Abstract
Objective
The NIH All of Us Research Program aims to advance personalized medicine by not only linking patient records, surveys, and genomic data but also engaging with participants, particularly from groups traditionally underrepresented in biomedical research (UBR). This study details how the dialogue between scientists and community members, including many from communities of color, shaped local research priorities.
Materials and Methods
We recruited area quantitative, basic, and clinical scientists as well as community members from our Community and Participant Advisory Boards with a predetermined interest in All of Us research as members of a Special Interest Group (SIG). An expert community engagement scientist facilitated 6 SIG meetings over the year, explicitly fostering openness and flexibility during conversations. We qualitatively analyzed discussions using a social movement framework tailored for community-based participatory research (CBPR) mobilization.
Results
The SIG evolved through CBPR stages of emergence, coalescence, momentum, and maintenance/integration. Researchers prioritized community needs above personal academic interests while community members kept discussions focused on tangible return of value to communities. One key outcome includes SIG-driven shifts in programmatic and research priorities of the All of Us Research Program in Southeastern Wisconsin. One major challenge was building equitable conversations that balanced scientific rigor and community understanding.
Discussion
Our approach allowed for a rich dialogue to emerge. Points of connection and disconnection between community members and scientists offered important guidance for emerging areas of genomic inquiry.
Conclusion
Our study presents a robust foundation for future efforts to engage diverse communities in CBPR, particularly on healthcare concerns affecting UBR communities.
Keywords: community-based participatory research, NIH All of Us Research Program, genomic research, ethnic and racial minorities
Background and significance
The NIH’s All of Us Research Program is a pioneering effort to advance personalized medicine by linking electronic health records, surveys, and genomic and other biologic data from a diverse population into a robust research database.1 All of Us makes this data available to a broad community of researchers using a cloud-based platform with tiered levels of access and statistical tools for collaborative data querying and analysis, the Researcher Workbench (RWB).2 The RWB allows the All of Us Research Program to balance wide accessibility with participant privacy and ensures that users have appropriate training in responsible data use.
Inclusion of individuals from communities traditionally underrepresented in biomedical research (UBR) is a stated priority for the All of Us Research Program; this extends to including such communities in decisions regarding research priorities carried out using this rich database. However, this commitment faces challenges due to the need for equitable partnerships between UBR communities and scientists with technical expertise in data use. Such partnerships must navigate community concerns about the intrusiveness of genetic sampling and historic mistrust of biomedical research. Furthermore, local researchers, community members, and All of Us staff held limited awareness of the strengths and weaknesses of the RWB.
The Medical College of Wisconsin (MCW) has enrolled more than 11 000 participants since 20183 and is committed to fostering research that addresses disease disparities impacting Southeast Wisconsin communities, including Milwaukee, one of the most racially and socioeconomically segregated cities in the United States.4 Notably, over 3000 enrollees come from communities of color. However, engaging these nonwhite racial/ethnic groups in the research process has lagged behind enrollment. To address this gap and in response to feedback from our Community and Participant Advisory Boards (CAB/PAB), we sought to increase genomics science literacy as a foundation for equitable dialogue with community members, focusing on Milwaukee’s communities of color.
We developed a Special Interest Group (SIG) based on the long-standing Science Shop model, which brings together community stakeholders and scientists in ways that deliberately balance power, emphasize the practical utility of understanding, and request that scientists work to pragmatically address community priorities.5 This approach emphasizes scientific exchange within a community-based participatory research (CBPR) framework.6 The composition of SIGs ranges from physicians and epidemiologists to trainees and community members. Collaborative research involving community members and scientists has demonstrated numerous benefits, including enhancing research outcomes and fostering meaningful partnerships between researchers and the community.6–10 Constructivist learning theory also underpins our approach—by engaging community members as active participants in the research process, we worked toward empowerment, ensuring that they not only contributed their unique perspectives but also shaped the research agenda.11 This approach fostered a deeper understanding of genomics and its potential impact on community health, while exposing scientists to community priorities.
To provide a structured framework for understanding the progression of our SIG meetings, we used the social movement framework of CBPR.12 Social movement theories offer valuable insights into the processes of collective action and community change, emphasizing the importance of partnership, resource mobilization, framing processes, and strategic action.12 This framework offers a structured lens through which to analyze the development of collective action initiatives.13–15 Using this framework, we aimed to gain insights into the SIG’s evolution, identify key turning points, and understand the factors that contributed to successful co-creation of knowledge by scientists and community members.
Objective
We describe key aspects of SIG formation and evolution, emphasizing its role in building community understanding of genomics and aligning researcher interests with the health priorities of Milwaukee communities, particularly communities of color that have been historically marginalized in research. Consistent with an evolving consensus among community-engaged researchers,16 we designated all participating community members as coauthors, recognizing their intellectual contributions in data generation during meetings and analysis, beyond mere manuscript preparation. This reflects CBPR’s inclusive approach to emphasize the involvement of all partners—from problem identification to dissemination—and the broader shift in scientific publishing toward recognizing critical yet nontraditional authors.
Methods
SIG formation
We worked with the established MCW All of Us Research Program CAB and PAB to identify community leaders who were interested in engaging further through a deep, science-oriented discussion of genomics with a particular focus on All of Us. The CAB included leaders from organizations serving diverse Southeastern Wisconsin communities, especially African American and Hispanic communities. The PAB consisted of All of Us enrollees who volunteered to assist the MCW site to optimize the experience of future participants. We invited researchers from institutions affiliated with the Clinical and Translational Science Institute of Southeast Wisconsin, which is the academic home of MCW’s All of Us Research Program.
The SIG included 5 community members and 7 researchers with consistent attendance (at least 4 of the 6 meetings), along with 4 facilitators from the MCW research team (Table S1). Community (CAB & PAB) participants included 1 African American, 1 Hispanic, 1 mixed-race individual identifying primarily as African American, and 2 White members. Although this group broadly reflected the region’s demographics, no Asian, Native American, or other racial/ethnic groups were represented, as they were not part of the CAB or PAB. One LGBT+ community leader from the CAB was unable to join due to time constraints. Researchers consisted of 6 Asians, 1 Hispanic, and 3 White members.
According to the 2020 Census, Milwaukee’s population was approximately 38% African American, 32% White (non-Hispanic), 20% Hispanic, 5% Asian, 3% mixed race, and 1% urban Native American.17 Our inclusion approach built on the existing social infrastructure of the CAB and PAB, where extensive efforts to elevate understanding of All of Us-related genomic considerations had already occurred. While the CAB and PAB were carefully recruited to represent Milwaukee, they were not fully representative of all subgroups in the region.
The CAB, PAB, and SIG brought together a wide array of perspectives across ages, races, ethnicities, and genders (Table S1). To ensure all SIG members started with a sense of trust and mutual respect, the facilitator team first met separately with community members and scientists, clarifying that the goal was not to proceed as rapidly as possible to a scientific hypothesis, but rather to build mutual understanding of various members’ priorities when identifying key questions leveraging the All of Us Research Program database.
Ethical considerations
This analysis of SIG evolution was approved by the MCW Institutional Review Board (IRB) (PRO00044833). At the beginning of the process, we presented an informational letter explaining the purpose of the SIG, and the overlapping roles as SIG members (eg, CAB/PAB membership, identifying research questions on one hand and serving as the primary data source for the present manuscript on the other). The IRB requested that All of Us staff involved directly in participant recruitment be excluded from serving as part of the SIG research team, and that specific guidance on navigating potentially conflicting roles and priorities be included in the informational letter. This allowed participants to choose their level of involvement as either participant-authors or simply participants. The entire informational letter is provided in the Supplementary File.
SIG meetings and data collection
We held 6 SIG meetings, which occurred virtually at approximately 2-month intervals over Zoom. With participants’ consent, recordings and transcripts were produced and stored within the MCW private server. Rather than using a static facilitator guide, we employed thematic content, framing, and small stories approaches to rapidly analyze meeting transcripts after each session. Thus, the agenda for following meetings was based in part on questions posed by participants, discontinuities in the conversation, and gaps in understanding for both community members and researchers. We also balanced these agendas with facilitator priorities like summarizing how a study using the All of Us RWB might unfold. This facilitation approach reflects strategies drawn from collaborative ethnography and community engagement.18 The resulting agendas for the meeting series are provided (Table S2).
Most meetings began with brief PowerPoint presentations exploring pertinent genomics topics and relevant data from All of Us or peer-reviewed sources. These presentations used visual storytelling to ensure the meeting was both informative and conducive to active participation from all members. To provide illustrative, locally relevant descriptive data, S.K.T. created an RWB workspace using the All of Us Research Program Controlled Tier Dataset v6 and all SIG members with access were invited to use this workspace.
Data analysis
Building on the intersession work using small stories to shape focus group agendas, further thematic content analysis allowed us to extract and interpret patterns across all sessions.19 Using a deductive approach to thematic analysis, we recognized that the SIG process partially aligned with the social movement framework stages of CBPR: (1) emergence, (2) coalescence, (3) momentum, and (4) maintenance and integration.12,13 In the emergence stage, the group defines its purpose and strategy. Coalescence sees increased organization and consensus-building. Momentum focuses on active implementation, in this case with particular emphasis on community-scientist collaborative hypothesis generation. Finally, maintenance and integration consolidate gains and foster ongoing collaboration.
Framing analysis enabled us to understand how different health issues were conceptualized and discussed by participants, and on the perspectives and biases influencing these conversations.20 The small stories approach permitted us to capture the meeting interactions, providing a deeper understanding of the community’s lived experiences and perceptions.21,22 K.M. and S.K.T. completed an initial review to identify broad themes. Following this initial set, S.K.T. did 2 additional reviews to code meeting-specific realizations and ensure that the perspectives of both community members and researchers were accurately and fairly represented.
Results
Table 1 presents a detailed mapping of the SIG-driven process using the same themes and headings outlined in Tremblay et al.’s social movement theory framework for CBPR.12 This table adapts the framework to our SIG research, integrating both methods and outcomes. The following sections explain how the SIG aligns with key stages of the social movement framework—emergence, coalescence, momentum, and maintenance. Each stage’s thematic analysis highlights important aspects such as disease priorities and the use of visual storytelling to engage participants.
Table 1.
Social movement framework-CBPR dimensions of the SIG-driven process.
| CBPR dimension | Description as related to this SIG process |
|---|---|
| Context | Regular meetings of the CAB and PAB under the MCW All of Us Research Program facilitate information dissemination to local communities and CTSI-affiliated researchers, promoting participation in the program and utilization of the RWB to further precision medicine. |
| Problem |
|
| Partnership | The MCW All of Us research team established the SIG, integrating researchers and community members to address shared concerns. |
| Cause | SIG’s goals included enhancing public understanding of science (especially genetics), setting research goals and at least one central hypothesis directly derived from community priorities, and conducting analysis using the All of Us Research Program dataset housed on the RWB. |
| Collective action strategy | By enhancing community understanding of science to create equitable conversation, we sought to develop a research hypothesis that addresses a community health priority by using the All of Us Research Program dataset. |
| Framing processes |
|
| Opportunities |
|
| Resources |
|
| Community and System Changes | The SIG process marked the beginning of ongoing collaborative dialogues between community members and researchers, laying the groundwork for continuous research initiatives and strengthening partnerships across the CTSI of Southeast Wisconsin. |
| Stage 1: Emergence | Following identification of the problems, the first SIG meeting served as the creation of the partnership and to develop the aims by building on resources and opportunities detailed above. |
| Stage 2: Coalescence | SIG meetings #2-#4 worked to develop the cause and collective action strategy using the framing strategies as detailed above. |
| Stage 3: Momentum | The fifth SIG meeting sought to implement the collective action strategy. |
| Stage 4: Maintenance and integration | SIG meeting #6 came as the last SIG meeting. While this was a formal end to the SIG, the ideas, values, and research priorities were incorporated into activities of the larger MCW All of Us Research Program site. |
Abbreviations: CAB, Community Advisory Board; CTSI, Clinical and Translational Science Institute of Southeastern Wisconsin; MCW, Medical College of Wisconsin; PAB, Participant Advisory Board; RWB, Researcher Workbench; SIG, Special Interest Group.
Emergence
The inaugural meeting explained the partnership and laid the groundwork for clarifying the group’s purpose, which are components of the emergence stage (Table 1).
Community health priorities and data exploration
Discussions on health conditions began broadly, informed by CAB input, covering diabetes, mental health, cancer, heart disease, Alzheimer’s/dementia, asthma/COPD, obesity, HIV/AIDS, pregnancy and childbirth, and other areas (men’s health, adverse childhood experiences, wellness/prevention). The group then focused on diabetes, kidney disease, heart disease, Alzheimer’s/dementia, obesity, hypertension, and health concerns related to environmental exposures. The meeting also featured a demonstration of the All of Us Research Program Data Browser (https://databrowser.researchallofus.org/), showcasing the range of data accessible on the RWB. Throughout, participants shared personal and community narratives that provided valuable insights into their lived experiences, embodying the ‘small stories’ approach. For instance, one community voice stressed the need to focus on diseases like kidney disease, highlighting the gaps in awareness in the community. The session concluded with an invitation for members to formulate 1 or 2 key research questions for future exploration.
Visual storytelling and participant dynamics
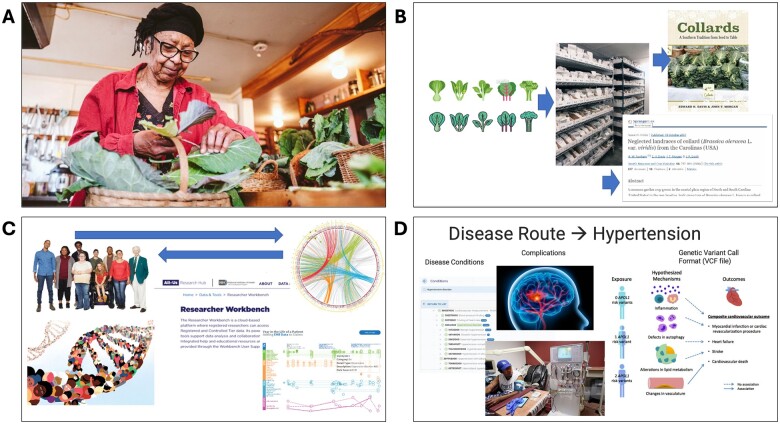
Elements in the PowerPoint presentations enabled the framing of health issues in the community experience. Visual storytelling techniques were used to make the concepts of precision medicine and genomics more accessible and relevant to the community (Figure 1).23,24 We described a prime example of how local communities, individual seed savers, and plant geneticists worked together to locate, monitor, and preserve the genetic diversity of heirloom collards (Figure 1A and B), a staple for many Black American families.25,26 This compelling story of preservation illustrated the importance of community involvement in scientific endeavors.
Figure 1.
Slides from SIG presentations demonstrating visual storytelling. Photo 1A courtesy of Ézé Amos.
Coalescence
The subsequent 3 SIG meetings focused on developing the cause and collective action strategy (Table 1), suggesting group coalescence.
The second meeting emphasized balancing personal research goals with collaborative SIG aims. We explored 3 methods for selecting a study population: by disease, demographic groups, or those affected by an environmental exposure. While researchers initially proposed a disease-specific approach to selecting a study population, focusing on conditions like diabetes and hypertension, community members emphasized concerns about the life-altering impact of complications from these conditions that they believed disproportionately affected their communities—amputations, retinopathy, and dialysis for diabetes, and stroke and dialysis for hypertension (Figure 1D). This exchange underscored the need for researchers to prioritize the lived healthcare concerns of community members.
The SIG also considered the appropriate geographic scope of research driven by their recommendations. Most members unequivocally restricted the area to Southeastern Wisconsin, especially Milwaukee. One community member inquired about the generational impacts of environmental exposures. A researcher highlighted the APOL1 gene’s role in kidney disease risk among hypertensive Black Americans.27,28 During the wide-ranging discussion of potential topics, one community member repeatedly emphasized the need for early agreement to hasten data exploration. This advocacy for a more focused hypothesis early in the dialogue served as a counterbalance to potential groupthink, enabling more comprehensive discussions.
This second meeting also showcased the need for local data to guide quantitative conversations. SIG members familiar with coding languages (R/Python) and tools like Jupyter Notebook branched to form a data analysis subgroup. By using the RWB, the group generated descriptive data to assess the feasibility of various ideas while also building analytic capacity.
Third meeting
During the third meeting, the discussion focused on identifying questions that could be addressed using the All of Us Research Program dataset. Facilitators first described the personal genetic results provided to participants: genetic return of results (GRoR). GRoR includes the “Hereditary Disease Risk” and “Medicine and Your DNA” reports.29 The former includes disease-associated genetic variants that are highly penetrant, clinically significant, and medically actionable (found in ∼1%-2% of participants) while the latter identifies variants in 7 genes that significantly affect medication metabolism.29
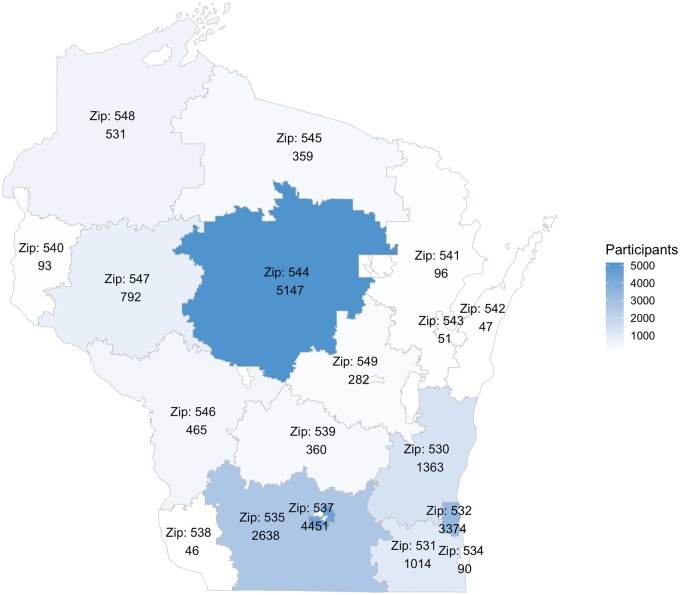
Next, facilitators reviewed participant counts in Wisconsin and nationally for obesity, hypertension, diabetes, chronic kidney disease, and attention-deficit hyperactivity disorder (Table 2). We noted a significant decrease in numbers after filtering for disease complications or whole-genome sequencing availability (Table 2). Additionally, we provided geographical context by presenting a map of Wisconsin with participant numbers by 3-digit zip codes (Figure 2).
Table 2.
All of Us research program database participant counts.
| Disease | National participant countsa | SE WI participant countsa | SE WI complication countsa | ||||
|---|---|---|---|---|---|---|---|
| Hypertension (SNOMED: 38341003) | Total: 103 632 | Total: 6090 | CKD: 373 Ischemic stroke: 9 Dialysis dependence: 99 | ||||
| 58 990 Female | 41 575 Male | 31 273 with WGS | 3497 Female | 2385 Male | 1985 with WGS | ||
| Diabetes (SNOMED: 73211009) | Total: 48 354 | Total: 2427 | CKD as well: 583 Dialysis dependence: 77 | ||||
| 28 039 Female | 18 937 Male | 14 211 with WGS | 1347 Female | 989 Male | 760 with WGS | ||
| Chronic Kidney Disease (SNOMED: 709044004) | Total: 22 120 | Total: 1374 | Dialysis dependence: 100 | ||||
| 10 669 Female | 10 751 Male | 6163 with WGS | 681 Female | 643 Male | 428 with WGS | ||
| Obesity (SNOMED: 414916001) | Total: 65 706 | Total: 4676 | |||||
| 44 659 Female | 19 213 Male | 19 935 with WGS | 3086 Female | 1445 Male | 1478 with WGS | ||
| ADHD (SNOMED: 406506008) | Total: 6648 | Total: 779 | |||||
| 3885 Female | 2546 Male | 2014 with WGS | 476 Female | 270 Male | 246 with WGS | ||
Participant counts as of 9 November 2022.
Abbreviations: CKD, chronic kidney disease; SE WI, Southeast Wisconsin; WGS, whole-genome sequencing; ADHD, attention-deficit hyperactivity disorder.
Figure 2.
All of Us Research Program participant counts shown by 3-digit zip codes on a map of Wisconsin as of 9 November 2022.
Reviewing specific GRoR-related examples and participant counts encouraged SIG members to brainstorm RWB-directed questions. Many community members asked about epigenetic research, reflecting the belief that trauma experienced by ancestors (ie, slavery) can influence genetic expression in later generations. However, the unavailability of epigenetics data on RWB led to a consensus summarized by one facilitator, “While we recognize the power of epigenetics in health outcomes, we must focus on the genetics we can analyze now, setting a stage for a more comprehensive future study.” SIG community members favored hypotheses related to GRoR; one community member described their rationale: “It makes sense to start with what we know our community has access to—their genetic results—and build from there.”
Visual storytelling to understand the All of Us research program dataset
Visual storytelling of quantitative data using maps and tables proved essential for distilling complex concepts and active community member engagement. The success of visualizations in promoting broad understanding led us to establish an information subgroup dedicated to communicating complex scientific ideas in ways easily understood by the community. One community member’s statement, “I think it’s really important…to put it in layman’s terms,” preceded a short diversion into the discussion of readily accessible platforms.
Community members also became aware of the importance of academic deliverables when conducting research. Following the third meeting, SIG members collaboratively drafted a conference abstract on the SIG process.30 As with the current manuscript, the abstract was collaboratively written, promoting inclusivity and shared ownership of our research endeavors.
Fourth meeting
The fourth meeting’s presentation focused on education regarding genetic research concepts, including the distinction between gene and variant and between correlation and causation. We also compared family linkage, candidate gene, and genome-wide association studies. Finally, we explained that all datasets had strengths and weaknesses—for example, one must consider selection bias in the volunteer All of Us participant cohort regardless of its size, breadth, and diversity.
Although we had hoped to identify candidate hypotheses during this meeting, the group could not settle on one research direction. However, the community members became more open to supplementing Wisconsin research with national comparisons to enhance generalizability and statistical power. We also found that SIG researchers lacked the coding expertise needed for certain RWB analyses. We acknowledged this skill gap and were eventually forced to focus on candidate gene methods due to resource and expertise constraints. This was communicated transparently to balance SIG expectations with feasibility.
Despite efforts to educate and engage, facilitators noted that community members were less vocal during technical discussions. Off-line individual conversations with community members to discuss the silence indicated discomfort with many of the genetic concepts. To decrease this knowledge discrepancy and hesitance with genetics, we convened a community-member-only meeting, which ran after the fifth SIG meeting (described below), to demystify genetic concepts and promote public understanding of science.
The SIG also demonstrated a commitment to disseminating findings and making SIG discussions accessible to the public. “We’re looking at platforms like Medium and even considering tools like Chat-GPT to help simplify complex scientific terms,” shared an information subgroup member. This desire to disseminate SIG discussions led to the publication of abbreviated meeting notes through Medium posts by Z.F. (https://medium.com/@zeno.franco/community-scientist-co-creation-of-a-testable-genetics-research-question-998c4f1215bf). The post featured AI-generated artwork symbolizing diversity in genetics (Figure 3A). Community partners actively participated in selecting the final image for each blog post and reviewed the blog text for clarity and approachability by nonscientists.
Figure 3.
AI-generated artwork (MidJourney AI) chosen in collaboration with the community to visually represent key concepts raised during SIG discussions and published with meeting notes on Medium.
After this fourth meeting, facilitators engaged with community members to affirm 3 main disease priorities: diabetes, kidney disease, and hypertension. After a literature review, we generated 10 hypotheses and worked with scientists to narrow down to 3 based on: (1) availability of a candidate gene variant for a complication associated with a disease priority; (2) inclusion of that variant in All of Us Research Program GRoR; (3) capacity for broad generalization; (4) novelty; and (5) alignment with prior SIG conversations.
Momentum
The fifth meeting aimed to build momentum by taking collective action toward a shared research agenda. Community/scientist collaborative hypothesis generation has often been viewed as one of the most difficult areas to navigate equity in CBPR. To address this, we presented 3 possible hypothesis focus areas (Table 3), each with a researcher-driven version and a community-driven version so that each group could both see that their own priorities were being addressed, while also viewing how the priorities of the other group changed the details and emphasis. Table 3 includes a summary of relevant literature on candidate variants in the “Review of Information” column, an indication of whether a genetic variant is included in the All of Us Research Program (a key priority for SIG community partners) in the “GROR” column, and the facilitators’ estimate of scientific novelty, based on available literature and publication volume (a priority for SIG researchers), in the “Novelty” column.
Table 3.
Three candidate gene hypotheses presented at the Fifth SIG Meeting.
| Researcher-driven hypotheses (with a focus on the genetic variants) | Community-driven hypotheses (with a focus on health condition and outcomes) | Review of information | GRoR | Novelty |
|---|---|---|---|---|
| Hypothesis #1: Diabetes and kidney disease | ||||
| Type 2 diabetic patients who carry either the UMOD or TENM3 variants are more likely to have chronic kidney disease. |
|
|
No | High |
| Hypothesis #2: Hypertension, kidney disease, and racial disparities | ||||
| Black and Hispanic Americans who carry the MYH9 variants associated with hypertension are at a higher risk of nondiabetic end-stage kidney disease. |
|
|
No | Mid |
| Hypothesis #3: Pharmacogenetics and racial disparities | ||||
| Black American patients with diabetes undergoing percutaneous coronary intervention not only have a higher prevalence of high on-treatment platelet reactivity to clopidogrel but also have higher CYP2C19 variant carrier status compared with Non-Hispanic White Americans. |
|
|
Yes | Low |
GRoR consists of variants assessed and returned to All of Us participants via the participant portal29; The “Review of Information” column provides a summarized literature review, and the “Novelty” column reflects scientific novelty as assessed by the facilitator team, based on available literature and publication volume.
Abbreviation: GRoR, genetic return of results.
Notably, where community members struggled with scientific terminology, the researchers struggled with community-driven language:
Researcher 1: [viewing slide of community-driven hypotheses] “It’s not a statement. It’s not a hypothesis. [viewing slide of researcher-driven hypotheses] I like this one, this is a hypothesis, right?”
Facilitator 1: “Right, but I wanted to make it simple enough to help everyone in the group pick one.”
Researcher 1: “I would think that’s confusing.”
Facilitator 2: [misunderstands Researcher 1’s statements as a criticism of the hypothesis itself, not the language used in the community-driven version]
Researcher 2: “I think it was a matter of just changing that slide before, that [community-driven version] was the background and now these [researcher-centered version] are your hypotheses.”
After significant discussion about the 2 versions of hypotheses, one community member expressed uncertainty about their implications:
Community Representative 1: “For me it’s still not clear what we are trying to figure Out … what are we going to do with it? Is it like verify if it’s true [comparing All of Us Research Program data to prior published studies] … or discover something, like it is not only for people of European decent, but it’s also for these other populations?”
We also noted a perspective shift regarding the studies that researchers found valuable:
Researcher 1: “We also need to think about—beyond the novelty—is relevance, or another word is impact. Or community impact. That’s another thing that has not appeared on the table [Table 3].”
Community member insights like “It’s about what affects us, our families, our neighbors” directed the research team’s focus toward conditions that both bear a genetic footprint AND resonate deeply within local narratives. The following Medium post by Z.F. summarized the fifth meeting: https://medium.com/@zeno.franco/collaborating-to-prioritize-hypotheses-for-genetics-research-7b67157c1e2b. The post again featured AI-generated artwork (Figure 3B), chosen in collaboration with community members to illustrate key themes.
While the fifth meeting did not result in a hypothesis selection, it concluded with a renewed dedication to the consensus-building process and being sure to anchor questions in real-world implications rather than abstract scientific curiosity. Looking ahead, the SIG acknowledged the evolving landscape of available data and prepared for the integration of epigenetic data when it becomes available. “We’re setting the groundwork for what’s to come,” one participant stated. This forward-looking perspective ensures a model that is both resilient and adaptable to emerging scientific opportunities.
Community member-only meeting
Individual conversations with community members following the fourth and fifth meetings indicated a need to decelerate the process without researchers present. One community member hosted this in-person session off campus at his organization’s offices and started by distributing paper copies of Chat-GPT-assisted simplifications of the scientific discussions in laymen’s terms and metaphors, enhancing comprehension and engagement. Facilitators provided additional details explaining genetic hypotheses and RWB constraints.
During this conversation, many of the community members openly shared specific health concerns they and their family members faced, renewing interests in disease priorities such as cancer, which were eliminated in earlier SIG meetings. This session also highlighted a divide between seeking novel research avenues appreciated by scientists versus focusing on tangible community impact—community members held greater interest in hypothesis #3 while scientists preferred hypothesis #2 (Table 3). Z.F.’s Medium post along with another AI-generated artwork (Figure 3C) captured this candid exchange of ideas and concerns: https://medium.com/@zeno.franco/community-discussion-to-narrow-options-for-candidate-gene-research-b8ed04f5840a
Maintenance and integration
This sixth meeting centered on the need to balance focused scientific questions with broader community-driven dialogues. One scientist highlighted, “It’s about capturing the essence of what the community is concerned about, be it clinical issues or broader health outcomes.” The group’s approach thus evolved into a community-driven research paradigm, prioritizing diseases like end-stage renal disease and cancers prevalent in Milwaukee.
“It’s not just about the data; it’s about what we can do with it,” a community member emphasized, underlining the importance of research relevancy and practicality. Community members understood the importance of statistical power and sample size in producing significant, accurate, and actionable findings that benefit the community. These example statements from a researcher and community member highlight the symbiotic relationship that developed between community members and researchers during the SIG process.
Moreover, community members clearly articulated their health concerns, identifying disease complications and heritability risk as having a major impact on their individual and familial quality of life. Ranking the actionability and relevance of a question higher than novelty, community members advocated for studies with tangible improvements in health disparities, screening practices, and targeted prescribing. This focus suggests that enhancing the practical utility of All of Us Research Program data may increase community interest in sustained participation in the program.
The sixth meeting marked the maintenance and integration stage (Table 1) as insights were integrated into the broader All of Us Research Program processes. A research team member explained: “We’re moving from hypergrassroots discussions to a more national perspective, where the community is not just participating but actively shaping our research strategy.” Recognizing the need for more equitable dialogue, the research team opted to hold separate discussions for community members and researchers, laying the groundwork for future unified collaboration. This temporary separation aims to address specific needs and strengthen future integrated discussions, aligning with the principles of CBPR. The insights and approaches developed will guide the next phases once the SIG reconvenes.
Discussion
Our SIG process offers a nuanced reflection on the intersection of community engagement and scientific inquiry. This research confirms the applicability of the social movement framework to CBPR efforts and reaffirms CBPR’s power in fostering meaningful research partnerships and combating mistrust in science.6,12,13 Our dialogue provided a forum for Southeastern Wisconsin community members to directly influence local All of Us Research Program priorities. While we did not identify a specific final hypothesis, the process improved community member understanding of genomics, yielded rich discussions, and informed future research priorities and participant recruitment strategies.
Reflecting on the SIG journey
Throughout the SIG meetings, the facilitators gleaned valuable lessons (Table 4) that shaped the discussions.
Table 4.
Lessons learned by SIG research team.
| SIG meeting # | Lessons learned by SIG research team |
|---|---|
| 1 |
Incorporating visual storytelling in CBPR presentations:
• Investing time and expertise in crafting informative stories that align community priorities with research goals proved highly beneficial. o The positive response to the successful community-academic collaboration around collard greens illustrated the value of visual storytelling23,24 in communicating complex scientific concepts and engaging a diverse group of participants (Figure 1A and B). • Facilitating interactive discussions fostered a collaborative atmosphere. o Presentations included more interactive elements to engage the audience, such as questions prompting participants to reflect on their interests and roles in the discussion. • Including visual elements from previous meetings in future presentations or the Medium posts maintained continuity and coherence. o The inclusion of AI-generated artwork (Figure 3) offered a venue to illustrate the nuanced differences between conversations. |
| 2 |
Outlier contributions:
• Diverse perspectives were crucial to shaping research hypotheses. o Some SIG members served as outliers, advocating for expediting analysis and addressing community needs promptly. o Although narrowing the research hypothesis earlier would have increased efficiency, later discussion demonstrated that many members needed more time to understand the data and research possibilities. o We actively moderated discussions to ensure inclusivity and balance power dynamics. Data analysis needs: • Realizing a crucial need for bioinformaticians to prosecute SIG-driven research questions, we established a data analysis subgroup and recruited pre- and postgraduate trainees to become acquainted with the Researcher Workbench. • A major barrier to progress was the complexity of Researcher Workbench itself. This required academic SIG members to develop competency with the platform and seek expert advice. • While this process slowed the initial timeline, it also pushed SIG members to develop data analysis skills. This newfound knowledge ultimately informed the presentation of research topics. |
| 3 |
Building community understanding of genetics:
• Initially, we focused on leveraging community knowledge of local health needs. However, we realized the importance of also enhancing the community understanding of genetic principles and their implications. • Community members wanted deeper knowledge of genetic nuances, including assumptions and potential errors in data interpretation. Deepening community knowledge around genetic interpretation is an important part of returning value to the community. Community members believe this will empower them to engage more fully in the research discourse. |
| 4 |
Community member engagement:
• We acknowledged the need for separate community member meetings to create more inclusive dialogues and set up community-member-specific meeting after the fifth SIG meeting. Technical assistance: • Bringing bioinformatics experts on board not only addressed the immediate need for technical skill but also laid the groundwork for long-term capacity building within the SIG. This critical step equipped the group to transform vast datasets into actionable health knowledge. |
| 5 |
Balancing scientific accuracy and accessibility:
• Conversations with community engagement experts led us to revise how we presented the initial 3 hypotheses, making them more accessible for community members (Table 3). • Although these hypotheses were more comprehensible, we faced challenges in maintaining scientific accuracy. This highlighted the dichotomy between scientific precision and community accessibility. |
| 6 |
Need for separate meetings for participation:
• Community members became more vocal and influential as the process progressed, particularly when leading discussions on healthcare priorities. • Researchers realized that prioritizing community-driven health concerns from the outset would have provided a stronger foundation for collaborative work. • To foster focused discussions, we planned to split future meetings between community members and researchers. This will allow space for identifying specific priorities before reconvening the full SIG. Building capacity: • In parallel with these discussions, efforts are underway to secure support for bioinformatics expertise and smaller grants. • The goal is to develop robust analysis capabilities to enable targeted research projects with bioinformatics experts. This ensures that communities contributing to the national dataset receive maximum value in return. |
Abbreviation: SIG, Special Interest Group.
Evolution of stakeholder perspectives
A notable outcome was the reciprocal evolution of both community members’ and scientists’ perspectives. As community members gained scientific understanding, their health priorities for research shifted from broad concerns to specific areas, reflecting a growing ability and willingness to articulate their research needs. Simultaneously, scientists underwent a paradigm shift, recognizing the importance of community-driven hypotheses and prioritizing research questions with tangible community benefits. This mutual understanding demonstrates the importance of longitudinal engagement and dialogue in fostering a truly collaborative research environment.37
Leveraging AI for communication
We were struck by the impact of enhancing meeting notes with AI-generated artwork (Figure 3). This innovative approach to presenting complex research concepts captured the spirit of our collaborative work. Community members actively participated in selecting these images, ensuring they resonated with the SIG discussion. Additionally, during the community-only meeting, one member creatively employed AI tools like ChatGPT to generate metaphors and analogies. This appeared to deepen their understanding of scientific terminology, empowering them to more fully engage in the research process. These examples illustrate how AI can enhance communication and bridge knowledge gaps within a CBPR framework.38
Challenges and solutions in CBPR
The success of our CBPR approach hinged on several key elements, including a multi-faceted communication strategy, skilled facilitation, and meticulous planning to foster transparency, trust, and meaningful engagement. A crucial aspect of our approach was the deliberate temporary suspension of self-interest, asking all parties to commit to a group process and work toward a single shared outcome of a testable hypothesis. This approach, rooted in the concept that collaboration often transcends individual interests,39 fostered a sense of unity and shared purpose among participants.
Numerous phone calls, emails, and texts between CAB/PAB members and research staff provided individual members with informal channels and a safe space for open dialogue to address questions and concerns. We also needed strong facilitation to guide the polyvocal discussions and drive toward consensus by focusing on solutions acceptable to all parties.40 The careful planning of discussions and the strategic invocation of expertise were essential in making complex scientific concepts accessible to all participants.40
One significant hurdle in the SIG process was managing the diversity of opinions and ensuring that every voice was heard. We tackled this by adopting flexible discussion formats and actively encouraging participation from quieter members. Another challenge was bridging the communication gap between scientists and community members. To address this, we used visual aids, simplified language, and community-only discussions to make complex scientific concepts more accessible.24 Furthermore, while the separation of researchers and community members discussions toward the end may seem to run counter to the ethos of collaboration in CBPR, structured capacity building, sometimes outside of the main partnership activities, is critical to ensuring community partners are able to engage more equitably.41
Impact on future research
The SIG discussions yielded benefits for all 3 stakeholders. Researchers gained a deeper understanding of community priorities, enabling them to incorporate these insights into their projects and enhance the relevance and impact of their work. Community members developed a deeper understanding of the scientific process, potentially bridging the local knowledge gap. The MCW All of Us research team gained critical insights into improving participant enrollment, research dissemination, and community outreach.
Notably, the tension between scientific novelty and community impact when deciding on a hypothesis proved particularly fertile. Balancing both considerations may meaningfully advance scientific knowledge and community health. For example, while studying heavily researched variants like CYP2C19 from hypothesis #3 (Table 3) to localize community concerns, separate exploration of additional variants that are more common in UBR populations can generate scientific novelty. For research using the RWB, our work underscored 2 specific needs: scientists who can effectively communicate genetic complexities to the community and robust bioinformatics support.
As the SIG progressed, we recognized the inherent challenges in converging on a single hypothesis and the importance of acknowledging individual research interests. This led to a broadening of our approach, culminating in the development of several promising research ideas that individual labs could pursue while incorporating the community’s priorities. This evolution has already yielded tangible outcomes, including increased researcher participation in training on the RWB and a successful grant collaboration between academic researchers and community partners focused on addressing SIG-directed health disparities.
Limitations of the study
This study’s limitations include the scalability of the SIG and potential representation biases. The intimate nature of our discussions may not be replicable in larger groups, and our findings may not fully capture the diversity of community opinions. Furthermore, data limitations restricted our ability to explore certain research questions, including epigenetics and environmental exposures, which could have impacted the study’s direction. The choice of virtual meetings, while inclusive and convenient, potentially lacked the depth of in-person interactions.
Conclusion
The implications of our study extend beyond the local context, offering insights into how equitable conversations are necessary for strong CBPR. This approach fosters a deeper public understanding of science, empowering communities of color and UBR populations to actively participate in and shape research endeavors. The lessons learned from our SIG process provide a valuable roadmap for effectively implementing RWB-related CBPR in other settings.
Our approach demonstrates the potential for community-driven research to contribute significantly to personalized medicine, particularly in ensuring that health interventions are tailored to meet the specific needs and priorities of diverse communities. This project contributes to a growing body of knowledge on how community engagement can enhance the applicability and impact of scientific research, bridging the gap between academic pursuits and community needs.
Supplementary Material
Acknowledgments
We are grateful to Jessica Olson, PhD, MPH for her expertise in ensuring that meeting presentations were accessible and engaging for community members as well as her mentorship of S.T. We would like to thank and acknowledge the All of Us staff members who made this project possible, including Jenna Koney, Kairee Larson, Kristyn Ertl, and Karen Dotson. We received permission to acknowledge those named in this section.
Contributor Information
Suma K Thareja, Medical College of Wisconsin, Milwaukee, WI 53226, United States; All of Us Wisconsin, Milwaukee, WI 53226, United States.
Xin Yang, All of Us Wisconsin, Milwaukee, WI 53226, United States.
Paramita Basak Upama, Marquette University, Milwaukee, WI 53233, United States.
Aziz Abdullah, Inpower, Milwaukee, WI 53212, United States.
Shary Pérez Torres, United Community Center, Milwaukee, WI 53204, United States.
Linda Jackson Cocroft, Black Women 50+ Health & Lifestyles Magazine, Milwaukee, WI 53007, United States.
Michael Bubolz, Eisen Dev, Milwaukee, WI 53207, United States.
Kari McGaughey, All of Us Wisconsin, Milwaukee, WI 53226, United States.
Xuelin Lou, Medical College of Wisconsin, Milwaukee, WI 53226, United States.
Sailaja Kamaraju, Medical College of Wisconsin, Milwaukee, WI 53226, United States.
Sheikh Iqbal Ahamed, Marquette University, Milwaukee, WI 53233, United States.
Praveen Madiraju, Marquette University, Milwaukee, WI 53233, United States.
Anne E Kwitek, Medical College of Wisconsin, Milwaukee, WI 53226, United States.
Jeffrey Whittle, Medical College of Wisconsin, Milwaukee, WI 53226, United States; All of Us Wisconsin, Milwaukee, WI 53226, United States; Clement J Zablocki Veterans Affairs Medical Center, Milwaukee, WI 53295, United States.
Zeno Franco, Medical College of Wisconsin, Milwaukee, WI 53226, United States; All of Us Wisconsin, Milwaukee, WI 53226, United States; Clement J Zablocki Veterans Affairs Medical Center, Milwaukee, WI 53295, United States.
Author contributions
All authors were part of the SIG. Suma K. Thareja: Conceptualization, Data curation, Formal Analysis, Investigation, Visualization, Writing—original draft, Writing—review & editing. Xin Yang: Data curation, Investigation, Visualization, Writing—review & editing. Paramita Basak Upama: Data curation, Writing—review & editing. Aziz Abdullah: Data curation, Writing—review & editing: Shary Pérez Torres: Data curation, Writing—review & editing. Linda Jackson Cocroft: Data curation, Writing—review & editing. Michael Bubolz: Data curation, Writing—review & editing. Kari McGaughey: Data curation, Formal Analysis, Writing—review & editing. Xuelin Lou: Data curation, Writing—review & editing. Sailaja Kamaraju: Data curation, Writing—review & editing. Sheikh Iqbal Ahamed: Data curation, Writing—review & editing. Praveen Madiraju: Data curation, Writing—review & editing. Anne E. Kwitek: Data curation, Writing—review & editing. Jeff Whittle: Conceptualization, Resources, Supervision, Data curation, Project administration, Funding acquisition, Writing—review & editing. Zeno Franco: Conceptualization, Resources, Supervision, Data curation, Project administration, Funding acquisition, Writing—review & editing.
Supplementary material
Supplementary material is available at Journal of the American Medical Informatics Association online.
Funding
This work is supported by All of Us Wisconsin (NIH 3OT2OD026555-01S4). S.K.T.’s postdoctoral fellowship was partially funded by the Robert D. and Patricia E. Kern Institute for the Transformation of Medical Education.
Conflicts of interest
None declared.
Data availability
The data are not publicly available to protect participant confidentiality surrounding specific text derived from members in this qualitative CBPR study. Questions can be addressed to the corresponding author.
Ethics approval
The Medical College of Wisconsin Institutional Review Board (IRB) reviewed and approved this project PRO00044833. The informational letter provided to all SIG participants is included in the Supplementary File. All the study procedures were conducted in accordance with US Federal Regulations for the Protection of Human Subjects.
References
- 1. National Institutes of Health. All of Us Research Program Overview. Accessed February 28, 2023. https://allofus.nih.gov/about/program-overview
- 2. National Institutes of Health. Researcher Workbench. Accessed June 28, 2024. https://www.researchallofus.org/data-tools/workbench/
- 3. Froedtert & the Medical College of Wisconsin All of Us Research Program. Home Page. Accessed February 28, 2023. https://mcw.joinallofus.org/
- 4. Levine MV. The State of Black Milwaukee in national perspective: racial inequality in the nation’s 50 largest metropolitan areas. In: 65 Charts and Tables. UWM Digital Commons. Center for Economic Development Publications; 2020:56. [Google Scholar]
- 5. Leydesdorff L, Ward J. Science shops: a kaleidoscope of science-society collaborations in Europe. Public Understand Sci. 2005;14:353-372. [Google Scholar]
- 6. CDC/ATSDR. Principles of Community Engagement (NIH Publication No. 11-7782). 2nd ed. US Department of Health and Human Services, National Institutes of Health; 2015. [Google Scholar]
- 7. Parry M, Owadally T, O’Hara A, et al. Community- and patient-partner engagement in women’s cardiovascular disease research: a rapid review of the evidence. CJC Open. 2024;6:485-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sheldon E, Ezaydi N, Ditmore M, et al. Patient and public involvement in the development of health services: engagement of underserved populations in a quality improvement programme for inflammatory bowel disease using a community-based participatory approach. Health Expect. 2024;27:e14004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Collins SE, Clifasefi SL, Stanton J, et al. Community-based participatory research (CBPR): towards equitable involvement of community in psychology research. Am Psychol. 2018;73:884-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Unertl KM, Schaefbauer CL, Campbell TR, et al. Integrating community-based participatory research and informatics approaches to improve the engagement and health of underserved populations. J Am Med Inform Assoc. 2016;23:60-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bada SO, Olusegun S. Constructivism learning theory: a paradigm for teaching and learning. J Res Method Educ. 2015;5:66-70. [Google Scholar]
- 12. Tremblay MC, Martin DH, Macaulay AC, et al. Can we build on social movement theories to develop and improve community-based participatory research? A framework synthesis review. Am J Community Psychol. 2017;59:333-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tremblay MC, Martin DH, McComber AM, et al. Understanding community-based participatory research through a social movement framework: a case study of the Kahnawake Schools Diabetes Prevention Project. BMC Public Health. 2018;18:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Toke S, Correa-Velez I, Riggs E. Exploring trauma- and violence-informed pregnancy care for Karen women of refugee background: a community-based participatory study. Int J Environ Res Public Health. 2024;21:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mosnier E, Hoyer M, Artigas F, et al. Enhancing sexual health and empowerment among migrant women sex workers: a community health worker-led intervention in Marseille, France. Front Public Health. 2024;12:1359363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coughlin SS. Coauthorship by patients and other stakeholders with limited knowledge of scientific publishing practices. Emerg Themes Epidemiol. 2021;18:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. U.S. Census Bureau. Milwaukee City, Wisconsin. 2020. Accessed September 13, 2024. https://data.census.gov/profile/Milwaukee_city,_Wisconsin?g=160XX00US5553000#race-and-ethnicity
- 18. Lassiter LE. The Chicago Guide to Collaborative Ethnography. University of Chicago Press; 2005. [Google Scholar]
- 19. Braun V, Clarke V. Using thematic analysis in psychology. Qual Res Psychol. 2006;3:77-101. [Google Scholar]
- 20. Borah P. Conceptual issues in framing theory: a systematic examination of a decade’s literature. J Commun. 2011;61:246-263. [Google Scholar]
- 21. Georgakopoulou A. Thinking big with small stories in narrative and identity analysis. Narrative Inq. 2006;16:122-130. [Google Scholar]
- 22. Bamberg M, Georgakopoulou A. Small stories as a new perspective in narrative and identity analysis. Text & Talk. 2008;28:377-396. [Google Scholar]
- 23. Villar ME. Community engagement and co-creation of strategic health and environmental communication: collaborative storytelling and game-building. J Sci Commun. 2021;20:C08. [Google Scholar]
- 24. Dahlstrom MF. Using narratives and storytelling to communicate science with nonexpert audiences. Proc Natl Acad Sci USA. 2014;111:13614-13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wood D. A Community of Seed Savers Has a Recipe to Revive Rare Varieties of Collard Greens. NPR; 2022.
- 26. Farnham MW, Davis EH, Morgan JT, et al. Neglected landraces of collard (Brassica oleracea L. var. viridis) from the Carolinas (USA). Genet Resour Crop Evol. 2008;55:797-801. [Google Scholar]
- 27. Nadkarni GN, Gignoux CR, Sorokin EP, et al. Worldwide frequencies of APOL1 renal risk variants. N Engl J Med. 2018;379:2571-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hung AM, Shah SC, Bick AG, et al. ; VA Million Veteran Program COVID-19 Science Initiative. APOL1 risk variants, acute kidney injury, and death in participants with African ancestry hospitalized with COVID-19 from the million veteran program. JAMA Intern Med. 2022;182:386-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Venner E, Muzny D, Smith JD, et al. ; All of Us Research Program Regulatory Working Group. Whole-genome sequencing as an investigational device for return of hereditary disease risk and pharmacogenomic results as part of the All of Us Research Program. Genome Med. 2022;14:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thareja S, Yang X, Mia MR, et al. 441 Southeastern Wisconsin community-based participatory research using the All of Us researcher workbench [abstract]. J Clin Transl Sci. 2023;7:131-131. [Google Scholar]
- 31. Vujkovic M, Keaton JM, Lynch JA, et al. ; VA Million Veteran Program. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat Genet. 2020;52:680-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Palmer ND, Ng MC, Hicks PJ, et al. Evaluation of candidate nephropathy susceptibility genes in a genome-wide association study of African American diabetic kidney disease. PLoS One. 2014;9:e88273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Behar DM, Rosset S, Tzur S, et al. African ancestry allelic variation at the MYH9 gene contributes to increased susceptibility to non-diabetic end-stage kidney disease in Hispanic Americans. Hum Mol Genet. 2010;19:1816-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pendyala LK, Torguson R, Loh JP, et al. Racial disparity with on-treatment platelet reactivity in patients undergoing percutaneous coronary intervention. Am Heart J. 2013;166:266-272. [DOI] [PubMed] [Google Scholar]
- 35. Thomas CD, Franchi F, Keeley EC, et al. Impact of the ABCD-GENE score on clopidogrel clinical effectiveness after PCI: a multi-site, real-world investigation. Clin Pharmacol Ther. 2022;112:146-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Angiolillo DJ, Capodanno D, Danchin N, et al. Derivation, validation, and prognostic utility of a prediction rule for nonresponse to clopidogrel: the ABCD-GENE score. JACC Cardiovasc Interv. 2020;13:606-617. [DOI] [PubMed] [Google Scholar]
- 37. Resnik DB, Kennedy CE. Balancing scientific and community interests in community-based participatory research. Account Res. 2010;17:198-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pillai M, Griffin AC, Kronk CA, et al. Toward community-based natural language processing (CBNLP): cocreating with communities. J Med Internet Res. 2023;25:e48498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Collins KR. Beyond self-interest: why the market rewards those who reject it? by Krzysztof Pelc Oxford University Press, 2022. World Trade Rev. 2023;22:708-711. [Google Scholar]
- 40. Israel BA, Schulz AJ, Parker EA, et al. Review of community-based research: assessing partnership approaches to improve public health. Annu Rev Public Health. 1998;19:173-202. [DOI] [PubMed] [Google Scholar]
- 41. Allen ML, Culhane-Pera KA, Pergament S, et al. A capacity building program to promote CBPR partnerships between academic researchers and community members. Clin Transl Sci. 2011;4:428-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are not publicly available to protect participant confidentiality surrounding specific text derived from members in this qualitative CBPR study. Questions can be addressed to the corresponding author.