ABSTRACT
The nonfermenting bacteria belonging to Acinetobacter spp. and Pseudomonas spp. are capable of colonizing both humans and animals and can also be opportunistic pathogens. More specifically, the species Acinetobacter baumannii and Pseudomonas aeruginosa have been recurrently reported as multidrug-resistant and even pandrug-resistant in clinical isolates. Both species were categorized among the ESKAPE pathogens, ESKAPE standing for Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter species. These six pathogens are the major cause of nosocomial infections in the United States and are a threat all over the world because of their capacity to become increasingly resistant to all available antibiotics. A. baumannii and P. aeruginosa are both intrinsically resistant to many antibiotics due to complementary mechanisms, the main ones being the low permeability of their outer membrane, the production of the AmpC beta-lactamase, and the production of several efflux systems belonging to the resistance-nodulation-cell division family. In addition, they are both capable of acquiring multiple resistance determinants, such as beta-lactamases or carbapenemases. Even if such enzymes have rarely been identified in bacteria of animal origin, they may sooner or later spread to this reservoir. The goal of this article is to give an overview of the resistance phenotypes described in these pathogens and to provide a comprehensive analysis of all data that have been reported on Acinetobacter spp. and Pseudomonas spp. from animal hosts.
ACINETOBACTER spp.
The Acinetobacter genus includes 50 species of nonmotile Gram-negative rods that are strictly aerobic, adapted to a wide range of temperatures, and able to survive on abiotic surfaces. Many species belonging to the Acinetobacter genus are able to cause infections, favored by the presence of indwelling devices, in immune-compromised human hosts (1). The lethality of Acinetobacter infections is elevated in more than 50% of cases (2). Among the Acinetobacter spp., A. baumannii is the most prevalent, responsible for 95% of infections and outbreaks in hospitals, followed by A. nosocomialis and A. pittii. The ability of A. baumannii to survive in the hospital environment promotes its diffusion by outbreaks and epidemics. To date, several global epidemics have occurred, sustained by a few strains belonging to successful lineages, namely, clonal complex I-III, as characterized by multilocus sequence typing (3). Recently, another lineage with the potential for global diffusion, delineated as sequence type (ST) 25, has emerged (4). Preventing the introduction of A. baumannii into hospital settings could contribute to preventing the further spread of multidrug-resistant isolates. Although its reservoir remains unknown, this organism has been found in soil, water, and food, including fish, milk, raw vegetables, and meat, which has earned it the definition of “ubiquitous.” The presence in retail meat samples of A. baumannii isolates belonging to a clonal complex commonly associated with multidrug-resistant clones invites the speculation that food may carry organisms into hospital settings. A highly selective pressure exerted by antimicrobial usage may positively select those isolates able to acquire and/or develop resistance mechanisms (5). Unfortunately, Acinetobacter spp. can also be pathogenic for animals. In the following paragraphs, an overview of the infections, the principal mechanisms of antibiotic resistance, and their epidemiology in Acinetobacter spp. among animals will be presented.
Acinetobacter Infections in Animal Hosts
Acinetobacter species are commensals of several body sites in many animal hosts. A. baumannii is frequently isolated from the eyes of horses. It is also isolated from the fecal flora of cattle, equids, and rabbits; from lice and ked of cattle, sheep and dogs; and from the mouths of dogs and cats, with a reported prevalence of 6.5% (9/138) in Reunion Island (6–12). Besides commensalism, the pathogenic role of Acinetobacter in animals cannot be neglected, with infections occurring that are similar to those observed in humans. The presence of foreign bodies in critically ill animals represents a risk factor for developing Acinetobacter spp. infections (13, 14). Furthermore, propagation of multidrug-resistant isolates may occur that are similar to the outbreaks generated in human clinics (15, 16). In the effort to understand the relevance of Acinetobacter as an animal pathogen, Mathewson and Simpson analyzed 347 animal specimens. Although the analysis was conducted on a phenotypic basis, they found Acinetobacter to be prevalent in as many as 14.5% (50/347) of isolates, principally from equine hosts (27%) followed by canine (17%), feline (2%), bovine (2%), and various other hosts (2%) (17). Acinetobacter spp. have also been associated with wound and respiratory tract infections in horses (18, 19) and with urinary tract and respiratory infections and sepsis in dogs and cats (20, 21). Less frequently, Acinetobacter spp. have been found in association with other animal diseases such as bovine mastitis (22, 23) and skin and mucous diseases in birds.
Besides the veterinary relevance of Acinetobacter spp. and, in particular, A. baumannii as an infective agent, many investigations have been conducted with an anthropocentric perspective, studying animals as a reservoir of antimicrobial-resistant bacteria and a source of infections for humans. Indeed, sporadic investigations of animals infected by multidrug-resistant isolates of Acinetobacter spp. have been reported and their epidemiology discussed. In the following section, the most common antimicrobial resistance mechanisms detected in Acinetobacter spp. will be described.
Antimicrobial Resistance in Acinetobacter spp.
A. baumannii poses a public health concern because of its propensity to develop multidrug resistance. In particular, the acquisition of carbapenem resistance poses a serious threat of therapeutic failures (1). The occurrence of Acinetobacter spp. infections in animal hosts poses principally two issues: first, treating such infections is challenging because Acinetobacter spp. isolates are often naturally resistant to many of the antibiotics authorized for use in veterinary medicine; second, the presence and/or the development of multidrug-resistant isolates in animal hosts may serve as reservoir of multidrug-resistant isolates for humans.
Intrinsic resistance
A. baumannii exhibits an intrinsic reduced susceptibility to several antibiotic classes, including beta-lactams, macrolides, trimethoprim, and fosfomycin (24). The mechanisms underlying such intrinsic resistances consist of natural membrane impermeability, basal efflux activity, and the presence of two chromosomally encoded beta-lactamases, an ADC cephalosporinase and an OXA-51 oxacillinase (25). To date, three efflux systems belonging to the resistance-nodulation-division family have been characterized in A. baumannii, encoded by the adeABC, adeFGH, and adeIJK operons (26). Homologs of these operons have been recovered in other Acinetobacter spp. such as A. calcoaceticus, A. nosocomialis, and A. pittii, among others (27–29). The AdeIJK efflux system is constitutively expressed and contributes to a basal resistance to beta-lactams, tetracyclines, macrolides and lincosamides, phenicols, fusidic acid, and fluoroquinolones.
Acquired resistance
The development of acquired resistance can occur by two processes: mutation in chromosomal structures and the acquisition of exogenous genes by horizontal gene transfer. Mutations in the two-component regulatory system AdeRS and in the regulators AdeL and AdeN have been shown to lead to the overproduction of the efflux pumps AdeABC, AdeFGH, and AdeIJK, respectively, and consequently to an increase in resistance. In particular, overproduction of AdeABC contributes to an increase of resistance to beta-lactams, aminoglycosides, fluoroquinolones, tetracyclines and tigecycline, macrolides and lincosamides, and chloramphenicol, whereas overproduction of AdeFGH contributes to resistance to quinolones, antifolates, and chloramphenicol (27).
Resistance to beta-lactams in A. baumannii
Certain insertion sequences, including ISAbaI among others, can provide a strong promoter for the overexpression of the genes located downstream. This phenomenon can be responsible for the overproduction of ADC and OXA-51, leading to the development of high-level resistance to third- and fourth-generation cephalosporins in the first case and of carbapenem resistance in the second case (30, 31). Other insertion sequences, such as ISAba125 and ISAba825, are able to insert into porin-encoding genes, causing the inactivation of the porins and subsequent resistance to carbapenems (32).
Resistance to third- and/or fourth-generation cephalosporins, other than penicillins and their derivatives, can also be mediated by the acquisition of genes coding for exogenous enzymes such as the class A beta-lactamases TEM and SHV in the extended-spectrum variants CTX-M, PER, GES, and VEB (33, 34). Among A. baumannii isolates from human infections, the most common mechanism of carbapenem resistance is mediated by the acquisition of OXAs hydrolyzing carbapenems. The enzymes OXA-23 and OXA-58 are frequently identified in clinical isolates, whereas OXA-24/40 and OXA-143 are rarer. The insertion sequence ISAbaI can mediate the overexpression of the acquired blaOXAs, leading to high-level resistance to carbapenems (35). The presence of class B metallo-beta-lactamases such as SIM, IMP, VIM, and NDM-1 has also been reported (36).
Resistance to aminoglycosides in A. baumannii
Frequently, resistance to carbapenems is associated with aminoglycoside resistance. This is classically mediated by aminoglycoside-modifying enzymes, which catalyze reactions of acetylation, phosphorylation, or O-nucleotidyl transfer. Among such enzymes, AAC(6′)-I is cryptic in several Acinetobacter spp. and confers, when the relative gene is expressed, resistance to netilmicin, tobramycin, gentamicin, and amikacin. Acquired aminoglycoside-modifying enzymes have been frequently detected in A. baumannii, with AAC(3)-I, APH(3′)-VI, and ANT(2″)-I being the most prevalent (37). More recently, 16S rRNA methylases have been described as another mechanism conferring resistance to aminoglycosides (38, 39). This mechanism confers high-level resistance to amikacin, gentamicin, netilmicin, tobramycin, and kanamycin. Among the known methylases, ArmA is the only one to be reported in A. baumannii clinical human isolates, whereas no report exists from animals (40, 41).
Resistance to fluoroquinolones in A. baumannii
In contrast to beta-lactam and aminoglycoside resistances, which are mostly based on the acquisition of exogenous determinants, the development of fluoroquinolone resistances is mainly due to point mutations of the gyrase and topoisomerase enzymes. Of particular importance for high-level resistance are GyrA Ser83Leu together with ParC Ser80Leu and Glu84Lys amino acid substitutions (42, 43).
Resistance to other antibiotics in A. baumannii
Certain antibiotic classes are of limited therapeutic interest for the treatment of A. baumannii infections, mainly because of their toxicity in humans. However, in certain circumstances some of these antibiotics’ properties are fundamental, as in the case of rifampicin, which is able to easily penetrate tissues. Resistance to this antibiotic occurs principally by mutation of the rpoB gene and acquisition of an enzyme that modifies the rifampicin, encoded by the arr-2 gene, and that is usually located on class I integrons (44). The development of multidrug resistance has forced intensified usage of “old antibiotics” such as colistin. Resistance to colistin is mediated by mutation in the proteins PmrAB, a two-component system in A. baumannii (45). Colistin resistance in animal isolates has never been reported. Tigecycline is considered a last-resort treatment of infections caused by multidrug-resistant Acinetobacter spp. Emergence of resistant isolates, mainly overexpressing efflux pumps, has been reported among human patients (46). Furthermore, coselection of tigecycline resistance by usage of other antibiotics, including tetracycline, has been demonstrated in enterococci (47). This is a concern, considering that tigecycline is not allowed in veterinary practice, whereas tetracycline could contribute to the development of a potential reservoir for human contamination. Recently, Ewers et al. reported two tigecycline-resistant A. baumannii isolates from two dogs in Germany (48).
Most common mobile genetic elements in A. baumannii
All the described acquired resistance mechanisms can be located on the chromosome or on plasmids, eventually associated with transposons. For instance, blaOXA-23 has been found located on several transposon structures containing ISAbaI, such as Tn2006, Tn2007, Tn2008, Tn2008B, and Tn2009 (49). A very successful strategy for A. baumannii to develop multidrug resistance is the acquisition of the so-called resistance islands, such as AbaR. The acquisition of such islands seems to be consecutive to a transposition event in a hot-spot sequence, the ATPase encoding gene. Several AbaR islands have been described, with AbaR1 containing as many as 25 genes encoding mechanisms conferring resistance to several antimicrobial classes (50, 51). This brief overview of resistance mechanisms encountered in A. baumannii is far from exhaustive but highlights the potential and the propensity for multidrug resistance development. Therefore, understanding the epidemiology of this species and the intersection of its different habitats and hosts is a high priority. In the following section, we will focus on reports concerning resistance to carbapenems, aminoglycosides, and fluoroquinolones in A. baumannii from animal settings.
Antimicrobial Resistance in Acinetobacter spp. from Food-Producing Animals
The first evidence of carbapenemase-producing Acinetobacter spp. of animal origin dates back to 2010, when Poirel et al. (52) investigated the carriage of carbapenem-resistant Gram-negative organisms in a dairy farm in France. In this investigation, nine isolates sampled from 50 cows were identified as Acinetobacter genomospecies 15TU, a close relative of the species Acinetobacter lwoffii, and all isolates were resistant to carbapenems, harboring a blaOXA-23 gene located on a Tn2008 transposon. The isolates demonstrated resistance not to fluoroquinolones but to kanamycin. Later, an A. lwoffii isolate producing NDM-1 was found in a chicken in China. The isolate was multidrug-resistant, and the blaNDM-1 gene was located on a conjugative plasmid (53). Later, a sporadic A. baumannii isolate was found in China in a survey conducted in 2011 to 2012 for carbapenem resistance in Gram-negative organisms from food-producing animals. The isolates harbored blaNDM-1 located on a plasmid that revealed similarity to plasmids found in isolates of human origin; furthermore, it demonstrated coresistance to aminoglycosides, with the exception of amikacin, and fluoroquinolones. Unfortunately, the sequence type of the isolate was not determined (54). The presence of A. baumannii isolates producing OXA-23 has been documented in wild fish from the Mediterranean Sea. In these isolates, also demonstrating multidrug resistance, the aac(6′)-Ib and aac(3′)-I genes coding for aminoglycoside-modifying enzymes were found. The isolates belonged to ST2, the most widely spread clone in human clinics with which multidrug-resistant isolates are associated. Further investigations revealed that the isolates found in fish were similar to the isolates found contemporaneously in human clinical infections (55). Most likely, the fish were colonized after exposure of water contaminated with clinical waste. Contemporaneously, Al Bayssari et al. (56) reported the presence of A. baumannii demonstrating high-level resistance to imipenem in livestock in Lebanon. The isolates (n = 5) were found in cattle, pigs, and fowls, and all of them harbored the blaOXA-23 gene. One isolate coharbored blaOXA-58. Sequence type determination revealed that among the isolates, one belonged to ST2 and another to ST20, the first being globally spread in human clinics and the second also found in dogs in Switzerland (13). Recently, a report from Pailhoriès et al. (57) revealed the presence of blaOXA-24 in A. baumannii in healthy cattle in Reunion Island. The blaOXA-24 gene occurred in an ST that had never been reported before, suggesting that carbapenem resistance can emerge and disseminate among animals independently from human cross-contamination. The presence of OXA-23 in species other than A. baumannii is quite infrequent, but Klotz et al. (58) has reported the emergence of blaOXA-23 located on Tn2008 in two isolates identified as Acinetobacter indicus colonizing two calves in Germany.
Antimicrobial Resistance in Acinetobacter spp. from Companion Animals
In 2011, a study conducted in Switzerland demonstrated the presence of A. baumannii isolates (n = 19) in infections of pets and horses (13). The majority of these isolates (n = 12) were resistant to fluoroquinolones and harbored the GyrA Ser83Leu and ParC Ser80Leu mutations. Seventeen isolates were resistant to aminoglycosides, and among those the genes aacC2, aacC1, and aadA1 were present. Three isolates in this study, identified in three diseased dogs, demonstrated reduced susceptibility to carbapenems and harbored an ISAbaI inserted upstream from the blaOXA-51 gene. These isolates belonged to ST12 and ST15, which are common among human isolates. An even more worrisome finding has been the detection of Acinetobacter spp. harboring blaOXA-23 in companion animals. The first report dates from 2012 from a screening of fecal carriage in hospitalized horses. On this occasion Smet et al. (59) found two multidrug-resistant Acinetobacter spp. harboring blaOXA-23 located on a Tn2008 transposon. The second report concerned a single isolate associated with a urinary tract infection in a cat in Portugal in 2009. In this isolate, blaOXA-23 was located on a Tn2006 transposon that was chromosomally located. Furthermore, an ISAbaI copy was located upstream from blaADC, and mutations conferring fluoroquinolone resistance were detected, as well (60). This isolate also belonged to ST2, reinforcing the hypothesis that a cross-transmission among humans and pets could be at the base of the animal colonization. In our recent study (21) conducted in the framework of Resapath, the French network for the surveillance of antimicrobial resistance in diseased animals, we analyzed 49 Acinetobacter spp. isolates collected from 2011 to 2015. Among those isolates, the majority were identified as A. baumannii (n = 41), three as A. lwoffii, and one each as A. haemolyticus, A. radioresistens, A. schindleri, A. johnsonii, and A. junii. Among the A. baumannii isolates, seven isolated from the urine of dogs and cats affected by urinary tract infections demonstrated multidrug resistance with high-level resistance to carbapenems. All these isolates harbored a blaOXA-23 located on a chromosomal Tn2008B-like transposon and belonged to ST25. This finding was quite surprising since all previously described A. baumannii isolates of animal origin that were resistant to carbapenems by OXA enzymes production have been reported as belonging to ST2. We also demonstrated that this clone was able to propagate in two regions of France and persist for at least two years among diseased pets. Our study was amplified by a contemporaneous report from the Nantes region, where two dogs were found to be colonized by ST25 A. baumannii harboring blaOXA-23 (61). During a 13-year (2000 to 2013) investigation conducted in Germany by Ewers et al. (48) on diagnostic veterinary samples, three out of 223 A. baumannii isolates harbored blaOXA-23 on a Tn2008 transposon located on a plasmid. These isolates belonged to ST10 and to ST1, two multidrug-resistant sequence types associated with isolates responsible for human infections. In Japan, two isolates of A. radioresistens have been isolated from a diseased cat and dog. The isolates were resistant to carbapenems and harbored a blaOXA-23 gene, which A. radioresistens is considered to be the source of, and a blaIMP-1 gene, together with genes encoding aminoglycoside-modifying enzymes (62). A summary of all the reports described at time of writing is provided in Figure 1.
FIGURE 1.
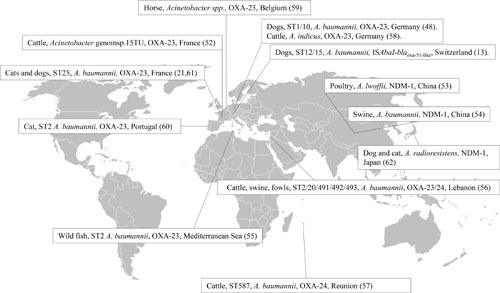
Overview of Acinetobacter spp., sequence types, and acquired carbapenem resistance mechanisms.
Overall, recovering carbapenem-resistant A. baumannii in animal hosts continues to be surprising when considering that usage of carbapenems is not allowed in veterinary medicine. However, coselective pressure on OXA enzymes by the usage of other beta-lactams can be speculated, similalry to the role of other environmental factors.
PSEUDOMONAS spp.
Pseudomonas spp. are Gram-negative bacteria comprising more than 200 species at the time of writing (http://www.bacterio.net/pseudomonas.html) that can be ubiquitously found in humans, animals, soil, and plants (63, 64). Pseudomonas spp. were extensively studied for their beneficial or deleterious associations with plants (P. putida, P. syringae, P. fluorescens, etc.) but also for their roles in soil bioremediation due to specific biodegradation properties (P. putida, P. stutzeri, P. alcaligenes, etc.) (65–67). Only a few species are of clinical interest in either humans or animals, and P. aeruginosa is by far the most frequently reported pathogen. For this reason, this section will focus on this unique species, which is also the only one in which antibiotic resistance was reported in animal hosts.
P. aeruginosa Infections in Animal Hosts
P. aeruginosa is a ubiquitous bacterium normally found in water and soil, but also an opportunistic pathogen of humans, animals, and plants (68). In humans, P. aeruginosa is mostly nosocomial, causing severe infections in patients with underlying conditions. Immunosuppressed or intubated-ventilated patients presenting compromised host defenses are particularly vulnerable to this pathogen. It is primarily associated with burn victims and cystic fibrosis patients (69, 70).
P. aeruginosa has not been extensively studied in infections of animal origin since this bacterium is more often considered an environmental contaminant rather than a true pathogen. Apart from sporadic descriptions, it has mostly been reported in cats and dogs, where it is an important pathogen causing otitis externa and otitis media (71–74). Together with Staphylococcus pseudintermedius, it is one of the two main ear pathogens, and its prevalence ranges from 6.5 to 27.8% depending on the study (71, 72, 75, 76). However, P. aeruginosa is also implicated in skin infections, including deep pyoderma, often in association with other bacterial pathogens (72, 77, 78).
More surprisingly, P. aeruginosa infections have been recurrently described in fur animals, where it seems to be particularly virulent (79, 80). The first victims are minks (Neovison vison). These mammalian carnivores of the Mustelidae family are raised for fur production, principally in Denmark, China, the Netherlands, Poland, and the United States. First described in 1953, the acute and fatal hemorrhagic pneumonia caused by P. aeruginosa can decimate farmed minks and lead to high economic losses (81). The second victims are chinchillas, a rodent species which is raised for both pets and laboratory animals—but not for fur. The infections, mainly otitis, are often due to uncleaned water and cage environment and are favored by a weak immunity of this animal species. Since P. aeruginosa has a particular capacity of dissemination between these animals, it is very important to rapidly isolate the diseased individuals.
P. aeruginosa is much less reported in livestock animals. It is an opportunistic pathogen that is very rarely reported in etiological surveys of bovine mastitis (82–86). However, several case reports suggest that, though unusual, outbreaks of P. aeruginosa can be severe and spread either clonally or nonclonally in different herds (87–90). The origin of the infection is often contaminated liquids, teat dips, or even a contaminated antibiotic preparation (91, 92). One study also reports its implication in 12% of the urinary tract infections in cattle in Israel (93).
Antimicrobial Resistance in P. aeruginosa
P. aeruginosa is one of the ESKAPE bacteria (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter spp.), for which the therapeutic options are increasingly limited (94). In addition to its capacity to form biofilms, P. aeruginosa is intrinsically resistant to many antibiotics, including beta-lactams (penicillin G, aminopenicillins alone or in combination with inhibitors, first- and second-generation cephalosporins, cefixime, cefuroxime, cefotaxime, ceftriaxone, ertapenem), kanamycin, tetracycline, chloramphenicol, trimethoprim, and quinolones (95). P. aeruginosa is also known for its capacity to rapidly acquire additional resistances, so that the combination of intrinsic and acquired resistances can lead to therapeutic failures (96).
Intrinsic resistance
It is commonly admitted that in P. aeruginosa, intrinsic resistance is mainly mediated by a combination of impermeability, production of the inducible AmpC cephalosporinase, and the presence of efflux pumps (97). On the one hand, the permeability of the outer membrane is up to 100-fold lower in P. aeruginosa than in Escherichia coli (97), and on the other hand, two constitutively expressed drug efflux systems, MexAB-OprM and MexXY-OprM, directly participate in intrinsic resistance. Both systems belong to the resistance-nodulation-division family and were identified in the laboratory strain PAO1 (98). MexAB-OprM confers resistance to beta-lactams (with the exception of imipenem), fluoroquinolones, trimethoprim-sulfonamides, chloramphenicol, and tetracyclines, while MexXY-OprM is involved in resistance to cefepime, aminoglycosides, fluoroquinolones, tetracyclines, chloramphenicol, trimethoprim-sulfonamides, and macrolides. Ten other efflux systems were also described in P. aeruginosa, none of which plays a role in intrinsic resistance. Finally, P. aeruginosa harbors two beta-lactamase encoding genes. The first one is the constitutively expressed blaOXA-50 oxacillinase, which only plays a minor role in beta-lactam resistance (99). The second is the inducible AmpC beta-lactamase, which confers resistance to beta-lactams, including cefuroxime and ceftriaxone, even though this mechanism may be redundant with the MexAB-OprM efflux system (100, 101).
Like many bacterial species, P. aeruginosa can live either as planktonic cells or as organized communities called biofilms (102). Quorum-sensing, the cell-to-cell communication mechanism involving the Las, Rhl, and PQS systems in P. aeruginosa, was shown to be involved in biofilm formation, particularly through the Rhl system (103, 104). An important characteristic of biofilms is their tolerance to different external stresses, including antibiotic treatment (105). Tolerance is a physiological state of the bacteria that does not involve any acquired mutation and cannot be transmitted to the progeny of a mother cell. For example, tolerance to aminoglycosides (gentamicin, tobramycin), tetracyclines, and colistin has been described (93, 106). Colistin targets different zones of the biofilm compared to other molecules, so that combined treatment with colistin/gentamicin or colistin/tetracycline is more appropriate to eradicate the majority of the cells composing the biofilm (106).
Acquired resistance
Besides intrinsic resistance, P. aeruginosa is capable of acquiring numerous additional resistances, either through point mutations in pre-existing genes or through horizontal transfer of resistance determinants (96). The use of specific antibiotics during treatment can readily select for point mutations which lead to the overexpression of one or another efflux system. Depending on the affected system (MexAB-OprM, MexXY-OprM, MexCD-OprJ, or MexEF-OprN), elevated resistance levels toward their specific antibiotic substrates are observed (98, 107). Overproduction of AmpC can also be obtained through mutations in regulatory genes. The porin OprD can also be modified, which is the preferential pathway toward carbapenem resistance in human clinical isolates of P. aeruginosa. And finally, mutations in the target genes gyrA/gyrB and parC/parD confer resistance to fluoroquinolones (see below).
In parallel, a large number of acquired enzymes conferring beta-lactam resistance were identified in P. aeruginosa (108). These include extended-spectrum beta-lactamases of the PER, SHV, PME, GES, and VEB families, as well as the CTX-M enzymes typically found in Enterobacteriaceae. Metallo-beta-lactamases conferring resistance to carbapenems were also reported, mainly IMP and VIM enzymes, even though carbapenem resistance in P. aeruginosa is mostly due to the oprD gene, which can be repressed, mutated, or deleted (108). These enzymes are increasingly found in human clinical isolates but have not been reported yet in animal isolates.
Antimicrobial Resistance in Pseudomonas spp. in Cats and Dogs
P. aeruginosa is one of the main pathogens causing otitis externa and otitis media (71–74) but is also implicated in skin infections (72, 77, 78). The treatment of such infections starts by a thorough cleaning (deep ear flush in the case of otitis) and a topical disinfection, which is often followed by an antibiotic treatment using mainly fluoroquinolones, aminoglycosides or polymyxins. In this respect, monitoring of antimicrobial resistance in P. aeruginosa is clearly needed. However, most of the clinically relevant antibiotics do not have referenced clinical breakpoints, which is a serious data gap for effective surveillance that will have to be filled in in the near future.
Resistance to fluoroquinolones
Ciprofloxacin is considered the most active fluoroquinolone against P. aeruginosa (109). The prevalence of resistance to this molecule in P. aeruginosa of animal origin ranges from very low to high rates (Table 1). Indeed, variable resistance rates have been reported, ranging from 3.7% in China in 2009 to 2010 and 8.7% in Croatia (2007 to 2009) to 16% in Canada (2003 to 2006), 4.8% and 20% in two Brazilian studies, and 21% in the United States (73, 74, 78, 110–113), and even reaching 63% in France between 2008 and 2011. These divergences may be due to the methodology (MICs versus disk diffusion), the levels of fluoroquinolone usage in the countries where strains were collected, or the type of sampling (otitis versus skin infection or mild versus severe infections).
TABLE 1.
Antimicrobial susceptibility to fluoroquinolones and aminoglycosides in P. aeruginosa isolates of animal origin
| Country | Year | Animal host | Pathology | No. of isolates | Method | Percentage (%) of resistanced | Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|
| CIP | ENR | GEN | AMI | |||||||
| US. | 1998–2003 | Dogs/cats | Otitis | 319 | Sensititre | –e | 38.0 | 15.0 | 11.0 | 71 |
| U.S. | 1992–2005 | Dogs | Pyoderma | 20 | Disk diffusion | 25.0 | 40.0 | 5.0 | 5.0 | 78 |
| Europea | 2008–2010 | Dogs | SSTIf | 160 | Agar dilution | – | 16.9 | 18.8 | – | 114 |
| Cats | SSTI | 11 | – | 18.2 | 9.0 | – | ||||
| Croatia | 2007–2009 | Dogs | Otitis | 104 | Etest | 8.7 | 51.9 | 43.3 | – | 74 |
| Croatia | 1998–2000 | Dogs | Otitis | 183 | Agar dilution | 3.8 | 26.2 | 10.9 | 7.6 | 116 |
| Canada | 2003–2006 | Dogs | SSTI | 106 | Sensititre | 16.0 | 31.0 | 7.0 | 3.0 | 73 |
| China | 2009–2010 | Dogs | SSTI | 27 | Broth microdilution | 14.8 | – | 14.8 | 11.1 | 110 |
| Germany | 2004–2006 | Dogs/cats | SSTI | 71 | Broth microdilution | – | 24.0 | 27.0 | – | 115 |
| Urinary/genital tract | 28 | – | 11.0 | 11.0 | – | |||||
| France | 2008–2011 | Dogs | SSTI | 46 | Agar dilution | 63.0 | – | 56.5 | 15.2 | 124 |
| Horses | Diverseb | 10 | 0.0 | – | 10.0 | 0.0 | ||||
| Cows | Diversec | 12 | 0.0 | – | 8.4 | 0.0 | ||||
| Brazil | 2010–2012 | Dogs | Otitis, pyoderma | 104 | Disk diffusion | 4.8 | 26.0 | 4.8 | 2.9 | 113 |
| Japan | 2005–2007 | Cows | Mastitis | 116 | Broth microdilution | 0.0 | 31.0 | 4.3 | 1.7 | 154 |
| Japan | Unknown | Chinchillas | Healthy | 22 | 4.5 | 81.0 | 0.0 | 0.0 | 150 | |
| Denmark | 2002–2005 | Minks | Hemorrhagic pneumonia | 39 | Sensititre | – | 5.1 | 0.0 | – | 144 |
| China | 2010–2011 | Minks | Hemorrhagic pneumonia | 30 | VITEK-2 | 13.3 | – | 0.0 | 0.0 | 146 |
Czech Republic, France, Germany, Hungary, Italy, the Netherlands, Poland, Spain, Sweden, and the United Kingdom.
Respiratory infections, skin or eye infections, metritis.
Mastitis, digestive and respiratory infections.
CIP, ciprofloxacin; ENR, enrofloxacin; GEN, gentamicin; AMI, amikacin.
–, Not performed.
SSTI, Skin and Soft Tissues Infections.
The importance of resistance to ciprofloxacin also has to be put into perspective here since this molecule is not used in veterinary medicine. Nevertheless, among the three main fluoroquinolones prescribed in animals (enrofloxacin, marbofloxacin, and the more recent pradofloxacin), the most frequently tested is enrofloxacin, which presents high rates of resistance (Table 1). Indeed, 18.2% of isolates collected throughout Europe were resistant (and 81.8% presented an intermediate phenotype), as were 24% in Germany (49% intermediately resistant), 26.2% in Croatia, 31% and 38% in two Canadian studies, 49% in the United States, and 26.0% and 70% in two Brazilian studies (71, 73, 76, 111, 112, 114–116). On the other hand, even though ciprofloxacin is the major active metabolite of enrofloxacin, divergences in the prevalence of resistance are observed when both antibiotics are tested on the same collection of isolates (116). This may be because of the different in vitro activities of enrofloxacin, marbofloxacin, and ciprofloxacin. Globally, there is a lack of harmonized studies to clarify the clinical relevance of those discrepancies in the resistance of P. aeruginosa to the major fluoroquinolones used in routine veterinary practice.
The targets of quinolones and fluoroquinolones are the DNA gyrase and the DNA topoisomerase IV, which are both constituted of two subunits, named GyrA/GyrB and ParC/ParE, respectively (117). The main resistance mechanism in both Gram-positive and Gram-negative bacteria involves mutations in these targets. In P. aeruginosa, GyrA and ParC mutations were identified (though not systematically) in ciprofloxacin-resistant clinical isolates, while GyrB mutations are thought to confer only moderate resistance (118). Such point mutations were also reported in veterinary isolates, and the most frequent ones, namely Thr83Ile in GyrA and Ser87Leu in ParC, were also reported in human isolates (73, 110, 118, 119). Efflux pumps have also been identified as a key mechanism in fluoroquinolone resistance, notably through the MexAB-OprM or MexF-OprN systems (108, 120–122). Only one study reported the overexpression of these efflux systems in veterinary isolates (123). However, this subject should definitely be further explored in P. aeruginosa of animal origin, since overproduction of efflux pumps is easily selected by usage of veterinary -licensed antibiotics, conferring resistance to several antibiotics including aminoglycosides and even a few carbapenems due to their wide substrate specificity (98).
Resistance to aminoglycosides
Resistances to gentamicin and amikacin are often reported, probably because they are used as first- and second-line antibiotics for the treatment of otitis but also of pyoderma and corneal ulcers in cats and dogs. Precautions need to be taken with this family of antibiotics because of their nephro- and ototoxicity.
The prevalence of resistance to gentamicin is systematically higher than resistance to amikacin (Table 1). Gentamicin resistance was reported from dogs in two studies of soft tissue infections in the United States (5% and 7%), in ophthalmic infections in Brazil (10%), in otitis in Canada (15%), in diverse infectious contexts in Germany (11%) and Brazil (4.8%), in soft tissue infections in China and in Europe (14.8% and 18.8%), in otitis in Croatia (16.9% and 43.3% in two studies, respectively), and in otitis in France (56.5%) (71, 73, 74, 78, 110–112, 114–116, 124). On the other hand, amikacin resistance rates observed in the same studies (when available) ranged from 5% and 3% in the United States, 10% (ophthalmic infections) and 2.9% (otitis and pyoderma) in Brazil, 11% in Canada, 11.1% in China, 12.6% in Croatia, and 15.2% in France. The divergences between the two antibiotics may have the same causes as cited above for fluoroquinolones.
Aminoglycoside resistance in P. aeruginosa is principally mediated by the MexXY-OprM multidrug efflux system. This system is constitutively expressed and implicated in the intrinsic resistance of P. aeruginosa to aminoglycosides (112). However, its overexpression can easily be induced by the use of its substrate antibiotics, thus conferring an elevated resistance to these very same antibiotics, including aminoglycosides (98, 125). The role of MexXY-OprM in animal strains was studied by Chuanchuen et al. in pets and bovine mastitis (see below) (126, 127). The role of the MexXY efflux pump in aminoglycoside resistance was evidenced in cats and dogs sampled in Thailand and the United States (123, 127), in addition to the presence of aminoglycoside-modifying enzymes, which have also been reported in isolates from the United States and Canada (73, 110). Aminoglycoside resistance can be achieved by inactivation of these antibiotics through specific modifications mediated by enzymes of the AAC, APH, and ANT families (128). The nature of the modifications and the spectrum of inactivated molecules depend on the modifying enzyme implicated (128). Finally, three methylases—which bind to the target site of the aminoglycosides and confer high-resistance phenotypes to several molecules, including gentamicin and amikacin—have been described in P. aeruginosa, namely ArmA, RmtA, and RmtD (129–131), none of which have been reported yet in veterinary isolates.
Resistance to polymyxins
Polymyxin B is one of the first-line antibiotic treatments in cases of otitis and eye infections in cats and dogs (132). Antimicrobial susceptibility data on this molecule are still rare, but when data are available, polymyxin B is always the most efficient antibiotic. Indeed, no resistant isolate was described in the United States (78), in Canada (71), or in Brazil (112). Polymyxin-resistant veterinary isolates were nonetheless reported in Germany—where four isolates (4/71, 5.6%) from soft tissue infections and two isolates (2/28, 7.1%) from urinary/genital tract infections presented an MIC to colistin of >2 mg/liter—and recently in Brazil, where 3/10 isolates from ophthalmic infections showed polymyxin B resistance (111, 115). However, the molecular basis of these resistant phenotypes remains unknown.
The discovery of a plasmidic gene, mcr-1, conferring resistance to Enterobacteriaceae has shed new light on colistin resistance (133). Interestingly, this gene has been successfully transferred in vitro to P. aeruginosa, but no field strain of mcr-1-carrying P. aeruginosa has been reported yet. Colistin-resistance can also be achieved under laboratory conditions in a reversible manner by repeated exposure to subinhibitory concentrations of colistin (134). Colistin is a last-resort antibiotic in cases of multidrug-resistant strains, but colistin resistance is fortunately still very rare in human clinical isolates (135). When studied molecularly, these resistant isolates mostly present modifications in the lipopolysaccharide (136–138).
Resistance to carbapenems
Carbapenem use is forbidden in veterinary medicine, including in companion animals. Consequently, the occurrence of carbapenem-resistant pathogens in animals has only sporadically been described. In 2014, an IMP-45-producing P. aeruginosa strain was detected in a dog during routine surveillance for carbapenem resistance (139). Recently, a study was performed in France on 30 isolates from cats and dogs (including one cattle isolate) presenting a decreased susceptibility to imipenem and/or meropenem (140). No carbapenemase gene was detected, and only a few isolates showed an altered OprD (6/30), which is a major cause of carbapenem resistance in humans. On the contrary, most of the isolates displayed alterations in efflux pumps (MexAB-OprM [n = 12], MexEF-OprN [n = 4], MexXY [n = 8], and CzcCBA [n = 3]). Since these efflux pumps also confer resistances to antibiotics that are used in veterinary medicine (notably fluoroquinolones and aminoglycosides), the observed decreased susceptibility to carbapenems is thus probably a consequence of noncarbapenem antibiotic use. In Brazil, carbapenem-resistant isolates were also reported (7.7% of isolates resistant to imipenem, 1.0% to meropenem), but no molecular characterization was performed (112).
Antimicrobial Resistance in Pseudomonas spp. in Minks
P. aeruginosa is especially virulent in minks, where it is a major cause of hemorrhagic pneumonia. This infection is decimating farmed minks (N. vison) and causes high economic losses (81). P. aeruginosa dissemination is due to local outbreaks of clonal strains, but clones vary between outbreaks (79, 141, 142). The origin of the contamination is mostly environmental, and clones spread in farms due to contaminated water containers or food, standing water, and uncleaned cages (79). Prevention of hemorrhagic pneumonia mostly relies on multivalent vaccines, but their expensive price and short protection period leads to innovative research such as research in phages (79, 142, 143).
Antibiotics are used to treat minks. Penicillins, aminoglycosides, and macrolides are the main families of molecules used to treat fur animals in Denmark, independent of the pathology and the pathogen identified. Their use steadily increased between 2001 and 2006 (144) and increased significantly (102% increase) from 2007 to 2012 (145). However, data on antimicrobial resistance in P. aeruginosa from hemorrhagic pneumonia were only reported in three studies, which showed an overall high susceptibility of most of the isolates. The first study included 39 isolates collected in Denmark between 2000 and 2005 (144), the second one comprised 30 isolates originating from China from 2010 to 2011 (146), and the third one included 41 isolates collected in Denmark between 2014 and 2016 (147). Danish isolates from the first sampling period (2000 to 2005) were susceptible to gentamicin and colistin, while 5.1% were resistant to enrofloxacin. Resistance to aminoglycosides was suspected in both collections, but no proportions can be inferred because of the lack of referenced breakpoints. Isolates from the same country but the second sampling period (2014 to 2016) were also susceptible to gentamicin and ciprofloxacin, while 17% were resistant to colistin. Unfortunately, colistin resistance was only inferred by MIC results, but no molecular characterization was performed. Chinese isolates were more resistant to fluoroquinolones (13.3%) and also presented resistance to ticarcillin/clavulanic acid. However, no resistance was observed to aminoglycosides. The differences in resistance may reflect local specificities in terms of antibiotic treatment.
Antimicrobial Resistance in Pseudomonas spp. in Chinchillas
P. aeruginosa is the main cause of infections in chinchillas, often due to uncleaned water and cages. Chinchillas have been extensively studied as models of middle ear infections (148), but studies dedicated to otitis media in this animal species are scarce (149).
Antibiotic susceptibility has only been reported once, in 67 chinchillas in Japan (150), of which 23 were raised as pets and 21 as laboratory animals. A total of 22 P. aeruginosa isolates were identified, which clustered in seven pulsed-field gel electrophoresis patterns. No resistance phenotype was observed for aminoglycosides, even though nine isolates presented a decreased susceptibility to gentamicin and one to amikacin. One isolate was resistant to ciprofloxacin, while MICs for enrofloxacin—the major veterinary fluoroquinolone—were much higher than those to ciprofloxacin. Finally, six isolates showed intermediate resistance to ceftazidime and five to imipenem. The number of isolates presenting reduced susceptibilities should undoubtedly prompt further studies in these animals which are in close contact with humans.
Antimicrobial Resistance in Pseudomonas spp. in Food-Producing Animals
Cattle are the only food-producing animals for which substantial data on antimicrobial resistance in P. aeruginosa are available. Data for chickens and pigs are very scarce and mostly describe Pseudomonas spp. as environmental contaminants (151, 152). Only two studies specifically designed for the detection of carbapenem-resistant isolates reported the presence of VIM-2 in P. aeruginosa in fowl in Lebanon and VIM-1 in Pseudomonas putida in chicken cloacal swabs as well as in their environment in China (56, 153).
In cattle, only three articles reported on resistance phenotypes in P. aeruginosa isolates from bovine mastitis. Ohnishi et al. studied 116 P. aeruginosa strains collected from the milk of 115 cows in Japan between 2005 and 2007 (154). P. aeruginosa was found in 0.65% of the milk isolates that had been under control and caused moderate to severe infections in half of the cases. Isolates presented high susceptibility rates toward piperacillin, ceftazidime, cefepime, imipenem, ciprofloxacin, amikacin, and tobramycin. Amikacin resistance was suspected in two isolates and carbapenem-resistance in two others, but this could not be confirmed molecularly. This is considerably different from what has been seen in Japan in human isolates, where multidrug-resistant and carbapenemase-producing strains were recurrently found. Haenni et al. also reported 12 isolates from cattle in France in 2010 (124), which all belonged to nonhuman clones. In parallel, P. aeruginosa was recovered in 0.61% of the bovine isolates collected the same year through the Resapath network (www.resapath.anses.fr). Isolates originated from mastitis and respiratory tract infections and were susceptible to the majority of antibiotics tested, except fosfomycin (9/12, 75%) and ticarcillin (3/12, 25%). Thus, these results confirm the low incidence of P. aeruginosa in bovine mastitis in both countries, suggest that clones circulating in animals differ from the ones isolated in humans, and prove their capacity to cause severe infections.
Chuanchuen et al. reported a molecular study of the MexXY efflux pumps in 18 P. aeruginosa isolates collected from bovine mastitis in Thailand (126). All of these field isolates presented decreased susceptibility to a variety of aminoglycosides, and three displayed an MIC to gentamicin higher than those of the PAO1 control strains. These decreased susceptibilities to aminoglycosides were partly attributed to overexpression of the MexXY efflux system but also to the presence of genes coding for aminoglycoside-modifying genes, such as aph(3′)-IIb and aac(6′)-IIb. Finally, two carbapenem-resistant isolates producing the VIM-2 enzyme were reported in Lebanon (56).
Antimicrobial Resistance in Pseudomonas spp. in Horses
P. aeruginosa is a rare pathogen in horses, sporadically causing skin or respiratory infections. It is more frequently associated with genital tract infections such as endometritis, which can lead to reduced fertility or even sterility. Horse-to-horse transmission is a potential source of transmission since nonpathogenic isolates may be incidentally introduced into the vagina of the mare during coitus (155). However, a wide variety of clones was identified in certain studies, suggesting transmission through contaminated material during artificial insemination or opportunistic growth of bacteria from environmental sources if conditions are favorable (156–158).
Only two limited studies from France and Brazil reported the antimicrobial susceptibility of P. aeruginosa isolates from horses (111, 124). No multiresistant isolates were reported in France among the 10 animals sampled, and only fosfomycin resistance was prevalent (6/10, 60%). Interestingly, two strains belonged to clones ST155 and ST27, which are associated with human outbreaks and sometimes display multiresistance phenotypes. In Brazil, only three animals were included in the study, and two out of the three P. aeruginosa isolates studied presented multiple resistances to fluoroquinolones and aminoglycosides.
The paucity of infections due to P. aeruginosa in horses may explain the lack of information on antimicrobial resistance in such isolates. However, the spread of resistance in all reservoirs (human, animal, and environmental) and the need for data on all potential niches will probably prompt scientists to explore this field.
CONCLUSION
A. baumannii and P. aeruginosa are two major nosocomial pathogens in humans, and increasingly resistant strains are being characterized all over the world. In contrast, they are more rarely found in animals, as is evidenced by the low number of publications in the veterinary field. However, when considering taking measures to avoid further spread of antimicrobial-resistant organisms or emergence of further resistance, the intersections of all the ecological domains must be explored. The crossroads of humans and animals is especially important—on the one hand for protection of professionals, as in the case of breeders and livestock farmers, and on the other hand, for physical proximity, adoption of pets in Europe being a growing phenomenon with 75 million pet-owing households. In this context, the emergence of multidrug-resistant bacteria from animals is worrisome: first, because the therapeutic options for animals are dramatically diminishing and, second, because the animal reservoir of multidrug-resistant bacteria is gaining in prevalence and complexity. The process contributing to such development is articulated, consisting of cross-contamination between human and animals, selective and coselective pressure by antimicrobial usage, and the spread of multidrug-resistant organisms in intensive breeding frameworks. Considering this context, studies dedicated to Acinetobacter spp. and Pseudomonas spp. on farms or generally in animal hosts are both limited in number and quite sparse in their geographical distribution, thus impeding the elaboration of a general picture of the modality of the spread of certain clones and the emergence of resistance mechanisms. Ideally, concerted investigations between human and veterinary clinics would provide useful keys to understanding such phenomena. To this end, global and vigilant surveys are priorities to preserve public health.
ACKNOWLEDGMENTS
We thank Katy Jeannot for her helpful proofreading.
REFERENCES
- 1.Fishbain J, Peleg AY. 2010. Treatment of Acinetobacter infections. Clin Infect Dis 51:79–84 10.1086/653120. [PubMed] [DOI] [PubMed] [Google Scholar]
- 2.Karaiskos I, Giamarellou H. 2014. Multidrug-resistant and extensively drug-resistant Gram-negative pathogens: current and emerging therapeutic approaches. Expert Opin Pharmacother 15:1351–1370 10.1517/14656566.2014.914172. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodríguez-Valera F. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol 43:4382–4390 10.1128/JCM.43.9.4382-4390.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahl JW, Del Franco M, Pournaras S, Colman RE, Karah N, Dijkshoorn L, Zarrilli R. 2015. Phylogenetic and genomic diversity in isolates from the globally distributed Acinetobacter baumannii ST25 lineage. Sci Rep 5:15188 10.1038/srep15188. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lupo A, Vogt D, Seiffert SN, Endimiani A, Perreten V. 2014. Antibiotic resistance and phylogenetic characterization of Acinetobacter baumannii strains isolated from commercial raw meat in Switzerland. J Food Prot 77:1976–1981 10.4315/0362-028X.JFP-14-073. [PubMed] [DOI] [PubMed] [Google Scholar]
- 6.Belmonte O, Pailhoriès H, Kempf M, Gaultier MP, Lemarié C, Ramont C, Joly-Guillou ML, Eveillard M. 2014. High prevalence of closely-related Acinetobacter baumannii in pets according to a multicentre study in veterinary clinics, Reunion Island. Vet Microbiol 170:446–450 10.1016/j.vetmic.2014.01.042. [DOI] [PubMed] [Google Scholar]
- 7.Cattabiani F, Cabassi E, Allodi C, Gianelli F. 1976. Bacterial flora of the conjunctival sac of the horse. Ann Sclavo 18:91–119. (In Italian.) [PubMed] [PubMed] [Google Scholar]
- 8.Johns IC, Baxter K, Booler H, Hicks C, Menzies-Gow N. 2011. Conjunctival bacterial and fungal flora in healthy horses in the UK. Vet Ophthalmol 14:195–199 10.1111/j.1463-5224.2010.00867.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 9.Kumsa B, Socolovschi C, Parola P, Rolain JM, Raoult D. 2012. Molecular detection of Acinetobacter species in lice and keds of domestic animals in Oromia Regional State, Ethiopia. PLoS One 7:e52377 10.1371/journal.pone.0052377. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore CP, Heller N, Majors LJ, Whitley RD, Burgess EC, Weber J. 1988. Prevalence of ocular microorganisms in hospitalized and stabled horses. Am J Vet Res 49:773–777. [PubMed] [PubMed] [Google Scholar]
- 11.Rudi K, Moen B, Sekelja M, Frisli T, Lee MR. 2012. An eight-year investigation of bovine livestock fecal microbiota. Vet Microbiol 160:369–377 10.1016/j.vetmic.2012.06.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 12.Saphir DA, Carter GR. 1976. Gingival flora of the dog with special reference to bacteria associated with bites. J Clin Microbiol 3:344–349. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endimiani A, Hujer KM, Hujer AM, Bertschy I, Rossano A, Koch C, Gerber V, Francey T, Bonomo RA, Perreten V. 2011. Acinetobacter baumannii isolates from pets and horses in Switzerland: molecular characterization and clinical data. J Antimicrob Chemother 66:2248–2254 10.1093/jac/dkr289. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaneechoutte M, Devriese LA, Dijkshoorn L, Lamote B, Deprez P, Verschraegen G, Haesebrouck F. 2000. Acinetobacter baumannii-infected vascular catheters collected from horses in an equine clinic. J Clin Microbiol 38:4280–4281. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boerlin P, Eugster S, Gaschen F, Straub R, Schawalder P. 2001. Transmission of opportunistic pathogens in a veterinary teaching hospital. Vet Microbiol 82:347–359 10.1016/S0378-1135(01)00396-0. [DOI] [PubMed] [Google Scholar]
- 16.Zordan S, Prenger-Berninghoff E, Weiss R, van der Reijden T, van den Broek P, Baljer G, Dijkshoorn L. 2011. Multidrug-resistant Acinetobacter baumannii in veterinary clinics, Germany. Emerg Infect Dis 17:1751–1754 10.3201/eid1709.101931. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathewson JJ, Simpson RB. 1982. Glucose-nonfermenting Gram-negative bacilli associated with clinical veterinary specimens. J Clin Microbiol 15:1016–1018. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abbott Y, O’Mahony R, Leonard N, Quinn PJ, van der Reijden T, Dijkshoorn L, Fanning S. 2005. Characterization of a 2.6 kbp variable region within a class 1 integron found in an Acinetobacter baumannii strain isolated from a horse. J Antimicrob Chemother 55:367–370 10.1093/jac/dkh543. [PubMed] [DOI] [PubMed] [Google Scholar]
- 19.Boguta L, Gradzki Z, Borges E, Maurin F, Kodjo A, Winiarczyk S. 2002. Bacterial flora in foals with upper respiratory tract infections in Poland. J Vet Med B Infect Dis Vet Public Health 49:294–297 10.1046/j.1439-0450.2002.00570.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 20.Francey T, Gaschen F, Nicolet J, Burnens AP. 2000. The role of Acinetobacter baumannii as a nosocomial pathogen for dogs and cats in an intensive care unit. J Vet Intern Med 14:177–183 10.1111/j.1939-1676.2000.tb02233.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 21.Lupo A, Châtre P, Ponsin C, Saras E, Boulouis HJ, Keck N, Haenni M, Madec JY. 2016. Clonal spread of Acinetobacter baumannii sequence type 25 carrying blaOXA-23 in companion animals in France. Antimicrob Agents Chemother 61:61. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malinowski E, Lassa H, Kłlossowska A, Smulski S, Markiewicz H, Kaczmarowski M. 2006. Etiological agents of dairy cows’ mastitis in western part of Poland. Pol J Vet Sci 9:191–194. [PubMed] [PubMed] [Google Scholar]
- 23.Nam HM, Lim SK, Kang HM, Kim JM, Moon JS, Jang KC, Kim JM, Joo YS, Jung SC. 2009. Prevalence and antimicrobial susceptibility of Gram-negative bacteria isolated from bovine mastitis between 2003 and 2008 in Korea. J Dairy Sci 92:2020–2026 10.3168/jds.2008-1739. [PubMed] [DOI] [PubMed] [Google Scholar]
- 24.Ruppé É, Woerther PL, Barbier F. 2015. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann Intensive Care 5:61 10.1186/s13613-015-0061-0. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonomo RA, Szabo D. 2006. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin Infect Dis 43(Suppl 2):S49–S56 10.1086/504477. [PubMed] [DOI] [PubMed] [Google Scholar]
- 26.Coyne S, Courvalin P, Périchon B. 2011. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother 55:947–953 10.1128/AAC.01388-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coyne S, Rosenfeld N, Lambert T, Courvalin P, Périchon B. 2010. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 54:4389–4393 10.1128/AAC.00155-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Damier-Piolle L, Magnet S, Brémont S, Lambert T, Courvalin P. 2008. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob Agents Chemother 52:557–562 10.1128/AAC.00732-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenfeld N, Bouchier C, Courvalin P, Périchon B. 2012. Expression of the resistance-nodulation-cell division pump AdeIJK in Acinetobacter baumannii is regulated by AdeN, a TetR-type regulator. Antimicrob Agents Chemother 56:2504–2510 10.1128/AAC.06422-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corvec S, Caroff N, Espaze E, Giraudeau C, Drugeon H, Reynaud A. 2003. AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J Antimicrob Chemother 52:629–635 10.1093/jac/dkg407. [PubMed] [DOI] [PubMed] [Google Scholar]
- 31.Mugnier PD, Poirel L, Nordmann P. 2009. Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii. J Bacteriol 191:2414–2418 10.1128/JB.01258-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mussi MA, Relling VM, Limansky AS, Viale AM. 2007. CarO, an Acinetobacter baumannii outer membrane protein involved in carbapenem resistance, is essential for l-ornithine uptake. FEBS Lett 581:5573–5578 10.1016/j.febslet.2007.10.063. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.Bonnin RA, Potron A, Poirel L, Lecuyer H, Neri R, Nordmann P. 2011. PER-7, an extended-spectrum beta-lactamase with increased activity toward broad-spectrum cephalosporins in Acinetobacter baumannii. Antimicrob Agents Chemother 55:2424–2427 10.1128/AAC.01795-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naas T, Coignard B, Carbonne A, Blanckaert K, Bajolet O, Bernet C, Verdeil X, Astagneau P, Desenclos JC, Nordmann P, French Nosocomial Infection Early Warning Investigation and Surveillance Network. 2006.VEB-1 Extended-spectrum beta-lactamase-producing Acinetobacter baumannii, France. Emerg Infect Dis 12:1214–1222 10.3201/eid1708.051547. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans BA, Amyes SG. 2014. OXA β-lactamases. Clin Microbiol Rev 27:241–263 10.1128/CMR.00117-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathlouthi N, Al-Bayssari C, Bakour S, Rolain JM, Chouchani C. 2017. Prevalence and emergence of carbapenemases-producing Gram-negative bacteria in Mediterranean basin. Crit Rev Microbiol 43:43–61 10.3109/1040841X.2016.1160867. [PubMed] [DOI] [PubMed] [Google Scholar]
- 37.Seward RJ, Lambert T, Towner KJ. 1998. Molecular epidemiology of aminoglycoside resistance in Acinetobacter spp. J Med Microbiol 47:455–462 10.1099/00222615-47-5-455. [PubMed] [DOI] [PubMed] [Google Scholar]
- 38.Liou GF, Yoshizawa S, Courvalin P, Galimand M. 2006. Aminoglycoside resistance by ArmA-mediated ribosomal 16S methylation in human bacterial pathogens. J Mol Biol 359:358–364 10.1016/j.jmb.2006.03.038. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Périchon B, Courvalin P, Galimand M. 2007. Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob Agents Chemother 51:2464–2469 10.1128/AAC.00143-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doi Y, Adams JM, Yamane K, Paterson DL. 2007. Identification of 16S rRNA methylase-producing Acinetobacter baumannii clinical strains in North America. Antimicrob Agents Chemother 51:4209–4210 10.1128/AAC.00560-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu YS, Zhou H, Yang Q, Chen YG, Li LJ. 2007. Widespread occurrence of aminoglycoside resistance due to ArmA methylase in imipenem-resistant Acinetobacter baumannii isolates in China. J Antimicrob Chemother 60:454–455 10.1093/jac/dkm208. [PubMed] [DOI] [PubMed] [Google Scholar]
- 42.Vila J, Ruiz J, Goñi P, Jimenez de Anta T. 1997. Quinolone-resistance mutations in the topoisomerase IV parC gene of Acinetobacter baumannii. J Antimicrob Chemother 39:757–762 10.1093/jac/39.6.757. [PubMed] [DOI] [PubMed] [Google Scholar]
- 43.Vila J, Ruiz J, Goñi P, Marcos A, Jimenez de Anta T. 1995. Mutation in the gyrA gene of quinolone-resistant clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 39:1201–1203 10.1128/AAC.39.5.1201. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houang ET, Chu YW, Lo WS, Chu KY, Cheng AF. 2003. Epidemiology of rifampin ADP-ribosyltransferase (arr-2) and metallo-beta-lactamase (blaIMP-4) gene cassettes in class 1 integrons in Acinetobacter strains isolated from blood cultures in 1997 to 2000. Antimicrob Agents Chemother 47:1382–1390 10.1128/AAC.47.4.1382-1390.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother 53:3628–3634 10.1128/AAC.00284-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peleg AY, Adams J, Paterson DL. 2007. Tigecycline efflux as a mechanism for nonsusceptibility in Acinetobacter baumannii. Antimicrob Agents Chemother 51:2065–2069 10.1128/AAC.01198-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freitas AR, Novais C, Correia R, Monteiro M, Coque TM, Peixe L. 2011. Non-susceptibility to tigecycline in enterococci from hospitalised patients, food products and community sources. Int J Antimicrob Agents 38:174–176 10.1016/j.ijantimicag.2011.04.014. [PubMed] [DOI] [PubMed] [Google Scholar]
- 48.Ewers C, Klotz P, Leidner U, Stamm I, Prenger-Berninghoff E, Göttig S, Semmler T, Scheufen S. 2017. OXA-23 and ISAba1-OXA-66 class D β-lactamases in Acinetobacter baumannii isolates from companion animals. Int J Antimicrob Agents 49:37–44 10.1016/j.ijantimicag.2016.09.033. [PubMed] [DOI] [PubMed] [Google Scholar]
- 49.Nigro SJ, Hall RM. 2016. Structure and context of Acinetobacter transposons carrying the oxa23 carbapenemase gene. J Antimicrob Chemother 71:1135–1147 10.1093/jac/dkv440. [PubMed] [DOI] [PubMed] [Google Scholar]
- 50.Iacono M, Villa L, Fortini D, Bordoni R, Imperi F, Bonnal RJ, Sicheritz-Ponten T, De Bellis G, Visca P, Cassone A, Carattoli A. 2008. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob Agents Chemother 52:2616–2625 10.1128/AAC.01643-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaikh F, Spence RP, Levi K, Ou HY, Deng Z, Towner KJ, Rajakumar K. 2009. ATPase genes of diverse multidrug-resistant Acinetobacter baumannii isolates frequently harbour integrated DNA. J Antimicrob Chemother 63:260–264 10.1093/jac/dkn481. [PubMed] [DOI] [PubMed] [Google Scholar]
- 52.Poirel L, Berçot B, Millemann Y, Bonnin RA, Pannaux G, Nordmann P. 2012. Carbapenemase-producing Acinetobacter spp. in cattle, France. Emerg Infect Dis 18:523–525 10.3201/eid1803.111330. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Wu C, Zhang Q, Qi J, Liu H, Wang Y, He T, Ma L, Lai J, Shen Z, Liu Y, Shen J. 2012. Identification of New Delhi metallo-β-lactamase 1 in Acinetobacter lwoffii of food animal origin. PLoS One 7:e37152 10.1371/journal.pone.0037152. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang WJ, Lu Z, Schwarz S, Zhang RM, Wang XM, Si W, Yu S, Chen L, Liu S. 2013. Complete sequence of the bla(NDM-1)-carrying plasmid pNDM-AB from Acinetobacter baumannii of food animal origin. J Antimicrob Chemother 68:1681–1682 10.1093/jac/dkt066. [PubMed] [DOI] [PubMed] [Google Scholar]
- 55.Brahmi S, Touati A, Cadière A, Djahmi N, Pantel A, Sotto A, Lavigne JP, Dunyach-Remy C. 2016. First description of two sequence type 2 Acinetobacter baumannii isolates carrying OXA-23 carbapenemase in Pagellus acarne fished from the Mediterranean Sea near Bejaia, Algeria. Antimicrob Agents Chemother 60:2513–2515 10.1128/AAC.02384-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al Bayssari C, Dabboussi F, Hamze M, Rolain JM. 2015. Emergence of carbapenemase-producing Pseudomonas aeruginosa and Acinetobacter baumannii in livestock animals in Lebanon. J Antimicrob Chemother 70:950–951 10.1093/jac/dku469. [PubMed] [DOI] [PubMed] [Google Scholar]
- 57.Pailhoriès H, Kempf M, Belmonte O, Joly-Guillou ML, Eveillard M. 2016. First case of OXA-24-producing Acinetobacter baumannii in cattle from Reunion Island, France. Int J Antimicrob Agents 48:763–764 10.1016/j.ijantimicag.2016.09.005. [PubMed] [DOI] [PubMed] [Google Scholar]
- 58.Klotz P, Göttig S, Leidner U, Semmler T, Scheufen S, Ewers C. 2017. Carbapenem-resistance and pathogenicity of bovine Acinetobacter indicus-like isolates. PLoS One 12:e0171986 10.1371/journal.pone.0171986. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smet A, Boyen F, Pasmans F, Butaye P, Martens A, Nemec A, Deschaght P, Vaneechoutte M, Haesebrouck F. 2012. OXA-23-producing Acinetobacter species from horses: a public health hazard? J Antimicrob Chemother 67:3009–3010 10.1093/jac/dks311. [PubMed] [DOI] [PubMed] [Google Scholar]
- 60.Pomba C, Endimiani A, Rossano A, Saial D, Couto N, Perreten V. 2014. First report of OXA-23-mediated carbapenem resistance in sequence type 2 multidrug-resistant Acinetobacter baumannii associated with urinary tract infection in a cat. Antimicrob Agents Chemother 58:1267–1268 10.1128/AAC.02527-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hérivaux A, Pailhoriès H, Quinqueneau C, Lemarié C, Joly-Guillou ML, Ruvoen N, Eveillard M, Kempf M. 2016. First report of carbapenemase-producing Acinetobacter baumannii carriage in pets from the community in France. Int J Antimicrob Agents 48:220–221 10.1016/j.ijantimicag.2016.03.012. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.Kimura Y, Miyamoto T, Aoki K, Ishii Y, Harada K, Watarai M, Hatoya S. 2017. Analysis of IMP-1 type metallo-β-lactamase-producing Acinetobacter radioresistens isolated from companion animals. J Infect Chemother 23:655–657 10.1016/j.jiac.2017.03.011. [PubMed] [DOI] [PubMed] [Google Scholar]
- 63.Silby MW, Winstanley C, Godfrey SAC, Levy SB, Jackson RW. 2011. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35:652–680 10.1111/j.1574-6976.2011.00269.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 64.Argudín MA, Deplano A, Meghraoui A, Dodémont M, Heinrichs A, Denis O, Nonhoff C, Roisin S. 2017. Bacteria from animals as a pool of antimicrobial resistance genes. Antibiotics (Basel) 6:6 10.3390/antibiotics6020012. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Brien HE, Desveaux D, Guttman DS. 2011. Next-generation genomics of Pseudomonas syringae. Curr Opin Microbiol 14:24–30 10.1016/j.mib.2010.12.007. [PubMed] [DOI] [PubMed] [Google Scholar]
- 66.Wu X, Monchy S, Taghavi S, Zhu W, Ramos J, van der Lelie D. 2011. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbiol Rev 35:299–323 10.1111/j.1574-6976.2010.00249.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lalucat J, Bennasar A, Bosch R, García-Valdés E, Palleroni NJ. 2006. Biology of Pseudomonas stutzeri. Microbiol Mol Biol Rev 70:510–547 10.1128/MMBR.00047-05. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walker TS, Bais HP, Déziel E, Schweizer HP, Rahme LG, Fall R, Vivanco JM. 2004. Pseudomonas aeruginosa-plant root interactions: pathogenicity, biofilm formation, and root exudation. Plant Physiol 134:320–331 10.1104/pp.103.027888. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davies JC. 2002. Pseudomonas aeruginosa in cystic fibrosis: pathogenesis and persistence. Paediatr Respir Rev 3:128–134 10.1016/S1526-0550(02)00003-3. [DOI] [PubMed] [Google Scholar]
- 70.Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2:1051–1060 10.1016/S1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 71.Hariharan H, Coles M, Poole D, Lund L, Page R. 2006. Update on antimicrobial susceptibilities of bacterial isolates from canine and feline otitis externa. Can Vet J 47:253–255. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 72.Petersen AD, Walker RD, Bowman MM, Schott HC II, Rosser EJ Jr. 2002. Frequency of isolation and antimicrobial susceptibility patterns of Staphylococcus intermedius and Pseudomonas aeruginosa isolates from canine skin and ear samples over a 6-year period (1992–1997). J Am Anim Hosp Assoc 38:407–413 10.5326/0380407. [PubMed] [DOI] [PubMed] [Google Scholar]
- 73.Rubin J, Walker RD, Blickenstaff K, Bodeis-Jones S, Zhao S. 2008. Antimicrobial resistance and genetic characterization of fluoroquinolone resistance of Pseudomonas aeruginosa isolated from canine infections. Vet Microbiol 131:164–172 10.1016/j.vetmic.2008.02.018. [PubMed] [DOI] [PubMed] [Google Scholar]
- 74.Mekić S, Matanović K, Šeol B. 2011. Antimicrobial susceptibility of Pseudomonas aeruginosa isolates from dogs with otitis externa. Vet Rec 169:125 10.1136/vr.d2393. [PubMed] [DOI] [PubMed] [Google Scholar]
- 75.ANSES. 2016. Résapath - Réseau d’épidémiosurveillance de l’antibiorésistance des bactéries pathogènes animales, bilan 2015, Lyon et Ploufragan-Plouzané, France, November 2016. [Google Scholar]
- 76.Colombini S, Merchant SR, Hosgood G. 2000. Microbial flora and antimicrobial susceptibility patterns from dogs with otitis media. Vet Dermatol 11:235–239 10.1046/j.1365-3164.2000.00191.x. [DOI] [Google Scholar]
- 77.Done SH. 1974. Pseudomonas aeruginosa infection in the skin of a dog: a case report. Br Vet J 130:lxviii–lxix 10.1016/S0007-1935(17)35852-9. [PubMed] [DOI] [PubMed] [Google Scholar]
- 78.Hillier A, Alcorn JR, Cole LK, Kowalski JJ. 2006. Pyoderma caused by Pseudomonas aeruginosa infection in dogs: 20 cases. Vet Dermatol 17:432–439 10.1111/j.1365-3164.2006.00550.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 79.Wilson DJ, Baldwin TJ, Whitehouse CH, Hullinger G. 2015. Causes of mortality in farmed mink in the Intermountain West, North America. J Vet Diagn Invest 27:470–475 10.1177/1040638715586438. [PubMed] [DOI] [PubMed] [Google Scholar]
- 80.Shimizu T, Homma JY, Aoyama T, Onodera T, Noda H. 1974. Virulence of Pseudomonas aeruginosa and spontaneous spread of pseudomonas pneumonia in a mink ranch. Infect Immun 10:16–20. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Farrell RK, Leader RW, Gorham JR. 1958. An outbreak of hemorrhagic pneumonia in mink; a case report. Cornell Vet 48:378–384. [PubMed] [PubMed] [Google Scholar]
- 82.Daniel RC, O’Boyle D, Marek MS, Frost AJ. 1982. A survey of clinical mastitis in South-East Queensland dairy herds. Aust Vet J 58:143–147 10.1111/j.1751-0813.1982.tb00625.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 83.Bradley AJ, Leach KA, Breen JE, Green LE, Green MJ. 2007. Survey of the incidence and aetiology of mastitis on dairy farms in England and Wales. Vet Rec 160:253–257 10.1136/vr.160.8.253. [PubMed] [DOI] [PubMed] [Google Scholar]
- 84.Persson Y, Nyman AK, Grönlund-Andersson U. 2011. Etiology and antimicrobial susceptibility of udder pathogens from cases of subclinical mastitis in dairy cows in Sweden. Acta Vet Scand 53:36 10.1186/1751-0147-53-36. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Botrel MA, Haenni M, Morignat E, Sulpice P, Madec JY, Calavas D. 2010. Distribution and antimicrobial resistance of clinical and subclinical mastitis pathogens in dairy cows in Rhône-Alpes, France. Foodborne Pathog Dis 7:479–487 10.1089/fpd.2009.0425. [DOI] [PubMed] [Google Scholar]
- 86.Nam HM, Kim JM, Lim SK, Jang KC, Jung SC. 2010. Infectious aetiologies of mastitis on Korean dairy farms during 2008. Res Vet Sci 88:372–374 10.1016/j.rvsc.2009.12.008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 87.Sela S, Hammer-Muntz O, Krifucks O, Pinto R, Weisblit L, Leitner G. 2007. Phenotypic and genotypic characterization of Pseudomonas aeruginosa strains isolated from mastitis outbreaks in dairy herds. J Dairy Res 74:425–429 10.1017/S0022029907002610. [PubMed] [DOI] [PubMed] [Google Scholar]
- 88.Daly M, Power E, Björkroth J, Sheehan P, O’Connell A, Colgan M, Korkeala H, Fanning S. 1999. Molecular analysis of Pseudomonas aeruginosa: epidemiological investigation of mastitis outbreaks in Irish dairy herds. Appl Environ Microbiol 65:2723–2729. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McLennan MW, Kelly WR, O’Boyle D. 1997. Pseudomonas mastitis in a dairy herd. Aust Vet J 75:790–792 10.1111/j.1751-0813.1997.tb15652.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.Osborne AD, Armstrong K, Catrysse NH, Butler G, Versavel L. 1981. An outbreak of Pseudomonas mastitis in dairy cows. Can Vet J 22:215–216. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 91.Anderson B, Barton M, Corbould A, Dunford PJ, Elliott J, Leis T, Nicholls TJ, Sharman M, Stephenson GM. 1979. Pseudomonas aeruginosa mastitis due to contamination of an antibiotic preparation used in dry-cow therapy. Aust Vet J 55:90–91 10.1111/j.1751-0813.1979.tb15179.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 92.Kirk J, Mellenberger R. 2016. Mastitis control program for Pseudomonas mastitis in dairy cows. Purdue Dairy Page. https://www.extension.purdue.edu/dairy/health/hlthpub_mastitis.htm.
- 93.Yeruham I, Elad D, Avidar Y, Goshen T, Asis E. 2004. Four-year survey of urinary tract infections in calves in Israel. Vet Rec 154:204–206 10.1136/vr.154.7.204. [PubMed] [DOI] [PubMed] [Google Scholar]
- 94.Rice LB. 2008. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis 197:1079–1081 10.1086/533452. [PubMed] [DOI] [PubMed] [Google Scholar]
- 95.CA-SFM/EUCAST. 2016. Comité de l’antibiogramme de la Société Française de Microbiologie. http://www.sfm-microbiologie.org/page/page/showpage/page_id/90.html.
- 96.Breidenstein EB, de la Fuente-Núñez C, Hancock RE. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426 10.1016/j.tim.2011.04.005. [PubMed] [DOI] [PubMed] [Google Scholar]
- 97.Hancock RE. 1998. Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative Gram-negative bacteria. Clin Infect Dis 27(Suppl 1):S93–S99 10.1086/514909. [PubMed] [DOI] [PubMed] [Google Scholar]
- 98.Li XZ, Plésiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28:337–418 10.1128/CMR.00117-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Girlich D, Naas T, Nordmann P. 2004. Biochemical characterization of the naturally occurring oxacillinase OXA-50 of Pseudomonas aeruginosa. Antimicrob Agents Chemother 48:2043–2048 10.1128/AAC.48.6.2043-2048.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Masuda N, Gotoh N, Ishii C, Sakagawa E, Ohya S, Nishino T. 1999. Interplay between chromosomal beta-lactamase and the MexAB-OprM efflux system in intrinsic resistance to beta-lactams in Pseudomonas aeruginosa. Antimicrob Agents Chemother 43:400–402. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lodge JM, Minchin SD, Piddock LJ, Busby SJ. 1990. Cloning, sequencing and analysis of the structural gene and regulatory region of the Pseudomonas aeruginosa chromosomal ampC beta-lactamase. Biochem J 272:627–631 10.1042/bj2720627. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Davey ME, O’toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64:847–867 10.1128/MMBR.64.4.847-867.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.de Kievit TR. 2009. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ Microbiol 11:279–288 10.1111/j.1462-2920.2008.01792.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 104.Lequette Y, Greenberg EP. 2005. Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J Bacteriol 187:37–44 10.1128/JB.187.1.37-44.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Harmsen M, Yang L, Pamp SJ, Tolker-Nielsen T. 2010. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol Med Microbiol 59:253–268 10.1111/j.1574-695X.2010.00690.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 106.Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol 68:223–240 10.1111/j.1365-2958.2008.06152.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 107.Jeannot K, Elsen S, Köhler T, Attree I, van Delden C, Plésiat P. 2008. Resistance and virulence of Pseudomonas aeruginosa clinical strains overproducing the MexCD-OprJ efflux pump. Antimicrob Agents Chemother 52:2455–2462 10.1128/AAC.01107-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Potron A, Poirel L, Nordmann P. 2015. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents 45:568–585 10.1016/j.ijantimicag.2015.03.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 109.Oliphant CM, Green GM. 2002. Quinolones: a comprehensive review. Am Fam Physician 65:455–464. [PubMed] [PubMed] [Google Scholar]
- 110.Lin D, Foley SL, Qi Y, Han J, Ji C, Li R, Wu C, Shen J, Wang Y. 2012. Characterization of antimicrobial resistance of Pseudomonas aeruginosa isolated from canine infections. J Appl Microbiol 113:16–23 10.1111/j.1365-2672.2012.05304.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 111.Leigue L, Montiani-Ferreira F, Moore BA. 2016. Antimicrobial susceptibility and minimal inhibitory concentration of Pseudomonas aeruginosa isolated from septic ocular surface disease in different animal species. Open Vet J 6:215–222 10.4314/ovj.v6i3.9. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Aires JR, Köhler T, Nikaido H, Plésiat P. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother 43:2624–2628. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Arais LR, Barbosa AV, Carvalho CA, Cerqueira AM. 2016. Antimicrobial resistance, integron carriage, and gyrA and gyrB mutations in Pseudomonas aeruginosa isolated from dogs with otitis externa and pyoderma in Brazil. Vet Dermatol 27:113-7e31. 10.1111/vde.12290. [PubMed] [DOI] [PubMed] [Google Scholar]
- 114.Ludwig C, de Jong A, Moyaert H, El Garch F, Janes R, Klein U, Morrissey I, Thiry J, Youala M. 2016. Antimicrobial susceptibility monitoring of dermatological bacterial pathogens isolated from diseased dogs and cats across Europe (ComPath results). J Appl Microbiol 121:1254–1267 10.1111/jam.13287. [PubMed] [DOI] [PubMed] [Google Scholar]
- 115.Werckenthin C, Alesík E, Grobbel M, Lübke-Becker A, Schwarz S, Wieler LH, Wallmann J. 2007. Antimicrobial susceptibility of Pseudomonas aeruginosa from dogs and cats as well as Arcanobacterium pyogenes from cattle and swine as determined in the BfT-GermVet monitoring program 2004–2006. Berl Munch Tierarztl Wochenschr 120:412–422. [PubMed] [PubMed] [Google Scholar]
- 116.Seol B, Naglić T, Madić J, Bedeković M. 2002. In vitro antimicrobial susceptibility of 183 Pseudomonas aeruginosa strains isolated from dogs to selected antipseudomonal agents. J Vet Med B Infect Dis Vet Public Health 49:188–192 10.1046/j.1439-0450.2002.00548.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 117.Jacoby GA. 2005. Mechanisms of resistance to quinolones. Clin Infect Dis 41(Suppl 2):S120–S126 10.1086/428052. [PubMed] [DOI] [PubMed] [Google Scholar]
- 118.Mouneimné H, Robert J, Jarlier V, Cambau E. 1999. Type II topoisomerase mutations in ciprofloxacin-resistant strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother 43:62–66. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Matsumoto M, Shigemura K, Shirakawa T, Nakano Y, Miyake H, Tanaka K, Kinoshita S, Arakawa S, Kawabata M, Fujisawa M. 2012. Mutations in the gyrA and parC genes and in vitro activities of fluoroquinolones in 114 clinical isolates of Pseudomonas aeruginosa derived from urinary tract infections and their rapid detection by denaturing high-performance liquid chromatography. Int J Antimicrob Agents 40:440–444 10.1016/j.ijantimicag.2012.06.021. [PubMed] [DOI] [PubMed] [Google Scholar]
- 120.Li XZ, Nikaido H, Poole K. 1995. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother 39:1948–1953 10.1128/AAC.39.9.1948. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. 2000. Contribution of the MexX-MexY-oprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:2242–2246 10.1128/AAC.44.9.2242-2246.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Le Thomas I, Couetdic G, Clermont O, Brahimi N, Plésiat P, Bingen E. 2001. In vivo selection of a target/efflux double mutant of Pseudomonas aeruginosa by ciprofloxacin therapy. J Antimicrob Chemother 48:553–555 10.1093/jac/48.4.553. [PubMed] [DOI] [PubMed] [Google Scholar]
- 123.Beinlich KL, Chuanchuen R, Schweizer HP. 2001. Contribution of multidrug efflux pumps to multiple antibiotic resistance in veterinary clinical isolates of Pseudomonas aeruginosa. FEMS Microbiol Lett 198:129–134 10.1111/j.1574-6968.2001.tb10631.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 124.Haenni M, Hocquet D, Ponsin C, Cholley P, Guyeux C, Madec JY, Bertrand X. 2015. Population structure and antimicrobial susceptibility of Pseudomonas aeruginosa from animal infections in France. BMC Vet Res 11:9 10.1186/s12917-015-0324-x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Morita Y, Tomida J, Kawamura Y. 2012. MexXY multidrug efflux system of Pseudomonas aeruginosa. Front Microbiol 3:408 10.3389/fmicb.2012.00408. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chuanchuen R, Wannaprasat W, Ajariyakhajorn K, Schweizer HP. 2008. Role of the MexXY multidrug efflux pump in moderate aminoglycoside resistance in Pseudomonas aeruginosa isolates from Pseudomonas mastitis. Microbiol Immunol 52:392–398 10.1111/j.1348-0421.2008.00051.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 127.Poonsuk K, Chuanchuen R. 2012. Contribution of the MexXY multidrug efflux pump and other chromosomal mechanisms on aminoglycoside resistance in Pseudomonas aeruginosa isolates from canine and feline infections. J Vet Med Sci 74:1575–1582 10.1292/jvms.12-0239. [PubMed] [DOI] [PubMed] [Google Scholar]
- 128.Poole K. 2005. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:479–487 10.1128/AAC.49.2.479-487.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yokoyama K, Doi Y, Yamane K, Kurokawa H, Shibata N, Shibayama K, Yagi T, Kato H, Arakawa Y. 2003. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 362:1888–1893 10.1016/S0140-6736(03)14959-8. [DOI] [PubMed] [Google Scholar]
- 130.Doi Y, de Oliveira Garcia D, Adams J, Paterson DL. 2007. Coproduction of novel 16S rRNA methylase RmtD and metallo-beta-lactamase SPM-1 in a panresistant Pseudomonas aeruginosa isolate from Brazil. Antimicrob Agents Chemother 51:852–856 10.1128/AAC.01345-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li J, Zou M, Dou Q, Hu Y, Wang H, Yan Q, Liu WE. 2016. Characterization of clinical extensively drug-resistant Pseudomonas aeruginosa in the Hunan province of China. Ann Clin Microbiol Antimicrob 15:35 10.1186/s12941-016-0148-y. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jeannot K, Bolard A, Plésiat P. 2017. Resistance to polymyxins in Gram-negative organisms. Int J Antimicrob Agents 49:526–535 10.1016/j.ijantimicag.2016.11.029. [PubMed] [DOI] [PubMed] [Google Scholar]
- 133.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 134.Lee JY, Park YK, Chung ES, Na IY, Ko KS. 2016. Evolved resistance to colistin and its loss due to genetic reversion in Pseudomonas aeruginosa. Sci Rep 6:25543 10.1038/srep25543. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Martis N, Leroy S, Blanc V. 2014. Colistin in multi-drug resistant Pseudomonas aeruginosa blood-stream infections: a narrative review for the clinician. J Infect 69:1–12 10.1016/j.jinf.2014.03.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 136.Muller C, Plésiat P, Jeannot K. 2011. A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and β-lactams in Pseudomonas aeruginosa. Antimicrob Agents Chemother 55:1211–1221 10.1128/AAC.01252-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Moskowitz SM, Brannon MK, Dasgupta N, Pier M, Sgambati N, Miller AK, Selgrade SE, Miller SI, Denton M, Conway SP, Johansen HK, Høiby N. 2012. PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 56:1019–1030 10.1128/AAC.05829-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gutu AD, Sgambati N, Strasbourger P, Brannon MK, Jacobs MA, Haugen E, Kaul RK, Johansen HK, Høiby N, Moskowitz SM. 2013. Polymyxin resistance of Pseudomonas aeruginosa phoQ mutants is dependent on additional two-component regulatory systems. Antimicrob Agents Chemother 57:2204–2215 10.1128/AAC.02353-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang Y, Wang X, Schwarz S, Zhang R, Lei L, Liu X, Lin D, Shen J. 2014. IMP-45-producing multidrug-resistant Pseudomonas aeruginosa of canine origin. J Antimicrob Chemother 69:2579–2581 10.1093/jac/dku133. [PubMed] [DOI] [PubMed] [Google Scholar]
- 140.Haenni M, Bour M, Châtre P, Madec JY, Plésiat P, Jeannot K. 2017. Resistance of animal strains of Pseudomonas aeruginosa to carbapenems. Front Microbiol 8:1847 10.3389/fmicb.2017.01847. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Salomonsen CM, Themudo GE, Jelsbak L, Molin S, Høiby N, Hammer AS. 2013. Typing of Pseudomonas aeruginosa from hemorrhagic pneumonia in mink (Neovison vison). Vet Microbiol 163:103–109 10.1016/j.vetmic.2012.12.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 142.Hammer AS, Pedersen K, Andersen TH, Jørgensen JC, Dietz HH. 2003. Comparison of Pseudomonas aeruginosa isolates from mink by serotyping and pulsed-field gel electrophoresis. Vet Microbiol 94:237–243 10.1016/S0378-1135(03)00103-2. [DOI] [PubMed] [Google Scholar]
- 143.Gu J, Li X, Yang M, Du C, Cui Z, Gong P, Xia F, Song J, Zhang L, Li J, Yu C, Sun C, Feng X, Lei L, Han W. 2016. Therapeutic effect of Pseudomonas aeruginosa phage YH30 on mink hemorrhagic pneumonia. Vet Microbiol 190:5–11 10.1016/j.vetmic.2016.03.016. [PubMed] [DOI] [PubMed] [Google Scholar]
- 144.Pedersen K, Hammer AS, Sørensen CM, Heuer OE. 2009. Usage of antimicrobials and occurrence of antimicrobial resistance among bacteria from mink. Vet Microbiol 133:115–122 10.1016/j.vetmic.2008.06.005. [PubMed] [DOI] [PubMed] [Google Scholar]
- 145.Jensen VF, Sommer HM, Struve T, Clausen J, Chriél M. 2016. Factors associated with usage of antimicrobials in commercial mink (Neovison vison) production in Denmark. Prev Vet Med 126:170–182 10.1016/j.prevetmed.2016.01.023. [PubMed] [DOI] [PubMed] [Google Scholar]
- 146.Qi J, Li L, Du Y, Wang S, Wang J, Luo Y, Che J, Lu J, Liu H, Hu G, Li J, Gong Y, Wang G, Hu M, Shiganyan, Liu Y. 2014. The identification, typing, and antimicrobial susceptibility of Pseudomonas aeruginosa isolated from mink with hemorrhagic pneumonia. Vet Microbiol 170:456–461 10.1016/j.vetmic.2014.02.025. [PubMed] [DOI] [PubMed] [Google Scholar]
- 147.Nikolaisen NK, Lassen DCK, Chriél M, Larsen G, Jensen VF, Pedersen K. 2017. Antimicrobial resistance among pathogenic bacteria from mink (Neovison vison) in Denmark. Acta Vet Scand 59:60 10.1186/s13028-017-0328-6. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Giebink GS. 1999. Otitis media: the chinchilla model. Microb Drug Resist 5:57–72 10.1089/mdr.1999.5.57. [PubMed] [DOI] [PubMed] [Google Scholar]
- 149.Wideman WL. 2006. Pseudomonas aeruginosa otitis media and interna in a chinchilla ranch. Can Vet J 47:799–800. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 150.Hirakawa Y, Sasaki H, Kawamoto E, Ishikawa H, Matsumoto T, Aoyama N, Kawasumi K, Amao H. 2010. Prevalence and analysis of Pseudomonas aeruginosa in chinchillas. BMC Vet Res 6:52 10.1186/1746-6148-6-52. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Agersø Y, Sandvang D. 2005. Class 1 integrons and tetracycline resistance genes in alcaligenes, arthrobacter, and Pseudomonas spp. isolated from pigsties and manured soil. Appl Environ Microbiol 71:7941–7947 10.1128/AEM.71.12.7941-7947.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.de Oliveira KM, dos S Júlio PD, Grisolia AB. 2013. Antimicrobial susceptibility profile of Pseudomonas spp. isolated from a swine slaughterhouse in Dourados, Mato Grosso do Sul State, Brazil. Rev Argent Microbiol 45:57–60. [PubMed] [PubMed] [Google Scholar]
- 153.Zhang R, Liu Z, Li J, Lei L, Yin W, Li M, Wu C, Walsh TR, Wang Y, Wang S, Wu Y. 2017. Presence of VIM-positive Pseudomonas species in chickens and their surrounding environment. Antimicrob Agents Chemother 61:61 10.1128/AAC.00167-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ohnishi M, Sawada T, Hirose K, Sato R, Hayashimoto M, Hata E, Yonezawa C, Kato H. 2011. Antimicrobial susceptibilities and bacteriological characteristics of bovine Pseudomonas aeruginosa and Serratia marcescens isolates from mastitis. Vet Microbiol 154:202–207 10.1016/j.vetmic.2011.06.023. [PubMed] [DOI] [PubMed] [Google Scholar]
- 155.Metcalf ES. 2001. The role of international transport of equine semen on disease transmission. Anim Reprod Sci 68:229–237 10.1016/S0378-4320(01)00159-2. [DOI] [PubMed] [Google Scholar]
- 156.Atherton JG, Pitt TL. 1982. Types of Pseudomonas aeruginosa isolated from horses. Equine Vet J 14:329–332 10.1111/j.2042-3306.1982.tb02446.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 157.Tazumi A, Maeda Y, Buckley T, Millar B, Goldsmith C, Dooley J, Elborn J, Matsuda M, Moore J. 2009. Molecular epidemiology of clinical isolates of Pseudomonas aeruginosa isolated from horses in Ireland. Ir Vet J 62:456–459 10.1186/2046-0481-62-7-456. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kidd TJ, Gibson JS, Moss S, Greer RM, Cobbold RN, Wright JD, Ramsay KA, Grimwood K, Bell SC. 2011. Clonal complex Pseudomonas aeruginosa in horses. Vet Microbiol 149:508–512 10.1016/j.vetmic.2010.11.030. [PubMed] [DOI] [PubMed] [Google Scholar]


