ABSTRACT
Bacteria of the genus Stenotrophomonas are found throughout the environment, in close association with soil, sewage, and plants. Stenotrophomonas maltophilia, the first member of this genus, is the predominant species, observed in soil, water, plants, animals, and humans. It is also an opportunistic pathogen associated with the increased number of infections in both humans and animals in recent years. In this article, we summarize all Stenotrophomonas species (mainly S. maltophilia) isolated from animals and food products of animal origin and further distinguish all isolates based on antimicrobial susceptibility and resistance phenotypes. The various mechanisms of both intrinsic and acquired antimicrobial resistance, which were mainly identified in S. maltophilia isolates of nosocomial infections, have been classified as follows: multidrug efflux pumps; resistance to β-lactams, aminoglycosides, quinolones, trimethoprim-sulfamethoxazole, and phenicols; and alteration of lipopolysaccharide and two-component regulatory systems. The dissemination, coselection, and persistence of resistance determinants among S. maltophilia isolates have also been elaborated.
INTRODUCTION
The genus Stenotrophomonas comprises 16 characterized species (Table 1), and 13 validated species are included in the List of Prokaryotic names with Standing in Nomenclature (1). The first Stenotrophomonas species—Stenotrophomonas maltophilia—was isolated in 1943 from human pleural fluid. It was classified as Bacterium bookeri and subsequently renamed Pseudomonas maltophilia/Xanthomonas maltophilia (1, 2). Another 12 Stenotrophomonas species were first identified residing in soil, sewage, or plants. Of the remaining three species, Stenotrophomonas sp. D-1 and Stenotrophomonas koreensis were first isolated from deer fur and animal compost, respectively, and Stenotrophomonas africana was initially isolated from a sample of cerebrospinal fluid from a human immunodeficiency virus seropositive Rwandan refugee with primary meningoencephalitis (3). S. maltophilia is the most widely distributed bacterium of the Stenotrophomonas spp. in the environment and is isolated from soil, water, plants, animals, and humans. Moreover, the number of nosocomial infections caused by this opportunistic pathogen is increasing (4). Therefore, various studies of Stenotrophomonas in both animals and humans focus on the emergence, infections, treatment, and antimicrobial resistance of S. maltophilia as an opportunistic pathogen (4, 5). The main purpose of this article is to describe the antimicrobial resistance of S. maltophilia isolated from animals.
TABLE 1.
Characterization of Stenotrophomonas species
| Species | Year of first identification/designation | Host when first identified | Characterization | Countries/continents | Ref. |
|---|---|---|---|---|---|
| S. maltophilia | 1943 | Human | S. maltophilia, a new bacterial genus for X. maltophilia, is first identified from a specimen of pleural fluid | England/Europe | 99 |
| S. africana | 1997 | Human | Opportunistic pathogen from cerebrospinal fluid | Rwanda/Africa | 3 |
| S. nitritireducens | 2000 | Ammonia-supplied biofilters | It reduced nitrite, but not nitrate, without production of nitrogen | Germany/Europe | 100 |
| S. sp. D-1 | 2002 | Animal (deer fur) | A keratin-degrading bacterium isolated from soil containing deer fur; 16S rDNA revealed it has only 90.6% homology with S. nitritireducens | Japan/Asia | 101 |
| S. acidaminiphila | 2002 | Upflow anaerobic sludge blanket (UASB) reactor | A strictly aerobic, mesophilic bacterium isolated from UASB reactor treating a petrochemical wastewater | Burkina Faso/Africa | 102 |
| S. rhizophila | 2002 | Environment (plant) | Plant-associated bacterium with antifungal properties | Germany/Europe | 103 |
| S. dokdonensis | 2006 | Environment (soil) | The levels of 16S rDNA sequence similarity between S. dokdonensis and the type strains of Stenotrophomonas species ranged from 95.5 to 97.5% | Korea/Asia | 104 |
| S. koreensis | 2006 | Environment (animal compost) | A Gram-negative, rod-shaped, non-spore-forming bacterium was isolated from compost near Daejeon city | Korea/Asia | 105 |
| S. humi | 2007 | Environment (soil) | The nitrate-reducing bacterium was isolated from soil | Belgium/Europe | 106 |
| S. terrae | 2007 | Environment (soil) | The nitrate-reducing bacterium was isolated from soil | Belgium/Europe | 106 |
| S. chelatiphaga | 2009 | Environment (sewage) | An EDTA-utilizing gammaproteobacterial strain was isolated from municipal sewage sludge | Russia/Europe | 107 |
| S. ginsengisoli | 2010 | Environment (soil) | A Gram-negative, non-spore-forming, rod-shaped bacterium was isolated from soil from a ginseng field | Korea/Asia | 108 |
| S. daejeonensis | 2011 | Environment (sewage) | Comparative 16S rDNA analysis showed it was related most closely to S. acidaminiphila (97.9% similarity) | Korea/Asia | 109 |
| S. pavanii | 2011 | Environment (plant) | A Gram-negative, rod-shaped, non-spore-forming, and nitrogen-fixing bacterium was isolated from stems of a Brazilian sugar cane variety | Brazil/South America | 110 |
| S. tumulicola | 2015 | Environment (spot and gels) | A major contaminant of the stone chamber interior in blackish moldy spots and viscous gels (biofilms) collected from both tumuli | Japan/Asia | 111 |
| S. sp. DDT-1 | 2016 | Environment (contaminated soil) | A novel bacterium capable of utilizing 1,1,1-trichloro-2,2-bis(p-chlorophenyl)ethane (DDT) as the sole carbon and energy source. | China/Asia | 112 |
The earliest study of S. maltophilia reported its isolation from sources associated with rabbits, raw milk, and frozen fish in 1961 (6). It is the predominant bacterial species in swine and chicken feces (7), as well as in composted swine manure (8). S. maltophilia isolates have been found to coexist with influenza virus in the oral, nasal, and tracheal tissues of pigs and horses (9, 10). S. maltophilia is a predominant bacterial species in raw milk, milk processing plants, and milk products such as cheese (11–13) and is likely a constituent of the normal microflora of the mouth and cloacae of squirrels and captive healthy snakes (14, 15). In aquaculture, Stenotrophomonas spp. are predominant members of bacterial communities found in the internal organs of cultured snow crabs (Chionoecetes) (16) and are commonly isolated from cultured yellowtail (17), shrimp (18), and samples taken from salmon farms (19, 20).
Although Stenotrophomonas spp. are less frequently considered as primary pathogens, S. maltophilia is the major cause of the bacteriospermia in porcine or bovine semen in the United States and United Kingdom (21–23), as well as the infection of Xenopus laevis oocytes (24). It was also found to be associated with an outbreak of lymphadenitis in Omani goats (25) and causes fleece rot in sheep (26). Closely related S. maltophilia strains were isolated from an outbreak of bovine mastitis (27), which may be explained by the higher adhesion of these isolates to bovine mammary gland epithelial cells (28). S. maltophilia was identified as a cause of pyogranulomatous hepatitis in a female buffalo (Bubalus bubalis) in a herd in Serres, Greece (29), as well as the cause of necrosis and friability of the nictitating membrane of the giant panda (Ailuropoda melanoleuca) (30). It is also associated with chronic respiratory disease among horses, dogs, and cats (31, 32), as well as septicemia in pigs and crocodiles (33, 34). Moreover, the DNA of S. maltophilia is identified most frequently in the knee joints of dogs with inflammatory arthritis (35).
ANTIMICROBIAL SUSCEPTIBILITY
The susceptibility testing methods for S. maltophilia include disk diffusion, agar/broth dilution, commercially available microdilution strips, and microtiter panels (Table 2). Although the Clinical Laboratory Standards Institute (CLSI) has not defined breakpoints for S. maltophilia isolated from animals, the breakpoints for human isolates of S. maltophilia for sulfamethoxazole/trimethoprim (SXT), minocycline, levofloxacin, ticarcillin-clavulanic acid, ceftazidime, and chloramphenicol have been commonly adopted (36). The breakpoints for Enterobacteriaceae and Pseudomonas spp. are also frequently employed to interpret the susceptibility data for S. maltophilia (29, 32). Other breakpoints, such as those specified by the National Reference Laboratory for Antibiotics (National Institute of Public Health, Prague, Czech Republic) and the Antibiogram Committee of the French Microbiology Society, have also been used (13, 15).
TABLE 2.
Antimicrobial resistance of S. maltophilia isolated from animals and animal products
| Origin | Year of identification | Country | Strain no. | Standards and methods of susceptibility testinga | Antimicrobial agents used for susceptibility testing (resistance rates, %)b | Ref. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-Lactams (penicillins, cephalosporins, carbapenems) | Macrolides | Phenicols | Aminoglycosides | Tetracyclines | Fluoroquinolones | Polymyxins | Lincosamides/glycopeptides | Sulfonamides | ||||||
| Swine semen | 2000 | USA | 6 | NCCLS M31-A, 1999; disk diffusion | AMP (100) | ERY (100) and TIL (100) | GEN and SPT (100) | OTC (100) | CL (0) | Triple sulfa (100) | 21 | |||
| Omani goats | 2003 | Oman | 15 | NCCLS M2-A4, 1992; disk diffusion | PEN, AMP, AMC, and TIC (100) CAZ, CTX, and CEP (100) | ERY (0) | CHL (0) | KAN, GEN, and AMK (0) | TET (0) | ENO (0) | SXT (0) | 25 | ||
| Salmon farm | 2003 | Chile | 1 | NCCLS M7-A5, 1998; agar dilution | OTC (R), DOX (R), MIN (S) | 19 | ||||||||
| Yellowtail (Seriola quinqueradiata) | 2005 | Japan | 6 | Sensi-Disks (Showa, Tokyo, Japan); disk diffusion | AMP (100)CTX and CAZ (100) | 17 | ||||||||
| 13-lined ground squirrel | 2007 | USA | 1 | Clinical Microbiology Procedures Handbook; broth microdilution | AMP and AMX (R) | CHL (R) | GEN and SPT (R) | TET (R) | 14, 113 | |||||
| Captive snakesc | 2007 | Czech Republic | 47 | NCCLS M2-A8, 2003; breakpoints from National Reference Laboratory for Antibiotics (National Institute of Public Health, Prague, Czech Republic); broth microdilution | AMP (87.2), ATM (89.4), CAZ (68.1), CFP (63.8), CFZ (95.7), CPS (51.1), CTX (85.1), CXM (95.7), FEP (80.9), FOX (95.7), MEM (74.5), PIP (48.3), SAM (68.1), TZP (36.2) | CHL (61.7) | AMK (31.9), GEN (25.5), TOB (57.4) | TET (89.4) | LVX (0), OFX (2.1), CIP (42.6) | CL (21.3) | SXT (2.1) | 15 | ||
| Horse, cat, dog, and python | 2009 | Germany | 7 | Automated susceptibility test strips ATB PSE 5 and ATB VET strips (BioMérieux); microdilution | TIC, PIP, IPM, and CAZ (100) | CHL (28.6) | AMK (42.9), GEN (71.4), TOB (57.1) | TET (100) | CIP and ENO (0) | CL (0) | SXT (14.3) | 31 | ||
| Giant panda | 2010 | USA | 1 | Unknown | AMC, AMP, CAZ, CTX, CEF, and CEP (R) | AZI (R) | CHL (S) | GEN, NEO, and TOB (R) | DOX (S) OTC and TET (R) | CIP and ENO (S) | CL (R) | CLI and VAN (R) | SXT (I) | 30 |
| Captive snakes | 2010 | Czech Republic | 45 | CLSI M100-S19, 2009; broth microdilution | CAZ (44.4) | CHL (28.9) | LVX (0) | SMX (2.2) | 37 | |||||
| Buffalo (Bubalus bubalis) | 2010 | Greece | 1 | CLSI M100-S15, 2005; breakpoints of Pseudomonas spp. used; broth microdilution | TIC and PIP (R)CAZ and IPM (R) | CHL (S) | AMK, GEN, and TOB (R) | TET (R) | CIP and ENO (S) | CL (S) | SXT (S) | (29) | ||
| Horse | 2010 | Denmark | 7 | CLSI M100-S13, 2003; broth microdilution | PEN, AMP, and AMC (100)CF, CPD, and IPM (100) | ERY (100) | GEN and AMK (100) | TET (0) | MAR and ENO (0) | SXT (0) | 32 | |||
| Oocytes of Xenopus laevis | 2011 | USA | 5 | Unknown; disk diffusion | AMX, AMC, and TIC (100), CZ, CF, CTX, CPD, CEF, CXM, and CN (100) CRO (80), CAZ and IPM (0) | CHL (100) | GEN and TOB (100) AMK (0) | TET (100) | CIP (0)DIF, ENO, OFX, and OBX (100) MAR (80) | 24 | ||||
| Milk and Cheese | 2012 | France | 3 | Antibiogram Committee of the French Microbiology Society (CA-SFM), 2008/2009; disk diffusion | AM, PIP, AMX, AMC, TIM, CTX, and CAZ (100)IPM (66.7) | CHL (100) | TET (100) | 13 | ||||||
| Pig | 2012 | China | 7 | Unknown; disk diffusion | AMP, AMX, and novobiocin (100) CTX and CAZ (100) | GEN and STR (100) | S3 and TMP (100) | 10 | ||||||
| Bovine mastitis | 2012 | Japan | 13 | CLSI M31-A3 (2008) and M100-S21 (2011); commercially prepared microtiter panel (Opt Panel MP) and disk diffusion | MOX (0), CAZ (92.3) | CHL (7.7) | CIP (7.7)ENO (0) | SXT (15.4) | 27 | |||||
| Pig | 2015 | China | 1 | CLSI VET01-A4 (2013) and M100-S24 (2014); broth microdilution | AMP, AMC, CEF, CAZ, and MEM (R) | ERY and AZI (R) | CHL and FFC (R) | GEN, STR, and SPT (R) | TET and DOX (R) | ENO (R), LVX (I), CIP (R) | CL (R) | SMX and SXT (R) | 33 | |
For more than one strain, the resistance rate was calculated, and the susceptibility results were interpreted as resistant/intermediate/susceptible (R/I/S) for single strains. CLSI breakpoints were only available for S. maltophilia from humans for SXT, MIN, LEV, TIM, CAZ, SXT, and CHL determined using disk diffusion or dilution methods. For other antimicrobials, the breakpoints for Enterobacteriaceae or Pseudomonas spp. were used to interpret the susceptibility results for S. maltophilia.
PEN, penicillin G; AMP, ampicillin; AMX, amoxicillin; PIP, piperacillin; TIC, ticarcillin; TIM, ticarcillin/clavulanic acid; AMC, amoxycillin/clavulanic acid; SAM, ampicillin/sulbactam; TZP, piperacillin/tazobactam; CAZ, ceftazidime; CTX, cefotaxime; CEF, ceftiofur; CEP, cephalothin; CFZ, cefazolin; CFP, cefoperazone; CN, cephalexin; CRO, ceftriaxone; CPS, cefoperazone/sulbactam; CF, cephalothin; CXM, cefuroxime; FEP, cefepime; FOX, cefoxitin; CPD, cefpodoxime; CZ, cefazolin; MOX, moxalactam; IPM, imipenem; MEM, meropenem; ERY, erythromycin; TIL, tilmicosin; AZI, azithromycin; CHL, chloramphenicol; FFC, florfenicol; GEN, gentamicin; KAN, kanamycin; AMK, amikacin; SPT, spectinomycin; STR, streptomycin; NEO, neomycin; TOB, tobramycin; TET, tetracycline; DOX, doxycycline; OTC, oxytetracycline; MIN, minocycline; CIP, ciprofloxacin; LVX, levofloxacin; OFX, ofloxacin; ENO, enrofloxacin; MAR, marbofloxacin; DIF, difloxacin; OFX, ofloxacin; OBX, orbifloxacin; CL, colistin; CLI, clindamycin; VAN, vancomycin; S3, sulfonamides; SMX, sulfamethoxazole; TMP, trimethoprim; SXT, trimethoprim-sulfamethoxazole.
Resistance rates varied with incubation temperature (30°C or 37°C) and time (24 h or 48 h). Susceptibility data presented here were determined when isolates were incubated at 37°C for 24 h.
Available data are limited for the antimicrobial susceptibility of S. maltophilia, because it is not considered as a major pathogen in animals. However, S. maltophilia isolates from animals are resistant to numerous antimicrobials that are commonly used in human and veterinary medicine, including β-lactams (penicillins and cephalosporins), aminoglycosides, macrolides, and tetracyclines (except minocyline) (Table 2). In contrast, they are often susceptible to fluoroquinolones, polymyxins (mainly including polymyxin B and polymyxin E [colistin]), and SXT. The antibiotic resistance of S. maltophilia varies among different animal species. For example, one isolate from swine in China showed high resistance to most antimicrobials, including fluoroquinolones, polymyxins, and SXT (33), whereas isolates from Omani goats were susceptible to all tested antimicrobials except β-lactams (25). Despite its intrinsic resistance to β-lactams, the resistance rates of S. maltophilia isolates from captive snakes to these antimicrobials range from 36.2 to 95.7% (15, 37). Moreover, antimicrobial resistance varies with the incubation temperature and time. For instance, the MICs at 37°C and 30°C (after 24 h or 48 h) of 24 antibiotics were determined (microdilution method) for S. maltophilia isolates from captive snakes, but resistance rates increased when the strains were incubated at 30°C or for 48 h (37). However, SXT and levofloxacin were the most effective drugs at both temperatures. In addition, the S. maltophilia isolates from animal products also exhibit a multidrug-resistant (MDR) phenotype. For example, S. maltophilia was the most frequently isolated species among a large collection of Gram-negative bacteria isolated from milk and cheese in France. These S. maltophilia isolates showed high resistance rates to β-lactams, chloramphenicol, and tetracycline (13), representing a potential risk to food safety and public health.
MOLECULAR MECHANISMS OF ANTIMICROBIAL RESISTANCE
S. maltophilia employs an array of mechanisms that singularly or collectively, intrinsic or acquired contribute to antimicrobial resistance (Table 3). The following subsections provide detailed descriptions of the major mechanisms.
TABLE 3.
Molecular mechanisms of antimicrobial resistance of S. maltophiliaa
| Resistance mechanisms and related genes | Products | Antibiotic resistance phenotype | Intrinsic/acquired resistance | Gene location | Ref. |
|---|---|---|---|---|---|
| Multidrug efflux pumps | |||||
| smrA | ABC-type efflux pump | Fluoroquinolones, tetracycline, doxorubicin | NK/yes | C | 38 |
| fuaABC | ABC-type efflux pump | Fusaric acid | Yes/no | C | 41 |
| macABCsm | ABC-type efflux pump | Macrolides, aminoglycosides, polymyxins | Yes/NK | C | 39 |
| emrCABsm | MFS-type efflux pump | Nalidixic acid, erythromycin | No/yes | C | 40 |
| mfsA | MFS-type efflux pump | Aminoglycosides, cephalosporins, fluorpquinolones, erythromycin, rifampicin, tetracycline, chloramphenicol | Yes/NK | C | 50 |
| smeABC | RND-type efflux pump | β-lactams, aminoglycosides and quinolones | No/yes | C | 42 |
| smeDEF | RND-type efflux pump | Quinolones, tetracyclines, macrolides, chloramphenicol, novobiocin, SXT | Yes/yes | C | 43, 53 |
| smeVWX | RND-type efflux pump | Chloramphenicol, quinolones, tetracyclines | No/yes | C | 44 |
| smeIJK | RND-type efflux pump | Aminoglycosides, tetracyclines, fluorpquinolones, leucomycin | Yes/yes | C | 46, 55 |
| smeYZ | RND-type efflux pump | Aminoglycosides, SXT | Yes/yes | C | 45, 46, 54 |
| smeOP-TolCSm | RND-type efflux pump | Nalidixic acid, doxycycline, aminoglycosides, macrolides | Yes/no | C | 47 |
| β-lactamases | |||||
| blaL1 | Metallo-β-lactamase | β-Lactams except monobactams | Yes/yes | C or P | 56, 97 |
| blaL2 | Cephalosporinase | Penicillins and cephalosporins | Yes/yes | C | 57, 97 |
| blaTEM-2, blaTEM-116, blaTEM-127, blaCTX-M-1, blaSHV-1 and blaCTX-M-15 | β-lactamase | Penicillins and/or cephalosporins | No/yes | P | 62–65 |
| blaNDM-1 | Metallo-β-lactamase | β-Lactams except monobactams | No/yes | C | 66 |
| Aminoglycoside-inactivating enzymes | |||||
| aac(6′)-Iz | Aminoglycoside acetyltransferase | Amikacin, netilmicin, sisomicin, tobramycin | Yes/no | C | 67 |
| aph(3′)-IIc | Aminoglycoside phosphotransferase | Kanamycin, neomycin, butirosin, paromomycin | Yes/no | C | 68 |
| aac(6′)-Iak | Aminoglycoside acetyltransferase | Amikacin, arbekacin, dibekacin, isepamicin, kanamycin, neomycin, netilmicin, sisomicin, tobramycin | Yes/no | C | 69 |
| aac(6′)-Iam | Aminoglycoside acetyltransferase | NK | NK | C | 45 |
| Qnr family | |||||
| Smqnr | Pentapeptide repeat proteins | Low-level quinolone resistance | Yes/no | C | 76–78 |
| SXT resistance | |||||
| sul1 and sul2 | Folate reductase enzyme | Trimethoprim/sulfamethoxazole | No/yes | C or P | 82–84 |
| dfrA1, dfrA5, dfrA12, dfrA17, and dfrA27 | Dihydrofolate reductase enzyme | Trimethoprim/sulfamethoxazole | No/yes | C or P | 85 |
| Phenicol exporters | |||||
| floR | MFS exporter protein | Chloramphenicol, florfenicol | No/yes | P | 83 |
| floRv | MFS exporter protein | Chloramphenicol, florfenicol | No/yes | GI in C | 33 |
| cmlA | MFS exporter protein | Chloramphenicol | No/yes | I-integron | 90 |
| Lipopolysaccharide | |||||
| spgM | Phosphoglucomutase | Polymyxin B/E, nalidixic acid, gentamicin | Yes/NK | C | 92 |
| phoPQ | Two-component regulatory system | Polymyxin B, chloramphenicol, ampicillin, aminoglycosides | Yes/yes | C | 96 |
NK, not known.
Multidrug Efflux Pumps
The genome of S. maltophilia encodes multidrug efflux pumps, which contribute to intrinsic or acquired antibiotic resistance, as follows: ATP-binding cassette (ABC)-type (SmrA, FuaABC, and MacABCsm), major facilitator superfamily (MFS)-type (EmrCABsm, MsfA), and eight predicted resistance nodulation cell division (RND)-type efflux systems with SmeABC, SmeDEF, SmeVWX, SmeIJK, SmeYZ, and SmeOP-TolCSm characterized (38–47) and SmeMN and SmeGH uncharacterized (45). Most of the efflux pumps are superficially quiescent or expressed at low levels (39, 42, 44), and their overexpression is associated with reduced antibiotic susceptibility. Acquired resistance may be due to mutations in regulatory genes of these efflux systems (43, 46, 48).
SmrA, the first ABC-type efflux pump identified in S. maltophilia, confers acquired resistance to fluoroquinolones, tetracycline, doxorubicin, and multiple dyes (38). FuaABC, a fusaric acid (5-butylpicolinic acid, a mycotoxin) efflux pump, which is classified as a member of a subfamily of the ABC-type family, is induced by fusaric acid and contributes to fusaric acid resistance when overexpressed (41). The MacABCsm efflux pump confers intrinsic resistance to aminoglycosides, macrolides, and polymyxins and contributes to oxidative and envelope stress tolerance as well as biofilm formation (39). The MFS-type pump EmrCABsm is involved in the extrusion of hydrophobic compounds, including the antibiotics nalidixic acid and erythromycin, as well as the uncoupling agents carbonyl cyanide 3-chlorophenylhydrazone, and tetrachlorosalicylanilide (40). A novel MFS efflux pump (MfsA) with 14 transmembrane domains plays an important role in mediating resistance to paraquat (49), as well as to antibiotics such as aminoglycosides (kanamycin, streptomycin, and neomycin), cephalosporins (cefazolin and cefalexin), fluoroquinolones (ciprofloxacin, norfloxacin, levofloxacin, and ofloxacin), the macrolide erythromycin, rifampicin, tetracycline, and chloramphenicol (50).
SmeABC is involved in acquired, but not intrinsic, resistance to β-lactams, aminoglycosides, and quinolones. The deletion of smeC (encoding a porin) affects susceptibility to certain antibiotics, suggesting the relationship of porin to other unidentified efflux pumps (42). SmeDEF is involved in intrinsic and acquired (in the condition of overexpression) resistance to quinolones, tetracyclines, macrolides, chloramphenicol, novobiocin, and SXT, as well as acquired resistance to triclosan (51–53). SmeVWX mediates acquired resistance to chloramphenicol, quinolones, and tetracyclines and when overexpressed, increases susceptibility to aminoglycosides (44). SmeYZ mediates intrinsic resistance to aminoglycosides and SXT (45, 54), while SmeIJK is involved in intrinsic reduced susceptibility to gentamicin, amikacin, tetracycline, minocycline, ciprofloxacin, and leucomycin (45, 55). SmeIJK also mediated acquired resistance to levofloxacin, when overexpressed alone or in coordinate hyperproduction with SmeYZ (46). The activity of the SmeOP-TolCSm efflux pump is associated with the decreases in susceptibility to nalidixic acid, doxycycline, aminoglycosides (amikacin and gentamicin), and macrolides (erythromycin and leucomycin), as well as several nonantibiotic compounds including carbonyl cyanide 3-chlorophenylhydrazone, crystal violet, sodium dodecyl sulfate, and tetrachlorosalicylanilide (47).
Resistance to β-Lactam Antibiotics
The S. maltophilia genome encodes the inducible β-lactamases L1 and L2. L1 is a class B Zn2+-dependent metallo-β-lactamase with substrate preference for penicillins, cephalosporins, and carbapenems, except for monobactams; and L2 is a class A clavulanic acid-sensitive cephalosporinase that hydrolyzes penicillins, cephalosporins, and monobactams (56, 57). The expression of L1 and L2 is simultaneously regulated by AmpR, a transcriptional regulator encoded by ampR, located upstream of blaL2, which acts as a weak repressor or activator of the blaL2 in the presence or absence of β-lactam antibiotics, respectively (58). The induction of β-lactamases is inhibited by the deletion of the ampN-ampG operon, which encodes a permease transporter (59). The hyperproduction of L1/L2 β-lactamases occurs when the transcription of mrcA or ampDI (encoding penicillin-binding protein 1a [PBP1a] and a cytoplasmic N-acetyl-muramyl-l-alanine amidase [AmpDI], respectively) is inhibited (60, 61). In addition, the β-lactamases TEM-2, TEM-116, TEM-127, CTX-M-1, SHV-1, and CTX-M-15 and the globally disseminated metallo-β-lactamase NDM-1 are present in human clinical and environmental isolates of S. maltophilia (62–66), suggesting that this pathogen may serve as a reservoir for mobile genes that encode β-lactamases.
Resistance to Aminoglycosides
The mechanisms employed by S. maltophilia that mediate resistance to aminoglycosides primarily involve aminoglycoside-modifying enzymes and multidrug efflux pumps. These enzymes include the aminoglycoside acetyltransferase AAC(6′)-Iz (67) and the aminoglycoside phosphotransferase APH(3′)-IIc (68), both of which confer low-level resistance to aminoglycosides, with the exception of gentamicin. The novel aminoglycoside acetyltransferase AAC(6′)-Iak, which exhibits 86.3% amino acid identity to AAC(6′)-Iz, is expressed by an MDR S. maltophilia strain isolated from Nepal and acetylates amikacin, arbekacin, dibekacin, isepamicin, kanamycin, neomycin, netilmicin, sisomicin, and tobramycin, but not apramycin, gentamicin, or lividomycin (69). Moreover, AAC(6′)-Iam [84.3% amino acid sequence identity to AAC(6′)-Iak], was detected in a clinical isolate of S. maltophilia (45). However, the resistance phenotype conferred by this enzyme is unknown. In addition, the efflux pumps SmeABC, SmeYZ, SmeOP-TolCsm, and MacABCsm are associated with aminoglycoside resistance (Table 3).
Resistance to Quinolones
Mutations in the quinolone-resistance-determining region of genes encoding topoisomerases (gyrA, gyrB, parC, and parE) are associated with the major mechanism of quinolone resistance employed by bacteria (70). So far, mutations have not been detected in the quinolone-resistance-determining region of gyrA of S. maltophilia (71, 72). Amino acid residue substitutions are present in the quinolone-resistance-determining region-encoding regions of gyrB, parC, and parE of clinical isolates of S. maltophilia that cause bacteremia; however, these alterations have not been directly associated with quinolone resistance (73). The specific mechanisms associated with the quinolone resistance of S. maltophilia are mediated by both the efflux pumps and the chromosomal qnr gene (Smqnr) that protects gyrase and topoisomerase IV from quinolones (74). Smqnr and its functional 12 variants belong to the qnr family (75) and contribute to low-level intrinsic quinolone resistance (76–78). Genes that encode efflux pumps that mediate quinolone resistance are as follows: smeDEF, smeIJK, smeABC, and smeVWX (Table 3). The most prevalent cause of quinolone resistance in S. maltophilia is the overproduction of multidrug efflux pumps, among which the SmeDEF plays the most important role (79). Furthermore, overexpression of smeVWX in clinical isolates of S. maltophilia is associated with high-level resistance to quinolones (80).
Resistance to Trimethoprim-Sulfamethoxazole
The resistance of Gram-negative bacteria to sulfonamides is mainly conferred by the acquisition of either sul1 or sul2, encoding dihydropteroate synthases (81). The sul1 gene carried by class 1 integrons and sul2, which is linked to insertion sequence common region (ISCR) elements, was identified in SXT-resistant S. maltophilia isolates (82–84). The resistance of S. maltophilia to trimethoprim is mainly conferred by the dihydrofolate reductase dfr genes. For instance, the dfrA variant genes (dfrA1, dfrA5, dfrA12, dfrA17, and dfrA27), which are located within class 1 integrons as part of various resistance gene cassettes, are associated with high-level trimethoprim resistance in S. maltophilia isolates. Both types of sul and dfr genes can occur together in high-level SXT-resistant isolates (85, 86). Moreover, the efflux pumps SmeDEF, TolCsm, and SmeYZ are associated with SXT resistance (54, 87, 88).
Resistance to Phenicols
The main phenicol resistance determinant in S. maltophilia is floR, which encodes an exporter protein of the MFS family that mediates resistance to chloramphenicol and florfenicol (83). Florfenicol is extensively used in livestock to prevent or cure bacterial infections (89). In addition, the MFS exporter gene cmlA1 and chloramphenicol acetyltransferase genes catB2 and catB8, which separately reside in a gene cassette of class 1 integrons, confer resistance to chloramphenicol in S. maltophilia (82, 85, 90). Reports of the prevalence of floR in S. maltophilia are rare. One report that investigated an international collection of 55 clinical isolates of S. maltophilia found that four strains harbored floR (83). The novel variant floRv was detected in one porcine S. maltophilia isolate in China. The floRv gene encodes an exporter protein of 404 amino acids, which is 84.1 to 91.8% identical to FloR sequences deposited in GenBank. This FloR variant mediates resistance to chloramphenicol and florfenicol (33).
Alteration of Lipopolysaccharide and Two-Component Regulatory Systems
As in other Gram-negative bacteria, lipopolysaccharide (LPS) is an important structural component of the outer membrane of S. maltophilia and forms an effective barrier to exogenous compounds (91). The spgM gene encodes a phosphoglucomutase that is associated with LPS biosynthesis in S. maltophilia (92). Mutants lacking spgM, which produce less LPS compared with the SpgM+ strain, synthesize shorter O-polysaccharide chains and exhibit modest increases in susceptibility to polymyxin B, colistin, nalidixic acid, and gentamicin but increased resistance to vancomycin (92). The mobile colistin resistance gene mcr-1, which encodes a phosphoethanolamine transferase, couples phosphoethanolamine to the lipid A domain of the LPS component of the outer membrane of Gram-negative bacteria, and negates the efficacy of polymixins (93), has not been detected in Stenotrophomonas spp. The two-component regulatory system PhoPQ is involved in the resistance of numerous Gram-negative bacteria, including S. maltophilia, to cationic antimicrobial polypeptides, i.e., polymyxin B (94–96). Mutation of S. maltophilia PhoP increases susceptibility to polymyxin B, chloramphenicol, ampicillin, gentamicin, kanamycin, streptomycin, and spectinomycin (96). Moreover, downregulation of the SmeZ efflux transporter expressed by a PhoP mutant contributes to increased drug susceptibility, particularly to aminoglycosides (96).
DISSEMINATION, COSELECTION, AND PERSISTANCE OF RESISTANCE DETERMINANTS
As described above, the reduced susceptibility of S. maltophilia to most antibiotics can be attributed to intrinsic and acquired resistance. The proteins mediating intrinsic resistance of S. maltophilia include chromosomally encoded multidrug efflux pumps, antibiotic-inactivating enzymes (L1/L2 β-lactamases and aminoglycoside-inactivating enzymes), and the chromosomally encoded Qnr pentapeptide repeat proteins (74), which are present in most, if not all, strains of S. maltophilia, suggesting they did not arise during the recent evolution of resistance caused by antibiotic therapy. In addition, S. maltophilia can acquire mechanisms to increase its resistance through horizontal gene transfer via integrons, transposons, plasmids, and genomic islands (GIs). The sul1 gene is always associated with the class 1 integron in S. maltophilia, indicating the role of the latter in the acquisition and dissemination of sul1 within this species (82–86, 90). The qacEΔ1 gene, which encodes resistance to quaternary amines, coexists with sul1 at the 3′-termini of class 1 integrons (83, 85, 90). The gene cassettes, which comprise the variable regions of integrons, integrate different combinations of drug-resistance genes donated by other Gram-negative bacteria, including those encoding resistance to aminoglycosides [aacA4, aacA7, aadA1, aadA2, aadA4, aadA5, aadB, aac(6′)-II, aac(6′)-Ib, aac(3′)-Ia, and ant(3″)-Ia], trimethoprim (dfrA1, dfrA5, dfrA12, dfrA17 and dfrA27), β-lactams (blaCARB-8), rifampicin (arr-3), and chloramphenicol (catB2, catB8, cmlA1) (82, 85, 90).
ISCR elements are frequently associated with antimicrobial resistance genes and are always linked to sul2 in S. maltophilia. For example, seven sul2-positive S. maltophilia isolates harbor ISCR elements (five ISCR2 and two ISCR3 elements) on a plasmid (83). Moreover, sul2 and floR are linked to ISCR2 in all sul2-positive S. maltophilia isolates. Constitutively expressed blaTEM-2 resides within a novel Tn1/Tn3-type transposon in the genome of S. maltophilia isolate J675Ia (65). The transposon could mobilize blaTEM-2 onto the broad host-range conjugative plasmid R388, which is then transferred to E. coli.
The genes encoding β-lactamases L1 and L2 are invariably chromosomal and reside on an approximately 200-kb plasmid present in 10 clinical isolates of S. maltophilia (97). However, the sequences of the L1 and L2 genes diverge from that of the published strain IID 1275, indicating that the presence of β-lactamase genes on a plasmid may lead to their relatively quick evolution (97).
A literature search identified only a single report of an MDR GI in the S. maltophilia isolate GZP-Sm1 in China (33). GZP-Sm1 was isolated from swine with septicemia, and susceptibility testing revealed that the isolate was resistant to most antimicrobials employed in human and veterinary clinical practice (33). Whole-genome sequencing identified a GI of 40,226 bp, which contains an MDR region (19,364 bp) and is flanked by IS26 in opposite orientations (Fig. 1). Furthermore, six resistance genes exist in this region, including floRv (phenicol resistance), tet(A)-tetR (tetracycline resistance), strA/strB (streptomycin resistance), sul1 (sulfonamide resistance), and aadA2 (streptomycin/spectinomycin resistance). The MDR region comprises several segments with sequence similarity to plasmids or chromosomal sequences of other Gram-negative bacteria. For example, the aadA2 cassette and the 3′-CS region (qacEΔ1-sul1-Δorf5), which form part of an integron structure identified in this GI, occur in diverse bacterial species such as Salmonella spp., Pseudomonas spp., and E. coli. The 4,766-bp segment of Δsul-floRv-lysR-traG is 86.3% identical to the corresponding region of plasmid pAB (accession no. HQ917128) detected in a clinical isolate of Acinetobacter baumannii from Chile. The composite transposon comprising IS26-tet(A)-tetR-IS26 flanked by a direct repeat of GC is 95.1% identical to the corresponding region of the plasmid pB12 from uncultivable bacteria (accession no. JX469826). Inverse PCR showed that the GI could be excised from the chromosome by recombination between the direct repeats to generate a circular extrachromosomal form (Fig. 1). The emerging resistance of S. maltophilia to numerous antimicrobials raises the concern that the presence of resistance genes in the novel MDR GI drastically limit therapeutic options and may enhance their coselection when antimicrobials are administered.
FIGURE 1.
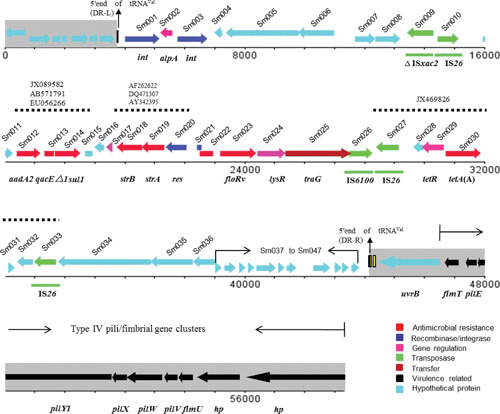
Linear representation of the complete GI and its flanking regions in S. maltophilia GZP-Sm1. The regions in gray represent the flanking regions of the GI when inserted into the bacterial chromosome. The arrows indicate the directions of gene transcription, and truncated genes are indicated by rectangles without arrowheads. Genes are depicted in different colors, and the regions of particular relevance (≥95% nucleotide sequence identity) are indicated by the dotted lines (33).
S. maltophilia could acquire antibiotic resistance from Gram-positive bacteria. For example, a gene cluster involved in resistance to antibiotics and heavy metals was detected in a clinical isolate of S. maltophilia (98). These genes encode a macrolide phosphotransferase (mphBM) and a cadmium efflux determinant (cadA), as well as its transcriptional regulator (cadC), encoding its cognate transcriptional regulator. The cadC-cadA region is flanked by a truncated IS257 sequence and a region coding for a bin3 invertase. The sequences of these genetic elements are highly similar to those of Staphylococcus aureus, indicating their Gram-positive origin.
CONCLUSION
S. maltophilia is the most widely distributed environmental species among Stenotrophomonas, and it is also an opportunistic pathogen associated with the increased number of infections in both humans and animals. S. maltophilia isolates from animals are resistant to most antimicrobials used in both human and veterinary medicine, which compromise the design of optimal therapeutic strategies in clinical chemotherapy. The antimicrobial resistances in S. maltophilia are conferred not only by intrinsic mechanisms, but also by multiple acquired resistance mechanisms, which are commonly associated with mobile genetic elements such as integrons, transposons, and plasmids. Moreover, for the first time, the transmission mechanism conferred by MDRGI was identified in a porcine S. maltophilia isolate. Therefore, continued surveillance of MDR S. maltophilia from animals is warranted for not only optimizing treatment of infections caused by this bacterium, but also tackling the transmission of antimicrobial resistance from animals to humans by either food-chain or environmental routes.
ACKNOWLEDGEMENT
This work was supported in part by the National Natural Science Foundation of China (grant no. 31422055) and the National Key Basic Research Program of China (grant no. 2013CB127200).
REFERENCES
- 1.http://www.bacterio.net/stenotrophomonas.html. [PubMed]
- 2.Swings J, De Vos P, Van den Mooter M, De Ley J. 1983. Transfer of Pseudomonas maltophilia Hugh 1981 to the genus Xanthomonas as Xanthomonas maltophilia. Int J Syst Bacteriol 33:409–413 10.1099/00207713-33-2-409. [DOI] [Google Scholar]
- 3.Drancourt M, Bollet C, Raoult D. 1997. Stenotrophomonas africana sp. nov., an opportunistic human pathogen in Africa. Int J Syst Bacteriol 47:160–163 10.1099/00207713-47-1-160. [PubMed] [DOI] [PubMed] [Google Scholar]
- 4.Brooke JS. 2012. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev 25:2–41 10.1128/CMR.00019-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan RP, Monchy S, Cardinale M, Taghavi S, Crossman L, Avison MB, Berg G, van der Lelie D, Dow JM. 2009. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat Rev Microbiol 7:514–525 10.1038/nrmicro2163. [PubMed] [DOI] [PubMed] [Google Scholar]
- 6.Hugh R, Ryschenkow E. 1961. Pseudomonas maltophilia, an alcaligenes-like species. J Gen Microbiol 26:123–132 10.1099/00221287-26-1-123. [PubMed] [DOI] [PubMed] [Google Scholar]
- 7.Chien YC, Chen CJ, Lin TH, Chen SH, Chien YC. 2011. Characteristics of microbial aerosols released from chicken and swine feces. J Air Waste Manag Assoc 61:882–889 10.3155/1047-3289.61.8.882. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Guo Y, Zhu N, Zhu S, Deng C. 2007. Molecular phylogenetic diversity of bacteria and its spatial distribution in composts. J Appl Microbiol 103:1344–1354 10.1111/j.1365-2672.2007.03367.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 9.Mancini DA, Mendonça RM, Dias AL, Mendonça RZ, Pinto JR. 2005. Co-infection between influenza virus and flagellated bacteria. Rev Inst Med Trop São Paulo 47:275–280 10.1590/S0036-46652005000500007. [PubMed] [DOI] [PubMed] [Google Scholar]
- 10.Hou D, Bi Y, Sun H, Yang J, Fu G, Sun Y, Liu J, Pu J. 2012. Identification of swine influenza A virus and Stenotrophomonas maltophilia co-infection in Chinese pigs. Virol J 9:169 10.1186/1743-422X-9-169. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munsch-Alatossava P, Alatossava T. 2006. Phenotypic characterization of raw milk-associated psychrotrophic bacteria. Microbiol Res 161:334–346 10.1016/j.micres.2005.12.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 12.Cleto S, Matos S, Kluskens L, Vieira MJ. 2012. Characterization of contaminants from a sanitized milk processing plant. PLoS One 7:e40189 10.1371/journal.pone.0040189. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coton M, Delbés-Paus C, Irlinger F, Desmasures N, Le Fleche A, Stahl V, Montel MC, Coton E. 2012. Diversity and assessment of potential risk factors of Gram-negative isolates associated with French cheeses. Food Microbiol 29:88–98 10.1016/j.fm.2011.08.020. [PubMed] [DOI] [PubMed] [Google Scholar]
- 14.Cloud-Hansen KA, Villiard KM, Handelsman J, Carey HV. 2007. Thirteen-lined ground squirrels (Spermophilus tridecemlineatus) harbor multiantibiotic-resistant bacteria. J Am Assoc Lab Anim Sci 46:21–23. [PubMed] [PubMed] [Google Scholar]
- 15.Hejnar P, Bardon J, Sauer P, Kolár M. 2007. Stenotrophomonas maltophilia as a part of normal oral bacterial flora in captive snakes and its susceptibility to antibiotics. Vet Microbiol 121:357–362 10.1016/j.vetmic.2006.12.026. [PubMed] [DOI] [PubMed] [Google Scholar]
- 16.Kim M, Kwon TH, Jung SM, Cho SH, Jin SY, Park NH, Kim CG, Kim JS. 2013. Antibiotic resistance of bacteria isolated from the internal organs of edible snow crabs. PLoS One 8:e70887 10.1371/journal.pone.0070887. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furushita M, Okamoto A, Maeda T, Ohta M, Shiba T. 2005. Isolation of multidrug-resistant Stenotrophomonas maltophilia from cultured yellowtail (Seriola quinqueradiata) from a marine fish farm. Appl Environ Microbiol 71:5598–5600 10.1128/AEM.71.9.5598-5600.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matyar F, Kaya A, Dinçer S. 2008. Antibacterial agents and heavy metal resistance in Gram-negative bacteria isolated from seawater, shrimp and sediment in Iskenderun Bay, Turkey. Sci Total Environ 407:279–285 10.1016/j.scitotenv.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 19.Miranda CD, Kehrenberg C, Ulep C, Schwarz S, Roberts MC. 2003. Diversity of tetracycline resistance genes in bacteria from Chilean salmon farms. Antimicrob Agents Chemother 47:883–888 10.1128/AAC.47.3.883-888.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miranda CD, Zemelman R. 2002. Antimicrobial multiresistance in bacteria isolated from freshwater Chilean salmon farms. Sci Total Environ 293:207–218 10.1016/S0048-9697(02)00022-0. [DOI] [PubMed] [Google Scholar]
- 21.Althouse GC, Kuster CE, Clark SG, Weisiger RM. 2000. Field investigations of bacterial contaminants and their effects on extended porcine semen. Theriogenology 53:1167–1176 10.1016/S0093-691X(00)00261-2. [DOI] [PubMed] [Google Scholar]
- 22.Althouse GC, Lu KG. 2005. Bacteriospermia in extended porcine semen. Theriogenology 63:573–584 10.1016/j.theriogenology.2004.09.031. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.Kilburn C, Rooks DJ, McCarthy AJ, Murray RD. 2013. Antimicrobial resistance in some Gram-negative bacteria isolated from the bovine ejaculate. Reprod Domest Anim 48:525–528 10.1111/rda.12127. [PubMed] [DOI] [PubMed] [Google Scholar]
- 24.O’Connell D, Mruk K, Rocheleau JM, Kobertz WR. 2011. Xenopus laevis oocytes infected with multi-drug-resistant bacteria: implications for electrical recordings. J Gen Physiol 138:271–277 10.1085/jgp.201110661. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson EH, Al-Busaidy R, Hameed MS. 2003. An outbreak of lymphadenitis associated with Stenotrophomonas (Xanthomonas) maltophilia in Omani goats. J Vet Med B Infect Dis Vet Public Health 50:102–104 10.1046/j.1439-0450.2003.00643.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 26.Macdiarmid JA, Burrell DH. 1986. Characterization of Pseudomonas maltophilia isolates from fleece rot. Appl Environ Microbiol 51:346–348. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohnishi M, Sawada T, Marumo K, Harada K, Hirose K, Shimizu A, Hayashimoto M, Sato R, Uchida N, Kato H. 2012. Antimicrobial susceptibility and genetic relatedness of bovine Stenotrophomonas maltophilia isolates from a mastitis outbreak. Lett Appl Microbiol 54:572–576 10.1111/j.1472-765X.2012.03246.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 28.Hagi T, Sasaki K, Aso H, Nomura M. 2013. Adhesive properties of predominant bacteria in raw cow’s milk to bovine mammary gland epithelial cells. Folia Microbiol (Praha) 58:515–522 10.1007/s12223-013-0240-z. [PubMed] [DOI] [PubMed] [Google Scholar]
- 29.Petridou E, Filioussis G, Karavanis E, Kritas SK. 2010. Stenotrophomonas maltophilia as a causal agent of pyogranulomatous hepatitis in a buffalo (Bubalus bubalis). J Vet Diagn Invest 22:772–774 10.1177/104063871002200522. [PubMed] [DOI] [PubMed] [Google Scholar]
- 30.Boedeker NC, Walsh T, Murray S, Bromberg N. 2010. Medical and surgical management of severe inflammation of the nictitating membrane in a giant panda (Ailuropoda melanoleuca). Vet Ophthalmol 13(Suppl):109–115 10.1111/j.1463-5224.2010.00802.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 31.Albini S, Abril C, Franchini M, Hüssy D, Filioussis G. 2009. Stenotrophomonas maltophilia isolated from the airways of animals with chronic respiratory disease. Schweiz Arch Tierheilkd 151:323–328 10.1024/0036-7281.151.7.323. [PubMed] [DOI] [PubMed] [Google Scholar]
- 32.Winther L, Andersen RM, Baptiste KE, Aalbæk B, Guardabassi L. 2010. Association of Stenotrophomonas maltophilia infection with lower airway disease in the horse: a retrospective case series. Vet J 186:358–363 10.1016/j.tvjl.2009.08.026. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.He T, Shen J, Schwarz S, Wu C, Wang Y. 2015. Characterization of a genomic island in Stenotrophomonas maltophilia that carries a novel floR gene variant. J Antimicrob Chemother 70:1031–1036. [PubMed] [DOI] [PubMed] [Google Scholar]
- 34.Harris NB, Rogers DG. 2001. Septicemia associated with Stenotrophomonas maltophilia in a West African dwarf crocodile (Osteolaemus tetraspis subsp. tetraspis). J Vet Diagn Invest 13:255–258 10.1177/104063870101300313. [PubMed] [DOI] [PubMed] [Google Scholar]
- 35.Muir P, Oldenhoff WE, Hudson AP, Manley PA, Schaefer SL, Markel MD, Hao Z. 2007. Detection of DNA from a range of bacterial species in the knee joints of dogs with inflammatory knee arthritis and associated degenerative anterior cruciate ligament rupture. Microb Pathog 42:47–55 10.1016/j.micpath.2006.10.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 36.Clinical and Laboratory Standards Institute. 2015. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Fifth Informational Supplement M-S, CLSI, Wayne, PA.
- 37.Hejnar P, Kolár M, Sauer P. 2010. Antibiotic resistance of Stenotrophomonas maltophilia strains isolated from captive snakes. Folia Microbiol (Praha) 55:83–87 10.1007/s12223-010-0014-9. [PubMed] [DOI] [PubMed] [Google Scholar]
- 38.Al-Hamad A, Upton M, Burnie J. 2009. Molecular cloning and characterization of SmrA, a novel ABC multidrug efflux pump from Stenotrophomonas maltophilia. J Antimicrob Chemother 64:731–734 10.1093/jac/dkp271. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Lin YT, Huang YW, Liou RS, Chang YC, Yang TC. 2014. MacABCsm, an ABC-type tripartite efflux pump of Stenotrophomonas maltophilia involved in drug resistance, oxidative and envelope stress tolerances and biofilm formation. J Antimicrob Chemother 69:3221–3226 10.1093/jac/dku317. [PubMed] [DOI] [PubMed] [Google Scholar]
- 40.Huang YW, Hu RM, Chu FY, Lin HR, Yang TC. 2013. Characterization of a major facilitator superfamily (MFS) tripartite efflux pump EmrCABsm from Stenotrophomonas maltophilia. J Antimicrob Chemother 68:2498–2505 10.1093/jac/dkt250. [PubMed] [DOI] [PubMed] [Google Scholar]
- 41.Hu RM, Liao ST, Huang CC, Huang YW, Yang TC. 2012. An inducible fusaric acid tripartite efflux pump contributes to the fusaric acid resistance in Stenotrophomonas maltophilia. PLoS One 7:e51053 10.1371/journal.pone.0051053. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li XZ, Zhang L, Poole K. 2002. SmeC, an outer membrane multidrug efflux protein of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 46:333–343 10.1128/AAC.46.2.333-343.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alonso A, Martínez JL. 2000. Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob Agents Chemother 44:3079–3086 10.1128/AAC.44.11.3079-3086.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen CH, Huang CC, Chung TC, Hu RM, Huang YW, Yang TC. 2011. Contribution of resistance-nodulation-division efflux pump operon smeU1-V-W-U2-X to multidrug resistance of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 55:5826–5833 10.1128/AAC.00317-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, Sebaihia M, Saunders D, Arrowsmith C, Carver T, Peters N, Adlem E, Kerhornou A, Lord A, Murphy L, Seeger K, Squares R, Rutter S, Quail MA, Rajandream MA, Harris D, Churcher C, Bentley SD, Parkhill J, Thomson NR, Avison MB. 2008. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol 9:R74 10.1186/gb-2008-9-4-r74. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gould VC, Okazaki A, Avison MB. 2013. Coordinate hyperproduction of SmeZ and SmeJK efflux pumps extends drug resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 57:655–657 10.1128/AAC.01020-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin CW, Huang YW, Hu RM, Yang TC. 2014. SmeOP-TolCSm efflux pump contributes to the multidrug resistance of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 58:2405–2408 10.1128/AAC.01974-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho HH, Sung JY, Kwon KC, Koo SH. 2012. Expression of Sme efflux pumps and multilocus sequence typing in clinical isolates of Stenotrophomonas maltophilia. Ann Lab Med 32:38–43 10.3343/alm.2012.32.1.38. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Srijaruskul K, Charoenlap N, Namchaiw P, Chattrakarn S, Giengkam S, Mongkolsuk S, Vattanaviboon P. 2015. Regulation by SoxR of mfsA, which encodes a major facilitator protein involved in paraquat resistance in Stenotrophomonas maltophilia. PLoS One 10:e0123699 10.1371/journal.pone.0123699. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dulyayangkul P, Charoenlap N, Srijaruskul K, Mongkolsuk S, Vattanaviboon P. 2016. Major facilitator superfamily MfsA contributes to multidrug resistance in emerging nosocomial pathogen Stenotrophomonas maltophilia. J Antimicrob Chemother 71:2990–2991 10.1093/jac/dkw233. [PubMed] [DOI] [PubMed] [Google Scholar]
- 51.Zhang L, Li XZ, Poole K. 2001. SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 45:3497–3503 10.1128/AAC.45.12.3497-3503.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hernández A, Ruiz FM, Romero A, Martínez JL. 2011. The binding of triclosan to SmeT, the repressor of the multidrug efflux pump SmeDEF, induces antibiotic resistance in Stenotrophomonas maltophilia. PLoS Pathog 7:e1002103 10.1371/journal.ppat.1002103. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sánchez MB, Martínez JL. 2015. The efflux pump SmeDEF contributes to trimethoprim-sulfamethoxazole resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 59:4347–4348 10.1128/AAC.00714-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin YT, Huang YW, Chen SJ, Chang CW, Yang TC. 2015. The SmeYZ efflux pump of Stenotrophomonas maltophilia contributes to drug resistance, virulence-related characteristics, and virulence in mice. Antimicrob Agents Chemother 59:4067–4073 10.1128/AAC.00372-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang YW, Liou RS, Lin YT, Huang HH, Yang TC. 2014. A linkage between SmeIJK efflux pump, cell envelope integrity, and σE-mediated envelope stress response in Stenotrophomonas maltophilia. PLoS One 9:e111784 10.1371/journal.pone.0111784. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crowder MW, Walsh TR, Banovic L, Pettit M, Spencer J. 1998. Overexpression, purification, and characterization of the cloned metallo-β-lactamase L1 from Stenotrophomonas maltophilia. Antimicrob Agents Chemother 42:921–926. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walsh TR, MacGowan AP, Bennett PM. 1997. Sequence analysis and enzyme kinetics of the L2 serine β-lactamase from Stenotrophomonas maltophilia. Antimicrob Agents Chemother 41:1460–1464. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okazaki A, Avison MB. 2008. Induction of L1 and L2 β-lactamase production in Stenotrophomonas maltophilia is dependent on an AmpR-type regulator. Antimicrob Agents Chemother 52:1525–1528 10.1128/AAC.01485-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang YW, Lin CW, Hu RM, Lin YT, Chung TC, Yang TC. 2010. AmpN-AmpG operon is essential for expression of L1 and L2 β-lactamases in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 54:2583–2589 10.1128/AAC.01283-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang TC, Huang YW, Hu RM, Huang SC, Lin YT. 2009. AmpDI is involved in expression of the chromosomal L1 and L2 β-lactamases of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 53:2902–2907 10.1128/AAC.01513-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin CW, Lin HC, Huang YW, Chung TC, Yang TC. 2011. Inactivation of mrcA gene derepresses the basal-level expression of L1 and L2 β-lactamases in Stenotrophomonas maltophilia. J Antimicrob Chemother 66:2033–2037 10.1093/jac/dkr276. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.al Naiemi N, Duim B, Bart A. 2006. A CTX-M extended-spectrum β-lactamase in Pseudomonas aeruginosa and Stenotrophomonas maltophilia. J Med Microbiol 55:1607–1608 10.1099/jmm.0.46704-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 63.Lavigne JP, Gaillard JB, Bourg G, Tichit C, Lecaillon E, Sotto A. 2008. Extended-spectrum β-lactamases-producing Stenotrophomonas maltophilia strains: CTX-M enzymes detection and virulence study. Pathol Biol (Paris) 56:447–453 10.1016/j.patbio.2008.07.013. (In French.) [PubMed] [DOI] [PubMed] [Google Scholar]
- 64.Maravić A, Skočibušić M, Fredotović Z, Cvjetan S, Samanić I, Puizina J. 2014. Characterization of environmental CTX-M-15-producing Stenotrophomonas maltophilia. Antimicrob Agents Chemother 58:6333–6334 10.1128/AAC.03601-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Avison MB, von Heldreich CJ, Higgins CS, Bennett PM, Walsh TR. 2000. A TEM-2 β-lactamase encoded on an active Tn1-like transposon in the genome of a clinical isolate of Stenotrophomonas maltophilia. J Antimicrob Chemother 46:879–884 10.1093/jac/46.6.879. [PubMed] [DOI] [PubMed] [Google Scholar]
- 66.Liu W, Zou D, Wang X, Li X, Zhu L, Yin Z, Yang Z, Wei X, Han L, Wang Y, Shao C, Wang S, He X, Liu D, Liu F, Wang J, Huang L, Yuan J. 2012. Proteomic analysis of clinical isolate of Stenotrophomonas maltophilia with blaNDM-1, blaL1 and blaL2 β-lactamase genes under imipenem treatment. J Proteome Res 11:4024–4033 10.1021/pr300062v. [PubMed] [DOI] [PubMed] [Google Scholar]
- 67.Li XZ, Zhang L, McKay GA, Poole K. 2003. Role of the acetyltransferase AAC(6′)-Iz modifying enzyme in aminoglycoside resistance in Stenotrophomonas maltophilia. J Antimicrob Chemother 51:803–811 10.1093/jac/dkg148. [PubMed] [DOI] [PubMed] [Google Scholar]
- 68.Okazaki A, Avison MB. 2007. Aph(3′)-IIc, an aminoglycoside resistance determinant from Stenotrophomonas maltophilia. Antimicrob Agents Chemother 51:359–360 10.1128/AAC.00795-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tada T, Miyoshi-Akiyama T, Dahal RK, Mishra SK, Shimada K, Ohara H, Kirikae T, Pokhrel BM. 2014. Identification of a novel 6′-N-aminoglycoside acetyltransferase, AAC(6′)-Iak, from a multidrug-resistant clinical isolate of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 58:6324–6327 10.1128/AAC.03354-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sánchez MB. 2015. Antibiotic resistance in the opportunistic pathogen Stenotrophomonas maltophilia. Front Microbiol 6:658 10.3389/fmicb.2015.00658. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ribera A, Doménech-Sanchez A, Ruiz J, Benedi VJ, Jimenez de Anta MT, Vila J. 2002. Mutations in gyrA and parC QRDRs are not relevant for quinolone resistance in epidemiological unrelated Stenotrophomonas maltophilia clinical isolates. Microb Drug Resist 8:245–251 10.1089/10766290260469499. [PubMed] [DOI] [PubMed] [Google Scholar]
- 72.Valdezate S, Vindel A, Echeita A, Baquero F, Cantó R. 2002. Topoisomerase II and IV quinolone resistance-determining regions in Stenotrophomonas maltophilia clinical isolates with different levels of quinolone susceptibility. Antimicrob Agents Chemother 46:665–671 10.1128/AAC.46.3.665-671.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cha MK, Kang CI, Kim SH, Cho SY, Ha YE, Chung DR, Peck KR, Song JH. 2016. Emergence of fluoroquinolone-resistant Stenotrophomonas maltophilia in blood isolates causing bacteremia: molecular epidemiology and microbiologic characteristics. Diagn Microbiol Infect Dis 85:210–212 10.1016/j.diagmicrobio.2016.02.020. [PubMed] [DOI] [PubMed] [Google Scholar]
- 74.Sanchez MB, Hernandez A, Martinez JL. 2009. Stenotrophomonas maltophilia drug resistance. Future Microbiol 4:655–660 10.2217/fmb.09.45. [PubMed] [DOI] [PubMed] [Google Scholar]
- 75.Gordon NC, Wareham DW. 2010. Novel variants of the Smqnr family of quinolone resistance genes in clinical isolates of Stenotrophomonas maltophilia. J Antimicrob Chemother 65:483–489 10.1093/jac/dkp476. [PubMed] [DOI] [PubMed] [Google Scholar]
- 76.Sánchez MB, Hernández A, Rodríguez-Martínez JM, Martínez-Martínez L, Martínez JL. 2008. Predictive analysis of transmissible quinolone resistance indicates Stenotrophomonas maltophilia as a potential source of a novel family of Qnr determinants. BMC Microbiol 8:148 10.1186/1471-2180-8-148. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sánchez MB, Martínez JL. 2010. SmQnr contributes to intrinsic resistance to quinolones in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 54:580–581 10.1128/AAC.00496-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shimizu K, Kikuchi K, Sasaki T, Takahashi N, Ohtsuka M, Ono Y, Hiramatsu K. 2008. Smqnr, a new chromosome-carried quinolone resistance gene in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 52:3823–3825 10.1128/AAC.00026-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garcia-Leon G, Salgado F, Oliveros JC, Sanchez MB, Martinez JL. 2014. Interplay between intrinsic and acquired resistance to quinolones in Stenotrophomonas maltophilia. Environ Microbiol 16:1282–1296. [PubMed] [DOI] [PubMed] [Google Scholar]
- 80.García-León G, Ruiz de Alegría Puig C, García de la Fuente C, Martínez-Martínez L, Martínez JL, Sánchez MB. 2015. High-level quinolone resistance is associated with the overexpression of smeVWX in Stenotrophomonas maltophilia clinical isolates. Clin Microbiol Infect 21:464–467 10.1016/j.cmi.2015.01.007. [PubMed] [DOI] [PubMed] [Google Scholar]
- 81.Rådström P, Swedberg G. 1988. RSF1010 and a conjugative plasmid contain sulII, one of two known genes for plasmid-borne sulfonamide resistance dihydropteroate synthase. Antimicrob Agents Chemother 32:1684–1692 10.1128/AAC.32.11.1684. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barbolla R, Catalano M, Orman BE, Famiglietti A, Vay C, Smayevsky J, Centrón D, Piñeiro SA. 2004. Class 1 integrons increase trimethoprim-sulfamethoxazole MICs against epidemiologically unrelated Stenotrophomonas maltophilia isolates. Antimicrob Agents Chemother 48:666–669 10.1128/AAC.48.2.666-669.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Toleman MA, Bennett PM, Bennett DM, Jones RN, Walsh TR. 2007. Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg Infect Dis 13:559–565 10.3201/eid1304.061378. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chung HS, Kim K, Hong SS, Hong SG, Lee K, Chong Y. 2015. The sul1 gene in Stenotrophomonas maltophilia with high-level resistance to trimethoprim/sulfamethoxazole. Ann Lab Med 35:246–249 10.3343/alm.2015.35.2.246. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hu LF, Chang X, Ye Y, Wang ZX, Shao YB, Shi W, Li X, Li JB. 2011. Stenotrophomonas maltophilia resistance to trimethoprim/sulfamethoxazole mediated by acquisition of sul and dfrA genes in a plasmid-mediated class 1 integron. Int J Antimicrob Agents 37:230–234 10.1016/j.ijantimicag.2010.10.025. [PubMed] [DOI] [PubMed] [Google Scholar]
- 86.Hu LF, Chen GS, Kong QX, Gao LP, Chen X, Ye Y, Li JB. 2016. Increase in the prevalence of resistance determinants to trimethoprim/sulfamethoxazole in clinical Stenotrophomonas maltophilia isolates in China. PLoS One 11:e0157693 10.1371/journal.pone.0157693. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang YW, Hu RM, Yang TC. 2013. Role of the pcm-tolCsm operon in the multidrug resistance of Stenotrophomonas maltophilia. J Antimicrob Chemother 68:1987–1993 10.1093/jac/dkt148. [PubMed] [DOI] [PubMed] [Google Scholar]
- 88.Sánchez MB, Martínez JL. 2015. The efflux pump SmeDEF contributes to trimethoprim-sulfamethoxazole resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 59:4347–4348 10.1128/AAC.00714-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He T, Shen Y, Schwarz S, Cai J, Lv Y, Li J, Feßler AT, Zhang R, Wu C, Shen J, Wang Y. 2016. Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J Antimicrob Chemother 71:1466–1473 10.1093/jac/dkw016. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.Chang LL, Lin HH, Chang CY, Lu PL. 2007. Increased incidence of class 1 integrons in trimethoprim/sulfamethoxazole-resistant clinical isolates of Stenotrophomonas maltophilia. J Antimicrob Chemother 59:1038–1039 10.1093/jac/dkm034. [PubMed] [DOI] [PubMed] [Google Scholar]
- 91.Rahmati-Bahram A, Magee JT, Jackson SK. 1995. Growth temperature-dependent variation of cell envelope lipids and antibiotic susceptibility in Stenotrophomonas (Xanthomonas) maltophilia. J Antimicrob Chemother 36:317–326 10.1093/jac/36.2.317. [PubMed] [DOI] [PubMed] [Google Scholar]
- 92.McKay GA, Woods DE, MacDonald KL, Poole K. 2003. Role of phosphoglucomutase of Stenotrophomonas maltophilia in lipopolysaccharide biosynthesis, virulence, and antibiotic resistance. Infect Immun 71:3068–3075 10.1128/IAI.71.6.3068-3075.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 94.Gooderham WJ, Hancock RE. 2009. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol Rev 33:279–294 10.1111/j.1574-6976.2008.00135.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 95.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461–472 10.1016/j.cell.2005.05.030. [PubMed] [DOI] [PubMed] [Google Scholar]
- 96.Liu MC, Tsai YL, Huang YW, Chen HY, Hsueh PR, Lai SY, Chen LC, Chou YH, Lin WY, Liaw SJ. 2016. Stenotrophomonas maltophilia PhoP, a two-component response regulator, involved in antimicrobial susceptibilities. PLoS One 11:e0153753 10.1371/journal.pone.0153753. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Avison MB, Higgins CS, von Heldreich CJ, Bennett PM, Walsh TR. 2001. Plasmid location and molecular heterogeneity of the L1 and L2 β-lactamase genes of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 45:413–419 10.1128/AAC.45.2.413-419.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alonso A, Sanchez P, Martínez JL. 2000. Stenotrophomonas maltophilia D457R contains a cluster of genes from Gram-positive bacteria involved in antibiotic and heavy metal resistance. Antimicrob Agents Chemother 44:1778–1782 10.1128/AAC.44.7.1778-1782.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Palleroni NJ, Bradbury JF. 1993. Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) Swings et al. 1983. Int J Syst Bacteriol 43:606–609 10.1099/00207713-43-3-606. [PubMed] [DOI] [PubMed] [Google Scholar]
- 100.Finkmann W, Altendorf K, Stackebrandt E, Lipski A. 2000. Characterization of N2O-producing Xanthomonas-like isolates from biofilters as Stenotrophomonas nitritireducens sp. nov., Luteimonas mephitis gen. nov., sp. nov. and Pseudoxanthomonas broegbernensis gen. nov., sp. nov. Int J Syst Evol Microbiol 50:273–282 10.1099/00207713-50-1-273. [PubMed] [DOI] [PubMed] [Google Scholar]
- 101.Yamamura S, Morita Y, Hasan Q, Rao SR, Murakami Y, Yokoyama K, Tamiya E. 2002. Characterization of a new keratin-degrading bacterium isolated from deer fur. J Biosci Bioeng 93:595–600 10.1016/S1389-1723(02)80243-2. [DOI] [PubMed] [Google Scholar]
- 102.Assih EA, Ouattara AS, Thierry S, Cayol JL, Labat M, Macarie H. 2002. Stenotrophomonas acidaminiphila sp. nov., a strictly aerobic bacterium isolated from an upflow anaerobic sludge blanket (UASB) reactor. Int J Syst Evol Microbiol 52:559–568 10.1099/00207713-52-2-559. [PubMed] [DOI] [PubMed] [Google Scholar]
- 103.Wolf A, Fritze A, Hagemann M, Berg G. 2002. Stenotrophomonas rhizophila sp. nov., a novel plant-associated bacterium with antifungal properties. Int J Syst Evol Microbiol 52:1937–1944. [PubMed] [DOI] [PubMed] [Google Scholar]
- 104.Yoon JH, Kang SJ, Oh HW, Oh TK. 2006. Stenotrophomonas dokdonensis sp. nov., isolated from soil. Int J Syst Evol Microbiol 56:1363–1367 10.1099/ijs.0.64091-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 105.Yang HC, Im WT, Kang MS, Shin DY, Lee ST. 2006. Stenotrophomonas koreensis sp. nov., isolated from compost in South Korea. Int J Syst Evol Microbiol 56:81–84 10.1099/ijs.0.63826-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 106.Heylen K, Vanparys B, Peirsegaele F, Lebbe L, De Vos P. 2007. Stenotrophomonas terrae sp. nov. and Stenotrophomonas humi sp. nov., two nitrate-reducing bacteria isolated from soil. Int J Syst Evol Microbiol 57:2056–2061 10.1099/ijs.0.65044-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 107.Kaparullina E, Doronina N, Chistyakova T, Trotsenko Y. 2009. Stenotrophomonas chelatiphaga sp. nov., a new aerobic EDTA-degrading bacterium. Syst Appl Microbiol 32:157–162 10.1016/j.syapm.2008.12.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 108.Kim HB, Srinivasan S, Sathiyaraj G, Quan LH, Kim SH, Bui TP, Liang ZQ, Kim YJ, Yang DC. 2010. Stenotrophomonas ginsengisoli sp. nov., isolated from a ginseng field. Int J Syst Evol Microbiol 60:1522–1526 10.1099/ijs.0.014662-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 109.Lee M, Woo SG, Chae M, Shin MC, Jung HM, Ten LN. 2011. Stenotrophomonas daejeonensis sp. nov., isolated from sewage. Int J Syst Evol Microbiol 61:598–604 10.1099/ijs.0.017780-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 110.Ramos PL, Van Trappen S, Thompson FL, Rocha RC, Barbosa HR, De Vos P, Moreira-Filho CA. 2011. Screening for endophytic nitrogen-fixing bacteria in Brazilian sugar cane varieties used in organic farming and description of Stenotrophomonas pavanii sp. nov. Int J Syst Evol Microbiol 61:926–931 10.1099/ijs.0.019372-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 111.Handa Y, Tazato N, Nagatsuka Y, Koide T, Kigawa R, Sano C, Sugiyama J. 2016. Stenotrophomonas tumulicola sp. nov., a major contaminant of the stone chamber interior in the Takamatsuzuka Tumulus. Int J Syst Evol Microbiol 66:1119–1124 10.1099/ijsem.0.000843. [PubMed] [DOI] [PubMed] [Google Scholar]
- 112.Pan X, Lin D, Zheng Y, Zhang Q, Yin Y, Cai L, Fang H, Yu Y. 2016. Biodegradation of DDT by Stenotrophomonas sp. DDT-1: characterization and genome functional analysis. Sci Rep 6:21332 10.1038/srep21332. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hindler JF, Tamashiro L. 2004. Broth microdilution MIC test, p 5.2.1–5.2.17. In Clarke L, Della-Latta P, Denys GA, Douglas SD, Garcia LS, Hazen KC, Hindler JF, Jenkins SG, Mangels JI, Miller JM, Nachamkin I, Pfaller MA, Snyder JW, Weissfeld AS, York MK (ed), Clinical Microbiology Procedures Handbook, 2nd ed. ASM Press, Washington, DC. [Google Scholar]


