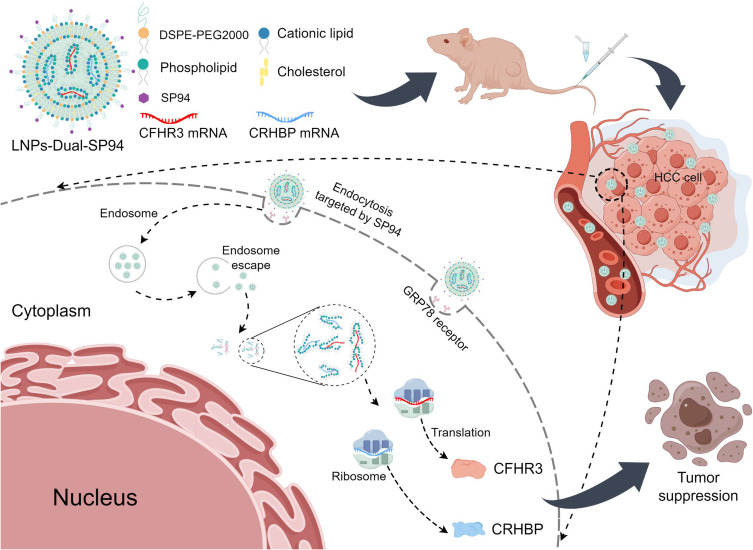
Abstract
Purpose
Hepatocellular carcinoma (HCC) is a deadly disease requiring the identification of new therapeutic targets and strategies.
Methods
This study identified genes linked to HCC progression via differential analysis. Key genes were identified through univariate and multivariate Cox regression analysis. The biological effects of co-expressed CRHBP and CFHR3 were evaluated in vitro. mRNAs encoding CRHBP and CFHR3 were encapsulated in lipid nanoparticles (LNPs), with the addition of SP94 peptide on the LNPs surface to enhance targeting. The therapeutic efficacy of dual-mRNA LNPs was evaluated in HCC cells and mouse models.
Results
CRHBP and CFHR3 were closely associated with HCC progression. Low expression of CRHBP (P < 0.01, HR = 1.931 [1.174–3.175]) and CFHR3 (P < 0.05, HR = 1.755 [1.066–2.890]) was identified as a poor prognostic factor for HCC. The risk score model combining CRHBP and CFHR3 demonstrated superior predictive power (P < 0.001, HR = 2.935 [1.768–4.872]). Co-expression of CRHBP and CFHR3 significantly inhibited the malignant biological functions of HCC cells. Treatment with SP94 peptide-modified dual-mRNA LNPs markedly suppressed HCC tumor growth and exhibited excellent biocompatibility and safety.
Conclusion
Our study proposes a dual-targeted therapeutic strategy for HCC, which may represent a promising treatment approach.
Keywords: CRHBP, CFHR3, Lipid nanoparticles, mRNA
Graphical Abstract
Introduction
Hepatocellular carcinoma (HCC) constitutes about 80% of primary liver cancers, with surgical resection as the main treatment, augmented by radiotherapy, targeted therapy, and immunotherapy.1–3 However, the prognosis for advanced HCC is poor due to its insidious onset and high recurrence and metastasis rates.4 Therefore, potential biomarkers associated with HCC progression must be further explored to develop new effective therapies.
As an emerging therapeutic tool, mRNA therapy has shown great potential, especially in the successful development of the COVID-19 mRNA vaccine.5,6 Its advantages include the ability to rapidly develop and formulate personalized therapeutic regimens for tumors, transient expression and non-integration, and the ability to achieve synergistic effects through the simultaneous delivery of multiple mRNAs.7 Despite these benefits, the application of mRNA therapeutics in cancer treatment faces several challenges, including poor stability in vivo.8 More effective delivery systems and targeting strategies are essential to enhance their efficacy.
Lipid nanoparticles (LNPs) are nanoscale particles composed of lipid molecules, which are commonly used to deliver nucleic acid drugs such as mRNA and protect them from degradation by enzymes in vivo.9 Due to the high delivery efficiency, stability and mass production of LNPs, their application in cancer therapy is promising. However, LNPs delivery of mRNA still has shortcomings, such as limited targeting and low delivery efficiency in certain tissues.10 In this regard, optimization of the physicochemical properties and surface ligands of LNPs could improve their aggregation in tumors, thereby enhancing the therapeutic effect and reducing the damage to normal tissues.11
Corticotropin-releasing hormone-binding protein (CRHBP) was a key component of the hypothalamic-pituitary-adrenal axis,12 primarily involved in cellular metabolism and stress responses by binding and regulating corticotropin-releasing hormone (CRH).13,14 Studies demonstrated that CRHBP inhibited the proliferation, metastasis, and angiogenesis of HCC by downregulating cyclin B2 expression and disrupting the cyclin B2-CDK1 complex.15 Complement factor H-related protein 3 (CFHR3) was an important member of the human factor H protein family, bound to heparin and involved in complement regulation.16 In HCC, CFHR3 suppressed cell proliferation and metastasis by promoting STAT3 phosphorylation and inhibiting p53 expression through miR-590-3p mediation, thereby exerting significant tumor-suppressive effects.17 Moreover, the low expression of CRHBP and CFHR3 was closely associated with HCC progression.18 Therefore, CRHBP and CFHR3 highlighted their potential as prognostic biomarkers and therapeutic targets for HCC.
This study identified CRHBP and CFHR3 as novel biomarkers linked to HCC through comprehensive patient cohort analysis. Subsequently, a dual-gene risk score model was developed, demonstrating the potential for predicting the progression of HCC. Co-amplified expression of these two genes exerts a synergistic effect, more effectively inhibiting the malignant progression of HCC cells. We designed mRNA-LNPs targeting HCC cells and evaluated their therapeutic efficacy in vitro and in vivo. These findings offered a potential new therapeutic strategy for HCC treatment.
Material and Methods
Data Acquisition and Preprocessing
Transcriptomic data and corresponding clinical information for HCC patients were sourced from The Cancer Genome Atlas (TCGA). Samples with complete clinical data were retained, with 343 tumor tissues and 50 normal tissues included in the study. Gene expression data were converted to Transcripts Per Million (TPM) values and log-transformed for analysis.
Identification and Evaluation of Genes Associated with HCC Progression
The Wilcoxon test identified differentially expressed genes (DEGs) linked to HCC progression. Genes were filtered using |logFC| > 1 and an adjusted P-value < 0.05. HCC progression-related genes were defined as the intersecting DEGs between HCC and normal tissues, as well as between advanced and early-stage HCC tissues. Cox proportional hazards analyses, both univariate and multivariate, were used to identify risk factors for overall survival (OS) in HCC patients. The LASSO Cox proportional hazards regression model was developed using the glmnet package in R. The model was developed with five-fold cross-validation. Receiver Operating Characteristic (ROC) curves assessed the model’s sensitivity and specificity. The survival analysis was further validated using Kaplan-Meier survival curves, stratified by the risk score. Log rank tests were performed to assess the statistical significance of the differences between groups.
Clinical Samples
Between 2014 and 2018, 136 tissue samples were collected from HCC-diagnosed patients at the First Affiliated Hospital of Gannan Medical University. Informed consent was obtained from all participants. Patients undergoing chemotherapy or other treatments before the therapeutic surgery were excluded from the study. The study protocol adhered to the Declaration of Helsinki and was approved by the Ethics Committee of the First Affiliated Hospital of Gannan Medical University.
Immunohistochemistry
Formalin-fixed, paraffin-embedded (FFPE) tissue sections of HCC were deparaffinized and rehydrated. Antigen retrieval was conducted using high-pressure heating in sodium citrate buffer (pH 6.0) or EDTA buffer (pH 9.0). Immunohistochemistry (IHC) was conducted according to standard protocols. Primary antibodies against CFHR3 (1:100, 16583-1-AP, Proteintech), CRHBP (1:100, A6568, ABclonal), and Ki67 (1:5000, HA721115, Huabio) were incubated overnight at 4°C. Staining was visualized using a DAB (3,3’-diaminobenzidine) solution. A pathologist assessed IHC scoring by evaluating the staining intensity and the proportion of stained cells.
Cell Culture
The HCC cell lines (HepG2, Hep3B) and the immortalized human liver epithelial cell line (THLE-2) were sourced from the Chinese Academy of Sciences (Shanghai, China). The HCCLM9 cell line was supplied by the Liver Cancer Institute, Fudan University, Shanghai, China. All cell lines were cultured in DMEM with 10% FBS, 100 μg/mL penicillin, and 100 μg/mL streptomycin. The cells were incubated at 37 °C with 5% CO2.
qRT-PCR
According to the manufacturer’s instructions, total RNA was extracted from the cells using the TransZol reagent (ET101-01-V2, TransGen Biotech). The extracted RNA was promptly reverse-transcribed into cDNA using the cDNA synthesis kit (AU341-02, TransGen Biotech). The synthesized cDNA was then mixed with ChamQ SYBR qPCR Master Mix (Q411, Vazyme) for quantitative PCR (qPCR) analysis. The primer sequences utilized were: CRHBP-F: 5’-tcttccgagtccatgaaccagg-3’; CRHBP-R: 5’-tcgatgctggtggtggaactacc-3’; CFHR3-F: 5’- agcagatggaaattcttcaggatc-3’; CFHR3-R: 5’- agtagaaaggaggtggtatcacc-3’; β-ACTIN-F: 5’-caccattggcaatgagcggttc-3’; β-ACTIN-R: 5’-aggtctttgcggatgtccacgt-3’. Gene expression levels were determined via the 2−ΔΔCT method.
Construction of Stably Transfected Cell Lines
The CRHBP overexpression plasmid and CFHR3 overexpression plasmid were constructed by Generay Technologies (Shanghai, China). A single positive bacterial colony was selected on LB agar plates containing ampicillin, and successful cloning was confirmed by colony PCR and sequencing. Cells were infected with lentiviral particles, and stably transfected HepG2 and HCCLM9 cells were obtained after puromycin selection. The HCC cells with simultaneous overexpression of CRHBP and CFHR3 were designated as Dual-OE.
Cell Viability Assay
Cell viability was evaluated using the Cell Counting Kit-8 (CCK-8; GK10001, Glpbio) and the EdU Assay Kit (C0075S, Beyotime) according to the manufacturer’s protocols. The cells were cultured in 96-well plates and incubated at intervals of 0, 24, 48, 72, and 96 hours. Optical density (OD) at 450 nm was measured using a microplate reader (Thermo Fisher Scientific) after incubation. The EdU assay: cells were treated with 10 μM EdU, followed by fixation, permeabilization, and incubation with Azide 555. Nuclei were then stained with Hoechst 33342 for 10 minutes. Cell viability was determined by calculating the percentage of EdU-positive cells.
Transwell Assay
Cells were seeded in a serum-free medium in the upper compartment of a Matrigel-coated Transwell chamber for invasion assay. The lower compartment contained medium with 10% FBS. The chamber was incubated at 37°C for a specified period to facilitate cell migration or invasion through the membrane. Following incubation, the migrated or invaded cells were fixed with methanol, stained with crystal violet, and imaged under a microscope for quantification.
Western Blotting
Proteins were extracted from the cells, size-separated via SDS-PAGE, and transferred to nitrocellulose membranes. The membranes were blocked with 10% skimmed milk powder to prevent non-specific binding, followed by overnight incubation with the primary antibody. The primary antibodies used: anti-CRHBP (1:1000, A6568, ABclonal), anti-CFHR3 (1:1000, AB302792, Proteintech), and anti-β-actin (1:40000, HA722023, Huabio).
mRNA Synthesis
The plasmid containing the open reading frame (ORF) of CRHBP with a T7 promoter was linearized using the restriction enzyme Mlu I. The purified linear DNA was then transcribed in vitro with the HiScribe T7 ARCA mRNA Kit (E2060S, NEB). Cy5-labeled UTP was added to the reaction mixture to synthesize Cy5-labeled mRNA during this process. Finally, the mRNA obtained was purified and stored at −80 °C. The plasmid containing the ORF of CFHR3 with a T7 promoter was linearized using the restriction enzyme Sap I. The remaining steps were as described above. In this study, naked mRNA referred to CRHBP-mRNA and naked CFHR3-mRNA.
Synthesis of LNPs-mRNA and LNPs-mRNA-SP94
Lipid nanoparticles were formulated by dissolving ALC-0315, DSPE, cholesterol, and DSPE-PEG2000-NH2 in ethanol at a molar ratio of 55:5:38.5:1.5. Then the lipid mixture and mRNA were mixed at nitrogen-to-phosphate (N/P) ratio of 6 through the microfluidic cartridge at a flow rate ratio of 3:1 (aqueous phase: organic phase). The resulting mixture was then subjected to dialysis against HEPE buffer at 4°C for 24 hours using a 10,000 Da ultrafiltration membrane (UFC910096, Millipore) to obtain the final mRNA-loaded lipid nanoparticles. In this study, LNPs encapsulating enhanced green fluorescent protein (EGFP) mRNA (abbreviated as LNPs-EGFP) were used as a negative control. LNPs-CRHBP mRNA (abbreviated as LNPs-CRHBP), LNPs-CFHR3 mRNA (abbreviated as LNPs-CFHR3), and LNPs-Dual mRNAs (abbreviated as LNPs-Dual).
The carboxyl group of the SP94 peptide was activated using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and then incubated for 4 hours with amino-functionalized lipid nanoparticles containing the mRNA. The unbound peptide was then removed using a dialysis membrane with a molecular weight cut-off of 100 kDa. The SP94-modified lipid nanoparticles containing mRNA were stored at 4°C.
Characterization of Lipid Nanoparticles
Nanoparticle size, polydispersity index (PDI), and zeta potential were measured using a Zetasizer (Malvern Instruments, UK). The morphology of nanoparticles was examined using transmission electron microscopy (TEM, Japan). mRNA encapsulation efficiency and concentration were measured using the RiboGreen assay ((R11490; Invitrogen).
Stability Assay
LNPs-Dual-SP94 was incubated in either PBS (0.01 M, pH 7.4) or PBS with 10% FBS at 37 °C. The size of the particles was monitored at specified time intervals using dynamic light scattering (DLS) technique.
Hemolysis Assay
Following three gentle PBS washes of anticoagulated mouse blood, the centrifuged erythrocyte suspension was added to PBS solution (negative control), deionized water (positive control), or various NPs solutions. After thorough mixing, samples were incubated at 37 °C for 2 hours. Subsequently, the samples were centrifuged at 12,000 rpm for 10 minutes, and hemolysis images were captured. The optical density at 570 nm of the supernatant in each tube was measured with a microplate reader.
Cellular Uptake
HCC cells were seeded in confocal dishes. After cell adhesion, they were exposed to specific concentrations of nanomaterials for 4 hours. Lyso-Tracker Green (C1047S, Beyotime) was then added to the cells and incubated for 15 minutes. Finally, Hoechst 33342 (diluted 1:1000) was added for nuclear staining. Images were captured using a ZEISS confocal laser scanning microscopy (Germany) after staining.
In vitro Transfection Efficiency
HCC cells were seeded in 6-well plates at a density of 5×104 cells per well.19 The cells were then transfected with LNPs-EGFP (EGFP mRNA concentrations: 0.125, 0.25, 0.5, and 1 μg/mL) for 24 hours. PBS and naked EGFP mRNA were used as negative controls, while Lipofectamine 3000 (Lip3000) was used as a positive control. The medium was replaced, followed by a 24-hour incubation of the cells. EGFP expression was subsequently assessed using flow cytometry.
Pharmacokinetics and Biodistribution Study
Male 5-week-old BALB/c nude mice were utilized for the in vivo pharmacokinetic study (n=5 per group). In the first part of the experiment, the mice were administered either naked Cy5-mRNA or Cy5-LNPs-Dual-SP94 via tail vein injection. Blood samples were collected from the orbital vein at 0, 0.25, 0.5, 1, 2, 4, 6, 8, and 10 hours post-injection. The fluorescence intensity of Cy5-mRNA in the blood samples was quantified using a fluorescence spectrophotometer to monitor the pharmacokinetics of the mRNA. In the biodistribution study, male BALB/c nude mice with subcutaneous tumor xenografts received an intravenous injection of either naked Cy5-mRNA or Cy5-LNPs-Dual-SP94 at an mRNA dose of 800 µg/kg body weight (n=5 per group). The MILabs U-OI imaging system (MILabs, Netherlands) was used to quantify fluorescence intensity in tumors and major organs 24 hours post-administration, evaluating mRNA tissue distribution.
Antitumor Effects in vivo
The HCCLM9 cells (5×106 cells) were injected subcutaneously into the dorsal region of healthy male BALB/c nude mice. Treatment was initiated when the average subcutaneous tumor size exceeded 100 mm³. The mice with established subcutaneous tumors were randomly assigned to five groups (n=5 per group), receiving PBS, LNPs, LNPs-EGFP, LNPs-Dual, or LNPs-Dual-SP94 every three days (mRNA dose of 800 µg/kg body weight) for a total of six cycles. The tumor volume and body weight of the mice were monitored daily. The average tumor volume (mm³) was determined using the formula: (length × width² × 0.5). Animal experiments received approval from the Animal Ethics Committee of the First Affiliated Hospital of Nanchang University.
In vivo Biosafety Studies
Tumors and major organs (liver, kidneys, lungs, spleen, and heart) were collected post-antitumor treatment. These tissues were then subjected to immunohistochemistry and hematoxylin-eosin (H&E) staining to examine histopathological alterations. Blood samples from the mice were analyzed for alanine aminotransferase (ALT), aspartate aminotransferase (AST), creatinine (Cr), and blood urea nitrogen (BUN) to assess hepatic and renal toxicity.
Statistical Analysis and Visualization
The D’Agostino-Pearson test assessed the data’s normality. Non-parametric tests were applied to data that did not pass the D’Agostino-Pearson normality test. Parametric or non-parametric t-tests were employed to investigate differences between two distinct groups. Experimental results were reported as mean ± standard deviation. Statistical analyses and visualizations were conducted using R (v4.2.3) or GraphPad Prism (v9.5). A P-value below 0.05 was deemed statistically significant.
Results
Identification of Genes Associated with HCC Progression
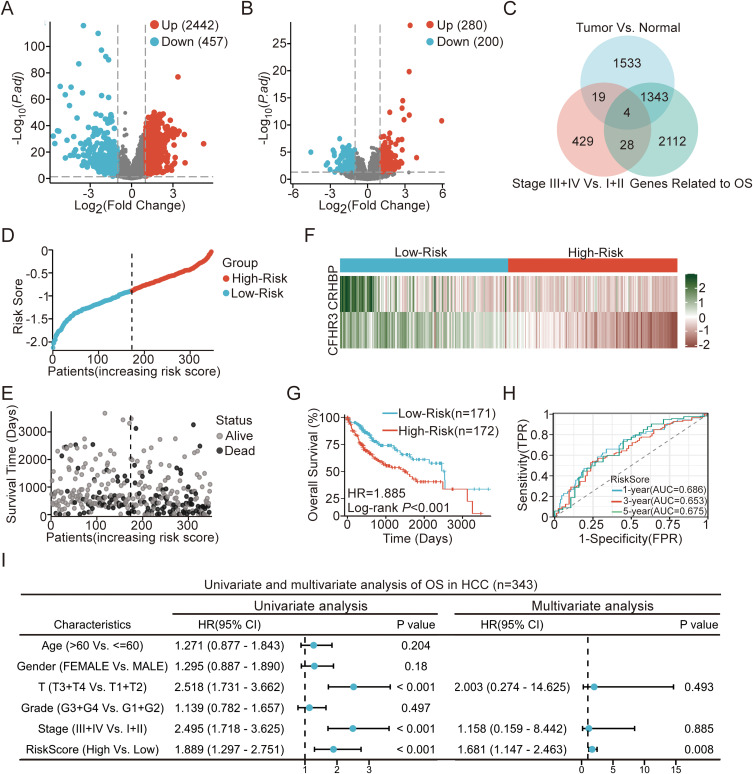
We compared 343 HCC tissue samples with 50 normal liver tissue samples through the TCGA database and found that 2442 genes were upregulated, while 457 genes were downregulated (Figure 1A). To investigate genes associated with HCC progression, we classified the 343 HCC tissue samples into early-stage (Stage I+II) and late-stage (Stage III+IV) groups according to pathological staging. Differential expression analysis between these two groups revealed that 280 genes were upregulated and 200 genes were downregulated in late-stage HCC tissues compared to early-stage tissues (Figure 1B). We identified 23 genes likely critical to HCC progression by intersecting DEGs between HCC and normal liver tissues with those between late-stage and early-stage HCC tissues (Figure 1C).
Figure 1.
Identification of the prognostic model for HCC based on TCGA cohort. (A) Volcano plot of DEGs between HCC and normal tissues. (B) Volcano plot of DEGs between HCC stages I+II and III+IV. (C) Venn diagram showing the intersections of DEGs in HCC vs normal tissues and HCC stages III+IV vs I+II. (D) Risk score of each patient with HCC. (E) Patient survival based on the risk score. (F) Heat map of CRHBP and CFHR3 in high- and low-risk groups. (G) Kaplan–Meier survival analysis of HCC patient OS by risk score. (H) ROC curve analysis for the prognostic model’s value over different years. (I) Forrest plot of univariate or multivariate Cox proportional hazard regression indicated the impact of different characteristics on OS.
The Construction of a Prognostic Model Based on CFHR3 and CRHBP
To identify genes associated with overall survival (OS) in HCC patients, we conducted a univariate Cox regression analysis on 23 differentially expressed genes (DEGs). This analysis identified four DEGs significantly associated with the OS of HCC patients (Figure 1C). We conducted a Lasso regression analysis of four OS-related genes to develop a prognostic model. This analysis led to the selection of two key genes, CFHR3 and CRHBP, which were utilized to form the final prognostic model (Figure S1A and B). The risk score for each sample was computed using the formula: Risk Score = (−1.371 × CFHR3 expression) + (−1.090 × CRHBP expression).
We validated the model by calculating the risk scores for 343 HCC patients using the specified formula and categorizing them into high-risk and low-risk groups (Figure 1D). An increase in risk score correlated with higher mortality in HCC patients (Figure 1E). The expression of two key DEGs also decreased (Figure 1F). Kaplan-Meier survival curves indicated significantly poorer OS in the high-risk group compared to the low-risk group (Figure 1G; P < 0.001). The ROC curve demonstrated AUC values of 0.686 (1 year), 0.653 (3 years), and 0.675 (5 years), suggesting the model’s effectiveness in predicting the OS of HCC patients (Figure 1H). Multivariate Cox regression analysis identified the risk score derived from CFHR3 and CRHBP expression levels as an independent prognostic factor for OS in HCC patients (Figure 1I). This finding highlighted the potential clinical utility of the two-gene risk score model in predicting patient outcomes.
Verification of Prognostic Models in Clinical Tissue Samples
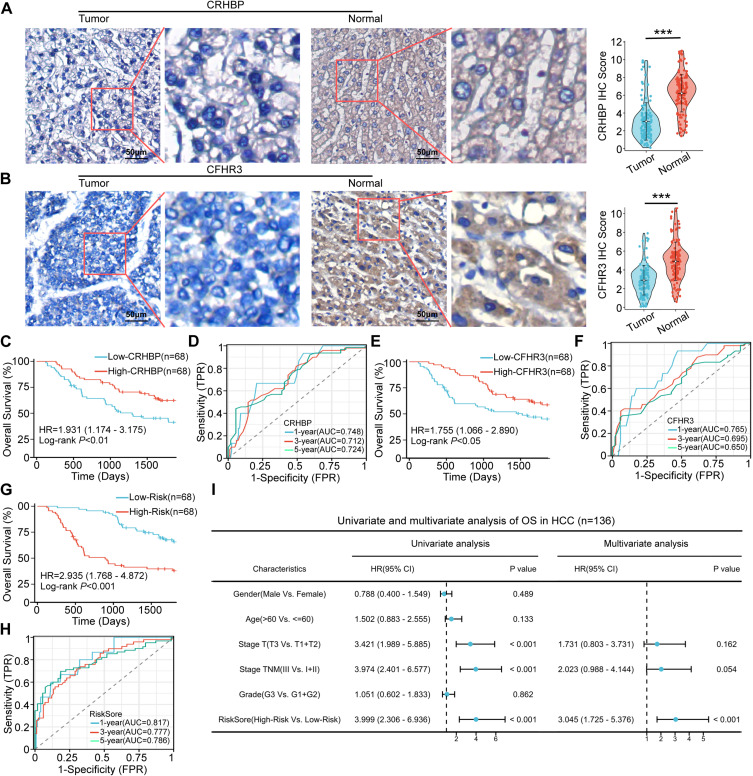
To validate the prognostic model’s predictive value, we assessed CFHR3 and CRHBP expression in 136 HCC patients using IHC staining. IHC staining results indicated that CFHR3 and CRHBP expression levels were lower in HCC tissues compared to adjacent normal liver tissues (Figure 2A-B). Subsequently, 136 patients were categorized into low-expression and high-expression groups based on the median IHC staining scores of CRHBP or CFHR3. Kaplan-Meier survival curves indicated significantly lower OS in the low-expression groups for CRHBP (P < 0.01) and CFHR3 (P < 0.05) compared to the high-expression groups (Figure 2C–E). The ROC curve demonstrated that CRHBP and CFHR3 expression effectively predict OS in HCC patients (Figure 2D–F). Furthermore, according to the above mentioned risk score formula, we calculated the risk scores of 136 patients with HCC based on the IHC staining score of CRHBP and CFHR3. Kaplan-Meier survival curves indicated significantly poorer OS in the high-risk group compared to the low-risk group (Figure 2G). The ROC curve demonstrated that the AUC values of the risk score were 0.817 (1 year), 0.777 (3 years), and 0.786 (5 years), surpassing those of CRHBP and CFHR3. This indicated that the predictive model offered superior predictive value for OS of HCC compared to CRHBP and CFHR3 (Figure 2H). Univariate and multivariate Cox regression analysis identified the risk score as an independent risk factor for OS of HCC in our clinical cohort (Figure 2I).
Figure 2.
Valuation of prognostic models in clinical tissue samples. (A and B) Representative images (left) and quantitative results (right) of immunohistochemistry staining of the protein levels of CRHBP (A) and CFHR3 (B) in clinical HCC samples. Scale bar, 50 μm. Statistical significance was determined using a two-tailed t-test (*** P < 0.001). (C) Kaplan–Meier survival analysis of the OS of patients with HCC according to CRHBP expression. (D) ROC curve analysis for the prognostic value of the CRHBP for different years. (E) Kaplan–Meier survival analysis of the OS of patients with HCC according to CFHR3 expression. (F) ROC curve analysis for the prognostic value of the CFHR3 for different years. (G) Kaplan–Meier analysis of the OS of patients with HCC according to risk score. (H) ROC curve analysis for the prognostic value of the prognostic model for different years. (I) Forrest plot of univariate or multivariate Cox proportional hazard regression indicated the impact of different characteristics on OS.
Overexpression of CRHBP and CFHR3 Inhibited HCC Cells Proliferation, Migration, and Invasion
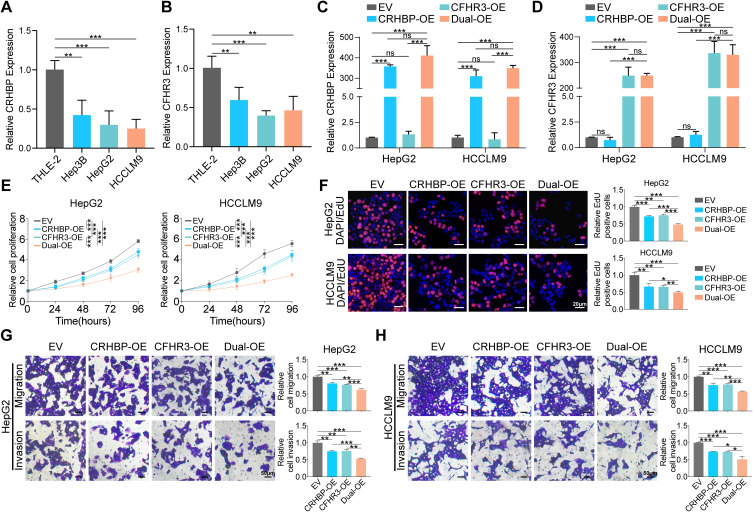
CRHBP and CFHR3 expression levels were analyzed in a normal liver cell line (THLE2) and three HCC cell lines. CRHBP and CFHR3 levels were significantly lower in HCC cell lines compared to the normal liver cell line (Figure 3A and B). To investigate the function of CRHBP and CFHR3, we independently or simultaneously overexpressed CRHBP and CFHR3 in HepG2 and HCCLM9 cells by transfecting with CRHBP-overexpressing (CRHBP-OE) or CFHR3-overexpressing (CFHR3-OE) plasmids, or by simultaneously transfecting both CRHBP-overexpressing and CFHR3-overexpressing plasmids (Dual-OE). As a result, a significant increase in CRHBP expression was observed in the CRHBP-OE and Dual-OE (Figures 3C, S2A). Similarly, CFHR3 expression was significantly increased in the CFHR3-OE and Dual-OE (Figures 3D, S2B), indicating the successful construction of single or dual gene overexpression cell lines. Functional assays, including CCK8 and EdU, demonstrated reduced proliferative capacity in HepG2 and HCCLM9 cells in the CRHBP-OE and CFHR3-OE compared to the control group. Notably, the proliferative capacity of HepG2 and HCCLM9 cells in the Dual-OE was significantly inhibited (Figure 3E–F). Moreover, transwell migration and invasion assays demonstrated that overexpression of CRHBP or CFHR3 inhibited cell migration and invasion abilities, with co-overexpression of CRHBP and CFHR3 further significantly suppressing these abilities (Figure 3G–H). The data suggested that co-overexpression of CRHBP and CFHR3 more effectively inhibits the proliferation, migration, and invasion of HCC cells in vitro.
Figure 3.
Overexpression of CRHBP and CFHR3 suppressed HCC cell proliferation, migration, and invasion in vitro. (A and B) RT-qPCR analysis of CRHBP (A) and CFHR3 (B) expression in three human HCC cell lines (Hep3B, HepG2, and HCCLM9) and normal liver cell line (THLE-2). (C and D) The efficiency of overexpression of CRHBP (C) and CFHR3 (D) was verified by RT-qPCR analysis in HepG2 and HCCLM9 cells as indicated treatment (EV, empty vector; CRHBP-OE, overexpression vector encoding CRHBP; CFHR3-OE, overexpression vector encoding CFHR3; Dual-OE, overexpression vector encoding CRHBP and overexpression vector encoding CFHR3). (E) CCK-8 assay in HepG2 and HCCLM9 cells as indicated treatment. (F) Representative images (left) and quantitative results (right) of EdU assay in HepG2 and HCCLM9 cells as indicated treatment. Scale bar, 20 μm. (G–H) Representative images (left) and quantitative results (right) of transwell migration and invasion assays in HepG2 (G) and HCCLM9 (H) cells as indicated treatment. Scale bar, 50 μm. * P < 0.05; ** P < 0. 01; *** P < 0.001; ns, no statistical difference.
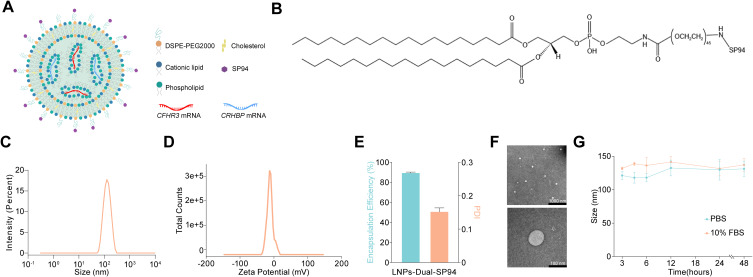
Preparation and Characterization of LNPs-Dual-SP94
To explore the potential of CRHBP and CFHR3 as therapeutic targets for HCC, we synthesized lipid nanoparticles encapsulating CRHBP and CFHR3 mRNA and modified them with the targeting peptide SP94, resulting in LNPs-Dual-SP94 (Figure 4A and B). As depicted in Figure 4C–F, LNPs-Dual-SP94 exhibited a spherical shape with a uniform particle size distribution and an average size of 129 nm. The encapsulation efficiency of the mRNA was 89.97 ± 0.21%, and the zeta potential of LNPs-Dual-SP94 was −11.5 mV. Moreover, the size of LNPs-Dual-SP94 did not change significantly in the presence of 10% serum for 48 hours, demonstrating its stability in serum (Figure 4G).
Figure 4.
Preparation and characterization of LNPs-Dual-SP94. (A) Schematic illustration of LNPs-Dual-SP94. (B) Chemical formula of DSPE-PEG2000-SP94. (C and D) Particle sizes (C) and zeta potentials (D) were measured by dynamic light scattering. (E) The encapsulation efficiency of mRNA (left) and the polydispersity index (PDI) of LNPs-Dual-SP94 (right). (F) Representative TEM images of two fields of view of LNPs-Dual-SP94. Scale bars, 1000 nm and 100 nm, respectively. (G) Stability of LNP-Dual-SP94 over 48 hours in PBS or PBS containing 10% serum at 37 °C.
mRNA Nanoparticles Demonstrated High Transfection Efficiency, Safety, and Enhanced SP94-mediated Cellular Uptake in vitro
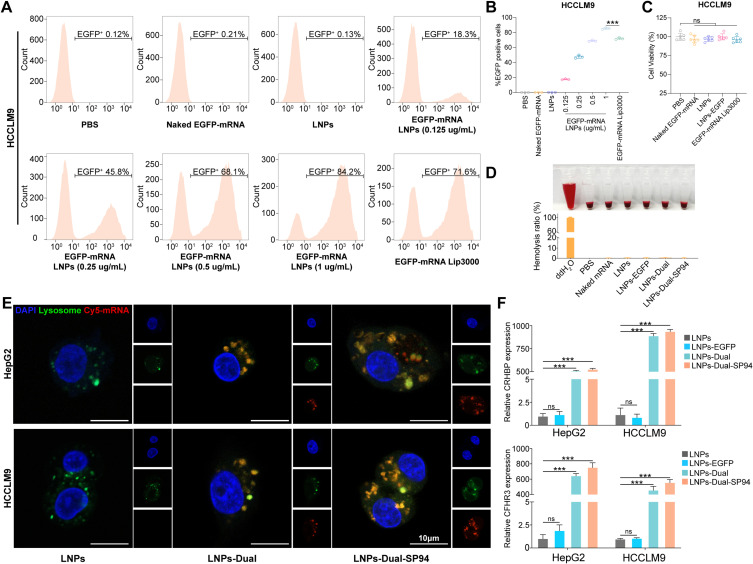
Flow cytometry was used to assess the transfection efficiency of mRNA NPs by analyzing EGFP expression in HCCLM9 and HepG2 cells. The results indicated a dose-dependent increase in EGFP expression, with concentrations of EGFP mRNA ranging from 0.125 to 1 μg/mL. Notably, a significantly higher percentage of EGFP-positive cells was observed in the LNPs-transfected group compared to the Lip3000-transfected group at the 1 μg/mL concentration (Figure 5A and B, S3A–B). The in vitro cytotoxicity of NPs was assessed by treating HCCLM9 and HepG2 cells with each NPs group for 48 hours. Cell viability remained unaffected in the LNPs and LNPs-EGFP compared to the control group. These experiments demonstrated that NPs exhibited high mRNA transfection efficiency with minimal cytotoxicity in HCCLM9 and HepG2 cells (Figures 5C, S3C).
Blood compatibility was a crucial parameter for assessing the biocompatibility of blood-contacting biomedical materials. As illustrated in Figure 5D, the positive control group exhibited significant hemolysis upon visual inspection, while the negative control group and all NPs groups showed no apparent hemolysis. Hemolysis was quantitatively evaluated by measuring the hemoglobin released into the supernatant at 540 nm using a microplate reader. Consistently, quantitative analysis using a microplate reader found that the hemolysis rates for all NPs groups remained below 5%. In conclusion, all tested NPs demonstrated excellent blood compatibility, indicating their suitability for intravenous injection in vivo.
Figure 5.
mRNA NPs demonstrated high transfection efficiency, safety, and enhanced SP94-mediated cellular uptake in vitro. (A and B) Representative images (A) and quantitative results (B) of the in vitro transfection efficiency (percentage of EGFP-positive cells) in the HCCLM9 cells were determined by flow cytometry. (C) Cell viability of the HCCLM9 cells after treatment with PBS, naked EGFP-mRNA (1 μg/mL), LNPs, LNPs-EGFP (1 μg/mL), or EGFP-mRNA Lip3000 (1 μg/mL). (D) Hemolysis of erythrocytes after incubation with PBS (negative control), deionized water (positive control), or various NPs for 2 hours. (Inset: corresponding photos of centrifuge tube containing different samples). (E) Confocal laser scanning microscopy images of HCCLM9 cells after incubation with naked mRNA, LNPs-Dual or LNPs-Dual-SP94 for 4 hours. Endosomes were stained by Lysotracker (green), nuclei were stained by 4′,6-diamidino-2-phenylindole (DAPI) (blue), and mRNA was labeled with Cy5. Scale bars: 10 μm. (F) The expression levels of CRHBP and CFHR3 in HepG2 and HCCLM9 cells treated with LNPs, LNPs-EGFP, LNPs-Dual, or LNPs-Dual-SP94 were verified by RT-qPCR analysis. The mRNA concentration was 1 μg/mL. *** P < 0.001; ns, no statistical difference.
High uptake levels and successful lysosomal release were prerequisites for efficient gene delivery and expression. After 4 hours of incubation, cells with naked mRNA exhibited negligible fluorescence. Significant cellular fluorescence was observed in cells incubated with both LNPs-Dual-SP94 and LNPs-Dual. Moreover, compared to LNPs-Dual, the uptake of LNPs-Dual-SP94 by HCCLM9 and HepG2 cells was markedly enhanced due to SP94 mediation (Figure 5E). These findings demonstrated that all NPs examined effectively entered the cytoplasm and contributed positively to the protection and transport of mRNA. Additionally, SP94 specifically targeted HCC cells, significantly enhancing the cellular uptake of mRNA, whereas naked mRNA could barely enter the cells.
We subsequently analyzed the expression levels of CRHBP and CFHR3 in HepG2 and HCCLM9 cells following treatment with the LNPs, LNPs-EGFP, LNPs-Dual, or LNPs-Dual-SP94. RT-qPCR and WB results showed high levels of CRHBP or CFHR3 expression in the LNPs-Dual-SP94 relative to non-targeted LNPs-Dual (Figures 5F and S4). These results suggested that nanoparticles functionalized with the HCC-targeting ligand SP94 had superior targeting capabilities.
LNPs-Dual-SP94 Suppressed HCC Cell Proliferation, Migration, and Invasion in vitro
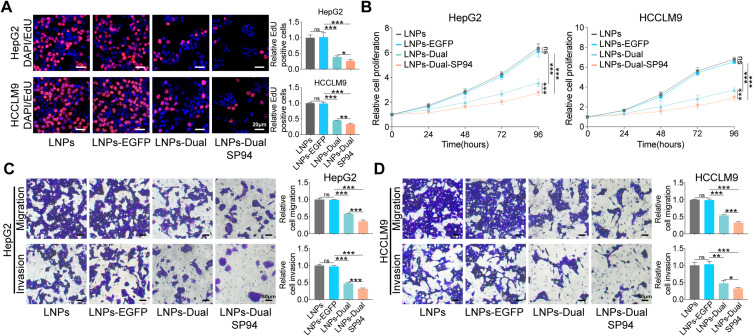
After confirming the targeting capability of mRNA NPs, we further evaluated their therapeutic functions on HCC cells in vitro. The results demonstrated that LNPs-Dual significantly inhibited cell proliferation, migration, and invasion compared to the single mRNA delivery group (Figure S5A–D). These suggested that dual-mRNA therapy held potential therapeutic value. Notably, compared to non-targeted LNPs-Dual, LNPs-Dual-SP94 treatment led to a significant reduction in cell proliferation, migration, and invasion, while no significant differences were observed between the LNPs and LNPs-EGFP groups (Figure 6A–D). These findings indicated that SP94-modified mRNA NPs enhanced association with HCC cells through a receptor-mediated mechanism and exhibited excellent antitumor effects in vitro.
Figure 6.
LNPs-Dual-SP94 inhibited the proliferation, migration, and invasion of HCC Cells in vitro. (A) Representative images (left) and quantitative results (right) of EdU assay in HepG2 and HCCLM9 cells treated with LNPs, LNPs-EGFP, LNPs-Dual, or LNPs-Dual-SP94. Scale bar, 20 μm. (B) CCK-8 assay in HepG2 and HCCLM9 cells as indicated treatment. C-D Representative images (left) and quantitative results (right) of transwell migration and invasion assays in HepG2 (C) and HCCLM9 (D) cells treated with LNPs, LNPs-EGFP, LNPs-Dual or LNPs-Dual-SP94. Scale bar, 50 μm. The mRNA concentration was 1 μg/mL. * P < 0.05; ** P < 0. 01; *** P < 0.001; ns, no statistical difference.
In vivo Evaluation of Therapeutic Efficacy and Toxicity of LNPs-Dual-SP94 in Xenograft Model
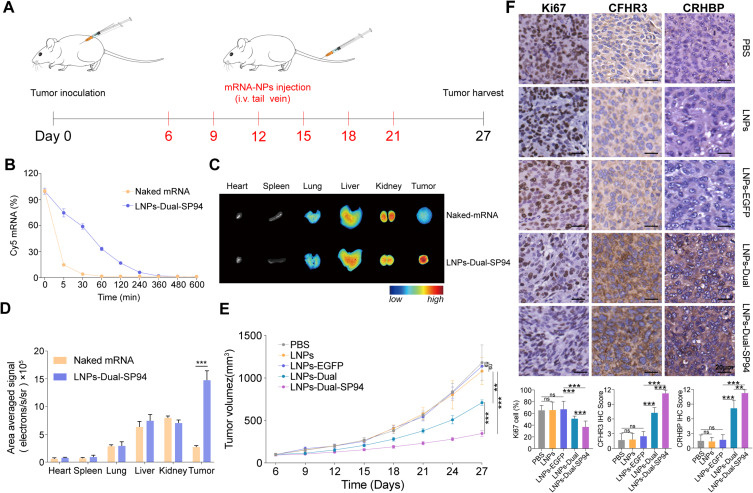
To predict the in vivo performance of mRNA NPs for systemic delivery, the pharmacokinetics were evaluated by measuring plasma levels of Cy5-labeled mRNA after injecting either naked mRNA or LNPs-Dual-SP94 into mice via the tail vein. Figure 7B demonstrated that while naked mRNA was quickly cleared from the bloodstream, LNPs-Dual-SP94 showed extended circulation with an elimination half-life exceeding 30 minutes. Notably, 17% of LNPs-Dual-SP94 remained detectable in the plasma at two hours post-injection (Figure 7B). To further investigate the biodistribution and tumor accumulation of NPs in the HCC xenograft model, we injected naked mRNA and LNPs-Dual-SP94 through the tail vein to BALB/c nude mice with subcutaneous HCCLM9 tumors. As revealed in Figure 7C and D, mice treated with LNPs-Dual-SP94 had significantly higher tumor site fluorescence intensity than those treated with naked mRNA.
Figure 7.
In vivo therapeutic efficacy and toxicity evaluation of LNPs-Dual-SP94 in HCCLM9 xenograft model. (A) Schematic illustration of HCCLM9 tumor inoculation and treatment of HCCLM9 tumor‐bearing mice. (B) In vivo pharmacokinetics of naked mRNA and LNPs-Dual-SP94. (C and D) Representative images (C) and quantitative results (D) illustrate the biodistribution of naked mRNA and LNPs-Dual-SP94 in various organs (including tumors) of HCCLM9 tumor-bearing mice. (E) Tumor growth of the tumor‐bearing mice treated with PBS, LNPs, LNPs-EGFP, LNPs-Dual, or LNPs-Dual-SP94. (F) IHC staining analysis of CRHBP, CFHR3, and Ki67 expression and quantitative results in tumor tissues after systemic treatment in various groups of mice. Scale bar, 20 μm. ** P < 0. 01; *** P < 0.001; ns, no significance.
Based on the observed antitumor effects of LNPs-Dual-SP94 in vitro, we further assessed its efficacy in vivo using a xenograft mouse model. Mice were administered LNPs-Dual-SP94 via intravenous (i.v.) tail vein injection every three days, totaling six systemic doses (Figure 7A). Tumor-bearing mice treated with PBS, LNPs, or LNPs-EGFP were used as control groups. In contrast to the rapid tumor growth observed in the control groups, treatment with LNPs-Dual-SP94 significantly inhibited tumor growth (Figure 7E). Notably, no significant changes in body weight were observed across all treatment groups, suggesting minimal systemic toxicity (Figure S6A). Immunohistochemical analysis of collected tumors revealed a significant elevation in CRHBP and CFHR3 expression in both the LNPs-Dual and LNPs-Dual-SP94 (Figure 7F). Additionally, there was a marked reduction in Ki67 protein expression, indicating a decrease in tumor proliferation. These results collectively demonstrated that LNPs-Dual-SP94 exerted potent antitumor effects in vivo.
H&E staining of cardiac, hepatic, splenic, pulmonary, and renal tissues revealed no significant damage, highlighting the efficacy and safety of these NPs (Figure S6B). Blood index analysis further confirmed the absence of significant hepatotoxicity and nephrotoxicity of mRNA NPs (Figure S6C). The NPs demonstrated good biocompatibility in mice, providing sufficient evidence for their potential clinical application.
Discussion
In this study, we developed a prognostic risk score model for HCC based on the two genes associated with malignant progression, CFHR3 and CRHBP. Multivariate Cox regression analysis confirmed that the risk score derived from CFHR3 and CRHBP was an independent prognostic factor for patients with HCC. The accuracy of this model was validated in an independent clinical cohort. In vitro studies demonstrated that the synergistic amplification effect of co-overexpressing CRHBP and CFHR3 significantly suppressed the malignant biological behaviors of HCC cells. We developed dual mRNAs-loaded LNPs based on these findings, modified with the HCC-specific ligand peptide SP94. Our research indicated that the simultaneous delivery of these dual mRNAs exhibited superior antitumor effects against HCC, providing a new perspective for therapeutic approaches to this disease.
In recent years, next-generation sequencing (NGS) technology has led to substantial genomic and molecular data generation.20 Leveraging these data, researchers have developed various models to guide clinical treatment for HCC. For instance, Gu et al developed a six-long non-coding RNA predictive signature for assessing HCC recurrence risk.21 Similarly, Xu et al constructed a nine-gene model for prognostic evaluation in HCC patients.22 However, the therapeutic value of these signatures remained inadequately validated. This study developed a potentially more practical and cost-effective dual-gene prognostic risk score model. Additionally, our dual-gene model was validated at the protein level compared to previous conventional prognostic models, enhancing its potential for clinical application. While this study contributed to the advancement of prognostic assessment systems for HCC, its reliability necessitated validation through large-scale, multicenter prospective studies. Identifying prognostically relevant gene signatures in this study highlighted their potential clinical significance, warranting further investigation into the biological function of these dual genes in HCC.
Previous studies have demonstrated a strong association between CRHBP and CFHR3 with disease progression.15,18,23,24 Yang et al found that CRHBP exerted antitumor effects in renal cell carcinoma by regulating the mitochondrial apoptosis pathway.25 Additionally, CRHBP influenced hematopoiesis independently of CRH, and its downregulation correlated with poor prognosis, increased metastasis, and enhanced tumor invasiveness in acute myeloid leukemia (AML), identifying it as a novel tumor suppressor in AML.26 Similarly, the Complement Factor H gene was crucial for regulating complement system activation, with its deletion or mutation contributing to various diseases.27–29 The complement system has been shown to provide immunosurveillance against tumors, inhibiting their growth.30,31 Abnormal expression of CFHR3, a complement factor H protein family member, allowed cancer cells to evade immune surveillance and reduced chemotherapy sensitivity.32,33 However, the therapeutic potential of CRHBP and CFHR3 for HCC remains uncertain.
The efficacy of gene therapy fundamentally hinged on the efficient delivery and expression of therapeutic genes.34 Nevertheless, due to the complexity of tumors, previous single-target therapies have not demonstrated superior therapeutic outcomes.35 From a novel perspective, gene therapy can be enhanced by increasing the number of therapeutic targets. In this study, the co-transmission of CRHBP and CFHR3 exhibited a stronger inhibitory effect on HCC cells than single-gene groups, indicating a synergistic effect when these genes were co-transmitted through physical mixing. Similarly, Chen et al demonstrated that the co-delivery of pVEGF and pNGF significantly improved therapeutic efficacy by promoting stable angiogenesis in ischemic hindlimbs.36 Additionally, dual mRNAs encoding PCCA and PCCB delivered via LNPs showed superior efficacy compared to single mRNA, as they produced higher PCC enzyme activity, restored liver function, and reduced disease-associated toxins in models of propionic acidemia.37 This synergistic effect may be due to the fact that tumors are often caused by the combined action of multiple genetic abnormalities. Therefore, multi-gene combination therapy offers a more rational and superior approach, representing the future direction of tumor therapy.38
mRNA therapy represents a significant breakthrough in modern medicine,39 enabling the intracellular synthesis of therapeutic proteins at high levels while minimizing the risk of unintended gene alterations or mutations associated with plasmid DNA.40 However, current delivery systems encounter some limitations, including low transfection efficiency, non-specific distribution, and mRNA degradation.41 Thus, the efficient and specific delivery of mRNA to targeted cells remains a significant challenge. LNPs demonstrated tremendous potential as mRNA delivery systems, as evidenced by their successful application in COVID-19 vaccines.42 By engineering the surface of LNPs to enhance targeting specificity, these nanoparticles more precisely delivered mRNA to specific cells, thereby improving therapeutic efficacy and safety.11 SP94 (SFSIIHTPILPL) was a targeting peptide identified through phage display technology,43 selectively bound to glucose-regulated protein 78 (GRP78), which was a receptor overexpressed on the surface of HCC cells.44 Conjugation of SP94 with LNPs enabled targeted delivery to HCC cells via receptor-mediated endocytosis.45 This approach minimized off-target effects by leveraging the specificity of SP94 for GRP78 and the enhanced permeability and retention (EPR) effect. He et al reported that nanocapsules coated with SP94 peptide selectively delivered the CRISPR/Cas9 system to HCC cells for antitumor effects.46 Furthermore, SP94-PEG-modified GDYO nanosheets efficiently delivered sorafenib and adriamycin, reducing the drug dose while increasing bioavailability. SP94-targeted peptide was considered as an ideal ligand for HCC due to its advantages of small size, low immunogenicity and economy, which significantly enhanced drug internalization in HCC and accumulation at the tumor site.47,48
In this study, we employed active targeting by modifying LNPs containing CRHBP mRNA and CFHR3 mRNA with the targeting peptide SP94. This modification facilitated the delivery of dual mRNAs into target cells, resulting in stable and efficient expression. Both in vivo and in vitro experiments confirmed the significant antitumor effects of LNPs modified with the SP94 targeting peptide, with no evident side effects on major organs. Therefore, dual mRNAs-targeted delivery based on LNPs showed significant promise for HCC therapy.
Conclusions
This study developed a prognostic model based on the HCC progression-related genes CRHBP and CFHR3, which effectively predicted the prognosis and survival rates of HCC patients. Notably, the co-overexpression of CRHBP and CFHR3 significantly outperformed single-gene expression in suppressing the malignant biological functions of HCC cells. Furthermore, we designed LNPs loaded with dual mRNAs and functionalized with the HCC-specific ligand SP94. Both in vitro and in vivo experiments confirmed that these nanoparticles exhibit excellent biocompatibility, low cytotoxicity, and significant cellular uptake and antitumor effects. In conclusion, this study provided a potential therapeutic strategy for HCC treatment.
Funding Statement
This work was supported by the major science and technology R & D projects of Jiangxi Province (20213AAG01013), Natural Science Foundation of China (82472225,32071223,32100729, training plan for academic and technical leaders of major disciplines in Jiangxi Province (20213BCJ22015), Jiangxi Province Science and Technology Innovation Talent Project (jxsq2023201037), science and technology innovation base plan of Jiangxi Province (20212BCD42006), Natural Science Foundation of Jiangxi Province Youth Project (20242BAB20404), Youth Talent Research Training Foundation of the First Affiliated Hospital of Nanchang University (YFYPY2023103), Full time Talent Introduction and Research Start up Fund of the First Affiliated Hospital of Nanchang University(RSC-0020).
Abbreviations
HCC, hepatocellular carcinoma; LNPs, lipid nanoparticles; TCGA, the cancer genome atlas; TPM, transcripts per million; DEGs, differentially expressed genes; LASSO, least absolute shrinkage and selection operator; ROC, receiver operating characteristic; FFPE, formalin-fixed, paraffin-embedded; ALT, alanine aminotransferase; AST, aspartate aminotransferase; Cr, creatinine; BUN, blood urea nitrogen; OS, overall survival; NGS, next-generation sequencing; AML, acute myeloid leukemia.
Data Sharing Statement
Upon reasonable request, requests for further inquiries may be sent to ndyfy00973@ncu.edu.cn.
Ethics Approval and Consent to Participate
The Ethics Committee of the First Affiliated Hospital of Gannan Medical University and the First Affiliated Hospital of Nanchang University approved this study (LLSC-2023245, IIT-2024614). All participants provided written informed consent before their involvement in the research. All animal experiments received approval from the Animal Ethics Committee of the First Affiliated Hospital of Nanchang University (CDYFY-IACUC-202309QR004). The experimental protocols were performed in strict accordance with the Guide for the Ethical Review of Laboratory Animal Welfare.
Disclosure
The authors declare no competing interests in this work.
References
- 1.Ladd AD, Duarte S, Sahin I, Zarrinpar A. Mechanisms of drug resistance in HCC. Hepatology. 2024;79(4):926–940. doi: 10.1097/hep.0000000000000237 [DOI] [PubMed] [Google Scholar]
- 2.Toh MR, Wong EYT, Wong SH, et al. Global Epidemiology and Genetics of Hepatocellular Carcinoma. Gastroenterology. 2023;164(5):766–782. doi: 10.1053/j.gastro.2023.01.033 [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Pinyol R, Yarchoan M, et al. Adjuvant and neoadjuvant immunotherapies in hepatocellular carcinoma. Nat Rev Clin Oncol. 2024;21(4):294–311. doi: 10.1038/s41571-024-00868-0 [DOI] [PubMed] [Google Scholar]
- 4.Tang Y, Xu L, Ren Y, et al. Identification and Validation of a Prognostic Model Based on Three MVI-Related Genes in Hepatocellular Carcinoma. Int J Biol Sci. 2022;18(1):261–275. doi: 10.7150/ijbs.66536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kowalski PS, Rudra A, Miao L, Anderson DG. Delivering the Messenger: advances in Technologies for Therapeutic mRNA Delivery. Mol Ther. 2019;27(4):710–728. doi: 10.1016/j.ymthe.2019.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu C, Shi Q, Huang X, Koo S, Kong N, Tao W. mRNA-based cancer therapeutics. Nat Rev Cancer. 2023;23(8):526–543. doi: 10.1038/s41568-023-00586-2 [DOI] [PubMed] [Google Scholar]
- 7.Qin S, Tang X, Chen Y, et al. mRNA-based therapeutics: powerful and versatile tools to combat diseases. Signal Transduct Target Ther. 2022;7(1):166. doi: 10.1038/s41392-022-01007-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan M, Han Z, Liang Y, et al. mRNA nanodelivery systems: targeting strategies and administration routes. Biomater Res. 2023;27(1):90. doi: 10.1186/s40824-023-00425-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zong Y, Lin Y, Wei T, Cheng Q. Lipid Nanoparticle (LNP) Enables mRNA Delivery for Cancer Therapy. Adv Mater. 2023;35(51):e2303261. doi: 10.1002/adma.202303261 [DOI] [PubMed] [Google Scholar]
- 10.Cullis PR, Hope MJ. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol Ther. 2017;25(7):1467–1475. doi: 10.1016/j.ymthe.2017.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kon E, Ad-El N, Hazan-Halevy I, Stotsky-Oterin L, Peer D. Targeting cancer with mRNA-lipid nanoparticles: key considerations and future prospects. Nat Rev Clin Oncol. 2023;20(11):739–754. doi: 10.1038/s41571-023-00811-9 [DOI] [PubMed] [Google Scholar]
- 12.Ketchesin KD, Stinnett GS, Seasholtz AF. Corticotropin-releasing hormone-binding protein and stress: from invertebrates to humans. Stress. 2017;20(5):449–464. doi: 10.1080/10253890.2017.1322575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Argentieri MA, Nagarajan S, Seddighzadeh B, Baccarelli AA, Shields AE. Epigenetic Pathways in Human Disease: the Impact of DNA Methylation on Stress-Related Pathogenesis and Current Challenges in Biomarker Development. EBioMedicine. 2017;18:327–350. doi: 10.1016/j.ebiom.2017.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tezval H, Dubrowinskaja N, Peters I, et al. Tumor Specific Epigenetic Silencing of Corticotropin Releasing Hormone -Binding Protein in Renal Cell Carcinoma: association of Hypermethylation and Metastasis. PLoS One. 2016;11(10):e0163873. doi: 10.1371/journal.pone.0163873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Li M, Liu Y, et al. CRHBP is degraded via autophagy and exerts anti-hepatocellular carcinoma effects by reducing cyclin B2 expression and dissociating cyclin B2-CDK1 complex. Cancer Gene Ther. 2022;29(8–9):1217–1227. doi: 10.1038/s41417-021-00423-4 [DOI] [PubMed] [Google Scholar]
- 16.Skerka C, Chen Q, Fremeaux-Bacchi V, Roumenina LT. Complement factor H related proteins (CFHRs). Mol Immunol. 2013;56(3):170–180. doi: 10.1016/j.molimm.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 17.Wan Z, Li X, Luo X, Wang B, Zhou X, Chen A. The miR-590-3p/CFHR3/STAT3 signaling pathway promotes cell proliferation and metastasis in hepatocellular carcinoma. Aging. 2022;14(14):5783–5799. doi: 10.18632/aging.204178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia HB, Wang HJ, Fu LQ, et al. Decreased CRHBP expression is predictive of poor prognosis in patients with hepatocellular carcinoma. Oncol Lett. 2018;16(3):3681–3689. doi: 10.3892/ol.2018.9073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong N, Tao W, Ling X, et al. Synthetic mRNA nanoparticle-mediated restoration of p53 tumor suppressor sensitizes p53-deficient cancers to mTOR inhibition. Sci Transl Med. 2019;11(523). doi: 10.1126/scitranslmed.aaw1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Craig AJ, von Felden J, Garcia-Lezana T, Sarcognato S, Villanueva A. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17(3):139–152. doi: 10.1038/s41575-019-0229-4 [DOI] [PubMed] [Google Scholar]
- 21.Gu JX, Zhang X, Miao RC, et al. Six-long non-coding RNA signature predicts recurrence-free survival in hepatocellular carcinoma. World J Gastroenterol. 2019;25(2):220–232. doi: 10.3748/wjg.v25.i2.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Z, Peng B, Liang Q, et al. Construction of a Ferroptosis-Related Nine-lncRNA Signature for Predicting Prognosis and Immune Response in Hepatocellular Carcinoma. Front Immunol. 2021;12:719175. doi: 10.3389/fimmu.2021.719175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hageman GS, Hancox LS, Taiber AJ, et al. Extended haplotypes in the complement factor H (CFH) and CFH-related (CFHR) family of genes protect against age-related macular degeneration: characterization, ethnic distribution and evolutionary implications. Ann Med. 2006;38(8):592–604. [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H, Zhang L, Wang P. Complement factor H‑related 3 overexpression affects hepatocellular carcinoma proliferation and apoptosis. Mol Med Rep. 2019;20(3):2694–2702. doi: 10.3892/mmr.2019.10514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang K, Xiao Y, Xu T, et al. Integrative analysis reveals CRHBP inhibits renal cell carcinoma progression by regulating inflammation and apoptosis. Cancer Gene Ther. 2020;27(7–8):607–618. doi: 10.1038/s41417-019-0138-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbieri DA, Telonis AG, Figueroa MKE. Corticotropin Releasing Hormone Binding Protein (CRHBP) Regulates Hematopoietic Function and Acts As a Novel AML Tumor Suppressor through a Crh-Independent Mechanism. Blood. 2022;140(Supplement 1):5840–5841. [Google Scholar]
- 27.Yang C, Chen J, Yu Z, et al. Mining of RNA Methylation-Related Genes and Elucidation of Their Molecular Biology in Gallbladder Carcinoma. Front Oncol. 2021;11:621806. doi: 10.3389/fonc.2021.621806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes AE, Orr N, Esfandiary H, Diaz-Torres M, Goodship T, Chakravarthy U. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet. 2006;38(10):1173–1177. doi: 10.1038/ng1890 [DOI] [PubMed] [Google Scholar]
- 29.Kavanagh D, Richards A, Atkinson J. Complement regulatory genes and hemolytic uremic syndromes. Annu Rev Med. 2008;59:293–309. doi: 10.1146/annurev.med.59.060106.185110 [DOI] [PubMed] [Google Scholar]
- 30.Carroll MC, Isenman DE. Regulation of humoral immunity by complement. Immunity. 2012;37(2):199–207. doi: 10.1016/j.immuni.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reis ES, Mastellos DC, Ricklin D, Mantovani A, Lambris JD. Complement in cancer: untangling an intricate relationship. Nat Rev Immunol. 2018;18(1):5–18. doi: 10.1038/nri.2017.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao X, Zhou L, Tey SK, et al. Tumour extracellular vesicle-derived Complement Factor H promotes tumorigenesis and metastasis by inhibiting complement-dependent cytotoxicity of tumour cells. J Extracell Vesicles. 2020;10(1):e12031. doi: 10.1002/jev2.12031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui L, Fu J, Pang JC, et al. Overexpression of IL-7 enhances cisplatin resistance in glioma. Cancer Biol Ther. 2012;13(7):496–503. doi: 10.4161/cbt.19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K, Kievit FM, Zhang M. Nanoparticles for cancer gene therapy: recent advances, challenges, and strategies. Pharmacol Res. 2016;114:56–66. doi: 10.1016/j.phrs.2016.10.016 [DOI] [PubMed] [Google Scholar]
- 35.Benedetti R, Conte M, Iside C, Altucci L. Epigenetic-based therapy: from single- to multi-target approaches. Int J Biochem Cell Biol. 2015;69:121–131. doi: 10.1016/j.biocel.2015.10.016 [DOI] [PubMed] [Google Scholar]
- 36.Chen Y, Chen Z, Duan J, et al. H(2)O(2)-responsive VEGF/NGF gene co-delivery nano-system achieves stable vascularization in ischemic hindlimbs. J Nanobiotechnology. 2022;20(1):145. doi: 10.1186/s12951-022-01328-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang L, Park JS, Yin L, et al. Dual mRNA therapy restores metabolic function in long-term studies in mice with propionic acidemia. Nature Communi. 2020;11(1):5339. doi: 10.1038/s41467-020-19156-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cáceres B, Ramirez A, Carrillo E, et al. Deciphering the Mechanism of Action Involved in Enhanced Suicide Gene Colon Cancer Cell Killer Effect Mediated by Gef and Apoptin. Cancers. 2019;11(2). doi: 10.3390/cancers11020264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang X, Kong N, Zhang X, Cao Y, Langer R, Tao W. The landscape of mRNA nanomedicine. Nat Med. 2022;28(11):2273–2287. doi: 10.1038/s41591-022-02061-1 [DOI] [PubMed] [Google Scholar]
- 40.Han J, Lim J, Wang CJ, et al. Lipid nanoparticle-based mRNA delivery systems for cancer immunotherapy. Nano Converg. 2023;10(1):36. doi: 10.1186/s40580-023-00385-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim SA, Cox A, Tung M, Chung EJ. Clinical progress of nanomedicine-based RNA therapies. Bioact Mater. 2022;12:203–213. doi: 10.1016/j.bioactmat.2021.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu MZ, Wang NN, Zhu JQ, Lin YX. The clinical progress and challenges of mRNA vaccines. Wiley Interdiscip Rev Nanomed Nanobiotec. 2023;15(5):e1894. doi: 10.1002/wnan.1894 [DOI] [PubMed] [Google Scholar]
- 43.Lo A, Lin CT, Wu HC. Hepatocellular carcinoma cell-specific peptide ligand for targeted drug delivery. Mol Cancer Ther. 2008;7(3):579–589. doi: 10.1158/1535-7163.Mct-07-2359 [DOI] [PubMed] [Google Scholar]
- 44.Jiang B, Zhang R, Zhang J, et al. GRP78-targeted ferritin nanocaged ultra-high dose of doxorubicin for hepatocellular carcinoma therapy. Theranostics. 2019;9(8):2167–2182. doi: 10.7150/thno.30867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Tong L, Xiong Z, et al. Ferroptosis-inducing nanomedicine and targeted short peptide for synergistic treatment of hepatocellular carcinoma. J Nanobiotechnology. 2024;22(1):533. doi: 10.1186/s12951-024-02808-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He L, Li Z, Su D, et al. Tumor Microenvironment-Responsive Nanocapsule Delivery CRISPR/Cas9 to Reprogram the Immunosuppressive Microenvironment in Hepatoma Carcinoma. Adv Sci. 2024;11(26):e2403858. doi: 10.1002/advs.202403858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao Y, Shi D, Shang M, et al. GRP78-targeted and doxorubicin-loaded nanodroplets combined with ultrasound: a potential novel theranostics for castration-resistant prostate cancer. Drug Deliv. 2022;29(1):203–213. doi: 10.1080/10717544.2021.2023698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raza F, Zheng M, Zhong H, et al. Engineered tumor microvesicles modified by SP94 peptide for arsenic trioxide targeting drug delivery in liver cancer therapy. Biomater Adv. 2023;155:213683. doi: 10.1016/j.bioadv.2023.213683 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Upon reasonable request, requests for further inquiries may be sent to ndyfy00973@ncu.edu.cn.