Abstract
In recent years, extracellular vesicles (EVs) have emerged as novel key players in plant–microbe interactions. While it is immensely useful to draw on the established “minimal information for studies of extracellular vesicles” (MISEV) guidelines and precedents in mammalian systems, working with plants and their associated microbes poses specific challenges. To navigate researchers through these obstacles, we offer detailed step‐by‐step suggestions for those embarking on EV research in the context of plant–microbe interactions. The advice is based on recent publications and our collective experience from the diverse plant and microbe systems studied in a dedicated research consortium. We provide considerations for experimental design, optimization, quality control, and recommendations on how to increase yield, purity, and reproducibility of EV isolation. With this perspective article, we aim not only to assist researchers in our field but also to promote discussions on plant and microbe EVs in the broader EV community.
Keywords: plant‐microbe interactions, EV isolation, EV size profile, EV marker, EV quality control, biological fluid, axenic culture, apoplastic wash fluid
1. BACKGROUND
Extracellular vesicles (EVs) are central mediators in inter‐cellular and inter‐organismal communication across diverse biological systems. These membranous structures can be generated and released via different cellular pathways and cell types to the extracellular space (Box 1; Colombo et al., 2014). In the interaction of plants with both pathogenic and beneficial microbes, EVs have garnered significant interest owing to their potential to modulate the relationship between both partners (Cai, Qiao, et al., 2018; Chalupowicz et al., 2023; Wang et al., 2016).
Particle types defined by the minimal information for studies of extracellular vesicles (MISEV) guideline and examples thereof from the field of plant–microbe interactions
Extracellular vesicles (EVs): “Particles that are released from cells, are delimited by a lipid bilayer, and cannot replicate on their own” (Welsh et al., 2024). Example: In samples derived from colonized plants, these can be of plant or microbe origin.
Non‐vesicular extracellular particles (NVEPs): “Multimolecular assemblies that are released from cells and do not have a lipid bilayer (non‐vesicular extracellular particle fraction” (Welsh et al., 2024). Example: If working with plant samples one might look out for RuBisCo complexes or lipoproteins. For samples originating from liquid cultures, particles from the medium might be contaminants.
Extracellular particles (EPs): “Umbrella term for all particles outside the cell, including EVs and NVEPs” (Welsh et al., 2024). Example: See above as for EVs and NVEPs.
EV mimetic: “EV‐like particles that are produced through direct artificial manipulation” (Welsh et al., 2024). Example: Unintentionally generated vesicles derived from cell lysis, for example, by infiltration or centrifugation during apoplastic wash fluid (AWF) isolation.
Artificial cell‐derived vesicles (ACDVs): “EV mimetics that are produced in the laboratory under conditions of induced cell disruption, such as extrusion” (Welsh et al., 2024). Example: In the plant field, vesicles sourced from juices or disrupted tissue have also been termed “plant‐derived nanovesicles” (PDNVs; Pinedo et al., 2021).
Research efforts discerning the composition of EV cargos of plants and associated microbes, at present, typically focus on proteins and various RNA classes, including small RNAs, messenger RNAs, long non‐coding RNAs, circular RNAs, and fragments of transfer as well as ribosomal RNAs (Kusch et al., 2023; Kwon et al., 2021; Ruf et al., 2022; Wang et al., 2023). In particular, the bidirectional RNA exchange between plants and microbes has been suggested to tune the interaction in several systems (Cai, Qiao, et al., 2018; Cheng et al., 2023; Dunker et al., 2020; Wang et al., 2016; Weiberg et al., 2013; Wong‐Bajracharya et al., 2022) and is serving as a blueprint for the development of novel types of pesticides (Cai, He, et al., 2018). Accordingly, there is an increasing interest in studying EVs and their cargos in the context of plant–microbe interactions, which necessitates suitable EV isolation protocols.
However, EV isolation procedures in plant, microbial, and mammalian systems vary due to differences in physiology (e.g., the presence/absence of a cell wall; Brown et al., 2015) and cultivation. The current lack of recommendations for experimental procedures and documentation standards regarding plant(‐microbe) systems in the MISEV (minimal information for studies of extracellular vesicles) guideline hinders reproducibility and comparability across such studies (Welsh et al., 2024). Consequently, the importance of well‐documented and reproducible workflows cannot be overstated, serving as the foundation for robust scientific conclusions. Previous reviews have sought to address these issues for EV isolations from (healthy) plants (Pinedo et al., 2021; Rutter & Innes, 2020). Nonetheless, further refinements and tailored protocols specific to this field are urgently needed, especially for scientists studying plant–microbe interactions.
In this perspective article, we attempt to provide comprehensive support for the establishment and optimization of EV isolation procedures in the context of plant–microbe interactions. Drawing upon recent advancements and novel insights from first‐hand experience gained in the context of a dedicated research consortium (Research Unit FOR5116 “exRNA” funded by the Deutsche Forschungsgemeinschaft [DFG]; https://www.biologie.uni‐hamburg.de/en/forschung/forschungsverbuende/dfg‐ru5116.html), we wish to supply researchers new to this burgeoning field with practical advice to master the complexities of EV isolation. These include hints regarding the cultivation of organisms and the retrieval of biological fluids for EV isolation, the actual EV isolation procedure, and measures for EV quality control. By this, we aim to empower scientists to elucidate the nuanced mechanisms governing potential cross‐kingdom plant–microbe communication mediated by EVs or to discover novel potential colonization strategies enabled by EVs.
2. CULTIVATION AND RETRIEVAL OF BIOLOGICAL FLUIDS FOR EV ISOLATION
Living cells can release EVs into their environment. Depending on the cell type and organism of interest, this environment can be very diverse in the plant–microbe field. Appropriate cultivation of the source organisms significantly influences the experimental EV isolation success. Isolation of EVs typically requires some kind of biological fluid as starting material, which is ideally fully devoid of cells and cellular debris. The latter is usually achieved through filtration (0.22 or 0.45 µm pore size) and low‐speed centrifugation of EV‐containing samples (reviewed in Pinedo et al., 2021; Rutter & Innes, 2020). Plant samples are typically centrifuged at 10,000 × g (Cai, He, et al. 2018; Cai, Qiao, et al., 2018; Regente et al., 2009; Rutter & Innes, 2017), fungal samples between 4000 × g and 15,000 × g (in some instances by two consecutive cleaning steps; Bleackley et al., 2020; Hill & Solomon, 2020; Kwon et al., 2021; Rutter et al., 2022), and bacterial samples between 4500 × g and 10,000 × g (Bahar et al., 2016; Janda et al., 2023; McMillan & Kuehn, 2023; Nascimento et al., 2016). In cases involving microbial or liquid plant cultures (e.g., plant cell suspension cultures, plant tissue cultures, or hydroponic systems), EVs can be directly isolated from culture supernatants (De Palma et al., 2020; Janda et al., 2023; Kocholata et al., 2022; Kwon et al., 2021). For whole plants grown in soil or other solid substrates (e.g., mineral composites such as vermiculite or solid media for in vitro cultivation), growth conditions need to be optimized to isolate apoplastic wash fluid (AWF)—a frequently used source for EV isolation from plants approximating the full repertoire of naturally secreted EVs. AWF is a liquid commonly obtained by infiltrating buffer into the intercellular space (apoplast) and subsequent centrifugation of the infiltrated plant specimens to collect the buffer along with (nano‐)particles and molecules present in the apoplast (O'Leary et al., 2014).
2.1. Plants
For plants grown on solid substrates, AWF can be isolated from entire seedlings or adult plants, isolated leaves, and potentially roots, as accomplished in many different plant species (Chen et al., 2022; Kusch et al., 2023; Regente et al., 2017; Rutter & Innes, 2017). Preceding growth conditions notably affect buffer infiltration. For instance, high humidity supports infiltration efficiency, because stomata are wide open (Chincinska, 2021; Rumyantseva et al., 2023). We, therefore, recommend increasing the humidity at least several hours before buffer infiltration by covering plants with a lid. However, it is crucial to note that adjusting humidity levels may alter plant gene expression and hence affect the interaction with any microbes under study (Yao et al., 2023).
Apart from AWF, EVs have also been isolated from the liquid medium of plant cell or tissue cultures (Boccia et al., 2022; Kocholata et al., 2022). On the one hand, this approach reduces potential contaminations due to cell damage through infiltration‐centrifugation steps during AWF isolation, and a direct comparison could be used to identify these in AWF samples. On the other hand, plant liquid culture experiments neglect the systemic context and are restricted to systems for which the microbe of study can be co‐cultivated. Hydroponic plant systems also offer the possibility to retrieve EVs from the medium but are not necessarily suitable for all types of plant–microbe interactions (De Palma et al., 2020).
Disruptive methods such as tissue blending are unsuitable for EV isolation due to associated contamination from cellular debris. This can lead to the generation of artificial cell‐derived vesicles, EV mimetics, which in the plant field, have been termed “plant‐derived nanovesicles” (Box 1; Pinedo et al., 2021). In general, the generation of such plant‐derived nanovesicles is undesirable in the context of EV isolation. However, comparing their characteristics with those of EVs can support the existence of unique EV profiles. Depending on the plant species, obtaining sufficient AWF volume for EV isolation can be challenging. To enhance AWF yield, we suggest employing young(er) plants with soft(er) tissue and optimising buffer infiltration with an efficient vacuum pump (employed vacuum typically between 25 and 45 kPa; Figueir et al., 2018; Regente et al., 2008). Vacuum infiltration is followed by a very low‐speed centrifugation step. Applied centrifugation forces range between 400 and 900 × g depending on the plant species (Cai, Qiao, et al., 2018; Regente et al., 2008; Rutter & Innes, 2017). We do not recommend exceeding 900 × g unless a higher force is necessary to obtain the fluid (Lohaus et al., 2001). Further, customized growth conditions are proposed to find the best balance between plant age and leaf size—larger and younger leaves will yield more AWF than smaller or older leaves (Chen et al., 2022). Significant scale‐up may be necessary for sufficient AWF (and thus EV) yield, especially when microbial EVs should be co‐enriched. In cases where infiltration is aggravated due to high lignin content or cuticular waxes, one might consider using low amounts of a non‐ionic detergent (e.g., Tween 20) in the infiltration buffer to break surface tension (Nouchi et al., 2012). If the EVs are stable, this treatment is unlikely to affect their integrity and might aid their stability during storage (van de Wakker et al., 2022). However, attention should be paid when performing measurements of the zeta potential (an indicator of particle surface charge) as it might be altered (Midekessa et al., 2020).
We further advise assessing cell viability of infiltrated plant tissue post‐AWF isolation, for example, using trypan blue staining to visualize potential damage (e.g., dead cells; Mulaosmanovic et al., 2020) and/or inspect the retrieved AWF by microscopy for organelle debris potentially released because of cell injury during its isolation. At the molecular level, immunoblotting of AWF samples with antibodies targeting abundant intracellular proteins (e.g., of endomembrane or cytoplasmic origin, e.g., chloroplast components) should complement these efforts to validate proper AWF isolation. In principle, such cellular content could also result from organelle secretion, which has been described recently for animal cells (Suh & Lee, 2024). However, although we cannot exclude the possibility that certain plant species or cell types may secrete organelles, this has not been reported to the best of our knowledge. Hence, we consider any organelle debris detected in AWF samples rather a putative byproduct of the unavoidably harsh conditions of the isolation process. Recurring cytoplasmic contamination is often indicated by green (chlorophyll‐based) coloration in the case of leaf‐derived AWF. In such instances, we recommend streamlining the AWF isolation to minimize the damage to the plant material. This may include selecting an infiltration buffer that does not compromise cell integrity, as do high detergent or non‐isotonic salt concentrations; opting for the lowest effective infiltration time; centrifuging as slowly and shortly as possible; and handling the plant samples with great care (avoid agitation and tissue damage). Special attention is warranted when working with plants infected by necrotrophic or hemibiotrophic pathogens, which cause cell damage and tissue lesions during pathogenesis. In these instances, AWF should be preferably collected at the early infection stage before necrotic lesions occur. The above‐outlined methods can be used to confirm that pathogen growth has not yet compromised plant cell integrity.
2.2. Culturable microorganisms
Several plant‐associated microbes can be cultivated in vitro. While EVs from microbial axenic cultures (i.e., cultures in the absence of the plant) are most likely not equivalent to those produced during interaction with their plant hosts, the relative ease of handling and scalability are advantageous for an initial survey of EV characteristics and EV‐associated molecules.
As outlined above for plants, the first step is choosing appropriate growth conditions since these can impact EV production and EV cargo (Jonca et al., 2021; McMillan & Kuehn, 2023; Welsh et al., 2024). It is further important to consider the microbial growth or developmental stage at the time point of EV isolation and any potential factor that may impinge on the physiology and cellular activities. EV biogenesis and the permeability of cellular barriers, such as the microbial cell wall, can vary greatly depending on the cellular morphology influenced by the growth conditions. Furthermore, the biological functions of EVs can differ depending on the microbial growth stage (Saad et al., 2024) and lifestyle (Johnston et al., 2023). The latter can be influenced by growing the microbes in liquid media or on agar plates for biofilm formation, and EVs can be isolated from both conditions (Janda et al., 2023). Multicellular growth forms such as bacterial biofilms or mycelia of fungi and oomycetes may increase the heterogeneity of EVs. For mycelia, EV diffusion into the medium can be hindered, for example, by the cell wall (Rutter et al., 2022). Because of the aforementioned variability, experimental details such as nutrient composition and pH of the culture medium, aeration/rotation, cultivation time, growth phase, and temperature should be well documented and reported to increase reproducibility (Welsh et al., 2024).
Opting for liquid culture over colonized plant materials is usually a compromise between relevance to the biological question and EV yield. In some systems, culture conditions can be adjusted to allow the microbial cells to simulate developmentally and transcriptionally certain stages of plant colonization, for example, by the addition of plant extracts or dedicated salts (Kwon et al., 2021; Li et al., 2022). Moreover, there are media known that resemble specific plant locations, such as the apoplast (Rico & Preston, 2008) or xylem (Hiery et al., 2013; Neumann & Dobinson, 2003), or that trigger virulence in phytobacteria (Wengelnik et al., 1996) or mimic symbiotic conditions (Li et al., 2022). It is helpful to understand, for example, based on existing transcriptomics or proteomics data, to what extent a given culture condition simulates the situation in planta. If ‐omics resources are limited for testing different conditions, one could use expression levels of hallmark genes for colonization that are specifically upregulated during certain infection stages as an indicator of whether the growth conditions mimic the in planta situation. We propose using defined media composition for microbial cultures instead of complex media with ill‐defined components of natural origin, such as yeast extract, which may contain EV‐like nanoparticles. Contamination from such components can be reduced by filtration or ultracentrifugation, similar to how blood sera are particle‐depleted before their addition to mammalian cell cultures (Lehrich et al., 2021). The potential effect of this procedure on the growth of the microorganisms should be, however, assessed carefully. If unavoidable, the unconditioned complex medium should be included in all experiments as an important negative control for comparison (Welsh et al., 2024).
Any optimization to increase the viability and the intactness of the EV‐secreting microbes during culture and harvesting of conditioned media would reduce the contamination by EV mimetics (Box 1). In general, high cell viability (commonly ≥95%, certified by vital staining) at the time point of EV isolation is advised (Shekari et al., 2023). However, a compromise may be necessary to increase the EV yield. For example, starvation (Debbi et al., 2022) or cell wall stress (Olicón‐Hernández et al., 2015) may increase the release of EV‐like particles (Box 1). In some fungi, such as Colletotrichum higginsianum, it may be necessary to use cell wall‐degrading enzymes to break apart the mycelium partially and to release EVs from the paramural space (Rutter et al., 2022). We recommend taking such steps with caution, because several studies have demonstrated that the vesicle cargo of some bacterial and fungal species is influenced by nutrient availability (Bahar et al., 2016; Dauros Hill & Solomon, 2020; Hong et al., 2019; McMillan & Kuehn, 2023; Singorenko et al., 2017). It must also be noted that many commercially available protoplasting enzyme preparations tend to be crude mixtures with additional proteinase and RNase activities and are often derived from fungi, which may confound any ‐omics analyses of fungal EVs. While such organism‐specific treatments may be necessary to yield sufficient EV quantities, it is important to address the potential consequences on the cellular status, the occurrence of putative contaminants, and the biological relevance of the EVs obtained.
Prior to EV isolation, microbial cells must be removed from the culture medium. This can be achieved by centrifugation and/or filtration. Centrifugation time and force, as well as filtration pressure, should be reduced as much as possible to prevent cell lysis and minimize changes in cell physiology. At this point, defined aliquots of the cells should be snap‐frozen for later comparison with the isolated EVs. This can be done by an immunoblot or gene expression analysis via quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR), for example. To address any putative contamination of the EV preparations with living cells, it is suggested to introduce streak controls of the samples on a solid medium. These should take place instantly after the removal of microbial cells from the liquid culture and after obtaining the crude EV suspension. A quick quality control via microscopy and, optionally, organelle staining, would be beneficial to address contamination from dead or lysed cells before proceeding with EV isolation. It should be noted, though, that certain organisms may secrete organelles, similar to various animal cell types that recently have been reported to secrete mitochondria (Suh & Lee, 2024), which would render such organelles unsuitable as markers for cell lysis. If cellular, non‐EV marker proteins are already known, an aliquot from each step in preparing the biological fluid and EV isolation can be tested for cellular contamination by immunoblotting.
3. EV ISOLATION
Once the biological source fluid (e.g., AWF or culture medium) has been obtained and cleared of cellular material, EV isolation can proceed. The methods published for the isolation of EVs from plants, plant‐colonizing microbes, and colonized plants include a combination of differential ultracentrifugation, ultrafiltration, size exclusion chromatography, and density gradient centrifugation (Bleackley et al., 2020; Cai, Qiao, et al., 2018; Janda et al., 2023; Kwon et al., 2021; Regente et al., 2017; Rutter & Innes, 2017). We encourage researchers to explore diverse methods used in the plant–microbe field, especially if generating sufficient quantities of biological source fluid is critical (Rutter et al., 2022). In these instances, scientists might consider a low specificity/high recovery approach such as a precipitation (polymer)‐based method to obtain more EVs in the crude pellet (Welsh et al., 2024). For detailed considerations on designing a suitable differential ultracentrifugation protocol, we refer to the review by Rutter & Innes (2020). It should be noted that the appropriate procedure depends on the size and density of EVs of the targeted organism(s), which may differ between biological fluids obtained from uncolonized and colonized plants. Thus, centrifugation force and rotors should be selected with great care as this might influence the accumulation of different EV subclasses or even contaminants (Box 1), making it necessary to establish the ideal rotor type for the given system. Fixed angle rotors have a higher pelleting efficiency (lower k‐factor) compared to swing‐out rotors that hold equivalent sample tube volumes and, therefore, may be more suitable for obtaining the crude EV pellets from a larger volume of starting biological fluid, while swing‐out rotors are essential for density gradients. For a detailed overview of the effect of rotor types on EV isolation, we refer the readers to a previous study (Cvjetkovic et al., 2014). Common centrifugation forces are between 40,000 and 100,000 × g in the case of plants and fungi (Bleackley et al., 2020; Cai, Qiao, et al., 2018; Hill & Solomon, 2020; Kwon et al., 2021; Regente et al., 2009; Rutter & Innes, 2017; Rutter et al., 2022), and between 31,000 and 150,000 × g in case of bacteria (Bahar et al., 2016; Janda et al., 2023; McMillan & Kuehn, 2023; Nascimento et al., 2016). In general, centrifugation time should be minimized as prolonged centrifugation time can lead to the accumulation of impurities (Box 1), cause artifacts, and might affect EV integrity (Cvjetkovic et al., 2014). Typical centrifugation times for an EV pelleting step (in some instances several runs are performed) are usually 60 min for plant AWF (Cai, Qiao, et al., 2018; Regente et al., 2009; Rutter & Innes, 2017), between 60 and 90 min for fungal cultures (Bleackley et al., 2020; Hill & Solomon, 2020; Kwon et al., 2021; Rutter et al., 2022), and between 90 and 180 min for bacterial cultures (Bahar et al., 2016; Janda et al., 2023; McMillan & Kuehn, 2023; Nascimento et al., 2016). Purification of crude EV pellets obtained by any of the abovementioned methods is recommended, particularly for explorative downstream analyses such as any ‐omics approaches and should exploit different physical and biochemical properties than the initial EV isolation procedure (Welsh et al., 2024).
A specific challenge when isolating EVs in the context of plant–microbe interactions is distinguishing between the EVs originating from the different organisms or even separating them physically. In the case of bacterial OMVs, researchers might benefit from recent advancements. For example, a fluorescent probe sensitive to outer membrane vesicles (OMVs) with aggregation‐induced emission (AIE) characteristics that could aid in distinguishing between plant and bacterial EVs has been reported lately (Wang et al., 2023). Alternatively, the separation of OMVs from plant EVs in a mixture could, at some point, be facilitated by targeting bacterial lipopolysaccharides exploiting a secreted effector protein of the bacterial species Cupriavidus necator (Hofer, 2021). If technically possible, the separate characterization of plant‐ and microbe‐derived EVs enables the assessment of changes that occur during plant colonization. This can be, for example, achieved by measuring the zeta potential, which in addition to EV size, can reveal differences in EV surface charge (see below; Janda et al., 2023). It is important to note that isolating EVs from the natural interaction site rather than from (co‐)cultivated plants and microbes may allow for conclusions with higher biological relevance. Allocation of EV‐associated molecules is possible if reference datasets are available for each organism, even when no specific EV markers (see below) have been identified yet. If desired, plant‐ and microbe‐derived EVs may be separated by immunoaffinity capture or fluorescence‐activated cell sorting (He et al., 2021; Kondratov et al., 2020). However, for these procedures, reliable EV biomarkers and suitable antibodies are required, which remain an exception for many species (as discussed below). Particularly for obligate biotrophic microbes, which cannot be cultivated without their host plant, the establishment of EV biomarkers would be highly desired to capture microbe‐derived EVs from the colonized plant tissue.
4. EV QUALITY CONTROL PARAMETERS
To characterize isolated particles and to verify if these are genuine EVs (see Box 1 for alternative particle categories), we recommend three types of analysis: (1) an estimation of EV particle size and concentration measurements, (2) visual inspection by electron microscopy, and (3) molecular analysis for the presence of EV biomarker proteins. In agreement with the MISEV guideline, we advise to use at least two of these independent yet complementing methods (Théry et al., 2018; Welsh et al., 2024).
4.1. EV size profiles and concentration
Common single particle‐based methods to analyse the size and concentration of nanoparticles in plant EV samples are nanoparticle tracking analysis (NTA) and dynamic light scattering (Welsh et al., 2024). To measure the sample concentration, NTA is a popular method since besides the concentration, it determines the size profile and, depending on the manufacturer, the zeta potential, which represents the overall electric charge of the EV's surface (Varga et al., 2020; Welsh et al., 2024). Both, changes in the zeta potential and the size profile in EV samples derived from infected plants as compared to uncolonized plants and microbial EVs can be a hint for the co‐presence of plant and microbial EVs (Janda et al., 2023). Nonetheless, NTA does not distinguish between EVs and other spherical particles (Box 1) such as bigger protein complexes. Further, the concentration of smaller particles such as EVs in polydisperse samples might be underrepresented due to the intense light scattering of larger particles, preventing smaller particles from being tracked (Filipe et al., 2010). Newer NTA generations include lasers to detect EV particles via a fluorescence light detector. If available, measuring EVs with fluorescently labelled biomarkers (see below) or staining of EVs with lipophilic dyes or probes prior to the measurement can enable a more precise estimation of the EV size profile and concentration, as fewer non‐EV or non‐lipid contaminants, respectively, will be measured. However, it is crucial to include appropriate controls, such as buffer only, to demonstrate that unbound dye has been removed from the sample. Of note, the binding of antibodies or the intercalation of membrane dyes could lead to a distortion of the actual particle size (Varga et al., 2020). One also has to consider that NTA and dynamic light scattering measure, in fact, the hydrodynamic diameter and provide an overestimation of the actual EV diameter, which has to be taken into account when comparing it with size estimates from electron micrographs (Varga et al., 2020). We, therefore, encourage researchers to explore also advanced single‐particle analysers (e.g. nanoflow cytometry or microfluidic resistive pulse sensing) that at present are not commonly used in the field.
The EV quantity can also be derived based on the total lipid, protein, or nucleic acid content (Bahar et al., 2016; McMillan & Kuehn, 2023; McMillan et al., 2021). For protein concentration measurements, commonly used methods are the Bradford assay or staining with bicinchoninic acid (BCA). However, the respective substances also react with reducing sugars and phospholipids and do not discriminate contaminants from EVs (Théry et al., 2018). The total lipid content is typically quantified by staining EVs with lipid dyes such as FM4‐64, DiOC6, and DiR, followed by fluorescence intensity measurements (Rutter & Innes, 2017). Some dyes stain nucleic acids, such as RiboGreen, or the RNA dye SYTOTM RNASelectTM (Fortunato et al., 2021). While the single‐particle‐based techniques can give further information on different EV (sub‐)populations by measuring size, such differences cannot be revealed by quantifying total lipid, nucleic acid, or protein concentrations (Welsh et al., 2024).
4.2. Morphology
Electron microscopic techniques are the method of choice to characterize the morphological features of EVs. In addition, electron microscopy can provide information about the purity of EV samples and the potential occurrence of different EV (sub‐)populations (Bahar et al., 2016; Janda et al., 2023; McMillan et al., 2021; Rutter & Innes, 2017). Currently, the most precise albeit tedious approach is cryo‐electron microscopy (Chernyshev et al., 2015; Skliar et al., 2018). Other frequently used electron microscopic techniques like transmission or scanning electron microscopy will underestimate the diameter of EVs as these will desiccate in the process of sample preparation (Bachurski et al., 2019; Chuo et al., 2018). In transmission electron micrographs, EVs typically appear as cup‐shaped structures (Panagopoulou et al., 2020; Rutter et al., 2020), whereas in scanning electron micrographs, EVs are usually spherical blebs (Chernyshev et al., 2015; Janda et al., 2023). Attention should be given when the electron micrographs reveal impurities such as flagellar structures when working with bacterial EVs (Janda et al., 2023) or organellar structures when working with plant or fungal samples as they are indicators of impurities. These contaminants can have severe effects on any downstream experiments and it might be necessary to optimize the purification process before proceeding with further analyses.
4.3. EV biomarkers and molecular cargo
There are ongoing efforts to establish suitable EV markers in the plant(‐microbe) field. EV biomarkers are molecules (in particular proteins) that are characteristic of EVs or EV sub‐types of a given organism (Welsh et al., 2024). They can significantly improve the EV quality and quantity assessment, might allow for EV purification via immunoaffinity capture, and increase the portfolio for experimental downstream analyses (He et al., 2021). We encourage following the MISEV guidelines regarding the recommendation to establish at least two different positive EV biomarkers. Ideally, one of these should be an integral membrane or glycosylphosphatidylinositol (GPI)‐anchored protein (or outer membrane for Gram‐negative bacteria) and the other a cytosolic (or periplasmic for Gram‐negative bacteria) protein with lipid‐ or membrane protein‐binding ability. It is advised to establish additionally one negative marker, which could be a common co‐isolated contaminant, for example, a constituent of non‐vesicular extracellular particles (Théry et al., 2018; Box 1). Moreover, a proper EV biomarker should be abundant in the EV fractions for easy and reliable detection.
For finding organism‐specific EV markers, there is a benefit in using organisms that can be grown in liquid culture, as EVs are obtained from a single species and it is easier to avoid contaminations from lysed cells due to handling. For obligate parasites or symbionts, the mixture of EVs obtained and the potential homology of proteins and nucleic acids in mixed samples may complicate the analyses. A first step to establish a new EV biomarker might be a proteomic survey, which can provide a list of candidate proteins. Such data can be used to select candidates that show potential relevance for plant–microbe interactions (e.g., association of a protein with a nucleic acid of interest; Cai, Qiao, et al., 2018), or that have homology to established marker proteins in other biological systems. Currently, there are only a handful of commonly tested EV markers in the field, including a homolog of human tetraspanin CD63 in the dicotyledonous reference plant Arabidopsis thaliana (TET8; Cai, He, et al., 2018; Cai, Qiao, et al., 2018; Box 2). Orthologs of well‐established mammalian EV markers such as tetraspanins are absent in certain fungi and bacteria, but other biomarker proteins have been suggested for these organisms (Box 2). Larger collections of EV proteomics data such as Vesiclepedia or EVpedia (Chitti et al., 2024; Kim et al., 2015), which also include established EV biomarkers, can help deciding which candidate protein(s) to select.
Examples of known and suggested EV markers for plants and plant‐colonizing microbes
Plants
-
‐
Arabidopsis thaliana: PENETRATION1 (PEN1) and PATELLIN1 (PATL1) (Rutter & Innes, 2017), TETRASPANIN8 (TET8) (Cai, Qiao, et al., 2018), EXOCYST COMPONENT OF 70 kDa PROTEIN E2 (EXO70E2) (Wang et al., 2010)
-
‐
Sorghum bicolor: A. thaliana PENETRATION (PEN1) orthologs (Chaya et al., 2024)
Phytopathogenic fungi
-
‐
Botrytis cinerea: PUNCHLESS1 (PLS1) (He et al., 2023)
-
‐
Colletotrichum higginsianum: Brain modulosignalin homolog1 (Bmh1) (Rutter et al., 2022)
-
‐
Fusarium graminearum: Suppressor of Rvs167 mutation (Sur7) (Garcia‐Ceron et al., 2021)
-
‐
Fusarium oxysporum f.sp. vasinfectum: Heat shock protein of 70 kDa (Hsp70) (Bleackley et al., 2020)
-
‐
Zymoseptoria tritici: Suppressor of Rvs167 mutation (Sur7) (Hill & Solomon, 2020)
Phytopathogenic oomycetes
-
‐
Phytophthora sojae: TETRASPANIN1 (TET1), TETRASPANIN3 (TET3) (Zhu et al., 2023)
Phytopathogenic bacteria
-
‐
Pseudomonas syringae pv. tomato DC3000: Outer membrane protein F (OprF), Ampicillin C (AmpC) (Janda et al., 2023)
-
‐
Xanthomonas oryzae: ELONGATION FACTOR‐Thermo unstable (EF‐Tu) (Bahar et al., 2016)
-
‐
Xylella fastidiosa: Lipase/esterase A (LesA), Motility protein B (MopB) (Nascimento et al., 2016)
If specific antibodies are available or epitope‐tagged protein variants can be expressed, the candidate list can be further narrowed down based on protease protection or immunoaffinity capture assays to gain information on which proteins are rather inside the EV lumen than only loosely associated with the EV surface (forming the so‐called EV corona; Heidarzadeh et al., 2023). Particularly useful would be an EV‐associated integral membrane protein with both an “extracellular” and “cytosolic” terminus. These termini could be labelled by genetic engineering with a reporter tag on the luminal side (e.g., for EV quantification) and with an epitope tag on the extra‐vesicular side (e.g. for immunoaffinity capture). However, such an approach requires the genetic manipulation of the organism(s) of interest to express labelled EV markers, which is not always possible.
Protease protection assays are commonly used to determine the localization of EV‐associated proteins. They are based on the proteolytic digestion of EV‐associated proteins in the presence or the absence of an EV‐disrupting detergent (Cvjetkovic et al., 2016). A typical protease protection assay would include EVs treated with buffer‐only, protease‐only, detergent‐only, and protease with detergent. Harsh protease treatments may compromise the integrity of EVs and could lead to false results where even intraluminal proteins are degraded (Foers et al., 2018). Therefore, we highly recommend including the detergent‐only control. If, for example, luminescence originating from EVs with luciferase‐tagged intraluminal proteins increases with the detergent‐only control but disappears upon protease treatment, the protease concentration should be reduced (Bonsergent et al., 2021). If there is already evidence suggesting that the protein of interest is a cargo, a less invasive alternative to determine the localization might be immunoaffinity capture or immunogold labelling. However, EVs will only be able to bind to the solid phase or be markable if the epitope for the antibody is present on the outside of the EV.
A similar approach could be used for EV‐associated nucleic acids (nuclease protection assay). Many EV‐associated nucleic acids are protected by proteins (He et al., 2021). Hence, addition of a protease might be necessary in addition to the nuclease to access the nucleic acid for digestion (Zand Karimi et al., 2022). This setup increases the complexity of required controls further. Protease and nuclease treatments should be carried out on fresh EVs and may require a washing step to remove the respective hydrolytic enzyme sufficiently. Therefore, these would be performed immediately after obtaining a crude EV preparation, before further purification. As some proteins can be associated with the EV corona but still be relevant for the plant–microbe interaction, it may be important to optimize protease treatment to suit the biological question at hand. If it is not possible to generate transgenic organisms that express tagged EV markers, surface labelling with membrane‐impermeant biotinylation reagents may help distinguishing internal and external cargo (Cvjetkovic et al., 2016).
4.4. Controls
Stepwise quality checks are necessary during optimization and troubleshooting of an EV isolation protocol. The effect of altering each step in the protocol should be assessed by the above‐mentioned approaches. While this may be a tedious process, it is necessary for maximising the EV yield (as opposed to other EV‐like particles, Box 1), assessing suitable storage conditions, or decreasing intracellular and other contaminants. During initial method establishment and troubleshooting for sources of contamination, it is useful to include the liquid medium (if any), buffers, or even filtered distilled water as negative controls to determine the extent of nanoparticle contamination derived from the materials, equipment, and handling. It is further important to assess for contaminants following the removal of cells from the medium or AWF, and also after subsequent centrifugation or filtration steps. If ultracentrifugation is used to pellet EVs, then it may be informative to examine the supernatant for EVs, especially if the EV pellet is loose or difficult to see. It is not uncommon to deal with EV pellets that are not visible to the naked eye. If ultrafiltration is used to concentrate the samples, it is worth testing for EVs bound to the membrane or eluting with the filtrate. The same ideas apply to immunoaffinity capture, where one would compare the input crude EVs, the supernatant after each wash step, the eluate, and the beads after elution.
In the mammalian field, EVs are typically stored for longer periods at −80°C; however, preserving EV integrity during storage remains a matter of debate (Görgens et al., 2022). Similarly, experience within the Research Unit FOR5116 has revealed that the suitability for EV storage greatly depends on the source organism and is generally improved when so‐called low‐bind tubes are used (Evtushenko et al., 2020). We recommend initially comparing stored EVs that have undergone freeze‐thawing and fresh EVs for their intactness (e.g., by transmission electron microscopy or NTA), content (e.g., by Bioanalyzer‐based nucleic acid profiling), and biological activity (e.g., by enzymatic or other functional assays). EV storage conditions are dependent on the purpose and the biological question at hand. Generally, measurements of morphology, biophysical size and concentration, for example, by NTA or electron microscopy, should be performed with fresh EVs, which we propose to store short‐term (e.g., overnight) at 4°C with gentle rotation, if necessary. However, it may be possible to snap‐freeze freshly prepared EV samples for subsequent protein and nucleic acid analyses, for example, in case it is challenging to extract EV content on the same day that the EVs are obtained. Ideally, freshly prepared EVs are directly used for protein or nucleic acid extraction up to a point where the samples are safe for storage.
5. CONCLUDING REMARKS
Here we present a practical proposal for establishing EV work in the field of plant–microbe interactions (see Figure 1 for a proposed workflow and Table 1 for a synopsis of questions and recommendations). While the existing MISEV guidelines and resources from the mammalian field are highly informative, the diversity and complexity of our systems, comprising two interacting organisms, present specific challenges. There are no one‐size‐fits‐all solutions for striking a balance between sufficient EV yield and purity and identifying specific molecular markers. We encourage scientists to be creative to bypass these obstacles. At the same time, adapting protocols to the EV research in the context of plant–microbe interactions, the thorough use of controls, and precise documentation are essential for creating new field‐specific standards. An open exchange of challenges and solutions can help the plant–microbe EV field to grow in the future.
FIGURE 1.
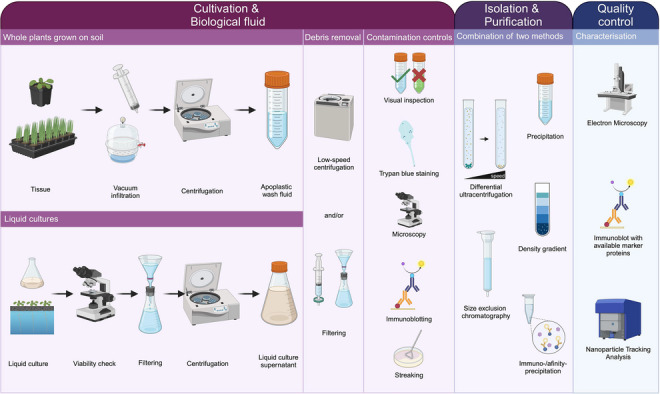
A workflow for designing EV isolation procedures from different sources in plant–microbe systems. The growth conditions of the source organism and the research question determine the downstream workflow for EV isolation. Cultivation and biological fluid: For plants grown on soil, the biological fluid, AWF, is commonly obtained by vacuum infiltration followed by very low‐speed centrifugation. Removal of cellular debris involves low‐speed centrifugation and filtering. Contamination controls for the tissue include visual inspection of the AWF colour (if applicable), viability stains (e.g., trypan blue), microscopy for organelle debris, and immunoblotting targeting intracellular proteins. When organisms are grown in liquid culture (e.g., microbes, plant cell cultures, hydroponic systems), the culture supernatant or medium serves as the biological fluid. For cell cultures, viability checks of source cells before harvesting the supernatant are advised. Removal of intact cells and larger debris involves filtering and centrifugation, followed by another step of low‐speed centrifugation and/or filtering for cell debris. Controls for contamination with live cells or cell debris are microscopy, immunoblotting with known non‐EV markers, and streaking of the cell‐free culture supernatant, where applicable. Isolation and purification of EVs can be achieved by precipitation via polymers, differential ultracentrifugation, density gradient centrifugation, size exclusion chromatography, or immuno‐/affinity‐purification. Details may vary based on research questions and the manageability of obtaining biological fluid. We refer to the MISEV 2023 guidelines for a detailed register of pros and cons for the different approaches (Welsh et al., 2024). Working with crude EV samples is acceptable for pilot studies, but purification with a second method exploiting different physical and biological properties should be performed for thorough examination. Quality control and initial characterization of isolated (and purified) EVs largely overlap. Two independent methods are recommended for EV characterization, with regular quality control suggested. Electron microscopy confirms the presence of typical structures, and nanoparticle tracking validates consistent particle size isolation, both independent of knowledge about biomarkers. If available, EVs can be tested for the presence of suitable marker proteins by immunoblotting. When EVs are isolated from unicellular organisms, streaking of the EV pellet aids in corroborating that no replicating entities are present. Created with BioRender.com. AWF, apoplastic wash fluid; EV, extracellular vesicles; MISEV, minimal information for studies of extracellular vesicles.
TABLE 1.
Synopsis of questions and recommendations for EV isolation in plant–microbe interactions.
| Question | Recommendation | Sample type | Reference |
| 1) Cultivation | |||
|---|---|---|---|
| a) How to obtain microbial EVs from axenic cultures that are biologically relevant for plant–microbe interactions? |
‐ Try different media mimicking in planta conditions. e.g., apoplast or xylem ‐ Compare the transcriptome and proteome of cultured cells to those from colonised plant material to determine how representative the cultures are. e.g., are the so‐called “plant‐specific” genes, or those important for plant colonization up‐regulated? Are the developmental stage, morphology, and physiology of the cultured cells matching the colonization stage of interest? |
Liquid culture | (Hiery et al., 2013; Jonca et al., 2021; Kwon et al., 2021; Li et al., 2022; McMillan & Kuehn, 2023; Rico & Preston, 2008; Wengelnik et al., 1996) |
| b) How to cultivate my organism for good EV yield? |
‐ Scale up It may be too laborious to harvest the biological fluid, isolate the EVs, and perform quality controls and extractions on the same day. To determine a stopping point, test for stability of the biological material from suitable steps in the procedure, when stored, e.g., biological fluid, EV pellet, or suspension |
All | – |
|
‐ Adjust conditions to open stomata to improve infiltration e.g., humidity or light conditions ‐ Use younger plants |
Plants grown on soil | (Chincinska, 2021; Nouchi et al., 2012; Rumyantseva et al., 2023) | |
|
‐ Optimise conditions for cultivation e.g., rich vs. minimal medium, pH etc. ‐ Increase surface area exposed to medium e.g., smaller mycelial clumps for fungi ‐ Degrade the cell wall to release paramural vesicles Stress conditions may increase vesiculation but can compromise the biological relevance of the EV cargos |
Liquid culture | (McMillan & Kuehn, 2023; Rutter et al., 2022) | |
| c) How to choose the right cultivation time? |
‐ Perform vesicle isolation in a time series to check for EV yield and relevant EV cargos if known ‐ Check for cell viability by staining (ideally ≥95%; see below) |
Liquid culture | (Janda et al., 2023; Shekari et al., 2023) |
| 2) Obtaining biological fluid and EV isolation | |||
|---|---|---|---|
| a) How to increase apoplastic fluid yield? |
‐ Optimise vacuum pressure for infiltration ‐ Optimise centrifugation time and force The goal is to find a balance between infiltration/extraction efficiency and cell integrity (see 2b) ‐ Use surfactants to decrease the leaf hydrophobicity Should only be considered in extreme cases when no AWF can be obtained otherwise ‐ Use more plants |
Plants grown on soil |
RU5116 (own experience) (Nouchi et al., 2012) |
| b) How and when to check for contamination with cells/cell debris? |
‐ Immunoblotting with antibodies directed against endomembrane/cytoplasmic contaminants ‐ Perform (light) microscopy (check for organelle contamination) Consider the possibility of bona fide secretion of organelles, especially for organisms other than plants |
All | (Delaunois et al., 2013) |
|
After infiltration/centrifugation: ‐ Green AWF and pellets indicate cell lysis ‐ Trypan blue staining |
Plants grown on soil | (Mulaosmanovic et al., 2020) RU5116 (own experience) | |
|
‐ Viability staining (e.g., propidium iodide) staining of cultures just prior to EV isolation (ideally ≥95% viable) ‐ Check via microscopy: ‐ Cell morphology and signs of lysis before and after removing them from culture supernatant ‐ Cell‐free culture supernatant for carried over cells and debris ‐ Streak cell‐free culture supernatant and EV suspension on agar plate to check for contamination with living cells It is possible to work as sterile as possible under the clean bench, with sterile filtered solutions and sterilised ultracentrifuge tubes |
Liquid culture |
RU5116 (own experience) |
|
| c) How to reduce contamination from cell lysis? |
‐ Settle with the lowest possible time for infiltration; find a balance between yield and purity (see above) ‐ Handle the leaves with great care when blotting dry and otherwise ‐ Choose infiltration buffer that does not compromise cell integrity |
Plants grown on soil | (O'Leary et al., 2014) |
|
‐ Minimise centrifugation force and time ‐ Apply gentle vacuum when filtering |
All | RU5116 (own experience) | |
| d) How to determine which EV subpopulations are relevant for my biological question? |
‐ Further purify and fractionate crude EV preparation according to their biophysical properties or known markers, e.g., size exclusion chromatography, density gradients, immunoaffinity capture, fluorescence‐activated cell sorting, or advanced single‐particle analysers ‐ Check resulting fractions for molecules of interest |
All | (Bleackley et al., 2020; Cai, Qiao, et al., 2018; Garcia‐Ceron et al., 2021; Rutter & Innes, 2017) |
| e) What is the best ultracentrifugation force/duration for isolating my vesicles? |
‐ Dependent on the size and density of the EVs, of the organism(s), and of the density of the used media/buffer ‐ Different pellets have to be compared if not much is known about the organism ‐ Centrifuge as short as possible |
All | (Rutter & Innes, 2020) |
| f) How to distinguish between plant vesicles and microbe vesicles? |
‐ Take advantage of specific plant and microbial vesicle markers if available ‐ Comparison of properties from EVs isolated from microbe, plant, and colonised plant |
All | (Janda et al., 2023) |
| 3) Quality check | |||
|---|---|---|---|
| a) How to determine if a protein or nucleic acid of interest is EV cargo? |
‐ Carry out immunoaffinity capture assay ‐ Carry out nuclease or protease protection assays: ‐ Check if luminal cargo is removed after treatment with detergent (Triton X‐100 etc.) and hydrolytic enzyme (e.g., protease or nuclease) One might check for sufficient EV disruption under the electron microscope |
All | (Bonsergent et al., 2021; Huang et al., 2021; Kwon et al., 2021; Zand Karimi et al., 2022) |
| b) How to identify the correct storage conditions? |
‐ Check EVs in NTA/transmission electron microscopy for aggregate formation ‐ Perform immunoblot analysis with antibodies directed against intraluminal cargo Protein integrity is used as a proxy for EV integrity ‐ Perform immunoaffinity capture Signal intensity in subsequent immunoblot analysis should decrease if EVs were disrupted and membrane fragments fused in random orientation ‐ Use low‐bind tubes ‐ Some EVs cannot be frozen In some cases, freezing can have a negative effect on the properties of the EVs. In these cases, short‐term storage of the EVs at 4°C with slight rotation can be considered. |
All |
FOR5116 (own experience) |
| c) How to differentiate EVs from other particles? |
‐ Perform NTA (size profile and zeta potential) ‐ Stain with a lipophilic dye (e.g., FM4‐64, DiOC6, DiR) ‐ Transmission electron microscopy (TEM) ‐ Comparison of the results obtained using the above methods with literature ‐ Use of advanced single‐particle analysers such as nanoflow cytometry or microfluidic resistive pulse sensing Try to compare recorded EV properties with the ones of closely related species or universal EV properties where applicable, e.g., NTA size profiles and zeta potentials or cup‐shaped structures in TEM micrographs, respectively. Consider testing differing experimental conditions until the ideal cultivation/isolation procedure is found. |
All | (Welsh et al., 2024) |
Abbreviation: EV, extracellular vesicles.
AUTHOR CONTRIBUTIONS
Hannah Thieron: Conceptualization (equal); visualization (lead); writing—original draft (equal); writing—review and editing (equal). Laura Krassini: Conceptualization (equal); writing—original draft (equal); writing—review and editing (equal). Seomun Kwon: Conceptualization (equal); writing—original draft (equal); writing—review and editing (equal). Sebastian Fricke: Conceptualization (supporting); writing—review and editing (supporting). Sabrine Nasfi: Conceptualization (supporting); writing—review and editing (supporting). Lorenz Oberkofler: Conceptualization (supporting); writing—review and editing (supporting). Alessa Ruf: Conceptualization (supporting); writing—review and editing (supporting). Julia Kehr: Funding acquisition (equal); supervision (equal); writing—review and editing (supporting). Karl‐Heinz Kogel: Funding acquisition (equal); supervision (equal); writing—review and editing (supporting). Arne Weiberg: Funding acquisition (equal); supervision (equal); writing—review and editing (equal). Michael Feldbrügge: Funding acquisition (equal); supervision (equal); writing—review and editing (supporting). Silke Robatzek: Funding acquisition (equal); supervision (equal); writing—review and editing (supporting). Ralph Panstruga: Conceptualization (supporting); funding acquisition (equal); project administration (lead); supervision (lead); writing—review and editing (lead).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interests.
ACKNOWLEDGEMENTS
We appreciate the usage of BioRender (https://www.biorender.com/) to create Figure 1. This work was enabled by grants within the research unit FOR5116 “exRNA” funded by the Deutsche Forschungsgemeinschaft (DFG; project number 433194101). Individual grants of this research unit comprise FE 448/15‐1 to M.F., KE 856/8‐1 to J.K., KO 1208/30‐1 to K.‐H.K., PA 861/22‐1 to R.P., RO 3550/16‐1 to S.R., and WE 5707/2‐1 to A.W.). Work in the lab of S.R. in the context of this article was further funded by a project within the DFG‐funded Collaborative Research Centre SFB924 “Yield” (SFB924/3TP15) and the Advanced Grant “MultiX” (grant number 884235) of the European Research Council (ERC). S.N. was supported by a Dr. Ernst‐Leopold Klipstein Foundation scholarship.
Open access funding enabled and organized by Projekt DEAL.
Thieron, H. , Krassini, L. , Kwon, S. , Fricke, S. , Nasfi, S. , Oberkofler, L. , Ruf, A. , Kehr, J. , Kogel, K.‐H. , Weiberg, A. , Feldbrügge, M. , Robatzek, S. , & Panstruga, R. (2024). Practical advice for extracellular vesicle isolation in plant–microbe interactions: Concerns, considerations, and conclusions. Journal of Extracellular Vesicles, 13, e70022. 10.1002/jev2.70022
Hannah Thieron, Laura Krassini, and Seomun Kwon contributed equally to the study.
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
REFERENCES
- Bachurski, D. , Schuldner, M. , Nguyen, P.‐H. , Malz, A. , Reiners, K. S. , Grenzi, P. C. , Babatz, F. , Schauss, A. C. , Hansen, H. P. , Hallek, M. , & Pogge von Strandmann, E. (2019). Extracellular vesicle measurements with nanoparticle tracking analysis—An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. Journal of Extracellular Vesicles, 8, 1596016. 10.1080/20013078.2019.1596016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar, O. , Mordukhovich, G. , Luu, D. D. , Schwessinger, B. , Daudi, A. , Jehle, A. K. , Felix, G. , & Ronald, P. C. (2016). Bacterial outer membrane vesicles induce plant immune responses. Molecular Plant‐Microbe Interactions, 29(5), 374–384. 10.1094/MPMI-12-15-0270-R [DOI] [PubMed] [Google Scholar]
- Bitto, N. J. , Petrovski, S. , Hill, A. F. , & Kaparakis‐Liaskos, M. (2023). Planktonic and biofilm‐derived Pseudomonas aeruginosa outer membrane vesicles facilitate horizontal gene transfer of plasmid DNA. Microbiology Spectrum, 11, e05179–e05122. 10.1128/spectrum.05179-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleackley, M. R. , Samuel, M. , Garcia‐Ceron, D. , McKenna, J. A. , Lowe, R. G. T. , Pathan, M. , Zhao, K. , Ang, C. S. , Mathivanan, S. , & Anderson, M. A. (2020). Extracellular vesicles from the cotton pathogen Fusarium oxysporum f. sp. vasinfectum induce a phytotoxic response in plants. Frontiers in Plant Science, 10, 1610. 10.3389/fpls.2019.01610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccia, E. , Alfieri, M. , Belvedere, R. , Santoro, V. , Colella, M. , Del Gaudio, P. , Moros, M. , Dal Piaz, F. , Petrella, A. , Leone, A. , & Ambrosone, A. (2022). Plant hairy roots for the production of extracellular vesicles with antitumor bioactivity. Communications Biology, 5(1), 848. 10.1038/s42003-022-03781-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsergent, E. , Grisard, E. , Buchrieser, J. , Schwartz, O. , Théry, C. , & Lavieu, G. (2021). Quantitative characterization of extracellular vesicle uptake and content delivery within mammalian cells. Nature Communications, 12, 1864. 10.1038/s41467-021-22126-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, L. , Wolf, J. M. , Prados‐Rosales, R. , & Casadevall, A. (2015). Through the wall: Extracellular vesicles in Gram‐positive bacteria, mycobacteria and fungi. Nature Reviews Microbiology, 13(10), 620–630. 10.1038/nrmicro3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Q. , He, B. , Kogel, K. H. , & Jin, H. (2018a). Cross‐kingdom RNA trafficking and environmental RNAi — nature's blueprint for modern crop protection strategies. Current Opinion in Microbiology, 46, 58–64. 10.1016/j.mib.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Q. , Qiao, L. , Wang, M. , He, B. , Lin, F. , Palmquist, J. , & Jin, H. (2018b). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science, 360(6393), 1126–1129. 10.1126/science.aar4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalupowicz, L. , Mordukhovich, G. , Assoline, N. , Katsir, L. , Sela, N. , & Bahar, O. (2023). Bacterial outer membrane vesicles induce a transcriptional shift in arabidopsis towards immune system activation leading to suppression of pathogen growth in planta. Journal of Extracellular Vesicles, 12(1), 12285. 10.1002/jev2.12285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaya, T. , Banerjee, A. , Rutter, B. D. , Adekanye, D. , Ross, J. , Hu, G. , Innes, R. W. , & Caplan, J. L. (2024). The extracellular vesicle proteomes of Sorghum bicolor and Arabidopsis thaliana are partially conserved. Plant Physiology, 194(3), 1481–1497. 10.1093/plphys/kiad644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, A. , He, B. , & Jin, H. (2022). Isolation of extracellular vesicles from arabidopsis. Current Protocols, 2(1), e352. 10.1002/cpz1.352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, A. P. , Lederer, B. , Oberkofler, L. , Huang, L. , Johnson, N. R. , Platten, F. , Dunker, F. , Tisserant, C. , & Weiberg, A. (2023). A fungal RNA‐dependent RNA polymerase is a novel player in plant infection and cross‐kingdom RNA interference. PLoS Pathogens, 19(12), e1011885. 10.1371/journal.ppat.1011885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyshev, V. S. , Rachamadugu, R. , Tseng, Y. H. , Belnap, D. M. , Jia, Y. , Branch, K. J. , Butterfield, A. E. , Pease, L. F. , Bernard, P. S. , & Skliar, M. (2015). Size and shape characterization of hydrated and desiccated exosomes. Analytical and Bioanalytical Chemistry, 407(12), 3285–3301. 10.1007/s00216-015-8535-3 [DOI] [PubMed] [Google Scholar]
- Chincinska, I. A. (2021). Leaf infiltration in plant science: Old method, new possibilities. Plant Methods, 17(1), 83. 10.1186/s13007-021-00782-x<./bib> [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitti, S. V. , Gummadi, S. , Kang, T. , Shahi, S. , Marzan, A. L. , Neveda, C. , Sanwlani, R. , Bramich, K. , Stewart, S. , Petrovska, M. , Sen, B. , Ozkan, A. , Akinfenwa, M. , Foneska, P. , & Mathivanan, S. (2024). Vesiclepedia 2024: An extracellular vesicles and extracellular particles repository. Nucleic Acids Research, 52(D1), D1694–1D698. 10.1093/nar/gkad1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuo, S. T. Y. , Chien, J. C. Y. , & Lai, C. P. K. (2018). Imaging extracellular vesicles: Current and emerging methods. Journal of Biomedical Science, 25, 91. 10.1186/s12929-018-0494-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, M. , Raposo, G. , & Théry, C. (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annual Review of Cell and Developmental Biology, 30, 255–289. 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- Cvjetkovic, A. , Jang, S. C. , Konečná, B. , Höög, J. L. , Sihlbom, C. , Lässer, C. , & Lötvall, J. (2016). Detailed analysis of protein topology of extracellular vesicles‐evidence of unconventional membrane protein orientation. Scientific Reports, 6, 36338. 10.1038/srep36338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvjetkovic, A. , Lötvall, J. , & Lässer, C. (2014). The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. Journal of Extracellular Vesicles, 3(1), 23111. 10.3402/jev.v3.23111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauros Singorenko, P. , Chang, V. , Whitcombe, A. , Simonov, D. , Hong, J. , Phillips, A. , Swift, S. , & Blenkiron, C. (2017). Isolation of membrane vesicles from prokaryotes: A technical and biological comparison reveals heterogeneity. Journal of Extracellular Vesicles, 6(1), 1324731. 10.1080/20013078.2017.1324731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbi, L. , Guo, S. , Safina, D. , & Levenberg, S. (2022). Boosting extracellular vesicle secretion. Biotechnology Advances 2, 59, 107983. 10.1016/j.biotechadv.2022.107983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunois, B. , Colby, T. , Belloy, N. , Conreux, A. , Harzen, A. , Baillieul, F. , Clément, C. , Schmidt, J. , Jeandet, P. , & Cordelier, S. (2013). Large‐scale proteomic analysis of the grapevine leaf apoplastic fluid reveals mainly stress‐related proteins and cell wall modifying enzymes. BMC Plant Biology, 13, 24. 10.1186/1471-2229-13-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma, M. , Ambrosone, A. , Leone, A. , Del Gaudio, P. , Ruocco, M. , Turi, L. , Bokka, R. , Fiume, I. , Tucci, M. , & Pocsfalvi, G. (2020). Plant roots release small extracellular vesicles with antifungal activity. Plants, 9(12), 1777. 10.3390/plants9121777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker, F. , Trutzenberg, A. , Rothenpieler, J. S. , Kuhn, S. , Pröls, R. , Schreiber, T. , Tissier, A. , Kemen, A. , Kemen, E. , Hückelhoven, R. , & Weiberg, A. (2020). Oomycete small RNAs bind to the plant RNA‐induced silencing complex for virulence. Elife, 9, e56096. 10.7554/eLife.56096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evtushenko, E. G. , Bagrov, D. V. , Lazarev, V. N. , Livshits, M. A. , & Khomyakova, E. (2020). Adsorption of extracellular vesicles onto the tube walls during storage in solution. PLoS ONE, 15(12), e0243738. 10.1371/journal.pone.0243738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo, J. , Sousa Silva, M. , & Figueiredo, A. (2018). Subtilisin‐like proteases in plant defence: The past, the present and beyond. Molecular Plant Pathology, 19(4), 1017–1028. 10.1111/mpp.12567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipe, V. , Hawe, A. , & Jiskoot, W. (2010). Critical evaluation of nanoparticle tracking analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharmaceutical Research, 27(5), 796–810. 10.1007/s11095-010-0073-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foers, A. D. , Chatfield, S. , Dagley, L. F. , Scicluna, B. J. , Webb, A. I. , Cheng, L. , Hill, A. F. , Wicks, I. P. , & Pang, K. C. (2018). Enrichment of extracellular vesicles from human synovial fluid using size exclusion chromatography. Journal of Extracellular Vesicles, 7(1), 1490145. 10.1080/20013078.2018.1490145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato, D. , Mladenović, D. , Criscuoli, M. , Loria, F. , Veiman, K. L. , Zocco, D. , Koort, K. , & Zarovni, N. (2021). Opportunities and pitfalls of fluorescent labeling methodologies for extracellular vesicle profiling on high‐resolution single‐particle platforms. International Journal of Molecular Sciences, 22(19), 10510. 10.3390/ijms221910510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Ceron, D. , Lowe, R. G. T. , McKenna, J. A. , Brain, L. M. , Dawson, C. S. , Clark, B. , Berkowitz, O. , Faou, P. , Whelan, J. , Bleackley, M. R. , & Anderson, M. A. (2021). Extracellular vesicles from Fusarium graminearum contain protein effectors expressed during infection of corn. Journal of Fungi, 7(11), 977. 10.3390/jof7110977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görgens, A. , Corso, G. , Hagey, D. W. , Jawad Wiklander, R. , Gustafsson, M. O. , Felldin, U. , Lee, Y. , Bostancioglu, R. B. , Sork, H. , Liang, X. , Zheng, W. , Mohammad, D. K. , van de Wakker, S. I. , Vader, P. , Zickler, A. M. , Mamand, D. R. , Ma, L. , Holme, M. N. , Stevens, M. M. , … El Andaloussi, S. (2022). Identification of storage conditions stabilizing extracellular vesicles preparations. Journal of Extracellular Vesicles, 11(6), 12238. 10.1002/jev2.12238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, B. , Cai, Q. , Qiao, L. , Huang, C. Y. , Wang, S. , Miao, W. , Ha, T. , Wang, Y. , & Jin, H. (2021). RNA‐binding proteins contribute to small RNA loading in plant extracellular vesicles. Nature Plants, 7(3), 342–352. 10.1038/s41477-021-00863-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, B. , Wang, H. , Liu, G. , Chen, A. , Calvo, A. , Cai, Q. , & Jin, H. (2023). Fungal small RNAs ride in extracellular vesicles to enter plant cells through clathrin‐mediated endocytosis. Nature Communications, 14(1), 1–15. 10.1038/s41467-023-40093-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidarzadeh, M. , Zarebkohan, A. , Rahbarghazi, R. , & Sokullu, E. (2023). Protein corona and exosomes: New challenges and prospects. Cell Communication and Signaling, 21, 64. 10.1186/s12964-023-01089-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiery, E. , Adam, S. , Reid, S. , Hofmann, J. , Sonnewald, S. , & Burkovski, A. (2013). Genome‐wide transcriptome analysis of Clavibacter michiganensis subsp. michiganensis grown in xylem mimicking medium. Journal of Biotechnology, 168(4), 348–354. 10.1016/j.jbiotec.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Hill, E. H. , & Solomon, P. S. (2020). Extracellular vesicles from the apoplastic fungal wheat pathogen Zymoseptoria tritici . Fungal Biology and Biotechnology, 7, 13. 10.1186/s40694-020-00103-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer, U. (2021). Lassoing OMVs with an LPS receptor. Nature Reviews Microbiology, 19(11), 682. 10.1038/s41579-021-00633-5 [DOI] [PubMed] [Google Scholar]
- Hong, J. , Dauros‐Singorenko, P. , Whitcombe, A. , Payne, L. , Blenkiron, C. , Phillips, A. , & Swift, S. (2019). Analysis of the Escherichia coli extracellular vesicle proteome identifies markers of purity and culture conditions. Journal of Extracellular Vesicles, 8(1), 1632099. 10.1080/20013078.2019.1632099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. , Wang, S. , Cai, Q. , & Jin, H. (2021). Effective methods for isolation and purification of extracellular vesicles from plants. Journal of Integrative Plant Biology, 63(12), 2020–2030. 10.1111/jipb.13181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda, M. , Rybak, K. , Krassini, L. , Meng, C. , Feitosa‐junior, O. , Stigliano, E. , Szulc, B. , Menke, F. L. H. , Malone, J. G. , Brachmann, A. , Klingl, A. , Ludwig, C. , & Robatzek, S. (2023). Biophysical and proteomic analyses of Pseudomonas syringae pv. tomato DC3000 extracellular vesicles suggest adaptive functions during plant infection. MBio, 14, e03589–e03522. 10.1128/mbio.03589-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonca, J. , Waleron, M. , Czaplewska, P. , Bogucka, A. , Steć, A. , Dziomba, S. , Jasiecki, J. , Rychłowski, M. , & Waleron, K. (2021). Membrane vesicles of pectobacterium as an effective protein secretion system. International Journal of Molecular Sciences, 22(22), 12574. 10.3390/ijms222212574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. K. , Lee, J. , Kim, S. R. , Choi, D. S. , Yoon, Y. J. , Kim, J. H. , Go, G. , Nhung, D. , Hong, K. , Jang, S. C. , Kim, S. H. , Park, K. S. , Kim, O. Y. , Park, H. T. , Seo, J. H. , Aikawa, E. , Baj‐Krzyworzeka, M. , van Balkom, B. W. , Belting, M. , … Gho, Y. S. (2015). EVpedia: A community web portal for extracellular vesicles research. Bioinformatics, 31(6), 933–939. 10.1093/bioinformatics/btu741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocholata, M. , Prusova, M. , Auer Malinska, H. , Maly, J. , & Janouskova, O. (2022). Comparison of two isolation methods of tobacco‐derived extracellular vesicles, their characterization and uptake by plant and rat cells. Scientific Reports, 12, 19896. 10.1038/s41598-022-23961-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov, K. , Nikitin, Y. , Fedorov, A. , Kostareva, A. , Mikhailovskii, V. , Isakov, D. , Ivanov, A. , & Golovkin, A. (2020). Heterogeneity of the nucleic acid repertoire of plasma extracellular vesicles demonstrated using high‐sensitivity fluorescence‐activated sorting. Journal of Extracellular Vesicles, 9(1), 1743139. 10.1080/20013078.2020.1743139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch, S. , Singh, M. , Thieron, H. , Spanu, P. D. , & Panstruga, R. (2023). Site‐specific analysis reveals candidate cross‐kingdom small RNAs, tRNA and rRNA fragments, and signs of fungal RNA 2 phasing in the barley‐powdery mildew interaction. Molecular Plant Pathology, 24(6), 570–587. 10.1111/mpp.13324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, S. , Rupp, O. , Brachmann, A. , Blum, C. F. , Kraege, A. , Goesmann, A. , & Feldbrügge, M. (2021). mRNA inventory of extracellular vesicles from Ustilago maydis . Journal of Fungi, 7(7), 562. 10.3390/jof7070562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrich, B. M. , Liang, Y. , & Fiandaca, M. S. (2021). Foetal bovine serum influence on in vitro extracellular vesicle analyses. Journal of Extracellular Vesicles, 10(3), e12061. 10.1002/jev2.12061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D. , Li, Z. , Wu, J. , Tang, Z. , Xie, F. , Chen, D. , Lin, H. , & Li, Y. (2022). Analysis of outer membrane vesicles indicates that glycerophospholipid metabolism contributes to early symbiosis between Sinorhizobium fredii HH103 and soybean. Molecular Plant‐Microbe Interactions, 35(4), 311–322. 10.1094/MPMI-11-21-0288-R [DOI] [PubMed] [Google Scholar]
- Lohaus, G. , Pennewiss, K. , Sattelmacher, B. , Hussmann, M. , & Hermann Muehling, K. (2001). Is the infiltration‐centrifugation technique appropriate for the isolation of apoplastic fluid? A critical evaluation with different plant species. Physiologia Plantarum, 111(4), 457–465. 10.1034/j.1399-3054.2001.1110405.x [DOI] [PubMed] [Google Scholar]
- McMillan, H. M. , & Kuehn, M. J. (2023). Proteomic profiling reveals distinct bacterial extracellular vesicle subpopulations with possibly unique functionality. Applied and Environmental Microbiology, 89(1), e01686–e01622. 10.1128/aem.01686-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan, H. M. , Rogers, N. , Wadle, A. , Hsu‐Kim, H. , Wiesner, M. R. , Kuehn, M. J. , & Hendren, C. O. (2021). Microbial vesicle‐mediated communication: Convergence to understand interactions within and between domains of life. Environmental Science: Processes and Impacts, 23(5), 664–677. 10.1039/d1em00022e [DOI] [PubMed] [Google Scholar]
- Midekessa, G. , Godakumara, K. , Ord, J. , Viil, J. , Lättekivi, F. , Dissanayake, K. , Kopanchuk, S. , Rinken, A. , Andronowska, A. , Bhattacharjee, S. , Rinken, T. , & Fazeli, A. (2020). Zeta potential of extracellular vesicles: Toward understanding the attributes that determine colloidal stability. ACS Omega, 5(27), 16701–16710. 10.1021/acsomega.0c01582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulaosmanovic, E. , Lindblom, T. U. T. , Bengtsson, M. , Windstam, S. T. , Mogren, L. , Marttila, S. , Stützel, H. , & Alsanius, B. W. (2020). High‐throughput method for detection and quantification of lesions on leaf scale based on trypan blue staining and digital image analysis. Plant Methods, 16, 62. 10.1186/s13007-020-00605-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento, R. , Gouran, H. , Chakraborty, S. , Gillespie, H. W. , Almeida‐Souza, H. O. , Tu, A. , Rao, B. J. , Feldstein, P. A. , Bruening, G. , Goulart, L. R. , & Dandekar, A. M. (2016). The type II secreted lipase/esterase LesA is a key virulence factor required for Xylella fastidiosa pathogenesis in grapevines. Scientific Reports, 6, 18598. 10.1038/srep18598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann, M. J. , & Dobinson, K. F. (2003). Sequence tag analysis of gene expression during pathogenic growth and microsclerotia development in the vascular wilt pathogen Verticillium dahliae . Fungal Genetics and Biology, 38(1), 54–62. 10.1016/S1087-1845(02)00507-8 [DOI] [PubMed] [Google Scholar]
- Nouchi, I. , Hayashi, K. , Hiradate, S. , Ishikawa, S. , Fukuoka, M. , Chen, C. P. , & Kobayashi, K. (2012). Overcoming the difficulties in collecting apoplastic fluid from rice leaves by the infiltration‐centrifugation method. Plant and Cell Physiology, 53(9), 1659–1668. 10.1093/pcp/pcs102 [DOI] [PubMed] [Google Scholar]
- O'Leary, B. M. , Rico, A. , McCraw, S. , Fones, H. N. , & Preston, G. M. (2014). The infiltration‐centrifugation technique for extraction of apoplastic fluid from plant leaves using Phaseolus vulgaris as an example. Journal of Visualized Experiments: JoVE, 19(94), e52113. 10.3791/52113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olicón‐Hernández, D. R. , Hernández‐Lauzardo, A. N. , Pardo, J. P. , Peña, A. , Velázquez‐del Valle, M. G. , & Guerra‐Sánchez, G. (2015). Influence of chitosan and its derivatives on cell development and physiology of Ustilago maydis . International Journal of Biological Macromolecules, 79, 654–660. 10.1016/j.ijbiomac.2015.05.057 [DOI] [PubMed] [Google Scholar]
- Panagopoulou, M. S. , Wark, A. W. , Birch, D. J. S. , & Gregory, C. D. (2020). Phenotypic analysis of extracellular vesicles: A review on the applications of fluorescence. Journal of Extracellular Vesicles, 9(1), 1710020. 10.1080/20013078.2019.1710020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinedo, M. , Canal, L. , & Marcos Lousa, C. (2021). A call for Rigor and standardization in plant extracellular vesicle research. Journal of Extracellular Vesicles, 10(6), e12048. 10.1002/jev2.12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regente, M. , Corti Monzón, G. , & De La Canal, L. (2008). Phospholipids are present in extracellular fluids of imbibing sunflower seeds and are modulated by hormonal treatments. Journal of Experimental Botany, 59(3), 553–562. 10.1093/jxb/erm329 [DOI] [PubMed] [Google Scholar]
- Regente, M. , Corti‐Monzón, G. , Maldonado, A. M. , Pinedo, M. , Jorrín, J. , & de la Canal, L. (2009). Vesicular fractions of sunflower apoplastic fluids are associated with potential exosome marker proteins. FEBS Letters, 583(20), 3363–3366. 10.1016/j.febslet.2009.09.041 [DOI] [PubMed] [Google Scholar]
- Regente, M. , Pinedo, M. , Clemente, H. S. , Balliau, T. , Jamet, E. , & De La Canal, L. (2017). Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. Journal of Experimental Botany, 68(20), 5485–5495. 10.1093/jxb/erx355 [DOI] [PubMed] [Google Scholar]
- Rico, A. , & Preston, G. M. (2008). Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast‐induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Molecular Plant‐Microbe Interactions, 21(2), 269–282. 10.1094/MPMI-21-2-0269 [DOI] [PubMed] [Google Scholar]
- Ruf, A. , Oberkofler, L. , Robatzek, S. , & Weiberg, A. (2022). Spotlight on plant RNA‐containing extracellular vesicles. Current Opinion in Plant Biology, 69, 102272. 10.1016/j.pbi.2022.102272 [DOI] [PubMed] [Google Scholar]
- Rumyantseva, N. I. , Valieva, A. I. , Kostyukova, Y. A. , & Ageeva, M. V. (2023). The effect of leaf plasticity on the isolation of apoplastic fluid from leaves of tartary buckwheat plants grown in vivo and in vitro. Plants, 12(23), 4048. 10.3390/plants12234048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter, B. D. , Chu, T. T. H. , Dallery, J. F. , Zajt, K. K. , O'Connell, R. J. , & Innes, R. W. (2022). The development of extracellular vesicle markers for the fungal phytopathogen Colletotrichum higginsianum . Journal of Extracellular Vesicles, 11(5), e12216. 10.1002/jev2.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter, B. D. , & Innes, R. W. (2017). Extracellular vesicles isolated from the leaf apoplast carry stress‐response proteins. Plant Physiology, 173(1), 728–741. 10.1104/pp.16.01253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter, B. D. , & Innes, R. W. (2020). Growing pains: Addressing the pitfalls of plant extracellular vesicle research. New Phytologist, 228(5), 1505–1510. 10.1111/nph.16725 [DOI] [PubMed] [Google Scholar]
- Saad, M. G. , Beyenal, H. , & Dong, W. J. (2024). Dual roles of the conditional extracellular vesicles derived from Pseudomonas aeruginosa biofilms: Promoting and inhibiting bacterial biofilm growth. Biofilm, 7, 100183. 10.1016/j.bioflm.2024.100183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekari, F. , Alibhai, F. J. , Baharvand, H. , Börger, V. , Bruno, S. , Davies, O. , Giebel, B. , Gimona, M. , Salekdeh, G. H. , Martin‐Jaular, L. , Mathivanan, S. , Nelissen, I. , Nolte‐'t Hoen, E. , O'Driscoll, L. , Perut, F. , Pluchino, S. , Pocsfalvi, G. , Salomon, C. , Soekmadji, C. , … Nieuwland , R. (2023). Cell culture‐derived extracellular vesicles: Considerations for reporting cell culturing parameters. Journal of Extracellular Biology, 2(10), e115. 10.1002/jex2.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skliar, M. , Chernyshev, V. S. , Belnap, D. M. , Sergey, G. V. , Al‐Hakami, S. M. , Bernard, P. S. , Stijleman, I. J. , & Rachamadugu, R. (2018). Membrane proteins significantly restrict exosome mobility. Biochemical and Biophysical Research Communications, 501(4), 1055–1059. 10.1016/j.bbrc.2018.05.107 [DOI] [PubMed] [Google Scholar]
- Suh, J. , & Lee, Y. S. (2024). Mitochondria as secretory organelles and therapeutic cargos. Experimental and Molecular Medicine, 56, 66–85. 10.1038/s12276-023-01141-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J. M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7(1), 1535750. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wakker, S. I. , van Oudheusden, J. , Mol, E. A. , Roefs, M. T. , Zheng, W. , Görgens, A. , El Andaloussi, S. , Sluijter, J. P. G. , & Vader, P. (2022). Influence of short term storage conditions, concentration methods and excipients on extracellular vesicle recovery and function. European Journal of Pharmaceutics and Biopharmaceutics, 170, 59–69. 10.1016/j.ejpb.2021.11.012 [DOI] [PubMed] [Google Scholar]
- Varga, Z. , Fehér, B. , Kitka, D. , Wacha, A. , Bóta, A. , Berényi, S. , Pipich, V. , & Fraikin, J. L. (2020). Size measurement of extracellular vesicles and synthetic liposomes: The impact of the hydration shell and the protein corona. Colloids and Surfaces B: Biointerfaces, 192, 111053. 10.1016/j.colsurfb.2020.111053 [DOI] [PubMed] [Google Scholar]
- Wang, F. , Ho, P. Y. , Kam, C. , Yang, Q. , Liu, J. , Wang, W. , Zhao, E. , & Chen, S. (2023). An AIE‐active probe for efficient detection and high‐throughput identification of outer membrane vesicles. Aggregate, 4(4), e312. 10.1002/agt2.312 [DOI] [Google Scholar]
- Wang, J. , Ding, Y. , Wang, J. , Hillmer, S. , Miao, Y. , Lo, S. W. , Wang, X. , Robinson, D. G. , & Jiang, L. (2010). EXPO, an exocyst‐positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell, 22(12), 4009–4030. 10.1105/tpc.110.080697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Weiberg, A. , Lin, F.‐M. , Thomma, B. P. H. J. , Huang, H.‐D. , & Jin, H. (2016). Bidirectional cross‐kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nature Plants, 2, 16151. 10.1038/nplants.2016.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , He, B. , Wu, H. , Cai, Q. , Ramírez‐Sánchez, O. , Abreu‐Goodger, C. , Birch, P. R. J. , & Jin, H. (2023). Plant mRNAs move into a fungal pathogen via extracellular vesicles to reduce infection. Cell Host & Microbe, 32(1), 93–105. 10.1016/j.chom.2023.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg, A. , Wang, M. , Lin, F.‐M. , Zhao, H. , Zhang, Z. , Kaloshian, I. , Huang, H.‐D. , & Jin, H. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science, 342(6154), 118–123. 10.1126/science.1239705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh, J. A. , Goberdhan, D. C. I. , O'Driscoll, L. , Buzas, E. I. , Blenkiron, C. , Bussolati, B. , Cai, H. , Di Vizio, D. , Driedonks, T. A. P. , Erdbrügger, U. , Falcon‐Perez, J. M. , Fu, Q. L. , Hill, A. F. , Lenassi, M. , Lim, S. K. , Mahoney, M. G. , Mohanty, S. , Möller, A. , Nieuwland, R. , … Witwer, K. W. (2024). Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. Journal of Extracellular Vesicles, 13(2), e12404. 10.1002/jev2.12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengelnik, K. , Marie, C. , Russel, M. , & Bonas, U. (1996). Expression and localization of HrpA1, a protein of Xanthomonas campestris pv. vesicatoria essential for pathogenicity and induction of the hypersensitive reaction. Journal of Bacteriology, 178(4), 1061–1069. 10.1128/jb.178.4.1061-1069.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong‐Bajracharya, J. , Singan, V. R. , Monti, R. , Plett, K. L. , Ng, V. , Grigoriev, I. V. , Martin, F. M. , Anderson, I. C. , & Plett, J. M. (2022). The ectomycorrhizal fungus Pisolithus microcarpus encodes a microRNA involved in cross‐kingdom gene silencing during symbiosis. Proceedings of the National Academy of Sciences of the United States of America, 119(3), e2103527119. 10.1073/pnas.2103527119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, L. , Jiang, Z. , Wang, Y. , Hu, Y. , Hao, G. , Zhong, W. , Wan, S. , & Xin, X. (2023). High air humidity dampens salicylic acid pathway and NPR1 function to promote plant disease. EMBO Journal, 42(21), e113499. 10.15252/embj.2023113499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zand Karimi, H. , Baldrich, P. , Rutter, B. D. , Borniego, L. , Zajt, K. K. , Meyers, B. C. , & Innes, R. W. (2022). Arabidopsis apoplastic fluid contains SRNA‐ and circular RNA‐protein complexes that are located outside extracellular vesicles. Plant Cell, 34(5), 1863–1881. 10.1093/plcell/koac043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J. , Qiao, Q. , Sun, Y. , Xu, Y. , Shu, H. , Zhang, Z. , Liu, F. , Wang, H. , Ye, W. , Dong, S. , Wang, Y. , Ma, Z. , & Wang, Y. (2023). Divergent sequences of tetraspanins enable plants to specifically recognize microbe‐derived extracellular vesicles. Nature Communications, 14, 4877. 10.1038/s41467-023-40623-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.


