Abstract
Ross River virus (RRV) is an indigenous Australian arthropod-borne alphavirus responsible for epidemic polyarthritis (EPA), myalgia, and lethargy in humans. Macrophages and monocytes have been associated with human RRV disease, and previous studies have shown that RRV is capable of infecting macrophages via both a natural virus receptor and by Fc receptor-mediated antibody-dependent enhancement (ADE). Similar to other viruses, such as human immunodeficiency virus and dengue virus, ADE infection results in dramatic RRV growth increases for in vitro macrophage cultures. This study demonstrates that RRV could resist lipopolysaccharide (LPS)-induced antiviral activity in macrophage cultures when infection was via the ADE pathway. Investigation of this infection pathway found that RRV was able to suppress the transcription and translation of key antiviral genes (tumor necrosis factor and inducible nitric oxide synthase) in LPS-stimulated macrophages by disrupting the transcription into mRNA of the genes coding for the associated transcription factors IRF-1 and NF-κB. The transcription of non-antiviral control genes was not perturbed by RRV-ADE infection, and de novo protein synthesis also was not significantly affected in RRV-ADE infected cells. The ADE pathway of infection allowed RRV to specifically target antiviral genes in macrophages, resulting in unrestricted virus replication. As ADE has been observed for several virus families and associated with disease and adverse vaccination outcomes, these findings may have broad relevance to viral disease formation and antiviral vaccination strategies.
Ross River virus (RRV) is an alphavirus responsible for the greatest incidence of arboviral disease in Australia. RRV is a notifiable infection that in recent years has caused an average of 4,800 cases of disease annually, ranging from 2,600 to 7,800 cases over the period surveyed (21). There are significant concerns that RRV poses an increasing health risk, particularly because RRV disease has been reported on the outskirts of major Australian population centers (1, 21). The virus is responsible for outbreaks of epidemic polyarthritis (EPA), as well as myalgia, arthralgia, rash, and lethargy, which in some patients can persist from months to years.
Synovial effusions from EPA sufferers have been shown to have a predominant monocyte infiltrate (7), and recent studies of a mouse model have shown macrophages to be the general agent of RRV disease, as well as the cell responsible for the severe muscle pathology observed in mice postinfection (19). In vitro studies with a range of human and mouse macrophages and monocytes (20) have shown that RRV can freely infect macrophage cells, but not T-cell and B-cell lines, and can infect monocytes if RRV is complexed with a subneutralizing concentration of anti-RRV antibody. In macrophages, RRV infectivity and growth were dramatically enhanced by anti-RRV antibodies (20), therefore displaying the capacity of RRV for antibody-dependent enhancement (ADE) of infection. ADE has been reported for a number of other viruses, including influenza virus, dengue virus, and human immunodeficiency virus (HIV) (25, 26, 29), as well as other alphaviruses (28). Studies have identified the Fc receptor of monocytes and macrophages as the cellular conduit for antibody-complexed virus entry (6, 28, 32), with complement receptors also recognized as having a role in HIV infection (8). While the in vivo implications of ADE have not been thoroughly identified, there has been considerable concern that ADE must be taken into account in vaccine design for viral diseases caused by dengue virus and HIV (16, 22, 25), particularly because it has been posited that the ADE mechanism is responsible for the life-threatening dengue hemorrhagic fever and dengue shock syndrome and can act as the cellular avenue for lymphocytotropic HIV strains to immediately adjust their tropism for macrophages, which is thought to predispose the host to persistent infection (25, 32).
Via studies of the infection of macrophages by RRV under lipopolysaccharide (LPS)-induced antiviral conditions in vitro, we have identified a mechanism of ADE which would explain why viruses are able to grow to high titers in macrophage cells which normally produce a number of potent antiviral proteins (e.g., tumor necrosis factor [TNF] and inducible nitric oxide synthase [NOS2]). From our evidence, antibody-facilitated virus entry into macrophages specifically inhibited the transcription and expression of key antiviral genes in macrophages, leading to the enhanced in vitro growth of RRV at LPS concentrations which normally restrict RRV growth after non-ADE virus entry into cells.
MATERIALS AND METHODS
Cells.
RAW 264.7 mouse macrophages (ATCC TIB-71) were cultured in EMEM (Life Technologies, Melbourne, Australia) medium supplemented with 100 mM glucose, 10 mM HEPES, 0.5% sodium bicarbonate, 2.0 mM l-glutamine, 50 IU of penicillin-streptomycin, and 5.0% heat-inactivated fetal calf serum (HI-FCS) (TRACE, Sydney, Australia). For 2 weeks prior to virus infection, cultures were treated with the antimycoplasma agent tylosin at 8.0 mg/ml (Sigma, St. Louis, Mo.). To reduce the background of LPS contamination in macrophage cultures, cells were grown in EMEM containing polymyxin B sulfate (Sigma) at 50 mg/liter for at least a week before virus infection. No polymyxin B sulfate was added to LPS-treated cultures. EMEM was used because it contained no nitrites or nitrates which would interfere with reactive nitrogen intermediate (RNI) measurements.
Vero cells were grown in M199 medium (Life Technologies) supplemented with 2.0 mM l-glutamine, 50 IU of penicillin-streptomycin, nonessential amino acids, and 5.0% HI-FCS.
Both cell lines were incubated at 37°C in a humidified atmosphere containing 5.0% CO2.
Virus.
Virus generated from the infectious clone of the mouse virulent RRV strain T48 (17), was used to infect RAW 264.7 cell cultures at a multiplicity of infection (MOI) of 0.01 or 0.1. For ADE of infection, RRV was incubated with a 10−3 dilution of polyclonal anti-RRV antibody-positive mouse serum (titer, 1/5,120) for 1 h at room temperature (RRV-ADE), while virus for control RRV infection (non-ADE) was incubated concurrently with a 10−3 dilution of normal mouse serum (NMS) prior to RAW 264.7 infection (both sera were heat inactivated at 56°C for 30 min). Twenty-four hours before infection, RAW cells were seeded into 24-well trays (Nunc, Roskilde, Denmark) at a density of 5.0 × 105 cells/ml with EMEM-FCS containing LPS (E. coli serotype O111:B4; Sigma) at a concentration of 1.0 or 250 ng/ml; LPS-reduced cultures were maintained in polymyxin B sulfate. Virus was placed on the confluent cell monolayer in phosphate-buffered saline (PBS) (TRACE) for 1 h at 37°C, after which the virus inoculum was removed and fresh medium (±LPS) was added. Infected cultures were incubated for 24 and 48 h postinfection. Culture medium was collected at the appropriate time points for RRV titration or TNF protein and RNI analyses, and the cells were harvested for RNA extraction.
TNF ELISA analysis.
TNF was measured in RAW 264.7 cell supernatants by a sandwich enzyme-linked immunosorbent assay (ELISA) method, which has been described previously (31). Briefly, Nunc (Roskilde, Denmark) maxisorp ELISA plates were coated with a hamster anti-mouse TNF (TN3-19.12) monoclonal antibody diluted in a pH 9.6 bicarbonate buffer. The plates were incubated overnight at 4°C followed by blocking for 2 h at room temperature (PBS plus 3% bovine serum albumin [BSA]), after which the standards and samples were added; sample supernatants were added undiluted. The plates were incubated overnight at 4°C, followed by the addition of polyclonal rabbit anti-mouse TNF antibody and incubation for 2 h at room temperature. Each well received a sheep anti-rabbit antibody–alkaline phosphatase conjugate, and the plate was incubated for 90 min at room temperature before treatment with a diethanolamine alkaline substrate buffer and further incubation for 30 to 60 min at room temperature; plates were then read at 410 nm (reference at 630 nm). All antibodies, standards, and samples were diluted in PBS containing 1% BSA and 0.1% Tween 20. The plates were washed six times between each step with PBS plus 0.1% Tween 20.
RNI assay.
RNIs were assessed in RAW 264.7 cell supernatants by the Greiss reagent-based methodology. Before the addition of the Greiss reagent, all samples, controls, standards and blanks were treated with NADPH and nitrate reductase. After addition of the Greiss reagent, all samples were trichloroacetic acid treated, and the A540s were read (reference, 630 nm). This gave the total nitrate concentration for the samples; for the total RNI concentration, nitrite levels were also determined, and this was achieved by repeating the nitrate methodology, but with the omission of NADPH and nitrate reductase. This method has been described in full previously (30).
Plaque assay.
RRV concentrations in RAW 264.7 cell supernatants were measured by plaque assay on confluent Vero cells. Serially diluted (10−1 to 10−6) samples were adsorbed to cell monolayers for 1 h at 37°C prior to a 48-h incubation (37°C, 5% CO2, humidified) under a semisolid overlay containing M199 medium, 2% HI-FCS, 4% HI-newborn serum (bovine [NBS]; TRACE), 0.02% DEAE-dextran (Pharmacia, Uppsala, Sweden), and 1.0% agar. The plates were stained with 0.02% neutral red, plaques were counted, and geometric mean virus concentrations were calculated and expressed as log10 virus titer (PFU per milliliter).
Cytokine neutralization assays.
RAW cells were grown in culture medium containing 1.0 ng of LPS per ml for at least a week before infection. Cells were seeded into 24-well trays, as described above, in medium containing the specific anticytokine antibodies. The cells were infected at an MOI of 0.1 for 1 h at 37°C, the excess virus was removed, and fresh medium was added which contained the relevant neutralizing antibody. Cultures were incubated for 48 h at 37°C, after which supernatants were collected for plaque assay. The following antimouse cytokine antibodies were used: TNF, TN3-19.12 (31); gamma interferon (IFN-γ), R4.6A2 (American Type Culture Collection, Rockville, Md.); IFN-αβ, Cytimmune rabbit anti-mouse IFN-αβ (Lee Biomolecular, San Diego, Calif.). Anti-TNF and IFN-γ antibodies, as well as species-specific control antibodies, were used at 2.0 mg/ml, and anti-IFN-αβ was used at 100 U/ml.
Determination of RRV infectivity.
RAW 264.7 cultures were infected with RRV exactly as described earlier. The percentage of infected cells was determined by immunofluorescence assays (IFA) and fluorescence-activated cell sorting (FACS; Becton-Dickinson FACSsort) at 24 h postinfection.
(i) IFA.
Confluent RRV-infected and noninfected control cells in glass chamber slides (Nunc, Inc., Naperville, Ill.) were fixed with acetone-methanol (1:1) for 1 min, after which they were left overnight at 4°C in PBS. The cells were then incubated for 2 h at 37°C with mouse anti-RRV hyperimmune ascitic fluid (HIAF) diluted to 10−3 in PBS containing 1% HI-FCS. This was followed by the addition of fluorescein isothiocyanate (FITC)-conjugated sheep anti-mouse immunoglobulin G (IgG) antibodies (Fab fragments only; Silenius, Melbourne, Australia) in PBS plus 1% HI-FCS for 1 h at 37°C, after which the cells were counted with a Zeiss fluorescent microscope. Between each antibody incubation and prior to viewing under the microscope, the cells were washed three times with sterile PBS.
(ii) FACS.
RAW cells (5 × 105) were scraped into solution and fixed with 4% paraformaldehyde (Sigma) for 5 min at room temperature. The cells were washed once in PBS and then once in PBS plus 0.1% saponin (ICN, Costa Mesa, Calif.), followed by incubation for 30 min at room temperature in PBS containing 5% HI-FCS and 0.1% saponin. Mouse anti-RRV HIAF was added to the cells at a 1/500 dilution in PBS plus saponin and incubated for 45 min in the dark at room temperature, followed by two washes with PBS-saponin. FITC-conjugated sheep anti-mouse IgG antibodies (Fab fragments only; Silenius) were added at 1/100 in PBS-saponin and incubated for 45 min in the dark at room temperature. This was followed by three washes with PBS alone before resuspension of the cells in 0.5 ml of PBS for FACS analysis.
Gene transcription studies.
RAW 264.7 cells were cultured and RRV infected, or mock infected, exactly as described earlier. At 24 h postinfection, total RNA was extracted from control and infected cells (5.0 × 105/extraction) by the method of Chomczynski and Sacchi (3). Isolated RNA was used for the synthesis of cDNA by using an oligo(dT) primer and avian myeloblastosis virus reverse transcriptase (Promega, Madison, Wis.). The cDNA was then used for specific PCR assays with Taq polymerase (Boehringer, Mannheim, Germany) and the appropriate primers. Primers for TNF and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were purchased from Clontech (Palo Alto, Calif.) and used in accordance with the manufacturer's instructions. The primer sequences for NOS2, IFN regulatory factor-1 (IRF-1), NF-κB, β-actin, and hypoxanthine phosphoribosyltransferase (HPRT) are shown in Table 1. The cDNA used in all PCRs was initially denatured at 94°C for 2 min, followed by 25 to 35 cycles of 94°C (30 s), 55 to 62°C (40 s), and 70°C (50 s) and 1 cycle at 70°C (5 min).
TABLE 1.
Primer sequences used in the RT-PCR studies of RRV-infected RAW 264.7 cellsa
| Gene | Sequenceb
|
|
|---|---|---|
| 5′ Primer | 3′ Primer | |
| NOS2 | CATGGCTTGCCCCTGGAAGTTTCTCTTCAAAG | GCAGCATCCCCTCTGATGGTGCCATCG |
| IRF-1 | AAGCACGGCTGGGACATCAACAAG | GTGGTGTAACTGCTGTGGTCATCA |
| NF-κB | GCACACCCCAGCATCTCCACTCCG | GCCATTGCAGGGCTCCTGA |
| HPRT | GTTGGATACAGGCCAGACTTTGTTG | GATTCAACTTGCGCTCATCTTAGGC |
| β-Actin | GCTCGGCCGTGGTGGTGAAGC | GTGGGGCGCCCCAGGCACCA |
TNF and GAPDH primers were purchased from Clontech (Palo Alto, Calif.).
All primer sequences are reported in the 5′ → 3′ orientation.
After the appropriate number of PCR cycles, the amplified DNA was analyzed by gel electrophoresis followed by ethidium bromide staining or Southern blotting and detected with the ECL enhanced chemiluminescence detection system as recommended by the manufacturer (Amersham Corporation, Arlington Heights, Ill.). Southern blotting was performed on the PCR products for NOS2 (probe, 5′ GGGACAGCACAGAATGGTCCATCCCTG 3′), IRF-1 (probe, 5′ CTTCCTCTTGGTTTTGCTCTTAGTGTCTCGGCTGG 3′), NF-κB (probe, 5′ GCACACCCCAGCATCTCCACTCCG 3′), and HPRT (probe, 5′ GTTGTTGGATTGCCCTTGAC 3′) to enhance the detection of the specific transcribed cellular sequences. The chemiluminescent signals were quantified with a gel scanner (Advanced Vision Research) calibrated with a densitometric step tablet (Kodak, Rochester, N.Y.). For each cytokine and factor, results were normalized for the relative quantity of total mRNA by comparison to β-actin for ethidium bromide staining or HPRT after Southern blotting. Values are expressed as fold increase or decrease in mRNA expression in infected RAW 264.7 cells over that of mock-infected cells at the same time postinfection, which were assigned an arbitrary value of 1 and represent the mean and standard deviation of cytokine or factor mRNA levels in RAW cells.
Study of de novo protein synthesis in RRV-infected RAW 264 cultures.
RAW 264.7 cultures were grown with 1.0 ng of LPS per ml for 24 h before infection with RRV (±anti-RRV antibody), as described previously, and incubated for 20 h at 37°C. All cultures were then starved by a 1.0-h incubation in PBS, after which the cells were pulsed with a tritiated (3H+)-amino acid mixture (Amersham, Little Chalfont, United Kingdom) at 50 mCi/well in amino acid-deficient EMEM plus 5% FCS, and incubated for 2.5 h at 37°C. The cells were then dissolved in Laemmli buffer (plus 2-mercaptoethanol) and boiled for 5 min prior to being loaded onto a 10% acrylamide gel (4% stacking gel). After running the gel at 135 V for 1.5 h, the gel was fixed in Coomassie destain solution (30% methanol plus 10% acetic acid) for 1 h at room temperature. The fixed gel was washed in distilled water and placed into Amersham Amplify solution for 25 min at room temperature. The gel was washed in nanopure water and dried under vacuum at 80°C for 1 h. The dried gel was then exposed to preflashed X-ray film and stored at −70°C for 1 month.
RESULTS
A low concentration of LPS significantly inhibits the growth of RRV in RAW 264.7 cells, but does not affect the proportion of cells infected.
Table 2 shows that incubating RAW 264.7 cells with a low concentration of LPS (1.0 ng/ml) leads to a significant restriction of the normal (non-ADE) in vitro growth of RRV at 24 h postinfection (1.0 ng of LPS per ml was found to stimulate the production of 5.2 ng of TNF per ml at 20 h and 5.8 ng of TNF per ml at 43 h posttreatment [data not shown]). The antiviral capacity of TNF for RRV in RAW cell cultures was demonstrated by the significant enhancement of RRV growth in LPS-treated (1.0 ng/ml) cultures containing specific antimouse TNF antibodies (1.2-log10 [PFU/ml] increase in RRV growth); the inclusion of anti-IFN-α/β or -IFN-γ antibodies also enhanced the growth of RRV in LPS-treated RAW cells, but not as effectively as anti-TNF (data not shown). However, these LPS-induced antiviral conditions did not affect the ability of RRV to initially infect macrophages (Table 2). By both IFA microscopy and FACS, a slight increase in normal RRV infectivity for RAW cells was demonstrated for cells cultured with 1.0 ng of LPS per ml compared to cells cultured under null-LPS conditions. Furthermore, for ADE cultures, infectivity at 24 h postinfection was similarly enhanced (compared to normal, non-ADE infection) under both LPS conditions tested (null LPS = 14% and 1.0 ng of LPS per ml = 12.3% by IFA). The inhibition of RRV growth in RAW cells observed due to LPS exposure, therefore, did not appear to be due to virus entry into the cell, but suggests that the postentry intracellular activities were of greater importance.
TABLE 2.
Comparison of in vitro RRV growth and percent RRV cell infectivity at 24 h postinfection in RAW 264.7 cells cultured with or without LPSa
| RAW 264.7 treatment | Log10 RRV titer (PFU/ml) | % RRV infectivityb
|
|
|---|---|---|---|
| IFA | FACS | ||
| LPS (1.0 ng/ml) | 4.91 ± 0.77 | 0.50 | 1.47 |
| No LPSc | 7.16 ± 0.02 | 0.13 | 0.46 |
The kinetics of control RRV and RRV-ADE growth under conditions in which no exogenous LPS was added to cultures and in cultures which were specifically treated to reduce LPS background (namely, with polymyxin B sulfate added) have been described previously (19, 20). In LPS-containing cultures, virus growth at 24 and 48 h post-RRV-ADE infection was found to be significantly greater than growth following control (non-ADE) RRV infection (P < 0.01). At 120 to 336 h postinfection, RRV-ADE growth was not significantly different (P > 0.10) from virus growth after control RRV infection.
Mock-infected control RAW 264.7 cultures analyzed by FACS showed between 0.11 and 0.18% fluorescence for null-LPS and 1.0-ng/ml LPS cultures, while no fluorescent cells were detected for mock-infected cultures by IFA analysis.
Cultures were treated with polymyxin B sulfate to reduce background LPS.
RRV was resistant to LPS-induced antiviral activity in macrophages when infection was antibody mediated via the Fc receptor pathway (RRV-ADE).
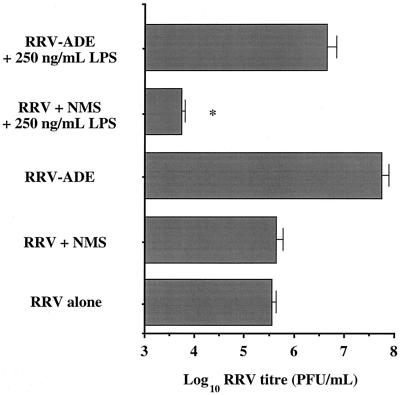
The inclusion of a high dose of LPS (250 ng/ml) in RRV-infected RAW cell cultures had the predicted effect of significantly (P < 0.05) restricting the growth of progeny RRV in vitro, after non-ADE control infection (Fig. 1). If RAW cell infection was done with anti-RRV antibodies (RRV-ADE), rather than with control sera, the antiviral impact of the high LPS dose was totally negated, with RRV growth exceeding that of non-LPS control macrophage cultures. This result demonstrated that normal RRV infection of macrophages was susceptible to the antiviral affects of LPS and resulted in restricted RRV growth, whereas RRV-ADE infection via the macrophage Fc receptor led to a perturbation of LPS-induced antiviral activity, allowing the virus to achieve very high levels of replication, although not as high as that observed for RRV-ADE infection of macrophages cultured without exogenous LPS (Fig. 1).
FIG. 1.
Growth of RRV at 24 h postinfection in RAW 264.7 cultures treated with 250 ng of LPS per ml. LPS and control (no LPS) cultures were RRV infected at an MOI of 0.01 after incubation for 1 h at 4°C with either normal mouse serum (RRV + NMS), serum containing polyclonal anti-RRV antibodies (RRV-ADE), or no serum (RRV alone); sera were diluted to 10−3 prior to incubation with RRV. ∗, significant (P < 0.05) decrease in RRV titer compared to RRV + NMS (as determined by unpaired Student's t test).
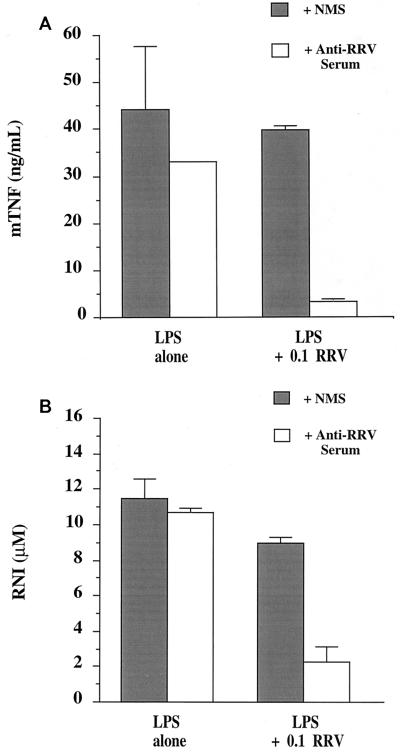
Production of TNF protein and RNIs was significantly inhibited for LPS-stimulated RAW 264.7 cultures after RRV-ADE infection.
Figure 2 shows the results of studies in which RAW cells were cultured with LPS (250 ng/ml) prior to and during a 48-h RRV infection, after which culture media were collected for TNF protein and RNI analysis. Noninfected (±anti-RRV antibody) cultures responded to LPS stimulation by producing significant levels of both TNF and RNIs in RAW cell supernatants. Control RRV infection (plus NMS) showed nonsignificant reductions in TNF and RNI production, whereas RRV-ADE infection resulted in a dramatic inhibition of TNF and RNI production in RAW cell cultures. The post-RRV-ADE infection reduction in TNF and RNIs coincided with the enhanced RRV growth found for identical LPS-stimulated and infected cultures, as shown in Fig. 1.
FIG. 2.
LPS-stimulated (250 ng/ml) TNF (A) and RNI (B) production in RAW 264.7 cultures infected with RRV at an MOI of 0.1. Cells were RRV infected after virus incubation with either NMS or serum containing polyclonal anti-RRV antibodies (RRV-ADE) for 1 h at 4°C; both sera were diluted to 10−3 prior to incubation with RRV. Noninfected, LPS-stimulated control cultures were mock infected with PBS containing either NMS or anti-RRV serum alone at a 10−3 dilution.
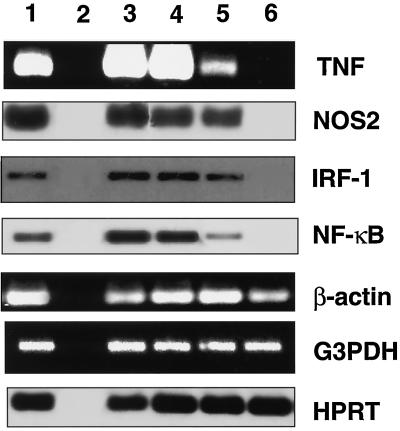
Transcription of the genes coding for TNF, NOS2, NF-κB, and IRF-1 was specifically inhibited in LPS-stimulated RAW 264.7 cultures infected via the RRV-ADE pathway.
Figure 3 shows the reverse transcriptase PCR (RT-PCR) analysis of gene transcription in LPS-treated (250 ng/ml) RAW cell cultures infected with RRV. The transcription of NOS2 message (as representative of RNI activity) was unaffected for control RRV-infected cultures in comparison to noninfected cultures. TNF transcription was clearly detected for control RRV-infected RAW cells, but was reduced by 30-fold compared to that in noninfected controls. Message for NOS2 (iNOS) and TNF, however, was completely ablated in RAW cell cultures following RRV-ADE infection (Fig. 3). Further investigation examined the transcription of IRF-1, which has been identified as the major transcription factor responsible for NOS2 regulation (11, 27), and NF-κB (Fig. 3). RT-PCR studies of RNA from concurrent LPS-stimulated RAW cell cultures found that IRF-1 message was completely ablated by RRV-ADE infection, whereas IRF-1 message was detected at similar levels for both control RRV-infected and noninfected RAW cell cultures. NF-κB transcription was reduced by 10-fold for control RRV-infected cultures compared to noninfected cultures, similar to what was observed for TNF transcription. As found for IRF-1, the transcription of NF-κB was completely ablated in RAW 264.7 cultures following RRV-ADE infection, and the ablation of message for both transcription factors corresponded with the absence of RNA message for NOS2 and TNF.
FIG. 3.
Transcription of NOS2 (RNI), TNF, IRF-1, NF-κB and three control genes (coding for β-actin, GAPDH, and HPRT) in RRV-infected (MOI of 0.1) and LPS-treated (250 ng/ml) RAW 264.7 cultures at 24 h postinfection. RRV infection was performed with a 10−3 dilution of either NMS (RRV + NMS) or anti-RRV serum (RRV-ADE). Total RAW 264.7 RNA was first reverse transcribed with an oligo(dT) primer, and the subsequent cDNA was used in specific PCRs with primer sets described in Table 1 or purchased from Clontech. NOS2, IRF-1, and HPRT results were enhanced by Southern hybridization after RT-PCR amplification. Lanes: 1, positive control; 2, negative control; 3, not infected plus NMS; 4, not infected plus anti-RRV serum; 5, RRV plus NMS; 6, RRV plus anti-RRV serum (RRV-ADE).
Three control genes coding for proteins with no antiviral involvement (HPRT, GAPDH, and β-actin) were also assessed in LPS-stimulated RAW cells (Fig. 3). The transcription of these genes was found to be unaffected by either control RRV infection or RRV-ADE infection compared to that in noninfected control cultures, indicating that the transcription of other cellular genes continued during antibody-enhanced RRV infection.
Control cultures rendered LPS free with polymyxin B sulfate were RRV-ADE infected, the RNA was extracted from these cells, and RT-PCR analysis was performed for GAPDH. Without LPS-induced protection, GAPDH transcription in infected cells was ablated within 48 h postinfection (data not shown), demonstrating that this control gene was sensitive to disruption by RRV infection.
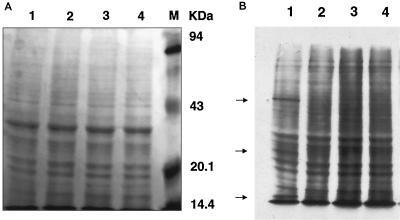
General de novo protein synthesis in RAW 264.7 cells was not significantly influenced by RRV-ADE infection.
A study was performed to assess whether the observed downregulation of antiviral protein production by RRV-ADE infection was simply due to the property of many virus families to nonspecifically shut down general cellular de novo protein synthesis. This was achieved by pulsing infected and LPS-treated (1.0 ng/ml) macrophage cultures with 3H+-amino acids followed by gel autoradiography.
Figure 4 shows that cellular de novo protein synthesis was not significantly down-regulated by either control RRV or RRV-ADE infection protocols in LPS-treated macrophages. In contrast, polymyxin B sulfate-treated (null-LPS) macrophage cultures showed significant reductions in de novo protein synthesis post-RRV and -RRV-ADE infection (as assessed by cpm; data not shown), indicating that unstimulated RAW cells were normally susceptible to RRV-induced host protein shutdown. The enhanced infection of RAW cells via ADE mechanisms was also confirmed by the detection of de novo RRV proteins at 50, 30, and 15 kDa in the RRV-ADE cell protein sample. The massive reduction in TNF protein and RNI production by LPS-stimulated RAW cells was not, therefore, a result of a general virus-mediated host protein shutdown, but was associated with the specific targeting of antiviral protein production by ADE-mediated entry of RRV into macrophages.
FIG. 4.
Analysis of general de novo protein synthesis in LPS-treated (1.0 ng/ml) RAW 264.7 cultures at 24 h post-RRV infection (MOI of 0.1; plus anti-RRV or NMS). The loading control (A) was stained with Coomassie blue and shows the molecular mass markers. (B) Efficiency of 3H+-amino acid utilization by LPS-treated and RRV-infected RAW 264.7 cells as a measure of de novo protein synthesis. Lanes: 1, RRV-ADE; 2, RRV plus NMS; 3, noninfected plus anti-RRV serum; 4, noninfected plus NMS. Enhanced infectivity by RRV-ADE in RAW cells is shown by the detection of viral proteins (arrows).
DISCUSSION
RRV can infect macrophages via a normal cell receptor, as well as utilize ADE mechanisms to significantly enhance its infectivity on Fc receptor-bearing cells, such as monocytes and macrophages (20). For RRV, the ADE mechanism is particularly useful, because the virus was found to be very sensitive to antiviral cytokine activity. As evidenced by these studies, ADE entry into macrophages afforded RRV with protection from LPS-induced antiviral activity in vitro, resulting in enhanced cell infectivity and dramatically increased RRV titers compared to those of control (non-ADE) RRV infection. RRV has been found to induce cytopathic effect in macrophages in vitro (20), which is a type of infection that has been suggested relies upon control via cytokine activity, rather than cytotoxicity (11, 34). Such a suggestion supports the view that cytokine expression needs to be circumvented by RRV because this poses a major threat to virus survival. The ability of viruses to interfere with the activity of related antiviral pathways has been shown previously with human cytomegalovirus, which inhibited class II major histocompatibility complex expression in endothelial cells and fibroblasts by disabling IFN-γ-stimulated Jak/STAT-mediated signal transduction (24). Antiviral intracellular pathway interference has also been implicated by this study, which found that RRV achieved protection from LPS-induced antiviral factors, post-ADE entry, by specifically perturbing the transcription of key antiviral genes through targeting the transcription factors IRF-1 and NF-κB.
Studies using constitutive transgene overexpression have identified IRF-1 as having a potent antiviral function for three virus families (27). Further investigations with IRF-1−/− mice have established the IRF-1 link to IFN-α/β-induced antiviral activity, although not all virus families tested were sensitive to IFN-α/β antiviral activity mediated via IRF-1 (13). The involvement of IRF-1 with NOS2 activity has also been demonstrated by studies of macrophages from IRF-1−/− mice (10). IRF-1−/− macrophages were shown to produce dramatically attenuated levels of the stable nitric oxide (NO) end product, nitrite, after in vitro stimulation by combinations of IFN-α/β and IFN-γ, TNF, and LPS. This study also revealed that the massive downregulation of NO activity was a result of the absence of NOS2 mRNA, which clearly connects IRF-1 with being crucial to NOS2 gene activation and, therefore, NO-mediated antiviral activity (12). NF-κB, which is solely responsible for TNF transcription, has been identified along with IRF-1 as being critical to the induction of NOS2 (33). The ADE-mediated disruption of IRF-1 and NF-κB by RRV, therefore, specifically neutralizes the downstream expression of NO and TNF while allowing non-antiviral proteins to be synthesized, further supporting viral growth.
Studies have identified activities associated with the impact of cytokines on Fc receptor function. For example, a study has found that IFN-γ augmented the Fc receptor-mediated infection of human monocytes by dengue virus, via the IFN-γ-induced increase in Fcγ receptor numbers on cells (15). Of particular relevance to our study has been previous work showing that the cross-linking of Fcγ receptors on monocytes with antibody (Fc domain binding only) resulted in the induction of TNF secretion (4, 5). These findings strongly suggest a link between the functional Fc receptor proteins on the cell surface and the intracellular pathways attached to TNF expression. It is possible, therefore, that RRV-antibody (ADE) infection via Fc receptors provides the virus with an efficient route for the disruption of normal cytokine activation post-Fc receptor binding. In fact, it has been shown that immune complex binding to Fcγ receptors on P388D1 cells can lead to the inhibition of IFN-γ-mediated Ia antigen expression through the manipulation of adenylate cyclase (9), indicating that under certain binding conditions cytokine activity can be inhibited via the Fc receptor.
Whether ADE is a factor in RRV disease is yet to be determined, although severe disease resulting from secondary infection by another arbovirus, dengue virus, is a well-studied example of ADE and human disease (14, 25). Also, a study of infection by another arbovirus, yellow fever virus (YF), showed that anti-YF monoclonal antibodies enhanced YF-induced mortality in mice, as reflected by significantly reduced survival times (2). An RRV study (20) has considered whether preexisting enhancing antibodies could be detected in EPA patients, or whether antibodies against the related alphavirus, Barmah Forest virus (BFV), could enhance RRV infection in vitro. This study found no simple correlation of in vitro enhancement of infection to clinical disease when anti-RRV IgGs from EPA patients and asymptomatic seropositive individuals were compared. Also, anti-BFV antibodies did not enhance RRV infection in vitro, suggesting that cross-reactive antibodies were not involved.
An animal model example of particular relevance to RRV disease is the infection of goats with caprine arthritis-encephalitis virus (CAEV). CAEV induces a gradual development of debilitating arthritis in goats, and a vaccine study (23) showed that goats vaccinated against CAEV developed a more severe and rapid arthritis than control vaccinated animals. A subsequent study (18) has shown that goat macrophages, which infiltrate joint tissues after CAEV infection, have an altered expression of cytokine and chemokine genes post-CAEV infection, with message for IL-8 and MCP-1 upregulated and that for TGF-β1 downregulated. Similar to our RRV study, investigation of cytokine expression in stimulated macrophage cultures found that message for TNF, IL-1β, IL-6, and IL-12 p40 was reduced. A role for NF-κB was also considered, but was found not to be affected by CAEV infection (18). This evidence, from both the vaccine and macrophage studies, provides very firm parallels between CAEV disease in the goat and RRV-induced EPA in humans. Macrophages have been shown to be the cellular agents of RRV disease and associated muscle pathology in a mouse model (19), and further studies are needed to define whether there is in situ alteration of the cytokine profile that correlates with disease. This study (19) also described a massive infiltration of macrophages into the muscle of RRV-infected mice, which strongly suggests that there was an upregulation of chemokine production by macrophages, as found in vitro for CAEV infection. Although in vivo ADE has not been described for RRV in either human studies or in mouse models, the CAEV evidence would advise that care be taken with the development of future RRV vaccines.
In conclusion, the ADE pathway of infection has been shown to protect RRV from LPS-stimulated antiviral activity in macrophages, whereas RRV infection via the natural virus receptor on macrophages resulted in the dramatic restriction of RRV growth in LPS-stimulated cultures. Further examination found that RRV-ADE infection significantly inhibited LPS-induced RNI and TNF protein production in macrophage cultures, with the transcription of the corresponding genes found to be ablated in these cultures. However, general de novo protein synthesis and the transcription of nonantiviral control genes were found to be unaffected after in vitro RRV-ADE infection. This specific inhibition of RNI and TNF activity in RRV-ADE-infected macrophages was found to result from the ability of Fc receptor-mediated RRV infection to effectively disrupt, at the mRNA level, the transcription factors IRF-1 and NF-κB. Because RRV is very sensitive to antiviral cytokine activity in vitro, ADE mechanisms provide a way for the virus to establish an infection cycle in macrophage cells, which we have shown previously (19) to be responsible for RRV disease in mice. Therefore, blocking the ADE mechanism of antiviral cytokine downregulation could be critical to the inhibition of RRV disease formation, because normal macrophage-mediated cytokine responses would restrict virus replication and spread.
ACKNOWLEDGMENTS
This study was funded in part by an Australian Research Council grant (awarded to B.A.L.) and a National Health and Medical Research Council of Australia grant held by L. Dalgarno and R. Weir.
B.A.L. thanks Lyn Dalgarno, Ron Weir, and Eva Lee (Division of Biochemistry & Molecular Biology, Faculty of Science, Australian National University) for supporting the early macrophage studies which gave impetus to this work.
REFERENCES
- 1.Amin J, Hueston L, Dwyer D E, Capon A. Ross River virus infection in the north-west outskirts of the Sydney basin. Commun Dis Intell. 1998;22:101–102. doi: 10.33321/cdi.1998.22.18. [DOI] [PubMed] [Google Scholar]
- 2.Barrett A D, Gould E A. Antibody-mediated early death in vivo after infection with yellow fever virus. J Gen Virol. 1986;67:2539–2542. doi: 10.1099/0022-1317-67-11-2539. [DOI] [PubMed] [Google Scholar]
- 3.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 4.Debets J M H, van der Linden C J, Dieteren I E, Leeuwenberg J F, Buurman W A. Fc-receptor cross-linking induces rapid secretion of tumor necrosis factor (cachectin) by human peripheral blood monocytes. J Immunol. 1988;141:1197–1201. [PubMed] [Google Scholar]
- 5.Debets J M H, Van de Winkel J G J, Ceuppens J L, Dieteren I E M, Buurman W A. Cross-linking of both FcRI and FcRII induces secretion of TNF by human monocytes, requiring high affinity Fc-FcR interactions. J Immunol. 1990;144:1304–1310. [PubMed] [Google Scholar]
- 6.Dworak L J, Wolfinbarger J B, Bloom M E. Aleutian mink disease parvovirus infection of K562 cells is antibody-dependent and is mediated via an Fc(gamma)RII receptor. Arch Virol. 1997;142:363–373. doi: 10.1007/s007050050082. [DOI] [PubMed] [Google Scholar]
- 7.Fraser J R E. Epidemic polyarthritis and Ross River virus disease. Clin Rheum Dis. 1986;12:369–388. [PubMed] [Google Scholar]
- 8.Fust G. Enhancing antibodies in HIV infection. Parasitology. 1997;115:S127–S140. doi: 10.1017/s0031182097001819. [DOI] [PubMed] [Google Scholar]
- 9.Hanaumi K, Gray P, Suzuki T. Fc gamma receptor-mediated suppression of gamma-interferon-induced Ia antigen expression on a murine macrophage-like cell line (P338D1) J Immunol. 1984;133:2852–2856. [PubMed] [Google Scholar]
- 10.Kägi D, Ledermann B, Bürki K, Zinkernagel R M, Hengartner H. Molecular mechanisms of lymphocyte-mediated cytotoxicity and their role in immunological protection and pathogenesis in vivo. Annu Rev Immunol. 1996;14:207–232. doi: 10.1146/annurev.immunol.14.1.207. [DOI] [PubMed] [Google Scholar]
- 11.Kamijo R, Harada H, Matsuyama T, et al. Requirement for transcription factor IRF-1 in NO synthase induction in macrophages. Science. 1994;263:612–615. doi: 10.1126/science.7510419. [DOI] [PubMed] [Google Scholar]
- 12.Karupiah G, Xie Q-W, Buller M L, Nathan C, Duarte C, MacMicking J D. Inhibition of viral replication by interferon-γ-induced nitric oxide synthase. Science. 1993;261:1445–1448. doi: 10.1126/science.7690156. [DOI] [PubMed] [Google Scholar]
- 13.Kimura T, Nakayama K, Penninger J, et al. Involvement of the IRF-1 transcription factor in antiviral responses to interferon. Science. 1994;264:1921–1924. doi: 10.1126/science.8009222. [DOI] [PubMed] [Google Scholar]
- 14.Kliks S C, Nisalak A, Brandt W E, Wahl L, Burke D S. Antibody-dependent enhancement of dengue virus growth in human monocytes as a risk factor for dengue haemorrhagic fever. Am J Trop Med Hyg. 1989;40:444–451. doi: 10.4269/ajtmh.1989.40.444. [DOI] [PubMed] [Google Scholar]
- 15.Kontny U, Kurane I, Ennis F A. Gamma interferon augments Fcγ receptor-mediated dengue virus infection of human monocytic cells. J Virol. 1988;62:3928–3933. doi: 10.1128/jvi.62.11.3928-3933.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozlowski P A, Black K P, Shen L, Jackson S. High prevalence of serum IgA HIV-1 infection-enhancing antibodies in HIV-infected persons. Masking by IgG. J Immunol. 1995;154:6163–6173. [PubMed] [Google Scholar]
- 17.Kuhn R J, Niesters H G, Hong Z, Strauss J H. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology. 1991;182:430–441. doi: 10.1016/0042-6822(91)90584-x. [DOI] [PubMed] [Google Scholar]
- 18.Lechner F, Machado J, Bertani G, Seow H F, Dobbelaere D A, Peterhans E. Caprine arthritis encephalitis virus dysregulates the expression of cytokines in macrophages. J Virol. 1997;71:7488–7497. doi: 10.1128/jvi.71.10.7488-7497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lidbury B A, Simeonovic C, Maxwell G E, Marshall I D, Hapel A J. Macrophage-induced muscle pathology results in morbidity and mortality for Ross River virus infected mice. J Infect Dis. 2000;181:27–34. doi: 10.1086/315164. [DOI] [PubMed] [Google Scholar]
- 20.Linn M L, Aaskov J, Suhrbier A. Antibody-dependent enhancement and persistence in macrophages of an arbovirus associated with arthritis. J Gen Virol. 1996;77:407–411. doi: 10.1099/0022-1317-77-3-407. [DOI] [PubMed] [Google Scholar]
- 21.Mackenzie J S, Broom A K, Hall R A, Johansen C A, Lindsay M D, Phillips D A, Ritchie S A, Russell R C, Smith D W. Arboviruses in the Australian region, 1990 to 1998. Commun Dis Intell. 1998;22:93–100. doi: 10.33321/cdi.1998.22.17. [DOI] [PubMed] [Google Scholar]
- 22.Mascola J R, Mathieson B J, Zack P M, Walker M C, Halstead S B, Burke D S. Summary report: workshop on the potential risks of antibody-dependent enhancement in human HIV vaccine trials. AIDS Res Hum Retrovir. 1993;9:1175–1184. doi: 10.1089/aid.1993.9.1175. [DOI] [PubMed] [Google Scholar]
- 23.McGuire T C, Adams D S, Johnson G C, Klevjer-Anderson P, Barbee D D, Gorman J R. Acute arthritis in caprine arthritis-encephalitis virus challenge exposure of vaccinated or persistently infected goats. Am J Vet Res. 1986;47:537–540. [PubMed] [Google Scholar]
- 24.Miller D M, Rahill B M, Boss J M, Lairmore M D, Durbin J E, Waldman W J, Sedmak D D. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J Exp Med. 1998;187:675–683. doi: 10.1084/jem.187.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morens D M, Halstead S B. Measurement of antibody-dependent infection enhancement of four dengue virus serotypes by monoclonal and polyclonal antibodies. J Gen Virol. 1990;71:2909–2914. doi: 10.1099/0022-1317-71-12-2909. [DOI] [PubMed] [Google Scholar]
- 26.Ochiai H, Kurokawa M, Matsui S, Yamamoto T, Kuroki Y, Kishimoto C, Shiraki K. Infection enhancement of influenza A NWS virus in primary murine macrophages by anti-hemagglutinin monoclonal antibody. J Med Virol. 1992;36:217–221. doi: 10.1002/jmv.1890360312. [DOI] [PubMed] [Google Scholar]
- 27.Pine R. Constitutive expression of an ISGF2/IRF1 transgene leads to interferon-independent activation of interferon-inducible genes and resistance to virus infection. J Virol. 1992;66:4470–4478. doi: 10.1128/jvi.66.7.4470-4478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porterfield J S. Antibody-dependent enhancement of viral infectivity. Adv Virus Res. 1986;31:335–355. doi: 10.1016/s0065-3527(08)60268-7. [DOI] [PubMed] [Google Scholar]
- 29.Robinson W E, Jr, Montefiori D C, Mitchell W M. Antibody-dependent enhancement of human immunodeficiency virus type 1 infection. Lancet. 1988;i:790–794. doi: 10.1016/s0140-6736(88)91657-1. [DOI] [PubMed] [Google Scholar]
- 30.Rockett K A, Awburn M M, Rockett E J, Cowden W B, Clark I A. Possible role of nitric oxide in malarial immunosuppression. Parasite Immunol. 1994;16:243–249. doi: 10.1111/j.1365-3024.1994.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 31.Sheehan K C, Ruddle N H, Schreiber R D. Generation and characterisation of hamster monoclonal antibodies that neutralise murine tumour necrosis factor. J Immunol. 1989;142:3884–3893. [PubMed] [Google Scholar]
- 32.Trischmann H, Davis D, Lachmann P J. Lymphocytotropic strains of HIV type 1 when complexed with enhancing antibodies can infect macrophages via Fc gamma RIII, independently of CD4. AIDS Res Hum Retrovir. 1995;11:343–352. doi: 10.1089/aid.1995.11.343. [DOI] [PubMed] [Google Scholar]
- 33.Xie Q W, Kashiwabara Y, Nathan C. Role of transcription factor NF-kappa B/Rel in induction of nitric oxide synthase. J Biol Chem. 1994;269:4705–4708. [PubMed] [Google Scholar]
- 34.Zinkernagel R M. Immunology taught by viruses. Science. 1996;271:173–178. doi: 10.1126/science.271.5246.173. [DOI] [PubMed] [Google Scholar]