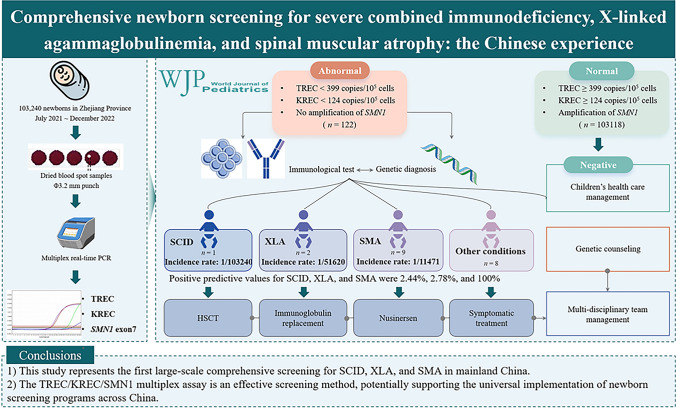
Abstract
Background
Newborn screening (NBS) for severe combined immunodeficiency (SCID), X-linked agammaglobulinemia (XLA), and spinal muscular atrophy (SMA) enables early diagnosis and intervention, significantly improving patient outcomes. Advances in real-time polymerase chain reaction (PCR) technology have been instrumental in facilitating their inclusion in NBS programs.
Methods
We employed multiplex real-time PCR to simultaneously detect T-cell receptor excision circles (TRECs), kappa-deleting recombination excision circles (KRECs), and the absence of the survival motor neuron (SMN) 1 gene in dried blood spots from 103,240 newborns in Zhejiang Province, China, between July 2021 and December 2022.
Results
Of all the samples, 122 were requested further evaluation. After flow cytometry evaluation and/or genetic diagnostics, we identified one patient with SCID, two patients with XLA, nine patients with SMA [one of whom also had Wiskott–Aldrich Syndrome (WAS)], and eight patients with other medical conditions. The positive predictive values (PPVs) of NBS for SCID, XLA, and SMA were 2.44%, 2.78%, and 100%, respectively. The estimated prevalence rates in the Chinese population were 1 in 103,240 for SCID, 1 in 51,620 for XLA, and 1 in 11,471 for SMA.
Conclusion
This study represents the first large-scale screening in mainland China using a TREC/KREC/SMN1 multiplex assay, providing valuable epidemiological data. Our findings suggest that this multiplex assay is an effective screening method for SCID, XLA, and SMA, potentially supporting the universal implementation of NBS programs across China.
Graphical abstract
Keywords: Genetic diagnosis, Newborn screening, Severe combined immunodeficiency, Spinal muscular atrophy, X-linked agammaglobulinemia
Introduction
Newborn screening (NBS) aims to detect severe congenital and hereditary diseases in infants, facilitating early diagnosis and treatment to significantly improve the prognosis of affected patients [1]. Since the implementation of NBS for congenital hypothyroidism (CH) and phenylketonuria (PKU) in the 1980s, followed by congenital adrenal hyperplasia (CAH) and glucose-6-phosphate dehydrogenase (G6PD) deficiency in the 2000s, the application of tandem mass spectrometry (MS/MS) to over 40 genetic metabolic diseases has revolutionized the field of NBS [2]. Currently, NBS in China faces the challenge of expanding the range of target diseases that can be integrated into routine programs.
Severe combined immunodeficiency (SCID), one of the most severe forms of primary immunodeficiency disorders (PIDs), is caused by a spectrum of genetic mutations that result in critically impaired cellular and humoral immunity [3]. Real-time polymerase chain reaction (PCR)-based quantification of T-cell receptor excision circles (TRECs), which serve as DNA biomarkers of normal T-cell development, is widely used in NBS for SCID and can also identify other T-cell deficiencies [4, 5]. Patients diagnosed with SCID through NBS typically undergo hematopoietic stem cell transplantation (HSCT) at a significantly younger median age than those diagnosed based on clinical symptoms [6]. In general, patients with SCID who undergo HSCT at an earlier age experience superior outcomes compared to those who undergo transplantation later in life [7]. Kappa-deleting recombination excision circles (KRECs) are DNA fragments produced during B-cell maturation in the bone marrow [8]. Nakagawa et al. [9] described a PCR-based method for detecting KRECs, which was subsequently validated and utilized in pilot studies to identify B-cell deficiencies, such as X-linked agammaglobulinemia (XLA) [10–13]. The combined TREC/KREC assay provides a more comprehensive NBS approach for diverse forms of PIDs, enabling the detection of conditions that might be overlooked when using TRECs alone [14].
Spinal muscular atrophy (SMA) is a genetic disorder characterized by progressive, symmetrical muscle weakness and atrophy [15]. It is a common genetic condition that can lead to severe outcomes in infants, including death. SMA affects about 1 in 6000–11,000 live births, with an estimated carrier frequency of about 1 in 35–50 individuals [16]. SMA is primarily caused by mutations in the survival motor neuron (SMN) 1 gene located on chromosome 5q13, with about 95% of cases identified in newborns through screening for the homozygous absence of SMN1 exon 7 [17]. The most profound therapeutic benefits of current treatments, such as onasemnogene abeparvovec [18], nusinersen [19], and risdiplam [20], are observed in patients who receive treatment prior to the manifestation of clinical symptoms. The American College of Medical Genetics and Genomics (ACMG) recommends population-wide screening for SMA, given the efficacy of these therapeutics and the additional benefits of early pre-symptomatic identification [21]. NBS for SMA using real-time PCR technology, similar to that employed in SCID-NBS, has been implemented in the United States of America (USA) and several other countries. In addition, determining the SMN1 and SMN2 copy numbers through multiplex ligation-dependent probe amplification (MLPA), the standard diagnostic method for SMA is crucial for clinical categorization and prognosis [22]. Previous studies have demonstrated the feasibility of using a multiplex qPCR assay to screen for both PID and SMA with the same real-time PCR technology [23, 24].
We conducted a study in Zhejiang Province to evaluate the effectiveness of incorporating TREC/KREC/SMN1 screening into our NBS programs. This approach employs multiplex real-time PCR assays to screen newborns for severe PIDs by assessing T- and B-cell levels and detecting deletions in exon 7 of the SMN1 gene. This study describes the NBS process and management of SCID, XLA, and SMA, based on the inaugural large-scale NBS implementation in the Mainland of China.
Methods
Dried blood spot samples
The NBS pilot trial for SMA, SCID, and XLA was conducted on newborns born in Zhejiang Province between July 2021 and December 2022. Dried blood spot (DBS) samples were collected 48 h after birth from adequately fed newborns. A 3.2-mm punch was extracted from the leftover DBS specimens collected during routine NBS at the Zhejiang Neonatal Screening Centre. The trial covered 303 maternity units across 75 counties in Zhejiang Province, excluding Ningbo City. This study was approved by the Research Ethics Committee of the Children’s Hospital of Zhejiang University School of Medicine (approval number: 2021-IRB-036) and compiled patient data without personal identifiers.
TREC/KREC/SMN1 screening assay
The TREC/KREC/SMN1 NBS assay was performed using multiplex real-time PCR. DNA was extracted from 3.2-mm diameter punches (NeoMDx DNA Extraction kit, Xinbo, Suzhou, China). The DBS discs (3.2 mm) were punched and washed once in 100 μL of DNA elution buffer, then shaken at 1500 rpm for 10 min. After removing the wash buffer, 40 μL of fresh DNA elution buffer was added, and the samples were heated at 95 °C for 30 min. The eluted DNA from the supernatant underwent multiplex real-time PCR analysis to simultaneously quantify TREC, KREC, SMN1 exon 7, and RPP30 in 96-well formats using a NeoMDx PCR kit (Xinbo, Suzhou, China) on a SLAN® 96S real-time PCR instrument (Hongshi, Shanghai, China). The cycling conditions were as follows: 37 °C for 2 min, then 94 °C for 5 min, followed by 40 cycles of 93 °C for 10 s, 60 °C for 30 s, and 69 °C for 40 s. RPP30 was used as an internal positive control for sample quantity. Individual cycle thresholds were determined by inspecting the amplification curves and automatically set using the instrument software. The quantities of TREC and KREC were calculated and expressed as copies per 105 nucleated cells using the following formula:
Based on the distribution data of 3400 normal newborns, the optimal cutoff values for TREC and KREC copies/105 cells were determined using the Youden index method, which was 0.005. The K–S test for these samples yielded a P value < 0.05, indicating that the distribution was almost normal. The cutoff values for TREC and KREC were set at < 399 copies/105 cells and < 124 copies/105 cells, respectively. Samples were classified as having a homozygous deletion of exon 7 of SMN1 if no amplification of SMN1 was detected. For TREC and KREC values, abnormal values were defined as those below the established cutoffs for TREC and/or KREC, provided there was no DNA amplification failure. All abnormal values and incomplete samples were confirmed using a repeat assay.
Clinical procedures
After obtaining informed consent, referred infants were examined to confirm the diagnosis. A complete blood count, flow cytometry, and immunoglobulin levels were assessed for infants with suspected PID. Infants with critically low TREC and/or KREC values, particularly those with a suspected family history of immunodeficiency, underwent trio-whole exome sequencing (trio-WES) for comprehensive genetic analysis. To obtain more accurate epidemiological data and reduce the risk of missed diagnoses, referred infants with moderately low TREC and/or KREC values underwent whole exome sequencing (WES) as probands using genomic DNA extracted from fresh whole blood. Unreferred infants with abnormal TREC and/or KREC values also underwent WES, with genomic DNA extracted from their DBS. To minimize the inherent limitations of whole exome variant detection, the average sequencing depth was ×150, with more than 98% of the exonic regions achieving a coverage of over ×50, and 99% achieving a coverage of at least ×30. The probe detection method was used to capture the exonic and flanking intronic regions of the genome, and the obtained DNA sequences were compared to the human genome hg19 reference sequence from the University of California, Los Angeles (UCLA) database. Variants with at least tenfold coverage were analyzed using bioinformatic tools for pathogenicity, following data interpretation guidelines provided by the ACMG. Variant nomenclature was based on the guidelines set by the Human Genome Variation Society (HGVS). Only variants that fit the inheritance model of the patient’s family and were potentially linked to the fetal phenotypes were reported. All suspected clinically significant variants were verified using Sanger sequencing. Copy number variations (CNVs) were identified using the sliding window method based on read depth algorithms with WES data. MLPA was performed using whole blood samples for patients suspected of having SMA.
Results
Overall screening results
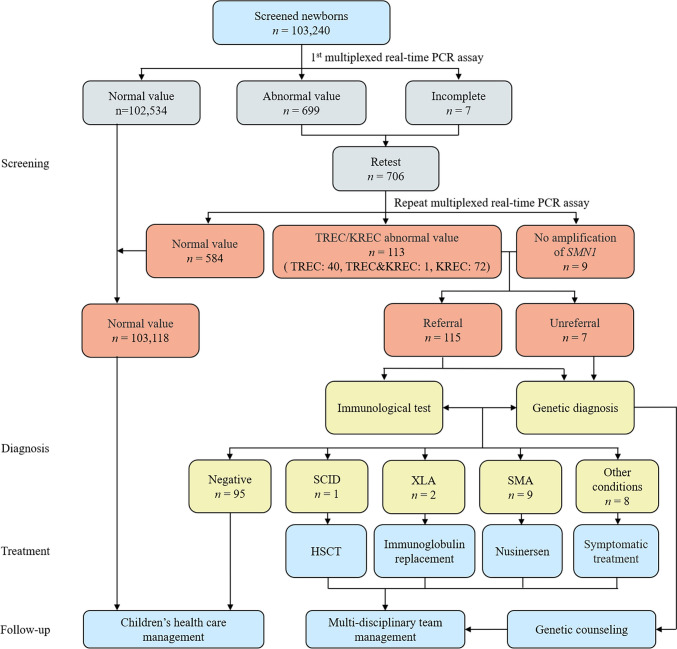
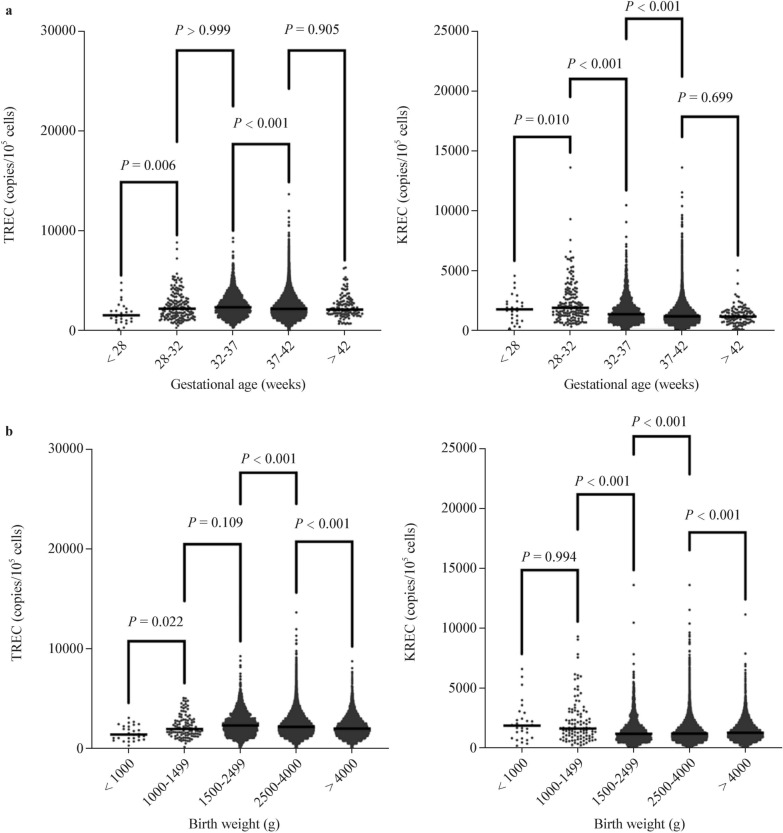
The screening diagnostic decision algorithm is illustrated in Fig. 1. Throughout the study period, comprehensive screening for SCID, XLA, and SMA was conducted on 103,240 newborns, covering 99.99% of all live births. Table 1 presents the baseline characteristics of the screened newborns, including sex, ethnicity, birth weight (BW), and gestational age (GA). The TREC and KREC values for these infants are shown in Fig. 2. The mean TREC levels were significantly lower in ultra-preterm infants (GA < 28 weeks) compared to other groups, with the highest values observed in very preterm infants (GA: 28–32 weeks). In contrast, the mean KREC levels peaked in very preterm infants and gradually declined with increasing GA, reaching their lowest in post-term infants (GA > 42 weeks). Similarly, the mean TREC levels were the lowest in very-low BW infants (BW < 1000 g) and highest in low BW infants (BW: 1500–2499 g). The mean KREC levels were highest in infants weighing 1000–1499 g, then rapidly declined with increasing weight, reaching their lowest in normal newborns (BW: 2500–4000 g).
Fig. 1.
Schematic diagram of the T-cell receptor excision circle (TREC)/kappa-deleting recombination excision circle (KREC)/survival motor neuron 1 (SMN1) newborn screening, diagnostic, and treatment integration system. PCR polymerase chain reaction, SCID severe combined immunodeficiency, XLA X-linked agammaglobulinemia, SMA spinal muscular atrophy, HSCT hematopoietic stem cell transplantation
Table 1.
Baseline characteristics of the screened newborns
| Characteristics | n | % |
|---|---|---|
| Gender | ||
| Male | 53,712 | 52.03 |
| Female | 49,528 | 47.97 |
| Gestational age (GA) | ||
| < 28 wk | 27 | 0.03 |
| ≥ 28 wk, < 32 wk | 162 | 0.16 |
| ≥ 32 wk, < 37 wk | 2215 | 2.15 |
| ≥ 37 wk, < 42 wk | 100,717 | 97.56 |
| ≥ 42 wk | 119 | 0.12 |
| Birth weight (BW), g | ||
| < 1000 | 29 | 0.03 |
| 1000–1499 | 110 | 0.11 |
| 1500–2499 | 2077 | 2.01 |
| 2500–4000 | 95,507 | 92.51 |
| > 4000 | 5517 | 5.34 |
| Total | 103,240 | 100.00 |
Fig. 2.
T-cell receptor excision circles (TRECs) and kappa-deleting recombination excision circles (KRECs) in newborns by a gestational age and b birth weight. TREC T-cell receptor excision circles, KREC kappa-deleting recombination excision circles
Among the newborns screened, 100,836 (97.67%) were born at full term, whereas 2,404 (2.33%) were delivered prematurely. A total of 699 samples with abnormal values and 7 incomplete samples from the initial analysis were subjected to repeated testing of the original DBS. After the retest, 122 samples (0.12%) had abnormal values: 40 with abnormal TREC, 72 with abnormal KREC, 1 with abnormalities in both TREC and KREC, and 9 with no amplification of SMN1. Of these, 117 were from full-term newborns (95.9%), while 5 were from premature newborns (4.1%). A total of 106 infants with abnormal TREC and/or KREC values underwent immunological testing, while the other 7 infants refused referral or were lost to follow-up. All nine infants who tested positive on SMA screening were referred for further evaluation. Overall, 115 of the 122 samples (94.26%) were successfully referred for the follow-up.
Screening performance
The 106 referred infants with abnormal TREC/KREC values underwent a series of diagnostic immunological tests, including flow cytometry, immunoglobulin level assessments, and WES. In contrast, the seven unreferred infants were tested with WES on DBS samples as probands. MLPA was performed on all infants with undetectable SMN1 cycle threshold (Ct) levels to assess the copy numbers of exons 7 and 8 in both SMN1 and SMN2. We identified one patient with SCID, two patients with XLA, and nine patients with SMA, one of whom also had Wiskott–Aldrich Syndrome (WAS). In addition, eight patients were diagnosed with other medical conditions, such as trisomy 21 (n = 1), 22q11.2 deletion syndrome (n = 4), Noonan syndrome (n = 1), 47,XYY syndrome (n = 1), and pancytopenia (n = 1). At the time of publication of this article, no new cases had been identified among those with negative screening results. Therefore, the positive predictive values (PPVs) of NBS were 2.44% (1/41) for SCID, 2.78% (2/72) for XLA, and 2.65% (3/113) for PID. The specificities for SCID and XLA were 99.96% and 99.93%, respectively, with both conditions demonstrating a sensitivity and negative predictive value (NPV) of 100%. The PPV, NPV, sensitivity, and specificity for SMA were all 100%. Our overall screening performance for SCID, XLA, and SMA yielded a combined PPV of 9.84%.
Confirmation and management
Primary immunodeficiency disorders
Table 2 presents comprehensive data on all individuals confirmed to have PIDs. Patient 1 (P1) exhibited abnormal TREC values in both the initial and repeat assays, while KREC values remained normal. Flow cytometric analysis of lymphocyte subsets revealed that the levels of CD3 + T cells (0.19×109/L), CD4 + T cells (0.11×109/L), CD19 + B cells (0.55×109/L), and CD16 + CD56 + natural killer (NK)-cells (0.38×109/L) were consistent with a T-B + NK + SCID phenotype. In addition, his serum immunoglobulin G (IgG) (5.0 g/L) and IgM (0.34 g/L) levels were within the normal ranges, whereas his serum IgA levels were low (0.01 g/L). Trio-WES identified a hemizygous mutation in intron 7 of the IL2RG gene (NM_000206.2: c.925-13 T > G) in both the patient and his mother. An identical mutation was detected in a DBS sample obtained from his deceased brother, who died from severe sepsis at 3 months of age. At 4 months old, P1 received a successful mismatched HSCT from his father. He is now nearly 2 years old and remains in excellent health.
Table 2.
Summary of patients with SCID and XLA
| Patients | Gender | GA (wk) | BW (g) | Age at DBS | TREC (copies/105 cells) | KREC (copies/105 cells) | Lymphocyte subsets (×109cells/L) | Immunoglobulin (g/L) | Mutation genes | Diagnosis |
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | M | 39+6 | 3830 | 3 d | 0 | 293 |
Total 1.12 CD3 + 0.19 (17.6%) CD4 + 0.11 (10.15%) CD8 + 0.01 (1.25%) CD19 + 0.55 (49.15%) CD56 + 0.38 (34.05%) |
IgG 5.0 IgA 0.01 IgM 0.34 |
IL2RG | T-B + NK + SCID |
| P2 | M | 40+3 | 3640 | 3 d | 2963 | 0 |
Total 4.80 CD3 + 4.49 (93.6%) CD19 + 0.03 (0.7%) |
IgG 7.2 IgA 0.01 IgM 0.02 |
BTK | XLA |
| P3 | M | 38+4 | 3100 | 4 d | 1799 | 0 |
Total 5.04 CD3 + 4.60 (91.2%) CD19 + 0.03 (0.55%) |
IgG 5.9 IgA 0.02 IgM 0.03 |
BTK | XLA |
M male, F female, GA gestational age, BW birth weight, DBS dried blood spot, TREC T-cell receptor excision circles, KREC kappa-deleting recombination excision circles, IgG immunoglobulin G, IgA immunoglobulin A, IgM immunoglobulin M, NK natural killer cell, SCID severe combined immunodeficiency, XLA X-linked agammaglobulinemia
P2 and P3 initially exhibited an absence of KRECs in the DBS samples, which persisted in subsequent tests. Immunological evaluations revealed a B-cell deficiency (P2: 0.7% CD19 + B cells; P3: 0.55% CD19 + B cells) and hypogammaglobulinemia, including deficiencies in IgM and IgA (P2: IgM 0.02 g/L, IgA 0.01 g/L; P3: IgM 0.03 g/L, IgA 0.01 g/L), while IgG levels were normal (P2: IgG 7.2 g/L, P3: IgG 5.9 g/L) (Table 2). The XLA diagnosis for the two patients was confirmed by detecting maternal semi-zygotic variants in the BTK gene: c.92 T > C (p.L31P) in one patient and c.599dup (p.P201Afs*5) in the other. At 6 months of age, both P2 and P3 began receiving immunological replacement therapy, consisting of intravenous immunoglobulin infusions (400–600 mg/kg every 3–4 weeks). This treatment was necessary due to a reduction in IgG levels caused by the depletion of maternal IgG and a severe decrease in the endogenous production of IgG (P2: IgG = 0.9 g/L, P3: IgG = 0.6 g/L).
Spinal muscular atrophy
Cases P4 to P12, all of whom had positive SMA-NBS results, received a confirmatory diagnosis and determination of the SMN2 copy number using MLPA. Among these patients, three had two copies of SMN2, while six had three copies, with no false-positive results observed.
P4, who had two copies of SMN2, was admitted to the newborn intensive care unit due to hypoxia at birth, along with hypotonia, joint contractures, and areflexia before the NBS results were available. Similarly, P5 and P6, who each had two copies of SMN2, exhibited hypotonia shortly after birth without any accompanying respiratory issues. Given the gravity of the illness, the projected outcome, and the financial burden of medical intervention, the families of P4, P5, and P6 opted to discontinue treatment, resulting in the tragic demise of all three infants within 2–6 months after birth.
At the time of diagnosis, none of the patients with three copies of SMN2 (P7–P12) showed any symptoms. P7 through P10 began nusinersen treatment at a median age of 85 days (range: 59–137 days) and have remained asymptomatic, achieving age-appropriate milestones at their most recent follow-ups. P11, a migrant, was referred to a local hospital for follow-up care but did not receive treatment due to financial constraints. At her 6-month follow-up, P11 exhibited signs of delayed motor development and slight muscle weakness.
It is worth noting that P12, who had three copies of SMN2, was symptomatic at the time of nusinersen treatment. P12 was diagnosed with both SMA and WAS. On day 2 after birth, he developed petechial rashes and severe eczema all over his body, along with thrombocytopenia and the presence of small platelets. Trio-WES revealed a hemizygous nonsense mutation in exon 7 of the WAS gene (NM_000377.3: c.629C > G, p.Ser210Ter), inherited from his mother. Despite these findings, he had normal lymphocyte and immunoglobulin levels, consistent with the normal TREC/KREC values. P12 underwent an umbilical cord blood stem cell transplantation from a human leukocyte antigen-matched unrelated donor at 12 months of age, and nusinersen treatment was started at 15 months of age. Unfortunately, despite these interventions, P12 exhibited delays and regression in motor development since 6 months of age. Although subsequent evaluations showed slight improvements in motor development, his motor milestones remained significantly delayed. At his most recent visit, he had not yet achieved the ability to sit independently (Table 3).
Table 3.
Summary of patients with SMA
| Patients | Gender | GA (wk) | BW (g) | Age at DBS | SMN1 Ct | SMN2 copies | Diagnosis | DMT | Age at treatment | Symptomatic at treatment | Age at last clinical | Symptomatic at last | Motor milestone achieved |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P4 | M | 39+3 | 3500 | 3 d | NA | 2 | SMA | None | / | / | Died at 2 mon | Y | None |
| P5 | M | 31+4 | 1525 | 11 d | NA | 2 | SMA | None | / | / | Died at 6 mon | Y | None |
| P6 | F | 39+2 | 3270 | 3 d | NA | 2 | SMA | None | / | / | Died at 3 mon | Y | None |
| P7 | F | 38+2 | 3350 | 5 d | NA | 3 | SMA | Nusinersen | 59 d | N | 1 y 7 mon | N | Walking independently (11 mon) |
| P8 | M | 40 | 3170 | 3 d | NA | 3 | SMA | Nusinersen | 137 d | N | 1 y 6 mon | N | Walking independently (12 mon) |
| P9 | M | 38+3 | 2860 | 2 d | NA | 3 | SMA | Nusinersen | 62 d | N | 1 y 4 mon | N | Walking independently (13 mon) |
| P10 | M | 39+3 | 3610 | 3 d | NA | 3 | SMA | Nusinersen | 82 d | N | 1 y 4 mon | N | Walking independently (13 mon) |
| P11 | M | 40 | 3320 | 3 d | NA | 3 | SMA | None | / | / | 6 mon | Y | Rolling; sitting with support |
| P12 | M | 39+6 | 3490 | 3 d | NA | 3 | SMA; WAS | Nusinersen | 1 y 3 mon | N | 2 y 3 mon | Y | Sitting with support |
NA no amplification, F female, M male, SMA spinal muscular atrophy, GA gestational age, BW birth weight, Ct cycle threshold, DMT disease-modifying treatment, Y yes, N no, WAS Wiskott–Aldrich Syndrome, NA not available
Other medical conditions
Four patients diagnosed with 22q11.2 deletion syndrome (P13–P16) were identified through the NBS program due to their low TREC levels. P17, a patient with trisomy 21, was born as a 31+4-week premature twin and conceived through in vitro fertilization. P18 was diagnosed with Noonan syndrome after trio-WES revealed a de novo heterozygous variant of the PTPN11 gene (NM_002834.5, c.922A > G). P19 initially presented with undetectable KREC copies, and further karyotyping confirmed a diagnosis of 47,XYY syndrome. Following referral, P19 was evaluated by a local immunologist, who found a normal immunophenotype along with normal lymphocyte and immunoglobulin levels. P20 was born with no measurable levels of TREC or KREC. Tragically, P20 died due to a combination of severe infection, respiratory failure, and pulmonary hemorrhage. Trio-WES did not reveal any genetic variations associated with immunodeficiency. Table 4 contains detailed information on all of these cases.
Table 4.
Summary of other medical conditions
| Patient | Gender | GA (wk) | BW (g) | Age at DBS | Diagnosis | TREC (copies/105 cells) | KREC (copies/105 cells) | Mutation genes |
|---|---|---|---|---|---|---|---|---|
| P13 | M | 41+1 | 2945 | 3 d | 22q11.2 deletion syndrome | 373 | 1463 | 2.68 Mb microdeletion in chromosome 22 long arm q11.2 (chr22:18,893,817–21,576,514×1) |
| P14 | F | 38+3 | 3250 | 7 d | 22q11.2 deletion syndrome | 287 | 185 | 2.92 Mb microdeletion in chromosome 22 long arm q11.2 (chr22:18,659,481–21,576,514×1) |
| P15 | F | 40 | 3450 | 4 d | 22q11.2 deletion syndrome | 60 | 1094 | 2.52 Mb microdeletion in chromosome 22 long arm q11.2 (chr22:18,893,817–21,414,846×1) |
| P16 | F | 39+6 | 3800 | 3 d | 22q11.2 deletion syndrome | 255 | 2988 | 2.52 Mb microdeletion in chromosome 22 long arm q11.2 (chr22:18,893,817–21,414,846×1) |
| P17 | F | 31+4 | 1470 | 3 d | Trisomy 21 | 1146 | 0 | Chromosome 21 triploid repeat |
| P18 | M | 40 | 3700 | 3 d | Noonan syndrome | 314 | 4054 | De novo variation of PTPN11, c.922A > G (p.N308D), LP |
| P19 | M | 40 | 3400 | 7 d | 47,XYY syndrome | 5017 | 0 | 47, XYY |
| P20 | F | 38+3 | 3450 | 4 d | Pancytopenia | 0 | 0 | No definite pathogenic variation associated with phenotype was detected |
M mail, F female, GA gestational age, BW birth weight, DBS dried blood spot, TREC T-cell receptor excision circles, KREC kappa-deleting recombination excision circles
In another case, the initial KREC score was slightly below the specified threshold. Subsequent investigations revealed that the infant’s mother had systemic lupus erythematosus (SLE) but was in remission after having undergone treatment. She was stable during pregnancy and did not receive any immunosuppressants. Immunological evaluation and follow-up reviews revealed a normal immunophenotype for the infant, and WES did not identify any gene variants associated with immunodeficiency. Consequently, this case is not included in Table 4.
Discussion
Early screening, accurate diagnosis, and timely intervention for significant birth abnormalities are critical for improving disease prognosis and lowering medical expenses [25]. Both SCID and SMA are included in the recommended list of essential disorders for newborn screening by the Health Resources & Services Administration [21]. Currently, an increasing number of countries are either implementing or considering the inclusion of PID and SMA screening in their universal NBS programs.
In this study, the prevalence rates of SCID, XLA, and SMA in the Chinese population were estimated from the screening data. The occurrence rate of SCID was determined to be 1 in 103,240, which is significantly lower than that reported in Israel (1 in 22,159) [26], Wisconsin, USA (1 in 41,539) [27], and Taiwan of China (1 in 53,195) [28]. The reported incidence of XLA varies considerably by region and ethnicity: 1 in 200,000 live births in Switzerland [29]; 1 in 20,000,000 to 1 in 10,000,000 in Spain [30]; 1 in 285,000 to 1 in 100,000 in Norway [31]; 1 in 160,000 to 1 in 27,000 in Israel [32]; and 1 in 379,000 in the USA [33]. However, our study suggests that the prevalence may be higher in Chinese individuals, with an estimated rate of 1 in 51,620. Due to the relatively low occurrence of both SCID and XLA, it is necessary to obtain more accurate prevalence data by implementing generalized screening programs. The incidence of SMA in this study was 1 in 11,471, which is comparable to prevalence observed in other regions: Australia (1 in 10,390) [34], Taiwan, China (1 in 17,181) [35], and New York, USA (1 in 19,117) [36]. In addition, this figure is marginally lower than the prevalence reported in one of our previous multicenter studies (1 in 9,788) [37]. Owing to the extended screening period, larger sample size, and more consistent management of the single-center screening process across the entire region, the incidence rates reported in this study are likely to be more accurate.
Based on the current cut-off values for TREC and KREC, 113 samples (0.11%) below the cut-off value were considered abnormal TREC/KREC values. Among these patients, P20, who had pancytopenia, exhibited abnormal TREC and KREC values. Therefore, the positive rate for the TREC-alone assay was 0.03% (41/103,240), while the positive rate for the KREC-alone assay was 0.07% (73/103,240). This rate is comparable to figures reported in Sweden (0.10%; 93/89,462) [10] and Seville (0.097%; 5/5,160) [11], and is slightly higher than that reported in Japan (0.037%; 39/105,419) [12]. The rates reported here do not indicate a significant increase compared to other TREC-alone screening rates [38, 39]. In this study, we identified one patient with SCID and two patients with XLA, yielding a PPV of 2.44% for SCID and 2.78% for XLA. The overall PPV for PIDs was 2.65%. Considering the incidence of XLA observed in this study, a slight increase in the positivity rate for KREC is considered acceptable. However, higher cut-off values may lead to lower positive rates. While this approach could help reduce false positives, it may also risk overlooking cases of SCID or XLA, although the cases with undetectable TREC or KREC levels in our study still can be identified. In addition, it may also fail to identify other immunological disorders. Previous research has identified maternal immunosuppression, preterm birth [40], and congenital heart problems [41] as the primary factors contributing to T-cell and/or B-cell lymphopenia in infants. We found that TREC levels increased most rapidly when BW increased from < 1000 g to 1000–2499 g and GA increased from < 28 weeks to 28–32 weeks, corresponding to periods of rapid thymic development and increased T-cell production [42]. Conversely, the population median KREC levels decreased most rapidly as BW increased from 1000–1499 g to 1500–2499 g, and as GA progressed from 28–32 weeks to 32–37 weeks. This decline in KREC levels is attributed to B-lymphocyte production by the liver during the fetal period. As the liver rapidly increases in size and weight, active B-cell production occurs, diluting KREC concentrations in the peripheral blood [43, 44]. Analyzing the factors influencing these correlations can provide a theoretical basis for optimizing cut-off values, thereby ensuring sensitivity, reducing the false-positive rate, and improving screening efficiency.
Studies have shown that KREC analysis is valuable for classifying SCID and identifying B-cell deficiencies. KRECs can also be useful for detecting other PIDs, such as late-onset adenosine deaminase deficiency deficiencies, Nijmegen breakage syndrome, and other XLA-like disorders [14, 45]. While we did not detect other types of PIDs, our analysis revealed one case of SCID with undetectable TRECs (TREC; 0 copies/105 cells) and two cases of XLA with undetectable KRECs (KREC; 0 copies/105 cells). For instance, P1, who exhibited the T-B + NK + SCID phenotype, had abnormal TREC values and normal KREC values, which corroborates the role of KREC in SCID classification. In addition to SCID and XLA, our screening program identified eight patients with other medical conditions, including 22q11.2 deletion syndrome, Noonan syndrome, trisomy 21, 47,XYY syndrome, and pancytopenia of unknown etiology. 22q11.2 deletion syndrome, which is associated with thymic stromal deficiency leading to T-cell immunodeficiency, may sometimes exhibit low TREC levels while generally maintaining normal KREC levels [46]. In our study, all four patients with this condition presented with abnormal TREC and normal KREC values. Noonan syndrome, identified by abnormal TREC levels in our study, may be associated with a common phenotype of lymphatic dysplasia [47]. Thus, multiplexed TREC/KREC assays are both feasible and sensitive for the detection of T- and/or B-cell lymphopenia. The addition of KREC to our program did not affect the overall screening performance; moreover, it played a valuable role in predicting SCID subtypes. In addition, KREC screening is essential for the identification of trisomy 21 syndrome [48], and may also be valuable for detecting rare diseases such as 47,XYY syndrome [49].
Despite advances in genetic screening technologies and therapeutic methodologies that have accelerated the implementation of pilot SMA screening and led to its incorporation into routine neonatal screening programs in certain areas, global implementation remains limited. As of 2021, newborn screening for SMA has been implemented in only nine countries, representing less than 2% of the global newborn population [50]. Several pilot trials have expanded SMA-NBS by incorporating it into existing SCID-NBS panels. Our study not only addresses the lack of data in mainland China but also establishes the incorporation of measuring TREC/KREC/SMN1 into our NBS program. Semi-quantitative detection of SMN1 exon 7 by real-time PCR, used in NBS for SMA due to the homozygous deletion of exon 7, is a genetic screening method with a theoretical PPV of 100%. However, due to various constraints, including sample quality and primer design, the detection of exon 7 of the SMN1 gene has been associated with a few false-positive results in prior investigations [34]. Our study did not identify any false positives, and SMN1 Ct values were undetectable in all patients. This resulted in a PPV, sensitivity, and specificity of 100%, consistent with findings from most independent SMA studies, providing further evidence of the reliability and feasibility of our multiplex TREC/KREC/SMN1 assays.
NBS provides a critical opportunity for pre-symptomatic diagnosis, allowing for early intervention and management of conditions. All three patients with PID in our study were promptly treated and closely followed up, including receiving genetic counseling. They were in excellent physical condition, had avoided recurrent infections, and experienced outcomes similar to those observed in P1’s brother. Previous studies have shown that children with SMA who begin treatment before symptoms appear have significantly higher survival rates, better respiratory and feeding support, and approved attainment of motor milestones compared to those who start treatment after the onset of symptoms [15]. In this study, the four children with three copies of SMN2 who received nusinersen treatment before the onset of symptoms remained asymptomatic during the follow-up period, achieving age-appropriate motor milestones. Both untreated cases with three SMN2 copies developed symptoms at 6 months of age and exhibited a tendency toward deterioration. P12 was a patient with combined SMA and WAS. During communication with the doctor, the parents were concerned that the subsequent HSCT would interfere with the effectiveness of the prior nusinersen treatment. Despite clear explanations from the medical team HSCT and nusinersen treatment could be administered concurrently without conflict, the parents insisted on prioritizing HSCT and postponing nusinersen treatment. P12 developed symptoms while waiting for a well-matched donor. Although his head control and muscle strength improved with symptomatic treatment using nusinersen, he remained incapable of sitting independently. We hypothesized that the clinical outcome of P12 was the result of multiple factors, including potential interference of WAS, implementation of HSCT, and the delayed initiation of nusinersen treatment. Despite the brief follow-up period and the potential influence of other medical conditions, the findings of this study align with prior SMA-NBS pilot research, demonstrating that children who received pre-symptomatic treatment through NBS showed substantial improvements in motor function [15, 36]. Unfortunately, all three infants with two SMN2 copies discontinued treatment, resulting in inconclusive evidence regarding the impact of NBS on their prognosis in this study.
Previous studies have suggested that SCID-NBS is cost-effective [52–54]. While the cost-effectiveness of NBS for SMA has been disputed owing to the high cost of treatment drugs, comprehensive economic analyses from various nations indicate that NBS for SMA is both more cost-effective and clinically beneficial, compared to scenarios without screening [55, 56]. Adding SMA-NBS to SCID-NBS incurs only a minimal additional cost when using a multiplex qPCR assay, which further reduces the overall cost of SMA screening. Economic analysis of NBS programs in Australia for SCID and SMA demonstrates significant health and financial benefits [57]. The combined screening approach is projected to yield 95 quality-adjusted life-years (QALYs) per 100,000 newborns screened, while simultaneously generating cost savings of $8.6 million. These data strongly support the cost-effectiveness of implementing a joint NBS program for SCID and SMA [51]. Tesorero et al. [58] integrated sickle cell disease screening into the original NBS panel, suggesting that a multiplex real-time PCR assay could be integrated with the current screening tests to include additional target diseases through the analysis of disease-associated DNA molecules or genes. In the present study, we used a multiplex real-time PCR assay to minimize the cost of NBS for SCID, SMA, and XLA. Given that disease incidence significantly impacts cost-effectiveness, the higher prevalence of XLA in the Chinese population identified in our study underscores the significance and potential cost of implementing NBS for this condition. The timely interventions made possible by our screening significantly improved the prognosis of affected infants. As the main factor in the cost-effectiveness of SMA-NBS, the recent reduction in the price of nusinersen in mainland China to 33,000 RMB has considerably lowered overall treatment costs. However, related studies have not yet been performed in China. Considering disease incidence, the reliability of screening techniques, treatment accessibility, and improved prognosis, we anticipate that combined TREC/KREC/SMN1 screening will not only provide long-term cost savings for the government but will also improve and save lives.
Despite limitations such as reduced specificity, the need for additional validation, and the potential for false-positive or false-negative results, molecular screening using multiplex real-time PCR assays offers a convenient and cost-effective complementary approach to traditional biochemical screening in public health NBS programs. WES is crucial for identifying children with other genetic diseases, such as 22q11.2 deletion syndrome and atypical biochemical markers. Recent advancements in genetic testing technology have led to increased attention on applying next-generation sequencing (NGS) to NBS. NGS offers significant advantages, including high-throughput capabilities and shortened diagnostic times for patients with complex medical conditions [59]. In our study, a subset of 26 neonates with abnormal TREC/KREC screening results were found to carry likely pathogenic mutations as identified by WES. However, because these patients did not exhibit phenotypic confirmation at the time of referral, they were excluded from the list of confirmed patients. These cases continue to be monitored, and this issue will be addressed in subsequent follow-ups. Therefore, employing NGS as a first-tier test for NBS could significantly increase the costs of both screening and confirmatory testing and increase the requirements for gene interpretation, genetic counseling, and long-term management [60, 61]. Building on insights from our previous multicenter study [62], we have begun implementing NGS-NBS as an optional component in our NBS program in Zhejiang Province. We believe that biochemical combined genetic screening represents a promising future trend in NBS.
In summary, this study employed real-time PCR to concurrently screen for SCID, SMA, and XLA in a single assay, demonstrating the clinical effectiveness of this combined screening approach in a large cohort of newborns. In addition to validating the diagnoses of the associated diseases and identifying patients with specific syndromes, we developed an integrated system for early neonatal screening, diagnosis, treatment, and follow-up that is ready for widespread adoption and implementation. This system encompasses the creation of a comprehensive epidemiological database in mainland China, the establishment of a disease cohort, the integration of multi-disciplinary team treatments, and the standardization of clinical pathways. Negative screening results should also be managed through comprehensive pediatric healthcare services established in China [63], which provide continuous healthcare services for all children from birth. For asymptomatic children, genetic counseling and health management can be offered based on genetic analysis and genotype–phenotype correlations. The establishment of a standardized screening system is expected to increase the utility of NBS.
Author contributions
CC: formal analysis, investigation, data curation, writing–original draft, writing–review and editing. ZC: methodology, software. WDW: validation. WBY: writing–review and editing. XR: data curation. HXL: investigation. YX: investigation. GZG: conceptualization, supervision. YRL: conceptualization, supervision, project administration.
Funding
This work was sponsored by the National Key Research and Development Program of China (No. 2022YFC2703401).
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
No financial or non-financial benefits have been received or will be received from any party related directly or indirectly to the subject of this article.
Ethical approval
This study was approved by the Institutional Review Board of the Ethics Committee in Children’s Hospital, Zhejiang University School of Medicine (approval number 2021-IRB-036). Informed consent to infants in the study has been obtained from their parent or legal guardian.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhi-Gang Gao, Email: ebwk@zju.edu.cn.
Ru-Lai Yang, Email: chsczx@zju.edu.cn.
References
- 1.Currier R, Puck JM. SCID newborn screening: what we’ve learned. J Allergy Clin Immunol. 2021;147:417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu X, Wang Z, Ye J, Han L, Qiu W. Newborn screening in China: phenylketonuria, congenital hypothyroidism and expanded screening. Ann Acad Med Singap. 2008;37:107–14. [PubMed] [Google Scholar]
- 3.Biggs CM, Haddad E, Issekutz TB, Roifman CM, Turvey SE. Newborn screening for severe combined immunodeficiency: a primer for clinicians. CMAJ. 2017;189:E1551–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazenberg MD, Otto SA, de Pauw ES, Roelofs H, Fibbe WE, Hamann D, et al. T-cell receptor excision circle and T-cell dynamics after allogeneic stem cell transplantation are related to clinical events. Blood. 2002;99:3449–53. [DOI] [PubMed] [Google Scholar]
- 5.McDonald-McGinn DM, Sullivan KE, Marino B, Philip N, Swillen A, Vorstman JA, et al. 22q11.2 deletion syndrome. Nat Rev Dis Primers. 2015;1:15071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dvorak CC, Cowan MJ, Logan BR, Notarangelo LD, Griffith LM, Puck JM, et al. The natural history of children with severe combined immunodeficiency: baseline features of the first fifty patients of the primary immune deficiency treatment consortium prospective study 6901. J Clin Immunol. 2013;33:1156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pai SY, Logan BR, Griffith LM, Buckley RH, Parrott RE, Dvorak CC, et al. Transplantation outcomes for severe combined immunodeficiency, 2000–2009. N Engl J Med. 2014;371:434–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Zelm MC, Szczepanski T, van der Burg M, van Dongen JJ. Replication history of B lymphocytes reveals homeostatic proliferation and extensive antigen-induced B cell expansion. J Exp Med. 2007;204:645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakagawa N, Imai K, Kanegane H, Sato H, Yamada M, Kondoh K, et al. Quantification of κ-deleting recombination excision circles in Guthrie cards for the identification of early B-cell maturation defects. J Allergy Clin Immunol. 2011;128:223-5.e2. [DOI] [PubMed] [Google Scholar]
- 10.Zetterström RH, Barbaro M, Ohlsson A, Borte S, Jonsson S, Winiarski J, et al. Newborn screening for primary immune deficiencies with a TREC/KREC/ACTB triplex assay—a three-year pilot study in Sweden. Int J Neonat Screen. 2017;3:11. [Google Scholar]
- 11.de Felipe B, Olbrich P, Lucenas JM, Delgado-Pecellin C, Pavon-Delgado A, Marquez J, et al. Prospective neonatal screening for severe T- and B-lymphocyte deficiencies in Seville. Pediatr Allergy Immunol. 2016;27:70–7. [DOI] [PubMed] [Google Scholar]
- 12.Kimizu T, Nozaki M, Okada Y, Sawada A, Morisaki M, Fujita H, et al. Multiplex real-time PCR-based newborn screening for severe primary immunodeficiency and spinal muscular Atrophy in Osaka, Japan: Our Results after 3 Years. Genes (Basel). 2024;15:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pal Singh S, Dammeijer F, Hendriks RW. Role of Bruton’s tyrosine kinase in B cells and malignancies. Mol Cancer. 2018;17:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King JR, Hammarström L. Newborn screening for primary immunodeficiency diseases: history, current and future practice. J Clin Immunol. 2018;38:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercuri E, Sumner CJ, Muntoni F, Darras BT, Finkel RS. Spinal muscular atrophy. Nat Rev Dis Primers. 2022;8:52. [DOI] [PubMed] [Google Scholar]
- 16.Keinath MC, Prior DE, Prior TW. Spinal muscular atrophy: mutations, testing, and clinical relevance. Appl Clin Genet. 2021;14:11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–65. [DOI] [PubMed] [Google Scholar]
- 18.Strauss KA, Farrar MA, Muntoni F, Saito K, Mendell JR, Servais L, et al. Onasemnogene abeparvovec for presymptomatic infants with three copies of SMN2 at risk for spinal muscular atrophy: the Phase III SPR1NT trial. Nat Med. 2022;28:1390–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corey DR. Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nat Neurosci. 2017;20:497–9. [DOI] [PubMed] [Google Scholar]
- 20.Paik J. Risdiplam: a review in spinal muscular atrophy. CNS Drugs. 2022;36:401–10. [DOI] [PubMed] [Google Scholar]
- 21.Health Resources & Services Administration (HRSA). Recommended uniform screening panel. In: Advisory Committee on Heritable Disorders in Newborns and Children. 2024. https://www.hrsa.gov/advisory-committees/ heritable-disorders/rusp. Accessed 10 Jan 2024.
- 22.Simard LR, Bélanger MC, Morissette S, Wride M, Prior TW, Swoboda KJ. Preclinical validation of a multiplex real-time assay to quantify SMN mRNA in patients with SMA. Neurology. 2007;68:451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor JL, Lee FK, Yazdanpanah GK, Staropoli JF, Liu M, Carulli JP, et al. Newborn blood spot screening test using multiplexed real-time PCR to simultaneously screen for spinal muscular atrophy and severe combined immunodeficiency. Clin Chem. 2015;61:412–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutierrez-Mateo C, Timonen A, Vaahtera K, Jaakkola M, Hougaard DM, Bybjerg-Grauholm J, et al. Development of a multiplex real-time PCR assay for the newborn screening of SCID, SMA, and XLA. Int J Neonat Screen. 2019;5:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCandless SE, Wright EJ. Mandatory newborn screening in the United States: history, current status, and existential challenges. Birth Defects Res. 2020;112:350–66. [DOI] [PubMed] [Google Scholar]
- 26.Rechavi E, Lev A, Simon AJ, Stauber T, Daas S, Saraf-Levy T, et al. First year of Israeli newborn screening for severe combined immunodeficiency-clinical achievements and insights. Front Immunol. 2017;8:1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verbsky JW, Baker MW, Grossman WJ, Hintermeyer M, Dasu T, Bonacci B, et al. Newborn screening for severe combined immunodeficiency; the Wisconsin experience (2008–2011). J Clin Immunol. 2012;32:82–8. [DOI] [PubMed] [Google Scholar]
- 28.Chien Y-H, Yu H-H, Lee N-C, Ho H-C, Kao S-M, Lu M-Y, et al. Newborn screening for severe combined immunodeficiency in Taiwan. Int J Neonat Screen. 2017;3:16. [Google Scholar]
- 29.Ryser O, Morell A, Hitzig WH. Primary immunodeficiencies in Switzerland: first report of the national registry in adults and children. J Clin Immunol. 1988;8:479–85. [DOI] [PubMed] [Google Scholar]
- 30.Matamoros Florí N, Mila Llambi J, Español Boren T, Raga Borja S, Fontan CG. Primary immunodeficiency syndrome in Spain: first report of the National Registry in Children and Adults. J Clin Immunol. 1997;17:333–9. [DOI] [PubMed] [Google Scholar]
- 31.Stray-Pedersen A, Abrahamsen TG, Frøland SS. Primary immunodeficiency diseases in Norway. J Clin Immunol. 2000;20:477–85. [DOI] [PubMed] [Google Scholar]
- 32.Broides A, Nahum A, Mandola AB, Rozner L, Pinsk V, Ling G, et al. Incidence of typically severe primary immunodeficiency diseases in consanguineous and non-consanguineous populations. J Clin Immunol. 2017;37:295–300. [DOI] [PubMed] [Google Scholar]
- 33.Winkelstein JA, Marino MC, Lederman HM, Jones SM, Sullivan K, Burks AW, et al. X-linked agammaglobulinemia: report on a United States registry of 201 patients. Medicine (Baltimore). 2006;85:193–202. [DOI] [PubMed] [Google Scholar]
- 34.Kariyawasam DST, Russell JS, Wiley V, Alexander IE, Farrar MA. The implementation of newborn screening for spinal muscular atrophy: the Australian experience. Genet Med. 2020;22:557–65. [DOI] [PubMed] [Google Scholar]
- 35.Chien YH, Chiang SC, Weng WC, Lee NC, Lin CJ, Hsieh WS, et al. Presymptomatic diagnosis of spinal muscular atrophy through newborn screening. J Pediatr. 2017;190:124-9.e1. [DOI] [PubMed] [Google Scholar]
- 36.Lee BH, Deng S, Chiriboga CA, Kay DM, Irumudomon O, Laureta E, et al. Newborn screening for spinal muscular atrophy in New York state: clinical outcomes from the first 3 years. Neurology. 2022;99:e1527–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Y, Lin CH, Yin X, Zhu L, Yang J, Shen Y, et al. Newborn screening for spinal muscular atrophy in China using DNA mass spectrometry. Front Genet. 2019;10:1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogel BH, Bonagura V, Weinberg GA, Ballow M, Isabelle J, DiAntonio L, et al. Newborn screening for SCID in New York State: experience from the first two years. J Clin Immunol. 2014;34:289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Audrain M, Thomas C, Mirallie S, Bourgeois N, Sebille V, Rabetrano H, et al. Evaluation of the T-cell receptor excision circle assay performances for severe combined immunodeficiency neonatal screening on Guthrie cards in a French single centre study. Clin Immunol. 2014;150:137–9. [DOI] [PubMed] [Google Scholar]
- 40.Melville JM, Moss TJ. The immune consequences of preterm birth. Front Neurosci. 2013;7:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelly B, Mohanakumar S, Hjortdal VE. Diagnosis and management of lymphatic disorders in congenital heart disease. Curr Cardiol Rep. 2020;22:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tangshewinsirikul C, Panburana P. Sonographic measurement of fetal thymus size in uncomplicated singleton pregnancies. J Clin Ultrasound. 2017;45:150–9. [DOI] [PubMed] [Google Scholar]
- 43.Duijts L, Bakker-Jonges LE, Labout JA, Jaddoe VW, Hofman A, Steegers EA, et al. Fetal growth influences lymphocyte subset counts at birth: the generation R Study. Neonatology. 2009;95:149–56. [DOI] [PubMed] [Google Scholar]
- 44.Srisupundit K, Piyamongkol W, Tongprasert F, Luewan S, Tongsong T. Reference range of fetal splenic circumference from 14 to 40 weeks of gestation. Arch Gynecol Obstet. 2011;283:449–53. [DOI] [PubMed] [Google Scholar]
- 45.Kwan A, Puck JM. History and current status of newborn screening for severe combined immunodeficiency. Semin Perinatol. 2015;39:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Froňková E, Klocperk A, Svatoň M, Nováková M, Kotrová M, Kayserová J, et al. The TREC/KREC assay for the diagnosis and monitoring of patients with DiGeorge syndrome. PLoS ONE. 2014;9:e114514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zenker M, Edouard T, Blair JC, Cappa M. Noonan syndrome: improving recognition and diagnosis. Arch Dis Child. 2022;107:1073–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eissa E, Afifi HH, Abo-Shanab AM, Thomas MM, Taher MB, Kandil R, et al. Importance of TREC and KREC as molecular markers for immunological evaluation of down syndrome children. Sci Rep. 2023;13:15445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gul Y, Kapaklı H, Aytekin SE, Guner ŞN, Keles S, Zamani AG, et al. Evaluation of immunological abnormalities in patients with rare syndromes. Cent Eur J Immunol. 2022;47:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dangouloff T, Vrščaj E, Servais L, Osredkar D. SMA NBS World Study Group. Newborn screening programs for spinal muscular atrophy worldwide: where we stand and where to go. Neuromuscul Disord. 2021;31:574–82. [DOI] [PubMed] [Google Scholar]
- 51.Shih STF, Keller E, Wiley V, Farrar MA, Wong M, Chambers GM. Modelling the cost-effectiveness and budget impact of a newborn screening program for spinal muscular atrophy and severe combined immunodeficiency. Int J Neonat Screen. 2022;8:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bessey A, Chilcott J, Leaviss J, de la Cruz C, Wong R. A cost-effectiveness analysis of newborn screening for severe combined immunodeficiency in the UK. Int J Neonat Screen. 2019;5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shih STF, Keller E, Wiley V, Wong M, Farrar MA, Chambers GM. Economic evaluation of newborn screening for severe combined immunodeficiency. Int J Neonat Screen. 2022;8:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding Y, Thompson JD, Kobrynski L, Ojodu J, Zarbalian G, Grosse SD. Cost-effectiveness/cost-benefit analysis of newborn screening for severe combined immune deficiency in Washington State. J Pediatr. 2016;172:127–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weidlich D, Servais L, Kausar I, Howells R, Bischof M. Cost-effectiveness of newborn screening for spinal muscular atrophy in England. Neurol Ther. 2023;12:1205–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jalali A, Rothwell E, Botkin JR, Anderson RA, Butterfield RJ, Nelson RE. Cost-effectiveness of nusinersen and universal newborn screening for spinal muscular atrophy. J Pediatr. 2020;227:274-80.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Velikanova R, van der Schans S, Bischof M, van Olden RW, Postma M, Boersma C. Cost-effectiveness of newborn screening for spinal muscular Atrophy in The Netherlands. Value Health. 2022;25:1696–704. [DOI] [PubMed] [Google Scholar]
- 58.Tesorero R, Janda J, Hörster F, Feyh P, Mütze U, Hauke J, et al. A high-throughput newborn screening approach for SCID, SMA, and SCD combining multiplex qPCR and tandem mass spectrometry. PLoS One. 2023;18:e0283024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smon A, Repic Lampret B, Groselj U, Zerjav Tansek M, Kovac J, Perko D, et al. Next generation sequencing as a follow-up test in an expanded newborn screening programme. Clin Biochem. 2018;52:48–55. [DOI] [PubMed] [Google Scholar]
- 60.Remec ZI, Trebusak Podkrajsek K, Repic Lampret B, Kovac J, Groselj U, Tesovnik T, et al. Next-generation sequencing in newborn screening: a review of current state. Front Genet. 2021;12:662254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang RL, Qian GL, Wu DW, Miao JK, Yang X, Wu BQ, et al. A multicenter prospective study of next-generation sequencing-based newborn screening for monogenic genetic diseases in China. World J Pediatr. 2023;19:663–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deng B, Hua J, Zhou Y, Zhan D, Zhu L, Zhan Y, et al. Legionella pneumonia complicated with rhabdomyolysis and acute kidney injury diagnosed by metagenomic next-generation sequencing: a case report. World J Emerg Med. 2023;14:322–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He Z, Xu T. China’s actions to achieve universal health coverage for children. China CDC Wkly. 2022;4:802–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.