Abstract
With the implementation of the policy of “centralized slaughtering and chilled to market” and the development of the livestock processing industry, numerous researchers have begun to focus on the selection and breeding of broilers bred for slaughter. The selection of breeds with excellent slaughtering performance and high meat production performance has become one of the most important selective breeding goals. In our previous study, we conducted transcriptome sequencing on chicken breast tissues with high and low breast muscle rates and found higher early growth response protein 1 (EGR1) expression in breast tissues with a low breast muscle ratio, thus hypothesizing that the EGR1 gene is involved in the growth and development process of chicken muscle tissues. Therefore, we analyzed the gene functions and polymorphisms of EGR1 to investigate its association with slaughter traits. We used various experimental methods, including RT-qPCR, Cell Counting Kit 8, 5-ethynyl-2′-deoxyuridine, western blot, flow cytometry, and immunofluorescence, to validate EGR1’s role in chicken primary myoblasts. The results of our functional validation experiments indicate that EGR1 is highly expressed in breast tissues with a low breast muscle content and plays a key role in regulating of muscle growth and development by promoting proliferation and inhibiting the differentiation of chicken primary myoblasts. In addition, we explored the relationship between the EGR1 gene polymorphisms and slaughter traits using mixed linear models for the first time. In a population of Jiangfeng M3 lineage partridge chickens, we identified 4 EGR1 single-nucleotide polymorphisms, 2 of which were significantly associated with slaughter traits, including live weight, slaughter weight, semi-eviscerated weight, eviscerated weight, leg weight, wing weight, and breast muscle rate. In summary, ectopic expression of EGR1 promotes the proliferation and differentiation of chicken primary myoblasts. In addition, polymorphisms in EGR1 were associated with slaughter performance, providing a potential basis for further utilization of EGR1 as a breeding marker.
Keywords: Early growth response protein 1 (EGR1), Polymorphism, Slaughter performance, Association analysis, Skeletal muscle
Introduction
The mechanisms regulating skeletal muscle growth and development are complex, and among these mechanisms molecular mechanisms, nutritional metabolism, and genetics play important roles. Studies on molecular mechanisms have focused on exploring signaling pathways in skeletal muscle cells, such as mammalian target of rapamycin complex 1, PI3K/Akt, and mitogen-activated protein kinase, which promote muscle synthesis and growth (Brennan et al., 2021; Goodman, 2019; Zhao et al., 2022). Studies on nutrient metabolism have focused primarily on the effects of protein intake and the type and regulation of metabolic pathways on skeletal muscle growth, revealing that the amount and type of protein intake have a significant effect on the rate and extent of muscle synthesis. In addition, regulation of metabolic pathways, such as adenosine monophosphate-activated protein kinase and peroxisome proliferator-activated receptor-γ coactivator-1α, can promote skeletal muscle synthesis and growth (Kong et al., 2022; Xu et al., 2020). Exploring the effects of genes on skeletal muscle growth has been a major focus of genetic research, and many genetic variants are associated with skeletal muscle mass and growth. For example, the relationship between genes such as insulin-like growth factor 1, myostatin, and ACTN3 and muscle growth has been extensively studied (Mohammadabadi et al., 2021; Seto et al., 2021; Yoshida and Delafontaine, 2020). Overall, an in-depth study of these influencing factors will help us to comprehensively understand the mechanisms of skeletal muscle growth and development and provide more effective approaches and strategies for the promotion of muscle health.
Owing to rapid advances in molecular biology and genomics technology in recent years, the study of the association between genetic polymorphisms and broiler slaughter performance has become a topic of interest in the field of broiler breeding. These genotypic variants can affect various traits such as growth performance, slaughter performance, and meat quality in broilers, thereby affecting the quality and yield of meat products. For example, the rs313277885 polymorphism of CRELD1 is associated with breast muscle, abdominal fat, and liver weight in Jiangfeng partridge chickens, and the rs144502152 polymorphism of DNAJC30 is significantly associated with abdominal fat and breast muscle weight (Zhou et al., 2023). PRKG2 is an important candidate gene for the traits of growth and meat quality in Chinese Ningdu Yellow chickens, and its rs16400745 polymorphism is significantly associated with 12 slaughter and meat quality traits (Xiong et al., 2022). Therefore, in-depth studies on the association between chicken slaughter performance and genetic polymorphisms can allow for better selection and optimization of the chicken genome and can help to improve the slaughter performance and production efficiency of chickens. In addition, understanding the genetic polymorphisms of chickens helps breeders better predict slaughter performance and other relevant characteristics of chickens for improved selective breeding planning and goals.
Early growth response protein 1 (EGR1) is a transcription factor with important biological functions which belongs to the EGR family of C2H2-type zinc finger proteins (Pei and Grishin, 2015; Sogut et al., 2021; Thiel and Rossler, 2023). The EGR family includes transcription factors such as EGR1, EGR2, EGR3, and EGR4, which share an N-terminal DNA-binding domain and a C-terminal activation domain. The DNA-binding domain consists of 3 C2H2 zinc-fingers, which can bind to DNA sequences of target genes to regulate gene transcription, and the activation domain consists of numerous common transcriptional activation regions, such as TA1 and TA2, which can enhance or attenuate transcription efficiency (O'Donovan et al., 1999; Russo et al., 1993; Svaren et al., 1996). Currently, there are few studies on the regulatory mechanisms of EGR1 in muscle growth and development. However, some previous studies have shown that EGR1 induces Sirtuin 1 expression in skeletal muscle cells and inhibits cellular responses to mechanical stimuli. Simultaneously, high Sirtuin 1 expression inhibits EGR1 expression, forming an autoregulatory loop (Pardo and Boriek, 2012). Furthermore, the transcription factor EGR1 could promote the differentiation of bovine skeletal muscle satellite cells by regulating myogenin (MyoG) expression. In addition, several studies have found that the upregulation of EGR1 expression promotes the proliferation and migration of vascular smooth muscle cells and vascular regeneration. In summary, EGR1 plays an important role in muscle growth and development, but its specific regulatory mechanism requires further in-depth study.
Materials and methods
Experimental animals and slaughter trait data
In this study, the M3 line of partridge chickens bred by Guangzhou Jiangfeng Breeding Science and Technology Co. were used as research subjects and reared under equivalent environmental and management conditions. Animal experiments in this study were approved by the Animal Protection Committee of South China Agricultural University. We randomly selected 130 males and 260 females at 77 days of age and sent them to Guangzhou Jiangfeng Industrial Co. Ltd. Zhuowei Poultry Processing Plant for slaughtering. During the 12 h prior to slaughter, the experimental partridge chickens were provided with drinking water only. We measured the live weight of the partridge chickens prior to slaughter. After carotid artery exsanguination, the partridge chickens were blanched in water at a temperature of about 60°C for 3 min to remove feathers, followed by drying and subjection to slaughter performance measurements, including carcass weight, semi-eviscerated weight, eviscerated weight, breast muscle weight, leg weight (including leg bones), wing weight, and abdominal fat weight. In addition, we calculated the slaughter rate, semi-evisceration rate, evisceration rate, breast muscle rate, leg rate, wing rate, and abdominal fat rate. All measurements and calculations were conducted in accordance with the agricultural industry standard “Terminology and Measurement Calculation Method for Poultry Performance” (NY/T823-2020).
HE staining
Tissue samples were collected and prepared into paraffin sections, which were then placed in an environmentally friendly dewaxing solution (Servicebio, China). This was followed by continuous soaking in anhydrous ethanol and 75 % alcohol (MACKLIN, China), and finally washed with tap water. The slices should then be dyed in the hematoxylin dye solution (Servicebio, China) for a period of between three and five minutes, after which they should be rinsed with water and returned to their original blue state. The slices are then dehydrated in 95 % alcohol (MACKLIN, China) for one minute and subsequently stained in eosin solution (Servicebio, China) for 15 seconds. Subsequently, the samples are dehydrated and sealed.
Transcriptome sequencing
We divided the M3 line partridge hens into high and low breast rate groups (n = 4) according to evisceration weight, breast weight and breast muscle rate (Table 1). Breast muscle tissue was taken and stored at -80°C. Tissue samples were sent to ANNOROAD GENE TECHNOLOGY for RNA extraction and transcriptome sequencing. All raw data was submitted to the NCBI SRA database (accession number: PRJNA1126171; link: https://www.ncbi.nlm.nih.gov/sra/PRJNA1126171).
Table 1.
Individual slaughtering performance of M3 partridge hens with high and low breast muscle rates.
| Slaughter Traits | Group |
|||||||
|---|---|---|---|---|---|---|---|---|
| High-1 | High-2 | High-3 | High-4 | Low-1 | Low-2 | Low-3 | Low-4 | |
| EW (g) | 966 | 946 | 949 | 974 | 951 | 980 | 954 | 972 |
| BMW (g) | 165.58 | 163.28 | 164.05 | 168.82 | 134.99 | 140.77 | 135.33 | 138.64 |
| BMP (%) | 17.14 | 17.26 | 17.29 | 17.33 | 14.19 | 14.36 | 14.19 | 14.26 |
High = high breast muscle rate group; Low = low breast muscle rate group; EW = evisceration weight; BMW = breast muscle weight; BMP = breast muscle percentage.
Isolation and culture of chicken primary myoblasts
The shells of 11-day-old eggs were cracked on an ultraclean bench, the embryos were removed, and the whole legs of the embryos were immediately separated and placed in phosphate-buffered solution. Forceps were used to separate the muscles of the whole leg from the leg bone, and the muscles were subsequently cut into pieces, a process that took about 10 min. Next, digestion was performed using trypsin in an incubator at 37°C for 15 - 20 min, and then digestion was stopped by adding 15 % complete medium. The digested mixture was filtered, and the filtrate was placed in a centrifuge and centrifuged at 1,000 × g for 5 min. The supernatant was discarded. The cell pellet was then suspended in 15 % complete medium, seeded into new cell culture dishes, and cultured in an incubator at 37°C with 5 % CO2. Finally, cells were purified by differential adhesion and cultured using culture medium containing 15 % fetal bovine serum, 0.2 % penicillin-streptomycin, and 1640 medium. Chicken primary myoblasts (CPMs) were induced to differentiate in 1640 medium containing 5 % horse serum and 0.2 % penicillin-streptomycin.
DNA extraction, PCR, and DNA sequencing
We collected blood samples from the subwing veins of 390 partridge chickens and performed DNA extraction according to the DNA extraction kit manufacturer's instructions (OMEGA, USA). We amplified all DNA samples using 2 × Taq MasterMix (Kangwei Century, China), which targets the double-stranded DNA of the 5′ untranslated region (UTR), 3′ UTR, and coding sequence (CDS) region of the EGR1 gene. The PCR reaction conditions were as follows: an initial 2-minute pre-denaturation at 94°C, followed by 30 cycles of 30 seconds 94°C amplification, a 30 seconds 60°C amplification, and a 1minute 72°C amplification. A final amplification was performed at 72°C for 2 min, and then samples were held at 4°C indefinitely. Finally, we sent the PCR products to Sangon Biotech for purification and sequencing.
RNA extraction, cDNA synthesis, and real-time fluorescence quantitative PCR
We extracted total the RNA from tissues or cells using the MagZol reagent (Guangzhou Meiji, China) according to the manufacturer's instructions. Subsequently, the total RNA was reverse transcribed into cDNA according to the MonScript RTase III reagent manufacturer's instructions (Suzhou Mona, China), and the PCR products were stored at −20°C. Next, we performed RT-qPCR experiments using the ChamQ Universal SYBR qPCR Master Mix (Nanjing Vazyme Biotechnology Co., Ltd., China) and a Bio-Rad CFX96 real-time detection system. Finally, we used chicken glyceraldehyde 3-phosphate dehydrogenase as a reference gene and analyzed the data using the 2−ΔΔCt method. RT-qPCR primers were designed using Primer-BLAST and are detailed in Supplementary Table 1.
Construction of overexpression vectors and synthesis of interfering fragments
We amplified the CDS region of the chicken EGR1 gene using NCBI-designed primers and cloned it into the Acc65I and XbaI sites of the pcDNA3.1-3 × FLAG plasmid. Subsequently, plasmids were extracted according to the manufacturer's instructions on the HiPure Plasmid/BAC EF Mini Kit (Guangzhou Magen Biotechnology Co., Ltd., China). The interfering fragment (5′-CATCAGTGCGCACAGTGTA-3′) was synthesized and supplied by Guangzhou RiboBio Co., Ltd.
Cell Counting Kit 8 (CCK-8) and flow cytometry assays
In the CCK-8 treatment assay, CPMs were seeded into 96-well plates and transfected after the cell density reached 60 %–70 %. Cells were processed according to the manufacturer's instructions on the Cell Counting Kit-8 (New Cell and Molecular Biotech Co., Ltd., China). At 12, 24, 36, and 48 h after transfection, the absorbance was measured with a filter wavelength of 450 nm using a Model 680 microplate reader, and experimental data were recorded and tallied.
In the flow cytometry detection assay, CPMs were seeded into 12-well plates and transfected after the cell density reached 70 % - 80 %. At 48 h after transfection, the medium was aspirated and the cells were digested with trypsin for 1 minute. Then, the complete medium was added to terminate the digestion. The cell suspension was transferred to a 1.5 mL centrifuge tube and centrifuged at 1,000 × g for 5 min, and the supernatant was discarded. Then, 1 mL of precooled phosphate-buffered saline (PBS) was added, and the cells were recentrifuged. Thereafter, 1 mL of precooled 70 % ethanol was added, and the cells were fixed at a temperature of 4°C for 12 h. The fixed cells were further processed according to the manufacturer's instructions on the Cell Cycle and Apoptosis Detection Kit (Beyontime Biotechnology Co. Ltd., China), and red fluorescence was detected at an excitation wavelength of 488 nm using a CytoFLEX flow cytometer (Beckman Coulter, USA) with simultaneous detection of light scattering. FlowJo software was used to analyze the DNA content and light scattering of cells.
Flow cytometry and 5-ethynyl-2′-deoxyuridine (EdU) fluorescence imaging assays
CPMs were seeded onto 6-well and 12-well plates, of which 6-well plates were used for EdU flow cytometry and 12-well plates were used for EdU fluorescence imaging assays. Transfection was performed when the cell density reached 70 % - 80 %. After 48 h of cell transfection, the medium was aspirated, 50 μM EdU medium was added, and the plates were incubated for 2 h at 37°C. Flow cytometry and the EdU fluorescence imaging assay were performed separately according to the manufacturer's instructions on the Cell-Light EdU Apollo In Vitro Kit (Guangzhou RiboBio Co., Ltd., China). After completing the staining process, the red fluorescence of the cells in the 6-well plates was detected using a CytoFLEX flow cytometer at an excitation wavelength of 488 nm and the experimental results were statistically analyzed using CytExpert software. Meanwhile, cells in 12-well plates were observed and photographed using a fluorescence inverted microscope, and the images were statistically analyzed using ImageJ software.
Immunofluorescence
CPMs were seeded into 12-well plates and transfected after the cell density reached 70 % - 80 %. At 48 h after cell transfection, the medium in each well was first aspirated and the cells were washed thrice with PBS for 5 min each. The cells were then fixed with 4 % paraformaldehyde for 20 min. After fixation was completed, the plates were washed thrice with PBS for 5 min each time. The cells were then permeabilized using a 0.1 % Triton X-100 reagent for 10 - 15 min. After 10 - 15 min of permeabilization, the cells were washed thrice with PBS for 5 min each time. The cells were then blocked for 30 min using goat serum, a 1:800 dilution of mouse anti-MyHC primary antibody was added, and the plates were incubated overnight at 4°C. The next day, the primary antibody was recovered and the plates were washed thrice with PBS for 5 min each time. Then, a 1:100 dilution of FITC-goat anti-mouse IgG secondary antibody was added, and the plates were incubated away from light for 2 h at room temperature before being washed thrice with PBS for 5 min at a time. Subsequently, DAPI staining solution was added for nuclei staining. After staining for 2 - 3 min, the cells were washed with PBS thrice for 5 min at a time. Finally, the images were observed and photographed using a fluorescence inverted microscope and statistically analyzed using ImageJ software.
Data statistics and analysis
The preliminary statistics and the processing of the experimental data were conducted using Excel 2019 software, and the results were expressed as “mean ± standard deviation.” GraphPad Prism and R Studio software were used for statistical analysis and graph plotting. A two-tailed unpaired t-test was used to assess the statistical significance between the 2 groups, and the least significant difference rule was used to assess significant differences for multiple comparisons. In addition, a linkage disequilibrium analysis was performed using the Haploview 4.2 software.
The association analysis of single-nucleotide polymorphism (SNP) loci and slaughter performance was performed using a mixed linear model in SPSS 26.0 analysis software. The formula is as follows:
where denotes observed value, denotes mean, denotes genotype fixed effect, denotes gender fixed effect, and denotes random error.
Genotype frequencies and gene frequency distributions for different alleles were calculated using Excel 2019 software. The polymorphism information content (PIC) of SNP loci was calculated as follows:
where and the frequencies of the “ith” and “jth” alleles, respectively, and n is the number of polymorphic alleles. > 0.5 indicates high polymorphism, < 0.25 indicates low polymorphism, and 0.25 < < 0.5 indicates moderate polymorphism.
Results
Transcriptome sequencing results for breast muscle tissues with high and low breast muscle rates
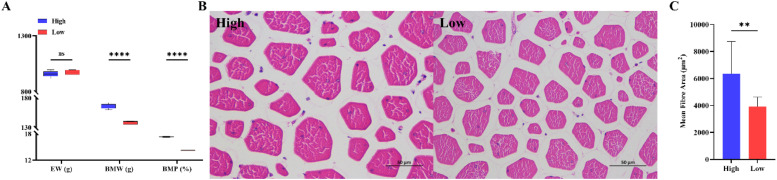
Table 1 demonstrates the evisceration weight, breast weight, and percentage of high and low breast muscle rates of M3 line partridge hens. The results showed that the difference in evisceration weight between the groups with high and low breast muscle rates was not significant (P > 0.05), but the breast muscle weight and rate of the high breast muscle rate group were significantly higher than those of the low breast muscle rate group (P < 0.05) (Fig. 1A).
Fig. 1.
Selection of individuals for transcriptome sequencing of high and low breast muscle rates. Note: A. Results of pairwise comparison of evisceration weight, breast muscle weight, and breast muscle rate for individuals with high and low breast muscle rates (n = 4); B. Hematoxylin–eosin staining plots of breast muscle tissues from individuals with high and low breast muscle rates (scale bar: 50 μm); and C. Statistical results of the average myofiber area of breast muscle tissues of individuals with high and low breast muscle rates (n = 4).
To gain more insight into the differing characteristics of the muscle fiber in the high and low breast muscle rate groups, this study used electron microscopy to observe Hematoxylin–eosin-stained sections and compare their muscle fiber microstructure. The results showed that the mean muscle fiber area was significantly greater in the high breast muscle rate group than in the low breast muscle rate group (Fig. 1B and C).
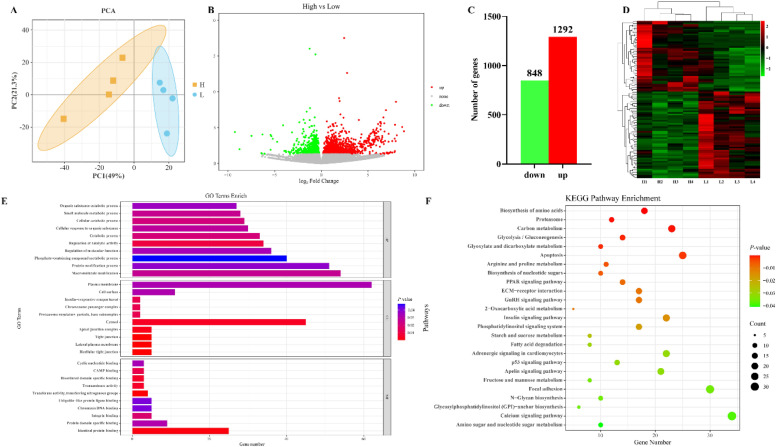
To investigate the genetic regulation mechanisms of muscle growth and development in chickens, breast muscle tissues from individual animals with high and low breast muscle rates were routinely transcriptome sequenced (n = 4). PCA analysis of the count values of each sample demonstrates that the 2 groups showed a trend of separation (Fig. 2A). A total of 2,140 differentially expressed genes (DEGs) were identified between the high and low breast muscle rate groups, with 1,292 genes upregulated and 848 genes downregulated, based on the log2 Fold Change = 0, P < 0.05 criteria (Fig. 2B and C). These DEGs were plotted as heat maps using R Studio software to better compare and analyze the differential expression between high and low breast muscle rate groups (Fig. 2D). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed to explore the biological functions of these 2,140 DEGs. According to the results of GO enrichment analysis, these DEGs were mainly involved in protein modification processes, regulation of molecular functions, chromatin DNA binding, and cyclic nucleotide binding (Fig. 2E). According to the results of the KEGG pathway enrichment analysis, these DEGs were mainly involved in the peroxisome proliferator-activated receptor, insulin, calcium, gonadotropin-releasing hormone, and p53 signaling pathways (Fig. 2F).
Fig. 2.
Transcriptome sequencing results of high and low breast muscle rate breast muscle tissues. Note: A. Principal component analysis (PCA) plot of transcriptome samples from breast tissue with high and low breast muscle rates; B. Volcano plot of differentially expressed genes (DEGs); C. Statistical plot of DEGs showing that 848 genes were upregulated and 1,292 genes were downregulated; D. Expression heatmap of the top 100 DEGs; E. Results of Gene Ontology (GO) enrichment analysis of DEGs; F. Results of Kyoto Encyclopedia of Genes and Genome (KEGG) pathway enrichment analysis of DEGs.
Table 2 presents the top 15 DEGs identified through transcriptome sequencing of breast muscle tissue from chickens with high and low breast muscle rates. The genes that may be associated with muscle growth and development include SP5, LONRF1, MRAS, DIO2, CAMK1G, GPR37L1, CUX2, and EGR1. In this study, EGR1 was selected as a candidate gene to better investigate its role and influence in skeletal muscle growth and development.
Table 2.
High and low breast muscle rates of breast muscle tissues via ordinary transcriptome sequencing of the top 15 DEGs.
| Gene ID | Gene name | FPKM_H | FPKM_L | log2FoldChange | P-value | P-adjust |
|---|---|---|---|---|---|---|
| 107052728 | RP1 | 0.0000 | 0.0383 | −6.2761 | 0.0000 | 0.0008 |
| 418892 | C4A | 0.4991 | 0.2393 | 1.3085 | 0.0000 | 0.0109 |
| 693261 | SP5 | 0.0457 | 0.0000 | 5.0230 | 0.0000 | 0.0304 |
| 422742 | LONRF1 | 8.8563 | 4.8000 | 1.1364 | 0.0002 | 0.1422 |
| 396359 | SYNPR | 0.2053 | 0.6161 | −1.3771 | 0.0002 | 0.1642 |
| 395149 | MRAS | 15.3147 | 40.2016 | −1.2794 | 0.0002 | 0.1811 |
| 373903 | DIO2 | 0.2035 | 0.5226 | −1.1085 | 0.0003 | 0.2140 |
| 419859 | CAMK1G | 0.0799 | 0.0107 | 2.9225 | 0.0005 | 0.2853 |
| 421176 | GPR37L1 | 0.0288 | 0.0820 | −1.2304 | 0.0007 | 0.3996 |
| 430848 | RPP25 | 0.0443 | 0.0043 | 3.7525 | 0.0013 | 0.5583 |
| 418120 | TMEM178B | 0.0598 | 0.2711 | −1.8778 | 0.0013 | 0.5583 |
| 416875 | CUX2 | 0.0091 | 0.0499 | −2.2657 | 0.0024 | 0.8072 |
| 768614 | IFA3L | 0.2491 | 0.0490 | 2.3230 | 0.0027 | 0.8407 |
| 428531 | CCDC178 | 0.0375 | 0.0118 | 1.8325 | 0.0029 | 0.8558 |
| 373931 | EGR1 | 6.9693 | 16.8984 | −1.0970 | 0.0030 | 0.8558 |
Expression pattern of EGR1
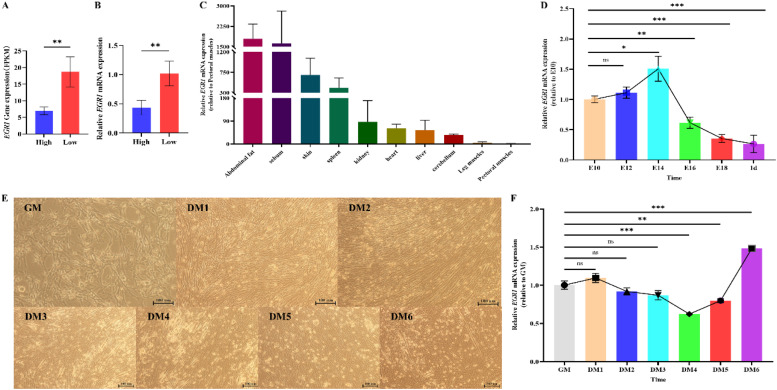
The transcriptome sequencing results of high and low breast muscle rates in breast muscle tissues were analyzed, and the transcription factor EGR1 was screened as a candidate gene. Sequencing results showed that EGR1 expression was higher in the breast muscle tissues of the low breast muscle rate group (Fig. 3A). Subsequent qRT-PCR validation was performed, and the results confirmed the differences in EGR1 expression in breast muscle tissues across the high and low breast muscle rate groups (Fig. 3B). A further investigation of EGR1 expression in different tissues of 77-day-old M3 partridge chickens was conducted via the qRT-PCR assay. The results demonstrated that EGR1 expression was higher in the abdominal adipose, subcutaneous adipose, skin, and spleen tissues than in breast muscle tissues (Fig. 3C). qRT-PCR was employed to investigate EGR1 expression in leg muscles at different embryonic periods. The results showed that EGR1 expression was highest at E14, followed by a gradual decrease (Fig. 3D). To explore the role of EGR1 in skeletal muscle development, CPMs were isolated in this study and the cells were observed via microscopy at each period. By the 3rd day of differentiation, thicker myotubes began to form and more myotubes were intertwined after the 5th day of differentiation, (Fig. 3E). The qRT-PCR results showed that EGR1 expression decreased and then increased with the time of differentiation (Fig. 3F).
Fig. 3.
Expression pattern of early growth response protein 1 (EGR1). Note: A. Fragments per kilobase of transcript per million mapped read (FPKM) values of EGR1 in the breast muscle tissues of individuals with high and low breast muscle rates from common transcriptome sequencing results; B. EGR1 expression in the breast tissues of individuals with high and low breast muscle rates (n = 4); C. EGR1 expression in a variety of different chicken tissues (n = 3); D. EGR1 expression in the development of skeletal muscle in chicken embryos (n = 3); E. Differentiation process of CPMs; F. EGR1 expression during differentiation of CPMs (n = 6).
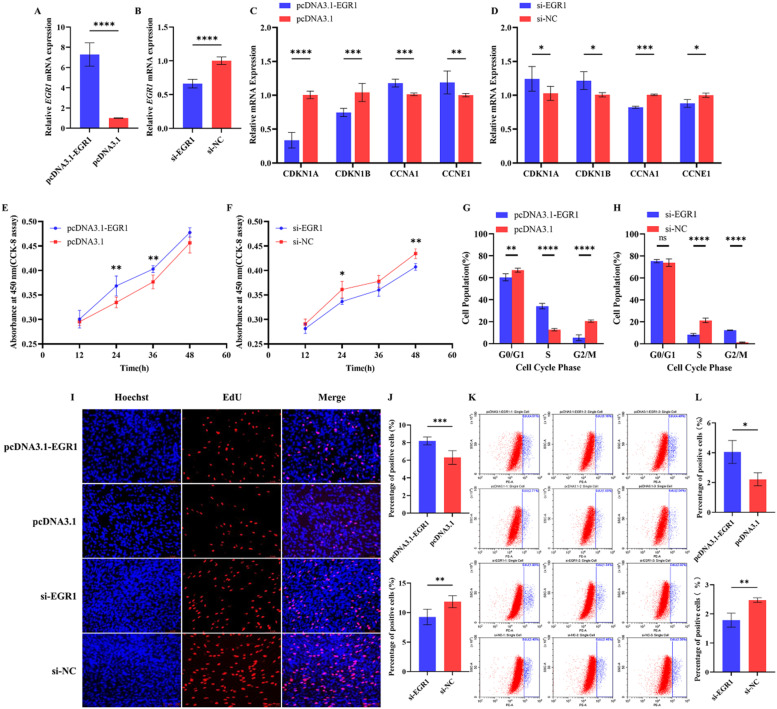
EGR1 promotes the proliferation of primary myoblasts
To further investigate the role of EGR1 in the proliferation of CPMs, this study used EGR1 overexpressed vectors and siRNA for transfection of proliferating CPMs. First, the expression efficiency of EGR1 overexpression or interference was examined using qRT-PCR. The results showed that the overexpression efficiency of EGR1 increased more than 7-fold, whereas the interference efficiency was found to be around 40 % (Fig. 4A and B). Next, the mRNA expression levels of several cell cycle-related genes (CDKN1A, CDKN1B, CCNA1, and CCNE1) were examined using qRT-PCR. The experimental results showed that EGR1 overexpression significantly decreased the expression levels of CDKN1A and CDKN1B and increased the expression levels of CCNA1 and CCNE1, whereas the opposite result was obtained by EGR1 interference (Fig. 4C and D). In addition, CCK-8 assays were performed to further investigate the effect of EGR1 on cell proliferation. The results showed that cell proliferation was significantly enhanced at 24 and 36 h in the presence of EGR1 overexpression, whereas it was significantly inhibited at 24 and 48 h in the presence of EGR1 interference (Fig. 4E and F). Next, the cell cycle was examined 48 h after transfection using flow cytometry, which showed a significant increase in the proportion of CPMs overexpressing EGR1 in the S phase and a significant increase in the proportion of CPMs in the G2M phase after EGR1 interference. Finally, flow cytometry and fluorescence imaging were used to detect EdU staining. The results of the fluorescence imaging assay method showed that EGR1 overexpression increased the proportion of CPMs in the proliferative phase, whereas EGR1 interference decreased this proportion (Fig. 4I and J). This conclusion was confirmed by the experimental results of flow cytometry (Fig. 4K and L). In summary, the above experimental results indicate that EGR1 can promote the proliferation of CPMs.
Fig. 4.
Early growth response protein 1 (EGR1) promotes the proliferation of CPMs. Note: A. Overexpression efficiency of the EGR1 overexpression vector in CPMs (n = 6); B. Interference efficiency of the EGR1 interference fragment in CPMs (n = 6); C and D. Changes in mRNA expression of cell proliferation marker genes in CPMs after transfection of the EGR1 overexpression vector or interference fragment for 48 h (n = 6); E and F. Results of the Cell Counting Kit 8 (CCK-8) cell viability assay in CPMs after transfection of EGR1 overexpression vector or interference fragments at 12, 24, 36, and 48 h (n = 6); G and H. The results of the cell cycle assay in CPMs after transfection with EGR1 overexpression vectors or interference fragments for 48 h (n = 6); I. The results of 5-ethynyl-2′-deoxyuridine (EdU) staining of CPMs 48 h after transfection with EGR1 overexpression vectors or interference fragments; J. The ratio of the number of cells in the proliferative stage to the total number of cells 48 after transfection with the EGR1 overexpression vector or interference fragment (n = 6); K. The results of the ratio of the number of cells in the proliferative stage to the total number of cells in CPMs 48 h after transfection with the EGR1 overexpression vector or interference fragment detected using flow cytometry (n = 6); L. The results of the ratio of the number of cells in the proliferative stage to the total number of cells in CPMs 48 h after transfection with the EGR1 overexpression vector or interference fragment detected using flow cytometry (n = 3).
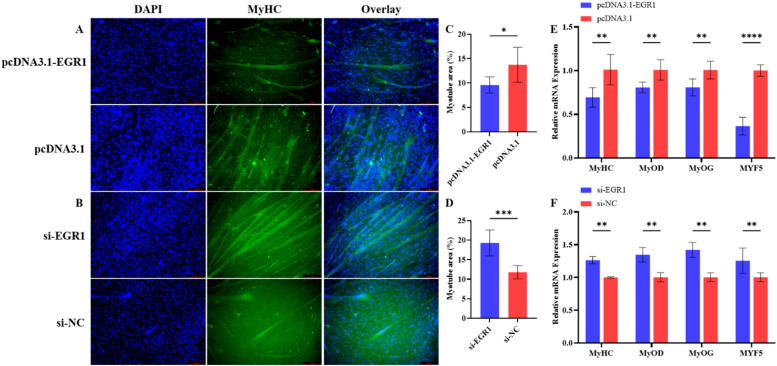
EGR1 inhibits the differentiation of chicken primary myoblasts
To investigate the potential role of EGR1 in the differentiation of CPMs, EGR1 overexpression vector and siRNA were used to transfect CPMs in this study. After 48 h of transfection with EGR1 overexpression vectors or interference fragments, cells were induced to differentiate for 3 days using a differentiation medium and myofibers were stained and visualized via the immunofluorescence assay. The results of this experiment showed that CPMs in which EGR1 expression interference was present differentiated into more mature and thicker myofibers, whereas cells overexpressing EGR1 differentiated into slenderer myofibers (Fig. 5A and B). The muscle fiber area in the images was counted using ImageJ, and the results showed that the area of muscle fibers with EGR1 expression interference was significantly larger than that of the control group, whereas the area of muscle fibers overexpressing EGR1 was significantly smaller than that of the control group (Fig. 5C and D). To further validate the effect of EGR1 on the differentiation of CPMs, this study examined the mRNA expression levels of several genes associated with muscle cell differentiation (MyHC, myoblast determination protein 1 [MyOD], MyOG, and MYF5) using qRT-PCR. The experimental results showed that EGR1 overexpression significantly decreased the expression of MyHC, MyOD, MyOG, and MYF5, whereas the opposite result was obtained by EGR1 interference (Fig. 5E and F). Taken together, the experiment results proved that EGR1 inhibits CPMs differentiation.
Fig. 5.
Early growth response protein 1 (EGR1) inhibits CPMs differentiation. Note: A and B. Immunofluorescence staining results of CPMs after overexpressing or interfering with EGR1 for 48 h and inducing differentiation for 3 days; C and D. Percentage of the myofiber area of the entire microscopic field of view of CPMs after overexpressing or interfering with EGR1 for 48 h and inducing differentiation for 3 days (n = 6); E and F. Changes in mRNA expression of muscle differentiation marker genes in CPMs after transfection of EGR1 overexpression vector or interfering fragment for 48 h and inducing differentiation for 3 days (n = 6).
EGR1 polymorphism analysis
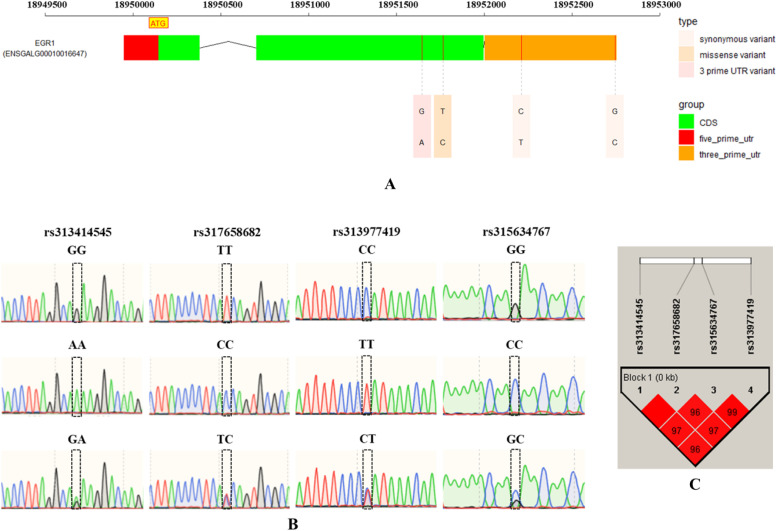
Supplementary Table 2 provides statistical data on the slaughter performance of pure lines of M3 partridge chickens, which will be used in the analysis of the association between polymorphisms in the EGR1 gene and slaughter performance. The full-length sequence of the EGR1 gene was amplified to screen for polymorphisms. The results revealed that 4 SNP loci were present in the pure lines of M3 partridge chickens, of which 2 SNPs (rs313414545 and rs317658682) were located in the CDS region and the other 2 (rs315634767 and rs313977419) were located in the 3′ UTR (Fig. 6A). All 4 SNP loci have 3 genotypes (Fig. 6B). The 4 SNPs were analyzed for linkage disequilibrium using Haploview software, revealing their location within the same haplotype block, and there was a high level of disequilibrium among them, as indicated by D' values ranging from 0.96 to 1.00 (Fig. 6C).
Fig. 6.
Early growth response protein 1 (EGR1) gene polymorphism analysis. Note: A. EGR1 gene polymorphism distribution sites; B. EGR1 gene polymorphism base mutation types; C. EGR1 gene polymorphism linkage disequilibrium analysis. The values in the box indicate the linkage disequilibrium values (D') for single nucleotide polymorphisms, which will not be displayed when D' = 1. The deeper red the box color, the stronger the linkage disequilibrium.
In this study, the chi-square test was used to analyze whether the allele frequencies conformed to the Hardy–Weinberg equilibrium. The results are shown in Table 3, and all of the above SNPs conformed to the Hardy–Weinberg equilibrium (P > 0.05). In addition, Table 3 lists parameters related to genetic polymorphisms, including PIC, observed heterozygosity, expected heterozygosity, and effective number of alleles, which can be used to assess the genetic diversity and information content of SNPs.
Table 3.
Genotype and allele frequencies and diversity parameters of EGR1 SNPs.
| SNP | Genotype frequency (n) |
Allele Gene frequency |
P-value | Genetic polymorphism |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | BB | AB | A | B | PIC | He | Ne | Ho | |||
| rs313414545 | 139(0.38) | 45(0.13) | 178(0.49) | 0.63 | 0.37 | 0.58 | 0.36 | 0.47 | 1.87 | 0.49 | |
| rs317658682 | 23(0.06) | 192(0.53) | 147(0.41) | 0.27 | 0.73 | 0.77 | 0.32 | 0.39 | 1.64 | 0.41 | |
| rs315634767 | 139(0.38) | 47(0.13) | 176(0.49) | 0.63 | 0.37 | 0.75 | 0.36 | 0.47 | 1.88 | 0.49 | |
| rs313977419 | 26(0.07) | 186(0.52) | 145(0.41) | 0.28 | 0.72 | 0.95 | 0.32 | 0.40 | 1.67 | 0.41 | |
EGR1 = early growth response protein 1; SNP = single-nucleotide polymorphism; PIC = polymorphism information content; Ho = observed heterozygosity; He = expected heterozygosity; Ne = effective number of alleles. P-value: Hardy–Weinberg equilibrium test, with P > 0.05 indicating that the population conforms to the Hardy–Weinberg equilibrium.
Table 4.
Data presenting the significant association between SNP locus rs317658682 of the EGR1 gene and slaughter performance.
| Trait | rs317658682 |
P-value | ||
|---|---|---|---|---|
| CC (189) | TC (144) | TT (23) | ||
| Live weight (g) | 1547.08±184.17A | 1570.14±199.13B | 1657.83±208.87AB | 0.029 |
| Carcass weight (g) | 1393.23±171.64A | 1399.38±215.70B | 1498.30±192.43AB | 0.046 |
| Semi-evisceration weight (g) | 1265.83±166.04A | 1285.23±176.63B | 1367.22±183.78AB | 0.026 |
| Evisceration weight (g) | 1049.69±138.94A | 1065.44±145.79B | 1133.52±147.96AB | 0.027 |
| Leg weight (g) | 375.88±58.73A | 381.84±61.56B | 408.91±62.12AB | 0.044 |
SNP = single-nucleotide polymorphism; EGR1 = early growth response protein 1. In the same row, the same letter indicates that there is no significant difference between the variables (P > 0.05), whereas different letters indicate that there is a significant difference between the variables (P < 0.05). The aforementioned convention is also applicable to the following table.
Detailed analysis of the association between different genotypes and slaughter performance was performed using the MLM model in SPSS software, and the results are presented in Table 5, Table 6 for SNP loci significantly associated with slaughter performance. Specifically, the TT genotype at rs317658682 was found to have significantly (P < 0.05) higher live weight, carcass weight, semi-evisceration weight, evisceration weight, and leg weight than the CC and TC genotypes. Furthermore, the GG genotype at rs313977419 exhibited significantly elevated (P < 0.05) live weight, semi-evisceration weight, evisceration weight, leg weight, and wing weight in comparison to the CC genotype. However, it demonstrated a notable reduction (P < 0.05) in breast muscle percentage when contrasted with both the CC and GC genotypes.
Table 5.
Data presenting the significant association between SNP locus rs313977419 the EGR1 gene and slaughtering performance.
| Trait | rs313977419 |
P-value | ||
|---|---|---|---|---|
| CC (191) | GC (145) | GG (26) | ||
| Live weight (g) | 1543.82±182.32A | 1574.07±201.55 | 1648.85±204.55A | 0.023 |
| Semi-evisceration weight (g) | 1263.37±164.58A | 1288.27±178.76 | 1358.15±180.32A | 0.025 |
| Evisceration weight (g) | 1047.39±138.20A | 1068.43±146.93 | 1125.31±145.98A | 0.026 |
| Leg weight (g) | 374.72±58.34A | 383.25±62.15 | 406.28±60.80A | 0.034 |
| Wing weight (g) | 137.56±19.73A | 141.22±20.34 | 148.44±20.72A | 0.020 |
| Breast muscle ratio (%) | 15.22±1.52B | 15.39±1.64A | 14.50±1.45AB | 0.028 |
SNP = single-nucleotide polymorphism; EGR1 = early growth response protein 1.
Table 6.
Association analysis of EGR1 haplotype combinations with slaughter performance.
| SNPs | Haplotype | Double Type | Semi-evisceration weight | P-value |
|---|---|---|---|---|
| rs313414545 rs317658682 rs315634767 rs313977419 |
H1:ACTC (0.620) H2:GTCG (0.260) H3:GCCC (0.094) |
H1H1 (132) | 1259.47±167.52B | 0.048 |
| H2H1 (122) | 1298.40±177.62D | |||
| H2H2 (22) | 1377.50±181.20ABCD | |||
| H2H3 (14) | 1231.21±182.73A | |||
| H3H1 (44) | 1288.39±167.12C | |||
| H3H3 (4) | 1278.00±168.04 |
EGR1 = early growth response protein 1; SNP = single-nucleotide polymorphism.
These haplotype blocks were correlated with slaughter performance using the MLM model to more accurately characterize the relationship between SNPs and slaughter performance. Table 6 lists the slaughter performance parameters that are significantly associated with haplotype blocks. The results showed that the H2H2 genotype had a significantly (P < 0.05) higher semi-evisceration weight than the H1H1, H2H1, H2H3, and H3H1 genotypes.
Discussion
Currently, transcriptome sequencing technology is widely used to detect DEGs under 2 or more different conditions. The technique has been useful for studying muscle growth and development in animals such as pigs (Xu et al., 2012), cows (Guo et al., 2015), and goats (Wang et al., 2015). Muscle growth and development is also one of the most important concerns in broiler production, and increasing broiler meat production is a major goal of broiler selective breeding. In this study, the breast muscle tissues of chickens with high and low breast muscle rates were studied intensively using transcriptome sequencing technology to reveal the differences in muscle growth and development between chickens with high and low breast muscle rates. In total, 2,140 DEGs were found between the breast muscle tissues of chickens with high and low breast muscle rates, of which 1,292 genes were upregulated and 848 genes were downregulated.
We further analyzed these DEGs for GO and KEGG pathway enrichment. GO enrichment analysis results revealed that these genes are mainly involved in biological processes such as protein modification, regulation of molecular function, chromatin DNA binding, and cyclic nucleotide binding, which play crucial roles in muscle growth and development. Protein modification refers to the alteration of protein structure and function during protein synthesis by chemical modifications such as phosphorylation, methylation, acetylation, ubiquitination, and glycosylation. These modifications affect protein function in muscle growth and development (Han and Martinage, 1992; Solis and Russell, 2021). In addition, molecular functional regulators are crucial for muscle growth and development. For example, insulin-like growth factor 1 promotes muscle cell proliferation and differentiation, which in turn promotes muscle growth and development (Philippou et al., 2007), and MyoD is an important transcription factor involved in the regulation of the expression of several genes and plays an important part in muscle cell differentiation and muscle fiber formation (Li and Olson, 1992). Thus, changes in such biological processes and molecular mechanisms may be an important reason for differences in the muscle growth and development of chickens with high and low breast muscle rates.
The results of KEGG pathway enrichment analysis show that these DEGs are mainly involved in the peroxisome proliferator-activated receptor, insulin, calcium, gonadotropin-releasing hormone, and p53 signaling pathways, which are also important factors in muscle growth and development. For example, insulin stimulates glucose uptake and utilization by muscle cells, as well as promotes muscle protein synthesis and inhibits protein breakdown, thereby promoting muscle growth and development (Duan et al., 2010). On the contrary, the p53 signaling pathway plays an important role in the regulation of the cell cycle and apoptosis (Cai et al., 2018). Collectively, these signaling pathways interact to synergistically regulate processes such as muscle cell proliferation, differentiation, and protein metabolism, which in turn affect muscle growth and development.
Transcription factors can bind to specific sequence regions on DNA and recruit other transcription factors or proteins, such as co-activators, transcriptional cofactors, and RNA polymerases, to form a complex transcriptional regulatory complex that activates or represses gene transcription (Latchman, 1997). As a transcription factor, EGR1 can bind to specific sequence regions on DNA, thereby regulating gene transcription. During muscle growth and development, EGR1 may play a regulatory role and be involved in the proliferation and differentiation of myogenic cells, affecting muscle cell development and function. The EGR1 protein has been demonstrated to promote the differentiation of bovine skeletal muscle satellite cells by regulating the expression of the MyoG (Zhang et al., 2018). The expression pattern of EGR1 was explored to gain insight into the regulatory mechanism of the candidate gene EGR1 in chicken skeletal muscle growth and development.
The results showed that the expression level of EGR1 was higher in the breast muscle tissues of partridge chickens with a low breast muscle rate, suggesting that EGR1 is closely related to the tissue differentiation and growth and muscle development. Compared with breast muscle tissues, the expression of EGR1 was higher in abdominal lipids, subcutaneous lipids, the skin, and splenic tissues of 77-day-old M3 lineage partridge chickens, indicating that EGR1 is expressed in multiple tissues and is not limited to muscle tissues. This phenomenon may be related to EGR1’s role in various biological processes, such as cell proliferation, differentiation, apoptosis, inflammation, and stress response (Havis and Duprez, 2020; Pagel and Deindl, 2012). It was also found that EGR1 expression showed significant temporal variations during the embryonic development of skeletal muscle in chickens. Specifically, EGR1 expression gradually increased during the E10 - E14 stage and gradually decreased during the E16 - 1d stage. Research has shown that during the E10 - E14 stage, muscle development occurs via myogenic cell formation, whereas during the E16 - 1d stage, muscle development occurs via the maturation and differentiation of muscle cells (Hayes and Hikida, 1976). This suggests that there are differences in the mechanisms by which EGR1 regulates muscle tissue organization at different developmental stages, with its primary role probably centered on promoting myogenic cell proliferation. This conclusion was also validated in experiments involving EGR1 overexpression and interference in CPMs. In addition, the expression level of EGR1 decreased and then increased during the differentiation of CPMs, suggesting that changes in EGR1 expression during muscle differentiation of chickens are linear, but there may be a regulatory equilibrium point. This phenomenon may be related to the complex role of EGR1 in regulating myocyte proliferation and differentiation.
The continuous scientific and technological advances that have occurred over time have led to the widespread application of molecular marker-assisted selective breeding techniques in the field of poultry production.
The continuous scientific and technological advances that have occurred over time have led to the widespread application of molecular marker-assisted selective breeding techniques in the field of poultry production. This technique employs molecular marker technology and genetic principles to screen poultry breeds with excellent genetic characteristics with speed and accuracy and avoids the tedious breeding process and long rearing cycles associated with traditional breeding methods (Cui et al., 2019). EGR1 is a transcription factor that plays an important regulatory role in several biological processes, including cell proliferation, differentiation, apoptosis, and stress response (Havis and Duprez, 2020; Pagel and Deindl, 2012). In addition, EGR1 plays important regulatory roles in multiple signaling pathways, including cell cycle, DNA damage repair, apoptosis, immune response, and neural development (Thiel and Cibelli, 2002; Weisz et al., 2004). However, there are no reports on the association of EGR1 polymorphisms with chicken slaughter performance. Four SNP loci in the CDS and 3′ UTR regions of EGR1 were identified in a study of a pure-line population of M3 partridge chickens from 10 generations of Jiangfeng partridge chickens. The CDS region comprises DNA sequences that encode proteins in eukaryotes, and the presence of SNPs within this region may affect the function or expression of genes, thereby increasing or decreasing disease risk (Chu and Wei, 2019). The 3′ UTR region is an important regulatory element of mRNAs as it controls mRNA stability, degradation rates, subcellular localization, translation levels, and the fate of specific mRNAs. 3′ UTR mutations may also lead to disease (Chabanon et al., 2004; Kuersten and Goodwin, 2003; Zhu et al., 2002). Among these SNPs, rs313414545 and rs317658682 are located in the CDS region, with rs313414545 representing a missense mutation resulting in the substitution of arginine for lysine. Additionally, rs315634767 and rs313977419 are located in the 3′ UTR region.
In genetic polymorphism analysis, the effective number of alleles, genetic heterozygosity, and polymorphic information content are crucial markers used to measure the degree of genetic variation in a given population (Botstein et al., 1980). Polymorphic information content is positively correlated with genetic variation. When polymorphic information content is higher, the population also has a higher degree of genetic variation, indicating greater genetic potential (Marshall et al., 1998). In this study, we found that 4 SNP loci in EGR1 in a pure population of M3 lines from 10 generations of Jiangfeng partridge chickens exhibited moderate polymorphism and conformed to the Hardy–Weinberg equilibrium. This implies that these loci have a high degree of polymorphism and genetic variation, providing great potential for the genetic improvement of Jiangfeng partridge chickens. After association analysis, 2 SNP loci were found to be significantly associated with slaughter performance in the pure-line population of M3 lines of Jiangfeng partridge chickens in the 10th generation. Specifically, the TT genotype at rs317658682 exhibited a significantly higher association (P < 0.05) with live weight, slaughter weight, semi-evisceration weight, evisceration weight, and leg weight than the CC and TC genotypes, whereas the GG genotype at rs313977419 exhibited a significantly higher association (P < 0.05) with live weight, semi-evisceration weight, evisceration weight, leg weight, and wing weight than the CC genotype, but its breast muscle rate was significantly lower (P < 0.05) than the CC and GC genotypes. These results suggest that these 2 SNP loci have a close relationship with the growth and slaughter performance of the pure Jiangfeng M3 line of partridge chickens. Individual SNP loci are often less informative than haplotype combinations because animal phenotypes can be affected by mutations in multiple genes (Georges et al., 2019). The linkage disequilibrium analysis of these 4 SNP loci revealed that they were located in the same haplotype block and showed a high degree of linkage disequilibrium. The results of association analysis showed that the H2H2 genotype had a significantly higher semi-evisceration weight than the H1H1, H2H1, H2H3, and H3H1 genotypes (P < 0.05). In summary, EGR1 may become a new SNP marker in the genetics and selective breeding of chickens, and this provides a reference for further optimization of chicken growth and slaughter performance.
In conclusion, analysis of the transcriptome sequencing results of breast muscle tissues from high and low breast muscle rate groups and validation of cellular functions revealed that EGR1 promotes the proliferation and inhibits the differentiation of primary adult myoblasts in chickens. The EGR1 polymorphisms rs317658682 and rs313977419 were found to be significantly associated with slaughter performance.
Declaration of competing interest
The authors claim that none of the results in this paper have been published or are under consideration for publication elsewhere. The authors have declared that no competing interests exist.
Acknowledgments
This work was supported by the Key-Area Research and Development Program of Guangdong Province, No. 2022B0202100002, Science and Technology Program of Guangzhou City No. 2024B03J1353.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2024.104533.
Appendix. Supplementary materials
References
- Hayes V.E., Hikida R.S. Naturally-occurring degeneration in chick muscle development: ultrastructure of the M. complexus. J. Anat. 1976;122:67–76. [PMC free article] [PubMed] [Google Scholar]
- Botstein D., White R.L., Skolnick M., Davis R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Han K.K., Martinage A. Post-translational chemical modification(s) of proteins. Int. J. Biochem. 1992;24:19–28. doi: 10.1016/0020-711x(92)90225-p. [DOI] [PubMed] [Google Scholar]
- Li L., Olson E.N. Regulation of muscle cell growth and differentiation by the MyoD family of helix-loop-helix proteins. Adv. Cancer Res. 1992;58:95–119. doi: 10.1016/s0065-230x(08)60292-4. [DOI] [PubMed] [Google Scholar]
- Russo M.W., Matheny C., Milbrandt J. Transcriptional activity of the zinc finger protein NGFI-A is influenced by its interaction with a cellular factor. Mol. Cell Biol. 1993;13:6858–6865. doi: 10.1128/mcb.13.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J., Sevetson B.R., Apel E.D., Zimonjic D.B., Popescu N.C., Milbrandt J. NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol. Cell Biol. 1996;16:3545–3553. doi: 10.1128/mcb.16.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchman D.S. Transcription factors: an overview. Int. J. Biochem. Cell Biol. 1997;29:1305–1312. doi: 10.1016/s1357-2725(97)00085-x. [DOI] [PubMed] [Google Scholar]
- Marshall T.C., Slate J., Kruuk L.E., Pemberton J.M. Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- O'Donovan K.J., Tourtellotte W.G., Millbrandt J., Baraban J.M. The EGR family of transcription-regulatory factors: progress at the interface of molecular and systems neuroscience. Trends. Neurosci. 1999;22:167–173. doi: 10.1016/s0166-2236(98)01343-5. [DOI] [PubMed] [Google Scholar]
- Thiel G., Cibelli G. Regulation of life and death by the zinc finger transcription factor Egr-1. J. Cell Physiol. 2002;193:287–292. doi: 10.1002/jcp.10178. [DOI] [PubMed] [Google Scholar]
- Zhu S., Yoon K., Sterneck E., Johnson P.F., Smart R.C. CCAAT/enhancer binding protein-beta is a mediator of keratinocyte survival and skin tumorigenesis involving oncogenic Ras signaling. Proc. Natl. Acad. Sci. u S. a. 2002;99:207–212. doi: 10.1073/pnas.012437299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten S., Goodwin E.B. The power of the 3′ UTR: translational control and development. Nat. Rev. Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- Chabanon H., Mickleburgh I., Hesketh J. Zipcodes and postage stamps: mRNA localisation signals and their trans-acting binding proteins. Brief. Funct. Genomic. Proteomic. 2004;3:240–256. doi: 10.1093/bfgp/3.3.240. [DOI] [PubMed] [Google Scholar]
- Weisz L., Zalcenstein A., Stambolsky P., Cohen Y., Goldfinger N., Oren M., Rotter V. Transactivation of the EGR1 gene contributes to mutant p53 gain of function. Cancer Res. 2004;64:8318–8327. doi: 10.1158/0008-5472.CAN-04-1145. [DOI] [PubMed] [Google Scholar]
- Philippou A., Maridaki M., Halapas A., Koutsilieris M. The role of the insulin-like growth factor 1 (IGF-1) in skeletal muscle physiology. In. Vivo. 2007;21:45–54. [PubMed] [Google Scholar]
- Duan C., Ren H., Gao S. Insulin-like growth factors (IGFs), IGF receptors, and IGF-binding proteins: roles in skeletal muscle growth and differentiation. Gen. Comp. Endocrinol. 2010;167:344–351. doi: 10.1016/j.ygcen.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Pagel J.I., Deindl E. Disease progression mediated by egr-1 associated signaling in response to oxidative stress. Int. J. Mol. Sci. 2012;13:13104–13117. doi: 10.3390/ijms131013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo P.S., Boriek A.M. An autoregulatory loop reverts the mechanosensitive Sirt1 induction by EGR1 in skeletal muscle cells. Aging. 2012;4:456–461. doi: 10.18632/aging.100470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Qian H., Feng X., Xiong Y., Lei M., Ren Z., Zuo B., Xu D., Ma Y., Yuan H. Differential proteome and transcriptome analysis of porcine skeletal muscle during development. J. Proteomics. 2012;75:2093–2108. doi: 10.1016/j.jprot.2012.01.013. [DOI] [PubMed] [Google Scholar]
- Guo B., Greenwood P.L., Cafe L.M., Zhou G., Zhang W., Dalrymple B.P. Transcriptome analysis of cattle muscle identifies potential markers for skeletal muscle growth rate and major cell types. BMC. Genomics. 2015;16:177. doi: 10.1186/s12864-015-1403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J., Grishin N.V. C2H2 zinc finger proteins of the SP/KLF, Wilms tumor, EGR, Huckebein, and Klumpfuss families in metazoans and beyond. Gene. 2015;573:91–99. doi: 10.1016/j.gene.2015.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.H., Zhang C.L., Plath M., Fang X.T., Lan X.Y., Zhou Y., Chen H. Global transcriptional profiling of longissimus thoracis muscle tissue in fetal and juvenile domestic goat using RNA sequencing. Anim. Genet. 2015;46:655–665. doi: 10.1111/age.12338. [DOI] [PubMed] [Google Scholar]
- Cai B., Ma M., Chen B., Li Z., Abdalla B.A., Nie Q., Zhang X. MiR-16-5p targets SESN1 to regulate the p53 signaling pathway, affecting myoblast proliferation and apoptosis, and is involved in myoblast differentiation. Cell Death. Dis. 2018;9:367. doi: 10.1038/s41419-018-0403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Tong H., Zhang Z., Shao S., Liu D., Li S., Yan Y. Transcription factor EGR1 promotes differentiation of bovine skeletal muscle satellite cells by regulating MyoG gene expression. J. Cell Physiol. 2018;233:350–362. doi: 10.1002/jcp.25883. [DOI] [PubMed] [Google Scholar]
- Chu D., Wei L. Nonsynonymous, synonymous and nonsense mutations in human cancer-related genes undergo stronger purifying selections than expectation. BMC. Cancer. 2019;19:359. doi: 10.1186/s12885-019-5572-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H.X., Shen Q.C., Zheng M.Q., Su Y.C., Cai R.C., Yu Y., Yang X.R., Chen Z.W., Wen J., Zhao G.P. A selection method of chickens with blue-eggshell and dwarf traits by molecular marker-assisted selection. Poult. Sci. 2019;98:3114–3118. doi: 10.3382/ps/pez069. [DOI] [PubMed] [Google Scholar]
- Georges M., Charlier C., Hayes B. Harnessing genomic information for livestock improvement. Nat. Rev. Genet. 2019;20:135–156. doi: 10.1038/s41576-018-0082-2. [DOI] [PubMed] [Google Scholar]
- Goodman C.A. Role of mTORC1 in mechanically induced increases in translation and skeletal muscle mass. J. Appl. Physiol. 2019;127:581–590. doi: 10.1152/japplphysiol.01011.2018. 1985. [DOI] [PubMed] [Google Scholar]
- Havis E., Duprez D. EGR1 Transcription factor is a multifaceted regulator of matrix production in tendons and other connective tissues. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Chen X., Huang Z., Chen D., Yu B., Chen H., Luo Y., Zheng P., Yu J., He J. Grape seed proanthocyanidin extract promotes skeletal muscle fiber type transformation via AMPK signaling pathway. J. Nutr. Biochem. 2020;84 doi: 10.1016/j.jnutbio.2020.108462. [DOI] [PubMed] [Google Scholar]
- Yoshida T., Delafontaine P. Mechanisms of IGF-1-mediated regulation of skeletal muscle hypertrophy and atrophy. Cells. 2020;9 doi: 10.3390/cells9091970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C.M., Emerson C.P., Jr., Owens J., Christoforou N. p38 MAPKs - roles in skeletal muscle physiology, disease mechanisms, and as potential therapeutic targets. JCI. Insight. 2021;6 doi: 10.1172/jci.insight.149915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadabadi M., Bordbar F., Jensen J., Du M., Guo W. Key genes regulating skeletal muscle development and growth in farm animals. Animals. (Basel) 2021:11. doi: 10.3390/ani11030835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto J.T., Roeszler K.N., Meehan L.R., Wood H.D., Tiong C., Bek L., Lee S.F., Shah M., Quinlan K.G.R., Gregorevic P., Houweling P.J., North K.N. ACTN3 genotype influences skeletal muscle mass regulation and response to dexamethasone. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abg0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogut M.S., Venugopal C., Kandemir B., Dag U., Mahendram S., Singh S., Gulfidan G., Arga K.Y., Yilmaz B., Kurnaz I.A. ETS-domain transcription factor Elk-1 regulates stemness genes in brain tumors and CD133+ BrainTumor-initiating cells. J. Pers. Med. 2021:11. doi: 10.3390/jpm11020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis C., Russell B. Striated muscle proteins are regulated both by mechanical deformation and by chemical post-translational modification. Biophys. Rev. 2021;13:679–695. doi: 10.1007/s12551-021-00835-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong S., Cai B., Nie Q. PGC-1alpha affects skeletal muscle and adipose tissue development by regulating mitochondrial biogenesis. Mol. Genet. Genomics. 2022;297:621–633. doi: 10.1007/s00438-022-01878-2. [DOI] [PubMed] [Google Scholar]
- Xiong X., Zhou M., Zhu X., Tan Y., Wang Z., Gong J., Xu J., Wen Y., Liu J., Tu X., Rao Y. RNA sequencing of the pituitary gland and association analyses reveal PRKG2 as a candidate gene for growth and carcass traits in Chinese Ningdu Yellow chickens. Front. Vet. Sci. 2022;9 doi: 10.3389/fvets.2022.892024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao X., Shen X., Zhang Y., Zhang Y., Ye L., Li D., Zhu Q., Yin H. CircCCDC91 regulates chicken skeletal muscle development by sponging miR-15 family via activating IGF1-PI3K/AKT signaling pathway. Poult. Sci. 2022;101 doi: 10.1016/j.psj.2022.101803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel G., Rossler O.G. Glucose homeostasis and pancreatic islet size are regulated by the transcription factors elk-1 and Egr-1 and the protein phosphatase calcineurin. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms24010815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Cai D., Wei G., Cai B., Kong S., Ma M., Zhang J., Nie Q. Polymorphisms of CRELD1 and DNAJC30 and their relationship with chicken carcass traits. Poult. Sci. 2023;102 doi: 10.1016/j.psj.2022.102324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.