ABSTRACT
There is broad consensus internationally that surveillance of the levels of antimicrobial resistance (AMR) occurring in various systems underpins strategies to address the issue. The key reasons for surveillance of resistance are to determine (i) the size of the problem, (ii) whether resistance is increasing, (iii) whether previously unknown types of resistance are emerging, (iv) whether a particular type of resistance is spreading, and (v) whether a particular type of resistance is associated with a particular outbreak. The implications of acquiring and utilizing this information need to be considered in the design of a surveillance system. AMR surveillance provides a foundation for assessing the burden of AMR and for providing the necessary evidence for developing efficient and effective control and prevention strategies. The codevelopment of AMR surveillance programs in humans and animals is essential, but there remain several key elements that make data comparisons between AMR monitoring programs, and between regions, difficult. Currently, AMR surveillance relies on uncomplicated in vitro antimicrobial susceptibility methods. However, the lack of harmonization across programs and the limitation of genetic information of AMR remain the major drawbacks of these phenotypic methods. The future of AMR surveillance is moving toward genotypic detection, and molecular analysis methods are expected to yield a wealth of information. However, the expectation that these molecular techniques will surpass phenotypic susceptibility testing in routine diagnosis and monitoring of AMR remains a distant reality, and phenotypic testing remains necessary in the detection of emerging resistant bacteria, new resistance mechanisms, and trends of AMR.
INTRODUCTION
There is broad consensus internationally that surveillance of the levels of antimicrobial resistance (AMR) occurring in various systems underpins strategies to address the issue. The key reasons for surveillance of resistance are to determine (i) the size of the problem, (ii) whether resistance is increasing, (iii) whether previously unknown types of resistance are emerging, (iv) whether a particular type of resistance is spreading, and (v) whether a particular type of resistance is associated with a particular outbreak. The implications of acquiring and utilizing this information need to be considered in the design of a surveillance system.
HISTORICAL BACKGROUND
Nearly 50 years ago Anderson (1) reported a spike in infections caused by multidrug-resistant Salmonella enterica serovar Typhimurium phage type 29 in calves in the United Kingdom. Such infections became prominent following the adoption of intensive farming practices such as profligate antibiotic use in feed without veterinary prescription. Many human infections resulted. Anderson (1) concluded that the Salmonella epidemic could be eliminated “not by the massive use of antibiotics but by improvement in conditions of animal husbandry and reduction in the opportunities for the initiation and spread of the disease.” In light of the public health implications of this finding, the Swann Report recommended restricting the use of antimicrobials in livestock production and imposing greater veterinary oversight (2). A national AMR monitoring program for Salmonella in animals commenced in the United Kingdom in 1970 (3), and surveys were conducted in other countries (4). Martel and Coudert (5) reported on the national surveillance of AMR in animal (mainly of bovine origin) isolates of Salmonella and Escherichia coli, which had been in operation in France since 1969. Although there is scant reference in the literature to national AMR surveillance in animals for the next 20 years, reviews by Cohen and Tauxe (6) and DuPont and Steele (7) confirmed that the use of antimicrobials in livestock contributed to increased AMR in foodborne Salmonella. DuPont and Steele (7) recommended national monitoring of the use of antimicrobials in food-producing animals. They also recommended molecular epidemiological investigation to determine whether antimicrobial-resistance determinants arising in food animals as a result of antibiotic use, either by direct cross-infection or through horizontal transfer, influences the pool of genetic resistance determinants that are important to human health. Drug resistance in Salmonella was once again identified as the most significant public health threat arising from the use of antibiotics in livestock.
In the late 1980s and 1990s, third-generation cephalosporins and fluoroquinolones became widely available for use in food-producing animals. In 1996, following the emergence of multidrug-resistant Salmonella Typhimurium DT104, S. enterica serovar Newport resistant to third-generation cephalosporins, and fluoroquinolone-resistant Campylobacter, the Centers for Disease Control (CDC), the U.S. Department of Agriculture, and the Food and Drug Administration (FDA) established the National Antimicrobial Resistance Monitoring System (NARMS). Its purpose was to monitor changes in antimicrobial susceptibilities of zoonotic pathogens from human and animal clinical specimens and carcasses of food-producing animals at slaughter plants over time to assess the effectiveness of intervention strategies (8). At the same time, in response to the emergence of vancomycin-resistant enterococci in poultry and pigs in Europe related to avoparcin use, The Danish Integrated Antimicrobial Resistance Monitoring Program (DANMAP) was launched and established baseline levels of resistance in pathogenic, zoonotic, and indicator bacteria of pigs, cattle, and broilers (9). The AMR surveillance strategies and recommendations of these foundational programs have since been adopted in many countries throughout the world (e.g., 10; selected case studies indicated below), and in many cases, linked with antimicrobial use monitoring and human AMR surveillance programs.
CODEVELOPMENT WITH AMR SURVEILLANCE IN HUMANS
Antimicrobial susceptibility testing (AST) for surveillance of AMR in animals essentially evolved from existing surveillance programs focused on human pathogens, with several key differentiating characteristics. While both programs utilize the same AST methodology, namely MIC testing utilizing agar dilution or microbroth dilution techniques, there are major differences in breakpoint determination, types of bacteria screened, and interpretation. Within the animal arena, application of different terminologies, techniques, and clinical breakpoints versus epidemiological cutoff values (ECOFFs) has meant that it has been difficult to compare data across national programs, and sometimes within programs over time as breakpoints are re-evaluated or created and methodological advances made.
SIMILARITIES AND DIFFERENCES BETWEEN HUMAN AND ANIMAL AMR SURVEILLANCE PROGRAMS
In human AMR surveillance programs, AST is performed on major bacterial pathogens, collected from primary accession clinical microbiology laboratories. As such, these isolates can be considered convenience samples obtained from either hospitalized patients or outpatients in the community for which a culture and susceptibility test has been requested by the referring clinician. The site of infection is important because the breakpoints used to classify the isolate as susceptible or resistant may differ between an isolate from the urinary tract and a skin or soft tissue infection depending on the pharmacokinetics of the antimicrobial agent. The main focus of programs also changes over time, often in response to international attempts to harmonize and integrate AST data. For example, in Australia, yearly AMR surveillance of human Gram-negative pathogens focused on hospital-acquired infections in odd-numbered years and community-acquired infections in even-numbered years. However, from 2013, surveillance only focused on sepsis or blood isolates, regardless of the source. In many cases, if the primary accession laboratories utilize the same testing methodology and quality assurance, testing indicates excellent agreement between laboratories (such as use of automated MIC determination technology), the AST data can be collected passively from clinical microbiology laboratory records, and surveillance activities can focus largely on the further characterization and molecular epidemiology of isolates, such as resistance mechanism screening and multilocus sequence typing. Many of these epidemiological characteristics can now be obtained through whole-genome sequence analysis (discussed below), with genomic epidemiology rapidly becoming an important feature of many global surveillance programs.
AMR surveillance in animal pathogens closely mirrors human surveillance programs in that it is focused on isolates obtained from clinical infections in animals. This includes both animal-only pathogens (e.g., Mannheimia haemolytica) and zooanthroponotic pathogens (e.g., methicillin-resistant Staphylococcus aureus [MRSA]). However, there is often much more variability in AST methodology applied to the isolates, with many veterinary diagnostic laboratories still being heavily reliant on disc diffusion methodology because of its rapidity and price. Automated MIC testing is increasingly available in veterinary laboratories but is often an additional cost for the client. Therefore, AMR surveillance focused on animal pathogens still often relies on centralized collection of isolates by an independent reference laboratory, which then conducts “gold standard” AST (MIC testing), making this a very expensive option for regular ongoing surveillance programs. Where veterinary diagnostic laboratory testing is well coordinated and methodologically similar, such as in France’s RESAPATH program, passive collection of animal pathogen AST data is possible and has generated some useful information for antimicrobial stewardship policy and procedures (see example below). Surveillance of animal pathogens directly relates to antimicrobial use in companion animals (defined here as cats and dogs), performance animals (mainly horses, though in some countries horses are food-producing animals), livestock predominantly raised for food and fiber production and, potentially, exotic animals in zoological collections.
AMR surveillance of animals diverges from human surveillance in the development of programs aimed at AMR monitoring of zoonotic foodborne and commensal bacteria that are present in the gastrointestinal tract of food-producing animals at slaughter. In these cases, an epidemiological sampling strategy is used to obtain representative samples postslaughter and isolate key bacteria that are either directly associated with foodborne illness or are indicators or potential reservoirs of AMR determinants. The direct emphasis is monitoring resistance in bacteria from a public health and food safety perspective rather than having a direct benefit to animal health and production. To be included in a targeted livestock surveillance program, the microorganism must either be a commensal of animals that has the potential to provide sentinel information on trends and emergence of AMR, or it is a pathogen of animals and/or humans that has the potential to develop or is known to have developed AMR that is of concern to human health, and its primary route of infection is foodborne.
PRESENT MONITORING OF AMR IN ANIMALS: PRINCIPLES AND PRACTICES
Recommendations for implementation of national surveillance of AMR in animals have been developed by the World Organization for Animal Health, the World Health Organization Advisory Group on Integrated Surveillance of Antimicrobial Resistance, and the EFSA Working Group on Developing Harmonised Schemes for Monitoring Antimicrobial Resistance in Zoonotic Agents (11). In summary, these recommendations cover how to design sampling strategies, target animal and bacterial species, determine which antimicrobials to test, conduct AST and interpret results, and present the data. Some of these points will briefly be covered below.
Scope: Animal Species
AMR in animal and zooanthroponotic pathogens
Major animal species targeted for AMR monitoring in animal pathogens are usually divided into companion animals (i.e., dogs and cats), performance animals, (i.e., horses), livestock including aquaculture species produced for food), and exotics and wildlife. Comparison among animal species can be problematic due to the differences between clinical breakpoints and site of infection (e.g., skin and soft tissue versus urine), the fact that there are no animal-specific breakpoints for some antimicrobial agents (in which case human breakpoints are used by default), and overall relevance to animal health versus public health.
AMR in zoonotic foodborne and commensal indicator bacteria
Cattle, pigs, and poultry provide the top three sources of meat; are critical in the maintenance of supplies of high-quality, low-cost food for human consumption, providing 13% of human calories and 30% of protein consumption; and produce around 40% of global gross domestic product (12). Most livestock-associated AMR surveillance programs focus on these three main sources. Cattle can be further subdivided into grass-fed (extensive) and lot-fed (intensive) beef cattle, cull dairy cattle, and veal (dairy calf) production systems. Antimicrobial use in each beef production sector varies considerably, from virtually no antimicrobial treatments in grass-fed cattle to a high proportion of animals receiving at least one treatment in feedlots. Poultry production includes both domestic broilers and turkeys, but commercial and free-range laying flocks are also of significant interest from an AMR standpoint given that eggs are a primary source of Salmonella food poisoning in many countries and there is only a restricted range of antimicrobial agents that can be used in egg-producing birds. The aquaculture industry, while not the largest in terms of food production in many countries, is the fastest-growing sector, with consumption expected to continue to increase in the coming decades (13). High antimicrobial use was a feature of the industry in its infancy, but as the industry has matured, control of endemic bacterial diseases by vaccination has seen antimicrobial use in some countries drop to very low levels (14). Other minor food animal species that are sometimes considered in country-specific AMR monitoring programs in livestock include horses, rabbits, and wild game species.
Scope: Antimicrobials
Inclusion of an antimicrobial drug in an AMR surveillance program depends upon whether it has been used, currently or historically, in animals to such a degree that development of AMR has been demonstrated or is of future concern to human health, particularly when the antimicrobial is within a class that is deemed to be of critical importance (Table 1). The final list of antimicrobials that are screened can vary considerably between countries and programs and can often change between annual reports as new resistance mechanisms are identified or more streamlined approaches are adopted. As an example, the DANMAP 2015 (15) report screened Salmonella and Escherichia coli isolates for susceptibility to the same 14 antimicrobial agents mainly used in human medicine, whereas in DANMAP 2011 (16) the numbers were 16 and 18 antimicrobials, respectively. The main differences were the historical inclusion of several animal-only antimicrobials such as apramycin and florfenicol, which are no longer included in screening programs. Furthermore, the recent identification of plasmid-mediated mechanisms for resistance to critically important antimicrobials in animal commensal isolates (e.g., Mcr-1 mediated transferable colistin resistance [17] and carbapenemases normally associated with human pathogens [18, 19]) has renewed efforts to ensure that livestock AMR surveillance programs include “last-resort” drug classes in their screening panels. This is particularly the case in countries where there has been reported use of colistin in livestock production, where the mcr-1 gene remained undetected for a number of years until its chance discovery in humans and livestock.
TABLE 1.
Antimicrobial classes and agents registered for human and veterinary use that are often screened in antimicrobial resistance surveillance programsa,b
| Antibacterial class and antibacterial (use in AMR surveillance) | Principal human use | Principal animal use |
|---|---|---|
| Narrow-spectrum penicillins | ||
| Benzylpenicillin (pen G) and phenoxymethylpenicillin (pen V) (AP) | Primary agents in pneumococcal and streptococcal infection | N/A |
| Procaine penicillin | Intramuscular—occasional substitute for benzylpenicillin | Primary agent for predominantly Gram-positive infections in a wide range of animals, mostly horses (often in combination with gentamicin) and livestock (intramuscular administration only) |
| Benzathine penicillin | Intramuscular—syphilis treatment and rheumatic fever prophylaxis | N/A |
| Penethemate hydriodide | N/A | Hydrolized to benzylpenicillin following injection for treatment of mastitis and respiratory and uterine infections, mainly in dairy cattle |
| Moderate-spectrum penicillins | ||
| Amoxycillin and ampicillin (AP, ZFP, AC) | Principal role in respiratory tract infections; widespread i.v. hospital use in combination for a range of moderate and serious infections; surgical and endocarditis prophylaxis | Broad-spectrum primary agent for a large range of infections in dogs and cats, horses, and livestock (oral or injectable) |
| Antistaphylococcal penicillin | ||
| Cloxacillin, dicloxacillin, and flucloxacillin (methicillin)Oxacillin (AP, ZP [MRSA/MRSP only]) | Standard treatment for Staphylococcus aureus infections (not MRSA)Surgical prophylaxis, especially orthopedics | Cloxacillin only: intramammary treatment of mastitis due to staphylococci and streptococci in dairy cattleOxacillin susceptibility used as a surrogate for MRSP identification in VDLs |
| Beta-lactamase inhibitor combinations | ||
| Amoxycillin-clavulanate (AP, ZP, ZFP, AC) | Second-line agent for respiratory tract infections; role in certain types of skin/soft tissue infections and mixed staphylococcal/Gram-negative infections and aerobic/anaerobic infections | Primary or second-line broad-spectrum agent in dogs and cats only (oral and injectable) for a wide range of infections (skin, soft tissue, and UTI)Intramammary formulation only for mastitis in dairy cattle |
| Piperacillin-tazobactam (AP) | Valuable agents for a range of severe mixed aerobic-anaerobic infections including intra-abdominal infections, aspiration pneumonia, skin/soft tissue infections.Neutropenic sepsis | N/A |
| First-generation cephalosporins | ||
| Cephalexin, cephalothin, and cephazolin (AP, ZP, ZFP, AC) | Treatment of minor and staphylococcal infections in penicillin-allergic patients Prophylaxis in orthopedic and other surgery | Primary agent for skin, soft tissue, and UTIs as well as surgical prophylaxis in dogs and cats only |
| Cephalonium/cephapirin | N/A | Intramammary treatment of mastitis due to staphylococci and streptococci in dairy cattle/intrauterine treatment for metritis in cattle |
| Second-generation cephalosporins and cephamycins | ||
| Cefaclor and cefuroxime-axetil | Treatment of respiratory infections in penicillin-allergic patients | Intramammary treatment of mastitis due to staphylococci and streptococci in dairy cattle |
| Cefoxitin (AP, ZP, ZFP, AC) | Useful antianaerobic activity, major role in surgical prophylaxis | N/A |
| Third-generation cephalosporins | ||
| Ceftriaxone (AP, ZP, ZFP, AC) | Major agent in severe pneumonia and meningitisUsed in selected cases for treatment of gonorrhea and alternative for prophylaxis of meningococcal infection | N/A |
| Cefotaxime (AP, ZP, ZFP, AC) | Major agent in severe pneumonia and meningitis | N/A |
| Ceftazidime (AP, ZP, ZFP, AC) | Restricted role in pseudomonal infection and neutropenic sepsis | N/A |
| Cefovecin (AP, ZP) | N/A | Reserve agent for skin, soft tissue, periodontal, and UTIs in dogs and cats only where compliance with oral medication is compromised (injection only) |
| Ceftiofur (AP, ZP, ZFP, AC) | N/A | Reserve agent for respiratory infections in cattleOff-label use for infections resistant to first-line therapies in individual food-producing animals (injection only) |
| Cefpodoxime (AP) | Broad-spectrum oral third-generation cephalosporin available in some countries | Reserve oral agent for skin, soft tissue, periodontal, and UTIs in dogs and cats only |
| Fourth-generation cephalosporins | ||
| Cefepime | Moderate-to-severe pneumonia, skin and soft tissue infections, complicated and uncomplicated UTIs (broad-spectrum) | N/A |
| Cefquinome (AP) | N/A | Reserve agent for respiratory infections in cattle and pigs, coliform mastitis in cattle |
| Carbapenems | ||
| Imipenem (AP, ZP, ZFP, AC), meropenem (AP, ZP, ZFP, AC), doripenem, and ertapenem | Very broad-spectrum reserve agents for multiresistant and serious Gram-negative and mixed infections | Use as a last resort option for multi-resistant Gram-negative infections in dogs has been reported |
| Tetracyclines/glycylcyclines | ||
| Doxycycline (AP), minocycline (AP), and demeclocycline | Major agents for minor respiratory tract infections and acneSupportive role in pneumonia for treating Mycoplasma and Chlamydia pneumoniae Malaria prophylaxis (doxycycline) | Doxycycline only: major primary agent for respiratory skin, soft tissue, urinary tract, and periodontal infections in dogs and cats including Mycoplasma and Chlamydia (oral only); occasional use of minocycline for MRSP |
| Chlortetracycline, oxytetracycline, and tetracycline (AP, ZP, ZFP, AC) | N/A | Major broad-spectrum primary agent for systemic infections in livestock |
| Tigecycline (ZFP, AC) | Reserve agent for multiresistant Gram-positive and some multiresistant Gram-negative infections | N/A |
| Glycopeptides | ||
| Vancomycin (AP, ZP, AC) | Drug of choice for serious methicillin-resistant staphylococcal infectionsReserve agent for enterococcal infection when there is resistance or penicillin allergy | N/A |
| Teicoplanin (AC) | Substitute for vancomycin if intolerance or outpatient i.v. therapyvanB vancomycin-resistant enterococcal infections | N/A |
| Aminoglycosides/aminocyclitols | ||
| Neomycin (including framycetin) (AP, AC) | Topical agent for skin infection and gut suppression. | Primary agent for enteric infections in livestock (oral form); broad-spectrum primary agent for a range of systemic infections in livestock and horses (parenteral form) |
| Gentamicin and tobramycin (AP, ZP, ZFP, AC) | Standard agents in combination for serious and pseudomonal infectionGentamicin used in combination for endocarditis | Gentamicin only: primary agent for broad-spectrum infections in horses (with penicillin); primary agent for short-term treatment of serious/life threatening infections in dogs and cats due to nephrotoxicityCannot be administered to livestock in Australia |
| Amikacin (AP, ZP, ZFP, AC) | Reserve agents for Gram-negatives resistant to gentamicin and tobramycin | Use as a last-resort option for multiresistant infections in companion animalsUse as a reserve agent for gentamicin-resistant infections in horses |
| Spectinomycin (AP, AC) | Spectinomycin only used for gonorrhea (infrequently) | Primary agent in combination with lincomycin for gastrointestinal and respiratory infections in pigs and broilers including mycoplasma (oral and injectable) |
| Streptomycin (AP, ZP, ZFP, AC) | Rare use in treatment of TB and enterococcal endocarditis | N/A |
| Apramycin (AP) | N/A | Primary agent for E. coli and Salmonella infections in calves, pigs, and broilers |
| Dihydrostreptomycin | N/A | Banned in livestock (except in oral or intramammary preparations) due to residue issues (apart from treatment of acute leptospirosis in cattle) |
| Sulfonamides and DHFR inhibitors | ||
| Trimethoprim (AP, ZP, ZFP, AC) | Treatment and prophylaxis of UTI | N/A |
| Trimethoprim-sulfamethoxazole (co-trimoxazole) (AP, ZP, ZFP, AC) | Minor infections, especially treatment and prophylaxis of UTIStandard for treatment and prophylaxis of Pneumocystis carinii infection and nocardiasisImportant for community-acquired MRSA infections | Trimethoprim-sulphonamide combinations are used as primary agents for broad-spectrum infections in livestock, horses, and dogs, including enteritis and pneumonia (oral and injectable) |
| Sulfadiazine, sulfadoxine, and sulfaquinoxaline | N/A | Oral sulfonamides (without trimethoprim) are also used for coccidiosis in poultry |
| Oxazolidinones | ||
| Linezolid (AP, ZP, AC) | Treatment of multiresistant Gram-positive infections, especially MRSA and VRE. | N/A |
| Macrolides | ||
| Azithromycin (ZFP) | Treatment of Chlamydia trachomatis infectionsMajor agent for treatment and suppression of atypical mycobacterial infection | Occasional use in dogs and cats for chlamydia/mycoplasma infection and foals for Rhodococcus infection (see erythromycin) |
| Clarithromycin | Treatment of minor Gram-positive infectionsMajor agent for treatment and suppression of atypical mycobacterial infection | Occasional use in dogs and cats for chlamydia/mycoplasma infection and foals for Rhodococcus infection (see erythromycin) |
| Erythromycin and roxithromycin (AP, ZFP, AC) | Treatment of minor Gram-positive, Chlamydia and Mycoplasma infections | Erythromycin only: livestock for respiratory infections and other serious systemic infections including mastitis; respiratory disease in broilers; administered to foals in combination with rifampicin for Rhodococcus infection |
| Spiramycin | Treatment of toxoplasmosis in pregnancy | Periodontal and other anaerobic infections in dogs and cats (with metronidazole) |
| Oleandomycin | N/A | Intramammary formulation in combination with neomycin and tetracycline for mastitis |
| Tulathromycin, gamithromycin, and tildipirosin (AP) | N/A | Primary agent for respiratory infections in cattle and pigs |
| Tilmicosin, tylosin, and kitasamycin (AP) | N/A | Primary agent for respiratory infections in cattleTreatment and prevention of enteritis and respiratory diseases in cattle, poultry, and pigs (especially Lawsonia infection)Growth promotion in pigs |
| Lincosamides | ||
| Clindamycin and lincomycin (AP) | Reserved for Gram-positive and anaerobic infections in penicillin-allergic patientsClindamycin topical used for acne | Clindamycin: Gram-positive and anaerobic infections in dogs and cats including osteomyelitisLincomycin: oral or injectable in livestock for respiratory and enteric infections (often in combination with spectinomycin) |
| Streptogramins | ||
| Quinupristin with dalfopristin (AC) | Reserve agent for multiresistant Gram-positive infections (MRSA and vancomycin-resistant Enterococcus faecium). | N/A |
| Pristinamycin | As for quinupristin-dalfopristin | N/A |
| Virginiamycin | N/A | Laminitis prevention in horses, rumen acidosis prevention in cattle, necrotic enteritis prevention in broilers |
| Quinolones/Fluoroquinolones | ||
| Naladixic acid (ZFP, AC)Ciprofloxacin (AP, ZP, ZFP, AC) | First-generation quinolone no longer used in human medicineMajor oral agent for the treatment of Gram-negative infections resistant to other agentsMinor role in meningococcal prophylaxis | N/A (often included in AMR surveillance as an indicator of reduced susceptibility to the quinolone class)N/A |
| Moxifloxacin | Restricted role in the management of serious respiratory infections, especially pneumonia in patients with severe penicillin allergy | N/A |
| Enrofloxacin, marbofloxacin, and pradofloxacin (AP, ZP) | N/A | Reserve agents for treatment of Gram-negative serious, chronic or life-threatening infections in dogs, cats, and occasionally horses and exotics, treatment of complicated pyoderma due to mixed infectionsRespiratory infections in feedlot cattle; historic use in poultryCannot be administered to food-producing animals in Australia |
| Ansamycins | ||
| Rifampicin (rifampin) (AP, ZP) | Meningococcal and H. influenzae type b prophylaxisStandard part of TB regimensImportant oral agent in combination for MRSA infections | Used in combination with a macrolide for treatment of Rhodococcus infection in foals |
| Bacitracin and gramicidin | Topical agents with Gram-positive activity | Treatment and prevention of necrotic enteritis in poultry, topical agents for mucocutaneous infections in companion animals (Gram-positive) |
| Polymyxins | ||
| Colistin (ZFP, AC) | Reserve agent for very multiresistant Gram-negative infection (both inhaled and intravenous) | N/A |
| Phenicols | ||
| Chloramphenicol (AP, ZP, ZFP, AC) | Usage largely as topical eye preparationOccasional need for the treatment of bacterial meningitis. | Reserve agent for multiresistant infections in companion animals (dogs and cats only), especially E. coli and MRSP |
| Florfenicol (AP, ZP, ZFP, AC) | N/A | Respiratory infections in cattle and pigsOff-label use for multiresistant E. coli in pigs |
| Nitrofurans | ||
| Nitrofurantoin (AP) | Treatment and prophylaxis of UTIs only | N/A |
| Lipopeptides | ||
| Daptomycin (AP, ZP, AC) | Reserve agent for serious MRSA and VRE infections | N/A |
The table focusses on animal pathogens (AP), zooanthroponotic pathogens (ZP), zoonotic foodborne pathogens (ZFP), and animal commensal indictor organisms (AC). Adapted from Shaban et al. (11).
N/A, not applicable; i.v., intravenous; MRSP, methicillin-resistant Staphylococcus pseudintermedius; VDL, veterinary diagnostic laboratories; DHFR, dihydrofolate reductase; UTI, urinary tract infection; VRE, vancomycin-resistant enterococci; MRSP, methicillin-resistant Staphylococcus pseudintermedius.
Scope: Microorganisms
There are four distinct categories of bacteria that can be isolated from animals for inclusion in AMR monitoring programs. This includes animal-only pathogens and/or zooanthroponotic pathogens usually isolated from sick companion/performance animals and livestock, as well as zoonotic foodborne and indicator commensal bacteria isolated from healthy livestock at slaughter. The main organisms targeted for AST in each category are described below and summarized in Table 2.
TABLE 2.
Microorganisms of interest in AMR monitoring programs focused on both zoonotic foodborne pathogens and commensals in healthy livestock and major animal pathogensa,b
| Organism | Animal context | Human context |
|---|---|---|
| Methicillin-resistant Staphylococcus pseudintermedius (primary animal pathogen) | Recently emerged and spread epidemically in companion animals | Infections in humans are rare, can be a reservoir of SCCmec-associated resistance genes |
| Enterotoxigenic Escherichia coli, Mannheimia haemolytica, Pasteurella multocida, Histophilus somnus, Actinobacillus pleuropneumoniae (primary animal pathogens) | Relevant to pig and veal production, may drive use of third-generation cephalosporins; major reason for antimicrobial use in feedlot cattle and pigs; resistance to first-choice antimicrobials could result in increased use of third-generation cephalosporins and fluoroquinolones | Multidrug-resistant strains coming through the food chain would drive use of broad-spectrum cephalosporins and carbapenems in humans; uncommon to rare human pathogens |
| MRSA (zooanthroponotic pathogen/commensal in animals) | Livestock-associated MRSA; some human MRSA subtypes now adapted to animal hosts (i.e., horses and dogs) | Major human AMR surveillance organism; veterinarians in clinical practice have a higher rate of MRSA nasal carriage than the general population |
| ExPEC (zooanthroponotic pathogen/commensal in animals) | Some human-associated multidrug-resistant subtypes (e.g., ST131, ST648, ST354) can colonize and cause infections in dogs | Major human AMR surveillance organism; similarity between canine, avian, and human strains carrying ESBLs suggests potential for cross-transmission |
| Multidrug-resistant Salmonella (e.g., S. enterica serovars Newport, Typhimurium) (zoonotic foodborne pathogens) | Propensity to develop AMR under antimicrobial selection pressure in livestock production; in particular, resistance to critically important antimicrobials | Eggs and meat often implicated in outbreaks; invasive disease in humans often treated with fluoroquinolones in adults and third-generation cephalosporins in children |
| Fluoroquinolone-resistant Campylobacter (zoonotic foodborne pathogen) | Major foodborne risk bacteria associated with poultry; fluoroquinolone use a major selection pressure in livestock systems | Undercooked poultry meat and cross-contamination of fresh food often implicated in outbreaks; macrolides (in children) and fluoroquinolones are the treatments of choice for complicated infections |
| Commensal Enterococcus spp. (commensal indicator organism in livestock)Commensal E. coli. (commensal indicator organism in livestock) | Gram-positive indicator organism in many surveillance programs; vancomycin resistance related to avoparcin use; streptogramin resistance related to virginiamycin use.Have been shown to be reservoirs of plasmid-associated resistance of public health significance (e.g., ESBL and plasmid-borne AmpC β-lactamases). | Historical use of avoparcin and other Gram-positive spectrum growth promoters linked to vanA type vancomycin resistance in human isolatesGram-negative indicator organisms, frequently harbour multidrug resistance on mobile genetic elements with potential for horizontal movement into human ExPEC |
Adapted from Shaban et al. (11).
ESBL, Extended spectrum β-lactamase; MRSA, methicillin-resistant Staphylococcus aureus; ExPEC, multidrug-resistant extraintestinal pathogenic E. coli.
Animal-only pathogens
While these are rarely involved in causing human infections, they are the main drivers of antibiotic use in either companion or production animals, particularly in the face of developing resistance. In companion animals, methicillin-resistant Staphylococcus pseudintermedius is the major bacterium requiring ongoing monitoring, due to its propensity to develop multidrug-resistant and extensive drug-resistant profiles. S. pseudintermedius is the resident skin microbiota of dogs and often causes secondary bacterial infections in animals with primary allergic skin disease. The organism is increasingly recognized as a cause of otitis externa and surgical site infections, with nosocomial transmission as an important feature. methicillin-resistant S. pseudintermedius epidemiology is characterized by widespread dissemination of resistant clones.
In livestock, the main drivers of antimicrobial use are the bacterial pathogens involved in bovine and porcine respiratory disease complexes. In cattle, M. haemolytica, Pasteurella multocida, and Histophilus somni have traditionally been susceptible to most registered antimicrobials (20). However, in 2012, the first integrative-conjugative element (ICE) containing multiple resistance genes was identified in a P. multocida isolate from a case of bovine respiratory disease in the United States (21, 22), and similar ICEs have subsequently been identified in M. haemolytica and H. somni. Some isolates with ICE elements have also become resistant to fluoroquinolones (23). Among the main respiratory pathogens of pigs, Actinobacillus pleuropneumoniae, Haemophilus parasuis, and P. multocida, AMR to traditional drugs of choice is also increasing, with the first ICE identified in 2016 in A. pleuropneumoniae (24).
Enterotoxigenic E. coli is a major pathogen of sucker and weaner pigs and neonatal calves. While controlled to some extent by vaccination and management, outbreaks—particularly of postweaning diarrhea in pigs—can be explosive and unpredictable. Resistance to multiple antimicrobials is common, including recent development of resistance to third-generation cephalosporins (e.g., 25) and fluoroquinolones (e.g., 26) in some countries. Salmonella that are isolated from cases of infection in animals are usually serotyped and archived by national reference laboratories and are often available for inclusion in more active AMR surveillance focused on carriage by healthy livestock at slaughter (see 3 below).
Zooanthroponotic pathogens
Bacteria that are naturally transmissible from vertebrate animals to humans and vice versa (zooanthroponotic transmission) present public health risks at the human-animal-ecosystem interface. The major organisms include MRSA and extraintestinal pathogenic E coli. In companion animals, distinct MRSA clones appear to colonize specific animal host species. For example, health care-associated MRSA clone ST22 (EMRSA-15) is most commonly isolated from dogs and cats, while community-associated MRSA CC 8 (ST8, ST612, and ST254) clones are host-adapted to horses (27). Livestock-associated MRSA (LA-MRSA) ST398 is now endemic in many animal production systems throughout the world and is mainly a risk for humans in direct contact with animals, such as farmers and veterinarians. Additional LA-MRSA sequence types have been identified in dairy cattle, pigs, and poultry. A novel hybrid LA-MRSA CC9/CC398 genotype identified as a cause of infection in Danish citizens without direct contact with livestock (and no reported livestock reservoir in Denmark) has implicated retail poultry meat produced in other European countries as a potential source of infection (28).
Highly similar strains of extraintestinal pathogenic E. coli cause clinical infections in both humans and companion animals (29). A number of broad-host-range sequence types that are associated with multidrug resistance, including resistance to both third-generation cephalosporins and fluoroquinolones, have been reported in both humans and animals. These include ST131 (30), ST648 (31), and ST354 (32).
Zoonotic foodborne pathogens
The two main foodborne zoonotic pathogens screened in AMR surveillance programs focused on healthy animals at slaughter are Salmonella spp. and Campylobacter spp. Together, these organisms comprise the most common causes of foodborne disease derived from the consumption or handling of animal products that may require antimicrobial chemotherapy for severe or invasive infections. It is therefore paramount to report trends in resistance to antimicrobial agents that are likely to be used as first-line treatments for these infections. Multidrug-resistant Salmonella strains that also possess resistance to third-generation cephalosporins such as S. enterica serovars Newport, Typhimurium, and Heidelberg have been associated with ground beef and chicken meat, respectively (33). Emerging resistance to fluoroquinolones has been documented in Salmonella isolates derived from pigs (e.g., 34) and poultry meat (35).
Commensals as indicator bacteria
Monitoring of resistance in indicator bacteria is undertaken in healthy livestock because these organisms are ubiquitous in nature, food, animals, and humans and reflect AMR characteristics arising from selective pressure across these environments. It has also been suggested that they represent potential reservoirs of transferrable resistance genes encoded on mobile genetic elements. Enterococcus spp. are included as an AMR indicator organism because they can potentially share mobile genetic elements with other Gram-positive organisms (36). Similarly, plasmid-mediated transfer of AMR genes among commensal E. coli in the gut of healthy animals is well documented (37). However, it is important to note that the resident gut commensal E. coli may also include phylogenetic groups with the right repertoire of virulence genes to cause extraintestinal infection. Avian pathogenic E. coli and other virulent E. coli lineages are carried in the gut of healthy poultry and have been implicated as sources of foodborne infection in humans, either through direct cross-transmission via meat and eggs or through indirect transmission of plasmids and other mobile genetic elements encoding extended-spectrum beta-lactamases and AMR determinants (38, 39). Broad-host-range commensal E. coli sequence types that have acquired resistance to third-generation cephalosporins and/or fluoroquinolones such as ST10 (common between humans and animals) have been isolated in distinct global regions (40, 41). However, a whole-genome sequencing (WGS) study suggested that cephalosporin resistance in human and animal isolates from matching STs was disseminated on different E. coli plasmids (42). The availability of rapid and inexpensive WGS platforms (discussed below) is therefore a welcome addition to AMR monitoring programs to provide context and risk assessment.
Laboratory Testing Methodologies for AMR Monitoring
AST results have historically been intended primarily to guide physicians and veterinarians regarding appropriate antimicrobial therapy. Results are generally reported as susceptible, intermediate, or resistant after applying relevant clinical breakpoints, and there has been little incentive to report quantitative AMR data. For the purposes of surveillance, quantitative results achieved using different laboratory methods or applying nonstandard breakpoints are of limited value for detecting trends or evaluating levels of resistance on a broader level, and comparison of the data is rarely useful. However, reporting and retaining quantitative MIC data provides a mechanism to detect shifts in MIC over time and facilitate early detection of emerging resistance. This approach supports comparison with surveillance data from other systems and allows data to be reinterpreted.
MIC testing is the gold standard technique for determining an isolate’s individual susceptibility to an antimicrobial agent (defined as the lowest concentration inhibiting growth of the organism using a series of 2-fold dilutions). While a number of techniques for determining MIC have been developed, including broth microdilution using 96-well plates, agar dilution, Etest graded strips, and automated systems, broth microdilution is most applicable to AMR surveillance (43). The Kirby-Bauer disk diffusion technique is often employed in veterinary diagnostic laboratories for AST on individual clinical isolates from animals; however, it is not readily amenable to AMR surveillance projects. The availability of broth microdilution methodologies, such as Sensititre, that utilize freeze-dried, predetermined antimicrobial ranges allows for automated or semiautomated testing, individual plate customization, or adoption of standardized plates (such as the NARMS panel). Nevertheless, there are often cases when additional antimicrobials not available in a standardized plate format need to be evaluated, and thorough knowledge and application of the most appropriate standards are therefore required. The Clinical Laboratory Standards Institute (CLSI) has developed clinical breakpoints for human and veterinary pathogens for both MIC broth microdilution and disk diffusion AST techniques. The European Committee on Antimicrobial Susceptibility Testing (EUCAST) has developed clinical breakpoints for human pathogens, but veterinary pathogen clinical breakpoints are still in development.
CLSI and clinical breakpoints
The CLSI (formerly the National Committee for Clinical Laboratory Standards) is a not-for-profit standards development organization formed in 1968. It has established clinical breakpoints for registered veterinary and human antimicrobial agents according to the label dosing regime when a standard method of testing performance is adopted. Clinical breakpoints are determined using a combination of in vitro and in vivo data to predict the likelihood of clinical cure based on pharmacokinetic-pharmacodynamic parameters. They do not, however, predict the likely presence of resistance mechanisms in isolates, are not available for all antimicrobials and all animal species (the default is to use human clinical breakpoints if veterinary-specific breakpoints are unavailable), and are subject to change as new pharmacokinetic-pharmacodynamic data are obtained. Based on clinical breakpoints, isolates are designated susceptible (bacterial infection may be appropriately treated with the dosage regimen recommended for that type of infection and infecting species), intermediate (bacterial infection may be appropriately treated in body sites where the drugs are physiologically concentrated or when a high dosage of drug can be used), or resistant (bacteria are not inhibited by the usually achievable concentrations of the agent with normal dosage schedules and/or fall in the range where specific microbial resistance mechanisms are likely and clinical outcome has not been predictable in in vivo-based studies) (44). Breakpoints are agreed upon by CLSI’s veterinary antimicrobial susceptibility subcommittee (first formed in 1982) after reviewing available data, which includes calculation of ECOFFs (note: the acronym favored by CLSI is ECV). An isolate may be described as nonsusceptible if its MIC is above the susceptible clinical breakpoint (i.e., it is in the intermediate or resistant range).
EUCAST and ECOFFs
EUCAST is a standing committee jointly organized in 1997 by ESCMID, ECDC, and European national breakpoint committees. EUCAST is funded by the European Union. EUCAST first developed the term “ECOFF” and publishes ECOFFs for specific antimicrobials and veterinary pathogen combinations based on MIC distributions of a large number of isolates. ECOFFs classify an organism as wild type or non-wild type based on the normal distribution of MICs for fully susceptible isolates that do not contain any resistance determinants that could influence the MIC phenotype (phenotypically detectable acquired resistance mechanisms). For example, a point mutation in the fluoroquinolone resistance-determining region of gyrA imparting reduced susceptibility to enrofloxacin could shift the MIC of an E. coli isolate from 0.06 μg/ml (within the wild-type range) to 0.25 μg/ml (the ECOFF). This isolate would be described as non-wild type but should not be described as resistant or be included in percentage resistant calculations for isolate collections, because it is still below the clinical resistance breakpoint of ≥4 μg/ml. MIC distributions are often bimodal, with clinically resistant isolates normally distributed around a high MIC and very few isolates falling within the interval range from the non-wild type ECOFF to the clinical resistance breakpoint. This can best be appreciated in an MIC distribution for a hypothetical antimicrobial (Fig. 1). ECOFF phenotypes can now also be more readily justified on the basis of whole-genome sequence data confirming the presence of specific resistance determinants (see below). However, ECOFFs should not be used to classify an isolate as clinically resistant or to calculate the percentage of isolates that are multidrug-resistant (resistant to at least one drug in three antimicrobial classes) or extensively drug resistant (resistant to all but one or two classes), because this cannot be verified from a statistical analysis of MIC distributions without relevant pharmacological data. It is also extremely important to measure the actual MIC; if it falls below a testing threshold, arbitrary “less than” values cannot be used to calculate an ECOFF (45, 46).
FIGURE 1.
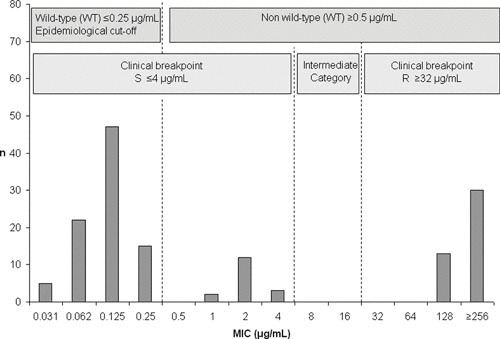
MIC distribution for a hypothetical bacterial species targeted in antimicrobial resistance surveillance programs. Arrows indicate the epidemiological cutoff value (ECOFF) established according to EUCAST recommendations, separating the wild type (no resistance determinants) from the non-wild type (presumed resistance determinants that could be verified by whole-genome sequencing analysis), and the clinical breakpoint. Susceptible, resistant, and intermediate value columns are indicated (45).
The advantage of measuring and reporting ECOFFs is that AST data, when viewed as an MIC distribution, can be more readily compared over time because the proportion of isolates shifting MICs (either higher or lower) can be directly linked to antimicrobial selection pressure or antimicrobial stewardship policies. Reporting and retaining quantitative MIC data provides a mechanism to detect shifts in the MIC over time and facilitate early detection of emerging resistance. This approach supports comparison with surveillance data from other systems and allows data to be reinterpreted if breakpoints or cutoff values change—from the perspective of animal clinical breakpoints (if available) versus human clinical breakpoints—if ECOFFs are applied, or if data from different laboratories are compared (47). The current difficulty in interpreting veterinary-specific AST data is where recent changes in clinical breakpoints established for veterinary species have shifted the CLSI clinical breakpoint to below the corresponding ECOFF. This can be seen with current amoxicillin-clavulanate breakpoints for dogs and cats for E. coli isolated from skin and soft tissue infections (≥1 μg/ml) and urinary tract infections (≥16 μg/ml) now being below the current ECOFF (≥32 μg/ml), which is also the human clinical breakpoint. Using the veterinary clinical breakpoints, isolates with MICs that do not indicate the presence of resistance mechanisms and are within the wild-type distribution could still be classified as resistant (but the same isolates would be classified as susceptible if they were zooanthroponotic and had human clinical breakpoints applied). At this time, displaying the MIC data distribution in tabular form with both the ECOFF and clinical breakpoint values clearly marked would appear to be the best way to present the data for interpretation. However, it is disingenuous to present percentage non-wild type as equivalent to percentage resistance if only ECOFFs have been used to define the MIC distribution.
Sampling Strategies for Healthy Livestock at Slaughter Surveys
The relationship between sample numbers and the sensitivity of a surveillance system to detect increases in resistance has been explored (48). For example, if a sample size of 200 yields a resistance rate of 5% to a particular antibiotic, the resistance level measured in a second set of 200 samples would need to rise above 11% before it can be stated that the level of resistance in the population has significantly increased. Sensitivity can be improved by increasing the number of samples. If 1,000 samples were included in each round, an increase from 5 to 7% is indicative of increasing resistance. It is possible that these numbers do not account for the nonrandom distribution (clustering) of resistance isolates, and where clustering occurs, the sample size requirements are much higher. Randomization of sample collection on an abattoir chain represents the best method of avoiding any potential clustering effects. The EFSA Working Group on Developing Harmonised Schemes for Monitoring Antimicrobial Resistance in Zoonotic Agents recommended that European Union member states should collect data on at least 170 isolates each year as the most accurate and achievable number for all possible outcomes (49). This number was determined based on a range of assumptions (95% confidence intervals with 80% power) and to achieve a desired level of accuracy for estimates of resistance. If resistance is already widespread (50% frequency), only a relatively large change in proportion of resistance is considered relevant (15% increase). For the detection of the initial emergence of resistance (such as to critically important human antimicrobials, for example, 0.1 to 5% frequency), an increase of a few percent should be also detectable.
The number of samples per flock or herd is also an important consideration. Current EFSA guidelines suggest that a single animal from a single farm provides enough precision at a national level. However, greater precision (and significant reductions in cost) of sampling for the same estimation of AMR prevalence was achieved in a study of Campylobacter isolates by reducing the number of birds per flock (n = 155) sampled from five to two (50). However, if only a single animal was sampled, the flock sample size needed to be increased to 250.
While a wide range of potential samples are listed in the various recommendations, the most appropriate samples (particularly for large countries with long distances between farms) are abattoir specimens in the form of cecal content samples for pigs and beef and carcass rinses or swabs for poultry.
Isolation of Zoonotic and Commensal Bacteria from Gastrointestinal Samples
International or national standardized methods of isolation of each bacterial species need to be employed and applied to each sample (ISO6579:2002 for Salmonella and ISO102721:2006 for Campylobacter). For Campylobacter and Salmonella this includes the use of suitable enrichment broth and selective media, preferably employing chromogenic agar. All isolates are required to be confirmed to the species level prior to AST, using appropriate biochemical or genetically based tests. However, matrix-assisted laser desorption ionization–time of flight mass mass spectroscopy is becoming the preferred method in many countries. Isolates need to be appropriately identified and stored for easy traceability.
Recommendations for Reporting and Database Management
An isolate-level database is at the core of any program for the surveillance of AMR. The database contains relevant details of demographic and microbiological characteristics derived from routine diagnostic samples, convenience samples, or targeted surveillance program samples. Data should be stored in secure databases that facilitate simple entry and retrieval, flexible reporting, and ad hoc analysis. Compatibility with similar national and international databases is important. Electronic transfer of data from other systems is highly recommended, rather than manual data entry, which is time- and resource-consuming and error-prone.
SELECTED CASE STUDIES FOR CURRENT AMR SURVEILLANCE IN ANIMALS PROGRAMS
Denmark: DANMAP
DANMAP was established by the Danish Ministry of Food, Agriculture, and Fisheries and the Danish Ministry of Health in 1995 to monitor antimicrobial use in the human and veterinary sectors and to monitor AMR in human and animal pathogens, zoonotic bacteria, and indicator bacteria. DANMAP had its genesis in the 1990s when Danish scientists established the link between avoparcin use in poultry and carriage and contamination of meat with vancomycin-resistant enterococci. It is the first national surveillance program to be initiated by a country and forms a successful blueprint that has been replicated, albeit with modifications, by several other countries. From the outset, DANMAP adopted a coordinated, One Health strategy; they developed a highly integrated, systematic, and continuous program that covered the entire food chain, relating antibiotic consumption with resistance, from “farm to fork to sickbed.” Unique methods of integrating data were developed that created outcomes for action through cross-sector collaboration between scientists and authorities. DANMAP has been highly successful due to adequate funding, excellent planning, and collaboration at all sectors, but also because Denmark is a small country with a large economic reliance on high-quality agricultural produce (approximately 80% of antimicrobials used in the animal sector are administered to pigs) and relatively short distances between farms, processing facilities, and laboratories. Susceptibility testing (one isolate per bacterial species per farm, meat sample, or patient) is performed with commercial Sensititre plates according to CLSI guidelines using ECOFFs validated by EUCAST when possible.
Data from DANMAP documenting the increasing prevalence of vancomycin-resistant enterococci in poultry and pig meat was instrumental in the Danish government’s decision to ban the use of antimicrobials for growth promotion in the 1990s. Steady increases in the amount of therapeutic use of antimicrobials in animals were recorded following the ban, concomitant with the increased detection of extended-spectrum beta-lactamases in commensal E. coli isolates from livestock. Despite the introduction of new guidelines governing the use of antimicrobials, consumption continued to rise. In a further attempt to reduce overall antimicrobial use, the Danish government instituted the “yellow card” system in 2010 for veterinarians and their clients, along with a voluntary withdrawal of the use of cephalosporins in pig production. These efforts resulted in a decrease in detection of extended-spectrum beta-lactamases in indicator E. coli from pigs (51).
France: RESAPATH
The French National Observatory for Epidemiology of Bacterial Resistance to Antibiotics (ONERBA) centralizes data from human and animal surveillance covering 17 surveillance networks. Created in 1997, ONERBA is an organization whose scientific and technical activities rely mainly on the networks for surveillance of resistance already established; only one of these networks (RESAPATH) is devoted to isolates obtained from animals. RESAPATH, operated by ANSES, the French Agency for Food, Environmental, and Occupational Health and Safety, coordinates the voluntary contribution of antimicrobial susceptibility data from isolates from diseased food-producing animals and companion animals obtained by 63 public and private diagnostic laboratories distributed throughout the country. It commenced in 1982 and dealt only with bovine isolates; it was expanded to include swine and poultry isolates in 2000, and other animal species including companion animals and horses were added in 2007. RESAPATH is a key component of the EcoAntibio 2017 plan to combat AMR in animals. The EcoAntibio 2017 plan aims to reduce antimicrobial use in the veterinary sector by 25% by 2017 by introducing/refining 40 broad measures. EcoAntibio 2017 supports the mission of EFSA and ESVAC. ANSES manages the Salmonella surveillance network and also publishes reports on antimicrobial sales data in the French animal sector (from 1999 onward).
ANSES antimicrobial use data demonstrated a 27.9% increase in the consumption of antimicrobials between 1999 and 2009, though data collected between 2009 and 2010 show a 12.2% decrease. However, during this time there was a concomitant increase in the use of critically important antimicrobials (third- and fourth-generation cephalosporins and fluoroquinolones). RESAPATH confirmed high rates of resistance to critical antimicrobials among E. coli isolates from cattle, horses, and companion animals concomitant with increased availability and prescribing of these drugs. However, they were able to demonstrate a drop in resistance frequency in their most recent report when EcoAntibio 2017 energies were focused on education and more appropriate therapeutic guidelines.
Canada: CIPARS
The Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) was established in 2002. A key feature of CIPARS is that reports are an amalgamation of human data with animal data on AMR and antimicrobial use. CIPARS aims to provide a unified approach for monitoring trends in antimicrobial use and resistance in humans and animals and for facilitating assessment of the public health impact of antimicrobials used in humans and agricultural sectors to enable accurate comparison with data from other countries that use similar surveillance systems.
Surveillance data from CIPARS have been instrumental in strengthening our understanding of how AMR in animals can have an adverse effect on public health. Presentation of human and animal data in an integrated fashion is useful for ensuring that animal surveillance and future interventions both have a focus on human health. Several examples of the impact of CIPARS have been reported, including demonstrating a link between an increasing frequency of detection of multidrug-resistant Salmonella Heidelberg in humans and the use of ceftiofur in poultry production in parts of Canada (52).
United States: NARMS
In 1996, collaboration was established between federal, state, and local agencies in the United States for performing surveillance of AMR in enteric bacteria from humans, retail meats, and animals (NARMS). An important feature of NARMS is that methodology in sampling and laboratories has been sufficiently stable since its inception to allow for sound comparison of results between years, thus demonstrating time-based trends in the emergence of resistance. The laboratory methodology is comparable across the three arms of NARMS (humans, food, and animals) as well, which provides a strong basis for One Health comparisons between these three sources. This provides a powerful mechanism for tracking the evolution of resistance in zoonotic enteric pathogens over time.
Data on the occurrence of fluoroquinolone resistance in Campylobacter spp. isolated from poultry have been used in regulatory and legal processes in the United States to reduce the availability of enrofloxacin in animal production. Applications to register new antimicrobial products for use in the animal industries are now evaluated within the context of NARMS findings and through human food safety risk assessments (53). Arguably, one of the most important outcomes of NARMS has been the demonstration of the widespread and increasing level of resistance to third-generation cephalosporins in nontyphoidal Salmonella from food animals. These data were clearly very influential in the FDA decision to introduce additional legal constraints on the use of cephalosporin drugs in food-producing animals. The most recent NARMS report (54) is the first to present WGS data in combination with AST. Summarizing the NARMS data, a major finding confirmed that 80% of Salmonella isolates from retail meats in the United States have remained susceptible to all antimicrobials tested for the past 10 years, and ceftriaxone resistance has decreased in both human and retail poultry isolates over the same time period (19).
FUTURE MONITORING OF AMR IN ANIMALS
Historically, infectious disease surveillance has been based on a number of laboratory processes that require specialized reagents and dedicated personnel with proficiency in a series of separate methods, each providing a distinct piece of microbiological information to aid the clinician in treating and tracking infectious diseases. These laboratory tests provide species identification based on metabolic byproducts, antibiotic susceptibility patterns derived from the in vitro growth response of organisms to different drug concentrations, antigenic structure (e.g., serotype) defined by an algorithm of agglutination reactions, and sometimes virulence properties such as toxin production or the presence of pathognomonic virulence traits (e.g., stx) usually revealed by PCR. Phylogenic relationships based on pulsed-field gel electrophoresis (PFGE) patterns, multilocus sequence typing, or other methods were used to investigate and respond to disease outbreaks and to attribute strain types to a source. This daunting assortment of tests is a major impediment to establishing integrated AMR surveillance in many countries. It is also the limiting factor in the design scope of existing surveillance systems, which are necessarily limited in the number and variety of samples and pathogens under surveillance.
In looking toward the future of AMR surveillance, it is clear that the main way in which it will change is through greater use of DNA sequencing technologies. The development of affordable WGS technologies, along with complementary advances in bioinformatics, provides a single, rapid, and comprehensive laboratory procedure by which to characterize bacterial strains. The power of WGS for public health surveillance has already been demonstrated. In a clinical setting, WGS technology is providing solutions to what were once intractable problems in the characterization of pathogens. For zoonotic foodborne infections in particular, WGS now offers a means to more accurately conduct outbreak investigations, to better understand the virulence traits of pathogenic bacteria and the factors that influence their adaptability to food animal environments, and to conduct genetic analysis of surveillance isolates. Moreover, data are growing that show the power of WGS to provide definitive data on AMR determinants regardless of the source of the sample, allowing direct confirmation of the correct ECOFF value separating wild-type strains from those carrying resistance determinants.
Since the array of phenotypic traits provided by traditional microbiology methods can theoretically be deduced from the genotype, and at lower costs, WGS is expected to remove the limitation on surveillance imposed by the need for multiple procedures. For example, DNA sequence analysis has demonstrated the specificity to identify more than 2,300 of the 2,600 Salmonella serotypes (SeqSero), to attribute foodborne pathogens to geographic origin (tuna scrape), and to catalogue the array of genes underlying traditional phenotypic testing, including antibiotic resistance genes (see below). For Listeria epidemiology, WGS has replaced PFGE for outbreak investigations and will soon replace PFGE and other typing tools for other pathogens. This has started a paradigm shift in the analytical approach to infectious disease by increasing the data that can be quickly extracted from an infectious agent, thereby transforming the laboratory science of pathogen identification, phylogenetic analysis, epidemiology, and surveillance. While the science is still developing, early studies show how WGS data can augment, and in some cases replace, in vitro AST for both surveillance and clinical purposes.
The surveillance of AMR is based on in vitro AST methods, which measure the growth response of bacteria to different drug concentrations in a defined incubation environment. These methods consist mainly of measuring the MIC of antibiotics arrayed in 2-fold serial dilutions, or the diameter of inhibition zones around disks containing standard amounts of antibiotic. These methods are an uncomplicated and proven way to predict the presence of acquired antibiotic resistance traits and to select appropriate therapy. Despite nearly 100 years of experience with this approach, there are several well-known limitations to methods based on an in vitro growth response. There is a lack of harmonization that hinders the interlaboratory comparison of data, an absence of validated methods for many pathogens, practical limitations on the number of agents that can be tested simultaneously, and shifting interpretive standards (55).
THE LIMITATIONS OF PCR FOR RESISTOMICS
In some cases, the results of in vitro AST alone are inadequate, and additional genetic information is needed. For example, methicillin resistance in S. aureus (MRSA) is conferred by the mecA gene. A key characteristic of this gene is its varied expression in vitro, which can lead to false-negative results. For this reason, the definition of MRSA includes detection of the mecA gene. Similarly, the very slow growth rate of Mycobacterium tuberculosis (MTB) in the laboratory causes long delays in the laboratory reporting of susceptibility data. PCR-based testing for rifampin resistance based on mutation in the rpoB gene correlates well with phenotypic methods and can be used to provide more timely information for treating infections. Furthermore, in AMR surveillance, phenotype information in the form of MICs or disk diffusion zone sizes usually does not reveal the underlying genetic mechanisms of which there are thousands of known alleles. This information can help attribute pathogens and resistances to different sources, allowing for targeted measures to combat the spread of resistant pathogens.
Since its development in 1983, PCR has been the most common method for detecting the presence of specific genes. Variations on the method have proven indispensable in genotyping bacteria and augmenting the phenotypic susceptibility data in surveillance. Despite the power and ease of PCR, it has several important limitations. In practice, it is limited largely to the detection of known DNA sequences. Even for genes with known sequences, an amplification product usually must be subjected to DNA sequencing to confirm the results, to identify the resistance allele, or to detect structural mutations conferring resistance. False negatives are a problem because amplification can fail due to the presence of a single mutation or to small variations in sample preparation procedures. False positives also can occur, usually through contamination as a result of carryover of DNA template from another specimen.
While it is possible to account for false test results, PCR is limited in the number of genes it can detect in a single assay, and its utility is incomplete without also determining the DNA sequence of the amplicon. DNA microarrays overcome some of the limitations of PCR-based resistance gene detection by enabling the simultaneous detection of thousands of gene alleles. However, microarray methods also rely on known DNA sequences, may give false-positive and -negative results, and are labor-intensive and expensive.
RESISTOME SURVEILLANCE
Since WGS does not depend on known prior sequence information, it circumvents the limitations of PCR and microarray resistance genotyping methods. While WGS reduces the time and resources needed to generate microbiological data, it requires powerful and sophisticated data analysis via bioinformatics to be useful. To facilitate resistome typing from WGS data, several resistance gene databases have been developed that contain all known resistance genes. These include ResFinder (56), CARD (57), Arg-ANNOT (58), and the CVM resistance gene database (59). These databases make analysis easier by reducing the time needed to compile the complement of resistance genes for a given isolate. As with differences in methods and interpretive criteria for in vitro susceptibility testing, the variety of resistance gene databases can lead to confusion in the scientific literature without some central reference for harmonized gene naming. The NCBI is attempting to overcome this challenge by resolving discrepancies in the nomenclature of resistance alleles in the GenBank repository, where inaccurate and duplicative annotation is a well-known problem. NCBI recently released the Bacterial Antimicrobial Resistance Reference Gene Database (http://www.ncbi.nlm.nih.gov/bioproject/313047), which is a curated and standardized collection of over 3,440 resistance genes and alleles. NCBI has also taken over curation of the Lahey Clinic’s Beta-Lactamase Allele Database, and routinely assigns allele designations for over two dozen beta-lactamase gene families (http://www.ncbi.nlm.nih.gov/pathogens/submit_beta_lactamase/). In addition, NCBI has developed a tool to automatically catalogue resistance genes in all submitted microbial genome sequences and ultimately will link resistome data automatically with phylogenetic trees and other data produced for the GenomeTrakr Project by NCBI’s Pathogen Detection Pipeline. These initiatives will help ensure harmonization of bioinformatics specific to resistome surveillance by providing an index for gene classification.
The effort to develop and curate comprehensive resistance gene databases was motivated to facilitate the use of “resistomics” both to augment AMR surveillance and as a means to guide therapy, related but distinct endeavors. To date, a few studies have examined the correlations between the presence of known resistance determinants and clinical resistance in zoonotic foodborne pathogens. The results show a very high correlation between genotypes and phenotypes for E. coli, Salmonella, and Campylobacter (59–63) for most antimicrobial agents. Some studies have performed similar analyses with Staphylococcus, Klebsiella, Pseudomonas, and Enterococcus, although these bacteria do not typically cause serious foodborne infections (60, 61, 64). While there are lower correlations for some antimicrobial agents, overall, these studies show that WGS analysis is very sensitive and specific for identifying acquired resistance determinants. In this way, WGS can provide a corollary to ECVs (ECOFFS) by its ability to identify non-wild-type strains.
In addition to correlating resistance genotypes and phenotypes, WGS-based analysis completes in one step what was unpractical with PCR strategies, namely, identifying new alleles conferring resistance to the same drug class. For example, in one study of gentamicin-resistant Campylobacter from human infections and from retail meats, PCR failed to detect aminoglycoside resistance genes in many of the human isolates (65). The use of WGS revealed the presence of six gentamicin-resistance genes that had not previously been found in Campylobacter. This included the aph(2′)-Ig gene, which has <30% amino acid identity to other aph(2′) genes and thus would not have been discovered easily without WGS.
In traditional AMR surveillance, the range of antimicrobial classes that are monitored is determined by various considerations intended to optimize the physical limitations of a 96-well panel format. An important advantage of WGS-based surveillance is the ability to detect resistances that are excluded from phenotypic monitoring for whatever reason. In an early study comparing Salmonella genomes, WGS revealed the colocalization of extended-spectrum cephalosporin resistance encoded by the blaCMY-2 gene on a plasmid with the sugE gene, which confers high-level MICs to cetylpyridinium chloride, a chemical used for carcass decontamination. This raises the possibility that chemicals used in processing might influence the resistance profiles of pathogens that reach the food supply. Similarly, WGS has revealed resistance determinants for innate immunity, heavy metals, and less commonly used antibiotics (66). Importantly, WGS supersedes PCR-based methods for plasmid typing by capturing plasmid replicon information, which may signify the animal origin of some resistances.
There is great interest in the potential of WGS to predict the likelihood that a pathogen will respond to anti-infective therapy and thereby serve as a clinical diagnostic test. This is distinct from the utility of WGS in public health surveillance, where a resistance gene is viewed in terms of hazard characterization. The use of WGS to guide clinical treatment options touches on the practice of medicine and standards of care and involves multiple datasets that include clinical outcome information. While DNA tests do serve this purpose for MRSA and MTB as noted above, the parameters for WGS-based susceptibility testing continue to develop for other pathogens. Genomic sequence data have been used to evaluate tentative laboratory breakpoints for streptomycin resistance in Salmonella and E. coli (67). Similarly, quinolone MICs in Salmonella can be distinguished by whether the underlying genetic mechanism for resistance is mutations in gyrA (MIC of >16 μg/ml) or qnr mutations (MIC of 8 to 16 μg/ml) (63). EUCAST established a subcommittee to explore the role of WGS in AST and released a document for public comment on the topic (http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Consultation/2016/Aerococcus_and_Kingella_BP_consultation_and_responses_20161212.pdf.) At the least, WGS can provide information on which antimicrobials are not likely to be effective based on resistance gene content.
RETROSPECTIVE RESISTANCE SURVEILLANCE
In addition to identifying known resistance determinants, WGS allows for rapid retrospective analysis of bacterial genomes as new resistance genes are discovered. An example of this is illustrated by the discovery in 2015 of a mobile colistin resistance (mcr-1) gene in E. coli from animals, retail meats, and humans in China (17). Colistin is considered a drug of last resort for treating multidrug-resistant Gram-negative infections (68). It has not been part of most routine surveillance, since its use in humans is rare due to toxicity issues. With increasing resistance to front-line drugs, colistin resistance has become more important, and plasmid-mediated resistance adds concern about horizontal spread (17, 69, 70). With WGS, it is now possible to examine historical isolates for the presence of mcr-1 and other new resistance genes with very little outlay of resources and time. Instead of reviving banked historical isolates and performing traditional susceptibility testing, it is now possible to examine all publicly available genomes to provide a rapid answer. As a result, bacteria with mcr-1 were detected in over 20 countries within weeks of the original publication (71–73). In the United States, over 55,000 genomes of bacteria from domestic sources were found to be negative for mcr-1. This illustrates the unprecedented power of WGS to quickly provide answers to important questions about emerging AMR threats that previously would have been difficult to resolve quickly.
Identifying Resistance Plasmids
One of the drawbacks of Illumina DNA sequencing technologies is the short read length, which results in genomes consisting of many contigs (74). This makes it difficult to localize a given resistance gene to the chromosome or a plasmid. Some databases, such as PlasmidFinder (75), were assembled to help make these determinations, but these databases are far from complete, and while they indicate the presence of plasmid origins of replication, the physical association of resistance genes to particular plasmids is not always obvious.
Technologies that provide longer reads of DNA, and therefore fewer contigs, make it easier to completely close plasmids and chromosomes and to determine the physical linkages of genes with mobile DNA elements (41, 76, 77). This information can be used to assess the phylogenetic relationships of plasmid incompatibility groups and the variations at MDR integration sites where MDR islands are often found. This information sheds light on the likely origins and drivers of resistance and the potential risk associated with the use of antimicrobial drugs where coselection may lead to cross-resistance. Plasmids from food isolates may carry resistance to antimicrobial drugs not used in food animals (76). Furthermore, heavy metals such as copper and zinc used in agriculture and livestock production and aquaculture as antibiotic replacements may coselect for AMR (78). The details of gene arrangements on plasmids and other mobile elements can only be fully understood with closed genomes. Affordable methods to routinely achieve this will significantly enhance surveillance of AMR.
METAGENOMICS AND MOVING BEYOND CULTURE
Although genomic sequencing reveals the panoply of resistance traits in an individual organism, metagenomic methods do not rely on classic cultivation techniques and thus are not limited by which microorganisms can be grown in culture on artificial media. This is due in part to changing laboratory practices and market forces driving the rapid expansion of culture-independent diagnostic tests for foodborne and other illnesses, which do not require or produce a bacterial isolate for analysis. This will impact public health surveillance in a number of ways, including the ability to track trends in antibiotic resistance.
Metagenomics methods provide the total DNA content of a complex biological sample by direct sequencing. They can identify many microorganisms present in a sample and have been used to identify new pathogens. More importantly for resistance surveillance, metagenomic sequencing reveals the overall resistance gene content of a sample, including genes from unknown bacteria, those that are not easy to culture (e.g., anaerobes), and those for which no culture methods exist. This ability to conduct “deep surveillance” into new environments provides a much more complete picture of the microbiome and resistome associated with a given source. It will be important to link resistance genes with their host bacterium in metagenomic assays but more important to understand which genes are traversing ecological bottlenecks to reach human pathogens and compromise treatment. This ability will also enable a more complete One Health approach to combating resistance. This paradigm emphasizes the interconnectedness of animal, human, and environmental systems when addressing sustainability and preventing disease. The cost savings afforded by next-generation sequencing (NGS) technologies will make it possible to expand routine surveillance to more types of environments and enhance our understanding of AMR from a broader ecological perspective.
LIMITATIONS OF GENOME-BASED AMR SURVEILLANCE
There are several limitations to WGS-based AMR surveillance. While the assay does not require a premeditated design based on known sequences, the interpretation of results requires an understanding of structure-function relationships, or at least sufficient sequence similarity to orthologs to allow inferences to be made. For AMR surveillance, this means that some form of phenotypic susceptibility testing will be necessary to detect novel genes, as will a closely curated gene database to add new genes as they are found. The design of phenotypic susceptibility testing may change, however, as certain agents selected especially for epidemiological purposes (e.g., streptomycin, chloramphenicol) in Salmonella may be tracked adequately by WGS alone. Beyond the quinolones, WGS has not been adequately vetted for its ability to detect resistance resulting from a combination of mutations, which would require straightforward bioinformatics pipelines to evaluate multiple acquired mechanisms working together. Nor can WGS easily detect changes in susceptibility caused by differential gene expression, such as the combination of loci in the Mar regulon that lead to MIC changes or to silent genes whose expression depends on the genetic background of the host.
Despite these limitations, NGS technologies will make it easier to collect larger data sets at lower costs. With freely available bioinformatics tools, along with efforts to automate informatics at GenBank and other places online, it is anticipated that this technology will become automated in the near future for microbial surveillance while replacing programs and processes that require separate laboratory processes. As data on the relationship between genotype and MIC increase, NGS methods may replace traditional susceptibility testing for more pathogens. WGS has already largely replaced PFGE in strain typing, PCR for speciation, and a number of other traditional microbiological techniques (79, 80). Thus, it can be expected that veterinarians, physicians, and other public health officials who make decisions based on the characteristics of microbial pathogens will come to rely more and more on what can be gleaned from the genome as NGS methods become routine.
SUMMARY AND CONCLUSION
AMR surveillance is the cornerstone to address many of the global aspects of AMR. It provides a foundation for assessing the burden of AMR and for providing the necessary evidence for developing efficient and effective control and prevention strategies. The codevelopment of AMR surveillance programs in humans and animals is essential, but there remain several key elements that make data comparisons between human and animal AMR monitoring programs difficult. Differences in AMR programs between regions and countries also create challenges for data comparison. Currently, AMR surveillance relies on uncomplicated in vitro antimicrobial susceptibility methods. However, the lack of harmonization across programs and the limitation of genetic information of AMR remain the major drawbacks of these phenotypic methods. The future of AMR surveillance is moving toward genotypic detection, and molecular analysis methods are expected to yield a wealth of information. However, the expectation that these molecular techniques will surpass phenotypic susceptibility testing in routine diagnosis and monitoring of AMR remains a distant reality, and phenotypic testing remains necessary in the detection of emerging resistant bacteria, new resistance mechanisms, and trends of AMR.
DISCLAIMER
The views expressed in this article are those of the authors and do not necessarily reflect the official policy of the Department of Health and Human Services, the U.S. Food and Drug Administration, or the U.S. government.
ACKNOWLEDGMENTS
The authors thank Ramon Shaban, Geoff Simons, David Jordan, and John Turnidge for critical review and intellectual contributions.
REFERENCES
- 1.Anderson ES. 1968. Drug resistance in Salmonella typhimurium and its implications. BMJ 3:333–339 10.1136/bmj.3.5614.333. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.UK Joint Committee of Houses of Parliament. 1969. Report on the use of antibiotics in animal husbandry and veterinary medicine (‘Swann Report’). Her Majesty’s Stationery Office, London. [PubMed] [Google Scholar]
- 3.Wray C, McLaren IM, Beedell YE. 1993. Bacterial resistance monitoring of salmonellas isolated from animals, national experience of surveillance schemes in the United Kingdom. Vet Microbiol 35:313–319 10.1016/0378-1135(93)90156-2. [DOI] [PubMed] [Google Scholar]
- 4.Pocurull DW, Gaines SA, Mercer HD. 1971. Survey of infectious multiple drug resistance among Salmonella isolated from animals in the United States. Appl Microbiol 21:358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martel JL, Coudert M. 1993. Bacterial resistance monitoring in animals: the French national experiences of surveillance schemes. Vet Microbiol 35:321–338 10.1016/0378-1135(93)90157-3. [DOI] [PubMed] [Google Scholar]
- 6.Cohen ML, Tauxe RV. 1986. Drug-resistant Salmonella in the United States: an epidemiologic perspective. Science 234:964–969 10.1126/science.3535069. [PubMed] [DOI] [PubMed] [Google Scholar]
- 7.DuPont HL, Steele JH. 1987. Use of antimicrobial agents in animal feeds: implications for human health. Rev Infect Dis 9:447–460 10.1093/clinids/9.3.447. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Tollefson L, Angulo FJ, Fedorka-Cray PJ. 1998. National surveillance for antibiotic resistance in zoonotic enteric pathogens. Vet Clin North Am Food Anim Pract 14:141–150 10.1016/S0749-0720(15)30285-1. [DOI] [PubMed] [Google Scholar]
- 9.Aarestrup FM, Bager F, Jensen NE, Madsen M, Meyling A, Wegener HC. 1998. Resistance to antimicrobial agents used for animal therapy in pathogenic-, zoonotic- and indicator bacteria isolated from different food animals in Denmark: a baseline study for the Danish Integrated Antimicrobial Resistance Monitoring Programme (DANMAP). APMIS 106:745–770 10.1111/j.1699-0463.1998.tb00222.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 10.Moyaert H, de Jong A, Simjee S, Thomas V. 2014. Antimicrobial resistance monitoring projects for zoonotic and indicator bacteria of animal origin: common aspects and differences between EASSA and EFSA. Vet Microbiol 171:279–283 10.1016/j.vetmic.2014.02.038. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Shaban RZ, Simon GI, Trott DJ, Turnidge J, Jordan D. 2014. Surveillance and reporting of antimicrobial resistance and antibiotic usage in animals and agriculture in Australia. Report to the Department of Agriculture, Griffith University and University of Adelaide, Australia. [Google Scholar]
- 12.Pagel SW, Gautier P. 2012. Use of antimicrobial agents in livestock. Rev Sci Tech 31:145–188 10.20506/rst.31.1.2106. [PubMed] [DOI] [PubMed] [Google Scholar]
- 13.Weir M, Rajić A, Dutil L, Uhland C, Bruneau N. 2012. Zoonotic bacteria and antimicrobial resistance in aquaculture: opportunities for surveillance in Canada. Can Vet J 53:619–622. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 14.Brudeseth BE, Wiulsrød R, Fredriksen BN, Lindmo K, Løkling KE, Bordevik M, Steine N, Klevan A, Gravningen K. 2013. Status and future perspectives of vaccines for industrialised fin-fish farming. Fish Shellfish Immunol 35:1759–1768 10.1016/j.fsi.2013.05.029. [PubMed] [DOI] [PubMed] [Google Scholar]
- 15.DANMAP. 2016. DANMAP 2015. Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans. https://www.danmap.org/.
- 16.DANMAP. 2012. DANMAP 2011. Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans. https://www.danmap.org/.
- 17.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 18.Guerra B, Fischer J, Helmuth R. 2014. An emerging public health problem: acquired carbapenemase-producing microorganisms are present in food-producing animals, their environment, companion animals and wild birds. Vet Microbiol 171:290–297 10.1016/j.vetmic.2014.02.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 19.Johnson TJ. 2017. Carbapenemase-producing Enterobacteriaceae in swine production in the United States: impact and opportunities. Antimicrob Agents Chemother 61:e02348-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Garch F, de Jong A, Simjee S, Moyaert H, Klein U, Ludwig C, Marion H, Haag-Diergarten S, Richard-Mazet A, Thomas V, Siegwart E. 2016. Monitoring of antimicrobial susceptibility of respiratory tract pathogens isolated from diseased cattle and pigs across Europe, 2009-2012: VetPath results. Vet Microbiol 194:11–22 10.1016/j.vetmic.2016.04.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 21.Michael GB, Kadlec K, Sweeney MT, Brzuszkiewicz E, Liesegang H, Daniel R, Murray RW, Watts JL, Schwarz S. 2012. ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: structure and transfer. J Antimicrob Chemother 67:91–100 10.1093/jac/dkr411. [PubMed] [DOI] [PubMed] [Google Scholar]
- 22.Michael GB, Kadlec K, Sweeney MT, Brzuszkiewicz E, Liesegang H, Daniel R, Murray RW, Watts JL, Schwarz S. 2012. ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: analysis of the regions that comprise 12 antimicrobial resistance genes. J Antimicrob Chemother 67:84–90 10.1093/jac/dkr406. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.Lubbers BV, Hanzlicek GA. 2013. Antimicrobial multidrug resistance and coresistance patterns of Mannheimia haemolytica isolated from bovine respiratory disease cases: a three-year (2009-2011) retrospective analysis. J Vet Diagn Invest 25:413–417 10.1177/1040638713485227. [PubMed] [DOI] [PubMed] [Google Scholar]
- 24.Bossé JT, Li Y, Fernandez Crespo R, Chaudhuri RR, Rogers J, Holden MT, Maskell DJ, Tucker AW, Wren BW, Rycroft AN, Langford PR, the BRaDP1T Consortium. 2016. ICEApl1, an integrative conjugative element related to ICEHin1056, identified in the pig pathogen Actinobacillus pleuropneumoniae. Front Microbiol 7:810 10.3389/fmicb.2016.00810. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jahanbakhsh S, Smith MG, Kohan-Ghadr HR, Letellier A, Abraham S, Trott DJ, Fairbrother JM. 2016. Dynamics of extended-spectrum cephalosporin resistance in pathogenic Escherichia coli isolated from diseased pigs in Quebec, Canada. Int J Antimicrob Agents 48:194–202 10.1016/j.ijantimicag.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Smith M, Do TN, Gibson JS, Jordan D, Cobbold RN, Trott DJ. 2014. Comparison of antimicrobial resistance phenotypes and genotypes in enterotoxigenic Escherichia coli isolated from Australian and Vietnamese pigs. J Glob Antimicrob Resist 2:162–167 10.1016/j.jgar.2014.03.008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 27.Walther B, Monecke S, Ruscher C, Friedrich AW, Ehricht R, Slickers P, Soba A, Wleklinski CG, Wieler LH, Lübke-Becker A. 2009. Comparative molecular analysis substantiates zoonotic potential of equine methicillin-resistant Staphylococcus aureus. J Clin Microbiol 47:704–710 10.1128/JCM.01626-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen J, Stegger M, Andersen PS, Petersen A, Larsen AR, Westh H, Agersø Y, Fetsch A, Kraushaar B, Käsbohrer A, Feβler AT, Schwarz S, Cuny C, Witte W, Butaye P, Denis O, Haenni M, Madec JY, Jouy E, Laurent F, Battisti A, Franco A, Alba P, Mammina C, Pantosti A, Monaco M, Wagenaar JA, de Boer E, van Duijkeren E, Heck M, Domínguez L, Torres C, Zarazaga M, Price LB, Skov RL. 2016. Evidence for human adaptation and foodborne transmission of livestock-associated methicillin-resistant Staphylococcus aureus. Clin Infect Dis 63:1349–1352 10.1093/cid/ciw532. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Platell JL, Johnson JR, Cobbold RN, Trott DJ. 2011. Multidrug-resistant extraintestinal pathogenic Escherichia coli of sequence type ST131 in animals and foods. Vet Microbiol 153:99–108. [PubMed] [DOI] [PubMed] [Google Scholar]
- 30.Platell JL, Cobbold RN, Johnson JR, Heisig A, Heisig P, Clabots C, Kuskowski MA, Trott DJ. 2011. Commonality among fluoroquinolone-resistant sequence type ST131 extraintestinal Escherichia coli isolates from humans and companion animals in Australia. Antimicrob Agents Chemother 55:3782–3787 10.1128/AAC.00306-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ewers C, Bethe A, Stamm I, Grobbel M, Kopp PA, Guerra B, Stubbe M, Doi Y, Zong Z, Kola A, Schaufler K, Semmler T, Fruth A, Wieler LH, Guenther S. 2014. CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: another pandemic clone combining multiresistance and extraintestinal virulence? J Antimicrob Chemother 69:1224–1230 10.1093/jac/dkt516. [PubMed] [DOI] [PubMed] [Google Scholar]
- 32.Vangchhia B, Abraham S, Bell JM, Collignon P, Gibson JS, Ingram PR, Johnson JR, Kennedy K, Trott DJ, Turnidge JD, Gordon DM. 2016. Phylogenetic diversity, antimicrobial susceptibility and virulence characteristics of phylogroup F Escherichia coli in Australia. Microbiology 162:1904–1912 10.1099/mic.0.000367. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.Iwamoto M, Reynolds J, Karp BE, Tate H, Fedorka-Cray PJ, Plumblee JR, Hoekstra RM, Whichard JM, Mahon BE. 2017. Ceftriaxone-resistant nontyphoidal Salmonella from humans, retail meats, and food animals in the United States, 1996-2013. Foodborne Pathog Dis 14:74–83. [PubMed] [DOI] [PubMed] [Google Scholar]
- 34.Jiu Y, Zhu S, Khan SB, Sun M, Zou G, Meng X, Wu B, Zhou R, Li S. 2016. Phenotypic and genotypic resistance of Salmonella isolates from healthy and diseased pigs in China during 2008-2015. Microb Drug Resist 23:651–659. [PubMed] [DOI] [PubMed] [Google Scholar]
- 35.Lin D, Chen K, Wai-Chi Chan E, Chen S. 2015. Increasing prevalence of ciprofloxacin-resistant food-borne Salmonella strains harboring multiple PMQR elements but not target gene mutations. Sci Rep 5:14754 10.1038/srep14754. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clewell DB, Weaver KE, Dunny GM, Coque TM, Francia MV, Hayes F. 2014. Extrachromosomal and mobile elements in Enterococci: transmission, maintenance, and epidemiology. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from Commensals to Leading Causes of Drug Resistant Infection. [Internet.] Massachusetts Eye and Ear Infirmary, Boston, MA. [PubMed] [Google Scholar]
- 37.Al-Tawfiq JA, Laxminarayan R, Mendelson M. 2017. How should we respond to the emergence of plasmid-mediated colistin resistance in humans and animals? Int J Infect Dis 54:77–84 10.1016/j.ijid.2016.11.415. [PubMed] [DOI] [PubMed] [Google Scholar]
- 38.Kluytmans JA, Overdevest IT, Willemsen I, Kluytmans-van den Bergh MF, van der Zwaluw K, Heck M, Rijnsburger M, Vandenbroucke-Grauls CM, Savelkoul PH, Johnston BD, Gordon D, Johnson JR. 2013. Extended-spectrum β-lactamase-producing Escherichia coli from retail chicken meat and humans: comparison of strains, plasmids, resistance genes, and virulence factors. Clin Infect Dis 56:478–487 10.1093/cid/cis929. [DOI] [PubMed] [Google Scholar]
- 39.Mitchell NM, Johnson JR, Johnston B, Curtiss R III, Mellata M. 2015. Zoonotic potential of Escherichia coli isolates from retail chicken meat products and eggs. Appl Environ Microbiol 81:1177–1187 10.1128/AEM.03524-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abraham S, Jordan D, Wong HS, Johnson JR, Toleman MA, Wakeham DL, Gordon DM, Turnidge JD, Mollinger JL, Gibson JS, Trott DJ. 2015. First detection of extended-spectrum cephalosporin- and fluoroquinolone-resistant Escherichia coli in Australian food-producing animals. J Glob Antimicrob Resist 3:273–277 10.1016/j.jgar.2015.08.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 41.Doi Y, Hazen TH, Boitano M, Tsai YC, Clark TA, Korlach J, Rasko DA, Chattaway MA, DoNascimento V, Wain J, Helmuth R, Guerra B, Schwarz S, Threlfall J, Woodward MJ, Coldham N, Doi Y, Hazen TH, Boitano M, Tsai YC, Clark TA, Korlach J, Rasko DA. 2014. Whole-genome assembly of Klebsiella pneumoniae coproducing NDM-1 and OXA-232 carbapenemases using single-molecule, real-time sequencing. Antimicrob Agents Chemother 58:5947–5953 10.1128/AAC.03180-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Been M, Lanza VF, de Toro M, Scharringa J, Dohmen W, Du Y, Hu J, Lei Y, Li N, Tooming-Klunderud A, Heederik DJ, Fluit AC, Bonten MJ, Willems RJ, de la Cruz F, van Schaik W. 2014. Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet 10:e1004776 10.1371/journal.pgen.1004776. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jorgensen JH, Ferraro MJ. 2009. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 49:1749–1755 10.1086/647952. [PubMed] [DOI] [PubMed] [Google Scholar]
- 44.Lubbers BV, Turnidge J. 2015. Antimicrobial susceptibility testing for bovine respiratory disease: getting more from diagnostic results. Vet J 203:149–154 10.1016/j.tvjl.2014.12.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 45.CLSI. 2011. Generation, presentation and application of antimicrobial susceptibility test data for bacteria of animal origin. https://clsi.org/standards/products/veterinary-medicine/documents/vet05/.
- 46.Silley P. 2012. Susceptibility testing methods, resistance and breakpoints: what do these terms really mean? Rev Sci Tech 31:33–41 10.20506/rst.31.1.2097. [PubMed] [DOI] [PubMed] [Google Scholar]
- 47.European Food Safety Authority. 2012. Technical specifications for the analysis and reporting of data on antimicrobial resistance (AMR) in the European Union Summary Report. Parma, Italy. [Google Scholar]
- 48.World Health Organization. 2002. Surveillance Standards for Antimicrobial Resistance. WHO, Geneva, Switzerland. [Google Scholar]
- 49.European Food Safety Authority--Working Group on Developing Harmonised Schemes for Monitoring Antimicrobial Resistance in Zoonotic Agents. 2008. Harmonised monitoring of antimicrobial resistance in Salmonella and Campylobacter isolates from food animals in the European Union. Clin Microbiol Infect 14:522–533. [PubMed] [DOI] [PubMed] [Google Scholar]
- 50.Regula G, Lo Fo Wong DMA, Ledergerber U, Stephan R, Danuser J, Bissig-Choisat B, Stärk KDC. 2005. Evaluation of an antimicrobial resistance monitoring program for Campylobacter in poultry by simulation. Prev Vet Med 70:29–43 10.1016/j.prevetmed.2005.02.017. [PubMed] [DOI] [PubMed] [Google Scholar]
- 51.Agersø Y, Aarestrup FM. 2013. Voluntary ban on cephalosporin use in Danish pig production has effectively reduced extended-spectrum cephalosporinase-producing Escherichia coli in slaughter pigs. J Antimicrob Chemother 68:569–572 10.1093/jac/dks427. [PubMed] [DOI] [PubMed] [Google Scholar]
- 52.Parmley EJ, Pintar K, Majowicz S, Avery B, Cook A, Jokinen C, Gannon V, Lapen DR, Topp E, Edge TA, Gilmour M, Pollari F, Reid-Smith R, Irwin R. 2013. A Canadian application of One Health: integration of Salmonella data from various Canadian surveillance programs (2005-2010). Foodborne Pathog Dis 10:747–756 10.1089/fpd.2012.1438. [PubMed] [DOI] [PubMed] [Google Scholar]
- 53.Gilbert JM, White DG, McDermott PF. 2007. The US national antimicrobial resistance monitoring system. Future Microbiol 2:493–500 10.2217/17460913.2.5.493. [PubMed] [DOI] [PubMed] [Google Scholar]
- 54.FDA 2016. National Antimicrobial Resistance Monitoring System - Enteric Bacteria (NARMS): NARMS Integrated Report 2014. Rockville, Maryland: U.S. Department of Health and Human Services, Food & Drug Administration. Available at: http://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/default.htm. [Google Scholar]
- 55.Kahlmeter G. 2014. Defining antibiotic resistance-towards international harmonization. Ups J Med Sci 119:78–86 10.3109/03009734.2014.901446. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644 10.1093/jac/dks261. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O’Brien JS, Pawlowski AC, Piddock LJV, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357 10.1128/AAC.00419-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain JM. 2014. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220 10.1128/AAC.01310-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tyson GH, McDermott PF, Li C, Chen Y, Tadesse DA, Mukherjee S, Bodeis-Jones S, Kabera C, Gaines SA, Loneragan GH, Edrington TS, Torrence M, Harhay DM, Zhao S. 2015. WGS accurately predicts antimicrobial resistance in Escherichia coli. J Antimicrob Chemother 70:2763–2769 10.1093/jac/dkv186. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stoesser N, Batty EM, Eyre DW, Morgan M, Wyllie DH, Del Ojo Elias C, Johnson JR, Walker AS, Peto TE, Crook DW. 2013. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother 68:2234–2244 10.1093/jac/dkt180. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agersø Y, Lund O, Larsen MV, Aarestrup FM. 2013. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother 68:771–777 10.1093/jac/dks496. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.Zhao S, Tyson GH, Chen Y, Li C, Mukherjee S, Young S, Lam C, Folster JP, Whichard JM, McDermott PF. 2015. Whole-genome sequencing analysis accurately predicts antimicrobial resistance phenotypes in Campylobacter spp. Appl Environ Microbiol 82:459–466 10.1128/AEM.02873-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mcdermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. 2016. The use of whole genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother. 60:5515–5520. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gordon NC, Price JR, Cole K, Everitt R, Morgan M, Finney J, Kearns AM, Pichon B, Young B, Wilson DJ, Llewelyn MJ, Paul J, Peto TE, Crook DW, Walker AS, Golubchik T. 2014. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol 52:1182–1191 10.1128/JCM.03117-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao S, Mukherjee S, Chen Y, Li C, Young S, Warren M, Abbott J, Friedman S, Kabera C, Karlsson M, McDermott PF. 2015. Novel gentamicin resistance genes in Campylobacter isolated from humans and retail meats in the USA. J Antimicrob Chemother 70:1314–1321 10.1093/jac/dkv001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pal C, Bengtsson-Palme J, Rensing C, Kristiansson E, Larsson DG. 2014. BacMet: antibacterial biocide and metal resistance genes database. Nucleic Acids Res 42(D1):D737–D743 10.1093/nar/gkt1252. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tyson GH, Li C, Ayers S, McDermott PF, Zhao S. 2016. Using whole-genome sequencing to determine appropriate streptomycin epidemiological cutoffs for Salmonella and Escherichia coli. FEMS Microbiol Lett 363:363 10.1093/femsle/fnw009. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaye KS, Pogue JM, Tran TB, Nation RL, Li J. 2016. Agents of last resort: polymyxin resistance. Infect Dis Clin North Am 30:391–414 10.1016/j.idc.2016.02.005. [PubMed] [DOI] [PubMed] [Google Scholar]
- 69.Hu Y, Liu F, Lin IY, Gao GF, Zhu B. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:146–147 10.1016/S1473-3099(15)00533-2. [DOI] [PubMed] [Google Scholar]
- 70.Stoesser N, Mathers AJ, Moore CE, Day NP, Crook DW. 2016. Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. Lancet Infect Dis 16:285–286 10.1016/S1473-3099(16)00010-4. [DOI] [PubMed] [Google Scholar]
- 71.Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, Zankari E, Leekitcharoenphon P, Stegger M, Kaas RS, Cavaco LM, Hansen DS, Aarestrup FM, Skov RL. 2015. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill 20:20 10.2807/1560-7917.ES.2015.20.49.30085. [PubMed] [DOI] [PubMed] [Google Scholar]
- 72.McGann P, Snesrud E, Maybank R, Corey B, Ong AC, Clifford R, Hinkle M, Whitman T, Lesho E, Schaecher KE. 2016. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob Agents Chemother 60:4420–4421 10.1128/AAC.01103-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suzuki S, Ohnishi M, Kawanishi M, Akiba M, Kuroda M. 2016. Investigation of a plasmid genome database for colistin-resistance gene mcr-1. Lancet Infect Dis 16:284–285 10.1016/S1473-3099(16)00008-6. [DOI] [PubMed] [Google Scholar]
- 74.Lesho E, Clifford R, Onmus-Leone F, Appalla L, Snesrud E, Kwak Y, Ong A, Maybank R, Waterman P, Rohrbeck P, Julius M, Roth A, Martinez J, Nielsen L, Steele E, McGann P, Hinkle M. 2016. The challenges of implementing next generation sequencing across a large healthcare system, and the molecular epidemiology and antibiotic susceptibilities of carbapenemase-producing bacteria in the healthcare system of the U.S. Department of Defense. PLoS One 11:e0155770 10.1371/journal.pone.0155770. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903 10.1128/AAC.02412-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Y, Mukherjee S, Hoffmann M, Kotewicz ML, Young S, Abbott J, Luo Y, Davidson MK, Allard M, McDermott P, Zhao S. 2013. Whole-genome sequencing of gentamicin-resistant Campylobacter coli isolated from U.S. retail meats reveals novel plasmid-mediated aminoglycoside resistance genes. Antimicrob Agents Chemother 57:5398–5405 10.1128/AAC.00669-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang J, Stephan R, Power K, Yan Q, Hächler H, Fanning S. 2014. Nucleotide sequences of 16 transmissible plasmids identified in nine multidrug-resistant Escherichia coli isolates expressing an ESBL phenotype isolated from food-producing animals and healthy humans. J Antimicrob Chemother 69:2658–2668 10.1093/jac/dku206. [PubMed] [DOI] [PubMed] [Google Scholar]
- 78.Seiler C, Berendonk TU. 2012. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front Microbiol 3:399 10.3389/fmicb.2012.00399. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Larsen MV, Cosentino S, Lukjancenko O, Saputra D, Rasmussen S, Hasman H, Sicheritz-Pontén T, Aarestrup FM, Ussery DW, Lund O. 2014. Benchmarking of methods for genomic taxonomy. J Clin Microbiol 52:1529–1539 10.1128/JCM.02981-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Salipante SJ, SenGupta DJ, Cummings LA, Land TA, Hoogestraat DR, Cookson BT. 2015. Application of whole-genome sequencing for bacterial strain typing in molecular epidemiology. J Clin Microbiol 53:1072–1079 10.1128/JCM.03385-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]


