ABSTRACT
Strains of Staphylococcus aureus, and to a lesser extent other staphylococcal species, are a significant cause of morbidity and mortality. An important factor in the notoriety of these organisms stems from their frequent resistance to many antimicrobial agents used for chemotherapy. This review catalogues the variety of mobile genetic elements that have been identified in staphylococci, with a primary focus on those associated with the recruitment and spread of antimicrobial resistance genes. These include plasmids, transposable elements such as insertion sequences and transposons, and integrative elements including ICE and SCC elements. In concert, these diverse entities facilitate the intra- and inter-cellular gene mobility that enables horizontal genetic exchange, and have also been found to play additional roles in modulating gene expression and genome rearrangement.
As has been the case for other bacterial genera, studies of the genetic basis of staphylococcal pathogenicity and antimicrobial resistance highlighted the presence of determinants not uniformly represented in the genomes of all staphylococcal strains. Although not a fundamental requirement for survival per se, such accessory DNA usually encodes functions that are advantageous in a particular environmental niche. The extent of the accessory component of the genome has been revealed by comparative analysis of whole Staphylococcus aureus genome sequences, which indicate that it can constitute in excess of 20% of a strain’s genetic makeup (1–5). It is clear that the acquisition, maintenance, and dissemination of accessory functions have been central to the ongoing success of staphylococci as pathogens. These processes are underpinned by interactions between mobile genetic elements, such as insertion sequences and transposons that mediate intracellular movement of DNA, and plasmids, bacteriophage and integrative and conjugative elements (ICEs) that facilitate intercellular DNA mobility.
MECHANISMS OF GENETIC EXCHANGE
DNA can be introduced into staphylococci by each of the three classical bacterial gene transfer mechanisms of transformation, transduction, and conjugation. Transformation is thought to be limited by extracellular nucleases and is usually very inefficient (6, 7). However, homologs of competence genes from other Gram-positive species are evident in the S. aureus chromosome, leading to the suggestion that natural competence might arise under suitable growth conditions and/or in subpopulations of cells (8). Staphylococci usually possess one or more prophages integrated within their genomes, and generalized transduction is likely to be a frequent mechanism of genetic exchange between strains (9, 10). Staphylococcal conjugative plasmids not only mediate their own self-transfer, but also facilitate the exchange of other coresident nonconjugative plasmids, either by mobilization if the other plasmid carries a relaxation system (11) or more generally via cointegrate formation, and potentially subsequent resolution, in a process termed “conduction” (12, 13). Conjugation is also thought to represent the most likely basis for the apparent transmission of large DNA segments that correspond to approximately 10 to 20% of the chromosome (14). In addition to their own transfer, chromosomally located ICEs in staphylococcal strains (15) might also mediate transmission of plasmid and chromosomal DNA (16). A novel but poorly understood mechanism of staphylococcal genetic exchange, termed mixed-culture transfer or phage-mediated conjugation, was described in the 1980s (6, 17, 18). Although phage, or perhaps components of phage, play a role, the process has been shown to be mechanistically distinct from transduction. More recently, another phage-mediated mechanism, termed autotransduction, has been defined (19), which like mixed-culture transfer, requires the DNA recipient to be lysogenic for specific prophage. It is possible that mixed-culture transfer and autotransduction are actually different perspectives on the same phenomenon. There have been several reports that describe enhanced DNA transfer between staphylococci, by conjugation and transduction, in response to subinhibitory levels of some antibiotics; however, in most cases the basis of this stimulation is yet to be delineated (10, 20–22). Notably, genetic exchange has also been observed to occur more efficiently in vivo than in vitro (23).
Identical or nearly identical accessory genes, elements, and plasmids have been detected in different staphylococcal species and other bacterial genera, such as enterococci and streptococci (24–28). Such observations suggest that, directly and/or indirectly, the gene transfer mechanisms operating in staphylococci facilitate not only intraspecific transfer, but also interspecific and intergeneric exchange and hence access to an extended and shared reservoir of determinants.
STAPHYLOCOCCAL PLASMIDS
One or more plasmids are usually found in clinical isolates of S. aureus and coagulase-negative staphylococci (7, 29). Historically, staphylococcal plasmids can be categorized into one of three main classes based on physical/genetic organization and functional characteristics (24, 25, 30). These correspond to small plasmids that utilize a rolling circle (RC) replication mechanism and usually carry a single resistance gene or are cryptic, larger multiresistance plasmids that often carry several resistance genes and conjugative plasmids that usually also contain multiple resistance determinants and are typically larger, again due to the conjugation-associated genes they contain; theta-mode replication is used by multiresistance and conjugative plasmids. However, categorization based on resistance genes is parochial and does not necessarily reflect relatedness between plasmids, since resistance determinants can be readily lost from, or acquired by, plasmid backbones. For example, there are plasmids closely related to multiresistance and conjugative plasmids that lack any recognized resistance genes. Toxins are also encoded by some plasmids (29, 31–34), and the functions of many genes carried by larger plasmids have not been investigated, let alone elucidated.
Analysis of fully sequenced S. aureus plasmids in the databases has illustrated that plasmid size correlates strongly with the mode of replication used and hence with the replication initiation gene possessed. Plasmids smaller than 10 kb use the RC mechanism, whereas those larger than 14.5 kb utilize theta-mode replication, with plasmids in the intermediate 10 to 14.5-kb size range using either of these mechanisms (35). Fifteen plasmid incompatibility groups (Inc1 to Inc15) were established for staphylococcal plasmids, incorporating both RC replicating (RCR) and theta replicating plasmids (24, 36). However, the rate of plasmid discovery through genome sequencing has rendered incompatibility testing impractical. A plasmid typing system for plasmids from Gram-positive bacteria (including staphylococci) based on replication region DNA sequence has been established (37) and incorporated into the PlasmidFinder database (38). Unfortunately, coincident identification of additional replicon types (39, 40) has subsequently resulted in discordant classification of some staphylococcal plasmids.
Small RCR Plasmids
The most thoroughly characterized staphylococcal plasmids are those that utilize asymmetric RC replication via a single-stranded DNA intermediate (for detailed reviews of RC plasmid replication, see Khan [41] and Ruiz-Maso et al. [44]). Probably reflecting constraints imposed by this replication strategy, RCR (sometimes alternatively called just RC or single-stranded DNA) plasmids are usually less than 5 kb in size and are rarely found to contain transposable elements (24). This restriction in RCR plasmid size probably reflects elevated production of high-molecular-weight DNA with increasing plasmid size, which impairs cell growth, leading to counterselection against host cells and hence the plasmid itself (42). RCR plasmids are frequently found cointegrated within larger theta replicating plasmids, commonly flanked by insertion sequences such as IS257; in such cases the replication system of the integrated RCR plasmid is often inactivated in some way (28, 43).
Staphylococcal RCR plasmids can be subdivided into four families, exemplified by pT181, pC194, pE194, and pSN2, based on replication region sequence similarity (24); representative plasmids are shown in Fig. 1. These families contain plasmids found in different staphylococcal species and are related to plasmids of other bacterial genera, such as Bacillus, Lactobacillus, and Streptococcus, attesting to the horizontal transmission of RCR plasmids (42). Plasmids belonging to each of these four families encode evolutionarily distinct replication initiator (Rep) proteins that possess the conserved domains Rep_trans, Rep_1, Rep_2, and RepL, respectively (35). However, despite the Rep proteins having unique evolutionary origins, they perform a remarkably similar role in recognizing, binding, and nicking specific DNA sequences in the double-strand origin (dso). In the RCR plasmids examined so far, the Rep proteins also perform cleavage and ligation reactions at the termination of leading strand synthesis (44). pT181 family Rep proteins recruit the host-encoded helicase, PcrA, which is essential for synthesis of the new, and displacement of the parental, leading strands (45, 46). The leading strand contains a single-strand origin of replication (sso) that, upon displacement, is recognized and primed for replication by host-encoded RNA polymerase (47). The inability of host RNA polymerase to recognize and prime the sso limits the host range of RCR plasmids. For example, the ssoA carried by pT181 only functions efficiently in staphylococci, whereas the ssoU carried by pE194 family plasmids allows stable replication in a broader range of Gram-positive hosts (48).
FIGURE 1.
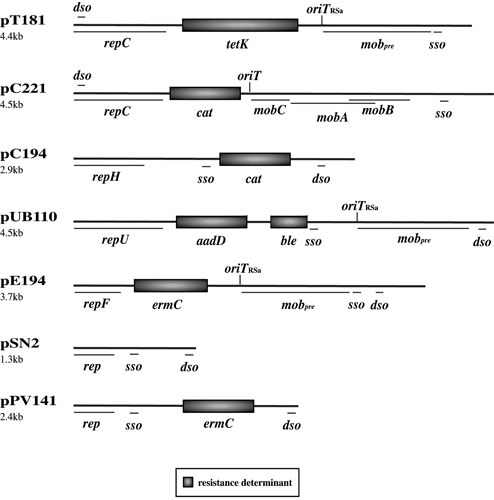
Maps of representative RCR plasmids: pT181, pC221, pC194, pUB110, pE194, pSN2, and pPV141 (25, 231); see text for additional references. Plasmid sizes are shown on the left. Resistance and plasmid maintenance genes/loci are shown: rep/repC/repF/repH/repU, initiation of replication; dso, double-stranded origin of DNA replication; sso, single-stranded origin of DNA replication; mobA/mobB/mobC/mobpre, mobilization; oriT/oriTRSa, origin of DNA transfer; cat, chloramphenicol resistance. Refer to Tables 1 and 2 for the antimicrobial resistance(s) conferred by other resistance determinants.
RCR plasmids are usually maintained at 10 to 60 copies per cell (7), and copy number control is achieved via an antisense RNA that is transcribed divergently to the replication initiation gene (35). In the case of pT181, binding of the antisense RNA causes transcriptional attenuation of repC mRNA (49), whereas for pC194 family plasmids antisense binding is believed to block translation of the Rep protein (50, 51). Antisense inhibition of Rep translation is also thought to occur in pE194 family plasmids, but transcriptional regulation mediated by a repressor protein may also play a role (35), as has been shown for the streptococcal RCR plasmid pMV158 (52, 53).
Some staphylococcal RCR plasmids contain mobilization systems that facilitate their horizontal transmission when coresident with a conjugative plasmid. Plasmids such as pC221 possess a multigene mobilization system (mobCAB and origin of transfer [oriT]) (Fig. 1) (11). The pC221 MobA relaxase and MobC relaxase accessory proteins are both required for nicking at oriT (54, 55). Other RCR plasmids possess a different mobilization locus that is homologous to the mobM relaxase gene and oriT of pMV158 (56, 57). This locus was originally identified as a site-specific recombination function on staphylococcal plasmids such as pT181 and pE194 (Fig. 1) (24), where the constituent sequences were named pre and RSA (corresponding to mobpre and oriTRSa, respectively) (Fig. 1). Additionally, the replication initiation protein and dso of several RCR plasmids have been shown to “moonlight” as a relaxase and oriT for mobilization mediated by the ICE Tn916 (see below) (58).
RCR plasmids, including those in different families, often share highly similar DNA segments, such that they appear to be mosaic structures consisting of discrete functional cassettes encoding replication, resistance, and sometimes recombination/mobilization functions (24, 59). Exchange of these segments between plasmids is thought to be promoted by the single-stranded intermediates generated by the RC replication strategy, which enhance the frequency of homologous recombination events between regions of flanking sequence similarity (60). DNA processing at replication and transfer origins also promotes the swapping of such cassettes (24, 61).
Theta Replicating Plasmids
Most staphylococcal plasmids larger than 10 kb employ theta-mode replication and are maintained at low copy numbers (7, 35, 62–65). The majority encode a replication initiation protein containing an N-terminal conserved RepA_N domain (29, 35, 66), which corresponds to a winged helix-turn-helix DNA binding domain that binds to origin Rep box repeat sequences characteristically located centrally within the rep gene coding sequence (67–69); the C-terminal domain of the Rep protein from the S. aureus conjugative plasmid pSK41 (see below) has been shown to interact with the host encoded DnaG primase (70). An interesting variation is seen in the plasmid pTZ2162, where the RepA_N-type initiation protein has been split into two smaller distinct polypeptides, expressed from two partially overlapping rep genes that have likely arisen from a frameshift (71). In staphylococcal plasmids encoding RepA_N type initiators, copy number is thought to be controlled by a small antisense RNA that binds to the leader region of the rep mRNA to block its translation (68, 72, 73).
Two groups of nonconjugative multiresistance plasmids were described previously in staphylococci: the β-lactamase/heavy-metal resistance plasmids and the pSK1 family plasmids; both utilize RepA_N initiation proteins (74). S. aureus strains from the 1960s and 1970s commonly contained plasmids that conferred resistance to β-lactam antibiotics and heavy metals or other inorganic ions. These plasmids characteristically carried a Tn552-like β-lactamase-encoding transposon or a derivative thereof (75, 76) and frequently possessed a composite structure designated Tn4004 conferring resistance to mercuric ions and operons encoding resistance to arsenical and/or cadmium ions (Fig. 2) (6, 77). Additionally, some β-lactamase/heavy-metal resistance plasmids carry the transposon Tn4001 or Tn551 conferring resistance to aminoglycosides (6, 78) and macrolide-lincosamide-streptogramin type B (MLS) antibiotics (79), respectively, and/or a qacA or qacB gene mediating multidrug resistance to antiseptics and disinfectants (Fig. 2) (6).
FIGURE 2.
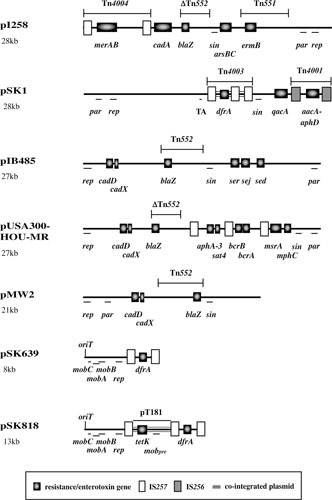
Maps of representative nonconjugative theta-replicating plasmids: pI258, pSK1, pIB485, pUSA300-HOU-MR, pMW2, pSK639, and pSK818 (3, 25, 29, 65, 85, 89, 232); see text for additional references. Resistance/enterotoxin genes, transposons, insertion sequences, and cointegrated plasmids are shown: arsBC, arsenic resistance; cadA, cadmium resistance; mphC, macrolide resistance; msrA, macrolide/streptogramin B resistance; qacA, antiseptic/disinfectant resistance; ser, sej, and sed, enterotoxins. Refer to Tables 1 and 2 for the antimicrobial resistance(s) conferred by other resistance determinants. Plasmid maintenance genes/loci are also shown: par, novel partitioning system; rep, initiation of replication; sin, multimer resolution; TA, Fst-like toxin-antitoxin system; mobA/mobB/mobC/mobpre, mobilization.
Plasmids related to the prototype, pSK1 (Fig. 2), were first identified in epidemic S. aureus and coagulase-negative staphylococcal strains isolated in Australia during the 1980s and later in isolates from Europe (25). In addition to a qacA gene mediating multidrug resistance to antiseptics/disinfectants carried ubiquitously by pSK1 family plasmids (80, 81), members of this family variously encode Tn4001, which confers resistance to gentamicin and other aminoglycosides (82), a Tn552-like β-lactamase transposon, Tn4002 (83), and/or a composite structure designated Tn4003 that mediates trimethoprim resistance (84). Tn4003 represents a vestige of a cointegrated pSK639-like plasmid (see below (26, 85).
As more plasmid sequences have accumulated through genome sequencing it has become clear that there is a broad diversity of plasmids with a repA_N-type initiation gene, including plasmids lacking resistance genes and/or containing toxin genes, such as pIB485 (Fig. 2) and related plasmids that have been isolated from strains of wide geographic distribution (29). Two other loci are nearly ubiquitous on RepA_N-encoding plasmids, suggesting that they are important for plasmid survival (29). The first is a multimer resolution system that includes a gene coding for a serine recombinase, which is usually annotated as sin or bin3 (86, 87). The second is homologous to the pSK1 par (formerly orf245) gene, which is usually located upstream of, and transcribed divergently from, the rep gene. This locus increases plasmid segregational stability and is thought to constitute a novel single-protein partitioning system (64, 88). In contrast, type I and II partitioning systems, comprising two protein-encoding genes, are found only occasionally on these plasmids (29). Finally, an Fst-like type I toxin-antitoxin system, which might contribute to plasmid segregational stability, is carried by pSK1 (89), and related loci have been identified on numerous other theta replicating staphylococcal plasmids (90, 91).
A minority of theta replicating staphylococcal plasmids code for a Rep protein containing a Rep_3 conserved domain (35); initiation proteins of this type from Gram-negative bacteria, encoded by iteron plasmids such as F, pPS10, and R6K, contain tandem winged helix-turn-helix domains (92). This type of replication system was first detected in the genus in plasmids from Staphylococcus epidermidis related to the 8-kb trimethoprim resistance plasmid pSK639 (Fig. 2), which also contains a mobilization system related to that of the RCR plasmid pC221 (65). More recently, rep_3-type genes have been found in addition to the usual repA_N-type gene on larger β-lactamase/heavy-metal resistance plasmids such as pN315 (1), whereas in the case of pMW2 (3) (Fig. 2), only a remnant of the repA_N gene remains. Finally, plasmids employing an initiation protein containing a PriCT_1 conserved domain, such as pETB, which encodes exfoliative toxin B (32), have also been found in staphylococci but appear to be quite rare; some also encode a RepA_N or Rep_3 type initiator (35).
Conjugative Plasmids
The largest staphylococcal plasmids (>30 kb) are those that encode their own conjugative transfer (7, 25). Three distinct families of conjugative plasmids have been identified in staphylococci: the well-known pSK41/pGO1-like plasmids and the recently characterized pWBG4 and pWBG749 families.
S. aureus plasmids such as pSK41 (43, 93, 94) (Fig. 3), pGO1 (95, 96), and pJE1 (20) were first identified in association with gentamicin resistance in the mid 1970s (13, 25). Structurally related conjugative plasmids were found in coagulase-negative staphylococci (97, 98), and interspecific transfer has been demonstrated (99, 100). In pSK41 and many relatives, the plasmid backbone consists of two IS257-flanked segments, termed the transfer (tra) region and region I (Fig. 3) (94). However, in other family members (e.g., pPR9) these segments are contiguous, and this likely represents the ancestral configuration prior to the incorporation of additional DNA segments (43, 101, 102).
FIGURE 3.
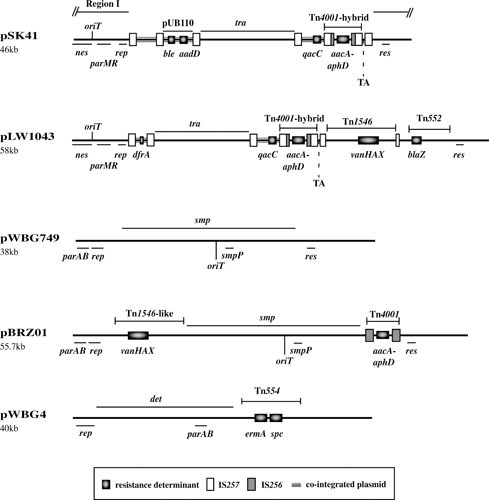
Maps of representative conjugative theta-replicating plasmids: pSK41, pLW1043, pWBG749, pBRZ01, and pWBG4 (27, 43, 128, 136, 137); see text for additional references. Resistance genes, transposons, insertion sequences, and cointegrated plasmids are shown: refer to Tables 1 and 2 for the antimicrobial resistance(s) conferred by the resistance determinants. Plasmid maintenance genes/loci (if known) are also shown: parAB, type I partitioning system; parMR, type II partitioning system; rep, initiation of replication; res, multimer resolution; TA, Fst-like toxin-antitoxin system; tra/smp/det genes, conjugative transfer; nes/smpP genes, relaxases; oriT, origin of transfer. Note that pLW1043 and pBRZ01 are members of the pSK41 and pWBG749 families, respectively.
Multiple copies of IS257, mostly in the same orientation, are a feature of many pSK41-like plasmids (Fig. 3). These usually delimit segments that carry resistance genes and often correspond to cointegrated copies of smaller plasmids (43, 103). Such determinants include the aminoglycoside and bleomycin resistance genes aadD and ble, respectively, on a copy of pUB110 and the small multidrug resistance determinant qacC on another RCR plasmid (Fig. 3). Plasmids such as pJE1 and pGO1 also contain a Tn4003-like structure derived from a pSK639-like trimethoprim-resistance plasmid (see above). The aacA-aphD gene encoded by Tn4001-IS257 hybrid structures is responsible for resistance to gentamicin and other aminoglycosides (104). Members of the family have also been found that contain IS257-flanked genes encoding resistance to tetracycline (tetK) (29), MLS antibiotics (ermC) (105), mupirocin (mupA/ileS2) (102, 106), and most recently, multidrug resistance to phenicols, lincosamides, oxazolidinones (e.g., linezolid), pleuromutilins, and streptogramin A compounds (PhLOPSA phenotype; cfr) (107). Plasmids of this type can also carry unit transposons, such as Tn552 (blaZ) (43) and the vanA glycopeptide resistance transposon Tn1546 (27). The pSK41-like plasmid pLW1043 (Fig. 3) was responsible for the first reported high-level vancomycin-resistant S. aureus strain, isolated in Michigan in 2002 (27), and resulted from the transposition of Tn1546 into a copy of IS257 within the pSK41-like plasmid pAM829 (27, 108). A Tn1546-containing Inc18 broad-host-range conjugative plasmid, pAM830, carried by a coisolated Enterococcus faecalis strain, is thought to have mediated the intergeneric transfer event (27, 109). Such enterococcal Inc18 vanA plasmids themselves have subsequently been detected in some vancomycin-resistant S. aureus strains, but their scarcity in S. aureus suggests some limitation on their establishment in this species, possibly inefficient replication, an incompatibility with staphylococcal cell wall biosynthesis, a restriction barrier, and/or high metabolic burden associated with vanA carriage (108). Cointegrates composed of Inc18 and pSK41-like plasmids are also thought to occur (110), and it has been suggested that carriage of pSK41-like plasmids might enhance the recipient ability of S. aureus cells for enterococcal donors containing Inc18 plasmids, but the mechanism has not been determined (111).
Conjugative transfer of pSK41 family plasmids occurs only on solid surfaces and does so at comparatively low frequencies, in the range of 10−5 to 10−7 transconjugants per donor cell (13). The majority of genes thought to be responsible for conjugative transfer are located in the tra region (Fig. 3) (93, 95). The tra genes are most similar to the transfer genes of the Lactococcus lactis plasmid pMRC01 (112) and the broad host range plasmid pIP501 (43), originally identified in Streptococcus agalactiae, exhibiting both synteny and product amino acid sequence similarity (43, 113, 114). Several deduced products show homology to proteins from the type IV secretion system superfamily, including to coupling proteins, peptidoglycan hydrolases, and several other core mating pair formation-type proteins (115, 116). Thus, the basic conjugative process is likely to resemble that of more extensively characterized Gram-negative counterparts, viz., transfer of a single strand of plasmid DNA via direct cell-cell contact. However, no pilus-like structure seems to be associated with Gram-positive conjugation (114), perhaps reflecting the distinction in the cell envelope organizations and accounting for the solid surface requirement for conjugative transfer. The origin of transfer, oriT, and the relaxase, Nes, that acts upon it, are encoded in region I (117, 118), as are a number of genes of unknown function that might also play a role in conjugative transfer (20, 43, 113).
As mentioned above, pSK41-like conjugative plasmids encode a RepA_N type replication initiation protein, whose translation is regulated by an antisense RNA called RNAI (67, 70, 72, 73). However, several members of the family, such as pPR9 and pUSA03, possess a novel chimeric rep gene where the sequences that usually encode the N-terminal RepA_N DNA-binding domain, and the origin Rep box repeats to which it binds, appear to have been replaced by sequences encoding a helix-turn-helix protein sequence related to phage replication proteins and distinct DNA sequence repeats (35, 102). The usual upstream antisense RNA control region and downstream in-frame end of the rep gene, encoding the conserved C-terminal end of the Rep protein that interacts with host DnaG primase (70), remain intact in these plasmids, but the variation is sufficient to allow compatibility with the pSK41 replication system (102).
All known pSK41 family plasmids encode a serine recombinase and a type II active partitioning system, the latter representing a significant difference from repA_N replicating nonconjugative staphylococcal plasmids, which usually contain an unusual par gene or occasionally a type I partitioning system (see above). The pSK41 res locus is functional, facilitating the conversion of plasmid multimers into monomers, with the Res protein shown to bind to a resolution site, comprising three subsites, located in the intergenic region upstream of the res gene; Res binding also autoregulates transcription of its own promoter (119). Functionality of the pSK41 partitioning system has also been demonstrated (120). It comprises a centromere-like site, parC, and two genes in an operon, parM and parR. The ParM protein is an actin-like motor protein that forms single-stranded helical filaments (121). ParC is a ribbon-helix-helix protein that binds to DNA repeat sequences within parC (120) and interacts with ParM to modulate its activity (122). ParR also autoregulates transcription from the partitioning operon promoter, which is also controlled by the plasmid-encoded ArtA protein (see below). Some pSK41-like plasmids encode an additional potential plasmid maintenance determinant in the form of an Fst-like toxin-antitoxin system, which is located on a small IS257-flanked DNA segment adjacent to Tn4001-hybrid elements in plasmids conferring gentamicin resistance (94).
In pSK41, transcription of the majority (21 out of 30) of the plasmid backbone genes is controlled by the product of the conserved artA gene (corresponding to trsN on pGO1) (101, 123). The ArtA protein is a ribbon-helix-helix DNA-binding protein that represses five promoters in addition to its own—including two operon promoters in the tra region and two more operon promoters in region 1 (Fig. 3). ArtA is therefore thought to coordinate basal-level transcription of most of the plasmid’s backbone genes (101).
The pWBG4 family of conjugative plasmids, also called diffusible pigment (DiP) plasmids, were first identified as a distinct family of S. aureus conjugative plasmids in the 1980s by the Grubb laboratory (124–127), but the complete sequence of pWBG4 (Fig. 3) was only reported in 2016 (128). The first example of this family, pWBG14, was isolated in Western Australia in 1968 and conferred resistance to MLS antibiotics (ermA) and spectinomycin (spc) and production of a diffusible orange pigment. Related plasmids were subsequently identified carrying resistance to aminoglycosides and trimethoprim (dfrD), and more recently several pWBG4-related plasmids were implicated in the conjugative transfer of linezolid and phenicol resistance (cfr and fexA) (128, 129) in America, China, and Germany (128, 129). pWBG4 family plasmids carry an ∼18-kb conjugation-gene cluster detA-detV which is distinct from that of pSK41/pGO1 family plasmids and uniquely includes a MobC-like conjugative relaxase (DetB) and T4CP (DetA) related to those of the mobilizable Enterobacter cloacae plasmid CloDF13 (128, 129). pWBG4 plasmids also encode a PriCT_1-domain replication-initiation protein common to enterococcal Inc18 plasmids (35). pWBG4 family plasmids have been isolated from both S. aureus and S. epidermidis, and transfer between and within these species has been demonstrated (125).
The pWBG749 family was also only recently sequenced and identified as being a distinct family of S. aureus conjugative plasmids. pWBG749 (Fig. 3) was originally isolated in 1995 (29, 125, 130, 131) from colonizing S. aureus present in remote indigenous communities in Western Australia, but more recent sequencing has identified variants isolated as early as 1985 (36, 128, 132–135). pWBG749 encodes a RepA_N initiation protein, possesses a type IB partitioning system (29), and carries an ∼23-kb conjugation gene cluster smpA-X (136) (Fig. 3). While most identified pWBG749 family plasmids lack antimicrobial-resistance determinants, members carrying resistance to penicillin (blaZ), aminoglycosides (aacA-aphD), and vancomycin (vanA) have been isolated (130–133, 137). Despite their frequent cryptic nature, pWBG749 family plasmids likely have an extensive impact on antimicrobial resistance dissemination through their so-far unique ability to horizontally mobilize large multiresistance plasmids in addition to smaller plasmids (131).
The mechanism by which pWBG749 mobilizes nonconjugative plasmids was only recently recognized as a naturally occurring phenomenon, potentially explaining why it was previously overlooked (128, 136). Most characterized plasmid mobilization systems described require a mobilizable plasmid to carry its own relaxase gene (138). Plasmids capable of being mobilized by pWBG749 need only carry a short “mimic” of the pWBG749 origin-of-transfer sequence (136). The conjugative relaxase of pWBG749, SmpP, along with an oriT specificity factor, SmpO, acts in trans on the oriT to pilot it to the mating apparatus. Approximately 50% of unique nonconjugative staphylococcal plasmids carry a pWBG749 family oriT mimic (136). Most common mimics are near-identical to those on pWBG749 or the closely related conjugative plasmid pWBG745 and its vancomycin resistance-encoding derivative pBRZ01 (Fig. 3) (137). Bioinformatic inspection of 397 sequenced clinical S. aureus isolates from the MRSA Orange County initiative of the Broad Institute (broadinstitute.org) identified just 24 contigs matching pWBG749 or pWBG745, but astonishingly, 736 oriT mimics were identified among the sequence contigs. This abundance is likely explained by the presence of up to three oriT mimic variants per plasmid on common large multidrug resistance plasmids such as pIB485, pMW2, and pUSA300-HOU-MR (Fig. 2) and on small RCR plasmids (128, 136, 138).
Analogous to pWBG749-like oriT mimics, multiple mimic types that resemble the pSK41 oriT sequence were also detected recently on numerous staphylococcal plasmids (139). Although pSK41 could not mobilize the variants tested, it would seem likely that other pSK41 family plasmids encoding relaxation systems with appropriate specificity might also mediate relaxase-in trans mobilization. Recent bioinformatic analyses have revealed that nearly all theta replicating S. aureus plasmids carry at least one oriT mimic sequence, with many possessing both pWBG749 and pSK41 types, suggesting that the capacity for conjugative mobilization via the relaxase in trans mechanism is widespread (138).
TRANSPOSABLE ELEMENTS
Insertion sequences and composite transposons detected in staphylococci are listed in Table 1, and unit transposons are presented in Table 2. For many, designation as a transposable element is based on possession of diagnostic characteristics, such as terminal inverted repeats, flanking insertion sequences and/or target duplications, presence of an open reading frame encoding a transposase homolog, and/or detection in varied genetic contexts, rather than formal demonstration of mobility in a recA recombination-defective host. As outlined for specific elements below, in addition to facilitating the translocation of genes between replicons, insertion sequence elements are increasingly being shown to play more subtle roles in phenotypic expression and genome evolution.
TABLE 1.
Insertion sequences and composite transposonsa
| ISb | Tnc | Associated resistance(s)/phenotype(s) | Determinant(s) | TDd (bp) |
|---|---|---|---|---|
| IS256 | Tn4001e | Gentamicin/kanamycin/tobramycin | aacA-aphD | 8 |
| Phenicols/lincosamides/oxazolidinones/pleuromutilins/streptogramin A | cfr | |||
| IS257f | Kanamycin/neomycin/paromomycin/tobramycin | aadD | 8 | |
| Kanamycin/neomycin | aphA-3 | |||
| Bacitracin | bcrAB | |||
| Bleomycin | ble | |||
| Cadmium | cadDX | |||
| Trimethoprim | dfrK | |||
| MLS | ermC | |||
| Fosfomycin | fosB5 | |||
| Fusidic acid | fusB | |||
| Mupirocin | ileS2 (mupA) | |||
| Lysostaphin immunity factor | lif | |||
| Preprolysostaphin | lss | |||
| Antiseptics/disinfectants | qacC | |||
| Streptothricin | sat4 | |||
| Tetracycline | tetK | |||
| Tetracycline | tetL | |||
| Streptogramin A | vat(A) | |||
| Streptogramin A/pleuromutilins/lincosamides | vga(A) | |||
| Streptogramin B | vgb(A) | |||
| Tn4003 | Trimethoprim | dfrA | ||
| Tn4004 | Mercury | merAB | ||
| Tn6072 | Gentamicin/kanamycin/tobramycin | aacA-aphD | ||
| Tn6072 | Spectinomycin | spc | ||
| IS1181 | 8 | |||
| IS1182 | Tn5405g | Streptomycin | aadE | 8 |
| Tn5405 | Kanamycin/neomycin | aphA-3 | ||
| Tn5405 | Streptothricin | sat4 | ||
| IS1272 | TnSha1 | Triclosan | fabI | Absent |
| TnSha2 | Triclosan | fabI | ||
| IS1293 | Lysostaphin immunity factor | lif | Unknown | |
| Preprolysostaphin | lss | |||
| IS21-558f | Phenicols/lincosamides/oxazolidinones/pleuromutilins/streptogramin A | cfr | 6 | |
| Lincosamides | lsa(B) | |||
| ISEnfa4 | Phenicols/lincosamides/oxazolidinones/pleuromutilins/streptogramin A | cfr | 8 | |
| ISSau10 | Kanamycin/neomycin/paromomycin/tobramycin | aadD | 8 | |
| Trimethoprim | dfrK | |||
| MLS | ermC | |||
| MLS | ermT | |||
| Tetracycline | tetL | |||
| ISSha1 | 8 |
See Paulsen et al. (25), Firth and Skurray (26), Fu et al. (237), Allignet and El Solh (238), Chen et al. (239), Kadlec and Schwarz (240), Highlander et al. (232), O’Neill and Chopra (241), de Vries et al. (242), and the text for references. Additional elements detected in genome sequences but not described here (e.g., ISSau1 through ISSau8 and ISSau11) are listed in ISfinder (http://www-is.biotoul.fr).
In the case of IS, association is based on probable involvement in the acquisition and dissemination of the resistance.
Associated composite transposons are shown.
TD, target duplication.
Tn3851 and Tn4031 are likely to be similar or identical to Tn4001.
IS257 is also known as IS431, and IS21-558 is also known as ISSau9.
Tn3854 may be similar to Tn5405.
TABLE 2.
Unit transposonsa
| Transposon | Family | Associated resistance(s) | Determinant(s) | TDb (bp) |
|---|---|---|---|---|
| Tn551 | Tn3 | MLS | ermB | 5 |
| Tn1546 | Vancomycin/teicoplanin | vanHAX | 5 | |
| Tn552c | Tn7 | Penicillins | blaZ | 6/7 |
| Tn5404 | Streptomycin | aadE | 6 | |
| Kanamycin/neomycin | aphA-3 | |||
| Streptothricin | sat4 | |||
| Tn554d | Tn554 | MLS | ermA | Absent |
| Spectinomycin | spc | |||
| ψTn554 | Cadmium | cadA | Absent | |
| Tn558 | Chloramphenicol/florfenicol | fexA | Absent | |
| Tn559 | Trimethoprim | dfrK | Absent | |
| Tn5406 | Streptogramin A/pleuromutilins/lincosamides | vga(A)v | Absent | |
| Tn6133 | MLS | ermA | Absent | |
| Spectinomycin | spc | |||
| Streptogramin A/pleuromutilins/lincosamides | vga(E) |
Inverted copies of IS256 flank the composite transposon Tn4001 (Figs. 2 and 3; Table 1) (82). Tn4001 and elements related to it are responsible for the emergence of linked resistance to the aminoglycosides, gentamicin, tobramycin, and kanamycin in staphylococci, having been found on the chromosome and theta replicating plasmids (Figs. 2 and 3) in clinical S. aureus and coagulase-negative staphylococcal isolates from around the world (6, 25). Indeed, some of the earliest reported gentamicin-resistant strains, dating from 1975, have been shown to carry a Tn4001-like structure on the chromosome (140). Resistance is mediated by the gene aacA-aphD, which encodes a bifunctional enzyme possessing both aminoglycoside acetyltransferase and phosphotransferase activities (75). Transposons closely related to Tn4001 are also evident in enterococci (141) and streptococci (142). In enterococci, IS256 and related insertion sequences are also associated with elements encoding resistance to vancomycin (143) and erythromycin (144). IS256 also appears to be contributing to the spread of the cfr multiresistance gene, forming a composite-like structure with a copy of the related enterococcal element ISEnfa4 (145, 146).
Roles in gene expression and regulation have been attributed to IS256 via both transcriptional promotion and insertional inactivation. Transcription of the aacA-aphD resistance gene of Tn4001 is directed by a complete promoter located within the end of the upstream copy of IS256, and it has also been found that a small flanking deletion can result in a −35-like sequence at the terminus of that element forming part of a hybrid promoter which mediates a higher level of aminoglycoside resistance (147). Additionally, roles for IS256 hybrid promoters have been demonstrated in the expression of methicillin resistance (148, 149). Insertional inactivation of genes by IS256 has been shown to modulate expression of several phenotypes, including virulence and glycopeptide resistance. Alternating IS256 insertions and excisions in the ica operon responsible for the synthesis of the polysaccharide intercellular adhesin mediate phase-variable biofilm formation in S. epidermidis strains responsible for indwelling device infections (150). Notably, biofilm phase variation has also been attributed to IS256 insertions that affect global regulatory circuits, including the sarA global regulator, the quorum-sensing agr system, and rsbU which positively regulates the σB stress response factor (151, 152). IS256 inactivation of the tcaA locus was found to increase the teicoplanin resistance of a glycopeptide intermediate-resistant S. aureus strain (153), and IS256 insertion into the promoter region of the walKR two-component regulator system similarly resulted in decreased glycopeptide susceptibility (154, 155). Insertion into the promoter repressor of toxins gene, rot, was also found to enhance the virulence of a USA500 strain (156).
IS256 utilizes a “copy-out-paste-in” transposition mechanism via an excised circular double-stranded DNA intermediate (157–159). Its transposase contains an N-terminal helix-turn-helix domain that mediates binding to the element’s terminal inverted repeats (160). The capacity of IS256 to mediate phase variability has been linked to a capacity for precise excision of the element and the target duplication generated during insertion (150). Interestingly, this excision behavior occurs independently of the element’s transposase and is instead thought to involve host-mediated illegitimate recombination acting on the flanking target duplications (161).
IS257 (also known as IS431) has been found in diverse genetic contexts in staphylococci, variously associated with a number of determinants (Table 1). The prevalence of resistance genes flanked by copies of this element resulted in the designation of several such composite structures as transposons, viz., Tn4003 and Tn4004. Although the possibility that these structures can behave as traditional composite transposons cannot be excluded, such an organization is probably a consequence of the transposition mechanism of IS257. This insertion sequence element is thought to undergo nonresolved replicative transposition (26, 103). The expected outcome of such an event involving two replicons is cointegration, so that the replicons become fused with a directly repeated copy of IS257 at each junction, as has been experimentally demonstrated by Needham et al. (162). The IS257-like element ISSau10 is also found in such configurations (163, 164). Many IS257-flanked segments represent plasmids cointegrated into larger plasmids (Figs. 2 and 3) or the chromosome (26, 103). Frequently observed deletion events adjacent to IS257 may result from intramolecular transposition of IS257. However, replicon fusions and sequence deletions have also been shown to have resulted from homologous recombination between pre-existing copies of IS257 (43).
IS257 is a member of the IS6 family of insertion sequences (159). Another member of this family, IS26, which is found flanking resistance genes with increasing prevalence in Gram-negative bacteria, has recently been shown to undergo a second “conservative” mode of transpositional movement (165, 166); the process was IS26 transposase dependent and recA independent in Escherichia coli. It involves the insertion of a single copy of the element and a flanking DNA fragment (often containing a resistance gene), termed a translocatable unit (TU), adjacent to an existing copy of IS26 in another replicon. The resulting composite structure can be equivalent to an IS26-mediated cointegration event into an “IS26-less” replicon, but importantly, movement of an IS26 TU was found to occur at a much higher frequency than cointegrate formation mediated by transposition of just an IS26 copy. As a consequence, the presence of IS26 in a replicon predisposes it to the acquisition of IS26 TUs. The capacity of IS257 to promote movement via comparable TUs is yet to be investigated, but the similar properties of these elements in their distinct hosts, particularly their prevalence in gene arrays punctuated by directly repeated insertion sequence copies, suggests it is a distinct possibility (165, 166).
The activities of IS257 are illustrated by the pSK41 family plasmids (Fig. 3). It would seem that IS257-mediated cointegration has enabled these conjugative plasmids to collect functions, in the form of smaller plasmids, as they move horizontally through bacterial populations. Insertions and flanking deletions mediated by this element provide a mechanism for the inactivation or removal of deleterious sequences, such as redundant replication function.
Interestingly, short (148-149 bp) elements, named ISLE39 and ISLE49, that lack any transposase coding sequence but possess terminal inverted repeats similar to those of IS257, have been identified in several staphylococcal species (167). These presumably represent non-autonomous miniature inverted repeat transposable elements (MITEs) that could be rendered mobile by transposase encoded by IS257 copies present in the same cell.
A central role for IS257 has been suggested in the evolution of staphylococcal trimethoprim resistance, including the probable capture of the resistance gene, dfrA, from the chromosome of an S. epidermidis strain (26, 103). Furthermore, an IS257-hybrid promoter is responsible for transcription of this determinant (103, 168). In some plasmids, the high-level trimethoprim resistance typically conferred by this determinant has been moderated as a consequence of IS257-associated flanking deletions. An analogous IS257-hybrid promoter transcribes the tetA(K) tetracycline resistance gene of a cointegrated copy of pT181 within type III SCCmec elements (see below), and several other resistance genes are likely to be similarly promoted (102, 169).
One or more copies of IS257 are carried by most types of staphylococcal cassette chromosome mec (SCCmec) elements (where it is commonly denoted IS431; see below). As well as flanking various resistance determinants within these elements, in several SCCmec types an IS257 copy has mediated deletion of the mecI and part of the mecR1 regulatory genes, resulting in derepressed transcription of the mecA methicillin resistance gene (170). Equivalent mecI-mecR1 deletions are associated with another insertion sequence in some other SCCmec types, which instead contain a remnant of IS1272, ψIS1272 (see below) (171–173). Multiple intact copies of IS1272 have been found on the chromosomes of S. aureus and coagulase-negative staphylococcal strains, and it is particularly prevalent in Staphylococcus haemolyticus (4, 172, 174). IS1272 is also associated with the elements TnSha1 and TnSha2, which encode the fabI determinant responsible for resistance to the biocide triclosan (175); these elements are believed to target palindromic sequences and not generate target duplications during transposition.
IS21-558 (also called ISSau9) and derivatives thereof have been linked with the cfr multiresistance gene in a range of contexts and hosts (145). Its name reflects the fact that it is a member of the IS21 family of insertion sequence elements and that it was initially detected within a variant of Tn558 (176), which itself carries a fexA determinant that mediates chloramphenicol and florfenicol efflux (177). Within the Tn558 variant, two directly repeated IS21-558 copies formed a composite configuration flanking both cfr and the lsa(B) lincosamide resistance gene (176).
In addition to the high-level glycopeptide resistance VanA element Tn1546 of enterococcal origin (27) described above, one other Tn3-type transposon, Tn551, has been detected in staphylococci. This MLS (ermB) transposon is closely related to Tn917 from E. faecalis but confers constitutive rather than inducible MLS resistance (7). Tn551 was originally identified on the β-lactamase/heavy-metal resistance plasmid, pI258 (Fig. 2) but has subsequently been shown to transpose to numerous sites in other large plasmids and the chromosome. Despite a tendency to “hot-spot,” it has served as a valuable tool for mapping and mutagenesis studies (7, 178).
Tn552 and Tn5404 are Tn7 family unit transposons. Tn552 appears to be restricted to a very limited set of insertion sites on the chromosome and in plasmids (25). In theta replicating plasmids (Figs. 2 and 3), the insertion sequence sites of Tn552 and Tn552-like elements are normally located within resolution sites upstream of resolvase (recombinase) genes encoded by the plasmids (43, 86, 87, 179). An invertible segment can be formed between resolution sites that are present in both the plasmid and transposon (180). In some ICE6013 elements (see below) Tn552 is also found proximal to the probable recombinase genes (181). This property is shared with Tn5035-like elements from Gram-negative bacteria, which are termed “res site hunters” (182). Despite such insertional specificity, Tn552-type transposons are thought to represent the source of all staphylococcal β-lactamase genes (76). However, in many plasmids, only remnants of such transposons remain, presumably as a consequence of multiple insertion events and/or rearrangements mediated by the recombinase systems present on both the transposon and plasmids.
Tn5404 is thought to have resulted from the transposition of a chromosomal element to a plasmid in a clinical S. aureus strain (183). This transposon shares an invertible segment (between resolution sites) with an adjacent copy of Tn552, such that an entire copy of one or the other of these elements is generated depending on the orientation of the segment (183). Another invertible segment within Tn5404 represents a composite structure designated Tn5405 (183), which encodes resistance to aminoglycosides (aadE, aphA) and streptothricin (sat4) (184). Tn5405 is bounded by copies of IS1182, an element related to IS1272 (see above) (185). Moreover, one of the IS1182 elements flanking Tn5405 is interrupted by a copy of IS1181, which is both prevalent and active in S. aureus (1, 185, 186).
An incomplete vestige of an element related to IS1181 has been detected on the plasmid pACK1 from Staphylococcus simulans biovar staphylolyticus ATCC1362. This element, designated IS1293, is located at one end of a segment that contains the genes encoding preprolysostaphin (lss) and lysostaphin immunity factor (lif) (187). It is likely that the lss-lif segment of this plasmid, which possesses a truncated remnant of IS257 at its other end, has been acquired through horizontal transfer (187). Numerous copies of another IS1181-like element, ISSha1, have been found in S. haemolyticus strains (188).
The MLS- and spectinomycin-resistance element Tn554 is unusual in that it lacks terminal inverted repeats and generates no target sequence duplications upon insertion (189). It inserts at an efficiency approaching 100% into a unique site in the S. aureus and S. epidermidis chromosomes, termed att554 (189), within the radC gene (190), with a preference for one orientation. Secondary insertions can be generated in the chromosome or plasmids, albeit at much lower frequency, if att554 is absent or occupied by a pre-existing copy of Tn554. However, natural isolates have been identified that carry this or related elements, such as ψTn554 encoding cadmium resistance, at different secondary sites, including some located in SCCmec regions (see below) (1, 179). Insertion of Tn554 is dependent on two tyrosine-based site-specific recombinases encoded by the tnpA and tnpB genes and the product of a third, tnpC, that influences efficiency and orientation specificity (191, 192). The recombinases of Tn554 are related to integrases of bacteriophage and ICEs such as Tn916 (see below). The Tn554-like elements, Tn558 (177), Tn559 (190), Tn5406 (193), and Tn6133 (194) exhibit equivalent insertion site specificity but encode additional or alternative resistance determinants.
INTEGRATIVE AND CONJUGATIVE ELEMENTS
ICEs are mobile genetic elements that share characteristics of both conjugative plasmids and chromosomally integrating elements such as transposons and prophage. ICEs primarily exist within the host chromosome but can excise, circularize, and transfer via conjugation. ICEs encode all genes necessary for mating-pair formation and DNA transfer. The broad impact of ICEs on an organism’s capacity for horizontal gene transfer and dissemination of antimicrobial resistance and virulence factors is significant. However, perhaps due to the often-changing classification of ICEs as genomic islands, transposons, or conjugative transposons and the evolving differences in naming conventions (195, 196), the roles of ICEs in S. aureus gene transfer are not yet as widely recognized as those of their plasmid and bacteriophage counterparts.
Identified S. aureus ICEs can be grouped into two families: those identical or closely related to the Tn916 ICE and those related to the distinct ICE6013 family (16). The conjugation gene clusters of Tn916 and ICE6013 family elements both carry conjugation genes related to those of Bacillus subtilis ICEBs1 (Fig. 4) and more distantly, plasmids such as Clostridium perfringens pCW3 and Enterococcus spp. pIP501 (197). S. aureus Tn916 and ICE6013 family elements vary in their integration mechanism, insertion site preference, and antimicrobial-resistance gene cargo.
FIGURE 4.
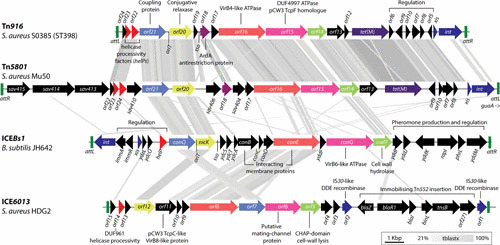
Genetic organization of Tn916, Tn5801, ICEBs1, and ICE6013. A comparison of Tn916, Tn5801, ICEBs1, and ICE6013 constructed using EasyFig (233). Amino acid similarity comparisons (20% lower cutoff) using tblastx and are shown as gray shading between regions on each ICE. Genes with likely common functions—regardless of sequence similarity—are similarly colored. Positions of the conjugative origin of transfer (oriT) and single-strand origin of replication (sso) are indicated if known. Recombinase attachment/target sites are indicated by green rectangles. Annotations were collated from previously published figures for each ICE (15, 181, 197, 204, 206, 234, 235).
Tn916 elements are extremely promiscuous in Gram-positive organisms, and those identified in S. aureus are often identical to those in other Gram-positive genera. Tn916 transfer from E. faecalis to S. aureus and transfer between S. aureus strains has been experimentally demonstrated (15, 198). The 18-kb Tn916 element carries genes for conjugation, integration, excision, and the tetracycline-minocycline-resistance gene tetM. Tn916 integration is mediated by the tyrosine recombinase Int, and excision is stimulated by Int and the excisionase Xis. Int preferentially targets pairs of poly-A and poly-T tracts separated by a 6-bp spacer for Tn916 insertion (199). Tn916 excision is stimulated in the presence of tetracycline and other ribosome-targeting antibiotics through an antiattenuation mechanism resulting in the derepression of xis; therefore, antibiotic exposure directly drives the spread of Tn916-mediated resistance (200–202).
Various distinct Tn916-like elements have been described in S. aureus, but the most well-documented example is the Tn5801 subfamily. Tn5801 (Fig. 4) was originally identified in S. aureus strain Mu50 (1) and carries conjugation and resistance genes (tetM) closely related to those of Tn916 in both sequence and organization. Tn5801 is larger (25 kb) than Tn916 and harbors several Tn5801-specific genes within the conjugation-gene cluster. int and xis genes of Tn5801 are distinct from those of Tn916, as is the integration preference for the 3′ end of the conserved GMP-synthase gene, guaA. Tn5801 transfer between S. aureus strains has also been demonstrated (15). Like Tn916 elements, Tn5801 elements have been identified in human and animal-associated S. aureus and are particularly prevalent in S. aureus clonal complex CC398 (15, 203, 204). Both elements are also abundant in other animal-associated Staphylococcus spp. (204, 205), suggesting that agricultural and companion animals may be reservoirs of Tn916 and Tn5801-carrying staphylococci.
ICE6013 is the most recently identified family of S. aureus ICEs, and although it is also related to Tn916 and Tn5801, it is more closely related to B. subtillis ICEBs1 than any other S. aureus element (206) (Fig. 4). Unlike Tn916 and Tn5801, ICE6013 family members generally lack antimicrobial-resistance determinants, although the first discovered ICE6013 element in S. aureus HDG2 carries a Tn552 insertion conferring penicillin resistance, and the insertion has immobilized this copy of ICE6013 (181, 206) (Fig. 4). As well as being one of the smallest known families of ICE, with a minimum size of 12.7 kb, ICE6013 is the only known ICE to utilize a DDE family transposase for its integration and excision and has an insertion preference for the sequence 5′-TTTTTANNNTAAAAA-3′ (181). ICE6013 was originally identified in the ST239 S. aureus strain HDG2, and related elements have been identified in 44% of distinct S. aureus sequence types, including the dominant American community-associated lineage USA300. Conjugation frequencies of ICE6013 approach rates similar to that of the conjugative plasmid pSK41/pG01. Some strains harbor various combinations of up to four unique ICE6013, Tn916, and Tn5801 family ICEs simultaneously (23, 181). The Tn916/Tn5801 orf18 gene encodes the ArdA antirestriction protein (Fig. 4), which inhibits type I restriction modification systems ubiquitous in S. aureus, so capture of Tn916/Tn5801 ICE may increase the propensity for horizontal acquisition of additional mobile elements (207–209).
In addition to their own conjugative transfer, ICEs can mobilize the transfer of compatible nonconjugative plasmids or other integrative elements (16, 210). ICEBs1 is able to mobilize the transfer of RCR plasmids encoding RepU replication initiators through an interaction dependent on RepU and the ICEBs1 T4CP, ConQ (58, 197). ICE6013, Tn916, and Tn5801-family ICEs share closely related ConQ homologs (Fig. 4), so it is plausible that S. aureus ICEs may facilitate mobilization of RCR plasmids. Indeed, ICEBs1- and Tn916-mediated mobilization of S. aureus antimicrobial-resistance plasmids pC194 and pUB110 has already been demonstrated, albeit in a B. subtilis host (58, 211). The impact of S. aureus ICEs on the capacity to facilitate horizontal gene transfer of other mobile genetic elements warrants further investigation (16, 138).
STAPHYLOCOCCAL CASSETTE CHROMOSOME
Most methicillin-resistant S. aureus isolates contain a 20- to 70-kb DNA segment, generically termed the mec region, that is not present in methicillin-sensitive strains. These segments are a type of genomic island (i.e., are bounded by terminal inverted repeats and subject to horizontal transfer) called staphylococcal cassette chromosome mec (SCCmec) (179). Methicillin resistance is mediated, at least in part, by the mecA gene or the more recently identified mecC gene (212, 213), both of which encode a related low-affinity penicillin-binding protein, termed PBP2a. Two regulatory loci, mecI and mecR1, are commonly found upstream of, and transcribed divergently to, mecA/mecC (however, see discussion of IS1272/IS257, above). In combination, the mecA/mecC, mecI, and mecR1 genes and associated insertion sequence(s) are referred to as the mec gene complex, of which several classes have been defined (A, B, C1, C2, D, and E) on the basis of structural diversity (http://www.sccmec.org).
Additionally, SCCmec contains the ccr gene complex, which comprises either a combination of ccrA and ccrB genes (one of each) or a single ccrC gene; different allotypes for these genes also exist, and several types of the ccr gene complex have been defined (http://www.sccmec.org) (214). Each ccr gene encodes a recombinase that mediates the excision and circularization of the cassette and its site- and orientation-specific insertion into the attBSCC site of the S. aureus chromosome and homologous sites in other staphylococcal species (179). Recently, a conserved gene located upstream of the ccr gene(s) has been shown to encode a DNA helicase (215) referred to as cch or cch2, and the latter is also preceded by a putative primase (polA). This implies that SCC elements might be able to replicate (postexcision), and multiple copies would likely facilitate the horizontal transfer process. Moreover, a gene located downstream of the ccr gene(s) encodes a uracil-DNA glycosylase inhibitor (SAUGI). S. aureus pathogenicity islands, which are specifically induced to excise and be packaged by invading bacteriophage, carry homologs of cch, and regulate their excision in response to phage-induced changes in uracil metabolism. These commonalities suggest that phage may also have a role in stimulating and/or facilitating SCCmec transfer (128, 215).
At present, SCCmec has been classified into 12 allotypes based on the organization of the mec and ccr gene complexes (Fig. 5) (216); subtypes also exist and are defined by the structure of the three joining (J) regions (previously called junkyard regions) located between these gene complexes and the chromosomal regions flanking SCCmec. The J regions also act as a chromosomal hot-spot for the insertion of additional antimicrobial resistance determinants, often in association with transposable elements (173). These include Tn554 or related elements encoding resistance to erythromycin and spectinomycin, or cadmium, and IS257-flanked segments conferring resistance to mercury, tetracycline, and/or aminoglycosides and bleomycin; the latter two segments are known to be cointegrated copies of the plasmids pT181 and pUB110, respectively (Fig. 5) (26, 103). Based on this classification system, an online tool, SCCmecFinder, has recently been developed for rapid whole-genome sequence-based SCCmec typing (217).
FIGURE 5.
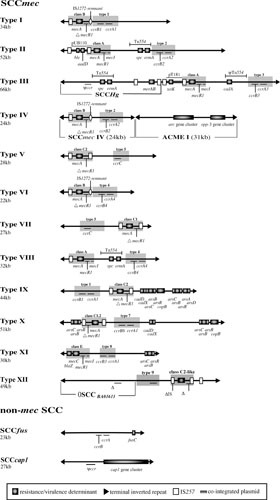
Maps of representative SCC elements (4, 216); see text for additional references. Resistance/virulence genes, transposons, insertion sequences, and cointegrated plasmids are shown: arsBC, arsenic resistance; cadA, cadmium resistance; cap1 genes, capsular polysaccharide (220); fusC (previously known as far), fusidic acid resistance (236); mecA/mecC, β-lactam resistance. Refer to Table 1 for the antimicrobial resistance(s) conferred by other resistance determinants. Cassette recombinase genes (ccrA, ccrB, and ccrC), mecA/mecC regulatory genes (mecI and mecR1), and an arginine catabolic mobile element (ACME I [227]) are also shown; mec classes and ccr types are denoted by gray shading. Note that cch genes, polA, and SAUGI are not shown and that IS257 is also known as IS431.
It is important to note that SCCmec elements are not exclusive to S. aureus and are frequently carried by coagulase-negative staphylococci. Moreover, the ubiquitous carriage of a mecA homolog by Staphylococcus sciuri has led to the suggestion that this or closely related species represent the origin of the mec determinants found in other species (218). In this regard, a recent study by Rolo et al. (219) indicates that Staphylococcus vitulinus and Staphylococcus fleurettii contributed to the evolution of the mec complex and that S. sciuri likely provided the mobile element into which this complex was later incorporated. In any case, it is clear that coagulase-negative staphylococci act as a reservoir from which methicillin-sensitive S. aureus can acquire SCCmec, contributing to the emergence of new methicillin-resistant S. aureus clones.
However, not all SCC elements encode resistance to methicillin, because related structures have also been identified (179, 220, 221). These include SCCcap1, which encodes a type I capsular polysaccharide (220), and SCCfur (also known as SCC476) and SCCHg (also known as SCCmercury), which encode resistance to fusidic acid and mercuric chloride, respectively (4, 222) (Fig. 5). Additionally, the arginine catabolic mobile element, which is broadly associated with certain coagulase-negative staphylococci (223–225), has also been identified in association with SCCmec allotypes, largely type IV (226). This element is a virulence/colonization determinant that utilizes SCCmec-encoded cassette recombinases and integrates into the attBSCC site (223, 227, 228) (Fig. 5). Arginine catabolic mobile element allotypes are defined by the presence or absence of the opp-3 and arc gene clusters, the latter of which encode a complete arginine deiminase pathway, and carriage of this element has been shown to enhance fitness and skin colonization (228–230).
PERSPECTIVES
Mobile genetic elements and the mechanisms of genetic exchange they facilitate/exploit act in concert to catalyze rapid microbial adaptation by providing access to an extended reservoir of niche-adaptive functions. Clinical staphylococci represent a salient illustration of the evolutionary potential this affords. Genomic technologies have greatly expanded our understanding of variability between strains (clinical and nonclinical) and the relationships between them and revealed a growing repertoire of mobile genetic elements, thereby illustrating the significance of horizontal genetic exchange. There is little doubt that the ongoing acceleration of genomics will yield further insights in the future.
ACKNOWLEDGMENTS
Our research on staphylococcal genetics is supported by National Health and Medical Research Council (Australia) project grants APP1081412 (to N.F., S.O.J., and S.M.K.) and APP1145697 (to N.F. and J.P.R.).
REFERENCES
- 1.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240 10.1016/S0140-6736(00)04403-2. [PubMed] [DOI] [PubMed] [Google Scholar]
- 2.Fitzgerald JR, Sturdevant DE, Mackie SM, Gill SR, Musser JM. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc Natl Acad Sci U S A 98:8821–8826 10.1073/pnas.161098098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827 10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 4.Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, Barron A, Bason N, Bentley SD, Chillingworth C, Chillingworth T, Churcher C, Clark L, Corton C, Cronin A, Doggett J, Dowd L, Feltwell T, Hance Z, Harris B, Hauser H, Holroyd S, Jagels K, James KD, Lennard N, Line A, Mayes R, Moule S, Mungall K, Ormond D, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Sharp S, Simmonds M, Stevens K, Whitehead S, Barrell BG, Spratt BG, Parkhill J. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A 101:9786–9791 10.1073/pnas.0402521101. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindsay JA, Holden MT. 2004. Staphylococcus aureus: superbug, super genome? Trends Microbiol 12:378–385 10.1016/j.tim.2004.06.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 6.Lyon BR, Skurray R. 1987. Antimicrobial resistance of Staphylococcus aureus: genetic basis. Microbiol Rev 51:88–134. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novick RP. 1990. The Staphylococcus as a molecular genetic system, p 1–37. In Novick RP (ed), Molecular Biology of the Staphylococci. VCH, New York, NY. [PubMed] [Google Scholar]
- 8.Morikawa K, Takemura AJ, Inose Y, Tsai M, Nguyen Thi T, Ohta T, Msadek T. 2012. Expression of a cryptic secondary sigma factor gene unveils natural competence for DNA transformation in Staphylococcus aureus. PLoS Pathog 8:e1003003 10.1371/journal.ppat.1003003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindsay JA. 2014. Staphylococcus aureus genomics and the impact of horizontal gene transfer. Int J Med Microbiol 304:103–109 10.1016/j.ijmm.2013.11.010. [PubMed] [PubMed] [DOI] [PubMed] [Google Scholar]
- 10.Stanczak-Mrozek KI, Laing KG, Lindsay JA. 2017. Resistance gene transfer: induction of transducing phage by sub-inhibitory concentrations of antimicrobials is not correlated to induction of lytic phage. J Antimicrob Chemother 72:1624–1631 10.1093/jac/dkx056. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Projan SJ, Archer GL. 1989. Mobilization of the relaxable Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J Bacteriol 171:1841–1845 10.1128/jb.171.4.1841-1845.1989. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Archer GL, Thomas WD Jr. 1990. Conjugative transfer of antimicrobial resistance genes between staphylococci, p 115–122. In Novick RP (ed), Molecular Biology of the Staphylococci. VCH, New York, NY. [Google Scholar]
- 13.Macrina FL, Archer GL. 1993. Conjugation and broad host range plasmids in streptococci and staphylococci, p 313–329. In Clewell DB (ed), Bacterial Conjugation. Plenum Press, New York, NY. 10.1007/978-1-4757-9357-4_12 [DOI] [Google Scholar]
- 14.Robinson DA, Enright MC. 2004. Evolution of Staphylococcus aureus by large chromosomal replacements. J Bacteriol 186:1060–1064 10.1128/JB.186.4.1060-1064.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Vries LE, Christensen H, Skov RL, Aarestrup FM, Agersø Y. 2009. Diversity of the tetracycline resistance gene tet(M) and identification of Tn916- and Tn5801-like (Tn6014) transposons in Staphylococcus aureus from humans and animals. J Antimicrob Chemother 64:490–500 10.1093/jac/dkp214. [PubMed] [DOI] [PubMed] [Google Scholar]
- 16.Sansevere EA, Robinson DA. 2017. Staphylococci on ICE: overlooked agents of horizontal gene transfer. Mob Genet Elements 7:1–10 10.1080/2159256X.2017.1368433. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birmingham VA, Pattee PA. 1981. Genetic transformation in Staphylococcus aureus: isolation and characterization of a competence-conferring factor from bacteriophage 80 alpha lysates. J Bacteriol 148:301–307. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacey RW. 1980. Evidence for two mechanisms of plasmid transfer in mixed cultures of Staphylococcus aureus. J Gen Microbiol 119:423–435. [DOI] [PubMed] [Google Scholar]
- 19.Haaber J, Leisner JJ, Cohn MT, Catalan-Moreno A, Nielsen JB, Westh H, Penadés JR, Ingmer H. 2016. Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat Commun 7:13333 10.1038/ncomms13333. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans J, Dyke KG. 1988. Characterization of the conjugation system associated with the Staphylococcus aureus plasmid pJE1. J Gen Microbiol 134:1–8. [DOI] [PubMed] [Google Scholar]
- 21.al-Masaudi SB, Day MJ, Russell AD. 1991. Effect of some antibiotics and biocides on plasmid transfer in Staphylococcus aureus. J Appl Bacteriol 71:239–243 10.1111/j.1365-2672.1991.tb04454.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 22.Barr V, Barr K, Millar MR, Lacey RW. 1986. Beta-lactam antibiotics increase the frequency of plasmid transfer in Staphylococcus aureus. J Antimicrob Chemother 17:409–413 10.1093/jac/17.4.409. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.McCarthy AJ, Loeffler A, Witney AA, Gould KA, Lloyd DH, Lindsay JA. 2014. Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol Evol 6:2697–2708 10.1093/gbe/evu214. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novick RP. 1989. Staphylococcal plasmids and their replication. Annu Rev Microbiol 43:537–565 10.1146/annurev.mi.43.100189.002541. [PubMed] [DOI] [PubMed] [Google Scholar]
- 25.Paulsen IT, Firth N, Skurray RA. 1997. Resistance to antimicrobial agents other than β-lactams, p 175–212. In Crossley KB, Archer GL (ed), The Staphylococci in Human Disease. Churchill Livingstone, London, UK. [Google Scholar]
- 26.Firth N, Skurray RA. 1998. Mobile elements in the evolution and spread of multiple-drug resistance in staphylococci. Drug Resist Updat 1:49–58 10.1016/S1368-7646(98)80214-8. [DOI] [PubMed] [Google Scholar]
- 27.Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569–1571 10.1126/science.1090956. [PubMed] [DOI] [PubMed] [Google Scholar]
- 28.Schwarz S, Shen J, Wendlandt S, Fessler AT, Wang Y, Kadlec K, Wu CM. 2014. Plasmid-mediated antimicrobial resistance in staphylococci and other firmicutes. Microbiol Spectr 2:PLAS-0020-2014. 10.1128/microbiolspec.PLAS-0020-2014. [DOI] [PubMed] [Google Scholar]
- 29.Shearer JE, Wireman J, Hostetler J, Forberger H, Borman J, Gill J, Sanchez S, Mankin A, Lamarre J, Lindsay JA, Bayles K, Nicholson A, O’Brien F, Jensen SO, Firth N, Skurray RA, Summers AO. 2011. Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. G3 (Bethesda) 1:581–591 10.1534/g3.111.000760. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malachowa N, DeLeo FR. 2010. Mobile genetic elements of Staphylococcus aureus. Cell Mol Life Sci 67:3057–3071 10.1007/s00018-010-0389-4. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S, Iandolo JJ, Stewart GC. 1998. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol Lett 168:227–233 10.1111/j.1574-6968.1998.tb13278.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi T, Hayashi T, Takami H, Ohnishi M, Murata T, Nakayama K, Asakawa K, Ohara M, Komatsuzawa H, Sugai M. 2001. Complete nucleotide sequence of a Staphylococcus aureus exfoliative toxin B plasmid and identification of a novel ADP-ribosyltransferase, EDIN-C. Infect Immun 69:7760–7771 10.1128/IAI.69.12.7760-7771.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayles KW, Iandolo JJ. 1989. Genetic and molecular analyses of the gene encoding staphylococcal enterotoxin D. J Bacteriol 171:4799–4806 10.1128/jb.171.9.4799-4806.1989. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omoe K, Hu DL, Takahashi-Omoe H, Nakane A, Shinagawa K. 2003. Identification and characterization of a new staphylococcal enterotoxin-related putative toxin encoded by two kinds of plasmids. Infect Immun 71:6088–6094 10.1128/IAI.71.10.6088-6094.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwong SM, Ramsay JP, Jensen SO, Firth N. 2017. Replication of staphylococcal resistance plasmids. Front Microbiol 8:2279 10.3389/fmicb.2017.02279. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Udo EE, Grubb WB. 1991. A new incompatibility group plasmid in Staphylococcus aureus. FEMS Microbiol Lett 62:33–36 10.1111/j.1574-6968.1991.tb04412.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 37.Jensen LB, Garcia-Migura L, Valenzuela AJ, Løhr M, Hasman H, Aarestrup FM. 2010. A classification system for plasmids from enterococci and other Gram-positive bacteria. J Microbiol Methods 80:25–43 10.1016/j.mimet.2009.10.012. [PubMed] [DOI] [PubMed] [Google Scholar]
- 38.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903 10.1128/AAC.02412-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lozano C, García-Migura L, Aspiroz C, Zarazaga M, Torres C, Aarestrup FM. 2012. Expansion of a plasmid classification system for Gram-positive bacteria and determination of the diversity of plasmids in Staphylococcus aureus strains of human, animal, and food origins. Appl Environ Microbiol 78:5948–5955 10.1128/AEM.00870-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCarthy AJ, Lindsay JA. 2012. The distribution of plasmids that carry virulence and resistance genes in Staphylococcus aureus is lineage associated. BMC Microbiol 12:104 10.1186/1471-2180-12-104. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan SA. 1997. Rolling-circle replication of bacterial plasmids. Microbiol Mol Biol Rev 61:442–455. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Helinski DR, Toukdarian AE, Novick RP. 1996. Replication control and other stable maintenance mechanisms of plasmids, p 2295–2324. In Neidhardt FC, Curtiss R, Ingraham JL, Lin ECC, Low KB Jr, Magasanik B, Reznikoff W, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC. [Google Scholar]
- 43.Berg T, Firth N, Apisiridej S, Hettiaratchi A, Leelaporn A, Skurray RA. 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J Bacteriol 180:4350–4359. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruiz-Maso JA, Macho NC, Bordanaba-Ruiseco L, Espinosa M, Coll M, Del Solar G. 2015. Plasmid rolling-circle replication. Microbiol Spectr 3:PLAS-0035-2014. 10.1128/microbiolspec.PLAS-0035-2014. [DOI] [PubMed] [Google Scholar]
- 45.Soultanas P, Dillingham MS, Papadopoulos F, Phillips SE, Thomas CD, Wigley DB. 1999. Plasmid replication initiator protein RepD increases the processivity of PcrA DNA helicase. Nucleic Acids Res 27:1421–1428 10.1093/nar/27.6.1421. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang TL, Naqvi A, Anand SP, Kramer MG, Munshi R, Khan SA. 2002. Biochemical characterization of the Staphylococcus aureus PcrA helicase and its role in plasmid rolling circle replication. J Biol Chem 277:45880–45886 10.1074/jbc.M207383200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 47.Kramer MG, Khan SA, Espinosa M. 1997. Plasmid rolling circle replication: identification of the RNA polymerase-directed primer RNA and requirement for DNA polymerase I for lagging strand synthesis. EMBO J 16:5784–5795 10.1093/emboj/16.18.5784. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kramer MG, Espinosa M, Misra TK, Khan SA. 1999. Characterization of a single-strand origin, ssoU, required for broad host range replication of rolling-circle plasmids. Mol Microbiol 33:466–475 10.1046/j.1365-2958.1999.01471.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 49.Novick RP, Iordanescu S, Projan SJ, Kornblum J, Edelman I. 1989. pT181 plasmid replication is regulated by a countertranscript-driven transcriptional attenuator. Cell 59:395–404 10.1016/0092-8674(89)90300-0. [DOI] [PubMed] [Google Scholar]
- 50.Alonso JC, Tailor RH. 1987. Initiation of plasmid pC194 replication and its control in Bacillus subtilis. Mol Gen Genet 210:476–484 10.1007/BF00327200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 51.Maciag IE, Viret JF, Alonso JC. 1988. Replication and incompatibility properties of plasmid pUB110 in Bacillus subtilis. Mol Gen Genet 212:232–240 10.1007/BF00334690. [PubMed] [DOI] [PubMed] [Google Scholar]
- 52.del Solar G, Espinosa M. 2000. Plasmid copy number control: an ever-growing story. Mol Microbiol 37:492–500 10.1046/j.1365-2958.2000.02005.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 53.López-Aguilar C, Romero-López C, Espinosa M, Berzal-Herranz A, Del Solar G. 2015. The 5′-tail of antisense RNAII of pMV158 plays a critical role in binding to the target mRNA and in translation inhibition of repB. Front Genet 6:225 10.3389/fgene.2015.00225. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caryl JA, Smith MC, Thomas CD. 2004. Reconstitution of a staphylococcal plasmid-protein relaxation complex in vitro. J Bacteriol 186:3374–3383 10.1128/JB.186.11.3374-3383.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caryl JA, Thomas CD. 2006. Investigating the basis of substrate recognition in the pC221 relaxosome. Mol Microbiol 60:1302–1318 10.1111/j.1365-2958.2006.05188.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grohmann E, Zechner EL, Espinosa M. 1997. Determination of specific DNA strand discontinuities with nucleotide resolution in exponentionally growing bacteria harboring rolling circle-replicating plasmids. FEMS Microbiol Lett 152:363–369 10.1111/j.1574-6968.1997.tb10453.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 57.Guzmán LM, Espinosa M. 1997. The mobilization protein, MobM, of the streptococcal plasmid pMV158 specifically cleaves supercoiled DNA at the plasmid oriT. J Mol Biol 266:688–702 10.1006/jmbi.1996.0824. [PubMed] [DOI] [PubMed] [Google Scholar]
- 58.Lee CA, Thomas J, Grossman AD. 2012. The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. J Bacteriol 194:3165–3172 10.1128/JB.00301-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Projan SJ, Novick R. 1988. Comparative analysis of five related staphylococcal plasmids. Plasmid 19:203–221 10.1016/0147-619X(88)90039-X. [DOI] [PubMed] [Google Scholar]
- 60.Gruss A, Ehrlich SD. 1989. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev 53:231–241. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wassenaar TM, Ussery DW, Ingmer H. 2016. The qacC gene has recently spread between rolling circle plasmids of Staphylococcus, indicative of a novel gene transfer mechanism. Front Microbiol 7:1528 10.3389/fmicb.2016.01528. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sheehy RJ, Novick RP. 1975. Studies on plasmid replication. V Replicative intermediates. J Mol Biol 93:237–253 10.1016/0022-2836(75)90130-8. [DOI] [PubMed] [Google Scholar]
- 63.Gering M, Götz F, Brückner R. 1996. Sequence and analysis of the replication region of the Staphylococcus xylosus plasmid pSX267. Gene 182:117–122 10.1016/S0378-1119(96)00526-4. [DOI] [PubMed] [Google Scholar]
- 64.Firth N, Apisiridej S, Berg T, O’Rourke BA, Curnock S, Dyke KGH, Skurray RA. 2000. Replication of staphylococcal multiresistance plasmids. J Bacteriol 182:2170–2178 10.1128/JB.182.8.2170-2178.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Apisiridej S, Leelaporn A, Scaramuzzi CD, Skurray RA, Firth N. 1997. Molecular analysis of a mobilizable theta-mode trimethoprim resistance plasmid from coagulase-negative staphylococci. Plasmid 38:13–24 10.1006/plas.1997.1292. [PubMed] [DOI] [PubMed] [Google Scholar]
- 66.Weaver KE, Kwong SM, Firth N, Francia MV. 2009. The RepA_N replicons of Gram-positive bacteria: a family of broadly distributed but narrow host range plasmids. Plasmid 61:94–109 10.1016/j.plasmid.2008.11.004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kwong SM, Skurray RA, Firth N. 2004. Staphylococcus aureus multiresistance plasmid pSK41: analysis of the replication region, initiator protein binding and antisense RNA regulation. Mol Microbiol 51:497–509 10.1046/j.1365-2958.2003.03843.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 68.Kwong SM, Lim R, Lebard RJ, Skurray RA, Firth N. 2008. Analysis of the pSK1 replicon, a prototype from the staphylococcal multiresistance plasmid family. Microbiology 154:3084–3094 10.1099/mic.0.2008/017418-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 69.Liu MA, Kwong SM, Pon CK, Skurray RA, Firth N. 2012. Genetic requirements for replication initiation of the staphylococcal multiresistance plasmid pSK41. Microbiology 158:1456–1467 10.1099/mic.0.057620-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 70.Schumacher MA, Tonthat NK, Kwong SM, Chinnam NB, Liu MA, Skurray RA, Firth N. 2014. Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein. Proc Natl Acad Sci U S A 111:9121–9126 10.1073/pnas.1406065111. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nakaminami H, Noguchi N, Nishijima S, Kurokawa I, Sasatsu M. 2008. Characterization of the pTZ2162 encoding multidrug efflux gene qacB from Staphylococcus aureus. Plasmid 60:108–117 10.1016/j.plasmid.2008.04.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 72.Kwong SM, Skurray RA, Firth N. 2006. Replication control of staphylococcal multiresistance plasmid pSK41: an antisense RNA mediates dual-level regulation of Rep expression. J Bacteriol 188:4404–4412 10.1128/JB.00030-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kwong SM, Firth N. 2015. Structural and sequence requirements for the antisense RNA regulating replication of staphylococcal multiresistance plasmid pSK41. Plasmid 78:17–25 10.1016/j.plasmid.2015.01.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 74.Firth N, Skurray RA. 2006. Genetics: accessory elements and genetic exchange, p 413–426. In Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI (ed), Gram-Positive Pathogens. ASM Press, Washington, DC. 10.1128/9781555816513.ch33 [DOI] [Google Scholar]
- 75.Rowland SJ, Dyke KGH. 1990. Tn552, a novel transposable element from Staphylococcus aureus. Mol Microbiol 4:961–975 10.1111/j.1365-2958.1990.tb00669.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 76.Dyke K, Gregory P. 1997. Resistance mediated by β-lactamase, p 139–157. In Crossley KB, Archer GL (ed), The Staphylococci in Human Disease. Churchill Livingstone, London, UK. [Google Scholar]
- 77.Shalita Z, Murphy E, Novick RP. 1980. Penicillinase plasmids of Staphylococcus aureus: structural and evolutionary relationships. Plasmid 3:291–311 10.1016/0147-619X(80)90042-6. [DOI] [PubMed] [Google Scholar]
- 78.Gillespie MT, Skurray RA. 1986. Plasmids in multiresistant Staphylococcus aureus. Microbiol Sci 3:53–58. [PubMed] [PubMed] [Google Scholar]
- 79.Novick RP, Edelman I, Schwesinger MD, Gruss AD, Swanson EC, Pattee PA. 1979. Genetic translocation in Staphylococcus aureus. Proc Natl Acad Sci U S A 76:400–404 10.1073/pnas.76.1.400. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tennent JM, Lyon BR, Midgley M, Jones IG, Purewal AS, Skurray RA. 1989. Physical and biochemical characterization of the qacA gene encoding antiseptic and disinfectant resistance in Staphylococcus aureus. J Gen Microbiol 135:1–10. [DOI] [PubMed] [Google Scholar]
- 81.Rouch DA, Cram DS, DiBerardino D, Littlejohn TG, Skurray RA. 1990. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol Microbiol 4:2051–2062 10.1111/j.1365-2958.1990.tb00565.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 82.Rouch DA, Byrne ME, Kong YC, Skurray RA. 1987. The aacA-aphD gentamicin and kanamycin resistance determinant of Tn4001 from Staphylococcus aureus: expression and nucleotide sequence analysis. J Gen Microbiol 133:3039–3052. [DOI] [PubMed] [Google Scholar]
- 83.Gillespie MT, Lyon BR, Skurray RA. 1988. Structural and evolutionary relationships of β-lactamase transposons from Staphylococcus aureus. J Gen Microbiol 134:2857–2866. [DOI] [PubMed] [Google Scholar]
- 84.Rouch DA, Messerotti LJ, Loo LS, Jackson CA, Skurray RA. 1989. Trimethoprim resistance transposon Tn4003 from Staphylococcus aureus encodes genes for a dihydrofolate reductase and thymidylate synthetase flanked by three copies of IS257. Mol Microbiol 3:161–175 10.1111/j.1365-2958.1989.tb01805.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 85.Leelaporn A, Firth N, Paulsen IT, Skurray RA. 1996. IS257-mediated cointegration in the evolution of a family of staphylococcal trimethoprim resistance plasmids. J Bacteriol 178:6070–6073 10.1128/jb.178.20.6070-6073.1996. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paulsen IT, Gillespie MT, Littlejohn TG, Hanvivatvong O, Rowland SJ, Dyke KG, Skurray RA. 1994. Characterisation of sin, a potential recombinase-encoding gene from Staphylococcus aureus. Gene 141:109–114 10.1016/0378-1119(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 87.Rowland S-J, Stark WM, Boocock MR. 2002. Sin recombinase from Staphylococcus aureus: synaptic complex architecture and transposon targeting. Mol Microbiol 44:607–619 10.1046/j.1365-2958.2002.02897.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 88.Simpson AE, Skurray RA, Firth N. 2003. A single gene on the staphylococcal multiresistance plasmid pSK1 encodes a novel partitioning system. J Bacteriol 185:2143–2152 10.1128/JB.185.7.2143-2152.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jensen SO, Apisiridej S, Kwong SM, Yang YH, Skurray RA, Firth N. 2010. Analysis of the prototypical Staphylococcus aureus multiresistance plasmid pSK1. Plasmid 64:135–142 10.1016/j.plasmid.2010.06.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.Kwong SM, Jensen SO, Firth N. 2010. Prevalence of Fst-like toxin-antitoxin systems. Microbiology 156:975–977, discussion 977 10.1099/mic.0.038323-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 91.Weaver KE, Reddy SG, Brinkman CL, Patel S, Bayles KW, Endres JL. 2009. Identification and characterization of a family of toxin-antitoxin systems related to the Enterococcus faecalis plasmid pAD1 par addiction module. Microbiology 155:2930–2940 10.1099/mic.0.030932-0. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Konieczny I, Bury K, Wawrzycka A, Wegrzyn K. 2014. Iteron plasmids. Microbiol Spectr 2:PLAS-0026-2014. 10.1128/microbiolspec.PLAS-0026-2014. [DOI] [PubMed] [Google Scholar]
- 93.Firth N, Ridgway KP, Byrne ME, Fink PD, Johnson L, Paulsen IT, Skurray RA. 1993. Analysis of a transfer region from the staphylococcal conjugative plasmid pSK41. Gene 136:13–25 10.1016/0378-1119(93)90442-6. [DOI] [PubMed] [Google Scholar]
- 94.Liu MA, Kwong SM, Jensen SO, Brzoska AJ, Firth N. 2013. Biology of the staphylococcal conjugative multiresistance plasmid pSK41. Plasmid 70:42–51 10.1016/j.plasmid.2013.02.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 95.Morton TM, Eaton DM, Johnston JL, Archer GL. 1993. DNA sequence and units of transcription of the conjugative transfer gene complex (trs) of Staphylococcus aureus plasmid pGO1. J Bacteriol 175:4436–4447 10.1128/jb.175.14.4436-4447.1993. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Caryl JA, O’Neill AJ. 2009. Complete nucleotide sequence of pGO1, the prototype conjugative plasmid from the staphylococci. Plasmid 62:35–38 10.1016/j.plasmid.2009.03.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 97.Jaffe HW, Sweeney HM, Weinstein RA, Kabins SA, Nathan C, Cohen S. 1982. Structural and phenotypic varieties of gentamicin resistance plasmids in hospital strains of Staphylococcus aureus and coagulase-negative staphylococci. Antimicrob Agents Chemother 21:773–779 10.1128/AAC.21.5.773. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Archer GL, Scott J. 1991. Conjugative transfer genes in staphylococcal isolates from the United States. Antimicrob Agents Chemother 35:2500–2504 10.1128/AAC.35.12.2500. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jaffe HW, Sweeney HM, Nathan C, Weinstein RA, Kabins SA, Cohen S. 1980. Identity and interspecific transfer of gentamicin-resistance plasmids in Staphylococcus aureus and Staphylococcus epidermidis. J Infect Dis 141:738–747 10.1093/infdis/141.6.738. [PubMed] [DOI] [PubMed] [Google Scholar]
- 100.Archer GL, Dietrick DR, Johnston JL. 1985. Molecular epidemiology of transmissible gentamicin resistance among coagulase-negative staphylococci in a cardiac surgery unit. J Infect Dis 151:243–251 10.1093/infdis/151.2.243. [PubMed] [DOI] [PubMed] [Google Scholar]
- 101.Ni L, Jensen SO, Ky Tonthat N, Berg T, Kwong SM, Guan FH, Brown MH, Skurray RA, Firth N, Schumacher MA. 2009. The Staphylococcus aureus pSK41 plasmid-encoded ArtA protein is a master regulator of plasmid transmission genes and contains a RHH motif used in alternate DNA-binding modes. Nucleic Acids Res 37:6970–6983 10.1093/nar/gkp756. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pérez-Roth E, Kwong SM, Alcoba-Florez J, Firth N, Méndez-Alvarez S. 2010. Complete nucleotide sequence and comparative analysis of pPR9, a 41.7-kilobase conjugative staphylococcal multiresistance plasmid conferring high-level mupirocin resistance. Antimicrob Agents Chemother 54:2252–2257 10.1128/AAC.01074-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Skurray RA, Firth N. 1997. Molecular evolution of multiply-antibiotic-resistant staphylococci. Ciba Found Symp 207:167–183, discussion 183–191. [PubMed] [DOI] [PubMed] [Google Scholar]
- 104.Byrne ME, Gillespie MT, Skurray RA. 1990. Molecular analysis of a gentamicin resistance transposonlike element on plasmids isolated from North American Staphylococcus aureus strains. Antimicrob Agents Chemother 34:2106–2113 10.1128/AAC.34.11.2106. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sasatsu M, Shima K, Shibata Y, Kono M. 1989. Nucleotide sequence of a gene that encodes resistance to ethidium bromide from a transferable plasmid in Staphylococcus aureus. Nucleic Acids Res 17:10103 10.1093/nar/17.23.10103. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Morton TM, Johnston JL, Patterson J, Archer GL. 1995. Characterization of a conjugative staphylococcal mupirocin resistance plasmid. Antimicrob Agents Chemother 39:1272–1280 10.1128/AAC.39.6.1272. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bender J, Strommenger B, Steglich M, Zimmermann O, Fenner I, Lensing C, Dagwadordsch U, Kekulé AS, Werner G, Layer F. 2015. Linezolid resistance in clinical isolates of Staphylococcus epidermidis from German hospitals and characterization of two cfr-carrying plasmids. J Antimicrob Chemother 70:1630–1638. [DOI] [PubMed] [Google Scholar]
- 108.Périchon B, Courvalin P. 2004. Heterologous expression of the enterococcal vanA operon in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 48:4281–4285 10.1128/AAC.48.11.4281-4285.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Flannagan SE, Chow JW, Donabedian SM, Brown WJ, Perri MB, Zervos MJ, Ozawa Y, Clewell DB. 2003. Plasmid content of a vancomycin-resistant Enterococcus faecalis isolate from a patient also colonized by Staphylococcus aureus with a VanA phenotype. Antimicrob Agents Chemother 47:3954–3959 10.1128/AAC.47.12.3954-3959.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tenover FC, Weigel LM, Appelbaum PC, McDougal LK, Chaitram J, McAllister S, Clark N, Killgore G, O’Hara CM, Jevitt L, Patel JB, Bozdogan B. 2004. Vancomycin-resistant Staphylococcus aureus isolate from a patient in Pennsylvania. Antimicrob Agents Chemother 48:275–280 10.1128/AAC.48.1.275-280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhu W, Clark N, Patel JB. 2013. pSK41-like plasmid is necessary for Inc18-like vanA plasmid transfer from Enterococcus faecalis to Staphylococcus aureusin vitro. Antimicrob Agents Chemother 57:212–219 10.1128/AAC.01587-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dougherty BA, Hill C, Weidman JF, Richardson DR, Venter JC, Ross RP. 1998. Sequence and analysis of the 60 kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactis DPC3147. Mol Microbiol 29:1029–1038 10.1046/j.1365-2958.1998.00988.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 113.Firth N, Berg T, Skurray RA. 1999. Evolution of conjugative plasmids from Gram-positive bacteria. Mol Microbiol 31:1598–1600. [PubMed] [PubMed] [Google Scholar]
- 114.Grohmann E, Muth G, Espinosa M. 2003. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol Mol Biol Rev 67:277–301 10.1128/MMBR.67.2.277-301.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guglielmini J, Néron B, Abby SS, Garcillán-Barcia MP, de la Cruz F, Rocha EP. 2014. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res 42:5715–5727 10.1093/nar/gku194. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73:775–808 10.1128/MMBR.00023-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Climo MW, Sharma VK, Archer GL. 1996. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J Bacteriol 178:4975–4983 10.1128/jb.178.16.4975-4983.1996. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Edwards JS, Betts L, Frazier ML, Pollet RM, Kwong SM, Walton WG, Ballentine WK III, Huang JJ, Habibi S, Del Campo M, Meier JL, Dervan PB, Firth N, Redinbo MR. 2013. Molecular basis of antibiotic multiresistance transfer in Staphylococcus aureus. Proc Natl Acad Sci U S A 110:2804–2809 10.1073/pnas.1219701110. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.LeBard RJ, Jensen SO, Arnaiz IA, Skurray RA, Firth N. 2008. A multimer resolution system contributes to segregational stability of the prototypical staphylococcal conjugative multiresistance plasmid pSK41. FEMS Microbiol Lett 284:58–67 10.1111/j.1574-6968.2008.01190.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 120.Schumacher MA, Glover TC, Brzoska AJ, Jensen SO, Dunham TD, Skurray RA, Firth N. 2007. Segrosome structure revealed by a complex of ParR with centromere DNA. Nature 450:1268–1271 10.1038/nature06392. [PubMed] [DOI] [PubMed] [Google Scholar]
- 121.Popp D, Xu W, Narita A, Brzoska AJ, Skurray RA, Firth N, Ghoshdastider U, Maéda Y, Robinson RC, Schumacher MA. 2010. Structure and filament dynamics of the pSK41 actin-like ParM protein: implications for plasmid DNA segregation. J Biol Chem 285:10130–10140 10.1074/jbc.M109.071613. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brzoska AJ, Jensen SO, Barton DA, Davies DS, Overall RL, Skurray RA, Firth N. 2016. Dynamic filament formation by a divergent bacterial actin-like ParM protein. PLoS One 11:e0156944 10.1371/journal.pone.0156944. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sharma VK, Johnston JL, Morton TM, Archer GL. 1994. Transcriptional regulation by TrsN of conjugative transfer genes on staphylococcal plasmid pGO1. J Bacteriol 176:3445–3454 10.1128/jb.176.12.3445-3454.1994. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Townsend DE, Ashdown N, Annear DI, Grubb WB. 1985. A conjugative plasmid encoding production of a diffusible pigment and resistance to aminoglycosides and macrolides in Staphylococcus aureus. Aust J Exp Biol Med Sci 63:573–586 10.1038/icb.1985.61. [PubMed] [DOI] [PubMed] [Google Scholar]
- 125.Townsend DE, Bolton S, Ashdown N, Annear DI, Grubb WB. 1986. Conjugative, staphylococcal plasmids carrying hitch-hiking transposons similar to Tn554: intra- and interspecies dissemination of erythromycin resistance. Aust J Exp Biol Med Sci 64:367–379 10.1038/icb.1986.39. [PubMed] [DOI] [PubMed] [Google Scholar]
- 126.Townsend DE, Grubb WB, Annear DI. 1985. A plasmid for diffusible pigment production in Staphylococcus aureus. Aust J Exp Biol Med Sci 63:463–472 10.1038/icb.1985.51. [PubMed] [DOI] [PubMed] [Google Scholar]
- 127.Udo EE, Wei MQ, Grubb WB. 1992. Conjugative trimethoprim resistance in Staphylococcus aureus. FEMS Microbiol Lett 76:243–248 10.1111/j.1574-6968.1992.tb05470.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 128.Ramsay JP, Kwong SM, Murphy RJ, Yui Eto K, Price KJ, Nguyen QT, O’Brien FG, Grubb WB, Coombs GW, Firth N. 2016. An updated view of plasmid conjugation and mobilization in Staphylococcus. Mob Genet Elements 6:e1208317 10.1080/2159256X.2016.1208317. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shore AC, Lazaris A, Kinnevey PM, Brennan OM, Brennan GI, O’Connell B, Feßler AT, Schwarz S, Coleman DC. 2016. First report of cfr-carrying plasmids in the pandemic sequence type 22 methicillin-resistant Staphylococcus aureus staphylococcal cassette chromosome mec type IV clone. Antimicrob Agents Chemother 60:3007–3015 10.1128/AAC.02949-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.O’Brien FG, Coombs GW, Pearman JW, Gracey M, Moss F, Christiansen KJ, Grubb WB. 2009. Population dynamics of methicillin-susceptible and -resistant Staphylococcus aureus in remote communities. J Antimicrob Chemother 64:684–693 10.1093/jac/dkp285. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.O’Brien FG, Ramsay JP, Monecke S, Coombs GW, Robinson OJ, Htet Z, Alshaikh FA, Grubb WB. 2015. Staphylococcus aureus plasmids without mobilization genes are mobilized by a novel conjugative plasmid from community isolates. J Antimicrob Chemother 70:649–652 10.1093/jac/dku454. [PubMed] [DOI] [PubMed] [Google Scholar]
- 132.Udo E, Townsend DE, Grubb WB. 1987. A conjugative staphylococcal plasmid with no resistance phenotype. FEMS Microbiol Lett 40:279–283 10.1111/j.1574-6968.1987.tb02039.x. [DOI] [Google Scholar]
- 133.Udo EE, Grubb WB. 1996. Molecular and phage typing of Staphylococcus aureus harbouring cryptic conjugative plasmids. Eur J Epidemiol 12:637–641 10.1007/BF00499464. [PubMed] [DOI] [PubMed] [Google Scholar]
- 134.Udo EE, Grubb WB. 1990. Excision of a conjugative plasmid from the staphylococcal chromosome. J Med Microbiol 33:227–234 10.1099/00222615-33-4-227. [PubMed] [DOI] [PubMed] [Google Scholar]
- 135.Udo EE, Grubb WB. 1990. A new class of conjugative plasmid in Staphylococcus aureus. J Med Microbiol 31:207–212 10.1099/00222615-31-3-207. [PubMed] [DOI] [PubMed] [Google Scholar]
- 136.O’Brien FG, Yui Eto K, Murphy RJ, Fairhurst HM, Coombs GW, Grubb WB, Ramsay JP. 2015. Origin-of-transfer sequences facilitate mobilisation of non-conjugative antimicrobial-resistance plasmids in Staphylococcus aureus. Nucleic Acids Res 43:7971–7983 10.1093/nar/gkv755. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rossi F, Diaz L, Wollam A, Panesso D, Zhou Y, Rincon S, Narechania A, Xing G, Di Gioia TS, Doi A, Tran TT, Reyes J, Munita JM, Carvajal LP, Hernandez-Roldan A, Brandão D, van der Heijden IM, Murray BE, Planet PJ, Weinstock GM, Arias CA. 2014. Transferable vancomycin resistance in a community-associated MRSA lineage. N Engl J Med 370:1524–1531 10.1056/NEJMoa1303359. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ramsay JP, Firth N. 2017. Diverse mobilization strategies facilitate transfer of non-conjugative mobile genetic elements. Curr Opin Microbiol 38:1–9 10.1016/j.mib.2017.03.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 139.Pollet RM, Ingle JD, Hymes JP, Eakes TC, Eto KY, Kwong SM, Ramsay JP, Firth N, Redinbo MR. 2016. Processing of nonconjugative resistance plasmids by conjugation nicking enzyme of staphylococci. J Bacteriol 198:888–897 10.1128/JB.00832-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wright CL, Byrne ME, Firth N, Skurray RA. 1998. A retrospective molecular analysis of gentamicin resistance in Staphylococcus aureus strains from UK hospitals. J Med Microbiol 47:173–178 10.1099/00222615-47-2-173. [PubMed] [DOI] [PubMed] [Google Scholar]
- 141.Hodel-Christian SL, Murray BE. 1992. Comparison of the gentamicin resistance transposon Tn5281 with regions encoding gentamicin resistance in Enterococcus faecalis isolates from diverse geographic locations. Antimicrob Agents Chemother 36:2259–2264 10.1128/AAC.36.10.2259. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Horaud T, de Céspèdes G, Trieu-Cuot P. 1996. Chromosomal gentamicin resistance transposon Tn3706 in Streptococcus agalactiae B128. Antimicrob Agents Chemother 40:1085–1090. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Quintiliani R Jr, Courvalin P. 1996. Characterization of Tn1547, a composite transposon flanked by the IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene 172:1–8 10.1016/0378-1119(96)00110-2. [DOI] [PubMed] [Google Scholar]
- 144.Rice LB, Carias LL, Marshall SH. 1995. Tn5384, a composite enterococcal mobile element conferring resistance to erythromycin and gentamicin whose ends are directly repeated copies of IS256. Antimicrob Agents Chemother 39:1147–1153 10.1128/AAC.39.5.1147. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shen J, Wang Y, Schwarz S. 2013. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J Antimicrob Chemother 68:1697–1706 10.1093/jac/dkt092. [PubMed] [DOI] [PubMed] [Google Scholar]
- 146.Li D, Wu C, Wang Y, Fan R, Schwarz S, Zhang S. 2015. Identification of multiresistance gene cfr in methicillin-resistant Staphylococcus aureus from pigs: plasmid location and integration into a staphylococcal cassette chromosome mec complex. Antimicrob Agents Chemother 59:3641–3644 10.1128/AAC.00500-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Matsuo H, Kobayashi M, Kumagai T, Kuwabara M, Sugiyama M. 2003. Molecular mechanism for the enhancement of arbekacin resistance in a methicillin-resistant Staphylococcus aureus. FEBS Lett 546:401–406 10.1016/S0014-5793(03)00644-6. [DOI] [PubMed] [Google Scholar]
- 148.Maki H, Murakami K. 1997. Formation of potent hybrid promoters of the mutant llm gene by IS256 transposition in methicillin-resistant Staphylococcus aureus. J Bacteriol 179:6944–6948 10.1128/jb.179.22.6944-6948.1997. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Couto I, Wu SW, Tomasz A, de Lencastre H. 2003. Development of methicillin resistance in clinical isolates of Staphylococcus sciuri by transcriptional activation of the mecA homologue native to the species. J Bacteriol 185:645–653 10.1128/JB.185.2.645-653.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ziebuhr W, Krimmer V, Rachid S, Lössner I, Götz F, Hacker J. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol 32:345–356 10.1046/j.1365-2958.1999.01353.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 151.Conlon KM, Humphreys H, O’Gara JP. 2004. Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J Bacteriol 186:6208–6219 10.1128/JB.186.18.6208-6219.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vuong C, Kocianova S, Yao Y, Carmody AB, Otto M. 2004. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidisin vivo. J Infect Dis 190:1498–1505 10.1086/424487. [PubMed] [DOI] [PubMed] [Google Scholar]
- 153.Maki H, McCallum N, Bischoff M, Wada A, Berger-Bächi B. 2004. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother 48:1953–1959 10.1128/AAC.48.6.1953-1959.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jansen A, Türck M, Szekat C, Nagel M, Clever I, Bierbaum G. 2007. Role of insertion elements and yycFG in the development of decreased susceptibility to vancomycin in Staphylococcus aureus. Int J Med Microbiol 297:205–215 10.1016/j.ijmm.2007.02.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 155.McEvoy CR, Tsuji B, Gao W, Seemann T, Porter JL, Doig K, Ngo D, Howden BP, Stinear TP. 2013. Decreased vancomycin susceptibility in Staphylococcus aureus caused by IS256 tempering of WalKR expression. Antimicrob Agents Chemother 57:3240–3249 10.1128/AAC.00279-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Benson MA, Ohneck EA, Ryan C, Alonzo F III, Smith H, Narechania A, Kolokotronis SO, Satola SW, Uhlemann AC, Sebra R, Deikus G, Shopsin B, Planet PJ, Torres VJ. 2014. Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Mol Microbiol 93:664–681 10.1111/mmi.12682. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Prudhomme M, Turlan C, Claverys JP, Chandler M. 2002. Diversity of Tn4001 transposition products: the flanking IS256 elements can form tandem dimers and IS circles. J Bacteriol 184:433–443 10.1128/JB.184.2.433-443.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Loessner I, Dietrich K, Dittrich D, Hacker J, Ziebuhr W. 2002. Transposase-dependent formation of circular IS256 derivatives in Staphylococcus epidermidis and Staphylococcus aureus. J Bacteriol 184:4709–4714 10.1128/JB.184.17.4709-4714.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Siguier P, Gourbeyre E, Varani A, Ton-Hoang B, Chandler M. 2015. Everyman’s guide to bacterial insertion sequences. Microbiol Spectr 3:MDNA3-0030-2014. 10.1128/microbiolspec.MDNA3-0030-2014. [DOI] [PubMed] [Google Scholar]
- 160.Hennig S, Ziebuhr W. 2010. Characterization of the transposase encoded by IS256, the prototype of a major family of bacterial insertion sequence elements. J Bacteriol 192:4153–4163 10.1128/JB.00226-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hennig S, Ziebuhr W. 2008. A transposase-independent mechanism gives rise to precise excision of IS256 from insertion sites in Staphylococcus epidermidis. J Bacteriol 190:1488–1490 10.1128/JB.01290-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Needham C, Noble WC, Dyke KGH. 1995. The staphylococcal insertion sequence IS257 is active. Plasmid 34:198–205 10.1006/plas.1995.0005. [PubMed] [DOI] [PubMed] [Google Scholar]
- 163.Kadlec K, Schwarz S. 2010. Identification of a plasmid-borne resistance gene cluster comprising the resistance genes erm(T), dfrK, and tet(L) in a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob Agents Chemother 54:915–918 10.1128/AAC.01091-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gómez-Sanz E, Kadlec K, Feßler AT, Zarazaga M, Torres C, Schwarz S. 2013. Novel erm(T)-carrying multiresistance plasmids from porcine and human isolates of methicillin-resistant Staphylococcus aureus ST398 that also harbor cadmium and copper resistance determinants. Antimicrob Agents Chemother 57:3275–3282 10.1128/AAC.00171-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Harmer CJ, Moran RA, Hall RM. 2014. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. MBio 5:e01801-14 10.1128/mBio.01801-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Harmer CJ, Hall RM. 2016. IS26-mediated formation of transposons carrying antibiotic resistance genes. MSphere 1:e00038-16 10.1128/mSphere.00038-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bjorland J, Steinum T, Sunde M, Waage S, Sviland S, Oppegaard H, Heir E. 2006. Deletion of pT181-like sequence in an smr-encoding mosaic plasmid harboured by a persistent bovine Staphylococcus warneri strain. J Antimicrob Chemother 57:46–51. [PubMed] [DOI] [PubMed] [Google Scholar]
- 168.Leelaporn A, Firth N, Byrne ME, Roper E, Skurray RA. 1994. Possible role of insertion sequence IS257 in dissemination and expression of high- and low-level trimethoprim resistance in staphylococci. Antimicrob Agents Chemother 38:2238–2244 10.1128/AAC.38.10.2238. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Simpson AE, Skurray RA, Firth N. 2000. An IS257-derived hybrid promoter directs transcription of a tetA(K) tetracycline resistance gene in the Staphylococcus aureus chromosomal mec region. J Bacteriol 182:3345–3352 10.1128/JB.182.12.3345-3352.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Katayama Y, Ito T, Hiramatsu K. 2001. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob Agents Chemother 45:1955–1963 10.1128/AAC.45.7.1955-1963.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Archer GL, Niemeyer DM, Thanassi JA, Pucci MJ. 1994. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob Agents Chemother 38:447–454 10.1128/AAC.38.3.447. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Archer GL, Thanassi JA, Niemeyer DM, Pucci MJ. 1996. Characterization of IS1272, an insertion sequence-like element from Staphylococcus haemolyticus. Antimicrob Agents Chemother 40:924–929. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hiramatsu K, Cui L, Kuroda M, Ito T. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol 9:486–493 10.1016/S0966-842X(01)02175-8. [DOI] [PubMed] [Google Scholar]
- 174.Bouchami O, de Lencastre H, Miragaia M. 2016. Impact of insertion sequences and recombination on the population structure of Staphylococcus haemolyticus. PLoS One 11:e0156653 10.1371/journal.pone.0156653. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Furi L, Haigh R, Al Jabri ZJ, Morrissey I, Ou HY, León-Sampedro R, Martinez JL, Coque TM, Oggioni MR. 2016. Dissemination of novel antimicrobial resistance mechanisms through the insertion sequence mediated spread of metabolic genes. Front Microbiol 7:1008 10.3389/fmicb.2016.01008. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kehrenberg C, Aarestrup FM, Schwarz S. 2007. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob Agents Chemother 51:483–487 10.1128/AAC.01340-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kehrenberg C, Schwarz S. 2005. Florfenicol-chloramphenicol exporter gene fexA is part of the novel transposon Tn558. Antimicrob Agents Chemother 49:813–815 10.1128/AAC.49.2.813-815.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Iandolo JJ, Bannantine JP, Stewart GC. 1997. Genetic and physical map of the chromosome of Staphylococcus aureus, p 39–53. In Crossley KB, Archer GL (ed), The Staphylococci in Human Disease. Churchill Livingstone, London, UK. [Google Scholar]
- 179.Ito T, Okuma K, Ma XX, Yuzawa H, Hiramatsu K. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist Updat 6:41–52 10.1016/S1368-7646(03)00003-7. [DOI] [PubMed] [Google Scholar]
- 180.Rowland SJ, Dyke KG. 1989. Characterization of the staphylococcal beta-lactamase transposon Tn552. EMBO J 8:2761–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sansevere EA, Luo X, Park JY, Yoon S, Seo KS, Robinson DA. 2017. Transposase-mediated excision, conjugative transfer, and diversity of ICE6013 elements in Staphylococcus aureus. J Bacteriol 199:199 10.1128/JB.00629-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Minakhina S, Kholodii G, Mindlin S, Yurieva O, Nikiforov V. 1999. Tn5053 family transposons are res site hunters sensing plasmidal res sites occupied by cognate resolvases. Mol Microbiol 33:1059–1068 10.1046/j.1365-2958.1999.01548.x. [DOI] [PubMed] [Google Scholar]
- 183.Derbise A, Dyke KGH, el Solh N. 1995. Rearrangements in the staphylococcal β-lactamase-encoding plasmid, pIP1066, including a DNA inversion that generates two alternative transposons. Mol Microbiol 17:769–779 10.1111/j.1365-2958.1995.mmi_17040769.x. [DOI] [PubMed] [Google Scholar]
- 184.Derbise A, Aubert S, El Solh N. 1997. Mapping the regions carrying the three contiguous antibiotic resistance genes aadE, sat4, and aphA-3 in the genomes of staphylococci. Antimicrob Agents Chemother 41:1024–1032. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Derbise A, Dyke KG, el Solh N. 1996. Characterization of a Staphylococcus aureus transposon, Tn5405, located within Tn5404 and carrying the aminoglycoside resistance genes, aphA-3 and aadE. Plasmid 35:174–188 10.1006/plas.1996.0020. [PubMed] [DOI] [PubMed] [Google Scholar]
- 186.Chesneau O, Lailler R, Derbise A, El Solh N. 1999. Transposition of IS1181 in the genomes of Staphylococcus and Listeria. FEMS Microbiol Lett 177:93–100 10.1111/j.1574-6968.1999.tb13718.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 187.Thumm G, Götz F. 1997. Studies on prolysostaphin processing and characterization of the lysostaphin immunity factor (Lif) of Staphylococcus simulans biovar staphylolyticus. Mol Microbiol 23:1251–1265 10.1046/j.1365-2958.1997.2911657.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 188.Takeuchi F, Watanabe S, Baba T, Yuzawa H, Ito T, Morimoto Y, Kuroda M, Cui L, Takahashi M, Ankai A, Baba S, Fukui S, Lee JC, Hiramatsu K. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J Bacteriol 187:7292–7308 10.1128/JB.187.21.7292-7308.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Murphy E. 1990. Properties of the site-specific transposable element Tn554, p 123–135. In Novick RP (ed), Molecular Biology of the Staphylococci. VCH, New York, NY. [Google Scholar]
- 190.Kadlec K, Schwarz S. 2010. Identification of the novel dfrK-carrying transposon Tn559 in a porcine methicillin-susceptible Staphylococcus aureus ST398 strain. Antimicrob Agents Chemother 54:3475–3477 10.1128/AAC.00464-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bastos MC, Murphy E. 1988. Transposon Tn554 encodes three products required for transposition. EMBO J 7:2935–2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Van Houdt R, Leplae R, Lima-Mendez G, Mergeay M, Toussaint A. 2012. Towards a more accurate annotation of tyrosine-based site-specific recombinases in bacterial genomes. Mob DNA 3:6 10.1186/1759-8753-3-6. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Haroche J, Allignet J, El Solh N. 2002. Tn5406, a new staphylococcal transposon conferring resistance to streptogramin a and related compounds including dalfopristin. Antimicrob Agents Chemother 46:2337–2343 10.1128/AAC.46.8.2337-2343.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Schwendener S, Perreten V. 2011. New transposon Tn6133 in methicillin-resistant Staphylococcus aureus ST398 contains vga(E), a novel streptogramin A, pleuromutilin, and lincosamide resistance gene. Antimicrob Agents Chemother 55:4900–4904 10.1128/AAC.00528-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hochhut B, Waldor MK. 1999. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol Microbiol 32:99–110 10.1046/j.1365-2958.1999.01330.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 196.Roberts AP, Chandler M, Courvalin P, Guédon G, Mullany P, Pembroke T, Rood JI, Smith CJ, Summers AO, Tsuda M, Berg DE. 2008. Revised nomenclature for transposable genetic elements. Plasmid 60:167–173 10.1016/j.plasmid.2008.08.001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Auchtung JM, Aleksanyan N, Bulku A, Berkmen MB. 2016. Biology of ICEBs1, an integrative and conjugative element in Bacillus subtilis. Plasmid 86:14–25 10.1016/j.plasmid.2016.07.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 198.Jones JM, Yost SC, Pattee PA. 1987. Transfer of the conjugal tetracycline resistance transposon Tn916 from Streptococcus faecalis to Staphylococcus aureus and identification of some insertion sites in the staphylococcal chromosome. J Bacteriol 169:2121–2131 10.1128/jb.169.5.2121-2131.1987. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Scott JR, Bringel F, Marra D, Van Alstine G, Rudy CK. 1994. Conjugative transposition of Tn916: preferred targets and evidence for conjugative transfer of a single strand and for a double-stranded circular intermediate. Mol Microbiol 11:1099–1108 10.1111/j.1365-2958.1994.tb00386.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 200.Mullany P, Roberts AP, Wang H. 2002. Mechanism of integration and excision in conjugative transposons. Cell Mol Life Sci 59:2017–2022 10.1007/s000180200001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Watson DA, Musher DM, Hamill RJ. 1993. Promotion of transposon Tn916 excision via exposure to streptomycin. J Infect Dis 168:1334–1335 10.1093/infdis/168.5.1334. [PubMed] [DOI] [PubMed] [Google Scholar]
- 202.Scornec H, Bellanger X, Guilloteau H, Groshenry G, Merlin C. 2017. Inducibility of Tn916 conjugative transfer in Enterococcus faecalis by subinhibitory concentrations of ribosome-targeting antibiotics. J Antimicrob Chemother 72:2722–2728 10.1093/jac/dkx202. [PubMed] [DOI] [PubMed] [Google Scholar]
- 203.León-Sampedro R, Novais C, Peixe L, Baquero F, Coque TM. 2016. Diversity and evolution of the Tn5801-tet(M)-like integrative and conjugative elements among Enterococcus, Streptococcus, and Staphylococcus. Antimicrob Agents Chemother 60:1736–1746 10.1128/AAC.01864-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.de Vries LE, Hasman H, Jurado Rabadán S, Agersø Y. 2016. Sequence-based characterization of Tn5801-like genomic islands in tetracycline-resistant Staphylococcus pseudintermedius and other Gram-positive bacteria from humans and animals. Front Microbiol 7:576 10.3389/fmicb.2016.00576. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Mingoia M, Morici E, Tili E, Giovanetti E, Montanari MP, Varaldo PE. 2013. Characterization of Tn5801.Sag, a variant of Staphylococcus aureus Tn916 family transposon Tn5801 that is widespread in clinical isolates of Streptococcus agalactiae. Antimicrob Agents Chemother 57:4570–4574 10.1128/AAC.00521-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Smyth DS, Robinson DA. 2009. Integrative and sequence characteristics of a novel genetic element, ICE6013, in Staphylococcus aureus. J Bacteriol 191:5964–5975 10.1128/JB.00352-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Serfiotis-Mitsa D, Roberts GA, Cooper LP, White JH, Nutley M, Cooper A, Blakely GW, Dryden DT. 2008. The Orf18 gene product from conjugative transposon Tn916 is an ArdA antirestriction protein that inhibits type I DNA restriction-modification systems. J Mol Biol 383:970–981 10.1016/j.jmb.2008.06.005. [PubMed] [DOI] [PubMed] [Google Scholar]
- 208.Chen K, Reuter M, Sanghvi B, Roberts GA, Cooper LP, Tilling M, Blakely GW, Dryden DT. 2014. ArdA proteins from different mobile genetic elements can bind to the EcoKI type I DNA methyltransferase of E. coli K12. Biochim Biophys Acta 1844:505–511 10.1016/j.bbapap.2013.12.008. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.McMahon SA, Roberts GA, Johnson KA, Cooper LP, Liu H, White JH, Carter LG, Sanghvi B, Oke M, Walkinshaw MD, Blakely GW, Naismith JH, Dryden DT. 2009. Extensive DNA mimicry by the ArdA anti-restriction protein and its role in the spread of antibiotic resistance. Nucleic Acids Res 37:4887–4897 10.1093/nar/gkp478. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Delavat F, Miyazaki R, Carraro N, Pradervand N, van der Meer JR. 2017. The hidden life of integrative and conjugative elements. FEMS Microbiol Rev 41:512–537 10.1093/femsre/fux008. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Naglich JG, Andrews RE Jr. 1988. Tn916-dependent conjugal transfer of PC194 and PUB110 from Bacillus subtilis into Bacillus thuringiensis subsp. israelensis. Plasmid 20:113–126 10.1016/0147-619X(88)90014-5. [DOI] [PubMed] [Google Scholar]
- 212.García-Álvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, Curran MD, Walpole E, Brooks K, Pickard DJ, Teale C, Parkhill J, Bentley SD, Edwards GF, Girvan EK, Kearns AM, Pichon B, Hill RL, Larsen AR, Skov RL, Peacock SJ, Maskell DJ, Holmes MA. 2011. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis 11:595–603 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ito T, Hiramatsu K, Tomasz A, de Lencastre H, Perreten V, Holden MT, Coleman DC, Goering R, Giffard PM, Skov RL, Zhang K, Westh H, O’Brien F, Tenover FC, Oliveira DC, Boyle-Vavra S, Laurent F, Kearns AM, Kreiswirth B, Ko KS, Grundmann H, Sollid JE, John JF Jr, Daum R, Soderquist B, Buist G, International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). 2012. Guidelines for reporting novel mecA gene homologues. Antimicrob Agents Chemother 56:4997–4999 10.1128/AAC.01199-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wu Z, Li F, Liu D, Xue H, Zhao X. 2015. Novel type XII staphylococcal cassette chromosome mec harboring a new cassette chromosome recombinase, CcrC2. Antimicrob Agents Chemother 59:7597–7601 10.1128/AAC.01692-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Mir-Sanchis I, Roman CA, Misiura A, Pigli YZ, Boyle-Vavra S, Rice PA. 2016. Staphylococcal SCCmec elements encode an active MCM-like helicase and thus may be replicative. Nat Struct Mol Biol 23:891–898 10.1038/nsmb.3286. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53:4961–4967 10.1128/AAC.00579-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kaya H, Hasman H, Larsen J, Stegger M, Johannesen TB, Allesøe RL, Lemvigh CK, Aarestrup FM, Lund O, Larsen AR. 2018. SCCmecFinder, a web-based tool for typing of staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. MSphere 3:e00612-17 10.1128/mSphere.00612-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wu S, Piscitelli C, de Lencastre H, Tomasz A. 1996. Tracking the evolutionary origin of the methicillin resistance gene: cloning and sequencing of a homologue of mecA from a methicillin susceptible strain of Staphylococcus sciuri. Microb Drug Resist 2:435–441 10.1089/mdr.1996.2.435. [PubMed] [DOI] [PubMed] [Google Scholar]
- 219.Rolo J, Worning P, Nielsen JB, Bowden R, Bouchami O, Damborg P, Guardabassi L, Perreten V, Tomasz A, Westh H, de Lencastre H, Miragaia M. 2017. Evolutionary origin of the staphylococcal cassette chromosome mec (SCCmec). Antimicrob Agents Chemother 61:61 10.1128/AAC.02302-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Luong TT, Ouyang S, Bush K, Lee CY. 2002. Type 1 capsule genes of Staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J Bacteriol 184:3623–3629 10.1128/JB.184.13.3623-3629.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Mongkolrattanothai K, Boyle S, Murphy TV, Daum RS. 2004. Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing pbp4 and tagF genes in a commensal staphylococcal species: a possible reservoir for antibiotic resistance islands in Staphylococcus aureus. Antimicrob Agents Chemother 48:1823–1836 10.1128/AAC.48.5.1823-1836.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Chongtrakool P, Ito T, Ma XX, Kondo Y, Trakulsomboon S, Tiensasitorn C, Jamklang M, Chavalit T, Song JH, Hiramatsu K. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob Agents Chemother 50:1001–1012 10.1128/AAC.50.3.1001-1012.2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Barbier F, Lebeaux D, Hernandez D, Delannoy AS, Caro V, François P, Schrenzel J, Ruppé E, Gaillard K, Wolff M, Brisse S, Andremont A, Ruimy R. 2011. High prevalence of the arginine catabolic mobile element in carriage isolates of methicillin-resistant Staphylococcus epidermidis. J Antimicrob Chemother 66:29–36 10.1093/jac/dkq410. [PubMed] [DOI] [PubMed] [Google Scholar]
- 224.Miragaia M, de Lencastre H, Perdreau-Remington F, Chambers HF, Higashi J, Sullam PM, Lin J, Wong KI, King KA, Otto M, Sensabaugh GF, Diep BA. 2009. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PLoS One 4:e7722 10.1371/journal.pone.0007722. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Pi B, Yu M, Chen Y, Yu Y, Li L. 2009. Distribution of the ACME-arcA gene among meticillin-resistant Staphylococcus haemolyticus and identification of a novel ccr allotype in ACME-arcA-positive isolates. J Med Microbiol 58:731–736 10.1099/jmm.0.007351-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 226.Sabat AJ, Köck R, Akkerboom V, Hendrix R, Skov RL, Becker K, Friedrich AW. 2013. Novel organization of the arginine catabolic mobile element and staphylococcal cassette chromosome mec composite island and its horizontal transfer between distinct Staphylococcus aureus genotypes. Antimicrob Agents Chemother 57:5774–5777 10.1128/AAC.01321-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 228.Diep BA, Stone GG, Basuino L, Graber CJ, Miller A, des Etages SA, Jones A, Palazzolo-Ballance AM, Perdreau-Remington F, Sensabaugh GF, DeLeo FR, Chambers HF. 2008. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis 197:1523–1530 10.1086/587907. [PubMed] [DOI] [PubMed] [Google Scholar]
- 229.Thurlow LR, Joshi GS, Clark JR, Spontak JS, Neely CJ, Maile R, Richardson AR. 2013. Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13:100–107 10.1016/j.chom.2012.11.012. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Joshi GS, Spontak JS, Klapper DG, Richardson AR. 2011. Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol Microbiol 82:9–20 10.1111/j.1365-2958.2011.07809.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Somkuti GA, Solaiman DK, Steinberg DH. 1997. Molecular properties of the erythromycin resistance plasmid pPV141 from Staphylococcus chromogenes. Plasmid 37:119–127 10.1006/plas.1997.1278. [PubMed] [DOI] [PubMed] [Google Scholar]
- 232.Highlander SK, Hultén KG, Qin X, Jiang H, Yerrapragada S, Mason EO Jr, Shang Y, Williams TM, Fortunov RM, Liu Y, Igboeli O, Petrosino J, Tirumalai M, Uzman A, Fox GE, Cardenas AM, Muzny DM, Hemphill L, Ding Y, Dugan S, Blyth PR, Buhay CJ, Dinh HH, Hawes AC, Holder M, Kovar CL, Lee SL, Liu W, Nazareth LV, Wang Q, Zhou J, Kaplan SL, Weinstock GM. 2007. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol 7:99 10.1186/1471-2180-7-99. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010 10.1093/bioinformatics/btr039. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wright LD, Grossman AD. 2016. Autonomous replication of the conjugative transposon Tn916. J Bacteriol 198:3355–3366 10.1128/JB.00639-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Wright LD, Johnson CM, Grossman AD. 2015. Identification of a single strand origin of replication in the integrative and conjugative element ICEBs1 of Bacillus subtilis. PLoS Genet 11:e1005556 10.1371/journal.pgen.1005556. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lannergård J, Norström T, Hughes D. 2009. Genetic determinants of resistance to fusidic acid among clinical bacteremia isolates of Staphylococcus aureus. Antimicrob Agents Chemother 53:2059–2065 10.1128/AAC.00871-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Fu Z, Liu Y, Chen C, Guo Y, Ma Y, Yang Y, Hu F, Xu X, Wang M. 2016. Characterization of fosfomycin resistance gene, fosB, in methicillin-resistant Staphylococcus aureus isolates. PLoS One 11:e0154829 10.1371/journal.pone.0154829. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Allignet J, El Solh N. 1999. Comparative analysis of staphylococcal plasmids carrying three streptogramin-resistance genes: vat-vgb-vga. Plasmid 42:134–138 10.1006/plas.1999.1412. [PubMed] [DOI] [PubMed] [Google Scholar]
- 239.Chen L, Mediavilla JR, Smyth DS, Chavda KD, Ionescu R, Roberts RB, Robinson DA, Kreiswirth BN. 2010. Identification of a novel transposon (Tn6072) and a truncated staphylococcal cassette chromosome mec element in methicillin-resistant Staphylococcus aureus ST239. Antimicrob Agents Chemother 54:3347–3354 10.1128/AAC.00001-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Kadlec K, Schwarz S. 2009. Identification of a novel trimethoprim resistance gene, dfrK, in a methicillin-resistant Staphylococcus aureus ST398 strain and its physical linkage to the tetracycline resistance gene tet(L). Antimicrob Agents Chemother 53:776–778 10.1128/AAC.01128-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.O’Neill AJ, Chopra I. 2006. Molecular basis of fusB-mediated resistance to fusidic acid in Staphylococcus aureus. Mol Microbiol 59:664–676 10.1111/j.1365-2958.2005.04971.x. [DOI] [PubMed] [Google Scholar]
- 242.de Vries LE, Christensen H, Agersø Y. 2012. The diversity of inducible and constitutively expressed erm(C) genes and association to different replicon types in staphylococci plasmids. Mob Genet Elements 2:72–80 http://dx.doi.org/10.4161/mge.20109. [DOI] [PMC free article] [PubMed] [Google Scholar]


