ABSTRACT
Members of the highly heterogeneous family Pasteurellaceae cause a wide variety of diseases in humans and animals. Antimicrobial agents are the most powerful tools to control such infections. However, the acquisition of resistance genes, as well as the development of resistance-mediating mutations, significantly reduces the efficacy of the antimicrobial agents. This article gives a brief description of the role of selected members of the family Pasteurellaceae in animal infections and of the most recent data on the susceptibility status of such members. Moreover, a review of the current knowledge of the genetic basis of resistance to antimicrobial agents is included, with particular reference to resistance to tetracyclines, β-lactam antibiotics, aminoglycosides/aminocyclitols, folate pathway inhibitors, macrolides, lincosamides, phenicols, and quinolones. This article focusses on the genera of veterinary importance for which sufficient data on antimicrobial susceptibility and the detection of resistance genes are currently available (Pasteurella, Mannheimia, Actinobacillus, Haemophilus, and Histophilus). Additionally, the role of plasmids, transposons, and integrative and conjugative elements in the spread of the resistance genes within and beyond the aforementioned genera is highlighted to provide insight into horizontal dissemination, coselection, and persistence of antimicrobial resistance genes. The article discusses the acquisition of diverse resistance genes by the selected Pasteurellaceae members from other Gram-negative or maybe even Gram-positive bacteria. Although the susceptibility status of these members still looks rather favorable, monitoring of their antimicrobial susceptibility is required for early detection of changes in the susceptibility status and the newly acquired/developed resistance mechanisms.
THE FAMILY PASTEURELLACEAE AND ITS ROLE IN ANIMAL INFECTIONS
The family Pasteurellaceae (order Pasteurellales, class Gammaproteobacteria) comprises a highly heterogeneous group of Gram-negative bacteria. Evaluation by sequence comparison of housekeeping genes, 16s rRNA gene sequence-based phylogenetic analysis, DNA-DNA hybridization, and analysis of the biochemical and physiological capacities has identified a number of distinct genetic and phenotypic groups (1, 2). As a consequence, the family Pasteurellaceae has undergone numerous reclassifications during the past years and currently (late 2017) contains 25 genera: Actinobacillus, Aggregatibacter, Avibacterium, Basfia, Bibersteinia, Bisgaardia, Chelonobacter, Cricetibacter, Frederiksenia, Gallibacterium, Haemophilus, Histophilus, Lonepinella, Mannheimia, Mesocricetibacter, Muribacter, Necropsobacter, Nicoletella, Otariodibacter, Pasteurella, Phocoenobacter, Testudinibacter, Ursidibacter, Vespertiliibacter, and Volucribacter (International Committee on Sytematics of Prokaryotes, http://www.the-icsp.org/taxa-covered-family-pasteurellaceae). A new genus, Rodentibacter, has recently been proposed. Rodentibacter pneumotropicus combinatio nova (comb. nov.), which will be reclassified from [Pasteurella] pneumotropica, with NCTC 8141T (also designated CCUG 12398T) as the type strain (3). The use of square brackets enclosing the genus name indicates that there is a proposal to reclassify the species to another genus or that the species has been shown not to be a member of the genus sensu stricto, as is the case for [Pasteurella] aerogenes, which is now excluded from Pasteurella sensu stricto based on genetic analysis (4). Additional information related to reclassification of genera may be found in references 1, 2, and 5. The use of quotation marks around “Actinobacillus porcitonsillarum” denotes that this is currently not a validated species name (5).
Whole-genome sequencing has become an important tool for the classification of members of the family Pasteurellaceae, providing a better understanding of the molecular evolution of isolates and their host specificities, niche preferences, pathogenic potential, and mechanisms of transfer and uptake of mobile genetic elements involved in antimicrobial (multi)resistance. Such knowledge can also be applied to improve/develop vaccines, diagnostic tests, disease control measures, and intervention strategies (2, 6–10).
This article deals with the genera which include pathogens of veterinary importance and will focus on the genera Pasteurella, Mannheimia, Actinobacillus, Haemophilus, and Histophilus, for which sufficient data on antimicrobial susceptibility and the detection of resistance genes are currently available. This article compiles the latest information on antimicrobial resistance, associated with the aforementioned genera, and the data published in previous book chapters (10, 11).
Many isolates of the genera Pasteurella, Mannheimia, Actinobacillus, Haemophilus, and Histophilus are commonly found on the mucous membranes of the respiratory tract and/or genital tract of reptiles, birds, and numerous mammals, including a wide variety of food-producing animals. Some species, e.g., Pasteurella multocida, may be found in many different hosts, including in humans, while others, such as Actinobacillus pleuropneumoniae and Mannheimia haemolytica (formerly [Pasteurella] haemolytica), have a narrow host range, being found primarily in pigs and ruminants, respectively (5).
P. multocida is the most relevant animal-pathogenic Pasteurella species. Various capsular types of P. multocida are known, some of which preferentially occur in connection with specific diseases in animals. For example, (i) capsular type A is the causative agent of pneumonia in several animal species including, but not limited to, cattle, sheep, and pigs; mastitis in sheep; snuffles in rabbits; and fowl cholera in poultry. (ii) Capsular types B and E are the causative agents of hemorrhagic septicemia of cattle and water buffaloes in Asia and Africa, respectively. (iii) Capsular type D isolates are mainly involved in atrophic rhinitis and pneumonia in swine (12, 13). (iv) Capsular type F isolates are mainly seen in poultry but may also be involved in fatal peritonitis in calves (14).
M. haemolytica comprises 12 capsular serotypes (A1, A2, A5 to A9, A12 to A14, A16, and A17). Serotypes A1 and A6 have been the ones most commonly associated with respiratory diseases in cattle (15) or in sheep (16), respectively.
A. pleuropneumoniae is the causative agent of porcine pleuropneumonia (13), which causes huge economic losses in the swine industry worldwide. Currently, two biovars (the NAD-dependent biovar 1 and the NAD-independent biovar 2) and 16 serovars of A. pleuropneumoniae are distinguished (17–19). Serovars 1, 5, 9, and 11 are considered to be more virulent than other serovars (20).
Several species of the genus Haemophilus also represent animal pathogens, with [Haemophilus] parasuis being of major economic importance. [H.] parasuis causes Glässer’s disease (also known as porcine polyserositis or infectious polyarthritis) in pigs, which is characterized by high fevers, polyserositis, polysynovitis, respiratory distress, and meningitis. More than 15 serotypes have been identified, with serotypes 1, 5, 12, 13, and 14 being thought to be the most virulent (21).
Histophilus somni (formerly [Haemophilus] somnus), Histophilus ovis, and Histophilus agni are also pathogens of cattle and sheep. H. somni is the etiological agent of thromboembolic meningoencephalitis in cattle. It has also been associated with various other diseases in sheep and diseases such as bronchopneumonia, necrotic laryngitis, myocarditis, arthritis, conjunctivitis, myositis, mastitis, and abortion (10).
All diseases in which Pasteurella, Mannheimia, Actinobacillus, Haemophilus, or Histophilus isolates act as primary pathogens commonly occur as peracute or acute forms and are accompanied by a high mortality rate, although subacute and chronic forms are also observed. As secondary pathogens, P. multocida and M. haemolytica play a major role in the final progression to severe bronchopneumonia and pleuropneumonia in cattle, sheep, and goats (22), as well as in enzootic pneumonia in calves (23) and progressive atrophic rhinitis of swine (24). Respiratory tract infections, in which bacteria such as P. multocida and M. haemolytica isolates are involved, are often multifactorial and polymicrobial diseases, with viruses and other bacteria, such as Mycoplasma spp. representing the primary pathogens (13, 22–24). Under certain environmental and/or management conditions (such as transport, marketing, change of feed, climate, or ventilation) which result in stress to the animals, especially in the presence of viruses and/or Mycoplasma spp. which may initiate damage to the host mucosal membranes, the bacterial pathogens can rapidly proliferate, resulting in high morbidity. Under conditions of low stress, the mortality rate may be low. As the amount of stress increases, however, the mortality rate also increases. Economic losses associated with acute pneumonic episodes are primarily due to increased costs in medications and retarded growth rates rather than mortality of the affected animals. Besides these major pathogens, various other members of the family Pasteurellaceae have been reported to be less frequently associated with diseases in humans and animals (10).
As seen for many other bacteria, members of the family Pasteurellaceae also respond to the selective pressure imposed by the use of antimicrobial agents by developing or acquiring resistance genes or resistance-mediating mutations. In this regard, mobile genetic elements, such as plasmids, transposons, or integrative and conjugative elements (ICEs) that carry resistance genes, play an important role in the dissemination of antimicrobial resistance, with members of the family Pasteurellaceae acting as donors and/or recipients. This article provides a brief description of the most recent data on the susceptibility status of selected members of the family Pasteurellaceae and an update on the current knowledge regarding genes and mutations conferring antimicrobial resistance among Pasteurellaceae of veterinary concern and on the dissemination, coselection, and persistence of such resistance genes.
ANTIMICROBIAL SUSCEPTIBILITY OF PASTEURELLA, MANNHEIMIA, ACTINOBACILLUS, HAEMOPHILUS, AND HISTOPHILUS
Although prudent use guidelines request identification of the causative pathogen and determination of its in vitro susceptibility prior to the onset of antimicrobial therapy, the generally acute nature of the diseases—and in veterinary medicine, the rapid spread of the causative pathogens within animal herds—often requires an immediate therapeutic intervention in which the initial choice of the antimicrobial agents may be revised on the basis of the results of the diagnostic tests. In this regard, two major aspects need to be considered: (i) the correct performance of in vitro susceptibility tests and (ii) representative data on the susceptibility status of members of the Pasteurellaceae.
Susceptibility Testing
In vitro antimicrobial susceptibility testing (AST) is performed to predict how a bacterium may respond to an antimicrobial agent in vivo (clinical response) or to monitor changes in susceptibility in relation to time and geographic location. In both instances, results may be reported qualitatively, e.g., susceptible, intermediate, or resistant (S-I-R), or quantitatively, e.g., as the minimal inhibitory concentration (MIC). When performing AST for surveillance purposes, the interpretive criteria are based on the bacterial population distributions relative to inhibition zone sizes and/or MIC values. Interpretive criteria for clinical consideration require the generation of a bacterium’s antibiogram in addition to knowledge of the pharmacokinetic parameters of the chosen drug in the target animal species and the pharmacodynamic parameters associated with the in vivo bacterium-antimicrobial agent-host interactions. In either situation, to ensure intra- and interlaboratory reproducibility, it is essential that standardized AST methods be used (25). Recently, it has been suggested that whole-genome sequencing for identification of resistance genes may be a useful adjunct to phenotypic testing, because it not only provides a reproducible means of analysis, but can provide added data for epidemiological purposes (9, 26, 27).
The results of in vitro AST ensure the most efficacious antimicrobial therapy by excluding antimicrobial agents to which the causative bacterial pathogen already shows resistance, or reduced susceptibility, under in vitro conditions. Correct performance of the tests is essential to most accurately predict the clinical response of the bacterium. Several members of the family Pasteurellaceae are classified as “fastidious organisms” since they require specific growth conditions, e.g., supplementation of the media with components essential for the growth of the bacteria. In this regard, performance of in vitro AST should follow standardized and internationally accepted rules. The in vitro AST of Pasteurella, Mannheimia, Actinobacillus, Haemophilus, and Histophilus is no exception. While many studies have reported in vitro susceptibility data for isolates of the these genera, obtained from animal sources in various parts of the world, there has been a notable absence of standardization in testing methods. In most cases, it is difficult to compare the results because of the use of different methods and breakpoints. The Clinical and Laboratory Standards Institute (CLSI) has published three documents, VET01-A4 (28), VET01-S (29), and VET06 (30), which provide the latest information on methods for in vitro susceptibility testing of P. multocida, Pasteurella spp. other than P. multocida, M. haemolytica, A. pleuropneumoniae, and H. somni. Because there is no CLSI-approved method for antimicrobial susceptibility testing of [H.] parasuis, methods recently developed and published should be followed when testing this organism (31, 32).
The CLSI documents VET01-S (29) and VET06 (30) also contain clinical breakpoints on the basis of which isolates are classified as susceptible, intermediate, or resistant. In contrast to epidemiological cutoff values, clinical breakpoints may predict the clinical outcome when the antimicrobial agent in question is dosed and administered as recommended (25, 33). Veterinary-specific clinical breakpoints applicable to bovine, canine, equine, feline, or porcine P. multocida, M. haemolytica, H. somni, and/or A. pleuropneumoniae are currently available for a number of antimicrobial agents, including ampicillin/amoxicillin, cefazolin, cefpodoxime, ceftiofur, chloramphenicol, danofloxacin, enrofloxacin, gamithromycin, florfenicol, gentamicin, penicillin G, pradofloxacin, spectinomycin, tetracycline, tiamulin, tildipirosin, tilmicosin, and/or tulathromycin (29, 34).
Susceptibility Status of Selected Pathogens of the Family Pasteurellaceae
In veterinary medicine, bacteria belonging to the family Pasteurellaceae are currently not included in most national antimicrobial susceptibility monitoring and surveillance programs. Only the German GERM-Vet program, which uses CLSI AST methodology and CLSI clinical breakpoints, included P. multocida and A. pleuropneumoniae from respiratory tract infections of swine (piglets, weaners, and adult swine), as well as P. multocida and M. haemolytica from cattle (calves and adult cattle) and M. haemolytica from sheep and goats (35) (Table 1). Moreover, P. multocida from dogs and cats suffering from either respiratory tract infections or infections of mouth, ear, or skin were collected between 2004 and 2006 in the BfT-GermVet monitoring program and analyzed for their in vitro susceptibility (36) (Table 1). In addition, Portis and coworkers (37) published susceptibility data collected in a large-scale surveillance program over a 10-year period (2000 to 2009) from the bovine respiratory disease pathogens M. haemolytica, P. multocida, and H. somni in the United States and Canada (Table 1). A comparison of the percentages of resistance between the respective P. multocida and M. haemolytica isolates from North America and Germany showed that among the German isolates resistance to newer antimicrobial agents, such as ceftiofur, florfenicol, enrofloxacin, and tulathromycin, is rarely detected, if at all. In contrast, the isolates from North America showed low levels of resistance (up to 11.6%) to florfenicol, enrofloxacin, and tulathromycin. However, all isolates were susceptible to ceftiofur. It should also be noted that none of the 72 P. multocida isolates from respiratory tract infections of dogs and cats investigated in the BfT-GermVet program exhibited resistance to any of the antimicrobial agents shown in Table 1. Only sulfonamide resistance was seen in 31 (43%) isolates (36).
TABLE 1.
Percentages of resistance of P. multocida, M. haemolytica, H. somni, and A. pleuropneumoniae isolates from different animal sources against selected antimicrobial agents
| Bacteria | Origin | Year(s) of isolation | No. of isolates | % resistant isolatesa | Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PEN | XNL | FLO | ENR | TET | TIL | TUL | |||||
| P. multocida | Cattle/USA, Canada | 2009 | 328 | 3.3 | 0.0 | 11.6 | 2.1 | 40.8 | 23.8 | 4.6 | 37 |
| Cattle/Germany | 2013 | 48 | 2.1 | 0.0 | 0.0 | 0.0 | 10.4 | 2.1 | 0.0 | 35 | |
| Adult swine/Germany | 2013 | 90 | 5.6 | 0.0 | 0.0 | 0.0 | 11.1 | 0.0 | 0.0 | 35 | |
| Weaner/Germany | 2013 | 25 | 0.0 | 0.0 | 0.0 | 0.0 | 8.0 | 0.0 | 0.0 | 35 | |
| Piglet/Germany | 2013 | 35 | 2.9 | 0.0 | 2.9 | 0.0 | 2.9 | 0.0 | 0.0 | 35 | |
| Dogs, cats/Germany | 2004–2006 | 72 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 36 | |
| M. haemolytica | Cattle/USA, Canada | 2009 | 304 | 27.3 | 0.0 | 8.6 | 6.6 | 43.7 | 27.3 | 8.9 | 37 |
| Calf/Germany | 2012–2013 | 63 | 11.1 | 0.0 | 1.6 | 0.0 | 22.2 | 1.6 | 0.0 | 35 | |
| Adult cattle/Germany | 2012–2013 | 35 | 8.8 | 0.0 | 0.0 | 0.0 | 0.0 | 2.9 | 2.9 | 35 | |
| Sheep, goat/Germany | 2012–2013 | 42 | 4.8 | 0.0 | 0.0 | 0.0 | 2.4 | 0.0 | 0.0 | 35 | |
| H. somni | Cattle/USA, Canada | 2009 | 174 | 4.5 | 0.0 | 1.7 | 7.4 | 42.5 | 18.4 | 10.9 | 37 |
| A. pleuropneumoniae | Swine/Germany | 2012 | 102 | 5.9 | 0.0 | 0.0 | 0.0 | 13.7 | 1.0 | 7.8 | 35 |
PEN, penicillin G; XNL, ceftiofur; FLO, florfenicol; ENR, enrofloxacin; TET, tetracycline; TIL, tilmicosin; TUL, tulathromycin.
In addition to these data from GERM-Vet (35), Bft-GermVet (36), and the aforementioned large-scale surveillance program (37), numerous other studies have dealt with the in vitro susceptibility of bovine, porcine, and avian Pasteurellaceae in different countries (e.g., 31, 38–43). Major problems arising from a comparison of the results of these studies are that (i) isolates have not been selected according to a defined sampling plan, (ii) different AST methods have been used, (iii) different interpretive criteria have been used for the evaluation of the results, and/or (iv) isolates may have not been investigated for their relatedness to prevent the inclusion of multiple members of the same clone in the test collection.
Additional studies that have determined the susceptibility to antimicrobial agents of Pasteurellaceae isolates from animal sources in different countries include the following. VetPath is a pan-European antibiotic susceptibility monitoring program collecting pathogens from diseased (but not antimicrobial-treated) cattle, pigs, and poultry. Two studies dealing with respiratory tract pathogens, including P. multocida and M. haemolytica from cattle and P. multocida and A. pleuropneumoniae from pigs, and covering the years 2002 to 2006 (38) and 2009 to 2012 (39) have been published. The results of these studies showed that, for most antimicrobial agents and pathogens, the percentages of resistance remained largely unchanged in the period from 2009 to 2012 compared to those of the period from 2002 to 2006. Moreover, these data also showed that resistance to amoxicillin/clavulanic acid, ceftiofur, enrofloxacin, florfenicol, tulathromycin, tiamulin, and tilmicosin was absent or <2%. A study conducted in Australia investigated P. multocida and A. pleuropneumoniae isolates from pigs with respiratory tract infections for their antimicrobial susceptibility (40). This study illustrated that Australian isolates of swine bacterial respiratory pathogens also exhibited low levels of resistance to antimicrobial agents commonly used in the pig industry. A study from Canada in which A. pleuropneumoniae isolates were tested for their resistance pheno- and genotypes revealed that all isolates were susceptible to ceftiofur, florfenicol, enrofloxacin, erythromycin, clindamycin, trimethoprim/sulfamethoxazole, and tilmicosin. A low level of resistance was observed toward tiamulin, penicillin, and ampicillin as well as danofloxacin, whereas the majority of the tested isolates were resistant to chlortetracycline (88.4%) and oxytetracycline (90.7%) (41). A large-scale study of 2,989 M. haemolytica isolates from feedlot cattle in Canada revealed that 87.8% of the isolates were pan-susceptible, whereas the percentages of resistant isolates varied between 0.0 and 4.5% for the antimicrobial agents tested (42). It should also be noted that a literature review of antimicrobial resistance in pathogens associated with bovine respiratory disease has recently been published (43).
MOLECULAR MECHANISMS OF ANTIMICROBIAL RESISTANCE IN PASTEURELLA, MANNHEIMIA, ACTINOBACILLUS, HAEMOPHILUS, AND HISTOPHILUS
The following subsections provide an update of the current status of resistance genes and resistance-mediating mutations known to occur in Pasteurellaceae. The focus will be on members of the genera Pasteurella, Mannheimia, Actinobacillus, Haemophilus, and Histophilus, which infect animals and for which data on the genetic basis of resistance are available (Table 2). As far as information is available, the association of the resistance genes with mobile genetic elements and their potential to spread across species and genus borders will be discussed.
TABLE 2.
Antimicrobial resistance genes and mutations identified in Pasteurella, Mannheimia, Actinobacillus, Haemophilus, and Histophilus isolates of veterinary importance
| Antimicrobial agent | Resistance gene or resistance-mediating mutation(s) | Protein specified by the resistance gene or mutation position | Detected ina | Referenceb/Accession numberc | ||||
|---|---|---|---|---|---|---|---|---|
| Pas | Man | Act | Hae | His | ||||
| Tetracycline | tet(A) | 12-TMS efflux protein | – | – | + | – | – | 72, 81 |
| tet(B) | 12-TMS efflux protein | + | + | + | + | – | 53, 66, 75, 81 | |
| tet(C) | 12-TMS efflux protein | – | – | + | – | – | 81 | |
| tet(G) | 12-TMS efflux protein | + | + | + | – | – | 64, 76, 81 | |
| tet(H) | 12-TMS efflux protein | + | + | + | – | + | 48, 49, 57, 61 | |
| tet(L) | 14-TMS efflux protein | + | + | + | – | – | 79, 81 | |
| tet(M) | Ribosome protective protein | + | – | + | – | – | 49, 81 | |
| tet(O) | Ribosome protective protein | + | – | + | – | – | 65, 81 | |
| Penicillins | bla CMY-2 | β-Lactmase | + | – | – | – | – | 90 |
| bla OXA-2 | β-Lactmase | + | + | – | – | + | 55, 57 | |
| bla PSE-1 | β-Lactmase | + | – | – | – | – | 76 | |
| bla ROB-1 | β-Lactmase | + | + | + | + | – | 85, 87, 88, 93 | |
| bla TEM-1 | β-Lactmase | + | – | – | – | – | 89 | |
| Streptomycin | strA | Aminoglycoside-3″-phosphotransferase | + | + | + | + | + | 55–57, 121, 194 |
| strB | Aminoglycoside-6-phosphotransferase | + | + | + | – | + | 55–57, 121 | |
| Streptomycin/spectinomycin | aadA1 | aminoglycoside-3″-adenyltransferase | + | – | – | – | – | 76 |
| aadA14 | aminoglycoside-3″-adenyltransferase | + | – | – | – | – | 132 | |
| aadA25 | aminoglycoside-3″-adenyltransferase | + | + | – | – | + | 55, 57 | |
| Spectinomycin | Mutation in 16S rRNA | C1192G | + | – | – | – | – | 143 |
| Mutation in rpsE coding for ribosomal protein S5 | 3-bp deletions resulting in S32I and loss of F33 or loss of K33 | + | – | – | – | – | 143 | |
| Kanamycin/neomycin | aphA1 | Aminoglycoside-3′-phosphotransferase | + | + | + | + | + | 55–57, 123; JN202624 |
| aphA3 | Aminoglycoside-3′-phosphotransferase | + | – | – | – | – | 135 | |
| Gentamicin | aacC2 | Aminoglycoside 3-N-acetyltransferase | – | – | + | + | – | 95; JN202624 |
| aacC4 | Aminoglycoside 3-N-acetyltransferase | – | – | + | – | – | 81 | |
| aadB | Aminoglycoside-2″-adenyltransferase | + | + | + | – | + | 55, 57, 81 | |
| Sulfonamides | sul2 | Sulfonamide-resistant dihydropteroate synthase | + | + | + | + | + | 55–57, 95; JN202624 |
| Trimethoprim | dfrA1 | Trimethoprim-resistant DHFR | + | – | – | – | – | 76 |
| dfrA14 | Trimethoprim-resistant DHFR | – | + | – | – | 122 | ||
| dfrA20 | Trimethoprim-resistant DHFR | + | – | – | – | – | 62 | |
| Macrolides | erm(T) | rRNA methylase | – | – | – | + | – | 154 |
| erm(A) | rRNA methylase | – | – | + | – | – | 70 | |
| erm(C) | rRNA methylase | – | – | + | – | – | 70 | |
| erm(42) | rRNA methylase | + | + | – | – | + | 57, 155, 157 | |
| Mutation in 23S rRNA | A2058G, A2059G | + | + | – | + | – | 153, 159 | |
| mrs(E)-mph(E) | Macrolide efflux protein and phospotransferase | + | + | – | – | + | 57, 155, 157 | |
| Lincosamides | lnu(C) | O-nucleotidyltransferase | – | – | – | + | – | 161 |
| Chloramphenicol | catA1 | Type A chloramphenicol acetyltransferase | + | + | – | – | – | 163 |
| catA3 | Type A chloramphenicol acetyltransferase | + | + | + | + | – | 95, 163; JN202624 | |
| catB2 | Type B chloramphenicol acetyltransferase | + | – | – | – | – | 76 | |
| Chloramphenicol/florfenicol | floR | 12-TMS efflux protein | + | + | + | + | + | 57, 73, 174, 179, 180 |
| (Fluoro)quinolones | Mutation in gyrA | G75S, S83F, S83I, S83R, S83V, S83Y, A84P, D87G, D87H, D87N, D87Y, D492G, D492V, G627E | + | + | + | + | – | 9, 55, 187, 189–192 |
| Mutation in gyrB | V211I, D254G | +d | – | – | + | – | 192 | |
| Mutation in parC | S73I, S73R, S80L, G83C, I84S, S85R, S85Y, E89K, Q227H, L379I, C578Y | + | + | + | + | – | 55, 187, 189, 190, 192 | |
| Mutation in parE | P440S, S459F, E461D, E461K, D479E, T551A | – | – | + | + | – | 190, 192 | |
| qnrA1 | Quinolone resistance protein | – | – | – | + | – | 191 | |
| qnrB6 | Quinolone resistance protein | – | – | – | + | – | 191 | |
| aac(6′)-Ib-cr | Quinolone resistance protein | – | – | – | + | – | 191 | |
| Streptothricin | sat2 | Streptothricin-acetyl-transferase | + | – | – | – | – | 130 |
Pas, Pasteurella; Man, Manheimia; Act, Actinobacillus; Hae, Haemophiluis; His, Histophilus; +, present; –, absent.
At least a reference or an accession number is included for each genus in which a specific gene or mutation is found. Additional information and references may be found in the text of this chapter.
Accession number was provided in case of unpblished data.
The mutations in gyrB were detected in Pasteurella isolates after passage on subinhibitory concentrations of fluoroquinolones.
Resistance to Tetracyclines
Tetracycline resistance is a highly heterogeneous property with more than 40 resistance genes known to date (http://faculty.washington.edu/marilynr/). In bacteria of the genera Pasteurella, Mannheimia, Actinobacillus, and Haemophilus, at least nine tetracycline resistance genes (tet genes), representing two resistance mechanisms (tetracycline exporters and ribosome protective proteins) have been detected (Table 2).
Tetracycline resistance mediated by specific exporters
Among the tet genes coding for membrane-associated proteins of the major facilitator superfamily which specifically export tetracyclines from the bacterial cell, the genes tet(A), tet(B), tet(C), tet(G), tet(H), tet(L), and tet(K) have been identified in bacteria of the aforementioned five genera, but tet(K) is commonly found on small plasmids only in human pathogens. Studies from the late 1970s and 1980s reported transferable tetracycline resistance among P. multocida and M. haemolytica isolates of animal origin (44–47), although the type of tet gene involved was not determined in any of these studies. In 1993, Hansen and coworkers identified a novel type of tet gene, designated tet(H) (48). The tet(H) gene was first detected on plasmid pVM111, which originated from a P. multocida isolate obtained from a turkey in the late 1970s in the United States. (44). In a subsequent study, Hansen et al. found the tet(H) gene to be the predominant tet gene among P. multocida and M. haemolytica from infections of cattle and pigs in North America (49). Since the tet(H) gene was located either in chromosomal DNA or on plasmids, they speculated about the involvement of a transposable element in the spread of tet(H) (49). The corresponding transposon, Tn5706, was identified in 1998 on plasmid pPMT1 from a bovine P. multocida isolate (50). Tn5706 is a small, nonconjugative composite transposon of 4,378 bp and represents the first known resistance-mediating transposon identified among members of the genus Pasteurella (51). The tetR-tet(H) gene region in Tn5706 is bracketed by inverted copies of the two closely related insertion sequences, IS1596 and IS1597 (50) (Fig. 1). Truncated Tn5706 elements, in which these insertion sequences were deleted in part or completely, have been found on small plasmids in isolates of M. haemolytica, P. multocida, and [P.] aerogenes (52, 53) and more recently as part of ICEs in isolates of P. multocida, H. somni, and M. haemolytica (54–57). The tet(H) gene has also been found in bacteria outside of the family Pasteurellaceae, namely in Moraxella spp. and Acinetobacter radioresistens (58), both obtained from salmon farms in Chile. The A. radioresistens isolate also harbored the insertion sequence IS1599 (58), which is closely related to the Tn5706-associated insertion sequences IS1596 and IS1597.
FIGURE 1.
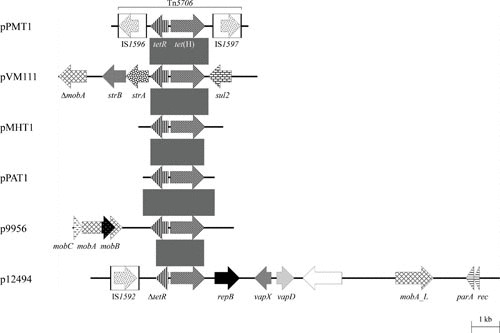
Schematic representation of the structure and organization of genes found in tet(H)-carrying plasmids from P. multocida, M. haemolytica, [P.] aerogenes, and A. pleuropneumoniae. Comparison of the maps of the partially sequenced plasmids pPMT1 (accession no. Y15510) and pVM111 (accession nos. AJ514834 and U00792), both from P. multocida, pMHT1 (accession no. Y16103) from M. haemolytica, and pPAT1 (accession no. AJ245947) from [P.] aerogenes (accession no. Z21724) and the completely sequenced plasmids p9956 (accession no. AY362554; 5,674 bp) and p12494 (accession no. DQ517426; 14,393 bp), both from A. pleuropneumoniae. Genes are shown as arrows, with the arrowhead indicating the direction of transcription. The following genes are involved in antimicrobial resistance: tetR-tet(H) (tetracycline resistance), sul2 (sulfonamide resistance), and strA and strB (streptomycin resistance); plasmid replication: repB; mobilization functions: mobA, mobB, mobC, and mobA_L; recombination functions: rec; DNA partition: par; virulence: vapD and vapX; unknown function: the open reading frame indicated by the white arrow. The Δ symbol indicates a truncated functionally inactive gene. The white boxes in the maps of pPMT1 and p12494 indicate the limits of the insertion sequences IS1592, IS1596, and IS1597; the arrows within these boxes indicate the reading frames of the corresponding transposase genes. Gray shaded areas indicate the tetR-tet(H) gene region common to all these plasmids with ≥95% nucleotide sequence identity. A distance scale in kilobases is shown at the bottom of the figure.
The tetR-tet(H) gene region of the P. multocida plasmid pVM111 is bracketed by the sul2 sulfonamide resistance gene and the strA and strB streptomycin resistance genes and thus represents part of a novel resistance gene cluster (59) (Fig. 1). The tet(H) gene is occasionally present in tetracycline-resistant A. pleuropneumoniae (9, 41, 60–63). Two tet(H)-carrying plasmids, p9956 and p12494 (5,674 bp and 14,393 bp, respectively), have been isolated from porcine A. pleuropneumoniae and sequenced completely (61). Structural analysis showed that they differed distinctly from one another and from the tet(H)-carrying plasmids previously found in Pasteurella spp. and Mannheimia spp. (Fig. 1). In plasmid p12494, a 100-bp insertion in the tetR gene was detected which resulted in the loss of 60 amino acids at the C-terminus. MIC determination of the corresponding A. pleuropneumoniae isolate, in the presence and absence of tetracycline, suggested that this tet(H) gene was constitutively expressed (61).
The tet(B) gene has been identified as the dominant tet gene among porcine [P.] aerogenes isolates (53) and has also been detected in porcine P. multocida isolates from the United States and Germany (64) and Spain (65), as well as in a bovine M. haemolytica isolate from France (66). In all but two cases, the tet(B) gene was located in one or two copies in the chromosome. The gene tet(B) is part of the nonconjugative transposon Tn10 (67, 68) and represents the most widely spread tet gene among Enterobacteriaceae (69). Hybridization studies using SfuI-digested whole cellular DNA of tet(B)-carrying P. multocida and [P.] aerogenes isolates suggested that there are complete copies of Tn10 in the majority of the isolates investigated (53, 64). In the 4.8-kb plasmid pPAT2, recovered from [P.] aerogenes in Germany, the tetR-tet(B) genes proved to be part of a largely truncated Tn10 element (53), whereas in the 5.1-kb P. multocida plasmid from Spain, pB1001, the tet(B) gene was found without tetR (65).
Studies of tetracycline-resistant A. pleuropneumoniae from pigs (9, 60, 70, 71) also revealed the presence of the gene tet(B), which appears to be the predominant tet gene in isolates, including those from Spain, Switzerland, Japan, Korea, Australia, and the United Kingdom (9, 60, 62, 63, 72, 73). This gene was also detected in the “A. porcitonsillarum” reference strain CCUG46996, and a single field isolate of this bacterium (62). Recently, whole-genome sequencing revealed that some United Kingdom isolates of A. pleuropneumoniae carry tet(B) in their chromosome as part of a transposon insertion disrupting the comM gene, whereas other isolates have Tn10 insertions carrying tet(B) as part of the 56-kb ICEApl1, located in a copy of tRNA-Leu (9, 74). Neither of the ICEs so far identified in P. multocida (ICEPmu1) and M. haemolytica (ICEMh1) carry tet(B) (54–56). It would be interesting to determine if the previously reported chromosomally encoded tet(B) genes in these species are part of ICEs related to ICEApl1 or other distinct ICEs. As more whole-genome sequences become available, it is likely that more ICEs will be identified in isolates of the Pasteurellaceae.
Among the Spanish A. pleuropneumoniae isolates, the tet(B) gene was found mainly on small plasmids, which were indistinguishable by their HindIII and DraI restriction patterns (60). One of these, p11745, a 5,486-bp plasmid, was sequenced completely. Again, as with pB1001 from P. multocida, neither the tetR repressor gene nor other parts of Tn10 were detectable in p11745 (60) (Fig. 2). Although both of these plasmids, isolated in Spain, encode a replication gene, rep, upstream of the tet(B) gene, there is little sequence similarity outside of the tet(B) gene (Fig. 2). In addition, genes for mobilization, mobC and mobA/L, are found in p11745, but not in pB1001. The 4,597-bp plasmid pHPS1019, isolated from [H.] parasuis in China (accession number HQ622101), encodes the same rep and tet(B) genes as pB1001 and also shares an extended region of identity downstream of the tet(B) gene (Fig. 2). In these two plasmids, there are direct repeats flanking the tet(B) gene, extending from within the 3′ terminus of rep for 484 bp in pB1001 and 198 bp in in pHPS1019. Another distinct tet(B) plasmid, pHS-Tet, was isolated from [H.] parasuis in Australia (75). This 5,147-bp plasmid, also without an accompanying tetR repressor gene, carries different mobilization genes than those in p11745 (Fig. 2).
FIGURE 2.
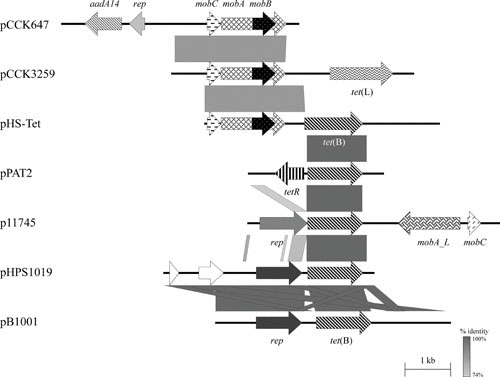
Schematic representation of the structure and organization of aadA14-, tet(L)-, and tet(B)-carrying plasmids from P. multocida, M. haemolytica, [H.] parasuis, [P.] aerogenes, and A. pleuropneumoniae. Comparison of the maps of the aadA14-carrying streptomycin/spectinomycin resistance plasmid pCCK647 (accession no. AJ884726; 5,198 bp) from P. multocida, the tet(L)-carrying tetracycline resistance plasmid pCCK3259 (accession no. AJ966516; 5,317 bp) from M. haemolytica, and the tet(B)-carrying tetracycline resistance plasmids pHS-Tet (accession no. AY862435; 5,147 bp) from [H.] parasuis, pPAT2 (accession no. AJ278685; partially sequenced) from [P.] aerogenes, p11745 (accession no. DQ176855; 5,486 bp) from A. pleuropneumoniae, pHPS1019 (accession no. HQ622101; 4,597 bp) from [H.] parasuis, and pB1001 (accession no. EU252517; 5,128 bp) from P. multocida. Genes are shown as arrows, with the arrowhead indicating the direction of transcription. The following genes are involved in antimicrobial resistance: tetR-tet(B), tet(B), and tet(L) (tetracycline resistance) and aadA14 (streptomycin/spectinomycin resistance); plasmid replication: rep; mobilization functions: mobA, mobB, and mobC; unknown function: the open reading frames indicated by white arrows. Gray-shaded areas indicate the regions common to plasmids, and the different shades of gray illustrate the percentages of nucleotide sequence identity between the plasmids, as indicated by the scale at the bottom of the figure. A distance scale in kilobases is shown.
The gene tet(G) has been found on the chromosome of six epidemiologically related M. haemolytica isolates from cattle (64) and on plasmid pJR1 from avian P. multocida (76). Surprisingly, the tet(G) structural gene in plasmid pJR1 was found in the absence of a corresponding tetR repressor gene, which is considered to be essential for the tetracycline-inducible expression of tet(G). It should be noted that plasmid pJR1 has not been transferred into susceptible recipient strains for phenotypic confirmation of the activity of the resistance genes found on this plasmid (76). Previously, tet(G) was shown to be part of the multiresistance gene cluster present in Salmonella enterica serovar Typhimurium DT104 (77), which has since been shown to be part of an integrative mobilizable element, SGI1 (78). It is currently not known whether a related integrative mobilizable element is present in the chromosomes of the six M. haemolytica isolates carrying tet(G).
The tet(L) gene, which is commonly found in Gram-positive cocci and Bacillus spp., was detected on small plasmids of M. haemolytica and Mannheimia glucosida, but also in the chromosomal DNA of single M. haemolytica and P. multocida isolates, all originating from cattle in Belgium (79). One such plasmid of 5,317 bp from M. haemolytica, designated pCCK3259, was sequenced completely (Fig. 2). Besides the tet(L) gene, it contains only the mobABC operon responsible for mobilization of the plasmid (79). In Gram-positive bacteria, the tet(L) gene is inducibly expressed via translational attenuation (80). The corresponding regulatory region, however, was absent in plasmid pCCK3259, whereas all elements required for constitutive expression of the tet(L) gene were detected in the upstream sequence (79). Two different-sized tet(L)-carrying plasmids (5.6 and >12 kb) have also been identified in porcine A. pleuropneumoniae (60). The smaller one of these plasmids, p9555, was sequenced completely and revealed striking structural similarities to plasmid pCCK3259 from M. haemolytica (60). Again, the regulatory region of the tet(L) gene was missing in plasmid p9555. While plasmid p9555 could be transferred into Escherichia coli, where it expressed tetracycline resistance (60), plasmid pCCK3259 did not replicate in E. coli but did in a P. multocida host (79).
More recently, isolates of A. pleuropneumoniae from Japan and South Korea were shown to harbor the tet(A) gene, as well as other tetracycline resistance genes (72, 81). In both reports, detection of resistance genes was by PCR, and localization to plasmid or chromosome was not determined. Of the 65 isolates tested by Kim et al. (81), 62 were resistant to tetracycline, with 21 of them carrying tet(A). Eleven of these isolates harboured two or more tetracycline resistance genes, as for example one isolate with tet(A), tet(B), tet(C), tet(G), and tet(L); another with tet(A), tet(B), tet(C), and tet(G); and another with tet(A), tet(B), and tet(M)/tet(O) (81).
Tetracycline resistance mediated by ribosome protective proteins
Another two tetracycline resistance genes, tet(M) and tet(O), both coding for ribosome protective proteins, have been identified in P. multocida and A. pleuropneumoniae. The gene tet(M) is associated with the conjugative transposon Tn916 (82). It is considered to be the most widespread tet gene among Gram-positive and Gram-negative bacteria (69; http://faculty.washington.edu/marilynr/). It has been detected by hybridization in the chromosomal DNA of two bovine P. multocida isolates (49, 66). The gene tet(O), previously identified in Campylobacter spp. and streptococci, was detected in chromosomal DNA of a porcine A. pleuropneumoniae isolate in 2004 (71). More recently, the tet(O) gene was also detected in eight porcine A. pleuropneumoniae isolates from Spain (60), five from Japan (72), four from Korea (73), and seven from Canada (41). In the isolates from Spain, the tet(O) gene was located on small plasmids of ca. 6 kb. Sequencing of a 2,489-bp region, including the complete tet(O) gene, of one of these plasmids (p13142) revealed 99% sequence identity to the tet(O) gene of Campylobacter jejuni (60).
Resistance to β-Lactam Antibiotics
In general, resistance of Pasteurellaceae to β-lactam antibiotics is based on the production of a β-lactamase or the presence of penicillin-binding proteins with low affinity to β-lactams (10, 11, 83). However, the latter mechanism has only been reported for the human pathogen Haemophilus influenzae (10, 11, 83). Other mechanisms, such as reduced outer membrane permeability or multidrug efflux systems that can efficiently export β-lactams from the bacterial cell (10, 11, 83), have rarely—if at all—been identified in Pasteurellaceae.
So far, five β-lactamase (bla) genes have been identified among Pasteurellaceae: blaROB-1 (84–88), blaTEM-1 (89), blaPSE-1 (76), blaCMY-2 (90), and blaOXA-2 (55, 57). It is interesting to note that the complete blaOXA-2 gene, identified as part of ICEPmu1, was found to be nonfunctional in P. multocida but functional in E. coli (55). According to the existing classification schemes of β-lactamases, the ROB-1 and TEM enzymes are assigned to the Ambler class A because of their structure and to the Bush class 2b on the basis of their substrate profile (91). Members of this class can hydrolyze penicillins and first-generation cephalosporins (narrow spectrum of activity) but are sensitive to inhibition by β-lactamase inhibitors such as clavulanic acid. The PSE-1 β-lactamase also belongs to the Ambler class A but to the Bush class 2c. This enzyme, also known as CARB-2 β-lactamase, can hydrolyze carbenicillin and is also inactivated by clavulanic acid.
Although initially identified in H. influenzae from a human infection (92), ROB-1 β-lactamases are widely distributed and have been detected in porcine A. pleuropneumoniae isolates (62, 93–95), porcine “A. porcitonsillarum” isolates (62), bovine and porcine P. multocida, [P.] aerogenes, and M. haemolytica isolates (84, 85, 96), as well as porcine [H.] parasuis isolates (88, 97). Comparisons confirmed that the similar sized blaROB-1-carrying plasmids from P. multocida and M. haemolytica (85) were structurally closely related. The sequences of the small blaROB-1-carrying plasmids pB1000 from [H.] parasuis and pB1002 from P. multocida (65, 88) are also structurally related to the blaROB-1-carrying plasmid pAB2 from bovine M. haemolytica (98) and the APP7_A plasmid from A. pleuropneumoniae (accession number CP001094) (Fig. 3), as well as to the tet(B)-carrying tetracycline resistance plasmid pHS-Tet from [H.] parasuis (75). Comparative analysis confirmed that all so far sequenced blaROB-1 genes from M. haemolytica, A. pleuropneumoniae, “A. porcitonsillarum,” and [H.] parasuis code for an identical β-lactamase protein of 305 amino acids. Mutations in the blaROB-1 gene, which resulted in resistance to extended-spectrum cephalosporins and β-lactamase inhibitors, have been produced in vitro (99). Only a single report has described the detection of a TEM-1 β-lactamase in a P. multocida isolate from a human dog-bite wound (89). Similarly, PSE-1 β-lactamase has been found in a single avian P. multocida isolate (76). In these studies, the TEM-1 β-lactamase was confirmed by isoelectric focusing and sequence analysis of part of the blaTEM-1 gene, whereas the blaPSE-1 gene was completely sequenced. The blaCMY-2 gene, detected by Chander and coworkers (90) in an apparently ceftiofur-resistant P. multocida isolate from a pig, was only detected by PCR. Since the blaCMY-2 gene was also detected in ceftiofur-susceptible isolates in the same study and no functional analysis has been conducted, the role of blaCMY-2 in ceftiofur resistance of P. multocida remains questionable.
FIGURE 3.
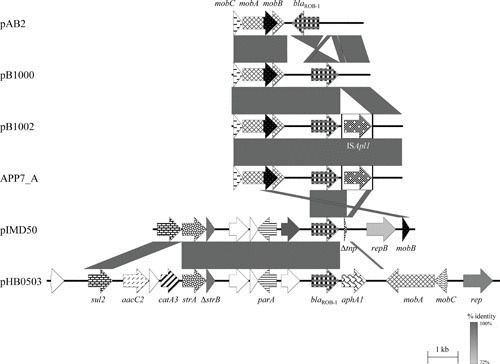
Schematic representation of the structure and organization of the blaROB-1-carrying resistance plasmids from M. haemolytica, [H.] parasuis, P. multocida, A. pleuropneumoniae, and “A. porcitonsillarum.” Comparison of the maps of blaROB-1-carrying resistance plasmids pAB2 (accession no. Z21724; 4,316 bp) from M. haemolytica, pB1000 (accession no. DQ840517; 4,613 bp) from [H.] parasuis, pB1002 (accession no. EU283341; 5,685 bp) from P. multocida, APP7_A (accession no. CP001094; 5,685 bp) from A. pleuropneumoniae, pIMD50 (accession no. AJ830711; 8,751 bp) from “A. porcitonsillarum,” and pHB0503 (accession no. EU715370; 15,079 bp) from A. pleuropneumoniae. It should be noted that another three pIMD50-related blaROB-1-carrying resistance plasmids from “A. porcitonsillarum” have been sequenced completely: pKMA5 (accession no. AM748705), pKMA202 (accession no. AM748706), and pKMA1467 (accession no. AJ830712). Genes are shown as arrows, with the arrowhead indicating the direction of transcription. The following genes are involved in antimicrobial resistance: sul2 (sulfonamide resistance), strA and strB (streptomycin resistance), blaROB-1 (β-lactam resistance), aacC2 (gentamicin resistance), catA3 (chloramphenicol resistance), and aphA1 (kanamycin/neomycin resistance); plasmid replication: rep; mobilization functions: mobA, mobB, and mobC; resolvase function: res; DNA partition: parA; unknown function: open reading frames indicated by white arrows. The prefix Δ indicates a truncated functionally inactive gene. Gray-shaded areas indicate the regions common to plasmids, and the different shades of gray illustrate the percentages of nucleotide sequence identity between the plasmids, as indicated by the scale at the bottom of the figure. A distance scale in kilobases is shown.
β-Lactam resistance among Pasteurellaceae is often associated with small plasmids. These range in size between 4.1 and 5.7 kb in P. multocida (65, 84, 89), 4.1 and 5.2 kb in M. haemolytica (85, 87, 98, 100–104), 2.5 and 15.1 kb in A. pleuropneumoniae (93–95, 105–107), 8.7 and 13.4 kb in “A. porcitonsillarum” (62, 108), and 2.7 and 4.6 kb in [H.] parasuis (88, 97). Complete sequences indicate that blaROB-1-carrying plasmids of ≥6 kb encode additional resistance genes, as shown in Table 3. Most of the β-lactam resistance plasmids detected among Pasteurellaceae have been identified phenotypically by transformation or conjugation experiments. Recently, some β-lactam-resistant A. pleuropneumoniae isolates from South Korea were found to be negative by PCR for both blaROB-1 and blaTEM-1 (73, 81). The mechanism of this resistance has not yet been investigated.
TABLE 3.
Subset of resistance plasmids identified in Pasteurella, Mannheimia, and Actinobacillus of veterinary importance
| Plasmid designation | Plamsid size (kb) | Plasmid sequencing | Resistance genotype | Bacterial source | Accession number | Reference |
|---|---|---|---|---|---|---|
| pARD3079 | 4.0 | Complete | sul2 | A. pleuropneumoniae | AM748707 | 108 |
| pKMA2425 | 3.1 | Complete | sul2 | A. pleuropneumoniae | AJ830714 | 108 |
| pKMA757 | 4.5 | Complete | sul2 | “A. porcitonsillarum” | AJ830713 | 108 |
| ABB7_B | 4.2 | Complete | sul2, strA | A. pleuropneumoniae | NC_010941 | Unpublished |
| pFZG1012 | Unknown | Partial | sul2, strA | [H.] parasuis | HQ015158 | Unpublished |
| pYC93 | 4.2 | Complete | sul2, strA | [H.] parasuis | HM486907 | 194 |
| pB1005 | 4.2 | Complete | sul2, strA | P. multocida | FJ197818 | 65 |
| pHN06 | 5.4 | Complete | sul2, strA | P. multocida | NC_017035 | 114 |
| Unnamed | 5.4 | Complete | sul2, strA | P. multocida | CP003314 | 114 |
| pCCK1900 | 10.2 | Complete | sul2, strA, strB, floR | P. multocida | FM179941 | 178 |
| pMS260 | 8.1 | Complete | sul2, strA, strB | A. pleuropneumoniae | AB109805 | 121 |
| pTYM1 | 4.2 | Complete | sul2, strA, ΔstrB | A. pleuropneumoniae | AF303375 | 94 |
| pPSAS1522 | 4.2 | Complete | sul2, strA, ΔstrB | A. pleuropneumoniae | AJ877041 | 108 |
| pYFC1 | 4.2 | Complete | sul2, strA, ΔstrB | M. haemolytica | M83717 | 103 |
| pPASS2 | 4.7 | Partial | sul2, strA, ΔstrB | [P.] aerogenes | Not provided | 125 |
| pPASS1 | 5.5 | Partial | sul2, strA, ΔstrB | [P.] aerogenes | Not provided | 125 |
| pIG1 | 5.4 | Complete | sul2, strA, ΔstrB | P. multocida | U57647 | 150 |
| pB1003 | 5.1 | Complete | sul2, strA, ΔstrB | P. multocida | EU360945 | 65 |
| pPMSS1 | 4.2 | Partial | sul2, strA, ΔstrB | P. multocida | Not provided | 125 |
| pB1003 | 5.0 | Complete | sul2, strA, ΔstrB | P. multocida | EU360945 | 65 |
| pKMA505 | 8.6 | Complete | sul2, strA, ΔstrB | “A. porcitonsillarum” | NC_007094 | 108 |
| pYMH5 | 5.0 | Complete | sul2, Δsul2, strA, ΔstrB, aphA1 | A. paragallinarum | EF015636 | 123 |
| pIMD50 | 8.8 | Complete | sul2, strA, ΔstrB, blaROB-1 | “A. porcitonsillarum” | NC_007095 | 108 |
| pKMA5 | 9.5 | Complete | sul2, strA, ΔstrB, blaROB-1 | “A. porcitonsillarum” | NC_009623 | 108 |
| pKMA1467 | 11.1 | Complete | sul2, strA, ΔstrB, blaROB-1 | “A. porcitonsillarum” | NC_007096 | 108 |
| pOV | 13.6 | Complete | sul2, strA, ΔstrB, blaROB-1 | P. multocida | JX827416 | Unpublished |
| pKMA202 | 13.4 | Complete | sul2, strA, ΔstrB, blaROB-1 | “A. porcitonsillarum” | AM748706 | 108 |
| pM3389T | 6.1 | Complete | sul2, ΔstrA, dfrA14 | A. pleuropneumoniae | KP197005 | 122 |
| pM3224T | 6.1 | Complete | sul2, ΔstrA, dfrA14, strB | A. pleuropneumoniae | KP197004 | 122 |
| pMHSCS1 | 5.0 | Complete | sul2, catA3, strA, ΔstrB | Mannheimia taxon 10a | AJ249249 | 125 |
| pMVSCS1 | 5.6 | Complete | sul2, catA3, strA, ΔstrB | M. varigena | NC_003411 | 116 |
| pPASCS1 | 5.6 | Partial | sul2, catA3, strA, ΔstrB | [P.] aerogenes | Not provided | 125 |
| pPASCS2 | 6.0 | Partial | sul2, catA3, strA, ΔstrB | [P.] aerogenes | Not provided | 125 |
| pPASCS3 | 6.1 | Partial | sul2, catA3, strA, ΔstrB | [P.] aerogenes | Not provided | 125 |
| pCCK13698 | 15.0 | Complete | sul2, catA3, ΔstrA, floR | B. trehalosi | AM183225 | 183 |
| pFZ51 | 15.7 | Complete | sul2, aacC2, catA3, strA, ΔstrB, blaROB-1, aphA1 | [H.] parasuis | JN202624 | Unpublished |
| pHB0503 | 15.1 | Complete | sul2, aacC2, catA3, strA, ΔstrB, blaROB-1, aphA1 | A. pleuropneumoniae | EU715370 | 95 |
| pVM111 | 9.8 | Partial | sul2, tetR-tet(H), strA, strB | P. multocida | AJ514834 | 49, 59 |
| pCCK154 | 11.0 | Partial | sul2, dfrA20 | P. multocida | AJ605332 | 146 |
| pJR1 | 6.8 | Complete | sul2, tet(G), catB2 | P. multocida | AY232670 | 76 |
| p9956 | 5.7 | Complete | tetR-tet(H) | A. pleuropneumoniae | AY362554 | 60 |
| pMHT1 | 4.4 | Partial | tetR-tet(H) | M. haemolytica | Y16103 | 64 |
| pPAT1 | 5.5 | Partial | tetR-tet(H) | [P.] aerogenes | AJ245947 | 52 |
| pPMT1 | 6.8 | Partial | tetR-tet(H) | P. multocida | Y15510 | 50 |
| pB1018 | 6.0 | Complete | tetR-tet(H) | P. multocida | JQ319774 | Unpublished |
| p12494 | 14.4 | Complete | ΔtetR-tet(H) | A. pleuropneumoniae | DQ517426 | 61 |
| p9555 | 5.7 | Complete | tet(L) | A. pleuropneumoniae | AY359464 | 60 |
| pCCK3259 | 5.3 | Complete | tet(L) | M. haemolytica | NC_006976 | 79 |
| p11745 | 5.5 | Complete | tet(B) | A. pleuropneumoniae | DQ176855 | 60 |
| pHS-Tet | 5.1 | Complete | tet(B) | [H.] parasuis | AY862435 | 75 |
| pTetHS016 | 3.4 | Complete | tet(B) | [H.] parasuis | KC818265 | 195 |
| pHPS1019 | 4.6 | Complete | tet(B) | [H.] parasuis | HQ622101 | Unpublished |
| pB1001 | 5.1 | Complete | tet(B) | P. multocida | EU252517 | 65 |
| pPAT2 | 4.8 | Partial | tetR-tet(B) | [P.] aerogenes | AJ278685 | 53 |
| p13142 | 6.0 | Partial | tet(O) | A. pleuropneumoniae | AY987963 | 60 |
| pB1006 | 6.0 | Complete | tet(O) | P. multocida | FJ234438 | 65 |
| pJR2 | 5.3 | Complete | aadA1, blaPSE-1 | P. multocida | AY232671 | 76 |
| pCCK343 | 5.4 | Complete | aadA1, dfrA1, sat2 | [P.] aerogenes | FR687372 | 130 |
| pCCK647 | 5.2 | Complete | aadA14 | P. multocida | NC_006868 | 132 |
| Unnamed | Unknown | Partial | bla ROB-1 | A. pleuropneumoniae | Not provided | 105 |
| APP7_A | 5.7 | Complete | bla ROB-1 | A. pleuropneumoniae | CP001094 | Unpublished |
| pB1000 | 4.6 | Complete | bla ROB-1 | [H.] parasuisP. multocida | DQ840517GU080062, GU080067 | 88 65 |
| pAB2 | 4.3 | Complete | bla ROB-1 | M. haemolytica | Z21724 | 98 |
| pPH51 | 4.1 | Partial | bla ROB-1 | M. haemolytica | X52872 | 85 |
| pYFC2 | 4.2 | Partial | bla ROB-1 | M. haemolytica | Not provided | 103 |
| pB1002 | 5.7 | Complete | bla ROB-1 | P. multocida | EU283341 | 65 |
| pJMA-1 | 2.7 | Complete | bla ROB-1 | [H.] parasuis | KP164834 | 97 |
| pCCK411 | 5.3 | Complete | blaROB-1, aphA3 | P. multocida | FR798946.1 | 135 |
| pFS39 | 7.6 | Complete | blaROB-1, erm(T) | [H.] parasuis | KC405064 | 154 |
| pFAB-1 | 4.3 | Partial | bla TEM-1 | P. multocida | Not provided | 89 |
| pHPSF1 | 6.3 | Complete | floR | [H.] parasuis | KR262062.1 | 180 |
| pM3446F | 7.7 | Complete | floR | A. pleuropneumoniae | KP696484 | 181 |
| pCCK381 | 10.8 | Complete | floR | P. multocida | NC_006994 | 174 |
| pMH1405 | 7.7 | Complete | floR | P. multocida | NC_0192601 | 179 |
| p518 | 3.9 | Complete | floR, strA, ΔstrB | A. pleuropneumoniae | KT355773 | 182 |
| pQY431 | 7.8 | Complete | aacA-aphD | [H.] parasuis | KC405065 | Unpublished |
| FJS5863 | 7.8 | Complete | aacA-aphD, blaROB-1 | [H.] parasuis | HQ015159 | Unpublished |
| pHN61 | 6.3 | Complete | lnu(C) | [H.] parasuis | FJ947048 | 161 |
Initially identified as M. haemolytica.
Resistance to Aminoglycosides and Aminocyclitols
Resistance to aminoglycoside and aminocyclitol antibiotics is usually mediated by enzymes that inactivate the drugs by adenylation, acetylation, or phosphorylation. Moreover, mutations in chromosomal genes have also been identified to mediate resistance to selected members of these classes of antimicrobial agents (109).
Resistance to aminoglycosides and aminocyclitols by enzymatic inactivation
Streptomycin and/or spectinomycin resistance mediated by enzymatic inactivation
The first aminoglycoside resistance genes detected in Pasteurella and Mannheimia were those mediating streptomycin resistance. In 1978, Berman and Hirsh published a report on plasmids coding for streptomycin resistance along with sulfonamide resistance, or sulfonamide and tetracycline resistance, in P. multocida from turkeys (44). Streptomycin resistance is commonly associated with small nonconjugative plasmids of less than 15 kb in P. multocida (44–47, 59, 65, 110–114), [P.] aerogenes (53), M. haemolytica (103, 115), Mannheimia varigena (116), “A. porcitonsillarum” (62), A. pleuropneumoniae (94, 95, 107, 117–122), and Avibacterium paragallinarum (123). In one case, streptomycin resistance was mediated by a conjugative multiresistance plasmid of approximately 113 kb in an avian P. multocida isolate, but the resistance genes were not investigated (47). Many of these plasmids carry additional resistance genes such as the sulfonamide resistance gene sul2, the kanamycin/neomycin resistance gene aphA1, the chloramphenicol resistance gene catA3 (Fig. 4), and/or the ampicillin resistance gene blaROB-1 (Fig. 3) (Table 3).
FIGURE 4.
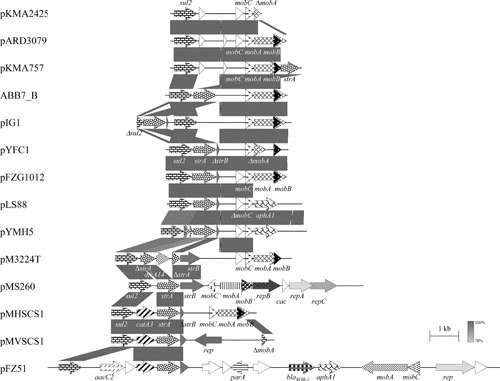
Schematic representation of the structure and organization of selected sul2-based (multi-)resistance plasmids from A. pleuropneumoniae, “A. porcitonsillarum,” A. paragallinarum, H. ducreyi, [H.] parasuis, M. haemolytica, Mannheimia unnamed taxon 10, M. varigena, and P. multocida. Comparison of the maps of the plasmids pKMA2425 (accession no. AJ830714; 3,156 bp) from A. pleuropneumoniae, pARD3079 (accession no. AM748707; 4,065 bp) from A. pleuropneumoniae, pKMA757 (accession no. AJ830713; 4,556 bp) from “A. porcitonsillarum,” ABB7_B (accession no. NC_010941; 4,236 bp) from A. pleuropneumoniae, pIG1 (accession no. U57647) from P. multocida, pYFC1 (accession no. M83717) from M. haemolytica, pFZG1012 (accession no. HQ015158; partially sequenced) from [H.] parasuis, pLS88 (accession no. L23118; 4,772 bp) from H. ducreyi, pYMH5 (accession no. EF015636; 4,772 bp) from A. paragallinarum, pM3224T (accession no. KP197004; 6,050 bp) from A. pleuropneumoniae, pMS260 (accession no. AB109805; 8,124 bp) from A. pleuropneumoniae, pMVSCS1 (accession no. AJ319822; 5,621 bp) from M. varigena, pMHSCS1 (accession no. AJ249249; 4,992 bp) from Mannheimia unnamed taxon 10, pFZ51 (accession no. JN202624; 15,672 bp) from [H.] parasuis, and pKMA757 (accession no. AJ830713; 4,556 bp) from “A. porcitonsillarum.” The map of another sul2-based multiresistance plasmid, pIMD50 (accession no. AJ830711) from “A. porcitonsillarum,” is displayed in Fig. 3. Genes are shown as arrows, with the arrowhead indicating the direction of transcription. The following genes are involved in antimicrobial resistance: sul2 (sulfonamide resistance), strA and strB (streptomycin resistance), catA3 (chloramphenicol resistance), aphA1 (kanamycin/neomycin resistance), and blaROB-1 (β-lactam resistance); plasmid replication: rep, repA, repB, and repC; mobilization functions: mobA, mobB, mobC, mobA′, mobB′, and mobC′; unknown function: open reading frames indicated by white arrows. The prefix Δ indicates a truncated functionally inactive gene. Gray-shaded areas indicate the regions common to plasmids and the different shades of gray illustrate the percentages of nucleotide sequence identity between the plasmids, as indicated by the scale at the bottom of the figure. A distance scale in kilobases is shown.
The predominant streptomycin resistance gene in bacteria of the genera Pasteurella, Mannheimia, and Actinobacillus is strA. It codes for an aminoglycoside-3″-phosphotransferase of 269 amino acids and is sometimes found together with the gene strB, which codes for an aminoglycoside-6-phosphotransferase of 278 amino acids. Both genes are part of transposon Tn5393 from Erwinia amylovora (124). In the streptomycin-resistant Pasteurella, Mannheimia, and Actinobacillus isolates, strA is usually complete, whereas various truncated strB genes have been identified (116, 121, 125). Isolates that carry a functionally active strA gene, but a largely truncated strB gene, have been shown to be highly resistant to streptomycin (125), suggesting that strA is the relevant gene for the expression of streptomycin resistance. Further support for this hypothesis comes from the observation that plasmid pSTOJO1, which carries an intact strB gene but a functionally inactive strA gene that is disrupted by the integration of a dfrA14 gene cassette, does not express streptomycin resistance in E. coli (126). Studies of the prevalence and distribution of the strA-strB genes showed that the strA gene—in combination with a complete or truncated copy of strB—occurs on plasmids or in the chromosomal DNA of a wide range of commensal and pathogenic bacteria from humans, animals, and plants (127–129).
An aadA1 gene coding for an aminoglycoside-3″-adenyltransferase that mediates resistance to both the aminoglycoside streptomycin and the aminocyclitol spectinomycin has been detected on the 5.2-kb plasmid pJR2 from avian P. multocida (76) and the 5.4-kb plasmid pCCK343 from porcine [P.] aerogenes (130). In pJR2, the aadA1 gene is part of a gene cassette, along with blaPSE-1, that is inserted into a relic of a class 1 integron. The intI1 gene coding for the integrase in the 5′-conserved segment of this integron is truncated, but without affecting the promotor, and the sul1 gene in the 3′ conserved fragment is missing completely (76). In plasmid pCCK343, a gene cassette array comprised of dfrA1-sat2-aadA1 is part of a truncated class 2 integron (130). In this case, the streptothricin acetyltransferase gene, sat2, conferred resistance to the streptothricin antibiotic nourseothricin (MIC, >256 mg/liter), in addition to the streptomycin and spectinomycin resistance (MICs of >256 mg/liter for both) conferred by the aminoglycoside adenyltransferase gene, aadA1. Attempts to identify the aadA genes in bovine isolates of P. multocida and M. haemolytica from Germany that are highly resistant to streptomycin and spectinomycin using PCR were unsuccessful (131). In all these isolates, streptomycin resistance was based on an strA gene, whereas a spectinomycin resistance gene could not be identified.
A novel streptomycin-spectinomycin resistance gene, designated aadA14, was identified on a small 5.2-kb plasmid from a bovine P. multocida capsular type F isolate from Belgium (132). The corresponding isolate was obtained from a case of fatal peritonitis in a calf (14). The 261-amino acid AadA14 adenyltransferase protein exhibited only 51.4 to 56.0% identity to the so far known AadA proteins and hence proved to be only distantly related to AadA proteins previously found in other bacteria.
Two ICEs from Pasteurellaceae, ICEPmu1 and ICEMh1, have been shown to contain genes associated with resistance to streptomycin and other aminoglycosides and aminocyclitols (55, 56). In addition to other resistance genes, the 92-kb ICEMh1 contains strA and strB, conferring resistance to streptomycin (MIC, 256 mg/liter), as well as aphA1, conferring resistance to kanamycin/neomycin (MICs, ≥512 mg/liter and 64 mg/liter, respectively) (56). The 82-kb ICEPmu1 carries 12 resistance genes, including strA and strB, aadA25, aadB, and aphA1, with the latter three genes conferring resistance to streptomycin (MIC, ≥256 mg/liter) and spectinomycin (MIC, ≥512 mg/liter), gentamicin (MIC, 128 mg/liter), and kanamycin/neomycin (MICs, ≥128 mg/liter and ≥32 mg/liter), respectively, in P. multocida strain 36950 (54, 55).
Kanamycin and neomycin resistance mediated by enzymatic inactivation
Kanamycin/neomycin resistance has been associated with the gene aphA1, also known as aph(3′)-Ia, which codes for an aminoglycoside-3′-phosphotransferase that mediates resistance to kanamycin and neomycin. This gene has been identified on transposon Tn903 (133). Subsequently, it was detected together with the streptomycin resistance genes strA-strB and the sulfonamide resistance gene sul2 on the broad-host-range plasmid pLS88 from human host-specific Haemophilus ducreyi (134). A pLS88-related plasmid has also been identified in the A. paragallinarum strain A14 (123) (Fig. 4). Further studies on kanamycin/neomycin-resistant [P.] aerogenes, P. multocida, and M. glucosida isolates identified the aphA1 gene mostly in chromosomal DNA (135). In a single P. multocida isolate, this gene was found on the 5,955-bp plasmid pCCK3152, along with complete strA and sul2 genes and a truncated strB gene (135). The 15.1-kb multiresistance plasmid pHB0503 (95) and the 15.7-kb plasmid pFZ51 (accession number JN202624), both carrying sul2, aacC2, catA3, strA, ΔstrB, and blaROB-1, along with aphA1, have been identified in Chinese A. pleuropneumoniae and [H.] parasuis isolates, respectively. Kang et al. (95) reported that the A. pleuropneumoniae isolate harboring pHB0503 was resistant to streptomycin (MIC, 512 mg/liter), kanamycin (MIC, 256 mg/liter), and gentamicin (MIC, 512 mg/liter), as well as to penicillin (MIC, 256 mg/liter), sulfadimidine (MIC, 1,024 mg/liter), and chloramphenicol (MIC, 16 mg/liter), whereas the [H.] parasuis isolate harboring pFZ51 has not been characterized (Fig. 4).
A second kanamycin/neomycin resistance gene, aphA3 [also known as aph(3′)-III], was detected on the 5.1-kb plasmid pCCK411 in single isolates of P. multocida and [P.] aerogenes (135). In addition to the aphA3 gene, this plasmid also carried a mobABC operon for mobilization. A not further specified aminoglycoside-3′-phosphotransferase gene mediating kanamycin resistance was also found, together with a blaROB-1 gene, on the 6-kb plasmid pTMY2 from A. pleuropneumoniae (94). Two virtually identical 7,777-bp plasmids from [H.] parasuis, FJS5863 (accession number HQ015159), and pQY431 (accession number KC405065) contain the aacA-aphD gene encoding a bifunctional aminoglycoside N-acetyltransferase and aminoglycoside phosphotransferase, in addition to the blaROB-1 gene, but no functional studies of these plasmids have been published.
Gentamicin resistance mediated by enzymatic inactivation
Although P. multocida, A. pleuropneumoniae, and “A. porcitonsillarum” isolates with MICs of gentamicin of ≥32 mg/liter have been detected, most attempts to detect specific resistance genes, such as aadB [ant(2″)-Ia], aacC2 [aac(3)-IIc], or aacC4 [aac(3)-IVa], have failed (62; Kehrenberg and Schwarz, unpublished data). Moreover, attempts to transfer gentamicin resistance from P. multocida donors were also unsuccessful (Kehrenberg and Schwarz, unpublished data). Recently, PCR analysis revealed that among 12 gentamicin-resistant A. pleuropneumoniae isolates from South Korea, 11 produced amplicons specific for aadB [ant(2″)-Ia], and one for aacC4 [aac(3)-IVa] (81). In addition, a 15.1-kb plasmid from A. pleuropneumoniae (pHB0503) has been found carrying aacC2, encoding an aminoglycoside-acetyltransferase (95). This plasmid conferred an MIC of gentamicin of 512 mg/liter in the original serovar 4 clinical isolate (from China) and an MIC of 256 mg/liter when transformed into M62, the reference strain of A. pleuropneumoniae serovar 4 (95). The 15.7-kb plasmid pFZ51 (accession number JN202624) from [H.] parasuis also carries aacC2 but has not been functionally characterized (Fig. 4).
Resistance to aminocyclitols by mutations
Ribosomal mutations conferring spectinomycin resistance have been described in a variety of bacteria (136–140). All these mutations were present in a specific region of helix 34 in 16S rRNA. This region encompassed the cross-linked positions 1063 to 1066 and 1090 to 1093, which are known to be involved in the binding of spectinomycin to the ribosome. Moreover, the rpsE gene coding for the ribosomal protein S5 is also relevant for the drug binding, and mutations in rpsE have been described to affect spectinomycin binding (141, 142). The analysis of high-level spectinomycin-resistant P. multocida isolates (MICs of spectinomycin ≥4,096 mg/liter), in which no enzymatic inactivation of the drug could be detected, revealed four types of mutations: (i) a C1192G transversion (no additional mutations in positions which have been associated with spectinomycin resistance were found) in 16S rRNA in all six or (ii) in five of the six rRNA operons, (iii) the aforementioned transversion in only two of the six operons accompanied by a 3-bp deletion in rpsE that resulted in a change of the amino acids 32-SF-33 to 32-I, and (iv) a 3-bp deletion in rpsE that resulted in the loss of 23-K without additional rRNA mutations (143). Molecular modeling suggested that both types of mutations in the S5 protein have a negative impact on spectinomycin binding to the ribosome.
Resistance to Folate Pathway Inhibitors
Sulfonamides and trimethoprim are competitive inhibitors of different enzymatic steps in folic acid metabolism. Sulfonamides represent structural analogs of p-aminobenzoic acid and inhibit the enzyme dihydropteroate synthase (DHPS), which—in the initial step of folic acid metabolism—catalyzes the synthesis of dihydropteroic acid from dihydropteridin and p-aminobenzoic acid. Trimethoprim inhibits the enzyme dihydrofolate reductase (DHFR), which—in a later step of folic acid metabolism—reduces dihydrofolic acid to tetrahydrofolic acid. Resistance to both drugs is commonly mediated by replacement of susceptible DHPS or DHFR enzymes by those with reduced affinity to sulfonamides or trimethoprim, respectively. Moreover, overproduction of susceptible targets, or mutations in chromosomal DHPS or DHFR genes, which alter substrate specificity, may also cause resistance (109). However, overproduction of a trimethoprim-sensitive DHFR, encoded by the gene folH (144), or a short insertion into the chromosomal gene folP encoding a DHPS (145), have so far only been reported to confer resistance to trimethoprim or sulfonamides, respectively, in human host-specific H. influenzae.
Sulfonamide resistance mediated by altered DHPSs
Sulfonamide resistance among Pasteurella, Mannheimia, Actinobacillus, and Haemophilus isolates is commonly mediated by a type 2 DHPS with reduced affinity to sulfonamides. The corresponding gene, sul2, is frequently found on small plasmids with sizes ranging from 3.1 to 15.7 kb (Table 3). The sul2-encoded DHPS proteins commonly consist of 271 amino acids. However, several variants ranging in size between 263 and 289 amino acids have also been reported (11, 55, 56, 125, 134, 146). Sequence analysis showed that single-base pair insertions downstream of codon 225 in the DHPS of the H. ducreyi plasmid pLS88 (134) resulted in a shortened C-terminus that differs from all so far known DHPS variants. A mutation in the translational stop codon of the sul2 gene from plasmids pYFC1 of M. haemolytica (103) and pTYM1 of A. pleuropneumoniae (94) led to an extension of 12 amino acids at the C-terminus. In plasmid pCCK154 from P. multocida, the loss of a single “A” at position 793 within the sul2 gene caused a frame-shift mutation which led to the substitution of 6 codons and extended the reading frame by 18 codons (146). Finally, a recombination in the 3′ end of the sul2 reading frame changed the final three codons and extended the sul2 reading frame by one codon in plasmid pVM111 from P. multocida (59). Plasmids carrying sul2 have also been identified in bacteria other than Pasteurellaceae, e.g., E. coli (147) and Photobacterium damselae subsp. piscicida (148). In addition to their location on small plasmids, sul2 genes have also been detected on broad-host-range conjugative (e.g., pGS05) or nonconjugative (e.g., RSF1010, pLS88) plasmids (134, 147, 149) and on the ICEPmu1 (55).
Various studies revealed that the sul2 gene is often linked to the strA-strB genes (65, 103, 108, 122, 125, 134, 150). PCR assays were developed to confirm the linkage of sul2 and strA in both orientations (125). In some cases, the strA gene followed by a truncated ΔstrB gene was detected upstream of sul2 (125, 150). However, in most P. multocida, [P.] aerogenes, and Mannheimia isolates studied, these genes were found in the orientation sul2-strA, whereas a truncated ΔstrA may also be found upstream of sul2 (65, 103, 125, 134). Detailed studies of the noncoding spacer between sul2 and strA revealed different lengths (121, 125) and showed that this region might represent a hot spot for recombination events. A catA3 gene, coding for chloramphenicol resistance, was found to be inserted between sul2 and strA via illegitimate recombination. Such sul2-catA3-strA clusters have been found on various plasmids as well as in the chromosomal DNA of [P.] aerogenes, M. haemolytica, M. varigena, and Mannheimia taxon 10 isolates (116, 125). Further insertion of aacC2 between sul2 and catA3 was identified in pHB0503 from A. pleuropneumoniae (95). In plasmid pVM111 from an avian P. multocida isolate (44, 48), a Tn5706-like tetR-tet(H) segment, responsible for tetracycline resistance, was also found to be inserted between sul2 and strA via illegitimate recombination (59). The resulting sul2-tetR-tet(H)-strA cluster, however, has not yet been detected on plasmids other than pVM111 or in the chromosomal DNA of Pasteurella, Mannheimia, or Actinobacillus isolates (Fig. 1). In addition to insertions between sul2 and strA, two recent A. pleuropneumoniae plasmids, pM3224T and pM3389T, were found to have insertions of dfrA14 (encoding trimethoprim resistance) disrupting the strA gene, located either downstream (pM3224T) (Fig. 4) or upstream (pM3389T) of sul2 (122).
Several small plasmids that carry only the sulfonamide resistance gene sul2, but no other resistance genes, have been sequenced completely. These include the plasmids pKMA2425 (3,156 bp) and pARD3079 (4,065 bp), both from A. pleuropneumoniae, as well as pKMA757 (4,556 bp) from “A. porcitonsillarum” (108) (Fig. 4). A 4,236-bp A. pleuropneumoniae plasmid, ABB7_B (accession number NC_010941), carries sul2 and strA. All of these plasmids share the same mobilization genes (mobABC), except pKM2425, which contains only mobC and a truncated copy of mobA (Fig. 4).
Other sul genes, such as sul1 and sul3, which also code for dihydropteroate synthetases with reduced affinity to sulfonamides, have not yet been detected among Pasteurellaceae species (62; Kehrenberg and Schwarz, unpublished data).
Trimethoprim Resistance Mediated by Altered DHFRs
A novel trimethoprim resistance gene was detected on the 11-kb plasmid pCCK154 from bovine P. multocida (146). This plasmid was transferable into E. coli, where it replicated and expressed high-level resistance to sulfonamides and trimethoprim. Sequence analysis identified the gene sul2 for sulfonamide resistance and a novel gene, designated dfrA20, for trimethoprim resistance. The dfrA20 gene codes for a trimethoprim-resistant DHFR of 169 amino acids, which is only distantly related to the DHFRs of Gram-negative bacteria, but upon cluster analysis appears to be related to those found in the Gram-positive genera Staphylococcus, Bacillus, and Listeria (146).
A different trimethoprim resistance gene, dfrA1, was identified on plasmid pCCK343 recovered from a porcine intestinal [P.] aerogenes isolate (130). This 5,415-bp plasmid contains a backbone sequence with homology to pHS-Tet, including the mobC gene (but not mobAB), with insertion of a partially truncated class 2 integron containing dfrA1-sat2-aadA1. In addition to conferring trimethoprim resistance (MIC, >256 mg/liter), the presence of the streptothricin acetyltransferase gene, sat2, and the aminoglycoside adenyltransferase gene, aadA1, conferred resistance to the streptothricin antibiotic nourseothricin (MIC, >256 mg/liter) and to streptomycin and spectinomycin (MICs of >256 mg/liter each), respectively. The truncated integron showed high identity with a sequence found in an E. coli ICE, AGI-5 (130).
Earlier studies of trimethoprim-resistant bovine M. haemolytica isolates from France revealed that this resistance was not associated with plasmids, and also was not transferable by conjugation. Hybridization experiments with gene probes specific for the genes dfrA1 to dfrA5 did not yield positive results (151). Similar negative PCR results were obtained for A. pleuropneumoniae and “A. porcitonsillarum” isolates in Switzerland, in which none of 27 different dfrA or dfrB genes—including dfrA20—could be identified using primers designed to detect groups of dfr genes (62). Recently, whole-genome sequencing was used to identify dfrA14 in trimethoprim-resistant A. pleuropneumoniae isolates from the United Kingdom (122). This gene may have been responsible for trimethoprim resistance in the Swiss isolates of “A. porcitonsillarum” and A. pleuropneumoniae (62), if there was a failure to amplify a product using the primers designed to detect dfrA5/dfrA14/dfrA25. Alternatively, these isolates possess another mechanism for trimethoprim resistance that remains to be elucidated. The study by Bossé et al. (122) highlights the value of whole-genome sequencing for determination of the genetic basis of resistance, not only providing information regarding the presence of specific resistance genes and/or mutations, but also facilitating localization of the gene(s) within the chromosomal DNA or on plasmids.
In the United Kingdom isolates, two distinct mobilizable trimethoprim resistance plasmids were identified (122) in which the dfrA14 gene was inserted into strA, as has been reported for enterobacterial plasmids pSTOJO1 and pCERC1 (126, 152). Differences in the gene order of flanking regions, with pM3224T carrying sul2-ΔstrA-dfrA14-ΔstrA-strB (Fig. 4) and pM3389T carrying ΔstrA-dfrA14-ΔstrA-sul2, suggest separate recombination of the ΔstrA-dfrA14-ΔstrA cassette (likely of enterobacterial origin) into different Pasteurellaceae plasmids.
Resistance to Macrolides
Many Gram-negative bacteria are believed to be innately resistant to macrolides due to permeability barriers or multidrug efflux pumps. However, chemical modification of the ribosomal target site by rRNA methylases or mutations in ribosomal proteins have also been described (109).
Macrolide Resistance Mediated by rRNA Methylases
Studies of the presence of macrolide resistance genes in bacteria of the genus Actinobacillus led to the identification of the rRNA methylase genes erm(A) and erm(C) in A. pleuropneumoniae (70). Mating experiments showed that these genes were transferred into Moraxella catarrhalis and/or Enterococcus faecalis (70). PCR-directed analysis of bovine P. multocida and M. haemolytica which exhibited MICs of erythromycin of ≥16 mg/liter did not detect any of the three genes erm(A), erm(B), or erm(C) (Kehrenberg and Schwarz, unpublished data). Matter and coworkers obtained similar results for A. pleuropneumoniae and “A. porcitonsillarum” isolates with MICs of tilmicosin of ≥32 mg/liter, in which none of the genes erm(A), erm(B), or erm(C) could be detected by PCR (62). Australian isolates of A. pleuropneumoniae with MICs of erythromycin of ≥16 mg/liter and/or MICs of tilmicosin of ≥32 mg/liter (40) also failed to amplify specific products for erm(A), erm(B), erm(C), erm(42), mph(E), mef(A), msr(A), or msr(E) (153). Furthermore, whole-genome sequencing of A. pleuropneumoniae HS 3572 (MIC of erythromycin of 16 mg/liter; MIC of tilmicosin of 32 mg/liter; MIC of tulathromycin of 16 mg/liter) revealed neither specific macrolide resistance genes nor any known point mutations previously associated with macrolide resistance (153). In [H.] parasuis, a 7,577-bp plasmid, pFS39, has been reported to carry the erm(T) gene, previously only identified in Gram-positive bacteria, along with blaROB-1 (154). The [H.] parasuis isolate harboring pFS39 had MICs of 64 mg/liter for both erythromycin and lincomycin (154). A novel monomethyltransferase gene, erm(42), has been identified in the chromosome of M. haemolytica and P. multocida isolates with high levels of resistance to multiple macrolides (155–157). The sequence of this monomethyltransferase gene was found to be divergent from previously reported erm genes and was only detected by whole-genome sequencing of the resistant isolates.
Macrolide Resistance Mediated by Mutations in Ribosomal Proteins and 23S rRNA
Mutations in ribosomal proteins L4 or L22 have been reported in human macrolide-resistant H. influenzae (158). In isolates of P. multocida and M. haemolytica reported to be highly resistant (MICs >64 mg/liter) to multiple macrolides including erythromycin, tilmicosin, tildipirosin, tulathromycin, and gamithromycin, mutations were found in one of two locations (either A2058G or A2059G) in all six copies of the 23S rRNA (159). Similarly, genome sequencing revealed an A2059G transition in all six copies of the 23S rRNA gene in one isolate of [H.] parasuis (HS 315) with MICs of erythromycin of 64 mg/liter, tilmicosin of ≥128 mg/liter, and tulathromycin of 64 mg/liter; however, other isolates with MICs of erythromycin of 64 mg/liter, but lower MICs of tilmicosin and tulathromycin compared to HS 315, did not show this, or any other known, mutation associated with macrolide resistance (153).
Macrolide Resistance Mediated by Active Efflux or Inactivation by Phosphotransferases
It was shown that some isolates of M. haemolytica, P. multocida, and H. somni had genes, mrs(E) and mph(E), encoding macrolide efflux and phospotransferase proteins, respectively (57, 155–157). These two genes, found in tandem and expressed from the same promoter, were also predicted to be chromosomally encoded (155, 156). When found in combination with erm(42), the presence of the mrs(E)-mph(E) operon resulted in the highest level of resistance to all macrolides tested (tulathromycin, gamithromycin, tilmicosin, and clindamycin), whereas those with just the mrs(E)-mph(E) genes were not resistant to clindamycin and had lower MICs for tilmicosin but higher MICs for gamithromycin and tulathromycin compared to isolates with only the erm(42) gene (155, 156, 160). All three genes [erm(42), mrs(E), and mph(E)] were identified as part of ICEPmu1 (55), which might explain their dissemination across strain, species, and genus boundaries.
Resistance to Lincosamides
Resistance mechanisms to lincosamides may involve 23S rRNA methyltransferase, efflux proteins, inactivation enzyme genes, and mutations (e.g., 23S rRNA, L4 and/or L22 genes). Lincosamide transferase genes have been commonly detected among Gram-positive bacteria, and the lnu(F) gene has been detected in Salmonella and E. coli (http://faculty.washington.edu/marilynr/ermweb4.pdf). However, an lnu(C) gene, located on the 6.3-kb plasmid pHN61, was identified in a porcine [H.] parasuis isolate from China (161). The high MICs of lincomycin (32 mg/liter) and clindamycin (8 mg/liter) and the low MIC of erythromycin (0.25 mg/liter) indicates that lnu(C) mediates resistance to lincosamides but not to macrolides. The replication and mobilization region of pHN61 shares sequence similarity with that in pHB0503 from an A. pleuropneumoniae isolate.
Resistance to Phenicols
Resistance to nonfluorinated phenicols may be due to enzymatic inactivation by chloramphenicol acetyltransferases, whereas resistance to fluorinated and/or nonfluorinated phenicols can be mediated by phenicol-specific exporters (109, 162). Other mechanisms, such as permeability barriers, have rarely been detected among Pasteurellaceae.
Chloramphenicol resistance mediated by enzymatic inactivation
Resistance to chloramphenicol is usually due to enzymatic inactivation of the drug by chloramphenicol acetyltransferases. Two types of chloramphenicol acetyltransferases, A and B, specified by a number of different catA and catB genes, are currently known (162). The catA and catB genes are often located on plasmids, transposons, or gene cassettes. Plasmids mediating chloramphenicol resistance have been identified in porcine P. multocida isolates (113), bovine P. multocida and M. haemolytica isolates (163), porcine [P.] aerogenes (125), bovine Mannheimia spp. (116, 125), and porcine A. pleuropneumoniae (95, 106, 117, 164, 165). As previously seen with other resistance plasmids, those mediating chloramphenicol resistance are commonly less than 15 kb in size and occasionally carry additional resistance genes (Table 3). Initial molecular studies of chloramphenicol resistance among P. multocida and M. haemolytica isolates included the detection of the three most frequently occurring cat genes among Gram-negative bacteria—catA1, catA2, and catA3 (formerly known as catI, catII, and catIII)—by specific PCR assays (163). The catA3 gene was detected on small plasmids of 5.1 kb, whereas the catA1 gene was located on plasmids of either 17.1 or 5.5 kb (163). In porcine [P.] aerogenes and bovine Mannheimia isolates, catA3 genes have been identified as parts of chromosomal or plasmid-borne resistance gene clusters (116, 125). In these gene clusters, the catA3 gene was always inserted between the sulfonamide resistance gene sul2 and the streptomycin resistance gene strA (116, 125) (Fig. 4). In pHB0503, a 15.1-kb plasmid isolated from A. pleuropneumoniae, this gene arrangement has been disrupted by insertion of aacC2 and a truncated copy of an ATPase gene between sul2 and catA3 (95). A similar 15.7-kb plasmid from [H.] parasuis, pFZ51, contains the same resistance gene cluster as pHB0503 (accession number JN202624; Fig. 4, Table 3).
Although the catA2 gene is commonly found in H. influenzae (166, 167), this gene has not yet been detected in chloramphenicol-resistant Pasteurella, Mannheimia, or Actinobacillus. However, a catB2 gene, which codes for a different type of chloramphenicol acetyltransferase than the aforementioned catA genes (162), was found on plasmid pJR1 from avian P. multocida (76). The catB2 gene is part of a gene cassette and thus needs the integron-associated promotor for its expression. In plasmid pJR1, the catB2 cassette was located outside of an integron structure. Gene cassettes located at secondary sites outside of integrons may be expressed if a suitable promotor is available (126). However, such a promotor has not been identified in pJR1 (76).
Chloramphenicol and florfenicol resistance mediated by active efflux
So far, resistance to florfenicol, a structural analogue of thiamphenicol, has rarely been detected among Pasteurellaceae, except in Korea, where 34% of A. pleuropneumoniae isolates collected between 2006 and 2010 and 43% of isolates collected between 2010 and 2013 were resistant to florfenicol (73, 81). All of the isolates characterized by Yoo et al. (73) had MICs of florfenicol of between 8 and ≤64 mg/liter and carried the floR gene, which codes for an exporter of the major facilitator family that mediates the efflux of phenicols from the bacterial cell. Monitoring studies to specifically determine the MIC values of florfenicol among bovine and porcine respiratory tract pathogens showed that all P. multocida, M. haemolytica, and A. pleuropneumoniae isolates collected between 1994 and 2005 were susceptible to florfenicol (168–171; Schwarz, unpublished data). Data from the monitoring programs GERM-Vet and BfT-GermVet, conducted in Germany, also confirmed that all P. multocida from pigs, poultry, dogs, and cats were susceptible to florfenicol (36, 172, 173), whereas 8.6% of M. haemolytica and 11.6% of P. multocida isolated from cattle in Canada and United States in 2009 were resistant (37).
The first report of a florfenicol-resistant bovine P. multocida isolate was published in 2005 (174). This isolate of capsular type A originated from a calf that had died from pneumonia in the United Kingdom. Florfenicol resistance was based on the presence of the gene floR. This gene was located on the 10,874-bp plasmid pCCK381 (Fig. 5). Interestingly, a florfenicol-resistant S. enterica subsp. enterica serovar Dublin isolate recovered from the same calf carried a plasmid indistinguishable from pCCK381. This observation, and the fact that plasmid pCCK381 was able to replicate and express florfenicol resistance in different E. coli hosts (174), strongly suggested that the plasmid pCCK381 can be exchanged between bacteria of different genera. Sequence analysis confirmed that this plasmid consists of three segments that show extended similarity to plasmids pDN1 from Dichelobacter nodosus (175), pMBSF1 from E. coli (176), and pRVS1 from Vibrio salmonicida (177). All these plasmids have been found either in bacteria such as E. coli from cattle and pigs, which have previously been shown to carry the floR gene along with a variable length ORF with homology to a lysR regulator gene, or in bacteria which cause diseases in fish and ruminants, such as cold-water vibriosis (V. salmonicida) or infectious pododermatitis (D. nodosus), for the control of which florfenicol is approved in several non-European Union countries. Although it is not possible to determine in retrospect where and when plasmid pCCK381 evolved, structural analysis suggested that this plasmid is most likely the result of interplasmid recombination processes (174). It is also interesting to note that pCCK381 has been isolated from both porcine and bovine isolates of P. multocida in Germany (178).
FIGURE 5.
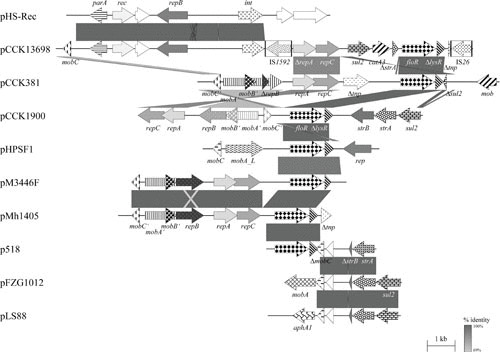
Schematic representation of the structure and organization of selected floR-based (multi-)resistance plasmids from B. trehalosi compared to an in-part-related plasmid from [H.] parasuis, and A. pleuropneumoniae, [H.] parasuis, P. multocida, and sul2-based (multi-)resistance plasmids from H. ducreyi and [H.] parasuis. Comparison of the maps of plasmids pCCK13698 (accession no. AM183225) from B. trehalosi and its in-part-related plasmid pHS-Rec (accession no. AY862436; 9,462 bp) from [H.] parasuis, pCCK381 (accession no. AJ871969; 10,874 bp) from P. multocida, pCCK1900 (accession no. FM179941; 10,226 bp) from P. multocida, pHPSF1 (accession no. KR262062; 6,328 bp) from [H.] parasuis, pM3446F (accession no. KP696484; 7,709 bp) from A. pleuropneumoniae, pMh1405 (accession no. NC_019260; 7,674 bp) from M. haemolytica, p518 (accession no. KT355773; 3,937 bp) from A. pleuropneumoniae, pFZG1012 (accession no. HQ015158; partially sequenced) from [H.] parasuis, and pLS88 (accession no. L23118; 4,772 bp) from H. ducreyi. Genes are shown as arrows, with the arrowhead indicating the direction of transcription. The following genes are involved in antimicrobial resistance: sul2 (sulfonamide resistance), strA and strB (streptomycin resistance), catA3 (chloramphenicol resistance), floR (chloramphenicol/florfenicol resistance), and aphA1 (kanamycin/neomycin resistance); plasmid replication: rep, repA, repB, and repC; mobilization functions: mobA, mobB, mobC, and mob; transposition functions: tnp; recombinase or integrase functions: rec and int; DNA partition: parA; unknown function: open reading frames indicated by white arrows. The prefix Δ indicates a truncated functionally inactive gene. The boxes in the map of pCCK13698 indicate the limits of the insertion sequences IS1592 and IS26; the arrows within these boxes indicate the reading frames of the corresponding transposase genes. Gray-shaded areas indicate the regions common to plasmids, and the different shades of gray illustrate the percentages of nucleotide sequence identity between the plasmids, as indicated by the scale at the bottom of the figure. A distance scale in kilobases is shown.
More recently, a number of smaller plasmids with floR as the only resistance gene have been identified in [H.] parasuis (pHPSF1), M. haemolytica (pMH1405), and A. pleuropneumoniae (pM3446F and p518) (179–182). Plasmids pMH1405 and pM3446F, both 7.7 kb, share extensive regions of identity with pCCK381, including the mobilization and replication genes, whereas the 6.3-kb pHPSF1 contains different mob and rep genes. The smallest plasmid, the 3.9-kb p518, shares the floR-lsyR region (high sequence identity) with pMh1405, and the remaining region, containing ΔmobC and strA-ΔstrB sequences, with [H.] parasuis plasmid pFZG1012 (Fig. 5).
The floR gene has also been found on multiresistance plasmids pCCK13698 and pCCK1900 (178, 183), and it is part of ICEPmu1 and ICEPmu1-related ICEs detected in M. haemolytica and H. somni (55). The 14,969-bp pCCK13698 plasmid was identified in a bovine isolate of Bibersteinia trehalosi (184) which originated from a calf that had died from a respiratory tract infection in France (183). The 10.2-kb pCCK1900 plasmid was recovered from a porcine P. multocida isolate in Germany (178). Both plasmids differ structurally from plasmid pCCK381 and from each other (Fig. 5). While in pCCK13698, the floR-lysR sequence is located downstream of the complete genes sul2 and catA3, and a truncated strA streptomycin resistance gene (183), in plasmid pCCK1900, the floR-lysR genes are found downstream of (and in the opposite orientation to) complete sul2, strA, and strB genes (Fig. 5). Moreover, plasmid pCCK13698 shares almost 6.8 kb of identity with the [H.] parasuis plasmid pHS-Rec (75) and harbors two insertion sequences, IS1592 and IS26 (183). In contrast, pCCK1900 appears to have arisen by insertion of the floR-lysR genes into the broad-host-range plasmid RSF1010 (178). Such analysis reveals a high level of involvement of the region floR-lysR in interplasmid recombination events. In ICEPmu1, floR is found as part of “resistance region 1,” a 15,711-bp sequence bracketed by copies of ISApl1, an insertion sequence initially identified in A. pleuropneumoniae (185). This region carries aphA1, strA, strB, sul2, floR, and erm(42), with the floR-lysR genes found to be bracketed by a truncated and a complete ISCR2 element (55).
Resistance to (Fluoro)quinolones
Quinolones are broad-spectrum antimicrobial agents that inhibit bacterial DNA gyrase and topoisomerase IV. Resistance is commonly due to mutational alterations in the genes coding for the different subunits of both enzymes but is also due to active efflux or protection of the enzymes by Qnr proteins (109).
(Fluoro)quinolone resistance mediated by target site mutations
Very little is known about (fluoro)quinolone resistance in Pasteurella, Mannheimia, Actinobacillus, or Haemophilus. Studies of Pasteurella spp. from humans and animals (36, 172, 173, 186) and A. pleuropneumoniae (62) identified virtually all isolates to be highly susceptible to the fluoroquinolones tested. The first report describing the analysis of the quinolone resistance determining region (QRDR) of the proteins encoded by the genes gyrA and parC in P. multocida isolates identified a Ser83Ile alteration in GyrA in an isolate that had a nalidixic acid MIC of 256 mg/liter, whereas Asp87Gly alterations were detected in isolates with nalidixic acid MICs of 4 and 12 mg/liter (187). None of these isolates exhibited resistance to fluoroquinolones. Whole-genome sequencing of the multiresistant bovine P. multocida isolate 36950, exhibiting an enrofloxacin MIC of 2 mg/liter, led to the identification of two base pair exchanges in the QRDR of gyrA, which resulted in amino acid alterations GGT → AGT (Gly75Ser) and AGC → AGA (Ser83Arg). In addition, a single base pair exchange in the QRDR of parC, TCA → TTA, which resulted in a Ser80Leu exchange, was also seen (55).
More recently, fluoroquinolone-resistant clinical isolates of P. multocida, with MICs of 0.5 mg/liter for both enrofloxacin and ciprofloxacin, were found to have Asp87Asn or Ala84Pro mutations in GyrA (188). In isolates further selected for resistance by passage on subinhibitory concentrations of fluoroquinolones, multiple mutations in gyrA, gyrB, and parC, but not parE, were found to be associated with high-level fluoroquinolone resistance (MICs, >4 mg/liter). Similarly, in M. haemolytica, resistance to nalidixic acid was associated with at least one amino acid substitution in one or both of GyrA and ParC, whereas all of the isolates with fluoroquinolone MICs ≥8.0 mg/liter had two mutations in GyrA and one additional change in ParC (189). A study of enrofloxacin-resistant A. pleuropneumoniae isolates in Taiwan (190) identified various mutations in gyrA, parC, and parE, with resistant isolates carrying at least one mutation in the QRDR of gyrA, resulting in amino acid changes at codon 83 or 87. Functional efflux pumps were also found to contribute to enrofloxacin resistance in these isolates, as demonstrated by addition of an efflux pump inhibitor (190). In a recent study comparing antimicrobial resistance profiles and the presence of resistance genes in whole-genome sequences of A. pleuropneumoniae (9), only a slightly elevated enrofloxacin MIC of 0.25 mg/liter was detected in a limited number of isolates, and in all cases this resistance was associated with the GyrA Ser83Phe substitution described by Wang et al. (190).
In a survey of 115 [H.] parasuis isolates collected from South China between 2008 and 2010, 20 were identified with high levels of resistance to nalidixic acid (MICs ≥128 mg/liter) as well as resistance to enrofloxacin and other fluoroquinolones (MICs ≥4 mg/liter), and these were shown to have at least one mutation in the gyrA gene causing changes at codon 83 or 87 (191). In another survey, of 138 [H.] parasuis isolates from China between 2002 and 2009, 60% were found to be resistant to enrofloxacin, and sequencing of PCR products demonstrated 10 point mutations in the quinolone QRDR regions of gyrA, gyrB, parC, and parE contributing to varying degrees of enrofloxacin resistance (192). However, the mutation causing the amino acid change Asp87Asn was common to all resistant isolates. This study further showed, by site-directed mutagenesis, that point mutations leading to three amino acid changes (Asp87Asn in GyrA, Ser73Arg in ParC, and Thr551Ala in ParE) were specifically involved in fluoroquinolone resistance (192).
(Fluoro)quinolone resistance mediated by other mechanisms
The AcrAB efflux pump has been shown in Enterobacteriaceae to be able to export fluoroquinolones from the cell (193). Although the use of efflux inhibitors has demonstrated a contribution of efflux pump(s) to fluoroquinolone resistance in A. pleuropneumoniae and P. multocida (188, 190), the specific proteins involved were not identified. A PCR survey of 115 Chinese [H.] parasuis isolates detected quinolone resistance genes qnrA1, qnrB6, and aac(6′)-Ib-cr in three, one, and three isolates, respectively (191). To date, this is the only report of acquired quinolone resistance genes in the Pasteurellaceae.
DISSEMINATION, COSELECTION, AND PERSISTENCE OF RESISTANCE GENES IN PASTEURELLA, MANNHEIMIA, ACTINOBACILLUS, HAEMOPHILUS, AND HISTOPHILUS
Molecular analysis of isolates of Pasteurella, Mannheimia, Actinobacillus, and Haemophilus revealed that, in many cases, antimicrobial resistance genes are associated with plasmids. Most of the resistance plasmids identified to date are <15 kb in size and are nonconjugative, though many carry mob genes (Table 3) (Fig. 1 to 5) and have been shown to be mobilizable. For these plasmids to be horizontally transferred by conjugation, the genes encoding the conjugation machinery must be supplied in trans. The only conjugative elements so far identified in members of the Pasteurellaceae are ICEs, including those recently described in P. multocida, M. haemolytica, H. somni, and A. pleuropneumoniae (55–57, 74). These ICEs are likely, along with others yet to be identified, be the source of the genes required for conjugal transfer of the smaller mobilizable plasmids identified in these species.
The number of plasmids for which complete sequences are available continues to grow. Currently, there are complete sequences for 60 plasmids carrying antimicrobial resistance genes (Table 3). In addition, the resistance gene regions of several other plasmids (also listed in Table 3) have been sequenced. Despite the relatively small size of these plasmids, many of them carry two or more resistance genes (Table 3, Fig. 3 to 5). To date, there are no sequences for resistance plasmids from Histophilus isolates in GenBank (last accessed October 27, 2017). Sequence analysis has provided insights into (i) the genes involved in antimicrobial resistance of Pasteurella, Mannheimia, Actinobacillus, and Haemophilus and their organization; (ii) the structural relationships between the resistance plasmids; and (iii) mechanisms resulting in the formation of multiresistance plasmids and plasmid-borne multiresistance gene clusters.
Some of the genes accounting for antimicrobial resistance in Pasteurella, Mannheimia, Haemophilus, and Actinobacillus, such as the tetracycline resistance gene tet(H) and the β-lactamase gene blaROB-1, appear to be more commonly found among Pasteurellaceae. In contrast, other resistance genes, such as the streptomycin resistance genes strA-strB, have been detected in a wide range of Gram-negative bacteria. The fact that these genes are associated with the transposon Tn5393 (124), and have also been found on two mobilizable broad-host-range plasmids, pLS88 (134) and RSF1010 (147), might explain their widespread occurrence, which is likely due to horizontal gene transfer.
The location of resistance genes on mobile genetic elements allows their spread into bacteria of other species and genera. This has been confirmed by the detection of highly similar plasmids in different host bacteria, such as the tet(H)-carrying plasmid pMHT1, which was found in P. multocida, M. haemolytica, M. varigena, and M. glucosida (64), and the sul2-strA-carrying plasmid pPMSS1, which was identified in P. multocida, [P.] aerogenes, and M. haemolytica (125). Several of these small plasmids carry mob genes which allow mobilization in the presence of a conjugative element. Mobilization has also been confirmed for the 8.1-kb sul2-strA-carrying plasmid pMS260 from A. pleuropneumoniae (121). This plasmid, which closely resembles the broad-host-range plasmid RSF1010 (147), proved to be mobilizable into a wide variety of respiratory tract pathogens including P. multocida, Bordetella bronchiseptica, and Pseudomonas aeruginosa, as well as other isolates of A. pleuropneumoniae (119). Moreover, most of the small resistance plasmids found in Pasteurella, Mannheimia, Actinobacillus, and Haemophilus are able to replicate and express their resistance properties in E. coli. On the other hand, most resistance plasmids originating from the Enterobacteriaceae and harboring either catA3, strA-strB, tet(B), or sul2, usually cannot replicate in Pasteurella, Mannheimia, Actinobacillus, or Haemophilus hosts. With the exception of the RSF1010-like plasmid pMS260 from A. pleuropneumoniae (121), analysis of the regions flanking the sul2 and strA-strB genes in plasmids of Pasteurella, Mannheimia, Actinobacillus, and Haemophilus revealed sequences similar not to those of Enterobacteriaceae plasmids, but rather to those of plasmids known to occur in Pasteurellaceae. This observation suggests that, most likely, recombination events between endogenous plasmids of Pasteurellaceae and resistance plasmids from other bacterial sources, which may be replication-deficient in Pasteurellaceae, have occurred. As a result, the horizontally acquired resistance genes became inserted into new plasmidic replicons which then have been stably maintained in Pasteurellaceae. Studies of the plasmid-borne sul2-strA gene cluster in Pasteurella and Mannheimia showed that the noncoding spacer region between the two resistance genes may represent the target site for further recombination events (116, 125). So far, a catA3 gene and the entire tetR-tet(H) region, respectively, have been found to be integrated between the genes sul2 and strA in different plasmids (59, 116).
Based on the observation that many of the resistance plasmids so far detected in Pasteurella, Mannheimia, and Actinobacillus carry more than one resistance gene, coselection of these resistance genes is likely. Once such a plasmid is transferred to a new host, all resistance genes located on it are also transferred. As a consequence, a new host bacterium gains resistance to two or more antimicrobial agents, or classes of antimicrobial agents, by the acquisition of a single small plasmid. In these cases, selective pressure imposed by the use of one such antimicrobial agent, e.g., tetracyclines, is sufficient to favor the exchange of the multiresistance plasmid. The location of different resistance genes on the same plasmid also enables their persistence, particularly if the resistance genes are organized in a cluster. A multiresistance gene cluster in which the genes sul2, catA3, and strA are organized as a transcriptional unit has been identified on several plasmids, as well as in chromosomal DNA of [P.] aerogenes and several Mannheimia spp. (116, 125). It is highly unlikely that individual genes from such a cluster are lost. A study of the location of chloramphenicol resistance genes in Pasteurella and Mannheimia isolates from Germany showed that the dominant chloramphenicol resistance gene catA3 was located in a sul2-catA3-strA cluster in all cases, except for a single bovine M. glucosida isolate (125). Although chloramphenicol has been prohibited for use in food-producing animals in the European Union and the United States for many years, aminoglycosides, including streptomycin, as well as sulfonamides are still used. Thus, streptomycin and/or sulfonamides might present the selective pressure that ensures the maintenance of the entire cluster. Thus, clusters such as that containing the genes sul2-catA3-strA may play an important role in maintaining resistance genes without direct selective pressure.
Analysis of the resistance genes, their location, and their organization provides insight into (i) the gene pool to which bacteria of the genera Pasteurella, Mannheimia, Actinobacillus, and Haemophilus have access; (ii) horizontal transfer processes which play a key role in the dissemination of the resistance genes between and beyond bacteria of the four aforementioned genera; and (iii) integration and recombination events which are of major importance for the development of novel resistance plasmids and the formation of multiresistance gene clusters in Pasteurella, Mannheimia, Actinobacillus, and Haemophilus isolates.
CONCLUDING REMARKS
Isolates of several genera in the family Pasteurellaceae cause a number of economically important diseases in cattle, swine, and other food-producing animals. Due to the multifactorial and polymicrobial nature of some of the infections in which Pasteurella, Mannheimia, Actinobacillus, Haemophilus, and Histophilus isolates are involved, prevention is—except in confined production systems with high biosecurity—a cumbersome task that often yields unsatisfying results. Despite hygienic measures, improved management and the use of vaccines, antimicrobial agents are indispensable tools for the control of infections in which bacteria of the family Pasteurellaceae are involved. Increasing numbers of resistant, or multiresistant, isolates are reducing the efficacy of antimicrobial agents currently approved for use in animals. It is anticipated that in the near future, there will be no new classes of antimicrobial agents approved for use in veterinary medicine. Thus, veterinarians will have to rely on currently available antimicrobial agents. To retain their efficacy, prescription and administration of antimicrobial agents should be undertaken with discretion supported by an accurate diagnosis, a careful choice of the antimicrobial agent(s), and the most appropriate dosing regimen. Imprudent use of antimicrobials bears a high risk of selecting resistant bacteria.
Many of the resistance genes known to be present in Pasteurella, Mannheimia, Actinobacillus, and Haemophilus are associated with plasmids or transposons and thus may be exchanged horizontally, not only between bacteria of the family Pasteurellaceae, but also with other Gram-negative bacteria. This update on the molecular basis of antimicrobial resistance illustrates that Pasteurella, Mannheimia, Actinobacillus, and Haemophilus have obviously acquired a number of resistance genes from other Gram-negative, or maybe even Gram-positive, bacteria. Knowledge of the location and colocation of the resistance genes on mobile genetic elements, as well as the conditions for their coselection and persistence, will be valuable information for veterinarians and will assist them in selecting the most efficacious antimicrobial agents for control of isolates of the family Pasteurellaceae. Even though the current susceptibility status of Pasteurellaceae from infections in animals looks rather favorable, continuous monitoring of the antimicrobial susceptibility of Pasteurellaceae is required for early detection of changes in the susceptibility status and for analyzing the respective isolates for newly acquired/developed resistance genes and resistance-mediating mutations.
ACKNOWLEDGMENTS
Funding for GBM and SS was provided by grant SCHW382/10-2 from the German Research Foundation (DFG). Funding for JB was provided by the Biotechnology and Biological Sciences Research Council (BB/K021109/1, BB/G018553, and BB/M023052/1), as well as through a CONFAP-the UK Academies Fellowship (FAPEMIG – APQ-00689-16).
REFERENCES
- 1.Christensen H, Kuhnert P, Busse HJ, Frederiksen WC, Bisgaard M. 2007. Proposed minimal standards for the description of genera, species and subspecies of the Pasteurellaceae. Int J Syst Evol Microbiol 57:166–178 10.1099/ijs.0.64838-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 2.Christensen H, Kuhnert P, Nørskov-Lauritsen N, Planet PJ, Bisgaard M. 2014. The family Pasteurellaceae, p 535–564. In DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The Prokaryotes. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 3.Adhikary S, Nicklas W, Bisgaard M, Boot R, Kuhnert P, Waberschek T, Aalbæk B, Korczak B, Christensen H. 2017. Rodentibacter gen. nov. including Rodentibacter pneumotropicus comb. nov., Rodentibacter heylii sp. nov., Rodentibacter myodis sp. nov., Rodentibacter ratti sp. nov., Rodentibacter heidelbergensis sp. nov., Rodentibacter trehalosifermentans sp. nov., Rodentibacter rarus sp. nov., Rodentibacter mrazii and two genomospecies. Int J Syst Evol Microbiol 67:1793–1806 10.1099/ijsem.0.001866. [DOI] [PubMed] [Google Scholar]
- 4.Christensen H, Kuhnert P, Bisgaard M, Mutters R, Dziva F, Olsen JE. 2005. Emended description of porcine [Pasteurella] aerogenes, [Pasteurella] mairii and [Actinobacillus] rossii. Int J Syst Evol Microbiol 55:209–223 10.1099/ijs.0.63119-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 5.Christensen H, Bisgaard M. 2008. Taxonomy and biodiversity of members of Pasteurellaceae, p 1–25. In Kuhnert P, Christensen H (ed), Pasteurellaceae: Biology, Genomics and Molecular Aspects. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 6.Bonaventura MP, Lee EK, Desalle R, Planet PJ. 2010. A whole-genome phylogeny of the family Pasteurellaceae. Mol Phylogenet Evol 54:950–956 10.1016/j.ympev.2009.08.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 7.Moustafa AM, Seemann T, Gladman S, Adler B, Harper M, Boyce JD, Bennett MD. 2015. Comparative genomic analysis of Asian haemorrhagic septicaemia-associated strains of Pasteurella multocida identifies more than 90 haemorrhagic septicaemia-specific genes. PLoS One 10:e0130296 10.1371/journal.pone.0130296. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clawson ML, Murray RW, Sweeney MT, Apley MD, DeDonder KD, Capik SF, Larson RL, Lubbers BV, White BJ, Kalbfleisch TS, Schuller G, Dickey AM, Harhay GP, Heaton MP, Chitko-McKown CG, Brichta-Harhay DM, Bono JL, Smith TP. 2016. Genomic signatures of Mannheimia haemolytica that associate with the lungs of cattle with respiratory disease, an integrative conjugative element, and antibiotic resistance genes. BMC Genomics 17:982 10.1186/s12864-016-3316-8. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bossé JT, Li Y, Rogers J, Fernandez Crespo R, Li Y, Chaudhuri RR, Holden MT, Maskell DJ, Tucker AW, Wren BW, Rycroft AN, Langford PR. 2017. Whole genome sequencing for surveillance of antimicrobial resistance in Actinobacillus pleuropneumoniae. Front Microbiol 8:311 10.3389/fmicb.2017.00311. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kehrenberg C, Walker RD, Wu CC, Schwarz S. 2006. Antimicrobial resistance in members of the family Pasteurellaceae, p 167–186. In Aarestrup FM (ed), Antimicrobial Resistance in Bacteria of Animal Origin. ASM Press, Washington, DC. [Google Scholar]
- 11.Schwarz S. 2008. Mechanisms of antimicrobial resistance in Pasteurellaceae, p 199–228. In Kuhnert P, Christensen H (ed), Pasteurellaceae: Biology, Genomics and Molecular Aspects. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 12.Quinn PJ, Markey BK, Carter ME, Donnelly WJ, Leonard FC. 2002. Veterinary Microbiology and Microbial Disease, 2nd ed. Blackwell Publishing, Ames, IA. [Google Scholar]
- 13.Radostits OM, Gay C, Blood DC, Hinchcliff KW. 2000. Diseases caused by bacteria III, p 779-908. In Radostits OM, Gay C, Blood DC, Hinchcliff KW (eds), Veterinary medicine: a textbook of the diseases of cattle, sheep, pigs, goats, and horses. 9th ed. Saunders, Philadelphia, London. [Google Scholar]
- 14.Catry B, Chiers K, Schwarz S, Kehrenberg C, Decostere A, de Kruif A. 2005. A case of fatal peritonitis in calves caused by Pasteurella multocida capsular type F. J Clin Microbiol 43:1480–1483 10.1128/JCM.43.3.1480-1483.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh K, Ritchey JW, Confer AW. 2011. Mannheimia haemolytica: bacterial-host interactions in bovine pneumonia. Vet Pathol 48:338–348 10.1177/0300985810377182. [PubMed] [DOI] [PubMed] [Google Scholar]
- 16.Rice JA, Carrasco-Medina L, Hodgins DC, Shewen PE. 2007. Mannheimia haemolytica and bovine respiratory disease. Anim Health Res Rev 8:117–128 10.1017/S1466252307001375. [PubMed] [DOI] [PubMed] [Google Scholar]
- 17.Maldonado J, Valls L, Martínez E, Riera P. 2009. Isolation rates, serovars, and toxin genotypes of nicotinamide adenine dinucleotide-independent Actinobacillus pleuropneumoniae among pigs suffering from pleuropneumonia in Spain. J Vet Diagn Invest 21:854–857 10.1177/104063870902100615. [PubMed] [DOI] [PubMed] [Google Scholar]
- 18.Sárközi R, Makrai L, Fodor L. 2015. Identification of a proposed new serovar of Actinobacillus pleuropneumoniae: serovar 16. Acta Vet Hung 63:444–450 10.1556/004.2015.041. [PubMed] [DOI] [PubMed] [Google Scholar]
- 19.Bossé JT, Li Y, Sárközi R, Gottschalk M, Angen Ø, Nedbalcova K, Rycroft AN, Fodor L, Langford PR. 2017. A unique capsule locus in the newly designated Actinobacillus pleuropneumoniae serovar 16 and development of a diagnostic PCR assay. J Clin Microbiol 55:902–907 10.1128/JCM.02166-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frey J. 1995. Virulence in Actinobacillus pleuropneumoniae and RTX toxins. Trends Microbiol 3:257–261 10.1016/S0966-842X(00)88939-8. [DOI] [PubMed] [Google Scholar]
- 21.Oliveira S, Blackall PJ, Pijoan C. 2003. Characterization of the diversity of Haemophilus parasuis field isolates by use of serotyping and genotyping. Am J Vet Res 64:435–442 10.2460/ajvr.2003.64.435. [PubMed] [DOI] [PubMed] [Google Scholar]
- 22.Hodgins DC, Conlon JA, Shewen PE. 2002. Respiratory viruses and bacteria in cattle, p 213–229. In Brogden KA, Guthmiller JM (ed), Polymicrobial Diseases. ASM Press, Washington, DC. 10.1128/9781555817947.ch12 [DOI] [PubMed] [Google Scholar]
- 23.Brockmeier SL, Halbur PG, Thacker EL. 2002. Porcine respiratory disease complex, p 231–258. In Brogden KA, Guthmiller JM (ed), Polymicrobial Diseases. ASM Press, Washington, DC. 10.1128/9781555817947.ch13 [DOI] [Google Scholar]
- 24.Magyar T, Lax AJ. 2002. Atrophic rhinitis, p 169–197. In Brogden KA, Guthmiller JM (ed), Polymicrobial Diseases. ASM Press, Washington, DC. 10.1128/9781555817947.ch10 [DOI] [PubMed] [Google Scholar]
- 25.Watts J, Sweeney MT, Lubbers B. 2017. Antimicrobial susceptibility testing of bacteria of veterinary origin. In Aarestrup F, Schwarz S, Shen J, Cavaco L (ed), Antimicrobial Resistance in Bacteria from Livestock and Companion Animals, ASM Press, Washington, DC. [Google Scholar]
- 26.Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agersø Y, Lund O, Larsen MV, Aarestrup FM. 2013. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother 68:771–777 10.1093/jac/dks496. [PubMed] [DOI] [PubMed] [Google Scholar]
- 27.Köser CU, Ellington MJ, Peacock SJ. 2014. Whole-genome sequencing to control antimicrobial resistance. Trends Genet 30:401–407 10.1016/j.tig.2014.07.003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clinical and Laboratory Standards Institute (CLSI). 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; Approved standard, 4th edition. CLSI document VET01-A4. Clinical and Laboratory Standards Institute; Wayne, PA. [Google Scholar]
- 29.Clinical and Laboratory Standards Institute (CLSI). 2015. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, 3rd ed. CLSI suplement VET01S. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 30.Clinical and Laboratory Standards Institute (CLSI). 2017. Methods for antimicrobial susceptibility testing of infrequently isolated or fastidious bacteria isolated from animals, 1st ed. CLSI document VET06-Ed1. Clinical and Laboratory Standards Institute; Wayne, PA. [Google Scholar]
- 31.Dayao DA, Kienzle M, Gibson JS, Blackall PJ, Turni C. 2014. Use of a proposed antimicrobial susceptibility testing method for Haemophilus parasuis. Vet Microbiol 172:586–589 10.1016/j.vetmic.2014.06.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 32.Prüller S, Turni C, Blackall PJ, Beyerbach M, Klein G, Kreienbrock L, Strutzberg-Minder K, Kaspar H, Meemken D, Kehrenberg C. 2016. Towards a standardized method for broth microdilution susceptibility testing of Haemophilus parasuis. J Clin Microbiol 55:264–273 10.1128/JCM.01403-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bywater R, Silley P, Simjee S. 2006. Antimicrobial breakpoints: definitions and conflicting requirements. Vet Microbiol 118:158–159 10.1016/j.vetmic.2006.09.005. [PubMed] [DOI] [PubMed] [Google Scholar]
- 34.Schwarz S, Böttner A, Goossens L, Hafez HM, Hartmann K, Kaske M, Kehrenberg C, Kietzmann M, Klarmann D, Klein G, Krabisch P, Luhofer G, Richter A, Schulz B, Sigge C, Waldmann KH, Wallmann J, Werckenthin C. 2008. A proposal of clinical breakpoints for amoxicillin applicable to porcine respiratory tract pathogens. Vet Microbiol 126:178–188 10.1016/j.vetmic.2007.06.023. [PubMed] [DOI] [PubMed] [Google Scholar]
- 35.Bundesamt für Verbraucherschutz und Lebensmittelsicherheit (BVL). 2016. Berichte zur Resistenzmonitoringstudie 2012/2013 - Resistenzsituation bei klinisch wichtigen tierpathogenen Bakterien. Springer Nature, Cham, Switzerland. 10.1007/978-3-319-31697-0. http://www.bvl.bund.de/SharedDocs/Downloads/09_Untersuchungen/Archiv_berichte_Resistenzmonitoring/Bericht_Resistenzmonitoring_2012_2013.pdf;jsessionid=05577AB30DA96869657351B75EAE9267.2_cid350?__blob=publicationFile&v=5. [DOI] [Google Scholar]
- 36.Schwarz S, Alesík E, Grobbel M, Lübke-Becker A, Werckenthin C, Wieler LH, Wallmann J. 2007. Antimicrobial susceptibility of Pasteurella multocida and Bordetella bronchiseptica from dogs and cats as determined in the BfT-GermVet monitoring program 2004–2006. Berl Munch Tierarztl Wochenschr 120:423–430. [PubMed] [PubMed] [Google Scholar]
- 37.Portis E, Lindeman C, Johansen L, Stoltman G. 2012. A ten-year (2000-2009) study of antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex—Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni—in the United States and Canada. J Vet Diagn Invest 24:932–944 10.1177/1040638712457559. [PubMed] [DOI] [PubMed] [Google Scholar]
- 38.de Jong A, Thomas V, Simjee S, Moyaert H, El Garch F, Maher K, Morrissey I, Butty P, Klein U, Marion H, Rigaut D, Vallé M. 2014. Antimicrobial susceptibility monitoring of respiratory tract pathogens isolated from diseased cattle and pigs across Europe: the VetPath study. Vet Microbiol 172:202–215 10.1016/j.vetmic.2014.04.008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.El Garch F, de Jong A, Simjee S, Moyaert H, Klein U, Ludwig C, Marion H, Haag-Diergarten S, Richard-Mazet A, Thomas V, Siegwart E. 2016. Monitoring of antimicrobial susceptibility of respiratory tract pathogens isolated from diseased cattle and pigs across Europe, 2009–2012: VetPath results. Vet Microbiol 194:11–22 10.1016/j.vetmic.2016.04.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 40.Dayao DA, Gibson JS, Blackall PJ, Turni C. 2014. Antimicrobial resistance in bacteria associated with porcine respiratory disease in Australia. Vet Microbiol 171:232–235 10.1016/j.vetmic.2014.03.014. [PubMed] [DOI] [PubMed] [Google Scholar]
- 41.Archambault M, Harel J, Gouré J, Tremblay YDN, Jacques M. 2012. Antimicrobial susceptibilities and resistance genes of Canadian isolates of Actinobacillus pleuropneumoniae. Microb Drug Resist 18:198–206 10.1089/mdr.2011.0150. [PubMed] [DOI] [PubMed] [Google Scholar]
- 42.Noyes NR, Benedict KM, Gow SP, Booker CW, Hannon SJ, McAllister TA, Morley PS. 2015. Mannheimia haemolytica in feedlot cattle: prevalence of recovery and associations with antimicrobial use, resistance, and health outcomes. J Vet Intern Med 29:705–713 10.1111/jvim.12547. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeDonder KD, Apley MD. 2015. A literature review of antimicrobial resistance in pathogens associated with bovine respiratory disease. Anim Health Res Rev 16:125–134 10.1017/S146625231500016X. [PubMed] [DOI] [PubMed] [Google Scholar]
- 44.Berman SM, Hirsh DC. 1978. Partial characterization of R-plasmids from Pasteurella multocida isolated from turkeys. Antimicrob Agents Chemother 14:348–352 10.1128/AAC.14.3.348. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirsh DC, Martin LD, Rhoades KR. 1981. Conjugal transfer of an R-plasmid in Pasteurella multocida. Antimicrob Agents Chemother 20:415–417 10.1128/AAC.20.3.415. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirsh DC, Martin LD, Rhoades KR. 1985. Resistance plasmids of Pasteurella multocida isolated from turkeys. Am J Vet Res 46:1490–1493. [PubMed] [PubMed] [Google Scholar]
- 47.Hirsh DC, Hansen LM, Dorfman LC, Snipes KP, Carpenter TE, Hird DW, McCapes RH. 1989. Resistance to antimicrobial agents and prevalence of R plasmids in Pasteurella multocida from turkeys. Antimicrob Agents Chemother 33:670–673 10.1128/AAC.33.5.670. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hansen LM, McMurry LM, Levy SB, Hirsh DC. 1993. A new tetracycline resistance determinant, Tet H, from Pasteurella multocida specifying active efflux of tetracycline. Antimicrob Agents Chemother 37:2699–2705 10.1128/AAC.37.12.2699. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen LM, Blanchard PC, Hirsh DC. 1996. Distribution of tet(H) among Pasteurella isolates from the United States and Canada. Antimicrob Agents Chemother 40:1558–1560. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kehrenberg C, Werckenthin C, Schwarz S. 1998. Tn5706, a transposon-like element from Pasteurella multocida mediating tetracycline resistance. Antimicrob Agents Chemother 42:2116–2118. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hunt ML, Adler B, Townsend KM. 2000. The molecular biology of pasteurella multocida. Vet Microbiol 72:3–25 10.1016/S0378-1135(99)00183-2. [DOI] [PubMed] [Google Scholar]
- 52.Kehrenberg C, Schwarz S. 2000. Identification of a truncated, but functionally active tet(H) tetracycline resistance gene in Pasteurella aerogenes and Pasteurella multocida. FEMS Microbiol Lett 188:191–195 10.1111/j.1574-6968.2000.tb09192.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 53.Kehrenberg C, Schwarz S. 2001. Molecular analysis of tetracycline resistance in Pasteurella aerogenes. Antimicrob Agents Chemother 45:2885–2890 10.1128/AAC.45.10.2885-2890.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Michael GB, Kadlec K, Sweeney MT, Brzuszkiewicz E, Liesegang H, Daniel R, Murray RW, Watts JL, Schwarz S. 2012. ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: structure and transfer. J Antimicrob Chemother 67:91–100 10.1093/jac/dkr411. [PubMed] [DOI] [PubMed] [Google Scholar]
- 55.Michael GB, Kadlec K, Sweeney MT, Brzuszkiewicz E, Liesegang H, Daniel R, Murray RW, Watts JL, Schwarz S. 2012. ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: analysis of the regions that comprise 12 antimicrobial resistance genes. J Antimicrob Chemother 67:84–90 10.1093/jac/dkr406. [PubMed] [DOI] [PubMed] [Google Scholar]
- 56.Eidam C, Poehlein A, Leimbach A, Michael GB, Kadlec K, Liesegang H, Daniel R, Sweeney MT, Murray RW, Watts JL, Schwarz S. 2015. Analysis and comparative genomics of ICEMh1, a novel integrative and conjugative element (ICE) of Mannheimia haemolytica. J Antimicrob Chemother 70:93–97 10.1093/jac/dku361. [PubMed] [DOI] [PubMed] [Google Scholar]
- 57.Klima CL, Zaheer R, Cook SR, Booker CW, Hendrick S, Alexander TW, McAllister TA, Onderdonk AB. 2014. Pathogens of bovine respiratory disease in North American feedlots conferring multidrug resistance via integrative conjugative elements. J Clin Microbiol 52:438–448 10.1128/JCM.02485-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miranda CD, Kehrenberg C, Ulep C, Schwarz S, Roberts MC. 2003. Diversity of tetracycline resistance genes in bacteria from Chilean salmon farms. Antimicrob Agents Chemother 47:883–888 10.1128/AAC.47.3.883-888.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kehrenberg C, Tham NTT, Schwarz S. 2003. New plasmid-borne antibiotic resistance gene cluster in Pasteurella multocida. Antimicrob Agents Chemother 47:2978–2980 10.1128/AAC.47.9.2978-2980.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blanco M, Gutiérrez-Martin CB, Rodríguez-Ferri EF, Roberts MC, Navas J. 2006. Distribution of tetracycline resistance genes in Actinobacillus pleuropneumoniae isolates from Spain. Antimicrob Agents Chemother 50:702–708 10.1128/AAC.50.2.702-708.2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blanco M, Kadlec K, Gutiérrez Martín CB, de la Fuente AJ, Schwarz S, Navas J. 2007. Nucleotide sequence and transfer properties of two novel types of Actinobacillus pleuropneumoniae plasmids carrying the tetracycline resistance gene tet(H). J Antimicrob Chemother 60:864–867 10.1093/jac/dkm293. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.Matter D, Rossano A, Limat S, Vorlet-Fawer L, Brodard I, Perreten V. 2007. Antimicrobial resistance profile of Actinobacillus pleuropneumoniae and Actinobacillus porcitonsillarum. Vet Microbiol 122:146–156 10.1016/j.vetmic.2007.01.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 63.Dayao D, Gibson JS, Blackall PJ, Turni C. 2016. Antimicrobial resistance genes in Actinobacillus pleuropneumoniae, Haemophilus parasuis and Pasteurella multocida isolated from Australian pigs. Aust Vet J 94:227–231 10.1111/avj.12458. [PubMed] [DOI] [PubMed] [Google Scholar]
- 64.Kehrenberg C, Salmon SA, Watts JL, Schwarz S. 2001. Tetracycline resistance genes in isolates of Pasteurella multocida, Mannheimia haemolytica, Mannheimia glucosida and Mannheimia varigena from bovine and swine respiratory disease: intergeneric spread of the tet(H) plasmid pMHT1. J Antimicrob Chemother 48:631–640 10.1093/jac/48.5.631. [PubMed] [DOI] [PubMed] [Google Scholar]
- 65.San Millan A, Escudero JA, Gutierrez B, Hidalgo L, Garcia N, Llagostera M, Dominguez L, Gonzalez-Zorn B. 2009. Multiresistance in Pasteurella multocida is mediated by coexistence of small plasmids. Antimicrob Agents Chemother 53:3399–3404 10.1128/AAC.01522-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chaslus-Dancla E, Lesage-Descauses M-C, Leroy-Sétrin S, Martel J-L, Lafont J-P. 1995. Tetracycline resistance determinants, Tet B and Tet M, detected in Pasteurella haemolytica and Pasteurella multocida from bovine herds. J Antimicrob Chemother 36:815–819 10.1093/jac/36.5.815. [PubMed] [DOI] [PubMed] [Google Scholar]
- 67.Chalmers R, Sewitz S, Lipkow K, Crellin P. 2000. Complete nucleotide sequence of Tn10.J Bacteriol 182:2970–2972 10.1128/JB.182.10.2970-2972.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lawley TD, Burland V, Taylor DE. 2000. Analysis of the complete nucleotide sequence of the tetracycline-resistance transposon Tn10. Plasmid 43:235–239 10.1006/plas.1999.1458. [PubMed] [DOI] [PubMed] [Google Scholar]
- 69.Chopra I, Roberts M. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65:232–260 10.1128/MMBR.65.2.232-260.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wasteson Y, Roe DE, Falk K, Roberts MC. 1996. Characterization of tetracycline and erythromycin resistance in Actinobacillus pleuropneumoniae. Vet Microbiol 48:41–50 10.1016/0378-1135(95)00130-1. [DOI] [PubMed] [Google Scholar]
- 71.Ouellet V, Forest A, Nadeau M, Sirois M. 2004. Characterization of tetracycline resistance determinants in Actinobacillus pleuropneumoniae. Abstr A-113, 104th ASM General Meeting, p 22. [Google Scholar]
- 72.Morioka A, Asai T, Nitta H, Yamamoto K, Ogikubo Y, Takahashi T, Suzuki S. 2008. Recent trends in antimicrobial susceptibility and the presence of the tetracycline resistance gene in Actinobacillus pleuropneumoniae isolates in Japan. J Vet Med Sci 70:1261–1264 10.1292/jvms.70.1261. [PubMed] [DOI] [PubMed] [Google Scholar]
- 73.Yoo AN, Cha SB, Shin MK, Won HK, Kim EH, Choi HW, Yoo HS. 2014. Serotypes and antimicrobial resistance patterns of the recent Korean Actinobacillus pleuropneumoniae isolates. Vet Rec 174:223 10.1136/vr.101863. [PubMed] [DOI] [PubMed] [Google Scholar]
- 74.Bossé JT, Li Y, Fernandez Crespo R, Chaudhuri RR, Rogers J, Holden MTG, Maskell DJ, Tucker AW, Wren BW, Rycroft AN, Langford PR, the BRaDP1T Consortium. 2016. ICEApl1, an integrative conjugative element related to ICEHin1056, identified in the pig pathogen Actinobacillus pleuropneumoniae. Front Microbiol 7:810 10.3389/fmicb.2016.00810. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lancashire JF, Terry TD, Blackall PJ, Jennings MP. 2005. Plasmid-encoded Tet B tetracycline resistance in Haemophilus parasuis. Antimicrob Agents Chemother 49:1927–1931 10.1128/AAC.49.5.1927-1931.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu J-R, Shieh HK, Shien J-H, Gong S-R, Chang P-C. 2003. Molecular characterization of plasmids with antimicrobial resistant genes in avian isolates of Pasteurella multocida. Avian Dis 47:1384–1392 10.1637/z7035. [PubMed] [DOI] [PubMed] [Google Scholar]
- 77.Briggs CE, Fratamico PM. 1999. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother 43:846–849. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doublet B, Boyd D, Mulvey MR, Cloeckaert A. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol Microbiol 55:1911–1924 10.1111/j.1365-2958.2005.04520.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 79.Kehrenberg C, Catry B, Haesebrouck F, de Kruif A, Schwarz S. 2005. tet(L)-mediated tetracycline resistance in bovine Mannheimia and Pasteurella isolates. J Antimicrob Chemother 56:403–406 10.1093/jac/dki210. [PubMed] [DOI] [PubMed] [Google Scholar]
- 80.Schwarz S, Cardoso M, Wegener HC. 1992. Nucleotide sequence and phylogeny of the tet(L) tetracycline resistance determinant encoded by plasmid pSTE1 from Staphylococcus hyicus. Antimicrob Agents Chemother 36:580–588 10.1128/AAC.36.3.580. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim B, Hur J, Lee JY, Choi Y, Lee JH. 2016. Molecular serotyping and antimicrobial resistance profiles of Actinobacillus pleuropneumoniae isolated from pigs in South Korea. Vet Q 36:137–144 10.1080/01652176.2016.1155241. [PubMed] [DOI] [PubMed] [Google Scholar]
- 82.Flannagan SE, Zitzow LA, Su YA, Clewell DB. 1994. Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid 32:350–354 10.1006/plas.1994.1077. [PubMed] [DOI] [PubMed] [Google Scholar]
- 83.Tristram S, Jacobs MR, Appelbaum PC. 2007. Antimicrobial resistance in Haemophilus influenzae. Clin Microbiol Rev 20:368–389 10.1128/CMR.00040-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Livrelli VO, Darfeuille-Richaud A, Rich CD, Joly BH, Martel J-L. 1988. Genetic determinant of the ROB-1 β-lactamase in bovine and porcine Pasteurella strains. Antimicrob Agents Chemother 32:1282–1284 10.1128/AAC.32.8.1282. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Livrelli V, Peduzzi J, Joly B. 1991. Sequence and molecular characterization of the ROB-1 β-lactamase gene from Pasteurella haemolytica. Antimicrob Agents Chemother 35:242–251 10.1128/AAC.35.2.242. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Juteau J-M, Levesque RC. 1990. Sequence analysis and evolutionary perspectives of ROB-1 β-lactamase. Antimicrob Agents Chemother 34:1354–1359 10.1128/AAC.34.7.1354. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Azad AK, Coote JG, Parton R. 1992. Distinct plasmid profiles of Pasteurella haemolytica serotypes and the characterization and amplification in Escherichia coli of ampicillin-resistance plasmids encoding ROB-1 β-lactamase. J Gen Microbiol 138:1185–1196 10.1099/00221287-138-6-1185. [PubMed] [DOI] [PubMed] [Google Scholar]
- 88.San Millan A, Escudero JA, Catalan A, Nieto S, Farelo F, Gibert M, Moreno MA, Dominguez L, Gonzalez-Zorn B. 2007. β-lactam resistance in Haemophilus parasuis is mediated by plasmid pB1000 bearing blaROB-1. Antimicrob Agents Chemother 51:2260–2264 10.1128/AAC.00242-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Naas T, Benaoudia F, Lebrun L, Nordmann P. 2001. Molecular identification of TEM-1 β-lactamase in a Pasteurella multocida isolate of human origin. Eur J Clin Microbiol Infect Dis 20:210–213 10.1007/PL00011254. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.Chander Y, Oliveira S, Goyal SM. 2011. Characterisation of ceftiofur resistance in swine bacterial pathogens. Vet J 187:139–141 10.1016/j.tvjl.2009.10.013. [PubMed] [DOI] [PubMed] [Google Scholar]
- 91.Bush K, Jacoby GA, Medeiros AA. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother 39:1211–1233 10.1128/AAC.39.6.1211. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Medeiros AA, Levesque R, Jacoby GA. 1986. An animal source for the ROB-1 β-lactamase of Haemophilus influenzae type b. Antimicrob Agents Chemother 29:212–215 10.1128/AAC.29.2.212. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Juteau J-M, Sirois M, Medeiros AA, Levesque RC. 1991. Molecular distribution of ROB-1 β-lactamase in Actinobacillus pleuropneumoniae. Antimicrob Agents Chemother 35:1397–1402 10.1128/AAC.35.7.1397. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chang C-F, Yeh T-M, Chou C-C, Chang Y-F, Chiang T-S. 2002. Antimicrobial susceptibility and plasmid analysis of Actinobacillus pleuropneumoniae isolated in Taiwan. Vet Microbiol 84:169–177 10.1016/S0378-1135(01)00459-X. [DOI] [PubMed] [Google Scholar]
- 95.Kang M, Zhou R, Liu L, Langford PR, Chen H. 2009. Analysis of an Actinobacillus pleuropneumoniae multi-resistance plasmid, pHB0503. Plasmid 61:135–139 10.1016/j.plasmid.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 96.Philippon A, Joly B, Reynaud D, Paul G, Martel J-L, Sirot D, Cluzel R, Névot P. 1986. Characterization of a β-lactamase from Pasteurella multocida. Ann Inst Pasteur Microbiol 1985 137A:153–158 10.1016/S0769-2609(86)80020-5. [DOI] [PubMed] [Google Scholar]
- 97.Moleres J, Santos-López A, Lázaro I, Labairu J, Prat C, Ardanuy C, González-Zorn B, Aragon V, Garmendia J. 2015. Novel blaROB-1-bearing plasmid conferring resistance to β-lactams in Haemophilus parasuis isolates from healthy weaning pigs. Appl Environ Microbiol 81:3255–3267 10.1128/AEM.03865-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wood AR, Lainson FA, Wright F, Baird GD, Donachie W. 1995. A native plasmid of Pasteurella haemolytica serotype A1: DNA sequence analysis and investigation of its potential as a vector. Res Vet Sci 58:163–168 10.1016/0034-5288(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 99.Galán JC, Morosini MI, Baquero MR, Reig M, Baquero F. 2003. Haemophilus influenzae bla(ROB-1) mutations in hypermutagenic deltaampC Escherichia coli conferring resistance to cefotaxime and β-lactamase inhibitors and increased susceptibility to cefaclor. Antimicrob Agents Chemother 47:2551–2557 10.1128/AAC.47.8.2551-2557.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Craig FF, Coote JG, Parton R, Freer JH, Gilmour NJ. 1989. A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J Gen Microbiol 135:2885–2890. [DOI] [PubMed] [Google Scholar]
- 101.Schwarz S, Spies U, Reitz B, Seyfert H-M, Lämmler C, Blobel H. 1989. Detection and interspecies-transformation of a β-lactamase-encoding plasmid from Pasteurella haemolytica. Zentralbl Bakteriol Mikrobiol Hyg [A] 270:462–469 10.1016/S0176-6724(89)80017-3. [DOI] [PubMed] [Google Scholar]
- 102.Rossmanith SER, Wilt GR, Wu G. 1991. Characterization and comparison of antimicrobial susceptibilities and outer membrane protein and plasmid DNA profiles of Pasteurella haemolytica and certain other members of the genus Pasteurella. Am J Vet Res 52:2016–2022. [PubMed] [PubMed] [Google Scholar]
- 103.Chang YF, Ma DP, Bai HQ, Young R, Struck DK, Shin SJ, Lein DH. 1992. Characterization of plasmids with antimicrobial resistant genes in Pasteurella haemolytica A1. DNA Seq 3:89–97 10.3109/10425179209034001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 104.Murphy GL, Robinson LC, Burrows GE. 1993. Restriction endonuclease analysis and ribotyping differentiate Pasteurella haemolytica serotype A1 isolates from cattle within a feedlot. J Clin Microbiol 31:2303–2308. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chang YF, Shi J, Shin SJ, Lein DH. 1992. Sequence analysis of the ROB-1 β-lactamase gene from Actinobacillus pleuropneumoniae. Vet Microbiol 32:319–325 10.1016/0378-1135(92)90154-L. [DOI] [PubMed] [Google Scholar]
- 106.Lalonde G, Miller JF, Tompkins LS, O’Hanley P. 1989. Transformation of Actinobacillus pleuropneumoniae and analysis of R factors by electroporation. Am J Vet Res 50:1957–1960. [PubMed] [PubMed] [Google Scholar]
- 107.Ishii H, Hayashi F, Iyobe S, Hashimoto H. 1991. Characterization and classification of Actinobacillus (Haemophilus) pleuropneumoniae plasmids. Am J Vet Res 52:1816–1820. [PubMed] [PubMed] [Google Scholar]
- 108.Matter D, Rossano A, Sieber S, Perreten V. 2008. Small multidrug resistance plasmids in Actinobacillus porcitonsillarum. Plasmid 59:144–152 10.1016/j.plasmid.2007.11.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 109.Schwarz S, Cloeckaert A, Roberts MC. 2006. Mechanisms and spread of bacterial resistance to antimicrobial agents, p 73–98. In Aarestrup FM (ed), Antimicrobial Resistance in Bacteria of Animal Origin. ASM Press, Washington, DC. [Google Scholar]
- 110.Schwarz S, Spies U, Schäfer F, Blobel H. 1989. Isolation and interspecies-transfer of a plasmid from Pasteurella multocida encoding for streptomycin resistance. Med Microbiol Immunol (Berl) 178:121–125 10.1007/BF00203308. [DOI] [PubMed] [Google Scholar]
- 111.Silver RP, Leming B, Garon CF, Hjerpe CA. 1979. R-plasmids in Pasteurella multocida. Plasmid 2:493–497 10.1016/0147-619X(79)90033-7. [DOI] [PubMed] [Google Scholar]
- 112.Coté S, Harel J, Higgins R, Jacques M. 1991. Resistance to antimicrobial agents and prevalence of R plasmids in Pasteurella multocida from swine. Am J Vet Res 52:1653–1657. [PubMed] [PubMed] [Google Scholar]
- 113.Yamamoto J, Sakano T, Shimizu M. 1990. Drug resistance and R plasmids in Pasteurella multocida isolates from swine. Microbiol Immunol 34:715–721 10.1111/j.1348-0421.1990.tb01049.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 114.Liu W, Yang M, Xu Z, Zheng H, Liang W, Zhou R, Wu B, Chen H. 2012. Complete genome sequence of Pasteurella multocida HN06, a toxigenic strain of serogroup D. J Bacteriol 194:3292–3293 10.1128/JB.00215-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zimmerman ML, Hirsh DC. 1980. Demonstration of an R plasmid in a strain of Pasteurella haemolytica isolated from feedlot cattle. Am J Vet Res 41:166–169. [PubMed] [PubMed] [Google Scholar]
- 116.Kehrenberg C, Schwarz S. 2002. Nucleotide sequence and organization of plasmid pMVSCS1 from Mannheimia varigena: identification of a multiresistance gene cluster. J Antimicrob Chemother 49:383–386 10.1093/jac/49.2.383. [PubMed] [DOI] [PubMed] [Google Scholar]
- 117.Gilbride KA, Rosendal S, Brunton JL. 1989. Plasmid mediated antimicrobial resistance in Ontario isolates of Actinobacillus (Haemophilus) pleuropneumoniae. Can J Vet Res 53:38–42. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 118.Willson PJ, Deneer HG, Potter A, Albritton W. 1989. Characterization of a streptomycin-sulfonamide resistance plasmid from Actinobacillus pleuropneumoniae. Antimicrob Agents Chemother 33:235–238 10.1128/AAC.33.2.235. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ishii H, Nakasone Y, Shigehara S, Honma K, Araki Y, Iyobe S, Hashimoto H. 1990. Drug-susceptibility and isolation of a plasmid in Haemophilus (Actinobacillus) pleuropneumoniae. Nippon Juigaku Zasshi 52:1–9 10.1292/jvms1939.52.1. [PubMed] [DOI] [PubMed] [Google Scholar]
- 120.Kiuchi A, Hara M, Tabuchi K. 1992. Drug resistant plasmid of Actinobacillus pleuropneumoniae isolated from swine pleuropneumonia in Thailand. Kansenshogaku Zasshi 66:1243–1247 10.11150/kansenshogakuzasshi1970.66.1243. [PubMed] [DOI] [PubMed] [Google Scholar]
- 121.Ito H, Ishii H, Akiba M. 2004. Analysis of the complete nucleotide sequence of an Actinobacillus pleuropneumoniae streptomycin-sulfonamide resistance plasmid, pMS260. Plasmid 51:41–47 10.1016/j.plasmid.2003.10.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 122.Bossé JT, Li Y, Walker S, Atherton T, Fernandez Crespo R, Williamson SM, Rogers J, Chaudhuri RR, Weinert LA, Oshota O, Holden MTG, Maskell DJ, Tucker AW, Wren BW, Rycroft AN, Langford PR, BRaDP1T Consortium. 2015. Identification of dfrA14 in two distinct plasmids conferring trimethoprim resistance in Actinobacillus pleuropneumoniae. J Antimicrob Chemother 70:2217–2222 10.1093/jac/dkv121. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hsu YM, Shieh HK, Chen WH, Sun TY, Shiang J-H. 2007. Antimicrobial susceptibility, plasmid profiles and haemocin activities of Avibacterium paragallinarum strains. Vet Microbiol 124:209–218 10.1016/j.vetmic.2007.04.024. [PubMed] [DOI] [PubMed] [Google Scholar]
- 124.Chiou C-S, Jones AL. 1993. Nucleotide sequence analysis of a transposon (Tn5393) carrying streptomycin resistance genes in Erwinia amylovora and other Gram-negative bacteria. J Bacteriol 175:732–740 10.1128/jb.175.3.732-740.1993. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kehrenberg C, Schwarz S. 2001. Occurrence and linkage of genes coding for resistance to sulfonamides, streptomycin and chloramphenicol in bacteria of the genera Pasteurella and Mannheimia. FEMS Microbiol Lett 205:283–290 10.1111/j.1574-6968.2001.tb10962.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 126.Ojo KK, Kehrenberg C, Schwarz S, Odelola HA. 2002. Identification of a complete dfrA14 gene cassette integrated at a secondary site in a resistance plasmid of uropathogenic Escherichia coli from Nigeria. Antimicrob Agents Chemother 46:2054–2055 10.1128/AAC.46.6.2054-2055.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sundin GW. 2000. Examination of base pair variants of the strA-strB streptomycin resistance genes from bacterial pathogens of humans, animals and plants. J Antimicrob Chemother 46:848–849 10.1093/jac/46.5.848. [DOI] [PubMed] [Google Scholar]
- 128.Sundin GW. 2002. Distinct recent lineages of the strA-strB streptomycin-resistance genes in clinical and environmental bacteria. Curr Microbiol 45:63–69 10.1007/s00284-001-0100-y. [PubMed] [DOI] [PubMed] [Google Scholar]
- 129.Sundin GW, Bender CL. 1996. Dissemination of the strA-strB streptomycin-resistance genes among commensal and pathogenic bacteria from humans, animals, and plants. Mol Ecol 5:133–143 10.1111/j.1365-294X.1996.tb00299.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 130.Kehrenberg C, Schwarz S. 2011. Trimethoprim resistance in a porcine Pasteurella aerogenes isolate is based on a dfrA1 gene cassette located in a partially truncated class 2 integron. J Antimicrob Chemother 66:450–452 10.1093/jac/dkq461. [PubMed] [DOI] [PubMed] [Google Scholar]
- 131.Schwarz S, Kehrenberg C, Salmon SA, Watts JL. 2004. In vitro activities of spectinomycin and comparator agents against Pasteurella multocida and Mannheimia haemolytica from respiratory tract infections of cattle. J Antimicrob Chemother 53:379–382 10.1093/jac/dkh059. [PubMed] [DOI] [PubMed] [Google Scholar]
- 132.Kehrenberg C, Catry B, Haesebrouck F, de Kruif A, Schwarz S. 2005. Novel spectinomycin/streptomycin resistance gene, aadA14, from Pasteurella multocida. Antimicrob Agents Chemother 49:3046–3049 10.1128/AAC.49.7.3046-3049.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Oka A, Sugisaki H, Takanami M. 1981. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol 147:217–226 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- 134.Dixon LG, Albritton WL, Willson PJ. 1994. An analysis of the complete nucleotide sequence of the Haemophilus ducreyi broad-host-range plasmid pLS88. Plasmid 32:228–232 10.1006/plas.1994.1060. [PubMed] [DOI] [PubMed] [Google Scholar]
- 135.Kehrenberg C, Schwarz S. 2005. Molecular basis of resistance to kanamycin and neomycin in Pasteurella and Mannheimia isolates of animal origin. Abstr A47, ASM Conference on Pasteurellaceae 2005, p. 55. [Google Scholar]
- 136.Makosky PC, Dahlberg AE. 1987. Spectinomycin resistance at site 1192 in 16S ribosomal RNA of E. coli: an analysis of three mutants. Biochimie 69:885–889 10.1016/0300-9084(87)90216-1. [DOI] [PubMed] [Google Scholar]
- 137.De Stasio EA, Moazed D, Noller HF, Dahlberg AE. 1989. Mutations in 16S ribosomal RNA disrupt antibiotic-RNA interactions. EMBO J 8:1213–1216. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brink MF, Brink G, Verbeet MP, de Boer HA. 1994. Spectinomycin interacts specifically with the residues G1064 and C1192 in 16S rRNA, thereby potentially freezing this molecule into an inactive conformation. Nucleic Acids Res 22:325–331 10.1093/nar/22.3.325. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Galimand M, Gerbaud G, Courvalin P. 2000. Spectinomycin resistance in Neisseria spp. due to mutations in 16S rRNA. Antimicrob Agents Chemother 44:1365–1366 10.1128/AAC.44.5.1365-1366.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.O’Connor M, Dahlberg AE. 2002. Isolation of spectinomycin resistance mutations in the 16S rRNA of Salmonella enterica serovar Typhimurium and expression in Escherichia coli and Salmonella. Curr Microbiol 45:429–433 10.1007/s00284-002-3684-y. [PubMed] [DOI] [PubMed] [Google Scholar]
- 141.Funatsu G, Schiltz E, Wittmann HG. 1972. Ribosomal proteins. XXVII. Localization of the amino acid exchanges in protein S5 from two Escherichia coli mutants resistant to spectinomycin. Mol Gen Genet 114:106–111 10.1007/BF00332781. [PubMed] [DOI] [PubMed] [Google Scholar]
- 142.Davies C, Bussiere DE, Golden BL, Porter SJ, Ramakrishnan V, White SW. 1998. Ribosomal proteins S5 and L6: high-resolution crystal structures and roles in protein synthesis and antibiotic resistance. J Mol Biol 279:873–888 10.1006/jmbi.1998.1780. [PubMed] [DOI] [PubMed] [Google Scholar]
- 143.Kehrenberg C, Schwarz S. 2007. Mutations in 16S rRNA and ribosomal protein S5 associated with high-level spectinomycin resistance in Pasteurella multocida. Antimicrob Agents Chemother 51:2244–2246 10.1128/AAC.00229-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.de Groot R, Sluijter M, de Bruyn A, Campos J, Goessens WH, Smith AL, Hermans PW. 1996. Genetic characterization of trimethoprim resistance in Haemophilus influenzae. Antimicrob Agents Chemother 40:2131–2136. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Enne VI, King A, Livermore DM, Hall LM. 2002. Sulfonamide resistance in Haemophilus influenzae mediated by acquisition of sul2 or a short insertion in chromosomal folP. Antimicrob Agents Chemother 46:1934–1939 10.1128/AAC.46.6.1934-1939.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kehrenberg C, Schwarz S. 2005. dfrA20, A novel trimethoprim resistance gene from Pasteurella multocida. Antimicrob Agents Chemother 49:414–417 10.1128/AAC.49.1.414-417.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Scholz P, Haring V, Wittmann-Liebold B, Ashman K, Bagdasarian M, Scherzinger E. 1989. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75:271–288 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- 148.Kim EH, Aoki T. 1996. Sulfonamide resistance gene in a transferable R plasmid of Pasteurella piscicida. Microbiol Immunol 40:397–399 10.1111/j.1348-0421.1996.tb01085.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 149.Rådström P, Swedberg G. 1988. RSF1010 and a conjugative plasmid contain sulII, one of two known genes for plasmid-borne sulfonamide resistance dihydropteroate synthase. Antimicrob Agents Chemother 32:1684–1692 10.1128/AAC.32.11.1684. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wright CL, Strugnell RA, Hodgson ALM. 1997. Characterization of a Pasteurella multocida plasmid and its use to express recombinant proteins in P. multocida. Plasmid 37:65–79 10.1006/plas.1996.1276. [PubMed] [DOI] [PubMed] [Google Scholar]
- 151.Escande F, Gerbaud G, Martel J-L, Courvalin P. 1991. Resistance to trimethoprim and 2,4-diamino-6,7-diisopropyl-pteridine (0/129) in Pasteurella haemolytica. Vet Microbiol 26:107–114 10.1016/0378-1135(91)90047-J. [DOI] [PubMed] [Google Scholar]
- 152.Anantham S, Hall RM. 2012. pCERC1, a small, globally disseminated plasmid carrying the dfrA14 cassette in the strA gene of the sul2-strA-strB gene cluster. Microb Drug Resist 18:364–371 10.1089/mdr.2012.0008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 153.Dayao DAE, Seddon JM, Gibson JS, Blackall PJ, Turni C. 2016. Whole genome sequence analysis of pig respiratory bacterial pathogens with elevated minimum inhibitory concentrations for macrolides. Microb Drug Resist 22:531–537 10.1089/mdr.2015.0214. [PubMed] [DOI] [PubMed] [Google Scholar]
- 154.Yang SS, Sun J, Liao XP, Liu BT, Li LL, Li L, Fang LX, Huang T, Liu YH. 2013. Co-location of the erm(T) gene and blaROB-1 gene on a small plasmid in Haemophilus parasuis of pig origin. J Antimicrob Chemother 68:1930–1932 10.1093/jac/dkt112. [PubMed] [DOI] [PubMed] [Google Scholar]
- 155.Kadlec K, Brenner Michael G, Sweeney MT, Brzuszkiewicz E, Liesegang H, Daniel R, Watts JL, Schwarz S. 2011. Molecular basis of macrolide, triamilide, and lincosamide resistance in Pasteurella multocida from bovine respiratory disease. Antimicrob Agents Chemother 55:2475–2477 10.1128/AAC.00092-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Desmolaize B, Rose S, Warrass R, Douthwaite S. 2011. A novel Erm monomethyltransferase in antibiotic-resistant isolates of Mannheimia haemolytica and Pasteurella multocida. Mol Microbiol 80:184–194 10.1111/j.1365-2958.2011.07567.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 157.Michael GB, Eidam C, Kadlec K, Meyer K, Sweeney MT, Murray RW, Watts JL, Schwarz S. 2012. Increased MICs of gamithromycin and tildipirosin in the presence of the genes erm(42) and msr(E)-mph(E) for bovine Pasteurella multocida and Mannheimia haemolytica. J Antimicrob Chemother 67:1555–1557 10.1093/jac/dks076. [PubMed] [DOI] [PubMed] [Google Scholar]
- 158.Peric M, Bozdogan B, Jacobs MR, Appelbaum PC. 2003. Effects of an efflux mechanism and ribosomal mutations on macrolide susceptibility of Haemophilus influenzae clinical isolates. Antimicrob Agents Chemother 47:1017–1022 10.1128/AAC.47.3.1017-1022.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Olsen AS, Warrass R, Douthwaite S. 2015. Macrolide resistance conferred by rRNA mutations in field isolates of Mannheimia haemolytica and Pasteurella multocida. J Antimicrob Chemother 70:420–423 10.1093/jac/dku385. [PubMed] [DOI] [PubMed] [Google Scholar]
- 160.Desmolaize B, Rose S, Wilhelm C, Warrass R, Douthwaite S. 2011. Combinations of macrolide resistance determinants in field isolates of Mannheimia haemolytica and Pasteurella multocida. Antimicrob Agents Chemother 55:4128–4133 10.1128/AAC.00450-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen LP, Cai XW, Wang XR, Zhou XL, Wu DF, Xu XJ, Chen HC. 2010. Characterization of plasmid-mediated lincosamide resistance in a field isolate of Haemophilus parasuis. J Antimicrob Chemother 65:2256–2258 10.1093/jac/dkq304. [PubMed] [DOI] [PubMed] [Google Scholar]
- 162.Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev 28:519–542 10.1016/j.femsre.2004.04.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 163.Vassort-Bruneau C, Lesage-Descauses MC, Martel J-L, Lafont J-P, Chaslus-Dancla E. 1996. CAT III chloramphenicol resistance in Pasteurella haemolytica and Pasteurella multocida isolated from calves. J Antimicrob Chemother 38:205–213 10.1093/jac/38.2.205. [PubMed] [DOI] [PubMed] [Google Scholar]
- 164.Kawahara K, Kawase H, Nakai T, Kume K, Danbara H. 1990. Drug resistance plasmids of Actinobacillus (Haemophilus) pleuropneumoniae serotype 2 strains isolated from swine. Kitasato Arch Exp Med 63:131–136. [PubMed] [PubMed] [Google Scholar]
- 165.Ishii H, Fukuyasu T, Iyobe S, Hashimoto H. 1993. Characterization of newly isolated plasmids from Actinobacillus pleuropneumoniae. Am J Vet Res 54:701–708. [PubMed] [PubMed] [Google Scholar]
- 166.Powell M, Livermore DM. 1988. Mechanisms of chloramphenicol resistance in Haemophilus influenzae in the United Kingdom. J Med Microbiol 27:89–93 10.1099/00222615-27-2-89. [PubMed] [DOI] [PubMed] [Google Scholar]
- 167.Murray IA, Martinez-Suarez JV, Close TJ, Shaw WV. 1990. Nucleotide sequences of genes encoding the type II chloramphenicol acetyltransferases of Escherichia coli and Haemophilus influenzae, which are sensitive to inhibition by thiol-reactive reagents. Biochem J 272:505–510 10.1042/bj2720505. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hörmansdorfer S, Bauer J. 1996. Resistance pattern of bovine Pasteurella. Berl Munch Tierarztl Wochenschr 109:168–171. (In German.) [PubMed] [PubMed] [Google Scholar]
- 169.Hörmansdorfer S, Bauer J. 1998. Resistance of bovine and porcine Pasteurella against florfenicol and other antibiotics. Berl Munch Tierarztl Wochenschr 111:422–426. (In German.) [PubMed] [PubMed] [Google Scholar]
- 170.Priebe S, Schwarz S. 2003. In vitro activities of florfenicol against bovine and porcine respiratory tract pathogens. Antimicrob Agents Chemother 47:2703–2705 10.1128/AAC.47.8.2703-2705.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kehrenberg C, Mumme J, Wallmann J, Verspohl J, Tegeler R, Kühn T, Schwarz S. 2004. Monitoring of florfenicol susceptibility among bovine and porcine respiratory tract pathogens collected in Germany during the years 2002 and 2003. J Antimicrob Chemother 54:572–574 10.1093/jac/dkh371. [PubMed] [DOI] [PubMed] [Google Scholar]
- 172.Kaspar H, Schröer U, Wallmann J. 2007. Quantitative resistance level (MIC) of Pasteurella multocida isolated from pigs between 2004 and 2006: national resistance monitoring by the BVL. Berl Munch Tierarztl Wochenschr 120:442–451. [PubMed] [PubMed] [Google Scholar]
- 173.Wallmann J, Schröer U, Kaspar H. 2007. Quantitative resistance level (MIC) of bacterial pathogens (Escherichia coli, Pasteurella multocida, Pseudomonas aeruginosa, Salmonella sp., Staphylococcus aureus) isolated from chickens and turkeys: national resistance monitoring by the BVL 2004/2005. Berl Munch Tierarztl Wochenschr 120:452–463. [PubMed] [PubMed] [Google Scholar]
- 174.Kehrenberg C, Schwarz S. 2005. Plasmid-borne florfenicol resistance in Pasteurella multocida. J Antimicrob Chemother 55:773–775 10.1093/jac/dki102. [PubMed] [DOI] [PubMed] [Google Scholar]
- 175.Whittle G, Katz ME, Clayton EH, Cheetham BF. 2000. Identification and characterization of a native Dichelobacter nodosus plasmid, pDN1. Plasmid 43:230–234 10.1006/plas.1999.1456. [DOI] [PubMed] [Google Scholar]
- 176.Blickwede M, Schwarz S. 2004. Molecular analysis of florfenicol-resistant Escherichia coli isolates from pigs. J Antimicrob Chemother 53:58–64 10.1093/jac/dkh007. [PubMed] [DOI] [PubMed] [Google Scholar]
- 177.Sørum H, Roberts MC, Crosa JH. 1992. Identification and cloning of a tetracycline resistance gene from the fish pathogen Vibrio salmonicida. Antimicrob Agents Chemother 36:611–615 10.1128/AAC.36.3.611. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kehrenberg C, Wallmann J, Schwarz S. 2008. Molecular analysis of florfenicol-resistant Pasteurella multocida isolates in Germany. J Antimicrob Chemother 62:951–955 10.1093/jac/dkn359. [PubMed] [DOI] [PubMed] [Google Scholar]
- 179.Katsuda K, Kohmoto M, Mikami O, Tamamura Y, Uchida I. 2012. Plasmid-mediated florfenicol resistance in Mannheimia haemolytica isolated from cattle. Vet Microbiol 155:444–447 10.1016/j.vetmic.2011.09.033. [PubMed] [DOI] [PubMed] [Google Scholar]
- 180.Li B, Zhang Y, Wei J, Shao D, Liu K, Shi Y, Qiu Y, Ma Z. 2015. Characterization of a novel small plasmid carrying the florfenicol resistance gene floR in Haemophilus parasuis. J Antimicrob Chemother 70:3159–3161 10.1093/jac/dkv230. [PubMed] [DOI] [PubMed] [Google Scholar]
- 181.Bossé JT, Li Y, Atherton TG, Walker S, Williamson SM, Rogers J, Chaudhuri RR, Weinert LA, Holden MTG, Maskell DJ, Tucker AW, Wren BW, Rycroft AN, Langford PR, BRaDP1T Consortium. 2015. Characterisation of a mobilisable plasmid conferring florfenicol and chloramphenicol resistance in Actinobacillus pleuropneumoniae. Vet Microbiol 178:279–282 10.1016/j.vetmic.2015.05.020. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.da Silva GC, Rossi CC, Santana MF, Langford PR, Bossé JT, Bazzolli DMS. 2017. p518, A small floR plasmid from a South American isolate of Actinobacillus pleuropneumoniae. Vet Microbiol 204:129–132 10.1016/j.vetmic.2017.04.019. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kehrenberg C, Meunier D, Targant H, Cloeckaert A, Schwarz S, Madec J-Y. 2006. Plasmid-mediated florfenicol resistance in Pasteurella trehalosi. J Antimicrob Chemother 58:13–17 10.1093/jac/dkl174. [PubMed] [DOI] [PubMed] [Google Scholar]
- 184.Blackall PJ, Bojesen AM, Christensen H, Bisgaard M. 2007. Reclassification of [Pasteurella] trehalosi as Bibersteinia trehalosi gen. nov., comb. nov. Int J Syst Evol Microbiol 57:666–674 10.1099/ijs.0.64521-0. [DOI] [PubMed] [Google Scholar]
- 185.Tegetmeyer HE, Jones SC, Langford PR, Baltes N. 2008. ISApl1, a novel insertion element of Actinobacillus pleuropneumoniae, prevents ApxIV-based serological detection of serotype 7 strain AP76. Vet Microbiol 128:342–353 10.1016/j.vetmic.2007.10.025. [PubMed] [DOI] [PubMed] [Google Scholar]
- 186.Goldstein EJC, Citron DM, Merriam CV, Warren YA, Tyrrell KL, Fernandez HT. 2002. In vitro activities of garenoxacin (BMS-284756) against 170 clinical isolates of nine Pasteurella species. Antimicrob Agents Chemother 46:3068–3070 10.1128/AAC.46.9.3068-3070.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cárdenas M, Barbé J, Llagostera M, Miró E, Navarro F, Mirelis B, Prats G, Badiola I. 2001. Quinolone resistance-determining regions of gyrA and parC in Pasteurella multocida strains with different levels of nalidixic acid resistance. Antimicrob Agents Chemother 45:990–991 10.1128/AAC.45.3.990-991.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kong LC, Gao D, Gao YH, Liu SM, Ma HX. 2014. Fluoroquinolone resistance mechanism of clinical isolates and selected mutants of Pasteurella multocida from bovine respiratory disease in China. J Vet Med Sci 76:1655–1657 10.1292/jvms.14-0240. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Katsuda K, Kohmoto M, Mikami O, Uchida I. 2009. Antimicrobial resistance and genetic characterization of fluoroquinolone-resistant Mannheimia haemolytica isolates from cattle with bovine pneumonia. Vet Microbiol 139:74–79 10.1016/j.vetmic.2009.04.020. [PubMed] [DOI] [PubMed] [Google Scholar]
- 190.Wang Y-C, Chan JP-W, Yeh K-S, Chang C-C, Hsuan S-L, Hsieh Y-M, Chang Y-C, Lai T-C, Lin W-H, Chen T-H. 2010. Molecular characterization of enrofloxacin resistant Actinobacillus pleuropneumoniae isolates. Vet Microbiol 142:309–312 10.1016/j.vetmic.2009.09.067. [PubMed] [DOI] [PubMed] [Google Scholar]
- 191.Guo L, Zhang J, Xu C, Zhao Y, Ren T, Zhang B, Fan H, Liao M. 2011. Molecular characterization of fluoroquinolone resistance in Haemophilus parasuis isolated from pigs in South China. J Antimicrob Chemother 66:539–542 10.1093/jac/dkq497. [PubMed] [DOI] [PubMed] [Google Scholar]
- 192.Zhang Q, Zhou M, Song D, Zhao J, Zhang A, Jin M. 2013. Molecular characterisation of resistance to fluoroquinolones in Haemophilus parasuis isolated from China. Int J Antimicrob Agents 42:87–89 10.1016/j.ijantimicag.2013.03.011. [PubMed] [DOI] [PubMed] [Google Scholar]
- 193.Poole K. 2005. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother 56:20–51 10.1093/jac/dki171. [PubMed] [DOI] [PubMed] [Google Scholar]
- 194.Chen L, Wu D, Cai X, Guo F, Blackall PJ, Xu X, Chen H. 2012. Electrotransformation of Haemophilus parasuis with in vitro modified DNA based on a novel shuttle vector. Vet Microbiol 155:310–316 10.1016/j.vetmic.2011.08.020. [PubMed] [DOI] [PubMed] [Google Scholar]
- 195.Luan SL, Chaudhuri RR, Peters SE, Mayho M, Weinert LA, Crowther SA, Wang J, Langford PR, Rycroft A, Wren BW, Tucker AW, Maskell DJ, BRaDP1T Consortium. 2013. Generation of a Tn5 transposon library in Haemophilus parasuis and analysis by transposon-directed insertion-site sequencing (TraDIS). Vet Microbiol 166:558–566 10.1016/j.vetmic.2013.07.008. [PubMed] [DOI] [PubMed] [Google Scholar]


