ABSTRACT
During the COVID-19 pandemic, Nepal, like other countries, faced emerging SARS-CoV-2 variants. To evaluate the circulating variants, 278 samples collected between September 2021 and March 2022 were sequenced in the country. From these, 229 high-quality genomes were obtained (82.97% Omicron and 17.03% Delta) highlighting the genomic diversity of SARS-CoV-2 in Nepal.
KEYWORDS: COVID-19, whole-genome sequencing, coronavirus, Illumina
ANNOUNCEMENT
During the COVID-19 pandemic caused by SARS-CoV-2, a positive-sense, single-stranded RNA virus of the family Coronaviridae and genus Betacoronavirus (1), the availability of complete, high-quality viral genome sequence data was limited in Nepal (2, 3).
A total of 278 nasopharyngeal swab samples from the government approved National Public Health laboratory (collected from different parts of the country) and KM-TU Biotech Covid Lab (collected from Kathmandu) from September 2021 to March 2022 were studied. The samples were stored at −80°C until use. RNA was extracted using Chemagic Prime Viral RNA kit (CMG-1449) on a Perkin Elmer Chemagic 360 instrument. Real Time Multiplex RT-PCR was performed using the Liferiver Novel Coronavirus (2019-nCoV) Real Time Multiplex RT-PCR Kit (Ref no.: RR-0479-02), and SARS-CoV-2-positive samples with Ct values less than 30 were used for sequencing.
Following Illumina COVIDSeq protocol (#1000000126053-04), the SARS-CoV-2 genome of each sample was amplified using CPP1 and CPP2 ARTIC v3 pools of primers (https://github.com/artic-network/artic-ncov2019/blob/master/primer_schemes/nCoV-2019/V3/nCoV-2019.tsv). Amplicons from each of the two PCR reactions per sample were pooled and indexed with “IDT for Illumina-PCR Indexes” (unique index per sample) using on-bead tagmentation. Each indexed sample was then pooled for the final library, followed by quality check with Agilent Bioanalyzer (Kit Cat. 50671504) and quantified with Qubit 3.0 (Kit Cat. Q32851). The final library was diluted to 4 nM and denatured. The sample was loaded on the sequencer at 10 pmol. Sequencing was done on Illumina MiSeq with the 2 × 300 bp paired-end protocol using MiSeqV3 reagent kit (MS-102–3003) at the IVDRL, Tribhuvan University, Nepal.
The 229 high-quality genomic sequences (Table 1) were analyzed using the CDC SARS-CoV-2 BFX-UT_ARTIC_Illumina pipeline (https://github.com/CDCgov/SARS-CoV-2_Sequencing/tree/master/protocols/BFX-UT_ARTIC_Illumina). The Illumina reads were mapped (bwa-mem version 0.7.17-r1188) to the SARS-CoV-2 reference genome (MN908947.3), sorted, and unmapped reads were removed using samtools (version 1.17 using htslib 1.18). The total number of reads and the average sequencing depth (201–4,766) per genome are listed in Table 1. Primers were trimmed, and the consensus genome (39 complete and 190 nearly complete coding sequence) was generated using iVar version 1.4.2 (4). Variants were called with iVar version 1.4.2 followed by LoFreq version 2.1.5 (5), then annotated with snpEff version 5.1f and snpSift version 4.3t (6). The sequences were concatenated, and lineages were assigned using Pangolin version 1.1.14 (7) and NextClade version 2.14.0 (8), combined in R version 4.2.2. Sequences with more than 3,000 Ns compared to the reference genome were excluded. Full genomes were aligned using MAFFT version 0.7 (9), and a phylogenetic tree was generated with Augur version 17.7.0 from Nextstrain (10). The tree was visualized and edited in iTOL version 5 (11), with clade labels added in Microsoft Preview. Default settings were used for all these software unless otherwise specified.
TABLE 1.
Details of SARS-CoV-2 samples, sequence analysis, and hyperlinks of the study
| S. No. | Sample name | Collection_Date | Hyperlinked SRA accession ID | GenBank accession ID | GC content (%) | Lineage (Nextclade) | clade_who | Length | qc.overallScore (Nextclade) | Genome coverage (%) (Nextclade) | Number of reads | Sequencing depth |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | BB466 | 2021-12 | SRR29872869 | PP825474 | 39 | BA.1.17.2 | Omicron | 29,760 | 60.61 | 91.6 | 139,487 | 605.13 |
| 2 | qc5158 | 2022-01 | SRR29872868 | PP825475 | 39.80 | BA.2 | Omicron | 29,805 | 11.56 | 95.7 | 236,261 | 1,470.6 |
| 3 | qc8160 | 2022-02 | SRR29872857 | PP825476 | 39.40 | BA.2 | Omicron | 29,648 | 18.46 | 94.4 | 141,771 | 725.48 |
| 4 | qc8213 | 2022-02 | SRR29872846 | PP825477 | 39 | BA.2.10 | Omicron | 29,636 | 14.08 | 94.8 | 278,256 | 1,550.1 |
| 5 | qc8215 | 2022-02 | SRR29872835 | PP825478 | 38.70 | BA.2 | Omicron | 29,797 | 36.76 | 93.3 | 255,107 | 1,525 |
| 6 | qc8219 | 2022-02 | SRR29872824 | PP825479 | 39.40 | BA.2.10 | Omicron | 29,621 | 19.43 | 94.2 | 245,897 | 1,388.8 |
| 7 | qc8220 | 2022-02 | SRR29872823 | PP825480 | 38.50 | BA.2.10 | Omicron | 29,781 | 5.58 | 96.6 | 182,429 | 1,242.1 |
| 8 | qc8221 | 2022-02 | SRR29872822 | PP825481 | 39 | BA.2 | Omicron | 29,640 | 4.92 | 96.2 | 256,586 | 1,565.4 |
| 9 | qc8228 | 2022-02 | SRR29872821 | PP825482 | 41.30 | BA.2 | Omicron | 29,755 | 64.59 | 91.3 | 176,481 | 905.09 |
| 10 | qc8249 | 2022-01 | SRR29872820 | PP825483 | 38.80 | BA.2 | Omicron | 29,708 | 94.81 | 89.6 | 169,426 | 925.52 |
| 11 | qc8258 | 2022-02 | SRR29872867 | PP825484 | 38.80 | BA.2 | Omicron | 29,778 | 13.55 | 95.3 | 190,765 | 1,127.2 |
| 12 | qc8260 | 2022-02 | SRR29872866 | PP825485 | 37.90 | BA.2.10 | Omicron | 29,602 | 54.21 | 91.4 | 225,175 | 1,652.3 |
| 13 | qc8261 | 2022-02 | SRR29872865 | PP825486 | 38.20 | BA.2.10 | Omicron | 29,703 | 15.12 | 94.9 | 284,906 | 1,597.7 |
| 14 | qc8273 | 2022-01 | SRR29872864 | PP825487 | 38.80 | BA.2 | Omicron | 29,678 | 16.84 | 94.6 | 177,154 | 1,098.1 |
| 15 | qc8274 | 2022-01 | SRR29872863 | PP825488 | 39.10 | BA.2 | Omicron | 29,747 | 44.49 | 92.5 | 134,298 | 751.64 |
| 16 | qc8280 | 2022-01 | SRR29872862 | PP825489 | 37.90 | BA.2.10 | Omicron | 29,638 | 16.72 | 94.5 | 281,714 | 1,847.9 |
| 17 | qc8281 | 2022-01 | SRR29872861 | PP825490 | 39.40 | BA.2.2 | Omicron | 29,805 | 12.99 | 95.5 | 321,001 | 1,749.7 |
| 18 | qc8317 | 2022-02 | SRR29872860 | PP825491 | 38.80 | BA.2.10 | Omicron | 29,637 | 17.36 | 94.4 | 130,560 | 714.6 |
| 19 | qc8319 | 2022-02 | SRR29872859 | PP825492 | 38.40 | BA.2 | Omicron | 29,774 | 17.48 | 95 | 175,423 | 1,061.8 |
| 20 | qc8321 | 2022-02 | SRR29872858 | PP825493 | 37.90 | BA.2 | Omicron | 29,799 | 60.72 | 91.7 | 238,459 | 1,453.1 |
| 21 | qc8322 | 2022-02 | SRR29872856 | PP825494 | 39 | BA.2.2 | Omicron | 29,631 | 17.53 | 94.6 | 162,709 | 1,035.8 |
| 22 | qc8323 | 2022-02 | SRR29872855 | PP825495 | 39.30 | BA.2 | Omicron | 29,697 | 18.02 | 94.6 | 169,050 | 951.75 |
| 23 | qc8324 | 2022-02 | SRR29872854 | PP825496 | 41.80 | BA.2 | Omicron | 29,647 | 9.86 | 95.4 | 347,504 | 1,853.2 |
| 24 | qc8326 | 2022-02 | SRR29872853 | PP825497 | 41.10 | BA.2 | Omicron | 29,642 | 8.09 | 95.6 | 398,228 | 2,255.1 |
| 25 | qc8327 | 2022-02 | SRR29872852 | PP825498 | 39 | BA.2 | Omicron | 29,660 | 71.68 | 90.6 | 247,540 | 1,428.3 |
| 26 | qc8338 | 2022-02 | SRR29872851 | PP825499 | 39.80 | BA.2.10 | Omicron | 29,713 | 61.01 | 91.4 | 239,720 | 1,205.3 |
| 27 | qc8342 | 2022-02 | SRR29872850 | PP825500 | 40.90 | BA.2 | Omicron | 29,578 | 49.26 | 91.7 | 236,197 | 1,134.4 |
| 28 | qc8344 | 2022-02 | SRR29872849 | PP825501 | 38.60 | BA.2 | Omicron | 29,634 | 8.13 | 95.6 | 271,237 | 1,608.3 |
| 29 | QC8512 | 2022-03 | SRR29872848 | PP825502 | 38.70 | BA.2 | Omicron | 29,729 | 0.69 | 99.6 | 154,684 | 1,217.7 |
| 30 | QC8514 | 2022-03 | SRR29872847 | PP825503 | 41.30 | BA.2 | Omicron | 29,722 | 30.41 | 93.6 | 77,197 | 456.37 |
| 31 | QC8516 | 2022-03 | SRR29872845 | PP825504 | 39.30 | BA.2.10 | Omicron | 29,729 | 0 | 98.7 | 152,397 | 1,006 |
| 32 | QC8532 | 2022-03 | SRR29872844 | PP825505 | 42.20 | BA.2 | Omicron | 29,774 | 14.1 | 95.4 | 170,158 | 999.32 |
| 33 | QC8534 | 2022-03 | SRR29872843 | PP825506 | 39.40 | BA.2.2 | Omicron | 29,776 | 0.98 | 97.9 | 108,161 | 718.66 |
| 34 | QC8538 | 2022-03 | SRR29872842 | PP825507 | 39.90 | BA.2 | Omicron | 29,729 | 36.22 | 93.2 | 171,852 | 1,127.5 |
| 35 | QC8598 | 2022-03 | SRR29872841 | PP825508 | 38.40 | BA.2 | Omicron | 29,775 | 0 | 99.4 | 140,043 | 1,101.9 |
| 36 | QC8629 | 2022-03 | SRR29872840 | PP825509 | 39.90 | BA.2 | Omicron | 29,781 | 1.56 | 99.7 | 139,410 | 956.51 |
| 37 | QC8630 | 2022-03 | SRR29872839 | PP825510 | 44.30 | BA.2.10 | Omicron | 29,441 | 80.47 | 89.5 | 88,085 | 381.77 |
| 38 | QC8636 | 2022-03 | SRR29872838 | PP825511 | 38.80 | BA.2 | Omicron | 29,779 | 0 | 99 | 124,409 | 956.03 |
| 39 | QC8638 | 2022-03 | SRR29872837 | PP825512 | 43.80 | BA.2 | Omicron | 29,635 | 70 | 90.7 | 83,769 | 398.32 |
| 40 | QC8641 | 2022-03 | SRR29872836 | PP825513 | 40.20 | BA.2.10 | Omicron | 29,698 | 9.96 | 95.6 | 73,226 | 497.95 |
| 41 | QC8644 | 2022-03 | SRR29872834 | PP825514 | 43.20 | BA.2 | Omicron | 29,682 | 17.39 | 94.7 | 117,215 | 591.27 |
| 42 | QC8651 | 2022-03 | SRR29872833 | PP825515 | 39.80 | BA.2 | Omicron | 29,771 | 38.81 | 93.1 | 131,622 | 945.49 |
| 43 | QC8661 | 2022-03 | SRR29872832 | PP825516 | 39.70 | BA.2 | Omicron | 29,776 | 5.02 | 98 | 83,146 | 577.5 |
| 44 | QC8663 | 2022-03 | SRR29872831 | PP825517 | 43.90 | BA.2 | Omicron | 29,741 | 39.46 | 92.9 | 70,942 | 291.18 |
| 45 | QC8664 | 2022-03 | SRR29872830 | PP825518 | 39.40 | BA.2 | Omicron | 29,723 | 2.48 | 97.4 | 145,775 | 1,031.2 |
| 46 | QC8665 | 2022-03 | SRR29872829 | PP825519 | 40.20 | BA.2 | Omicron | 29,729 | 1.02 | 97.7 | 137,181 | 929.15 |
| 47 | QC8667 | 2022-03 | SRR29872828 | PP825520 | 44.40 | BA.2.10.1 | Omicron | 29,755 | 31.57 | 93.5 | 112,736 | 433.42 |
| 48 | QC8668 | 2022-03 | SRR29872827 | PP825521 | 40.50 | BA.2.10 | Omicron | 29,739 | 0 | 99.5 | 100,208 | 657.97 |
| 49 | QC8710 | 2022-03 | SRR29872826 | PP825522 | 41 | BA.2 | Omicron | 29,779 | 51.57 | 92.3 | 106,539 | 740.48 |
| 50 | QC8711 | 2022-03 | SRR29872825 | PP825523 | 40.20 | BA.2 | Omicron | 29,781 | 1.37 | 97.7 | 137,732 | 998.91 |
| 51 | QC8712 | 2022-03 | SRR29874965 | PP825524 | 39.90 | BA.2 | Omicron | 29,709 | 55.25 | 91.8 | 183,189 | 1,347.3 |
| 52 | QC8714 | 2022-03 | SRR29874964 | PP825525 | 44 | BA.2 | Omicron | 29,568 | 58.27 | 91.2 | 88,203 | 345.46 |
| 53 | QC8731 | 2022-03 | SRR29874934 | PP825526 | 38.60 | BA.2.10 | Omicron | 29,789 | 0 | 99.8 | 180,103 | 1,353.3 |
| 54 | QC8732 | 2022-03 | SRR29874923 | PP825527 | 39.10 | BA.2 | Omicron | 29,773 | 0 | 99.2 | 134,732 | 998.56 |
| 55 | QC8733 | 2022-03 | SRR29874959 | PP825528 | 39.70 | BA.2 | Omicron | 29,744 | 0.02 | 98.5 | 62,387 | 437.29 |
| 56 | QC8734 | 2022-03 | SRR29874948 | PP825529 | 43.40 | BA.2 | Omicron | 29,779 | 1.6 | 97.6 | 63,998 | 361.41 |
| 57 | QC8735 | 2022-03 | SRR29874947 | PP825530 | 50.20 | BA.2 | Omicron | 29,755 | 77.51 | 90.6 | 61,243 | 276.08 |
| 58 | QC8736 | 2022-03 | SRR29874946 | PP825531 | 40.50 | BA.2 | Omicron | 29,779 | 0 | 99.7 | 110,527 | 745.68 |
| 59 | QC8737 | 2022-03 | SRR29874945 | PP825532 | 38.40 | BA.2.3 | Omicron | 29,729 | 0 | 99.2 | 106,569 | 831.41 |
| 60 | QC8738 | 2022-03 | SRR29874943 | PP825533 | 38.20 | BA.2 | Omicron | 29,776 | 0.09 | 98.5 | 97,165 | 770.57 |
| 61 | QC8768 | 2022-03 | SRR29874963 | PP825534 | 38.80 | BA.2 | Omicron | 29,773 | 13.36 | 95.4 | 83,925 | 639.71 |
| 62 | QC8803 | 2022-02 | SRR29874944 | PP825535 | 38.20 | BA.2 | Omicron | 29,729 | 0 | 98.8 | 150,622 | 1,178.4 |
| 63 | QC8812 | 2022-02 | SRR29874942 | PP825536 | 39.10 | BA.2 | Omicron | 29,726 | 22.54 | 94.3 | 103,461 | 742.99 |
| 64 | QC8837 | 2022-02 | SRR29874941 | PP825537 | 38.50 | BA.2 | Omicron | 29,745 | 0 | 98.8 | 106,001 | 808.3 |
| 65 | QC8844 | 2022-03 | SRR29874940 | PP825538 | 38.70 | BA.2 | Omicron | 29,769 | 5.57 | 96.6 | 60,147 | 455.91 |
| 66 | QC8845 | 2022-03 | SRR29874939 | PP825539 | 41.50 | BA.2 | Omicron | 29,729 | 11.64 | 95.5 | 76,449 | 440.91 |
| 67 | QC8847 | 2022-03 | SRR29874938 | PP825540 | 40 | BA.2.10 | Omicron | 29,790 | 7.23 | 96.4 | 79,020 | 542.74 |
| 68 | QC8849 | 2022-03 | SRR29874937 | PP825541 | 40.10 | BA.2 | Omicron | 29,729 | 8.11 | 96 | 122,295 | 813.73 |
| 69 | QC8850 | 2022-03 | SRR29874936 | PP825542 | 38.20 | BA.2 | Omicron | 29,766 | 2.75 | 97.2 | 97,401 | 769.71 |
| 70 | QC8851 | 2022-03 | SRR29874935 | PP825543 | 38.40 | BA.2.10 | Omicron | 29,792 | 3.72 | 97.1 | 80,651 | 648.05 |
| 71 | QC8852 | 2022-03 | SRR29874933 | PP825544 | 39.40 | BA.2 | Omicron | 29,777 | 2.81 | 97.2 | 163,295 | 1,178.1 |
| 72 | QC8853 | 2022-03 | SRR29874932 | PP825545 | 38.10 | BA.2 | Omicron | 29,805 | 85.87 | 90.4 | 83,370 | 630.61 |
| 73 | QC8854 | 2022-03 | SRR29874931 | PP825546 | 38.50 | BA.2 | Omicron | 29,781 | 40.68 | 93 | 112,793 | 856.61 |
| 74 | QC8856 | 2022-03 | SRR29874930 | PP825547 | 38.40 | BA.2 | Omicron | 29,778 | 93.59 | 90 | 66,083 | 500.9 |
| 75 | QC8858 | 2022-03 | SRR29874929 | PP825548 | 38.90 | BA.2 | Omicron | 29,781 | 91.52 | 90.1 | 97,493 | 710.69 |
| 76 | QC8868 | 2022-03 | SRR29874928 | PP825549 | 41.20 | BA.2 | Omicron | 29,779 | 7.8 | 96.2 | 147,903 | 909.59 |
| 77 | QC8870 | 2022-03 | SRR29874927 | PP825550 | 38.40 | BA.2 | Omicron | 29,781 | 13.31 | 95.5 | 106,570 | 770.01 |
| 78 | QC8911 | 2022-03 | SRR29874926 | PP825551 | 39.40 | BA.2 | Omicron | 29,781 | 8.28 | 96.2 | 127,597 | 948.84 |
| 79 | QC8912 | 2022-03 | SRR29874925 | PP825552 | 42.60 | BA.2 | Omicron | 29,744 | 94.02 | 89.9 | 63,355 | 366.15 |
| 80 | QC8913 | 2022-03 | SRR29874924 | PP825553 | 40.70 | BA.2 | Omicron | 29,729 | 80.67 | 90.5 | 82,204 | 584.12 |
| 81 | QC8914 | 2022-03 | SRR29874922 | PP825554 | 40.50 | BA.2 | Omicron | 29,781 | 3.78 | 97 | 89,511 | 637.07 |
| 82 | QC8916 | 2022-03 | SRR29874921 | PP825555 | 39.30 | BA.2 | Omicron | 29,805 | 42.35 | 92.9 | 106,977 | 761.39 |
| 83 | QC8917 | 2022-03 | SRR29874920 | PP825556 | 42.50 | BA.2 | Omicron | 29,770 | 69.31 | 91.1 | 114,848 | 615.2 |
| 84 | QC8920 | 2022-03 | SRR29874919 | PP825557 | 41.30 | BA.2 | Omicron | 29,773 | 71.31 | 91.1 | 112,709 | 765.84 |
| 85 | QC8927 | 2022-03 | SRR29874918 | PP825558 | 38.20 | BA.2 | Omicron | 29,755 | 78.81 | 90.6 | 95,313 | 751.59 |
| 86 | QC8938 | 2022-03 | SRR29874917 | PP825559 | 41 | BA.2 | Omicron | 29,768 | 30.54 | 93.7 | 103,909 | 672.93 |
| 87 | QC8944 | 2022-03 | SRR29874916 | PP825560 | 39.80 | BA.2.31.1 | Omicron | 29,771 | 30.54 | 93.7 | 131,260 | 973.09 |
| 88 | QC8946 | 2022-03 | SRR29874962 | PP825561 | 41 | BA.2 | Omicron | 29,779 | 7.7 | 96.3 | 150,787 | 1,019 |
| 89 | QC8948 | 2022-03 | SRR29874961 | PP825562 | 40.30 | BA.2 | Omicron | 29,770 | 91.03 | 90 | 66,787 | 490.69 |
| 90 | QC8957 | 2022-03 | SRR29874960 | PP825563 | 40.50 | BA.2 | Omicron | 29,781 | 3.93 | 97 | 251,044 | 1,876.1 |
| 91 | QC8958 | 2022-03 | SRR29874958 | PP825564 | 42.60 | BA.2 | Omicron | 29,771 | 22.33 | 94.5 | 252,585 | 1,414 |
| 92 | QC8965 | 2022-03 | SRR29874957 | PP825565 | 39.20 | BA.2 | Omicron | 29,787 | 4.43 | 98.5 | 276,743 | 2,243.1 |
| 93 | QC8966 | 2022-03 | SRR29874956 | PP825566 | 40.50 | BA.2.50 | Omicron | 29,814 | 0.02 | 98.7 | 237,570 | 1,813.3 |
| 94 | QC8967 | 2022-03 | SRR29874955 | PP825567 | 40.50 | BA.2.12.1 | Omicron | 29,781 | 6.7 | 96.4 | 343,467 | 2,534.5 |
| 95 | QC8968 | 2022-03 | SRR29874954 | PP825568 | 40.70 | BA.2.50 | Omicron | 29,781 | 12.46 | 95.6 | 110,853 | 818.2 |
| 96 | QC8969 | 2022-03 | SRR29874953 | PP825569 | 40.40 | BA.2 | Omicron | 29,781 | 0 | 98.9 | 175,990 | 1,358 |
| 97 | QC8975 | 2022-03 | SRR29874952 | PP825570 | 41.20 | BA.2 | Omicron | 29,779 | 19.43 | 94.8 | 124,140 | 897.66 |
| 98 | QC8979 | 2022-03 | SRR29874951 | PP825571 | 38.90 | BA.2 | Omicron | 29,760 | 82.54 | 90.4 | 110,570 | 895.92 |
| 99 | QC8981 | 2022-03 | SRR29874950 | PP825572 | 39.60 | BA.2.3 | Omicron | 29,744 | 19.62 | 94.6 | 213,190 | 1,735.1 |
| 100 | QC9031 | 2022-03 | SRR29874949 | PP825573 | 38.40 | BA.2.3 | Omicron | 29,776 | 1.49 | 97.6 | 120,816 | 1,087.3 |
| 101 | QC9032 | 2022-03 | SRR29875192 | PP825574 | 38.30 | BA.2.3 | Omicron | 29,763 | 10.7 | 95.7 | 139,954 | 1,237.8 |
| 102 | QC9033 | 2022-03 | SRR29875191 | PP825575 | 38.60 | BA.2.3 | Omicron | 29,781 | 0 | 98.8 | 139,818 | 1,228.1 |
| 103 | QC9034 | 2022-03 | SRR29875180 | PP825576 | 38.30 | BA.2.9 | Omicron | 29,799 | 36.58 | 93.3 | 164,730 | 1,474.4 |
| 104 | QC9035 | 2022-03 | SRR29875169 | PP825577 | 39.20 | BA.2 | Omicron | 29,779 | 9.66 | 96 | 174,932 | 1,504.9 |
| 105 | QC9036 | 2022-03 | SRR29875158 | PP825578 | 40.50 | BA.2 | Omicron | 29,781 | 0.01 | 98.7 | 190,449 | 1,516.8 |
| 106 | QC9037 | 2022-03 | SRR29875147 | PP825579 | 42.10 | BA.2 | Omicron | 29,779 | 12.22 | 95.6 | 258,998 | 1,845.1 |
| 107 | QC9038 | 2022-03 | SRR29875146 | PP825580 | 39.90 | BA.2 | Omicron | 29,781 | 2.74 | 97.3 | 237,240 | 1,990.9 |
| 108 | QC9039 | 2022-03 | SRR29875145 | PP825581 | 40.80 | BA.2 | Omicron | 29,775 | 0.1 | 98.5 | 154,325 | 1,211.5 |
| 109 | QC9051 | 2022-03 | SRR29875144 | PP825582 | 38.20 | BA.2 | Omicron | 29,734 | 0.17 | 99.6 | 183,635 | 1,857.9 |
| 110 | QC9052 | 2022-03 | SRR29875143 | PP825583 | 38.40 | BA.2 | Omicron | 29,734 | 0.54 | 97.9 | 256,284 | 2,459.7 |
| 111 | QC9053 | 2022-03 | SRR29875190 | PP825584 | 40.20 | BA.2 | Omicron | 29,729 | 0 | 99.1 | 218,645 | 1,753.8 |
| 112 | QC9055 | 2022-03 | SRR29875189 | PP825585 | 42.60 | BA.2 | Omicron | 29,729 | 0.42 | 98 | 190,770 | 1,236.4 |
| 113 | QC9056 | 2022-03 | SRR29875188 | PP825586 | 40.20 | BA.2 | Omicron | 29,786 | 17.36 | 99.6 | 271,489 | 2,311 |
| 114 | QC9057 | 2022-03 | SRR29875187 | PP825587 | 39.30 | BA.2 | Omicron | 29,729 | 0.68 | 97.8 | 187,485 | 1,645.5 |
| 115 | QC9058 | 2022-03 | SRR29875186 | PP825588 | 41.80 | BA.2 | Omicron | 29,729 | 19.16 | 94.6 | 204,331 | 1,099.9 |
| 116 | QC9059 | 2022-03 | SRR29875185 | PP825589 | 40.90 | BA.4.2 | Omicron | 29,714 | 22.06 | 94.4 | 101,473 | 604.09 |
| 117 | QC9081 | 2022-03 | SRR29875184 | PP825590 | 38.20 | BA.2 | Omicron | 29,729 | 0.04 | 98.4 | 159,177 | 1,571.5 |
| 118 | QC9097 | 2022-03 | SRR29875183 | PP825591 | 38.50 | BA.2 | Omicron | 29,675 | 9.77 | 95.6 | 229,436 | 1,808.7 |
| 119 | QC9116 | 2022-03 | SRR29875182 | PP825592 | 39.30 | BA.2 | Omicron | 29,809 | 0.62 | 98.1 | 234,880 | 2,084.3 |
| 120 | QC9117 | 2022-03 | SRR29875181 | PP825593 | 40.70 | BA.2.10 | Omicron | 29,781 | 13.23 | 95.5 | 132,163 | 1,049 |
| 121 | SQ1513 | 2021-11 | SRR29875179 | PP825594 | 38.90 | B.1.617.2 | Delta | 29,821 | 68.38 | 91.4 | 177,005 | 789.19 |
| 122 | SQ1529 | 2021-12 | SRR29875178 | PP825595 | 39 | B.1.617.2 | Delta | 29,855 | 46.04 | 94.4 | 518,055 | 2,193.2 |
| 123 | SQ1537 | 2021-12 | SRR29875177 | PP825596 | 38.60 | B.1.617.2 | Delta | 29,806 | 16.6 | 95.2 | 198,401 | 891.36 |
| 124 | sq1539 | 2021-12 | SRR29875176 | PP825597 | 38.40 | B.1.617.2 | Delta | 29,454 | 51.05 | 91.1 | 140,714 | 597.45 |
| 125 | SQ1580 | 2021-12 | SRR29875175 | PP825598 | 38.60 | B.1.617.2 | Delta | 29,821 | 60.8 | 91.8 | 455,614 | 2,032.7 |
| 126 | SQ1635 | 2021-12 | SRR29875174 | PP825599 | 39 | B.1.617.2 | Delta | 29,614 | 56.11 | 91.4 | 113,343 | 498.8 |
| 127 | sq1637 | 2021-12 | SRR29875173 | PP825600 | 38.40 | B.1.617.2 | Delta | 29,819 | 1.72 | 97.6 | 171,968 | 766.33 |
| 128 | sq1640 | 2021-12 | SRR29875172 | PP825601 | 38.40 | B.1.617.2 | Delta | 29,860 | 58.85 | 92 | 313,925 | 1,327.5 |
| 129 | SQ1651 | 2021-12 | SRR29875171 | PP825602 | 39.50 | B.1.617.2 | Delta | 29,785 | 40.97 | 93.2 | 179,134 | 769.07 |
| 130 | sq1654 | 2021-12 | SRR29875170 | PP825603 | 38.70 | B.1.617.2 | Delta | 29,814 | 16.75 | 96.2 | 145,600 | 1,070.6 |
| 131 | sq1655 | 2021-12 | SRR29875168 | PP825604 | 39.40 | B.1.617.2 | Delta | 29,794 | 3.39 | 97.5 | 254,578 | 1,110.5 |
| 132 | SQ1665 | 2021-12 | SRR29875167 | PP825605 | 38.50 | B.1.617.2 | Delta | 29,819 | 10.78 | 96.5 | 133,604 | 603.22 |
| 133 | SQ1667 | 2021-12 | SRR29875166 | PP825606 | 39.20 | B.1.617.2 | Delta | 29,806 | 23.02 | 96 | 115,505 | 495.66 |
| 134 | sq1672 | 2021-12 | SRR29875165 | PP825607 | 38 | B.1.617.2 | Delta | 29,492 | 1.64 | 97.4 | 207,546 | 912.01 |
| 135 | SQ1680 | 2021-11 | SRR29875164 | PP825608 | 39.90 | B.1.617.2 | Delta | 29,806 | 42.7 | 93.3 | 139,687 | 585.89 |
| 136 | SQ1682 | 2021-11 | SRR29875163 | PP825609 | 39.70 | B.1.617.2 | Delta | 29,798 | 72.56 | 91.1 | 207,047 | 873.18 |
| 137 | SQ1689 | 2021-11 | SRR29875162 | PP825610 | 39 | B.1.617.2 | Delta | 29,820 | 21.81 | 94.8 | 206,476 | 900.04 |
| 138 | sq1690 | 2021-11 | SRR29875161 | PP825611 | 38.40 | AY.4 | Delta | 29,691 | 88.05 | 90.2 | 81,881 | 348.81 |
| 139 | sq1702 | 2021-11 | SRR29875160 | PP825612 | 38.80 | AY.75 | Delta | 29,817 | 30.92 | 96.6 | 346,627 | 1,501.6 |
| 140 | sq1703 | 2021-11 | SRR29875159 | PP825613 | 38.50 | B.1.617.2 | Delta | 29,735 | 81.46 | 90.4 | 81,599 | 348.63 |
| 141 | sq1705 | 2021-11 | SRR29875157 | PP825614 | 38.80 | B.1.617.2 | Delta | 29,821 | 22.86 | 94.7 | 185,534 | 810.47 |
| 142 | SQ1707 | 2021-11 | SRR29875156 | PP825615 | 39.80 | B.1.617.2 | Delta | 29,811 | 38.93 | 95.9 | 145,128 | 616.64 |
| 143 | SQ1713 | 2021-11 | SRR29875155 | PP825616 | 41.20 | B.1.617.2 | Delta | 29,785 | 52.98 | 92.2 | 190,191 | 749.53 |
| 144 | SQ1720 | 2021-12 | SRR29875154 | PP825617 | 39.30 | B.1.617.2 | Delta | 29,889 | 12.46 | 97.2 | 1,097,224 | 4,765.7 |
| 145 | sq1723 | 2021-12 | SRR29875153 | PP825618 | 38.80 | B.1.617.2 | Delta | 29,827 | 58.95 | 91.8 | 223,543 | 954.62 |
| 146 | sq1729 | 2021-12 | SRR29875152 | PP825619 | 38.30 | B.1.617.2 | Delta | 29,738 | 91.99 | 92.6 | 117,549 | 499.5 |
| 147 | sq1742 | 2021-12 | SRR29875151 | PP825620 | 38.20 | B.1.617.2 | Delta | 29,636 | 13.05 | 95.1 | 265,739 | 1,152.6 |
| 148 | SQ1752 | 2021-12 | SRR29875150 | PP825621 | 38.90 | B.1.617.2 | Delta | 29,878 | 76.47 | 94.9 | 355,941 | 1,565.4 |
| 149 | SQ1755 | 2021-12 | SRR29875149 | PP825622 | 39.20 | B.1.617.2 | Delta | 29,885 | 10.48 | 96.1 | 808,264 | 3,431.5 |
| 150 | sq1780 | 2021-12 | SRR29875148 | PP825623 | 38.10 | B.1.617.2 | Delta | 29,315 | 90.31 | 88.9 | 110,417 | 470.77 |
| 151 | sq1819 | 2021-12 | SRR29876922 | PP825624 | 38.20 | B.1.617.2 | Delta | 29,573 | 70.81 | 90.5 | 46,018 | 200.75 |
| 152 | SQ1841 | 2021-12 | SRR29876921 | PP825625 | 38.90 | B.1.617.2 | Delta | 29,887 | 21.63 | 95.4 | 960,625 | 4,271 |
| 153 | SQ1868 | 2021-12 | SRR29876862 | PP825626 | 38.90 | B.1.617.2 | Delta | 29,748 | 58.66 | 91.9 | 138,073 | 599.77 |
| 154 | SQ1871 | 2021-12 | SRR29876851 | PP825627 | 42.60 | B.1.617.2 | Delta | 29,871 | 67.18 | 91.5 | 546,357 | 1,719.4 |
| 155 | SQ1874 | 2021-12 | SRR29876916 | PP825628 | 38.70 | B.1.617.2 | Delta | 29,819 | 47.9 | 93.8 | 154,688 | 680.96 |
| 156 | SQ5043 | 2021-09 | SRR29876905 | PP825629 | 38.50 | B.1.617.2 | Delta | 29,819 | 3.82 | 97.4 | 974,178 | 4,394.8 |
| 157 | sq5149 | 2022-01 | SRR29876894 | PP825630 | 41.40 | BA.2 | Omicron | 29,584 | 88.01 | 89.5 | 163,335 | 744.84 |
| 158 | sq5150 | 2022-01 | SRR29876883 | PP825631 | 39 | BA.2 | Omicron | 29,755 | 23.47 | 94.2 | 218,801 | 1,082.5 |
| 159 | sq5151 | 2022-01 | SRR29876873 | PP825632 | 39.40 | BA.2.2 | Omicron | 29,781 | 5.98 | 96.6 | 173,514 | 1,000.1 |
| 160 | sq5152 | 2022-01 | SRR29876871 | PP825633 | 38.90 | BA.2.2 | Omicron | 29,802 | 6.61 | 96.4 | 210,124 | 1,004 |
| 161 | sq5153 | 2022-01 | SRR29876920 | PP825634 | 39.10 | AY.76 | Delta | 29,598 | 160.68 | 89.1 | 182,923 | 914.95 |
| 162 | sq5154 | 2022-01 | SRR29876872 | PP825635 | 39.40 | BA.2 | Omicron | 29,755 | 25.15 | 94.1 | 154,101 | 892.92 |
| 163 | sq5157 | 2022-01 | SRR29876870 | PP825636 | 39.60 | BA.2.10 | Omicron | 29,755 | 38.85 | 93 | 208,367 | 869.91 |
| 164 | sq5207 | 2022-01 | SRR29876869 | PP825637 | 38.60 | BA.1.1 | Omicron | 29,752 | 69.2 | 91.1 | 237,055 | 1,295.5 |
| 165 | sq5208 | 2022-01 | SRR29876868 | PP825638 | 38.60 | BA.1.1 | Omicron | 29,727 | 74.73 | 90.7 | 126,607 | 583.6 |
| 166 | sq5209 | 2022-01 | SRR29876867 | PP825639 | 38.10 | BA.1.1 | Omicron | 29,752 | 46.34 | 92.4 | 253,489 | 1,285.2 |
| 167 | sq5215 | 2022-01 | SRR29876866 | PP825640 | 38.50 | BA.2.10 | Omicron | 29,632 | 31.53 | 93.1 | 210,497 | 1,052.7 |
| 168 | sq5218 | 2022-01 | SRR29876865 | PP825641 | 38.40 | BA.2.10 | Omicron | 29,807 | 4.39 | 96.9 | 278,136 | 1,430.4 |
| 169 | sq5220 | 2022-01 | SRR29876864 | PP825642 | 38.10 | BA.1.1 | Omicron | 29,752 | 23.83 | 94.3 | 253,216 | 1,516.1 |
| 170 | sq5224 | 2022-01 | SRR29876863 | PP825643 | 38 | BA.2.10 | Omicron | 29,792 | 14.72 | 95.3 | 229,726 | 1,252.5 |
| 171 | sq5226 | 2022-01 | SRR29876861 | PP825644 | 38.60 | BA.2.10 | Omicron | 29,772 | 15.97 | 95 | 348,822 | 2,029.8 |
| 172 | sq5230 | 2022-01 | SRR29876860 | PP825645 | 39 | BA.2.10 | Omicron | 29,704 | 28.13 | 93.6 | 347,876 | 1,944.6 |
| 173 | sq5234 | 2022-01 | SRR29876859 | PP825646 | 38 | BA.2.10 | Omicron | 29,772 | 6.07 | 96.4 | 289,439 | 1,610.9 |
| 174 | sq5236 | 2022-01 | SRR29876858 | PP825647 | 39 | BA.2.10 | Omicron | 29,755 | 32.79 | 93.4 | 146,310 | 768.35 |
| 175 | sq5240 | 2022-01 | SRR29876857 | PP825648 | 38.10 | BA.2.10 | Omicron | 29,755 | 24.41 | 94.1 | 266,493 | 1,470.9 |
| 176 | sq5248 | 2022-01 | SRR29876856 | PP825649 | 38.50 | BA.2.10 | Omicron | 29,699 | 4.71 | 96.4 | 233,513 | 1,462.2 |
| 177 | sq5254 | 2022-01 | SRR29876855 | PP825650 | 39 | BA.2 | Omicron | 29,742 | 11.56 | 95.5 | 200,688 | 1,111.9 |
| 178 | sq5257 | 2022-01 | SRR29876854 | PP825651 | 38.90 | BA.2.10 | Omicron | 29,797 | 30.99 | 93.7 | 131,641 | 824.97 |
| 179 | sq5260 | 2022-01 | SRR29876853 | PP825652 | 38.60 | BA.2.10 | Omicron | 29,584 | 25.15 | 93.5 | 271,286 | 1,550.4 |
| 180 | sq5303 | 2022-01 | SRR29876852 | PP825653 | 38.40 | BA.2 | Omicron | 29,773 | 1.11 | 97.7 | 384,010 | 2,044.3 |
| 181 | sq5304 | 2022-01 | SRR29876850 | PP825654 | 38.50 | BA.1.1 | Omicron | 29,752 | 8.26 | 96 | 267,500 | 1,395.2 |
| 182 | sq5373 | 2022-01 | SRR29876849 | PP825655 | 38.70 | BA.1 | Omicron | 29,798 | 632.37 | 96.3 | 259,655 | 1,661 |
| 183 | sq5379 | 2022-01 | SRR29876848 | PP825656 | 39.30 | BA.2 | Omicron | 2,9652 | 28.48 | 93.4 | 281,980 | 1,290.9 |
| 184 | sq5392 | 2022-01 | SRR29876847 | PP825657 | 39 | BA.2 | Omicron | 29,781 | 0.15 | 98.4 | 308,318 | 1,936.6 |
| 185 | sq5400 | 2022-01 | SRR29876846 | PP825658 | 38.20 | BA.2 | Omicron | 29,805 | 0.68 | 98 | 220,174 | 1,302.4 |
| 186 | sq5406 | 2022-01 | SRR29876845 | PP825659 | 39 | BA.2 | Omicron | 29,805 | 5.32 | 96.7 | 322,155 | 1,746.6 |
| 187 | sq5408 | 2022-01 | SRR29876844 | PP825660 | 38.70 | BA.2 | Omicron | 29,803 | 11.53 | 95.7 | 215,785 | 1,091.3 |
| 188 | sq5452 | 2022-01 | SRR29876919 | PP825661 | 38.50 | BA.1.1 | Omicron | 29,804 | 0.67 | 98 | 293,342 | 1,820 |
| 189 | sq5476 | 2022-01 | SRR29876918 | PP825662 | 38.30 | BA.2.10 | Omicron | 29,664 | 6.74 | 95.9 | 166,667 | 864.34 |
| 190 | sq5489 | 2022-01 | SRR29876917 | PP825663 | 37.90 | BA.2 | Omicron | 29,807 | 2.85 | 97.2 | 243,357 | 1,375.1 |
| 191 | sq5498 | 2022-01 | SRR29876915 | PP825664 | 38.60 | BA.2 | Omicron | 29,755 | 34.81 | 93.3 | 308,560 | 1,624.5 |
| 192 | sq5502 | 2022-01 | SRR29876914 | PP825665 | 38.90 | BA.1 | Omicron | 29,611 | 32.75 | 93 | 193,930 | 1,104.5 |
| 193 | sq5509 | 2022-01 | SRR29876913 | PP825666 | 37.90 | B.1.617.2 | Delta | 29,769 | 13.83 | 97.1 | 239,876 | 1,447.8 |
| 194 | sq5514 | 2022-01 | SRR29876912 | PP825667 | 38.40 | BA.2 | Omicron | 29,799 | 3.53 | 97 | 367,112 | 2,013.3 |
| 195 | sq5519 | 2022-01 | SRR29876911 | PP825668 | 38.50 | BA.2.10 | Omicron | 29,755 | 81.47 | 90.4 | 249,932 | 1,436.6 |
| 196 | sq5526 | 2022-01 | SRR29876910 | PP825669 | 39.60 | BA.2 | Omicron | 29,755 | 32.79 | 93.4 | 216,286 | 990.24 |
| 197 | sq6155 | 2022-02 | SRR29876909 | PP825670 | 38.90 | BA.2 | Omicron | 29,626 | 14.64 | 94.7 | 230,171 | 1,574.4 |
| 198 | sq6157 | 2022-02 | SRR29876908 | PP825671 | 39.70 | BA.2.10 | Omicron | 29,660 | 15.04 | 94.8 | 266,996 | 1,628.6 |
| 199 | sq6158 | 2022-02 | SRR29876907 | PP825672 | 41 | BA.2.10 | Omicron | 29,653 | 15.38 | 94.7 | 171,948 | 939.71 |
| 200 | sq6164 | 2022-02 | SRR29876906 | PP825673 | 41.30 | BA.2.10 | Omicron | 29,701 | 9.16 | 95.7 | 228,609 | 1,208.5 |
| 201 | sq6167 | 2022-02 | SRR29876904 | PP825674 | 39.20 | BA.2 | Omicron | 29,704 | 12.46 | 95.2 | 232,901 | 1,288.5 |
| 202 | sq6168 | 2022-02 | SRR29876903 | PP825675 | 39.30 | BA.2.10 | Omicron | 29,806 | 12.17 | 95.6 | 309,913 | 1,877.6 |
| 203 | sq6218 | 2022-02 | SRR29876902 | PP825676 | 38.80 | BA.2 | Omicron | 29,628 | 12.04 | 95 | 307,585 | 1,864.9 |
| 204 | sq6220 | 2022-02 | SRR29876901 | PP825677 | 38.30 | BA.2 | Omicron | 29,584 | 13.31 | 94.7 | 175,806 | 1,171.7 |
| 205 | sq6224 | 2022-02 | SRR29876900 | PP825678 | 39.10 | BA.2 | Omicron | 29,805 | 11.76 | 95.7 | 345,097 | 2,066.6 |
| 206 | sq6227 | 2022-02 | SRR29876899 | PP825679 | 38.80 | BA.2 | Omicron | 29,779 | 19.85 | 94.6 | 257,662 | 1,601.1 |
| 207 | SQ6292 | 2022-03 | SRR29876898 | PP825680 | 38.90 | BA.2.12.1 | Omicron | 29,780 | 4.06 | 96.9 | 234,714 | 1,846 |
| 208 | SQ6294 | 2022-04 | SRR29876897 | PP825681 | 39.50 | BA.2 | Omicron | 29,780 | 13.34 | 95.5 | 265,917 | 1,967 |
| 209 | SQ6296 | 2022-04 | SRR29876896 | PP825682 | 41.50 | BA.2.43 | Omicron | 29,728 | 35.51 | 93.2 | 96,169 | 609.25 |
| 210 | SQ6297 | 2022-04 | SRR29876895 | PP825683 | 40.50 | BA.2.78 | Omicron | 29,779 | 6.38 | 96.5 | 222,689 | 1,663.5 |
| 211 | SQ6298 | 2022-04 | SRR29876893 | PP825684 | 38.50 | BA.2.43 | Omicron | 29,775 | 2.64 | 97.3 | 222,151 | 1,798.6 |
| 212 | SQ6299 | 2022-04 | SRR29876892 | PP825685 | 39.70 | BA.2.43 | Omicron | 29,771 | 0 | 99.3 | 366,263 | 2,817.3 |
| 213 | SQ6301 | 2022-04 | SRR29876891 | PP825686 | 39.10 | BA.2 | Omicron | 29,772 | 20.85 | 94.6 | 327,061 | 2,558.5 |
| 214 | SQ6303 | 2022-04 | SRR29876890 | PP825687 | 41.20 | BA.2 | Omicron | 29,734 | 28.76 | 93.8 | 238,759 | 1,743.1 |
| 215 | SQ6307 | 2022-04 | SRR29876889 | PP825688 | 38.30 | B.1.617.2 | Delta | 29,821 | 25 | 99.5 | 196,880 | 1,737.3 |
| 216 | SQ6308 | 2022-04 | SRR29876888 | PP825689 | 43.30 | BA.2.3 | Omicron | 29,576 | 46.49 | 91.9 | 154,097 | 789.96 |
| 217 | SQ6310 | 2022-04 | SRR29876887 | PP825690 | 45.60 | BA.2.3 | Omicron | 29,738 | 87.73 | 90.2 | 229,620 | 893.29 |
| 218 | SQ6311 | 2022-04 | SRR29876886 | PP825691 | 40.80 | BA.1.1 | Omicron | 29,819 | 7.47 | 97.8 | 324,671 | 2,308.9 |
| 219 | SQ6316 | 2022-01 | SRR29876885 | PP825692 | 39.10 | BA.2.38 | Omicron | 29,775 | 1.53 | 97.9 | 313,996 | 2,449.5 |
| 220 | SQ6321 | 2022-05 | SRR29876884 | PP825693 | 44.40 | BA.2 | Omicron | 29,426 | 65.25 | 90.3 | 132,006 | 498.47 |
| 221 | SQ6324 | 2022-04 | SRR29876882 | PP825694 | 39.80 | BA.2 | Omicron | 29,679 | 18.49 | 94.5 | 289,957 | 2,361.7 |
| 222 | SQ6325 | 2022-04 | SRR29876881 | PP825695 | 38.80 | BA.2 | Omicron | 29,729 | 5.85 | 96.4 | 210,095 | 1,540.1 |
| 223 | SQ6330 | 2022-05 | SRR29876880 | PP825696 | 40 | BA.2.12.1 | Omicron | 29,665 | 8.82 | 95.7 | 207,454 | 1,653.4 |
| 224 | SQ6339 | 2022-05 | SRR29876879 | PP825697 | 38.40 | BA.2 | Omicron | 29,789 | 12.72 | 96.5 | 361,239 | 3,407.8 |
| 225 | SQ6360 | 2022-05 | SRR29876878 | PP825698 | 39.20 | BA.2 | Omicron | 29,441 | 41.44 | 91.8 | 189,082 | 1,360.9 |
| 226 | SQ6364 | 2022-05 | SRR29876877 | PP825699 | 39.70 | BA.2.9 | Omicron | 29,441 | 52.91 | 91.1 | 208,067 | 1,797.8 |
| 227 | SQ6366 | 2022-05 | SRR29876876 | PP825700 | 39.30 | BA.2 | Omicron | 29,435 | 84.23 | 89.4 | 170,110 | 1,201.8 |
| 228 | SQ6400 | 2022-02 | SRR29876875 | PP825701 | 43 | BA.2.38 | Omicron | 29,657 | 17.08 | 94.6 | 566,559 | 3,000.6 |
| 229 | ssq5500 | 2022-01 | SRR29876874 | PP825702 | 38.30 | BA.1.17.2 | Omicron | 29,804 | 44.2 | 92.8 | 244,823 | 1,697.3 |
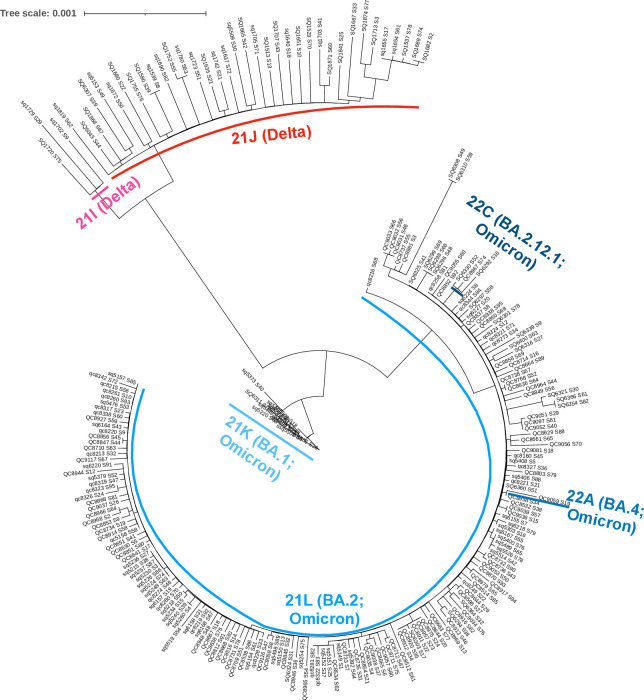
Among the samples, 82.97% were Omicron with BA.2 and its sub-lineages (94.21%, n = 179/190) and 17.03% were Delta with B.1.617.2 (92.3%, n = 36/39). The phylogenetic tree showed the SARS-CoV-2 genomic diversity in Nepal (Fig. 1). This study provides insights and contributes valuable data for genomic surveillance in understanding and monitoring SARS-CoV-2 variants in Nepal.
Fig 1.
Phylogenetic analysis of complete SARS-CoV-2 genome sequences using GTR substitution model. The phylogenetic analysis showed a distinct clustering pattern among the circulating variants where the tree shows distinct clustering of Delta (red) and Omicron (blue).
ACKNOWLEDGMENTS
This work was funded by the United States National Institutes of Health (grant U01AI151810).
Contributor Information
Krishna D. Manandhar, Email: krishna.manandhar@gmail.com.
John J. Dennehy, Queens College Department of Biology, Queens, New York, USA
DATA AVAILABILITY
The GenBank accession numbers of the 229 sequences from this study are provided in Table 1. The raw reads are available in the SRA database under the BioProject number PRJNA1137145, and the accession IDs for these raw reads are also available in Table 1.
ETHICS APPROVAL
Ethical approval was obtained from Nepal Health Research Council (NHRC) (Regd.: 274/2020).
REFERENCES
- 1. Dhama K, Khan S, Tiwari R, Sircar S, Bhat S, Malik YS, Singh KP, Chaicumpa W, Bonilla-Aldana DK, Rodriguez-Morales AJ. 2020. Coronavirus disease 2019-COVID-19. Clin Microbiol Rev 33:e00028-20. doi: 10.1128/CMR.00028-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rayamajhee B, Pokhrel A, Syangtan G, Khadka S, Lama B, Rawal LB, Mehata S, Mishra SK, Pokhrel R, Yadav UN. 2021. How well the government of Nepal is responding to COVID-19? An experience from a resource-limited country to confront unprecedented pandemic. Front Public Health 9:597808. doi: 10.3389/fpubh.2021.597808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bastola A, Sah R, Rodriguez-Morales AJ, Lal BK, Jha R, Ojha HC, Shrestha B, Chu DKW, Poon LLM, Costello A, Morita K, Pandey BD. 2020. The first 2019 novel coronavirus case in Nepal. Lancet Infect Dis 20:279–280. doi: 10.1016/S1473-3099(20)30067-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H. 2021. Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. doi: 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wilm A, Aw PPK, Bertrand D, Yeo GHT, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N. 2012. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 40:11189–11201. doi: 10.1093/nar/gks918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 6:80–92. doi: 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. O’Toole Á, Scher E, Underwood A, Jackson B, Hill V, McCrone JT, Colquhoun R, Ruis C, Abu-Dahab K, Taylor B, Yeats C, du Plessis L, Maloney D, Medd N, Attwood SW, Aanensen DM, Holmes EC, Pybus OG, Rambaut A. 2021. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol 7:veab064. doi: 10.1093/ve/veab064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aksamentov I, Roemer C, Hodcroft E, Neher RA. 2021. Nextclade: clade assignment, mutation calling and quality control for viral genomes. JOSS 6:3773. doi: 10.21105/joss.03773 [DOI] [Google Scholar]
- 9. Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34:4121–4123. doi: 10.1093/bioinformatics/bty407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Letunic I, Bork P. 2021. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. doi: 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The GenBank accession numbers of the 229 sequences from this study are provided in Table 1. The raw reads are available in the SRA database under the BioProject number PRJNA1137145, and the accession IDs for these raw reads are also available in Table 1.