ABSTRACT
Conjugative plasmids are the main carriers of transmissible antibiotic resistance (AbR) genes. For that reason, strategies to control plasmid transmission have been proposed as potential solutions to prevent AbR dissemination. Natural mechanisms that bacteria employ as defense barriers against invading genomes, such as restriction-modification or CRISPR-Cas systems, could be exploited to control conjugation. Besides, conjugative plasmids themselves display mechanisms to minimize their associated burden or to compete with related or unrelated plasmids. Thus, FinOP systems, composed of FinO repressor protein and FinP antisense RNA, aid plasmids to regulate their own transfer; exclusion systems avoid conjugative transfer of related plasmids to the same recipient bacteria; and fertility inhibition systems block transmission of unrelated plasmids from the same donor cell. Artificial strategies have also been designed to control bacterial conjugation. For instance, intrabodies against R388 relaxase expressed in recipient cells inhibit plasmid R388 conjugative transfer; pIII protein of bacteriophage M13 inhibits plasmid F transmission by obstructing conjugative pili; and unsaturated fatty acids prevent transfer of clinically relevant plasmids in different hosts, promoting plasmid extinction in bacterial populations. Overall, a number of exogenous and endogenous factors have an effect on the sophisticated process of bacterial conjugation. This review puts them together in an effort to offer a wide picture and inform research to control plasmid transmission, focusing on Gram-negative bacteria.
INTRODUCTION
Antibiotics have saved the lives of countless people suffering from bacterial infections since Alexander Fleming discovered penicillin in 1928 (1). Nevertheless, this success was accompanied by the emergence of antibiotic resistance (AbR). It is thought that AbR arose originally as a self-protection mechanism of producer organisms (2). AbR genes rapidly disseminated through the biosphere as a result of the selection pressure established by human application of antibiotics (3). Resistance mechanisms capable of rendering newly discovered drugs ineffective emerged with astonishing speed, rapidly reaching human pathogens and increasingly invalidating newer antimicrobial therapies (4). Altogether, >20,000 potential resistance genes of nearly 400 types have been predicted from bacterial genome sequences (5). The danger created by the ever-increasing number of pathogens resistant to conventional antibiotics is further increased by a significant drop in the development of new antimicrobial compounds (6). This situation demands solutions to prevent the hundreds of thousands of people dying each year as a result of AbR from becoming millions (7). Proposed strategies include more-accurate prescription policies and a controlled use and release of antibiotics in animal husbandry and agriculture, restrictions difficult to implement on a global scale (3).
Alternatives to conventional antibiotics are emerging to treat this global crisis. For example, inhibitors of bacterial virulence are promising alternatives with an advantage over antibiosis in that selection for resistance might not occur because pathogen growth would not be impaired (8). Additional lines of attack under development are vaccines (9), phage therapy (10), predatory bacteria (11), and antiplasmid strategies (12–14), among others. In this context, this review focuses on natural and artificial strategies that could be employed in Gram-negative bacteria to control the transmission of conjugative plasmids, the main propagation devices involved in AbR dissemination.
HORIZONTAL GENE TRANSFER
AbR genes are transferred either vertically when bacteria divide or laterally from one bacterium to another through horizontal gene transfer (HGT), an important source of bacterial variability (15). HGT is mediated by mobile genetic elements (MGEs), that is, DNA devices for the intra- or intercellular movement of DNA (16). Intracellular mobility is produced by transposons, DNA fragments with the ability to move from one genome location to another, including different replicons of the same cell. Intercellular mobility occurs by one of three main processes: transformation, conjugation, or transduction. Transformation involves extracellular DNA uptake, integration, and functional expression. Bacteria must be in a physiological state of competence to acquire exogenous DNA, which could be natural or artificially induced. Most naturally transformable bacteria develop competence in response to specific environmental conditions, such as altered growth conditions, nutrient access, cell density, or starvation (13). Conjugation requires genetic elements encoding the apparatus needed for their transfer from a donor to a recipient cell through direct contact (16). Transduction is mediated by bacteriophages when they accidentally pack segments of host DNA and inject them into a new host. Transduction may be generalized or specialized, depending on whether any gene may be transferred or only those located near the site of prophage integration (17).
BACTERIAL CONJUGATION
Conjugation is arguably the most common mechanism of HGT (18), and that with the broadest host range (19). Encoded either in autonomously replicating conjugative plasmids or in integrative and conjugative elements (ICEs) inserted in the bacterial chromosome, conjugation systems allow the transfer of large DNA fragments containing diverse adaptive traits (20). Indeed, they are major vehicles for the spread of AbR genes (21, 22).
Either double-stranded DNA (dsDNA) or single-stranded DNA (ssDNA) molecules can be transported from donor to recipient cells. dsDNA conjugation was described in Actinobacteria. The translocation mechanism involves a single protein, a plasmid-encoded septal DNA translocase similar to the segregation ATPase FtsK, unlike the complex machinery needed for “classic” ssDNA conjugation (23). Conjugative systems involved in ssDNA conjugation carry two sets of genetic components: mobility (MOB) for conjugative DNA processing, and mating-pair formation (MPF) for DNA delivery through the membranes of donor and recipient bacteria. The MOB component includes an origin of transfer (oriT), a short DNA sequence required in cis for plasmid transfer (24); a relaxase to initiate conjugation; and a type IV coupling protein (T4CP) to interconnect DNA processing with DNA transport. MPF genes code for a complex of proteins that build the type IV secretion system (T4SS).
Plasmids can be classified into three mobility categories: conjugative, mobilizable, and nonmobilizable. A conjugative plasmid contains the two sets of components necessary for its own transfer, whereas a mobilizable plasmid lacks MPF genes and uses the T4SS of a coresident self-transmissible element, thus escaping from pilus synthesis burden (20). In general, conjugative plasmids are large (>30 kb) and of low copy number, while mobilizable plasmids are small (<15 kb) and have relatively higher copy number. Plasmids unable to transfer by conjugation or mobilization are called nonmobilizable (20). Nevertheless, nonmobilizable plasmids may be transferred by physical association with a transmissible plasmid (the process is called cointegration, if the resulting plasmid maintains the physical association, or conduction, if the two plasmids resolve in recipient cells) (25).
Conjugative Transfer, the Process
The initial requirement for bacterial conjugation is the expression of MPF genes in donor cells. Four MPF classes are found in conjugative systems from Proteobacteria: MPFT (whose prototype is T-DNA transfer system of Agrobacterium tumefaciens pTi plasmid), MPFF (exemplified by conjugative plasmid F), MPFI (exemplified by IncI plasmid R64), and MPFG (related to a broad family of ICEs whose prototype is ICEHin1056 of Haemophilus influenzae) (20). The MPFT class encodes the simplest T4SS, consisting of 11 proteins called VirB1 to VirB11 from A. tumefaciens T4SS (26). The T4SS complex can be divided in four parts: the pilus, the core channel complex, the inner membrane platform, and the cytoplasmic ATPases that supply the energy for pilus biogenesis and substrate transport (27). The conjugative pilus is the appendage that extends from the donor cell to reach the recipient cells within its proximity and subsequently retracts it to facilitate cell-to-cell contact (28). Retraction has not been demonstrated for all types of pili (29). Pilus morphology determines the ability of plasmids to transfer in liquid media or on solid surfaces (such as biofilms). Plasmids that determine rigid pili (Inc groups M, N, P, and W) or thick flexible pili (Inc groups C, D, F, H, J, T, V, and X) transfer better on solid media, while plasmids encoding thin flexible pili (Inc groups I, B, and K) transfer equally well in both situations (30, 31). This feature, added to plasmid host range (32), and the contribution of pili to establishing bacterial biofilms (33), are important determinants for plasmid dissemination in the environment (22). Once donor-recipient contact is established, the next step in conjugation is DNA processing (Fig. 1), driven by the MOB proteins. Based on MOB sequences and DNA-processing mechanism, transmissible plasmids are classified into six MOB families: MOBF, MOBH, MOBQ, MOBC, MOBP, and MOBV (34). The key protein for DNA transfer initiation, present in all transmissible plasmids, is the relaxase. Together with specific auxiliary factors, the relaxase assembles a nucleoprotein complex on the oriT called the relaxosome. The relaxase is directed to the nic site within the oriT by auxiliary factors, such as TrwA and the chromosomally encoded integration host factor (IHF) in the case of plasmid R388 (35), where the relaxase cleaves the phosphodiester bond of the DNA strand to be transferred (T-strand) (36). The transesterification reaction results in a covalent link between relaxase and ssDNA (37), followed by DNA replication from the 3′ end of the cleaved strand, using the complementary circular strand as a template. A helicase domain, usually present at the C terminus of the relaxase domain (34), unwinds DNA to displace the T-strand (38). Then, the relaxase produces a second cleavage at the nic site to release the T-strand from the newly formed strand (39).
FIGURE 1.
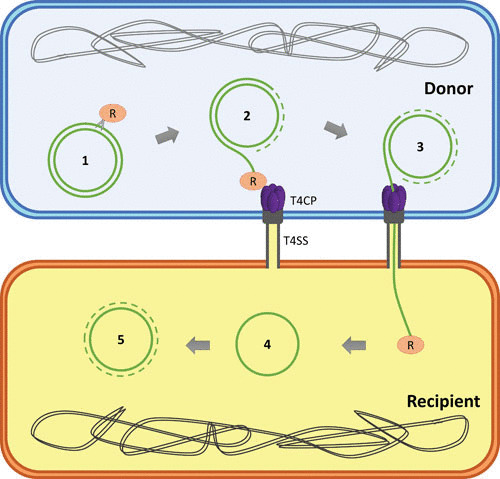
DNA processing during bacterial conjugation. (1) The relaxase (R) cleaves plasmid DNA at the nic site and forms a covalent intermediate with the 5′ end of the oriT. (2) The T4SS protein machinery recruits the relaxosome through interaction with the T4CP, while the donor DNA is replicated using the uncleaved DNA strand as a template. (3) The relaxase releases the T-strand by a second cleavage reaction at the nic site and acts as pilot protein for the ssDNA to be transferred through the T4SS, helped by the T4CP pumping activity. (4) In the recipient cell, the relaxase carries out the reverse nicking reaction to recircularize the T-strand. (5) The transferred ssDNA is replicated to generate a complete copy of the original plasmid.
After the nicking reaction, the T4CP recruits the relaxosome to the T4SS (40) in order to start the DNA transfer process. Then, the nucleoprotein complex is delivered to the inner membrane platform at the base of the T4SS to cross the channel that connects donor with recipient cells (29). According to the shoot-and-pump model (40), once the relaxase is shot through the channel acting as a pilot protein for the T-strand, T4CP pumps remaining ssDNA using the energy derived from ATP hydrolysis. When a complete copy of plasmid ssDNA reaches the recipient cell, the relaxase recognizes the nic site as a termination site and carries out the reverse nicking reaction, resolving the covalent intermediate relaxase-DNA and resulting in recircularization of the T-strand in the recipient cell (41, 42). Finally, a second strand is synthesized by rolling-circle replication to generate a copy of the original conjugative plasmid in the recipient cell, thus turning it into a new donor.
NATURAL STRATEGIES THAT CONTROL CONJUGATION
Several strategies, called eco-evo (based on an ecological and evolutionary perspective), have been explored with the aim of restoring antibiotic susceptibility in the environment (14). Since conjugation is a key mechanism involved in AbR dissemination (18, 22), this review focuses on both natural and artificial strategies to control this process (Fig. 2). Natural strategies can be defined as mechanisms that bacteria already employ in the environment to prevent conjugation (for instance, exclusion systems), while artificial strategies are human-based strategies not used by nature for this purpose (for example, antibodies targeting conjugative relaxases). It is worth noting that environmental factors like temperature, pH, chemical and physical composition, redox status, or moisture, as well as anthropogenic factors (e.g., organic or inorganic pollutants), significantly influence conjugation rates (43–45). Besides, the type of environment (human and animal microbiota, rhizosphere, manure, soil, wastewater treatment plants, aquatic environments that receive waste streams, etc.) is also an essential factor determining conjugation dynamics (17, 46). However, these factors are out of this review’s scope due to the variability of effects in different conjugative systems and the difficulty of designing a strategy to control conjugation based on environmental factors. The genetic determinants that are the basis of natural strategies can be located in the host chromosome (host strategies) or in the plasmid genome (plasmid strategies). Among host strategies, restriction-modification (RM) and CRISPR-Cas systems are the most common mechanisms to prevent stable acquisition of foreign DNA in bacteria. Conjugative plasmids display mechanisms that regulate their own transfer, block the entry of related plasmids into the same cell, or inhibit conjugative transfer of plasmids present in the same donor bacteria. Regulatory networks for bacterial conjugation comprise a set of complex responses to maximize DNA transport and minimize the burden to cells carrying the conjugative machinery (47). However, plasmid regulatory factors are diverse between different groups of conjugative systems. To give an example, the regulatory proteins encoded by prototype plasmids F, RP4, and R388 bear no homology relationship whatsoever. Another example of this diversity could be the specific antagonistic signaling of the pair cCF10/iCF10 pheromone-inhibitor peptides in the regulation of the conjugative plasmid pCF10 of Enterococcus faecalis (48). Therefore, our analysis of plasmid barriers will focus on exclusion and fertility inhibition systems, which are more conserved between different conjugative plasmids.
FIGURE 2.
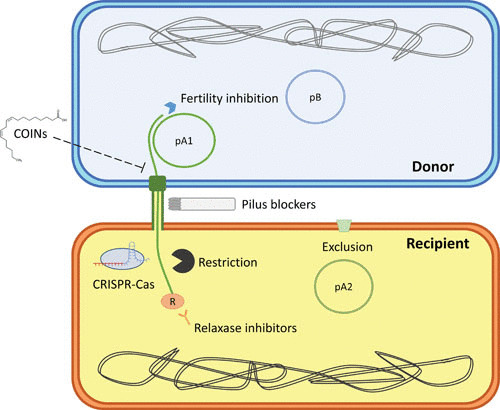
Natural and artificial mechanisms that control the transmission of conjugative plasmids. Natural mechanisms include RM and CRISPR-Cas systems (encoded by the recipient chromosome), exclusion systems (used to prevent the entrance of related plasmids in the same recipient), and fertility inhibition systems (encoded by plasmids in donor bacteria). Artificial mechanisms interfere with key components of the conjugative process, such as the relaxase, the pilus, or conjugation-related ATPases.
Host Strategies: Restriction
Restriction was first observed in the 1950s when bacteriophage λ, propagated in Escherichia coli B, was found to grow poorly on E. coli K-12 (49). RM systems code for a diverse group of enzymes, ubiquitous among prokaryotes, involved in defense against invading genomes, such as phages or plasmids (50). They comprise two opposing enzymatic activities, restriction endonuclease (REase) and methyltransferase (MTase) (51). The REase recognizes and cleaves foreign DNA at a specific site, whereas the MTase confers protection from cleavage to host genome by methylating a defined adenine or cytosine residue within the specificity site. Due to their ability to recognize self from nonself DNA, RM systems are considered a primitive, innate immune system (52). They are classified in four types, based on molecular structure, sequence recognition, cleavage position, and cofactor requirements (53).
RM systems are major players in the coevolutionary interaction between MGEs and their hosts (54). RM systems may have additional roles (51). For example, MGE-encoded RM systems act as toxin-antitoxin stability systems. During cell division, the failure to segregate RM systems efficiently results in postsegregational killing of the progeny lacking the RM-containing plasmids. This is due to the higher stability of the REase (toxin), which attacks the unmodified host genome of the progeny lacking the MTase (antitoxin) (55). Thus, the MGE is stabilized by the RM system and the RM system acquires the ability to be transferred. Although this role contributes to the stability of RM-containing plasmids instead of being a barrier to conjugation, it seems to be a minor role, since only 10% of the plasmids encode RM systems, whereas 69% of the chromosomes do so (54).
While host defense against bacteriophage infection has been extensively described (56), inhibition of bacterial conjugation by RM systems has been reported to a lesser extent. Several reports revealed that inactivation of restriction systems in recipient cells (57–62) or deletion of methylation systems in donor cells (63) increases conjugation frequency, while others showed a reduction in conjugative transfer when the number of restriction sites in the donor plasmid was increased (64–66). Accordingly, the ability of phages and plasmids to escape restriction highlights the importance of RM systems as defense devices against foreign DNA. The mechanisms used in this coevolutionary arms race between bacteria and parasitic DNA molecules to avoid restriction include four different strategies (Table 1). A number of conjugative plasmids encode antirestriction proteins, named Ard (alleviation of restriction of DNA). ArdA and ArdB, encoded by conjugative transposons and plasmids of the IncN, IncI, and IncF groups, are examples of direct inhibitors of REases that mimic DNA after their rapid expression in recipient cells (67, 68). ArdC protein from IncW plasmid pSa protects incoming T-strand by transient occlusion of restriction sites after being pumped into recipient cells (69). Another strategy is the selection of plasmid variants that lost restriction sites, as seems to happen in the case of plasmid RP4 (70). A combination of more than one antirestriction strategy is exemplified by the case of the E. faecalis Tn916-like conjugative transposons, which confer antimicrobial resistance to both Gram-positive and Gram-negative bacteria. Two reasons for Tn916’s broad host range are the presence within the element of ardA antirestriction systems and few restriction sites (71). This observation highlights the importance of antirestriction strategies to counteract RM systems of potential hosts, thus increasing the ability of conjugative elements to spread to a greater variability of bacteria.
TABLE 1.
Antirestriction strategies
| Mechanism | Antirestriction strategy | Examples | Reference(s) |
|---|---|---|---|
| Incoming genome modification | Reduction or reorientation of restriction sites | T3 or T7 phages, RP4 plasmid | 70, 254, 255 |
| Incorporation of unusual bases or methylation | Mu or SPβ phages | 256 | |
| Restriction site occlusion | Transient occlusion of restriction sites by proteins cotransported with the DNA | P1 phage DarA/DarB, IncW plasmids ArdC | 69, 257 |
| Host RM system alteration | MTase stimulation to modify incoming DNA | λ phage Ral protein | 258 |
| Destruction of REase cofactors | T3 phage SAMase | 259 | |
| REase inhibition | Direct inhibition of REases through mimicking DNA size, shape, and electric charge | T7 phage Ocr protein, IncN, IncF, and IncI plasmids ArdA/ArdB | 68, 260 |
Host Strategies: CRISPR-Cas
Additional defense systems, sometimes operating synergistically with RM systems, are CRISPR-Cas systems (72). Unlike RM systems, which provide a primitive innate immunity, CRISPR-Cas systems can be thought of as providing adaptive immunity, sequence-directed against foreign elements (73). CRISPR loci, present in 45% of bacterial and 84% of archaeal sequenced genomes (74), consist of an array of repetitive sequences of 30 to 40 bp, partially palindromic, and interspersed by equally short spacer sequences of viral or plasmid origin (75).
The CRISPR-Cas defense mechanism can be divided in two phases: immunization and immunity (76). In the immunization phase, also known as adaptation or spacer acquisition, sequences from the invading genome integrate into the CRISPR array. The acquisition of new spacers provides an efficient response against phages that escape immunity by mutating the target site (77). In the immunity phase, immunity is accomplished in two steps: guide RNA biogenesis, where a CRISPR array is transcribed and processed to generate small CRISPR RNAs (crRNAs); and targeting, in which the spacer in the crRNA serves as a guide to direct cleavage of the complementary sequence at the invading DNA (protospacer) by the Cas nucleases.
Bacteria must distinguish between protospacers of invading genomes and spacers of their CRISPR arrays to avoid cleavage of their own chromosome (78). CRISPR-Cas systems can be classified into three types, based on their Cas content, crRNA biogenesis mechanism, and targeting requirements (79). In type I and II systems, autoimmunity is prevented through a sequence called protospacer adjacent motif (PAM), only present in the invading DNA, upstream of the protospacer. The presence of this sequence is essential for foreign DNA cleavage by Cas nucleases (80). No PAM requirements have been described in type III systems, where autoimmunity inhibition is thought to occur through differential base pairing between crRNA and protospacer, preventing cleavage when full complementarity is detected (81). In addition, Chi sites (8-nucleotide motifs highly enriched in bacterial genomes) limit the acquisition of chromosomal fragments, favoring the acquisition of foreign elements, also more likely fragmented during replication (82). A failure in autoimmunity prevention leads to host death, a consequence that is being exploited for the use of CRISPR-Cas systems as genome-editing tools in both prokaryotes and eukaryotes (83, 84).
Among the numerous emerging applications of CRISPR-Cas systems (85), their ability to attack plasmid DNA during conjugation provides new weapons against AbR dissemination. In their first work, Marraffini and Sontheimer showed that a spacer from a clinical isolate of Staphylococcus epidermidis, which matched a region of the relaxase gene of staphylococcal conjugative plasmids, prevented transfer of plasmids containing this sequence by conjugation and transformation (78). Moreover, the CRISPR-Cas target was shown to be DNA instead of RNA by placing a self-splicing intron in the relaxase target sequence. In this line of research, the analysis of CRISPR spacers related to conjugative plasmids revealed that protospacers are not randomly distributed but display a MOB family-dependent bias. Whereas MOBP plasmids are usually targeted within the lagging regions, protospacers of the MOBF family are mostly located in the leading region (the first plasmid section entering the recipient cell). Nevertheless, when conjugative transfer of the MOBF plasmid F was inhibited using a type I CRISPR-Cas system, the level of protection was independent of the protospacer position and the DNA strand, suggesting that the observed bias depends either on the spacer acquisition phase or on the first regions becoming double-stranded (86). Additional studies demonstrate the conjugation-interfering role of CRISPR-Cas in different bacteria (87) and highlight the importance of these systems in preventing the acquisition of MGEs carrying AbR genes (88). In addition to plasmid transfer inhibition, spacers of plasmid origin could target AbR genes to induce plasmid loss (89) or even trigger AbR pathogen death (90, 91), among other interesting alternatives with countless possibilities.
Other Host Factors Involved in the Control of Conjugation
Recently discovered defense systems against phage infection and bacterial transformation might also be involved in protection against bacterial conjugation. This is the case for prokaryotic Argonaute proteins, homologs to the eukaryotic nucleases involved in RNA interference (92, 93), or bacteriophage exclusion, a mechanism that protects bacteria from phage replication (94, 95).
Besides the previously described defense barriers against incoming DNA, several studies aimed to find additional host barriers to conjugation or potential targets to control the process. Early studies demonstrated the contribution of the basic cellular machinery (replication, protein synthesis, or energy supply) in bacterial conjugation (96). In particular, DNA polymerase III was shown to be required in recipient cells for the synthesis of the transferred complementary strand (97), as well as in donors to replace the transferred strand (98). Another example is helicase PcrA of Bacillus subtilis, needed for ICEBs1 DNA unwinding after nicking (99). Although its E. coli homolog, UvrD, is not essential for growth, PcrA is a second helicase essential for B. subtilis viability (100). Nevertheless, targeting essential enzymes as a barrier to conjugation would kill the host, acting therefore like a conventional antibiotic (101). To avoid the selective pressure that increases the probability of AbR emergence, nonessential functions are preferred to control bacterial conjugation. This may be the case of the stationary-phase sigma factor RpoS, which regulates ICEclc excision in Pseudomonas knackmussii and is required for its conjugative transfer (102).
A mechanism potentially deleterious for conjugation as well as for recipient cell viability is the SOS response. The SOS response is stimulated by the appearance of ssDNA and its interaction with the RecA protein, which inactivates the LexA repressor, thereby inducing several genes involved in DNA repair, recombination, and mutagenesis (103). Some conjugative plasmids are adapted to counteract the SOS response through a plasmid SOS interference (psi) system that inhibits RecA binding to ssDNA (104). Similarly, the SOS response to DNA damage inactivates the LexA repressor homolog, present in several ICEs, that controls integrase expression and ICE propagation (105). Therefore, the SOS response can be a positive or a negative regulator of bacterial conjugation.
Host factors involved in regulation of bacterial conjugation are exemplified by the case of plasmid F, a narrow-host-range plasmid well adapted to E. coli (106). While broad-host-range plasmids regulate their transfer mostly through plasmid-encoded repressors (107), narrow-host-range plasmids rely on several host-encoded regulatory factors that act at DNA, RNA, or protein levels (Table 2).
TABLE 2.
Host-encoded factors involved in conjugative transfer of IncF plasmids
| Level | Host factor | Regulatory function | Reference(s) |
|---|---|---|---|
| DNA | ArcA/ArcB | Two-component regulatory system that activates transfer in response to oxygen levels | 261 |
| SdhABCD | Succinate dehydrogenase that has a repressive effect under aerobic conditions, probably by regulating transcription of the activator TraJ | 261 | |
| Dam | Methylase that modifies certain promoter regions, changing their sensitivity to binding of activators, such as the leucine regulator Lrp to traJ promoter | 262 | |
| H-NS | Global repressor that silences newly acquired DNA, including transfer genes | 263 | |
| RpoS/RpoH | Alternate sigma factors that stimulate transcription from H-NS silenced promoters | 264, 265 | |
| FIS | Activator or repressor, depending on whether it acts alone or in competition with H-NS | 266 | |
| IHF | Transcriptional activator of transfer genes, besides its primary role as part of the relaxosome architecture | 35, 267 | |
| CRP | The cyclic AMP receptor is also a positive regulator of traJ expression in response to glucose levels | 268 | |
| Unknown | Host-encoded regulator involved in F transfer repression during stationary phase | 269 | |
| RNA | RNase E | Ribonuclease that cleaves the antisense RNA FinP (downregulates the translation of the activator TraJ) | 139 |
| Hfq | Global regulator that binds traJ mRNA, promoting its degradation | 270 | |
| Protein | HslV/HslU | Heat shock protease-chaperone pair involved in TraJ degradation mediated by the two-component system CpxAR in response to extracytoplasmic stress | 271 |
| GroEL | Chaperonin that interacts with TraJ, promoting its proteolysis | 272 |
The first systematic screening for host genes involved in conjugation was carried out by Pérez-Mendoza and de la Cruz (108) using two collections of E. coli mutants as recipient cells: the Keio collection of 3,908 single-gene deletion mutants and a collection of 20,000 random transposon insertion mutants, which covered >99% of the E. coli nonessential genome. They studied the transfer of the broad-host-range IncW plasmid R388 on solid media through an automated conjugation assay based on the emission of luminescence by transconjugant bacteria. The work indicated that no nonessential recipient genes play a crucial role in conjugation. Therefore, required genes can be either essential for cell growth or redundant. The latter could be the case for the uvrD mutant, which showed 41% of wild-type conjugative transfer. UvrD is DNA helicase II, involved in rolling-circle replication of many plasmids (109). Despite being an interesting candidate, its barely significant effect suggested the involvement of an alternative helicase. Besides uvrD mutation, only mutations in lipopolysaccharide (LPS) biosynthesis showed a significant but modest decrease in R388 transfer (6 to 32% of wild type). A more drastic effect was observed on F plasmid liquid transfer, suggesting a role for LPS in mating-pair stabilization. Accordingly, several mutants were described that affect membrane integrity and were defective in recipient ability while increasing susceptibility to antibiotics, detergents, or phages. Among them were particular mutants of LPS biosynthesis or the outer membrane protein OmpA (110–117). Other reports characterized the effect of rfa (LPS synthesis) and ompA mutants on conjugation, proposing an adhesin at the F pilus tip as the receptor of its specific LPS group (118). More recently, recipient LPS was established as the specific receptor for the PilV adhesin of IncI plasmid R64 during liquid conjugation (119), while OmpA was shown to interact with F plasmid TraN for mating-pair stabilization (120).
Additional approaches using transposon mutagenesis revealed the nitrogen-related phosphotransferase system as the responsible mechanism for conjugative transfer inhibition of IncP-9 naphthalene catabolic plasmid pNAH7 from E. coli to Pseudomonas putida (121). Besides, combining transposon mutagenesis and massive sequencing (122), Johnson and Grossman found that there were no nonessential genes crucial for B. subtilis ICEBs1 conjugation (123). Functions slightly affecting the process were associated with membrane composition, agreeing with previous reports.
Although this review focuses on conjugative plasmids from Gram-negative bacteria, Gram-positive hosts also provide useful data on conjugation control. Gram-negative bacteria display a complex T4SS spanning two membranes with a cell-surface-attached filamentous pilus. In contrast, Gram-positive systems display a simpler T4SS for ssDNA translocation across their single cytoplasmic membrane, with a peptidoglycan hydrolase for local digestion of the cell wall, and adhesins that mediate cell contact (124). The signal for initiating conjugal transfer remains unknown in Gram-negative bacteria (125). On the contrary, many plasmids from Gram-positive bacteria rely on secreted signaling peptides called pheromones to initiate conjugation (for instance, E. faecalis plasmids pAD1 and pCF10). These pheromones, and the machinery needed for their processing and secretion, are encoded by the chromosome of recipient bacteria (126). The previously mentioned ICEBs1 from B. subtilis uses an opposite mechanism that requires the uptake of inhibitory peptides by recipient cells, using a host-encoded oligopeptide permease (127).
Plasmid Strategies: Exclusion
The exclusion phenomenon was first observed when exponentially growing cells harboring plasmid F acted as poor conjugation recipients (128). This phenotype, later called “superinfection immunity” (129), was the combination of two independent mechanisms, plasmid incompatibility and exclusion. Both phenomena refer to an interference between related sex factors, associated with replication and conjugation, respectively (130). Plasmid F contains two exclusion systems, surface exclusion and entry exclusion, later considered as prototypes for all others. Surface exclusion acts through the outer membrane protein TraT, by reducing the ability of recipient cells (∼10-fold) to form stable mating aggregates, whereas entry exclusion involves the recipient inner membrane protein TraS, which inhibits DNA transfer (∼100-fold) after mating pairs have stabilized (131, 132). The precise mechanism of action remains unclear in both cases. Some hypotheses proposed candidates for the TraT receptor in donor cells, including pilins, a hypothetical adhesin at the pilus tip, or the mating-pair stabilization protein TraN. However, none was confirmed (118, 133). The mechanism of TraS exclusion involves the inner membrane protein TraG in donor cells (134). TraG-TraS recognition was later confirmed, suggesting that TraG is translocated into recipient cells for transfer initiation, a process blocked by TraS (135). However, the interacting partner in conjugative plasmids not related to F is unknown, although TraG-VirB6 similarities point to VirB6 as the TraS counterpart (136).
All conjugative plasmids contain at least one exclusion gene, usually TraS-like, indicating their importance for the conjugative element. Exclusion systems may be used to prevent competition among identical plasmid backbones, for donor cells to avoid uneconomical excess of DNA transfer, or for recipient cells to prevent death by lethal zygosis (an excess of conjugative cell contacts causing membrane damage). Interestingly, only IncF and IncH plasmids, which produce pili that are firmly attached to the donor cell, encode both types of exclusion systems, while plasmids whose pili detach easily from the cell express only entry exclusion (136).
Plasmid Strategies: Fertility Inhibition
Fertility inhibition was discovered when certain plasmids carrying multiple AbR determinants were introduced in cells containing plasmid F (137). These R plasmids were IncFII plasmids that produced protein FinO. FinO reduced F transfer by increasing intracellular levels of the antisense RNA FinP (138). FinP RNA specifically downregulates traJ mRNA translation, whose product is a transcriptional activator of the transfer region. FinO binds FinP and traJ mRNA helping duplex formation, which triggers traJ mRNA cleavage by RNase III and protects FinP from degradation by RNase E (139, 140). The F plasmid is naturally derepressed due to finO insertional inactivation by insertion sequence IS3, resulting in low levels of FinP (141). Therefore, the FinOP system results in a small fraction of cells being transfer competent, contributing to regulate the balance between conjugative transfer and plasmid burden (including metabolic overhead of constitutive expression and vulnerability to pilus-specific phages) in IncF plasmids. The absence of FinOP regulation in early transconjugant cells produces a transient epidemic spread that ensures infection of the recipient cell population (106).
Besides the FinOP autoregulatory mechanism, which also affects other IncF plasmids due to FinO trans activity, additional fertility inhibition systems were identified that reduce conjugative transfer of unrelated coresident plasmids (142). These mechanisms may play a role as competition tools for colonization of new hosts (143). Eleven functions from different plasmid groups have been associated with fertility inhibition of IncF, IncW, IncP, and T-DNA of A. tumefaciens pTi plasmid, as schematized in Fig. 3.
FIGURE 3.
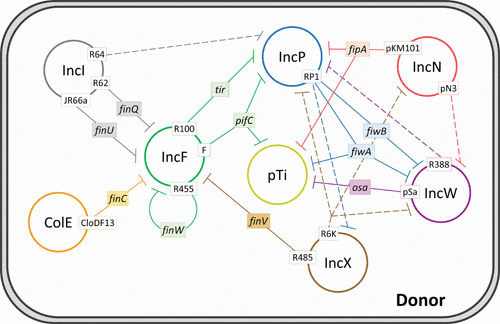
Network of interactions between conjugative plasmids that affects their conjugation capacity. Plasmid incompatibility groups are represented by colored circles. Continuous lines show fertility inhibition systems caused by genes in colored rectangles from plasmids in white boxes. Dashed lines show fertility inhibition systems caused by unidentified genes from plasmids in white boxes. See text for further details.
Transfer Inhibition of IncF Plasmids
Similar to the FinOP system, the FinQ and FinW systems act at the RNA level but independently of the main regulator TraJ. FinQ is encoded by IncI1 plasmids and acts via Rho-independent transcription termination at several sites of the tra operon (142–146). FinW is present in IncFI plasmids such as R455 and reduces transcription of TraM (142, 143, 145), a regulator activated by TraJ and essential for DNA processing during F transfer (147). FinC, FinU, and FinV fertility inhibition systems act posttranscriptionally. FinC is expressed by the mobilizable plasmid CloDF13 (which uses its own T4CP), probably to inhibit the function of the helper F T4CP during CloDF13 transfer (148). FinU and FinV are encoded by IncI1 plasmid JR66a and IncX plasmid R485, respectively. Since FinU inhibited both pilus assembly and entry exclusion, it was suggested to affect transcription of the tra operon (143). Since transcription reduction was not proportional to the observed effect, the primary target of FinU was suggested to be the translation or function of one or more transfer proteins (145). FinV reduced pilus formation but did not produce an effect on surface exclusion (143). Therefore, it was suggested to act posttranscriptionally, affecting the activity of one of the proteins required for pilus assembly (145).
Transfer Inhibition of IncW Plasmids
The elements fiwA and fiwB, encoded by IncP1α plasmids such as RP1, inhibit transfer of IncW plasmids (149–151). When acting together, they reduce R388 conjugation 1 million times. While fiwA affects only R388 transferability, fiwB also affects pilus production, conferring resistance to PR4 bacteriophage (150). Unidentified genes in the IncX plasmid R6K also inhibited the fertility of IncW plasmid R388 (149) and IncN plasmid R46 (152). Similarly, IncP plasmid RP4 reduced conjugal transfer of the rhizobial plasmids pRmeGR4a and pRmeGR4b (153).
Transfer Inhibition of IncP Plasmids
IncP plasmids are targets of fertility inhibition as well. The IncI plasmid R64 encodes a function that inhibits IncP plasmid RP4 conjugative transfer up to 100-fold (154). IncX plasmid R6K and IncP plasmid RP1 showed reciprocal fertility inhibition through an unknown mechanism that resembled the FinOP regulation system (149). The first IncN plasmids reported to inhibit IncP plasmids’ fertility were pN3 (149) and R390, which also inhibited transfer of IncW plasmid pSa (155). Fertility inhibition of IncP plasmids (or fip) was localized in IncN plasmid pKM101, which reduced RP1 transfer by 10,000-fold (156). The absence of effect in pilus synthesis or entry exclusion suggested that fip acted in a different way than the FinOP system. An apparently independent function was found in F plasmid that inhibits plasmid RP4 conjugative transfer 1,000-fold (157). It was identified as pifC (or repC), a gene involved in the initiation of F replication (158) and in the regulation of pif operon expression (phage interference function) (159). PifC inhibited both RP4 conjugation and RP4-assisted mobilization (160). As occurred with FipA of pKM101, PifC inhibition did not affect exclusion or pilus synthesis. A pifC functional homolog named tir (transfer inhibition of RP4) was discovered in the replication region of IncF plasmid R100 (161). The target of FipA and PifC in IncP plasmids was TraG, the T4CP that connects the relaxosome with the T4SS. Both proteins inhibited RSF1010 mobilization (which uses TraG of RP1), while CloDF13 mobilization (which codes for its own T4CP) was not affected. In addition, IncN-assisted RSF1010 mobilization enhanced by overexpression of traG was lost in the presence of fipA or pifC (162).
Transfer Inhibition of pTi Plasmid’s T-DNA
IncW plasmid pSa abolished the plant tumor-inducing activity of the pTi’s T-DNA of A. tumefaciens (163, 164). This suppressive activity was attributed to the osa gene (oncogenic suppression activity) (165). In contrast, the oncogenic suppression caused by IncQ mobilizable plasmids seemed to act by recruiting T-DNA’s MPF for mobilization and competing for transfer more efficiently than T-DNA (166–169). Osa protein shows homology to FiwA from RP1 (170), responsible for IncW fertility inhibition (151), and was located at the bacterial inner membrane (171). Osa mode of action was related to export inhibition of VirE2 (172), a protein involved in T-DNA endonuclease protection and transport (173). Export of VirE3 and VirF virulence proteins was blocked by Osa (174). Afterwards, IncQ plasmid RSF1010 mobilization by T-DNA’s transfer system was confirmed to be reduced by Osa (169). By the use of a transfer DNA immunoprecipitation assay, Cascales et al. (175) discovered that both IncQ mobilizable plasmids (which use the same pathway as T-DNA) and Osa fertility inhibitor suppressed plant tumorigenesis through inhibition of T-DNA and VirE2 binding to the T4CP (VirD4) receptor, blocking their passage through A. tumefaciens T4SS. In contrast to IncQ plasmids, which were proposed to block T-DNA’s T4CP as a competing substrate with higher copy number and affinity for T4SS than T-DNA (167), Osa exerted its effects by modulating VirD4 receptor activity through direct protein-protein interaction. As occurred with FipA and PifC, Osa only inhibited mobilization of plasmids lacking their own T4CP (such as RSF1010), whereas mobilization of plasmids carrying their own T4CP (such as CloDF13) was not affected, thus confirming previous results (175). Recently, Osa crystal structure was solved and shown to belong to the ParB/Srx superfamily (176). ATPase and DNase activities were discovered within its active site, activities that were common to their homologs (fertility inhibition protein ICE1056Fin of H. influenzae ICEHin1056 and partition system elements KorB from IncP1α plasmid RK2 and ParB from bacteriophage P1). In addition, it was shown that T-DNA transfer was inhibited by Osa homologs ICE1056Fin and FiwA, and even by the unrelated fertility inhibition factors FipA and PifC. Immunoprecipitation and Western blot analysis showed Osa interaction with two other T4SS components, VirB4 and VirB11 ATPases. By in vitro reconstitution of a partial T4SS (comprising VirB4, VirB11, and Osa), degradation of T-DNA covalently bound to VirD2 relaxase was observed. This observation has placed Osa DNase activity as a key function of a fertility inhibition mechanistic model (176).
ARTIFICIAL STRATEGIES TO CONTROL CONJUGATION
Conjugation inhibitors (COINs) have been employed to target specific components of the conjugation machinery, such as conjugative relaxases or the pilus tip. Other compounds considered to be COINs inhibited conjugative transfer of several plasmids in different hosts either indirectly, by affecting bacterial growth, or directly, by targeting a common conjugative function, such as T4SS.
Relaxase Inhibitors
Garcillán-Barcia et al. obtained intracellular antibodies (intrabodies) able to inhibit conjugal transfer of plasmid R388 by blocking relaxase activity in recipient cells (42). After mice were immunized with TrwC relaxase, immunoglobulin variable sequences were PCR-amplified from mice mRNA, assembled as scFv (single-chain variable fragment) antibodies, and cloned into an M13 phagemid vector fused to pIII protein. The resulting library of phagemids was submitted to rounds of panning against the purified relaxase immobilized onto ELISA plates, to select antibodies that bind the relaxase more efficiently. The antibody with higher affinity for TrwC was expressed in an E. coli strain carrying mutations in the major disulfide bond reduction systems to allow intrabody folding and stability in the reducing cytoplasm of bacteria. It was used as a recipient in an R388 mating experiment, obtaining a 20-fold reduction in transfer frequency compared to a control expressing an unrelated intrabody. To search in vivo for more-potent intrabodies able to directly inhibit R388 conjugation, a second library from the first round of panning was expressed in recipient cells. The screening was performed by the use of a high-throughput conjugation assay that relies on the emission of luminescence in transconjugant cells (177). Several intrabodies inhibited R388 conjugation from 40- to 10,000-fold, confirming that R388 relaxase carried out an important function in recipient cells that could be blocked as a viable strategy to prevent plasmid transmission. When CloDF13 mobilization by R388 was tested, no change was detected, due to the usage of its own relaxase (MobC). An interesting broad-range intrabody was discovered that also reduced conjugative transfer of MOBF plasmids pKM101 and F, whose relaxase domains are 51 and 37% identical to TrwC, respectively. In addition, by mapping the epitopes recognized by one of the intrabodies, the R388 mechanism to terminate conjugative DNA processing was clarified, establishing the second catalytic tyrosine of the relaxase as an important player in this reaction. Although intrabodies are not the most suitable therapeutic candidates for conjugation control due to the pharmacokinetic problems of any macromolecule as a drug, the recognized epitopes of the relaxase could be targeted by other means in order to generate better applicable COINs.
Relaxase activity of F plasmid TraI was targeted in vitro by bisphosphonates, a set of small molecules that could apparently interfere with the relaxase active site by mimicking a covalent phosphotyrosine intermediate (178). Two of the most potent compounds were promising hits because they had already been approved for bone loss treatment, a fact that would facilitate their inclusion into the market. However, besides inhibition of F transfer in vivo, they also caused unexpected selective death of bacteria containing both a catalytically active TraI and F plasmid. Moreover, it was later concluded that bisphosphonates act as mere chelating agents that could affect any metal-dependent cellular process (179).
Pilus Blockers
Donor-recipient contact, the first step for plasmid conjugative transmission, can be inhibited by interfering with the function of the conjugative pilus (Fig. 4). The best-known inhibitors of conjugation by blocking conjugative pili are bacteriophages. Some DNA and RNA bacteriophages use conjugative pili as receptors to infect bacteria containing certain plasmids. The attachment of these phages to conjugative pili can obstruct potential donor-recipient contacts. This section will focus on COIN activity of male-specific bacteriophages without lytic activity, although phages inducing bacterial lysis possess high potential as antimicrobials against AbR bacteria. In fact, the male-specific phage PRD1, with lytic activity against bacteria containing IncP plasmids, was described as an effective plasmid-curing agent. It reduced the frequency of AbR bacteria even under selective pressure for plasmid maintenance, which promoted the emergence of conjugation-deficient mutants resistant to PRD1 (180, 181). Furthermore, lytic phages cause faster extinction of conjugative plasmids in bacterial populations, probably due to selection for phage resistance mutations that increase genetic burden, indirectly affecting plasmid stability (182). Although lytic phages have been amply studied for their use in phage therapy, nonlytic bacteriophages infecting Pseudomonas aeruginosa, such as the filamentous Pf3 and Pf1 phages, could also be effective as antimicrobial adjuvants thanks to their ability to increase susceptibility to antibiotics (183). A similar effect was observed when Boeke et al. (184) expressed the pIII protein of F-specific filamentous phages, involved in pilus recognition. pIII causes pleiotropic effects in bacterial membranes, including increased sensitivity to detergents, antibiotics, and colicins and even a reduction in F conjugation and male-specific phage infection, probably by blocking pilus retraction.
FIGURE 4.
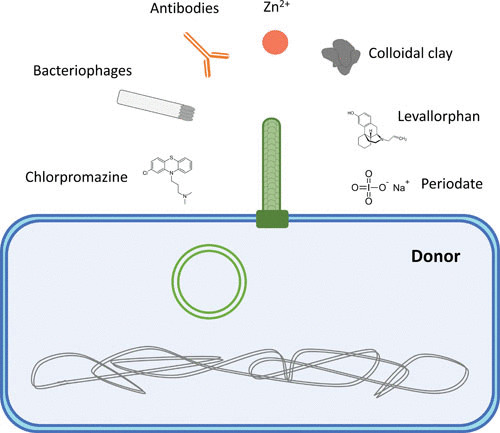
Artificial inhibitors of donor-recipient contact (pilus blockers). The tranquilizer chlorpromazine prevents plasmid conjugation and phage infection, possibly by modifying membrane topology. Male-specific bacteriophages bind the pilus tip through their pIII protein, blocking MPF and biofilm formation. Antibodies against conjugative pilus inhibit conjugation of specific plasmids. Zn2+ in the mating medium blocks F pilus contact with Zn2+-containing receptor sites. Colloidal clay forms a coating on bacterial cells preventing liquid mating, phage infection, and predation. The opioid levallorphan inhibits MPF and adsorption of male-specific bacteriophages, probably by damaging pilus or bacterial membrane. Sodium periodate alters F pili, inhibiting donor fertility and bacteriophage infection. See the section on pilus blockers in the text for additional information.
The first report about the COIN activity of bacteriophages originates from an investigation on pilus function. After Brinton et al. (185) suggested an association between RNA phage receptors and transport of genetic material, Knolle (186) found that an inactivated RNA phage (called fr) interfered with F conjugation in the same way that mating partially prevented phage invasion. Similar results were obtained with phages f1 and f2 as mating inhibitors, which attach to the tip and the sides of the F pilus, respectively (187). F-specific DNA and RNA phages (M13 and R17) were employed by Novotny et al. (188) to prevent the formation of mating pairs, providing evidence that supports F pilus as the common element involved in an early step of both phage infection and conjugation. To discard nonspecific inhibition of bacterial growth caused by phages, transfer of IncF or IncI plasmids from the same donor cell was blocked by inactivated F- or I-specific bacteriophages, respectively (189). Using a cell counter to measure mating pairs, Ou (190) demonstrated that phage f1 inhibited MPF function completely, while MS2 did so only partially. Since the filamentous DNA phage f1 attaches to the F pilus tip while the RNA bacteriophage MS2 attaches laterally along the pilus, the pilus tip was established as the specific attachment mating site. Schreil and Christensen (191) confirmed that MS2 interfered with F conjugation, but not due to competition for a common transport channel. Moreover, they disagreed with the reverse effect stated by Knolle (186), noticing that conjugation did not affect MS2 invasion.
A more recent study (192) revealed that M13 inhibition of F conjugation involved physical occlusion of the conjugative pilus by phage particles. Exogenous addition of pIII soluble fragment inhibited conjugation at nanomolar concentrations, whereas addition of the nonspecific protein bovine serum albumin did not. This result suggested that the effect was mediated by the phage coat protein pIII, known to interact with the F pilus (193). The concentration of pIII needed to inhibit F conjugation was 1,000-fold higher than the number of nonreplicating phage particles. The apparent higher affinity of phage particles when compared to isolated pIII protein could be due to cooperation between more than one pIII protein monomer to bind the pilus when they are attached to the phage structure, or to the irreversibility of the binding reaction in the case of phage particles. Lin et al. (192) also observed a 5-fold reduction in donor ability when bacteria were infected with replicating phages, probably due to decreased pilus elaboration. This effect could be important at low phage concentrations, when physical occlusion is less relevant. By constructing a chimeric phage in which the M13 N-terminal domain of pIII was substituted by the homologous sequence of If1 phage, M13 binding specificity was changed from F pilus to I pilus. Consequently, the chimeric phage inhibited conjugative transfer of IncI plasmids instead of F. They also presented a quantitative model for conjugation in the presence of phages that accurately described their COIN effect. Unlike other COINs, bacteriophages have the advantage of potential coevolution in case resistant bacteria appear.
A kinetic competition study between conjugation and M13 infection suggested that phage multiplicity of infection has to be high for the phages to act as effective antagonists to conjugation. At lower phage concentrations, conjugation persists despite phage inhibition, even in the absence of selective pressure (194). In spatially structured populations, such as surface-associated colonies and biofilms, M13 protein pIII effectively inhibited F conjugation. Moreover, spatial structure itself suppressed F conjugation due to isolation of donor and recipient populations, restricting conjugation to boundaries between them (195).
Besides conjugation and phage infection, conjugative pili are involved in the elaboration of biofilms, important targets in the battle against resistance (33). Therefore, bacteriophages affecting F conjugation could also prevent biofilm formation. Actually, male-specific filamentous DNA bacteriophage f1 prevented early biofilm formation by E. coli carrying F plasmid. Additionally, the fact that the RNA bacteriophage MS2 did not cause an inhibitory effect suggested that the pilus tip, not the sides, was important for early biofilm formation (196).
Antibodies directed against conjugative pili that are able to inhibit transfer of plasmids even more specifically than bacteriophages have been used to identify closely related resistance factors by analyzing the degree of inhibition (197). The results of this work agreed with previous serological analysis of sex pili detected through antigen-antibody reactions and observed by electron microscopy (198).
Other COINs could interfere with elaboration of mating pairs, either by blocking pilus tip in donors or pilus receptor in recipients or through nonspecific disorganization of bacterial membranes. A case in point is Zn2+, which seemed contradictory at the start. First, Zn2+ prevented phage M13 adsorption to F pilus (199). Then, a reduction in F donor fertility was shown, probably by blocking pilus tip, thus inhibiting its interaction with recipient cells (200). Conversely, incubation with Zn2+ before mating enhanced the ability of recipients to form mating pairs (201). These paradoxical effects were explained by the use of the Zn2+ chelator orthophenanthroline (202). Zn2+ is probably involved in the formation of receptor sites on the recipient surface, and the initial contact could occur between the pilus tip and Zn2+ of receptor sites. Therefore, pretreatment of recipient cells increased their fertility through Zn2+ incorporation. However, an excess of Zn2+ in the mating medium would compete for the tips of F pili, hindering their access to receptor sites. The reduction of Zn2+ availability by the mentioned chelating agent drastically decreased conjugation, mainly acting during MPF.
Unlike Zn2+, the effect caused by periodate in F donors was irreversible (203). After donor pretreatment, the number of transconjugant cells was greatly reduced, whereas treatment of recipient cells had no significant effect. Perborate and persulfate also decreased donor fertility, but to a lesser extent. The fact that addition of periodate to a mating in progress did not prevent conjugation between mating pairs already formed suggested an effect on MPF, probably by altering the surface of donor cells via polysaccharide oxidation. Consistent with this observation, Dettori et al. (204) showed that periodate also inhibits the adsorption of RNA bacteriophages to the sides of F pili. Consequently, an alteration of F pili seems to inhibit both donor fertility and bacteriophage infection (200).
The morphine derivative levallorphan, like Zn2+ and periodate, inhibited adsorption of phage MS2 to F pili (205). This inhibition was comparable to inhibition of MPF during R-factor transfer from Proteus rettgeri (now Providencia rettgeri) to E. coli, since both effects can be observed at the same concentration of levallorphan. Both inhibitory effects, on phage adsorption and conjugation, could be caused by damage of F pilus or the whole bacterial membrane (206). The tranquilizer chlorpromazine also reduced both IncF plasmid conjugative transfer and adsorption of male-specific bacteriophages (207). Since this is a cationic amphipathic molecule, it could act by modifying membrane topology through its insertion in the lipid bilayer. Another COIN probably affecting MPF is ammonium (153). It inhibited conjugative transfer of the rhizobial plasmid pRmeGR4a and pRmeGR4a-assisted mobilization of pRmeGR4b between Rhizobium meliloti strains. However, ammonium did not affect transmission of IncP plasmid RP4 or the rhizobial plasmids to A. tumefaciens. Thus, its effect seemed to take place on R. meliloti recipient cells, probably on their surface, but not on the transfer machinery.
An inert barrier between donors and recipients is an additional possibility to control bacterial conjugation. Colloidal clay, typically present in natural waters, prevented the transfer of IncF plasmid R1drd19 by forming a coating on bacterial cells (208). This clay envelope was also responsible for E. coli protection from bacteriophage infection and bacterial predation (209, 210). In contrast, plasmids that promote conjugation less efficiently in liquid, such as IncP plasmid RP4, enhanced their transfer in water containing nanoalumina particles. In the presence of these particles, RP4 upregulated the expression of genes required for MPF (211). An unspecified component of E. coli cell wall was also described as an inhibitor of conjugal transfer. In this case, the inhibitory mechanism presumably involved competence between cell wall components and actual partners on the surface of cells, thereby preventing MPF (212, 213).
Nonspecific COINs
Bisphosphonates were first described as relaxase-specific inhibitors (178) but then reappraised as chelating agents (179). Similarly, other reported COINs were later revealed as inhibitors of different cellular processes. For instance, the ability of some plasmid-curing agents to inhibit conjugative transfer is easily attributable to their antiplasmid effect, which favors the growth of plasmid-free cells (214). The increased sensitivity of E. coli containing F plasmid to bile salts and SDS represents another example of this effect. While plasmid-free cells are resistant to these toxic detergents, cells with an active system for pilin secretion are more susceptible to their entry through the T4SS pore (215). A similar behavior was found by overexpressing RP4 genes, which caused enhanced cell permeability (216). Another interesting antiplasmid effect is mediated by the type VI secretion system (T6SS). T6SSs are produced by Gram-negative bacteria to kill prokaryotic and eukaryotic cells through contact-dependent delivery of toxic effectors (217). P. aeruginosa T6SS is assembled in response to T6SS attacks by competing bacteria in microbial communities (218). Besides T6SS, T4SS structural proteins of plasmid RP4 triggered P. aeruginosa attacks by T6SS (219). The work suggested that these donor-directed counterattacks are induced at MPF-mediated membrane perturbations in P. aeruginosa recipients to potentially block the acquisition of foreign DNA. Thus, T6SS would represent a new type of immune system against HGT, through a mechanism that indirectly inhibits conjugative transfer by killing donor cells.
Several antimicrobial drugs, even at subinhibitory concentrations, act as inhibitors of plasmid conjugative transfer. However, their lethal effects in donors and recipients, or the absence of COIN activity in nongrowing bacteria, suggested that these compounds interfere with essential bacterial functions rather than recognizing a specific plasmid target (220, 221). In fact, most of these antibiotics act on cellular functions, such as DNA replication, transcription, translation, or membrane integrity, which are also involved in conjugation (43). Similarly, COIN activity of other compounds could be related to their antibacterial activity. This is the case with nitrofurans (222) and pipemidic acid (223), which inhibited transfer of several plasmids in different hosts by interfering with DNA replication. Moreover, copper surfaces inhibit conjugal transfer indirectly (224), presumably by killing bacteria through DNA and membrane damage (225–227). Epigallocatechin gallate, an antimicrobial component of tea, inhibited conjugative transfer of plasmid R100 in E. coli (228). In addition, the phenolic compounds rottlerin and “the red compound” extracted from the plant Mallotus philippensis inhibit transfer of several plasmids at subinhibitory concentrations (229). Likewise, Carica papaya seed macerate, containing a previously detected antibacterial substance (230), was considered a COIN for a Salmonella enterica serovar Typhimurium conjugative plasmid in the mouse digestive tract at nonlethal concentrations (231). Another example could be sodium propionate, produced by intestinal bacteria and abundant in the large intestine. It was found to reduce the transfer frequency of IncF plasmid pSLT in the mouse intestine (232). It also presented antibacterial properties against several microorganisms (233).
On the contrary, subinhibitory concentrations of certain antimicrobial agents can indirectly promote conjugation (234). For example, DNA damage caused by ciprofloxacin or mitomycin C induced SOS response, which is responsible for upregulating the excision and transfer of SXT ICE from Vibrio cholerae (235). In an SOS-independent manner, conjugative transposons from Bacteroides and E. faecalis increased their transfer when exposed to low concentrations of tetracycline (236, 237). Similarly, β-lactams stimulated the formation of bacterial aggregates, thus increasing conjugative transfer of a plasmid from S. aureus (238).
Unsaturated Fatty Acids
The first systematic search for COINs used conjugative plasmid R388 in E. coli as a model system (177). A luminescence-based high-throughput assay was used to measure R388 conjugation in the presence of >12,000 microbial extracts containing a variety of bioactive compounds. A control assay discarded compounds affecting bacterial growth, plasmid stability, or light emission. The first hits were oleic and linoleic acids, C18 unsaturated fatty acids (uFAs) containing one or two double bonds, respectively. The most potent compound was an atypical fatty acid named dehydrocrepenynic acid (DHCA), identified in an extract from the fungus Sistotrema sernanderi. DHCA is a C18 fatty acid with double bonds at positions 9 and 14 and a triple bond at position 12. Conjugation analysis using related compounds (including saturated fatty acids), suggested that a carboxylic group, a long carbon chain, and a double bond position were important features of COINs. Plasmids affected by linoleic acid and DHCA were R388 and pOX38, whereas RP4 or R6K were not. The fact that some plasmids were not affected argues against general metabolic disturbances as a cause. In addition, these results suggested that the inhibition target was involved in DNA processing (MOB), more similar between R388 and pOX38 than RP4 and R6K (239). However, the absence of effect on IncN plasmid pKM101, with a MOB module more similar to R388 and pOX38 than to RP4, weakened this hypothesis.
Recently, a novel set of natural COINs was discovered by analyzing a collection of bioactive compounds isolated from marine microorganisms (240). These compounds, called tanzawaic acids, are fungal polyketides more complex than uFAs. They are carboxylic acids with two aromatic rings at the end of an unsaturated aliphatic chain, thus confirming the importance of these two chemical characteristics for COIN activity.
2-Alkynoic Fatty Acids
A better understanding of COIN action in relevant conjugative systems is essential as a first step to treat complex environments. In order to chemically and biologically characterize the previously reported COIN activity, a set of 2-alkynoic fatty acids (2-AFAs) was synthesized (241). 2-Hexadecynoic acid (2-HDA) was identified as the most effective synthetic COIN, with similar potency to natural uFAs (177). A clinically representative set of conjugative plasmids was tested in the presence of 2-HDA to determine its activity range. Similarly to natural uFAs, 2-HDA inhibited transfer of IncF plasmids, the most common carriers of AbR genes in pathogenic Enterobacteriaceae (242). Transfer of IncW, IncF, and IncH plasmids was strongly inhibited by 2-HDA, while IncI, IncX, and IncL/M plasmid transfer was only moderately inhibited. On the other hand, IncN and IncP plasmids were resistant to COIN action. Also interesting for future applications was the fact that conjugation was inhibited irrespective of the bacterial host used as donor. The most remarkable result was obtained through a liquid mating experiment using the multiresistant plasmid R1drd19. In the absence of COINs, the IncF plasmid invaded the entire recipient population after just four generations. This was due to the high transmissibility of the plasmid, which caused plasmid dissemination even though plasmid-containing cells had slower growth rates than plasmid-free cells. Conversely, the presence of 2-HDA in the mating medium prevented plasmid conjugative transfer, flipping over the balance between plasmid transmission and burden, thus favoring colonization by plasmid-free cells. Consequently, 2-HDA was able to block plasmid invasiveness and reduce the prevalence of plasmid-containing cells in the bacterial population. Reversion from plasmid invasion to plasmid loss occurred at 50 μM 2-HDA, comparable to the observed 50% inhibitory concentration in R388 transfer. This suggested that 50% inhibition of conjugation was sufficient to prevent plasmid spread in the absence of selective pressure. These observations highlight the potential application of COINs to prevent AbR dissemination.
A recent study analyzed the activity of conjugative ATPases in the presence of uFAs (243). The component of R388 MPF system TrwD (VirB11 homolog), involved in pilus synthesis and DNA translocation (244, 245), was identified as the potential target. Conjugation frequency correlated with TrwD ATPase activity in the presence of different compounds, including saturated fatty acids, uFAs (177), synthetic AFAs, and AFA inactive analogs. Nevertheless, the absence of known TrwD homologs in IncF plasmids (246) still leaves unanswered questions. Given that transfer of mobilizable plasmids by IncF MPF is also affected by these COINs (241), the target in these plasmids, well adapted to E. coli after a long history of coevolution, could be an unidentified chromosomal ATPase involved in F conjugation and F-helped mobilization. Alternatively, another plasmid ATPase, such as the TrwK homolog TraC (VirB4), the only ATPase of plasmid F known to be required for MPF biogenesis (28), could be responsible.
T4SS Inhibitors Discovered by Biochemical Analysis
Other approaches to discover COINs take advantage of newly generated biochemical knowledge concerning related processes, such as T4SS-related virulence inhibition. For example, VirB11 ATPase was used as a target for the development of inhibitors, with the aim of preventing Helicobacter pylori virulence. The first described inhibitors targeted the H. pylori VirB11-type ATPase Cagα, blocking CagA toxin transport to host cells. The most active compound (CHIR-1, identified by a high-throughput screening that measured ATPase activity in the presence of small compound libraries) reduced H. pylori pathogenic effects in gastric cells and the ability of treated bacteria to colonize gastric mucosa in mice (247). Docking analysis using Cagα allowed the identification of a series of competitive inhibitors with potential as antibacterial agents (248). These antivirulence compounds could be tested in conjugative VirB11 homologs, such as TrwD of IncW plasmid R388.
Another example of how a T4SS inhibitor involved in protein secretion can extrapolate its activity to inhibit conjugative T4SS was reported by Shaffer et al. (249). They started from the structure of pilicides, small peptidomimetic molecules that target pilin chaperones and thereby inhibit assembly of type I pili, which mediate adhesion of uropathogenic E. coli (250). By screening a collection of pilicide derivatives with a central 2-pyridone scaffold, they found that compounds C10 and KSK85 disrupted H. pylori cag T4SS, thus inhibiting translocation of the oncogenic protein CagA and peptidoglycan to gastric cells. In addition, these molecules, in particular C10, effectively inhibited conjugative transfer of A. tumefaciens T-DNA to plant cells, and transmission of plasmids pKM101 and R1-16 between E. coli strains.
Type III secretion systems (T3SSs) of several pathogens were also targeted by new antivirulence drugs. They generally act as needles to inject virulence effectors into host cells (251). Several compounds were found to block T3SSs in different pathogenic bacteria (252). An interesting work developed a whole-cell high-throughput screening of T3SS inhibitors based in S. Typhimurium. A compound was identified that inhibited both T3SSs and T2SSs, probably by targeting an outer membrane component conserved between these two secretion systems (253). These results provide a proof of concept that compounds with a broad spectrum of activity against different bacterial secretion systems could be developed.
CONCLUSIONS
We have reviewed the multiple mechanisms by which conjugative transmission of plasmids can be affected. Given that conjugation is a sophisticated multistep process involving complexes of >15 proteins, it is reasonable that diverse mechanisms have been identified that disrupt one or more of the various steps. Up to now, the most efficient blocking mechanisms are the natural ones, such as RM, CRISPR, etc., that bacteria have used and perfected for millions of years to avoid "excessive" plasmid transmission. Artificial mechanisms found by scientific research are not yet as efficient but have the advantage over natural mechanisms of being less discriminating, which in our specific case is a valuable characteristic. The fight against AbR requires blocking mechanisms that impede its dissemination irrespective of the plasmid platform in which the resistance gene lies. Thus, COINs such as 2-HDA or other uFAs are broad-range inhibitors that can be used as lead compounds on which the pharmacological industry could work to obtain effective anticonjugation drugs. Alternative compounds and genetic devices have been reviewed that offer potentially different approaches to achieve the same goal. We hope that this review can inspire additional work that helps us win the fight against AbR transmission.
ACKNOWLEDGMENTS
Work at the F.D.L.C. laboratory was financed by the Spanish Ministry of Economy and Competitiveness (BFU2014-55534-C2-1-P and RTC-2015-3184-1), as well as by the European Seventh Framework Programme (612146/FP7-ICT-2013-10). M.G.’s work was supported by a Ph.D. fellowship from the University of Cantabria (Spain).
REFERENCES
- 1.Fleming A. 1929. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. Influenzae. Br J Exp Pathol 10:226–236. [PubMed] [Google Scholar]
- 2.D’Costa VM, McGrann KM, Hughes DW, Wright GD. 2006. Sampling the antibiotic resistome. Science 311:374–377. 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 3.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. 10.1128/MMBR.00016-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clatworthy AE, Pierson E, Hung DT. 2007. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol 3:541–548. 10.1038/nchembio.2007.24. [PubMed] [DOI] [PubMed] [Google Scholar]
- 5.Liu B, Pop M. 2009. ARDB—Antibiotic Resistance Genes Database. Nucleic Acids Res 37(Database issue):D443–D447. 10.1093/nar/gkn656. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper MA, Shlaes D. 2011. Fix the antibiotics pipeline. Nature 472:32. 10.1038/472032a. [PubMed] [DOI] [PubMed] [Google Scholar]
- 7.O’Neill J. 2014. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. Review on Antimicrobial Resistance, London, United Kingdom. https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf.
- 8.Heras B, Scanlon MJ, Martin JL. 2015. Targeting virulence not viability in the search for future antibacterials. Br J Clin Pharmacol 79:208–215. 10.1111/bcp.12356. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scully IL, Swanson K, Green L, Jansen KU, Anderson AS. 2015. Anti-infective vaccination in the 21st century—new horizons for personal and public health. Curr Opin Microbiol 27:96–102. 10.1016/j.mib.2015.07.006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 10.Nobrega FL, Costa AR, Kluskens LD, Azeredo J. 2015. Revisiting phage therapy: new applications for old resources. Trends Microbiol 23:185–191. 10.1016/j.tim.2015.01.006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Dwidar M, Monnappa AK, Mitchell RJ. 2012. The dual probiotic and antibiotic nature of Bdellovibrio bacteriovorus. BMB Rep 45:71–78. 10.5483/BMBRep.2012.45.2.71. [PubMed] [DOI] [PubMed] [Google Scholar]
- 12.Williams JJ, Hergenrother PJ. 2008. Exposing plasmids as the Achilles’ heel of drug-resistant bacteria. Curr Opin Chem Biol 12:389–399. 10.1016/j.cbpa.2008.06.015. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721. 10.1038/nrmicro1234. [PubMed] [DOI] [PubMed] [Google Scholar]
- 14.Baquero F, Coque TM, de la Cruz F. 2011. Ecology and evolution as targets: the need for novel eco-evo drugs and strategies to fight antibiotic resistance. Antimicrob Agents Chemother 55:3649–3660. 10.1128/AAC.00013-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrence JG, Retchless AC. 2009. The interplay of homologous recombination and horizontal gene transfer in bacterial speciation. Methods Mol Biol 532:29–53. 10.1007/978-1-60327-853-9_3. [PubMed] [DOI] [PubMed] [Google Scholar]
- 16.Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3:722–732. 10.1038/nrmicro1235. [PubMed] [DOI] [PubMed] [Google Scholar]
- 17.Davison J. 1999. Genetic exchange between bacteria in the environment. Plasmid 42:73–91. 10.1006/plas.1999.1421. [PubMed] [DOI] [PubMed] [Google Scholar]
- 18.Halary S, Leigh JW, Cheaib B, Lopez P, Bapteste E. 2010. Network analyses structure genetic diversity in independent genetic worlds. Proc Natl Acad Sci U S A 107:127–132. 10.1073/pnas.0908978107. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amábile-Cuevas CF, Chicurel ME. 1992. Bacterial plasmids and gene flux. Cell 70:189–199. 10.1016/0092-8674(92)90095-T. [PubMed] [DOI] [PubMed] [Google Scholar]
- 20.Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EP, de la Cruz F. 2010. Mobility of plasmids. Microbiol Mol Biol Rev 74:434–452. 10.1128/MMBR.00020-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waters VL. 1999. Conjugative transfer in the dissemination of beta-lactam and aminoglycoside resistance. Front Biosci 4:D433–D456. 10.2741/A439. [PubMed] [DOI] [PubMed] [Google Scholar]
- 22.Norman A, Hansen LH, Sørensen SJ. 2009. Conjugative plasmids: vessels of the communal gene pool. Philos Trans R Soc Lond B Biol Sci 364:2275–2289. 10.1098/rstb.2009.0037. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thoma L, Muth G. 2015. The conjugative DNA-transfer apparatus of Streptomyces. Int J Med Microbiol 305:224–229. 10.1016/j.ijmm.2014.12.020. [PubMed] [DOI] [PubMed] [Google Scholar]
- 24.Fürste JP, Pansegrau W, Ziegelin G, Kröger M, Lanka E. 1989. Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid-encoded products with the transfer origin. Proc Natl Acad Sci U S A 86:1771–1775. 10.1073/pnas.86.6.1771. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark AJ, Adelberg EA. 1962. Bacterial conjugation. Annu Rev Microbiol 16:289–319. 10.1146/annurev.mi.16.100162.001445. [PubMed] [DOI] [PubMed] [Google Scholar]
- 26.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol 59:451–485. 10.1146/annurev.micro.58.030603.123630. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, Lu F, Redzej A, Fronzes R, Orlova EV, Waksman G. 2014. Structure of a type IV secretion system. Nature 508:550–553. 10.1038/nature13081. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke M, Maddera L, Harris RL, Silverman PM. 2008. F-pili dynamics by live-cell imaging. Proc Natl Acad Sci U S A 105:17978–17981. 10.1073/pnas.0806786105. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cabezón E, Ripoll-Rozada J, Peña A, de la Cruz F, Arechaga I. 2015. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev 39:81–95. 10.1111/1574-6976.12085. [PubMed] [DOI] [PubMed] [Google Scholar]
- 30.Bradley DE. 1980. Morphological and serological relationships of conjugative pili. Plasmid 4:155–169. 10.1016/0147-619X(80)90005-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 31.Bradley DE, Taylor DE, Cohen DR. 1980. Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J Bacteriol 143:1466–1470. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.del Solar G, Alonso JC, Espinosa M, Díaz-Orejas R. 1996. Broad-host-range plasmid replication: an open question. Mol Microbiol 21:661–666. 10.1046/j.1365-2958.1996.6611376.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.Ghigo JM. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442–445. 10.1038/35086581. [PubMed] [DOI] [PubMed] [Google Scholar]
- 34.Garcillán-Barcia MP, Francia MV, de la Cruz F. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev 33:657–687. 10.1111/j.1574-6976.2009.00168.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 35.de la Cruz F, Frost LS, Meyer RJ, Zechner EL. 2010. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev 34:18–40. 10.1111/j.1574-6976.2009.00195.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 36.Byrd DR, Matson SW. 1997. Nicking by transesterification: the reaction catalysed by a relaxase. Mol Microbiol 25:1011–1022. 10.1046/j.1365-2958.1997.5241885.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 37.Guasch A, Lucas M, Moncalián G, Cabezas M, Pérez-Luque R, Gomis-Rüth FX, de la Cruz F, Coll M. 2003. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat Struct Biol 10:1002–1010. 10.1038/nsb1017. [PubMed] [DOI] [PubMed] [Google Scholar]
- 38.Llosa M, Grandoso G, Hernando MA, de la Cruz F. 1996. Functional domains in protein TrwC of plasmid R388: dissected DNA strand transferase and DNA helicase activities reconstitute protein function. J Mol Biol 264:56–67. 10.1006/jmbi.1996.0623. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Pansegrau W, Lanka E. 1996. Mechanisms of initiation and termination reactions in conjugative DNA processing. Independence of tight substrate binding and catalytic activity of relaxase (TraI) of IncPα plasmid RP4. J Biol Chem 271:13068–13076. 10.1074/jbc.271.22.13068. [DOI] [PubMed] [Google Scholar]
- 40.Llosa M, Gomis-Rüth FX, Coll M, de la Cruz Fd F. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol 45:1–8. 10.1046/j.1365-2958.2002.03014.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 41.Draper O, César CE, Machón C, de la Cruz F, Llosa M. 2005. Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. Proc Natl Acad Sci U S A 102:16385–16390. 10.1073/pnas.0506081102. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcillán-Barcia MP, Jurado P, González-Pérez B, Moncalián G, Fernández LA, de la Cruz F. 2007. Conjugative transfer can be inhibited by blocking relaxase activity within recipient cells with intrabodies. Mol Microbiol 63:404–416. 10.1111/j.1365-2958.2006.05523.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 43.Viljanen P, Boratynski J. 1991. The susceptibility of conjugative resistance transfer in gram-negative bacteria to physicochemical and biochemical agents. FEMS Microbiol Rev 8:43–54. [PubMed] [DOI] [PubMed] [Google Scholar]
- 44.van Elsas JD, Fry JC, Hirsch P, Molin S. 2000. Ecology of plasmid transfer and spread, p 175–206. In Thomas CM (ed), The Horizontal Gene Pool: Bacterial Plasmids and Gene Spread. Harwood Academic Publishers, Williston, VT. [Google Scholar]
- 45.Aminov RI. 2011. Horizontal gene exchange in environmental microbiota. Front Microbiol 2:158. 10.3389/fmicb.2011.00158. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Elsas JD, Bailey MJ. 2002. The ecology of transfer of mobile genetic elements. FEMS Microbiol Ecol 42:187–197. 10.1111/j.1574-6941.2002.tb01008.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 47.Koraimann G, Wagner MA. 2014. Social behavior and decision making in bacterial conjugation. Front Cell Infect Microbiol 4:54. 10.3389/fcimb.2014.00054. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chatterjee A, Cook LC, Shu CC, Chen Y, Manias DA, Ramkrishna D, Dunny GM, Hu WS. 2013. Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer. Proc Natl Acad Sci U S A 110:7086–7090. 10.1073/pnas.1212256110. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bertani G, Weigle JJ. 1953. Host controlled variation in bacterial viruses. J Bacteriol 65:113–121. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts RJ, Vincze T, Posfai J, Macelis D. 2015. REBAS—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 43(D1):D298–D299. 10.1093/nar/gku1046. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vasu K, Nagaraja V. 2013. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev 77:53–72. 10.1128/MMBR.00044-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bickle TA. 2004. Restricting restriction. Mol Microbiol 51:3–5. 10.1046/j.1365-2958.2003.03846.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 53.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev SK, Dryden DT, Dybvig K, Firman K, Gromova ES, Gumport RI, Halford SE, Hattman S, Heitman J, Hornby DP, Janulaitis A, Jeltsch A, Josephsen J, Kiss A, Klaenhammer TR, Kobayashi I, Kong H, Krüger DH, Lacks S, Marinus MG, Miyahara M, Morgan RD, Murray NE, Nagaraja V, Piekarowicz A, Pingoud A, Raleigh E, Rao DN, Reich N, Repin VE, Selker EU, Shaw PC, Stein DC, Stoddard BL, Szybalski W, Trautner TA, Van Etten JL, Vitor JM, Wilson GG, Xu SY. 2003. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res 31:1805–1812. 10.1093/nar/gkg274. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oliveira PH, Touchon M, Rocha EP. 2014. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res 42:10618–10631. 10.1093/nar/gku734. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kobayashi I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res 29:3742–3756. 10.1093/nar/29.18.3742. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samson JE, Magadán AH, Sabri M, Moineau S. 2013. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 11:675–687. 10.1038/nrmicro3096. [PubMed] [DOI] [PubMed] [Google Scholar]
- 57.Roer L, Aarestrup FM, Hasman H. 2015. The EcoKI type I restriction-modification system in Escherichia coli affects but is not an absolute barrier for conjugation. J Bacteriol 197:337–342. 10.1128/JB.02418-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waldron DE, Lindsay JA. 2006. Sau1: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J Bacteriol 188:5578–5585. 10.1128/JB.00418-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pinedo CA, Smets BF. 2005. Conjugal TOL transfer from Pseudomonas putida to Pseudomonas aeruginosa: effects of restriction proficiency, toxicant exposure, cell density ratios, and conjugation detection method on observed transfer efficiencies. Appl Environ Microbiol 71:51–57. 10.1128/AEM.71.1.51-57.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geisenberger O, Ammendola A, Christensen BB, Molin S, Schleifer KH, Eberl L. 1999. Monitoring the conjugal transfer of plasmid RP4 in activated sludge and in situ identification of the transconjugants. FEMS Microbiol Lett 174:9–17. 10.1111/j.1574-6968.1999.tb13543.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 61.Schäfer A, Kalinowski J, Pühler A. 1994. Increased fertility of Corynebacterium glutamicum recipients in intergeneric matings with Escherichia coli after stress exposure. Appl Environ Microbiol 60:756–759. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P. 1991. Shuttle vectors containing a multiple cloning site and a lacZα gene for conjugal transfer of DNA from Escherichia coli to Gram-positive bacteria. Gene 102:99–104. 10.1016/0378-1119(91)90546-N. [DOI] [PubMed] [Google Scholar]
- 63.Zhou H, Wang Y, Yu Y, Bai T, Chen L, Liu P, Guo H, Zhu C, Tao M, Deng Z. 2012. A non-restricting and non-methylating Escherichia coli strain for DNA cloning and high-throughput conjugation to Streptomyces coelicolor. Curr Microbiol 64:185–190. 10.1007/s00284-011-0048-5. [PubMed] [DOI] [PubMed] [Google Scholar]
- 64.Ohtani N, Sato M, Tomita M, Itaya M. 2008. Restriction on conjugational transfer of pLS20 in Bacillus subtilis 168. Biosci Biotechnol Biochem 72:2472–2475. 10.1271/bbb.80315. [PubMed] [DOI] [PubMed] [Google Scholar]
- 65.Purdy D, O’Keeffe TA, Elmore M, Herbert M, McLeod A, Bokori-Brown M, Ostrowski A, Minton NP. 2002. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol Microbiol 46:439–452. 10.1046/j.1365-2958.2002.03134.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 66.Elhai J, Vepritskiy A, Muro-Pastor AM, Flores E, Wolk CP. 1997. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J Bacteriol 179:1998–2005. 10.1128/jb.179.6.1998-2005.1997. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McMahon SA, Roberts GA, Johnson KA, Cooper LP, Liu H, White JH, Carter LG, Sanghvi B, Oke M, Walkinshaw MD, Blakely GW, Naismith JH, Dryden DT. 2009. Extensive DNA mimicry by the ArdA anti-restriction protein and its role in the spread of antibiotic resistance. Nucleic Acids Res 37:4887–4897. 10.1093/nar/gkp478. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilkins BM. 2002. Plasmid promiscuity: meeting the challenge of DNA immigration control. Environ Microbiol 4:495–500. 10.1046/j.1462-2920.2002.00332.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 69.Belogurov AA, Delver EP, Agafonova OV, Belogurova NG, Lee LY, Kado CI. 2000. Antirestriction protein Ard (type C) encoded by IncW plasmid pSa has a high similarity to the “protein transport” domain of TraC1 primase of promiscuous plasmid RP4. J Mol Biol 296:969–977. 10.1006/jmbi.1999.3493. [PubMed] [DOI] [PubMed] [Google Scholar]
- 70.Wilkins BM, Chilley PM, Thomas AT, Pocklington MJ. 1996. Distribution of restriction enzyme recognition sequences on broad host range plasmid RP4: molecular and evolutionary implications. J Mol Biol 258:447–456. 10.1006/jmbi.1996.0261. [PubMed] [DOI] [PubMed] [Google Scholar]
- 71.Clewell DB, Weaver KE, Dunny GM, Coque TM, Francia MV, Hayes F. 2014. Extrachromosomal and mobile elements in enterococci: transmission, maintenance, and epidemiology, pp. 309–420. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Boston: Massachusetts Eye and Ear Infirmary. [PubMed] [Google Scholar]
- 72.Dupuis ME, Villion M, Magadán AH, Moineau S. 2013. CRISPR-Cas and restriction-modification systems are compatible and increase phage resistance. Nat Commun 4:2087. 10.1038/ncomms3087. [PubMed] [DOI] [PubMed] [Google Scholar]
- 73.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. 10.1126/science.1138140. [PubMed] [DOI] [PubMed] [Google Scholar]
- 74.Grissa I, Vergnaud G, Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. 10.1186/1471-2105-8-172. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jansen R, Embden JD, Gaastra W, Schouls LM. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43:1565–1575. 10.1046/j.1365-2958.2002.02839.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 76.Marraffini LA. 2015. CRISPR-Cas immunity in prokaryotes. Nature 526:55–61. 10.1038/nature15386. [PubMed] [DOI] [PubMed] [Google Scholar]
- 77.Andersson AF, Banfield JF. 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320:1047–1050. 10.1126/science.1157358. [PubMed] [DOI] [PubMed] [Google Scholar]
- 78.Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845. 10.1126/science.1165771. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. 10.1038/nrmicro2577. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sashital DG, Wiedenheft B, Doudna JA. 2012. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell 46:606–615. 10.1016/j.molcel.2012.03.020. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marraffini LA, Sontheimer EJ. 2010. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463:568–571. 10.1038/nature08703. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Levy A, Goren MG, Yosef I, Auster O, Manor M, Amitai G, Edgar R, Qimron U, Sorek R. 2015. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520:505–510. 10.1038/nature14302. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sontheimer EJ, Barrangou R. 2015. The bacterial origins of the CRISPR genome-editing revolution. Hum Gene Ther 26:413–424. 10.1089/hum.2015.091. [PubMed] [DOI] [PubMed] [Google Scholar]
- 84.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239. 10.1038/nbt.2508. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pennisi E. 2013. The CRISPR craze. Science 341:833–836. 10.1126/science.341.6148.833. [PubMed] [DOI] [PubMed] [Google Scholar]
- 86.Westra ER, Staals RH, Gort G, Høgh S, Neumann S, de la Cruz F, Fineran PC, Brouns SJ. 2013. CRISPR-Cas systems preferentially target the leading regions of MOBF conjugative plasmids. RNA Biol 10:749–761. 10.4161/rna.24202. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Samson JE, Magadan AH, Moineau S. 2015. The CRISPR-Cas immune system and genetic transfers: reaching an equilibrium. Microbiol Spectr 3:PLAS-0034-2014. 10.1128/microbiolspec.PLAS-0034-2014. [PubMed] [DOI] [PubMed] [Google Scholar]
- 88.Price VJ, Huo W, Sharifi A, Palmer KL. 2016. CRISPR-Cas and restriction-modification act additively against conjugative antibiotic resistance plasmid transfer in Enterococcus faecalis. mSphere 1:e00064-16. 10.1128/mSphere.00064-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71. 10.1038/nature09523. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA. 2014. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol 32:1146–1150. 10.1038/nbt.3043. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yosef I, Manor M, Kiro R, Qimron U. 2015. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci U S A 112:7267–7272. 10.1073/pnas.1500107112. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Makarova KS, Wolf YI, van der Oost J, Koonin EV. 2009. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol Direct 4:29. 10.1186/1745-6150-4-29. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Swarts DC, Jore MM, Westra ER, Zhu Y, Janssen JH, Snijders AP, Wang Y, Patel DJ, Berenguer J, Brouns SJ, van der Oost J. 2014. DNA-guided DNA interference by a prokaryotic Argonaute. Nature 507:258–261. 10.1038/nature12971. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barrangou R, van der Oost J. 2015. Bacteriophage exclusion, a new defense system. EMBO J 34:134–135. 10.15252/embj.201490620. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Goldfarb T, Sberro H, Weinstock E, Cohen O, Doron S, Charpak-Amikam Y, Afik S, Ofir G, Sorek R. 2015. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J 34:169–183. 10.15252/embj.201489455. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Curtiss R, III, Charamella LJ, Stallions DR, Mays JA. 1968. Parental functions during conjugation in Escherichia coli K-12. Bacteriol Rev 32:320–348. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 97.Wilkins BM, Hollom SE. 1974. Conjugational synthesis of F lac+ and Col I DNA in the presence of rifampicin and in Escherichia coli K12 mutants defective in DNA synthesis. Mol Gen Genet 134:143–156. 10.1007/BF00268416. [DOI] [PubMed] [Google Scholar]
- 98.Kingsman A, Willetts N. 1978. The requirements for conjugal DNA synthesis in the donor strain during Flac transfer. J Mol Biol 122:287–300. 10.1016/0022-2836(78)90191-2. [PubMed] [DOI] [PubMed] [Google Scholar]
- 99.Lee CA, Babic A, Grossman AD. 2010. Autonomous plasmid-like replication of a conjugative transposon. Mol Microbiol 75:268–279. 10.1111/j.1365-2958.2009.06985.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Petit MA, Dervyn E, Rose M, Entian KD, McGovern S, Ehrlich SD, Bruand C. 1998. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol Microbiol 29:261–273. 10.1046/j.1365-2958.1998.00927.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 101.Tarantino PM, Jr, Zhi C, Wright GE, Brown NC. 1999. Inhibitors of DNA polymerase III as novel antimicrobial agents against gram-positive eubacteria. Antimicrob Agents Chemother 43:1982–1987. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Miyazaki R, Minoia M, Pradervand N, Sulser S, Reinhard F, van der Meer JR. 2012. Cellular variability of RpoS expression underlies subpopulation activation of an integrative and conjugative element. PLoS Genet 8:e1002818. 10.1371/journal.pgen.1002818. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roca AI, Cox MM. 1997. RecA protein: structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol 56:129–223. 10.1016/S0079-6603(08)61005-3. [PubMed] [DOI] [PubMed] [Google Scholar]
- 104.Petrova V, Chitteni-Pattu S, Drees JC, Inman RB, Cox MM. 2009. An SOS inhibitor that binds to free RecA protein: the PsiB protein. Mol Cell 36:121–130. 10.1016/j.molcel.2009.07.026. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Baharoglu Z, Bikard D, Mazel D. 2010. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet 6:e1001165. 10.1371/journal.pgen.1001165. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Frost LS, Koraimann G. 2010. Regulation of bacterial conjugation: balancing opportunity with adversity. Future Microbiol 5:1057–1071. 10.2217/fmb.10.70. [PubMed] [DOI] [PubMed] [Google Scholar]
- 107.Fernandez-Lopez R, Del Campo I, Revilla C, Cuevas A, de la Cruz F. 2014. Negative feedback and transcriptional overshooting in a regulatory network for horizontal gene transfer. PLoS Genet 10:e1004171. 10.1371/journal.pgen.1004171. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pérez-Mendoza D, de la Cruz F. 2009. Escherichia coli genes affecting recipient ability in plasmid conjugation: are there any? BMC Genomics 10:71. 10.1186/1471-2164-10-71. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bruand C, Ehrlich SD. 2000. UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol Microbiol 35:204–210. 10.1046/j.1365-2958.2000.01700.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 110.Watanabe T, Arai T, Hattori T. 1970. Effects of cell wall polysaccharide on the mating ability of Salmonella typhimurium. Nature 225:70–71. 10.1038/225070a0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 111.Monner DA, Jonsson S, Boman HG. 1971. Ampicillin-resistant mutants of Escherichia coli K-12 with lipopolysaccharide alterations affecting mating ability and susceptibility to sex-specific bacteriophages. J Bacteriol 107:420–432. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Skurray RA, Hancock RE, Reeves P. 1974. Con– mutants: class of mutants in Escherichia coli K-12 lacking a major cell wall protein and defective in conjugation and adsorption of a bacteriophage. J Bacteriol 119:726–735. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Havekes L, Tommassen J, Hoekstra W, Lugtenberg B. 1977. Isolation and characterization of Escherichia coli K-12 F– mutants defective in conjugation with an I-type donor. J Bacteriol 129:1–8. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hoekstra WP, Havekes AM. 1979. On the role of the recipient cell during conjugation in Escherichia coli. Antonie van Leeuwenhoek 45:13–18. 10.1007/BF00400773. [PubMed] [DOI] [PubMed] [Google Scholar]
- 115.Sanderson KE, Janzer J, Head J. 1981. Influence of lipopolysaccharide and protein in the cell envelope on recipient capacity in conjugation of Salmonella typhimurium. J Bacteriol 148:283–293. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Manoil C, Rosenbusch JP. 1982. Conjugation-deficient mutants of Escherichia coli distinguish classes of functions of the outer membrane OmpA protein. Mol Gen Genet 187:148–156. 10.1007/BF00384398. [PubMed] [DOI] [PubMed] [Google Scholar]
- 117.Duke J, Guiney DG, Jr. 1983. The role of lipopolysaccharide structure in the recipient cell during plasmid-mediated bacterial conjugation. Plasmid 9:222–226. 10.1016/0147-619X(83)90024-0. [DOI] [PubMed] [Google Scholar]
- 118.Anthony KG, Sherburne C, Sherburne R, Frost LS. 1994. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol Microbiol 13:939–953. 10.1111/j.1365-2958.1994.tb00486.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 119.Ishiwa A, Komano T. 2000. The lipopolysaccharide of recipient cells is a specific receptor for PilV proteins, selected by shufflon DNA rearrangement, in liquid matings with donors bearing the R64 plasmid. Mol Gen Genet 263:159–164. 10.1007/s004380050043. [PubMed] [DOI] [PubMed] [Google Scholar]
- 120.Klimke WA, Rypien CD, Klinger B, Kennedy RA, Rodriguez-Maillard JM, Frost LS. 2005. The mating pair stabilization protein, TraN, of the F plasmid is an outer-membrane protein with two regions that are important for its function in conjugation. Microbiology 151:3527–3540. 10.1099/mic.0.28025-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 121.Inoue K, Miyazaki R, Ohtsubo Y, Nagata Y, Tsuda M. 2013. Inhibitory effect of Pseudomonas putida nitrogen-related phosphotransferase system on conjugative transfer of IncP-9 plasmid from Escherichia coli. FEMS Microbiol Lett 345:102–109. 10.1111/1574-6968.12188. [PubMed] [DOI] [PubMed] [Google Scholar]
- 122.van Opijnen T, Camilli A. 2013. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol 11:435–442. 10.1038/nrmicro3033. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Johnson CM, Grossman AD. 2014. Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis. Mol Microbiol 93:1284–1301. 10.1111/mmi.12736. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bhatty M, Laverde Gomez JA, Christie PJ. 2013. The expanding bacterial type IV secretion lexicon. Res Microbiol 164:620–639. 10.1016/j.resmic.2013.03.012. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lang S, Kirchberger PC, Gruber CJ, Redzej A, Raffl S, Zellnig G, Zangger K, Zechner EL. 2011. An activation domain of plasmid R1 TraI protein delineates stages of gene transfer initiation. Mol Microbiol 82:1071–1085. 10.1111/j.1365-2958.2011.07872.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dunny GM. 2013. Enterococcal sex pheromones: signaling, social behavior, and evolution. Annu Rev Genet 47:457–482. 10.1146/annurev-genet-111212-133449. [PubMed] [DOI] [PubMed] [Google Scholar]
- 127.Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. 2005. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci U S A 102:12554–12559. 10.1073/pnas.0505835102. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lederberg J, Cavalli LL, Lederberg EM. 1952. Sex compatibility in Escherichia coli. Genetics 37:720–730. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Watanabe T. 1963. Infective heredity of multiple drug resistance in bacteria. Bacteriol Rev 27:87–115. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Novick RP. 1969. Extrachromosomal inheritance in bacteria. Bacteriol Rev 33:210–263. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Achtman M, Kennedy N, Skurray R. 1977. Cell–cell interactions in conjugating Escherichia coli: role of traT protein in surface exclusion. Proc Natl Acad Sci U S A 74:5104–5108. 10.1073/pnas.74.11.5104. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Achtman M, Manning PA, Edelbluth C, Herrlich P. 1979. Export without proteolytic processing of inner and outer membrane proteins encoded by F sex factor tra cistrons in Escherichia coli minicells. Proc Natl Acad Sci U S A 76:4837–4841. 10.1073/pnas.76.10.4837. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Klimke WA, Frost LS. 1998. Genetic analysis of the role of the transfer gene, traN, of the F and R100-1 plasmids in mating pair stabilization during conjugation. J Bacteriol 180:4036–4043. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Anthony KG, Klimke WA, Manchak J, Frost LS. 1999. Comparison of proteins involved in pilus synthesis and mating pair stabilization from the related plasmids F and R100-1: insights into the mechanism of conjugation. J Bacteriol 181:5149–5159. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Audette GF, Manchak J, Beatty P, Klimke WA, Frost LS. 2007. Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology 153:442–451. 10.1099/mic.0.2006/001917-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 136.Garcillán-Barcia MP, de la Cruz F. 2008. Why is entry exclusion an essential feature of conjugative plasmids? Plasmid 60:1–18. 10.1016/j.plasmid.2008.03.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 137.Watanabe T, Fukasawa T. 1962. Episome-mediated transfer of drug resistance in Enterobacteriaceae. IV. Interactions between resistance transfer factor and F-factor in Escherichia coli K-12. J Bacteriol 83:727–735. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Koraimann G, Teferle K, Markolin G, Woger W, Högenauer G. 1996. The FinOP repressor system of plasmid R1: analysis of the antisense RNA control of traJ expression and conjugative DNA transfer. Mol Microbiol 21:811–821. 10.1046/j.1365-2958.1996.361401.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 139.Jerome LJ, van Biesen T, Frost LS. 1999. Degradation of FinP antisense RNA from F-like plasmids: the RNA-binding protein, FinO, protects FinP from ribonuclease E. J Mol Biol 285:1457–1473. 10.1006/jmbi.1998.2404. [PubMed] [DOI] [PubMed] [Google Scholar]
- 140.Jerome LJ, Frost LS. 1999. In vitro analysis of the interaction between the FinO protein and FinP antisense RNA of F-like conjugative plasmids. J Biol Chem 274:10356–10362. 10.1074/jbc.274.15.10356. [PubMed] [DOI] [PubMed] [Google Scholar]
- 141.Frost LS, Ippen-Ihler K, Skurray RA. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev 58:162–210. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gasson MJ, Willetts NS. 1977. Further characterization of the F fertility inhibition systems of “unusual” Fin+ plasmids. J Bacteriol 131:413–420. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gasson MJ, Willetts NS. 1975. Five control systems preventing transfer of Escherichia coli K-12 sex factor F. J Bacteriol 122:518–525. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gasson M, Willetts N. 1976. Transfer gene expression during fertility inhibition of the Escherichia coli K12 sex factor F by the I-like plasmid R62. Mol Gen Genet 149:329–333. 10.1007/BF00268535. [PubMed] [DOI] [PubMed] [Google Scholar]
- 145.Gaffney D, Skurray R, Willetts N, Brenner S. 1983. Regulation of the F conjugation genes studied by hybridization and tra-lacZ fusion. J Mol Biol 168:103–122. 10.1016/S0022-2836(83)80325-8. [PubMed] [DOI] [PubMed] [Google Scholar]
- 146.Ham LM, Skurray R. 1989. Molecular analysis and nucleotide sequence of finQ, a transcriptional inhibitor of the F plasmid transfer genes. Mol Gen Genet 216:99–105. 10.1007/BF00332236. [DOI] [PubMed] [Google Scholar]
- 147.Penfold SS, Simon J, Frost LS. 1996. Regulation of the expression of the traM gene of the F sex factor of Escherichia coli. Mol Microbiol 20:549–558. 10.1046/j.1365-2958.1996.5361059.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 148.Willetts N. 1980. Interactions between the F conjugal transfer system and CloDF13::Tna plasmids. Mol Gen Genet 180:213–217. 10.1007/BF00267372. [PubMed] [DOI] [PubMed] [Google Scholar]
- 149.Olsen RH, Shipley PL. 1975. RP1 properties and fertility inhibition among P, N, W, and X incompatibility group plasmids. J Bacteriol 123:28–35. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yusoff K, Stanisich VA. 1984. Location of a function on RP1 that fertility inhibits Inc W plasmids. Plasmid 11:178–181. 10.1016/0147-619X(84)90022-2. [DOI] [PubMed] [Google Scholar]
- 151.Fong ST, Stanisich VA. 1989. Location and characterization of two functions on RP1 that inhibit the fertility of the IncW plasmid R388. J Gen Microbiol 135:499–502. [PubMed] [DOI] [PubMed] [Google Scholar]
- 152.Pinney RJ, Smith JT. 1974. Fertility inhibition of an N group R factor by a group X R factor, R6K. J Gen Microbiol 82:415–418. 10.1099/00221287-82-2-415. [PubMed] [DOI] [PubMed] [Google Scholar]
- 153.Herrera-Cervera JA, Olivares J, Sanjuan J. 1996. Ammonia inhibition of plasmid pRmeGR4a conjugal transfer between Rhizobium meliloti strains. Appl Environ Microbiol 62:1145–1150. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Datta N, Hedges RW, Shaw EJ, Sykes RB, Richmond MH. 1971. Properties of an R factor from Pseudomonas aeruginosa. J Bacteriol 108:1244–1249. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Coetzee JN, Datta N, Hedges RW. 1972. R factors from Proteus rettgeri. J Gen Microbiol 72:543–552. 10.1099/00221287-72-3-543. [PubMed] [DOI] [PubMed] [Google Scholar]
- 156.Winans SC, Walker GC. 1985. Fertility inhibition of RP1 by IncN plasmid pKM101. J Bacteriol 161:425–427. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tanimoto K, Iino T. 1983. Transfer inhibition of RP4 by F factor. Mol Gen Genet 192:104–109. 10.1007/BF00327654. [PubMed] [DOI] [PubMed] [Google Scholar]
- 158.Tanimoto K, Iino T. 1984. An essential gene for replication of the mini-F plasmid from origin I. Mol Gen Genet 196:59–63. 10.1007/BF00334092. [PubMed] [DOI] [PubMed] [Google Scholar]
- 159.Miller JF, Malamy MH. 1983. Identification of the pifC gene and its role in negative control of F factor pif gene expression. J Bacteriol 156:338–347. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Miller JF, Lanka E, Malamy MH. 1985. F factor inhibition of conjugal transfer of broad-host-range plasmid RP4: requirement for the protein product of pif operon regulatory gene pifC. J Bacteriol 163:1067–1073. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tanimoto K, Iino T, Ohtsubo H, Ohtsubo E. 1985. Identification of a gene, tir of R100, functionally homologous to the F3 gene of F in the inhibition of RP4 transfer. Mol Gen Genet 198:356–357. 10.1007/BF00383019. [PubMed] [DOI] [PubMed] [Google Scholar]
- 162.Santini JM, Stanisich VA. 1998. Both the fipA gene of pKM101 and the pifC gene of F inhibit conjugal transfer of RP1 by an effect on traG. J Bacteriol 180:4093–4101. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Loper JE, Kado CI. 1979. Host range conferred by the virulence-specifying plasmid of Agrobacterium tumefaciens. J Bacteriol 139:591–596. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Farrand S, Kado C, Ireland C. 1981. Suppression of tumorigenicity by the IncW R plasmid pSa in Agrobacterium tumefaciens. Mol Gen Genet 181:44–51. 10.1007/BF00339003. [DOI] [Google Scholar]
- 165.Close SM, Kado CI. 1991. The osa gene of pSa encodes a 21.1-kilodalton protein that suppresses Agrobacterium tumefaciens oncogenicity. J Bacteriol 173:5449–5456. 10.1128/jb.173.17.5449-5456.1991. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ward JE, Jr, Dale EM, Binns AN. 1991. Activity of the Agrobacterium T-DNA transfer machinery is affected by virB gene products. Proc Natl Acad Sci U S A 88:9350–9354. 10.1073/pnas.88.20.9350. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Binns AN, Beaupré CE, Dale EM. 1995. Inhibition of VirB-mediated transfer of diverse substrates from Agrobacterium tumefaciens by the IncQ plasmid RSF1010. J Bacteriol 177:4890–4899. 10.1128/jb.177.17.4890-4899.1995. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stahl LE, Jacobs A, Binns AN. 1998. The conjugal intermediate of plasmid RSF1010 inhibits Agrobacterium tumefaciens virulence and VirB-dependent export of VirE2. J Bacteriol 180:3933–3939. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lee LY, Gelvin SB. 2004. Osa protein constitutes a strong oncogenic suppression system that can block vir-dependent transfer of IncQ plasmids between Agrobacterium cells and the establishment of IncQ plasmids in plant cells. J Bacteriol 186:7254–7261. 10.1128/JB.186.21.7254-7261.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen CY, Kado CI. 1994. Inhibition of Agrobacterium tumefaciens oncogenicity by the osa gene of pSa. J Bacteriol 176:5697–5703. 10.1128/jb.176.18.5697-5703.1994. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chen CY, Kado CI. 1996. Osa protein encoded by plasmid pSa is located at the inner membrane but does not inhibit membrane association of VirB and VirD virulence proteins in Agrobacterium tumefaciens. FEMS Microbiol Lett 135:85–92. 10.1111/j.1574-6968.1996.tb07970.x. [DOI] [PubMed] [Google Scholar]
- 172.Lee LY, Gelvin SB, Kado CI. 1999. pSa causes oncogenic suppression of Agrobacterium by inhibiting VirE2 protein export. J Bacteriol 181:186–196. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chumakov MI. 2013. Protein apparatus for horizontal transfer of agrobacterial T-DNA to eukaryotic cells. Biochemistry (Mosc) 78:1321–1332. 10.1134/S000629791312002X. [PubMed] [DOI] [PubMed] [Google Scholar]
- 174.Schrammeijer B, den Dulk-Ras A, Vergunst AC, Jurado Jácome E, Hooykaas PJ. 2003. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res 31:860–868. 10.1093/nar/gkg179. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cascales E, Atmakuri K, Liu Z, Binns AN, Christie PJ. 2005. Agrobacterium tumefaciens oncogenic suppressors inhibit T-DNA and VirE2 protein substrate binding to the VirD4 coupling protein. Mol Microbiol 58:565–579. 10.1111/j.1365-2958.2005.04852.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Maindola P, Raina R, Goyal P, Atmakuri K, Ojha A, Gupta S, Christie PJ, Iyer LM, Aravind L, Arockiasamy A. 2014. Multiple enzymatic activities of ParB/Srx superfamily mediate sexual conflict among conjugative plasmids. Nat Commun 5:5322. 10.1038/ncomms6322. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fernandez-Lopez R, Machón C, Longshaw CM, Martin S, Molin S, Zechner EL, Espinosa M, Lanka E, de la Cruz F. 2005. Unsaturated fatty acids are inhibitors of bacterial conjugation. Microbiology 151:3517–3526. 10.1099/mic.0.28216-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 178.Lujan SA, Guogas LM, Ragonese H, Matson SW, Redinbo MR. 2007. Disrupting antibiotic resistance propagation by inhibiting the conjugative DNA relaxase. Proc Natl Acad Sci U S A 104:12282–12287. 10.1073/pnas.0702760104. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nash RP, McNamara DE, Ballentine WK, III, Matson SW, Redinbo MR. 2012. Investigating the impact of bisphosphonates and structurally related compounds on bacteria containing conjugative plasmids. Biochem Biophys Res Commun 424:697–703. 10.1016/j.bbrc.2012.07.012. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Jalasvuori M, Friman VP, Nieminen A, Bamford JK, Buckling A. 2011. Bacteriophage selection against a plasmid-encoded sex apparatus leads to the loss of antibiotic-resistance plasmids. Biol Lett 7:902–905. 10.1098/rsbl.2011.0384. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ojala V, Laitalainen J, Jalasvuori M. 2013. Fight evolution with evolution: plasmid-dependent phages with a wide host range prevent the spread of antibiotic resistance. Evol Appl 6:925–932. 10.1111/eva.12076. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Harrison E, Wood AJ, Dytham C, Pitchford JW, Truman J, Spiers A, Paterson S, Brockhurst MA. 2015. Bacteriophages limit the existence conditions for conjugative plasmids. mBio 6:e00586. 10.1128/mBio.00586-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hagens S, Habel A, Bläsi U. 2006. Augmentation of the antimicrobial efficacy of antibiotics by filamentous phage. Microb Drug Resist 12:164–168. 10.1089/mdr.2006.12.164. [PubMed] [DOI] [PubMed] [Google Scholar]
- 184.Boeke JD, Model P, Zinder ND. 1982. Effects of bacteriophage f1 gene III protein on the host cell membrane. Mol Gen Genet 186:185–192. 10.1007/BF00331849. [PubMed] [DOI] [PubMed] [Google Scholar]
- 185.Brinton CC, Jr, Gemski P, Jr, Carnahan J. 1964. A new type of bacterial pilus genetically controlled by the fertility factor of E. coli K 12 and its role in chromosome transfer. Proc Natl Acad Sci U S A 52:776–783. 10.1073/pnas.52.3.776. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Knolle P. 1967. Evidence for the identity of the mating-specific site of male cells of Escherichia coli with the receptor site of an RNA phage. Biochem Biophys Res Commun 27:81–87. 10.1016/S0006-291X(67)80043-3. [DOI] [PubMed] [Google Scholar]
- 187.Ippen KA, Valentine RC. 1967. The sex hair of E. coli as sensory fiber, conjugation tube, or mating arm? Biochem Biophys Res Commun 27:674–680. 10.1016/S0006-291X(67)80088-3. [PubMed] [DOI] [PubMed] [Google Scholar]
- 188.Novotny C, Knight WS, Brinton CC, Jr. 1968. Inhibition of bacterial conjugation by ribonucleic acid and deoxyribonucleic acid male-specific bacteriophages. J Bacteriol 95:314–326. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Salzman TC. 1971. Coordination of sex pili with their specifying R factors. Nat New Biol 230:278–279. 10.1038/newbio230278a0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 190.Ou JT. 1973. Inhibition of formation of Escherichia coli mating pairs by f1 and MS2 bacteriophages as determined with a Coulter counter. J Bacteriol 114:1108–1115. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Schreil W, Christensen JR. 1974. Conjugation and phage MS 2 infection with Hfr Escherichia coli. Arch Mikrobiol 95:19–28. 10.1007/BF02451744. [PubMed] [DOI] [PubMed] [Google Scholar]
- 192.Lin A, Jimenez J, Derr J, Vera P, Manapat ML, Esvelt KM, Villanueva L, Liu DR, Chen IA. 2011. Inhibition of bacterial conjugation by phage M13 and its protein g3p: quantitative analysis and model. PLoS One 6:e19991. 10.1371/journal.pone.0019991. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lubkowski J, Hennecke F, Plückthun A, Wlodawer A. 1999. Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA. Structure 7:711–722. 10.1016/S0969-2126(99)80092-6. [DOI] [PubMed] [Google Scholar]
- 194.Wan Z, Goddard NL. 2012. Competition between conjugation and M13 phage infection in Escherichia coli in the absence of selection pressure: a kinetic study. G3 (Bethesda) 2:1137–1144. 10.1534/g3.112.003418. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Freese PD, Korolev KS, Jiménez JI, Chen IA. 2014. Genetic drift suppresses bacterial conjugation in spatially structured populations. Biophys J 106:944–954. 10.1016/j.bpj.2014.01.012. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.May T, Tsuruta K, Okabe S. 2011. Exposure of conjugative plasmid carrying Escherichia coli biofilms to male-specific bacteriophages. ISME J 5:771–775. 10.1038/ismej.2010.158. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Harden V, Meynell E. 1972. Inhibition of gene transfer by antiserum and identification of serotypes of sex pili. J Bacteriol 109:1067–1074. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lawn AM, Meynell E. 1970. Serotypes of sex pili. J Hyg (Lond) 68:683–694. 10.1017/S0022172400042625. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Tzagoloff H, Pratt D. 1964. The initial steps in infection with coliphage M13. Virology 24:372–380. 10.1016/0042-6822(64)90174-6. [PubMed] [DOI] [PubMed] [Google Scholar]
- 200.Ou JT, Anderson TF. 1972. Effect of Zn2+ on bacterial conjugation: inhibition of mating pair formation. J Bacteriol 111:177–185. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ou JT. 1973. Effect of Zn2+ on bacterial conjugation: increase in ability of F– cells to form mating pairs. J Bacteriol 115:648–654. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ou JT, Reim R. 1976. Effect of 1,10-phenanthroline on bacterial conjugation in Escherichia coli K-12: inhibition of maturation from preliminary mates into effective mates. J Bacteriol 128:363–371. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sneath PH, Lederberg J. 1961. Inhibition by periodate of mating in Escherichia coli K-12. Proc Natl Acad Sci U S A 47:86–90. 10.1073/pnas.47.1.86. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Dettori R, Maccacaro G, Piccinin G. 1961. Sex-specific bacteriophages of Escherichia coli K12. G Microbiol 9:141–150. [Google Scholar]
- 205.Raab C, Röschenthaler R. 1970. Inhibition of adsorption and replication of the RNA-phage MS-2 in Escherichia coli C 3000 by levallorphan. Biochem Biophys Res Commun 41:1429–1436. 10.1016/0006-291X(70)90546-2. [DOI] [PubMed] [Google Scholar]
- 206.Löser R, Boquet PL, Röschenthaler R. 1971. Inhibition of R-factor transfer by levallorphan. Biochem Biophys Res Commun 45:204–211. 10.1016/0006-291X(71)90070-2. [DOI] [PubMed] [Google Scholar]
- 207.Mándi Y, Molnár J. 1981. Effect of chlorpromazine on conjugal plasmid transfer and sex pili. Acta Microbiol Acad Sci Hung 28:205–210. [PubMed] [PubMed] [Google Scholar]
- 208.Singleton P. 1983. Colloidal clay inhibits conjugal transfer of R-plasmid R1drd-19 in Escherichia coli. Appl Environ Microbiol 46:756–757. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Roper MM, Marshall KC. 1977. Effects of a clay mineral on microbial predation and parasitism of Escherichia coli. Microb Ecol 4:279–289. 10.1007/BF02013272. [PubMed] [DOI] [PubMed] [Google Scholar]
- 210.Roper MM, Marshall KC. 1978. Effect of clay particle size on clay-Escherichia coli-bacteriophage interactions. Microbiology 106:187–189. [Google Scholar]
- 211.Qiu Z, Yu Y, Chen Z, Jin M, Yang D, Zhao Z, Wang J, Shen Z, Wang X, Qian D, Huang A, Zhang B, Li JW. 2012. Nanoalumina promotes the horizontal transfer of multiresistance genes mediated by plasmids across genera. Proc Natl Acad Sci U S A 109:4944–4949. 10.1073/pnas.1107254109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Schwartz GH, Eiler D, Kern M. 1965. Solubilization of the conjugation inhibitor from Escherichia coli cell wall. J Bacteriol 89:89–94. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Lancaster JH, Goldschmidt EP, Wyss O. 1965. Characterization of conjugation factors in Escherichia coli cell walls. I. Inhibition of recombination by cell walls and cell extracts. J Bacteriol 89:1478–1481. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Spengler G, Molnár A, Schelz Z, Amaral L, Sharples D, Molnár J. 2006. The mechanism of plasmid curing in bacteria. Curr Drug Targets 7:823–841. 10.2174/138945006777709601. [PubMed] [DOI] [PubMed] [Google Scholar]
- 215.Bidlack JE, Silverman PM. 2004. An active type IV secretion system encoded by the F plasmid sensitizes Escherichia coli to bile salts. J Bacteriol 186:5202–5209. 10.1128/JB.186.16.5202-5209.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Daugelavicius R, Bamford JK, Grahn AM, Lanka E, Bamford DH. 1997. The IncP plasmid-encoded cell envelope-associated DNA transfer complex increases cell permeability. J Bacteriol 179:5195–5202. 10.1128/jb.179.16.5195-5202.1997. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Russell AB, Peterson SB, Mougous JD. 2014. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol 12:137–148. 10.1038/nrmicro3185. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Basler M, Ho BT, Mekalanos JJ. 2013. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152:884–894. 10.1016/j.cell.2013.01.042. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ho BT, Basler M, Mekalanos JJ. 2013. Type 6 secretion system-mediated immunity to type 4 secretion system-mediated gene transfer. Science 342:250–253. 10.1126/science.1243745. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Weisser J, Wiedemann B. 1987. Inhibition of R-plasmid transfer in Escherichia coli by 4-quinolones. Antimicrob Agents Chemother 31:531–534. 10.1128/AAC.31.4.531. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Debbia EA, Massaro S, Campora U, Schito GC. 1994. Inhibition of F’lac transfer by various antibacterial drugs in Escherichia coli. New Microbiol 17:65–68. [PubMed] [PubMed] [Google Scholar]
- 222.Michel-Briand Y, Laporte JM. 1985. Inhibition of conjugal transfer of R plasmids by nitrofurans. J Gen Microbiol 131:2281–2284. [DOI] [PubMed] [Google Scholar]
- 223.Nakamura S, Inoue S, Shimizu M, Iyobe S, Mitsuhashi S. 1976. Inhibition of conjugal transfer of R plasmids by pipemidic acid and related compounds. Antimicrob Agents Chemother 10:779–785. 10.1128/AAC.10.5.779. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Warnes SL, Highmore CJ, Keevil CW. 2012. Horizontal transfer of antibiotic resistance genes on abiotic touch surfaces: implications for public health. mBio 3:e00489-12. 10.1128/mBio.00489-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Warnes SL, Green SM, Michels HT, Keevil CW. 2010. Biocidal efficacy of copper alloys against pathogenic enterococci involves degradation of genomic and plasmid DNAs. Appl Environ Microbiol 76:5390–5401. 10.1128/AEM.03050-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Warnes SL, Caves V, Keevil CW. 2012. Mechanism of copper surface toxicity in Escherichia coli O157:H7 and Salmonella involves immediate membrane depolarization followed by slower rate of DNA destruction which differs from that observed for Gram-positive bacteria. Environ Microbiol 14:1730–1743. 10.1111/j.1462-2920.2011.02677.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 227.Hong R, Kang TY, Michels CA, Gadura N. 2012. Membrane lipid peroxidation in copper alloy-mediated contact killing of Escherichia coli. Appl Environ Microbiol 78:1776–1784. 10.1128/AEM.07068-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zhao WH, Hu ZQ, Hara Y, Shimamura T. 2001. Inhibition by epigallocatechin gallate (EGCg) of conjugative R plasmid transfer in Escherichia coli. J Infect Chemother 7:195–197. 10.1007/s101560100035. [PubMed] [DOI] [PubMed] [Google Scholar]
- 229.Oyedemi BO, Shinde V, Shinde K, Kakalou D, Stapleton PD, Gibbons S. 2016. Novel R-plasmid conjugal transfer inhibitory and antibacterial activities of phenolic compounds from Mallotus philippensis (Lam.) Mull. Arg. J Glob Antimicrob Resist 5:15–21. 10.1016/j.jgar.2016.01.011. [PubMed] [DOI] [PubMed] [Google Scholar]
- 230.Emeruwa AC. 1982. Antibacterial substance from Carica papaya fruit extract. J Nat Prod 45:123–127. 10.1021/np50020a002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 231.Leite AA, Nardi RM, Nicoli JR, Chartone-Souza E, Nascimento AM. 2005. Carica papaya seed macerate as inhibitor of conjugative R plasmid transfer from Salmonella typhimurium to Escherichia coli in vitro and in the digestive tract of gnotobiotic mice. J Gen Appl Microbiol 51:21–26. 10.2323/jgam.51.21. [PubMed] [DOI] [PubMed] [Google Scholar]
- 232.García-Quintanilla M, Ramos-Morales F, Casadesús J. 2008. Conjugal transfer of the Salmonella enterica virulence plasmid in the mouse intestine. J Bacteriol 190:1922–1927. 10.1128/JB.01626-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Heseltine WW, Galloway LD. 1951. Some antibacterial properties of sodium propionate. J Pharm Pharmacol 3:581–585. 10.1111/j.2042-7158.1951.tb13103.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 234.Couce A, Blázquez J. 2009. Side effects of antibiotics on genetic variability. FEMS Microbiol Rev 33:531–538. 10.1111/j.1574-6976.2009.00165.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 235.Beaber JW, Hochhut B, Waldor MK. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72–74. 10.1038/nature02241. [PubMed] [DOI] [PubMed] [Google Scholar]
- 236.Stevens AM, Shoemaker NB, Li LY, Salyers AA. 1993. Tetracycline regulation of genes on Bacteroides conjugative transposons. J Bacteriol 175:6134–6141. 10.1128/jb.175.19.6134-6141.1993. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Torres OR, Korman RZ, Zahler SA, Dunny GM. 1991. The conjugative transposon Tn925: enhancement of conjugal transfer by tetracycline in Enterococcus faecalis and mobilization of chromosomal genes in Bacillus subtilis and E. faecalis. Mol Gen Genet 225:395–400. 10.1007/BF00261679. [PubMed] [DOI] [PubMed] [Google Scholar]
- 238.Barr V, Barr K, Millar MR, Lacey RW. 1986. Beta-lactam antibiotics increase the frequency of plasmid transfer in Staphylococcus aureus. J Antimicrob Chemother 17:409–413. 10.1093/jac/17.4.409. [PubMed] [DOI] [PubMed] [Google Scholar]
- 239.Francia MV, Varsaki A, Garcillán-Barcia MP, Latorre A, Drainas C, de la Cruz F. 2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev 28:79–100. 10.1016/j.femsre.2003.09.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 240.Getino M, Fernández-López R, Palencia-Gándara C, Campos-Gómez J, Sánchez-López JM, Martínez M, Fernández A, de la Cruz F. 2016. Tanzawaic acids, a chemically novel set of bacterial conjugation inhibitors. PLoS One 11:e0148098. 10.1371/journal.pone.0148098. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Getino M, Sanabria-Ríos DJ, Fernández-López R, Campos-Gómez J, Sánchez-López JM, Fernández A, Carballeira NM, de la Cruz F. 2015. Synthetic fatty acids prevent plasmid-mediated horizontal gene transfer. mBio 6:e01032-e15. 10.1128/mBio.01032-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. 10.1128/AAC.01707-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ripoll-Rozada J, García-Cazorla Y, Getino M, Machón C, Sanabria-Ríos D, de la Cruz F, Cabezón E, Arechaga I. 2016. Type IV traffic ATPase TrwD as molecular target to inhibit bacterial conjugation. Mol Microbiol 100:912–921. 10.1111/mmi.13359. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kerr JE, Christie PJ. 2010. Evidence for VirB4-mediated dislocation of membrane-integrated VirB2 pilin during biogenesis of the Agrobacterium VirB/VirD4 type IV secretion system. J Bacteriol 192:4923–4934. 10.1128/JB.00557-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Atmakuri K, Cascales E, Christie PJ. 2004. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol 54:1199–1211. 10.1111/j.1365-2958.2004.04345.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Guglielmini J, Néron B, Abby SS, Garcillán-Barcia MP, de la Cruz F, Rocha EP. 2014. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res 42:5715–5727. 10.1093/nar/gku194. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Hilleringmann M, Pansegrau W, Doyle M, Kaufman S, MacKichan ML, Gianfaldoni C, Ruggiero P, Covacci A. 2006. Inhibitors of Helicobacter pylori ATPase Cagα block CagA transport and cag virulence. Microbiology 152:2919–2930. 10.1099/mic.0.28984-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 248.Sayer JR, Walldén K, Pesnot T, Campbell F, Gane PJ, Simone M, Koss H, Buelens F, Boyle TP, Selwood DL, Waksman G, Tabor AB. 2014. 2- and 3-substituted imidazo[1,2-a]pyrazines as inhibitors of bacterial type IV secretion. Bioorg Med Chem 22:6459–6470. 10.1016/j.bmc.2014.09.036. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Shaffer CL, Good JA, Kumar S, Krishnan KS, Gaddy JA, Loh JT, Chappell J, Almqvist F, Cover TL, Hadjifrangiskou M. 2016. Peptidomimetic small molecules disrupt type IV secretion system activity in diverse bacterial pathogens. mBio 7:e00221-e16. 10.1128/mBio.00221-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Pinkner JS, Remaut H, Buelens F, Miller E, Aberg V, Pemberton N, Hedenström M, Larsson A, Seed P, Waksman G, Hultgren SJ, Almqvist F. 2006. Rationally designed small compounds inhibit pilus biogenesis in uropathogenic bacteria. Proc Natl Acad Sci U S A 103:17897–17902. 10.1073/pnas.0606795103. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Galán JE, Lara-Tejero M, Marlovits TC, Wagner S. 2014. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol 68:415–438. 10.1146/annurev-micro-092412-155725. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Charro N, Mota LJ. 2015. Approaches targeting the type III secretion system to treat or prevent bacterial infections. Expert Opin Drug Discov 10:373–387. 10.1517/17460441.2015.1019860. [PubMed] [DOI] [PubMed] [Google Scholar]
- 253.Felise HB, Nguyen HV, Pfuetzner RA, Barry KC, Jackson SR, Blanc MP, Bronstein PA, Kline T, Miller SI. 2008. An inhibitor of Gram-negative bacterial virulence protein secretion. Cell Host Microbe 4:325–336. 10.1016/j.chom.2008.08.001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Krüger DH, Barcak GJ, Reuter M, Smith HO. 1988. EcoRII can be activated to cleave refractory DNA recognition sites. Nucleic Acids Res 16:3997–4008. 10.1093/nar/16.9.3997. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Meisel A, Bickle TA, Krüger DH, Schroeder C. 1992. Type III restriction enzymes need two inversely oriented recognition sites for DNA cleavage. Nature 355:467–469. 10.1038/355467a0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 256.Warren RA. 1980. Modified bases in bacteriophage DNAs. Annu Rev Microbiol 34:137–158. 10.1146/annurev.mi.34.100180.001033. [PubMed] [DOI] [PubMed] [Google Scholar]
- 257.Iida S, Streiff MB, Bickle TA, Arber W. 1987. Two DNA antirestriction systems of bacteriophage P1, darA, and darB: characterization of darA– phages. Virology 157:156–166. 10.1016/0042-6822(87)90324-2. [DOI] [PubMed] [Google Scholar]
- 258.Zabeau M, Friedman S, Van Montagu M, Schell J. 1980. The ral gene of phage lambda. I. Identification of a non-essential gene that modulates restriction and modification in E. coli. Mol Gen Genet 179:63–73. 10.1007/BF00268447. [PubMed] [DOI] [PubMed] [Google Scholar]
- 259.Studier FW, Movva NR. 1976. SAMase gene of bacteriophage T3 is responsible for overcoming host restriction. J Virol 19:136–145. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Walkinshaw MD, Taylor P, Sturrock SS, Atanasiu C, Berge T, Henderson RM, Edwardson JM, Dryden DT. 2002. Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol Cell 9:187–194. 10.1016/S1097-2765(02)00435-5. [DOI] [PubMed] [Google Scholar]
- 261.Serna A, Espinosa E, Camacho EM, Casadesús J. 2010. Regulation of bacterial conjugation in microaerobiosis by host-encoded functions ArcAB and sdhABCD. Genetics 184:947–958. 10.1534/genetics.109.109918. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Camacho EM, Serna A, Casadesús J. 2005. Regulation of conjugal transfer by Lrp and Dam methylation in plasmid R100. Int Microbiol 8:279–285. [PubMed] [PubMed] [Google Scholar]
- 263.Will WR, Lu J, Frost LS. 2004. The role of H-NS in silencing F transfer gene expression during entry into stationary phase. Mol Microbiol 54:769–782. 10.1111/j.1365-2958.2004.04303.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 264.Shin M, Song M, Rhee JH, Hong Y, Kim YJ, Seok YJ, Ha KS, Jung SH, Choy HE. 2005. DNA looping-mediated repression by histone-like protein H-NS: specific requirement of Eσ70 as a cofactor for looping. Genes Dev 19:2388–2398. 10.1101/gad.1316305. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wada C, Imai M, Yura T. 1987. Host control of plasmid replication: requirement for the σ factor σ32 in transcription of mini-F replication initiator gene. Proc Natl Acad Sci U S A 84:8849–8853. 10.1073/pnas.84.24.8849. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Dorman CJ. 2009. Nucleoid-associated proteins and bacterial physiology. Adv Appl Microbiol 67:47–64. 10.1016/S0065-2164(08)01002-2. [PubMed] [DOI] [PubMed] [Google Scholar]
- 267.Gamas P, Caro L, Galas D, Chandler M. 1987. Expression of F transfer functions depends on the Escherichia coli integration host factor. Mol Gen Genet 207:302–305. 10.1007/BF00331593. [PubMed] [DOI] [PubMed] [Google Scholar]
- 268.Starcic M, Zgur-Bertok D, Jordi BJ, Wösten MM, Gaastra W, van Putten JP. 2003. The cyclic AMP-cyclic AMP receptor protein complex regulates activity of the traJ promoter of the Escherichia coli conjugative plasmid pRK100. J Bacteriol 185:1616–1623. 10.1128/JB.185.5.1616-1623.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Frost LS, Manchak J. 1998. F– phenocopies: characterization of expression of the F transfer region in stationary phase. Microbiology 144:2579–2587. 10.1099/00221287-144-9-2579. [PubMed] [DOI] [PubMed] [Google Scholar]
- 270.Will WR, Frost LS. 2006. Hfq is a regulator of F-plasmid TraJ and TraM synthesis in Escherichia coli. J Bacteriol 188:124–131. 10.1128/JB.188.1.124-131.2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Lau-Wong IC, Locke T, Ellison MJ, Raivio TL, Frost LS. 2008. Activation of the Cpx regulon destabilizes the F plasmid transfer activator, TraJ, via the HslVU protease in Escherichia coli. Mol Microbiol 67:516–527. 10.1111/j.1365-2958.2007.06055.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 272.Zahrl D, Wagner A, Tscherner M, Koraimann G. 2007. GroEL plays a central role in stress-induced negative regulation of bacterial conjugation by promoting proteolytic degradation of the activator protein TraJ. J Bacteriol 189:5885–5894. 10.1128/JB.00005-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]


