Abstract
Introduction:
Pharmacovigilance programs protect patient health and safety by identifying adverse event signals through postmarketing surveillance of claims data and spontaneous reports. Electronic health records (EHRs) provide new opportunities to address limitations of traditional approaches and promote discovery-oriented pharmacovigilance.
Methods:
To evaluate the current state of EHR-based medication safety signal identification, we conducted a scoping literature review of studies aimed at identifying safety signals from routinely collected patient-level EHR data. We extracted information on study design; EHR data elements utilized; analytic methods employed; drugs and outcomes evaluated; and key statistical and data analysis choices.
Results:
We identified 81 eligible studies. Disproportionality methods were the predominant analytic approach, followed by data mining and regression. Variability in study design makes direct comparisons difficult. Studies varied widely in terms of data, confounding adjustment, and statistical considerations.
Conclusion:
Despite broad interest in utilizing EHRs for safety signal identification, current efforts fail to leverage the full breadth and depth of available data or to rigorously control for confounding. The development of best practices and application of common data models would promote the expansion of EHR-based pharmacovigilance.
1. Introduction
Pharmacovigilance programs protect the health and safety of patients by identifying signals of adverse events through postmarketing surveillance after medications and vaccines become available in routine care [1]. A signal, in this instance, refers to “information that arises from one or multiple sources, which suggests a new potentially causal association, or a new aspect of a known association, between an intervention and an event or set of related events” [2]. While many adverse events are recognized during clinical trials of new medications, these trials are, by definition, tightly controlled and do not reflect the variability and complexity of the general population in terms of demographics, concomitant medication, and comorbid conditions [3]. Further, premarketing trials are limited in terms of sample size and duration and may not capture rare events. Once approved for widespread use, previously unidentified adverse events may be recognized through postmarketing surveillance and require adjustment of clinical recommendations or even withdrawal of a medication from the market.
Postmarketing surveillance has traditionally relied on spontaneous reporting of suspected adverse drug events by concerned healthcare professionals, consumers, and pharmaceutical manufacturers [3]. These reports are collected and maintained in national and international databases, such as the FDA’s Adverse Event Reporting System (FAERS) [4], the European Medicines Agency’s (EMA) EudraVigilance [5], and the WHO’s VigiBase [6]. However, data in spontaneous reporting systems are limited to voluntary reports of suspected adverse events directly related to a medication exposure, are subject to reporting bias, and provide limited information on patient characteristics [3]. With the expansion of longitudinal sources of healthcare data, including health insurance claims and electronic health records (EHRs), new opportunities are emerging to address these limitations and promote discovery-oriented pharmacovigilance [7].
The US FDA’s active medical product safety surveillance system, Sentinel, serves as an important complement to FAERS passive surveillance approach. Sentinel uses health insurance claims as the primary vehicle for signal identification [8]. Claims data are well-suited for longitudinally following patients after medication exposures owing to their capture of pharmacy dispensing records, outpatient encounters, and hospitalizations during well-defined periods of health plan enrollment. However, such data lack clinical granularity and often under-capture subtle events that do not trigger formal coding. In contrast, EHRs contain comprehensive structured and semi-structured data as well as rich, unstructured clinical narratives that could be leveraged to improve safety signal identification. The integration of EHR data as a resource for safety surveillance is a priority of Sentinel, but current signal identification methods used with spontaneous reporting systems and claims data may not fully leverage the breadth and depth of EHR data [7]. New signal identification methods may be required to incorporate the variable information in EHRs, control the impact of confounding, and address the variability in terms of data collection processes, data gaps, and data quality across health systems.
In addition to Sentinel, international pharmacovigilance efforts would also benefit from advances in this area. Prior work by the EU-ADR project highlighted many of the difficulties around using EHR data for signal identification. The EMA’s Data Analysis and Real World Interrogation Network (DARWIN) [9], which collects real world health data for regulatory decision making, will need to address these issues, as well. Additionally, large scale health database efforts, such as the All of Us Research Program [10] in the US and the European Health Data Evidence Network (EHDEN) [11] might also benefit from methods to improve signal identification using EHR data.
To support the expansion of EHR-based pharmacovigilance, we conducted a scoping literature review of current practices in the use of routinely collected EHR data for medication safety signal identification. We included studies that performed completely hypothesis-free discovery, as well as those that attempted to identify evidence of a signal between specific drug-event pairs or among sets of related exposures and events. While there are a number of excellent reviews of the pharmacovigilance literature, few have as their primary focus the application of signal identification methods to EHR data. Most discuss signal identification more broadly, focus on methods applied to spontaneous reporting systems, or concentrate on recognition of known adverse drug effects (ADEs) for reporting or patient care tools [12–19]. Those reviews that do emphasize EHR-based signal identification are often smaller narrative reviews or focus on important related concerns, such as extracting documented ADEs from clinical texts or the use of pharmacogenomic data, which is beyond the scope of this review [3,20–28]. Herein, we focus on analytic methods using routinely-collected EHR data for medication safety signal identification and describe how both structured and unstructured EHR data are being utilized.
2. Methods
We conducted a scoping review of the literature documenting adverse event signal identification in longitudinal EHR data, characterizing the analytics methods employed, handling of statistical challenges, and integration of diverse clinical data. For the purposes of this review, we define signal identification as a statistical or algorithmic approach to identify an excess burden of adverse events following drug exposure, whether or not the exposure was documented as associated with the outcome by the patient or provider at the time of the event. A protocol was not preregistered for this review.
2.1. Data sources and search strategy
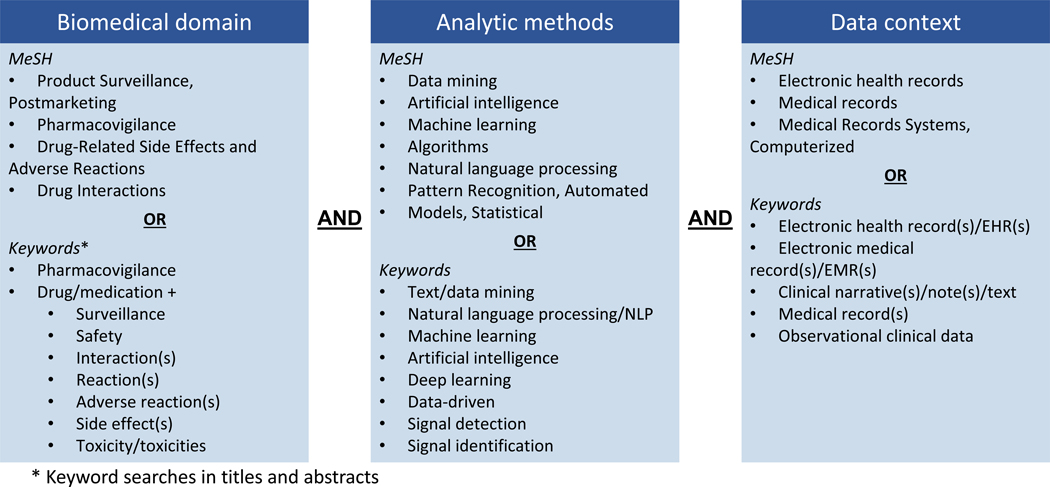
We queried MEDLINE and Embase, the two largest bibliographic databases of published biomedical literature, for all publications indexed as of April 14, 2023.We identified potentially relevant articles in both databases using both MeSH (Medical Subject Headings) terms and keywords. Our search strategy was designed to capture all citations encompassing three concepts of interest: biomedical domain, analytic methods, and data context (see Figure 1). Search queries were crafted for each of these concepts and results were combined to identify publications of interest. The biomedical domain search terms required studies to focus on pharmacovigilance biomedical research (e.g., drug-related side effects and post-marketing surveillance). The analytic methods search terms required studies to report on methods used for signal identification (e.g., natural language processing and machine learning). The data context search terms required studies to analyze EHR-based data with or without integration with spontaneous reporting systems. We subsequently excluded non-English language publications. The full search query is provided in Table 1. We also reviewed the reference lists of eligible articles for relevant studies and searched for full text publications associated with eligible conference abstracts.
Fig 1.
Search criteria used in OvidSP to identify publications of interest for the literature review. For each primary query, MeSH terms are listed along with keywords from titles and abstracts.
Table 1.
Full search query
| # | Subqueries |
|---|---|
| 1 | “Drug-Related Side Effects and Adverse Reactions”/ or Pharmacovigilance/ or Product Surveillance, Postmarketing/ or Drug Interactions/ |
| 2 | (((drug or medication) adj3 (reaction* or interaction* or safety or “side effect*” or toxicit* or surveillance or “adverse effect*”)) or pharmacovigilance).ti,ab. |
| 3 | 1 or 2 |
| 4 | limit 3 to English language |
| 5 | Data Mining/ or Artificial Intelligence/ or Machine Learning/ or Algorithms/ or Natural Language Processing/ or Pattern Recognition, Automated/ or Models, Statistical/ |
| 6 | (“text mining” or “data mining” or “NLP” or “natural language processing” or “machine learning” or “artificial intelligence” or “deep learning” or “signal detection” or “signal identification” or “data-driven” or “data driven”).ti,ab. |
| 7 | 5 or 6 |
| 8 | limit 7 to English language |
| 9 | Electronic Health Records/ or Medical Records/ or Medical Records Systems, Computerized/ |
| 10 | (“electronic health record*” or “electronic medical record*” or “EHR*” or “EMR*” or “clinical narrative*” or “clinical note*” or “clinical text*” or “observational clinical data” or “medical record*”).ti,ab. |
| 11 | 9 or 10 |
| 12 | limit 11 to English language |
| 13 | 4 and 8 and 12 |
| 14 | remove duplicates from 13 |
2.2. Study selection
Studies were eligible for inclusion in our review if they 1) analyzed patient-level EHR data collected through routine clinical care; and 2) implemented, evaluated, or proposed analytic methods for identifying or discovering adverse event signals. We focused on original research studies and excluded commentaries and prior literature reviews. Abstracts were independently screened for inclusion by at least two members of the study team (Smith, Davis, Coughlin, or Zabotka). Those cases in which reviewers disagreed on study eligibility were adjudicated by the two primary reviewers (Davis, Smith).
2.3. Data extraction
For each eligible study, we extracted key information on the clinical population; study design; data sources; EHR features extracted; analytic methods used; drugs and outcomes evaluated; handling of temporality of exposures and outcomes; adjustment for confounding; and consideration of multiple testing. We noted any direct comparisons of methods, as well as strengths and limitations of analytic methods.
3. Results
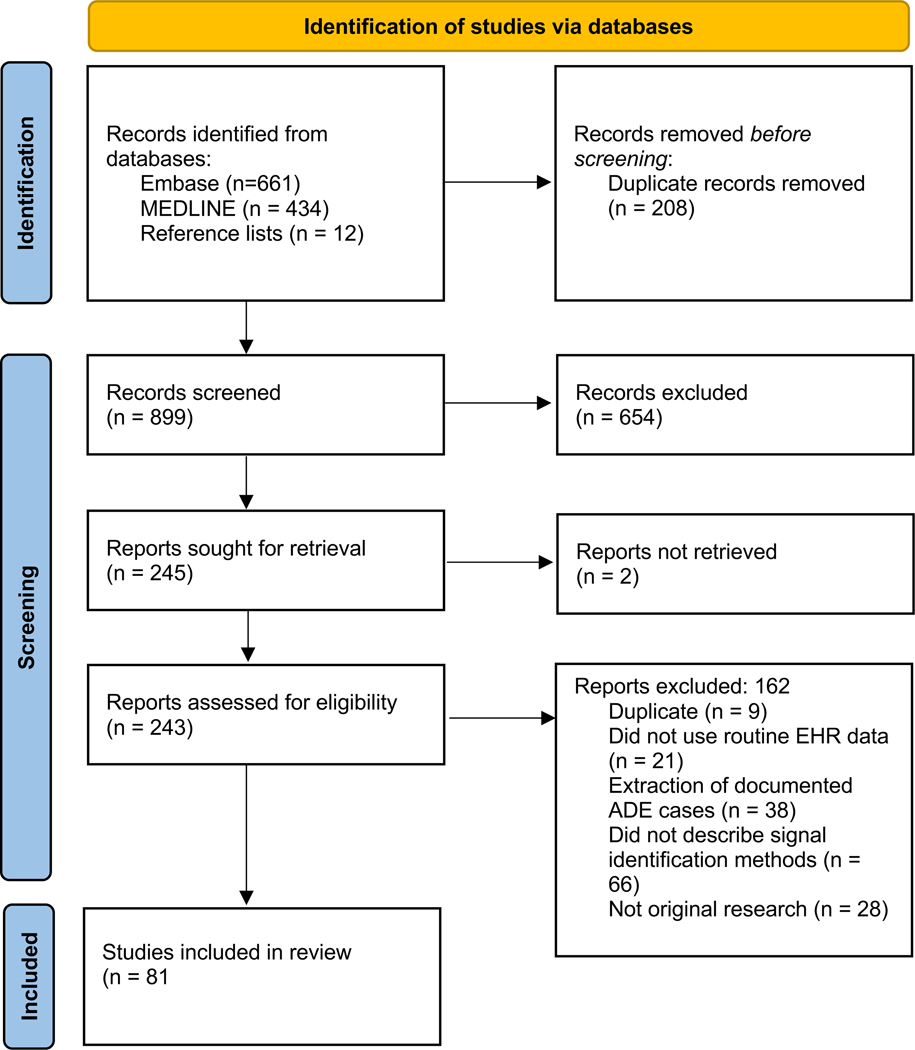
Figure 2 details our search results and disposition of each study using a PRISMA flow chart. Our search returned 1095 publications and we included an additional 12 studies retrieved from reference lists of relevant studies. After deduplication, 899 study abstracts were screened and 245 were selected for full review. During full review, we excluded an additional 164 publications. Most studies excluded at this stage were not original research, did not conduct adverse event signal identification, failed to provide sufficient detail about methods used for signal identification, or sought to extract documented ADEs from clinical notes using natural language processing (NLP). After full text review, 81 publications were eligible for inclusion.
Fig 2.
PRISMA flow chart of article disposition
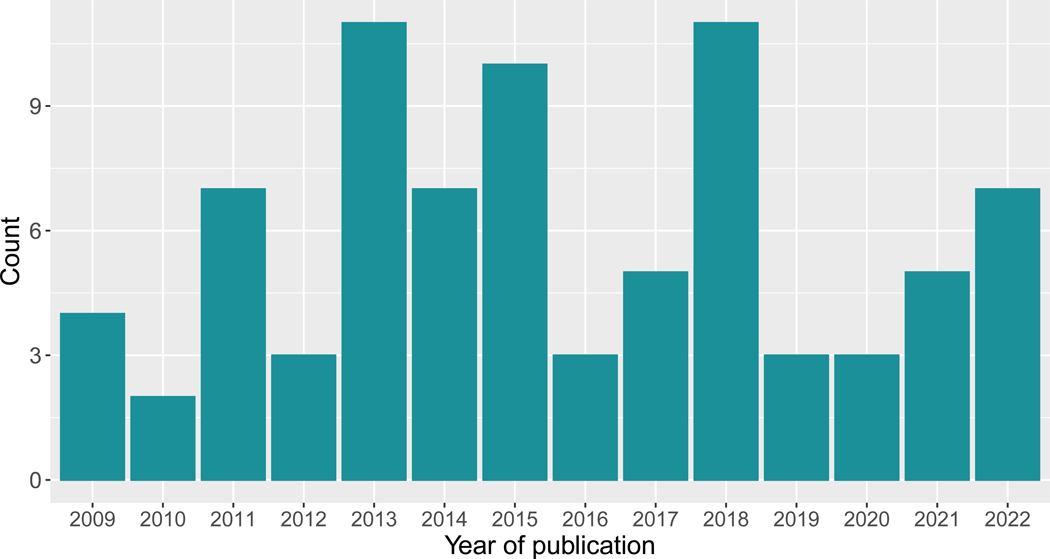
Table 2 provides an overview of the 81 original research studies included in our review and Table 3 summarizes details extracted from these studies along with relevant citations. For 12 studies, only the abstract was available and served as the full text for review, limiting the information we were able to extract. Figure 3 highlights sustained interest in methods for EHR-based signal identification over the past two decades. Studies were conducted on EHR data from Europe (n=26, 32%), Asia (n=14, 17%), and the United States (n=38, 47%). Studies from Asia and the United States primarily considered EHR data from academic medical centers, while those in Europe often included data collected from more diverse clinical settings, such as community health centers contributing EHR data to The Health Improvement Network (THIN) [29] or the General Practice Research Database (GPRD) [30] in the UK. Sample sizes were not consistently reported, with some studies reporting the number of observations used for analysis and others reporting the number of patients in a data resource. When reported, sample sizes varied widely, from just a few hundred patients to millions of patients. Single-drug adverse events were the primary focus, with 14 (17%) studies aimed at identifying drug-drug interactions (DDIs).
Table 2.
Summary of original research studies eligible for review. Sample sizes represent actual sample size used for analysis, if available, and data resource sample size otherwise.
| Study | Setting | Disproportionality analysis | Regression | Machine learning and data mining | Sequential analyses |
|---|---|---|---|---|---|
| Chazard et al. 2009 [32] | France and the Netherlands, variable settings | X | |||
| Edwards R.I. 2009 [33] | International database | X | |||
| Ryan P.B. et al. 2009 [34] | Not reported | X | |||
| Wang X. et al. 2009 [35] | US, single academic medical center | X | |||
| Brownstein J.S. et al. 2010 [36] | US, single academic medical center | X | X | ||
| Harpaz et al. 2010 [37] | US, single academic medical center | X | |||
| Brown J.S. et al. 2011 [38] | US, variable settings | X | |||
| Chazard et al. 2011 [39] | Europe variable settings | X | |||
| Coloma et al. 2011 [40] | Europe, variable settings | X | |||
| Coloma P.M. et al. 2011 [41] | Europe, variable settings | X | X | ||
| Ferrajolo C. et al. 2011 [42] | Europe, variable settings | X | |||
| Ji et al. 2011 [43] | US, single VA medical center | X | X | ||
| Park M.Y. et al. 2011 [44] | Korea, single academic medical center | X | |||
| Trifiro G. et al. 2011 [45] | Europe, variable settings | X | |||
| LePendu P. et al. 2012 [46] | US, variable settings | X | |||
| Star K. et al. 2012 [47] | United Kingdom, variable settings | X | |||
| Yoon et al. 2012 [48] | Korea | X | |||
| Afzal Z. et al. 2013 [49] | Denmark, variable settings | X | |||
| An L. et al. 2013 [50] | US, variable settings | X | |||
| Harpaz R. et al. 2013 [51] | New York, single academic medical center | X | |||
| Kulldorff M. et al. 2013 [52] | US, variable settings | X | |||
| Lependu et al. 2013 [53] | US, single academic medical center | X | |||
| LePendu P. et al. 2013 [54] | US, single academic medical center | X | X | ||
| Lian Duan et al. 2013 [55] | Simulated data | X | X | ||
| Liu et al. 2013 [56] | US, single academic medical center | X | X | ||
| Reps J. et al. 2013 [57] | United Kingdom, variable settings | X | X | ||
| Ryan et al. 2013 [31] | US, variable settings | X | X | X | |
| Sauzet O. et al. 2013 [58] | United Kingdom, variable settings | X | |||
| Eriksson et al. 2014 [59] | The Netherlands, single academic medical center | X | |||
| Ferrajolo et al. 2014 [60] | Europe, variable settings | X | |||
| Iyer et al. 2014 [61] | US, single academic medical center | X | |||
| Ji et al. 2014 [62] | US, single VA medical center | X | |||
| Li Y. et al. 2014 [63] | US, single academic medical center | X | X | ||
| Patel and Kaelber 2014 [64] | US, variable settings | X | |||
| Roitmann E. et al. 2014 [65] | Denmark, single medical center | X | |||
| Cederholm S. et al. 2015 [66] | United Kingdom, variable settings | X | X | ||
| Du L. et al. 2015 [67] | US, variable settings | X | |||
| Girardeau Y. et al. 2015 [68] | France, single academic medical center | X | |||
| Li Y. et al. 2015 [69] | US, variable settings | X | X | ||
| Pacurariu A.C. et al. 2015 [70] | Europe, variable settings | X | |||
| Patadia V.K. et al. 2015 [71] | Europe, variable settings | X | |||
| Reps J. et al. 2015 [72] | United Kingdom, variable settings | X | |||
| Star K. et al. 2015 [73] | United Kingdom, variable settings | X | |||
| Wang G. et al. 2015 [74] | US, single academic medical center | X | X | ||
| Zhang P. et al. 2015 [75] | US, variable settings | X | |||
| Hauben M. et al. 2016 [76] | United Kingdom, variable settings | X | X | ||
| Lorberbaum T. et al. 2016 [77] | US, single academic medical center | X | |||
| Lorberbaum T. et al. 2016 [78] | US, academic medical centers | X | |||
| Boland, M.R. et al. 2017 [79] | US, single academic medical center | X | X | ||
| Fan Y. et al. 2017 [80] | US, single academic medical center | X | |||
| Lee S. et al. 2017 [81] | Korea, single academic medical center | X | |||
| Personeni et al. 2017 [82] | US, single academic medical center | X | |||
| Wang et al. 2017 [83] | US, single academic medical center | X | |||
| Chen W. et al. 2018 [84] | China, single academic medical center | X | |||
| Choi L. et al. 2018 [85] | US, single academic medical center | X | |||
| Jeong E. et al. 2018 [86] | Korea, single academic medical center | X | X | X | |
| Patadia V.K. et al. 2018 [87] | Europe, variable settings | X | X | ||
| Shimai Y. et al. 2018 [88] | Japan, single academic medical center | X | |||
| Tham M.Y. et al. 2018 [89] | Singapore, single academic medical center | X | |||
| Vajravelu et al. 2018 [90] | United Kingdom, variable settings | X | X | ||
| Wang L. et al. 2018 [91] | US, single academic medical center | X | |||
| Wang X. et al. 2018 [92] | US, variable settings | X | |||
| Whalen E. et al. 2018 [93] | United Kingdom, variable settings | X | X | ||
| Zhou et al. 2018 [94] | United Kingdom, variable settings | X | |||
| Dang T.-T. et al. 2019 [95] | US, single academic medical center | X | X | ||
| Davazdahemami B. and Delen D. 2019 [96] | US, variable settings | X | |||
| Duan R. et al. 2019 [97] | US, single academic medical center | X | |||
| Yu et al. 2020 [98] | US, single academic medical center | X | |||
| Yu Y. et al. 2020 [99] | China, single academic medical center | X | |||
| Zhang et al. 2020 [100] | US, single academic medical center | X | |||
| Akimoto H. et al. 2021 [101] | Japan, single academic medical center | X | |||
| Nie X. et al. 2021 [102] | China, single academic medical center | X | X | ||
| Shin H. and Lee S. 2021 [103] | Korea, single academic medical center | X | |||
| Shin H. et al. 2021 [104] | Korea, single academic medical center | X | |||
| Wu et al. 2021 [105] | US, single academic medical center | X | |||
| Challa A.P. et al 2022 [106] | US, single academic medical center | X | |||
| Kaas-Hansen B.S. et al. 2022 [107] | Europe, variable settings | X | |||
| Kundrot S. et al. 2022 [108] | Multinational, variable settings | X | |||
| Mower J. et al. 2022 [109] | US, single academic medical center | X | X | ||
| Nie X. et al. 2022 [110] | China, single academic medical center | X | X | ||
| Sauzet O. and Cornelius V. 2022 [111] | United Kingdom, variable settings | X | |||
| Yu Y. et al. 2022 [112] | China, single academic medical center | X | X | ||
| 48 (59%) | 25 (31%) | 30 (37%) | 3 (4%) |
Abbreviations: electronic health record (EHR)
Table 3.
Characteristics of original research studies reviewed. Studies may be counted in multiple subcategories within each group.
| N | % | Relevant studies | |
|---|---|---|---|
| Total original research studies | 81 | ||
| Study aim | |||
| Signal identification for specified associations | 27 | 33.3% | [34–36,40–42,48,49,53,58,68,69,71,76,78,80,87,88,91,93–95,98,100,103,109,111] |
| Signal identification across many exposures | 44 | 54.3% | [31,32,37,39,44,45,47,51,54–57,59–61,65–67,70,72–75,77,79,81,82,84–86,89,90,92,96,97,99,101,102,104–108,110,112] |
| Signal identification across many outcomes | 34 | 42.0% | [31,32,37–39,43,44,50,52,54,56,57,59,61–66,73,74,81–86,89,93,104–108] |
| Type of event investigated | |||
| Single-drug adverse event | 68 | 84.0% | [31–49,51–60,63–66,69–74,76,79,81–91,93–95,98–104,106–112] |
| Drug-drug interaction | 14 | 17.3% | [50,54,61,62,67,68,75,77,78,80,92,96,105,107] |
| Analysis frame | |||
| Retrospective | 75 | 92.6% | [31–35,37–45,47–86,88–93,95–97,99–112] |
| Prospective | 6 | 7.4% | [36,46,54,87,94,98] |
| Study design | |||
| Cohort | 54 | 66.7% | [31,32,34–43,45,47,50–52,55,57,59,61–65,68–74,76–80,82–85,87,95,97–99,101,104–111] |
| Case-control | 22 | 27.2% | [31,46,48,53,54,56,58,59,61,67,75,81,85,90–92,96,99,102,103,110,112] |
| Self-controlled | 12 | 14.8% | [31,34,44,49,60,66,86,89,91,93,94,100] |
| Outcome type | |||
| Binary | 70 | 86.4% | [31–46,48,49,51–56,59–61,63–65,67,68,70,71,73,74,76,79,81–92,94–101,104,105] |
| Continuous | 4 | 4.9% | [50,77,78,89] |
| Time-to-event | 10 | 12.3% | [47,58,62,66,73,80,81,93,103,111] |
| Methods | |||
| Disproportionality | 48 | 59.3% | [31,34,35,40–45,49–51,53,55–61,63,64,66,67,69–71,76–78,83,86–91,93–96,98,99,102,104,109–112] |
| Regression | 25 | 30.9% | [31,36,37,48,54,63,68,69,74,75,79–81,85,86,90,92,97,101–103,105,106,110,112] |
| Machine learning and data mining | 30 | 37.0% | [31–33,38,39,41,43,46,47,52,55–57,62,65,66,72–74,76,79,82,84,86,93,95,100,107–109] |
| Sequential analyses | 3 | 3.7% | [36,54,87] |
| EHR components accessed | |||
| Demographics | 46 | 56.8% | [31,32,36,38–40,42,43,48–50,52–54,56–63,66,68,69,72,77,78,80,81,84,85,90,94,96,97,99,101–105,107,110–112] |
| Medication orders | 75 | 92.6% | [31–34,36–43,45–52,54–60,62–68,70–90,92–108,110–112] |
| Diagnostic/procedural codes | 60 | 74.1% | [31–34,36,38–43,45–49,52,54–58,60,62,64–67,69–73,75,76,78–85,87,90,92–94,96–104,108,110–112] |
| Laboratory results | 28 | 34.6% | [32,36,37,39,43,44,48,50,51,56,60,62–64,68,70,81,83,84,86,88,89,94,99,101,102,110,112] |
| Vital signs | 3 | 3.7% | [50,64,77] |
| Clinical text | 21 | 25.9% | [35,37,39,46,51,53,54,59,61,63,65,69,70,74,80,85,91,95,106,107,109] |
| Data sources* | |||
| EHR only | 66 | 81.5% | [31,32,34–43,46,48–50,52–60,62–68,70,72,74–76,79–81,84–90,92–97,99,100,102,105–112] |
| EHR and spontaneous reports | 15 | 18.5% | [45,51,61,69,71,73,77,78,82,83,87,91,98,101,103,104] |
| Control for confounding | |||
| None reported | 32 | 39.5% | [35,41,43,45,47,50,51,55,57,62,64,65,67,70,71,73–76,79,82–84,87,88,92,98,104,106–109,111] |
| Demographics | 35 | 43.2% | [31,32,36,38–40,42,48,49,52–54,56,58–61,68,72,77,78,80,81,85,90,96,97,99,101–103,105,110,112] |
| Drug exposures | 14 | 17.3% | [31,36,39,46,53,54,61,68,72,78,95,96,101,105] |
| Comorbidities | 22 | 27.2% | [31,32,36,37,39,46,48,53,54,56,61,63,69,80,81,85,96,99,101,102,110,112] |
| Statistical considerations | |||
| Explicit consideration of temporal constraints | 74 | 91.4% | [31,32,34–36,38–46,48–64,66–83,85–87,89–107,110–112] |
| Discussion of missing data | 6 | 7.4% | [79,84,86,90,99,103] |
| Adjustment for multiple testing | 20 | 24.7% | [38,51,52,54,59,63,65,68,75,77,78,81,86,87,90,92,98,105,106,108] |
Note, no studies only accessed spontaneous reports as our inclusion criteria limited the review to studies using EHR data. Abbreviations: electronic health record (EHR)
Fig 3.
Included studies of EHR-based signal identification by year
3.1. Study designs
Most studies employed a cohort (n=54, 67%) or a case-control design (n=22, 27%). Case-control studies included random matching, simple demographic matching (e.g., matching on age and sex only), matching on medical history, and propensity score matching. Self-controlled designs were less common (n=12, 15%). Ryan and colleagues [31] directly compared study designs and observed self-controlled designs to be more accurate in their identification of known ADEs and negative controls, yet all designs underperformed in estimating signal strength.
3.2. Analytic Methods
Table 4 provides a description of the major analytic approaches, including common specific methods reported and both benefits and limitations of how the methods were applied. The most common methods applied were various disproportionality analysis (n=48, 59%), machine learning/data mining algorithms (n=30, 37%), and regression-based modeling (n=25, 31%). Disproportionality analysis, generally speaking, refers to methods that identify combinations of exposures and adverse effects that occur more frequently than expected by using information on all drugs and effects in the available sample population [14]. Specific methods included the reporting odds ratio (ROR), proportional reporting ratio (PRR), gamma Poisson shrinker (GPS) and associated variants, and empirical Bayes geometric mean (EBGM), among others. The most frequent regression-based approaches were logistic and Cox regression. Studies using machine learning and data mining algorithms used a variety of supervised and unsupervised methods, including association-rule mining, clustering, random forests, the tree-based scan statistic, and neural networks. Most studies using sequential analyses applied the maximized sequential probability ratio test (MaxSPRT). Studies generally took a retrospective approach, seeking to identify adverse events from historical extracts of EHR data. However, 6 studies (7%) implemented a prospective analysis framework which could be used for prospective surveillance for emerging evidence of adverse event signals. These studies used statistical process control, regression, and sequence symmetry analysis.
Table 4.
Summary of methods employed in reviewed studies, highlighting advantages, challenges, and opportunities to advance EHR-based safety signal identification.
| Methodological class and specific examples | Advantages | Challenges and limitations | Opportunities for advancing signal identification in EHR data |
|---|---|---|---|
| Disproportionality analysis • Reporting odds ratios • Proportional reporting ratio • Gamma Poisson shrinker • Empirical Bayes geometric mean • Relative risks and rate ratios |
• Methods familiar in pharmacovigilance programs using spontaneous reports • Simple and fast to implement • Many methods could be conducted via distributed analyses |
• Current implementations often neglect to control for confounding or control for a limited number of confounders • Current implementations typically do not control for multiple testing or use conservative multiple testing adjustment |
• Increased use of propensity score methods could leverage the breadth of EHR data to address high-dimensional confounding • Distributed analyses across health systems could leverage larger datasets while protecting privacy • Simple approaches could provide initial screening and hypothesis generation to motivate more detailed, complex investigations |
| Regression-based modeling • Logistic regression • Poisson regression • Cox regression |
• Methods familiar in biomedical applications, including pharmacovigilance and pharmacoepidemiology • Simple and fast to implement • Many methods could be conducted via distributed analyses • Current implementations typically control for some confounding |
• Requires parameterization of all associations, including interactions and nonlinearity • Current implementations often control for a limited number of patient characteristics • Current implementations typically do not control for multiple testing or use conservative multiple testing adjustment |
• Increased use of regularization could leverage the breadth of EHR data to address high-dimensional confounding • Distributed analyses across health systems could leverage larger datasets while protecting privacy • Recent advances (e.g., DDI-WAS) highlight potential for hypothesis-free discovery applications |
| Machine learning and data mining • Random forests • Neural networks • Clustering • Temporal pattern discovery • Association rules • Tree-based Scan Statistic (TreeScan) |
• Data-driven approach to learning • Supervised and unsupervised methods enable hypothesis-driven and discovery applications • Methods address high-dimensional confounding by enabling nonlinear associations and interactions |
• Few studies have considered the same algorithms, providing limited evidence of comparative performance • Higher complexity and computation costs compared to regression and disproportionality analyses • Limitations of model interpretability and concerns about transportability may reduce acceptance by key stakeholders • Data hungry methods may require larger datasets than simpler, traditional approaches |
• Support use of the breadth of EHR data to address high-dimensional confounding and integration of diverse data types • Opportunities to leverage inherent hierarchical relationships among clinical concepts through approaches such as TreeScan • Opportunities for hypothesis-free discovery applications |
| Sequential analyses • Statistical process control • Sequential probability ratio tests |
• Designed for active, ongoing surveillance • Evidence from existing studies that prospective monitoring may identify adverse event signals in EHR data earlier than in spontaneous reporting data • Multiple testing control is inherent in method designs and parameterizations • Existing methods support confounding adjustment and binary or continuous outcomes |
• Few studies currently explore these methods for EHR-based signal identification or implement these methods prospectively in EHRs • Prospective surveillance requires different data infrastructure than traditional retrospective analyses • New protocols and staffing resources would be required for responding to surveillance-based alerts and following-up on potential safety signals |
• Prior applications in claims-based data provide roadmap for wider implementation in EHRs • Enable ongoing monitoring for signals starting at drug approval • Dashboards monitoring multiple potential drug safety signals could provide insight and early warning • Frequency of evaluation can be matched to event and exposure volumes for each drug |
Abbreviations: electronic health record (EHR)
Some studies aimed to evaluate the performance of different methods through signal identification applied to specified drug-outcome associations (n=27, 33%). Others focused on signal discovery across many drug exposures linked to a specific outcome (n=44, 54%) or across many outcomes after exposure to a specific drug (n=34, 42%). Methods were evaluated by comparing results to previously reported adverse events and known negative controls to document false negatives and false positives. Novel associations were also documented, with several studies including additional steps to filter out false positives and prioritize potential novel adverse events. This filtering and prioritization were accomplished by ranking events by measures of signal strength [37,43,57,57,61,63,69,71,82,82,99,102,106,107,109,112], comparing findings to parallel analyses of spontaneous reporting data [61,69,71,82,87,91], incorporating external knowledge sources [32,43,66,102,105–108,110,112], or assessing protopathic bias using Longitudinal Evaluation of Observational Profiles of Adverse events Related to Drugs (LEOPARD) [42,60,70,71,87]. Three studies [54,63,90] prescreened potential adverse events using disproportionality analyses and conducted more in-depth investigation into the potential signals using regression-based models. These studies found such a step-wise approach improved precision of signal identification and reduced false positives.
Few studies directly compared adverse event signal identification across methods. Wang and colleagues [74] found a random forest model based on data extracted from clinical notes outperformed disproportionality analyses on the same data, as well as disproportionality analyses on spontaneous reporting data. Jeong and colleagues [86] compared disproportionality analyses to machine learning models—including random forests, L1 regression, support vector machines, and neural networks—that used the summary statistics from disproportionality analyses of laboratory values as inputs, finding random forests to have the highest discrimination and all machine learning models outperforming the disproportionality analyses.
3.3. EHR Components
Table 3 includes a summary of those portions of the EHR used by analyses described in each study. Structured medication data were accessed in the vast majority of studies (n=75, 93%). Studies that did not use structured medication data identified drug exposures by using NLP to extract information from clinical notes [35,53,61,91,109]. Medication information typically captured drug names and timing of exposure. Dosing and route information was rarely considered. Most reviewed studies accessed diagnostic/procedural codes (n=60, 74%), with those not accessing diagnostic/procedural codes using laboratory data or vital signs to determine the presence of adverse events. Laboratory data was accessed in 28 (35%) studies for outcome determination or confounding adjustment. Use of demographic data was noted in 46 (57%) studies for population definition, stratification, or confounding adjustment. Few studies used vital sign data or health care utilization metrics.
Unstructured data from clinical notes were accessed in 21 studies (26%). Studies generally relied on a combination of NLP extracted event or medication information alongside structured EHR. However, three studies relied solely on clinical notes, extracting drug exposure, comorbidities, and adverse events using NLP without mentioning access to other components of the EHR for signal identification analyses [74,106,109]. The types of clinical notes accessed also varied, but included admission history and physical exam notes, discharge summaries, clinic visit notes, and nursing documentation. Methods and tools used to extract potential adverse events from unstructured data varied widely, from simple regular expression text matching to more sophisticated NLP systems that mapped text mentions to concepts in clinical ontologies. An in-depth discussion of specific NLP methods for clinical text is beyond the scope of this review, but we direct the interested reader to several high-quality reviews on the subject [20,24,28,113].
One study by LePendu and colleagues [54] described methods to transform unstructured clinical notes into a deidentified patient-feature matrix encoded using medical terminologies. The matrix forms a timeline, noting when events occurred (or were recorded) including drug exposures and outcomes of interest. They demonstrated its utility for identifying both single-drug adverse events and drug-drug interactions earlier than official alerts by finding signals on retrospective data. It also allowed filtering of spurious signals by adjusting for potential confounding and could be used to compile prevalence information and estimate performance.
3.4. Integration with Spontaneous Reporting Data
Several studies (n=15, 19%) utilized both spontaneous reporting and EHR data. Associations identified in spontaneous reports either directed subsequent EHR investigations of specific drug-event pairs of interest or filtered EHR-identified signals. This sequential approach was aimed at reducing the risk of false positive signals from observational EHR data while replicating signals from spontaneous reports in a more diverse population. A study by Li and colleagues [69] found a combination of analyses using both spontaneous reporting and EHR data more accurately identified ADEs than analyses in either resource alone. Prospective studies compared the time to signal recognition between EHR-based analyses and parallel analyses in spontaneous reporting data. These studies reported EHR-based methods were able to identify adverse event signals sooner than they would have been identified in spontaneous reporting databases [87,98]. A study by Patadia and colleagues [71] highlighted considerations of the potential interplay between current spontaneous reporting systems and ADE identification in EHR data. They found signals were detectable in EHR data earlier than spontaneous reporting systems; however, applying the same methods to EHR data collected after initial warnings were issued changed practice patterns and reduced the utility of subsequent EHR data for signal identification.
3.5. Statistical considerations
Control for confounding was often discussed as a limitation, yet 40% of studies reported no specific steps to control for confounding (n=32). When confounding was addressed, this typically took the form of matching, stratification, or statistical adjustment for a small number of patient features. Ten studies (12%) limited confounding control to simple demographic characteristics and 24 (30%) studies controlled for some combination of demographics, indications, other drug exposures, and comorbidities. Self-controlled series were used to adjust for time-invariant confounders in 12 studies (15%) and several studies undertook propensity score matching/adjustment (n=12, 15%). Limited details of propensity scoring methods were reported and few studies provided specific justification for the selection of confounders.
Most studies (n=74, 91%) explicitly documented temporal considerations to ensure drug exposures occurred within a specified time window prior to adverse event indication or documentation. Several studies considered time more specifically by conducting survival analyses [80,81,103] or using chronographs of temporal event patterns [47,66,73,76,93]. Temporality was further explicitly considered by the five studies [42,60,70,71,87] that used LEOPARD to explore the rate of drug initiation before and after adverse events to filter out ADEs that may be due to confounding by indication.
As most studies controlled for few, if any patient features, missing data was rarely mentioned and studies implicitly assumed no bias in drug exposure or outcome ascertainment. Six studies mentioned concerns about missing data and conducted complete case analyses [79,84,86,90,99,103]. No studies considered imputation.
While all studies evaluated multiple pre-specified ADEs or many potential drug-event pairs, most did not discuss concerns regarding adjustments for multiple comparisons (n=61, 75%). When mentioned, Bonferroni correction and false discovery rates were the most common approaches to handling multiple comparisons.
4. Discussion
The body of work discussed in this review, along with prior reviews of pharmacovigilance studies and methods, make it clear that EHR data can make an important contribution to medication safety signal identification. However, there are challenges remaining. Studies varied widely in terms of the methods implemented, the data utilized, temporal and statistical considerations, and other limitations. Below we discuss each of these areas and provide recommendations for future research.
4.1. Analytic Methods
The most commonly used methods were the same approaches popular in spontaneous reporting systems, such as disproportionality analysis and regression-based modeling. While 37% of studies employed machine learning or data mining methods, the selected algorithms were highly variable and do not provide sufficient information to compare specific algorithms. Fewer studies applied sequential analysis methods. Each of these approaches have benefits and limitations (see Table 4).
Disproportionality methods, such as ROR, PRR, and GPS, are commonly applied to spontaneous reporting data despite concerns about underreporting and the inability to provide a true incidence rate when the number of outcomes is known and the number of exposures is not [3]. While EHRs do not necessarily capture all instances of exposure and outcomes (due to missing data and patients visiting multiple health systems), they can provide for more-complete capture of numerator and denominator than spontaneous reports [14]. This can potentially improve estimates of drug utilization and condition incidence. However, as currently applied to EHR data, disproportionality methods are not leveraging the breadth of EHR data nor controlling for key confounding. Integration of more advanced propensity score matching taking into account the rich patient-level information in the EHR may improve the use of these methods for EHR-based pharmacovigilance.
Similarly, regression-based methods also allow for confounding adjustment. EHRs can provide rich clinical detail for such adjustments, but nearly half of studies either reported no specific steps to control for confounding or only controlled for simple demographic features. When utilized for signal identification (as opposed to confirmation), regression methods must be adjusted for multiple testing, however many studies did not report taking this into consideration.
While machine learning, data mining, and sequence analysis are promising, there are only a few examples of each method and thus limited evidence of performance. The tree-based scan statistic, which was applied in two coordinated studies [38,52], highlights the potential of these methods for adverse event signal identification. TreeScan, the data mining tool that implements the tree-based scan statistic, can simultaneously evaluate a number of potential adverse events (and groups of related adverse events) to determine if any occur with higher probability among exposed patients [52,114]. Simultaneously, it evaluates if those outcomes occur with increased risk among patients exposed to individual drugs or groups of related drugs, automatically adjusting for the inherent multiple testing. This approach, normalizing drugs to classes and specific outcomes to broader categories of related outcomes, makes use of the hierarchical structure of the terminologies used for both exposures and outcomes, and can aggregate multiple weaker signals into significant ones.
As described above, there were also very few studies comparing the performance of different signal identification methods to one another. This prevents us from commenting on which methods may best identify adverse event signals within EHR data. Future efforts should develop large, standardized datasets with established drug-event associations against which methods can be compared more directly within and across studies.
4.2. Data
Access to large longitudinal claims databases have somewhat alleviated the volume issues with traditional surveillance methods, but such databases do not have the depth, or granularity, of patient information available in structured and unstructured EHR data. The breadth of EHR information lies in the many different types of data available – demographics, diagnoses, laboratory results, vital signs, problem lists, and unstructured clinical text – and, unfortunately, our review highlights that most EHR-based signal identification studies have yet to take full advantage of these complementary data types. For example, while many studies accessed diagnosis codes for outcome ascertainment, few took advantage of the other recorded diagnoses to control for confounding. Studies also frequently operationalized outcomes using only diagnosis codes, only laboratory values, or only NLP-processed unstructured clinical notes. While these studies serve to validate each method with different data types, methods combining different EHR data types could lead to a richer abstraction of the patient and better capture of a wide variety of adverse events. This increase in data dimensionality, however, poses unique challenges, including cases in which critical features may be sparsely populated. Possible methodological considerations to help address these difficulties might include mapping of differing feature types to a common terminology, such as MedDRA, or developing methods that can utilize both binary outcomes and continuous outcomes, such as lab values.
4.3. Temporal and Statistical Considerations
Many studies developed interesting approaches with significant opportunity for extension, while often neglecting to address a common set of limitations. These include lack of control for confounding, adjustment for multiple testing, and attention to missing data. In terms of confounding control, most studies either did not report any confounding control or controlled for only a few factors, most commonly only age and sex. Some, however, took advantage of the rich EHR data available to control for factors such as medications, comorbid conditions, and utilization of healthcare resources. Methods such as propensity score matching and self-controlled study designs are effective ways to address confounding that could have been integrated into many of the methods reviewed. Given the breadth of potential confounders in the EHR, further research and evaluation of methods using published literature to identify confounders [115] are warranted.
Few studies took common statistical approaches to preempt false positive signals. As noted above, most studies minimally controlled for confounding, despite the ability of proper confounding control to reduce false positives. Our review also highlighted a lack of adherence to recommendations to adjust for multiple comparisons when evaluating many potential adverse event signals, with 75% of studies not addressing this statistical concern. Given these limitations, many studies required additional analytic steps to filter out false positives and determine which drug-event associations highlighted by their analyses warranted further investigation. Common filtering approaches included ranking drug-event pairs by measures of signal strength, comparing findings to parallel analyses in spontaneous reporting data, incorporating external knowledge sources, or assessing protopathic bias using LEOPARD. All of these approaches can reduce false positives, but it is also important to empirically estimate the false discovery rate so that methods can be accurately evaluated and compared.
Careful attention to temporal relationships within data are also critical when performing EHR-based signal identification. Most studies were explicit in their methods to ensure documented exposures occurred before the suspected adverse events. However, studies were not consistent in how they determined initial exposures, minimum unexposed time before an index exposure, or relevant follow-up requirements. Best practices for establishing a minimum length of medical history in EHR data prior to exposures would help more consistently ensure patients were not exposed earlier than suspected. Similarly, standard practices should be established for handling cases when exposure and outcome are found in the same note but the temporal ordering of events is not explicitly defined or when mentions of an outcome in a clinical note does not necessarily mean that it began on the day the note was recorded. Evaluations by Patadia and colleagues [71] revealed another temporality concern regarding the interaction between early suspicion of potential ADE signals and changes in prescribing patterns. They found EHR data collected after media coverage or regulatory warnings of a newly detected ADE were less likely to correctly identify the ADE signal compared to EHR data collected prior to the dissemination of information about the suspected signal. To avoid biasing results, studies must consider announcements of any preliminary safety concerns when defining temporal start and end points for studies in retrospective data. Further research on how to handle the temporality of longitudinal EHR data is required.
While common in routinely collected EHR data, missing data was rarely discussed in the articles we reviewed. Missingness was likely not discussed for studies using structured medications and diagnoses because the lack of recording is typically presumed to be the absence of a diagnosis or medication. However, undercoding – when structured codes do not adequately represent a patient’s condition or the full scope of work being performed – is a limitation of structured EHR data that all studies should consider. In studies utilizing other EHR features, missingness may become more of a concern and should be addressed. For example, missingness may be of particular importance when considering data on social determinants of health, which may not be well-recorded in certain subpopulations. Furthermore, given the decentralized nature of healthcare in the United States, patient data is often scattered across multiple EHRs and health systems. As mentioned above, efforts to link longitudinal claims data with EHRs, as well as efforts promoting Health Information Exchange, could somewhat alleviate such concern.
4.4. Other Limitations of Current Work
During our analyses, we noticed a number of additional limitations of current research in EHR-based signal identification. Medication dosing, for instance was almost never considered in the studies we reviewed and few studies focused on exposures that may be better-captured in EHRs compared to claims data. For example, the EHR can be a source for identifying adverse events due to blood products and contrast media, as well as herbals and other non-conventional medications. EHR note type was also rarely mentioned; cardiology notes reveal different information than rheumatology notes, for example. Most studies implemented analysis at only a single site, which can result in inadequate cases to identify signals, particularly for rare events.
While many different studies used spontaneous reports data in combination with EHR data, it remains unclear how the two data sources can best complement one another to improve sensitivity and reduce false positives safety signals. Additionally, while some studies used derivatives of FAERS, such as TWOSIDES [116], it is probable that such derivatives could be helpful in more studies. Integration of other data sources may be helpful, as well; disproportionality methods, for example, suffer from confounding and frequent non-causal associations with indications and comorbidities [117]. Databases such as SIDER [118] contain known indications and adverse effects from drug product labels and could be used to identify confounding by indication. EHR data itself could be used to identify common comorbidities. The combination of these data resources can be used to prioritize unexplained signals or discount those signals with other likely explanations.
Finally, we note the lack of studies focused on children and pregnancy. A review by McMahon and colleagues [18] noted the lack of robust pediatric-focused post-marking surveillance. While a body of work in claims-based maternal-fetal outcomes research exists, there is widespread recognition regarding the shortcoming of pharmacovigilance as it relates to pregnancy [119–121]. Studies of these populations are critical to drug safety efforts and would likely benefit from the addition of EHR data.
4.5. Recommendations
The current state of research involving EHR-based signal identification is promising but would benefit from a more systematic approach to methods evaluation and the development of best practices. Methods and data models should also take advantage of the full breadth and depth of EHR data. Most reviewed studies focused on a limited number of EHR features, many simply accessing specific diagnoses and medication orders of interest. Accessing a broader set of features captured in EHRs could allow more thorough control for confounding, a unique advantage of EHR data over spontaneous reports. Further advancement of prospective approaches should also be prioritized as the existing studies indicate such approaches may speed identification of safety concerns. Newer methods utilizing recent advances in deep learning, symbolic artificial intelligence, and large language models should also be further explored.
Similarly, development of a common data model for tailoring longitudinal EHR datasets for pharmacovigilance studies would allow consistent application of methods and simplify evaluation. A common EHR transformation for adverse event signal identification could be accomplished by extending the work of LePendu and colleagues [54] using patient feature matrices. While their study exclusively used features extracted from clinical text, structured data could be also represented using the same framework and terminology (e.g., MedDRA). Further extensions could allow the incorporation of both continuous features and the integration of claims data to address concerns when relevant data is collected from multiple health care organizations.
A standardized data model for pharmacovigilance would also support distributed analyses that could be performed at multiple sites and foster greater collaboration. While the sensitive nature of EHR data makes large repositories difficult, methods for federated machine learning and distributed analysis like those currently employed by Sentinel, DARWIN, and EDHEN initiatives would enable larger-scale EHR-based data analysis. This is particularly important for identifying safety signals involving less common medications or rare events that may not have sufficient data for detection at any individual site.
Finally, it is important to both define success metrics and estimate performance to better understand the types of signals that would be captured poorly in EHR data. Development of a common resources of known adverse event signals and control drug-event associations would support comparative evaluations across signal identification methods and enable reproducibility. Results for existing and future novel approaches to signal identification could be reported against this standard reference to provide more comparable baseline comparisons and support the collection of evidence needed to establish best practices.
5. Conclusion
The current state of drug safety signal identification with EHR data is promising. However, comparing signal identification methods to one another based upon available research is difficult due to differences in study designs and populations; EHR data models and components utilized; and diverse combinations of exposures and health outcomes evaluated. In addition, published studies differ substantially in their treatment of confounders, temporal considerations, and adjustments for multiple testing. Future efforts to evaluate available methods, create a common data model for EHR data, build shared reference sets for validation, and develop best practices for signal discovery and confirmation are necessary.
Key Points.
Detailed electronic health record data could enrich pharmacovigilance programs that have traditionally relied on voluntary spontaneous reporting systems, with hopes of enhancing and speeding our understanding of medication safety signal concerns.
Our review of electronic health record-based signal identification studies highlights great variability in study design and limited application of methods for handling confounders, temporal considerations, and adjustments for multiple testing.
While current research is promising, the community of pharmacovigilance researchers and practitioners would benefit from a more systematic approach to methods evaluation, comparison, and benchmarking to develop best practices for implementation and expansion of electronic health record-based pharmacovigilance.
Funding:
This project was supported by Task Order 7540119F19002 under Master Agreement 75F40119D10037 from the US Food and Drug Administration (FDA). The FDA reviewed and approved this manuscript. The FDA had no role in data collection, management, or analysis. The views expressed represent those of the authors and do not necessarily represent the official views of the FDA.
Footnotes
Statements and Declarations
Conflicts of interest: The authors declare no competing interests.
Ethics approval: Not applicable
Consent to participate: Not applicable
Consent for publication: Not applicable
Code availability: Not applicable
Availability of data and materials:
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
6. References
- 1.World Health Organization. What is Pharmacovigilance? [Internet]. [cited 2022 Jul 22]. Available from: https://www.who.int/teams/regulation-prequalification/regulation-and-safety/pharmacovigilance
- 2.Hauben M, Aronson JK. Defining “signal” and its subtypes in pharmacovigilance based on a systematic review of previous definitions. Drug Saf. 2009;32:99–110. [DOI] [PubMed] [Google Scholar]
- 3.Coloma PM, Trifiro G, Patadia V, Sturkenboom M Postmarketing safety surveillance: Where does signal detection using electronic healthcare records fit into the big picture? Drug Saf. 2013;36:183–97. [DOI] [PubMed] [Google Scholar]
- 4.U.S. Food & Drug Administration. Questions and Answers on FDA’s Adverse Event Reporting System (FAERS) [Internet]. 2018. Available from: https://www.fda.gov/drugs/surveillance/questions-and-answers-fdas-adverse-event-reporting-system-faers
- 5.European Medicines Agency. EudraVigilance system overview [Internet]. [cited 2022 Aug 25]. Available from: https://www.ema.europa.eu/en/human-regulatory/research-development/pharmacovigilance/eudravigilance/eudravigilance-system-overview
- 6.World Health Organization Uppsala Monitoring Centre. What is VigiBase? [Internet]. [cited 2022 Aug 25]. Available from: https://who-umc.org/vigibase/
- 7.Desai RJ, Matheny ME, Johnson K, Marsolo K, Curtis LH, Nelson JC, et al. Broadening the reach of the FDA Sentinel system: A roadmap for integrating electronic health record data in a causal analysis framework. NPJ Digit Med. 2021;4:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Platt R, Brown JS, Robb M, McClellan M, Ball R, Nguyen MD, et al. The FDA Sentinel Initiative - An Evolving National Resource. N Engl J Med. 2018;379:2091–3. [DOI] [PubMed] [Google Scholar]
- 9.European Medicines Agency. Data Analysis and Real World Interrogation Network (DARWIN EU) [Internet]. [cited 2023 May 14]. Available from: https://www.darwin-eu.org [Google Scholar]
- 10.National Institutes of Health. All of Us Research Program [Internet]. [cited 2023 May 13]. Available from: https://allofus.nih.gov [Google Scholar]
- 11.European Health Data & Evidence Network [Internet]. [cited 2023 May 13]. Available from: https://www.ehden.eu [Google Scholar]
- 12.Vilar S, Friedman C, Hripcsak G Detection of drug-drug interactions through data mining studies using clinical sources, scientific literature and social media. Brief Bioinform. 2018;19:863–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson AM, Thabane L, Holbrook A Application of data mining techniques in pharmacovigilance. Br J Clin Pharmacol. 2004;57:127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Almenoff JS, Pattishall EN, Gibbs TG, DuMouchel W, Evans SJW, Yuen N Novel statistical tools for monitoring the safety of marketed drugs. Clin Pharmacol Ther. 2007;82:157–66. [DOI] [PubMed] [Google Scholar]
- 15.Lu Z Information technology in pharmacovigilance: Benefits, challenges, and future directions from industry perspectives. Drug Healthc Patient Saf. 2009;1:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho T-B, Le L, Thai DT, Taewijit S Data-driven approach to detect and predict adverse drug reactions. Curr Pharm Des. 2016;22:3498–526. [DOI] [PubMed] [Google Scholar]
- 17.Bates DW, Evans RS, Murff H, Stetson PD, Pizziferri L, Hripcsak G. Detecting adverse events using information technology. J Am Med Inform Assoc JAMIA. 2003;10:115–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMahon AW, Wharton GT, Bonnel R, DeCelle M, Swank K, Testoni D, et al. Pediatric post-marketing safety systems in North America: assessment of the current status. Pharmacoepidemiol Drug Saf. 2015;24:785–92. [DOI] [PubMed] [Google Scholar]
- 19.Izem R, Sanchez-Kam M, Ma H, Zink R, Zhao Y Sources of Safety Data and Statistical Strategies for Design and Analysis: Postmarket Surveillance. Ther Innov Regul Sci. 2018;52:159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong A, Plasek JM, Montecalvo SP, Zhou L Natural Language Processing and Its Implications for the Future of Medication Safety: A Narrative Review of Recent Advances and Challenges. Pharmacotherapy. 2018;38:822–41. [DOI] [PubMed] [Google Scholar]
- 21.Zarrinpar A, David Cheng T-Y, Huo Z. What Can We Learn About Drug Safety and Other Effects in the Era of Electronic Health Records and Big Data That We Would Not Be Able to Learn From Classic Epidemiology?. J Surg Res. 2020;246:599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalinin AA, Higgins GA, Reamaroon N, Soroushmehr S, Allyn-Feuer A, Dinov ID, et al. Deep learning in pharmacogenomics: From gene regulation to patient stratification. Pharmacogenomics. 2018;19:629–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warrer P, Hansen EH, Juhl-Jensen L, Aagaard L. Using text-mining techniques in electronic patient records to identify ADRs from medicine use. Br J Clin Pharmacol. 2012;73:674–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo Y, Thompson WK, Herr TM, Zeng Z, Berendsen MA, Jonnalagadda SR, et al. Natural Language Processing for EHR-Based Pharmacovigilance: A Structured Review. Drug Saf. 2017;40:1075–89. [DOI] [PubMed] [Google Scholar]
- 25.Moore TJ, Furberg CD. Electronic Health Data for Postmarket Surveillance: A Vision Not Realized. Drug Saf. 2015;38:601–10. [DOI] [PubMed] [Google Scholar]
- 26.Wisniewski AFZ, Bate A, Bousquet C, Brueckner A, Candore G, Juhlin K, et al. Good Signal Detection Practices: Evidence from IMI PROTECT. Drug Saf. 2016;39:469–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Ventola C Big data and pharmacovigilance: Data mining for adverse drug events and interactions. P T. 2018;43:340–51. [PMC free article] [PubMed] [Google Scholar]
- 28.Harpaz R, Callahan A, Tamang S, Low Y, Odgers D, Finlayson S, et al. Text Mining for Adverse Drug Events: the Promise, Challenges, and State of the Art. Drug Saf. 2014;37:777–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The Health Improvement Network [Internet]. [cited 2022 Aug 25]. Available from: https://www.the-health-improvement-network.com [Google Scholar]
- 30.García Rodríguez LA, Pérez Gutthann S. Use of the UK General Practice Research Database for pharmacoepidemiology. Br J Clin Pharmacol. 1998;45:419–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan PB, Stang PE, Overhage JM, Suchard MA, Hartzema AG, DuMouchel W, et al. A comparison of the empirical performance of methods for a risk identification system. Drug Saf. 2013;36 Suppl 1:S143–158. [DOI] [PubMed] [Google Scholar]
- 32.Chazard E, Ficheur G, Merlin B, Genin M, Preda C, PSIP consortium, et al. Detection of adverse drug events detection: data aggregation and data mining. Stud Health Technol Inform. 2009;148:75–84. [PubMed] [Google Scholar]
- 33.Edwards RI Advanced methods in pharmacovigilance and toxicosurveillance. Clin Toxicol. 2009;47:485. [Google Scholar]
- 34.Ryan PB, Powell GE, Pattishall EN, Beach KJ Performance of screening multiple observational databases for active drug safety surveillance. Pharmacoepidemiol Drug Saf PDS. 2009;18:S78. [Google Scholar]
- 35.Wang X, Hripcsak G, Markatou M, Friedman C Active Computerized Pharmacovigilance Using Natural Language Processing, Statistics, and Electronic Health Records: A Feasibility Study. J Am Med Inform Assoc. 2009;16:328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brownstein JS, Murphy SN, Goldfine AB, Grant RW, Sordo M, Gainer V, et al. Rapid identification of myocardial infarction risk associated with diabetes medications using electronic medical records. Diabetes Care. 2010;33:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harpaz R, Haerian K, Chase HS, Friedman C. Mining electronic health records for adverse drug effects using regression based methods. Proc ACM Int Conf Health Inform - IHI 10 [Internet]. Arlington, Virginia, USA: ACM Press; 2010. [cited 2022 Jul 22]. p. 100. Available from: http://portal.acm.org/citation.cfm?doid=1882992.1883008 [Google Scholar]
- 38.Brown JS, Dashevsky I, Fireman B, Herrinton L, McClure D, Murphy M, et al. Data mining with a tree-based scan statistic. Pharmacoepidemiol Drug Saf. 2011;20:S331. [DOI] [PubMed] [Google Scholar]
- 39.Chazard E, Ficheur G, Bernonville S, Luyckx M, Beuscart R. Data mining to generate adverse drug events detection rules. IEEE Trans Inf Technol Biomed Publ IEEE Eng Med Biol Soc. 2011;15:823–30. [DOI] [PubMed] [Google Scholar]
- 40.Coloma PM, Schuemie MJ, Trifirò G, Gini R, Herings R, Hippisley-Cox J, et al. Combining electronic healthcare databases in Europe to allow for large-scale drug safety monitoring: the EU-ADR Project. Pharmacoepidemiol Drug Saf. 2011;20:1–11. [DOI] [PubMed] [Google Scholar]
- 41.Coloma PM, Trifiro G, Gini R, Herings R, Mazzaglia G, Giaquinto C, et al. Comparison of methods for drug safety signal detection using electronic healthcare record (EHR) databases: The added value of longitudinal, Time-stamped patient information. Pharmacoepidemiol Drug Saf. 2011;20:S142. [Google Scholar]
- 42.Ferrajolo C, Trifiro G, Coloma PM, Schuemie MJ, Gini R, Herings R, et al. Drug use and acute liver injury in children: Signal detection using multiple healthcare databases. Drug Saf. 2011;34:983–4. [Google Scholar]
- 43.Ji Y, Ying H, Dews P, Mansour A, Tran J, Miller RE, et al. A potential causal association mining algorithm for screening adverse drug reactions in postmarketing surveillance. IEEE Trans Inf Technol Biomed Publ IEEE Eng Med Biol Soc. 2011;15:428–37. [DOI] [PubMed] [Google Scholar]
- 44.Park MY, Yoon D, Lee K, Kang SY, Park I, Lee S-H, et al. A novel algorithm for detection of adverse drug reaction signals using a hospital electronic medical record database. Pharmacoepidemiol Drug Saf. 2011;20:598–607. [DOI] [PubMed] [Google Scholar]
- 45.Trifiro G, Patadia V, Schuemie MJ, Coloma PM, Gini R, Herings R, et al. EU-ADR healthcare database network vs. spontaneous reporting system database: preliminary comparison of signal detection. Stud Health Technol Inform. 2011;166:25–30. [PubMed] [Google Scholar]
- 46.LePendu P, Bauer-Mehren A, Iyer S, Shah NH Analyzing unstructured clinical notes for phase IV drug safety surveillance. Circulation. 2012;126. [Google Scholar]
- 47.Star K, Strandell J, Friden S, Sallstedt L, Johansson J, Edwards RI Temporal pattern discovery on electronic health records-a source of reference in signal detectionwork. Pharmacoepidemiol Drug Saf. 2012;21:347. [Google Scholar]
- 48.Yoon D, Park MY, Choi NK, Park BJ, Kim JH, Park RW. Detection of adverse drug reaction signals using an electronic health records database: Comparison of the Laboratory Extreme Abnormality Ratio (CLEAR) algorithm. Clin Pharmacol Ther. 2012;91:467–74. [DOI] [PubMed] [Google Scholar]
- 49.Afzal Z, Kors JA, Sturkenboom MC, Schuemie MJ Identifying drug-safety signals in electronic health records: An evaluation of automated case-detection algorithms with different sensitivity and specificity. Pharmacoepidemiol Drug Saf. 2013;22:285–6. [Google Scholar]
- 50.An L, Ravindran PP, Renukunta S, Denduluri S Co-medication of pravastatin and paroxetine-a categorical study. J Clin Pharmacol. 2013;53:1212–9. [DOI] [PubMed] [Google Scholar]
- 51.Harpaz R, Vilar S, DuMouchel W, Salmasian H, Haerian K, Shah NH, et al. Combing signals from spontaneous reports and electronic health records for detection of adverse drug reactions. J Am Med Inform Assoc. 2013;20:413–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kulldorff M, Dashevsky I, Avery TR, Chan AK, Davis RL, Graham D, et al. Drug safety data mining with a tree-based scan statistic. Pharmacoepidemiol Drug Saf. 2013;22:517–23. [DOI] [PubMed] [Google Scholar]
- 53.Lependu P, Iyer SV, Bauer-Mehren A, Harpaz R, Ghebremariam YT, Cooke JP, et al. Pharmacovigilance using Clinical Text. AMIA Jt Summits Transl Sci Proc AMIA Jt Summits Transl Sci. 2013;2013:109. [PMC free article] [PubMed] [Google Scholar]
- 54.LePendu P, Iyer SV, Bauer-Mehren A, Harpaz R, Mortensen JM, Podchiyska T, et al. Pharmacovigilance using clinical notes. Clin Pharmacol Ther. 2013;93:547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lian Duan L, Khoshneshin M, Street WN, Liu M. Adverse drug effect detection. IEEE J Biomed Health Inform. 2013;17:305–11. [DOI] [PubMed] [Google Scholar]
- 56.Liu M, McPeek Hinz ER, Matheny ME, Denny JC, Schildcrout JS, Miller RA, et al. Comparative analysis of pharmacovigilance methods in the detection of adverse drug reactions using electronic medical records. J Am Med Inform Assoc JAMIA. 2013;20:420–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reps JM, Garibaldi JM, Aickelin U, Soria D, Gibson J, Hubbard R. Comparison of algorithms that detect drug side effects using electronic healthcare databases. Soft Comput. 2013;17:2381–97. [Google Scholar]
- 58.Sauzet O, Carvajal A, Escudero A, Molokhia M, Cornelius VR Illustration of the weibull shape parameter signal detection tool using electronic healthcare record data. Drug Saf. 2013;36:995–1006. [DOI] [PubMed] [Google Scholar]
- 59.Eriksson R, Werge T, Jensen LJ, Brunak S. Dose-specific adverse drug reaction identification in electronic patient records: temporal data mining in an inpatient psychiatric population. Drug Saf. 2014;37:237–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferrajolo C, Coloma PM, Verhamme KMC, Schuemie MJ, de Bie S, Gini R, et al. Signal detection of potentially drug-induced acute liver injury in children using a multi-country healthcare database network. Drug Saf. 2014;37:99–108. [DOI] [PubMed] [Google Scholar]
- 61.Iyer SV, Harpaz R, LePendu P, Bauer-Mehren A, Shah NH. Mining clinical text for signals of adverse drug-drug interactions. J Am Med Inform Assoc JAMIA. 2014;21:353–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji Y, Ying H, Tran J, Dews P, Mansour A, Massanari RM. A temporal interestingness measure for drug interaction signal detection in post-marketing surveillance. Annu Int Conf IEEE Eng Med Biol Soc IEEE Eng Med Biol Soc Annu Int Conf. 2014;2014:2722–5. [DOI] [PubMed] [Google Scholar]
- 63.Li Y, Salmasian H, Vilar S, Chase H, Friedman C, Wei Y A method for controlling complex confounding effects in the detection of adverse drug reactions using electronic health records. J Am Med Inform Assoc. 2014;21:308–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patel VN, Kaelber DC. Using aggregated, de-identified electronic health record data for multivariate pharmacosurveillance: a case study of azathioprine. J Biomed Inform. 2014;52:36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roitmann E, Eriksson R, Brunak S Patient stratification and identification of adverse event correlations in the space of 1190 drug related adverse events. Front Physiol. 2014;5:Article 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cederholm S, Hill G, Asiimwe A, Bate A, Bhayat F, Persson Brobert G, et al. Structured Assessment for Prospective Identification of Safety Signals in Electronic Medical Records: Evaluation in the Health Improvement Network. Drug Saf. 2015;38:87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Du L, Chakraborty A, Chiang C-W, Cheng L, Quinney SK, Wu H, et al. Graphic Mining of High-Order Drug Interactions and Their Directional Effects on Myopathy Using Electronic Medical Records. CPT Pharmacomet Syst Pharmacol. 2015;4:481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Girardeau Y, Trivin C, Durieux P, Le Beller C, Louet Agnes L-L, Neuraz A, et al. Detection of Drug-Drug Interactions Inducing Acute Kidney Injury by Electronic Health Records Mining. Drug Saf. 2015;38:799–809. [DOI] [PubMed] [Google Scholar]
- 69.Li Y, Ryan PB, Wei Y, Friedman C. A Method to Combine Signals from Spontaneous Reporting Systems and Observational Healthcare Data to Detect Adverse Drug Reactions. Drug Saf. 2015;38:895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pacurariu AC, Straus SM, Trifiro G, Schuemie MJ, Gini R, Herings R, et al. Useful Interplay Between Spontaneous ADR Reports and Electronic Healthcare Records in Signal Detection. Drug Saf. 2015;38:1201–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patadia VK, Schuemie MJ, Coloma P, Herings R, van der Lei J, Straus S, et al. Evaluating performance of electronic healthcare records and spontaneous reporting data in drug safety signal detection. Int J Clin Pharm. 2015;37:94–104. [DOI] [PubMed] [Google Scholar]
- 72.Reps JM, Garibaldi JM, Aickelin U, Gibson JE, Hubbard RB. A supervised adverse drug reaction signalling framework imitating Bradford Hill’s causality considerations. J Biomed Inform. 2015;56:356–68. [DOI] [PubMed] [Google Scholar]
- 73.Star K, Watson S, Sandberg L, Johansson J, Edwards IR Longitudinal medical records as a complement to routine drug safety signal analysis. Pharmacoepidemiol Drug Saf. 2015;24:486–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang G, Jung K, Winnenburg R, Shah NH A method for systematic discovery of adverse drug events from clinical notes. J Am Med Inform Assoc. 2015;22:1196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang P, Du L, Wang L, Liu M, Cheng L, Chiang C-W, et al. A Mixture Dose–Response Model for Identifying High-Dimensional Drug Interaction Effects on Myopathy Using Electronic Medical Record Databases. CPT Pharmacomet Syst Pharmacol. 2015;4:474–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hauben M, Liu Q, Hung E, Blackwell W, Fram D, Bate A Signal detection using Temporal Pattern Discovery (TPD) in Electronic Health Records (EHRs) - Lessons from statins and rhabdomyolysis. Pharmacoepidemiol Drug Saf. 2016;25:441–2. [Google Scholar]
- 77.Lorberbaum T, Sampson KJ, Chang JB, Iyer V, Woosley RL, Kass RS, et al. Coupling Data Mining and Laboratory Experiments to Discover Drug Interactions Causing QT Prolongation. J Am Coll Cardiol. 2016;68:1756–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lorberbaum T, Sampson KJ, Woosley RL, Kass RS, Tatonetti NP An Integrative Data Science Pipeline to Identify Novel Drug Interactions that Prolong the QT Interval. Drug Saf. 2016;39:433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boland MR, Polubriaginof F, Tatonetti NP. Development of A Machine Learning Algorithm to Classify Drugs Of Unknown Fetal Effect. Sci Rep. 2017;7:12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fan Y, Adam TJ, McEwan R, Pakhomov SV, Melton GB, Zhang R Detecting Signals of Interactions Between Warfarin and Dietary Supplements in Electronic Health Records. Stud Health Technol Inform. 2017;245:370–4. [PMC free article] [PubMed] [Google Scholar]
- 81.Lee S, Choi J, Kim H-S, Kim GJ, Lee KH, Park CH, et al. Standard-based comprehensive detection of adverse drug reaction signals from nursing statements and laboratory results in electronic health records. J Am Med Inform Assoc. 2017;24:697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Personeni G, Bresso E, Devignes M-D, Dumontier M, Smail-Tabbone M, Coulet A. Discovering associations between adverse drug events using pattern structures and ontologies. J Biomed Semant. 2017;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang L, Rastegar-Mojarad M, Liu S, Zhang H, Liu H. Discovering adverse drug events combining spontaneous reports with electronic medical records: a case study of conventional DMARDs and biologics for rheumatoid arthritis. AMIA Jt Summits Transl Sci Proc AMIA Jt Summits Transl Sci. 2017;2017:95–103. [PMC free article] [PubMed] [Google Scholar]
- 84.Chen W, Yang J, Wang H-L, Shi Y-F, Tang H, Li G-H Discovering Associations of Adverse Events with Pharmacotherapy in Patients with Non-Small Cell Lung Cancer Using Modified Apriori Algorithm. BioMed Res Int. 2018;2018:1245616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Choi L, Carroll RJ, Beck C, Mosley JD, Roden DM, Denny JC, et al. Evaluating statistical approaches to leverage large clinical datasets for uncovering therapeutic and adverse medication effects. Bioinforma Oxf Engl. 2018;34:2988–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jeong E, Park N, Choi Y, Park RW, Yoon D Machine learning model combining features from algorithms with different analytical methodologies to detect laboratory-event-related adverse drug reaction signals. PLoS ONE. 2018;13:e0207749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Patadia VK, Schuemie MJ, Coloma PM, Herings R, van der Lei J, Sturkenboom M, et al. Can electronic health records databases complement spontaneous reporting system databases? A historical-reconstruction of the association of rofecoxib and acute myocardial infarction. Front Pharmacol. 2018;9:594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shimai Y, Takeda T, Okada K, Manabe S, Teramoto K, Mihara N, et al. Screening of anticancer drugs to detect drug-induced interstitial pneumonia using the accumulated data in the electronic medical record. Pharmacol Res Perspect. 2018;6:e00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tham MY, Ye Q, Ang PS, Fan LY, Yoon D, Park RW, et al. Application and optimisation of the Comparison on Extreme Laboratory Tests (CERT) algorithm for detection of adverse drug reactions: Transferability across national boundaries. Pharmacoepidemiol Drug Saf. 2018;27:87–94. [DOI] [PubMed] [Google Scholar]
- 90.Vajravelu RK, Scott FI, Mamtani R, Li H, Moore JH, Lewis JD. Medication class enrichment analysis: a novel algorithm to analyze multiple pharmacologic exposures simultaneously using electronic health record data. J Am Med Inform Assoc JAMIA. 2018;25:780–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang L, Rastegar-Mojarad M, Ji Z, Liu S, Liu K, Moon S, et al. Detecting pharmacovigilance signals combining electronic medical records with spontaneous reports: A case study of conventional disease-modifying antirheumatic drugs for rheumatoid arthritis. Front Pharmacol. 2018;9:875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang X, Zhang P, Chiang C-W, Wu H, Shen L, Ning X, et al. Mixture drug-count response model for the high-dimensional drug combinatory effect on myopathy. Stat Med. 2018;37:673–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Whalen E, Hauben M, Bate A Time Series Disturbance Detection for Hypothesis-Free Signal Detection in Longitudinal Observational Databases. Drug Saf. 2018;41:565–77. [DOI] [PubMed] [Google Scholar]
- 94.Zhou X, Douglas IJ, Shen R, Bate A. Signal Detection for Recently Approved Products: Adapting and Evaluating Self-Controlled Case Series Method Using a US Claims and UK Electronic Medical Records Database. Drug Saf. 2018;41:523–36. [DOI] [PubMed] [Google Scholar]
- 95.Dang T-T, Nguyen T-H, Ho T-B Causality assessment of adverse drug reaction: Controlling confounding induced by polypharmacy. Curr Pharm Des. 2019;25:1134–43. [DOI] [PubMed] [Google Scholar]
- 96.Davazdahemami B, Delen D Examining the effect of prescription sequence on developing adverse drug reactions: The case of renal failure in diabetic patients. Int J Med Inf. 2019;125:62–70. [DOI] [PubMed] [Google Scholar]
- 97.Duan R, Boland MR, Moore JH, Chen Y ODAL: A one-shot distributed algorithm to perform logistic regressions on electronic health records data from multiple clinical sites. Pac Symp Biocomput Pac Symp Biocomput. 2019;24:30–41. [PMC free article] [PubMed] [Google Scholar]
- 98.Yu Y, Ruddy KJ, Wen A, Zong N, Tsuji S, Chen J, et al. Integrating Electronic Health Record Data into the ADEpedia-on-OHDSI Platform for Improved Signal Detection: A Case Study of Immune-related Adverse Events. AMIA Jt Summits Transl Sci Proc AMIA Jt Summits Transl Sci. 2020;2020:710–9. [PMC free article] [PubMed] [Google Scholar]
- 99.Yu Y, Nie X, Song Z, Xie Y, Zhang X, Du Z, et al. Signal Detection of Potentially Drug-Induced Liver Injury in Children Using Electronic Health Records. Front Pediatr. 2020;8:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang W, Peissig P, Kuang Z, Page D. Adverse Drug Reaction Discovery from Electronic Health Records with Deep Neural Networks. Proc ACM Conf Health Inference Learn. 2020;2020:30–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Akimoto H, Nagashima T, Minagawa K, Hayakawa T, Takahashi Y, Asai S Signal Detection of Potential Hepatotoxic Drugs: Case-Control Study Using Both a Spontaneous Reporting System and Electronic Medical Records. Biol Pharm Bull. 2021;44:1514–23. [DOI] [PubMed] [Google Scholar]
- 102.Nie X, Jia L, Peng X, Zhao H, Yu Y, Chen Z, et al. Detection of Drug-Induced Thrombocytopenia Signals in Children Using Routine Electronic Medical Records. Front Pharmacol [Internet]. 2021. [cited 2023 May 2];12. Available from: https://www.frontiersin.org/articles/10.3389/fphar.2021.756207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shin H, Lee S An OMOP-CDM based pharmacovigilance data-processing pipeline (PDP) providing active surveillance for ADR signal detection from real-world data sources. BMC Med Inform Decis Mak. 2021;21:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shin H, Cha J, Lee Y, Kim J-Y, Lee S Real-world data-based adverse drug reactions detection from the Korea Adverse Event Reporting System databases with electronic health records-based detection algorithm. Health Informatics J. 2021;27:14604582211033014. [DOI] [PubMed] [Google Scholar]
- 105.Wu P, Nelson SD, Zhao J, Stone CA, Feng Q, Chen Q, et al. DDIWAS: High-throughput electronic health record-based screening of drug-drug interactions. J Am Med Inform Assoc JAMIA. 2021;28:1421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Challa AP, Niu X, Garrison EA, Van Driest SL, Bastarache LM, Lippmann ES, et al. Medication history-wide association studies for pharmacovigilance of pregnant patients. Commun Med. 2022;2:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kaas-Hansen BS, Placido D, Rodríguez CL, Thorsen-Meyer H-C, Gentile S, Nielsen AP, et al. Language-agnostic pharmacovigilant text mining to elicit side effects from clinical notes and hospital medication records. Basic Clin Pharmacol Toxicol. 2022;131:282–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kundrot S, Warnick J, Erdman C, Robert K, Brown J. Demonstration of TreeScan Techniques on a Federated Real-World Data Network. DRUG Saf. ADIS INT LTD 5 THE WAREHOUSE WAY, NORTHCOTE 0627, AUCKLAND, NEW ZEALAND; 2022. p. 1238–9. [Google Scholar]
- 109.Mower J, Bernstam E, Xu H, Myneni S, Subramanian D, Cohen T. Improving Pharmacovigilance Signal Detection from Clinical Notes with Locality Sensitive Neural Concept Embeddings. AMIA Annu Symp Proc AMIA Symp. 2022;2022:349–58. [PMC free article] [PubMed] [Google Scholar]
- 110.Nie X, Yu Y, Jia L, Zhao H, Chen Z, Zhang L, et al. Signal Detection of Pediatric Drug-Induced Coagulopathy Using Routine Electronic Health Records. Front Pharmacol. 2022;13:935627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sauzet O, Cornelius V. Generalised weibull model-based approaches to detect non-constant hazard to signal adverse drug reactions in longitudinal data. Front Pharmacol [Internet]. 2022. [cited 2023 May 2];13. Available from: https://www.frontiersin.org/articles/10.3389/fphar.2022.889088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu Y, Nie X, Zhao Y, Cao W, Xie Y, Peng X, et al. Detection of pediatric drug-induced kidney injury signals using a hospital electronic medical record database. Front Pharmacol [Internet]. 2022. [cited 2023 May 2];13. Available from: https://www.frontiersin.org/articles/10.3389/fphar.2022.957980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Murphy RM, Klopotowska JE, de Keizer NF, Jager KJ, Leopold JH, Dongelmans DA, et al. Adverse drug event detection using natural language processing: A scoping review of supervised learning methods. PloS One. 2023;18:e0279842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kulldorff M TreeScan Software for the Tree-Based Scan Statistic [Internet]. Available from: https://www.treescan.org [Google Scholar]
- 115.Malec SA, Wei P, Bernstam EV, Boyce RD, Cohen T Using computable knowledge mined from the literature to elucidate confounders for EHR-based pharmacovigilance. J Biomed Inform. 2021;117:103719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tatonetti NP, Ye PP, Daneshjou R, Altman RB Data-driven prediction of drug effects and interactions. Sci Transl Med. 2012;4:125ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Harpaz R, DuMouchel W, LePendu P, Bauer-Mehren A, Ryan P, Shah NH. Performance of pharmacovigilance signal-detection algorithms for the FDA adverse event reporting system. Clin Pharmacol Ther. 2013;93:539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kuhn M, Letunic I, Jensen LJ, Bork P. The SIDER database of drugs and side effects. Nucleic Acids Res. 2016;44:D1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dathe K, Schaefer C. The Use of Medication in Pregnancy. Dtsch Arzteblatt Int. 2019;116:783–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stock SJ, Norman JE. Medicines in pregnancy. F1000Research. 2019;8:F1000 Faculty Rev-911. [Google Scholar]
- 121.Braillon A, Bewley S. Prescribing in pregnancy shows the weaknesses in pharmacovigilance. BMJ. 2018;361:k2334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.