ABSTRACT
Non-typhoidal Salmonella is the most common foodborne bacterial pathogen in most countries. It is widely present in food animal species, and therefore blocking its transmission through the food supply is a prominent focus of food safety activities worldwide. Antibiotic resistance in non-typhoidal Salmonella arises in large part because of antibiotic use in animal husbandry. Tracking resistance in Salmonella is required to design targeted interventions to contain or diminish resistance and refine use practices in production. Many countries have established systems to monitor antibiotic resistance in Salmonella and other bacteria, the earliest ones appearing the Europe and the US. In this chapter, we compare recent Salmonella antibiotic susceptibility data from Europe and the US. In addition, we summarize the state of known resistance genes that have been identified in the genus. The advent of routine whole genome sequencing has made it possible to conduct genomic surveillance of resistance based on DNA sequences alone. This points to a new model of surveillance in the future that will provide more definitive information on the sources of resistant Salmonella, the specific types of resistance genes involved, and information on how resistance spreads.
BACKGROUND
Nontyphoidal Salmonella enterica (NTS) is a ubiquitous, motile, Gram-negative bacillus that is one of the most common bacterial causes of gastrointestinal disease worldwide. Salmonellae colonize the intestinal tract of a wide range of animal hosts, including pigs, cattle, poultry (1), and wildlife, as well as companion animals such as dogs, cats, birds, and reptiles (2). Humans acquire infection from the ingestion of contaminated foods. In the United States, most illnesses are associated with seeded vegetables, eggs, poultry, beef, and pork. Other sources include dairy, fruits, sprouts, and fish (3). Its ubiquity in nature and the variety of vectors mediating fecal-oral spread have made salmonellosis the most important foodborne bacterial zoonosis. For many decades, the cornerstone of Salmonella epidemiology has been the Kaufmann-White serotyping scheme (4). Based on antibodies to the three major surface antigens (somatic O, flagellar H, and capsular Vi antigens), over 2,500 distinct serovars are currently recognized. A comprehensive body of scientific information on Salmonella developed over the years makes it one of the best-understood bacterial pathogens. A great deal is known about Salmonella epidemiology and genetics, the various virulence factors, interaction of the bacterium with host cells, the host range of serovars, and the causes of antibiotic resistance.
Most human cases of acute NTS diarrhea are self-limiting and do not require treatment with antibiotics unless they are severe, invasive, or occur in the elderly, children, or those with underlying comorbidities (5). Cases that become invasive may result in life-threatening bloodstream infections. Antimicrobial resistance is especially problematic in these systemic infections, where antibiotic therapy can be life-saving. Efforts to limit the public health burden of salmonellosis, and the pressures leading to antibiotic resistance, have focused on farm practices, interventions in processing facilities, and consumer education on safe food handling practices.
In animals, Salmonella may cause overt signs of disease or may be carried asymptomatically and be shed into the environment (6). In food-producing animals, Salmonella is a continuous threat to animal health, especially in cattle, where infection typically presents as diarrhea with fever, anorexia, and dehydration (2). Less commonly, infection results in respiratory disease and death. Salmonella infections in dairy herds, where they may become endemic (7), result in decreased milk production and increased production costs, including the use of antibiotics. In addition to overt salmonellosis, a chronic asymptomatic carrier state may exist in food-producing animals. In both the carrier state and in cases of overt infection, exposure to antimicrobials promotes the evolution of resistant serovars that may be transmitted to humans. In the case of foodborne zoonotic bacteria, resistance spreads from food-producing animals to humans via the consumption of meat products derived either from treated animals or from foods cross-contaminated at processing or retail (8). Resistance also can spread by direct animal contact (as with pets) or environmental routes such as water or wildlife. The U.S. Centers for Disease Control and Prevention (CDC) classifies antibiotic-resistant Salmonella as a serious public health threat (9).
Bacterial resistance to antibiotics is a naturally occurring phenomenon that can be accelerated by selection pressures exerted through the use of antibiotics in medical and veterinary practice (10). Resistance is mediated by a limited number of mechanisms common to other pathogens. These include enzymatic modification, target protection, energy-dependent efflux, and permeability changes in the cell wall. The use and misuse of antimicrobials in both humans and animals result in antimicrobial resistance in both pathogenic and commensal bacteria within the treated host. Many classes of antimicrobial agents used in food-producing and companion animals are the same as those used in human medicine (11). Therefore, resistance developing in one drug use environment can compromise the efficacy of drugs used in other settings. For this reason, many countries are attempting to implement antimicrobial drug use monitoring.
The relationship between resistances in food animal bacteria and antibiotic use in animal agriculture is poorly defined. Very few countries can collect detailed information on the amounts and indications of antimicrobial compounds in different animal species over time. Only in Denmark is it mandatory for veterinarians to report (via VetStat) medicines used in their own practices. Like most countries, the United States and Japan collect data on total annual sales (in kilograms of active ingredient) by drug class. Japan collects sales data only for drugs used therapeutically (12). Most European Union countries also mandate collection of sales data (13). While usually the only practicable approach, bulk annual amounts of active ingredient sold is of limited utility and is not a reliable surrogate for actual drug use in the production environment (14). In July 2016, the U.S. FDA mandated that pharmaceutical companies report antimicrobial sales by the four major animal species: cattle, chickens, turkey, and swine, along with a combined “other” category (15). The goal of this provision is to better monitor drug use and better understand the drivers of antimicrobial resistance in Salmonella and other foodborne microorganisms.
While there are numerous small targeted surveys in the scientific literature describing antimicrobial resistance in Salmonella from different places and sources, an ongoing integrated national surveillance system is necessary to combat antibiotic resistance (16). The WHO defines integrated surveillance as “the coordinated sampling and testing of bacteria from food animals, foods, and clinically ill humans; and the subsequent evaluation of antimicrobial resistance trends throughout the food production and supply chain using harmonized methods” (1). It provides necessary data to identify emerging hazards, assess risks, monitor trends, measure interventions, and inform mitigation policies. The generation of robust and consistent data is important to the development and assessment of response measures used to combat antimicrobial resistance. In the United States, this work is conducted by the National Antimicrobial Resistance Monitoring System (NARMS). Similar systems are in place across the European Union, in Canada, across Latin America, and in some Asian and African countries. Most of these programs are focused on susceptibility surveillance of human clinical isolates. Active surveillance of animal and retail meat isolates operates to varying degrees in different countries (1). This article will review the latest available information on resistance in Salmonella (through test year 2014), with an emphasis on the situation in the United States and Europe.
THE BURDEN OF SALMONELLOSIS
NTS is recognized as one of the most common bacterial causes of foodborne diarrheal disease worldwide. Foodborne NTS is estimated to cause over 93 million cases of gastroenteritis annually and 155,000 deaths globally (8), resulting in about 4 million disability-adjusted life years (17). At-risk populations include children <1 year of age and adults ≥60, who are most vulnerable to infection and tend to have more severe disease (18). Extraintestinal or invasive NTS infections, often associated with certain serovars or phylogenetic clades, add to the global burden of illness, especially in resource-poor countries and immunocompromised patients (19). An estimated 3.4 million invasive infections occur annually, resulting in 681,000 deaths, with the young and old in Africa being most affected (20). The relatively high prevalence of highly invasive strain subtypes, such as those that have been identified among S. enterica serovars Enteritidis (21), Typhimurium (22), and Kentucky (23) may be expected to drive different antibiotic use practices to control disease in countries where invasive subtypes are endemic (24).
Despite sustained efforts to control NTS in animals and to educate the public on safe food-handling practices, the incidence of human salmonellosis has not changed significantly in the United States (25). The latest data from the CDC on microbial causes of zoonotic foodborne diseases ranked Salmonella first in incidence, at 15.5 cases per 100,000 inhabitants, resulting in over 2,100 hospitalizations and 32 deaths (18). In the European Union in 2014, the incidence of Salmonella infections (23 per 100,000) ranked well behind Campylobacter (71 per 100,000). In 2014, a total of 88,715 confirmed cases of salmonellosis were reported by 28 European Union member states and 4 nonmember states (26). In both the United States and the European Union (and in many other countries), S. enterica serovars Typhimurium and Enteritidis are consistently the most frequently isolated among confirmed human cases of disease (18, 26). In the United States and the European Union, poultry meat is the most commonly contaminated animal-derived food commodity (26, 27).
TREATMENT OF SALMONELLOSIS
The extent of antibiotic resistance in Salmonella varies by country and is influenced by antimicrobial use practices in humans and animals, as well as geographical differences in the epidemiology of Salmonella and regional serovar differences. In developed countries, drug resistance in Salmonella is driven largely by the use of antimicrobials in food-producing animals (28). In general, resistance profiles reflect the length of time an agent has been in use. Thus, irrespective of isolation source (humans, foods, food animals), the most frequent types of resistances are usually for older antimicrobials such as tetracycline, sulfamethoxazole, and streptomycin (26, 27, 29, 30).
Antimicrobial therapy is usually not indicated for uncomplicated infection. Therapy should be considered for populations at increased risk for invasive infection such as people >50 years of age with atherosclerosis, the immunocompromised, and those with cardiac disease. In these patients, the recommended antimicrobials include a fluoroquinolone such as ciprofloxacin, a third-generation cephalosporin such as ceftriaxone, trimethoprim/sulfamethoxazole, or amoxicillin. The recommended empiric antimicrobial therapy for bloody diarrhea in immunocompetent adults is either a fluoroquinolone or azithromycin, both of which show potent in vitro activity against Salmonella in the United States and the European Union. Additionally, most highly resistant strains of Salmonella are susceptible to carbapenem drugs (31, 32), making it a drug of last resort. Recommended empiric therapy for children includes a third-generation cephalosporin for those <3 months of age or azithromycin. Because fluoroquinolones, extended-spectrum cephalosporins, azithromycin, and carbapenems are critically important antibiotics for the management of salmonellosis, emerging resistance to these drug classes is a paramount concern (33).
ANTIMICROBIAL RESISTANCE IN SALMONELLA FROM THE UNITED STATES
The United States has extensive data back to 1996 on antimicrobial resistance in Salmonella, which is collected through NARMS (34). The NARMS program generates susceptibility data on Salmonella representing 5% of the nationally reported human clinical cases. These results are compared with data on Salmonella from 13 other sources that include (i) samples of retail chicken, turkey, pork, and beef collected in 14 states; (ii) cecal specimens from chickens, turkeys, cattle (dairy and beef), and pigs (hogs and sows) at slaughter; and (iii) carcass swabs, carcass rinses, and ground product from chickens, turkeys, cattle, and pigs at slaughter. A line listing of these data is freely available online (35), where information on nearly 160,000 isolates dating back to the start of NARMS can be downloaded and analyzed using standard spreadsheet and database software programs.
Few data are available from before 1996 to document the historical trends in Salmonella resistance. To address this gap, Tadesse et al. (36) conducted a retrospective study of 2,149 banked human clinical Salmonella strains and documented changing resistance patterns in strains dating back to 1948. Comparing data from pre-1960 with those from post-1989 for S. Typhimurium (which constituted most of the banked isolates) showed that resistance rose from 0% to 33% for ampicillin, 0% to 26% for chloramphenicol, 0% to 43% for streptomycin, 20% to 43% for tetracycline, and 0% to 37% for sulfamethoxazole. While recognizing the inherent limitations in directly comparing two disparate data sets, it is striking how the historical and modern data in juxtaposition show a monotonic increasing trend in resistance to older antimicrobial compounds (ampicillin, chloramphenicol, streptomycin, sulfonamide, tetracycline) from pre-1960s up until the 1990s, followed by a decline to current levels approximating those of the 1970s (Fig. 1). This study examined just over 2,100 human clinical isolates collected over 6 decades, a number now tested annually in NARMS.
FIGURE 1.
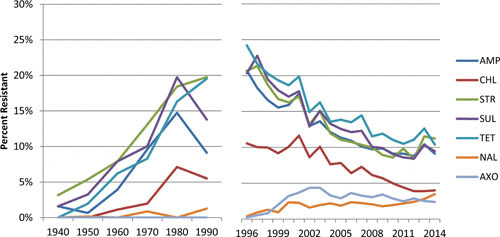
Temporal changes in resistance of clinical nontyphoidal Salmonella from the 1940s to 2014. AMP, ampicillin; CHL, chloramphenicol; STR, streptomycin; SUL, sulfonamides; TET, tetracycline; NAL, nalidixic acid; AXO, ceftriaxone.
In NARMS, antimicrobial susceptibility testing is centralized at government laboratories of the FDA, the CDC, and the U.S. Department of Agriculture. All three laboratories employ identical media, methods, quality control parameters, and repeat testing criteria, along with a common drug panel from a single manufacturer. The compounds tested have many commonalities with the European Union testing design (see below) except that ceftriaxone is used in the United States as a class representative for extended-spectrum cephalosporins (Table 1), while in Europe either ceftazidime or cefotaxime is used. The United States currently also includes amoxicillin-clavulanate, cefoxitin, streptomycin, and trimethoprim/sulfamethoxazole, while the European Union member states test trimethoprim alone. Some European Union countries report resistance data for tigecycline and colistin.
TABLE 1.
Antimicrobials tested in the European Union and United States and criteria used to interpret microbiological and clinical resistance
| Breakpoints (μg/ml) | |||
|---|---|---|---|
| European Uniona | U.S.a | ||
| Antimicrobial | Microbiological | Clinical | Clinical |
| Tetracycline (TET) | >8 | >4b | >8 |
| Sulfonamide (SUL) | >256 | >256b | >256 |
| Ampicillin (AMP) | >8 | >8 | >16 |
| Chloramphenicol (CHL) | >16 | >8 | >16 |
| Nalidixic acid (NAL) | >16 | >16b | >16 |
| Third-generation cephalosporin (CEP)c | >0.5 | >1 | >2 |
| Gentamicin (GEN) | >2 | >2 | >8 |
| Trimethoprim-sulfa (COT)d | >2 | >2 | >2 |
| Ciprofloxacin (CIP) | >0.064 | >0.064 | >0.5 |
| Azithromycin (AZI) | >16e | >16e | >16f |
| Colistin (COL) | >2 | N/A | |
European Union breakpoints are from EUCAST, and U.S. breakpoints are from CLSI unless otherwise noted.
Derived from CLSI.
In the European Union, isolates are tested against ceftazidime and cefotaxime.
In the European Union, non-human isolates are tested against trimethoprim and sulfonamide separately. In the United States, isolates are tested against trimethoprim-sulfamethoxazole and sulfamethoxazole.
Reference 149.
Different breakpoints are used that result in more conservative interpretations in the European Union compared with the United States (Table 1). The European Union uses EUCAST epidemiological cutoff values (ECOFFs) as breakpoints where available (37), which are based on the highest MIC of the wild-type population. In contrast, the United States currently interprets susceptibility results in Salmonella based on the Clinical and Laboratory Standards Institute (CLSI) clinical breakpoints (for all agents except streptomycin). Clinical breakpoints also rely on data from MIC distributions, which are combined with pharmacological information and clinical outcome trials to set clinical breakpoints. In accord with WHO recommendations (1), NARMS MIC data are also presented as MIC distributions so that other interpretive criteria can be applied to allow direct comparisons (27). The presentation of MIC data also allows for increased power in detecting slight shifts in bacterial susceptibility to some antibiotics, giving users of the data increased ability to create predictive models of resistance based on policy interventions (38).
The latest U.S. surveillance data (test year 2014) from NARMS comprised antimicrobial susceptibility results for 5,043 NTS isolates, including 2,127 from humans, 262 from retail meats, 1,579 from hazard analysis and critical control point samples (39), and 1,075 from animal cecal samples (27). The prevalence of Salmonella in U.S. retail meats in 2014 was 9.1% in chicken, 5.5% in ground turkey, 1.3% in pork, and 0.8% in beef.
A general overview of key antibiotic resistance trends in U.S. clinical isolates of Salmonella is shown in Fig. 2. Approximately 82% of Salmonella isolated from humans in 2014 had no resistance to any of the antimicrobial drugs tested, a trend that has improved since NARMS testing began in 1996. Multidrug resistance (MDR, resistant to ≥3 antimicrobial classes) was present in around 10% of isolates on average, appearing in 9.3% of human isolates in 2014. In 2014, resistance in human strains was most frequent to streptomycin (11.2%), tetracycline (10.4%), sulfamethoxazole (9.4%), and ampicillin (9.1%), followed by lower levels of resistance to chloramphenicol (4.0%), nalidixic acid (3.5%), ceftriaxone (2.4%), cefoxitin (2.2%), amoxicillin-clavulanate (2.1%), gentamicin (1.4%), trimethoprim-sulfamethoxazole (1.3%), ciprofloxacin (0.4%), and azithromycin (<0.1%) (27). Resistance to the three critically important drugs ceftriaxone, azithromycin, and ciprofloxacin was below 3% (Fig. 2). These three major findings were largely unchanged from the previous 10 years. The most common Salmonella serovars infecting humans in the United States in 2014 were Enteritidis (21%), Typhimurium (12%), and Newport (11%), followed by Javiana (6%), I 4,[5],12:i:– (5%), and Infantis (3.4%) (27).
FIGURE 2.
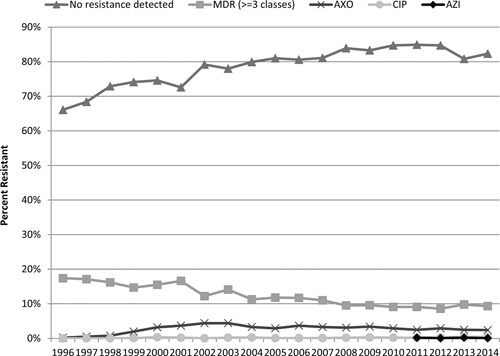
Resistances to critically important antimicrobials in human clinical Salmonella isolates from the United States. MDR, multidrug resistant; AXO, ceftriaxone; AZI, azithromycin; CIP, ciprofloxacin.
Multidrug resistance
In general, resistance in human isolates of Salmonella has been fairly low and stable over the past decade in the United States. Because a substantial proportion of human infections are acquired from non-food-animal sources (3), however, it is not unexpected that resistance in animal isolates tends to be higher. By animal species, resistance to any of the 14 tested compounds was most frequent in turkey sources (approximately 60% to 80% resistant to ≥1 agent), followed by broilers (about 30% to 60%), and cattle (30% to 40%). These findings are consistent with other studies that show that Salmonella from poultry sources tend to be more resistant than Salmonella from cattle sources (38) and that swine isolates tend to be more resistant than those from cattle (40).
In human clinical strains, MDR is most frequent in S. enterica serovars Typhimurium (28.9%), I 4,[5],12:i:– (27.9%), and Heidelberg (7.6%). While MDR is declining in S. Typhimurium (see below), it has risen in S. 4,[5],12:i:–, from 5.5% in 2007 to 50% in 2014, with 47.3% exhibiting resistance to more than four drug classes. In animals at slaughter, MDR was most frequent in isolates from turkeys (47%) and hogs (20%), followed by broilers (15%), dairy cattle (10%), beef cattle (6%), and sows (6%) (27). In 2014, MDR was detected in 36% of retail ground turkey isolates and 20% of retail chicken meat. MDR is disproportionately high in broilers among serovars Kentucky (33% hazard analysis and critical control point to 58% cecal isolates) and Typhimurium (16% to 53% among hazard analysis and critical control point and cecal isolates, respectively).
Measuring MDR by the number of resistance phenotypes without regard to the importance of the drug classes involved is of limited value. To help overcome this limitation, NARMS has published new tools online that allow the user to investigate any combination of specific MDR patterns and their changes over time (31). This permits a more refined analysis of MDR patterns in assessing risks, for identifying specific resistance trends of higher public health importance, and to better understand the specific drivers of resistance where coresistances are involved.
The percentage of Salmonella strains that are resistant to ampicillin, chloramphenicol, streptomycin, sulfonamide, and tetracycline (the ACSSuT penta-resistant phenotype) has been tracked as a hallmark of the globally disseminated Salmonella Typhimurium DT104 for decades. The ACSSuT resistance pattern has declined steadily in all U.S. salmonellae from 34% in 1996 to 14.5% in 2014. Declining levels of both S. Typhimurium (27) and MDR S. Typhimurium (41) are the main drivers behind overall declining levels of MDR Salmonella in human isolates. The ACSSuT resistance in cattle isolates of S. Typhimurium declined sharply from 67% in 2009 to 7% in 2014. This highlights an important feature of Salmonella resistance, namely, the serovar-specific nature of some resistance patterns whose ascendancy and decline may be temporally associated with the prevalence of specific strains.
In the United States in recent years, the ASSuT (ampicillin, streptomycin, sulfonamide, tetracycline) MDR pattern has increased among human isolates of S. I 4,[5],12:i:–, where it climbed from 1.4% in 2009 to 43% in 2014. Outbreak investigations of this strain have pointed to swine as a possible source (42), backed by findings of increased S. I 4,[5],12:i:– in diseased pigs in Minnesota (40), one of the top five pig-producing states in the United States.
While MDR is not common in S. Enteritidis, it is driven by a higher resistance to nalidixic acid compared with other serovars. MDR is common in S. Newport, where the MDR-AmpC phenotype on an IncA/C or IncI backbone is a common feature of resistance (43).
Quinolone Resistance
Overall, fluoroquinolone resistance has been consistently low among Salmonella isolated from all U.S. surveillance sources. In human isolates, it predominantly presents in serovar Enteritidis (47%) and is associated with travel (44). Since 2007, ciprofloxacin resistance has been detected in a total of only four cattle and six swine isolates in the United States. Ciprofloxacin resistance is not present (using CLSI breakpoints) in isolates from U.S. poultry Salmonella, where fluoroquinolones have not been used since 2005. In 2014, the first instance of ciprofloxacin-resistant Salmonella in meat was a single isolate from a retail pork sample which carried the qnrS gene. This was the first report of qnr genes present in retail meat Salmonella isolated in the United States (27).
While ciprofloxacin resistance is rare in U.S. salmonellae in general, decreased susceptibility to ciprofloxacin (DSC, MIC ≥ 0.125 μg/ml) has increased in humans and cattle strains as well as in swine strains. (Strains with this MIC would be considered microbiologically resistant according to EUCAST breakpoints.) Studies have reported extremely low quinolone resistance among Salmonella isolated from feedlot and beef cows as well as dairy cows (45, 46). However, decreased susceptibility is rarely assessed. Distribution of extrachromosomal qnr genes is thought to be the main reason why this phenotype is increasing in frequency. Because fluoroquinolones are widely, and often repeatedly, used to treat bovine respiratory disease, a common illness in cattle herds, there is a possibility that fluoroquinolone use may help to propagate an acquired resistance gene. Particularly concerning is the emergence of DSC in S. enterica serovar Dublin isolates from humans and cattle, where increasing cephalosporin resistance is also occurring. The incidence of human S. Dublin infections is relatively low, but it can cause invasive disease with more severe outcomes. S. Dublin also causes severe disease in cattle, mainly respiratory infections, and ranks among the top four serovars isolated from retail ground beef and cattle samples in NARMS and the top serovar among isolates derived from clinical specimens (40). As of the 2014 NARMS testing year, 57% (4/7) of DSC S. Dublin isolates from humans and 40% (18/42) of DSC S. Dublin isolates from cattle were also resistant to ceftriaxone. This combination of DSC and ceftriaxone resistance puts significant limitations on possible treatment options for human illness.
Cephalosporin Resistance
Among critically important human antibiotics, the temporal association of ceftiofur use in food animals and the emergence of ceftriaxone resistance in both animals and humans has been an area of concern in the United States, the European Union, and elsewhere. In the United States, ceftriaxone resistance was not detected in Salmonella prior to the approval of ceftiofur (36) and rose following its approval for use in the United States in livestock and poultry (Fig. 3). From 1996 through 2009, the percentage of NTS human isolates resistant to ceftriaxone increased from 0.2% to 3.4% (41). These rising trends have caused several countries, including the United States, to limit certain uses of cephalosporins in animal agriculture, with positive effects. A well-known example occurred in Canada when voluntary restrictions on ceftiofur use were followed by a rapid and significant decline in ceftriaxone-resistant S. Heidelberg (and Escherichia coli) in chickens, retail chicken meats, and human clinical isolates (47). In the United States, the FDA announced plans in 2008 to restrict the use of some cephalosporins, which went into effect in 2012 (48). As of 2014, it appears that the restrictions may be producing the desired effect of reducing cephalosporin resistance in humans and select animal species. Ceftriaxone resistance has declined since 2009 in human (3.4% to 2.4%) and retail chicken (38% to 18%) Salmonella isolates (Fig. 3).
FIGURE 3.
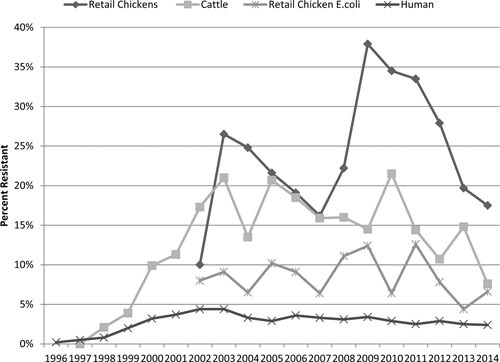
Trends in third-generation cephalosporin resistance in Salmonella from the United States.
Changes in ceftriaxone resistance were most notable for S. Heidelberg, where resistance in human isolates declined to 8.5% in 2014, down from a peak of 24% in 2010 (27). Ceftriaxone resistance in retail chicken S. Heidelberg remained at 0% from 2011 to 2013 (down from a peak of 32% in 2009), but reappeared in 3/24 isolates in 2014. In retail ground turkey isolates in 2014, resistance continued to decline to 7% after peaking at 22% in 2011. In cattle Salmonella isolates in 2014, ceftriaxone resistance reached its lowest level (7.6%) since 1999 (31). Studies of diagnostic cattle isolates show varied results. While some studies show demonstrable decreases in cephalosporin resistance among dairy cattle since 2012 (46), others show continued increases (40) or even no resistance at all among healthy feedlot cattle (49). Many of these disparities are due to regionalization of serovar frequencies, as well as the types of samples analyzed (diseased versus healthy animals).
Ceftriaxone resistance in human strains is most common in the same serovars in which MDR prevails, namely, Typhimurium, Newport, Heidelberg, and I 4,[5],12:i:–, with the addition of Dublin, 11.5% of which were ceftriaxone resistant. Ceftriaxone resistance was also high in S. Dublin from cattle (34.6%) and ground beef (60%). In other nonhuman sources, ceftriaxone was disproportionately high in serovars Newport from cattle (66.7%), Kentucky from broilers (66%), Typhimurium from broiler meat (72%), and Heidelberg from fattening turkeys (60%).
NARMS data show that extended-spectrum cephalosporin resistance among U.S. isolates of Salmonella (and E. coli) is usually mediated by blaCMY genes, whereas extended-spectrum beta-lactamases (ESBLs) have been rare (50). This appears to be changing. Examining NARMS strains from 2012 to 2014, 26 instances of ESBLs occurred, mainly conferred by members of the CTX-M family, along with 3 instances of blaSHV-12 and one case of blaSHV-30. Finding blaCTX-M-65 in a 2014 retail chicken sample led to an expanded examination of U.S. human and animal strains and revealed that the gene had become widespread in a strain of serovar Infantis previously identified in Europe and South America (51).
ANTIMICROBIAL RESISTANCE IN EUROPE
Denmark established the world’s first integrated antimicrobial resistance surveillance program in 1995, and other members of the European Union have followed with their own national programs. In the European Union, antimicrobial resistance in Salmonella is tracked in data submitted by European Union member states and Norway and is reported jointly by the European Food Safety Authority (EFSA) and the European Centre for Disease Prevention and Control (ECDC).
The latest European Union data are from 2014 (26), when for the first time, all 28 member states along with Iceland and Norway submitted isolate-level data on poultry and poultry meat products. Therefore, the European Union report focuses on resistance in human and poultry sources of isolates. Countries submit both MIC data, which is interpreted using published ECOFFS, and susceptibility categories interpreted from disk diffusion. In 2014, 21 member states and Norway provided data on human Salmonella isolates. Twelve countries (Austria, Denmark, Estonia, Finland, Greece, Ireland, Italy, Luxembourg, the Netherlands, Norway, Portugal, and Romania) reported isolate-level results in the form of inhibition zone diameters or MICs, which allows for improved comparability between human and animal/food isolates. Ten countries reported categorical interpretations of susceptible (S), intermediate (I), or resistant (R) according to the clinical breakpoints. The number and types of antimicrobials reported varied by country, from 2 countries testing only three antimicrobials to 13 countries testing all 10 antimicrobials in the priority panel. This mixture of breakpoints and testing methods, sampling strategies, differences in serovar by country, and the incomplete nature of the isolate-level data mean that the results must be interpreted, and the country differences compared, with caution.
In 2014 in the European Union, a total of 14,412 Salmonella isolates from human infections were tested, constituting 16% of all confirmed human cases of illness. When examining all serovars (n = 247) and countries (n = 22) combined, the most common resistances in human Salmonella isolates were to tetracyclines (30.3%), sulfonamides (28.6%), and ampicillin (28.2%), followed by lower levels of resistance to trimethoprim-sulfamethoxazole (9.2%), ciprofloxacin (8.8%), chloramphenicol (6%), gentamicin (2.7%), and cefotaxime (1.1%) (26). The top three resistances mirror the order of resistance profiles in the United States (except for streptomycin, which is not reported by EFSA).
Although the United States and the European Union employ different criteria for surveillance reporting, these differences are absent or minor for most drugs. Small discrepancies are evident for extended-spectrum cephalosporins and gentamicin that have nugatory effect on reported rates of resistance. Breakpoints for ciprofloxacin have the largest affect, where application of the EUCAST criteria changes the U.S. resistance percentage in human isolate data from 0.4% to 4.3%. The EUCAST breakpoints were applied for the purposes of comparison below.
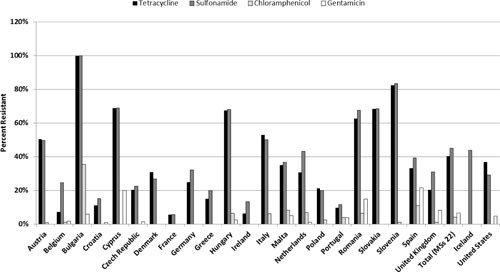
Monitoring data from food sources show some commonalities with the U.S. situation. An overall comparison between the European Union and the United States of resistance to the “older” antimicrobials of chloramphenicol, tetracycline, gentamicin, and ampicillin is shown for human isolates (Fig. 4) and broiler isolates (Fig. 5) of Salmonella spp. For human data, the U.S. surveillance data are comparable to Slovenia, which reports the lowest resistance levels for these drugs among the member states. Resistance data for broiler strains of Salmonella spp. show that the United States is slightly below the European Union average.
FIGURE 4.
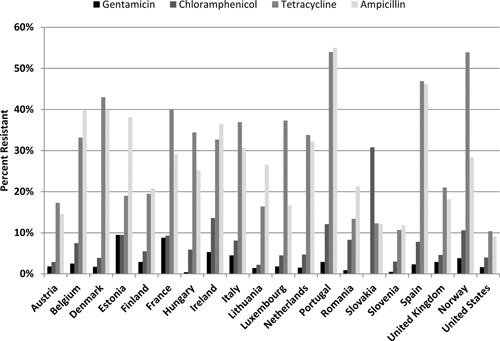
Resistance to gentamicin, chloramphenicol, tetracycline, and ampicillin in human Salmonella isolates from select European Union countries, Norway, and the United States. Breakpoints used for interpreting MICs were derived from the EUCAST.
FIGURE 5.
Resistance to gentamicin, chloramphenicol, tetracycline, and ampicillin in broiler Salmonella isolates from select European Union countries, Iceland, and the United States. Breakpoints used for interpreting MICs were derived from the EUCAST.
Other general features of resistance in both the European Union and the United States are evident. With some exceptions, Salmonella recovered from poultry meats is generally more resistant than isolates from human infections in both regions (26, 31). Among poultry isolates, resistance levels in the European Union were generally higher in turkeys than chickens. This is comparable to what was observed in the United States, where turkey isolates tended to be the most resistant, followed by those from chicken, swine, and cattle (27). One might presume that this pattern is the result of higher antimicrobial use in turkeys than in broilers, but there have been no on-farm studies that confirm a causal relationship (52). In general, fluoroquinolone resistance is higher in the European Union, and third-generation cephalosporin resistance is higher in the United States. Other resistance patterns are compared below.
Multidrug Resistance
In 2014, 10 member states tested Salmonella against 9 classes of antimicrobials (26). Overall, only 54.8% of human isolates were susceptible to all agents tested. MDR was high overall (26.0%) in human isolates from the European Union, with very high prevalence in some countries. Among poultry sources, MDR was low in laying hens; high in broiler meat, turkey meat, and broilers; and very high in turkeys. As with other types of resistance, MDR tends to be associated with certain serovars, generally being rare in S. Enteritidis. In Europe, MDR variants of S. Kentucky (74.6%), along with monophasic S. Typhimurium I 4,[5],12:i:– (69.4%) and S. Infantis (61.9%), are especially problematic in humans. As noted above, the United States also is witnessing a rise in MDR S. Typhimurium I 4,[5],12:i:–. While S. Kentucky rarely causes human disease in the United States, it commonly exhibits MDR.
Extended-spectrum cephalosporin-resistant S. Infantis (carrying the blaCTX-M65 gene) has emerged in the United States as a public health concern. The circulating clone is similar to an Italian strain of S. Infantis carrying the blaCTX-M65 gene (53). While MDR S. Infantis has not increased in poultry isolates collected for NARMS surveillance, EFSA reports that S. Infantis isolates from European Union broiler meat express very high levels of MDR (>70.0%). Isolates from Italian broilers exhibit exceptionally high levels of resistance to third-generation cephalosporins, which may be characteristic of the CTX-M clone. Particularly concerning was the detection of high-level resistance to ciprofloxacin in these isolates. The clone may be limited to chickens, because no resistance to third-generation cephalosporin was detected in fattening turkeys (26).
Much of the difference in resistance among poultry isolates from the United States and the European Union is likely due to the variation in serovar profiles of poultry isolates between the two regions. S. Infantis and S. Enteritidis are among the top three serovars in chickens and chicken meats in the European Union, but in the United States, serovar Kentucky predominates. Likewise, in European Union fattening turkeys, serovars Derby, Kentucky, and Newport account for 30% of Salmonella isolates, but in the United States, serovars Hadar and Reading round out the top two in turkeys and turkey meats (31).
Quinolone Resistance
A comparison of human isolates from European countries that reported data for both fluoroquinolones and extended-spectrum cephalosporins is shown alongside the U.S. data in Fig. 6. The average resistance to extended-spectrum cephalosporins is low (1.1%) in the European Union data, while ciprofloxacin resistance was found in 8.8% of all human isolates, ranging from 0% to 19% among reporting countries compared with 4.3% in the United States. The European Union resistance to ciprofloxacin is driven in part by a high prevalence of ciprofloxacin resistance in serovar Kentucky, where 84% were resistant to ciprofloxacin. This aligns with previous findings on the establishment of ST198 S. Kentucky in humans and poultry flocks throughout Europe and other areas (23). While this strain has been found in the United States, it has predominated in travelers and imported foods (54).
FIGURE 6.
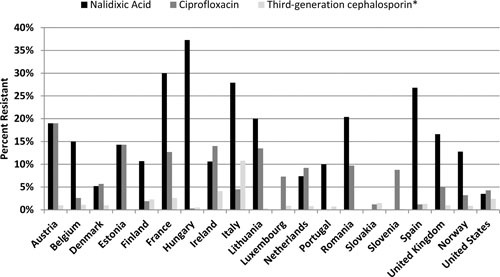
Resistance to quinolones and extended-spectrum cephalosporins in human isolates of Salmonella from select European Union countries, Norway, and the United States. Breakpoints used for interpreting MICs were derived from the EUCAST. Among critically important drugs (defined here as macrolides, fluoroquinolones, extended-spectrum cephalosporins, and carbapenems), azithromycin, meropenem, and colistin resistances were very rare and not reported in most countries. *Percentage based on reporting of either cefotaxime or ceftazidime resistance from the European Union or ceftriaxone from the United States.
Differences between Europe and the United States for critically important resistances are evident in isolates from poultry and poultry meat (Fig. 7). In the European Union, a very high proportion of Salmonella from broilers (average = 53.5%; n = 23 countries) was resistant to ciprofloxacin, ranging from 0% (Denmark and Ireland) to over 80% (Bulgaria, Hungary, and Slovenia). Similarly, ciprofloxacin resistance in broiler meats (average = 42.6%; n = 11 countries) ranged from 0% (Ireland) to 97.9% (Hungary). In meat from fattening turkeys, for which only three European Union countries reported, ciprofloxacin resistance rates varied from 6.9% in France to 74.2% in Germany and 91% in Hungary. Some of the ciprofloxacin-resistant isolates were not resistant to nalidixic acid, which is common for plasmid-mediated quinolone-resistance (PMQR) mechanisms (55). In contrast, the U.S. data show that 1/143 retail chicken meat isolates in 2014 was nonsusceptible to ciprofloxacin (MIC = 0.125 μg/ml), and no other ciprofloxacin resistance was detected from any poultry sources.
FIGURE 7.
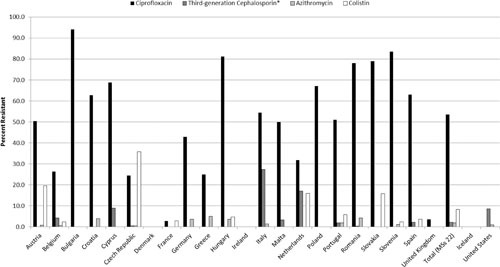
Resistance to quinolones, extended-spectrum cephalosporins, macrolides, and colistin in broiler strains of Salmonella from select European Union countries, Iceland, and the United States. Breakpoints used for interpreting MICs were derived from the EUCAST. Among critically important drugs (defined here as macrolides, fluoroquinolones, extended-spectrum cephalosporins, and carbapenems), azithromycin, meropenem, and colistin resistances were very rare and not reported in most countries. *Percentage based on reporting of either cefotaxime or ceftazidime resistance from the European Union or ceftriaxone from United States.
Cephalosporin Resistance
Resistance to third-generation cephalosporins ranged from 0% to 10.8% in the European Union (26) and was present in 2.4% of U.S. human clinical isolates (31). Among broiler isolates, the European Union averaged 2.3% resistance to third-generation cephalosporin (cefotaxime), with resistance entirely absent from 14 of the 23 reporting countries. Resistance was higher in the United States, where ceftriaxone resistance was detected in 8.7% of broiler isolates collected at slaughter. Only Italy (27.3%) and Cyprus (8.9%) were higher. Among the 11 member states that monitor meat derived from broilers, rare resistance was reported from Belgium (3.7%) and Spain (0.8%), compared with NARMS data which showed 17.5% resistance in 2014 isolates, down from a peak of 37.9% in 2009 (Fig. 7).
CTX-M is just one of several ESBL enzymes that can confer resistance to extended-spectrum cephalosporins and most other beta-lactam antimicrobials. In 2014, EFSA reported that serovar Infantis comprised the highest proportion of broiler isolates exhibiting an ESBL phenotype (18/30, 60%), followed by serovar Paratyphi B L(+) tartrate positive (6/30, 20%). Member states did not report molecular characterization of isolates, so the determination of ESBL positivity is presumptive; however, other studies do show that CTX-M is the most prevalent ESBL in Europe (56), dominating over the TEM and SHV enzyme families. Though they predominate in the United States, AmpC enzymes, which confer resistance to most beta-lactams and beta-lactamase inhibitors, are less frequently encountered in Europe. Genes encoding AmpC enzymes are typically carried on large plasmids that harbor other antimicrobial resistance genes but can also be chromosomally located (57). Of the 18 broiler isolates with an AmpC phenotype, 9 (50%) were serovar Heidelberg. Eleven of the eighteen isolates were from the Netherlands.
Colistin Resistance
Following the discovery of transmissible colistin resistance mediated by the mcr-1 gene in E. coli in China, (58) colistin-resistant Salmonella isolates were found in several countries (59). Colistin resistance was reported in The Netherlands (21.5%) and Denmark (5.9%) in 2014 (26). Overall, EFSA reports colistin resistance in Salmonella recovered from broilers (8.3%), laying hens (13.5%), and turkeys (1.8%) (Fig. 7). Most of these (72% of broilers and 80% of laying hens) were serovar Enteritidis, which may reflect a level of intrinsic resistance. Numerous separate studies have detected the mcr-1 gene in food animal and human Salmonella strains from several European countries (60–62) and from human strains in the United States (63). Additionally, mcr-1 has been found in multidrug-resistant isolates, raising the possibility of coselection (64). While colistin has been used routinely in livestock in several countries, it is not marketed or available for use in food animals in the United States. Therefore, NARMS does not test for phenotypic colistin resistance in routine surveillance. In addition, the mcr-1 gene was not found in whole-genome sequence data on over 6,500 U.S. Salmonella genomes, mostly from retail meats (65).
Other Resistances
In regard to other critically important antibiotics, azithromycin resistance remains rare in Salmonella from both continents, ranging in detection from <0.1% of U.S. isolates to 1.7% of strains from Denmark. Similarly, azithromycin resistance in Salmonella from chickens and turkeys was identified but was not frequent (Fig. 7).
Very few cases of carbapenem-resistant Salmonella infections have been reported in humans in the European Union and the United States (66–69), and resistance has only occasionally been found in Salmonella and other Gram-negative bacteria from food animals (70, 71). The impact of these sporadic findings on public health is of paramount concern. While currently, this impact appears to be small, there is speculation that carbapenemase genes can transfer between humans, animals, and the environment and that the use of third-generation cephalosporins may maintain bacterial populations that express these genes (70). Regardless, as of the writing of this article, carbapenem resistance had not been reported in any retail meat or food isolates of Salmonella.
Tigecycline also is not licensed for veterinary use, is not targeted in NARMS, and is an optional drug for susceptibility testing in the European Union. The 2014 EFSA data show microbiological resistance to tigecycline in 9.3% of all Salmonella spp. from broilers and 8% from turkeys. Most tigecycline resistance was associated with serovar Infantis in poultry, and most resistant strains had MICs just above the ECOFF breakpoint at 2 or 4 μg/ml. Resistance to tigecycline in Salmonella is thought to be mediated by regulatory gene mutations leading to increased efflux.
RESISTANCE IN OTHER REGIONS
Few countries have ongoing surveillance of antimicrobial susceptibility in Salmonella from food-producing or companion animals. Most data on antimicrobial resistance in Salmonella is derived from cases of human clinical illness. Sustained integrated surveillance of the food chain exists to different degrees outside the European Union and North America. In South America, for example, the Colombian Integrated Program for Antimicrobial Resistance Surveillance (COIPARS) monitors Salmonella from poultry farms, slaughterhouses, and retail poultry for resistance (72). In Southeast Asia, some Salmonella resistance data are systematically collected and published in periodic summary reports. The Japanese Veterinary Antimicrobial Resistance Monitoring System (JVARM) (73) began in 2000 and tracks antimicrobial resistance in food animal Salmonella throughout Japan.
In 2013, JVARM reported Salmonella resistance data collected from 2008 to 2011 on isolates from diagnostic samples from livestock and poultry infections (73). MICs were determined for a total of 688 Salmonella isolates: 301 from cattle, 236 from pigs, and 151 from chickens. Most were serovars Typhimurium (244 isolates, 35.5%), Choleraesuis (85 isolates, 12.4%), and Infantis (48 isolates, 7%).
JVARM reports that resistance rates against most antimicrobials studied were largely unchanged during the 2008 to 2011 sampling interval, with some slight declines (73). For compounds tested in both 2008 and 2011, resistances were as follows: tetracycline, 61% and 46%; ampicillin, 37% and 24%; kanamycin, 19% and 13%; chloramphenicol, 19% and 11%; nalidixic acid, 11% and 9%; gentamicin, 6% and 3%; cefazolin, 1.8% and 3.6%. Among critically important antimicrobial agents, resistance to cefotaxime was found in 1.7% of pig strains and 3% of chicken strains in 2010. In 2011, cefotaxime resistance was present in 10% of cattle isolates. Resistance to nalidixic acid was most common in pig isolates (16% to 21%). No resistance to colistin or ciprofloxacin was detected (73).
In 2012, JVARM added sampling of bacteria from animals at slaughter (12). The top serovars again were Typhimurium (37.5%), Choleraesuis (12.6%), and Infantis (6%). Between 2012 and 2013, a total of 365 Salmonella isolates (140 from cattle, 143 from pigs, and 82 from chickens) were subjected to antimicrobial susceptibility testing (12). Resistance was most frequent in Salmonella from cattle and swine for streptomycin (67.9% and 70.0% respectively), tetracycline (34.5% to 66.1% and 53.0% to 66.7%, respectively), and ampicillin (34.5% to 60.7% and 25.3% to 45.0%, respectively). Resistance to cefazolin and cefotaxime was rare in Salmonella isolates from cattle and chickens (0% to 8.9%). Resistance to colistin was found in a few isolates from pigs and chickens (0% to 8.9%). When comparing resistance between 2008 and 2009 with data from 2012 to 2013, overall resistance to most antimicrobials was stable in Japan except for nalidixic acid, which increased significantly (0.6% to 5.0%) in Salmonella from cattle, and kanamycin, tetracycline, and chloramphenicol resistance, which decreased significantly in isolates from pigs. Among the 212 Salmonella isolates tested between 2012 and 2013 from broilers at slaughter, resistance was common for streptomycin (77.7% to 84.7%), tetracycline (74.5% to 82.2%), ampicillin (22.9% to 31.9%), kanamycin (31.9% to 42.4%), and trimethoprim-sulfamethoxazole (31.9% to 48.3%). Resistance to nalidixic acid was high (29.8% to 19.5%), but resistance was infrequent for cefazolin (5.9% to 7.4%), cefotaxime (5.1% to 7.4%), and chloramphenicol (0% to 0.8%) (12). The JVARM program is planning to add retail meat testing in the near future.
China has one of the largest food animal production economies of any country. It is estimated that there are about 30 million annual cases of salmonellosis in China (74). One study estimated that between 1994 and 2005, about 22% of foodborne diseases in China were caused by Salmonella (75). While China works to implement an integrated national antimicrobial resistance monitoring system suited to its annual production volume, the status of resistance in Salmonella can be assessed only from a limited number of targeted studies, most of which target retail meats or human cases of salmonellosis or focus on specific resistance traits.
A 2005 survey by Yan et al. (76) examined raw retail samples of pork (n = 45), chicken (n =120), beef (n = 45), mutton (n = 45), seafood (n = 96), and milk powder (n = 36) in nine cities in Hebei province in northern China. The most common serovars were Agona (13.6%), Senftenberg (9.9%), Meleagridis (8.6%), and Derby (8.6%). MDR was most common in serovars Derby, Indiana, and Saintpaul. Among critical antimicrobial agents, nalidixic acid resistance was found in 31% of isolates, most commonly in isolates recovered from chicken (14/16, 74%). Ceftriaxone resistance was found in two (2.5%) isolates, and ciprofloxacin resistance was detected in eight (9.9%) isolates. Two years later, a study by Yang et al. (77) examined retail meats consisting of 515 chicken, 91 pork, 78 beef, and 80 lamb samples from nearby Shanxi Province in 2007 to 2008. The most common serovars were Enteritidis (31.5%), Typhimurium (13.4%), Shubra (10.0%), Indiana (9.7%), and Derby (9.5%). In addition to the common resistance to sulfamethoxazole (67%), trimethoprim/sulfamethoxazole (58%), and tetracycline (56%), a very high proportion of isolates were resistant to nalidixic acid (35%), ciprofloxacin (21%), and ceftriaxone (16%). Of particular interest, nearly all isolates of serovars Shubra (89%) and Indiana (88%) were resistant to more than antimicrobials, much higher than in other serovars (77).
A 2011 study by Bai et al. (78) examined resistance in Salmonella from large-scale chicken and swine abattoirs in Henan, China, where 128/283 (45.2%) chicken samples and 70/240 (29.2%) pig samples yielded Salmonella isolates for antimicrobial susceptibility. The most common resistance was to nalidixic acid (91%), followed by ampicillin (66%) and tetracycline (47%). Ciprofloxacin resistance was detected in 11 (8.6%) poultry strains, all of which were serovar Indiana and 6 of which also carried an ESBL. In the pig strains, the most common resistance was to tetracycline (67%), followed by nalidixic acid (64%) and chloramphenicol (60%). Ciprofloxacin resistance was found in 10% (7/70) of the isolates, 5 of which also carried an ESBL. This study shed light on the nature of MDR in serovar Indiana in China, where 11 of 198 isolates were coresistant to ciprofloxacin and cefotaxime and harbored blaCTX-M-65 and aac(6′)-Ib-cr genes, typically carried by plasmids and conferring resistance to the front-line drugs for salmonellosis. Another important study, by Lai et al., examined resistance in Salmonella in the Shandong province of China from 2009 to 2012. This survey showed a general rise in resistance during the sampling interval, with ciprofloxacin resistance appearing in 42% of salmonellae in 2012 (79).
Various later reports described the MDR traits found in serovar Indiana in China (77, 80, 81). In a study of ESBL-producing Salmonella from production environments in China, Zhang et al. (82) explored the genetic constituents of resistance in CTX-M-producing Salmonella isolates from chickens and pig facilities. They found that serovars Typhimurium and Indiana most commonly carried various transmissible CTX-M alleles, which in some cases was coupled to PMQR along with other resistance determinants. Serovar Indiana showed higher resistance levels than serovar Typhimurium. This combination of ESBL and PMQR in Salmonella has been noted in other studies from China (83, 84).
Despite that fact that Salmonella serovars and resistance can display distinct geographic characteristics, Salmonella clones have shown a remarkable ability to spread worldwide. This presents a perpetual public health issue concerning the international spread of new resistant clones as has been seen in the past. The striking example of global dissemination illustrated by MDR DT104 is a case in point. This strain spread to almost all countries and became a major driver of resistance reported in many regions (85–87). However, there has been a decrease in reported MDR DT104 in the past 10 years (41). Recent phylogenetic analysis indicates that the strain may have initially emerged in Europe in the early 1970s, followed by multiple transmission events between countries and hosts (88), likely related to the trade in breeding animals, human travel, and international sales of food products. The case of DT104 exemplifies the need to consider salmonellosis in the context of a global One Health paradigm, where resistance gaining ascendancy in animals can gain a foothold and spread around the world to become a major cause of resistant human infections.
ANTIMICROBIAL-RESISTANT SALMONELLA FROM COMPANION ANIMALS
Companion animals pose a public health risk for human salmonellosis in many countries (2, 89, 90). Although the prevalence of Salmonella in companion animals, especially in dogs and cats, varies greatly (ranging from 0% and 70% [2, 91, 92]), a number of serovars important to human health have been found in pet animals. Hoelzer (2) reported that among the top 20 most common human Salmonella serovars, 15 have been isolated from domestic dogs and cats, including Typhimurium, Enteritidis, Newport, Heidelberg, Montevideo, Muenchen, Oranienburg, Braederup, Agona, Infantis, Thompson, Paratpyhi B, Stanley, Tennessee, and Hadar. According to a 2006 annual report of the National Veterinary Services Laboratories of the U.S. Department of Agriculture (USDA), serovars Newport, Typhimurium, Montevideo, and Enteritidis were the most common serovars isolated from sick dogs and cats in the United States (93).
Because companion animals, especially dogs and cats, are considered family members, many antimicrobials that are important to human health have been used in veterinary practice to treat infections of dogs and cats, including category I antimicrobial agents, such as third/fourth-generation cephalosporins, fluoroquinolones, nitroimidazoles, penicillin beta-lactam inhibitors; category II agents, such as first/second-generation cephalosporins, penicillin, lincosamides, macrolides, and trimethoprim-sulfonamides; and category III agents, such as chloramphenicol, sulfonamides, and tetracycline (94–97). There is an increased concern about the rapid emergence and spread of MDR bacteria from pets to humans due to the extensive use of antimicrobial agents in these animals and their close contact with humans (98). Various MDR bacteria such as E. coli, Salmonella, Staphylococcus, Pseudomonas, Streptococcus, Klebsiella, Proteus, and Enterococcus have been isolated from diseased dogs and cats (95–100).
Currently, there is no surveillance program in the United States for antimicrobial resistance in bacteria from companion animals, and antimicrobial use in these animals is not routinely monitored. Therefore, no reliable data are available to assess the trend of antimicrobial resistance in bacteria isolated from companion animals. Several reports have shown that the increased use of antimicrobials to treat diseased dogs and cats is associated with the emergence of antimicrobial resistance in Salmonella. Among these reports are early U.S. NARMS Animal Arm reports (1997 to 2004), which feature data on antimicrobial resistance in Salmonella isolated from sick dogs and cats. The Salmonella isolates were submitted to the USDA National Veterinary Services Laboratory in Ames, IA, and tested for susceptibilities to 16 antimicrobials. Salmonella isolated from diseased dogs and cats had varied rates of resistance to different antimicrobials: ampicillin (24.6% to 58.8%), streptomycin (23.7% to 58.8%), tetracycline (22.7% to 53.8%), sulfamethoxazole (6.3% to 58.8%), and chloramphenicol (9.4% to 43.8%) (101). When surveillance first began in 1997, Salmonella isolates from sick dogs (n = 38) were susceptible to amoxicillin/clavulanic acid, ceftiofur, ceftriaxone, cephalothin, gentamicin, nalidixic acid, and trimethoprim/sulfamethoxazole, but 1 year later, isolates appeared resistant to all of these drugs except nalidixic acid. By 2002, resistance to amoxicillin/clavulanic reached 30.3%, ceftiofur 29.5%, ceftriaxone 29.5%, cephalothin 31.2%, and trimethoprim/sulfamethoxazole 12.3%. While dog isolates from 1997 were susceptible to most drugs tested, cat isolates (n = 28) from that same year were resistant to amoxicillin/clavulanic acid (10.7%) and ceftiofur, ceftriaxone, and cephalothin (10.7%) but were susceptible to gentamicin, nalidixic acid, and trimethoprim/sulfamethoxazole. All Salmonella isolates from sick dogs and cats from 1997 to 2004 were susceptible to amikacin and ciprofloxacin (Table 2). Overall, these NARMS data show that Salmonella isolates from sick dogs and cats have increased in their resistance to amoxicillin/clavulanic acid, ceftiofur, gentamicin, and nalidixic acid over the years, with nalidixic acid resistance first appearing in dog isolates in 2000 and in cat isolates in 2004. Analysis of all 5,709 Salmonella isolates from domestic food and companion animals collected for NARMS in 1997 and 1998 showed that extended-spectrum cephalosporin-resistance levels differed significantly among host animal species, with higher resistances found in isolates from turkeys, horses, cats, and dogs. All Salmonella isolates resistant to extended-spectrum cephalosporins carried the blaCMY gene (102).
TABLE 2.
Percentage resistance of Salmonella isolated from clinical companion animalsa
| Antimicrobialsb | 1997 | 1998 | 1999 | 2000 | 2001c | 2002 | 2003 | 2004 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dogn = 38 | Catn = 28 | Dogn = 57 | Catn = 29 | Dogn = 57 | Catn = 25 | Dogn = 44 | Catn = 17 | Dogn = 64 | Dogn = 92 | Catn = 19 | Dogn = 68 | Catn = 32 | Dogn = 53 | Catn = 22 | |
| Amoxicillin/clavulanic acid | 0 | 10.7 | 7.0 | 6.9 | 10.5 | 8.0 | 13.6 | 17.6 | 29.7 | 38.0 | 10.5 | 20.6 | 3.1 | 26.4 | 18.2 |
| Ampicillin | 31.6 | 53.6 | 24.6 | 48.3 | 26.3 | 40.0 | 34.1 | 58.8 | 35.9 | 43.5 | 26.3 | 29.4 | 25.0 | 30.2 | 31.8 |
| Apramycin | 0 | 3.6 | 1.8 | 0 | 0 | 4.0 | 4.5 | 0 | 1.6 | ||||||
| Cefoxitin | 11.4 | 17.6 | 29.7 | 37.0 | 10.5 | 20.6 | 3.1 | 22.6 | 13.6 | ||||||
| Ceftiofur | 0 | 10.7 | 7.0 | 6.9 | 8.8 | 8.0 | 11.4 | 17.6 | 29.7 | 37.0 | 10.5 | 20.6 | 3.1 | 22.6 | 13.6 |
| Cephalothin | 0 | 10.7 | 8.8 | 13.8 | 14.0 | 8.0 | 11.4 | 23.5 | 29.7 | 39.1 | 10.5 | 26.5 | 9.4 | ||
| Chloramphenicol | 13.2 | 28.6 | 21.1 | 20.7 | 17.5 | 36.0 | 25.0 | 35.3 | 29.7 | 43.5 | 18.5 | 26.5 | 9.4 | 28.3 | 22.7 |
| Gentamicin | 0 | 0 | 10.5 | 6.9 | 1.8 | 12.0 | 6.8 | 11.8 | 9.4 | 5.4 | 5.3 | 17.6 | 3.1 | 1.9 | 9.1 |
| Kanamycin | 18.4 | 32.2 | 12.3 | 27.6 | 12.3 | 24.0 | 9.1 | 35.3 | 10.9 | 20.7 | 5.3 | 19.1 | 12.5 | 9.4 | 0 |
| Nalidixic acid | 0 | 0 | 0 | 0 | 0 | 0 | 4.5 | 0 | 1.6 | 15.2 | 0 | 4.4 | 0 | 5.7 | 4.5 |
| Streptomycin | 23.7 | 35.7 | 33.3 | 51.7 | 31.6 | 48.0 | 31.8 | 58.8 | 39.1 | 44.6 | 21.1 | 32.4 | 21.9 | 32.1 | 27.3 |
| Sulfisoxazole | 31.6 | 50 | 31.6 | 48.3 | 29.8 | 44.0 | 34.1 | 58.8 | 40.6 | 38.0 | 21.1 | 30.9 | 6.3 | 30.2 | 31.8 |
| Tetracycline | 36.8 | 57.1 | 31.6 | 44.8 | 33.3 | 48.0 | 31.8 | 52.9 | 45.3 | 45.7 | 26.3 | 38.2 | 25.0 | 30.2 | 22.7 |
| Trimethoprim/sulfamethoxazole | 0 | 0 | 5.3 | 3.4 | 10.5 | 4.0 | 2.3 | 11.8 | 1.6 | 16.3 | 5.3 | 7.4 | 3.1 | 0 | 0 |
The data were obtained from the animal arm of NARMS report (http://www.ars.usda.gov/Main/docs.htm?docid=18034). All isolates were obtained from the National Veterinary Services Laboratories.
The resistant breakpoints were adopted from early CLSI guidelines as descripted in the NARMS 1997 to 2004 reports. All isolates were susceptible to amikacin, ceftriaxone, and ciprofloxacin (resistant breakpoints of 64 ug/ml and 4 ug/ml were used for ceftriaxone and ciprofloxacin, respectively).
No cat isolates were tested in 2001.
Other countries have also reported antimicrobial resistance in Salmonella from companion animals. A 2005 to 2006 Canadian survey of pet dog feces showed that 23.2% of the dogs carried Salmonella, and 20% of the Salmonella isolates were resistant to at least one antimicrobial, while 14% were resistant to multiple antimicrobials. The most common pattern, found in 13.3% of isolates, was resistance to amoxicillin/clavulanic acid, ampicillin, cefoxitin, ceftiofur, and ceftriaxone (103). Among the resistant Salmonella isolates, 79.2% were S. Heidelberg, 12.5% were S. Kentucky, and 8.3% were S. Indiana (103). A Belgian study by Van Immerseel et al. (104) showed that the prevalence of Salmonella in different cat groups (healthy house cats, group-housed cats, and sick cats) ranged from 0.36% to 51.4%. In this study, S. Typhimurium, S. Enteritidis, S. Bovismorbificans, and S. 4:i:– were identified. Most S. Typhimurium isolates from group-housed cats were resistant to ampicillin, chloramphenicol, and tetracycline; S. 4:i:– from diseased cats was resistant to ampicillin, chloramphenicol, sulfonamides, tetracycline, and trimethoprim/sulfamethoxazole. The resistance genes blaTEM, cat, sul2, tet(A), and dfrA were identified in this S. 4:i:– isolate. Another study in the Netherlands found that 1% of diarrheic dogs were positive for Salmonella and that 53% of the Salmonella isolates were resistant to cephalexin, 37% to tetracycline, 14% to amoxicillin/clavulanic acid, 6% to trimethoprim/sulfonamides, and 4% to enrofloxacin (98).
S. Typhimurium DT104 with the ACSSuT resistance profile was first isolated in the United Kingdom from sea gulls in the mid-1980s, but not from humans until 1989. During the past 30 years, the strain has become prevalent in humans, food animals, and companion animals in many countries (98, 105). S. Typhimurium DT104 in humans significantly increased in England and Wales from 1991 to 1995, from 259 in 1990 to 2,873 in 1994 and 3,837 in 1995. Most of the human S. Typhimurium DT104 isolates had R-type ACSSuT. Further epidemiologic studies indicated that cats played an important role in the epidemic spread of this organism through many populations (106). A similar report from Scotland during the same time period showed that S. Typhimurium DT104 was isolated in 1% to 2% of cat fecal samples, and the cats shed the organism for at least 7 to 14 weeks (107). MDR Typhimurium var. Copenhagen DT104 also was isolated from horses, dogs, and cats in Germany (108). The dog and cat isolates had R-type ACSSuT. Two horse isolates showed additional resistance to florfenicol, gentamicin, kanamycin, and trimethoprim. In the United States, several human outbreaks of MDR S. Typhimurium DT104 with R-type ACSSuT or ACKSSuT were associated with small-animal veterinary clinics and animal shelters in Idaho, Minnesota, and Washington in 1999 to 2000. During these outbreaks, cats were confirmed as a source of infections (109, 110). A number of MDR S. Typhimurium outbreaks associated with pet rodents were also reported in the United States from 2003 to 2004 (111). The S. Typhimurium isolated from patients and pet hamsters showed the same R-type ACSSuT resistance profile and an indistinguishable pulsed field gel electrophoresis profile.
Multidrug-resistant S. Typhimurium with high-level fluoroquinolone resistance has been isolated from dogs and cats in Japan (112). These isolates had MICs of >256 μg/ml for nalidixic acid, 24 to 32 μg/ml for ciprofloxacin, and 24 to 32 μg/ml for norfloxacin. Two of the isolates had R-type ACSSuT in addition to resistance to nalidixic acid and ciprofloxacin (ACSSuTNCp). Another isolate from a dog showed additional resistance to trimethoprim/sulfamethoxazole, trimethoprim, and gentamicin (112).
Contaminated dog and cat foods are increasingly recognized as a risk factor for Salmonella infections in pets. Several surveys conducted in the United States and Canada showed that the prevalence of Salmonella in pet foods and treats ranged from 21% to 50% (2). A survey was conducted by the U.S. FDA to investigate the prevalence and antimicrobial resistance of Salmonella in dog treats sold in the U.S. market (113). Investigators found that 41% of animal-derived pet treats were contaminated with Salmonella and 26% of the isolates were resistant to tetracycline, 23% to streptomycin, 19% to sulfamethoxazole, 8% to chloramphenicol, and 8% to ampicillin. More than one third (36%) of the Salmonella isolates were resistant to at least one antimicrobial, and 13% of isolates displayed resistance to four or more antimicrobials. Two isolates were identified as S. Typhimurium DT104, with the characteristic R-type ACSSuT. One S. Typhimurium isolate was resistant to kanamycin in addition to the above five antimicrobials. One S. Brandenburg isolate was resistant to eight antimicrobials, including ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, tetracycline, gentamicin, apramycin, and cephalothin. Pet foods are also a risk factor for human infections. A multistate outbreak of tetracycline-resistant Salmonella serovar I 4,[5],12:i– was attributed to frozen feeder rodents. This outbreak caused over 500 clinical illnesses between 2008 and 2010 (114).
Salmonella has also been isolated from other types of pets, such as lizards, snakes, turtles, birds, and fish. Unfortunately, most reports do not contain antimicrobial resistance information for these isolates (115–117). Reptiles are considered a natural reservoir of Salmonella and constitute a significant source of human salmonellosis (2, 89, 90). The U.S. NARMS animal testing component reports from 1997 to 2004 feature antimicrobial susceptibility data on Salmonella isolated from exotic animals, including lizard, snakes, iguanas, and other reptiles and turtles. Those isolates showed low levels of resistance to most antimicrobials tested: a little over 10% resistance to ampicillin, streptomycin, sulfamethoxazole, and tetracycline but <10% resistance to amoxicillin/clavulanic acid, cefoxitin, ceftiofur, ceftriaxone, cephalothin, chloramphenicol, gentamicin, kanamycin, nalidixic acid, and trimethoprim/sulfamethoxazole. They were all susceptible to amikacin and ciprofloxacin (101).
These data indicate that companion animals are important reservoirs of Salmonella and that many Salmonella strains isolated from companion animals have developed resistance. Furthermore, some Salmonella isolates showed high resistance to medically important antimicrobials. To protect public health, there is a need to establish an antimicrobial resistance surveillance program for companion animals. There are ongoing discussions on how existing U.S. surveillance programs can fill this void.
ANTIMICROBIAL RESISTANCE GENES
To fully interpret the antimicrobial resistance data, it is important to characterize the underlying genetic mechanisms in bacteria from humans, animals, and food (16). Acquired resistance in Salmonella results from mutations in chromosomal genes (both structural and regulatory sequences) and by acquisition of preformed, exogenous genes transmitted on mobile elements such as plasmids, integrons, and transposons. While both mech anisms can lead to rapid changes in bacterial populations, horizontal gene transfer is more consequential in the evolution of resistance in Salmonella, where a single plasmid conjugation event can confer resistance to seven or more agents (118). Following conjugative transfer, mobile DNA elements can be maintained as extrachromosomal plasmids or be incorporated wholly or partially into the chromosome as genomic islands. Detailed information on the genes and their context provides insights into the evolution of resistance and its sources.
THE IMPACT OF ROUTINE WHOLE-GENOME SEQUENCING
The DNA sequence information on the number and types of different resistance genes in Salmonella is growing at a very rapid rate, perhaps faster than for any other pathogen at this time. This is due to a push in the public health arena for early adoption of whole-genome sequencing (WGS) as a tool for food safety monitoring and outbreak investigations. Specialists in food safety have spearheaded initiatives to set data quality standards, provide proficiency testing, explore analytical approaches, and foster data sharing arrangements to deploy WGS globally to combat infectious diseases (119). As of this writing, the National Center for Biotechnology Information (NCBI) pathogen detection web portal (https://www.ncbi.nlm.nih.gov/pathogens/) lists Salmonella first among genera with completed genomic sequence (>100,000 genomes), followed by E. coli/Shigella (>37,000), Listeria (>16,000), and Campylobacter (>14,000), with the majority of each belonging to environmental (including food) isolates and with several hundred new genomes being uploaded weekly. Currently, most of the Salmonella genomes are generated by the U.S. FDA’s GenomeTrakr program, which submitted an average of 2,400 Salmonella genomic sequences per month in 2015 (120). The unprecedented stream of WGS data on current strains of Salmonella will soon expand greatly when the CDC PulseNet program and other food safety monitoring systems worldwide shift away from pulsed field gel electrophoresis to WGS for routine surveillance and outbreak investigations. Based on past PusleNet testing volumes in the United States alone, this will add approximately 45,000 more Salmonella genomic sequences annually to public databases.
Beginning in test year 2014, the U.S. NARMS program began including WGS analysis of Salmonella in annual reports and uploading the raw WGS data to the NCBI. In addition, the WGS has been determined for all the historical Salmonella (currently over 6,500 isolates) recovered from retail meat sources since testing began in 2002. These genomes also are available in the public domain at NCBI and have accompanying susceptibility phenotypes (Bioproject number PRJNA290865). As WGS data accumulate in NARMS and other surveillance programs, methods for analyzing and reporting changes in the resistome will augment traditional susceptibility information (see Chapter 28). Studies show a very high correlation between the presence of known resistance genes and clinical resistance in Salmonella (121, 122) and other foodborne bacteria (123, 124). Thus, it is a simple process to predict resistance in Salmonella with a high degree of accuracy for most major drug classes based on WGS data alone. The NARMS web page (35) provides simple and powerful tools to explore the Salmonella resistome in U.S. national surveillance. Resistome Tracker (125) is one publicly available tool that provides visually informative displays of antibiotic resistance genes in Salmonella. Similar tools to quickly identify the resistance (and other) genes in sequence reads have been incorporated into automated analytical processes at NCBI (https://www.ncbi.nlm.nih.gov/pathogens/) or can be applied locally using software applications and resistance gene databases freely available on the web (e.g., ResFinder, CARD). The development of simple bioinformatics tools will enable a comprehensive, near-real-time monitoring of the resistome in Salmonella from foods, the environment, animals, and human infections that will be accessible by all interested parties.
After many years of surveillance and research, and especially with the large amounts of WGS data now being generated through surveillance programs, much is known about the specific alleles underlying resistance in different Salmonella serovars. Michael and Schwarz (126–128) have published three comprehensive reviews on this topic since 2006. Table 3 shows the WGS-based resistome of the U.S. Salmonella isolates that are deposited at NCBI. While the canon of resistance genes is changing very rapidly, the current status in Salmonella is summarized below.
TABLE 3.
Acquired antimicrobial resistance genes in nontyphoidal Salmonellaa
| Antimicrobial class | Resistance genes |
|---|---|
| Rifampicin | arr-2, arr-3 |
| Fluoroquinolone | qnrA1, qnrA2, qnrB1, qnrB11, qnrB12, qnrB17, qnrB19, qnrB2, qnrB25, qnrB26, qnrB3, qnrB32, qnrB34, qnrB35, qnrB37, qnrB38, qnrB4, qnrB40, qnrB4, qnrB47, qnrB48, qnrB51, qnrB6, qnrB69, qnrB7, qnrB9, qnrD, qnrS1, qnrS2, qnrS3, qnrS4, qnrVC4, aac(6′)Ib-cr, norA, oqxA, oqxB, qepA |
| Aminoglycoside | aac(2′)-Ia, aac(2′)-Ib, aac(2′)-Ic, aac(3)-IIa, aac(3)-IId, aac(3)-IIe, aac(3)-IVa, aac(3)-Ia, aac(3)-Id, aac(3)-Ik, aac(3)-VIa, aac(6′)-33, aac(6′)-IIa, aac(6′)-IIc, aac(6′)-Ib, aac(6′)-Ic, aac(6′)-If, aac(6′)-Ii, aac(6′)-Im, aac(6′)-Iz, aac(6′)-aph(2″), aacA4, aadA1, aadA10, aadA11, aadA12, aadA13, aadA14, aadA15, aadA16, aadA17, aadA2, aadA22, aadA23, aadA24, aadA3, aadA4, aadA5, aadA6, aadA7, aadA8, aadA8b, aadB, aadD, ant(6)-Ia, aph(2″)-Ib, aph(3′)-III, aph(3′)-IIa, aph(3′)-IIb, aph(3′)-IIc, aph(3′)-Ia, aph(3′)-Ic, aph(3′)-Id, aph(3′)-VIa, aph(4)-Ia, aph(6)-Ic, armA, rmtB, rmtE, spc, sph, strA, strB |
| Beta-lactam (bla genes) | ACT-4, ACT-5, ACT-6, ACT-7, CARB-1, CARB-2, CARB-3, CARB-5, CARB-6, CEPH-A, CKO-1, CMG, CMY-15, CMY-16, CMY-17, CMY-18, CMY-2, CMY- 20, CMY-22, CMY-23, CMY-24, CMY-3, CMY-30, CMY-32, CMY-33, CMY-36, CMY-4, CMY-41, CMY-42, CMY-44, CMY-46, CMY-47, CMY-48, CMY-5, CMY-53, CMY-54, CMY-56, CMY-58, CMY-59, CMY-6, CMY-61, CMY-64, CMY-68 ,CMY-70, CMY-83, CMY-87, CMY-98, CTX-M-1, CTX-M-11, CTX-M-14, CTX-M-14b, CTX-M-15, CTX-M-2, CTX-M-24, CTX-M-27, CTX-M-3, CTX-M-5, CTX-M-55, CTX-M-65, CTX-M-8, CTX-M-9, DHA-1, DHA-2, DHA- 3, DHA-5, HERA-3, HERA-5, HERA-6, KPC-2, LEN1, LEN11, LEN9, MAL-1, MIR-3, MIR-5, MOR-1, MOR-2, NDM-1, OKP-B-12, OXA-1, OXA-10, OXA- 114, OXA-129, OXA-134, OXA-17, OXA-2, OXA-21, OXA-23, OXA-27, OXA-278, OXA-335, OXA-34, OXA-36, OXA-4, OXA-48, OXA-50, OXA-58, OXA-61, OXA-66, OXA-9, OXA-90, OXY-1-4, OXY-1-5, OXY-2, OXY-2-7, OXY-2-8, OXY-5-1, OXY-6, OXY-6-1, OXY-6-2, PAO, PER-2, SED1, SHV-1, SHV-100, SHV-105, SHV-11, SHV-12, SHV-122, SHV-129, SHV-2, SHV-25, SHV-28, SHV-39, SHV-45, SHV-99, TEM-1, TEM-10, TEM-104, TEM-105, TEM-106, TEM-116, TEM-12 ,TEM-123, TEM-124, TEM-126, TEM-127, TEM-135, TEM- 141 ,TEM-143, TEM-144, TEM-148, TEM-154, TEM-155, TEM-156, TEM-157, TEM-159, TEM-162, TEM-166, TEM-169, TEM-171, TEM-176, TEM-183, TEM-199, TEM-1A, TEM-1B, TEM-1C, TEM-1D, TEM-2 ,TEM-205, TEM-213, TEM-22, TEM-30, TEM-33, TEM-42, TEM-52, TEM-52B, TEM-57, TEM-59, TEM-63, TEM-67, TEM-7, TEM-70, TEM-76, TEM-79, TEM-90, TEM-95, VEB- 5, VIM-1, Z, ZEG-1 |
| Beta-lactam (other) | cepA-29, cfxA, cfxA3, cphA1, mecA, hugA |
| Phenicol | cat(pC221), catA1, catA2, catA3, catB2, catB3, catB7, catQ, cml, cmlA1, fexA, floR |
| Trimethoprim | dfrA10, dfrA12, dfrA14, dfrA15, dfrA16, dfrA17, dfrA18, dfrA21, dfrA23, dfrA24, dfrA25, dfrA27, dfrA29, dfrA31, dfrA32, dfrA5, dfrA7, dfrA8, dfrB1, dfrB3, dfrB5, dfrG |
| Macrolides/lincosamides | ere(A), erm(42), erm(A), erm(B), erm(C), erm(D), erm(F), erm(G), lnu(A), lnu(C), lnu(F), lsa(A), mef(A), mef(B), mph(A), mph(B), mph(C), mph(E), msr(A), msr(C), msr(D), msr(E), vga(A) |
| Polymyxin | mcr-1 |
| Sulfonamide | sul1, sul2, sul3 |
| Tetracycline | tet(32), tet(38), tet(39), tet(41), tet(A), tet(B), tet(C), tet(D), tet(E), tet(G), tet(H), tet(J), tet(K), tet(L), tet(M), tet(O), tet(P), tet(Q), tet(W), tet(X) |
| Fosfomycin | fosA |
| Fusidic acid | fusA, fusB3 |
NCBI last accessed 4 March 2017. Total number of genomes, N = 81,936: U.S., n = 38,725; non-U.S., n = 27,095; unknown origin, n = 16,116.
Fluoroquinolone Resistance
Fluoroquinolone resistance has been well characterized in Salmonella and other bacterial pathogens. It has long been known that a combination of mutations in specific regions of the topoisomerase genes encoded by gyrA, gyrB, parC, and parE confers resistance to fluoroquinolones in Salmonella and other enterics. More recently, PMQR determinants have been identified including multiple alleles of qnrA, qnrB, qnrD, and qnrS, which function by protecting the topoisomerase targets from inhibition resulting in decreased fluoroquinolone susceptibility (129). A total of 32 qnr genes have been identified in Salmonella (Table 3). The qnrB19 allele has emerged to become the predominant one in Salmonella in the United States, where it is present in 0.5% of resistant human strains. The quinolone efflux pump encoded by qepA (130, 131), a bifuntional enzyme encoded by aac(60)-Ib-cr (132), and the oqxA gene (133) are known also to affect quinolone MICs in Salmonella isolates. The PMQR gene, oqxAB, also mediates resistance to nalidixic acid and chloramphenicol, as well as olaquindox. It has been found in multiple serovars in China (134) and in S. Typhimurium strains in Europe (135). It appears to augment the development of fluoroquinolone resistance in Salmonella (136) and can be selected by florfenicol exposure (137).
Beta-lactamase Resistance
Resistance to the β-lactam class of compounds is conferred by at least 1,177 beta-lactamase genes. Michael and Schwarz report that 13 beta-lactamase families have been identified in Salmonella, including ACC, CMY, CTX-M, DHA, PER, PSE, SCO, SHV, and TEM, as well as 4 carbapenemases of types KPC, NDM, OXA, and VIM (126). To this list can be added FOX (in a U.S. clinical strain of serovar Newport), multiple HERA alleles (prevalent in U.S. turkey isolates), and LAP (in a U.S. clinical strain of S. Saintpaul) (121), as well as HUGA and CEP (in United Kingdom clinical isolates), VEB (linked to armA in a U.S. clinical isolate), and ZEG (in a U.S. riverine isolate) (NCBI Bioproject number PRJNA290865). In the United States, the blaCMY gene is present in most strains exhibiting resistance to extended-spectrum cephalosporins. Carbapenems are not used in U.S. agriculture, nor are they approved for food-producing animals in any country. No carbapenemases producing salmonellae have been found in NARMS testing, and two cases have been associated with travel (66, 69).
Macrolide Resistance
Among the critically important macrolide class of drugs, only azithromycin has been explored as a treatment for salmonellosis, mainly for S. Typhi. Resistance to azithromycin (MIC, ≥16 μg/ml) in NTS has been associated with the presence of mphA (121, 138) and mphE (unpublished data), although other macrolide resistance genes have been found in Salmonella, including ere(A), lnu(F), mef(B), and msr(E) (121, 126, 138). The full complement of known macrolide-lincosamide resistance genes in Salmonella is shown in Table 3.
Trimethoprim-Sulfonamide Resistance
Trimethoprim-sulfonamide has long been a recommended treatment for salmonellosis. At least 42 trimethoprim resistance genes have been identified in bacteria, 19 of which have been detected in Salmonella. These include dfrA1, dfrA3, dfrA5, dfrA7, dfrA8, dfrA10, dfrA12, dfrA13, dfrA14, dfrA15, dfrA16, dfrA17, dfrA18, dfrA19, dfrA21, dfrA23, dfrA24, dfrA25, dfrA27, dfrA29, dfrA31, and dfrA32, along with dfrB1, dfrB3, dfrB5, dfrB6, and dfrG. At least eight of these have been found in U.S. isolates (dfrA1, dfrA5, dfrA7, dfrA8, dfrA12, dfrA14, dfrA15, dfrA17, and dfrA18). Sulfonamide resistance mediated by sul1, sul2, and sul3 are common in Salmonella.
Tetracycline Resistance
Among the 46 tetracycline resistance genes identified in bacteria to date (139), 20 have been reported in Salmonella. These include energy efflux pumps encoded by tet(A), tet(B), tet(C), tet(D), tet(E), tet(G), tet(H), tet(J), tet(K), and tet(L). The tet(A) and tet(B) genes are consistently the most common ones identified. Ribosomal protection mechanisms conferred by tet(M) and tet(O), as well as rare instances of tet(X), which encodes a tetracycline-degrading enzyme, have been found in U.S. isolates of Salmonella (121), usually along with other tet genes.
Aminoglycoside Resistance
Aminoglycoside resistance is coded by a wide range of alleles, which is complicated by the use of two different naming systems in the literature. More than 146 alleles have been found in bacteria. Resistance occurs by ribosomal modification, efflux, and enzymatic inactivation. Among these mechanisms, those based on enzymatic modification of the drug are most common in the clinical setting. These enzymes function through three general reactions resulting in phosphorylation, adenylation, or acelylation. The streptomycin phosphotransferases encoded by strA and strB (also designated aph) are the most common overall, present in about 27% of resistant U.S. strains (Table 3) (121). A total of 23 3′O-adenyltransferase (aad) genes have been identified in Salmonella, including aadA1, aadA2, aadA3, aadA4, aadA5, aadA6, aadA7, aadA8, aadA8b, aadA10, aadA11, aadA12, aadA13, aadA14, aadA15, aadA16, aadA17, aadA21, aadA22, aadA23, aadA24, aadA26, and aadA27 (126). At least 10 of these have been found in U.S. isolates. Various forms of the aac gene family conferring gentamicin resistance are present in Salmonella. These include aac(2′)-Ia, aac(2′)-Ib, aac(2′)-Ic, aac(3)-IIa, aac(3)-IId, aac(3)-IIe, aac(3)-Iva, aac(3)-Ia, aac(3)-Id, aac(3)-Ik, aac(3)-Via, aac(6′)-33, aac(6′)-IIa, aac(6′)-IIc, aac(6′)-Ib, aac(6′)-Ic, aac(6′)-If, aac(6′)-Ii, aac(6′)-Im, aac(6′)-Iz, aac(6′)-aph(2″) (bifunctional enzyme conferring coresistance to low levels of fluoroquinolones), and aacA4.
At least six RNA methylase enzymes (ArmA, RmtA, RmtB, RmtC, RmtD, and NpmA) are known, which reside on mobile elements, often with other resistance genes, and confer very high levels of resistance to various aminoglycosides (140). Among the Enterobacteriaceae, ArmA is the most frequently identified, and both armA and rmtC have been found in Salmonella (141, 142). An interesting feature of aminoglycoside resistance revealed by WGS is the propensity for Salmonella to harbor numerous aminoglycoside resistance genes (and often other resistance gene classes) within a single bacterium (121). In fact, a small minority of aminoglycoside-resistant strains sequenced to date in the United States was found to carry just one resistance gene, and many carry five or more alleles. For example, one U.S. isolate of S. I 4,5,12:i:– has the aminoglycoside genotype aph(3′)-Ic, aac(6′)-IIc, rmtE, aph(3′)-Ia, strA, strB, aph(3′)-Ic along with unrelated resistance determinants coded by blaSHV-12, blaTEM-1B, ere(A), anrB19, sul1, sul2, tet(B), tet(A), and tet(M). The advantage of carrying so many resistance determinants all conferring resistance to the same drug class is not obvious but may partly reflect the fact that aminoglycosides have been used longer than most drug classes in food animal production and that individual strains have adapted to accommodate the array of genes enriched by decades of selective pressures.
Phenicol Resistance
Chloramphenicol was the first broad-spectrum antibiotic to be used both orally and systemically, and it has been used around the world to treat Salmonella infections. Resistance in Salmonella is associated with the presence of chloramphenicol acetyltransferase genes encoded by a small number of variants of catA (catA1, catA2, catA3) and catB (catB2, catB3, catB7, catB8), as well as efflux by cmlA (cmlA1, cmlA4, cmlA9) and floR. Acetylation blocks binding of the drug to the ribosome target. The fluorine atom of florfenicol prevents acetylation, circumventing modification by the cat genes. Florfenicol efflux resistance also results from expression of floR and fexA.
Colistin Resistance
Colistin (polymyxin E) is now considered a reserve drug for treating MDR Gram-negative bacteria, including carbapenem-resistant strains. The first report of mobile colistin resistance gene mcr-1 was reported in late 2015 in E. coli in China (58). Soon thereafter, several instances of mcr-1 in Salmonella were reported (143–148). In the NCBI database, there are now at least five alleles of mcr designated mcr-1 through mcr-5.
CONCLUDING REMARKS
The global challenge of antimicrobial resistance is being addressed on many fronts. The nature and magnitude of resistance in the agriculture/food sector is perhaps best evaluated by examining resistance in the principal foodborne bacterial pathogen Salmonella. The situations in North America and Europe, where surveillance data are extensive, provide a good backdrop to a global understanding of resistance in this important pathogen, including resistance data to measure the impact of drug use restrictions designed to limit resistance.
Overall, antimicrobial resistance surveillance data for Salmonella show common and unique features in different regions, where resistance to older drugs is generally highest and serovar differences impact overall trends. Differences in resistance by serovar often illustrate the most salient trends, as demonstrated by fluoroquinolone-resistant S. Kentucky in Europe or MDR S. Dublin in the United States.
The contribution of zoonotic infections from companion animals is not well understood or adequately addressed by surveillance programs. However, the relative ease with which the resistome of Salmonella can be evaluated by examination of WGS data heralds a new era in antimicrobial resistance monitoring where the resistance allele will become more prominent as the hazard under surveillance. As more is learned about the array of resistant strain types infecting humans, the range of sources causing human infections will become clearer, including the proportion of those arising from companion animals and other sources. Along with more detailed drug use information, this will lead to a better understanding of the sources and drivers of resistance and better interventions to protect public health.
ACKNOWLEDGEMENTS
We acknowledge our FDA colleagues Claudine Kabera and Chih-Hao Hsu for help with the tables and graphs, and John Graham and Adrienne Ivory for review of the manuscript.
REFERENCES
- 1.WHO. 2013. Integrated Surveillance of Antimicrobial Resistance: Guidance from a WHO Advisory Group. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Hoelzer K, Moreno Switt AI, Wiedmann M. 2011. Animal contact as a source of human non-typhoidal salmonellosis. Vet Res (Faisalabad) 42:34 10.1186/1297-9716-42-34. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Interagency Food Safety Analytics Collaboration (IFSAC). 2015. Foodborne illness source attribution estimates for Salmonella, Escherichia coli O157 (E. coli O157), Listeria monocytogenes (Lm), and Campylobacter using outbreak surveillance data. U.S. Department of Health and Human Services, CDC, FDA, USDA-FSIS.
- 4.CDC. 2014. National Enteric Disease Surveillance: Salmonella Annual Report, 2014. CDC, Atlanta, GA. [Google Scholar]
- 5.Rabsch W, Tschäpe H, Bäumler AJ. 2001. Non-typhoidal salmonellosis: emerging problems. Microbes Infect 3:237–247 10.1016/S1286-4579(01)01375-2. [DOI] [PubMed] [Google Scholar]
- 6.Patterson SK, Kim HB, Borewicz K, Isaacson RE. 2016. Towards an understanding of Salmonella enterica serovar Typhimurium persistence in swine. Anim Health Res Rev 17:159–168 10.1017/S1466252316000165. [PubMed] [DOI] [PubMed] [Google Scholar]
- 7.Cobbold RN, Rice DH, Davis MA, Besser TE, Hancock DD. 2006. Long-term persistence of multi-drug-resistant Salmonella enterica serovar Newport in two dairy herds. J Am Vet Med Assoc 228:585–591 10.2460/javma.228.4.585. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease ‘Burden of Illness’ Studies. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889 10.1086/650733. [PubMed] [DOI] [PubMed] [Google Scholar]
- 9.CDC. 2013. Antibiotic resistance threats in the United States, 2013. http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf#page=112.
- 10.McDermott PF, Zhao S, Wagner DD, Simjee S, Walker RD, White DG. 2002. The food safety perspective of antibiotic resistance. Anim Biotechnol 13:71–84 10.1081/ABIO-120005771. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.FDA. 2012. Guidance for Industry No. 209: The judicious use of medically important antimicrobials drugs in food-producing animals. HHS, Food and Drug Administration, Rockville, MD. https://www.fda.gov/downloads/AnimalVeterinary/GuidanceComplianceEnforcement/GuidanceforIndustry/UCM216936.pdf.
- 12.Ministry of Agriculture FaF, National Veterinary Assay Laboratory. 2016. JVARM: Report on the Japanese Veterinary Antimicrobial Resistance Monitoring System, 2012–2013. Ministry of Agriculture, Forestry, and Fisheries, Tokyo, Japan. [Google Scholar]
- 13.EMA. 2016. Sales of veterinary antimicrobial agents in 29 European countries in 2014. Sixth ESVAC Report. European Medicines Agency, Agency EM, London, United Kingdom.
- 14.Bondt N, Jensen VF, Puister-Jansen LF, van Geijlswijk IM. 2013. Comparing antimicrobial exposure based on sales data. Prev Vet Med 108:10–20 10.1016/j.prevetmed.2012.07.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 15.Federal Register. 2016. Antimicrobial animal drug sales and distribution reporting. Docket no. FDA-2012-N-0447.
- 16.WHO. 2014. Antimicrobial Resistance Global Report on Surveillance. WHO, Geneva, Switzerland. [PubMed] [Google Scholar]
- 17.Havelaar AH, Kirk MD, Torgerson PR, Gibb HJ, Hald T, Lake RJ, Praet N, Bellinger DC, de Silva NR, Gargouri N, Speybroeck N, Cawthorne A, Mathers C, Stein C, Angulo FJ, Devleesschauwer B, World Health Organization Foodborne Disease Burden Epidemiology Reference Group. 2015. World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med 12:e1001923 10.1371/journal.pmed.1001923. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.CDC. 2014. Foodborne Diseases Active Surveillance Network (FoodNet): FoodNet Surveillance Report for 2014 (Final Report). U.S. Department of Health and Human Services, CDC, Atlanta, GA. [Google Scholar]
- 19.Reddy EA, Shaw AV, Crump JA. 2010. Community-acquired bloodstream infections in Africa: a systematic review and meta-analysis. Lancet Infect Dis 10:417–432 10.1016/S1473-3099(10)70072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ao TT, Feasey NA, Gordon MA, Keddy KH, Angulo FJ, Crump JA. 2015. Global burden of invasive nontyphoidal Salmonella disease, 2010(1). Emerg Infect Dis 21:21 10.3201/eid2106.140999. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feasey NA, Hadfield J, Keddy KH, Dallman TJ, Jacobs J, Deng X, Wigley P, Barquist L, Langridge GC, Feltwell T, Harris SR, Mather AE, Fookes M, Aslett M, Msefula C, Kariuki S, Maclennan CA, Onsare RS, Weill FX, Le Hello S, Smith AM, McClelland M, Desai P, Parry CM, Cheesbrough J, French N, Campos J, Chabalgoity JA, Betancor L, Hopkins KL, Nair S, Humphrey TJ, Lunguya O, Cogan TA, Tapia MD, Sow SO, Tennant SM, Bornstein K, Levine MM, Lacharme-Lora L, Everett DB, Kingsley RA, Parkhill J, Heyderman RS, Dougan G, Gordon MA, Thomson NR. 2016. Distinct Salmonella Enteritidis lineages associated with enterocolitis in high-income settings and invasive disease in low-income settings. Nat Genet 48:1211–1217 10.1038/ng.3644. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingsley RA, Msefula CL, Thomson NR, Kariuki S, Holt KE, Gordon MA, Harris D, Clarke L, Whitehead S, Sangal V, Marsh K, Achtman M, Molyneux ME, Cormican M, Parkhill J, MacLennan CA, Heyderman RS, Dougan G. 2009. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res 19:2279–2287 10.1101/gr.091017.109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Hello S, Bekhit A, Granier SA, Barua H, Beutlich J, Zając M, Münch S, Sintchenko V, Bouchrif B, Fashae K, Pinsard JL, Sontag L, Fabre L, Garnier M, Guibert V, Howard P, Hendriksen RS, Christensen JP, Biswas PK, Cloeckaert A, Rabsch W, Wasyl D, Doublet B, Weill FX. 2013. The global establishment of a highly-fluoroquinolone resistant Salmonella enterica serotype Kentucky ST198 strain. Front Microbiol 4:395 10.3389/fmicb.2013.00395. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kariuki S, Gordon MA, Feasey N, Parry CM. 2015. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine 33(Suppl 3):C21–C29 10.1016/j.vaccine.2015.03.102. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marder EPC, Cieslak PR, Cronquist AB, Dunn J, Lathrop S, Rabatsky-Ehr T, Ryan P, Smith K, Tobin-D’Angelo M, Vugia DJ, Zansky S, Holt KG, Wolpert BJ, Lynch M, Tauxe R, Geissler AL. 2017. Incidence and trends of infections with pathogens transmitted commonly through food and the effect of increasing use of culture-independent diagnostic tests on surveillance: Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2013–2016. MMWR Morb Mortal Wkly Rep 66:397–403 10.15585/mmwr.mm6615a1. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.EFSA-ECDC. 2016. EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control), 2016. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals, and food in 2014. EFSA J 14:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.FDA. 2016. National Antimicrobial Resistance Monitoring System - Enteric Bacteria (NARMS): NARMS Integrated Report 2014. U.S. Department of Health and Human Services, Food & Drug Administration, Rockville, MD. http://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/default.htm.
- 28.Anderson AD, Nelson JM, Rossiter S, Angulo FJ. 2003. Public health consequences of use of antimicrobial agents in food animals in the United States. Microb Drug Resist 9:373–379 10.1089/107662903322762815. [PubMed] [DOI] [PubMed] [Google Scholar]
- 29.CIPARS. 2016. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS), Annual Report, 2014.
- 30.DANMAP. 2015. DANMAP 2015: use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, foods and humans in Denmark. http://www.danmap.org.
- 31.FDA. 2017. The 2015 NARMS Integrated Report. U.S. Department of Health and Human Services, FDA, Rockville, MD. [Google Scholar]
- 32.Miriagou V, Carattoli A, Fanning S. 2006. Antimicrobial resistance islands: resistance gene clusters in Salmonella chromosome and plasmids. Microbes Infect 8:1923–1930 10.1016/j.micinf.2005.12.027. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.WHO. 2017. WHO Guidelines on Use of Medically Important Antimicrobials in Food-Producing Animals. World Health Organization, Geneva, Switzerland. [PubMed] [PubMed] [Google Scholar]
- 34.Karp BE, Tate H, Plumblee JR, Dessai U, Whichard JM, Thacker EL, Hale KR, Wilson W, Friedman CR, Griffin PM, McDermott PF. 2017. National Antimicrobial Resistance Monitoring System: two decades of advancing public health through integrated surveillance of antimicrobial resistance. Foodborne Pathog Dis 14:545–557 10.1089/fpd.2017.2283. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.FDA. 2016. NARMS now: integrated data. http://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm416741.htm. Accessed 24 November 2017.
- 36.Tadesse DA, Singh A, Zhao S, Bartholomew M, Womack N, Ayers S, Fields PI, McDermott PF. 2016. Antimicrobial resistance in Salmonella in the United States from 1948 to 1995. Antimicrob Agents Chemother 60:2567–2571 10.1128/AAC.02536-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.EUCAST. European Committee on Antimicrobial Susceptibility Testing. http://www.eucast.org. Accessed 25 January 2017.
- 38.Bjork KE, Kopral CA, Wagner BA, Dargatz DA. 2015. Comparison of mixed effects models of antimicrobial resistance metrics of livestock and poultry Salmonella isolates from a national monitoring system. Prev Vet Med 122:265–272 10.1016/j.prevetmed.2015.10.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Billy TJ, Wachsmuth IK. 1997. Hazard analysis and critical control point systems in the United States Department of Agriculture regulatory policy. Rev Sci Tech 16:342–348 10.20506/rst.16.2.1029. [PubMed] [DOI] [PubMed] [Google Scholar]
- 40.Hong S, Rovira A, Davies P, Ahlstrom C, Muellner P, Rendahl A, Olsen K, Bender JB, Wells S, Perez A, Alvarez J. 2016. Serotypes and antimicrobial resistance in Salmonella enterica recovered from clinical samples from cattle and swine in Minnesota, 2006 to 2015. PLoS One 11:e0168016 10.1371/journal.pone.0168016. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medalla F, Hoekstra RM, Whichard JM, Barzilay EJ, Chiller TM, Joyce K, Rickert R, Krueger A, Stuart A, Griffin PM. 2013. Increase in resistance to ceftriaxone and nonsusceptibility to ciprofloxacin and decrease in multidrug resistance among Salmonella strains, United States, 1996–2009. Foodborne Pathog Dis 10:302–309 10.1089/fpd.2012.1336. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawakami VM, Bottichio L, Angelo K, Linton N, Kissler B, Basler C, Lloyd J, Inouye W, Gonzales E, Rietberg K, Melius B, Oltean H, Wise M, Sinatra J, Marsland P, Li Z, Meek R, Kay M, Duchin J, Lindquist S. 2016. Notes from the field: outbreak of multidrug-resistant Salmonella infections linked to pork--Washington, 2015. MMWR Morb Mortal Wkly Rep 65:379–381 10.15585/mmwr.mm6514a4. [PubMed] [DOI] [PubMed] [Google Scholar]
- 43.Folster JP, Grass JE, Bicknese A, Taylor J, Friedman CR, Whichard JM. 2016. Characterization of resistance genes and plasmids from outbreaks and illness clusters caused by Salmonella resistant to ceftriaxone in the United States, 2011–2012. Microb Drug Resist. 10.1089/mdr.2016.0080. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Donnell AT, Vieira AR, Huang JY, Whichard J, Cole D, Karp BE. 2014. Quinolone-resistant Salmonella enterica serotype Enteritidis infections associated with international travel. Clin Infect Dis 59:e139–e141 10.1093/cid/ciu505. [PubMed] [DOI] [PubMed] [Google Scholar]
- 45.Smith AB, Renter DG, Cernicchiaro N, Shi X, Nagaraja TG. 2016. Prevalence and quinolone susceptibilities of Salmonella isolated from the feces of preharvest cattle within feedlots that used a fluoroquinolone to treat bovine respiratory disease. Foodborne Pathog Dis 13:303–308 10.1089/fpd.2015.2081. [PubMed] [DOI] [PubMed] [Google Scholar]
- 46.Valenzuela JR, Sethi AK, Aulik NA, Poulsen KP. 2017. Antimicrobial resistance patterns of bovine Salmonella enterica isolates submitted to the Wisconsin Veterinary Diagnostic Laboratory: 2006–2015. J Dairy Sci 100:1319–1330 10.3168/jds.2016-11419. [PubMed] [DOI] [PubMed] [Google Scholar]
- 47.Government of Canada. 2016. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2014 Annual Report Summary. Public Health Agency of Canada, Guelph, ON. [Google Scholar]
- 48.Federal Register. 2008. Cephalosporin Drugs; Extralabel Animal Drug Use; Order of Prohibition. https://www.federalregister.gov/documents/2012/01/06/2012-35/new-animal-drugs-cephalosporin-drugs-extralabel-animal-drug-use-order-of-prohibition
- 49.Khaitsa ML, Kegode RB, Bauer ML, Gibbs PS, Lardy GP, Doetkott DK. 2007. A longitudinal study of Salmonella shedding and antimicrobial resistance patterns in North Dakota feedlot cattle. J Food Prot 70:476–481 10.4315/0362-028X-70.2.476. [PubMed] [DOI] [PubMed] [Google Scholar]
- 50.Sjölund-Karlsson M, Howie RL, Blickenstaff K, Boerlin P, Ball T, Chalmers G, Duval B, Haro J, Rickert R, Zhao S, Fedorka-Cray PJ, Whichard JM. 2013. Occurrence of β-lactamase genes among non-Typhi Salmonella enterica isolated from humans, food animals, and retail meats in the United States and Canada. Microb Drug Resist 19:191–197 10.1089/mdr.2012.0178. [PubMed] [DOI] [PubMed] [Google Scholar]
- 51.Tate H, Folster JP, Hsu CH, Chen J, Hoffmann M, Li C, Morales C, Tyson GH, Mukherjee S, Brown AC, Green A, Wilson W, Dessai U, Abbott J, Joseph L, Haro J, Ayers S, McDermott PF, Zhao S. 2017. Comparative analysis of extended-spectrum-β-lactamase CTX-M-65-producing Salmonella enterica serovar Infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob Agents Chemother 61:61 10.1128/AAC.00488-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Helke KL, McCrackin MA, Galloway AM, Poole AZ, Salgado CD, Marriott BP. 2017. Effects of antimicrobial use in agricultural animals on drug-resistant foodborne salmonellosis in humans: a systematic literature review. Crit Rev Food Sci Nutr 57:472–488 10.1080/10408398.2016.1230088. [PubMed] [DOI] [PubMed] [Google Scholar]
- 53.Franco A, Leekitcharoenphon P, Feltrin F, Alba P, Cordaro G, Iurescia M, Tolli R, D’Incau M, Staffolani M, Di Giannatale E, Hendriksen RS, Battisti A. 2015. Emergence of a clonal lineage of multidrug-resistant ESBL-producing Salmonella Infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS One 10:e0144802 10.1371/journal.pone.0144802. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rickert-Hartman R, Folster JP. 2014. Ciprofloxacin-resistant Salmonella enterica serotype Kentucky sequence type 198. Emerg Infect Dis 20:910–911 10.3201/eid2005.131575. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacoby GA, Strahilevitz J, Hooper DC. 2014. Plasmid-mediated quinolone resistance. Microbiol Spectr 2:PLAS-0006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cantón R, González-Alba JM, Galán JC. 2012. CTX-M enzymes: origin and diffusion. Front Microbiol 3:110 10.3389/fmicb.2012.00110. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fey PD, Safranek TJ, Rupp ME, Dunne EF, Ribot E, Iwen PC, Bradford PA, Angulo FJ, Hinrichs SH. 2000. Ceftriaxone-resistant Salmonella infection acquired by a child from cattle. N Engl J Med 342:1242–1249 10.1056/NEJM200004273421703. [PubMed] [DOI] [PubMed] [Google Scholar]
- 58.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 59.Liakopoulos A, Mevius DJ, Olsen B, Bonnedahl J. 2016. The colistin resistance mcr-1 gene is going wild. J Antimicrob Chemother 71:2335–2336 10.1093/jac/dkw262. [PubMed] [DOI] [PubMed] [Google Scholar]
- 60.Arcilla MS, van Hattem JM, Matamoros S, Melles DC, Penders J, de Jong MD, Schultsz C, COMBAT Consortium. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:147–149 10.1016/S1473-3099(15)00541-1. [PubMed] [DOI] [PubMed] [Google Scholar]
- 61.Olaitan AO, Chabou S, Okdah L, Morand S, Rolain JM. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:147 10.1016/S1473-3099(15)00540-X. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, Zankari E, Leekitcharoenphon P, Stegger M, Kaas RS, Cavaco LM, Hansen DS, Aarestrup FM, Skov RL. 2015. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill 20:pii=30085 10.2807/1560-7917.ES.2015.20.49.30085. [PubMed] [DOI] [PubMed] [Google Scholar]
- 63.Watkins LF, Folster J, Chen J, Karlsson MS, Boyd S, Leung V, McNutt A, Medus C, Wang X, Hanna S, Smith N, Colón A, Barringer A, Dunbar-Manley C, Balk J, Friedman C. 2017. Emergence of mcr-1 among nontyphoidal Salmonella isolates in the United States. Open Forum Infect Dis 4(suppl_1):S129–S130 10.1093/ofid/ofx163.182. [DOI] [Google Scholar]
- 64.Anjum MF, Duggett NA, AbuOun M, Randall L, Nunez-Garcia J, Ellis RJ, Rogers J, Horton R, Brena C, Williamson S, Martelli F, Davies R, Teale C. 2016. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J Antimicrob Chemother 71:2306–2313 10.1093/jac/dkw149. [PubMed] [DOI] [PubMed] [Google Scholar]
- 65.CDC. 2016. Newly reported gene, mcr -1, threatens last-resort antibiotics. https://www.cdc.gov/drugresistance/mcr1.html.
- 66.Savard P, Gopinath R, Zhu W, Kitchel B, Rasheed JK, Tekle T, Roberts A, Ross T, Razeq J, Landrum BM, Wilson LE, Limbago B, Perl TM, Carroll KC. 2011. First NDM-positive Salmonella sp. strain identified in the United States. Antimicrob Agents Chemother 55:5957–5958 10.1128/AAC.05719-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rasheed JK, Kitchel B, Zhu W, Anderson KF, Clark NC, Ferraro MJ, Savard P, Humphries RM, Kallen AJ, Limbago BM. 2013. New Delhi metallo-β-lactamase-producing Enterobacteriaceae, United States. Emerg Infect Dis 19:870–878 10.3201/eid1906.121515. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Day MR, Meunier D, Doumith M, de Pinna E, Woodford N, Hopkins KL. 2015. Carbapenemase-producing Salmonella enterica isolates in the UK. J Antimicrob Chemother 70:2165–2167. [DOI] [PubMed] [Google Scholar]
- 69.Miriagou V, Tzouvelekis LS, Rossiter S, Tzelepi E, Angulo FJ, Whichard JM. 2003. Imipenem resistance in a Salmonella clinical strain due to plasmid-mediated class A carbapenemase KPC-2. Antimicrob Agents Chemother 47:1297–1300 10.1128/AAC.47.4.1297-1300.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mollenkopf DF, Stull JW, Mathys DA, Bowman AS, Feicht SM, Grooters SV, Daniels JB, Wittum TE. 2016. Carbapenemase-producing Enterobacteriaceae recovered from the environment of a swine farrow-to-finish operation in the United States. Antimicrob Agents Chemother AAC.01298-16 10.1128/AAC.01298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woodford N, Wareham DW, Guerra B, Teale C. 2014. Carbapenemase-producing Enterobacteriaceae and non-Enterobacteriaceae from animals and the environment: an emerging public health risk of our own making? J Antimicrob Chemother 69:287–291 10.1093/jac/dkt392. [PubMed] [DOI] [PubMed] [Google Scholar]
- 72.Donado-Godoy P, Castellanos R, León M, Arevalo A, Clavijo V, Bernal J, León D, Tafur MA, Byrne BA, Smith WA, Perez-Gutierrez E. 2015. The establishment of the Colombian Integrated Program for Antimicrobial Resistance Surveillance (COIPARS): a pilot project on poultry farms, slaughterhouses and retail market. Zoonoses Public Health 62(Suppl 1):58–69 10.1111/zph.12192. [PubMed] [DOI] [PubMed] [Google Scholar]
- 73.Ministry of Agriculture, Forestry and Fisheries, National Veterinary Assay Laboratory. 2013. JVARM: A Report on the Japanese Veterinary Antimicrobial Resistance Monitoring System, 2008–2011. Ministry of Agriculture, Forestry and Fisheries, Tokyo, Japan. [Google Scholar]
- 74.Wu H, Xia X, Cui Y, Hu Y, Xi M, Wang X, Shi X, Wang D, Meng J, Yang B. 2013. Prevalence of extended-spectrum β-lactamase-producing Salmonella on retail chicken in six provinces and two national cities in the People’s Republic of China. J Food Prot 76:2040–2044 10.4315/0362-028X.JFP-13-224. [PubMed] [DOI] [PubMed] [Google Scholar]
- 75.Wang S, Duan H, Zhang W, Li JW. 2007. Analysis of bacterial foodborne disease outbreaks in China between 1994 and 2005. FEMS Immunol Med Microbiol 51:8–13 10.1111/j.1574-695X.2007.00305.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 76.Yan H, Li L, Alam MJ, Shinoda S, Miyoshi S, Shi L. 2010. Prevalence and antimicrobial resistance of Salmonella in retail foods in northern China. Int J Food Microbiol 143:230–234 10.1016/j.ijfoodmicro.2010.07.034. [PubMed] [DOI] [PubMed] [Google Scholar]
- 77.Yang B, Qu D, Zhang X, Shen J, Cui S, Shi Y, Xi M, Sheng M, Zhi S, Meng J. 2010. Prevalence and characterization of Salmonella serovars in retail meats of marketplace in Shaanxi, China. Int J Food Microbiol 141:63–72 10.1016/j.ijfoodmicro.2010.04.015. [PubMed] [DOI] [PubMed] [Google Scholar]
- 78.Bai L, Lan R, Zhang X, Cui S, Xu J, Guo Y, Li F, Zhang D. 2015. Prevalence of Salmonella isolates from chicken and pig slaughterhouses and emergence of ciprofloxacin and cefotaxime co-resistant S. enterica serovar Indiana in Henan, China. PLoS One 10:e0144532 10.1371/journal.pone.0144532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lai J, Wu C, Wu C, Qi J, Wang Y, Wang H, Liu Y, Shen J. 2014. Serotype distribution and antibiotic resistance of Salmonella in food-producing animals in Shandong province of China, 2009 and 2012. Int J Food Microbiol 180:30–38 10.1016/j.ijfoodmicro.2014.03.030. [PubMed] [DOI] [PubMed] [Google Scholar]
- 80.Xia S, Hendriksen RS, Xie Z, Huang L, Zhang J, Guo W, Xu B, Ran L, Aarestrup FM. 2009. Molecular characterization and antimicrobial susceptibility of Salmonella isolates from infections in humans in Henan Province, China. J Clin Microbiol 47:401–409 10.1128/JCM.01099-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu Y, Zhao H, Liu Y, Zhou X, Wang J, Liu T, Beier RC, Hou X. 2015. Characterization of quinolone resistance in Salmonella enterica serovar Indiana from chickens in China. Poult Sci 94:454–460 10.3382/ps/peu133. [PubMed] [DOI] [PubMed] [Google Scholar]
- 82.Zhang WH, Lin XY, Xu L, Gu XX, Yang L, Li W, Ren SQ, Liu YH, Zeng ZL, Jiang HX. 2016. CTX-M-27 producing Salmonella enterica serotypes Typhimurium and Indiana are prevalent among food-producing animals in China. Front Microbiol 7:436 10.3389/fcimb.2017.00436. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiang HX, Song L, Liu J, Zhang XH, Ren YN, Zhang WH, Zhang JY, Liu YH, Webber MA, Ogbolu DO, Zeng ZL, Piddock LJ. 2014. Multiple transmissible genes encoding fluoroquinolone and third-generation cephalosporin resistance co-located in non-typhoidal Salmonella isolated from food-producing animals in China. Int J Antimicrob Agents 43:242–247 10.1016/j.ijantimicag.2013.12.005. [PubMed] [DOI] [PubMed] [Google Scholar]
- 84.Li L, Liao XP, Liu ZZ, Huang T, Li X, Sun J, Liu BT, Zhang Q, Liu YH. 2014. Co-spread of oqxAB and blaCTX-M-9G in non-Typhi Salmonella enterica isolates mediated by ST2-IncHI2 plasmids. Int J Antimicrob Agents 44:263–268 10.1016/j.ijantimicag.2014.05.014. [PubMed] [DOI] [PubMed] [Google Scholar]
- 85.Baggesen DL, Sandvang D, Aarestrup FM. 2000. Characterization of Salmonella enterica serovar Typhimurium DT104 isolated from Denmark and comparison with isolates from Europe and the United States. J Clin Microbiol 38:1581–1586. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Davis MA, Hancock DD, Besser TE. 2002. Multiresistant clones of Salmonella enterica: the importance of dissemination. J Lab Clin Med 140:135–141 10.1067/mlc.2002.126411. [PubMed] [DOI] [PubMed] [Google Scholar]
- 87.Threlfall EJ, Ward LR, Hampton MD, Ridley AM, Rowe B, Roberts D, Gilbert RJ, Van Someren P, Wall PG, Grimont P. 1998. Molecular fingerprinting defines a strain of Salmonella enterica serotype Anatum responsible for an international outbreak associated with formula-dried milk. Epidemiol Infect 121:289–293 10.1017/S0950268898001149. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leekitcharoenphon P, Hendriksen RS, Le Hello S, Weill FX, Baggesen DL, Jun SR, Ussery DW, Lund O, Crook DW, Wilson DJ, Aarestrup FM. 2016. Global genomic epidemiology of Salmonella enterica serovar Typhimurium DT104. Appl Environ Microbiol 82:2516–2526 10.1128/AEM.03821-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Damborg P, Broens EM, Chomel BB, Guenther S, Pasmans F, Wagenaar JA, Weese JS, Wieler LH, Windahl U, Vanrompay D, Guardabassi L. 2015. Bacterial zoonoses transmitted by household pets: state-of-the-art and future perspectives for targeted research and policy actions. J Comp Pathol 155(1 Suppl 1):S27–S40. 10.1016/j.jcpa.2015.03.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.Rijks JM, Cito F, Cunningham AA, Rantsios AT, Giovannini A. 2015. Disease risk assessments involving companion animals: an overview for 15 selected pathogens taking a European perspective. J Comp Pathol 155(1 Suppl 1):S75–S97. 10.1016/j.jcpa.2015.08.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 91.Jay-Russell MT, Hake AF, Bengson Y, Thiptara A, Nguyen T. 2014. Prevalence and characterization of Escherichia coli and Salmonella strains isolated from stray dog and coyote feces in a major leafy greens production region at the United States-Mexico border. PLoS One 9:e113433 10.1371/journal.pone.0113433. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lowden P, Wallis C, Gee N, Hilton A. 2015. Investigating the prevalence of Salmonella in dogs within the Midlands region of the United Kingdom. BMC Vet Res 11:239 10.1186/s12917-015-0553-z. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.USAHA. 2006. Report of the Committee on Salmonella. http://www.usaha.org/upload/Committee/Salmonella/report-sal-2008.pdf.
- 94.Anholt RM, Berezowski J, Ribble CS, Russell ML, Stephen C. 2014. Using informatics and the electronic medical record to describe antimicrobial use in the clinical management of diarrhea cases at 12 companion animal practices. PLoS One 9:e103190 10.1371/journal.pone.0103190. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Summers JF, Hendricks A, Brodbelt DC. 2014. Prescribing practices of primary-care veterinary practitioners in dogs diagnosed with bacterial pyoderma. BMC Vet Res 10:240 10.1186/s12917-014-0240-5. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Prescott JF, Hanna WJ, Reid-Smith R, Drost K. 2002. Antimicrobial drug use and resistance in dogs. Can Vet J 43:107–116. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 97.Hölsö K, Rantala M, Lillas A, Eerikäinen S, Huovinen P, Kaartinen L. 2005. Prescribing antimicrobial agents for dogs and cats via university pharmacies in Finland: patterns and quality of information. Acta Vet Scand 46:87–93 10.1186/1751-0147-46-87. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guardabassi L, Schwarz S, Lloyd DH. 2004. Pet animals as reservoirs of antimicrobial-resistant bacteria. J Antimicrob Chemother 54:321–332 10.1093/jac/dkh332. [PubMed] [DOI] [PubMed] [Google Scholar]
- 99.Rzewuska M, Czopowicz M, Kizerwetter-Świda M, Chrobak D, Błaszczak B, Binek M. 2015. Multidrug resistance in Escherichia coli strains isolated from infections in dogs and cats in Poland (2007–2013). Sci World J 2015:408205 10.1155/2015/408205. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cummings KJ, Aprea VA, Altier C. 2015. Antimicrobial resistance trends among canine Escherichia coli isolates obtained from clinical samples in the northeastern USA, 2004–2011. Can Vet J 56:393–398. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 101.USDA-ARS. NARMS animal arm summary tables and reports. http://www.ars.usda.gov/Main/docs.htm?docid=18034. Accessed 17 October 2017.
- 102.Eaves DJ, Randall L, Gray DT, Buckley A, Woodward MJ, White AP, Piddock LJ. 2004. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob Agents Chemother 48:4012–4015 10.1128/AAC.48.10.4012-4015.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Leonard EK, Pearl DL, Finley RL, Janecko N, Reid-Smith RJ, Peregrine AS, Weese JS. 2012. Comparison of antimicrobial resistance patterns of Salmonella spp. and Escherichia coli recovered from pet dogs from volunteer households in Ontario (2005–06). J Antimicrob Chemother 67:174–181 10.1093/jac/dkr430. [PubMed] [DOI] [PubMed] [Google Scholar]
- 104.Van Immerseel F, Pasmans F, De Buck J, Rychlik I, Hradecka H, Collard JM, Wildemauwe C, Heyndrickx M, Ducatelle R, Haesebrouck F. 2004. Cats as a risk for transmission of antimicrobial drug-resistant Salmonella. Emerg Infect Dis 10:2169–2174 10.3201/eid1012.040904. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Poppe C, Smart N, Khakhria R, Johnson W, Spika J, Prescott J. 1998. Salmonella typhimurium DT104: a virulent and drug-resistant pathogen. Can Vet J 39:559–565. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 106.Wall PG, Threllfall EJ, Ward LR, Rowe B. 1996. Multiresistant Salmonella Typhimurium DT104 in cats: a public health risk. Lancet 348:471 10.1016/S0140-6736(96)24033-4. [PubMed] [DOI] [PubMed] [Google Scholar]
- 107.Low JC, Tennant B, Munro D. 1996. Multiple-resistant Salmonella Typhimurium DT104 in cats. Lancet 348:1391 10.1016/S0140-6736(05)65465-7. [PubMed] [DOI] [PubMed] [Google Scholar]
- 108.Frech G, Kehrenberg C, Schwarz S. 2003. Resistance phenotypes and genotypes of multiresistant Salmonella enterica subsp. enterica serovar Typhimurium var. Copenhagen isolates from animal sources. J Antimicrob Chemother 51:180–182 10.1093/jac/dkg058. [PubMed] [DOI] [PubMed] [Google Scholar]
- 109.Centers for Disease Control and Prevention (CDC). 2001. Outbreaks of multidrug-resistant Salmonella Typhimurium associated with veterinary facilities: Idaho, Minnesota, and Washington, 1999. MMWR Morb Mortal Wkly Rep 50:701–704. [PubMed] [PubMed] [Google Scholar]
- 110.Wright JG, Tengelsen LA, Smith KE, Bender JB, Frank RK, Grendon JH, Rice DH, Thiessen AM, Gilbertson CJ, Sivapalasingam S, Barrett TJ, Besser TE, Hancock DD, Angulo FJ. 2005. Multidrug-resistant Salmonella Typhimurium in four animal facilities. Emerg Infect Dis 11:1235–1241 10.3201/eid1108.050111. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Swanson SJ, Snider C, Braden CR, Boxrud D, Wünschmann A, Rudroff JA, Lockett J, Smith KE. 2007. Multidrug-resistant Salmonella enterica serotype Typhimurium associated with pet rodents. N Engl J Med 356:21–28 10.1056/NEJMoa060465. [PubMed] [DOI] [PubMed] [Google Scholar]
- 112.Izumiya H, Mori K, Kurazono T, Yamaguchi M, Higashide M, Konishi N, Kai A, Morita K, Terajima J, Watanabe H. 2005. Characterization of isolates of Salmonella enterica serovar Typhimurium displaying high-level fluoroquinolone resistance in Japan. J Clin Microbiol 43:5074–5079 10.1128/JCM.43.10.5074-5079.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.White DG, Datta A, McDermott P, Friedman S, Qaiyumi S, Ayers S, English L, McDermott S, Wagner DD, Zhao S. 2003. Antimicrobial susceptibility and genetic relatedness of Salmonella serovars isolated from animal-derived dog treats in the USA. J Antimicrob Chemother 52:860–863 10.1093/jac/dkg441. [PubMed] [DOI] [PubMed] [Google Scholar]
- 114.Cartwright EJ, Nguyen T, Melluso C, Ayers T, Lane C, Hodges A, Li X, Quammen J, Yendell SJ, Adams J, Mitchell J, Rickert R, Klos R, Williams IT, Barton Behravesh C, Wright J. 2016. A multistate investigation of antibiotic-resistant Salmonella enterica serotype I 4,[5],12:i:- infections as part of an international outbreak associated with frozen feeder rodents. Zoonoses Public Health 63:62–71 10.1111/zph.12205. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Centers for Disease Control and Prevention (CDC). 2003. Reptile-associated salmonellosis: selected states, 1998–2002. MMWR Morb Mortal Wkly Rep 52:1206–1209. [PubMed] [PubMed] [Google Scholar]
- 116.Centers for Disease Control and Prevention (CDC). 2005. Salmonellosis associated with pet turtles: Wisconsin and Wyoming, 2004. MMWR Morb Mortal Wkly Rep 54:223–226. [PubMed] [PubMed] [Google Scholar]
- 117.Centers for Disease Control and Prevention (CDC). 2007. Turtle-associated salmonellosis in humans:United States, 2006–2007. MMWR Morb Mortal Wkly Rep 56:649–652. [PubMed] [PubMed] [Google Scholar]
- 118.Welch TJ, Fricke WF, McDermott PF, White DG, Rosso ML, Rasko DA, Mammel MK, Eppinger M, Rosovitz MJ, Wagner D, Rahalison L, Leclerc JE, Hinshaw JM, Lindler LE, Cebula TA, Carniel E, Ravel J. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309 10.1371/journal.pone.0000309. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.GMI. The Global Microbial Identifier. http://www.globalmicrobialidentifier.org/. [PubMed]
- 120.FDA. 2017. GenomeTrakr Network. https://www.fda.gov/Food/FoodScienceResearch/WholeGenomeSequencingProgramWGS/ucm363134.htm. Accessed 24 November 2017.
- 121.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. 2016. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 60:5515–5520 10.1128/AAC.01030-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agersø Y, Lund O, Larsen MV, Aarestrup FM. 2013. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother 68:771–777 10.1093/jac/dks496. [PubMed] [DOI] [PubMed] [Google Scholar]
- 123.Tyson GH, McDermott PF, Li C, Chen Y, Tadesse DA, Mukherjee S, Bodeis-Jones S, Kabera C, Gaines SA, Loneragan GH, Edrington TS, Torrence M, Harhay DM, Zhao S. 2015. WGS accurately predicts antimicrobial resistance in Escherichia coli. J Antimicrob Chemother 70:2763–2769 10.1093/jac/dkv186. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao S, Tyson GH, Chen Y, Li C, Mukherjee S, Young S, Lam C, Folster JP, Whichard JM, McDermott PF. 2015. Whole-genome sequencing analysis accurately predicts antimicrobial resistance phenotypes in Campylobacter spp. Appl Environ Microbiol 82:459–466 10.1128/AEM.02873-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.FDA. 2017. Resistome Trakcer. https://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm570694.htm.
- 126.Michael GB, Schwarz S. 2016. Antimicrobial resistance in zoonotic nontyphoidal Salmonella: an alarming trend? Clin Microbiol Infect 22:968–974 10.1016/j.cmi.2016.07.033. [PubMed] [DOI] [PubMed] [Google Scholar]
- 127.Michael GB, Butaye P, Cloeckaert A, Schwarz S. 2006. Genes and mutations conferring antimicrobial resistance in Salmonella: an update. Microbes Infect 8:1898–1914 10.1016/j.micinf.2005.12.019. [PubMed] [DOI] [PubMed] [Google Scholar]
- 128.Michael GB, Schwarz S. 2013. Antimicrobial resistance in Salmonella, p 120–135. In Methner U (ed), Salmonella in Domestic Animals. CAB eBooks, Barrow, PA. 10.1079/9781845939021.0120 [DOI] [Google Scholar]
- 129.Tran JH, Jacoby GA. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc Natl Acad Sci USA 99:5638–5642 10.1073/pnas.082092899. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Périchon B, Courvalin P, Galimand M. 2007. Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob Agents Chemother 51:2464–2469 10.1128/AAC.00143-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yamane K, Wachino J, Suzuki S, Kimura K, Shibata N, Kato H, Shibayama K, Konda T, Arakawa Y. 2007. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother 51:3354–3360 10.1128/AAC.00339-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, Bush K, Hooper DC. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med 12:83–88 10.1038/nm1347. [PubMed] [DOI] [PubMed] [Google Scholar]
- 133.Hansen LH, Johannesen E, Burmølle M, Sørensen AH, Sørensen SJ. 2004. Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob Agents Chemother 48:3332–3337 10.1128/AAC.48.9.3332-3337.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bai L, Zhao J, Gan X, Wang J, Zhang X, Cui S, Xia S, Hu Y, Yan S, Wang J, Li F, Fanning S, Xu J. 2016. Emergence and diversity of Salmonella enterica serovar Indiana isolates with concurrent resistance to ciprofloxacin and cefotaxime from patients and food-producing animals in China. Antimicrob Agents Chemother 60:3365–3371 10.1128/AAC.02849-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Campos J, Mourão J, Marçal S, Machado J, Novais C, Peixe L, Antunes P. 2016. Clinical Salmonella Typhimurium ST34 with metal tolerance genes and an IncHI2 plasmid carrying oqxAB-aac(6′)-Ib-cr from Europe. J Antimicrob Chemother 71:843–845 10.1093/jac/dkv409. [PubMed] [DOI] [PubMed] [Google Scholar]
- 136.Wong MH, Chan EW, Liu LZ, Chen S. 2014. PMQR genes oqxAB and aac(6′)Ib-cr accelerate the development of fluoroquinolone resistance in Salmonella typhimurium. Front Microbiol 5:521 10.3389/fmicb.2014.00521. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen Y, Sun J, Liao XP, Shao Y, Li L, Fang LX, Liu YH. 2016. Impact of enrofloxacin and florfenicol therapy on the spread of OqxAB gene and intestinal microbiota in chickens. Vet Microbiol 192:1–9 10.1016/j.vetmic.2016.05.014. [PubMed] [DOI] [PubMed] [Google Scholar]
- 138.Nair S, Ashton P, Doumith M, Connell S, Painset A, Mwaigwisya S, Langridge G, de Pinna E, Godbole G, Day M. 2016. WGS for surveillance of antimicrobial resistance: a pilot study to detect the prevalence and mechanism of resistance to azithromycin in a UK population of non-typhoidal Salmonella. J Antimicrob Chemother 71:3400–3408 10.1093/jac/dkw318. [PubMed] [DOI] [PubMed] [Google Scholar]
- 139.Nguyen F, Starosta AL, Arenz S, Sohmen D, Dönhöfer A, Wilson DN. 2014. Tetracycline antibiotics and resistance mechanisms. Biol Chem 395:559–575 10.1515/hsz-2013-0292. [PubMed] [DOI] [PubMed] [Google Scholar]
- 140.Wachino J, Arakawa Y. 2012. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist Updat 15:133–148 10.1016/j.drup.2012.05.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 141.Folster JP, Rickert R, Barzilay EJ, Whichard JM. 2009. Identification of the aminoglycoside resistance determinants armA and rmtC among non-Typhi Salmonella isolates from humans in the United States. Antimicrob Agents Chemother 53:4563–4564 10.1128/AAC.00656-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Naas T, Bentchouala C, Lima S, Lezzar A, Smati F, Scheftel JM, Nordmann P. 2009. Plasmid-mediated 16S rRNA methylases among extended-spectrum-beta-lactamase-producing Salmonella enterica Senftenberg isolates from Algeria. J Antimicrob Chemother 64:866–868 10.1093/jac/dkp312. [PubMed] [DOI] [PubMed] [Google Scholar]
- 143.Campos J, Cristino L, Peixe L, Antunes P. 2016. MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1,4,[5],12:i:- and S. Rissen clones in Portugal, 2011 to 2015. Euro Surveill 21:pii=30270 10.2807/1560-7917.ES.2016.21.26.30270. [PubMed] [DOI] [PubMed] [Google Scholar]
- 144.Carnevali C, Morganti M, Scaltriti E, Bolzoni L, Pongolini S, Casadei G. 2016. Occurrence of mcr-1 in colistin-resistant Salmonella enterica isolates recovered from humans and animals in Italy, 2012 to 2015. Antimicrob Agents Chemother 60:7532–7534. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, Day M, Muller-Pebody B, Ellington MJ, de Pinna E, Johnson AP, Hopkins KL, Woodford N. 2016. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother 71:2300–2305 10.1093/jac/dkw093. [PubMed] [DOI] [PubMed] [Google Scholar]
- 146.Quesada A, Ugarte-Ruiz M, Iglesias MR, Porrero MC, Martínez R, Florez-Cuadrado D, Campos MJ, García M, Píriz S, Sáez JL, Domínguez L. 2016. Detection of plasmid mediated colistin resistance (MCR-1) in Escherichia coli and Salmonella enterica isolated from poultry and swine in Spain. Res Vet Sci 105:134–135 10.1016/j.rvsc.2016.02.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 147.Rau RB, de Lima-Morales D, Wink PL, Ribeiro AR, Martins AF, Barth AL. 2017. Emergence of mcr-1 producing Salmonella enterica serovar Typhimurium from retail meat first detection in Brazil. Foodborne Pathog Dis 15(1):58–59. 10.1089/fpd.2017.2346. [PubMed] [DOI] [PubMed] [Google Scholar]
- 148.Torpdahl M, Hasman H, Litrup E, Skov RL, Nielsen EM, Hammerum AM. 2017. Detection of mcr-1-encoding plasmid-mediated colistin-resistant Salmonella isolates from human infection in Denmark. Int J Antimicrob Agents 49:261–262 10.1016/j.ijantimicag.2016.11.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 149.Sjölund-Karlsson M, Joyce K, Blickenstaff K, Ball T, Haro J, Medalla FM, Fedorka-Cray P, Zhao S, Crump JA, Whichard JM. 2011. Antimicrobial susceptibility to azithromycin among Salmonella enterica isolates from the United States. Antimicrob Agents Chemother 55(9):3985–3989. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Seidman JC, Coles CL, Silbergeld EK, Levens J, Mkocha H, Johnson LB, Muñoz B, West SK. 2014. Increased carriage of macrolide-resistant fecal E. coli following mass distribution of azithromycin for trachoma control. Int J Epidemiol 43(4):1105–1113. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Schmidt JW, Agga GE, Bosilevac JM, Brichta-Harhay DM, Shackelford SD, Wang R, Wheeler TL, Arthur TM. 2015. Occurrence of Antimicrobial-Resistant Escherichia coli and Salmonella enterica in the Beef Cattle Production and Processing Continuum. Appl Environ Microbiol 81(2):713–725. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]


