ABSTRACT
We report the complete genome sequences of nine double recombinant vaccine-derived novel oral poliovirus type 2 genomes from acute flaccid paralysis (AFP) cases (n = 3), AFP case contacts (n = 4), and environmental surveillance sampling (n = 2) in Nigeria.
KEYWORDS: poliovirus, nOPV2, next generation sequencing
ANNOUNCEMENT
Poliovirus, from the family Picornaviridae, is a single-stranded, positive-sense RNA (+ssRNA) virus with a genome of approximately 7,500 nucleotides and a causative agent of poliomyelitis. To better address the global evolving risk of type 2 circulating vaccine-derived poliovirus (cVDPV2), a novel oral poliovirus type 2 (nOPV2) vaccine was genetically engineered and first distributed to countries in March 2021 (1, 2). The nOPV2 vaccine is a modified version of the preexisting type 2 monovalent OPV (mOPV2) vaccine and provides comparable protection against poliovirus while being more genetically stable and less likely to revert to a form that can cause paralysis (1). The nOPV2 vaccine’s increased genetic stability should result in a reduced risk of seeding new cVDPV2 emergences compared to the mOPV2 (Sabin 2) vaccine.
The nine samples were received at the Centers for Disease Control and Prevention via FTA cards from the WHO National Polio Laboratory (polioeradication.org), University of Maiduguri Teaching Hospital, Maiduguri, Nigeria (see Table 1). Viral RNA was extracted using the QIAamp Viral RNA Mini Kit (Cat. No. 52906; Qiagen), and cDNA was generated by using SuperScript IV First-Strand Sequencing System (Cat. No. 18091200; Invitrogen) using random primers. Klenow Fragment (3′−5′ exo-) (Cat. No. M0212S, NEB) was used for cDNA second-strand synthesis. For Illumina sequencing, the library was prepared using the Nextera XT Library Kit (Cat. No. FC-131-1096; Illumina) and sequenced on the Illumina MiSeq platform (2 × 250 bp).
TABLE 1.
Sequencing summary of nine double recombinant circulating vaccine-derived nOPV2 genomes from Nigeria
| GenBank accession |
SRA accession |
Sample name |
Collection year |
Type | Country | State | Surveillance type |
Genome length (nt) |
Average coverage |
SNP count when compared to VP1 reference (AY184220) |
Total no. of pre- processed reads |
Total no. of post- processed reads |
Total no. of mapped reads |
GC content (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PQ059262 | SRR30089174 | NIE23-001 | 2023 | nOPV2 | Nigeria | Katsina State | AFP case contact | 7,442 | 1,262.7 | 12 | 2,224,238 | 439,508 | 68,963 | 46.2 |
| PQ059263 | SRR30089173 | NIE23-002 | 2023 | nOPV2 | Nigeria | Kano State | AFP case | 7,442 | 366.0 | 13 | 1,600,498 | 302,501 | 20,323 | 46.0 |
| PQ059264 | SRR30089172 | NIE23-003 | 2023 | nOPV2 | Nigeria | Kano State | Environmental | 7,442 | 867.8 | 16 | 1,881,262 | 372,247 | 44,987 | 46.0 |
| PQ059265 | SRR30089171 | NIE23-004 | 2023 | nOPV2 | Nigeria | Kano State | Environmental | 7,442 | 78.3 | 16 | 713,470 | 32,591 | 4,058 | 46.0 |
| PQ059266 | SRR30089170 | NIE24-005 | 2024 | nOPV2 | Nigeria | Kano State | AFP case | 7,442 | 403.5 | 13 | 2,029,418 | 757,739 | 25,357 | 46.2 |
| PQ059267 | SRR30089169 | NIE24-006 | 2024 | nOPV2 | Nigeria | Kano State | AFP case contact | 7,442 | 1,031.2 | 16 | 2,159,638 | 407,361 | 59,321 | 46.2 |
| PQ059268 | SRR30089168 | NIE24-007 | 2024 | nOPV2 | Nigeria | Kano State | AFP case contact | 7,442 | 503.1 | 15 | 1,406,028 | 494,068 | 30,176 | 46.1 |
| PQ059269 | SRR30089167 | NIE24-008 | 2024 | nOPV2 | Nigeria | Kano State | AFP case | 7,442 | 104.7 | 17 | 208,442 | 100,142 | 6,089 | 46.0 |
| PQ059270 | SRR30089166 | NIE24-009 | 2024 | nOPV2 | Nigeria | Kano State | AFP case contact | 7,442 | 139.5 | 12 | 862,648 | 197,435 | 8,816 | 46.0 |
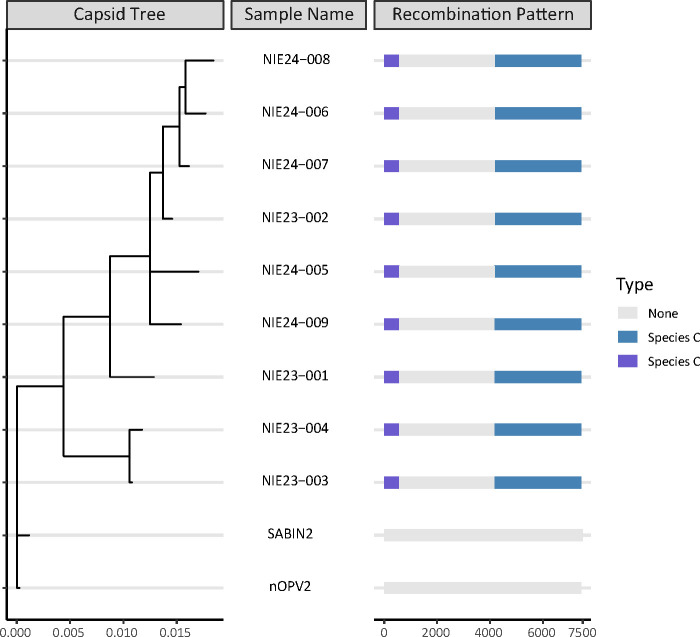
Raw read data were processed using VPipe version 1.0, our in-house pipeline for processing viral specimens (3), using default parameters. Additionally, a reference-based assembly was performed in Geneious Prime version 2023.1.1 using default parameters and a medium sensitivity. Genome alignments were performed using MAFFT version 7.490 to compare with the reference genome (GenBank accession AY184220) and verify results and identify recombination events (see Table 1). The MrBayes version 3.2.6 plugin in Geneious Prime using an HKY substitution model and a gamma rate variation was used to make the capsid tree (see Fig. 1), and regions of recombination events were identified as Enterovirus species C.
Fig 1.
Double recombinant cVDPV nOPV2 Nigeria samples showing a capsid tree and the recombination pattern of their full-length genomes. All samples are in the NIE-KTS-1 emergence group and show similar double recombination events. Sabin2 (AY184220) and nOPV2 (MZ245455) sequences are shown for reference. “None” delineates areas where no recombination was observed. Purple shows the 5′ untranslated region recombinant area matching an Enterovirus species C. Blue shows the P2/P3 recombinant area of the genome matching a different Enterovirus species C. All nine full-length genomes show a similar recombinant pattern, and all genomes show a 98%–99.8% sequence pairwise percent identity to each other.
We isolated the full nOPV2 genomes from patients, contacts, and environmental sampling in Nigeria that have undergone double recombination events and were identified as VDPV2s (4). The entire 5′ untranslated region and the non-structural region (P2/P3) of the genome showed evidence of recombination (see Fig. 1), essentially removing the nOPV2 modifications except three nucleotide markers within the capsid (P1) region. The identification of these double recombinant genomes is responsible for classifying a new cVDPV2 emergence group from nOPV2 origin in Nigeria called NIE-KTS-1, first detected in an AFP contact in 2023. This emergence group in Nigeria is in addition to the previously reported cVDPV nOPV2 emergence group, NIE-KBS-1 previously detected in 2023 as well (5), yet distinctly different cVDPV nOPV2 strains. Identifying these nine double recombinant genomes from AFP case patients is noteworthy for tracking the genetic characterization of all circulating nOPV2 strains at the current stage of nOPV2 vaccine use.
ACKNOWLEDGMENTS
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Contributor Information
Jaume Jorba, Email: jjorba@cdc.gov.
Jelle Matthijnssens, Katholieke Universiteit Leuven, Leuven, Belgium.
DATA AVAILABILITY
The genome sequences have been deposited in GenBank with the accession numbers PQ059262-PQ059270. The postprocessed FASTQ reads have been deposited in the Sequence Read Archive with the run accession numbers SRR30089166-SRR30089174.
REFERENCES
- 1. Yeh MT, Bujaki E, Dolan PT, Smith M, Wahid R, Konz J, Weiner AJ, Bandyopadhyay AS, Van Damme P, De Coster I, Revets H, Macadam A, Andino R. 2020. Engineering the live-attenuated polio vaccine to prevent reversion to virulence. Cell Host Microbe 27:736–751. doi: 10.1016/j.chom.2020.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Macklin GR, Peak C, Eisenhawer M, Kurji F, Mach O, Konz J, Gast C, Bachtiar NS, Bandyopadhyay AS, Zipursky S, nOPV2 Working Group . 2023. Enabling accelerated vaccine roll-out for Public Health Emergencies of International Concern (PHEICs): novel oral polio vaccine type 2 (nOPV2) experience. Vaccine (Auckl) 41:A122–A127. doi: 10.1016/j.vaccine.2022.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wagner DD, Marine RL, Ramos E, Ng TFF, Castro CJ, Okomo-Adhiambo M, Harvey K, Doho G, Kelly R, Jain Y, Tatusov RL, Silva H, Rota PA, Khan AN, Oberste MS. 2022. VPipe: an automated bioinformatics platform for assembly and management of viral next-generation sequencing data. Microbiol Spectr 10:e0256421. doi: 10.1128/spectrum.02564-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Global Polio Eradication Initiative WHO . 2016. Classification and reporting of vaccine-derived polioviruses (VDPV) - GPEI guidelines 2016. World Health Organization. Available from: https://polioeradication.org/wp-content/uploads/2016/09/Reporting-and-Classification-of-VDPVs_Aug2016_EN.pdf [Google Scholar]
- 5. Davlantes E, Jorba J, Henderson E, Bullard K, Deka MA, Kfutwah A, Martin J, Bessaud M, Shulman LM, Hawes K, Diop OM, Bandyopadhyay AS, Zipursky S, Burns CC. 2023. Notes from the field: circulating vaccine-derived poliovirus type 2 emergences linked to novel oral poliovirus vaccine type 2 use - six African Countries, 2021-2023. MMWR Morb Mortal Wkly Rep 72:1041–1042. doi: 10.15585/mmwr.mm7238a4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequences have been deposited in GenBank with the accession numbers PQ059262-PQ059270. The postprocessed FASTQ reads have been deposited in the Sequence Read Archive with the run accession numbers SRR30089166-SRR30089174.