ABSTRACT
Bordetella bronchiseptica is involved in respiratory tract infections mainly in dogs and pigs but may also cause infections in humans. Valid and representative data on antimicrobial susceptibility of B. bronchiseptica is rare. Approved antimicrobial susceptibility testing methods have been published, but very few clinical breakpoints are available. The MIC values are low for most agents but high for β-lactam antibiotics and macrolides. Information on the genetic basis of resistance is scarce. For a small number of isolates that are resistant or show elevated MICs, the molecular basis of resistance was identified. Three tetracycline resistance genes, tet(A), tet(C), and tet(31), coding for major facilitator superfamily efflux pumps, were identified. Two other major facilitator superfamily exporter genes confer resistance to chloramphenicol (cmlB1) or to chloramphenicol and florfenicol (floR). Two class B chloramphenicol acetyltransferase genes (catB1 and catB3), which confer resistance to nonfluorinated phenicols by enzymatic inactivation, have been identified in B. bronchiseptica. Like the trimethoprim resistance genes dfrA1 and dfrB1, which code for trimethoprim-insensitive dihydrofolate reductases, the genes catB1 and catB3 were located on gene cassettes and found in class 1 integrons also harboring the sulfonamide resistance gene sul1. In addition, the gene sul2 has also been detected. Both sul1 and sul2 code for sulfonamide-insensitive dihydropteroate synthases. A gene cassette harboring the β-lactamase gene blaOXA-2 was also identified, whereas β-lactam resistance in B. bronchiseptica seems to be more likely due to reduced influx in combination with the species-specific β-lactamase encoded by blaBOR-1. The resistance genes were mostly located on conjugative plasmids.
BORDETELLA BRONCHISEPTICA
B. bronchiseptica is a bacterium within the phylum Proteobacteria and the class Betaproteobacteria. It belongs to the order Burkholderiales and the family Alcaligenaceae. In the genus Bordetella, B. bronchiseptica is one of 14 approved species (http://www.bacterio.net/bordetella.html). B. bronchiseptica is a small, coccoid-shaped Gram-negative bacterium with a size of about 0.2 to 0.5 µm by 0.5 to 2 µm. It is motile due to peritrichous flagella. In comparison to other Bordetella spp., its nutritional requirements are simple, and it grows on blood agar plates at 35 to 37°C overnight. The colonies are small, grayish-white, smooth, and shiny, usually without or only with a small zone of hemolysis.
B. bronchiseptica is a commensal of the upper respiratory tract of diverse animal species, including mammals and birds. In veterinary medicine, it also plays an important role as a primary and secondary pathogen of the upper respiratory tract in several mammals but is most important and best described in dogs and in pigs. In contrast, Bordetella pertussis, the causative agent of whooping cough in humans, has rarely been reported in other mammals. Experimental infections showed that rhesus macaques and baboons can develop clinical disease (1), and at least one case of an epizootic of whooping cough among chimpanzees in a zoo has been described (2). Bordetella avium is commonly identified in birds, although B. avium infections have also been described in single human patients with cystic fibrosis (3, 4).
Clinical Relevance
B. bronchiseptica is a facultative respiratory tract pathogen and causes respiratory tract infections in mammals (5). In general, clinical infections caused by B. bronchiseptica require additional factors (infectious or noninfectious stressors) and can be seen as multifactorial diseases (6). In addition to other bacteria or viruses, for example, transport and crowding are accompanying factors in the porcine respiratory disease complex. In the clinical scenario, B. bronchiseptica may be a primary pathogen and pave the way for other respiratory tract pathogens such as Pasteurella multocida. This is commonly the case in one of the major diseases associated with B. bronchiseptica in pigs: atrophic rhinitis. A mild form of atrophic rhinitis is seen when B. bronchiseptica is the only pathogen, whereas a progressive and much more severe form is seen when P. multocida is involved (7, 8). In dogs, B. bronchiseptica may also act as a secondary pathogen in the kennel cough complex, also known as canine infectious tracheobronchitis. In kennel cough, canine parainfluenza virus is considered the major pathogen and may pave the way for a subsequent B. bronchiseptica infection.
Zoonotic Potential
The vast majority of patients suffering from a clinical infection with B. bronchiseptica are either very young or old. Rarely, reports can be found with patients in the age group of 10 to 50 years; commonly, people in that age group and infected by B. bronchiseptica are immunocompromised, such as a 43-year-old man who was HIV-positive (9, 10) and an 11-year-old girl suffering from cystic fibrosis (11). However, contact with infected animals may also play a role in human B. bronchiseptica infections (12). The patients show respiratory symptoms, such as sinusitis, tracheobronchitis, or a pertussis-like cough (13). Septicemia and meningitis have been also described (14, 15).
Prophylaxis and Therapy
On the one hand, vaccination is available for small animals, especially for dogs. Kennel cough vaccines comprising either B. bronchiseptica alone or B. bronchiseptica and canine parainfluenza virus type 2 are commercially available, the former available as an injectable vaccine and the latter as an injectable vaccine or a vaccine for intranasal application. For cats, only an intranasal vaccine against B. bronchiseptica is available. For rabbits, an injectable vaccine against B. bronchiseptica and P. multocida is on the market. In pigs, autogenous vaccination is used.
On the other hand, in addition to symptomatic treatment, a treatment with antimicrobial agents is a good and successful option to treat B. bronchiseptica infections. This prevents additional complications or additional secondary bacterial infections but does not help against other components of the multifactorial disease complexes. Thus, it is important to also reduce viral and environmental stressors. Because these respiratory diseases are highly contagious, it is also helpful to avoid contact of diseased animals with healthy animals. Such a quarantine is likely possible for pets but difficult if not impossible for pigs and rabbit breeding units, because B. bronchiseptica has already spread between animals before they show the first clinical signs of disease.
In human patients also, B. bronchiseptica infections can be treated with antimicrobial agents. However, in human medicine, the correct identification of B. bronchiseptica is the major problem, because this bacterium is not a common human pathogen. Of note, the very common use of β-lactams as first-choice antibiotics does not lead to therapeutic success in B. bronchiseptica infections. In contrast, in combination with a β-lactamase inhibitor, this treatment was successful (16). However, most patients with clinical infections have other severe underlying diseases hampering the treatment and leading to the critical situations described in the few case reports available (13, 17).
ANTIMICROBIAL SUSCEPTIBILITY OF B. BRONCHISEPTICA
Antimicrobial susceptibility testing prior to the treatment of clinical B. bronchiseptica infections is of major relevance in both human and veterinary medicine. To predict the success or failure of an antimicrobial therapy, the correct in vitro determination of the antimicrobial susceptibility of the B. bronchiseptica isolates is of utmost importance.
Antimicrobial Susceptibility Testing Methods
An internationally accepted testing procedure is available from the Clinical and Laboratory Standards Institute (CLSI) (18, 19). B. bronchiseptica isolates can be tested by agar disk diffusion or by determining the MIC by agar dilution or by broth micro- or macrodilution. For this, the standard procedure as described for fast-growing aerobic bacteria in CLSI document VET01-S (19) should be applied. The inoculum can be prepared by either the growth method or the direct colony suspension method and should be equivalent to a 0.5 McFarland standard. Incubation should be for 16 to 20 h at 35°C ± 2°C in ambient air. The CLSI-approved media are Mueller-Hinton agar for disk diffusion and agar dilution as well as cation-adjusted Mueller-Hinton broth for broth dilution assays. Escherichia coli ATCC 25922 or Staphylococcus aureus ATCC 25923 (disk diffusion)/ATCC 29213 (MIC determination) are recommended as quality controls (19). However, it has been reported that an increase of the incubation time to 24 h may be advantageous. The authors of this study showed that the MIC values of ten isolates determined in five replicates were more stable when read after 24 h incubation time, although the classification of the isolates as susceptible, intermediate, or resistant did not change (20).
Clinical Breakpoints
CLSI document VET01-S is currently the only antimicrobial susceptibility testing document that contains approved clinical breakpoints specific to B. bronchiseptica (19). However, breakpoints are only available for a few agents, namely, for ampicillin (test result can be extrapolated to amoxicillin and hetacillin), florfenicol, tildipirosin, and tulathromycin. For tulathromycin, it is worthwhile to mention that it is absolutely essential to stick to the prescribed pH value to end up with correct results for B. bronchiseptica as well as for other bacteria. Breakpoints are available for disk diffusion and for MICs determined by broth dilution or agar dilution for florfenicol, tildipirosin, and tulathromycin (19). In contrast, there are only MIC breakpoints for ampicillin (19, 21). Since B. bronchiseptica is commonly resistant to ampicillin, these breakpoints serve the diagnostic laboratory mainly to exclude ampicillin and related antimicrobial agents of the β-lactam class from treatment recommendations.
Epidemiological Cutoff Values
An interpretation of the susceptibility testing results by using epidemiological cutoff values is even more difficult. Epidemiological cutoff values for B. bronchiseptica had been available solely for trimethoprim-sulfamethoxazole on the EUCAST homepage and have been removed in the meantime (https://mic.eucast.org/Eucast2/). The only data still shown on the website are tetracycline MICs without giving an epidemiological cutoff value (https://mic.eucast.org/Eucast2/SearchController/search.jsp?action=performSearch&BeginIndex=0&Micdif=mic&NumberIndex=50&Antib=-1&Specium=791). In contrast to other MIC values, the MICs for trimethoprim-sulfamethoxazole show a very wide range for B. bronchiseptica isolates, for example, comprising more than all 12 dilution steps tested from ≤0.03 mg/liter to ≥64 mg/liter (22). In comparison, tetracycline MICs of the same 349 isolates were distributed from ≤0.12 mg/liter to 2 mg/liter representing the wild-type population, with the vast majority of isolates (n = 227) having a MIC of 0.25 mg/liter (22). Three isolates showed a distinctly higher MIC of 64 mg/liter and were considered non-wild type and were later shown to harbor a specific tetracycline resistance gene (23). A very similar situation is seen on the EUCAST website, with 443 isolates distributed normally from 0.12 mg/liter to 4 mg/liter and 4 isolates showing higher MICs of 64 mg/liter (https://mic.eucast.org/Eucast2/SearchController/search.jsp?action=performSearch&BeginIndex=0&Micdif=mic&NumberIndex=50&Antib=-1&Specium=791). Tetracycline MIC distributions are compared in Table 1.
TABLE 1.
Tetracycline MIC distributions of B. bronchiseptica isolates
| Origin | Country | Year of isolation | No. of isolates | No. of isolates with an MIC of … mg/litera | Reference | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | |||||
| Pigs | Germany | 2011/2012 | 90 | n.t. | n.t. | 0 | 1 | 52 | 27 | 5 | 3 | 0 | 0 | 0 | 2 | 0 | 27 |
| Companion animalsb | Germany | 2010–2012 | 43 | n.t. | n.t. | 1 | 14 | 22 | 4 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 24 |
| Pigs | Germany | 2010-2012 | 107 | n.t. | n.t. | 2 | 51 | 37 | 4 | 5 | 0 | 0 | 0 | 1 | 4 | 3 | 24 |
| Pigs | Europec | 2010–2012 | 118 | n.t. | 0 | 0 | 42 | 52 | 9 | 7 | 4 | 1 | 0 | 0 | 0 | 3 | 25 |
| Pigs | Germany | 2010 | 43 | n.t. | n.t. | 0 | 4 | 24 | 10 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 26 |
| Dogs (n = 8), cats (n = 5) | Germany | 2010 | 13 | n.t. | n.t. | 0 | 0 | 9 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 60 |
| Pigs | Germany | 2009 | 69 | n.t. | n.t. | 0 | 9 | 51 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 3 | 29 |
| Pigs | Germany | 2008 | 93 | n.t. | n.t. | 0 | 49 | 35 | 5 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 30 |
| Dogs (n = 34), cats ( = 8) | Germany | 2004–2006 | 42 | 0 | 0 | 0 | 13 | 18 | 3 | 6 | 1 | 0 | 0 | 0 | 1 | 0 | 39 |
| Pigs | Germany | 2003 | 82 | n.t. | n.t. | 8 | 60 | 11 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 22 |
| Pigs | Germany | 2002/2003 | 138 | n.t. | n.t. | 9 | 99 | 23 | 3 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 58 |
| Pigs | Germany | 2002 | 91 | n.t. | n.t. | 6 | 63 | 17 | 1 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 22 |
| Pigs | Germany | 2001 | 98 | n.t. | n.t. | 5 | 65 | 27 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 22 |
| Pigs | Germany | 2000 | 78 | n.t. | n.t. | 29 | 39 | 7 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 22 |
n.t., not tested.
Horses (n = 24), dogs (n = 8), rabbits (n = 8), cats (n = 2), ferret (n = 1).
Belgium (n = 24), Denmark (n = 9), France (n = 12), Germany (n = 14), The Netherlands (n = 22), Poland (n = 14), Spain (n = 21), United Kingdom (n = 2).
Published Monitoring Studies
Collected information about antimicrobial susceptibility testing studies in a PhD thesis (23) shows that a direct comparison of susceptibility data is very difficult to accomplish due to different methodologies used. In the corresponding study, MIC determination followed the CLSI-approved antimicrobial susceptibility testing protocol (22). This study showed an overall favorable situation with low MIC values and no change in MICs over a period of 4 years among 349 porcine B. bronchiseptica isolates from pigs. More recent publications, however, were performed basically—but not exactly—according to the CLSI standard (24, 25). Commonly, in routine diagnostics as well as in some publications, such as a study of B. bronchiseptica isolates from Poland (26), disk diffusion is performed. While in the case of MIC determination, MIC distributions are often shown, the distribution of inhibitory zone diameters is not provided, and results are only given as percentages of isolates classified as susceptible, resistant, or (if available) intermediate. Due to the lack of approved breakpoints, these results have to be used with caution. Distributions of MIC values for florfenicol are shown in Table 2. While clinical breakpoints are available for florfenicol and often the testing range is reduced to the dilution steps of clinical interest, for tetracycline, a wider test range is applied. The tetracycline MICs show a clear bimodal distribution, with most of the isolates having an MIC value around 0.25 or 0.5 mg/liter and single isolates showing MICs of 16 mg/liter or higher (Table 1). For the florfenicol MICs, a bimodal distribution is not so clear, which is not due to the shorter testing ranges (Table 2). Isolates with an MIC in the upper range of the normal distribution around the MIC values of 1 to 4 mg/liter have to be classified as intermediate or even as resistant according to the clinical breakpoints.
TABLE 2.
Florfenicol MIC distributions of B. bronchiseptica isolates
| Origin | Country | Year of isolation | No. of isolates | No. of isolates with an MIC of … mg/litera | Isolates in %b | Reference | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | S | I | R | |||||
| Companion animalsc | Germany | 2010–2012 | 43 | n.t. | n.t. | 0 | 0 | 1 | 16 | 11 | 12 | 2 | 1 | 0 | 0 | 24 | |||
| Pigs | Germany | 2010–2012 | 107 | n.t. | n.t. | 0 | 1 | 3 | 39 | 49 | 14 | 1 | 0 | 0 | 0 | 86.0 | 13.1 | 0.9 | 24 |
| Pigs | Germany | 2011/2012 | 90 | n.t. | n.t. | 0 | 0 | 0 | 1 | 8 | 79 | 2 | 0 | 0 | 0 | 10.0 | 87.8 | 2.2 | 27 |
| Pigs | Europed | 2010–2012 | 118 | n.t. | n.t. | n.t. | n.t. | n.t. | 10 | 52 | 50 | 1 | 0 | 0 | 5 | 52.5 | 42.4 | 5.1 | 25 |
| Pigs | Germany | 2010 | 43 | n.t. | n.t. | 0 | 0 | 0 | 3 | 4 | 32 | 1 | 1 | 2 | 0 | 16.3 | 74.4 | 9.3 | 28 |
| Pigs | Germany | 2009 | 69 | n.t. | n.t. | 0 | 0 | 0 | 2 | 15 | 46 | 4 | 0 | 1 | 1 | 24.6 | 66.7 | 8.7 | 29 |
| Pigs | Germany | 2008 | 93 | n.t. | n.t. | 0 | 0 | 1 | 18 | 41 | 31 | 1 | 0 | 1 | 0 | 64.5 | 33.3 | 2.2 | 30 |
| Dogs (n = 34), cats (n = 8) | Germany | 2004–2006 | 42 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 32 | 0 | 0 | 1 | 0 | 39 | |||
| Pigs | Germany | 2003 | 82 | n.t. | n.t. | n.t. | n.t. | n.t. | n.t. | 67 | 11 | 0 | 0 | 4 | n.t. | 81.7 | 13.4 | 4.9 | 22 |
| Pigs | Germany | 2003 | 51 | n.t. | n.t. | 0 | 0 | 0 | 4 | 40 | 7 | 0 | 0 | 0 | 0 | 72.5 | 26.5 | 1.0 | 61 |
| Pigs | Germany | 2002/2003 | 138 | n.t. | n.t. | n.t. | n.t. | n.t. | n.t. | 111 | 25 | 0 | 0 | 2 | n.t. | 80.4 | 18.2 | 1.4 | 58 |
| Pigs | Korea | 1998–2003 | 70 | n.t. | n.t. | 0 | 0 | 0 | remaining 67 isolates | 3 | 0 | 0 | 0 | 0 | 95.7 | 4.3 | 0.0 | 59 | |
| Pigs | Germany | 2002 | 91 | n.t. | n.t. | n.t. | n.t. | n.t. | n.t. | 73 | 17 | 0 | 1 | 0 | n.t. | 80.2 | 18.7 | 1.1 | 22 |
| Pigs | Germany | 2002 | 80 | n.t. | n.t. | 0 | 0 | 2 | 17 | 59 | 1 | 0 | 0 | 1 | 0 | 97.4 | 1.3 | 1.3 | 61 |
| Pigs | Germany | 2001 | 73 | n.t. | n.t. | 0 | 0 | 0 | 9 | 17 | 38 | 5 | 0 | 4 | 0 | 35.6 | 52.1 | 12.3 | 62 |
| Pigs | Germany | 2001 | 98 | n.t. | n.t. | n.t. | n.t. | n.t. | n.t. | 71 | 26 | 1 | 0 | 0 | n.t. | 72.5 | 26.5 | 1.0 | 22 |
| Pigs | Germany | 2000 | 87 | n.t. | n.t. | 0 | 0 | 0 | 11 | 18 | 26 | 26 | 6 | 0 | 0 | 33.3 | 29.9 | 36.8 | 62 |
| Pigs | Germany | 2000 | 78 | n.t. | n.t. | n.t. | n.t. | n.t. | n.t. | 67 | 7 | 2 | 2 | 0 | n.t. | 85.9 | 9.0 | 5.1 | 22 |
n.t., not tested.
Given for the porcine isolates, for which clinical breakpoints are available from the CLSI: S, susceptible; I, intermediate; R, resistant.
Horses (n = 24), dogs (n = 8), rabbits (n = 8), cats (n = 2), ferret (n = 1).
Belgium (n = 24), Denmark (n = 9), France (n = 12), Germany (n = 14), The Netherlands (n = 22), Poland (n = 14), Spain (n = 21), United Kingdom (n = 2).
Another fact reducing the information on susceptibility of B. bronchiseptica is that often several agents of the same class are tested (24, 25, 27–30). Moreover, two classes licensed and used to treat respiratory tract infections are most commonly included in the panel for B. bronchiseptica: β-lactams and macrolides. Both classes are not useful against B. bronchiseptica, and in vitro susceptibility testing revealed high MIC values and—when breakpoints were available—100% resistant isolates (24, 25, 27–30). For other antimicrobial agents, the studies shown in Table 1 confirm the favorable situation with respect to susceptibility and resistance of B. bronchiseptica.
ANTIMICROBIAL RESISTANCE IN B. BRONCHISEPTICA
In general, little information concerning antimicrobial resistance in B. bronchiseptica is available from the published literature. As a facultative pathogen in a genus of bacteria that harbors human pathogens (B. pertussis) as well as apathogenic species (e.g., Bordetella tumbae), several publications focused on virulence factors and the pathogenicity of bordetellae including B. bronchiseptica (31). Other studies dealt with immunity and vaccination strategies (32). In fact, a B. bronchiseptica vaccine was one of the first antibacterial vaccines and started as a temperature-sensitive vaccine for intranasal application (33). Treatment of clinical B. bronchiseptica infections is easily possible with the appropriate antimicrobial agents licensed for food-producing animals. Commonly, tetracyclines are used, and more than 90% of all B. bronchiseptica isolates show low tetracycline MIC values (https://mic.eucast.org/Eucast2/SearchController/search.jsp?action=performSearch&BeginIndex=0&Micdif=mic&NumberIndex=50&Antib=-1&Specium=791; 22, 34, 35). Overall, due to the favorable situation in terms of antimicrobial susceptibility, treatment problems are rare, and a search for alternative agents is often not necessary.
All three antimicrobial resistance mechanisms— enzymatic inactivation of the antimicrobial agent, reduced intracellular accumulation, and target site modifications—have been identified and described in B. bronchiseptica isolates.
Tetracycline Resistance
High tetracycline MIC values are seen in single isolates of virtually all publications and throughout all years. In contrast to other respiratory tract pathogens, such as Pasteurellaceae with about 30% tetracycline-resistant isolates, only about 1% of the B. bronchiseptica isolates are tetracycline-resistant, e.g., 3 of 349 German porcine isolates (22) and 4 of 447 isolates (https://mic.eucast.org/Eucast2/SearchController/search.jsp?action=performSearch&BeginIndex=0&Micdif=mic&NumberIndex=50&Antib=-1&Specium=791). Isolates collected from pretreated animals, because tetracyclines are commonly used, might show a higher resistance rate.
The first two B. bronchiseptica isolates considered tetracycline-resistant were isolated in the United Kingdom from cats (35). The gene tet(C) was identified in these feline isolates. This gene codes for a specific efflux protein of the major facilitator superfamily (MFS) conferring resistance in various Gram-negative bacteria by active efflux of tetracyclines. To date, the tet(C) gene has been also identified in a porcine isolate (Kadlec and Schwarz, unpublished). Another MFS tet gene, tet(A), was identified for the first time in B. bronchiseptica in porcine isolates from Germany (23). Later, tet(A) was also identified in all eight tetracycline-resistant isolates from another study of German isolates (24). A third MFS gene, tet(31), has been confirmed in B. bronchiseptica (36). All three genes have also been described in other Gram-negative bacteria but not in other common respiratory tract pathogens, except tet(C) in Chlamydia suis (37).
Sulfonamide Resistance
Among the three sulfonamide resistance genes sul1, sul2, and sul3, so far only sul1 and sul2 have been identified in B. bronchiseptica. All sul genes code for an alternative dihydropteroate synthase that is insensitive to sulfonamides. As in other bacteria, sul1 was in B. bronchiseptica a part of the 3′-conserved segment of class 1 integrons that were located on plasmids (38). The gene sul2 has also been identified in B. bronchiseptica isolates obtained from dogs and cats in the BfT-GermVet study (39). This gene is commonly seen in close proximity to strA and strB (40). In a study by Prüller and coworkers (24), the sul2-positive B. bronchiseptica isolates were also strA and strB positive, but an analysis of the linkage of these genes was not performed. Complete class 1 integrons are common in other Gram-negative bacteria but have not been described in respiratory tract pathogens of the family Pasteurellaceae so far. In contrast, the gene cluster sul2-strA-strB has been seen in Pasteurellaceae and is also common in several other Gram-negative bacteria (40–42).
Trimethoprim Resistance
Up to now, three trimethoprim resistance genes have been described in B. bronchiseptica (24, 38). All three genes (dfrA1, dfrA7, and dfrB1) code for alternative dihydrofolate reductases. Among the various dfr genes that have been described in Gram-negative bacteria, about 30 genes code for class A dihydrofolate reductases, such as the genes dfrA1 and dfrA7, and only seven code for class B dihydrofolate reductases, namely, dfrB1, dfrB2, dfrB3, dfrB4, dfrB5, dfrB6, and dfrB7 (43). The acquisition of such a resistance gene and thereby the replacement of the naturally occurring trimethoprim-sensitive dihydrofolate reductase by an alternative trimethoprim-insensitive enzyme leads to very high trimethoprim MIC values (>256 mg/liter) compared to wild-type isolates with low MIC values of <0.12 mg/liter. This observation has also been made in B. bronchiseptica (38). However, trimethoprim alone is usually neither tested nor used for treatment. These dfr genes are commonly located on gene cassettes, a fact that has been also described for dfrA1 and dfrB1 in B. bronchiseptica (38). The dfr-carrying gene cassettes described in B. bronchiseptica are shown in Fig. 1. In the study that described the identification of dfrA7, the authors also detected a sul1 gene, a hint about the presence of a class 1 integron, but did not confirm the location of dfrA7 in a gene cassette (24).
FIGURE 1.
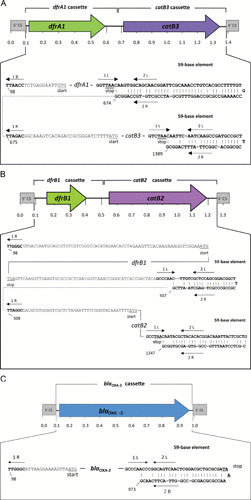
Schematic presentation of the class 1 integrons described so far in B. bronchiseptica isolates. The reading frames of the antimicrobial resistance genes are shown as arrows, and the conserved segments of the class 1 integron are shown as boxes. The beginning and the end of the integrated cassettes are shown in detail below. The translational start and stop codons are underlined. The 59-base elements are shown in bold type, and the putative IntI1 integrase binding domains 1L, 2L, 2R, and 1R are indicated by arrows. The numbers refer to the positions of the bases in the EMBL database entries with the following accession numbers: (a) AJ844287, (b) AJ879564, and (c) AJ877267 (41, 50).
Aminoglycoside and Aminocyclitol Resistance
In general, the streptomycin MIC values of B. bronchiseptica isolates are high, as described for 150 isolates, 132 of which had MICs of 32 to 128 mg/liter and the remaining 18 of which had distinctly higher MICs of ≥1,024 mg/liter. In 17/18 streptomycin-resistant isolates (tentatively classified as resistant by MICs of ≥1,024 mg/liter), the genes strA and strB were detected (24). These genes often occur together and code for phosphotransferases, namely for the aminoglycoside-3′-phosphotransferase and the aminoglycoside-6′-phosphotransferase, respectively. Thus, they confer resistance by the inactivation of streptomycin. For neomycin, the majority of B. bronchiseptica isolates showed MICs of 1 to 8 mg/liter (22, 24). In four isolates with distinctly higher MICs of ≥128 mg/liter, no resistance gene was detected (24). MICs of gentamicin are commonly around 2 mg/liter (22, 24), and isolates exhibiting high MICs have not yet been observed. For the aminocyclitol spectinomycin, data from the German BfT-GermVet study revealed that all 42 isolates from cats and dogs had very high MICs of ≥512 mg/liter (39).
Phenicol Resistance
B. bronchiseptica isolates resistant to florfenicol that also exhibited high MICs to chloramphenicol, but also isolates that were susceptible to florfenicol but had high chloramphenicol MIC values have been described (22, 44). Among the latter isolates, two phenicol resistance mechanisms were identified. The catB1 and catB3 genes code for class B chloramphenicol acetyltransferases which inactivate only nonfluorinated phenicols, such as chloramphenicol. As described for other Gram-negative bacteria, these genes were located on gene cassettes and integrated into class 1 integrons (Fig. 1) (38). Another chloramphenicol resistance mechanism was identified in one B. bronchiseptica isolate. The isolate harbored a novel gene, cmlB1, coding for an MFS exporter. Database searches revealed that the gene is still very rare. Only one additional database entry was found which described the cmlB1 gene in the whole-genome sequence of an Acinetobacter pittii isolate (45). No phenotype was described for this A. pittii isolate. In contrast to cmlA genes, cmlB1 was not part of a gene cassette. It was located on a large nonconjugative plasmid and also conferred chloramphenicol resistance after transfer to E. coli (44).
Most of the B. bronchiseptica isolates that were resistant to florfenicol and had high chloramphenicol MICs harbored the widely distributed resistance gene floR. This gene also codes for an MFS exporter and was located in the chromosomal DNA of 7/10 florfenicol-resistant B. bronchiseptica isolates (44). The remaining three isolates showed distinctly lower florfenicol and chloramphenicol MIC values and were no longer classified as florfenicol-resistant when an efflux inhibitor was added. The inhibitor PaβNA indicates the presence of a not further specified exporter of the resistance-nodulation-cell division type (44).
β-Lactam Resistance
In B. bronchiseptica, the species-specific β-lactamase gene blaBOR-1 has been described (46). Involvement of a β-lactam hydrolyzing enzyme in the decreased susceptibility of B. bronchiseptica to β-lactam antibiotics is underlined by the fact that β-lactam MICs are lower in the presence of the β-lactamase inhibitor clavulanic acid. Among 150 isolates from pigs, cats, and dogs, 147 were positive in a PCR for blaBOR-1 (24). In addition to this class A β-lactamase gene, the class D β-lactamase gene blaOXA-2 has been described in B. bronchiseptica (47). As previously reported in Enterobacteriaceae, blaOXA-2 was located on a gene cassette and integrated into a class 1 integron (Fig. 1). In addition, it was shown that low membrane permeability could contribute to the β-lactam resistance of B. bronchiseptica (47).
Fluoroquinolone Resistance
Fluoroquinolones are usually highly active against B. bronchiseptica. An early study in which fluoroquinolones were evaluated for their activity against porcine respiratory bacterial pathogens revealed that ciprofloxacin was the most active quinolone against nine strains of B. bronchiseptica with mean MICs of 0.58 mg/liter (48). In another study, all 78 canine B. bronchiseptica isolates were reported to be susceptible to enrofloxacin (49). In a study of feline B. bronchiseptica isolates, all 43 strains tested were susceptible to marbofloxacin and enrofloxacin (MIC90, 0.5 mg/liter), while 93% and 84% of the strains were susceptible, respectively, to ciprofloxacin and difloxacin, with MIC90 values of 1 and 8 mg/liter, respectively (50). Testing of 42 B. bronchiseptica isolates from dogs and cats for their susceptibility to pradofloxacin revealed MICs in the range between 0.12 mg/liter and 1 mg/liter with both MIC50 and MIC90 values of 0.25 mg/liter (51). Porcine B. bronchiseptica isolates (n = 349; 2000 to 2003) ranged in their MICs between ≤0.015 mg/liter and 2 mg/liter with MIC50 and MIC90 values of 0.25 and 0.5 mg/liter, respectively (22). The marbofloxacin MIC values of 504 B. bronchiseptica isolates collected in various European countries between 1994 and 2013 ranged between 0.06 mg/liter and 2 mg/liter with both MIC50 and MIC90 values of 0.5 mg/liter (52).
LOCATION OF RESISTANCE GENES ON MOBILE GENETIC ELEMENTS
Most of the resistance genes described so far were located on plasmids. In all cases, the corresponding resistance plasmid was the only plasmid harbored by the respective field isolates. Most of these plasmids were conjugative and could be successfully transferred to E. coli (23, 35, 38, 44). The easy transfer into E. coli and also the good maintenance in E. coli is in contrast to the transfer of plasmids isolated from other respiratory tract pathogens, namely Pasteurellaceae.
In addition to plasmids, B. bronchiseptica makes use of gene cassettes as mobile genetic elements. Trimethoprim, chloramphenicol, and β-lactam resistance genes have been already described as part of gene cassettes in B. bronchiseptica (Fig. 1). While very common in Enterobacteriaceae, other respiratory tract pathogens do not often carry class 1 integrons.
Moreover, for the tetracycline resistance gene tet(A), remnants of the small nonconjugative transposon Tn1721 occurring commonly in E. coli and other Enterobacteriaceae were identified by sequence analysis (Fig. 2). The fact that different parts of transposon Tn1721 were present on the two further analyzed plasmids indicates that different genetic events led to the final plasmid structure and that, very likely, a Tn1721-located gene tet(A) was acquired more than once by B. bronchiseptica. Tn1721 has been described in E. coli but not in Pasteurellaceae. In Pasteurella, the genes tet(B) and tet(H) are the most common tetracycline resistance genes (41, 53). The streptomycin resistance genes strA and strB are often located on plasmids and are associated with the transposon Tn5393 (54). Although not described so far, it is very likely that such a location is also present in B. bronchiseptica. The genes strA and strB are also found in Pasteurellaceae: plasmids and integrative and conjugative elements carrying strA, strB, and/or sul2 have been described (41, 42, 53, 55, 56). However, these genes seem to be ancient and have also been found in streptomycin-resistant bacteria from permafrost (57). Thus, it is not astonishing that these genes are present in a wide variety of bacterial genera.
FIGURE 2.
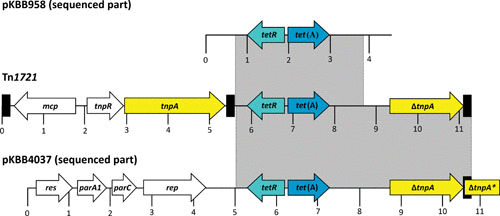
Comparison of Tn1721 (GenBank accession no. X61367) and the sequenced parts of the resistance plasmids pKBB958 (GenBank accession no. AM183165) and pKBB4037 (GenBank accession no. AJ877266) from B. bronchiseptica. A distance scale in kb is given below each map. The genes tetR, tet(A), mcp, tnpR, tnpA, ΔtnpA, res, parA1, parC, and ΔtnpA* are presented as arrows, with the arrowhead indicating the direction of transcription. The Δ symbol indicates a truncated, functionally inactive gene. The black boxes represent the terminal or internal 38-bp repeats of Tn1721. The gray shaded areas indicate the homologous parts between the B. bronchiseptica plasmids and Tn1721 (26).
CONCLUDING REMARKS
B. bronchiseptica is in general susceptible to most antimicrobial agents, which therefore can be used to treat clinical infections. In addition to taxonomy and the identification of novel species, as well as pathogenicity and immunization, in which B. bronchiseptica offers a lot of lessons to learn, antimicrobial resistance in B. bronchiseptica appears to be of less interest judging from the number of published studies. B. bronchiseptica has proved to be able to acquire resistance genes from other bacterial genera, especially from E. coli. The future will show whether B. bronchiseptica will gain further resistance genes directed against important or critically important antimicrobial agents.
ACKNOWLEDGMENTS
We acknowledge support for the work done on B. bronchiseptica from Heike Kaspar, Federal Office of Consumer Protection and Food Safety (BVL), Berlin, Germany.
REFERENCES
- 1.Warfel JM, Beren J, Kelly VK, Lee G, Merkel TJ. 2012. Nonhuman primate model of pertussis. Infect Immun 80:1530–1536 10.1128/IAI.06310-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gustavsson OE, Röken BO, Serrander R. 1990. An epizootic of whooping cough among chimpanzees in a zoo. Folia Primatol (Basel) 55:45–50 10.1159/000156498. [PubMed] [DOI] [PubMed] [Google Scholar]
- 3.Spilker T, Liwienski AA, LiPuma JJ. 2008. Identification of Bordetella spp. in respiratory specimens from individuals with cystic fibrosis. Clin Microbiol Infect 14:504–506 10.1111/j.1469-0691.2008.01968.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 4.Harrington AT, Castellanos JA, Ziedalski TM, Clarridge JE III, Cookson BT. 2009. Isolation of Bordetella avium and novel Bordetella strain from patients with respiratory disease. Emerg Infect Dis 15:72–74 10.3201/eid1501.071677. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodnow RA. 1980. Biology of Bordetella bronchiseptica. Microbiol Rev 44:722–738. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brockmeier SL, Halbur PG, Thacker EL. 2002. Porcine respiratory disease complex, p 231–258. In Brogden KA, Guthmiller JM (ed), Polymicrobial Diseases. ASM Press, Washington, DC. 10.1128/9781555817947.ch13 [DOI] [Google Scholar]
- 7.Magyar T, Lax AJ. 2002. Atrophic rhinitis, p 169–197. In Brogden KA, Guthmiller JM (ed), Polymicrobial Diseases. ASM Press, Washington, DC. 10.1128/9781555817947.ch10 [DOI] [PubMed] [Google Scholar]
- 8.OIE. 2012. Atrophic rhinitis of swine, chapter 2.8.2. In Terrestrial Manual 2012. OIE, Paris, France. [Google Scholar]
- 9.Rampelotto RF, Hörner A, Hörner C, Righi R, Hörner R. 2016. Pneumonia caused by Bordetella bronchiseptica in two HIV-positive patients. Sao Paulo Med J 134:268–272 10.1590/1516-3180.2015.02492701. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazumder SA, Cleveland KO. 2010. Bordetella bronchiseptica bacteremia in a patient with AIDS. South Med J 103:934–935 10.1097/SMJ.0b013e3181ebcdbc. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Register KB, Sukumar N, Palavecino EL, Rubin BK, Deora R. 2012. Bordetella bronchiseptica in a paediatric cystic fibrosis patient: possible transmission from a household cat. Zoonoses Public Health 59:246–250 10.1111/j.1863-2378.2011.01446.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gueirard P, Weber C, Le Coustumier A, Guiso N. 1995. Human Bordetella bronchiseptica infection related to contact with infected animals: persistence of bacteria in host. J Clin Microbiol 33:2002–2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woolfrey BF, Moody JA. 1991. Human infections associated with Bordetella bronchiseptica. Clin Microbiol Rev 4:243–255 10.1128/CMR.4.3.243. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dworkin MS, Sullivan PS, Buskin SE, Harrington RD, Olliffe J, MacArthur RD, Lopez CE. 1999. Bordetella bronchiseptica infection in human immunodeficiency virus-infected patients. Clin Infect Dis 28:1095–1099 10.1086/514761. [PubMed] [DOI] [PubMed] [Google Scholar]
- 15.Belen O, Campos JM, Cogen PH, Jantausch BA. 2003. Postsurgical meningitis caused by Bordetella bronchiseptica. Pediatr Infect Dis J 22:380–381 10.1097/01.inf.0000059766.51912.e8. [PubMed] [DOI] [PubMed] [Google Scholar]
- 16.Wernli D, Emonet S, Schrenzel J, Harbarth S. 2011. Evaluation of eight cases of confirmed Bordetella bronchiseptica infection and colonization over a 15-year period. Clin Microbiol Infect 17:201–203 10.1111/j.1469-0691.2010.03258.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 17.Mattoo S, Cherry JD. 2005. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev 18:326–382 10.1128/CMR.18.2.326-382.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.CLSI. 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard, 4th ed. CLSI document VET01-A4. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 19.CLSI. 2015. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; CLSI supplement VET01-S3. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 20.Prüller S, Frömke C, Kaspar H, Klein G, Kreienbrock L, Kehrenberg C. 2015. Recommendation for a standardised method of broth microdilution susceptibility testing for porcine Bordetella bronchiseptica. PLoS One 10:e0123883 10.1371/journal.pone.0123883. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarz S, Böttner A, Goossens L, Hafez HM, Hartmann K, Kaske M, Kehrenberg C, Kietzmann M, Klarmann D, Klein G, Krabisch P, Luhofer G, Richter A, Schulz B, Sigge C, Waldmann KH, Wallmann J, Werckenthin C. 2008. A proposal of clinical breakpoints for amoxicillin applicable to porcine respiratory tract pathogens. Vet Microbiol 126:178–188 10.1016/j.vetmic.2007.06.023. [PubMed] [DOI] [PubMed] [Google Scholar]
- 22.Kadlec K, Kehrenberg C, Wallmann J, Schwarz S. 2004. Antimicrobial susceptibility of Bordetella bronchiseptica isolates from porcine respiratory tract infections. Antimicrob Agents Chemother 48:4903–4906 10.1128/AAC.48.12.4903-4906.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadlec K, Kehrenberg C, Schwarz S. 2006. tet(A)-mediated tetracycline resistance in porcine Bordetella bronchiseptica isolates is based on plasmid-borne Tn1721 relics. J Antimicrob Chemother 58:225–227 10.1093/jac/dkl149. [PubMed] [DOI] [PubMed] [Google Scholar]
- 24.Prüller S, Rensch U, Meemken D, Kaspar H, Kopp PA, Klein G, Kehrenberg C. 2015. Antimicrobial susceptibility of Bordetella bronchiseptica isolates from swine and companion animals and detection of resistance genes. PLoS One 10:e0135703 10.1371/journal.pone.0135703. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El Garch F, de Jong A, Simjee S, Moyaert H, Klein U, Ludwig C, Marion H, Haag-Diergarten S, Richard-Mazet A, Thomas V, Siegwart E. 2016. Monitoring of antimicrobial susceptibility of respiratory tract pathogens isolated from diseased cattle and pigs across Europe, 2009-2012: VetPath results. Vet Microbiol 194:11–22 10.1016/j.vetmic.2016.04.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 26.Stepniewska K, Urbaniak K, Markowska-Daniel I. 2014. Phenotypic and genotypic characterization of Bordetella bronchiseptica strains isolated from pigs in Poland. Pol J Vet Sci 17:71–77 10.2478/pjvs-2014-0009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 27.GERM-Vet. 2016. BVL-Report · 10.5 Berichte zur Resistenzmonitoringstudie. ISBN 978-3-319-31696-3. Springer, Basel, Switzerland. (in German). [Google Scholar]
- 28.GERM-Vet. 2014. BVL-Report · 8.6 Berichte zur Resistenzmonitoringstudie. ISBN 978-3-319-05995-2. Springer, Basel, Switzerland. (in German). [Google Scholar]
- 29.GERM-Vet. 2012. Berichte zur Resistenzmonitoringstudie 2009. ISBN 978-3-0348-0504-9. Springer, Basel, Switzerland. (in German). [Google Scholar]
- 30.GERM-Vet. 2012. Berichte zur Resistenzmonitoringstudie 2008. ISBN 978-3-0348-0422-6. Springer, Basel, Switzerland. (in German). [Google Scholar]
- 31.Gerlach G, von Wintzingerode F, Middendorf B, Gross R. 2001. Evolutionary trends in the genus Bordetella. Microbes Infect 3:61–72 10.1016/S1286-4579(00)01353-8. [PubMed] [DOI] [PubMed] [Google Scholar]
- 32.Carbonetti NH, Wirsing von König CH, Lan R, Jacob-Dubuisson F, Cotter PA, Deora R, Merkel TJ, van Els CA, Locht C, Hozbor D, Rodriguez ME. 2016. Highlights of the 11th International Bordetella Symposium: from basic biology to vaccine development. Clin Vaccine Immunol 23:842–850 10.1128/CVI.00388-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimizu T. 1978. Prophylaxis of Bordetella bronchiseptica infection in guinea pigs by intranasal vaccination with live strain ts-S34. Infect Immun 22:318–321. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortensen JE, Brumbach A, Shryock TR. 1989. Antimicrobial susceptibility of Bordetella avium and Bordetella bronchiseptica isolates. Antimicrob Agents Chemother 33:771–772 10.1128/AAC.33.5.771. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Speakman AJ, Binns SH, Osborn AM, Corkill JE, Kariuki S, Saunders JR, Dawson S, Gaskell RM, Hart CA. 1997. Characterization of antibiotic resistance plasmids from Bordetella bronchiseptica. J Antimicrob Chemother 40:811–816 10.1093/jac/40.6.811. [PubMed] [DOI] [PubMed] [Google Scholar]
- 36.Kadlec K, Kaspar H, Mankertz J, Schwarz. 2012. First identification of tet(31) among tetracycline-resistant porcine Bordetella bronchiseptica. Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC) 16–19 June 2012, San Francisco, CA. [Google Scholar]
- 37.Dugan J, Rockey DD, Jones L, Andersen AA. 2004. Tetracycline resistance in Chlamydia suis mediated by genomic islands inserted into the chlamydial inv-like gene. Antimicrob Agents Chemother 48:3989–3995 10.1128/AAC.48.10.3989-3995.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadlec K, Kehrenberg C, Schwarz S. 2005. Molecular basis of resistance to trimethoprim, chloramphenicol and sulphonamides in Bordetella bronchiseptica. J Antimicrob Chemother 56:485–490 10.1093/jac/dki262. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Schwarz S, Alesík E, Grobbel M, Lübke-Becker A, Werckenthin C, Wieler LH, Wallmann J. 2007. Antimicrobial susceptibility of Pasteurella multocida and Bordetella bronchiseptica from dogs and cats as determined in the BfT-GermVet monitoring program 2004-2006. Berl Munch Tierarztl Wochenschr 120:423–430. [PubMed] [PubMed] [Google Scholar]
- 40.Sundin GW, Bender CL. 1996. Dissemination of the strA-strB streptomycin-resistance genes among commensal and pathogenic bacteria from humans, animals, and plants. Mol Ecol 5:133–143 10.1111/j.1365-294X.1996.tb00299.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 41.Kehrenberg C, Tham NT, Schwarz S. 2003. New plasmid-borne antibiotic resistance gene cluster in Pasteurella multocida. Antimicrob Agents Chemother 47:2978–2980 10.1128/AAC.47.9.2978-2980.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michael GB, Kadlec K, Sweeney MT, Brzuszkiewicz E, Liesegang H, Daniel R, Murray RW, Watts JL, Schwarz S. 2012. ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: analysis of the regions that comprise 12 antimicrobial resistance genes. J Antimicrob Chemother 67:84–90 10.1093/jac/dkr406. [PubMed] [DOI] [PubMed] [Google Scholar]
- 43.Toulouse JL, Edens TJ, Alejaldre L, Manges AR, Pelletier JN. 2017. Integron-associated DfrB4, a previously uncharacterized member of the trimethoprim-resistant dihydrofolate reductase B family, is a clinically identified emergent source of antibiotic resistance. Antimicrob Agents Chemother 61:5 10.1128/AAC.02665-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kadlec K, Kehrenberg C, Schwarz S. 2007. Efflux-mediated resistance to florfenicol and/or chloramphenicol in Bordetella bronchiseptica: identification of a novel chloramphenicol exporter. J Antimicrob Chemother 59:191–196 10.1093/jac/dkl498. [PubMed] [DOI] [PubMed] [Google Scholar]
- 45.Barreto-Hernández E, Falquet L, Reguero MT, Mantilla JR, Valenzuela EM, González E, Cepeda A, Escalante A. 2013. Draft genome sequences of multidrug-resistant Acinetobacter sp. strains from Colombian hospitals. Genome Announc 1:e00868-e13 10.1128/genomeA.00868-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lartigue MF, Poirel L, Fortineau N, Nordmann P. 2005. Chromosome-borne class A BOR-1 β-lactamase of Bordetella bronchiseptica and Bordetella parapertussis. Antimicrob Agents Chemother 49:2565–2567 10.1128/AAC.49.6.2565-2567.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kadlec K, Wiegand I, Kehrenberg C, Schwarz S. 2007. Studies on the mechanisms of β-lactam resistance in Bordetella bronchiseptica. J Antimicrob Chemother 59:396–402 10.1093/jac/dkl515. [PubMed] [DOI] [PubMed] [Google Scholar]
- 48.Hannan PC, O’Hanlon PJ, Rogers NH. 1989. In vitro evaluation of various quinolone antibacterial agents against veterinary mycoplasmas and porcine respiratory bacterial pathogens. Res Vet Sci 46:202–211. [PubMed] [PubMed] [Google Scholar]
- 49.Speakman AJ, Dawson S, Corkill JE, Binns SH, Hart CA, Gaskell RM. 2000. Antibiotic susceptibility of canine Bordetella bronchiseptica isolates. Vet Microbiol 71:193–200 10.1016/S0378-1135(99)00171-6. [PubMed] [DOI] [PubMed] [Google Scholar]
- 50.Carbone M, Pennisi MG, Masucci M, De Sarro A, Giannone M, Fera MT. 2001. Activity and postantibiotic effect of marbofloxacin, enrofloxacin, difloxacin and ciprofloxacin against feline Bordetella bronchiseptica isolates. Vet Microbiol 81:79–84 10.1016/S0378-1135(01)00349-2. [PubMed] [DOI] [PubMed] [Google Scholar]
- 51.Schink AK, Kadlec K, Hauschild T, Brenner Michael G, Dörner JC, Ludwig C, Werckenthin C, Hehnen HR, Stephan B, Schwarz S. 2013. Susceptibility of canine and feline bacterial pathogens to pradofloxacin and comparison with other fluoroquinolones approved for companion animals. Vet Microbiol 162:119–126 10.1016/j.vetmic.2012.08.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 52.El Garch F, Kroemer S, Galland D, Morrissey I, Woehrle F. 2017. Survey of susceptibility to marbofloxacin in bacteria isolated from diseased pigs in Europe. Vet Rec 180:591 10.1136/vr.103954. [PubMed] [DOI] [PubMed] [Google Scholar]
- 53.Kehrenberg C, Schwarz S. 2002. Nucleotide sequence and organization of plasmid pMVSCS1 from Mannheimia varigena: identification of a multiresistance gene cluster. J Antimicrob Chemother 49:383–386 10.1093/jac/49.2.383. [PubMed] [DOI] [PubMed] [Google Scholar]
- 54.Chiou CS, Jones AL. 1993. Nucleotide sequence analysis of a transposon (Tn5393) carrying streptomycin resistance genes in Erwinia amylovora and other Gram-negative bacteria. J Bacteriol 175:732–740 10.1128/jb.175.3.732-740.1993. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.San Millan A, Escudero JA, Gutierrez B, Hidalgo L, Garcia N, Llagostera M, Dominguez L, Gonzalez-Zorn B. 2009. Multiresistance in Pasteurella multocida is mediated by coexistence of small plasmids. Antimicrob Agents Chemother 53:3399–3404 10.1128/AAC.01522-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eidam C, Poehlein A, Leimbach A, Michael GB, Kadlec K, Liesegang H, Daniel R, Sweeney MT, Murray RW, Watts JL, Schwarz S. 2015. Analysis and comparative genomics of ICEMh1, a novel integrative and conjugative element (ICE) of Mannheimia haemolytica. J Antimicrob Chemother 70:93–97 10.1093/jac/dku361. [PubMed] [DOI] [PubMed] [Google Scholar]
- 57.Petrova M, Kurakov A, Shcherbatova N, Mindlin S. 2014. Genetic structure and biological properties of the first ancient multiresistance plasmid pKLH80 isolated from a permafrost bacterium. Microbiology 160:2253–2263 10.1099/mic.0.079335-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 58.Wallmann J, Kaspar H, Kroker R. 2004. [The prevalence of antimicrobial susceptibility of veterinary pathogens isolated from cattle and pigs: national antibiotic resistance monitoring 2002/2003 of the BVL]. Berl Munch Tierarztl Wochenschr 117:480–492. The prevalence of antimicrobial susceptibility of veterinary pathogens isolated from cattle and pigs: national reistance monitoring 2002/2003 of the BVL. [PubMed] [Google Scholar]
- 59.Shin SJ, Kang SG, Nabin R, Kang ML, Yoo HS. 2005. Evaluation of the antimicrobial activity of florfenicol against bacteria isolated from bovine and porcine respiratory disease. Vet Microbiol 106:73–77 10.1016/j.vetmic.2004.11.015. [PubMed] [DOI] [PubMed] [Google Scholar]
- 60.Schwarz S, Kadlec K, Silley P. 2013. Antimicrobial Resistance in Bacteria of Animal Origin. ZETT-Verlag, Steinen, Germany. [Google Scholar]
- 61.Kehrenberg C, Mumme J, Wallmann J, Verspohl J, Tegeler R, Kühn T, Schwarz S. 2004. Monitoring of florfenicol susceptibility among bovine and porcine respiratory tract pathogens collected in Germany during the years 2002 and 2003. J Antimicrob Chemother 54:572–574 10.1093/jac/dkh371. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.Priebe S, Schwarz S. 2003. In vitro activities of florfenicol against bovine and porcine respiratory tract pathogens. Antimicrob Agents Chemother 47:2703–2705 10.1128/AAC.47.8.2703-2705.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]


