Abstract
Background
Coxiella burnetii is a zoonotic pathogen that causes Q fever and is found worldwide. Ticks serve as the primary reservoir, playing an important role in maintaining the natural cycle of C. burnetii. C. burnetii is transmitted to animals when ticks feed on their blood. However, information on C. burnetii infection in ticks remains limited, despite the widespread prevalence of the infection in humans and animals across China.
Methods
In this study, 192 engorged ticks were collected from Baiyin City of Gansu Province, China. The presence of Coxiella burnetii in ticks was specifically identified by detecting the IS1111 gene using nested polymerase chain reaction (nPCR). In addition, the 16S rRNA gene of C. burnetii was molecularly characterized using nPCR. A total of 10 spacer sequences (Cox 2, 5, 18, 20, 22, 37, 51, 56, 57, and 61) were amplified using PCR against positive specimens for MST analysis.
Results
All collected ticks were identified as Hyalomma marginatum, and 90 of them tested positive for C. burnetii, with a positive rate of 46.9% (90/192). The 16S rRNA gene analysis showed that the novel C. burnetii variants detected in this study were closely related to other C. burnetii strains in the world. The allele codes found in the present study for loci Cox2-Cox5-Cox18-Cox20-Cox22-Cox37-Cox51-Cox56-Cox57-Cox61 were 8-4-9-5-7-5-2-3-11-6. This represents a novel combination of allele values, similar to MST28, currently designated as MST85 in the Multi Spacers Typing (MST) database.
Conclusion
Our results revealed the circulation of a novel MST genotype of C. burnetii in Baiyin City, Gansu Province, China. The detection of C. burnetii in ticks suggests a potential public health risk to the local human population.
Keywords: ticks, Coxiella burnetii, IS1111, 16S rRNA, genotyping, MST
Introduction
Q fever, caused by the obligate intracellular bacterium Coxiella burnetii, is a worldwide disease that can infect both animals and humans (1). It was first reported in Australia in 1935 (2), with the Netherlands having the highest prevalence (3). Q fever has spread to almost all countries worldwide (4). It is now one of the most widely distributed zoonotic diseases, affecting the health of both humans and animals (5, 6). Humans become infected mainly from animals through infected aerosols and the ingestion of raw milk or dairy products (6). Human infection can manifest with chills, fever, and headache (6, 7). At the same time, severe cases of Q fever presenting complications such as hepatitis, endocarditis, rare spinal infections, prosthetic joint infections, and even death have been reported (6–9). Meanwhile, since Q fever is not a legally reported infectious disease in China, its clinical symptoms are atypical and not emphasized and thus difficult to diagnose (10). The rate of clinical misdiagnosis and underdiagnosis is high, and the disease is also easily neglected (11).
Ticks can transmit C. burnetii to animals while feeding on their blood, and these animals can subsequently transmit the agent to humans (12). In addition, C. burnetii direct transmission to humans by ticks through biting has been reported, though it is rare (13); hence, its potential risk to humans should be considered. To date, more than 40 hard ticks from genera Haemaphysalis, Amblyomma, Rhipicephalus, Hyalomma, and Dermacentor and at least 14 soft ticks from Ornithodoros have been documented as vectors for C. burnetii (14–17). In China, C. burnetii has been identified in Hyalomma (18–20), Dermacentor (19–22), Rhipicephalus (19, 23), and Haemaphysalis (19–22, 24).
Multi Spacers Typing (MST), utilized for genotyping C. Burnetii (5, 25), not only reflects the predominant genotypes in each region but also enables strain sequence typing comparisons, thereby facilitating traceability to the source of C. burnetii infection (5, 25, 26). MST can be directly applied to DNA extracted from specimens without the need to culture pathogen isolates, offering the advantage of high reproducibility between laboratories (27). To date, 79 MST types have been identified based on the MST database, and only MST16 has been found in rats from Yunnan, China (28). Furthermore, four potential novel MST types were identified, including two in rats from Yunnan (28) and another two in hedgehogs from Hubei (29).
Baiyin City is located in the central part of Gansu Province, near Lanzhou, and at least two species of ticks have been identified within its territory. Although C. burnetii has been identified in D. nuttalli, D. silvarum, Ha. japonica, and Hy. asiaticum from Gansu Province (20, 22), no studies on C. burnetii infection in ticks have been reported in Baiyin City, Gansu. In the present study, C. burnetii was screened in ticks from Baiyin City, and the MST types of C. burnetii were identified to determine its prevalence in ticks within the region.
Materials and methods
Collection and identification of ticks and DNA extraction
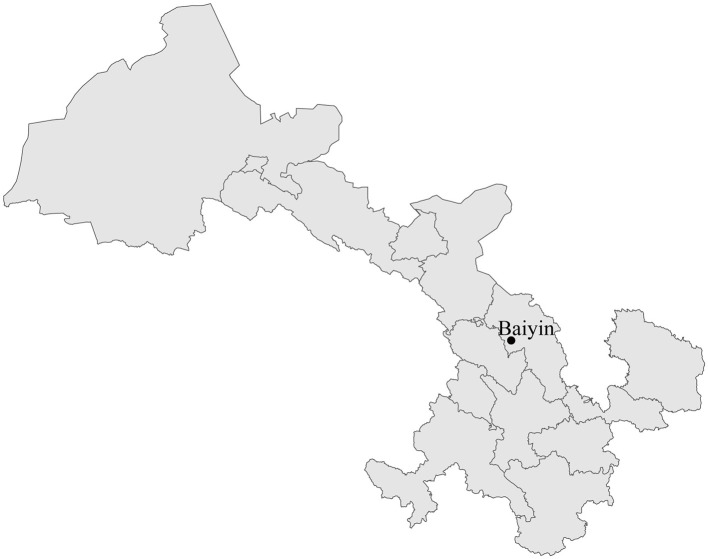
In August 2019, one adult tick was collected from the body of each goat in Baiyin City, Gansu Province (Figure 1). Ticks were initially identified at the species level based on morphological characteristics under a stereoscopic microscope. The taxonomical keys, mainly including the shape of the basis capitulum, palp, scutum, coxae I, anal groove, eyes, festoons, adanal plates, spiracle, and hypostomal teeth, were used for the identification of tick species. In addition, tick species were confirmed by analyzing the cytochrome c oxidase I (COI) gene sequence obtained using polymerase chain reaction (PCR) (30). All collected tick specimens were washed twice with 75% alcohol and then washed twice with phosphate-buffered saline (PBS). Following the manufacturer's instructions, total DNA was extracted from each tick using the tissue DNA extraction kit (Omega, Norcross, GA, USA). The extracted DNA sample was eluted into 80 μl ddH2O and stored at −80°C before screening C. burnetii.
Figure 1.
Map with the location of the collection site of ticks (•) in Baiyin City, Gansu Province, China.
Molecular identification and characterization of C. burnetii
Coxiella burnetii was screened by amplifying the IS1111 gene using nested polymerase chain reaction (nPCR). Primer pair QBT1/QBT2 was used for the first round of nPCR (31), and QBTN3/QBTN4 was used for the second round of nPCR (32), yielding a 440-bp amplicon.
To better understand the genetic characteristics, a partial 16S rRNA gene (624–627 bp) was amplified from the samples positive for C. burnetii using nPCR. Primer pairs Cox16S-F1/16S-R and 16S-F/16S-R were used as the first and second rounds, respectively (33). All primers used in this study are shown in Table 1.
Table 1.
Primer sequences used in this study.
| Target gene | Primer | Oligonucleotide sequences (5′- 3′) | Amplicon size (bp) | References |
|---|---|---|---|---|
| IS1111 | QBT1 | TATGTATCCACCGTAGCCAGTC | 687 | (31) |
| QBT2 | CCCAACAACACCTCCTTATTC | |||
| QBTN3 | AAGCGTGTGGAGGAGCGAACC | 440 | (32) | |
| QBTN4 | CTCGTAATCACCAATCGCTTCGTC | |||
| 16S rRNA | 16S-F1 | CGTAGGAATCTACCTTRTAGWGG | 624-627 | (33) |
| 16S-F | TGAGAACTAGCTGTTGGRRAGT | |||
| 16S-R | GCCTACCCGCTTCTGGTACAATT | |||
| COX2 | COX2F | CAACCCTGAATACCCAAGGA | 397 | (34) |
| COX2R | GAAGCTTCTGATAGGCGGGA | |||
| COX5 | COX5F | CAGGAGCAAGCTTGAATGCG | 395 | |
| COX5R | TGGTATGACAACCCGTCATG | |||
| COX18 | COX18F | CGCAGACGAATTAGCCAATC | 557 | |
| COX18R | TTCGATGATCCGATGGCCTT | |||
| COX20 | COX20F | GATATTTATCAGCGTCAAAGCAA | 631 | |
| COX20R | TCTATTATTGCAATGCAAGTGG | |||
| COX22 | COX22F | GGGAATAAGAGAGTTAGCTCA | 383 | |
| COX22R | CGCAAATTTCGGCACAGACC | |||
| COX37 | COX37F | GGCTTGTCTGGTGTAACTGT | 463 | |
| COX37R | ATTCCGGGACCTTCGTTAAC | |||
| COX51 | COX51F | TAACGCCCGAGAGCTCAGAA | 674 | |
| COX51R | GCGAGAACCGAATTGCTATC | |||
| COX56 | COX56F | CCAAGCTCTCTGTGCCCAAT | 479 | |
| COX56R | ATGCGCCAGAAACGCATAGG | |||
| COX57 | COX57F | TGGAAATGGAAGGCGGATTC | 617 | |
| COX57R | GGTGGAAGGCGTAAGCCTTT | |||
| COX61 | COX61F | GAAGATAGAGCGGCAAGGAT | 611 | |
| COX61R | GGGATTTCAACTTCCGATAGA |
MST genotype of C. burnetii
MST was performed to determine the genotypes of C. burnetii using PCR to target 10 spacers with the highest variability, as previously described (34). These spacers include Cox2, Cox5, Cox18, Cox20, Cox22, Cox37, Cox51, Cox56, Cox57, and Cox61. All primers used in this study are shown in Table 1.
Sequencing and nucleotide sequence analysis
The PCR products were analyzed using electrophoresis on a 1% agarose gel, and the spacer sequence PCR products were analyzed on a 1.2% agarose gel. All the PCR products of the expected size were purified and cloned into pMD19-T vectors (Takara, Dalian, China) for sequencing with the universal primers (Sangon, Beijing, China).
Bioedit v. 7. 1. 11 was used to edit all newly generated sequences in this study (35). The obtained IS1111 and 16S rRNA genes were analyzed using BLAST comparison on the NCBI website. The nucleotide sequence identities were calculated using the MegAlign program available within the Lasergene software package (36). To better understand the relationship between the C. burnetii identified in this study and other strains, the maximum-likelihood (ML) tree was reconstructed based on the 16S rRNA gene sequence using MEGA 6.0.6 software (37). The optimal nucleotide substitution model General Time Reversible (GTR) nucleotide substitution model as well as the gamma (G)-distribution and proportion of invariable sites (i.e., GTR+G+ I) were determined using the MEGA 6.0.6 (37). Bootstrap values were calculated from 1,000 replicates, and the phylogenetic trees were rooted at the midpoint for clarity.
Individual spacer sequences of C. burnetii obtained in this study were concatenated. The MST genotype was determined by comparing the results with the MST database of C. burnetii (https://ifr48.timone.Univ-mrs.Fr/mst/coxiella_burnetii/ accessed on 24 April 2024). A phylogenetic tree of the MST genotypes was built using the unweighted pair group method with the arithmetic mean method (UPGMA) using MEGA 6.0.6 (37). A minimum spanning tree was generated using the software GrapeTree with parameters implemented in MSTree v2 (http://localhost:8000/) for the 10 alleles from all STs (38).
Results
Identification of C. burnetii and ticks
A total of 192 ticks were collected from the body of goats, and all ticks were identified as Hy. marginatum based on the morphology. Subsequently, the COI gene sequence obtained from all ticks showed 99.4–100% nucleotide identity with each other and exhibited 97.7–98.7% nucleotide identity with known sequences of this tick species deposited in the GenBank database (GenBank numbers: OQ799122, PP330223, and KX000648). Furthermore, in the phylogenetic tree based on the COI gene, all newly generated sequences in this study had the closest relationship with those of Hy. marginatum (Figure 2). Therefore, all these ticks were confirmed to be Hy. marginatum.
Figure 2.
Molecular identification of ticks based on the phylogenetic analysis with the COI gene. The maximum-likelihood (ML) tree was reconstructed using the MEGA 6.0.6 under the GTR+G+I model with 1,000 replicates. The numbers at each node indicated bootstrap values, and only bootstrap values >70% are shown at appropriate nodes. Taxa marked by circles depict representative sequences obtained in this study.
Gel electrophoresis analysis showed that the size of the 90 PCR products was in accordance with the expected size. Sequencing of the PCR products and further BLAST showed that all these newly generated sequences most closely resembled those of C. burnetii and shared the highest nucleotide identity of 97.2–100% nucleotide identity with known IS1111 gene sequences of C. burnetii. The positive rate of C. burnetii infection in Hy. marginatum ticks was 46.9% (90/192). Moreover, all these 90 IS1111 gene sequences presented 99.5–100% nucleotide identity with each other. All the IS1111 gene sequences obtained in this study have been submitted to GenBank under the accession numbers PP929917–PP930006.
Molecular characterization of 16s rRNA gene of C. burnetii
To better understand the genetic characteristic, a partial 16S rRNA gene was successfully amplified from 56 out of 90 C. burnetii-positive tick specimens. After sequencing, 56 partial 16S rRNA gene sequences showed 99.1–100% nucleotide identity with known those of C. burnetii from the GenBank database. Furthermore, all these 56 partial 16S rRNA gene sequences presented 99.8–100% nucleotide identity with each other. All the 16S rRNA gene sequences obtained in this study have been submitted to the GenBank database under the accession numbers PP930513–PP930568.
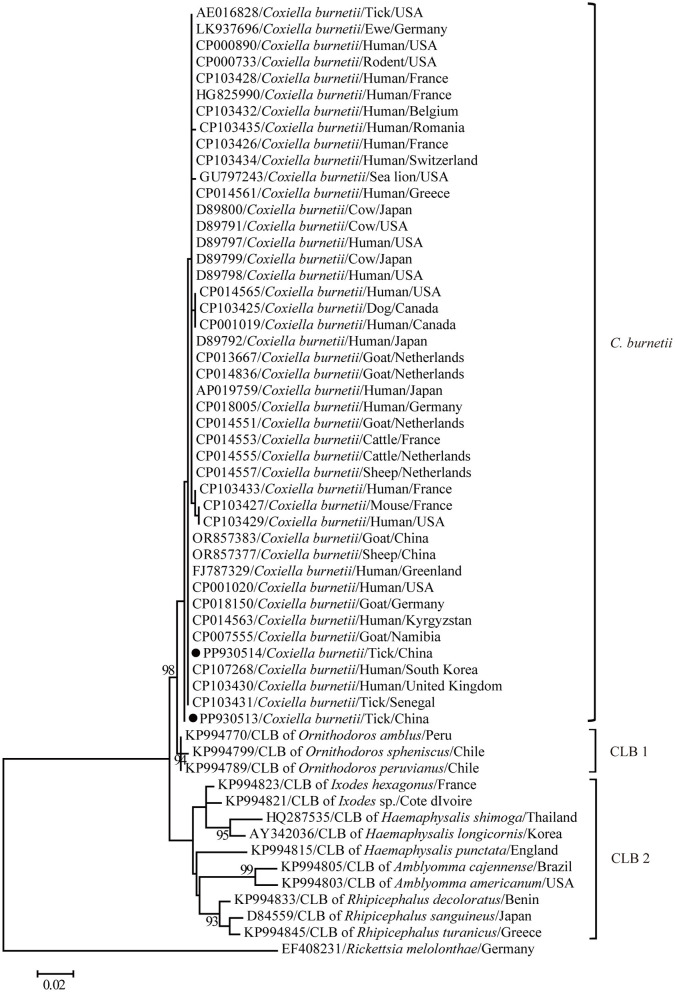
The maximum-likelihood tree based on the partial 16S rRNA gene sequences was reconstructed to get a better understanding of the relationships between the C. burnetii variants determined in this study and other known strains. In general, clear segregation into three clusters was observed in the partial 16S rRNA gene tree in this study: C. burnetii, CLB1, and CLB2 (Figure 3). Coxiella burnetii variants identified in this study clustered together with other known C. burnetii strains including those identified from humans and separated from two groups of Coxiella endosymbiont (CLB) (Figure 3). Furthermore, C. burnetii was closely related to CLB1 and distantly related to CLB2. Consistently, C. burnetii shared 98.4–99.3% and 93.8–97.7% nucleotide identities with CLB1 and CLB2 for the partial 16S rRNA gene, respectively. In addition, CLB1 was distantly related to CLB2 and only shared 93.3–96.9% nucleotide identity.
Figure 3.
A phylogenetic tree based on the 16S rRNA gene. The numbers at each node indicate bootstrap values, and only bootstrap values >70% are shown at appropriate nodes. Taxa marked by circles depict representative sequences obtained in this study.
MST genotyping of C. burnetii
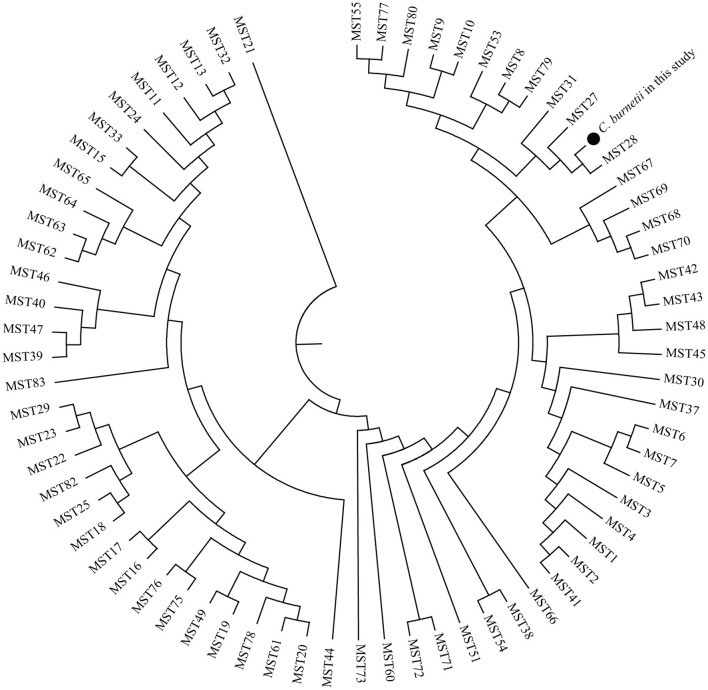
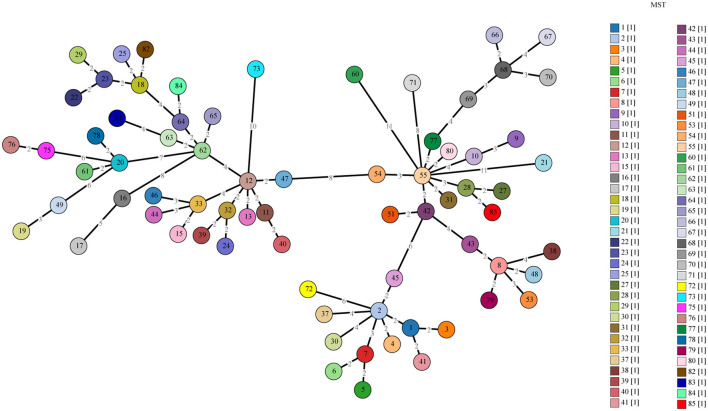
Allelic loci were successfully obtained at 10 allelic intervals from 55 tick samples (Table 2). The allele codes found in the present study for loci Cox2-Cox5-Cox18-Cox20-Cox22-Cox37-Cox51-Cox56-Cox57-Cox61 were 8-4-9-5-7-5-2-3-11-6. The allele values of the single spacer sequences of C. burnetii in different specimens are shown in Table 2. The 10 successfully amplified spacer sequences were combined and compared to the sequences in the MST database, and the results showed that the allele values identified in this study were a novel combination of allele values similar to MST28 found in sheep, cattle, ticks, and humans from Kazakhstan, Central Asia (Figure 4). Compared to MST28, which has an allele value of 4 for the Cox57 spacer, this combination of allele values showed a value of 11, which has been found in MST types 66–70, indicating a unique MST genotype. This novel MST genotype has been submitted to the MST database, currently defined in the database as MST85. The minimum spanning tree constructed from 10 alleles of all STs showed that MST28 was a putative ancestral genotype for MST27 and MST85 (Figure 5).
Table 2.
Coxiella burnetii genotyping based on multiple spacer sequence typing.
| Sample category | Sample number | Intergenic spacer | MST genotype | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COX2 | COX5 | COX18 | COX20 | COX22 | COX37 | COX51 | COX56 | COX57 | COX61 | |||
| 1 | 55 | 8 | 4 | 9 | 5 | 7 | 5 | 2 | 3 | 11 | 6 | Novel |
| 2 | 2 | 8 | 4 | 9 | 5 | 7 | 5 | 2 | 3 | NA | 6 | NI |
| 3 | 1 | 8 | 4 | 9 | 5 | 7 | 5 | 2 | NA | 11 | 6 | NI |
| 4 | 4 | 8 | 4 | 9 | NA | 7 | 5 | 2 | 3 | 11 | 6 | NI |
| 5 | 7 | 8 | 4 | NA | 5 | 7 | 5 | 2 | 3 | 11 | 6 | NI |
| 6 | 1 | 8 | 4 | NA | 5 | 7 | 5 | 2 | NA | 11 | 6 | NI |
| 7 | 1 | 8 | 4 | NA | 5 | 7 | 5 | 2 | 3 | 11 | NA | NI |
| 8 | 2 | 8 | 4 | NA | NA | 7 | 5 | 2 | 3 | 11 | 6 | NI |
| 9 | 1 | 8 | NA | 9 | 5 | 7 | 5 | 2 | 3 | 11 | 6 | NI |
| 10 | 1 | 8 | NA | 9 | NA | NA | 5 | 2 | NA | NA | 6 | NI |
| 11 | 1 | 8 | NA | 9 | NA | NA | NA | 2 | 3 | 11 | 6 | NI |
| 12 | 1 | 8 | NA | NA | 5 | 7 | 5 | 2 | 3 | NA | 6 | NI |
| 13 | 1 | 8 | NA | NA | NA | 7 | 5 | 2 | 3 | 11 | 6 | NI |
| 14 | 1 | 8 | NA | NA | NA | NA | NA | 2 | NA | 11 | NA | NI |
| 15 | 1 | 8 | NA | NA | 5 | 7 | 5 | 2 | 3 | 11 | 6 | NI |
| 16 | 4 | NA | 4 | 9 | 5 | 7 | 5 | 2 | 3 | 11 | 6 | NI |
| 17 | 1 | NA | 4 | 9 | 5 | 7 | NA | NA | 3 | NA | 6 | NI |
| 18 | 1 | NA | 4 | 9 | NA | 7 | 5 | 2 | 3 | 11 | 6 | NI |
| 19 | 1 | NA | 4 | 9 | NA | NA | 5 | NA | 3 | NA | NA | NI |
| 20 | 2 | NA | NA | 9 | 5 | 7 | 5 | 2 | 3 | 11 | 6 | NI |
| 21 | 1 | NA | NA | NA | 5 | NA | NA | 2 | NA | NA | NA | NI |
NA, not amplified; NI, not identified.
Figure 4.
Phylogenetic tree of C. burnetii MST genotypes identified in this study with known genotypes. The MST genotype identified in this study and known MST genotypes from the MST database (https://ifr48.timone.univ-mrs.fr/mst/coxiella_burnetii/) were used. Phylogenetic analysis was performed using the unweighted pair group method with the arithmetic mean (UPGMA) method. Taxa marked by circles depict the sequences obtained in this study.
Figure 5.
A minimum spanning tree for the ten allele profiles with all STs.
Discussion
Q fever has always been a public health problem of concern in the international community (4, 39). Worldwide, Q fever epidemics have occurred in recent years in some countries, including Chile (40), Ethiopia (41), Iran (42), and the Netherlands (43). Q fever was first reported in China in 1950, and the first isolation of C. burnetii was performed in 1962 from a patient with chronic Q fever (44). In Chinese history, small outbreaks of Q fever have occurred in Xizang, Xinjiang, and Inner Mongolia, and sporadic cases of Q fever have been reported in 64 cities/municipalities across 19 provinces (19, 44). Although direct transmission from ticks to humans is scarce, C. burnetii infection in ticks can reflect its threat to local domestic animals and further reflect its risk to local populations. However, there is insufficient information on tick-borne C. burnetii in China. Therefore, a better understanding of the epidemiology of C. burnetii infection in ticks would be helpful for the prevention and control of Q fever in humans. In this study, C. burnetii was identified in Hy. marginatum ticks from Baiyin City of Gansu Province, China. This finding is consistent with previous reports of C. burnetii in several species within the genus Hyalomma. The positive rate of C. burnetii infection in Hy. marginatum ticks in this study was 46.9%, which was higher than that in D. Nuttalli, Hy. asiaticum, D. silvarum, and Ha.japonica collected from other areas of Gansu Province (20, 22). The different positive rates may be related to the detection method, collection site, and ecological environment.
It is well-known that rRNA operons are weakly affected by horizontal gene transfer (45–47), and no recombination occurred in these regions (48). Thus, the 16S rRNA gene plays a crucial role in species identification and construction of phylogenetic relationships of prokaryotes, including Coxiella. The 16S rRNA gene sequences obtained in this study had the highest homology and clustered with other known C. burnetii sequences, suggesting that the pathogen detected in this study should be considered as C. burnetii. The 16S rRNA gene obtained in this study had 100% nucleotide identity and presented a close genetic relationship with C. burnetii variants from humans including Ammassalik (FJ787329) (49), CbuK_Q154 (CP107268) (50), and Schperling (CP014563) (51), suggesting a high risk of its infection in the local population.
MST is a well-established genotyping method for C. burnetii, which is of great significance for the traceability of geographical and natural host sources for Q fever (52). Currently, 80 C. burnetii MST genotypes worldwide are stored in the MST database. In recent years, the application of MST genotyping technology for C. burnetii has also been reported in China. MST16 was found in wild rats from Yunnan (28), and two potential novel MSTs were identified in hedgehogs from Hubei Province (29). However, MST genotyping of C. burnetii identified in ticks has yet to be reported in China. In this study, a novel MST was identified and characterized by a novel combination of known allele values. This novel MST was identified from the majority of C. burnetii-positive tick samples, suggesting that it is the predominant MST genotype in Baiyin City.
The main limitation of this study is that we cannot rule out the possibility that C. burnetii may have originated from the blood meal, as the ticks were collected from goats. Therefore, it is still unclear whether Hy. marginatum ticks can serve as the effective vector of C. burnetii. However, only Hy. marginatum ticks were collected in the endemic areas of C. burnetii in this study; therefore, Hy. marginatum may be the vector of C. burnetii in the local area, which should be confirmed in future studies.
Conclusion
In this study, C. burnetii was found in Hy. Marginatum in Gansu Province, China. A novel MST defined as MST85, similar to MST28, was identified. It is necessary to study the C. burnetii carried by ticks to provide a theoretical basis for the prevention and control of Q fever in China.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Hebei Natural Science Foundation (No. C2022406003), the Key Research and Development Program of Hebei Province (No. 213777109D), and the Scientific Research Foundation for High-level Talents of Chengde Medical University (No. 202001).
Data availability statement
The data presented in the study are deposited in the GenBank repository, accession number PP929917-PP930006 and PP930513-PP930568.
Ethics statement
The animal study was approved by Scientific Ethics Committee of Chengde Medical University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
Z-YX: Data curation, Investigation, Methodology, Resources, Validation, Writing – original draft. F-NW: Data curation, Investigation, Methodology, Resources, Writing – original draft. RJ: Data curation, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft. JX: Data curation, Investigation, Methodology, Resources, Validation, Writing – original draft. Y-CG: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. W-PG: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1470242/full#supplementary-material
References
- 1.Newton P, Thomas DR, Reed SCO, Lau N, Xu B, Ong SY, et al. Lysosomal degradation products induce Coxiella burnetii virulence. Proc Natl Acad Sci USA. (2020) 117:6801–10. 10.1073/pnas.1921344117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derrick EH. “Q” fever, a new fever entity: clinical features, diagnosis and laboratory investigation. Rev Infect Dis. (1983) 5:790–800. 10.1093/clinids/5.4.790 [DOI] [PubMed] [Google Scholar]
- 3.Broertjes J, Franz E, Friesema IHM, Jansen H-J, Reubsaet FAG, Rutjes SA, et al. Epidemiology of pathogens listed as potential bioterrorism agents, the Netherlands, 2009–2019. Emerg Infect Dis. (2023) 29:1–9. 10.3201/eid2907.221769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anastácio S, Sousa SRD, Saavedra MJ, Silva GJD. Role of goats in the epidemiology of Coxiella burnetii. Biology. (2022) 11:1703. 10.3390/biology11121703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mobarez AM, Baseri N, Khalili M, Mostafavi E, Stenos J, Esmaeili S. Genetic diversity of Coxiella burnetii in Iran by multi-spacer sequence typing. Pathogens. (2022) 11:1175. 10.3390/pathogens11101175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.España PP, Uranga A, Cillóniz C, Torres A. Q fever (Coxiella burnetii). Semin Respir Crit Care Med. (2020) 41:509–21. 10.1055/s-0040-1710594 [DOI] [PubMed] [Google Scholar]
- 7.Melenotte C, Million M, Raoult D. New insights in Coxiella burnetii infection: diagnosis and therapeutic update. Expert Rev Anti Infect Ther. (2020) 18:75–86. 10.1080/14787210.2020.1699055 [DOI] [PubMed] [Google Scholar]
- 8.Lee J, Choi Y-J, Yong HS, Baek M-J, Na JO, Choi JY, et al. Fever endocarditis combined with thrombus and antiphospholipid syndrome. Korean Circ J. (2023) 53:106. 10.4070/kcj.2022.0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang S, Xue B, Hu X, Zhou W, Zhang M, Zhao M. Spinal infection caused by Coxiella burnetii. BMC Infect Dis. (2023) 23:6. 10.1186/s12879-022-07938-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Souza EARD, André MR, Labruna MB, Horta MC. Q fever and coxiellosis in Brazil: an underestimated disease? A brief review. Rev Bras Parasitol Veterinária. (2022) 31:e009822. 10.1590/s1984-29612022051 [DOI] [PubMed] [Google Scholar]
- 11.Ullah Q, Jamil T, Saqib M, Iqbal M, Neubauer H. Q fever—a neglected zoonosis. Microorganisms. (2022) 10:1530. 10.3390/microorganisms10081530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duron O, Sidi-Boumedine K, Rousset E, Moutailler S, Jourdain E. The importance of ticks in Q fever transmission: What has (and has not) been demonstrated? Trends Parasitol. (2015) 31:536–52. 10.1016/j.pt.2015.06.014 [DOI] [PubMed] [Google Scholar]
- 13.Borawski K, Dunaj J, Czupryna P, Pancewicz S, Swierzbińska R, Żebrowska A, et al. Assessment of Coxiella burnetii presence after tick bite in north-eastern Poland. Infection. (2020) 48:85–90. 10.1007/s15010-019-01355-w [DOI] [PubMed] [Google Scholar]
- 14.Zhao G-P, Wang Y-X, Fan Z-W, Ji Y, Liu M, Zhang W-H, et al. Mapping ticks and tick-borne pathogens in China. Nat Commun. (2021) 12:1075. 10.1038/s41467-021-21375-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Celina SS, Cerný J. Coxiella burnetii in ticks, livestock, pets and wildlife: a mini-review. Front Vet Sci. (2022) 9:1068129. 10.3389/fvets.2022.1068129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Körner S, Makert GR, Ulbert S, Pfeffer M, Mertens-Scholz K. The prevalence of Coxiella burnetii in hard ticks in europe and their role in Q fever transmission revisited-a systematic review. Front Vet Sci. (2021) 8:655715. 10.3389/fvets.2021.655715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yessinou RE, Katja MS, Heinrich N, Farougou S. Prevalence of Coxiella-infections in ticks - review and meta-analysis. Ticks Tick Borne Dis. (2022) 13:101926. 10.1016/j.ttbdis.2022.101926 [DOI] [PubMed] [Google Scholar]
- 18.Batu N, Wang Y, Liu Z, Huang T, Bao W, He H, et al. Molecular epidemiology of Rickettsia sp. and Coxiella burnetii collected from Hyalomma asiaticum in bactrian camels (Camelus bactrianus) in inner Mongolia of China. Ticks Tick Borne Dis. (2020) 11:101548. 10.1016/j.ttbdis.2020.101548 [DOI] [PubMed] [Google Scholar]
- 19.Ni J, Lin H, Xu X, Ren Q, Aizezi M, Luo J, et al. Coxiella burnetii is widespread in ticks (Ixodidae) in the Xinjiang areas of China. BMC Vet Res. (2020) 16:317. 10.1186/s12917-020-02538-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang F, Wang XH, Yu GW. Study on Q termal medium and molecular characteristics along Hexi corridor in Gansu Province. J Pathog Biol. (2023) 18: 158–61 (in Chinese). 10.13350/j.cjpb.230207 [DOI] [Google Scholar]
- 21.Luo J, Li J, Zhang GF, Liu PW, Luo WY, Su XL, et al. Infection and molecular analysis of Coxiella burnetii in ticks from three areas of Heilongjiang Province of China. Chin Vet Sci. (2021) 5: 1385–91 (in Chinese). 10.16656/j.issn.1673-4696.2021.0186 [DOI] [Google Scholar]
- 22.Zhang F, Liu ZH. Molecular epidemiological studies on Coxiella burnetii from northwestern China. J Pathog Biol. (2011) 6: 183–5 (in Chinese). 10.13350/j.cjpb.2011.03.007 [DOI] [Google Scholar]
- 23.Jiao J, Zhang J, He P, OuYang X, Yu Y, Wen B, et al. Identification of tick-borne pathogens and genotyping of Coxiella burnetii in Rhipicephalus microplus in Yunnan Province, China. Front Microbiol. (2021) 12:736484. 10.3389/fmicb.2021.736484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi Y, Ai L, Zhu C, Ye F, Lv R, Wang J, et al. Wild hedgehogs and their parasitic ticks coinfected with multiple tick-borne pathogens in Jiangsu Province, eastern China. Microbiol Spectr. (2022) 10:e02138–22. 10.1128/spectrum.02138-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Truong A-T, Youn SY, Yoo M-S, Lim J-Y, Yoon S-S, Cho YS. Genotyping of Coxiella burnetii from cattle by multispacer sequence typing and multiple locus variable number of tandem repeat analysis in the republic of Korea. Genes. (2022) 13:1927. 10.3390/genes13111927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eldin C, Mélenotte C, Mediannikov O, Ghigo E, Million M, Edouard S, et al. From Q fever to Coxiella burnetii infection: a paradigm change. Clin Microbiol Rev. (2017) 30:115–90. 10.1128/CMR.00045-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemsley CM, Essex-Lopresti A, Norville IH, Titball RW. Correlating genotyping data of Coxiella burnetii with genomic groups. Pathogens. (2021) 10:604. 10.3390/pathogens10050604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu M, He P, OuYang X, Yu Y, Wen B, Zhou D, et al. Novel genotypes of Coxiella burnetii circulating in rats in Yunnan Province, China. BMC Vet Res. (2022) 18:204. 10.1186/s12917-022-03310-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong X-Q, Xiao X, Liu J-W, Han H-J, Qin X-R, Lei S-C, et al. Occurrence and genotyping of Coxiella burnetii in hedgehogs in China. Vect Borne Zoon Dis. (2020) 20:580–5. 10.1089/vbz.2019.2589 [DOI] [PubMed] [Google Scholar]
- 30.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. (1994) 3:294–9. [PubMed] [Google Scholar]
- 31.Hoover TA, Vodkin MH, Williams JC. A Coxiella bumetii repeated DNA element resembling a bacterial insertion sequence. J Bacteriol. (1992) 174:5540–8. 10.1128/jb.174.17.5540-5548.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mares-Guia MAMM, Guterres A, Rozental T, Ferreira M dos S, Lemos ERS. Clinical and epidemiological use of nested PCR targeting the repetitive element IS 1111 associated with the transposase gene from Coxiella burnetii. Braz J Microbiol. (2018) 49:138–43. 10.1016/j.bjm.2017.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo M-G, Lee S-H, Ouh I-O, Lee GH, Goo Y-K, Kim S, et al. Molecular detection and genotyping of Coxiella-like endosymbionts in ticks that infest horses in south Korea. PLoS ONE. (2016) 11:e0165784. 10.1371/journal.pone.0165784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glazunova O, Roux V, Freylikman O, Sekeyova Z, Fournous G, Tyczka J, et al. Coxiella burnetii genotyping. Emerg Infect Dis. (2005) 11:1211–7. 10.3201/eid1108.041354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall TA. BioEdit : a user-friendly biological sequence alignment editor and analysis. Nuclc Acids Symp. (1999) 41:95–8. [Google Scholar]
- 36.Burland TG. DNASTAR's Lasergene sequence analysis software. Methods Mol Biol. (2000) 132:71–91. 10.1385/1-59259-192-2:71 [DOI] [PubMed] [Google Scholar]
- 37.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 60. Mol Biol Evol. (2013) 30:2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Z, Alikhan N-F, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. (2018) 28:1395–404. 10.1101/gr.232397.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dijkstra F, Hoek WVD, Wijers N, Schimmer B, Rietveld A, Wijkmans CJ, et al. The 2007–2010 Q fever epidemic in the Netherlands: characteristics of notified acute Q fever patients and the association with dairy goat farming. FEMS Immunol Med Microbiol. (2012) 64:3–12. 10.1111/j.1574-695X.2011.00876.x [DOI] [PubMed] [Google Scholar]
- 40.Tapia T, Stenos J, Flores R, Duery O, Iglesias R, Olivares MF, et al. Evidence of Q fever and rickettsial disease in Chile. Trop Med Infect Dis. (2020) 5:99. 10.3390/tropicalmed5020099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robi DT, Demissie W, Temteme S. Coxiellosis in livestock: epidemiology, public health significance, and prevalence of Coxiella burnetii infection in ethiopia. Vet Med Res Rep. (2023) 14:145–58. 10.2147/VMRR.S418346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rahravani M, Moravedji M, Mostafavi E, Mohammadi M, Seyfi H, Baseri N, et al. The epidemiological survey of Coxiella burnetii in small ruminants and their ticks in western Iran. BMC Vet Res. (2022) 18:292. 10.1186/s12917-022-03396-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klaassen CHW, Nabuurs-Franssen MH, Tilburg JJ, Hamans MA, Horrevorts AM. Multigenotype Q fever outbreak, the Netherlands. Emerg Infect Dis. (2009) 15:613–4. 10.3201/eid1504.081612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El-Mahallawy HS, Lu G, Kelly P, Xu D, Li Y, Fan W, et al. Q fever in China: a systematic review, 1989–2013. Epidemiol Infect. (2015) 143:673–81. 10.1017/S0950268814002593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rivera MC, Jain R, Moore JE, Lake JA. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci USA. (1998) 95:6239–44. 10.1073/pnas.95.11.6239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daubin V, Moran NA, Ochman H. Phylogenetics and the cohesion of bacterial genomes. Sci New Ser. (2003) 301:829–32. 10.1126/science.1086568 [DOI] [PubMed] [Google Scholar]
- 47.Větrovský T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS ONe. (2013) 8:e57923. 10.1371/journal.pone.0057923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hashimoto JG, Stevenson BS, Schmidt TM. Rates and consequences of recombination between rRNA operons. J Bacteriol. (2003) 185:966–72. 10.1128/JB.185.3.966-972.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koch A, Svendsen CB, Christensen JJ, Bundgaard H, Vindfeld L, Christiansen CB, et al. Q fever in Greenland. Emerg Infect Dis. (2010) 16:511–3. 10.3201/eid1603.091220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beare PA, Unsworth N, Andoh M, Voth DE, Omsland A, Gilk SD, et al. Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect Immun. (2009) 77:642–56. 10.1128/IAI.01141-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuley R, Kuijt E, Smits MA, Roest HIJ, Smith HE, Bossers A. Genome plasticity and polymorphisms in critical genes correlate with increased virulence of Dutch outbreak-related Coxiella burnetii strains. Front Microbiol. (2017) 8:1526. 10.3389/fmicb.2017.01526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chochlakis D, Santos AS, Giadinis ND, Papadopoulos D, Boubaris L, Kalaitzakis E, et al. Genotyping of Coxiella burnetii in sheep and goat abortion samples. BMC Microbiol. (2018) 18:204. 10.1186/s12866-018-1353-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in the study are deposited in the GenBank repository, accession number PP929917-PP930006 and PP930513-PP930568.