Abstract
The Mexican fruit fly, Anastrepha ludens, is a polyphagous true fruit fly (Diptera: Tephritidae) considered 1 of the most serious insect pests in Central and North America to various economically relevant fruits. Despite its agricultural relevance, a high-quality genome assembly has not been reported. Here, we described the generation of a chromosome-level genome for the A. ludens using a combination of PacBio high fidelity long-reads and chromatin conformation capture sequencing data. The final assembly consisted of 140 scaffolds (821 Mb, N50 = 131 Mb), containing 99.27% complete conserved orthologs (BUSCO) for Diptera. We identified the sex chromosomes using 3 strategies: (1) visual inspection of Hi-C contact map and coverage analysis using the HiFi reads, (2) synteny with Drosophila melanogaster, and (3) the difference in the average read depth of autosomal vs sex chromosomal scaffolds. The X chromosome was found in 1 major scaffold (100 Mb) and 8 smaller contigs (1.8 Mb), and the Y chromosome was recovered in 1 large scaffold (6.1 Mb) and 35 smaller contigs (4.3 Mb). Sex chromosomes and autosomes showed considerable differences of transposable elements and gene content. Moreover, evolutionary rates of orthologs of A. ludens and Anastrepha obliqua revealed a faster evolution of X-linked, compared with autosome-linked, genes, consistent with the faster-X effect, leading us to new insights on the evolution of sex chromosomes in this diverse group of flies. This genome assembly provides a valuable resource for future evolutionary, genetic, and genomic translational research supporting the management of this important agricultural pest.
Keywords: Mexican fruit fly (mexfly), fraterculus group, genome assembly, Hi-C, PacBio HiFi, long-read sequencing, sex chromosomes, faster-X effect
Introduction
Anastrepha is a highly diverse genus of the Tephritidae family (true fruit flies) distributed in the tropics and subtropics of the Americas (Norrbom and Kim 1988). Some of the Anastrepha species are of major economic importance due to their ability to infest a wide range of fleshy fruits in which larvae will complete development (Aluja 1994). Among them, Anastrepha ludens (Loew), commonly known as the Mexican fruit fly, is a highly polyphagous pest that has been documented to attack at least 50 fruit species including important mango and citrus commercial cultivars (Baker et al. 1944; Norrbom and Kim 1988; Norrbom 2022). The geographical range of the Mexican fruit fly extends from the southern United States through Central America (Foote et al. 1993; Hernandez-Ortiz and Aluja 1993; Ruiz-Arce et al. 2015; Dupuis et al. 2019). However, studies have indicated that its geographical distribution could likely increase due to climate change (Hill et al. 2016; Skendžić et al. 2021), which may seriously impact the economy and food security of newly affected countries. Notably, 2 of the most important regulatory agencies in the world, the Animal and Plant Health Inspection Service (APHIS), from the United States of America, and the European Food Safety Authority (EFSA), from the European Union, consider that Mexican fruit fly satisfy the criteria for imposing quarantine on products infested with this pest (EFSA PLH Panel et al. 2020; APHIS 2023). This presents unique challenges for implementing effective bio-surveillance and pest control strategies in countries that both export and import crops susceptible to Mexican fruit fly infestations.
Genomic information opens new avenues for developing innovative identification and control techniques for pest species. A. ludens along with other 32 species, including other important pests such as Anastrepha obliqua, and the Anastrepha fraterculus complex, make up the fraterculus group (Norrbom et al. 2012). In this group, species diagnosis is mostly challenging due to morphological and genetic similarities among closely related species, resulting from recent divergence and hybridization (Zucchi 2000; Norrbom et al. 2012; Perre et al. 2014; Scally et al. 2016; Díaz et al. 2018; Congrains et al. 2021). Genome-scale methods have been employed to improve species-level identification of tephritid pests and have enabled to discriminate closely related species with complex evolutionary histories (Dupuis et al. 2018; Doellman et al. 2020; Congrains et al. 2023; Doorenweerd et al. 2024). Likewise, large-scale sequencing data has been used to track the geographic origin of potential invasive pests in this family, which can be applied to detect vulnerabilities in border inspection procedures (Dupuis et al. 2019; Deschepper et al. 2023; Zhang et al. 2023).
Control techniques that can benefit from the availability of complete genomes include the Sterile Insect Technique (SIT). The SIT requires mass rearing, sterilization through radiation, and subsequent release of large numbers of sterile males from the target species to suppress the growth of wild pest populations (Hendrichs and Robinson 2009). For the Mexican fruit fly, genetic sexing strains (GSS) based on the black pupae (bp) marker have been developed to selectively remove females before mass releases (Zepeda-Cisneros et al. 2014; Ramírez-Santos et al. 2021), but the genetic basis for bp remains unknown. A high-quality genome for A. ludens will facilitate modern functional genomics to characterize the bp trait. Combined with adequate protocols for targeted gene disruption using CRISPR/Cas9 (Sim et al. 2019; Choo et al. 2022; Paulo et al. 2022), the effort could lead to the development of new, more stable GSS for this and other closely related species. A reference genome also opens horizons to explore strategies based on transgenic genomic modifications, like homing-based gene drives (Meccariello et al. 2024) and precision-guided SIT (Kandul et al. 2019), which are promising tools to mitigate the negative effects associated to radiation-based sterilization (Barry et al. 2003; Orozco-Dávila et al. 2015; Landeta-Escamilla et al. 2016).
Assembly of sex chromosomes have been particularly challenging due to the high content of repetitive regions (Bachtrog 2003; Kejnovsky et al. 2009; Tomaszkiewicz et al. 2017), the haploid nature of Y or W (in XX/XY and ZZ/ZW systems, respectively) (Tomaszkiewicz et al. 2017), and in some taxa, complex systems with multiple sex chromosomes (Carey et al. 2022). Recent improvements in sequencing technology and assembly algorithms have provided the tools to generate sex chromosomes assemblies with higher accuracy and contiguity (Carey et al. 2022). For instance, long-read sequencing (i.e. PacBio and Nanopore sequencing approaches) has been used to generate telomere-to-telomere human sex chromosomes (Miga et al. 2020; Rhie et al. 2023). Furthermore, chromatin conformation capture (Hi-C) sequencing has been applied to accurately assign and order contigs into scaffolds at the chromosome-scale (Burton et al. 2013), which can be beneficial for assembling sex chromosomes (Xue et al. 2021; Carey et al. 2022). Despite these advances, there is still no standard method for identifying sex chromosomes in a genome assembly (Carey et al. 2022). In addition, some genome sequencing initiatives have overlooked the particularities of sex chromosomes, especially the sex-limited chromosomes (i.e. chromosomes present in haploid form in only one sex, such as Y and W) (Tomaszkiewicz et al. 2017; Deakin et al. 2019). For example, from a total of 13 chromosome scale genome assemblies of Tephritidae species deposited in the NCBI Genome database (https://www.ncbi.nlm.nih.gov/datasets/genome/? taxon=7211, accessed on 2023 November 2), only 3 (including this study) were able to properly identify at least 1 sex chromosome.
Here, we present a high-quality, chromosome-scale genome assembly of A. ludens generated using PacBio high-fidelity (HiFi) reads coupled with chromatin conformation capture (Hi-C) data. We further performed comparative genomics analysis of transposable elements (TEs) between A. ludens and other tephritid species to determine the extent of conservation of TE composition across the Family. The highly contiguous and complete (in terms of gene content) genome generated here, also allowed us to identify and characterize scaffolds assigned to both sex chromosomes (X and Y) of A. ludens. Using this data, we estimated the evolutionary rates between autosomal and sex-linked orthologous genes in A. ludens and a close relative, A. obliqua (both species belong to the fraterculus group), which in turn will contribute to understanding of the evolution of sex chromosomes in this group.
Materials and methods
Sample collection
Unmated A. ludens males were sampled from the wild-type Willacy strain, which is routinely maintained at the United State Department of Agriculture (USDA) APHIS Plant Protection and Quarantine Sterile Mexican Fruit Fly Production Facility in Edinburg, Texas, USA (Dupuis et al. 2019). The specimens were flash frozen in liquid nitrogen and transported on dry ice to the USDA—Agricultural Research Service (ARS)—Daniel K. Inouye U.S. Pacific Basin Agricultural Research Center in Hilo, HI, USA. Specimens were stored at −80°C until further processing.
DNA extraction, library preparation, and sequencing
For PacBio HiFi sequencing high molecular weight genomic DNA (HMW gDNA) was extracted from the thorax of a single male fly using the Qiagen MagAttract HMW DNA Kit (Qiagen, Hilden, Germany), followed by 2× bead clean-up. DNA concentration was quantified using Qubit dsDNA high sensitivity assay (Thermo Fisher Scientific) and a DS-11 Spectrophotometer and Fluorometer (DeNovix Inc., Wilmington, DE, USA). DNA purity was evaluated using ratios of absorbance at 260/280 and 260/230 obtained using a DS-11 Spectrophotometer and Fluorometer (DeNovix Inc.). The fragment size distribution was assessed using Agilent Femto Pulse analyzer (Agilent Technologies). Extracted HMW gDNA was sheared using a Diagenode Megaruptor 2 (Denville, NJ, USA) to a mean size of ∼15 kb, as further confirmed by Femto Pulse analyzer (Agilent Technologies, Ankeny, IA, USA). The sequencing library was prepared using the SMRTbell Express Template Prep Kit 2.0 (PacBio, Menlo Park, CA, USA) and sequenced on a Sequel II system (Pacific Biosciences, Menlo Park, CA, USA) using a Pacific Biosciences 8M SMRT Cell. The PacBio subreads were processed to generate HiFi reads using circular consensus sequencing mode on the SMRTLink v8.0 software. The Hi-C library was prepared from the head and abdomen of the same individual using the Arima HiC 2.0 kit (Arima Genomics, San Diego, CA, USA). The sample was sheared to generate an average size of ∼450 bp using a Bioruptor Pico (Diagenode, Denville, NJ, USA). A short-read library was prepared using NEBNext Ultra DNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA), and sequenced with 150-bp paired-end run on an Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA).
Genome assembly
The HiFi reads were composed with 3 or more passes (subreads) and had a minimum read quality of 0.99 using default HiFi settings in the SMART Link software. Raw HiFi reads were filtered to remove adapter containing reads using HiFiAdapterFilt v2.0 (Sim et al. 2022). Remaining reads were used to generate a haplotype aware contig assembly with hifiasm v0.16.1-r375 (Cheng et al. 2021). This program generates 2 assemblies: the first with longer pseudohaploid contigs (primary assembly) and the second with the heterozygous regions within this assembly (alternate assembly). The primary assembly were used for subsequent analysis. Potential contaminant sequences (i.e. genomic fragments of Bacteria) were identified using BlobTools v2.6.1 (Challis et al. 2020). This assessment consisted of a taxon-based annotation, the analysis of GC content and read coverage. Taxonomic annotation was carried out by locally aligning all assembled contigs to the NCBI nucleotide database (downloaded on 2022 February 14) using an e-value cutoff of 10−25 and the MegaBLAST algorithm implemented in BLAST + v2.7.1 (Camacho et al. 2009). Likewise, contigs shorter than 10 Mb were aligned against reference proteomes from the UniProt database (March 2020) using an e-value cutoff of 10−25 and the BLASTX algorithm in DAIMOND v2.0.9.147 (Buchfink et al. 2021). Genome coverage was estimated by mapping cleaned PacBio reads to the primary assembly using Minimap v2.22-r1101 (Li 2018, 2021). The estimated genome size and heterozygosity were calculated using GenomeScope v2 (Ranallo-Benavidez et al. 2020). Additionally, the consensus quality value (QV) of the final assembly was calculated using YAK v0.1-r69 (https://github.com/lh3/yak) based on the k-mer counts of the filtered HiFi reads.
To generate a chromosome-level genome assembly, we used Hi-C sequencing data and the primary assembly following the steps implemented in the Arima-HiC Mapping Pipeline (Arima Genomics Inc., https://github.com/ArimaGenomics/mapping_pipeline). First, the paired-end reads were independently mapped to the primary assembly using BWA-MEM2 (Vasimuddin et al. 2019). The reads with a ligation junction were considered chimeric, and their 3′-ends were trimmed using the filter_five_end.pl script included in the Arima-HiC Mapping Pipeline. Trimmed pair-end reads were then combined using the script two_read_bam_combiner.pl included in the Arima-HiC Mapping Pipeline. Reads with mapping quality <10 were removed using SAMtools v1.3.1 (Li et al. 2009). PCR duplicates were excluded using MarkDuplicates option in Picard Tools v2.26.10 (http://broadinstitute.github.io/picard/). We used YaHS v1.1 (Zhou et al. 2023) with no contig error correction option to generate the Hi-C scaffolds. Visualization and manual curation were performed using Juicebox v1.11 (Durand et al. 2016). We used BlobTools v2.6.1 (Challis et al. 2020) once again to generate plots, assess assembly statistics, and further contamination removal.
Assessment of the genome assembly
Completeness in terms of gene content was evaluated on the contig and scaffold level assemblies, as well as the predicted proteins (see details below) with Benchmarking Universal Single-Copy Orthologs (BUSCO) v5.4.4 (Manni et al. 2021) using the 3,285 conserved orthologs for Diptera in orthoDB v10 database (Kriventseva et al. 2019). For the assemblies, the gene prediction was performed using the pre-trained set for Drosophila melanogaster in Augustus v3.5 (Stanke et al. 2006).
Genome annotation
The genome assembly was submitted to the GenBank, and the annotation was performed internally using the NCBI Eukaryotic Genome Annotation Pipeline v10.1 (Thibaud-Nissen et al. 2016). A full report of the A. ludens genome annotation can be found at https://www.ncbi.nlm.nih.gov/refseq/annotation_euk/Anastrepha_ludens/GCF_028408465.1-RS_2023_03/. This method provided a set of standardized annotated features, which facilitates comparisons between studies. We extracted all the coding sequences (CDS) using gffread v0.12.1 (Pertea and Pertea 2020) from the general feature format (GFF) file and the set of longest CDS per gene was obtained using the script get_longest_CDS_per_gene.py (https://github.com/popphylotools/get_longest_CDS_per_gene/). Functional annotation was performed on the protein set (translated longest CDS per gene) using the default parameters in eggNOG-mapper v2 implemented in eggNOG v5.0 (Huerta-Cepas et al. 2019; Cantalapiedra et al. 2021). The functional annotation of eggNOG includes the assignment of Clusters of Orthologous Groups of proteins (COG/KOG) (Tatusov et al. 1997, 2003), gene ontology (GO) terms (The Gene Ontology Consortium et al. 2023) and to protein families using the Pfam database (Mistry et al. 2021). The distribution of the GO terms with a percentage higher than 2% was visualized using the Gene Ontology file of 01-Nov-2018 in WEGO v. 2 (Ye et al. 2006, 2018).
TEs were annotated using the Extensive de novo TE Annotator software (Ou et al. 2019). For this analysis, regions correspond to genes were excluded (option –exclude), and fasta files with CDS (option –cds) of the species and a manually curated TE database developed for D. melanogaster (Rech 2022) (option –curatedlib) were provided. To enable comparisons in the Tephritidae family, the TEs of the following species were also predicted using the same approach: A. obliqua (GenBank: GCF_027943255.1) (Sim et al. 2024), Rhagoletis zephyria (GenBank: GCF_001687245.2), Ceratitis capitata (GenBank: GCF_000347755.3) (Papanicolaou et al. 2016), Bactrocera dorsalis (GenBank: GCF_023373825.1) (Jiang et al. 2022), and Zeugodacus cucurbitae (GenBank: GCF_028554725.1). Retrotransposons were classified in the TE class I and DNA transposons in the TE class II (Wicker et al. 2007). The frequency of TE classes and gene content was estimated in windows of 500 kb across the A. ludens genome, and visualized in ChromoMap v.0.4.1 (Anand and Rodriguez Lopez 2022) using R v4.2.2 (R Core Team 2022).
Identification and characterization of sex chromosomes
We used 3 strategies to identify contigs and scaffolds linked to sex chromosomes in the final A. ludens genome assembly, including: (1) visualization of the Hi-C contact map and coverage of HiFi reads across the genome, (2) synteny with D. melanogaster, and (3) the difference in the average read depth of autosomal vs sex chromosomal scaffolds (known as the average depth (AD)-ratio or chromosome quotient approach) (Hall et al. 2013; Bidon et al. 2015). In the first approach, we expect heteromorphic sex chromosomes (XY for a male individual) to show approximate half of the coverage observed in the autosomes of the Hi-C reads. Consequently, the contact map would display them with less intensity than the other chromosomes (autosomes). Additionally, we calculated the coverage of the HiFi reads in windows of 500 kb across the A. ludens genome using a mapping quality threshold of 20 in mosdepth (Pedersen and Quinlan 2018).
Cytogenetic and genomic studies have shown conservation at chromosome level in many species across the order Diptera, which exhibit high levels of synteny in 6 chromosomes arms, which are referred to as Muller elements from A to F (White 1949; Holt et al. 2002; Vicoso and Bachtrog 2013, 2015). The X chromosome of groups of ancient divergence in Diptera such as the Tephritidae family corresponds to the Muller element F, different to what was found in D. melanogaster, which X chromosome corresponds to the Muller element A (Vicoso and Bachtrog 2013). To confirm this pattern, the synteny between A. ludens and D. melanogaster (GCF_000001215.4) (Hoskins et al. 2015) genomes was conducted using the NCBI assembly-assembly alignment pipeline. The visualization was performed in the Comparative Genome Viewer tool of the NCBI (https://www.ncbi.nlm.nih.gov/genome/cgv/).
For the AD-ratio approach, whole-genome sequencing (WGS) data from 25 males and 25 females of A. ludens, previously described in Paulo et al. (2024), were used to estimate differential coverage of chromosomes between sexes. Briefly, WGS reads were filtered using fastp (Chen et al. 2018) and mapped against the Mexican fruit fly genome using BWA-MEM (Li and Durbin 2009; Li 2013). Duplicates were marked with the SAMBLASTER tool (Faust and Hall 2014) and removed from alignments along with non-mapped reads and low-quality aligned reads using the SAMtools v1.3.1 (Li et al. 2009) view module (parameters: -q 15 -F 1028) (Li et al. 2009). Filtered alignments were sorted and merged into a single BAM file per sex using SAMtools v1.3.1 (Li et al. 2009). After mapping, male and female read depths were calculated for each scaffold using a mapping quality (Q) threshold of 30 in mosdepth (Pedersen and Quinlan 2018). The AD-ratio of each scaffold was calculated by dividing the average read depth in the female group by the average read depth in the male group: AD-ratio = ADfemale/(ADmale × norm). A normalization factor (norm) was used to account for differences in sequencing coverage between groups and was calculated by dividing the total number of reads in the female BAM file by the total number of reads in the male BAM file: norm = BAMfemale/BAMmale. The AD-ratio is expected to be close to zero for perfectly mapped Y-linked scaffolds, 1 for autosomal, and 2 for X-linked scaffolds. We used relaxed interval cutoffs, classifying scaffolds as Y-linked if AD-ratio ≤ 0.3 (3.33 times as many alignments from male data than from female data) as suggested by Hall et al. (2013). We further classified scaffolds as autosomal if AD-ratio ranges between 0.7 and 1.3, or X-linked if AD-ratio ≥ 1.7 (Bidon et al. 2015).
Evolutionary rates
We calculated the nonsynonymous to synonymous substitution rates ratio (Ka/Ks) of orthologous CDSs from A. ludens and a close relative A. obliqua (GenBank: GCF_027943255.1) (Sim et al. 2024). Annotation files (GFF) of A. obliqua and A. ludens genomes were used to extract all complete CDS in gffread v0.12.1 (Pertea and Pertea 2020). The set of longest CDS per gene was extracted using the script get_longest_CDS_per_gene.py (https://github.com/popphylotools/get_longest_CDS_per_gene). The pair of orthologs was inferred using best reciprocal blast strategy. For that, 2 BLASTN searches were performed using an e-value cut-off of 10−6 in BLAST + v2.7.1 (Camacho et al. 2009). The first search involved using CDS of A. obliqua as query and the CDS of A. ludens as subject, and in the second, the species in the query and subject were interchanged. The pairs of CDSs that resulted as hit with the highest bit score in both BLAST searches were retained for further analysis. The Ka/Ks ratio was calculated using KaKs_Calculator v3.0 (Zhang 2022). To ensure the accuracy of our analysis, we performed a filtering step to remove potential non-orthologous genes, excluding genes with a difference in length >30% of the longest CDS, without variation, sequences with over 25% of variable sites, and Ks exceeding 2. We estimated the average values of Ka/Ks, Ka and Ks in windows of 500 kb across the A. ludens genome and results were visualized using ChromoMap v.0.4.1 (Anand and Rodriguez Lopez 2022) in R v4.2.2 (R Core Team 2022). We tested if the evolutionary rates (Ka, Ks, and Ka/Ks) for X-linked genes were significantly different than the observed for autosome-linked genes using the Wilcoxon sum rank test with Bonferroni-corrected P-values in R v4.2.2 (R Core Team 2022). Additionally, we compared the proportion of rapidly evolving genes (Ka/Ks > 1) in the X chromosome and autosomes using the Fisher's exact test in R v4.2.2 (R Core Team 2022).
Gene content of the sex chromosomes
In addition to the characterization of the sex chromosome-linked genes predicted using the NCBI Eukaryotic Genome Annotation Pipeline v10.1 (Thibaud-Nissen et al. 2016) and GO assignment (Huerta-Cepas et al. 2019; Cantalapiedra et al. 2021), we conducted similarity searches to find previously identified known sex-linked genes in tephritid fruit flies: Maleness-on-the-Y (MoY) and Gigyf (gyf). MoY is a male-specific gene located in the Y chromosome, and plays a key role in sex determination in Tephritidae (Meccariello et al. 2019). In contrast, the gyf gene has been described on the X chromosome of Bactrocera tryoni (Teprhitidae) (Choo et al. 2019). Although truncated paralogs of gyf may exist on the Y chromosome, we anticipated finding the complete CDS only on the X-linked scaffold. For this analysis, we aligned the MoY of B. dorsalis (GenBank: MK165749.1) and gyf of B. tryoni (GenBank: XM_040111897.1) against the final A. ludens genome assembly using 2 e-value cutoffs (10−5 and 10) and 2 searching algorithms (BLASTn and tBLASTn) implemented in BLAST + v2.7.1 (Camacho et al. 2009).
Results and discussion
Genome assembly
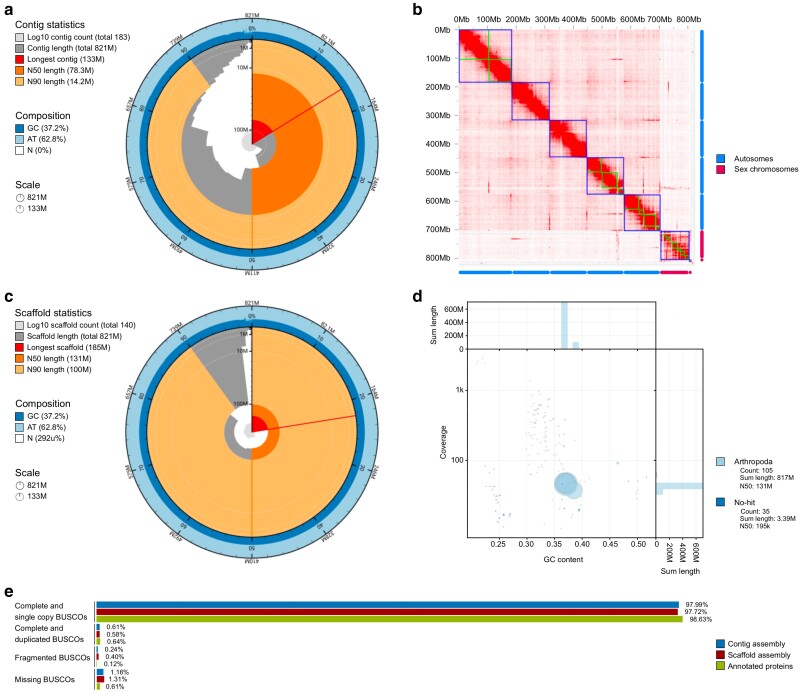
The contig assembly was generated based on 4.8 million PacBio HiFi reads with an average length of 8.7 kb (totaling 41.3 Gb of raw HiFi data). The taxon-based annotation approach implemented in BlobTools (Challis et al. 2020) identified 19 contigs (454 kb) suspected to originate from Bacteria, which were excluded for further analysis. The filtered assembly comprised 183 contigs with a total length of 821 Mb with N50 of 78.3 Mb and GC content of 37.2% (Fig. 1a). The estimated genome size was 753 Mb, which is less than the total number of assembled bases. The heterozygosity calculated based on k-mer counts was 1.5%, which is high when compared with other animal taxa (Gan et al. 2019; Papa et al. 2023; Supple et al. 2024; Wu et al. 2024), but moderate for an insect (Deng et al. 2024). The alternate assembly was substantially less contiguous than the primary, which included 15,663 contigs with N50 of 181.426 kb (Supplementary Table 1 in File 1). The differences in contiguity between the primary and the alternate assembly is due to the heterozygosity found in this genome, with the alternate assembly representing each heterozygous region in the genome graph. The scaffolding of the primary assembly was performed based on Hi-C data containing 93.2 million pair-end reads (Fig. 1b). The Hi-C contact map revealed some signals of interchromosomal interactions near putative centromere and telomere regions of the chromosomes. This is an indication of centromere-centromere and interchromosomal telomere-telomere interactions happening within the nuclei, which has been documented in other Diptera species (Hoencamp et al. 2021; Lukyanchikova et al. 2022). A pattern that is concordant with the Rabl-like chromatin conformation (Rabl 1885). Their locations in the Hi-C map suggest a Rabl-like chromatin conformation in A. ludens, extending this phenomenon to this species as well.
Fig. 1.
Statistics of the Anastrepha ludens (Mexican fruit fly) genome assembly. a) Snail plot visualization of the contig assembly. b) Hi-C contact map indicates that contigs (green squares) can be grouped in 7 major scaffolds (blue squares). c) Snail plot visualization of the scaffold assembly. d) Coverage and GC content of taxon-annotated scaffolds, after removing potential contaminant sequences from Bacteria. e) BUSCO assessment of the contig and scaffold assemblies, as well as the set of annotated proteins (1 per gene) of the scaffold assembly using Diptera conserved orthologs.
The final scaffold assembly comprised of 140 scaffolds with N50 of 131 Mb, which represents a 67% increase compared with the N50 of the contig assembly (Fig. 1a and c). Cytogenetic analysis of A. ludens shows that males have 5 pairs of autosomes and 1 pair of sex chromosomes (namely XY) (Garcia-Martinez et al. 2009). This observation is consistent with the presence of the 7 longest scaffolds, which collectively constitute 98.9% of the genome assembly and with the coverage pattern visualized in the Hi-C contact map (Fig. 1b). This assembly showed an adjusted QV of 61.259, which implies approximately only 1 error every million of bases. Moreover, BlobTools analysis (Challis et al. 2020) showed no signals for potential contaminants remained in the final assembly (Fig. 1d).
An evaluation of the assembly's completeness revealed that the A. ludens genome contains most of the BUSCO gene set expected for the Diptera lineage. Specifically, 99.27% of these genes were found to be complete in the annotated genes of the scaffold assembly (98.63% single-copy and 0.64% duplicated), only 0.12% fragmented, and 0.61% were missing (Fig. 1e).
Genome annotation
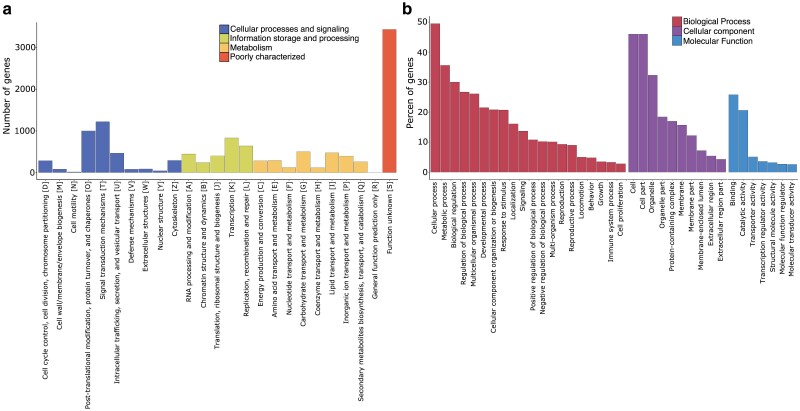
The genome annotation revealed a total of 16,617 predicted genes, of which 87.78% were protein coding genes (PCGs) (Table 1). Notably, 99% of the predicted PCGs were complete sequences (i.e. initiating with a Methionine start codon and finishing with a stop codon). Eggnog annotated 13,028 complete PCGs. Furthermore, functional categorization revealed that 90% of these genes were assigned to COG/KOG categories, 73% to protein families (Pfam), and 70% to GO terms. The most common COG/KOG class was cellular processes and signaling class (3,740 genes, 28.6%), and the second was the poorly characterized class (3,570 genes, 27.4%) (Fig. 2a). In terms of GO, the most prevalent GO terms per category were cell (46%) for cellular component, binding (25.8%) for molecular function, and cellular process (49.5%) for biological process (Fig. 2b).
Table 1.
Statistics of the annotation of the Anastrepha ludens (Mexican fruit fly) genome assembly.
| Feature | Number | |
|---|---|---|
| Genes | 16,617 | |
| Protein coding genes | 14,586 | |
| Non-protein coding genes | lncRNA | 824 |
| rRNA | 695 | |
| snoRNA | 45 | |
| snRNA | 90 | |
| tRNA | 377 | |
| Pseudogenes | 688 | |
| mRNA | 25,247 |
Fig. 2.
Functional annotation summary of the Anastrepha ludens genome assembly. a) Distribution of Clusters of Orthologous Groups of proteins (COG/KOG) functional categories. b) Distribution of gene ontology (GO) classification including GO terms with percentages >2%.
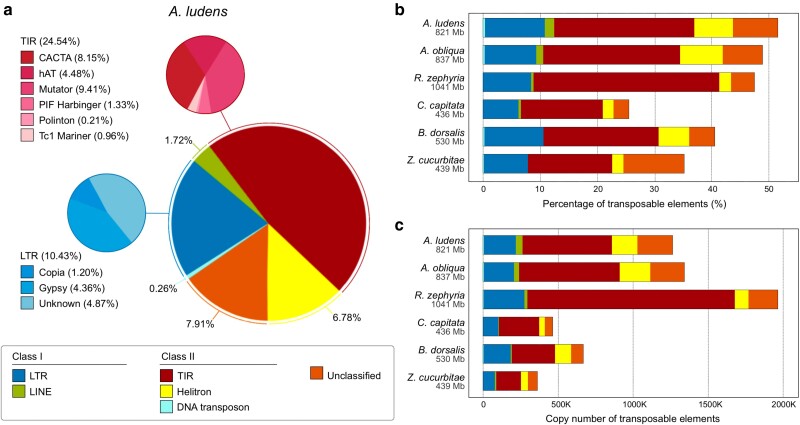
Repetitive elements encompassed 424 Mb (51.69%) of the A. ludens genome (Fig. 3). Among these, TEs class II (DNA transposons, Helitron and TIR: CACTA, Mutator, PIF Harbinger, Tc1 Mariner, hAT and Polinton) were the most abundant (31.59%), with TIR being the most prevalent order (24.55%). TEs class I (LTRs, and non-LTRs) represented 12.19% of the genome (Fig. 3a). It's worth mentioning that TE content may substantially vary among taxa (Wells and Feschotte 2020) and even between closely related species (Feschotte and Pritham 2007). For instance, the proportion of LTRs varies from ∼2 to 17% across Drosophila species (Sessegolo et al. 2016; Mérel et al. 2020). In the case of Tephritidae genomes, TE content ranged from 25.5% (C. capitata) to 51.7% (A. ludens) (Fig. 3b). Among the Tephritidae species compared, the top 3 most prevalent TEs (excluding unclassified TEs) were TIR, LTR, and Helitron, with average proportions of 20.5, 9.7, and 4.6%, respectively. R. zephyria genome exhibited the highest proportion (32.5%) and copy number (1,384,103) of TIR elements (Fig. 3b and c). Furthermore, both species of Anastrepha showed similar genomic proportion and copy number of the annotated TE categories, and the largest proportion and copy number of LINE and Helitron (Fig. 3b and c).
Fig. 3.
TEs classification found in the genomes of Anastrepha ludens and 5 other Tephritidae species. a) Genomic proportions of TEs classified in orders and superfamilies annotated in the A. ludens genome. TE categories with genomic proportions exceeding 0.1% are shown. Genomic proportion b) and copy number c) of TEs classified in orders found in the genomes of A. ludens, Anastrepha obliqua (GCF_027943255.1), Rhagoletis zephyria (GCF_001687245.2), Ceratitis capitata (GCF_000347755.3), Bactrocera dorsalis (GCF_023373825.1) and Zeugodacus cucurbitae (GCF_028554725.1). Genome sizes in megabases (Mb) are shown in gray below the species names.
Identification of sex chromosomes
Hi-C contact map revealed that 5 scaffolds (NC_071498.1, NC_071499.1, NC_071500.1, NC_071501.1, and NC_071502.1) had approximately double the coverage relative to the remaining 2 main scaffolds (scaffolds larger than 5 Mb) and a group of smaller contigs (Fig. 1b). The coverage analysis using the HiFi reads also showed a similar pattern (Supplementary Fig. 1 in File 1). The average of the HiFi coverage for the scaffolds NC_071498.1, NC_071499.1, NC_071500.1, NC_071501.1, and NC_071502.1 was 42.6×, which is approximately double of the coverage of NC_071503.1 (22.1×) and NC_071504.1 (18.7×) (Supplementary Fig. 1 in File 1). These results suggest that NC_071498.1, NC_071499.1, NC_071500.1, NC_071501.1, and NC_071502.1 are autosomes, while NC_071502.1 and NC_071504.1 are linked to the sex chromosomes.
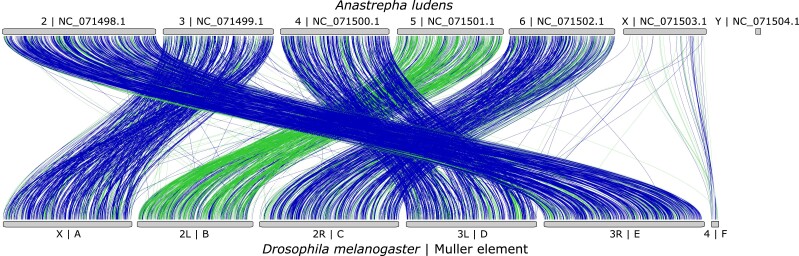
We found substantial level of synteny and homology between A. ludens and D. melanogaster genomes, especially for scaffolds from NC_071498.1 to NC_071502.1 (Fig. 4). This high degree of conservation suggests that these scaffolds correspond to autosomes, which agrees with the results of the coverage analysis of the Hi-C and HiFi data. Given the high level of synteny across Diptera (White 1949; Holt et al. 2002; Vicoso and Bachtrog 2013, 2015; Sved et al. 2016), we established the correspondence between scaffolds NC_071498.1, NC_071499.1, NC_071500.1, NC_071501.1, and NC_071502.1 with the Muller elements E, A, D, B, and C, respectively. The scaffold NC_071503.1 displayed relatively low levels of homology and synteny with the element F, aligning with the pattern expected for the X chromosome in taxa with ancient divergence in higher Diptera, such as the Tephritidae family (Vicoso and Bachtrog 2013). Conversely, the scaffold NC_071504.1 did not show homology with D. melanogaster genome, making it a potential candidate for the male-specific Y chromosome, which is expected to have low degree of conservation across taxa including Diptera (Mahajan and Bachtrog 2017; Tomaszkiewicz et al. 2017; Vicoso 2019).
Fig. 4.
Synteny analysis of Anastrepha ludens and Drosophila melanogaster (GCF_000001215.4) genomes. Muller elements are shown next to the arm chromosome names of the D. melanogaster genome. Forward and reverse alignments are shown in green and purple, respectively. An interactive version of this figure is available at https://www.ncbi.nlm.nih.gov/genome/cgv/browse/GCF_028408465.1/GCF_000001215.4/36815/28586.
Scaffold-to-chromosome classification using the AD-ratio approach (Table 2 and Supplementary Table 2 in File 1) largely corroborates with the previous analysis, assigning the chromosome-level scaffolds NC_071498.1, NC_071499.1, NC_071500.1, NC_071501.1, and NC_071502.1 to autosomes (AD-ratio = 0.93–0.94), the scaffold NC_071503.1 to the X chromosome (AD-ratio = 1.73), and the scaffold NC_071504.1 to the Y chromosome (AD-ratio = 0.05). We also detected 8 additional contigs assigned to the X chromosome (AD-ratio ranges between 1.75 and 2.17) with a total of 1.8 Mb (∼1.8% of the total X chromosome length). Moreover, we identified 35 additional contigs with an AD-ratio close to zero (AD-ratio ranges between 0.0 and 0.27), which is expected for Y-linked sequences, with a combined length of 4.3 Mb (Supplementary Table 2 in File 1). Together with scaffold NC_071504.1 (the longest Y-linked scaffold with 6.1 Mb), we estimate the Y chromosome of A. ludens to be 10.4 Mb in length.
Table 2.
Scaffolds identification in the Anastrepha ludens genome assembly by the AD-ratio approach.
| Scaffold ID | Length (bp) | AD-female | AD-male | AD-ratio | Normalized AD-ratio | Designation |
|---|---|---|---|---|---|---|
| NC_071498.1 | 185,090,897 | 26.05 | 15.16 | 1.72 | 0.93 | A |
| NC_071499.1 | 133,334,115 | 25.58 | 14.89 | 1.72 | 0.93 | A |
| NC_071500.1 | 131,385,309 | 26.63 | 15.49 | 1.72 | 0.93 | A |
| NC_071501.1 | 128,169,930 | 26.26 | 15.3 | 1.72 | 0.93 | A |
| NC_071502.1 | 127,816,056 | 26.17 | 15.18 | 1.72 | 0.94 | A |
| NC_071503.1 | 99,992,179 | 32.06 | 10.05 | 3.19 | 1.73 | X |
| NC_071504.1 | 6,146,282 | 0.3 | 3.56 | 0.08 | 0.05 | Y |
A, autosome; X, X chromosome; Y, Y chromosome.
Overall, the coverage analysis using the Hi-C contact map and HiFi reads, along with synteny, and the AD-ratio results, all agree that the chromosome-level scaffolds NC_071498.1, NC_071499.1, NC_071500.1, NC_071501.1, and NC_071502.1 are autosomes (hereafter referred to as mitotic chromosomes 2, 3, 4, 5, and 6, respectively); NC_071503.1 is the X chromosome; and NC_071504.1 is the largest fragment of the Y chromosome.
Characterization TE and gene content of sex chromosomes
The sex chromosomes showed ∼50% reduction in gene density (genes per Mb) and an ∼10% increase in the proportion of TEs, when compared with the autosomes (Fig. 5 and Supplementary Tables 3 and 4 in File 1). Both sex chromosomes had substantially higher TE content of class I (4.5× for X chromosome and 3× for Y chromosome) relative to the autosomes (Supplementary Table 3 in File 1). The X chromosome had an increase of both class I orders (LTR and LINE), while the Y chromosome only increased the LTR elements (Supplementary Table 3 in File 1). The increment of LINE element in the X and Z chromosome has been reported in other metazoans (Bellott et al. 2010) and in chromosome 4 [homolog to the ancestral X in Diptera (Vicoso and Bachtrog 2013)] in D. melanogaster (Kaminker et al. 2002). In humans, LINE elements may play a role in X chromosome inactivation (Bailey et al. 2000; Chow et al. 2010; Barros de Andrade e Sousa et al. 2019), but the role of these TEs in other taxa such insects is still poorly understood.
Fig. 5.
Heatmap of frequency of TEs and PCGs across the Anastrepha ludens genome assembly. The frequencies of the TE classes and PCGs were calculated in windows of 500 kb. TEs class I are composed of LTRs (Copia, Gypsy, and unclassified LTRs) and non-LTRs (DIRS, LINE and unclassified non-LTR elements). TEs class II are composed of DNA transposons, TIR (CACTA, Mutator, PIF Harbinger, Tc1 Mariner, hAT, and Polinton), and non-TIR element Helitron.
Evolutionary rates of sex chromosome-linked genes
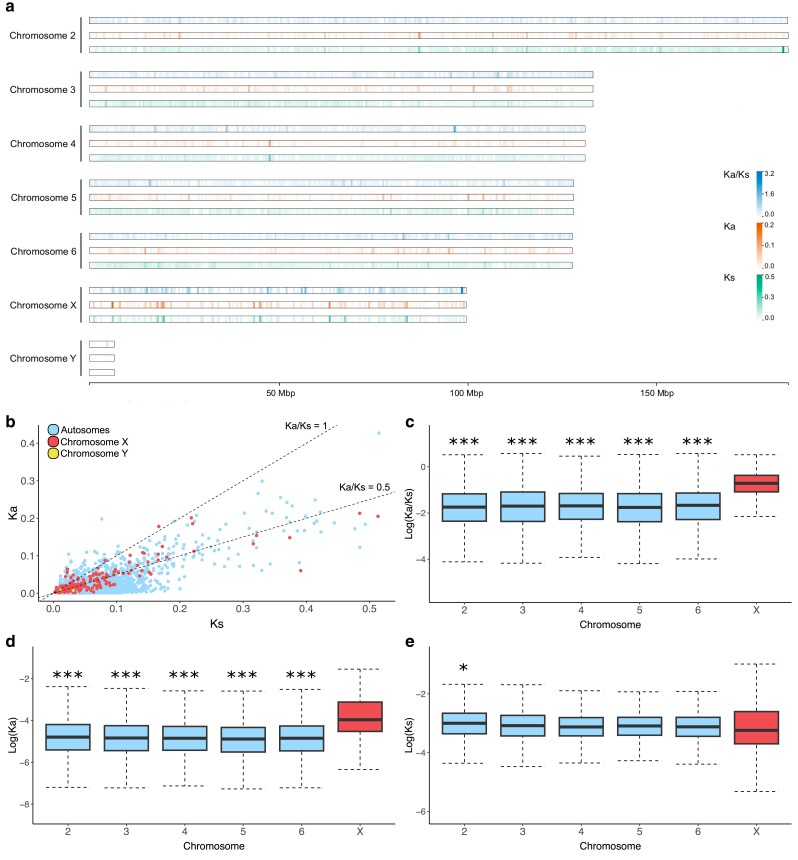
We observed a lower proportion of orthologous genes of A. ludens and A. obliqua in the sex chromosomes than in the autosomes (Supplementary Table 4 in File 1 and Table 5 in File 2), which was 3 times fewer for X chromosome (χ2P-value < 0.01) and 15 times fewer for Y chromosome (χ2P-value < 0.01), suggesting higher genetic divergence in sex chromosome-linked genes. Furthermore, orthologous genes in the X chromosome showed higher median of Ka and Ka/Ks compared with orthologs located in the autosomes (Fig. 6). These differences were significant for the comparison of Ka/Ks of genes located in autosomes (compared independently) and X-linked genes (Bonferroni corrected P-value of Wilcoxon sum rank test < 0.001). This elevated rate of evolution is concordant with the faster-X (or Z) effect (Charlesworth et al. 1987; Meisel and Connallon 2013). This phenomenon is highly widespread across animal taxa, it has been found in insects (Begun et al. 2007; Hu et al. 2013; Mongue et al. 2022), birds (Mank et al. 2007; Hayes et al. 2020), mammals (Hvilsom et al. 2012; Kousathanas et al. 2014), and fishes (Darolti et al. 2023). However, exceptions to the faster-X evolution have also been reported (Rousselle et al. 2016; Whittle et al. 2020; Darolti et al. 2023), which reflects the complexity of the evolutionary trajectories of the sex chromosomes.
Fig. 6.
Evolutionary rates of 9,325 orthologous genes of Anastrepha ludens and Anastrepha obliqua. a) A heatmap of the average of nonsynonymous to synonymous rate ratio (Ka/Ks), nonsynonymous rate (Ka) and synonymous rate (Ks) calculated in windows of 500 kb and plotted across the A. ludens genome. b) Scatter plot of Ka and Ks calculated for each orthologous gene. Colors indicate location of the gene in the A. ludens genome: light blue for autosomes, red for the X chromosome and yellow for the Y chromosome. Box plots of evolutionary rates: Log(Ka/Ks) c), Log(Ka) d), and Log(Ks) e). These parameters were estimated for each pair of ortholog and plotted according to the respective gene chromosome location. Box plot of Y-linked genes was not included due to the low number of orthologous genes (only 2). Asterisks above the boxplots in the autosomes indicate significant difference of evolutionary rates between autosome-linked relative to X-linked genes using Bonferroni corrected P-value of Wilcoxon sum rank test (*P-value < 0.05 and ***P-value < 0.001). The boxes indicate the interquartile range (IQR), the medians are shown as black lines inside the boxes and the whiskers were estimated at 1.5 IQR. Evolutionary rates for each pair of orthologs are available on Supplementary Table 5 in File S2.
Two non-mutually exclusive evolutionary processes have been postulated to explain this effect: relaxed purifying selection and positive selection (Charlesworth et al. 1987; Vicoso and Charlesworth 2009; Parker et al. 2022). We found significant differences not only for Ka/Ks, but also for Ka (Bonferroni corrected P-value of Wilcoxon sum rank test < 0.001) in all the comparisons between autosomes and the X chromosome and only 1 comparison (median higher in chromosome 2 than in the X) for Ks (Bonferroni corrected P-value of Wilcoxon sum rank test < 0.05) (Fig. 6 and Supplementary Table 5 in File 2). Additionally, the proportion of genes with Ka/Ks higher than 1 (i.e. genes probably evolving under positive selection) was 10 times higher in the X chromosome than in the autosomes (Fisher's exact test P value < 0.01). Although we cannot rule out the possibility of relaxed selection, these results support that, in some extent, the faster-X effect is due to adaptive evolution in these taxa.
Gene content of the sex chromosomes
The NCBI eukaryotic genome annotation pipeline predicted 1,140 genes (1,011 PCGs, 98 long non-coding RNA, 7 small nuclear RNA, and 24 ribosomal RNA) located in the X chromosome (scaffolds NC_071503.1, NW_026530029.1, and NW_026530059.1) of A. ludens. In contrast, only 54 (3 long non-coding RNA and 51 PCGs) Y-linked genes were predicted, all of them located in the major scaffold (NC_071504.1). The majority of these Y-linked genes were annotated as uncharacterized proteins and only 4 genes had GO terms assigned, 1 of those genes (protein accession number: XP_053969199.1) was associated with GO terms related to the nervous system (Supplementary Table 6 in File 1). This result suggests that at least some of these genes represent evolutionary novelties, making them potential targets for studies of sex-specific genes and sex determination.
The similarity searches revealed a putative homolog to the gyf in the X chromosome of A. ludens (alignment coordinates in the X chromosome: 11,460,704–11,456,115, e-value of 0.0, and identity of 58%), so this gene is in synteny between A. ludens and B. tryoni. We failed to find any paralog of this gene in A. ludens genome, which is consistent with the origin of the Y-linked typo-gyf in a clade of the Bactrocera genus (Choo et al. 2019). The annotation pipeline predicted 5 isoforms (GenBank: XM_054113069.1—XM_054113073.1) for the gyf gene, all containing 8 exons and with a range of sizes from 5,670 to 5,864 bp. Although the function of this gene has not been experimentally validated in Tephritidae, the gyf homolog in Drosophila has a role in the regulation of autophagy (Kim et al. 2015). Additionally, we could not identify any MoY-like sequences in the Mexican fruit fly genome assembly, even when applying relaxed set of BLAST parameters (e-value = 10−5 to 10) for comparisons based on nucleotide and protein sequences of the gene described for B. dorsalis. MoY is a Y-specific gene in tephritid fruit flies, encoding for a small protein necessary for normal male development (Meccariello et al. 2019). In the context of Tephritidae phylogeny (Han and Ro 2016; Congrains et al. 2023; Sim et al. 2024), this result suggests that MoY has been lost or diverged rapidly during the evolution of Anastrepha lineage. Alternatively, it is possible that this gene is specific to the subfamily Dacinae, as previously proposed (Meccariello et al. 2019). Additionally, given the apparent fragmentation of the Y chromosome into 36 pieces in our assembly, it is also possible that the genomic region for MoY has not been covered by our sequencing approach. A more contiguous assembly of the Y chromosome of an Anastrepha species may help to clarify the origin of the MoY gene.
Conclusion
In this study, we present a highly contiguous (N50 = 131 Mb) and complete (98.9% of complete BUSCOs) chromosome-scale genome assembly of the polyphagous pest A. ludens, the Mexican fruit fly. This high-quality genome assembly enables us to identify a major scaffold (∼100 Mb) and 8 contigs (totaling of 1.8 Mb) linked to the X chromosome, as well as a substantial part of the Y chromosome (1 scaffold and 35 contigs totaling 10.4 Mb in length). Our findings revealed that these sex chromosomes have distinctive TE and gene content when compared with the autosomes. Furthermore, an analysis of evolutionary rates between A. ludens and A. obliqua indicates that X-linked genes have evolved faster than autosome-linked genes, which is concordant with the faster-X effect. In addition to the valuable contribution with new insights into the evolution of X chromosome within the Tephritidae family, the reported genome assembly is a relevant resource for developing new molecular tools to manage this economically important fruit pest.
Supplementary Material
Acknowledgments
This research used resources provided by the SCINet project of the USDA Agricultural Research Service, ARS project number 2040-22430-028-000-D. This material was made possible, in part, by a Cooperative Agreement from the APHIS. It may not necessarily express APHIS’ views. USDA is an equal opportunity employer. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. We thank Norman B. Barr and Terrance N. Todd for contributing to this study.
Contributor Information
Carlos Congrains, U.S. Department of Agriculture-Agricultural Research Service, Daniel K. Inouye U.S. Pacific Basin Agricultural Research Center, Tropical Pest Genetics and Molecular Biology Research Unit, Hilo, HI 96720, USA; Department of Plant and Environmental Protection Services, University of Hawaii at Manoa, Honolulu, HI 96822, USA.
Sheina B Sim, U.S. Department of Agriculture-Agricultural Research Service, Daniel K. Inouye U.S. Pacific Basin Agricultural Research Center, Tropical Pest Genetics and Molecular Biology Research Unit, Hilo, HI 96720, USA.
Daniel F Paulo, U.S. Department of Agriculture-Agricultural Research Service, Daniel K. Inouye U.S. Pacific Basin Agricultural Research Center, Tropical Pest Genetics and Molecular Biology Research Unit, Hilo, HI 96720, USA; Department of Plant and Environmental Protection Services, University of Hawaii at Manoa, Honolulu, HI 96822, USA.
Renee L Corpuz, U.S. Department of Agriculture-Agricultural Research Service, Daniel K. Inouye U.S. Pacific Basin Agricultural Research Center, Tropical Pest Genetics and Molecular Biology Research Unit, Hilo, HI 96720, USA.
Angela N Kauwe, U.S. Department of Agriculture-Agricultural Research Service, Daniel K. Inouye U.S. Pacific Basin Agricultural Research Center, Tropical Pest Genetics and Molecular Biology Research Unit, Hilo, HI 96720, USA.
Tyler J Simmonds, U.S. Department of Agriculture-Agricultural Research Service, Daniel K. Inouye U.S. Pacific Basin Agricultural Research Center, Tropical Pest Genetics and Molecular Biology Research Unit, Hilo, HI 96720, USA.
Sheron A Simpson, U.S. Department of Agriculture-Agricultural Research Service, Genomics and Bioinformatics Research Unit, Stoneville, MS 38776, USA.
Brian E Scheffler, U.S. Department of Agriculture-Agricultural Research Service, Genomics and Bioinformatics Research Unit, Stoneville, MS 38776, USA.
Scott M Geib, U.S. Department of Agriculture-Agricultural Research Service, Daniel K. Inouye U.S. Pacific Basin Agricultural Research Center, Tropical Pest Genetics and Molecular Biology Research Unit, Hilo, HI 96720, USA.
Data availability
Raw data of PacBio long-reads and Hi-C reads were deposited in the Sequence Read Archive of the NCBI under accession numbers SRX25519850 and SRX14205169, respectively. The A. ludens genome assembly is available under the accession number GCF_028408465.1 in the NCBI Assembly database. The annotated features can be accessed at https://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/28586/GCF_028408465.1-RS_2023_03/. The NCBI accession numbers of the WGS data used for the AD-ratio analysis can be found in the Supplementary Table 7 in File 1. Scripts and commands used to assemble the genome can be found at https://doi.org/10.15482/USDA.ADC/25762509.v1.
Supplemental material available at G3 online.
Funding
This study was funded by the USDA Agricultural Research Service (ARS) with project “Advancing Molecular Pest Management, Diagnostics, and Eradication of Fruit Flies and Invasive Species” (2040-22430-028-000-D) as well as USDA-APHIS Plant Protection Act 7721 with projects “Developing molecular diagnostic tools to determine strain and mating status of fruit fly incursions” (3.0271) and “Integrative identification methods for Bactrocera fruit flies” (3.0624). This study was also partially supported by a USDA-National Institute of Food and Agriculture (NIFA) Agriculture and Food Research Initiative (AFRI) Competitive grant awarded to SBS (CALW-2016-04601). This research used resources provided by the SCINet project and/or the AI Center of Excellence of the USDA Agricultural Research Service, ARS project numbers 0201-88888-003-000D and 0201-88888-002-000D.
Literature Cited
- The Gene Ontology Consortium; Aleksander SA, Balhoff J, Carbon S, Cherry JM, Drabkin HJ, Ebert D, Feuermann M, Gaudet P, Harris NL, et al. 2023. The gene ontology knowledgebase in 2023. Genetics. 224(1):iyad031. doi: 10.1093/genetics/iyad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluja M. 1994. Bionomics and management of Anastrepha. Annu Rev Entomol. 39(1):155–178. doi: 10.1146/annurev.en.39.010194.001103. [DOI] [Google Scholar]
- Anand L, Rodriguez Lopez CM. 2022. ChromoMap: an R package for interactive visualization of multi-omics data and annotation of chromosomes. BMC Bioinformatics. 23(1):33. doi: 10.1186/s12859-021-04556-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APHIS . 2023. Regulated plant pest table. [accessed 2023 Apr 28]. https://www.aphis.usda.gov/aphis/ourfocus/planthealth/import-information/rppl/rppl-table.
- Bachtrog D. 2003. Adaptation shapes patterns of genome evolution on sexual and asexual chromosomes in Drosophila. Nat Genet. 34(2):215–219. doi: 10.1038/ng1164. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Carrel L, Chakravarti A, Eichler EE. 2000. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci U S A. 97(12):6634–6639. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AC, Stone WE, Plummer CC, McPhail M.. 1944. A Review of Studies on the Mexican Fruit Fly and Related Mexican Species. USDA Miscellaneous Publications 531. Washington (DC): U.S. Government Printing Office. p. 155. [Google Scholar]
- Barros de Andrade e Sousa L, Jonkers I, Syx L, Dunkel I, Chaumeil J, Picard C, Foret B, Chen C-J, Lis JT, Heard E, et al. 2019. Kinetics of Xist-induced gene silencing can be predicted from combinations of epigenetic and genomic features. Genome Res. 29(7):1087–1099. doi: 10.1101/gr.245027.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry JD, McInnis DO, Gates D, Morse JG. 2003. Effects of irradiation on Mediterranean fruit flies (Diptera: Tephritidae): emergence, survivorship, lure attraction, and mating competition. J Econ Entomol. 96(3):615–622. doi: 10.1093/jee/96.3.615. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, Hillier LW, Poh Y-P, Hahn MW, Nista PM, Jones CD, Kern AD, Dewey CN, et al. 2007. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 5(11):e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellott DW, Skaletsky H, Pyntikova T, Mardis ER, Graves T, Kremitzki C, Brown LG, Rozen S, Warren WC, Wilson RK, et al. 2010. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature. 466(7306):612–616. doi: 10.1038/nature09172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidon T, Schreck N, Hailer F, Nilsson MA, Janke A. 2015. Genome-wide search identifies 1.9 Mb from the polar bear Y chromosome for evolutionary analyses. Genome Biol Evol. 7(7):2010–2022. doi: 10.1093/gbe/evv103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA PLH Panel; Bragard C, Dehnen-Schmutz K, Di Serio F, Gonthier P, Jacques M-A, Jaques Miret JA, Justesen AF, Magnusson CS, Milonas P, et al. 2020. Pest categorisation of non-EU Tephritidae. EFSA J. 18(1):e05931. doi: 10.2903/j.efsa.2020.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Reuter K, Drost H-G. 2021. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 18(4):366–368. doi: 10.1038/s41592-021-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JN, Adey A, Patwardhan RP, Qiu R, Kitzman JO, Shendure J. 2013. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. 31(12):1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics. 10(1):421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. 2021. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol. 38(12):5825–5829. doi: 10.1093/molbev/msab293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey SB, Lovell JT, Jenkins J, Leebens-Mack J, Schmutz J, Wilson MA, Harkess A. 2022. Representing sex chromosomes in genome assemblies. Cell Genom. 2(5):100132. doi: 10.1016/j.xgen.2022.100132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis R, Richards E, Rajan J, Cochrane G, Blaxter M. 2020. BlobToolKit—interactive quality assessment of genome assemblies. G3 (Bethesda). 10(4):1361–1374. doi: 10.1534/g3.119.400908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. 1987. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 130(1):113–146. doi: 10.1086/284701. [DOI] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Concepcion GT, Feng X, Zhang H, Li H. 2021. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 18(2):170–175. doi: 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo A, Fung E, Nguyen TNM, Okada A, Crisp P.. 2022. CRISPR/Cas9 mutagenesis to generate novel traits in Bactrocera tryoni for sterile insect technique. In: Verma PJ, Sumer H, Liu J, editors. Applications of Genome Modulation and Editing. New York (NY): Springer. p. 151–171. [Google Scholar]
- Choo A, Nguyen TNM, Ward CM, Chen IY, Sved J, Shearman D, Gilchrist AS, Crisp P, Baxter SW. 2019. Identification of Y-chromosome scaffolds of the Queensland fruit fly reveals a duplicated gyf gene paralogue common to many Bactrocera pest species. Insect Mol Biol. 28(6):873–886. doi: 10.1111/imb.12602. [DOI] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, et al. 2010. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 141(6):956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- Congrains C, Dupuis JR, Rodriguez EJ, Norrbom AL, Steck G, Sutton B, Nolazco N, de Brito RA, Geib SM. 2023. Phylogenomic analysis provides diagnostic tools for the identification of Anastrepha fraterculus (Diptera: Tephritidae) species complex. Evol Appl. 16(9):1598–1618. doi: 10.1111/eva.13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congrains C, Zucchi RA, de Brito RA. 2021. Phylogenomic approach reveals strong signatures of introgression in the rapid diversification of neotropical true fruit flies (Anastrepha: Tephritidae). Mol Phylogenet Evol. 162:107200. doi: 10.1016/j.ympev.2021.107200. [DOI] [PubMed] [Google Scholar]
- Darolti I, Fong LJM, Sandkam BA, Metzger DCH, Mank JE. 2023. Sex chromosome heteromorphism and the Fast-X effect in poeciliids. Mol Ecol. 32(16):4599–4609. doi: 10.1111/mec.17048. [DOI] [PubMed] [Google Scholar]
- Deakin JE, Potter S, O’Neill R, Ruiz-Herrera A, Cioffi MB, Eldridge MDB, Fukui K, Marshall Graves JA, Griffin D, Grutzner F, et al. 2019. Chromosomics: bridging the gap between genomes and chromosomes. Genes (Basel). 10(8):627. doi: 10.3390/genes10080627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Ren S, Liu Q, Zhou D, Zhong C, Jin Y, Xie L, Gu J, Xiao C. 2024. A high heterozygosity genome assembly of Aedes albopictus enables the discovery of the association of PGANT3 with blood-feeding behavior. BMC Genomics. 25(1):336. doi: 10.1186/s12864-024-10133-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschepper P, Vanbergen S, Zhang Y, Li Z, Hassani IM, Patel NA, Rasolofoarivao H, Singh S, Wee SL, De Meyer M, et al. 2023. Bactrocera dorsalis in the Indian Ocean: a tale of two invasions. Evol Appl. 16(1):48–61. doi: 10.1111/eva.13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz F, Luís A, Lima A, Nakamura AM, Fernandes F, Sobrinho I, de Brito RA. 2018. Evidence for introgression among three species of the Anastrepha fraterculus group, a radiating species complex of fruit flies. Front Genet. 9:359. doi: 10.3389/fgene.2018.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doellman MM, Hood GR, Gersfeld J, Driscoe A, Xu CCY, Sheehy RN, Holmes N, Yee WL, Feder JL. 2020. Identifying diagnostic genetic markers for a cryptic invasive agricultural pest: a test case using the apple maggot fly (Diptera: Tephritidae). Ann Entomol Soc Am. 113(4):246–256. doi: 10.1093/aesa/saz069. [DOI] [Google Scholar]
- Doorenweerd C, San Jose M, Geib S, Barr N, Rubinoff D. 2024. Genomic data reveal new species and the limits of mtDNA barcode diagnostics to contain a global pest species complex (Diptera: Tephritidae: Dacinae). Syst Entomol. 49(2):279–293. doi: 10.1111/syen.12616. [DOI] [Google Scholar]
- Dupuis JR, Bremer FT, Kauwe A, San Jose M, Leblanc L, Rubinoff D, Geib SM. 2018. HiMAP: robust phylogenomics from highly multiplexed amplicon sequencing. Mol Ecol Resour. 18(5):1000–1019. doi: 10.1111/1755-0998.12783. [DOI] [PubMed] [Google Scholar]
- Dupuis JR, Ruiz-Arce R, Barr NB, Thomas DB, Geib SM. 2019. Range-wide population genomics of the Mexican fruit fly: toward development of pathway analysis tools. Evol Appl. 12(8):1641–1660. doi: 10.1111/eva.12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, Aiden EL. 2016. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3(1):99–101. doi: 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust GG, Hall IM. 2014. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics. 30(17):2503–2505. doi: 10.1093/bioinformatics/btu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. 2007. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 41(1):331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote RH, Blanc FL, Norrbom AL. 1993. Handbook of the Fruit Flies (Diptera: Tephritidae) of America North of Mexico. Ithaca: Comstock Publishing Associates. [Google Scholar]
- Gan HM, Falk S, Morales HE, Austin CM, Sunnucks P, Pavlova A. 2019. Genomic evidence of neo-sex chromosomes in the eastern yellow robin. GigaScience. 8(9):giz111. doi: 10.1093/gigascience/giz111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez V, Hernandez-Ortiz E, Zepeta-Cisneros CS, Robinson AS, Zacharopoulou A, Franz G. 2009. Mitotic and polytene chromosome analysis in the Mexican fruit fly, Anastrepha ludens (Loew) (Diptera: Tephritidae). Genome. 52(1):20–30. doi: 10.1139/G08-099. [DOI] [PubMed] [Google Scholar]
- Hall AB, Qi Y, Timoshevskiy V, Sharakhova MV, Sharakhov IV, Tu Z. 2013. Six novel Y chromosome genes in Anopheles mosquitoes discovered by independently sequencing males and females. BMC Genomics. 14(1):273. doi: 10.1186/1471-2164-14-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H-Y, Ro K-E. 2016. Molecular phylogeny of the superfamily Tephritoidea (Insecta: Diptera) reanalysed based on expanded taxon sampling and sequence data. J Zool Syst Evol Res. 54(4):276–288. doi: 10.1111/jzs.12139. [DOI] [Google Scholar]
- Hayes K, Barton HJ, Zeng K. 2020. A study of faster-Z evolution in the great tit (Parus major). Genome Biol Evol. 12(3):210–222. doi: 10.1093/gbe/evaa044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrichs J, Robinson A. 2009. Sterile insect technique. In: Resh VH, Cardé RT, editors. Encyclopedia of Insects. 2nd ed. San Diego: Elsevier Science & Technology. p. 953–957. [Google Scholar]
- Hernandez-Ortiz V, Aluja M. 1993. Listado de especies del género neotropical Anastrepha (Diptera: Tephritidae) con notas sobre su distribución y plantas hospederas. Folia Entomol Mex. 88:89–105. [Google Scholar]
- Hill MP, Bertelsmeier C, Clusella-Trullas S, Garnas J, Robertson MP, Terblanche JS. 2016. Predicted decrease in global climate suitability masks regional complexity of invasive fruit fly species response to climate change. Biol Invasions. 18(4):1105–1119. doi: 10.1007/s10530-016-1078-5. [DOI] [Google Scholar]
- Hoencamp C, Dudchenko O, Elbatsh AMO, Brahmachari S, Raaijmakers JA, van Schaik T, Sedeño Cacciatore Á, Contessoto VG, van Heesbeen RGHP, van den Broek B, et al. 2021. 3D genomics across the tree of life reveals condensin II as a determinant of architecture type. Science 372(6545):984–989. doi: 10.1126/science.abe2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro José MC, Wides R, et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 298(5591):129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Hoskins RA, Carlson JW, Wan KH, Park S, Mendez I, Galle SE, Booth BW, Pfeiffer BD, George RA, Svirskas R, et al. 2015. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 25(3):445–458. doi: 10.1101/gr.185579.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Eisen MB, Thornton KR, Andolfatto P. 2013. A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence. Genome Res. 23(1):89–98. doi: 10.1101/gr.141689.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, et al. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47(D1):D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvilsom C, Qian Y, Bataillon T, Li Y, Mailund T, Sallé B, Carlsen F, Li R, Zheng H, Jiang T, et al. 2012. Extensive X-linked adaptive evolution in central chimpanzees. Proc Natl Acad Sci U S A. 109(6):2054–2059. doi: 10.1073/pnas.1106877109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Liang L, Wang J, Zhu S. 2022. Chromosome-level genome assembly of Bactrocera dorsalis reveals its adaptation and invasion mechanisms. Commun Biol. 5(1):25. doi: 10.1038/s42003-021-02966-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, et al. 2002. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 3(12):research0084. doi: 10.1186/gb-2002-3-12-research0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandul NP, Liu J, Sanchez C HM, Wu SL, Marshall JM, Akbari OS. 2019. Transforming insect population control with precision guided sterile males with demonstration in flies. Nat Commun. 10(1):84. doi: 10.1038/s41467-018-07964-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kejnovsky E, Hobza R, Cermak T, Kubat Z, Vyskot B. 2009. The role of repetitive DNA in structure and evolution of sex chromosomes in plants. Heredity (Edinb). 102(6):533–541. doi: 10.1038/hdy.2009.17. [DOI] [PubMed] [Google Scholar]
- Kim M, Semple I, Kim B, Kiers A, Nam S, Park H-W, Park H, Ro S-H, Kim J-S, Juhász G, et al. 2015. Drosophila Gyf/GRB10 interacting GYF protein is an autophagy regulator that controls neuron and muscle homeostasis. Autophagy. 11(8):1358–1372. doi: 10.1080/15548627.2015.1063766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousathanas A, Halligan DL, Keightley PD. 2014. Faster-X adaptive protein evolution in house mice. Genetics. 196(4):1131–1143. doi: 10.1534/genetics.113.158246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriventseva EV, Kuznetsov D, Tegenfeldt F, Manni M, Dias R, Simão FA, Zdobnov EM. 2019. OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 47(D1):D807–D811. doi: 10.1093/nar/gky1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeta-Escamilla A, Hernández E, Arredondo J, Díaz-Fleischer F, Pérez-Staples D. 2016. Male irradiation affects female remating behavior in Anastrepha serpentina (Diptera: Tephritidae). J Insect Physiol. 85:17–22. doi: 10.1016/j.jinsphys.2015.11.011. [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 1303.3997. http://arxiv.org/abs/1303.3997.
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34(18):3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2021. New strategies to improve minimap2 alignment accuracy. Bioinformatics. 37(23):4572–4574. doi: 10.1093/bioinformatics/btab705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukyanchikova V, Nuriddinov M, Belokopytova P, Taskina A, Liang J, Reijnders MJMF, Ruzzante L, Feron R, Waterhouse RM, Wu Y, et al. 2022. Anopheles mosquitoes reveal new principles of 3D genome organization in insects. Nat Commun. 13(1):1960. doi: 10.1038/s41467-022-29599-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S, Bachtrog D. 2017. Convergent evolution of Y chromosome gene content in flies. Nat Commun. 8(1):785. doi: 10.1038/s41467-017-00653-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Axelsson E, Ellegren H. 2007. Fast-X on the Z: rapid evolution of sex-linked genes in birds. Genome Res. 17(5):618–624. doi: 10.1101/gr.6031907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of Eukaryotic, Prokaryotic, and Viral genomes. Mol Biol Evol. 38(10):4647–4654. doi: 10.1093/molbev/msab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meccariello A, Hou S, Davydova S, Fawcett JD, Siddall A, Leftwich PT, Krsticevic F, Papathanos PA, Windbichler N. 2024. Gene drive and genetic sex conversion in the global agricultural pest Ceratitis capitata. Nat Commun. 15(1):372. doi: 10.1038/s41467-023-44399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meccariello A, Salvemini M, Primo P, Hall B, Koskinioti P, Dalíková M, Gravina A, Gucciardino MA, Forlenza F, Gregoriou M-E, et al. 2019. Maleness-on-the-Y (MoY) orchestrates male sex determination in major agricultural fruit fly pests. Science. 365(6460):1457–1460. doi: 10.1126/science.aax1318. [DOI] [PubMed] [Google Scholar]
- Meisel RP, Connallon T. 2013. The faster-X effect: integrating theory and data. Trends Genet. 29(9):537–544. doi: 10.1016/j.tig.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérel V, Boulesteix M, Fablet M, Vieira C. 2020. Transposable elements in Drosophila. Mob DNA. 11(1):23. doi: 10.1186/s13100-020-00213-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga KH, Koren S, Rhie A, Vollger MR, Gershman A, Bzikadze A, Brooks S, Howe E, Porubsky D, Logsdon GA, et al. 2020. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 585(7823):79–84. doi: 10.1038/s41586-020-2547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, et al. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res. 49(D1):D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongue AJ, Hansen ME, Walters JR. 2022. Support for faster and more adaptive Z chromosome evolution in two divergent lepidopteran lineages*. Evolution. 76(2):332–345. doi: 10.1111/evo.14341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrbom A. 2022. Anastrepha ludens (Mexican fruit fly). CABI Compendium. Vol. Pest, Natural enemy, Invasive species. Wallingford (UK): CAB International. [Google Scholar]
- Norrbom AL, Kim KC. 1988. A List of the Reported Host Plants of the species of Anastrepha (Diptera: Tephritidae). Riverdale (MD): U.S. Department of Agriculture, Animal and Plant Health Inspection Service, Plant Protection and Quarantine. [Google Scholar]
- Norrbom AL, Korytkowski CA, Zucchi RA, Uramoto K, Venable GL, McCormick J, Dallwitz MJ.. 2012. Anastrepha and Toxotrypana: descriptions, illustrations, and interactive keys. [accessed 2021 Nov 15]. http://delta-intkey.com/anatox/indext.htm.
- Orozco-Dávila D, de Lourdes Adriano-Anaya M, Quintero-Fong L, Salvador-Figueroa M. 2015. Sterility and sexual competitiveness of Tapachula-7 Anastrepha ludens males irradiated at different doses. PLoS One. 10(8):e0135759. doi: 10.1371/journal.pone.0135759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Su W, Liao Y, Chougule K, Agda JRA, Hellinga AJ, Lugo CSB, Elliott TA, Ware D, Peterson T, et al. 2019. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 20(1):275. doi: 10.1186/s13059-019-1905-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa Y, Wellenreuther M, Morrison MA, Ritchie PA. 2023. Genome assembly and isoform analysis of a highly heterozygous New Zealand fisheries species, the tarakihi (Nemadactylus macropterus). G3 (Bethesda). 13(2):jkac315. doi: 10.1093/g3journal/jkac315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanicolaou A, Schetelig MF, Arensburger P, Atkinson PW, Benoit JB, Bourtzis K, Castañera P, Cavanaugh JP, Chao H, Childers C, et al. 2016. The whole genome sequence of the Mediterranean fruit fly, Ceratitis capitata (Wiedemann), reveals insights into the biology and adaptive evolution of a highly invasive pest species. Genome Biol. 17(1):192. doi: 10.1186/s13059-016-1049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker DJ, Jaron KS, Dumas Z, Robinson-Rechavi M, Schwander T. 2022. X chromosomes show relaxed selection and complete somatic dosage compensation across Timema stick insect species. J Evol Biol. 35(12):1734–1750. doi: 10.1111/jeb.14075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo DF, Cha AY, Kauwe AN, Curbelo K, Corpuz RL, Simmonds TJ, Sim SB, Geib SM. 2022. A unified protocol for CRISPR/Cas9-mediated gene knockout in tephritid fruit flies Led to the recreation of white eye and white puparium phenotypes in the melon fly. J Econ Entomol. 115(6):2110–2115. doi: 10.1093/jee/toac166. [DOI] [PubMed] [Google Scholar]
- Paulo DF, Nguyen TNM, Corpuz RL, Kauwe AN, Rendon P, Ruano REY, Cardoso AAS, Gouvi G, Fung E, Crisp P, et al. 2024. The genetic basis of the black pupae phenotype in tephritid fruit flies. bioRxiv 597636. doi: 10.1101/2024.06.07.597636, preprint: not peer reviewed. [DOI]
- Pedersen BS, Quinlan AR. 2018. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics. 34(5):867–868. doi: 10.1093/bioinformatics/btx699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perre P, Jorge LR, Lewinsohn TM, Zucchi RA. 2014. Morphometric differentiation of fruit fly pest species of the Anastrepha fraterculus group (Diptera: Tephritidae). Ann Entomol Soc Am. 107(2):490–495. doi: 10.1603/AN13122. [DOI] [Google Scholar]
- Pertea G, Pertea M. 2020. GFF Utilities: GffRead and GffCompare. F1000Res. 9:ISCB Comm J-304. doi: 10.12688/f1000research.23297.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl C. 1885. Uber Zelltheilung. Morphol Jahrb. 10:214–330. [Google Scholar]
- Ramírez-Santos E, Rendon P, Gouvi G, Zacharopoulou A, Bourtzis K, Cáceres C, Bloem K. 2021. A novel genetic sexing strain of Anastrepha ludens for cost-effective sterile insect technique applications: improved genetic stability and rearing efficiency. Insects. 12(6):499. doi: 10.3390/insects12060499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo-Benavidez TR, Jaron KS, Schatz MC. 2020. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 11(1):1432. doi: 10.1038/s41467-020-14998-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2022. R: A Language and Environment for Statistical Computing. https://www.R-project.org/.
- Rech GE, Radío S, Guirao-Rico S, Aguilera L, Horvath V, Green L, Lindstadt H, Jamilloux V, Quesneville H, González J. 2022. Population-scale long-read sequencing uncovers transposable elements associated with gene expression variation and adaptive signatures in Drosophila. Nat Commun. 13:1948. doi: 10.1038/s41467-022-29518-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, Nurk S, Cechova M, Hoyt SJ, Taylor DJ, Altemose N, Hook PW, Koren S, Rautiainen M, Alexandrov IA, et al. 2023. The complete sequence of a human Y chromosome. Nature. 621(7978):344–354. doi: 10.1038/s41586-023-06457-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousselle M, Faivre N, Ballenghien M, Galtier N, Nabholz B. 2016. Hemizygosity enhances purifying selection: lack of fast-Z evolution in two Satyrine butterflies. Genome Biol Evol. 8(10):3108–3119. doi: 10.1093/gbe/evw214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Arce R, Owen CL, Thomas DB, Barr NB, McPheron BA. 2015. Phylogeographic structure in Anastrepha ludens (Diptera: Tephritidae) populations inferred with mtDNA sequencing. J Econ Entomol. 108(3):1324–1336. doi: 10.1093/jee/tov082. [DOI] [PubMed] [Google Scholar]
- Scally M, Into F, Thomas DB, Ruiz-Arce R, Barr NB, Schuenzel EL. 2016. Resolution of inter and intra-species relationships of the West Indian fruit fly Anastrepha obliqua. Mol Phylogenet Evol. 101:286–293. doi: 10.1016/j.ympev.2016.04.020. [DOI] [PubMed] [Google Scholar]
- Sessegolo C, Burlet N, Haudry A. 2016. Strong phylogenetic inertia on genome size and transposable element content among 26 species of flies. Biol Lett. 12(8):20160407. doi: 10.1098/rsbl.2016.0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim SB, Congrains C, Velasco-Cuervo S, Corpuz RL, Kauwe AN, Scheffler B, Geib SM. 2024. Genome report: chromosome-scale genome assembly of the West Indian fruit fly Anastrepha obliqua (Diptera: Tephritidae). G3 (Bethesda) GenesGenomesGenetics. 14(4):jkae024. doi: 10.1093/g3journal/jkae024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim SB, Corpuz RL, Simmonds TJ, Geib SM. 2022. HiFiAdapterFilt, a memory efficient read processing pipeline, prevents occurrence of adapter sequence in PacBio HiFi reads and their negative impacts on genome assembly. BMC Genomics. 23(1):157. doi: 10.1186/s12864-022-08375-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim SB, Kauwe AN, Ruano REY, Rendon P, Geib SM. 2019. The ABCs of CRISPR in Tephritidae: developing methods for inducing heritable mutations in the genera Anastrepha, Bactrocera and Ceratitis. Insect Mol Biol. 28(2):277–289. doi: 10.1111/imb.12550. [DOI] [PubMed] [Google Scholar]
- Skendžić S, Zovko M, Živković IP, Lešić V, Lemić D. 2021. The impact of climate change on agricultural insect pests. Insects. 12(5):440. doi: 10.3390/insects12050440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34(Web Server):W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supple MA, Escalona M, Adkins J, Buchalski MR, Alexandre N, Sahasrabudhe RM, Nguyen O, Sacco S, Fairbairn C, Beraut E, et al. 2024. A genome assembly of the American black bear, Ursus americanus, from California. J Hered. 115(5):498–506. doi: 10.1093/jhered/esae037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved JA, Chen Y, Shearman D, Frommer M, Gilchrist AS, Sherwin WB. 2016. Extraordinary conservation of entire chromosomes in insects over long evolutionary periods. Evolution. 70(1):229–234. doi: 10.1111/evo.12831. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 4(1):41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science. 278(5338):631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Thibaud-Nissen F, DiCuccio M, Hlavina W, Kimchi A, Kitts PA, Murphy TD, Pruitt KD, Souvorov A. 2016. P8008 The NCBI Eukaryotic genome annotation pipeline. J Anim Sci. suppl_4(94):184. doi: 10.2527/jas2016.94supplement4184x. [DOI] [Google Scholar]
- Tomaszkiewicz M, Medvedev P, Makova KD. 2017. Y and W chromosome assemblies: approaches and discoveries. Trends Genet. 33(4):266–282. doi: 10.1016/j.tig.2017.01.008. [DOI] [PubMed] [Google Scholar]
- Vasimuddin M, Misra S, Li H, Aluru S.. 2019. Efficient architecture-aware acceleration of BWA-MEM for multicore systems. In: 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS). Rio de Janeiro (Brazil). p. 314–324.
- Vicoso B. 2019. Molecular and evolutionary dynamics of animal sex-chromosome turnover. Nat Ecol Evol. 3(12):1632–1641. doi: 10.1038/s41559-019-1050-8. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D. 2013. Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature. 499(7458):332–335. doi: 10.1038/nature12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D. 2015. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. 13(4):e1002078. doi: 10.1371/journal.pbio.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. 2009. Effective population size and the faster-X effect: an extended model. Evolution. 63(9):2413–2426. doi: 10.1111/j.1558-5646.2009.00719.x. [DOI] [PubMed] [Google Scholar]
- Wells JN, Feschotte C. 2020. A field guide to Eukaryotic transposable elements. Annu Rev Genet. 54(1):539–561. doi: 10.1146/annurev-genet-040620-022145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJD. 1949. Cytological evidence on the phylogeny and classification of the Diptera. Evolution. 3(3):252–261. doi: 10.2307/2405562. [DOI] [PubMed] [Google Scholar]
- Whittle CA, Kulkarni A, Extavour CG. 2020. Absence of a faster-X effect in beetles (Tribolium, Coleoptera). G3 (Bethesda). 10(3):1125–1136. doi: 10.1534/g3.120.401074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 8(12):973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Wu S, Wang K, Dou T, Yuan S, Yan S, Xu Z, Liu Y, Jian Z, Zhao J, Zhao R, et al. 2024. High quality assemblies of four indigenous chicken genomes and related functional data resources. Sci Data. 11(1):300. doi: 10.1038/s41597-024-03126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Gao Y, Wu M, Tian T, Fan H, Huang Y, Huang Z, Li D, Xu L. 2021. Telomere-to-telomere assembly of a fish Y chromosome reveals the origin of a young sex chromosome pair. Genome Biol. 22(1):203. doi: 10.1186/s13059-021-02430-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Jing W, Li S, Li R, Bolund L, et al. 2006. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34(Web Server):W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Zhang Y, Cui H, Liu J, Wu Y, Cheng Y, Xu H, Huang X, Li S, Zhou A, et al. 2018. WEGO 2.0: a web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 46(W1):W71–W75. doi: 10.1093/nar/gky400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepeda-Cisneros CS, Meza Hernández JS, García-Martínez V, Ibañez-Palacios J, Zacharopoulou A, Franz G.. 2014. Development, genetic and cytogenetic analyses of genetic sexing strains of the Mexican fruit fly, Anastrepha ludens Loew (Diptera: Tephritidae). BMC Genom Data. 15(S2):S1. doi: 10.1186/1471-2156-15-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. 2022. Kaks_Calculator 3.0: calculating selective pressure on coding and non-coding sequences. Genomics Proteomics Bioinformatics. 20(3):536–540. doi: 10.1016/j.gpb.2021.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu S, Meyer D, Liao M, Zhao Z, Virgilio Y, Feng M, Qin S, Singh Y, Wee S, et al. 2023. Genomes of the cosmopolitan fruit pest Bactrocera dorsalis (Diptera: Tephritidae) reveal its global invasion history and thermal adaptation. J Adv Res. 53:61–74. doi: 10.1016/j.jare.2022.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, McCarthy SA, Durbin R. 2023. YaHS: yet another Hi-C scaffolding tool. Bioinformatics. 39(1):btac808. doi: 10.1093/bioinformatics/btac808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchi RA. 2000. Taxonomia. In: Malavasi A, Zucchi RA, editors. Moscas-das-Frutas de Importância Econômica no Brasil: Conhecimento Básico e Aplicado. Ribeirão Preto (Brazil): Holos. p. 13–24. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data of PacBio long-reads and Hi-C reads were deposited in the Sequence Read Archive of the NCBI under accession numbers SRX25519850 and SRX14205169, respectively. The A. ludens genome assembly is available under the accession number GCF_028408465.1 in the NCBI Assembly database. The annotated features can be accessed at https://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/28586/GCF_028408465.1-RS_2023_03/. The NCBI accession numbers of the WGS data used for the AD-ratio analysis can be found in the Supplementary Table 7 in File 1. Scripts and commands used to assemble the genome can be found at https://doi.org/10.15482/USDA.ADC/25762509.v1.
Supplemental material available at G3 online.