Abstract
Previous studies have described poliovirus genomes in which the internal ribosome entry (IRES) for encephalomyocarditis virus (EMCV) is positioned between the P1 and P2-P3 open reading frames of the poliovirus genome. Although these dicistronic poliovirus genomes were replication competent, most exhibited evidence of genetic instability, and the EMCV IRES was deleted upon serial passage. One possible reason for instability of the genome is that the dicistronic genome was at least 108% larger than the wild-type poliovirus genome, which could reduce the efficiency of encapsidation. To address this possibility, we have constructed dicistronic poliovirus replicons by substituting the EMCV IRES and the gene encoding luciferase in place of the poliovirus P1 region; the resulting dicistronic replicons are smaller than the wild-type poliovirus genome. One dicistronic genome was constructed in which the poliovirus 5′ nontranslated region was fused to the gene encoding luciferase, followed by the complete EMCV IRES fused to the P2-P3 region of the poliovirus genome (PV-Luc-EMCV). A second dicistronic genome, EMCV-Luc-PV, was constructed with the first 108 nucleotides of the poliovirus genome fused to the EMCV IRES, followed by the gene encoding luciferase and the poliovirus IRES fused to the remaining P2-P3 region of the poliovirus genome. Both dicistronic replicons expressed abundant luciferase following transfection of in vitro-transcribed RNA into HeLa cells at 30, 33, or 37°C. The luciferase activity detected from PV-Luc-EMCV increased rapidly during the first 4 h following transfection and then plateaued, peaking after approximately 24 h. In contrast, the luciferase activity detected from EMCV-Luc-PV increased for approximately 12 h following transfection; by 24 h posttransfection, the overall levels of luciferase activity were similar to that of PV-Luc-EMCV. To analyze encapsidation of the dicistronic replicons, we used a system in which the capsid protein (P1) is provided in trans from a recombinant vaccinia virus (VV-P1). The PV-Luc-EMCV replicon was unstable upon serial passage in the presence of VV-P1, with deletions of the EMCV IRES region detected even during the initial transfection at 37°C. Following serial passage in the presence of VV-P1 at 33 or 30°C, we detected deleted genomes in which the luciferase gene was fused with the P2-P3 genes of the poliovirus genome so as to maintain the translational reading frame. In contrast, the EMCV-Luc-PV replicon was genetically stable during passage with VV-P1 at 33 or 30°C. The encapsidation of EMCV-Luc-PV was compared to that of monocistronic replicons encoding luciferase with either a poliovirus or EMCV IRES. Analysis of the encapsidated replicons after four serial passages with VV-P1 revealed that the dicistronic replicon was encapsidated more efficiently than the monocistronic replicon with the EMCV IRES but less efficiently than the monicistronic replicon with the poliovirus IRES. The results of this study suggest a genetic predisposition for picornavirus genomes to contain a single IRES region and are discussed with respect to a role of the IRES in encapsidation.
Poliovirus (PV) is a small, nonenveloped RNA virus of the Picornaviridae family. The poliovirus genome is a single-stranded RNA approximately 7,440 nucleotides in length. The plus-strand, genomic RNA functions as mRNA for viral protein expression and serves as a template for negative-strand RNA synthesis. The genomic RNA is initially translated as a long single polyprotein, which has been subdivided into three regions: the P1 region encodes the structural capsid proteins, while the P2 and P3 regions encode nonstructural proteins (18, 30, 31). The individual proteins are released from the polyprotein by virus-encoded protease 2Apro, which cleaves the P1 from the P2-P3 region in an autocatalytic reaction (32). The protease 3Cpro processes the remaining P2-P3 region proteins (16, 34). P1 is processed in trans by a 3Cpro fusion with the viral RNA-dependent RNA polymerase (3Dpol), which is referred to as 3CDpro (8).
One of the first steps in a PV infection is translation of the genomic RNA. The initiation of translation occurs in a cap-independent manner in the 5′ nontranslated region (5′NTR), at a region referred to as the internal ribosome entry site (IRES) (12, 13, 25–27). The IRES region contains a high degree of RNA secondary structure encompassing nucleotides 134 to 556 of the PV 5′NTR (10, 19, 22). All members of the family Picornaviridae contain IRES elements in the 5′NTR. Encephalomyocarditis virus (EMCV) contains an IRES element encompassing approximately 500 nucleotides (nucleotides 335 to 836) (14). Although the EMCV IRES performs the same function as the PV IRES, there is little sequence or secondary structure homology (33). This fact has been exploited in previous studies to construct PV genomes that contain both the PV and EMCV IRES elements (dicistronic genomes). Dicistronic genomes were constructed to encode the EMCV IRES between the P1 and P2-P3 regions of PV (1, 7, 20, 21). Previous studies by Molla et al. (20, 21) and Paul et al. (24) have described the construction and characterization of dicistronic PVs in which the EMCV IRES was inserted into the PV infectious clone. Constructs which contained the EMCV IRES between the P1 and P2-P3 junction or the 2A-2B junction generated viable viruses which retained the EMCV IRES, although it was not clear for how many serial passages. Alexander et al. (1) characterized a dicistronic PV which contained the EMCV IRES positioned between the PV IRES and P1. Although this virus retained the EMCV IRES for a single passage, analysis after five serial passages revealed that the genome had lost the EMCV IRES (1). One possibility for genetic instability with the dicistronic genomes could be that all were larger than the wild-type PV genome, which might result in constraints in encapsidation and facilitate deletion upon serial passage.
To explore the reasons for the instability of the dicistronic genomes, we have used a system in which the P1 coding region of PV can be removed and replaced with a foreign gene (replicon) (6). Removal of the P1 gene allows for the insertion of approximately 2.5 kb into the PV genome without increasing the overall genome size. Using this system, we have constructed dicistronic genomes containing the IRES elements from both PV and EMCV and the gene encoding luciferase. One replicon contains the PV IRES followed by the luciferase gene, EMCV IRES, and the remaining P2-P3 region genes (PV-Luc-EMCV); this genome was similar to the previously described dicistronic PV genome (20, 21). The second replicon (EMCV-Luc-PV), which contains the IRES elements in the reciprocal order, has not been previously described. Encapsidation and serial passage of the dicistronic replicons in the presence of recombinant vaccinia virus VV-P1 revealed a difference in genetic stability between the two genomes. The results of this study are discussed with respect to an as yet unappreciated role for the IRES in enhancing encapsidation.
MATERIALS AND METHODS
Unless otherwise noted, all chemicals and reagents were purchased from Sigma Chemical Company. The tissue culture media and supplements, as well as synthetic DNA primers, restriction enzymes, Taq polymerase, Superscript preamplification system, and reagents for PCR, were purchased from Gibco BRL. The luciferase assay kits and pGEM-T Easy vector cloning kits were purchased from Promega (Madison, Wis.). Tri-reagent was purchased from Molecular Research Center, Inc. QIAquick gel extraction kits were purchased from Qiagen. MAXIscript T7 kits and RPA III kits were purchased from Ambion.
Construction of dicistronic genomes.
Plasmids pT7-IC-Gag1 (6, 28), pT7-IC-CEAsig− (3), pGEM-Luc (Promega), and pCITE-4a(+) (Novagen) were used for construction of the dicistronic genome pPV-Luc-EMCV. An EcoRI-to-SnaBI fragment of pT7-IC-CEAsig− was cloned into pT7-IC-Gag1, creating the pT7-XhoI-SnaBI cloning vector. The luciferase gene from pGEM-Luc was amplified by PCR, creating an XhoI site 5′ to the start codon and an HpaI site 3′ of the stop codon (primers, 5′-CTCGAGGAAGACGCCAAAAACATAAG-3′ and 5′-GTTAACCAATTTGGACTTTCCGCC-3′). The EMCV IRES was amplified from pCITE-4a(+) and encoded both XhoI and SnaBI sites at the 5′ end of the fragment and an HpaI site at the 3′ end, following the AUG start codon (primers, 5′-CTCGAGAAGCTTTACGTAGGTTATTTTCCACCATATTGC-3′ and 5′-GTTAACGGCCATATTATCATCGTG-3′). The EMCV IRES fragment (XhoI to HpaI) was cloned into the XhoI-SnaBI region of pT7-XhoI-SnaBI. After confirmation, the resulting plasmid was digested with XhoI and SnaBI and the luciferase fragment (XhoI to HpaI) was subcloned, resulting in plasmid pPV-Luc-EMCV. The plasmid sequence was confirmed by DNA sequencing.
For construction of the second dicistronic genome, we used plasmids pEMCV-Luc and pPV-Luc (15; see Fig. 4). A PCR fragment containing the T7 promoter, first 108 nucleotides of PV, EMCV IRES, and luciferase gene from pEMCV-Luc was amplified, creating a SacI site 3′ of the luciferase stop codon (primers, 5′-CCAGTGAATTCCTAATACGACTCACTATAGGTTAAAACAGC-3′ and 5′-GAGCTCTTACAATTTGGACTTTCCGCCC-3′). The PV IRES from pPV-Luc was amplified with a SacI site at the 5′ of the region and a SnaBI site at the 3′ end (primers, 5′-GAGCTCGACGCACAAAACCAAGTTCAATAG-3′ and 5′-TACGTACATTATGATACAATTGTCTG-3′). These two fragments, along with a SnaBI-to-EcoRI fragment from pPV-Luc, which contains the remaining PV genome and plasmid, were subcloned together in a three-way ligation. The resulting plasmid, pEMCV-Luc-PV, was characterized by restriction enzyme analysis, and the sequence was confirmed by DNA sequencing.
FIG. 4.
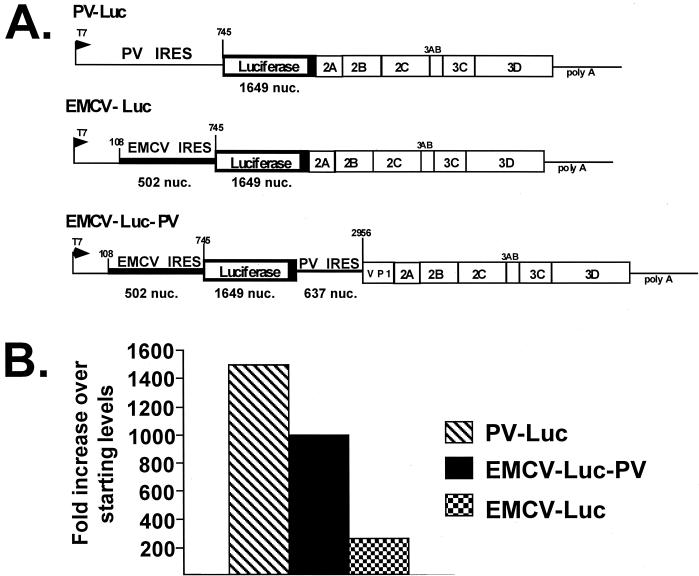
Encapsidation of the dicistronic EMCV-Luc-PV compared with that of monocistronic replicons. (A) Schematic of the three replicons used to compare encapsidation efficiency. Encapsidated dicistronic replicon EMCV-Luc-PV was obtained from serial passage in the presence of VV-P1 at 33°C. Two additional monocistronic encapsidated replicons were also used; PV-Luc encodes the luciferase gene substituted for the P1 gene of PV, and EMCV-Luc contains the first 108 nucleotides (nuc.) of the PV 5′NTR fused with the EMCV IRES followed by the luciferase gene. Both monocistronic replicons were encapsidated and propagated at 33°C. (B) The passages were initiated using replicons at approximately 0.5 infectious unit per cell. Luciferase activity was determined and designated pass 1. Three additional serial passages of the replicons were performed under identical conditions. Equal amounts of each passage were used to infect cells, and luciferase activity was determined. The fold increase in luciferase expression over starting amounts was determined for each replicon.
In vitro transcription and transfections.
Plasmids pPV-Luc-EMCV and pEMCV-Luc-PV were linearized with the restriction enzyme SalI. RNA was generated by in vitro transcription reactions using T7 RNA polymerase as previously described (6). Relative levels of in vitro-transcribed RNA were determined by spot densitometry using the Alphaimager 3.2 program. For transfection, HeLa H1 cells were first infected with recombinant vaccinia virus VV-P1 (4, 28) at 10 PFU/cell for 3 h. Equal amounts of RNA were transfected by the DEAE-dextran method as previously described (6). Transfection and subsequent incubations were all carried out at 37, 33, or 30°C.
Encapsidation of replicons.
The encapsidation and serial passage of poliovirus replicons using VV-P1 have been described previously (4, 28). Briefly, in vitro-transcribed RNA was transfected into HeLa H1 cells previously infected for 3 h with VV-P1 as described above. The cultures were harvested 48 h after transfection by three freeze-thaw cycles, overlaid on a 30% sucrose cushion (30% sucrose, 30 mM Tris [pH 8.0], 0.1 mM NaCl), and centrifuged in an SW41 rotor (Beckman) at 40,000 rpm (4°C) overnight. Supernatants were discarded; the pellets were resuspended in serum-free Dulbecco modified Eagle medium and used for serial passage.
For serial passage of the encapsidated replicons, HeLa H1 cells were infected with 10 PFU of VV-P1 per cell (at the designated temperature). After 3 h, the cells were infected with the supernatant containing the encapsidated replicons. Cultures were harvested 48 h postinfection by three successive freeze-thaw cycles and clarified by centrifugation at 14,000 rpm for 20 min. These supernatants were stored at −70°C or used for additional passages.
Luciferase assay.
For analysis of luciferase protein expression, HeLa H1 cells were transfected or infected and harvested at the designated times by being washed once with phosphate-buffered saline (PBS) and then resuspended in 1 ml of PBS. The cells were pelleted for 6 min at 14,000 rpm, and the PBS was removed; 100 μl of 1× lysis buffer (Promega) was used to resuspend the cell pellet. The samples were assayed for light detection using a luminometer (Moonlight 2010; Analytical Luminescence Laboratories) (29). The raw relative light unit (RLU) data were used to calculate the total RLU in a sample. All infections for the determination of luciferase activity were carried out overnight (15 h) at 37°C unless otherwise specified. The luciferase activities presented were representative of three different experiments.
RNA isolation and RT-PCR analysis.
Tri-reagent-LS was used to isolate total RNA from HeLa H1 cells infected overnight with passages of encapsidated replicon according to the manufacturer's instructions as modified for the Superscript kit (Gibco BRL). The total RNA was resuspended in 100 μl of RNase-free water, and 5 μl was used in the reverse transcription (RT) reaction. Two minus-sense primers, spanning PV nucleotides 3068 to 3086 (primer c; 5′-TCGAATACCAACATACGG-3′) or nucleotides 3481 to 3495 (primer d; 5′-CTACTCCACATGACG-3′), were used. The two positive-sense primers used for PCR spanned nucleotides 1541 to 1560 of luciferase (primer b; 5′-CGACGCGGGCGTGGCAGGTC-3′) or nucleotides 1 to 30 of PV (primer a; 5′-TTAAAACAGCTCTGGGGTTGTACCCACCCC-3′). The minus-sense and positive-sense primers were used in all possible combinations in the PCRs. The RT-PCR products were gel isolated, purified using a QIAquick gel extraction kit, and cloned into the pGEM-T Easy vector system. Individual colonies from the ligations were grown; plasmid DNA was extracted and used for DNA sequencing.
Comparison of the encapsidation of dicistronic and monocistronic replicons.
The monocistronic replicons PV-Luc and EMCV-Luc are described elsewhere (15; see Fig. 4). EMCV-Luc contains the nucleotides corresponding to the EMCV IRES substituted for the PV IRES (13). EMCV-Luc, PV-Luc, and EMCV-Luc-PV were encapsidated at 33°C in the presence of VV-P1. After four serial passages under identical conditions at 33°C, the amount of encapsidated replicons was estimated by luciferase expression. The amount of luciferase detected following the four serial passages was divided by the amount obtained using the starting inoculum to determine the fold increase of replicon obtained following serial passage.
RESULTS
Construction of dicistronic replicon genomes.
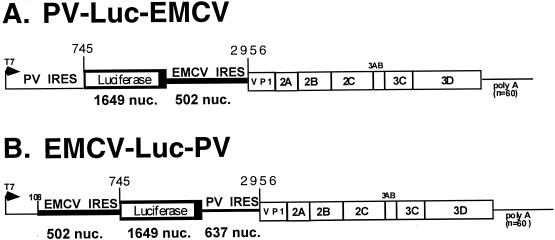
In this study, we have constructed two dicistronic replicons that encode the luciferase gene in place of the PV P1 gene (Fig. 1). The first replicon, PV-Luc-EMCV, contains the complete 5′NTR of PV followed by the gene encoding luciferase. The stop codon of the carboxy terminus of luciferase is immediately followed by the EMCV IRES (nucleotides 335 to 836 of the EMCV genome). The EMCV IRES is fused to nucleotide 2956 of the PV genome so that the AUG codon contributed by the EMCV IRES is followed, in frame, by the codons from the remaining VP1 gene. Note that the EMCV IRES does not contain the upstream poly(C) tract as found in the previously described dicistronic genomes (20, 21). Translation promoted by the EMCV IRES in this construct would be predicted to include the region encoding the final 143 amino acids of the VP1 protein, followed by the intact P2-P3 proteins of PV.
FIG. 1.
Construction of dicistronic replicons PV-Luc-EMCV and EMCV-Luc-PV. (A) PV-Luc-EMCV. The gene encoding luciferase was cloned following the complete PV 5′NTR. Following the termination codon for luciferase, the IRES from EMCV was fused to nucleotide (nuc.) 2956 of the PV genome, maintaining the translational reading frame of the VP1-P2-P3 region of the genome. The plasmid containing the dicistronic genome (pPV-Luc-EMCV) was linearized with SalI, followed by in vitro transcription to generate the replicon RNA used for transfection. The predicted genome size is 7,381 nucleotides. (B) EMCV-Luc-PV. The EMCV IRES (502 nucleotides) was cloned 3′ to the first 108 nucleotides of the PV genome. The gene encoding luciferase was cloned after the EMCV IRES. Following the termination codon for luciferase, the 637-nucleotide region of the PV 5′NTR was cloned in frame with the VP1-P2-P3 region. The plasmid containing the replicon, pEMCV-Luc-PV, was linearized with SalI prior to in vitro transcription to generate the replicon RNA.
In the second dicistronic genome constructed, the IRES of EMCV was positioned 5′ to the luciferase gene and the IRES from the PV genome was 3′ to the luciferase gene. This dicistronic genome contained the first 108 nucleotides of PV at the extreme 5′ end of the genome, as this region has been shown to be important for viral replication (2). For the PV IRES region, nucleotides 109 to 745 of the PV 5′NTR were fused in frame with the PV VP1 gene at nucleotide 2956. As with the previous dicistronic genome, the PV IRES would promote synthesis of a truncated VP1 protein followed by the complete P2-P3 region proteins. To our knowledge, this arrangement of IRES elements has not been previously described in a complete PV genome. Note that the two dicistronic genomes are identical in size and approximately 60 nucleotides smaller than the complete PV genome.
Analysis of luciferase expression from dicistronic replicons.
Luciferase protein expression was first analyzed from the two dicistronic replicons following RNA transfection. In preliminary experiments, we established that the increase in luciferase expression directly correlates with the capacity of the genome to undergo self-amplification following transfection into cells. Genomes in which the open reading frame was disrupted so that a truncated 3Dpol was expressed did not produce increased levels of luciferase after the first 2 h following transfection (data not shown). Thus, following transfection, both translation and replication of the dicistronic genomes can be ascertained by measurement of luciferase. Three different temperatures, 37, 33, and 30°C, were used for the transfections. HeLa cells were first infected with VV-P1, and 3 h later in vitro-transcribed replicon RNA was transfected into the cells. At 2, 4, 6, 11, and 24 h after transfection, luciferase activity was determined (Fig. 2). We chose to analyze translation and replication in the presence of VV-P1 since subsequent experiments would use these experimental conditions for encapsidation. Similar expression profiles were generated for each replicon at the different temperatures in the absence of VV-P1 (data not shown).
FIG. 2.
Kinetics of luciferase gene expression from dicistronic replicons PV-Luc-EMCV and EMCV-Luc-PV. Equivalent amounts of in vitro-transcribed RNA from pPV-Luc-EMCV (A) or pEMCV-Luc-PV (B) were transfected into HeLa cells previously infected with VV-P1 for 3 h. The cells were then incubated at the designated temperatures for the specified times. Cell lysates were prepared and analyzed for luciferase expression. Total RLU was plotted versus the same time posttransfection for each temperature.
Expression of luciferase from the PV-Luc-EMCV replicon increased rapidly during the first 4 h, and then leveled off, reaching a peak at 24 h. The kinetics of luciferase expression correlated with that observed for production of viral proteins from previously described dicistronic PV genomes (20, 21). The kinetics for expression of luciferase from the EMCV-Luc-PV replicon was slightly different from the PV-Luc-EMCV replicon. Luciferase activity increased in a linear fashion during the first 12 h following transfection and then plateaued, reaching a level similar to that for the replicon PV-Luc-EMCV at approximately 24 h.
Encapsidation of dicistronic replicons.
In previous studies, we have demonstrated that replicons can be encapsidated by serial passage in the presence of a recombinant vaccinia virus, VV-P1, which provides capsids proteins in trans. To monitor encapsidation, luciferase activity was analyzed following a single round of encapsidation (transfection) and after two, four, and eight serial passages in the presence of VV-P1. Analysis of luciferase expression from the replicon PV-Luc-EMCV following transfection at 37°C revealed luciferase activity from the transfection and through two serial passages ranging from 100- to 1,000-fold over background (Table 1). By serial passage 4, in the presence of VV-P1, we did not detect any luciferase activity over the background levels. Furthermore, no 3Dpol-related proteins (i.e., 3CD) were detected following metabolic labeling and immunoprecipitation. Additional analysis using RT-PCR (see below) confirmed the absence of replicon genomes. In contrast, luciferase activity was detected following transfection and all subsequent serial passages at 33 or 30°C for PV-Luc-EMCV.
TABLE 1.
Analysis of the genetic stability of PV-Luc-EMCV following encapsidation and serial passage at three different temperatures
| Passage | 37°C
|
33°C
|
30°C
|
|||
|---|---|---|---|---|---|---|
| Luciferase activity?b | Genomea | Luciferase activity? | Genome | Luciferase activity? | Genome | |
| Transfection | Yes | Deleted | Yes | Dicis | Yes | Dicis |
| 2 | Yes | Deleted | Yes | Dicis | Yes | Dicis |
| 4 | No | — | Yes | Deleted | Yes | Mixed |
| 8 | — | — | Yes | Deleted | Yes | Deleted |
Deleted, deleted genome detected by RT-PCR (see Fig. 3 for description of genomes); dicis, starting dicistronic genome sequence detected by RT-PCR; mixed, multiple bands from RT-PCR, including both dicistronic genomes and deleted genomes; —, not done.
Luciferase activity from an infection with replicons from a transfection or designated passages. Yes, 100-fold over background luciferase activity obtained from uninfected HeLa cells; No, background luciferase activity. Loss of the replicon was confirmed by no 3Dpol-related proteins immunoprecipitated from metabolically labeled cells infected with the designated passage. Note that in passages 4 and 8 at 33°C, the levels of luciferase activity were 104- to 106-fold over background. See text for details.
RT-PCR was used to analyze the genome stability of PV-Luc-EMCV during the transfection and serial passages. PCR primers were chosen to amplify critical regions of the dicistronic replicon spanning the 5′NTR of PV, the luciferase gene, the EMCV IRES, and the VP1 and 2A genes of PV. RT-PCR from PV-Luc-EMCV encapsidated at 37°C revealed the presence of smaller than predicted amplified DNA even during the first round of encapsidation (Table 1 and Fig. 3). Similarly, RT-PCR analysis of PV-Luc-EMCV passaged at 33°C revealed amplified DNA smaller than expected. In contrast to the samples passaged at 37°C, at passage 4, abundant levels of luciferase activity were still detected from the 33°C-passaged replicons; the levels at passages 4 and 8 at 33°C were 104- to 106-fold over background (Table 1). RT-PCR of PV-Luc-EMCV following transfection and serial passage at 30°C revealed that during transfection and through passage 4, DNA was amplified at the appropriate size for the intact PV IRES-luciferase-EMCV IRES genes. However, analysis of the passage 4 samples revealed amplified DNA that was full length or smaller than expected; by passage 8, only the truncated DNA was amplified by RT-PCR.
FIG. 3.
Analysis of PV-Luc-EMCV genomes following transfection and serial passage. (A) Schematic of the starting dicistronic genome. The boxed numbers refer to nucleotides in the PV genome. The underlined numbers refer to positions in the gene encoding luciferase. Nucleotides 335 to 836 of the EMCV genome correspond to the IRES [without poly(C) tract]. Primers used for RT-PCR (a to d) are marked by arrows and described in Materials and Methods. (B) Schematic of the major genomes recovered by RT-PCR from transfections and serial passages. The RT-PCR products recovered from transfections (Trfxn) or serial passages in the presence of VV-P1 (p2, p4, and p8) were cloned and sequenced. The major deleted genomes recovered at all temperatures consisted of a fusion between the luciferase (nucleotide 1798) and VP1 (nucleotide 3034) genes to maintain the translational reading frame. The original dicistronic genome was also recovered from early passages at lower temperatures. (C) Nucleotide and predicted amino acid sequences of the luciferase-VP1 gene fusion. The entire EMCV IRES was deleted from the original dicistronic construct, along with 78 nucleotides of the PV VP1 coding region (from nucleotides 2956 to 3033). The PV nucleotides are boxed, and the luciferase nucleotides are underlined. The first two nucleotides of the luciferase stop codon are in bold; the last two luciferase nucleotides and the first two PV nucleotides are identical. Note that although a Y-G amino acid dipeptide is present a few amino acids from the 3′ end of the luciferase gene, the P4 amino acid is F rather than the L required for optimal 2Apro cleavage (9).
To further characterize the nature of the deletions of the replicon genome, we cloned the RT-PCR DNA and determined the DNA sequence. The major deleted genome observed consisted of an in-frame fusion between nucleotide 1798 of the luciferase gene and nucleotide 3034 of the PV VP1 gene (Fig. 3C). This deletion was found after several independent transfections and serial passage trials at 37, 33, and 30°C. The resulting gene fusion reestablished the translational reading frame between luciferase and the VP1-P2-P3 region of the PV genome. The entire EMCV IRES element was deleted. The fusion genome was stable for at least 18 additional serial passages at 33°C. Other deletion products were found which also contained fusions between the luciferase gene and VP1 (luciferase nucleotide 1795 and nucleotide 3040 of PV or nucleotide 1453 of luciferase and 2955 of PV [this fusion also contained 13 random nucleotides between the two sites]). These two fusion products were recovered from individual RT-PCR clones and were each detected only once. Thus, most (98%) of the deleted genomes cloned contained the 1798–3034 in-frame deletion. At present, it is not clear why we did not detect similar deleted genomes from the passages at 37°C.
In contrast, the dicistronic replicon EMCV-Luc-PV appeared stable at all three temperatures examined for serial passage in the presence of VV-P1 (Table 2). Luciferase activity was detected following transfection and up to two serial passages at 37°C (at least 100-fold over background). However, by passage 4 at 37°C, we did not detect luciferase activity above background. We confirmed that there were no replicon genomes in these cultures by using metabolic labeling and immunoprecipitation for 3CD. The dicistronic replicon, EMCV-Luc-PV, appeared stable following transfection and serial passage at 33 or 30°C. Analysis of these encapsidated replicons following eight serial passages revealed abundant luciferase activity at least 100-fold over background. Furthermore, using RT-PCR, we did not amplify smaller DNA products for the EMCV IRES-luciferase-PV IRES genes, indications of gene deletion. DNA sequence of the RT-PCR product confirmed the presence of the starting dicistronic genome (data not shown).
TABLE 2.
Analysis of the genetic stability of EMCV-Luc-PV following encapsidation and serial passagea
| Passage | 37°C
|
33°C
|
30°C
|
|||
|---|---|---|---|---|---|---|
| Luciferase activity? | Genome | Luciferase activity? | Genome | Luciferase activity? | Genome | |
| Transfection | Yes | Dicis | Yes | Dicis | Yes | Dicis |
| 2 | Yes | Dicis | Yes | Dicis | Yes | Dicis |
| 4 | No | — | Yes | Dicis | Yes | Dicis |
| 8 | — | — | Yes | Dicis | Yes | Dicis |
For details, see the footnotes to Table 1.
Comparison of the encapsidation of dicistronic EMCV-Luc-PV and monocistronic replicons.
To further characterize the dicistronic replicon EMCV-Luc-PV, we compared the encapsidation of EMCV-Luc-PV with that of two previously constructed monocistronic replicons (Fig. 4A). In one replicon, EMCV-Luc, the EMCV IRES is substituted for the PV IRES; this replicon contains the first 108 nucleotides of the PV genome fused with the EMCV IRES followed by the gene encoding luciferase (15). The second replicon, PV-Luc, contains the intact PV 5′NTR fused to luciferase. All three replicons produce luciferase and have genomes that are smaller than the wild-type PV genome. We compared the relative encapsidation efficiency of each of these replicons following four serial passages. In preliminary studies, we found that the amount of luciferase activity detected following infection from the individual replicons reflected the amount of encapsidated replicon (29). The luciferase activity obtained following the fourth serial passage was compared with the luciferase activity used to initiate the serial passages (pass 1). Passage of all replicons was carried out under identical conditions at 33°C. The results are represented as fold increase over the starting levels of encapsidated replicon (Fig. 4B). The monocistronic replicon, PV-Luc, was most efficiently amplified after four serial passages, with an approximately 1,500-fold increase over the starting amount of encapsidated replicon. In contrast, the monocistronic replicon with the EMCV IRES substituted for the PV IRES (EMCV-Luc) was amplified only 200-fold following the four serial passages, a result consistent with our previous studies (15). Surprisingly, we found that the dicistronic replicon EMCV-Luc-PV was amplified approximately 1,000-fold over the starting amount, clearly greater than that observed for the monocistronic EMCV-Luc replicon but less than that found for PV-Luc.
DISCUSSION
In this study, we investigated the genetic stability and encapsidation of dicistronic PV replicons. We constructed two replicons which contained the luciferase reporter gene and the IRES regions from both PV and EMCV. The replicon PV-Luc-EMCV was genetically unstable at three different temperatures used for encapsidation. A replicon genome was identified from these passages which contained a fusion between the luciferase gene and the remaining PV polyprotein. The replicon containing this deletion was stable and could be passaged following growth at 33°C. In contrast, the replicon EMCV-Luc-PV was stable following propagation at 33 and 30°C. Comparison of the encapsidation efficiency between a monocistronic replicon, which contains the EMCV IRES substituted for the poliovirus IRES, with the dicistronic replicon EMCV-Luc-PV revealed that the dicistronic replicon was more efficiently encapsidated following serial passage in the presence of VV-P1.
Previous studies have described the characterization of dicistronic PV genomes containing the EMCV IRES positioned between the P1 and P2-P3 open reading frames or the 2A and 2B proteins. Although both genomes produced infectious viruses upon transfection, it was not clear if these viruses were genetically stable following serial passages (20, 21, 24). Genomes with the EMCV IRES positioned between the PV IRES and P1 were unstable after five serial passages, resulting in a deletion of the EMCV IRES (1). Similarly, a dicistronic genome with the gene encoding chloramphenicol acetyltransferase and the EMCV IRES was unstable following five serial passages (1). The reason for the genetic instability was not clear, since previous studies established that little sequence homology exists between the EMCV and PV IRES elements, making it unlikely that there would be genetic recombination between the two (11, 13). One possibility for the instability of these dicistronic genomes was they were all approximately 108 to 117% larger than the wild-type PV genome (1, 20, 21, 24). It was not possible, though, to address the question of whether the genome size had any relationship to the subsequent genetic instability because all of the PV genes present were required for infectivity. Replicons do not have this limitation, as the capsids are provided in trans. The dicistronic replicons then were constructed so that the overall size was similar to that of the PV genome; thus, genome size would not be the major reason for the instability of these dicistronic genomes. However, the results of our analysis of PV-Luc-EMCV demonstrated genome instability, indicating a genetic predisposition for PV to contain a single IRES element. Analysis of the replicon gene deletion found during the propagation of PV-Luc-EMCV provides some insights into the constraints that the PV genome has overcome in order to maintain a monocistronic genome. The major deleted genome recovered contained a fusion between the luciferase gene and the VP1 gene, so that the translational reading frame was maintained. The reason for the fusion at nucleotides 3034 of the PV genome is not evident. Based on what is known about PV recombination, we would speculate that the recombination occurred during minus-strand synthesis (17). It is possible that the 78 nucleotides of VP1 not found in the deleted genome might have been undergoing translation and thus blocked for access by the replicase synthesizing minus strands. In other words, the presence of ribosomes near the first 78 nucleotides of the VP1 gene might have disrupted the PV RNA-dependent RNA polymerase synthesizing a minus strand from the opposite direction, thus facilitating a translocation of the replicase to the luciferase gene. The fact that the complete luciferase gene was recovered is somewhat perplexing. One explanation might be that the polymerase had landed many times within the EMCV IRES, followed by continued RNA transcription to make a complete minus strand. Plus strand from this minus-strand RNA might not be infectious because it would lack a complete IRES for translation initiation of the P2 and P3 genes and/or the translational reading frame would not be conserved between the luciferase and remaining P2-P3 region. Thus, the first site where the PV replicase might land during recombination and maintain the translational reading frame would be the 3′ end of the luciferase gene. Once this deleted genome had been made, there was a selective advantage for this genome over the dicistronic genome. Most probably, a combination of enhanced translation/replication and encapsidation facilitated by the PV IRES contributed to the advantage of the monocistronic over the dicistronic replicon. In support of this, we found greater amplification in the encapsidation of monocistronic PV-Luc compared with any other mono- or dicistronic replicons (Fig. 4).
An extension of this rationale could be applied to understanding the genome stability of the dicistronic replicon EMCV-Luc-PV following serial passage. In this case, one deletion that might be predicted for the dicistronic EMCV-Luc-PV would be removal of the complete luciferase gene and EMCV IRES to form a smaller replicon consisting of a complete PV 5′NTR fused with the remaining VP1 gene. If this replicon occurred though, the overall genome size would be approximately 70% of the wild-type PV genome. Previous studies analyzing the lengths of PV defective interfering genomes have reported that a genome 27% the size of PV could be encapsidated (5); smaller size genomes were not tested. We have found that genomes 70% or smaller were not efficiently encapsidated using our complementation system (W. S. Choi and C. D. Morrow, unpublished data). A second deleted genome which might occur would result in the recombinant 5′NTR, including the first 108 nucleotides of the PV genome, and the EMCV IRES followed by the luciferase gene fused to the VP1-P2-P3 region of the PV genome; this replicon would have deleted the PV IRES. The overall gene structure of this replicon would be similar to that observed for the major deletion product found following passage of the dicistronic PV-Luc-EMCV. To determine why this replicon was not found, we compared the relative encapsidation efficiency of a similar replicon EMCV-Luc with that of the dicistronic replicon, EMCV-Luc-PV. We found that the dicistronic replicon, EMCV-Luc-PV, was more efficiently amplified than EMCV-Luc under identical experimental conditions. The dicistronic replicon then would have a selective advantage in encapsidation compared with the monocistronic EMCV-Luc replicon. Why the dicistronic replicon was amplified more efficiently than the monocistronic containing the EMCV IRES is not clear. Translation/replication of the monocistronic EMCV-Luc was similar, if not greater, than that for the dicistronic EMCV-Luc-PV replicon (Johansen and Morrow, unpublished). One possibility is the poliovirus IRES plays a role in enhancing encapsidation. In support of this idea, recent studies have suggested a coupling of RNA replication/translation and encapsidation (23). Since the newly synthesized plus-strand PV genome RNA could either be translated or encapsidated, it is possible that the interaction of viral proteins, possibly P1, with the IRES might be involved in the switch from translation to encapsidation. Future studies using the complementation system in combination with the monocistronic and dicistronic replicons will allow the opportunity to address this possibility.
ACKNOWLEDGMENTS
We thank Miroslav J. Novak for helpful discussions and Dee Martin for preparation of the manuscript.
We thank Sylvia McPherson, UAB AIDS Center Molecular Biology Core, for construction of dicistronic replicons (supported by grant AI27767). The UAB AIDS Center Sequencing Core Facilities (AI27767) carried out DNA sequencing. L.K.J. was supported in part by training grant T32GM08111. This work was supported by grants AI25005 and AI28147 to C.D.M.
REFERENCES
- 1.Alexander L, Lu H H, Wimmer E. Poliovirus containing picornavirus type 1 and/or type 2 internal ribosomal entry site elements: genetic hybrids and the expression of a foreign gene. Proc Natl Acad Sci USA. 1994;91:1406–1410. doi: 10.1073/pnas.91.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andino R, Rieckhof E, Achacosp P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansardi D C, Moldoveanu Z, Porter D C, Walker D E, Conry R M, LoBuglio A F, McPherson S, Morrow C D. Characterization of poliovirus replicons encoding carcinoembryonic antigens. Cancer Res. 1994;54:6359–6363. [PubMed] [Google Scholar]
- 4.Ansardi D C, Porter D C, Morrow C D. Complementation of a poliovirus defective genome by a recombinant vaccinia virus which provides P1 capsid precursor in trans. J Virol. 1993;67:3684–3690. doi: 10.1128/jvi.67.6.3684-3690.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barclay W, Li Q, Hutchinson G, Moon D, Richardson A, Percy N, Almond J W, Evans D J. Encapsidation studies of poliovirus subgenomic replicons. J Gen Virol. 1998;79:1725–1734. doi: 10.1099/0022-1317-79-7-1725. [DOI] [PubMed] [Google Scholar]
- 6.Choi W S, Pal-Ghosh R, Morrow C D. Expression of human immunodeficiency virus type 1 (HIV-1) Gag, Pol, and Env proteins from chimeric HIV-1–poliovirus minireplicons. J Virol. 1991;65:2875–2883. doi: 10.1128/jvi.65.6.2875-2883.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gromeier M, Alexander L, Wimmer E. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneic poliovirus recombinants. Proc Natl Acad Sci USA. 1996;93:2370–2375. doi: 10.1073/pnas.93.6.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris K S, Reddigari S R, Nicklin J H, Wimmer E. Purification and characterization of poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J Virol. 1992;66:7481–7489. doi: 10.1128/jvi.66.12.7481-7489.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellen C U T, Lee C K, Wimmer E. Determinants of substrate recognition by poliovirus 2A proteinase. J Virol. 1992;66:3330–3338. doi: 10.1128/jvi.66.6.3330-3338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iizuka N, Kohara M, Hagino-Yamagishi K, Abe S, Komatsu T. Construction of less neurovirulent polioviruses by introducing deletions into the 5′ noncoding sequence of the genome. J Virol. 1989;63:5354–5363. doi: 10.1128/jvi.63.12.5354-5363.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 12.Jang S K, Davies M V, Kaufman R J, Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989;63:1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jang S K, Krausslich H G, Nicklin J H, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jang S K, Pestova T V, Hellen C U T, Witherell G W, Wimmer E. Cap-independent translations of picornavirus RNAs: structure and function of the internal ribosomal entry site. Enzyme. 1990;44:292–309. doi: 10.1159/000468766. [DOI] [PubMed] [Google Scholar]
- 15.Johansen, L., and C. D. Morrow. The RNA encompassing the internal ribosome entry site in the poliovirus 5′ non-translational region enhances the encapsidation of genomic RNA. Virology, in press. [DOI] [PubMed]
- 16.Jore J, Degues B, Jackson R J, Pouwels P H, Enger-Valk B E. Poliovirus protein 3CD is the active protease for processing of the precursor protein P1 in vitro. J Gen Virol. 1988;69:1627–1636. doi: 10.1099/0022-1317-69-7-1627. [DOI] [PubMed] [Google Scholar]
- 17.Kirkegaard K, Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitamura N, Semler B L, Rothberg P G, Larsen G R, Adler C J, Dorner A J, Emini E A, Hanecak R, Lee J J, van der Werf S, Anderson C W, Wimmer E. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981;291:547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- 19.Kuge S, Nomoto A. Construction of a viable deletion and insertion mutants of the Sabin strain type 1 poliovirus: function of the 5′ noncoding sequence in viral replication. J Virol. 1987;61:1478–1487. doi: 10.1128/jvi.61.5.1478-1487.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molla A, Jang S, Paul A B, Reuer Q, Wimmer E. Cardioviral internal ribosome entry site is functional in a genetically engineered dicistronic poliovirus. Nature. 1992;356:255–257. doi: 10.1038/356255a0. [DOI] [PubMed] [Google Scholar]
- 21.Molla A A, Paul V, Schmid M, Jang S K, Wimmer E. Studies on dicistronic poliovirus implicate viral proteinase 2Apro in RNA replication. Virology. 1993;196:739–747. doi: 10.1006/viro.1993.1531. [DOI] [PubMed] [Google Scholar]
- 22.Nicholson R, Pelletier J, Le S-Y, Sonenberg N. Structural and functional analysis of the ribosome landing pad of poliovirus type 2: in vitro translation studies. J Virol. 1991;63:5886–5894. doi: 10.1128/jvi.65.11.5886-5894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nugent C I, Johnson K L, Sarnow P, Kirkegaard K. Functional coupling between replication and packaging of poliovirus replicon RNA. J Virol. 1999;73:427–435. doi: 10.1128/jvi.73.1.427-435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul V, Mugavero J, Molla A A, Wimmer E. Internal ribosomal entry site scanning of the poliovirus polyprotein: implications for proteolytic processing. Virology. 1998;250:241–253. doi: 10.1006/viro.1998.9376. [DOI] [PubMed] [Google Scholar]
- 25.Pelletier J, Kaplan G, Racaniello V R, Sonenberg N. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol Cell Biol. 1988;8:1103–1112. doi: 10.1128/mcb.8.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelletier J, Sonenberg N. Internal binding of eukaryotic ribosomes on poliovirus RNA: translation in HeLa cell extracts. J Virol. 1989;63:441–444. doi: 10.1128/jvi.63.1.441-444.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 28.Porter D C, Ansardi D C, Choi W S, Morrow C D. Encapsidation of genetically engineered poliovirus minireplicons which express human immunodeficiency virus type 1 Gag and Pol proteins upon infection. J Virol. 1993;67:3712–3719. doi: 10.1128/jvi.67.7.3712-3719.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porter D C, Ansardi D C, Wang J, McPherson S, Moldoveanu Z, Morrow C D. Demonstration of the specificity of poliovirus encapsidation using novel replicons which encode enzymatically active firefly luciferase. Virology. 1998;243:1–11. doi: 10.1006/viro.1998.9046. [DOI] [PubMed] [Google Scholar]
- 30.Rueckert R, Wimmer E. Systematic nomenclature for picornavirus proteins. J Virol. 1984;50:957–959. doi: 10.1128/jvi.50.3.957-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Semler B L, Hanecak R, Anderson C W, Wimmer E. Cleavage sites in the polypeptide precursors of poliovirus protein P2-X. Virology. 1981;114:589–594. doi: 10.1016/0042-6822(81)90242-7. [DOI] [PubMed] [Google Scholar]
- 32.Toyoda H, Nicklin M J H, Murray M G, Anderson C W, Dunn J J, Studier F W, Wimmer E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986;45:761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- 33.Wimmer E, Hellen C U T, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 34.Ypma-Wong M F, Dewalt P G, Johnson V H, Lamb J G, Semler B L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988;166:265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]