Abstract
Triple-negative breast cancer (TNBC) is a highly invasive breast cancer subtype that is challenging to treat due to inherent heterogeneity and absence of estrogen, progesterone, and human epidermal growth factor 2 receptors. Kinase signaling networks drive cancer growth and development, and kinase inhibitors are promising anti-cancer strategies in diverse cancer subtypes. Kinase inhibitor screens are an efficient, valuable means of identifying compounds that suppress cancer cell growth in vitro, facilitating the identification of kinase vulnerabilities to target therapeutically. The Kinase Chemogenomic Set is a well-annotated library of 187 kinase inhibitor compounds that indexes 215 kinases of the 518 in the known human kinome representing various kinase networks and signaling pathways, several of which are understudied. Our screen revealed 14 kinase inhibitor compounds effectively inhibited TNBC cell growth and proliferation. Upon further testing, three compounds, THZ531, THZ1, and PFE-PKIS 29, had the most significant and consistent effects across a range of TNBC cell lines. These cyclin-dependent kinase (CDK)12/CDK13, CDK7, and phosphoinositide 3-kinase inhibitors, respectively, decreased metabolic activity in TNBC cell lines and promote a gene expression profile consistent with the reversal of the epithelial-to-mesenchymal transition, indicating these kinase networks potentially mediate metastatic behavior. These data identified novel kinase targets and kinase signaling pathways that drive metastasis in TNBC.
Keywords: breast cancer, compound screen, inhibitor, kinase
Introduction
Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer and has a propensity to metastasize. Compared to other breast cancer subtypes, TNBC often has worse clinical outcomes and has high early recurrence rates [1]. TNBC is a highly invasive, metastatic subtype known for its lack of expression of estrogen, progesterone receptors, and human epidermal growth factor receptor 2 (HER2) [2], making it very difficult to treat with hormone receptor-specific therapies like tamoxifen [3]. Therefore, there is a need to develop drugs that more effectively target TNBC.
Metastasis contributes significantly to worse outcomes. The TNBC 5-year relative survival rate drops to less than 15% after metastasizing to distant regions of the body, compared to a nearly 32% survival rate for all breast cancers combined. In contrast, localized TNBC has a 92% survival rate [4], demonstrating the urgent need to identify novel therapies that prevent metastasis.
Epithelial-to-mesenchymal transition (EMT) is the process by which epithelial cells adopt a more mesenchymal phenotype. While necessary in the context of embryonic development, EMT is utilized by cancer cells to invade and migrate to other sites during metastasis [5]. When cancer cells upregulate mesenchymal genes, they acquire greater potential for invasion and migration. In contrast, the upregulation of epithelial genes results in a more benign phenotype, indicating that the cells may be less likely to migrate and progress [6].
Several factors influence TNBC development and progression, including environmental factors and genetic mutations [7]. While kinases have key roles in cell growth and survival, dysregulated expression or activity of kinases results in aberrant cell growth and proliferation and can promote tumor development. However, kinases are clinically relevant due to their targetability [8]. For example, hormone receptor-positive/HER2− breast cancer is treated with a cyclin-dependent kinase (CDK)4/CDK6 inhibitor combination therapy [9,10] which drives cell cycle arrest [11]. Multiple kinase inhibitors are used for the treatment of other breast cancer subtypes as well, including lapatinib, alpelisib, and everolimus, which target HER2, phosphoinositide 3-kinase (PI3K), and mechanistic target of rapamycin (mTOR), respectively [12]. Kinase inhibition has proven to be an effective means of therapy for multiple cancer types, including breast cancer [13,14]. Currently, the only kinase inhibitors being used to treat TNBC are tumor agnostic in nature [15,16], but there are currently no kinase inhibitor compounds FDA-approved for use specifically against TNBC [17].
While kinase inhibitors show great potential as therapeutics, some kinases are vastly understudied, and therefore, their value as targets is unknown. There are 518 known kinases in the human kinome, and 164 are encoded near gene regions involved in tumor formation [18]. Understanding the role of all kinases is necessary to understand diseases and how to treat them effectively. In order to help identify potential kinase vulnerabilities, the Kinase Chemogenomic Set (KCGS), a library of 187 kinase inhibitors indexing 215 human kinases [19], was developed. Screening the KCGS for activity on TNBC cells may provide insight into druggable nodes and shed light on the roles of studied and understudied ‘dark’ kinases.
In this study, a patient-derived xenograft (PDX) model of TNBC was used. PDX models are generated from a primary tumor isolated from a cancer patient that is transplanted and propagated in immunodeficient mice [20]. PDX cancer models often maintain the attributes of the original tumor, including metastatic potential and cell morphology [21,22]. Therefore, primary cell lines derived from PDXs are more translational models than previously established cell lines.
Here, we used a PDX-derived TNBC cell line, TU-BcX-4IC cells (4IC), and evaluated cell growth, migration, and EMT expression after exposure to the KCGS kinase inhibitors. After initial screening for morphological changes, more in-depth analyses were performed on 4IC and additional TNBC cell lines: BT-20, BT-549 (549), MDA-MB-231 (231), and MDA-MB-468 (468). An estrogen receptor-positive cell line, MCF-7, was included as a hormone receptor-positive cell line comparison. Morphological imaging and follow-up analyses indicated that three compounds (THZ531, THZ1, and PFE-PKIS 29) were the most effective at altering TNBC morphology, survival, and migration.
Methods
Roswell Park Memorial Institute (RPMI) 1640 medium (72400-047), fetal bovine serum (FBS) (10437-028), minimum essential medium (MEM) amino acids solution (11130-051), trypan blue (15250-061), 0.25% trypsin-EDTA (1×) (25200-056), and antibiotic-antimycotic (15240-062) were purchased from Gibco (Thermo Fisher Scientific, Waltham, Massachusetts, USA). MEM nonessential amino acid solution (M1745) and dimethyl sulfoxide (D2650) were purchased from Sigma-Aldrich (St. Louis, Missouri, USA). Sterile PBS was purchased from Cytiva (Marlborough, Massachusetts, USA).
Cell culture
Dr. Burow’s laboratory at the Tulane University School of Medicine generated the human PDX-derived TNBC cell line used in these studies, TU-BcX-4IC cells [23].
BT-20, BT-549, MCF-7, MDA-MB-231, and MDA-MB-468 cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, Virginia, USA). BT-20, BT-549, MDA-MB-231, MDA-MB-468, and TU-BcX-4IC cells were maintained in RPMI (Gibco 72400-047) supplemented with 10% FBS (Gibco 10437-028), 1% antibiotic-antimycotic (Gibco 15240-062), 1% MEM nonessential amino acids (Sigma M1745), and 1% MEM essential amino acid solution (Gibco 11130-051). MCF-7 cells were maintained in low-glucose DMEM (Gibco 11885-084) supplemented with 10% heat-inactivated FBS (HyClone SH30396.03) and 1% penicillin-streptomycin solution (HyClone SV30010, Logan, Utah, USA).
Kinase Chemogenomic Set
The KCGS library was created by the Structural Genomics Consortium at The University of North Carolina at Chapel Hill and purchased from CancerTools (cancertools.org, London, England, UK). The 187 kinase inhibitors were provided in a 384-well plate with 1 µL aliquots of 10 mM compound in DMSO. Upon receiving the plate, each well was diluted to provide a compound concentration of 1 mM in DMSO (Sigma D2650). Before treating the cells, the kinase inhibitors were diluted into complete RPMI media to a final concentration of 1 µM.
Crystal violet staining
TU-BcX-4IC cells were seeded overnight at 8000 cells/cm2 in 24-well tissue culture plates. The following day, the cells were treated with the kinase inhibitors at a concentration of 1 µM; 72 h after initial treatment, compounds were removed, and cells were washed with 1× sterile PBS. The cells were stained with 3% crystal violet (Sigma C0775) in 100% methanol (Spectrum 67-56-1) for 30 min. The stain was removed, and wells were washed with DI water until clear. The stain dried overnight.
The same protocol was followed for MCF-7, MDA-MB-231, and BT-20 cell lines, which were all seeded at 18,000 cells/cm2 in 48-well plates 1 day before treatment with the 14 hits from our initial screen on the TU-BcX-4IC cells (Table 1).
Table 1.
Fourteen kinase inhibitor compounds that were further tested on subsequent cell lines and their targeted kinases
| Compound | Kinase targets |
|---|---|
| GW810372X | GSK3B, CLK2, HIPK1 |
| GSK461364 | PLK1, NEK2 |
| PFE-PKIS 40 | PIK3CG, PIK3CA, PIK3C2G, PIK3CD, PIK3CB, PIK3C2B, MTOR, PI4KB |
| GSK237701A | NEK9, PDGFRB, PDGFRA, PLK1 |
| GW440146 | MAP2K3, BLK |
| GSK579289A | NEK2, NEK9, PLK1 |
| XMD-17-51 | NUAK1, ABL1, AURKA |
| THZ531 | CDK12, CDK13 |
| TPKI-26 | PLK1, PLK2, PLK3 |
| BI2536 | PLK1, PLK2, PLK3, RPS6KA4, CAMKK1, CAMKK2, MYLK |
| THZ1 | CDK7, IGF1R, INSR, MAP3K11, MAPK8, TBK1, TYK2 |
| TPKI-24 | PLK1, PLK2, PLK3, MYLK, DAPK3 |
| SB-772077-B | ROCK1, ROCK2, RPS6KA5, PIM1, RSK1 |
| PFE-PKIS 29 | PIK3CG, PIK3CA, PIK3C2G, PIK3C2B, PIK3CD, PIK3CB, MTOR, PI4KB |
The cells were imaged at 4× and 20× magnification using an inverted Agilent Cytation 10 confocal imaging system (Santa Clara, California, USA) and quantified using ImageJ software (National Institutes of Health, Bethesda, Maryland, USA; http://imagej.nih.gov/ij/).
Immunofluorescence staining
TU-BcX-4IC, MDA-MB-231, BT-549, and MCF-7 cells were seeded overnight in 8-well chamber slides (Nunc Lab Tek 154534; Rochester, New York, USA) at a density of 3500, 7000, 4500, and 4500 cells/cm2, respectively. Cells were treated with 1 µM kinase inhibitor for 72 h. The cells were fixed in 10% formalin and then stained with Phalloidin-iFluor 488 Reagent (Abcam ab176753; Waltham, Boston, Massachusetts, USA) according to the manufacturer’s protocol. Counterstaining with DAPI-AntiFade was performed. The cells were imaged at 40× magnification using an inverted Agilent Cytation 10 confocal imaging system.
Quantitative reverse transcription-PCR
Total RNA from vehicle-treated cancer cells (DMSO) and compound-treated cancer cells were isolated using an RNeasy Mini RNA extraction kit (Qiagen, Germantown, Maryland, USA). A total of 500 ng of RNA was used for cDNA synthesis with an Applied Bioscience purification kit (Thermo Fisher Scientific, USA). qRT-PCR was performed using the SYBR Green qPCR SuperMix (Bio-Rad, Hercules, California, USA) according to the manufacturer’s protocol. Oligonucleotide primers were designed using the vendor’s software (IDT, Coralville, Iowa, USA). Table 2 lists the primer sequences used for qRT-PCR. PCR conditions were 2 min at 95 °C, 40 cycles of 15 s at 95 °C, and 30 s at 60 °C. The target and reference genes were amplified in separate wells. All reactions were performed in duplicate. The 2−∆∆CT method was used to quantify gene expressions and normalized data to glyceraldehyde-3-phosphate dehydrogenase, which was used as an internal control.
Table 2.
List of primers used for qRT-PCR
| Gene | Forward (5'–3') | Reverse (5'–3') |
|---|---|---|
| CDH1 | GTTAAGCACAACAGCAACA | GCATCAGCATCAGTCACT |
| CD24 | TGCTCCTACCCACGCAGATT | GGCCAACCCAGAGTTGGAA |
| VIM | GAGAACTTTGCCGTTGAAGC | GCTTCCTGTAGGTGGCAATC |
| JAG1 | GACTCATCAGCCGTGTCTCA | TGGGGAACACTCACACTCAA |
| SNAI1 | TTCTCCTCTACTTCAGTCTCT | GGCTGAGGTATTCCTTGTT |
| GAPDH | CGCTGAGTACGTCGTGGAGTC | GCAGGAGGCATTGCAGATGA |
CDH1, epithelial cadherin; CD24, cluster of differentiation 24; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; JAG1, Jagged1; qRT-PCR, Quantitative reverse transcription-PCR; SNAI1, snail family transcriptional repressor 1; VIM, Vimentin.
Cell Titer Glo assay
TU-BcX-4IC, MCF-7, and MDA-MB-231 cells were seeded in a 96-well plate at 45,000 cells/cm2 density. The cells were treated with THZ531, THZ1, and PFE-PKIS 29 for 72 h at a concentration of 1 µM. After treatment, the Cell Titer Glo (CTG; Promega G7570; Madison, Wisconsin, USA) was performed according to the manufacturer’s protocol.
XTT assay
MDA-MB-231, TU-BcX-4IC, MDA-MB-468, BT-20, BT-549, and MCF-7 cells were seeded in a 96-well plate at 45,000 cells/cm2 density. The cells were treated with THZ531, THZ1, and PFE-PKIS 29 for 72 h at a concentration of 1 µM. After treatment, the CyQUANT XTT Cell Viability Assay (Thermo Fisher X12223) was performed according to the manufacturer’s protocol.
Boyden chamber migration assay
TU-BCX-4IC cells were seeded in a Boyden chamber in Opti-MEM (Gibco 353182) containing 1 µM THZ531, THZ1, or PFE-PKIS 29 at a density of 25,000 cells per chamber. DMSO-containing Opti-MEM was used as a vehicle control. Complete RPMI media was added below the Boyden chamber as a chemoattractant. Cells were incubated for 24 h and stained with 3% crystal violet in methanol. After washing, cells remaining in the top of the Boyden chamber were carefully removed using a cotton swab. Cells were imaged and quantified using ImageJ analysis software.
Statistical analysis
GraphPad PRISM 8 (GraphPad Software, Boston, Massachusetts, USA) was used for all statistical analyses. Paired t-test was used to determine the differences between the control and treated groups. Asterisks (*) indicate statistical significance: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Results
Significant morphological changes detected in breast cancer cells treated with kinase inhibitors
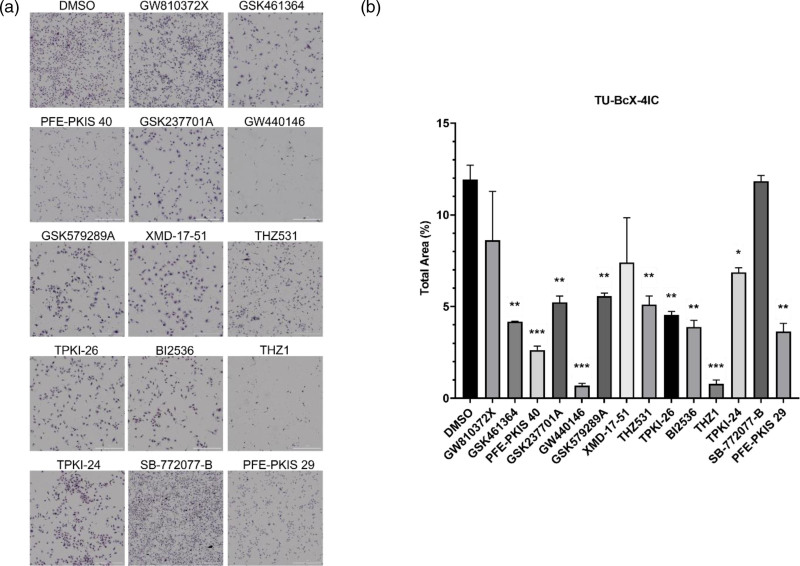
TU-BcX-4IC cells were treated with the KCGS compound library at a concentration of 1 µM and, after 72 h, stained the cells with crystal violet (see Supplementary data 1, Supplemental digital content 1, http://links.lww.com/ACD/A564). The crystal violet dye stains proteins, DNA, and lipids. During EMT, mesenchymal cells have more fibroblastic shapes, while epithelial cells are more round [24], and morphological changes may give insight into major gene and protein expression alterations [25]. Therefore, this technique provided preliminary insight into morphological changes and suggested EMT transition changes that could be observed after treatment. A total of 14 compounds caused noticeable morphological changes in vitro (Table 1), including but not limited to decreased cell number and spread cytoplasm (Fig. 1a). However, only 11 of these compounds (GSK461364, PFE-PKIS 40, GSK237701A, GW440146, GSK579289A, THZ531, TPKI-26, BI2536, THZ1, TPKI-24, and PFE-PKIS 29) caused a significant, quantifiable decrease in cell number compared to the vehicle-treated control suggesting these kinase inhibitors affected cell growth or survival with inhibitor treatment (Fig. 1b).
Fig. 1.
Crystal violet staining of TU-BcX-4IC cells after treatment with KCGS library for 72 h. (a) Morphological changes were observed in treated cells kinase inhibitor drugs (GW810372X, GSK461364, PFE-PKIS 40, GSK237701A, GW440146, GSK579289A, XMD-17-51, THZ531, TPKI-26, BI2536, THZ1, TPKI-24, SB-772022-B, and PFE-PKIS) at 4× magnification. (b) Quantification of the total cell area in each treatment well (4× magnification) using ImageJ analysis software. The values are the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. KCGS, Kinase Chemogenomic Set.
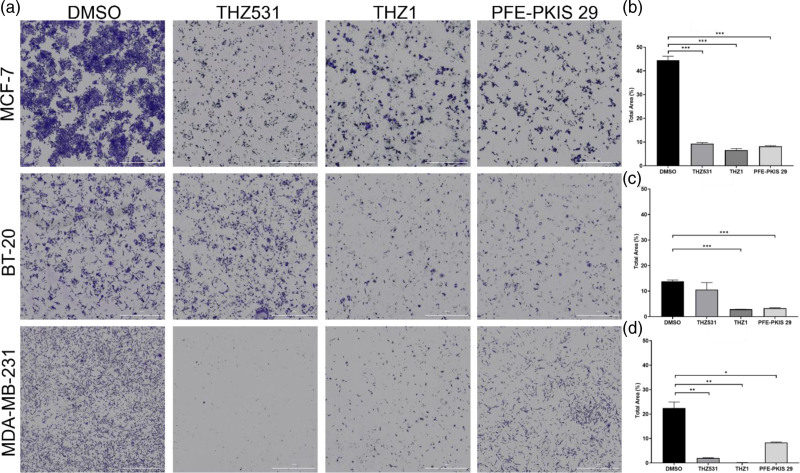
To assess whether the changes in morphology and cell growth induced by the KCGS compounds could be generally applied to TNBC, the 14 inhibitors were screened in three additional breast cancer cell lines: MCF-7, BT-20, and MDA-MB-231, and stained with crystal violet. Three compounds demonstrated consistent effects on cellular morphology across TNBC cell lines: THZ531, THZ1, and PFE-PKIS 29 (Fig. 2a). These three inhibitors cause visible and quantifiable reductions in cell number compared to vehicle-treated control groups (Fig. 2b). Screening the remaining 11 compounds of the 14 original hits in the MCF-7, BT-20, and MDA-MB-231 cell lines revealed less consistent across cell lines compared to the three lead compounds (see Supplementary data 2, Supplemental digital content 2, http://links.lww.com/ACD/A565).
Fig. 2.
THZ531, THZ1, and PFE-PKIS 29 result in significant morphological changes to other triple-negative and hormone-dependent breast cancer cell lines. (a) MCF-7, BT-20, and MDA-MB-231 breast cancer cell lines indicate severely altered morphologies after treatment with three kinase inhibitor drugs from the KCGS compound library. The total cell area in each treatment well for MCF-7 (b), BT-20 (c), and MDA-MB-231 (d) cell lines was quantified and compared to the vehicle-treated control group. The values are the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. KCGS, Kinase Chemogenomic Set.
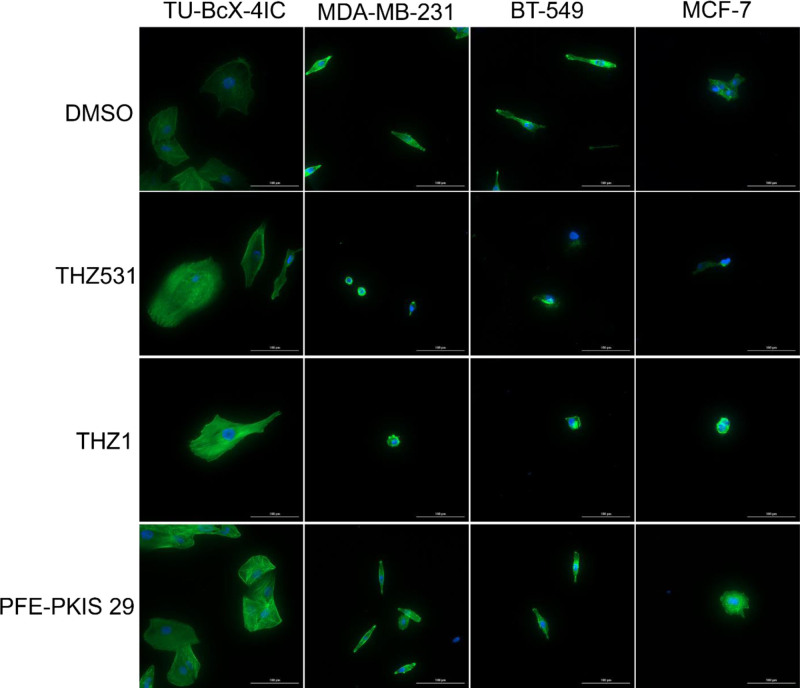
Lead compounds altered cytoskeletal actin filaments in MCF-7, TU-BcX-4IC, MDA-MB-231, and BT-549 cell lines
To further evaluate actin cytoskeletal morphology changes after treatment with the KCGS library compounds, we performed a phalloidin stain after 72 h of treatment at 1 µM on MCF-7, TU-BcX-4IC, MDA-MB-231, and BT-549 cell lines. Distinct morphological changes were observed after exposure to the kinase inhibitors (Fig. 3), including but not limited to cytoskeletal shrinkage and altered cell shape.
Fig. 3.
THZ531, THZ1, and PFE-PKIS 29 cause changes to cytoskeletal actin filaments in other triple-negative and hormone-dependent breast cancer cell lines. Phalloidin staining of actin filaments and counterstaining with DAPI further indicate morphological changes induced by compound treatment at 40× magnification.
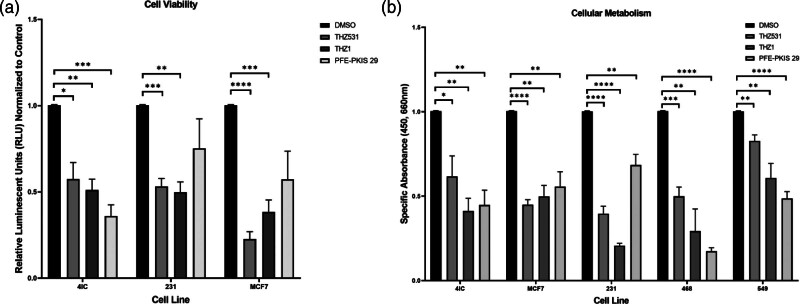
Kinase inhibition significantly decreased cellular proliferation and metabolism of breast cancer cells
In order to understand how the kinase inhibitor compounds affected viability, a CTG viability assay was performed. THZ531 and THZ1 decreased the viability of TU-BcX-4IC, MDA-MB-231, and MCF-7 breast cancer cell lines, while PFE-PKIS 29 only altered viability in the TU-BcX-4IC cell line (Fig. 4a). To validate the changes in viability seen in the CTG assay, the orthogonal XTT cell metabolic assay was performed. THZ531, THZ1, and PFE-PKIS 29 all caused a significant decrease in cellular metabolism across the five breast cancer cell lines evaluated regardless of hormone-receptor status (Fig. 4b).
Fig. 4.
THZ531, THZ1, and PFE-PKIS 29 significantly reduce metabolism and cell viability of triple-negative and estrogen-receptor-positive breast cancer cell lines. (a) Cell Titer Glo viability assay shows significantly downregulated viability after 72 h of treatment with novel kinase inhibitor compounds in vitro (n = 3). (b) XTT metabolic assay shows significant downregulation of metabolic activity after 72 h of treatment with novel kinase inhibitor drugs obtained from the KCGS compound library (n = 3). Values normalized to vehicle-treated control. The values are the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. KCGS, Kinase Chemogenomic Set.
THZ531, THZ1, and PFE-PKIS 29 significantly changed epithelial-to-mesenchymal transition marker gene expression
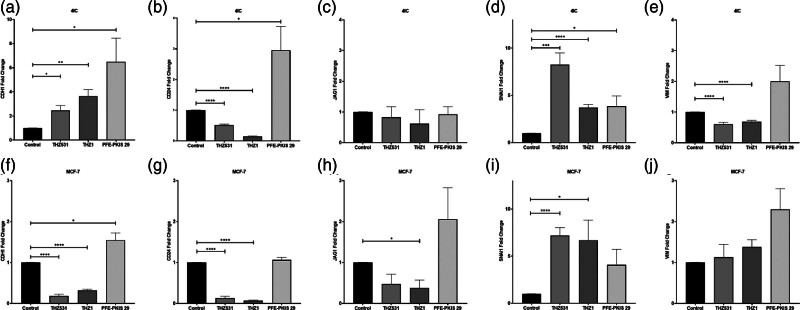
To determine if exposure to the kinase inhibitors impacted transcription of genes that drive the EMT suggested by changes in morphology, qRT-PCR was performed after 1 µM THZ531, THZ1, and PFE-PKIS 29 treatments on TU-BcX-4IC and MCF-7 cells (Fig. 5). In both cell lines, THZ531 and THZ1 decrease the expression of cluster of differentiation 24 (CD24) and Jagged1 (JAG1), suggesting a reversal of EMT. Snail family transcriptional repressor 1 (SNAI1) expression is upregulated in both cell lines treated with all three lead compounds, suggesting that EMT is likely. However, upregulated epithelial cadherin (CDH1) paired with downregulated CD24 and Vimentin (VIM) in the THZ-treated TU-BcX-4IC is potentially indicative of another process being involved.
Fig. 5.
Expression of epithelial-to-mesenchymal transition markers after 72 h of treatment of MCF-7 (a–e) and 4IC (f–j) cell lines. qRT-PCR showing the expression of CDH1 (a and f), CD24 (b and g), JAG1 (c and h), SNAI1 (d and i), and VIM (e and j). The values are the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (n = 3). CDH1, epithelial cadherin; CD24, cluster of differentiation 24; JAG1, Jagged1; qRT-PCR, Quantitative reverse transcription-PCR; SNAI1, snail family transcriptional repressor 1; VIM, Vimentin.
However, other cell-specific gene expression changes are associated with THZ531, THZ1, and PFE-PKIS 29 treatment. Upregulation of CDH1 and downregulation of VIM in the 4IC cell line treated with the THZ compounds may suggest that the morphologic changes are less likely due to a process like EMT after exposure to these specific kinase inhibitors. The PFE-PKIS 29-treated MCF-7 cells upregulated CDH1, CD24, and JAG1, which was not seen during treatment with THZ compounds. Overall, downregulation of some mesenchymal-representing genes with an increased expression of some genes representing an epithelial phenotype after THZ531 and THZ1 treatment in the TU-BcX-4IC cell line suggests reversal of EMT or a more epithelial phenotype. The effects of compound treatment on MCF-7 cells were less convincing, as reduced expression of CDH1 and amplified expression of VIM were associated with THZ531 and THZ1 treatment. However, lower expression of CD24 and JAG1 suggests that THZ531 and THZ1 treatment may affect the ability of MCF-7 cells to undergo EMT; however, other processes may be involved.
THZ531, THZ1, and PFE-PKIS 29 suppressed triple-negative breast cancer cell migration
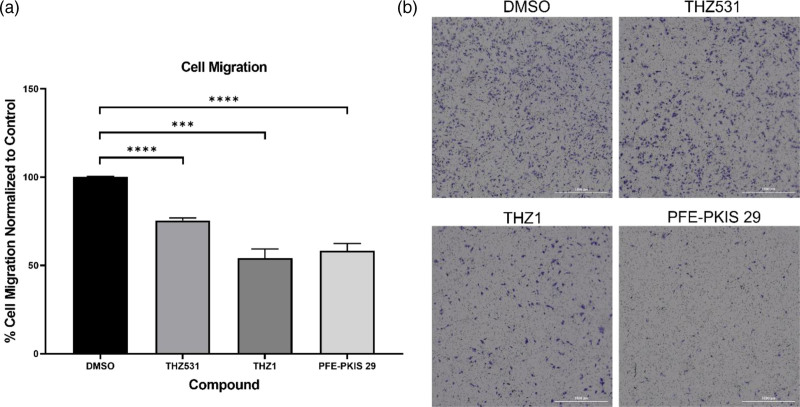
During EMT, cancer cells acquire more migratory and metastatic capabilities. To determine if changes to EMT marker expression subsequently affected migration potential, we assessed 4IC cell migration and invasion post-inhibitor treatment. Migration was reduced to 75.5%, 54.2%, and 58.3% compared to the vehicle-treated control (100%) when treated with THZ531, THZ1, and PFE-PKIS 29, respectively (Fig. 6). These data suggest that THZ531, THZ1, and PFE-PKIS 29 impair migration of TNBC cells.
Fig. 6.
(a) Migration of TU-BcX-4IC cell line during treatment with THZ531, THZ1, and PFE-PKIS 29. (b) Images were quantified using ImageJ analysis software and normalized to the control. The values are the mean ± SEM. ***P < 0.001; ****P < 0.0001 (n = 4).
Discussion
Kinases have been studied for their important roles in the progression of cancers [26] including in TNBC. AMP-activated protein kinase is upregulated in TNBC compared to non-TNBC [27]. Tumor protein p53 is upregulated in TNBC subtypes but not in all non-TNBC lines [28], indicating the potential role of an upstream kinase dysregulator. The FDA has also approved several kinase inhibitor drugs for clinical use in some breast cancers. For example, HER inhibitors are clinically approved against HER-positive breast cancers [10]. Currently, 80 FDA-approved kinase inhibitors target 22 kinases in the known human kinome [17], and about a quarter of all kinases are considered understudied [29]. Understudied kinases may be essential future targets for cancer therapeutics, and through the use of medicinal chemistry, targeting understudied kinases may be achievable [30].
In this study, we treated several breast cancer cell lines with the kinase inhibitors of the KCGS and evaluated the effects of compound treatment on cell growth and viability, metabolism, morphology, EMT marker expression, and migration. Our aim was to identify novel kinase pathways that affected breast cancer cell proliferation and metastatic capabilities to find new targets for breast cancer treatment. We identified three compounds (THZ531, THZ1, and PFE-PKIS 29) that resulted in significant and consistent morphologic alterations across cell lines and inhibited cell proliferation and growth.
EMT is associated with metastasis and drug resistance in cancer [31]. Therefore, monitoring EMT status using EMT markers and functional Boyden chamber assays is crucial to determining the ability of a compound to reduce the risk of EMT-related progression and cancer cell migration. In TU-BcX-4IC cells, upregulated CDH1 and downregulated JAG1, paired with decreased migration and invasion during treatment with all three leading compounds, indicate the potential reversal of EMT. THZ531 and THZ1 treatment caused decreased CD24 and VIM gene expression, suggesting decreased EMT capacity and potentially reversing the mesenchymal phenotype. However, upregulation of SNAI1 during treatment with all three compounds would suggest EMT progression, indicating another pathway may be involved. In MCF-7 cells, downregulation of CDH1 during treatment with THZ531 and THZ1 suggests that EMT could occur during treatment despite decreased expression of CD24 and JAG1 in the same treatment groups. Overall, qRT-PCR is a good indicator of changes at the gene level but does not provide a full story, as gene expression does not necessarily correlate with protein expression and, therefore, functional changes in the cell. All three compounds significantly inhibited the invasion and migration of TU-BcX-4IC cells through a Boyden chamber, consistent with the increased CDH1 expression and decreased CD24, JAG1, and VIM expression in these cells.
The annotation of THZ531, THZ1, and PFE-PKIS 29 suggests these compounds inhibit CDK12/CDK13, CDK7, and PI3K kinases, respectively. Higher expression of CDK13 and CDK7 is correlated with worse prognoses in breast cancers [32,33], and higher expression of CDK12 is related to later-stage breast cancers [34], suggesting the potential utility of targeting these kinases for breast cancer treatment. However, no FDA-approved compounds currently target these kinases against TNBC specifically [17], despite promising clinical trial outcomes [35].
THZ531 inhibits both CDK12 and CDK13 [36]. CDK12 plays a part in a range of processes throughout the cell, including but not limited to DNA repair, cell cycle, transcription, and translation [37]. This DNA repair regulator has implications for various cancers, including breast cancer. Its overexpression promotes breast tumor development [38]. CDK13 regulates transcript splicing [39] and may be necessary for metastatic behavior in cancer cells [40]. THZ531 promotes apoptosis through the inhibition of DNA repair [36]. Consistent with our studies, another CDK12/13 inhibitor, SR-4835, also induces TNBC death and alters gene expression pathways [38].
THZ1 mainly targets CDK7 with limited activity towards CDK12 and CDK13 [41,42]. CDK7 phosphorylates a range of downstream kinases to promote cell cycle progression under normal conditions [43,44]. Inhibitors of CDK7 have been evaluated in TNBC cells specifically, as TNBC lines appear especially sensitive to the inhibition of transcription [45]. This may provide a partial explanation of the observation that the THZ1, THZ531, and PFE-PKIS 29 all had a similar effect on the MCF-7 cell line, whereas the TNBC cell lines were much more potently affected by THZ1 than THZ531 and PFE-PKIS 29. Despite THZ1’s potency in TNBC cell lines, CDK7’s important role in transcriptional regulation is evident when inhibited in estrogen receptor-positive MCF-7 cells, as these cells become less resistant to therapy [46]. Therefore, CDK7 inhibitors may even be successful in hormone receptor-positive breast cancer. Currently, the CDK7 inhibitor Samuraciclib is in clinical trials for the treatment of hormone receptor-positive and HER2-negative breast cancers, as well as castration-resistant prostate cancer [47].
While data on the use of PFE-PKIS 29 are limited, there are studies on the kinases inhibited by PFE-PKIS 29: the family of PI3Ks (PIK3CG, PIK3CA, PIK3C2G, PIK3C2B, PIK3CD, PIK3CB), PI4KB, and mTOR. PI3K is a direct upstream activator of CDK7 [48], which may partially explain the evident and overwhelming phenotypic changes seen in the cells treated with PFE-PKIS 29. There is evidence that inhibition of several kinases simultaneously, including mTOR, significantly affects TNBC [49], warranting further investigation of PFE-PKIS 29 and its targets in TNBC. Additionally, the roles of the PI3K kinases are crucial to cell growth and division, highlighting the importance of inhibiting their activity as potential future therapeutics.
Our screen of well-annotated kinase inhibitors has identified THZ531, THZ1, and PFE-PKIS 29 as compounds that elicit strong phenotypic effects in several breast cancer cell lines, both triple-negative and estrogen receptor-positive. In conclusion, screening well-annotated compounds in disease-relevant phenotypic assays can provide insight into lead kinase targets. More in-depth examination of specific cancer cell biological pathways altered by lead compounds of interest may result in promising future therapeutics for breast cancer in general, specifically TNBC, which urgently requires improved targeted therapies.
Acknowledgements
The Structural Genomics Consortium (SGC) is a registered charity (no. 1097737) that receives funds from Bayer AG, Boehringer Ingelheim, Bristol Myers Squibb, Genentech, Genome Canada through Ontario Genomics Institute [OGI-196], EU/EFPIA/OICR/McGill/KTH/Diamond Innovative Medicines Initiative 2 Joint Undertaking [EUbOPEN grant 875510], Janssen, Merck KGaA (aka EMD in Canada and USA), Pfizer, and Takeda.
We would like to acknowledge the Cancer Prevention and Research Institute of Texas (Award # RP210046) for funding and support.
We would like to acknowledge the Burow laboratory at Tulane University for their intellectual contributions and support. We thank the Drewry laboratory at The University of North Carolina Chapel Hill for providing guidance, as well as the compounds used for our screening and subsequent treatment experiments.
Conflicts of interest
There are no conflicts of interest.
Supplementary Material
Footnotes
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website, www.anti-cancerdrugs.com.
References
- 1.Ovcaricek T, Frkovic SG, Matos E, Mozina B, Borstnar S. Triple negative breast cancer – prognostic factors and survival. Radiol Oncol 2011; 45:46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 2007; 13:4429–4434. [DOI] [PubMed] [Google Scholar]
- 3.Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol 2005; 23:7350–7360. [DOI] [PubMed] [Google Scholar]
- 4.SEER Cancer Stat Facts: Female Breast Cancer Subtypes. Bethesda, MD: National Cancer Institute. https://seer.cancer.gov/statfacts/html/breast-subtypes.html. [Accessed 1 July 2024] [Google Scholar]
- 5.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009; 119:1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest 2009; 119:1429–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene 2007; 26:2126–2132. [DOI] [PubMed] [Google Scholar]
- 8.Sarkar N, Singh A, Kumar P, Kaushik M. Protein kinases: role of their dysregulation in carcinogenesis, identification and inhibition. Drug Res (Stuttg). 2023; 73:189–199. [DOI] [PubMed] [Google Scholar]
- 9.Shah M, Nunes MR, Stearns V. CDK4/6 inhibitors: game changers in the management of hormone receptor–positive advanced breast cancer? Oncology (Williston Park) 2018; 32:216–222. [PMC free article] [PubMed] [Google Scholar]
- 10.Kannaiyan R, Mahadevan D. A comprehensive review of protein kinase inhibitors for cancer therapy. Expert Rev Anticancer Ther 2018; 18:1249–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol 2016; 13:417–430. [DOI] [PubMed] [Google Scholar]
- 12.Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Rev Dis Primers 2019; 5:66. [DOI] [PubMed] [Google Scholar]
- 13.GIancu G, Serban D, Badiu CD, Tanasescu C, Tudosie MS, Tudor C, et al. Tyrosine kinase inhibitors in breast cancer (Review). Exp. Ther. Med 2021; 23:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Zhao S, Xin Q, Zhang Y, Wang K, Li M. Recent progress of CDK4/6 inhibitors’ current practice in breast cancer. Cancer Gene Ther 2024; 2. doi: 10.1038/s41417-024-00747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subbiah V, Wolf J, Konda B, Kang H, Spira A, Weiss J, et al. Tumour-agnostic efficacy and safety of selpercatinib in patients with RET fusion-positive solid tumours other than lung or thyroid tumours (LIBRETTO-001): a phase 1/2, open-label, basket trial. Lancet Oncol 2022; 23:1261–1273. [DOI] [PubMed] [Google Scholar]
- 16.Drilon A, Siena S, Ou SI, Patel M, Ahn MJ, Lee J, et al. Safety and antitumor activity of the multi-targeted Pan-TRK, ROS1, and ALK inhibitor entrectinib (RXDX-101): combined results from two phase 1 trials (ALKA-372-001 and STARTRK-1). Cancer Discov. 2017; 7:400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roskoski R, Jr. Properties of FDA-approved small molecule protein kinase inhibitors: a 2024 update. Pharmacol Res 2024; 200:107059. [DOI] [PubMed] [Google Scholar]
- 18.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science 2002; 298:1912–1934. [DOI] [PubMed] [Google Scholar]
- 19.Wells CI, Al-Ali H, Andrews DM, Asquith CRM, Axtman AD, Dikic I, et al. The kinase chemogenomic set (KCGS): an open science resource for kinase vulnerability identification. Int J Mol Sci 2021; 22:566–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai Y, Wei X, Lin S, Qin L, Cheng L, Li P. Current status and perspectives of patient-derived xenograft models in cancer research. J. Hematol. Oncol 2017; 10:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med 2011; 17:1514–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida GJ. Applications of patient-derived tumor xenograft models and tumor organoids. J. Hematol. Oncol 2020; 13:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matossian MD, Chang T, Wright MK, Burks HE, Elliott S, Sabol RA, et al. In-depth characterization of a new patient-derived xenograft model for metaplastic breast carcinoma to identify viable biologic targets and patterns of matrix evolution within rare tumor types. Clin Transl Oncol 2022; 24:127–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nurmagambetova A, Mustyatsa V, Saidova A, Vorobjev I. Morphological and cytoskeleton changes in cells after EMT. Sci Rep 2023; 13:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haghighi M, Caicedo JC, Cimini BA, Carpenter AE, Singh S. High-dimensional gene expression and morphology profiles of cells across 28,000 genetic and chemical perturbations. Nat Methods 2022; 19:1550–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cicenas J, Zalyte E, Bairoch A, Gaudet P. Kinases and cancer. Cancers (Basel) 2018; 10:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang X, Li X, Xie X, Ye F, Chen B, Song C, et al. High expressions of LDHA and AMPK as prognostic biomarkers for breast cancer. Breast 2016; 30:39–46. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence RT, Perez EM, Hernández D, Miller CP, Haas KM, Irie HY, et al. The proteomic landscape of triple-negative breast cancer. Cell Rep 2015; 11:630–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Essegian D, Khurana R, Stathias V, Schürer SC. The clinical kinase index: a method to prioritize understudied kinases as drug targets for the treatment of cancer. Cell Rep Med. 2020; 1:100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson B, Rosston P, Ong HW, Hossain MA, Davis-Gilbert ZW, Drewry DH. How many kinases are druggable? A review of our current understanding. Biochem J 2023; 480:1331–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du B, Shim JS. Targeting epithelial-mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules 2016; 21:965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang L, Zhu C, Jin J, Wang X, Yu L, Guan X. Expression of CDK7 correlates with molecular subtypes and predicts clinical outcomes in breast cancer. Transl Cancer Res. 2021; 10:669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu XL, Zhan R, Zhao GM, Qian ZH, Gong CC, Li YQ. Expression of CDK13 was associated with prognosis and expression of HIF-1α and beclin1 in breast cancer patients. J Invest Surg 2022; 35:442–447. [DOI] [PubMed] [Google Scholar]
- 34.Lu KQ, Li ZL, Zhang Q, Yin Q, Zhang YL, Ni WJ, et al. CDK12 is a potential biomarker for diagnosis, prognosis and immunomodulation in pan-cancer. Sci Rep 2024; 14:6574. doi: 10.1038/s41598-024-56831-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Batalini F, Xiong N, Tayob N, Polak M, Eismann J, Cantley LC, et al. Phase 1b clinical trial with alpelisib plus olaparib for patients with advanced triple-negative breast cancer. Clin Cancer Res 2022; 28:1493–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang T, Kwiatkowski N, Olson CM, Dixon-Clarke SE, Abraham BJ, Greifenberg AK, et al. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat Chem Biol 2016; 12:876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang S, Hu L, Wu Z, Chen Z, Liu S, Xu X, Qian A. CDK12: a potent target and biomarker for human cancer therapy. Cells 2020; 9:1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Filippone MG, Gaglio D, Bonfanti R, Tucci FA, Ceccacci E, Pennisi R, et al. CDK12 promotes tumorigenesis but induces vulnerability to therapies inhibiting folate one-carbon metabolism in breast cancer. Nat Commun 2022; 13:2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berro R, Pedati C, Kehn-Hall K, Wu W, Klase Z, Even Y, et al. CDK13, a new potential human immunodeficiency virus type 1 inhibitory factor regulating viral mRNA splicing. J Virol 2008; 82:7155–7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Even Y, Escande ML, Fayet C, Genevière AM. CDK13, a kinase involved in pre-mRNA splicing, is a component of the perinucleolar compartment. PLoS One 2016; 11:e0149184–e0149114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sava GP, Fan H, Coombes RC, Buluwela L, Ali S. CDK7 inhibitors as anticancer drugs. Cancer Metastasis Rev 2020; 39:805–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014; 511:616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stelzer G, Rosen R, et al. The GeneCards Suite: From Gene Data UNIT 1.30 Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics 2016; 54:1.30.1–1.30.33. [DOI] [PubMed] [Google Scholar]
- 44.Larochelle S, Merrick KA, Terret ME, Wohlbold L, Barboza NM, Zhang C, et al. Requirements for Cdk7 in the assembly of Cdk1/cyclin B and activation of Cdk2 revealed by chemical genetics in human cells. Mol Cell 2007; 25:839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Zhang T, Kwiatkowski N, Abraham BJ, Lee TI, Xie S, et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell 2015; 163:174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun B, Mason S, Wilson RC, Hazard SE, Wang Y, Fang R, et al. Inhibition of the transcriptional kinase CDK7 overcomes therapeutic resistance in HER2-positive breast cancers. Oncogene 2020; 39:50–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coombes RC, Howell S, Lord SR, Kenny L, Mansi J, Mitri Z, et al. Dose escalation and expansion cohorts in patients with advanced breast cancer in a Phase I study of the CDK7-inhibitor samuraciclib. Nat Commun 2023; 14:4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Desai SR, Pillai PP, Patel RS, McCray AN, Win-Piazza HY, Acevedo-Duncan ME. Regulation of Cdk7 activity through a phosphatidylinositol (3)-kinase/PKC-ι-mediated signaling cascade in glioblastoma. Carcinogenesis 2012; 33:10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He J, McLaughlin RP, van der Noord V, Foekens JA, Martens JWM, van Westen G, et al. Multi-targeted kinase inhibition alleviates mTOR inhibitor resistance in triple-negative breast cancer. Breast Cancer Res Treat 2019; 178:263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]