ABSTRACT
Campylobacter is a major foodborne pathogen and has become increasingly resistant to clinically important antimicrobials. To cope with the selection pressure from antimicrobial use in both veterinary and human medicine, Campylobacter has developed multiple mechanisms for antibiotic resistance, including modification or mutation of antimicrobial targets, modification or inactivation of antibiotics, and reduced drug accumulation by drug efflux pumps. Some of these mechanisms confer resistance to a specific class of antimicrobials, while others give rise to multidrug resistance. Notably, new antibiotic resistance mechanisms continuously emerge in Campylobacter, and some examples include the recently discovered multidrug resistance genomic islands harboring multiple genes involved in the resistance to aminoglycosides and macrolides, a novel Cfr(C) conferring resistance to phenicols and other drugs, and a potent multidrug efflux pump CmeABC variant (RE-CmeABC) that shows a significantly enhanced function in multidrug resistance and is associated with exceedingly high-level resistance to fluoroquinolones. These newly emerged resistance mechanisms are horizontally transferable and greatly facilitate the adaptation of Campylobacter in the food-producing environments where antibiotics are frequently used. In this article, we will discuss how Campylobacter resists the action of various classes of antimicrobials, with an emphasis on newly discovered mechanisms.
INTRODUCTION
Campylobacter, a foodborne bacterial pathogen, is the leading cause of human gastroenteritis worldwide. According to data from the World Health Organization, the estimated incidence of gastroenteritis due to Campylobacter spp. in high-income countries is between 4.4 and 9.3 per 1,000 people (1). Most Campylobacter infections are mild and self-limiting and may not require antimicrobial therapy; however, antibiotic treatment is required for severe or prolonged infections. In clinical settings, fluoroquinolones and macrolides are the drugs of choice to treat campylobacteriosis (2–6), but in some cases, tetracyclines and gentamicin are used to treat systemic infection with Campylobacter (5, 6). In a report from the Centers for Disease Control and Prevention (CDC) on antibiotic resistance threats in the United States in 2013, drug-resistant Campylobacter was listed under “microorganisms with a threat level of serious” (http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf). The CDC indicated that almost 24% of Campylobacter strains tested were resistant to ciprofloxacin (fluoroquinolone) or azithromycin (macrolide), indicating that approximately 310,000 Campylobacter infections are caused by drug-resistant Campylobacter each year in the United States. Although contaminated undercooked poultry meat is a main source of infection for human campylobacteriosis (2, 7), ruminant Campylobacter is also a significant contributor for foodborne illnesses (8–15).
As a foodborne pathogen transmitted via foodborne routes, Campylobacter is constantly exposed to antimicrobials used for food production. In dealing with antimicrobial selection, Campylobacter has evolved various mechanisms of resistance to antimicrobials. Some of the mechanisms confer resistance to a specific class of antimicrobials, while others may confer multidrug resistance. Previous publications have provided excellent reviews on antibiotic resistance in Campylobacter (5, 16–19). However, several new antibiotic resistance mechanisms have emerged in Campylobacter in recent years. Examples include the rRNA methylase Erm(B) (mediating macrolide resistance), a functionally enhanced multidrug efflux pump variant (RE-CmeABC), methylarsenite efflux permease ArsP conferring resistance to organoarsenicals, a novel fosXCC gene conferring fosfomycin resistance, and the rRNA methyltransferase Cfr(C) mediating multidrug resistance. In this review, we will summarize the current state of antibiotic resistance in Campylobacter, with an emphasis on the newly emerged mechanisms.
RESISTANCE TO FLUOROQUINOLONES
The fluoroquinolones (e.g., ciprofloxacin, enrofloxacin, etc.) are a family of synthetic broad-spectrum antibacterial agents that are active against a wide range of Gram-positive and Gram-negative organisms (20, 21). To date, they are one of the drugs of choice to treat campylobacteriosis in humans as well as other bacterial diseases in both animals and humans (21–23). Fluoroquinolones target two essential enzymes, DNA gyrase and topoisomerase IV, and impair DNA replication (21, 24). Generally, mutations in the genes encoding the subunits of DNA gyrase (GyrA and GyrB), topoisomerase IV (ParC and ParE), or both are responsible for the resistance of bacteria to fluoroquinolones (25, 26). In Campylobacter, the main resistance mechanism to fluoroquinolones is mediated by point mutations in the quinolone resistance-determining region of GyrA (4, 5). To date, mutations in GyrB have not been associated with fluoroquinolone resistance in Campylobacter (27–29). The absence of genes encoding ParC and ParE implies that they are not involved in fluoroquinolone action and resistance in Campylobacter (27–32). Notably, a single point mutation in the quinolone resistance-determining region of gyrA is sufficient to substantially reduce the susceptibility of Campylobacter to fluoroquinolone antimicrobials (5, 30, 33, 34). Multiple resistance-associated mutations, including T86I, T86K, A70T, and D90N, have been reported in Campylobacter (4, 5, 33, 35). The C257T change in the gyrA gene, which leads to the T86I substitution in gyrase, is the most frequently observed mutation conferring resistance to fluoroquinolones in Campylobacter (4, 5, 36). In addition to mutations in GyrA, the functional multidrug efflux pump, CmeABC, is also required for fluoroquinolone resistance in Campylobacter. Inactivation of cmeABC in fluoroquinolone-resistant mutants (carrying specific GyrA mutations) made the resistant mutants susceptible to fluoroquinolones (30). Until now, mutations in GyrA together with CmeABC have been the only identified mechanisms of fluoroquinolone resistance in Campylobacter. Plasmid-mediated quinolone-resistance determinants, such as qnr, aac(6′)-Ib-cr, and qepA, have not been reported in Campylobacter.
RESISTANCE TO MACROLIDES
The macrolide antibiotics (azithromycin, clarithromycin, erythromycin, telithromycin, etc.) are a class of drugs for the treatment of gastric diseases caused by Helicobacter pylori and Campylobacter and for respiratory tract infections in humans (37). Antibiotics in this class, including erythromycin, tylosin, spiramycin, tilmicosin, and roxithromycin, are also approved for growth promotion and therapeutic purposes in animals (38). Macrolides target the 50S subunit of the bacterial ribosome and inhibit protein synthesis through interference with the peptide translocation step (39, 40). Generally, bacterial resistance to macrolides is mediated by three mechanisms: enzymatic inactivation of macrolides, modification or point mutations in the target, and enhanced drug efflux (5, 41). In Campylobacter, modification of the ribosomal target, leading to macrolide resistance, can occur either by enzyme-mediated methylation or by point mutation in the 23S rRNA and/or ribosomal proteins L4 and L22 (4, 5, 41). Although an early report suggested the presence of rRNA methylation genes in Campylobacter rectus isolates based on the result of Southern hybridization (42), an rRNA methylating enzyme was not formally identified until recently, when Erm(B) was identified in both Campylobacter jejuni and Campylobacter coli from various sources, including swine, chicken, ducks, and humans (43–45). The erm(B) gene was either located in the chromosomal DNA or carried by a plasmid (43). This gene alone is able to confer high-level resistance to macrolides (44). It is worth noting that the erm(B) gene is associated with multidrug resistance genomic islands (MDRGIs), which include several resistance genes [aacA-aphD, sat4, aphA-3, fosXCC, aad9, and tet(O)] and mediate resistance to multiple classes of antibiotics (43, 44) (Fig. 1). Generally, MDRGIs are located in the region between cadF and CCO1582, nfsB and cinA, or cj0168c and sodB (43–45) and are transferrable among different Campylobacter spp. by natural transformation under laboratory conditions (43).
FIGURE 1.
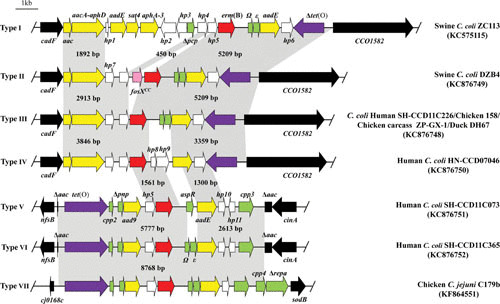
Chromosomal organization and comparison of seven types (I to VII) of MDRGIs containing the erm(B) gene (modified from references 43–45). erm(B) is in red, aminoglycoside resistance genes are in yellow, the streptothricin resistance gene (sat4) is in blue, the tetracycline resistance gene [tet(O)] is in purple, genes with predicted functions are in green, and genes coding hypothetical proteins are in white. The tet(O) gene is intact in types V and VI but is truncated in other types. The border genes of the MDRGIs are depicted by black box arrows. The gray shading indicates regions sharing more than 98% DNA identity. A representative strain for each type of MDRGI is indicated on the right side of the panel.
Point mutations in domain V of the 23S rRNA have been recognized as the most common mechanism for macrolide resistance in Campylobacter (4, 5, 41, 46). These point mutations occur at positions 2074 and 2075 of the 23S rRNA in Campylobacter, which correspond to positions 2058 and 2059, respectively, in Escherichia coli. Among the reported resistance-associated mutations, the A2074C, A2074G, and A2075G mutations confer high-level resistance to macrolide antibiotics (erythromycin MIC >128 µg/ml) in Campylobacter (46–50), with A2075G being the predominant mutation in clinical and field isolates (4, 5, 41, 46). Campylobacter contains three copies of 23S rRNA genes (51), and usually, macrolide resistance-associated mutations occur in all three copies for most Campylobacter isolates with high-level resistance (47, 48, 52).
In addition to target modification, active efflux also contributes to macrolide resistance in Campylobacter (48–50, 53–56). The CmeABC efflux system functions synergistically with target mutations, and inactivation of CmeABC significantly reduces the resistance to macrolide antibiotics in isolates with high-, intermediate-, or low-level macrolide resistance (19). Additionally, the synergy between the CmeABC efflux pump and mutations in the ribosomal proteins L4 (G74D) and L22 (inserted at position 86 or 98) also confers macrolide resistance in Campylobacter (50, 53).
RESISTANCE TO TETRACYCLINES
Tetracyclines, discovered in the 1940s, are an important class of antibiotics that are widely used in both human and animal medicine. Tetracyclines have broad-spectrum activity against Gram-positive and Gram-negative bacteria, as well as chlamydiae, mycoplasmas, rickettsiae, and protozoan parasites (57). It is well established that tetracyclines inhibit bacterial protein synthesis by preventing the attachment of aminoacyl-tRNA to the ribosomal acceptor (A) site (58, 59). Because of the long history and widespread use of tetracyclines, a number of resistance determinants to this class of drugs have been observed in a variety of bacteria (60, 61). Generally, the resistance to tetracyclines is mediated by one of four mechanisms: efflux pumps, chemical modification of tetracyclines, ribosomal protection proteins, and mutations in rRNA (60).
To date, resistance to tetracyclines in Campylobacter is conferred by the ribosomal protection protein Tet(O) and efflux pumps (CmeABC and CmeG). Tet(O) belongs to one of the characterized ribosomal protection proteins (61), several of which are paralogs of the translational GTPase EF-G and actively remove tetracycline from the ribosome in a GTP hydrolysis-dependent fashion (62–64). The well-documented action mode is that Tet(O) recognizes and binds to an open A site on the bacterial ribosome and then induces a conformational change that results in the sequential release of the bound tetracycline molecule (60, 64). This conformational change is able to persist and allows the A site to function in protein elongation (60, 64). A recent study indicated that several critical residues located in the three loops of Tet(O) disrupt the binding of tetracycline to the ribosome complex (59). The tet(O) gene, which is widely present in C. jejuni and C. coli (65, 66), can be located either in the chromosomal DNA or on a plasmid (e.g., pTet and pCC31) (67–69). Based on the G-C content, sequence homology, codon usage, and hybridization analysis, it appears that the Campylobacter tet(O) gene was probably acquired from a Gram-positive origin by horizontal gene transfer (66, 68). The CmeABC and CmeG multidrug efflux pumps contribute to both intrinsic and acquired resistance to tetracycline in Campylobacter (55, 70–72). CmeABC functions synergistically with Tet(O) to confer high-level resistance to tetracycline (70). Inactivation of either CmeABC or CmeG increases the susceptibility of Campylobacter to tetracyclines (70, 72).
RESISTANCE TO AMINOGLYCOSIDES
Aminoglycosides are bactericidal antibiotics that bind to ribosomes and inhibit protein synthesis (73). They are structurally characterized by an aminocyclitol ring bound to one or more amino sugars by pseudoglycosidic bonds (74–76). This class of antimicrobials is generally considered to have broad-spectrum bacteriocidal activity and is clinically used to treat acute and systemic Campylobacter infections (77, 78), although they have limited activity in anaerobic environment. On the basis of the in vitro susceptibility of many Campylobacter strains to aminoglycosides (79, 80), oxygen levels in the microaerophilic environments preferred by Campylobacter are sufficient to allow the transport of compounds into the intracellular environment (18). A total of five mechanisms of aminoglycoside resistance in bacteria have been described (74, 76, 81): (i) reduced accumulation of the drug in the intracellular environment, conferred by a multidrug efflux pump that transports the drug back into the extracellular environment or due to decreased permeability of the bacterial cellular membrane to the drug (74), (ii) methylation of 16S rRNA in sites that interfere with drug binding (75, 82), (iii) mutations in the binding sites of rRNA, especially in Mycobacterium spp. with a single copy of the ribosomal operon (75), (iv) active swarming, a nonspecific mechanism in P. aeruginosa cells that exhibits adaptive antibiotic resistance against several antibiotics, including gentamicin (83), and (v) enzymatic modification at the -OH or -NH2 groups of the 2-deoxystreptamine nucleus or sugar moieties of the antibiotic, which is considered the most important mechanism (81). Among the known mechanisms of aminoglycoside resistance, modification of the aminoglycoside structure by enzymes such as aminoglycoside acetyltransferases, aminoglycoside phosphotransferases, and aminoglycoside nucleotidyltransferases is the most significant and prevalent in several bacterial species, including Campylobacter spp. (16, 78, 84). In Campylobacter, each of the above-mentioned aminoglycoside-modifying enzymes has been detected.
Aminoglycoside phosphotransferases constitute the majority of aminoglycoside-modifying enzymes identified in Campylobacter spp. and are responsible for phosphorylation of the 3′ hydroxyl group of aminoglycosides. They also mediate kanamycin and neomycin resistance. Aminoglycoside phosphotransferases are divided into eight groups according to the resistance of additional specific aminoglycosides (I to VIII) (76, 81). To date, only types I, III, IV, and VII, which mainly mediate kanamycin resistance, have been detected in Campylobacter. The aphA-1 gene, also known as aph(3′)-Ia, was identified adjacent to the insertion sequence IS15-delta commonly found in Gram-negative bacteria, suggesting that it may have originated from the Enterobacteriaceae family of organisms (85). Sequence analysis showed identical homology to the kanamycin resistance gene of the Tn903 transposon in E. coli (85). The aphA-1 gene is also commonly used as a resistance marker gene in cloning vehicles (81). Different from aphA-1, the aphA-3 gene is commonly detected in Gram-positive bacteria, such as Staphylococcus, and has been identified in clinical Campylobacter isolates. It is located on plasmids or chromosomes (80, 84). Some plasmids in C. jejuni harbor the aphA-3 gene as part of the resistance cluster that includes the aadE and sat4 genes, which originated from Gram-positive bacteria (86). Subsequently, the aadE-sat4-aphA-3 cluster together with additional aminoglycoside resistance genes, including aacA-aphD and aac, was identified in a genomic island on a chromosome of C. coli (87). Clonal expansion and horizontal transmission have been involved in dissemination of this novel aminoglycoside resistance genomic island (87). The identification of the aphA-3 gene in Campylobacter provides another piece of evidence suggesting the transfer of antibiotic resistance genes from Gram-positive bacteria to Gram-negative bacteria. The plasmid-encoded aphA-7 gene, mediating kanamycin resistance, may be an indigenous gene of Campylobacter based on its G+C content at 32.8%, which is similar to that of the Campylobacter genome (88). The aphA-7 gene was found on small plasmids of 9.5 and 11.5 kb in C. jejuni (89), and the presence of this gene in C. coli has also been documented (87).
The aacA4 gene encodes aminoglycoside 6′-N-acetyltransferase, AAC(6′)-Ib7, conferring resistance to tobramycin, kanamycin, and neomycin (90). Additionally, the aacA4 gene was associated with class 1 integron and found in C. jejuni isolated from the water lines of a broiler chicken house environment (91). The plasmid-borne gene aac(6′)-Ie/aph(2″)-Ia (also named aacA/aphD and encoding a bifunctional enzyme) was described in a clinical isolate of C. jejuni from a U.S. soldier deployed to Thailand. It was found to encode phosphotransferase activity and was named aph(2″)-If (92). Gentamicin resistance, conferred by aacA/aphD that is associated with an aminoglycoside resistance genomic island, was reported in C. coli from China, and clonal expansion may be involved in dissemination of this entire resistance island (87). Subsequently, the gentamicin resistance-related gene aph(2″)-Ig (encoding a phosphotransferase) was detected in a C. coli isolate from retail chicken meat (93). Recently, several variants of gentamicin resistance genes [aph(2″)-Ib, Ic, If1, If3, Ih, and aac(6′)-Ie/aph(2″)-If2] were identified in Campylobacter isolates from humans and retail meats in the United States. The same resistance profile and similar pulsed-field gel electrophoresis patterns shared by isolates from human and retail chicken indicated that retail chicken is a potential source for gentamicin-resistant C. coli that causes infections in humans (94). The increasing prevalence and emergence of novel genes of gentamicin resistance has led to an increasing number of studies on gentamicin resistance mechanisms.
The sat4 gene encoding a streptothricin acetyltransferase is present either as a single gene or in the aadE-sat4-aphA-3 cluster in streptothricin-resistant Campylobacter spp. (87, 95). The aminoglycoside 3-adenyltransferase gene (aadA) confers resistance to streptomycin and spectinomycin, while the aminoglycoside 6-adenyltransferase gene (aadE) only confers resistance to streptomycin. The aadA gene was identified in the multidrug resistance plasmid pCG8245, which contains various aminoglycoside resistance genes in C. jejuni (95). In contrast, aadE was commonly associated with the aadE-sat4-aphA-3 gene cluster that was detected on the plasmid or chromosome of C. jejuni and C. coli (87, 94, 95). The 286-amino-acid streptomycin resistance protein, ANT(6)-Ib, encoded by ant(6)-Ib, belongs to a family of aminoglycoside nucleotidyltransferases and was identified in Campylobacter fetus subsp. fetus (96). Recently, a novel streptomycin resistance gene was described, and its widespread presence among C. coli isolates may partly account for the prevalence of streptomycin resistance in C. coli (97).
RESISTANCE TO β-LACTAMS
β-Lactam antibiotics are a class of broad-spectrum antibiotics that inhibit bacterial cell wall biosynthesis. This class of antibiotic agents contains a β-lactam ring in their molecular structures. β-lactam antibiotics are the most widely used antibiotics and account for more than half of the total antibiotic market worldwide (98). For the past decades, the prevalence of β-lactam-resistant bacteria has greatly increased (99, 100). To date, three mechanisms contributing to β-lactam resistance in Campylobacter have been identified: enzymatic inactivation, reduced uptake, and efflux pump. A previous study showed that β-lactamase-positive Campylobacter were more resistant than β-lactamase-negative Campylobacter to amoxicillin, ampicillin, and ticarcillin (101). OXA-61 (Cj0299) is the only identified and characterized β-lactamase in C. jejuni (102–104). Notably, almost half of OXA-61-carrying Campylobacter are susceptible to ampicillin, suggesting that the expression level of OXA-61 modulates the resistance phenotype (102). Indeed, a recent study indicated that a G → T transversion in the OXA-61 promoter enhances the expression of β-lactamase and is linked to high-level β-lactam resistance in C. jejuni isolates (104). The porins of C. jejuni and C. coli form a relatively small cation-selective pore that may contribute to intrinsic resistance to antimicrobial agents. These cation-selective pores in C. jejuni and C. coli are able to exclude most β-lactams with a molecular weight greater than 360 or that are anionic (105). The CmeABC and CmeDEF efflux pumps may also contribute to β-lactam resistance. Inactivation of these efflux pumps results in increased susceptibility to ampicillin (70, 106, 107).
RESISTANCE TO PHENICOLS
Nonfluorinated (chloramphenicol) or fluorinated phenicols (florfenicol) are highly effective against a wide variety of Gram-positive and Gram-negative bacteria. Phenicols were once widely applied in both human and veterinary practices for the prevention and treatment of many bacterial infections. Resistance to phenicols in Campylobacter is mediated by enzymatic inactivation via chloramphenicol acetyltransferases, target site mutations in 23S rRNA, target site modification in 23S rRNA via the rRNA methyltransferase Cfr(C), or enhanced extrusion by efflux pumps. Acetylation of the drug by chloramphenicol acetyltransferases (encoded by cat) confers resistance to chloramphenicol but not to florfenicol (108). The G2073A mutation in the 23S rRNA gene of Campylobacter (corresponding to position 2057 in the 23S rRNA gene of E. coli) is associated with chloramphenicol and florfenicol resistance (109). The first cfr gene was discovered in a bovine Staphylococcus sciuri isolate in 2000 (110). It encodes an rRNA methyltransferase that methylates the adenine at position 2503 in the 23S rRNA, resulting in resistance to five chemically unrelated antimicrobial classes: phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A (known as the PhLOPSA phenotype) (111). Since its discovery, the cfr gene has been detected in a variety of Gram-positive and Gram-negative bacteria (110, 112–116). A recent study identified a novel plasmid-borne cfr-like gene, designated cfr(C), in multidrug-resistant C. coli isolates of cattle origin. Similar to cfr and cfr(B), the cfr(C) gene was found to confer transferable resistance to phenicols and oxazolidinones (linezolid) as well as lincosamides and pleuromutilins (Campylobacter is naturally resistant to streptogramin) in both C. jejuni and C. coli (117). Additionally, the recently identified multidrug efflux pump variant RE-CmeABC alone can confer elevated resistance to phenicols (see details in “CmeABC” below) (118).
RESISTANCE TO FOSFOMYCIN
Fosfomycin is a broad-spectrum antibiotic with bactericidal activity against both Gram-positive and Gram-negative bacteria (119). Fosfomycin inhibits bacterial cell wall synthesis by inactivating the essential enzyme for the catalysis of bacterial peptidoglycan biosynthesis (120). Campylobacter resistance to fosfomycin is rare, and the resistance rate has remained low (121, 122). To date, the only mechanism of fosfomycin resistance identified in Campylobacter is the fosXCC gene, which encodes a protein that shares 63.9% identity to fosfomycin resistance determinant FosX, found in Listeria monocytogenes. FosX inactivates fosfomycin by catalyzing the addition of groups to its epoxide (120, 123). The fosXCC gene is contained in the MDRGI in C. coli, and is transferrable to C. jejuni by natural transformation (43, 123).
RESISTANCE TO ARSENICS
Arsenic compounds have been commonly used in the poultry industry for promoting growth and controlling diseases. Due to their potential risk to human health and the environment, they were recently withdrawn from poultry use in the United States. However, the organic form of arsenic, roxarsone, is still used as a feed additive in other countries. To survive in the poultry production environment, Campylobacter has developed ways to resist the action of arsenic compounds. Campylobacter isolates from conventional poultry products showed significantly higher levels of arsenic resistance than those from antimicrobial-free poultry products (124). Recently, several arsenic detoxification mechanisms have been identified in C. jejuni, including arsenate reductase ArsC, arsenite efflux transporters Acr3 and ArsB, and methylarsenite efflux permease ArsP (125–128). The two arsenite transporters (Acr3 and ArsB) belong to different families (129) and extrude toxic AS(III) out of bacterial cells. The ArsB family has been found only in bacteria and archaea, while the Acr3 family exists in prokaryotes and fungi, as well as in plant genomes (129–132). As an arsenate reductase, ArsC converts As(V) to AS(III) in the cytoplasm (130, 133), which is subsequently extruded by Acr3 or ArsB transporters (125, 130). Acr3 in C. jejuni consists of 347 amino acids and contains 10 predicted transmembrane helices. The presence of the acr3-containing operon is significantly associated with elevated resistance to arsenite and arsenate in Campylobacter. Furthermore, inactivation of acr3 leads to reductions in the MICs of both arsenite and arsenate. Acr3 is not involved in the resistance to other classes of antibiotics in Campylobacter (125). ArsB in C. jejuni consists of 428 amino acids and contains 11 probable transmembrane helices. The amino acid sequence of ArsB is homologous to ArsB in Shewanella sp. ANA-3 (134), S. aureus (135), E. coli (136–138), and Acidithiobacillus caldus (139). Inactivation of arsB resulted in increased susceptibility of Campylobacter to both arsenite and arsenate, but not to other heavy metals and antibiotics (126). Interestingly, analysis of various Campylobacter isolates of different animal origins for the distribution of arsB and acr3 genes indicated that all of the tested strains contained at least one of the two genes (126). ArsP in C. jejuni consists of 315 amino acids and contains 8 probable transmembrane helices. arsP is the first gene in the four-gene ars operon, which contains arsP, arsR, arsC, and acr3. The presence of ArsP is associated with elevated MIC of roxarsone. Inactivation of arsP results in reduced resistance to several organic arsenics including arsanilic acid, nitarsone, and roxarsone (127). It was also revealed that ArsP is an efflux permease for trivalent organoarsenicals including methylarsenite and trivalent forms of aromatic arsenicals (128). ArsP does not play a role in the resistance to inorganic arsenic.
MULTIDRUG EFFLUX PUMPS
The antibiotic efflux transporters play an essential role in the intrinsic and acquired resistance to structurally diverse antimicrobials. In Campylobacter, several multidrug efflux pumps (CmeABC, CmeDEF, and CmeG) have been functionally characterized for their contributions to antimicrobial resistance.
CmeABC
CmeABC is the predominant antibiotic efflux system in C. jejuni and belongs to the resistance-nodulation-cell division superfamily of multidrug efflux transporters. This efflux system is encoded by a three-gene operon comprising cmeA, cmeB, and cmeC (70) and consists of a membrane fusion protein (CmeA), an inner membrane transporter (CmeB), and an outer membrane protein (CmeC) (70). CmeABC extrudes toxic compounds and contributes to Campylobacter resistance to structurally diverse antimicrobials (70, 71). It should be noted that every component of the CmeABC system is required for its full function as an efflux pump. As mentioned above, CmeABC functions synergistically with other mechanisms in conferring high-level resistance to antibiotics (30, 48, 50, 53, 55, 70, 140, 141). These examples illustrate the important role of CmeABC in conferring resistance to clinically important antibiotics such as macrolide and fluoroquinolone. Interestingly, CmeABC also contributes to resistance to bacteriocins, antimicrobial peptides produced by bacteria (142, 143). As the predominant efflux system in Campylobacter, cmeABC is conserved among different Campylobacter spp. and is widely distributed in Campylobacter isolates (144). This efflux system has been functionally characterized in C. jejuni (70, 71, 145), C. coli (33, 141), Campylobacter lari, C. fetus, and Campylobacter hyointestinals (144) and has been shown to contribute to antibiotic resistance in all examined species. In general, the sequences of cmeABC are highly conserved within a species, but significant sequence polymorphisms are observed in the cmeABC genes among different Campylobacter spp. (144, 146–148). The expression of cmeABC is modulated by a transcriptional regulator called CmeR (149) that functions as a repressor for cmeABC. The cmeABC operon is inducible by bile salts and salicylate (150, 151), and the induction by bile is due to conformational changes in the DNA binding motif of CmeR, releasing its repression on the cmeABC promoter (152–154).
Notably, a potent variant of CmeABC, named RE-CmeABC, has recently emerged in C. jejuni (118). This variant CmeABC is much more powerful in conferring multidrug resistance and is especially potent to florfenicol and chloramphenicol. The RE-CmeABC operon has a unique CmeB sequence that shows only ∼80% amino acid sequence identity to CmeB in other C. jejuni strains. The sequence variation in CmeB contributed mostly to the enhanced function of RE-CmeABC. In addition to the enhanced resistance to various antibiotics, RE-CmeABC also promotes the emergence of fluoroquinolone-resistant mutants under selection pressure. In the presence of GyrA mutations, RE-CmeABC confers exceedingly high-level resistance (ciprofloxacin MIC ≥ 128 µg/ml) to fluoroquinolone (118). Additionally, Re-CmeABC is increasingly prevalent in C. jejuni isolates in China, suggesting that it facilitates the adaptation of Campylobacter to antibiotic selection pressure.
CmeDEF
CmeDEF is another resistance-nodulation-cell division-type efflux pump identified in C. jejuni. CmeD (Cj1031) is an outer membrane protein of 424 amino acids which shares low but significant sequence homology to HefA of H. pylori and TolC of E. coli, the outer membrane components of antibiotic efflux systems (1). CmeE (Cj1032) is a membrane fusion protein composed of 246 amino acids, which shares significant homology with the membrane fusion protein of HefB in H. pylori. CmeF is an inner membrane transporter and is predicted to contain a 12-transmembrane helical domain structure. The sequence of CmeF (1,005 amino acids) shares certain homology with many other resistance-nodulation-cell division-type efflux transporters such as HefC of H. pylori and AcrB, AcrD, and AcrF of E. coli (1, 155). The low sequence identity between CmeDEF and CmeABC suggests that these two efflux systems may have different functions and abilities to extrude antibiotics and other toxic compounds. Several studies have determined the contribution of cmeDEF to antimicrobial resistance. Pumbwe et al. (106) reported that the insertional mutation of cmeF in Campylobacter resulted in increased susceptibility to structurally unrelated antimicrobial compounds, including ampicillin, ethidium bromide, acridine orange, SDS, sodium deoxycholate, bile, detrimide, and triclosan. Akiba et al. (107) also reported that the cmeF mutant of C. jejuni NCTC 11168 showed a 2-fold decrease in resistance to ampicillin and ethidium bromide, but the authors did not observe any changes in the susceptibility to other tested antimicrobials, including bile salts. Another study, by Ge et al. (33), found that inactivation of cmeF in C. jejuni 81-176 had no effects on susceptibility to ciprofloxaxin, erythromycin, tetracycline, and chloramphenicol. In general, CmeDEF appears to play a modest role in antibiotic resistance in a strain-dependent manner, and its natural function in Campylobacter physiology remains unknown.
CmeG
CmeG (Cj1375) is one of the predicted MFS (major facilitator superfamily) transporters and is present in all the C. jejuni strains sequenced to date. Analysis of its amino acid sequence revealed that CmeG is a homolog of Bmr of B. subtilis and NorA of S. aureus (72), both of which contribute to multidrug resistance in bacteria (72). In addition, CmeG is predicated to be an inner membrane protein and possesses 12 transmembrane domains. Inactivation of cmeG significantly reduced resistance to various classes of antimicrobials, including ciprofloxacin, erythromycin, tetracycline, gentamicin, ethidium bromide, and cholic acid, while overexpression of cmeG enhanced the resistance to various fluoroquinolone antimicrobials, including ciprofloxacin, enrofloxacin, norfloxacin, and moxifloxacin but not to the other antibiotics tested in the study (72). Accumulation assays demonstrated that the cmeG mutant accumulated more ethidium bromide and ciprofloxacin than the wild-type strain (72). These results indicate that CmeG is a functional efflux transporter in Campylobacter. The expression of cmeG appears to be regulated by the Fur protein and iron concentrations, because inactivation of Fur or iron depletion resulted in the upregulation of cmeG (156–158). The detailed mechanism underlying cmeG regulation remains to be determined.
SUMMARY AND PERSPECTIVES
Campylobacter is a major foodborne pathogen, and its resistance to clinically important antibiotics is increasingly prevalent. Particularly, rising fluoroquinolone resistance in Campylobacter has been reported in many countries (6), limiting its usage for the treatment of campylobacteriosis. Campylobacter is highly mutable to fluoroquinolone treatment, and acquisition of resistance does not incur a fitness cost, contributing to the rapid development and persistence of fluoroquinolone-resistant Campylobacter (159). In contrast, development of macrolide resistance in Campylobacter occurs slowly and incurs a significant fitness cost in the absence of selection pressure, contributing to the overall low prevalence of macrolide-resistant Campylobacter. However, a horizontally transferable erm(B) has recently emerged in Campylobacter (43–45), which may significantly influence the epidemiology of macrolide-resistant Campylobacter. This possibility warrants enhanced efforts to monitor its further spread in Campylobacter isolates. Importantly, several new multidrug resistance mechanisms, including MDRGIs, Cfr(C), and RE-CmeABC, have been detected in Campylobacter, which greatly increases its ability to cope with selection pressure from multiple antibiotics. These examples illustrate the extraordinary ability of Campylobacter to acquire new mechanisms for adaptation to antimicrobial usage. With that said, it is likely that new antibiotic resistance mechanisms will continue to emerge in Campylobacter. Thus, innovative strategies are needed to curb the rise and spread of antibiotic-resistant Campylobacter.
ACKNOWLEDGMENTS
We apologize to all the investigators whose research could not be appropriately cited owing to space limitations.
The work of Zhangqi Shen, Yang Wang, and Jianzhong Shen on antimicrobial resistance genes in Campylobacter during 2013–2017 was financially supported by the National Basic Research Program of China (2013CB127200). The work of Qijing Zhang is supported by grant R01AI118283 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.WHO. 2013. The global view of campylobacteriosis. Report of an expert consultation. World Health Organization, Utrecht, The Netherlands, 9–11 July 2012. [PubMed] [Google Scholar]
- 2.Allos BM. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis 32:1201–1206 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 3.Engberg J, Aarestrup FM, Taylor DE, Gerner-Smidt P, Nachamkin I. 2001. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg Infect Dis 7:24–34 10.3201/eid0701.010104. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Payot S, Bolla JM, Corcoran D, Fanning S, Mégraud F, Zhang Q. 2006. Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect 8:1967–1971 10.1016/j.micinf.2005.12.032. [PubMed] [DOI] [PubMed] [Google Scholar]
- 5.Luangtongkum T, Jeon B, Han J, Plummer P, Logue CM, Zhang Q. 2009. Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol 4:189–200 10.2217/17460913.4.2.189. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaser M, Engberg J. 2008. Clinical aspects of Campylobacter jejuni and Campylobacter coli infections, p 99–121. In Nachamkin I, Szymanski CM, Blaser MJ (eds), Campylobacter, 3rd ed. ASM Press, Washington, DC. 10.1128/9781555815554.ch6 [DOI] [Google Scholar]
- 7.Altekruse SF, Tollefson LK. 2003. Human campylobacteriosis: a challenge for the veterinary profession. J Am Vet Med Assoc 223:445–452 10.2460/javma.2003.223.445. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Wieczorek K, Denis E, Lynch O, Osek J. 2013. Molecular characterization and antibiotic resistance profiling of Campylobacter isolated from cattle in Polish slaughterhouses. Food Microbiol 34:130–136 10.1016/j.fm.2012.12.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 9.Sanad YM, Closs G Jr, Kumar A, LeJeune JT, Rajashekara G. 2013. Molecular epidemiology and public health relevance of Campylobacter isolated from dairy cattle and European starlings in Ohio, USA. Foodborne Pathog Dis 10:229–236 10.1089/fpd.2012.1293. [PubMed] [DOI] [PubMed] [Google Scholar]
- 10.Châtre P, Haenni M, Meunier D, Botrel MA, Calavas D, Madec JY. 2010. Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from cattle between 2002 and 2006 inFrance. J Food Prot 73:825–831 10.4315/0362-028X-73.5.825. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Hakkinen M, Heiska H, Hänninen ML. 2007. Prevalence of Campylobacter spp. in cattle in Finland and antimicrobial susceptibilities of bovine Campylobacter jejuni strains. Appl Environ Microbiol 73:3232–3238 10.1128/AEM.02579-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Englen MD, Hill AE, Dargatz DA, Ladely SR, Fedorka-Cray PJ. 2007. Prevalence and antimicrobial resistance of Campylobacter in US dairy cattle. J Appl Microbiol 102:1570–1577 10.1111/j.1365-2672.2006.03189.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 13.Jesse TW, Englen MD, Pittenger-Alley LG, Fedorka-Cray PJ. 2006. Two distinct mutations in gyrA lead to ciprofloxacin and nalidixic acid resistance in Campylobacter coli and Campylobacter jejuni isolated from chickens and beef cattle. J Appl Microbiol 100:682–688 10.1111/j.1365-2672.2005.02796.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 14.Inglis GD, Morck DW, McAllister TA, Entz T, Olson ME, Yanke LJ, Read RR. 2006. Temporal prevalence of antimicrobial resistance in Campylobacter spp. from beef cattle in Alberta feedlots. Appl Environ Microbiol 72:4088–4095 10.1128/AEM.02830-05. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bae W, Kaya KN, Hancock DD, Call DR, Park YH, Besser TE. 2005. Prevalence and antimicrobial resistance of thermophilic Campylobacter spp. from cattle farms in Washington State. Appl Environ Microbiol 71:169–174 10.1128/AEM.71.1.169-174.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iovine NM. 2013. Resistance mechanisms in Campylobacter jejuni. Virulence 4:230–240 10.4161/viru.23753. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wieczorek K, Osek J. 2013. Antimicrobial resistance mechanisms among Campylobacter. BioMed Res Int 2013:1 10.1155/2013/340605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q, Plummer P. 2008. Mechanisms of antibiotic resistance in Campylobacter, p 263–276. In Nachamkin I, Szymanski CM, Blaser MJ (eds), Campylobacter, 3rd ed. ASM Press, Washington, DC. [Google Scholar]
- 19.Shen Z, Su C, Yu E, Zhang Q. 2013. Multidrug efflux transporters in Campylobacter, p 223–235. In Yu EW, Zhang Q, Brown MH (eds), Microbial Efflux Pumps: Current Research. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 20.Appelbaum PC, Hunter PA. 2000. The fluoroquinolone antibacterials: past, present and future perspectives. Int J Antimicrob Agents 16:5–15 10.1016/S0924-8579(00)00192-8. [PubMed] [DOI] [PubMed] [Google Scholar]
- 21.Hooper DC. 1998. Clinical applications of quinolones. Biochim Biophys Acta 1400:45–61 10.1016/S0167-4781(98)00127-4. [PubMed] [DOI] [PubMed] [Google Scholar]
- 22.Martinez M, McDermott P, Walker R. 2006. Pharmacology of the fluoroquinolones: a perspective for the use in domestic animals. Vet J 172:10–28 10.1016/j.tvjl.2005.07.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.Oliphant CM, Green GM. 2002. Quinolones: a comprehensive review. Am Fam Physician 65:455–464. [PubMed] [PubMed] [Google Scholar]
- 24.Redgrave LS, Sutton SB, Webber MA, Piddock LJ. 2014. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol 22:438–445 10.1016/j.tim.2014.04.007. [PubMed] [DOI] [PubMed] [Google Scholar]
- 25.Hooper DC. 1999. Mechanisms of fluoroquinolone resistance. Drug Resist Updat 2:38–55 10.1054/drup.1998.0068. [PubMed] [DOI] [PubMed] [Google Scholar]
- 26.Hooper DC. 2001. Emerging mechanisms of fluoroquinolone resistance. Emerg Infect Dis 7:337–341 10.3201/eid0702.010239. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Payot S, Cloeckaert A, Chaslus-Dancla E. 2002. Selection and characterization of fluoroquinolone-resistant mutants of Campylobacter jejuni using enrofloxacin. Microb Drug Resist 8:335–343 10.1089/10766290260469606. [PubMed] [DOI] [PubMed] [Google Scholar]
- 28.Piddock LJ, Ricci V, Pumbwe L, Everett MJ, Griggs DJ. 2003. Fluoroquinolone resistance in Campylobacter species from man and animals: detection of mutations in topoisomerase genes. J Antimicrob Chemother 51:19–26 10.1093/jac/dkg033. [PubMed] [DOI] [PubMed] [Google Scholar]
- 29.Bachoual R, Ouabdesselam S, Mory F, Lascols C, Soussy CJ, Tankovic J. 2001. Single or double mutational alterations of gyrA associated with fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Microb Drug Resist 7:257–261 10.1089/10766290152652800. [PubMed] [DOI] [PubMed] [Google Scholar]
- 30.Luo N, Sahin O, Lin J, Michel LO, Zhang Q. 2003. In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob Agents Chemother 47:390–394 10.1128/AAC.47.1.390-394.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper R, Segal H, Lastovica AJ, Elisha BG. 2002. Genetic basis of quinolone resistance and epidemiology of resistant and susceptible isolates of porcine Campylobacter coli strains. J Appl Microbiol 93:241–249 10.1046/j.1365-2672.2002.01650.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 32.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668 10.1038/35001088. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.Ge B, McDermott PF, White DG, Meng J. 2005. Role of efflux pumps and topoisomerase mutations in fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother 49:3347–3354 10.1128/AAC.49.8.3347-3354.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Q, Lin J, Pereira S. 2003. Fluoroquinolone-resistant Campylobacter in animal reservoirs: dynamics of development, resistance mechanisms and ecological fitness. Anim Health Res Rev 4:63–71 10.1079/AHR200356. [PubMed] [DOI] [PubMed] [Google Scholar]
- 35.Hänninen ML, Hannula M. 2007. Spontaneous mutation frequency and emergence of ciprofloxacin resistance in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother 60:1251–1257 10.1093/jac/dkm345. [PubMed] [DOI] [PubMed] [Google Scholar]
- 36.Smith JL, Fratamico PM. 2010. Fluoroquinolone resistance in campylobacter. J Food Prot 73:1141–1152 10.4315/0362-028X-73.6.1141. [PubMed] [DOI] [PubMed] [Google Scholar]
- 37.Chu DT. 1999. Recent developments in macrolides and ketolides. Curr Opin Microbiol 2:467–474 10.1016/S1369-5274(99)00002-8. [DOI] [PubMed] [Google Scholar]
- 38.McEwen SA, Fedorka-Cray PJ. 2002. Antimicrobial use and resistance in animals. Clin Infect Dis 34(Suppl 3):S93–S106 10.1086/340246. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Brisson-Noel A, Trieu-Cuot P, Courvalin P. 1988. Mechanism of action of spiramycin and other macrolides. J Antimicrob Chemother 22(Suppl B):13–23. [PubMed] [DOI] [PubMed] [Google Scholar]
- 40.Poehlsgaard J, Douthwaite S. 2005. The bacterial ribosome as a target for antibiotics. Nat Rev Microbiol 3:870–881 10.1038/nrmicro1265. [PubMed] [DOI] [PubMed] [Google Scholar]
- 41.Gibreel A, Taylor DE. 2006. Macrolide resistance in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother 58:243–255 10.1093/jac/dkl210. [PubMed] [DOI] [PubMed] [Google Scholar]
- 42.Roe DE, Weinberg A, Roberts MC. 1995. Mobile rRNA methylase genes in Campylobacter (Wolinella) rectus. J Antimicrob Chemother 36:738–740 10.1093/jac/36.4.738. [PubMed] [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Zhang M, Deng F, Shen Z, Wu C, Zhang J, Zhang Q, Shen J. 2014. Emergence of multidrug-resistant Campylobacter species isolates with a horizontally acquired rRNA methylase. Antimicrob Agents Chemother 58:5405–5412 10.1128/AAC.03039-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin S, Wang Y, Zhang Q, Zhang M, Deng F, Shen Z, Wu C, Wang S, Zhang J, Shen J. 2014. Report of ribosomal RNA methylase gene erm(B) in multidrug-resistant Campylobacter coli. J Antimicrob Chemother 69:964–968 10.1093/jac/dkt492. [PubMed] [DOI] [PubMed] [Google Scholar]
- 45.Deng F, Wang Y, Zhang Y, Shen Z. 2015. Characterization of the genetic environment of the ribosomal RNA methylase gene erm(B) in Campylobacter jejuni. J Antimicrob Chemother 70:613–615 10.1093/jac/dku418. [PubMed] [DOI] [PubMed] [Google Scholar]
- 46.Corcoran D, Quinn T, Cotter L, Fanning S. 2006. An investigation of the molecular mechanisms contributing to high-level erythromycin resistance in Campylobacter. Int J Antimicrob Agents 27:40–45 10.1016/j.ijantimicag.2005.08.019. [PubMed] [DOI] [PubMed] [Google Scholar]
- 47.Gibreel A, Kos VN, Keelan M, Trieber CA, Levesque S, Michaud S, Taylor DE. 2005. Macrolide resistance in Campylobacter jejuni and Campylobacter coli: molecular mechanism and stability of the resistance phenotype. Antimicrob Agents Chemother 49:2753–2759 10.1128/AAC.49.7.2753-2759.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin J, Yan M, Sahin O, Pereira S, Chang YJ, Zhang Q. 2007. Effect of macrolide usage on emergence of erythromycin-resistant Campylobacter isolates in chickens. Antimicrob Agents Chemother 51:1678–1686 10.1128/AAC.01411-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mamelli L, Prouzet-Mauléon V, Pagès JM, Mégraud F, Bolla JM. 2005. Molecular basis of macrolide resistance in Campylobacter: role of efflux pumps and target mutations. J Antimicrob Chemother 56:491–497 10.1093/jac/dki253. [PubMed] [DOI] [PubMed] [Google Scholar]
- 50.Caldwell DB, Wang Y, Lin J. 2008. Development, stability, and molecular mechanisms of macrolide resistance in Campylobacter jejuni. Antimicrob Agents Chemother 52:3947–3954 10.1128/AAC.00450-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, Brinkac LM, DeBoy RT, Parker CT, Daugherty SC, Dodson RJ, Durkin AS, Madupu R, Sullivan SA, Shetty JU, Ayodeji MA, Shvartsbeyn A, Schatz MC, Badger JH, Fraser CM, Nelson KE. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol 3:e15 10.1371/journal.pbio.0030015. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vacher S, Menard A, Bernard E, Santos A, Megraud F. 2005. Detection of mutations associated with macrolide resistance in thermophilic Campylobacter spp. by real-time PCR. Microb Drug Resist 11:40–47 10.1089/mdr.2005.11.40. [PubMed] [DOI] [PubMed] [Google Scholar]
- 53.Cagliero C, Mouline C, Cloeckaert A, Payot S. 2006. Synergy between efflux pump CmeABC and modifications in ribosomal proteins L4 and L22 in conferring macrolide resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother 50:3893–3896 10.1128/AAC.00616-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kurincic M, Botteldoorn N, Herman L, Smole Mozina S. 2007. Mechanisms of erythromycin resistance of Campylobacter spp. isolated from food, animals and humans. Int J Food Microbiol 120:186–190 10.1016/j.ijfoodmicro.2007.03.012. [PubMed] [DOI] [PubMed] [Google Scholar]
- 55.Gibreel A, Wetsch NM, Taylor DE. 2007. Contribution of the CmeABC efflux pump to macrolide and tetracycline resistance in Campylobacter jejuni. Antimicrob Agents Chemother 51:3212–3216 10.1128/AAC.01592-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Payot S, Avrain L, Magras C, Praud K, Cloeckaert A, Chaslus-Dancla E. 2004. Relative contribution of target gene mutation and efflux to fluoroquinolone and erythromycin resistance, in French poultry and pig isolates of Campylobacter coli. Int J Antimicrob Agents 23:468–472 10.1016/j.ijantimicag.2003.12.008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 57.Chopra I, Roberts M. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65:232–260 10.1128/MMBR.65.2.232-260.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Epe B, Woolley P, Hornig H. 1987. Competition between tetracycline and tRNA at both P and A sites of the ribosome of Escherichia coli. FEBS Lett 213:443–447 10.1016/0014-5793(87)81539-9. [DOI] [PubMed] [Google Scholar]
- 59.Li W, Atkinson GC, Thakor NS, Allas U, Lu CC, Chan KY, Tenson T, Schulten K, Wilson KS, Hauryliuk V, Frank J. 2013. Mechanism of tetracycline resistance by ribosomal protection protein Tet(O). Nat Commun 4:1477 10.1038/ncomms2470. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Connell SR, Tracz DM, Nierhaus KH, Taylor DE. 2003. Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob Agents Chemother 47:3675–3681 10.1128/AAC.47.12.3675-3681.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roberts MC. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett 245:195–203 10.1016/j.femsle.2005.02.034. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.Dönhöfer A, Franckenberg S, Wickles S, Berninghausen O, Beckmann R, Wilson DN. 2012. Structural basis for TetM-mediated tetracycline resistance. Proc Natl Acad Sci USA 109:16900–16905 10.1073/pnas.1208037109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Margus T, Remm M, Tenson T. 2007. Phylogenetic distribution of translational GTPases in bacteria. BMC Genomics 8:15 10.1186/1471-2164-8-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Connell SR, Trieber CA, Dinos GP, Einfeldt E, Taylor DE, Nierhaus KH. 2003. Mechanism of Tet(O)-mediated tetracycline resistance. EMBO J 22:945–953 10.1093/emboj/cdg093. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taylor DE, Courvalin P. 1988. Mechanisms of antibiotic resistance in Campylobacter species. Antimicrob Agents Chemother 32:1107–1112 10.1128/AAC.32.8.1107. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor DE, Garner RS, Allan BJ. 1983. Characterization of tetracycline resistance plasmids from Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother 24:930–935 10.1128/AAC.24.6.930. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taylor DE. 1986. Plasmid-mediated tetracycline resistance in Campylobacter jejuni: expression in Escherichia coli and identification of homology with streptococcal class M determinant. J Bacteriol 165:1037–1039 10.1128/jb.165.3.1037-1039.1986. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Batchelor RA, Pearson BM, Friis LM, Guerry P, Wells JM. 2004. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 150:3507–3517 10.1099/mic.0.27112-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 69.Sahin O, Plummer PJ, Jordan DM, Sulaj K, Pereira S, Robbe-Austerman S, Wang L, Yaeger MJ, Hoffman LJ, Zhang Q. 2008. Emergence of a tetracycline-resistant Campylobacter jejuni clone associated with outbreaks of ovine abortion in the United States. J Clin Microbiol 46:1663–1671 10.1128/JCM.00031-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin J, Michel LO, Zhang Q. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother 46:2124–2131 10.1128/AAC.46.7.2124-2131.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pumbwe L, Piddock LJ. 2002. Identification and molecular characterisation of CmeB, a Campylobacter jejuni multidrug efflux pump. FEMS Microbiol Lett 206:185–189 10.1111/j.1574-6968.2002.tb11007.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 72.Jeon B, Wang Y, Hao H, Barton YW, Zhang Q. 2011. Contribution of CmeG to antibiotic and oxidative stress resistance in Campylobacter jejuni. J Antimicrob Chemother 66:79–85 10.1093/jac/dkq418. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spahn CM, Schäfer MA, Krayevsky AA, Nierhaus KH. 1996. Conserved nucleotides of 23 S rRNA located at the ribosomal peptidyltransferase center. J Biol Chem 271:32857–32862 10.1074/jbc.271.51.32857. [PubMed] [DOI] [PubMed] [Google Scholar]
- 74.Jana S, Deb JK. 2006. Molecular understanding of aminoglycoside action and resistance. Appl Microbiol Biotechnol 70:140–150 10.1007/s00253-005-0279-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 75.Magnet S, Blanchard JS. 2005. Molecular insights into aminoglycoside action and resistance. Chem Rev 105:477–498 10.1021/cr0301088. [PubMed] [DOI] [PubMed] [Google Scholar]
- 76.Smith CA, Baker EN. 2002. Aminoglycoside antibiotic resistance by enzymatic deactivation. Curr Drug Targets Infect Disord 2:143–160 10.2174/1568005023342533. [PubMed] [DOI] [PubMed] [Google Scholar]
- 77.Lawrence PK, Kittichotirat W, McDermott JE, Bumgarner RE. 2010. A three-way comparative genomic analysis of Mannheimia haemolytica isolates. BMC Genomics 11:535 10.1186/1471-2164-11-535. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vakulenko SB, Mobashery S. 2003. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev 16:430–450 10.1128/CMR.16.3.430-450.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tenover FC, Elvrum PM. 1988. Detection of two different kanamycin resistance genes in naturally occurring isolates of Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother 32:1170–1173 10.1128/AAC.32.8.1170. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gibreel A, Sköld O, Taylor DE. 2004. Characterization of plasmid-mediated aphA-3 kanamycin resistance in Campylobacter jejuni. Microb Drug Resist 10:98–105 10.1089/1076629041310127. [PubMed] [DOI] [PubMed] [Google Scholar]
- 81.Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist Updat 13:151–171 10.1016/j.drup.2010.08.003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Doi Y, Arakawa Y. 2007. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin Infect Dis 45:88–94 10.1086/518605. [PubMed] [DOI] [PubMed] [Google Scholar]
- 83.Overhage J, Bains M, Brazas MD, Hancock RE. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J Bacteriol 190:2671–2679 10.1128/JB.01659-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lambert T, Gerbaud G, Trieu-Cuot P, Courvalin P. 1985. Structural relationship between the genes encoding 3′-aminoglycoside phosphotransferases in Campylobacter and in Gram-positive cocci. Ann Inst Pasteur Microbiol 1985 136B:135–150 10.1016/S0769-2609(85)80040-5. [DOI] [PubMed] [Google Scholar]
- 85.Ouellette M, Gerbaud G, Lambert T, Courvalin P. 1987. Acquisition by a Campylobacter-like strain of aphA-1, a kanamycin resistance determinant from members of the family Enterobacteriaceae. Antimicrob Agents Chemother 31:1021–1026 10.1128/AAC.31.7.1021. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gibreel A, Tracz DM, Nonaka L, Ngo TM, Connell SR, Taylor DE. 2004. Incidence of antibiotic resistance in Campylobacter jejuni isolated in Alberta, Canada, from 1999 to 2002, with special reference to tet(O)-mediated tetracycline resistance. Antimicrob Agents Chemother 48:3442–3450 10.1128/AAC.48.9.3442-3450.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qin S, Wang Y, Zhang Q, Chen X, Shen Z, Deng F, Wu C, Shen J. 2012. Identification of a novel genomic island conferring resistance to multiple aminoglycoside antibiotics in Campylobacter coli. Antimicrob Agents Chemother 56:5332–5339 10.1128/AAC.00809-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tenover FC, Gilbert T, O’Hara P. 1989. Nucleotide sequence of a novel kanamycin resistance gene, aphA-7, from Campylobacter jejuni and comparison to other kanamycin phosphotransferase genes. Plasmid 22:52–58 10.1016/0147-619X(89)90035-8. [DOI] [PubMed] [Google Scholar]
- 89.Tenover FC, Fennell CL, Lee L, LeBlanc DJ. 1992. Characterization of two plasmids from Campylobacter jejuni isolates that carry the aphA-7 kanamycin resistance determinant. Antimicrob Agents Chemother 36:712–716 10.1128/AAC.36.4.712. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rather PN, Munayyer H, Mann PA, Hare RS, Miller GH, Shaw KJ. 1992. Genetic analysis of bacterial acetyltransferases: identification of amino acids determining the specificities of the aminoglycoside 6′-N-acetyltransferase Ib and IIa proteins. J Bacteriol 174:3196–3203 10.1128/jb.174.10.3196-3203.1992. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee MD, Sanchez S, Zimmer M, Idris U, Berrang ME, McDermott PF. 2002. Class 1 integron-associated tobramycin-gentamicin resistance in Campylobacter jejuni isolated from the broiler chicken house environment. Antimicrob Agents Chemother 46:3660–3664 10.1128/AAC.46.11.3660-3664.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Toth M, Frase H, Antunes NT, Vakulenko SB. 2013. Novel aminoglycoside 2″phosphotransferase identified in a Gram-negative pathogen. Antimicrob Agents Chemother 57:452–457 10.1128/AAC.02049-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen Y, Mukherjee S, Hoffmann M, Kotewicz ML, Young S, Abbott J, Luo Y, Davidson MK, Allard M, McDermott P, Zhao S. 2013. Whole-genome sequencing of gentamicin-resistant Campylobacter coli isolated from U.S. retail meats reveals novel plasmid-mediated aminoglycoside resistance genes. Antimicrob Agents Chemother 57:5398–5405 10.1128/AAC.00669-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao S, Mukherjee S, Chen Y, Li C, Young S, Warren M, Abbott J, Friedman S, Kabera C, Karlsson M, McDermott PF. 2015. Novel gentamicin resistance genes in Campylobacter isolated from humans and retail meats in the USA. J Antimicrob Chemother 70:1314–1321 10.1093/jac/dkv001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nirdnoy W, Mason CJ, Guerry P. 2005. Mosaic structure of a multiple-drug-resistant, conjugative plasmid from Campylobacter jejuni. Antimicrob Agents Chemother 49:2454–2459 10.1128/AAC.49.6.2454-2459.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Abril C, Brodard I, Perreten V. 2010. Two novel antibiotic resistance genes, tet(44) and ant(6)-Ib, are located within a transferable pathogenicity island in Campylobacter fetus subsp. fetus. Antimicrob Agents Chemother 54:3052–3055 10.1128/AAC.00304-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Olkkola S, Culebro A, Juntunen P, Hänninen ML, Rossi M. 2016. Functional genomics in Campylobacter coli identified a novel streptomycin resistance gene located in a hypervariable genomic region. Microbiology 162:1157–1166 10.1099/mic.0.000304 [PubMed] [DOI] [PubMed] [Google Scholar]
- 98.Hamad B. 2010. The antibiotics market. Nat Rev Drug Discov 9:675–676 10.1038/nrd3267. [PubMed] [DOI] [PubMed] [Google Scholar]
- 99.Wise R. 2002. Antimicrobial resistance: priorities for action. J Antimicrob Chemother 49:585–586 10.1093/jac/49.4.585. [PubMed] [DOI] [PubMed] [Google Scholar]
- 100.Jovetic S, Zhu Y, Marcone GL, Marinelli F, Tramper J. 2010. β-Lactam and glycopeptide antibiotics: first and last line of defense? Trends Biotechnol 28:596–604 10.1016/j.tibtech.2010.09.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 101.Lachance N, Gaudreau C, Lamothe F, Larivière LA. 1991. Role of the beta-lactamase of Campylobacter jejuni in resistance to beta-lactam agents. Antimicrob Agents Chemother 35:813–818 10.1128/AAC.35.5.813. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Griggs DJ, Peake L, Johnson MM, Ghori S, Mott A, Piddock LJ. 2009. Beta-lactamase-mediated beta-lactam resistance in Campylobacter species: prevalence of Cj0299 (bla OXA-61) and evidence for a novel beta-lactamase in C. jejuni. Antimicrob Agents Chemother 53:3357–3364 10.1128/AAC.01655-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alfredson DA, Korolik V. 2005. Isolation and expression of a novel molecular class D beta-lactamase, OXA-61, from Campylobacter jejuni. Antimicrob Agents Chemother 49:2515–2518 10.1128/AAC.49.6.2515-2518.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zeng X, Brown S, Gillespie B, Lin J. 2014. A single nucleotide in the promoter region modulates the expression of the β-lactamase OXA-61 in Campylobacter jejuni. J Antimicrob Chemother 69:1215–1223 10.1093/jac/dkt515. [PubMed] [DOI] [PubMed] [Google Scholar]
- 105.Page WJ, Huyer G, Huyer M, Worobec EA. 1989. Characterization of the porins of Campylobacter jejuni and Campylobacter coli and implications for antibiotic susceptibility. Antimicrob Agents Chemother 33:297–303 10.1128/AAC.33.3.297. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pumbwe L, Randall LP, Woodward MJ, Piddock LJ. 2004. Expression of the efflux pump genes cmeB, cmeF and the porin gene porA in multiple-antibiotic-resistant Campylobacter jejuni. J Antimicrob Chemother 54:341–347 10.1093/jac/dkh331. [PubMed] [DOI] [PubMed] [Google Scholar]
- 107.Akiba M, Lin J, Barton YW, Zhang Q. 2006. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J Antimicrob Chemother 57:52–60 10.1093/jac/dki419. [PubMed] [DOI] [PubMed] [Google Scholar]
- 108.Wang Y, Taylor DE. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23–28 10.1016/0378-1119(90)90463-2. [DOI] [PubMed] [Google Scholar]
- 109.Ma L, Shen Z, Naren G, Li H, Xia X, Wu C, Shen J, Zhang Q, Wang Y. 2014. Identification of a novel G2073A mutation in 23S rRNA in amphenicol-selected mutants of Campylobacter jejuni. PLoS One 9:e94503 10.1371/journal.pone.0094503. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schwarz S, Werckenthin C, Kehrenberg C. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob Agents Chemother 44:2530–2533 10.1128/AAC.44.9.2530-2533.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob Agents Chemother 50:2500–2505 10.1128/AAC.00131-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu Y, Wang Y, Schwarz S, Li Y, Shen Z, Zhang Q, Wu C, Shen J. 2013. Transferable multiresistance plasmids carrying cfr in Enterococcus spp. from swine and farm environment. Antimicrob Agents Chemother 57:42–48 10.1128/AAC.01605-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dai L, Wu CM, Wang MG, Wang Y, Wang Y, Huang SY, Xia LN, Li BB, Shen JZ. 2010. First report of the multidrug resistance gene cfr and the phenicol resistance gene fexA in a Bacillus strain from swine feces. Antimicrob Agents Chemother 54:3953–3955 10.1128/AAC.00169-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang Y, Wang Y, Schwarz S, Shen Z, Zhou N, Lin J, Wu C, Shen J. 2012. Detection of the staphylococcal multiresistance gene cfr in Macrococcus caseolyticus and Jeotgalicoccus pinnipedialis. J Antimicrob Chemother 67:1824–1827 10.1093/jac/dks163. [PubMed] [DOI] [PubMed] [Google Scholar]
- 115.Wang Y, He T, Schwarz S, Zhou D, Shen Z, Wu C, Wang Y, Ma L, Zhang Q, Shen J. 2012. Detection of the staphylococcal multiresistance gene cfr in Escherichia coli of domestic-animal origin. J Antimicrob Chemother 67:1094–1098 10.1093/jac/dks020. [PubMed] [DOI] [PubMed] [Google Scholar]
- 116.Shen J, Wang Y, Schwarz S. 2013. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J Antimicrob Chemother 68:1697–1706 10.1093/jac/dkt092. [PubMed] [DOI] [PubMed] [Google Scholar]
- 117.Tang Y, Dai L, Sahin O, Wu Z, Liu M, Zhang Q. 2017. Emergence of a plasmid-borne multidrug resistance gene cfr(C) in foodborne pathogen Campylobacter. J Antimicrob Chemother 72:1581–1588 10.1093/jac/dkx023. [PubMed] [DOI] [PubMed] [Google Scholar]
- 118.Yao H, Shen Z, Wang Y, Deng F, Liu D, Naren G, Dai L, Su CC, Wang B, Wang S, Wu C, Yu EW, Zhang Q, Shen J. 2016. Emergence of a potent multidrug efflux pump variant that enhances Campylobacter resistance to multiple antibiotics. MBio 7:e01543–e16 10.1128/mBio.01543-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Forsgren A, Walder M. 1983. Antimicrobial activity of fosfomycin in vitro. J Antimicrob Chemother 11:467–471 10.1093/jac/11.5.467. [PubMed] [DOI] [PubMed] [Google Scholar]
- 120.Fillgrove KL, Pakhomova S, Newcomer ME, Armstrong RN. 2003. Mechanistic diversity of fosfomycin resistance in pathogenic microorganisms. J Am Chem Soc 125:15730–15731 10.1021/ja039307z. [PubMed] [DOI] [PubMed] [Google Scholar]
- 121.Schwaiger K, Schmied EM, Bauer J. 2008. Comparative analysis of antibiotic resistance characteristics of Gram-negative bacteria isolated from laying hens and eggs in conventional and organic keeping systems in Bavaria, Germany. Zoonoses Public Health 55:331–341 10.1111/j.1863-2378.2008.01151.x. [DOI] [PubMed] [Google Scholar]
- 122.Gomez-Garces JL, Cogollos R, Alos JL. 1995. Susceptibilities of fluoroquinolone-resistant strains of Campylobacter jejuni to 11 oral antimicrobial agents. Antimicrob Agents Chemother 39:542–544 10.1128/AAC.39.2.542. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang Y, Yao H, Deng F, Liu D, Zhang Y, Shen Z. 2015. Identification of a novel fosXCC gene conferring fosfomycin resistance in Campylobacter. J Antimicrob Chemother 70:1261–1263. [PubMed] [DOI] [PubMed] [Google Scholar]
- 124.Sapkota AR, Price LB, Silbergeld EK, Schwab KJ. 2006. Arsenic resistance in Campylobacter spp. isolated from retail poultry products. Appl Environ Microbiol 72:3069–3071 10.1128/AEM.72.4.3069-3071.2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang L, Jeon B, Sahin O, Zhang Q. 2009. Identification of an arsenic resistance and arsenic-sensing system in Campylobacter jejuni. Appl Environ Microbiol 75:5064–5073 10.1128/AEM.00149-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shen Z, Han J, Wang Y, Sahin O, Zhang Q. 2013. The contribution of ArsB to arsenic resistance in Campylobacter jejuni. PLoS One 8:e58894 10.1371/journal.pone.0058894. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shen Z, Luangtongkum T, Qiang Z, Jeon B, Wang L, Zhang Q. 2014. Identification of a novel membrane transporter mediating resistance to organic arsenic in Campylobacter jejuni. Antimicrob Agents Chemother 58:2021–2029 10.1128/AAC.02137-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen J, Madegowda M, Bhattacharjee H, Rosen BP. 2015. ArsP: a methylarsenite efflux permease. Mol Microbiol 98:625–635 10.1111/mmi.13145. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rosen BP. 1999. Families of arsenic transporters. Trends Microbiol 7:207–212 10.1016/S0966-842X(99)01494-8. [PubMed] [DOI] [PubMed] [Google Scholar]
- 130.Rosen BP. 2002. Biochemistry of arsenic detoxification. FEBS Lett 529:86–92 10.1016/S0014-5793(02)03186-1. [PubMed] [DOI] [PubMed] [Google Scholar]
- 131.Indriolo E, Na G, Ellis D, Salt DE, Banks JA. 2010. A vacuolar arsenite transporter necessary for arsenic tolerance in the arsenic hyperaccumulating fern Pteris vittata is missing in flowering plants. Plant Cell 22:2045–2057 10.1105/tpc.109.069773. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fu HL, Meng Y, Ordóñez E, Villadangos AF, Bhattacharjee H, Gil JA, Mateos LM, Rosen BP. 2009. Properties of arsenite efflux permeases (Acr3) from Alkaliphilus metalliredigens and Corynebacterium glutamicum. J Biol Chem 284:19887–19895 10.1074/jbc.M109.011882. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ji G, Silver S. 1992. Reduction of arsenate to arsenite by the ArsC protein of the arsenic resistance operon of Staphylococcus aureus plasmid pI258. Proc Natl Acad Sci USA 89:9474–9478 10.1073/pnas.89.20.9474. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Saltikov CW, Cifuentes A, Venkateswaran K, Newman DK. 2003. The ars detoxification system is advantageous but not required for As(V) respiration by the genetically tractable Shewanella species strain ANA-3. Appl Environ Microbiol 69:2800–2809 10.1128/AEM.69.5.2800-2809.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ji G, Silver S. 1992. Regulation and expression of the arsenic resistance operon from Staphylococcus aureus plasmid pI258. J Bacteriol 174:3684–3694 10.1128/jb.174.11.3684-3694.1992. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen CM, Misra TK, Silver S, Rosen BP. 1986. Nucleotide sequence of the structural genes for an anion pump. The plasmid-encoded arsenical resistance operon. J Biol Chem 261:15030–15038. [PubMed] [PubMed] [Google Scholar]
- 137.San Francisco MJ, Tisa LS, Rosen BP. 1989. Identification of the membrane component of the anion pump encoded by the arsenical resistance operon of R-factor R773. Mol Microbiol 3:15–21 10.1111/j.1365-2958.1989.tb00098.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 138.Tisa LS, Rosen BP. 1990. Molecular characterization of an anion pump. The ArsB protein is the membrane anchor for the ArsA protein. J Biol Chem 265:190–194. [PubMed] [PubMed] [Google Scholar]
- 139.Tuffin IM, de Groot P, Deane SM, Rawlings DE. 2005. An unusual Tn21-like transposon containing an ars operon is present in highly arsenic-resistant strains of the biomining bacterium Acidithiobacillus caldus. Microbiology 151:3027–3039 10.1099/mic.0.28131-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 140.Piddock LJ, Griggs D, Johnson MM, Ricci V, Elviss NC, Williams LK, Jørgensen F, Chisholm SA, Lawson AJ, Swift C, Humphrey TJ, Owen RJ. 2008. Persistence of Campylobacter species, strain types, antibiotic resistance and mechanisms of tetracycline resistance in poultry flocks treated with chlortetracycline. J Antimicrob Chemother 62:303–315 10.1093/jac/dkn190. [PubMed] [DOI] [PubMed] [Google Scholar]
- 141.Cagliero C, Mouline C, Payot S, Cloeckaert A. 2005. Involvement of the CmeABC efflux pump in the macrolide resistance of Campylobacter coli. J Antimicrob Chemother 56:948–950 10.1093/jac/dki292. [PubMed] [DOI] [PubMed] [Google Scholar]
- 142.Hoang KV, Stern NJ, Lin J. 2011. Development and stability of bacteriocin resistance in Campylobacter spp. J Appl Microbiol 111:1544–1550 10.1111/j.1365-2672.2011.05163.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hoang KV, Stern NJ, Saxton AM, Xu F, Zeng X, Lin J. 2011. Prevalence, development, and molecular mechanisms of bacteriocin resistance in Campylobacter. Appl Environ Microbiol 77:2309–2316 10.1128/AEM.02094-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Guo B, Lin J, Reynolds DL, Zhang Q. 2010. Contribution of the multidrug efflux transporter CmeABC to antibiotic resistance in different Campylobacter species. Foodborne Pathog Dis 7:77–83 10.1089/fpd.2009.0354. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lin J, Sahin O, Michel LO, Zhang Q. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect Immun 71:4250–4259 10.1128/IAI.71.8.4250-4259.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Olah PA, Doetkott C, Fakhr MK, Logue CM. 2006. Prevalence of the Campylobacter multi-drug efflux pump (CmeABC) in Campylobacter spp. Isolated from freshly processed turkeys. Food Microbiol 23:453–460 10.1016/j.fm.2005.06.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 147.Cagliero C, Cloix L, Cloeckaert A, Payot S. 2006. High genetic variation in the multidrug transporter cmeB gene in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother 58:168–172 10.1093/jac/dkl212. [PubMed] [DOI] [PubMed] [Google Scholar]
- 148.Fakhr MK, Logue CM. 2007. Sequence variation in the outer membrane protein-encoding gene cmeC, conferring multidrug resistance among Campylobacter jejuni and Campylobacter coli strains isolated from different hosts. J Clin Microbiol 45:3381–3383 10.1128/JCM.01208-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lin J, Akiba M, Sahin O, Zhang Q. 2005. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob Agents Chemother 49:1067–1075 10.1128/AAC.49.3.1067-1075.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lin J, Cagliero C, Guo B, Barton YW, Maurel MC, Payot S, Zhang Q. 2005. Bile salts modulate expression of the CmeABC multidrug efflux pump in Campylobacter jejuni. J Bacteriol 187:7417–7424 10.1128/JB.187.21.7417-7424.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shen Z, Pu XY, Zhang Q. 2011. Salicylate functions as an efflux pump inducer and promotes the emergence of fluoroquinolone-resistant Campylobacter jejuni mutants. Appl Environ Microbiol 77:7128–7133 10.1128/AEM.00763-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gu R, Su CC, Shi F, Li M, McDermott G, Zhang Q, Yu EW. 2007. Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni. J Mol Biol 372:583–593 10.1016/j.jmb.2007.06.072. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lei HT, Shen Z, Surana P, Routh MD, Su CC, Zhang Q, Yu EW. 2011. Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni. Protein Sci 20:712–723 10.1002/pro.602. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Routh MD, Su CC, Zhang Q, Yu EW. 2009. Structures of AcrR and CmeR: insight into the mechanisms of transcriptional repression and multi-drug recognition in the TetR family of regulators. Biochim Biophys Acta 1794:844–851 10.1016/j.bbapap.2008.12.001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pumbwe L, Randall LP, Woodward MJ, Piddock LJ. 2005. Evidence for multiple-antibiotic resistance in Campylobacter jejuni not mediated by CmeB or CmeF. Antimicrob Agents Chemother 49:1289–1293 10.1128/AAC.49.4.1289-1293.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Palyada K, Sun YQ, Flint A, Butcher J, Naikare H, Stintzi A. 2009. Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genomics 10:481 10.1186/1471-2164-10-481. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Holmes K, Mulholland F, Pearson BM, Pin C, McNicholl-Kennedy J, Ketley JM, Wells JM. 2005. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151:243–257 10.1099/mic.0.27412-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 158.Palyada K, Threadgill D, Stintzi A. 2004. Iron acquisition and regulation in Campylobacter jejuni. J Bacteriol 186:4714–4729 10.1128/JB.186.14.4714-4729.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Luo N, Pereira S, Sahin O, Lin J, Huang S, Michel L, Zhang Q. 2005. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc Natl Acad Sci USA 102:541–546 10.1073/pnas.0408966102. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]


