Abstract
Interspecies transmission of influenza A viruses circulating in wild aquatic birds occasionally results in influenza outbreaks in mammals, including humans. To identify early changes in the receptor binding properties of the avian virus hemagglutinin (HA) after interspecies transmission and to determine the amino acid substitutions responsible for these alterations, we studied the HAs of the initial isolates from the human pandemics of 1957 (H2N2) and 1968 (H3N2), the European swine epizootic of 1979 (H1N1), and the seal epizootic of 1992 (H3N3), all of which were caused by the introduction of avian virus HAs into these species. The viruses were assayed for their ability to bind the synthetic sialylglycopolymers 3′SL-PAA and 6′SLN-PAA, which contained, respectively, 3′-sialyllactose (the receptor determinant preferentially recognized by avian influenza viruses) and 6′-sialyl(N-acetyllactosamine) (the receptor determinant for human viruses). Avian and seal viruses bound 6′SLN-PAA very weakly, whereas the earliest available human and swine epidemic viruses bound this polymer with a higher affinity. For the H2 and H3 strains, a single mutation, 226Q→L, increased binding to 6′SLN-PAA, while among H1 swine viruses, the 190E→D and 225G→E mutations in the HA appeared important for the increased affinity of the viruses for 6′SLN-PAA. Amino acid substitutions at positions 190 and 225 with respect to the avian virus consensus sequence are also present in H1 human viruses, including those that circulated in 1918, suggesting that substitutions at these positions are important for the generation of H1 human pandemic strains. These results show that the receptor-binding specificity of the HA is altered early after the transmission of an avian virus to humans and pigs and, therefore, may be a prerequisite for the highly effective replication and spread which characterize epidemic strains.
Wild aquatic birds are the natural reservoirs for influenza A viruses of all known hemagglutinin (HA) and neuraminidase (NA) subtypes. Such viruses are occasionally transmitted to other species, including domestic poultry, sea mammals, pigs, horses, and humans, where they can cause severe outbreaks of influenza (reviewed in reference 40). For example, the 1957 and 1968 influenza pandemics originated from avian-human reassortant viruses (H2N2 “Asian” pandemic virus in 1957; H3N2 “Hong Kong” pandemic virus in 1968). Recent introduction of an H1N1 avian virus into pigs led to the emergence of a so-called avian-like swine virus lineage (36). Although progenitors of H1N1 human viruses and H1N1 “classical” swine viruses have not been unambiguously identified, they too are thought to have originated from avian virus precursors.
All influenza A viruses bind to cellular glycoconjugates containing terminal sialic acid, but the exact molecules (glycoproteins or glycolipids) that serve as biological receptors of influenza viruses in birds and other species remain to be determined. Cellular receptors of influenza viruses appear to differ among distinct animal species because the binding specificity of viruses varies considerably depending on the host animals from which the viruses are isolated. Namely, human influenza A and B strains and swine influenza viruses preferentially bind receptors that contain terminal 6′-sialyl(N-acetyllactosamine) residues (6′SLN; Neu5Acα2-6Galβ1-4GlcNAc), whereas avian and equine viruses bind poorly to 6′SLN, preferring instead the terminal 3′-sialylgalactose (Neu5Acα2-3Gal) moiety (8, 13, 23, 33; see also reference 28 for a review of earlier data). The molecular mechanisms by which influenza viruses distinguish between these sialyloligosaccharide determinants are poorly defined.
A comparison of the amino acid sequences of influenza A viruses from different hosts revealed six amino acids in the HA receptor-binding site, which are highly conserved among avian viruses (138A, 190E, 194L, 225G, 226Q, and 228G), but bear substitutions in human viruses (23). This finding suggested that mutations at these positions are required for adaptation of the avian virus HA to human hosts. However, the role of individual mutations at most of these positions in the alteration of the HA receptor-binding properties remains undefined.
Both H2 and H3 human viruses bear the same substitutions, 226Q→L and 228G→S, with respect to the avian consensus sequence (8). The single mutation 226L→Q in the H3 human virus HA changes its specificity from preferential Neu5Acα2-6Gal recognition to preferential Neu5Acα2-3Gal binding (31, 32). As was shown by site-directed mutagenesis, mutations at position 228 of the human H3 HA affect HA binding to erythrocytes (19, 39); however, the effects of such mutations on the ability of the HA to recognize the type of Neu5Ac-Gal linkage were not clearly determined. Interestingly, some H2N2 viruses isolated from humans during the first year of the 1957 pandemic contain 228G (17) as do most avian viruses, whereas some avian H3 viruses contain “human” 228S (2, 16). The receptor-binding properties of such atypical avian and human viruses have not been characterized. Thus, the contribution of substitutions at position 228 to the adaptation of avian viruses to human receptors remains unknown.
The currently circulating human H1N1 viruses are thought to have originated from an avian virus that was transmitted to humans at the beginning of this century and gave rise to the so-called Spanish influenza pandemic. At least four mutations in the conserved positions of the avian receptor-binding site (138, 190, 194, and 225) separate contemporary H1 human viruses from avian strains (23, 33), and those in positions 190 and 225 are also present in the recently sequenced H1N1 human viruses from 1918 (30). Effects of substitutions at these positions on the HA receptor-binding properties and their possible contribution to the adaptation of the H1 avian HA to human hosts are unclear.
Previous studies compared the receptor-binding specificities of influenza viruses that were already well adapted to their hosts, making it difficult to define minimal changes in the specificity of the avian HA required for efficient replication in a new species. Furthermore, because many mutations have been introduced into the HA of these viruses since transmission from birds, it has been difficult to determine the contribution of each mutation to the binding specificity of this protein. We compare here the receptor-binding properties of the earliest available influenza viruses isolated during the 1957 and 1968 human pandemics and during swine and seal epizootics with those of closely related avian viruses of the H1, H2, and H3 subtypes. Two major questions were addressed: what are the earliest detectable alterations in the receptor-binding specificity of the avian HA after introduction into a new host, and what are the most critical mutations in the HA that account for these changes?
MATERIALS AND METHODS
Viruses.
The viruses used in this study and their abbreviations are listed in Tables 1 to 3. H2N2 human virus isolates were from the repository of the Centers for Disease Control and Prevention, Atlanta, Ga. H1N1 avian-like swine viruses were from the repository of the Instituto Superiore di Sanita, and seal viruses were from the repository of the University of Wisconsin-Madison. The R4 reassortant virus, which carries the HA of A/Udorn/307/72 (H3N2) with the mutation 226L→Q, with all other genes from A/mallard/New York/6750/78 (H2N2), was described previously (24). The R2 reassortant with two mutations (226L→Q and 228S→G) in the HA of A/Udorn/307/72 was generated by reverse genetics (39). The other viruses were from the repository of St. Jude Children's Research Hospital. All viruses were grown in 10- to 11-day-old chicken eggs. Allantoic fluids were clarified by low-speed centrifugation, and virus was pelleted by high-speed centrifugation, followed by resuspension in 0.1 M Tris buffer (pH 7.2) containing 50% glycerol, and stored at −20°C.
TABLE 1.
Binding of sialylglycopolymers to H3 influenza viruses
| Virus strain | Abbreviation | Apparent association constant (Kass, μM−1 Neu5Ac)a
|
Relative affinityb | |
|---|---|---|---|---|
| 3′SL-PAA | 6′SLN-PAA | |||
| Avian | ||||
| Duck/Hokkaido/10/85 | dkHok10/85 | 60 | 3 | 20 |
| Duck/Hokkaido/9/85 | dkHok9/85 | 60 | 3 | 20 |
| Duck/Hokkaido/7/82 | dkHok7/82 | 60 | 2 | 30 |
| Duck/Hokkaido/33/80 | dkHok33/80 | 60 | 3 | 20 |
| Duck/Hokkaido/8/80 | dkHok8/80 | 20 | 1 | 20 |
| Duck/Ukraine/1/63 | dkUkr/63 | 25 | 1.5 | 17 |
| Mallard/NY/6874/78 | maNY6874/78 | 30 | 1.5 | 20 |
| Duck/Memphis/928/74 | dkMem928/74 | 70 | <0.5 | >140 |
| Human | ||||
| Aichi/2/68 | Aichi2/68 | 2 | 10 | 0.2 |
| Memphis/102/72 | Mem102/72 | 3 | 20 | 0.15 |
| Los Angeles/2/87 | LA2/87 | 2 | 30 | 0.07 |
| Shanghai/11/89 | Shang11/89 | 2 | 20 | 0.1 |
| Udorn/72 and variantsc | ||||
| Udorn/307/72 (226L, 228S) | Udorn/72 | 4 | 13 | 0.3 |
| R4 (226Q, 228S) | Udorn/72/R4 | 30 | 2 | 15 |
| R2 (226Q, 228G) | Udorn/72/R2 | 65 | 2 | 33 |
| Seal | ||||
| Seal/MA/3908/92 | se3908/92 | 50 | 1 | 50 |
| Seal/MA/3911/92 | se3911/92 | 40 | 1.5 | 27 |
| Seal/MA/3984/92 | se3984/92 | 5.5 | 1 | 5.5 |
| Seal/MA/4007/93 | se4007/93 | 50 | 1 | 50 |
The binding assay was performed and the apparent association constants of virus complexes with sialylglycopolymers were calculated as described in Materials and Methods and in the text.
Calculated as Kass[3′SL-PAA]/Kass[6′SLN-PAA].
TABLE 3.
Binding of sialylglycopolymers to H1N1 influenza viruses
| Virus strain | Abbreviation | Apparent association constant (Kass, μM−1 Neu5Ac)a
|
Relative affinityb | |
|---|---|---|---|---|
| 3′SL-PAA | 6′SLN-PAA | |||
| Avian | ||||
| Turkey/MN/1661/81 | tyMN/81 | 60 | 1 | 60 |
| Mallard/TN/11464/85 | maTN/85 | 60 | 1 | 60 |
| Duck/Alberta/35/76 | dkAlb/76 | 70 | 1 | 70 |
| Duck/Bavaria/1/77 | dkBav/77 | 70 | 1 | 70 |
| Duck/Australia/749/80 | dkAus/80 | 45 | 1 | 45 |
| Avian-like swine | ||||
| Swine/Arnsberg/6554/79 | swArn/79 | 25 | 15 | 1.7 |
| Swine/Netherlands/3/80 | swNet/80 | 45 | 10 | 4.5 |
| Swine/Germany/2/81 | swGer/81 | 35 | 10 | 3.5 |
| Swine/Belgium/1/83 | swBel/83 | 35 | 10 | 3.5 |
| Swine/Netherlands/12/85 | swNet/85 | 30 | 8 | 3.8 |
| Swine/Italy/671/87 | swIta/87 | 9 | 15 | 0.6 |
| Swine/Germany/8533/91 | swGer/91 | 15 | 20 | 0.8 |
| Swine/Schles.-Holstein/1/92 | swHol/92 | 15 | 30 | 0.5 |
| Classical swine | ||||
| Turkey/MO/1/81 | tyMO/81 | 8 | 25 | 0.3 |
| Swine/Hokkaido/2/81 | swHok/81 | 4 | 14 | 0.3 |
| Swine/Italy/437/76 | swIta/76 | 10 | 25 | 0.4 |
| Turkey/NC/1/88 | tyNC/88 | 6 | 11 | 0.6 |
Sialosides and sialylglycopolymers.
Free N-acetylneuraminic acid (Neu5Ac), α-methyl glycoside of N-acetylneuraminic acid (MN; Neu5Acα2Me), and 3′-sialyllactose (3′SL; Neu5Acα2-3Galβ1-4Glc) were purchased from Sigma. 6′-Sialyl(N-acetyllactosamine) (6′SLN) was a gift from V. E. Piskarev, Nesmeyanov Institute of Organoelement Compounds, Moscow, Russia. The sialylglycopolymers 3′SL-PAA and 6′SLN-PAA, containing 20 mol% of 3′SL and 6′SLN, respectively, and 5 mol% of biotin attached to a soluble polyacrylamide carrier, were synthesized as described previously (3). The sialidase inhibitor zanamivir (2,3-didehydro-2,4-dideoxy-4-guanidino-N-acetyl-d-neuraminic acid; GG167) was kindly provided by R. Bethell, Glaxo Wellcome R&D.
Assay of virus-binding affinity for sialylglycopolymers.
The general protocol for this solid-phase receptor-binding assay was described previously (12). Modifications of the assay for the present study were limited to the use of synthetic sialylglycopolymers and of a biotin-streptavidin detection system. In brief, 50-μl aliquots of bovine fetuin solution in phosphate-buffered saline (PBS; 5 μg/ml) were incubated in 96-well polyvinyl chloride microplates (Costar) at 4°C overnight. The plates were washed with water and air dried. Concentrated virus stocks were diluted with PBS to an HA titer of 20 to 100. Then, 40 μl of virus solution was added to each well of the fetuin-coated microplates. After incubation at 4°C overnight, the plates were washed with ice-cold washing buffer (WB; 0.01% Tween 80 in 0.2× PBS). Serial twofold dilutions of sialylglycopolymers in the reaction buffer (RB; 0.02% Tween 80, 0.02% bovine serum albumin, 1 μM sialidase inhibitor GG167 in PBS) were added into the wells (20 μl/well), and the plates were incubated at 4°C for 2 h. After washing, streptavidin-peroxidase solution in RB (1/2,000) was added at 25 μl/well, and the plates were incubated at 4°C for 1 h. After washing, the peroxidase activity in the wells was assayed with o-phenylenediamine substrate solution. The absorbancies (at 490 nm) were determined with a model 3550 microplate reader (Bio-Rad), transferred to a PC using Microplate Manager 4.0 software (Bio-Rad), and processed with Microsoft Excel software. The data were converted to Scatchard plots (A490/C versus A490, where C is the concentration of the sialylglycopolymer and A490 is the absorbency in the corresponding well; see Fig. 1). The apparent association constants of virus complexes with sialylglycopolymers (Kass) were determined from the slopes of these plots and are expressed in micromolar amounts of Neu5Ac (with respect to concentration of sialic acid residues present in the solution). Because the assay includes extensive washing steps and is based on a complex multivalent binding of sialylglycopolymers to a virus with numerous HAs, these constants are not true equilibrium association constants. However, they provided a reliable and convenient measure of the relative affinity of viruses for the sialylglycopolymers. Thus, although the absolute values of the apparent association constants determined on different days varied up to fourfold, the relative affinities of the viruses were highly reproducible. The data presented in the tables are average values of two to three independent experiments performed on different days.
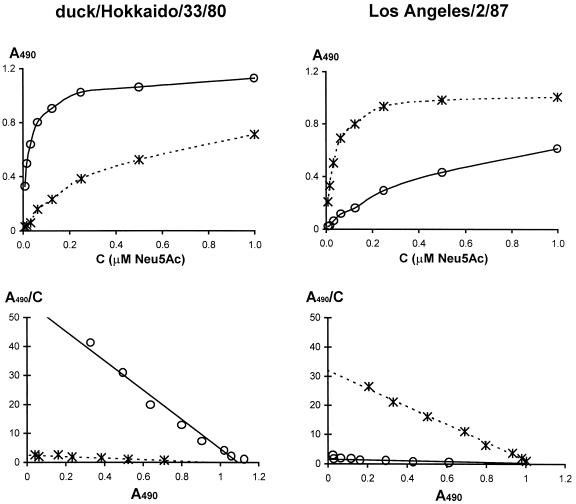
FIG. 1.
Binding of sialylglycopolymers 3′SL-PAA (solid lines) and 6′SLN-PAA (dotted lines) by H3 subtype influenza virus strains A/duck/Hokkaido/33/80 and A/Los Angeles/2/87. The binding assay is described in Materials and Methods. The upper panels represent the primary data (dependence of absorbancy in the wells, A490, versus concentration of the polymer); the lower panels show the corresponding Scatchard plots.
Assay of virus binding of sialic acid and sialosides.
The apparent association constants of the influenza virus complexes with low-molecular-mass receptor analogs were determined in a fetuin-binding inhibition assay, as previously described (12, 22). This assay is based on the competition between the receptor analog under study and a standard preparation of peroxidase-labeled fetuin for the binding sites on a solid-phase immobilized virus. The competitive reaction was performed for 1 h at 2 to 4°C in the presence of 1 μM GG167 to block the activity of viral neuraminidase.
Analysis of HA amino acid sequences.
The HA amino acid sequences were obtained from GenBank. The sequences were analyzed with the use of GeneDoc 2.3 software (25; Karl B. Nicholas and Hugh B. Nicholas, Jr. [GeneDoc, http://www.cris.com/∼Ketchup/genedoc.shtml]). Only partial sequences that covered the region of the receptor-binding site of the HA (amino acids 130 to 230 of HA1) were analyzed. This region was arbitrarily defined by a sphere with a 15-Å radius around the glycosidic oxygen atom of the bound sialic acid in the X31 HA complex with 3′-sialyllactose (1HGG structure; Protein DataBank). The H3 numbering system, in accord with the alignment of Nobusawa et al. (26), is used throughout this study.
Phylogenetic relationships among the H1N1 viruses from different hosts were estimated with the PHYLIP 3.572 software package (11; J. Felsenstein, Phylogeny Inference Package, version 3.5c [1993], Department of Genetics, University of Washington, Seattle [http://evolution.genetics.washington.edu/phylip.html]). The tree was obtained for the amino acid sequences of the whole HA1 by using a neighbor-joining algorithm and the Dayhoff PAM matrix; H2 HA was used as an outgroup. The TREEVIEW 1.5.2 program (27) was used to draw the tree.
The positions of distinct amino acid residues with respect to the virus receptor-binding site and bound sialyloligosaccharides were analyzed with the RasMol 2.6 (34) and WebLab ViewerPro 3.10 (Molecular Simulations, Inc., San Diego, Calif.) computer programs. The crystallographic structure of the X31 HA complex with pentasaccharide LSTc (10) was kindly provided by M. B. Eisen.
RESULTS
Assay of virus receptor-binding specificity.
To determine the receptor-binding specificity of influenza viruses in previous studies, we used two synthetic sialylglycopolymers, 3′SL-PAA and 6′SLN-PAA (carrying the Neu5Acα2-3Gal and Neu5Acα2-6Gal moieties, respectively), and a competitive binding assay (13). In the present study, we developed a direct binding assay by using biotin-labeled polymers. The viruses were immobilized in the wells of microtiter plates and were then incubated with variable concentrations of biotinylated sialylglycopolymers. The amount of bound polymer was determined with a streptavidin-peroxidase detection system. This assay is technically simpler than competitive assays, and the Scatchard transformation of the binding data enables easy determination of apparent association constants from the slopes of binding plots. Figure 1 shows examples of primary binding data and corresponding Scatchard plots. The 3′SL-containing polymer bound strongly to the duck virus duck/Hokkaido/33/80 (H3N8) and bound substantially more weakly to the human strain Los Angeles/2/87 (H3N2). By contrast, 6′SLN-PAA bound much better to the human strain than it did to the avian strain. This binding pattern agrees with results obtained for nonbiotinylated polymers in a competitive binding assay (13).
Alterations of the avian H3 HA at the beginning of the 1968 Hong Kong pandemic.
The reassortant virus possessing an avian H3 HA and all other genes from a previously circulating human virus caused the 1968 pandemic. A few mutations in the HA, including those at positions 144, 193, 226, and 228, were found to separate the earliest human isolate Aichi/2/68 from its putative avian virus precursor (2) (see Fig. 3 for the location of these mutations on the HA molecule). To determine the contribution of these mutations to the receptor-binding properties, we compared sialylglycopolymer-binding activities of viruses described by Bean et al. (2). All H3 avian viruses exhibited the same binding specificity, that is, they bound 3′SL-PAA more than an order of magnitude better than they did 6′SLN-PAA (Table 1). Three of the avian H3 viruses tested (duck/Hokkaido/8/80, mallard/NY/6874/78, and duck/Hokkaido/7/82) differed from other avian viruses by having arginine or serine instead of glycine in HA position 228 (Fig. 2, H3). The first two strains and duck/Ukraine/63 virus reproducibly showed two- to threefold lower affinities for 3′SL-PAA compared to duck/Hokkaido/7/82 and other H3 avian strains. Although duck/Ukraine/63 bears “avian” 226Q and 228G (amino acids conserved among the majority of avian HAs), it also has a nonconservative substitution, 227S→P, in the HA that can affect the structure of the region surrounding this residue. Because dk/Hokkaido/7/82 was reported to contain 228S (16) but did not differ in our experiments from H3 viruses with 228G, the HA sequence of the preparation of the virus we used in the binding studies was determined. The HA was found to contain glycine at position 228. These findings indicated that the presence of an “atypical” amino acid at position 228 (and 227) correlated with a lower affinity for 3′SL-PAA. Mutations 228G→R and 227S→P did not substantially increase the affinity for 6′SLN-PAA.
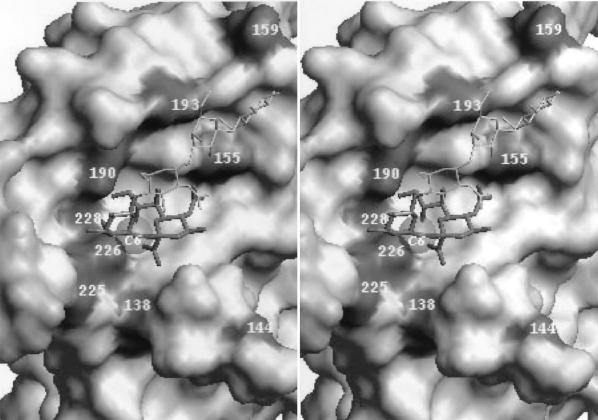
FIG. 3.
Positions of amino acids in the HA receptor-binding site that differ between early epidemic human and swine viruses and their closely related avian counterparts (stereo view). The figure is based on the crystallographic model of the X31 HA complex with the Neu5Acα2-6Gal-containing receptor analog LSTc (Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4Glc) (10). The solvent-accessible molecular surface of the protein is shown. The sialic acid residue and penultimate galactose ring of LSTc are displayed as thick stick bonds; the rest of the molecule is shown as a thin white line. The gray transparent sphere (C6) in close proximity to amino acid 226 represents the van der Waals surface of the C6′-carbon atom of Gal. The figure was generated with WebLab ViewerPro 3.10 (Molecular Simulations, Inc., San Diego, Calif.).
FIG. 2.
Partial HA amino acid sequences (HA1 positions 130 to 230) of influenza viruses that were tested for their binding to sialylglycopolymers. Full names of the virus strains are listed in Tables 1 to 3. Differences with respect to the top sequence are shown. The H3 numbering system is used; the position in the H1 HA that is absent from the H3 HA is indicated by an underscore. The RBS line shows the position of the amino acid with respect to the HA receptor-binding site. The “R” designates that the amino acid residue contacts either sialic acid or penultimate galactose in the X31 virus HA complex with 3′-sialyllactose (1HGG structure; Brookhaven Protein Database). The star indicates that an amino acid is within 15 Å of the C2 carbon atom of the sialic acid in the 1HGG structure. The figure was generated with GeneDoc 2.3 software (25).
All human H3 isolates, including the earliest, Aichi/2/68, bound to 6′SLN-PAA with a higher affinity and to 3′SL-PAA with a lower affinity than did all of the avian viruses tested. The Aichi/2/68 HA bears substitutions 144G, 182I, 193S, 226L, and 228S with respect to the avian HA consensus sequence (Fig. 2). The 182V→I mutation of Aichi/2/68 is unique and is absent in other human virus strains; hence, it is unlikely to be critical for the “human” receptor-binding phenotype. Because 144G is present in the HA of the avian virus mallard/NY/6874/78 that does not bind to 6′SLN-PAA any better than other avian viruses, it is probably not essential for the increased affinity of human viruses for 6′SLN-PAA. The duck/Memphis/74 strain reproducibly bound to 6′SLN-PAA more weakly than any other avian virus. This feature correlates with the presence of 193D in this HA receptor-binding site (RBS), whereas other avian viruses bear 193N, and the early human strains bear 193S. Residue 193 is relatively far from the region of the RBS, which accommodates the Neu5Acα2-6Gal moiety (10), but it can potentially interact with more distant saccharide rings of the Neu5Acα2-6Gal-terminated receptors and with the polymeric part of 6′SLN-PAA (Fig. 3). Although these findings suggest that mutations at position 193 can modulate the recognition of 6′SLN-containing receptors, substitutions in positions 226 and 228 appear to be primarily responsible for the differences in the receptor specificity between avian viruses and Aichi/2/68.
To further evaluate the effects of mutations 226Q→L and 228G→S on changes of the receptor-binding properties of avian HA, we analyzed laboratory variants of the human virus Udorn/307/72, which possessed distinct amino acids at these positions (Table 1). The R2 variant, for example, bears “avian” amino acids at both positions and is indistinguishable from the natural avian H3 viruses with respect to its binding to sialylglycopolymers. Thus, in the presence of 226Q/228G, other amino acids that differ from avian viruses (i.e., at positions 144, 155, 188, and 193) do not appear to substantially affect the avian virus-like receptor-binding specificity of the R2 virus. The R4 variant has a single mutation, 228G→S, with respect to R2. This substitution has no effect on the affinity of the HA for 6′SLN-PAA but reduces its affinity for 3′SL-PAA, similar to the effect of substitution 228G→R in the avian viruses described above. By contrast, the substitution 226Q→L (compare the R4 variant with its parent, Udorn/72) markedly changes the receptor-binding properties of the virus by both substantially increasing its affinity for 6′SLN-PAA and decreasing its affinity for 3′SL-PAA. These data show that the 226Q→L mutation primarily determines the change in recognition of the Neu5Ac-Gal linkage during interspecies transfer, whereas substitutions of glycine at position 228 of the avian HA have only limited effect on the Neu5Ac-Gal linkage specificity.
Alterations of the avian H2 HA at the beginning of the Asian pandemic.
The “Asian” pandemic of 1957 was caused by a reassortant virus that contained the genes of the surface glycoproteins and the PB1 protein from an H2N2 avian strain and the remainder from a human virus. We analyzed the receptor-binding properties (Table 2) and amino acid sequences (Fig. 2, H2) of the panel of H2 avian viruses (35) and the earliest known H2 human strains (17).
TABLE 2.
Binding of sialylglycopolymers to H2 influenza viruses
| Virus strain | Amino acid at position:
|
Abbreviation | Apparent association constant (Kass, μM−1 Neu5Ac)a
|
Relative affinityb | ||
|---|---|---|---|---|---|---|
| 226 | 228 | 3′SL-PAA | 6′SLN-PAA | |||
| Avian | ||||||
| Mallard/Potsdam/178/83 | Q | G | maPot/83 | 90 | 1.5 | 60 |
| Peking dk/Potsdam/1689/85 | Q | G | pdkPot/86 | 65 | 1 | 65 |
| Pintail/Praimorie/625/76 | Q | G | pinPraim/76 | 100 | 1 | 100 |
| Duck/Hong Kong/273/78 | Q | G | dkHK/78 | 50 | 1 | 50 |
| Mallard/MT/61 | Q | G | maMT/61 | 100 | 2 | 50 |
| Herring gull/DE/677/88 | Q | G | hguDE/88 | 110 | 2 | 55 |
| Gull/MD/19/77 | Q | G | guMD/77 | 40 | 1 | 40 |
| Mallard/ALB/353/88 | Q | G | maALB/88 | 95 | 1 | 95 |
| Guinea Fowl/NJ/3070/91 | Q | G | gfNJ/91 | 70 | 1 | 70 |
| Mallard/ONT/56/76 | Q | G | maONT/76 | 40 | 1.2 | 33 |
| Mallard/NY/6750/78 | Q | G | maNY/78 | 60 | 2 | 30 |
| Human | ||||||
| El Salvador/2/57 | Q | G | Sal/57 | 50 | 1 | 50 |
| Leningrad/134/57 | Q | G | Len/57 | 50 | 1 | 50 |
| RI/5−/57 | Q | G | RI5−/57 | 40 | 0.5 | 80 |
| Davis/1/57 | L | G | Dav/57 | 1.5 | 12 | 0.13 |
| Albany/7/57 | L | G | Alb/57 | 1 | 12 | 0.08 |
| RI/5+/57 | L | S | RI5+/57 | 1 | 25 | 0.04 |
| Albany/6/58 | L | S | Alb/58 | 1 | 30 | 0.03 |
| Malaya/16/58 | L | S | Mal/58 | 1 | 25 | 0.04 |
| Sao Paolo/3/59 | L | S | SP3/59 | 1 | 25 | 0.04 |
| Victoria/15681/59 | L | S | Vic/59 | 1.5 | 20 | 0.075 |
| Ohio/2/59 | L | S | Ohio/59 | 2 | 25 | 0.08 |
| Ann Arbor/6/60 | L | S | AA6/60 | 25 | 2.5 | 10 |
| Berlin/3/64 | L | S | Ber/64 | 5 | 30 | 0.17 |
All H2 avian viruses contained 226Q and 228G and bound strongly to 3′SL-PAA and weakly to 6′SLN-PAA. Among the H2N2 viruses isolated from humans in the first year of the pandemic, three strains (El Salvador/57, Leningrad/57, and RI/5−/57) carried the amino acids typical for avian viruses, 226Q and 228G, and displayed the avian virus-like receptor-binding phenotype. In contrast to these strains, Davis/57 contains only one amino acid substitution in the region of the receptor-binding site, 226Q→L, whereas Albany/57 contains two substitutions, 226Q→L and 186N→I. Both of these variants with 226L displayed a dramatic shift in receptor-binding activity, with their affinity for 6′SLN-PAA increasing ca. 10-fold and that for 3′SL-PAA decreasing >10-fold. Thus, these two strains are similar to the earliest human H3 pandemic strain, Aichi/2/68, with respect to their affinity for sialylglycopolymers. The RI/5+/57 strain and other H2 human viruses isolated in later years displayed a further twofold increase in affinity for 6′SLN-PAA; this feature correlates with the presence of serine at position 228 of the HA. Among the human H2 viruses, the Ann Arbor/6/60 strain showed the most unusual receptor-binding properties. Despite the presence of 226L/228S, the receptor-binding specificity of this strain is closer to that of the avian viruses. Three substitutions in the RBS region (137R→Q, 158G→E, and 186N→I; Fig. 2, H2), which separate this strain from the human consensus sequence, could be responsible for the altered binding specificity. Mutations in HA positions 137 and 186 were previously found to be involved in the adaptation of human H3N2 virus to growth in the presence of animal sera, the receptor specificity of the serum-resistant variants being similar to that of Ann Arbor/60 (20).
Receptor-binding specificity of H3 viruses from seals.
In 1991, H3N3 avian virus-like viruses were isolated from lung tissues of seals that died of pneumonia along the Cape Cod peninsula of Massachusetts (5). We examined four seal viruses isolated at different times during the outbreak (Table 1); the HA sequences of two had been determined earlier (Fig. 2, H3). Three of the four isolates displayed a typical avian virus-like pattern of binding to sialylglycopolymers, that is, an affinity for 3′SL-PAA of about 50 μM−1 and an affinity for 6′SLN-PAA of about 1 μM−1. This finding suggests that an avian influenza virus can infect seals without substantial changes in its receptor-binding specificity. By comparison with three other H3 seal viruses, seal/MA/3984/92 showed a lower binding affinity for 3′SL-PAA with no increase in binding to 6′SLN-PAA. The reasons for this discrepancy are not presently clear. Two amino acid substitutions in the upper portion of the HA globular head, 138A→S and 220R→S, separate this strain from the seal/MA/3911/92 virus. Both substitutions involve amino acid residues that are conserved among influenza virus HAs of all antigenic subtypes (23, 26). Position 220 is located on the boundary between the HA monomers relatively distant from the RBS. Amino acid 138 resides in the receptor-binding pocket (Fig. 3) and is much more likely to be responsible for the decreased binding to 3′SL-PAA.
Alterations of avian H1N1 viruses during their replication in pigs.
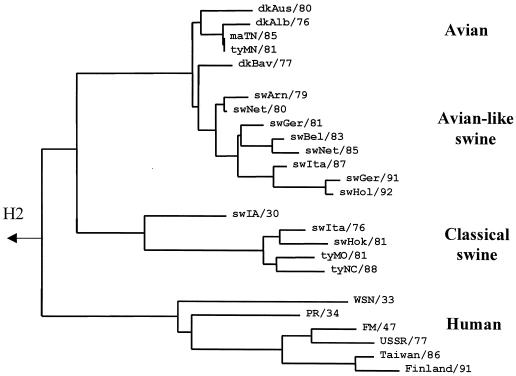
In 1979, an avian virus-like virus was isolated from pigs (36). This virus continues to circulate in European pigs (4, 18) and provides a rare opportunity for investigation of the receptor-binding specificity changes during adaptation of an avian virus to a new host. Besides so-called avian-like swine viruses, “classical” H1N1 swine influenza virus strains were included in this study to represent the receptor-binding properties of viruses that have circulated in pigs for about 80 years and are highly adapted to this host (Fig. 4 shows phylogenetic relationships among H1 avian, swine, and human viruses).
FIG. 4.
Phylogenetic tree for H1 subtype influenza viruses from different hosts, as determined based on the amino acid sequences of the HA1. The tree was constructed as described in Materials and Methods using the HA sequences of viruses listed in Fig. 2 and the following sequences of classical swine and human viruses from GenBank: sw/Iowa/15/30, WSN/33, PR/8/34 (Cambridge strain), FM/1/47, USSR/90/77, Taiwan/1/86, and Finland/168/91. The figure was generated with the TREEVIEW 1.5.2 program (27).
We found that the receptor-binding characteristics of H1N1 avian strains are similar to those of H2 and H3 avian viruses (Table 3). By contrast, the early avian-like swine isolates swine/Arnsberg/79 and swine/Netherlands/80 showed a substantial increase in affinity for 6′SLN-PAA. Notably, these swine viruses bound 6′SLN-PAA with the same affinity as do the early human pandemic strains Albany/57 (H2N2), Davis/57 (H2N2), and Aichi/68 (H3N2). Unlike H2 and H3 human viruses, the early avian-like swine viruses retained the ability of their avian predecessors to bind 3′SL-PAA. However, the affinity for 3′SL-PAA shown by swine isolates from subsequent years was gradually reduced, reflecting continued evolution of the virus in swine. The three more recent avian-like swine viruses isolated in 1987, 1991, and 1992 displayed binding affinities that were similar to those of classical swine viruses.
Four amino acid substitutions affecting the avian virus consensus sequence are present in the region of the receptor-binding site of the early avian-like swine viruses swine/Arnsberg/79 and swine/Netherlands/80 (positions 155, 159, 190, and 225; see Fig. 2, H1, and Fig. 3). One or more of these substitutions should be thus responsible for the increase in the affinity of avian H1 HA for the 6′SLN-PAA-containing glycopolymer. We therefore compared amino acid residues at these positions among the H1 HAs of classical swine and human viruses (Table 4). These positions were conserved in all 13 H1 avian HAs analyzed. Although classical swine viruses and avian-like swine viruses belong to distinct phylogenetic lineages (Fig. 3), most strains of both lineages carry the same substitutions, 155T→V, 159T→N, and 190E→D, suggesting that these changes are essential for adaptation of the avian HA to swine hosts. The mutation 225G→E is present in the HA of two early avian-like swine viruses but is absent in later isolates (Fig. 2) and in the classical swine strains. Thus, although this substitution could be involved in the early stages of adaptation of the avian H1 HA to swine, it was not preserved during subsequent replication of the virus in this host.
TABLE 4.
Amino acids at positions 155, 159, 190, and 225 of the HA of H1 viruses from birds, swine, and humansa
| Virus type (no. of sequences analyzed) | Amino acid residue at position:
|
|||
|---|---|---|---|---|
| 155 | 159 | 190 | 225 | |
| H1 | ||||
| Avian (13) | T* | T* | E* | G* |
| Classical swine (22) | V* | N[19], D[2], S[1] | D[21], E[1] | G[20], D[1], N[1] |
| Avian-like swine (8) | V[6], I[2] | N* | D[7], E[1] | G[6], E[2] |
| Human (42) | T* | G* | D[33], E[5], N[4] | D[22], G[19], N[1] |
| Human, 1918 (3)b | T* | S* | D* | D[2], G[1] |
| H2-H15 avian (14)c | Variable: T, V, I, L | Variable: G, A, S, T, N, Q, D | E* | G[13], N[1] |
The analysis was performed with sequences obtained from the GenBank and from the literature (29, 41). An asterisk next to an amino acid indicates that it was conserved in all sequences. Otherwise, the number of sequences for a particular amino acid is given in brackets.
Reid et al. (30).
The analysis was performed with a previously described set of avian virus HA sequences representing each of the 15 HA subtypes (23).
Among the human viruses, no changes in the avian sequence were detected at position 155, whereas a substitution at position 159 (T→G) differed from that in swine viruses (T→N or T→S). Because, similar to swine viruses, H1N1 human viruses display a high binding affinity for 6′SLN-PAA (13), one can conclude that amino acids at these two positions are not of primary importance for recognition of the 6′SLN determinants. What, then, can be the role of these substitutions for HA adaptation to swine? The side chain of the amino acid at position 155 participates in the formation of the pocket that accommodates the acyl substituent at 5-N of Neu5Ac (see Fig. 3). Thus, we hypothesize that the substitution 155T→I/V in the swine virus HA increases the affinity of the virus for 5N-glycolyl analog of the sialic acid that is abundant in pigs but absent in birds and humans (see reference 38 and references therein). Amino acid 159 is at the tip of the HA (Fig. 3) and therefore could potentially affect interactions of viruses with the distant parts of the sialyloligosaccharides or with protein parts of the receptors.
Most human viruses bear the same substitution as swine viruses at position 190 (E→D) and carry a similar substitution at position 225 (G→D). In fact, every swine and human H1 HA analyzed in this study differed from the avian consensus at least in one of these positions (Table 4). Thus, mutations at positions 190 and 225 correlate most closely with the ability of the virus to bind 6′SLN and appear then to be primarily involved in the adaptation of the H1 avian virus HA to swine and humans.
Probing of the HA receptor-binding specificity with monovalent sialosides.
To gain insight into the molecular mechanisms by which mutations in key HA positions alter the receptor-binding specificity of the avian virus HA during adaptation to other hosts, we compared the binding affinities of the test viruses for low-molecular-mass receptor analogs: free N-acetylneuraminic acid, methyl sialoside (MN), 3′SL, and 6′SLN. The absolute values of association constants and the relative affinities with respect to the affinity for the α-anomer of sialic acid (Krel) are presented in Table 5.
TABLE 5.
Binding of monovalent receptor analogs, free α-anomer of Neu5Ac, α-methylglycoside of Neu5Ac (MN), 3′SL, and 6′SLN by influenza virusesa
| Virus group and strain | Host species | HA subtype | Apparent association constant (Kass [mM−1])
|
Relative affinity with respect to Neu5Ac (Krel)b
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Neu5Ac | MN | 3′SL | 6′SLN | 3′SL | 6′SLN | MN | |||
| Group 1 | |||||||||
| Mal/TN/11464/85 (226Q, 228G) | Avian | H1 | 1.3 | 0.1 | 11 | 0.5 | 8.5 | 0.4 | 0.08 |
| Dk/Hokkaido/7/82 (226Q, 228G) | Avian | H3 | 2 | 0.15 | 18 | 0.55 | 9 | 0.3 | 0.08 |
| Leningrad/134/57 (226Q, 228G) | Human | H2 | 0.7 | 0.05 | 14 | 0.3 | 20 | 0.4 | 0.07 |
| Udorn/72/R2 (226Q, 228G) | Human | H3 | 2.5 | 0.2 | 33 | 0.5 | 13 | 0.2 | 0.08 |
| Udorn/72/R4 (226Q, 228S) | Human | H3 | 2.3 | 0.25 | 19 | 0.6 | 8.3 | 0.3 | 0.1 |
| Group 2 | |||||||||
| Albany/7/57 (226L, 228G) | Human | H2 | 1.5 | 1.2 | 1.3 | 1.1 | 0.9 | 0.7 | 0.8 |
| RI/5+/57 (226L, 228S) | Human | H2 | 1.2 | 0.7 | 0.5 | 2.3 | 0.4 | 1.9 | 0.6 |
| Aichi/2/68 (226L, 228S) | Human | H3 | 1.4 | 0.5 | 1 | 0.8 | 0.7 | 0.6 | 0.4 |
| Udorn/72 (226L, 228S) | Human | H3 | 4.8 | 1.3 | 2.8 | 1.8 | 0.6 | 0.4 | 0.3 |
| Group 3 | |||||||||
| Sw/Netherlands/3/80 (225E, 155I)c | Swine | 5.7 | 0.2 | 21 | 1.2 | 3.4 | 0.2 | 0.04 | |
| Sw/Germany/2/81 (190D, 155V)c | Swine | 5 | 0.3 | 8 | 1.3 | 1.6 | 0.3 | 0.06 | |
| Seal viruses | |||||||||
| Seal/3911/92 | H3 | 1.6 | 14 | 9 | |||||
| Seal/3984/92 (138S)c | H3 | 1.4 | 1.3 | 0.9 | |||||
Binding was studied, and apparent association constants were determined in solution in a competitive assay, as described in Materials and Methods.
The relative affinities reflect the ratio of the Kass values for a given sialoside and that for Neu5Ac (Krel = K[X]/K[Neu5Ac]). The higher the relative affinity, the better the sialosides bind (compared to free αNeu5Ac).
226Q and 228G are present in the HA of these viruses. Amino acids that differ from the avian virus consensus sequence in the other “key positions” of the HA RBS (23) are given in parentheses.
Comparison of binding patterns indicated three distinct groups of viruses. The first included authentic avian viruses (mallard/Tennessee/85 and duck/Hokkaido/82), human strains with the “avian” consensus sequence 226Q/228G (R2 and Leningrad/57), and R4. These viruses bound 3′SL ca. 10-fold more avidly than free sialic acid (Krel from 8.3 to 20), indicating significant energetically favorable interactions between the avian virus HA and the 3-linked lactose moiety of 3′SL. They also showed weak relative binding to 6′SLN and methyl sialoside by comparison to free Neu5Ac. Thus, the asialic parts of 6′SLN and MN do not appear to fit into the receptor-binding site of the HA without significant energy losses. Although R4 differed from other strains in this group by virtue of a 228G→S substitution, it displayed a binding pattern typical of other avian viruses. This finding supports the notion that a single 228G→S mutation in the avian HA does not markedly alter its receptor-binding characteristics as detected in this assay (see above and Tables 1 and 2).
The viruses of the second group (Albany/57, RI/5+/57, Aichi/68, and Udorn/72) are human isolates of the H2 and H3 subtype that share a common substitution, 226Q→L from the avian consensus sequence. Among them, Albany/57 carries the “avian” 228G but displays a binding pattern similar to that of other viruses from this group. Thus, 226L must be primarily responsible for their receptor-binding characteristics, regardless of the amino acid at position 228. Comparison between viruses with 226L and those of the first group (226Q) revealed several changes in receptor-binding properties. First, the substitution Q→L increased the affinity for 6′SLN and MN, an effect that correlated with an increase in Krel. Thus, the improved binding of these viruses to 6′SLN and MN appears to stem from a better fit of their asialic portions, N-acetyllactosamine and methyl moieties, due to the 226Q→L mutation. These two moieties share one structural feature, the hydrocarbon group, which participates in the formation of the glycosidic bond with sialic acid (6′-CH2 group of Gal in 6′SLN and 2-methyl group of MN). According to a model of the human virus HA complexed with Neu5Acα2-6Gal-containing sialyloligosaccharide LSTc (Fig. 3), this hydrocarbon group is positioned in immediate proximity to the amino acid at position 226. Hence, there may be direct steric interference in the avian HA between 226Q and the 6-hydrocarbon group of galactose, and a mutation from glutamine to leucine may create more room to accommodate this group. Another characteristic effect of the 226Q→L mutation is an appreciable decrease in affinity for 3′SL, rendering it comparable to the affinity for free N-acetylneuraminic acid (Krel = 0.4 to 0.9). Thus, the leucine at position 226 of the HA appears to abrogate favorable interactions between the 3-linked lactose and the protein that are typical for the avian virus HA.
Swine viruses belong to a third receptor-binding group, which resembles human H2 and H3 viruses with their higher affinity for 6′SLN compared to that of avian virus strains. However, the detailed molecular mechanism responsible for the increased affinity for the 6′SLN moiety appears to differ between the swine and human viruses. That is, H1 swine strains show no enhancement of their relative affinities for 6′SLN and MN but do have a substantially higher affinity for free Neu5Ac compared to the avian viruses. We conclude, therefore, that the HAs of swine viruses have a higher absolute affinity for 6′SLN and MN because the affinity of the HA for the sialic acid moiety of these analogs is increased. Another difference between the human and swine viruses is a higher absolute and relative affinity of swine viruses for 3′SL (and, as a consequence, for 3′SL-PAA; see Table 3). This feature indicates that amino acid substitutions in the receptor-binding pocket of H1 swine viruses (most likely, those in positions 190 and 225) decrease HA interactions with the 3-linked galactose moiety of 3′SL less markedly than does the 226Q→L mutation in the human H2 and H3 HAs.
To find a molecular basis for the decreased affinity of seal/MA/3984/92 isolate for 3′SL-PAA, we analyzed this virus and the seal/MA/3911/92 strain for their binding of Neu5Ac and 3′SL. The results presented in Table 5 indicate that the former virus has a markedly decreased affinity for 3′SL because it does not bind to the asialic portion of the 3′SL (Krel[3′SL] = 0.9). Thus, the 138A→S substitution in the HA of seal/3984/92 abolishes the fit of the 3-linked Gal moiety to the avian receptor-binding site.
DISCUSSION
The intent of this study was to gain insight into the initial events allowing transmission of avian virus HA to humans and other species by analyzing alterations of viral receptor-binding properties early after introduction of virus from birds to humans, pigs, or seals. We found that the majority of avian viruses, irrespective of their HA subtype, displayed very similar patterns of binding to 3′SL-PAA and 6′SLN-PAA polymers, with affinities ranging from 50 to 100 μM−1 (3′SL-PAA) and 1 to 2 μM−1 (6′SLN-PAA). These data indicate that the receptor-binding properties of viruses of wild aquatic birds are relatively conserved. The earliest available isolates of pandemic human H2 and H3 viruses and of epizootic avian-like swine H1 viruses bound 6′SLN-PAA with at least a fourfold-higher affinity than did the avian viruses. It can be concluded, therefore, that the ability to recognize 6′SLN was acquired relatively early after the introduction of an avian virus into humans and pigs, at least by the time influenza epidemics became apparent and viral isolates were obtained. The affinity of human and pig viruses for 6′SLN-PAA somewhat increased during consecutive years of epidemics, suggesting continued refinement of receptor-binding properties in these new hosts.
Our results confirm previous conclusions of Connor et al. (8) that the receptor-binding properties of H2 human virus strains with 226Q/228G are similar to those of avian H2 viruses. One of these strains (A/RI/5−/57) is a laboratory mutant selected by passage of parental human virus (226L/228S) in the presence of horse serum (6). The exact passage history of the two other strains is unknown. However, because any virus from 1957 must have been isolated and undergone multiple passages in eggs, these strains, similar to RI5−/57, may be receptor-binding mutants derived in the laboratory. Indeed, a shift toward an avian virus-like receptor specificity has been well documented upon passage in eggs of human influenza A and B viruses (13, 15, 30a) and of H2 isolates from 1957 in particular (see reference 8 and references therein). For these reasons, there is no clear evidence that H2N2 viruses with avian virus-like receptor specificity replicated in humans. By contrast, “human virus-like” H2N2 isolates certainly circulated among humans during the first year of the 1957 pandemic, because egg adaptation never favors selection of mutants with human virus-like specificity (15, 30a).
Besides their increased affinity for 6′SLN, another characteristic of the earliest available H2 and H3 subtype human viral HAs was their substantially decreased affinity for 3′SL-PAA compared to that of avian viruses. This feature may indicate that there is negative selective pressure in humans against viruses with this specificity. The mechanism for such pressure could be neutralization of the virus by human respiratory mucins, which are known to carry predominantly Neu5Acα2-3Gal moieties (references 1 and 9 and references therein). Alternatively, a decreased affinity for Neu5Acα2-3Gal could merely reflect effects of HA mutations that became fixed under selective pressure for stronger binding to 6′SLN.
In contrast to human and swine viruses, there were no changes in the receptor-binding specificities among three of the four viral isolates from seals. Thus, either the selective pressure against viruses with avian receptor specificity is low (or absent) in these species or else the animals were only recently infected with avian viruses so that there was not enough time for the selection to occur.
Previously, we identified six key amino acid positions in the HA receptor-binding site that are highly conserved in the avian influenza viruses but can change during adaptation of the avian HA to humans after interspecies transfer (23). In this study, we further defined roles of five of these mutations in the changes of the receptor-binding properties of the HA. First of all, we found that a single mutation at position 138, 190, 225, 226, or 228 of the avian HA decreases the affinity of the virus for receptor analogs 3′SL and 3′SL-PAA and, therefore, most likely, for cellular receptors on the target cells of avian intestinal epithelia. This finding can explain previous reports concerning the inability of H3 viruses with a single 228G→S substitution in the HA to replicate in the intestinal tract of ducks (24, 39). Binding data indicate that the affinity for Neu5Acα2-3Gal determinants decreases because mutations at these positions lower (positions 190, 225, and 228) or completely abrogate (positions 138 and 226) the favorable interactions of viruses with the 3-linked galactose moiety of the receptor analog (see Table 5, relative binding to 3′SL). These data support our hypothesis about a tight fit of the Gal moiety of the Neu5Acα2-3Gal determinant to the avian receptor-binding site and its destruction by these mutations (23, 39).
In apparent conflict with this interpretation is the presence of mutations at position 228 and the lowered affinity for 3′SL-PAA of some H3 viral isolates from wild ducks (2, 16) (see Fig. 2, H3). It cannot be excluded, however, that these viruses, as well as seal/MA/3984/92 (138S), are laboratory variants. As shown recently for the receptor-binding variants of swine influenza virus A/NJ/11/76, the virus with the lower affinity for cellular receptors can readily outgrow the high-affinity variant in laboratory passages because of more-rapid release from cells and, consequently, more-rapid spread (14). In natural infection, by contrast, a higher affinity may be required for efficient transmission of the virus from one infected host to another (a task performed in laboratory passages by the researcher).
The available data strongly suggest that the avian HA in humans and swine is subjected to selective pressure toward mutants with increased affinity for the 6′SLN determinant and decreased affinity for Neu5Acα2-3Gal. Our data show that the substitution 226Q→L in the H2 and H3 HA is most critical for this change, whereas an additional mutation, 228G→S, makes only a marginal contribution. It is tempting to speculate in this view that the human viral isolates Davis/57 and Chile/57 with 226L and 228G (17) represent the earliest steps of adaptation of the avian H2 HA to humans.
In the case of the H1 HA, we could not compare receptor-binding properties of early human and avian viruses because such isolates are not available. However, a good correlation between mutations in the avian H1 HA in swine and substitutions in the same positions in human viruses suggests either similar molecular mechanisms of H1 HA adaptation to these hosts or origination of H1 human viruses and classical swine viruses from a common precursor virus that carried mutations in the RBS. The binding data (Table 3) suggest that mutations 190E→D and/or 225G→E/D are most critical for enhancement of the affinity of the avian H1 HA affinity for 6′SLN and decrease in binding to Neu5Acα2-3Gal. This conclusion is supported by the fact that substitutions in these two positions (often with reversion to the avian consensus sequence) are commonly observed during the egg adaptation of contemporary human H1N1 influenza viruses and have the most profound effect on the virus recognition of the type of Neu5Ac-Gal linkage (15).
Substitutions at positions 190, 225, and 226 and some others in the receptor-binding site of avian virus-like viruses isolated from other species could serve as markers of the virus host and potential to cause epidemics. For example, the amino acid sequences of three 1918 H1N1 virus strains from the “Spanish” influenza pandemic have been reported recently (30). These sequences were derived directly from viral RNA present in tissues of infected humans and would be therefore free from artifacts of laboratory adaptation. All three variants bear 190D, and two of them additionally carry 225D, suggesting that 1918 pandemic viruses had a swine virus-like receptor-binding specificity recognizing both 6′SLN- and Neu5Acα2-3Gal-containing receptors. However, these viruses have 155S, which is typical of avian and human viruses but not of swine viruses. The latter notion seems to be more consistent with the theory that the 1918 influenza virus spread from humans to swine (30) rather than in the opposite direction. Another example is the H5N1 viruses isolated from humans during the 1997 outbreak in Hong Kong that carry no substitutions in positions 190, 225, and 226 (7, 37) and display avian virus-like receptor-binding specificity (21). These properties are consistent with the direct introduction of these viruses from chickens to humans and with the fact that viruses were not transmitted from human to human.
In summary, our data suggest that a shift of the receptor-binding specificity of the avian virus HA from Neu5Acα2-3Gal recognition to Neu5Acα2-6Gal recognition is a prerequisite for the generation of human pandemic viruses. The results also indicate that one or two amino acid mutations in the avian virus HA are sufficient for this shift; hence, a limited number of replications of an avian virus in humans appears to be sufficient for such changes.
ACKNOWLEDGMENTS
We thank Peng Gao for sequencing the HA1 gene of dk/Hokkaido/7/82 virus, V. E. Piskarev for the generous gift of 6′SLN, and M. B. Eisen for crystallographic structures of influenza virus HA complexes with sialyloligosaccharides. We are grateful to R. Bethell of GlaxoWellcome R&D for providing the NA inhibitor zanamivir (GG167). We also thank Krisna Wells and Martha McGregor for technical assistance and J. Gilbert for editing the manuscript.
This work was supported by Public Health Service research grants from the National Institute of Allergy and Infectious Diseases, by Cancer Center Support (CORE) grant CA-21765, and by the American Lebanese Syrian Associated Charities. Mikhail Matrosovich was supported by a Karnofsky fellowship from St. Jude Children's Research Hospital.
REFERENCES
- 1.Baum L G, Paulson J C. Sialyloligosaccharides of the respiratory epithelium in the selection of human influenza virus receptor specificity. Acta Histochem Suppl Band. 1990;XL:35–38. [PubMed] [Google Scholar]
- 2.Bean W J, Schell M, Katz J, Kawaoka Y, Naeve C, Gorman O, Webster R G. Evolution of the H3 influenza virus hemagglutinin from human and nonhuman hosts. J Virol. 1992;66:1129–1138. doi: 10.1128/jvi.66.2.1129-1138.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bovin N V, Korchagina E Y, Zemlyanukhina T V, Byramova N E, Galanina O E, Zemlyakov A E, Ivanov A E, Zubov V P, Mochalova L V. Synthesis of polymeric neoglycoconjugates based on N-substituted polyacrylamides. Glycoconj J. 1993;10:142–151. doi: 10.1007/BF00737711. [DOI] [PubMed] [Google Scholar]
- 4.Brown I H, Ludwig S, Olsen C W, Hannoun C, Scholtissek C, Hinshaw V S, Harris P A, McCauley J W, Strong I, Alexander D J. Antigenic and genetic analyses of H1N1 influenza A viruses from European pigs. J Gen Virol. 1997;78:553–562. doi: 10.1099/0022-1317-78-3-553. [DOI] [PubMed] [Google Scholar]
- 5.Callan R J, Early G, Kida H, Hinshaw V S. The appearance of H3 influenza viruses in seals. J Gen Virol. 1995;76:199–203. doi: 10.1099/0022-1317-76-1-199. [DOI] [PubMed] [Google Scholar]
- 6.Choppin P W, Tamm I. Two kinds of particles with contrasting properties in influenza A virus strains from the 1957 pandemic. Virology. 1959;8:539–542. doi: 10.1016/0042-6822(59)90059-5. [DOI] [PubMed] [Google Scholar]
- 7.Claas E C J, Osterhaus A D M E, Vanbeek R, Dejong J C, Rimmelzwaan G F, Senne D A, Krauss S, Shortridge K F, Webster R G. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 8.Connor R J, Kawaoka Y, Webster R G, Paulson J C. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 9.Couceiro J N, Paulson J C, Baum L G. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993;29:155–165. doi: 10.1016/0168-1702(93)90056-s. [DOI] [PubMed] [Google Scholar]
- 10.Eisen M B, Sabesan S, Skehel J J, Wiley D C. Binding of the influenza A virus to cell-surface receptors: structures of five hemagglutinin-sialyloligosaccharide complexes determined by X-ray crystallography. Virology. 1997;232:19–31. doi: 10.1006/viro.1997.8526. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 12.Gambaryan A S, Matrosovich M N. A solid-phase enzyme-linked assay for influenza virus receptor-binding activity. J Virol Methods. 1992;39:111–123. doi: 10.1016/0166-0934(92)90130-6. [DOI] [PubMed] [Google Scholar]
- 13.Gambaryan A S, Tuzikov A B, Piskarev V E, Yamnikova S S, Lvov D K, Robertson J S, Bovin N V, Matrosovich M N. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: non-egg-adapted human H1 and H3 influenza A and influenza B viruses share a common high binding affinity for 6′-sialyl(N-acetyllactosamine) Virology. 1997;232:345–350. doi: 10.1006/viro.1997.8572. [DOI] [PubMed] [Google Scholar]
- 14.Gambaryan A S, Matrosovich M N, Bender C A, Kilbourne E D. Differences in the biological phenotype of low-yielding (L) and high-yielding (H) variants of swine influenza virus A/NJ/11/76 are associated with their different receptor-binding activity. Virology. 1998;247:223–231. doi: 10.1006/viro.1998.9274. [DOI] [PubMed] [Google Scholar]
- 15.Gambaryan A S, Robertson J S, Matrosovich M N. Effects of egg-adaptation on the receptor-binding properties of human influenza A and B viruses. Virology. 1999;258:232–239. doi: 10.1006/viro.1999.9732. [DOI] [PubMed] [Google Scholar]
- 16.Kida H, Kawaoka Y, Naeve C W, Webster R G. Antigenic and genetic conservation of H3 influenza virus in wild ducks. Virology. 1987;159:109–119. doi: 10.1016/0042-6822(87)90353-9. [DOI] [PubMed] [Google Scholar]
- 17.Klimov A I, Bender C A, Hall H E, Cox N J. Evolution of human influenza A (H2N2) viruses. In: Brown L E, Hampson A W, Webster R G, editors. Options for the control of influenza III. Amsterdam, The Netherlands: Elsevier Science B.V.; 1996. pp. 546–552. [Google Scholar]
- 18.Ludwig S, Stitz L, Planz O, Van H, Fitch W M, Scholtissek C. European swine virus as a possible source for the next influenza pandemic? Virology. 1995;212:555–561. doi: 10.1006/viro.1995.1513. [DOI] [PubMed] [Google Scholar]
- 19.Martin J, Wharton S A, Lin Y P, Takemoto D K, Skehel J J, Wiley D C, Steinhauer D A. Studies of the binding properties of influenza hemagglutinin receptor-site mutants. Virology. 1998;241:101–111. doi: 10.1006/viro.1997.8958. [DOI] [PubMed] [Google Scholar]
- 20.Matrosovich M, Gao P, Kawaoka Y. Molecular mechanisms of serum resistance of human influenza H3N2 virus and their involvement in virus adaptation in a new host. J Virol. 1998;72:6373–6380. doi: 10.1128/jvi.72.8.6373-6380.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matrosovich M, Zhou N, Kawaoka Y, Webster R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J Virol. 1999;73:1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matrosovich M N, Gambaryan A S, Tuzikov A B, Byramova N E, Mochalova L V, Golbraikh A A, Shenderovich M D, Finne J, Bovin N V. Probing of the receptor-binding sites of the H1 and H3 influenza A and influenza B virus hemagglutinins by synthetic and natural sialosides. Virology. 1993;196:111–121. doi: 10.1006/viro.1993.1459. [DOI] [PubMed] [Google Scholar]
- 23.Matrosovich M N, Gambaryan A S, Teneberg S, Piskarev V E, Yamnikova S S, Lvov D K, Robertson J S, Karlsson K A. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology. 1997;233:224–234. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- 24.Naeve C W, Hinshaw V S, Webster R G. Mutations in the hemagglutinin receptor-binding site can change the biological properties of an influenza virus. J Virol. 1984;51:567–569. doi: 10.1128/jvi.51.2.567-569.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholas K B, Nicholas H B, Jr, Deerfield D W. GeneDoc: analysis and visualization of genetic variation. EMBNEW News. 1997;4:14–14. [Google Scholar]
- 26.Nobusawa E, Aoyama T, Kato H, Suzuki Y, Tateno Y, Nakajima K. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 27.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 28.Paulson J C. Interactions of animal viruses with cell surface receptors. In: Conn M, editor. The receptors. Vol. 2. Orlando, Fla: Academic Press, Inc.; 1985. pp. 131–219. [Google Scholar]
- 29.Raymond F L, Caton A J, Cox N J, Kendal A P, Brownlee G G. The antigenicity and evolution of influenza H1 haemagglutinin, from 1950–1957 and 1977–1983: two pathways from one gene. Virology. 1986;148:275–287. doi: 10.1016/0042-6822(86)90325-9. [DOI] [PubMed] [Google Scholar]
- 30.Reid A H, Fanning T G, Hultin J V, Taubenberger J K. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc Natl Acad Sci USA. 1999;96:1651–1656. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Robertson J S. Clinical influenza virus and the embryonated hen's eggs. Rev Med Virol. 1993;3:97–106. [Google Scholar]
- 31.Rogers G N, Paulson J C, Daniels R S, Skehel J J, Wilson I A, Wiley D C. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 32.Rogers G N, Pritchett T J, Lane J L, Paulson J C. Differential sensitivity of human, avian, and equine influenza A viruses to a glycoprotein inhibitor of infection: selection of receptor specific variants. Virology. 1983;131:394–408. doi: 10.1016/0042-6822(83)90507-x. [DOI] [PubMed] [Google Scholar]
- 33.Rogers G N, D'Souza B L. Receptor binding properties of human and animal H1 influenza virus isolates. Virology. 1989;173:317–322. doi: 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 34.Sayle R, Milner-White E J. RasMol: biomolecular graphics for all. Trends Biochem Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 35.Schafer J R, Kawaoka Y, Bean W J, Suss J, Senne D, Webster R G. Origin of the pandemic 1957 H2 influenza A virus and the persistence of its possible progenitors in the avian reservoir. Virology. 1993;194:781–788. doi: 10.1006/viro.1993.1319. [DOI] [PubMed] [Google Scholar]
- 36.Scholtissek C, Burger H, Bachmann P A, Hannoun C. Genetic relatedness of hemagglutinins of the H1 subtype of influenza A viruses isolated from swine and birds. Virology. 1983;129:521–523. doi: 10.1016/0042-6822(83)90194-0. [DOI] [PubMed] [Google Scholar]
- 37.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X Y, Fukuda K, Cox N. Characterization of an avian influenza (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki T, Horiike G, Yamazaki Y, Kawabe K, Masuda H, Miyamoto D, Matsuda M, Nishimura S I, Yamagata T, Ito T, Kida H, Kawaoka Y, Suzuki Y. Swine influenza virus strains recognize sialylsugar chains containing the molecular species of sialic acid predominantly present in the swine tracheal epithelium. FEBS Lett. 1997;404:192–196. doi: 10.1016/s0014-5793(97)00127-0. [DOI] [PubMed] [Google Scholar]
- 39.Vines A, Wells K, Matrosovich M, Castrucci M R, Ito T, Kawaoka Y. The role of influenza A virus hemagglutinin residues 226 and 228 in receptor specificity and host range restriction. J Virol. 1998;72:7626–7631. doi: 10.1128/jvi.72.9.7626-7631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu X, Rocha E P, Regenery H L, Kendal A P, Cox N J. Genetic and antigenic analyses of influenza A (H1N1) viruses, 1986–1991. Virus Res. 1993;28:37–55. doi: 10.1016/0168-1702(93)90088-5. [DOI] [PubMed] [Google Scholar]