Abstract
Tumour innervation associates with worse patient outcomes in multiple cancers1,2, suggesting that it may regulate metastasis. We observed that highly metastatic murine mammary tumours acquired more innervation than less metastatic tumours. This enhanced innervation was driven by expression of the axon guidance molecule SLIT2 in tumour vasculature. Breast cancer cells induced spontaneous calcium activity in sensory neurons and elicited release of the neuropeptide substance-P (SP). Using three-dimensional co-cultures and in vivo models, we found that neuronal SP promoted breast tumour growth, invasion, and metastasis. Moreover, patient tumours with elevated SP exhibited enhanced lymph node metastatic spread. SP acted on tumoral tachykinin receptors (TACR1) to drive death of a small population of TACR1-high cancer cells. Single-stranded RNAs (ssRNAs) released from dying cells acted on neighbouring tumoral toll-like receptors 7 (TLR7) to activate a non-canonical pro-metastatic gene expression program, including PI3K-AKT signalling. This SP/ssRNA-induced Tlr7 gene expression signature associated with reduced breast cancer survival outcomes. Therapeutic targeting of this neuro-cancer axis with the TACR1 antagonist aprepitant, an approved anti-nausea drug, suppressed breast cancer growth and metastasis in multiple models. Our findings reveal that hyperactivation of sensory neurons regulates multiple aspects of metastatic progression in breast cancer via a therapeutically targetable neuropeptide/extracellular ssRNA sensing axis.
Nerve fibres have been detected within malignant tissues for decades3. Most solid tumours are innervated by the peripheral nervous system, receiving input from autonomic (sympathetic and parasympathetic) and/or sensory nerves. Primary tumours have been shown to secrete neurotrophic factors including members of the neurotrophin family4 or axon guidance molecules5 as a means of recruiting such innervation. However, the involvement of other stromal cells in regulating tumour innervation is unexplored. Tumour innervation has been increasingly implicated in experimental tumorigenesis. Glioma cells form functional synapses with neurons to promote tumour growth and invasion6–8. The autonomic nervous system has been shown to promote initiation of gastric9 and prostate10 tumours. Immunomodulatory roles have recently been reported for autonomic and sensory nerves in melanoma11,12. Healthy breast tissue receives abundant sensory innervation13,14, and there is pathological evidence of breast tumour innervation2. However, the role of sensory innervation in breast cancer metastasis remains poorly understood. In this study, we used 3D co-culture, genetic, and in vivo metastasis assays to define an intricate paracrine mechanism by which sensory nerves act directly on cancer cells to drive breast cancer metastasis.
Tumour endothelium regulates innervation
We recently identified that SLIT2, an axon guidance molecule, was expressed in the endothelium of metastatic tumours15. Consistent with this, we observed that SLIT2 was overexpressed in the endothelium of highly metastatic breast tumours (4T1, EO771 LM2, HCC1806 LM2) relative to isogenic, poorly metastatic tumours (Extended Data Fig. 1a, b). Given its established role in axon guidance16, we hypothesized that ecSLIT2 may regulate innervation of primary breast tumours. To test this, we performed orthotopic transplantations of 4T1 breast cancer cells into an inducible knockout model (Cdh5(PAC)-creERT2) in which tamoxifen was used to drive endothelial-specific deletion of Slit2. Control mice harboured Slit2-floxed alleles but were Cre-negative and received equal amounts of tamoxifen. ecSLIT2 loss strongly reduced tumoral innervation, including a significant reduction in sensory innervation (Extended Data Fig. 1c, d). In contrast, loss of SLIT2 in the tumour compartment did not alter tumour innervation (Extended Data Fig. 1e). These data implicate SLIT2 originating from the tumour vasculature as a driver of breast tumour innervation.
Innervation predicts metastatic capacity
Our prior work established ecSLIT2 as a driver of metastasis. Given its ability to regulate tumour innervation, we hypothesized that innervation may regulate metastatic progression. In a survey of publicly available patient data, we observed that breast tumours expressing higher levels of pan-neuronal markers (βIII-tubulin or PGP9.5), exhibited higher rates of metastatic recurrence (Extended Data Fig. 1f). In murine models, immunofluorescence staining and western blotting for βIII-tubulin revealed that highly metastatic breast tumours (4T1, EO771 LM2, HCC1806 LM2) were more innervated than their respective less metastatic isogenic tumours (67NR/4T07, EO771, HCC1806 respectively; Extended Data Fig. 1g, h). Importantly, βIII-tubulin expression arose from the stromal compartment, consistent with its neuronal expression (Extended Data Fig. 1h). Metastatic mammary tumours at distant organ sites were also innervated (Extended Data Fig. 1i). Moreover, across two independent patient cohorts totalling 30 breast tumours that we analysed, increased innervation associated with increased lymph node dissemination (Fig. 1a, b). Collectively, these findings reveal that the extent of tumoral innervation correlates with metastatic propensity.
Figure 1: Sensory neurons promote invasion, proliferation, and metastasis in breast cancer.
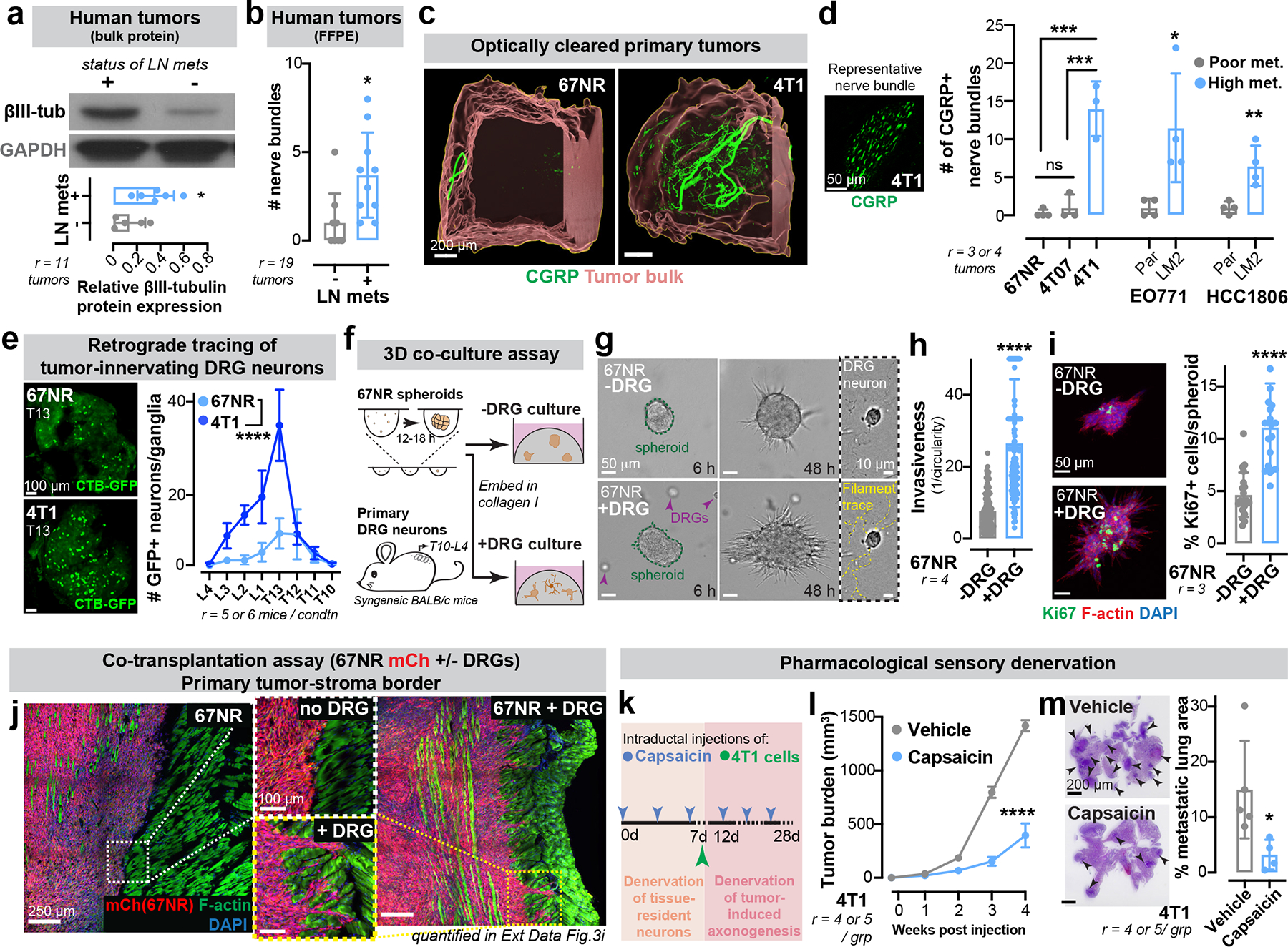
(a-b) Breast tumours from patients with lymph node spread express higher βIII-tubulin than those with localized disease. Cohort 1 (a), r=11; *p = 0.0329. Cohort 2 (b), r=19; *p = 0.0119. t-test, Mean±SD. MW: βIII-tubulin (50 kDa).
(c) CGRP+ sensory innervation in poorly (67NR) or highly (4T1) metastatic optically cleared mammary tumours. r = 3 tumours/ group.
(d) Left: CGRP+ sensory nerve bundle. Right: CGRP+ innervation in highly metastatic primary tumours relative to corresponding isogenic poorly metastatic tumours. ***p (67NR/4T1) = 0.0002, ***p (4T07/4T1) = 0.0003, nsp (67NR/4T07) = 0.6636 by one-way ANOVA; *p (EO771 Par/LM2) = 0.0272, **p (HCC1806 Par/LM2) = 0.0073, Mann-Whitney test. Mean ± SD.
(e) Retrograde tracing of DRG neurons innervating 67NR/4T1 tumours using GFP-tagged cholera toxin β (CTB-GFP). Scale, 100 μm. ****p < 0.0001, t-test of AUC. Mean±SEM.
(f-i) Co-culture of 67NR cancer cell spheroids and DRG neurons isolated from syngeneic mice in 3D collagen I. (f) Schematic. (g) Time-series of 67NR cancer cell spheroids and DRG neurons. Quantification of spheroid invasion (h) and proliferation (i). ****p < 0.0001, Mann-Whitney test, Mean ± SD.
(j) Representative tile scans of primary tumours arising from mCh+ 67NR cancer cells transplanted with or without DRG neurons; enlarged insets of the tumour-stroma border are shown. Quantified in Extended Data Fig. 3e.
(k-m) Sensory nerve-specific denervation of 4T1 mammary tumours. (k) Schematic. (l) Tumour growth. ****p < 0.0001, t-test. Mean ± SEM. (m) Metastatic area quantified by H&E. *p = 0.0385, t-test. Mean ± SD.
To identify the source of peripheral innervation for breast tumours, GFP-tagged cholera-toxin β (CTB-GFP) was injected intraductally to retrogradely label neurons innervating the abdominal mammary glands of mice (Extended Data Fig. 1j). We identified dorsal root ganglia (DRG; T10-L4) sensory neurons as a predominant source of innervation for these mammary glands (Extended Data Fig. 1j, k). Importantly, we also observed abundant sensory innervation marked by calcitonin gene-related peptide (CGRP) in syngeneic (4T1 and EO771) and genetically initiated (MMTV-PyMT and C3(1)-TAg) mouse breast cancer models, a patient-derived xenograft (PDX) model of HER2+ breast cancer, and three independent primary human breast tumours (Extended Data Fig. 1l). Further supporting sensory innervation of metastatic breast cancer, we observed a significantly higher degree of sensory innervation in optically cleared 4T1 tumours relative to isogenic poorly metastatic 67NR tumours (Fig. 1c). Immunofluorescence-based quantification of CGRP+ nerve bundles revealed increased sensory innervation in highly metastatic tumours relative to isogenic, poorly metastatic tumours (Fig. 1d). Additionally, intra-tumoral nerve-tracing using CTB-GFP revealed far greater neurons within the dorsal root ganglia innervating highly metastatic 4T1 tumours than 67NR tumours (Fig. 1e). These findings reveal that sensory innervation is enhanced in breast tumours with higher metastatic propensity.
Sensory innervation drives metastasis
To determine if sensory neurons impact metastatic phenotypes, we developed a 3D co-culture system comprised of 67NR (poorly metastatic) cancer cell spheroids and primary sensory neurons dissected from the DRGs of syngeneic naïve mice (Fig. 1f). Spheroids and neurons were embedded in 3D collagen I, a protein highly enriched in breast cancer stroma17. During the culture, DRG neurons cultured with cancer cells exhibited enhanced viability relative to those cultured alone (Extended Data Fig. 2a), and extended axons into the surrounding matrix (Fig. 1g, Extended Data Fig. 2b). Upon co-culture with DRG neurons, 67NR breast cancer spheroids exhibited significantly more invasion (Fig. 1 g, h, Extended Data Fig. 2c) and proliferation (Fig. 1i). Consistent with these in vitro findings, cancer cells adjacent to nerve bundles in 4T1 primary tumours were more proliferative than cancer cells from a distant region of the same tumour (Extended Data Fig. 2d). DRG neurons enhanced the colony forming capacity of 67NR breast cancer cells within Matrigel in a cell-dose dependent manner (Extended Data Fig. 2e, f). DRGs similarly increased the invasiveness and colony formation capacity of MMTV-PyMT derived organoids and cancer cell clusters, respectively (Extended Data Fig. 2 g–i). We next investigated the effect of DRGs on primary human tumour organoids; organoids were isolated from four distinct patients’ breast tumours (3/4 ER+ PR+ HER2- and 1/4 ER+ PR+ HER2+; Extended Data Fig. 2m) and co-cultured with murine DRG neurons (Extended Data Fig. 2j). Consistent with our findings in mouse models, DRG neurons strongly increased invasion and proliferation of organoids from all four human tumours (Extended Data Fig. 2k, l). These findings reveal that DRG neurons enhance the proliferative and invasive phenotypes of murine and human breast cancer cells in vitro.
Next, we assessed the impact of sensory innervation on breast cancer metastasis in vivo. To do this, we sought to experimentally enhance innervation of poorly metastatic 67NR tumours and conversely to reduce innervation of highly metastatic 4T1 tumours. We first co-transplanted a small number of DRG neurons along with mCherry-labelled 67NR cancer cells into syngeneic BALB/cJ hosts (Extended Data Fig. 3a). Tumoral mCherry expression allowed us to track cancer cells within non-fluorescent hosts as they progressed through the metastatic cascade. Consistent with neuronal viability and functionality upon co-injection with cancer cells in vivo, we observed a significant increase in the extent of sensory innervation (Extended Data Fig. 3b) and neurotransmitter expression in these tumours (Extended Data Fig. 3c). Importantly, co-transplantation with sensory neurons significantly enhanced the growth of 67NR primary tumours (Extended Data Fig. 3d) in a neuronal cell-dose dependent manner (Extended Data Fig. 3i). DRGs significantly promoted the collective invasion of 67NR cancer cells into the surrounding muscle (Fig. 1j, Extended Data Fig. 3e). It is widely believed that 67NR cancer cells fail to metastasize due to their inability to intravasate18. However, the use of a fluorescent reporter and imaging at single-cell resolution revealed the presence of hundreds of 67NR cancer cells within the lungs of mice (Extended Data Fig. 3f). A vast majority of these cells were associated with the lung endothelium, consistent with metastatic failure due to impaired extravasation (Extended Data Fig. 3f). A minority of 67NR cells had exited the lung endothelium to form micro-metastases. This contrasted with isogeneic 4T07 cells, which efficiently formed micro-metastases, and 4T1 cells, which efficiently formed macro-metastases18. Mice co-transplanted with 67NR cells and DRGs harboured a significantly higher number of circulating mCherry+ tumour cells and micro-metastases (Extended Data Fig. 3g, h respectively). Additionally, we co-injected 67NR cancer cells with DRG neurons into the tail veins of mice (Extended Data Fig. 4a) and observed neuronal cell bodies within the lung epithelium, consistent with neuronal viability upon injection in vivo (Extended Data Fig. 4b). We observed that DRG neurons significantly enhanced metastatic colonization by 67NR cancer cells in an experimental metastasis assay as well (Extended Data Fig. 4c). Consistently, we also observed enhanced tumour growth and micro-metastases in co-transplantation experiments performed with EO771 breast cancer cells (Extended Data Fig. 4 d–f). These findings reveal that DRG neurons are sufficient to enhance the metastatic capacity of breast cancer cells in co-transplantation experiments in vivo.
We next performed sensory-specific denervation of 4T1 tumours using capsaicin, a TRPV1 agonist and neurotoxin that degenerates sensory nerves. We first injected capsaicin into the mammary ductal tree to remove tissue-resident sensory innervation, then intraductally injected 4T1 cancer cells while continuing to administer capsaicin to maintain a sustained denervated state (Fig. 1k). By performing capsaicin injections intraductally, we restricted its effects to the mammary ductal epithelium. Immunofluorescence analysis confirmed successful denervation of sensory nerves within the ipsilateral mammary gland (Extended Data Fig. 5a, b). We discovered that sensory-specific denervation greatly reduced tumour growth (Fig. 1l) and suppressed the number of macro-metastases (quantified in mice bearing size-matched primary tumours; Fig. 1m). Importantly, capsaicin had no effect on the invasiveness or proliferation of 4T1 cancer cells in vitro, consistent with the impact of capsaicin on these phenotypes occurring via its effects on neurons, rather than on tumour cells (Extended Data Fig. 5 c–d). To further control for possible confounding effects of TRPV1 agonism in immune cells19, we repeated these experiments in NOD-SCID gamma (NSG) mice bearing 4T1 tumours and observed similar anti-tumour effects of capsaicin (Extended Data Fig. 5 e–f). Collectively, these findings demonstrate the requirement for sensory innervation in breast cancer progression and metastasis.
Neuronal substance-P drives metastasis
Cancer cells are known to migrate along nerves in a process called perineural invasion20, requiring physical contact. In our co-culture models, however, we did not detect physical interactions between neurons and cancer cells (Extended Data Fig. 2b). Therefore, we hypothesized that DRG neurons may mediate pro-metastatic effects via secreted molecules. To test this, we harvested conditioned medium from 67NR tumour-DRG co-cultures (DRG-CM; Fig. 2a). We used conditioned medium from 67NR tumour only cultures as control (tumour-CM). Treatment of 67NR cancer cells with DRG-CM phenocopied the invasion and proliferation effects of DRG neurons (Figure 2 b,c), consistent with a secreted mediator.
Figure 2: Neuronal substance-P promotes breast cancer metastasis.
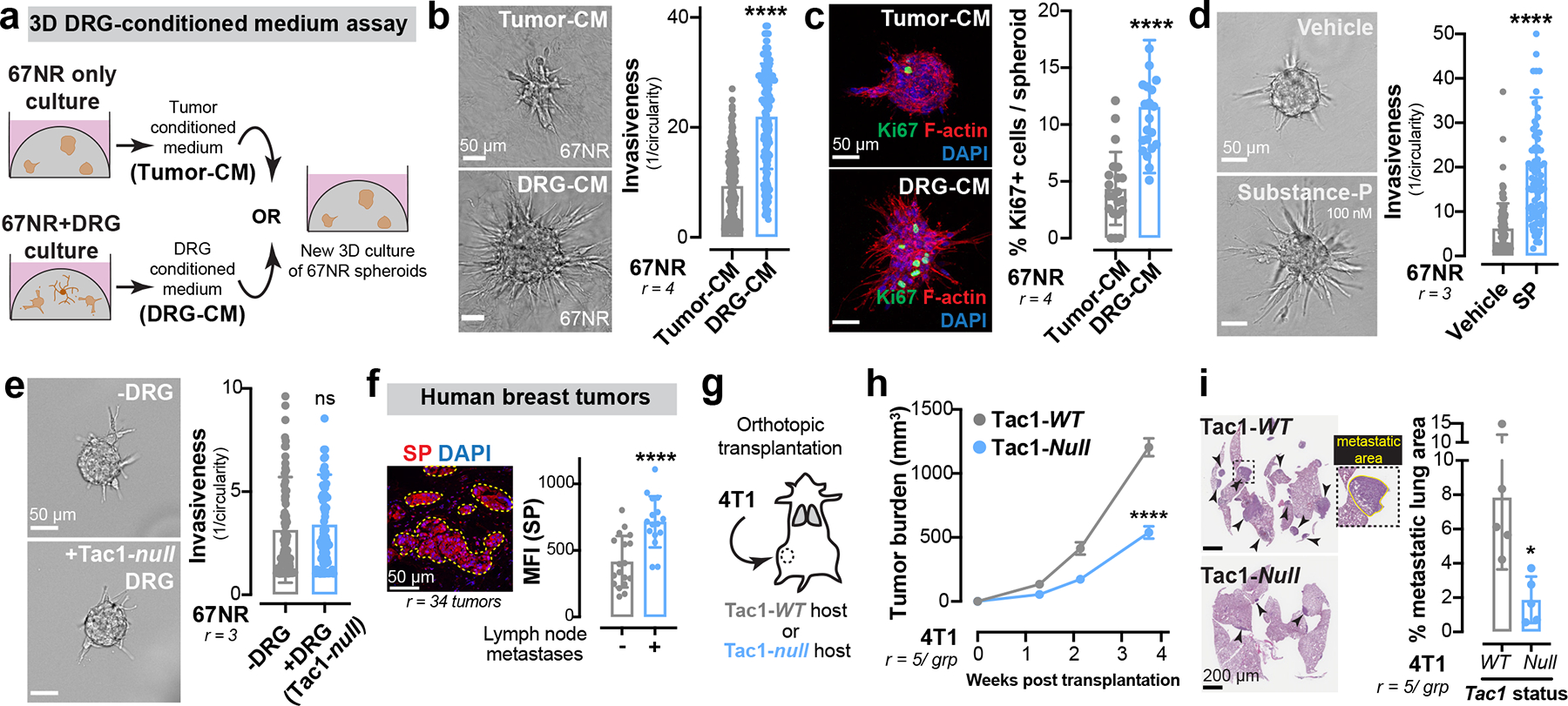
(a-c) Tumour-CM was isolated from a 3D culture of 67NR spheroids. DRG-CM was isolated from a 3D co-culture of 67NR spheroids and DRG neurons. (a) Schematic. Quantification of spheroid invasion (b) and proliferation (c). ****p < 0.0001, Mann-Whitney test. Mean ± SD.
(d) Invasion quantification of 67NR spheroids cultured with/without substance-P (SP). ****p < 0.0001, Mann-Whitney test. Mean ± SD.
(e) Invasion quantification of 67NR spheroids co-cultured with/without DRG neurons isolated from Tac1-null mice. nsp = 0.3070, Mann-Whitney test. Mean ± SD.
(f) Left: A primary human breast tumour immuno-stained for SP with the highlighted tumour region (ROI) used for mean fluorescence intensity (MFI) measurements. Right: MFI of SP in breast tumours of patients with lymphatic spread compared to those with localized disease. ****p < 0.0001, t-test. Mean ± SD.
(g-i) Orthotopic transplantation of 4T1 cells into the abdominal mammary glands of Tac1-WT and Tac1-null host mice. (g) Schematic. (h) Tumour growth. ****p < 0.0001, t-test. Mean ± SEM. (i) Metastatic area quantified by H&E. *p = 0.0165, t-test. Mean ± SD.
Sensory DRG neurons secrete neuropeptides that stimulate various biological processes. We tested the ability of three common DRG-secreted neuropeptides21, CGRP, substance-P (SP), and galanin to promote invasion of 67NR spheroids into collagen I. We found that SP and galanin significantly increased 67NR spheroid invasion and proliferation (Extended Data Fig. 6 a–c), with SP exerting the largest magnitude effect, while CGRP exhibited a modest effect (Fig. 2d, Extended Data Fig. 6b). Importantly, DRG-CM contained higher levels of SP than tumour-CM (Extended Data Fig. 6d). As an orthogonal approach, we performed a co-culture containing SP-deficient DRG neurons isolated from Tac1-null mice, which lack the Tac1 gene that encodes the precursor protein for SP). We found that Tac1−/− DRG neurons had no impact on the invasiveness of 67NR spheroids (Fig. 2e), demonstrating that neuropeptide SP is required for the pro-invasive effects of DRG neurons on breast cancer cells.
Mice bearing highly metastatic 4T1 tumours exhibited higher SP expression in the tumour compartment and in plasma (Extended Data Fig. 6e, f) relative to mice with poorly metastatic 67NR tumours. This SP expression was innervation-dependent since intraductal treatment of 4T1 tumours with capsaicin, a sensory neurotoxin, significantly reduced tumoral SP levels (Extended Data Fig. 6g). Importantly, SP expression was higher in primary tumours of patients with lymph node metastases relative to those with localized disease (Fig. 2f). Inhibition of extracellular SP, through intraductal administration of an anti-SP neutralizing antibody (Extended Data Fig. 6h, i), strongly reduced tumour growth and metastasis (quantified in mice bearing size-matched primary tumours; Extended Data Fig. 6j, k). Significantly, mice lacking host-derived SP (Tac1-null) exhibited significantly reduced orthotopic tumour growth and metastasis across multiple syngenetic breast cancer models (Fig. 2 g–i, Extended Data Fig. 6 l–o). These data identify neuronal substance-P as a driver of breast tumour growth and metastasis.
Biological activity of SP is mediated through tachykinin receptor 1 (TACR1 or NK1R)21. To determine if SP mediates metastasis via tumoral TACR1, we used lentiviral shRNA-mediated silencing to deplete Tacr1 in 4T1 cancer cells (Extended Data Fig. 6p). TACR1 depletion significantly reduced 4T1 spheroid invasion and proliferation in vitro (Extended Data Fig. 6q, r) and 4T1 tumour growth and metastasis (quantified in mice bearing size-matched primary tumours; Extended Data Fig. 6 s–u) in vivo.
SP causes ssRNA release from cancer cells
We observed that conditioned medium from DRG neurons alone (DRGonly-CM) was not sufficient to increase the invasiveness of 67NR spheroids (Extended Data Fig. 7 a, b) and contained less SP than conditioned medium from the 67NR+DRG co-culture described above (DRG-CM, Extended Data Fig. 6d). Release of SP from DRG neurons is triggered by an increase in intracellular calcium22. This suggests that exposure to cancer cells alters the calcium activity of DRG neurons. To test this hypothesis, we quantified calcium levels with the Fluo-4 calcium dye (Fig. 3a). As previously reported23, DRG neurons cultured in isolation showed minimal change in calcium levels (Fig. 3b, Extended Data Fig. 7c). In contrast, when cultured with cancer cells, DRG neurons exhibited spontaneous calcium spikes (Fig. 3b, Extended Data Fig. 7c). Consistently, we observed increased SP release from DRG neurons cultured with tumour-CM (Extended Data Fig. 6d). Furthermore, DRG neurons were unable to drive invasiveness of cancer cell spheroids upon treatment with tetrodotoxin, a sodium channel blocker that inhibits action potentials (Extended Data Fig. 7d). These findings reveal a bi-directional crosstalk between cancer cells and sensory neurons, whereby cancer cells enhance sensory neuron activity, driving SP release, which then acts on tumoral TACR1 to drive metastasis.
Figure 3: Neuronal SP promotes the release of metastatic ssRNAs from cancer cells.
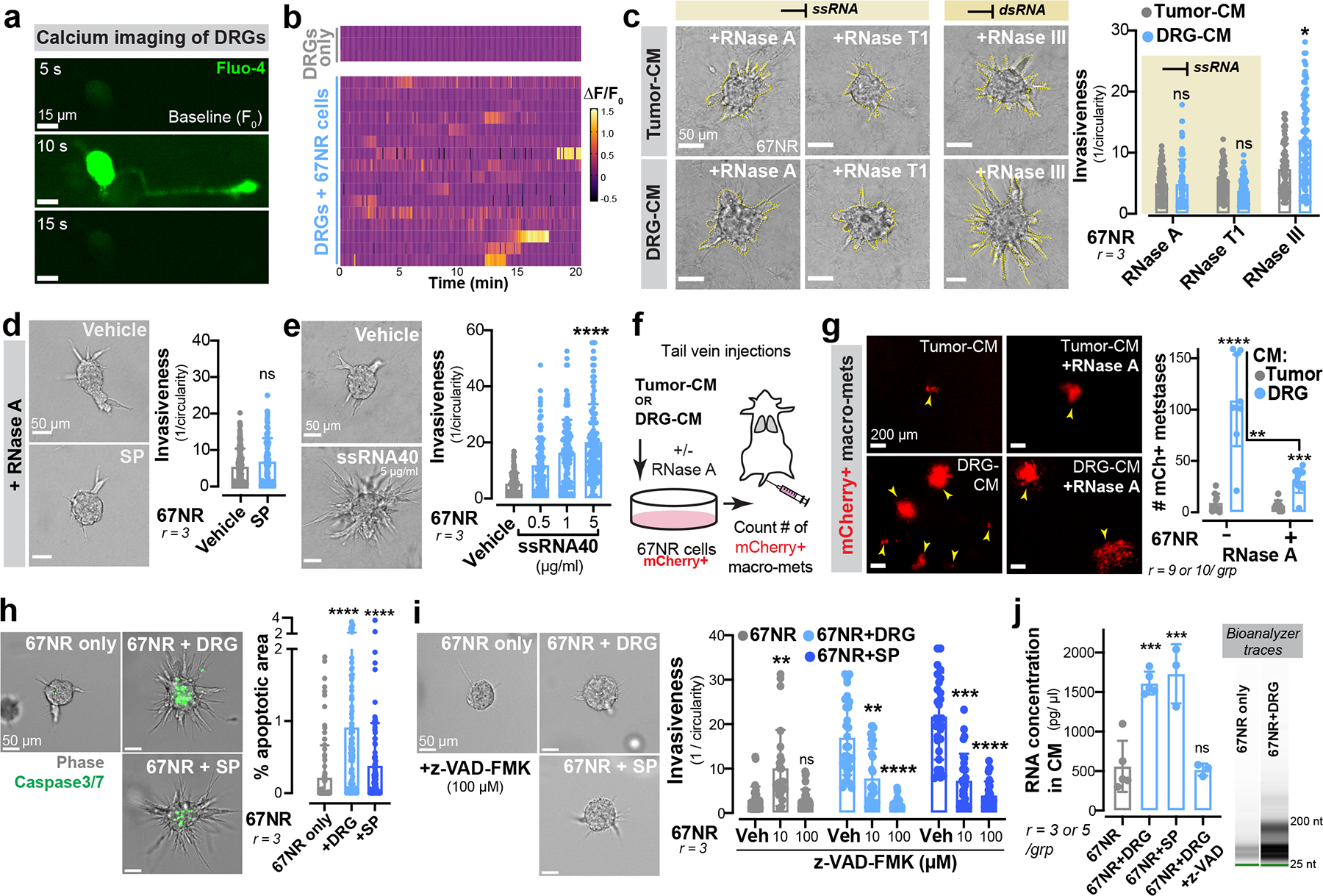
(a) Time series of a DRG neuron co-cultured with 67NR cancer cells in the presence of the calcium dye Fluo-4.
(b) Spontaneous calcium events (ΔF/F0) in DRG neurons cultured with or without 67NR cancer cells.
(c) 67NR spheroids cultured in tumour-CM or DRG-CM in the presence of either ssRNA- or dsRNA-specific RNases. Left: Representative images. Right: Invasion quantification. nsp > 0.9999, *p = 0.0215, Kruskal-Wallis test. Mean ± SD.
(d) Invasion quantification of 67NR spheroids cultured with SP and RNase A. Vehicle-treated 67NR spheroids (+/−RNase A) are plotted in Extended Data Fig. 6b since the experiments were conducted together. nsp = 0.2416, Mann-Whitney test. Mean ± SD.
(e) Invasion quantification of 67NR spheroids cultured with a ssRNA mimetic. ****p < 0.0001, Kruskal-Wallis test. Mean ± SD.
(f-g) mCherry+ 67NR cancer cells were pre-treated with tumour-CM or DRG-CM with or without RNase A prior to tail vein injections into syngeneic hosts. (f) Schematic. (g) Quantification of mCherry+ metastases. ****p < 0.0001, ***p = 0.0005, **p = 0.0015, Mann-Whitney test. Mean ± SD.
(h) Percent apoptotic area within 67NR cancer cell spheroids when cultured alone, with DRG neurons, or with SP. ****p < 0.0001, Kruskal-Wallis test. Mean ± SD.
(i) Invasion quantification of 67NR spheroids cultured alone, with DRG neurons, or with SP – all in the presence of z-VAD-FMK, a pan-caspase inhibitor. Veh = vehicle. 67NR only group: **p = 0.0056, nsp > 0.9999; 67NR+DRG group: **p = 0.0083, ****p < 0.0001; 67NR + SP group: ***p = 0.0004, ****p < 0.0001. Kruskal-Wallis test.
(j) RNA concentration within conditioned medium of 67NR only, 67NR+DRG (+/−z-VAD-FMK), 67NR+SP cultures. Extracellular RNA species detected are <200 nucleotides. ***p = 0.0002, nsp = 0.8248, ANOVA. Mean ± SD.
To systematically dissect the molecular basis by which SP (within DRG-CM) drives metastasis, we treated DRG-CM with DNase, RNase A, or heat inactivation to degrade proteins. Treatment of DRG-CM with RNAse A, but not DNase or heat inactivation, significantly impaired the ability of DRG-CM to promote invasion of 67NR spheroids (Fig. 3c; Extended Data Fig. 7e). Using RNases specific for single-stranded RNA (ssRNA, RNase T124) or double stranded RNA (dsRNA, RNase III25), we implicated ssRNAs as mediators of the pro-invasive effects of DRG-CM on breast cancer cells (Fig. 3c, Extended Data Fig. 7f).
We next sought to determine if SP and ssRNAs signal through a common pathway that drives breast cancer metastasis. We observed that treatment of 67NR spheroids with SP in the presence of RNase A completely abolished the invasion-promoting effects of SP (Fig. 3d) but did not alter the effects of CGRP or galanin (Extended Data Fig. 6b). Because the above experiments were performed using exogenously added SP and in the absence of sensory neurons, we propose a model wherein sensory neurons secrete SP, which in turn promotes the release of ssRNA molecules from breast cancer cells that drive invasion and proliferation.
Cancer cell dissemination is associated with increased levels of circulating nucleic acids26 and decreased nuclease activity in the plasma of cancer patients27. The use of RNases as anti-tumour agents dates to the 1950s, often with contradictory effects28,29. To define the role of ssRNA in breast cancer metastasis, we treated 67NR spheroids with a ssRNA mimetic, ssRNA40. Treatment with ssRNA40, but not dsRNA mimetic Poly I:C significantly increased spheroid invasion and proliferation (Fig. 3e; Extended Data Fig. 7 g, h). Moreover, intraductal injections of two independently generated recombinant RNase A enzymes significantly impaired tumour growth and metastasis of 4T1 cancer cells without any observed impact on dsRNA levels (Extended Data Fig. 7 i–l).
Our in vitro studies revealed ssRNAs within DRG-CM as promoters of tumour invasion. To assess the impact of DRG-CM on metastasis in vivo, we pre-treated 67NR cancer cells with DRG-CM or tumour-CM for two days prior to tail vein injections (Fig. 3f). Pre-treatment with DRG-CM was sufficient to confer metastasis-forming capacity onto the poorly metastatic 67NR cells, demonstrated by the formation of hundreds of macro-metastases (Fig. 3g). 67NR cancer cells pre-treated with DRG-CM also formed significantly more metastases to the lung, liver, brain, and ovaries upon intra-cardiac injection (Extended Data Fig. 7 m–o). Pre-treatment of 67NR cancer cells with DRG-CM supplemented with RNase inhibited the ability of DRG-CM to confer a metastasis promoting phenotype (Fig. 3g). In further support of the pro-metastatic effects of this axis, we found that SP and ssRNA40 promoted the invasiveness of spheroids from two additional murine breast cancer cell lines (4T1 and Py8119 cells) and a human breast cancer cell line (MDA-MB-231; Extended Data Fig. 8a, b).
Neuronal SP could induce ssRNA release by cancer cells either via a regulated secretory pathway or by cell death. In support of the latter model, we identified a small (<1% of spheroid area) but significant increase in cancer cell apoptosis upon co-culture with neurons or upon treatment with SP (Fig. 3h). In contrast, the neuropeptide galanin did not promote cancer cell death (Extended Data Fig. 9a). Importantly, addition of a pan-caspase inhibitor (z-VAD-FMK) blocked the pro-invasive effects of both DRG co-culture and SP treatment (Fig. 3i). While co-culture with DRG neurons and SP treatment increased the amount of RNA released into the conditioned medium, addition of z-VAD-FMK prevented RNA release (Fig. 3j). Collectively, these data support a model whereby DRG neurons release SP, which induces apoptosis in a subset of cancer cells and subsequent ssRNA release.
Using immunofluorescence and flow cytometry analyses, we found that a small sub-population of cancer cells expressed high levels of the SP receptor TACR1 (Extended Data Fig. 9b, c). We hypothesized that these TACR1-high cells may be most susceptible to cell death upon SP treatment. Consistent with this, we found a ~40% reduction in the number of TACR1-high cancer cells after co-culture with DRG neurons or SP treatment (Extended Data Fig. 9b, c), suggesting that heterogeneous expression of tumoral TACR1 underlies heterogeneous cell death upon SP exposure. In further support of this, depletion of tumoral TACR1 suppressed the ability of SP to induce cell death (Extended Data Fig. 9d, e) and invasion (Extended Data Fig. 9f). Taken together, these data demonstrate that neuronal SP causes cell death in a sub-population of TACR1-high cancer cells, causing release of ssRNAs that drive metastasis.
Sensory nerves signal via tumoral TLR7
We next sought to define the mechanism by which extracellular ssRNAs promote breast cancer metastasis. In mice, ssRNAs are detected by the TLR7 receptor, which mediates innate immune RNA sensing and effector responses30. To determine if tumoral TLR7 mediates the downstream pro-metastatic effects of ssRNAs, we depleted Tlr7 in 67NR cancer cells prior to co-culture with primary DRG neurons (Extended Data Fig. 9g). TLR7-depleted breast cancer spheroids did not exhibit enhanced invasiveness or proliferation upon co-culture with DRG neurons (Fig. 4a, b). In contrast, tumour spheroids lacking the dsRNA sensor Tlr3 co-cultured with DRGs retained their invasiveness (Extended Data Fig. 9h, i). Moreover, SP was unable to promote invasion of Tlr7-depleted 67NR spheroids (Fig. 4c), suggesting signalling through a common pathway. Treatment of 67NR spheroids with synthetic TLR7 agonists such a R837 (imiquimod, TLR7-specific agonist) or R848 (resiquimod, TLR7/8 dual-agonist) also significantly increased spheroid invasion (Extended Data Fig. 9j). Additionally, lentiviral shRNA-mediated depletion of Tlr7 in 4T1 cancer cells reduced spheroid invasion in vitro (Extended Data Fig. 9k, l), and tumour growth and metastasis in vivo (Fig. 4 d–f). The anti-metastatic effects of Tlr7 depletion were immune-independent since (i) we did not observe changes in the abundance of tumour-infiltrating CD45+, CD3+, or NK1.1+ immune cells upon TLR7 loss (Extended Data Fig. 9m), and (ii) loss of Tlr7 significantly reduced tumour growth and metastasis in immune-compromised NSG mice (Extended Data Fig. 9 n–p). Importantly, RNase A treatment of TLR7-depleted 4T1 tumours did not further reduce tumour growth (Extended Data Fig. 9 q–s), consistent with a common pathway comprising ssRNA and Tlr7. Taken together, these data reveal that neuron-dependent release of ssRNA species from cancer cells signals in a paracrine manner via TLR7 to drive invasion, growth, and metastasis.
Figure 4: Sensory neurons signal via tumoral TLR7 to promote metastasis.
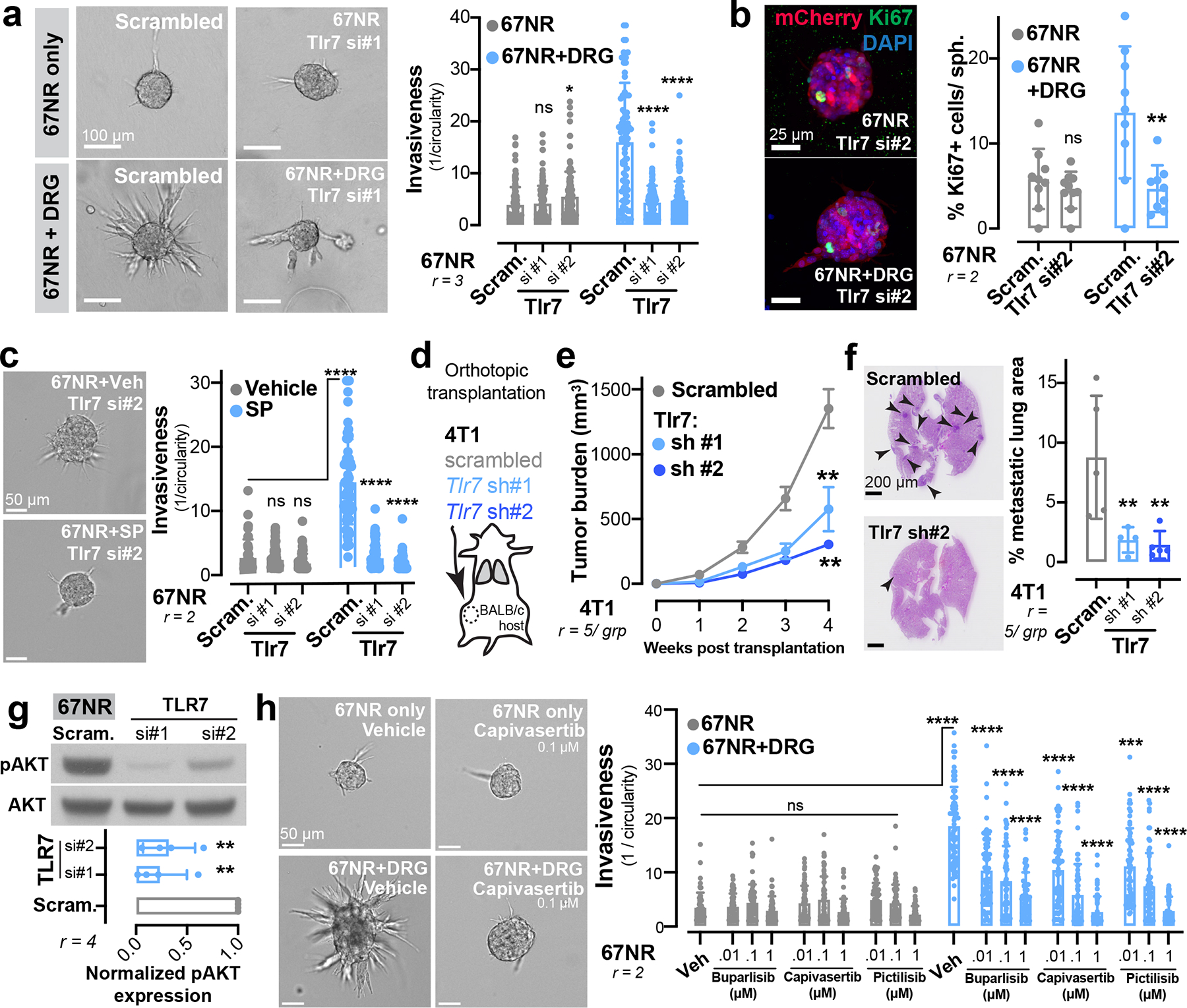
(a-b) 67NR cancer cell spheroids depleted for TLR7 and cultured with or without DRG neurons. (a) Invasion quantification. nsp = 0.5787, *p = 0.031, ****p < 0.0001, ANOVA. (b) Proliferation quantification. Sph. = spheroid. nsp = 0.8751, **p = 0.0048, t-test. Mean ± SD.
(c) Invasion quantification of 67NR cancer cell spheroids depleted for TLR7 and cultured with or without SP. nsp = 0.07 (scram. vs si#1), 0.926 (scram. vs si#2), ****p < 0.0001, Kruskal-Wallis test. Mean ± SD.
(d-f) Orthotopic transplantation of 4T1 cancer cells depleted for TLR7. (d) Schematic. (e) Tumour growth. **p = 0.0070 (scrambled vs sh#1), 0017 (scrambled vs sh#2), ANOVA. Mean ± SEM. (f) Metastatic area quantified by H&E. **p = 0.0086 (scram. vs sh#1), 0.0085 (scram. vs sh#2), ANOVA. Mean ± SD.
(g) Western blotting of phosphorylated (pAKT) and total AKT levels in 67NR cancer cells depleted of Tlr7. **p = 0.0013 (scram. vs si#1), **p = 0.0015 (scram. vs si#2), ANOVA. Mean ± SD. MW: AKT (60 kDa).
(h) Invasion quantification of 67NR cancer cell spheroids co-cultured with DRG neurons in the presence of small molecule inhibitors of PI3K signalling (10 nM, 100 nM, or 1 μM). ***p = 0.0005, ****p < 0.0001, Kruskal-Wallis test. Mean ± SD.
To define the tumoural gene expression response downstream of Tlr7 activation, we performed mRNA sequencing of control and TLR7-depleted 4T1 spheroids (Extended Data Fig. 10a). Gene-set enrichment analysis (GSEA) revealed that TLR7 depletion repressed expression of genes implicated in cancer cell invasion, proliferation, and metastasis—most notably focal adhesion genes, ECM-receptors, and PI3K-Akt signalling pathway components (Extended Data Fig. 10a). Tlr7 is the predominant sensor for pathogen-derived ssRNAs and canonically activates a MyD88-dependent inflammatory cytokine response in immune cells31. Given the absence of an inflammatory gene signature upon Tlr7 depletion in cancer cells, we hypothesized that Tlr7 was signalling through a non-canonical, MyD88-independent pathway. Consistent with this, depletion of MyD88 from 67NR cancer cell spheroids did not alter the ability of DRG neurons to promote invasion (Extended Data Fig. 10b, c).
Non-canonical PI3K signalling downstream of TLR activation has been observed in certain cell types (such as macrophages32) or certain disease contexts (such as myocardial ischemia33). Given that PI3K-Akt signalling was one of the top downregulated pathways upon tumoral TLR7 depletion (Extended Data Fig. 10a), we hypothesized that PI3K may be activated downstream of neuronal activation of tumoral TLR7 receptors. Consistent with this, TLR7 depletion in cancer cells significantly reduced the levels of phosphorylated AKT (pAKT) (Fig. 4g). Importantly, three independent small molecule inhibitors of PI3K signalling (buparlisib, capivasertib, and pictilisib) prevented DRG neuron-induced cancer spheroid invasiveness in a dose-dependent manner (Fig. 4h). All three PI3K inhibitors also prevented SP-induced cancer spheroid invasiveness (Extended Data Fig. 10d). Taken together, these data implicate PI3K signalling as a MyD88-independent mechanism downstream of TLR7 as a pathway that confers pro-invasive phenotypes in breast cancer cells.
To further assess the role of this signalling axis in human metastatic disease, we calculated a gene expression signature based on transcripts that were differentially expressed upon Tlr7 depletion and used it to stratify breast cancer patient survival. In multivariate analyses conducted across both the TCGA and the METABRIC cohorts, we found that the Tlr7-high signature correlated with reduced overall survival (METABRIC: HR 1.616, p = 0.024; TCGA: HR 2.730, p = 0.001, Extended Data Fig. 10e, f). These findings reveal that TLR7 promotes a pro-metastatic gene expression program that associates with poor breast cancer patient survival.
Aprepitant inhibits this neuro-cancer axis
Our results demonstrate a crucial role for neuronal SP and its receptor TACR1 in driving breast cancer metastatic progression. Aprepitant is a small molecule therapeutic drug that acts as an antagonist of TACR1 and is used clinically to treat nausea34. The effect of TACR1 antagonism on cancer cell proliferation and invasion in traditional 2D culture models has yielded conflicting results35–38. In our in vitro 3D culture models, aprepitant significantly decreased invasiveness of 4T1, Py8119, and MDA-MB-231 spheroids in a dose-dependent manner (Extended Data Fig. 11a). Administration of clinically relevant doses of aprepitant to mice significantly inhibited tumour growth and metastatic progression in multiple models including 4T1 cancer cells (Fig. 5 a–c), Py8119 cancer cells (Extended Data Fig. 11b, c), and MMTV-PyMT organoids (Extended Data Fig. 11b, d). Aprepitant also significantly impaired tumour growth in two independent PDX models (Fig. 5d, e). Taken together, our findings identify a neuronal activity-dependent, metastasis-promoting axis comprising of SP/TACR1/ssRNA/Tlr7. We also provide proof-of-concept for therapeutic targeting of multiple nodes in this axis, including an approved anti-nausea medication.
Figure 5: The antiemetic aprepitant targets this neuro-cancer axis to inhibit metastasis.
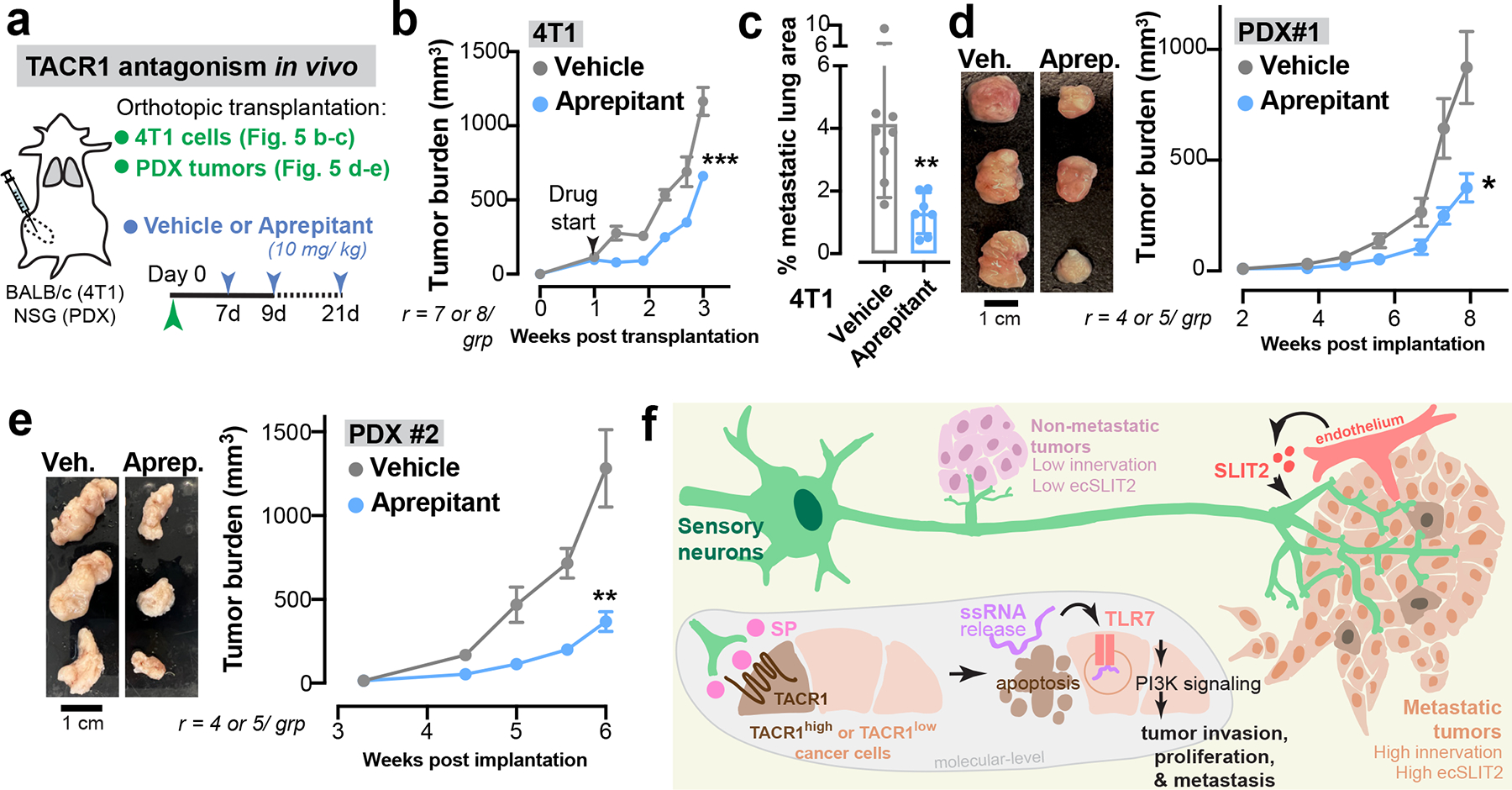
(a-e) A clinically used anti-nausea medication, aprepitant was evaluated for its potential in inhibiting breast cancer progression and metastasis. (a) Schematic. 4T1: orthotopic tumour, aprepitant delivered intra-tumorally. Patient-derived xenografts (PDXs): subcutaneous tumour, aprepitant delivered intraperitoneally. (b) 4T1 tumour growth. ***p = 0.0003, Mann-Whitney test. Mean ± SEM. (c) 4T1 metastatic area quantified by H&E. **p = 0.0086, t-test. Mean ± SD. r = 7–8 mice/ grp. (d,e) Tumour growth for two independent PDX models. *p = 0.0116, **p = 0.0037, t-test. Mean ± SEM.
(f) Model: Breast tumours are frequently innervated by sensory nerves. Metastatic breast tumours have more sensory innervation than isogeneic tumours with lower metastatic potential. In the tumour stroma, neuronal SP causes cell death in a subset of TACR1-high cancer cells, causing ssRNA release that acts in a paracrine manner on tumoral TLR7 receptors of neighbouring cancer cells to drive breast cancer metastasis.
Progression of cancers from neoplastic lesions is associated with an increase in nerve density1. Cancer cells are known to express neurotrophic factors that drive neuriteogenesis1,39,40. In this study, we demonstrate a novel role for stromal cells in regulating the extent of tumour innervation. We find that an axon guidance molecule, SLIT2, expressed by the tumour endothelium drives innervation. Within tumours, nerves have been recognized as critical signalling structures that can elicit tumorigenic4,9,10,41,42 or tumour suppressive41,43,44 effects. The mammary gland receives abundant sensory fibre input13,14, yet the impact of this sensory innervation on breast cancer progression remained poorly characterized. Conceptually, our work reveals the critical dependency of breast tumours on sensory innervation for tumour growth, invasion, and metastatic progression. We found that neuronal substance-P elicits the release of ssRNAs from a minority of TACR1-high apoptotic cancer cells. The released ssRNA acts in a paracrine manner on tumoral TLR7 receptors to promote metastasis (Fig. 5f)—thus linking neuropeptide secretion to RNA release and subsequent innate RNA sensing in the tumour compartment. We observed that neuropeptide-mediated activation of TLR7 in cancer cells drives a non-canonical PI3K-dependent gene expression signature that associates with worse survival outcomes in breast cancer patients. This work provides direct molecular evidence that links neuronal activity to extracellular RNA mediated activation of a key oncogenic signalling pathway in an epithelial tumour.
Neuronal activity within the central and peripheral nervous system regulates various aspects of tissue homeostasis, plasticity, and immune cell function45. Gliomas, for example, have been found to integrate into neuronal circuits in the brain and depend on neuronal activity for tumour growth7,8. Certain head and neck cancers can reprogram innervating sensory nerves into a more adrenergic state to support tumour growth. Our data reveals an instructive role for cancer cells of an epithelial origin in altering neuronal activity. Breast cancer cells in-turn take advantage of this increased neuronal activity to enhance multiple metastatic phenotypes in an activity-dependent manner.
Neurons are evolutionarily conserved modulators of immunity and their immune-regulatory functions have been shown to be critical in tumorigenesis11,12,41,46,47. In contrast to these previously reported indirect effects on the tumour stroma, we observed pro-tumorigenic effects of innervation directly on cancer cells via the activation of tumoral TLR7 signalling. While TLR7 canonically functions as a ssRNA pathogen sensor in innate-immunity31, we uncovered a non-canonical, immune-independent, tumour-intrinsic, and metastasis-associated transcriptional response upon neuronally-induced TLR7 activation in these cancer cells. Such ability of cancer cells to selectively exploit tumorigenic responses downstream of innate immune receptors while repressing immune activating responses has been previously described15,48. These findings also raise the possibility that pathogenic RNAs that emerge during viral infections may trigger or accelerate metastatic progression via tumoral TLR7 activation.
This work focused on the cancer-cell intrinsic mechanisms by which sensory neurons regulate breast cancer metastasis and led to the identification of aprepitant as a disruptor of the functional interactions between sensory nerves and breast cancer cells. Given the safety and tolerability of aprepitant, our findings warrant investigations into the clinical efficacy of this agent upon its prolonged use in breast cancer in combination with standard of care regimens as an anti-metastatic therapeutic approach.
MATERIALS AND METHODS
Animal studies.
All animal experiments were conducted in accordance with the Institutional Animal Care and Use Committee (IACUC) at The Rockefeller University. All animals were housed at ambient temperature in a 12 h light/dark cycle with unrestricted access to food and water. Female mice were age-matched; litter mates were used when possible and animals were randomized to each experimental cohort. No statistical methods were used to predetermine sample sizes. All tumour growth studies were performed in accordance with IACUC guidelines and mice were euthanized when the tumour volume exceeded 1500 mm3. Tumor measurements and metastasis quantification was performed in a blinded manner. Wild-type BALB/cJ, C57BL/6J, FVB, Tac1-null49 (JAX, 004103), and Nav1.8-Cre (JAX, 036564) animals were obtained from Jackson laboratories. The endothelial-specific inducible Cre line Cdh5(PAC)-creERT2 was obtained from R. Adams50. Cdh5(PAC)-creERT2; Slit2fl/fl mice were crossed for at least six generations with pure wild-type BALB/c mice. To activate Cre, mice were injected intraperitoneally with 150 μl of tamoxifen diluted in corn oil (10 mg/ml) for 2 consecutive days. Concomitantly, mouse chow was replaced with tamoxifen-supplemented food (250 mg/kg) (Envigo TD.130856). MMTV-PyMT51 mice were a generous gift from Kevin J. Cheung. Fixed C3(1)-Tag52 tumours were donated by Andrew J. Ewald. Patient-derived xenografts (PDXs) used in this study were obtained from The Jackson Laboratories (TNM00096 and TNM00098) were maintained subcutaneously in NSG mice. PDX tumours were implanted by Jackson Laboratories prior to shipment to Rockefeller University; tumours were <100 mm3 when aprepitant53 treatments were initiated.
Human samples.
All research involving human samples at Rockefeller University is supervised by the Institutional Review Board (IRB). This study was found to be IRB-exempt. All human tumour samples used in the study were received from the Cooperative Human Tissue Network (CHTN). Samples used in Fig. 1a, and Extended Data Fig. 2 j–m were received in basal medium the day after surgery. Samples used for Western blotting (n = 11) were snap-frozen upon arrival and stored at −80 °C until use. Samples used in Fig. 1b and Fig. 2f (n = 19) were received as FFPE blocks or sections and were processed for immunofluorescence analysis. De-identified pathology information for each of these samples is used to determine their lymph node status.
Primary DRG culture.
Dorsal root ganglia (DRG) neurons were isolated from 5–8-week-old female syngeneic mice (BALB/cJ, FVB, or C57BL/6J). Each mouse was perfused with 20 ml of cold HBSS (Sigma-Aldrich, 55021C). Ganglia were collected into cold HBSS, pelleted by spinning at 100 g for 2 minutes. Ganglia were then digested in papain (Worthington LS003126) and collagenase (Worthington LS004176) at 37°C. The digested neurons were triturated with fire-polished Pasteur pipettes. DRG neurons were further purified using a Percoll (GE Healthcare, 17-0891-01) gradient (12.5% and 28%). Pelleted neurons were resuspended in neuronal growth media (DMEM-F12 with HEPES (Fisher Scientific 11–330-032), penicillin-streptomycin (ThermoFischer Scientific 15140–122), bovine serum albumin (BSA; Sigma-Aldrich, A9576), and fetal bovine serum (FBS; Sigma-Aldrich, F4135)) at a concentration of 50–100 neurons/ μl. For all co-culture experiments, the DRG: cancer cell ratio of 1:200 was used unless otherwise specified.
Isolation of primary tumour organoids.
Human or mouse mammary tumours were processed into organoids as previously described54. Samples were mechanically disrupted with a scalpel, and enzymatically (collagenase-based) digested. The digested tissue was centrifuged for 10 minutes at 1500 rpm. The pelleted tissue was then treated with DNAse (Sigma, D4263). Differential centrifugations (quick spins at 400 g for 3–7 seconds) were used to separate out single/ stromal cells from epithelial organoids.
Tissue culture and spheroid generation.
4T1, EO771, HEK293T, Py8119 and MDA-MB-231 cells were obtained from American Tissue Type collection (ATCC). 4T07 and 67NR cells were a generous gift from W. P. Schiemann. EO771 cells were maintained in RPMI with 10% FBS and 10 mM HEPES (ThermoFisher Scientific 15-630-080). 67NR, 4T1, and MDA-MB-231 cancer cells were maintained in DMEM (Gibco, 11995065) with 10% FBS. Py8119 cells were cultured in Ham’s F-12K (Kaighn’s) medium (ThermoFisher Scientific, 21127022) with 10% FBS. Contamination with mycoplasma was ruled out on a quarterly basis using PCR-based protocols55.
Spheroids were formed using the hanging drop method56. 50, 3000, and 1500 cells from 4T1, 67NR, and Py8119 respectively were resuspended in 25 μl of media and plated into a lid of a tissue-culture treated dish. Droplets were placed sufficiently apart to not merge when lid was inverted and placed back on the PBS-filled reservoir. Spheroids were harvested after an overnight incubation. For MDA-MB-231 spheroids 5000 cells were resuspended in 25 μl of media containing 5% Matrigel (Corning, 354320).
Three-dimensional cultures.
For invasion assays, organoids/ spheroids were embedded in neutralized, cold-polymerized, fibrillar rat-tail collagen I (Corning, 354236). Invasion assays in the presence of DRG neurons consisted of 100 neurons/ well in a 24-well plate. The endpoint for 3D invasion assays involving cancer cells lines was ~48 hours, while that for cultures involving MMTV-PyMT or primary human tumor organoids was ~5 days. For colony formation assays using MMTV-PyMT mouse model, organoids were briefly dissociated into small cell clusters and embedded in Matrigel (Corning, 354320; 5,000 clusters/ well in a 24-well plate). For cell lines, colony formation assays were performed using 67NR cancer cells (10,000 cells/ well in a 24-well plate) and DRG neurons (100 neurons/ well in a 24-well plate). The base medium for all organoid cultures (and co-cultures) included DMEM (Gibco, 11995065), L-glutamine (ThermoFisher, 35050061), 1% penicillin-streptomycin (Gibco, 15070063), 30% BSA (Sigma-Aldrich, A9576), FBS (Sigma-Aldrich, F4135), N2 and B27 supplements (ThermoFisher Scientific, 17502048 and 17504001 respectively). Human organoids were additionally supplemented with hEGF (Sigma-Aldrich, E9644) while murine cultures were supplemented with FGF2 (Sigma, F0291). Conditioned medium was collected 24 and 48 hours after the setup of the original 3D culture in collagen I. Conditioned medium was collected and replaced with fresh neuronal growth medium. For conditioned medium experiments, RNase A (ThermoFisher Scientific, EN0531) was used at a concentration of 25 μg/ ml, RNase T1 (ThermoFisher Scientific, EN0541) was used at a concentration of 2000 U/ ml, RNase III (ThermoFisher Scientific, AM2290) was used at a concentration of 2.5 U/ ml, DNase I was used at a concentration of 2 U/ ml, and heat inactivation was performed at 100°C for 20 minutes. For ssRNA/ dsRNA mimetic experiments, ssRNA4057 (InvivoGen, tlrl-lrna40) and Poly(I:C)58 (InvivoGen, tlrl-pic) were used at concentrations of 0.5, 1 and 5 μg/ ml. ssRNA40 is complexed with the cationic lipid LyoVec to facilitate its uptake. Furthermore, the two imidazoquinoline amines, Imiquimod59 (R837, TLR7 agonist, Selleckchem, S1211) and Resiquimod60 (R848, TLR 7/8 agonist, Selleckchem, S8133) were used at concentrations of 1 and 10 μg/ ml for TLR7 stimulation. For apoptosis blockage the pan-caspase inhibitor z-VAD-FMK (Selleckchem, S7023) was used at a concentration of 100 μM. PI3K/Akt signalling was inhibited using buparlisib61, capivasertib62, or pictilisib63 at 0.01–1 μM concentrations as noted in the figures.
Matrigel transwell invasion assay:
67NR cancer cells were serum-starved in 0.2% FBS DMEM overnight. The following day, Matrigel Invasion Chambers (Corning, 354483) were rehydrated with 0.2% FBS DMEM for 2 hours at 37°C. Meanwhile, 750 μl of neuronal growth medium (NGM) was added to wells of a 24-well plate and seeded with either 1000 DRG neurons or 100 nM SP. The starved cancer cells were resuspended at 2.5 × 104 cells/ml in 0.1% BSA DMEM. Matrigel invasion chambers were emptied following rehydration, transferred to wells containing NGM, and 500 μl of cells (1.25 × 10^4) were added to the chamber. Cells were allowed to invade for 24 hours at 37°C. Media was then removed from Matrigel inserts and the inserts were washed with PBS to remove cancer cells that failed to invade. Inserts were fixed in 4% paraformaldehyde for 30 minutes followed by a PBS wash. The inserts were stained with 0.1% crystal violet, washed 3 times in PBS, and invaded cells were counted in five fields of view using the 10x objective on an inverted Primo Vert (Zeiss) microscope.
Lentiviral production and transduction.
Third generation lentivirus system was used to produce virus from HEK293T cells grown in 10 cm plates. HEK293T cells were plated at 70% confluency the night before transfection and cultured in DMEM (Gibco, 11995065) with 10% FBS (Sigma-Aldrich, F4135). Cells were transfected with 2.5 μg of Gag-Pol, 5 μg of VSV-G, 2.5 μg of RSV-Rev (Cell Biolabs, VPK-205) and 7.5 μg of pLKO.puro1 vector (Sigma) cloned to contain the appropriate shRNA, using 60 μl of Lipofectamine 2000 (ThermoFisher Scientific, 11668019). After an overnight incubation, the medium was replaced with fresh medium. Virus-containing medium was collected 48- and 72-hours post-transfection. The supernatant was filtered through a 0.45 μm filter (Pall, 4614), mixed 1:1 with fresh medium and substituted with 8 μg/ ml polybrene (Sigma-Aldrich, TR-1003-G) to transduce pre-plated 4T1 cells at 50% confluency. Selection with 3 μg of puromycin (ThermoFisher Scientific, A1113803) was conducted 36 hours post-transduction. Knockdown was validated using qPCR and Western Blotting.
Tlr7 shRNA#1 (mouse):
F 5’-CCGGTCTATGGAGAGCCGGTGATAACTCGAGTTATCACCGGCTCTCCATAGATTTTTG-3’
R 5’AATTCAAAAATCTATGGAGAGCCGGTGATAACTCGAGTTATCACCGGCTCTCCATAGA-3’
Tlr7 shRNA#2 (mouse):
F 5’-CCGGACCACTTTGCCACCTAATTTACTCGAGTAAATTAGGTGGCAAAGTGGTTTTTTG-3’
R 5’AATTCAAAAAACCACTTTGCCACCTAATTTACTCGAGTAAATTAGGTGGCAAAGTGGT-3’
Tacr1 shRNA#1 (mouse):
F 5’- CGGGAGGACAGTGACCAATTATTTCTCGAGAAATAATTGGTCACTGTCCTCTTTTTG-3’
R 5’AATTCAAAAAGAGGACAGTGACCAATTATTTCTCGAGAAATAATTGGTCACTGTCCTC-3’
Tacr1 shRNA#2 (mouse):
F 5’- CCGGTGGAGAAGGCAAGCGTTATATCTCGAGATATAACGCTTGCCTTCTCCATTTTTG-3’
R 5’AATTCAAAAATGGAGAAGGCAAGCGTTATATCTCGAGATATAACGCTTGCCTTCTCCA-3’
siRNA Transfection.
Transient knockdown of Tlr7 was performed using at least two independent siRNAs against these genes in 67NR cells (sequences below). The siRNA and Lipofectamine™ RNAiMAX transfection reagent (ThermoFisher Scientific, 13778150) were diluted in Opti-MEM (ThermoFisher Scientific, 31985070) and added to 67NR cells at ~60% confluency. After an overnight incubation, the culture medium was replaced with fresh medium. Successful knockdown was validated using qPCR and Western Blotting.
Tlr7 (mouse) siRNA sequences:
5’-GCAGAAGGAAUUACUCACAUGCUAA-3’
5’-CCUUUCAGUGAAUAAGAUAUCUCCT-3’
MyD88 (mouse) siRNA sequences:
5’- UGGUUGUUUCUGACGAUUAUCUACA-3’
5’- UGUUAGACCGUGAGGAUAUACUGAA-3’
5’- AGGUUUGCAUCUUCUUAUUCCUUTC-3’
Protein Isolation.
Cell lines were washed with and scraped into ice-cold PBS (without any Ca2+ or Mg2+), centrifuged for 5 minutes at 300 g (4°C). The cell pellet was resuspended in an appropriate volume of RIPA-based lysis buffer, vortexed, and left on ice for 5 to 10 minutes. For primary tumours, ~10 mg of tumour tissue was homogenized in ~100 μl ice-cold RIPA lysis buffer using an electric homogenizer (Bel-Art Homogenizer System, Fisher Scientific, 03-421-215; Bel-Art Pestles and Tubes, Fisher Scientific, 03-421-221). Samples were centrifuged at 10,000 g (4°C) for 10 minutes and the supernatants were transferred to new, prechilled Eppendorf tubes. A BCA assay kit (Thermo Scientific, PI23225) was used to quantify the amount of protein in each sample. Samples were stored at −80°C.
Western blotting.
Whole cell protein lysates were thawed on ice for 30 minutes before use. Samples were diluted with 4x protein loading buffer (Li-Cor, 928–40004) and 10x sample reducing agent (ThermoFisher Scientific, NP0009). Equal amounts of protein were loaded in 4–12% Bis-tris Mini Protein Gels (Fisher Scientific, NP0336PK2). SDS-Page was performed in MES-SDS running buffer (20x, ThermoFisher Scientific, NP0002) at 140 V for ~1 hour or until the dye front had run off the gel. The gels were transferred at 100 V for 1.5 hours at 4°C in transfer buffer (ThermoFisher Scientific, NP0006) onto PVDF membranes (ThermoFisher Scientific, 88520). Membranes were blocked in 5% nonfat dry milk (BioRad, 1706404) in TBS-T (TBS (Cell Signalling Technology, 12498S) + 0.1% Tween (Millipore, P2287)) for 1 hour at room temperature. Primary antibodies were diluted in 3% BSA/ TBS-T (Millipore, A2153) and incubated overnight at 4°C. Membranes were washed thrice with 0.1% TBS-T and incubated for 1 hour at room temperature with corresponding HRP-linked secondary antibodies, added at a dilution of 1:10,000. Membranes were then washed three times with 0.1% TBS-T and activated by an ECL blotting solution (ThermoFisher Scientific, 32106). X-ray films (Imaging Solutions Company, 110102) were exposed to the membrane from 10 seconds up to 2 minutes and developed afterwards. ImageJ was used to quantify band intensity. Primary antibodies used included mouse βIII-tubulin (1:1000; Biolegend, MMS-435P, Clone TUJ1), rabbit GAPDH (1:1000; Cell Signalling 2118, 14C10), mouse HSP-60 (1:1000; Santa Cruz Biotechnology, sc-13115), and rabbit TACR1 (1:1000; ThermoFisher, PA1–32229). Uncropped Western blots are provided in Supplementary Figure 1.
Immunofluorescence.
Tumours, lungs, and mammary glands harvested from mice were fixed in 4% paraformaldehyde (Fisher Scientific, AA433689M) overnight at 4°C and then transferred into 25% sucrose (Sigma-Aldrich, S0389), 0.1% sodium azide (Fisher Scientific, 71448–16) in PBS for 24 hours at 4°C. Tissues were embedded in Tissue-Tek O.C.T. Compound (VWR, 4583) and frozen at −80°C. Sections (20–50 μm thick) were cut using a cryostat set to −20°C cutting temperature for lungs and tumours and −27°C for mammary glands. The sections were collected onto Superfrost Plus microscope slides (Fisher Scientific, 12-550-15). Prior to immunofluorescent staining, O.C.T. was removed by incubating the slides in PBS for 1 hour.
Formalin-fixed paraffin-embedded tissue sections were dewaxed and rehydrated using xylene and descending concentrations of ethanol. Antigen retrieval was performed by boiling samples in citrate buffer (Sigma-Aldrich, C9999) for 30 minutes. Sections were then permeabilized using 0.5% Triton X-100 (Millipore, T9284). Slides were blocked in 10% FBS (Sigma-Aldrich, F4135), 1% BSA (Millipore, A2153), 0.1% Tween-20 (Millipore, P2287) in PBS for up to 2 hours at room temperature. Incubation with primary and secondary antibodies (diluted in 1% FBS, 1% BSA, 0.1% Tween-20) was performed overnight at 4°C or for 2–3 hours at room temperature. Slides were mounted with Prolong Gold (Fisher Scientific, P36930). Primary antibodies used in the study include rabbit anti-CGRP (1:250, Cell Signalling, mAb#14959), mouse anti-βIII-tubulin (1:200, Biolegend, MMS-435P, Clone TUJ1), rabbit anti-Ki-67 (1:250, abcam, ab15580), rabbit anti-substance P (1:200, Sigma-Aldrich, AB1566), mouse anti-neurofilament-L (1:400, NFL, Cell Signalling, #2837), mouse anti-dsRNA (1:200, Axxora, JBS-RNT-SCI-10010200), and phalloidin (1:200, ThermoFisher Scientific, A12379). Nuclei were counterstained using Hoechst (1:500, ThermoFisher Scientific, H3579). All secondary antibodies were AlexaFluor conjugates (1:200, ThermoFisher Scientific, A21206, A21202, A31573, A31572, A31571, A31570, or A21208).
qPCR.
RNA was extracted from cells using the RNA purification kit (Norgen Biotek, 37500), according to the manufacturer’s protocol. Reverse transcription (RT-PCR) was performed on 2–5 μg of RNA using the SuperScript III First-Strand Synthesis System (ThermoFisher Scientific, 18080051). Real time qPCR was conducted in a 384-well PCR microplate (Fisher scientific, AB-1384) performed on an Applied Biosystem StepOne Real-Time PCR System using Fast SYBR Green master mix (Fisher Scientific, 43-856-18). ΔCt values were used to calculate the relative levels of the mRNA of interest which were normalized to the mRNA levels of GAPDH. Primers used in the studies include:
GAPDH (mouse):
Forward – 5’-AGGTCGGTGTGAACGGATTTG-3’
Reverse – 5’-TGTAGACCATGTAGTTGAGGTCA-3’
Tlr7 (mouse):
Forward – 5’-CACCACCAATCTTACCCTTACC-3’
Reverse – 5’-CAGATGGTTCAGCCTACGGAA-3’
Tacr1 (mouse):
Forward – 5’-CTCCACCAACACTTCTGAGTC-3’
Reverse – 5’-TCACCACTGTATTGAATGCAGC-3’
Tlr3 (mouse):
Forward – 5’- GTGAGATACAACGTAGCTGACTG-3’
Reverse – 5’- TCCTGCATCCAAGATAGCAAGT-3’
Flow cytometry.
All steps were performed on ice and under protection from light. Fresh flow buffer containing 25 mM HEPES (Fisher Scientific 11-330-032), 1% BSA (Sigma-Aldrich, A9576), 1% FBS (Sigma-Aldrich, F4135), and 1% penicillin-streptomycin (ThermoFischer Scientific 15140–122) in PBS was prepared. 67NR cancer cells were detached from culture dishes, strained through a 70 μm filter, pelleted, washed with ice-cold PBS, and incubated in 50 μl of staining buffer (anti-TACR1, 1:200 in flow buffer, alamone labs, ATR-001) for 20 minutes. Cells were washed three times with 1 ml of flow buffer. The last step was repeated for secondary antibody as well (1:200, Thermo Fischer Scientific, A31572). DAPI (Thomas Scientific, C756W66) was used as a live/ dead stain. Flow cytometry data was obtained using the Attune NxT cytometer and data was analyzed using FlowJo. Gating strategy is depicted in Supplementary Fig. 2.
Mammary-fat-pad injections.
Cancer cells (67NR, 4T1, EO771, or Py8119) were resuspended in a 50:50 mix of PBS: Matrigel (Corning, 354320) at a concentration of 106 cells/ ml. For DRG co-transplantations, 100 DRGs were injected per animal (added to the cancer cell suspension just prior to injections into the animal). Orthotopic transplantations were conducted in 5–7-week-old syngeneic female mice as previously described64. Mice were anesthetized, immobilized, and the surgical site was shaved and sterilized using povidone iodine (Abcam, ab143439). A midline incision was made to expose the abdominal mammary gland. 50 μl of the cell suspension was injected into the mammary gland. The surgical wound was closed using 9 mm autoclips (Fisher Scientific, NC9938480). Tumours measurements were taken using digital calipers twice a week and tumour volume was calculated according to r × h2/2. Tumours and lungs were typically harvested 3 to 4 weeks after transplantation when the tumour volume reached ~1500 mm3, in accordance with our IACUC protocols. Macro-metastases (>100 cells) within the lungs were quantified using hematoxylin and eosin (H&E) staining of 5 μm thick tumour sections. Micro-metastases were counted manually in lung sections and typically consist of < 50 cells. For circulating tumour cell (CTC) enumeration, cardiac puncture was used to obtain 750 μl – 1 ml of blood from animals with late-stage tumours. Following lysis of red blood cells, the remainder of the sample was smeared on a glass microscope slide and examined under a confocal microscope for mCherry+ expression. Cells with intact nuclei and mCherry expression were identified as a CTC.
Tail-vein injections.
67NR cancer cells were resuspended in PBS at a concentration of 750,000 cells/ ml and stored on ice. Tail vein injections were performed in 6–9-week-old syngeneic female mice. Mice were placed in a heating chamber set to 37°C for 10 minutes and immobilized in a restraining device. 200 μl of the cell suspension was injected via the tail vein. Lungs from these mice were harvested ~1 week later and the number of macro-metastases were examined under the dissection microscope.
Intracardiac injections.
100,000 mCherry+ 67NR cancer were injected into the left ventricle in 100 μL of PBS using a 26 g needle while mice were under 2.5% isoflurane anesthesia. Two weeks later, the brain, liver, lung, femur, and ovaries were collected and number of mCherry+ macro-metastases were examined under a dissection microscope.
Intraductal injections.
Intraductal injections of cancer cells or reagents/ drugs were performed to restrict delivery within the ductal epithelium. Injections were performed under a dissection microscope as previously described65. Using a pair of micro-dissecting tweezers, the dead skin around the nipple is removed. A 33-gauge micro-syringe (needle: CAL7637–01, syringe: 89221–012) is used to inject no more than 20 μl into the ductal epithelium. 4T1 cancer cells were resuspended at a concentration of 106 cells/ ml in PBS. Capsaicin66 was dissolved in 10% ethanol, 10% Tween-80, and 80% saline and injected into mice (20 μl/ mouse) at a concentration of 75 mg/ kg. Anti-SP (BioGeneX AR069GP) or IgG control (BioXCell BE0095) antibodies were injected into 4T1 tumour-bearing NOD-SCID gamma mice at a concentration of 20 μg/ mouse. RNase A (two sources: ThermoFisher Scientific EN0531 and 12091021) was used at a concentration of 5 mg/ ml.
Genotyping of transgenic mouse lines
Tail snips collected from 2.5-week-old mice were lysed in DirectPCR Lysis Reagent (Viagen Biotech, 102-T) and Proteinase K (Millipore Sigma, 3115828001) to extract DNA. PCR reactions using the primers listed below were used to detect the presence of a transgene.
MMTV-PyMT:
F: 5’- GGAAGCAAGTACTTCACAAGGG-3’
R: 5’-GGAAAGTCACTAGGAGCAGGG-3’
Internal positive control F: 5’-CAAATGTTGCTTGTCTGGTG-3’
Internal positive control R: 5’-GTCAGTCGAGTGCACAGTTT-3’
Positive control runs at 200 bp while the transgene runs at 556 bp.
Tac1-KO
F: 5’- AGAATTTAAAGCTCTTTTGCC-3’
R: 5’- GCTCATCAGTATGTGACATAGAAA-3’
Mutant allele runs at 175 bp while the wild type runs at 190 bp.
Nav1.8 Cre
F: 5’- GGTTCGCAAGAACCTGATGG-3’
R: 5’- GCCTTCTCTACACCTGCGG-3’
This is a generic Cre primer. The transgene runs at 570 bp.
Optical tissue clearing.
Clearing of murine mammary tumours was performed using a previously described AdipoCLEAR protocol67. Tumours were fixed in 4% paraformaldehyde (ThermoFisher Scientific, AA433689M) and washed in increasing concentrations of methanol (20–100%) in B1N buffer (water, 0.1% Triton X-100 (Sigma-Aldrich, T9284), 0.3 M glycine (Sigma-Aldrich, G5417)). Delipidation was performed using 100% dicholoromethane (Sigma-Aldrich, 270997). Samples were washed in decreasing concentrations of methanol (100–20%) in B1N buffer. Tumours were incubated with primary antibodies (anti-CRP, 1:200, Cell Signalling, mAb#14959) for 4 days with continuous shaking, washed, and incubated with secondary antibodies (AlexaFluor conjugates) for another 4 days. After immunolabelling, samples were dehydrated in an increasing series of methanol/ water (25–100%). Samples were washed in dicholoromethane overnight and cleared in dibenzyl ether (Sigma-Aldrich, 33630).
Retrograde tracing.
GFP-tagged cholera toxin-β68 was used for retrograde tracing experiments at a concentration of 0.1% in a total injection volume of 20 μl. For the normal mammary gland. CTB was injected intraductally. For tracing experiments in 67NR, 4T1 tumors, CTB was injected intratumorally 2–3 weeks post-transplantation. DRGs were isolated from these animals 1 week after CTB injections, fixed in 4% PFA, and mounted onto a coverslip.
Imaging and image analysis.
Optically cleared tissues were imaged on a light sheet microscope (LaVision BioTec) with a 4x/0.3 LVMI-Fluar objective with 5.6–6 mm WD. Images were imported into Imaris and rendered in 3D. Confocal microscopy was performed using either the LSM 780 laser scanning confocal microscope (Zeiss) or A1R confocal microscope (Nikon). Phase contrast/ brightfield images were acquired using the CellDiscoverer7 (Zeiss) microscope. For experiments in which immunofluorescence was used to infer relative protein expression levels, all images were collected and processed using identical parameters. All image analysis was performed using Fiji/ ImageJ2, unless otherwise stated below.
Extent of innervation in tissue sections was estimated by manually counting the number of nerve bundles in size-matched tumors, positively identified by βIII-tubulin/ neurofilament-L for total innervation analysis, or CGRP for sensory-specific innervation. Two sections per tumor were averaged as a single datapoint in the graphs. Any innervation in the surrounding tissue was not included in the analysis.
For cleared 3D samples, Imaris (version 10) was used for rendering the maximum intensity projection image. Autofluorescence (images collected at the 488 nm wavelength) was used to define the tumor boundary.
Proliferation in cancer cell spheroids was assessed as percentage of Ki67+ nuclei in respect to total number of cells per organoid/ spheroid identified by DAPI.
Invasiveness was reported as an inverse of the circularity of the manually-traced organoid/ spheroid boundary. Circularity is defined as the ratio of the square of the perimeter to 4π times the area. While a circle has a circularity of one, invasive spheroids/ organoids have lower circularity scores. Circularity scores are the standard measurement for invasion in 3D cultures since they account for both the number and the length of invasion strands.
Metastatic area was measured by manually tracing the area of metastases as seen by H&E staining of the lung. Total lung area was measured using auto-threshold. The percentage of metastatic area, which accounts for number and size of metastases, was reported in the manuscript. The percent metastatic area averaged across 3–4 lung sections per mouse was average and plotted as a single data point in the graphs.
For in vivo invasion quantifications, we imaged ~20 ROIs within 67NR or 67NR+DRG tumors across 3 independent tumors per condition. We classified it as invasive if there was >1 invasion strand within the ROI.
For SP expression in mouse tumors, the mean fluorescence intensity for the entire ROI was measured. For substance-P expression levels in human breast tumours, fluorescence intensity was measured within an ROI drawn around any cancer-cell rich areas of the tumour. In all cases, multiple ROIs were measured per tumour, each dot in the plot corresponds to data from a single ROI. r = independent tumours.
For SLIT2 expression analysis within the tumour endothelium, the mean fluorescence intensity for SLIT2 along the endothelium (marked by endomucin) was measured. multiple ROIs were measured per tumour, each dot in the plot corresponds to data from a single ROI. r = independent tumours.
NucView 488 Caspase-3 substrate (Biotium, 30029-T) was used to visualize apoptosis within spheroids. The green fluorescence was used to measure the apoptotic area (thresholded in ImageJ) and a manually traced spheroid boundary was used to measure total area.
Calcium imaging was performed using the Fluo-4 calcium dye69 (excitation 488 nm/ emission 525 nm; ThermoFischer Scientific, F14201). DRG neurons were cultured on a collagen I layer either alone or in the presence of 67NR cancer cells for at least 48 hours prior to loading with the calcium dye. Cells were incubated with Fluo-4 (5 μM) for 60 minutes at 37 °C. Neurons were washed thrice, and medium was replaced with the BrainPhys Imaging Optimized Medium (StemCell Technologies, 05796). Live-cell imaging was performed for ~1 hour to capture spontaneous calcium spikes using the Nikon TiE inverted microscope with Perfect Focus mechanism. Images were collected every 3–5 seconds. Imaris (version 10) was used to measure the mean fluorescence intensity within the soma of neurons within each ROI, at each time point. A rolling baseline was defined as the 10th percentile of the fluorescence intensity every 35 image frames. ΔF/F0 values are represented in the heatmap (Fig. 3b) and individual calcium traces (Extended Data Fig. 7c).
Statistical analysis.
For all experiments, independent biological replicates are listed as “r”. For 3D invasion/ proliferation assays used in the study, each dot in the plot represents quantification for a single spheroid/ organoid (or “n”). Groups were compared using tests for significance as indicated in the figure legends. A significant difference was concluded at p < 0.05. Unless otherwise noted: (1) all data are expressed as mean ± standard deviation. (2) all statistical tests were two-sided.
RNA-sequencing.
Libraries were constructed from 500 ng of total RNAs isolated from 4T1 (scrambled vs Tlr7 shRNA) using the TruSeq RNA Library Prep Kit (Illumina). Constructed libraries were sequenced using Illumina NextSeq (High Output, 75 SR) at the Rockefeller University Genomics Center. Sequencing reads were mapped to the mouse genome (assembly GRCm38) using STAR aligner (v2.7.5) at default settings. STAR was also used for counting reads mapping to genes. Further analyses were performed using RStudio (v4.1.0). Differentially expressed genes were determined using DESeq2 (v1.32.0). For gene set enrichment analysis, genes were ranked according to log fold changes shrunken with the àshr` method and enriched pathways of the Kegg database were identified using the `gseKEGG` function of the clusterProfiler package for RStudio (v4.0.0).
Analysis of the TCGA and METABRIC studies.
To assess the association of TLR7-dependent signalling with outcome in breast cancer patients, we analysed the association of a TLR7 (orthologous to human TLR8)-dependent gene signature with survival in two large independent datasets. Specifically, we generated a signature of genes downregulated in cancer spheroids in which Tlr7 expression was silenced (see section above) and calculated the TLR7 signature score by summing the scaled expression of the genes contained in the signature and using the ranked signature score as a covariate in a Cox proportional hazards model including tumour stage, age, and the ranked signature score using the Survival (v3.2–11) and forestmodel (v0.6.2) packages for RStudio. For the TCGA study, we used clinical data as recently curated and included only primary tumour samples. For the METABRIC70 study we used the validation cohort.
Extended Data
Extended Data Figure 1: Breast tumours are frequently innervated by sensory nerves.
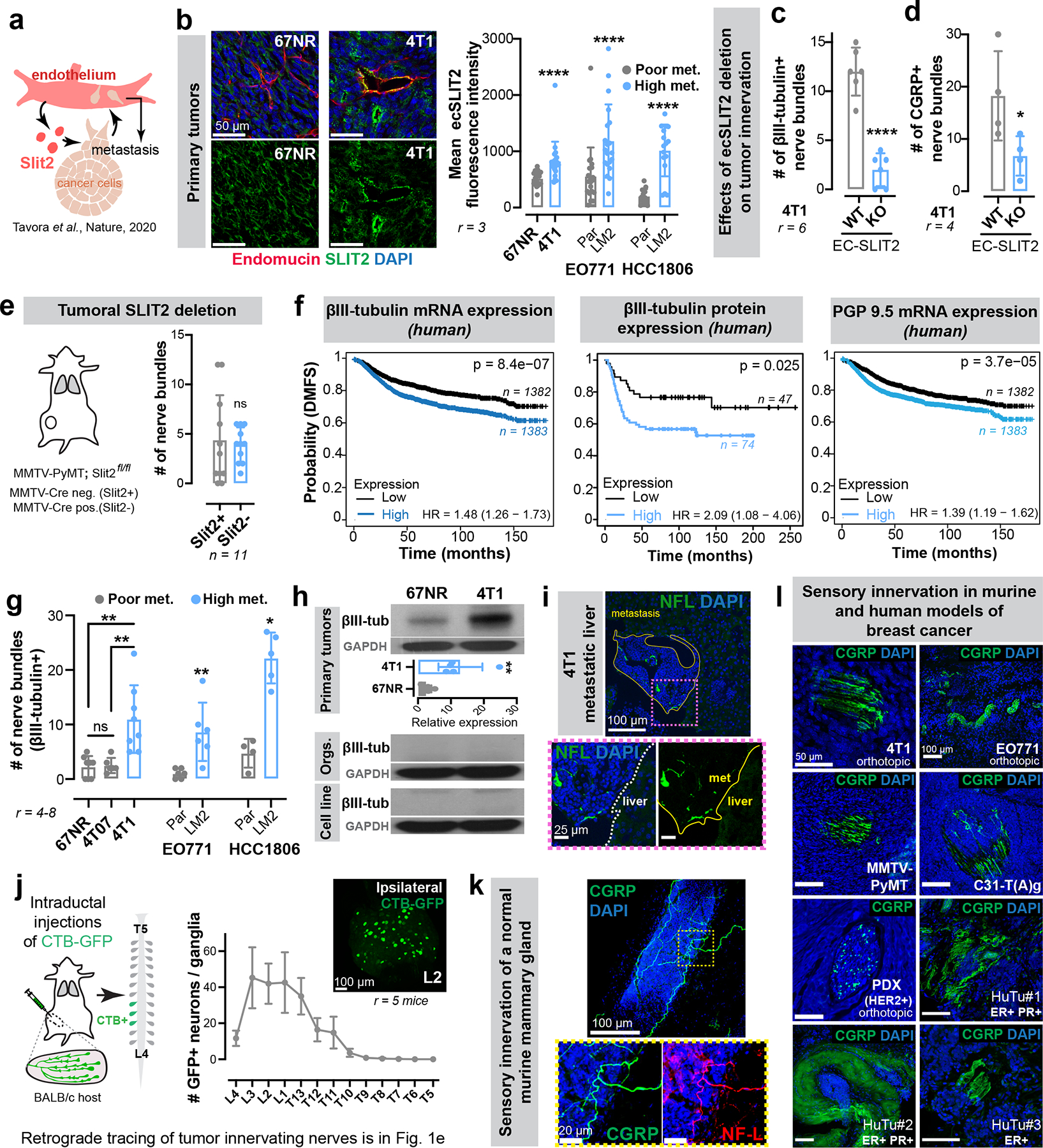
(a) We previously uncovered a requirement for endothelial-derived SLIT2 (ecSLIT2) in metastasis15.
(b) Quantification of ecSLIT2 expression in poorly vs highly metastatic breast tumour models. ****p < 0.0001, Mann-Whitney test. Mean ± SD.
(c-d) Quantification of total (c) and CGRP+ sensory (d) innervation in 4T1 tumours grown in ecSLIT2 knockout (endothelial-specific depletion of SLIT2) mice. **** p < 0.0001, *p = 0.0489, t-test. Mean ± SD.
(e) Quantification of innervation in SLIT2-knockout mammary tumours in Slit2fl/fl; MMTV-PyMT; MMTV-Cre mice. nsp = 0.6414, Mann-Whitney test. Mean ± SD.
(f) Kaplan-Meier plots showing distant metastasis-free survival (DMFS) of breast cancer patients sorted by the median expression level of βIII-tubulin (mRNA, protein) or PGP9.5 (mRNA) of their tumours. The x-axes were set to have >10 surviving patients in each arm.
(f) Nerve bundle abundance in highly metastatic primary tumours relative to corresponding isogenic poorly metastatic tumours. Between 4 and 8 tumours were analysed per group. **p (67NR vs 4T1) = 0.001, **p (4T07 vs 4T1) = 0.0019, nsp (67NR vs 4T07) = 0.9095, ANOVA; **p (EO771 Par vs LM2) = 0.0065, *p (HCC1806 Par vs LM2) = 0.0159, Mann-Whitney test. Mean ± SD.
(g) Expression of pan-neuronal βIII-tubulin in 67NR vs 4T1 primary tumours, stroma-depleted primary tumour organoids, or cell lines. **p = 0.0048, t-test. Mean ± SD. MW: βIII-tubulin (50 kDa).
(h) Liver metastasis from a 4T1 tumour-bearing mouse immuno-stained for nerve fibres. r = 3/group.
(i) Retrograde tracing of DRG neurons innervating abdominal mammary glands of wild-type mice. Mean ± SEM.
(k-l) Sensory, CGRP+ innervation observed within the normal murine mammary gland (k), murine models of breast cancer (l), and primary human tumours (l). r = 3 tumours/ group.
Extended Data Figure 2: Sensory neurons promote cancer invasion and proliferation across multiple ex vivo murine and human models of breast cancer.
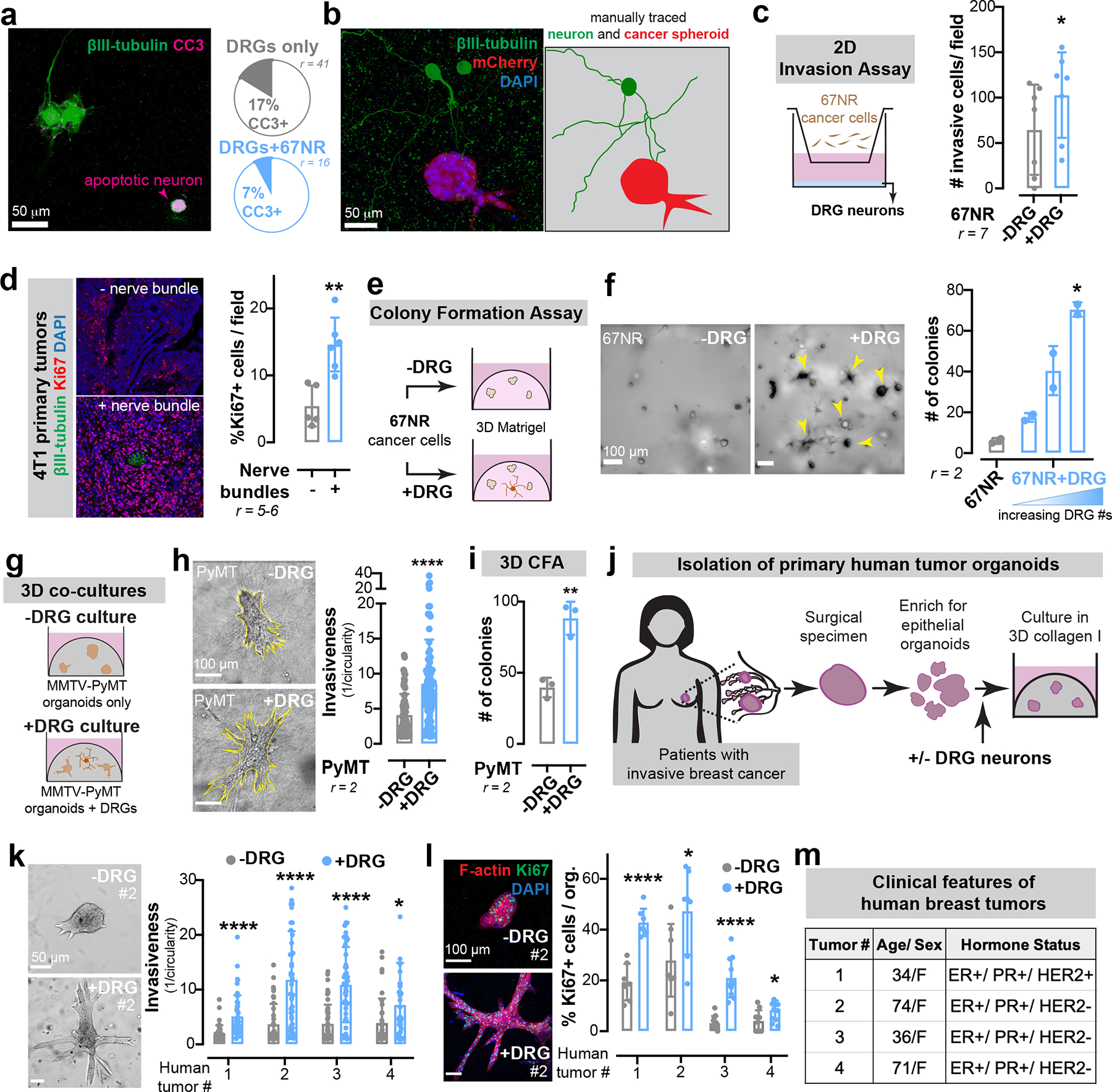
(a) Frequency of apoptosis (cleaved caspase 3, CC3+) in DRG neurons cultured alone or in the presence of 67NR cancer cells.
(b) Micrographs of mCherry+ 67NR spheroids co-cultured with βIII-tubulin+ DRG neurons. Neuron and spheroid boundaries are manually traced to illustrate little physical contact. n = 10 ROIs/ group, r = 4.
(c) 2D invasion assay of 67NR cancer cells that were cultured alone or in the presence of primary DRG neurons. *p = 0.0432, t-test. Mean ± SD.
(d) Quantification of the number of mitotically active cells within regions of 4T1 primary tumours adjacent to or far away from a nerve bundle. **p = 0.0043, Mann-Whitney test. Mean ± SD.
(e-f) 3D colony formation assay. 67NR cancer cells were cultured alone or in the presence of DRG neurons in 3D Matrigel. (e) Schematic. (f) Quantification. **p = 0.0067, Kruskal-Wallis test. Mean ± SD.
(g-i) 3D co-culture assays of MMTV-PyMT mammary tumour organoids and primary DRG neurons. (g) Schematic. (h) Invasion assay. ****p < 0.0001, Mann-Whitney test. (i) Colony formation assay. **p = 0.0033, t-test. Mean ± SD.
(j-m) 3D co-culture assays of 4 independent primary human breast tumours and DRG neurons. (j) Schematic of organoid isolation. (k) Invasion quantification. ****p < 0.0001, *p = 0.0266, Mann-Whitney test. Mean ± SD. (l) Proliferation quantification. ****p < 0.0001, *p = 0.0401 (#2), 0.0267 (#4), t-test. Mean ± SD. (m) Basic de-identified clinical information for human tumour samples cultured.
Extended Data Figure 3: Co-transplantation of breast cancer cells with DRG neurons drives metastasis.
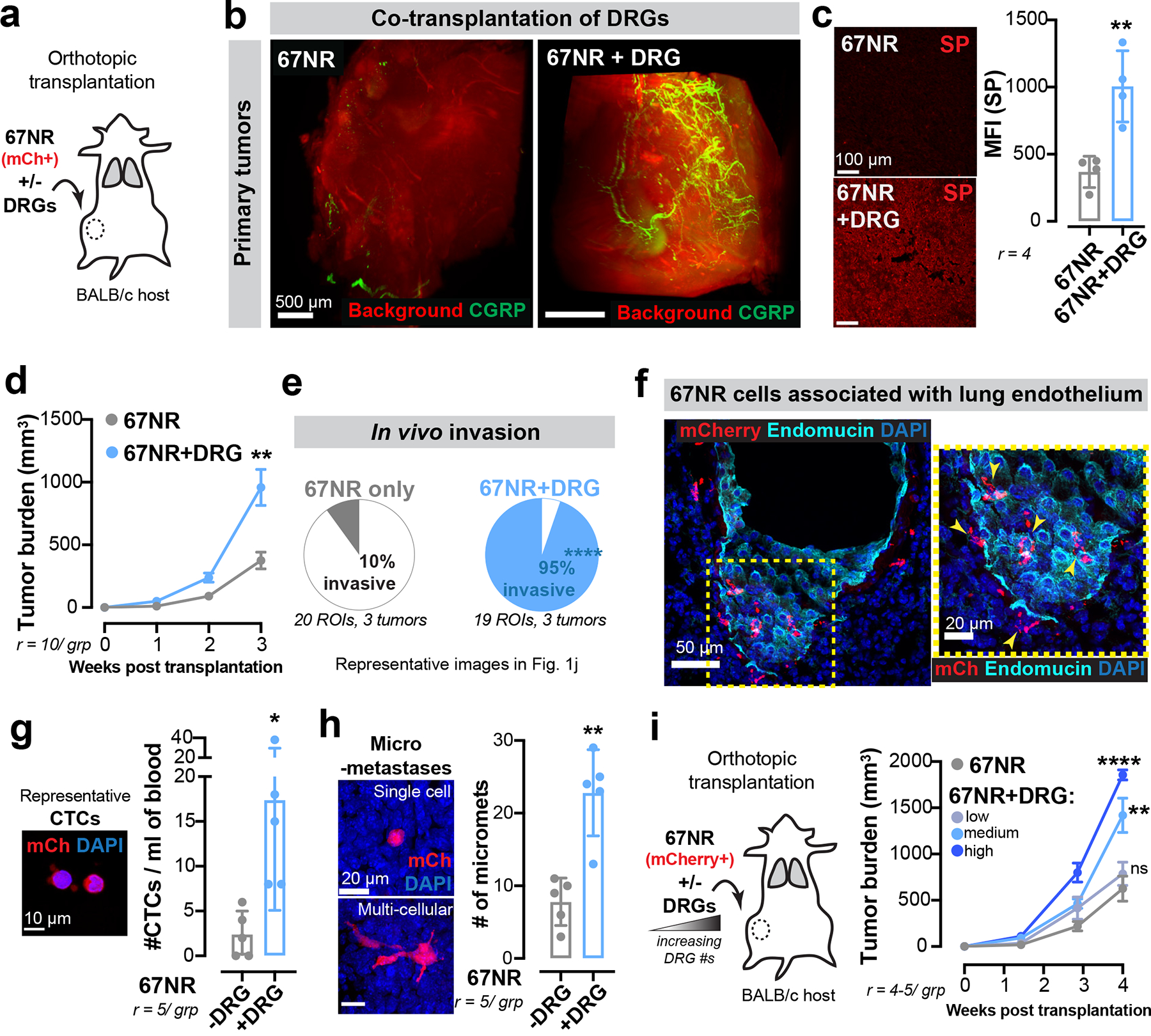
(a-h) Co-transplantation of mCherry+ (mCh) 67NR cancer cells and DRG neurons. (a) Schematic. (b) Optically cleared 67NR tumours transplanted with or without DRG neurons and immunostained for CGRP+ sensory nerves. r = 2 tumours/ group. (c) Mean fluorescence intensity (MFI) for SP in 67NR primary tumours transplanted with or without DRG neurons. **p = 0.0046, t-test. Mean ± SD. (d) Tumour growth. **p = 0.0017, t-test. Mean ± SEM. (e) Percentage of the tumour-stroma boundary with a pushing vs invasive morphology. ****p < 0.0001, Chi-square test. (f) Association of mCh+ 67NR cancer cells with the lung endothelium in mice transplanted with orthotopic 67NR tumours. r = 3 lungs/ group. (g) CTC enumeration in mice transplanted with mCh+ 67NR cancer cells with or without DRG neurons. *p = 0.0287, t-test. Mean ± SD. (h) mCh+ micro-metastases. **p = 0.0079, Mann-Whitney test. Mean ± SD.
(i) Co-transplantation of 67NR cancer cells with increasing numbers of DRG neurons. nsp = 0.4124, **p = 0.0026, ****p < 0.0001, ANOVA. Mean ± SEM.
Extended Data Figure 4: Sensory neurons drive metastatic colonization in multiple breast cancer models.
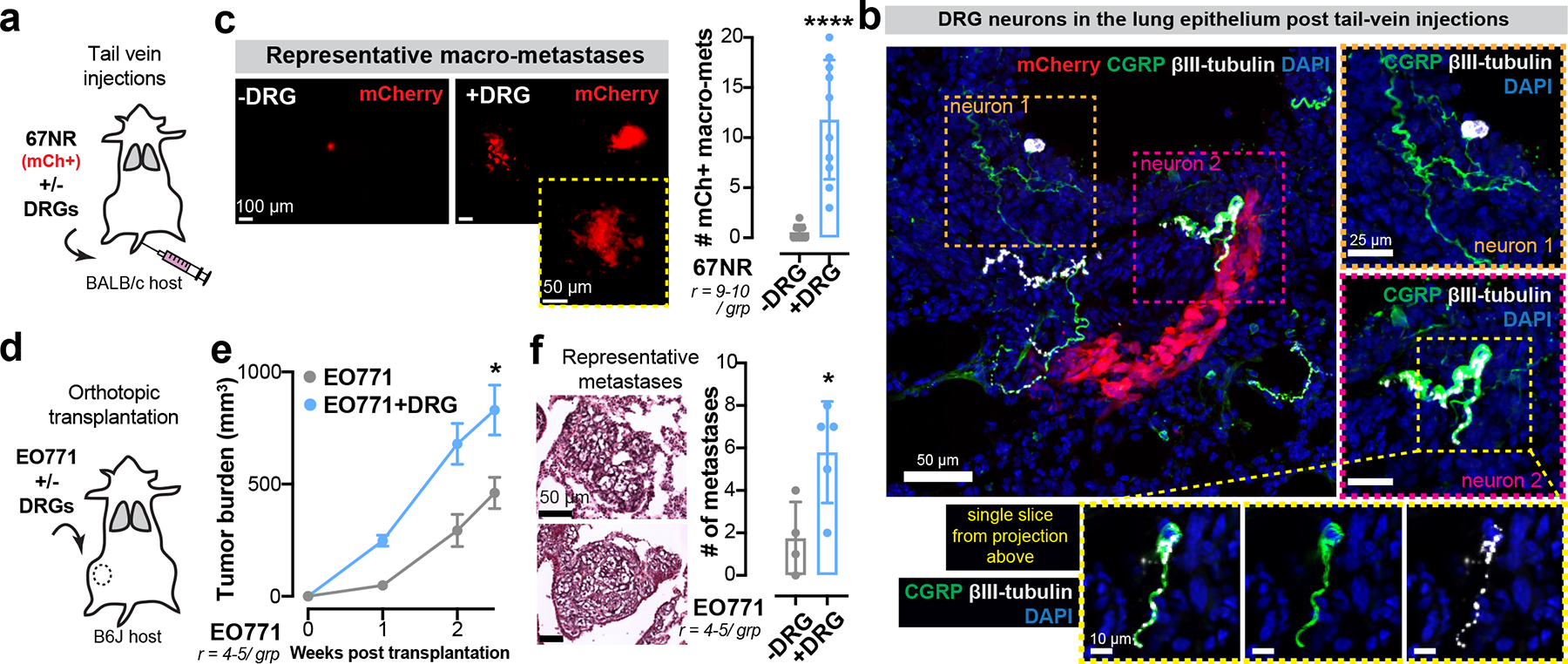
(a-c) Tail vein injections of mCh+ 67NR cancer cells with or without DRG neurons. (a) Schematic. (b) CGRP+/ βIII-tubulin+ neuronal cell bodies in mice co-injected with DRG neurons. r = 3 lungs/ group. (c) mCh+ metastases. ****p < 0.0001, Mann-Whitney test. Mean ± SD.
(d-f) Co-transplantation of EO771 cancer cells and DRG neurons. (d) Schematic. (e) Tumour growth. *p = 0.0345, t-test. Mean ± SEM. (f) Number of micro-metastases counted by H&E. *p = 0.0249, t-test. Mean ± SD.
Extended Data Figure 5: Capsaicin does not alter growth or invasion of cancer cells in vitro and reduces tumour growth in immune-compromised mice in vivo.
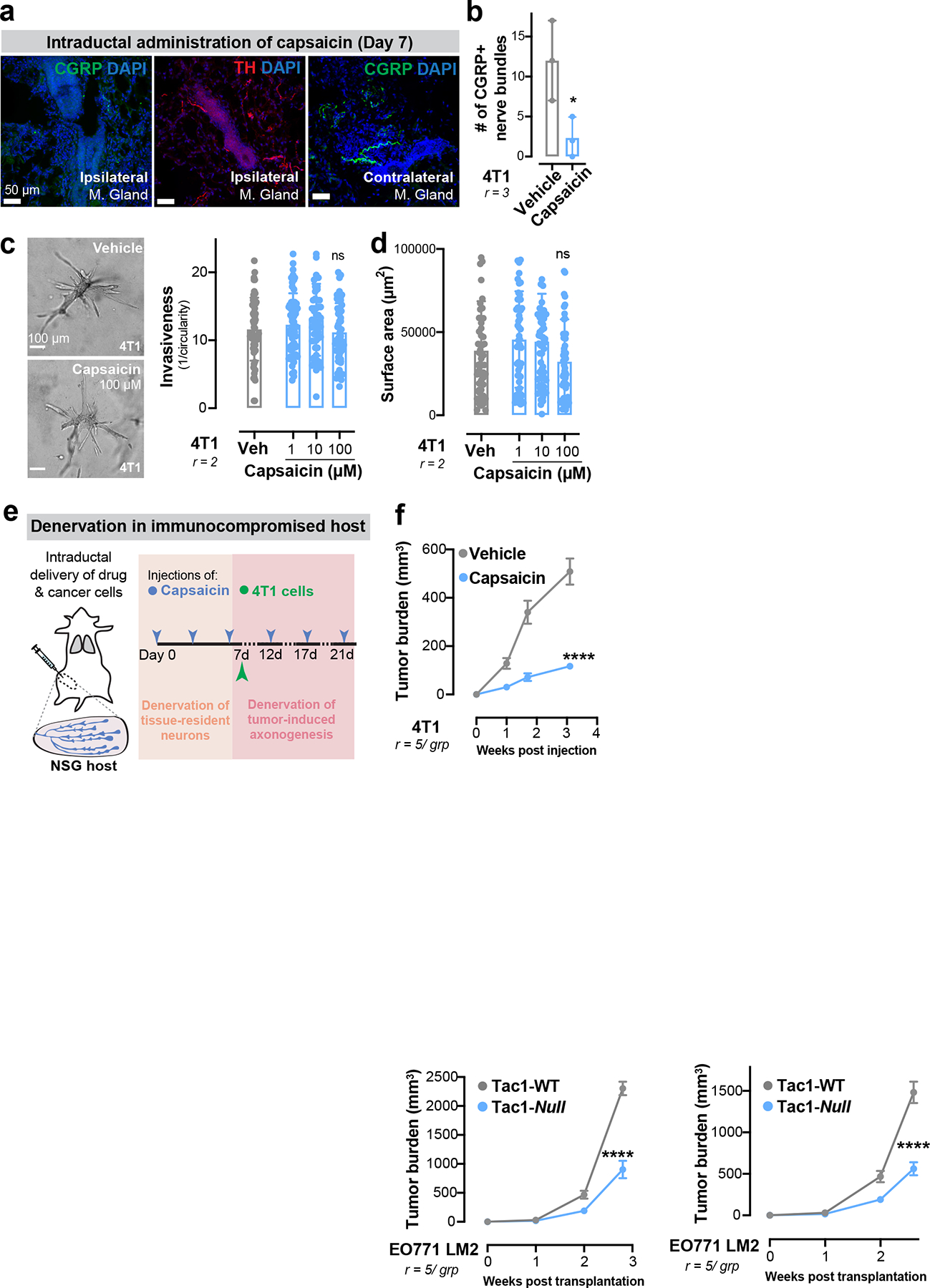
(a) Sensory (CGRP+) and sympathetic (TH+) innervation in ipsilateral or contralateral mammary glands 7 days after capsaicin administration.
(b) Quantification of CGRP+ nerve bundles post-capsaicin administration. *p = 0.0403, t-test. Mean ± SD.
(c-d) 3D culture of 4T1 spheroids in the presence of capsaicin (1–100 μM). (c) Invasion quantification. nsp > 0.9999, Kruskal-Wallis test. (d) Spheroid surface area. nsp = 0.7108, Kruskal-Wallis test. Mean ± SD.
(e-f) Sensory-specific denervation of 4T1 tumours grown in NSG mice. (e) Schematic. (f) Tumour growth. . ****p < 0.0001, t-test. Mean ± SEM.
Extended Data Figure 6: Neuronal substance-P drives breast cancer metastasis via the activation of the tumoral TACR1 receptor.
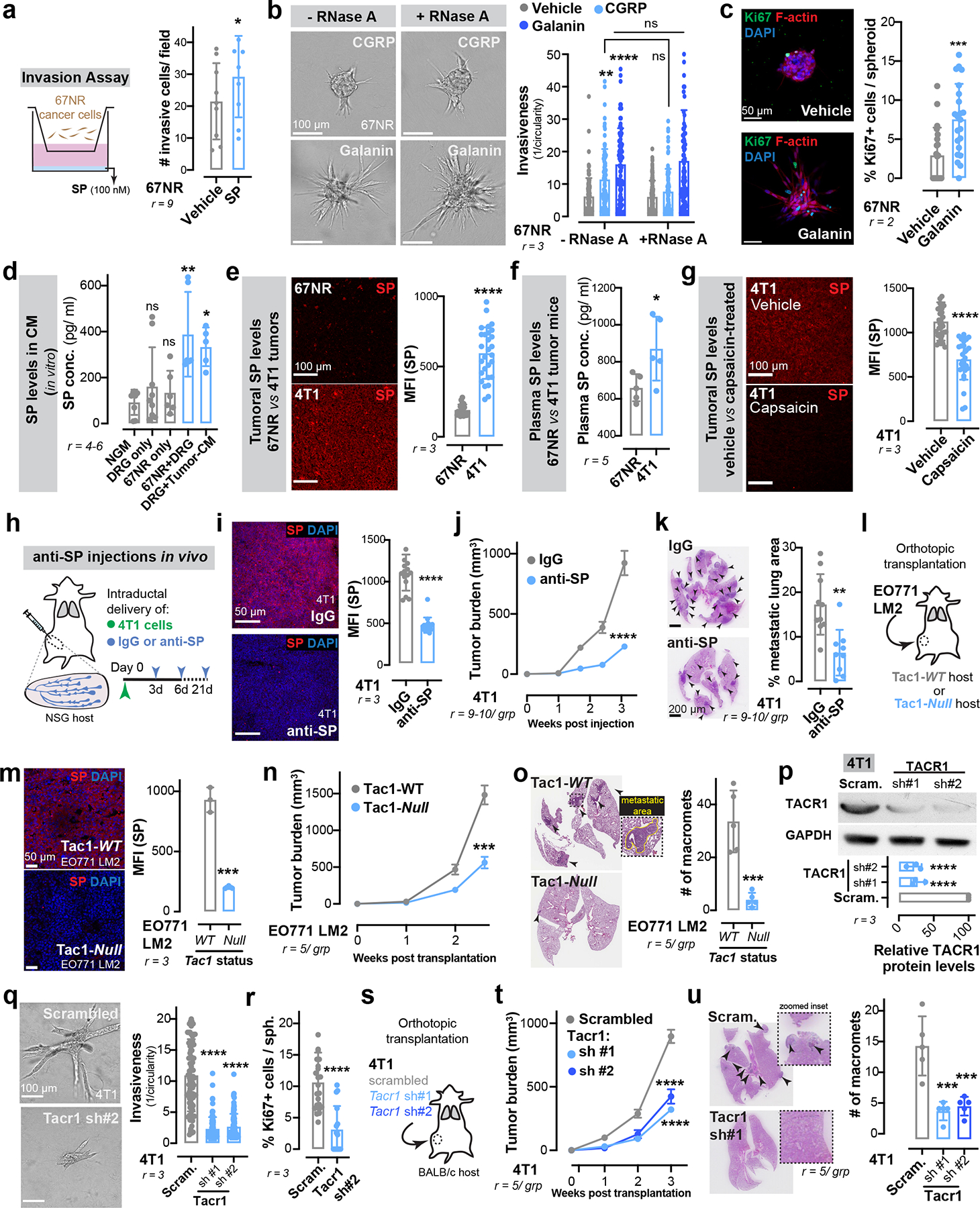
(a) Invasion assay of 67NR cancer cells in response to SP. *p = 0.0254, t-test. Mean ± SD.
(b-c) 67NR cancer cell spheroids cultured with neuropeptides galanin or CGRP, with or without RNase A. (b) Invasion quantification. **p = 0.0011, ****p < 0.0001, nsp (CGRP) = 0.1066, nsp (galanin) > 0.9999, Kruskal-Wallis test. Vehicle-treated 67NR spheroids (−/+RNase A) are also plotted in Fig. 2d and Fig. 3d respectively since the experiments were conducted together. (c) Proliferation quantification. ***p = 0.0006, Mann-Whitney test. Mean ± SD.
(d) ELISA-based quantification for SP levels in conditioned medium. p values are based on comparisons with base medium (NGM). nsp = 0.7426 (vs DRG only), 0.8724 (vs 67NR only); *p = 0.0213; **p = 0.0024, ANOVA. Mean ± SD.
(e-g) In vivo measurements of SP expression. (e) Mean fluorescence intensity (MFI) of SP in 4T1 vs 67NR primary tumours. ****p < 0.0001, t-test. (f) Plasma SP levels in 67NR vs 4T1 tumour-bearing mice. *p = 0.0359, t-test. (g) MFI of SP in 4T1 vehicle vs capsaicin-treated primary tumours. ****p < 0.0001, t-test. Mean ± SD.
(h-k) Effects of an SP-blocking antibody on 4T1 tumour growth and metastasis. (h) Schematic. (i) MFI of SP in IgG vs anti-SP treated 4T1 tumours. ****p < 0.0001, Mann-Whitney test. Mean ± SD. (j) Tumour growth. ****p < 0.0001, Mann-Whitney test. Mean ± SEM. (k) Metastatic area quantified by H&E. **p = 0.0012, t-test. Mean ± SD.
(l-o) Orthotopic transplantation of EO771 LM2 cells into the abdominal mammary glands of Tac1-WT and Tac1-null host mice. (l) Schematic. (m) MFI of SP in primary tumours. ***p = 0.0003, t-test. Mean ± SD. (n) Tumour growth. ***p = 0.0003, t-test. Mean ± SEM. (o) Number of macro-metastases quantified by H&E. ***p = 0.0006, t-test. Mean ± SD.
(p-r) Depletion of SP’s receptor, TACR1 in 4T1 cancer cell spheroids. (p) Validation of knockdown. ****p < 0.0001, ANOVA. MW: TACR1 (46 kDa). Quantification of spheroid invasion (q) and proliferation (r). ****p < 0.0001, Mann-Whitney test. Mean ± SD.
(s-u) Orthotopic transplantation of 4T1 cancer cells depleted for TACR1. (s) Schematic. (t) Tumour growth. ****p < 0.0001, ANOVA. Mean ± SEM. (u) Metastatic area quantified by H&E. ***p = 0.0003, ANOVA. Mean ± SD.
Extended Data Figure 7: Neuronal SP drives metastasis in a ssRNA-dependent manner.
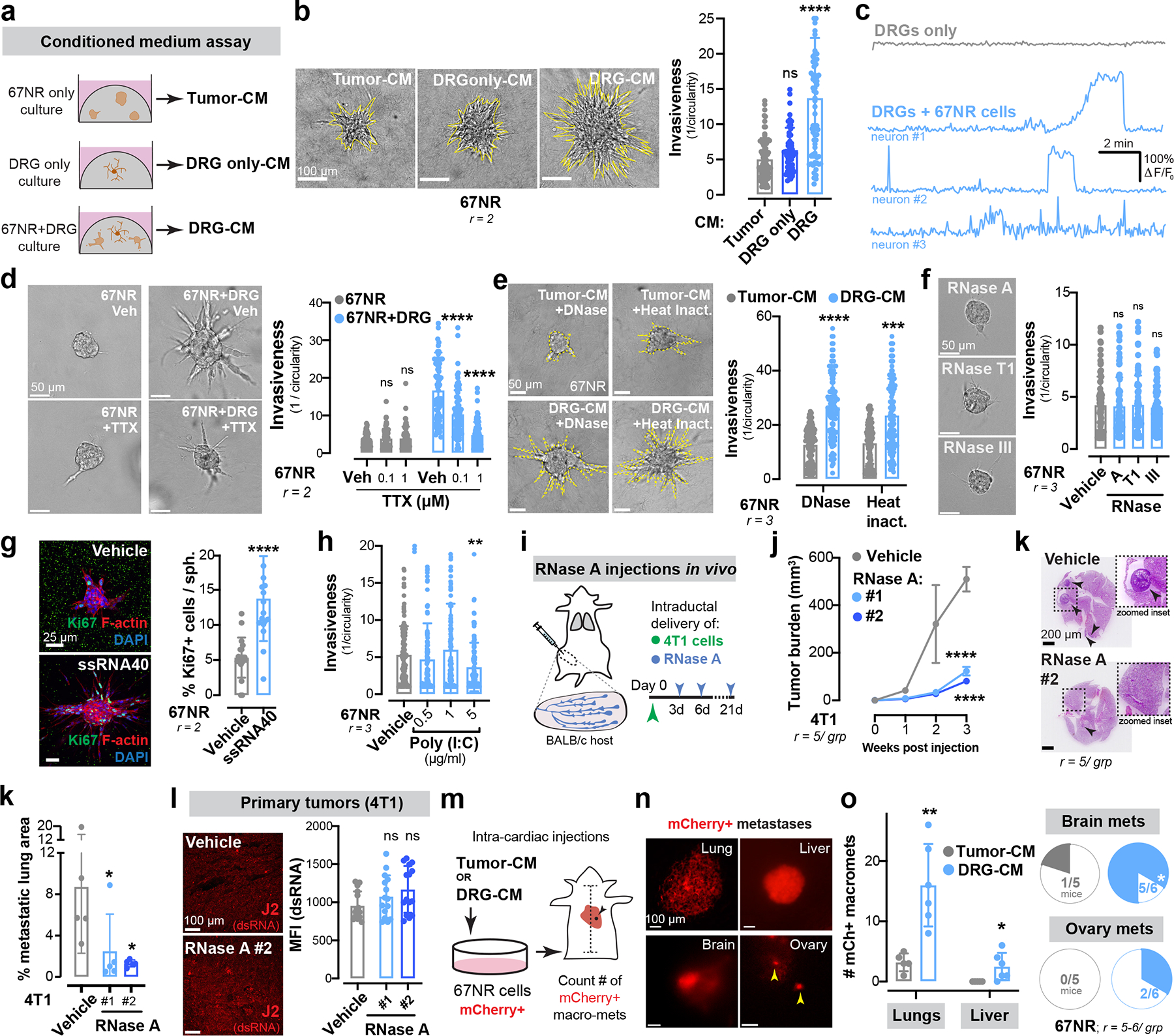
(a-b) Schematic for isolation of conditioned medium from tumour only, DRG only, or tumour-DRG cultures (a). Invasion quantification, nsp = 0.1463, ****p < 0.0001, Kruskal-Wallis test (b). Mean ± SD.
(c) Calcium fluorescence traces (ΔF/F0) of DRG neurons cultured alone or in the presence of 67NR cancer cells. F = measured fluorescence, F0 = baseline fluorescence.
(d) Invasion quantification of 67NR spheroids co-cultured with DRG neurons in the presence of a sodium channel blocker, tetrodotoxin (TTX). nsp > 0.9999, ****p < 0.0001, Kruskal-Wallis test. Mean ± SD.
(e) Invasion quantification of 67NR spheroids cultured with DNase-treated or heat inactivated tumour-CM or DRG-CM. ***p = 0.001, ****p < 0.0001, Kruskal-Wallis test. Mean ± SD.
(f) Invasion quantification of 67NR spheroids cultured with solely RNase A, RNase T1 or RNase III. nsp > 0.9999, Kruskal-Wallis test. Mean ± SD.
(g) Proliferation quantification of 67NR spheroids cultured with or without ssRNA40. ****p < 0.0001, Mann-Whitney test. Mean ± SD.
(h) Invasion quantification of 67NR spheroids cultured with a dsRNA mimetic, Poly (I:C). **p = 0.0072, Kruskal-Wallis test. Mean ± SD.
(i-k) Effect of RNase A treatment on 4T1 tumour growth and metastasis. (i) Schematic. (j) Tumour growth. ****p < 0.0001, ANOVA. Mean ± SEM. (k) Metastatic area quantified by H&E. *p = 0.0392 (vehicle vs RNase A #1), *p = 0.0347 (vehicle vs RNase A #2), ANOVA. Mean ± SD.
(l) MFI of dsRNA (measured using an anti-dsRNA antibody, J2) in vehicle vs RNase A treated 4T1 primary tumours. nsp = 0.22 (RNase A #1), 0.0625 (RNase A #2), ANOVA. Mean ± SD.
(m-o) Intra-cardiac injections of mCherry+ 67NR cancer cells pre-treated with tumour-CM or DRG-CM. (m) Schematic. (n) Representative mCherry+ metastases. (o) Quantification of mCherry+ metastases. **p = 0.0028, *p = 0.0366 (liver), *p = 0.0313 (brain), t-test. Mean ± SD.
Extended Data Figure 8: SP-driven activation of TACR1 and ssRNA-driven activation of TLR7 promote breast cancer invasiveness across multiple models of breast cancer.
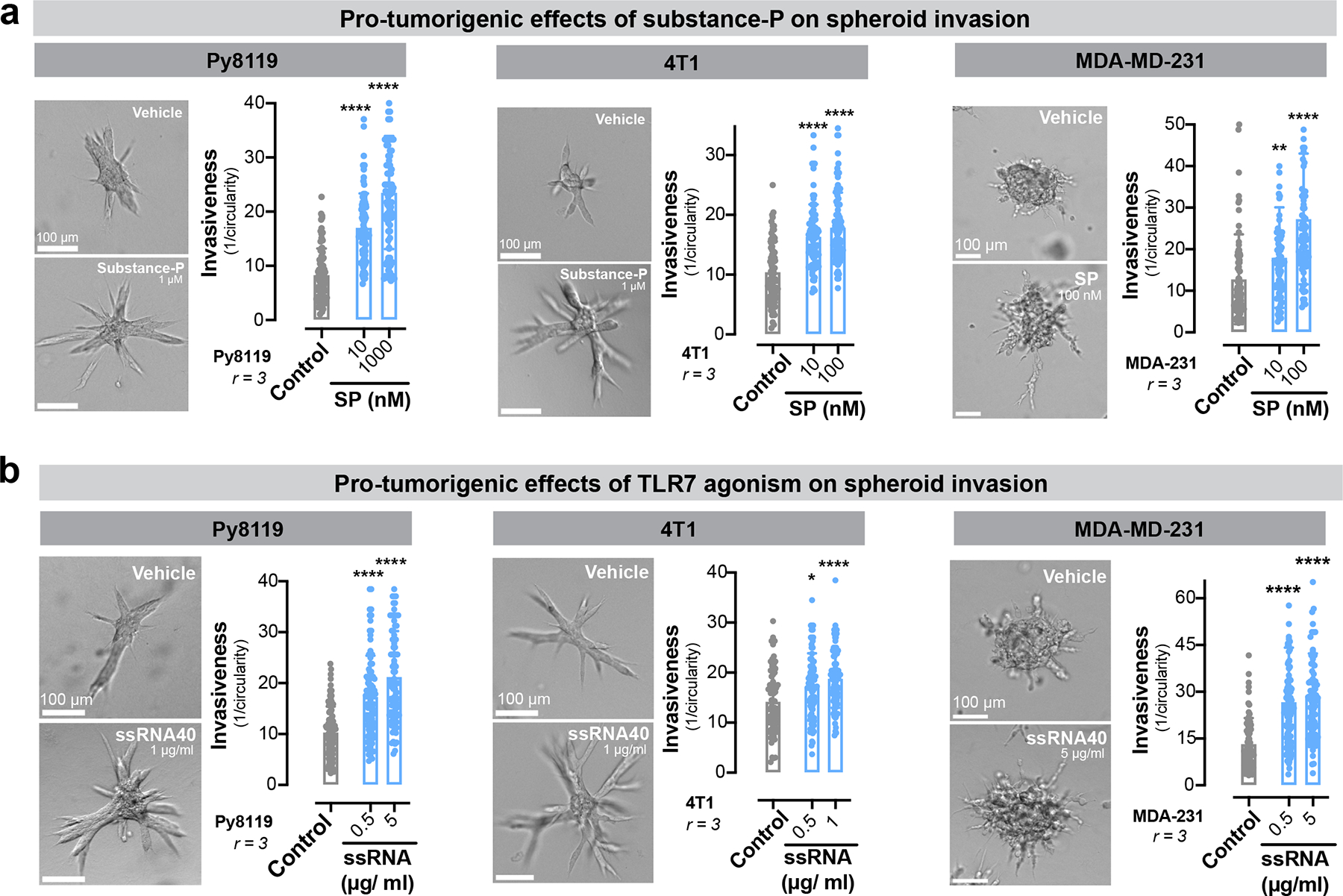
(a) Invasion quantification of Py8119, 4T1, and MDA-MB-231 breast cancer spheroids in the presence of SP. **p = 0.0013, ****p < 0.0001, Kruskal-Wallis test. Mean ± SD.
(b) Invasion quantification of Py8119, 4T1, and MDA-MB-231 breast cancer spheroids in the presence of ssRNA40. *p = 0.0235, ****p < 0.0001, Kruskal-Wallis test. Mean ± SD.
Extended Data Figure 9: Neuronal SP signals via tumoral TLR7 receptors to drive metastasis.
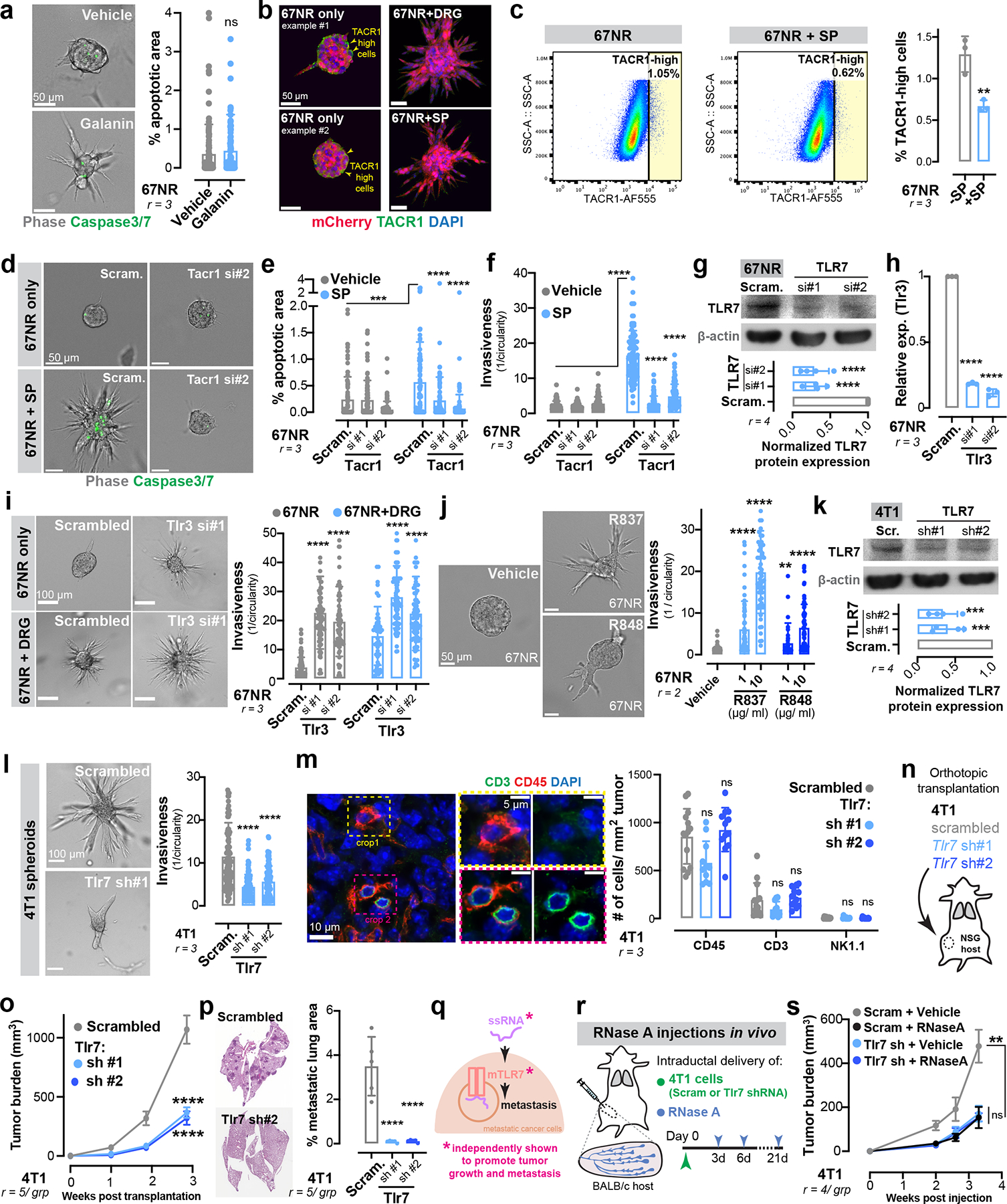
(a) Percent apoptotic area within 67NR spheroids cultured with galanin. nsp = 0.0807, Mann-Whitney test. Mean ± SD.
(b) 67NR spheroids cultured with or without DRG neurons and immunostained for TACR1. n = 10 spheroids/ group.
(c) Flow cytometry analysis of TACR1 expression in 67NR cancer cells. **p = 0.0089, t-test. Mean ± SD.
(d-f) 67NR spheroids depleted for TACR1 and cultured with SP. (e) Percent apoptotic area. ***p = 0.0003, Mann-Whitney test, ****p < 0.0001, Kruskal-Wallis test. (f) Invasion quantification. ****p < 0.0001, Kruskal-Wallis test. Mean ± SD.
(g) Validation of Tlr7 knockdown in 67NR cancer cells. ****p < 0.0001, ANOVA. Mean ± SD. MW: TLR7 (135 kDa).
(h) Validation of Tlr3 knockdown in 67NR cancer cells. ****p < 0.0001, ANOVA. Mean ± SD.
(i) Invasion quantification of 67NR spheroids depleted for TLR3 and cultured with or without DRG neurons. ****p < 0.0001, Kruskal-Wallis test. Mean ± SD.
(j) Invasion quantification of 67NR spheroids cultured in the presence of TLR7 agonists, R837 or R848. ****p < 0.0001, **p = 0.0011, Kruskal-Wallis test. Mean ± SD.
(k) Validation of Tlr7 knockdown in 4T1 cancer cells. ***p = 0.0006, ANOVA. Mean ± SD. MW: TLR7 (135 kDa).
(l) Invasion quantification of 4T1 spheroids depleted of TLR7. ****p < 0.0001, Kruskal-Wallis test. Mean ± SD.
(m) Immunostaining for CD45 (all immune), CD3 (T cells), or NK1.1 (NK cells) on 4T1 primary tumours depleted of TLR7. nsp > 0.9999 (all conditions), Kruskal-Wallis test. Mean ± SD.
(n-p) Orthotopic transplantations of 4T1 cancer cells depleted for TLR7 in NSG mice. (n) Schematic. (o) Tumour growth. ****p < 0.0001, ANOVA. Mean ± SEM. (p) Metastatic area quantified by H&E. ****p < 0.0001, ANOVA. Mean ± SD.
(q-s) Orthotopic transplantation of 4T1 cancer cells depleted for TLR7, followed by periodic injections of RNase A. (q,r) Schematics. (s) Tumour growth. **p = 0.0033, nsp = 0.9923, ANOVA. Mean ± SD.
Extended Data Figure 10: Sensory neurons activate a non-canonical TLR7 signalling axis in cancer cells which correlates with poor patient outcome.
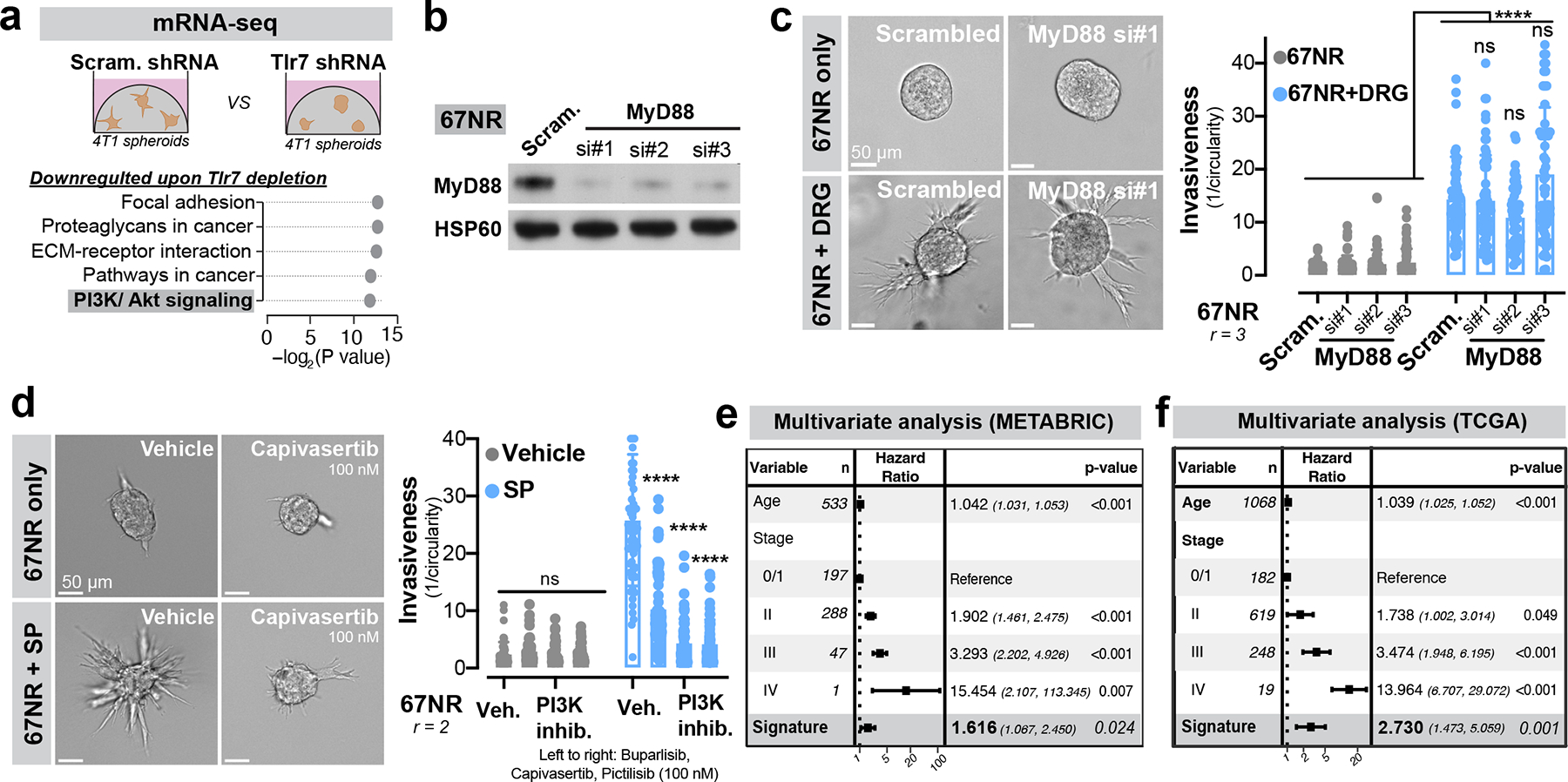
(a) mRNA sequencing of control vs Tlr7 depleted 4T1 spheroids. Top five downregulated pathways in 4T1 spheroids depleted for Tlr7 as assessed by gene set enrichment analysis (p values according to permutation testing).
(b) Validation of MyD88 knockdown in 67NR cancer cells. MW: MyD88 (33 kDa). r = 3.
(c) Invasion quantification of 67NR spheroids depleted for MyD88 and cultured with or without DRG neurons. . ****p < 0.0001, Mann-Whitney test. nsp > 0.9999 (si#1), nsp = 0.0783 (si#2), nsp = 0.4140 (si#3), Kruskal-Wallis test. Mean ± SD.
(d) Invasion quantification of 67NR spheroids cultured with or without SP and a PI3K inhibitor (left to right: buparlisib, capivasertib, or pictilisib). nsp (Veh vs buparlisib) = 0.1273, nsp (Veh vs capivasertib) = 0.6967, nsp (Veh vs pictilisib) > 0.9999, ****p < 0.0001, Kruskal-Wallis test. Mean ± SD.
(e-f) Multivariate analysis of the association of age, tumour stage and a Tlr7-dependent gene signature with survival in breast cancer patients from the METABRIC (e) and TCGA (f) datasets (p values according to multivariate Cox proportional hazard models, error bars indicate 95% confidence intervals).
Extended Data Figure 11: Aprepitant impairs tumour growth and metastasis of Py8119 and MMTV-PyMT models of breast cancer.
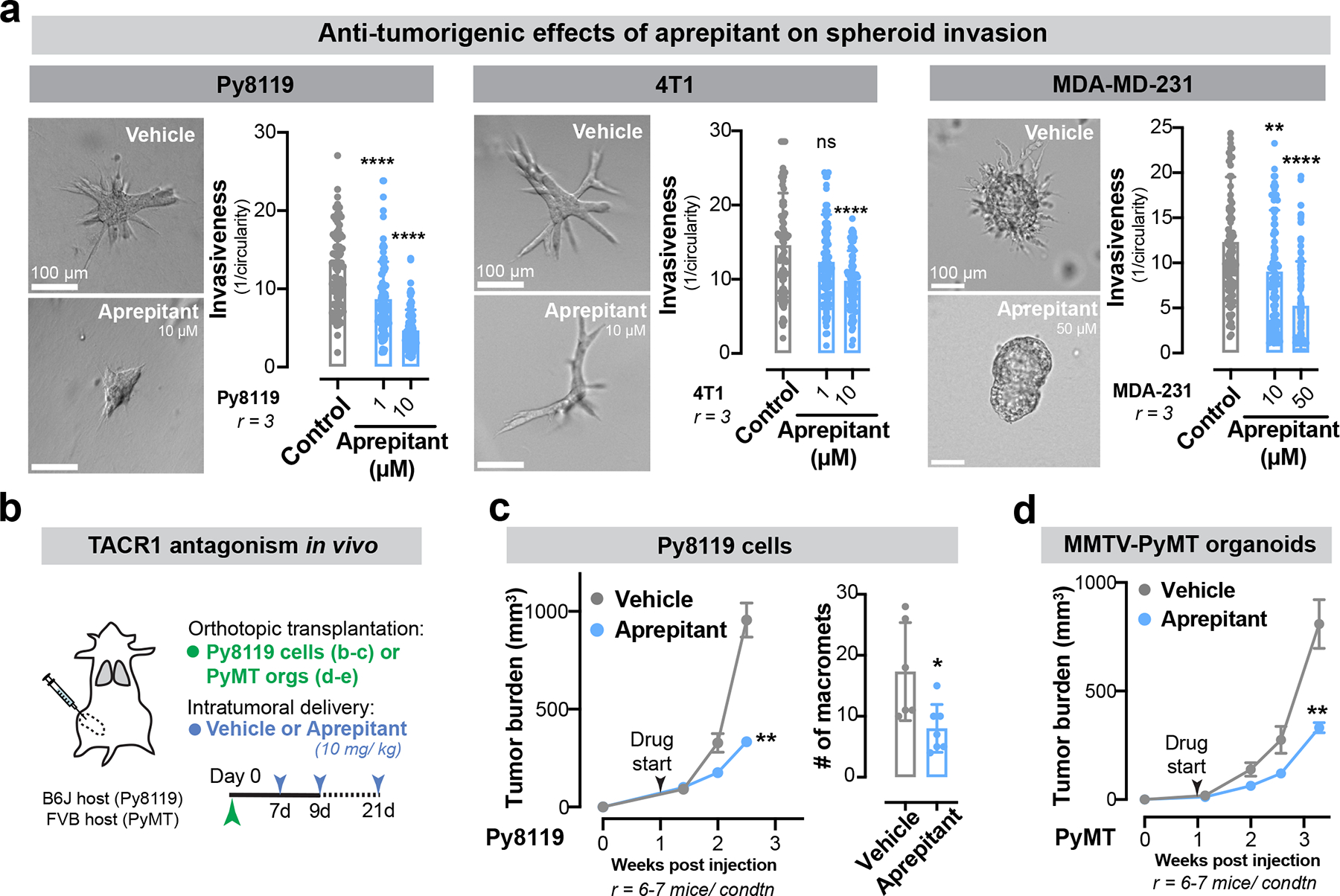
(a) Invasion quantification of Py8119, 4T1, and MDA-MB-231 breast cancer spheroids cultured in the presence of aprepitant. ****p < 0.0001, nsp = 0.9621, **p = 0.0013, Kruskal-Wallis test. Mean ± SD
(b-d) Aprepitant was evaluated for its potential in inhibiting breast cancer progression and metastasis. (b) Schematic. (c) Py8119 tumour growth and metastasis count. **p = 0.0012, *p = 0.0117, Mann-Whitney test. (d) MMTV-PyMT tumor growth. **p = 0.0047, Mann-Whitney test. Mean ± SEM.
Supplementary Material
ACKOWLEDGEMENTS
We are grateful to members of our laboratory for critical discussions and feedback on the manuscript text. We are thankful for P. Rajasethupathy’s advice on calcium imaging analysis; M. Klatt’s technical help with several animal experiments. We thank the various resource centers at Rockefeller University: A. North, C. Pyrgaki, Banerjee P., and other staff of the Bio Imaging Resource Center, C. Zhao and staff of the Genomics Resource Center, and S. Mazel and staff of the Flow Cytometry Resource Center. The results published here are in part based upon data generated by the TCGA Research Network. This work was supported by U54CA261701, R35CA274446, the Black Family Metastasis Center, the Breast Cancer Research Foundation and the Reem Kayden award. V.P. was supported by the Hope Funds for Cancer Research postdoctoral fellowship. I.K. is member of the German Academic Scholarship Foundation (Studienstiftung des deutschen Volkes) and was awarded a fellowship from Boehringer Ingelheim Fonds (BIF). B.N.O. was supported by a Max Eder grant of the German Cancer Aid (reference 70114327) and is a fellow of the digital clinician scientist program at BIH-Charité.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
Code availability. All custom computer code is publicly available under https://github.com/benostendorf/padmanaban_etal_2024.
Data availability.
Raw sequencing data and count tables for transcriptional profiling of 4T1-derived spheroids have been deposited at the Gene Expression Omnibus under accession number GSE267958. Reads were mapped to the mouse genome assembly GRCm38. Data for the METABRIC study are publicly available under the EGA accession number EGAS00000000083; data from the TCGA study are publicly available from https://portal.gdc.cancer.gov.
REFERENCES
- 1.Ayala GE et al. Cancer-related axonogenesis and neurogenesis in prostate cancer. Clin Cancer Res 14, 7593–7603 (2008). 10.1158/1078-0432.CCR-08-1164 [DOI] [PubMed] [Google Scholar]
- 2.Huang D et al. Nerve fibers in breast cancer tissues indicate aggressive tumor progression. Medicine (Baltimore) 93, e172 (2014). 10.1097/MD.0000000000000172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oertel H Innervation and Tumour Growth: A Preliminary Report. Can Med Assoc J 18, 135–139 (1928). [PMC free article] [PubMed] [Google Scholar]
- 4.Renz BW et al. beta2 Adrenergic-Neurotrophin Feedforward Loop Promotes Pancreatic Cancer. Cancer Cell 34, 863–867 (2018). 10.1016/j.ccell.2018.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Latil A et al. Quantification of expression of netrins, slits and their receptors in human prostate tumors. Int J Cancer 103, 306–315 (2003). 10.1002/ijc.10821 [DOI] [PubMed] [Google Scholar]
- 6.Osswald M et al. Brain tumour cells interconnect to a functional and resistant network. Nature 528, 93–98 (2015). 10.1038/nature16071 [DOI] [PubMed] [Google Scholar]
- 7.Venkatesh HS et al. Neuronal Activity Promotes Glioma Growth through Neuroligin-3 Secretion. Cell 161, 803–816 (2015). 10.1016/j.cell.2015.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venkatesh HS et al. Electrical and synaptic integration of glioma into neural circuits. Nature 573, 539–545 (2019). 10.1038/s41586-019-1563-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao CM et al. Denervation suppresses gastric tumorigenesis. Sci Transl Med 6, 250ra115 (2014). 10.1126/scitranslmed.3009569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magnon C et al. Autonomic nerve development contributes to prostate cancer progression. Science 341, 1236361 (2013). 10.1126/science.1236361 [DOI] [PubMed] [Google Scholar]
- 11.Balood M et al. Nociceptor neurons affect cancer immunosurveillance. Nature 611, 405–412 (2022). 10.1038/s41586-022-05374-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Globig AM et al. The beta(1)-adrenergic receptor links sympathetic nerves to T cell exhaustion. Nature (2023). 10.1038/s41586-023-06568-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerendai I et al. Transneuronal labelling of nerve cells in the CNS of female rat from the mammary gland by viral tracing technique. Neuroscience 108, 103–118 (2001). 10.1016/s0306-4522(01)00399-2 [DOI] [PubMed] [Google Scholar]
- 14.Hebb C & Linzell JL Innervation of the mammary gland. A histochemical study in the rabbit. Histochem J 2, 491–505 (1970). 10.1007/BF01003127 [DOI] [PubMed] [Google Scholar]
- 15.Tavora B et al. Tumoural activation of TLR3-SLIT2 axis in endothelium drives metastasis. Nature 586, 299–304 (2020). 10.1038/s41586-020-2774-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brose K et al. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell 96, 795–806 (1999). 10.1016/s0092-8674(00)80590-5 [DOI] [PubMed] [Google Scholar]
- 17.Nguyen-Ngoc KV et al. ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proc Natl Acad Sci U S A 109, E2595–2604 (2012). 10.1073/pnas.1212834109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aslakson CJ & Miller FR Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res 52, 1399–1405 (1992). [PubMed] [Google Scholar]
- 19.Bujak JK, Kosmala D, Szopa IM, Majchrzak K & Bednarczyk P Inflammation, Cancer and Immunity-Implication of TRPV1 Channel. Front Oncol 9, 1087 (2019). 10.3389/fonc.2019.01087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liebig C, Ayala G, Wilks JA, Berger DH & Albo D Perineural invasion in cancer: a review of the literature. Cancer 115, 3379–3391 (2009). 10.1002/cncr.24396 [DOI] [PubMed] [Google Scholar]
- 21.Kastin A Handbook of biologically active peptides. (Academic press, 2013). [Google Scholar]
- 22.Otsuka M & Konishi S Release of substance P-like immunoreactivity from isolated spinal cord of newborn rat. Nature 264, 83–84 (1976). 10.1038/264083a0 [DOI] [PubMed] [Google Scholar]
- 23.Eshete F & Fields RD Spike frequency decoding and autonomous activation of Ca2+-calmodulin-dependent protein kinase II in dorsal root ganglion neurons. J Neurosci 21, 6694–6705 (2001). 10.1523/JNEUROSCI.21-17-06694.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishikawa S et al. Two histidine residues are essential for ribonuclease T1 activity as is the case for ribonuclease A. Biochemistry 26, 8620–8624 (1987). 10.1021/bi00400a019 [DOI] [PubMed] [Google Scholar]
- 25.Robertson HD, Webster RE & Zinder ND Purification and properties of ribonuclease III from Escherichia coli. J Biol Chem 243, 82–91 (1968). [PubMed] [Google Scholar]
- 26.Bremnes RM, Sirera R & Camps C Circulating tumour-derived DNA and RNA markers in blood: a tool for early detection, diagnostics, and follow-up? Lung Cancer 49, 1–12 (2005). 10.1016/j.lungcan.2004.12.008 [DOI] [PubMed] [Google Scholar]
- 27.Huang W et al. Site-specific RNase A activity was dramatically reduced in serum from multiple types of cancer patients. PLoS One 9, e96490 (2014). 10.1371/journal.pone.0096490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Lamirande G Action of deoxyribonuclease and ribonuclease on the growth of Ehrlich ascites carcinoma in mice. Nature 192, 52–54 (1961). 10.1038/192052a0 [DOI] [PubMed] [Google Scholar]
- 29.Ledoux L Action of ribonuclease on two solid tumours in vivo. Nature 176, 36–37 (1955). 10.1038/176036a0 [DOI] [PubMed] [Google Scholar]
- 30.Lund JM et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A 101, 5598–5603 (2004). 10.1073/pnas.0400937101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawasaki T & Kawai T Toll-like receptor signaling pathways. Front Immunol 5, 461 (2014). 10.3389/fimmu.2014.00461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ojaniemi M et al. Phosphatidylinositol 3-kinase is involved in Toll-like receptor 4-mediated cytokine expression in mouse macrophages. Eur J Immunol 33, 597–605 (2003). 10.1002/eji.200323376 [DOI] [PubMed] [Google Scholar]
- 33.Ha T et al. TLR2 ligands induce cardioprotection against ischaemia/reperfusion injury through a PI3K/Akt-dependent mechanism. Cardiovasc Res 87, 694–703 (2010). 10.1093/cvr/cvq116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hesketh PJ et al. The oral neurokinin-1 antagonist aprepitant for the prevention of chemotherapy-induced nausea and vomiting: a multinational, randomized, double-blind, placebo-controlled trial in patients receiving high-dose cisplatin--the Aprepitant Protocol 052 Study Group. J Clin Oncol 21, 4112–4119 (2003). 10.1200/JCO.2003.01.095 [DOI] [PubMed] [Google Scholar]
- 35.Rosso M, Robles-Frias MJ, Covenas R, Salinas-Martin MV & Munoz M The NK-1 receptor is expressed in human primary gastric and colon adenocarcinomas and is involved in the antitumor action of L-733,060 and the mitogenic action of substance P on human gastrointestinal cancer cell lines. Tumour Biol 29, 245–254 (2008). 10.1159/000152942 [DOI] [PubMed] [Google Scholar]
- 36.Munoz M & Rosso M The NK-1 receptor antagonist aprepitant as a broad spectrum antitumor drug. Invest New Drugs 28, 187–193 (2010). 10.1007/s10637-009-9218-8 [DOI] [PubMed] [Google Scholar]
- 37.Nagakawa O et al. Effect of prostatic neuropeptides on invasion and migration of PC-3 prostate cancer cells. Cancer Lett 133, 27–33 (1998). 10.1016/s0304-3835(98)00186-4 [DOI] [PubMed] [Google Scholar]
- 38.Nizam E & Erin N Differential consequences of neurokinin receptor 1 and 2 antagonists in metastatic breast carcinoma cells; Effects independent of Substance P. Biomed Pharmacother 108, 263–270 (2018). 10.1016/j.biopha.2018.09.013 [DOI] [PubMed] [Google Scholar]
- 39.Austin M, Elliott L, Nicolaou N, Grabowska A & Hulse RP Breast cancer induced nociceptor aberrant growth and collateral sensory axonal branching. Oncotarget 8, 76606–76621 (2017). 10.18632/oncotarget.20609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jurcak NR et al. Axon Guidance Molecules Promote Perineural Invasion and Metastasis of Orthotopic Pancreatic Tumors in Mice. Gastroenterology 157, 838–850 e836 (2019). 10.1053/j.gastro.2019.05.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamiya A et al. Genetic manipulation of autonomic nerve fiber innervation and activity and its effect on breast cancer progression. Nat Neurosci 22, 1289–1305 (2019). 10.1038/s41593-019-0430-3 [DOI] [PubMed] [Google Scholar]
- 42.Zahalka AH et al. Adrenergic nerves activate an angio-metabolic switch in prostate cancer. Science 358, 321–326 (2017). 10.1126/science.aah5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Partecke LI et al. Subdiaphragmatic vagotomy promotes tumor growth and reduces survival via TNFalpha in a murine pancreatic cancer model. Oncotarget 8, 22501–22512 (2017). 10.18632/oncotarget.15019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Renz BW et al. Cholinergic Signaling via Muscarinic Receptors Directly and Indirectly Suppresses Pancreatic Tumorigenesis and Cancer Stemness. Cancer Discov 8, 1458–1473 (2018). 10.1158/2159-8290.CD-18-0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boilly B, Faulkner S, Jobling P & Hondermarck H Nerve Dependence: From Regeneration to Cancer. Cancer Cell 31, 342–354 (2017). 10.1016/j.ccell.2017.02.005 [DOI] [PubMed] [Google Scholar]
- 46.Kalinichenko VV, Mokyr MB, Graf LH Jr., Cohen RL & Chambers DA Norepinephrine-mediated inhibition of antitumor cytotoxic T lymphocyte generation involves a beta-adrenergic receptor mechanism and decreased TNF-alpha gene expression. J Immunol 163, 2492–2499 (1999). [PubMed] [Google Scholar]
- 47.Mohammadpour H et al. beta2 adrenergic receptor-mediated signaling regulates the immunosuppressive potential of myeloid-derived suppressor cells. J Clin Invest 129, 5537–5552 (2019). 10.1172/JCI129502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benci JL et al. Opposing Functions of Interferon Coordinate Adaptive and Innate Immune Responses to Cancer Immune Checkpoint Blockade. Cell 178, 933–948 e914 (2019). 10.1016/j.cell.2019.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao YQ et al. Primary afferent tachykinins are required to experience moderate to intense pain. Nature 392, 390–394 (1998). 10.1038/32897 [DOI] [PubMed] [Google Scholar]
- 50.Wang Y et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 465, 483–486 (2010). 10.1038/nature09002 [DOI] [PubMed] [Google Scholar]
- 51.Guy CT, Cardiff RD & Muller WJ Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol 12, 954–961 (1992). 10.1128/mcb.12.3.954-961.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maroulakou IG, Anver M, Garrett L & Green JE Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc Natl Acad Sci U S A 91, 11236–11240 (1994). 10.1073/pnas.91.23.11236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hale JJ et al. Structural optimization affording 2-(R)-(1-(R)-3, 5-bis(trifluoromethyl)phenylethoxy)-3-(S)-(4-fluoro)phenyl-4- (3-oxo-1,2,4-triazol-5-yl)methylmorpholine, a potent, orally active, long-acting morpholine acetal human NK-1 receptor antagonist. J Med Chem 41, 4607–4614 (1998). 10.1021/jm980299k [DOI] [PubMed] [Google Scholar]
- 54.Padmanaban V et al. Organotypic culture assays for murine and human primary and metastatic-site tumors. Nat Protoc 15, 2413–2442 (2020). 10.1038/s41596-020-0335-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young L, Sung J, Stacey G & Masters JR Detection of Mycoplasma in cell cultures. Nat Protoc 5, 929–934 (2010). 10.1038/nprot.2010.43 [DOI] [PubMed] [Google Scholar]
- 56.Foty R A simple hanging drop cell culture protocol for generation of 3D spheroids. J Vis Exp (2011). 10.3791/2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heil F et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303, 1526–1529 (2004). 10.1126/science.1093620 [DOI] [PubMed] [Google Scholar]
- 58.Alexopoulou L, Holt AC, Medzhitov R & Flavell RA Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413, 732–738 (2001). 10.1038/35099560 [DOI] [PubMed] [Google Scholar]
- 59.Hemmi H et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol 3, 196–200 (2002). 10.1038/ni758 [DOI] [PubMed] [Google Scholar]
- 60.Jurk M et al. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat Immunol 3, 499 (2002). 10.1038/ni0602-499 [DOI] [PubMed] [Google Scholar]
- 61.Maira SM et al. Identification and characterization of NVP-BKM120, an orally available pan-class I PI3-kinase inhibitor. Mol Cancer Ther 11, 317–328 (2012). 10.1158/1535-7163.MCT-11-0474 [DOI] [PubMed] [Google Scholar]
- 62.Davies BR et al. Preclinical pharmacology of AZD5363, an inhibitor of AKT: pharmacodynamics, antitumor activity, and correlation of monotherapy activity with genetic background. Mol Cancer Ther 11, 873–887 (2012). 10.1158/1535-7163.MCT-11-0824-T [DOI] [PubMed] [Google Scholar]
- 63.Folkes AJ et al. The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. J Med Chem 51, 5522–5532 (2008). 10.1021/jm800295d [DOI] [PubMed] [Google Scholar]
- 64.Padmanaban V et al. E-cadherin is required for metastasis in multiple models of breast cancer. Nature 573, 439–444 (2019). 10.1038/s41586-019-1526-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krause S, Brock A & Ingber DE Intraductal injection for localized drug delivery to the mouse mammary gland. J Vis Exp (2013). 10.3791/50692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jancso G, Kiraly E, Such G, Joo F & Nagy A Neurotoxic effect of capsaicin in mammals. Acta Physiol Hung 69, 295–313 (1987). [PubMed] [Google Scholar]
- 67.Chi J et al. Three-Dimensional Adipose Tissue Imaging Reveals Regional Variation in Beige Fat Biogenesis and PRDM16-Dependent Sympathetic Neurite Density. Cell Metab 27, 226–236 e223 (2018). 10.1016/j.cmet.2017.12.011 [DOI] [PubMed] [Google Scholar]
- 68.Luppi PH, Fort P & Jouvet M Iontophoretic application of unconjugated cholera toxin B subunit (CTb) combined with immunohistochemistry of neurochemical substances: a method for transmitter identification of retrogradely labeled neurons. Brain Res 534, 209–224 (1990). 10.1016/0006-8993(90)90131-t [DOI] [PubMed] [Google Scholar]
- 69.Gee KR et al. Chemical and physiological characterization of fluo-4 Ca(2+)-indicator dyes. Cell Calcium 27, 97–106 (2000). 10.1054/ceca.1999.0095 [DOI] [PubMed] [Google Scholar]
- 70.Curtis C et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012). 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data and count tables for transcriptional profiling of 4T1-derived spheroids have been deposited at the Gene Expression Omnibus under accession number GSE267958. Reads were mapped to the mouse genome assembly GRCm38. Data for the METABRIC study are publicly available under the EGA accession number EGAS00000000083; data from the TCGA study are publicly available from https://portal.gdc.cancer.gov.


