Abstract
T-20 is a synthetic peptide that potently inhibits replication of human immunodeficiency virus type 1 by interfering with the transition of the transmembrane protein, gp41, to a fusion active state following interactions of the surface glycoprotein, gp120, with CD4 and coreceptor molecules displayed on the target cell surface. Although T-20 is postulated to interact with an N-terminal heptad repeat within gp41 in a trans-dominant manner, we show here that sensitivity to T-20 is strongly influenced by coreceptor specificity. When 14 T-20-naive primary isolates were analyzed for sensitivity to T-20, the mean 50% inhibitory concentration (IC50) for isolates that utilize CCR5 for entry (R5 viruses) was 0.8 log10 higher than the mean IC50 for CXCR4 (X4) isolates (P = 0.0055). Using NL4.3-based envelope chimeras that contain combinations of envelope sequences derived from R5 and X4 viruses, we found that determinants of coreceptor specificity contained within the gp120 V3 loop modulate this sensitivity to T-20. The IC50 for all chimeric envelope viruses containing R5 V3 sequences was 0.6 to 0.8 log10 higher than that for viruses containing X4 V3 sequences. In addition, we confirmed that the N-terminal heptad repeat of gp41 determines the baseline sensitivity to T-20 and that the IC50 for viruses containing GIV at amino acid residues 36 to 38 was 1.0 log10 lower than the IC50 for viruses containing a G-to-D substitution. The results of this study show that gp120-coreceptor interactions and the gp41 N-terminal heptad repeat independently contribute to sensitivity to T-20. These results have important implications for the therapeutic uses of T-20 as well as for unraveling the complex mechanisms of virus fusion and entry.
Human immunodeficiency virus type 1 (HIV-1) binds the CD4 molecule, normally expressed on a subset of T lymphocytes, monocyte/macrophages, and dendritic cells, as the initial step of viral entry into the host cell (42). All naturally occurring isolates of HIV-1 require in addition to the CD4 molecule a chemokine receptor, usually CXCR4 or CCR5, for viral entry to occur (reviewed in references 18 and 31). Viruses that use CXCR4 (X4 viruses) or both coreceptors (X4R5 viruses) are frequently associated with CD4 T-cell depletion and disease progression in vivo, while viruses that use CCR5 (R5 viruses) usually predominate during transmission and the asymptomatic stages of HIV-1 infection (reviewed in reference 2). Entry of HIV-1 into the target cell is mediated by two envelope glycoproteins, gp120 and gp41, that are modified by extensive glycosylation and proteolytically processed from a precursor molecule, gp160 (21, 33). Mature gp41 and gp120 are noncovalently associated into oligomeric complexes that facilitate entry of the virion into the host cell (34, 44, 56). The efficiency of gp120 dissociation from gp41, neutralization by soluble CD4, and binding to CD4 have been linked to the stability of the gp120-gp41 complex (28, 34, 40, 43, 57).
The surface glycoprotein, gp120, mediates CD4 and coreceptor binding (29). The third variable (V3) loop of gp120 contains determinants of coreceptor specificity and tropism (3, 10, 19, 20, 22, 38, 45, 47, 49, 50, 52, 59). The viral transmembrane glycoprotein, gp41, contains an N-terminal extracellular domain, a membrane-spanning domain (MSD), and a C-terminal cytoplasmic tail (21, 33). The function of gp41 is to anchor the glycoprotein complex within the host-derived viral membrane and mediate membrane fusion. Conformational changes that occur in gp41 following gp120 interactions with CD4 and coreceptor promote fusion between the viral envelope and the target cell membrane (6). These conformational changes in gp41 expose a putative hydrophobic fusion peptide located near the N terminus (7), which is believed to insert into the target cell membrane. It is postulated that the bridged target cell and viral membranes are brought together as two heptad repeats within gp41 (one located C terminal to the fusion peptide [HR1] and the other located N terminal to the MSD [HR2]) associate to form a coiled-coil bundle (6, 32, 48, 51). Functional studies have shown that HR1 and HR2 are essential for virus-cell membrane fusion to occur (15, 53). Although crystallographic and biophysical data suggest that the fusion process of HIV-1 may be analogous to that described for the hemagglutinin glycoprotein of influenza virus (4, 5), the specific events remain to be defined.
Short synthetic peptides that interact with sequences within HR1 and HR2 have been used effectively to inhibit viral entry and cell to cell fusion in vitro (35, 41, 54, 55) and viral replication in vivo (25). One peptide, T-20 (formerly DP-178), corresponds to a linear 36-amino-acid sequence within HR2 and potently inhibits entry of HIV-1 into host cells most likely by interfering with the transition of gp41 into its fusion-active state (17, 26, 55). T-20 peptides are proposed to interact with a target sequence within HR1, inhibiting association with native HR2 and preventing apposition of the viral and cellular membranes (7, 25). In support of this model, Rimsky et al. identified a contiguous three-amino-acid residue sequence within HR1 (GIV at positions 36 to 38) that is critical for inhibition of HIV-1 HXB3 entry by T-20 and for efficient association between HR1 and T-20 peptides (41). The generation of viral resistance to T-20 in vitro was associated with substitutions at two positions within the GIV sequence (G→S or D and V→M). Even though the GIV sequence is highly conserved among isolates of HIV-1 (36), a commonly studied, laboratory-adapted X4 strain, NL4.3, contains a G→D substitution at position 36 within HR1. Despite this G→D substitution, NL4.3 remains sensitive to high concentrations of T-20 (41).
In this report, we show that higher concentrations of T-20 were required to inhibit primary R5 isolates than X4 isolates. By analyzing chimeric viruses containing combinations of envelope sequences derived from X4 and R5 viruses, we show that coreceptor specificity defined by the gp120 V3 loop substantially modulates sensitivity to inhibition by T-20. The results presented here show that the process of fusion initiated by interactions with CCR5 or CXCR4 is differentially susceptible to inhibitors that target intramolecular interactions of gp41. These findings have important implications for the therapeutic uses of T-20 as well as for unraveling the complex mechanisms of virus-cell membrane fusion and understanding viral resistance to peptide inhibitors and the evolution of coreceptor-specific viral quasispecies.
MATERIALS AND METHODS
Primary HIV-1 isolates.
Primary isolates of HIV-1 were obtained from infected patients with Institutional Review Board-approved informed consent at the UAB 1917 Clinic. Peripheral blood mononuclear cells (PBMC) were isolated from whole blood by Ficoll-Paque (Pharmacia, Piscataway, N.J.) gradient centrifugation and then cocultured with normal donor PBMC that had been cultured for 1 to 3 days in the presence of 3 μg of phytohemagglutinin (PHA) per ml to activate T cells. Cultures were maintained in RPMI 1640 supplemented with 15% fetal bovine serum (HyClone, Logan, Utah), 2 mM glutamine, 1× nonessential amino acids, 1× penicillin-streptomycin, and 20 or 30 U of recombinant human interleukin-2 (Roche, Indianapolis, Ind.) per ml for 7 days. At day 7, one half of the medium was removed and replaced with fresh interleukin-2-containing medium containing PHA-activated blasts. Production of virus was monitored by enzyme-linked immunosorbent assay (ELISA) (Coulter, Miami, Fla.) to detect HIV-1 p24 in the culture supernatant. At day 14, the virus-containing supernatant was collected, clarified by low-speed centrifugation, and passed through a 0.45-μm-pore-size filter to produce cell-free virus stocks. Stocks were stored in 1-ml aliquots at −70°C.
Determination of SI/NSI phenotypes and coreceptor specificity of primary isolates.
Primary isolates were used to infect MT-2 cells to determine their syncytium-inducing (SI) or noninducing (NSI) phenotype, using the AIDS Clinical Trials Group (ACTG) virology manual MT-2 assay (23). Briefly, 5 × 104 MT-2 cells were plated into 96-well plates, and 50 μl of cell-free virus stock was added. Cultures were monitored for the presence of syncytia at days 3, 6, 9, 12, and 14 and scored positive (SI) if syncytia were present or negative (NSI) if no syncytia were observed during the 14-day period. All virus stocks were also tested for infectivity in PBMC. Coreceptor specificity was determined using GHOST4 HIV indicator cells expressing CCR5, CXCR4, or no coreceptor (11). Briefly, 2 × 104 cells were plated into 12-well plates in Dulbecco modified Eagle medium (DMEM) containing selective antibiotics puromycin (except the parental line), hygromycin, and G418. The next day, the cells were infected with 0.3 ml of virus stock for 2 h at 37°C, and the culture volume was adjusted to 1 ml with complete DMEM with antibiotics. Coreceptor utilization was indicated by detection of long terminal repeat (LTR)-driven green fluorescence protein by flow cytometry at 48 or 72 h postinfection over background fluorescence in the coreceptor-negative parental cells.
T-20 phenotypic sensitivity assays.
JC53-BL HIV-1 indicator cells are a derivative of HeLa cells that express high levels of CD4 and the HIV-1 coreceptors CCR5 and CXCR4 and were kindly supplied by Tranzyme Inc., Birmingham, Ala. (X. Wu et al., unpublished data). JC53-BL cells contain reporter cassettes of luciferase and β-galactosidase that are each expressed from an HIV-1 LTR. Expression of the reporter genes is dependent on production of HIV-1 tat. JC53-BL cells were routinely subcultured every 3 to 4 days by trypsinization and were maintained in DMEM supplemented with 10% fetal bovine serum and 1× penicillin-streptomycin (cDMEM). The infectious titer of all virus stocks was determined on JC53-BL cells by direct counting of blue foci prior to analysis of T-20 inhibition (data not shown). For each set of analyses, 5 × 104 JC53-BL cells were plated per well into 24-well tissue culture plates 1 day prior to infection. An equivalent amount of each virus stock (2,000 infectious units) was added to the cell monolayer in the presence of DEAE-dextran (40 μg/ml) in DMEM in a final volume of 0.25 ml. After adsorption at 37°C for 2 h, 0.75 ml of cDMEM was added to each well. The infected cultures were incubated at 37°C with 5% CO2 in a humidified incubator for 2 days. Infections were performed and maintained in the absence of T-20 (control) or in the presence of 0.008, 0.04, 0.2, 1, and 5 μg of T-20 per ml in duplicate wells. To measure luciferase activity at 2 days postinfection, the supernatant was removed and the cells were lysed using a Promega (Madison, Wis.) luciferase assay system kit. The light intensity of each cell lysate was measured on a BMG luminometer using Lumistar version 2.04 software. Mock-infected wells were used to determine background luminescence, which was subtracted from the sample wells. Mock-infected wells were also cultured with each dose of T-20 to control for cell viability (data not shown). Two luciferase-positive control wells were included with each analysis. Virus infectivity was calculated by dividing the luciferase units (LU) produced at each concentration of T-20 by the LU produced by the control infection (no T-20). The mean 50% inhibitory concentration (IC50) was calculated for each virus using the predicted exponential growth function in Microsoft Excel, which uses existing x-y data to estimate the corresponding T-20 concentration (x) from a known value (y), which in this case was 50% infectivity. Mean IC50s were calculated using all replicates for each virus and are expressed ± the standard deviation. The mean coefficient of variation for all sets of replicate analyses was 25% (range, 9 to 42%). The Wilcoxon rank sum test was applied to pairwise comparisons to determine whether the observed differences between IC50 for individual chimeric viruses or the mean IC50s for X4 and R5 primary isolates were statistically significant.
To identify β-galactosidase-expressing cells, the supernatant was removed and the cells were fixed in 0.25% glutaraldehyde–0.8% formaldehyde in phosphate-buffered saline (PBS) for 5 min at room temperature, washed three times with PBS, and then stained using a solution containing 400 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per ml, 4 mM MgCl2, 4 mM potassium ferrocyanide, and 4 mM potassium ferricyanide in PBS for 2 h at 37°C. The staining solution was then removed; cells were washed one time with PBS and then overlaid with 0.5 ml of PBS for microscopic counting of blue foci. Counts for mock-infected wells, used to determine background, were subtracted from counts for the sample wells. The results of all luciferase experiments were confirmed by direct counting of blue foci in parallel infections (data not shown).
For infections performed in PBMC, each virus stock was individually titered on cells from a CCR5 wild-type donor by measuring p24 production at day 7 in wells containing serial dilutions of virus and calculating the 50% tissue culture infectious dose (TCID50) using the Spearman-Karber formula. PBMC from the same donor were used for the T-20 inhibition assay, in which 1,000 TCID50 of each virus stock was used to infect 106 PHA-activated PBMC in the presence or absence of 0.0016 to 1 μg of T-20 per ml, using a modification of the ACTG virology manual HIV drug susceptibility assay. Supernatant p24 was measured at day 7 postinfection for each virus and used to calculate the virus infectivity and IC50 of T-20.
HIV-1 envelope chimeras.
Plasmids containing envelope sequences derived from HIV-1 strain NL4.3 or JRFL in an NL4.3 proviral background have been previously described (37–39). Briefly, each plasmid was constructed into the backbone of the HIV-1 NL4.3 provirus by subcloning regions of the JRFL envelope gene into restriction site-modified pNL4.3. Plasmids containing gp120 V3 loops from HIV-1 strains ADA, SF162, and SF2 in an NLHX (NL4.3 containing the HXB2 envelope) proviral background have been previously described (20). The nucleotide positions of restriction sites used for subcloning (see Fig. 2), numbered according to NL4.3, are as follows: SalI, 5785; KpnI, 6343; StuI, 6822; MstII, 7305; PvuI, 7655; BamHI, 8474; and XhoI, 8887. The envelope coding region begins at position 6224, the V3 loop spans positions 7103 to 7267, the gp120/gp41 cleavage site is at 7747, and the coding region ends at 8785. To make infectious viral stocks representing each proviral construct, each plasmid was transformed into competent Escherichia coli cells and grown on Luria-Bertani agar containing ampicillin (100 μg/ml) overnight. Well-isolated colonies were used for overnight cultures, from which ampicillin-resistant plasmids were isolated. Each plasmid construct was analyzed by restriction digest to verify that all HIV-1 sequences were maintained. Each plasmid was then transfected into a 100-mm-diameter tissue culture plate containing an 80% confluent monolayer of 293T cells in cDMEM using Fugene 6 (Roche/Boehringer Mannheim, Indianapolis, Ind.) at a 3:1 ratio (Fugene volume to microgram of DNA) as specified by the manufacturer. Transfected 293T cells were incubated at 37°C with 5% CO2 in a humidified incubator for 2 days; then the virus-containing supernatant was collected, clarified by low-speed centrifugation, and passed through a 0.45-μm-pore-size filter to produce cell-free virus stocks. Stocks were stored in 0.5-ml aliquots at −70°C.
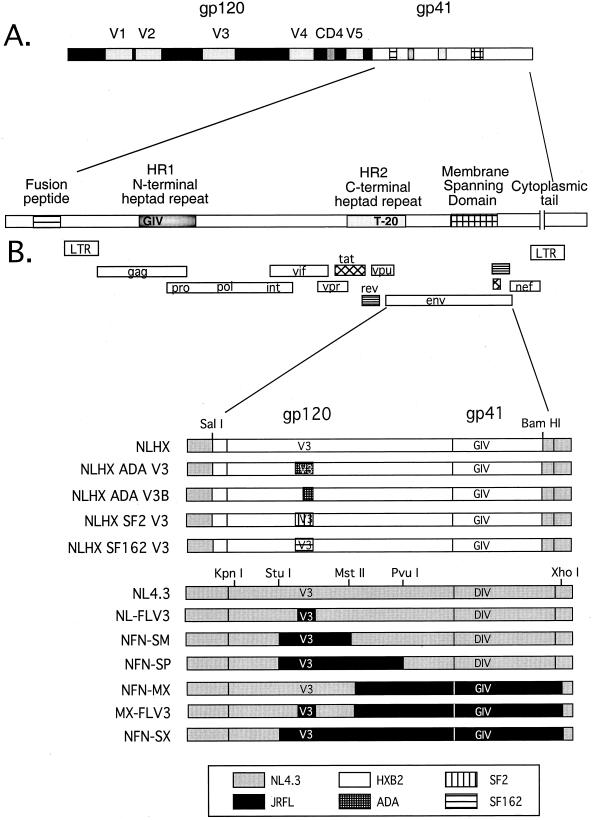
FIG. 2.
Organization of the HIV-1 envelope gene and the panel of chimeric envelope constructs used to define determinants of T-20 sensitivity. (A) Two envelope glycoproteins, gp120 and gp41, are proteolytically processed from a precursor molecule, gp160. V3, one of five variable loops in gp120, contains the major determinants of coreceptor specificity. “CD4” indicates a conserved region involved in binding to the CD4 receptor. gp41 contains an extracellular domain, an MSD, and an intracellular cytoplasmic tail. The extracellular domain contains several conserved regions that are important for fusion and entry: the fusion peptide, the N-terminal heptad repeat (HR1), and the C-terminal heptad repeat (HR2). The 36-amino-acid T-20 peptide sequence corresponds to residues within the C-terminal half of HR2. (B) All chimeras were constructed in an NL4.3 proviral background. “V3” indicates the gp120 V3 loop; “GIV or DIV” represents the critical amino acid sequence of the T-20 interaction site in the N-terminal heptad repeat (HR1). NLHX-based constructs contain the HXB2-derived envelope in an NL4.3 proviral background. Wild-type NL4.3 represents two constructs: NL4.3 and NL(Stu)Xba. NL4.3 sequences are represented by gray boxes, and the chimeric envelope sequences of JRFL, HXB2, ADA, SF2, and SF162 are indicated in the key.
RESULTS
Primary X4 isolates are more sensitive than R5 isolates to inhibition by T-20.
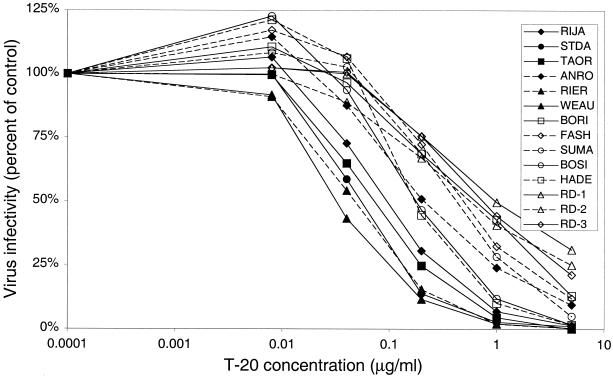
T-20 has been previously shown to inhibit cell-to-cell fusion and cell-free virus infectivity in vitro (35, 41, 54, 55) and effectively block viral replication in vivo (25). However, phenotypic resistance to T-20 can readily develop under selective pressure in both environments, especially when suboptimal doses of T-20 are used (41; G. M. Shaw, unpublished data). In this study, we analyzed the concentrations of T-20 required to inhibit a panel of primary isolates derived from HIV-1-infected subjects that were T-20 naive and represented various clinical settings (Table 1). We used a derivative of HeLa cells, JC53-BL indicator cells, which have been engineered to express surface levels of CD4, CCR5, and CXCR4 comparable to levels produced by PBMC (data not shown). This cell line is highly susceptible to infection by diverse primary isolates, including both X4 and R5 strains (Wu et al., unpublished data). These cells produce LTR-driven reporter gene products, β-galactosidase and luciferase, when infected with HIV-1 and allow quantitative measurement of virus infectivity. We obtained 14 viral isolates from HIV-1-infected individuals by patient PBMC coculture with donor PHA-activated blasts. The ability of each primary isolate to induce syncytia in cultures of MT-2 cells (23) and infect GHOST cells expressing CCR5 or CXCR4 (11) was determined (Table 1). To evaluate their sensitivity to T-20, equivalent infectious units of each viral isolate were used to infect JC53-BL cells (Fig. 1). Infections were performed and maintained in the presence of increasing doses of T-20 (0.008, 0.04, 0.2, 1, and 5 μg/ml) for 48 h. The relative level of virus infection for each isolate was calculated by the ratio of LU produced at each T-20 concentration to the control (no T-20). The mean IC50 of T-20 for all 14 isolates varied over 1.5 log10, ranging from 0.029 to 0.982 μg/ml (mean = 0.257 ± 0.272). As shown in Fig. 1, the majority of X4 viral isolates were more sensitive than the R5 isolates to T-20. The mean IC50 of T-20 for the X4 and X4R5 isolates was 0.067 ± 0.054 μg/ml (n = 6), while the mean IC50 for the R5 isolates was 0.400 ± 0.285 μg/ml (n = 8), a statistically significant difference of about 0.8 log10 (P = 0.0055). Although the X4 isolates were more sensitive to the inhibitory action of T-20 in this analysis, it was not possible to attribute this observation solely to coreceptor usage because of the inherent genetic diversity in other regions of the HIV-1 genome.
TABLE 1.
HIV-1 primary isolates obtained from T-20-naive subjects
| Viral isolatea | Patient information
|
Coreceptord | Phenotypee | |
|---|---|---|---|---|
| CD4 countb | Viral loadc | |||
| RIJA | 233 | <50 | X4 | SI |
| STDA | 218 | 33,327 | X4R5 | SI |
| TAOR | 132 | 1,057 | X4R5 | SI |
| ANRO | 220 | 155,355 | X4R5 | SI |
| RIER | 42 | 440,827 | X4R5 | SI |
| WEAU | 358 | 12,402 | X4R5 | SI |
| BORI | 931 | 171,804 | R5 | NSI |
| FASH | 262 | 1,283,820 | R5 | NSI |
| SUMA | 853 | 4,033,370 | R5 | NSI |
| BOSI | 13 | 145,986 | R5 | NSI |
| HADE | 75 | 4,012,275 | R5 | NSI |
| RD-1 | 820 | <200 | R5 | NSI |
| RD-2 | 820 | <200 | R5 | NSI |
| RD-3 | 820 | <200 | R5 | NSI |
HIV-1 primary isolates obtained by patient PBMC coculture.
CD4+ cells per microliter of blood, determined by flow cytometry.
HIV-1 RNA copies per milliliter of plasma, determined by Roche Amplicor assay.
Determined by green fluorescent protein expression in infected GHOST, GHOST-CCR5, or GHOST-CXCR4 cell lines.
Determined on MT-2 cells.
FIG. 1.
Sensitivity of HIV-1 primary isolates to inhibition by T-20. JC53-BL HIV-1 indicator cells were infected with 2,000 infectious units of each primary isolate in the absence (control) or presence of increasing doses of T-20. Cells were lysed at 48 h postinfection, and luciferase activity was measured. Virus infectivity, calculated by dividing the infectivity at each concentration of T-20 by the infectivity of the control, is plotted along the vertical axis on a linear scale; T-20 concentration is plotted along the horizontal axis on a log10 scale. Each curve represents the inhibition profile of an individual viral isolate. X4 viruses are represented by filled symbols, and R5 viruses are represented by open symbols. Each point represents the mean infectivity calculated from two independent experiments, each with duplicate wells. The range of T-20 IC50 for all isolates was 0.029 to 0.982 μg/ml (1.5 log10).
Substitution of R5 V3 loops into an X4 envelope background decreases sensitivity to T-20.
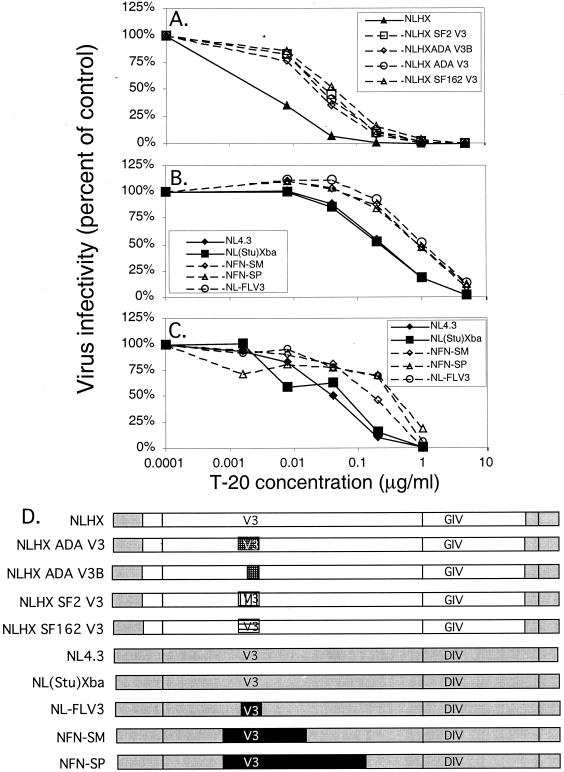
To determine whether coreceptor usage influenced sensitivity to T-20, we analyzed a panel of chimeric proviruses in which V3 loop sequences derived from a series of R5 viruses were inserted into an X4 envelope background (Fig. 2). Figure 2A shows the organization of the HIV-1 envelope gene, and Fig. 2B shows the envelope sequence combinations present in the chimeras used in this study. The first panel of chimeric viruses was constructed in an NL4.3 proviral background, and all contain gp120 and gp41 envelope sequences derived from HXB2, an X4 strain adapted to replication in T-cell lines (NLHX) (20). The V3 loop sequences (Fig. 3A) were derived from two R5 viruses, SF162 and ADA, and a T-cell line-adapted X4R5 virus, SF2, that predominantly uses CCR5 as its coreceptor (8, 20). These chimeras have been described previously and were originally constructed to analyze the determinants of CCR5 utilization (20). When the relative virus infectivity of each construct was analyzed in the presence of T-20, all R5 V3 loop substitutions into NLHX resulted in decreased sensitivity to T-20 (Fig. 4A). The NLHX virus that contains R5 V3 loop sequences derived from SF162 required 0.8 log10 more T-20 to inhibit 50% of its infectivity than NLHX (IC50 increased from 0.005 to 0.032 μg/ml, P = 0.028). Similarly, the virus that contains R5 V3 loop sequences derived from ADA demonstrated an IC50 of 0.024 μg/ml, which was 0.7 log10 higher than that for NLHX (P = 0.028). A virus that contains only the C-terminal sequences of the ADA V3 loop, NLHX ADA V3B, also had an increased IC50 of 0.019 μg/ml, about 0.6 log10 higher than that for NLHX (P = 0.028). Finally, the IC50 for the virus that contains the SF2-derived R5 V3 loop was 0.025 μg/ml, about 0.7 log10 higher than that for NLHX (P = 0.028). These results showed clearly that determinants of coreceptor specificity within this panel of chimeras modulated the response to T-20 and that the mean IC50 for the four R5 chimeras was 0.7 log10 higher than that for the X4 virus NLHX.
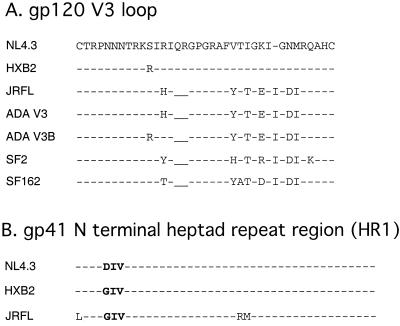
FIG. 3.
V3 loop and HR1 amino acid sequence alignments. HIV-1 NL4.3 is used as the reference sequence. Conserved amino acids are indicated as dashed lines, and residues that differ from the NL4.3 sequence are indicated by amino acid letter. Underscores represent gaps in the sequence. (A) Predicted amino acid sequences for the V3 loops (residues 294 to 329 in NL4.3 gp120) used in this study. HXB2 and NL4.3 are X4 specific; ADA, ADA V3B, JFRL, SF162, and SF2 are R5 specific. (B) Differences in the predicted amino acid sequences for the highly conserved HIV-1 HR1 regions (residues 32 to 75 of NL4.3 gp41) used in this study are shown. The critical T-20 interaction site (residues 36 to 38) is shown in boldface.
FIG. 4.
Contribution of coreceptor specificity to inhibition by T-20. Cells were infected with 2,000 infectious units of each chimeric virus stock in the absence (control) or presence of increasing doses of T-20. Virus infectivity is plotted along the vertical axis on a linear scale, and T-20 concentration is plotted along the horizontal axis on a log10 scale. Each curve represents the inhibition profile of an individual virus. (A) Luciferase activity was measured at 48 h in infected JC53-BL cells. Each point represents the mean relative infectivity calculated from two independent experiments, each with duplicate wells. The results were confirmed by one experiment in which blue foci were counted (data not shown). The range of IC50s of T-20 for the NLHX-based chimeras was 0.005 to 0.0.032 μg/ml. (B) Luciferase activity was measured at 48 h in infected JC53-BL cells. Each point represents the mean infectivity calculated from four independent experiments, each with duplicate wells. The results were confirmed by two experiments in which blue foci were counted (data not shown). The range of IC50s of T-20 for the NL4.3/JRFL V3 loop chimeras was 0.120 to 0.443 μg/ml. (C) PBMC were infected with a subset of the NL4.3/JRFL envelope chimeras, and supernatant p24 was measured on day 7 by ELISA. Each point represents the mean infectivity calculated from one experiment with duplicate wells. The range of IC50s of T-20 for the NL4.3/JRFL V3 loop chimeras was 0.024 to 0.109 μg/ml. (D) Chimeric constructs used in this analysis.
To validate the effect of coreceptor specificity on inhibition by T-20, we analyzed a second set of chimeras that were constructed in an NL4.3 proviral and envelope background (Fig. 2B). HIV-1 NL4.3 is a T-cell line-adapted strain that is highly related to HXB2 and is specific for CXCR4 (1, 36). These NL4.3-based chimeric constructs contain different subsets of envelope sequences derived from HIV-1 JRFL, an R5 isolate obtained from the brain of a patient who died of AIDS (27, 38, 46). The NL4.3/JRFL constructs were originally designed to define determinants of macrophage tropism and resistance to neutralization by soluble CD4 (37–39). To determine the baseline sensitivity of NL4.3 to T-20 inhibition, viruses produced from two infectious proviral clones of NL4.3 that differ only in the modification of a StuI restriction site to XbaI in cellular flanking sequences upstream of the 5′ LTR [NL4.3 and NL(Stu)Xba] were analyzed (Fig. 4B). The NL(Stu)Xba provirus is the parental proviral clone in which all of the JRFL envelope chimeras were constructed (37–39). The IC50s for the two NL4.3 viruses were almost identical (0.120 and 0.130 μg/ml, mean = 0.125 ± 0.008). When the R5 V3 loop sequence derived from JRFL was inserted into NL4.3 (NL-FLV3), the IC50 of T-20 for that virus increased 0.6 log10, from 0.125 to 0.443 μg/ml (P < 0.001). Similar increases in the IC50 were observed when the JRFL V3 loop and flanking sequences were included (0.397 μg/ml for NFN-SM and 0.423 μg/ml for NFN-SP, P < 0.001 for both comparisons). Thus, the minimal requirements for a 0.5- to 0.6-log10 increase in IC50 involved only substitution of the R5 V3 loop and was consistent with the NLHX chimeras described above, confirming that coreceptor specificity affects susceptibility to the inhibitory effect of T-20 in vitro.
To determine whether our observations could be extended from the JC53-BL cell line to primary cells, we repeated the analysis of the NL4.3/JRFL chimeras in PBMC, using measurement of supernatant p24 by ELISA at day 7 postinfection instead of LU to calculate the IC50s. The IC50 for each virus was four to six times lower on PBMC than on JC53-BL cells (Fig. 4C). For example, the mean IC50 calculated for the NL4.3 viruses in PBMC was 0.024 μg/ml, compared to 0.125 μg/ml in JC53-BL cells. The mean IC50 for the viruses that contained the R5 V3 loop sequence (0.060, 0.094, and 0.109 μg/ml, mean = 0.088 ± 0.025) was 0.6 log10 higher than that for the NL4.3 viruses, which was consistent with our analyses performed on JC53-BL cells. Based on these results, we conclude that coreceptor modulation of T-20 inhibition also occurs in primary target cells in vitro.
Comparison of the baseline sensitivity to T-20 determined by a target sequence within HR1.
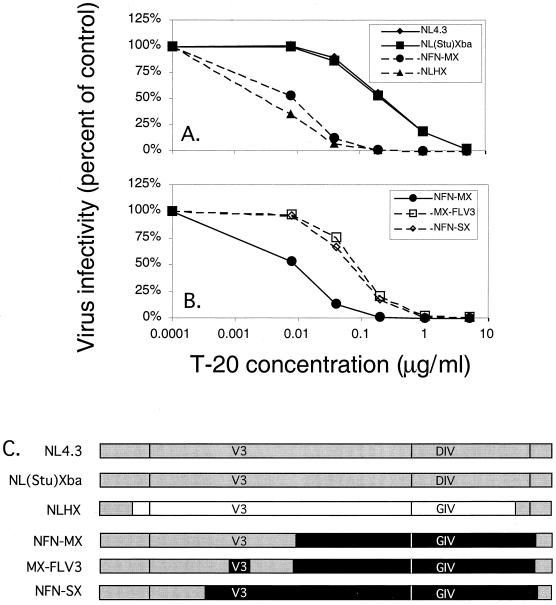
Rimsky et al. reported that a contiguous three-amino-acid sequence present at positions 36 to 38 within HR1 is a major determinant of sensitivity to T-20 (41). Here we investigated the effect of this sequence on sensitivity to T-20 in the context of different coreceptor specificities using envelope chimeras (Fig. 2). HXB2 and NL4.3 contain similar amino acid sequences within gp41 and differ by only one amino acid residue within HR1: a naturally occurring G-to-D substitution at position 36 in NL4.3 (Fig. 3B). The baseline sensitivity of NL4.3 and NLHX differed dramatically, even though their envelope sequences are highly related and their coreceptor specificities are the same (Fig. 5A). When the HXB2 envelope, which contains GIV within HR1, was inserted into NL4.3, the IC50 was about 1.4 log10 lower, from a mean of 0.125 to 0.005 μg/ml (P = 0.002). To validate the contribution of the HR1 sequence, we compared an X4 virus that contains the GIV sequence in HR1 derived from JRFL, NFN-MX, to NL4.3 and NLHX (Fig. 5A). The IC50 for NFN-MX was about 1.2 log10 lower than that for NL4.3 (0.008 versus 0.125 μg/ml of T-20, P < 0.001). We next determined whether placement of the JRFL-derived R5 V3 loop sequence back into the X4 envelope background of NFN-MX would result in an increase in the IC50. We compared NFN-MX to MX-FLV3, which is identical to NFN-MX except that it contains JRFL-derived R5 V3 loop sequences (Fig. 5C). Figure 5B shows that substitution of the JRFL V3 loop sequence in NFM-MX resulted an increase in the IC50 of T-20 of about 0.8 log10, from a baseline of 0.008 to 0.056 μg/ml (P < 0.001). The IC50 for a similar construct, NFN-SX, was comparable to that for NFN-MX (0.047 versus 0.056 μg/ml).
FIG. 5.
Contribution of HR1 to T-20 sensitivity. JC53-BL indicator cells were infected with 2,000 infectious units of each chimeric virus stock in the absence (control) or presence of increasing doses of T-20. At 48 h postinfection, cells were lysed and luciferase activity was measured. Virus infectivity is plotted along the vertical axis on a linear scale, and T-20 concentration is plotted along the horizontal axis on a log10 scale. (A) Each point represents the mean infectivity calculated from two independent experiments, each with duplicate wells. The results were confirmed by one experiment in which blue foci were counted (data not shown). The range of IC50s of T-20 for the NL4.3/HXB2/JRFL chimeras was 0.005 to 0.130 μg/ml. (B) Each point represents the mean infectivity calculated from four independent experiments, each with duplicate wells. The results were confirmed by two experiments in which blue foci were counted (data not shown). The range of IC50s of T-20 for these NL4.3/JRFL chimeras was 0.008 to 0.056 μg/ml. (C) Chimeric constructs used in this analysis.
Coreceptor specificity and HR1 independently modulate susceptibility to T-20.
The IC50s for all envelope chimeras analyzed in this study are shown in Table 2 in relation to their coreceptor specificity and the critical amino acid sequence present in HR1. The mean IC50s and standard deviations were calculated for each group of viruses based on their coreceptor specificity and the sequence present in HR1. The constructs that required the highest concentrations of T-20 to inhibit infectivity contained a combination of R5 V3 loop sequences and the DIV sequence in HR1 (R5/DIV). Substituting an X4 V3 loop sequence for the R5 V3 loop sequence (X4/DIV) resulted in a ∼0.5-log10 increase in sensitivity to T-20 relative to the R5/DIV viruses. Constructs containing an R5 V3 loop sequence combined with the GIV sequence (R5/GIV) were ∼1.1 log10 more sensitive to T-20 than the R5/DIV viruses. Finally, the most sensitive combination was an X4 V3 loop sequence combined with the GIV sequence in HR1 (X4/GIV). These viruses were ∼1.8 log10 more sensitive to T-20 than the R5/DIV viruses. These results show that coreceptor specificity and the critical target sequence present within HR1 produce additive effects that independently modulate sensitivity to T-20.
TABLE 2.
Independent effects of coreceptor specificity and T-20 target site in HR1
| Virus | Coreceptora | HR1b | IC50 (μg/ml) | Mean IC50c (μg/ml) | SD | Changed |
|---|---|---|---|---|---|---|
| NL-FLV3 | R5 | DIV | 0.443 | 0.421 | 0.02 | 0.0 |
| NFN-SM | R5 | DIV | 0.397 | |||
| NFN-SP | R5 | DIV | 0.423 | |||
| NL4.3 | X4 | DIV | 0.120 | 0.125 | 0.01 | −0.5 |
| NL(Stu)Xba | X4 | DIV | 0.130 | |||
| MX-FLV3 | R5 | GIV | 0.056 | 0.032 | 0.01 | −1.1 |
| NFN-SX | R5 | GIV | 0.047 | |||
| NLHX SF2 V3 | R5 | GIV | 0.025 | |||
| NLHX ADA V3B | R5 | GIV | 0.019 | |||
| NLHX ADA V3 | R5 | GIV | 0.024 | |||
| NLHX SF162 V3 | R5 | GIV | 0.032 | |||
| NFN-MX | X4 | GIV | 0.008 | 0.007 | 0.00 | −1.8 |
| NLHX | X4 | GIV | 0.005 |
Coreceptor specificity of each chimeric virus.
Amino acids 36 to 38 in the gp41 N-terminal heptad repeat that comprise a critical T-20 target site.
Calculated for chimeric viruses grouped by coreceptor specificity and T-20 target site.
Independent effects of coreceptor specificity and T-20 target site represented on a log10 scale using group mean IC50s.
DISCUSSION
Using chimeric HIV-1 constructs containing identical proviral backgrounds and defined envelope sequence combinations, we confirmed that sequences contained within gp41 determine the baseline sensitivity of a virus to T-20 independently of coreceptor specificity. Our studies showed that both X4 and R5 viruses containing the DIV variant sequence in HR1 display >1.0-log10-higher IC50s than similar viruses containing the GIV motif. For example, viruses that contain an X4 V3 loop sequence within the NL4.3 envelope background showed IC50s 1.0 log10 higher than those for similar constructs within the HXB2 or JRFL envelope background, which both contain GIV (Fig. 5A). These results, combined with the absence of other amino acid substitutions within envelope sequences derived from T-20-resistant viruses (41), suggest that this site interacts directly with T-20 peptides and is critical to its mode of action. Since the GIV sequence is highly conserved among isolates of HIV-1 (36), natural variants probably occur infrequently in the absence of selection and were not likely to have been a major factor in our analysis of primary isolates. Nevertheless, this highly conserved sequence can be altered without diminishing infectivity when viruses are exposed to the selective pressures of T-20 in vitro (41). Moreover, T-20-resistant viruses generated at suboptimal doses of T-20 in vivo also contain substitutions within the GIV region (Shaw, unpublished data).
A major finding in our study is the novel observation that coreceptor specificity, defined by V3 loop sequences, independently modulates sensitivity to inhibition by T-20. The insertion of four different R5 virus-derived V3 loop sequences into the NL4.3 or HXB2 X4 envelope background increased the IC50 an average of 0.7 log10 (range, 0.6 to 0.8 log10). Although most of the analyses were performed in a cell line, the results were confirmed for a subset of the chimeras in PBMC by analyzing p24 production in the supernatant. Even though the IC50 for each virus analyzed was about fivefold lower in PBMC, a 0.6-log10 increase in mean IC50 was observed for the three viruses containing R5 V3 loop sequences. Moreover, our analysis of R5 and X4 primary isolates was completely consistent with the results observed with the chimeric viruses containing defined envelope sequences. The mean IC50 for R5 primary isolates was about 0.8-log10 higher than the mean IC50 for X4 isolates. It is likely that coreceptor specificity is a strong modulator of sensitivity to T-20 in primary isolates of HIV-1, even though other regions of the envelope and viral genome may influence overall sensitivity. The findings reported here have several important implications. First, T-20 may more effectively inhibit viral strains that use CXCR4 in vivo and is the first clinically effective antiretroviral therapy that exhibits this additional potential. Heavily pretreated patients with advanced disease that harbor X4 viruses may experience additional benefits from treatment regimens containing T-20. Second, administration of T-20 as part of an antiretroviral regimen in patients with early disease might delay the emergence of more pathogenic strains of HIV-1 that are associated with CXCR4 specificity if viral replication is not completely suppressed (9, 12, 16, 58). Our data also suggest that R5 viruses may be able to generate resistance more rapidly than X4 viruses in vivo in the presence of suboptimal doses of T-20, since R5 viruses remain infectious at higher concentrations of T-20 in vitro. However, clinical studies of subjects undergoing T-20-containing regimens are necessary to confirm these hypotheses.
It is not immediately clear why coreceptor specificity would influence susceptibility of a viral isolate to inhibition by T-20. The mechanism of T-20 action probably involves competitive binding of the peptides to the HR1 domain in a dominant negative manner that inhibits its association with the native HR2 after gp120 binding to CD4 and coreceptor molecules (7, 17, 35). With T-20 bound, the conformation of gp41 would be held in an intermediate state, viral and cellular membranes would remain separated, and fusion would be prevented (35). One possible explanation for the effects of coreceptor specificity on the mechanism of T-20 inhibition is that gp120 binding to CD4 and CCR5 induces conformational changes that hinder T-20 peptide interactions with HR1, while gp120 binding to CD4 and CXCR4 induces a conformation more conducive to T-20 association with HR1. The stability of the gp120-gp41 association has been shown to be a major determinant of susceptibility to the nonpeptidic inhibitor RPR103611, whose target sequence maps to critical residues within gp41 (30). Similarly, the bis-azo compound inhibitor FP-21339, which modulates stability of the gp120-gp41 complex, inhibited a virus containing an X4 envelope at lower concentrations than an isogenic virus containing an R5 envelope (60). Furuta et al. showed that the presence of soluble CD4 alone was sufficient to trigger the conformational changes in gp41 that promote access of hemagglutinin epitope-tagged T-20 peptides for the envelope derived from HXB2, but not SF162 or JRFL, which required interaction with both CD4 and coreceptor molecules (17). Thus, the greater sensitivity of X4 viruses to T-20 inhibition in this study could be explained by more efficient dissociation of gp120 from gp41 upon contact with CD4/CXCR4 than with CD4/CCR5, leading to conformational changes that promote T-20 interaction with HR1.
It is also possible that the kinetics of the conformational changes within gp41 induced by gp120 binding to CD4 and coreceptor may differ for CCR5 and CXCR4. Doranz and colleagues reported that the binding of X4 gp120s to CXCR4 in the presence of CD4 is not as robust as binding of R5 gp120s to CCR5 (13, 14). The CD4-induced conformational changes in gp120-gp41 occur rapidly after binding (13, 24), but the kinetics of subsequent events have not been described. One study investigating the time dependence of T-20 inhibition using the X4 HIV-1 LAI envelope showed that cell-cell fusion was completely inhibited if T-20 was added within the first 15 min of cell mixing (35). T-20 added between 15 and 75 min resulted in partial inhibition of fusion (10 to 90%), and T-20 added after 75 min had no effect. This finding suggests that there is a distinct window of opportunity for T-20 to interact with HR1 within a transient structure that forms after CD4-coreceptor binding but before fusion is complete. After this time, it is likely that HR1 and HR2 have already complexed, so that T-20-mediated inhibition is no longer possible. If R5 gp120s bind to CD4/CCR5 with higher affinity and association constants than X4 gp120s bind to CD4/CXCR4, it is possible the gp120-CD4-CCR5 complex may trigger the conformational changes of gp41 that lead to fusion more rapidly than gp120-CD4-CXCR4. In contrast, the X4 gp120-CD4-CXCR4 complex may produce an intermediate structure that exposes the T-20 interaction for a longer period of time.
A third possible explanation is that CCR5 and CXCR4 have different rates of internalization after gp120 binding. Although gp120-bound coreceptor internalization is not required for HIV-1 entry, the rate at which this occurs may influence the exposure of complexed viruses to T-20 and thus cause differential inhibition of X4 and R5 viruses. Although we have presented alternative hypotheses that may explain our observations, these are not mutually exclusive and may contribute in concert to the differential inhibition of CXCR4 and CCR5-mediated fusion.
The results presented in this study provide strong evidence that the CD4-coreceptor-envelope interaction differs structurally and/or kinetically between X4 and R5 viruses. The coreceptor-specific modulation of T-20 inhibition suggests that although CCR5 and CXCR4 both serve as coreceptors for HIV-1, they may provide functional or kinetic differences in viral entry. Whether these differences play a role in pathogenesis and viral evolution in vivo is unclear, but uncovering the complex events and structures that define fusion will be important for a complete understanding of the HIV-1 entry into target cells.
ACKNOWLEDGMENTS
We thank Trimeris, Inc., Durham, N.C., for providing T-20, Jeannette Lee and the UAB Comprehensive Cancer Center Biostatistics Unit for performing statistical analysis of the data, and Robert Blumenthal at NCI/NIH, Frederick, Md., for participating in helpful discussions.
This work was supported by NIH grants R37A133319 (E.H.) and R37AI24745 (L.R.) and the Howard Hughes Medical Institute (G.M.S.). The experiments were performed in the Central Virus Core of the UAB Center for AIDS Research supported by grant P30-A1-27767.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 3.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S, Caron M G, Cullen B R. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 5.Carr C K. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 6.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 7.Chan D C, Kim P S. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 8.Cheng-Mayer C, Liu R, Landau N R, Stamatatos L. Macrophage tropism of human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. J Virol. 1997;71:1657–1661. doi: 10.1128/jvi.71.2.1657-1661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun T W, Stuyver L, Mizell S B, Ehler L A, Mican J A, Baseler M, Lloyd A L, Nowak M A, Fauci A S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 11.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 12.Derdeyn C A, Kilby J M, Miralles G D, Li L F, Sfakianos G, Saag M S, Hockett R D, Bucy R P. Evaluation of distinct blood lymphocyte populations in human immunodeficiency virus type 1-infected subjects in the absence or presence of effective therapy. J Infect Dis. 1999;180:1851–1862. doi: 10.1086/315117. [DOI] [PubMed] [Google Scholar]
- 13.Doranz B J, Baik S S, Doms R W. Use of a gp120 binding assay to dissect the requirements and kinetics of human immunodeficiency virus fusion events. J Virol. 1999;73:10346–10358. doi: 10.1128/jvi.73.12.10346-10358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doranz B J, Orsini M J, Turner J D, Hoffman T L, Berson J F, Hoxie J A, Peiper S C, Brass L F, Doms R W. Identification of CXCR4 domains that support coreceptor and chemokine receptor functions. J Virol. 1999;73:2752–2761. doi: 10.1128/jvi.73.4.2752-2761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubay J W, Roberts S J, Brody B, Hunter E. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J Virol. 1992;66:4748–4756. doi: 10.1128/jvi.66.8.4748-4756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 17.Furuta R A, Wild C T, Weng Y, Weiss C D. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman T L, Doms R W. Chemokines and coreceptors in HIV/SIV-host interactions. AIDS. 1998;12:S17–S26. [PubMed] [Google Scholar]
- 19.Hoffman T L, Doms R W. HIV-1 envelope determinants for cell tropism and chemokine receptor use. Mol Membr Biol. 1999;16:57–65. doi: 10.1080/096876899294760. [DOI] [PubMed] [Google Scholar]
- 20.Hung C S, Vander Heyden N, Ratner L. Analysis of the critical domain in the V3 loop of human immunodeficiency virus type 1 gp120 involved in CCR5 utilization. J Virol. 1999;73:8216–8226. doi: 10.1128/jvi.73.10.8216-8226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunter E. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 22.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 23.Japour A J, Fiscus S A, Arduino J M, Mayers D L, Reichelderfer P S, Kuritzkes D R. Standardized microtiter assay for determination of syncytium-inducing phenotypes of clinical human immunodeficiency virus type 1 isolates. J Clin Microbiol. 1994;32:2291–2294. doi: 10.1128/jcm.32.9.2291-2294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones P L, Korte T, Blumenthal R. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J Biol Chem. 1998;273:404–409. doi: 10.1074/jbc.273.1.404. [DOI] [PubMed] [Google Scholar]
- 25.Kilby J M, Hopkins S, Venetta T M, DiMassimo B, Cloud G A, Lee J Y, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson M R, Nowak M A, Shaw G M, Saag M S. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 26.Kliger Y, Shai Y. Inhibition of HIV-1 entry before gp41 folds into its fusion-active conformation. J Mol Biol. 2000;295:163–168. doi: 10.1006/jmbi.1999.3368. [DOI] [PubMed] [Google Scholar]
- 27.Koyanagi Y, Miles S, Mitsuyasu R T, Merrill J E, Vinters H V, Chen I S. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 28.Kozak S L, Platt E J, Madani N, Ferro F E, Jr, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labrosse B, Treboute C, Alizon M. Sensitivity to a nonpeptidic compound (RPR103611) blocking human immunodeficiency virus type 1 env-mediated fusion depends on sequence and accessibility of the gp41 loop region. J Virol. 2000;74:2142–2150. doi: 10.1128/jvi.74.5.2142-2150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Littman D R. Chemokine receptors: keys to AIDS pathogenesis? Cell. 1998;93:677–680. doi: 10.1016/s0092-8674(00)81429-4. [DOI] [PubMed] [Google Scholar]
- 32.Lu M, Blacklow S C, Kim P S. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 33.Luciw P A. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. [Google Scholar]
- 34.Moore J P, McKeating J A, Weiss R A, Sattentau Q J. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 35.Munoz-Barroso I, Durell S, Sakaguchi K, Appella E, Blumenthal R. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J Cell Biol. 1998;140:315–323. doi: 10.1083/jcb.140.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myers G, Korber B, Hahn B H, Jeang K, Mellors J W, McCutchan F E, Henderson L E, Pavlakis G N. Human retroviruses and AIDS. Los Alamos, N.Mex: Theoretical Biology and Biophysics, Los Alamos National Laboratory; 1995. [Google Scholar]
- 37.O'Brien W A, Chen I S, Ho D D, Daar E S. Mapping genetic determinants for human immunodeficiency virus type 1 resistance to soluble CD4. J Virol. 1992;66:3125–3130. doi: 10.1128/jvi.66.5.3125-3130.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Brien W A, Koyanagi Y, Namazie A, Zhao J Q, Diagne A, Idler K, Zack J A, Chen I S. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 39.O'Brien W A, Sumner-Smith M, Mao S H, Sadeghi S, Zhao J Q, Chen I S. Anti-human immunodeficiency virus type 1 activity of an oligocationic compound mediated via gp120 V3 interactions. J Virol. 1996;70:2825–2831. doi: 10.1128/jvi.70.5.2825-2831.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poignard P, Fouts T, Naniche D, Moore J P, Sattentau Q J. Neutralizing antibodies to human immunodeficiency virus type-1 gp120 induce envelope glycoprotein subunit dissociation. J Exp Med. 1996;183:473–484. doi: 10.1084/jem.183.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rimsky L T, Shugars D C, Matthews T J. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J Virol. 1998;72:986–993. doi: 10.1128/jvi.72.2.986-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sattentau Q J, Moore J P. The role of CD4 in HIV binding and entry. Philos Trans R Soc Lond B. 1993;342:59–66. doi: 10.1098/rstb.1993.0136. [DOI] [PubMed] [Google Scholar]
- 43.Sattentau Q J, Moore J P, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sattentau Q J, Zolla-Pazner S, Poignard P. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology. 1995;206:713–717. doi: 10.1016/s0042-6822(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 45.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 46.Smyth R J, Yi Y, Singh A, Collman R G. Determinants of entry cofactor utilization and tropism in a dualtropic human immunodeficiency virus type 1 primary isolate. J Virol. 1998;72:4478–4484. doi: 10.1128/jvi.72.5.4478-4484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan K, Liu J, Wang J, Shen S, Lu M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 50.Trujillo J R, Wang W K, Lee T H, Essex M. Identification of the envelope V3 loop as a determinant of a CD4-negative neuronal cell tropism for HIV-1. Virology. 1996;217:613–617. doi: 10.1006/viro.1996.0158. [DOI] [PubMed] [Google Scholar]
- 51.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 52.Westervelt P, Trowbridge D B, Epstein L G, Blumberg B M, Li Y, Hahn B H, Shaw G M, Price R W, Ratner L. Macrophage tropism determinants of human immunodeficiency virus type 1 in vivo. J Virol. 1992;66:2577–2582. doi: 10.1128/jvi.66.4.2577-2582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wild C, Dubay J W, Greenwell T, Baird T, Jr, Oas T G, McDanal C, Hunter E, Matthews T. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc Natl Acad Sci USA. 1994;91:12676–12680. doi: 10.1073/pnas.91.26.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wild C, Oas T, McDanal C, Bolognesi D, Matthews T. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc Natl Acad Sci USA. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wild C T, Shugars D C, Greenwell T K, McDanal C B, Matthews T J. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willey R L, Martin M A. Association of human immunodeficiency virus type 1 envelope glycoprotein with particles depends on interactions between the third variable and conserved regions of gp120. J Virol. 1993;67:3639–643. doi: 10.1128/jvi.67.6.3639-3643.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willey R L, Martin M A, Peden K W. Increase in soluble CD4 binding to and CD4-induced dissociation of gp120 from virions correlates with infectivity of human immunodeficiency virus type 1. J Virol. 1994;68:1029–1039. doi: 10.1128/jvi.68.2.1029-1039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong J K, Hezareh M, Gunthard H F, Havlir D V, Ignacio C C, Spina C A, Richman D D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 59.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 60.Zhang J L, Choe H, Dezube B J, Farzan M, Sharma P L, Zhou X C, Chen L B, Ono M, Gillies S, Wu Y, Sodroski J G, Crumpacker C S. The bis-azo compound FP-21399 inhibits HIV-1 replication by preventing viral entry. Virology. 1998;244:530–541. doi: 10.1006/viro.1998.9115. [DOI] [PubMed] [Google Scholar]