Abstract
Current studies of biological tissues require visualizing diverse cell types and molecular interactions, creating a growing need for versatile techniques to simultaneously probe numerous targets. Traditional multiplexed imaging is limited to around five targets at once. Emerging methods using sequential rounds of staining, imaging, and signal removal can probe tens of targets but require specialized hardware and time-consuming workflows and face challenges with sample distortion and artifacts. We present a highly multiplexed fluorescence microscopy method using semiconducting polymer dots (Pdots) in a single round of staining and imaging. Pdots are small, bright, and photostable fluorescent probes with a wide range of tunable Stokes shifts (20 to 450 nanometers). Multiple series of Pdots with varying excitation wavelengths allow for fast (<1 minute) and single-round imaging of up to 21 targets in the brain and kidney. This method is based on a simple immunofluorescence workflow, efficient use of spectral space, standard hardware, and straightforward analysis, making it widely applicable for bioimaging laboratories.
Fluorescent semiconducting polymer dots facilitate a single round of staining and imaging of 21 targets in mouse tissues.
INTRODUCTION
Biological tissues are complex networks of cells that are arranged within intricate extracellular environments and that adapt during development, aging, and disease. Fluorescence microscopy is heavily used for the study of these complex specimens and particularly benefits from the high molecular specificity of immunofluorescence and related staining methods due to their powerful ability to quantify the distributions of multiple molecules of interest within the specimen. Commonly used fluorophores in microscopy have absorption and emission spectral bandwidths of 30 to 60 nm and a Stokes shift of 20 to 30 nm between the absorption and emission maxima. Because each fluorophore occupies a combined ~70 nm of spectral bandwidth, the number of fluorophores concurrently visualized across visible and near-infrared wavelengths (400 to 750 nm) is commonly limited to ~5.
Prior work has sought to increase fluorescence multiplexing capabilities using various immunolabeling strategies and detection techniques (1). One family of techniques uses sequential cycles of staining and imaging and avoids spectral overlap by chemically inactivating or removing fluorescent dyes after each cycle [e.g., MxIF (2), t-CyCIF (3), IBEX (4), and SWITCH (5)]. A second family of sequential multiplexing techniques uses a single step to immunostain tissues with DNA-barcoded antibodies, followed by a sequential readout of barcodes using complementary fluorescent oligonucleotides [e.g., CODEX (6) and Immuno-SABER (7)]. Multiplexed transcriptomics methods are a third family of techniques that map mRNA distributions across cells and tissues [e.g., MERFISH (8) and seqFISH (9)]. These methods also use a single round of staining with barcoded probes for gene panels, followed by sequential rounds of reagent delivery and imaging to read out the barcodes and spatially localize the transcripts. Despite the large number of targets that can be probed, all of these sequential approaches encounter major limitations: extended times for probe delivery and incubation (usually 20 to 120 min or longer per cycle and on order of a day or more per experiment) that restrict the application to imaging of thin tissues; the potential for tissue distortion between rounds and for unsuccessful inactivation of probes; complex and computationally intensive probe design and analysis; and the requirement for specialized and automated fluidic handling equipment and dedicated imaging instruments.
Instead of multiple rounds of staining and imaging, a single round of highly multiplexed imaging would be desirable in many circumstances. Laser or ion beam ablation followed by mass spectroscopy can achieve higher plex imaging in a single round (10, 11), but this approach involves a tedious workflow with low throughput (e.g., pump-down of the sample chamber), is destructive, and requires expensive instrumentation. A fluorescence-based technique would alleviate some of these requirements, in principle. However, commonly used microscopy fluorophores have a small Stokes shift of 20 to 30 nm that, together with the 30- to 60-nm excitation and emission bandwidths, limits the number of fluorophores that may be used. In contrast, recently developed spectrally tunable semiconducting polymer dots (Pdots) (12–23) exhibit a variable Stokes shift of 20 to 450 nm, which can be tuned by varying the composition of the emitting Pdots (17, 18, 20, 21). This tunability of Stokes shift in Pdots provides much more spectral real estate for multiplexing than is possible using conventional fluorescent dyes. The related field of flow cytometry has led the way in pushing the limits of fluorescence multiplexing. However, many of the long Stokes shift fluorophores that made high multiplex flow cytometry possible, such as phycoerythrin tandem dyes and the BD Brilliant series of polymer dyes, can be prone to photobleaching and are thus unsuitable for imaging. In contrast, Pdots are photostable and can have similar or better photostability as water-soluble quantum dots (Qdots) (12–14, 20). (A comparison of the photostability of Pdots and BD Brilliant dyes is shown in fig. S1.) Pdots are also brighter than small-molecule organic dyes or water-soluble Qdots due to their larger absorption cross section (14, 16, 17, 23).
Here, we demonstrate highly multiplexed imaging of up to 21 targets in a single round of immunostaining and imaging by using only three series of Pdots (a Pdot series is defined by its excitation peak, such as a 355-nm excitable series of Pdots comprising over 10 Pdots—each with a distinct emission peak from below 400 nm to over 800 nm). With computational processing using linear unmixing (24) after a single round of image acquisition, overlapping spectral signals of multiple Pdots can be unmixed and reassigned to their corresponding channels to provide reliable signal separation and distinct multitarget imaging. We demonstrate the versatility of our technique by using both direct and indirect immunolabeling of tissues (mouse kidney and brain) and by making both separate and concurrent use of excitation-multiplexed and emission-multiplexed imaging.
RESULTS
Semiconducting Pdots with spectrally tunable properties
Pdots are semiconducting nanoparticles prepared by collapsing hydrophobic semiconducting polymers into a compact “dot,” the size of which can be tuned from ~5 nm to over ~20 nm (12, 13, 15, 18, 19). The brightness of Pdots is generally very high, owing to their large absorption cross section. By incorporating a Qdot into a Pdot, the brightness of the Qdot can be increased by up to an order of magnitude in comparison with a water-soluble Qdot alone (16). The photophysical property of Pdots that is of particular relevance to the current study is amplified energy transfer, which enables the Stokes shift to be tuned from ~20 nm to over 400 nm (20, 21). This characteristic, together with narrow emission bands, allows for the design and creation of a large color panel (e.g., 10) for a single wavelength excitation. Unlike Qdots, the absorption peak of Pdots can also be tuned to enable excitation multiplexing. Combining excitation and emission multiplexing enables the creation of a very large color panel that numbers in the many tens. These favorable properties of Pdots have now facilitated highly multiplexed flow cytometry analysis of single cells, and Pdots are commercially available as the StarBright series from Bio-Rad, with over 30 colors launched thus far. Unlike many of the tandem dyes used traditionally in flow cytometry, most Pdots have good photostability and may enable highly multiplexed fluorescence imaging. In addition, Pdots can be designed with further improved photostability or used under conditions that improve their photostability (22).
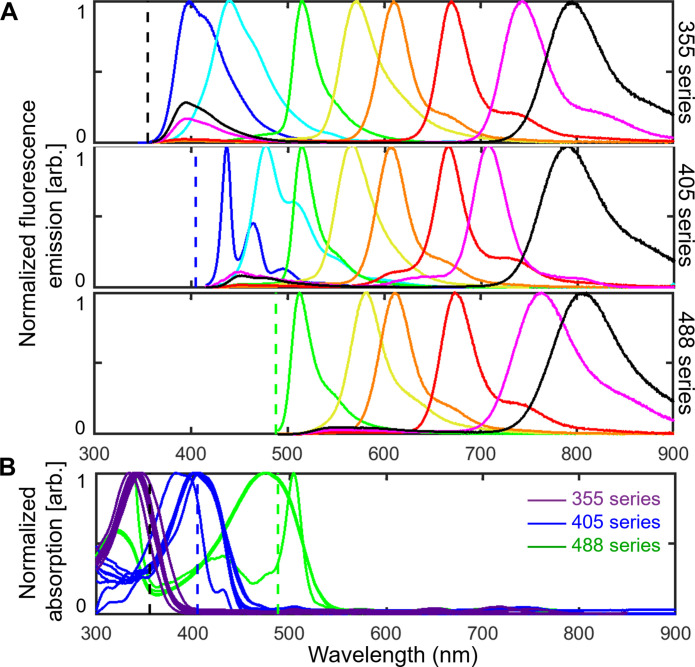
For multiplexed imaging in this study, we used three series of spectrally distinguishable Pdots with absorption maxima near 355, 405, or 488 nm for optimal excitation with commonly available laser lines. For each series, we used six to eight Pdots with a range of emissions covering the visible and near-infrared spectrum, from below 400 to over 800 nm. The excitation and emission spectra of this palette of 22 Pdots are shown in Fig. 1.
Fig. 1. Normalized fluorescence spectra of 22 Pdots.
Emission (A) and absorption (B) spectra of three series of Pdots. Each series contains six to eight Pdots with emission wavelengths that span the visible and near-infrared spectrum from below 400 to over 800 nm. Dashed lines indicate the excitation wavelengths used for imaging experiments (365, 405, and 488 nm).
To measure fluorescence from about 400 to 800 nm for our panels of Pdots, we sequentially recorded images using a set of bandpass emission filters (fig. S2) for a given excitation wavelength and then repeated the process for up to three excitation wavelengths. While the emission filters were selected to match each Pdot, some emission bleedthrough was present due to spectral overlap of the Pdots within each Pdot series (Fig. 1A). This emission bleedthrough is most notable for Pdots in adjacent spectral bands and for Pdots with a principal emission peak of 700 to 800 nm that exhibit a minor emission peak with a small Stokes shift of 50 to 100 nm. In addition, the excitation spectra of the three Pdot series partly overlap at the excitation wavelengths, leading to some cross-excitation (Fig. 1B). For instance, at 365-nm excitation, 355ex-Pdots (the 355-nm excitation series of Pdots) are excited at ~80% of their excitation maximum while the 405ex-Pdots and 488ex-Pdots are also cross-excited at ~20 to 30% of their excitation maximum. To address emission bleedthrough and cross-excitation, we used a linear unmixing algorithm in MATLAB (source code publicly available: see Data and materials availability) to reassign signals to their corresponding channels based on calibration data measured for individual Pdots. This procedure allowed us to generate unmixed images that correct for cross-talk.
Emission multiplexing: Visualizing eight targets using a single excitation wavelength in a single round
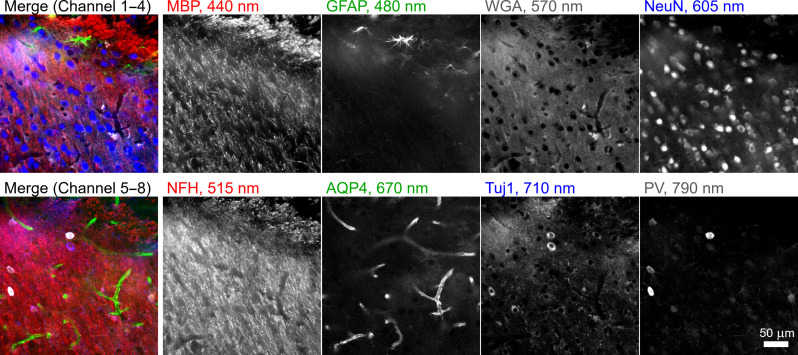
To concurrently label and visualize eight key structures in mouse brain using emission multiplexing with Pdots excited at 405 nm, we first selected and validated seven primary antibodies and the lectin wheat germ agglutinin (WGA) with distinct staining patterns in mouse brain. The panel of seven primary antibodies was selected to also be antigenically distinct [i.e., mouse immunoglobulin G1 (IgG1), mouse IgG2a, mouse IgG2b, mouse IgG3, rabbit, rat, and chicken] so that they could each be uniquely targeted by specific secondary antibodies (i.e., goat anti-mouse IgG1, goat anti-mouse IgG2a, goat anti-mouse IgG2b, goat anti-mouse IgG3, donkey anti-rabbit, donkey anti-rat, and donkey anti-chicken; see table S1 for details). Next, eight Pdots from the 405-nm series with emission peaks ranging from 430 to 800 nm were conjugated to secondary antibodies or directly to WGA. Figure 2 shows a 100-μm-thick brain tissue section stained for eight targets using these probes and then imaged within a few seconds in a single round of automated imaging using matched filter sets. The images displayed in Fig. 2 were linearly unmixed from raw data to correctly reassign bleedthrough from each spectral band to the corresponding stain (see Materials and Methods, Supplementary Text, and fig. S3 for details on the linear unmixing procedure and for a comparison of the raw and unmixed images). The secondary antibody-Pdot conjugates bind to primary antibodies accurately with a low background signal. Anti–myelin basic protein labeled oligodendrocytes, anti–glial fibrillary acidic protein labeled astrocytes/glial cells, WGA labeled N-acetylglucosamine, anti-NeuN labeled neuronal nuclei, anti–neurofilament H labeled heavy neurofilaments, anti–aquaporin 4 (AQP4) labeled AQP4 water channel protein, and anti-Tuj1 labeled class III β-tubulin on neural stem cells.
Fig. 2. Emission multiplexing with the 405ex-Pdot series.
A 100-μm-thick brain section was stained with eight different Pdots conjugated to a set of seven secondary antibodies and the lectin WGA. The specimen was imaged at the amygdalar and piriform area by illuminating with 405-nm light on a wide-field microscope. The signal measured through different bandpass filters was linearly unmixed. MBP, myelin basic protein; GFAP, glial fibrillary acidic protein; WGA, wheat germ agglutinin; NeuN, neuronal nuclei; NFH, neurofilament H; AQP4, aquaporin 4; Tuj1, class III β-tubulin; PV, parvalbumin.
In a related experiment for eight-channel emission multiplexing with the 405ex-Pdot series, we used a set of five secondary antibody-Pdot conjugates (anti-mouse IgG1, anti-mouse IgG2a, anti-mouse IgG2b, anti-rabbit, and anti-rat), a direct antibody-Pdot conjugate, and two Pdot-lectin conjugates to label and image a mouse kidney tissue section (fig. S4). We also performed seven-channel emission multiplexing with the 355ex-Pdot series in mouse brain and mouse kidney using a mixture of Pdot-secondary antibody conjugates, Pdot-primary conjugates, and Pdot-lectin conjugates (figs. S5 and S6). Together, this collection of experiments demonstrates the feasibility of using emission multiplexing with Pdots to achieve up to eight-channel imaging with a single excitation wavelength.
Excitation multiplexing: Visualizing three targets using a single emission filter in a single round
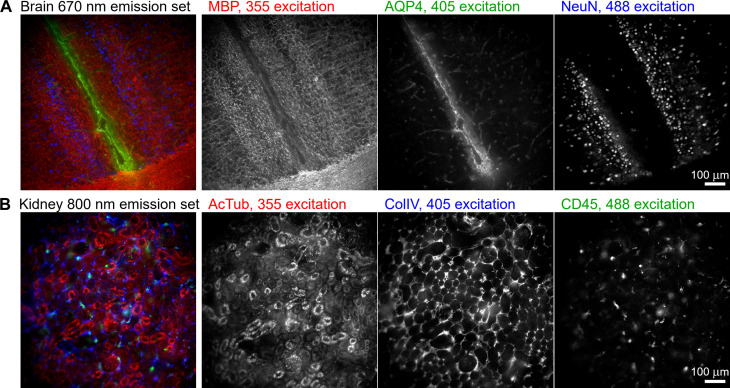
In addition to emission multiplexing, in which one excitation wavelength excites a set of Pdots that fluoresce across a range of emission wavelengths, Pdots can also be used for excitation multiplexing, in which Pdots that emit at the same wavelength are selectively excited using different illumination wavelengths. For example, in Fig. 3, we demonstrate three-channel excitation-multiplexed imaging of mouse brain tissue using Pdots that emit at 670 nm and that of mouse kidney tissue using Pdots that emit at 800 nm. In each case, the Pdots are selectively excited via illumination at 365, 405, and 488 nm. Unlike emission multiplexing, in which bleedthrough into neighboring emission channels is observed, here, we encounter some cross-excitation of Pdots that produces channel cross-talk (e.g., 405-nm illumination undesirably cross-excites the 488ex-Pdots at ~20%; see Fig. 1B). We corrected this cross-excitation in a similar manner as before using linear unmixing (see fig. S7 for a comparison of the raw and unmixed data). This ability to perform excitation multiplexing is unavailable to Qdots because they do not have distinct absorption peaks.
Fig. 3. Excitation multiplexing.
Three-channel excitation-multiplexed imaging of 100-μm-thick tissue sections of (A) mouse brain, imaged above the medial corpus callosum region, and (B) mouse kidney, imaged at the cortex region. Each set of three Pdots was illuminated at 365, 405, and 488 nm; a 667/30-nm emission filter was used for the brain, and an 800/60-nm emission filter for the kidney. MBP, myelin basic protein; AQP4, aquaporin 4; NeuN, neuronal nuclei; AcTub, acetylated tubulin; ColIV, collagen 4; CD45, leukocyte common antigen.
Excitation and emission multiplexing: Up to 21-plex imaging in a single round
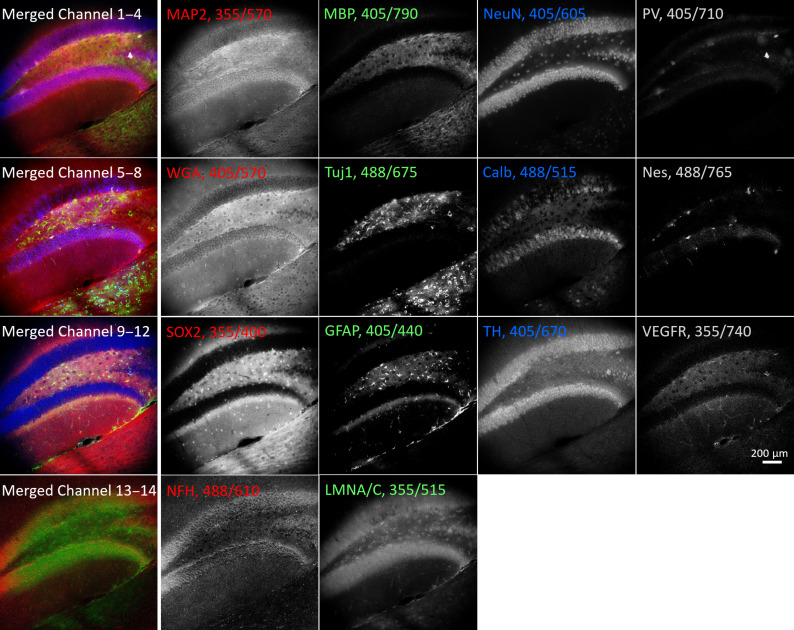
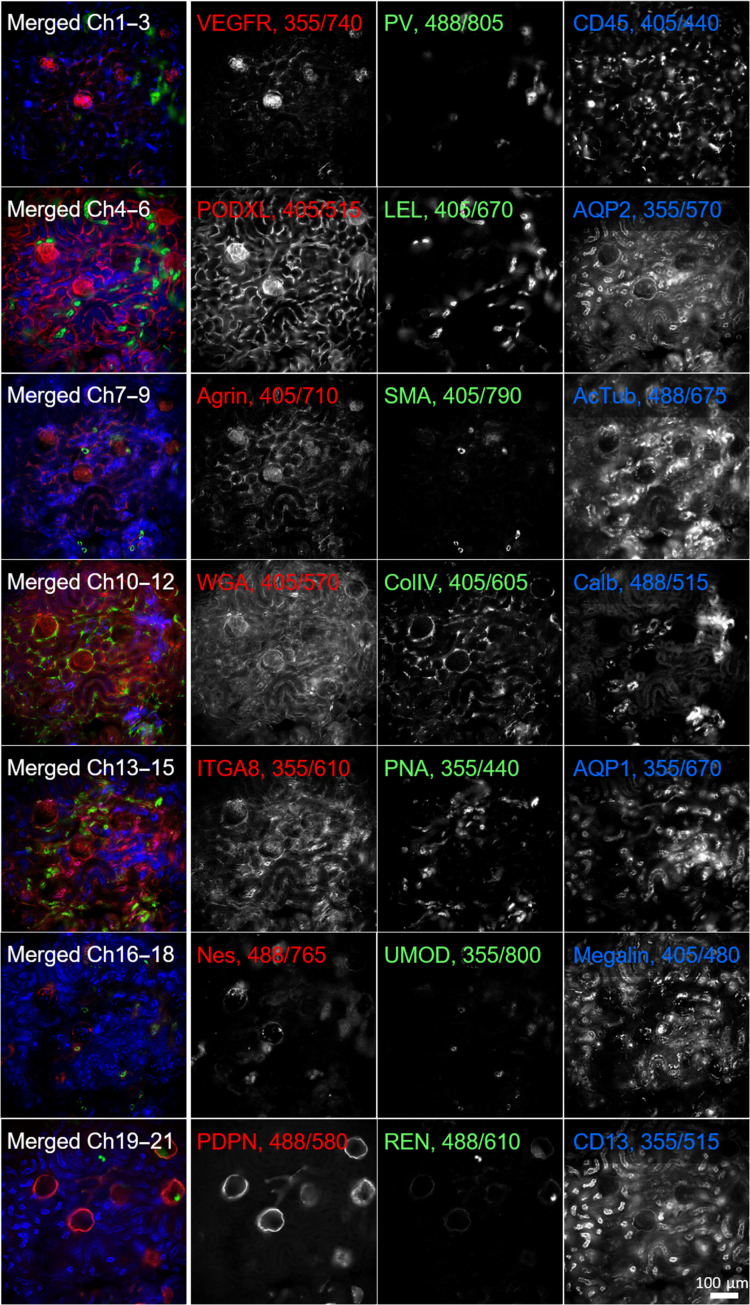
Having separately demonstrated the possibilities of using Pdots for excitation-multiplexed and emission-multiplexed imaging, we sought to combine them using our three series of Pdots to stain and visualize up to 21 targets in a single round. Using indirect immunofluorescence to stain a large number of targets in a single round is impractical due to limitations in the number of independent primary/secondary antibody combinations. Therefore, when creating larger palettes of probes, we chose to conjugate the Pdots directly to primary antibodies or lectins (see table S3). We validated independent antibodies or lectins as labeling their respective targets of interest, conjugated each antibody or lectin to a Pdot, revalidated the binding of antibody-Pdot conjugates to the original targets, and performed careful cross-talk calibration measurements for each Pdot. Using this procedure, we were able to perform 14-plex imaging of a mouse brain section and 21-plex imaging of a mouse kidney section, with the data acquisition for each being performed in under a minute (Figs. 4 and 5). As before, details of filters, antibodies, and Pdot conjugations are listed in tables S1 to S3, and the linear unmixing procedure is described in Materials and Methods.
Fig. 4. Excitation and emission multiplexing in brain tissue.
A 50-μm-thick mouse brain section showing the dentate gyrus of the hippocampus stained with 14 Pdots conjugated to primary antibodies and lectins. The tissue was imaged using a combination of 3 excitation wavelengths and 11 emission filters. The left column shows four partial four-channel composite images; the four columns to the right show corresponding grayscale images. Labels indicate the protein target or lectin (WGA) together with Pdot excitation/emission wavelengths. MAP2, microtubule-associated protein 2; MBP, myelin basic protein; NeuN, neuronal nuclei; PV, parvalbumin; WGA, wheat germ agglutinin; Tuj1, class III β-tubulin; Calb, calbindin; Nes, neuroepithelial stem cell protein; SOX2, sex-determining region Y-box 2; GFAP, glial fibrillary acidic protein; TH, tyrosine hydroxylase; VEGFR, vascular endothelial growth factor receptor 3; NFH, neurofilament H; LMNA/C, lamin A/C.
Fig. 5. Excitation and emission multiplexing in kidney tissue.
A 50-μm-thick mouse kidney section stained with 21 Pdots conjugated to primary antibodies and lectins. The tissue was imaged at the cortex region using a combination of 3 excitation wavelengths and 11 emission filters. The left column shows seven three-channel composite images of the same tissue region; the three columns to the right show corresponding grayscale images. Labels indicate the protein targets or lectins (LEL, WGA, and PNA) together with Pdot excitation/emission wavelengths. VEGFR, vascular endothelial growth factor receptor 3; PV, parvalbumin; CD45, leukocyte common antigen; PODXL, podocalyxin; LEL, Lycopersicon esculentum tomato lectin; AQP2, aquaporin 2; SMA = α–smooth muscle actin; AcTub, acetylated tubulin; WGA, wheat germ agglutinin; ColIV, collagen 4; Calb, calbindin; ITGA8, integrin α8; PNA, peanut agglutinin; AQP1, aquaporin 1; Nes, neuroepithelial stem cell protein; UMOD, uromodulin; Megalin, low-density lipoprotein–related protein 2, PDPN, podoplanin; REN, renin; CD13, aminopeptidase N.
We also investigated the compatibility of our workflow with formalin-fixed paraffin-embedded (FFPE) tissues in addition to the fresh fixed tissues shown above. This labeling was performed without any modifications to our brain and kidney panels of preconjugated Pdots and without any optimization of concentrations, cross-talk, or antigen retrieval. We chose 10 Pdot conjugated antibodies or lectins against targets that show broad expression in many organs, and we successfully stained and visualized targets in seven to nine channels each in the brain, kidney, thymus, prostate, and larynx (figs. S8 to S12). As before, these images were obtained using just three excitation wavelengths. In one to three channels per organ, we did not see a positive signal as the antibody targets were either not expressed (e.g., NeuN is only present in the brain) or in low abundance in those organs.
DISCUSSION
Multiplexed imaging is increasingly important to probe the interactions of various cell types in disease and in the context of discovery efforts such as tissue atlases. Our technique provides broad applicability because Pdots can be bound to commercially available conventional antibodies, which offers flexibility in the choice of cellular markers, antibody-Pdot combinations, etc. Compared to iterative staining methods (2–7), our method is simpler and faster and does not require probe inactivation reagents or suffer from tissue distortion. We make use of standard imaging hardware available in most bioimaging labs, unlike other multiplexing approaches that use more sophisticated hardware or may need dedicated instruments for automated fluidics or multiday acquisitions (6–11, 25). We have demonstrated that, in a single round of highly multiplexed imaging, we can concurrently investigate up to 21 targets in a single 50-μm-thick tissue specimen using three series of Pdots with excitation at 365, 405, and 488 nm. Additional 560- and 647-nm series of Pdots would enable excitation multiplexing (26) to a larger extent than we demonstrated here, and the number of targets could be increased further to ~30 in a single round through a combination of excitation and emission multiplexing with five series of Pdots. Combining this approach with antibody stripping or Pdot inactivation (e.g., via quenching or bleaching) may make it possible to measure over 100 targets in only four to five rounds of imaging.
In contrast to the above benefits, the use of Pdots for highly multiplexed imaging does face some challenges. The process of conjugating antibodies to Pdots places some restrictions on the choice of antibodies and requires careful pre- and postvalidation of antibody performance. For this study, we tested 54 Pdot-primary antibody (or Pdot-lectin) conjugates (table S3). Of these 54 conjugates, 31 consistently produced strong staining of the correct molecular targets, 10 produced weak staining, and 13 showed no staining or unsuitably high background indicative of nonspecific binding (see fig. S13 for the process of validation of Pdot-antibody conjugates). These results show an estimated ~60% success rate for Pdot conjugation. The failure rate of Pdot-primary antibody conjugates (~40% not binding their targets efficiently) is similar to that of DNA-conjugated primary antibodies and other approaches involving custom primary conjugates to conventional dyes (3). Purchasing different primary antibodies against the same target and repeating the conjugation often led to identifying a successful conjugate.
As part of our image processing workflow, we used linear unmixing—a simple and popular technique in the field of multiplexed imaging that uses raw/mixed image data and a reference/calibration matrix to generate unmixed spectral images (24, 27). Despite the technique’s simplicity, it requires a premeasurement of all the Pdots individually in each imaging channel before the unmixing can be performed. However, this calibration can be performed once for all subsequent experiments where the same imaging settings are used. Techniques like hyperspectral phasors (HySP) and blind source separation methods can unmix signals without prior measurements of reference spectra but generally perform worse when there are larger numbers of mixed channels (28–31). For instance, PICASSO (31) uses a sophisticated unmixing algorithm that seeks to maximize the differences between multiple channels but has some potential limitations in cases where channels may be innately similar to one another. In another approach, Lin et al. used a customized microscope with a seven-laser system and a continuously tunable emission filter together with variable Stokes shift fluorescent probes to achieve up to 19-channel immunofluorescence (25).
We also noticed higher levels of background when illuminating the 355-nm series of Pdots; at these shorter excitation wavelengths, biological tissues exhibit strong autofluorescence that can mask the specific staining or affect the unmixing calculation, generating images with autofluorescence contamination in the short wavelength channels. The use of thinner tissue sections or incorporating corrections for autofluorescence may improve the multiplexing performance of these Pdots. While most Pdots are bright and photostable, we noticed that some Pdots with larger Stokes shifts exhibit blue-shifting of their spectrum in a power density–dependent manner when exposed to bright focused light (e.g., when using a confocal microscope with a high–numerical aperture objective lens). This spectral shift can introduce errors during linear unmixing as signals tend to be weaker for Pdots with larger Stokes shift (i.e., redder emission channels). This may limit the current applications of Pdot multiplexing to microscopies that use lower power densities, such as wide-field and light sheet microscopy. In the future, engineering Pdots that do not exhibit spectral shifts would make them more broadly applicable and better suited to microscopy modes that involve very high excitation power densities.
With the availability of additional sets of Pdots excitable at 561 and 640 nm, together with increasing emission multiplexing and better compensation methods, it will be possible to further increase the number of targets that can be studied in a single round of staining and imaging. While fewer emission bands can be accommodated for these redder excitation wavelengths, incorporating these additional Pdot series could allow for probing ~30 targets in a single round. We are also investigating approaches to engineer these Pdots to have narrower excitation and emission bands to reduce bleedthrough and cross-excitation, which will further increase the number of targets that can be probed in a single round of imaging.
MATERIALS AND METHODS
Experimental design
This study aims to develop an approach to highly multiplexed fluorescence microscopy with a simple workflow and wide applicability in bioimaging laboratories with standard imaging hardware. The experimental design is based on the use of fluorescent semiconducting nanoparticles (Pdots) that have a large range of Stokes shifts and are spectrally distinct from each other, thus enabling combinations of excitation and emission multiplexing. Pdots are conjugated to commercially available antibodies of interest. By designing all experiments to be conducted in a single round of staining and imaging, our approach emphasizes simplicity, speed, and applicability to routine microscopy procedures.
Absorption and emission spectra
UV-Vis absorption spectra were recorded in a 1-cm quartz cuvette with a DU 720 scanning spectrophotometer (Beckman Coulter, Inc., CA USA). Fluorescence spectra were obtained using a Fluorolog-3 fluorometer (HORIBA Jobin Yvon, NJ USA).
Mouse brain and kidney sample preparation
All procedures used in this study were approved by the University of Washington Institutional Animal Care and Use Committee under protocol number 4442-01. Three mice were used in this study: mouse 1, a 6-month-old C57BL/6 Cre-ERT2 female; mouse 2, a 2-month-old C57BL/6 wild-type female; and mouse 3, a 5.5-month-old B6*P14 transgenic female. Tissues from these mice did not exhibit any fluorescent reporters. Immediately after CO2 euthanasia, mice were cardiac perfused with PBS-azide and 4% paraformaldehyde (PFA), each for 5 min. The kidneys, with renal capsule removed, were harvested and fixed in 10 ml of 4% PFA for 1 hour. The brain was fixed overnight in PFA. After fixation, the brain was embedded in agarose gel and sliced into 50- and 100-μm-thick sections on a vibratome along the coronal plane. The kidneys were sliced without embedding into 50- and 100-μm sections along the coronal plane. All sections were stored in PBS-azide at 4°C until further use.
Screening and validation of antibodies
The full list of primary and secondary antibodies used in this study is provided in tables S2 and S3. We validated the performance of primary antibodies by staining tissues with the primary antibody and then a prevalidated secondary antibody conjugated to conventional fluorescent dyes (such as ATTO 488, Alexa Fluor 568, or ATTO 647N). We confirmed that the antibodies bound the correct targets in desired tissues by comparing our images with the literature. Secondary antibodies were validated in a similar manner but in combination with a prevalidated primary antibody. After this first round of validation, the antibodies (primary or secondary) were conjugated to Pdots. In some cases, the conjugation could change the antibody’s structure, antigenicity, or binding efficiency. We therefore revalidated each antibody after conjugation to Pdots to confirm target binding via imaging. The process of conjugating antibodies to Pdots places some restrictions on the choice of antibodies. Some components typically found in the antibody buffer solution, such as bovine serum albumin, gelatin, albumin, and Tris buffer, can interfere with the conjugation reaction and make the yield harder to quantify. We chose to use antibodies without these components, which sometimes limited us to using antibodies that were not used widely in other papers.
Conjugation of antibodies to Pdots
The preparation of Pdot-conjugates has been described in detail in prior publications (12–14, 17, 21). Briefly, Pdots were conjugated to antibodies or lectin via a 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) hydrochloride–catalyzed reaction, in which 0.25 ml of a Pdot solution was mixed with 3 μl of EDC and 60 μl of an antibody solution and reacted for 3 hours. The final solution was then loaded onto a gel filtration column to purify the Pdot-antibodies from free unreacted antibodies.
Immunostaining
Tissues were incubated in a block/permeabilization solution (3% bovine serum albumin and 0.5% Triton-X 100) for at least 4 hours at room temperature (~23°C) on a shaker. All tissues were stained in a single round. For indirect immunofluorescence, tissues were stained overnight with a primary antibody and overnight with a Pdot-conjugated secondary antibody. For direct immunofluorescence, tissues were stained for 24 hours. Tissues were washed three times with PBS-azide for 10 min each and were imaged in a single round with the proper filter combinations (table S1). Luminescence tends to be dimmer for Pdots with larger Stokes shifts, and the quantum efficiency of standard scientific complementary metal-oxide semiconductor (sCMOS) cameras also declines toward the near-infrared region. To counter these issues, we paired the brightest immunostains with the dimmest Pdots and increased the concentration of the dimmest stains to achieve similar levels of signal intensity for each channel.
Immunostaining of FFPE tissue microarray
FFPE tissue microarray slides containing multiple organ sections of 5-μm thickness were purchased from TissueArray.com (MON621: from C57BL/6N mouse). The slides were baked at 60°C for 2 hours. Tissues were deparaffinized by incubation in 100% Histo-Clear (Electron Microscopy Sciences) for 30 min, followed by 50% Histo-Clear:50% ethanol for 5 min. Tissues were rehydrated by incubating in serial dilutions of ethanol in Milli-Q water: 100, 90, 80, 70, 60, and 50% ethanol for 5 min each, then lastly in Milli-Q water for 10 min. Antigen retrieval was performed with a sodium citrate buffer [10 mM sodium citrate and 0.05% Tween 20 (pH 6.0)], which was heated in the microwave for 35 s at high power. Slides were then inserted in the hot buffer, microwaved at low power for 2 to 3 min, cooled at room temperature (~23°C) for 50 min, and washed with PBS for 10 min before storing in PBS. The slides were then placed in a 50-ml conical tube with a block/permeabilization solution at room temperature for 2 hours on a shaker. A PAP pen was used to form a hydrophobic layer around the tissues. Tissues were stained with 10 Pdot-primary antibody conjugates in a single round of overnight staining in 250 μl of a block/perm solution. Staining was performed in a humidity chamber in the dark, and tissues were washed with PBS a few times the next day before imaging.
Imaging
All Pdot imaging was performed on a Nikon wide field microscope equipped with a motorized objective z-drive for focusing (Ti2-E, Nikon Instruments Inc.), automated XY stage (MS2000, Applied Scientific Instrumentation), motorized filter cube turrets, an assembly of motorized filter wheels (FW102C, Thorlabs), and a sCMOS camera (Orca Flash 4.0v3, Hamamatsu). All parts were controlled using the open-source Micromanager software (v2.0). Excitation was achieved by filtering the white light output of a SOLA U-nIR light engine (Lumencor) using three excitation filters. Emission filters were optimized to capture a band of 20 to 60 nm at the emission peaks of each Pdot.
For a few validation experiments, another Nikon wide field scope (Ti-S, Nikon Instruments Inc.) with four-channel light-emitting diode illumination (Thorlabs DC4104), quad-band dichroic, emission filters, and an Andor Zyla 5.5 sCMOS camera (Andor) was used. All dichroic, excitation, and emission filters are listed in table S1 and were purchased from Semrock or Chroma.
Linear unmixing
Mixed signals due to spectral overlap in each channel were computationally unmixed using a custom analysis script in MATLAB (v2024a). The emission profiles of each conjugated Pdot in solution were measured across the available emission filter channels to record the proportion of cross-talk and signal bleedthrough to neighboring channels. During initial stages of the project when excitation and emission multiplexing were performed separately, we used a simpler calibration procedure for just the few necessary channels. As more Pdots were conjugated and the full palette of 22 distinct Pdots was available, we performed a full calibration for each pure Pdot in solution (i.e., using each combination of illumination wavelength and emission channel for each Pdot) to produce a 22 × 22 calibration matrix. The inverse of the complex conjugate of this calibration matrix was multiplied to the raw mixed datasets to generate unmixed datasets using nonnegative least-squares fitting on a pixel-by-pixel basis.
Statistical analysis
Image data were processed using established software tools (e.g., ImageJ and MATLAB 2024a). Quantitative measurements, such as signal intensities, were extracted from regions of interest. Example raw data, including images and measurement values, are available in the Supplementary Materials and archived in Zenodo.
Acknowledgments
We gratefully acknowledge assistance from C. Mao in the initial stages of this project.
Funding: This work was supported by funding from the following: University of Washington (J.C.V. and D.T.C.), National Institutes of Health grant R01MH115767 (D.T.C. and J.C.V.), NIDDK Diabetic Complications Consortium grants DK076169 and DK115255 (J.C.V.), and Washington Research Foundation Postdoctoral Fellowship (C.P.)
Author contributions: Conceptualization: Z.G., C.P., J.Y., D.T.C., and J.C.V. Methodology: Z.G., C.P., J.Y., D.T.C., and J.C.V. Investigation: Z.G., C.P., and J.Y. Resources: Z.G., C.P., J.Y., M.W., M.C.S., D.T.C., and J.C.V. Data curation: Z.G., C.P., and J.Y. Validation: Z.G., C.P., J.Y., D.T.C., and J.C.V. Visualization: Z.G., C.P., D.T.C., and J.C.V. Formal analysis: Z.G., C.P., J.Y., and J.C.V. Software: Z.G., C.P., and J.C.V. Supervision: D.T.C. and J.C.V. Funding acquisition: D.T.C. and J.C.V. Project administration: D.T.C. and J.C.V. Writing—original draft: Z.G., C.P., J.Y., and D.T.C. Writing—review and editing: Z.G., C.P., M.C.S., J.Y., D.T.C., and J.C.V.
Competing interests: D.T.C., J.Y., and M.C.S. have a financial interest in Lamprogen Inc., which has licensed the Pdot technology from the University of Washington. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Two example datasets and the MATLAB source code used for linear unmixing of mixed signals have been deposited in Github (https://github.com/chetan-poudel/Pdot-linearunmixing) and archived in Zenodo (https://doi.org/10.5281/zenodo.11100240).
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S13
Tables S1 to S4
REFERENCES AND NOTES
- 1.Hickey J. W., Neumann E. K., Radtke A. J., Camarillo J. M., Beuschel R. T., Albanese A., McDonough E., Hatler J., Wiblin A. E., Fisher J., Croteau J., Small E. C., Sood A., Caprioli R. M., Angelo R. M., Nolan G. P., Chung K., Hewitt S. M., Germain R. N., Spraggins J. M., Lundberg E., Snyder M. P., Kelleher N. L., Saka S. K., Spatial mapping of protein composition and tissue organization: A primer for multiplexed antibody-based imaging. Nat. Methods 19, 284–295 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerdes M. J., Sevinsky C. J., Sood A., Adak S., Bello M. O., Bordwell A., Can A., Corwin A., Dinn S., Filkins R. J., Hollman D., Kamath V., Kaanumalle S., Kenny K., Larsen M., Lazare M., Li Q., Lowes C., Culloch C. C. M., Donough E. M., Montalto M. C., Pang Z., Rittscher J., Santamaria-Pang A., Sarachan B. D., Seel M. L., Seppo A., Shaikh K., Sui Y., Zhang J., Ginty F., Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl. Acad. Sci. U.S.A. 110, 11982–11987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin J.-R., Izar B., Wang S., Yapp C., Mei S., Shah P. M., Santagata S., Sorger P. K., Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife 7, e31657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radtke A. J., Kandov E., Lowekamp B., Speranza E., Chu C. J., Gola A., Thakur N., Shih R., Yao L., Yaniv Z. R., Beuschel R. T., Kabat J., Croteau J., Davis J., Hernandez J. M., Germain R. N., IBEX: A versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues. Proc. Natl. Acad. Sci. U.S.A. 117, 33455–33465 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray E., Cho J. H., Goodwin D., Ku T., Swaney J., Kim S.-Y., Choi H., Park Y.-G., Park J.-Y., Hubbert A., McCue M., Vassallo S., Bakh N., Frosch M. P., Wedeen V. J., Seung H. S., Chung K., Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell 163, 1500–1514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goltsev Y., Samusik N., Kennedy-Darling J., Bhate S., Hale M., Vazquez G., Black S., Nolan G. P., Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saka S. K., Wang Y., Kishi J. Y., Zhu A., Zeng Y., Xie W., Kirli K., Yapp C., Cicconet M., Beliveau B. J., Lapan S. W., Yin S., Lin M., Boyden E. S., Kaeser P. S., Pihan G., Church G. M., Yin P., Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat. Biotechnol. 37, 1080–1090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen K. H., Boettiger A. N., Moffitt J. R., Wang S., Zhuang X., Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah S., Lubeck E., Zhou W., Cai L., In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron 92, 342–357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giesen C., Wang H. A. O., Schapiro D., Zivanovic N., Jacobs A., Hattendorf B., Schüffler P. J., Grolimund D., Buhmann J. M., Brandt S., Varga Z., Wild P. J., Günther D., Bodenmiller B., Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Angelo M., Bendall S. C., Finck R., Hale M. B., Hitzman C., Borowsky A. D., Levenson R. M., Lowe J. B., Liu S. D., Zhao S., Natkunam Y., Nolan G. P., Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–442 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu C., Chiu D. T., Highly fluorescent semiconducting polymer dots for biology and medicine. Angew. Chem. Int. Ed. Engl. 52, 3086–3109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J., Rong Y., Kuo C.-T., Zhou X.-H., Chiu D. T., Recent advances in the development of highly luminescent semiconducting polymer dots and nanoparticles for biological imaging and medicine. Anal. Chem. 89, 42–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C., Schneider T., Zeigler M., Yu J., Schiro P. G., Burnham D. R., McNeill J. D., Chiu D. T., Bioconjugation of ultrabright semiconducting polymer dots for specific cellular targeting. J. Am. Chem. Soc. 132, 15410–15417 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye F., Wu C., Jin Y., Wang M., Chan Y.-H., Yu J., Sun W., Hayden S., Chiu D. T., A compact and highly fluorescent orange-emitting polymer dot for specific subcellular imaging. Chem. Commun. 48, 1778–1780 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan Y.-H., Ye F., Gallina M. E., Zhang X., Jin Y., Wu I.-C., Chiu D. T., Hybrid semiconducting polymer dot–quantum dot with narrow-band emission, near-infrared fluorescence, and high brightness. J. Am. Chem. Soc. 134, 7309–7312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rong Y., Wu C., Yu J., Zhang X., Ye F., Zeigler M., Gallina M. E., Wu I.-C., Zhang Y., Chan Y.-H., Sun W., Uvdal K., Chiu D. T., Multicolor fluorescent semiconducting polymer dots with narrow emissions and high brightness. ACS Nano 7, 376–384 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X., Yu J., Rong Y., Ye F., Chiu D. T., Uvdal K., High-intensity near-IR fluorescence in semiconducting polymer dots achieved by cascade FRET strategy. Chem. Sci. 4, 2143–2151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye F., Sun W., Zhang Y., Wu C., Zhang X., Yu J., Rong Y., Zhang M., Chiu D. T., Single-chain semiconducting polymer dots. Langmuir 31, 499–505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu I.-C., Yu J., Ye F., Rong Y., Gallina M. E., Fujimoto B. S., Zhang Y., Chan Y.-H., Sun W., Zhou X.-H., Wu C., Chiu D. T., Squaraine-based polymer dots with narrow, bright near-infrared fluorescence for biological applications. J. Am. Chem. Soc. 137, 173–178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen D., Wu I.-C., Liu Z., Tang Y., Chen H., Yu J., Wu C., Chiu D. T., Semiconducting polymer dots with bright narrow-band emission at 800 nm for biological applications. Chem. Sci. 8, 3390–3398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuo C.-T., Wu I.-C., Chen L., Yu J., Wu L., Chiu D. T., Improving the photostability of semiconducting polymer dots using buffers. Anal. Chem. 90, 11785–11790 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Zhang J., Yu J., Jiang Y., Chiu D. T., Ultrabright Pdots with a large absorbance cross section and high quantum yield. ACS Appl. Mater. Interfaces 14, 13631–13637 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.T. Zimmermann, J. Marrison, K. Hogg, P. O’Toole, “Clearing Up the Signal: Spectral Imaging and Linear Unmixing in Fluorescence Microscopy” in Confocal Microscopy, S. W. Paddock, Ed., vol. 1075 of Methods in Molecular Biology (Springer, 2014), pp. 129–148; https://link.springer.com/10.1007/978-1-60761-847-8_5. [DOI] [PubMed] [Google Scholar]
- 25.Lin J.-R., Chen Y.-A., Campton D., Cooper J., Coy S., Yapp C., Tefft J. B., McCarty E., Ligon K. L., Rodig S. J., Reese S., George T., Santagata S., Sorger P. K., High-plex immunofluorescence imaging and traditional histology of the same tissue section for discovering image-based biomarkers. Nat. Cancer 4, 1036–1052 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen K., Yan R., Xiang L., Xu K., Excitation spectral microscopy for highly multiplexed fluorescence imaging and quantitative biosensing. Light Sci. Appl. 10, 97 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.M. Alterman, Y. Y. Schechner, A. Weiss, Multiplexed fluorescence unmixing, in 2010 IEEE International Conference on Computational Photography (ICCP) (IEEE, 2010), pp. 1–8; http://ieeexplore.ieee.org/document/5585093/. [Google Scholar]
- 28.Fereidouni F., Bader A. N., Gerritsen H. C., Spectral phasor analysis allows rapid and reliable unmixing of fluorescence microscopy spectral images. Opt. Express 20, 12729–12741 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Cutrale F., Trivedi V., Trinh L. A., Chiu C.-L., Choi J. M., Artiga M. S., Fraser S. E., Hyperspectral phasor analysis enables multiplexed 5D in vivo imaging. Nat. Methods 14, 149–152 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Neher R. A., Mitkovski M., Kirchhoff F., Neher E., Theis F. J., Zeug A., Blind source separation techniques for the decomposition of multiply labeled fluorescence images. Biophys. J. 96, 3791–3800 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seo J., Sim Y., Kim J., Kim H., Cho I., Nam H., Yoon Y.-G., Chang J.-B., PICASSO allows ultra-multiplexed fluorescence imaging of spatially overlapping proteins without reference spectra measurements. Nat. Commun. 13, 2475 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S13
Tables S1 to S4