Abstract
Cardiac diseases represent common highly morbid conditions for which molecular mechanisms remain incompletely understood. Here we report the analysis of 1,459 protein measurements in 44,313 UK Biobank participants to characterize the circulating proteome associated with incident coronary artery disease, heart failure, atrial fibrillation and aortic stenosis. Multivariable-adjusted Cox regression identified 820 protein–disease associations—including 441 proteins—at Bonferroni-adjusted P < 8.6 × 10−6. Cis-Mendelian randomization suggested causal roles aligning with epidemiological findings for 4% of proteins identified in primary analyses, prioritizing therapeutic targets across cardiac diseases (for example, spondin-1 for atrial fibrillation and the Kunitz-type protease inhibitor 1 for coronary artery disease). Interaction analyses identified seven protein–disease associations that differed Bonferroni-significantly by sex. Models incorporating proteomic data (versus clinical risk factors alone) improved prediction for coronary artery disease, heart failure and atrial fibrillation. These results lay a foundation for future investigations to uncover disease mechanisms and assess the utility of protein-based prevention strategies for cardiac diseases.
Subject terms: Cardiovascular diseases, Molecular medicine, Proteomics
Schuermans et al. identify a causal relationship between the circulating proteins spondin-1 and atrial fibrillation and SPINT1 and coronary artery disease and show that adding proteomic data improves clinical risk factor-based cardiovascular risk prediction.
Main
Cardiac diseases represent the leading global cause of morbidity and mortality1, with coronary artery disease, heart failure, atrial fibrillation and aortic stenosis collectively accounting for more than 90% of cardiac deaths1,2. The prevention of these diseases typically relies on accurate risk prediction and pharmacotherapy of modifiable risk factors, which represent two complementary aspects of cardiovascular care that remain incompletely optimized. For instance, most high-risk individuals remain undetected until experiencing their first clinical event3,4, and even under currently optimal treatment conditions, there remains substantial residual risk that is incompletely captured by traditional risk factors5,6. Both the development of effective prediction tools and the discovery of therapeutic targets could considerably improve treatment outcomes and enhance early detection for different cardiac diseases.
The circulating proteome—a dynamic network that reflects genetic background as well as external factors such as environmental exposures and lifestyle alterations—may be leveraged for both risk prediction and disease risk modification. For instance, smaller studies have demonstrated that sparse protein-based risk scores can improve the prediction of cardiovascular events in certain populations7–11. Furthermore, targeted analyses of specific biomarkers suggest that integrating proteomic and genetic data can nominate causal protein–disease associations and reveal actionable drug targets in the bloodstream12,13. Nevertheless, protein–disease associations do not need to be causal to be usefully predictive and, conversely, associations that are causal and may generate therapeutic targets are not necessarily useful in the prediction of incident events. Whether population-scale, agnostic analyses of the circulating proteome can provide insights into both aspects of clinical care, that is, improve the prediction of first clinical events and reveal causal mediators for a range of cardiac disease subtypes, remains unclear.
Here, we performed a proteomic analysis of cardiac diseases in 44,313 unrelated individuals from the UK Biobank (UKB) Pharma Proteomics Project (PPP) (Table 1)14. The UKB-PPP is a population-based cohort with high-throughput proteomic profiling (using the Olink Explore 1536 platform) at study baseline and longitudinal follow-up for incident cardiac events (Fig. 1). In primary analyses, we used multivariable-adjusted time-to-event models to test the associations of 1,459 circulating proteins with incident coronary artery disease, heart failure, atrial fibrillation and aortic stenosis. While these models can identify single protein–disease associations, they cannot be used to infer causal protein–disease associations and do not provide useful measures of predictive performance. Therefore, in a first set of downstream analyses, we leveraged Mendelian randomization (MR) to infer causal roles among the identified proteins to prioritize therapeutic targets. Next, in a separate set of downstream analyses, we constructed protein-based risk scores and evaluated whether these could improve disease prediction beyond the use of traditional risk factors. Other downstream analyses assessed sex differences in protein–disease associations and tested enrichment of certain biological pathways in proteins associated with different cardiac diseases.
Table 1.
Baseline characteristics of UK Biobank Pharma Proteomics Project participants included in the present study (n = 44,313)
| UKB-PPP participants (n = 44,313) | |
|---|---|
| Age at blood draw, years | 56.4 ± 8.2 |
| Female, n | 24,701 (55.7%) |
| Race/ethnicity, n | – |
| Asian | 942 (2.1%) |
| Black | 1,051 (2.4%) |
| White | 41,481 (93.6%) |
| Mixed | 300 (0.7%) |
| Other | 539 (1.2%) |
| Smoking status, n | – |
| Never | 24,709 (55.8%) |
| Previous | 14,932 (33.7%) |
| Current | 4,672 (10.5%) |
| BMI, kg m−² | 27.3 ± 4.7 |
| Blood pressure, mm Hg | – |
| Systolic blood pressure | 139.5 ± 19.6 |
| Diastolic blood pressure | 82.3 ± 10.6 |
| Blood biochemistry, mg dl−1 | – |
| Total cholesterol | 221.4 ± 43.8 |
| LDL cholesterol | 138.3 ± 33.4 |
| HDL cholesterol | 56.4 ± 14.7 |
| Triglycerides | 129.8 (92.0 to 188.1) |
| Creatinine | 0.82 ± 0.19 |
| Type 2 diabetes mellitus, n | 1,218 (2.7%) |
| Medication use, n | – |
| Cholesterol-lowering medication use | 6,544 (14.8%) |
| Antihypertensive medication use | 6,214 (14.0%) |
| Townsend deprivation index | −2.08 (−3.63 to 0.70) |
Continuous variables are summarized as mean ± s.d. or median (IQR), as appropriate. Categorical variables are summarized as n (%).
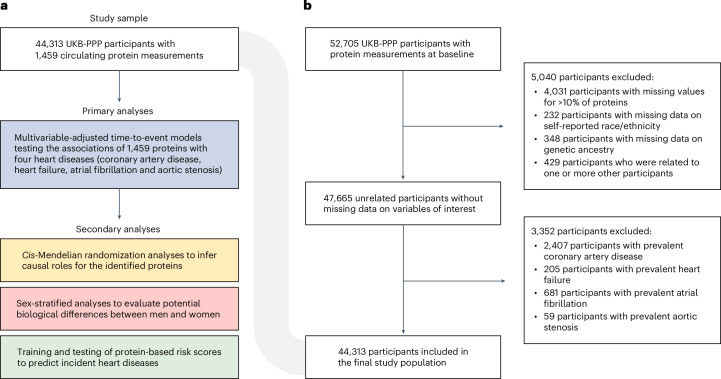
Fig. 1. Visual representation of the study design and participant inclusion and exclusion criteria.
The present study tested the associations of circulating proteins with common cardiac diseases (coronary artery disease, heart failure, atrial fibrillation and aortic stenosis) in the UKB-PPP. Primary analyses tested the epidemiological associations of 1,459 circulating proteins with cardiac diseases in 44,313 UKB-PPP participants without these diseases at baseline. Secondary analyses performed cis-MR analyses, tested for sex-specific effects and trained and tested protein-based risk scores. a, Study design. b, Participant inclusion and exclusion criteria.
Results
Associations of circulating proteins with heart diseases
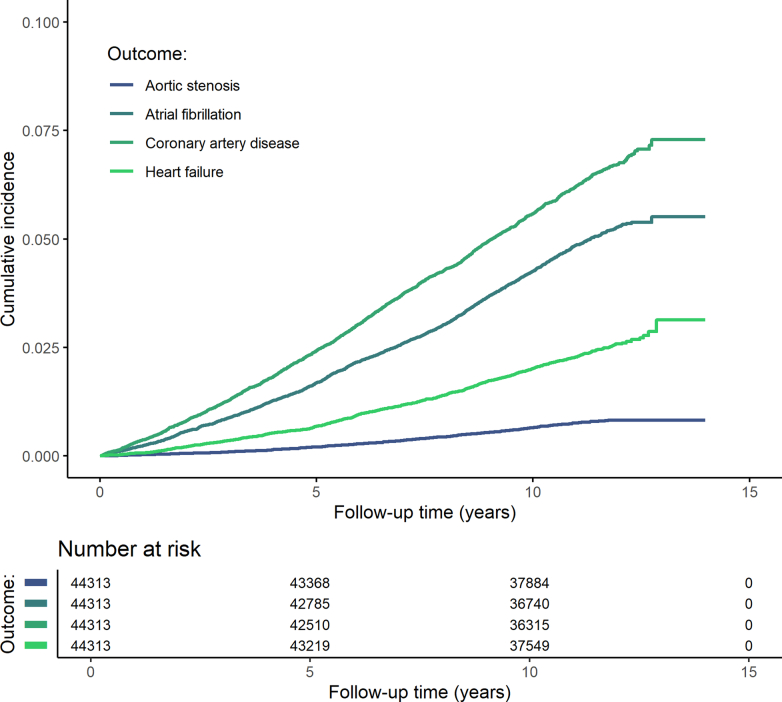
Of 44,313 UKB-PPP participants, 4,610 (10.4%) experienced at least one cardiac event over a median (interquartile range (IQR)) follow-up of 11.1 (10.4–11.8) years. Coronary artery disease had the highest cumulative incidence (6.2% (n = 2,729 of 44,313)), followed by atrial fibrillation (4.8% (n = 2,107 of 44,313)), heart failure (2.3% (n = 1,014 of 44,313)) and aortic stenosis (0.7% (n = 326 of 44,313)) (Extended Data Fig. 1).
Extended Data Fig. 1. Cumulative incidence of coronary artery disease, heart failure, atrial fibrillation, and aotic stenosis during follow-up.
Cumulative incidence plots were constructed using the Kaplan–Meier method. Participants were followed for a median (interquartile range) follow-up of 11.1 (10.4–11.8) years.
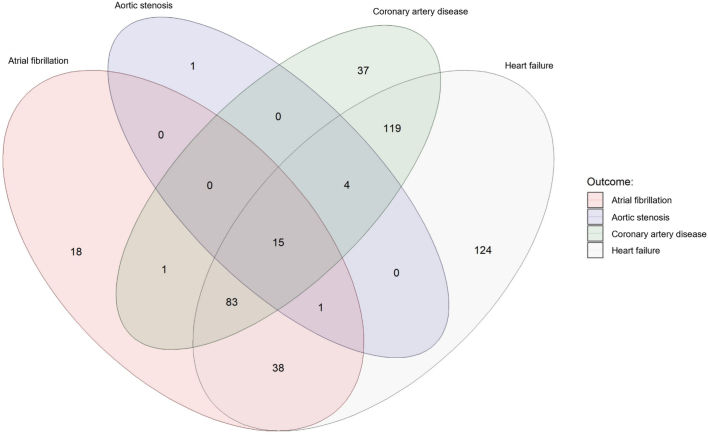
Primary analyses tested the associations of 1,459 circulating proteins (Supplementary Tables 1 and 2) with each of the incident heart diseases (coronary artery disease, heart failure, atrial fibrillation and aortic stenosis) using multivariable-adjusted Cox regression models. The correlation matrix of these circulating proteins is provided in Extended Data Fig. 2 and Supplementary Table 3. Primary analyses identified 820 protein–disease associations—reflecting 441 unique proteins—at Bonferroni-corrected P < 0.05/5,836 (P < 0.05/(1,459 tested proteins × 4 tested outcomes)) (Fig. 2 and Supplementary Table 4). Heart failure had the highest number of proteomic associations (n = 384), followed by coronary artery disease (n = 259), atrial fibrillation (n = 156) and aortic stenosis (n = 21). Among proteins with one or more significant associations, 261 (59.2%) were shared across multiple outcomes and 15 (3.4%) were shared across all four outcomes (Extended Data Fig. 3).
Extended Data Fig. 2. Correlations among circulating proteins measured at baseline.
All colored boxes represent Pearson correlation coefficients (r) indicating the correlations between the proteins that were measured in the final study cohort (N = 44,313). Red boxes indicate positive correlations between proteins (r > 0), whereas blue boxes indicate negative correlations between proteins (r < 0). Pearson correlation coefficients are provided in Supplementary Table 3. Each row and each column each represent one circulating protein. Proteins were clustered using a hierarchical cluster analysis based on the “complete linkage method”. Hierarchical clustering was performed using the hclust() function in R. The heat plot was constructed using the pheatmap() function (pheatmap package21 in R).
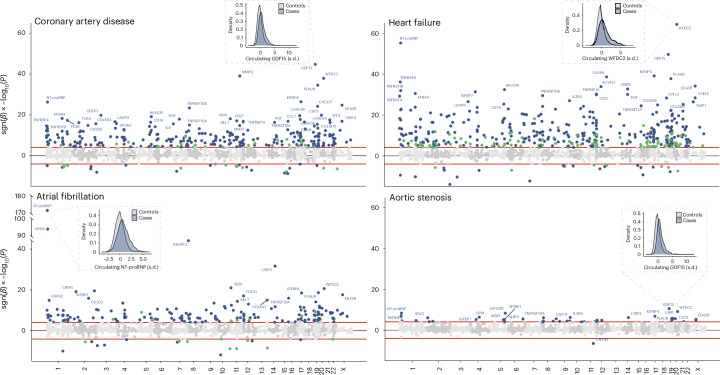
Fig. 2. Associations of circulating protein levels with incident coronary artery disease, heart failure, atrial fibrillation and aortic stenosis.
Miami plots visualize the associations of all 1,459 Olink proteins with coronary artery disease, heart failure, atrial fibrillation and aortic stenosis, tested using multivariable-adjusted Cox proportional hazards models (Methods). The y axis indicates the −log10(P) value for each association, multiplied by 1 if the association was positive (β > 0) or −1 if the association was negative (β < 0). The x axis indicates the genetic position of each protein’s encoding gene. Protein–disease associations with Bonferroni-corrected two-sided P < 0.05 (P < 0.05/5,836 or ~8.6 × 10−6) are shown in blue (if the protein was associated with more than one outcome) or green (if the protein was not associated with more than one outcome). The probability density functions show the distributions of the strongest protein–disease associations in cases (dark blue) versus controls (light blue) for each outcome. These analyses included 44,313 UKB-PPP participants, among whom 2,729 experienced coronary artery disease, 2,107 heart failure, 1,014 atrial fibrillation and 326 aortic stenosis events during follow-up.
Extended Data Fig. 3. Venn diagram showing the number of distinct and shared protein associations across outcomes.
All 441 proteins that were associated with one or more outcomes at Bonferroni-corrected P < 0.05 are represented in this graph.
The strongest protein–disease associations (by P value) were observed for atrial fibrillation, with N-terminal pro-B-type natriuretic peptide (NT-proBNP) and B-type natriuretic peptide (NPPB, also known as BNP) yielding hazard ratios (HRs) of 1.74 (95% confidence interval (CI) 1.67–1.81; P = 8.7 × 10−173) and 1.62 (95% CI 1.54–1.69; P = 5.6 × 10−95), respectively, for each s.d. increase in circulating protein levels (which were log2-transformed before analysis). NT-proBNP was also the second strongest for association with heart failure, with an HR of 1.57 (95% CI 1.48–1.66; P = 5.4 × 10−56) per s.d. The biomarker most strongly associated with heart failure was WAP four-disulfide core domain protein 2 (WFDC2), a fibroblast-derived mediator of fibrosis also known as human epididymis protein 4 (HE4)15, with an HR of 1.62 (95% CI 1.54–1.72; P = 4.1 × 10−65) per s.d. The proteins most strongly associated with incident coronary artery disease were growth differentiation factor 15 (GDF15; HR 1.31 (95% CI 1.26–1.36) per s.d.; P = 2.0 × 10−45) and matrix metalloproteinase-12 (MMP12; HR 1.29 (95% CI 1.24–1.34) per s.d.; P = 1.1 × 10−39); those most strongly associated with aortic stenosis were GDF15 (HR 1.44 (95% CI 1.29–1.60) per s.d.; P = 2.7 × 10−11) and WFDC2 (HR 1.40 (95% CI 1.26–1.55) per s.d.; P = 7.3 × 10−10). The distributions of the proteins with the strongest associations, stratified by incident cases versus controls, are shown in Fig. 1.
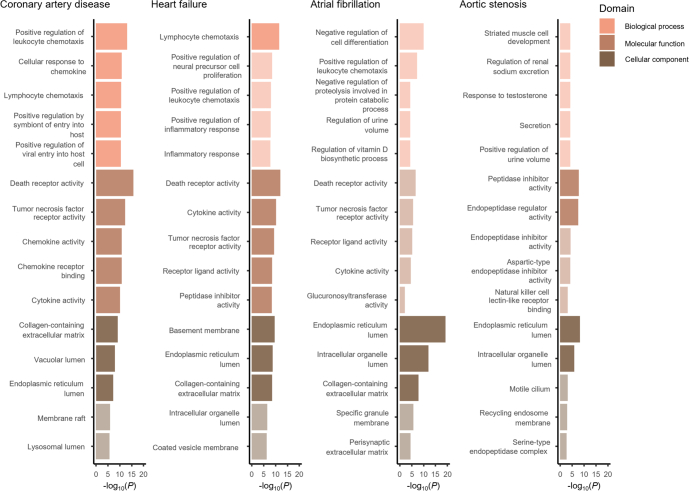
To gain insights into biological pathways associated with the identified proteins, we carried the 820 observed protein–disease associations forward for pathway enrichment analysis using the Gene Ontology resource16 via Enrichr17. The highest-scoring pathways for coronary artery disease and heart failure included inflammatory and immune-related processes involving leukocyte/lymphocyte chemotaxis and cellular response to cytokines (Extended Data Fig. 4 and Supplementary Table 5). Participants with coronary artery disease or heart failure during follow-up were also enriched for apoptosis-related proteins such as those from the tumor necrosis factor (TNF) receptor family. Proteins associated with aortic stenosis demonstrated enrichment for peptidase inhibitor activity, consistent with recent work suggesting an important role for certain peptidases in the progression of calcific aortic stenosis18. Furthermore, according to data from the Human Protein Atlas19, proteins associated with at least one cardiac outcome were more often actively secreted to the bloodstream (34.0% (n = 150 of 441 proteins)) than those without any significant associations (16.2% (n = 165 of 1,018); chi-squared test, P = 5.4 × 10−14).
Extended Data Fig. 4. Top biological processes, molecular functions, and cellular components enriched among proteins associated with coronary artery disease, heart failure, atrial fibrillation, and aortic stenosis.
Top biological functions, molecular pathways, and cellular components were queried using the Gene Ontology resource22,23 via Enrichr24. Enrichment tests were performed against a background gene set that included the genes corresponding to all 1,459 proteins tested in primary analyses. Gene sets with a false discovery rate-adjusted two-sided P < 0.05 were considered statistically significant. Bright colors indicate statistical significance, whereas dull colors indicate no statistical significance. All P-values shown were unadjusted for multiple comparisons.
Cis-Mendelian randomization analyses
Next, we performed MR analyses to infer causal effects of the identified proteins on coronary artery disease, heart failure, atrial fibrillation and aortic stenosis. As the use of cis-protein quantitative trait loci (cis-pQTLs; genetic variants associated with circulating protein levels that map near the protein-encoding gene) facilitates adherence to the assumptions of MR20,21, we only used variants within a 200-kilobase range of the protein-encoding gene to construct our genetic instruments. Of 441 unique Bonferroni-significant proteins in primary analyses, 430 (97.5%; corresponding to 802 protein–disease associations) had at least one valid cis-pQTL (±200 kilobases, P < 5 × 10−6, R2 < 0.1) (Supplementary Table 6). F-statistics were >10 for all proteins other than myoglobin, which was excluded from cis-MR analyses to minimize the risk of weak instrument bias. Median (IQR) F-statistics and R2 estimates (representing phenotypic variance explained by genetic instruments) were 1,515 (454–4,050) and 3.2% (1.0–7.9%), respectively (Supplementary Table 7). Consistent with the use of cis-pQTLs21, Steiger filtering did not identify any variants explaining more variance in the outcome than the exposure (Supplementary Table 8).
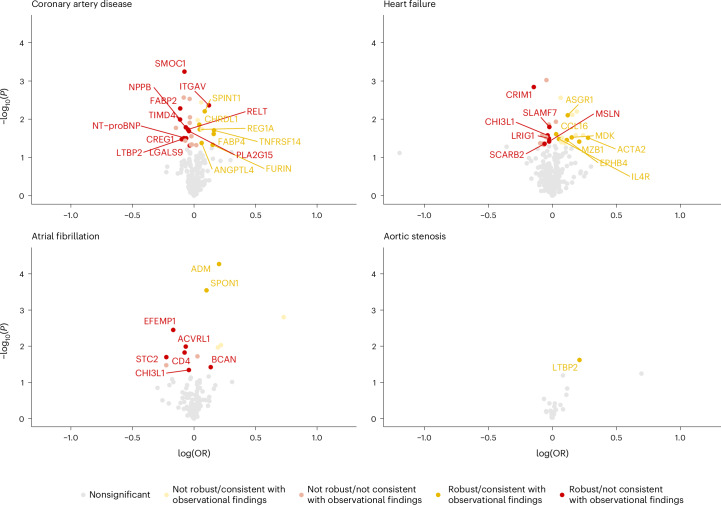
Of 801 protein–disease associations examined in cis-MR analyses, 76 (9.5%; representing 69 of 429 (16.1%) proteins) showed suggestive evidence of causality with P < 0.05 (Fig. 3 and Supplementary Table 9). Because it is routinely recommended to evaluate cis-MR findings across P value and R2 thresholds13, we performed multiple sensitivity analyses (using genetic variants at P < 5 × 10−4/< 5 × 10−6/< 5 × 10−8 and R2 < 0.001/< 0.01/< 0.1/< 0.2) to evaluate the robustness of the observed genetic associations (Supplementary Table 10). We further performed MR-Egger (Supplementary Table 10), one-sample MR (Supplementary Table 11) and multivariable-adjusted MR adjusting for proteins with shared pQTLs (Supplementary Tables 12 and13). A total of 40 of 76 (52.6%) genetic protein–disease associations were robust across all sensitivity analyses (directionally consistent across all MR models without evidence of horizontal pleiotropy). Genetic and observational analyses showed directional consistency for 17 of 40 (42.5%) robust genetic associations, corresponding to 2.1% of all protein–disease pairs and 4.0% of unique proteins tested in cis-MR analyses. These protein–disease associations all had positive effect estimates, implying that increased protein concentrations may promote cardiac disease risk and lowering would reduce risk. Furthermore, proteins were considered to be druggable for 14 of 17 (82.4%) robustly and directionally consistent protein–disease associations (Supplementary Table 14)22.
Fig. 3. Associations of genetically predicted protein levels with coronary artery disease, heart failure, atrial fibrillation and aortic stenosis.
The volcano plots visualize the genetic associations of all proteins identified in primary analyses with their corresponding outcomes, by plotting each association’s −log10(P) against the corresponding log(OR) per s.d. increase in genetically predicted protein levels. All analyses represent cis-MR analyses using the IVW (for instruments with two or more variants) or Wald ratio method (for instruments with one variant). Genetic instruments were constructed using cis-variants associated with circulating protein levels at P < 1 × 10−4 clumped at R2 < 0.1. Associations with two-sided P < 0.05 (not corrected for multiple comparisons) are shown in yellow (if the primary cis-MR analysis was directionally consistent with the observational analysis) or red (if the primary cis-MR analysis was not directionally consistent with the observational analysis). Bright colors and protein labels indicate robustness against sensitivity analyses (Methods), whereas dull colors indicate no robustness against sensitivity analyses. OR, odds ratio.
Because proprotein convertase subtilisin/kexin type 9 (PCSK9) is an established causal biomarker and therapeutic target for coronary artery disease23, we used this protein as a positive control for our cis-MR analyses (despite PCSK9 not reaching Bonferroni significance for coronary artery disease in epidemiological analyses). Each s.d. increase in genetically predicted PCSK9 was associated with 1.23-fold odds of coronary artery disease (95% CI 1.17–1.30; P = 6.0 × 10−16), supporting a cis-MR strategy for the identification of potential causal protein–disease associations. The proteins with the strongest epidemiological associations did not generally show strong genetic associations with cardiac diseases. For instance, neither genetic associations for WFDC2 nor GDF15 reached nominal significance (Supplementary Table 9). Genetically predicted MMP12, which was among the most strongly associated proteins for incident coronary artery disease in epidemiological analyses, was only modestly associated with a protective effect on coronary artery disease risk in primary cis-MR analyses (OR 0.97 (95% CI 0.95–1.00) per s.d.; P = 0.046).
The strongest robust genetic associations were observed for spondin-1 (SPON1) and adrenomedullin (ADM) with atrial fibrillation. Consistent with their observational associations, higher genetically predicted levels of SPON1 (OR 1.11 (95% CI 1.05–1.17) per s.d.; P = 2.9 × 10−4) and ADM (OR 1.23 (95% CI 1.11–1.35) per s.d.; P = 5.4 × 10−5) were associated with a greater risk of atrial fibrillation. Notably, colocalization analyses suggested shared causal genetic variants between these two proteins and atrial fibrillation (posterior probability for shared causal variants [H4] > 0.80; Supplementary Table 15). The Kunitz-type protease inhibitor 1 (SPINT1; also known as hepatocyte growth factor activator inhibitor type 1) and asialoglycoprotein receptor 1 (ASGR1) had the strongest directionally concordant and robust genetic associations for coronary artery disease (OR 1.09 (95% CI 1.03–1.23) per s.d.; P = 7.9 × 10−3) and heart failure (OR, 1.13 (95% CI 1.03–1.49) per s.d.; P = 2.4 × 10−2), respectively. For aortic stenosis, the latent-transforming growth factor β-binding protein 2 (LTBP2) was the only protein with a robust and directionally consistent genetic association (OR 1.24 (95% CI 1.03–1.49) per s.d.; P = 2.4 × 10−2).
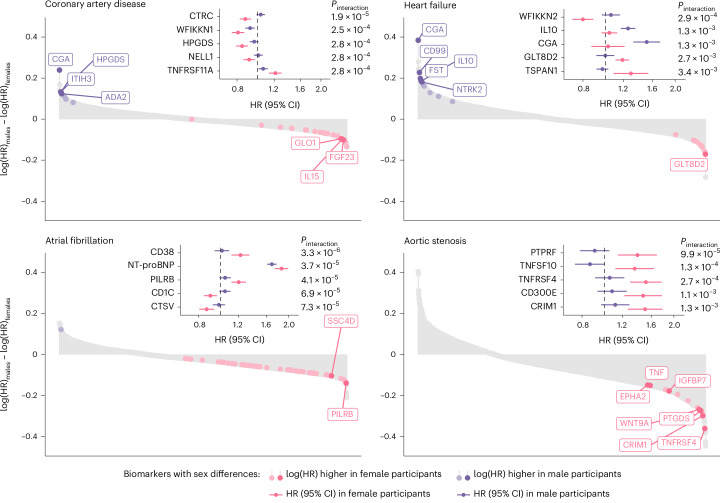
Sex-specific protein–disease associations
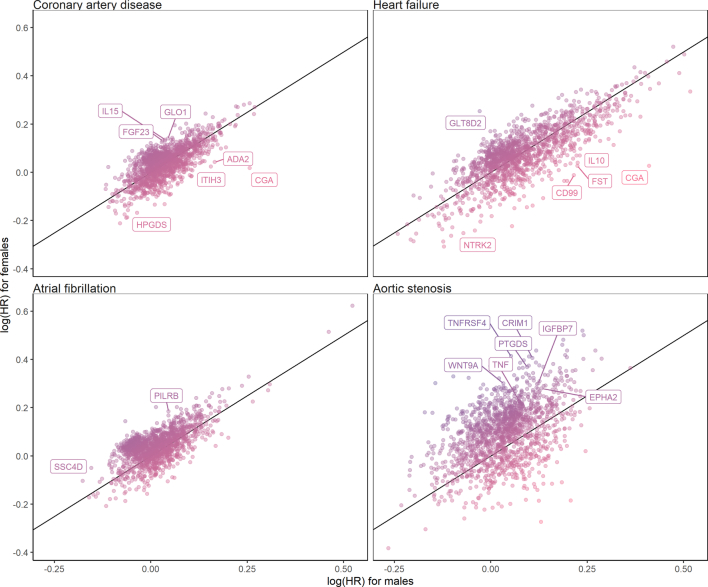
Because previous work suggested sex differences in the concentrations of cardiovascular biomarkers24, we hypothesized (a priori) that certain proteins were differentially associated with cardiac disease risk in men versus women. Therefore, we tested the multivariable-adjusted associations of all 1,459 proteins with cardiac diseases in men (n = 19,612) versus women (n = 24,701). A total of 467 protein–disease associations met the primary significance threshold (P < 0.05/5,836) for men versus 314 for women (Supplementary Table 16). Protein–disease associations (for all 1,459 tested biomarkers) showed strong correlation between sexes, indicated by a Pearson correlation coefficient (r) of 0.71 (Extended Data Fig. 5). The correlation between sexes was strongest for heart failure (r = 0.79), whereas it was comparatively weaker for aortic stenosis (r = 0.47).
Extended Data Fig. 5. Correlation between the effect sizes of protein–disease associations in male vs. female participants.
The scatter plots depict the correlation between the protein–disease associations’ effect sizes (that is, log[HR]) in female vs. male participants. HR indicates hazard ratio. All estimates were calculated using multivariable-adjusted Cox proportional hazards models, adjusted for age, age², self-reported race/ethnicity, the first ten principal components of genetic ancestry, smoking, normalized Townsend deprivation index, body mass index, systolic blood pressure, antihypertensive medication use, total cholesterol, high-density lipoprotein cholesterol, cholesterol-lowering medication use, serum creatinine, and prevalent type 2 diabetes. In addition, we included the cardiac outcomes that were not tested (for example, heart failure, atrial fibrillation, and aortic stenosis for incident coronary artery disease models) as time-varying covariates. The labeled protein–disease represent proteins that were associated with the indicated outcome at two-sided P < 0.05/5,836 (that is, Bonferroni-adjusted) in one sex without nominal significance (two-sided unadjusted P > 0.05) in the other sex. In addition, all proteins indicated in color had suggestive evidence for interaction by sex (two-sided unadjusted Pinteraction < 0.05). HR indicates hazard ratio.
We formally tested for sex interactions across all protein–disease associations reaching significance (P < 0.05/5,836) in at least one sex (n = 566) (Fig. 4 and Supplementary Table 16). Six protein–disease associations had a Bonferroni-significant (P < 0.05/566) sex-differential effect for atrial fibrillation, including T cell surface glycoprotein CD1c (CD1C; Pinteraction = 6.9 × 10−5), cyclic ADP-ribose hydrolase (CD38; Pinteraction = 3.3 × 10−6), cathepsin L2 (CTSV; Pinteraction = 7.3 × 10−5), NT-proBNP (Pinteraction = 3.7 × 10−5), paired immunoglobulin-like type 2 receptor β (PILRB; Pinteraction = 4.1 × 10−5) and WFDC2 (Pinteraction = 7.7 × 10−5). We also observed a sex-differential effect for chymotrypsin C (CTRC; Pinteraction = 1.9 × 10−5) on coronary artery disease. To test whether these associations differed between premenopausal and postmenopausal women, we performed association analyses in female participants stratified by menopause status and tested the interaction between these proteins and menopause status on cardiac outcome risk (Supplementary Fig. 1). These analyses revealed that the association of CD38 with atrial fibrillation was stronger in premenopausal than in postmenopausal women (Pinteraction = 4.4 × 10−2), although CD38 was positively associated with atrial fibrillation risk in both groups. There were no other significant interactions between circulating proteins and menopause status on cardiac disease risk, suggesting that the identified sex-differential effects are not strongly affected by menopause status.
Fig. 4. Sex-specific protein–disease associations and protein-by-sex interactions for coronary artery disease, heart failure, atrial fibrillation and aortic stenosis.
Lollipop plots depict the differences in effect sizes between male and female participants (log(HR)males − log(HR)females) for all tested protein–disease associations. Bright colors with labels represent proteins with two-sided P < 0.05/5,836 (Bonferroni-corrected) in one sex without nominal significance (two-sided P > 0.05) in the other sex; dull colors represent proteins with P < 0.05/5,836 in one sex and at least nominal significance (two-sided P < 0.05) in the other sex. In addition, all proteins indicated in color had suggestive evidence for interaction by sex (two-sided Pinteraction < 0.05). Forest plots depict the sex-stratified protein–disease associations (purple for men, pink for women) for the five proteins with the strongest sex–protein interactions. In these forest plots, central points indicate the HR of the indicated protein (per s.d.) with the indicated outcome stratified by sex (with corresponding 95% CIs). Pinteraction indicates the P value for the interaction term between ‘sex’ and the indicated protein on the corresponding outcome. All associations were tested using multivariable-adjusted Cox proportional hazards models (Methods) in 19,612 male and 24,701 female participants.
Protein-based prediction of cardiac diseases
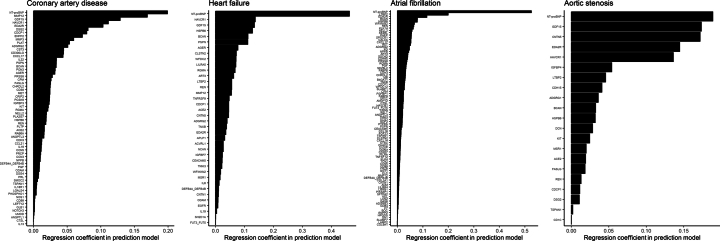
We next derived and tested the predictive accuracy of protein-based risk scores in addition to clinical risk factors in the UKB-PPP. We constructed protein-based, clinical, and combined (using proteomic and clinical variables) risk scores in the training set (80%; n = 35,450) using least absolute shrinkage and selection operator (LASSO) regression with tenfold cross-validation. Protein-based risk scores (using all 1,459 tested proteins as input) included 64 proteins for coronary artery disease, 38 for heart failure, 92 for atrial fibrillation and 21 for aortic stenosis (Supplementary Table 17 and Extended Data Fig. 6). The prediction models’ highest-weighted biomarkers were largely overlapping with those showing the strongest associations in primary analyses.
Extended Data Fig. 6. Protein weights for the primary protein-based prediction models of coronary artery disease, heart failure, atrial fibrillation, and aortic stenosis.
Each bar indicates the protein weights (that is, the absolute value of the corresponding regression coefficients).
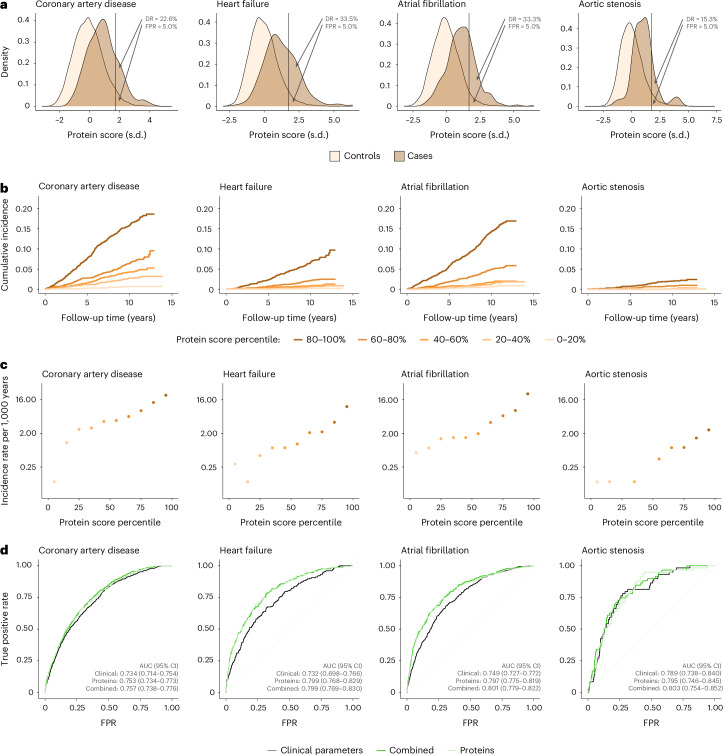
Analyses in the testing cohort (20%; n = 8,863) revealed that protein-based risk scores effectively stratified the risk of incident events across outcomes (Fig. 5a–c). The protein-based risk scores were strong independent predictors of incident events in multivariable-adjusted Cox regression models, with HRs of 2.19 (95% CI 1.87–2.55; P = 3.1 × 10−23) per s.d. increase for coronary artery disease, 2.49 (95% CI 2.10–2.95; P = 1.4 × 10−25) for heart failure, 2.39 (95% CI 2.13–2.69; P = 7.5 × 10−48) for atrial fibrillation and 2.70 (95% CI 1.65–4.42; P = 7.5 × 10−5) for aortic stenosis. The top versus bottom quintile of protein-based risk scores was associated with HRs of 8.15 (95% CI 4.07–16.30; P = 3.04 × 10−9) for coronary artery disease, 12.85 (95% CI 3.90–42.31; P = 2.67 × 10−5) for heart failure, 6.85 (95% CI, 3.40–13.80; P = 7.52 × 10−8) for atrial fibrillation and 2.70 (95% CI 0.45–16.13; P = 0.28) for aortic stenosis.
Fig. 5. Risk stratification and prediction of incident coronary artery disease, heart failure, atrial fibrillation and aortic stenosis by protein-based risk scores in the UKB-PPP.
a, Distributions of protein-based risk scores in cases and controls. b, Cumulative incidence of each outcome (calculated using the Kaplan–Meier method) by protein-based score quintiles. c, Incidence rate estimates according to protein-based score deciles on a logarithmically scaled y axis. d, Accuracies of the clinical, proteomic and combined risk scores in predicting the indicated outcomes (quantified using the ROC AUC) with corresponding 95% CI. For a, the vertical lines indicate the protein-based risk score values corresponding to an FPR of 5.0%; the DRs indicate ‘exact’ detection rates, calculated as the unadjusted proportions of cases with a positive test result at the corresponding protein-based risk score threshold. For b, incidence rate estimates are not displayed if the incidence in a protein score percentile bin was zero. All analyses were performed in the UKB-PPP testing set (n = 8,863). During a median (IQR) follow-up of 11.1 (10.4–11.8) years, 566 participants in the UKB-PPP testing set experienced coronary artery disease events, 203 experienced heart failure, 432 atrial fibrillation and 59 aortic stenosis.
Distributions of protein-based risk scores in individuals who did and did not experience an event during follow-up are shown in Fig. 5a. ROC curve analyses revealed that adding proteomic data improved the prediction of incident coronary artery disease, heart failure and atrial fibrillation (Fig. 5d). The increment in predictive accuracy compared to the clinical model—quantified using the area under the ROC curve (AUC)—was most pronounced for atrial fibrillation (AUC 0.801 (95% CI 0.779–0.822) versus 0.749 (95% CI 0.727–0.772); DeLong test: P = 2.0 × 10−10) and heart failure (AUC 0.799 (95% CI 0.769–0.830) versus 0.732 (95% CI 0.698–0.766); P = 1.7 × 10−6), followed by coronary artery disease (AUC 0.757 (95% CI 0.738–0.776) versus 0.734 (95% CI 0.714–0.754); P = 1.4 × 10−4). There was no statistically significant difference for aortic stenosis (AUC 0.803 (95% CI 0.754–0.852) versus 0.789 (95% CI 0.738–0.840); P = 0.35). For a false positive rate of 5% (where a test score incorrectly classifies 5% of controls as positive), the protein-based risk scores for coronary artery disease, heart failure, atrial fibrillation and aortic stenosis yielded exact detection rates (the proportions of cases that were correctly classified as positive; also known as true positive rates or sensitivities) of 22.6%, 33.5%, 33.3% and 15.3%, respectively (Fig. 5a). The corresponding exact detection rates of the combined risk scores for a false positive rate of 5% were 21.0%, 35.0%, 35.9% and 18.6%. Using a more stringent false positive rate cutoff of 1%, the corresponding true positive rates were 8.1%, 9.4%, 10.6% and 5.1%; the probabilities of experiencing an event during follow-up (given a positive test result) for these were 34.8%, 34.7%, 18.0% and 3.6%, respectively (Supplementary Table 18).
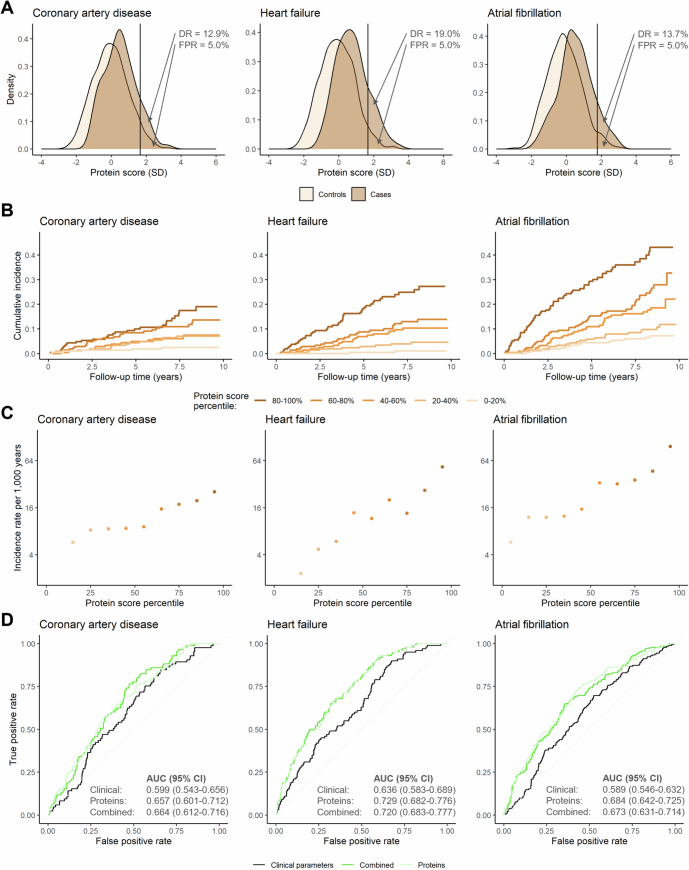
To evaluate the performance of the protein-based scores for coronary artery disease, heart failure and atrial fibrillation in an external cohort, we tested the accuracies of these scores in the Women’s Health Initiative (WHI). A total of 1,083 WHI participants who provided blood samples at the WHI-Long Life Study (LLS) visit, with data on 552 circulating protein analytes (measured using six Olink Target 96 assays), were included (Extended Data Fig. 7 and Supplementary Table 19). Among the 552 available protein analytes, there were 518 unique proteins that were also measured the UKB-PPP (Supplementary Table 20) and were used to retrain the proteomic models in the UKB-PPP training set (Supplementary Table 21). ROC curve analyses in the UKB-PPP testing set demonstrated that the retrained proteomic scores (based on the proteins that were overlapping between the UKB-PPP and the WHI-LLS) improved the prediction of incident events with increments that were similar to those observed using the scores that were constructed using the full protein set (based on all 1,459 circulating proteins measured in the UKB-PPP) (Supplementary Fig. 2). Similarly, analyses in the WHI-LLS showed that the combined models (based on both clinical and proteomic variables) were associated with a significantly better detection of coronary artery disease (AUC 0.664 (95% CI 0.612–0.716) versus 0.599 (95% CI 0.543–0.656); P = 1.4 × 10−2), heart failure (AUC 0.720 (95% CI 0.683–0.777) versus 0.636 (95% CI 0.583–0.689); P = 6.6 × 10−4) and atrial fibrillation (AUC 0.673 (95% CI 0.631–0.714) versus 0.589 (95% CI 0.546–0.632); P = 6.7 × 10−7) compared to the clinical models (Extended Data Fig. 8).
Extended Data Fig. 7. WHI-LLS participant inclusion and exclusion criteria for external validation analyses.
External validation analyses tested the performance of protein-based risk scores to predict incident coronary artery disease, heart failure, and atrial fibrillation in 1,083 participants from the Women’s Health Initiative who attended the Long Life Study (WHI-LLS).
Extended Data Fig. 8. Risk (A–C) stratification and (D) prediction of incident coronary artery disease, heart failure, atrial fibrillation, and aortic stenosis by protein-based risk scores in the WHI-LLS.
The indicated plots depict (A) the distributions of protein-based risk scores in cases and controls; (B) the cumulative incidence of each outcome (calculated using the Kaplan–Meier method) by protein-based score quintiles; (C) incidence rate estimates according to protein-based score deciles on a logarithmically scaled Y axis; and (D) the accuracies of the clinical, proteomic, and combined risk scores in predicting the indicated outcomes (quantified using the area under the receiver-operating characteristic curve [AUC] with corresponding 95% confidence intervals [CIs]). For (A), the vertical lines indicate the protein-based risk score values corresponding to a false positive rate (FPR) of 5.0%; the detection rates (DRs) indicate the “exact” detection rates, calculated as the unadjusted proportions of cases with a positive test result at the corresponding protein-based risk score threshold. For (C), incidence rate estimates are not displayed if the incidence of the indicated outcome in a protein score percentile bin was zero. All analyses were performed in the Women’s Health Initiative Long Life Study (WHI-LLS; n = 1,083). During a median (interquartile range) follow-up of 8.3 (5.6-8.9) years, 85 participants in the WHI-LLS cohort experienced coronary artery disease events, 100 experienced heart failure, and 182 atrial fibrillation.
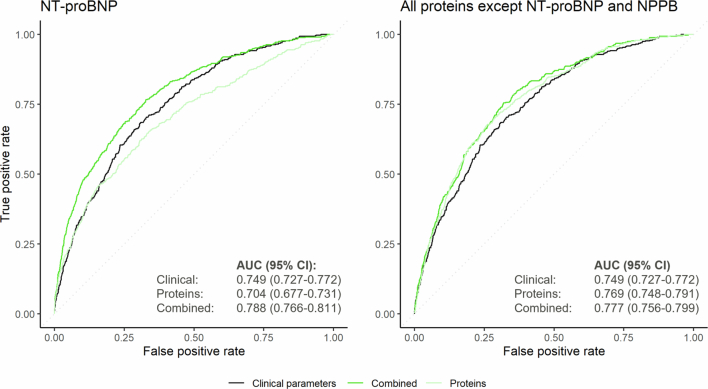
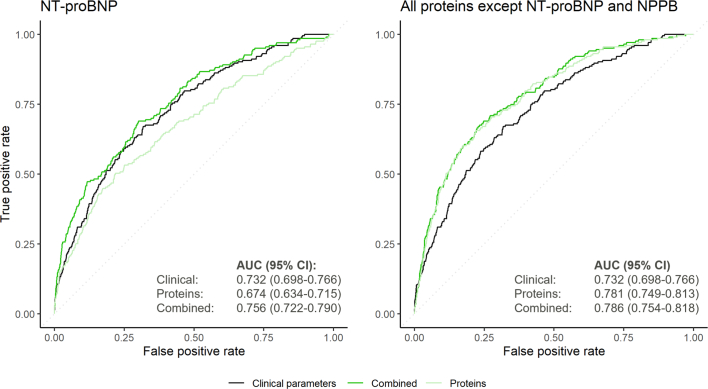
Given the disproportionately high weights for NT-proBNP in the protein-based risk scores for atrial fibrillation and heart failure (Supplementary Table 17), we further evaluated the performance of models including NT-proBNP alone versus those incorporating all other biomarkers in predicting these outcomes in the UKB-PPP testing set. We also excluded NPPB from the latter set of protein-based risk scores as NPPB and NT-proBNP are encoded by the same gene and released in the circulation in equimolar quantities25. Compared to the score based on clinical factors alone (0.749 (95% CI 0.727–0.772)), inclusion of NT-proBNP significantly improved the prediction of atrial fibrillation (AUC 0.788 (95% CI 0.766–0.811); P = 1.1× 10−6), resulting in a greater increment in predictive accuracy than the score incorporating all proteins other than NT-proBNP and NPPB (AUC 0.777 (95% CI 0.756–0.799); P = 9.2 × 10−7) (Extended Data Fig. 9). In contrast, for heart failure, the score incorporating all proteins except NT-proBNP and NPPB was associated with a greater improvement in predictive accuracy versus the clinical score (AUC 0.786 (95% CI, 0.754–0.818) versus 0.732 (95% CI 0.698–0.766); P = 1.4 × 10−5) than the score incorporating NT-proBNP alone (AUC 0.756 (95% CI 0.722–0.790); P = 0.07) (Extended Data Fig. 10).
Extended Data Fig. 9. Risk prediction of incident atrial fibrillation by risk scores incorporating NT-proBNP and all proteins except NT-proBNP and NPPB.
The receiver-operating characteristics curves depict the accuracy of the clinical, proteomic, and combined risk scores in predicting atrial fibrillation events in the UKB-PPP testing set (n = 8,863). Areas under the curve (AUCs) and corresponding 95% confidence intervals (95% CIs) quantify the performance of each model. Models with multiple candidate features were constructed using logistic least absolute shrinkage and selection operator (LASSO) models; the combined models included all clinical predictors (see Methods) as well as the indicated biomarkers (that is, NT-proBNP or all proteins except NT-proBNP and NPPB) as potential covariates in the final model. Participants were followed for a median (interquartile range) follow-up of 11.1 (10.4–11.8) years.
Extended Data Fig. 10. Risk prediction of incident heart failure by risk scores incorporating NT-proBNP and all proteins except NT-proBNP and NPPB.
The receiver-operating characteristics curves depict the accuracy of the clinical, proteomic, and combined risk scores in predicting heart failure events in the UKB-PPP testing set (n = 8,863). Areas under the curve (AUCs) and corresponding 95% confidence intervals (95% CIs) quantify the performance of each model. Models with multiple candidate features were constructed were constructed using logistic least absolute shrinkage and selection operator (LASSO) models; the combined models included all clinical predictors (see Methods) as well as the indicated biomarkers (that is, NT-proBNP or all proteins except NT-proBNP and NPPB) as potential covariates in the final model. Participants were followed for a median (interquartile range) follow-up of 11.1 (10.4–11.8) years.
Discussion
In a population-based cohort of ~45,000 middle-aged adults with circulating protein measurements and longitudinal follow-up, we characterized the proteomic architecture of incident coronary artery disease, heart failure, atrial fibrillation and aortic stenosis. We identified 820 significant protein–disease associations with important roles (potentially mediating or marking disease presence) for natriuretic peptides (for example, NT-proBNP), inflammatory mediators (for example, MMP12) and apoptosis-related factors (for example, GDF15) as predictors of cardiac diseases. Genetic analyses suggested causal or mediating roles—either protective or deleterious—for a substantial proportion of biomarkers identified in observational analyses. Sex-based analyses suggested generally preserved associations between men and women, albeit with varying weights of prediction including several biomarkers with strong sex interactions. Finally, we constructed sparse protein-based risk scores that improved the prediction of cardiac disease development in the general population. Our findings provide insights into the biology of cardiac diseases, with implications for the prediction of incident cardiovascular diagnoses and potential for targeted prevention and treatment of these conditions.
The findings from this study offer insights into potential causal roles of proteins associated with incident cardiac diseases. We found that 4% of proteins identified in primary analyses (and tested in cis-MR analyses) had putative causal associations that were directionally concordant with those derived from epidemiological models (primary analyses); however, we also identified many proteins (more than 5%) with genetic associations that were robust across sensitivity analyses yet directionally discordant with epidemiological estimates (‘opposite’). By systematically integrating observational and genetic data, our study corroborates and extends previous studies reporting similar discrepancies between genetic and epidemiological associations for selected proteins13,26. For instance, consistent with previous research13,26, primary cis-MR analyses revealed a protective effect of genetically predicted MMP12 on coronary artery disease, although observational analyses indicated strong associations of higher MMP12 levels with the same outcome. Whether these seemingly discordant observations reflect inherent differences between disease onset versus progression or compensatory response to subclinical disease (where protein levels increase before the onset of symptoms, potentially acting as adaptive or compensatory mechanisms to mitigate damage caused by the underlying disease) requires further investigation. Nevertheless, several proteins had consistent genetic and observational effects. One example of concordant observations was SPON1, for which higher levels (both measured and genetically predicted) were associated with increased atrial fibrillation risk. SPON1 is an extracellular protein expressed in tissues such as the heart and brain that has been implicated in Alzheimer’s dementia through its role in amyloid-β precursor protein processing27. Previous protein-focused analyses in patients with heart failure showed that the presence of atrial fibrillation was associated with activation of amyloid-β-related pathways, with SPON1 as one of the most strongly upregulated proteins in those with atrial fibrillation28. These data, together with colocalization findings indicating shared causal variants for SPON1 and atrial fibrillation, collectively suggest that SPON1 not only marks presence of a pro-arrhythmic substrate, but could also represent an upstream therapeutic target for preventing and/or treating atrial fibrillation. Given the paucity of identified robust biomarkers mediating atrial fibrillation risk, more data are needed regarding SPON1 and its role in arrhythmogenesis.
Biomarkers identified in proteomic analyses are often markers of already established disease, rather than mediators of disease biology. In this regard, our analyses identified several inflammation- and apoptosis-related proteins as strong predictors marking risk for cardiac disease but were unlikely causal biomarkers. WFDC2 (also known as HE4)—a profibrotic protease inhibitor with a potential role in natural immunity15—emerged as the strongest proteomic predictor of heart failure. Previous research in hospitalized heart failure patients demonstrated associations of circulating WFDC2 with disease severity as well as kidney function29. As WFDC2 is expressed exclusively in noncardiovascular tissues such as the respiratory tract, male and female genitourinary system and kidneys30, it is likely that the strong associations of WFDC2 with cardiac outcomes stem from peripheral organ responses rather than indicating direct cardiac dysfunction or vascular damage. Similarly, GDF15—another pleiotropic protein expressed across multiple organ systems31—was the strongest biomarker for coronary artery disease. As a member of the transforming growth factor-β (TGF-β) superfamily, GDF15 is upregulated in response to external stressors (for example, inflammation, hypoxia and oxidative stress) and is believed to reflect the cumulative impact of both acute and chronic exposure to cellular stressors31. Recent data suggest GDF15 as an independent prognostic biomarker for individuals with established atherosclerotic cardiovascular disease32. Nevertheless, cis-MR analyses detected no evidence of causality in the associations of these proteins with cardiac diseases. Assuming that these analyses had adequately strong genetic instruments and sufficient power, our findings collectively suggest that inflammation- and apoptosis-related biomarkers such as WFDC2 and GDF15 represent early disease markers without causal involvement in the pathogenesis of cardiac diseases, consistent with their pleiotropic and nonspecific effects in response to tissue damage across organs.
In addition, our findings demonstrate that circulating proteins can provide information beyond clinical risk factors to predict cardiac events. Risk scores integrating proteomic and clinical data led to better prediction of coronary artery disease, heart failure and atrial fibrillation than those based on clinical parameters alone, both internally in the UKB-PPP and externally in the WHI-LLS. Nevertheless, the clinical-proteomic scores yielded detection rates ranging 35–50% for these conditions, for a false positive rate of 10%. As cardiovascular prevention (for example, through statins) is offered at progressively lower cardiovascular risk thresholds, it is unlikely that proteomic scores will be an effective standalone test to screen for allocation of primary prevention therapies in people without known risk drivers33,34. Nevertheless, our analyses demonstrate that protein-based risk scores confer information that is not captured by clinical risk factors and may also provide biological insights. For example, in addition to confirming the established association of natriuretic peptide elevation with so-called ‘pre-heart failure’35, ROC analyses revealed that NT-proBNP was a better predictor of atrial fibrillation than all other proteins together (except NPPB or BNP). These results extend previous work demonstrating strong associations of circulating NT-proBNP with incident atrial fibrillation36 and align with recent data from the LOOP trial, suggesting that individuals with elevated NT-proBNP levels may derive more clinical benefit from atrial fibrillation screening than those with lower levels37. Collectively, these findings provide support for the use of NT-proBNP as a biomarker for atrial fibrillation in the general population.
Another finding from this study was evidence for biological sex differences underlying cardiac disease risk in men and women. The strongest sex interaction across all tested proteins was observed for CD38, which was significantly more strongly associated with incident atrial fibrillation in female than in male participants. CD38 is a glycoprotein expressed across various immune cells including lymphocytes and plasma cells38. Previous research suggests that CD38 is causally implicated in autoimmune diseases such as rheumatoid arthritis and systemic lupus erythematosus38,39. As a history of autoimmune diseases represents a risk factor for atrial fibrillation that affects women more strongly than men40, it could be possible that the observed sex differences for CD38 reflect a more important role for immunity-related pathways in women. Furthermore, some of the largest differences in protein–disease associations between sexes were observed for aortic stenosis. For instance, we identified several sex-specific senescence-associated biomarkers (for example, IGFBP7 and TNF) associated with incident aortic stenosis in female, but not male, participants. Previous histological work in aortic stenosis patients revealed distinct tissue composition differences between men and women, with women showing less valvular calcification but more fibrosis than men41. These findings indicate that fibrosis-related proteins are likely stronger markers for aortic stenosis in women than in men.
While this study benefits from a large sample size and the use of state-of-the-art proteomic profiling methods, findings must be interpreted in the context of limitations. First, the strength and quantity of protein–disease associations for each outcome were influenced by statistical power and, consequently, the number of cases per outcome. Conditions with lower incidence rates during follow-up (such as aortic stenosis) had fewer proteomic associations. Second, the study population was predominantly white, precluding generalization to other races/ethnicities. Third, causal inference using MR relies on the validity of the underlying instrumental variable assumptions. This study utilized a robust cis-MR framework (facilitating the adherence to these assumptions20,21) and tested the robustness of the genetic associations through many sensitivity analyses. Nevertheless, prioritized therapeutic targets remain to be evaluated in animal experiments and eventually human trials. Fourth, not all proteins identified in primary analyses had strong cis-pQTLs, precluding adequate cis-MR analyses. Genetic instrument strength also varied across proteins. Instruments with more variants have greater power to detect statistically significant genetic protein–disease associations, potentially leading to an underestimation of associations for instruments with fewer variants. Additionally, there are no established power calculation methods for two-sample MR analyses with binary outcomes (such as those performed in this study) beyond the use of F-statistics to evaluate genetic instrument strength. Nevertheless, we minimized type II error by adopting a lenient P value threshold (P < 0.05) to indicate statistical significance for primary cis-MR analyses and prioritizing genetic protein–disease associations that were robust to many sensitivity analyses. Finally, external validation of protein-based risk scores was performed using a restricted set of proteins that only included those that were measured in both the UKB-PPP and WHI-LLS. Therefore, the external validation results do not fully reflect the predictive ability of protein-based risk scores constructed using the full set of proteins evaluated in primary analyses. This limitation partially reflects the rapid evolution of large-scale proteomics research, characterized by increasingly extensive but imperfectly overlapping proteomic assays across platforms used in different cohorts. Nevertheless, our external validation approach still found that a limited panel of proteins measured in both the UKB-PPP and WHI-LLS significantly improved the prediction of incident coronary artery disease, heart failure and atrial fibrillation in both cohorts.
Leveraging a population-based cohort of ~45,000 participants, this study characterized the circulating proteome associated with incident coronary artery disease, heart failure, atrial fibrillation and aortic stenosis. The study findings support new applications for established biomarkers (for example, atrial fibrillation surveillance using NT-proBNP) and identify strong and potentially useful predictors of cardiac diseases (for example, WFDC2 for heart failure). These results lay a foundation for future investigations to uncover disease mechanisms and assess the clinical utility of protein-based prevention strategies for cardiac diseases.
Methods
Study design and participants
The study design is illustrated in Fig. 1. The UKB is a population-based cohort of ~500,000 volunteers aged 40–69 years at the time of study enrollment, recruited from 22 assessment centers across the United Kingdom during 2006–201042. At enrollment, participants provided informed consent; underwent physical examination; provided details on sociodemographic characteristics, lifestyle factors, medical history, and medication use; and donated blood samples. Follow-up for incident events occurred via linkage to electronic health records through March 2020.
The UKB-PPP is a precompetitive consortium of 13 biopharmaceutical companies funding the generation of blood-based proteomic data in a subset of UKB participants14,43. Upon release, the sponsors have no direct role in research activities of these features as is the case for the present work. The UKB-PPP includes 54,306 participants, of whom 46,673 (85.9%) were randomly selected from baseline, 6,385 (11.8%) were preselected by UKB-PPP consortium members based on certain characteristics of interest (for example, disease status or genetic ancestry) and 1,268 (2.3%) were selected because they attended multiple visits of the COVID-19 case–control imaging study14. We considered 52,705 participants with baseline proteomic data passing quality control for inclusion in the present study (Fig. 1). Participants were excluded if they had missing data for >10% of assay measurements or if they had missing data on self-reported race/ethnicity or genetic ancestry. We also excluded individuals inferred to be related (closer than third degree; kinship coefficient >0.0884) and those with self-reported or physician-ascertained coronary artery disease, heart failure, atrial fibrillation or aortic stenosis at baseline (see below for disease definitions).
The UKB was approved by the North West Multi-center Research Ethics Committee. All analyses were conducted under UKB application no. 7089. The Mass General Brigham Institutional Review Board approved the secondary use of these data.
Protein measurements and proteomic data processing
Blood samples donated by UKB-PPP study participants underwent proteomic profiling using the Olink Explore 1536 platform (Olink Proteomics), which measures 1,472 protein analytes across four different panels (the Cardiometabolic, Inflammation, Neurology and Oncology panels) representing 1,463 unique proteins (Supplementary Table 1)44. In brief, Olink uses proximity extension assay technology, whereby antibody pairs with conjugated oligonucleotides bind their target proteins in a pairwise manner. When an antibody pair has bound its target, complementary oligonucleotides undergo hybridization and, subsequently, extension by DNA polymerase. These DNA sequences—or tags—are then amplified through PCR amplification, which can be quantified using next-generation sequencing. For each assay and each sample, normalized protein expression values are calculated as the log2-transformed ratio of sequence read counts to the counts of the extension control, corrected for plate and batch effects14,43.
For proteins that were measured by multiple panels (TNF, IL-6 and CXCL8), we only evaluated data from the panel with the highest detectability per protein and, if necessary, the largest number of protein measurements exceeding the respective limit of detection (the Cardiometabolic panel for TNF and Oncology panel for IL-6 and CXCL8). We further excluded proteins with >10% missingness in the final study cohort (CTSS and NPM1 from the Neurology panel, PCOLCE from the Cardiometabolic panel and TACSTD2 from the Oncology panel; Supplementary Table 2) and imputed the remaining 1.1% of missing protein values using k-nearest neighbors (k = 10) via the impute.knn() function (impute package45 in R)43. The remaining 1,459 protein markers underwent z-score transformation before analysis.
Covariate ascertainment
Demographic characteristics, medical history, medication use and health behaviors were systematically assessed upon enrollment in the UKB. Self-reported-race/ethnicity was collected at baseline and used as a binary variable (white versus nonwhite) in analyses. Smoking was dichotomized as ever (current or past) smoking versus no history of smoking. Type 2 diabetes was defined by self-report or qualifying International Classification of Diseases (ICD) codes. The Townsend deprivation index—an area-level score that incorporates data on home ownership, automobile ownership, employment and household overcrowding—was used as a composite measure of material deprivation46. Townsend deprivation index scores were inverse-rank normalized and z-score transformed before analysis.
Anthropometric data, physical measurements and blood samples were obtained by trained study staff42. Body mass index (BMI) was calculated from standing height and weight measured at baseline. After a 5-min period of seated rest, blood pressure was measured using an electronic monitor (Omron 705 IT, OMRON Healthcare) on two separate occasions with a 1-min interval in between; the mean was calculated and used for analysis when both measurements were available. Total cholesterol, high-density lipoprotein (HDL) cholesterol and creatinine concentrations were quantified in baseline blood samples (AU5800, Beckman Coulter).
Missing values for BMI (missing for n = 717; 1.6%), systolic blood pressure (n = 2,210; 5.0%), total cholesterol (n = 1,995; 4.5%), HDL cholesterol (n = 5,591; 12.6%), serum creatinine (n = 2,010; 4.5%) and normalized Townsend deprivation index (n = 53; 0.1%) were imputed using linear regression models incorporating sex, age, race/ethnicity and the first ten principal components of genetic ancestry as predictors.
Outcome ascertainment
Follow-up for incident outcomes occurred through linkage to national health records until March 2020. Incident events were defined by the occurrence of (1) at least one qualifying ICD-9 or ICD-10 code for a corresponding in- or outpatient diagnosis (as either a primary or secondary disease diagnosis); or (2) at least one Office of Population Censuses and Surveys Classification of Surgical Operations and Procedures (OPCS) code for a qualifying procedure (for example, coronary artery revascularization for coronary artery disease). The specific codes used to define each outcome are listed in Supplementary Table 22 (refs. 47,48).
Proteomic association analyses
Primary analyses tested the associations of circulating protein levels with incident cardiac events using Cox proportional hazards models adjusted for age, age², sex, self-reported race/ethnicity, the first ten principal components of genetic ancestry, smoking, normalized Townsend deprivation index, BMI, systolic blood pressure, antihypertensive medication use, total cholesterol, HDL cholesterol, cholesterol-lowering medication use, serum creatinine (as a measure of kidney function) and prevalent type 2 diabetes. In addition, to increase the specificity of the detected protein associations for a given disease (for example, coronary artery disease), we included the other cardiac outcomes (for example, heart failure, atrial fibrillation and aortic stenosis) as time-varying covariates using the tmerge() function in R (survival package)49. Bonferroni-corrected P < 0.05/5,836 (P < 0.05/(1,459 tested proteins × 4 tested outcomes) or ~8.6 × 10−6) indicated statistical significance for the primary analyses. To illustrate the distributions of the most strongly associated proteins in individuals who experienced incident events versus those who did not, we constructed probability density functions showing the distributions of the strongest protein–disease associations in cases versus controls for each outcome using the ggplot2 package in R50.
Pathway enrichment analyses
Pathway enrichment analyses evaluated whether certain protein groups representing biologically distinct pathways were disproportionately up- or downregulated in individuals with incident cardiac events. Top biological functions, molecular pathways and cellular components were queried for each outcome using the Gene Ontology resource16 via Enrichr17. Enrichment tests were performed against a background gene set including the genes corresponding to all 1,459 proteins tested in primary analyses. Gene sets with a false discovery rate-adjusted P < 0.05 were considered statistically significant.
Main cis-Mendelian randomization analyses
We performed two-sample MR analyses to explore the causal roles of proteins that were statistically significantly associated with one or more cardiac outcomes in epidemiological models (primary analyses). These analyses tested the associations of protein quantitative trait loci (pQTLs; genetic variants associated with circulating protein levels) with coronary artery disease, heart failure, atrial fibrillation and aortic stenosis. We obtained pQTL data from 35,571 UKB-PPP participants who had their circulating proteomes profiled using the Olink Explore 1536 platform14. FinnGen (freeze 9; https://r9.finngen.fi/) was used for genetic association data for coronary artery disease (cases of total participants: n = 43,518 of 377,277), heart failure (n = 27,304 of 377,277), atrial fibrillation/flutter (n = 45,766 of 237,690) and operated calcific aortic stenosis (n = 9,153 of 377,277). Genetic association data were obtained from FinnGen rather than larger meta-GWASs (which often included the UKB) to avoid sample overlap between the exposure and outcome cohorts, which increases the risk of weak instrument bias in two-sample MR leading to inflated type I error rates51. All genetic data were derived from individuals of European ancestry, and there was no overlap between the exposure and outcome study cohorts.
Because the use of cis-pQTLs (pQTLs that map near the protein-encoding gene) facilitates adherence to the assumptions of MR20,21, we only used variants within a 200-kilobase range of the protein-encoding gene to construct our genetic instruments. We used a relaxed P value threshold for instrument selection (P < 5 × 10−6) relative to the conventional genome-wide threshold (P < 5 × 10−8) to increase the number of genetic instruments as the cis-regions for the assayed proteins represent only a small fraction of the genome, are expected to be enriched for associations, and to optimize power. All cis-pQTLs with P < 5 × 10−6 were clumped into largely independent loci (linkage disequilibrium R2 < 0.1) using PLINK52. Linkage disequilibrium information was obtained from the European panel of phase 3 of the 1000 Genomes Project53.
To minimize the risk of weak instrument bias, we only performed cis-MR analyses for genetic instruments with F-statistics >10. F-statistics were obtained by performing linear regression analyses of a protein’s genetic risk score (as the independent variable) against the measured levels of that protein (as the dependent variable) in the UKB-PPP. Genetic risk scores were calculated as weighted allele scores using the ‘clumping and thresholding’ method, applying the same P value and linkage disequilibrium R2 thresholds as those used in our primary cis-MR analyses (P < 5 × 10−6 and R2 < 0.1). All scores were calculated using genotype array data; for proteins where genetic risk score calculation failed, F-statistics were estimated using summary statistics as equation (1):
| 1 |
where n indicates the sample size of the original genome-wide association study, k the number of variants included in the genetic instrument and R2 the variance in the exposure explained by the genetic variants54.
Depending on the number of cis-pQTLs included in a protein’s genetic instrument, we used different MR methods to infer causal effects13. The inverse-variance-weighted (IVW) method was used with fixed effects for genetic instruments with two to three cis-pQTLs and with multiplicative random effects for those with more than three cis-pQTLs. The Wald ratio estimator was used for genetic instruments with only one cis-pQTL. We adjusted for between-variant correlation structure in all IVW models to avoid inflated estimates caused by residual correlation55,56.
Sensitivity cis-Mendelian randomization analyses
Because it is routinely recommended to evaluate the robustness of cis-MR estimates using multiple sensitivity analyses57, we performed additional analyses using different MR approaches and instrument selection parameters. First, we evaluated the possibility of reverse causation affecting our analyses by performing Steiger filtering to exclude variants explaining more variance in the outcome (cardiac diseases) than the exposure (circulating protein levels). Second, as cis-MR analyses often rely on pQTLs that may be residually correlated with each other, we carried out sensitivity analyses testing genetic instruments that were constructed using a range of linkage disequilibrium R2 thresholds (R2 < 0.001, R2 < 0.01, R2 < 0.1 and R2 < 0.2). Third, because the primary genetic instruments were constructed using subgenome-wide significant cis-pQTLs, we verified the robustness of our genetic associations against different P value thresholds (P < 5 × 10−4, P < 5 × 10−6 and P < 5 × 10−8). Fourth, we calculated effect estimates using the MR-Egger method to account for horizontal pleiotropy (effects of the genetic instruments on the outcome through pathways other than the exposure of interest). Fifth, we performed one-sample cis-MR analyses to test the associations of the prioritized proteins’ genetic risk scores with cardiac diseases in an external UKB sample (see below). Sixth, to account for the possibility that a certain protein’s genetic instrument could affect the outcomes through one or more other proteins, we calculated multivariable-adjusted cis-MR estimates that were adjusted for the genetic instruments of all prioritized proteins significantly associated with the tested protein’s genetic risk score (see below).
Genetic risk scores were calculated from genetic association data from the UKB-PPP as weighted allele scores using the ‘clumping and thresholding’ method, applying the same P value and linkage disequilibrium R2 thresholds as those used in the primary cis-MR analyses (P < 5 × 10−6 and R2 < 0.1). One-sample cis-MR was performed in UKB participants who were not included in the UKB-PPP, were free of cardiac diseases at baseline, and had no missing covariates (n = 407,230). Associations of the circulating proteins’ genetic risk scores with cardiac outcomes were tested using Cox regression models adjusted for age, age², sex, race/ethnicity and the first ten principal components of genetic ancestry. In addition, we interrogated the possibility that a certain protein’s genetic instrument was also associated with other proteins’ circulating levels (proteins with shared pQTLs). To investigate this, we tested the associations of genetic risk scores for all proteins with putative causal associations in the primary cis-MR analyses. Linear regression models adjusted for age, age², sex, race/ethnicity and the first ten principal components of genetic ancestry were employed for these analyses. For proteins with genetic instruments that were significantly associated with one or more other proteins (‘correlated proteins’), we then calculated multivariable-adjusted cis-MR estimates in the independent UKB sample (n = 407,230) using Cox regression models adjusted for age, age², sex, race/ethnicity, the first ten principal components of genetic ancestry and the genetic risk scores of all ‘correlated’ proteins.
Sensitivity analyses were performed for all genetic protein–disease associations with at least nominal significance (unadjusted P < 0.05) in primary cis-MR analysis. Genetic associations were considered robust if (1) the effect estimates were directionally consistent across all primary and sensitivity analyses and (2) MR-Egger suggested no horizontal pleiotropy (P ≥ 0.05 for the intercept test or P < 0.05 for the intercept test with P < 0.05 for the causal test). MR analyses were performed using the TwoSampleMR and MendelianRandomization packages in R58,59. Druggability profiles of proteins with robust genetic associations were extracted from a published list of druggable genes22.
Colocalization analyses
We performed colocalization analyses to test for shared causal variants between the prioritized proteins’ cis loci (from the UKB-PPP) and corresponding cardiac outcomes (from FinnGen). Analyses considered all variants that were present in the protein and outcome summary statistics within ±200 kilobases of each biomarker’s protein-encoding region. Colocalization analyses were performed using the coloc.abf() function (coloc package60 in R). All colocalization analysis results were expressed as test statistics representing the posterior probabilities of five hypotheses: H0, neither trait has an association with a genetic variant in the region; H1, only the indicated protein has an association with a genetic variant in the region; H2, only the indicated cardiac disease has an association with a genetic variant in the region; H3, both traits are associated but with different causal variants; and H4, both traits are associated and share a single causal variant. A posterior probability for H4 > 0.80 indicated strong colocalization evidence.
Sex-stratified association analyses
Sex-stratified analyses tested the associations of circulating protein levels with incident coronary artery disease, heart failure, atrial fibrillation and aortic stenosis in self-reported female and male participants separately. These analyses were performed using Cox proportional hazards models adjusted for age, age², self-reported race/ethnicity, the first ten principal components of genetic ancestry, smoking, normalized Townsend deprivation index, BMI, systolic blood pressure, antihypertensive medication use, total cholesterol, HDL cholesterol, cholesterol-lowering medication use, serum creatinine and prevalent type 2 diabetes. Coronary artery disease, heart failure, atrial fibrillation and aortic stenosis were included as time-varying covariates. The difference in effect size for the protein–disease associations was quantified by subtracting the natural logarithm of the HR in females from the natural logarithm of the HR in males (log(HR)males − log(HR)females).
We tested all protein–disease association reaching significance (P < 0.05/5,836) in at least one sex for protein-by-sex interactions. These analyses were performed by fitting an interaction term (sex × circulating protein levels) in Cox proportional hazards models adjusted for sex, age, age², self-reported race/ethnicity, the first ten principal components of genetic ancestry, smoking, normalized Townsend deprivation index, BMI, systolic blood pressure, antihypertensive medication use, total cholesterol, HDL cholesterol, cholesterol-lowering medication use, serum creatinine, prevalent type 2 diabetes and circulating levels of the tested protein.
Construction of protein-based prediction models
We constructed protein-based risk scores to predict incident cardiac events in the UKB-PPP. We created three risk scores for each cardiac outcome using logistic LASSO regression, including (1) a score based on clinical risk factors; (2) a score based on circulating proteins; and (3) a combined score (that is, using clinical risk factors and circulating proteins). The clinically evaluable variables used as covariates in primary analyses (age, sex, self-reported race/ethnicity, smoking, BMI, systolic blood pressure, antihypertensive medication use, total cholesterol, HDL cholesterol, cholesterol-lowering medication use, serum creatinine and type 2 diabetes) were fed into LASSO models for the clinical risk scores. Circulating levels of the 1,459 proteins tested in primary analyses were fed into LASSO models for the protein-based risk scores.
The study cohort was randomly divided into a training (80%; n = 35,450) and testing (20%; n = 8,863) set. All clinical, proteomic and combined prediction scores were constructed using LASSO regression for variable selection and regularization. In brief, LASSO is a regularized regression method that selects informative variables (for example, proteins or clinical risk factors) from high-dimensional and correlated datasets while shrinking the regression coefficients of less informative variables to zero. We used tenfold cross-validation to tune the regularization parameter (λ; the parameter that controls the strength of shrinkage and variable selection) for each LASSO model. During the cross-validation procedure, multiple LASSO models are iteratively constructed for each set of predictors (clinical risk factors, proteins or both) using different values for λ, with each λ corresponding to a certain number of variables included in the prediction model. A higher λ value corresponds to fewer predictive variables in the regression model. The accuracy of each LASSO model (with its respective λ value) was quantified using the ROC AUC.
We used the ‘one standard error rule’ to determine the optimal λ for all proteomic and combined prediction models. This approach reduces the complexity of prediction models that are derived from high-dimensional datasets by selecting the largest λ (which corresponds to the smallest number of predictive covariates) for which the AUC is within one standard error of the highest AUC value during the cross-validation process. For models based solely on clinical risk factors, the λ corresponding to the highest AUC was used, considering that these risk scores were derived from a specific set of risk factors rather than a high-dimensional dataset.
Evaluation of protein-based prediction models
The performance of each prediction model was evaluated internally in the testing set of the UKB-PPP cohort (see above) and externally in the WHI-LLS dataset (see below). in the testing set by ROC curve analysis, and the DeLong test was used to evaluate differences between AUCs. We also calculated the detection rates for each model at false positive rates of 0.01, 0.05 and 0.1. ‘Exact’ detection rates were calculated as the proportion of affected individuals with positive test results (the number of true positives divided by the number of true positives plus false negatives). ‘Approximated’ detection rates were calculated using equation (2):
| 2 |
where DR is the detection rate; Φ the cumulative distribution function of the normal distribution with 0 as mean and (σcases/σcontrols) as s.d.; Φ−1 the inverse cumulative distribution function of the standard normal distribution; μcases the mean of the cases; μcontrols the mean of the controls; σcases the s.d. of the cases; σcontrols the s.d. of the controls; and FPR the false positive rate.
In addition, we constructed Kaplan–Meier plots to visualize the cumulative incidence of each outcome during follow-up according to proteomic risk score quintiles. We also tested the multivariable-adjusted association of each risk score (as a continuous variable) with their corresponding outcome using multivariable-adjusted Cox regression models adjusted for age, age², self-reported race/ethnicity, the first ten principal components of genetic ancestry, smoking, normalized Townsend deprivation index, BMI, systolic blood pressure, antihypertensive medication use, total cholesterol, HDL cholesterol, cholesterol-lowering medication use, serum creatinine and prevalent type 2 diabetes. Coronary artery disease, heart failure, atrial fibrillation and aortic stenosis were included as time-varying covariates. We used the glmnet61 and pROC62 packages in R to construct and test all risk scores.
External validation of protein-based prediction models
External validation analyses were performed in the WHI63—a prospective study of women recruited at 40 centers across the United States from 1993 to 1998—for coronary artery disease, heart failure and atrial fibrillation (all outcomes for which proteomic data significantly improved prediction in the UKB-PPP). A subset of WHI participants were invited for the LLS, which consisted of a one-time in-person study visit (between March 2012 and May 2013) including a blood draw, clinical evaluation and assessment of functional status. A total of 1,333 WHI-LLS participants underwent proteomic profiling. After excluding participants with missing values for >10% of measured proteins, missing data on time between enrollment and time of blood donation, or a history of heart disease, we included data from 1,083 WHI-LLS participants (Extended Data Fig. 7 and Supplementary Table 19).
Proteomic profiling was performed using six Olink Target 96 platforms (the Cardiometabolic, Cardiovascular II, Cardiovascular III, Inflammation, Neurology and Oncology III panels), measuring a total of 552 protein analytes representing 518 unique proteins that were also measured the UKB-PPP (Supplementary Table 20). Because only a subset of proteins was measured in both the WHI-LLS and the UKB-PPP, all proteomic and combined models were retrained in the UKB-PPP using only the subset of proteins that was measured in both the WHI-LLS and the UKB-PPP.
WHI-LLS participants underwent follow-up for coronary artery disease (defined as a composite of fatal and nonfatal myocardial infarction using a standardized adjudication process64), heart failure (defined as probable or definite congestive heart failure hospitalization using a standardized adjudication process65) and atrial fibrillation (defined as a composite of hospitalized and outpatient atrial fibrillation using self-report). WHI-LLS participants underwent follow-up through February 2022, resulting in a median (IQR) follow-up duration of 8.4 (6.1–8.9) years after blood draw. The performance of each prediction model was evaluated by ROC curve analysis.
Statistical analyses
All tests were two-sided. Data analysis was performed using R (v.4.1.0; R Project for Statistical Computing) unless otherwise specified.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Supplementary Figs. 1 and 2.
Source data
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Acknowledgements
All UKB analyses were performed under application no. 7089. This work was supported by funding from the Belgian American Educational Foundation (A.S.), the US National Heart, Lung and Blood Institute (K08HL166687, M.C.H.; R01HL127564, P.N.), the US National Human Genome Research Institute (R00HG012956, Z.Y.), the American Heart Association (940166 and 979465, M.C.H.), the Adolph M. Hutter MD Professorship (J.L.J.) and the Paul & Phyllis Fireman Endowed Chair in Vascular Medicine from the Massachusetts General Hospital (P.N.).
Extended data
Author contributions
A.S., A.B.P., M.C.H. and P.N. designed the study. A.S. and A.B.P. performed data analysis. A.S., A.B.P., J.L., R.B., S.G., N.D., A.M.S., Z.Y., W.H., A.P.R., J.L.L., M.C.H. and P.N. interpreted the study findings. C.K. and A.P.R. contributed data from the WHI. A.S., M.C.H. and P.N. drafted the paper. All authors reviewed the paper and provided critical feedback.
Peer review
Peer review information
Nature Cardiovascular Research thanks Aroon Hingorani and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
Data supporting the results of the present study are available from the UKB (https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access) to bona fide researchers with institutional review board and UKB approval. These analyses were performed using the UKB resource under application no. 7089. The secondary use of these data was approved by the Mass General Brigham institutional review board. Pathway enrichment analyses were performed using the Gene Ontology resource via Enrichr (https://maayanlab.cloud/Enrichr/). The UKB-PPP was used for genetic association data for circulating proteins (protein quantitative trait locus data) through Synapse (10.7303/syn51364943). FinnGen (freeze 9) was used for genetic association data for coronary artery disease (https://r9.finngen.fi/pheno/I9_CHD), heart failure (https://r9.finngen.fi/pheno/I9_HEARTFAIL), atrial fibrillation/flutter (https://r9.finngen.fi/pheno/I9_AF), and operated calcific aortic stenosis (https://r9.finngen.fi/pheno/I9_CAVS_OPERATED). The Human Protein Atlas was used for functional characterization of proteins (https://www.proteinatlas.org/). The WHI was used for external validation analyses for the clinical, protein-based and combined prediction models. Data from the WHI (https://www.whi.org/) can be accessed by researchers who meet the criteria for access to confidential data. Source data are provided with this paper.
Code availability
Code used for the main analyses of this study can be accessed at https://github.com/aschuerm/ukbppp_cardiac_diseases.
Inclusion and ethics
Inclusion and ethics standards have been reviewed where applicable.
Competing interests
J.L.J. reports board membership of Imbria Pharmaceuticals; grant support from Abbott Diagnostics, AstraZeneca, BMS, HeartFlow and Novartis; previous consulting income from Abbott Diagnostics, AstraZeneca, Bayer, Beckman Coulter, Jana Care, Janssen, Novartis, Quidel, Roche Diagnostics and Siemens; and clinical end point committee/data safety monitoring board membership for Abbott, Bayer, AbbVie, CVRx, Pfizer, Roche Diagnostics and Takeda. M.C.H. reports consulting fees from CRISPR Therapeutics and Comanche Biopharma; advisory board service for Miga Health; and grant support from Genentech. P.N. reports research grants from Allelica, Amgen, Apple, Boston Scientific, Genentech/Roche and Novartis; personal fees from Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Creative Education Concepts, CRISPR Therapeutics, Eli Lilly & Co, Esperion Therapeutics, Foresite Capital, Foresite Labs, Genentech / Roche, GV, HeartFlow, Magnet Biomedicine, Merck, Novartis, TenSixteen Bio and Tourmaline Bio; equity in Bolt, Candela, Mercury, MyOme, Parameter Health, Preciseli and TenSixteen Bio; and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Art Schuermans, Ashley B. Pournamdari.
These authors jointly supervised this work: Michael C. Honigberg, Pradeep Natarajan.
Contributor Information
Michael C. Honigberg, Email: mhonigberg@mgh.harvard.edu
Pradeep Natarajan, Email: pnatarajan@mgh.harvard.edu.
Extended data
is available for this paper at 10.1038/s44161-024-00567-0.
Supplementary information
The online version contains supplementary material available at 10.1038/s44161-024-00567-0.
References
- 1.GBD 2019 Diseases and Injuries Collaborators. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet396, 1204–1222 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaduganathan, M., Mensah, G. A., Turco, J. V., Fuster, V. & Roth, G. A. The Global Burden of Cardiovascular Diseases and Risk: a compass for future health. J. Am. Coll. Cardiol.80, 2361–2371 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Mortensen, M. B., Nordestgaard, B. G., Afzal, S. & Falk, E. ACC/AHA guidelines superior to ESC/EAS guidelines for primary prevention with statins in non-diabetic Europeans: the Copenhagen General Population Study. Eur. Heart J.38, 586–594 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nurmohamed, N. S. et al. Proteomics and lipidomics in atherosclerotic cardiovascular disease risk prediction. Eur. Heart J.44, 1594–1607 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fruchart, J. C. et al. The residual risk reduction initiative: a call to action to reduce residual vascular risk in patients with dyslipidemia. Am. J. Cardiol.102, 1K–34K (2008). [DOI] [PubMed] [Google Scholar]
- 6.Figtree, G. A. et al. Mortality in STEMI patients without standard modifiable risk factors: a sex-disaggregated analysis of SWEDEHEART registry data. Lancet397, 1085–1094 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Nurmohamed, N. S. et al. Targeted proteomics improves cardiovascular risk prediction in secondary prevention. Eur. Heart J.43, 1569–1577 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganz, P. et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA315, 2532–2541 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Williams, S. A. et al. Plasma protein patterns as comprehensive indicators of health. Nat. Med.25, 1851–1857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deo, R. et al. Proteomic cardiovascular risk assessment in chronic kidney disease. Eur. Heart J.44, 2095–2110 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams, S. A. et al. A proteomic surrogate for cardiovascular outcomes that is sensitive to multiple mechanisms of change in risk. Sci. Transl. Med.14, 9625 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Folkersen, L. et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat. Metab.2, 1135–1148 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henry, A. et al. Therapeutic targets for heart failure identified using proteomics and mendelian randomization. Circulation145, 1205–1217 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun, B. B. et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature622, 329–338 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeBleu, V. S. et al. Identification of human epididymis protein-4 as a fibroblast-derived mediator of fibrosis. Nat. Med.19, 227–231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carbon, S. et al. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res.49, D325–D334 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res.44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi, B. et al. Dipeptidyl peptidase-4 induces aortic valve calcifcation by inhibiting insulin-like growth factor-1 signaling in valvular interstitial cells. Circulation135, 1935–1950 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Uhlén, M. et al. Tissue-based map of the human proteome. Science347, 6220 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Schmidt, A. F. et al. Genetic drug target validation using Mendelian randomisation. Nat. Commun.11, 1–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swerdlow, D. I. et al. Selecting instruments for Mendelian randomization in the wake of genome-wide association studies. Int. J. Epidemiol.45, 1600–1616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finan, C. et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med.9, eaag1166 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabatine, M. S. PCSK9 inhibitors: clinical evidence and implementation. Nat. Rev. Cardiol.16, 155–165 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Lau, E. S. et al. Sex differences in circulating biomarkers of cardiovascular disease. J. Am. Coll. Cardiol.74, 1543–1553 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, H. N. & Januzzi, J. L. Natriuretic peptide testing in heart failure. Circulation123, 2015–2019 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Sun, B. B. et al. Genomic atlas of the human plasma proteome. Nature558, 73–79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho, A. & Südhof, T. C. Binding of F-spondin to amyloid-β precursor protein: a candidate amyloid-β precursor protein ligand that modulates amyloid-β precursor protein cleavage. Proc. Natl Acad. Sci. USA101, 2548–2553 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santema, B. T. et al. Pathophysiological pathways in patients with heart failure and atrial fibrillation. Cardiovasc. Res.118, 2478–2487 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Boer, R. A. et al. The WAP four-disulfide core domain protein HE4: a novel biomarker for heart failure. JACC Heart Fail.1, 164–169 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Galgano, M. T., Hampton, G. M. & Frierson, H. F. Comprehensive analysis of HE4 expression in normal and malignant human tissues. Mod. Pathol.19, 847–853 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Wollert, K. C., Kempf, T. & Wallentin, L. Growth differentiation factor 15 as a biomarker in cardiovascular disease. Clin. Chem.63, 140–151 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Kato, E. T. et al. Growth differentiation factor 15 and cardiovascular risk: individual patient meta-analysis. Eur. Heart J.44, 293–300 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kivimäki, M., Hingorani, A. D. & Lindbohm, J. V. Comment on ‘A proteomic surrogate for cardiovascular outcomes that is sensitive to multiple mechanisms of change in risk’. Sci. Transl. Med.14, 4810 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Williams, S. A. & Ganz, P. Response to comment on “A proteomic surrogate for cardiovascular outcomes that is sensitive to multiple mechanisms of change in risk”. Sci. Transl. Med.14, 1355 (2022). [DOI] [PubMed] [Google Scholar]
- 35.Bozkurt, B. et al. Universal definition and classification of heart failure: a report of the Heart Failure Society of America, Heart Failure Association of the European Society of Cardiology, Japanese Heart Failure Society and Writing Committee of the Universal Definition of Heart Failure. J. Card. Fail.27, 387–413 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Patton, K. K. et al. N-terminal pro-B-type natriuretic peptide is a major predictor of the development of atrial fibrillation. Circulation120, 1768–1774 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xing, L. Y. et al. Effects of atrial fibrillation screening according to N-terminal pro-B-Type natriuretic peptide: a secondary analysis of the randomized LOOP study. Circulation147, 1788–1797 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cole, S. et al. Integrative analysis reveals CD38 as a therapeutic target for plasma cell-rich pre-disease and established rheumatoid arthritis and systemic lupus erythematosus. Arthritis Res. Ther.20, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostendorf, L. et al. Targeting CD38 with daratumumab in refractory systemic lupus erythematosus. N. Engl. J. Med.383, 1149–1155 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Tilly, M. J. et al. Autoimmune diseases and new-onset atrial fibrillation: a UK Biobank study. EP Europace25, 804–811 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simard, L. et al. Sex-related discordance between aortic valve calcification and hemodynamic severity of aortic stenosis. Circ. Res.120, 681–691 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gadd, D. et al. Blood protein assessment of leading incident diseases and mortality in the UK Biobank. Nat. Aging4, 939–948 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong, W. et al. Next generation plasma proteome profiling to monitor health and disease. Nat. Commun.12, 1–12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hastie, T., Tibshirani, R., Narasimhan, B. & Chu, G. impute: Imputation for microarray data. Bioconductorhttps://www.bioconductor.org/packages/devel/bioc/manuals/impute/man/impute.pdf (2023).
- 46.Jordan, H., Roderick, P. & Martin, D. The Index of Multiple Deprivation 2000 and accessibility effects on health. J. Epidemiol. Community Health58, 250–257 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pirruccello, J. P. et al. Genetic analysis of right heart structure and function in 40,000 people. Nat. Genet.54, 792–803 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Honigberg, M. C. et al. Association of premature natural and surgical menopause with incident cardiovascular disease. JAMA322, 2411–2421 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Therneau, T. M., Lumley, T., Atkinson, E. & Crowson, C. Package ‘survival’. CRANhttps://cran.r-project.org/web/packages/survival/survival.pdf (2023).
- 50.Wickham, H. Ggplot2: Elegant Graphics for Data Analysis (Springer, 2009); 10.1007/978-0-387-98141-3
- 51.Burgess, S., Davies, N. M. & Thompson, S. G. Bias due to participant overlap in two-sample Mendelian randomization. Genet. Epidemiol.40, 597–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet.81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ardissino, M. et al. Birth weight influences cardiac structure, function, and disease risk: evidence of a causal association. Eur. Heart J.10.1093/eurheartj/ehad631 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burgess, S., Dudbridge, F. & Thompson, S. G. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat. Med.35, 1880–1906 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burgess, S., Zuber, V., Valdes-Marquez, E., Sun, B. B. & Hopewell, J. C. Mendelian randomization with fine‐mapped genetic data: choosing from large numbers of correlated instrumental variables. Genet. Epidemiol.41, 714 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gkatzionis, A., Burgess, S. & Newcombe, P. J. Statistical methods for cis-Mendelian randomization with two-sample summary-level data. Genet. Epidemiol.47, 3–25 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hemani, G. et al. The MR-base platform supports systematic causal inference across the human phenome. eLife7, e34408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yavorska, O. O. & Burgess, S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 46, 1734–1739 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet.10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw.33, 1–22 (2010). [PMC free article] [PubMed] [Google Scholar]
- 62.Robin, X. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform.12, 1–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson, G. et al. Design of the Women’s Health Initiative Clinical Trial and Observational Study. Control. Clin. Trials19, 61–109 (1998). [DOI] [PubMed] [Google Scholar]
- 64.Curb, J. D. et al. Outcomes ascertainment and adjudication methods in the women’s health initiative. Ann. Epidemiol.13, S122–S128 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Hall, P. S. et al. Reproductive factors and incidence of heart failure hospitalization in the Women’s Health Initiative. J. Am. Coll. Cardiol.69, 2517–2526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs. 1 and 2.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Statistical source data.
Data Availability Statement
Data supporting the results of the present study are available from the UKB (https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access) to bona fide researchers with institutional review board and UKB approval. These analyses were performed using the UKB resource under application no. 7089. The secondary use of these data was approved by the Mass General Brigham institutional review board. Pathway enrichment analyses were performed using the Gene Ontology resource via Enrichr (https://maayanlab.cloud/Enrichr/). The UKB-PPP was used for genetic association data for circulating proteins (protein quantitative trait locus data) through Synapse (10.7303/syn51364943). FinnGen (freeze 9) was used for genetic association data for coronary artery disease (https://r9.finngen.fi/pheno/I9_CHD), heart failure (https://r9.finngen.fi/pheno/I9_HEARTFAIL), atrial fibrillation/flutter (https://r9.finngen.fi/pheno/I9_AF), and operated calcific aortic stenosis (https://r9.finngen.fi/pheno/I9_CAVS_OPERATED). The Human Protein Atlas was used for functional characterization of proteins (https://www.proteinatlas.org/). The WHI was used for external validation analyses for the clinical, protein-based and combined prediction models. Data from the WHI (https://www.whi.org/) can be accessed by researchers who meet the criteria for access to confidential data. Source data are provided with this paper.
Code used for the main analyses of this study can be accessed at https://github.com/aschuerm/ukbppp_cardiac_diseases.