Abstract
In a previous study of poliovirus vaccine-derived strains isolated from patients with vaccine-associated paralytic poliomyelitis (VAPP) (9, 11), we reported that a high proportion (over 50%) of viruses had a recombinant genome. Most were intertypic vaccine/vaccine recombinants. However, some had restriction fragment length polymorphism (RFLP) profiles different from those of poliovirus vaccine strains. We demonstrate here that five such recombinants, of 88 VAPP strains examined, carried sequences of wild (nonvaccine) origin. To identify the parental wild donor of these sequences, we used RFLP profiles and nucleotide sequencing to look for similarity in the 3D polymerase-coding region of 61 wild, cocirculating poliovirus isolates (43 type 1, 16 type 2, and 2 type 3 isolates). In only one case was the donor identified, and it was a wild type 1 poliovirus. For the other four vaccine/wild recombinants, the wild parent could not be identified. The possibility that the wild sequences were of a non-poliovirus-enterovirus origin could not be excluded. Another vaccine/wild recombinant, isolated in Belarus from a VAPP case, indicated that the poliovirus vaccine/wild recombination is not an isolated phenomenon. We also found wild polioviruses (2 of 15) carrying vaccine-derived sequences in the 3′ moiety of their genome. All these results suggest that genetic exchanges with wild poliovirus and perhaps with nonpoliovirus enteroviruses, are also a natural means of evolution for poliovirus vaccine strains.
Poliovirus (PV) is a human enterovirus belonging to the Picornaviridae family. The poliovirion is an icosahedral particle incorporating 60 copies each of four virus-specified polypeptides: VP1, VP2, VP3, and VP4. This protein coat encloses a single-stranded RNA genome of positive polarity, 7,500 nucleotides in length. The single large open reading frame is flanked by 5′ and 3′ extremities consisting of untranslated sequences.
PV, the causal agent of poliomyelitis, has three serotypes that do not cross-react in neutralization tests with type-specific sera. If panels of neutralizing monoclonal antibodies are used, antigenic differences are observed between the strains of the three serotypes. However, these differences are invariably confined to the limits of each serotype (5, 17, 23). This has made possible the efficient prophylaxis of poliomyelitis using two vaccines: the inactivated poliovirus vaccine (IPV) and the oral poliovirus vaccine (OPV), both of which contain all three PV serotypes.
The OPV consists of live attenuated Sabin strains obtained from wild strains (29). The OPV strains actively multiply in the gut of vaccinees, thereby eliciting a strong, long-lasting immune response (30). The digestive immunity induced by OPV prevents or limits reinfection in humans, thereby preventing natural PV transmission (13). This property has made OPV the main tool for poliomyelitis eradication (39).
Sabin PV strains have a generally good safety record in vaccine use. However, the selection of variants with increased neurovirulence constitutes a real problem with respect to vaccine safety. There is a low but constant risk of vaccine-assisted paralytic poliomyelitis (VAPP) with the use of OPV. The overall incidence of VAPP in the United States was found to be one case per 2.4 million doses distributed; for immunocompetent children receiving their first dose of OPV, the incidence was estimated at one case per 750,000 doses (28, 35). VAPP is most frequently associated with type 2 and 3 vaccine strains and rarely with type 1 vaccine strains (2, 24, 35). The risk of VAPP has increased by a factor of 10 to 14 in Romania over the last three decades (6, 34). To reduce this risk and to facilitate the transition to the exclusive use of IPV, a sequential vaccination schedule in which IPV is followed by OPV was recently introduced in the United States (1).
Sabin vaccine strains are subject to genetic variation during their multiplication in humans. Indeed, following their administration to humans, mutants bearing specific nucleotide changes are rapidly isolated, probably due to selection pressure exerted by factors such as temperature, target host cells, neutralizing antibodies, and other unknown factors (reviewed in reference 23). Loss of the original attenuated phenotype of OPV strains has been attributed to single- or multiple-nucleotide substitutions at precise sites in the genome. Natural genetic recombination in PV, another mechanism of variation in Sabin strains, was demonstrated many years ago (14, 32, 33) (for reviews, see references 4 and 19). Sabin vaccine PV-derived strains with recombinant intertypic genomes have been found to occur naturally (17, 22) and to be selected frequently in the gut of VAPP patients (9, 11, 21). In some cases, vaccine/wild (V/W) PV recombinants have been found, in which vaccine-specific segments of the Sabin virus genome have been replaced by nonvaccine sequences derived from wild PVs or, perhaps, from non-polio enteroviruses (NPEVs) (10, 19, 21, 30).
We previously used a combined analysis of two distant polymorphic segments of the viral genome to analyze a large number of OPV-derived strains isolated from VAPP cases in Romania. We detected a large number of vaccine/vaccine (V/V) recombinant strains (9). Most were easily recognized as intertypic V/V recombinants in restriction fragment length polymorphism (RFLP) tests, but we also observed many vaccine strains with genomic segments that gave atypical restriction patterns. These segments were thought to be derived from modified Sabin strains, wild PV, or from NPEVs. One of these recombinants was found to be a double recombinant containing wild sequences throughout the genome except in the capsid-encoding region (10). In this study, we further analyzed the genetic characteristics of these modified OPV-derived strains and assessed the occurrence of such strains in cases of VAPP. Nonvaccine sequences were found to be present in 6% of VAPP strains (V/W recombinants). We analyzed wild cocirculating PV strains in the search for the nonvaccine parentals of the V/W recombinants. We also detected OPV-derived sequences in the genomes of these wild strains (9% of cases). These results suggest that genetic exchanges with wild PVs (and perhaps with NPEVs) are a common mechanism of evolution for vaccine PV. We present here a detailed analysis of the five V/W Romanian recombinants and a sixth strain isolated in Belarus.
MATERIALS AND METHODS
Cells and viruses.
HEp-2c and simian Vero cells were grown in monolayers in Dulbecco modified Eagle medium supplemented with 5% fetal calf serum. The PV vaccine viruses, Sabin 1, 2, and 3, were obtained from the World Health Organization [Behringwerke (S0+1)] “master seeds.” The second passage at 34°C in HEp-2c cells of the original seed was used to prepare viral stocks. The reference virulent strains, PV1/Mahoney, PV2/MEF1, and PV3/Leon/37, were maintained in our laboratory and grown at 37°C in HEp-2c cells. The nomenclature used for strains in this study includes the PV serotype (P1, P2, or P3), origin (W, wild; V, vaccine-derived), laboratory index number (three digits), and year of isolation (two digits). Most of the PV strains studied here were isolated in Romania between 1980 and 1990.
Viruses were isolated from clinical specimens (generally stool specimens, but also nasopharyngeal swabs, samples of cerebrospinal fluid, or central nervous system tissue obtained at autopsy) from patients with poliomyelitis by using primary cultures of monkey kidney cells. The viruses were cultured further and titrated with HEp-2c cells. Viral stocks were prepared by clarifying the supernatant of infected cells by centrifugation for 20 min at 1,000 × g and were stored at −70°C. For neurovirulence tests, viruses were concentrated by ultracentrifugation at 30,000 rpm in a Kontron 50.38 rotor for 2 h and resuspended in phosphate-buffered saline supplemented with Ca++ and Mg++.
Virus identification.
Viruses were identified and typed by using a standard neutralizing assay with type-specific neutralizing antisera, as previously described (9, 11). Intratypic differentiation between W and V strains was carried out with strain-specific monoclonal antibodies (5). These antibodies were also used to separate homotypic strains from samples containing mixtures of V and W PVs. Virus origin (W or V) was systematically confirmed by RFLP assay (see below) (3). All strains analyzed in detail in this study were cloned by plaque purification. Virus stocks were prepared by one or two passages in vitro after cloning. If necessary, viruses were separated from mixtures by endpoint dilution in the presence of heterotype-specific neutralizing antisera and by plaque purification.
RFLP assay.
Recombinant genomes were detected with a double RFLP assay that has been described elsewhere (9). Briefly, two distant regions of the viral genome were subjected to PCR amplification followed by restriction enzyme digestion. The first region corresponded to nucleotides (nt) 2402 to 2881 (RFLP 1 assay) in the VP1 capsid-encoding region, and the second corresponded to nt 6086 to 6376 in the 3D polymerase-coding region (RFLP 3D1 assay). The RFLP profiles obtained with three different restriction enzymes from strains isolated in the field were compared with those of the original Sabin strains. If they were found to differ, the RFLP profiles of the field strains were compared with those of the wild strains tested.
We developed new RFLP assays to screen the entire genome for the recombination junction in the recombinant strains. Restriction enzymes were selected to differentiate the three Sabin strains over the entire length of the genome. The pairs of primers and the restriction enzymes used in RFLP assays are listed in Table 1 (some of the primers presented have been used in previous studies [3, 9, 11, 27]).
TABLE 1.
Primer pairs and restriction enzymes used for RFLP assays
| Region | Genomic primer: nucleotide positiona | Complementary primer: nucleotide positiona | Length (bp) | RFLP assay | Restriction enzymes |
|---|---|---|---|---|---|
| 5′UTR | UG52: (162)-CAAGCACTTCTGTTTCCCCGG-(182) | UC53: (595)-TTGTCACCATAAGCAGCCA-(577) | 434 | 5′ | HaeIII, AluI, HpaII |
| VP4 | UG53: (577)-TGGCTGCTTATGGTGACAAT-(595) | UC21: (1200)-TCAGGTAATTTCCACCACCA-(1181) | 624 | 4 | AluI, HpaII, DdeI |
| VP2 | UG21: (1172)-TCGAGAGGGTGGTGGTGGAA-(1191) | UC24: (1923)-ATCATGGTGTCTATTTCTGC-(1904) | 752 | 2 | AluI, DdeI, AluI |
| VP3 | UG24: (1913)-GACACCATGATTCCCCTTAA-(1932) | UC20: (2421)-TCATTACACGCTGACACAAA-(2402) | 509 | 3 | HaeIII, RsaI, TaqI |
| VP1 | UG1: (2402)-TTTGTGTCAGCGTGTAATGA-(2421) | UC1: (2881)-GAATTCCATGTCAAATCTAGA-(2862) | 480 | 1 | HaeIII, DdeI, HpaII |
| VP1-2A | UG19: (2870)-GACATGGAATTCACCTTTGTGG-(2891) | UC13: (3648)-TAGTACTTAGCTTCCATGTA-(3629) | 779 | 1-2A | HinfI, RsaI, Bsp1286 |
| P2 | UG13: (3617)-CCCACCTTCCAGTACATGGA-(3636) | UC14: (4842)-CTGGCCAGTGCATCACTGTG-(4823) | 1,226 | P2 | HaeIII, HinfI, DdeI |
| 2C | UG23: (4169)-AAGGGATTGGAGTGGGTGTC-(4188) | UC15: (4965)-CATCTCTTGAAGTTTGCTGG-(4946) | 797 | 2C | AluI, HinfI, RsaI |
| 3AB-C | UG15: (4936)-CTGTCACCAACCAGCAAACTT-(4956) | UC16: (5940)-TGTGAACCGTTCCCACCAAC-(5921) | 1,005 | 3A-C | HaeIII, HinfI, RsaI |
| 3D | UG7: (6086)-TTTGAAGGGGTGAAGGAACCAGC-(6108) | UC8: (6376)-GATGTCTCTCTTCTTCTTTCCC-(6355) | 291 | 3D1 | HaeIII, DdeI, RsaI |
| 3C-D | UG16: (5921)-GTTGGTGGGAACGGTTCACA-(5940) | UC12: (6516)-TCAATTAGTCTGGATTTTCCCTG-(6494) | 596 | 3D3 | HinfI, RsaI, Dde |
| 3D-3′UTR | UG17: (6536)-TCAGTGGCCATGAGAATGGC-(6555) | UC10: (7464)-TTTTTTTTTTTTTTTTTTTTTTTC-(7441) | 929 | 3D-3′ | HinfI, DdeI, HpaII |
Numbers refer to the nucleotide position of the primer on the Sabin 1 PV strain.
Viral RNA was reverse transcribed, and the resulting cDNA was amplified by PCR as previously described (3) but with a few modifications. Viral supernatant (1 μl), RNasin (20 U), and antisense primer (10 pmol) were heated at 80°C for 5 min and annealed by incubation at 42°C for 5 min. A mixture of 4 μl of transcriptase buffer (5×), 1 μl of deoxynucleoside triphosphates (10 mM each), 1 U of avian myeloblastosis virus reverse transcriptase, and distilled water to make a final volume of 20 μl was added to the annealed template-RNA solution, and transcription was allowed to occur at 42°C for 30 min. The reverse transcription (RT) products were heated at 95°C for 5 min for denaturation and placed immediately on ice. For primers UC14, UC16, and UC10, the temperature for annealing and RT was increased to 50°C.
The PCR products were digested with restriction enzymes, and the digestion products were resolved by electrophoresis as previously described (3, 9). The amplified fragment was named according to the corresponding region of the PV genome.
Nucleotide sequence analysis.
PCR products were sequenced with the primers used for the RFLP tests. Before being sequenced, PCR products were purified either directly after amplification on Qiaquick spin columns (Qiagen) or by isolating bands from low-melting-point agarose gels after electrophoresis by standard techniques (31).
PCR products were sequenced by the dideoxynucleotide method with the Sequenase kit, version 2.0 (U.S. Biochemicals) as previously described (9). We also used the BigDye Terminator Cycle Sequencing Ready Reaction kit, following the procedure recommended by Applied Biosystems, Perkin-Elmer. Sequences were aligned with the CLUSTAL W program, version 1.6 (37). The GenBank DNA sequence library was screened for similar sequences with the FastA program, version 3.0 (25). Dendrograms were constructed based on sequence similarity, using the programs included in the PHYLIP package, version 3.5 (7). The distance matrix was calculated by the Kimura two-parameter method, using DNADIST. Three reconstruction was performed with the KITSCH program of the PHYLIP package. In the dendrogram, the distance along the abscissa to the node connecting any two strains is a measure of the sequence divergence between those two strains. The tree was drawn using NJplot (26).
Neurovirulence.
The neurovirulence of field isolates was compared with that of reference Sabin strains and wild reference viruses using mice transgenic for the human PV receptor gene (PVR-Tg mice) (kindly provided by Akio Nomoto and T. Nomura). The intraperitoneal inoculation-mean healthy time (IP-MHT) test was used as previously described (11), except that each virus strain was tested in 12 mice instead of 6. Briefly, each mouse was inoculated IP with 108 PFU of virus strain in 500 μl of phosphate-buffered saline. The MHT was calculated for each virus as the mean number of days for which inoculated mice presented no clinical neurological signs (paresis, paralysis or prominent walking instability, or death) during the 14 days of observation. In most cases, the disease index ratio (or percentage) of diseased mice versus inoculated mice was also calculated.
RESULTS
Detection of recombinant strains.
We carried out an exhaustive search for recombinant genomes in 88 vaccine-derived PV strains isolated in Romania from patients with VAPP (9, 11). All patients from whom OPV-derived strains were isolated presented a poliomyelitis syndrome with persistent acute flaccid paralysis and were classified as having VAPP (all had received vaccine or had been in contact with vaccinees) according to World Health Organization case classification criteria (38). For each strain, two distant regions of the genome were studied by RT-PCR and comparison of restriction digestion profiles were studied by RFLP: the RFLP 1 and RFLP 3D1 assays were used to analyze the 5′ third of the VP1-coding region and the 5′ part of the 3D polymerase-coding region, respectively (11). For some strains, RFLP analysis was extended to two other genomic regions: the RFLP P2 assay analyzes almost all of the 2C-coding region and the RFLP 3D-3′ assay analyzes the 3′ end of the genome (see Materials and Methods). In this way, it was also possible to detect a multiple recombinant. The results of the screening of 88 V strains isolated from VAPP cases in Romania between 1980 and 1990, including those previously published (9–11), are presented in Table 2. None of the serotype 1 strains was found to have a recombinant genome. The proportion of recombinants was 81% for serotype 2 and 80% for serotype 3. Serotype 2 intertypic V/V recombinants showed preferential recombination with Sabin 1-derived sequences in the 3D polymerase-coding region and 3′ untranslated region (3′UTR) extremity of the genome (64%), whereas serotype 3 recombinants showed preferential recombination with Sabin 2 partners (68%).
TABLE 2.
Proportion of strains with recombinant genomes among VAPP case isolates in Romania, 1980 to 1990
| Polio-vaccine serotypea | RFLP assayb
|
Category of recombinant (genotype)c | No. of strains (% of recombinants) | Total no. of strains | % Recombinants in each serotype | |
|---|---|---|---|---|---|---|
| 1 + P2 | 3D1 + 3D-3′ | |||||
| S1 | S1 | S1 | ∼∼S1∼∼ | 5 (0) | 5 | 0 |
| S2 | S2 | ∼∼S2∼∼ | 9 (0) | 9 | ||
| S2 | S3 | S2/S3 | 9 (23) | |||
| S2 | S2 | S1 | S2/S1 | 24 (62) | ||
| S2/S3 | S1 | S2/S3/S1 | 1 (2) | 39 | 81 | |
| S2 | W | S2/W | 5 (13) | |||
| S3 | S3 | ∼∼S3∼∼ | 7 (0) | 7 | ||
| S3 | S3 | S1 | S3/S1 | 9 (32) | 28 | 80 |
| S3 | S2 | S3/S2 | 19 (68) | |||
| Total | Sn/Sn + S2/W | 67 (76) | 88d | 76 | ||
S1, S2, S3, Sabin PV vaccine-derived type 1, 2, and 3, respectively.
Characteristics of the genomes according to RFLP assays 1 and 3D1. In some cases, RFLP P2 and 3D-3′ assays were also used (see Materials and Methods). S2/S3, intertypic recombinant genome in which the 5′ part is derived from S2 and the 3′ part is derived from S3; W, wild (non-vaccine).
Sn, Sabin strain of any serotype; ∼∼S(n)∼∼, nonrecombinant Sabin strain; S2/W, V/W recombinants.
The analysis is based only on the 88 strains isolated in Romania during an 11-year period (1980 to 1990).
Most recombinants were found to be intertypic V/V recombinants, but five Sabin 2 strain genomes contained nonvaccine (W) sequences, as shown by sequencing (see details below) and were therefore identified as V/W recombinants. For the 88 strains isolated in Romania, V/W recombinants accounted for 13% of the type 2 recombinants and 5.7% of all vaccine-derived strains tested (Table 2). Four of the five V/W recombinants were isolated during a period in which wild PV was circulating in the region, but one (P2-V/057/87) was isolated 5 years (59 months) after the last recorded isolation of wild PV in Romania. We also analyzed a PV strain (P2-V/236/66) isolated in January 1966 in Moghilev, Belarus, from a 2-year-old unvaccinated patient with paralytic poliomyelitis. This Sabin 2 strain also had a nonvaccine sequence in the 3′ part of the genome, as shown by sequencing. Clinical and epidemiological data for patients from whom V/W recombinant strains and putative wild donor strains were isolated are presented in Table 3.
TABLE 3.
Clinical and epidemiological data on poliomyelitis cases from which V/W recombinant and cocirculating wild strains were isolated
| Strain | Patient age (mo) | OPV historya | Case classificationb | Serologyc
|
||
|---|---|---|---|---|---|---|
| P1 | P2 | P3 | ||||
| P2-V/406/80 | 19 | 1 T | VAPP-R | <4 | 512 | <4 |
| P2-V/553/80 | 24 | 1 T | VAPP-R | <4 | 180 | 32 |
| P2-V/598/80 | 25 | NV | VAPP-C | 190 | 512 | <4 |
| P2-V/146/81 | 18 | 1 T | VAPP-C | <8 | 256 | <8 |
| P2-V/057/87 | 6 | NV | VAPP-C | <4 | 350 | 10 |
| P2-V/263/66 | 24 | NV | VAPP-C | NK | NK | NK |
| P1-W/377/80 | 14 | NV | P1-Epid | 128 | <4 | <4 |
| P1-W/635/80 | 5 | NV | P1-Epid | <4 | <4 | <4 |
| P1-W/639/80 | 5 | 1 M1 | P1-Epid | 32 | <4 | <4 |
| P1-W/640/80 | 5 | 1 M1 | P1-Epid | 22 | <4 | <4 |
| P1-W/728/80 | 30 | NV | P1-Epid | 32 | >512 | >512 |
| P1-W/004/81 | 7 | NV | P1-Epid | <4 | <4 | <4 |
| P1-W/032/81 | 9 | NV | P1-Epid | 6 | <4 | <4 |
| P1-W/257/81 | 12 | NV | P1-Epid | 32 | <4 | <4 |
| P1-W/469/81 | 18 | 1 T | P1-Epid | 128 | <4 | <4 |
| P1-W/086/82 | 6 | NV | P1-Epid | 160 | <4 | <4 |
| P1-W/522/81 | 7 | 1 T | P1-Epid | 32 | <4 | <4 |
| P2-W/078/80 | 6 | NV | P2-Epid | <4 | <4 | <4 |
| P2-W/277/80 | 21 | 1 T | P2-Epid | <4 | <4 | <4 |
| P3-W/80/452 | 7 | 1 M1 | P3-Epid | 128 | <4 | <4 |
| P3-W/80/455 | 16 | 1 M1 | P3-Epid | 256 | <4 | <4 |
Number of doses of trivalent (T) or type 1 monovalent (M1) OPV. NV, nonvaccinated.
VAPP-R, VAPP recipient case; VAPP, VAPP contact case; Epid, epidemic poliomyelitis case with PV type 1 (P1-Epid), type 2 (P2-Epid), or type 3 (P3-Epid).
Reciprocal of the serum dilution neutralizing 100 TCID50 of homotypic PV type 1 (P1), type 2 (P2), and type 3 (P3). NK, not known.
Genomic structure of V/W recombinants.
All six V/W recombinants examined in this study were derived from Sabin 2 virus, as shown by their reaction with Sabin 2-specific neutralizing monoclonal antibodies (data not shown). In the RFLP 1 assay, Sabin 2-specific restriction patterns were detected in the VP1-coding region.
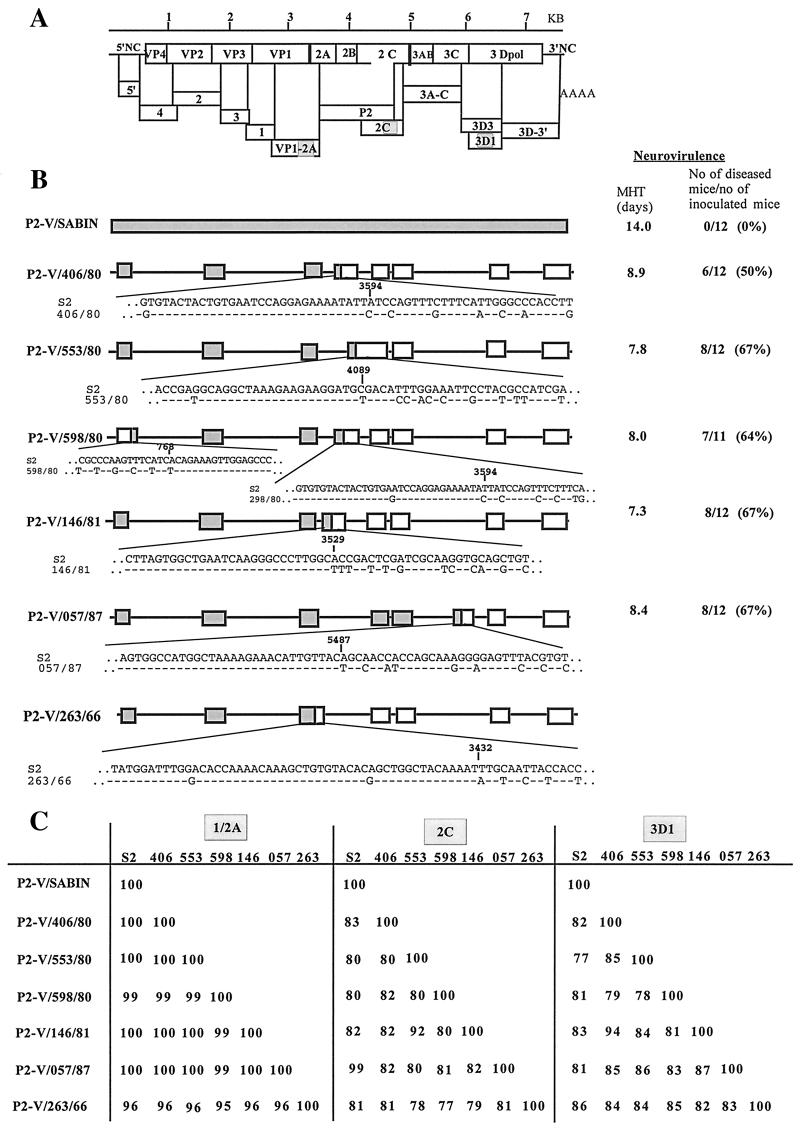
To locate the recombination junction, we first scanned almost all of the genome of these recombinant strains by a multiple RFLP technique (see Materials and Methods). Once the region containing the junction had been determined, it was sequenced to define the recombination site more precisely. The junction was located to within a few nucleotides, depending on the nucleotide differences between the vaccine and the wild strains. The recombination junctions were found to be different for each V/W strain (Fig. 1).
FIG. 1.
Genomic structure and neurovirulence of the V/W recombinant strains. (A) The genetic organization of the PV genome with the structural capsid proteins (VP1 through VP4) and the nonstructural proteins (2A through 3D) is shown. The size of the genome in kilobases is also indicated. Genomic fragments, obtained by RT-PCR and analyzed by RFLP, are indicated by boxes, and the name of each RFLP assay is given. (B) Schematic diagrams of the genomes of the Sabin 2 and recombinant V/W strains are shown. The structure of each genome was deduced from the RFLP assays shown in panel A and from partial DNA sequencing. Sequenced regions are indicated in boxes.  , Sabin 2 sequence; □, nonvaccine sequence. Sequences at the recombination junctions are given and compared with Sabin 2 sequences. The results of neurovirulence tests are indicated: MHT and the numbers and percentages of diseased transgenic mice after inoculation are given for six of the seven strains shown in panel B. (C) Percent nucleotide identity between the six V/W recombinants for three different regions of the genome. The sequences of the genome compared are indicated. VP1-2A, nt 3295 to 3444; 2C, nt 4546 to 4895; 3D1, nt 6172 to 6371 (nucleotide positions are numbered according to the Sabin 2 sequence). Sequences were compared pairwise using the CLUSTAL W program, and the percent nucleotide identity values for the pairs of strains are shown.
, Sabin 2 sequence; □, nonvaccine sequence. Sequences at the recombination junctions are given and compared with Sabin 2 sequences. The results of neurovirulence tests are indicated: MHT and the numbers and percentages of diseased transgenic mice after inoculation are given for six of the seven strains shown in panel B. (C) Percent nucleotide identity between the six V/W recombinants for three different regions of the genome. The sequences of the genome compared are indicated. VP1-2A, nt 3295 to 3444; 2C, nt 4546 to 4895; 3D1, nt 6172 to 6371 (nucleotide positions are numbered according to the Sabin 2 sequence). Sequences were compared pairwise using the CLUSTAL W program, and the percent nucleotide identity values for the pairs of strains are shown.
No recombination event was detected in the capsid-encoding region, except in a small segment at the 3′ end of the VP4-coding region in strain P2-V/598/80 (10). The recombination site of strain P2-V/263/66 was located between nt 3393 and 3432, near the junction between the VP1- and 2A-coding regions (nt 3385). This accounts for the lower-percentage nucleotide identity in the sequenced VP1-2A region (positions 3295 to 3444) between P2-V/263/66 and the other strains, including Sabin 2 (Fig. 1). In most cases, sequence comparisons for the 2C and 3D nonstructural protein-coding regions indicated that the W sequences in the 3′ moiety of the V/W recombinants were different in each strain (Fig. 1). This suggested that the V/W recombinants resulted from recombination with different wild donors. However, the nucleotide sequence of strain P2-V/553/80 was found to be 92% identical to that of strain P2-V/146/81 in the 2C region and 94% identical to that of strain P2-V/057/87 in the 3D1 region. This suggests that these sequences may have a common ancestor. The different partners and/or different sites of recombination indicated that the various V/W strains resulted from independent recombination events.
We estimated the circulation of the recombinant strains by determining the number of mutations accumulated in the Sabin 2-specific region of their genomes. A total of 550 to 1,045 nt were sequenced for each of the V/W strains, including the VP1-2A, the VP2, and, in some cases, the 2B (strain P2-V/553/80) or 3A-C (strain P2-V/057/87) regions (Table 4). Only 0 to 2 substitutions were found in each of the six V/W strains examined. This indicates that the recombinant viruses had circulated or multiplied for only short periods of time before isolation (see Discussion).
TABLE 4.
Percent nucleotide identities between parental Sabin 2 PV sequences and the vaccine-derived sequences present in V/W recombinants
| Strain | Target region | Sequence length (nt) | No. of mutations | % Nucleotide identity |
|---|---|---|---|---|
| P2-V/408/80 | VP2 + VP1-2A | 550 | 0 | 100.0 |
| P2-V/553/80 | VP2 + VP1-2A + P2 | 750 | 0 | 100.0 |
| P2-V/598/80 | VP2 + VP1-2A | 550 | 1 | 99.8 |
| P2-V/146/81 | VP2 + VP1-2A | 610 | 1 | 99.8 |
| P2-V/057/87 | VP2 + VP1-2A + 3AC | 1,045 | 2 | 99.8 |
| P2-V/263/66 | VP2 + VP1-2A | 522 | 1 | 99.8 |
Neurovirulence of V/W recombinants.
We assessed the pathogenicity of the V/W recombinants isolated from VAPP patients by comparing the neurovirulence of these strains with that of their Sabin 2 parent. We used mice transgenic for the human PV receptor gene, which have been shown to be susceptible to all three PV serotypes following parenteral inoculation. The IP-MHT assay, which involves daily follow-up of the clinical signs appearing in mice inoculated IP with virus, was used as described in Materials and Methods. The MHT recorded for each strain shown in Fig. 1 indicates that the five V/W Romanian recombinant strains tested had lost their attenuated phenotype. An MHT of 7.0 to 8.0, with a disease index (percentage of diseased mice) of 50 to 67% corresponds to moderate neurovirulence. In this test, Sabin 2 had an MHT of 14 days (with a disease index of 0%). This is the minimum value for neurovirulence (maximum attenuation). The MHT and disease indices (in parentheses) obtained in several experiments with neurovirulent reference strains were 3.3 (95%) for PV1/Mahoney, 5.8 (80%) for PV2/MEF1, and 2.0 (100%) for PV3/Leon/37.
Wild recombination partner.
We tried to identify the donor strains for sequences in the 3′ moiety of V/W recombinant strains by aligning the W sequences (2C and 3D1 segments) with homologous PV or NPEV sequences from a nucleotide sequence database (GenBank). The results of this search are presented in Table 5. In all cases, for the 2C-coding region, the highest percentage of nucleotide sequence identity was that with PVs (79.4% to 84.0%). Coxsackieviruses A21 (CA21) and A24 (CA24) appeared to be the most similar NPEVs (70.0 to 74.1% nucleotide identity). Comparison of the nucleotide and amino acid sequences corresponding to the 2C region suggested that the W sequences were derived from wild PVs and not from NPEVs. For the 3D polymerase-coding region, both nucleotide (80.0 to 87%) and amino acid (95.5 to 98.5%) sequence identities were too similar for significant differences to be detected between the W sequences and PV, CA21, and CA24 sequences. However, no CA21 or CA24 strains were isolated in Romania between 1970 and 1995.
TABLE 5.
Nucleotide and amino acid identitiesa
| Strain | Similarity in genomic regionb
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 2C
|
3D1
|
|||||||
| Nucleotide
|
Amino acid
|
Nucleotide
|
Amino acid
|
|||||
| Virus | % | Virus | % | Virus | % | Virus | % | |
| P3/119 | 84.0 | P3/119 | 100.0 | P3/Fin | 84.0 | P2/W-2 | 98.5 | |
| P3/Leon | 84.0 | P3/Leon | 100.0 | P1/Mah | 83.5 | CA21 | 98.5 | |
| P3/Sab | 83.7 | P3/Sab | 99.1 | P1/Sab | 83.0 | P2/Lan | 98.5 | |
| P2-V/406/80 | P3/Fin | 83.1 | P3/Fin | 99.1 | P3/Sab | 83.0 | P1/Mah | 97.0 |
| P2/Sab | 83.1 | P2/Sab | 98.3 | P3/Leon | 83.0 | P3/Fin | 97.0 | |
| CA24 | 73.8 | CA21 | 78.6 | CA24 | 83.0 | P1/Sab | 97.0 | |
| CA21 | 72.0 | CB5 | 70.9 | CA21 | 83.0 | P3/Sab | 97.0 | |
| P3/119 | 81.1 | P3/Fin | 100.0 | P3/Sab | 86.5 | P2/Lan | 98.5 | |
| P3/Leon | 81.1 | P3/Leon | 100.0 | P3/Fin | 86.5 | CA21 | 98.5 | |
| P2/Lan | 81.1 | P3/Sab | 99.0 | P3/Leon | 86.5 | P1/Mah | 97.0 | |
| P2-V/553/80 | P3/Sab | 81.1 | P2/Lan | 98.3 | P1/Mah | 86.5 | P3/Fin | 97.0 |
| P3/Fin | 80.9 | P2/Sab | 98.3 | P1/Sab | 84.0 | P1/Sab | 97.0 | |
| CA24 | 74.1 | CA21 | 78.6 | CA21 | 83.5 | P3/Sab | 97.0 | |
| CA21 | 71.8 | CB5 | 70.9 | CA24 | 80.5 | P3/Leon | 97.0 | |
| P3/119 | 80.9 | P3/119 | 98.3 | P1/Mah | 84.9 | P2/Lan | 98.5 | |
| P3/Leon | 80.9 | P3/Leon | 98.3 | P1/Sab | 84.4 | CA21 | 98.5 | |
| P2/Sab | 80.9 | P3/Sab | 97.4 | CA24 | 82.4 | P1/Mah | 97.0 | |
| P2-V/598/80 | P3/Sab | 80.9 | P2/Sab | 96.6 | P3/Sab | 81.4 | P1/Sab | 97.0 |
| P2/Lan | 79.4 | P2/Lan | 96.6 | P3/Leon | 81.4 | P3/Fin | 97.0 | |
| CA24 | 72.0 | CA21 | 78.6 | P2/Sab | 81.4 | P3/Sab | 97.0 | |
| CA21 | 70.0 | CB5 | 69.2 | CA21 | 81.4 | CB4 | ||
| P2/Lan | 82.6 | P3/Fin | 100.0 | CA21 | 85.0 | P2/Lan | 97.0 | |
| P2/Sab | 82.3 | P3/Leon | 100.0 | P2/Lan | 84.5 | CA21 | 97.0 | |
| P3/Fin | 82.0 | P3/Sab | 99.0 | P3/Fin | 83.0 | P1/Mah | 97.0 | |
| P2-V/146/81 | P3/Leon | 82.0 | P2/Lan | 99.0 | P1/Mah | 80.5 | P1/Sab | 95.5 |
| P1/Sab | 79.4 | P2/Sab | 98.3 | P3/Sab | 80.5 | P3/Fin | 95.5 | |
| CA24 | 72.0 | CA21 | 78.6 | P1/Sab | 80.0 | P3-Sab | 95.5 | |
| CA21 | 70.0 | CB5 | 70.9 | CA24 | 79.0 | P3/Leon | 95.5 | |
| P3/Sab | 84.5 | P2/Lan | 97.0 | |||||
| P3/Fin | 84.5 | CA21 | 97.0 | |||||
| P3/Leon | 84.5 | P1/Mah | 97.0 | |||||
| P2-V/057/87 | P2-Sab | 100.0 | P2-Sab | 100.0 | P1/Mah | 83.5 | P1/Sab | 95.5 |
| P1-Sab | 83.5 | P3/Fin | 95.5 | |||||
| CA21 | 82.5 | P3/Sab | 95.5 | |||||
| CA24 | 82.0 | P3/Leon | 95.5 | |||||
| P3/Fin | 82.0 | P3/119 | 99.1 | P1/Mah | 87.0 | P2/Lan | 98.5 | |
| P2/Sab | 81.4 | P3/Leon | 99.1 | P1/Sab | 86.5 | CA21 | 98.5 | |
| P2/W-2 | 80.2 | P3/Sab | 98.3 | CA21 | 85.5 | P1/Mah | 97.0 | |
| P2-V/263/66 | P2/Lan | 79.9 | P1/Mah | 96.6 | P2/Lan | 84.5 | P3/119 | 96.9 |
| P3/Leon | 79.9 | P3/Fin | 96.6 | CA24 | 84.5 | P3/Fin | 96.9 | |
| CA24 | 73.9 | CA21 | 79.8 | P3/Sab | 82.0 | P3/Leon | 96.9 | |
| CA21 | 71.7 | CB5 | 70.7 | P3/Leon | 82.0 | P3/Sab | 97.0 | |
Nucleotide and amino acid identities between the 2C and 3D genomic regions of the V/W recombinants and those of the most similar enteroviruses strains found in the GenBank are shown.
For each V/W recombinant, the nearest polio strains and NEPVs are listed. Nucleotides corresponding to positions 4546 to 4895 (2C) and 6172 to 6371 (3D1) of the Sabin 2 PV have been considered for this analysis. Virus strains (GenBank accession number) are as follows: P1/Mah, PV1/Mahoney (V01148); P1/Sab, P1/Sabin (V01150); P2/Lan, P2/Lansing (M12197); P2/Sab, P2/Sabin (X00595); P2/W-2, (D00625); P3/Fin, P3/Finland/23127/84 (X 04468); P3/Leon, P3/Leon/37 (K01392); P3/Sab, P3/Sabin (X00595); P3/119, P3/119 (X01076); CA24, coxsackievirus A24 (D90457); CA21, coxsackievirus A21 (D00538); CB4, coxsackievirus B4 (X05690); CB5, coxsackievirus B5 (X67706).
We investigated whether the wild sequences present in the V/W recombinants were typical of the cocirculating wild strains by analyzing the wild PVs isolated in Romania during the period in which the V/W recombinants were isolated.
Five of the V/W recombinant strains, all Sabin type 2-derived polioviruses, were isolated in Romania in 1980, 1981, or 1987. In 1980 to 1982, two overlapping poliomyelitis outbreaks occurred: one with a wild type 2 (P2-W) PV, which occurred from March to April 1980 (15 virologically confirmed cases), and the other with a wild type 1 (P1-W) PV, which occurred from March 1980 to July 1982 (133 virologically confirmed cases). Of a total of 148 cases, 52 occurred in 1980, 85 in 1981, and 11 in 1982. Two sporadic cases were recorded in 1980 in which wild type 3 poliovirus (P3-W) was isolated. A total of about 650 PV isolates were obtained from epidemic cases and their contacts in 1980 to 1982 in Romania. No wild PV was isolated in Romania during the 10-year period from August 1982 to March 1991, despite a high standard of virological surveillance. During this period, one case of acute flaccid paralysis was reported per 100,000 children under the age of 15 years; 997 children were examined, and the mean annual isolation rate for NPEV was 7.8%. Curiously, the V/W strain P2-V/057/87 was isolated in the middle of this 10-year period.
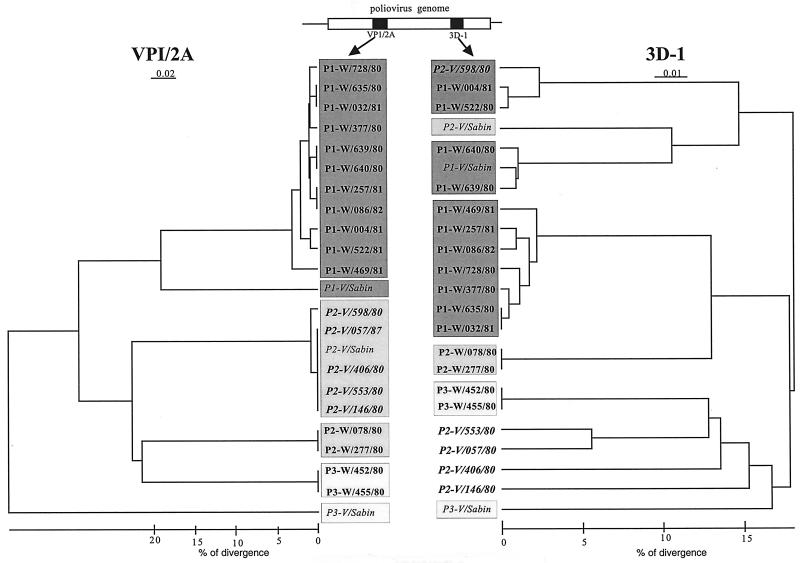
We tried to find the W partners from the sequences of the V/W recombinants. We first screened 61 strains randomly chosen from the wild cocirculating PVs for similarities in RFLP profile in the 3D polymerase-coding region (RFLP 3D1 assay). These strains, comprising 43 type 1 (P1-W), 16 type 2 (P2-W), and 2 type 3 (P3-W) strains, were grouped on the basis of similarity of RFLP 3D1 pattern (see Materials and Methods). Thus, the wild type 1 PV strains were classified into four groups, and the wild type 2 PV strains were assigned to a single group. The restriction profiles of two wild type 1 and two wild type 3 strains were found to be similar to those of the Sabin 1 and Sabin 2 strains, respectively. The RFLP 3D1 profiles of strains P1-W/004/81 and P1-W/522/81 were similar to that of the V/W recombinant, P2-V/598/80. No similarity in RFLP 3D1 profile was found between any of the other four V/W recombinants and the wild PVs tested.
The 3D1- and VP1-2A-coding regions were sequenced for representative members of each of the RFLP 3D1 groups. To increase the probability of finding the W parents of V/W recombinants, a careful case-by-case study was carried out. We also sequenced the 3D1 and VP1-2A regions of strains known to have been present in the same geographical area and at the same time as the patients from whom V/W recombinants were isolated. Phylogenetic trees were constructed from all these sequences, including those of the five V/W recombinants and the three OPV strains. The dendrograms are shown in Fig. 2. With the exception of strain P2-V/598/80, no significant similarity in the 3D1 genomic region was found between the V/W recombinants and the wild cocirculating PV strains tested. This suggests that the wild donor of four of the V/W recombinants may not be a wild PV. However, we cannot exclude the possibility that the wild partners escaped at the first step in the screening process, which involved grouping strains according to their RFLP 3D1 pattern.
FIG. 2.
Dendrogram showing the relatedness of strains based on the nucleotide sequences of the VP1-2A genomic region and those of the 3D polymerase-coding region. A schematic diagram of the PV genome (open box) and the sequenced genomic regions (black boxes) is shown; VP1-2A (positions 3295 to 3444) and 3D1 (positions 6172 to 6371) sequences were used for this analysis. Sequencing data were analyzed using DNASIS software, and the dendrogram was generated using the KITSCH program from the PHYLIP package. The VP1-2A sequences are grouped according to serotype: type 1,  ; type 2,
; type 2,  ; type 3,
; type 3,  . The same code is used for the 3D1 sequences. Vaccine, wild, and V/W recombinant strains are shown in italics, boldface, and outlined capitals, respectively. Percent nucleotide identity was estimated from the values obtained for nucleotide identity (CLUSTAL W) and from the genetic distances (KITCH) used for building the dendrogram.
. The same code is used for the 3D1 sequences. Vaccine, wild, and V/W recombinant strains are shown in italics, boldface, and outlined capitals, respectively. Percent nucleotide identity was estimated from the values obtained for nucleotide identity (CLUSTAL W) and from the genetic distances (KITCH) used for building the dendrogram.
The dendrograms shown in Fig. 2 also confirm that two P1-W strains (P1-W/639/80 and P1-W/640/80) contain Sabin 1-specific sequences in the 3D1 segment of their genome (W/V recombinants). The recombination junction between wild and vaccine sequences were located for both strains around nt 5285. The vaccine-derived nucleotide sequences of these strains diverged by 1.0 and 2.0% from those of the original Sabin 1 strain, respectively, indicating that either that donor Sabin strain or the recombinant virus must have been circulating for several months before isolation.
Wild PV partner of the V/W recombinant P2-V/598/80.
Strain P2-V/598/80 has been shown to be a type 2 V/W recombinant in which the only conserved vaccine-derived sequence is the capsid-coding region studied (10). Almost all of the 5′UTR, the nonstructural protein-coding region, and the 3′UTR have been replaced by wild sequences. In this study, we looked for the donor of the W nucleotide sequences in the 3D1 region and found a high degree of sequence similarity to two wild PV strains, P1-W/004/81 and P1-W/522/81, isolated at the beginning and end of 1981, respectively (Fig. 3). These two wild PV strains were isolated from distant regions of Romania. This strongly suggests that one of these P1-W epidemic strains or one of their ancestors was the donor of the 3D1 sequence present in the P2-V/598/80 V/W recombinant strain. With 97, 94, and 99% sequence identity in the 5′UTR, 2C, and 3D regions, respectively, the two P1-W strains appear to have closely related genomes. The low degree of nucleotide identity between the V/W recombinant and the two P1-W strains in the 5′UTR (87 to 88%) and in the 2C (77 to 78%) regions indicates that at least one other virus donated these sequences. Strain P2-V/598/80 therefore probably has a four-part recombinant genome resulting from genetic exchanges between the Sabin 2 strain (capsid), at least one wild (PV or NPEV) partner (5′UTR- and 2C-coding regions), and another wild PV (3Dpol region).
FIG. 3.
Sequence similarities between the P2-V598/80 V/W recombinant strain and two cocirculating wild PVs. A schematic diagram of the genetic organization of the PV genome is shown. The 5′UTR and 3′UTR regions, the genomic regions encoding structural capsid proteins (VP1 through VP4), and those encoding nonstructural proteins (2A through 3D) are shown. The size of the genome in kilobases is also indicated. A schematic diagram of the genome of P2-V/598/80, of the cocirculating strains, and of P2-V/Sabin is given. Sequenced regions are indicated: Sabin 2,  ; nonvaccine sequence, □. The sequenced parts of the genome compared are indicated: 5′ UTR, positions 220 to 468; VPI-2A, positions 3295 to 3444; 2C, positions 4546 to 4895; 3D1, positions 6172 to 6371. The percent nucleotide identity between P2-V/598/80, the two epidemic wild PVs, and the P2-V Sabin sequences was calculated after alignment with CLUSTAL W.
; nonvaccine sequence, □. The sequenced parts of the genome compared are indicated: 5′ UTR, positions 220 to 468; VPI-2A, positions 3295 to 3444; 2C, positions 4546 to 4895; 3D1, positions 6172 to 6371. The percent nucleotide identity between P2-V/598/80, the two epidemic wild PVs, and the P2-V Sabin sequences was calculated after alignment with CLUSTAL W.
The common ancestor of the closely related P1-W/004/81 and P1-W/522/81 or of another P1-W strain probably evolved by recombination. Indeed, the VP1-2A regions of P1-W/004/81 and P1-W/522/81 are closely related to all the other wild type 1 strains isolated during the outbreak. However, they fall into a different group if the 3Dpol region is considered (Fig. 2).
DISCUSSION
The analysis of PV strains isolated from vaccinees is an excellent way of studying the evolution of enteroviruses in humans. Studies of PVs isolated from patients with VAPP have demonstrated a high frequency of genetic recombination between OPV strains (9–11, 17, 21, 22). A high frequency of intertypic vaccine-derived recombinants was also found in this study by analyzing a total of 88 vaccine-derived PV strains isolated from VAPP cases in Romania. Sequencing combined with RFLP analysis for four different genomic fragments in the regions encoding proteins VP1, 2C, and the 3D polymerase and for the 3′ extremity of the genome showed that 76% of the strains implicated in these cases of VAPP (mostly types 2 and 3) had recombinant genomes. Similar results have been obtained for OPV strains isolated from healthy vaccinees (N. Cuervo et al., unpublished results).
Most of these recombinants were V/V recombinants, but five were shown to contain nonvaccine genomic segments and were classified as V/W recombinants. For one, the nonvaccine parent was identified among the known cocirculating wild PVs. For the others, the wild parent has not yet been identified and the possibility that the wild sequences originate from an NPEV cannot be excluded. Wild PV or NEPV sequences were detected in 6% of the VAPP strains studied. We analyzed wild cocirculating PV strains in an attempt to identify the nonvaccine parents of the V/W recombinants. We detected vaccine-derived sequences in 2 of 15 wild strains. Our findings strongly suggest that there is frequent interchange of genetic material between enteroviruses, whether PVs or NPEVs. Vaccination with OPV creates ideal conditions for this exchange by infecting the gut of the child with three different enterovirus genotypes. In some cases there may already be infection, or superinfection, with another enterovirus genotype.
Given the high frequency of recombinant genomes in the OPV strains excreted by healthy vaccinees (22; N. Cuervo et al., unpublished results), their contacts in the community, and patients with VAPP (9, 11, 21; this study), it seems highly likely that genetic recombination is involved in the natural evolution of Sabin strains. OPV strains are reputed to grow less efficiently in the human gut than wild epidemic PVs. Due to the quasispecies properties of PV populations, a large number of variants are likely to arise during the multiplication of vaccine strains in the human digestive tract. Two mechanisms may be responsible for this variation: mutations and recombination. Theoretically, recombination is a more powerful mechanism of variation than mutation because it may transfer a number of properties to the original virus in a single event. The presence of variant populations of PV in the human gut provides a wealth of material for the selection of variants that grow more efficiently.
We determined the neurovirulence of V/W recombinants in PVR-Tg mice and found that none retained the attenuated phenotype of the original Sabin 2 PV. However, the degree of neurovirulence acquired was intermediate between attenuation and the full neurovirulence of wild reference strains. This suggests that recombination alone or in combination with reverse mutations at the attenuating sites of Sabin 2 is not sufficient to render the original vaccine virus highly neurovirulent. This moderate increase in neurovirulence is, however, sufficient to allow Sabin viruses to cause poliomyelitis in man, as demonstrated by the identification of such strains as the etiological agent in certain VAPP cases (12).
To estimate the length of time for which the V/W recombinant strains had been circulating, we determined the number of mutations (as a percentage of the total number of bases) accumulated in the vaccine-derived segments. A large number of such mutations probably indicates the circulation of vaccine viruses before and/or after the recombination event. Very small numbers of mutations were found to have accumulated in the Sabin 2-specific regions of the genomes of the six V/W recombinants (from 0 to 0.2% mutated nucleotides). This indicates that recombination probably occurred in the gut of the vaccine recipient or in that of a contact before the disease. The number of mutations accumulated in the Sabin 1 genome during multiplication in humans, with or without disease transmission, has been estimated to be close to 1% per year for the VP1-coding region (16, 18). However, the frequency of variation may depend on genomic segment and strain. Our results for the Sabin sequences suggest that the W/V recombinants circulated or multiplied for several months before or after recombination with wild PVs.
We cannot determine whether the W sequences in the 3D1 region originated from wild PVs or from NPEVs, directly or via a wild PV, until the donor virus has been positively identified. Four of the five Romanian V/W recombinants were isolated during a period (1980 to 1982) of active circulation of wild PV in Romania. In an attempt to determine the origin of the wild sequences in V/W recombinants, we compared them with homologous sequences from a nucleotide sequence database (GenBank). PV rather than NPEV sequences were selected as the closest match, favoring the hypothesis that silent circulating wild PVs rather than NPEVs were the donors of the wild sequences of the V/W recombinants. This led us to try to identify the wild parent among the cocirculating epidemic PV strains. We looked for similarities in the sequence of the 3D1 region between the recombinants and 61 of 148 wild type 1, 2, and 3 poliovirus isolates. The W sequence in the 3D1 region of strain P2-V/598/80 appeared to be very similar to those of two type 1 wild PVs, isolated several months later in Romania (Fig. 3). Phylogenetic analysis of the sequences suggested that the two wild PVs and the donor of the sequence present in the V/W recombinant probably have a common ancestor. A similar situation was found in China (20), where two P1-V/W recombinants were found in which the W donor was identified as a wild PV circulating in a geographically narrow region. The demonstration that, during their natural multiplication, the PV strains could “trap” sequences from cocirculating wild PV may be used for an active detection of silent wild PV transmission by screening for the V/W recombinants.
However, the donors of the other V/W recombinants described here have not yet been identified among the 61 cocirculating wild PVs tested. Surprisingly, one of the V/W recombinant strains (P2-V/057/87) was isolated in the middle of a 9-year period (1983 to 1991) during which no wild PV was isolated in the surrounding region, under conditions of high-quality virological surveillance. Similarly, in Brazil, a V/W recombinant was isolated from a VAPP case 2 years after the last isolation of a wild PV on the American continent (8). This suggests that a NPEV rather than a wild PV may have been the donor of the W sequence of the recombinant. Although searches of GenBank mostly showed the most similar sequence to be that of a wild PV, this may be due to the small number of sequences from enteroviruses in the database corresponding to the genomic regions studied here. In the case of the W segments of the V/W recombinants, only the positive identification of an NPEV donor of these sequences could completely exonerate wild PV from being the recombination partner. There is no experimental evidence that recombinants between PV and NPEV are viable. However, there are findings suggesting that such recombinants circulate in nature, as the remarkable similarity to PV of the 3′ end of the CA21 or the CA24 reference strains shows (15, 36).
Thus, it is clear that OPV-derived PVs can acquire highly modified genomes not only by mutation but also by genetic exchanges with vaccine or wild PV, or even with NPEV. These PV vaccine-derived strains can survive for long periods of time by prolonged excretion and/or by natural transmission and may cause poliomyelitis in man. If PV eradication is to be successful after OPV is discontinued, we must hope that PV vaccine-derived strains will disappear before they can spread in the growing nonimmune population. This should be carefully studied before a general policy of discontinuing OPV immunization is implemented.
ACKNOWLEDGMENTS
This work was partly supported by grants to R.C. from the World Health Organization (V26/181/107) and from the European Commission (Copernicus-CIPA CT94-0123 and Inco-Copernicus ERBIC 15 CT96-0912).
REFERENCES
- 1.Anonymous. Poliomyelitis prevention in the United States: introduction of a sequential vaccination schedule of inactivated poliovirus vaccine followed by oral poliovirus vaccine. Recommendations of the Advisory Committee on Immunization Practices (ACIP) Morb Mortal Wkly Rep. 1997;46:1–25. [PubMed] [Google Scholar]
- 2.Assaad F, Cockburn W C. The relation between acute persisting paralysis and poliomyelitis vaccine—results of a ten-year inquiry. Bull W H O. 1982;60:231–238. [PMC free article] [PubMed] [Google Scholar]
- 3.Balanant J, Guillot S, Candrea A, Delpeyroux F, Crainic R. The natural genomic variability of poliovirus analyzed by a restriction fragment length polymorphism assay. Virology. 1991;184:645–654. doi: 10.1016/0042-6822(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 4.Cooper P D. Genetics of picornaviruses. Comp Virol. 1977;9:133–207. [Google Scholar]
- 5.Crainic R, Couillin P, Blondel B, Cabau N, Boue A, Horodniceanu F. Natural variation of poliovirus neutralization epitopes. Infect Immun. 1983;41:1217–1225. doi: 10.1128/iai.41.3.1217-1225.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crainic R, Furione M, Otelea D, Guillot S, Balanant J, Aubert-Combiescu A, Combiescu M, Candrea A. Natural evolution of oral vaccine poliovirus strains. In: Kurstak E, editor. Measles and poliomyelitis. Vienna, Austria: Springer-Verlag; 1993. pp. 371–390. [Google Scholar]
- 7.Felsenstein J. PHYLIP (Phylogeny Inference Package), version 3.5c. Seattle, Washington: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 8.Friedrich F, daSilva E F, Schatzmayr H G. Type 2 poliovirus recombinants isolated from vaccine-associated cases and from healthy contacts in Brazil. Acta Virol. 1996;40:27–33. [PubMed] [Google Scholar]
- 9.Furione M, Guillot S, Otelea D, Balanant J, Candrea A, Crainic R. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology. 1993;196:199–208. doi: 10.1006/viro.1993.1468. [DOI] [PubMed] [Google Scholar]
- 10.Georgescu M M, Delpeyroux F, Crainic R. Tripartite genome organization of a natural type 2 vaccine/nonvaccine recombinant poliovirus. J Gen Virol. 1995;76:2343–2348. doi: 10.1099/0022-1317-76-9-2343. [DOI] [PubMed] [Google Scholar]
- 11.Georgescu M-M, Delpeyroux F, Tardy-Panit M, Balanant J, Combiescu M, Combiescu A A, Guillot S, Crainic R. High diversity of poliovirus strains isolated from the central nervous system from patients with vaccine-associated paralytic poliomyelitis. J Virol. 1994;68:8089–8101. doi: 10.1128/jvi.68.12.8089-8101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgescu M M, Balanant J, Macadam A, Otelea D, Combiescu M, Combiescu A A, Crainic R, Delpeyroux F. Evolution of the Sabin type 1 poliovirus in humans: characterization of strains isolated from patients with vaccine-associated paralytic poliomyelitis. J Virol. 1997;71:7758–7768. doi: 10.1128/jvi.71.10.7758-7768.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghendon Y, Robertson S E. Interrupting the transmission of wild polioviruses with vaccines: immunological considerations. Bull W H O. 1994;72:973–983. [PMC free article] [PubMed] [Google Scholar]
- 14.Hirst G K. Genetic recombination with Newcastle virus, polioviruses and influenza. Cold Spring Harbor Symp Quant Biol. 1962;27:303–308. doi: 10.1101/sqb.1962.027.001.028. [DOI] [PubMed] [Google Scholar]
- 15.Hughes P J, North C, Minor P D, Stanway G. The complete nucleotide sequence of coxsackievirus A21. J Gen Virol. 1989;70:2943–2952. doi: 10.1099/0022-1317-70-11-2943. [DOI] [PubMed] [Google Scholar]
- 16.Kew O M, Mulders M N, Lipskaya G Y, daSylva E E, Pallansch M A. Molecular epidemiology of polioviruses. Semin Virol. 1995;6:401–414. [Google Scholar]
- 17.Kew O M, Nottay B K. Evolution of the oral poliovaccine strains in humans occurs by both mutation and intramolecular recombination. In: Chanock R, Lerner R, editors. Modern approaches to vaccines. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1984. pp. 357–367. [Google Scholar]
- 18.Kew O M, Sutter R W, Nottay B K, McDonough M J, Prevots D R, Quick L, Pallansch M A. Prolonged replication of a type 1 vaccine-derived poliovirus in an immunodeficient patient. J Clin Microbiol. 1998;36:2893–2899. doi: 10.1128/jcm.36.10.2893-2899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai M M C. RNA recombination in animal and plant viruses. Microbiol Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Zhang L B, Yoneyama T, Yoshida H, Shimizu H, Yoshii K, Hara M, Nomura T, Yoshikura H, Miyamura T, Hagiwara A. Genetic basis of the neurovirulence of type 1 polioviruses isolated from vaccine-associated paralytic patients. Arch Virol. 1996;141:1047–1054. doi: 10.1007/BF01718608. [DOI] [PubMed] [Google Scholar]
- 21.Lipskaya G Y, Muzychenko A R, Kutitova O K, Maslova S V, Equestre M, Drozdov S G, Bercoff R P, Agol V I. Frequent isolation of intertypic poliovirus recombinants with serotype 2 specificity from vaccine-associated polio cases. J Med Virol. 1991;35:290–296. doi: 10.1002/jmv.1890350415. [DOI] [PubMed] [Google Scholar]
- 22.Macadam A J, Arnold C, Howlett J, John A, Marsden S, Taffs F, Reeve P, Hamada N, Wareham K, Almond J, Cammack N, Minor P D. Reversion of the attenuated and temperature-sensitive phenotypes of the Sabin type 3 strain of poliovirus in vaccinees. Virology. 1989;172:408–414. doi: 10.1016/0042-6822(89)90183-9. [DOI] [PubMed] [Google Scholar]
- 23.Minor P D. The molecular biology of poliovaccines. J Gen Virol. 1992;73:3065–3077. doi: 10.1099/0022-1317-73-12-3065. [DOI] [PubMed] [Google Scholar]
- 24.Otelea D, Guillot S, Furione M, Combiescu A A, Balanant J, Candrea A, Crainic R. Genomic modifications in naturally occurring neurovirulent revertants of Sabin 1 polioviruses. Dev Biol Stand. 1993;78:33–38. [PubMed] [Google Scholar]
- 25.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perrière G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 27.Petitjean J, Quibriac M, Freymuth F, Fuchs F, Laconche N, Aymard M, Kopecka H. Specific detection of enteroviruses in clinical samples by molecular hybridization using poliovirus subgenomic riboprobes. J Clin Microbiol. 1990;28:307–311. doi: 10.1128/jcm.28.2.307-311.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prevots D R, Sutter R W, Strebel P M, Weibel R E, Cochi S L. Completeness of reporting for paralytic poliomyelitis, United States, 1980 through 1991. Implications for estimating the risk of vaccine-associated disease. Arch Pediatr Adolesc Med. 1994;148:479–485. doi: 10.1001/archpedi.1994.02170050037007. [DOI] [PubMed] [Google Scholar]
- 29.Sabin A B, Boulger L R. History of Sabin attenuated poliovirus oral live vaccine strains. J Biol Stand. 1973;1:115–118. [Google Scholar]
- 30.Sabin A B. Oral poliovirus vaccine: history of its development and use and current challenge to eliminate poliomyelitis from the world. J Infect Dis. 1985;151:420–436. doi: 10.1093/infdis/151.3.420. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Sergiescu D, Aubert C A, Crainic R. Recombination between guanidine-resistant and dextran sulfate-resistant mutants of type 1 poliovirus. J Virol. 1969;3:326–330. doi: 10.1128/jvi.3.3.326-330.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sergiescu D, Horodniceanu F, Klein R, Crainic R. Genetic transfer of guanidine resistance from type 2 to type 1 poliovirus. Arch Gesamte Virusforsch. 1966;18:231–243. doi: 10.1007/BF01241844. [DOI] [PubMed] [Google Scholar]
- 34.Strebel P M, Aubert C A, Ion N N, Biberi M S, Combiescu M, Sutter R W, Kew O M, Pallansch M A, Patriarca P A, Cochi S L. Paralytic poliomyelitis in Romania, 1984–1992. Evidence for a high risk of vaccine-associated disease and reintroduction of wild-virus infection. Am J Epidemiol. 1994;140:1111–1124. doi: 10.1093/oxfordjournals.aje.a117211. [DOI] [PubMed] [Google Scholar]
- 35.Strebel P M, Sutter R W, Cochi S L, Biellik R J, Brink E W, Kew O M, Pallansch M A, Orenstein W A, Hinman A R. Epidemiology of poliomyelitis in the United States one decade after the last reported case of indigenous wild virus-associated disease. Clin Infect Dis. 1992;14:568–579. doi: 10.1093/clinids/14.2.568. [DOI] [PubMed] [Google Scholar]
- 36.Supanaranond K, Takeda N, Yamazaki S. The complete nucleotide sequence of a variant of Coxsackievirus A24, an agent causing acute hemorraghic cunjunctivitis. Virus Genes. 1992;6:149–158. doi: 10.1007/BF01703064. [DOI] [PubMed] [Google Scholar]
- 37.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Health Organization. Manual for poliovirus surveillance. WHO/EPI/GEN/97.01. Geneva, Switzerland: World Health Organization; 1997. [Google Scholar]
- 39.Wright P F, Kim-Farley R J, de Quadros C A, Robertson S E, Scott R M, Ward N A, Henderson R H. Strategies for the global eradication of poliomyelitis by the year 2000. N Engl J Med. 1991;325:1774–1779. doi: 10.1056/NEJM199112193252504. [DOI] [PubMed] [Google Scholar]