Abstract
Objective
The All of Us Evenings with Genetics (EwG) Research Program at Baylor College of Medicine (BCM), funded to engage research scholars to work with the All of Us data, developed a training curriculum for the Researcher Workbench, the platform to access and analyze All of Us data. All of Us EwG developed the curriculum so that it could teach scholars regardless of their skills and background in programming languages and cloud computing. All of Us EwG delivered this curriculum at the first annual All of Us EwG Faculty Summit in May 2022. The curriculum was evaluated both during and after the Faculty Summit so that it could be improved for future training.
Materials and Methods
Surveys were administered to assess scholars’ familiarity with the programming languages and computational tools required to use the Researcher Workbench. The curriculum was developed using backward design and was informed by the survey results, a review of available resources for training users on the Researcher Workbench, and All of Us EwG members’ collective experience training students. The curriculum was evaluated using feedback surveys during the Faculty Summit as well as virtual meetings and emails following the Faculty Summit.
Results
The evaluation results demonstrated the success of the curriculum and identified areas for improvement.
Discussion and Conclusion
The curriculum has been adapted and improved in response to evaluations and in response to changes to the All of Us data and infrastructure to train more researchers through this program and other scholarly programs.
Keywords: curriculum, continuing education, biomedical research, medical informatics, genomics
Background and significance
The All of Us Research Program
The Precision Medicine Initiative from the White House includes the Department of Health and Human Services supported All of Us Research Program that has developed one of the most expansive and diverse health data repositories.1,2 The All of Us data are projected to include one million participants from various races, ethnicities, age groups, geographic locations, and more.3 These participants include groups historically underrepresented in biomedical research (UBR) and groups typically with poor access to healthcare.1,3 The data include demographic, survey, electronic health record (EHR), physical measurement, Fitbit, and genomic data from participants. EHR data include laboratory test results, disease diagnoses, procedures, and medications. The survey data include self-reported answers on topics, such as lifestyle, family health history, healthcare access, and social determinants of health.4 Genomic data include whole genome sequencing and genotyping array data, both of which are generated from participant-donated biosamples. The All of Us Research Program offers multiple enrollment options for participants: partnering health care provider organizations, participant centers, or online (https://joinallofus.org).
The All of Us Researcher Workbench platform
To ensure the privacy of participants sharing their data, the All of Us data can only be accessed and analyzed by researchers using the cloud-based All of Us Researcher Workbench platform.5 Individual participant-level data cannot be downloaded or exported.6 Therefore, the tools of the Researcher Workbench must be used for research with the All of Us data. These tools developed by All of Us help users query the data, select data subsets, and analyze the datasets in the Google Cloud via All of Us Jupyter Notebooks.7–9 The platform supports the Python and R programming languages, limited bash commands, and genomic analysis software, including Hail10 and PLINK.11 Researchers affiliated with institutions that have contractual access to the All of Us data can register to use the Researcher Workbench.12
The All of Us Evenings with Genetics Research Program
The All of Us Evenings with Genetics (EwG) Research Program at the Department of Molecular and Human Genetics (DMHG) at Baylor College of Medicine (BCM) is funded as a community engagement partner by the National Institutes of Health (NIH) to recruit and train research scholars from diverse backgrounds to use the All of Us data for projects through its annual All of Us Biomedical Researcher (BR) Scholars Program (NIH Award #: OT2 OD031932).13 All of Us EwG functions as a complementary but separate program from the All of Us Research Program. While the work of All of Us EwG is independent, NIH All of Us program officers are informed of their progress and milestones. This communication ensures alignment with the broader All of Us goals of promoting researcher engagement with the All of Us data for precision medicine advancements.
The All of Us BR Scholars Program focuses on mentorship, programming languages (Python and R), data analysis techniques (statistical and genomic), and the execution of interdisciplinary group research projects that leverage All of Us data. The annual program begins with a Faculty Summit, an intensive four-day boot camp introducing the program, mentors, All of Us data, and its analysis on the Researcher Workbench. At the Faculty Summit, scholars form research teams and plan group research projects using the All of Us data that they complete during the program year.
Scholars trained in the All of Us BR Scholars Program are postdoctoral fellows or early-career faculty recruited from across the United States, including from Minority Serving Institutions (MSIs) and from UBR groups. Scholars have interests aligned with the All of Us Research Program to foster research that advances health equity. Scholars have diverse career goals, ranging from research to teaching to clinical care, and this diversity supports the formation of cross-disciplinary projects that may not occur in traditional university settings. Scholars who join the program are typically new to utilizing the All of Us data for research and are unfamiliar with the Researcher Workbench, including its unique tools. Many scholars have a foundation in statistics using software such as Statistical Analysis System (SAS) or Statistical Package for the Social Sciences (SPSS). However, few are familiar with basic programming and scripting, and fewer still are familiar with cloud computing environments, such as used in the Researcher Workbench.
The life sciences are rapidly transforming into data-driven fields. The rise of genome sequencing, concomitant functional genomics, and systems biology necessitates a growing need for computational expertise.14 This trend is highlighted by various global surveys that identify the demand for more programming, scripting, and other computational training for life scientists.15 Large databases, such as that offered by the All of Us Research Program, are indispensable to researchers in this new era. However, the biological life sciences higher education curricula have been slow to adopt bioinformatics skills and competencies training despite the growing need in recent years.16,17 To bridge this gap, beginner training is crucial for researchers to analyze the All of Us data.
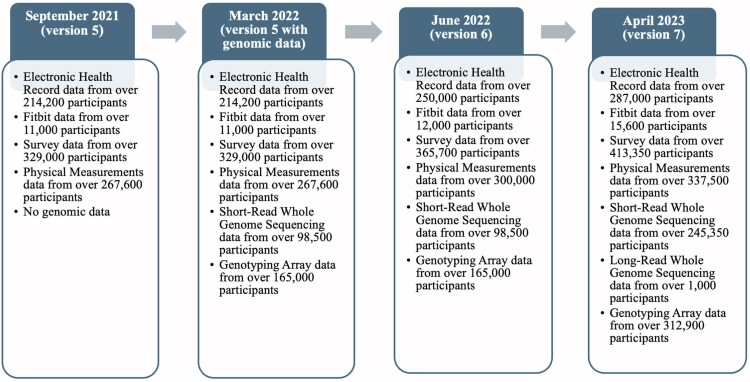
The All of Us data offer unique advantages and challenges compared to other popular databases, such as Gene Expression Omnibus, which houses gene expression data. The All of Us Research Program specifically focuses on utilizing data for precision medicine research. The data are incredibly valuable because of its unmatched diversity in participants, allowing researchers to explore the influence of individual variations across different populations. Additionally, the variety of data, including genomic, EHR, and lifestyle information, all collected from the same participants, enables research across these data sources. Figure 1 provides a summary of the types and quantity of data available across recent releases of the data.18 Since the late 1990’s, nationwide biobanks that provide genomic data linked to EHR data have emerged in some countries.19 Training researchers to access and analyze the EHR-linked biobank data from All of Us will advance precision medicine but also teach skills on handling “big data” that are increasingly becoming valuable with the rapidly growing amount of data in the life sciences.20
Figure 1.
Data types and their numbers available to researchers for recent releases of the All of Us data. Genomic data were first released in March 2022, two months before the Faculty Summit of the first All of Us BR Scholars Program in May 2022. Additional data were released shortly after the Faculty Summit in June 2022.
Objective
To enable rapid use of the Researcher Workbench for team projects, one of the main tasks of All of Us EwG is to teach scholars at the Faculty Summit how to access and analyze the All of Us data, regardless of scholars having limited skills and background in programming languages and cloud computing. This task was achieved by developing a training curriculum and delivering it over four one-hour sessions during the first annual Faculty Summit on May 19-21, 2022. This curriculum was evaluated for its effectiveness during and after the Faculty Summit so that the curriculum could be improved for future delivery at the Faculty Summit and other scholarly programs.
Materials and methods
Programming languages and computational tools survey
To assess the comfort and familiarity of incoming scholars with the programming languages and computational tools used in the Researcher Workbench, the All of Us EwG team prepared a survey included in the application for the All of Us BR Scholars Program. The results of this survey were used to inform the content of the training curriculum being created, ensuring that it matched the skill levels of incoming scholars.
Review of All of Us resources
Many resources from All of Us exist to train and help users on the Researcher Workbench. These resources include YouTube videos (https://www.youtube.com/@researcherworkbench4940/videos), the User Support Hub (https://support.researchallofus.org), and example projects within the Researcher Workbench. The All of Us EwG team evaluated and curated these resources into a synopsis to highlight those that are most useful in different anticipated research contexts, avoiding redundancy and ensuring that the training curriculum complemented and did not duplicate existing content.
Backward design of curriculum
To ensure effective training for the scholars on accessing and analyzing the All of Us data on the Researcher Workbench, the curriculum for the four Faculty Summit training sessions was developed using Understanding by Design by Wiggins and McTighe.21 The framework emphasizes backward design through a three-stage approach: (1) “Identify desired results,” such as what a student should know, understand, and be able to do; (2) “Determine acceptable evidence” that a student has understanding and proficiency; and (3) “Plan learning experiences and instructions.” An example of the backward design of the first session is provided in Supplementary Appendix S1.
The curriculum was also informed by All of Us EwG members’ collective decades of experience training biomedical students from the undergraduate to postgraduate level. PowerPoint slides were prepared using the All of Us Research Program’s Registered and Controlled Tier Curated Data Repository (CDR) version 5.
Additional resources selected and curated
In addition to the live training materials, the All of Us EwG team selected and curated resources to give scholars for self-paced, asynchronous learning after the Faculty Summit. The resources were chosen to provide support to scholars new or wanting to grow their programming and data analysis skills, thereby becoming more productive on the Researcher Workbench. These materials included books and an internally developed resource, the “Summit Binder.” The Summit Binder focused on topics deemed too time-intensive for live training but valuable for completing projects on the Researcher Workbench.
Costs and personnel for training
The primary costs for the training sessions were buying the books and printing the Summit Binder, which amounted to approximately $6000, and hiring and paying additional personnel, which amounted to approximately $7000, excluding the salary of the instructors themselves. Ten BCM graduate students, postdoctoral fellows, and research associates with strong programming and data analysis skills were recruited as coaches to help scholars attending live training sessions navigate the Researcher Workbench. The coaches were thoroughly trained in the curriculum prior to the Faculty Summit.
The instructors for the training sessions were All of Us EwG staff and included those who helped develop the curriculum. Instructors had doctoral degrees in fields such as human genetics, evolution and population, statistical genetics, or medicine. Most instructors had experience in biomedical research using programming languages and statistics. There were three instructors for the first training session, two for the second, two for the third, and one for the fourth.
Researcher Workbench costs for the training sessions were kept to a minimum. The costs within a Google Cloud project (workspace) on the Researcher Workbench are associated with data retrieval, analysis, and storage.22 Every new user on the Researcher Workbench receives a free $300 initial credit to get started. Scholars were expected to use up to $10 of this initial credit to complete all four training sessions at the Faculty Summit. The training examples and exercises at the Faculty Summit were intentionally designed to minimize data retrieval and analysis costs to less than $10, while still providing a scholar with a comprehensive and efficient training experience. Once a scholar started their own project outside the Faculty Summit, they continued to accrue additional costs beyond $10.
Evaluation and assessment
The curriculum was evaluated using feedback surveys that were collected at the end of each training session at the Faculty Summit. After the Faculty Summit, the All of Us EwG staff provided ongoing support to scholars embarking on their team project on the Researcher Workbench through virtual meetings and emails to answer questions. These communications identified gaps in knowledge or areas where scholars needed further reinforcement for a better or more timely understanding. At least two one-hour virtual meetings were held within seven months of the Faculty Summit with each research team. Additionally, two teams requested a third virtual meeting. All teams initiated anywhere from one to four email conversations with staff by the end of the program year.
Results
Programming languages and computational tools survey results
Table 1 summarizes the survey results for programming languages and computational tools used by scholars. The data revealed that fewer than 15% of the scholars reported prior experience or expertise in any specific language or tool.
Table 1.
Programming languages and computational tools survey and results.
| ||||||
| Please rank (1-5 stars) your familiarity with the following: | ||||||
| Five stars | Four stars | Three stars | Two stars | One star | Average response | |
|
|
||||||
| Jupyter Notebook | 0 | 3 | 3 | 8 | 21 | 1.66 |
| R programming language | 1 | 4 | 11 | 15 | 4 | 2.51 |
| Python programming language | 0 | 1 | 11 | 14 | 9 | 2.11 |
| All of Us Researcher Workbench | 0 | 0 | 4 | 12 | 19 | 1.57 |
| Plink | 1 | 1 | 3 | 5 | 25 | 1.51 |
| Hail | 0 | 0 | 1 | 5 | 29 | 1.20 |
Synopsis of All of Us resources
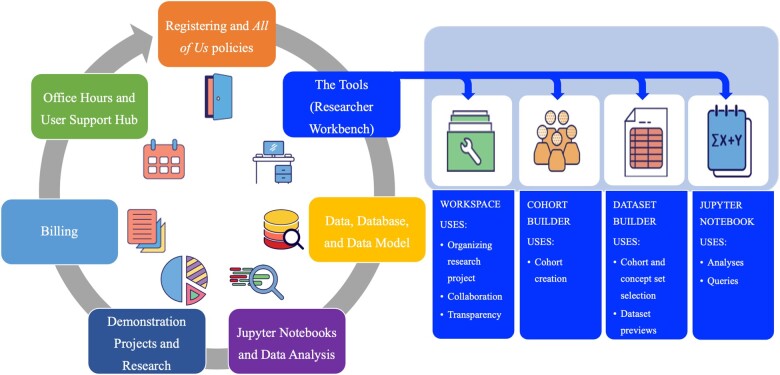
The synopsis of existing All of Us training and support resources is visualized in Figure 2. The synopsis was divided into topics, each offering a comprehensive list of resources.
Figure 2.
Visualization of the synopsis of available All of Us resources. The synopsis detailed resources on registering to become a new user on the All of Us Researcher Workbench and the All of Us policies. These topics and their resources were also detailed: Researcher Workbench Tools; Data, Database, and Data Model; Jupyter Notebooks and Data Analysis; Demonstration Projects and Research; Billing; and Office Hours and User Support Hub.
Training curriculum outline, exercises, and slides
Table 2 outlines the curriculum for the four training sessions, including the session titles, detailed outlines, and corresponding exercises. This curriculum was designed based on insights from the programming languages and computational tools survey (Table 1), along with the comprehensive review of existing All of Us resources (Figure 2). The training slides and exercises delivered at the Faculty Summit can be viewed online.23–26
Table 2.
Titles, outlines, and exercises of the four training sessions
| Training session titles, outlines, and exercises |
| Session 1: The All of Us Researcher Workbench and creating a project |
|
| Session 2: Creating an All of Us dataset |
|
| Session 3: Analyzing an All of Us dataset and other helpful tools |
|
| Session 4: Genomics overview and analyzing All of Us genomic data |
|
Scholar reaction to the training sessions
During the training sessions, scholars listened attentively and asked questions about the material presented to them, seeking clarification from instructors. Questions were more frequent during the fourth training session when analyzing genomic data. During the first three training sessions, most scholars were successful in completing the exercises provided but with considerable assistance from the coaches to provide technical support on the Researcher Workbench. The instructors also provided additional assistance to scholars when necessary.
Additional resources
Scholars received books about analyzing biological data with Python or R.27–31 Scholars also received the internally developed Summit Binder that included: (1) a curated list of online tutorials for Python, R, and Hail programming languages; (2) comprehensive code examples in both Python and R for common statistical analyses; (3) annotations of previous publications using All of Us data, categorized by the type of statistical analysis employed; and (4) a recommended list of example projects on the Researcher Workbench for practical application. A PDF file of the 2024 version of the Summit Binder can be viewed online.32
Evaluation results
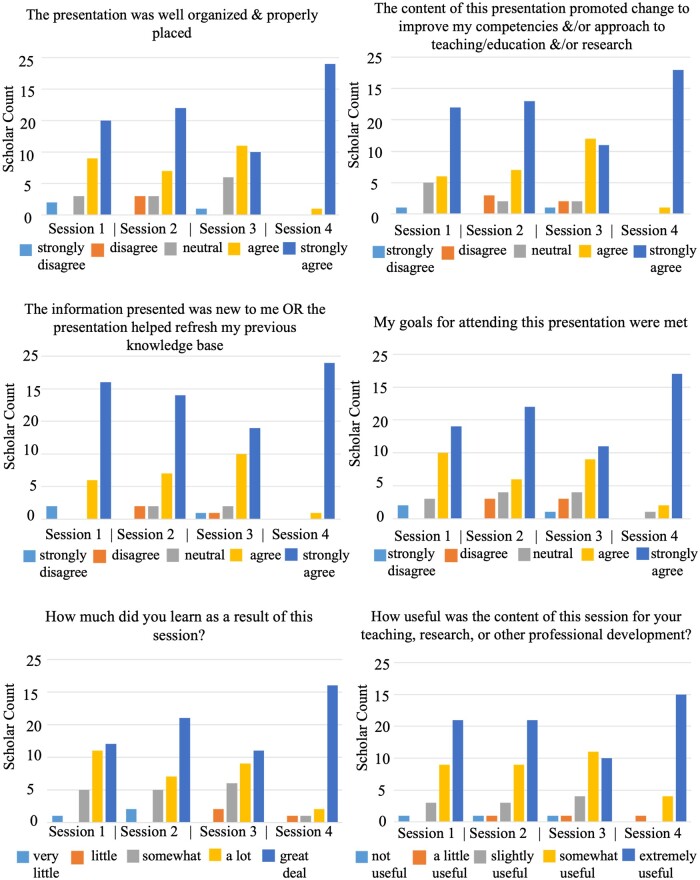
Evaluation results indicated that the curriculum successfully introduced scholars to the Researcher Workbench but also highlighted areas where scholars needed further training. Figure 3 summarizes the results of the six Likert scale questions in the feedback surveys. Table 3 presents the results, including comments and their common themes, from the feedback surveys when scholars were asked whether they could identify opportunities to incorporate improvements into their teaching and research. Supplementary Appendix S2 presents further comments grouped by subject from the feedback surveys when the scholars were asked for “positive comments,” “constructive comments,” or “what future topics would meet their educational needs.”
Figure 3.
Results from the six Likert scale questions in the feedback surveys for the four training sessions.
Table 3.
Results from the feedback surveys when the scholars were asked if they could identify opportunities to incorporate improvements into their teaching and research based on each session.
| Question | Session | Response |
|---|---|---|
| Based on what you have discussed today, have you identified opportunities to incorporate improvements into your teaching or research? | Session 1 |
|
| Session 2 |
|
|
| Session 3 |
|
|
| Session 4 |
|
|
| If yes, please explain what improvements you intend to make | Session 1 |
|
| Session 2 |
|
|
| Session 3 |
Improvement of exploring the Researcher Workbench and learning how to use it
|
|
| Session 4 |
|
Listed are the scholars’ explanations of these improvements as well as common themes identified across these explanations.
Table 4 captures the most common questions scholars asked following the Faculty Summit in virtual meetings and emails. These questions highlighted areas in which scholars required additional support.
Table 4.
Most frequently asked questions by scholars during virtual meetings and emails after the Faculty Summit
| Category | Questions |
|---|---|
| Billing | When am I being billed as I do research on the Researcher Workbench? |
| Can we pool our initial credits on the Researcher Workbench together on a project? | |
| Examples | Are there comprehensive examples of a research project with All of Us data from its beginning to publication? |
| Dataset creation | How do I create a control cohort for my trait of interest? |
| How can I restrict the size of my cohorts randomly? | |
| Can you show an example of creating a dataset from beginning to end? | |
| Data wrangling | How do I join datasets together? |
| How do I create a new dataset column based on other data? | |
| How do I filter my data? | |
| Storage | How do I copy files between workspaces [projects]? |
| How do I share files I created in a workspace, such as VCF files, with other team members in that same workspace? | |
| Genomics | How do I access the whole genome sequencing (WGS) and genotyping array data on the Researcher Workbench? |
| How do you get the VCF files and how much does it cost? | |
| What quality controls have been applied to the genomic data? | |
| What quality controls should I now apply to the genomic data? | |
| How do I access annotations from All of Us for the genomic data? | |
| How do you add the phenotypic data to the genomic data? | |
| How do you filter the genomic data to your variants of interest? | |
| Where can I get more information on using Hail? | |
| How do I convert the Hail MatrixTable to other file formats? |
Discussion
Achievements of the Faculty Summit training
All of Us EwG successfully trained scholars to access and analyze the All of Us data. This success was confirmed by the high feedback scores and positive comments at the Faculty Summit as well as the initiation and execution of team research projects on the Researcher Workbench. The training curriculum continues to be refined based on feedback.
Session strengths and usefulness highlighted by themes found in survey feedback
As shown in Figure 3, the session ratings varied but were generally positive, with the highest ratings for the fourth session.
As shown in Table 3, three common themes emerged of improvements scholars intended to make to their personal research and courses based on the sessions. First and most importantly, across all sessions, scholars were equipped to explore and further learn the Researcher Workbench after the Faculty Summit, such as creating workspaces, creating datasets, and using Jupyter Notebooks for data analysis. Comments such as “learned how to create workspaces [projects] in All of Us and use the notebook” were common.
Second, after the first two sessions, some scholars intended to improve student courses by using the Researcher Workbench. The first session introduced the Research Hub (https://researchallofus.org), a public website providing an overview of the All of Us Research Program and its Researcher Workbench. It includes a “Data Browser” where one can explore the data at an aggregate-level. Scholars saw opportunities for using this public website in their courses from comments such as “can use the public tier to help students design research questions.” The second session introduced creating datasets and viewing them in Jupyter Notebooks on the Researcher Workbench. Scholars saw opportunities to use these tools to introduce programming to students from comments such as “introducing coding to the biochemistry students.”
Third, after the first, second, and fourth sessions, some scholars intended to improve their research using the Researcher Workbench. From the first session, scholars saw opportunities to use the Researcher Workbench to generate research project ideas from comments such as “I plan on using the All of Us research workbench to generate project ideas and hypothesis.” From the second session, scholars saw opportunities to use the new skills they learned for research from comments such as “gave a great overview to create a dataset that'll later be foundation experiment.” From the fourth session, scholars saw opportunities to use the genomic analysis they learned for research from comments such as “looking forward to running HAIL on our data.”
Positive feedback on sessions
The positive comments received in the feedback survey (Supplementary Appendix S2) highlighted the helpful nature of the sessions. Scholars highly praised the support received from coaches. Some scholars commended the instructors and found the handouts to be useful. Scholars appreciated the fourth session’s teaching approach where they worked alongside the instructor on the Researcher Workbench. Comments such as “…I really appreciated how [the speaker] explained everything and we clicked through the exercises together” were common for the fourth session.
Constructive feedback on sessions
The constructive comments received in the feedback survey (Supplementary Appendix S2) highlighted that the sessions were too fast-paced and could benefit from live demonstrations. Comments such as “the presentation itself was fast paced and hard to follow” and “live demonstration would be helpful…” were noted. Scholars requested more in-depth training to further learn the Researcher Workbench. Other scholar suggestions included hearing the speaker better, giving presentation slides beforehand, moving all the training to earlier in the Faculty Summit, and learning in small groups.
Limitations indicated by questions
Post-Faculty Summit questions (Table 4) highlighted additional limitations. Scholars needed a more detailed explanation of billing costs and more emphasis on example projects with matching publications. Although six example projects were mentioned in the first session, more content on these example projects was needed. Scholars also needed more curated examples of handling phenotypic data and dataset creation, as well as guidance on file storage and transfer.
Post-Faculty Summit questions also identified the need for more in-depth discussion of genomic data analysis. Notably, some questions raised, such as adding phenotypic data and filtering variants, were already addressed in the fourth session. This need might be attributed to scholars lacking familiarity with Hail, the cloud-native language recommended for All of Us genomic data analysis due to its scalability and efficiency.33 Since Hail was a new tool for most scholars (Table 1), additional practice or targeted discussions focused on data manipulation techniques with Hail would be beneficial.
Team projects started on the Researcher Workbench and their funding
The training sessions and team-building activities at the Faculty Summit successfully led to scholars forming research teams and initiating several research projects on the Researcher Workbench: “Biological determinants in opioid use, lupus and cardiovascular disease” (genomic and non-genomic), “Evaluation of prostate cancer genetic variants using All of Us” (genomic), “Prevalence of pathogenic BRCA1 and BRCA2 mutations [for breast cancer] among Latinas in the All of Us cohort” (genomic), “Dissecting the influence of perceived discrimination and genetic liability on mental health” (genomic and non-genomic), “Pathways to Science Identity (PSI): Ready to apply pressure” (educational), and “Identify risk factors for IBD using the All of Us database” (genomic and non-genomic).
Project themes fell into two main categories: educational and genomic research. The educational project aimed to introduce undergraduate students to the Researcher Workbench and its functionality. Three of the genomic projects also incorporated non-genomic data analysis of EHR condition, drug, or survey data.
To pay for the Google Cloud costs of these projects on the Researcher Workbench after the Faculty Summit, the research teams applied for and received seed grants funded by the All of Us EwG Research Program’s NIH award. Grants of up to $30 000 were provided until the end of the program year to cover the Google Cloud costs of projects as well as various other costs, including travel, community outreach, and laboratory supplies. Scholars had spent up to $10 on the Google Cloud to complete training on the Researcher Workbench at the Faculty Summit, but to complete a whole research project on the Researcher Workbench after the Faculty Summit, teams spent anywhere from about $50-$1000 depending on the quantity of data and type of analysis used. A non-genomic educational project cost about $50, but a genomic project with a couple genome-wide association studies in addition to other analyses cost about $1000.
Research teams were encouraged to acquire other funding resources to pay their ongoing costs while using the Researcher Workbench by applying for NIH or other federal, private, or institutional grants. Also available to research teams was an additional $300 credit from the Google Cloud Platform (GCP), which can be used temporarily.34,35 It is recommended that researchers apply for these credits when they are close to needing a permanent billing solution.
Return of value to communities
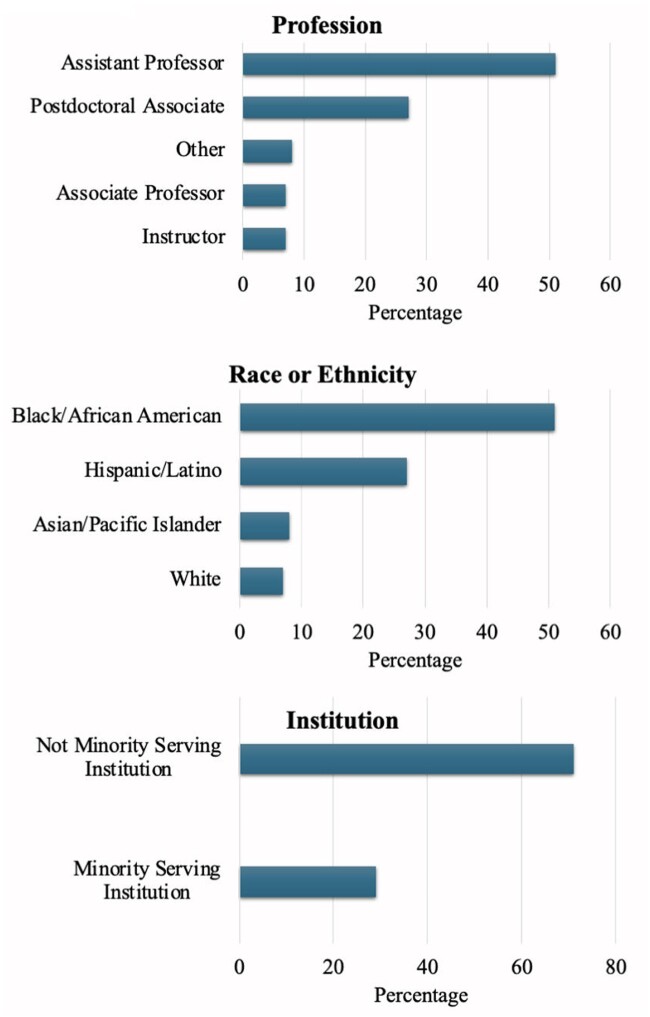
Scholar recruitment included those from MSIs and UBR groups. Any postdoctoral fellow or early-career faculty whose research or teaching aligned with the mission of the All of Us Research Program was eligible to apply and be a scholar. Figure 4 presents the data for various attributes, including race or ethnicity, of the 118 scholars from the past three years of the All of Us BR Scholars Program. There were 35 scholars in the 2022-2023 All of Us BR Scholars Program, 40 in the 2023-2024 All of Us BR Scholars Program, and 43 in the 2024-2025 All of Us BR Scholars Program. Most scholars were from racial or ethnic minorities or from an MSI.
Figure 4.
Attributes of all 118 scholars trained on the Researcher Workbench at the Faculty Summit for three years (2022-2023, 2023-2024, and 2024-2025) of the All of Us BR Scholars Program.
For the first year of the All of Us BR Scholars Program, nearly all research teams chose projects focused on UBR populations and health concerns where disparities among UBR populations exist. Four projects focused on studying cohorts of racial or ethnic minorities in the areas of breast cancer, prostate cancer, inflammatory bowel disease, and mental health. A fifth project studied lupus across all populations, but the disease is known to disproportionally afflict racial and ethnic minority women.36 A sixth educational project implemented the Researcher Workbench into the curriculum for an undergraduate class at an MSI.
The research team studying breast cancer, using a seed grant from All of Us EwG, held a special public community event for Latina women at their MSI institution that brought awareness of breast cancer risks, including genetic risk, and prevention.37 This event complemented the mission of their project on the Researcher Workbench for studying breast cancer in Latina women. The research teams studying breast cancer and prostate cancer, as well as the educational team, presented their research and educational outcomes at various scientific conferences to audiences of undergraduate students, graduate students, and faculty.
The curriculum developed can continue to be a valuable tool to train researchers on the Researcher Workbench by being adapted and implemented by other institutions. The curriculum was designed to help researchers who register on the Researcher Workbench but fail to make progress on projects because of a lack of familiarity with programming, cloud computing, and the unique tools of the Researcher Workbench. Overall, the All of Us BR Scholars Program and its training curriculum for the Researcher Workbench were catalysts for raising awareness and training researchers on the Researcher Workbench that led to projects largely addressing health concerns in UBR populations, including in subsequent years of the program beyond the first year.
Summary of curriculum changes based on feedback
Several changes have been implemented in the training curriculum since its first year. Only one or two instructors now teach each training session. All sessions now have scholars working on the Researcher Workbench simultaneously with the instructor. This approach had not been implemented in some sessions because of the exercise time given for practice. Exercises are still included in sessions so that after scholars work on the Researcher Workbench with the instructor, they still get further practice with the exercises that reinforce and assess what they have learned, as recommended in stage two of the backward design approach of Wiggins and McTighe.21
To manage pace, some content from the second session is extended to the third session, with backing up Jupyter Notebooks moved to materials available after the Faculty Summit. By prioritizing teaching, only the essential skills for using the Researcher Workbench in the Faculty Summit sessions, a manageable pace that optimizes learning and retaining knowledge is ensured. Exercises are now standardized to facilitate peer support. One more Faculty Summit session is added that covers frequently asked genomics topics, such as file types, quality control filters, and annotations with Hail, so that there are now five sessions instead of four. To accommodate, the first session is moved to a virtual session before the Faculty Summit. While more Python and R training was requested, it is not included in the Faculty Summit because of time constraints. Instead, four virtual sessions on Python and four virtual sessions on R are delivered after the Faculty Summit. Additional content on example projects, billing costs, and dataset wrangling are also delivered as virtual sessions after the Faculty Summit. Combined, the Faculty Summit training sessions and subsequent virtual sessions provide robust training to use the Researcher Workbench.
Conclusions
The training curriculum for the Researcher Workbench was successful in its inaugural year. Developed through careful assessment and backward design, the curriculum was delivered over four one-hour sessions. Feedback from the Faculty Summit and subsequent interactions continue to guide further improvements, making the curriculum more effective.
The training successfully introduced researchers to the Researcher Workbench and facilitated project initiation. This introduction works especially well in the context of a team environment that leverages diversity in technical expertise and disciplinary knowledge, as shown by the experiences of scholars in the first year of the All of Us BR Scholars Program. During 2024, more tools, such as RStudio and SAS, became available for data analysis on the Researcher Workbench, broadening researchers’ options. Scholars attending the 2023 and 2024 iterations of the Faculty Summit were trained via new versions of the curriculum, which has led to other projects focusing on health concerns in UBR populations.
In its first year, the All of Us BR Scholars Program achieved a significant milestone when one research team secured a one-year $50 000 grant from Prostate Cancer Research to support their prostate cancer project. Since 2022, multiple research teams have presented their research at various conferences across the United States. The breast cancer research team gave oral presentations at the 2022 Annual Biomedical Research Conference for Minoritized Students (ABRCMS), 2023 ABRCMS, and 2023 Society for Advancing Chicanos/Hispanics and Native Americans in Science (SACNAS) National Diversity in STEM (NDiSTEM) Conference.38–40 They also gave a poster presentation at the 2023 16th American Association of Cancer Research Conference on the Science of Cancer Health Disparities in Racial/Ethnic Minorities and the Medically Underserved.41 The prostate cancer research team gave oral presentations at the 2023 ABRCMS and 2023 SACNAS NDiSTEM Conference.39–40 They also gave a poster presentation at the 2023 International Genetic Epidemiology Society Annual Meeting.42 The educational outreach team gave an oral presentation at the 2022 ABRCMS.38 A pregnancy loss research team formed in year two of the program gave a poster presentation at the 2024 Society for Reproductive Investigation Annual Scientific Meeting.43 A hypertensive heart disease team also formed in year two of the program gave a poster presentation at the 2024 Pulmonary Hypertension Association International PH Conference.44
Outside the All of Us BR Scholars Program, the training curriculum for the Researcher Workbench has been adapted and used for other scholarly programs and audiences. Another All of Us EwG team adapted it into a year-round Train-the-Trainer Series for researchers learning the Research Workbench.45,46 Hundreds of users on the Researcher Workbench have attended this virtual series. Building on the core curriculum, a condensed version was delivered for the 2023 ASHG virtual interactive workshop47 and will be modified and delivered again for the upcoming 2024 ASHG in-person workshop.
Overall, this curriculum sets a strong foundation for training researchers to access and analyze All of Us data. As All of Us continues to update the data and the Researcher Workbench, the curriculum will also be adapted, ensuring its continued relevance and impact. The latest curriculum slides, using the All of Us Research Program’s Controlled Tier CDR version 7, are available online for others to utilize with proper acknowledgement.48 This versatile curriculum will continue to be used to empower a broad range of researchers to effectively analyze All of Us data on the Researcher Workbench.
Supplementary Material
Acknowledgments
The All of Us Research Program would not be possible without the partnership of its participants. We gratefully acknowledge All of Us participants for their contributions, without whom the training curriculum for the All of Us Researcher Workbench would not have been possible. We also thank the NIH’s All of Us Research Program for providing the available participant data.
Contributor Information
Julie R Coleman, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Jasmine N Baker, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Shamika Ketkar, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Ashley M Butler, Department of Pediatrics, Baylor College of Medicine, Houston, TX 77030, United States.
LaTerrica Williams, Department of Pediatrics, Baylor College of Medicine, Houston, TX 77030, United States.
Latanya Hammonds-Odie, Department of Biological Sciences, Georgia Gwinnett College, Lawrenceville, GA 30043, United States.
Elizabeth G Atkinson, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States; Jan and Dan Duncan Neurological Research Institute, Texas Children’s Hospital, Houston, TX 77030, United States.
Debra D Murray, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Brendan Lee, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Kim C Worley, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, United States.
Author contributions
The authors confirm their contributions to the article as follows: study conception and design: Debra D. Murray, Kim C. Worley, Brendan Lee; curriculum design and data science staff: Julie R. Coleman, Jasmine N. Baker, Elizabeth G. Atkinson, Shamika Ketkar, Kim C. Worley; data collection: Julie R. Coleman, Jasmine N. Baker, Shamika Ketkar, Ashley M. Butler, LaTerrica Williams; analysis and interpretation of results: Julie R. Coleman, Jasmine N. Baker, Shamika Ketkar, Elizabeth G. Atkinson. All authors assisted with draft manuscript preparation, reviewed the results, approved the final version of the manuscript, and agree to be accountable for all aspects of the work.
Supplementary material
Supplementary material is available at Journal of the American Medical Informatics Association online.
Funding
This work was supported by a National Institutes of Health grant number OT2 OD031932 to Brendan Lee and Debra D. Murray. The All of Us Research Program is supported by the National Institutes of Health, Office of the Director: Regional Medical Centers: 1 OT2 OD026549; 1 OT2 OD026554; 1 OT2 OD026557; 1 OT2 OD026556; 1 OT2 OD026550; 1 OT2 OD 026552; 1 OT2 OD026553; 1 OT2 OD026548; 1 OT2 OD026551; 1 OT2 OD026555; IAA #: AOD 16037; Federally Qualified Health Centers: HHSN 263201600085U; Data and Research Center: 5 U2C OD023196; Biobank: 1 U24 OD023121; The Participant Center: U24 OD023176; Participant Technology Systems Center: 1 U24 OD023163; Communications and Engagement: 3 OT2 OD023205; 3 OT2 OD023206; and Community Partners: 1 OT2 OD025277; 3 OT2 OD025315; 1 OT2 OD025337; 1 OT2 OD025276.
Conflicts of interest
The authors declare no competing interests directly or indirectly related to the work reported here. All authors benefit indirectly from funding that BCM and DMHG receive from a joint venture with Baylor Genetics, but the activities reported here are not related to that funding.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. Bianchi DW, Brennan PF, Chiang MF, et al. The All of Us research program is an opportunity to enhance the diversity of US biomedical research. Nat Med. 2024;30(2):330-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372(9):793-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. All of Us Research Program Investigators. The "All of Us" research program. N Engl J Med. 2019;381(7):668-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cronin RM, Jerome RN, Mapes B, et al. Development of the initial surveys for the All of Us research program. Epidemiology. 2019;30(4):597-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. All of Us Research Program. Researcher workbench. Accessed June 19, 2024. https://www.researchallofus.org/data-tools/workbench
- 6. All of Us Research Program. What data are available for download in the Researcher Workbench? Accessed June 6, 2024. https://www.researchallofus.org/faq/what-data-is-available-for-download-in-the-researcher-workbench
- 7. All of Us Research Program. Cohort Builder & Cohort Review [Video]. Accessed March 1, 2024. https://youtu.be/G6_GG2CJ9mA
- 8. All of Us Research Program. Dataset Builder & Concept Sets [Video]. Accessed March 1, 2024. https://youtu.be/cUuDKUxjQoo
- 9. All of Us Research Program. Notebooks & Code Snippets [Video]. Accessed March 4, 2024. https://youtu.be/NvMWBlVyyUU
- 10. Hail Team. Hail. Accessed February 3, 2024. https://github.com/hail-is/hail
- 11. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. All of Us Research Program. Institutional Agreements. Accessed March 5, 2024. https://www.researchallofus.org/institutional-agreements
- 13. Baylor College of Medicine. NIH award supports diverse researchers in All of Us Research Program. Accessed April 5, 2024. https://www.bcm.edu/news/nih-award-supports-diverse-researchers-in-all-of-us-research-program.
- 14. National Research Council. A New Biology for the 21st Century. Washington, DC: National Academies Press; 2009. [Google Scholar]
- 15. Attwood TK, Blackford S, Brazas MD, et al. A global perspective on evolving bioinformatics and data science training needs. Briefings Bioinf. 2019;20(2):398-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Işık EB, Brazas MD, Schwartz R, et al. Grand challenges in bioinformatics education and training. Nat Biotechnol. 2023;41(8):1171-1174. [DOI] [PubMed] [Google Scholar]
- 17. Pevzner P, Shamir R. Computing has changed biology—biology education must catch up. Science. 2009;325(5940):541-542. [DOI] [PubMed] [Google Scholar]
- 18. All of Us Research Program. Release Notes. Accessed August 21, 2024. https://support.researchallofus.org/hc/en-us/sections/360006773231-Release-Notes
- 19. Abul-Husn NS, Kenny EE. Personalized medicine and the power of electronic health records. Cell. 2019;177(1):58-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dall'Alba G, Casa PL, Abreu FP, Notari DL, de Avila ESS. A survey of biological data in a big data perspective. Big Data. 2022;10(4):279-297. [DOI] [PubMed] [Google Scholar]
- 21. Wiggins GP, McTighe J. Understanding by Design. Expanded 2nd ed. Alexandria, VA: Association for Supervision and Curriculum Development; 2005. [Google Scholar]
- 22. All of Us Research Program. What exactly am I paying for?. Accessed June 6, 2024. https://support.researchallofus.org/hc/en-us/articles/360040040852-What-exactly-am-I-paying-for
- 23. Coleman J, Baker J, Ketkar S, et al. Training I: the All of Us researcher workbench and creating a project [version 1; not peer reviewed]. F1000Research. 2023;12:1284 (slides). 10.7490/f1000research.1119636.1 [DOI] [Google Scholar]
- 24. Coleman J, Baker J, Ketkar S, et al. Training II: creating an All of Us dataset [version 1; not peer reviewed]. F1000Research. 2023;12:1298 (slides). 10.7490/f1000research.1119638.1 [DOI] [Google Scholar]
- 25. Coleman J, Baker J, Ketkar S, et al. Training III: analyzing an All of Us dataset and other helpful tools [version 1; not peer reviewed]. F1000Research. 2023;12:1346 (slides). 10.7490/f1000research.1119642.1 [DOI] [Google Scholar]
- 26. Coleman J, Baker J, Ketkar S, et al. Training IV: genomics overview and analyzing All of Us genomic data [version 1; not peer reviewed]. F1000Research. 2023;12:1406 (slides). 10.7490/f1000research.1119650.1 [DOI] [Google Scholar]
- 27. Allesina S, Wilmes M. Computing Skills for Biologists: A Toolbox. Princeton, NJ: Princeton University Press; 2019. [Google Scholar]
- 28. Libeskind-Hadas R, Bush EC. Computing for Biologists: Python Programming and Principles. Cambridge; New York: Cambridge University Press; 2014. [Google Scholar]
- 29. Haddock SHD, Dunn CW. Practical Computing for Biologists. Sunderland, MA: Sinauer Associates, Inc.; 2010. [Google Scholar]
- 30. Quicke DLJ, Butcher BA, Welton RAK. Practical R for Biologists: An Introduction. Wallingford, Oxfordshire; Boston, MA: CAB International; 2021. [Google Scholar]
- 31. Mills M, Barban N, Tropf FC. An Introduction to Statistical Genetic Data Analysis. Cambridge, MA: The MIT Press; 2020. [Google Scholar]
- 32. Coleman JR, Baker JN, Ketkar S, et al. Resources for the All of Us researcher workbench. Zenodo. Accessed August 23, 2024. 10.5281/zenodo.12007807 [DOI]
- 33. Hail Team. Scalable Genetic Analysis. Accessed February 4, 2024. https://www.broadinstitute.org/videos/broade-hail-introduction-hail-tool-scalable-genomic-analysis
- 34. All of Us Research Program. Utilizing Free GCP Credits on the Workbench While Setting up a Long-term Billing Solution. Accessed June 6, 2024. https://support.researchallofus.org/hc/en-us/articles/7505124745364-Utilizing-Free-GCP-Credits-on-the-Workbench-While-Setting-Up-a-Long-term-Billing-Solution
- 35. Google Cloud. Free Cloud features and trial offer. Accessed June 6, 2024. https://cloud.google.com/free/docs/free-cloud-features#free-trial
- 36. Reid MR, Danguecan AN, Colindres I, et al. An ecological approach to understanding and addressing health inequities of systemic lupus erythematosus. Lupus. 2023;32(5):612-624. [DOI] [PubMed] [Google Scholar]
- 37. Visbal AP. Cafecito Para La Salud UHD 2023. [Video]. Accessed August 19, 2024. https://youtu.be/LidrqxVzvoQ
- 38. Murray DD, Sander CT, Visbal AP. What Can We Do with the Largest, Diverse Dataset?: Two Approaches Using the All of Us Workbench. Presented at: Annual Biomedical Research Conference for Minoritized Students. Anaheim, CA; November 9-12, 2022.
- 39. Lloyd SM, Gavile CM, Katsonis P. What Can We Do with the Largest, Diverse Dataset?: Two Approaches using the All of Us Workbench. Presented at: Annual Biomedical Research Conference for Minoritized Students. Phoenix, AZ; November 15-18, 2023. https://vimeo.com/883479533
- 40. Samayoa C, Krieger KL, Murray DD. Early career faculty research projects using All of Us data: two approaches using the all of us workbench. Presented at: Society for Advancing Chicanos/Hispanics and Native Americans in Science National Diversity in STEM Conference. Portland, OR; October 26-28, 2023.
- 41. Samayoa C. Prevalence of pathogenic BRCA1/2 variants among Latinas in the All of Us cohort. Poster Presented at: 16th AACR Conference on the Science of Cancer Health Disparities in Racial/Ethnic Minorities and the Medically Underserved. Orlando, FL; September 29-October 2, 2023.
- 42. Lewis DD. Evolutionary Action analysis of ultra-rare genetic variants in African American men with prostate cancer from the All of Us Research Program. Poster presented at: International Genetic Epidemiology Society Annual Meeting. Nashville, TN; November 5-6, 2023.
- 43. Cope D, Carreon SA, Moon CI, et al. Perceived medical discrimination among pregnant Black and Latina participants in the All of Us database. Poster Presented at: Society for Reproductive Investigation Annual Scientific Meeting. Vancouver, BC; March 12-16, 2024.
- 44. Chakafana G, Xuan M, Winston-Gray C, et al. Identification of key genetic factors associated with hypertensive heart disease in African Americans in the United States. Poster presented at: Pulmonary Hypertension Association International PH Conference. Indianapolis, IN; August 15-18, 2024.
- 45. Ritter DI, Byun J, Wang J, et al. Experiences in providing a community educational resource for the All of Us Researcher Workbench. J Am Med Inform Assoc. 2024;31(12):2952-2957. 10.1093/jamia/ocae226 [DOI] [PMC free article] [PubMed]
- 46. Ritter DI, Byun J, Wang J, et al. All of Us/Evenings with Genetics researcher workbench support train-the-trainer slide decks. Zenodo. Accessed August 23, 2024. 10.5281/zenodo.11453503 [DOI]
- 47. Coleman JR, Ketkar S, Byun J. Workshop: accessing and analyzing All of Us biomedical and genomic data via the researcher workbench. American Society of Human Genetics; September 21, 2023. Accessed August 22, 2024. https://learning.ashg.org/products/workshop-accessing-and-analyzing-all-of-us-biomedical-and-genomic-data-via-the-researcher-workbench
- 48. Coleman JR, Baker JN, Ketkar S, et al. All of Us Evenings with Genetics data trainings for the 2024 biomedical researchers faculty summit. Zenodo. Accessed August 23, 2024. 10.5281/zenodo.12009041 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.