ABSTRACT
Streptococcus agalactiae (Group B Streptococcus) strain COH1 is a representative strain of serotype III, multi-locus sequence type 17, which is disproportionately associated with neonatal meningitis. Here we report the transcriptome of COH1 when interacting with human brain endothelial cells compared with COH1 alone.
KEYWORDS: Group B Streptococcus, transcriptome, blood-brain barrier
ANNOUNCEMENT
To cause bacterial meningitis, bacteria must interact with and penetrate the blood-brain barrier (BBB) (1). The BBB is composed of highly specialized brain endothelial cells (BECs) that serve to maintain barrier integrity and prevent pathogens from exiting the circulation and entering the central nervous system (2). The surface of BECs represents an environment that meningitis-causing bacteria must interact with. Streptococcus agalactiae (Group B Streptococcus [GBS]) is the leading cause of neonatal meningitis, and presently, there is no vaccine available (1, 3, 4). GBS serotype III, multi-locus sequence type (MLST) 17, is disproportionately associated with neonatal meningitis (1, 5, 6). Much work has been conducted to identify how the BBB responds to bacteria such as GBS (1, 7–11). However, little is known about how GBS responds to association with BECs.
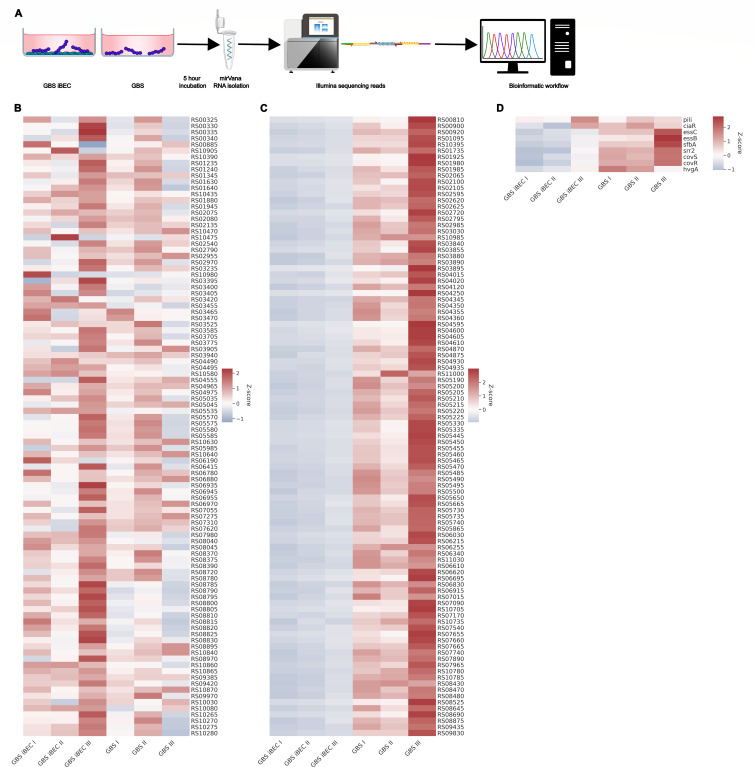
GBS strain COH1 (serotype III, MLST-17, a clinical isolate obtained from a newborn) was either incubated alone in wells containing endothelial cell (EC) media (hESFM + B27) or in wells containing EC media and human induced pluripotent stem cell-derived brain-like endothelial cells (iBECs) for 5 h at 37°C + 5% CO2 (multiplicity of infection of 10) (10, 12–17). In the publication originating the data set presented here, more details are available on the methods for iBEC obtention (18). After washing away non-cell-associated bacteria with phosphate-buffered saline, total RNA was immediately collected using the mirVana RNA (Thermo Scientific) isolation kit (Fig. 1A) (18). This process was repeated three times to collect individual biological replicates. The quality of the extracted total RNA samples was checked using the Agilent RNA 6000 Nano Kit, assay mode Prokaryote Total RNA Nano, on a Bio Analyzer. cDNA libraries were prepared as previously described (19). For cDNA library preparation, the Illumina Stranded Total RNA Prep Ligation with Ribo-Zero Plus protocol was used according to the manufacturer’s recommendations. Libraries were pooled in equimolar concentrations and fractionated using the Agencourt AMPure kit and sequenced on an Illumina NextSeq500 platform in single-end mode for 75 cycles. Obtained reads (Illumina read length 75 bp) were trimmed using Cutadapt (version 1.15), and Illumina TruSeq adapters were removed from the 3′ end. Reads with a Phred quality score less than 20 and following downstream bases were cut. READemption (version 0.4.3, DOI: 10.5281/zenodo.250598) was used for downstream mapping (segemehl version 0.2.0 associated with READemption) and analysis (20–23). Streptococcus agalactiae COH1 genome and annotation from National Center for Biotechnology Information GenBank (accession number HG939456.1, assembly accession number GCA_000689235.1) were extended with an existing sRNA annotation (24). Differential gene expression was performed with R package DESeq2 (version 1.18.1, R version 3.4.4) based on raw read counts (25). As differentially expressed, were genes defined with an adjusted (Benjamini-Hochberg-corrected) P value of 0.05. The complete bioinformatic workflow and number of reads of the individual conditions and replicates can be found under DOI https://zenodo.org/records/12652566. These methods are expanded versions of descriptions in our related work (18).
Fig 1.
Heatmaps of COH1 transcriptome when interacting with human brain endothelial cells compared with COH1 alone. (A) Schematic of the workflow: Group B Streptococcus (GBS) and induced pluripotent stem cell-derived brain-like endothelial cells (iBECs). (B–D) Heatmap of differentially regulated genes: (B) top upregulated genes, (C) top downregulated genes, and (D) selection of known GBS adhesins.
Compared with GBS in media alone, 430 of the 2,068 total annotated genes were found to be differentially regulated in GBS associated with iBECs, with 360 genes exhibiting downregulation (18). These data suggest that GBS can sense host cell association and accordingly adapts its gene expression pattern (Fig. 1B and C). Examination of known GBS adhesins that have been demonstrated to facilitate interaction with BECs does not show differential regulation at the transcript level, suggesting that these factors are not upregulated when interacting with the BBB (Fig. 1D) (6, 7, 26–28). Our data suggest that other contributing factors may facilitate GBS intimate interaction with BECs, and presently unknown mechanisms of BBB dysfunction could be discovered by examining the differentially expressed bacterial genes uncovered here (18).
ACKNOWLEDGMENTS
We thank our funding sources, National Institutes of Health/National Institute of Neurological Disorders and Stroke (R15NS131921 to B.J.K.), the Alexander von Humboldt Postdoctoral Scholarship (to N.V.), and Deutsche Forschungsgemeinschaft (German Research Foundation, GRK2157 to A.S.-U.). Additionally, we acknowledge Sarah Reichardt from the Helmholtz Institute for RNA and Infection for her support in RNA sample processing and quality check.
Contributor Information
Brandon J. Kim, Email: bjkim4@ua.edu.
André O. Hudson, Rochester Institute of Technology, Rochester, New York, USA
DATA AVAILABILITY
Data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus under accession number GSE197489.
REFERENCES
- 1. Doran KS, Fulde M, Gratz N, Kim BJ, Nau R, Prasadarao N, Schubert-Unkmeir A, Tuomanen EI, Valentin-Weigand P. 2016. Host-pathogen interactions in bacterial meningitis. Acta Neuropathol 131:185–209. doi: 10.1007/s00401-015-1531-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abbott NJ, Patabendige AAK, Dolman DEM, Yusof SR, Begley DJ. 2010. Structure and function of the blood–brain barrier. Neurobiol Dis 37:13–25. doi: 10.1016/j.nbd.2009.07.030 [DOI] [PubMed] [Google Scholar]
- 3. Bjerkhaug AU, Ramalingham S, Mboizi R, Le Doare K, Klingenberg C. 2024. The immunogenicity and safety of Group B streptococcal maternal vaccines: a systematic review. Vaccine (Auckl) 42:84–98. doi: 10.1016/j.vaccine.2023.11.056 [DOI] [PubMed] [Google Scholar]
- 4. Nuccitelli A, Rinaudo CD, Maione D. 2015. Group B Streptococcus vaccine: state of the art. Ther Adv Vaccines 3:76–90. doi: 10.1177/2051013615579869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bianchi-Jassir F, Paul P, To KN, Carreras-Abad C, Seale AC, Jauneikaite E, Madhi SA, Russell NJ, Hall J, Madrid L, Bassat Q, Kwatra G, Le Doare K, Lawn JE. 2020. Systematic review of Group B streptococcal capsular types, sequence types and surface proteins as potential vaccine candidates. Vaccine (Auckl) 38:6682–6694. doi: 10.1016/j.vaccine.2020.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tazi A, Disson O, Bellais S, Bouaboud A, Dmytruk N, Dramsi S, Mistou MY, Khun H, Mechler C, Tardieux I, Trieu-Cuot P, Lecuit M, Poyart C. 2010. The surface protein HvgA mediates group B Streptococcus hypervirulence and meningeal tropism in neonates. J Exp Med 207:2313–2322. doi: 10.1084/jem.20092594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Banerjee A, Kim BJ, Carmona EM, Cutting AS, Gurney MA, Carlos C, Feuer R, Prasadarao NV, Doran KS. 2011. Bacterial pili exploit integrin machinery to promote immune activation and efficient blood-brain barrier penetration. Nat Commun 2:462. doi: 10.1038/ncomms1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doran KS, Engelson EJ, Khosravi A, Maisey HC, Fedtke I, Equils O, Michelsen KS, Arditi M, Peschel A, Nizet V. 2005. Blood-brain barrier invasion by group B Streptococcus depends upon proper cell-surface anchoring of lipoteichoic acid. J Clin Invest 115:2499–2507. doi: 10.1172/JCI23829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Doran KS, Liu GY, Nizet V. 2003. Group B streptococcal beta-hemolysin/cytolysin activates neutrophil signaling pathways in brain endothelium and contributes to development of meningitis. J Clin Invest 112:736–744. doi: 10.1172/JCI17335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim BJ, Bee OB, McDonagh MA, Stebbins MJ, Palecek SP, Doran KS, Shusta EV. 2017. Modeling Group B Streptococcus and blood-brain barrier interaction by using induced pluripotent stem cell-derived brain endothelial cells. mSphere 2:e00398-17. doi: 10.1128/mSphere.00398-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim BJ, Hancock BM, Bermudez A, Del Cid N, Reyes E, van Sorge NM, Lauth X, Smurthwaite CA, Hilton BJ, Stotland A, Banerjee A, Buchanan J, Wolkowicz R, Traver D, Doran KS. 2015. Bacterial induction of Snail1 contributes to blood-brain barrier disruption. J Clin Invest 125:2473–2483. doi: 10.1172/JCI74159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Espinal ER, Matthews T, Holder BM, Bee OB, Humber GM, Brook CE, Divyapicigil M, Sharp J, Kim BJ. 2022. Group B Streptococcus-induced macropinocytosis contributes to bacterial invasion of brain endothelial cells. Pathogens 11:474. doi: 10.3390/pathogens11040474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim B.J, McDonagh MA, Deng L, Gastfriend BD, Schubert-Unkmeir A, Doran KS, Shusta EV. 2019. Streptococcus agalactiae disrupts P-glycoprotein function in brain endothelial cells. Fluids Barriers CNS 16:26. doi: 10.1186/s12987-019-0146-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim B.J, Shusta EV, Doran KS. 2019. Past and current perspectives in modeling bacteria and blood-brain barrier interactions. Front Microbiol 10:1336. doi: 10.3389/fmicb.2019.01336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lippmann ES, Azarin SM, Kay JE, Nessler RA, Wilson HK, Al-Ahmad A, Palecek SP, Shusta EV. 2012. Derivation of blood-brain barrier endothelial cells from human pluripotent stem cells. Nat Biotechnol 30:783–791. doi: 10.1038/nbt.2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilson CB, Weaver WM. 1985. Comparative susceptibility of group B streptococci and Staphylococcus aureus to killing by oxygen metabolites. J Infect Dis 152:323–329. doi: 10.1093/infdis/152.2.323 [DOI] [PubMed] [Google Scholar]
- 17. Rubens CE, Wessels MR, Heggen LM, Kasper DL. 1987. Transposon mutagenesis of type III group B Streptococcus: correlation of capsule expression with virulence. Proc Natl Acad Sci U S A 84:7208–7212. doi: 10.1073/pnas.84.20.7208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vollmuth N, Bridgers BE, Armstrong ML, Wood JF, Gildea AR, Espinal ER, Hooven TA, Barbieri G, Westermann AJ, Sauerwein T, Foerstner KU, Schubert-Unkmeir A, Kim BJ. 2024. Group B Streptococcus transcriptome when interacting with brain endothelial cells. J Bacteriol 206:e0008724. doi: 10.1128/jb.00087-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Westermann AJ, Förstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Müller L, Reinhardt R, Stadler PF, Vogel J. 2016. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature New Biol 529:496–501. doi: 10.1038/nature16547 [DOI] [PubMed] [Google Scholar]
- 20. Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 21. Förstner KU, Vogel J, Sharma CM. 2014. READemption-a tool for the computational analysis of deep-sequencing-based transcriptome data. Bioinformatics 30:3421–3423. doi: 10.1093/bioinformatics/btu533 [DOI] [PubMed] [Google Scholar]
- 22. Hoffmann S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, Stadler PF, Hackermüller J. 2009. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput Biol 5:e1000502. doi: 10.1371/journal.pcbi.1000502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Otto C, Stadler PF, Hoffmann S. 2014. Lacking alignments? The next-generation sequencing mapper segemehl revisited. Bioinformatics 30:1837–1843. doi: 10.1093/bioinformatics/btu146 [DOI] [PubMed] [Google Scholar]
- 24. Keogh RA, Spencer BL, Sorensen HM, Zapf RL, Briaud P, Bonsall AE, Doran KS, Carroll RK. 2021. Global annotation, expression analysis, and stability of candidate sRNAs in group B Streptococcus. MBio 12:e0280321. doi: 10.1128/mBio.02803-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maisey HC, Hensler M, Nizet V, Doran KS. 2007. Group B streptococcal pilus proteins contribute to adherence to and invasion of brain microvascular endothelial cells. J Bacteriol 189:1464–1467. doi: 10.1128/JB.01153-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seo HS, Mu R, Kim BJ, Doran KS, Sullam PM. 2012. Binding of glycoprotein Srr1 of Streptococcus agalactiae to fibrinogen promotes attachment to brain endothelium and the development of meningitis. PLoS Pathog 8:e1002947. doi: 10.1371/journal.ppat.1002947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Sorge NM, Quach D, Gurney MA, Sullam PM, Nizet V, Doran KS. 2009. The group B streptococcal serine-rich repeat 1 glycoprotein mediates penetration of the blood-brain barrier. J Infect Dis 199:1479–1487. doi: 10.1086/598217 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus under accession number GSE197489.