ABSTRACT
For nearly a century the use of antibiotics to treat infectious diseases has benefited human and animal health. In recent years there has been an increase in the emergence of antibiotic-resistant bacteria, in part attributed to the overuse of compounds in clinical and farming settings. The genus Listeria currently comprises 17 recognized species found throughout the environment. Listeria monocytogenes is the etiological agent of listeriosis in humans and many vertebrate species, including birds, whereas Listeria ivanovii causes infections mainly in ruminants. L. monocytogenes is the third-most-common cause of death from food poisoning in humans, and infection occurs in at-risk groups, including pregnant women, newborns, the elderly, and immunocompromised individuals.
Currently, multidrug resistance is not a common feature encountered in Listeria species. However, as has been observed with other pathogens of importance to humans, Listeria species have the ability to rapidly develop resistance to any antimicrobial agent, a feature that represents an emerging and increasing threat to human and animal health. Isolates of Listeria have been reported with varying degrees of resistance to commonly used antibiotics, and the first multidrug-resistant Listeria isolate was identified in France in 1988. These isolates developed resistance through a number of well-known mechanisms, including target gene mutations, such as within genes encoding efflux pumps, together with the acquisition of mobile genetic elements. This article will focus, in particular, on describing the mechanisms that confer resistance in Listeria species to antibiotics, biocides, and heavy metals.
INTRODUCTION TO THE GENUS LISTERIA AND SPECIES
Listeria species are small rod-shaped bacteria, typically 0.5 μm in diameter and 1 to 2 μm in length. They are Gram-positive bacteria, facultative anaerobes, and nonsporulating. They are catalase-positive, oxidase-negative, and motile at low temperatures. They grow at temperatures in the range of 4 to 45°C and pH values of 4.7 to 9.6, and have a low GC content. The genus Listeria comprises 17 recognized species: L. monocytogenes, L. ivanovii, L. grayi, L. innocua, L. seeligeri, L. welshimeri, L. marthii, L. fleischmannii, L. floridensis, L. aquatica, L. newyorkensis, L. cornellensis, L. rocourtiae, L. weihenstephanensis, L. grandensis, L. riparia, and L. booriae (Fig. 1) (1). Listeria species can be classified into Listeria sensu stricto (L. monocytogenes, L. ivanovii, L. innocua, L. seeligeri, L. welshimeri, and L. marthii) and Listeria sensu lato (L. grayi, L. fleischmannii, L. floridensis, L. aquatica, L. newyorkensis, L. cornellensis, L. rocourtiae, L. weihenstephanensis, L. grandensis, L. riparia, and L. booriae) (1).
FIGURE 1.
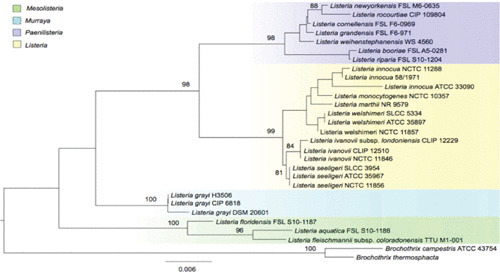
Listeria species maximum likelihood phylogenetic tree based on concatenated nucleotide sequences of the 16S rRNA genes from all Listeria species. Values on branches represent bootstrap values based on 500 bootstrap replicates; bootstrap values >80% are not displayed. Listeria species are color coded according the new genera classification proposed by Orsi et al. (1).
Phylogenetic studies indicate that the most closely related genus is Brochothrix, and there is a relatedness to other Gram-positive bacteria with low GC content, such as Bacillus, Clostridium, Enterococcus, Staphylococcus, and Streptococcus (2, 3). Orsi and Wiedmann (1) proposed a reclassification of the Listeria genus into four genera, Listeria, Mesolisteria, Paenilisteria, and Murraya (Fig. 1). The species that belong to the Mesolisteria genus are unable to grow at temperatures below 7°C. Listeria sensu stricto species and L. grayi possess motility genes that enable them to move, in contrast to the rest of the Listeria species.
Listeria species can be classified into serotypes based on the serological reactions of listerial somatic antigen (O-antigen) and flagellar antigen (H-antigen) with specific antisera. L. monocytogenes can be differentiated into at least 13 serotypes, and these are divided into 4 lineages, I to IV (Table 1) (4). The majority of L. monocytogenes isolates cluster into lineages I and II, described by Piffaretti et al. (5). Lineage IV is the most recently identified, and currently few isolates belong to this lineage (6, 7).
TABLE 1.
Listeria monocytogenes lineages and serotype distribution
| Lineage | Serotypes | Distribution | Genetic characteristics |
|---|---|---|---|
| I | 1/2b, 3b, 4b | Commonly isolated from various sources; overrepresented among human isolates | Lowest diversity among the lineages; lowest levels of recombination among the lineages |
| II | 1/2a, 1/2c, 3a, 3c | Commonly isolated from various sources; overrepresented among food and food-related as well as natural environments | Most diverse, highest recombination levels |
| III | 4a, 4b, 4c | Most isolates obtained from ruminants | Very diverse; recombination levels between those for lineage I and lineage II |
| IV | 4a, 4b, 4c | Most isolates obtained from ruminants | Few isolates analyzed to date |
L. monocytogenes is an important facultative human foodborne pathogen, and it is the third leading cause of deaths due to foodborne bacteria the United States (8). Listeria outbreaks are distributed globally, causing a significant economic impact on the food industry and public health. This bacterium was first isolated in rabbits and guinea pigs with pronounced monocytosis (9). Listeria species are ubiquitous and widespread in the environment. The pathogenicity of Listeria species is associated with intercellular replication, and its reservoir extends from the environment to humans and animals, providing an ecological niche (10).
LISTERIA SPECIES IN DOMESTIC ANIMALS AND THE FARM ENVIRONMENT
Listeria species have been isolated from a wide range of domestic and wild animals including mammals, birds, fish, and crustaceans (11, 12). The clinical disease is usually observed in domestic ruminants and humans, though occasionally also in poultry, pigs, rabbits, and other species. Clinical listeriosis in birds is rare, but birds tend to be asymptomatic carriers, because they can ingest this bacterium through contaminated food, water, bedding, or soil (13). Younger cohorts of bird populations seem to be more susceptible (14). Like rabbits, guinea pigs are also susceptible, and L. monocytogenes has also been isolated in canine cutaneous infection (15). There have been reports of symptoms such as miscarriages and septicemia associated with L. monocytogenes equine infections (16). The prevalence of Listeria in wildlife species ranges from 1 to 60%, although species were mainly captive or domestic (17–19). Less than 1% prevalence was recorded in wild birds and mammals in their natural habitats in Japan (20), while Listeria species were recorded in 5% of sampled red fox, beech marten, and raccoon populations in Poland (21). Contact with contaminated silage is believed to be an important transmission vector for livestock infection (22).
Listeria can be classified as a saprophytic bacterium, because its reservoirs are the soil and the intestinal tracts of asymptomatic animals, with soil and fecal contamination resulting in the presence of these bacteria on plants and fodder, particularly silage, along with walls, floors, and drains in the agricultural environment (23, 24).
Evidence indicates that farm systems with cattle species have a higher prevalence of L. monocytogenes, including the species that cause human listeriosis, compared to other animals (25). In the addition, it appears that cattle play an important part in the amplification and spread of L. monocytogenes in agricultural environments (Fig. 2), while there is evidence of a difference in epidemiology and transmission between ruminant species (25, 26). L. monocytogenes can be isolated from cattle farms more frequently during the spring, a feature that may be related to variables including housing with silage feeding during the winter and the application of slurry (27). This would indicate that farmyard environments are reservoirs. The latter feature is not surprising since healthy bovine species with fecal carriage of L. monocytogenes as high as 46.3% have been reported (28, 29).
FIGURE 2.
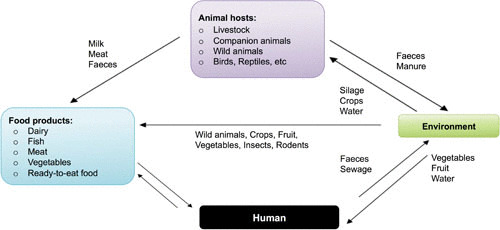
Transmission dynamics of listeriosis involving human and animal hosts. Potential transmission pathways of Listeria species are indicated by arrows, and vehicles are represented by colored boxes.
However, the disease ecology caused by Listeria species is not fully understood, and fundamental questions regarding the interaction between the hosts, the pathogen, and environment remain to be answered. Specifically, the determinants and mechanisms of transmission from the principal reservoirs, i.e., ruminants, within agricultural ecosystems merit further investigations (30).
OVERVIEW OF THE PATHOGENESIS OF LISTERIOSIS
The genus Listeria contains two pathogenic species, L. monocytogenes and L. ivanovii. However, on rare occasions infections caused by L. innocua and L. seeligeri have occurred (31, 32). Listeriosis in humans and many vertebrate species, including birds, is caused by L. monocytogenes, with L. ivanovii infections being specific to ruminants (33). Most infections caused by Listeria result from the ingestion of contaminated food such as raw milk, meat, and fish; ready-to-eat meals; unpasteurized dairy products; and uncooked vegetables in humans and from contaminated silage or other sources of feed in animals. The cycle of infection is completed with the liberation of Listeria into the environment in the feces. Listeria is a saprophyte when it is found in the environment but makes a transition to a physiological state that promotes bacterial survival and replication in host cells (24). Listeria has evolved a number of mechanisms to invade and exploit host processes to multiply, spread from cell-to-cell, and evade the immune system without damaging the host cell. The intracellular infectious cycle consists of distinct stages: (i) invasion of the eukaryotic host cell, (ii) escape from the intracellular vacuole, (iii) intercellular proliferation, (iv) actin filament-based intercellular spread, and (v) dissemination to adjacent cells (Fig. 3) (34).
FIGURE 3.
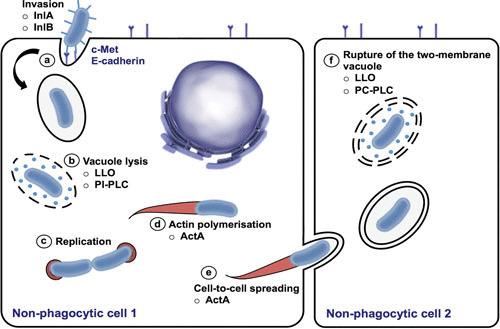
L. monocytogenes intracellular life cycle. (a) Listeria invades the host cells via a zipper mechanism, by the interaction of surface internalins InlA and InlB with the host cell surface receptors E-cadherin and Met, respectively. (b) Listeria escapes from the phagosome before the fusion with the lysosome occurs, by the action of the secreted proteins, the pore-forming toxin LLO, and phosphatidylinositide phospholipase C (PI-PLC). (c) Listeria may replicate in the cytosol, and (d) it spreads by actin polymerization, which propels the bacteria unidirectionally, (e) promoting cell-to-cell spreading of Listeria. (f) Rupture of the two-membrane vacuole is mainly mediated by the action of LLO and phosphatidylcholine-specific phospholipase C (PC-PLC).
As indicated, Listeria is an intracellular pathogen that spreads from cell to cell, and L. monocytogenes has evolved mechanisms to evade exposure to the host innate immune defenses. One example of immune evasion mechanisms in L. monocytogenes is the O-acetyltransferase (oatA), which encodes for O-acetylation of muramic residues contained in the peptidoglycan. OatA increases the bacterial cell wall resistance to antimicrobial compounds such as lysozyme, thereby favoring bacterial persistence in macrophages and virulence in vivo (34). However, the host has evolved innate and acquired immunity mechanisms, including the induction of apoptosis and the generation of cytotoxic T-cells that recognize and lyse listeria-infected cells, releasing bacteria into the extracellular space and rendering them susceptible to infiltrating phagocytes (35). (For a more complete description of the bacterial mechanisms underpinning the pathogenesis and virulence of Listeria, the following references 33 and 36–39 provide additional information.)
LISTERIOSIS IN FOOD-PRODUCING AND OTHER ANIMALS AND IN HUMANS
Listeria species have been isolated from more than 50 animal species, both domestic and wild (Table 2), with presentation of disease varying according to the host (Table 3). Most infections in animals are subclinical, but invasive listeriosis can occur either sporadically or as part of an outbreak (12). Clinical disease is seen most often in domestic ruminants and humans, though occasional cases have been reported in poultry, pigs, rabbits, and other species.
TABLE 2.
Mammals, birds, and other species from which Listeria species have been isolated
| Mammals | Cats, cattle, chinchillas, deer, dogs, ferrets, foxes, goats, guinea pigs, horses, humans, jackals, lemmings, mice, mink, monkeys, moose, pigs, rabbits, raccoons, rats, sheep, skunks, and voles |
| Birds | Canaries, chaffinches, chickens, cranes, doves, ducks, eagles, geese, hawks, lorikeets, owls, parrots, partridges, pheasants, pigeons, seagulls, turkeys, white grouse, whitethroat grouse, and wood grouse |
| Other | Ants, crustaceans, lizards, fish, flies, frogs, snails, ticks and tortoises |
TABLE 3.
Listeria species, hosts, and forms of disease
| Species | Host | Forms of disease |
|---|---|---|
| L. monocytogenes |
|
|
| L. ivanovii | Cattle, sheep | Abortion |
| L. innocua | Sheep | Encephalitis (rare) |
Listeriosis in Food-Producing Domestic Ruminants
In ruminants, infection with Listeria and disease are predominantly due to the consumption of spoiled silage contaminated with L. ivanovii or L. monocytogenes. Listeria species can replicate in poorly fermented silage with pH values above 5.5, and these bacteria can reach numbers of up to 107 colony-forming units/kg. Other contamination sources apart from silage include barn equipment such as bedding, water, and feeding troughs (40), where bacteria can survive within biofilms. Depending on the number of bacteria ingested, the pathogenic properties of the individual bacterial isolate, and the immune status of the host, listeriosis may present as meningoencephalitis, abortion, or septicemia. It is uncommon for different clinical manifestations to be observed during the same outbreak. The following subsections provide a brief synopsis of the clinical features associated with these presentations.
Meningoencephalitis
Meningoencephalitis (rhombencephalitis) is caused by L. monocytogenes and is the most characteristic clinical manifestation of listeriosis in adult ruminants. Indeed, it is among the most common causes of neurological disease in ruminants. A naturally occurring case of ovine meningoencephalitis due to L. innocua has also been reported (41). The incubation period is usually 2 to 3 weeks, and the course of the infection is more acute and frequently fatal in sheep and goats (death may occur within 2 to 3 days after the onset of symptoms) but is subacute to chronic in cattle. Once these bacteria cross the intestinal barrier they can gain access to the central nervous system (CNS) via the blood stream. An alternative route to the CNS is through damaged oral, nasal, or ocular mucosas, where the bacterium can track along the trigeminal nerve terminals with subsequent intraneural direct spread to the brain stem and to further areas of the brain via axonal pathways (42). Clinical presentation depends on the localization and distribution of lesions in the nervous system (30). Early and nonspecific symptoms such as depression and anorexia are followed by neurologic signs, often unilateral, such as facial and tongue paralysis with profuse salivation, nystagmus, absence of palpebral reflex, and dropped ear. The syndrome is known as “circling disease” because it is characterized by a lack of coordination, leaning against objects, head deviation sometimes with head tilt, involuntary torticollis, and walking aimlessly in circles as a result of brainstem lesions. In a further development of the infection, prostration is followed by paddling movements and convulsions before death. The course of the infection is short, extending from 1 to 4 days in sheep and from 1 to 2 weeks in cattle.
Abortion
L. monocytogenes and L. ivanovii can gain access to the developing fetus via hematogenous penetration of the placental barrier. Vaginal transmission can also occur. The depression of cell-mediated immunity during advanced pregnancy is thought to play an important role in the development of listeriosis (33). The incubation period can be as short as 1 day, and the outcome of the fetal infection is mainly abortion, though cases of stillbirths have been reported. Abortion occurs in the last trimester of gestation (after 7 months in cattle [approximately 9 months gestation] and 12 weeks in sheep and goats [approximately 20 weeks gestation]), and abortion storms are more commonly reported in sheep and goats. In both species, the rates of abortion in a group are low but may reach as high as 15%. Mothers have no other clinical signs, with the exception of fever and anorexia, and it is not known yet why these animals never develop CNS infection or overt septicemic disease (33). If the placenta is retained, metritis may result.
Septicemia
The septicemic form of listeriosis is relatively uncommon in adult ruminants and generally, but not invariably, occurs in the neonate as a consequence of in utero infection. It is marked by weakness, depression, inappetence, fever, and death. In this clinical form, multiple foci of necrosis in the liver and, less frequently, the spleen may be noted at necropsy.
Other presentations
Mastitis has rarely been associated with L. monocytogenes infection and is not as frequent as with Brucella species, Mycobacterium bovis, Escherichia coli, Staphylococcus species, or Streptococcus species. However, subclinical L. monocytogenes mastitis is also reported (43). Gastrointestinal infections can occasionally occur in sheep and goats (44), with dullness, inappetence, pyrexia, diarrhea, and death within 24 hours of clinical signs. Unilateral uveitis and keratoconjunctivitis (ocular listeriosis) are also reported in cattle (45) and are attributed to direct ocular contact with contaminated silage.
In addition to the clinical cases of listeriosis, a significant number of animals within a herd may be subclinically infected and excrete Listeria species in their feces, a feature that represents a strong risk factor for contamination of the environment and a means by which the bacterium could gain access to the slaughterhouse (46).
Listeriosis in Other Animals
Birds
Birds are usually subclinical carriers of L. monocytogenes. Secretions and excretions from colonized birds are rich sources of these bacteria, and they can play a major role in transmission and spread of Listeria species to both humans and animals via the ingestion of contaminated food, feed, water, litter, and soil (13). Outbreaks of listeriosis are occasionally observed in the young animals. Predisposing factors include immunosuppression, wet/damp conditions, moist litters, and cold (47). The most frequent form is septicemia occurring with symptoms including depression, listlessness, diarrhea, and emaciation. Peracute deaths occur sometimes without other clinical signs (13). The encephalitic form of listeriosis in birds, meningoencephalitis, is a rare occurrence. However, an outbreak of encephalitic listeriosis in red-legged partridges between 8 and 28 days of age has been recently documented (48). Meningoencephalitis in birds is characterized by incoordination, torticollis, stupor, and paresis or paralysis (49). In young geese, both meningoencephalitis and septicaemia can be seen concurrently. Salpingitis is observed in hens during the acute systemic phase (49).
Rabbits
In rabbits, L. monocytogenes usually causes abortion during late pregnancy or sudden death. Encephalitis is rare. L. monocytogenes was first isolated by Murray et al. as the etiological agent of septicemia in laboratory rabbits and guinea pigs in England in 1926 (9).
Swine
Swine listeriosis is caused by L. monocytogenes, but it is uncommon. The primary manifestation is septicemia in young piglets, with death within 3 to 4 days. Encephalitis in adults and abortions are also seen occasionally.
Others
Septicemia and neurologic signs resembling rabies have been reported in dogs, and L. monocytogenes has also been isolated in canine cutaneous infection (15). Rare cases of encephalitis or septicemia occur in cats, with typical nonspecific symptoms including depression, inappetence, abdominal pain, vomiting, and diarrhea. In horses, there have been reports of meningoencephalitis, abortions, intrauterine infection, septicemia, and kerato-conjunctivitis associated with L. monocytogenes (16). Occasionally, septicemia disease is the usual form of listeriosis in other species, but abortions can also occur.
Listeriosis in Humans
L. monocytogenes is the most commonly reported etiological agent in cases of human listeriosis, though adult meningitis caused by L. seeligeri (31), a fatal bacteremia caused by L. innocua (32), and various cases of listeriosis caused by L. ivanovii (50) have also been reported. The disease mainly affects pregnant women and immunocompromised individuals by either physiological means—due to an immature (in newborns and young children) or suppressed (in the elderly) immune system—by infectious means (arising from immunosuppressive viruses and advanced systemic pathologies), or by iatrogenic means (associated with postchemotherapy or pretransplant immunosuppression). Currently, listeriosis is regarded as a foodborne disease of serious public health concern with a high mortality rate (25 to 30%) and a variable incubation period from 3 to 70 days.
In pregnant women, listeriosis is most commonly noted in the third trimester, but cases have been reported at all stages of pregnancy. The main clinical symptoms documented include mild flu-like symptoms, bloody vaginal discharge, and septicemia, and these may result in premature delivery, abortion, or stillbirth (33). A perinatal listeriosis infection occurs when L. monocytogenes crosses the placenta and elicits an infection in the fetus, but cross-infection during delivery is also possible (51). Perinatal infection is classified on the basis of the clinical symptoms observed: an early onset of listeriosis occurs in the fetus or neonate within the first week after delivery and is characterized by a serious septicemia (granulomatosis infantisepticum). Late onset occurs within the 2nd to 4th week of life and is manifested as meningitis with hydrocephalus as sequela (52). In immunocompromised adults, listeriosis can manifest as a rare but severe form of infection including septicemia or encephalitis (meningoencephalitis or rhombencephalitis), with symptoms appearing after 2 to 10 weeks of infection (33, 53).
Healthy individuals (who are immunocompetent) rarely develop clinical signs after infection, though a milder gastroenteritis with fever, abdominal and back pain, headache, and myalgia can occur. The incubation period is reported to extend from 8 to 48 h, and symptoms are typically self-limiting and resolve within 1 to 3 days. Direct transmission of L. monocytogenes from animals to humans is reported among farmers, veterinarians, and animal handlers who are in frequent direct contact with colonized animals or healthy carriers. A cutaneous form characterized by a nonpainful, nonpruritic pyogranulomatous rash has been reported in individuals who handle infected newborns, fetuses, or aborted cows or who perform necropsies on septicemic animals (54), while cases of conjunctivitis have been reported in workers in poultry processing plants who handled listeria-positive chickens (13).
CHEMOTHERAPEUTIC OPTIONS FOR THE TREATMENT OF ANIMAL AND HUMAN LISTERIOSIS
Cell-mediated immunity is the main host defense strategy against Listeria species and is largely dependent on the action of cytotoxic T-cells—hence the association between listeriosis and conditions involving impairments of cell-mediated immunity (55). The intracellular lifecycle of this bacterium confounds the efficacy of antimicrobial chemotherapy together with the lack of development of anti-Listeria immunity by the host postinfection. However, an anti-listeriolysin O (LLO) neutralizing monoclonal antibody has been reported to increase host resistance to infection in mouse models (56). Anti-LLO antibodies have been shown to help arrest the growth of Listeria species within macrophages, and this approach can be used for the serodiagnosis of these infections in humans and animals (57, 58), though it cannot discriminate between a current or previous infection (59), and furthermore, it provides no indication of the duration of this immunity (46).
Antimicrobial chemotherapy is the only viable option for the treatment of listeriosis. For an antibiotic such as a penicillin-based agent to be effective against this bacterium, it must penetrate into the host cell, maintain a high intracellular concentration, and bind to the penicillin-binding protein 3 (PBP3) expressed by Listeria species (60). High drug doses and early treatment are essential to treat animals with encephalitis, though if signs of encephalitis are severe, death usually occurs despite the intervention. Treatment in sheep and goats usually has little value soon after the appearance of neurological signs or in chronic cases. According to The Merck Veterinary Manual (54), in cattle, chlortetracycline given at a dose of 10 mg/kg body weight per day and administered for 5 days intravenously is effective in the treatment of encephalitis cases. If penicillin G is used, it should be given intramuscularly at 44,000 U/kg body weight per day for 1 to 2 weeks, and the first injection should be accompanied by the same dose given intravenously. Time durations for antimicrobial treatment may vary according to the level of infection. In severe cases, it is often recommended to continue treatment for up to 1 week after the clinical symptoms have disappeared. Supportive care, including the provision of clean housing along with fluids and electrolytes, is an important part of therapy. High-dose dexamethasone (1 mg/kg, given intravenously) at first examination is considered beneficial by some but is controversial and will cause abortion during the last two trimesters in cattle and after day 135 in sheep. Gentamicin has been found to be effective in the treatment of bovine genital listeriosis (61). Other drugs of choice to treat cases of listeriosis in livestock include erythromycin and trimethoprim/sulfamethoxazole. In birds, penicillin, tetracycline, erythromycin, gentamicin, and trimethoprim/sulfamethoxazole may be used successfully to treat the septicemic forms of infection, while treatment of the encephalitic form is usually unsuccessful.
Strategies for vaccine development are currently under study, and these are urgently required to tackle the infection in sheep, though advances in these protocols remain to be validated (12).
In humans, there are specific antimicrobial chemotherapeutic strategies to treat listeriosis depending on the individual and the corresponding diagnosis. Ampicillin or penicillin G combined with an aminoglycoside, classically gentamicin, is the most commonly prescribed treatment for listeriosis (62, 63). However, the combination of trimethoprim/sulfamethoxazole can be used as an alternative treatment. A review by Janakiraman et al. (63) provides information on different treatment options for listeriosis in humans.
RESISTANCE OF LISTERIA SPECIES TO DIFFERENT ANTIMICROBIAL AGENTS
Antimicrobial agents have been used in a wide range of settings to eliminate or inhibit bacterial growth. Most of these compounds target unique bacterial cell features including cell wall synthesis, the bacterial membrane, particular stages of protein synthesis, DNA and RNA synthesis, and folic acid metabolism, and depending on the nature of the drug, these can result in bacterial cell death or the inhibition of growth (64).
Bacteria possess two types of resistance: intrinsic, or naturally occurring, resistance and acquired resistance via mutations in chromosomal genes and by horizontal gene transfer (65). Intrinsic resistance usually arises as a result of inherent structural or functional characteristics of the microorganism. An example of intrinsic resistance arises due to the lack of affinity of the antimicrobial compound for its bacterial target in L. monocytogenes, and this is found in two cases of β-lactam-based compounds, monobactams and broad-spectrum cephalosporins. In these cases, intrinsic resistance is caused by the low affinity of theses drugs for PBP3, the enzyme that catalyzes the final step during cell wall synthesis. Although a few intrinsic resistance mechanisms in Listeria species have been described, most cases of resistance to antimicrobial compounds in this bacterium are due to acquired mechanisms, such as mobile genetic elements including self-transferable plasmids and conjugative transposons (66, 67).
Listeria species are well known for their susceptibility to a wide range of antimicrobial agents. Nonetheless, they possess intrinsic or natural resistance to a select number of antimicrobial compounds. Although natural resistance differs among members of the genus Listeria, all of the Listeria species tested, including L. monocytogenes, L. innocua, L. welshimeri, L. ivanovii, L. grayi, and L. seeligeri, were susceptible or intermediately resistant to aminoglycosides, carbapenems, cefotiam, cefoperazone, first- and second-generation cephalosporins (cefaclor, cefazolin, loracarbef), chloramphenicol, dalfopristin/quinupristin, glycopeptides, lincosamides, macrolides, penicillins (except for oxacillin), and tetracyclines (68). Troxler et al. (68) demonstrated that L. monocytogenes and L. innocua were naturally resistant to fosfomycin and fusidic acid, whereas other Listeria species were found to be naturally resistant only to fusidic acid (Table 4) (68). Listeria species differ in their natural susceptibility to co-trimoxazole, fluoroquinolones, fosfomycin, fusidic acid, rifampicin, and trimethoprim (Table 4), while L. innocua, L. ivanovii, and L. seeligeri elaborated a natural resistance to the fluoroquinolones, enoxacin, and sparfloxacin (Table 4) (68).
TABLE 4.
Intrinsic or natural antibiotic susceptibility and resistance of Listeria species
| Listeria spp. | Naturally susceptible | Naturally resistant |
| L. monocytogenes | Trimethoprim | Fosfomycin |
| Co-trimoxazole | Fusidic acid | |
| L. innocua | Trimethoprim | Fosfomycin |
| Co-trimoxazole | Fusidic acid | |
| Enoxacin | ||
| Sparfloxacin | ||
| L. welshimeri | Trimethoprim | Fusidic acid |
| Co-trimoxazole | ||
| L. ivanovii | Fosfomycin | Enoxacin |
| Fusidic acida | Fusidic acida | |
| Trimethoprim | Fleroxacin | |
| Co-trimoxazole | Pefloxacin | |
| Sparfloxacin | ||
| L. seeligeri | Trimethoprim | Enoxacin |
| Co-trimoxazole | Fleroxacin | |
| Pefloxacin | ||
| Sparfloxacin | ||
| Fusidic acid | ||
| L. grayi | All Fluoroquinolones | Fusidic acid |
| Trimethoprim | ||
| Co-trimoxazole |
Both fusidic acid-susceptible and -resistant L. ivanovii isolates occur naturally in the environment.
Listeria species are rarely reported to develop acquired resistance to antimicrobial compounds. Nonetheless, selective pressure imposed in the past following the overuse use of various antimicrobial agents has resulted in members of this genus acquiring target gene mutations and resistance-encoding genes on mobile genetic elements (69). Transfer of resistance genes between Listeria species and other bacteria, such as Enterococcus and Streptococcus, by self-transferable plasmids has also been demonstrated (70).
Multidrug resistance was first documented in L. monocytogenes in 1988 in France (71). Since then, other Listeria species that are resistant to one or more compounds have been isolated, and several antimicrobial-resistance genes have been identified and characterized (62).
The following section provides a summary of the resistance-encoding genes that have been identified and mutations known to be associated with resistance to different classes of antibiotics, biocides, and heavy metals in Listeria species.
Antibiotics
Since the discovery of penicillin in 1929, antibiotics have been commercially used for the treatment of a wide variety of clinical diseases and have also been used for animal production purposes as additives in animal feed (72, 73). Over this period of time, selective pressure arising from the overuse of antimicrobial compounds has led to the emergence of bacteria expressing resistance to one or more of these agents. Similarly, members of the genus Listeria have developed mutations in the chromosome and/or acquired resistance genes carried on mobile genetic elements. In contrast and in comparison to other foodborne pathogens of human health importance, Listeria species do not exhibit these same features. Thus, data describing antimicrobial resistance in Listeria species is limited. Similar to other bacteria, Listeria species can express innate resistance to certain classes of antimicrobial compounds. In addition, several resistance genes have been reported to be located on mobile genetic elements such as conjugative transposons and plasmids. Therefore, members of the genus Listeria may play a role in the dissemination of resistance genes among bacteria.
Multidrug resistance has been documented in various isolates of Listeria, and several antimicrobial-resistance-encoding genes have been identified by means of molecular-based approaches. In the subsections below, a summary of the known mutations and genes associated with resistance to the different classes of antimicrobial agents is provided. The location of resistance genes on mobile genetic elements and the colocation of other markers is discussed.
Resistance to quinolones and fluoroquinolones
Quinolones and fluoroquinolones represent a class of antimicrobial agent which is important for the treatment of severe and invasive infections in animals and humans, with special interest for public and animal health. Quinolones were introduced into clinical use in 1962 in the form of nalidixic acid, and fluoroquinolones were first licensed for veterinary use in several countries in the 1980s (74). Subsequently, their use was followed by the reported emergence of antimicrobial resistance in bacteria. These bacteria were isolated from humans and food-producing animals (69, 75). Resistance to ciprofloxacin is mainly attributed to target gene mutations within the quinolone-resistance-determining regions of genes encoding the bacterial topoisomerase enzymes (76). The presence of plasmid-mediated quinolone-resistance (PMQR)-encoding genes can also contribute to the ciprofloxacin-resistance phenotype (77). Although these PMQR genes confer only low-level resistance to fluoroquinolones, the presence of PMQR (particularly qnr genes) may provide a selective advantage for bacteria exposed to fluoroquinolones and facilitate the subsequent development of high-level chromosomal quinolone resistance. In some Listeria isolates, the quinolone-resistance-determining region of DNA gyrase subunit A was altered; the deduced amino acid sequences revealed substitutions including Ser84 → Thr and Asp/Glu88 → Phe, both representing amino acid changes at hot spots commonly associated with resistance to these agents (78, 79). Another mechanism, involving efflux pumps such as Lde, MdrL, and FepA (see “Concluding Remarks”), has also been associated with resistance to fluoroquinolones in Listeria species (67, 80, 81). Macrolide-based antibiotics and cefotaxime, as well as heavy metals and ethidium bromide, are also exported by MdrL (80).
Resistance to penicillins and cephalosporins
Penicillins and cephalosporins are β-lactam-based antibiotics that inhibit bacterial cell wall synthesis. Listeria species are usually susceptible to penicillins, with the exception of oxacillin, monobactams, and broad-spectrum cephalosporins including cefetamet, cefixime, ceftibuten, ceftazidime, cefdinir, cefpodoxime, cefotaxime, ceftriaxone, and cefuroxime, to which this bacterium is naturally resistant (68). Only, the penA-encoding gene, a known PBP first identified from Neisseria meningitides, has been associated with resistance to penicillin G in L. monocytogenes (82). Some of the previously described mechanisms associated with the innate or natural resistance to cephalosporins most often include cell wall-acting gene products, two-component systems, and efflux pumps. The PBPs are enzymes that are responsible for extending the glycan chains in peptidoglycan and cross-linking the peptides between chains. Several PBPs were identified in Listeria and function to confer a natural resistance phenotype to modern cephalosporins (83). The gene oatA encodes an O-acetyltransferase, which catalyzes the acetylation of muramic acid in peptidoglycan, thus conferring resistance to cefotaxime and gallidermin. OatA is important for pathogenesis in mice, probably via protection from macrophage killing (84). Other mechanisms also confer reduced susceptibility to cephalosporins, including the two-component systems CesR and LiaSR, and efflux pumps (MdrL and AnrAB) (85).
Resistance to aminoglycosides
Aminoglycosides inhibit protein synthesis by binding to the 30S ribosomal subunit. Although more than 170 aminoglycoside-resistance-encoding genes have been described in bacteria, these can be grouped into three major classes: acetyltransferase-acting modifying enzymes, nucleotidyltransferases, and phosphotransferases (86). The streptomycin-resistance gene, aad6, which encodes for 6-N-streptomycin adenylyltransferase, has been identified in L. monocytogenes and L. innocua (69, 70). To date, no other aminoglycoside-resistance genes have been described in Listeria species.
Resistance to tetracyclines
Tetracycline resistance is the most frequent resistance phenotype detected in Listeria species. Tetracyclines inhibit protein synthesis by binding to the 30S ribosomal subunit. More than 50 tetracycline-resistance-encoding genes have been described, and five of these—tet(A), tet(K), tet(L), tet(M), and tet(S)—have been reported in Listeria species. Two known mechanisms of resistance to tetracyclines have been reported in Listeria species. These are mechanisms that involve efflux of the drug mediated by proton antiporters, conferring resistance to tetracycline only [including the genes tet(A), tet(K), and tet(L)] and ribosome protection proteins, conferring resistance to both tetracycline and minocycline [encoded by tet(M) and tet(S)]. Two types of mobile genetic elements, conjugative plasmids and transposons originating from Enterococcus-Streptococcus, are thought to be responsible for the emergence of resistance to tetracycline in Listeria. The tet(M) gene is often associated with the conjugative transposon Tn916, while tet(S) and tet(L) are more often carried on plasmids (87).
Resistance to phenicols
The molecular basis of bacterial resistance to chloramphenicol and its fluorinated derivative florfenicol has been reviewed by van Hoek et al. and Schwarz et al. (88, 89). In Listeria species, enzymatic inactivation of this drug by type A chloramphenicol acetyltransferases (Cat) and the export of chloramphenicol/florfenicol by specific efflux proteins are the dominant resistance mechanisms. A cat gene (type A-8) has been detected in Listeria, located on a plasmid (89, 90). The floR gene is associated with the export of florfenicol in L. monocytogenes, and 50% of floR-positive isolates may also confer resistance to chloramphenicol (82). Florfenicol and chloramphenicol coresistance conferred by floR has also been reported in Gram-negative bacteria such as Pasteurella piscicida and E. coli (82).
Resistance to macrolides (macrolides-lincosamides-streptogramin)
The macrolides (macrolides-lincosamides-streptogramin B [MLSB]) inhibit protein synthesis through binding to the 50S ribosomal subunit of bacteria. The resistance determinants responsible include rRNA methylases that modify the 23S ribosomal target sites, ATP-binding cassette (ABC) transporters, and efflux proteins of the major facilitator superfamily (MFS), as well as factors such as the ere-encoding genes that function as inactivating enzymes. The most common mechanism of MLSB resistance is due to the presence of rRNA methylases, encoded by the erm genes. These enzymes methylate an adenine base, which prevents drug binding to the 50S ribosomal subunit, resulting in MLSB resistance. There are currently 92 MLSB resistance genes recognized in bacteria (88), but only erm(A), erm(B), and erm(C) have been reported in Listeria species (91, 92). Plasmid pDB2011 from L. innocua, isolated from prepackaged sprouts in Switzerland, contained three antibiotic genes: spc (spectinomycin-adenyltransferase), erm(A) (erythromycin-methylase), and dfrD (trimethoprim-dihydrofolate); spc was located together with erm(A) on the transposon Tn554 (93). In Gram-positive bacteria, resistance to 14- and 15-membered-ring macrolides, such as erythromycin, can also be mediated by efflux pumps belonging to the ABC transporter family, such as msr(A) found in Staphylococcus species, or to the MFS mef(A) found in Streptococcus pneumoniae (69, 92). Although several studies have investigated the possible presence of mef(A) and msr(A) in Listeria species, further studies are necessary to extend our knowledge of the possible role of these efflux pumps in Listeria.
Resistance to trimethoprim
Trimethoprim is a folate pathway inhibitor, and folic acid is essential for the synthesis of adenine and thymine, two of the four bases that are involved as structural components of nucleic acids. Trimethoprim resistance has been described in L. monocytogenes due to the dfrD resistance-encoding gene. High-level trimethoprim resistance in Listeria is mainly due to the replacement of a trimethoprim-sensitive dihydrofolate reductase by a plasmid-, transposon-, or cassette-borne version. To date, more than 40 trimethoprim-resistant dihydrofolate resistance-mediating reductase (dfr) genes have been identified (94). Initially, the dfr genes were classified into two major families according to their amino acid structure, dfrA and dfrB, and currently, six plasmid-mediated families can be distinguished in Gram-positive bacteria, including dfrC, dfrD, dfrG, and dfrK (88).
In Listeria, only two of these—dfrG and dfrD—have been reported. The dfrD gene was found to be encoded on the plasmid pIP823 (95), while the dfrG gene was recently found in Tn6198, a Tn916-like element associated with a trimethoprim-resistance gene (96).
Biocides
Biocides have been widely used to control the microbial ecology of various niches for centuries. European Union regulation no. 528/2102, covering biocidal products (97), divides biocides into four categories: disinfectants, preservatives, pest control agents, and other biocidal products. These compounds are used in a wide variety of settings including the food and cosmetic industries, for personal hygiene, and for applications in veterinary practice and farming (98). Currently, there is a diverse range of chemicals including alcohols, biguanides, bisphenols, chlorine-releasing agents, iodophors, peroxides, and quaternary ammonium compounds (QACs), among others, that constitute biocides. Reduced susceptibility to these agents has been recognized for decades and is increasing (98, 99).
This section provides a summary of the known mechanisms of resistance or tolerance of Listeria species.
Quaternary ammonium compounds
QACs are amphoteric surfactants with a broad spectrum of antimicrobial activity and are widely used as disinfectants in clinical, domestic, and environmental settings (100). The most commonly used QAC is benzalkonium chloride (BC), also known as alkyl dimethyl benzyl ammonium chloride, which targets cytoplasmic membrane permeability functions, causing potassium leakage, osmoregulation disruption, enzyme inactivation, and protein denaturation (101, 102). Tolerance to BC was first described in L. monocytogenes and L. innocua isolates in 1998. BC tolerance may be conferred by transposons or efflux pumps encoded on the chromosome or on plasmids. One such tolerance mechanism previously described in L. monocytogenes was associated with the bcrABC operon, which can be encoded in the chromosome or on a putative IS1216 composite transposon harbored by the large plasmid pLM80 (103, 104). The bcrABC cassette is composed of a TetR-like transcriptional regulator (bcrA) and two small multidrug resistance (SMR) efflux pump-encoding genes (bcrB and bcrC) (103, 104). Another resistance mechanism identified in L. monocytogenes is the chromosomally integrated transposon Tn6188, which is related to other Tn554-like transposons. Transposon Tn6188 is a 5,117-bp structure found integrated within the radC gene, and it consists of three transposase genes (tnpABC), along with genes encoding a putative transcriptional regulator and QacH, an SMR transmembrane protein associated with the export of BC (105, 106). The qacH gene also confers tolerance to a wide range of QACs including BC, benzethonium chloride, cetylpyridinium chloride monohydrate, cetyltrimethylammonium bromide, domiphen bromide, dodecyltrimethylammonium bromide, ethidium bromide, and the sanitizer Weiquat, resulting in higher MIC values being recorded as well as in increased expression of the transporter (106). Although Tn6188 has been widely identified in L. monocytogenes, Tn6188 has not been identified in other Listeria species to date. The putative EmrE SMR-related efflux pump located within Listeria genomic island 1 (LGI1) in L. monocytogenes can also confer tolerance to BC and QAC-based sanitizers (107). There are two MFS efflux pumps, MdrL and Lde, that have also been reported to be partially responsible for BC tolerance in L. monocytogenes (108–110).
Triclosan
Triclosan is a chlorinated aromatic compound with a broad spectrum of antibacterial and antifungal activity, which has been used for over 40 years as an antiseptic, disinfectant, and preservative in clinical and cosmetics settings (111, 112). It is bacteriostatic in nature at low or sublethal concentrations, acting to inhibit a specific enzyme, FabI, an enoyl-acyl carrier protein reductase linked to fatty acid biosynthesis, (113–116). Inhibition of FabI results in reductions in lipid biosynthesis (117). At high or bactericidal concentrations, triclosan has a more general membrane-disrupting action, consistent with potassium leakage from the cell (118, 119). It has been shown that point mutations and overexpression of the fabI-encoding gene conferred tolerance to triclosan in some bacteria, but not in Listeria species (116, 120, 121). Exposure of L. monocytogenes to sublethal concentrations of triclosan can cause cross-resistance to several aminoglycosides and exhibit two types of colony morphology: normal-size and pinpoint colonies. This study demonstrated that adaptation to triclosan may be due to point mutations in several genes including ferrochelatase (hemH) or glutamyl tRNA reductase (hemA) (122).
Heavy Metals
From a saprophytic bacterium (all Listeria species) to intracellular pathogen (L. monocytogenes and L. ivanovii), maintaining heavy metal homeostasis is central to the survival of Listeria species across a broad range of environmental niches. Many heavy metals, such as iron and zinc, are an essential component of cellular function, and bacteria compete to sequester them using a variety of mechanisms such as siderophores and ATP-transporters. As heavy metal concentrations increase, associated toxicity can lead to cell death. Many factors can lead to an increase in heavy metal concentration in niches associated with Listeria species. The use of fertilizers or industrial processing can contaminate soil or water environments with heavy metals to levels which can be toxic to bacteria. Similarly, for pathogenic Listeria species, both heavy metal scavenging and resistance are necessary for survival and infection. It is becoming clear that heavy metals are an integral part of the host immune defense, either through host sequestering to deny availability or by exploiting their toxicity to kill invading pathogens (123–125).
Balancing the intracellular concentration of heavy metals is thus a matter of life or death for bacteria such as Listeria species. Studies investigating heavy metal resistance (HMR) in Listeria largely focus on L. monocytogenes, although these resistance mechanisms are often found in other Listeria species (126, 127). While much progress has been made in identifying mechanisms utilized by Listeria in resistance to certain heavy metals (e.g., cadmium and arsenic), less is known about many others (e.g., mercury and lead).
Many genes for HMR in bacteria are contained on mobile genetic elements such as plasmids and transposons (127–129), and as such, they are often found in association with a variety of other genetic markers, some of which may confer resistance to other antimicrobial agents or environmental stressors. Perhaps the most significant implication of this is the possibility of one resistance phenotype coselecting for another (130). Although mobile genetic element-mediated antibiotic resistance has not been frequently reported in Listeria, the potential coselection of Listeria plasmids containing multiple resistance markers has been reported (127, 131). In particular, heavy metal-mediated coselection of environmental stress resistance markers may contribute to survival and persistence of Listeria species in various environmental niches. Coselection of HMR plasmids may be elicited by a number of factors, including the use of heavy metals in agriculture in fertilizers or supplemented into animal diets as growth promoters or therapeutics (132, 133), heavy metal discharge in industrial wastewater effluent (134), and global mining activities (135). In addition, genetic markers of HMR may be associated with genes coding for disinfectant-resistance determinants (136); as a result, the use of disinfectants in food processing environments, which are frequently colonized by Listeria species (137, 138), may also serve to coselect for HMR (139). A multipronged approach will thus be necessary in any effort designed to limit the maintenance and spread of HMR-mediating mobile genetic elements in bacterial populations including Listeria species and others.
Cadmium resistance
Cadmium is not essential for the growth or survival of Listeria species, and it can be toxic to bacteria at low levels (140). Of all the heavy metals, cadmium-resistance phenotypes and mechanisms are among the most extensively studied in L. monocytogenes (136, 139, 141, 142). Resistance to cadmium was first correlated with plasmid carriage, with a cadAC operon being subsequently identified as the resistance determinant (143, 144). Efflux of Cd2+ is driven by the cadA-encoded energy-dependent pump, while cadC serves as a negatively acting regulatory protein of the operon (145, 146). The Listeria cadAC genes were first identified in a plasmid-borne Tn5422 transposon and were initially thought to be more prevalent among serotype 1/2 isolates (147). Recent studies have shown a more uniform distribution across serotype 1/2 and 4 isolates (148). Based on the frequency of plasmid carriage, plasmid profiling along with cadmium and arsenic-resistance phenotyping and serotyping has been suggested as a simpler means of subtyping L. monocytogenes isolates with acceptable discriminatory power (147). Variations of cadA described in Listeria include cadA1, the Tn5422 determinant described above, the cadA2 gene identified on plasmid pLM80 (149), cadA3 harbored by the EGD-e strain (3, 150), and cadA4 located on the chromosome of the outbreak strain ScottA (Fig. 4) (151).
FIGURE 4.
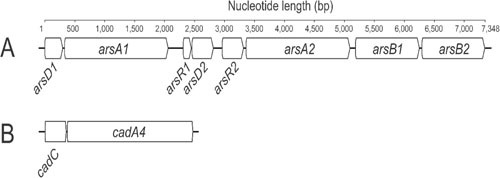
Heavy metal resistance operons in the L. monocytogenes strain ScottA. (A) Arsenic resistance operon. (B) cadAC cadmium resistance operon.
Arsenic resistance
Like cadmium, arsenic is also a nonessential heavy metal for bacterial growth and metabolism but becomes toxic as concentrations increase. Arsenic contamination of soil and water environments can arise from a number of sources including mining, industrial processing, and geothermal activity (152). Organic arsenic is less toxic than the inorganic form, and the use of arsenic has applications in medicine, pest control, and organic arsenic-based drugs. Organoarsenates have also been used as growth enhancers in food production systems (153, 154). Arsenic resistance in Listeria has been well characterized, and an arsenic-resistance cassette initially identified in L. innocua CLIP11262 on plasmid pLI100 has subsequently been identified in a number of L. monocytogenes isolates (Fig. 4) (127, 141, 151). Although harbored on plasmid pLI100, arsenic resistance in L. monocytogenes isolates to date typically show chromosomal association (141, 147). In contrast to cadmium resistance, which does not appear to have serotype-specific association, arsenic resistance has been primarily associated with serotype 4b isolates (139, 147). The use of organoarsenates in intensive poultry food production systems has also been implicated in selecting for arsenic resistance in Listeria species (139).
Zinc resistance
Unlike cadmium and arsenic, zinc is an essential micronutrient for Listeria species and is involved in diverse functions such as metabolism, cell division, and stress resistance (155). Two principle transport mechanisms responsible for sequestering zinc have been characterized in Listeria: the zurLAM and the zinABC operons (155). As with other heavy metals, however, zinc becomes toxic to Listeria species as the concentration increases. Elevated zinc levels are utilized in the host response to pathogen infection, and previous work has shown that this is associated with a bacteriostatic effect on L. monocytogenes (124, 156). Unlike other bacteria, cadA has not been shown to contribute to zinc resistance in Listeria species (128, 144).
Copper resistance
Although copper plays an important role in cellular function, Listeria species must maintain copper homeostasis to combat cytotoxic effects due to elevated concentrations (155). The csoR-copA-copZ operon confers copper resistance in L. monocytogenes (157). Other genetic markers implicated in copper resistance include the copper transporter CptA and a CutC homolog encoded by lmo1018 (155, 157). Copper is used as a growth promoter in food animals and, as such, has implications for selection of copper-resistant Listeria species; future work should address the implications of this selection pressure on coselection of other resistance markers.
MOBILE GENETIC ELEMENTS ASSOCIATED WITH ANTIMICROBIAL RESISTANCE IN LISTERIA SPECIES
DNA elements that can transfer within a genome or to another genome are generally referred to as mobile genetic elements. Transposons and plasmids are among the most commonly described mobile genetic elements in Listeria species and have been identified as one of the most important driving forces underlying the evolution of the species (158). As knowledge of the diversity of these mobile genetic elements increases, it is becoming apparent that they are frequently associated with a plethora of resistance phenotypes. Their dissemination through the Listeria population raises a number of concerns for food safety and public health; mobile genetic elements contributing to resistance of Listeria strains to disinfection regimes utilized in food processing have already been described, and reports are now emerging of resistance to clinically relevant antibiotics (96, 103, 105).
Transposons
Transposons are an important mechanism for the transfer of resistance markers through bacterial populations. A number of transposons and their insert sites (both chromosomal and plasmid) have been identified among Listeria species (129, 158), harboring resistance markers to different classes of antimicrobial agents, some of which include the transposon Tn917, which is implicated in cadmium resistance and is closely related to another plasmid-borne Tn5422 cadmium-resistance transposon (144, 159), the Tn554-like transposon, which carries an arsenic-resistance operon (158), a Tn6188 transposon (also a Tn554-like transposon), which carries the qacH gene and confers increased resistance to QACs, a bcrABC resistance cassette, which harbors an IS1216 composite transposon, a Tn916 to Tn1545 family conjugative transposon associated with tetracycline resistance and which contains tet(M) (160), and a Tn6198 multidrug-resistance transposon that confers resistance to tetracycline and trimethoprim (96).
Previous studies have noted the capacity for the transfer of chromosomal-located resistance determinants between Listeria species as well as other genera (96, 161), highlighting the potential for proliferation of antimicrobial resistance in Listeria species populations. The recent increase in whole-genome sequencing of Listeria genomes will undoubtedly expand our understanding of the repertoire of transposable elements present in Listeria species and provide insights into their putative transfer patterns. Such knowledge will be crucial in directing future control strategies aimed at tackling the issue of horizontal gene transfers among Listeria species.
Plasmids
Maintaining plasmids can impose a fitness cost on the host bacterium, which could lead to that host being outcompeted by other bacteria, in the absence of a selective pressure (162). However, resistance plasmid carriage is widespread in bacterial species, including subgroups of Listeria species (147, 163). Recent advances in understanding the factors underlying their ubiquity among bacterial populations have shown that very low levels of antimicrobial agent, often significantly below the MIC, can select for their carriage (164). This is further exacerbated by coselection, because plasmids often confer resistance to many different antimicrobial compounds, such as antibiotics, heavy metals, or biocides, and thus are maintained in bacterial populations by a multitude of selective pressures (103, 165).
Plasmids sharing replication mechanisms generally cannot exist together in the same host, which is often referred to as plasmid incomparability, and this feature forms the basis of incompatibility (Inc) grouping. Although commonly used for classification of Gram-negative plasmids, its use is less common among Gram-positive bacteria (166, 167). Studies relating to Listeria species plasmids suggest two distinct groupings based on their associated replicon and genetic diversity (127). Few instances of Listeria species harboring more than one plasmid have been reported (168), but more research is needed to understand the dynamics of plasmid carriage and incompatibility in Listeria species.
HMR markers are among the most commonly described with regard to Listeria plasmids, particularly cadmium-resistance genes (129, 144, 169). These plasmids frequently harbor other resistance markers associated with biocide and/or stress resistance, as well as efflux systems (127, 129, 136). These markers may contribute to the survival and persistence of Listeria species in food processing environments and, as such, present a significant concern for food safety and public health (127, 131). Another intriguing observation is the conservation of identical or similar plasmids among isolates from geographically diverse locations, suggesting that strong selective pressures are maintaining plasmid carriage among Listeria species (131).
Although the frequency of antibiotic resistance among Listeria species has been reported to be low (69, 170), plasmid carriage of resistance-encoding mechanisms to antibiotics including chloramphenicol, erythromycin, and tetracycline has been reported (71, 171, 172). In addition, a recently identified multidrug-resistance plasmid in L. innocua conferring resistance to both trimethoprim and spectinomycin exhibited a broad host range (93). The emergence of such resistance mechanisms highlights the need for increased surveillance efforts to attempt to minimize the risk of emergence of clinically relevant antimicrobial resistance in Listeria species and in L. monocytogenes in particular.
EFFLUX PUMPS ASSOCIATED WITH ANTIMICROBIAL RESISTANCE IN LISTERIA SPECIES
Efflux pumps are transmembrane-spanning protein complexes capable of transporting a broad range of chemically and structurally unrelated substrate molecules from inside the bacterial cell to the extracellular matrix (173). Bacterial efflux pumps have been classified into five families: the ABC superfamily, the multidrug and toxic compound extrusion family (MATE), the MFS, the SMR family, and the resistance-nodulation-division superfamily (174). The most commonly identified efflux pumps found in Gram-positive bacteria are those represented by the MFS family, in contrast to those of the resistance-nodulation-division family being found in Gram-negative bacteria. The latter family is rarely reported in Gram-positive bacteria. All of these systems harness the proton motive force as an energy source, with the exception of the ABC transporters, which use direct ATP hydrolysis to drive the transport of substrates (Fig. 5) (175). In the past few years, several efflux pumps have been described in L. monocytogenes.
FIGURE 5.
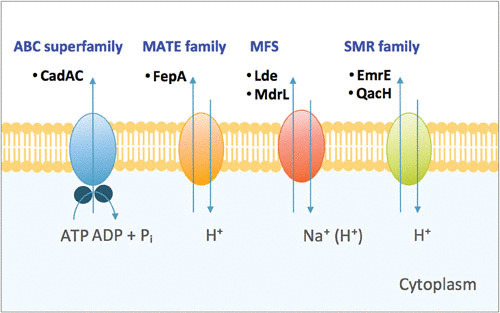
Diagrammatic representation of the four families of efflux pumps in L. monocytogenes. The ATP-binding cassette (ABC) superfamily, the major facilitator superfamily (MFS), the multidrug and toxic-compound extrusion (MATE) family, and the small multidrug resistance (SMR) family. Common examples of the individual proteins that form each class of efflux pump are shown.
The first efflux pump to be described in L. monocytogenes was cadAC. Shortly thereafter, an MFS efflux pump, denoted MdrL, was described in L. monocytogenes, and it was reported to export cefotaxime, ethidium bromide, heavy metals, and macrolides (Table 5) (80). Several subsequent studies have also reported on the role of MdrL in the adaptation of L. monocytogenes to BC (108, 109). L. monocytogenes encodes two efflux pumps that are highly similar to QacA in Staphylococcus aureus, and they were identified as MdrT and MdrM. Both MFS efflux pumps confer resistance to cholic acid (176, 177). MrdT and MdrM also play a role during bacterial replication within the cytosol of infected cells, by secreting cyclic-di-AMP, which triggers the production of type I interferons, including beta interferon, which promotes L. monocytogenes virulence (178).
TABLE 5.
Multidrug efflux transporters characterzsed in L. monocytogenes
| Gene | Efflux pump | Regulator | Substrates |
|---|---|---|---|
| lmo1409 | MdrL | LadR |
|
| lmo1617 | MdrM | MarR |
|
| lmo2588 | MdrT | BrtAa |
|
| lmo2741 | Lde | Unknown |
|
| lmo1851 | EmrE | Unknown | Benzalkonium chloride |
| lmo2089 | FepA | FepRa |
|
| lmo2115 | AnrB | VirR |
|
| cadAC | CadAC | CadC |
|
| bcrABC | BcrBC | BcrAa | Benzalkonium chloride |
| qacH | QacH | TetR |
|
Belongs to the TetR family.
Two efflux pumps that confer resistance to fluoroquinolones have been characterized in L. monocytogenes, Lde and FepA. The former is encoded by the lde gene, whose protein product is a 12-segment transmembrane-spanning putative MFS efflux pump with 44% amino acid identity with PmrA from S. pneumoniae. Lde was characterized in L. monocytogenes CLIP 21369 by insertional inactivation of the lde gene, and these data demonstrated that this pump conferred resistance to ciprofloxacin and norfloxacin together with a reduced susceptibility to the dyes acridine orange and ethidium bromide (Table 5) (67, 179). Other studies have also suggested that lde may be partly responsible for BC tolerance (110). Lde has other interesting functions, because it may also cooperate with a eukaryotic multidrug resistance-related protein (MRP)-like efflux transporter to reduce the activity of ciprofloxacin, a substrate for both pumps, in J774 macrophages infected with L. monocytogenes (174, 180). Furthermore, the L. monocytogenes in vivo-induced virulence factor Hpt mediates uptake of fosfomycin in L. monocytogenes, thereby conferring a resistant phenotype in vitro and thus constituting an antibacterial in vitro-in vivo paradox, since the bacteria are resistant in vitro but susceptible to the drug in vivo (174, 181).
Another mechanism that may confer resistance to fluoroquinolones in L. monocytogenes is linked with the overexpression of a multidrug and toxic compound extrusion efflux pump, FepA. It has been demonstrated that a single point mutation in the FepA transcriptional regulator fepR, which belongs to the TetR family, produces a frameshift mutation that causes the introduction of a premature stop codon, resulting in an inactive truncated protein. This single point mutation was responsible for the overexpression of fepA, which confers resistance to ciprofloxacin, ethidium bromide, and norfloxacin, thereby confirming the role of FepR as a local repressor of fepA (Table 5) (81).
AnrB, an ABC transporter, confers innate resistance of L. monocytogenes to the lantibiotic nisin. The AnrB function was demonstrated by a nonpolar deletion mutation in the lmo2115 gene, and the loss of this multidrug transporter increased susceptibility to bacitracin, β-lactams, gallidermin, and nisin (Table 5) (182).
In 2015, a novel putative efflux pump, EmrE (324 bp), was described within LGI1 in L. monocytogenes (107). An emrE deletion mutant demonstrated an increase in the susceptibility to BC along with QAC-based sanitizers but exhibited no effect on the MICs for acriflavine, chloramphenicol, ciprofloxacin, erythromycin, gentamicin, tetracycline, and triclosan (Table 5) (107).
CONCLUDING REMARKS
Listeria species are ubiquitous throughout the food chain and the natural environment, and these microorganisms have been isolated from a wide variety of domestic and wild animals. Since 2009, nine new Listeria species have been discovered, and these remain to be further characterized.
Listeriosis may manifest in a mild form, presenting with gastroenteritis-like symptoms, or in a more severe form where Listeria crosses the blood-brain barrier and placenta, invading the CNS and the fetus. Antibiotic treatment strategies for listeriosis vary according to the infection type and the nature of the infected host. In humans, ampicillin or penicillin G in combination with an aminoglycoside is regarded as the front-line treatment (63). The primary antibiotics used to treat listeriosis in animals are chlortetracycline, penicillin, tetracycline, erythromycin, gentamicin, and trimethoprim/sulfamethoxazole (61). Although the incidence of antibiotic resistance in Listeria species remains low, multidrug-resistant Listeria species strains have been isolated from numerous sources including clinical isolates, retail foods, and the environment (69, 92, 183). Furthermore, the range of antibiotics to which resistance has been acquired is broad and includes compounds currently used to treat listeriosis, such as ampicillin, penicillin, trimethoprim, tetracycline, and erythromycin, among others (88, 183). Resistance to erythromycin, rifampicin, trimethoprim, and especially, to tetracycline has been commonly reported in Listeria species from Europe and North America and is consistent with the identification of tet-, erm-, aad-, and dfr-encoding genes (79). This development may be problematic for patients who are allergic to penicillin because the combination of trimethoprim and sulfamethoxazole is used for the treatment of listeriosis (63).
Multidrug resistance in Listeria species has also been linked to the presence of efflux pumps, which confer resistance to a wide variety of antimicrobials, including antibiotics, biocides, heavy metals, and other antimicrobial compounds. Lde and AnrB confer resistance to fluoroquinolones, whereas EmrE, BcrABC, and QacH confer tolerance to BC. Tolerance to biocides is of special concern in food-producing plants and farms, because these agents are used as sanitizers and their decreasing efficacy hinders the elimination of Listeria from these environments, increasing the risk of cross-contamination to the final product or animals. Efflux pumps and antimicrobial-resistance genes can be encoded chromosomally or on mobile genetic elements, e.g., plasmid pDB2011 from L. innocua, which contains three antibiotic resistant genes, spc, erm(A), and dfrD, and confers resistance to spectinomycin, erythromycin, and trimethoprim, and Tn6188, which encodes for the qacH gene and confers resistance to QACs. Furthermore, Listeria can acquire antimicrobial genes from foreign sources by self-transferable plasmids or conjugative transposons from other bacterial species such as Enterococcus and Streptococcus (62, 170, 183).
Despite the ever-increasing threat of antimicrobial resistance to human health and the fact that Listeria is widespread in the environment and agriculture, relatively little research has been focused on this topic to date.
ACKNOWLEDGMENTS
This work was supported by grant 11/F/008 from the Irish Department of Agriculture and Food and the Marine (DAFM) under the Food Institutional Research Measure (FIRM) Network.
REFERENCES
- 1.Orsi RH, Wiedmann M. 2016. Characteristics and distribution of Listeria spp., including Listeria species newly described since 2009. Appl Microbiol Biotechnol 100:5273–5287 10.1007/s00253-016-7552-2. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feresu SB, Jones D. 1988. Taxonomic studies on Brochothrix, Erysipelothrix, Listeria and atypical lactobacilli. J Gen Microbiol 134:1165–1183. [PubMed] [DOI] [PubMed] [Google Scholar]
- 3.Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couvé E, de Daruvar A, Dehoux P, Domann E, Domínguez-Bernal G, Duchaud E, Durant L, Dussurget O, Entian KD, Fsihi H, García-del Portillo F, Garrido P, Gautier L, Goebel W, Gómez-López N, Hain T, Hauf J, Jackson D, Jones LM, Kaerst U, Kreft J, Kuhn M, Kunst F, Kurapkat G, Madueno E, Maitournam A, Vicente JM, Ng E, Nedjari H, Nordsiek G, Novella S, de Pablos B, Pérez-Diaz JC, Purcell R, Remmel B, Rose M, Schlueter T, Simoes N, et al. 2001. Comparative genomics of Listeria species. Science 294:849–852. [PubMed] [DOI] [PubMed] [Google Scholar]
- 4.Orsi RH, den Bakker HC, Wiedmann M. 2011. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol 301:79–96 10.1016/j.ijmm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Piffaretti JC, Kressebuch H, Aeschbacher M, Bille J, Bannerman E, Musser JM, Selander RK, Rocourt J. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc Natl Acad Sci USA 86:3818–3822 10.1073/pnas.86.10.3818. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts A, Nightingale K, Jeffers G, Fortes E, Kongo JM, Wiedmann M. 2006. Genetic and phenotypic characterization of Listeria monocytogenes lineage III. Microbiology 152:685–693 10.1099/mic.0.28503-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 7.Ward TJ, Ducey TF, Usgaard T, Dunn KA, Bielawski JP. 2008. Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl Environ Microbiol 74:7629–7642 10.1128/AEM.01127-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States: major pathogens. Emerg Infect Dis 17:7–15 10.3201/eid1701.P11101. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray EGD, Webb RA, Swann MBR. 1926. A disease of rabbits characterised by a large mononuclear leucocytosis, caused by a hitherto undescribed bacillus Bacterium monocytogenes (n.sp.). J Pathol Bacteriol 29:407–439 10.1002/path.1700290409. [DOI] [Google Scholar]
- 10.Walland J, Lauper J, Frey J, Imhof R, Stephan R, Seuberlich T, Oevermann A. 2015. Listeria monocytogenes infection in ruminants: is there a link to the environment, food and human health? A review. Schweiz Arch Tierheilkd 157:319–328 10.17236/sat00022. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Lyautey E, Hartmann A, Pagotto F, Tyler K, Lapen DR, Wilkes G, Piveteau P, Rieu A, Robertson WJ, Medeiros DT, Edge TA, Gannon V, Topp E. 2007. Characteristics and frequency of detection of fecal Listeria monocytogenes shed by livestock, wildlife, and humans. Can J Microbiol 53:1158–1167 10.1139/W07-084. [PubMed] [DOI] [PubMed] [Google Scholar]
- 12.Lopez J. 2008. Listeria monocytogenes, p. 1238–1254. In OIE Biological Standards Commission (ed), Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, vol 2, 6th ed. World Organisation for Animal Health (OIE), Paris, France. [Google Scholar]
- 13.Dhama K, Karthik K, Tiwari R, Shabbir MZ, Barbuddhe S, Malik SVS, Singh RK. 2015. Listeriosis in animals, its public health significance (food-borne zoonosis) and advances in diagnosis and control: a comprehensive review. Vet Q 35:211–235 10.1080/01652176.2015.1063023. [PubMed] [DOI] [PubMed] [Google Scholar]
- 14.Gray ML. 1958. Listeriosis in fowls: a review. Avian Dis 2:296 10.2307/1587530. [DOI] [Google Scholar]
- 15.Loncarevic A, Artursson, Johansson. 1999. A case of canine cutaneous listeriosis. Vet Dermatol 10:69–71 10.1046/j.1365-3164.1999.00127.x. [DOI] [PubMed] [Google Scholar]
- 16.Revold T, Abayneh T, Brun-Hansen H, Kleppe SL, Ropstad E-O, Hellings RA, Sørum H. 2015. Listeria monocytogenes associated kerato-conjunctivitis in four horses in Norway. Acta Vet Scand 57:76 10.1186/s13028-015-0167-2. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber A, Prell A, Potel J, Schäfer R. 1993. Occurrence of Listeria monocytogenes in snakes, tortoises, lizards and amphibians raised as pets. Berl Munch Tierarztl Wochenschr 106:293–295. (In German.) [PubMed] [PubMed] [Google Scholar]
- 18.Arumugaswamy R, Gibson LF. 1999. Listeria in zoo animals and rivers. Aust Vet J 77:819–820 10.1111/j.1751-0813.1999.tb12955.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 19.Bauwens L, Vercammen F, Hertsens A. 2003. Detection of pathogenic Listeria spp. in zoo animal faeces: use of immunomagnetic separation and a chromogenic isolation medium. Vet Microbiol 91:115–123 10.1016/S0378-1135(02)00265-1. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida T, Sugimoto T, Sato M, Hirai K. 2000. Incidence of Listeria monocytogenes in wild animals in Japan. J Vet Med Sci 62:673–675 10.1292/jvms.62.673. [PubMed] [DOI] [PubMed] [Google Scholar]
- 21.Nowakiewicz A, Zięba P, Ziółkowska G, Gnat S, Muszyńska M, Tomczuk K, Majer Dziedzic B, Ulbrych Ł, Trościańczyk A. 2016. Free-living species of carnivorous mammals in Poland: red fox, beech marten, and raccoon as a potential reservoir of Salmonella, Yersinia, Listeria spp. and coagulase-positive Staphylococcus.PLoS One 11:e0155533 10.1371/journal.pone.0155533. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenlon DR, Wilson J, Donachie W. 1996. The incidence and level of Listeria monocytogenes contamination of food sources at primary production and initial processing. J Appl Bacteriol 81:641–650 10.1111/j.1365-2672.1996.tb03559.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.Fieseler L, Doyscher D, Loessner MJ, Schuppler M. 2014. Acanthamoeba release compounds which promote growth of Listeria monocytogenes and other bacteria. Appl Microbiol Biotechnol 98:3091–3097 10.1007/s00253-014-5534-9. [PubMed] [DOI] [PubMed] [Google Scholar]
- 24.Freitag NE, Port GC, Miner MD. 2009. Listeria monocytogenes: from saprophyte to intracellular pathogen. Nat Rev Microbiol 7:623–628 10.1038/nrmicro2171. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nightingale KK, Schukken YH, Nightingale CR, Fortes ED, Ho AJ, Her Z, Grohn YT, McDonough PL, Wiedmann M. 2004. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl Environ Microbiol 70:4458–4467 10.1128/AEM.70.8.4458-4467.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver SP, Jayarao BM, Almeida RA. 2005. Foodborne pathogens in milk and the dairy farm environment: food safety and public health implications. Foodborne Pathog Dis 2:115–129 10.1089/fpd.2005.2.115. [PubMed] [DOI] [PubMed] [Google Scholar]
- 27.Wilkes G, Edge TA, Gannon VPJ, Jokinen C, Lyautey E, Neumann NF, Ruecker N, Scott A, Sunohara M, Topp E, Lapen DR. 2011. Associations among pathogenic bacteria, parasites, and environmental and land use factors in multiple mixed-use watersheds. Water Res 45:5807–5825 10.1016/j.watres.2011.06.021. [PubMed] [DOI] [PubMed] [Google Scholar]
- 28.Esteban JI, Oporto B, Aduriz G, Juste RA, Hurtado A, Roberts A, Wiedmann M, Buncic S, Iida T, Kanzaki M, Nakama A, Kokubo Y, Maruyama T, Kaneuchi C, Skovgaard N, Norrung B, de Valk H, Vaillant V, Jacquet C, Rocourt J, Le Querrec F, Stainer F, Quelquejeu N, Pierre O, Pierre V, Desenclos J, Goulet V, McLauchlin J, Hall S, Velani S, Gilbert R, Giovannacci I, Ragimbeau C, Queguiner S, Salvat G, Vendeuvre J, Carlier V, Ermel G, Nightingale K, Schukken Y, Nightingale C, Fortes E, Ho A, Her Z, Grohn Y, McDonough P, Wiedmann M, Gandhi M, Chikindas M, Fenlon D, et al. 2009. Faecal shedding and strain diversity of Listeria monocytogenes in healthy ruminants and swine in Northern Spain. BMC Vet Res 5:2. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber A, Potel J, Schäfer-Schmidt R, Prell A, Datzmann C. 1995. Studies on the occurrence of Listeria monocytogenes in fecal samples of domestic and companion animals. Zentralbl Hyg Umweltmed 198:117–123. (In German.) [PubMed] [PubMed] [Google Scholar]
- 30.Walland J, Lauper J, Frey J, Imhof R, Stephan R, Seuberlich T, Oevermann A. 2015. Listeria monocytogenes infection in ruminants: is there a link to the environment, food and human health? A review. Schweiz Arch Tierheilkd 157:319–328 10.17236/sat00022. [PubMed] [DOI] [PubMed] [Google Scholar]
- 31.Rocourt J, Hof H, Schrettenbrunner A, Malinverni R, Bille J. 1986. Acute purulent Listeria seelingeri meningitis in an immunocompetent adult. Schweiz Med Wochenschr 116:248–251. (In French.) [PubMed] [PubMed] [Google Scholar]
- 32.Perrin M, Bemer M, Delamare C. 2003. Fatal case of Listeria innocua bacteremia. J Clin Microbiol 41:5308–5309 10.1128/JCM.41.11.5308-5309.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vázquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Domínguez-Bernal G, Goebel W, González-Zorn B, Wehland J, Kreft J. 2001. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev 14:584–640 10.1128/CMR.14.3.584-640.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cossart P, Lebreton A. 2014. A trip in the “New Microbiology” with the bacterial pathogen Listeria monocytogenes. FEBS Lett 588:2437–2445 10.1016/j.febslet.2014.05.051. [PubMed] [DOI] [PubMed] [Google Scholar]
- 35.Appelberg R, Castro AG, Silva MT. 1994. Neutrophils as effector cells of T-cell-mediated, acquired immunity in murine listeriosis. Immunology 83:302–307. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 36.Carvalho F, Sousa S, Cabanes D. 2014. How Listeria monocytogenes organizes its surface for virulence. Front Cell Infect Microbiol 4:48 10.3389/fcimb.2014.00048. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cossart P, Lecuit M. 1998. Interactions of Listeria monocytogenes with mammalian cells during entry and actin-based movement: bacterial factors, cellular ligands and signaling. EMBO J 17:3797–3806 10.1093/emboj/17.14.3797. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pizarro-Cerdá J, Kühbacher A, Cossart P. 2012. Entry of Listeria monocytogenes in mammalian epithelial cells: an updated view. Cold Spring Harb Perspect Med 2:a010009 10.1101/cshperspect.a010009. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camejo A, Carvalho F, Reis O, Leitão E, Sousa S, Cabanes D. 2011. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2:379–394 10.4161/viru.2.5.17703. [PubMed] [DOI] [PubMed] [Google Scholar]
- 40.Mohammed HO, Stipetic K, McDonough PL, Gonzalez RN, Nydam DV, Atwill ER. 2009. Identification of potential on-farm sources of Listeria monocytogenes in herds of dairy cattle. Am J Vet Res 70:383–388 10.2460/ajvr.70.3.383. [PubMed] [DOI] [PubMed] [Google Scholar]
- 41.Walker JK, Morgan JH, McLauchlin J, Grant KA, Shallcross JA. 1994. Listeria innocua isolated from a case of ovine meningoencephalitis. Vet Microbiol 42:245–253 10.1016/0378-1135(94)90023-X. [PubMed] [DOI] [PubMed] [Google Scholar]
- 42.Oevermann A, Di Palma S, Doherr MG, Abril C, Zurbriggen A, Vandevelde M. 2010. Neuropathogenesis of naturally occurring encephalitis caused by Listeria monocytogenes in ruminants. Brain Pathol 20:378–390 10.1111/j.1750-3639.2009.00292.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hird DW, Genigeorgis C. 1990. Listeriosis in food animals: clinical signs and livestock as a potential source of direct (nonfoodborne) infection for humans, p 31–39. In Miller, AJ, Smith, JL, Somkuti, GA (ed), Foodborne Listeriosis: Topics in Industrial Microbiology. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 44.Clark RG, Gill JM, Swanney S. 2004. Listeria monocytogenes gastroenteritis in sheep. N Z Vet J 52:46–47 10.1080/00480169.2004.36391. [PubMed] [DOI] [PubMed] [Google Scholar]
- 45.Starič J, Križanec F, Zadnik T. 2008. Listeria monocytogenes keratoconjunctivitis and uveitis in dairy cattle. Bull Vet Inst Pulawy 52:351–355. [Google Scholar]
- 46.Ivanek R, Gröhn YT, Wiedmann M. 2006. Listeria monocytogenes in multiple habitats and host populations: review of available data for mathematical modeling. Foodborne Pathog Dis 3:319–336 10.1089/fpd.2006.3.319. [PubMed] [DOI] [PubMed] [Google Scholar]
- 47.Kahn CM. 2005. Listeriosis, p. 2240–2241. In Kahn CN, Line Scott (ed), The Merck Veterinary Manual, 9th ed. Merck Publishing Group, Rahway, NJ. [Google Scholar]
- 48.Jeckel S, Wood A, Grant K, Amar C, King SA, Whatmore AM, Koylass M, Anjum M, James J, Welchman DB. 2015. Outbreak of encephalitic listeriosis in red-legged partridges (Alectoris rufa). Avian Pathol 44:269–277 10.1080/03079457.2015.1042427. [PubMed] [DOI] [PubMed] [Google Scholar]
- 49.Kurazono M, Nakamura K, Yamada M, Yonemaru T, Sakoda T. 2003. Pathology of listerial encephalitis in chickens in Japan. Avian Dis 47:1496–1502 10.1637/7066. [PubMed] [DOI] [PubMed] [Google Scholar]
- 50.Guillet C, Join-Lambert O, Le Monnier A, Leclercq A, Mechaï F, Mamzer-Bruneel M-F, Bielecka MK, Scortti M, Disson O, Berche P, Vazquez-Boland J, Lortholary O, Lecuit M. 2010. Human listeriosis caused by Listeria ivanovii. Emerg Infect Dis 16:136–138 10.3201/eid1601.091155. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lecuit M, Nelson DM, Smith SD, Khun H, Huerre M, Vacher-Lavenu M-C, Gordon JI, Cossart P. 2004. Targeting and crossing of the human maternofetal barrier by Listeria monocytogenes: role of internalin interaction with trophoblast E-cadherin. Proc Natl Acad Sci USA 101:6152–6157 10.1073/pnas.0401434101. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLauchlin J. 1992. Listeriosis. BMJ 304:1583–1584 10.1136/bmj.304.6842.1583. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goulet V, King LA, Vaillant V, de Valk H. 2013. What is the incubation period for listeriosis? BMC Infect Dis 13:11 10.1186/1471-2334-13-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wesley IV. 1999. Listeriosis in animals, p. 39–73. In Ryser ET, Marth EH (ed), Listeria, Listeriosis, and Food Safety, 2nd ed. Marcel Dekker Inc., New York, NY. [Google Scholar]
- 55.Portnoy DA, Auerbuch V, Glomski IJ. 2002. The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J Cell Biol 158:409–414 10.1083/jcb.200205009. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edelson BT, Cossart P, Unanue ER. 1999. Cutting edge: paradigm revisited: antibody provides resistance to Listeria infection. J Immunol 163:4087–4090. [PubMed] [PubMed] [Google Scholar]
- 57.Berche P, Bonnichon M, Beretti JL, Raveneau J, Gaillard JL, Veron M, Kreis H, Reich KA, Cossart P, Geoffroy C, Geslin P. 1990. Detection of anti-listeriolysin O for serodiagnosis of human listeriosis. Lancet 335:624–627 10.1016/0140-6736(90)90411-W. [PubMed] [DOI] [PubMed] [Google Scholar]
- 58.Low JC, Davies RC, Donachie W. 1992. Purification of listeriolysin O and development of an immunoassay for diagnosis of listeric infections in sheep. J Clin Microbiol 30:2705–2708. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baetz AL, Wesley IV, Stevens MG. 1996. The use of listeriolysin O in an ELISA, a skin test and a lymphocyte blastogenesis assay on sheep experimentally infected with Listeria monocytogenes, Listeria ivanovii, or Listeria innocua. Vet Microbiol 51:151–159 10.1016/0378-1135(96)00033-8. [DOI] [PubMed] [Google Scholar]
- 60.Hof H, Nichterlein T, Kretschmar M. 1997. Management of listeriosis. Clin Microbiol Rev 10:345–357. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chopra S, Sharma V, Shukla S, Nayak A. 2015. Antibiogram of Listeria spp. isolated from reproductive disorders and livestock products of ruminants. J Anim Res 2:187–190. [Google Scholar]
- 62.Charpentier E, Courvalin P. 1999. Antibiotic resistance in Listeria spp. Antimicrob Agents Chemother 43:2103–2108. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Janakiraman V. 2008. Listeriosis in pregnancy: diagnosis, treatment, and prevention. Rev Obstet Gynecol 1:179–185. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 64.Wright GD. 2010. Q&A: Antibiotic resistance: where does it come from and what can we do about it? BMC Biol 8:123 10.1186/1741-7007-8-123. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. 2015. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51 10.1038/nrmicro3380. [PubMed] [DOI] [PubMed] [Google Scholar]
- 66.Charpentier E, Gerbaud G, Courvalin P. 1999. Conjugative mobilization of the rolling-circle plasmid pIP823 from Listeria monocytogenes BM4293 among Gram-positive and Gram-negative bacteria. J Bacteriol 181:3368–3374. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Godreuil S, Galimand M, Gerbaud G, Jacquet C, Courvalin P. 2003. Efflux pump Lde is associated with fluoroquinolone resistance in Listeria monocytogenes. Antimicrob Agents Chemother 47:704–708 10.1128/AAC.47.2.704-708.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Troxler R, von Graevenitz A, Funke G, Wiedemann B, Stock I. 2000. Natural antibiotic susceptibility of Listeria species: L. grayi, L. innocua, L. ivanovii, L. monocytogenes, L. seeligeri and L. welshimeri strains. Clin Microbiol Infect 6:525–535 10.1046/j.1469-0691.2000.00168.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 69.Morvan A, Moubareck C, Leclercq A, Hervé-Bazin M, Bremont S, Lecuit M, Courvalin P, Le Monnier A. 2010. Antimicrobial resistance of Listeria monocytogenes strains isolated from humans in France. Antimicrob Agents Chemother 54:2728–2731 10.1128/AAC.01557-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lungu B, O’Bryan CA, Muthaiyan A, Milillo SR, Johnson MG, Crandall PG, Ricke SC. 2011. Listeria monocytogenes: antibiotic resistance in food production. Foodborne Pathog Dis 8:569–578 10.1089/fpd.2010.0718. [PubMed] [DOI] [PubMed] [Google Scholar]
- 71.Poyart-Salmeron C, Carlier C, Trieu-Cuot P, Courtieu AL, Courvalin P. 1990. Transferable plasmid-mediated antibiotic resistance in Listeria monocytogenes. Lancet 335:1422–1426 10.1016/0140-6736(90)91447-I. [DOI] [PubMed] [Google Scholar]
- 72.Jones FT, Ricke SC. 2003. Observations on the history of the development of antimicrobials and their use in poultry feeds. Poult Sci 82:613–617 10.1093/ps/82.4.613. [PubMed] [DOI] [PubMed] [Google Scholar]
- 73.Castanon JIR. 2007. History of the use of antibiotic as growth promoters in European poultry feeds. Poult Sci 86:2466–2471 10.3382/ps.2007-00249. [PubMed] [DOI] [PubMed] [Google Scholar]
- 74.Robicsek A, Jacoby GA, Hooper DC. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis 6:629–640 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 75.Lyon SA, Berrang ME, Fedorka-Cray PJ, Fletcher DL, Meinersmann RJ. 2008. Antimicrobial resistance of Listeria monocytogenes isolated from a poultry further processing plant. Foodborne Pathog Dis 5:253–259 10.1089/fpd.2007.0070. [PubMed] [DOI] [PubMed] [Google Scholar]
- 76.Chen S, Cui S, McDermott PF, Zhao S, White DG, Paulsen I, Meng J. 2007. Contribution of target gene mutations and efflux to decreased susceptibility of Salmonella enterica serovar typhimurium to fluoroquinolones and other antimicrobials. Antimicrob Agents Chemother 51:535–542 10.1128/AAC.00600-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu J-H, Deng Y-T, Zeng Z-L, Gao J-H, Chen L, Arakawa Y, Chen Z-L. 2008. Coprevalence of plasmid-mediated quinolone resistance determinants QepA, Qnr, and AAC(6′)-Ib-cr among 16S rRNA methylase RmtB-producing Escherichia coli isolates from pigs. Antimicrob Agents Chemother 52:2992–2993 10.1128/AAC.01686-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lampidis R, Kostrewa D, Hof H. 2002. Molecular characterization of the genes encoding DNA gyrase and topoisomerase IV of Listeria monocytogenes. J Antimicrob Chemother 49:917–924 10.1093/jac/dkf065. [PubMed] [DOI] [PubMed] [Google Scholar]
- 79.Moreno LZ, Paixão R, Gobbi DDS, Raimundo DC, Ferreira TP, Moreno AM, Hofer E, Reis CMF, Matté GR, Matté MH. 2014. Characterization of antibiotic resistance in Listeria spp. isolated from slaughterhouse environments, pork and human infections. J Infect Dev Ctries 8:416–423 10.3855/jidc.4188. [PubMed] [DOI] [PubMed] [Google Scholar]
- 80.Mata MT, Baquero F, Pérez-Díaz JC. 2000. A multidrug efflux transporter in Listeria monocytogenes. FEMS Microbiol Lett 187:185–188 10.1111/j.1574-6968.2000.tb09158.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 81.Guérin F, Galimand M, Tuambilangana F, Courvalin P, Cattoir V. 2014. Overexpression of the novel MATE fluoroquinolone efflux pump FepA in Listeria monocytogenes is driven by inactivation of its local repressor FepR. PLoS One 9:e106340 10.1371/journal.pone.0106340. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Srinivasan V, Nam HM, Nguyen LT, Tamilselvam B, Murinda SE, Oliver SP. 2005. Prevalence of antimicrobial resistance genes in Listeria monocytogenes isolated from dairy farms. Foodborne Pathog Dis 2:201–211 10.1089/fpd.2005.2.201. [PubMed] [DOI] [PubMed] [Google Scholar]
- 83.Zawadzka-Skomial J, Markiewicz Z, Nguyen-Distèche M, Devreese B, Frère J-M, Terrak M. 2006. Characterization of the bifunctional glycosyltransferase/acyltransferase penicillin-binding protein 4 of Listeria monocytogenes. J Bacteriol 188:1875–1881 10.1128/JB.188.5.1875-1881.2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aubry C, Goulard C, Nahori M-A, Cayet N, Decalf J, Sachse M, Boneca IG, Cossart P, Dussurget O. 2011. OatA, a peptidoglycan O-acetyltransferase involved in Listeria monocytogenes immune escape, is critical for virulence. J Infect Dis 204:731–740 10.1093/infdis/jir396. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Allen KJ, Wałecka-Zacharska E, Chen JC, Katarzyna K-P, Devlieghere F, Van Meervenne E, Osek J, Wieczorek K, Bania J. 2016. Listeria monocytogenes: an examination of food chain factors potentially contributing to antimicrobial resistance. Food Microbiol 54:178–189 10.1016/j.fm.2014.08.006. [DOI] [Google Scholar]
- 86.Shaw KJ, Rather PN, Hare RS, Miller GH. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev 57:138–163. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lancaster H, Roberts AP, Bedi R, Wilson M, Mullany P. 2004. Characterization of Tn916S, a Tn916-like element containing the tetracycline resistance determinant tet(S). J Bacteriol 186:4395–4398 10.1128/JB.186.13.4395-4398.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van Hoek AHAM, Mevius D, Guerra B, Mullany P, Roberts AP, Aarts HJM. 2011. Acquired antibiotic resistance genes: an overview. Front Microbiol 2:203 10.3389/fmicb.2011.00203. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev 28:519–542 10.1016/j.femsre.2004.04.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.Li L, Olsen RH, Shi L, Ye L, He J, Meng H. 2016. Characterization of a plasmid carrying cat, ermB and tetS genes in a foodborne Listeria monocytogenes strain and uptake of the plasmid by cariogenic Streptococcus mutans. Int J Food Microbiol 238:68–71 10.1016/j.ijfoodmicro.2016.08.038. [PubMed] [DOI] [PubMed] [Google Scholar]
- 91.Roberts MC, Facinelli B, Giovanetti E, Varaldo PE. 1996. Transferable erythromycin resistance in Listeria spp. isolated from food. Appl Environ Microbiol 62:269–270. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Granier SA, Moubareck C, Colaneri C, Lemire A, Roussel S, Dao T-T, Courvalin P, Brisabois A. 2011. Antimicrobial resistance of Listeria monocytogenes isolates from food and the environment in France over a 10-year period. Appl Environ Microbiol 77:2788–2790 10.1128/AEM.01381-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bertsch D, Anderegg J, Lacroix C, Meile L, Stevens MJA. 2013. pDB2011, a 7.6 kb multidrug resistance plasmid from Listeria innocua replicating in Gram-positive and Gram-negative hosts. Plasmid 70:284–287 10.1016/j.plasmid.2013.06.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 94.Huovinen P, Huovinen P. 2001. Resistance to trimethoprim-sulfamethoxazole. Clin Infect Dis 32:1608–1614 10.1086/320532. [PubMed] [DOI] [PubMed] [Google Scholar]
- 95.Charpentier E, Courvalin P. 1997. Emergence of the trimethoprim resistance gene dfrD in Listeria monocytogenes BM4293. Antimicrob Agents Chemother 41:1134–1136. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bertsch D, Uruty A, Anderegg J, Lacroix C, Perreten V, Meile L. 2013. Tn6198, a novel transposon containing the trimethoprim resistance gene dfrG embedded into a Tn916 element in Listeria monocytogenes. J Antimicrob Chemother 68:986–991 10.1093/jac/dks531. [PubMed] [DOI] [PubMed] [Google Scholar]
- 97.European Union. 2012. Regulation (EU) 528/2012 of 22 May 2012 concerning the making available on the market and use of biocidal products. European Union Regul.
- 98.Harbarth S, Tuan Soh S, Horner C, Wilcox MH. 2014. Is reduced susceptibility to disinfectants and antiseptics a risk in healthcare settings? A point/counterpoint review. J Hosp Infect 87:194–202 10.1016/j.jhin.2014.04.012. [PubMed] [DOI] [PubMed] [Google Scholar]
- 99.Ortega Morente E, Fernández-Fuentes MA, Grande Burgos MJ, Abriouel H, Pérez Pulido R, Gálvez A. 2013. Biocide tolerance in bacteria. Int J Food Microbiol 162:13–25 10.1016/j.ijfoodmicro.2012.12.028. [PubMed] [DOI] [PubMed] [Google Scholar]
- 100.McBain AJ, Ledder RG, Moore LE, Catrenich CE, Gilbert P. 2004. Effects of quaternary-ammonium-based formulations on bacterial community dynamics and antimicrobial susceptibility. Appl Environ Microbiol 70:3449–3456 10.1128/AEM.70.6.3449-3456.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McDonnell G, Russell AD. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev 12:147–179. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goddard P, McCue K. 2001. Disinfectants and Antiseptics. Disinfection, Sterilization, and Preservation. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 103.Elhanafi D, Dutta V, Kathariou S. 2010. Genetic characterization of plasmid-associated benzalkonium chloride resistance determinants in a Listeria monocytogenes strain from the 1998-1999 outbreak. Appl Environ Microbiol 76:8231–8238 10.1128/AEM.02056-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dutta V, Elhanafi D, Kathariou S. 2013. Conservation and distribution of the benzalkonium chloride resistance cassette bcrABC in Listeria monocytogenes. Appl Environ Microbiol 79:6067–6074 10.1128/AEM.01751-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Müller A, Rychli K, Muhterem-Uyar M, Zaiser A, Stessl B, Guinane CM, Cotter PD, Wagner M, Schmitz-Esser S. 2013. Tn6188: a novel transposon in Listeria monocytogenes responsible for tolerance to benzalkonium chloride. PLoS One 8:e76835 10.1371/journal.pone.0076835. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Müller A, Rychli K, Zaiser A, Wieser C, Wagner M, Schmitz-Esser S. 2014. The Listeria monocytogenes transposon Tn6188 provides increased tolerance to various quaternary ammonium compounds and ethidium bromide. FEMS Microbiol Lett 361:166–173 10.1111/1574-6968.12626. [PubMed] [DOI] [PubMed] [Google Scholar]
- 107.Kovacevic J, Ziegler J, Wałecka-Zacharska E, Reimer A, Kitts DD, Gilmour MW. 2015. Tolerance of Listeria monocytogenes to quaternary ammonium sanitizers is mediated by a novel efflux pump encoded by emrE. Appl Environ Microbiol 82:939–953 10.1128/AEM.03741-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tamburro M, Ripabelli G, Vitullo M, Dallman TJ, Pontello M, Amar CFL, Sammarco ML. 2015. Gene expression in Listeria monocytogenes exposed to sublethal concentration of benzalkonium chloride. Comp Immunol Microbiol Infect Dis 40:31–39 10.1016/j.cimid.2015.03.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 109.Mereghetti L, Quentin R, Marquet-Van Der Mee N, Audurier A. 2000. Low sensitivity of Listeria monocytogenes to quaternary ammonium compounds. Appl Environ Microbiol 66:5083–5086 10.1128/AEM.66.11.5083-5086.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Romanova NA, Wolffs PFG, Brovko LY, Griffiths MW. 2006. Role of efflux pumps in adaptation and resistance of Listeria monocytogenes to benzalkonium chloride. Appl Environ Microbiol 72:3498–3503 10.1128/AEM.72.5.3498-3503.2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jones RD, Jampani HB, Newman JL, Lee AS. 2000. Triclosan: a review of effectiveness and safety in health care settings. Am J Infect Control 28:184–196 10.1067/mic.2000.102378. [PubMed] [DOI] [PubMed] [Google Scholar]
- 112.Schweizer HP. 2001. Triclosan: a widely used biocide and its link to antibiotics. FEMS Microbiol Lett 202:1–7 10.1111/j.1574-6968.2001.tb10772.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 113.Heath RJ, Yu YT, Shapiro MA, Olson E, Rock CO. 1998. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J Biol Chem 273:30316–30320 10.1074/jbc.273.46.30316. [PubMed] [DOI] [PubMed] [Google Scholar]
- 114.Heath RJ, Rubin JR, Holland DR, Zhang E, Snow ME, Rock CO. 1999. Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J Biol Chem 274:11110–11114 10.1074/jbc.274.16.11110. [PubMed] [DOI] [PubMed] [Google Scholar]
- 115.Slater-Radosti C, Van Aller G, Greenwood R, Nicholas R, Keller PM, DeWolf WE Jr, Fan F, Payne DJ, Jaworski DD. 2001. Biochemical and genetic characterization of the action of triclosan on Staphylococcus aureus. J Antimicrob Chemother 48:1–6 10.1093/jac/48.1.1. [PubMed] [DOI] [PubMed] [Google Scholar]
- 116.Fan F, Yan K, Wallis NG, Reed S, Moore TD, Rittenhouse SF, DeWolf WE Jr, Huang J, McDevitt D, Miller WH, Seefeld MA, Newlander KA, Jakas DR, Head MS, Payne DJ. 2002. Defining and combating the mechanisms of triclosan resistance in clinical isolates of Staphylococcus aureus. Antimicrob Agents Chemother 46:3343–3347 10.1128/AAC.46.11.3343-3347.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kampf G, Kramer A. 2004. Epidemiologic background of hand hygiene and evaluation of the most important agents for scrubs and rubs. Clin Microbiol Rev 17:863–893 10.1128/CMR.17.4.863-893.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Villalaín J, Mateo CR, Aranda FJ, Shapiro S, Micol V. 2001. Membranotropic effects of the antibacterial agent triclosan. Arch Biochem Biophys 390:128–136 10.1006/abbi.2001.2356. [PubMed] [DOI] [PubMed] [Google Scholar]
- 119.Escalada MG, Russell AD, Maillard J-Y, Ochs D. 2005. Triclosan-bacteria interactions: single or multiple target sites? Lett Appl Microbiol 41:476–481 10.1111/j.1472-765X.2005.01790.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 120.Bailey AM, Constantinidou C, Ivens A, Garvey MI, Webber MA, Coldham N, Hobman JL, Wain J, Woodward MJ, Piddock LJV. 2009. Exposure of Escherichia coli and Salmonella enterica serovar Typhimurium to triclosan induces a species-specific response, including drug detoxification. J Antimicrob Chemother 64:973–985 10.1093/jac/dkp320. [PubMed] [DOI] [PubMed] [Google Scholar]
- 121.Nielsen LN, Larsen MH, Skovgaard S, Kastbjerg V, Westh H, Gram L, Ingmer H. 2013. Staphylococcus aureus but not Listeria monocytogenes adapt to triclosan and adaptation correlates with increased fabI expression and agr deficiency. BMC Microbiol 13:177 10.1186/1471-2180-13-177. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kastbjerg VG, Hein-Kristensen L, Gram L. 2014. Triclosan-induced aminoglycoside-tolerant Listeria monocytogenes isolates can appear as small-colony variants. Antimicrob Agents Chemother 58:3124–3132 10.1128/AAC.02266-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Appelberg R. 2006. Macrophage nutriprive antimicrobial mechanisms. J Leukoc Biol 79:1117–1128 10.1189/jlb.0206079. [PubMed] [DOI] [PubMed] [Google Scholar]
- 124.Botella H, Peyron P, Levillain F, Poincloux R, Poquet Y, Brandli I, Wang C, Tailleux L, Tilleul S, Charrière GM, Waddell SJ, Foti M, Lugo-Villarino G, Gao Q, Maridonneau-Parini I, Butcher PD, Castagnoli PR, Gicquel B, de Chastellier C, Neyrolles O. 2011. Mycobacterial p(1)-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe 10:248–259 10.1016/j.chom.2011.08.006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.White C, Lee J, Kambe T, Fritsche K, Petris MJ. 2009. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem 284:33949–33956 10.1074/jbc.M109.070201. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.den Bakker HC, Cummings CA, Ferreira V, Vatta P, Orsi RH, Degoricija L, Barker M, Petrauskene O, Furtado MR, Wiedmann M. 2010. Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics 11:688 10.1186/1471-2164-11-688. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kuenne C, Voget S, Pischimarov J, Oehm S, Goesmann A, Daniel R, Hain T, Chakraborty T. 2010. Comparative analysis of plasmids in the genus Listeria. PLoS One 5:5 10.1371/journal.pone.0012511. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yoon KP, Silver S. 1991. A second gene in the Staphylococcus aureus cadA cadmium resistance determinant of plasmid pI258. J Bacteriol 173:7636–7642 10.1128/jb.173.23.7636-7642.1991. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Allnutt TR, Bradbury MI, Fanning S, Chandry PS, Fox EM. 2016. Draft genome sequences of 15 isolates of Listeria monocytogenes serotype 1/2a, subgroup ST204. Genome Announc 4:4 10.1128/genomeA.00935-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wales AD, Davies RH. 2015. Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics (Basel) 4:567–604 10.3390/antibiotics4040567. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schmitz-Esser S, Müller A, Stessl B, Wagner M. 2015. Genomes of sequence type 121 Listeria monocytogenes strains harbor highly conserved plasmids and prophages. Front Microbiol 6:380 10.3389/fmicb.2015.00380. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mortvedt JJ. 1996. Heavy metal contaminants in inorganic and organic fertilizers. Fert Res 43:55–61 10.1007/BF00747683. [DOI] [Google Scholar]
- 133.Yazdankhah S, Rudi K, Bernhoft A. 2014. Zinc and copper in animal feed: development of resistance and co-resistance to antimicrobial agents in bacteria of animal origin. Microb Ecol Health Dis 25:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Barakat MA. 2011. New trends in removing heavy metals from industrial wastewater. Arab J Chem 4:361–377 10.1016/j.arabjc.2010.07.019. [DOI] [Google Scholar]
- 135.Guan Y, Shao C, Ju M. 2014. Heavy metal contamination assessment and partition for industrial and mining gathering areas. Int J Environ Res Public Health 11:7286–7303 10.3390/ijerph110707286. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xu D, Nie Q, Wang W, Shi L, Yan H. 2016. Characterization of a transferable bcrABC and cadAC genes-harboring plasmid in Listeria monocytogenes strain isolated from food products of animal origin. Int J Food Microbiol 217:117–122 10.1016/j.ijfoodmicro.2015.10.021. [PubMed] [DOI] [PubMed] [Google Scholar]
- 137.Pritchard TJ, Flanders KJ, Donnelly CW. 1995. Comparison of the incidence of Listeria on equipment versus environmental sites within dairy processing plants. Int J Food Microbiol 26:375–384 10.1016/0168-1605(94)00130-X. [DOI] [PubMed] [Google Scholar]
- 138.Fox EM, Wall PG, Fanning S. 2015. Control of Listeria species food safety at a poultry food production facility. Food Microbiol 51:81–86 10.1016/j.fm.2015.05.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 139.Mullapudi S, Siletzky RM, Kathariou S. 2008. Heavy-metal and benzalkonium chloride resistance of Listeria monocytogenes isolates from the environment of turkey-processing plants. Appl Environ Microbiol 74:1464–1468 10.1128/AEM.02426-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Khan Z, Rehman A, Hussain SZ, Nisar MA, Zulfiqar S, Shakoori AR. 2016. Cadmium resistance and uptake by bacterium, Salmonella enterica 43C, isolated from industrial effluent. AMB Express 6:54 10.1186/s13568-016-0225-9. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lee S, Rakic-Martinez M, Graves LM, Ward TJ, Siletzky RM, Kathariou S. 2013. Genetic determinants for cadmium and arsenic resistance among Listeria monocytogenes serotype 4b isolates from sporadic human listeriosis patients. Appl Environ Microbiol 79:2471–2476 10.1128/AEM.03551-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xu D, Li Y, Zahid MSH, Yamasaki S, Shi L, Li JR, Yan H. 2014. Benzalkonium chloride and heavy-metal tolerance in Listeria monocytogenes from retail foods. Int J Food Microbiol 190:24–30 10.1016/j.ijfoodmicro.2014.08.017. [PubMed] [DOI] [PubMed] [Google Scholar]
- 143.Lebrun M, Loulergue J, Chaslus-Dancla E, Audurier A. 1992. Plasmids in Listeria monocytogenes in relation to cadmium resistance. Appl Environ Microbiol 58:3183–3186. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lebrun M, Audurier A, Cossart P. 1994. Plasmid-borne cadmium resistance genes in Listeria monocytogenes are similar to cadA and cadC of Staphylococcus aureus and are induced by cadmium. J Bacteriol 176:3040–3048 10.1128/jb.176.10.3040-3048.1994. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tynecka Z, Gos Z, Zajac J. 1981. Energy-dependent efflux of cadmium coded by a plasmid resistance determinant in Staphylococcus aureus. J Bacteriol 147:313–319. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Endo G, Silver S. 1995. CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. J Bacteriol 177:4437–4441 10.1128/jb.177.15.4437-4441.1995. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McLauchlin J, Hampton MD, Shah S, Threlfall EJ, Wieneke AA, Curtis GD. 1997. Subtyping of Listeria monocytogenes on the basis of plasmid profiles and arsenic and cadmium susceptibility. J Appl Microbiol 83:381–388 10.1046/j.1365-2672.1997.00238.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 148.Ratani SS, Siletzky RM, Dutta V, Yildirim S, Osborne JA, Lin W, Hitchins AD, Ward TJ, Kathariou S. 2012. Heavy metal and disinfectant resistance of Listeria monocytogenes from foods and food processing plants. Appl Environ Microbiol 78:6938–6945 10.1128/AEM.01553-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nelson KE, Fouts DE, Mongodin EF, Ravel J, DeBoy RT, Kolonay JF, Rasko DA, Angiuoli SV, Gill SR, Paulsen IT, Peterson J, White O, Nelson WC, Nierman W, Beanan MJ, Brinkac LM, Daugherty SC, Dodson RJ, Durkin AS, Madupu R, Haft DH, Selengut J, Van Aken S, Khouri H, Fedorova N, Forberger H, Tran B, Kathariou S, Wonderling LD, Uhlich GA, Bayles DO, Luchansky JB, Fraser CM. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res 32:2386–2395 10.1093/nar/gkh562. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mullapudi S, Siletzky RM, Kathariou S. 2010. Diverse cadmium resistance determinants in Listeria monocytogenes isolates from the turkey processing plant environment. Appl Environ Microbiol 76:627–630 10.1128/AEM.01751-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Briers Y, Klumpp J, Schuppler M, Loessner MJ. 2011. Genome sequence of Listeria monocytogenes Scott A, a clinical isolate from a food-borne listeriosis outbreak. J Bacteriol 193:4284–4285 10.1128/JB.05328-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sarkar A, Paul B. 2016. The global menace of arsenic and its conventional remediation: a critical review. Chemosphere 158:37–49 10.1016/j.chemosphere.2016.05.043. [PubMed] [DOI] [PubMed] [Google Scholar]
- 153.Henke KR, Atwood DA. Arsenic in human history and modern societies, p 277–302. In Arsenic. John Wiley & Sons, Ltd., Chichester, United Kingdom. 10.1002/9780470741122.ch5 [DOI] [Google Scholar]
- 154.Chapman HD, Johnson ZB. 2002. Use of antibiotics and roxarsone in broiler chickens in the USA: analysis for the years 1995 to 2000. Poult Sci 81:356–364 10.1093/ps/81.3.356. [PubMed] [DOI] [PubMed] [Google Scholar]
- 155.Jesse HE, Roberts IS, Cavet JS. 2014. Metal ion homeostasis in Listeria monocytogenes and importance in host-pathogen interactions. Adv Microb Physiol 65:83–123 10.1016/bs.ampbs.2014.08.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 156.Castillo Y, Tachibana M, Nakatsu Y, Watanabe K, Shimizu T, Watarai M. 2015. Combination of zinc and all-trans retinoic acid promotes protection against Listeria monocytogenes infection. PLoS One 10:e0137463 10.1371/journal.pone.0137463. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Corbett D, Schuler S, Glenn S, Andrew PW, Cavet JS, Roberts IS. 2011. The combined actions of the copper-responsive repressor CsoR and copper-metallochaperone CopZ modulate CopA-mediated copper efflux in the intracellular pathogen Listeria monocytogenes. Mol Microbiol 81:457–472 10.1111/j.1365-2958.2011.07705.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 158.Kuenne C, Billion A, Mraheil MA, Strittmatter A, Daniel R, Goesmann A, Barbuddhe S, Hain T, Chakraborty T. 2013. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 14:47 10.1186/1471-2164-14-47. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Camejo A, Buchrieser C, Couvé E, Carvalho F, Reis O, Ferreira P, Sousa S, Cossart P, Cabanes D. 2009. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog 5:e1000449 10.1371/journal.ppat.1000449. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bertrand S, Huys G, Yde M, D’Haene K, Tardy F, Vrints M, Swings J, Collard J-M. 2005. Detection and characterization of tet(M) in tetracycline-resistant Listeria strains from human and food-processing origins in Belgium and France. J Med Microbiol 54:1151–1156 10.1099/jmm.0.46142-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 161.Pourshaban M, Ferrini AM, Mannoni V, Oliva B, Aureli P. 2002. Transferable tetracycline resistance in Listeria monocytogenes from food in Italy. J Med Microbiol 51:564–566 10.1099/0022-1317-51-7-564. [PubMed] [DOI] [PubMed] [Google Scholar]
- 162.Humphrey B, Thomson NR, Thomas CM, Brooks K, Sanders M, Delsol AA, Roe JM, Bennett PM, Enne VI. 2012. Fitness of Escherichia coli strains carrying expressed and partially silent IncN and IncP1 plasmids. BMC Microbiol 12:53 10.1186/1471-2180-12-53. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Clewell DB, Weaver KE, Dunny GM, Coque TM, Francia MV, Hayes F. 2014. Extrachromosomal and mobile elements in enterococci: transmission, maintenance, and epidemiology. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Massachusetts Eye and Ear Infirmary, Boston, MA. Available from https://www.ncbi.nlm.nih.gov/books/NBK190430/. [PubMed] [Google Scholar]
- 164.Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, Andersson DI. 2011. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog 7:e1002158 10.1371/journal.ppat.1002158. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gullberg E, Albrecht LM, Karlsson C, Sandegren L, Andersson DI. 2014. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. MBio 5:e01918–e14 10.1128/mBio.01918-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jensen LB, Garcia-Migura L, Valenzuela AJS, Løhr M, Hasman H, Aarestrup FM. 2010. A classification system for plasmids from enterococci and other Gram-positive bacteria. J Microbiol Methods 80:25–43 10.1016/j.mimet.2009.10.012. [PubMed] [DOI] [PubMed] [Google Scholar]
- 167.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238 10.1128/AAC.01707-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Romanova N, Favrin S, Griffiths MW. 2002. Sensitivity of Listeria monocytogenes to sanitizers used in the meat processing industry. Appl Environ Microbiol 68:6405–6409 10.1128/AEM.68.12.6405-6409.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang H, Zhou Y, Bao H, Zhang L, Wang R, Zhou X. 2015. Plasmid-borne cadmium resistant determinants are associated with the susceptibility of Listeria monocytogenes to bacteriophage. Microbiol Res 172:1–6 10.1016/j.micres.2015.01.008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 170.Bertsch D, Muelli M, Weller M, Uruty A, Lacroix C, Meile L. 2014. Antimicrobial susceptibility and antibiotic resistance gene transfer analysis of foodborne, clinical, and environmental Listeria spp. isolates including Listeria monocytogenes. MicrobiologyOpen 3:118–127 10.1002/mbo3.155. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Poyart-Salmeron C, Trieu-Cuot P, Carlier C, MacGowan A, McLauchlin J, Courvalin P. 1992. Genetic basis of tetracycline resistance in clinical isolates of Listeria monocytogenes. Antimicrob Agents Chemother 36:463–466 10.1128/AAC.36.2.463. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hadorn K, Hächler H, Schaffner A, Kayser FH. 1993. Genetic characterization of plasmid-encoded multiple antibiotic resistance in a strain of Listeria monocytogenes causing endocarditis. Eur J Clin Microbiol Infect Dis 12:928–937 10.1007/BF01992167. [PubMed] [DOI] [PubMed] [Google Scholar]
- 173.Piddock LJV. 2006. Multidrug-resistance efflux pumps: not just for resistance. Nat Rev Microbiol 4:629–636 10.1038/nrmicro1464. [PubMed] [DOI] [PubMed] [Google Scholar]
- 174.Li X-Z, Nikaido H. 2009. Efflux-mediated drug resistance in bacteria: an update. Drugs 69:1555–1623 10.2165/11317030-000000000-00000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Webber MA, Piddock LJV. 2003. The importance of efflux pumps in bacterial antibiotic resistance. J Antimicrob Chemother 51:9–11 10.1093/jac/dkg050. [PubMed] [DOI] [PubMed] [Google Scholar]
- 176.Crimmins GT, Herskovits AA, Rehder K, Sivick KE, Lauer P, Dubensky TW Jr, Portnoy DA. 2008. Listeria monocytogenes multidrug resistance transporters activate a cytosolic surveillance pathway of innate immunity. Proc Natl Acad Sci USA 105:10191–10196 10.1073/pnas.0804170105. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kaplan Zeevi M, Shafir NS, Shaham S, Friedman S, Sigal N, Nir Paz R, Boneca IG, Herskovits AA. 2013. Listeria monocytogenes multidrug resistance transporters and cyclic di-AMP, which contribute to type I interferon induction, play a role in cell wall stress. J Bacteriol 195:5250–5261 10.1128/JB.00794-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Schwartz KT, Carleton JD, Quillin SJ, Rollins SD, Portnoy DA, Leber JH. 2012. Hyperinduction of host beta interferon by a Listeria monocytogenes strain naturally overexpressing the multidrug efflux pump MdrT. Infect Immun 80:1537–1545 10.1128/IAI.06286-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Marquez B, Pourcelle V, Vallet CM, Mingeot-Leclercq M-P, Tulkens PM, Marchand-Bruynaert J, Van Bambeke F. 2014. Pharmacological characterization of 7-(4-(Piperazin-1-yl)) ciprofloxacin derivatives: antibacterial activity, cellular accumulation, susceptibility to efflux transporters, and intracellular activity. Pharm Res 31:1290–1301 10.1007/s11095-013-1250-x. [DOI] [PubMed] [Google Scholar]
- 180.Lismond A, Tulkens PM, Mingeot-Leclercq M-P, Courvalin P, Van Bambeke F. 2008. Cooperation between prokaryotic (Lde) and eukaryotic (MRP) efflux transporters in J774 macrophages infected with Listeria monocytogenes: studies with ciprofloxacin and moxifloxacin. Antimicrob Agents Chemother 52:3040–3046 10.1128/AAC.00105-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Scortti M, Lacharme-Lora L, Wagner M, Chico-Calero I, Losito P, Vázquez-Boland JA. 2006. Coexpression of virulence and fosfomycin susceptibility in Listeria: molecular basis of an antimicrobial in vitro-in vivo paradox. Nat Med 12:515–517 10.1038/nm1396. [PubMed] [DOI] [PubMed] [Google Scholar]
- 182.Collins B, Curtis N, Cotter PD, Hill C, Ross RP. 2010. The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various beta-lactam antibiotics. Antimicrob Agents Chemother 54:4416–4423 10.1128/AAC.00503-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Walsh D, Duffy G, Sheridan JJ, Blair IS, McDowell DA. 2001. Antibiotic resistance among Listeria, including Listeria monocytogenes, in retail foods. J Appl Microbiol 90:517–522 10.1046/j.1365-2672.2001.01273.x. [PubMed] [DOI] [PubMed] [Google Scholar]


