Abstract
Virion infectivity factor (Vif) is a protein encoded by human immunodeficiency virus types 1 and 2 (HIV-1 and -2) and simian immunodeficiency virus, plus other lentiviruses, and is essential for viral replication either in vivo or in culture for nonpermissive cells such as peripheral blood lymphoid cells, macrophages, and H9 T cells. Defects in the vif gene affect virion morphology and reverse transcription but not the expression of viral components. It has been shown that Vif colocalizes with Gag in cells and Vif binds to the NCp7 domain of Gag in vitro. However, it seems that Vif is not specifically packaged into virions. The molecular mechanism(s) for Vif remains unknown. In this report, we demonstrate that HIV-1 Vif is an RNA-binding protein and specifically binds to HIV-1 genomic RNA in vitro. Further, Vif binds to HIV-1 RNA in the cytoplasm of virus-producing cells to form a 40S mRNP complex. Coimmunoprecipitation and in vivo UV cross-linking assays indicated that Vif directly interact with HIV-1 RNA in the virus-producing cells. Vif-RNA binding could be displaced by Gag-RNA binding, suggesting that Vif protein in the mRNP complex may mediate viral RNA interaction with HIV-1 Gag precursors. Furthermore, we have demonstrated that these Vif mutants that lose the RNA binding activity in vitro do not support vif-deficient HIV-1 replication in H9 T cells, suggesting that the RNA binding capacity of Vif is important for its function. Further studies regarding Vif-RNA interaction in virus-producing cells will be important for studying the function of Vif in the HIV-1 life cycle.
Virion infectivity factor (Vif) protein of human immunodeficiency virus type 1 (HIV-1) is a highly basic, 23-kDa protein composed of 192 amino acids. Sequence analysis of viral DNA from HIV-1-infected-individuals has shown that the open reading frame of Vif remains intact (58, 66, 67). Deletion of the vif gene will dramatically decrease the replication of simian immunodeficiency virus (SIV) in macaques and HIV-1 replication in SCID-hu mice (3, 22). These studies indicate that the vif gene is required for viral replication in vivo. In cell culture systems, vif-deficient (vif−) HIV-1 is incapable of establishing infection in certain cells, such as H9 T cells, peripheral blood mononuclear cells, and monocyte-derived macrophages. This has led to classification of these cells as nonpermissive. However, in some cells, such as C8166, Jurkat, SupT1, and HeLa-T4 cells, the vif gene is not required, and these cells have been classified as permissive (29, 32, 33, 60, 64).
Extensive studies have been performed to identify the role of Vif in the viral life cycle. A defective vif gene can be complemented by wild-type Vif protein expressed in the virus-producing cells but not in the target cells, indicating that Vif functions in the virus-producing cells or within cell-free virions (7, 32, 64). Defects of the vif gene do not have detectable effects on viral transcription and translation or on virion production. HIV-1 variants with a defective vif gene are able to bind and penetrate the target cells but are not able to complete intracellular reverse transcription and endogenous reverse transcription (ERT) in cell-free virions (17, 36, 59, 64). Conversely, it has also been reported that the stability of newly synthesized viral DNA in the target cells is impaired (54). Recently, we demonstrated that defects in the vif gene have much less of an effect on ERT if detergent is not used. When ERT was driven by addition of deoxyribonucleoside triphosphates at high concentrations, certain levels of plus-strand viral DNA could also be completed. Interestingly, if vif− viruses, generated from nonpermissive cells and harboring larger quantities of viral DNA generated by ERT, were allowed to infect permissive cells, they could partially bypass the block at intracellular reverse transcription, through which vif− viruses without deoxynucleoside triphosphate (dNTP) treatment could not pass. Consequently, viral infectivity can be partially rescued from the vif− phenotype (24). Most of the studies indicated that the expression of viral components, including viral proteins and nucleic acids, is not altered in the virions produced from nonpermissive cells (31, 32, 64). However, deletion of the vif gene will result in alterations of virion morphology (8, 10, 38). It has been shown that the quantity of Vif protein in the HIV-1 virions generated from chronically infected cells is approximately 7 to 28 molecules per virion (13, 31, 55). As the virion-associated Vif proteins do not depend on the expression of viral components and the amount of Vif in the virus-producing cells, it seems that Vif proteins are not specifically incorporated into virions (13, 55). Recently, it was reported that Vif was absent from virions when highly purified virions were used for quantitative analysis (23).
Based on these investigations, it has been proposed that Vif functions in the virus-producing cells and could affect viral assembly. The expression of Vif in infected cells is quite high, and the majority of Vif in the virus-producing cells is in the cytoplasmic fraction; some are associated with the cellular membrane. The molar ratio of Vif to Gag precursors in the infected cells is 1:1.7, suggesting that Vif may play a structural rather than a regulatory role in the virus-producing cells (35, 55). As Vif is required by nonpermissive but not the permissive cells for HIV-1 replication, two possibilities exist. In permissive cells, there may be a Vif cellular homologue which can replace Vif function in the virus-producing cells; alternatively, there may be an inhibitor(s) for viral replication in nonpermissive cells which require Vif to counteract their effects (62). Recently, it was proposed that Vif protein is required to counteract an unknown endogenous inhibitor(s) in the virus-producing cells (42, 53). HIV-1 Vif can complement the function of HIV-1 Vif and SIVAGM Vif in human nonpermissive cells, whereas it cannot complement the function of HIV-1 and SIVAGM Vif in simian cells. However, SIVAGM can complement the function of HIV-1 Vif and SIVAGM Vif in simian cells but not the function of HIV-1 Vif and SIVAGM Vif in human cells. This work indicates that a cellular cofactor(s) is involved in the action of Vif protein (56). Conversely, as a Vif mutant (Vif from HIV-1F12) can inhibit wild-type HIV-1 replication in the permissive cells, a Vif homologue in the permissive cells may also exist (18). Interestingly, although it seems that Vif is not specifically incorporated into virions, Vif is able to bind to the NCp7 domain of Gag precursors (9, 39). Vif protein is found to colocalize with Gag precursors in the cytoplasm of HIV-1-infected cells (52). This Vif-Gag interaction in vivo, however, could be indirect (51).
Overall, Vif may directly or indirectly be involved in the viral assembly process. Uncovering the molecular mechanism(s) of Vif will be extremely important for understanding HIV-1 pathogenesis and in identifying new targets for HIV-1 treatment. In this report, we attempt to evaluate Vif-binding proteins or nucleic acids in virus-producing cells. We show herein that Vif is an RNA-binding protein and is an integral component of an mRNP complex of viral RNA in the cytoplasm of virus-producing cells. The Vif protein in this mRNP complex may protect viral RNA from various endogenous inhibitors and could mediate viral RNA engagement with HIV-1 Gag precursors. As such, the interaction between Vif and HIV-1 RNA may play an important role in the late events of the HIV-1 life cycle.
MATERIALS AND METHODS
Plasmids, mutagenesis, and protein expression.
The vif gene and its fragments of HIV-1 were amplified by PCR with pNL4-3 as the template. The PCR fragments were then cloned into pGEM-T vector (Promega). HIV-1 gag, NCp7, and SIVmac251 vif genes were also cloned into pGEM-T by the same procedure. A PCR-based mutagenesis method, as described previously (68), was adapted to create point mutations in the vif gene. The PCR fragments were inserted into pGEM-T. All of the clones were confirmed by DNA sequencing. The cloned genes were then inserted into pGEX-KG for glutathione S-transferase (GST) fusion protein expression, into pCITE-4a vector for 35S-labeled in vitro protein synthesis, and into pSLX-CMV-RRE for retrovirus-mediated expression in specific cells. Of note, pSLX-CMV-RRE was constructed by inserting the HIV-1 Rev response element (RRE) into the 3′ portion of pSLX-CMV to allow for Vif expression to be Rev dependent. An HIV-1 infectious clone lacking the vif gene, pNL4-3Δvif, was constructed by replacing the EagI-EcoRI fragment of pNL4-3 with the EagI-EcoRI fragment of p197-1 (5′-half mutant of pNL4-3 with vif deletion) (34). To generate HIV-1 riboprobes, HIV-1 DNA fragments in various regions of pNL4-3 were amplified by PCR and inserted into pGEM-T.
To generate GST, GST-Gag, GST-p7, GST-Vif, and GST-Vif mutant fusions, pGEX-KG only and pGEX-KG harboring gag, NCp7, vif, and vif mutants, respectively, were transformed into Escherichia coli BL21 competent cells (Novagen). After the expression of GST or GST fusion proteins induced by 1 mM isopropylthio-β-d-galactoside, the bacterial cells were lysed by adding bacterial lysing buffer (cold phosphate-buffered saline [PBS] containing 0.5% Triton X-100, 0.2 mg of lysozyme/ml, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 2 μg of leupeptin/ml, and 1 μg of aprotinin/ml), followed by sonication. The insoluble fraction was pelleted at 10,000 × g for 10 min, and the supernatant was applied to a glutathione-conjugated agarose bead (Sigma) column. After washing, the bound GST or GST fusion proteins were eluted with 5 mM glutathione. The free glutathione was removed by dialysis assay. The protein concentration was measured by the Bradford method, and the samples were frozen at −70°C.
In vitro transcription and translation.
The HIV-1 and γ-actin RNAs (including riboprobes) were synthesized by in vitro transcription with linearized pGEM-T harboring HIV-1 DNA fragments or the γ-actin DNA fragment as the template. A standard protocol (Novagen) was followed. To synthesize 35S-labeled Vif protein or Vif fragments, the Novagen single-tube protein system 3 was used for in vitro translation, following the protocol supplied by the manufacturer. The translation efficiency was determined by measuring the trichloroacetic acid (TCA)-insoluble radioactive counts.
In vitro RNA-protein binding assays.
Several methods were used to study in vitro RNA-Vif binding. First, polynucleotide homopolymer-conjugated beads (Sigma) were mixed with in vitro-translated, 35S-labeled Vif or Vif fragments (50,000 cpm) in washing/binding buffer (150 mM NaCl, 10 mM Tris-HCl [pH 8.0], 0.1% Triton X-100). Binding was allowed to proceed at 23°C for 20 min and then at 4°C for 1 h. The beads were then washed with washing/binding buffer three times, and the bead-bound proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For gel mobility shift assays, GST or GST fusion proteins (10 pmol of each) were allowed to bind to riboprobes (105 cpm) in RNA-protein buffer modified from a previous description (65) [30 mM Tris-HCl (pH 8.0), 12% glycerol, 70 mM KCl, 1.3 mM dithiothreitol, 2.5 mM MgCl2, 0.01% Triton X-100, 20 U of RNasin RNase inhibitor (Promega), 750 ng of poly(dI-dC) (Sigma) or tRNA]. Binding was allowed to continue at 23°C for 15 min. The mixtures were then fractionated in native 5% Tris-borate-EDTA (TBE) gels. For filter binding assays, the GST and GST fusion proteins (10 pmol) were allowed to bind with riboprobes (105 or 2 × 105 cpm) in RNA-protein buffer [containing 750 ng of poly(dI-dC) as the inhibitor] at 23°C for 15 min. The mixtures were then slowly loaded onto presoaked HAWP nitrocellulose filters (0.45-μm pore size; Millipore). After washing with ice-cold PBS three times, the radioactivity remaining on the filters was determined by liquid scintillation counting (12). Finally, for UV light-induced cross-linking assays, GST or GST fusion proteins (10 pmol) were mixed with riboprobes (105 cpm) in the presence of RNA-protein binding buffer at 23°C for 15 min. The mixtures were then irradiated in a 300-nm UV light source for 10 min. RNase A (1 μg/ml) was added to digest the unprotected riboprobes. The mixtures were then heated at 95°C for 5 min and fractionated by SDS-PAGE.
Rate-zonal sedimentation.
HIV-1NL4-3-infected H9 T cells were harvested, washed twice with cold PBS, and then lysed with cell/virus lysing buffer (10 mM Tris-HCl, 100 mM NaCl, 1.5 mM MgCl2, 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin/ml, 1 μg of aprotinin/ml, 1 μg of benzamidine/ml). The nuclear fraction was removed by centrifugation at 1,400 × g for 3 min. The cytoplasmic portion was then placed onto a 15 to 30% sucrose gradient. After centrifugation at 250,000 × g for 3 h at 4°C, the gradient was collected in 12 parts. Vif protein in each fraction was then analyzed by Western blotting; HIV-1 unspliced and spliced RNAs and γ-actin RNA were extracted and detected by reverse transcriptase-mediated PCR (RT-PCR) as described previously (69, 70). The primer pair for HIV-1 RNA at the ψ site (unspliced RNA) was 5′-AGCAGTGGCGCCCGAACAGGGA-3′ (sense) plus 5′-TGCCCATACTATATGTTTTA-3′ (antisense); the probe was 5′-ATGGGTGCGAGAGCGTCGGTA-3′. The primer and probe for HIV-1 spliced RNA were described previously (49). The primer pair for γ-actin RNA was 5′-AAGAGATCGCCGCGCTGGTC-3′ (sense) plus 5′-GTACTTCAGGGTCAGGATGC-3′ (antisense); the probe was 5′-GTGTTTCCTTCCATCGTCGG-3′.
Coimmunoprecipitation.
Coimmunoprecipitation was used as described previously (47), with some modification. Briefly, the Vif protein-enriched fraction was mixed with anti-Vif antibody and protein A-conjugated Sepharose. As a control, preimmune rabbit serum was also mixed with the same sample and protein A-conjugated Sepharose. Binding was allowed to proceed at 4°C for 2 h. After washing three times, the Sepharose-bound Gag protein was subjected to SDS-PAGE followed by Western blot detection with anti-p7 antibody; the Sepharose-bound HIV-1 unspliced RNA and γ-actin RNA were subjected to RT-PCR analysis.
In vivo UV light-induced cross-linking and oligo(dT)-cellulose chromatography.
The protocol of Adam et al. (1) was followed, with some modification. Briefly, HIV-1NL4-3-infected H9 T cells were washed and exposed to a 300-nm UV light source for 10 min. The cells were then lysed with cell/virus lysing buffer. The postnuclear fraction was adjusted to 1 mM EDTA and 0.5% SDS. After heating at 65°C for 5 min, rapid chilling, and addition of high-salt buffer, the postnuclear fraction was allowed to pass through an oligo(dT)-conjugated cellulose column which was obtained from the mRNA purification kit (Pharmacia). After washing with high-salt buffer, the mRNA was eluted from the cellulose column and ethanol precipitated. The protocol supplied by the manufacturer was followed. After suspension in TN buffer, RNase A (1 μg/ml) was added to digest unprotected RNA. The mixtures were then analyzed by SDS-PAGE and Western blotting.
Viral infectivity assay.
Infectious clone pNL4-3Δvif was transfected into RD (rhabdomyosarcoma) cells to generate HIV-1NL4-3Δvif as described previously (68). Conversely, murine retroviral vectors (pSLX-CMV-RRE) containing HIV-1 vif and vif mutant genes were transfected into PA317 cells. The G418-resistant PA317 cells were then cocultured with H9 cells for 2 days to allow the vif or vif mutant-containing recombinant amphotropic murine leukemia virus (MLV) to infect H9 T cells. Subsequently, G418-resistant H9 T cells were selected. The H9 T cells (106) harboring vif or vif mutant genes were then infected with HIV-1NL4-3Δvif (1 ng of p24 antigen equivalents) as described previously (69, 70). Viral growth was monitored by detecting HIV-1 p24 antigen in the supernatant via enzyme-linked immunosorbent assay (ELISA) (Du Pont).
RESULTS
Vif specifically binds to HIV-1 RNA in vitro.
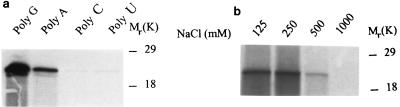
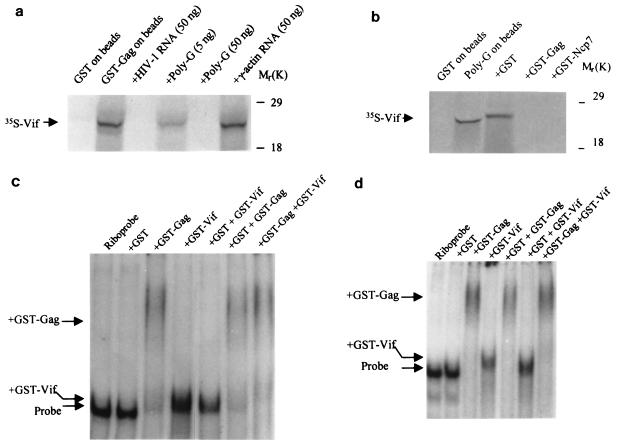
Our studies were initiated by searching for Vif-associated proteins or nucleic acids. As HIV-1 Vif binds to the NCp7 domain of the Gag protein (9, 39; our unpublished data), we investigated whether Vif binds to nucleic acids. With ribonucleotide homopolymer-conjugated agarose beads, which have been used to study other RNA-protein interactions (61), we found that Vif can bind to ribonucleotide homopolymers (Fig. 1a). Vif preferentially binds to poly(G), indicating that the binding is sequence specific, as described for the NCp7 protein of HIV-1 (30). This binding was quite strong, as it still occurred when the NaCl concentration reached 500 mM (Fig. 1b). Moreover, the Vif protein of SIVmac251 also bound to poly(G)-conjugated beads (data not shown).
FIG. 1.
HIV-1 Vif binds to polynucleotide homopolymers in vitro. (a) In vitro-translated, 35S-labeled HIV-1 Vif was mixed with polynucleotide homopolymers [poly(G), poly(A), poly(C), and poly(U)]-conjugated agarose beads, respectively. (b) In vitro-translated, 35S-labeled HIV-1 Vif was mixed with poly(G)-conjugated agarose beads in the presence of NaCl at various concentrations. After a binding and washing procedure, the bead-associated Vif protein was fractionated by SDS-PAGE.
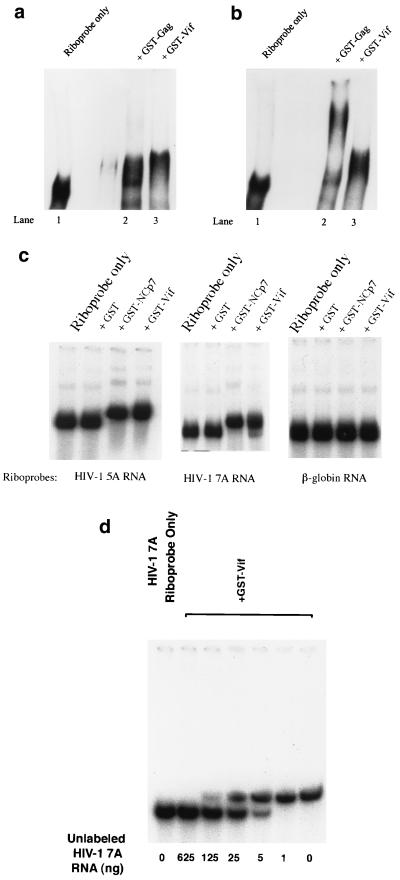
Further, GST-Vif protein was allowed to bind with HIV-1 riboprobes, and the mixtures were fractionated in native gels. Various inhibitors were tested in the binding buffer. Interestingly, the location of GST-Vif (or GST-Gag)-RNA complex in the gel shift was mainly affected by the inhibitors. The mechanism behind this phenomenon is unknown. Figure 2a indicates that GST-Vif-RNA complex and GST-Gag-RNA complex located at almost the same place in the gel when tRNA was used as the competitor. However, the location of GST-Gag-RNA complex and GST-Vif-RNA complex could be clearly separated on the gel when poly(dI-dC) was used as the competitor (Fig. 2b). Thus, poly(dI-dC), rather than tRNA, was used as the inhibitor for the rest of this work. Further, the gel mobility shift assay indicated that Vif can strongly bind to HIV-1 RNA at various regions but not to γ-actin RNA, suggesting that Vif specifically binds to HIV-1 RNA (Fig. 2c). Neither free poly(dI-dC) or tRNA can inhibit the HIV-1 RNA-Vif binding (Fig. 2a to c). This binding is similar to the binding between NCp7 and RNA, which is also HIV-1 RNA but not actin RNA specific and cannot be inhibited by free tRNA (5). To further verify the Vif-RNA shift, unlabeled HIV-1 RNA, at various concentrations, was added into the solutions for Vif-RNA binding. The mixture was then subject to gel shifting. Figure 2d demonstrates that the binding between Vif and HIV-1 riboprobe could be competitively inhibited by unlabeled HIV-1 RNA, in a concentration-dependent way. This result also indicated that the Vif-RNA shift, even though relatively minor, was substantial.
FIG. 2.
Gel shift assay to analyze the interaction between Vif and RNA. (a and b) GST-Gag and GST-Vif (10 pmol) were allowed to bind with the HIV-1 riboprobe 7A (located in the HIV-1 RNA genome at nucleotides 5104 to 5287) in the presence of 750 ng of tRNA (a) or poly(dI-dC) (b). (c) GST and GST-fusion proteins (GST-NCp7 and GST-Vif) (10 pmol) were allowed to bind with the HIV-1 riboprobe 5A (located in the HIV-1 RNA genome at nucleotides 3677 to 3925) and riboprobe 7A, respectively. As a control, a riboprobe generated from the γ-actin gene (nucleotides 81 to 280) was also allowed to bind with GST and GST-Vif. The RNA-protein mixtures were then fractionated on a 5% native TBE gel. (d) GST-Vif protein (10 pmol) were allowed to bind with HIV-1 riboprobe 7A in the presence of in vitro-transcribed, unlabeled HIV-1 7A at various concentrations. The mixtures were then fractionated on a 5% native TBE gel, followed by autoradiography.
Alternatively, filter binding assays, described by others for the analysis of NCp7 of HIV-1 and RNA binding, were used (15, 19). After binding in vitro, the mixture of GST-Vif and HIV-1 RNA was placed onto a nitrocellulose filter, and the TCA-insoluble radioactivity remaining on the filter was quantitated. This assay also demonstrated that GST-Vif strongly binds to HIV-1 RNA (Table 1). Interestingly, the binding between Vif and HIV-1 RNA is stronger than the binding between the Gag precursor and HIV-1 RNA at many regions of HIV-1 genomic RNA but not at the ψ site (Table 1 and data not shown). This result also indicated that the distance of protein-RNA shifting on the native gel is not correlated with the binding ability.
TABLE 1.
Interaction between Vif and HIV-1 RNA analyzed by filter binding assaya
| Assay | Radioactivity remaining on filter (cpm)
|
||
|---|---|---|---|
| 1Bb | 5Ac | 7Ac | |
| Riboprobe only | 726 | 744 | 711 |
| Riboprobe plus: | |||
| GST | 892 | 923 | 721 |
| GST-Gag | 46,727 | 13,236 | 26,847 |
| GST-NCp7 | 49,263 | 18,162 | 62,415 |
| GST-Vif | 36,223 | 38,215 | 51,886 |
HIV-1 riboprobes 1B (177 to 446 [ψ site]), 5A, and 7A were allowed to bind with GST or GST fusion proteins. The RNA-protein mixtures were then placed onto a nitrocellulose filter. After washing, the radioactivity remaining on the filter was quantitated. The values reported are the means of duplicated experiments.
Input radioactivity was 105 cpm.
Input radioactivity was 2 × 105 cpm.
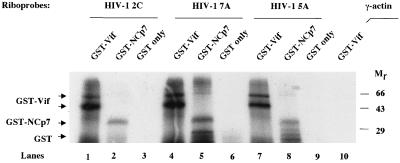
The in vitro UV cross-linking method was also used to study the interaction between Vif and RNA. HIV-1 riboprobes were mixed individually with GST-Vif, GST-NCp7, and GST proteins, followed by UV light-induced cross-linking. After RNase A digestion and heating, GST-Vif proteins that were cross-linked with 32P-labeled nucleotides were fractionated by SDS-PAGE (Fig. 3). This experiment demonstrated that the HIV-1 riboprobes could bind to HIV-1 Vif protein. However, the γ-actin riboprobe did not bind to GST-Vif under the same conditions (Fig. 3, lane 10), further indicating that binding between Vif and HIV-1 RNA is sequence specific. Of note, the smear covering the bottom half of lanes 1, 2, 4, 5, 7, and 8 may be due to the degraded Vif and NCp7 protein, as the smear does not appear in the lanes with GST (lanes 3, 6, and 9).
FIG. 3.
In vitro UV light-induced cross-linking assay. GST-Vif was mixed with HIV-1 riboprobes 2C (located in the HIV-1 RNA genome at nucleotides 1467 to 1677), 5A, and 7A. As controls, GST and GST-NCp7 were also mixed with HIV-1 riboprobes, while GST-Vif was mixed with an γ-actin riboprobe (lane 10). The mixtures were irradiated with UV light, followed by digestion with RNase A. The samples were then fractionated by SDS-PAGE, and protein-associated 32P-labeled RNA was visualized via autoradiography.
Vif binds to HIV-1 RNA and forms a 40S mRNP complex in the virus-producing cells.
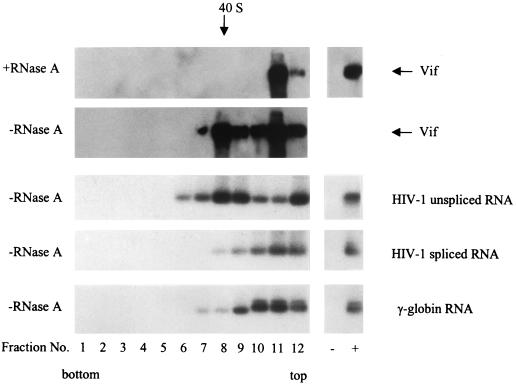
It has been shown that Vif does not specifically incorporate into virions and localizes mainly in the cytoplasmic rather than the nuclear fraction (13, 23, 35, 55). If Vif truly interacts with HIV-1 RNA in vivo, Vif could bind to HIV-1 RNA in the cytoplasmic fraction to form an mRNP complex. Similar to other retroviral genomic RNAs, the size of HIV-1 genomic RNA may be 34S to 38S (16). As such, Vif could also be associated with a particle which is larger or equal to 34S to 38S. To examine this possibility, the postnuclear fractions of HIV-1-infected cells were further fractionated by rate-zonal sedimentation, with or without RNase A treatment. As shown in Fig. 4, Vif is associated with an RNase A-sensitive particle (fraction 8). The sedimentation coefficient for this particle is approximately 40S. As expected, the unspliced HIV-1 RNA was enriched in this fraction, while spliced HIV-1 RNA and a cellular RNA, γ-actin RNA, did not occur in this fraction. Of note, the quantity of particle-associated Vif (fraction 8) is still lower than that of particle-free Vif (<10S) (fractions 11 and 12), possibly because of particle degradation by contaminating RNase, particle-free Vif proteins in the cytoplasm, or association with membranous structures.
FIG. 4.
Copurification of unspliced HIV-1 RNA with Vif by rate-zonal sedimentation. HIV-1NL4-3-infected H9 cells were lysed, and the postnuclear fraction was divided into two parts; one part was treated with RNase A (1 μg/ml), and the other part was treated with the RNase inhibitor RNasin (320 U/ml). Both portions were then placed onto a 15 to 30% sucrose gradient for ultracentrifugation. Twelve fractions were collected, and Vif protein in all fractions was detected via Western blotting. The sedimentation coefficient was calculated as described previously (44, 70). HIV-1 unspliced and spliced RNAs and γ-globin RNA were detected by RT-PCR.
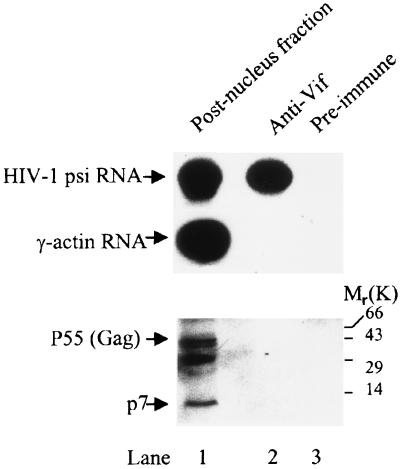
To examine the components of this RNase-sensitive particle, we used an anti-Vif antibody to capture Vif and its potential partner(s) (Fig. 4, −RNase A, fraction 8). As shown in Fig. 5, Vif-associated components included HIV-1 unspliced RNA but not γ-actin RNA. As well, NCp7 and Gag proteins were also not detected in the Vif-associated particle by coimmunoprecipitation assay, indicating that the Vif-associated particle is not the budding complex associated with the cellular membrane. Conversely, the interaction between Gag and Vif, as described by others, may not be in this fraction (40S) (9, 39, 52). The complex that both Vif and Gag are associated with may be detergent sensitive; alternatively, if it is detergent resistant, the proteins may be too small to reach the 40S fraction in rate-zonal sedimentation (250,000 × g for 3 h).
FIG. 5.
Binding between Vif and HIV-1 RNA analyzed by coimmunoprecipitation assay. Fraction 8 in the -RNase A panel in Fig. 4 was mixed with anti-Vif antibody or preimmune rabbit serum, as well as protein A-conjugated Sepharose beads. The bead-associated HIV-1 unspliced RNA and host cell γ-actin RNA were then detected by RT-PCR, while the bead-associated HIV-1 Gag protein was detected with an anti-p7 antibody via Western blotting.
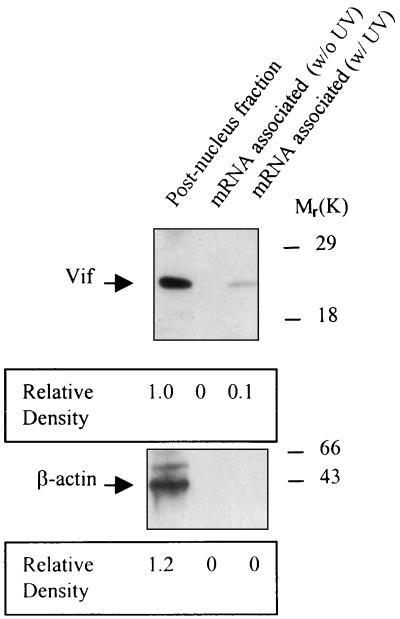
To further verify that Vif is an mRNA-associated protein, an in vivo UV light-induced cross-linking assay was used (1). After UV irradiation, mRNA and associated proteins in the postnuclear fraction of HIV-1-infected H9 T cells were purified by oligo(dT)-cellulose chromatography. As shown in Fig. 6, Vif but not β-actin protein was copurified with mRNA. These in vivo data demonstrated that Vif can specifically and directly coat HIV-1 RNA in the cytoplasm of HIV-1-infected cells to form an mRNP complex. However, as the complex was still sensitive to RNase A digestion (Fig. 4), Vif protein may not completely coat HIV-1 RNA.
FIG. 6.
Interaction between Vif and mRNA analyzed by in vivo UV cross-linking assay. HIV-1NL4-3-infected H9 cells were washed and irradiated with UV light. The cells were lysed, and mRNA and associated proteins were then isolated from the postnuclear fraction by oligo(dT)-cellulose chromatography. After digestion with RNase A, the mRNA-associated proteins were detected via Western blotting with either anti-Vif or anti-β-actin antibody.
Vif-RNA binding can be displaced by Gag-RNA binding in vitro.
It has been demonstrated that Vif binds to the NCp7 domain of the Gag protein and colocalizes with Gag protein in virus-producing cells (9, 39, 52; data not shown). However, Vif does not incorporate into virions specifically (13, 23, 35, 55). The molar ratio of Vif to Gag in HIV-1 infected cells is 1:1.7, while the ratio in the purified virions is 1:100 (55). Further, as genomic RNA in the cytoplasm will be packaged into virions, why would these Vif proteins that coat the genomic RNA and bind to Gag precursors not specifically be packaged into virions with genomic RNA? To study the interactions between Gag, RNA, and Vif, we first examined the effect of free RNA on the binding activities of 35S-labeled Vif to GST-Gag protein. In the presence of free ribonucleotide homopolymers [poly(G)] or in vitro-synthesized HIV-1 RNA, the binding between GST-Gag and Vif, and the binding between GST-NCp7 and Vif, significantly decreased, suggesting that the binding between Vif and Gag can be inhibited by viral RNA [poly(G)] rather than γ-actin RNA (Fig. 7a and data not shown). Conversely, the binding between Vif and poly(G) was inhibited by GST-Gag and GST-NCp7, respectively, but not GST, indicating that the binding affinity of Vif to RNA will significantly decrease in the presence of HIV-1 Gag protein (Fig. 7b). This phenomenon was further demonstrated by gel mobility shift assays. When the HIV-1 riboprobe (5A or 7A) was mixed with both GST-Vif and GST-Gag, the binding of GST-Vif to HIV-1 RNA significantly decreased in the presence of GST-Gag. However, the binding between GST-Gag and HIV-1 RNA remained the same (Fig. 7c and d). It is notable that the binding ability of Vif to these two HIV-1 riboprobes is higher than that of Gag to RNA, as shown with a filter binding assay (Table 1). As such, the decreased binding of Vif to RNA in the presence of Gag is unlikely to be due to competitive inhibition.
FIG. 7.
Interaction between Vif, RNA, and Gag in vitro. (a) In vitro-translated, 35S-labeled Vif protein was subjected to binding with GST-Gag-conjugated agarose beads in the presence of free poly(G), HIV-1 7A RNA, or γ-actin RNA. After washing, the bead-associated Vif was fractionated by SDS-PAGE. (b) In vitro-translated, 35S-labeled Vif protein was subjected to binding with poly(G)-conjugated agarose beads or in the presence of GST, GST-Gag, and GST-p7. The bead-associated Vif was fractionated by SDS-PAGE. (c and d) HIV-1 riboprobes 5A (c) and 7A (d) were subjected to binding with various GST and GST fusion proteins (10 pmol of each), respectively. The mixtures were then fractionated in a native 5% TBE gel.
N terminus of Vif protein contains RNA binding sites, which are required for Vif function in the HIV-1 life cycle.
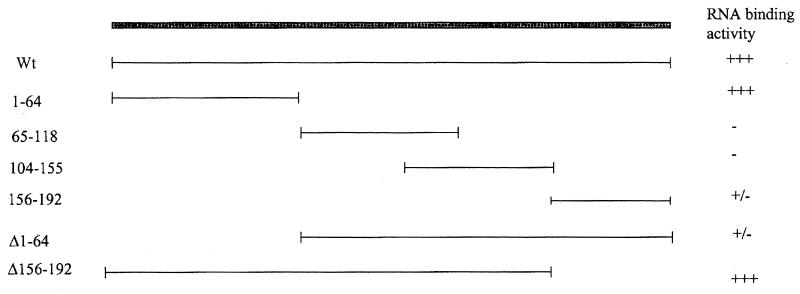
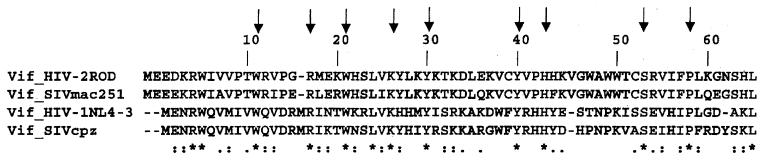
To analyze the RNA binding domain(s), Vif protein fragments were generated and labeled with [35S]methionine by in vitro translation. Via in vitro binding with poly(G)-conjugated beads, we found that the fragment from amino acids 1 to 64, which is located in the N terminus of the Vif, contains a strong RNA binding activity. Deletion of this region from Vif protein significantly decreased the RNA binding activity. The C terminus of Vif harboring many positive charged amino acids also had weak RNA binding activity (Fig. 8). It has been demonstrated that Vif proteins from several strains of HIV-1, HIV-2, and SIV are composed of heterogeneous sequences, yet they can functionally complement each other (46, 57). Multiple sequence alignments of the N termini of Vif proteins from several primate lentivirus strains can easily identify the conserved amino acids (Fig. 9). HIV-1 vif mutants at these amino acids, conserved in various strains of primate lentiviruses, were then generated by site-directed mutagenesis. In vitro binding demonstrated that Vif mutants at W11, Y30, and Y40 significantly decreased RNA binding activity (Table 2). It is notable that no identified RNA binding domains are similar to the sequence of the N terminus of Vif (11). As such, the molecular mechanism of Vif-RNA binding remains to be further clarified.
FIG. 8.
Localization of RNA binding sites in the Vif protein. The vif gene was truncated into several fragments and then cloned into the pCITE-4a vector. The 35S-labeled Vif fragments were then in vitro translated and allowed to bind with poly(G)-conjugated agarose beads. The bead-associated Vif fragments were fractionated by SDS-PAGE. Wt, wild type.
FIG. 9.
Alignment of the sequences from the N termini of various Vif proteins. ∗, conserved in all strains; :, not completely conserved but highly homologous; ., with some homology. Arrows indicate the amino acids that were selected for site-directed mutagenesis.
TABLE 2.
Effects of Vif mutations in the N terminus upon RNA binding activity and viral infectivity
| Vif construct | RNA binding activity (%)
|
Viral infectivityc | |
|---|---|---|---|
| Poly(G)a | HIV-1 RNA (7A)b | ||
| Wild type | 100 | 100 | ++ |
| W11A | 20 | 45 | − |
| R17A | 75 | NDd | ND |
| W21A | 68 | ND | + |
| K26A | 59 | ND | − |
| Y30A | 16 | 20 | − |
| Y40A | 8 | 28 | − |
| H43A, Y44A | 55 | ND | ND |
| S53A | 98 | ND | ND |
| P58A | 93 | 91 | ++ |
The in vitro-translated Vif mutants were allowed to bind with poly(G)-conjugated agarose beads, and binding activity was calculated by comparison with the input sample labeled with [35S]methionine. The binding between wild-type Vif and poly(G) was set at 100%. The others were calculated thereafter.
Vif and its mutants were expressed as GST fusion proteins from bacterial cells and isolated via glutathione-coated agarose beads. The purified proteins (10 pmol) were then subjected to binding with HIV-1 riboprobe 7A (105 cpm). The mixture was placed onto a nitrocellulose filter, and the TCA-insoluble radioactivity remaining on the filter was quantitated. The binding between wild-type Vif and HIV-1 RNA was set at 100%. The others were calculated thereafter. In most cases, the data reflect at least three independent experiments.
To examine the effects on HIV-1 infectivity in H9 T cells utilizing these Vif mutants, the mutants were cloned into a retroviral vector (pSLX-CMV-RRE). The recombinant MLVs were generated and allowed to infect H9 T cells. The G418-resistant H9 T cells were then infected with HIV-1NL4-3Δvif. The culture was considered to be positive (+) or strongly positive (++) when the quantity of HIV-1 p24 antigen in the supernatant was greater than 60 or 240 pg/ml on day 11 postinfection.
ND, not determined.
To investigate biological significance of Vif-RNA binding, the Vif mutants that lose the ability for in vitro Vif-RNA binding were constructed into an MLV vector containing an RRE. The recombinant retroviruses were generated from PA317 cells and allowed to infect H9 T cells. After G418 screening, the H9 T cells that harbored Vif mutants were infected with HIV-1NL4-3Δvif. The growth of HIV-1NL4-3Δvif was quite inefficient in these Vif mutants, indicating that the RNA binding activity of Vif is required for Vif function in the viral life cycle (Table 2).
DISCUSSION
In this work, we have studied Vif-RNA interactions with various in vitro assays including homopolymer binding, gel shifting, filter binding, and UV cross-linking plus various in vivo assays including copurification by rate-zonal sedimentation plus RNase digestion, UV cross-linking, and immunocoprecipitation. We demonstrated that in vitro (i) Vif preferentially binds to poly(G) rather than binding equally to all homopolymers (Fig. 1), indicating that Vif-RNA binding is sequence specific; (ii) GST-Vif is able to bind to HIV-1 RNA rather than γ-actin RNA in gel shift assays (Fig. 2); (iii) the binding between Vif and HIV-1 RNA cannot be inhibited by tRNA or poly(dI-dC) in a high quantity (750 ng) but can be inhibited by unlabeled HIV-1 RNA in a concentration-dependent way (Fig. 2 and Table 1); and (iv) UV cross-linking indicates that HIV-1 riboprobes, but not an γ-actin riboprobe, can bind to GST-Vif protein (Fig. 3). Further, we demonstrated that in vivo (i) Vif protein is copurified with an RNase-sensitive particle which contain HIV-1 unspliced RNA rather than spliced RNA and γ-actin RNA; and (ii) HIV-1 unspliced RNA, but not γ-actin RNA, was coimmunoprecipitated with Vif protein.
Overall, the present data indicated that Vif strongly and specifically bound to HIV-1 RNA. The specific sequence for Vif-RNA binding remains to be determined. It could be an RNA-specific structure, such as trans-activation-responsive (TAR) element for Tat and Rev-responsive element (RRE) for Rev. It could also be certain short sequences in HIV-1 RNA which can specifically bind to the Vif protein. These sequences may be selected by systematic evolution of ligands by exponential enrichment assay, as described previously for the hnRNP proteins (12, 63). It is well known that many hnRNP and mRNP proteins specifically bind to certain mRNA molecules by binding to select short sequences (12, 25, 26). Vif binds to viral RNA through its N terminus rather than the positively charged amino acid-enriched C terminus, indicating that this interaction does not simply depend on the clusters of positively charged amino acids in Vif (Fig. 8). Importantly, we have preliminarily demonstrated that these Vif mutants that lose RNA binding activity in vitro do not support HIV-1 vif− replication in nonpermissive cells, suggesting that the RNA binding ability of Vif is important for its function in the virus-producing cells (Table 2). As such, these data strongly suggest that Vif-RNA binding is not trivial and should play a role in the viral life cycle.
Our data have also demonstrated that HIV-1, a member of the lentivirus family, can encode an RNA-binding protein, Vif, to coat temporarily and specifically its genomic RNA, and form an mRNP complex in the cytoplasm of the virus-producing cells (Fig. 4 to 6). It has been recognized that posttranscriptional regulation of mRNA plays an important role in gene expression. The eukaryotic mRNAs are associated with select cellular proteins to form a particle (mRNP), which mainly exists in the cytoplasm. The proteins in the mRNP complex can regulate the translation, degradation, and localization of mRNA in the cytoplasm (25, 26, 40). Some mRNA-associated proteins are able to shuttle between the nucleus and the cytoplasm (2). Two major core proteins of mRNP, p50 and p70, have been found in the cytoplasm of different somatic mammalian cells (25). p50 has been characterized as a member of the Y-box-binding transcription factor family of proteins by both high structural homology and ability to bind specifically the Y-box sequence in double-stranded DNA. Further, this protein can melt the RNA secondary structure, change the RNA conformation, and promote the initiation of protein biosynthesis in vitro (27, 28). Other RNA-binding proteins, such as hnRNP A1, La autoantigen, and pyrimidine tract-binding protein, can also render translation cap-dependent in rabbit reticulocyte lysate (27). However, Xenopus Y-box protein FRGY2, an mRNA-binding protein, can inhibit translation (43). Of importance, it has been reported that protein N of vescular stomatitis virus, which is extensively packaged into virions, can bind to viral RNA to form an mRNP complex and inhibit ribosomal function in the host cell (1, 48).
The regulation of retroviral RNA may be more complicated than that of host mRNAs. Similar to host mRNAs, unspliced retroviral RNAs traffic from nucleus to cytoplasm, serve as the template in ribosomes for the synthesis of their structural proteins, and undergo degradation in the cytoplasm. Moreover, unspliced retroviral RNA is selectively packaged into virions for transmission through the packaging signal in its 5′ region. So far, however, little is known about this complicated regulatory process. Whether a cellular mRNA-associated protein(s) is involved in this process remains unknown. An early study showed that the nucleocapsid protein of Rous sarcoma virus might inhibit protein synthesis in vitro (21). Moreover, it has been proposed that dimerization of retroviral RNA plays a role in selecting packaging rather than translation processes (6). Our work demonstrated that in addition to Gag and Gag-Pol structural proteins, lentiviruses could encode another protein to potentially regulate this complicated process. Vif in the mRNP complex of viral RNA may play multiple roles for the intracellular trafficking and packaging of HIV-1 genomic RNA. It may maintain the HIV-1 RNA in a properly folded structure or prevent improper engagement with ribosomes. Our recent data indicated that Vif may have RNA chaperone activity whereby it performs these functions (37; unpublished data).
Conversely, Vif in mRNP complexes may prevent the interactions of HIV-1 RNA with various endogenous inhibitors, as described by others (42, 53). For instance, it may prevent activation of interferon or mRNA degradation systems, or it may prevent the engagement with the Gag proteins of endogenous retroviruses. In these circumstances, there may be no significant component difference of 40S mRNP complexes in permissive or nonpermissive cells. As such, a comparative analysis for the components of mRNP complexes in permissive or nonpermissive cells may be more informative, but not essential, for understanding the physiological significance of Vif-RNA binding.
Moreover, an interesting phenomenon has been demonstrated in this study. In the presence of Gag precursors, the ability of Vif to bind to RNA decreases. However, in contrast to decreased Vif-RNA binding, RNA will continue binding to Gag precursors (Fig. 7c and d). The precise mechanism of this displacement is unknown, and more in vivo evidence is required to confirm this phenomenon. The characteristics of the Vif, Gag, and RNA interactions are compatible with the fact that Vif does not specifically incorporate into virions, even though it strongly binds to viral RNA or Gag protein (Fig. 1, 2, and 7; Table 1) (9, 39). Based on these observations, we suggest that the binding between Vif and RNA in the mRNP complex could decrease and Gag-RNA binding would dominate at the site of aggregated Gag precursors (budding complex). If this is the case, Vif could be directly involved in the viral RNA folding and packaging process.
Retroviral RNA packaging is a complicated process composed of multiple steps: recognition, selection, dimerization, folding, and condensation. The viral RNA packaging signal (ψ site) and dimerization signal are both located in the same region of the 5′ terminus of retroviral RNA, separated by a short distance (50). Deletion of this region will significantly affect the RNA packaging into HIV-1 virions and the virion morphology (14, 20, 41). The nucleocapsid domains of Gag precursors, which harbors two zinc finger structures, directly binds to viral RNA in the packaging process. The first zinc finger structure and its flanking positively charged amino acids play a key role in the RNA binding activity (4, 20). It has been shown that Gag precursors and NCp7 of HIV-1 can specifically bind to HIV-1 RNA at the ψ site in vitro (5, 15, 19). However, several questions remain to be further investigated. How many Gag monomers are involved in the binding to the RNA packaging signal in viral unspliced RNA? Besides the binding between NCp7 and viral RNA at the ψ site, what is the sequential process for the condensation and folding of the rest of viral genomic RNA? Is any host cellular factor involved in this process? In recent studies, a double-stranded RNA-binding protein, Staufen, has been found within HIV-1 virions. Its incorporation into HIV-1 virions is dependent on genomic RNA packaging (45). This result suggests that RNA packaging could be quite complicated process and may not be facilitated only by Gag protein.
Based on our data, we propose that Vif may play roles in the lentiviral RNA folding and packaging process. Vif proteins in mRNP complexes of viral RNA could initially bind to NCp7 domains of the Gag precursor to mediate the engagement of HIV-1 genomic RNA to HIV-1 Gag precursors. As the entire viral RNA molecule starts to bind to the NCp7 domains of the Gag precursor, the affinity of Vif to RNA and Vif to Gag will decrease and Vif protein will gradually dissociate from this budding complex composed of Gag, Gag-Pol, and genomic RNA. This displacement process may guide the proper folding and condensation of viral RNA during packaging. If Vif is not expressed in the nonpermissive cells, HIV-1 genomic RNA will still be packaged into virions through the strong binding between the ψ site of viral RNA and certain NCp7 domains of Gag precursors, as shown by in vitro studies (5, 15, 19). However, the genomic viral RNA, which is not coated by Vif proteins, may have been altered or even damaged by the hostile environment of the cytoplasm, or the interaction between genomic RNA and the NCp7 domain of Gag precursor, and thus the folding and condensation of genomic RNA would proceed improperly. As a result, a virion generated from such a producing cell will have morphologic alterations and not complete reverse transcription, as previously demonstrated for HIV-1 lacking Vif from nonpermissive cell types (8, 10, 38). Further studies are required to confirm these hypotheses.
In summary, our studies demonstrate that Vif protein is an RNA-binding protein and binds to HIV-1 genomic RNA to form an mRNP complex in the cytoplasm of virus-producing cells. Vif in this complex may assist HIV-1 RNA to maintain proper folding and trafficking in the cytoplasm, or it may prevent cellular inhibitors from altering HIV-1 RNA. Further, Vif in this complex may mediate HIV-1 genomic RNA in association with the NCp7 domains of the HIV-1 Gag precursor. It remains to be clarified whether the defect of the vif gene will result in a detectable impairment in the RNA packaging/folding process. Comparative analysis of the nucleocapsid complex in the vif− virions, generated from permissive cells or nonpermissive cells, will further prove our hypothesis. Conversely, if an endogenous inhibitor exists in nonpermissive cells, what is the molecular mechanism for its impairment of genomic RNA, and how does Vif protein prevent it? Further, it is also of interest to dissect the molecular mechanisms of Vif-RNA binding and its regulation. We believe that understanding these questions will lead to further analysis of other molecular mechanisms involving Vif protein and thus generate a novel accessory protein target for rational design of HIV-1 therapeutics.
ACKNOWLEDGMENTS
We gratefully acknowledge D. H. Gabuzda for providing the anti-Vif antibody; R. C. Desrosiers for providing the p197-1 clone (5′ half of pNL4-3 with vif deletion) through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH; and L. Henderson for providing the anti-p7 antibody.
This work was supported by Thomas Jefferson University funds (H.Z.).
REFERENCES
- 1.Adam S A, Choi Y D, Dreyfuss G. Interaction of mRNA with proteins in vesicular stomatitis virus-infected cells. J Virol. 1986;57:614–622. doi: 10.1128/jvi.57.2.614-622.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonina E, Stauber R, Pavlakis G N. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J Biol Chem. 1998;273:13015–13021. doi: 10.1074/jbc.273.21.13015. [DOI] [PubMed] [Google Scholar]
- 3.Aldrovandi G M, Zack J A. Replication and pathogenicity of human immunodeficiency virus type 1 accessory gene mutants in SCID-hu mice. J Virol. 1996;70:1505–1511. doi: 10.1128/jvi.70.3.1505-1511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 5.Berkowitz R D, Luban J, Goff S P. Specific binding of human immunodeficiency virus type 1 Gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bieth E, Gabus C, Darlix J L. A study of the dimer formation of Rous sarcoma virus RNA and of its effect on viral protein synthesis in vitro. Nucleic Acids Res. 1990;18:119–127. doi: 10.1093/nar/18.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanc D, Patience C, Schulz T F, Weiss R, Spire B. Transcomplementation of VIF- HIV-1 mutants in CEM cells suggests that VIF affects late steps of the viral life cycle. Virology. 1993;193:186–192. doi: 10.1006/viro.1993.1114. [DOI] [PubMed] [Google Scholar]
- 8.Borman A M, Quillent C, Charneau P, Dauguet C, Clavel F. Human immunodeficiency virus type 1 Vif− mutant particles from restrictive cells: role of Vif in correct particle assembly and infectivity. J Virol. 1995;69:2058–2067. doi: 10.1128/jvi.69.4.2058-2067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouyac M, Courcoul M, Bertoia G, Baudat Y, Gabuzda D, Blanc D, Chazal N, Boulanger P, Sire J, Vigne R, Spire B. Human immunodeficiency virus type 1 Vif protein binds to the Pr55Gag precursor. J Virol. 1997;71:9358–9365. doi: 10.1128/jvi.71.12.9358-9365.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouyac M, Rey F, Nascimbeni M, Courcoul M, Sire J, Blanc D, Clavel F, Vigne R, Spire B. Phenotypically Vif− human immunodeficiency virus type 1 is produced by chronically infected restrictive cells. J Virol. 1997;71:2473–2477. doi: 10.1128/jvi.71.3.2473-2477.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 12.Burd C G, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camaur D, Trono D. Characterization of human immunodeficiency virus type 1 Vif particle incorporation. J Virol. 1996;70:6106–6111. doi: 10.1128/jvi.70.9.6106-6111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clavel F, Orenstein J M. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J Virol. 1990;64:5230–5234. doi: 10.1128/jvi.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coffin J. Structure of the retroviral genome. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. pp. 261–369. [Google Scholar]
- 17.Courcoul M, Patience C, Rey F, Blanc D, Harmache A, Sire J, Vigne R, Spire B. Peripheral blood mononuclear cells produce normal amounts of defective Vif− human immunodeficiency virus type 1 particles which are restricted for the preretrotranscription steps. J Virol. 1995;69:2068–2074. doi: 10.1128/jvi.69.4.2068-2074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Aloja P, Olivetta E, Bona R, Nappi F, Pedacchia D, Pugliese K, Ferrari G, Verani P, Federico M. gag, vif, and nef genes contribute to the homologous viral interference induced by a nonproducer human immunodeficiency virus type 1 (HIV-1) variant: identification of novel HIV-1-inhibiting viral protein mutants. J Virol. 1998;72:4308–4319. doi: 10.1128/jvi.72.5.4308-4319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darlix J L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 21.Darlix J L, Spahr P F. Binding sites of viral protein P19 onto Rous sarcoma virus RNA and possible controls of viral functions. J Mol Biol. 1982;160:147–161. doi: 10.1016/0022-2836(82)90172-3. [DOI] [PubMed] [Google Scholar]
- 22.Desrosiers R C, Lifson J D, Gibbs J S, Czajak S C, Howe A Y, Arthur L O, Johnson R P. Identification of highly attenuated mutants of simian immunodeficiency virus. J Virol. 1998;72:1431–1437. doi: 10.1128/jvi.72.2.1431-1437.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dettenhofer M, Yu X F. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J Virol. 1999;73:1460–1467. doi: 10.1128/jvi.73.2.1460-1467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dornadula G, Yang S, Pomerantz R J, Zhang H. Partial rescue of the Vif-negative phenotype of mutant human immunodeficiency virus type 1 strains from nonpermissive cells by intravirion reverse transcription. J Virol. 2000;74:2594–2602. doi: 10.1128/jvi.74.6.2594-2602.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dreyfuss G. Structure and function of nuclear and cytoplasmic ribonucleoprotein particles. Annu Rev Cell Biol. 1986;2:459–498. doi: 10.1146/annurev.cb.02.110186.002331. [DOI] [PubMed] [Google Scholar]
- 26.Dreyfuss G, Matunis M J, Pinol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 27.Evdokimova V M, Kovrigina E A, Nashchekin D V, Davydova E K, Hershey J W, Ovchinnikov L P. The major core protein of messenger ribonucleoprotein particles (p50) promotes initiation of protein biosynthesis in vitro. J Biol Chem. 1998;273:3574–3581. doi: 10.1074/jbc.273.6.3574. [DOI] [PubMed] [Google Scholar]
- 28.Evdokimova V M, Wei C L, Sitikov A S, Simonenko P N, Lazarev O A, Vasilenko K S, Ustinov V A, Hershey J W, Ovchinnikov L P. The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. J Biol Chem. 1995;270:3186–3192. doi: 10.1074/jbc.270.7.3186. [DOI] [PubMed] [Google Scholar]
- 29.Fisher A G, Ensoli B, Ivanoff L, Chamberlain M, Petteway S, Ratner L, Gallo R C, Wong-Staal F. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science. 1987;237:888–893. doi: 10.1126/science.3497453. [DOI] [PubMed] [Google Scholar]
- 30.Fisher R J, Rein A, Fivash M, Urbaneja M A, Casas-Finet J R, Medaglia M, Henderson L E. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fouchier R A, Simon J H, Jaffe A B, Malim M H. Human immunodeficiency virus type 1 Vif does not influence expression or virion incorporation of gag-, pol-, and env-encoded proteins. J Virol. 1996;70:8263–8269. doi: 10.1128/jvi.70.12.8263-8269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gabuzda D H, Lawrence K, Langhoff E, Terwilliger E, Dorfman T, Haseltine W A, Sodroski J. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabuzda D H, Li H, Lawrence K, Vasir B S, Crawford K, Langhoff E. Essential role of vif in establishing productive HIV-1 infection in peripheral blood T lymphocytes and monocyte/macrophages. J Acquir Immune Defic Syndr. 1994;7:908–915. [PubMed] [Google Scholar]
- 34.Gibbs J S, Regier D A, Desrosiers R C. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res Hum Retroviruses. 1994;10:343–350. doi: 10.1089/aid.1994.10.343. [DOI] [PubMed] [Google Scholar]
- 35.Goncalves J, Jallepalli P, Gabuzda D H. Subcellular localization of the Vif protein of human immunodeficiency virus type 1. J Virol. 1994;68:704–712. doi: 10.1128/jvi.68.2.704-712.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goncalves J, Korin Y, Zack J, Gabuzda D. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J Virol. 1996;70:8701–8709. doi: 10.1128/jvi.70.12.8701-8709.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 38.Hoglund S, Ohagen A, Lawrence K, Gabuzda D. Role of vif during packing of the core of HIV-1. Virology. 1994;201:349–355. doi: 10.1006/viro.1994.1300. [DOI] [PubMed] [Google Scholar]
- 39.Huvent I, Hong S S, Fournier C, Gay B, Tournier J, Carriere C, Courcoul M, Vigne R, Spire B, Boulanger P. Interaction and co-encapsidation of human immunodeficiency virus type 1 Gag and Vif recombinant proteins. J Gen Virol. 1998;79:1069–1081. doi: 10.1099/0022-1317-79-5-1069. [DOI] [PubMed] [Google Scholar]
- 40.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 41.Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madani N, Kabat D. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J Virol. 1998;72:10251–10255. doi: 10.1128/jvi.72.12.10251-10255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsumoto K, Meric F, Wolffe A P. Translational repression dependent on the interaction of the Xenopus Y-box protein FRGY2 with mRNA. Role of the cold shock domain, tail domain, and selective RNA sequence recognition. J Biol Chem. 1996;271:22706–22712. doi: 10.1074/jbc.271.37.22706. [DOI] [PubMed] [Google Scholar]
- 44.McEwen C R. Tables for estimating sedimentation through linear concentration gradients of sucrose solution. Anal Biochem. 1967;20:114–149. doi: 10.1016/0003-2697(67)90271-0. [DOI] [PubMed] [Google Scholar]
- 45.Mouland A J, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen E A. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J Virol. 2000;74:5441–5451. doi: 10.1128/jvi.74.12.5441-5451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reddy T R, Kraus G, Yamada O, Looney D J, Suhasini M, Wong-Staal F. Comparative analyses of human immunodeficiency virus type 1 (HIV-1) and HIV-2 Vif mutants. J Virol. 1995;69:3549–3553. doi: 10.1128/jvi.69.6.3549-3553.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reed R, Griffith J, Maniatis T. Purification and visualization of native spliceosomes. Cell. 1988;53:949–961. doi: 10.1016/s0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 48.Rosen C A, Siekierka J, Ennis H L, Cohen P S. Inhibition of protein synthesis in vesicular stomatitis virus infected Chinese hamster ovary cells: role of virus mRNA-ribonucleoprotein particle. Biochemistry. 1984;23:2407–2411. doi: 10.1021/bi00306a014. [DOI] [PubMed] [Google Scholar]
- 49.Seshamma T, Bagasra O, Oakes J W, Pomerantz R J. A quantitative reverse transcriptase-polymerase chain reaction for HIV-1-specific RNA species. J Virol Methods. 1992;40:331–345. doi: 10.1016/0166-0934(92)90091-q. [DOI] [PubMed] [Google Scholar]
- 50.Shen N, Jette L, Liang C, Wainberg M A, Laughrea M. Impact of human immunodeficiency virus type 1 RNA dimerization on viral infectivity and of stem-loop B on RNA dimerization and reverse transcription and dissociation of dimerization from packaging. J Virol. 2000;74:5729–5735. doi: 10.1128/jvi.74.12.5729-5735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon J H, Carpenter E A, Fouchier R A, Malim M H. Vif and the p55Gag polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J Virol. 1999;73:2667–2674. doi: 10.1128/jvi.73.4.2667-2674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simon J H, Fouchier R A, Southerling T E, Guerra C B, Grant C K, Malim M H. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J Virol. 1997;71:5259–5267. doi: 10.1128/jvi.71.7.5259-5267.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon J H, Gaddis N C, Fouchier R A, Malim M H. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat Med. 1998;4:1397–1400. doi: 10.1038/3987. [DOI] [PubMed] [Google Scholar]
- 54.Simon J H, Malim M H. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J Virol. 1996;70:5297–5305. doi: 10.1128/jvi.70.8.5297-5305.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simon J H, Miller D L, Fouchier R A, Malim M H. Virion incorporation of human immunodeficiency virus type-1 Vif is determined by intracellular expression level and may not be necessary for function. Virology. 1998;248:182–187. doi: 10.1006/viro.1998.9296. [DOI] [PubMed] [Google Scholar]
- 56.Simon J H, Miller D L, Fouchier R A, Soares M A, Peden K W, Malim M H. The regulation of primate immunodeficiency virus infectivity by Vif is cell species restricted: a role for Vif in determining virus host range and cross-species transmission. EMBO J. 1998;17:1259–1267. doi: 10.1093/emboj/17.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon J H, Southerling T E, Peterson J C, Meyer B E, Malim M H. Complementation of vif-defective human immunodeficiency virus type 1 by primate, but not nonprimate, lentivirus vif genes. J Virol. 1995;69:4166–4172. doi: 10.1128/jvi.69.7.4166-4172.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sova P, van Ranst M, Gupta P, Balachandran R, Chao W, Itescu S, McKinley G, Volsky D J. Conservation of an intact human immunodeficiency virus type 1 vif gene in vitro and in vivo. J Virol. 1995;69:2557–2564. doi: 10.1128/jvi.69.4.2557-2564.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sova P, Volsky D J. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. J Virol. 1993;67:6322–6326. doi: 10.1128/jvi.67.10.6322-6326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strebel K, Daugherty D, Clouse K, Cohen D, Folks T, Martin M A. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature. 1987;328:728–730. doi: 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- 61.Swanson M S, Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trono D. HIV accessory proteins: leading roles for the supporting cast. Cell. 1995;82:189–192. doi: 10.1016/0092-8674(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 63.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 64.von Schwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 66.Wieland U, Hartmann J, Suhr H, Salzberger B, Eggers H J, Kuhn J E. In vivo genetic variability of the HIV-1 vif gene. Virology. 1994;203:43–51. doi: 10.1006/viro.1994.1453. [DOI] [PubMed] [Google Scholar]
- 67.Wieland U, Seelhoff A, Hofmann A, Kuhn J E, Eggers H J, Mugyenyi P, Schwander S. Diversity of the vif gene of human immunodeficiency virus type 1 in Uganda. J Gen Virol. 1997;78:393–400. doi: 10.1099/0022-1317-78-2-393. [DOI] [PubMed] [Google Scholar]
- 68.Zhang H, Dornadula G, Alur P, Laughlin M A, Pomerantz R J. Amphipathic domains in the C terminus of the transmembrane protein (gp41) permeabilize HIV-1 virions: a molecular mechanism underlying natural endogenous reverse transcription. Proc Natl Acad Sci USA. 1996;93:12519–12524. doi: 10.1073/pnas.93.22.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang H, Dornadula G, Pomerantz R J. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenvironments: an important stage for viral infection of nondividing cells. J Virol. 1996;70:2809–2824. doi: 10.1128/jvi.70.5.2809-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang H, Zhang Y, Spicer T P, Abbott L Z, Abbott M, Poiesz B J. Reverse transcription takes place within extracellular HIV-1 virions: potential biological significance. AIDS Res Hum Retroviruses. 1993;9:1287–1296. doi: 10.1089/aid.1993.9.1287. [DOI] [PubMed] [Google Scholar]