Abstract
The complete RNA genome of the avian nephritis virus (ANV) associated with acute nephritis in chickens has been molecularly cloned and sequenced. Excluding the poly(A) tail, the genome comprises 6,927 nucleotides and contains three sequential open reading frames (ORFs). The first ORF (ORF 1a) contains a sequence encoding a serine protease motif, and the second ORF (ORF 1b) has a sequence encoding an RNA-dependent RNA polymerase. ORF 1a may be linked to the second ORF by a ribosomal frameshifting mechanism. The third ORF (ORF 2) may encode the virion structural proteins as a polyprotein precursor. Two RNAs, probably genonic and subgenonic RNA (7.5 and 3.0 kb), were detected in the cytoplasm of ANV-infected cells. ANV and human astroviruses have the same genonic organization, and both are characterized by the presence of two RNA bands. The amino acid homologies of the products of ORF 1a, 1b, and 2 were 20.3, 41.9, and 25.8% to products of the corresponding ORFs of human astrovirus serotype 1 (A/88 Newcastle strain). We have constructed a genonic-length cDNA clone of ANV to test whether the in vitro transcript is infectious. When a chicken kidney cell culture was transfected with RNA transcribed in vitro and the cDNA clone, infectious virus was produced with cytopathic effects in the absence of trypsin. These observations suggested that the ANV (G-4260 strain) is a new genus of the family Astroviridae.
Avian nephritis virus (ANV) acts as an etiological agent of growth retardation of young chickens by causing interstitial nephritis (15). The virus was first isolated from young chickens with chicken kidney (CK) cells in 1976 by our group (52). ANV is widely distributed in chickens worldwide, and turkeys may also be susceptible (5, 16). Field viruses of ANV exhibit different degrees of pathogenicity in chickens, producing results ranging from subclinical infection to death, and there are at least two serotypes (7, 14, 43).
ANV is an unclassified small round nonenveloped virus 28 nm in diameter and stable at pH 3.0; however, the buoyant density has not been determined because ANV is fragile in cesium chloride (CsCl). Growth inhibition of ANV by adding the nucleoside analog 5-bromo-2-deoxyuridine indicated that it has an RNA genome (52). The virus can replicate in the kidneys and intestine in vivo. Similar to picornaviruses, ANV does not require trypsin for in vitro cultivation. Based on pathological and immunologic data, however, ANV is distinct from avian encephalomyelitis virus and duck hepatitis viruses, which are members of avian enterovirus-like viruses of the family Picornaviridae (5, 28, 30, 31). These data led investigators to believe that ANV may belong to the genus Enterovirus of the family Picornaviridae, although neither the nucleotide nor the amino acid sequences had been reported yet (14). Small rounded nonenveloped RNA viruses (approximately 30 nm in diameter) are causative agents of gastroenteritis in animals and humans (23). Not only picornaviruses but also astroviruses are included as the agents (31, 33, 35, 53).
Astroviruses were first identified in diarrheal stools of children with gastroenteritis by electron microscopy (27). Astroviruses are small nonenveloped RNA viruses, approximately 28 nm in diameter, characterized by a star-like morphology (26). Astroviruses require trypsin to grow in cell culture (3, 42, 47, 50), which may have some implications for gastroenteritis. There are at least eight serotypes of human astroviruses (HAst). The genome consists of a positive-strand RNA of approximately 6,800 nucleotides (nt) with three open reading frames (ORFs). The first ORF (ORF 1a), encodes a 3C-like serine protease motif, and the second ORF (ORF 1b) encodes an RNA-dependent RNA polymerase (RdRp) motif; however, the astrovirus lacks a helicase motif typical of other positive-strand RNA viruses. ORF 1a is linked to ORF 1b by a ribosomal frameshifting motif, and both ORFs are probably translated from the genonic RNA. The third ORF (ORF 2) encodes a precursor polyprotein which is proteolytically processed to viral structural proteins. ORF 2 is likely expressed from a subgenonic RNA which is coterminal with the genonic RNA at the 3′ end (3, 33, 34).
Definitive classification of ANV requires genetic information. In this study, the complete RNA genome was molecularly cloned and sequenced. Sequence analysis data indicated that ANV is a new member of the family Astroviridae. ANV is the first avian astrovirus whose genome has been completely sequenced. The infectious cDNA clone developed here is a tool to identify the critical regions for pathology and serology on the ANV genome.
MATERIALS AND METHODS
Cells and virus.
Primary CK cell cultures were prepared from specific-pathogen-free (SPF) chickens (line PDL-1) (8) and were maintained in Eagle's minimal essential medium (EMEM) supplemented with antibiotics and 5% newborn bovine serum (15). Baby hamster kidney (BHK) cells were grown in EMEM containing 10% fetal calf serum. ANV (G-4260 strain) was plaque-purified through three consecutive cycles before use. Chicken antiserum to the G-4260 strain obtained by immunization of SPF chicks with the plaque-purified virus was used for fluorescent antibody (FA) tests (15).
EM observation.
Viral particles purified from the supernatants of infected CK cells and an ultrathin section of infected cells were observed with an electron microscope (EM; JEM-100CX; Nippon Denshi Co., Tokyo, Japan). Infected cell culture fluid was harvested when the cells exhibited a maximum cytopathic effect (CPE) at 36 to 48 h postinoculation and were concentrated with polyethylene glycol 6000 (9). After centrifugation (8,000 × g for 30 min), the pellet was suspended in phosphate-buffered saline and loaded onto a 10 to 45% continuous linear sucrose gradient prepared in a polyalomer SW40 ultracentrifuge tube. The sample was ultracentrifuged on the sucrose gradient for 3.5 h at 36,000 rpm (SW-40Ti; Beckman) at 4°C; settings for acceleration and decelation were both “7.” The fraction with the highest infectivity was obtained and examined for virus with an EM, after it was stained with 3% uranyl acetate (pH 7.0). For the preparation of ultrathin sections, infected CK cells were collected before the maximum CPE appeared (20 h p.i.), fixed with 2% glutaraldehyde in phosphate buffer, postfixed with 1% osmium tetroxide in phosphate buffer, dehydrated in a graded ethanol series, and embedded in an Epon mixture. Ultrathin sections were cut with a glass knife, stained with a lead citrate-uranyl acetate solution, and observed with an EM.
Isolation of ANV RNA.
To clone the ANV genome, the culture fluid of ANV-infected cells was purified by sucrose gradient ultracentrifugation because several attempts to clone the virus from cesium chloride gradient-purified viral particles were unsuccessful. The viral antigen was monitored at each step with the EM. The virion RNA was extracted by using ISOGEN-LS total RNA isolation reagent (Nippon Gene, Tokyo, Japan) according to the manufacturer's recommendation.
Cloning the genome of ANV and sequencing.
Standard molecular biology procedures were used to clone ANV cDNAs (41). Briefly, cDNA synthesis was performed using a Time Saver cDNA synthesis kit (Amersham Pharmacia Biotech), the reverse transcription of ANV RNA being primed by oligo(dT)12–18 or by random six-residue primers. Second-strand synthesis produced double-stranded cDNA, which was subsequently ligated into the EcoRI site of the pBluescript II (pBSII) KS+ vector (Stratagene, La Jolla, Calif.). This was used to transform the Escherichia coli DH5α strain. The remainder of the genonic cDNA was cloned by three successive rounds of primer extension using synthetic primers derived from the sequence at the termini of the preceding clones. Clones containing the 5′ end of the genome were obtained with the 5′-Full rapid amplification of cDNA ends (RACE) core kit (Takara, Tokyo, Japan). The cycle sequencing reaction was performed with a dye terminator cycle sequencing FS Ready Reaction kit (Perkin-Elmer Cetus) using M13 forward, M13 reverse, and custom primers and was analyzed with an ABI 373 automatic sequencer. The sequences of all clones derived from each round of primer extension and 5′-Full RACE were determined for both strands of the cDNA.
RNA blot hybridization.
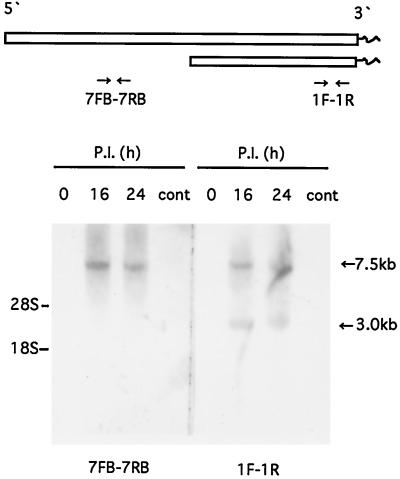
Two probes, one the 5′-end side (7FB-7RB; 302 bp at ORF 1a) and the other the 3′-end side (1F-1R; 280 bp at ORF 2 and the 3′-end nontranslated region [NTR]) (Table 1; Fig. 3), were synthesized with PCR DIG probe synthesis kits (Boehringer Mannheim). Total cytoplasmic RNA isolated from ANV-infected and uninfected cells at 16 and 24 h p.i. was resolved in a 1.0% agarose gel containing 0.1% sodium dodecyl sulfate, transferred to a nylon membrane, and probed with digoxigenin (DIG)-labeled DNAs. DIG nucleic acid detection kits (Boehringer Mannheim) were used according to the manufacturer's recommendations.
TABLE 1.
PCR primers used to generate PCR clones of ANV cDNA and the DIG-labeled probes for Northern blot hybridization
| Primer | Location (nt) | Sequencea |
|---|---|---|
| Sense | ||
| BT 7 | 1–22 | cgg cgg gat cct aat acg act cac tat agC CGAATA GAT GGG ATG GCT TCG |
| S 1 | 539–561 | GAG AAC GAG AGA ACC AAA CAC GG |
| S 2 | 583–604 | GTG CCT CTG GAT CTT TCT TGC G |
| 7FB | 1897–1916 | GGA CGG GAG GAT GCT TGG TG |
| 1F | 6596–6615 | CTA GAC CTC CGG CGC TTT CA |
| Antisense | ||
| A 2 | 382–356 | CAG GAG CGT GCG GGT GAG AAC AAC |
| A 1 | 473–450 | CTT TCA CTT TTT CAC GCG CCA GGT |
| P-12R | 661–638 | AAC ATC GAC AAG GGT ACA CGC TGC |
| 7RB | 2198–2179 | CAC GAA CGG TCC TCT TAG TC |
| 1R | 6879–6859 | CGG GAG AAC CAA CTT TAG CGC |
Uppercase letters, primer sequence corresponding to (5′ primer) or complementary to (3′ primer) the ANV genonic sequence; lowercase letters, sequence used to introduce restriction site and T7 promoter.
FIG. 3.
RNA blot hybridization analysis. Total cytoplasmic RNA isolated from ANV-infected cells and uninfected cells at the indicated hours p.i. were resolved in a 1.0% agarose gel which contained 0.1% sodium dodecyl sulfate, transferred to a nylon membrane, and probed with DIG-labeled ANV probes (7FB-7RB, 5′ end; 1F-1R, 3′ end). The positions of the 28S and 18S rRNAs are indicated on the left; those of genonic (7.5-kb) and subgenonic (3.0-kb) RNAs are indicated on the right. cont, control.
Comparative sequence analysis.
Both nucleotide and deduced amino acid sequences were compared, the multiple alignments were done, the phylogenetic trees of ORF 1b and ORF 2 were constructed, and the potential secondary structures of the ANV genome were examined using the software of GENETYX-WIN, version 4 (Software Development Co., Ltd., Tokyo, Japan). The following nucleotide sequences were obtained from GenBank/EMBL/DDBJ: HAst serotype 1 (HAst 1), Z25771 and L23513; HAst 2, L13745; feline astrovirus, AF056197; porcine astrovirus, AB037272; turkey astrovirus, AF206663.
Construction of full-length cDNA clone for ANV.
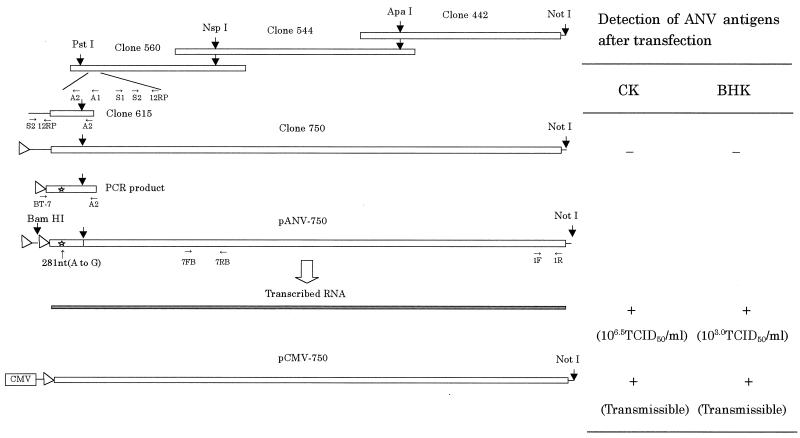
A full-length cDNA copy of ANV (G-4260 strain) was assembled in pBSII by sequentially linking overlapping fragments of the ANV cDNA at unique restriction sites (Fig. 1). These overlapping cDNA fragments were selected from the clones generated by cloning the genome of ANV for sequencing. Table 1 lists the PCR primers used for cloning clone 615 and the 5′-most primer on the viral genome comprising a T7 promoter as well as a BamHI site. The pBSII clone containing the full-length ANV cDNA between the BamHI and the NotI sites was named pANV-750. This plasmid harbors two T7 promoters organized in the same direction, one derived from the pBSII vector and the other derived from the 5′-most PCR primer. In the process of construction of pANV-750 from clone 750 and the PCR product (Fig. 1), a point mutation (A to G at nt 281; amino acid change Thr to Ala) was introduced by PCR. Finally, the vector plasmid was changed to pCMV vector (Clontech Laboratories, Inc., Palo Alto, Calif.) from pBSII at the NotI site and named pCMV-750.
FIG. 1.
Construction of the full-length ANV cDNA clone and infectivity. Open boxes, partial and full-length viral cDNA; solid box, RNA transcribed in vitro from pANV-750; arrowheads, T7 promoter; CMV, human cytomegalovirus immediate-early gene promoter; star, point mutation introduced by PCR; small arrow, primer direction and position (see Table 1); −, negative for infectious virus; +, positive for infectious virus (see Fig. 7). In vitro transcripts prepared from the full-length ANV cDNA clone or a plasmid which can transcribe the genonic-size ANV RNA under the control of the CMV promoter were transfected onto CK or BHK cells using Lipofectin reagent (Bethesda Research Laboratories). The cells were tested for the presence of ANV antigens by FA tests 2 days after transfection.
Demonstrating infectivity of RNA transcribed from the cDNA clone (pANV-750) and cDNA of ANV.
The ANV cDNA was positioned downstream of a viral T7 DNA-dependent RNA polymerase promoter, and the NotI restriction site allowed the linearization of the DNA immediately downstream of the viral poly(A) sequence. In vitro transcription with T7 polymerase (Promega, Madison, Wis.) was performed according to the manufacturer's recommendations in the presence of an m7G(5′)ppp(5′)G RNA cap structure analog and was followed by treatment with an RNase-free DNase (10).
CK and BHK cells were transfected with RNA (1 μg/dish) transcribed in vitro from pANV-750 and cDNAs (4 μg/dish) using Lipofectin reagent (Bethesda Research Laboratories, Gaithersburg, Md.) according to the manufacturer's protocol. After transfection, cells were incubated in medium supplemented with 10% serum at 37°C for 24 to 96 h, and the cells were observed every day for CPE and expression of viral antigens, as detected by an indirect immunofluorescence assay with the antiserum (16). To recover infectious virus for passing from the transfected cells, the cells and media were harvested after 96 h of incubation. Every transfection experiment included a negative control where the cells were mock-transfected in the absence of RNA or cDNA. Lysates from the mock-transfected cells were used as a negative control for subsequent infection of cells.
Nucleotide sequence accession number.
The complete nucleotide sequence of ANV has been submitted to the DDBJ, EMBL, and GenBank databases under accession no. AB033998.
RESULTS
Sequence analysis.
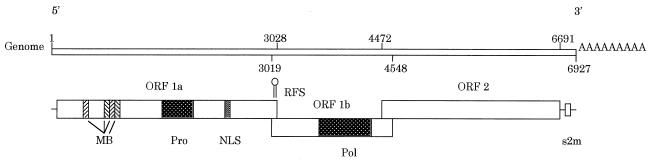
The genonic RNA of ANV was 6,927 nt in length, excluding the poly(A) tail. The genome possesses three overlapping ORFs, ORF 1a, 1b, and 2 (Fig. 2; Table 2). When the virus uses the first methionine, ORF 1a (nt 14 to 3028) may encode a polypeptide of 1,005 amino acids (aa). ORF 1b (nt 3019 to 4548) may encode a polypeptide of 483 aa; it overlaps ORF 1a by 10 nt and was in reading frame +1, and its first AUG codon was located 68 nt downstream of the ORF 1a termination codon. ORF 2 (nt 4472 to 6619), which may be present also in the subgenonic RNA (Fig. 3), began with an initiation codon at nt 4571 and ended with a stop codon 308 bases upstream from the 3′ end. ORF 2 was 2,148 nt in length and may encode a polypeptide of 683 aa.
FIG. 2.
Schematic representation of the ANV genome. Open boxes, ORFs. The locations of three ORFs, predicted transmembrane helices (MB), protease (Pro), nuclear localization signal (NLS), ribosomal frameshift structure (RFS), RNA-dependent RNA polymerase (Pol), and stem-loop II-like motif (s2m) are indicated. Numbering is according to the ANV genonic sequence (accession no. AB033998).
TABLE 2.
Structure of 5′- and 3′-end NTRs and three ORFs of astroviruses
| Virusa | No. of nucleotides in:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 5′ NTR | ORF 1a | ORF 1a–1b overlap | ORF 1b | ORF 1b–2 overlap | ORF 2 | 3′ NTR | Total | |
| ANV | 13 | 3,027 | 10 | 1,530 | 77 | 2,148 | 305 | 6,927 |
| HAst 1 | 85 | 2,889 | 70 | 1,557 | 221 | 2,574 | 80 | 6,813 |
| HAst 2 | 82 | 2,841 | 70 | 1,557 | 113 | 2,496 | 82 | 6,797 |
A potential ribosomal frameshift signal was identified in the small overlapping region between ORF 1a and 1b, consisting of the “shifty” heptanucleotide (AAAAAAC) from nt 3022 to 3028, followed by a stem-loop structure from nt 3035 to 3052 (Fig. 2 and 5).
FIG. 5.
Nucleotide sequence and predicted RNA secondary structure of the overlap region of ANV ORFs 1a and 1b. The deduced amino acid sequences encoded by ORF 1a, 1b, and 1a-1b surrounding the frameshift site are shown (17). The putative frameshift site (shifty heptanucleotide sequence) is underlined, and the termination codon of ORF 1a is boxed.
RNA blot hybridization.
By using a 3′-end probe in the Northern blot hybridization analysis, two RNA bands of 7.5 and 3.0 kb were identified in the ANV-infected cells at 16 and 24 h p.i. On the other hand, only one large RNA (7.5 kb) was observed with a 5′-end probe (Fig. 3).
Comparison of the sequence.
Comparison of the nucleotide sequence and amino acid sequence of ANV with sequences of other positive-strand RNA viruses identified a region of similarity with HAst that included the putative RdRp, serine protease, and 3′-end motif (17, 18, 21, 32, 33, 48). The nucleotide sequences of the ANV genonic RNA of ORF 1a, 1b, and 2 were compared with those of the HAst (Table 3). Between ANV (G-4260 strain) and HAst, the genonic composition was very similar except for the 5′- and 3′-end NTRs, but the homologies of ORFs 1a, 1b, and 2 were not very high. The highest amino acid homology was found in the product of ORF 1b (41.9%) (Table 3). Although the homologies were low, the typical amino acid motifs of serine protease and RdRp encoded by ORF 1a and 1b, respectively, were conserved (Fig. 4) (21).
TABLE 3.
Nucleotide and amino acid homology of three ORFs between ANV and human astroviruses
| Virusesa | % Nucleotide (amino acid) homology for:
|
||
|---|---|---|---|
| ORF 1a | ORF 1b | ORF 2 | |
| ANV/HAst 1 | 45.5 (20.3) | 53.7 (41.9) | 46.0 (25.8) |
| ANV/HAst 2 | 45.7 (20.3) | 53.3 (41.9) | 44.5 (26.9) |
| HAst 1/HAst 2 | 85.9 (93.8) | 93.2 (97.7) | 69.0 (68.3) |
Accession numbers of viruses are given in the footnote of Table 2.
FIG. 4.
Amino acid sequence homology of the predicted functional domains of ANV (G-4260) with those of HAst 1 (A/88 Newcastle). (A) Putative 3C-like serine protease domains (ORF 1a). ∗∗, putative catalytic residues; !, residues implicated in substrate binding (16); ∗, identical residues; colons, similar residues. (B) Putative RNA-dependent RNA polymerase domains (ORF 1b). Consensus 1 shows amino acid residues that are conserved in at least 80% of the polymerases of supergroup I (16, 19). U, bulky aliphatic residue (I, L, M, or V); @, aromatic residue, (F, Y, or W); &, bulky hydrophobic residue (aliphatic or aromatic); ·, any residue. Residues conserved in the (putative) polymerases of all positive-strand RNA viruses of eukaryotes are in boldface. Distances between the aligned conserved motifs and from the protein termini are indicated.
ORF 1a of ANV encoded a putative serine protease motif which was very similar to those of astroviruses, especially in having a serine at the active site (Fig. 4A), four transmembrane helices (aa 190 to 206, 300 to 316, 324 to 340, and 370 to 386), and a nuclear localization signal (aa 719 to 735) (Fig. 2). However, like those of HAst, it did not have a classical helicase-like motif (17, 48).
The characteristic YGDD motif (Fig. 4B) found in the RdRps (17, 20, 24, 36) was encoded by ORF 1b of ANV beginning at nt 4132. This region in which the YGDD motif was located contained the eight conserved motifs typical of the positive-strand RNA virus RdRps, indicating that it belongs to the so-called supergroup I, which includes the polymerases of picornaviruses, caliciviruses, and several other groups of plant viruses (17, 21).
The putative frameshifting signal of ANV was composed of 31 nt (Fig. 5) and was smaller than that of HAst (37 nt) (25). It showed a resemblance to that at the Gag-Pro junction of mouse mammary tumor virus rather than those of some viruses such as HAst, infectious bronchitis virus (IBV), and human immunodeficiency virus type 1, and it fit perfectly the simultaneous tRNA slippage model of −1 frameshifting described for the synthesis of Gag-related polyproteins (4). The ribosomal frameshifting is a common expression mechanism of some positive-strand RNA viruses (2, 6, 25, 38, 51).
ORF 2, which may encode the structural proteins, encoded a product with 25.8% amino acid homology to that of HAst 1 (Table 3).
A common RNA motif (35 nt) in the 3′ end of the genome of astroviruses was identified at nt 6707 to 6739 (90.9% homology) (Fig. 2) (18, 32).
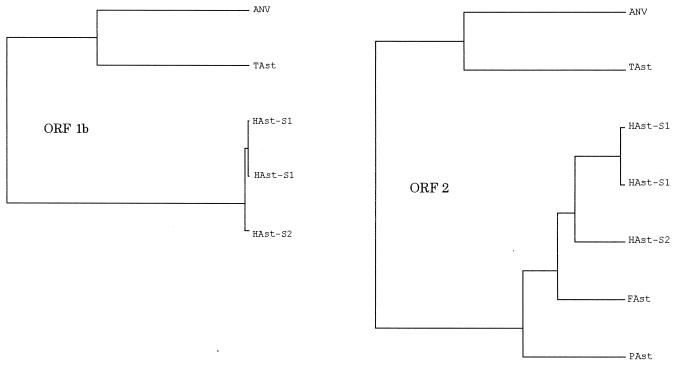
To gain further insight into the evolutionary relationship of ANV with other astroviruses, we generated a tentative phylogenetic tree by the amino acid sequence encoded by ORF 1b and ORF 2 of the astroviruses. The results showed that ANV constitutes a distinct evolutionary lineage not closely associated with mammalian astroviruses and that turkey astrovirus is the closest virus to ANV (Fig. 6).
FIG. 6.
Phylogenetic relationship for ORF 1b (RdRp) and ORF 2 (structural polyprotein) of astroviruses. Amino acid sequences were analyzed using GENETYX-WIN, version 4 (Software Development Co., Ltd.). The phylogenetic trees were constructed by the unweighted pair group method by arithmetic averaging. The following nucleotide sequences were obtained from GenBank/EMBL/DDBJ: HAst 1, Z25771 and L23513; HAst 2, L13745; FAst (feline astrovirus), AF056197; PAst (porcine astrovirus), AB037272; TAst (turkey astrovirus), AF206663.
Morphology and physicochemical properties of ANV.
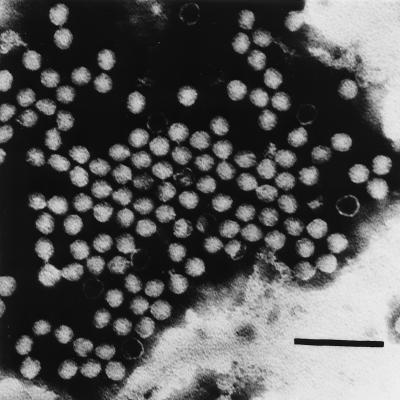
In ultrathin sections, ANV particles were visible in the cytoplasm of infected cells as large aggregates (14). Negative-staining observation of purified ANV demonstrated that the particles were spherical and about 30 nm in diameter, but the five- or six-pointed star-like structure, a characteristic of the astroviruses (13, 26, 39), was not observed, although some empty particles were also observed (Fig. 7). The unstable character of ANV in CsCl solution was reproduced.
FIG. 7.
Electron micrograph of purified ANV (G-4260 strain) particles. Bar = 100 nm.
Infectivity in vitro of RNA transcribed from cDNA and cDNA clones of ANV (G-4260 strain) in CK and BHK cells.
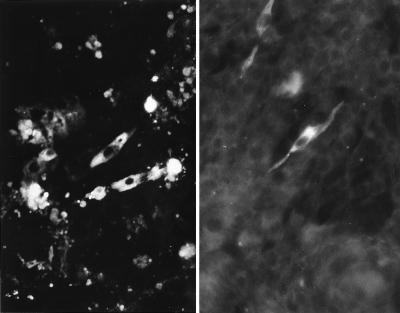
Beginning 48 h posttransfection (p.t.), ANV antigens were detected in the cytoplasm of the transfected CK cells by FA tests using anti-ANV chicken serum (Fig. 8). Typical ANV CPEs were observed at 48 h p.t. for RNA-transfected CK cells and at 72 h p.t. for cDNA-transfected CK cells. The culture supernatant contained infectious ANV at a titer of 106.5 50% tissue culture infective doses (TCID50)/ml at 96 h after RNA transfection.
FIG. 8.
Immunofluorescence of CK (left) and BHK (right) cells transfected with in vitro-transcribed RNA from pANV-750 or a plasmid which can transcribe the full-length ANV cDNA under the control of a cytomegalovirus promoter. Fluorescing fine granules are disseminated in the cytoplasm of the cells. The viruses produced in these cells were transmissible to CK cells 96 h p.t. Magnification, ×360.
The in vitro RNA transcript and the cDNA of ANV were also transfected onto a rodent cell line, BHK cells, which were largely refractory to ANV infection. ANV antigens were also detected in the transfected BHK cells from 48 to 96 h p.t. without CPE (Fig. 8). The culture fluid contained 103.0 TCID50/ml for CK cells, but recovered viruses from transfected BHK cells could not be repeatedly passaged on BHK cells.
To further confirm the specificity of the full-length cDNA clone (pANV-750) (Fig. 1), we compared the nucleotide sequences of the 5′-end regions of ANV recovered from the transfected CK cells with those of the wild-type G-4260 strain, because pANV-750 contained a point mutation at nt 281 (A to G). The 5′ ends of both the recovered and wild-type G-4260 strains were amplified by reverse transcription-PCR, and the PCR products were directly sequenced. This experiment confirmed that the recovered virus had indeed originated from pANV-750 and that this cDNA clone contained the full-length ANV genome.
DISCUSSION
The genome structure of ANV, which is essential for determining the taxonomy of viruses, had not been shown since the first isolation in 1976 even though the virus had been believed to be a member of the family Picornaviridae by morphological (Fig. 7) and physicochemical analyses (31, 52). This study indicated that the ANV genome consists of approximately 7,000 nt with three ORFs, ORF 1a, 1b, and 2 (Fig. 2). ORF 1a encoded a 3C-like serine protease motif, whereas the ORF 1b encoded a viral RdRp motif, and a ribosomal frameshift motif was also present between the two ORFs. On the other hand, ORF 2 of ANV, which may encode the capsid precursor polyprotein, encoded a product with 26% amino acid homology to those of HAst (17, 34, 48). As shown for the HAst, ORF 2 of ANV is likely expressed from subgenonic-size RNA (29, 34), which was detected in the ANV-infected cells, as well as genonic-size RNA (Fig. 3). The genome organization of ANV is apparently identical to that of HAst (Fig. 2) (3). The sequence analyses clearly demonstrated that ANV is not a Picornavirus but a member of the family Astroviridae (31, 33, 35).
ANV differs from previously described mammalian astroviruses in that trypsin is not required for growth in tissue culture (52). Trypsin-dependent replication of astroviruses in vitro is a very specific feature (3, 42, 47, 50). Virus particles produced in the absence of trypsin are minimally infectious. Monroe et al. demonstrated that the trypsin-free virus particles were composed of the precursor capsid protein (34). Bass and Qiu clearly proved that conversion of the large precursor protein (79 kDa) to three small mature capsid proteins (34, 29, and 26 kDa) correlated with enhanced infectivity by trypsin treatment (1). It is not clear why ANV does not require trypsin to replicate in CK cell culture in spite of being an astrovirus. One of the reasons might be that the CK cell is not a lined cell but a primary culture cell.
Picornaviruses can usually be purified with CsCl ultracentrifugation, but ANV cannot. The unstable property of ANV in CsCl solution was also described for HAst 1 but not for HAst 2 (29). Some animal astroviruses of bovine, ovine, porcine, feline, turkey, and duck origin have already been reported, and some of them are classified as Astroviridae mainly from their morphological properties (11, 12, 33, 40, 42, 50; R. E. Gough, M. S. Collins, E. Borland, and L. F. Keymer, Letter, Vet. Rec. 114:279, 1984). ANV particles did not show the typical star-like motif (Fig. 7); however, it is known that only minor populations of virions exhibit this star-like morphology, and they are visualized in stool specimens as star-like particles (26, 33, 39).
ANV is the first astrovirus which was not identified by the morphological character but by genonic analyses. The first AUG codon of ORF 1a remains to be identified because there are not typical motifs. The 5′-end NTR of the ANV G-4260 strain (13 nt) was short if the virus was using the first methionine (GXXAUGG) (22) in comparison with those of HAst (80 to 82 nt). When the virus uses the first methionine (AXXAUGG) (22), ORF 2 may encode a capsid protein precursor of 683 aa with a predicted molecular mass of 73.9 kDa, which is smaller than those of HAst (88 to 90 kDa) (17, 34). On the other hand, although the 3′-end NTR of ANV was more than three or four times the lengths of those of other astroviruses (Table 2), the common RNA motif (35 nt) of astroviruses was identified at nt 6707 to 6739 (90.9% homology) (18, 32). This common RNA motif is also found in the 3′ ends of the genomes of avian IBV and an equine rhinovirus (18). However, the position of the motif, which was named a stem-loop II-like motif (s2m) (Fig. 2), was different somehow in different viruses. In HAst, this motif contains the stop codon of ORF (18). IBV is a coronavirus with a 27.6-kb plus-strand RNA genome containing a 0.3- to 0.5-kb 3′ NTR and a poly(A) tail (49). Different strains have different tissue tropisms, infecting respiratory, reproductive, and gastrointestinal organs as well as the kidneys of chickens (44, 45). We also suggest that this conserved RNA motif in these different viruses might have resulted from a recombinational event during coinfection, because both ANV and IBV replicate in the chicken kidneys and gut (14, 18, 45). Of interest, astrovirus-like particles have been reported (Gough et al., Vet. Rec. 114:279) in association with fatal hepatitis in ducklings, suggesting a possible hepatic tropism for this virus.
Our study showed that ANV was genetically distinct from other astroviruses (Fig. 6). The RdRp polyprotein sequence of ANV (ORF 1b) had a 41.9% overall amino acid homology with that of HAst, while the polyprotein sequences encoded by ANV ORF 1a and 2 had 20 and 26% amino acid homology, respectively (Table 3). Turkey astrovirus was the closest virus to ANV. ORF 1a, 1b, and 2 of ANV encoded products with 29.9, 53.8, and 32.3% overall amino acid homology to those of turkey astrovirus (AF206663). Although the antigenic studies remain, from the comparison of the sequence and biological properties of these two viruses, they seem different (46). In the current taxonomy of picornaviruses, an amino acid homology of the RdRp lower than 60% is one of the important parameters to assign a new genus along with the genome structure and the biological character (19, 37).
The present findings strongly support the classification of ANV as a new genus of the family Astroviridae, and the virus group of Astroviridae can cause nephritis in chickens, in addition to gastroenteritis.
The ANV infectious cDNA clone described here should prove useful for defining the minimal and optimal requirements for the use of astrovirus groups as an expression vector and for developing a packaging system for vector RNA (10), and this cDNA clone would greatly facilitate and enhance studies on the replication strategy and the pathogenesis of the astroviruses in vitro and in the original host.
REFERENCES
- 1.Bass D M, Qiu S. Proteolytic processing of the astrovirus capsid. J Virol. 2000;74:1810–1814. doi: 10.1128/jvi.74.4.1810-1814.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brierley I, Digard P, Inglis S C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter M J, Willcocks M M. The molecular biology of astroviruses. Arch Virol Suppl. 1996;12:277–285. doi: 10.1007/978-3-7091-6553-9_30. [DOI] [PubMed] [Google Scholar]
- 4.Chamorro M, Parkin N, Varmus H E. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc Natl Acad Sci USA. 1992;89:713–717. doi: 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connor T J, McNeilly F, McFerran J B, McNulty M S. A survey of avian sera from Northern Ireland for antibody to avian nephritis virus. Avian Pathol. 1987;16:15–20. doi: 10.1080/03079458708436348. [DOI] [PubMed] [Google Scholar]
- 6.den Boon J A, Snijder E J, Chirnside E D, de Vries A A, Horzinek M C, Spaan W J. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frazier J A, Howes K, Reece R L, Kidd A W, Cavanagh D. Isolation of noncytopathic viruses implicated in the aetiology of nephritis and baby chick nephropathy and serologically related to avian nephritis virus. Avian Pathol. 1990;19:139–160. doi: 10.1080/03079459008418663. [DOI] [PubMed] [Google Scholar]
- 8.Furuta K, Ohashi H, Obana J, Sato S. Performance of 3 successive generations of specified-pathogen-free chickens maintained as a closed flock. Lab Anim. 1980;14:107–112. doi: 10.1258/002367780780942773. [DOI] [PubMed] [Google Scholar]
- 9.Garrett J K, Davis R B, Ragland W L. Enzyme-linked immunosorbent assay for detection of antibody to avian encephalomyelitis virus in chickens. Avian Dis. 1984;28:117–130. [PubMed] [Google Scholar]
- 10.Geigenmuller U, Ginzton N H, Matsui S M. Construction of a genome-length cDNA clone for human astrovirus serotype 1 and synthesis of infectious RNA transcripts. J Virol. 1997;71:1713–1717. doi: 10.1128/jvi.71.2.1713-1717.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harbour D A, Ashley C R, Williams P D, Gruffydd-Jones T J. Natural and experimental astrovirus infection of cats. Vet Rec. 1987;120:555–557. doi: 10.1136/vr.120.23.555. [DOI] [PubMed] [Google Scholar]
- 12.Herring A J, Gray E W, Snodgrass D R. Purification and characterization of ovine astrovirus. J Gen Virol. 1981;53:47–55. doi: 10.1099/0022-1317-53-1-47. [DOI] [PubMed] [Google Scholar]
- 13.Imada T. Avian nephritis virus infection. In: McFerran J B, McNulty M S, editors. Virus infections of birds. Vol. 4. Amsterdam, The Netherlands: Elsevier Science Publishers; 1993. pp. 479–483. [Google Scholar]
- 14.Imada T, Kawamura H. Infectious nephritis. In: Calnek B W, Barnes H J, Beard C W, McDougald L R, Saif Y M, editors. Diseases of poultry. 10th ed. Ames: Iowa State University Press; 1997. pp. 761–765. [Google Scholar]
- 15.Imada T, Yamaguchi S, Kawamura H. Pathogenicity for baby chicks of the G-4260 strain of the picornavirus “avian nephritis virus.”. Avian Dis. 1979;23:582–588. [PubMed] [Google Scholar]
- 16.Imada T, Yamaguchi S, Miura N, Kawamura H. Antibody survey against avian nephritis virus among chickens in Japan. Natl Inst Anim Health Q (Tokyo) 1980;20:79–80. [PubMed] [Google Scholar]
- 17.Jiang B, Monroe S S, Koonin E V, Stine S E, Glass R I. RNA sequence of astrovirus: distinctive genonic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc Natl Acad Sci USA. 1993;90:10539–10543. doi: 10.1073/pnas.90.22.10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jonassen C M, Jonassen T O, Grinde B. A common RNA motif in the 3′ end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus. J Gen Virol. 1998;79:715–718. doi: 10.1099/0022-1317-79-4-715. [DOI] [PubMed] [Google Scholar]
- 19.Kaku Y, Yamada S, Murakami Y. Sequence determination and phylogenetic analysis of RNA-dependent RNA polymerase (RdRp) of the porcine enterovirus 1 (PEV-1) Talfan strain. Arch Virol. 1999;144:1845–1852. doi: 10.1007/s007050050709. [DOI] [PubMed] [Google Scholar]
- 20.Kamer G, Argos P. Primary structural comparison of RNA-dependent RNA polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984;12:7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA virus: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 22.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurtz J B, Lee T W. Astroviruses: human and animal. Ciba Found Symp. 1987;128:92–107. doi: 10.1002/9780470513460.ch6. [DOI] [PubMed] [Google Scholar]
- 24.Lewis T L, Greenberg H B, Herrmann J E, Smith L S, Matsui S M. Analysis of astrovirus serotype 1 RNA, identification of the viral RNA-dependent RNA polymerase motif, and expression of a viral structural protein. J Virol. 1994;68:77–83. doi: 10.1128/jvi.68.1.77-83.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis T L, Matsui S M. Astrovirus ribosomal frameshifting in an infection-transfection transient expression system. J Virol. 1996;70:2869–2875. doi: 10.1128/jvi.70.5.2869-2875.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madeley C R, Cosgrove B P. 28nm particles in faeces in infantile gastroenteritis. Lancet. 1975;ii:451–452. doi: 10.1016/s0140-6736(75)90858-2. [DOI] [PubMed] [Google Scholar]
- 27.Madeley C R, Cosgrove B P. Viruses in infantile gastroenteritis. Lancet. 1975;ii:124. doi: 10.1016/s0140-6736(75)90020-3. [DOI] [PubMed] [Google Scholar]
- 28.Marvi P, Knowles N J, Mockett A P A, Britton P, Brown T D K, Cavanagh D. Avian encephalomyelitis virus is a picornavirus and is most closely related to hepatitis A virus. J Gen Virol. 1999;80:653–662. doi: 10.1099/0022-1317-80-3-653. [DOI] [PubMed] [Google Scholar]
- 29.Matsui S M, Kim J P, Greenberg H B, Young L M, Smith L S, Lewis T L, Herrmann J E, Blacklow N R, Dupuis K, Reyes G R. Cloning and characterization of human astrovirus immunoreactive epitopes. J Virol. 1993;67:1712–1715. doi: 10.1128/jvi.67.3.1712-1715.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNulty M S, Connor T J, McNeilly F, McFerran J B. Biological characterisation of avian enteroviruses and enterovirus-like viruses. Avian Pathol. 1990;19:75–87. doi: 10.1080/03079459008418658. [DOI] [PubMed] [Google Scholar]
- 31.Minor P D, Brown F, Domingo E, Hoey E, King A, Knowles N, Lemon S, Palmenberg A, Rueckert R, Stanway G, Wimmer E, Yin-Murphy M. Picornaviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy: sixth report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer-Verlag; 1995. pp. 329–336. [Google Scholar]
- 32.Monceyron C, Grinde B, Jonassen T O. Molecular characterisation of the 3′-end of the astrovirus genome. Arch Virol. 1997;142:699–706. doi: 10.1007/s007050050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monroe S S, Carter M J, Herrmann J E, Kurtz J B, Matsui S M. Family astroviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Classification and nomenclature of viruses: sixth report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer-Verlag; 1995. pp. 364–367. [Google Scholar]
- 34.Monroe S S, Jiang B, Stine S E, Koopmans M, Glass R I. Subgenonic RNA sequence of human astrovirus supports classification of Astroviridae as a new family of RNA viruses. J Virol. 1993;67:3611–3614. doi: 10.1128/jvi.67.6.3611-3614.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy F A. Virus taxonomy. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 15–57. [Google Scholar]
- 36.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pringle C R. Virus taxonomy at the XIth International Congress of Virology, Sydney, Australia, 1999. Arch Virol. 1999;144:2065–2070. doi: 10.1007/s007050050728. [DOI] [PubMed] [Google Scholar]
- 38.Prufer D, Tacke E, Schmitz J, Kull B, Kaufmann A, Rohde W. Ribosomal frameshifting in plants: a novel signal directs the −1 frameshift in the synthesis of the putative viral replicase of potato leafroll luteovirus. EMBO J. 1992;11:1111–1117. doi: 10.1002/j.1460-2075.1992.tb05151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Risco C, Carrascosa J L, Pedregosa A M, Humphrey C D, Sanchez-Fauquier A. Ultrastructure of human astrovirus serotype 2. J Gen Virol. 1995;76:2075–2080. doi: 10.1099/0022-1317-76-8-2075. [DOI] [PubMed] [Google Scholar]
- 40.Saif Y M, Saif L J, Hofacre C L, Hayhow C, Swayne D E, Dearth R N. A small round virus associated with enteritis in turkey poults. Avian Dis. 1990;34:762–764. [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Shimizu M, Shirai J, Narita M, Yamane T. Cytopathic astrovirus isolated from porcine acute gastroenteritis in an established cell line derived from porcine embryonic kidney. J Clin Microbiol. 1990;28:201–206. doi: 10.1128/jcm.28.2.201-206.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shirai J, Nakamura K, Shinohara K, Kawamura H. Pathogenicity and antigenicity of avian nephritis isolates. Avian Dis. 1991;35:49–54. [PubMed] [Google Scholar]
- 44.Siddell S, Wege H, ter Meulen V. The biology of coronaviruses. J Gen Virol. 1983;64:761–776. doi: 10.1099/0022-1317-64-4-761. [DOI] [PubMed] [Google Scholar]
- 45.Siller W G. Renal pathology of the fowl—a review. Avian Pathol. 1981;10:187–262. doi: 10.1080/03079458108418474. [DOI] [PubMed] [Google Scholar]
- 46.Thouvenelle M L, Haynes J S, Reynolds D L. Astrovirus infection in hatchling turkeys: histologic, morphometric, and ultrastructural findings. Avian Dis. 1995;39:328–336. [PubMed] [Google Scholar]
- 47.Willcocks M M, Ashton N, Kurtz J B, Cubitt W D, Carter M J. Cell culture adaptation of astrovirus involves a deletion. J Virol. 1994;68:6057–6058. doi: 10.1128/jvi.68.9.6057-6058.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willcocks M M, Brown T D, Madeley C R, Carter M J. The complete sequence of a human astrovirus. J Gen Virol. 1994;75:1785–1788. doi: 10.1099/0022-1317-75-7-1785. [DOI] [PubMed] [Google Scholar]
- 49.Williams A K, Wang L, Sneed L W, Collisson E W. Analysis of a hypervariable region in the 3′ non-coding end of the infectious bronchitis virus genome. Virus Res. 1993;28:19–27. doi: 10.1016/0168-1702(93)90086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woode G N, Gourley N E, Pohlenz J F, Liebler E M, Mathews S L, Hutchinson M P. Serotypes of bovine astrovirus. J Clin Microbiol. 1985;22:668–670. doi: 10.1128/jcm.22.4.668-670.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiong Z, Kim K H, Kendall T L, Lommel S A. Synthesis of the putative red clover necrotic mosaic virus RNA polymerase by ribosomal frameshifting in vitro. Virology. 1993;193:213–221. doi: 10.1006/viro.1993.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamaguchi S, Imada T, Kawamura H. Characterisation of a picornavirus isolated from broiler chicks. Avian Dis. 1979;23:571–581. [PubMed] [Google Scholar]
- 53.Yamashita T, Sakae K, Tsuzuki H, Suzuki Y, Ishikawa N, Takeda N, Miyamura T, Yamazaki S. Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans. J Virol. 1998;72:8408–8412. doi: 10.1128/jvi.72.10.8408-8412.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]