Abstract
Background
We retrospectively evaluated radiomics as a predictor of the tumor microenvironment (TME) and efficacy with anti‐PD‐1 mAb (IO) in R/M HNSCC.
Methods
Radiomic feature extraction was performed on pre‐treatment CT scans segmented using 3D slicer v4.10.2 and key features were selected using LASSO regularization method to build classification models with XGBoost algorithm by incorporating cross‐validation techniques to calculate accuracy, sensitivity, and specificity. Outcome measures evaluated were disease control rate (DCR) by RECIST 1.1, PFS, and OS and hypoxia and CD8 T cells in the TME.
Results
Radiomics features predicted DCR with accuracy, sensitivity, and specificity of 76%, 73%, and 83%, for OS 77%, 86%, 70%, PFS 82%, 75%, 89%, and in the TME, for high hypoxia 80%, 88%, and 72% and high CD8 T cells 91%, 83%, and 100%, respectively.
Conclusion
Radiomics accurately predicted the efficacy of IO and features of the TME in R/M HNSCC. Further study in a larger patient population is warranted.
Keywords: artificial intelligence, head and neck squamous cell carcinoma, imaging, immunotherapy, machine learning, pembrolizumab, radiomics, treatment response
1. INTRODUCTION
Recurrent/metastatic squamous cell carcinoma of the head and neck (R/M HNSCC) is associated with poor outcomes. 1 However, progress has been made in systemic therapy with the advent of anti‐PD‐1 mAb immunotherapy. First, in 2016, nivolumab significantly improved overall survival (OS) compared to standard chemotherapy in the platinum failure setting. 2 Then, more recently in 2019, in the frontline setting, pembrolizumab monotherapy improved survival compared to the EXTREME regimen for PD‐L1 expressors with CPS ≥1 and chemotherapy plus pembrolizumab for PD‐L1 expressors, as well as the total population. 3 Still, response rates to anti‐PD‐1 monotherapy remain low at 16%–20%. 2 , 3 , 4 As new therapeutic combinations make their way through clinical trials and with chemotherapy as a choice to add to pembrolizumab in the frontline setting, there is an even greater need for predictive biomarkers for anti‐PD‐1 mAb monotherapy.
In R/M HNSCC, PD‐L1 expression, immune gene expression (GEP), and tumor mutational burden (TMB) have been found to be predictive of efficacy. 5 , 6 , 7 However, even with two favorable biomarkers (TMB high/PD‐L1 high or GEP high/TMB high), the response rate to pembrolizumab was only 35%. 7 In addition, these biomarkers need an invasive procedure and reflect a small static “snapshot” sample of the tumor which can be heterogeneous. This speaks to the need for better biomarkers that can robustly and non‐invasively identify and predict those patients who will benefit from immunotherapy, inclusive of tumor heterogeneity.
Radiomics is a method for extraction of quantitative features from standard modality imaging with the goal of deeper understanding of cancer biology, genomics, and spatial heterogeneity. 8 , 9 , 10 Radiomics has been used to evaluate the efficacy of immunotherapy treatment and found to be predictive of efficacy in studies with combining advanced solid tumors or isolated to tumor types such as melanoma and non‐small cell lung cancer. 11 However, to the best of our knowledge, no study has been published evaluating radiomics as a biomarker for predicting immunotherapy efficacy in R/M HNSCC. 11 , 12 Radiomics has a number of benefits; it is non‐invasive and features are obtained from standard medical scans that makes it ideal for smoother clinical translation as a non‐invasive tool to determine response. Therefore, we evaluated the predictive value of radiomics based on pretreatment CT scans in anti‐PD‐1 mAb therapy treated R/M HNSCC patients and also its correlation with the tumor immune microenvironment.
2. MATERIALS AND METHODS
2.1. Study design and patient population
We conducted a single‐center retrospective analysis of patients with R/M HNSCC treated with anti‐PD‐1 mAb therapy between 2015 and 2020 at the UPMC Hillman Cancer Center, that had consented to the UPMC Hillman tissue banking protocol (HCC 99‐069). Clinical data was obtained via chart review, including baseline demographics, clinical characteristics, and treatments received. Our in vivo imaging core facility at UPMC Hillman Center determined treatment response by RECIST 1.1. Additional endpoints obtained were progression‐free survival (PFS) and overall survival (OS). Thirty‐five out of the 61 patients in this cohort had previously undergone tumor microenvironment analysis, including analysis for intratumoral hypoxia, CD8+ T cells (CD8) and Foxp3+ T cells (Treg). 13 As previously published, 14 to conduct these analyses, antigen retrieval was done on formalin‐fixed paraffin‐embedded sections from patient samples. Tissue sections were then stained with Pan cytokeratin eFluor 570 (Life Technologies), CD8 Alexa Fluor 647 (Biolegend), Foxp3 Alexa Fluor 488 (Life Technologies), and DAPI, and mounted with ProLong Diamond Antifade Mountant (Life Technologies). These sections were then imaged with an Olympus IX83 microscope with analysis via ImageJ software. 13 PD‐L1 Immunohistochemistry (IHC) scoring was obtained and characterized using the combined positive score (CPS).
All 61 patients were identified for the purpose of prediction analysis to identify the radiomics signatures that can predict OS in months, PFS in months, and Response. And 35 out of the 61 patients were identified to carry out prediction analysis to identify the radiomics signatures that can predict Hypoxia, CD8/TREGs, and Average CD8/panck Area. We had previously reported on these 35 patients in abstract form at the ASCO Annual meeting in 2021 (DOI: 10.1200/JCO.2021.39.15_suppl.6045 Journal of Clinical Oncology 39, no. 15_suppl (May 20, 2021) 6045); however, this analysis just evaluated disease control rate and hypoxia.
2.2. Lesion segmentation and feature extraction
Feature extraction was performed on the pre‐immunotherapy baseline CT scans. The lesions were segmented using 3D slicer v4.10.2 to create a volume of interest (VOI) for radiomics texture analysis. For each patient, we segmented radiomics texture features from three different lesions: Primary Recurrent Lesion (PL_RL), Distant Recurrent Lesion (D_RL), and Lymph Node Lesion (LN) (Figure 1). For normalization, sternocleidomastoid muscle was also segmented.
FIGURE 1.
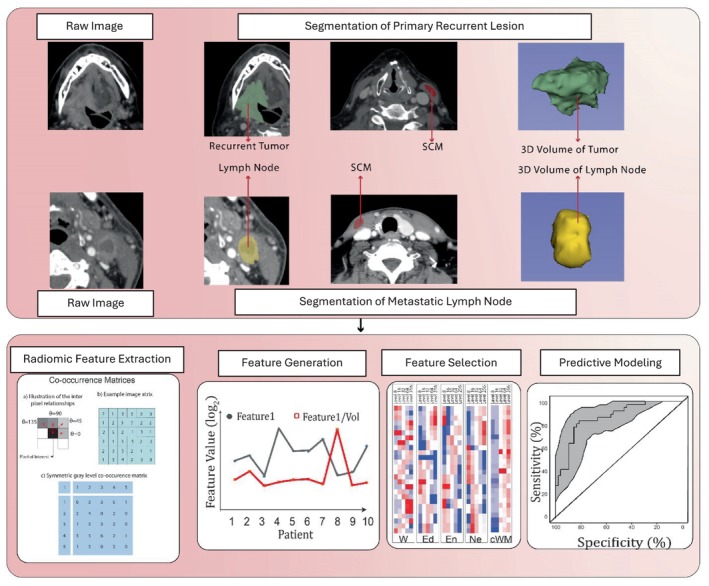
Workflow for radiomics analysis.
In the domain of radiomics feature extraction, intensity‐level histograms and gray‐level co‐occurrence matrices (GLCMs) were used. The extraction process encompasses two primary categories: first‐order features and second‐order features.
First‐order features are derived directly from the pixel intensity values of an image without considering the spatial relationships between the pixels. In our study for first‐order features, a comprehensive set of 10 metrics, informed by the theoretical framework of Papoulis et al., is utilized. 15 These metrics encompass the minimum, maximum, mean, standard deviation (SD), skewness, kurtosis, and four percentile values (1%, 5%, 95%, 99%).
GLCM (Gray‐Level Co‐occurrence Matrix) features represent a statistical approach widely employed in texture analysis to evaluate the spatial relationship of pixel pairs within an image. This method quantifies the occurrence frequency of pixel pairs with designated values and orientations, culminating in the creation of a matrix. This matrix is then used to extract features that describe the image's texture, offering insights into its structural variations and spatial patterns.
In this study, we computed a total of 195 features using established methodologies by Haralick et al., Clausi, and Soh et al. 16 , 17 , 18 These texture features include angular second moment, variance, contrast, correlation, inverse difference moment, entropy, sum average, sum variance, sum entropy, difference variance, difference entropy, information measure of correlation (I and II). Each feature was evaluated at four distinct orientations (0°, 45°, 90°, and 135°), representing the angle between the voxel of interest and its in‐plane neighbor.
To ensure rotation invariance, we computed average, median, and range values, resulting in 39 rotation‐invariant features. Additionally, we quantized the original images into five gray‐level categories (8, 16, 32, 64, and 256 gray levels) to enhance signal‐to‐noise ratio. At each quantization level, the 39 rotation‐invariant features were extracted, enabling robust analysis across varying levels of detail. This feature extraction pipeline was programmed using Python Programming Language (version: 3.10.8).
A total of 205 texture features were extracted from each lesion's VOI, including 10 histogram‐based first‐order features and 195 second‐order features using gray‐level co‐occurrence matrices. After feature normalization, we divided the 195 second‐level features by the volumes of each VOI to generate another 195 volume‐independent features per VOI. A total of 400 features (10 histogram‐based and 390 second‐order texture features) were calculated from each extracted volume of interest (VOI).
2.3. Feature selection
Given the large multi‐dimensional dataset, there was a need to perform feature selection in order to reduce the dimension of the dataset and only retain those features that were most meaningful and important for the prediction of the target outcome variable. Hence, for this purpose, we used the feature selection method, Least Absolute Shrinkage and Selection Operator (LASSO) technique. 13 This regularization method simultaneously performs regression and regularization analysis and identifies the most significant and relevant features that are needed for the prediction of outcome variables.
2.4. Bioinformatic analysis—Radiomics model building
Machine learning supervised analysis using binary logistic regression through eXtreme Gradient Boosting (XGBoost) algorithm was used for the purpose of radiomics model building. 14 XGBoost is a machine learning algorithm that employs a tree‐boosting technique to perform parallel computation, regularization, cross‐validation, and tree pruning simultaneously. For radiomic model building to predict OS, PFS, and treatment response, we have divided our dataset of 61 patients into training and testing with 70:30 split, where 70% of the dataset was used as the training set (n = 44) and the remaining 30% of the dataset was used as a separate independent hold‐out testing set (n = 17) for evaluating the performance of the final trained model. While training the model, to assess its performance, the training set was further divided into 10 folds using a 10‐fold cross‐validation technique. Therefore, care was taken to ensure that the hold‐out testing set (n = 17) was kept independent and the process of feature selection using LASSO regularization and fitting of the training model was performed exclusively on the training set alone. By doing so, this allowed us to evaluate the generalizability of our training model beyond the training set. Given the small sample size (n = 35) patients, for radiomic model building to predict Hypoxia, CD8/TREGs, and Average CD8/panck Area, we used Leave‐One‐Out Cross‐Validation or LOOCV, a configuration of k‐fold cross‐validation in which each patient/sample is hold‐out as the validation set and the reaming n‐1 patients/samples are used as the training set. To address and avoid overfitting of our radiomic models, especially given the characteristics of our dataset, all the training models were built by implementing hyper‐parameter tuning using the XGBoost algorithm's booster parameters; “eta,” a parameter used to control the learning rate (range: 0.001–0.3), and “max_depth,” a parameter used to control over‐fitting (range: 1–6).The final selected radiomics model was the one with the least number of LASSO features that yielded high prediction accuracy and required less computation time. Each of the radiomics model's accuracy was evaluated using the area under the receiver operating characteristic curve (AUC), sensitivity, and specificity. R software (version 3.4.0, R Foundation for Statistical Computing, Vienna, Austria) was used for all the analysis: XGBoost package (version 0.6.4.1) and the Machine Learning package mlr (version 2.11) were used for feature selection and model building. Finally, ROC analysis was performed using pROC package (version 1.9.1).
2.5. Statistical consideration
Survival analysis for OS and PFS was performed through the Kaplan–Meier method. The mean, median and 95% confidence interval (CI) for OS and PFS in months were calculated for 61 patients in the study.
3. RESULTS
We identified 61 R/M HNSCC patients treated with anti‐PD‐1 mAb monotherapy between 2015 and 2020. Baseline characteristics are shown in Table 1. The median age of our cohort was 61 years and was predominantly male (77%) and Caucasian (92%). The oral cavity was the most common primary site (36%), and of the 20 oropharyngeal cancer patients, 70% were HPV positive. The majority of patients received anti‐PD‐1 for platinum failure (61%), with the remainder being treated in the frontline setting. Platinum failure was defined as progression within 6 months of platinum based chemoradiation for locally advanced disease or after progression on platinum based chemotherapy given for R/M HNSCC. The median PFS and OS of the entire cohort were 3.3 [95% CI (1.4–5.2)] and 9.7 months [95% CI (8.3–11.1)], respectively. The overall response rate was 18% (3CR, 8 PR), with SD achieved in an additional 18% of patients for a disease control rate (CR/PR plus SD) of 36%.
TABLE 1.
Baseline characteristics of patients.
| Category | N (%) |
|---|---|
| Age | Median 61 (range 40–80) |
| Sex |
Female: 14 (23) Male: 47 (77) |
| Race |
White: 56 (92) Black: 5 (8) |
| Smoking |
Yes: 38 (62) No: 23 (38) |
| Primary site |
Oral cavity: 22 (36) Oropharynx: 20 (33) HPV a (+): 14/20 (70) Larynx/hypopharynx: 12(20) Other: 7 (11) |
| Type of recurrence |
Locoregional only: 26 (43) Distant only: 12 (20) Locoregional + distant: 23 (37) |
| Indication for anti‐PD‐1 |
Platinum failure: 37 (61%) Frontline: 24 (39%) |
| PD‐L1 by CPS b |
Positive: 17 (29) Negative: 17 (29) Unknown: 27 (44) |
HPV by p16 or HPV ISH for oropharynx primary.
PD‐L1 expression by combined positive score. Positive defined as ≥1.
We evaluated the texture features from the pre‐treatment CT scan including all applicable lesions (primary, distant, and/or lymph nodes), either alone or in combination. For OS, LASSO feature selection identified 37 features and our model based on the top 8 features (Table 2) via XGBoost machine‐learning method predicted OS with high accuracy, sensitivity, and specificity (77%, 86%, 70%) (Figure 2A). For PFS, 10 out of 22 LASSO features (Table 3) predicted PFS with 82%, 75%, and 89% accuracy, sensitivity, and specificity, respectively (Figure 2B). Our predictive model for disease control showed accuracy, sensitivity, and specificity of 76%, 73%, and 83%, respectively, using the top 8 out 55 LASSO features (Table 4, Figure 2C). Comparing the significant LASSO features from these three models for OS, PFS, and DCR did not show any overlapping features. For DCR and OS, the aforementioned best models used primary recurrent lesions, distant lesions and/or lymph nodes, while for PFS, the best model was with just the primary lesion and/or lymph node.
TABLE 2.
Top 8 features of 37 extracted from 61 patients for prediction of OS.
| Order no. | Feature | Gray‐level | Feature name |
|---|---|---|---|
| 1 | PL_RL_FV59 | 16 | Range of sum variance |
| 2 | PL_RL_FOF9 | 16 | First order: Skewness |
| 3 | PL_RL_FV50 | 16 | Average of difference entropy |
| 4 | LN_F142 | 64 | Range of information measure of correlation |
| 5 | D_RL_F125 | 64 | Average of sum entropy |
| 6 | D_RL_F25 | 8 | Range of information measure of correlation |
| 7 | PL_RL_FV1 | 8 | Average of angular second moment |
| 8 | PL_RL_FV83 | 32 | Average of inverse difference moment |
FIGURE 2.
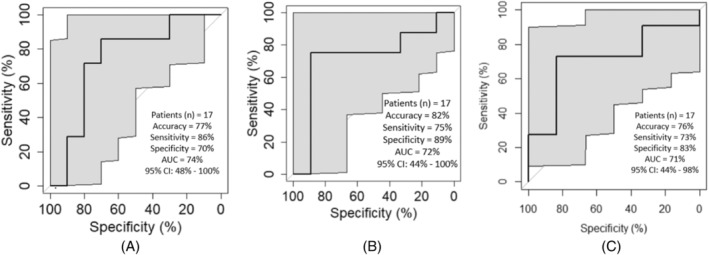
(A) ROC curve representing the performance of the predictive model when using top 8 of 24 LASSO features to predict overall survival. (B) ROC curve representing the performance of the predictive model when using top 10 of 22 LASSO features to predict progression‐free survival. (C) ROC curve representing the performance of the predictive model when using top 8 of 55 LASSO features to predict disease control (CR/PR/SD vs. PD).
TABLE 3.
Top 10 features of 22 extracted from 61 patients for prediction of PFS.
| Order no. | Feature | Gray‐level | Feature name |
|---|---|---|---|
| 1 | PL_RL_FV26 | 8 | Range of information measure of correlation |
| 2 | PL_RL_F11 | 8 | Average of difference entropy |
| 3 | PL_RL_F117 | 32 | Variance of information measure of correlation 2 |
| 4 | LN_F4 | 8 | Average of sum of squares |
| 5 | LN_F127 | 64 | Average of difference variance |
| 6 | LN_FV176 | 256 | Range of sum variance |
| 7 | LN_F3 | 8 | Average of correlation |
| 8 | PL_RL_F30 | 8 | Variance of sum of squares |
| 9 | PL_RL_FV39 | 8 | Variance of information measure of correlation 2 |
| 10 | PL_RL_FV173 | 256 | Range of sum of squares |
TABLE 4.
Top 8 features of 55 extracted from 61 patients for prediction of response.
| Order no. | Feature | Gray‐level | Feature name |
|---|---|---|---|
| 1 | PL_RL_FOF4 | First order: Standard deviation | |
| 2 | LN_FOF4 | First order: Standard deviation | |
| 3 | LN_FV52 | 16 | Average of information measure of correlation 2 |
| 4 | LN_F21 | 8 | Range of sum entropy |
| 5 | LN_FOF9 | First order: Skewness | |
| 6 | PL_RL_FV88 | 32 | Average of difference variance |
| 7 | PL_RL_FV78 | 16 | Variance of information measure of correlation 2 |
| 8 | D_RL_FOF7 | First order: Percentile 95 |
Previous studies have shown the predictive value of Neutrophil/lymphocyte (N/L) ratio and HPV status for anti‐PD‐1 mAb treatment. 19 , 20 Therefore we compared these variables to radiomics as well in combination with radiomics. For DCR, PFS, and OS, the AUC with radiomics was higher than that of N/L ration and HPV status as well as the combination of either N/L ratio or HPV status plus radiomics (Figure 3 and Figures S1 and S2). For example, the AUC for DCR was 71.21% with radiomics compared to 50.03% and 46.97% with N/L ratio and HPV status respectively (Figure 3).
FIGURE 3.
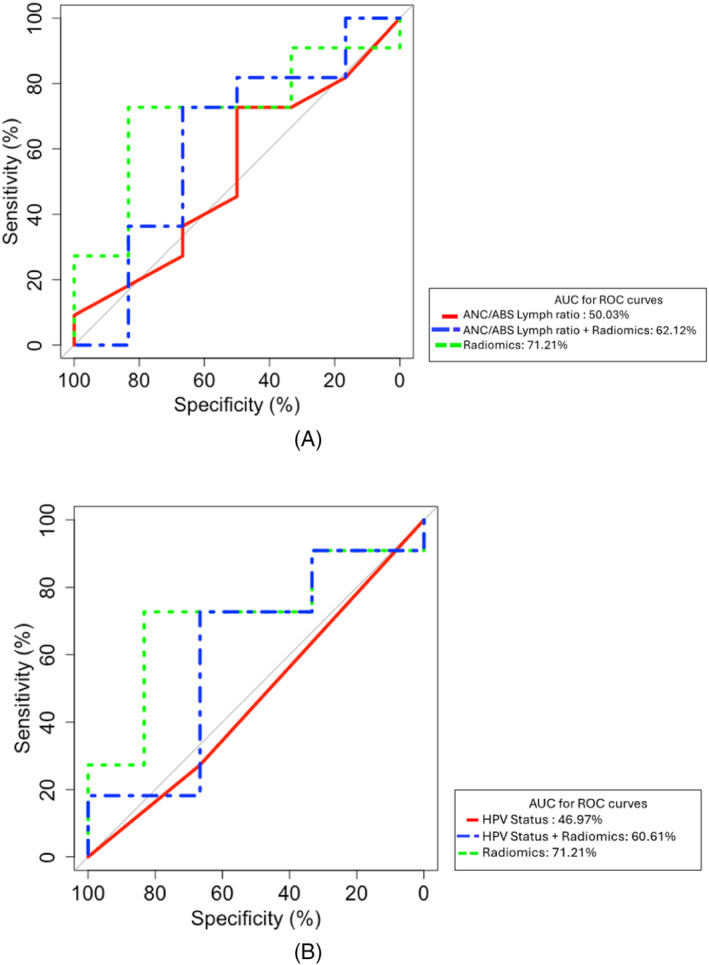
(A) ROC curve representing the performance of the radiomics predictive model compared to Neutrophil/Lymphocyte ratio for disease control (CR/PR + SD vs. PD). (B) ROC curve representing the performance of the radiomics predictive model compared to HPV status (positive vs. negative) for disease control (CR/PR + SD vs. PD).
We had previously published that both increased hypoxia and low CD8 tumor‐infiltrating lymphocytes were predictive of worse efficacy with anti‐PD‐1 mAb treatment in R/M HNSCC, with each independently associated with efficacy. 13 We applied our radiomics model to the 35 patients in our current cohort that also had intratumoral hypoxia and CD8 analysis as per our prior published analysis. Using 4 out of 29 LASSO features, our model predicted high hypoxia in the tumor microenvironment with accuracy, sensitivity, and specificity of 80%, 88%, and 72%, respectively (Figure 4A, Table S1). Our model with the top 7 of 25 LASSO features was also successful in predicting high CD8 T cells in the tumor microenvironment with 91%, 83%, and 100% accuracy, sensitivity, and specificity, respectively (Figure 4B, Table S2). Using 5 of 26 LASSO features, the accuracy, sensitivity, and specificity were 85%, 83%, and 88%, respectively, for the prediction of increased CD8/Treg ratio (Figure 4C, Table S3). There was no overlap in top features for hypoxia, CD8, or CD8/Treg prediction.
FIGURE 4.
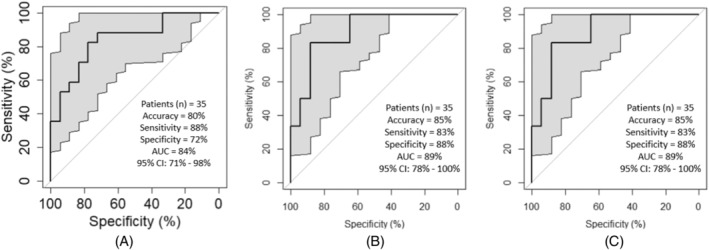
(A) ROC curve representing the performance of the predictive model when using top 4 of 29 LASSO features to predict hypoxia (high vs. low). (B) ROC curve representing the performance of the predictive model when using top 7 of 25 LASSO features to predict average CD8/PanCK (high vs. low). (C) ROC curve representing the performance of the predictive model when using top 5 of 26 LASSO features to predict CD8/Treg (high vs. low).
4. DISCUSSION
There is a great need for better predictive biomarkers for the efficacy of anti‐PD‐1 mAb therapy, especially non‐invasive biomarkers. In our analysis, we found that radiomics applied to pretreatment CT scans showed high accuracy, sensitivity, and specificity for predicting the efficacy of anti‐PD‐1 mAb therapy as well as intratumoral hypoxia, CD8 T cells, and CD8/Treg ratio. To our knowledge this is the first evaluation of the predictive value of radiomics in anti‐Pd‐1 mAb treated R/M HNSCC patients.
Better predictive biomarkers have been a constant need in solid tumors, including R/M HNSCC, since the first studies with this class of therapeutics. Reliable prediction of what patient is likely to benefit from anti‐PD‐1 monotherapy is even more important when there is an alternative choice of therapy for the same indication. For PD‐L1 expressors (CPS ≥1) with R/M HNSCC, there is the option of adding chemotherapy to pembrolizumab or just using pembrolizumab alone in the frontline setting. Exploratory subgroup analysis of KEYNOTE 048 suggests that Pembrolizumab monotherapy may be of less benefit in PD‐L1 CPS 1–19 patients, and while not compared directly, chemotherapy plus pembrolizumab has a numerically higher response rate compared to pembrolizumab monotherapy (36% vs. 19%), albeit at the cost of higher toxicity. 3 , 21 Therefore, a biomarker that can predict with high accuracy those that would benefit from anti‐PD‐1 mAb monotherapy would significantly benefit clinical decision making. Importantly, radiomics analysis of pre‐treatment CT scans is non‐invasive and practical, given all patients undergo pre‐treatment imaging.
Comparison of the predictive value of our radiomics model to HPV status and N/L ratio showed a higher AUC for the radiomics model. An important question is how radiomics compares to other established tumor biomarkers such as PD‐L1, TMB, and immune gene expression. There are limitations to cross comparison of trials, and most clinical trials report numeric efficacy with high or low biomarkers. However, an analysis of 258 R/M HNSCC patients treated with pembrolizumab in Keynote 055 and 012, where ROC analysis was used, showed an AUC for response with PD‐L1, TMB, and immune gene expression of 64%, 63%, and 71%, respectively, compared to 71.21% in our study. 7 A noted limitation of our study is that do to PD‐L1 not being standard in platinum failure patients, and lack of tissue availability, we were not able to compare the predictive value of radiomics directly to PD‐L1, TMB, or GEP in our population. Further analysis is needed to compare radiomics to these and other predictive biomarkers including whether there is additive and/or independent predictive value.
Additionally important is the correlation between radiomics and the tumor immune microenvironment. Previously we showed that hypoxia is independently associated with worse efficacy of anti‐PD‐1 mAb therapy in R/M HNSCC patients. 13 Here, we show that radiomics can predict hypoxia with high accuracy. There is much ongoing work in the field, including in R/M HNSCC, on modulating metabolism and pathways involved in hypoxia and therefore non‐invasive prediction could prove important in selecting patients most likely to benefit from these therapeutics.
Previous radiomics studies in patients with various diseases provide support for the potential clinical translation of our radiomics‐based model to predict immunotherapy response. Sun et al. used machine learning based radiomics score for predicting immunotherapy response. They discovered that they could predict CD8 gene expression based on their initial radiomics score. And patients with higher radiomics score had higher CD8 gene expression and better response to immunotherapy in comparison to patients with progressive or stable disease. Also, when compared to patients with progressive disease, patients with controlled disease (stable disease, partial response, complete response) did not have a significantly higher baseline radiomics score. 22 In another radiomics study, Mu et al. described a multi‐parametric radiomics model based on positron emission tomography (PET)/CT images to predict durable clinical benefit of immunotherapy response in patients with advanced NSCLC (AUC ranging from 0.81 to 0.86). They showed that compared with that of the multi‐parametric radiomics signature, the AUCs of the radiomics model generated by CT‐only‐based features to predict response were lower (AUCs ranging from 0.64 to 0.69). 23 These AUCs are also lower in comparison to our study most likely because the CT images used from PET/CT studies have lower resolution than the diagnostic CT studies as used in the present study and were non‐contrast enhanced CTs. Despite downsides, these results are consistent with our study in highlighting the possibility of radiomics models in predicting response to immunotherapy.
Our paper has a number of limitations. Primarily, it is the smaller sample size and there will be a need to validate these results in a larger cohort. Another limitation is that it is retrospective, albeit importantly, our in vivo imaging core facility performed and verified response assessments making our results more accurate than other retrospective studies that just dependent on chart review. Currently, we are recruiting prospectively into this cohort of anti‐PD‐1 mAb treated R/M HNSCC patients.
In summary, our study shows that radiomics analysis of pretreatment CT scans in R/M HNSCC anti‐PD‐1 treated patients is predictive of efficacy as well as intratumoral hypoxia and CD8 T cells with high accuracy. Further study is needed to validate radiomics as a predictor, but this technology has the potential to make a significant impact as a non‐invasive predictive biomarker in R/M HNSCC.
FUNDING INFORMATION
This research was partially funded by the University of Pittsburgh Hillman Cancer Center NCI Cancer Center Support Grant (CCSG) (RRC), The University of Pittsburgh Hillman Cancer Center startup funding (RRC), and the NIH/NCI SPORE in Head and Neck Cancer (P50CA097190).
Supporting information
Data S1. Supporting information.
Zandberg DP, Zenkin S, Ak M, et al. Evaluation of radiomics as a predictor of efficacy and the tumor immune microenvironment in anti‐PD‐1 mAb treated recurrent/metastatic squamous cell carcinoma of the head and neck patients. Head & Neck. 2025;47(1):129‐138. doi: 10.1002/hed.27878
Section Editor: Steven Chinn
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Hsieh RW, Borson S, Tsagianni A, Zandberg DP. Immunotherapy in recurrent/metastatic squamous cell carcinoma of the head and neck. Front Oncol. 2021;11:705614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ferris RL, Blumenschein G Jr, Fayette J, et al. Nivolumab for recurrent squamous‐cell carcinoma of the head and neck. N Engl J Med. 2016;375(19):1856‐1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burtness B, Harrington KJ, Greil R, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE‐048): a randomised, open‐label, phase 3 study. Lancet. 2019;394(10212):1915‐1928. [DOI] [PubMed] [Google Scholar]
- 4. Cohen EEW, Soulieres D, Le Tourneau C, et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head‐and‐neck squamous cell carcinoma (KEYNOTE‐040): a randomised, open‐label, phase 3 study. Lancet. 2019;393(10167):156‐167. [DOI] [PubMed] [Google Scholar]
- 5. Chow LQM, Haddad R, Gupta S, et al. Antitumor activity of pembrolizumab in biomarker‐unselected patients with recurrent and/or metastatic head and neck squamous cell carcinoma: results from the phase Ib KEYNOTE‐012 expansion cohort. J Clin Oncol. 2016;34(32):3838‐3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seiwert TY, Burtness B, Mehra R, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE‐012): an open‐label, multicentre, phase 1b trial. Lancet Oncol. 2016;17(7):956‐965. [DOI] [PubMed] [Google Scholar]
- 7. Seiwert TY, Haddad R, Bauml J, et al. Biomarkers predictive of response to pembrolizumab in head and neck cancer (HNSCC). Abstract LB‐339 Presented at: American Association for Cancer Research Annual Meeting; April 14–18, 2018; Chicago. 2018.
- 8. Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Braman N, Prasanna P, Whitney J, et al. Association of peritumoral radiomics with tumor biology and pathologic response to preoperative targeted therapy for HER2 (ERBB2)‐positive breast cancer. JAMA Netw Open. 2019;2(4):e192561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zinn PO, Singh SK, Kotrotsou A, et al. A coclinical radiogenomic validation study: conserved magnetic resonance radiomic appearance of periostin‐expressing glioblastoma in patients and xenograft models. Clin Cancer Res. 2018;24(24):6288‐6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Trebeschi S, Drago SG, Birkbak NJ, et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann Oncol. 2019;30(6):998‐1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khorrami M, Prasanna P, Gupta A, et al. Changes in CT radiomic features associated with lymphocyte distribution predict overall survival and response to immunotherapy in non‐small cell lung cancer. Cancer Immunol Res. 2020;8(1):108‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zandberg DP, Menk AV, Velez M, et al. Tumor hypoxia is associated with resistance to PD‐1 blockade in squamous cell carcinoma of the head and neck. J Immunother Cancer. 2021;9(5):e002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zandberg DP, Velez M, Menk AV, et al. The impact of tumor hypoxia on the clinical efficacy of anti‐PD‐1 mAb treatment in recurrent/metastatic HNSCC patients (R/M). J Clin Oncol. 2020;38(Suppl 15):6546. [Google Scholar]
- 15. Papoulis A, Pillai SU. Probability, Random Variables, and Stochastic Processes; 1965. McGraw‐Hill Higher Education. [Google Scholar]
- 16. Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern. 1973;SMC‐3(6):610‐621. doi: 10.1109/TSMC.1973.4309314 [DOI] [Google Scholar]
- 17. Clausi DA. An analysis of co‐occurrence texture statistics as a function of grey level quantization. CJoRS. 2014;28(1):45‐62. doi: 10.5589/m02-004 [DOI] [Google Scholar]
- 18. Soh L‐K, Tsatsoulis C. Texture analysis of SAR sea ice imagery using gray level co‐occurrence matrices. IEEE Trans Geosci Rem Sens. 1999;37(2):780‐795. doi: 10.1109/36.752194 [DOI] [Google Scholar]
- 19. Valero C, Lee M, Hoen D, et al. Pretreatment neutrophil‐to‐lymphocyte ratio and mutational burden as biomarkers of tumor response to immune checkpoint inhibitors. Nat Commun. 2021;12(1):729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xu Y, Zhu G, Maroun CA, et al. Programmed death‐1/programmed death‐ligand 1‐axis blockade in recurrent or metastatic head and neck squamous cell carcinoma stratified by human papillomavirus status: a systematic review and meta‐analysis. Front Immunol. 2021;12:645170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burtness B, Rischin D, Greil R, et al. Efficacy of first‐line (1L) pembrolizumab by PD‐L1 combined positive score <1, 1–19, and ≥20 in recurrent and/or metastatic (R/M) head and neck squamous cell carcinoma (HNSCC): KEYNOTE‐048 subgroup analysis [abstract]. In: Proceedings of the Annual Meeting of the American Association for Cancer Research 2020; 2020 Apr 27–28 and Jun 22–24. Philadelphia (PA): AACR. Cancer Res. 2020;80(16 Suppl):LB‐258. [Google Scholar]
- 22. Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour‐infiltrating CD8 cells and response to anti‐PD‐1 or anti‐PD‐L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19(9):1180‐1191. [DOI] [PubMed] [Google Scholar]
- 23. Mu W, Tunali I, Gray JE, Qi J, Schabath MB, Gillies RJ. Radiomics of (18)F‐FDG PET/CT images predicts clinical benefit of advanced NSCLC patients to checkpoint blockade immunotherapy. Eur J Nucl Med Mol Imaging. 2020;47(5):1168‐1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting information.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


