Abstract
Sex chromosomes of some closely related species are not homologous, and sex chromosome turnover is often attributed to mechanisms that involve linkage to or recombination arrest around sex-determining loci. We examined sex chromosome turnover and recombination landscapes in African clawed frogs (genus Xenopus) with reduced representation genome sequences from 929 individuals from 19 species. We recovered extensive variation in sex chromosomes, including at least eight nonhomologous sex-associated regions—five newly reported here, with most maintaining female heterogamety, but two independent origins of Y chromosomes. Seven of these regions are found in allopolyploid species in the subgenus Xenopus, and all of these reside in one of their two subgenomes, which highlights functional asymmetry between subgenomes. In three species with chromosome-scale genome assemblies (Xenopus borealis, Xenopus laevis, and Xenopus tropicalis), sex-specific recombination landscapes have similar patterns of sex differences in rates and locations of recombination. Across these Xenopus species, sex-associated regions are significantly nearer chromosome ends than expected by chance, even though this is where the ancestral recombination rate is highest in both sexes before the regions became sex associated. As well, expansions of sex-associated recombination arrest occurred multiple times. New information on sex linkage along with among-species variation in female specificity of the sex-determining gene dm-w argues against a “jumping gene” model, where dm-w moves around the genome. The diversity of sex chromosomes in Xenopus raises questions about the roles of natural and sexual selection, polyploidy, the recombination landscape, and neutral processes in driving sex chromosome turnover in animal groups with mostly heterogametic females.
Keywords: sex determination, genetic linkage, recombination landscape, allopolyploidization, sex chromosome turnover
Introduction
In animals, sexual differentiation generally involves a developmental turning point where genetic or environmental queues guide a bipotential larva to develop into one or the other adult sexual phenotypes. Mutations that alter sexual differentiation have fitness costs if they reduce fertility or cause sterility. Nonetheless, biological mechanisms that orchestrate sexual differentiation often evolve rapidly (for example in vertebrates; reviewed in Stoeck et al. 2021). This is especially true for genes that trigger sexual differentiation (“master sex-determining genes”; reviewed in Adolfi et al. 2021; Pan et al. 2021), even though these biological innovations can have pronounced genomic repercussions associated with the establishment of new sex chromosomes and changes in the modes of inheritance and rates and locations of recombination (hereafter the “recombination landscape”) of these chromosomes. Key questions in functional genomics ask how and why the genetic basis of crucial phenotypes—such as sexual differentiation—evolve rapidly when genetic changes that affect these traits have disruptive and potentially devastating consequences.
In species with genetic sex determination, sex-determining genes are frequently found in genomic locations that lack recombination, which reduces the efficacy of natural selection in sex-linked regions in several ways (e.g. McVean and Charlesworth 2000). Consequences of recombination suppression are evident in numerous independently evolved sex-linked regions that have lost genes and accumulated repetitive elements following recombination arrest (e.g. Skaletsky et al. 2003; Bachtrog 2013). Recombination arrest in genomic regions harboring sex-determining genes could either be a preexisting condition if sex-determining genes arise in genomic areas that already have low recombination (reviewed in Sardell and Kirkpatrick 2020), or it could evolve after a sex-determining gene appears because of neutral evolution (Ironside 2010; Jeffries et al. 2021) or nonneutral evolution (Charlesworth and Charlesworth 1980; Rice 1987; Charlesworth and Wall 1999; Zou et al. 2021). Neutral evolution might lead to recombination suppression due to the origin of mutations in linkage disequilibrium with the sex-determining allele that reduces the probability of recombination (Jeffries et al. 2021). Natural selection might favor recombination suppression, for example, if it resolves genomic conflict associated with mutations with sexually antagonistic fitness effects (Charlesworth and Charlesworth 1980), or under scenarios involving heterozygote advantage (Charlesworth and Wall 1999). Inversions in sex-determining regions also might be favored because they fix heterozygosity where one allele is recessive and disadvantageous (Jay et al. 2022). Recombination suppression could additionally be favored if inversions are followed by sex-chromosome-specific cis-regulatory evolution and the origin of dosage compensation (Lenormand and Roze 2022).
In many animals (including humans), the rates and genomic locations of genetic recombination within non-sex-linked genomic regions differ between oogenesis and spermatogenesis—a phenomenon known as heterochiasmy. At the extreme, recombination may be absent in one sex (achiasmy), which is the case in Drosophila melanogaster males (Morgan 1910). Explanations for why heterochiasmy exists include neutral evolution, mechanistic differences between oogenesis and spermatogenesis, and adaptation (reviewed in Sardell and Kirkpatrick 2020). In species with heterochiasmy, natural selection may favor the origin of sex-determining genes in (or translocation to) genomic regions that already have a low rate of recombination in the sex in which the sex-determining gene resides (Sardell and Kirkpatrick 2020). Low recombination could be favored by natural selection to ensure faithful inheritance of a nonrecombined sex-determining gene. It could also increase the chance that a sex-determining gene becomes genetically linked to and resolves genetic conflict associated with a mutation with sexually antagonistic fitness effects, which could favor the initial establishment of a nascent sex chromosome. Alternatively, heterochiasmy could contribute to the origin of sex-linked recombination arrest through neutral processes. In some ranid frogs, for example, extreme heterochiasmy exists where recombination during spermatogenesis is mostly limited to chromosome tips (Rodrigues et al. 2013). In these frogs, newly established Y chromosomes may immediately have large nonrecombining regions that span chromosome centers (Jeffries et al. 2018). In therian mammals, recombination arrest between nonpseudoautosomal portions of the X and Y chromosomes probably appeared after the origin of a male-determining allele (Sry); expansion of recombination arrest occurred in association with several inversions, and ultimately led to heteromorphic sex chromosomes, including profound differences in sequence and gene content between the X and Y chromosome (Ross et al. 2005).
In species with genetic sex determination, two predominant and nonexclusive hypotheses have been proposed to drive sex chromosome turnover, and both involve linkage to or recombination arrest of sex-determining regions of the sex chromosomes. In (i), the mutational load model, degeneration from recombination arrest causes the fitness of the sex-specific sex chromosome (Y or W) to decline, which favors the origin of new sex chromosomes elsewhere in the genome, and then allows for the loss of the loaded sex-specific sex chromosome (Blaser et al. 2013, 2014). In (ii), the sexual antagonism model, genomic conflict caused by sexually antagonistic mutations (which are beneficial in females but deleterious in males or vice versa) is resolved by linkage to a new or translocated sex-determining gene (Fisher 1930; Charlesworth and Charlesworth 1980; Rice 1987; van Doorn and Kirkpatrick 2007, 2010).
Under the mutational load model, an ancestral shared sex chromosome (the Z chromosome with female heterogamety or the X chromosome with male heterogamety) transitions to autosomal segregation, and the (degenerate) ancestral sex-specific sex chromosome (the W or Y, respectively) goes extinct when a new sex-specific sex chromosome arises elsewhere in the genome. Under this model, if the newly emerged sex-determining gene is dominant, then the heterogametic sex must remain unchanged if the ancestral degenerate sex chromosome is to be purged (Bull and Charnov 1977). However, if the newly emerged sex-determining gene is recessive, a change in the heterogametic sex is possible while also purging the (ancestral) loaded sex-specific sex chromosome (Bull and Charnov 1977). Under the sexual antagonism model, the heterogametic sex may change, but does not have to. Sex chromosomes might also turnover because of genetic drift (Saunders et al. 2018). Under a drift model (and similar to the mutational load model when dominance is assumed), transitions in heterogamety are neutral and predicted to be relatively rare because the ancestral shared sex chromosome has a higher frequency, and therefore is more likely to fix than the sex-specific sex chromosome.
The Sex Chromosomes of African Clawed Frogs (Xenopus)
One step toward understanding why sex chromosomes sometimes evolve rapidly is to characterize natural variation, and then to evaluate evidence for alternative explanations in species that have experienced rapid sex chromosome turnover. African clawed frogs (genus Xenopus) offer a compelling system with which to accomplish this. Amphibians in general have a remarkably high rate of sex chromosome evolution (Evans et al. 2012; Jeffries et al. 2018; Pennell et al. 2018; Ma and Veltsos 2021). All but one of the 29 species of Xenopus are allopolyploid—species with duplicated genomes that are derived from two or more ancestral species (Evans et al. 2015). Genomic redundancy associated with polyploidy has the potential to catalyze rapid evolution of sex chromosomes through genetic changes associated with gene duplication (subfunctionalization, neofunctionalization, redundancy, and gene loss), and possible structural changes following genome duplication (Force et al. 1999; Lynch et al. 2001; Hufton and Panopoulou 2009; Simakov et al. 2020).
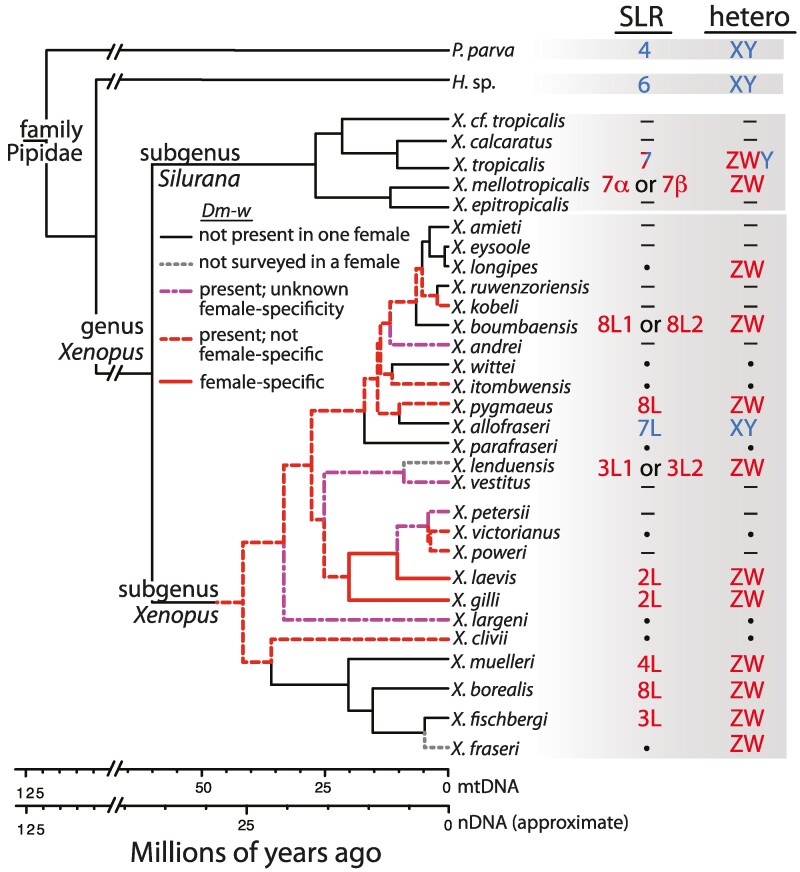
Xenopus includes two subgenera—Silurana and Xenopus—that diverged from one another ∼34 to 58 Ma (Feng et al. 2017) and whose diploid ancestors had different chromosome numbers (2n = 20 for the subgenus Silurana; 2n = 18 for the subgenus Xenopus, where n is the number of chromosomes in a gamete). Previous work demonstrates independent evolution of three nonhomologous sex-associated regions in four Xenopus species (Xenopus tropicalis, Xenopus mellotropicalis, Xenopus laevis, and Xenopus borealis on chromosomes 7, 7α or 7β, 2L, and 8L, respectively; Uno et al. 2008; Yoshimoto et al. 2008; Matsuda et al. 2015; Roco et al. 2015; Furman and Evans 2016; Mitros et al. 2019). Xenopus borealis, X. laevis, and X. mellotropicalis (all are allotetraploid species) have heterogametic females, whereas in X. tropicalis, there is a trio of sex chromosomes (W, Z, Y) rather than the typical pair (Roco et al. 2015; Furman et al. 2020), and both sexes can be heterogametic or homogametic (females: WW, WZ; males: ZZ, ZY, WY). The female-determining regions on the W chromosomes of X. tropicalis and X. mellotropicalis are homologous (Cauret et al. 2020), and the Y chromosome of X. tropicalis evolved from the Z chromosome after divergence of this species from X. mellotropicalis (Furman et al. 2020). Intraspecific variation in sex chromosomes also exists in X. borealis in that there is extensive variation in the size of the sex-associated region in different populations (>50 Mb in one population, <1 Mb in another; Evans et al. 2022).
In at least one Xenopus species (X. laevis), a gene called dm-w triggers female differentiation (Fig. 1; Yoshimoto et al. 2008, 2010; Cauret et al. 2020, 2023). Surveys of six other species detected dm-w in some males, and often in only a subset of females (Xenopus kobeli, Xenopus pygmaeus, Xenopus victorianus, Xenopus clivii, and Xenopus poweri), which suggests that dm-w is not the sex-determining gene in these species. In one species (Xenopus itombwensis) dm-w was found in all individuals, which suggests autosomal segregation (Cauret et al. 2020). Dm-w was lost in several other Xenopus species, such as X. borealis, Xenopus muelleri, Xenopus fischbergi, and others (Fig. 1; Bewick et al. 2011; Cauret et al. 2020, 2023; Song et al. 2021; Evans et al. 2022). In at least two species (X. clivii and Xenopus vestitus), dm-w may lack exon 4 (Cauret et al. 2023), which encodes a functionally important component of this protein (Hayashi et al. 2022), and the absence of this exon may incapacitate the female-determining function of dm-w in those species. Adding to this recent dynamic functional evolution, analysis of phylogenetic relationships of dm-w and its paralogs (dmrt1) demonstrates a recent origin of dm-w in the ancestor of subgenus Xenopus after divergence from the ancestor of the subgenus Silurana (Bewick et al. 2011). Sex chromosome variation is also evident in other genera that are in the same family as Xenopus (the family Pipidae): males are the heterogametic sex in Pipa parva and Hymenochirus sp. (sensu Gvoždík et al. 2024) and the sex-associated regions of these species are homologous to chromosome 6 (Chr6) and 4 (Chr4) of X. tropicalis, respectively (Cauret et al. 2020).
Fig. 1.
Phylogenetic affinities of at least eight nonhomologous sex-linked regions exist in Xenopus. The sex-linked regions (SLR) are on chromosomes that are indicated with numbers, and letters after these numbers refer to subgenomes (L or S for allotetraploids in subgenus Xenopus, L1, L2, S1, or S2 for allooctoploids in subgenus Xenopus, α or β for allotetraploids in subgenus Silurana; Evans et al. 2005, 2015). The heterogametic sex (hetero) is indicated with ZW for female heterogamety and XY for male heterogamety, or ZWY for X. tropicalis (see main text). The time-calibrated phylogeny was estimated from complete mitochondrial genomes (Evans et al. 2019) that do not reflect reticulating relationships among allopolyploid species. Scale bars illustrate variation in date estimates when analyses are performed using mitochondrial (Evans et al. 2019) and nuclear DNA (Evans et al. 2015). Black dots indicate unknown information from some species that were surveyed using RRGS data in this study; dashes indicate species for which RRGS data are not currently available. Results from X. laevis, X. borealis, X. tropicalis, X. mellotropicalis, P. parva, and Hymenochirus sp. (H. sp. as Hymenochirus boettgeri) were previously reported (Yoshimoto et al. 2008; Olmstead et al. 2010; Roco et al. 2015; Furman and Evans 2016; Cauret et al. 2020, 2023).
Goals
We set out to further characterize variation in sex-associated genomic regions in Xenopus and to evaluate sex-specific recombination landscapes within which these regions evolved. To this end, we used reduced representation genome sequencing (RRGS) to survey sex linkage in 929 individuals from 19 out of the 29 extant Xenopus species, including the only diploid species, 13 allotetraploid species, 4 allooctoploid species, and 1 allododecaploid species. To interpret our findings in the context of proposed mechanisms for sex chromosome turnover, we independently characterized the recombination landscape of both sexes using RRGS data from one family in each of three species for which chromosome-scale genome assemblies were available (X. borealis, X. laevis, and X. tropicalis). As detailed below, our results reveal extensive variation in the locations of sex-associated genomic regions (that are partially or completely linked to the sex-determining gene), heterogamety, and recover differences in the rates and genomic locations of recombination during oogenesis and spermatogenesis. We also found evidence for rapid evolution of reduction or arrest of recombination following the origin of new sex-associated genomic regions. These findings provide insights into several aspects of sex chromosome evolution in animal species, where females are usually the heterogametic sex, including the applicability of proposed mechanisms that drive turnover, and the origins of sex-linked recombination arrest.
Results
Frequent Sex Chromosome Turnover in Xenopus
We attempted to identify sex-associated genomic regions in 929 individuals from 19 Xenopus species using two approaches to analyze RRGS data (see Materials and Methods; Table 1, supplementary table S1, Supplementary Material online). The first tested for sex-associated single nucleotide polymorphisms (SNPs) in genotypes of RRGS reads mapped to a reference genome. Sex-associated SNPs are variants that occur more frequently in one sex than expected by chance. The second approach (RADsex; see Materials and Methods) did not require genotyping of mapped reads and instead tested for biased abundance or exclusivity of RRGS reads in one sex.
Table 1.
Summary of samples used in this study including species, ploidy, the total number of individuals sampled (total), the number of wild females and males (wild F, wild M), non-kin lab individuals of each sex (other F, other M), and information on lab families including the family name, origin of parents (lab or wild), and the number of offspring of each sex (daughters, sons)
| Species | Ploidy | Total | Wild F | Wild M | Other F | Other M | Name | Parents | Daughters | Sons |
|---|---|---|---|---|---|---|---|---|---|---|
| Subgenus Silurana | ||||||||||
| X. tropicalis | Diploid | 190 | 6 | 20 | 7 | 6 | Family 1 | Wild | 22 | 21 |
| Family 2 | Wild | 7 | 5 | |||||||
| C659 | Lab | 19 | 18 | |||||||
| C660 | Lab | 27 | 32 | |||||||
| X. mellotropicalis | Allotetraploid | 20 | 1 | 1 | ND | ND | Family 1 | Lab | 9 | 9 |
| Subgenus Xenopus | ||||||||||
| X. allofraseri | Allotetraploid | 77 | 8 | 15 | ND | ND | Family 0 | Wild/wilda | 5 | 3 |
| Family 1 | Wild | 24 | 11 | |||||||
| Family 2 | Wild/wilda | 5 | 6 | |||||||
| X. borealis | Allotetraploid | 96 | 24 | 24 | 1 | 1 | Family 1 | Lab | 24 | 22 |
| X. clivii | Allotetraploid | 62 | 27 | 20 | 1 | 14 | ND | ND | ND | ND |
| X. fischbergi | Allotetraploid | 31 | ND | ND | 2 | 2 | Family 1 | Lab | 18 | 9 |
| X. fraseri | Allotetraploid | 36 | 24 | 12 | ND | ND | ND | ND | ND | ND |
| X. gilli | Allotetraploid | 11 | 8 | 3 | ND | ND | ND | ND | ND | ND |
| X. laevis | Allotetraploid | 74 | 23 | 20 | 1 | 1 | Family 1 | Lab | 10 | 19 |
| X. largeni | Allotetraploid | 9 | 4 | 5 | ND | ND | ND | ND | ND | ND |
| X. muelleri | Allotetraploid | 24 | ND | ND | 1 | 3 | Family 1 | Lab | 11 | 9 |
| X. parafraseri | Allotetraploid | 16 | 7 | 9 | ND | ND | ND | ND | ND | ND |
| X. pygmaeus | Allotetraploid | 58 | 6 | 2 | ND | ND | Family 1 | Lab | 38 | 12 |
| X. victorianus | Allotetraploid | 34 | 15 | 19 | ND | ND | ND | ND | ND | ND |
| X. boumbaensis | Allooctoploid | 26 | 6 | 5 | ND | ND | Family 1 | Lab | 5 | 10 |
| X. itombwensis | Allooctoploid | 41 | 13 | 28 | ND | ND | ND | ND | ND | ND |
| X. lenduensis | Allooctoploid | 62 | 28 | 34 | ND | ND | ND | ND | ND | ND |
| X. wittei | Allooctoploid | 28 | 14 | 14 | ND | ND | ND | ND | ND | ND |
| X. longipes | Allododecaploid | 34 | 23 | 11 | ND | ND | ND | ND | ND | ND |
ND - Not Defined.
aThe same lab-reared individual was the father of X. allofraseri family 0 and family 2.
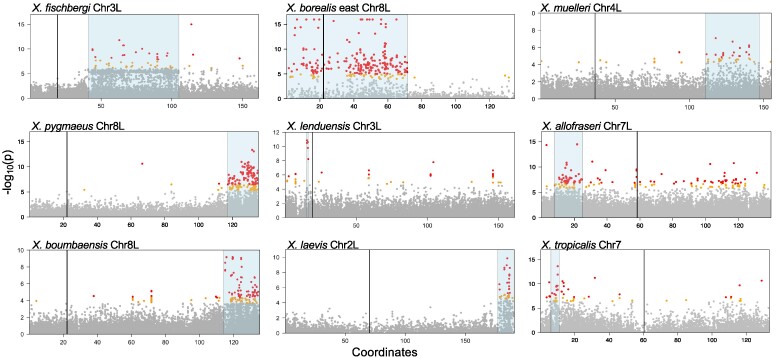
Using these approaches, we discovered previously unidentified sex-determining regions in six species, including four allotetraploids: Xenopus allofraseri (Chr7L), X. fischbergi (Chr3L), X. muelleri (Chr4L), X. pygmaeus (Chr8L), and two allooctoploids: Xenopus lenduensis (one of the two homeologs of Chr3L—Chr3L1 or Chr3L2) and Xenopus boumbaensis (one of the two homeologs of Chr8L—Chr8L1 or Chr8L2; Figs. 1 and 2, supplementary figs. S1 and S2, Supplementary Material online). The five newly identified sex-associated regions of recombination reduction or arrest are homologous to the following positions in the X. laevis reference genome: X. allofraseri: 10.8 to 19.8 Mb on Chr7L; X. fischbergi: 41 to 105 Mb on Chr3L; X. muelleri: 111 to 147 Mb on Chr4L; X. pygmaeus and X. boumbaensis: 117 to 135 Mb on Chr8L, X. lenduensis: 15 to 16.2 Mb on Chr3L (summarized in Table 2). These regions have 3 to ∼1,100 annotated genes (supplementary results, Supplementary Material online) but do not stand out as particularly gene rich or poor (supplementary figs. S3 and S4, Supplementary Material online).
Fig. 2.
Eight nonhomologous sex-associated regions in nine Xenopus species based on analyses of genetic association and sex specificity (RADsex; see main text), including five newly described here. Gray, orange, and red dots indicate the −log10 transformed probability (P) for a test of whether each SNP is associated with sex, and correspond to >0.1%, 0.1–0.05%, or the <0.05% percentiles, respectively, across genome-wide RRGS variants for each species. Sex-associated regions supported by association tests and RADsex analysis are highlighted in blue; centromere locations of the reference genomes are indicated with vertical lines. These plots show only the sex chromosomes for each species; plots of the whole genome for each species are provided in supplementary fig. S1, Supplementary Material online. Except for X. lenduensis, data from all species depicted here are at least partially from lab-bred families.
Table 2.
Sex-associated regions and heterogametic sex in Xenopus species identified or confirmed in this study
| Subgenus | Species | Heterogametic sex | Chr | Start | Stop | Evidence |
|---|---|---|---|---|---|---|
| Silurana | X. tropicalis | F/M | Chr7 | 0 | 11000000 | Sex association |
| Silurana | X. mellotropicalis | F | Chr7 | 0 | 11000000 | Both |
| Xenopus | X. allofraseri | M | Chr7L | 10800000 | 19800000 | Both |
| Xenopus | X. borealis | F | Chr8L | 0 | 54100000 | Both |
| Xenopus | X. boumbaensis | F | Chr8L | 117000000 | 135000000 | Sex association |
| Xenopus | X. fischbergi | F | Chr3L | 41000000 | 105000000 | Both |
| Xenopus | X. fraseri | F | ND | ND | ND | RADsex |
| Xenopus | X. gilli | F | Chr2L | 177000000 | 190000000 | RADsex |
| Xenopus | X. laevis | F | Chr2L | 177000000 | 190000000 | Both |
| Xenopus | X. muelleri | F | Chr4L | 111000000 | 147000000 | Both |
| Xenopus | X. pygmaeus | F | Chr8L | 117000000 | 135000000 | Both |
| Xenopus | X. lenduensis | F | Chr3L | 15000000 | 16200000 | Both |
| Xenopus | X. longipes | F | ND | ND | ND | RADsex |
ND - Not Defined.
Chromosome (Chr) and coordinates (Start, Stop) correspond to the reference genome for each subgenus (X. tropicalis for the subgenus Silurana, X. laevis for the subgenus Xenopus). Evidence for sex association is based on sex-association tests, RADsex analysis, or both.
We confirmed the locations of three previously reported nonhomologous sex chromosomes in two allotetraploids: X. laevis (Chr2L) and X. borealis west (Chr8L), and homologous sex chromosomes in the diploid X. tropicalis (Chr7) and the allotetraploid X. mellotropicalis (one of the two homeologs of Chr7; Fig. 2, supplementary fig. S1 and table S2, Supplementary Material online). Based on the analysis that does not use a reference genome—RADsex (see Materials and Methods; supplementary fig. S2 and table S3, Supplementary Material online), we recovered evidence for sex chromosomes in the allotetraploid Xenopus gilli that are homologous to those of X. laevis (on Chr2L, supplementary results, Supplementary Material online), which is consistent with a previous finding that dm-w is female-specific in X. gilli (Cauret et al. 2020).
The sex-determining regions of X. pygmaeus and X. boumbaensis reside in a homologous region of Chr8L, and these regions may be derived from their most recent common ancestor. Three other groups of species have sex-associated regions on the same chromosome but these are probably nonhomologous. For two of these groups—(X. pygmaeus and X. boumbaensis) + X. borealis and X. lenduensis + X. fischbergi, the genomic locations of these sex-associated regions do not overlap, which indicates independent origins or movement of the genomic location of a sex-determining gene. For the third group—X. allofraseri + (X. tropicalis, X. mellotropicalis)—their sex-associated regions overlap slightly. However, these species are highly diverged (and in different subgenera), and the ancestral heterogametic sex is female in X. tropicalis + X. mellotropicalis (Furman et al. 2020) but male in X. allofraseri (discussed below). It is therefore probable that their sex-association regions also arose independently. Thus, when combined with previous findings, a total of eleven Xenopus species have eight sex-determining regions that are not homologous to each other. Additional details about sex-associated regions of several Xenopus species are provided in supplementary results, Supplementary Material online.
Three pairs of closely related species were identified with homologous sex-associated regions: X. tropicalis + X. mellotropicalis, X. laevis + X. gilli, X. pygmaeus + X. boumbaensis, and all are closely related. For this reason, the available data provides very limited statistical power with which to accurately estimate a rate of sex chromosome turnover during the entirety of Xenopus diversification which transpired over dozens of millions of years. We instead calculated a minimum rate of sex chromosome turnover using the strong (and unlikely) assumptions that (i) we have observed all sex chromosome turnover events that ever occurred during Xenopus diversification of species with known sex-associated regions (n = 8), and (ii) that one of these states is the ancestral sex chromosome. Using a time-calibrated mitochondrial phylogeny that may overestimate divergence times (as discussed below) and therefore yield an even more underestimated minimum rate, seven changes occurred on a phylogeny with a total branch length of 321 My, yielding a minimum rate of 0.022 turnover events per million years (one turnover event every ∼46 My). As discussed below, the actual rate of sex chromosome turnover almost certainly is much higher than this.
Sex-Associated Recombination Suppression
Because all of these sex-associated regions except that of X. lenduensis were identified using linkage in families, the size of the area of complete sex-linked recombination arrest is probably smaller than estimates recovered from our analysis of families. This is likely the case in X. laevis where the bounds of the sex-associated region from a family are much larger than the sizes of the W- and Z-specific portions of Chr2L (Mawaribuchi et al. 2017). Other factors also contribute to noise in the signal of sex association, including repetitive sequences, and (for allotetraploids, allooctoploids, and allododecaploids mapped to X. laevis) mis-mapping of reads from one subgenome to the other subgenome, and (for allooctoploids and allododecaploids mapped to X. laevis and reads from the allotetraploid X. mellotropicalis mapped to the diploid X. tropicalis) reads from multiple closely related subgenomes co-mapping to one subgenome of X. laevis. For these reasons, we consider the estimated boundaries of the sex-associated regions to be approximate.
One line of evidence that would support recombination arrest would be divergence between the sex chromosomes. To explore this possibility, we calculated FST between females and males in 5 Mb genomic windows on the sex chromosomes of each species for which we were able to identify a sex-associated region, with the expectation that recombination arrest between the sex chromosomes would lead to atypically high FST between females and males in sex-associated compared with non-sex-associated windows. A caveat to this expectation is that recent sex-associated recombination arrest would have little or no signal of divergence. We found qualitative support for divergence due to recombination arrest in the sex-associated region of non-kin X. fischbergi and portions of this region in X. borealis (supplementary fig. S5, Supplementary Material online). More subtle signatures were evident in X. pygmaeus, X. tropicalis, and among kin of X. allofraseri, X. boumbaensis, and X. laevis. Atypically high FST between females and males was not evident in the small sex-associated regions of non-kin X. allofraseri, X. laevis, or X. lenduensis, kin of X. mellotropicalis, or in the large sex-associated region of non-kin X. muelleri (supplementary figs. S5 and S6, Supplementary Material online).
Subgenome Bias
All known sex-associated regions in the subgenus Xenopus—including those previously identified in X. laevis (Chr2L) and X. borealis (Chr8L)—are in the long (“L”) subgenome. All known sex-determining systems in Xenopus have heterogametic females except X. allofraseri—newly reported here to have heterogametic males—and the previously reported complex sex-determining system of X. tropicalis (discussed above; Roco et al. 2015; Furman et al. 2020). In the 17 species we analyzed from the subgenus Xenopus, the observed number of mapped reads was significantly higher in the L subgenome for 12 species and significantly higher in the short (“S”) subgenome for five species (P < 0.00001, χ2 test, 1 degree of freedom), but the magnitudes of these differences were modest (supplementary table S2, Supplementary Material online). This suggests that the finding that all sex-associated regions are in the L subgenome is unlikely to be a technical artifact of biased read mapping. Additional details about mapped reads for each species are provided in the Supplementary material.
Male and Female Heterogamety
We also recovered evidence for female heterogamety in the allotetraploid Xenopus fraseri and the allododecaploid Xenopus longipes, but were unable to discern the genomic locations of the female-linked regions in these two species because female-specific reads from each species mapped to repetitive elements in the X. laevis genome (supplementary results, Supplementary Material online).
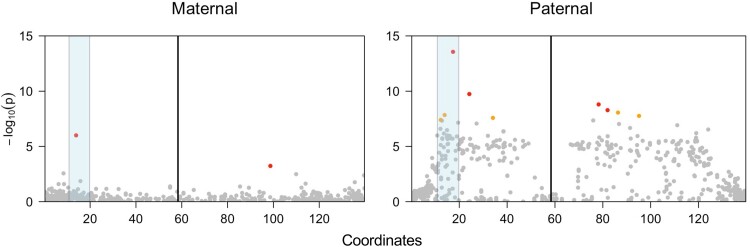
When genetic variation from parents and offspring is available, female or male heterogamety can be distinguished by separately examining the genetic associations of variants from each parent (i.e. variants that are heterozygous in the mother but homozygous in the father, and vice versa). Analyses of sex-association of maternal and paternal genetic variation in families of X. borealis, X. fischbergi, X. laevis, X. muelleri, and X. pygmaeus evidence female heterogamety (supplementary figs. S7 and S8, Supplementary Material online); female heterogamety in X. laevis and X. borealis has been previously reported (Chang and Witschi 1955; Furman and Evans 2016). Male heterogamety in X. allofraseri is evidenced by a strong signal of sex-linkage in paternal but not maternal variants—especially in the largest family we studied (Fig. 3), but also in two smaller families (supplementary fig. S8, Supplementary Material online).
Fig. 3.
Male heterogamety in X. allofraseri is evidenced by multiple strongly sex-associated paternal SNPs (right) but far fewer sex-associated maternal SNPs (left) on the sex chromosome (Chr7L). Labeling follows Fig. 2. Gray, orange, and red dots indicate that the −log10 transformed probability (P) that SNPs are associated with sex, and correspond to >0.1%, 0.1–0.05%, or the <0.05% percentiles, respectively, across maternal (left) or paternal (right) genome-wide RRGS variants. These data are from X. allofraseri family 1; data from the other two families are plotted in supplementary fig. S8, Supplementary Material online.
Analysis of maternal and paternal variants evidences a YZ sex chromosome genotype in the father of one X. tropicalis family (family 2) but reveals more complexity in three other X. tropicalis families where signatures of sex association were observed in both parents (most prominently in family C659; supplementary fig. S9, Supplementary Material online). A plausible explanation is that the sex chromosome genotypes of the mother and father of these three X. tropicalis families were WZ and ZY, respectively, in which case W- and Y-linked variation are both expected to be associated with sex.
Data from parents were not available for the X. boumbaensis family; to assess heterogamety, we instead calculated pairwise nucleotide diversity (π) for daughters and sons of the significantly sex-associated variants (P < 0.001) that mapped to the sex-associated region (117 to 135 Mb on Chr8L). We reasoned that π of these sex-associated variants should be higher in the heterogametic sex due to the presence of a shared and sex-specific genomic region (ZW for female heterogamety or XY for male heterogamety) when compared with the homogametic sex (ZZ for female heterogamety or XX for male heterogamety). The π of these positions was almost three times higher in females (π = 0.314, SE = 0.016, n = 265 positions) than males (π = 0.114, SE = 0.011, n = 261 positions), with the difference in the number of positions due to missing genotypes. This is consistent with the mother being the heterogametic sex in the X. boumbaensis family. As a proof of concept, we were able to use this approach to confirm female heterogamety in offspring of families of X. fischbergi, X. muelleri, and X. pygmaeus, though this method failed to provide strong evidence for heterogamety using a small number of sex-associated positions in X. lenduensis and X. allofraseri (supplementary Results, Supplementary Material online). As explained in further detail below (see Materials and Methods), because these diversity estimates are restricted to only sex-associated (variable) positions, they do not provide information about per-site variation.
Clear signatures of sex association were not detected in six other Xenopus species (X. clivii, X. itombwensis, Xenopus largeni, Xenopus parafraseri, X. victorianus, and Xenopus wittei), including some with reasonably large sample sizes (>50 individuals; supplementary Figs. S1 and S2, Supplementary Material online). For all six of these species, only wild-caught individuals were analyzed. This may indicate that these species have small regions of recombination arrest and, for some species, that sample sizes or sequencing depth were insufficient to detect significant sex linkage.
dm-w
We used Sanger sequencing to test in additional individuals (supplementary tables S4 and S5, Supplementary Material online) whether (i) one or both X. pygmaeus parents and their offspring carried dm-w, and (ii) a polymerase chain reaction (PCR)-amplified sex-associated region identified in the X. pygmaeus family (which originated from the Democratic Republic of the Congo, DRC) was also sex associated in non-kin individuals that are also from the DRC. We found (i) both X. pygmaeus parents and at least five of their sons and five of their daughters carried dm-w (supplementary fig. S10, Supplementary Material online), and that (ii) the PCR-amplified sex-associated regions identified in the X. pygmaeus family was not sex associated in the non-kin individuals we tested. A lack of female specificity of dm-w in X. pygmaeus is consistent with previous findings from other samples (Cauret et al. 2020).
Together, these efforts bring the total number of mostly or entirely nonhomologous sex-determining regions in the genus Xenopus to a total of (at least) eight, up from three that were previously known. The genomic locations of sex-associated regions are now known in a total of 11 Xenopus species and 2 additional species (including one dodecaploid) are established here to have heterogametic females (Fig. 1). Most or all of this sex chromosome diversity evolved during the diversification of extant species in the genus Xenopus from their most recent common ancestor beginning ∼50 Ma, or more recently (Fig. 1; Evans et al. 2015; Feng et al. 2017; Portik et al. 2023). This demonstrates a remarkably rapid rate of sex chromosome evolution in closely related amphibian species that have mostly heterogametic females.
Heterochiasmy
At the time of this study, three Xenopus species had chromosome-scale genome assemblies: X. tropicalis (Hellsten et al. 2010; Mitros et al. 2019; Bredeson et al. 2024), X. laevis (Session et al. 2016), and X. borealis (Evans et al. 2022), that allowed us to further refine estimates of sex-specific recombination landscapes using RRGS data. In all three species, we observed significantly more recombination during oogenesis than spermatogenesis, resulting in longer maternal than paternal map lengths (heterochiasmy; Table 3). The female-to-male ratio of the rate of recombination per base pair was only slightly higher in female than male X. tropicalis (1.32) and X. laevis (1.50) but substantially higher in X. borealis (2.66; Table 3). These genetic maps covered similar genomic regions (supplementary fig. S11, Supplementary Material online), which suggests that the differences in map length are not a consequence of genome-wide variation in recombination rates (for example, if, by chance, the maternal linkage groups were more frequently in regions where recombination was high in females, but the opposite for the paternal linkage groups).
Table 3.
Physical (in millions of base pairs, Mb) and genetic distances (in centimorgans, cM) that were covered by the largest linkage group in each chromosome, by species
| L subgenome | S subgenome | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Sex | Mb | cM | cM/Mb | F/M cM/Mb ratio | Mb | cM | cM/Mb | F/M cM/Mb ratio | Mb | cM | cM/Mb | F/M cM/Mb fm ratio |
| X. tropicalis | F | 1401 | 1191 | 0.85 (0.07) | 1.32a | ND | ND | ND | ND | ND | ND | ND | ND |
| M | 1342 | 867 | 0.65 (0.12) | ND | ND | ND | ND | ND | ND | ND | ND | ||
| X. laevis | F | 2450 | 1964 | 0.80 (0.08) | 1.50a | 1355 | 992 | 0.73 (0.05) | 1.31a | 1095 | 973 | 0.89 (0.16) | 1.75 |
| M | 2549 | 1367 | 0.54 (0.04) | 1405 | 785 | 0.56 (0.05) | 1144 | 582 | 0.51 (0.07) | ||||
| X. borealis | F | 2324 | 1739 | 0.75 (0.07) | 2.66a | 1261 | 910 | 0.72 (0.12) | 3.07a | 1063 | 829 | 0.78 (0.09) | 2.29a |
| M | 2402 | 675 | 0.28 (0.04) | 1342 | 315 | 0.23 (0.03) | 1060 | 360 | 0.34 (0.07) | ||||
ND - Not Defined.
The ratio of these distances (cM/Mb) and the female:male ratio of these ratios (F/M cM/Mb ratio) allows for comparisons of the average genome-wide recombination rate during oogenesis (F) and spermatogenesis (M). These statistics were also calculated for the L and S subgenomes (L and S subgenome, respectively) of the allotetraploids X. laevis and X. borealis. The weighted standard error of the mean is given in the parentheses. aSignificant departure of the F/M cM/Mb ratio from the null expectation of no bias (see Materials and Methods).
Xenopus laevis and X. borealis are derived from a common allotetraploid ancestor (Evans et al. 2005) and their L subgenomes are orthologous to each other and their S subgenomes are also orthologous to each other. We observed significantly more maternal than paternal recombination per base pair in both subgenomes of X. borealis, and also in the L subgenome of X. laevis (Table 3).
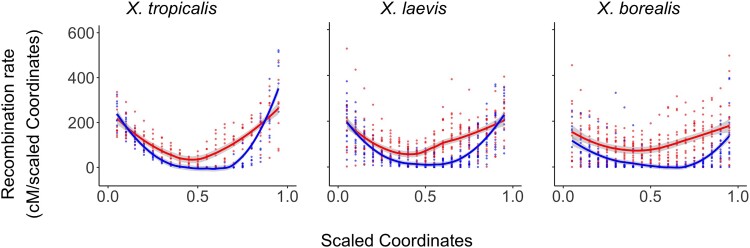
In all three species, there is pronounced heterogeneity in the rate of recombination which is evident along individual chromosomes (supplementary fig. S12, Supplementary Material online) and in fitted estimates across all chromosomes within each species (Fig. 4). Recombination in both sexes tends to be more frequent toward the tips than centers of chromosomes, but the bias toward the tips is more pronounced in spermatogenesis than oogenesis. This is evidenced by a more concave fit for male recombination of the smoothed recombination rates across scaled chromosomes and at individual chromosomes, including very low or absent recombination in the centers of chromosomes during spermatogenesis (fig. 4, supplementary fig. S12, Supplementary Material online).
Fig. 4.
Heterochiasmy is evidenced in three Xenopus species: X. tropicalis (left), X. laevis (center), and X. borealis (right) by maternal (red) and paternal (blue) recombination landscapes. Lines indicate smoothed estimates of recombination rates during oogenesis (red) and spermatogenesis (blue) across linkage groups from all intraspecific chromosomes, with standard errors of these estimates in gray. Chromosomes were scaled to be one unit long (x-axis); recombination rates (y-axis) were estimated from the first derivative of predicted values of a spline with a monotonic increase that was fitted to the largest maternal and paternal linkage group from each chromosome after scaling (red and blue dots, respectively). Plots of genetic versus physical map distances and corresponding estimates of recombination rate for individual chromosomes in each species are provided in supplementary figs. S11 and S12, Supplementary Material online.
These analyses also highlight sex differences in recombination within sex-associated regions. For example, in X. borealis, there is a relatively high rate of paternal recombination between the sex-associated portions of the two Z chromosomes within the first 54.1 Mb of Chr8L—especially in the first ∼25 Mb—but no observed maternal recombination between the sex-associated portions of the W and Z chromosomes in this region (supplementary figs. S11 and S12, Supplementary Material online). Moreover, the paternal map length of this region was 36.4 centimorgans based on 46 variable positions (centimorgams, cM), whereas the maternal map length was 0 cM based on 222 variable positions. In the sex-associated regions of X. tropicalis (<11 Mb on Chr7) which has a complex system for sex determination that includes three sex chromosomes (Roco et al. 2015), maternal and paternal recombination was detected between 9.6 to 9.9 Mb (3.0 cM, 4 markers) and 2.6 to 4.9 Mb (11.8 cM, 3 markers), respectively, but marker density was insufficient to quantify recombination in other portions of this sex-associated region. In the small sex-associated regions of X. laevis (∼177 to 190 Mb on Chr2L), there was insufficient marker density to estimate maternal or paternal recombination rates.
We performed stringent quality filtering before recombination analysis (see Materials and Methods), but genotyping errors may still be present in our data, for example because of technical sources such as allelic dropout, wherein one allele is not sequenced due to a heterozygous polymorphism that affects a restriction enzyme site. One concern is that genotype errors could artificially increase genetic map lengths in the parent with more markers (there were more markers in the mother than the father of the X. tropicalis and X. borealis families, and more in the father than the mother for the X. laevis family; see Materials and Methods). It is also possible that a higher density of markers increased power to detect recombination events. However, we did not observe a strong positive correlation between genetic distance (in cM) and marker density for maternal linkage groups, even though the maternal maps were significantly longer than the paternal maps (supplementary fig. S13, Supplementary Material online; Table 3). Compared with the paternal maps, the longer maternal maps in these species are therefore not likely to be due to genotyping errors.
Sex-Associated Regions Usually not Near Chromosome Centers
Except for X. fischbergi (Fig. 2), all known sex-associated regions in Xenopus are either entirely situated near a chromosome telomere, or span large regions where one end is near a chromosome telomere. Only one sex-associated region (of the population of X. borealis from East Kenya) mapped to a region that spanned a centromere in the X. laevis reference genome (Fig. 2). The small sex-associated region of X. lenduensis was on the short arm of Chr3L, which is a subtelocentric chromosome in X. laevis, resulting in this region mapping near (<5 Mb) to a centromere in the reference genome, and also near to the telomere of this chromosome. Comparative cytogenetic analysis of X. tropicalis and X. laevis suggests that the positions of genes relative to centromeres are frequently but not always conserved (Session et al. 2016); thus, there may be among-species variation in centromere locations that we are unable to detect.
Permutation tests indicate that Xenopus sex-associated regions tend to be significantly farther from the centers of chromosomes than expected by chance (P < 0.001, test statistic = 1.14, 95% confidence interval of permutations: 0.19 to 0.79), with the caveat that we assumed that relative chromosome lengths within all species in the subgenus Xenopus match those of X. laevis. This permutation result is inconsistent with a strong effect of natural selection favoring the establishment of sex-determining regions in genomic areas with low rates of recombination (which in Xenopus are in the centers of chromosomes in both sexes, Fig. 4, supplementary fig. S10, Supplementary Material online). However, it is notable that the largest sex-associated region detected here—on the W chromosome of X. fischbergi—is in the center of a chromosome (Fig. 2), where recombination is lowest during oogenesis (Fig. 4). The large sex-associated region of one population of X. borealis also spans a portion of a chromosome center. The size of this region may also be influenced by the ancestral recombination landscape because recombination in this region was probably rare during oogenesis even before the region became sex linked.
Discussion
No Evidence for dm-w Being a Jumping Sex-Determining Gene in Xenopus
Clear signatures of sex-associated genetic variation exist in thirteen Xenopus species (X. allofraseri, X. borealis, X. boumbaensis, X. fischbergi, X. fraseri, X. gilli, X. laevis, X. lenduensis, X. longipes, X. mellotropicalis, X. muelleri, X. pygmaeus, and X. tropicalis) and at least 8 sex-associated regions in 11 of these species are mostly or entirely nonhomologous. These observations rule out scenarios where sex determination in these species is random (Perrin 2016) or environmental.
One way that variation in the genomic locations of sex-linked regions could arise is via a “jumping” sex-determining gene, such as the sex-determining region cassette in strawberries (Tennessen et al. 2018) and sdY in salmonids (Yano et al. 2012). In X. pygmaeus, dm-w was previously detected in 6 of 9 females and 2 of 11 males (Cauret et al. 2020), and we report here a strong signature of sex association on Chr8L in one X. pygmaeus family where both parents and all offspring we surveyed of both sexes had dm-w. This illustrates that a species with dm-w has a genetic mechanism for sex determination that is not triggered by this gene. Consistent with this, the sex determination region of X. boumbaensis may be homologous to that of X. pygmaeus, but X. boumbaensis may lack dm-w (based on targeted capture sequencing of one female; Cauret et al. 2020, 2023). In other species that carry dm-w (X. clivii, X. itombwensis, X. kobeli, X. poweri, and X. victorianus), dm-w is also not female specific (Fig. 1; Cauret et al. 2020, 2023). Interestingly, available information from X. victorianus suggests that the absence of female specificity of dm-w may be restricted to the population in the region on and near the Lendu Plateau in the DRC; in populations from other portions of East Africa, dm-w may be female specific (Cauret et al. 2020). In a recently released chromosome-scale genome assembly of X. petersii (https://www.ncbi.nlm.nih.gov/datasets/genome/GCA_038501915.1), dm-w is in a homologous genomic location on Chr2L to X. laevis. In several other Xenopus species, dm-w is absent (X. borealis, Xenopus calcaratus, Xenopus epitropicalis, X. mellotropicalis, and X. tropicalis) or may be absent (X. allofraseri, Xenopus eysoole, X. fischbergi, X. longipes, X. muelleri, X. parafraseri, Xenopus ruwenzoriensis, and X. wittei; Cauret et al. 2020). For these reasons, a “jumping gene” scenario involving dm-w cannot account for the high diversity of nonhomologous sex-associated regions in Xenopus. Because we do not know what the sex-determining genes are in almost all Xenopus species, we are unable to rule out the possibility that there is a mobile sex-determining locus other than dm-w. It is conceivable that some among-species variation in the locations of sex-associated regions could stem from undetected structural variation because RRGS data were mapped to reference genomes from only two species.
Rapid Evolution of Reduced Recombination After the Birth of New Sex Chromosomes
In both sexes of three Xenopus species (X. tropicalis, X. laevis, and X. borealis), patterns of heterochiasmy are similar, with recombination tending to be proportionately higher on the tips of chromosomes compared with the centers in both sexes (Fig. 4, supplementary fig. S10, Supplementary Material online), and with the rate of recombination being significantly higher in females than males (Table 3). Recombination in these three species occurs in both sexes in genomic regions that are now largely sex-associated regions in other species, such as X. fischbergi (supplementary figs. S11 and S12, Supplementary Material online). Moreover, eight nonhomologous sex-associated regions are more frequently located on the tips of chromosomes than expected by chance (P < 0.001, permutation test). If the ancestral recombination landscape of these species matches X. tropicalis, X. laevis, and X. borealis—which seems plausible—then the locations of known sex-associated regions in Xenopus do not support a “recombination suppression first” scenario where natural selection favors the origin of new sex-determining regions in areas of the genome that already have low recombination rates. In contrast, it seems that rapidly recombining chromosomal tips are more likely to become sex-associated regions compared with the centers of chromosomes. This could reflect variation in the density of sex-related genes, which may be more frequently recruited to become triggers for sex determination than non-sex-related genes (Jeffries et al. 2018; Adolfi et al. 2021). Frequent origins of sex linkage at the tips of chromosomes could also be related to the high density of repetitive elements at chromosome tips (Bredeson et al. 2024) that could make these regions more susceptible to duplication, insertion, or translocation. Another possibility is that the extent of sex-linked recombination arrest is influenced by patterns of heterochiasmy, even though these patterns do not directly influence where a master sex-determining gene originates. The large regions of sex-linked recombination arrest in or near the centers of the sex chromosomes of X. borealis and of X. fischbergi are consistent with this possibility.
Several sex chromosomes were identified here using RRGS data from one or more families with limited or no data from non-kin (X. allofraseri, X. boumbaensis, X. fischbergi, X. mellotropicalis, X. muelleri, X. pygmaeus, and X. tropicalis). Because only one generation of recombination is represented in a family, the completely sex-linked region of recombination arrest is almost certainly smaller than the entire sex-associated region inferred from families. In X. pygmaeus, for example, Sanger sequencing identified sex-associated SNPs in a family, but not in wild-caught individuals. This also could reflect variation in the sex location within each species, and whole-genome sequencing could distinguish between these possibilities.
It is nonetheless clear that large regions (up to ∼34 to 64 Mb) of recombination reduction or arrest evolved rapidly after independent origins or movement of new sex-determining regions in several closely related species (X. muelleri, X. fischbergi, and one population of X. borealis); other smaller regions (up to ∼3 Mb or more) also arose in several species (X. allofraseri, X. laevis, X. lenduensis, X. pygmaeus + X. boumbaensis; Fig. 2). Perhaps most striking is the previously reported intraspecific variation in the size of the sex-associated region in X. borealis, which is ∼54.1 Mb in a population from eastern Kenya, but unidentified using RRGS of a population from western Kenya (supplementary fig. S1, Supplementary Material online; Evans et al. 2022). In captive X. borealis derived from eastern Kenya, we observed recombination in the sex-associated portion of Chr8L in males but not females, which suggests reduced or arrested recombination. There is also extensive allele-specific expression along the entire sex-associated portion of the W chromosome in X. borealis from eastern Kenya (Song et al. 2021), which is consistent with recombination arrest.
In the sex-associated region of X. fischbergi, FST between females and males is elevated (supplementary fig. S5, Supplementary Material online), which is consistent with divergence due to recombination arrest. Atypically high FST between females and males is also observed in portions of the sex-associated regions of other species—including X. borealis, which is closely related to X. fischbergi, and evidences the evolution of sex-associated recombination reduction or arrest in each species following their divergence. Together these observations suggest that the rate of recombination rapidly decreased multiple times when new sex-associated genomic regions evolved. Mechanisms behind sex-linked recombination reduction or arrest in Xenopus remain unknown, but possibilities such as inversions could be readily explored using cytogenetic and genomic approaches.
There are caveats to our inferences with respect to the recombination landscape and sex-associated recombination reduction or arrest. Errors in genome assembly, mismapped reads, and divergence in genome structure could generate markers in the genetic map that did not match the order of their genomic coordinates. While we have no reason to suspect a systematic bias by sex or genomic location of genotype errors or misplaced markers, their net effect would be to overestimate map lengths and recombination rates.
Functional Dominance of the L Subgenome
Except X. tropicalis, all extant species in Xenopus are allopolyploid, and their genomes have compartments called “subgenomes” that are derived from lower ploidy ancestors. Asymmetric evolution of subgenomes in the subgenus Xenopus is suggested by multiple lines of evidence, including differences in the length of the chromosomes in each subgenome (longer in the L than the S subgenome), and differences in gene expression (higher in the L), rates of pseudogenization (lower in the L), gene length (longer in the L), and there are differences in the compositions of transposable elements (Session et al. 2016; Elurbe et al. 2017; Furman et al. 2018). However, in natural populations of X. laevis, patterns of population structure and gene flow in each subgenome are similar (Premachandra et al. 2023). All known triggers for sex determination in the subgenus Xenopus allotetraploids are in the L subgenome (this study; Yoshimoto et al. 2008; Furman and Evans 2016), which further underscores the functional primacy (“subgenome dominance”) of the L compared with the S subgenome.
A plausible explanation for subgenome dominance is that it stems from differences in gene expression that were present immediately or soon after the origin of the ancestral allopolyploid (e.g. Schnable et al. 2011; Edger et al. 2017). Differences in genome size of the ancestral diploid species creates more potential transcription factor binding sites in the subgenome derived from the ancestor with the larger genome (An et al. 2024). Expression and functional divergence between subgenomes therefore may be an immediate consequence of allopolyploidization when the subgenomes are differently sized (An et al. 2024). It is also possible that selection to maintain cytonuclear interactions might favor retention of the maternally inherited subgenome, as has been proposed for various allopolyploid teleost fishes (Xu et al. 2023). Within the subgenus Xenopus, the maternally inherited subgenome has not been identified because diploid descendants of both diploid ancestors have not been identified and may be extinct. Interestingly, a partial gene duplication that gave rise to dm-w involved dmrt1.S, which is one of two homeologous copies of the male-related gene dmrt1 that resides in the (recessive) S subgenome (Bewick et al. 2011), even though dm-w resides in the (dominant) L subgenome. Of note is that our findings in Xenopus contrast with those from at least one example from pitcher plants, where the sex-determining locus resides in the “recessive” subgenome (Saul et al. 2023).
Whatever the cause of divergent subgenome evolution, it is clear that genome duplication coupled with pseudogenization and expression divergence between homeologs contributes to biological complexity in Xenopus. In Xenopus, sex-specific alleles such as dm-w are not duplicated by polyploidization, but must interface with autosomal sex-related genes such as dmrt1 that vary extensively in copy number and (presumably) relative expression levels among allotetraploid, allooctoploid, and allododecaploid species (Bewick et al. 2011). It is thus conceivable that genome duplication contributes to sex chromosome turnover. One example of this is known from weeping willows (Salix babylonica) where allopolyploidization was coupled with a transition from male to female heterogamety despite retention of the sex-associated region from one of the diploid ancestors (He et al. 2024). In Xenopus, however, the available evidence argues against this possibility because the sex-determining system was conserved across differing ploidy levels in X. tropicalis (a diploid) and X. mellotropicalis (a tetraploid) and in X. pygmaeus (a tetraploid) and X. boumbaensis (an octoploid). Sex chromosomes were also maintained through polyploidization in the plant genus Mercurialis (Toups et al. 2022).
Mutational Load and Sexual Antagonism
Under the mutational load model, recombination arrest leads to degeneration of sex-linked portions of a sex-specific sex chromosome (W or Y). In species with female heterogamety, increased mutational load on the nonrecombining portion of the W chromosome favors the establishment of a new sex-determining region elsewhere in the genome, extinction of the ancestral W chromosome, and concomitant autosomal segregation of the ancestral (nondegenerate) Z chromosome (Blaser et al. 2013, 2014). There are several ways that this can happen (Bull and Charnov 1977) that depend on whether transition between sex-determining systems happens within the same pair of sex chromosomes (a “homologous” transition sensu van Doorn and Kirkpatrick 2010) or between different pairs of chromosomes including an ancestral sex chromosome and an ancestral autosome (a “nonhomologous” transition sensu van Doorn and Kirkpatrick 2010).
In Xenopus, most known sex chromosome turnovers resulted in no change from female heterogamety. If newly emerged triggers for sex determination are dominant, then preservation of the heterogametic sex is consistent with expectations of the mutational load model. However, nonhomologous change in the heterogametic sex is still possible under the mutational load model if the newly emerged sex-determining locus is recessive (Table 2B in Bull and Charnov 1977). Thus, a switch of female to male heterogamety in X. allofraseri and the newly evolved Y chromosome in X. tropicalis (Roco et al. 2015; Furman et al. 2020) could allow the ancestral W chromosome to go extinct, and therefore still be consistent with the mutational load model. If this were to happen in X. tropicalis, it would be a homologous sex chromosome transition, whereas the turnover in X. allofraseri was nonhomologous.
Similar to the mutational load model, the sexual antagonism model can be difficult to detect and distinguish from other phenomena that result in similar genomic signatures (Ponnikas et al. 2018). However, some insights into the sexual antagonism model for sex chromosome turnover can be gleaned by comparing the recombination landscape to the positions of sex-associated regions in Xenopus. With female-higher recombination, a new male-determining gene would more likely be linked to a locus with sexually antagonistic effects than a new female-determining gene. This leads to the expectation that more turnover events should occur in species with male heterogamety if males have a lower rate of recombination than females (Prediction 1 in Sardell and Kirkpatrick 2020). In line with this expectation, at least ten sex chromosome turnover events occurred in ranid frogs with mostly male heterogamety and extreme female-higher recombination bias (in at least some species; Jeffries et al. 2018). However, almost as many (and maybe more) turnover events occurred in mostly female heterogametic species of Xenopus over a similar timespan, even though recombination in females is higher than in males (Table 1). This demonstrates that extreme heterochiasmy is not required for rapid sex chromosome turnover, and fails to support the expectation (when recombination is female biased) that the rate of sex chromosome turnover in species with female heterogamety should be more modest compared with species with male heterogamety. However, the comparison between the roles of heterochiasmy in ranids and Xenopus is confounded by polyploidy, which is far more common in Xenopus (Schmid et al. 2015), and clearly, more information is needed from other groups.
Even without heterochiasmy, if sexual antagonism was driving sex chromosome turnover, sex-linked regions should more frequently reside in areas with low rates of recombination where new sex-determining genes are more likely to be linked to mutations with sexually antagonistic fitness effects. Here, again this prediction is not borne out in Xenopus because most sex-associated regions occur on the ends of chromosomes where recombination is highest in both sexes (Fig. 4). However, if genes that are prone to sexually antagonistic mutations tend to occur on chromosome ends, this could favor the origin of sex-associated regions despite high rates of recombination.
Implications for Rapid Evolution of Sex Determination
The minimum rate of sex chromosome turnover calculated here (1 turnover every ∼46 My) is more than an order of magnitude slower than, for example, a rate recently estimated in cichlid fish (El Taher et al. 2021), but our estimate is extremely conservative because it is based on the unlikely assumption that all turnover events have been detected. Further, the mitochondrial diversification times in Fig. 1 (Evans et al. 2019) were constrained to match an analysis that used fossil calibration points (Feng et al. 2017), and are similar to times recovered from an analysis that uses a combination of fossil and geological calibration points (Evans et al. 2004). However, these divergence times are substantially older than those estimated from analysis of mitochondrial or nuclear DNA when different calibrations were used (Cannatella 2015; Evans et al. 2015). It is notable that other divergence estimates are even more recent, such as an estimate of 17 to 18 Ma for the age of the most recent common ancestor of species in the subgenus Xenopus (Session et al. 2016). More recent diversification of species in subgenus Xenopus would indicate an even more rapid pace of sex chromosome diversification. Over a large phylogenetic scale, changes in heterogamety point to a high rate of sex chromosome turnover in amphibians that also underestimate the true rate because turnovers that do not change the heterogametic sex are not counted (Evans et al. 2012; Pennell et al. 2018; Ma and Veltsos 2021). Results reported here and elsewhere (Dufresnes et al. 2015; Jeffries et al. 2018) open the possibility that very rapid sex chromosome evolution—even in very recent diversifications—is a widespread but poorly documented phenomenon in frogs (or amphibians in general).
The high diversity in nonhomologous sex-associated regions in Xenopus is remarkable in its own right and allows us to conclude that (i) sex determination has a genetic basis in many (or all) Xenopus, (ii) heterochiasmy and variation within the genome in the rate of recombination seem to be poor prognosticators for locations of sex-associated regions in Xenopus, (iii) extreme heterochiasmy is not necessary for rapid sex chromosome turnover, and (iv) sex-associated recombination reduction or arrest evolved rapidly in several Xenopus species after the origin of sex-determining regions.
Because the mutational load and sexual antagonism models make overlapping predictions with respect to changes in the heterogametic sex, we are unable to rule out or confirm the roles of these mechanisms. But caveats are necessary for the application of each one, specifically (i) that new sex-determining genes are occasionally recessive to account for changes from female to male heterogamety under the mutational load model, and that (ii) genes with potentially sexually antagonistic function must occur at the ends of chromosomes to account for these regions repeatedly becoming sex associated despite being where recombination is highest in both sexes under the sexual antagonism model. Our observations in Xenopus also fail to rule out a role of genetic drift in sex chromosome turnover because this model also allows for occasional changes in the heterogametic sex (Saunders et al. 2018). Although simulation studies have assumed otherwise (Saunders et al. 2019), caveat (i) does not seem far fetched, because this could arise via loss of function mutations at sex-related genes where the wild-type allele is haplosufficient. This is a major cause of autosomal recessive diseases in humans, which have a disease burden that is similar to autosomal dominant diseases (Baird et al. 1988; Xiao and Lauschke 2021).
New sex-determining regions may arise through mechanisms such as allelic divergence, such as Sry in eutherian mammals (Koopman et al. 1990), gene duplication, such as dm-w in Xenopus and dm-Y in medaka fish (Matsuda et al. 2007; Yoshimoto et al. 2008), gene recruitment, such as sdY in salmonids (Yano et al. 2012), and haploinsufficiency, such as dmrt1 in birds and the half-smooth tongue sole (Smith et al. 2009; Chen et al. 2014). Currently, only one genetic trigger for sex determination has been identified in amphibians: dm-w in X. laevis (Yoshimoto et al. 2008); another strong candidate has been identified in Bufotes viridis (Kuhl et al. 2024). An exciting implication of these discoveries in Xenopus is the opportunities they provide to identify and test other newly evolved triggers for sex determination. This can be accomplished in several ways that take advantage of currently available tools such as gene editing, chromosome-scale genome assemblies, and relative ease of animal husbandry. Characterization of more triggers for sex determination in amphibians promises to inform us about the relative importance of alternative mechanisms by which novel gene function and genetic pathways form (e.g. Adolfi et al. 2021), and the types of genes that evolution co-opts to become sex-determining genes (Pan et al. 2021).
Materials and Methods
Samples and Data
Species identifications were made by sequencing a portion of the 16S rRNA gene in the mitochondrial genome using primers 16sc-L and 16sd-H (Evans et al. 2003), and then comparing these sequences to complete mitochondrial genome sequences from all Xenopus species (Evans et al. 2019). For each wild-caught sample and each parent of each cross (except two X. tropicalis families), the sequenced region was identical or almost identical to the homologous region of only one Xenopus mitochondrial genome. Mitochondrial sequencing was not performed for two X. tropicalis families (PRJNA526297), but the group that generated these data also did the complete genome sequence of X. tropicalis (Mitros et al. 2019) and the species identification of those samples is thus not in question. Mitochondrial DNA sequences rarely introgress between Xenopus species that hybridize (e.g. Evans et al. 1998; Furman et al. 2017) or that are sympatric (e.g. Evans et al. 2011, 2015), are thus typically diagnostic of species.
We searched for sex-associated SNPs with RRGS of wild caught and lab reared individuals from a total of 929 individuals from 19 species (median: 34 individuals/species; range: 9 to 190 individuals/species; Table 1, supplementary table S1, Supplementary Material online). Samples from some species (n = 10) were exclusively from wild-caught individuals, whereas others were a combination of wild-caught and lab-reared individuals (n = 7), and some species (n = 2) were exclusively from lab-reared individuals. In almost all samples, the sex of the individual was determined by dissection after euthanasia. In a small number of individuals (<5%), sex was based on external characteristics (females: large size, cloacal extensions; males: nuptial pads, smaller size). Individuals with ambiguous sex were not included. RRGS was performed on these samples using methods described elsewhere for previously collected data (Furman and Evans 2016, 2018; Mitros et al. 2019; Cauret et al. 2020; Furman et al. 2020; Evans et al. 2022; Premachandra et al. 2023), and the same methodology as Premachandra et al. (2023) for all new RRGS data reported here.
Quality Control, Mapping, Genetic Association with Sex, Subgenome Mapping Bias
Raw data were demultiplexed using sabre version 1.0 (Joshi 2011) and trimmed using cutadapt (Martin 2011) and trimmomatic version 0.39 (Bolger et al. 2014). For analysis of genetic association, data from the two species in subgenus Silurana (X. tropicalis and X. mellotropicalis) were mapped to the X. tropicalis genome assembly version 10.0 (Bredeson et al. 2024) and data from the other species (all in subgenus Xenopus) were mapped to the X. laevis genome assembly version 10.1 (Session et al. 2016). Genetic associations between single SNPs and sex were calculated using angsd version 0.939 (Korneliussen et al. 2014) using binary alignment map (.bam) files as input and enforcing a minimum depth of two and a maximum depth of 100 in order a variant in each individual to be included, and we removed variants with a minor allele frequency below 10%.
For one randomly selected sample from each species in the subgenus Xenopus, we also quantified the number of mapped reads in each subgenome to evaluate whether there was some sort of systematic bias that could influence our inferences concerning sex linkage. We established an expectation for the number of mapped reads based on the size of each subgenome (the L subgenome is ∼54% of the whole genome).
Female and Male Heterogamety
For species with newly reported sex chromosomes, we evaluated the evidence for female versus male heterogamety in two ways. When genetic data from parents were available, we evaluated sex associations of maternally and paternally inherited SNPs with the expectation that sex association would only be detected in SNPs from the heterogametic parent. For these families and for the X. boumbaensis family (where parental data were not available) and wild-caught individuals (from X. lenduensis), we compared pairwise nucleotide diversity of sex-associated positions within the sex-determining region in females and males. We expected higher diversity in these sex-associated positions in the heterogametic sex as a consequence of divergence between the sex-specific (W or Y) and shared (Z or X) sex chromosomes. This analysis did not consider pairwise diversity per site because data from two subgenomes from the octoploid species are expected to map to each subgenome of the tetraploid (X. laevis) reference genome, and for this reason, diversity per-site estimates are inaccurate (specifically, overestimates). Variable sites were genotyped using the Genome Analysis Toolkit (McKenna et al. 2010) as detailed below (but without filtering), and the diversity of significantly sex-associated positions (P < 0.001) within the sex-associated region was calculated using vcftools version 0.1.16 (Danecek et al. 2011).
RADsex Analysis
Mapping of RRGS data from a polyploid to a heterospecific reference genome with a lower ploidy level causes multiple subgenomes to map to orthologous regions of the reference genome. This is an issue for our data from X. mellotropicalis (an allotetraploid) that was mapped to the X. tropicalis reference genome (which is diploid) and for the allooctoploids (X. boumbaensis, X. itombwensis, X. lenduensis, and X. wittei) and the allododecaploid (X. longipes) which were mapped to the X. laevis reference genome (which is an allotetraploid). To work around this limitation, we complemented the association tests with a reference genome-free approach (RADsex; Feron et al. 2021). This approach identifies sequence reads in the RRGS data that are found exclusively or almost exclusively in one sex. The heterogametic sex is expected to be enriched for sex-specific and sex-biased sequences that are within or linked to the sex-specific portion of the sex-specific sex chromosome. For some species, RADsex analysis identified a significant signal of sex bias (P < 0.001). We further assessed the RADsex results by using read mapping and BLAST (Altschul et al. 1997) to assess where these putatively sex-specific or sex-biased reads mapped in the X. tropicalis genome version 10.0 (for X. mellotropicalis and X. tropicalis) or X. laevis genome version 10.1 (for species in subgenus Xenopus). Multiple reads that mapped to the same genomic region (with ∼1 Mb) were inferred to be true positives; reads that mapped to scattered genomic regions were inferred to be false positives.
Rates of Sex Chromosome Turnover
We estimated the minimum rate of sex chromosome turnover in the genus Xenopus using a time-calibrated mitochondrial phylogeny (Evans et al. 2019) and assuming that sex-associated regions on different portions of the same sex chromosomes evolved independently. The total branch lengths of Xenopus species with known sex-associated regions were calculated after removing branches that subtend species with unknown sex-associated regions using the R package ape version 5.8 (Paradis et al. 2004).
F ST in Genomic Windows
We used angsd version 0.939 (Korneliussen et al. 2014) to calculate FST between females and males in nonoverlapping 5 Mb genomic windows on the sex chromosomes of each species for which we were able to identify a sex-associated region. We required a minimum depth of 5 and a maximum depth of 30 for a variant in each individual to be included, and a mapping quality of at least 30. Analyses were performed on kin (parents and offspring) and non-kin separately.
Recombination
We assessed the genomic locations of female- and male-specific recombination based on RRGS data from one family from each of three species. This included a family from X. tropicalis (family 1: 45 individuals including both wild-caught parents, 22 daughters, 21 sons), X. laevis (31 individuals including both parents, 10 daughters, 19 sons), and X. borealis (48 individuals including both parents, 24 daughters, 22 sons). For recombination analyses, data from X. tropicalis were mapped to the X. tropicalis genome assembly version 10.0 (Bredeson et al. 2024); data from X. laevis were mapped to the X. laevis genome assembly version 10.1 (Session et al. 2016); data from X. borealis were mapped to the version 1.0 genome assembly for this species (Evans et al. 2022). For X. tropicalis and X. laevis, the average coverage of mapped reads was >20 for all samples. For X. borealis, we excluded 13 samples because coverage of mapped reads was <9X, leaving both parents, 21 daughters and 9 sons.
Prior to linkage analysis, the VariantFiltration and SelectVariants functions of the Genome Analysis Toolkit (McKenna et al. 2010) were used to filter low-quality genotypes after calling genotypes with the HaplotypeCaller and GenotypeGVCFs functions. Positions with the following attributes were removed: QD > 2.0, QUAL < 20, SOR > 3.0, FS > 60.0, MQ < 30.0, where these acronyms respectively refer to variant confidence/quality by depth (QD), genotype quality (QUAL), symmetric odds ratio of 2 × 2 contingency table to detect strand bias (SOR), Fisher exact test for strand bias (FS), and map quality (MQ). We then used vcftools version 0.1.16 (Danecek et al. 2011) to remove positions where any sample had a missing genotype, and to ensure that for all sites, that the mean depth across samples was >30X. Genotype (vcf) files were then converted to JoinMap input files (loc) using a python script (vcf2loc.py; https://github.com/tomkurowski/vcf2loc).
Before calculating genetic distances, positions with short-range double crossover events that were supported by only one genotype or by multiple genotypes within <5 Mb were set to missing. The rationale for this conservative filtering is that these genotypes are most likely genotype errors, such as “under-called” heterozygous positions that were inferred to be homozygous (Furman and Evans 2018). We excluded positions that were heterozygous in both parents because these positions are minimally informative about linkage, and generated missing genotypes for some offspring. We used the regression algorithm and Kosambi mapping function as implemented by JoinMap version 5.0 (Van Ooijen 2019), following Bredeson et al. (2024), and the “cross pollination” population type, which allowed us to separately analyze recombination events that occurred during oogenesis and spermatogenesis. We removed markers with segregation distortion (P < 0.001), used a “Grouping tree” to select markers for analysis, and used map positions in the genome assemblies to force the order of markers for each species. Following Bredeson et al. (2024), a small number of markers were excluded (3, 37, and 0 markers for X. tropicalis, X. laevis, and X. borealis, respectively) whose genetic distances were discontinuous with respect to the coordinates of flanking markers. The number of maternal and paternal markers incorporated into linkage groups for X. tropicalis, X. laevis, and X. borealis were 598 and 272, 660 and 742, and 4,316 and 2,496, respectively. The actual number of positions that were analyzed was greater than this, but many sites were excluded prior to the final recombination analysis due to the way that JoinMap handles site patterns that have missing genotypes.
The weighted standard error of the ratio of the genetic distance in cM to physical distance in Mb was calculated using the physical distance in Mb of each linkage group in each chromosome as weights. Departure of the female:male cM/Mb ratio from an expectation of no bias was tested using a weighted one-sample t-test to test the null hypothesis that the differences between maternal and paternal cM/Mb across all chromosomes was zero, with the average of the physical distances of the maternal and paternal linkage groups on each chromosome used as weights.
Permutation Tests
We used permutations to test whether the distribution of sex-associated regions of sex chromosomes was random with respect to chromosomal location. We considered only the largest confirmed sex-associated region for each of eight species, and to ensure phylogenetic independence, we only included species for each nonhomologous sex-associated regions (X. boumbaensis, X. gilli, and X. mellotropicalis were excluded because the sex-associated regions of these species are homologous to those of X. pygmaeus, X. laevis, and X. tropicalis, respectively). Because the genomes of most Xenopus are not well characterized, we further assumed that chromosome lengths of X. laevis were conserved for species in subgenus Xenopus (Session et al. 2016; Smith et al. 2021); for X. tropicalis (which is in the subgenus Silurana), we used information from that species (Bredeson et al. 2024). We used as a test statistic the sum across eight species (X. allofraseri, X. borealis, X. fischbergi, X. laevis, X. lenduensis, X. muelleri, X. pygmaeus, and X. tropicalis) with clearly defined sex-associated regions (Fig. 1) of squared proportions of the sex chromosome that is between the centers of the sex-associated region and the sex chromosome:
where s is the coordinate (in base pairs) of the center of each sex-associated region, d is the length of each sex chromosome (in base pairs), and this value is summed for each of n = eight species.
This test statistic was compared with those from 1,000 permutations where the locations of sex-associated regions for each species were randomly placed along their respective sex chromosomes and the test statistic was recalculated for each permutation. We then compared the observed test statistic to the distribution of the permutated ones, with the one-sided expectation that the observed test statistics would be more extreme. This expectation was based on previous studies that suggested that recombination during oogenesis was lower at chromosome ends than centers of the chromosomes (Furman and Evans 2018; Furman et al. 2020). This result was not confirmed here, though we found the disparity between recombination rates in the tips and the centers of chromosomes to be more modest in females compared with males (see Results). A limitation of this approach is that it does not consider sex-associated regions that do not map contiguously to the largest sex-associated region due to rearrangements that occurred during divergence of the reference genome from the focal species. However, for most species, there was no strong signal of substantially sized sex-associated regions apart from the largest one, and for this reason, we expect this would have a modest effect.
Supplementary Material
Acknowledgments
The authors thank staff at the Central Animal Facility at McMaster University for assistance with animal care and Ben Bolker, Johan Van Ooijen, and Jessen Bredeson for helpful discussions. The authors thank C. Ofori-Boateng, D.V. Wasonga, B.L.S. Furman, C. Kusamba, W.M. Muninga, M.M. Aristote, M. Zigabe, A.M. Marcel, M. Luhumyo, and J.F. Akuku for assistance with fieldwork.
Contributor Information
Ben J Evans, Department of Biology, Life Sciences Building Room 328, McMaster University, 1280 Main Street West, Hamilton, ON Canada L8S4K1.
Václav Gvoždík, Institute of Vertebrate Biology of the Czech Academy of Sciences, Brno, Czech Republic; Department of Zoology, National Museum of the Czech Republic, Prague, Czech Republic.
Martin Knytl, Department of Biology, Life Sciences Building Room 328, McMaster University, 1280 Main Street West, Hamilton, ON Canada L8S4K1; Department of Cell Biology, Charles University, Viničná 7, Prague 12843, Czech Republic.
Caroline M S Cauret, Department of Biology, Life Sciences Building Room 328, McMaster University, 1280 Main Street West, Hamilton, ON Canada L8S4K1; Department of Botany and Plant Pathology, Oregon State University, Cordley Hall 4605, 2701 SW Campus Way, Corvallis, OR 97331, USA.
Anthony Herrel, UMR 7179, Mécanismes Adaptatifs et Evolution, Muséum national d'Histoire naturelle CNRS, Paris, France; Department of Biology, Evolutionary Morphology of Vertebrates, Ghent University, Ghent, Belgium; Department of Biology, University of Antwerp, Wilrijk, Belgium; Naturhistorisches Museum Bern, Bern, Switzerland.
Eli Greenbaum, Department of Biological Sciences, The University of Texas at El Paso, El Paso, TX 79968, USA.
Jay Patel, Department of Biology, Life Sciences Building Room 328, McMaster University, 1280 Main Street West, Hamilton, ON Canada L8S4K1.
Tharindu Premachandra, Department of Biology, Life Sciences Building Room 328, McMaster University, 1280 Main Street West, Hamilton, ON Canada L8S4K1.
Theodore J Papenfuss, Museum of Vertebrate Zoology, University of California, Berkeley, CA 94720, USA.
James Parente, Eugene Bell Center for Regenerative Biology and Tissue Engineering and National Xenopus Resource, Marine Biological Laboratory, Woods Hole, MA, USA.
Marko E Horb, Eugene Bell Center for Regenerative Biology and Tissue Engineering and National Xenopus Resource, Marine Biological Laboratory, Woods Hole, MA, USA.
John Measey, UMR 7179, Mécanismes Adaptatifs et Evolution, Muséum national d'Histoire naturelle CNRS, Paris, France; Centre for Invasion Biology, Department of Botany and Zoology, Stellenbosch University, Stellenbosch 7602, South Africa; Centre for Invasion Biology, Institute of Biodiversity, Yunnan University in Kunming, Yunnan Province, China; School of Biological Sciences, University of Portsmouth, Portsmouth, UK.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online.
Author Contributions
B.J.E. conceived of the study, reared several species (X. muelleri, X. fischbergi, and X. allofraseri), collected individuals from several species with collaborators (X. allofraseri, X. clivii, X. fraseri, X. gilli, X. itombwensis, X. laevis, X. largeni, X. lenduensis, X. parafraseri, X. tropicalis, and X. wittei), performed all computational analyses, and wrote the paper. V.G. collected and reared the X. pygmaeus family. M.K., J.P., and B.J.E. generated Sanger sequences. C.M.S.C. assisted with rearing of X. muelleri and X. fischbergi and collection of X. tropicalis and X. fraseri. A.H. collected X. allofraseri. E.G. collected several species (X. itombwensis, X. pygmaeus, X. victorianus, and X. wittei). J.P. and M.E.H. reared X. boumbaensis. Th.P. performed DNA extractions. Te.P. collected X. clivii. J.M. collected several species (X. gilli and X. laevis). All authors provided comments on the manuscript.
Funding
This work was supported by the Natural Science and Engineering Research Council of Canada (RGPIN-2017-05770 and RGPIN-2024-05290 to B.J.E.), a Resource Allocation Competition awards from Compute Canada (to B.J.E.), the Museum of Comparative Zoology (to B.J.E.), Czech Science Foundation (23-07331S to V.G.), Ministry of Culture of the Czech Republic (DKRVO 2024–2028/6.I.a, National Museum of the Czech Republic, 00023272 to V.G.), Marie Skłodowska-Curie Actions Fellowship CZ - UK (P JAC project CZ.02.01.01/00/22_010/0002902 to M.K.), the National Institute of Health (NIH R24OD030008 and P40OD010997 to MEH) and the National Research Foundation of South Africa (NRF grant no. 87759 to J.M.). Fieldwork by E.G. in DRC was funded by the Percy Sladen Memorial Fund, an IUCN/SSC Amphibian Specialist Group Seed Grant, K. Reed, MD, research funds from the Department of Biology at Villanova University, National Geographic Grants (nos. 8556 to 8508 and WW-R018-17), UTEP, and the National Science Foundation of the United States (DEB-1145459).
Data Availability
This study used RRGS data from previous accessions and newly collected data. Accessions for all RRGS data for individual samples are provided in supplementary table S1, Supplementary Material online. Previous accessions include X. laevis lab bred (single end): PRJNA421148, X. laevis wild caught (paired end): PRJNA906487, X. mellotropicalis lab bred (single end): PRJNA549161, X. borealis lab bred (single end): PRJNA319044, X. borealis wild caught (singe end): PRJNA616217, X. tropicalis wild caught (single end): PRJNA627066, X. tropicalis lab bred (single end): PRJNA526297. New RRGS data from the following species are available in BioProject PRJNA1086962: X. allofraseri (paired end), X. borealis (single end), X. boumbaensis (paired end), X. clivii (paired end), X. fischbergi (paired end), X. fraseri (paired end), X. gilli (single end), X. itombwensis (paired end), X. largeni (paired end), X. lenduensis (paired end), X. longipes (paired end), X. muelleri (paired end), X. parafraseri (paired end), X. pygmaeus (paired end), X. victorianus (single end), X. wittei (paired end), and BioProject PRJNA1095558: X. boumbaensis (paired end). Sanger sequences of a sex-associated locus of X. pygmaeus are available in GenBank (accession numbers PQ424455-PQ424482).
References
- Adolfi MC, Herpin A, Schartl M. The replaceable master of sex determination: bottom-up hypothesis revisited. Philos Trans R Soc B. 2021:376(1832):20200090. 10.1098/rstb.2020.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997:25(17):3389–3402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Pires JC, Conant GC. Gene expression bias between the subgenomes of allopolyploid hybrids is an emergent property of the kinetics of expression. PLoS Comput Biol. 2024:20(1):e1011803. 10.1371/journal.pcbi.1011803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 2013:14(2):113–124. 10.1038/nrg3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird PA, Anderson TW, Newcombe HB, Lowry RB. Genetic disorders in children and young adults—a population study. Am J Hum Genet. 1988:42:677–693. [PMC free article] [PubMed] [Google Scholar]
- Bewick AJ, Anderson DW, Evans BJ. Evolution of the closely related, sex-related genes DM-W and DMRT1 in African clawed frogs (Xenopus). Evolution. 2011:65(3):698–712. 10.1111/j.1558-5646.2010.01163.x. [DOI] [PubMed] [Google Scholar]
- Blaser O, Grossen C, Neuenschwander S, Perrin N. Sex-chromosome turnovers induced by deleterious mutation load. Evolution. 2013:67(3):635–645. 10.1111/j.1558-5646.2012.01810.x. [DOI] [PubMed] [Google Scholar]
- Blaser O, Neuenschwander S, Perrin N. Sex-chromosome turnovers: the hot-potato model. Am Nat. 2014:183(1):140–146. 10.1086/674026. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014:30(15):2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredeson JV, Mudd AB, Medina-Ruiz S, Mitros T, Smith OK, Miller KE, Lyons JB, Batra SS, Park J, Berkoff KC, et al. Conserved chromatin and repetitive patterns reveal slow genome evolution in frogs. Nat Commun. 2024:15(1):579. 10.1038/s41467-023-43012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Charnov EL. Changes in the heterogametic mechanism of sex determination. Heredity (Edinb). 1977:39(1):1–14. 10.1038/hdy.1977.38. [DOI] [PubMed] [Google Scholar]
- Cannatella DC. Xenopus in space and time: fossils, node calibrations, tip-dating, and paleobiography. Cytogenet Genome Res. 2015:145(3–4):283–301. 10.1159/000438910. [DOI] [PubMed] [Google Scholar]
- Cauret CM, Gansauge M-TT, Tupper A, Furman BLS, Knytl M, Song X, Greenbaum E, Meyer M, Evans BJ. Developmental systems drift and the drivers of sex chromosome evolution. Mol Biol Evol. 2020:37(3):799–810. 10.1093/molbev/msz268. [DOI] [PubMed] [Google Scholar]
- Cauret CMS, Jordan DC, Kukoly L, Burton S, Anele EU, Kwiecien JM, Gansauge M-T, Senthillmohan S, Greenbaum E, Meyer M, et al. Functional dissection and assembly of a small, newly evolved, W chromosome-specific genomic region of the African clawed frog Xenopus laevis. PLoS Genet. 2023:19(10):e1010990. 10.1371/journal.pgen.1010990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CY, Witschi E. Breeding of sex-reversed males of Xenopus laevis Daudin. Proc Soc Exp Biol Med. 1955:89(1):150–152. 10.3181/00379727-89-21742. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Wall JD. Inbreeding, heterozygote advantage and the evolution of neo-X and neo-Y sex chromosomes. Proc R Soc Biol Sci Series B. 1999:266(1414):51–56. 10.1098/rspb.1999.0603. [DOI] [Google Scholar]
- Charlesworth D, Charlesworth B. Sex differences in fitness and selection for centric fusions between sex chromosomes and autosomes. Genet Res. 1980:35(2):205–214. 10.1017/S0016672300014051. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhang G, Shao C, Huang Q, Liu G, Zhang P, Song W, An N, Chalopin D, Volff JN, et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet. 2014:46(3):253–260. 10.1038/ng.2890. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker R, Lunter G, Marth G, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011:27(15):2156–2158. 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresnes C, Borzée A, Horn A, Stöck M, Ostini M, Sermier R, Wassef J, Litvinchuck SN, Kosch TA, Waldman B, et al. Sex-chromosome homomorphy in Palearctic tree frogs results from both turnovers and X–Y recombination. Mol Biol Evol. 2015:32(9):2328–2337. 10.1093/molbev/msv113. [DOI] [PubMed] [Google Scholar]
- Edger PP, Smith R, McKain MR, Cooley AM, Vallejo-Marin M, Yuan Y, Bewick AJ, Ji L, Platts AE, Bowman MJ, et al. Subgenome dominance in an interspecific hybrid, synthetic allopolyploid, and a 140-year-old naturally established neo-allopolyploid monkeyflower. Plant Cell. 2017:29(9):2150–2167. 10.1105/tpc.17.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Taher A, Ronco F, Matschiner M, Salzburger W, Böhne A. Dynamics of sex chromosome evolution in a rapid radiation of cichlid fishes. Sci Adv. 2021:7:eabe8215. 10.1126/sciadv.abe8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elurbe DM, Paranjpe SS, Georgiou G, Van Kruijsbergen I, Bogdanovic O, Gibeaux R, Heald R, Lister R, Huynen MA, Van Heeringen SJ, et al. Regulatory remodeling in the allo-tetraploid frog Xenopus laevis. Genome Biol. 2017:18(1):198. 10.1186/s13059-017-1335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans BJ, Bliss SM, Mendel SA, Tinsley RC. The Rift Valley is a major barrier to dispersal of African clawed frogs (Xenopus) in Ethiopia. Mol Ecol. 2011:20(20):4216–4230. 10.1111/j.1365-294X.2011.05262.x. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Brown RM, McGuire JA, Supriatna J, Andayani N, Diesmos A, Iskandar DT, Melnick DJ, Cannatella DC. Phylogenetics of fanged frogs (Anura; Ranidae; Limnonectes): testing biogeographical hypotheses at the Asian-Australian faunal zone interface. Syst Biol. 2003:52:794–819. 10.1080/10635150390251063. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Carter TF, Greenbaum E, Gvoždík V, Kelley DB, McLaughlin PJ, Pauwels OSG, Portik DM, Stanley EL, Tinsley RC, et al. Genetics, morphology, advertisement calls, and historical records distinguish six new polyploid species of African clawed frog (Xenopus, Pipidae) from West and Central Africa. PLoS One. 2015:10(12):e0142823. 10.1371/journal.pone.0142823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans BJ, Gansauge MT, Stanley EL, Furman BLS, Cauret CMS, Ofori-Boateng C, Gvozdik V, Streicher JW, Greenbaum E, Tinsley RC, et al. Xenopus fraseri: Mr. Fraser, where did your frog come from? PLoS One. 2019:14(9):e0220892. 10.1371/journal.pone.0220892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans BJ, Kelley DB, Melnick DJ, Cannatella DC. Evolution of RAG-1 in polyploid clawed frogs. Mol Biol Evol. 2005:22(5):1193–1207. 10.1093/molbev/msi104. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Kelley DB, Tinsley RC, Melnick DJ, Cannatella DC. A mitochondrial DNA phylogeny of clawed frogs: phylogeography on sub-Saharan Africa and implications for polyploid evolution. Mol Phylogenet Evol. 2004:33(1):197–213. 10.1016/j.ympev.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Morales JC, Picker MD, Kelley DB, Melnick DJ. Absence of extensive introgression between Xenopus gilli and Xenopus laevis laevis (Anura: Pipidae), in southwestern Cape Province, South Africa. Copeia. 1998:1998(2):504–509. 10.2307/1447452. [DOI] [Google Scholar]
- Evans BJ, Mudd AB, Bredeson JV, Furman BLS, Wasonga DV, Lyons JB, Harland RM, Rokhsar DS. New insights into Xenopus sex chromosome genomics from the Marsabit clawed frog X. borealis. J Evol Biol. 2022:35(12):1777–1790. 10.1111/jeb.14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans BJ, Pyron RA, Wiens JJ. Polyploidization and sex chromosome evolution in amphibians. In: Soltis PS, Soltis DE, editors. Polyploidy and genome evolution. Berlin: Springer Verlag; 2012. p. 385–410. [Google Scholar]
- Feng YJ, Blackburn DC, Liang D, Hillis DM, Wake DB, Cannatella DC, Zhang P. Phylogenomics reveals rapid, simultaneous diversification of three major clades of Gondwanan frogs at the cretaceous-paleogene boundary. Proc Natl Acad Sci U S A. 2017:114(29):E5864–E5870. 10.1073/pnas.1704632114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feron R, Pan Q, Wen M, Imarazene B, Jouanno E, Anderson J, Herpin A, Journot L, Parrinello H, Klopp C, et al. RADSex: a computational workflow to study sex determination using restriction site-associated DNA sequencing data. Mol Ecol Resour. 2021:21(5):1715–1731. 10.1111/1755-0998.13360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. Oxford: Clarendon; 1930. [Google Scholar]
- Force A, Lynch M, Pickett B, Amores A, Yan YL, Postlethwait JH. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999:151(4):1531–1545. 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman BLS, Cauret CM, Colby G, Measey J, Evans BJ. Limited genomic consequences of hybridization between two African clawed frogs, Xenopus gilli and X. laevis (Anura: Pipidae). Sci Rep. 2017:7(1):1091. 10.1038/s41598-017-01104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman BLS, Cauret CMS, Knytl M, Song XY, Premachandra T, Ofori-Boateng C, Jordan DC, Horb ME, Evans BJ. A frog with three sex chromosomes that co-mingle together in nature: Xenopus tropicalis has a degenerate W and a Y that evolved from a Z chromosome. PLoS Genet. 2020:16(11):e1009121. 10.1371/journal.pgen.1009121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman BLS, Dang UJ, Evans BJ, Golding GB. Divergent subgenome evolution after allopolyploidization in African clawed frogs (Xenopus). J Evol Biol. 2018:31(12):1945–1958. 10.1111/jeb.13391. [DOI] [PubMed] [Google Scholar]
- Furman BLS, Evans BJ. Sequential turnovers of sex chromosomes in African clawed frogs (Xenopus) suggest some genomic regions are good at sex determination. G3. 2016:6:3625–3633. 10.1534/g3.116.033423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman BLS, Evans BJ. Divergent evolutionary trajectories of two young, homomorphic, and closely related sex chromosome systems. Genome Biol Evol. 2018:10(3):742–755. 10.1093/gbe/evy045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gvoždík V, Knytl M, Zassi-Boulou AG, Fornaini NR, Bergelová B. Tetraploidy in the Boettger's dwarf clawed frog (Pipidae: Hymenochirus boettgeri) from the Congo indicates non-conspecificity with the captive population. Zool J Linn Soc. 2024:200(4):1034–1047. 10.1093/zoolinnean/zlad119. [DOI] [Google Scholar]
- Hayashi S, Suda K, Fujimura F, Fujikawa M, Tamura K, Tsukamoto D, Evans BJ, Takamatsu N, Ito M. Neofunctionalization of a noncoding portion of a DNA transposon in the coding region of the chimerical sex-determining gene dm-W in Xenopus frogs. Mol Biol Evol. 2022:39(7):msac138. 10.1093/molbev/msac138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Wang Y, Wang Y, Zhang RG, Wang Y, Hörandl E, Mank JE, Ming R. Allopolyploidization from two dioecious ancestors leads to recurrent evolution of sex chromosomes and reversion to autosomes. Nat Commun. 2024:15. 10.1038/s41467-024-51158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kaptonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, et al. The genome of the western clawed frog Xenopus tropicalis. Science. 2010:328(5978):633–636. 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufton AL, Panopoulou G. Polyploidy and genome restructuring: a variety of outcomes. Curr Opin Genet Dev. 2009:19(6):600–606. 10.1016/j.gde.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Ironside JE. No amicable divorce? Challenging the notion that sexual antagonism drives sex chromosome evolution. BioEssays. 2010:32(8):718–726. 10.1002/bies.200900124. [DOI] [PubMed] [Google Scholar]
- Jay P, Tezenas E, Véber A, Giraud T. Sheltering of deleterious mutations explains the stepwise extension of recombination suppression on sex chromosomes and other supergenes. PLoS Biol. 2022:20(7):e3001698. 10.1371/journal.pbio.3001698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries DL, Gerchen JF, Scharmann M, Pannell JR. A neutral model for the loss of recombination on sex chromosomes. Philos Trans R Soc Lond B Biol Sci. 2021:376(1832):20200096. 10.1098/rstb.2020.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries DL, Lavanchy G, Sermier R, Sredl MJ, Miura I, Borzée A, Barrow LN, Canestrelli D, Crochet P-A, Dufresnes C, et al. A rapid rate of sex-chromosome turnover and non-random transitions in true frogs. Nat Commun. 2018:9(1):4088. 10.1038/s41467-018-06517-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N. Sabre - a barcode demultiplexing and trimming tool for FastQ files. GitHub: San Francisco, CA; 2011. [Google Scholar]
- Koopman P, Munsterberg A, Capel B, Vivian N, Lovellbadge R. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature. 1990:348(6300):450–452. 10.1038/348450a0. [DOI] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics. 2014:15(1):356. 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl H, Tan WH, Klopp C, Kleiner W, Koyun B, Ciorpac M, Feron R, Knytl M, Kloas W, Schartl M, et al. A candidate sex determination locus in amphibians which evolved by structural variation between X- and Y-chromosomes. Nat Commun. 2024:15(1):4781. 10.1038/s41467-024-49025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T, Roze D. Y recombination arrest and degeneration in the absence of sexual dimorphism. Science. 2022:375(6581):663–666. 10.1126/science.abj1813. [DOI] [PubMed] [Google Scholar]
- Lynch M, O'Hely M, Walsh B, Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001:159(4):1789–1804. 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W-J, Veltsos P. The diversity and evolution of sex chromosomes in frogs. Genes (Basel). 2021:12(4):483. 10.3390/genes12040483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011:17(1):10–12. 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Matsuda M, Shinomiya A, Kinoshita M, Suzuki A, Kobayashi T, Paul-Prasanth B, Lau E-I, Hamaguchi S, Sakaizumi M, Nagahama Y. DMY gene induces male development in genetically female (XX) medaka fish. Proc Natl Acad Sci U S A. 2007:104(10):3865–3870. 10.1073/pnas.0611707104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Uno Y, Kondo M, Gilchrist MJ, Zorn AM, Rokhsar DS, Schmid M, Taira M. A new nomenclature of Xenopus laevis chromosomes based on the phylogenetic relationship to Silurana/Xenopus tropicalis. Cytogenet Genome Res. 2015:145(3–4):187–191. 10.1159/000381292. [DOI] [PubMed] [Google Scholar]
- Mawaribuchi S, Takahashi S, Wada M, Uno Y, Matsuda Y, Kondo M, Fukui A, Takamatsu N, Taira M, Ito M. Sex chromosome differentiation and the W- and Z-specific loci in Xenopus laevis. Dev Biol. 2017:426(2):393–400. 10.1016/j.ydbio.2016.06.015. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010:20(9):1297–1303. 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean GAT, Charlesworth B. The effects of Hill-Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics. 2000:155(2):929–944. 10.1093/genetics/155.2.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitros T, Lyons JB, Session AM, Jenkins J, Shu S, Kwon T, Lane M, Ng C, Grammer TC, Khokha MK, et al. A chromosome-scale genome assembly and dense genetic map for Xenopus tropicalis. Dev Biol. 2019:452(1):8–20. 10.1016/j.ydbio.2019.03.015. [DOI] [PubMed] [Google Scholar]
- Morgan TH. Sex limited inheritance in Drosophila. Science. 1910:32(812):120–122. 10.1126/science.32.812.120. [DOI] [PubMed] [Google Scholar]
- Olmstead AW, Lindberg-Livingston A, Degitz SJ. Genotyping sex in the amphibian, Xenopus (Silurana) tropicalis, for endocrine disruptor bioassays. Aquat Toxicol. 2010:98(1):60–66. 10.1016/j.aquatox.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Pan Q, Kay T, Depincé A, Adolfi M, Schartl M, Guiguen Y, Herpin A. Evolution of master sex determiners: TGF-β signalling pathways at regulatory crossroads. Philos Trans R Soc Lond B Biol Sci. 2021:376(1832):20200091. 10.1098/rstb.2020.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analysis of phylogenetics and evolution in R language. Bioinformatics. 2004:20(2):289–290. 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Pennell MW, Mank JE, Peichel CL. Transitions in sex determination and sex chromosomes across vertebrate species. Mol Ecol. 2018:27(19):3950–3963. 10.1111/mec.14540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin N. Random sex determination: when developmental noise tips the sex balance. BioEssays. 2016:38(12):1218–1226. 10.1002/bies.201600093. [DOI] [PubMed] [Google Scholar]
- Ponnikas S, Sigeman H, Abbott JK, Hansson B. Why do sex chromosomes stop recombining? Trends Genet. 2018:34(7):492–503. 10.1016/j.tig.2018.04.001. [DOI] [PubMed] [Google Scholar]
- Portik DM, Streicher JW, Wiens JJ. Frog phylogeny: a time-calibrated, species-level tree based on hundreds of loci and 5,242 species. Mol Phylogenet Evol. 2023:188:107907. 10.1016/j.ympev.2023.107907. [DOI] [PubMed] [Google Scholar]
- Premachandra T, Cauret CMS, Conradie W, Measey J, Evans BJ. Population genomics and subgenome evolution of the allotetraploid frog Xenopus laevis in southern Africa. G3 (Bethesda). 2023:13(2):jkac325. 10.1093/g3journal/jkac325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution. 1987:41(4):911–914. 10.2307/2408899. [DOI] [PubMed] [Google Scholar]
- Roco ÁS, Olmstead AW, Degitz SJ, Amano T, Zimmerman LB, Bullejos M. Coexistence of Y, W, and Z sex chromosomes in Xenopus tropicalis. Proc Natl Acad Sci U S A. 2015:112(34):E4752–E4761. 10.1073/pnas.1505291112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues N, Betto-Colliard C, Jourdan-Pineau H, Perrin N. Within-population polymorphism of sex-determination systems in the common frog (Rana temporaria). J Evol Biol. 2013:26(7):1569–1577. 10.1111/jeb.12163. [DOI] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, et al. The DNA sequence of the human X chromosome. Nature. 2005:434(7031):325–337. 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardell JM, Kirkpatrick M. Sex differences in the recombination landscape. Am Nat. 2020:195(2):361–379. 10.1086/704943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul F, Scharmann M, Wakatake T, Rajaraman S, Marques A, Freund M, Bringmann G, Channon L, Becker D, Carroll E, et al. Subgenome dominance shapes novel gene evolution in the decaploid pitcher plant Nepenthes gracilis. Nat Plants. 2023:9(12):2000–2015. 10.1038/s41477-023-01562-2. [DOI] [PubMed] [Google Scholar]
- Saunders PA, Neuenschwander S, Perrin N. Sex chromosome turnovers and genetic drift: a simulation study. J Evol Biol. 2018:31(9):1413–1419. 10.1111/jeb.13336. [DOI] [PubMed] [Google Scholar]
- Saunders PA, Neuenschwander S, Perrin N. Impact of deleterious mutations, sexually antagonistic selection, and mode of recombination suppression on transitions between male and female heterogamety. Heredity (Edinb). 2019:123(3):419–428. 10.1038/s41437-019-0225-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Evans B, Bogart JP. Polyploidy in Amphibia. Cytogenet Genome Res. 2015:145(3–4):315–330. 10.1159/000431388. [DOI] [PubMed] [Google Scholar]
- Schnable JC, Springer NM, Freeling M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci U S A. 2011:108(10):4069–4074. 10.1073/pnas.1101368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016:538(7625):336–343. 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simakov O, Marletaz F, Yue J-X, O'Connell B, Jenkins J, Brandt A, Calef R, Tung C-H, Huang T-K, Schmutz J, et al. Deeply conserved synteny resolves early events in vertebrate evolution. Nat Ecol Evol. 2020:4(6):820–830. 10.1038/s41559-020-1156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003:423(6942):825–837. 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, Sinclair AH. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009:461(7261):267–271. 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- Smith OK, Limouse C, Fryer KA, Teran NA, Sundararajan K, Heald R, Straight AF. Identification and characterization of centromeric sequences in Xenopus laevis. Genome Res. 2021:31(6):958–967. 10.1101/gr.267781.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X-Y, Furman BLS, Premachandra T, Knytl M, Cauret CMS, Wasonga DV, Measey J, Dworkin I, Evans BJ. Sex chromosome degeneration, turnover, and sex-biased expression of sex-linked transcripts in African clawed frogs (Xenopus). Philos Trans R Soc Lond B Biol Sci. 2021:376(1832):20200095. 10.1098/rstb.2020.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck M, Kratochvil L, Kuhl H, Rovatsos M, Evans BJ, Suh A, Valenzuela N, Veyrunes F, Zhou Q, Gamble T, et al. A brief review of vertebrate sex evolution with a pledge for integrative research: towards ‘sexomics’. Philos Trans R Soc Lond B Biol Sci. 2021:376(1832):20200426. 10.1098/rstb.2020.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Wei N, Straub SCK, Govindarajulu R, Liston A, Ashman T-L. Repeated translocation of a gene cassette drives sex-chromosome turnover in strawberries. PLoS Biol. 2018:16(8):e2006062. 10.1371/journal.pbio.2006062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toups MA, Vicoso B, Pannell JR. Dioecy and chromosomal sex determination are maintained through allopolyploid speciation in the plant genus Mercurialis. PLoS Genet. 2022:18(7):e1010226. 10.1371/journal.pgen.1010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Nishida C, Yoshimoto S, Ito M, Oshima Y, Yokoyama S, Nakamura M, Matsuda Y. Diversity in the origins of sex chromosomes in anurans inferred from comparative mapping of sexual differentiation genes for three species of the Ranidae and Xenopodinae. Chromosome Res. 2008:16(7):999–1011. 10.1007/s10577-008-1257-z. [DOI] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M. Turnover of sex chromosomes induced by sexual conflict. Nature. 2007:449(7164):909–912. 10.1038/nature06178. [DOI] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M. Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics. 2010:186(2):629. 10.1534/genetics.110.118596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen JW. Joinmap 5, software for the calculation of genetic linkage maps in experimental populations. The Netherlands: Wageningen; 2019. [Google Scholar]
- Xiao Q, Lauschke V. The prevalence, genetic complexity and population-specific founder effects of human autosomal recessive disorders. NPJ Genom Med. 2021:6(1):41. 10.1038/s41525-021-00203-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Liao Z, Brock J, Du K, Li G, Chen Z, Wang Y, Gao Z, Agarwal G, Wei K, et al. Maternal dominance contributes to subgenome differentiation in allopolyploid fishes. Nat Commun. 2023:14(1):8357. 10.1038/s41467-023-43740-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano A, Guyomard R, Nicol B, Jouanno E, Quillet E, Klopp C, Cabau C, Bouchez O, Fostier A, Guiguen Y. An immune-related gene evolved into the master sex-determining gene in rainbow trout, oncorhynchus mykiss. Curr Biol. 2012:22(15):1423–1428. 10.1016/j.cub.2012.05.045. [DOI] [PubMed] [Google Scholar]
- Yoshimoto S, Ikeda K, Izutsu Y, Shiba T, Takamatsu N, Ito M. Opposite roles of DMRT1 and its W-linked paralog, DM-W, in sexual dimorphism of Xenopus laevis: implications of a ZZ/ZW-type sex-determining system. Development. 2010:137(15):2519–2526. 10.1242/dev.048751. [DOI] [PubMed] [Google Scholar]
- Yoshimoto S, Okada E, Umemoto H, Tamura K, Uno Y, Nishida-Umehara C, Matsuda Y, Takamatsu N, Shiba T, Ito M. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci U S A. 2008:105(7):2469–2474. 10.1073/pnas.0712244105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C, Massonnet M, Minio A, Patel S, Llaca V, Karn A, Gouker F, Cadle-Davidson L, Reisch B, Fennell A, et al. Multiple independent recombinations led to hermaphroditism in grapevine. Proc Natl Acad Sci U S A. 2021:118(15):15. 10.1073/pnas.2023548118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study used RRGS data from previous accessions and newly collected data. Accessions for all RRGS data for individual samples are provided in supplementary table S1, Supplementary Material online. Previous accessions include X. laevis lab bred (single end): PRJNA421148, X. laevis wild caught (paired end): PRJNA906487, X. mellotropicalis lab bred (single end): PRJNA549161, X. borealis lab bred (single end): PRJNA319044, X. borealis wild caught (singe end): PRJNA616217, X. tropicalis wild caught (single end): PRJNA627066, X. tropicalis lab bred (single end): PRJNA526297. New RRGS data from the following species are available in BioProject PRJNA1086962: X. allofraseri (paired end), X. borealis (single end), X. boumbaensis (paired end), X. clivii (paired end), X. fischbergi (paired end), X. fraseri (paired end), X. gilli (single end), X. itombwensis (paired end), X. largeni (paired end), X. lenduensis (paired end), X. longipes (paired end), X. muelleri (paired end), X. parafraseri (paired end), X. pygmaeus (paired end), X. victorianus (single end), X. wittei (paired end), and BioProject PRJNA1095558: X. boumbaensis (paired end). Sanger sequences of a sex-associated locus of X. pygmaeus are available in GenBank (accession numbers PQ424455-PQ424482).