ABSTRACT
The genus Streptococcus includes Gram-positive organisms shaped in cocci and organized in chains. They are commensals, pathogens, and opportunistic pathogens for humans and animals. Most Streptococcus species of veterinary relevance have a specific ecological niche, such as S. uberis, which is almost exclusively an environmental pathogen causing bovine mastitis. In contrast, S. suis can be considered as a true zoonotic pathogen, causing specific diseases in humans after contact with infected animals or derived food products. Finally, Streptococcus species such as S. agalactiae can be sporadically zoonotic, even though they are pathogens of both humans and animals independently. For clarification, a short taxonomical overview will be given here to highlight the diversity of streptococci that infect animals. Several families of antibiotics are used to treat animals for streptococcal infections. First-line treatments are penicillins (alone or in combination with aminoglycosides), macrolides and lincosamides, fluoroquinolones, and tetracyclines. Because of the selecting role of antibiotics, resistance phenotypes have been reported in streptococci isolated from animals worldwide. Globally, the dynamic of resistance acquisition in streptococci is slower than what is experienced in Enterobacteriaceae, probably due to the much more limited horizontal spread of resistance genes. Nonetheless, transposons or integrative and conjugative elements can disseminate resistance determinants among streptococci. Besides providing key elements on the prevalence of resistance in streptococci from animals, this article will also largely consider the mechanisms and molecular epidemiology of the major types of resistance to antimicrobials encountered in the most important streptococcal species in veterinary medicine.
TAXONOMIC OVERVIEW OF STREPTOCOCCI
More than 60 Streptococcus species have been recognized so far. Some of these, such as S. pyogenes, S. agalactiae, S. equi, S. canis, and S. iniae, produce hemolytic factors and, when cultivated on solid media containing blood, can be classified as beta-hemolytic. However, nonhemolytic variants can also be observed (1). Isolates belonging to other species, such as S. dysgalactiae subsp. dysgalactiae, S. pneumoniae, S. mutans, S. salivarius, S. sanguinis, S. gordonii, S. mitis, and S. oralis, produce hydrogen peroxide that partially lyses the erythrocytes, with the subsequent oxidation of the heme group resulting in a greenish pigment in the medium that is often interpreted as alpha-hemolysis. This oxidation process is influenced by several cultivation conditions and is variably evident. For this reason, it is preferable to consider those latter-mentioned species as nonhemolytic. The truly nonhemolytic species, mainly encompassing S. gallolyticus (formerly S. bovis), were also named gamma-hemolytic. A classification of Streptococcus species proposed by Rebecca Lancefield in the 1930s was based on the antigenic reaction of the cell wall-associated carbohydrates and remains classically used (2). On the basis of this approach, streptococci are distributed into groups ranging from A to W, depending on the antibodies recognizing the specific carbohydrates of a definite streptococcal species. Nevertheless, the whole picture is sometimes complicated by the fact that several antibodies can react with isolates belonging to the same species. For instance, depending on the isolates, S. dysgalactiae subsp. equisimilis may be classified as belonging to the C or G group, while it may also be classified, even though less commonly, as group A or L (3); isolates from S. phocae may belong to either the C or G group; isolates from S. infantarius are sporadically considered as group D; isolates from S. anginosus are indifferently classified as group A, C, G, F, or N; isolates from S. constellatus subsp. constellatus belong to either group F or N; sporadic isolates belonging to S. constellatus subsp. pharyngis can be considered as group C; isolates from the S. intermedius species can be considered as group N; and finally, isolates belonging to S. porcinus are classified in either group P, U, or V.
In the following sections, the most relevant Streptococcus species responsible for diseases in animals and/or humans will be summarized. The relative resistances to selected antibiotics will be discussed in the sections on macrolides-lincosamides-streptogramins B tetracyclines, beta-lactam resistance, fluoroquinolone resistance, and integrative and conjugative elements (ICEs).
Group A
Streptococci have diverse ecological origins, and certain species are exclusively adapted to a unique host as exemplified by the beta-hemolytic S. pyogenes, which is considered as the most pathogenic type of streptococcus for humans, together with S. pneumoniae, and is responsible for pharyngitis, erysipelas, and other invasive diseases such as soft tissue infection, rheumatic fever, glomerulonephritis, and streptococcal toxic shock syndrome (STSS) (4, 5). The finding of S. pyogenes in animals has been debated. According to Copperman (6), pets could have been the source of contagious pharyngitis (6), but no expansion of these findings has been reported, supporting the hypothesis that humans are the exclusive reservoir of S. pyogenes.
Group B
The beta-hemolytic S. agalactiae, or group B streptococci, according to the Lancefield’s classification, is a commensal of the human intestinal and urogenital tract and is infamous as a human pathogen causing severe diseases such as pneumonia, sepsis, and meningitis in newborns and pregnant women; recently, its pathogenic importance in elderly and immunocompromised patients has been re-evaluated (7). S. agalactiae is also an animal pathogen and has been reported from a variety of hosts such as fish with meningoencephalitis (8), camels with mastitis and joints infections (9), and horses with unspecified disease or death (10). Classically, S. agalactiae has been associated with mastitis in cows (11), and the zoonotic potential of S. agalactiae is debated. On one side, genomic comparative approaches highlight a specific host adaptation of S. agalactiae isolates causing infections (12); on the other side, infections of humans from consumption of fish infected by S. agalactiae has recently been documented (13). Also, experimental infections of fish with S. agalactiae isolates of human origin have resulted in fish death (14). Globally, the hygienic control measures implemented for controlling contagious bovine mastitis have contributed to a sharply decreased prevalence of S. agalactiae in the veterinary sector (15, 16).
Group D
Group D streptococci were divided into two diverging groups in the early 1980s: S. feacalis and S. faecium, which were renamed Enterococcus faecalis and Enterococcus faecium (17). Since then, several new species have been added to the Enterococcus genus (18), which will not be discussed in this review.
Formerly, S. bovis was included in the viridans group of streptococci, and its taxonomy has been reviewed in several studies. Overall, several previously identified S. bovis isolates were classified as group D according to Lancefield’s reaction, as shown in Table 1. Molecular evidence has provided the basis for the classification of the former S. bovis isolates into five species: S. gallolyticus subsp. gallolyticus, S. gallolyticus subsp. pasteurianus, S. gallolyticus subsp. macedonicus, S. infantarius subsp. infantarius, and S. lutetiensis (19). Isolates belonging to S. gallolyticus subsp. gallolyticus have been found as commensals of the gastrointestinal tract of humans and animals but also cause invasive diseases such as sepsis, endocarditis, arthritis, and meningitis in both humans and animals (Table 1). The transmission of S. gallolyticus subsp. gallolyticus between animals and humans has been reported (20), highlighting the zoonotic potential of this species. S. gallolyticus subsp. pasteurianus is an emergent infective agent in human medicine that is responsible for sepsis, bone and joint infections, and meningitis (21–23). In birds, this bacterium is responsible for similar diseases (24, 25). S. lutetiensis has rarely been associated with infective endocarditis and sepsis (26).
TABLE 1.
Overview of streptococci causative of infections in humans and animalsa
| Lancefield group | Hemolysin | Species | Host | Associated disease | References |
|---|---|---|---|---|---|
| B | Beta | S. agalactiae | Human | Sepsis, meningitis, pneumonia, joint and urinary tract infections | 7, 11, 199, 200 |
| Cows, camels, horses, dolphins, fish | Mastitis, joint infection, meningitis, death | ||||
| C | Alpha | S. dysgalactiae subsp. dysgalactiae | Humans | Endocarditis, joint infection, cellulitis | 48, 201, 202 |
| Fish, cows | Tissue necrosis, mastitis | ||||
| Beta | S. dysgalactiae subsp. equisimilis | Humans | STSS, sepsis, soft tissue infections, pneumonia, pharyngitis | 33, 203–210 | |
| Swine, seals, horses | Arthritis, endocarditis, lymphadenitis, joint infection, strangles-like disease, respiratory tract infection | ||||
| Beta | S. equi subsp. equi | Horses | Strangles disease | 32 | |
| Beta | S. equi subsp. zooepidemicus | Humans | Nephritis, STSS | 33 | |
| Sheep, horses | Mastitis, lymphadenitis, joint and respiratory infections, endometritis | 31, 32, 211 | |||
| Beta | S. phocae | Fish, seals | Respiratory infections, abortions, sepsis | 212 | |
| D | S. gallolyticus spp. gallolyticus | Humans | Sepsis, endocarditis, arthritis, meningitis | 213–216 | |
| Koalas, birds | Intestinal colonizer, endocarditis, sepsis | ||||
| S. gallolyticus spp. pasteurianus | Humans | Sepsis, bone and joint infections, meningitis | 21–25 | ||
| Birds | Sepsis, meningitis | ||||
| E | Beta | S. porcinus | Humans | Urinary tract, placenta, and wound infections, sepsis | 27, 28, 217, 218 |
| Swine | Endocarditis, respiratory tract infection, sepsis | ||||
| G | Beta | S. canis | Humans | Skin, soft-tissue and respiratory infections, sepsis | 34, 35, 219, 220 |
| Dogs, cats | Skin, soft-tissue, and urinary tract infections, otitis, arthritis, STSS | ||||
| R | Nonhemolytic/beta | S. suis | Humans | Sepsis, meningitis, endocarditis, STSS | 37, 221–225 |
| Swine, boars, rabbits | Sepsis, meningitis, pneumonia, arthritis | ||||
| Undefined | Beta | S. iniae | Humans | Soft-tissue infection | 48, 49 |
| Dolphins, fish | Abscess, streptococcosis, sepsis | 226–231 | |||
| S. uberis/S. parauberis | Cows, horses | Mastitis | 11, 16, 30, 43 |
Isolates belonging to S. dysgalactiae subsp. equisimilis react with antigen G with comparable prevalence of antigen C reaction, whereas reaction with antigens A and L are less common; isolates belonging to S. phocae react also with antigen G; isolates belonging to S. porcinus react also with antigens P, U, and V. Nonhemolytic variant can be recovered among isolates of S. agalactiae and S. dysgalactiae subsp. equisimilis.
Group E
In Lancefield’s group E, S. porcinus is typically associated with sepsis, endocarditis, pneumonia, and lymphadenitis in swine (27). Infections sustained by S. porcinus have also occurred in humans (28); however, S. porcinus isolates infecting humans seem to have a different origin when compared to S. porcinus isolates of nonhuman sources (29).
Groups C and G
S. dysgalactiae subsp. dysgalactiae belongs to Lancefield's group C and G and plays a major role in mastitis (30). On rare occasions, it has also been found in necrotic tissues of fish and in humans, as responsible for various diseases (Table 1). In the same Lancefield’s groups, S. dysgalactiae subsp. equisimilis is a beta-hemolytic bacterium found associated with strangles-like diseases in horses and with arthritis and endocarditis in swine. Unfortunately, this species is also associated with invasive diseases in humans, such as STSS and sepsis (Table 1). S. equi subsp. zooepidemicus organisms react with group C and G Lancefield’s antigens as well. This organism is commonly found in bovine mastitis (90, 94, 137; also see Table and Table 3) and has sporadically been found associated with mastitis in sheep (31) and is most prevalent in equine infective diseases as the causative agent of joint and respiratory tract infections (32). This species also causes severe infections in humans in association with the consumption of contaminated dairy products (33). Transmission of S. canis isolates belonging to Lancefield’s group C and G between pets and humans seems conceivable (34). This bacterium is responsible for arthritis in cats and humans (35) and endocarditis and skin, soft tissue, and urinary tract infections in dogs and humans. Skin lesions seem to represent the entry portal for establishing infections in humans (34, 36). In contrast, S. equi subsp. equi, belonging to Lancefield’s groups C and G and responsible for strangles disease in horses, is exclusively animal adapted (32).
TABLE 3.
Distribution of the tetracycline resistance genes in streptococci in animal hosts
| Animal host | Country | Year | Bacterial species | No. of isolates | Tetracycline genes | No. of TetR isolatesb | Percentage of resistance (%) | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Efflux | Ribosomal protection | |||||||||||
| tet(K) | tet(L) | tet(M) | tet(O) | tet(S) | ||||||||
| Cattle | USA | 1990 | S. agalactiae | 39 | 0 | 0 | NTa | 7 | NT | 10 | 25.6 | 137 |
| S. dysgalactiae | 21 | 1 | 1 | NT | 1 | NT | 9 | 42.9 | ||||
| S. uberis | 11 | 1 | NT | 1 | NT | 2 | 18.2 | |||||
| Cattle | France | 1984–2008 | S. agalactiae | 76 | NT | NT | 16 | 13 | 1 | 30 | 39.5 | 94 |
| S. dysgalactiae | 32 | NT | NT | 5 | 4 | 4 | 32 | 100.0 | ||||
| S. uberis | 101 | NT | NT | 23 | 36 | 3 | 62 | 61.4 | ||||
| Cattle | Portugal | 2002–2003 | S. agalactiae | 60 | 34 | NT | 13 | 20 | 0 | 34 | 56.7 | 90 |
| S. dysgalactiae | 18 | 0 | NT | 6 | 6 | 0 | 18 | 100.0 | ||||
| S. uberis | 30 | 0 | NT | 2 | 9 | 8 | 18 | 60.0 | ||||
| Ovine | Italy | 2004–2014 | S. uberis | 51 | 9 | NT | 12 | 12 | NT | 18 | 35.3 | 138 |
| Pig | USA | 1986 | S. suis | 21 | 0 | 0 | 5 | NT | NT | 14 | 66.7 | 103 |
| Pig | Denmark | 1989–2002 | S. suis | 103 | NT | 0 | 11 | 6 | 0 | 25 | 24.3 | 237 |
| Pig | Italy | 2003–2007 | S. suis | 57 | 0 | 0 | 2 | 38 | 0 | 51 | 89.5 | 140 |
| Pig | China | 2005–2012 | S. suis | 62 | NT | NT | 53 | 42 | NT | 57 | 91.9 | 170 |
| 34 | NT | NT | 24 | 9 | NT | 28 | 82.4 | |||||
| Pig | China | 2008–2010 | S. suis | 106 | NT | 2 | 16 | 86 | 1 | 105 | 99.1 | 146 |
| Dog/cat | France | 2010 | S. canis | 112 | NT | 1 | 31 | 16 | 5 | 36 | 32.1 | 148 |
| Dog/cat | Japan | 2015 | S. canis | 68 | 0 | 0 | 13 | 10 | NT | 16 | 23.5 | 149 |
| Dog/cat/horse/humanc | Portugal | 2000–2010 | S. canis | 85 | NT | 1 | 11 | 8 | 1 | 23 | 27.0 | 34 |
NT, not tested.
Discrepancies between the number of tetracycline-resistant isolates and the genes identified may be due to either unidentified genes or to isolates presenting an association of two or three tet genes.
Human isolates could not be individualized.
Group R
S. suis is a major pathogen for swine and causes meningitis, endocarditis, sepsis, arthritis, and pneumonia. In humans, S. suis is mostly responsible for meningitis and STSS. It is a well-recognized zoonotic agent, and indeed, human exposure to swine and swine-derived food products is a risk factor for infection by S. suis (37, 38). The production of a capsule seems to have a major role in pathogenesis, and capsular types 2 and 14 are the most prevalent among S. suis isolates that cause disease in humans (39).
Non-Lancefield Streptococci
All other streptococci lacking the Lancefield antigens are thus considered non-Lancefield (or nontypable) streptococci. Several are frequently encountered in animals and are detailed below.
S. uberis
If hygienic measures have been effective to control the dissemination of S. agalactiae, the same cannot be said for S. uberis, which remains a major animal pathogen and a leading cause of mastitis in cattle (15, 16). This nonserotypable organism has an environmental origin, possesses a flexible metabolism, and is almost exclusively adapted to cattle (40). Infections can occur by a variety of strains, which in some cases are able to persist and propagate among different cows within a herd (41, 42). Rarely, it has been responsible for mastitis in heifers, and even more rarely it has been isolated from shrimps (43–45). The control of S. uberis propagation is more challenging than that of other Streptococcus spp. probably because of its ability, among others, to survive on bedding material (46, 47).
S. iniae
Another nonserotypable species is S. iniae, which was primarily isolated from diseased dolphins and was subsequently confirmed as a major fish pathogen and as responsible for soft tissue infections in humans with zoonotic features (Table 1) (48–52).
S. pneumoniae
S. pneumoniae is a major streptococcal pathogen of humans and is responsible for serious infections such as pneumonia and meningitis; reports of infections in animals are extremely rare and concern horses with respiratory tract infections (53). A recent publication also reported S. pneumoniae, probably of human origin, in wild and captive chimpanzees (54). The spread of this pathogen due to the migration of infected animals to other communities or the reintroduction into wild populations of formerly captive animals might be a real danger.
Viridans streptococci
These organisms were defined as viridans because of the hemolytic features described above that produce a greenish pigmentation on blood agar, their absence of Lancefield antigens, and their resistance to the chemical compound optochin. Generally, viridans streptococci are implicated in the establishment of dental caries, arthritis, and infective endocarditis in humans (50–52).
Certain Streptococcus species, including S. sobrinus and S. mutans, the first representative of the mutans group; S. salivarius, S. vestibularis, and S. infantarius from the salivarius group; the anginosus group, including S. anginosus, S. constellatus, and S. intermedius; the sanguinus group, including S. sanguinis, S. parasanguinis, and S. gordonii; and most species of the mitis group, including S. mitis, S. oralis, S. cristatus, S. infantis, and S. peroris, have exclusive human adaptation; others have been found only in animal hosts, such as S. macacae from monkeys, S. ferus from wild rodents, S. orisratti from rats, and S. hyointestinalis and S. hyovaginalis from swine, with no associated diseases (55–59). Three species of the viridans group, namely S. sobrinus, S. criceti, and S. ratti, have been reported from humans and experimental rats, whereas S. alactolyticus has been found in swine, dogs, and humans (56, 60, 61); S. downei was isolated first from a monkey and more recently from human dental plaque (62, 63). Another species of the viridans group, S. pluranimalium, has been reported rarely. It was first reported in 1999 from bovine mastitis, and few clinical human cases have been reported since then (64, 65).
This brief introduction does not pretend to be exhaustive. The reviews by Póntigo et al. and Facklam provide a comprehensive nomenclature and classification of streptococci based on molecular and phenotypic features, respectively (1, 66).
EVOLUTION OF ANTIMICROBIAL RESISTANCE IN VETERINARY STREPTOCOCCI
Monitoring Programs
Prevalence data on antimicrobial resistance were principally obtained through dedicated studies performed at the scale of a region, a country, or a consortium of countries. However, only monitoring programs can give an evolutionary picture of antimicrobial resistance rates over time. Consequently, surveillance systems are highly valuable to follow trends and detect emergent resistant phenotypes.
Several national surveillance and monitoring programs in veterinary medicine exist in Europe (67), including the Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands program, the Swedish Veterinary Antimicrobial Resistance Monitoring program (SVARM), the Danish Integrated Antimicrobial Resistance Monitoring and Research Program, the German Resistance Monitoring in Veterinary Medicine program, and the French surveillance network for antimicrobial resistance in pathogenic bacteria of animal origin (RESAPATH). Two similar programs cover the North American continent, the National Antimicrobial Resistance Monitoring System for Enteric Bacteria in the United States and the Canadian Integrated Program for Antimicrobial Resistance Surveillance, and one reports Japanese data (the Japanese Veterinary Antimicrobial Resistance Monitoring program). An additional industry-based pan-European monitoring program commissioned by the Executive Animal Health Study Center investigates pathogens from farm (VetPath) and companion animals (ComPath). In addition to the recurrently criticized lack of harmonization (67, 68), a major feature of most programs is their main focus on bacteria of animal origin but of relevance for human health, such as zoonotics and commensal indicators. Accordingly, streptococci of animal origin were poorly included, and data on their resistance to antimicrobials remain limited at a global scale. Indeed, only two monitoring programs reported data on a long-term basis: the Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands program from 2002 to 2008 and RESAPATH from 2006 until today. Other programs such as the Swedish Veterinary Antimicrobial Resistance Monitoring program (in 2002), the German Resistance Monitoring in Veterinary Medicine program, and ComPath/VetPath have also documented antimicrobial resistance in streptococci, but on a sporadic basis.
Different Methodologies to Determine Antibiotic Resistance
Standard surveillance programs rely on phenotypic methods that are used in routine diagnostic laboratories. The most frequently used methods are antibiograms performed by disc diffusion and MIC performed by broth microdilution. These techniques generate qualitative or quantitative results, respectively, that are then interpreted according to official guidelines (EUCAST, CLSI, Antibiogram Committee of the French Microbiology Society (CA-SFM), etc.) so that the studied isolates can be classified as susceptible, intermediate, or resistant to the tested antibiotics. In the surveillance systems, the genotypic techniques are only optionally implemented as a second-line characterization.
This traditional approach may be disrupted by the democratization of next-generation sequencing methodologies. Recently, several publications proved the usefulness of large-scale genomic analyses for the efficient detection of resistance mechanisms and capsular types (69), for the sequence-based prediction of beta-lactam resistance using the penicillin-binding protein (PBP) transpeptidase signatures (70), and for the prediction of the antimicrobial profile and its potential evolution toward resistance over time (71). This is, of course, not an exhaustive list of publications using next-generation sequencing, especially in a field that is progressing very rapidly. There are still a couple of drawbacks to the direct implementation of next-generation sequencing in diagnostic laboratories, including the time needed to generate results (which exceeds the 48 hours traditionally needed for phenotypic testing) and the price. However, this methodology is so powerful that it will undoubtedly be used in monitoring programs, at least for long-term surveillance purposes.
Evolution of Resistance in Streptococcus spp.
The main Streptococcus species studied through monitoring programs were S. uberis and S. dysgalactiae isolated from bovine mastitis. S. agalactiae is also still considered a major streptococcal pathogen associated with bovine subclinical and mild to moderate clinical mastitis, but its incidence has drastically fallen in the past 20 years due to hygiene measures and guidelines for good practices. Consequently, S. agalactiae is now only rarely isolated from cattle mastitis, and the numbers are too small to be reliably reported. The evolution of antimicrobial resistance thus focuses on S. uberis and S. dysgalactiae. Here, we review data on resistance to the main antibiotics used in the treatment of animal infections due to S. uberis and S. dysgalactiae, i.e., penicillin G, tetracyclines, erythromycin, lincomycin, enrofloxacin, and streptomycin.
Evolution of Antimicrobial Resistance in the Netherlands
The Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands program reported antimicrobial resistance data on streptococci isolates collected from milk in the context of intramammary infections, but the monitoring of these pathogens stopped in 2008. From 2002 to 2008, the overall prevalence of resistance was quite stable for most antibiotics, with a seemingly upward trend for a few (72). Resistance to beta-lactams (represented by penicillin G and cefalotin) was only detected in rare cases of S. uberis and was absent in S. dysgalactiae. The highest rates of resistance were for tetracyclines (around 40% for S. uberis and 70% for S. dysgalactiae) and lincomycin (also around 40% for S. uberis and 25% for S. dysgalactiae). The discrepancy between the prevalence of tetracycline resistance in S. dysgalactiae compared to S. uberis is in accordance with what has been reported in other monitoring programs (see below) and numerous studies (see “Prevalence of Resistance to Macrolides among Streptococci of Bovine Origin” below). Resistance to erythromycin (around 20% for S. uberis and 10% for S. dysgalactiae) was systematically lower than for lincomycin, suggesting a significant prevalence of non-erm-mediated mechanisms of resistance.
RESAPATH, the Ongoing French Monitoring Program
RESAPATH is the only ongoing and long-term monitoring program for, among other topics, resistance to antimicrobials in streptococci in Europe. For S. uberis, from 2006 to 2015, antimicrobial resistance was tested on 600 to 1,500 isolates, depending on the nature of the antibiotics, and the global trend was quite stable for all antibiotics (Fig. 1). The highest prevalence of resistance was observed for enrofloxacin, which may be explained by the intrinsic low-level resistance of streptococci to fluoroquinolones. Indeed, in the RESAPATH network, resistance is defined as the addition of both resistant and intermediate phenotypes, which may lead to the over-estimation of the prevalence of resistance in the case of fluoroquinolones. For erythromycin and lincomycin, the resistance rates decreased from 24% to 17% between 2006 and 2007, were stable during next 6 years until 2013, and increased again in 2014 to 2015 up to around 22% of resistant isolates. Both curves matched perfectly, indicating a cross-resistance to macrolides-lincosamides involving erm genes (see “Resistance to Macrolides, Lincosamides, and Streptogramin B” below). Tetracycline resistance is following a very slow upward trend (from 14% in 2006 to 21% in 2015), to be confirmed in the coming years. Finally, streptomycin is the antibiotic presenting the lowest prevalence of resistance (from 11 to 16% over the 10-year period of 2006 to 2015), albeit the highest among resistance to aminoglycosides. Indeed, rates of resistance to kanamycin and gentamicin only reached 5% and 3%, respectively, in 2015. However, these resistances are of major importance since they may result in the loss of synergy between aminoglycosides and beta-lactams, which is a frequently used combination in veterinary practice. Taken together, these data show globally high basal levels of resistance of streptococci to antimicrobials in 2006 and a slight but increasing prevalence, which will have to be monitored in the near future.
FIGURE 1.
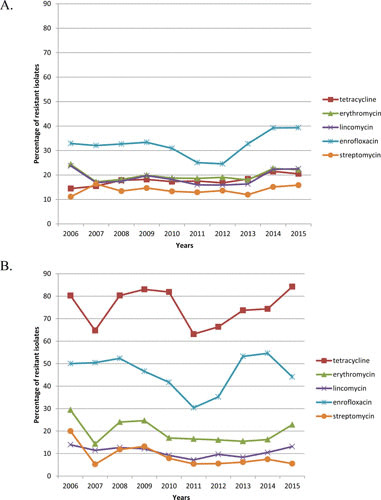
Ten-year evolution of resistance in France in (A) S. uberis and (B) S. dysgalactiae.
The number of S. dysgalactiae isolates that can be considered for global trends in prevalence is around 10 times lower than for S. uberis (25 compared to 235 isolates), so that the prevalence rates are subject to wider variations. However, the rates observed are different though more stable than for S. uberis. The antibiotic with the highest prevalence of resistance by far was tetracyclines. Up to 85% of the S. dysgalactiae isolates were resistant to this drug, which is in accordance with what has been reported in other studies (see “Prevalence of Resistance to Macrolides among Streptococci of Bovine Origin” below). Enrofloxacin presents fluctuating resistance rates of around 50%. Finally, erythromycin, lincomycin, and streptomycin present a stable prevalence of around 22%, 12%, and 6%, respectively. The 10% discrepancy between erythromycin and lincomycin resistance rates deserves special attention since it may signal a divergence in the resistance mechanisms involved compared to S. uberis.
Though originating from two countries only, these evolution rates constitute a starting point to address the issue of resistance in Streptococcus spp. in veterinary medicine. In line with these trends, the next sections will detail the epidemiology and mechanisms of resistance of the antibiotics mentioned above.
RESISTANCE TO MACROLIDES, LINCOSAMIDES, AND STREPTOGRAMIN B
Erythromycin was the first macrolide discovered in 1952 by McGuire as a natural product originating from Streptomyces erythreus. The other macrolides were derived from erythromycin by semi-synthesis, and their core unit consists of a lactone ring that can be constituted by 14, 15, and 16 carbon atoms. These molecules are bacteriostatic in staphylococci and bactericidal in streptococci, inhibiting the protein translocation by binding to the 23S or 50S ribosomal subunit at peculiar residues (i.e., the guanine 2505, the uridine 2609, and the adenines 2058, 2059, and 2062) (73). Macrolides have a broad spectrum of action, being effective against Gram-positive and Gram-negative bacteria and intracellular pathogens, and are reputed as valuable agents for their good pharmacodynamics properties, their relatively few side effects, and their good penetration in tissues. Lincosamides, including lincomycin and clindamycin, together with streptogramin B, including pristinamycin and quinupristin, are structurally unrelated to macrolides, but they have a common mechanism of action and, as a consequence, resistance to all these classes of antibiotics is crossed. In addition to the treatment of infections caused by intracellular pathogens, the usage of macrolides and lincosamides in human clinics is principally dedicated to the treatment of uncomplicated infections in patients who are allergic to beta-lactams. In veterinary medicine, macrolides and lincosamides are available as in-feed and injectable formulations and are used for the treatment of a variety of diseases ranging from respiratory tract infections to infective mastitis in food-producing animals, especially swine and cattle (74, 75). Frequently, macrolides and lincosamides are used in combination with other drugs such as aminoglycosides, ampicillin, colistin, tetracyclines, sulfonamides, and trimethoprim (76). Certain macrolides were also used as growth promoters (council regulation EC2821/98, 17 December 1998). Unfortunately, shortly after the introduction of macrolides in human therapy, resistant isolates were recovered; the emergence of resistant isolates has also occurred in animals. In the following subsections we will describe the most common mechanisms of resistance to macrolides, lincosamides, and streptogramins B found in streptococci of animal origin.
Macrolides, Lincosamides, and Streptogramins B Resistance Determinants
Target modification
Ribosomal mutation in the residues crucial to the binding of macrolides results in cross-resistance to all macrolides, lincosamides, and streptogramins B conferring the so-called MLSb phenotype. Human clinical isolates with such mutations have been sporadically observed, probably because this mechanism requires mutations in all the operon copies encoding the ribosomal subunits.
Target protection
The methylation of the adenine at position 2058 is enough to confer an MLSb resistance phenotype. This reaction is mediated by the methylases encoded by the erm (erythromycin ribosome methylation) gene family, originated by the natural producers of macrolides. In the presence of an Erm methylase, resistance to lincosamides and streptogramin B can be either constitutive or induced by the presence of erythromycin (73). Overall, this mechanism is the most prevalent method conferring resistance to macrolides in human and veterinary clinical isolates. Currently, about 40 variants of the erm gene have been reported, with erm(B) gene being the most prevalent (77). It is often located on mobile genetic elements (MGEs) and associated with genes conferring resistance to tetracyclines (see below). These two factors have consistently contributed to the dissemination of resistance to macrolides (78).
Efflux
The second most common mechanism of resistance to macrolides in streptococci is represented by the efflux mediated by the Mef efflux pumps. The mefA gene was described for the first time in S. pyogenes, and other variants have been reported since then that are principally represented by mefE and mefI (79). These efflux systems confer resistance to 14- and 15-carbon atom macrolides only, determining an M phenotype. Transcription of mef genes is coupled with the expression of msr genes encoding for an ATP-dependent efflux system. The presence of mef genes seems to be necessary to confer macrolide resistance (80). Genes of the mef family are often located on genetic units that can transfer by transformation, such as the MEGA (macrolide efflux genetic assembly) element harboring the mefE gene and conjugative transposons such as the Tn1207.3 harboring the mefA variant and the 5216IQ composite element harboring the mefI gene (81–83). With a lower prevalence, the mreA efflux pump has been reported as well (84). Finally, genes of the lsa family have been described, conferring cross-resistance to lincosamides, streptogramin A and pleuromutilins. These genes, encoding ATP-binding proteins, most likely promote the efflux of the antibiotics (192).
Drug modification
Lincosamides can be inactivated by adenylation in position 3 by a 3-lincosamide-O-nucleotidyltransferase encoded by the linB gene conferring an L phenotype (85). The linB gene was first found in a clinical isolate of E. faecium. Its origin remains unknown because no similar sequence has been found in natural lincosamide producers. The linB gene was reported sporadically; however, its transfer to S. agalactiae has occurred and has been reported from a human clinical isolate in Canada (86). After the first description, the linB gene was renamed lnuB. Later, Achard et al. unveiled the mechanism behind the lincomycin resistance in an S. agalactiae isolate that was surprisingly susceptible to clindamycin. It consisted of a novel nucleotidyl-transferase encoded by the lnuC gene (87). The LnuD-adenylating clindamycin was later discovered in S. uberis (88).
Inactivation of macrolides can also be caused by phosphotransferases encoded by the mph gene families with differential affinity for the different macrolide types and lincosamides according to the variant of the mph gene expressed (89).
Prevalence of Resistance to Macrolides among Streptococci of Bovine Origin
Studies of the resistance to macrolides in streptococci of bovine origin have focused on isolates responsible for clinical and subclinical mastitis. During an investigation of different streptococcal species from bovine mastitis during 2002 to 2003 in Portugal, Rato et al. found a constitutive MLSb phenotype in 11/60 S. agalactiae isolates; among those, 10 isolates were positive for the presence of an erm(B) gene and one harbored an erm(A) gene (90). Among S. dysgalactiae subsp. dysgalactiae isolates, 4/18 demonstrated a constitutive MLSb phenotype, with the presence of erm(A) and erm(B) in 1 and 3 isolates, respectively, whereas all the MLSb-resistant S. uberis isolates (8/30) harbored an erm(B) gene. An L phenotype was demonstrated in 11 isolates, 3 belonging to S. dysgalactiae subsp. dysgalactiae and 8 to S. uberis species, all of them harboring an lnuB gene. In 2005, Duarte et al. found that 8.5% (9/38) of S. agalactiae isolates from Brazil were resistant to erythromycin with a constitutive MLSb phenotype. All of the isolates harbored an erm(B) and an mreA gene; six of these coharbored erm(A) and were co-resistant to tetracyclines (29). Contemporaneously, Dogan et al. reported a 3.6% prevalence of erythromycin-resistant isolates among 83 S. agalactiae from bovines in New York. The resistance was mediated by Erm(B) in all isolates, and coresistance to tetracyclines was observed as well (91). A study from China was published in 2012 on 55 S. agalactiae isolates recovered from bovine mastitis during an undetermined period and reported a 23.5% rate of resistance to erythromycin. All isolates harbored the erm(B) gene (92). Pinto et al. analyzed 29 S. agalactiae isolates collected between 1980 and 2006 in Brazil. They found that 27% of the isolates were resistant to erythromycin, with erm(B) as the most prevalent gene, followed by the erm(A) variant. In this study, the presence of a mef allele was reported as well (93).
More recently, our investigation of 76 S. agalactiae isolates demonstrated an MLSb phenotype in eight isolates, including four isolates with a constitutive phenotype and four with an inducible phenotype. Six isolates harbored an erm(B) gene, and the two remaining ones harbored the erm(A) variant. In the same study, 4/32 isolates of S. dysgalactiae demonstrated the presence of an erm(B) gene; in this case also, the constitutive or inducible phenotypes were equally distributed (94). The erm(B) gene has also been confirmed as the most common gene conferring resistance to macrolides among S. dysgalactiae isolates; for instance, it was present in 4/4 isolates detected in dairy herds in the southwestern United States (95). We conducted a study to characterize the erythromycin resistance of 125 isolates of S. uberis collected from bovine mastitis during 2007 to 2008 in France. Overall, 111/125 isolates demonstrated an MLSb phenotype, constitutive in 42.3% of the isolates and inducible in the remaining ones. An erm(B) gene was present in all isolates. In this collection, 14 isolates demonstrated an L phenotype and harbored an lnuB gene. In one isolate, the less common lnuD gene was found as well (96). Contemporaneously, another study, conducted on dairy cows in Mayenne, France, confirmed MLSb resistance in 12 (12/55, 22%) S. uberis isolates, which all were positive for the erm(B) gene. An lnuB gene was present in four isolates with the L phenotype (97). In S. uberis, emergence of the mphB gene was documented in 2008 (98), but propagation of this mechanism has not occurred in a large scale.
Among others, the German Resistance Monitoring in Veterinary Medicine program has provided extensive data from Germany from 2007 to 2010 (Table 2). Several other studies from different parts of the world and based on the phenotypic characterization of mastitis isolates provided a comprehensive picture of the problematic link to the rise of resistance to macrolides in streptococci causing mastitis. An overview is provided in Table 2, and for exhaustive reports on temporal and geographical evolution of macrolide and lincosamide resistance in streptococci, we suggest the reports from Hendriksen et al. in 2008 and Thomas et al. in 2015 (99, 100). Overall, the lowest prevalence of macrolide and lincosamide resistance was observed in Sweden (101), and large differences in prevalence were observed among countries. However, no major variation was observed from one year to another in a single country.
TABLE 2.
Erythromycin and lincosamide resistance in streptococci in animal hostsa
| Animal host | Country | Year | Bacterial species | No. of isolates | Genetic determinants | Percentage of resistance (%) to | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mefA | mefE | msr | ermA | ermB | lnuB | lnuD | mph | mreA | M | L | ||||||
| Cattle | USA | ND | S. dysgalactiae | 152 | 10 | 25.6 | 136 | |||||||||
| S. uberis | 133 | 9 | 42.9 | |||||||||||||
| Cattle | France | S. uberis | 55 | 0 | 12 | 4 | 22 | 97 | ||||||||
| Cattle | USA | ND | S. dysgalactiae | 4 | 0 | 0 | 0 | 4 | 100 | 95 | ||||||
| S. uberis | 20 | 0 | 0 | 0 | 12 | 60 | ||||||||||
| Cattle | China | ND | S. agalactiae | 55 | 0 | 0 | 13 | 23.5 | 92 | |||||||
| Cattle | Brazil | 1980–2006 | S. agalactiae | 29 | 5 | 4 | 7 | 0 | 31 | 20.7 | 93 | |||||
| Cattle | France | 1984–2008 | S. agalactiae | 76 | 0 | 2 | 6 | 10.1 | 94 | |||||||
| S. dysgalactiae | 32 | 0 | 0 | 4 | ND | |||||||||||
| S. uberis | 101 | 0 | 0 | 75 | 5 | 1 | ND | |||||||||
| Cattle | Brazil | 1995–2000 | S. agalactiae | 38 | 0 | 6 | 9 | 0 | 9 | 8.5 | 8.5 | 29 | ||||
| Cattle | France | 1995–2000 | S. agalactiae | 8 | 0 | 0 | 134 | |||||||||
| S. dysgalactiae | 41 | 16.7 | 11.9 | |||||||||||||
| S. uberis | 50 | 28 | 36 | |||||||||||||
| Cattle | Argentina | 1999–2003 | S. agalactiae | 36 | 16.7 | 19.4 | 232 | |||||||||
| S. dysgalactiae | 8 | 12.5 | 12.5 | |||||||||||||
| Cattle | USA | 2000–2002 | S. agalactiae | 83 | 0 | 0 | 3 | 3.6 | 91 | |||||||
| Cattle | Portugal | 2002–2003 | S. agalactiae | 60 | 0 | 1 | 10 | 0 | 18.3 | 18.3 | 90 | |||||
| S. dysgalactiae | 18 | 0 | 1 | 3 | 3 | 22.2 | 38.9 | |||||||||
| S. uberis | 30 | 0 | 0 | 8 | 8 | 26.7 | 26.7 | |||||||||
| Cattle | Sweden | 2002–2003 | S. agalactiae | 6 | 16.7 | 16.7 | 133 | |||||||||
| S. dysgalactiae | 152 | 0 | 0.7 | |||||||||||||
| S. uberis | 113 | 0 | 0 | |||||||||||||
| Cattle | Korea | 2004–2008 | S. agalactiae | 5 | 0 | 60 | 233 | |||||||||
| S. bovis group | 24 | 12.5 | 33.3 | |||||||||||||
| S. uberis | 99 | 34.3 | 41.4 | |||||||||||||
| S. oralis | 30 | 36.7 | 36.7 | |||||||||||||
| S. salivarius | 13 | 0 | 69.2 | |||||||||||||
| S. intermedius | 7 | 42.8 | 71.4 | |||||||||||||
| Cattle | Turkey | ND | S. agalactiae | 5 | 0 | 40 | 169 | |||||||||
| S. uberis | 18 | 11 | 17 | |||||||||||||
| Cattle | France | 2007–2008 | S. uberis | 125 | 0 | 0 | 111 | 3 | 0 | ND | ND | 96 | ||||
| Cattle | Estonia | 2007–2009 | S. agalactiae | 1.3 | 6.2 | 168 | ||||||||||
| S. dysgalactiae | 6.7 | 7.8 | ||||||||||||||
| S. uberis | 8.2 | 6.6 | ||||||||||||||
| Cattle | Germany | 2007–2010 | S. agalactiae | 101 | 2 | 13 | 0 | 15 | 16.8 | ND | 234 | |||||
| S. dysgalactiae | 100 | 2 | 1 | 0 | 10 | 11 | ND | |||||||||
| S. uberis | 102 | 2 | 2 | 0 | 5 | 17.6 | ND | |||||||||
| Cattle | Switzerland | 2010–2012 | S. dysgalactiae | 46 | 2.2 | 2.2 | 238 | |||||||||
| S. uberis | 208 | 10.6 | 10.6 | |||||||||||||
| Cattle | Switzerland | 2011–2013 | S. dysgalactiae | 213 | ND | 37.4 | 167 | |||||||||
| S. uberis | 1,228 | ND | 49.7 | |||||||||||||
| Swine | EU | 1987–1997 | S. suis | 404 | 55.3 | 139 | ||||||||||
| Swine | Denmark | 1989–2002 | S. suis | 103 | 39 | 40.8 | 237 | |||||||||
| Swine | France | 1996–2000 | S. suis | 110 | 78.2 | 78.2 | 235 | |||||||||
| Swine | Belgium | 1999–2000 | S. suis | 87 | 0 | 62 | 71 | 71 | 236 | |||||||
| Swine | Spain | 1999–2001 | S. suis | 151 | 90.7 | 87.4 | 143 | |||||||||
| Swine | Italy | 2003–2007 | S. suis | 57 | 0 | 44 | 81 | 81 | 140 | |||||||
| Swine | China | 2005–2007 | S. suis | 421 | 67.2 | 68.4 | 142 | |||||||||
| Swine | China | 2005–2012 | S. suis | 96 | 18 | 0 | 35 | 0 | 38.5 | 38.5 | 170 | |||||
| Swine | China | 2008–2010 | S. suis | 106 | 51 | 51 | 70 | 67.9 | 67.9 | 146 | ||||||
| Swine | Brazil | 2009–2010 | S. suis | 260 | 46.5 | 84.6 | 144 | |||||||||
| Swine | Korea | 2010–2013 | S. suis | 227 | 39 | 218 | 94 | 95.6 | 104 | |||||||
ND, not determined; M, macrolides; L, lincosamides.
Prevalence of Resistance to Macrolides among Streptococci of Porcine Origin
Macrolides, lincosamides, and streptogramins B are widely used for the treatment of infections in swine. Unfortunately, it appears that the usage of these drugs has influenced the emergence of resistance in S. suis (102). High rates of resistance to these drugs were observed over time, ranging from 52% (11/21 isolates) in the first observation in Norway in 1986 (103) to 94% (216/226 isolates) in Korea during 2010 to 2013 (104). The most prevalent genetic determinant is the erm(B) gene, whereas mefA/E were sporadically detected in human isolates (105). Often, such resistances occur together with tetracycline resistance (see below). S. suis may also act as a reservoir of lincosamide resistance genes, as exemplified by the emergence of the lnuE gene, previously identified in S. suis, in staphylococcal isolates (106). The report from Hendriksen et al. shows a certain variability of erythromycin resistance in S. suis among European countries during 2002 to 2004 (107). In addition, other streptococcal species were rarely found to cause diseases in swine. Within the framework of the BfT-GermVET program, Lüthje and Schwarz reported the presence of S. dysgalactiae subsp. equisimilis in diseased swine, with 21 isolates demonstrating resistance to macrolides. Among those, 13 harbored an erm(B) gene, one an erm(B) gene together with mefA and msrD, and one a lnuB gene (108).
In all, such an alarming prevalence of macrolide resistance in a relevant zoonotic pathogen such as S. suis highlights the need to prevent infections through appropriate hygienic measures.
Prevalence of Resistance to Macrolides among Streptococci from Non-Food-Producing Animals
In Brazil, six isolates of S. dysgalactiae subsp. equisimilis from horses were included in a study of antimicrobial resistance in humans, and prevalence data on resistance to macrolides were similar in isolates from the two sectors (109). In Germany, an S. equi subsp. zooepidemicus isolate with an M phenotype was recovered from a horse and harbored a mefA and an msrD gene. S. canis resistant to macrolides, mostly with an MLSb phenotype, has been reported from diseased dogs in Denmark and Germany (110). From dogs and cats, six S. dysgalactiae subsp. equisimilis macrolide-resistant isolates were found in Germany with five isolates harboring an erm(B) gene and one harboring a mefA and a msrD gene (108).
TETRACYCLINES
Tetracyclines, which were discovered in the late 1940s, are bacteriostatic antibiotics that block bacterial protein synthesis by preventing the attachment of aminoacyl-tRNA to the ribosomal acceptor A site (111). Because of their broad-spectrum activity against both Gram-positive and Gram-negative bacteria, they rapidly became one of the most widely used antibiotics (112), and consequently, the first resistant isolate was reported in 1953 in Shigella dysenteriae (113). Tetracycline resistance rapidly and broadly disseminated in bacteria of human, animal, and environmental origin and is now considered one of the most frequently seen resistances to antimicrobials (114). In humans, tetracyclines have largely been supplanted by beta-lactams, while they remain one of the main classes of antibiotics used in veterinary medicine (115–117). In animals, tetracyclines are also considered as growth-promoting factors when mixed with food at subtherapeutic levels, in order for food-producing animals to gain weight more quickly (111). This practice was banned in Europe at the latest on 1 January 2006 since it can promote resistance selection, as exemplified by the increase of vancomycin-resistant enterococci in animals through the use of the glycopeptide avoparcin (118). However, growth promoters are still authorized in many countries worldwide, such as in the United States, and tetracycline is again the most frequently used antibiotic class (117). The past and present excessive use of tetracyclines first of all hampers the efficacy of these molecules, but may also have unexpected side effects such as the selection of hypervirulent S. agalactiae clones worldwide and increasing numbers of neonate infections (119).
Tetracycline Resistance Determinants
A total of 46 tet or otr genes have been identified as tetracycline resistance determinants in 126 genera (120, 121). They are commonly divided into two main groups characterized by their mode of action: the genes coding for efflux proteins and those coding for ribosomal protection enzymes (120). In Streptococcus spp., tet(K), tet(L), tet(M), tet(O), tet(Q), and tet(T) are the most frequently reported genes (111, 113, 121). Tet(K) and Tet(L) code for membrane-associated efflux systems that share nearly 60% amino acid identity (111) and confer resistance to tetracycline but not to minocycline. The tet(K) gene was discovered on a pT181 plasmid in Staphylococcus aureus (122), whereas tet(L) was found associated with small non-conjugative plasmids in streptococci (123). In contrast, Tet(M), Tet(O), Tet(S), Tet(Q), Tet(T), and the more recently identified Tet(W) are enzymes that protect the ribosome from the action of tetracycline, a mechanism conferring resistance to all available antibiotics of the tetracycline family. The tet(M) gene was concomitantly identified with tet(L) on streptococcal plasmids (123). It has now been extensively detected and studied in both Gram-negative and Gram-positive species and is often found on ICEs of the Tn916-Tn1545 family (124–127). Since the Tn916-Tn1545 elements also encode, among others, resistance to erythromycin and kanamycin, these mobile determinants promote the emergence of multiresistant isolates, as exemplified by their frequent association with the tet(M) and erm(B) resistance genes in streptococci isolates (120). The dissemination of the remaining protecting enzymes—Tet(O) (which was first discovered in Campylobacter coli [128]), Tet(S) (discovered in Listeria monocytogenes [129]), Tet(Q) (first described in Bacteroides species [130]), and the closely related Tet(T) (first detected in S. pyogenes [131])—was less efficient than the diffusion of Tet(M), probably because of the localization of the corresponding genes, which have never been reported on conjugative transposons such as Tn916 and Tn1545, mentioned above. Of note, Tet(M), Tet(O), Tet(S), and Tet(Q) are closely related since they share around 78% sequence identity (113), even though they can easily be differentiated using specific primers. Tet(W) is the latest protection enzyme detected in streptococci and was first identified in Butyrivibrio fibrisolvens (132).
Prevalence of Tetracycline Resistance among Streptococci of Bovine or Ovine Origin
Most studies reporting tetracycline resistance in isolates of bovine or ovine origin were performed on S. uberis, S. dysgalactiae, and S. agalactiae in the context of clinical or subclinical mastitis. Tetracycline is often the antibiotic presenting the highest prevalence of resistance. When considering the CLSI breakpoints which categorize as tetracycline resistant all isolates presenting an MIC of >4 mg/liter, resistance rates in S. uberis ranged from 1.8 to 4% in Sweden between 2002 and 2009 up to 60% in Portugal in 2003 (90, 101, 133), with intermediate prevalence of 12.9 to 22% in France, 15% in England, 27.1% in the United States (1997 to 1999), and 44% in Italy (99, 134–136). In S. dysgalactiae prevalence is overall higher, ranging from 6% in Sweden to 76.6% in the Netherlands and 100% in Portugal and France (90, 94). Prevalence rate figures are less frequently available for S. agalactiae but also suggest a very high frequency of tetracycline resistance, with 33.4% in Sweden in 2002 and 37.5% in France in 2000 (133, 134).
Only a few studies have reported the molecular characterization of tet resistance genes (Table 3) in streptococci of bovine origin. When reported, the tet genes did not have a bacterial specificity and were often described in combination, such as tet(M)/tet(O), tet(M)/tet(K), or tet(O)/tet(K) (90, 94, 137, 138).
Most studies detailed here were performed in Europe in the 2000s. Having a better and updated view of the evolution of resistance in veterinary streptococci would require monitoring of tetracycline resistance in bacteria from animal origins at a larger scale.
Prevalence of Tetracycline Resistance among Streptococci of Porcine Origin
In line with S. suis being the major Streptococcus spp. in swine, numerous publications have reported antimicrobial resistance in this pathogen, mostly in diseased animals. Tetracycline, as already noted for streptococci of bovine or ovine origin, is often the antibiotic presenting the highest prevalence of resistance. The lowest rates were reported in the oldest European isolates studied (no resistance in Danish isolates collected in 1967 to 1981 and 7.7% in Swedish isolates collected in 1992 to 1997) and in the context of the ARBAO-II study performed in 2002 to 2004 (48% resistance in the Netherlands and 52.2% in Denmark in 2003) (102, 107). Higher rates were then reported in the United States (66.7% in 1986), Spain (68.0% in 2004), Poland (64.0% in 2004), Japan (86.9%), Italy (89.5%), and a pan-European study (75.1%) (107, 139–141). Several studies showed resistance rates greater than 90%, with 91.7%, 95.4% and 97.9% in China, Spain, and Brazil, respectively (142–144).
A few molecular studies detailed the tet genes responsible for the phenotypic resistance detected. Tet(O) is by far the most commonly detected enzyme in S. suis, while Tet(M) has also been reported (Table 3), either alone or in association with other Tet determinants, in particluar Tet(O). The distribution of these tet genes may also vary depending on the serotype, but further work on larger cohorts is needed to have statistically relevant data. The tet(W) gene was repeatedly reported in S. suis, initially in a human patient in Italy, but then also in pig isolates (140, 145, 146). Of note, the only occurrence of tet(B) in streptococci was reported in 17 S. suis isolates (17/111, 15%) in the United States (147).
Tetracycline Resistance among Other Streptococci
Tetracycline resistance in S. canis was reported from diseased cats and dogs, with a prevalence of 32.1% in France, 23.5% in Japan, and 27% in Portugal (including a few isolates from horses and humans) (34, 148, 149). In these studies, tetracycline resistance was due principally to the presence of the tet(M) and tet(O) genes, alone or in combination. A study performed in Belgium on healthy individually owned cats and groups of cats noted a higher prevalence of tetracycline resistance in a cattery (52%) compared to individual animals (22.2%), most likely due to the clonal transmission of resistant strains in the cattery (150).
BETA-LACTAM RESISTANCE
Beta-lactams are the largest family of antibiotics available in both human and veterinary medicine. All members of this family act on the bacterial cell wall by covalently blocking the PBPs and thus impairing the continuous building of this protecting structure. Currently, bacterial resistance to the last generations of beta-lactams is one of the most challenging issues in both human and animal medicine. The key threats are the worldwide emergence and dissemination of inactivating enzymes such as extended-spectrum beta-lactamases, cephalosporinases (AmpCs), and carbapenemases in Gram-negative bacteria. All these resistance determinants are carried by plasmids and thus display a high capacity to efficiently disseminate in an intra- or interspecies manner. In Gram-positive bacteria, methicillin-resistant S. aureus—which possesses an additional PBP2A presenting a decreased affinity to beta-lactams—remains an issue in human medicine, despite the fact that its prevalence in hospitals has been considerably reduced in the past decades with improvements of hygiene measures. In veterinary medicine, methicillin-resistant Staphylococcus pseudintermedius is known to cause serious treatment challenges because of its associated multiresistance. However, the success of both methicillin-resistant S. aureus and methicillin-resistant S. pseudintermedius is more due to epidemic bursts of successful clones than to the mobilization of the mecA-carrying cassette.
Streptococci are unique among the major pathogens in the sense that they are incapable of acquiring any exogenous beta-lactam resistance genes. However, they can progressively mutate their own PBPs. Indeed, no isolate carrying a beta-lactamase (such as Gram-negative bacteria) or a new PBP (such as staphylococci) has been described yet, and a few species, including S. pyogenes, are even unable to develop decreased susceptibility to beta-lactams in vitro (151).
Beta-Lactam Resistance in Streptococci
To achieve beta-lactam resistance, streptococci sequentially modify their PBPs, specifically the class B PBP2B and PBP2X (and the class A PBP1A in the more resistant isolates). This was particularly exemplified in S. pneumoniae, the only Streptococcus spp. for which penicillin-resistance was successfully achieved and widely disseminated, where both mutated and mosaic PBPs were reported. Other less-documented genes were sporadically reported as PBP-independent penicillin-resistance mechanisms. These include the MurMN operon encoding enzymes that are responsible for the biosynthesis of branched muropeptide components (152), the ciaRH operon, a two-component signal-transducing system (153), the adr gene coding for a peptidoglycan O-acetyltransferase (154), the stkP gene encoding a serine/threonine kinase (155), the pstS gene encoding a subunit of a phosphate ABC transporter (156), and the spr1178 gene encoding for a putative iron permease (157). Penicillin-resistant S. pneumoniae has widely disseminated through the success of a limited number of serotypes, selected mainly by the excessive use of antibiotics (158), but the prevalence of this resistance has considerably decreased since the early 2000s by both the reduced consumption of antibiotics and the marketing of efficient vaccines (159, 160). The presence and characterization of mutated PBPs in isolates presenting decreased susceptibilities to beta-lactam were also reported in S. agalactiae of human origin (69, 161, 162). Recently, a classification of S. agalactiae was proposed which takes into account the different mutations in the PBPs (163).
In veterinary medicine, Streptococcus isolates presenting full penicillin resistance have only rarely been reported, and only a few molecular studies were conducted either on laboratory strains or on field isolates presenting reduced susceptibility to beta-lactams. The presence of three groups of PBPs (PBP 1, PBP 2, and PBP 3) was demonstrated in S. suis, and PBP modifications were strongly suggested to be responsible for penicillin G-resistant phenotypes in both in vitro mutants and field isolates (164). In S. uberis, Haenni et al. (165) showed that both a quality control strain and field strains were capable of developing a 60-fold MIC increase after 30 cycles of exposure to penicillin G. This increase was due to the accumulation of mutations in the class B (PBP 2B and PBP 2X) and A (PBP 1A) enzymes, including the systematic presence of the two specific E381K and Q554E mutations in the PBP 2X. Interestingly, PBP analysis of seven field strains collected in Switzerland, France, and Holland and presenting MICs of 0.25 to 0.5 mg/liter also revealed the systematic presence of these two key mutations (165). However, none of the tested strains (selected either in vitro or by treatment on farms) could achieve full resistance, since their MICs only reached 0.25 to 2 mg/liter, which is still considered intermediately resistant.
Phenotypic Reports on Beta-Lactam Activity in Bovine Mastitis
In veterinary medicine, beta-lactam resistance has mostly been documented in bovine streptococci, namely S. uberis, S. dysgalactiae, and S. agalactiae. Data were gathered in cases of clinical and subclinical mastitis, a pathology for which the first-line treatment is beta-lactams. Comparison between studies can be difficult because of the heterogeneity of the methods (disc diffusion, agar diffusion, broth microdilution) and the guidelines used. If no interpretation was inferred, an isolate was considered resistant when the MIC was ≥4 mg/liter according to the CLSI breakpoints.
Most studies based on the determination of MICs report the absence of penicillin G resistance in S. uberis and S. dysgalactiae, even though isolates presenting decreased susceptibilities (0.25 to 0.5 mg/liter) were regularly reported. In France, such nonsusceptible isolates were reported in 2002, with 14.0% of the S. uberis showing an MIC of 0.25 mg/liter (134), and a shift toward decreased susceptibilities was suggested in 2010 based on the comparison of disk diffusion and MIC results (166). In Sweden, two studies performed successively in 2003 (133) and 2008 to 2009 (101) showed a slight shift over the years toward decreased susceptibility, with 6.0% of the S. uberis isolates displaying an MIC of 0.25 mg/liter and 10.0% of the S. dysgalactiae isolates displaying an MIC of 0.12 mg/liter in 2009, whereas only 0.9% of the S. uberis isolates had an MIC of 0.25 mg/liter, and all the S. dysgalactiae isolates were fully susceptible (MIC, <0.06 mg/liter) in 2003. In the United States, one true resistant S. dysgalactiae isolate (MIC, 4 mg/liter) was detected over 152 strains tested, whereas 6.8% of the S. uberis isolates presented MICs of 0.5 mg/liter to penicillin, and one strain displayed an MIC of 1 mg/liter (136). The VetPath data (multicenter European data) collected between 2002 and 2006 showed no true resistance, but 29.8% of isolates presented decreased susceptibility (MICs ranging from 0.25 mg/liter to 1 mg/liter) (100). Though without any MIC values, other studies also reported data on beta-lactam resistance in veterinary streptococci, often with a very low prevalence of resistance. One pan-European study performed in 2002 to 2004 showed very low levels (0 to 3.9%) of penicillin resistance (99). A Swiss study detected susceptibilities to ampicillin of 92.3% for S. uberis and 94.8% for S. dysgalactiae from cows sampled in 2011 to 2013 (167). Kalmus et al. showed 0 to 0.4% resistance to penicillin G, ampicillin, and cefalotin in Estonia between 2007 and 2009 (168). In France, 12.9% of the S. uberis and 1.4% of the S. dysgalactiae presented resistance patterns to oxacillin, but these were not confirmed by MIC determination (135). In Turkey, 94% of the S. uberis isolates showed susceptibility to penicillin G (169).
These different data confirm beta-lactams as efficient antibiotics against streptococci isolated from bovine mastitis. However, this slow but clear shift of strains from full toward decreased susceptibility will have to be surveyed in the future. Beta-lactam resistance development in streptococci surely does not present the same dynamic as in Gram-negative bacteria, where plasmids play a major role. However, this should not hide rampant and silent acquisition of beta-lactam resistance in streptococci of animal origin, which may one day limit the therapeutic arsenal available for veterinarians.
Beta-Lactam Resistance Outside the Context of Cattle Mastitis
Antibiotic resistance was also monitored in S. suis isolated from pigs, and several studies reported the absence of resistance to any beta-lactams. Nevertheless, isolates presenting particularly high MICs to penicillin G were also recurrently reported, with unfortunately, no concomitant molecular work on the underlying mechanisms of resistance. Indeed, 4% of the Spanish isolates displayed MICs ranging from 4 to 16 mg/liter, whereas two Chinese studies reported 2.1% and 9.5% of isolates with an MIC of ≥4 mg/liter (142, 143, 170). In the Netherlands, 0.5% and 0.3% of over 1,163 isolates of S. suis tested were considered resistant to penicillin G and ampicillin between 2013 and 2015 (171). In Japan, Poland, and Portugal, resistance was reported in 0.9%, 8.1%, and 13.0% of the isolates, respectively (107, 141). However, despite these cases, beta-lactams can still be recommended as first-line antibiotics for the treatment of S. suis.
Aside from S. suis and streptococci isolated from bovine mastitis, reports of veterinary streptococci are quite rare. Beta-lactam resistance was described once in S. dysgalactiae subspecies equisimilis isolated from swine in Brazil and in four studies of S. canis isolated from pets and horses in France, Japan, Belgium, and Portugal (34, 148–150, 172), and these five studies found full susceptibility of all isolates to penicillin G.
FLUOROQUINOLONE RESISTANCE
Quinolones are not active against streptococci, because of their intrinsic resistance. However, fluoroquinolones may be an alternative to beta-lactam antibiotics to treat streptococcal infections. The main agents used in veterinary medicine are enrofloxacin, marbofloxacin, danofloxacin, and the more recent pradofloxacin.
Resistance to fluoroquinolones is generally mediated by point mutations in the quinolone resistance-determinants regions of the gyrA and parC genes (173). Furthermore, plasmidic qnr genes participate in the dissemination of low-level resistance, but they have never been reported in streptococci. Efflux pumps also play a role in fluoroquinolone resistance, as has been proved for the SatAB, an ABC transporter, in S. suis (174).
Fluoroquinolone-Resistance Phenotypes
Resistance to fluoroquinolones has rarely been reported in veterinary streptococci. Moreover, in the fluoroquinolone family, there is a wide variability of the agents tested (enrofloxacin and ciprofloxacin are the most frequently used), thus making comparisons among studies difficult.
A 1.5% prevalence of resistance to enrofloxacin in S. uberis and 5.5% in S. dysgalactiae was reported in France in 2010 (135). In the same bacterial species, MICs ranging from 0.5 to 2 mg/liter were observed in Sweden (101). These apparently elevated MICs are constitutive of streptococci, which have a basal MIC higher than that of Enterobacteriaceae or staphylococci. In a study performed between 1994 and 2001, the same range of MICs was reported, and no increase in the resistance rate was observed over the years (175). Except for three resistant strains (MIC, 4 mg/liter), all S. uberis and S. dysgalactiae isolates collected in a multicenter European study presented MICs to marbofloxacin ranging from 0.25 to 2 mg/liter (176). In S. suis, the resistance rates to enrofloxacin determined in a pan-European study performed in 2009 to 2012 and to ciprofloxacin determined in Japan in 1987 to 1996 were very similar: 0.7% and 0.3%, respectively (177). Enrofloxacin resistance was also observed in S. canis in France, where MICs ranged from 0.25 to 2 mg/liter (148). Streptococcus spp. isolated from cats and dogs were studied through the ComPath European network: no resistance was reported in dermatological samples, whereas 1.8% of the cats and 4.0% of the dogs presenting with a respiratory tract infection carried enrofloxacin-resistant streptococci (178, 179).
ROLE OF THE ICES IN THE EVOLUTION OF RESISTANCE
MGEs play a major role in the dissemination of antibiotic resistance genes. MGEs mostly comprise conjugative plasmids, transposons, phages, and ICEs (initially named conjugative transposons). ICEs are chromosomal, self-transmissible MGEs that are capable of promoting their excision, conjugation, and site-specific integration in a recipient cell (180, 181). One of the most emblematic members of the ICEs is the Tn916-Tn1545 family, which carries tet(M) and other antibiotic resistance genes (126, 127, 182, 183).
ICEs have been widely reported in streptococci, and they were recently detected in all Streptococcus spp. for which at least one complete genome was available, with S. suis being the most “colonized” species (184, 185). In human clinical streptococcal isolates, erm and tet resistance genes were recurrently reported on ICEs, such as erm(B) on ICESp1116 and erm(TR)-tet(O) on ICESp2905 in S. pyogenes (186, 187), erm(TR) on ICESagTR7 in S. agalactiae (188), and erm(B) and tet(O) on ICESsD9 in S. suis (189). Interestingly, resistance genes can also be mobilized by coresident ICEs, as demonstrated by the mobilization of an erm(T)-carrying plasmid in S. dysgalactiae subsp. equisimilis (190). Other antibiotic resistance determinants can also be found on ICEs, as exemplified by the presence of tet(M) and a chloramphenicol acetyl-transferase on ICESp23FST81 from S. agalactiae (191), lincosamide resistance (lsa genes) on different ICEs in S. agalactiae (192), and a multidrug resistance cluster on ICESsuNC28 carried by S. suis (193). Moreover, ICEs originating from different streptococcal species may form hybrids that can further transfer in vitro to a third streptococcal species (194). This illustrates the wide distribution and the plasticity of these MGEs and thus their role in the dissemination of resistance genes. Finally, resistance determinants may be adjacent to—and not inside—an ICE. This is exemplified by the first vancomycin-resistance determinants in streptococci, the vanG operons, which were identified in one S. agalactiae and two S. anginosus isolates (195). These vanG operons were immediately followed by a large chromosomal element named ICE-r (ICE-like sequences). A plausible hypothesis is that the integration of ICE-r in the streptococcal chromosome may have favored the subsequent integration of the vanG element.
Except for elements from the Tn916-Tn1545 family which were broadly reported in different streptococcal species originating from diverse animal hosts (9, 94, 170, 196), ICEs carrying resistance genes have rarely been reported in streptococci from animal origin. Indeed, only the mosaic tet(O/W/32/O) carried on ICESsu32457 in S. suis, which could then be transferred to S. pneumoniae, S. pyogenes, and S. agalactiae (197), as well as the lincomycin-resistance gene lsa (192) were described on such MGEs. This will likely change in the near future, since new research perspectives are emerging with the democratized access to high-throughput sequencing technologies and the subsequent databases (185, 198).
CONCLUSION
In veterinary medicine streptococci are frequent pathogens not only in food-producing but also in companion animals. However, as far as public health is concerned, there are few situations in which streptococci of animal origin may cause risk for humans, and vice-versa. Among those, S. suis and, to a lesser extent, S. agalactiae are likely the most relevant examples, and are both considered zoonotic pathogens. Studies of resistance to antimicrobials in streptococci of animal origin have largely focused on tetracyclines and macrolides/lincosamides, which are widely used in the animal sector globally. Accordingly, high resistance rates to these molecules have frequently been observed, which is also in line with specific niches covering major animal diseases, such as cattle mastitis of streptococcal origin. Molecular investigations highlighted the diversity of the resistance genes of the tet and erm families, together with the pivotal role of the ICEs. Nonetheless, most data originate from Europe, and there is a need for larger prevalence and molecular studies on a global scale. Of note, despite the wide use of penicillins to treat streptococcal infections, resistance to beta-lactams does not appear to be a crucial issue in veterinary streptococci, in contrast to what has been observed with the human-specific S. pneumoniae. In all, as in humans, antimicrobial resistance in Streptococcus of animal origin may largely differ depending on the Streptococcus sp. and therefore should not be considered as a whole.
REFERENCES
- 1.Póntigo F, Moraga M, Flores SV. 2015. Molecular phylogeny and a taxonomic proposal for the genus Streptococcus. Genet Mol Res 14:10905–10918 10.4238/2015.September.21.1. [PubMed] [DOI] [PubMed] [Google Scholar]
- 2.Lancefield RC, Freimer EH. 1966. Type-specific polysaccharide antigens of group B streptococci. J Hyg (Lond) 64:191–203 10.1017/S0022172400040456. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka D, Isobe J, Watahiki M, Nagai Y, Katsukawa C, Kawahara R, Endoh M, Okuno R, Kumagai N, Matsumoto M, Morikawa Y, Ikebe T, Watanabe H, Working Group for Group A Streptococci in Japan. 2008. Genetic features of clinical isolates of Streptococcus dysgalactiae subsp. equisimilis possessing Lancefield’s group A antigen. J Clin Microbiol 46:1526–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brouwer S, Barnett TC, Rivera-Hernandez T, Rohde M, Walker MJ. 2016. Streptococcus pyogenes adhesion and colonization. FEBS Lett 590:3739–3757 10.1002/1873-3468.12254. [PubMed] [DOI] [PubMed] [Google Scholar]
- 5.Carapetis JR, Steer AC, Mulholland EK, Weber M. 2005. The global burden of group A streptococcal diseases. Lancet Infect Dis 5:685–694 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 6.Copperman SM. 1982. Cherchez le chien: household pets as reservoirs of persistent or recurrent streptococcal sore throats in children. N Y State J Med 82:1685–1687. [PubMed] [PubMed] [Google Scholar]
- 7.Farley MM. 2001. Group B streptococcal disease in nonpregnant adults. Clin Infect Dis 33:556–561 10.1086/322696. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Olivares-Fuster O, Klesius PH, Evans J, Arias CR. 2008. Molecular typing of Streptococcus agalactiae isolates from fish. J Fish Dis 31:277–283 10.1111/j.1365-2761.2007.00900.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 9.Fischer A, Liljander A, Kaspar H, Muriuki C, Fuxelius HH, Bongcam-Rudloff E, de Villiers EP, Huber CA, Frey J, Daubenberger C, Bishop R, Younan M, Jores J. 2013. Camel Streptococcus agalactiae populations are associated with specific disease complexes and acquired the tetracycline resistance gene tetM via a Tn916-like element. Vet Res (Faisalabad) 44:86 10.1186/1297-9716-44-86. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yildirim AO, Lämmler C, Weiss R. 2002. Identification and characterization of Streptococcus agalactiae isolated from horses. Vet Microbiol 85:31–35 10.1016/S0378-1135(01)00481-3. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.McDonald TJ, McDonald JS. 1976. Streptococci isolated from bovine intramammary infections. Am J Vet Res 37:377–381. [PubMed] [PubMed] [Google Scholar]
- 12.Brochet M, Couvé E, Zouine M, Vallaeys T, Rusniok C, Lamy MC, Buchrieser C, Trieu-Cuot P, Kunst F, Poyart C, Glaser P. 2006. Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes Infect 8:1227–1243 10.1016/j.micinf.2005.11.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 13.Rajendram P, Mar Kyaw W, Leo YS, Ho H, Chen WK, Lin R, Pratim P, Badaruddin H, Ang B, Barkham T, Chow A. 2016. Group B streptococcus sequence type 283 disease linked to consumption of raw fish, Singapore. Emerg Infect Dis 22:1974–1977 10.3201/eid2211.160252. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans JJ, Klesius PH, Pasnik DJ, Bohnsack JF. 2009. Human Streptococcus agalactiae isolate in Nile tilapia (Oreochromis niloticus). Emerg Infect Dis 15:774–776 10.3201/eid1505.080222. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradley A. 2002. Bovine mastitis: an evolving disease. Vet J 164:116–128 10.1053/tvjl.2002.0724. [PubMed] [DOI] [PubMed] [Google Scholar]
- 16.Leigh JA. 1999. Streptococcus uberis: a permanent barrier to the control of bovine mastitis? Vet J 157:225–238 10.1053/tvjl.1998.0298. [PubMed] [DOI] [PubMed] [Google Scholar]
- 17.Schleifer KH, Kilpper-Bälz R. 1984. Transfer of Streptococcus faecalis and Streptococcus faecium to the genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. Int J Syst Evol Microbiol 34:31–34. [Google Scholar]
- 18.Köhler W. 2007. The present state of species within the genera Streptococcus and Enterococcus. Int J Med Microbiol 297:133–150 10.1016/j.ijmm.2006.11.008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 19.Ben-Chetrit E, Wiener-Well Y, Kashat L, Yinnon AM, Assous MV. 2016. Streptococcus bovis new taxonomy: does subspecies distinction matter? Eur J Clin Microbiol Infect Dis 36(2):387. [PubMed] [DOI] [PubMed] [Google Scholar]
- 20.Dumke J, Hinse D, Vollmer T, Schulz J, Knabbe C, Dreier J. 2015. Potential transmission pathways of Streptococcus gallolyticus subsp. gallolyticus. PLoS One 10:e0126507 10.1371/journal.pone.0126507. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gherardi G, Palmieri C, Marini E, Pompilio A, Crocetta V, Di Bonaventura G, Creti R, Facinelli B. 2016. Identification, antimicrobial resistance and molecular characterization of the human emerging pathogen Streptococcus gallolyticus subsp. pasteurianus. Diagn Microbiol Infect Dis 86:329–335 10.1016/j.diagmicrobio.2016.09.019. [PubMed] [DOI] [PubMed] [Google Scholar]
- 22.Sheng WH, Chuang YC, Teng LJ, Hsueh PR. 2014. Bacteraemia due to Streptococcus gallolyticus subspecies pasteurianus is associated with digestive tract malignancies and resistance to macrolides and clindamycin. J Infect 69:145–153 10.1016/j.jinf.2014.03.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.Sturt AS, Yang L, Sandhu K, Pei Z, Cassai N, Blaser MJ. 2010. Streptococcus gallolyticus subspecies pasteurianus (biotype II/2), a newly reported cause of adult meningitis. J Clin Microbiol 48:2247–2249 10.1128/JCM.00081-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnett J, Ainsworth H, Boon JD, Twomey DF. 2008. Streptococcus gallolyticus subsp. pasteurianus septicaemia in goslings. Vet J 176:251–253 10.1016/j.tvjl.2007.02.011. [PubMed] [DOI] [PubMed] [Google Scholar]
- 25.Li M, Gu C, Zhang W, Li S, Liu J, Qin C, Su J, Cheng G, Hu X. 2013. Isolation and characterization of Streptococcus gallolyticus subsp. pasteurianus causing meningitis in ducklings. Vet Microbiol 162:930–936 10.1016/j.vetmic.2012.11.038. [PubMed] [DOI] [PubMed] [Google Scholar]
- 26.Marmolin ES, Hartmeyer GN, Christensen JJ, Nielsen XC, Dargis R, Skov MN, Knudsen E, Kemp M, Justesen US. 2016. Bacteremia with the bovis group streptococci: species identification and association with infective endocarditis and with gastrointestinal disease. Diagn Microbiol Infect Dis 85:239–242 10.1016/j.diagmicrobio.2016.02.019. [PubMed] [DOI] [PubMed] [Google Scholar]
- 27.Wessman GE. 1986. Biology of the group E streptococci: a review. Vet Microbiol 12:297–328 10.1016/0378-1135(86)90081-7. [DOI] [PubMed] [Google Scholar]
- 28.Facklam R, Elliott J, Pigott N, Franklin AR. 1995. Identification of Streptococcus porcinus from human sources. J Clin Microbiol 33:385–388. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duarte RS, Barros RR, Facklam RR, Teixeira LM. 2005. Phenotypic and genotypic characteristics of Streptococcus porcinus isolated from human sources. J Clin Microbiol 43:4592–4601 10.1128/JCM.43.9.4592-4601.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jonsson P, Olsson SO, Olofson AS, Fälth C, Holmberg O, Funke H. 1991. Bacteriological investigations of clinical mastitis in heifers in Sweden. J Dairy Res 58:179–185 10.1017/S0022029900029721. [PubMed] [DOI] [PubMed] [Google Scholar]
- 31.Cuccuru C, Meloni M, Sala E, Scaccabarozzi L, Locatelli C, Moroni P, Bronzo V. 2011. Effects of intramammary infections on somatic cell score and milk yield in Sarda sheep. N Z Vet J 59:128–131 10.1080/00480169.2011.562862. [PubMed] [DOI] [PubMed] [Google Scholar]
- 32.Timoney JF. 2004. The pathogenic equine streptococci. Vet Res 35:397–409 10.1051/vetres:2004025. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.Hashikawa S, Iinuma Y, Furushita M, Ohkura T, Nada T, Torii K, Hasegawa T, Ohta M. 2004. Characterization of group C and G streptococcal strains that cause streptococcal toxic shock syndrome. J Clin Microbiol 42:186–192 10.1128/JCM.42.1.186-192.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinho MD, Matos SC, Pomba C, Lübke-Becker A, Wieler LH, Preziuso S, Melo-Cristino J, Ramirez M. 2013. Multilocus sequence analysis of Streptococcus canis confirms the zoonotic origin of human infections and reveals genetic exchange with Streptococcus dysgalactiae subsp. equisimilis. J Clin Microbiol 51:1099–1109 10.1128/JCM.02912-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iglauer F, Kunstýr I, Mörstedt R, Farouq H, Wullenweber M, Damsch S. 1991. Streptococcus canis arthritis in a cat breeding colony. J Exp Anim Sci 34:59–65. [PubMed] [PubMed] [Google Scholar]
- 36.Lacave G, Coutard A, Troché G, Augusto S, Pons S, Zuber B, Laurent V, Amara M, Couzon B, Bédos JP, Pangon B, Grimaldi D. 2016. Endocarditis caused by Streptococcus canis: an emerging zoonosis? Infection 44:111–114 10.1007/s15010-015-0809-3. [PubMed] [DOI] [PubMed] [Google Scholar]
- 37.Goyette-Desjardins G, Auger JP, Xu J, Segura M, Gottschalk M. 2014. Streptococcus suis, an important pig pathogen and emerging zoonotic agent: an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microbes Infect 3:e45 10.1038/emi.2014.45. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segura M, Calzas C, Grenier D, Gottschalk M. 2016. Initial steps of the pathogenesis of the infection caused by Streptococcus suis: fighting against nonspecific defenses. FEBS Lett 590:3772–3799 10.1002/1873-3468.12364. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Taniyama D, Sakurai M, Sakai T, Kikuchi T, Takahashi T. 2016. Human case of bacteremia due to Streptococcus suis serotype 5 in Japan: the first report and literature review. IDCases 6:36–38 10.1016/j.idcr.2016.09.011. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward PN, Holden MT, Leigh JA, Lennard N, Bignell A, Barron A, Clark L, Quail MA, Woodward J, Barrell BG, Egan SA, Field TR, Maskell D, Kehoe M, Dowson CG, Chanter N, Whatmore AM, Bentley SD, Parkhill J. 2009. Evidence for niche adaptation in the genome of the bovine pathogen Streptococcus uberis. BMC Genomics 10:54–71 10.1186/1471-2164-10-54. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phuektes P, Mansell PD, Dyson RS, Hooper ND, Dick JS, Browning GF. 2001. Molecular epidemiology of Streptococcus uberis isolates from dairy cows with mastitis. J Clin Microbiol 39:1460–1466 10.1128/JCM.39.4.1460-1466.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilesmith JW, Francis PG, Wilson CD. 1986. Incidence of clinical mastitis in a cohort of British dairy herds. Vet Rec 118:199–204 10.1136/vr.118.8.199. [PubMed] [DOI] [PubMed] [Google Scholar]
- 43.Compton CW, Heuer C, Parker K, McDougall S. 2007. Epidemiology of mastitis in pasture-grazed peripartum dairy heifers and its effects on productivity. J Dairy Sci 90:4157–4170 10.3168/jds.2006-880. [PubMed] [DOI] [PubMed] [Google Scholar]
- 44.Hasson KW, Wyld EM, Fan Y, Lingsweiller SW, Weaver SJ, Cheng J, Varner PW. 2009. Streptococcosis in farmed Litopenaeus vannamei: a new emerging bacterial disease of penaeid shrimp. Dis Aquat Organ 86:93–106 10.3354/dao02132. [PubMed] [DOI] [PubMed] [Google Scholar]
- 45.Parker KI, Compton C, Anniss FM, Weir A, Heuer C, McDougall S. 2007. Subclinical and clinical mastitis in heifers following the use of a teat sealant precalving. J Dairy Sci 90:207–218 10.3168/jds.S0022-0302(07)72622-X. [DOI] [PubMed] [Google Scholar]
- 46.Dodd FH. 1983. Mastitis: progress on control. J Dairy Sci 66:1773–1780 10.3168/jds.S0022-0302(83)82005-0. [DOI] [PubMed] [Google Scholar]
- 47.Zehner MM, Farnsworth RJ, Appleman RD, Larntz K, Springer JA. 1986. Growth of environmental mastitis pathogens in various bedding materials. J Dairy Sci 69:1932–1941 10.3168/jds.S0022-0302(86)80620-8. [DOI] [PubMed] [Google Scholar]
- 48.Koh TH, Sng LH, Yuen SM, Thomas CK, Tan PL, Tan SH, Wong NS. 2009. Streptococcal cellulitis following preparation of fresh raw seafood. Zoonoses Public Health 56:206–208 10.1111/j.1863-2378.2008.01213.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 49.Weinstein MR, Litt M, Kertesz DA, Wyper P, Rose D, Coulter M, McGeer A, Facklam R, Ostach C, Willey BM, Borczyk A, Low DE. 1997. Invasive infections due to a fish pathogen, Streptococcus iniae. S. iniae Study Group. N Engl J Med 337:589–594 10.1056/NEJM199708283370902. [PubMed] [DOI] [PubMed] [Google Scholar]
- 50.Nilson B, Olaison L, Rasmussen M. 2016. Clinical presentation of infective endocarditis caused by different groups of non-beta haemolytic streptococci. Eur J Clin Microbiol Infect Dis 35:215–218 10.1007/s10096-015-2532-5. [PubMed] [DOI] [PubMed] [Google Scholar]
- 51.Simón-Soro A, Mira A. 2015. Solving the etiology of dental caries. Trends Microbiol 23:76–82 10.1016/j.tim.2014.10.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 52.Yombi J, Belkhir L, Jonckheere S, Wilmes D, Cornu O, Vandercam B, Rodriguez-Villalobos H. 2012. Streptococcus gordonii septic arthritis: two cases and review of literature. BMC Infect Dis 12:215–220 10.1186/1471-2334-12-215. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burrell MH, Mackintosh ME, Taylor CE. 1986. Isolation of Streptococcus pneumoniae from the respiratory tract of horses. Equine Vet J 18:183–186 10.1111/j.2042-3306.1986.tb03591.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 54.Köndgen S, Calvignac-Spencer S, Grützmacher K, Keil V, Mätz-Rensing K, Nowak K, Metzger S, Kiyang J, Becker AL, Deschner T, Wittig RM, Lankester F, Leendertz FH. 2017. Evidence for human Streptococcus pneumoniae in wild and captive chimpanzees: a potential threat to wild populations. Sci Rep 7:14581 10.1038/s41598-017-14769-z. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beighton D, Hayday H. 1982. The streptococcal flora of the tongue of the monkey Macaca fascicularis. Arch Oral Biol 27:331–335 10.1016/0003-9969(82)90163-7. [PubMed] [DOI] [PubMed] [Google Scholar]
- 56.Devriese LA, Hommez J, Pot B, Haesebrouck F. 1994. Identification and composition of the streptococcal and enterococcal flora of tonsils, intestines and faeces of pigs. J Appl Bacteriol 77:31–36 10.1111/j.1365-2672.1994.tb03040.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 57.Devriese LA, Pot B, Vandamme P, Kersters K, Collins MD, Alvarez N, Haesebrouck F, Hommez J. 1997. Streptococcus hyovaginalis sp. nov. and Streptococcus thoraltensis sp. nov., from the genital tract of sows. Int J Syst Bacteriol 47:1073–1077 10.1099/00207713-47-4-1073. [PubMed] [DOI] [PubMed] [Google Scholar]
- 58.Freedman ML, Coykendall AL, O’Neill EM. 1982. Physiology of “mutans-like” Streptococcus ferus from wild rats. Infect Immun 35:476–482. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu H, Willcox MD, Knox KW. 2000. A new species of oral Streptococcus isolated from Sprague-Dawley rats, Streptococcus orisratti sp. nov. Int J Syst Evol Microbiol 50:55–61 10.1099/00207713-50-1-55. [PubMed] [DOI] [PubMed] [Google Scholar]
- 60.Rinkinen ML, Koort JM, Ouwehand AC, Westermarck E, Björkroth KJ. 2004. Streptococcus alactolyticus is the dominating culturable lactic acid bacterium species in canine jejunum and feces of four fistulated dogs. FEMS Microbiol Lett 230:35–39 10.1016/S0378-1097(03)00851-6. [DOI] [PubMed] [Google Scholar]
- 61.Toepfner N, Shetty S, Kunze M, Orlowska-Volk M, Krüger M, Berner R, Hentschel R. 2014. Fulminant neonatal sepsis due to Streptococcus alactolyticus: a case report and review. APMIS 122:654–656 10.1111/apm.12219. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.Beighton D, Russell RR, Hayday H. 1981. The isolation of characterization of Streptococcus mutans serotype h from dental plaque of monkeys (Macaca fascicularis). J Gen Microbiol 124:271–279. [DOI] [PubMed] [Google Scholar]
- 63.Yoo SY, Kim KJ, Lim SH, Kim KW, Hwang HK, Min BM, Choe SJ, Kook JK. 2005. First isolation of Streptococcus downei from human dental plaques. FEMS Microbiol Lett 249:323–326 10.1016/j.femsle.2005.06.020. [PubMed] [DOI] [PubMed] [Google Scholar]
- 64.Aryasinghe L, Sabbar S, Kazim Y, Awan LM, Khan HK. 2014. Streptococcus pluranimalium: a novel human pathogen? Int J Surg Case Rep 5:1242–1246 10.1016/j.ijscr.2014.11.029. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Devriese LA, Vandamme P, Collins MD, Alvarez N, Pot B, Hommez J, Butaye P, Haesebrouck F. 1999. Streptococcus pluranimalium sp. nov., from cattle and other animals. Int J Syst Bacteriol 49:1221–1226 10.1099/00207713-49-3-1221. [PubMed] [DOI] [PubMed] [Google Scholar]
- 66.Facklam R. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev 15:613–630 10.1128/CMR.15.4.613-630.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Silley P, Simjee S, Schwarz S. 2012. Surveillance and monitoring ofantimicrobial resistance and antibiotic consumption in humans and animals. Rev Sci Tech 31:105–120 10.20506/rst.31.1.2100. [PubMed] [DOI] [PubMed] [Google Scholar]
- 68.de Jong A, Thomas V, Klein U, Marion H, Moyaert H, Simjee S, Vallé M. 2013. Pan-European resistance monitoring programmes encompassing food-borne bacteria and target pathogens of food-producing and companion animals. Int J Antimicrob Agents 41:403–409 10.1016/j.ijantimicag.2012.11.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 69.Metcalf BJ, Chochua S, Gertz RE Jr, Hawkins PA, Ricaldi J, Li Z, Walker H, Tran T, Rivers J, Mathis S, Jackson D, Glennen A, Lynfield R, McGee L, Beall B, Active Bacterial Core surveillance team. 2017. Short-read whole genome sequencing for determination of antimicrobial resistance mechanisms and capsular serotypes of current invasive Streptococcus agalactiae recovered in the USA. Clin Microbiol Infect 23:574e577–574e514. [DOI] [PubMed] [Google Scholar]
- 70.Li Y, Metcalf BJ, Chochua S, Li Z, Gertz RE Jr, Walker H, Hawkins PA, Tran T, Whitney CG, McGee L, Beall BW. 2016. Penicillin-binding protein transpeptidase signatures for tracking and predicting beta-lactam resistance levels in Streptococcus pneumoniae. MBio 7:pii e00756-16 10.1128/mBio.00756-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mobegi FM, Cremers AJ, de Jonge MI, Bentley SD, van Hijum SA, Zomer A. 2017. Deciphering the distance to antibiotic resistance for the pneumococcus using genome sequencing data. Sci Rep 7:42808 10.1038/srep42808. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.MARAN. 2009. Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in The Netherlands in 2008. CIDC-Lelystad, Lelystad, The Netherlands. [Google Scholar]
- 73.Leclercq R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin Infect Dis 34:482–492 10.1086/324626. [PubMed] [DOI] [PubMed] [Google Scholar]
- 74.Collignon P, Powers JH, Chiller TM, Aidara-Kane A, Aarestrup FM. 2009. World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin Infect Dis 49:132–141 10.1086/599374. [PubMed] [DOI] [PubMed] [Google Scholar]
- 75.Edmondson PW. 1989. An economic justification of “blitz” therapy to eradicate Streptococcus agalactiae from a dairy herd. Vet Rec 125:591–593. [PubMed] [PubMed] [Google Scholar]
- 76.Pyörälä S, Baptiste KE, Catry B, van Duijkeren E, Greko C, Moreno MA, Pomba MC, Rantala M, Ružauskas M, Sanders P, Threlfall EJ, Torren-Edo J, Törneke K. 2014. Macrolides and lincosamides in cattle and pigs: use and development of antimicrobial resistance. Vet J 200:230–239 10.1016/j.tvjl.2014.02.028. [PubMed] [DOI] [PubMed] [Google Scholar]
- 77.De Mouy D, Cavallo JD, Leclercq R, Fabre R, AFICORPI-BIO Network. Association de Formation Continue en Pathologie Infectieuse des Biologistes. 2001. Antibiotic susceptibility and mechanisms of erythromycin resistance in clinical isolates of Streptococcus agalactiae: French multicenter study. Antimicrob Agents Chemother 45:2400–2402 10.1128/AAC.45.8.2400-2402.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Puopolo KM, Klinzing DC, Lin MP, Yesucevitz DL, Cieslewicz MJ. 2007. A composite transposon associated with erythromycin and clindamycin resistance in group B Streptococcus. J Med Microbiol 56:947–955 10.1099/jmm.0.47131-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 79.Cai Y, Kong F, Gilbert GL. 2007. Three new macrolide efflux (mef) gene variants in Streptococcus agalactiae. J Clin Microbiol 45:2754–2755 10.1128/JCM.00579-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ambrose KD, Nisbet R, Stephens DS. 2005. Macrolide efflux in Streptococcus pneumoniae is mediated by a dual efflux pump (mel and mef) and is erythromycin inducible. Antimicrob Agents Chemother 49:4203–4209 10.1128/AAC.49.10.4203-4209.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gay K, Stephens DS. 2001. Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J Infect Dis 184:56–65 10.1086/321001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 82.Mingoia M, Vecchi M, Cochetti I, Tili E, Vitali LA, Manzin A, Varaldo PE, Montanari MP. 2007. Composite structure of Streptococcus pneumoniae containing the erythromycin efflux resistance gene mefI and the chloramphenicol resistance gene catQ. Antimicrob Agents Chemother 51:3983–3987 10.1128/AAC.00790-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Santagati M, Iannelli F, Cascone C, Campanile F, Oggioni MR, Stefani S, Pozzi G. 2003. The novel conjugative transposon tn1207.3 carries the macrolide efflux gene mef(A) in Streptococcus pyogenes. Microb Drug Resist 9:243–247 10.1089/107662903322286445. [PubMed] [DOI] [PubMed] [Google Scholar]
- 84.Clancy J, Dib-Hajj F, Petitpas JW, Yuan W. 1997. Cloning and characterization of a novel macrolide efflux gene, mreA, from Streptococcus agalactiae. Antimicrob Agents Chemother 41:2719–2723. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bozdogan B, Berrezouga L, Kuo MS, Yurek DA, Farley KA, Stockman BJ, Leclercq R. 1999. A new resistance gene, linB, conferring resistance to lincosamides by nucleotidylation in Enterococcus faecium HM1025. Antimicrob Agents Chemother 43:925–929. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.de Azavedo JC, McGavin M, Duncan C, Low DE, McGeer A. 2001. Prevalence and mechanisms of macrolide resistance in invasive and noninvasive group B streptococcus isolates from Ontario, Canada. Antimicrob Agents Chemother 45:3504–3508 10.1128/AAC.45.12.3504-3508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Achard A, Villers C, Pichereau V, Leclercq R. 2005. New lnu(C) gene conferring resistance to lincomycin by nucleotidylation in Streptococcus agalactiae UCN36. Antimicrob Agents Chemother 49:2716–2719 10.1128/AAC.49.7.2716-2719.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Petinaki E, Guérin-Faublée V, Pichereau V, Villers C, Achard A, Malbruny B, Leclercq R. 2008. Lincomycin resistance gene lnu(D) in Streptococcus uberis. Antimicrob Agents Chemother 52:626–630 10.1128/AAC.01126-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chesneau O, Tsvetkova K, Courvalin P. 2007. Resistance phenotypes conferred by macrolide phosphotransferases. FEMS Microbiol Lett 269:317–322 10.1111/j.1574-6968.2007.00643.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.Rato MG, Bexiga R, Florindo C, Cavaco LM, Vilela CL, Santos-Sanches I. 2013. Antimicrobial resistance and molecular epidemiology of streptococci from bovine mastitis. Vet Microbiol 161:286–294 10.1016/j.vetmic.2012.07.043. [PubMed] [DOI] [PubMed] [Google Scholar]
- 91.Dogan B, Schukken YH, Santisteban C, Boor KJ. 2005. Distribution of serotypes and antimicrobial resistance genes among Streptococcus agalactiae isolates from bovine and human hosts. J Clin Microbiol 43:5899–5906 10.1128/JCM.43.12.5899-5906.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gao J, Yu FQ, Luo LP, He JZ, Hou RG, Zhang HQ, Li SM, Su JL, Han B. 2012. Antibiotic resistance of Streptococcus agalactiae from cows with mastitis. Vet J 194:423–424 10.1016/j.tvjl.2012.04.020. [PubMed] [DOI] [PubMed] [Google Scholar]
- 93.Pinto TC, Costa NS, Vianna Souza AR, Silva LG, Corrêa AB, Fernandes FG, Oliveira IC, Mattos MC, Rosado AS, Benchetrit LC. 2013. Distribution of serotypes and evaluation of antimicrobial susceptibility among human and bovine Streptococcus agalactiae strains isolated in Brazil between 1980 and 2006. Braz J Infect Dis 17:131–136 10.1016/j.bjid.2012.09.006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haenni M, Saras E, Bertin S, Leblond P, Madec JY, Payot S. 2010. Diversity and mobility of integrative and conjugative elements in bovine isolates of Streptococcus agalactiae, S. dysgalactiae subsp. dysgalactiae, and S. uberis. Appl Environ Microbiol 76:7957–7965 10.1128/AEM.00805-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Loch IM, Glenn K, Zadoks RN. 2005. Macrolide and lincosamide resistance genes of environmental streptococci from bovine milk. Vet Microbiol 111:133–138 10.1016/j.vetmic.2005.09.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 96.Haenni M, Saras E, Chaussière S, Treilles M, Madec JY. 2011. ermB-mediated erythromycin resistance in Streptococcus uberis from bovine mastitis. Vet J 189:356–358 10.1016/j.tvjl.2010.06.021. [PubMed] [DOI] [PubMed] [Google Scholar]
- 97.Schmitt-Van de Leemput E, Zadoks RN. 2007. Genotypic and phenotypic detection of macrolide and lincosamide resistance in Streptococcus uberis. J Dairy Sci 90:5089–5096 10.3168/jds.2007-0101. [PubMed] [DOI] [PubMed] [Google Scholar]
- 98.Achard A, Guérin-Faublée V, Pichereau V, Villers C, Leclercq R. 2008. Emergence of macrolide resistance gene mph(B) in Streptococcus uberis and cooperative effects with rdmC-like gene. Antimicrob Agents Chemother 52:2767–2770 10.1128/AAC.00481-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hendriksen RS, Mevius DJ, Schroeter A, Teale C, Meunier D, Butaye P, Franco A, Utinane A, Amado A, Moreno M, Greko C, Stärk K, Berghold C, Myllyniemi AL, Wasyl D, Sunde M, Aarestrup FM. 2008. Prevalence of antimicrobial resistance among bacterial pathogens isolated from cattle in different European countries: 2002–2004. Acta Vet Scand 50:28–38 10.1186/1751-0147-50-28. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thomas V, de Jong A, Moyaert H, Simjee S, El Garch F, Morrissey I, Marion H, Vallé M. 2015. Antimicrobial susceptibility monitoring of mastitis pathogens isolated from acute cases of clinical mastitis in dairy cows across Europe: VetPath results. Int J Antimicrob Agents 46:13–20 10.1016/j.ijantimicag.2015.03.013. [PubMed] [DOI] [PubMed] [Google Scholar]
- 101.Persson Y, Nyman AK, Grönlund-Andersson U. 2011. Etiology and antimicrobial susceptibility of udder pathogens from cases of subclinical mastitis in dairy cows in Sweden. Acta Vet Scand 53:36 10.1186/1751-0147-53-36. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Aarestrup FM, Rasmussen SR, Artursson K, Jensen NE. 1998. Trends in the resistance to antimicrobial agents of Streptococcus suis isolates from Denmark and Sweden. Vet Microbiol 63:71–80 10.1016/S0378-1135(98)00228-4. [PubMed] [DOI] [PubMed] [Google Scholar]
- 103.Wasteson Y, Høie S, Roberts MC. 1994. Characterization of antibiotic resistance in Streptococcus suis. Vet Microbiol 41:41–49 10.1016/0378-1135(94)90134-1. [PubMed] [DOI] [PubMed] [Google Scholar]
- 104.Gurung M, Tamang MD, Moon DC, Kim SR, Jeong JH, Jang GC, Jung SC, Park YH, Lim SK. 2015. Molecular basis of resistance to selected antimicrobial agents in the emerging zoonotic pathogen Streptococcus suis. J Clin Microbiol 53:2332–2336 10.1128/JCM.00123-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Palmieri C, Varaldo PE, Facinelli B. 2011. Streptococcus suis, an emerging drug-resistant animal and human pathogen. Front Microbiol 2:235–241 10.3389/fmicb.2011.00235. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao Q, Wendlandt S, Li H, Li J, Wu C, Shen J, Schwarz S, Wang Y. 2014. Identification of the novel lincosamide resistance gene lnu(E) truncated by ISEnfa5-cfr-ISEnfa5 insertion in Streptococcus suis: de novo synthesis and confirmation of functional activity in Staphylococcus aureus. Antimicrob Agents Chemother 58:1785–1788 10.1128/AAC.02007-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hendriksen RS, Mevius DJ, Schroeter A, Teale C, Jouy E, Butaye P, Franco A, Utinane A, Amado A, Moreno M, Greko C, Stärk KD, Berghold C, Myllyniemi AL, Hoszowski A, Sunde M, Aarestrup FM. 2008. Occurrence of antimicrobial resistance among bacterial pathogens and indicator bacteria in pigs in different European countries from year 2002–2004: the ARBAO-II study. Acta Vet Scand 50:19–29 10.1186/1751-0147-50-19. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lüthje P, Schwarz S. 2007. Molecular basis of resistance to macrolides and lincosamides among staphylococci and streptococci from various animal sources collected in the resistance monitoring program BfT-GermVet. Int J Antimicrob Agents 29:528–535 10.1016/j.ijantimicag.2006.12.016. [PubMed] [DOI] [PubMed] [Google Scholar]
- 109.Silva LG, Genteluci GL, Corrêa de Mattos M, Glatthardt T, Sá Figueiredo AM, Ferreira-Carvalho BT. 2015. Group C Streptococcus dysgalactiae subsp. equisimilis in south-east Brazil: genetic diversity, resistance profile and the first report of human and equine isolates belonging to the same multilocus sequence typing lineage. J Med Microbiol 64:551–558 10.1099/jmm.0.000052. [PubMed] [DOI] [PubMed] [Google Scholar]
- 110.Pedersen K, Pedersen K, Jensen H, Finster K, Jensen VF, Heuer OE. 2007. Occurrence of antimicrobial resistance in bacteria from diagnostic samples from dogs. J Antimicrob Chemother 60:775–781 10.1093/jac/dkm269. [PubMed] [DOI] [PubMed] [Google Scholar]
- 111.Chopra I, Roberts M. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65:232–260 10.1128/MMBR.65.2.232-260.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Speer BS, Shoemaker NB, Salyers AA. 1992. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin Microbiol Rev 5:387–399 10.1128/CMR.5.4.387. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Roberts MC. 1996. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Rev 19:1–24 10.1111/j.1574-6976.1996.tb00251.x. [DOI] [PubMed] [Google Scholar]
- 114.Kazimierczak KA, Rincon MT, Patterson AJ, Martin JC, Young P, Flint HJ, Scott KP. 2008. A new tetracycline efflux gene, tet(40), is located in tandem with tet(O/32/O) in a human gut firmicute bacterium and in metagenomic library clones. Antimicrob Agents Chemother 52:4001–4009 10.1128/AAC.00308-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Moulin G, Cavalié P, Pellanne I, Chevance A, Laval A, Millemann Y, Colin P, Chauvin C, Antimicrobial Resistance ad hoc Group of the French Food Safety Agency. 2008. A comparison of antimicrobial usage in human and veterinary medicine in France from 1999 to 2005. J Antimicrob Chemother 62:617–625 10.1093/jac/dkn213. [PubMed] [DOI] [PubMed] [Google Scholar]
- 116.Report UOH. 2015. Joint report on human and animal antibiotic use, sales and resistance, 2013. https://www.gov.uk/government/collections/antimicrobial-resistance-amr-information-and-resources.
- 117.FDA. 2014. 2012 Summary report on antimicrobials sold or distributed for use in food-producing animals. FDA, Washington, DC. http://www.fda.gov/downloads/ForIndustry/UserFees/AnimalDrugUserFeeActADUFA/UCM416983.pdf. [Google Scholar]
- 118.van den Bogaard AE, Bruinsma N, Stobberingh EE. 2000. The effect of banning avoparcin on VRE carriage in The Netherlands. J Antimicrob Chemother 46:146–148 10.1093/jac/46.1.146. [PubMed] [DOI] [PubMed] [Google Scholar]
- 119.Da Cunha V, Davies MR, Douarre PE, Rosinski-Chupin I, Margarit I, Spinali S, Perkins T, Lechat P, Dmytruk N, Sauvage E, Ma L, Romi B, Tichit M, Lopez-Sanchez MJ, Descorps-Declere S, Souche E, Buchrieser C, Trieu-Cuot P, Moszer I, Clermont D, Maione D, Bouchier C, McMillan DJ, Parkhill J, Telford JL, Dougan G, Walker MJ, Holden MTG, Poyart C, Glaser P, Glaser P, DEVANI Consortium. 2014. Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline. Nat Commun 5:4544 10.1038/ncomms5544. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Roberts MC. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett 245:195–203 10.1016/j.femsle.2005.02.034. [PubMed] [DOI] [PubMed] [Google Scholar]
- 121.Roberts MC, Schwarz S. 2016. Tetracycline and phenicol resistance genes and mechanisms: importance for agriculture, the environment, and humans. J Environ Qual 45:576–592 10.2134/jeq2015.04.0207. [PubMed] [DOI] [PubMed] [Google Scholar]
- 122.Khan SA, Novick RP. 1983. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid 10:251–259 10.1016/0147-619X(83)90039-2. [DOI] [PubMed] [Google Scholar]
- 123.Burdett V, Inamine J, Rajagopalan S. 1982. Heterogeneity of tetracycline resistance determinants in Streptococcus. J Bacteriol 149:995–1004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hartley DL, Jones KR, Tobian JA, LeBlanc DJ, Macrina FL. 1984. Disseminated tetracycline resistance in oral streptococci: implication of a conjugative transposon. Infect Immun 45:13–17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Courvalin P, Carlier C. 1987. Tn1545: a conjugative shuttle transposon. Mol Gen Genet 206:259–264 10.1007/BF00333582. [PubMed] [DOI] [PubMed] [Google Scholar]
- 126.Clewell DB, Flannagan SE, Jaworski DD, Clewell DB. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol 3:229–236 10.1016/S0966-842X(00)88930-1. [DOI] [PubMed] [Google Scholar]
- 127.Rice LB. 1998. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob Agents Chemother 42:1871–1877. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sougakoff W, Papadopoulou B, Nordmann P, Courvalin P. 1987. Nucleotide sequence and distribution of gene tetO encoding tetracycline resistance in Campylobacter coli. FEMS Microbiol Lett 44:153–159 10.1111/j.1574-6968.1987.tb02260.x. [DOI] [Google Scholar]
- 129.Charpentier E, Gerbaud G, Courvalin P. 1993. Characterization of a new class of tetracycline-resistance gene tet(S) in Listeria monocytogenes BM4210. Gene 131:27–34 10.1016/0378-1119(93)90665-P. [PubMed] [DOI] [PubMed] [Google Scholar]
- 130.Fletcher HM, Macrina FL. 1991. Molecular survey of clindamycin and tetracycline resistance determinants in Bacteroides species. Antimicrob Agents Chemother 35:2415–2418 10.1128/AAC.35.11.2415. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Clermont D, Chesneau O, De Cespédès G, Horaud T. 1997. New tetracycline resistance determinants coding for ribosomal protection in streptococci and nucleotide sequence of tet(T) isolated from Streptococcus pyogenes A498. Antimicrob Agents Chemother 41:112–116. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Barbosa TM, Scott KP, Flint HJ. 1999. Evidence for recent intergeneric transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O) in ruminal bacteria. Environ Microbiol 1:53–64 10.1046/j.1462-2920.1999.00004.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 133.Bengtsson B, Unnerstad HE, Ekman T, Artursson K, Nilsson-Ost M, Waller KP. 2009. Antimicrobial susceptibility of udder pathogens from cases of acute clinical mastitis in dairy cows. Vet Microbiol 136:142–149 10.1016/j.vetmic.2008.10.024. [PubMed] [DOI] [PubMed] [Google Scholar]
- 134.Guérin-Faublée V, Tardy F, Bouveron C, Carret G. 2002. Antimicrobial susceptibility of Streptococcus species isolated from clinical mastitis in dairy cows. Int J Antimicrob Agents 19:219–226 10.1016/S0924-8579(01)00485-X. [PubMed] [DOI] [PubMed] [Google Scholar]
- 135.Botrel MA, Haenni M, Morignat E, Sulpice P, Madec JY, Calavas D. 2010. Distribution and antimicrobial resistance of clinical and subclinical mastitis pathogens in dairy cows in Rhône-Alpes, France. Foodborne Pathog Dis 7:479–487 10.1089/fpd.2009.0425. [DOI] [PubMed] [Google Scholar]
- 136.Rossitto PV, Ruiz L, Kikuchi Y, Glenn K, Luiz K, Watts JL, Cullor JS. 2002. Antibiotic susceptibility patterns for environmental streptococci isolated from bovine mastitis in central California dairies. J Dairy Sci 85:132–138 10.3168/jds.S0022-0302(02)74061-7. [DOI] [PubMed] [Google Scholar]
- 137.Brown MB, Roberts MC. 1991. Tetracycline resistance determinants in streptococcal species isolated from the bovine mammary gland. Vet Microbiol 29:173–180 10.1016/0378-1135(91)90124-X. [DOI] [PubMed] [Google Scholar]
- 138.Lollai SA, Ziccheddu M, Duprè I, Piras D. 2016. Characterization of resistance to tetracyclines and aminoglycosides of sheep mastitis pathogens: study of the effect of gene content on resistance. J Appl Microbiol 121:941–951 10.1111/jam.13229. [PubMed] [DOI] [PubMed] [Google Scholar]
- 139.Wisselink HJ, Veldman KT, Van den Eede C, Salmon SA, Mevius DJ. 2006. Quantitative susceptibility of Streptococcus suis strains isolated from diseased pigs in seven European countries to antimicrobial agents licensed in veterinary medicine. Vet Microbiol 113:73–82 10.1016/j.vetmic.2005.10.035. [PubMed] [DOI] [PubMed] [Google Scholar]
- 140.Princivalli MS, Palmieri C, Magi G, Vignaroli C, Manzin A, Camporese A, Barocci S, Magistrali C, Facinelli B. 2009. Genetic diversity of Streptococcus suis clinical isolates from pigs and humans in Italy (2003–2007). Euro Surveill 14:pii=19310. http://www.eurosurveillance.org/content/10.2807/ese.14.33.19310-en 10.2807/ese.14.33.19310-en. [PubMed] [DOI] [PubMed] [Google Scholar]
- 141.Kataoka Y, Yoshida T, Sawada T. 2000. A 10-year survey of antimicrobial susceptibility of Streptococcus suis isolates from swine in Japan. J Vet Med Sci 62:1053–1057 10.1292/jvms.62.1053. [PubMed] [DOI] [PubMed] [Google Scholar]
- 142.Zhang C, Ning Y, Zhang Z, Song L, Qiu H, Gao H. 2008. In vitro antimicrobial susceptibility of Streptococcus suis strains isolated from clinically healthy sows in China. Vet Microbiol 131:386–392 10.1016/j.vetmic.2008.04.005. [PubMed] [DOI] [PubMed] [Google Scholar]
- 143.Vela AI, Moreno MA, Cebolla JA, González S, Latre MV, Domínguez L, Fernández-Garayzábal JF. 2005. Antimicrobial susceptibility of clinical strains of Streptococcus suis isolated from pigs in Spain. Vet Microbiol 105:143–147 10.1016/j.vetmic.2004.10.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 144.Soares TC, Paes AC, Megid J, Ribolla PE, Paduan KS, Gottschalk M. 2014. Antimicrobial susceptibility of Streptococcus suis isolated from clinically healthy swine in Brazil. Can J Vet Res 78:145–149. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 145.Manzin A, Palmieri C, Serra C, Saddi B, Princivalli MS, Loi G, Angioni G, Tiddia F, Varaldo PE, Facinelli B. 2008. Streptococcus suis meningitis without history of animal contact, Italy. Emerg Infect Dis 14:1946–1948 10.3201/eid1412.080679. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen L, Song Y, Wei Z, He H, Zhang A, Jin M. 2013. Antimicrobial susceptibility, tetracycline and erythromycin resistance genes, and multilocus sequence typing of Streptococcus suis isolates from diseased pigs in China. J Vet Med Sci 75:583–587 10.1292/jvms.12-0279. [PubMed] [DOI] [PubMed] [Google Scholar]
- 147.Chander Y, Oliveira SR, Goyal SM. 2011. Identification of the tet(B) resistance gene in Streptococcus suis. Vet J 189:359–360 10.1016/j.tvjl.2010.07.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 148.Haenni M, Hourquet C, Saras E, Madec JY. 2015. Genetic determinants of antimicrobial resistance in Streptococcus canis in France. J Glob Antimicrob Resist 3:142–143 10.1016/j.jgar.2015.02.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 149.Tsuyuki Y, Kurita G, Murata Y, Goto M, Takahashi T. 2017. Identification of group G streptococci isolates from companion animals in Japan and their antimicrobial resistance. Jpn J Infect Dis 70:394–398 10.7883/yoken.JJID.2016.375. [PubMed] [DOI] [PubMed] [Google Scholar]
- 150.Moyaert H, De Graef EM, Haesebrouck F, Decostere A. 2006. Acquired antimicrobial resistance in the intestinal microbiota of diverse cat populations. Res Vet Sci 81:1–7 10.1016/j.rvsc.2005.10.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 151.Pérez-Trallero E, Fernández-Mazarrasa C, García-Rey C, Bouza E, Aguilar L, García-de-Lomas J, Baquero F, Spanish Surveillance Group for Respiratory Pathogens. 2001. Antimicrobial susceptibilities of 1,684 Streptococcus pneumoniae and 2,039 Streptococcus pyogenes isolates and their ecological relationships: results of a 1-year (1998–1999) multicenter surveillance study in Spain. Antimicrob Agents Chemother 45:3334–3340 10.1128/AAC.45.12.3334-3340.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Filipe SR, Tomasz A. 2000. Inhibition of the expression of penicillin resistance in Streptococcus pneumoniae by inactivation of cell wall muropeptide branching genes. Proc Natl Acad Sci USA 97:4891–4896 10.1073/pnas.080067697. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Guenzi E, Gasc AM, Sicard MA, Hakenbeck R. 1994. A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol Microbiol 12:505–515 10.1111/j.1365-2958.1994.tb01038.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 154.Crisóstomo MI, Vollmer W, Kharat AS, Inhülsen S, Gehre F, Buckenmaier S, Tomasz A. 2006. Attenuation of penicillin resistance in a peptidoglycan O-acetyl transferase mutant of Streptococcus pneumoniae. Mol Microbiol 61:1497–1509 10.1111/j.1365-2958.2006.05340.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 155.Dias R, Félix D, Caniça M, Trombe MC. 2009. The highly conserved serine threonine kinase StkP of Streptococcus pneumoniae contributes to penicillin susceptibility independently from genes encoding penicillin-binding proteins. BMC Microbiol 9:121 10.1186/1471-2180-9-121. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Soualhine H, Brochu V, Ménard F, Papadopoulou B, Weiss K, Bergeron MG, Légaré D, Drummelsmith J, Ouellette M. 2005. A proteomic analysis of penicillin resistance in Streptococcus pneumoniae reveals a novel role for PstS, a subunit of the phosphate ABC transporter. Mol Microbiol 58:1430–1440 10.1111/j.1365-2958.2005.04914.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 157.Fani F, Leprohon P, Légaré D, Ouellette M. 2011. Whole genome sequencing of penicillin-resistant Streptococcus pneumoniae reveals mutations in penicillin-binding proteins and in a putative iron permease. Genome Biol 12:R115 10.1186/gb-2011-12-11-r115. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Goldstein FW. 1999. Penicillin-resistant Streptococcus pneumoniae: selection by both beta-lactam and non-beta-lactam antibiotics. J Antimicrob Chemother 44:141–144 10.1093/jac/44.2.141. [PubMed] [DOI] [PubMed] [Google Scholar]
- 159.Anonymous. 2008. Recent trends in antimicrobial resistance among Streptococcus pneumoniae and Staphylococcus aureus isolates: the French experience. Euro Surveill 13:pii=19035. http://eurosurveillance.org/content/10.2807/ese.13.46.19035-en. [PubMed] [Google Scholar]
- 160.Dagan R, Klugman KP. 2008. Impact of conjugate pneumococcal vaccines on antibiotic resistance. Lancet Infect Dis 8:785–795 10.1016/S1473-3099(08)70281-0. [DOI] [PubMed] [Google Scholar]
- 161.Kimura K, Suzuki S, Wachino J, Kurokawa H, Yamane K, Shibata N, Nagano N, Kato H, Shibayama K, Arakawa Y. 2008. First molecular characterization of group B streptococci with reduced penicillin susceptibility. Antimicrob Agents Chemother 52:2890–2897 10.1128/AAC.00185-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nagano N, Nagano Y, Kimura K, Tamai K, Yanagisawa H, Arakawa Y. 2008. Genetic heterogeneity in pbp genes among clinically isolated group B streptococci with reduced penicillin susceptibility. Antimicrob Agents Chemother 52:4258–4267 10.1128/AAC.00596-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kimura K, Nagano N, Arakawa Y. 2015. Classification of group B streptococci with reduced β-lactam susceptibility (GBS-RBS) based on the amino acid substitutions in PBPs. J Antimicrob Chemother 70:1601–1603. [DOI] [PubMed] [Google Scholar]
- 164.Cain D, Malouin F, Dargis M, Harel J, Gottschalk M. 1995. Alterations in penicillin-binding proteins in strains of Streptococcus suis possessing moderate and high levels of resistance to penicillin. FEMS Microbiol Lett 130:121–127 10.1111/j.1574-6968.1995.tb07708.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 165.Haenni M, Galofaro L, Ythier M, Giddey M, Majcherczyk P, Moreillon P, Madec JY. 2010. Penicillin-binding protein gene alterations in Streptococcus uberis isolates presenting decreased susceptibility to penicillin. Antimicrob Agents Chemother 54:1140–1145 10.1128/AAC.00915-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Haenni M, Saras E, Madec JY. 2010. Demonstration of a shift towards penicillin resistance in the Streptococcus uberis population. J Med Microbiol 59:993–995 10.1099/jmm.0.018978-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 167.Rüegsegger F, Ruf J, Tschuor A, Sigrist Y, Rosskopf M, Hässig M. 2014. Antimicrobial susceptibility of mastitis pathogens of dairy cows in Switzerland. Schweiz Arch Tierheilkd 156:483–488 10.1024/0036-7281/a000635. [PubMed] [DOI] [PubMed] [Google Scholar]
- 168.Kalmus P, Aasmäe B, Kärssin A, Orro T, Kask K. 2011. Udder pathogens and their resistance to antimicrobial agents in dairy cows in Estonia. Acta Vet Scand 53:4 10.1186/1751-0147-53-4. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bal EB, Bayar S, Bal MA. 2010. Antimicrobial susceptibilities of coagulase-negative staphylococci (CNS) and streptococci from bovine subclinical mastitis cases. J Microbiol 48:267–274 10.1007/s12275-010-9373-9. [PubMed] [DOI] [PubMed] [Google Scholar]
- 170.Zhang C, Zhang Z, Song L, Fan X, Wen F, Xu S, Ning Y. 2015. Antimicrobial resistance profile and genotypic characteristics of Streptococcus suis capsular type 2 isolated from clinical carrier sows and diseased pigs in China. BioMed Res Int 2015:284303. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.van Hout J, Heuvelink A, Gonggrijp M. 2016. Monitoring of antimicrobial susceptibility of Streptococcus suis in the Netherlands, 2013–2015. Vet Microbiol 194:5–10 10.1016/j.vetmic.2016.03.014. [PubMed] [DOI] [PubMed] [Google Scholar]
- 172.Moreno LZ, da Costa BL, Matajira CE, Gomes VT, Mesquita RE, Silva AP, Moreno AM. 2016. Molecular and antimicrobial susceptibility profiling of Streptococcus dysgalactiae isolated from swine. Diagn Microbiol Infect Dis 86:178–180 10.1016/j.diagmicrobio.2016.07.020. [PubMed] [DOI] [PubMed] [Google Scholar]
- 173.Drlica K, Zhao X. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev 61:377–392. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Escudero JA, San Millan A, Gutierrez B, Hidalgo L, La Ragione RM, AbuOun M, Galimand M, Ferrándiz MJ, Domínguez L, de la Campa AG, Gonzalez-Zorn B. 2011. Fluoroquinolone efflux in Streptococcus suis is mediated by SatAB and not by SmrA. Antimicrob Agents Chemother 55:5850–5860 10.1128/AAC.00498-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Meunier D, Acar JF, Martel JL, Kroemer S, Vallé M. 2004. Seven years survey of susceptibility to marbofloxacin of bovine pathogenic strains from eight European countries. Int J Antimicrob Agents 24:268–278 10.1016/j.ijantimicag.2003.12.011. [PubMed] [DOI] [PubMed] [Google Scholar]
- 176.Kroemer S, Galland D, Guérin-Faublée V, Giboin H, Woehrlé-Fontaine F. 2012. Survey of marbofloxacin susceptibility of bacteria isolated from cattle with respiratory disease and mastitis in Europe. Vet Rec 170:53 10.1136/vr.100246. [PubMed] [DOI] [PubMed] [Google Scholar]
- 177.El Garch F, de Jong A, Simjee S, Moyaert H, Klein U, Ludwig C, Marion H, Haag-Diergarten S, Richard-Mazet A, Thomas V, Siegwart E. 2016. Monitoring of antimicrobial susceptibility of respiratory tract pathogens isolated from diseased cattle and pigs across Europe, 2009–2012: VetPath results. Vet Microbiol 194:11–22 10.1016/j.vetmic.2016.04.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 178.Morrissey I, Moyaert H, de Jong A, El Garch F, Klein U, Ludwig C, Thiry J, Youala M. 2016. Antimicrobial susceptibility monitoring of bacterial pathogens isolated from respiratory tract infections in dogs and cats across Europe: ComPath results. Vet Microbiol 191:44–51 10.1016/j.vetmic.2016.05.020. [PubMed] [DOI] [PubMed] [Google Scholar]
- 179.Ludwig C, de Jong A, Moyaert H, El Garch F, Janes R, Klein U, Morrissey I, Thiry J, Youala M. 2016. Antimicrobial susceptibility monitoring of dermatological bacterial pathogens isolated from diseased dogs and cats across Europe (ComPath results). J Appl Microbiol 121:1254–1267 10.1111/jam.13287. [PubMed] [DOI] [PubMed] [Google Scholar]
- 180.Burrus V, Pavlovic G, Decaris B, Guédon G. 2002. Conjugative transposons: the tip of the iceberg. Mol Microbiol 46:601–610 10.1046/j.1365-2958.2002.03191.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 181.Wozniak RA, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8:552–563 10.1038/nrmicro2382. [PubMed] [DOI] [PubMed] [Google Scholar]
- 182.Santoro F, Vianna ME, Roberts AP. 2014. Variation on a theme; an overview of the Tn916/Tn1545 family of mobile genetic elements in the oral and nasopharyngeal streptococci. Front Microbiol 5:535–545 10.3389/fmicb.2014.00535. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Roberts AP, Mullany P. 2011. Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol Rev 35:856–871 10.1111/j.1574-6976.2011.00283.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 184.Ambroset C, Coluzzi C, Guédon G, Devignes MD, Loux V, Lacroix T, Payot S, Leblond-Bourget N. 2016. New insights into the classification and integration specificity of Streptococcus integrative conjugative elements through extensive genome exploration. Front Microbiol 6:1483–1504 10.3389/fmicb.2015.01483. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Huang J, Ma J, Shang K, Hu X, Liang Y, Li D, Wu Z, Dai L, Chen L, Wang L. 2016. Evolution and diversity of the antimicrobial resistance associated mobilome in Streptococcus suis: a probable mobile genetic elements reservoir for other streptococci. Front Cell Infect Microbiol 6:118 10.3389/fcimb.2016.00118. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Brenciani A, Tiberi E, Morici E, Oryasin E, Giovanetti E, Varaldo PE. 2012. ICESp1116, the genetic element responsible for erm(B)-mediated, inducible resistance to erythromycin in Streptococcus pyogenes. Antimicrob Agents Chemother 56:6425–6429 10.1128/AAC.01494-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Giovanetti E, Brenciani A, Tiberi E, Bacciaglia A, Varaldo PE. 2012. ICESp2905, the erm(TR)-tet(O) element of Streptococcus pyogenes, is formed by two independent integrative and conjugative elements. Antimicrob Agents Chemother 56:591–594 10.1128/AAC.05352-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Mingoia M, Morici E, Marini E, Brenciani A, Giovanetti E, Varaldo PE. 2016. Macrolide resistance gene erm(TR) and erm(TR)-carrying genetic elements in Streptococcus agalactiae: characterization of ICESagTR7, a new composite element containing IMESp2907. J Antimicrob Chemother 71:593–600 10.1093/jac/dkv408. [PubMed] [DOI] [PubMed] [Google Scholar]
- 189.Huang K, Song Y, Zhang Q, Zhang A, Jin M. 2016. Characterisation of a novel integrative and conjugative element ICESsD9 carrying erm(B) and tet(O) resistance determinants in Streptococcus suis, and the distribution of ICESsD9-like elements in clinical isolates. J Glob Antimicrob Resist 7:13–18 10.1016/j.jgar.2016.05.008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 190.Palmieri C, Magi G, Creti R, Baldassarri L, Imperi M, Gherardi G, Facinelli B. 2013. Interspecies mobilization of an ermT-carrying plasmid of Streptococcus dysgalactiae subsp. equisimilis by a coresident ICE of the ICESa2603 family. J Antimicrob Chemother 68:23–26 10.1093/jac/dks352. [PubMed] [DOI] [PubMed] [Google Scholar]
- 191.Croucher NJ, Walker D, Romero P, Lennard N, Paterson GK, Bason NC, Mitchell AM, Quail MA, Andrew PW, Parkhill J, Bentley SD, Mitchell TJ. 2009. Role of conjugative elements in the evolution of the multidrug-resistant pandemic clone Streptococcus pneumoniaeSpain23F ST81. J Bacteriol 191:1480–1489 10.1128/JB.01343-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Douarre PE, Sauvage E, Poyart C, Glaser P. 2015. Host specificity in the diversity and transfer of lsa resistance genes in group B Streptococcus. J Antimicrob Chemother 70:3205–3213. [PubMed] [DOI] [PubMed] [Google Scholar]
- 193.Huang K, Zhang Q, Song Y, Zhang Z, Zhang A, Xiao J, Jin M. 2016. Characterization of spectinomycin resistance in Streptococcus suis leads to two novel insights into drug resistance formation and dissemination mechanism. Antimicrob Agents Chemother 60:6390–6392 10.1128/AAC.01157-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Marini E, Palmieri C, Magi G, Facinelli B. 2015. Recombination between Streptococcus suis ICESsu32457 and Streptococcus agalactiae ICESa2603 yields a hybrid ICE transferable to Streptococcus pyogenes. Vet Microbiol 178:99–104 10.1016/j.vetmic.2015.04.013. [PubMed] [DOI] [PubMed] [Google Scholar]
- 195.Srinivasan V, Metcalf BJ, Knipe KM, Ouattara M, McGee L, Shewmaker PL, Glennen A, Nichols M, Harris C, Brimmage M, Ostrowsky B, Park CJ, Schrag SJ, Frace MA, Sammons SA, Beall B. 2014. vanG element insertions within a conserved chromosomal site conferring vancomycin resistance to Streptococcus agalactiae and Streptococcus anginosus. MBio 5:e01386-14 10.1128/mBio.01386-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Meng F, Kanai K, Yoshikoshi K. 2009. Structural characterization of Tn916-like element in Streptococcus parauberis serotype II strains isolated from diseased Japanese flounder. Lett Appl Microbiol 48:770–776. [PubMed] [DOI] [PubMed] [Google Scholar]
- 197.Palmieri C, Magi G, Mingoia M, Bagnarelli P, Ripa S, Varaldo PE, Facinelli B. 2012. Characterization of a Streptococcus suis tet(O/W/32/O)-carrying element transferable to major streptococcal pathogens. Antimicrob Agents Chemother 56:4697–4702 10.1128/AAC.00629-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Richards VP, Zadoks RN, Pavinski Bitar PD, Lefébure T, Lang P, Werner B, Tikofsky L, Moroni P, Stanhope MJ. 2012. Genome characterization and population genetic structure of the zoonotic pathogen, Streptococcus canis. BMC Microbiol 12:293–309 10.1186/1471-2180-12-293. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bekele T, Molla B. 2001. Mastitis in lactating camels (Camelus dromedarius) in Afar Region, north-eastern Ethiopia. Berl Munch Tierarztl Wochenschr 114:169–172. [PubMed] [PubMed] [Google Scholar]
- 200.Evans JJ, Pasnik DJ, Klesius PH, Al-Ablani S. 2006. First report of Streptococcus agalactiae and Lactococcus garvieae from a wild bottlenose dolphin (Tursiops truncatus). J Wildl Dis 42:561–569 10.7589/0090-3558-42.3.561. [PubMed] [DOI] [PubMed] [Google Scholar]
- 201.Jordal S, Glambek M, Oppegaard O, Kittang BR. 2015. New tricks from an old cow: infective endocarditis caused by Streptococcus dysgalactiae subsp. dysgalactiae. J Clin Microbiol 53:731–734 10.1128/JCM.02437-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nomoto R, Munasinghe LI, Jin DH, Shimahara Y, Yasuda H, Nakamura A, Misawa N, Itami T, Yoshida T. 2004. Lancefield group C Streptococcus dysgalactiae infection responsible for fish mortalities in Japan. J Fish Dis 27:679–686 10.1111/j.1365-2761.2004.00591.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 203.Hilmarsdóttir I, Valsdóttir F. 2007. Molecular typing of beta-hemolytic streptococci from two patients with lower-limb cellulitis: identical isolates from toe web and blood specimens. J Clin Microbiol 45:3131–3132 10.1128/JCM.00532-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kawata K, Minakami T, Mori Y, Katsumi M, Kataoka Y, Ezawa A, Kikuchi N, Takahashi T. 2003. rDNA sequence analyses of Streptococcus dysgalactiae subsp. equisimilis isolates from pigs. Int J Syst Evol Microbiol 53:1941–1946 10.1099/ijs.0.02666-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 205.Lopardo HA, Vidal P, Sparo M, Jeric P, Centron D, Facklam RR, Paganini H, Pagniez NG, Lovgren M, Beall B. 2005. Six-month multicenter study on invasive infections due to Streptococcus pyogenes and Streptococcus dysgalactiae subsp. equisimilis in Argentina. J Clin Microbiol 43:802–807 10.1128/JCM.43.2.802-807.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Nei T, Akutsu K, Shima A, Tsuboi I, Suzuki H, Yamamoto T, Tanaka K, Shinoyama A, Kojima Y, Washio Y, Okawa S, Sonobe K, Norose Y, Saito R. 2012. A case of streptococcal toxic shock syndrome due to group G streptococci identified as Streptococcus dysgalactiae subsp. equisimilis. J Infect Chemother 18:919–924 10.1007/s10156-012-0375-x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 207.Savini V, Catavitello C, Talia M, Manna A, Pompetti F, Di Bonaventura G, Di Giuseppe N, Febbo F, Balbinot A, Di Zacomo S, Esattore F, D’Antonio D. 2008. Beta-lactam failure in treatment of two group G Streptococcus dysgalactiae subsp. equisimilis pharyngitis patients. J Clin Microbiol 46:814–816 10.1128/JCM.00985-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Siljander T, Karppelin M, Vähäkuopus S, Syrjänen J, Toropainen M, Kere J, Vuento R, Jussila T, Vuopio-Varkila J. 2008. Acute bacterial, nonnecrotizing cellulitis in Finland: microbiological findings. Clin Infect Dis 46:855–861 10.1086/527388. [PubMed] [DOI] [PubMed] [Google Scholar]
- 209.Wajima T, Morozumi M, Hanada S, Sunaoshi K, Chiba N, Iwata S, Ubukata K. 2016. Molecular characterization of invasive Streptococcus dysgalactiae subsp. equisimilis, Japan. Emerg Infect Dis 22:247–254 10.3201/eid2202.141732. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zoric M, Nilsson E, Lundeheim N, Wallgren P. 2009. Incidence of lameness and abrasions in piglets in identical farrowing pens with four different types of floor. Acta Vet Scand 51:23–32 10.1186/1751-0147-51-23. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Casagrande Proietti P, Bietta A, Coppola G, Felicetti M, Cook RF, Coletti M, Marenzoni ML, Passamonti F. 2011. Isolation and characterization of β-haemolytic-streptococci from endometritis in mares. Vet Microbiol 152:126–130 10.1016/j.vetmic.2011.04.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 212.Imai D, Jang S, Miller M, Conrad PA. 2009. Characterization of beta-hemolytic streptococci isolated from southern sea otters (Enhydra lutris nereis) stranded along the California coast. Vet Microbiol 136:378–381 10.1016/j.vetmic.2008.11.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 213.García-País MJ, Rabuñal R, Armesto V, López-Reboiro M, García-Garrote F, Coira A, Pita J, Rodríguez-Macías AI, López-Álvarez MJ, Alonso MP, Corredoira J. 2016. Streptococcus bovis septic arthritis and osteomyelitis: a report of 21 cases and a literature review. Semin Arthritis Rheum 45:738–746 10.1016/j.semarthrit.2016.02.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 214.Osawa R, Sly LI. 1991. Phenotypic characterization of CO2-requiring strains of Streptococcus bovis from koalas. Appl Environ Microbiol 57:3037–3039. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sekizaki T, Nishiya H, Nakajima S, Nishizono M, Kawano M, Okura M, Takamatsu D, Nishino H, Ishiji T, Osawa R. 2008. Endocarditis in chickens caused by subclinical infection of Streptococcus gallolyticus subsp. gallolyticus. Avian Dis 52:183–186 10.1637/8048-070307-Case. [PubMed] [DOI] [PubMed] [Google Scholar]
- 216.van Samkar A, Brouwer MC, Pannekoek Y, van der Ende A, van de Beek D. 2015. Streptococcus gallolyticus meningitis in adults: report of five cases and review of the literature. Clin Microbiol Infect 21:1077–1083 10.1016/j.cmi.2015.08.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 217.Katsumi M, Kataoka Y, Takahashi T, Kikuchi N, Hiramune T. 1997. Bacterial isolation from slaughtered pigs associated with endocarditis, especially the isolation of Streptococcus suis. J Vet Med Sci 59:75–78 10.1292/jvms.59.75. [PubMed] [DOI] [PubMed] [Google Scholar]
- 218.O’Sullivan T, Friendship R, Blackwell T, Pearl D, McEwen B, Carman S, Slavić D, Dewey C. 2011. Microbiological identification and analysis of swine tonsils collected from carcasses at slaughter. Can J Vet Res 75:106–111. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 219.Miller CW, Prescott JF, Mathews KA, Betschel SD, Yager JA, Guru V, DeWinter L, Low DE. 1996. Streptococcal toxic shock syndrome in dogs. J Am Vet Med Assoc 209:1421–1426. [PubMed] [PubMed] [Google Scholar]
- 220.Reissmann S, Friedrichs C, Rajkumari R, Itzek A, Fulde M, Rodloff AC, Brahmadathan KN, Chhatwal GS, Nitsche-Schmitz DP. 2010. Contribution of Streptococcus anginosus to infections caused by groups C and G streptococci, southern India. Emerg Infect Dis 16:656–663 10.3201/eid1604.090448. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Hatrongjit R, Kerdsin A, Gottschalk M, Takeuchi D, Hamada S, Oishi K, Akeda Y. 2015. First human case report of sepsis due to infection with Streptococcus suis serotype 31 in Thailand. BMC Infect Dis 15:392–399 10.1186/s12879-015-1136-0. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Mancini F, Adamo F, Creti R, Monaco M, Alfarone G, Pantosti A, Ciervo A. 2016. A fatal case of streptococcal toxic shock syndrome caused by Streptococcus suis carrying tet (40) and tet (O/W/32/O), Italy. J Infect Chemother 22:774–776 10.1016/j.jiac.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 223.Sánchez del Rey V, Fernández-Garayzábal JF, Briones V, Iriso A, Domínguez L, Gottschalk M, Vela AI. 2013. Genetic analysis of Streptococcus suis isolates from wild rabbits. Vet Microbiol 165:483–486 10.1016/j.vetmic.2013.04.025. [PubMed] [DOI] [PubMed] [Google Scholar]
- 224.Sánchez del Rey V, Fernández-Garayzábal JF, Domínguez L, Gottschalk M, Vela AI. 2016. Screening of virulence-associated genes as a molecular typing method for characterization of Streptococcus suis isolates recovered from wild boars and pigs. Vet J 209:108–112 10.1016/j.tvjl.2015.11.007. [PubMed] [DOI] [PubMed] [Google Scholar]
- 225.Staats JJ, Feder I, Okwumabua O, Chengappa MM. 1997. Streptococcus suis: past and present. Vet Res Commun 21:381–407 10.1023/A:1005870317757. [PubMed] [DOI] [PubMed] [Google Scholar]
- 226.Anshary H, Kurniawan RA, Sriwulan S, Ramli R, Baxa DV. 2014. Isolation and molecular identification of the etiological agents of streptococcosis in Nile tilapia (Oreochromis niloticus) cultured in net cages in Lake Sentani, Papua, Indonesia. Springerplus 3:627–638 10.1186/2193-1801-3-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Bonar CJ, Wagner RA. 2003. A third report of “golf ball disease” in an Amazon River dolphin (Inia geoffrensis) associated with Streptococcus iniae. J Zoo Wildl Med 34:296–301 10.1638/1042-7260(2003)034[0296:ATROGB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 228.Chou L, Griffin MJ, Fraites T, Ware C, Ferguson H, Keirstead N, Brake J, Wiles J, Hawke JP, Kearney MT, Getchell RG, Gaunt P, Soto E. 2014. Phenotypic and genotypic heterogeneity among Streptococcus iniae isolates recovered from cultured and wild fish in North America, Central America and the Caribbean islands. J Aquat Anim Health 26:263–271 10.1080/08997659.2014.945048. [PubMed] [DOI] [PubMed] [Google Scholar]
- 229.El Aamri F, Padilla D, Acosta F, Caballero M, Roo J, Bravo J, Vivas J, Real F. 2010. First report of Streptococcus iniae in red porgy (Pagrus pagrus, L.). J Fish Dis 33:901–905 10.1111/j.1365-2761.2010.01191.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 230.Figueiredo HC, Netto LN, Leal CA, Pereira UP, Mian GF. 2012. Streptococcus iniae outbreaks in Brazilian Nile tilapia (Oreochromis niloticus L:) farms. Braz J Microbiol 43:576–580 10.1590/S1517-83822012000200019. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Keirstead ND, Brake JW, Griffin MJ, Halliday-Simmonds I, Thrall MA, Soto E. 2014. Fatal septicemia caused by the zoonotic bacterium Streptococcus iniae during an outbreak in Caribbean reef fish. Vet Pathol 51:1035–1041 10.1177/0300985813505876. [PubMed] [DOI] [PubMed] [Google Scholar]
- 232.Denamiel G, Llorente P, Carabella M, Rebuelto M, Gentilini E. 2005. Anti-microbial susceptibility of Streptococcus spp. isolated from bovine mastitis in Argentina. J Vet Med B Infect Dis Vet Public Health 52:125–128 10.1111/j.1439-0450.2005.00830.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 233.Nam HM, Lim SK, Kang HM, Kim JM, Moon JS, Jang KC, Joo YS, Kang MI, Jung SC. 2009. Antimicrobial resistance of streptococci isolated from mastitic bovine milk samples in Korea. J Vet Diagn Invest 21:698–701 10.1177/104063870902100517. [PubMed] [DOI] [PubMed] [Google Scholar]
- 234.Entorf M, Feßler AT, Kaspar H, Kadlec K, Peters T, Schwarz S. 2016. Comparative erythromycin and tylosin susceptibility testing of streptococci from bovine mastitis. Vet Microbiol 194:36–42 10.1016/j.vetmic.2015.12.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 235.Marie J, Morvan H, Berthelot-Hérault F, Sanders P, Kempf I, Gautier-Bouchardon AV, Jouy E, Kobisch M. 2002. Antimicrobial susceptibility of Streptococcus suis isolated from swine in France and from humans in different countries between 1996 and 2000. J Antimicrob Chemother 50:201–209 10.1093/jac/dkf099. [PubMed] [DOI] [PubMed] [Google Scholar]
- 236.Martel A, Baele M, Devriese LA, Goossens H, Wisselink HJ, Decostere A, Haesebrouck F. 2001. Prevalence and mechanism of resistance against macrolides and lincosamides in Streptococcus suis isolates. Vet Microbiol 83:287–297 10.1016/S0378-1135(01)00426-6. [DOI] [PubMed] [Google Scholar]
- 237.Tian Y, Aarestrup FM, Lu CP. 2004. Characterization of Streptococcus suis serotype 7 isolates from diseased pigs in Denmark. Vet Microbiol 103:55–62 10.1016/j.vetmic.2004.07.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 238.Overesch G, Stephan R, Perreten V. 2013. Antimicrobial susceptibility of gram-positive udder pathogens from bovine mastitis milk in Switzerland. Schweiz Arch Tierheilkd 155:339–350. [PubMed] [DOI] [PubMed] [Google Scholar]


