ABSTRACT
Previously, leaderless mRNAs (lmRNAs) were perceived to make up only a minor fraction of the transcriptome in bacteria. However, advancements in RNA sequencing technology are uncovering vast numbers of lmRNAs, particularly in archaea, Actinobacteria, and extremophiles and thus underline their significance in cellular physiology and regulation. Due to the absence of conventional ribosome binding signals, lmRNA translation initiation is distinct from canonical mRNAs and can therefore be differentially regulated. The ribosome’s inherent ability to bind a 5′-terminal AUG can stabilize and protect the lmRNA from degradation or allow ribosomal loading for downstream initiation events. As a result, lmRNAs remain translationally competent during a variety of physiological conditions, allowing them to contribute to multiple regulatory mechanisms. Furthermore, the abundance of lmRNAs can increase during adverse conditions through the upregulation of lmRNA transcription from alternative promoters or by the generation of lmRNAs from canonical mRNAs cleaved by an endonucleolytic toxin. In these ways, lmRNA translation can continue during stress and contribute to regulation, illustrating their importance in the cell. Due to their presence in all domains of life and their ability to be translated by heterologous hosts, lmRNAs appear further to represent ancestral transcripts that might allow us to study the evolution of the ribosome and the translational process.
In bacteria and archaea, translation initiates with a 30S ribosomal subunit interacting with an initiator tRNA at the ribosome binding site on a canonical mRNA to form a stable translation initiation complex that is primed for elongation. Canonical mRNAs contain both 5′ and 3′ untranslated regions (UTRs) containing information that will influence the stability and translation efficiency of the mRNA. Within the 5′ UTR, these signals can include ribosome recognition regions such as purine-rich Shine-Dalgarno (SD) sequences that are complementary to the anti-SD (aSD) sequence near the 16S rRNA 3′ terminus (1), AU-rich sequences that interact with ribosomal protein (r-protein) bS1 (2, 3) and prevent the formation of secondary structures, and enhancer regions. Additionally, 5′ UTRs may contain sequences that can be bound by trans-acting elements (i.e., proteins, antisense and small regulatory RNAs, or low-molecular-weight effectors) to change secondary structures or block translation initiation regions. Therefore, the regulatory and translation initiation signals are primarily contained within the 5′ UTR. Despite this functional importance of the 5′ UTR, there exists a class of mRNAs that are completely devoid of 5′ UTRs or possess very short 5′ UTRs. These mRNAs lack the SD sequence and any other translational signals and are so named leaderless mRNAs (lmRNAs). Thus, the mechanism underlying their recognition and binding by the translational apparatus is still not entirely elucidated.
lmRNAs can be naturally encoded on the chromosome and occur as a result of transcription and translation initiating at the same position or as a result of posttranscriptional or cotranslational processing events (Fig. 1). Intriguingly, lmRNAs are found in every domain of life and are universally translatable by heterologous hosts (4), signifying a fundamental lmRNA recognition capability of all translation systems. These results suggest that they represent ancestral RNA remnants. Hence, understanding their translatability could provide insight into the evolution of the ribosome.
FIGURE 1.
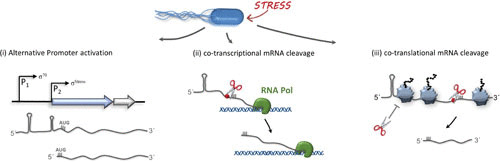
Mechanisms leading to the generation of lmRNAs in bacteria. Besides genes that are generally transcribed as lmRNAs, bacteria can generate lmRNAs in response to adverse environmental conditions (i) by activation of alternative promoters, where the transcriptional start point coincides with the A of the AUG start codon; (ii) by cotranscriptional cleavage, when the 5′ UTR is removed by RNases during the process of transcription; or (iii) cotranslationally. Here, the cleavage can be regulated by translating ribosomes that might either protect mRNAs from cleavage or expose specific sites for the processing event by RNases. Cleavage sites and potential RNases are indicated by red spheres and scissors, respectively.
WIDESPREAD OCCURRENCE OF lmRNAs
As the number of annotated genomes has drastically increased due to advancements in sequencing technology, we are beginning to uncover the vast number of lmRNAs. Recent in silico analyses examining translation initiation regions have made predictions as to the prevalence of lmRNAs across numerous bacterial and archaeal genomes (5–7). However, these studies can only estimate quantities based on promoter sequence logos and analysis of intergenic regions. Therefore, while they provide useful information regarding the high probability of lmRNAs, they cannot determine exact reproducible numbers of leaderless transcripts. Rather, transcriptome sequencing (RNA-seq)-based techniques, while limited in the number of transcriptomes analyzed in one study, can provide precise measurements of lmRNAs, which have been directly sequenced with high confidence. Such studies have uncovered a wide variety of lmRNAs in bacteria and archaea ranging from <1% up to ∼70% of primary and secondary transcripts (Table 1). lmRNAs are not confined to bacteria and archaea either. They are also found in eukaryotes, with all mammalian mitochondrial mRNAs being leaderless (8, 9).
TABLE 1.
Compilation of published transcriptome analyses outlining the number of leaderless mRNAs in a variety of bacterial and archaeal genomesa
| Nucleotides upstream of start codon | Total no. of transcription start sites or mRNAs identified | No. of lmRNAs | % of lmRNAs | Reference | |
|---|---|---|---|---|---|
| Bacteria | |||||
| Alphaproteobacteria | |||||
| Caulobacter crescentus NA1000 | 0 | 2,201 | 375 | 17 | 84 |
| Sinorhizobium meliloti 1021 | 0 | 4,430 | 171 | 4 | 119 |
| Deltaproteobacteria | |||||
| Geobacter sulfurreducens ATCC 51573 | ≤5 | 3,487 | 52 | 1.5 | 120 |
| Epsilonproteobacteria | |||||
| Campylobacter jejuni NCTC 11168 | 0 | 992 | 12 | 1.2 | 121 |
| Campylobacter jejuni | 0 | 3,241 | 48 | 1.48 | 122 |
| Gammaproteobacteria | |||||
| Escherichia coli BW25113 | 0 | 728 | 5 | 0.7 | 123 |
| Escherichia coli MG1655 | 0 | 4,261 | 14 | 124 | |
| Klebsiella pneumoniae MGH 78578 | ≤5 | 5,194 | 83 | 1.6 | 125 |
| Salmonella enterica serovar Typhimurium SL1344 | 0 | 1,873 | 23 | 1.2 | 126 |
| Xanthomonas campestris pv. campestris B100 | ≤2 | 3,067 | 130 | 8.4 | 127 |
| Legionella pneumophila strain Paris | 0 | 1,905 | 12 | 0.63 | 128 |
| Deinococcus-Thermus | |||||
| Deinococcus deserti RD19 | 0 | 1,958 | 916 | 47 | 91 |
| Firmicutes | |||||
| Clostridium acetobutylicum ATCC 824 | 0 | 616 | 212 | 34.4 | 129 |
| Bacillus amyloliquefaciens XH7 | ≤5 | 1,064 | 57 | 5.4 | 130 |
| Actinobacteria | |||||
| Mycobacterium tuberculosis H37Rv | ≤5 | 2,524 | 505 | 22 | 106 |
| Mycobacterium tuberculosis H37Rv | ≤5 | 4,979 | 1,098 | 22 | 79 |
| Mycobacterium smegmatis mc2155 | 0 | 1,098 | 206 | 19 | 79 |
| Mycobacterium avium TMC724 | ≤3 | 844 | 278 | 33 | 131 |
| Actinoplanes sp. SE50/110 | 0 | 661 | 135 | 20 | 132 |
| Corynebacterium glutamicum ATCC 13032 | 0 | 2,454 | 546 | 22 | 133 |
| Corynebacterium glutamicum ATCC 13032 | 0 | 2,147 | 707 | 33 | 134 |
| Streptomyces tsukubaensis NRRL 18488 | 0 | 3,678 | 581 | 15.8 | 135 |
| Cyanobacteria | |||||
| Prochlorococcus MED4 | 0 | 4,126 | 30 | 0.7 | 136 |
| Prochlorococcus MIT913 | 0 | 8,587 | 41 | 0.5 | 136 |
| Archaea | |||||
| Haloferax volcanii H26 | ≤5 | 4,749 | 1,329 | 72 | 137 |
| Thermococcus onnurineus NA1 | ≤5 | 1,082 | 117 | 10.8 | 13 |
| Pyrococcus abyssi GE5 | ≤5 | 1,893 | 27 | 1.4 | 138 |
| Sulfolobus solfataricus P2 | ≤3 | 1,040 | 718 | 69 | 139 |
The total number of lmRNAs identified in the different studies as well as their percentage (%) are given. mRNAs harboring up to 5 nucleotides upstream of the AUG start codon are included for completeness.
lmRNAs may represent a functional relic of an earlier evolutionary age, as bacteria and archaea closer to the root of the phylogenetic tree commonly have a higher proportion of lmRNAs. As determined by an in silico analysis in Gammaproteobacteria, lmRNA genes are most found in Xanthomonadales and Legionellales, which are located near the root of Gammaproteobacteria (6). Furthermore, phylogenetically close species tend to have a relatively equal proportion of lmRNA genes. Analysis of 16S rRNA revealed that archaeal genomes with the highest level of leaderless transcripts clustered together (10).
Additionally, in archaea, the majority of single genes or the proximal cistrons in operons encode lmRNAs (11–13). Operon structure appears to have coevolved with translation initiation signals, requiring additional SD sequences for an increasing number of cistrons. The proportion of operon distal genes shows positive correlation with the proportion of SD-led genes in both bacteria and archaea, supporting this notion (6). Furthermore, a transcriptome analysis in Escherichia coli uncovered previously uncharacterized 5′-AUGs at the end of canonical SD-led mRNAs (5′-uAUGs) (14). This genetic structure in which the first open reading frame (ORF) in a polycistronic mRNA is leaderless, parallels the operon structure in archaea (11). In E. coli, these 5′-uAUGs were shown to bind 70S ribosomes and were translated at biologically relevant levels, leading to the conclusion that these transcripts should also be classified as lmRNAs and are likely to exist in many bacterial species (14). This notion is supported by the characterization of the acuR-acuI-dddL operon in the Gram-negative alphaproteobacterium Rhodobacter sphaeroides (15) and the sptA mRNA in the haloarchaeal Natrinema sp. strain J7-2 (16), in which these mRNAs were found to be regulated by their ability to be alternatively transcribed as an lmRNA. The regulation ascribed to each of these leaderless transcripts is described below.
lmRNAs are not only naturally encoded on the chromosome but can also be a result of posttranscriptional processing (Fig. 1). In one such instance, leadered mRNAs are cleaved to generate a subset of lmRNAs that are specialized for translation during suboptimal growth conditions (17, 18). This phenomenon involves a conditionally active toxin whose endonucleolytic capability transforms the RNA landscape to allow for certain mRNAs to maintain translatability. This illustrates the presence of previously overlooked lmRNAs that are not evident from genome analysis but play a role in regulation. The exact mechanism of this processing is detailed below.
Therefore, with the advances in global RNA sequencing and classification of the translatome, the prevalence of lmRNAs is becoming apparent, demonstrating the need to understand their mechanism of translation and the role they play in cellular physiology and regulation.
MAIN PATHWAYS OF TRANSLATION INITIATION
Translation Initiation on Canonical mRNAs
Despite the fact that lmRNAs are rather infrequent in E. coli, the majority of studies addressing the pathway of translation initiation on these peculiar mRNAs have been performed in this model organism. In E. coli, the canonical pathway of translation initiation (Fig. 2A) entails first the initiation factor-assisted recognition and binding of the mRNA and the recruitment of the fMet-tRNAfMet to the 30S subunit. Next, the codon-anticodon interaction in the ribosomal P-site is established to form the 30S initiation complex. The efficiency of this step is affected by the strength of the SD-aSD interaction, secondary structures within the ribosome binding site, and the nature of the start codon. The three initiation factors (IF1, IF2, and IF3) contribute synergistically to the kinetics and fidelity of translation initiation (19–22). IF1 binds at the ribosomal A-site. Thus, it prevents elongator tRNA binding but also controls the conformational dynamics of the 30S subunit (21). Further, by providing the anchoring point for IF2 and IF3 on the 30S subunit, IF1 enhances their activity (23). IF2 selects for the correct initiator tRNA by directly binding to the N-formyl-methionine and aminoacyl acceptor stem of fMet-tRNAfMet (24). Hence, IF2 increases the affinity of fMet-tRNAfMet to the 30S subunit, whereas IF3 reduces the affinity of the fMet-tRNAfMet (25) and discriminates against noncognate initiation complexes by introducing conformational dynamics during complex formation (19). With respect to subunit joining, IF2 and IF3 likewise show opposite activities. Due to the localization of the C-domain of IF3 at the interface side of the platform of the 30S subunit, the factor sterically blocks subunit joining at the B2 bridge between helix 69 of the 23S rRNA and the 16S rRNA (26). After release of IF3, a dynamic conformational switch introduced by binding of GTP to IF2 stimulates the rate of subunit joining (20). Finally, binding of the 50S subunit triggers GTP hydrolysis, which causes a conformational change in IF2 and drives an intersubunit rotation. Thereby, IF2 is released and the 70S complex is stabilized and transitions into the elongation-competent state (27).
FIGURE 2.
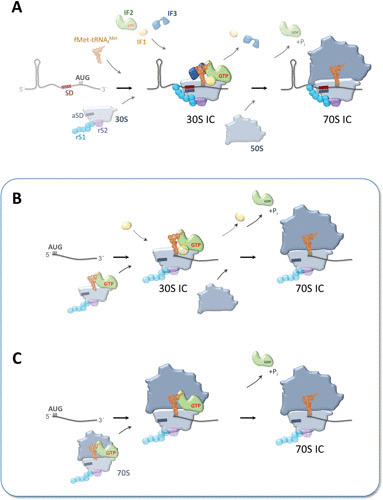
Potential pathways for translation initiation complex (IC) formation on lmRNAs. (A) Schematic showing the main steps during canonical initiation. (B and C) Potential steps during translation initiation on lmRNAs via 30S subunits and 70S monosomes, respectively. r-Proteins bS1 and uS2 are transparent, indicating their dispensability during this process. See text for details.
In addition to the initiation factors, some r-proteins were shown to play key roles during translation initiation complex formation. The multidomain r-protein bS1 is essential for canonical initiation (28). It interacts via the N-terminal helix with r-protein uS2 on the 30S subunit (Fig. 1) (29). The flexible and dynamic protein is suggested to scan the vicinity of the ribosome for mRNAs with its C-terminal RNA-binding domains (30). Further, protein bS1 promotes RNA unwinding by binding to single-stranded RNA during thermal breathing (31). Together, these activities suggest that the protein promotes mRNA recruitment and facilitates positioning of structured mRNA in the RNA track of the ribosome by unwinding secondary structures. r-Protein bS21, which is likewise essential for ribosome binding during initiation of MS2 RNA and canonical E. coli mRNA translation, is localized on the platform of the 30S subunit close to the 16S rRNA 3′ terminus. It is suggested to stimulate the base-pairing potential of the SD-aSD interaction by exposing the 16S rRNA 3′ terminus (32–34).
Factors and r-Proteins That Affect Translation Initiation on lmRNAs
In contrast to the established pathway for translation initiation on canonical mRNAs in bacteria, initiation on lmRNAs can occur with a reduced set of factors and/or r-proteins (Fig. 2B). Several lines of evidence indicate that for translation initiation on lmRNA r-proteins bS1 and uS2 are both dispensable, as the cI lmRNA is more efficiently translated in their absence, suggesting they negatively impact lmRNA translation (35–37). Upon bS1 depletion, lmRNAs become more abundant and more stable and their translation is stimulated, whereas translation of canonical mRNAs is greatly reduced, an effect that can be reverted by the addition of exogenous bS1 (38, 39). This positive impact of the lack of bS1 on lmRNA translation is likely due to the loss of competition with canonical mRNAs. As structured, leadered mRNAs can no longer compete for bS1-depleted ribosomes, this allows the ribosomes to selectively translate only unstructured or leaderless transcripts. Additionally, loss of bS1 will eliminate the negative impact of IF3 because bS1 is required for the discriminatory activity of IF3 against lmRNA translation (35). Interestingly, bS1 has been shown to be dispensable for the formation of the initiation complex on leadered mRNAs if they contain a strong SD sequence and a weakly structured ribosome binding site (40). Moreover, some Gram-positive bacteria with low G+C content, the majority of which contain relatively high levels of lmRNA (6) (Table 1), either lack a bS1 homolog or contain a homolog missing the first domain, rendering it unable to bind to the ribosome (41). The lack of a functional bS1 homolog in archaea also correlates with their high number of leaderless transcripts. Together, these data support the notion that bS1 is in general not essential for translation.
The role of r-protein uS2 in lmRNA translation might be rather indirect, as it represents the primary anchoring point for bS1 on the ribosome (29). Therefore, the effects seen as a result of uS2 depletion are the same as deleting bS1 alone (38, 42), as only translation of canonical mRNAs is affected while the translational efficiency of lmRNAs increases 8- to 10-fold (43). Additionally, bS2 mutants contain increased levels of 70S ribosomes, in line with the stimulation of lmRNA expression (44).
In addition to bS1 and uS2, r-protein bS21 is also expendable for lmRNA translation. Here, promoting the base-pairing potential of the SD-aSD interaction (33) is irrelevant for ribosome binding. However, the absence of bS21 did not affect fMet-tRNAfMet binding necessary for lmRNA translation, or poly(U) translation (32), which resembles lmRNAs in that it is translated under conditions in which 70S ribosomes are prevalent (44).
As the 5′-AUG initiation codon appears to act as the unique ribosome binding signal on lmRNAs, their translation is more dependent on the presence of fMet-tRNAfMet than of canonical mRNA. This notion is supported by the observation that IF2 promotes lmRNA translation, which can effectively compete with canonical mRNAs for 30S occupation when IF2 levels are increased (4, 45). In contrast, IF3, which decreases the affinity of the fMet-tRNAfMet (25), discriminates against lmRNA translation by recognizing it as “noncanonical” and destabilizing the initiation complex (45–49). This discrimination could be attributed to conformational dynamics introduced in the ribosome by IF3 during initiation complex formation rather than direct scanning of the initiation codon (19, 50) and requires the presence of r-protein bS1 (35). Therefore, the ratio between IF2 and IF3 can modulate the efficiency of lmRNA translation (45). These ratios are equal at a steady state (51) but can vary in response to physiological changes, as IF3 concentration increases during increased growth rate (52), while IF2 levels increase under cold shock (53). This suggests that the efficiency of lmRNA translation might also fluctuate in response to environmental changes.
Together, these observations propose the first pathway for initiation on lmRNAs in bacteria, where a 30S subunit with a prebound IF2-fMet-tRNAfMet complex binds to the 5′ terminus of the lmRNA in the absence of IF3. Here, the 5′-terminal AUG initiation codon and the 5′ triphosphate appear to act as ribosome binding signals (Fig. 2B). Intriguingly, this process is analogous to the eukaryotic initiation pathway in which the 40S subunit must first be loaded with tRNAi to scan for the start codon (54).
The second model suggests that the lmRNA is directly bound by an intact 70S ribosome through codon-anticodon base pairing with a prebound IF2-fMet-tRNAfMet complex (Fig. 2C). 70S ribosomes show a high preference for a 5′-terminal AUG (55) and bind to lmRNAs more strongly than 30S subunits (46). Furthermore, 70S initiation complexes are intrinsically more stable on lmRNAs, with a 5- to 10-fold-higher retention with the addition of fMet-tRNAfMet compared to 30S subunits (56), and chemically cross-linked 70S complexes, unable to dissociate into 30S and 50S subunits, retained the ability to translate leaderless but not canonical mRNAs (44). Also, inactivation of ribosome recycling factor to prevent subunit dissociation resulted in increased lmRNA translation, while bulk mRNA translation greatly diminished (44). Considering that IF3 prevents subunit association (26), the 70S initiation pathway on lmRNA can also be reconciled with the reduced translation initiation when IF3 levels are increased (35, 45). Finally, conditions that create a prevalence of 70S monosomes in vivo correlate with preferential translation of lmRNA and loss of canonical translation (44). While evidence suggests that the use of the 70S pathway is more likely, the two models are not mutually exclusive, making it possible that both the 30S and 70S initiation mechanisms are operating in vivo depending on the physiological conditions.
Although lmRNAs are generally more prominent in archaea, less is known about their initiation mechanism. It is considered to closely parallel the proposed bacterial pathway, with 70S-bound a/eIF2 recruiting Met-tRNAiMet and directing it to bind specifically to the initiation codon (Fig. 2C) (57, 58). However, there are likely additional auxiliary factors that contribute to archaeal lmRNA initiation, as they contain >10 genes encoding proteins homologous to eukaryotic initiation factors, including aIF1, aIF1A, aIF2, a/eIF2, and aIF6 (59–62). Moreover, archaea utilize a hybrid system where most factors are more similar to the eukaryotic mechanism; however, they follow the basic bacterial mechanism for initiation codon selection (61). Nonetheless, in both bacteria and archaea, the initiator tRNA is absolutely essential for stable initiation complex formation on lmRNA (4, 44, 56, 57).
Together, the working model for lmRNA translation in archaea suggests that a 70S-aIF1-aIF1A-a/eIF2-GTP-Met-tRNAiMet complex binds the mRNA (58). Although the exact order of events to form this complex is unknown, it is anticipated that a/eIF2 initially binds to the ribosome. The binding is stimulated by the concerted activities of aIF1 and aIF1A, which are hypothesized to induce structural changes in the 30S to facilitate a/eIF2-GTP binding to, in turn, recruit the Met-tRNAiMet (58, 63, 64). a/eIF2 performs distinct functions depending on whether or not it is bound to the ribosome. Off the ribosome it has a high affinity for 5′-triphosphorylated ends to protect mRNA from degradation (62, 65). However, this binding negatively impacts lmRNA, as it will block translation when a/eIF2 is bound. Once bound by the ribosome, a/eIF2 has a higher affinity for Met-tRNAiMet and instead positively contributes to translation initiation (66). This is especially important for lmRNAs, which are dependent on Met-tRNAiMet to be prebound to the 70S ribosome for efficient translation initiation complex formation.
Additional initiation factors aIF2 and aIF6, common only between archaea and eukaryotes, play indirect roles in archaeal lmRNA translation. aIF2 acts as a ribosome-dependent thermophilic GTPase that interacts preferentially with 50S subunits and 70S monosomes (67). It stimulates the binding of Met-tRNAiMet to the ribosome and thereby increases the translation of both leadered and leaderless mRNAs (67, 68). aIF6 localizes at the nucleation core of the 50S subunit to prevent formation of the 70S ribosome. It is upregulated during cold and heat shock to act as translational repressor under unfavorable conditions (62, 69). Therefore, it is possible that the ratios between archaeal initiation factors vary in response to external conditions, thereby similarly modulating lmRNA expression.
As demonstrated, the various initiation factors can modulate lmRNA translation in distinct ways when compared to canonical translation. Nevertheless, the reduced number of translational factors needed for lmRNA translation further illustrates their ancestral nature by following a simpler mechanism of initiation established prior to the addition of certain r-proteins and initiation factors to increase complexity.
Signals Intrinsic to the lmRNA That Affect Translation Initiation
Due to the absence of 5′-UTR signals in lmRNA to direct ribosome binding, it was hypothesized that sequences downstream of the start codon may instead fulfill this role. However, extensive examination has failed to produce a consensus sequence that may be responsible and has disproven the existence of a potential “downstream box” that was thought to interact with the 16S rRNA (70, 71). The presence of CA repeats or A-richness has been shown to increase the translational efficiency on both lmRNAs and leadered mRNAs (72, 73), and these sequences are thought to contribute to the reduction of secondary structures, resulting in a more open complex and therefore fewer constraints on ribosome binding.
The remaining factors present on every lmRNA are then reduced to only the initiation codon and the 5′ triphosphate, both of which play a role in lmRNA initiation. The 5′ triphosphate was found to influence ternary complex formation and stability on lmRNAs, with a 5′ hydroxyl abolishing translation of the naturally leaderless cI mRNA in E. coli but not the SD leadered version (74). Brock et al. demonstrated that in E. coli, the 70S ribosome can distinguish mRNAs by the presence or absence of a 5′-AUG and that this signal is both necessary and sufficient for ribosome binding and subsequent translation (75). Most convincing was the demonstration that the addition of an AUG triplet to the 5′ end of an internal fragment of the lacZ mRNA restored its ability to be bound by ribosomes and successfully translated in vivo (75).
It is the 5′-AUG itself, and not the codon-anticodon base pairing with the fMet-tRNAfMet, that contributes to the initial ribosome interaction with lmRNAs. The codon-anticodon interaction further stabilizes the complex (75, 76). To support the notion that the 5′-AUG itself is important, in many organisms, mutation of the AUG to another initiation codon (GUG, UUG, CUG, AUU) completely abolishes lmRNA translation (77, 78). In mycobacteria, a 5′-AUG or 5′-GUG is necessary and sufficient for lmRNA translation, with no other initiation codon enriched in the leaderless transcripts (79). Therefore, an AUG may be highly preferred but not absolutely required. The proximity of the AUG to the 5′ terminus also affects translation efficiency, as a further recessed AUG results in reduced translation (80). It has been proposed that the ribosome is able to examine 5′ termini for the presence of a start codon even on mRNAs that are not naturally leaderless (81, 82). This may occur as a result of coupling transcription and translation through interaction of the RNA polymerase (Pol) and the ribosome (83). Thereby the ribosome may directly scan each mRNA as it exits the RNA Pol. However, this would only account for the first round of ribosome binding on each lmRNA, after which free ribosomes would be responsible for lmRNA recognition. Without the benefit of direct binding facilitated through RNA Pol-ribosome interactions, the ribosome binding efficiency to lmRNA is likely decreased, accounting for the less efficient translation of lmRNA in general.
REGULATION UNDER VARIOUS PHYSIOLOGICAL CONDITIONS
Due to the lack of a 5′ UTR and canonical initiation signals, lmRNAs are typically regarded to be less efficiently translated during exponential growth. This observation may be an artifact of using E. coli as the model organism for the majority of lmRNA translation studies despite its relatively low number of lmRNAs. Conversely, the use of ribosome profiling data and reporter assays has shown that lmRNAs are translated with equal efficiency to non-SD or SD-led mRNAs in Caulobacter crescentus (84) and Mycobacterium spp. (79). Also, in both bacterial and archaeal systems, in some cases the translational efficiency is higher when deleting the entire 5′ UTR compared to mutations that disrupt the SD sequence (76, 85, 86). However, one has to carefully consider the impact of secondary structures introduced by addition or removal of 5′ UTRs.
Continued Translation of lmRNAs during Stress Conditions
Regardless of whether lmRNAs are less efficiently translated during exponential growth, lmRNA translation appears to be adapted to adverse physiological conditions. They can be translated during stress (17, 18, 87), without any initiation factors (55, 88), at low temperatures (89), and their translation is immune to various antibiotics (90). A higher occurrence of lmRNA genes also appears in many extremophiles such as radiation-resistant Deinococcus radiodurans, thermophilic Thermus thermophilus, and many archaea (6). In Deinococcus deserti, 60% of the transcriptome is leaderless and proteomics have confirmed that the lmRNAs are efficiently translated and encode the most abundant proteins present (91) (Table 1). Some of these lmRNAs code for proteins that are essential for viability, such as the HU proteins used for nucleoid compaction (92). The HU proteins in D. deserti were shown to be more efficiently synthesized from lmRNAs in contrast to the leadered variants, which resulted in protein levels that were insufficient to provide their essential function (92). Many of the leaderless transcripts code for small peptides that have homologs in other Deinococcus species (91). These small peptides are hypothesized to act as antioxidants, resulting in the radiation and desiccation tolerance displayed by Deinococcus species (91).
Numerous genes encoding resistance mechanisms against antibiotics targeting the protein synthesis machinery are transcribed as lmRNAs (82, 93–95). It is tempting to speculate that this mechanism is required for continued translation even in the presence of the antibiotic. The aminoglycoside antibiotic kasugamycin (Ksg) targets the ribosome in a position overlapping the kink between the P- and E-site codons and therefore interferes with the path of the mRNA immediately upstream of the start codon (96, 97). However, Ksg only affects 30S subunits without impairing fMet-tRNAfMet binding to the P-site in 70S ribosomes due to 50S stabilization (96). Therefore, lmRNA translation is resistant to Ksg treatment due to the lack of competition between the mRNA and Ksg for binding sites (90, 98). This mechanism further supports the 70S initiation pathway for lmRNA translation containing no 30S intermediate steps (98). Intriguingly, the mRNAs encoding the ABC transporter that exports Ksg in the producing actinobacterium Streptomyces kasugaensis correspondingly are leaderless or translationally coupled (99). By this means the synthesis of the export mechanism continues even when the intracellular concentration of Ksg increases, ensuring the survival of the producer cell.
Generation of lmRNAs during Stress
During adverse physiological conditions, lmRNAs are even produced via endoribonucleolytic cleavage, allowing for their continued translation. Stress conditions such as treatment with antibiotics affecting transcription or translation (100–102) or amino acid starvation (103) cause a reduction in the expression of the toxin-antitoxin system mazEF in E. coli. The labile antitoxin is quickly degraded and therefore no longer represses the stable MazF toxin. The active sequence-specific endoribonuclease MazF subsequently cleaves at single-stranded ACA sites to cause bulk mRNA degradation (Fig. 1). However, in some cases, the only available ACA site resides within the 5′ UTR just upstream of the initiation codon (17). This cleavage therefore generates a subset of mRNAs including both lmRNA and mRNA with shortened leaders that specifically contain a 5′-terminal hydroxyl group (104).
Concurrently, MazF also cleaves the 16S rRNA within mature 70S ribosomes upstream of position A1500, removing 43 nucleotides from the 3′ terminus, including the aSD sequence (17). Therefore, MazF cleavage results not only in a subset of short leadered/leaderless mRNAs but also in a subpopulation of so-called 70SΔ43 ribosomes that is specialized for lmRNA translation. Subsequent ribosome profiling has revealed that the transcripts that are processed by MazF upstream of the start codon are bound and selectively translated by the modified 70SΔ43 ribosomes (18). Whether the ribosome is able to select for the mRNAs in the MazF regulon via the absence of a complex leader sequence or through recognition of the 5′ hydroxyl remains to be elucidated. However, recent data suggest that the 70SΔ43 ribosome, in contrast to the 70S, is more promiscuous, as it binds both 5′-triphosphate and 5′-hydroxyl termini (H. J. Beck, M. Sauert, and I. Moll, unpublished data), presenting a new potential pathway for lmRNA initiation.
The ∼330 mRNAs that make up the MazF regulon do not appear to be functionally clustered, but there is an overrepresentation of essential genes, suggesting that the mRNAs that are not degraded by MazF are important for cell survival during stress and likely post-stress recovery (18). During recovery, the RNA ligase RtcB catalyzes the religation of the 43-nucleotide cleaved fragment to the 3′ end of the 16S rRNA, successfully restoring the ribosomes’ ability to translate canonical leadered mRNAs (105). Therefore, the cell has processes in place to not only reduce its translatome to move into dormancy but also to transition back to steady state by utilizing the lmRNA initiation mechanism.
Differential Expression in Response to Stress
Other organisms also exploit the use of lmRNAs to adapt to stress. In Mycobacterium tuberculosis the majority of toxin-antitoxin genes is transcribed as lmRNA (106). A transcriptome analysis revealed a significant increase in lmRNAs during starvation, and notably, lmRNAs were the most strongly upregulated. Although lmRNAs are typically thought to be less efficiently expressed, in M. tuberculosis they were shown to have longer half-lives than the SD-led mRNAs, resulting in nearly equivalent protein production. These results suggest that lmRNAs, while playing a secondary role during exponential growth, are key in the physiology of nonreplicating cells (106). Mycobacterium spp. also produce a MazF toxin shown to target both the 23S rRNA helix loop 70, to completely inactivate translation, and the 3′ end of the 16S rRNA, to alter ribosomal specificity by removing the aSD sequence (107, 108). Mycobacterial MazF recognizes a different specific sequence than E. coli MazF but targets similar functional regions of the translational machinery, suggesting that the orthologous toxins have evolved their relative specificities in an effort to exploit essential and accessible ribosomal regions, which could lead to an adaptive response to various conditions (108).
Shigella spp. utilize alternative promoters within one gene to transcribe a leadered and a leaderless variant of the virF mRNA, translation of which results in the major full-length and an N-terminally truncated VirF protein, respectively (109). By directly binding to consensus virF promoter elements, the truncated protein negatively regulates the levels of full-length VirF that triggers a regulatory cascade, leading to virulence and subsequent invasion of the intestinal epithelial barrier in humans (109). The conditions triggering the transcription of virF lmRNA from a secondary promoter within the canonical virF coding sequence remain elusive, but it has been speculated that environmental cues signaling unsuitable conditions for an invasive response may play a role. Alternative transcription in response to butanol and butyrate stress also leads to the generation of lmRNA in Clostridium acetobutylicum (110). It is thus conceivable that numerous organisms employ leaderless transcripts to ensure continuous translation, albeit with lower efficiency, during adverse physiological conditions.
Further Benefits of 5′-Terminal AUGs
The ability of ribosomes to interact with the 5′-AUG can additionally allow for indirect regulation of downstream ORFs in polycistronic mRNAs. As previously mentioned, a recent transcriptome analysis brought to light the prevalence of 5′-uAUGs, which were able to bind ribosomes and in some cases allow for efficient translation (14). Similar 5′-uAUGs were also discovered in D. deserti and in some cases were found to be involved in transcriptional attenuation (91). In the acuR-acuI-dddL operon in R. sphaeroides, the first cistron encodes a regulatory protein that represses the entire operon but is translated at a much lower rate than the downstream cistrons because it lacks a ribosome binding site (15). This genetic makeup generates equal transcriptional rates but disproportionate translational rates, resulting in the desired regulation. The pairwise arrangement of acuR-acuI, lacking a 5′ UTR, was observed in several individual strains of alpha-, beta-, and gammaproteobacteria (15), suggesting that utilizing an lmRNA to control protein expression is a widespread phenomenon.
In some cases, the additional upstream start codon is in frame with the canonical start codon, which could result in two independently translated protein isoforms from one transcript. Such cases of alternative translation initiation have been well documented in eukaryotes (reviewed in reference 110) and haloarchaea (16). In the haloarchaeon Natrinema sp. strain J7-2, the alternative initiation site on the sptA mRNA is 5 nucleotides from the 5′ terminus, thereby characterizing the transcript as both leadered and leaderless depending on the start codon utilized. Here, greater translation initiation efficiency occurs at the 5′-terminal AUG, with the internal AUG serving as a remedial initiation site in case the 5′-terminal AUG is disrupted (16). This observation is not surprising, as lmRNAs form the majority of transcripts in haloarchaea and are known to be efficiently translated (111). However, the SptA protein is processed posttranslationally to remove the N terminus when translated from the leaderless mRNA; thus, regardless of the initiation codon used, the final protein product remains the same (111). The presence of both AUGs does provide benefits, positively affecting mRNA stability to enhance expression. Redundancy in initiation as either a leadered or leaderless mRNA ensures its translation in response to different cellular signals or stress. Alternative translation initiation also occurs in M. tuberculosis, where the extracytoplasmatic sigma factor σE (sigE) is differentially translated to produce different isoforms depending on environmental cues. In response to surface stress or an alkaline pH, transcription of the leaderless variant of the sigE transcript is upregulated while the leadered variant, usually selected for during normal growth, is repressed (112). This genome structure is conserved in other mycobacteria, suggesting that this regulatory mechanism evolved early in mycobacteria history.
The translation of the upstream ORF can similarly affect downstream expression through translational coupling (113). A high percentage of leaderless small proteins are coupled to downstream genes in mycobacteria (79). Since translation of the first ORF can be initiated with a 70S ribosome, this may allow for increased expression of the entire operon under conditions of increased 70S concentration. 70S ribosomes can reinitiate downstream without dissociating by scanning using unidimensional diffusion to search for an SD sequence (114). This scanning can take place in the absence of initiation factors if fMet-tRNAfMet is present. Therefore, under conditions selecting for lmRNAs, translational coupling through reinitiation could allow the entire operon to maintain efficient expression.
One example in which the 70S scanning mechanism appears to be essential for downstream expression occurs in E. coli ptrB mRNA encoding a serine protease. The ptrB transcript contains a 5′-uAUG upstream of its canonical start site, and mutation of this 5′-uAUG drastically reduces downstream ptrB expression by >90% (115). The 5′-uAUG does not act as an initiation codon for translation, but rather as a 70S loading site, necessary during optimal growth conditions due to ptrB’s weak SD sequence and also to ensure continued translation during stress. In this case, regulation of the mRNA is dependent upon the inherent ability of a 70S ribosome to recognize a 5′-AUG via the 70S lmRNA initiation pathway (115).
Apart from initiating translation, an upstream leaderless AUG can elicit indirect effects from a bound ribosome. 70S ribosome binding at the 5′ terminus can stabilize the mRNA (2); block the action of exonucleases (116); prevent premature transcriptional termination (117); or block the binding of other ribosomes or low-molecular-weight effectors such as amino acids, coenzymes, vitamins, or cyclic di-GMP whose binding could affect the mRNA secondary structure. Increasing the mRNA’s half-life or protecting it from degradation will have a positive impact on surrounding ORFs. However, prohibiting ribosomal loading through steric hindrance will ultimately reduce downstream expression. Furthermore, occupying an area on the mRNA can also affect secondary structure, which in turn can lead to opening or occluding downstream ribosomal binding regions. Therefore, upstream ribosomal binding through recognition of the 5′-AUG can indirectly modulate the expression of the other cistrons in the operon in a variety of ways.
These regulation mechanisms are able to utilize an intrinsic property that may be an ancestral remnant used to recognize lmRNAs for their own specialized purposes. This “recycling” of inherent interaction mechanisms has already been shown: aside from assisting in translation initiation, the aSD sequence of the ribosome can interact with the SD sequence within coding sequences in archaea to induce pausing (113) and in bacteria for programmed frameshifting (114). In this way, intrinsic properties of the ribosome are used not only for initiation but during elongation as well.
PERSPECTIVE
Although it is impossible to definitively conclude the evolutionary origins of any biological process, there is substantial evidence to suggest that lmRNAs are ancestral in nature. From their ability to be translated by ribosomes of any domain of life, the utilization of a scaled-down mechanism of initiation, and their prevalence in organisms close to the root of the phylogenetic tree and in extremophiles, which are resistant to conditions reflecting the traits of prehistoric times, lmRNAs appear to predate SD-led mRNAs. lmRNAs can provide insight into the minimal requirements for translation. More-evolved prokaryotes and eukaryotes appear to have built upon the primitive method by increasing complexity of mRNA structure and number of initiation factors and other ancillary components. However, ribosomes have retained the intrinsic ability to recognize and bind the 5′-AUG signature of lmRNAs, allowing them to remain active in today’s cells. This suggests that ribosomes may inspect the 5′ termini of all mRNAs as a remnant of an ancestral mechanism that has evolved into the eukaryotic mechanism of 5′-cap binding. The existence of capped lmRNAs in the protozoan parasite Giardia lamblia that use the lmRNA recognition mechanism without ribosome scanning might provide a bridge between prokaryotic and eukaryotic mechanisms with parallels to archaea (118). Furthermore, 5′-end recognition may continue to be utilized in bacteria and archaea for ribosomal loading for downstream translation or to protect the mRNA from degradation. These observations illustrate the importance of studying lmRNAs to provide insight into the evolution and function of the ribosome.
lmRNAs are not simply a functional relic of ancient times. They are still being used to control translational efficiency and to modulate the stress response. The ability to withstand adverse conditions makes lmRNAs beneficial in combating stress and transitioning into dormancy. The use of lmRNAs provides a way to control protein production without the need for auxiliary factors, such as small RNAs or RNA-binding proteins. Therefore, no further energy expenditure is required to modulate translational efficiency, making lmRNAs a valuable tool during cellular shutdown. Considering that the threat of antibiotic resistance and persistence formation is increasing, elucidating the role of lmRNA and the mechanisms underlying its translation will be crucial in controlling bacterial infections. As the vast numbers of lmRNAs throughout bacteria and archaea will be identified, further examples of their various roles in multiple cellular processes are expected to be elucidated, which may provide insights into the intrinsic abilities of the ribosome.
REFERENCES
- 1.Shine J, Dalgarno L. 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A 71:1342–1346. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komarova AV, Tchufistova LS, Dreyfus M, Boni IV. 2005. AU-rich sequences within 5′ untranslated leaders enhance translation and stabilize mRNA in Escherichia coli. J Bacteriol 187:1344–1349. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boni IV, Isaeva DM, Musychenko ML, Tzareva NV. 1991. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res 19:155–162. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grill S, Gualerzi CO, Londei P, Bläsi U. 2000. Selective stimulation of translation of leaderless mRNA by initiation factor 2: evolutionary implications for translation. EMBO J 19:4101–4110. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang B, Halgamuge S, Tang SL. 2006. Analysis of SD sequences in completed microbial genomes: non-SD-led genes are as common as SD-led genes. Gene 373:90–99. [PubMed] [DOI] [PubMed] [Google Scholar]
- 6.Zheng X, Hu GQ, She ZS, Zhu H. 2011. Leaderless genes in bacteria: clue to the evolution of translation initiation mechanisms in prokaryotes. BMC Genomics 12:361. 10.1186/1471-2164-12-361. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srivastava A, Gogoi P, Deka B, Goswami S, Kanaujia SP. 2016. In silico analysis of 5′-UTRs highlights the prevalence of Shine-Dalgarno and leaderless-dependent mechanisms of translation initiation in bacteria and archaea, respectively. J Theor Biol 402:54–61. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Montoya J, Ojala D, Attardi G. 1981. Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature 290:465–470. [PubMed] [DOI] [PubMed] [Google Scholar]
- 9.Jones CN, Wilkinson KA, Hung KT, Weeks KM, Spremulli LL. 2008. Lack of secondary structure characterizes the 5′ ends of mammalian mitochondrial mRNAs. RNA 14:862–871. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torarinsson E, Klenk HP, Garrett RA. 2005. Divergent transcriptional and translational signals in Archaea. Environ Microbiol 7:47–54. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Tolstrup N, Sensen CW, Garrett RA, Clausen IG. 2000. Two different and highly organized mechanisms of translation initiation in the archaeon Sulfolobus solfataricus. Extremophiles 4:175–179. [PubMed] [DOI] [PubMed] [Google Scholar]
- 12.Slupska MM, King AG, Fitz-Gibbon S, Besemer J, Borodovsky M, Miller JH. 2001. Leaderless transcripts of the crenarchaeal hyperthermophile Pyrobaculum aerophilum. J Mol Biol 309:347–360. [PubMed] [DOI] [PubMed] [Google Scholar]
- 13.Cho S, Kim MS, Jeong Y, Lee BR, Lee JH, Kang SG, Cho BK. 2017. Genome-wide primary transcriptome analysis of H2-producing archaeon Thermococcus onnurineus NA1. Sci Rep 7:43044. 10.1038/srep43044. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beck HJ, Fleming IMC, Janssen GR. 2016. 5′-Terminal AUGs in Escherichia coli mRNAs with Shine-Dalgarno sequences: identification and analysis of their roles in non-canonical translation initiation. PLoS One 11:e0160144. 10.1371/journal.pone.0160144. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sullivan MJ, Curson AR, Shearer N, Todd JD, Green RT, Johnston AW. 2011. Unusual regulation of a leaderless operon involved in the catabolism of dimethylsulfoniopropionate in Rhodobacter sphaeroides. PLoS One 6:e15972. 10.1371/journal.pone.0015972. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang W, Wu Y, Li M, Wang J, Mei S, Tang B, Tang XF. 2016. Alternative translation initiation of a haloarchaeal serine protease transcript containing two in-frame start codons. J Bacteriol 198:1892–1901. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. 2011. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 147:147–157. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauert M, Wolfinger MT, Vesper O, Müller C, Byrgazov K, Moll I. 2016. The MazF-regulon: a toolbox for the post-transcriptional stress response in Escherichia coli. Nucleic Acids Res 44:6660–6675. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussain T, Llácer JL, Wimberly BT, Kieft JS, Ramakrishnan V. 2016. Large-scale movements of IF3 and tRNA during bacterial translation initiation. Cell 167:133–144.e13. 10.1016/j.cell.2016.08.074. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caban K, Pavlov M, Ehrenberg M, Gonzalez RL, Jr. 2017. A conformational switch in initiation factor 2 controls the fidelity of translation initiation in bacteria. Nat Commun 8:1475. 10.1038/s41467-017-01492-6. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milón P, Rodnina MV. 2012. Kinetic control of translation initiation in bacteria. Crit Rev Biochem Mol Biol 47:334–348. [PubMed] [DOI] [PubMed] [Google Scholar]
- 22.Milón P, Maracci C, Filonava L, Gualerzi CO, Rodnina MV. 2012. Real-time assembly landscape of bacterial 30S translation initiation complex. Nat Struct Mol Biol 19:609–615. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. 2006. How initiation factors maximize the accuracy of tRNA selection in initiation of bacterial protein synthesis. Mol Cell 23:183–193. [PubMed] [DOI] [PubMed] [Google Scholar]
- 24.Simonetti A, Marzi S, Jenner L, Myasnikov A, Romby P, Yusupova G, Klaholz BP, Yusupov M. 2009. A structural view of translation initiation in bacteria. Cell Mol Life Sci 66:423–436. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. 2006. How initiation factors tune the rate of initiation of protein synthesis in bacteria. EMBO J 25:2539–2550. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dallas A, Noller HF. 2001. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell 8:855–864. [DOI] [PubMed] [Google Scholar]
- 27.Marshall RA, Aitken CE, Puglisi JD. 2009. GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol Cell 35:37–47. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sørensen MA, Fricke J, Pedersen S. 1998. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo. J Mol Biol 280:561–569. [PubMed] [DOI] [PubMed] [Google Scholar]
- 29.Byrgazov K, Grishkovskaya I, Arenz S, Coudevylle N, Temmel H, Wilson DN, Djinovic-Carugo K, Moll I. 2015. Structural basis for the interaction of protein S1 with the Escherichia coli ribosome. Nucleic Acids Res 43:661–673. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lauber MA, Rappsilber J, Reilly JP. 2012. Dynamics of ribosomal protein S1 on a bacterial ribosome with cross-linking and mass spectrometry. Mol Cell Proteomics 11:1965–1976. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qu X, Lancaster L, Noller HF, Bustamante C, Tinoco I, Jr. 2012. Ribosomal protein S1 unwinds double-stranded RNA in multiple steps. Proc Natl Acad Sci U S A 109:14458–14463. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Duin J, Wijnands R. 1981. The function of ribosomal protein S21 in protein synthesis. Eur J Biochem 118:615–619. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.Backendorf C, Ravensbergen CJ, Van der Plas J, van Boom JH, Veeneman G, Van Duin J. 1981. Basepairing potential of the 3′ terminus of 16S RNA: dependence on the functional state of the 30S subunit and the presence of protein S21. Nucleic Acids Res 9:1425–1444. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Odom OW, Stöffler G, Hardesty B. 1984. Movement of the 3′-end of 16 S RNA towards S21 during activation of 30 S ribosomal subunits. FEBS Lett 173:155–158. [DOI] [PubMed] [Google Scholar]
- 35.Moll I, Resch A, Bläsi U. 1998. Discrimination of 5′-terminal start codons by translation initiation factor 3 is mediated by ribosomal protein S1. FEBS Lett 436:213–217. [DOI] [PubMed] [Google Scholar]
- 36.Tedin K, Resch A, Bläsi U. 1997. Requirements for ribosomal protein S1 for translation initiation of mRNAs with and without a 5′ leader sequence. Mol Microbiol 25:189–199. [PubMed] [DOI] [PubMed] [Google Scholar]
- 37.Shean CS, Gottesman ME. 1992. Translation of the prophage λ cl transcript. Cell 70:513–522. [DOI] [PubMed] [Google Scholar]
- 38.Moll I, Grill S, Gründling A, Bläsi U. 2002. Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD-box helicase on translation of leaderless and canonical mRNAs in Escherichia coli. Mol Microbiol 44:1387–1396. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Delvillani F, Papiani G, Dehò G, Briani F. 2011. S1 ribosomal protein and the interplay between translation and mRNA decay. Nucleic Acids Res 39:7702–7715. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duval M, Korepanov A, Fuchsbauer O, Fechter P, Haller A, Fabbretti A, Choulier L, Micura R, Klaholz BP, Romby P, Springer M, Marzi S. 2013. Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLoS Biol 11:e1001731. 10.1371/journal.pbio.1001731. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salah P, Bisaglia M, Aliprandi P, Uzan M, Sizun C, Bontems F. 2009. Probing the relationship between Gram-negative and Gram-positive S1 proteins by sequence analysis. Nucleic Acids Res 37:5578–5588. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Byrgazov K, Manoharadas S, Kaberdina AC, Vesper O, Moll I. 2012. Direct interaction of the N-terminal domain of ribosomal protein S1 with protein S2 in Escherichia coli. PLoS One 7:e32702. 10.1371/journal.pone.0032702 [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aseev LV, Chugunov AO, Efremov RG, Boni IV. 2013. A single missense mutation in a coiled-coil domain of Escherichia coli ribosomal protein S2 confers a thermosensitive phenotype that can be suppressed by ribosomal protein S1. J Bacteriol 195:95–104. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moll I, Hirokawa G, Kiel MC, Kaji A, Bläsi U. 2004. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res 32:3354–3363. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grill S, Moll I, Hasenöhrl D, Gualerzi CO, Bläsi U. 2001. Modulation of ribosomal recruitment to 5′-terminal start codons by translation initiation factors IF2 and IF3. FEBS Lett 495:167–171. [DOI] [PubMed] [Google Scholar]
- 46.O’Donnell SM, Janssen GR. 2002. Leaderless mRNAs bind 70S ribosomes more strongly than 30S ribosomal subunits in Escherichia coli. J Bacteriol 184:6730–6733. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tedin K, Moll I, Grill S, Resch A, Graschopf A, Gualerzi CO, Bläsi U. 1999. Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol Microbiol 31:67–77. [PubMed] [DOI] [PubMed] [Google Scholar]
- 48.Maar D, Liveris D, Sussman JK, Ringquist S, Moll I, Heredia N, Kil A, Bläsi U, Schwartz I, Simons RW. 2008. A single mutation in the IF3 N-terminal domain perturbs the fidelity of translation initiation at three levels. J Mol Biol 383:937–944. [PubMed] [DOI] [PubMed] [Google Scholar]
- 49.O’Connor M, Gregory ST, Rajbhandary UL, Dahlberg AE. 2001. Altered discrimination of start codons and initiator tRNAs by mutant initiation factor 3. RNA 7:969–978. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moazed D, Samaha RR, Gualerzi C, Noller HF. 1995. Specific protection of 16 S rRNA by translational initiation factors. J Mol Biol 248:207–210. [DOI] [PubMed] [Google Scholar]
- 51.Howe JG, Hershey JW. 1983. Initiation factor and ribosome levels are coordinately controlled in Escherichia coli growing at different rates. J Biol Chem 258:1954–1959. [PubMed] [PubMed] [Google Scholar]
- 52.Liveris D, Klotsky RA, Schwartz I. 1991. Growth rate regulation of translation initiation factor IF3 biosynthesis in Escherichia coli. J Bacteriol 173:3888–3893. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones PG, VanBogelen RA, Neidhardt FC. 1987. Induction of proteins in response to low temperature in Escherichia coli. J Bacteriol 169:2092–2095. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hinnebusch AG. 2017. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem Sci 42:589–611. [PubMed] [DOI] [PubMed] [Google Scholar]
- 55.Balakin AG, Skripkin EA, Shatsky IN, Bogdanov AA, Belozersky AN. 1992. Unusual ribosome binding properties of mRNA encoding bacteriophage λ repressor. Nucleic Acids Res 20:563–571. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Udagawa T, Shimizu Y, Ueda T. 2004. Evidence for the translation initiation of leaderless mRNAs by the intact 70 S ribosome without its dissociation into subunits in eubacteria. J Biol Chem 279:8539–8546. [PubMed] [DOI] [PubMed] [Google Scholar]
- 57.Benelli D, Maone E, Londei P. 2003. Two different mechanisms for ribosome/mRNA interaction in archaeal translation initiation. Mol Microbiol 50:635–643. [PubMed] [DOI] [PubMed] [Google Scholar]
- 58.Hasenöhrl D, Fabbretti A, Londei P, Gualerzi CO, Bläsi U. 2009. Translation initiation complex formation in the crenarchaeon Sulfolobus solfataricus. RNA 15:2288–2298. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bell SD, Jackson SP. 1998. Transcription and translation in Archaea: a mosaic of eukaryal and bacterial features. Trends Microbiol 6:222–228. [DOI] [PubMed] [Google Scholar]
- 60.Dennis PP. 1997. Ancient ciphers: translation in Archaea. Cell 89:1007–1010. [DOI] [PubMed] [Google Scholar]
- 61.La Teana A, Benelli D, Londei P, Bläsi U. 2013. Translation initiation in the crenarchaeon Sulfolobus solfataricus: eukaryotic features but bacterial route. Biochem Soc Trans 41:350–355. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.Benelli D, Londei P. 2011. Translation initiation in Archaea: conserved and domain-specific features. Biochem Soc Trans 39:89–93. [PubMed] [DOI] [PubMed] [Google Scholar]
- 63.Hasenöhrl D, Benelli D, Barbazza A, Londei P, Bläsi U. 2006. Sulfolobus solfataricus translation initiation factor 1 stimulates translation initiation complex formation. RNA 12:674–682. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pedullà N, Palermo R, Hasenöhrl D, Bläsi U, Cammarano P, Londei P. 2005. The archaeal eIF2 homologue: functional properties of an ancient translation initiation factor. Nucleic Acids Res 33:1804–1812. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arkhipova V, Stolboushkina E, Kravchenko O, Kljashtorny V, Gabdulkhakov A, Garber M, Nikonov S, Märtens B, Bläsi U, Nikonov O. 2015. Binding of the 5′-triphosphate end of mRNA to the γ-subunit of translation initiation factor 2 of the crenarchaeon Sulfolobus solfataricus. J Mol Biol 427:3086–3095. [PubMed] [DOI] [PubMed] [Google Scholar]
- 66.Hasenöhrl D, Lombo T, Kaberdin V, Londei P, Bläsi U. 2008. Translation initiation factor a/eIF2(-γ) counteracts 5′ to 3′ mRNA decay in the archaeon Sulfolobus solfataricus. Proc Natl Acad Sci U S A 105:2146–2150. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Londei P. 2005. Evolution of translational initiation: new insights from the archaea. FEMS Microbiol Rev 29:185–200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 68.Yatime L, Schmitt E, Blanquet S, Mechulam Y. 2004. Functional molecular mapping of archaeal translation initiation factor 2. J Biol Chem 279:15984–15993. [PubMed] [DOI] [PubMed] [Google Scholar]
- 69.Benelli D, Marzi S, Mancone C, Alonzi T, la Teana A, Londei P. 2009. Function and ribosomal localization of aIF6, a translational regulator shared by archaea and eukarya. Nucleic Acids Res 37:256–267. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Resch A, Tedin K, Gründling A, Mündlein A, Bläsi U. 1996. Downstream box-anti-downstream box interactions are dispensable for translation initiation of leaderless mRNAs. EMBO J 15:4740–4748. [PubMed] [PMC free article] [PubMed] [Google Scholar]
- 71.Moll I, Huber M, Grill S, Sairafi P, Mueller F, Brimacombe R, Londei P, Bläsi U. 2001. Evidence against an interaction between the mRNA downstream box and 16S rRNA in translation initiation. J Bacteriol 183:3499–3505. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martin-Farmer J, Janssen GR. 1999. A downstream CA repeat sequence increases translation from leadered and unleadered mRNA in Escherichia coli. Mol Microbiol 31:1025–1038. [PubMed] [DOI] [PubMed] [Google Scholar]
- 73.Brock JE, Paz RL, Cottle P, Janssen GR. 2007. Naturally occurring adenines within mRNA coding sequences affect ribosome binding and expression in Escherichia coli. J Bacteriol 189:501–510. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Giliberti J, O’Donnell S, Etten WJ, Janssen GR, Van Etten WJ, Janssen GR. 2012. A 5′-terminal phosphate is required for stable ternary complex formation and translation of leaderless mRNA in Escherichia coli. RNA 18:508–518. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brock JE, Pourshahian S, Giliberti J, Limbach PA, Janssen GR. 2008. Ribosomes bind leaderless mRNA in Escherichia coli through recognition of their 5′-terminal AUG. RNA 14:2159–2169. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Etten WJ, Janssen GR. 1998. An AUG initiation codon, not codon-anticodon complementarity, is required for the translation of unleadered mRNA in Escherichia coli. Mol Microbiol 27:987–1001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 77.O’Donnell SM, Janssen GR. 2001. The initiation codon affects ribosome binding and translational efficiency in Escherichia coli of cI mRNA with or without the 5′ untranslated leader. J Bacteriol 183:1277–1283. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hering O, Brenneis M, Beer J, Suess B, Soppa J. 2009. A novel mechanism for translation initiation operates in haloarchaea. Mol Microbiol 71:1451–1463. [PubMed] [DOI] [PubMed] [Google Scholar]
- 79.Shell SS, Wang J, Lapierre P, Mir M, Chase MR, Pyle MM, Gawande R, Ahmad R, Sarracino DA, Ioerger TR, Fortune SM, Derbyshire KM, Wade JT, Gray TA. 2015. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet 11:e1005641. 10.1371/journal.pgen.1005641. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krishnan KM, Van Etten WJ, III, Janssen GR. 2010. Proximity of the start codon to a leaderless mRNA’s 5′ terminus is a strong positive determinant of ribosome binding and expression in Escherichia coli. J Bacteriol 192:6482–6485. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu CJ, Janssen GR. 1996. Translation of vph mRNA in Streptomyces lividans and Escherichia coli after removal of the 5′ untranslated leader. Mol Microbiol 22:339–355. [DOI] [PubMed] [Google Scholar]
- 82.Wu CJ, Janssen GR. 1997. Expression of a streptomycete leaderless mRNA encoding chloramphenicol acetyltransferase in Escherichia coli. J Bacteriol 179:6824–6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kohler R, Mooney RA, Mills DJ, Landick R, Cramer P. 2017. Architecture of a transcribing-translating expressome. Science 356:194–197. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schrader JM, Zhou B, Li GW, Lasker K, Childers WS, Williams B, Long T, Crosson S, McAdams HH, Weissman JS, Shapiro L. 2014. The coding and noncoding architecture of the Caulobacter crescentus genome. PLoS Genet 10:e1004463. 10.1371/journal.pgen.1004463. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sartorius-Neef S, Pfeifer F. 2004. In vivo studies on putative Shine-Dalgarno sequences of the halophilic archaeon Halobacterium salinarum. Mol Microbiol 51:579–588. [PubMed] [DOI] [PubMed] [Google Scholar]
- 86.Condò I, Ciammaruconi A, Benelli D, Ruggero D, Londei P. 1999. cis-Acting signals controlling translational initiation in the thermophilic archaeon Sulfolobus solfataricus. Mol Microbiol 34:377–384. [PubMed] [DOI] [PubMed] [Google Scholar]
- 87.Amitai S, Kolodkin-Gal I, Hananya-Meltabashi M, Sacher A, Engelberg-Kulka H. 2009. Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins.” PLoS Genet 5:e1000390. 10.1371/journal.pgen.1000390. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Andreev DE, Terenin IM, Dunaevsky YE, Dmitriev SE, Shatsky IN. 2006. A leaderless mRNA can bind to mammalian 80S ribosomes and direct polypeptide synthesis in the absence of translation initiation factors. Mol Cell Biol 26:3164–3169. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grill S, Moll I, Giuliodori AM, Gualerzi CO, Bläsi U. 2002. Temperature-dependent translation of leaderless and canonical mRNAs in Escherichia coli. FEMS Microbiol Lett 211:161–167. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.Chin K, Shean CS, Gottesman ME. 1993. Resistance of λ cI translation to antibiotics that inhibit translation initiation. J Bacteriol 175:7471–7473. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de Groot A, Roche D, Fernandez B, Ludanyi M, Cruveiller S, Pignol D, Vallenet D, Armengaud J, Blanchard L. 2014. RNA sequencing and proteogenomics reveal the importance of leaderless mRNAs in the radiation-tolerant bacterium Deinococcus deserti. Genome Biol Evol 6:932–948. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bouthier de la Tour C, Blanchard L, Dulermo R, Ludanyi M, Devigne A, Armengaud J, Sommer S, de Groot A. 2015. The abundant and essential HU proteins in Deinococcus deserti and Deinococcus radiodurans are translated from leaderless mRNA. Microbiology 161:2410–2422. [PubMed] [DOI] [PubMed] [Google Scholar]
- 93.Baumeister R, Flache P, Melefors O, von Gabain A, Hillen W. 1991. Lack of a 5′ non-coding region in Tn1721 encoded tetR mRNA is associated with a low efficiency of translation and a short half-life in Escherichia coli. Nucleic Acids Res 19:4595–4600. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jones RL, III, Jaskula JC, Janssen GR. 1992. In vivo translational start site selection on leaderless mRNA transcribed from the Streptomyces fradiae aph gene. J Bacteriol 174:4753–4760. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.August PR, Flickinger MC, Sherman DH. 1994. Cloning and analysis of a locus (mcr) involved in mitomycin C resistance in Streptomyces lavendulae. J Bacteriol 176:4448–4454. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schluenzen F, Takemoto C, Wilson DN, Kaminishi T, Harms JM, Hanawa-Suetsugu K, Szaflarski W, Kawazoe M, Shirouzu M, Nierhaus KH, Yokoyama S, Fucini P. 2006. The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation. Nat Struct Mol Biol 13:871–878. [PubMed] [DOI] [PubMed] [Google Scholar]
- 97.Schuwirth BS, Day JM, Hau CW, Janssen GR, Dahlberg AE, Cate JH, Vila-Sanjurjo A. 2006. Structural analysis of kasugamycin inhibition of translation. Nat Struct Mol Biol 13:879–886. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moll I, Bläsi U. 2002. Differential inhibition of 30S and 70S translation initiation complexes on leaderless mRNA by kasugamycin. Biochem Biophys Res Commun 297:1021–1026. [DOI] [PubMed] [Google Scholar]
- 99.Ikeno S, Yamane Y, Ohishi Y, Kinoshita N, Hamada M, Tsuchiya KS, Hori M. 2000. ABC transporter genes, kasKLM, responsible for self-resistance of a kasugamycin producer strain. J Antibiot (Tokyo) 53:373–384. [DOI] [PubMed] [Google Scholar]
- 100.Müller C, Sokol L, Vesper O, Sauert M, Moll I. 2016. Insights into the stress response triggered by kasugamycin in Escherichia coli. Antibiotics (Basel) 5:E19. 10.3390/antibiotics5020019. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hazan R, Sat B, Engelberg-Kulka H. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J Bacteriol 186:3663–3669. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sat B, Hazan R, Fisher T, Khaner H, Glaser G, Engelberg-Kulka H. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J Bacteriol 183:2041–2045. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Aizenman E, Engelberg-Kulka H, Glaser G. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci U S A 93:6059–6063. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang Y, Zhang J, Hara H, Kato I, Inouye M. 2005. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J Biol Chem 280:3143–3150. [PubMed] [DOI] [PubMed] [Google Scholar]
- 105.Temmel H, Müller C, Sauert M, Vesper O, Reiss A, Popow J, Martinez J, Moll I. 2017. The RNA ligase RtcB reverses MazF-induced ribosome heterogeneity in Escherichia coli. Nucleic Acids Res 45:4708–4721. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cortes T, Schubert OT, Rose G, Arnvig KB, Comas I, Aebersold R, Young DB. 2013. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep 5:1121–1131. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schifano JM, Edifor R, Sharp JD, Ouyang M, Konkimalla A, Husson RN, Woychik NA. 2013. Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc Natl Acad Sci U S A 110:8501–8506. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schifano JM, Vvedenskaya IO, Knoblauch JG, Ouyang M, Nickels BE, Woychik NA. 2014. An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat Commun 5:3538. 10.1038/ncomms4538. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Di Martino ML, Romilly C, Wagner EG, Colonna B, Prosseda G. 2016. One gene and two proteins: a leaderless mRNA supports the translation of a shorter form of the Shigella VirF regulator. mBio 7:e01860-16. 10.1128/mBio.01860-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kochetov AV. 2008. Alternative translation start sites and hidden coding potential of eukaryotic mRNAs. BioEssays 30:683–691. [PubMed] [DOI] [PubMed] [Google Scholar]
- 111.Brenneis M, Hering O, Lange C, Soppa J. 2007. Experimental characterization of cis-acting elements important for translation and transcription in halophilic archaea. PLoS Genet 3:e229. 10.1371/journal.pgen.0030229. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Donà V, Rodrigue S, Dainese E, Palù G, Gaudreau L, Manganelli R, Provvedi R. 2008. Evidence of complex transcriptional, translational, and posttranslational regulation of the extracytoplasmic function sigma factor σE in Mycobacterium tuberculosis. J Bacteriol 190:5963–5971. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rex G, Surin B, Besse G, Schneppe B, McCarthy JE. 1994. The mechanism of translational coupling in Escherichia coli. Higher order structure in the atpHA mRNA acts as a conformational switch regulating the access of de novo initiating ribosomes. J Biol Chem 269:18118–18127. [PubMed] [PubMed] [Google Scholar]
- 114.Yamamoto H, Wittek D, Gupta R, Qin B, Ueda T, Krause R, Yamamoto K, Albrecht R, Pech M, Nierhaus KH. 2016. 70S-scanning initiation is a novel and frequent initiation mode of ribosomal translation in bacteria. Proc Natl Acad Sci U S A 113:E1180–E1189. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Beck HJ, Janssen GR. 2017. Novel translation initiation regulation mechanism in Escherichia coli ptrB mediated by a 5′-terminal AUG. J Bacteriol 199:e00091-17. 10.1128/JB.00091-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Deana A, Belasco JG. 2005. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev 19:2526–2533. [PubMed] [DOI] [PubMed] [Google Scholar]
- 117.Eriksen M, Sneppen K, Pedersen S, Mitarai N. 2017. Occlusion of the ribosome binding site connects the translational initiation frequency, mRNA stability and premature transcription termination. Front Microbiol 8:362. 10.3389/fmicb.2017.00362. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li L, Wang CC. 2004. Capped mRNA with a single nucleotide leader is optimally translated in a primitive eukaryote, Giardia lamblia. J Biol Chem 279:14656–14664. [PubMed] [DOI] [PubMed] [Google Scholar]
- 119.Schlüter JP, Reinkensmeier J, Barnett MJ, Lang C, Krol E, Giegerich R, Long SR, Becker A. 2013. Global mapping of transcription start sites and promoter motifs in the symbiotic α-proteobacterium Sinorhizobium meliloti 1021. BMC Genomics 14:156. 10.1186/1471-2164-14-156. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Qiu Y, Cho BK, Park YS, Lovley D, Palsson BØ, Zengler K. 2010. Structural and operational complexity of the Geobacter sulfurreducens genome. Genome Res 20:1304–1311. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Porcelli I, Reuter M, Pearson BM, Wilhelm T, van Vliet AH. 2013. Parallel evolution of genome structure and transcriptional landscape in the Epsilonproteobacteria. BMC Genomics 14:616. 10.1186/1471-2164-14-616. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dugar G, Herbig A, Förstner KU, Heidrich N, Reinhardt R, Nieselt K, Sharma CM. 2013. High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet 9:e1003495. 10.1371/journal.pgen.1003495. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Romero DA, Hasan AH, Lin YF, Kime L, Ruiz-Larrabeiti O, Urem M, Bucca G, Mamanova L, Laing EE, van Wezel GP, Smith CP, Kaberdin VR, McDowall KJ. 2014. A comparison of key aspects of gene regulation in Streptomyces coelicolor and Escherichia coli using nucleotide-resolution transcription maps produced in parallel by global and differential RNA sequencing. Mol Microbiol 94:963–987. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Thomason MK, Bischler T, Eisenbart SK, Förstner KU, Zhang A, Herbig A, Nieselt K, Sharma CM, Storz G. 2015. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J Bacteriol 197:18–28. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Seo JH, Hong JS, Kim D, Cho BK, Huang TW, Tsai SF, Palsson BO, Charusanti P. 2012. Multiple-omic data analysis of Klebsiella pneumoniae MGH 78578 reveals its transcriptional architecture and regulatory features. BMC Genomics 13:679. 10.1186/1471-2164-13-679. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kröger C, Dillon SC, Cameron ADS, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hébrard M, Händler K, Colgan A, Leekitcharoenphon P, Langridge GC, Lohan AJ, Loftus B, Lucchini S, Ussery DW, Dorman CJ, Thomson NR, Vogel J, Hinton JC. 2012. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A 109:E1277–E1286. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Alkhateeb RS, Vorhölter FJ, Rückert C, Mentz A, Wibberg D, Hublik G, Niehaus K, Pühler A. 2016. Genome wide transcription start sites analysis of Xanthomonas campestris pv. campestris B100 with insights into the gum gene cluster directing the biosynthesis of the exopolysaccharide xanthan. J Biotechnol 225:18–28. [PubMed] [DOI] [PubMed] [Google Scholar]
- 128.Sahr T, Rusniok C, Dervins-Ravault D, Sismeiro O, Coppee JY, Buchrieser C. 2012. Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biol 9:503–519. [PubMed] [DOI] [PubMed] [Google Scholar]
- 129.Venkataramanan KP, Min L, Hou S, Jones SW, Ralston MT, Lee KH, Papoutsakis ET. 2015. Complex and extensive post-transcriptional regulation revealed by integrative proteomic and transcriptomic analysis of metabolite stress response in Clostridium acetobutylicum. Biotechnol Biofuels 8:81. 10.1186/s13068-015-0260-9. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liao Y, Huang L, Wang B, Zhou F, Pan L. 2015. The global transcriptional landscape of Bacillus amyloliquefaciens XH7 and high-throughput screening of strong promoters based on RNA-seq data. Gene 571:252–262. [PubMed] [DOI] [PubMed] [Google Scholar]
- 131.Ignatov D, Malakho S, Majorov K, Skvortsov T, Apt A, Azhikina T. 2013. RNA-Seq analysis of Mycobacterium avium non-coding transcriptome. PLoS One 8:e74209. 10.1371/journal.pone.0074209. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schwientek P, Neshat A, Kalinowski J, Klein A, Rückert C, Schneiker-Bekel S, Wendler S, Stoye J, Pühler A. 2014. Improving the genome annotation of the acarbose producer Actinoplanes sp. SE50/110 by sequencing enriched 5′-ends of primary transcripts. J Biotechnol 190:85–95. [PubMed] [DOI] [PubMed] [Google Scholar]
- 133.Albersmeier A, Pfeifer-Sancar K, Rückert C, Kalinowski J. 2017. Genome-wide determination of transcription start sites reveals new insights into promoter structures in the actinomycete Corynebacterium glutamicum. J Biotechnol 257:99–109. [PubMed] [DOI] [PubMed] [Google Scholar]
- 134.Pfeifer-Sancar K, Mentz A, Rückert C, Kalinowski J. 2013. Comprehensive analysis of the Corynebacterium glutamicum transcriptome using an improved RNAseq technique. BMC Genomics 14:888. 10.1186/1471-2164-14-888. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bauer JS, Fillinger S, Förstner K, Herbig A, Jones AC, Flinspach K, Sharma C, Gross H, Nieselt K, Apel AK. 2017. dRNA-seq transcriptional profiling of the FK506 biosynthetic gene cluster in Streptomyces tsukubaensis NRRL18488 and general analysis of the transcriptome. RNA Biol 14:1617–1626. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Voigt K, Sharma CM, Mitschke J, Lambrecht SJ, Voß B, Hess WR, Steglich C. 2014. Comparative transcriptomics of two environmentally relevant cyanobacteria reveals unexpected transcriptome diversity. ISME J 8:2056–2068. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Babski J, Haas KA, Näther-Schindler D, Pfeiffer F, Förstner KU, Hammelmann M, Hilker R, Becker A, Sharma CM, Marchfelder A, Soppa J. 2016. Genome-wide identification of transcriptional start sites in the haloarchaeon Haloferax volcanii based on differential RNA-Seq (dRNA-Seq). BMC Genomics 17:629. 10.1186/s12864-016-2920-y. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Toffano-Nioche C, Ott A, Crozat E, Nguyen AN, Zytnicki M, Leclerc F, Forterre P, Bouloc P, Gautheret D. 2013. RNA at 92°C: the non-coding transcriptome of the hyperthermophilic archaeon Pyrococcus abyssi. RNA Biol 10:1211–1220. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wurtzel O, Sapra R, Chen F, Zhu Y, Simmons BA, Sorek R. 2010. A single-base resolution map of an archaeal transcriptome. Genome Res 20:133–141. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]


