Abstract
Background
Claudin-3 (CLDN3) participates in the formation of the tight-junctions (TJs) that regulate intercellular permeability. Altered CLDN3 expression has been linked to tumor progression in multiple tumor types. Despite its widespread expression in normal epithelial cells, CLDN3 is considered an attractive drug target candidate, since it may be more accessible in cancer cells than in normal cells due to their less orchestrated cell growth.
Methods
To comprehensively determine the prevalence of CLDN3 expression in cancer, a tissue microarray containing 14,966 samples from 133 different tumor types and subtypes as well as 608 samples of 76 different normal tissue types was analyzed by immunohistochemistry.
Results
CLDN3 immunostaining was observed in 8,479 (68.9%) of 12,314 analyzable tumors, including 11.6% with weak, 6.2% with moderate, and 51.1% with strong positivity. CLDN3 staining was found in 96 of 133 tumor categories, 80 of which contained at least one strongly positive case. CLDN3 positivity was most seen in neuroendocrine neoplasms (92–100%) and in adenocarcinomas (67–100%), tumors of the female genital tract, including various subtypes of ovarian and endometrial carcinoma (up to 100%), as well as different subtypes of breast cancer (95.3–100%). CLDN3 positivity was less common in squamous cell carcinomas (0–43.2%) and mainly absent in melanoma, mesenchymal, and hematolymphatic neoplasms. In clear cell renal cell carcinoma (ccRCC), low CLDN3 was strongly linked to poor ISUP (p < 0.0001), Fuhrman (p < 0.0001), and Thoenes (p < 0.0001) grades, advanced pT category (p < 0.0001), high UICC stage (p = 0.0006) and distant metastasis (p = 0.0011), as well as shortened overall (p = 0.0118) and recurrence-free (p < 0.0001) survival. In papillary RCC (pRCC), low CLDN3 was associated with poor grade (p < 0.05), high pT (p = 0.0273) and distant metastasis (p = 0.0357). In urothelial carcinoma high CLDN3 was linked to high grade (p < 0.0001) and nodal metastasis (p = 0.0111). The level of CLDN3 staining was unrelated to parameters of tumor aggressiveness in pancreatic, gastric, and breast cancer.
Conclusion
In conclusion, our data demonstrate significant levels of CLDN3 expression in many different tumor entities and identify reduced CLDN3 expression as a potential prognostic marker in RCC.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40364-024-00702-w.
Keywords: CLDN3, Tissue microarray, Cancer, Renal cell carcinoma, Biomarker
Introduction
Claudin-3 (CLDN3) is one of 27 known members of the claudin family [1]. Together with occludin and other junctional adhesion molecules, the claudins form the tight-junctions (TJs) that regulate intercellular permeability [2]. Claudins can be distinguished into paracellular barrier forming and pore forming claudins allowing for controlled diffusion of ions and water through TJs [3]. TJs display characteristic individual compositions and ratios of different claudins which define individual “penetrability properties” in different tissues and cell types [2, 4]. CLDN3 is a rather ubiquitously expressed barrier forming claudin which occurs in the intestine and many other epithelial tissues [5, 6].
Despite their widespread expression in normal cells, TJ components are considered attractive drug target candidates, since they may be more accessible in cancer cells than in normal cells. In normal epithelia, the accessibility of TJ proteins is limited by the orchestrated cell growth, the protection of individual TJ proteins by intact TJ structures, and the predominant expression of TJs at apical surfaces [7–10]. The misorientation of the cell division in cancerous tissues results in a markedly higher exposure of TJ components [7, 10, 11]. The expression of CLDN3 in cancer has been analyzed in more than 45 studies using immunohistochemistry (IHC). Aberrations of CLDN3 expression have been reported to occur in colorectal [12], breast [13–15], ovarian [16, 17], prostatic [18, 19], gastric [20–22], hepatic [23] and pulmonary cancers [24]. Several of these studies have found a link between either elevated [14, 18, 25] or reduced [19, 23, 24] CLDN3 expression levels and poor prognosis of cancer patients. It is of note that the reported rates of CLDN3 positivity varied considerably between studies. For example, the range of reported CLDN3 positive cases ranged from 25 to 73.6% in gastric cancer [20, 21], from 32 to 95% in breast cancer of no special type [26, 27], and from 41.4 to 97.0% in pulmonary adenocarcinoma [28, 29]. Such conflicting data between studies are typically caused by the use of different antibodies, IHC protocols, and criteria to define CLDN3 positivity.
To better understand the prevalence and potential clinical significance of CLDN3 expression in cancer, a comprehensive study analyzing a large number of neoplastic and non-neoplastic tissues under highly standardized conditions is needed. Therefore, CLDN3 expression was analyzed in more than 14,500 tumor tissue samples from 133 different tumor types and subtypes as well as 76 non-neoplastic tissue categories by IHC in a tissue microarray (TMA) format in this study.
Material and methods
Tissue Microarrays (TMAs)
The normal tissue TMA was composed of 8 samples from 8 different donors for each of 76 different normal tissue types (608 samples on one slide). The cancer TMAs contained a total of 14,966 primary tumors from 133 tumor types and subtypes. Detailed histopathological data on grade, pathological tumor stage (pT) or pathological lymph node status (pN) were available from breast cancers (n = 600), urothelial carcinomas (n = 829), ovarian cancers (n = 344), endometroid endometrial cancers (n = 182), thyroid (n = 518), gastric (n = 327), and pancreatic carcinomas (n = 598) as well as clear cell (n = 1,224) and papillary (n = 310) renal cell carcinomas (ccRCC, pRCC). Clinical follow up data were available from 789 patients with ccRCC and from 177 patients with pRCC with a median follow-up time of 48.0 and 50.5 months (range 1–250 and 1–247). The composition of both normal and cancer TMAs is described in detail in the results section. All samples were from the archives of the Institute of Pathology, University Medical Center Hamburg, Germany, the Institute of Patholgy, Clinical Center Osnabrueck, Germany, and Department of Pathology, Academic Hospital Fuerth, Germany. Tissues were fixed in 4% buffered formalin and then embedded in paraffin. The TMA manufacturing process was described earlier in detail [30, 31]. In brief, one tissue spot (diameter: 0.6 mm) per patient was used. The use of archived remnants of diagnostic tissues for TMA manufacturing, their analysis for research purposes, and the use of patient data were according to local laws (HmbKHG, §12) and analysis had been approved by the local ethics committee (Ethics commission Hamburg, WF-049/09). All work has been carried out in compliance with the Helsinki Declaration.
Immunohistochemistry (IHC)
Freshly cut TMA sections were immunostained on one day and in one experiment. Slides were deparaffinized with xylol, rehydrated through a graded alcohol series and exposed to heat-induced antigen retrieval for 5 min in an autoclave at 121 °C in pH 7.8 Tris–EDTA-Citrat (TEC) puffer. Endogenous peroxidase activity was blocked with Dako REAL Peroxidase-Blocking Solution (Agilent Technologies, Santa Clara, CA, USA; #S2023) for 10 min. Primary antibody specific against CLDN3 protein (rabbit recombinant monoclonal, HMV-309, ardoci GmbH, Hamburg, Germany) was applied at 37 °C for 60 min at a dilution of 1:150. For the purpose of antibody validation, the normal tissue TMA was also analyzed by the rabbit recombinant monoclonal CLDN3 antibody EPR19971 (Abcam Limited, Cambridge, GB) at a dilution of 1:40 and an otherwise identical protocol. Bound antibody was then visualized using the Dako REAL EnVision Detection System Peroxidase/DAB + , Rabbit/Mouse kit (Agilent Technologies, Santa Clara, CA, USA; #K5007) according to the manufacturer’s directions. The sections were counterstained with hemalaun. IHC scoring was predefined and has been used in multiple previous studies [32–34]. For tumor tissues, the percentage of positive neoplastic cells was estimated, and the staining intensity was semi-quantitatively recorded (0, 1 + , 2 + , 3 +) [35]. For statistical analyses, the staining results were categorized into four groups. Tumors without any staining were considered negative. Tumors with 1 + staining intensity in ≤ 70% of tumor cells and 2 + intensity in ≤ 30% of tumor cells were considered weakly positive. Tumors with 1 + staining intensity in > 70% of tumor cells, 2 + intensity in 31–70%, or 3 + intensity in ≤ 30% of tumor cells were considered moderately positive. Tumors with 2 + intensity in > 70% or 3 + intensity in > 30% of tumor cells were considered strongly positive. The analysis by one pathologist enables the best possible consistency of interpretation within the study. A possible impact of interobserver variation was excluded as much as possible by a four-tier categorization of tumor staining. Although interobserver variation is common in TMA studies between 1 + and 2 + there is little discrepancies between 0 + and 3 + .
Statistics
Statistical calculations were performed with JMP17® software (SAS®, Cary, NC, USA). Contingency tables and the chi2-test were performed to search for associations between CLDN3 immunostaining and tumor phenotype. Survival curves were calculated according to Kaplan–Meier. The Log-Rank test was applied to detect significant differences between groups.
Results
Technical issues
A total of 12,314 (82.3%) of 14,966 tumor samples were interpretable in our TMA analysis. Non-interpretable samples demonstrated lack of unequivocal tumor cells or lack of entire tissue spots. A sufficient number of samples (≥ 4) of each normal tissue type was evaluable.
CLDN3 immunostaining in normal tissues
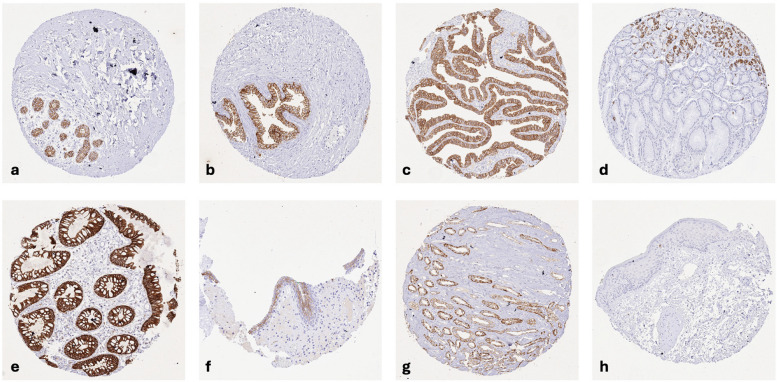
CLDN3 immunostaining was predominantly membranous. CLDN3 staining was particularly strong in luminal cells of breast glands, prostate, and seminal vesicle, follicular cells of the thyroid, respiratory epithelial cells, glandular cells of salivary glands, a small subset of gastric epithelial cells in the neck and in glandular pits, all epithelial cells of the small intestine and the colorectum, bile ducts in the liver and gallbladder epithelium, acinar cells of the pancreas, collecting ducts of the kidney, most epithelial cells in the cauda epididymis, epithelial cells of endometrium glands, the fallopian tube, and the endocervix (predominantly basolateral), megakaryocytes of the bone marrow, subsets of high endothelial venules and of monocytic cells in germinal centers of lymph nodes, as well as in squamous epithelial cells of tonsil crypts and corpuscles of Hassall’s in the thymus. A less intense, weak to moderate membranous CLDN3 staining was observed in the urothelium (predominantly in the upper half), epithelial cells of the parathyroidal gland, few epithelial cells of the adrenal gland, hepatocytes (predominantly at the bile secreting apical membrane), excretory ducts of salivary glands, islets cells of the pancreas, chief cells in the corpus epididymis, some renal tubular cells, hepatocytes, a large subset of corpus luteum cells of the ovary, pneumocytes, a subset of cells in the white pulp of the spleen, and the syncytiotrophoblast (surface membrane) of the first trimenon placenta. CLDN3 staining was absent in squamous epithelial cells of the epidermis, the ectocervix, and the esophagus, amnion, chorion, all muscle cells, and the brain. Representative images are shown in Fig. 1. All cell types identified as CLDN3 positive by HMV-309 were also positive by using EPR19971, although the signal was less intense for EPR19971 even at a dilution of 1:40 (Supplementary Fig. 1).
Fig. 1.
CLDN3 immunostaining of normal tissues. The panels show a strong membranous CLDN3 immunostaining of the luminal cells of breast glands (a) and of the prostate (b), epithelial cells of the fallopian tube (c), a small subset of gastric epithelial cells in the neck and in glandular pits (d), epithelial cells of the colorectum (e), the upper half of urothelial cells (f), and in collecting ducts of the kidney medulla (g) while CLDN3 staining is absent in squamous epithelial cells of the epidermis (h)
CLDN3 immunostaining in neoplastic tissues
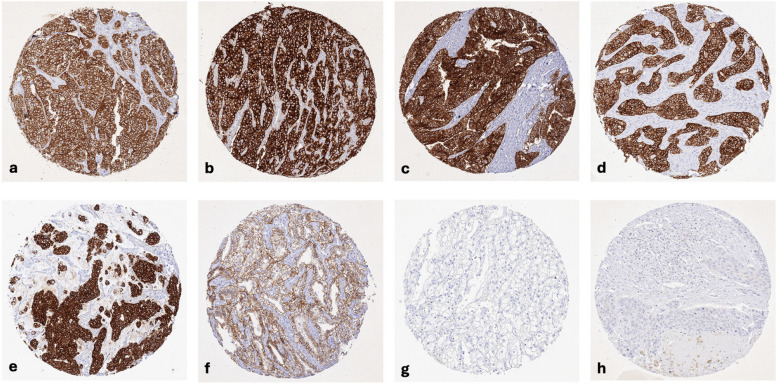
CLDN3 staining was observed in 8,479 (68.9%) of 12,314 analyzable tumors, including 11.6% with weak, 6.2% with moderate, and 51.1% with strong staining intensity. CLDN3 staining varied both in intensity and in its pattern between samples. Most CLDN3 positive tumors showed a purely membranous staining pattern but some tumors showed an additional cytoplasmic positivity. Representative images are shown in Fig. 2.
Fig. 2.
CLDN3 immunostaining in cancer. CLDN3 immunostaining was purely membranous in most tumors, with some showing an additional cytoplasmic positivity. The panels show a strong CLDN3 positivity in cancer cells of a neuroendocrine tumor of the appendix (a), an adenocarcinoma of the prostate (b), an endometrioid endometrial carcinoma (c), an invasive breast cancer of no special type (d), a muscle-invasive urothelial carcinoma (e), and clear cell renal cell carcinoma (f). CLDN3 staining is lacking in another clear cell renal cell carcinoma (g) and squamous cell carcinoma of the lung (h)
At least an occasional weak CLDN3 positivity was detected in 96 of 133 tumor categories and 80 categories included at least one case with strong CLDN3 positivity (Table 1).
Table 1.
CLDN3 immunostaining in human tumors
| CLDN3 immunostaining | |||||||
|---|---|---|---|---|---|---|---|
| Tumor entity | on TMA (n) | analyzable (n) | negative (%) | weak (%) | moderate (%) | strong (%) | |
| Tumors of the skin | Basal cell carcinoma of the skin | 41 | 13 | 100.0 | 0.0 | 0.0 | 0.0 |
| Squamous cell carcinoma of the skin | 95 | 80 | 93.8 | 6.3 | 0.0 | 0.0 | |
| Malignant melanoma | 19 | 19 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Malignant melanoma lymph node metastasis | 86 | 82 | 98.8 | 1.2 | 0.0 | 0.0 | |
| Merkel cell carcinoma | 2 | 2 | 50.0 | 0.0 | 50.0 | 0.0 | |
| Tumors of the head and neck | Squamous cell carcinoma of the larynx | 109 | 89 | 78.7 | 11.2 | 6.7 | 3.4 |
| Squamous cell carcinoma of the pharynx | 60 | 41 | 65.9 | 24.4 | 0.0 | 9.8 | |
| Oral squamous cell carcinoma (floor of the mouth) | 130 | 102 | 84.3 | 4.9 | 4.9 | 5.9 | |
| Pleomorphic adenoma of the parotid gland | 50 | 45 | 37.8 | 31.1 | 20.0 | 11.1 | |
| Warthin tumor of the parotid gland | 49 | 48 | 2.1 | 31.3 | 43.8 | 22.9 | |
| Basal cell adenoma of the salivary gland | 15 | 15 | 20.0 | 13.3 | 20.0 | 46.7 | |
| Tumors of the lung, pleura and thymus | Adenocarcinoma of the lung | 196 | 130 | 3.8 | 3.8 | 4.6 | 87.7 |
| Squamous cell carcinoma of the lung | 80 | 45 | 71.1 | 17.8 | 2.2 | 8.9 | |
| Mesothelioma, epithelioid | 40 | 24 | 95.8 | 4.2 | 0.0 | 0.0 | |
| Mesothelioma, biphasic | 29 | 17 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Thymoma | 29 | 29 | 69.0 | 6.9 | 13.8 | 10.3 | |
| Lung, neuroendocrine tumor (NET) | 29 | 25 | 8.0 | 0.0 | 8.0 | 84.0 | |
| Tumors of the female genital tract | Squamous cell carcinoma of the vagina | 30 | 26 | 76.9 | 11.5 | 3.8 | 7.7 |
| Squamous cell carcinoma of the vulva | 107 | 87 | 90.8 | 3.4 | 2.3 | 3.4 | |
| Squamous cell carcinoma of the cervix | 88 | 74 | 56.8 | 31.1 | 5.4 | 6.8 | |
| Adenocarcinoma of the cervix | 23 | 23 | 4.3 | 4.3 | 4.3 | 87.0 | |
| Endometrioid endometrial carcinoma | 288 | 261 | 0.4 | 5.7 | 10.3 | 83.5 | |
| Endometrial serous carcinoma | 36 | 32 | 0.0 | 0.0 | 6.3 | 93.8 | |
| Carcinosarcoma of the uterus | 57 | 46 | 21.7 | 17.4 | 4.3 | 56.5 | |
| Endometrial carcinoma, high grade, G3 | 13 | 13 | 30.8 | 7.7 | 15.4 | 46.2 | |
| Endometrial clear cell carcinoma | 9 | 8 | 0.0 | 12.5 | 0.0 | 87.5 | |
| Endometrioid carcinoma of the ovary | 93 | 71 | 0.0 | 4.2 | 4.2 | 91.5 | |
| Serous carcinoma of the ovary | 530 | 445 | 0.2 | 1.3 | 2.7 | 95.7 | |
| Mucinous carcinoma of the ovary | 75 | 51 | 19.6 | 19.6 | 7.8 | 52.9 | |
| Clear cell carcinoma of the ovary | 51 | 40 | 0.0 | 5.0 | 0.0 | 95.0 | |
| Carcinosarcoma of the ovary | 47 | 36 | 19.4 | 11.1 | 5.6 | 63.9 | |
| Granulosa cell tumor of the ovary | 44 | 42 | 90.5 | 4.8 | 2.4 | 2.4 | |
| Leydig cell tumor of the ovary | 4 | 4 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sertoli cell tumor of the ovary | 1 | 1 | 0.0 | 100.0 | 0.0 | 0.0 | |
| Sertoli Leydig cell tumor of the ovary | 3 | 3 | 66.7 | 0.0 | 33.3 | 0.0 | |
| Steroid cell tumor of the ovary | 3 | 3 | 66.7 | 0.0 | 0.0 | 33.3 | |
| Brenner tumor | 32 | 26 | 88.5 | 7.7 | 0.0 | 3.8 | |
| Tumors of the breast | Invasive breast carcinoma of no special type | 499 | 345 | 0.3 | 13.9 | 9.6 | 76.2 |
| Lobular carcinoma of the breast | 150 | 107 | 4.7 | 17.8 | 16.8 | 60.7 | |
| Medullary carcinoma of the breast | 8 | 7 | 0.0 | 14.3 | 0.0 | 85.7 | |
| Tubular carcinoma of the breast | 2 | 1 | 0.0 | 0.0 | 100.0 | 0.0 | |
| Mucinous carcinoma of the breast | 7 | 4 | 0.0 | 0.0 | 0.0 | 100.0 | |
| Tumors of the digestive system | Adenomatous polyp, low-grade dysplasia | 50 | 25 | 0.0 | 0.0 | 0.0 | 100.0 |
| Adenomatous polyp, high-grade dysplasia | 50 | 38 | 0.0 | 0.0 | 0.0 | 100.0 | |
| Adenocarcinoma of the colon | 2483 | 2219 | 0.1 | 0.9 | 1.0 | 98.0 | |
| Gastric adenocarcinoma, diffuse type | 215 | 189 | 23.3 | 9.5 | 2.6 | 64.6 | |
| Gastric adenocarcinoma, intestinal type | 215 | 184 | 9.8 | 8.7 | 10.9 | 70.7 | |
| Gastric adenocarcinoma, mixed type | 62 | 50 | 8.0 | 14.0 | 8.0 | 70.0 | |
| Adenocarcinoma of the esophagus | 83 | 76 | 6.6 | 7.9 | 10.5 | 75.0 | |
| Squamous cell carcinoma of the esophagus | 76 | 69 | 68.1 | 17.4 | 4.3 | 10.1 | |
| Squamous cell carcinoma of the anal canal | 91 | 65 | 83.1 | 10.8 | 0.0 | 6.2 | |
| Cholangiocarcinoma | 58 | 46 | 15.2 | 21.7 | 23.9 | 39.1 | |
| Gallbladder adenocarcinoma | 51 | 44 | 6.8 | 20.5 | 11.4 | 61.4 | |
| Gallbladder Klatskin tumor | 42 | 27 | 22.2 | 33.3 | 25.9 | 18.5 | |
| Hepatocellular carcinoma | 312 | 274 | 11.7 | 56.6 | 14.6 | 17.2 | |
| Ductal adenocarcinoma of the pancreas | 659 | 415 | 33.0 | 33.3 | 12.5 | 21.2 | |
| Pancreatic/Ampullary adenocarcinoma | 98 | 65 | 12.3 | 7.7 | 10.8 | 69.2 | |
| Acinar cell carcinoma of the pancreas | 18 | 17 | 5.9 | 17.6 | 11.8 | 64.7 | |
| Gastrointestinal stromal tumor (GIST) | 62 | 58 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Appendix, neuroendocrine tumor (NET) | 25 | 14 | 0.0 | 0.0 | 0.0 | 100.0 | |
| Colorectal, neuroendocrine tumor (NET) | 12 | 9 | 0.0 | 0.0 | 0.0 | 100.0 | |
| Ileum, neuroendocrine tumor (NET) | 53 | 45 | 0.0 | 0.0 | 0.0 | 100.0 | |
| Pancreas, neuroendocrine tumor (NET) | 101 | 76 | 2.6 | 0.0 | 3.9 | 93.4 | |
| Colorectal, neuroendocrine carcinoma (NEC) | 14 | 12 | 8.3 | 8.3 | 8.3 | 75.0 | |
| Ileum, neuroendocrine carcinoma (NEC) | 8 | 7 | 0.0 | 0.0 | 0.0 | 100.0 | |
| Gallbladder, neuroendocrine carcinoma (NEC) | 4 | 4 | 0.0 | 0.0 | 75.0 | 25.0 | |
| Pancreas, neuroendocrine carcinoma (NEC) | 14 | 9 | 0.0 | 22.2 | 11.1 | 66.7 | |
| Tumors of the urinary system | Non-invasive papillary urothelial carcinoma, pTa G2 low grade | 87 | 79 | 77.2 | 16.5 | 5.1 | 1.3 |
| Non-invasive papillary urothelial carcinoma, pTa G2 high grade | 80 | 73 | 63.0 | 26.0 | 8.2 | 2.7 | |
| Non-invasive papillary urothelial carcinoma, pTa G3 | 126 | 116 | 36.2 | 35.3 | 6.9 | 21.6 | |
| Urothelial carcinoma, pT2-4 G3 | 735 | 520 | 46.0 | 25.2 | 11.9 | 16.9 | |
| Squamous cell carcinoma of the bladder | 22 | 21 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Small cell neuroendocrine carcinoma of the bladder | 5 | 5 | 0.0 | 20.0 | 0.0 | 80.0 | |
| Sarcomatoid urothelial carcinoma | 25 | 12 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Urothelial carcinoma of the kidney pelvis | 62 | 55 | 65.5 | 25.5 | 1.8 | 7.3 | |
| Clear cell renal cell carcinoma | 1287 | 1045 | 10.1 | 25.6 | 16.6 | 47.7 | |
| Papillary renal cell carcinoma | 368 | 275 | 2.9 | 5.1 | 8.7 | 83.3 | |
| Clear cell (tubulo) papillary renal cell carcinoma | 26 | 18 | 5.6 | 5.6 | 11.1 | 77.8 | |
| Chromophobe renal cell carcinoma | 170 | 133 | 30.1 | 43.6 | 12.8 | 13.5 | |
| Oncocytoma of the kidney | 257 | 191 | 22.0 | 61.3 | 13.6 | 3.1 | |
| Tumors of the male genital organs | Adenocarcinoma of the prostate, Gleason 3 + 3 | 83 | 83 | 0.0 | 1.2 | 2.4 | 96.4 |
| Adenocarcinoma of the prostate, Gleason 4 + 4 | 80 | 80 | 0.0 | 0.0 | 1.3 | 98.8 | |
| Adenocarcinoma of the prostate, Gleason 5 + 5 | 85 | 85 | 0.0 | 0.0 | 1.2 | 98.8 | |
| Adenocarcinoma of the prostate (recurrence) | 258 | 237 | 0.0 | 1.3 | 0.8 | 97.9 | |
| Small cell neuroendocrine carcinoma of the prostate | 2 | 2 | 0.0 | 0.0 | 0.0 | 100.0 | |
| Seminoma | 682 | 640 | 95.5 | 3.9 | 0.6 | 0.0 | |
| Embryonal carcinoma of the testis | 54 | 41 | 82.9 | 14.6 | 2.4 | 0.0 | |
| Leydig cell tumor of the testis | 31 | 30 | 96.7 | 0.0 | 3.3 | 0.0 | |
| Sertoli cell tumor of the testis | 2 | 2 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Spermatocytic tumor of the testis | 1 | 1 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Yolk sac tumor | 53 | 40 | 60.0 | 37.5 | 2.5 | 0.0 | |
| Teratoma | 53 | 40 | 72.5 | 10.0 | 7.5 | 10.0 | |
| Squamous cell carcinoma of the penis | 92 | 67 | 95.5 | 3.0 | 1.5 | 0.0 | |
| Tumors of endocrine organs | Adenoma of the thyroid gland | 63 | 63 | 0.0 | 4.8 | 12.7 | 82.5 |
| Papillary thyroid carcinoma | 341 | 329 | 0.0 | 4.6 | 6.7 | 88.8 | |
| Follicular thyroid carcinoma | 109 | 106 | 0.0 | 7.5 | 8.5 | 84.0 | |
| Medullary thyroid carcinoma | 57 | 57 | 3.5 | 0.0 | 1.8 | 94.7 | |
| Parathyroid gland adenoma | 43 | 41 | 9.8 | 31.7 | 19.5 | 39.0 | |
| Anaplastic thyroid carcinoma | 19 | 19 | 84.2 | 5.3 | 0.0 | 10.5 | |
| Adrenal cortical adenoma | 48 | 43 | 95.3 | 2.3 | 2.3 | 0.0 | |
| Adrenal cortical carcinoma | 27 | 23 | 60.9 | 17.4 | 4.3 | 17.4 | |
| Pheochromocytoma | 51 | 48 | 97.9 | 2.1 | 0.0 | 0.0 | |
| Tumors of hematopoetic and lymphoid tissues | Hodgkin's lymphoma | 103 | 91 | 100.0 | 0.0 | 0.0 | 0.0 |
| Small lymphocytic lymphoma, B-cell type (B-SLL/B-CLL) | 50 | 39 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Diffuse large B cell lymphoma (DLBCL) | 113 | 101 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Follicular lymphoma | 88 | 77 | 100.0 | 0.0 | 0.0 | 0.0 | |
| T-cell non-Hodgkin's lymphoma | 25 | 19 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Mantle cell lymphoma | 18 | 15 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Marginal zone lymphoma | 16 | 12 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Diffuse large B-cell lymphoma (DLBCL) in the testis | 16 | 15 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Burkitt lymphoma | 5 | 2 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of soft tissue and bone | Granular cell tumor | 23 | 13 | 100.0 | 0.0 | 0.0 | 0.0 |
| Leiomyoma | 50 | 49 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Leiomyosarcoma | 94 | 87 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Liposarcoma | 96 | 82 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Malignant peripheral nerve sheath tumor (MPNST) | 15 | 13 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Myofibrosarcoma | 26 | 25 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Angiosarcoma | 42 | 28 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Angiomyolipoma | 91 | 69 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Dermatofibrosarcoma protuberans | 21 | 11 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Ganglioneuroma | 14 | 11 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Kaposi sarcoma | 8 | 2 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Neurofibroma | 117 | 84 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sarcoma, not otherwise specified (NOS) | 74 | 63 | 98.4 | 0.0 | 1.6 | 0.0 | |
| Paraganglioma | 41 | 27 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Ewing sarcoma | 23 | 12 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Rhabdomyosarcoma | 7 | 6 | 83.3 | 16.7 | 0.0 | 0.0 | |
| Schwannoma | 122 | 93 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Synovial sarcoma | 12 | 9 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Osteosarcoma | 19 | 10 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Chondrosarcoma | 15 | 8 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Rhabdoid tumor | 5 | 5 | 60.0 | 0.0 | 20.0 | 20.0 | |
| Solitary fibrous tumor | 17 | 17 | 100.0 | 0.0 | 0.0 | 0.0 | |
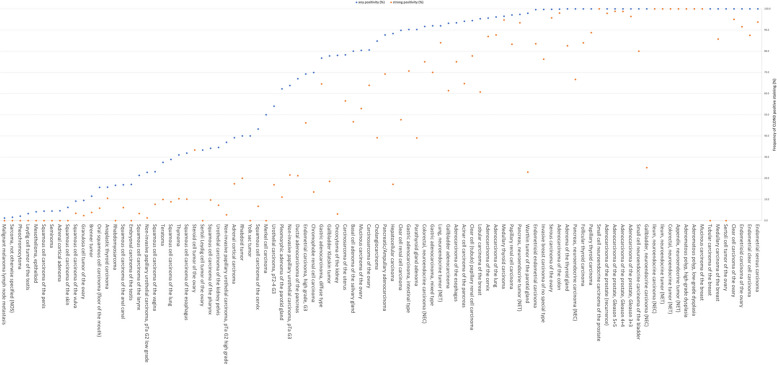
CLDN3 positivity was most seen in adenocarcinomas (67–100%) and neuroendocrine neoplasms (92–100%) from various organs as well as in other tumors of the female genital tract such as in various subtypes of ovarian and endometrial carcinoma (up to 100%) and different subtypes of breast cancer (95.3–100%). CLDN3 was less common in squamous cell carcinomas (0–43.2%) and mainly absent in melanoma, mesenchymal neoplasia, and in tumors of hematopoetic and lymphoid tissues. A graphical representation of a ranking order of CLDN3 positive and strongly positive cancers is given in Fig. 3.
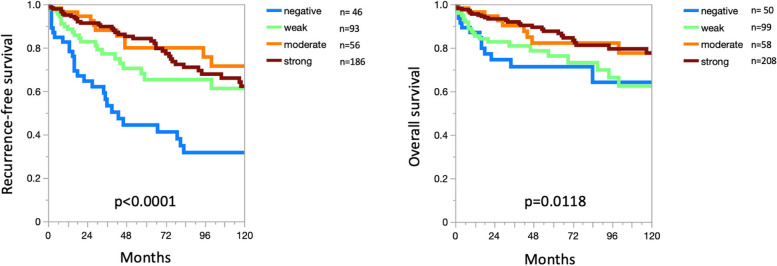
Fig. 4.
CLDN3 immunostaining and recurrence-free (A) and overall survival (B) in clear cell renal cell carcinoma
Fig. 3.
Ranking order of CLDN3 positive immunostaining in different human tumors.In ccRCC, low CLDN3 staining was strongly linked to poor ISUP (p < 0.0001), Fuhrman (p < 0.0001), and Thoenes (p < 0.0001) grades, advanced pT stage (p < 0.0001), high UICC stage (p = 0.0006), distant metastasis (p = 0.0011), as well as shortened overall (p = 0.0118; Fig. 4a) and recurrence-free (p < 0.0001; Fig. 4b) survival
In pRCC, low CLDN3 staining was associated with poor ISUP (p = 0.0019), Fuhrman (p = 0.0064), and Thoenes (p = 0.0315) grades, high pT (p = 0.0273), and distant metastasis (p = 0.0357). In urothelial carcinoma high CLDN3 staining was associated with high grade in non-invasive carcinomas (p < 0.0001), tumor invasiveness (pTa vs. pT2-4; p < 0.0001) as well as with nodal metastasis (p = 0.0111) and lymphovascular invasion (L1 status; p = 0.0062) in the subset of muscle-invasive carcinomas. The level of CLDN3 immunostaining was unrelated to parameters of tumor aggressiveness in ductal adenocarcinoma of the pancreas, gastric cancer and breast cancer. Associations with tumor phenotype are summarized in Table 2.
Table 2.
CLDN3 immunostaining and tumor phenotype
| CLDN3 immunostaining | |||||||
|---|---|---|---|---|---|---|---|
| n | negative (%) | weak (%) | moderate (%) | strong (%) | p | ||
| Invasive breast cancer of no special type | All cancers | 294 | 0.3 | 13.3 | 9.9 | 76.5 | |
| pT1 | 115 | 0.0 | 10.4 | 9.6 | 80.0 | 0.1192 | |
| pT2 | 147 | 0.7 | 11.6 | 11.6 | 76.2 | ||
| pT3-4 | 29 | 0.0 | 31.0 | 3.4 | 65.5 | ||
| G1 | 8 | 0.0 | 0.0 | 12.5 | 87.5 | 0.6289 | |
| G2 | 160 | 0.0 | 13.1 | 10.6 | 76.3 | ||
| G3 | 126 | 0.8 | 14.3 | 8.7 | 76.2 | ||
| pN0 | 148 | 0.7 | 12.8 | 9.5 | 77.0 | 0.6641 | |
| pN + | 117 | 0.0 | 12.8 | 12.0 | 75.2 | ||
| Endometrioid endometrial carcinoma | pT1 | 105 | 0.0 | 6.7 | 4.8 | 88.6 | 0.2436 |
| pT2 | 23 | 0.0 | 8.7 | 17.4 | 73.9 | ||
| pT3-4 | 37 | 0.0 | 8.1 | 13.5 | 78.4 | ||
| pN0 | 50 | 0.0 | 4.0 | 14.0 | 82.0 | 0.7811 | |
| pN + | 29 | 0.0 | 6.9 | 10.3 | 82.8 | ||
| Clear cell renal cell carcinoma | all cancers | 999 | 10.0 | 25.9 | 16.5 | 47.5 | |
| ISUP | |||||||
| 1 | 229 | 5.2 | 18.3 | 13.1 | 63.3 | < 0.0001 | |
| 2 | 355 | 5.9 | 25.4 | 16.9 | 51.8 | ||
| 3 | 221 | 10.4 | 34.4 | 17.6 | 37.6 | ||
| 4 | 64 | 48.4 | 23.4 | 14.1 | 14.1 | ||
| Fuhrman | |||||||
| 1 | 54 | 1.9 | 11.1 | 5.6 | 81.5 | < 0.0001 | |
| 2 | 598 | 5.7 | 24.4 | 17.4 | 52.5 | ||
| 3 | 252 | 11.9 | 32.1 | 18.7 | 37.3 | ||
| 4 | 78 | 43.6 | 28.2 | 11.5 | 16.7 | ||
| Thoenes | |||||||
| 1 | 300 | 6.0 | 19.7 | 15.3 | 59.0 | < 0.0001 | |
| 2 | 418 | 9.6 | 30.4 | 19.6 | 40.0 | ||
| 3 | 90 | 31.1 | 33.3 | 7.8 | 27.8 | ||
| UICC | |||||||
| 1 | 261 | 6.9 | 24.9 | 14.6 | 53.6 | 0.0006 | |
| 2 | 31 | 12.9 | 32.3 | 9.7 | 45.2 | ||
| 3 | 83 | 21.7 | 26.5 | 10.8 | 41.0 | ||
| 4 | 65 | 26.2 | 32.3 | 7.7 | 33.8 | ||
| pT1 | 574 | 5.1 | 23.7 | 17.1 | 54.2 | < 0.0001 | |
| pT2 | 111 | 6.3 | 22.5 | 20.7 | 50.5 | ||
| pT3-4 | 303 | 20.8 | 31.0 | 14.2 | 34.0 | ||
| pN0 | 151 | 15.2 | 27.2 | 11.9 | 45.7 | 0.1249 | |
| pN + | 22 | 27.3 | 27.3 | 22.7 | 22.7 | ||
| pM0 | 88 | 9.1 | 19.3 | 12.5 | 59.1 | 0.0011 | |
| pM + | 83 | 25.3 | 31.3 | 10.8 | 32.5 | ||
| Papillary renal cell carcinoma | All cancers | 239 | 2.9 | 5.9 | 9.2 | 82.0 | |
| ISUP | |||||||
| 1 | 29 | 0.0 | 3.4 | 13.8 | 82.8 | 0.0019 | |
| 2 | 113 | 0.9 | 4.4 | 3.5 | 91.2 | ||
| 3 | 62 | 6.5 | 8.1 | 17.7 | 67.7 | ||
| 4 | 4 | 25.0 | 25.0 | 25.0 | 25.0 | ||
| Fuhrman | |||||||
| 1 | 2 | 0.0 | 0.0 | 0.0 | 100 | 0.0064 | |
| 2 | 153 | 0.7 | 3.9 | 5.9 | 89.5 | ||
| 3 | 63 | 6.3 | 7.9 | 15.9 | 69.8 | ||
| 4 | 8 | 12.5 | 12.5 | 37.5 | 37.5 | ||
| Thoenes | |||||||
| 1 | 45 | 0.0 | 4.4 | 13.3 | 82.2 | 0.0315 | |
| 2 | 128 | 2.3 | 6.3 | 7.0 | 84.4 | ||
| 3 | 14 | 14.3 | 0.0 | 28.6 | 57.1 | ||
| UICC | |||||||
| 1 | 77 | 1.3 | 3.9 | 9.1 | 85.7 | 0.0625 | |
| 2 | 9 | 0.0 | 22.2 | 22.2 | 55.6 | ||
| 3 | 3 | 0.0 | 0.0 | 0.0 | 100.0 | ||
| 4 | 9 | 22.2 | 22.2 | 11.1 | 44.4 | ||
| pT1 | 169 | 1.8 | 5.3 | 5.3 | 87.6 | 0.0273 | |
| pT2 | 39 | 2.6 | 7.7 | 17.9 | 71.8 | ||
| pT3-4 | 25 | 12.0 | 8.0 | 16.0 | 64.0 | ||
| pN0 | 20 | 0.0 | 10.0 | 15.0 | 75.0 | 0.2338 | |
| pN + | 12 | 16.7 | 8.3 | 16.7 | 58.3 | ||
| pM0 | 23 | 0.0 | 4.3 | 17.4 | 78.3 | 0.0357 | |
| pM + | 11 | 18.2 | 18.2 | 27.3 | 36.4 | ||
| Ductal adenocarcinoma of the pancreas | pT1 | 8 | 50 | 25 | 0 | 25 | 0.662 |
| pT2 | 43 | 37.2 | 39.5 | 9.3 | 14 | ||
| pT3 | 242 | 35.5 | 33.9 | 12.8 | 17.8 | ||
| pT4 | 17 | 52.9 | 29.4 | 11.8 | 5.9 | ||
| G1 | 10 | 50 | 20 | 0 | 30 | 0.092 | |
| G2 | 218 | 35.3 | 33 | 11.9 | 19.7 | ||
| G3 | 68 | 41.2 | 38.2 | 13.2 | 7.4 | ||
| pN0 | 65 | 40 | 29.2 | 12.3 | 18.5 | 0.8041 | |
| pN + | 244 | 36.1 | 35.7 | 11.9 | 16.4 | ||
| Gastric adenocarcinoma | All cancers | 316 | 13.9 | 13.9 | 9.8 | 62.7 | |
| pT1-2 | 42 | 7.1 | 7.1 | 4.8 | 81.0 | 0.2596 | |
| pT3 | 103 | 15.5 | 11.7 | 12.6 | 60.2 | ||
| pT4 | 105 | 15.2 | 14.3 | 10.5 | 60.0 | ||
| pN0 | 61 | 13.1 | 8.2 | 8.2 | 70.5 | 0.5204 | |
| pN + | 189 | 13.8 | 13.8 | 11.1 | 61.4 | ||
| Urothelial carcinoma | All cancers | 619 | 50.7 | 27.6 | 10.3 | 11.3 | |
| pTa G2 low | 79 | 77.2 | 16.5 | 5.1 | 1.3 | < 0.0001 | |
| pTa G2 high | 73 | 63.0 | 26.0 | 8.2 | 2.7 | ||
| pTa G3 | 86 | 36.0 | 37.2 | 9.3 | 17.4 | ||
| pT2 | 97 | 50.5 | 20.6 | 14.4 | 14.4 | 0.5546 | |
| pT3 | 183 | 46.4 | 31.1 | 10.4 | 12.0 | ||
| pT4 | 92 | 43.5 | 28.3 | 12.0 | 16.3 | ||
| G2 | 18 | 55.6 | 27.8 | 11.1 | 5.6 | 0.6878 | |
| G3 | 354 | 46.3 | 27.7 | 11.9 | 14.1 | ||
| pN0 | 203 | 51.2 | 27.1 | 12.3 | 9.4 | 0.0111 | |
| pN + | 143 | 38.5 | 29.4 | 11.2 | 21.0 | ||
| L0 | 146 | 52.7 | 27.4 | 11.0 | 8.9 | 0.0062 | |
| L1 | 132 | 36.4 | 30.3 | 11.4 | 22.0 | ||
Abbreviations: G grade, pM pathologic status of distant metastasis, pN pathologic lymph node status, pT pathologic tumor stage, L Lymphatic invasion status, ISUP International Society of Urologic Pathologists, UICC Union for International Cancer Control
Discussion
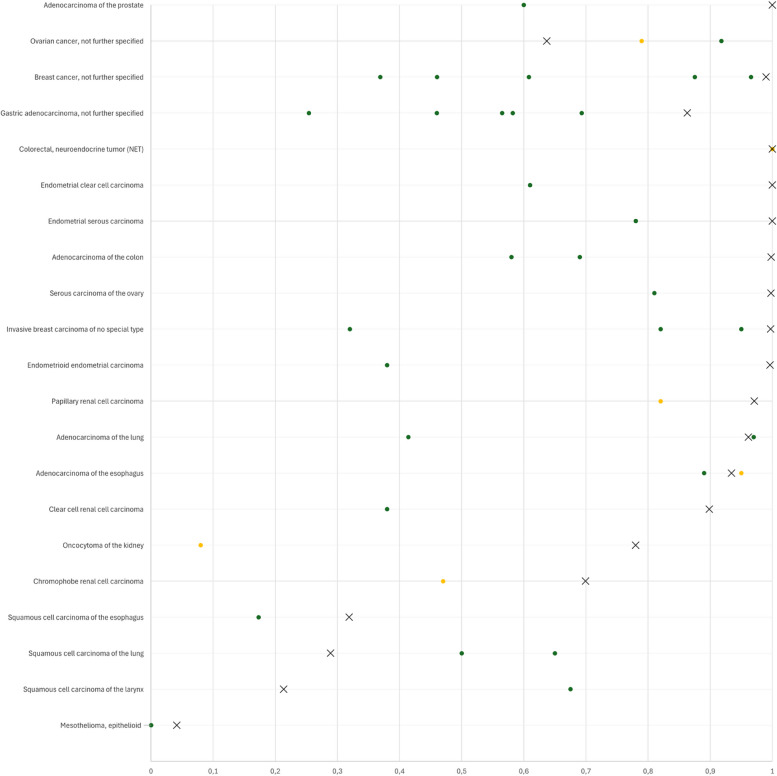
The results of our successful analysis of 14,966 tumors from 133 different tumor categories provide a comprehensive overview of CLDN3 expression in cancer. Although CLDN3 expression could be found in a wide range of tumor entities, it showed that CLDN3 positivity was most seen in neuroendocrine neoplasms and adenocarcinomas, as well as in tumors of the female genital tract and various subtypes of breast cancer. CLDN3 positivity was less frequent in squamous cell carcinomas and, as described by others [36], only rarely seen in hematolymphoid and in most mesenchymal neoplasms. Although previous studies on CLDN3 expression in cancer were limited in number and had provided partially conflicting data (summarized in Fig. 5), several earlier results are in agreement with our data. For example, CLDN3 positivity was described in 95% of 20 [37] and in 89% of 57 [38] esophageal adenocarcinomas (our study: 93.4%), 100% of 16 colorectal neuroendocrine tumors [39] (our study: 100%), and in 97% of 34 pulmonary adenocarcinomas [29] (our study: 96.2%).
Fig. 5.
CLDN3 protein expression in cancer (own findings vs. literature data). Graphical representation of CLDN3 data from this study (X) compared to the previous literature. The colors of the dots represent the number of tumors analyzed in these studies: red: n ≤ 20; yellow: n = 21 to 100; green: n > 100. For raw data and references, see suppl. Tab. 1
Claudins, which are essential for the formation of TJs in human epithelial and endothelial cells, are altered in a variety of tumors. Because both downregulation and upregulation of CLDN3 have been found in different tumor entities and both alterations have been associated with aggressive tumor characteristics, a tissue type dependency of CLDN3 function has been assumed [40]. The striking associations between a reduced CLDN3 expression and unfavorable histopathological tumor parameters and poor prognosis in ccRCC represents a key finding of our study. As adjuvant systemic therapies are increasingly being administered in high and intermediate risk RCC, there is a need for a better assessment of the individual risk of progression in these tumors [41, 42]. In the future, CLDN3 IHC could evolve towards a clinically useful prognostic marker in RCC, optimally in combination with other markers. A link between reduced CLDN3 expression and poor patient outcome or unfavorable tumor characteristics was previously also found in other cancer types. Jung et al. [21] described an association between reduced CLDN3 expression and L1 status as well as advanced T-stage in a study on 72 gastric adenocarcinomas. Che et al. [24] reported low CLDN3 expression in squamous cell carcinomas of the lung with high pT stage, nodal metastasis, and disease recurrence. Orea et al. [19] found lower disease-free survival and time to clinical progression in prostatic adenocarcinomas with low CLDN3 expression. Jiang et al. [23] found a shortened overall survival in hepatocellular carcinomas with reduced CLDN3 mRNA expression. Functional studies on cell line models identified associations between reduced CLDN3 expression and various cancer driving mechanisms such as a decrease in epithelial barrier function [43], invasiveness [43], dedifferentiation [43], proliferative potential [44], and reduced adhesion [40]. Alternatively, it cannot be excluded that reduced CLDN3 expression in tumors derived from CLDN3 expressing normal cells merely reflects tumor cell dedifferentiation which always parallels cancer progression.
That not only downregulation but also upregulation can be associated with tumor progression in a tumor type dependent manner is demonstrated in our study by the strong relationship between CLDN3 upregulation and grade and stage progression in urothelial carcinomas. This is in line with data from an earlier study by Nakanishi et al. showing a link between high CLDN3 expression and advanced stage, high grade and poor overall survival in a cohort of 129 urothelial cancers of the upper urinary tract [45]. A significant association between CLDN3 upregulation and tumor progression had also been reported for breast [25] and ovarian cancer [17]. Mechanisms that were suggested to explain a tumor promoting role of CLDN3 in cancer include a regulatory impact on cancer stemness [46] and increased chemoresistance [46]. In ovarian cancer cell lines, Agarwal et al. [47] found associations between CLDN3 upregulation and increased cell survival, invasion and motility. Again, it cannot be excluded that CLDN3 neo-expression can be caused by random alterations occurring during dedifferentiation in tumors cell derived from CLDN3 non-expressing normal cells.
Claudins represent potential therapeutic cancer drug targets for several cancer types due to their membranous localization [48]. Initial evidence for druggability of CLDN3 came from experiments with Clostridium perfringens enterotoxin (CPE), which causes food poisoning, and selectively binds to the ECL2 motive of CLDN3 [49, 50]. Non-cytotoxic CPE fragments have therefore been interrogated for their therapeutic potential in cancer. They showed anti-tumor efficacy in prostate [50], breast [51], and ovarian cancer cells [52]. Moreover, C-terminal fragment of CPE increased the efficacy of chemotherapy in ovarian cancer [53] and were also successfully used as a carrier to specifically deliver therapeutic drugs to ovarian cancer cells [54]. While CPE is not specific for CLDN3 but also binds to other claudins, specific antibodies have been developed for treating cancer [7, 55, 56]. Human monoclonal antibodies such as KM390755, IgGH6 [57], and h4G3 [7] been developed against the CLDN3 ECL1 and ECL2 domains. These antibodies were shown to induce antibody-dependent cellular cytotoxicity (ADCC) and in case of KM3907 also a complement-dependent cytotoxicity (CDC) [48, 55].
Considering the large scale of our study, our assay was extensively validated by comparing our IHC findings in normal tissues with data obtained by another independent anti-CLDN3 antibody and CLDN3 RNA data derived from three different publicly accessible databases [58–61]. This validation procedure was suggested by the international working group of antibody validation (IWGAV) [62]. To ensure an as broad as possible range of proteins to be tested for cross-reactivity, 76 different normal tissues categories were included in this analysis. Validity of our assay was supported by the detection of strongest claudin-3 immunostaining in tissues with highest RNA expression (intestine, thyroid, pancreas, and the prostate). True CLDN3 expression in tissues and cell types found to be CLDN3 positive by HMV-309 but lacking documented RNA expression (germinal center cells in lymphatic tissues, megakaryocytes in the bone marrow, squamous epithelium positivity in the thymus and the tonsil crypts, gallbladder, urothelium, placenta, epididymis, gastric mucosa, adrenal gland, and the parathyroidal gland) as well as in tissues with only very low CLDN3 RNA levels (endometrium) were confirmed by identical stainings seen by the independent antibody EPR19971 (Suppl. Figure 1). Given that these CLDN3 positive cell types constituted very small subpopulations of the respective organs, we assume that CLDN3 RNA had not been detected due to a massive dilution if RNAs from total organs were analyzed. Overall, these data document a high specificity of our IHC assay for CLDN3 detection.
Conclusion
Our data demonstrate significant levels of CLDN3 expression in many different tumor entities, and show that both increased and decreased levels of CLDN3 can occur during tumor progression in a cancer type dependent manner. The strong association between low CLDN3 expression and unfavorable prognostic tumor features may suggest a clinically useful role of CLDN3 expression measurement in ccRCC.
Supplementary Information
Acknowledgements
We are grateful to Laura Behm, Melanie Steurer, Inge Brandt, Maren Eisenberg, and Sünje Seekamp for excellent technical assistance.
Abbreviations
- CLDN3
Claudin 3
- TJ
Tight junction
- RCC
Renal cell carcinoma
- ccRCC
Clear cell renal cell carcinoma
- pRCC
Papillary renal cell carcinoma
- IHC
Immunohistochemistry
- TMA
Tissue microarray
- CPE
Clostridium perfringes enterotoxin
- ADCC
Antibody-dependent cellular cytotoxicity
- CDC
Complement-dependent cytotoxicity
- IWGAV
International working group of antibody validation
- G
Grade
- pM
Pathologic status of distant metastasis
- pN
Pathologic lymph node status
- pT
Pathologic tumor stage
- L
Lymphatic invasion status
- ISUP
International Society of Urologic Pathologists
- UICC
Union for International Cancer Control
Authors’ contributions
SB, NS, RS, GS, MF: contributed to conception, design, data collection, data analysis and manuscript writing. AM, FL, VC, FV, DD, RS, AH, CF, CB, KM, VR, AML, PL, SW, ML, FJ, TC, AHM, SS, EB, NG, SM, TK: participated in pathology data analysis, data interpretation, and collection of samples RS, MK, CHM: data analysis RS, GS, MF: study supervision All authors agree to be accountable for the content of the work.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
The use of archived remnants of diagnostic tissues for manufacturing of TMAs and their analysis for research purposes as well as patient data analysis has been approved by local laws (HmbKHG, §12) and by the local ethics committee (Ethics commission Hamburg, WF-049/09). All work has been carried out in compliance with the Helsinki Declaration. Patient consent was waived due to local laws (HmbKHG, §12,1) that permit research with anonymized diagnostic left-over tissue samples.
Consent for publication
Not applicable.
Competing interests
Conflict of interests The CLDN3 antibody clone HMV-309 was provided from ardoci GmbH (owned by a family member of GS).
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tsukita S, Tanaka H, Tamura A. The Claudins: from tight junctions to biological systems. Trends Biochem Sci. 2019;44:141–52. [DOI] [PubMed] [Google Scholar]
- 2.Günzel D, Yu ASL. Claudins and the modulation of tight junction permeability. Physiol Rev. 2013;93:525–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J. Context-dependent roles of claudins in tumorigenesis. Front Oncol. 2021;11:676781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber CR. Dynamic properties of the tight junction barrier. Ann N Y Acad Sci. 2012;1257:77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milatz S, et al. Claudin-3 acts as a sealing component of the tight junction for ions of either charge and uncharged solutes. Biochim Biophys Acta. 2010;1798:2048–57. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Hernandez V, Quiros M, Nusrat A. Intestinal epithelial claudins: expression and regulation in homeostasis and inflammation. Ann N Y Acad Sci. 2017;1397:66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H, et al. Development of human monoclonal antibody for Claudin-3 overexpressing carcinoma targeting. Biomolecules. 2019;10:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinemann U, Schuetz A. Structural features of tight-junction proteins. Int J Mol Sci. 2019;20:6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nehme Z, Roehlen N, Dhawan P, Baumert TF. Tight junction protein signaling and cancer biology. Cells. 2023;12:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Díaz-Coránguez M, Liu X, Antonetti DA. Tight junctions in cell proliferation. Int J Mol Sci. 2019;20:5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahin U, et al. FAST: a randomised phase II study of zolbetuximab (IMAB362) plus EOX versus EOX alone for first-line treatment of advanced CLDN18.2-positive gastric and gastro-oesophageal adenocarcinoma. Ann Oncol. 2021;32:609–19. [DOI] [PubMed] [Google Scholar]
- 12.Tokuhara Y, et al. Nuclear expression of claudin-3 in human colorectal adenocarcinoma cell lines and tissues. Oncol Lett. 2018;15:99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danzinger S, et al. Differential Claudin 3 and EGFR expression predicts BRCA1 mutation in triple-negative breast cancer. Cancer Invest. 2018;36:378–88. [DOI] [PubMed] [Google Scholar]
- 14.Kolokytha P, et al. Claudin-3 and claudin-4: distinct prognostic significance in triple-negative and luminal breast cancer. Appl Immunohistochem Mol Morphol. 2014;22:125–31. [DOI] [PubMed] [Google Scholar]
- 15.Tokés A-M, et al. Claudin-1, -3 and -4 proteins and mRNA expression in benign and malignant breast lesions: a research study. Breast Cancer Res. 2005;7:R296-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu KH, et al. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin Cancer Res. 2004;10:3291–300. [DOI] [PubMed] [Google Scholar]
- 17.Choi Y-L, et al. Expression profile of tight junction protein claudin 3 and claudin 4 in ovarian serous adenocarcinoma with prognostic correlation. Histol Histopathol. 2007;22:1185–95. [DOI] [PubMed] [Google Scholar]
- 18.Bartholow TL, Chandran UR, Becich MJ, Parwani AV. Immunohistochemical profiles of claudin-3 in primary and metastatic prostatic adenocarcinoma. Diagn Pathol. 2011;6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orea MJ, et al. Claudin-3 loss of expression is a prognostic marker in castration-resistant prostate cancer. Int J Mol Sci. 2023;24:803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jun K-H, Kim J-H, Jung J-H, Choi H-J, Chin H-M. Expression of claudin-7 and loss of claudin-18 correlate with poor prognosis in gastric cancer. Int J Surg. 2014;12:156–62. [DOI] [PubMed] [Google Scholar]
- 21.Jung H, Jun KH, Jung JH, Chin HM, Park WB. The expression of claudin-1, claudin-2, claudin-3, and claudin-4 in gastric cancer tissue. J Surg Res. 2011;167:e185-191. [DOI] [PubMed] [Google Scholar]
- 22.Soini Y, Tommola S, Helin H, Martikainen P. Claudins 1, 3, 4 and 5 in gastric carcinoma, loss of claudin expression associates with the diffuse subtype. Virchows Arch. 2006;448:52–8. [DOI] [PubMed] [Google Scholar]
- 23.Jiang L, et al. CLDN3 inhibits cancer aggressiveness via Wnt-EMT signaling and is a potential prognostic biomarker for hepatocellular carcinoma. Oncotarget. 2014;5:7663–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Che J, et al. Decreased expression of claudin-3 is associated with a poor prognosis and EMT in completely resected squamous cell lung carcinoma. Tumour Biol. 2015;36:6559–68. [DOI] [PubMed] [Google Scholar]
- 25.Jääskeläinen A, et al. High-level cytoplasmic claudin 3 expression is an independent predictor of poor survival in triple-negative breast cancer. BMC Cancer. 2018;18:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu S, et al. Claudin expression in high-grade invasive ductal carcinoma of the breast: correlation with the molecular subtype. Mod Pathol. 2013;26:485–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skálová H, et al. Impact of chemotherapy on the expression of claudins and cadherins in invasive breast cancer. Exp Ther Med. 2019;18:3014–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung JH, et al. Diagnostic utility of expression of claudins in non-small cell lung cancer: different expression profiles in squamous cell carcinomas and adenocarcinomas. Pathol Res Pract. 2009;205:409–16. [DOI] [PubMed] [Google Scholar]
- 29.Chaouche-Mazouni S, et al. Claudin 3, 4, and 15 expression in solid tumors of lung adenocarcinoma versus malignant pleural mesothelioma. Ann Diagn Pathol. 2015;19:193–7. [DOI] [PubMed] [Google Scholar]
- 30.Dancau A-M, Simon R, Mirlacher M, Sauter G. Tissue microarrays. Methods Mol Biol. 2016;1381:53–65. [DOI] [PubMed] [Google Scholar]
- 31.Kononen J, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–7. [DOI] [PubMed] [Google Scholar]
- 32.Lennartz M, et al. TRPS1 is a Highly Sensitive Marker for breast cancer: a tissue microarray study evaluating more than 19,000 tumors from 152 different tumor entities. Am J Surg Pathol. 2024;48:637–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schraps N, et al. Prevalence and significance of AGR2 expression in human cancer. Cancer Med. 2024;13:e70407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kind S, et al. KLK7 expression in human tumors: a tissue microarray study on 13,447 tumors. BMC Cancer. 2024;24:794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon R, Mirlacher M, Sauter G. Immunohistochemical analysis of tissue microarrays. Methods Mol Biol. 2010;664:113–26. [DOI] [PubMed] [Google Scholar]
- 36.Soini Y. Expression of claudins 1, 2, 3, 4, 5 and 7 in various types of tumours. Histopathology. 2005;46:551–60. [DOI] [PubMed] [Google Scholar]
- 37.Takala H, Saarnio J, Wiik H, Soini Y. Claudins 1, 3, 4, 5 and 7 in esophageal cancer: loss of claudin 3 and 4 expression is associated with metastatic behavior. APMIS. 2007;115:838–47. [DOI] [PubMed] [Google Scholar]
- 38.Montgomery E, et al. Overexpression of claudin proteins in esophageal adenocarcinoma and its precursor lesions. Appl Immunohistochem Mol Morphol. 2006;14:24–30. [DOI] [PubMed] [Google Scholar]
- 39.Ishida M, Kushima R, Okabe H. Claudin expression in rectal well-differentiated endocrine neoplasms (carcinoid tumors). Oncol Rep. 2009;21:113–7. [PubMed] [Google Scholar]
- 40.Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65:9603–6. [DOI] [PubMed] [Google Scholar]
- 41.Klatte T, Rossi SH, Stewart GD. Prognostic factors and prognostic models for renal cell carcinoma: a literature review. World J Urol. 2018;36:1943–52. [DOI] [PubMed] [Google Scholar]
- 42.Rosellini M, et al. Prognostic and predictive biomarkers for immunotherapy in advanced renal cell carcinoma. Nat Rev Urol. 2023;20:133–57. [DOI] [PubMed] [Google Scholar]
- 43.Ahmad R, et al. Loss of claudin-3 expression induces IL6/gp130/Stat3 signaling to promote colon cancer malignancy by hyperactivating Wnt/β-catenin signaling. Oncogene. 2017;36:6592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okugawa T, et al. Down-regulation of claudin-3 is associated with proliferative potential in early gastric cancers. Dig Dis Sci. 2012;57:1562–7. [DOI] [PubMed] [Google Scholar]
- 45.Nakanishi K, et al. Expression of occludin and claudins 1, 3, 4, and 7 in urothelial carcinoma of the upper urinary tract. Am J Clin Pathol. 2008;130:43–9. [DOI] [PubMed] [Google Scholar]
- 46.Ma L, Yin W, Ma H, Elshoura I, Wang L. Targeting claudin-3 suppresses stem cell-like phenotype in nonsquamous non-small-cell lung carcinoma. Lung Cancer Manag. 2019;8(1):LMT04. [DOI] [PMC free article] [PubMed]
- 47.Agarwal R, D’Souza T, Morin PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res. 2005;65:7378–85. [DOI] [PubMed] [Google Scholar]
- 48.Vonniessen B, Tabariès S, Siegel PM. Antibody-mediated targeting of Claudins in cancer. Front Oncol. 2024;14:1320766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao Z, McClane BA. Use of Clostridium perfringens Enterotoxin and the Enterotoxin Receptor-Binding Domain (C-CPE) for cancer treatment: opportunities and challenges. J Toxicol. 2012;2012: 981626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romanov V, Whyard TC, Waltzer WC, Gabig TG. A claudin 3 and claudin 4-targeted Clostridium perfringens protoxin is selectively cytotoxic to PSA-producing prostate cancer cells. Cancer Lett. 2014;351:260–4. [DOI] [PubMed] [Google Scholar]
- 51.Kominsky SL, et al. Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am J Pathol. 2004;164:1627–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santin AD, et al. Treatment of chemotherapy-resistant human ovarian cancer xenografts in C.B-17/SCID mice by intraperitoneal administration of Clostridium perfringens enterotoxin. Cancer Res. 2005;65:4334–42. [DOI] [PubMed] [Google Scholar]
- 53.Gao Z, et al. C terminus of Clostridium perfringens enterotoxin downregulates CLDN4 and sensitizes ovarian cancer cells to Taxol and Carboplatin. Clin Cancer Res. 2011;17:1065–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cocco E, et al. Clostridium perfringens enterotoxin carboxy-terminal fragment is a novel tumor-homing peptide for human ovarian cancer. BMC Cancer. 2010;10:349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kato-Nakano M, et al. Characterization and evaluation of the antitumour activity of a dual-targeting monoclonal antibody against claudin-3 and claudin-4. Anticancer Res. 2010;30:4555–62. [PubMed] [Google Scholar]
- 56.Li X, et al. Development of an anti-claudin-3 and -4 bispecific monoclonal antibody for cancer diagnosis and therapy. J Pharmacol Exp Ther. 2014;351:206–13. [DOI] [PubMed] [Google Scholar]
- 57.Romani C, et al. Evaluation of a novel human IgG1 anti-claudin3 antibody that specifically recognizes its aberrantly localized antigen in ovarian cancer cells and that is suitable for selective drug delivery. Oncotarget. 2015;6:34617–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thul PJ, et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. [DOI] [PubMed] [Google Scholar]
- 59.Lizio M, et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015;16:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 20013;45:580–585. [DOI] [PMC free article] [PubMed]
- 61.Lizio M, et al. Update of the FANTOM web resource: expansion to provide additional transcriptome atlases. Nucleic Acids Res. 2019;47:D752–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uhlen M, et al. A proposal for validation of antibodies. Nat Methods. 2016;13:823–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.