Abstract
The early evolution of eukaryotes and their adaptations to low-oxygen environments are fascinating open questions in biology. Genome-scale data from novel eukaryotes, and particularly from free-living lineages, are the key to answering these questions. The Parabasalia are a major group of anaerobic eukaryotes that form the most speciose lineage of Metamonada. The most well-studied are parasitic parabasalids, including Trichomonas vaginalis and Tritrichomonas foetus, but very little genome-scale data are available for free-living members of the group. Here, we sequenced the transcriptome of Pseudotrichomonas keilini, a free-living parabasalian. Comparative genomic analysis indicated that P. keilini possesses a metabolism and gene complement that are in many respects similar to its parasitic relative T. vaginalis and that in the time since their most recent common ancestor, it is the T. vaginalis lineage that has experienced more genomic change, likely due to the transition to a parasitic lifestyle. Features shared between P. keilini and T. vaginalis include a hydrogenosome (anaerobic mitochondrial homolog) that we predict to function much as in T. vaginalis and a complete glycolytic pathway that is likely to represent one of the primary means by which P. keilini obtains ATP. Phylogenomic analysis indicates that P. keilini branches within a clade of endobiotic parabasalids, consistent with the hypothesis that different parabasalid lineages evolved toward parasitic or free-living lifestyles from an endobiotic, anaerobic, or microaerophilic common ancestor.
Keywords: anaerobic eukaryotes, eukaryotic evolution, protist transcriptome, hydrogenosome
Significance.
Eukaryotes are enormously diverse, from animals, plants, and fungi to a range of single-celled forms. But many eukaryotic lineages are poorly sampled by genome-scale sequencing projects, resulting in a paucity of data with which to study eukaryotic evolution. Here, we sequenced and analyzed the transcriptome of P. keilini, a free-living, anaerobic eukaryote that is related to the important parasite Trichomonas vaginalis. Comparing their gene complements enables us to distinguish features that are common to the broader group—the Parabasalians—from those specific to Trichomonas during the evolution of parasitism. The new data also provide a basis for further studies of divergent and fascinating eukaryotic lineages.
Introduction
Animals, plants, and fungi are well-studied by biologists, and genomes for many lineages are now available. However, most eukaryotic diversity is microbial, and many groups are poorly sampled by genomics and transcriptomics (Sibbald and Archibald 2017). Among unicellular lineages, parasites are best represented by sequencing efforts. These include the parabasalid Trichomonas vaginalis, the causative agent of the most common nonviral sexually transmitted disease, trichomoniasis (Donné 1836). T. vaginalis infects the genitourinary tracts of 187 million people every year around the world (Menezes et al. 2016) and increases the transmission rate of human immunodeficiency virus (Petrin et al. 1998). There is therefore great interest in the biology of T. vaginalis and also in the evolution of parasitism in the whole Parabasalia.
There are over 450 described species of parabasalids. The majority are endobiotic, including parasites such as T. vaginalis and its relatives that infect other animals (Yamin and Ma 1979; Brugerolle and Lee 2000; Adl et al. 2007; Cepicka et al. 2010). The first free-living parabasalid to be discovered was P. keilini (Bishop 1935, 1939). P. keilini was originally isolated from pond water in Lincolnshire, UK, and later isolated from mangrove sediments in Japan, a lake in Cyprus (Yubuki et al. 2010); from freshwater sediments in Azerbaijan, sulfurous freshwater spring, and brackish sediments in Greece; and from inland salt marshes in Spain (Céza et al. 2022). As parabasalids are thought to be ancestrally endobiotic (Čepička et al. 2017), several questions naturally arise about a free-living member of the group: Did the free-living lifestyle of P. keilini evolve secondarily, or are transitions between free-living and host-associated lifestyles more common than anticipated in parabasalids? Answering these questions fully will require genome-scale data from closely related free-living and host-associated members of the group, alongside further study of their biology. As a step toward providing this knowledge base, we carried out transcriptome sequencing and bioinformatics analyses.
Here, we present a largely complete transcriptome dataset of P. keilini, a free-living anaerobic parabasalid isolated from salt marsh sediment in Spain. We perform phylogenetic analyses to place Pseudotrichomonas in the eukaryotic tree and use comparative genomics to trace gene content and hydrogenosome evolution within Parabasalia. We envisage that the P. keilini transcriptome will be of use in further analyses of early eukaryote genomic and metabolic evolution.
Transcriptome of P. keilini
We isolated and cultured P. keilini (supplementary fig. S1), then extracted RNA and sequenced it on the Illumina MiSeq and NovaSeq6000 platforms (see supplementary Methods, Supplementary Material online); reads were deposited at the NCBI Short Read Archive (PRJNA884676). Transcriptome reads were assembled into 137,389 transcripts using Trinity RNA-Seq (Haas et al. 2013), and predicted proteins were obtained using TransDecoder. We performed basic local alignment search tool-based filtering to remove contaminants from the assembly (see supplementary Methods, Supplementary Material online, supplementary table S1, Supplementary Material online), and clustered identical overlapping proteins to obtain 18,851 nonredundant predicted proteins, of which functions could be predicted for 12,811.
To evaluate the completeness of the P. keilini transcriptome, we conducted a benchmarking universal single copy orthologs (BUSCO) analysis (Simão et al. 2015) of P. keilini using a set of conserved eukaryotic genes and compared it to BUSCO values for other published Metamonada (supplementary table S2, Supplementary Material online). These datasets (a mixture of transcriptomes and genomes) had a median completeness of 47% of BUSCOs, with a 95% confidence interval ranging from 35.29 to 46.71. Our P. keilini transcriptome includes complete protein-coding genes for 47% of BUSCOs, the median value, and the same as some other free-living metamonads such as Anaeramoeba flamelloides. By comparison, the completely sequenced T. vaginalis genome encodes 53% of BUSCOs. Overall, we concluded that we had produced a near-complete transcriptome that is likely to be informative about the evolution and metabolism of P. keilini.
P. keilini branches within a clade of parasitic parabasalids
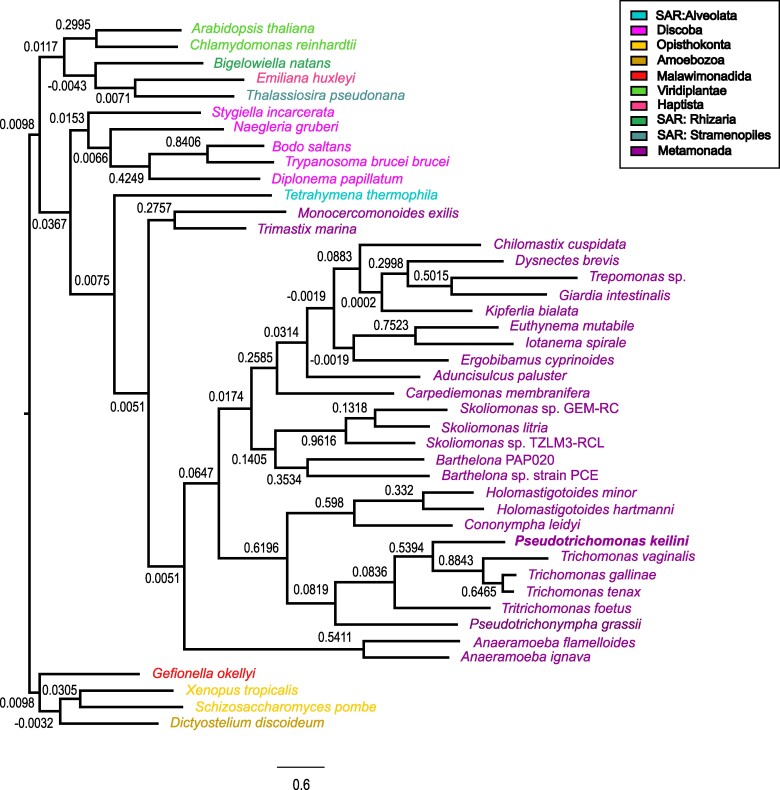
To investigate the relationship of P. keilini to other parabasalids and metamonads, we used SpeciesRax (Morel et al. 2022) to infer a species tree including P. keilini, its closest parabasalid relatives, and a representative sample of other eukaryotic lineages, using 13,346 gene families clustered from 43 genomes (Fig. 1) using Broccoli (Derelle et al. 2020; supplementary Methods, Supplementary Material online). Interestingly, the phylogeny indicates that P. keilini branches within a clade of parasitic parabasalids with very good support, as the sister lineage to a clade comprising T. vaginalis, Trichomonas gallinae, and Trichomonas tenax (extended quadripartition internode certainty [EQPIC] 0.53, reflecting good agreement between this species tree node and the underlying quartets of the input gene family trees). The cattle and feline parasite T. foetus branches sister to this P. keilini-T. vaginalis clade (Fig. 1). The phylogeny suggests either that P. keilini evolved a free-living lifestyle from a parasitic ancestor, or alternatively that there have been at least two transitions to parasitism within this clade: once in the ancestor of T. foetus and once in the ancestor of the Trichomonas clade. However, our phylogeny lacks many free-living and commensal endobiotic Parabasalia due to the lack of molecular data, and definitively testing between these hypotheses will require denser genomic sampling of related lineages. In 18S rRNA phylogenies, Pseudotrichomonas and Lacusteria are the two deepest branches within the order Trichomonadida. The position of Trichomonadida within Parabasalia is not resolved, but it might be the sister clade of Honigbergiellida, which contains several free-living species. The endobiotic Trichomitus batrachorum branches distantly and belongs to yet another order, the Hypotrichomonadida (Céza et al. 2022). Parasitic and free-living species seem to have arisen multiple times each within the predominantly commensal endobiotic Parabasalia.
Fig. 1.
Species tree of P. keilini among parabasalids and metamonads. SpeciesRax estimated species tree from 13,346 gene family trees, clustered from 43 genomes. Branch supports are EQPIC scores (Morel et al. 2022), reflecting the degree of agreement between quartets in the input gene trees and the inferred maximum likelihood species tree; >0.5 denotes high support). P. keilini groups robustly with the parasitic parabasalids T. vaginalis, T. gallinae, and T. tenax. Branch lengths in units of mean expected substitutions per site. Species are colored based on their taxonomic groups as indicated in the color code box on the right.
More broadly, the maximum likelihood SpeciesRax topology recovered the monophyly of Metamonada (including Parabasalia, Fornicata, Preaxostyla, Barthelona, Skoliomonas, and Anaeramoebae), although the deep relationships between these lineages were poorly resolved, with EQPIC scores close to 0 for the deepest splits within Metamonada, representing conflicting support from gene family trees. Alternative approaches such as concatenation (Stairs et al. 2021; Williamson et al. 2024) or reconciliation methods that model gene tree uncertainty (Cerón-Romero et al. 2022; Morel et al. 2024) may be required to resolve these deep branches of the eukaryotic tree.
Gene content evolution in P. keilini and its relatives
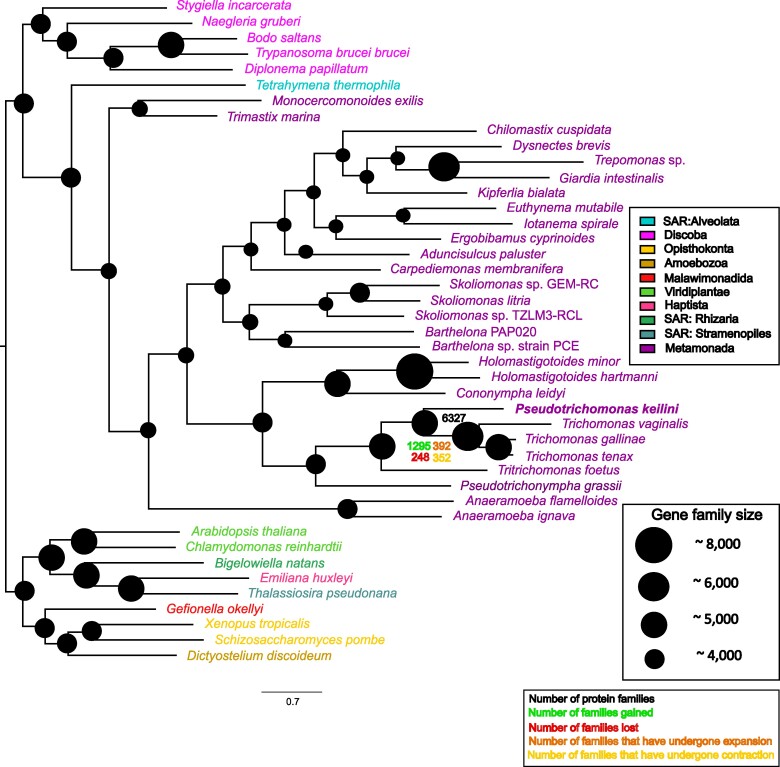
To investigate the evolutionary origins of the P. keilini genome, we mapped gene family evolution on the inferred eukaryotic species tree using a phylogenetic birth–death model implemented in Count (Csűös 2010), summarized in Fig. 2 (see also supplementary Methods, Supplementary Material online). The analysis suggested that the common ancestor of Parabasalia and Fornicata had a relatively small genome (2,975 gene families), with extensive gene gain in the parabasalid lineage after its divergence from the common ancestor with Fornicata. This is consistent with previous reports of gene family expansions in Parabasalia (Oyhenart and Breccia 2014; Handrich et al. 2019; Maciejowski et al. 2023). The common ancestor of P. keilini and T. vaginalis was inferred to have a gene repertoire comparable in size to that of P. keilini (5,280 gene families), with additional gene gains in the T. vaginalis lineage after its divergence from T. gallinae and T. tenax. Functional annotation of genes gained in the T. vaginalis lineage highlighted a role for binding to cellular components of the urogenital tract, a crucial step for extracellular parasite survival (Pereira-Neves and Benchimol 2007) (supplementary table S5, Supplementary Material online). Another key parasitic function can be seen through the presence of genes binding to spectrin, a protein found on the host cell surface that acts as the main gateway for target cell degradation (Fiori et al. 1997). These results are consistent with the view that the T. vaginalis lineage adapted to a parasitic lifestyle after divergence from its common ancestor with P. keilini, in part through the gene gains mapped here.
Fig. 2.
Gene family evolution in parabasalids and metamonads. We used a phylogenetic birth–death model implemented in Count (Csűös 2010) to map gene family evolution onto the inferred species tree. Numbers and the diameter of circles indicate gene family repertoire size at ancestral nodes, while family gains, losses, expansions, and contractions are plotted for the T. vaginalis, T. gallinae, and T. tenax lineages after their divergence from P. keilini. The analysis was also performed on a species tree manually edited to reflect the consensus view of deep eukaryotic relationships (supplementary fig. S2, Supplementary Material online), with closely similar results for gene content evolution within metamonads.
The maximum likelihood species tree contained some heterodox features that were poorly supported by EQPIC scores, including the branching of the ciliate Tetrahymena thermophila with metamonads rather than stramenopiles, alveolates and rhizarians (EQPIC 0.0075), and a deep-branching position of Anaeramoebae within metamonads (0.0051) rather than as sister to Parabasalia (Stairs et al. 2021). We therefore performed a sensitivity analysis in which we fit the phylogenetic birth–death model to a species tree edited to more closely reflect these current views of eukaryotic relationships (reviewed in Burki et al. 2020); see supplementary fig. S2, Supplementary Material online for the alternative tree. The results were closely similar.
The hydrogenosome of P. keilini
Metamonads are ancestrally anaerobic, and characterized metamonads have a range of reduced mitochondrial homologs or mitochondria-related organelles (MROs; Stairs et al. 2015). One of the best-characterized MROs is the hydrogenosome of T. vaginalis, and so we sought to investigate the MRO of P. keilini and to compare it with that of its close anaerobic, parasitic relative. To do so, we searched the P. keilini protein set for proteins previously implicated in MRO function and metabolism (Stairs et al. 2015), proteins that have been localized to the hydrogenosome in T. vaginalis (569 proteins, Schneider et al. 2011) and other hallmark mitochondrial proteins, including the mitochondrial carrier family of transporters.
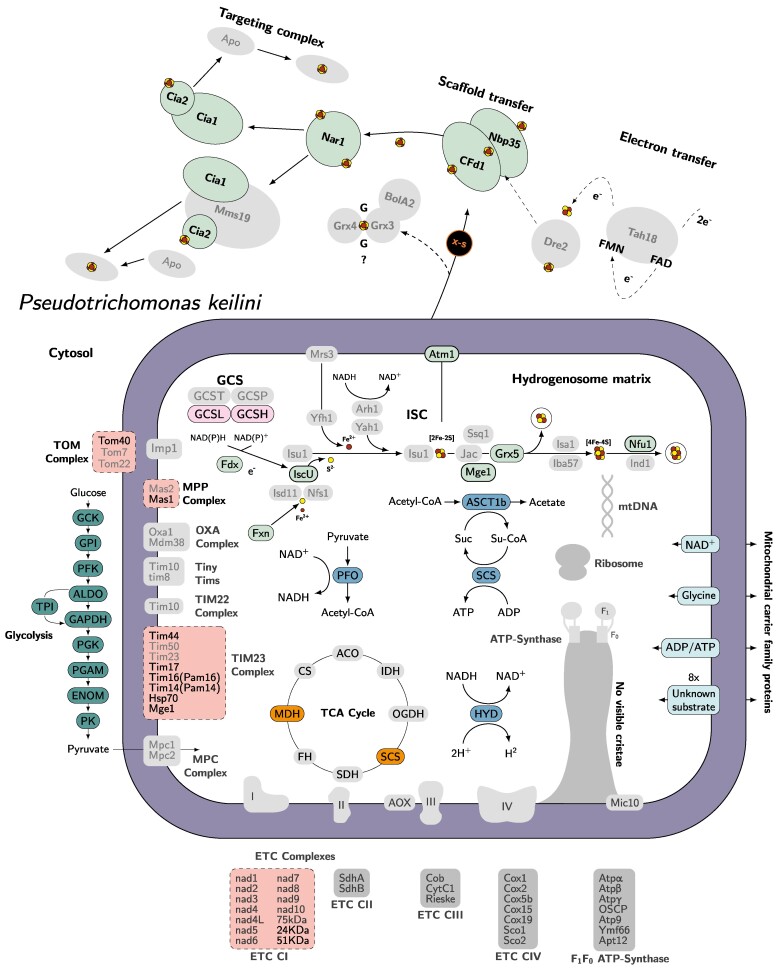
This analysis confirmed that P. keilini possesses a hydrogenosome that, in many respects, is similar to that of T. vaginalis: both organisms encode the same subset of 12 hallmark proteins drawn from a larger set found in a wide range of MROs across the eukaryotic tree of life (Stairs et al. 2015), including the key enzymes needed to reduce protons to molecular hydrogen via the oxidative decarboxylation of pyruvate (pyruvate-ferredoxin oxidoreductase, [FeFe]-hydrogenase, and the associated maturases) (supplementary fig. S4, Supplementary Material online, supplementary tables S2 to S5, Supplementary Material online). Overall, our analyses suggest that P. keilini has all the enzymes needed to carry out glycolysis, with the resulting pyruvate imported into the hydrogenosome for further catabolism (Fig. 3).
Fig. 3.
Predicted metabolic pathways in the P. keilini hydrogenosome. Key enzymatic reactions are depicted, including glycolysis, the main pathway used to produce pyruvate, which is then oxidized by the tricarboxylic acid (TCA) cycle in the hydrogenosome. The translocases of the outer and inner mitochondrial membranes (TOM and TIM, respectively) and the electron transport chain (ETC) subunits that were detected in complex I are also indicated; note that the subunits identified (NuoE and NuoF) are common in anaerobes, and are involved in other processes beyond oxidative phosphorylation (Hrdy et al. 2004; Stairs et al. 2021). Boxes with solid outlines represent complexes for which all subunits were identified, dashed outlines represent complexes with some subunits identified, and boxes in gray with no outline indicate complexes with no subunits identified. Figure design is adapted from Peña-Diaz and Lukeš (2018) and Lewis et al. (2019).
The transition from mitochondrion to hydrogenosome was previously inferred to have occurred in the common ancestor of all metamonads (Leger et al. 2017). This is consistent with our findings, in that phylogenetic analysis of the key hydrogenosomal enzymes recovered a monophyletic clade of parabasalids in each of the trees (supplementary figs. S3 to S14, Supplementary Material online). In total, the P. keilini transcriptome encodes orthologues of 487/569 T. vaginalis proteins localized to the hydrogenosome by proteomics (Schneider et al. 2011). Both organisms also encode the same complement of six paralogous mitochondrial carrier family proteins, suggesting that the requirements for transport into and out of the MRO are similar in each case (see supplementary Supplementary Material for more discussion on hydrogenosomal import in P. keilini).
The biosynthesis of iron–sulfur (Fe–S) clusters is perhaps the most widely (although not universally) conserved function of mitochondria and MROs (Stairs et al. 2015). The P. keilini hydrogenosome carries out Fe–S cluster biosynthesis using the ISC pathway, as in T. vaginalis. We detected orthologues of all the ISC enzymes present in T. vaginalis and Naegleria gruberi in the P. keilini transcriptome. Interestingly, P. keilini shares an iron–sulfur flavoprotein of bacterial origin used in the detoxification of reactive oxygen species with T. vaginalis and one other eukaryotic anaerobe, Entamoeba histolytica (Peña-Diaz and Lukeš 2018).
In sum, we sequenced the transcriptome of a free-living anaerobic parabasalid, P. keilini. Although free-living, the gene content, metabolism, and hydrogenosome of P. keilini are in many ways similar to its parasitic relative, T. vaginalis. The phylogenetic position of P. keilini in the species tree inferred from a large sample of protein-coding genes is consistent with the numerous transitions between free-living and host-associated lifestyles that have been suggested by 18S rRNA gene trees. Based on the analysis of BUSCO gene content, the transcriptome is likely to be largely complete and will represent a useful resource for future comparative analyses of parabasalids and eukaryotic evolution more broadly.
Supplementary Material
Acknowledgments
We thank František Šťáhlavský for providing the salt marsh sediment from which this strain of P. keilini was isolated, Pierre Lafont for help with figure design, and Will Maciejowski for technical assistance.
Contributor Information
Hend Abu-Elmakarem, Institute of Ecology and Evolution, University of Edinburgh, Edinburgh EH9 3FL, UK.
Stephen J Taerum, School of Life Sciences, Arizona State University, Tempe, AZ 85287, USA.
Celine Petitjean, School of Biological Sciences, University of Bristol, Bristol BS8 1TH, UK.
Michael Kotyk, Department of Zoology, Faculty of Science, Charles University, 128 00 Prague, Czech Republic; Institute of Vertebrate Biology, Czech Academy of Sciences, 603 00 Brno, Czech Republic.
Christopher Kay, School of Biological Sciences, University of Bristol, Bristol BS8 1TH, UK.
Ivan Čepička, Department of Zoology, Faculty of Science, Charles University, 128 00 Prague, Czech Republic.
David Bass, Centre for Environment, Fisheries and Aquaculture Science (Cefas), Lowestoft, UK.
Gillian H Gile, School of Life Sciences, Arizona State University, Tempe, AZ 85287, USA.
Tom A Williams, School of Biological Sciences, University of Bristol, Bristol BS8 1TH, UK.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online.
Funding
I.Č. and M.K. were supported by the Czech Science Foundation project no. 22-22538S. This work was supported in part by a grant from the US National Science Foundation, DEB-2045329, to G.H.G. This work was supported by the Gordon and Betty Moore Foundation through grant GBMF9741 to T.A.W.
Data Availability
Read data has been deposited in the NCBI SRA (accession number PRJNA884676). The transcriptome assembly, gene models, and annotations are available in a FigShare repository: https://doi.org/10.6084/m9.figshare.26528119.v1.
Literature Cited
- Adl SM, Leander BS, Simpson AG, Archibald JM, Anderson OR, Bass D, Bowser SS, Brugerolle G, Farmer MA, Karpov S, et al. Diversity, nomenclature, and taxonomy of protists. Syst Biol. 2007:56(4):684–689. 10.1080/10635150701494127. [DOI] [PubMed] [Google Scholar]
- Bishop A. Observations upon a “Trichomonas” from pond water. Parasitology. 1935:27(2):246–256. 10.1017/S0031182000015110. [DOI] [Google Scholar]
- Bishop A. A note upon the systematic position of “Trichomonas” keilini (Bishop, 1935). Parasitology. 1939:31(4):469–472. 10.1017/S0031182000012993. [DOI] [Google Scholar]
- Brugerolle G, Lee J. Phylum Parabasalia. In: Lee JJ, Leedale GF, Bradbury PC, editors. An illustrated guide to the protozoa: organisms traditionally referred to as protozoa, or newly discovered groups. Lawrence, Kansas, USA: Society of Protozoologists; 2000. p. 1196–1250. [Google Scholar]
- Burki F, Roger AJ, Brown MW, Simpson AGB. The new tree of eukaryotes. Trends Ecol Evol. 2020:35(1):43–55. 10.1016/j.tree.2019.08.008. [DOI] [PubMed] [Google Scholar]
- Čepička I, Dolan MF, Gile GH. Parabasalia. In: Archibald JM, Simpson AGB, Slamovits CH, Margulis L, Melkonian M, Chapman DJ, Corliss JO, editors. Handbook of the protists. Cham: Springer International Publishing; 2017. p. 1–44. [Google Scholar]
- Cepicka I, Hampl V, Kulda J. Critical taxonomic revision of parabasalids with description of one new genus and three new species. Protist. 2010:161(3):400–433. 10.1016/j.protis.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Cerón-Romero MA, Fonseca MM, de Oliveira Martins L, Posada D, Katz LA. Phylogenomic analyses of 2,786 genes in 158 lineages support a root of the eukaryotic tree of life between opisthokonts and all other lineages. Genome Biol Evol. 2022:14(8):evac119. 10.1093/gbe/evac119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Céza V, Kotyk M, Kubánková A, Yubuki N, Šťáhlavský F, Silberman JD, Čepička I. Free-living trichomonads are unexpectedly diverse. Protist. 2022:173(4):125883. 10.1016/j.protis.2022.125883. [DOI] [PubMed] [Google Scholar]
- Csűös M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics. 2010:26(15):1910–1912. 10.1093/bioinformatics/btq315. [DOI] [PubMed] [Google Scholar]
- Derelle R, Philippe H, Colbourne JK. Broccoli: combining phylogenetic and network analyses for orthology assignment. Mol Biol Evol. 2020:37(11):3389–3396. 10.1093/molbev/msaa159. [DOI] [PubMed] [Google Scholar]
- Donné A. Animalcules observés dans les matières purulentes et le produit des sécrétions des organes génitaux de l'homme et da la femme. CR Acad Sci. 1836:3:385–386. [Google Scholar]
- Fiori PL, Rappelli P, Addis MF, Mannu F, Cappuccinelli P. Contact-dependent disruption of the host cell membrane skeleton induced by Trichomonas vaginalis. Infect Immun. 1997:65(12):5142–5148. 10.1128/iai.65.12.5142-5148.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity. Nat Protoc. 2013:8(8):1494–1512. 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handrich MR, Garg SG, Sommerville EW, Hirt RP, Gould SB. Characterization of the BspA and Pmp protein family of trichomonads. Parasit Vectors. 2019:12(1):406. 10.1186/s13071-019-3660-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdy I, Hirt RP, Dolezal P, Bardonová L, Foster PG, Tachezy J, Martin Embley T. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004:432(7017):618–622. 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- Leger MM, Kolisko M, Kamikawa R, Stairs CW, Kume K, Čepička I, Silberman JD, Andersson JO, Xu F, Yabuki A, et al. Organelles that illuminate the origins of Trichomonas hydrogenosomes and Giardia mitosomes. Nat Ecol Evol. 2017:1(4):0092. 10.1038/s41559-017-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis WH, Lind AE, Sendra KM, Onsbring H, Williams TA, Esteban GF, Hirt RP, Ettema TJG, Embley TM. Convergent evolution of hydrogenosomes from mitochondria by gene transfer and loss. Mol Biol Evol. 2019:37(2):524–539. 10.1093/molbev/msz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski WJ, Gile GH, Jerlström-Hultqvist J, Dacks JB. Ancient and pervasive expansion of adaptin-related vesicle coat machinery across Parabasalia. Int J Parasitol. 2023:53(4):233–245. 10.1016/j.ijpara.2023.01.002. [DOI] [PubMed] [Google Scholar]
- Menezes CB, Frasson AP, Tasca T. Trichomoniasis-are we giving the deserved attention to the most common non-viral sexually transmitted disease worldwide? Microb Cell. 2016:3:404. 10.15698/mic2016.09.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel B, Schade P, Lutteropp S, Williams TA, Szöllősi GJ, Stamatakis A. SpeciesRax: a tool for maximum likelihood species tree inference from gene family trees under duplication, transfer, and loss. Mol Biol Evol. 2022:39(2):msab365. 10.1093/molbev/msab365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel B, Williams TA, Stamatakis A, Szöllősi GJ. AleRax: a tool for gene and species tree co-estimation and reconciliation under a probabilistic model of gene duplication, transfer, and loss. Bioinformatics. 2024:40(4):btae162. 10.1093/bioinformatics/btae162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyhenart J, Breccia JD. Evidence for repeated gene duplications in Tritrichomonas foetus supported by EST analysis and comparison with the Trichomonas vaginalis genome. Vet Parasitol. 2014:206(3-4):267–276. 10.1016/j.vetpar.2014.09.024. [DOI] [PubMed] [Google Scholar]
- Peña-Diaz P, Lukeš J. Fe–S cluster assembly in the supergroup Excavata. J Biol Inorg Chem. 2018:23(4):521–541. 10.1007/s00775-018-1556-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Neves A, Benchimol M. Phagocytosis by Trichomonas vaginalis: new insights. Biol Cell. 2007:99(2):87–101. 10.1042/BC20060084. [DOI] [PubMed] [Google Scholar]
- Petrin D, Delgaty K, Bhatt R, Garber G. Clinical and microbiological aspects of Trichomonas vaginalis. Clin Microbiol Rev. 1998:11(2):300–317. 10.1128/CMR.11.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider RE, Brown MT, Shiflett AM, Dyall SD, Hayes RD, Xie Y, Loo JA, Johnson PJ. The Trichomonas vaginalis hydrogenosome proteome is highly reduced relative to mitochondria, yet complex compared with mitosomes. Int J Parasitol. 2011:41(13-14):1421–1434. 10.1016/j.ijpara.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbald SJ, Archibald JM. More protist genomes needed. Nat Ecol Evol. 2017:1(5):145. 10.1038/s41559-017-0145. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single–copy orthologs. Bioinform. 2015:31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Stairs CW, Leger MM, Roger AJ. Diversity and origins of anaerobic metabolism in mitochondria and related organelles. Philos Trans R Soc B Biol Sci. 2015:370(1678):20140326. 10.1098/rstb.2014.0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stairs CW, Táborský P, Salomaki ED, Kolisko M, Pánek T, Eme L, Hradilová M, Vlček Č, Jerlström-Hultqvist J, Roger AJ, et al. Anaeramoebae are a divergent lineage of eukaryotes that shed light on the transition from anaerobic mitochondria to hydrogenosomes. Curr Biol. 2021:31(24):5605–5612.e5. 10.1016/j.cub.2021.10.010. [DOI] [PubMed] [Google Scholar]
- Williamson K, Eme L, Baños H, McCarthy C, Susko E, Kamikawa R, Orr RJS, Muñoz-Gómez SA, Simpson AGB, Roger AJ. A robustly rooted tree of eukaryotes reveals their excavate ancestry. bioRxiv 611237. 10.1101/2024.09.04.611237, 16 September 2024, preprint: not peer reviewed. [DOI]
- Yamin MA, Ma Y. Flagellates of the orders Trichomonadida Kirby, Oxymonadida Grassé, and Hypermastigida Grassi & Foà reported from lower termites (Isoptera families Mastotermitidae, Kalotermitidae, Hodotermitidae, Termopsidae, Rhinotermitidae, and Serritermitidae) and from the wood-feeding roach Cryptocercus (Dictyoptera: Cryptocercidae). Sociobiol. 1979:4(1):5–119. [Google Scholar]
- Yubuki N, Céza V, Cepicka I, Yabuki A, Inagaki Y, Nakayama T, Inouye I, Leander BS. Cryptic diversity of free-living parabasalids, Pseudotrichomonas keilini and Lacusteria cypriaca n. g., n. sp., as inferred from small subunit rDNA sequences. J Eukaryot Microbiol. 2010:57(6):554–561. 10.1111/j.1550-7408.2010.00509.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Read data has been deposited in the NCBI SRA (accession number PRJNA884676). The transcriptome assembly, gene models, and annotations are available in a FigShare repository: https://doi.org/10.6084/m9.figshare.26528119.v1.