Abstract
Loeys–Dietz syndrome (LDS) is a connective tissue disorder caused by mutations that decrease transforming growth factor-β signaling. LDS-causing mutations increase the risk of aneurysm throughout the arterial tree, yet the aortic root is a site of heightened susceptibility. Here we investigate the heterogeneity of vascular smooth muscle cells (VSMCs) in the aorta of Tgfbr1M318R/+ LDS mice by single-cell transcriptomics to identify molecular determinants of this vulnerability. Reduced expression of components of the extracellular matrix–receptor apparatus and upregulation of stress and inflammatory pathways were observed in all LDS VSMCs. However, regardless of genotype, a subset of Gata4-expressing VSMCs predominantly located in the aortic root intrinsically displayed a less differentiated, proinflammatory profile. A similar population was also identified among aortic VSMCs in a human single-cell RNA sequencing dataset. Postnatal VSMC-specific Gata4 deletion reduced aortic root dilation in LDS mice, suggesting that this factor sensitizes the aortic root to the effects of impaired transforming growth factor-β signaling.
Subject terms: Genetics, Transcriptomics
Bramel et al. identify a population of GATA4+ vascular smooth muscle cells enriched in the human and mouse aortic root that is intrinsically more susceptible to Loeys–Dietz-syndrome-causing mutations and demonstrate that postnatal deletion of Gata4 in vascular smooth muscle cells reduces aortic root dilation in a mouse model of Loeys–Dietz syndrome.
Main
Thoracic aortic aneurysms are localized vascular dilations that increase the risk of fatal dissections and/or rupture of the vessel wall1. Effective medical therapies to prevent life-threatening aortic events remain elusive1. Loeys–Dietz syndrome (LDS) is a hereditary connective tissue disorder that presents with highly penetrant aortic aneurysms2. LDS is caused by heterozygous, loss-of-function mutations in genes coding for positive effectors of the transforming growth factor-β (TGF-β) signaling pathway, including receptors, ligands and intracellular signaling mediators3–7. All of these mutations result in reduced phosphorylation/activation of Smad2 and Smad3, leading to defective Smad-dependent transcriptional regulation. Secondary compensatory mechanisms, including upregulation of angiotensin II type I receptor (AT1R) signaling and increased expression of TGF-β ligands and Smad proteins, ultimately elevate levels of Smad2/Smad3 activity at diseased aortic sites, with outcomes ranging from adaptive to maladaptive, depending on the disease progression and cellular context3,5,6,8–10. While LDS-causing mutations heighten aneurysm risk in all arteries, the aortic root is especially vulnerable to disease11–13. Several laboratories have highlighted how the cellular composition and/or the mechanical stresses may contribute to the increased risk of disease in this location; however, the molecular determinants of this susceptibility remain unclear10,14,15. Vascular smooth muscle cells (VSMCs) are the primary cellular component of the aortic wall, but the heterogeneity of VSMCs within the aorta and its implications for aneurysm are not fully understood. In this study, we investigate the transcriptional heterogeneity of VSMCs in the normal and diseased murine aorta leveraging both single-cell RNA sequencing (scRNA-seq) and spatial analysis via high throughput in situ hybridization. We identify Gata4 as a regional factor whose expression is intrinsically elevated in the aortic root and further upregulated in LDS samples. We also show that postnatal deletion of Gata4 in VSMCs ameliorates aortic root dilation in a murine model of LDS harboring a Tgfbr1M318R/+ genotype.
Results
Perturbed transcriptional profiles in Tgfbr1M318R/+ VSMCs
LDS mouse models expressing a heterozygous missense mutation in Tgfbr1 (Tgfbr1M318R/+) develop highly penetrant aortic root dilation9,10. To assess transcriptomic changes associated with vascular pathology in this model, we performed scRNA-seq on the aortic root and ascending aorta of control (Tgfbr1+/+) and LDS mice at 16 weeks of age, a time when dilation is consistently observed but before a decrease in survival below 90% (refs. 9,10).
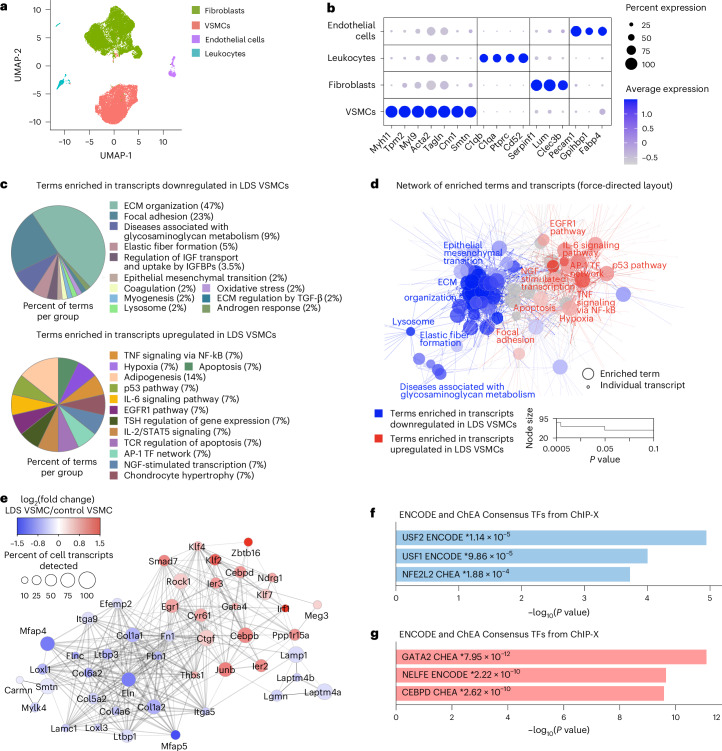
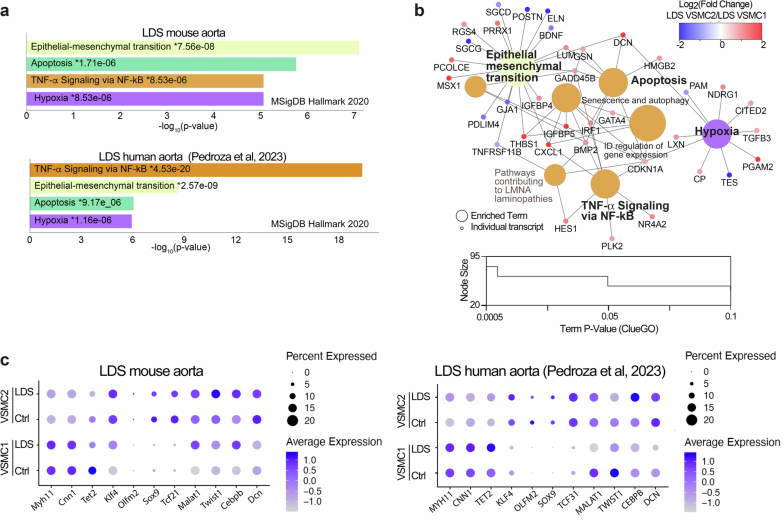
This analysis resulted in the identification of all the expected cell types according to well-established expression profiles16 (Fig. 1a,b and Extended Data Fig. 1). In consideration of the critical role of VSMCs in the pathogenesis of aortic aneurysm17, we focused the downstream analysis of LDS-driven transcriptional alterations on this cell type (Supplementary Table 1). Using the Cytoscape18 ClueGO19 plug-in to leverage gene set enrichment information from multiple databases, we produced a network of functionally related terms and pathways that are differentially enriched among downregulated and upregulated transcripts (Fig. 1c,d and Supplementary Table 2). The Tgfbr1M318R/+ LDS mutation caused broad downregulation of transcripts related to the maintenance of extracellular matrix (ECM)–receptor interactions and integrity of the elastic and contractile function of the aortic wall (Fig. 1c–e and Supplementary Table 2). Concurrently, the pathways involved in stress responses (that is, hypoxia and p53-dependent pathways), inflammation, senescence and cell death were enriched among transcripts upregulated in Tgfbr1M318R/+ VSMCs (Fig. 1c–e and Supplementary Table 2). We also noted the upregulation of mechanosensitive transcripts, such as Thbs1, Cyr61/CCN1 and Ctgf/CCN2(ref. 20) (Fig. 1e and Supplementary Table 1), suggesting that defective ECM–integrin receptor interactions contribute to induction of mechanical stress responses. Notably, as previously observed in other models of hereditary aortopathy21,22, loss of connections between VSMCs and elastic lamellae could be observed in LDS aortas as early as 6 weeks of age, before severe dilation9 (Extended Data Fig. 2).
Fig. 1. Downregulation of transcripts associated with ECM–receptor interactions and upregulation of stress and inflammation pathways in Tgfbr1M318R/+ LDS VSMCs.
a, A UMAP of aortic cells from control (Tgfbr1+/+) and LDS (Tgfbr1M318R/+) mice. b, A dot plot of cluster-defining transcripts used to identify endothelial cells, leukocytes, fibroblasts and VSMCs. The color of the dot represents a scaled average expression, while the size indicates the percentage of cells in which the transcript was detected. c, A ClueGO analysis of terms enriched among transcripts up- and downregulated in LDS VSMCs relative to the controls (IGF, insulin growth factor; IGFBP, IGF binding protein; TCR, T cell receptor; NGF, nerve growth factor; and TSH; thyroid stimulating hormone). d, A ClueGO network, in which each node represents a term/pathway or individual gene associated with that term. The terms enriched among transcripts downregulated in LDS VSMCs are highlighted in blue, while those enriched among transcripts upregulated in LDS VSMCs are highlighted in red. The size of the node indicates significance of the enrichment calculated by the ClueGO algorithm. e, A network of selected transcripts significantly dysregulated in LDS VSMCs. The size of the node is proportional to the percent of LDS cells in which the transcript was detected. The scale indicates the average log2 fold change in expression in LDS VSMCs relative to control VSMCs. The transcripts in blue are decreased in LDS VSMCs, while those in red are increased. f,g, EnrichR gene over-representation analysis for the ENCODE and ChEA Consensus transcription factors (TF) library showing the top three most significant terms associated with transcripts that are downregulated (f) or upregulated (g) in LDS VSMCs. The P values (*) refer to the significance of enrichment.
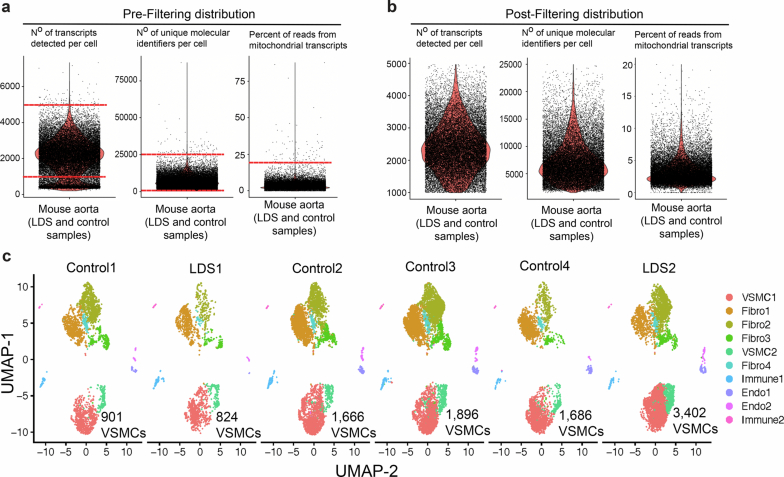
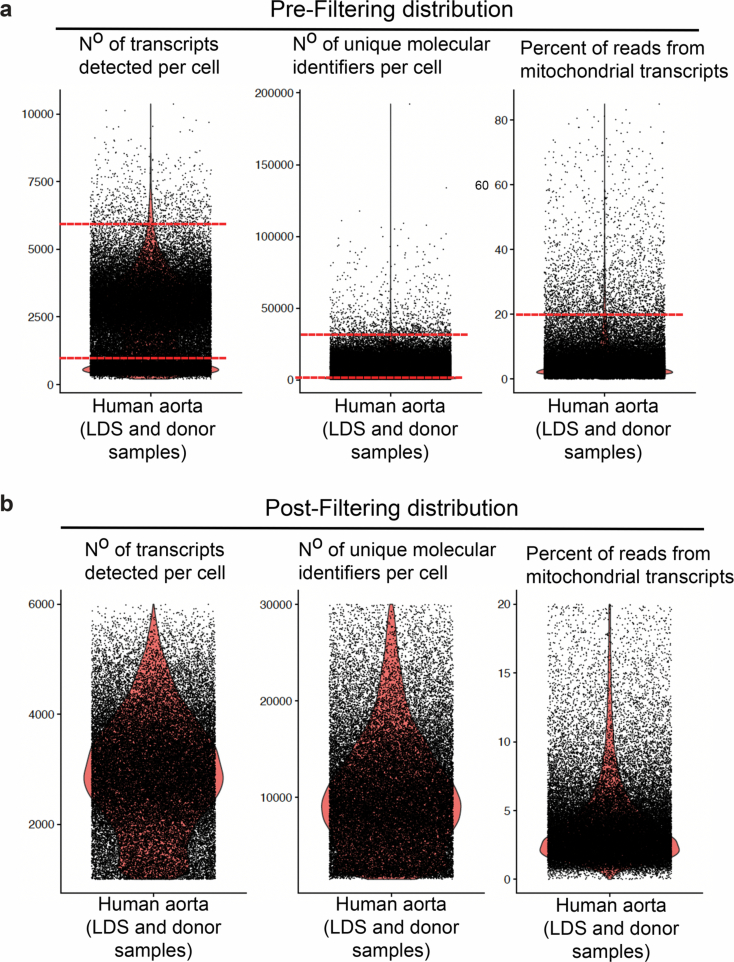
Extended Data Fig. 1. Quality control and filtering of scRNAseq of murine aorta.
ScRNAseq datasets from female control and Tgfbr1M318R/+ LDS mice were filtered by the number of transcripts per cell (nFeature), number of unique molecular identifiers per cell (nCounts), and the percent of reads from mitochondrial transcripts. Density plots showing the distribution of aortic cells from control and Tgfbr1M318R/+ LDS mice pre-filtering (a) and post-filtering (b) based on the following cutoffs which are indicated in the figure by horizontal bars: >1000 nFeature <5000, >1500 nCounts <25000, and <20% mitochondrial transcripts per cell. (c) UMAP of murine scRNAseq data split by mouse ID, showing the distribution of cells from each independent biological replicate (n = 6, 2 LDS and 4 control). Samples are labeled by genotype and the number of VSMCs per sample is indicated. A total of 4,226 LDS VSMCs and 6,149 control VSMCs were included in subsequent analyses.
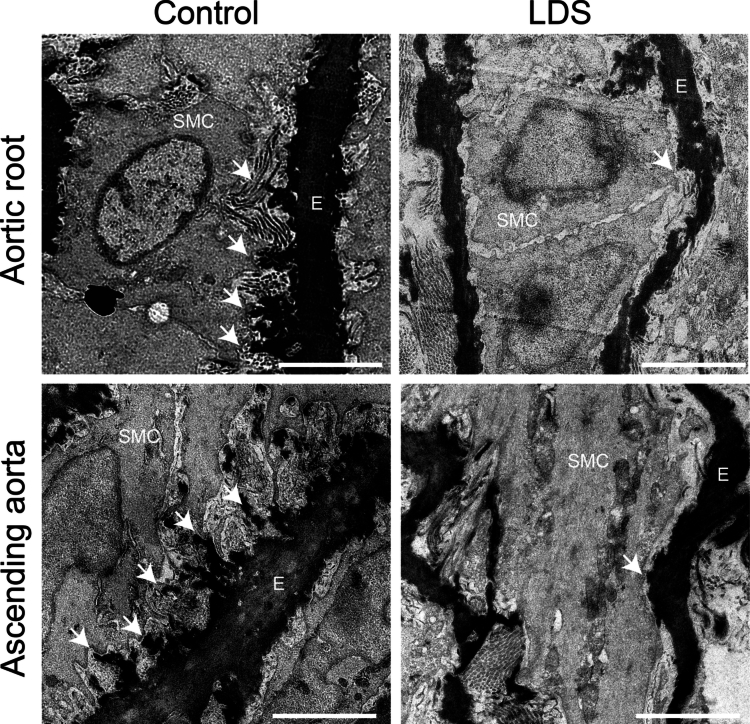
Extended Data Fig. 2. Defective connections between smooth muscle cells and elastic lamellae in the aorta of LDS mice.
Electron micrographs of longitudinal sections of the aortic root and ascending aorta of control and LDS mice at 6 weeks of age. Elastic fibers are stained dark (E, elastic lamellae; SMC, smooth muscle cell). Arrows indicate examples of dense plaques, which mark connections between elastic lamellae and smooth muscle cells. Image representative of three independent biological replicates. Scale bar is 2 microns.
Additional analysis of transcription factor target databases (ENCODE23 and Chromatin Immunoprecipitation Enrichment Analysis (ChEA) via EnrichR24,25) showed that the LDS-downregulated transcripts were enriched in targets of nuclear factor erythroid 2-related factor 2, also known as Nrf2, a transcription factor that activates expression of cytoprotective genes and suppresses expression of proinflammatory mediators26 (Fig. 1f and Supplementary Table 2). The targets of the upstream stimulatory factor family, which can modulate the expression of smooth muscle specific genes, were also enriched among downregulated transcripts27,28 (Fig. 1f and Supplementary Table 2). Conversely, the target genes for GATA transcription factors and CCAAT enhancer binding protein delta, also known as CEBPD, a positive transcriptional regulator of inflammatory responses mediated by interleukin-1 and interleukin-6 (refs. 29,30), were enriched among transcripts upregulated in LDS VSMCs (Fig. 1g and Supplementary Table 2).
Region- and disease-specific VSMC transcriptional patterns
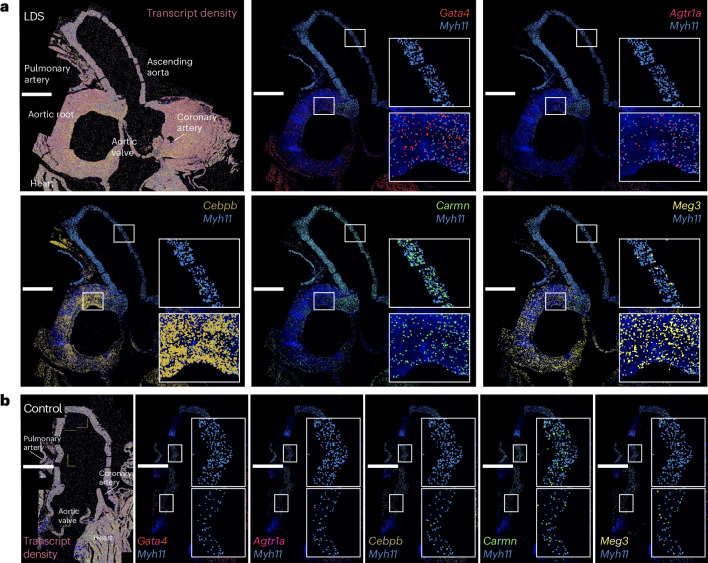
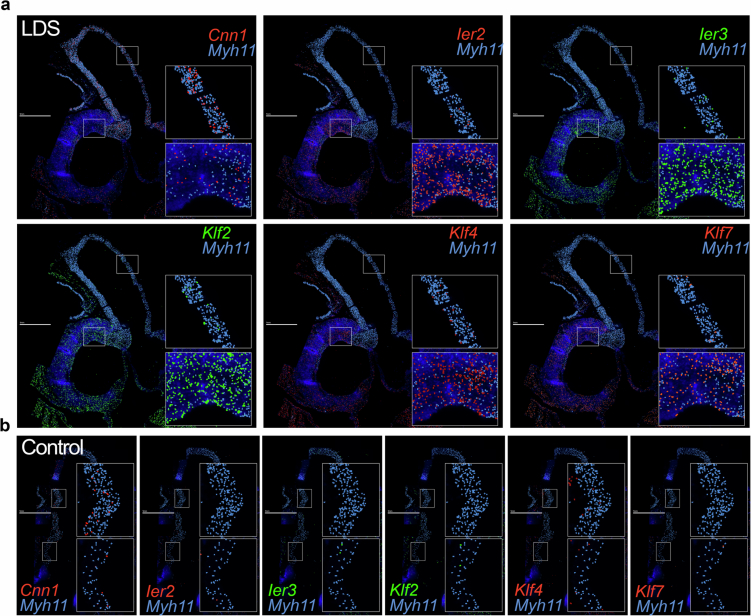
Given the regional vulnerability observed in LDS aortas, we leveraged insight gained from the literature and scRNA-seq analysis of the aorta of control and Tgfbr1M318R/+ mice to design a custom panel for high throughput in situ hybridization using the Multiplexed error-robust fluorescence in situ hybridization (MERFISH) spatial transcriptomics platform (Supplementary Table 3). The analysis of a longitudinal section of the proximal aorta of 16-week-old control and LDS mice showed regionally defined expression of several transcripts involved in the modulation of vascular phenotypes (Fig. 2 and Extended Data Fig. 3). The transcripts more highly detected in the aortic root of LDS mice relative to the ascending aorta included Agtr1a, which codes for angiotensin II receptor type 1a, a known contributor to LDS pathogenesis, and Gata4, which codes for a transcription factor known to positively regulate Agtr1a expression in the heart31,32. The transcripts coding for CCAAT enhancer binding protein beta (Cebpb), a proinflammatory mediator33, and maternally expressed gene 3 (Meg3), a long noncoding RNA that negatively regulates TGF-β signaling and promotes VSMC proliferation34, were also enriched in this region. In contrast, expression of cardiac mesoderm enhancer-associated noncoding RNA (Carmn), a positive regulator of VSMC contractile function that is downregulated in vascular disease, and expression of Myh11, coding for a marker of differentiated VSMCs, was enriched in the distal ascending aorta, a region that is only mildly affected in LDS mouse models35.
Fig. 2. MERFISH reveals spatially heterogeneous transcription profiles in LDS VSMCs.
a,b, MERFISH images of the proximal aorta of male LDS (a) and male control (b) mice at 16 weeks of age. Scale bars, 1 mm. The experiment was conducted once on one pair of age- and sex-matched samples. All the detected transcripts across the aortic tissue, with the key anatomic landmarks indicated, are displayed in a, top left. The depiction of the colocalization of Myh11 and the transcripts of interest are displayed in a, top middle, top right, bottom left, bottom middle and bottom right. The insets denote the regions of the ascending aorta and aortic root that are presented at higher magnification.
Extended Data Fig. 3. MERFISH reveals spatially heterogeneous expression of transcripts coding for Calponin-1, Immediate Early Response genes, and Kruppel-like Factor genes in LDS VSMCs.
MERFISH images of the proximal aorta of male LDS (a) and male control (b) mice at 16 weeks of age, scale bar is 1 mm. Experiment was conducted once on one pair of age and sex-matched samples. Scale bar is 1 mm. Panels depict the colocalization of Myh11 and transcripts of interest in the aorta of LDS (a) and control (b) mice. Insets denote regions of the ascending aorta and aortic root that are presented at higher magnification.
Spatial distribution of VSMC1/VSMC2-defining transcripts
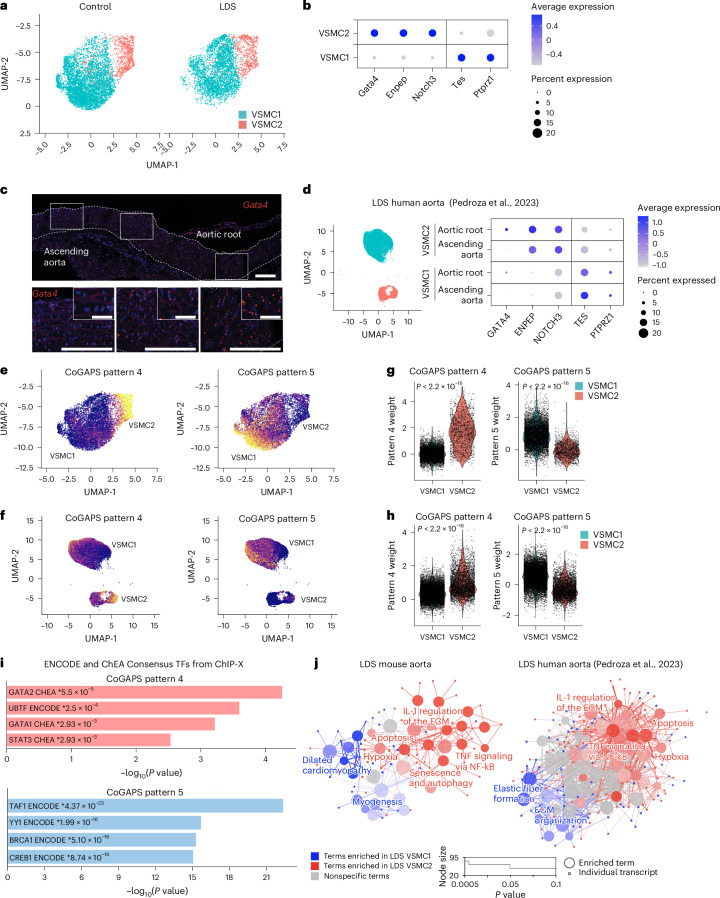
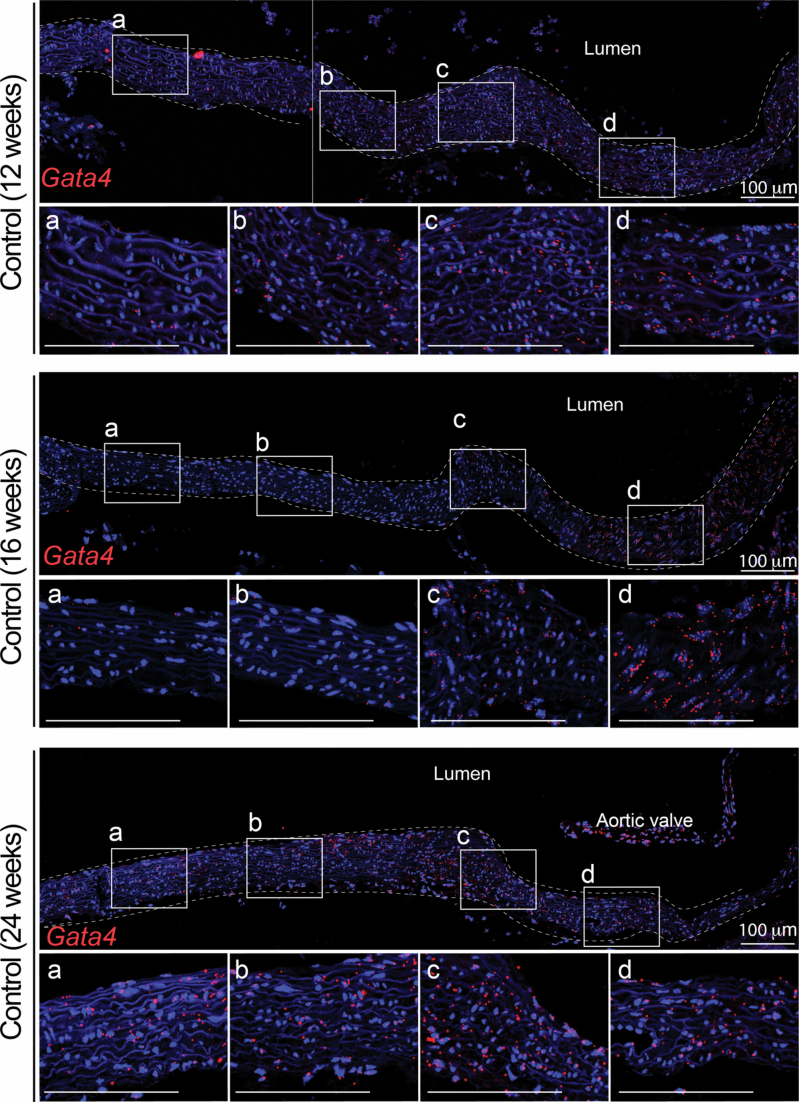
To examine if the spatial VSMC heterogeneity observed with MERFISH could be captured by scRNA-seq, we increased the clustering resolution for VSMCs, thus obtaining two subclusters, VSMC1 and VSMC2. We then examined the cluster-defining transcripts for these two VSMC subclusters (Supplementary Table 4), for transcripts our laboratory has previously shown to progressively increase (that is, Tes and Ptprz1) and decrease (that is, Enpep and Notch3) along the proximal-to-distal axis in the mouse ascending aorta22. Regardless of genotype, VSMC1 and VSMC2 showed differential expression of transcripts whose expression is intrinsically enriched in the ascending aorta and the aortic root, respectively22 (Fig. 3a,b and Supplementary Table 4). Gata4 was also noted among the transcripts that defined the VSMC2 subcluster and whose expression was highest in the aortic root, progressively diminishing along the proximal-to-distal axis in the ascending aorta, in both control and LDS aortas (Fig. 3c and Extended Data Figs. 4 and 5).
Fig. 3. Transcriptionally and spatially defined VSMC subclusters with distinct responses to LDS-causing mutations can be identified in both murine and human aortas.
a, A UMAP of VSMCs from control (Tgfbr1+/+) and LDS (Tgfbr1M318R/+) mice shown split by genotype. b, A dot plot showing enrichment of cluster-defining transcripts in VSMC1 and VSMC2. For a given transcript, the color of the dot represents a scaled average expression, while the size indicates the percentage of cells in which it was detected. c, RNA in situ hybridization showing the expression of GATA4 along the length of the murine aorta in a 16-week-old control animal, representative of five independent biological replicates. Bottom: the insets identify the location shown at a higher magnification. The dashed line identifies the approximate boundaries of the aortic wall. Scale bars, 100 mm. d, A UMAP of control and LDS VSMCs from human patients and a dot plot of cluster-defining markers in this dataset split by aortic region (Pedroza et al.38). e,f, A UMAP overlayed with weights for CoGAPS patterns 4 and 5, in mouse (e) and human (f) scRNA-seq datasets. g,h, Violin plots showing the distribution of pattern 4 and 5 weights in VSMC subclusters from mouse (g) and human (h) scRNA-seq datasets. The P values refer to Wilcoxon test. i, An EnrichR gene over-representation analysis for the ENCODE and ChEA Consensus transcription factor (TF) library showing the top four most significant terms associated with transcripts that define CoGAPS patterns 4 and 5. The P values (*) refer to the significance of enrichment. j, ClueGO network of terms differentially enriched in mouse and human LDS VSMC2 relative to VSMC1. The terms highlighted in blue are enriched in VSMC1, while those highlighted in red are enriched in VSMC2.
Extended Data Fig. 4. Expression of Gata4 transcript correlates with location along the proximal-to-distal axis in the mouse aorta.
RNA in situ hybridization showing the expression of Gata4 along the length of the murine aorta in 12-, 16- and 24-week-old control animals (three independent biological samples are shown; representative of two experiments showing similar results). Insets identify the location shown at higher magnification in the subsequent panel. The dashed line identifies the approximate boundaries of the aortic wall. Scale bar is 100 microns.
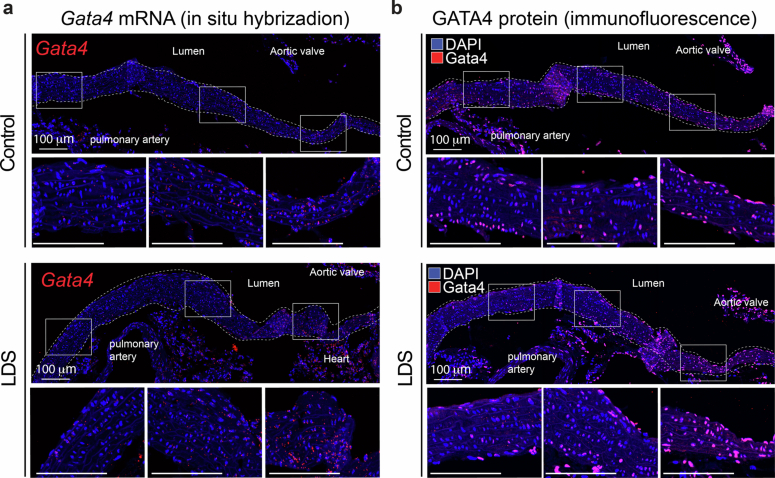
Extended Data Fig. 5. Gata4 mRNA and protein are upregulated in the aortic root of LDS mice.
(a) RNA in situ hybridization for Gata4 in the aortic root and ascending aorta of male control and LDS (Tgfbr1M318R/+) mice, representative of 5 independent biological replicates. Insets identify the location shown at higher magnification in the subsequent panel. Scale bar is 100 microns. (b) Immunofluorescence for Gata4 in the aortic root and ascending aorta of male control and LDS mice. Insets identify the location shown at higher magnification in the subsequent panel, representative of 4 independent biological replicates. Scale bar is 100 microns. The dashed line identifies the approximate boundaries of the aortic wall.
Considering previous work highlighting how cell lineage modulates the effect of LDS-causing mutations10,36, we explore the relationship between the VSMC2 and VSMC1 subclusters to the secondary heart field (SHF) and cardiac neural crest (CNC) lineage of origin (Extended Data Fig. 3). We found that VSMCs lineage-traced with a fluorescent reporter identifying CNC-derived cells were over represented in the VSMC1 subcluster (Extended Data Fig. 6a). However, reanalysis of a previously published dataset of SHF- and CNC-traced VSMCs (Supplementary Table 5) showed that VSMC1 and VSMC2 were not defined by lineage of origin, with VSMCs of both lineages found in either VSMC subcluster37 (Extended Data Fig. 6b). Nevertheless, as would be expected based on the known proximal-to-distal distribution of SHF- and CNC-derived VSMCs, there was overlap between VSMC2-defining and SHF-enriched transcripts (Extended Data Fig. 6b,c and Supplementary Tables 4 and 5). To assess if the VSMC substructure identified in murine models was relevant in the context of human aortic disease, we also reanalyzed a recently published scRNA-seq dataset of aortic tissue from patients with LDS and donor aortas in which the ascending aorta and aortic root were separately sequenced (Fig. 3d and Extended Data Fig. 7)38. The subpopulations of VSMCs expressing cluster-defining transcripts analogous to those found in VSMC1 and VSMC2 in mouse aortas could be identified in the human dataset (Fig. 3d and Supplementary Table 6). Although both VSMC1 and VSMC2 were present in human aortic root and ascending aorta, GATA4 expression was highest in the VSMC2 cluster from the aortic root, with no detectable expression in the ascending aorta (Fig. 3d).
Extended Data Fig. 6. VSMC1 and VSMC2 subclusters correlate but do not overlap with lineage-of-origin.
(a) A subset of cells included in the analysis derived from mice carrying the Rosa26-flox-STOP-flox-EGFP-L10a transgene and a Cre recombinase expressed under the control of a CNC-specific Wnt2 promoter (Wnt2-Cre). Dot plot shows detection of the Gfp reporter in VSMC1 and VSMC2. The color of the dot represents a scaled average expression while the size indicates the percentage of cells in which Gfp was detected. (b) UMAP of SHF-traced and non-SHF traced VSMCs and dot plot showing the SHF and non-SHF, and therefore CNC, enriched transcripts from Pedroza et al., 2022 in VSMC1 and VSMC2. (c) Dot plot of VSMC1 and VSMC2 cluster defining transcripts in SHF-traced and non-traced VSMCs from Pedroza et al., 2022.
Extended Data Fig. 7. Quality control and filtering of scRNAseq of human aorta.
Published scRNAseq dataset of donor and LDS patient aortic cells (Pedroza et al., 2023) was re-analyzed and filtered by the number of transcripts per cell (nFeature), number of unique molecular identifiers per cell (nCounts), and the percent of reads from mitochondrial transcripts. Density plots show the distribution of aortic cells from donor and LDS patients from pre-filtering (a) and post-filtering (b) based on the following cutoffs which are indicated in the figure by horizontal bars: >1000 nFeature < 6000, > nCounts < 30000, and < 20% mitochondrial transcripts per cell.
GATA4+ VSMC2 are poised toward a noncontractile phenotype
To examine the biological features of VSMC1 and VSMC2 and whether they were recapitulated in both murine and patient-derived LDS VSMCs, we used the Coordinated Gene Activity in Pattern Sets (CoGAPS) algorithm to identify latent patterns of coordinated gene expression in the Tgfbr1M318R/+ VSMC mouse dataset39. Two patterns, transcriptional patterns 4 and 5, were found to be enriched in the VSMC2 and VSMC1 subclusters, respectively, in the Tgfbr1M318R/+ VSMC mouse dataset (Fig. 3e,g and Supplementary Table 4). These same patterns were then projected onto the scRNA-seq data of VSMCs from the aorta of patients with LDS using ProjectR40, revealing a similar enrichment of pattern 4 in VSMC2 and pattern 5 in VSMC1 (Fig. 3e–h, Supplementary Table 4).
As previously observed for transcripts upregulated in Tgfbr1M318R/+ LDS VSMCs, pattern 4-associated transcripts were enriched for transcriptional targets of GATA family members (ENCODE23 and ChEA dataset, analyzed with EnrichR24,25,41,42; Fig. 3i). Differential gene set enrichment analysis using ClueGO19 to compare cluster-defining transcripts for VSMC1 and VSMC2 also showed that, in both mouse and human datasets, VSMC2-defining transcripts were enriched for pathways involved in inflammation, senescence and cellular stress (Fig. 3j and Supplementary Tables 7 and 8). In contrast, VSMC1 expressed higher levels of transcripts related to ECM–receptor interactions and contractile function (Fig. 3j, Extended Data Fig. 8 and Supplementary Tables 7 and 8). Network visualization of Molecular Signatures Database (MSigDB) VSMC2-enriched pathways shared by both mouse and human samples (probed with EnrichR25,41–44) (Extended Data Fig. 8a) and biological terms with shared ClueGO grouping (Fig. 3j and Supplementary Tables 7 and 8), highlighted the biological connections between these pathways and genes over-expressed in VSMC2 relative to VSMC1 (that is, Cxcl1(ref. 45), Irf1(ref. 46), Thbs1and Gata4(ref. 47)) (Extended Data Fig. 8b). Overall, in both mouse and human samples, the transcriptional profile of VSMC2 relative to VSMC1 resembled that of less-differentiated VSMCs and included lower expression of Myh11, Cnn1 and Tet2 and higher expression of transcripts associated with noncontractile VSMC phenotypes, including Klf4, Olfm2, Sox9, Tcf21, Malat1, Twist1 and Dcn48.
Extended Data Fig. 8. VSMC2-defining transcripts are enriched in modulators of inflammation and cellular stress in both mouse and human aortas.
(a) EnrichR bar plot of the top four significantly enriched pathways in the MSigDB Hallmark 2020 database in LDS VSMC2 relative to LDS VSMC1 in both mouse and human datasets. P-values refer to significance of enrichment. (b) Network visualization of MSigDB VSMC2-enriched pathways and biological terms with shared ClueGO grouping. The color of term nodes identifies ClueGO biological grouping. Individual genes are colored by the Log2 fold change of expression in LDS VSMC2 relative to LDS VSMC1 (blue genes are downregulated while red genes are upregulated in VSMC2 relative to VSMC1) (c) Dot plot of transcripts involved in modulation of VSMC contractile and non-contractile phenotypes in VSMC1 and VSMC2 in the mouse and human scRNAseq datasets. Color of the dot represents a scaled average expression, while the size indicates the percentage of cells in which the transcript was detected.
GATA4 is upregulated in the aortic root of LDS mice
Based on the analysis described above, including enrichment in GATA-dependent transcription in LDS upregulated transcripts (Fig. 1g) and its known role in driving the upregulation of propathogenic mediators of aneurysm progression, including Agtr1a/AT1R31, GATA4 emerged as a potential molecular determinant of increased risk of dilation of the aortic root in LDS.
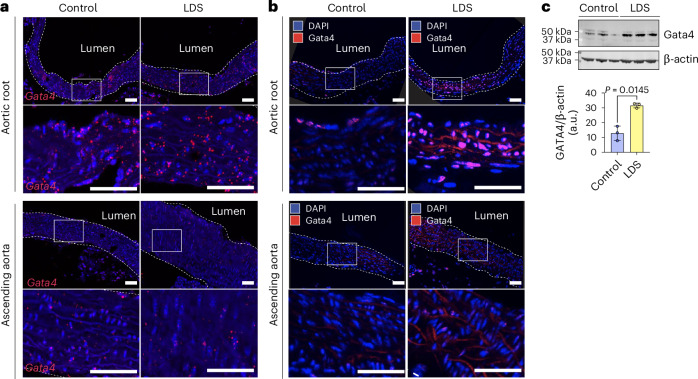
We speculated that the expression of Gata4 messenger RNA in a subset of VSMC (VSMC2) that are inherently enriched in this region may render these cells more vulnerable to the effects of LDS mutations (Fig. 3c and Extended Data Figs. 4 and 5). Expression of GATA4 mRNA was further upregulated in VSMCs in the LDS aorta, as assessed both by scRNA-seq (Supplementary Table 1) and RNA in situ hybridization (Fig. 4a and Extended Data Fig. 5). Given that expression of GATA4 protein is highly regulated at the post-transcriptional level through targeted degradation47,49, including via mechanosensitive pathways49, we also examined expression of GATA4 protein in control and LDS aortic samples and found that protein expression was increased in LDS aortic root, both by immunofluorescence and immunoblot assays (Fig. 4b,c, Extended Data Fig. 5b and Fig. 5).
Fig. 4. GATA4 mRNA and protein are upregulated in the aortic root of LDS mice.
a, An RNA in situ hybridization for Gata4 in the aortic root and ascending aorta of control and LDS (Tgfbr1M318R/+) male mice; the images are representative of five independent biological replicates. Bottom: the insets identify the location shown at higher magnification. Scale bars, 50 and 200 µm, respectively. b, Immunofluorescence for GATA4 in the aortic root and ascending aorta of control and LDS mice; the images are representative of five independent biological replicates. Bottom: the insets identify the location shown at higher magnification. Scale bars, 50 and 200 µm, respectively. The dashed line approximates the boundary of the aorta. c, An immunoblot for GATA4 expression relative to β-actin in aortic root lysates of control (n = 3 independent biological replicates), LDS mice (n = 3 independent biological replicates) and the related quantification of the immunoblot. The P value refers to a two-tailed Welch’s t-test (t = 6.32, d.f. = 2.43). Each symbol represents an independent biological replicate, and the bar graph shows the mean ± standard deviation.
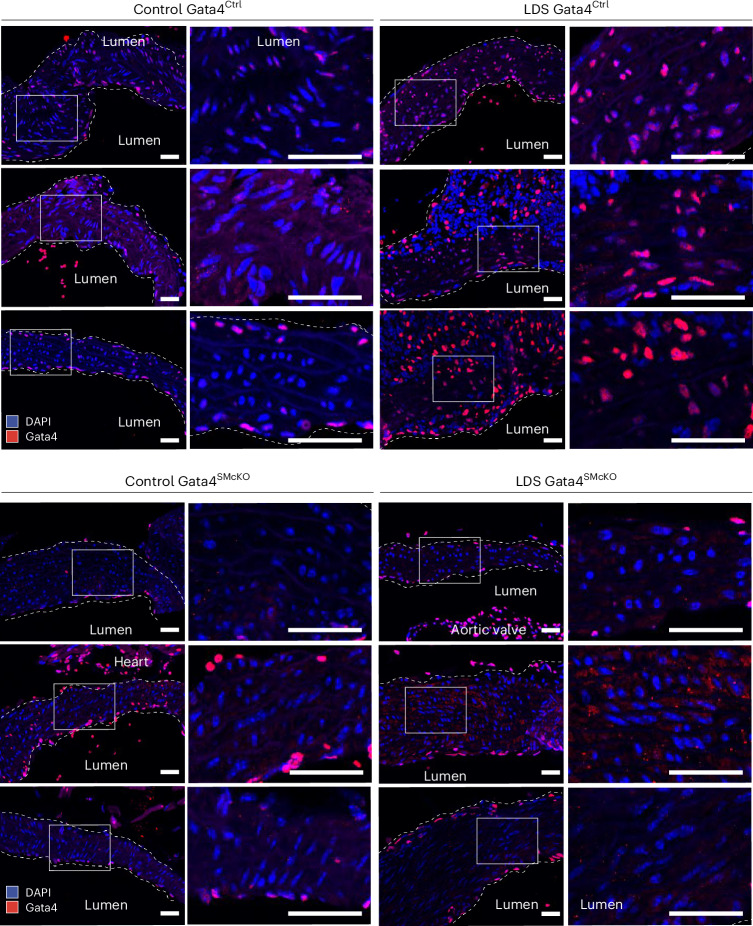
Fig. 5. GATA4 protein is upregulated in LDS aortic root of GATA4Ctrl and effectively ablated in GATA4SMcKO mice.
The immunofluorescence for GATA4 in the aorta of male mice of the indicated genotype at 16 weeks of age is shown. Three independent biological replicates are shown per genotype, abbreviated as follows: control (Tgfbr1+/+) and LDS (Tgfbr1M318R/+) with (GATA4SMcKO) or without (GATA4Ctrl) smooth muscle specific deletion of GATA4. The insets identify the location shown at a higher magnification. The images were acquired at 20× magnification. Scale bars, 50 and 200 µm, respectively. The dashed line approximates the boundary of the aorta.
Deletion of Gata4 in VSMC reduces aortic root dilation
To assess whether increased GATA4 levels in aortic root of LDS mouse models promoted dilation in this location, we crossed conditional Gata4fl°x/fl°x mice50 to LDS mice also expressing a transgenic, tamoxifen-inducible Cre recombinase under the control of a VSMC-specific promoter (Myh11-CreER)51. GATA4 deletion was induced starting at 6 weeks of age to bypass any potential role of this transcription factor during perinatal aortic development, during which rapid ECM remodeling occurs in conjunction with an increase in systolic pressure and cardiac output52 (Fig. 5). As Myh11-CreER is integrated on the Y chromosome, only male mice were used for this set of experiments.
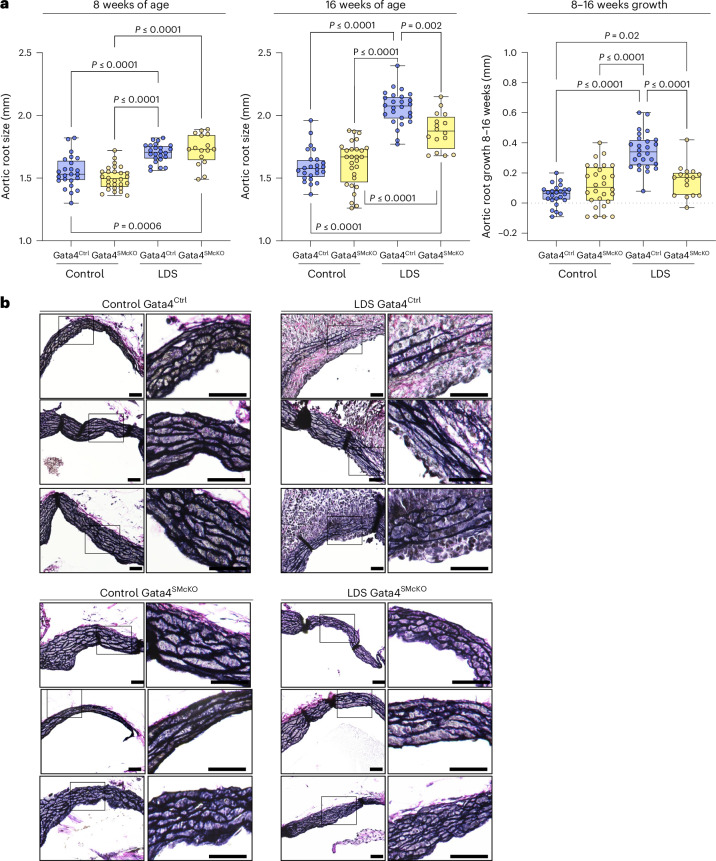
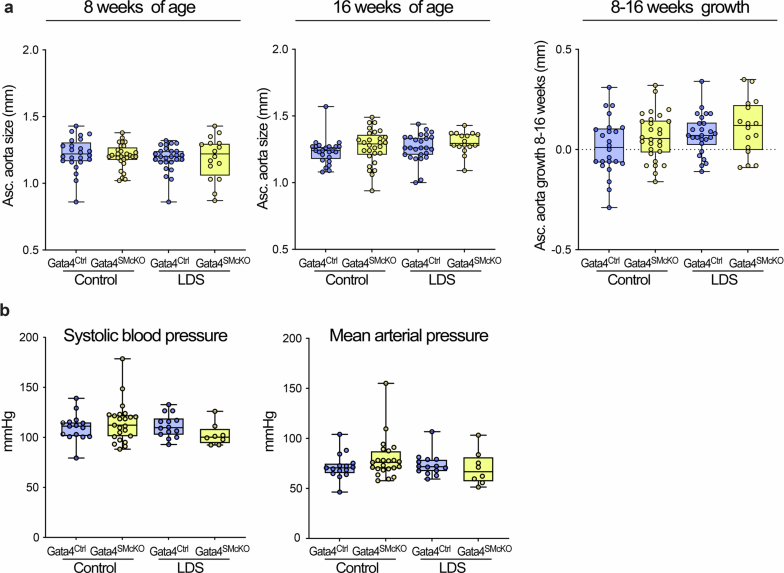
As expected, control LDS animals (Tgfbr1M318R/+; GATA4Ctrl) developed progressive focal dilation of the aortic root (albeit not reaching the 150% threshold of the normal diameter required for a definition of aneurysm) (Fig. 6a). This dilation was reduced in LDS mice with VSMC-specific postnatal deletion of GATA4 (Tgfbr1M318R/+; GATA4SMcKO) (Fig. 6a and Supplementary Table 9). The reduced aortic diameters also correlated with amelioration of aortic root medial architecture relative to control LDS aortas at 16 weeks of age (Fig. 6b). No notable dilation was observed in the ascending aorta of Tgfbr1M318R/+ mice at 16 weeks of age, and GATA4 deletion had no effect on the diameter of this aortic segment (Extended Data Fig. 9a). GATA4 deletion in VSMCs also did not associate with changes in blood pressure (Extended Data Fig. 9b).
Fig. 6. Smooth-muscle-specific deletion of GATA4 (GATA4SMcKO) reduces aortic root size and growth and improves aortic root media architecture in LDS mice.
a, Aortic root diameter of male control (Tgfbr1+/+) and LDS (Tgfbr1M318R/+) with (GATA4SMcKO) or without (GATA4Ctrl) smooth muscle specific deletion of GATA4, as measured by echocardiography at 8 and 16 weeks of age and aortic root growth from 8 to 16 weeks of age. The box plots show the upper quartile, median and lower quartile; each symbol represents an independent biological replicate. The whiskers identify the minimum to maximum range (control GATA4Ctrl n = 24; control GATA4SMcKO n = 28; LDS GATA4Ctrl n = 26; LDS GATA4SMcKO n = 16). The P values refer to Brown–Forsythe ANOVA (8 weeks: F* (F-statistic) = 25.98, DFn (degrees of freedom numerator) of 3, DFd (degrees of freedom denominator) of 66.69; 16 weeks: F* = 55.54, DFn of 3, DFd of 82.30; 8–16 weeks: F* = 33.39, DFn of 3, DFd of 78.63). b, VVG-stained aortic root sections from three independent biological replicates per genotype. The insets identify the area shown at a higher magnification. Scale bars, 50 and 200 µm, respectively.
Extended Data Fig. 9. Gata4 deletion does not affect size or growth of the ascending aorta or blood pressure in LDS mice.
(a) Ascending aortic diameter of male Ctrl (Tgfbr1+/+) and LDS (Tgfbr1M318R/+) mice with (Gata4SMcKO) or without (Gata4Ctrl) smooth muscle specific deletion of Gata4 as measured by echocardiography at 8 and 16 weeks of age, and aortic root growth from 8 to16 weeks of age. The box plots show the upper quartile, median and lower quartile; each symbol represents an independent biological replicate; whiskers identify the minimum to maximum range (Ctrl Gata4Ctrl n = 24; Ctrl Gata4SMcKO n = 28; LDS Gata4Ctrl n = 26; LDS Gata4SMcKO n = 16). No significant differences were detected (Brown-Forsythe ANOVA: 8 weeks F* = 0.31, DFn = 3, DFd = 52.88, P value = 0.8142; 16 weeks F* = 1.39, DFn = 3, DFd = 87.78, P value = 0.2491). (b) Systolic blood pressure and mean arterial pressure for male mice of the indicated genotypes. The box plots show the upper quartile, median and lower quartile; each symbol represents an independent biological replicate; whiskers identify the minimum to maximum range (Ctrl Gata4Ctrl n = 15; Ctrl Gata4SMcKO n = 23; LDS Gata4Ctrl n = 14; LDS Gata4SMcKO n = 8). No significant differences were detected (Brown-Forsythe ANOVA: Systolic pressure F* = 1.465, DFn= 3, DFd= 53.86, P value = 0.24; mean arterial F* = 1.055, DFn= 3, DFd= 38.74, P value = 0.379).
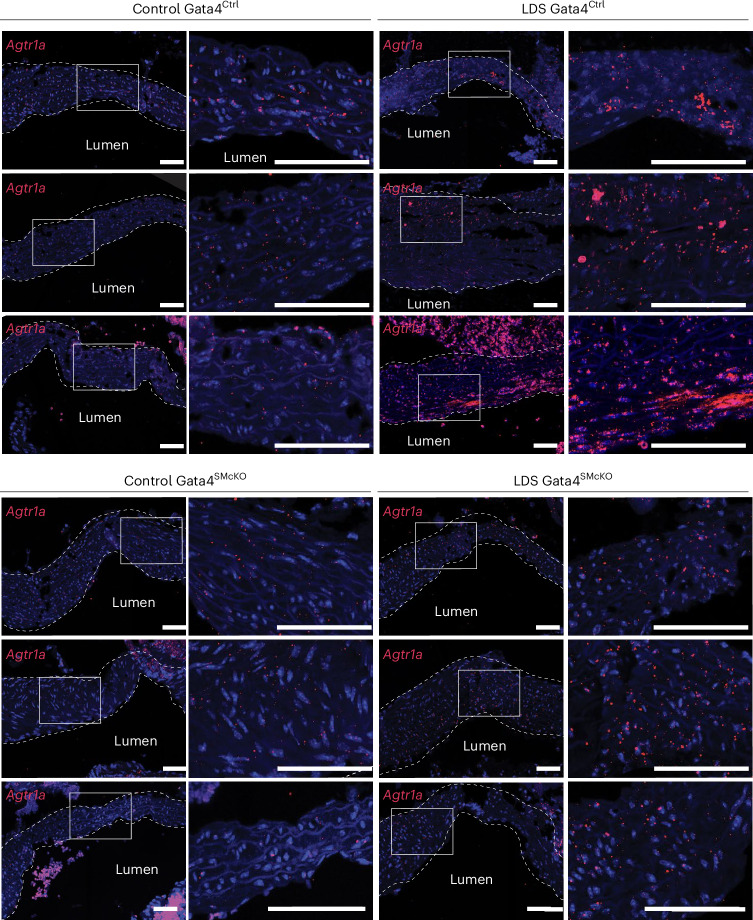
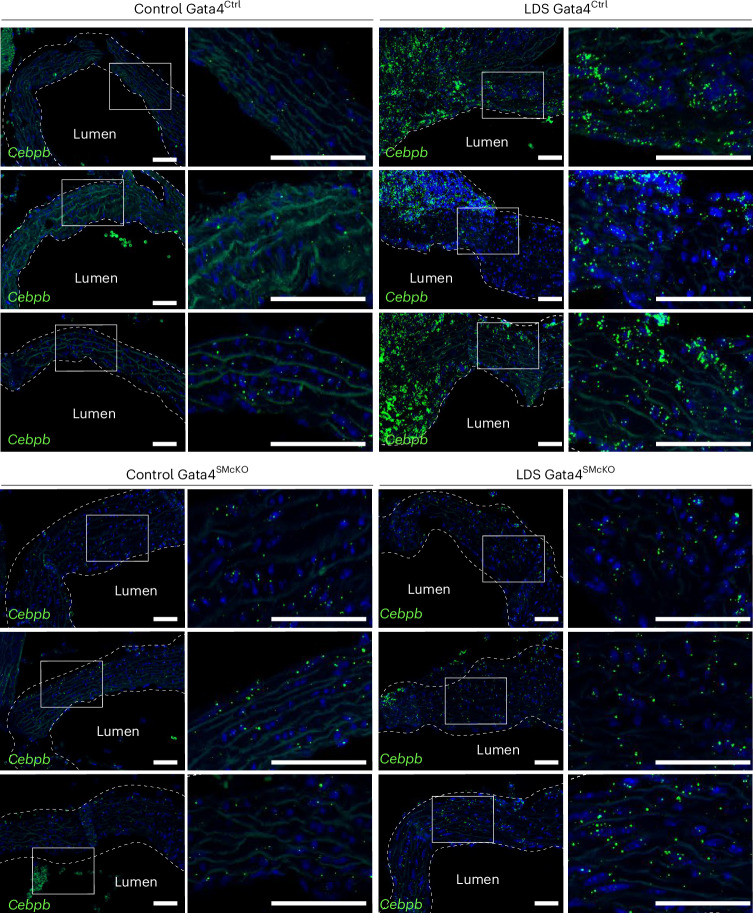
Previous work has shown that GATA4 binds to the Agtr1a promoter, inducing its expression in heart tissue31,32, and that Agtr1a is transcriptionally upregulated in the aortic root of LDS mice, resulting in upregulation of AT1R, which exacerbates LDS vascular pathology9,10,32. Accordingly, GATA4 deletion associated with reduced expression of Agtr1a in the aortic root of LDS mice (Fig. 7). Similarly, deletion of GATA4 reduced the expression of Cebpb and Cebpd (Fig. 8 and Extended Data Fig. 10), which code for proinflammatory transcription factors regulated by and/or interacting with GATA4 in other contexts30,33, which were upregulated in VSMCs in the presence of LDS mutations (Figs. 1 and 2 and Supplementary Table 1).
Fig. 7. Smooth muscle-specific deletion of GATA4 results in reduced expression of Agtr1a.
RNA in situ hybridization for Agtr1a in the aortic root of mice of the indicated genotype at 16 weeks of age. Three independent biological replicates are shown per genotype, abbreviated as follows: control (Tgfbr1+/+) and LDS (Tgfbr1M318R/+) with (GATA4SMcKO) or without (GATA4Ctrl) smooth-muscle-specific deletion of GATA4. The insets identify the location shown at a higher magnification. The images were acquired at 20× magnification. Scale bars, 50 and 200 µm, respectively. The dashed line approximates the boundary of the aorta.
Fig. 8. Smooth-muscle-specific deletion of GATA4 results in reduced expression of Cebpb.
RNA in situ hybridization for Cebpb in the aortic root of mice of the indicated genotype at 16 weeks of age. Three independent biological replicates are shown per genotype, abbreviated as follows: control (Tgfbr1+/+) and LDS (Tgfbr1M318R/+) with (GATA4SMcKO) or without (GATA4Ctrl) smooth-muscle-specific deletion of GATA4. The insets identify the location shown at a higher magnification. The images were acquired at 20× magnification. Scale bars, 50 and 200 µm, respectively. The dashed line approximates the boundary of the aorta.
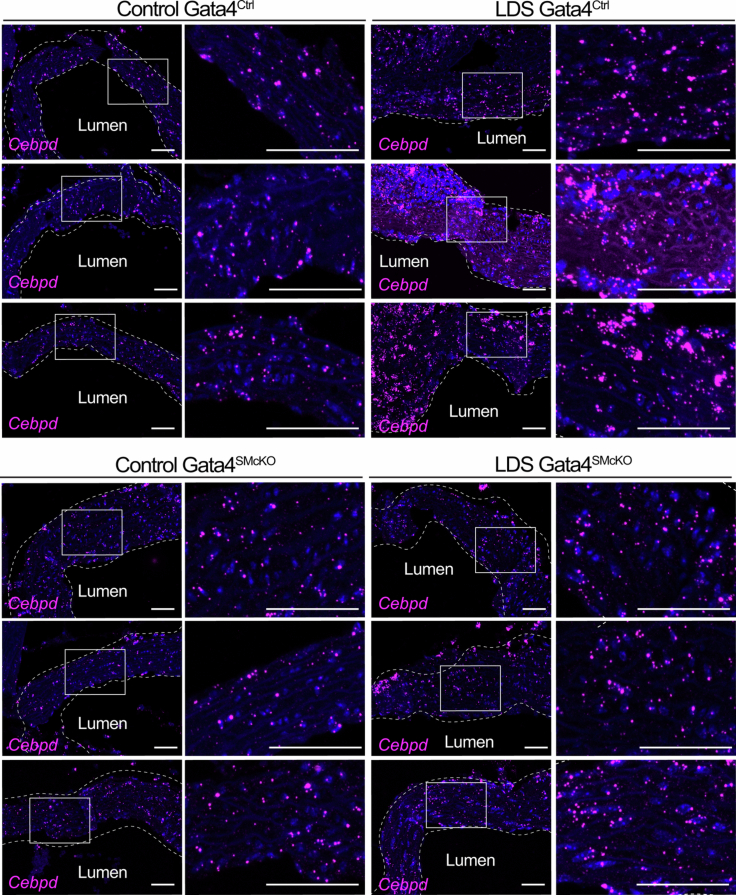
Extended Data Fig. 10. Smooth muscle-specific deletion of Gata4 results in reduced expression of Cebpd.
RNA in situ hybridization for Cebpd in the aortic root of male mice of the indicated genotype at 16 weeks of age. Three independent biological replicates are shown per genotype, abbreviated as follows Control (Tgfbr1+/+) and LDS (Tgfbr1M318R/+) with (Gata4SMcKO) or without (Gata4Ctrl) smooth muscle specific deletion of Gata4. Insets identify location shown at higher magnification in subsequent panels. Images were acquired at 20x magnification. Scale bars are 50 and 200 microns, respectively. The dashed line identifies the approximate boundaries of the aortic wall.
Discussion
LDS is a hereditary connective tissue disorder characterized by skeletal, craniofacial, cutaneous, immunological and vascular manifestations, including a high risk for aggressive arterial aneurysms2. It is caused by mutations that impair the signaling output of the TGF-β pathway, leading to defective transcriptional regulation of its target genes3–7. Although loss-of-signaling initiates vascular pathology, compensatory upregulation of positive modulators of the pathway results in a ‘paradoxical’ increase in activation of TGF-β-signaling mediators (that is, phosphorylated Smad2 and Smad3) and increased expression of target genes in diseased aortic tissue of both patients with LDS and mouse models3,5,6,8–10. This secondary upregulation depends, in part, on increased activation of angiotensin II signaling via AT1R, which positively modulates the expression of TGF-β ligands and TGF-β receptors53. Whereas upregulation of the TGF-β pathway can have both adaptive and maladaptive consequences depending on disease stage and cellular context53, upregulation of AT1R signaling has consistently been shown to be detrimental to vascular health, and both pharmacological (that is, with angiotensin receptor blockers) and genetic antagonism of this pathway ameliorates vascular pathology in LDS mouse models53,54.
Even though LDS-causing mutations confer an increased risk of disease across all arterial segments, the aortic root is one of the sites that is particularly susceptible to aneurysm development11–13. In this study, we leverage scRNA-seq in conjunction with spatial analysis via high throughput in situ hybridization to investigate the heterogeneity of VSMCs in an LDS mouse model, with the ultimate goal of identifying regional mediators that may drive upregulation of propathogenic signaling in this region. We identify distinct subpopulations of VSMCs characterized by expression patterns that preferentially map to the ascending aorta (VSMC1) and aortic root (VSMC2) in mouse aorta. We also show that the regional vulnerability of the aortic root depends, in part, on higher levels of Gata4 expression in a subset of VSMCs (VSMC2), which is intrinsically more vulnerable to the effect of an LDS-causing mutation.
Before the advent of single-cell analysis tools, which allow precise and unbiased unraveling of cellular identity, the ability to investigate VSMC heterogeneity in the proximal aorta was limited by the availability of experimental approaches to investigate known or expected diversity. In consideration of the mixed embryological origin of the aortic root and distal ascending aorta, earlier work, thus, focused on understanding how the effect of LDS mutations on VSMCs was modified by the SHF and CNC lineage of origin. In both mouse models and in induced pluripotent stem cell-derived in vitro models, signaling defects caused by LDS mutations were found to be more pronounced in VSMC derived from SHF (or cardiac mesoderm) progenitors relative to CNC-derived VSMCs10,36.
Similar to SHF-derived VSMCs, Gata4-expressing VSMC2 are enriched in the aortic root and are also more vulnerable to the effects of an LDS-causing mutation. They also express a transcriptional signature similar to that of SHF-derived VSMCs (Extended Data Fig. 3). Reciprocally, SHF-derived cells are over represented in the VSMC2 cluster in our dataset (Extended Data Fig. 3). However, the identity of VSMC2 and VSMC1 is not defined by lineage of origin, and the SHF- or CNC-derived origin is only an imperfect approximation of the VSMC heterogeneity that can now be assessed via scRNA-seq.
Heterogeneity beyond that imposed by lineage of origin was also shown by scRNA-seq analysis of the aorta of the Fbn1C1041G/+ Marfan syndrome mouse model, which revealed the existence of an aneurysm-specific population of transcriptionally modified smooth muscle cells at a later stage of aneurysmal disease and which could emerge from modulation of both SHF- and non-SHF (presumably CNC)-derived progenitors37,55. These cells, which could also be identified in the aneurysmal tissue derived from the aortic root of patients with Marfan syndrome, showed a transcriptional signature marked by a gradual upregulation of ECM genes and downregulation of VSMC contractile genes37,55. We were not able to identify this population of modified smooth muscle cells in the aorta of Tgfbr1M318R/+ LDS mouse models, even though it was shown to exist in the aorta of patients with LDS40.
Similar to the early effect of Smad3-inactivation, the Tgfbr1M318R/+ LDS mutation caused broad downregulation of gene programs required for ECM homeostasis and those favoring a differentiated VSMC phenotype22 (Fig. 1); conversely, proinflammatory transcriptional repertoires, with an enrichment in terms related to hypoxia, p53-dependent pathways and mechanical stress, were observed among upregulated transcripts. We speculate that this latter profile represents a response to loss of connections between VSMCs and elastic lamellae, which can be observed as early as 6 weeks of age (Extended Data Fig. 2) and is probably caused by decreased expression of ECM components whose expression requires TGF-β/Smad activity17.
We also noted downregulation of several components of the lysosome, whose function is required for cellular homeostasis and degradation of protein targets via selective autophagy56 (Fig. 1). GATA4 levels are regulated via p62-mediated selective autophagy47 and by mechanosensitive proteasome-mediated degradation49,57. The aortic root would be especially vulnerable to a defect in either of these processes given increased baseline levels of Gata4 mRNA expression in VSMC2. Increased levels of GATA4 may contribute to vascular pathogenesis by several potential mechanisms. In other cellular contexts, GATA4 has been shown to promote induction of the proinflammatory senescence-associated secretory phenotype, as well as transcription of the long noncoding RNA Malat1, which promotes aneurysm development in other mouse models58. GATA4 is also a negative regulator of contractile gene expression in Sertoli and Leydig cells59. In addition, GATA4 binds the promoter and activates the expression of Agtr1a31, which is known to drive propathogenic signaling in LDS aorta32. Accordingly, we find that GATA4 deletion downregulates expression of Agtr1a in the aortic media of LDS mouse models (Fig. 7).
Reanalysis of a scRNA-seq dataset of human aortic samples from patients with LDS, which included both the aortic root and the ascending aorta, shows that a population of Gata4-expressing VSMCs, similar to that found in mice, can also be identified in patients with LDS. In addition, patterns of coordinated gene expression identifying VSMC1 and VSMC2, which were learned from the scRNA-seq analysis of mouse aorta, could be projected onto the human dataset, suggesting that these two subsets of VSMCs are conserved across species and that the existence of a Gata4-expressing VSMC2 population may underlie increased risk in the aortic root of patients with LDS as well. Our study supports the notion that GATA4 is a factor sensitizing the aortic root to the effects of LDS mutations and that its inhibition early in the disease process reduces the rate of dilation. However, it is likely that, as for treatment with AT1R signaling inhibitors60, the beneficial effect of inhibiting this transcription factor will diminish at later stages of disease.
GATA4 is an important factor in aortic valve development and noncoding and missense variants in GATA4 have been implicated in patients with bicuspid aortic valve disease61,62. It also plays critical roles in regulation of numerous biological processes in nonvascular tissues63,64, making its direct targeting for therapeutic purposes unfeasible. However, this work highlights how the investigation of factors that increase or decrease the regional risk of aortic aneurysm may lead to a better understanding of adaptive and maladaptive pathways activated in response to a given aneurysm-causing mutation. This knowledge may be leveraged to develop therapeutic strategies that target the vulnerabilities of specific arterial segments.
Methods
All animal experiments were conducted according to protocols (approval number MO22M307) approved by the Johns Hopkins University School of Medicine Animal Care and Use Committee.
Animal experiments
All mice were maintained in an animal facility at 72 oF and 40–60% humidity. The mice were provided unlimited access to standard chow and water and a light–dark cycle of 10–14 h. Tgfbr1+/+ and Tgfbr1M318R/+(ref. 9) (The Jackson Laboratory, strain no. 036511) mice, some bearing the EGFP-L10a (The Jackson Laboratory, strain no. 024750) conditional tracer allele and a CNC-specific Cre recombinase expressed under the control of Wnt2 promoter (The Jackson Laboratory, strain no. 003829) were used for scRNA-seq as described below. All mice were maintained on a 129-background strain (Taconic, 129SVE). Tgfbr1+/+ and Tgfbr1M318R/+ mice were bred to Gata4flox/flox (The Jackson Laboratory, strain no. 008194) and mice carrying the Myh11-CreER transgene (The Jackson Laboratory, strain no. 019079). Myh11-CreER is integrated on the Y chromosome; therefore, only male mice were used for this set of experiments. Tgfbr1+/+ and Tgfbr1M318R/+ bearing Gata4flox/flox and Myh11-CreER are referred to as GATA4SMcKO. Tgfbr1+/+ and Tgfbr1M318R/+ bearing Gata4+/+ with or without Myh11-CreER or Gata4flox/flox or Gata4flox/+ without Myh11-CreER are referred to as GATA4Ctrl. All GATA4SMcKO and GATA4Ctrl mice were injected with 2 mg per day of tamoxifen (MilliporeSigma, T5648) starting at 6 weeks of age for five consecutive days. The mice were genotyped according to The Jackson Laboratory protocols for these models. Serial echocardiography was performed using the Visual Sonics Vivo 2100 machine and a 30 MHz probe. As there is some variability in the onset of aortic dilation in Tgfbr1M318R/+ mice and starting aortic size will affect final measurements, an aortic root diameter of 1.9 mm and above at baseline (8 weeks of age) was defined a priori as an exclusion criterion.
Molecular validation techniques
Aortic sample preparation
All mice were euthanized by halothane inhalation at a 4% concentration, 0.2 ml per liter of container volume (MilliporeSigma, H0150000). The heart and thoracic aorta were dissected en bloc and fixed in 4% paraformaldehyde (Electron Microscopy Sciences, 15710) in PBS at 4 oC overnight. The samples were subsequently incubated in 70% ethanol at 4 oC overnight before embedding in paraffin. Paraffin-embedded tissues were cut into 5 µm sections to expose a longitudinal section of the thoracic aorta. The sections were then stained with Verhoeff–van Gieson (StatLab, STVGI) to visualize elastic fiber morphology or to assess protein and RNA abundance by immunofluorescence or fluorescence in situ hybridization.
Immunofluorescence
Immunofluorescence was performed following a protocol adapted from Cell Signaling Technology (CST) for formaldehyde-fixed tissues. The paraffin-embedded sections were baked at 60 °C for 15 min, then deparaffinized in xylene followed by rehydration via a graded alcohol series: 100% ethanol, 95% ethanol, 70% ethanol and 1× PBS, each for 3 min. An antigen retrieval was performed in 10 mM sodium citrate buffer (0.05% Tween, pH 6.0) at 90 °C for 15 min. The slides were allowed to cool to room temperature before the sections were treated with 10 mg ml−1sodium borohydride in PBS (Sigma-Aldrich, 452882) for 20 min. The slides were permeabilized in Tris-buffered saline with Triton X-100 (TBT) (1× Tris-buffered saline (TBS) (Quality Biological, 351086101) with 0.1% Triton X-100 (Sigma-Aldrich, T9284) and 0.1 M glycine (Sigma-Aldrich, G8898)) for 20 min, followed by blocking with Fc Receptor Blocker (Innovex, NB309) and Background Buster (Innovex, NB306), each for 20 min at room temperature. The slides were then incubated with GATA4 antibody (CST, CST36966) diluted 1:100 in TBT overnight at 4 °C in a humid chamber. Following two washes with TBT, the slides were treated with donkey anti-rabbit Alexa Fluor 555 (ThermoFisher, A32794) at 1:100 for 1 h. The slides were washed again twice with TBT and once with TBS before mounting with Hard Set Mounting Media with 4,6-diamidino-2-phenylindole (DAPI) (VECTASHIELD, H-1500). The images were taken using a Zeiss LSM880 Airyscan FAST confocal microscope at 20× magnification and are presented as maximal intensity projection.
RNAscope fluorescence in situ hybridization
RNA in situ hybridization was performed using the RNAscope Multiplex Fluorescent Reagent Kit v2 Assay (ACD Biosciences, 323100) according to the manufacturer’s protocol with the following probes Mm-Gata4 (417881), Mm-Agtr1a (481161), Mm-Cebpd (556661) and Mm-Cebpb (547471). The images were taken using a Zeiss LSM880 Airyscan FAST confocal microscope at 20× magnification and are presented as maximal intensity projection.
Immunoblotting
Aortic root tissue was flash-frozen immediately upon dissection and stored at −80 oC until protein extraction. The protein was extracted using Full Moon Lysis Buffer (Full Moon Biosystems, EXB1000) with added phosphatase and protease inhibitors (MilliporeSigma, 11836170001 and 4906845001) and Full Moon lysis beads (Full Moon Biosystems, LB020) using an MP Biomedicals FastPrep 24 5G automatic bead homogenizer. After homogenization, the cell debris was pelleted, and the supernatant was collected. the samples were run on a 4–12% Bis–Tris Criterion XT gel (BioRad, 3450123) for 1.5 h at 100 V. The BioRad Turbo Blot standard 30 min protocol was used for transferring to a polyvinylidene difluoride membrane (BioRad, 1704273). The membrane was blocked for 45 min in diluted blocking buffer in PBS (LI-COR, 927-70003) for 1 h at room temperature. The primary antibodies for GATA4 (CST, CST36966) diluted 1:1,000 in blocking buffer and β-actin diluted 1:5,000 in blocking buffer (CST, CST3700) were incubated overnight at 4 °C. The membrane was washed with PBST (1× PBS + 0.05% Tween20) three times for 5 min each. The membrane was incubated with secondary antibodies (LI-COR 926-68072 and 926-32213) diluted 1:10,000 in blocking buffer. The membrane was washed three more times in PBST, then twice with 1× PBS and imaged on the LI-COR Odyssey machine.
Electron microscopy
The heart and thoracic aorta were prepared en bloc for electron microscopy. The samples were fixed in a solution of sodium cacodylate (0.1 M) and 4% paraformaldehyde (Electron Microscopy Sciences, 15710) overnight at 4 oC. They were then incubated in a 30% sucrose solution overnight at 4 oC. The samples were embedded in optimal cutting temperature compound then stored at −80 oC to freeze completely. The blocks were cut into 20 µm sections using a cryostat and mounted on slides. The clearest longitudinal cross sections of the thoracic aorta were selected and fixed with 2.5% glutaraldehyde solution, followed by a solution of 2% osmium in cacodylate buffer. These sections were then embedded in Epoxy resin (EPON), and the regions of interest—the aortic root and ascending aorta—were cut out. The sections of these regions, 60 nm, were then triple stained with tannic acid, uranyl acetate and lead citrate. The images were acquired by Johns Hopkins School of Medicine microscope facility staff on a Thermofisher Talos L120C G2 Electron microscope with a 16 megapixel CMOS camera at 120 Kv.
Transcriptomic analyses
scRNA-seq and analysis
The single-cell suspensions from each mouse were processed separately using the 10x Genomics 3′ v3 platform and sequenced on an Illumina NovaSeq. A total of 30,704 aortic cells were sequenced from six female mice. The raw data were processed, aligned to the mouse genome (mm10) and aggregated using 10x Genomics Cell Ranger V6. The data were then filtered using the Seurat V5 package based on the following criteria: >1,000 transcripts detected per cell but <5,000 and >1,500 total molecules detected per cell but <25,000 and <20% mitochondrial transcripts per cell. Filtering reduced this dataset from 30,704 aortic cells to 24,971 cells for further analysis. The data were then normalized using the function SCTransform v2. As samples were prepared on multiple days, the data were integrated across batches using reciprocal principal component analysis (RPCAIntegration). A principal component analysis and uniform manifold approximation and projection (UMAP) were performed, followed by the FindNeighbors and FindClusters functions. We opted to cluster at a low resolution (0.25) to differentiate aortic cell types and to identify only major subpopulations of smooth muscle cells that vary by a large number of differentially expressed genes. FindMarkers was used to identify cluster-defining transcripts and differentially expression genes between control and diseased cell populations based on a Wilcoxon rank sum test.
Reanalysis of human aortic cells from Pedroza et al. (2023)
For reanalysis of the ascending aorta and aortic root samples from a recently published scRNA-seq dataset of the donors and aortas of patients with LDS38, we used the following criteria: >1,000 transcripts detected per cell but <6,000 and >1,500 total molecules detected per cell <30,000 and <20% mitochondrial transcripts per cell. This reduces this dataset from 58,947 aortic cells to 43,349 for further analysis. We analyzed this dataset as described above with the FindClusters resolution parameter set to 0.15.
CoGAPS and projectR
CoGAPS39 (v3.22), an R package that utilizes non-negative matrix factorization to uncover latent patterns of coordinated gene expression representative of shared biological functions, was used to identify transcriptional patterns associated with VSMC subpopulations, with the npatterns parameter set to 8, in scRNA-seq analysis of murine aortas. ProjectR40 (v1.2), an R package that enables integration and analysis of multiple scRNA-seq datasets by identifying transcriptional patterns shared among datasets, was used to project these patterns into scRNA-seq analysis of the human aortic root and ascending aorta.
Gene over-representation analyses
ClueGO19 was used for gene over-representation analysis and visualization of enriched functional terms for transcripts globally dysregulated in all VSMCs as well as VSMC subsets.
The transcripts were filtered on the basis of an adjusted P value less than 0.05 and an average absolute log2 fold change of 0.25 or greater, as well as detection in at least 20% of either the control or LDS VSMCs. The resulting list of 502 downregulated and 200 upregulated genes was compared against five gene ontology databases (MSigDB Hallmark, KEGG, WikiPathways, Bioplanet and Reactome). The list of transcripts and ClueGO log files are provided in Supplementary Tables 2, 7 and 8. Differentially expressed gene lists were also analyzed using the online gene list enrichment analysis tool EnrichR25,41,42 (https://maayanlab.cloud/Enrichr/) for pathways using MSigDB43,44 and for transcription factors target enrichment using the ENCODE23 and ChEA24 databases.
MERFISH spatial transcriptomics
MERFISH spatial transcriptomics using a custom panel was performed on 5 µm formalin-fixed paraffin-embedded sections of control and LDS aortas according to manufacturer’s protocols (MERSCOPE Formalin-Fixed Paraffin-Embedded Tissue Sample Preparation User Guide Rev B, Vizgen). The slides were processed and imaged on a MERSCOPE instrument platform according to the manufacturer’s protocols (MERSCOPE Instrument User Guide Rev G, Vizgen). The raw images were processed by the instrument software to generate a matrix of spatial genomics measurements and associated image files that were analyzed using the MERSCOPE visualizer software.
Statistics
GraphPad Prism 10.0 was used for data visualization and statistical analysis. The data were tested for normality using the Shapiro–Wilk test and upon verification of normal distribution were analyzed using a Brown–Forsythe analysis of variance (ANOVA) test. For echocardiographic and blood pressure measurements, the data are presented as a box and whisker plot with the whiskers indicating the maximum and minimum values and a horizontal bar indicating the median. All individual data points are shown as dots. The figures indicating statistical significance include the statistical tests used in the figure caption.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
List of all detected transcripts comparing expression in mouse Tgfbr1M318R/+ VSMC relative to Tgfbr1+/+ VSMC.
Gene enrichment analysis using ClueGo and EnrichR for transcripts differentially detected in mouse Tgfbr1M318R/+ VSMC relative to Tgfbr1+/+ VSMC.
List of transcripts included in the custom MERFISH panel.
List of cluster-defining transcripts for VSMC1 and VSMC2 subcluster in Tgfbr1M318R/+ VSMCs, CoGAPS pattern 4 and pattern 5 gene lists and EnrichR results for CoGAPS patterns.
List of differentially expressed transcripts in mouse SHF-traced VSMC relative to non SHF-traced VSMC, reanalyzed from Pedroza et al.37.
List of cluster-defining transcripts for VSMC1 and VSMC2 subcluster in human LDS VSMCs, reanalyzed from Pedroza et al.38.
Differentially expressed transcripts in mouse Tgfbr1M318R/+ VSMC1 relative to VSMC2 Tgfbr1+/+ VSMC and gene enrichment analysis using ClueGo and EnrichR.
Differentially expressed transcripts in in human LDS VSMC1 relative to human LDS VSMC2 and Gene enrichment analysis using ClueGo and EnrichR.
Echocardiographic measurements of the aortic root and ascending aorta. Tgfbr1+/+ and Tgfbr1M318R/+ bearing Gata4fl°x/fl°x and Myh11-CreER are referred to as GATA4SMcKO. Tgfbr1+/+ and Tgfbr1M318R/+ bearing Gata4+/+ with or without Myh11-CreER or Gata4fl°x/fl°x or Gata4fl°x/+ without Myh11-CreER are referred to as GATA4Ctrl. An average measurement is provided for each animal in millimeters. This average is calculated from at least three measurements each taken from a separate echocardiographic image.
Source data
Unprocessed western gel images.
Western blot quantification data.
Blood pressure measurements, as ascending aortic echocardiography measurements are already provided in Supplementary Table 9.
Acknowledgements
Research in this publication was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under award numbers R01HL147947 to E.G.M. and F31HL163924 to E.E.B., as well as a generous gift from the Loeys–Dietz Foundation. Fluorescence Microscopy imaging was also supported by NIH award number S10OD023548 to the School of Medicine Microscope Facility.
Extended data
Author contributions
E.G.M. and E.E.B. conceptualized the study, designed the experiments, interpreted data, and prepared the manuscript. E.E.B. and T.J.C. generated and processed the scRNA-seq sequencing data. E.E.B. conducted the primary analysis of the scRNA-seq data and performed a reanalysis of published scRNA-seq datasets, with input from W.A.E.C., T.J.C., L.R. and J.T.M. E.G.M. conducted gene over-representation analysis and visualization. E.E.B., E.G.M., W.A.E. and L.R. were involved in sample preparation and processing for MERFISH. E.E.B. conducted in situ hybridization, immunofluorescence and immunoblotting experiments. E.E.B. was responsible for echocardiography, blood pressure measurements, genotyping and animal husbandry with support from T.J.C., M.S., W.A.E.C., L.R. and R.B. A.Z. performed histological staining and imaging. G.L.S.O. provided support for CoGAPS analysis and MERFISH high throughput in situ hybridization. A.J.P. and M.P.F. provided human scRNA-seq data and offered valuable insight on interpretation of the analysis. H.C.D. provided valuable input on the study design. E.G.M. and E.E.B. wrote the manuscript, and all authors contributed to its revision.
Peer review
Peer review information
Nature Cardiovascular Research thanks Helle Jorgensen, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
All scRNA-seq data, both raw fastq files and aggregated matrixes, is available in the Gene Expression Omnibus repository under accession number GSE267204. MERFISH data are available on Dataverse via https://dataverse.harvard.edu/dataverse/Gata4LDSAorticDilation (ref. 65).
Code availability
R Scripts for scRNA-seq analysis using the Seurat, CoGAPS and ProjectR packages (as described in Methods) are available as a GitHub repository at https://github.com/emilybramel/SingleCellRNAseqMurineAorta.git.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
is available for this paper at 10.1038/s44161-024-00562-5.
Supplementary information
The online version contains supplementary material available at 10.1038/s44161-024-00562-5.
References
- 1.Chou, E., Pirruccello, J. P., Ellinor, P. T. & Lindsay, M. E. Genetics and mechanisms of thoracic aortic disease. Nat. Rev. Cardiol.20, 168–180 (2023). [DOI] [PubMed] [Google Scholar]
- 2.MacCarrick, G. et al. Loeys–Dietz syndrome: a primer for diagnosis and management. Genet. Med.16, 576–587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loeys, B. L. et al. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat. Genet.37, 275–281 (2005). [DOI] [PubMed] [Google Scholar]
- 4.van de Laar, I. M. et al. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat. Genet.43, 121–126 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Lindsay, M. E. et al. Loss-of-function mutations in TGFB2 cause a syndromic presentation of thoracic aortic aneurysm. Nat. Genet.44, 922–927 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertoli-Avella, A. M. et al. Mutations in a TGF-beta ligand, TGFB3, cause syndromic aortic aneurysms and dissections. J. Am. Coll. Cardiol.65, 1324–1336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Micha, D. et al. SMAD2 mutations are associated with arterial aneurysms and dissections. Hum. Mutat.36, 1145–1149 (2015). [DOI] [PubMed] [Google Scholar]
- 8.van de Laar, I. M. et al. Phenotypic spectrum of the SMAD3-related aneurysms-osteoarthritis syndrome. J. Med. Genet.49, 47–57 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Gallo, E. M. et al. Angiotensin II-dependent TGF-β signaling contributes to Loeys–Dietz syndrome vascular pathogenesis. J. Clin. Invest.124, 448–460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacFarlane, E. G. et al. Lineage-specific events underlie aortic root aneurysm pathogenesis in Loeys–Dietz syndrome. J. Clin. Invest.129, 659–675 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams, J. A. et al. Early surgical experience with Loeys–Dietz: a new syndrome of aggressive thoracic aortic aneurysm disease. Ann. Thorac. Surg.83, S757–S763 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Hughes, G. C. Aggressive aortic replacement for Loeys–Dietz syndrome. Tex. Heart Inst. J.38, 663–666 (2011). [PMC free article] [PubMed] [Google Scholar]
- 13.Patel, N. D. et al. Aortic root replacement for children with Loeys–Dietz Syndrome. Ann. Thorac. Surg.103, 1513–1518 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Bersi, M. R., Bellini, C., Humphrey, J. D. & Avril, S. Local variations in material and structural properties characterize murine thoracic aortic aneurysm mechanics. Biomech. Model. Mechanobiol.18, 203–218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong, J. et al. In vitro lineage-specific differentiation of vascular smooth muscle cells in response to SMAD3 deficiency: implications for SMAD3-related thoracic aortic aneurysm. Arterioscler. Thromb. Vasc. Biol.40, 1651–1663 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalluri, A. S. et al. Single-cell analysis of the normal mouse aorta reveals functionally distinct endothelial cell populations. Circulation140, 147–163 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michel, J. B., Jondeau, G. & Milewicz, D. M. From genetics to response to injury: vascular smooth muscle cells in aneurysms and dissections of the ascending aorta. Cardiovasc. Res.114, 578–589 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bindea, G. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics25, 1091–1093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamashiro, Y. et al. Role of thrombospondin-1 in mechanotransduction and development of thoracic aortic aneurysm in mouse and humans. Circ. Res.123, 660–672 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bunton, T. E. et al. Phenotypic alteration of vascular smooth muscle cells precedes elastolysis in a mouse model of Marfan syndrome. Circ. Res.88, 37–43 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Bramel, E. E. et al. Postnatal Smad3 inactivation in murine smooth muscle cells elicits a temporally and regionally distinct transcriptional response. Front. Cardiovasc. Med.9, 826495 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo, Y. et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res.48, D882–D889 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lachmann, A. et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics26, 2438–2444 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie, Z. et al. Gene set knowledge discovery with enrichr. Curr. Protoc.1, e90 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashino, T., Yamamoto, M., Yoshida, T. & Numazawa, S. Redox-sensitive transcription factor Nrf2 regulates vascular smooth muscle cell migration and neointimal hyperplasia. Arterioscler. Thromb. Vasc. Biol.33, 760–768 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Chen, Y. H., Layne, M. D., Watanabe, M., Yet, S. F. & Perrella, M. A. Upstream stimulatory factors regulate aortic preferentially expressed gene-1 expression in vascular smooth muscle cells. J. Biol. Chem.276, 47658–47663 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Sellak, H., Choi, C., Browner, N. & Lincoln, T. M. Upstream stimulatory factors (USF-1/USF-2) regulate human cGMP-dependent protein kinase I gene expression in vascular smooth muscle cells. J. Biol. Chem.280, 18425–18433 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Wang, Q. et al. A hierarchical and collaborative BRD4/CEBPD partnership governs vascular smooth muscle cell inflammation. Mol. Ther. Methods Clin. Dev.21, 54–66 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ko, C. Y., Chang, W. C. & Wang, J. M. Biological roles of CCAAT/enhancer-binding protein delta during inflammation. J. Biomed. Sci.22, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herzig, T. C. et al. Angiotensin II type1a receptor gene expression in the heart: AP-1 and GATA-4 participate in the response to pressure overload. Proc. Natl Acad. Sci. USA94, 7543–7548 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bramel, E. E. et al. Distinct contribution of global and regional angiotensin II type 1a receptor inactivation to amelioration of aortopathy in Tgfbr1M318R/+ mice. Front. Cardiovasc. Med.9, 936142 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren, Q. et al. C/EBPβ: the structure, regulation, and its roles in inflammation-related diseases. Biomed. Pharmacother.169, 115938 (2023). [DOI] [PubMed] [Google Scholar]
- 34.Mondal, T. et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA–DNA triplex structures. Nat. Commun.6, 7743 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, S. et al. LncRNA CARMN inhibits abdominal aortic aneurysm formation and vascular smooth muscle cell phenotypic transformation by interacting with SRF. Cell. Mol. Life Sci.81, 175 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou, D. et al. hiPSC modeling of lineage-specific smooth muscle cell defects caused by TGFBR1A230T variant, and its therapeutic implications for Loeys–Dietz syndrome. Circulation144, 1145–1159 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pedroza, A. J. et al. Embryologic origin influences smooth muscle cell phenotypic modulation signatures in murine marfan syndrome aortic aneurysm. Arterioscler. Thromb. Vasc. Biol.42, 1154–1168 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pedroza, A. J. et al. Early clinical outcomes and molecular smooth muscle cell phenotyping using a prophylactic aortic arch replacement strategy in Loeys–Dietz syndrome. J. Thorac. Cardiovasc. Surg.166, e332–e376 (2023). [DOI] [PubMed] [Google Scholar]
- 39.Johnson, J. A. I. et al. Inferring cellular and molecular processes in single-cell data with non-negative matrix factorization using Python, R and GenePattern Notebook implementations of CoGAPS. Nat. Protoc.18, 3690–3731 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma, G., Colantuoni, C., Goff, L. A., Fertig, E. J. & Stein-O’Brien, G. projectR: an R/Bioconductor package for transfer learning via PCA, NMF, correlation and clustering. Bioinformatics36, 3592–3593 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf.14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res.44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst.1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castanza, A. S. et al. Extending support for mouse data in the Molecular Signatures Database (MSigDB). Nat. Methods20, 1619–1620 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korbecki, J., Maruszewska, A., Bosiacki, M., Chlubek, D. & Baranowska-Bosiacka, I. The potential importance of CXCL1 in the physiological state and in noncancer diseases of the cardiovascular system, respiratory system and skin. Int. J. Mol. Sci.10.3390/ijms24010205 (2022). [DOI] [PMC free article] [PubMed]
- 46.Shen, Y. et al. IRF-1 contributes to the pathological phenotype of VSMCs during atherogenesis by increasing CCL19 transcription. Aging13, 933–943 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang, C. et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science349, aaa5612 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yap, C., Mieremet, A., de Vries, C. J. M., Micha, D. & de Waard, V. Six shades of vascular smooth muscle cells illuminated by KLF4 (Kruppel-like factor 4). Arterioscler. Thromb. Vasc. Biol.41, 2693–2707 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeong, K. et al. Nuclear focal adhesion kinase controls vascular smooth muscle cell proliferation and neointimal hyperplasia through GATA4-mediated cyclin D1 transcription. Circ. Res.125, 152–166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watt, A. J., Battle, M. A., Li, J. & Duncan, S. A. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc. Natl Acad. Sci. USA101, 12573–12578 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wirth, A. et al. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat. Med.14, 64–68 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Wagenseil, J. E. & Mecham, R. P. Vascular extracellular matrix and arterial mechanics. Physiol. Rev.89, 957–989 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Dorst, D. C. H. et al. Transforming growth factor-β and the renin–angiotensin system in syndromic thoracic aortic aneurysms: implications for treatment. Cardiovasc. Drugs Ther.10.1007/s10557-020-07116-4 (2020). [DOI] [PMC free article] [PubMed]
- 54.Daugherty, A., Sawada, H., Sheppard, M. B. & Lu, H. S. Angiotensinogen as a therapeutic target for cardiovascular and metabolic diseases. Arterioscler. Thromb. Vasc. Biol.44, 1021–1030 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pedroza, A. J. et al. Single-cell transcriptomic profiling of vascular smooth muscle cell phenotype modulation in Marfan syndrome aortic aneurysm. Arterioscler. Thromb. Vasc. Biol.10.1161/ATVBAHA.120.314670 (2020). [DOI] [PMC free article] [PubMed]
- 56.Clement, M. et al. Vascular smooth muscle cell plasticity and autophagy in dissecting aortic aneurysms. Arterioscler. Thromb. Vasc. Biol.39, 1149–1159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pikkarainen, S. et al. GATA-4 is a nuclear mediator of mechanical stretch-activated hypertrophic program. J. Biol. Chem.278, 23807–23816 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Lino Cardenas, C. L. et al. An HDAC9-MALAT1-BRG1 complex mediates smooth muscle dysfunction in thoracic aortic aneurysm. Nat. Commun.9, 1009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, Y. Q., Batool, A., Chen, S. R. & Liu, Y. X. GATA4 is a negative regulator of contractility in mouse testicular peritubular myoid cells. Reproduction156, 343–351 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Smith, J. D., Chen, J. Z., Phillips, R., Daugherty, A. & Sheppard, M. B. Losartan Increases Survival of the Fbn1mgR/mgR mouse model of Marfan syndrome in an age-dependent manner. Preprint at bioRxiv10.1101/2021.02.19.429438 (2021).
- 61.Yang, B. et al. Protein-altering and regulatory genetic variants near GATA4 implicated in bicuspid aortic valve. Nat. Commun.8, 15481 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garg, V. et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature424, 443–447 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Lepage, D. et al. Gata4 is critical to maintain gut barrier function and mucosal integrity following epithelial injury. Sci. Rep.6, 36776 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou, P., He, A. & Pu, W. T. Regulation of GATA4 transcriptional activity in cardiovascular development and disease. Curr. Top. Dev. Biol.100, 143–169 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Bramel, E. E. MERFISH data for thoracic aorta of control and Loeys–Dietz syndrome mice. V1. Harvard Dataverse10.7910/DVN/KZWIFX (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of all detected transcripts comparing expression in mouse Tgfbr1M318R/+ VSMC relative to Tgfbr1+/+ VSMC.
Gene enrichment analysis using ClueGo and EnrichR for transcripts differentially detected in mouse Tgfbr1M318R/+ VSMC relative to Tgfbr1+/+ VSMC.
List of transcripts included in the custom MERFISH panel.
List of cluster-defining transcripts for VSMC1 and VSMC2 subcluster in Tgfbr1M318R/+ VSMCs, CoGAPS pattern 4 and pattern 5 gene lists and EnrichR results for CoGAPS patterns.
List of differentially expressed transcripts in mouse SHF-traced VSMC relative to non SHF-traced VSMC, reanalyzed from Pedroza et al.37.
List of cluster-defining transcripts for VSMC1 and VSMC2 subcluster in human LDS VSMCs, reanalyzed from Pedroza et al.38.
Differentially expressed transcripts in mouse Tgfbr1M318R/+ VSMC1 relative to VSMC2 Tgfbr1+/+ VSMC and gene enrichment analysis using ClueGo and EnrichR.
Differentially expressed transcripts in in human LDS VSMC1 relative to human LDS VSMC2 and Gene enrichment analysis using ClueGo and EnrichR.
Echocardiographic measurements of the aortic root and ascending aorta. Tgfbr1+/+ and Tgfbr1M318R/+ bearing Gata4fl°x/fl°x and Myh11-CreER are referred to as GATA4SMcKO. Tgfbr1+/+ and Tgfbr1M318R/+ bearing Gata4+/+ with or without Myh11-CreER or Gata4fl°x/fl°x or Gata4fl°x/+ without Myh11-CreER are referred to as GATA4Ctrl. An average measurement is provided for each animal in millimeters. This average is calculated from at least three measurements each taken from a separate echocardiographic image.
Unprocessed western gel images.
Western blot quantification data.
Blood pressure measurements, as ascending aortic echocardiography measurements are already provided in Supplementary Table 9.
Data Availability Statement
All scRNA-seq data, both raw fastq files and aggregated matrixes, is available in the Gene Expression Omnibus repository under accession number GSE267204. MERFISH data are available on Dataverse via https://dataverse.harvard.edu/dataverse/Gata4LDSAorticDilation (ref. 65).
R Scripts for scRNA-seq analysis using the Seurat, CoGAPS and ProjectR packages (as described in Methods) are available as a GitHub repository at https://github.com/emilybramel/SingleCellRNAseqMurineAorta.git.