ABSTRACT
In this chapter, we update our 2004 review of “The Life of Commensal Escherichia coli in the Mammalian Intestine” (https://doi.org/10.1128/ecosalplus.8.3.1.2), with a change of title that reflects the current focus on “Nutrition of E. coli within the Intestinal Microbiome.” The earlier part of the previous two decades saw incremental improvements in understanding the carbon and energy sources that E. coli and Salmonella use to support intestinal colonization. Along with these investigations of electron donors came a better understanding of the electron acceptors that support the respiration of these facultative anaerobes in the gastrointestinal tract. Hundreds of recent papers add to what was known about the nutrition of commensal and pathogenic enteric bacteria. The fact that each biotype or pathotype grows on a different subset of the available nutrients suggested a mechanism for succession of commensal colonizers and invasion by enteric pathogens. Competition for nutrients in the intestine has also come to be recognized as one basis for colonization resistance, in which colonized strain(s) prevent colonization by a challenger. In the past decade, detailed investigations of fiber- and mucin-degrading anaerobes added greatly to our understanding of how complex polysaccharides support the hundreds of intestinal microbiome species. It is now clear that facultative anaerobes, which usually cannot degrade complex polysaccharides, live in symbiosis with the anaerobic degraders. This concept led to the “restaurant hypothesis,” which emphasizes that facultative bacteria, such as E. coli, colonize the intestine as members of mixed biofilms and obtain the sugars they need for growth locally through cross-feeding from polysaccharide-degrading anaerobes. Each restaurant represents an intestinal niche. Competition for those niches determines whether or not invaders are able to overcome colonization resistance and become established. Topics centered on the nutritional basis of intestinal colonization and gastrointestinal health are explored here in detail.
KEYWORDS: microbiome, E. coli, colonization, nutrition, intestine, colonization resistance
INTRODUCTION
Escherichia coli is arguably the best understood of model organisms; the species comprised commensal and pathogenic strains (1). As a commensal, E. coli is a persistent colonizer of the vertebrate gut (2). It is estimated there are 1021 E. coli cells among the human population (3). As a pathogen, E. coli strains can cause both intestinal and extra-intestinal infections (2). E. coli infections are a significant health problem which is associated with increased mortality, morbidity, and health-care-related costs around the world (4). E. coli is responsible for a million deaths per year around the globe (5). Colonized commensal E. coli acts as a barrier to prevent colonization by invading pathogens (6). Furthermore, invading pathogens almost always need to colonize the intestine before initiating a successful infection (3, 7, 8). However, how E. coli colonizes the mammalian intestine is not completely understood. A better understanding of colonization mechanisms can be exploited to prevent pathogens from causing gastrointestinal diseases. The colonic mucus layer is the primary site of E. coli colonization, where it competes with other organisms for limiting nutrients (3). The mucus layer covering the intestinal epithelium not only provides nutrients but also provides attachment sites (9, 10). Innate and adaptive immunity, bacterial cell-to-cell communication, etc., also play important roles in the colonization of the intestine; these topics are discussed elsewhere (11–14). In this article, we focus discussion on the nutrients available in the mucus layer, competition for nutrients that support E. coli colonization, colonization resistance, and how invaders overcome colonization resistance.
MUCOSAL HABITAT OF THE LARGE INTESTINE
Since commensal Enterobacteriaceae generally colonize the mucus layer of the mammalian intestine, we begin by considering the mucus layer as a microbial habitat. Very few microorganisms can survive the drastic acidic pH of gastric juices in the stomach. Analysis of stomach mucosal biopsies and gastric juices revealed there are 102–104 cultivable bacteria per gram of stomach contents. Isolates from the stomach include Propionibacterium, Lactobacillus, Streptococcus, and Staphylococcus (15). In the small intestine, the bacterial population in the duodenum increases from 104–105 CFU/mL to 107–108 CFU/mL in the distal ileum. Bacteria in the small intestine must respond to changing conditions such as rapid transit of the luminal contents and influx of digestive enzymes and bile. Representative genera in the small intestine include Bacteroides, Clostridium, Lactobacillus, Staphylococcus, Streptococcus, etc. (16).
In contrast to the small intestine, the large intestine is colonized by 1011–1012 CFU/mL (17). The mammalian large intestine is divided into three sections—the cecum, the colon, and the rectum. The epithelium of the large intestine, which consists of goblet cells and enterocytes, is renewable, and in humans, as many as 2–5 × 106 epithelial cells are shed per minute (17).
The epithelium of the large intestine is covered by a mucus layer secreted by goblet cells lining the epithelium (10, 18). The mucus coat protects the epithelial cells from bacteria, digestive enzymes, and toxic substances (10). The major component of the mucus layer is the gel-forming glycoprotein, MUC2 mucin, which forms a large net-like polymer (18, 19). Structurally, mucin domains have a central protein core characterized by abundant proline, threonine, and serine, with glycans projecting in all directions (20). Sugars commonly found in the glycan chains include galactose, N-acetylglucosamine (NAG), N-acetylgalactosamine (GalNAc), fucose, and sialic acid [N-acetylneuraminic acid (NANA)] (21). The O-glycan repertoire on MUC2 mucin varies between host species, and the glycosylation of mucin makes it non-degradable by host digestive enzymes; however, the glycans can be degraded by bacterial species (20). Study of the O-glycosylation of MUC2 mucin from the human colon showed more than 100 MUC2 O-glycans of chain lengths between 2 and 12 residues, including some that were identified for the first time (21). Mouse colonic mucus consists of two layers totaling about 150 µm in depth compared to about 800 µm in humans (18, 22). The tightly packed inner layer is firmly attached to the epithelial lining and contains few, if any bacteria (18, 19). The firm inner mucus layer is renewed constantly and is continuously converted into the soft outer layer. The outer mucus layer proximal to the intestinal lumen is looser because of the proteolytic degradation of mucin (18, 19). The thickness of the outer layer is highly variable depending on what bacteria are present (23). The outer layer of mucus and the digesta in the lumen is colonized by over 500 species of microorganisms, mostly obligate anaerobes. Representative bacteria present in the colon include Bacteroides spp., Bifidobacterium spp., Clostridium spp., Peptococcus spp., Streptococcus spp., Enterococcus spp., Lactobacillus spp., members of the family Enterobacteriaceae, etc. (24–27).
Mucosal glycans, besides serving as attachment sites for bacterial adhesins (28), also serve as a source of nutrients for the gastrointestinal microbiota (29). Since the glycans on MUC2 mucin vary between host species, bacteria colonizing a specific host carry a distinct set of enzymes to degrade the glycans (20, 29). However, not all bacteria in the gut have the required glycan-degrading enzymes, and therefore, they rely on cross-feeding by other bacteria in the community (20). Cross-feeding of mucin non-degraders by mucin-degrading species contributes to the stability of the microbial community (30). Several members of Verrucomicrobia, Bacteroidetes, and Firmicutes harbor one or multiple copies of mucin-degrading glycoside hydrolases (31). Commensal E. coli is dispersed in the mucus layer but is not attached to the epithelium (32, 33). However, E. coli and other enteric bacteria do not usually produce extracellular enzymes for degrading mucin oligosaccharides (34). E. coli grows primarily on simple monosaccharides and disaccharides, which are released by anaerobes in the gut (35, 36). Henderson and colleagues (37) identified and characterized a secreted serine protease protein, Pic (for “protein involved in intestinal colonization”), in enteroaggregative E. coli (EAEC), which had mucinolytic activity. Homologs of Pic have been reported in some isolates of uropathogenic E. coli (UPEC), enteroinvasive E. coli, and enteropathogenic E. coli (EPEC) (38–42). However, none of the isolates of non-pathogenic E. coli tested had Pic (42). Pic promotes intestinal colonization of EAEC in streptomycin-treated mice and its growth in cecal mucus, cecal contents, and in minimal media containing mucin but not in the absence of mucin (43). Recently, using a human colonic organoid model, it was shown that purified Pic degrades the mucin layer, and EAEC producing Pic showed increased colonization of the colonic epithelium compared to a Pic deletion mutant (44). Thus, Pic plays an important role in MUC2 barrier disruption in the intestine (44), promoting mucus layer penetration and attachment to the epithelium, and hence, is a virulence factor; however, whether Pic plays a role in increasing the availability of any nutrients from mucus is not known.
Since the population of E. coli in the large intestine remains constant, its growth rate in the mucus layer must exceed the rate it is sloughed into the lumen to prevent being washed out of the mucus layer (45). It was estimated that E. coli needs to divide roughly every 2 hours to maintain a stable population of 108 CFU/g of feces, which was consistent with the generation time calculated for E. coli BJ4, a rat isolate (45–47). In these studies, growth rates were determined by 23S rRNA-targeted fluorescent in situ hybridization to estimate ribosomal counts, which correlate with the growth rate. E. coli BJ4 grows in the mucus layer of the mouse large intestine with a generation time of 40–80 min (46). The growth rates were similar in streptomycin-treated and germ-free mice (45). In conventionalized streptomycin-treated mice and conventionalized germ-free mice, the estimated generation time increased to 116–130 min following 9 days of conventionalization. This contrasts with the rapid growth (30-min generation time) of E. coli BJ4 in intestinal mucus in vitro (48, 49). Since the generation time of E. coli was higher in conventionalized mice as compared to streptomycin-treated and germ-free mice, it is thought that conventionalization possibly leads to a decrease in the rate of mucus sloughing (45). E. coli, however, does not grow on intestinal luminal contents in vitro (23, 48, 49), and the mucus layer hosts a distinct intestinal microbial niche compared to the luminal contents (23, 48, 49). This suggests that nutrients secreted by the host in the form of mucus are more important than the nutrients ingested by the host (47), or some inhibitors present in the luminal contents inhibit the growth of E. coli in the contents (48).
NUTRIENT SOURCES IN THE INTESTINE
The mammalian intestine is a highly complex ecosystem that is shaped by multiple factors: the host, complex microbial community, and interactions with the environment such as food intake and pathogens (50). Limiting nutrients are major determinants of species composition in an ecosystem, including the mammalian intestine (3, 51). The microbial community in the intestine competes for carbon and energy sources, terminal electron acceptors, nitrogen sources, and iron (52–56). Dietary nutrients, endogenously secreted nutrients, shed epithelial cells, microbial metabolites, and components of mucus are the nutrient sources for bacteria in the intestine (3, 17, 57–62). Absorption of nutrients by the host determines the availability of dietary nutrients to the gut microbiota (57). Animals have developed numerous physiological strategies to absorb nutrients in the small intestine, so most digestible nutrients are absorbed there and are not available to the microbiota of the large intestine (55). However, undigested carbohydrates such as dietary fiber and some of the escaped digestible carbohydrates enter the colon and are available to the microbiota (63). In addition to the dietary nutrients entering the gastrointestinal tract, other substances from endogenous sources, such as saliva, bile, urea, glutathione, etc., simultaneously enter into the lumen where they can be metabolized by the microbiota (59). Human saliva, for example, contains approximately 3,000 mg/L of total proteins in the form of amylases, mucins MUC5B (MG1) and MUC7 (MG2), cystatin, histatins, statherin, etc., the majority of which are glycosylated (64). In pigs weighing 30–40 kg, 500 g of saliva flows per day which has been calculated to contribute 400 mg of nitrogen daily (59). Since the epithelium of the colon is renewable and cells are constantly shed, the dead epithelial cells are also an important source of nutrition for the microbiota (17, 65).
Goblet cells in the intestine continuously synthesize and secrete mucin which is the dominant glycoprotein component of mucus in the intestine (66). Mucin comprised 80% polysaccharide and 20% protein and is highly viscous (65). In addition to mucin, mucus also contains many other glycoproteins, proteins, sugars, lipids, and glycolipids, most of which can serve as nutrient sources for the microbiota (65). Indigenous intestinal microbes encode enzymes required to degrade the mucus layer and utilize it. As the mucus is degraded, the components are also shed into the lumen which forms part of the luminal contents that are excreted in the feces (65).
NUTRIENT CONSUMPTION AND CROSS-FEEDING OF THE INTESTINAL MICROBIOTA
Dietary carbohydrates and proteins are primarily absorbed in the proximal part of the small intestine (67). Mass spectrometric analysis of free amino acids from the proximal small intestine and colon revealed that >90% of amino acids are absorbed in the small intestine (68). Since facultative anaerobes generally cannot degrade oligosaccharides or polysaccharides [rare exceptions include dextrin (3) by E. coli and L-arabinose liberated from plant polysaccharides by Salmonella Typhimurium (69)], they must rely on sugars that are liberated by obligate anaerobes upon hydrolysis of complex polysaccharides during their expansion in the gut (70, 71) (Fig. 1). Ng and co-workers (70) demonstrated that Bacteroides thetaiotamicron (B. thetaiotamicron) enhanced the levels of free sialic acids in the ceca of B. thetaiotamicron-mono-associated mice as compared to germ-free mice. While B. thetaiotamicron prefers dietary fiber, it can switch to mucus-derived glycans in the cecum when dietary polysaccharides are absent (72). Under conditions of chronic or intermittent dietary fiber deficiency, mucus-eroding microbiota promote access to the epithelium and increase susceptibility to pathogens (73). S. Typhimurium utilizes the monosaccharides, fucose and sialic acid, liberated by the microbiota; when those catabolic pathways were knocked out, competitive fitness of S. Typhimurium was reduced. Clostridium difficile also utilized sialic acid liberated by B. thetaiotamicron, and reduction of free sialic acid impaired C. difficile expansion in the gut (70). It is known that B. thetaiotamicron lacks the catabolic pathway required to consume sialic acid but has enzymes that release sialic acid from the mucus (70, 74). Jimenez and co-workers showed that B. thetaiotamicron digests dietary pectin and makes galacturonic acid available in the gut (75). Pectin, which is rich in D-galacturonic acid, is present in plant cell walls and can be digested by saccharolytic members of the microbiota such as B. thetaiotamicron (75, 76). Galacturonic acid released by the gut microbiota in the lumen is used by enterohemorrhagic E. coli (EHEC) and Citrobacter rodentium as a carbon source and as a signal to aid in the colonization and initiate infection (75).
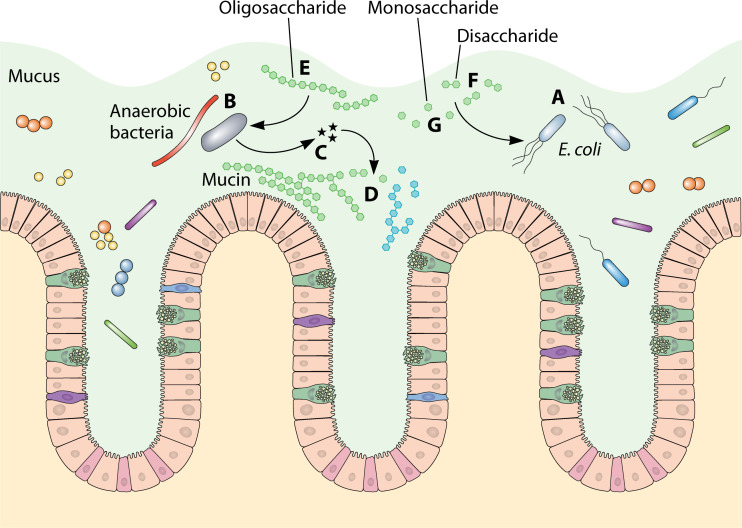
Fig 1.
Illustration of how E. coli (A) obtains nutrients in the mucosal layer of the large intestine. Anaerobes (B) produce glycosyl hydrolases (C) which degrade complex polysaccharides in dietary fibers and mucin (D) to release oligosaccharides (E), disaccharides (F), and monosaccharides (G). Oligosaccharides are preferred by anaerobes, while monosaccharides and disaccharides are preferred by E. coli.
B. thetaiotamicron has polysaccharide utilization loci (PULs) which encode glycolytic enzymes needed to utilize host as well as plant glycans (74). During colonization of germ-free mice with B. thetaiotamicron and Eubacterium rectale, B. thetaiotamicron upregulated several PULs and signaled the host to produce mucosal glycans which it can utilize. E. rectale responded to B. thetaiotamicron by downregulating glycan-hydrolyzing enzymes while upregulating sugar and amino acid transporters (77). In response to the signal from B. thetaiotamicron, the colonic epithelium upregulated genes involved in the synthesis of mucosal glycans (glycosphingolipids and O-glycans), suggesting the possibility that B. thetaiotamicron increases host production of glycans and benefits by utilizing those glycans. However, no follow-up studies of this possibility have been reported.
Studies of microbial interactions demonstrate functional redundancy among members of different phyla, and this metabolic flexibility helps to ensure the stability of the intestinal ecosystem (3, 77). More than 50% of its glycan hydrolase enzymes are predicted to be secreted by B. thetaiotamicron into the periplasm or to the cell exterior, thus liberating monosaccharides and oligosaccharides from complex polysaccharides (72). B. thetaiotamicron appears to utilize liberated monosaccharides, such as arabinose, xylose, fucose, galacturonate, β-hexosamines, fructose, glucose, galactose, etc., as genes involved in their transport and utilization are fitness determinants (78). Previously, it was also demonstrated that the colonization of germ-free mice by B. thetaiotamicron significantly reduced the concentrations of abundant monosaccharides present in the cecum (72). Many bacteria other than E. coli also rely on simple sugars released by anaerobic digestion (79, 80). Li and co-workers (23) provided evidence that genes for maltose utilization were upregulated in E. coli when colonizing germ-free mice together with B. thetaiotamicron as compared to E. coli colonizing germ-free mice alone, which is consistent with cross-feeding.
E. coli is usually grown in the laboratory in batch cultures on a high concentration of a single carbon source. When a mixture of two carbon sources at high concentration is present in a batch culture, the carbon source that supports the highest growth rate is typically utilized by E. coli first, resulting in diauxic growth (81). However, natural environments such as the intestine may contain mixtures of many carbon sources present at low concentrations (82). When E. coli was fed a mixture of six sugars—glucose, galactose, maltose, ribose, arabinose, and fructose, all six sugars were utilized simultaneously (81). When commensal E. coli MG1655 was grown in vitro in a minimal medium containing 13 different sugars which are known to be present in mucus, gluconate, NANA, galactose, mannose, N-acetylglucosamine, N-acetylgalactosamine, glucosamine, glucuronate, galacturonate, fucose, ribose, L-arabinose, and maltose, all nutrients except GalNAc were used (36). Catabolite-repressing sugars, i.e., NAG, gluconate, and galactose, were used first, and before those sugars were completely exhausted, the remaining sugars were metabolized simultaneously (36). Interestingly, the in vitro order of nutrient preference of commensal E. coli MG1655 and pathogenic EHEC EDL933 was almost identical except that EHEC EDL933 also used GalNAc (36). To determine which of those carbon sources supported colonization, a series of knockout mutants defective in carbon catabolism pathways were constructed and competed against the wild-type E. coli (36) using the streptomycin-treated mouse model (49, 83–86). Commensal E. coli MG1655 mutants defective in the utilization of arabinose, gluconate, fucose, N-acetylglucosamine, and N- acetylneuraminic acid showed colonization defects when competed against the wild-type commensal E. coli MG1655 (36, 87), indicating these carbon sources support colonization of the mouse intestine.
Gluconate, glucuronate, and galacturonate feed into the Entner-Doudoroff pathway and from there into the bottom half of glycolysis. It was known since the late 1990s that the Entner-Doudoroff pathway is critical for E. coli to colonize the mouse intestine (47). Eriksson and colleagues implicated the Entner-Doudoroff pathway as being important for the growth of Salmonella inside macrophages (88). The Thompson group went on to prove that glycolysis is required by Salmonella to replicate in macrophages and infect mice (89).
NUTRIENT LIMITATION IN THE INTESTINE
In nature, nutrients for bacterial growth are almost always limiting and are rarely available continuously. Since E. coli MG1655 uses up to seven different sugars during colonization (36, 87), this suggests that E. coli leads a scavenger lifestyle. It is likely these preferred nutrients are limiting in the mammalian intestine. In such nutrient-limiting conditions, E. coli relies on intracellularly stored carbon and energy (90). Glycogen is the primary carbon and energy storage molecule for enteric bacteria, which is synthesized when carbon is abundant and mobilized when needed to offset nutrient limitation (90–92). Knockout mutants unable to synthesize or degrade glycogen had significant colonization defects when competed against the wild type, suggesting that E. coli encounters famine conditions in the mouse intestine, and therefore, glycogen reserves are required for efficient colonization (90). The dependence of E. coli on glycogen during colonization was confirmed by providing an excess of gluconate (2% wt/vol) in the drinking water, which rescued the colonization defects of the glycogen metabolism mutants. Gluconate is not absorbed by animals and when it is provided to mice in excess, it therefore provides energy for E. coli to overcome mutational loss of glycogen stores. The conclusion is E. coli leads a feast and famine existence in the intestine that is mitigated by glycogen as a readily mobilizable energy source during intermittent famine.
NUTRIENT CONSUMPTION BY E. COLI PATHOGENS
Research on nutrient use by E. coli was not limited to commensal E. coli MG1655; nutrient use by other commensal strains E. coli Nissle 1917 and E. coli HS and some pathogenic strains EHEC EDL933, UPEC strain CFT073, and EPEC strain E2348/69 has also been explored. E. coli Nissle 1917 was isolated during World War I from a soldier who resisted infection during a Shigella outbreak (93). Since the early 1920s, Nissle 1917 has been used for the treatment of gastrointestinal diseases and is marketed under the trade name “Mutaflor” (94). Mutaflor is known to be effective in causing remission of ulcerative colitis without any side effects (94–96). Studies showed that E. coli Nissle 1917 uses seven different carbon sources during colonization of the intestine, including arabinose, fucose, galactose, gluconate, N-acetylgalactosamine, N-acetylneuraminate, and mannose (6). E. coli HS was isolated from a healthy human in 1958 and is considered a true human commensal as it colonizes humans at 1010 CFU/g of feces without causing disease (6, 97, 98). Although E. coli HS can use many carbon sources in vitro, it uses only six of those for colonizing the intestine, including galactose, arabinose, gluconate, N-acetylglucosamine, lactose, and ribose (6).
EHEC EDL933 caused the first major hamburger-borne outbreak of hemorrhagic colitis and hemolytic uremic syndrome in the United States (99). As described above, laboratory cultures of E. coli MG1655 and E. coli EDL933 utilize 13 monosaccharides equally well and in the same order (36). However, E. coli EDL933 occupies a distinct niche in the intestine which is defined by the utilization of seven different sugars: arabinose, galactose, hexuronates, mannose, N-acetylglucosamine, ribose, and sucrose (36). This indicates that E. coli strains with nearly identical catabolic potential vary significantly in the sugars that support their colonization. Studies of bovine intestinal contents (BIC) showed that E. coli EDL933 uses mannose, N-acetylglucosamine, N-acetylneuraminic acid, and galactose as its preferred carbohydrates during growth in bovine small intestinal contents with live endogenous microbiota (100). Although N-acetylneuraminic acid was not a preferred nutrient source used by E. coli EDL933 during colonization of the mouse large intestine (36), its utilization during colonization of bovine small intestinal contents (100) suggests that the nutritional requirements of E. coli strains may vary when it colonizes the small intestine as compared to the large intestine.
UPEC CFT073, a human urinary tract pathogen isolated from a patient with pyelonephritis, uses arabinose, galactose, mannose, N- acetylglucosamine, and ribose while colonizing the intestine, whereas it needs to import peptides and relies on gluconeogenesis for fitness during urinary tract infection (3, 101). Carbon sources utilized by EPEC E2348/69, the first diarrheal strain to be tested in humans (102), are also known to some extent (3). EPEC E2348/69 utilizes at least arabinose, galactose, mannose, N-acetylglucosamine, and ribose to colonize the intestine (3). In murine models of non-infectious colitis, the release of lactate by intestinal epithelial cells increased lactate availability in the gut lumen, and commensal E. coli and pathogenic Salmonella utilized lactate to increase their fitness (103). Garzetti and co-workers demonstrated that E. coli Mt1B1, which was isolated from a conventional mouse (104), utilizes galactitol while colonizing the mouse intestine. Galactitol is contained in several plants and is generated by galactose oxidation in the intestine; ~50% of E. coli strains can utilize it (71). Thus, the pathogens that have been investigated each utilize different carbon and energy sources to colonize.
NITROGEN ASSIMILATION IN THE INTESTINE
Until now, most research into the nutrition of enteric bacteria focused on carbon and energy sources and electron acceptors. Recently, nutrient sources other than sugars used by E. coli during colonization of the intestine have gained attention. Schubert and co-workers (68) showed that aspartate serves as the source of fumarate used for anaerobic respiration by E. coli in the intestine. Aspartase (L-aspartate ammonia-lyase) deaminates aspartate to produce fumarate and ammonia (105). The Schubert group also showed that E. coli assimilates the ammonia released by aspartase enzyme, which serves as the sole source of nitrogen under aerobic and anaerobic conditions in laboratory cultures (68, 106). It has been shown that, in an inflamed gut, pathogenic E. coli LF82 reprograms its metabolism to utilize L-serine, while the genes involved in the catabolism of simple sugars were significantly downregulated (107). When L-serine utilization genes were knocked out, it reduced the competitive fitness of the strain during inflammation. When L-serine was removed from the diet, its availability in the lumen was reduced, and this diminished the bloom of E. coli LF82 in the inflamed gut (107). However, it is important to note that serine use by commensal E. coli has not been studied in the intestine. The eut (ethanolamine utilization) gene cluster was upregulated in EHEC O157:H7 grown in bovine intestinal contents, and ethanolamine present in BIC was shown to be utilized by E. coli (108). EHEC can use ethanolamine as a nitrogen source in vitro but not as a carbon source (108). Taken together, these findings suggest that EHEC might use ethanolamine as a nitrogen source in the intestine. Interestingly, ethanolamine utilization genes are also upregulated in commensal E. coli MG1655 when grown in cecal mucus in vitro (87) and when colonizing the mouse intestine (109). However, an ethanolamine utilization pathway mutant (E. coli MG1655 ∆eutBC) was not defective in colonization (87), suggesting that ethanolamine is not used by commensal E. coli in the intestine. One conclusion from these findings is that different E. coli commensal and pathogenic strains utilize diverse nutrients while colonizing the mammalian intestine (Table 1), and nutrient choice may vary according to niche occupation, diet, and host physiology.
TABLE 1.
Nutrients used by different E. coli bio- and pathotypes to colonize the mammalian intestine
| Strain | Limiting nutrientsa in the intestine known so far |
|---|---|
| E. coli MG1655 | Arabinose, fucose, gluconate, N-acetylglucosamine, N-acetylneuraminic acid, maltose, glycogen |
| E. coli Nissle 1917 | Arabinose, fucose, galactose, gluconate, N-acetylgalactosamine, N- acetylneuraminic acid, and mannose |
| E. coli HS | Arabinose, galactose, gluconate, lactose, N-acetylglucosamine, and ribose |
| E. coli CFT073 | Arabinose, galactose, mannose, N-acetylglucosamine, and ribose |
| E. coli E2348/69 | Arabinose, galactose, mannose, N-acetylglucosamine, and ribose |
| E. coli EDL933 | Arabinose, fucose, galactose, hexuronates, mannose, N-acetylglucosamine, ribose, maltose, glycogen, and ethanolamine |
“Limiting nutrients” indicates mutants in the pathways involved in the utilization of the compound are defective in competitive colonization against the wild type by >0.8 log10 fold and Student’s t-test value of <0.05.
COMPETITION FOR NUTRIENTS IN THE MOUSE INTESTINE
The source of nutrients in the large intestine and some of the specific nutrients used by some of the resident microorganisms are known. However, not much is known about the mechanisms used by resident microbes to compete for the available nutrients. There are at least four different ways by which E. coli can compete for nutrients in the intestine (Fig. 2).
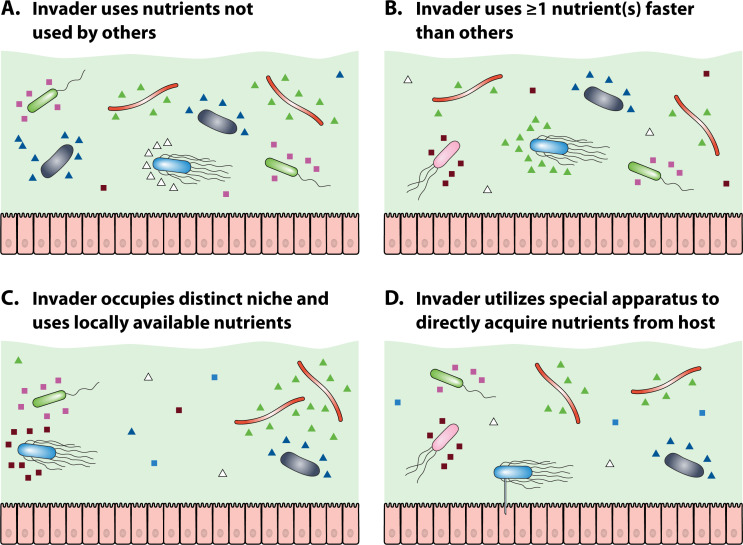
Fig 2.
Four strategies used by an invading bacterium (blue) to compete for nutrients (represented by colored triangles and squares) with resident microbiota; anaerobes are shown in green and black, and facultative anaerobes are shown in red and pink. (A) Invader utilizes nutrients (white squares) not used by other members of the microbiota, and hence, does not have to compete for the nutrients. (B) Invader utilizes at least one nutrient (green triangles) faster than other members of the microbiota [in this case, another facultative anaerobe (red)]; this provides an advantage over the competing microbiota. (C) Invader occupies a distinct niche in the intestine where it uses a nutrient (magenta squares) that is locally available. Although the invader may fail to compete with the microbiota in major niches, it will thrive on a locally available nutrient. (D) Invader acquires some nutrients directly from the host cell using a nanotube, so it does not have to compete with other bacteria for nutrients (created with BioRender.com).
First, as discussed above, since different E. coli strains have different in vivo nutritional preferences (3, 6, 36, 87), they can utilize nutrients not used by competitors in the intestine. To identify the nutrients used by each strain in the intestine, mutations in genes involved in the utilization of 13 different nutrients known to be present in the mucus were constructed in each strain, and the mutants were competed against the isogenic wild type using the streptomycin-treated mouse model. Based on competitive colonization experiments, if the mutant in a particular metabolic pathway had a significant defect in colonization when competing against its isogenic parent strain (≥0.8 log10 fold difference and P-value <0.05), it was concluded that the strain uses that nutrient for colonization. It was found that E. coli MG1655 uses five nutrients present in mucus: arabinose, fucose, gluconate, N-acetylglucosamine, and N-acetylneuraminic acid (sialic acid) (36, 87). E. coli Nissle 1917 used a different set of seven carbon sources to colonize the intestine: arabinose, fucose, galactose, gluconate, N-acetylgalactosamine, N-acetylneuraminate, and mannose. Another commensal strain, E. coli HS, used six different sugars, including arabinose, galactose, gluconate, N-acetylglucosamine, lactose, and ribose (6). Each of the three commensal strains, E. coli MG1655, E. coli Nissle 1917, and E. coli HS, utilized at least one sugar not used by the other strains in the intestine (6, 36, 87). These results suggest that each commensal can colonize the intestine in the presence of other strains by occupying distinct nutrient-defined niches. Thus, it is not surprising that mice pre-colonized with one of the human commensal E. coli strains allowed subsequent colonization by another human commensal E. coli strain (110). For example, when mice were pre-colonized for 10 days with E. coli MG1655 and subsequently fed E. coli HS or E. coli Nissle 1917, either competing strain grew from low to higher numbers, suggesting they occupied distinct niches in the intestine (110). Similar results were obtained when mice were pre-colonized with E. coli HS or E. coli Nissle 1917 and then challenged with the other commensals (110). These results are discussed in more detail below in the section entitled “Overcoming Colonization Resistance.”
The nutrient niches occupied by E. coli EDL933 differed from E. coli MG1655, in that the former strain used galactose, hexuronates, mannose, and ribose which were not used by the latter strain. Conversely, E. coli MG1655 used N-acetylneuraminic acid and gluconate which were not used by E. coli EDL933 (36). E. coli EDL933 also appeared to utilize ethanolamine, possibly as a nitrogen and carbon source, whereas E. coli MG1655 did not appear to utilize it (87, 108, 111). Similarly, E. coli EDL933 displayed a different nutritional program compared to E. coli Nissle 1917 or E. coli HS while colonizing the mouse intestine (6, 36). Furthermore, E. coli EDL933 demonstrated metabolic flexibility by switching to gluconeogenic substrates from glycolytic substrates when competing against commensal E. coli strains (112, 113). Bertin et al. (114), also demonstrated the importance of gluconeogenesis for maintaining colonization of enterohemorrhagic E. coli in the bovine intestine.
Since none of the commensal strains alone can saturate all the niches occupied by E. coli EDL933 in the intestine, they cannot prevent E. coli EDL933 from colonizing. In mice pre-colonized with E. coli MG1655 and E. coli HS individually, E. coli EDL933 grew from 105 CFU fed to 106–107 CFU/g of feces, showing that these commensal strains cannot prevent E. coli EDL933 from colonizing (110). In the case of E. coli Nissle 1917 pre-colonized mice, colonization of E. coli EDL933 was limited and reduced to 103 CFU/g of feces, but the pathogen was not eliminated (110). However, when E. coli EDL933 was fed to mice pre-colonized with three commensal strains, E. coli MG1655, E. coli Nissle 1917, and E. coli HS, it was eliminated from the intestine (110). Since together these commensal strains consumed all the nutrients utilized by E. coli EDL933 to colonize the intestine, these strains could exert colonization resistance based on nutrient limitation (110). In fact, just two pre-colonized strains fed together, E. coli Nissle 1917 and E. coli HS, utilized five sugars which are important for E. coli EDL933 colonization and thus prevented colonization of E. coli EDL933 (6).
While it seems reasonable to find a set of commensal E. coli strains which could be used to prevent colonization by any single pathogenic E. coli strain, it is unlikely to find a small set of commensal E. coli strains that will be effective in preventing colonization of all E. coli pathogens. This is understandable since E. coli pathotypes each occupy distinct intestinal niches (86). Uropathogenic E. coli CFT073 and enteropathogenic E. coli E2348/69 occupy niches distinct from those occupied by E. coli EDL933, as E. coli CFT073 and E. coli E2348/69 grew from low to higher numbers in E. coli EDL933 pre-colonized mice (86). When uropathogenic E. coli CFT073 pre-colonized mice were challenged with E. coli EDL933, E. coli EDL933 failed to grow to higher numbers but was not eliminated, suggesting that E. coli CFT073 outcompetes E. coli EDL933 in its preferred niche(s) (86). As expected, when E. coli Nissle 1917 and E. coli HS were pre-colonized together, a combination that conferred colonization resistance against E. coli EDL933, they failed to prevent colonization of E. coli CFT073 or E. coli E2348/69, indicating these pathotypes occupy intestinal niches not occupied by commensals E. coli Nissle 1917 and E. coli HS (86). The examples described above in this section indicate the six strains examined each use different nutrients to compete in distinct intestinal niches.
The second way in which E. coli can compete for nutrients is by growing faster on nutrients that are used by other strains, thereby outcompeting them (115). Spontaneous mutants of E. coli MG1655 with deletions in the flhDC operon were obtained during passage through the streptomycin-treated mouse intestine. These mutants grew 15%–30% faster than the wild type on several carbon sources (115). The flhDC operon encodes FlhD4C2 transcriptional regulator which controls expression of flagellar genes in E. coli (116). Those mutants were non-motile and were better colonizers of the mouse intestine than the wild type (115). During intestinal colonization by E. coli MG1655, it was found that 90% became non-motile by day 15 after feeding; all the non-motile mutants had 4–500 bp deletions in the flhDC promoter region (117). Colonization of germ-free mice with E. coli also led to the rapid selection of non-motile mutants having deletions in the flhDC operon (118). E. coli M1655 flhDC mutants had a competitive advantage in two ways: (i) genes involved in carbon and energy metabolism were upregulated, leading to more rapid growth, and (ii) energy conserved by being non-motile was directed to other cellular activities (115, 117). Thus, E. coli can gain a competitive advantage by outcompeting other microorganisms for their preferred nutrients.
The third strategy employed by E. coli to compete for nutrients is to occupy a distinct niche by entering a symbiotic association with anaerobes that release its preferred nutrients (83). When streptomycin-treated mice were fed E. coli MG1655 wild type, 90% of the cells became non-motile as a result of deletions in flhDC operon (117). The remaining 10% retained motility yet were better colonizers than the wild type during competition experiments (83). Those motile strains were envZ missense mutants; one of them (envZP41L) was more resistant to colicin V and bile salts and grew faster than the wild type on several sugars (83). The E. coli MG1655 envZP41L mutant isolated from mice had higher levels of OmpR~P and displayed different outer membrane protein profiles (higher OmpC and lower OmpF) than the wild type (84). When germ-free mice were fed E. coli MG1655, the germ-free mouse intestine also selected for mutants in the EnvZ/OmpR two-component system that had lower OmpF production and increased OmpC production compared to the wild type (119). In response to the osmolarity of the environment, EnvZ, the histidine kinase of the EnvZ/OmpR two-component system, phosphorylates response regulator OmpR which then regulates over 100 genes including those encoding outer membrane proteins OmpC and OmpF (120). As compared to flhDC deletion mutants, E. coli MG1655 envZP41L grew 15% slower in vitro in mouse cecal mucus and several sugars present in the mucus. However, the mutant colonized the mouse intestine better than the wild-type E. coli MG1655 and equally well as the E. coli MG1655 flhDC deletion mutant (83). Furthermore, E. coli MG1655 envZP41L appeared to occupy a minor galactose-defined intestinal niche that was not colonized or poorly colonized by an E. coli MG1655 flhDC mutant. When the envZP41L gene was transferred from the E. coli MG1655 background into E. coli Nissle 1917, the newly constructed E. coli Nissle 1917 envZP41L also produced higher levels of phosphorylated OmpR, more OmpC, and less OmpF than the parent strain E. coli Nissle 1917 (84). Like E. coli MG1655 envZP41L, it was more resistant to bile salts and colicin V and also grew slower than the parent strain in cecal mucus and on several sugars present in mucus, yet it colonized the mouse intestine better than the parent strain by 10-fold (84). Although E. coli Nissle 1917 envZP41L did not grow as fast as the parent strain on galactose as the sole carbon source in vitro, it utilized galactose to colonize a minor intestinal niche not colonized or poorly colonized by E. coli Nissle 1917 (84). These results suggest that hyper-activation of OmpR caused the envZP41L mutants to occupy a niche that is distinct from those occupied by the parent strains.
Since E. coli envZP41L colonized better by occupying a galactose-defined niche and had higher levels of OmpC and lower levels of OmpF, we hypothesized that outer membrane proteins played a role in colonizing distinct nutritional niches in the intestine. In a recent study (109), we found that changes in outer membrane protein profiles contributed to bile tolerance in the intestine and conferred better colonizing ability. An ompC deletion mutant was outcompeted by the wild type because it took time to adapt to the bile salt concentration in the intestine. Furthermore, we found an ompF deletion mutant outcompeted the wild type during colonization. Higher production of OmpC in the ompF mutant made it a better colonizer. However, OmpC overproduction via a constitutive promoter alone was not sufficient for better colonization except when ompF was deleted. A strain which overproduced OmpC and lacked OmpF was a better colonizer than the parent strain, suggesting that fine-tuning of OmpC and OmpF is crucial for colonization. We provided evidence that OmpC is important for E. coli to colonize the intestine because its smaller pore size excludes bile salts, while OmpF is deleterious because its larger pore size allows bile salts to enter the periplasm. However, the ompF mutant competed against the wild-type E. coli MG1655 in the same niche and did not grow faster than the parent strain with galactose as the sole carbon source. This suggests the galactose niche occupied by E. coli MG1655 envZP41L was not due to the alteration in outer membrane protein composition, and some other functions regulated by EnvZ/OmpR must be responsible for better galactose utilization. Interestingly, there is a putative OmpR-binding site upstream of galR promoter between galR and omrB (83, 121). Galactose transport and metabolism in E. coli are under the control of the GalR repressor (122). However, it is not yet known whether OmpR regulates galR transcription. Negative regulation of GalR by phosphorylated OmpR could explain why E. coli MG1655 envZP41L occupies a galactose-specific niche not occupied by wild-type E. coli MG1655.
Fourth, recently, a novel strategy used by EPEC to acquire nutrients from infected host cells was described by Pal and co-workers (123). In a process termed host-nutrient extraction, the authors showed that EPEC uses the CORE complex composed of integral inner membrane proteins to form tubular outer membrane extensions connecting the bacteria to the infected host cells. The nanotubes serve as a channel to import nutrients from the host cell cytoplasm (123). The authors showed that the infected bacteria acquired amino acids through the nanotubes, which suggests the possibility that other nutrient sources also could be acquired by the pathogens using this strategy (123). Since the attached pathogen is getting nutrients directly from the host cells without releasing nutrients to the competing microbiota, this could be a furtive strategy employed by pathogens to compete for nutrients and colonize the intestine.
NON-NUTRITIONAL ASPECTS OF COLONIZATION RESISTANCE
When intestinal pathogens enter the intestine, they must compete with the host microbiota to colonize and successfully establish an infection. Thus, the intestinal microbiota serves as a barrier to colonization by many invading pathogens (17, 50, 124, 125). Potential mechanisms of colonization resistance are not fully understood but are thought to include nutritional and non-nutritional aspects (126). Regarding non-nutritional causes, susceptibility to invading pathogens can be enhanced by perturbations of the microbiota caused by diverse factors such as diet, inflammation, or antibiotics (127–131). For instance, Sekirov and co-workers (130) demonstrated that clinically relevant doses of streptomycin and vancomycin in drinking water had a dose-dependent alteration on intestinal bacteria composition of C57BL/6 mice. They went on to show that greater alterations in the intestinal microbiota resulted in increased susceptibility to Salmonella Typhimurium colonization and more severe intestinal pathology. Thus, colonization resistance describes direct and indirect interactions between the microbiota, host, and pathogens that prevent pathogens from invading (132, 133). However, the mechanisms by which the resident microbiota confers colonization resistance are not completely understood. In addition to direct competition between the microbiota and pathogens for essential nutrients (6, 70, 71, 134), several factors are known to contribute to colonization resistance, including inhibition of pathogen growth by microbiota-derived substances including short-chain fatty acids (SCFA) and bacteriocins (135–138), ability to tolerate environmental stressors (109, 127), type VI secretion system-mediated killing of invading bacteria (139), and microbiota-induced stimulation of innate and adaptive immune responses (50, 140).
Although exploring the nutritional basis of colonization resistance is the focus of this review, it is worth discussing, in brief, some other non-nutritional mechanisms of colonization resistance. Osbelt et al. (135) demonstrated that the presence of SCFA (especially butyrate)-producing bacteria belonging to the phylum Firmicutes inhibited the growth of C. rodentium in mice. Supplementation of mice with butyrate was sufficient to reduce C. rodentium growth in vivo, suggesting the role of microbiota-derived metabolites in colonization resistance (135). SCFAs are known to exert diverse effects on pathogenic bacteria (141). For instance, SCFAs inhibit the growth of Enterobacteriaceae by acidifying the proximal colon and triggering intracellular acidification (136). Bacteriocins produced by many bacterial species, including lactic acid bacteria, have potent antimicrobial activity (142). Lactobacillus salivarius UCC118 has been shown to produce a bacteriocin Abp118 which significantly enhances resistance to infection by food-borne pathogen Listeria monocytogenes (137). Clostridium scindens, which converts primary bile acids to secondary bile acids, inhibits C. difficile infection in secondary bile acid-dependent fashion (143). E. coli Nissle 1917, which is an excellent biofilm former, outcompetes pathogenic E. coli strains during biofilm formation (144). E. coli Nissle 1917 has been shown to secrete a bifunctional (protease and chaperone) periplasmic protein DegP extracellularly and control biofilm formation by pathogenic strains (138). However, whether DegP has any role in colonization resistance conferred by E. coli Nissle 1917 is not yet known. Besides the microbiota-derived metabolites, environmental stressors like bile acids can also play a key role in colonization resistance. E. coli and Salmonella demonstrate enhanced bile resistance compared to other members of the microbiota when switched from plant-based maintenance diet to a high-fat diet (western diet), which promoted colonization by E. coli and Salmonella (127). Since a high-fat diet requires bile salts to be released into the gut lumen, high fat elevates the level of bile salts which inhibits other bacterial species but not E. coli or Salmonella spp., leading to a loss of colonization resistance against these species (127). While S. Typhimurium colonization was boosted by diet shifts and fat-elicited bile in mice lacking E. coli in their microbiota, competitive E. coli strains (mixture of three strains) provided protection against S. Typhimurium in the fat-challenged gut, suggesting that E. coli might limit the blooms of pathogens promoted by fat (127).
We recently showed that mice pre-colonized with E. coli MG1655 ∆ompC, which is sensitive to physiological bile salt concentration in the intestine, failed to prevent colonization of E. coli MG1655, further supporting the finding that environmental stressors play important roles in colonization resistance (109). Hecht et al. (139) demonstrated the role of the type VI secretion system in colonization resistance in Bacteroides fragilis. When mice were colonized with a symbiotic non-toxigenic B. fragilis, it limited the colonization of pathogenic enterotoxigenic B. fragilis. The strain-specific competition was attributed to the type VI secretion system of non-toxigenic B. fragilis, and the colonization resistance conferred by non-toxigenic B. fragilis protected the host from enterotoxigenic B. fragilis-induced colitis (139). E. coli has also been shown to utilize a type VI secretion system to fight against the invading competitor C. rodentium and limit its colonization. Interestingly, C. rodentium also utilizes the same strategy of contact-dependent inhibition to compete for intestinal niches with other bacteria (145).The resident microbiota can also stimulate host immune responses resulting in the production of antimicrobial peptides. These antimicrobial peptides can effectively inhibit intestinal pathogens and confer colonization resistance (50). For instance, the colonization of germ-free mice with intestinal microbiota triggers the expression of RegIIIγ from intestinal epithelial cells (140). C-type lectin RegIIIγ is an antimicrobial peptide which targets the peptidoglycan of Gram-positive bacterial cell walls, inhibiting pathogens such as Enterococci and Listeria monocytogenes in the intestine (140, 146).
NUTRIENT-DEPENDENT COLONIZATION RESISTANCE
Competition for available nutrients in the gut is profound, and the complexity of the gut microbiota ensures maximum utilization of the available substrates (147). Therefore, in a healthy, fully developed microbiota, it is unlikely that an incoming species finds an uncontested niche because it has to outcompete the established microbiota for nutrients to become established (148). This apparently explains why only a fraction of persons exposed to an enteric pathogen get infected during an outbreak. For instance, of the total 47,643 Japanese school children exposed to white radish sprout-associated E. coli 0157:H7 in Sakai City, Osaka, Japan, only 398 were hospitalized, and 8,355 were symptomatic (149). Experiments with conventional mice have also demonstrated that the attack rate of different E. coli strains was less than 20% (7). In conventional mice, the feces contains 105–106 CFU of E. coli per gram of feces (32), which might exert colonization resistance against introduced E. coli strains depending on their nutritional programs. It has been shown that antibiotic-treated mice and germ-free mice have increased amounts of nutrients in the gut, including carbohydrates and amino acids, which lead to reduction in colonization resistance (150, 151). Thus, susceptibility to an invading pathogen can be determined by the availability to invaders of nutrients that are not consumed by the resident microbiota.
The fact that different E. coli strains have different nutritional programs in the intestine could explain why some humans are susceptible to infection, while in others, the microbiota exerts colonization resistance (149, 152). Previous studies in our laboratory revealed the nutrients used by commensal and pathogenic E. coli to colonize. Different E. coli strains demonstrated distinct preferences in the utilization of sugars present in mucus, and a defect in utilizing one or more of those sugars led to a competitive disadvantage against the wild type (6, 36, 86, 87, 90, 110, 112, 113). When mice pre-colonized with any of three commensal E. coli strains (E. coli MG1655, E. coli HS, or E. coli Nissle 1917) were challenged with isogenic strains, the pre-colonized strain eliminated the isogenic challenge strain (110). Since isogenic strains compete equally well for the same nutrients, the pre-colonized strain conferred colonization resistance to the isogenic challenge strain by consuming nutrients (110). This supports the previous finding that when E. coli strains isolated from the feces of healthy human volunteers were fed to the same individual, they could not colonize (153). This is also true for pathogenic E. coli. It was demonstrated that two closely related O157:H7 EHEC strains, E. coli EDL933 and E. coli Sakai, exert colonization resistance against each other and compete in the same niches (86). These two strains have similar physiology, including an identical nutrient utilization spectrum, as they have >99.9% sequence identity in their orthologous protein-coding genes (154). The minor genomic differences reflect a difference in the annotation strategies that were used for the two genomes (155). The nearly identical physiology and metabolism of these two O157:H7 strains explain how they are able to exert colonization resistance against each other.
OVERCOMING COLONIZATION RESISTANCE
Having discussed many potential mechanisms of colonization resistance, we turn our attention to how invading pathogens overcome colonization resistance. When mice pre-colonized with one human E. coli commensal strain (E. coli MG1655, E. coli HS, or E. coli Nissle 1917) were challenged with a different commensal strain, the challenge strains were able to overcome colonization resistance, and the two strains co-colonized the intestine (110). Since each of the three human E. coli commensal strains utilizes at least one sugar not used by the other strain, these commensal strains are able to occupy distinct nutrient-defined niches not occupied by the other commensal strains (36, 87, 110). Prior colonization of any one of three commensal E. coli strains cannot prevent pathogenic E. coli EDL933 from colonizing the mouse intestine, although the population of the E. coli EDL933 challenge strain varied depending on the pre-colonized commensal strain. For example, in mice pre-colonized with E. coli Nissle 1917, E. coli EDL933 dropped from 105 CFU/g of feces to 5 × 103 CFU/g of feces (110), whereas in mice pre-colonized with E. coli MG1655 or E. coli HS, E. coli EDL933 grew to higher populations. Likely, the varying responses to E. coli EDL933 challenge are due to the different nutritional programs of the commensal strains (6, 36, 87). E. coli Nissle 1917 is a superior colonizer compared to E. coli MG1655 and E. coli HS and most likely fills more niches than the other commensal strains (110). Although E. coli EDL933 could overcome colonization resistance to any individual commensal strain, when mice were pre-colonized with mixtures containing E. coli Nissle 1917, E. coli HS, and E. coli MG1655 the commensal strains conferred colonization resistance (110). Thus, it appears that an approach to prevent or treat enteric infections would be to pre-colonize humans with strains that would occupy all nutritional niches such that the pathogens would be eliminated from the intestine. This strategy could be one mechanism by which Mutaflor mitigates traveler’s diarrhea. This approach, however, would not work if the invading species grows faster on the same nutrients and outcompetes the members of the microbiota. For instance, mice pre-colonized with E. coli MG1655 could not exert colonization resistance to E. coli MG1655 flhDC mutants as the mutants grew 15%–30% faster than the wild type on several carbon sources and, therefore, colonized better than the wild type (115).
E. coli competes for niches not only with other E. coli but also with other bacteria which utilize identical nutrients in the intestine. In germ-free mice infected and stably colonized with C. rodentium, challenge with E. coli decreased the C. rodentium population by almost 500-fold within 14 days of challenge (134). Both C. rodentium and E. coli grew well on monosaccharides, suggesting that the depletion of simple sugars by E. coli allowed it to outcompete the pathogen (134). Utilization of the limiting nutrient galactitol by E. coli prevented niche invasion by Salmonella Typhimurium which could otherwise use galactitol for colonization (71). Klebsiella michiganensis conferred colonization resistance against E. coli via superior sugar utilization; provision of a carbon source (galactitol) that is accessible to E. coli but not to K. michiganensis mitigated colonization resistance (156). This suggests that strategies can be developed where commensal strains selectively prevent colonization of pathogenic E. coli and other enteric pathogens. To this end, scientists have created a minimal bacterial community that provides colonization resistance to enteric pathogens. A bacterial consortium comprised 12 bacterial isolates also known as Oligo-Mouse-Consortia (Oligo-OMM12), providing partial protection against S. Typhimurium (157). When three more facultative anaerobes including E. coli Mt1B1 were added to Oligo-OMM12, the mixture conferred complete protection against S. Typhimurium (157). Microbial strains in the Oligo-MM12 could be tailored (expanded or reduced) to provide colonization resistance to specific pathogens (157). Although the focus of this article is on the nutritional basis of colonization resistance by different E. coli strains, this concept should hold true for other members of the resident microbiota and invading pathogens (133, 158).
RESTAURANT HYPOTHESIS
The microbial community structure within the intestine is determined by competition for resources. According to Rolf Freter, competition for nutrients is the most important factor for success in the intestinal ecosystem, although several other factors could contribute to an organism’s ability to colonize (159). Freter and colleagues developed an anerobic continuous-flow culture which demonstrated that stable multispecies communities similar to those in the large intestine of mice could be established (160). Freter postulated that, for many species to co-colonize, each species must use at least one limiting nutrient better than all others (7, 159). Thus, if the resident microbiota in the intestine consumes all the nutrients that an invading species needs to colonize, the invading species will not find the nutrients it requires and will fail to become established (62). While Freter’s nutrient-niche hypothesis has been widely accepted and is consistent with ecological succession during conventionalization of mice and microbiota-generated nutrient utilization by pathogens (161–163), its validity has been challenged by findings which demonstrate mixed-substrate utilization, metabolic flexibility, and heterogeneity of nutrients in time and space (reviewed extensively by Pereira and Berry)(148).
Some of the findings in studies conducted in our laboratory are also inconsistent with Freter’s nutrient niche hypothesis (83, 84, 117). For example, while Freter’s hypothesis can easily explain why the streptomycin-treated mouse intestine selects E. coli MG1655 flhDC mutants (115, 117), it cannot explain the selection of motile E. coli MG1655 envZ missense mutants by the streptomycin-treated mouse intestine (83, 117). E. coli MG1655 flhDC mutants grow 15%–30% faster on several sugars present in cecal mucus than E. coli MG1655 wild type, which explains why they were selected, consistent with the nutrient-niche hypothesis (115, 117). However, E. coli MG1655 envZP41L grew ~15% slower in vitro than the E. coli MG1655 flhDC mutant on mouse cecal mucus and several sugars present in mucus but was a much better colonizer than the wild-type E. coli MG1655 like the E. coli MG1655 flhDC mutant (83). Slower growth of envZP41L mutants that colonize equally well as the E. coli MG1655 flhDC mutants during competitive colonization is inconsistent with the nutrient-niche hypothesis (83). In follow-up studies in our laboratory, E. coli Nissle 1917 envZP41L, like E. coli MG1655 envZP41L, appeared to use galactose to colonize a second intestinal niche not colonized or poorly colonized by E. coli Nissle 1917 wild type (84). However, E. coli Nissle 1917 envZP41L grows slower than E. coli Nissle 1917 on galactose as the sole carbon source (84). Interestingly, E. coli Nissle 1917 envZP41L appeared to be worse than its parent strain at preventing EHEC EDL933 colonization despite being a better colonizer (84). These results, which are inconsistent with the nutrient-niche hypothesis, led to the development of the “restaurant hypothesis,” which emphasizes that organisms colonize the intestine as members of mixed biofilms and obtain the sugars they need for growth locally through cross-feeding from polysaccharide-degrading anaerobes (3, 83, 84). This contrasts with the nutrient-niche hypothesis, which assumes that nutrients are perfectly mixed in the intestine and are equally available to all species present (7).
Biofilms form in the mucus layers of the mammalian large intestine (164, 165) and our previous research demonstrated that commensal E. coli resides in mixed biofilms in the mouse intestine (83, 84). As discussed previously, E. coli growth in the intestine depends on the anaerobes, which degrade polysaccharides releasing monosaccharides and disaccharides (70, 71). Thus, anaerobes in the mixed biofilms release sugars that E. coli uses, in contrast to using nutrients from a perfectly mixed pool as assumed by the nutrient-niche hypothesis (3, 7). Those mixed biofilms feed E. coli, and the restaurant hypothesis proposes that different E. coli strains reside in different restaurants and interact with different anaerobes physically and metabolically. Hence, different E. coli strains may enter different restaurants which serve different nutrients. This hypothesis explains why different E. coli strains, despite utilizing the same nutrients in identical order in vitro, display different nutritional programs in the mouse intestine (3, 6, 36, 87).
The restaurant hypothesis can explain why E. coli MG1655 envZP41L and E. coli Nissle 1917 envZP41L are better colonizers than their parent strains, although they grow more slowly in cecal mucus and on several sugars present in mucus (83, 84). First, the mutants appear to enter a different biofilm (niche) consisting of different groups of anaerobes, which serve galactose in a niche that is not colonized or poorly colonized by the parent strains. Second, E. coli MG1655 envZP41L and E. coli Nissle 1917 envZP41L have decreased motility and different outer membrane protein profiles such as higher OmpC and lower OmpF than the parent strains, which could increase affinities to mixed biofilms. The higher phosphorylated OmpR levels present in envZ missense mutants also differentially regulate the expression of several other outer membrane protein genes cirA, fecA, fepA, and ompT via omrA and omrB small RNA expression (121). OmpC has been shown to function as an adhesin (166) and is highly expressed in biofilms compared to planktonic cells (167), suggesting that OmpC might contribute to biofilm formation. Interestingly, in our recently published study (109), we showed that an E. coli MG1655 ompC mutant colonizes the mouse intestine poorly compared to E. coli MG1655 wild type primarily for two reasons. First, the strain lacking ompC is more sensitive to physiological bile salt concentrations than the parent strain and is outcompeted by the parent strain in vitro and in vivo in the presence of bile salts. The poor colonization of E. coli MG1655 ompC is attributed to the mutant taking time to adapt to bile salts in the intestine. Second, we showed that mice pre-colonized with E. coli MG1655 wild type exerted colonization resistance to an E. coli MG1655 ompC mutant and eliminated it from the intestine. However, in pre-colonized mice, an E. coli MG1655 ompC mutant does not exert complete colonization resistance against E. coli MG1655 wild type, and the wild type persists at 102–103 CFU/g of feces in the intestine. Although the E. coli MG1655 ompC mutant and E. coli MG1655 wild type compete primarily in the same niche, there is a minor intestinal niche not saturated by the E. coli MG1655 ompC mutant in which E. coli MG1655 wild type thrives. This can also be explained by the restaurant hypothesis: E. coli MG1655 wild type enters a minor “restaurant” not colonized or poorly colonized by E. coli MG1655 ompC, in addition to a major “restaurant” in which E. coli MG1655 wild type and E. coli MG1655 ompC compete and differ in their tolerance to an environmental stressor, i.e., bile salts, and have different binding affinities in the mixed biofilms.
The restaurant hypothesis also explains why E. coli Nissle 1917 envZP41L, despite being a better colonizer than E. coli Nissle 1917 wild type, was worse than its parent strain at preventing EHEC EDL933 colonization (84). This result suggests that when EHEC EDL933 enters the mouse intestine, it grows in mucus planktonically before entering a biofilm. According to the restaurant hypothesis, when E. coli Nissle 1917 wild type and E. coli Nissle 1917 envZP41L grow in different biofilms utilizing different sugars, some sugars might escape the biofilms and become available to planktonic E. coli cells. Some planktonic anaerobic bacteria may also release sugars that become available to planktonic E. coli. EHEC EDL933 may then compete planktonically with escaped planktonic E. coli Nissle 1917 wild type or E. coli Nissle 1917 envZP41L for the available nutrients, and the outcome of this competition depends on the nature of available nutrients and the nutritional program of the two competitors. This could explain why a better grower in cecal mucus in vitro, i.e., E. coli Nissle 1917, appears to prevent colonization of EHEC EDL933 in the intestine more than E. coli Nissle 1917 envZP41L does (3, 84).
OVERCOMING COLONIZATION RESISTANCE AS A MECHANISM FOR INVASION
Under steady-state conditions, the intestinal microbiota can successfully exert colonization resistance to many invading pathogens (17, 50, 124, 125). When the microbiota is perturbed by different factors such as diet, inflammation, or antibiotics, expansion of invading pathogens can occur (127–131). Besides the external factors that perturb the microbiota, the invaders may also utilize strategies to exploit gaps in colonization resistance and promote their expansion in the gut (147).
Some invaders utilize their metabolic versatility to overcome colonization resistance in the gut. When EHEC EDL933 is the only E. coli strain in the intestine, it does not use gluconeogenic substrates and relies on glycolytic substrates; only when it is competing against E. coli MG1655 or E. coli Nissle 1917 does EHEC appear to switch to gluconeogenic substrates (112). This metabolic flexibility and its different nutritional programs from commensal E. coli allows EHEC EDL933 to overcome colonization resistance exerted by E. coli MG1655 or E. coli Nissle 1917 (6, 36, 110, 112). S. Typhimurium overcomes the inhibitory effects of propionate produced by gut microbiota by using it as a carbon source (168). This short-chain fatty acid, which is produced by anaerobic fermentation of sugars, appears to exert colonization resistance against S. Typhimurium by acidifying the intracellular space of S. Typhimurium and producing toxic by-products during its metabolism (169–172). Klebsiella pneumoniae also demonstrated metabolic flexibility, utilizing L-fucose to bypass nutritional competition with the resident microbiota and overcome colonization resistance (173). C. rodentium turned on the biosynthesis of amino acids to overcome colonization resistance exerted by the microbiota. Biosynthesis of amino acids was required for the colonization of specific pathogen-free mice, which have an intact microbiota, but not in germ-free mice (150). This is understandable since the concentrations of several amino acids were reduced in the intestinal contents of specific pathogen free (SPF) mice compared to those in germ free (GF) mice (150). Thus, metabolic versatility appears to be a key mechanism used by pathogens to overcome colonization resistance in the gut.
The other widely employed mechanism to overcome colonization resistance by invading pathogens is by inducing host-driven inflammation and utilizing nutrients uniquely present in those conditions (147). Stecher and colleagues (128) showed that inflammation induced by S. Typhimurium is sufficient to overcome colonization resistance by altering the microbiota composition. Winter et al. (174) showed, using models of colitis and intestinal injury, that nitrate generated by the host during inflammation conferred a growth advantage to E. coli in the inflamed gut of mice. We had previously shown that, in addition to aerobic respiration, E. coli utilizes fumarate and nitrate to respire anaerobically. However, fumarate is more important to E. coli than nitrate since nitrate is limiting in the intestine (53, 54), while aspartate is abundant and readily converted to fumarate (68). Similarly, during inflammation, the release of lactate by colonocytes increases the level of lactate in the gut lumen and bacteria like E. coli and Salmonella utilize the lactate to promote colonization (103). In addition to nitrate and lactate, further research demonstrated that the inflammatory host response induced by S. Typhimurium causes release of exudates that contain additional energy sources (electron donors) and anaerobic electron acceptors that promote pathogenic blooms in the gut (175, 176).
Some enteric pathogens also utilize “counter-attack” mechanisms such as bacteriocin production and use of secretion systems to antagonize the normal microbiota and replicate in the gut. For example, S. Typhimurium produces colicin Ib, which conferred a competitive advantage against commensal E. coli strains in the inflamed gut (177). Vibrio cholerae uses a type VI secretion system to kill commensal bacteria (178, 179). Other members of Enterobacteriaceae family, such as C. rodentium and S. Typhimurium, also utilize type VI secretion systems that provide a competitive advantage against the resident microbiota and promote their replication in the gut (145, 180). C. rodentium directly targeted and outcompeted commensal E. coli using its type VI secretion system to successfully establish itself within the murine gut (145). However, normally, enteric pathogens enter stationary phase upon excretion by a host and are typically dormant or viable but not culturable in the presence of various stressors in the environment (181, 182). Therefore, once an invading pathogen enters the intestine, it must exit stationary phase and enter the growth phase, utilizing the nutrients that provide energy, before inducing inflammation or antagonizing the microbiota. Thus, it appears that metabolic flexibility may be more important in overcoming colonization resistance than other mechanisms used by pathogens.
CONCLUDING REMARKS AND FUTURE DIRECTIONS
In this review, we focused on the nutritional aspects of intestinal colonization by E. coli. It is increasingly clear that colonized E. coli grows on free monosaccharides and disaccharides liberated by anaerobes in the intestine. Furthermore, different E. coli strains have different nutritional programs in the intestine despite using identical carbon and energy sources in vitro. There is fierce competition for nutrients in the intestine between invading species and the resident microbiota such that invaders can colonize only if they can outcompete the resident microbiota. While multiple factors can contribute to colonization and subsequent infection by invaders, it appears that competition for nutrients and the ability of pathogens to demonstrate metabolic flexibility are key. If the invading species can utilize a different nutrient not utilized by commensals or if the invading species can grow faster than the resident microbiota on a particular nutrient, it can grow, first planktonically, and then enter a restaurant where it can thrive. However, too little is known about the mechanisms used by those microbes to compete for nutrients. Direct evidence for the availability of nutrients in the intestine and the use of those nutrients by competing E. coli strains in the intestine is lacking. A metabolomic approach to determine the nutrients available and used by commensals and nutrients available post-colonization for invading pathogens can provide more information regarding the nutritional basis for overcoming colonization resistance. Metabolomic analysis using high-throughput LC-MS has revealed that there is an abundance of sugars, gluconeogenic substrates, amino acids, and their derivatives in streptomycin-treated mouse cecal mucus, which explains why E. coli strains can grow to ~109 CFU/g of feces in streptomycin-treated mice (unpublished work). Further work in this area is indicated to better understand the mechanisms used by microbes to colonize the intestine. Understanding the nutritional program of pathogens in the intestine and developing strategies to prevent pathogens from acquiring essential nutrients in the intestine could prove valuable in the fight to prevent gastrointestinal diseases.
ACKNOWLEDGMENTS
Research in the corresponding author’s laboratory is supported by Public Health Service grant GM117324.
Biographies

Sudhir Doranga is currently a post-doctoral fellow in the department of Cardiovascular and Metabolic Sciences at Cleveland Clinic. His post-doctoral work is focused on understanding the physiology and metabolism of Aggregatibacter and Fusobacterium in relation to their role in periodontitis. Sudhir received his BS and MS in Microbiology from Tribhuvan University, Nepal and Ph.D. in Microbiology/Cell and Molecular Biology from Oklahoma State University under the mentorship of Dr. Tyrrell Conway. His Ph.D. work focused on understanding the role of outer membrane proteins in colonization of the mammalian intestine by E. coli. He also led a project on understanding the nitrogen nutrition of E. coli in the intestine. He has presented in several national and regional conferences and has received numerous awards, including Robberson summer dissertation fellowship provided by the Oklahoma State University graduate college, and Microbiology graduate fellowship provided by the Microbiology department during his Ph.D.

Tyrrell Conway is a native Oklahoman who received his BS and PhD degrees in Microbiology from Oklahoma State University. He previously held faculty appointments at the University of Florida, University of Nebraska, Ohio State University and University of Oklahoma. Currently, Conway is a Regents Professor at Oklahoma State University. He is co-inventor of U.S. Patent number 5,000,000. His research team was first to genetically characterize the Entner-Doudoroff pathway, discovered the pathway for idonic acid catabolism, published the first transcriptome analysis of E. coli, first to characterize the carbon and respiratory metabolism of colonized E. coli, discovered the GadE regulon governing acid tolerance, and first to characterize the bacterial stringent response genome-wide. His current research is on the nutrition of E. coli in the intestinal microbiome. Conway is a fellow of the American Academy of Microbiology.
Contributor Information
Tyrrell Conway, Email: tconway@okstate.edu.
Susan T. Lovett, Brandeis University, Waltham, Massachusetts, USA
REFERENCES
- 1. Leimbach A, Hacker J, Dobrindt U. 2013. E. coli as an all-rounder: the thin line between commensalism and pathogenicity. Curr Top Microbiol Immunol 358:3–32. doi: 10.1007/82_2012_303 [DOI] [PubMed] [Google Scholar]
- 2. Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8:207–217. doi: 10.1038/nrmicro2298 [DOI] [PubMed] [Google Scholar]
- 3. Conway T, Cohen PS. 2015. Commensal and pathogenic Escherichia coli metabolism in the gut. Microbiol Spectr 3. doi: 10.1128/microbiolspec.MBP-0006-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Russo TA, Johnson JR. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect 5:449–456. doi: 10.1016/s1286-4579(03)00049-2 [DOI] [PubMed] [Google Scholar]
- 5. GBD 2019 Antimicrobial Resistance Collaborators . 2022. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the global burden of disease study 2019. Lancet 400:2221–2248. doi: 10.1016/S0140-6736(22)02185-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. 2013. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One 8:e53957. doi: 10.1371/journal.pone.0053957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Freter R, Brickner H, Fekete J, Vickerman MM, Carey KE. 1983. Survival and implantation of Escherichia coli in the intestinal tract. Infect Immun 39:686–703. doi: 10.1128/iai.39.2.686-703.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- 9. Jakobsson HE, Rodríguez-Piñeiro AM, Schütte A, Ermund A, Boysen P, Bemark M, Sommer F, Bäckhed F, Hansson GC, Johansson MEV. 2015. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep 16:164–177. doi: 10.15252/embr.201439263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Paone P, Cani PD. 2020. Mucus barrier, mucins and gut microbiota: the expected slimy partners? Gut 69:2232–2243. doi: 10.1136/gutjnl-2020-322260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lysenko ES, Ratner AJ, Nelson AL, Weiser JN. 2005. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog 1:e1. doi: 10.1371/journal.ppat.0010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pasetti MF, Salerno‐Gonçalves R, Sztein MB. 2005. Mechanisms of adaptive immunity that prevent colonization at mucosal surfaces. Colonization of mucosal surf:35–47. doi: 10.1128/9781555817619 [DOI] [Google Scholar]
- 13. Kaper JB, Sperandio V. 2005. Bacterial cell-to-cell signaling in the gastrointestinal tract. Infect Immun 73:3197–3209. doi: 10.1128/IAI.73.6.3197-3209.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clarke MB, Sperandio V. 2005. Events at the host-microbial interface of the gastrointestinal tract III. cell-to-cell signaling among microbial flora, host, and pathogens: there is a whole lot of talking going on. Am J Physiol Gastrointest Liver Physiol 288:G1105–9. doi: 10.1152/ajpgi.00572.2004 [DOI] [PubMed] [Google Scholar]
- 15. Delgado S, Cabrera-Rubio R, Mira A, Suárez A, Mayo B. 2013. Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microb Ecol 65:763–772. doi: 10.1007/s00248-013-0192-5 [DOI] [PubMed] [Google Scholar]
- 16. Kastl AJ, Terry NA, Wu GD, Albenberg LG. 2020. The structure and function of the human small intestinal microbiota: current understanding and future directions. Cell Mol Gastroenterol Hepatol 9:33–45. doi: 10.1016/j.jcmgh.2019.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Conway T, Krogfelt KA, Cohen PS. 2004. The life of commensal Escherichia coli in the mammalian intestine. EcoSal Plus 1. doi: 10.1128/ecosalplus.8.3.1.2 [DOI] [PubMed] [Google Scholar]
- 18. Johansson MEV, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. 2008. The inner of the two MUC2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105:15064–15069. doi: 10.1073/pnas.0803124105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johansson MEV, Ambort D, Pelaseyed T, Schütte A, Gustafsson JK, Ermund A, Subramani DB, Holmén-Larsson JM, Thomsson KA, Bergström JH, van der Post S, Rodriguez-Piñeiro AM, Sjövall H, Bäckström M, Hansson GC. 2011. Composition and functional role of the mucus layers in the intestine. Cell Mol Life Sci 68:3635–3641. doi: 10.1007/s00018-011-0822-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arike L, Hansson GC. 2016. The densely O-glycosylated MUC2 mucin protects the intestine and provides food for the commensal bacteria. J Mol Biol 428:3221–3229. doi: 10.1016/j.jmb.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Larsson JMH, Karlsson H, Sjövall H, Hansson GC. 2009. A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology 19:756–766. doi: 10.1093/glycob/cwp048 [DOI] [PubMed] [Google Scholar]
- 22. Gustafsson JK, Ermund A, Johansson MEV, Schütte A, Hansson GC, Sjövall H. 2012. An ex vivo method for studying mucus formation, properties, and thickness in human colonic biopsies and mouse small and large intestinal explants. Am J Physiol Gastrointest Liver Physiol 302:G430–8. doi: 10.1152/ajpgi.00405.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H, Limenitakis JP, Fuhrer T, Geuking MB, Lawson MA, Wyss M, Brugiroux S, Keller I, Macpherson JA, Rupp S, Stolp B, Stein JV, Stecher B, Sauer U, McCoy KD, Macpherson AJ. 2015. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun 6:8292. doi: 10.1038/ncomms9292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Simon GL, Gorbach SL. 1984. Intestinal flora in health and disease. Gastroenterology 86:174–193. [PubMed] [Google Scholar]
- 25. Borrelio SP. 1986. Microbial flora of the gastro-intestinal tract, p 2–16. In Hill MJ (ed), Microbial metabolism in the digestive tract. CRC Press, Inc, Boca Raton, Fla. [Google Scholar]
- 26. Tannock GW. 1997. Normal microbiota of the gastrointestinal tract of rodents, p 187–215. In Mackie RI, White BA, Isaacson RE (ed), Gastrointestinal microbiology. Chapman & Hall, London, UK. [Google Scholar]
- 27. Donaldson GP, Lee SM, Mazmanian SK. 2016. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 14:20–32. doi: 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Juge N. 2012. Microbial adhesins to gastrointestinal mucus. Trends Microbiol 20:30–39. doi: 10.1016/j.tim.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 29. Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10:323–335. doi: 10.1038/nrmicro2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Crouch LI, Liberato MV, Urbanowicz PA, Baslé A, Lamb CA, Stewart CJ, Cooke K, Doona M, Needham S, Brady RR, Berrington JE, Madunic K, Wuhrer M, Chater P, Pearson JP, Glowacki R, Martens EC, Zhang F, Linhardt RJ, Spencer DIR, Bolam DN. 2020. Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown. Nat Commun 11:4017. doi: 10.1038/s41467-020-17847-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Glover JS, Ticer TD, Engevik MA. 2022. Characterizing the mucin-degrading capacity of the human gut microbiota. Sci Rep 12:8456. doi: 10.1038/s41598-022-11819-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poulsen LK, Lan F, Kristensen CS, Hobolth P, Molin S, Krogfelt KA. 1994. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect Immun 62:5191–5194. doi: 10.1128/iai.62.11.5191-5194.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Møller AK, Leatham MP, Conway T, Nuijten PJM, de Haan LAM, Krogfelt KA, Cohen PS. 2003. An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect Immun 71:2142–2152. doi: 10.1128/IAI.71.4.2142-2152.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoskins LC, Agustines M, McKee WB, Boulding ET, Kriaris M, Niedermeyer G. 1985. Mucin degradation in human colon ecosystems. isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J Clin Invest 75:944–953. doi: 10.1172/JCI111795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ihssen J, Egli T. 2005. Global physiological analysis of carbon- and energy-limited growing Escherichia coli confirms a high degree of catabolic flexibility and preparedness for mixed substrate utilization. Environ Microbiol 7:1568–1581. doi: 10.1111/j.1462-2920.2005.00846.x [DOI] [PubMed] [Google Scholar]
- 36. Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, Leatham MP, Lins JJ, Allen RL, Laux DC, Cohen PS, Conway T. 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun 76:1143–1152. doi: 10.1128/IAI.01386-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Henderson IR, Czeczulin J, Eslava C, Noriega F, Nataro JP. 1999. Characterization of PIC, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect Immun 67:5587–5596. doi: 10.1128/IAI.67.11.5587-5596.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Parham NJ, Srinivasan U, Desvaux M, Foxman B, Marrs CF, Henderson IR. 2004. PicU, a second serine protease autotransporter of uropathogenic Escherichia coli. FEMS Microbiol Lett 230:73–83. doi: 10.1016/S0378-1097(03)00862-0 [DOI] [PubMed] [Google Scholar]
- 39. Heimer SR, Rasko DA, Lockatell CV, Johnson DE, Mobley HLT. 2004. Autotransporter genes PIC and TSH are associated with Escherichia coli strains that cause acute pyelonephritis and are expressed during urinary tract infection. Infect Immun 72:593–597. doi: 10.1128/IAI.72.1.593-597.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Abreu AG, Abe CM, Nunes KO, Moraes CTP, Chavez-Dueñas L, Navarro-Garcia F, Barbosa AS, Piazza RMF, Elias WP. 2016. The serine protease PIC as a virulence factor of atypical enteropathogenic Escherichia coli. Gut Microbes 7:115–125. doi: 10.1080/19490976.2015.1136775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hosseini Nave H, Mansouri S, Taati Moghadam M, Moradi M. 2016. Virulence gene profile and multilocus variable-number tandem-repeat analysis (MLVA) of enteroinvasive Escherichia coli (EIEC) isolates from patients with diarrhea in Kerman, Iran. Jundishapur J Microbiol 9:e33529. doi: 10.5812/jjm.33529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boisen N, Ruiz-Perez F, Scheutz F, Krogfelt KA, Nataro JP. 2009. Short report: high prevalence of serine protease autotransporter cytotoxins among strains of enteroaggregative Escherichia coli. Am J Trop Med Hyg 80:294–301. [PMC free article] [PubMed] [Google Scholar]
- 43. Harrington SM, Sheikh J, Henderson IR, Ruiz-Perez F, Cohen PS, Nataro JP. 2009. The PIC protease of enteroaggregative Escherichia coli promotes intestinal colonization and growth in the presence of mucin. Infect Immun 77:2465–2473. doi: 10.1128/IAI.01494-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu L, Saitz-Rojas W, Smith R, Gonyar L, In JG, Kovbasnjuk O, Zachos NC, Donowitz M, Nataro JP, Ruiz-Perez F. 2020. Mucus layer modeling of human colonoids during infection with enteroaggragative E. coli. Sci Rep 10:10533. doi: 10.1038/s41598-020-67104-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rang CU, Licht TR, Midtvedt T, Conway PL, Chao L, Krogfelt KA, Cohen PS, Molin S. 1999. Estimation of growth rates of Escherichia coli BJ4 in streptomycin-treated and previously germfree mice by in situ rRNA hybridization. Clin Diagn Lab Immunol 6:434–436. doi: 10.1128/CDLI.6.3.434-436.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Poulsen LK, Licht TR, Rang C, Krogfelt KA, Molin S. 1995. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. J Bacteriol 177:5840–5845. doi: 10.1128/jb.177.20.5840-5845.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peekhaus N, Conway T. 1998. What’s for dinner?: Entner-Doudoroff metabolism in Escherichia coli. J Bacteriol 180:3495–3502. doi: 10.1128/JB.180.14.3495-3502.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Licht TR, Tolker-Nielsen T, Holmstrøm K, Krogfelt KA, Molin S. 1999. Inhibition of Escherichia coli precursor-16S rRNA processing by mouse intestinal contents. Environ Microbiol 1:23–32. doi: 10.1046/j.1462-2920.1999.00001.x [DOI] [PubMed] [Google Scholar]
- 49. Wadolkowski EA, Laux DC, Cohen PS. 1988. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus. Infect Immun 56:1030–1035. doi: 10.1128/iai.56.5.1030-1035.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stecher B, Hardt WD. 2011. Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol 14:82–91. doi: 10.1016/j.mib.2010.10.003 [DOI] [PubMed] [Google Scholar]
- 51. Burson A, Stomp M, Greenwell E, Grosse J, Huisman J. 2018. Competition for nutrients and light: testing advances in resource competition with a natural phytoplankton community. Ecology 99:1108–1118. doi: 10.1002/ecy.2187 [DOI] [PubMed] [Google Scholar]
- 52. Freter R, Brickner H, Botney M, Cleven D, Aranki A. 1983. Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect Immun 39:676–685. doi: 10.1128/iai.39.2.676-685.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jones SA, Chowdhury FZ, Fabich AJ, Anderson A, Schreiner DM, House AL, Autieri SM, Leatham MP, Lins JJ, Jorgensen M, Cohen PS, Conway T. 2007. Respiration of Escherichia coli in the mouse intestine. Infect Immun 75:4891–4899. doi: 10.1128/IAI.00484-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jones SA, Gibson T, Maltby RC, Chowdhury FZ, Stewart V, Cohen PS, Conway T. 2011. Anaerobic respiration of Escherichia coli in the mouse intestine. Infect Immun 79:4218–4226. doi: 10.1128/IAI.05395-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reese AT, Pereira FC, Schintlmeister A, Berry D, Wagner M, Hale LP, Wu A, Jiang S, Durand HK, Zhou X, Premont RT, Diehl AM, O’Connell TM, Alberts SC, Kartzinel TR, Pringle RM, Dunn RR, Wright JP, David LA. 2018. Microbial nitrogen limitation in the mammalian large intestine. Nat Microbiol 3:1441–1450. doi: 10.1038/s41564-018-0267-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Das NK, Schwartz AJ, Barthel G, Inohara N, Liu Q, Sankar A, Hill DR, Ma X, Lamberg O, Schnizlein MK, Arqués JL, Spence JR, Nunez G, Patterson AD, Sun D, Young VB, Shah YM. 2020. Microbial metabolite signaling is required for systemic iron homeostasis. Cell Metab 31:115–130. doi: 10.1016/j.cmet.2019.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zeng X, Xing X, Gupta M, Keber FC, Lopez JG, Lee Y-C, Roichman A, Wang L, Neinast MD, Donia MS, Wühr M, Jang C, Rabinowitz JD. 2022. Gut bacterial nutrient preferences quantified in vivo. Cell 185:3441–3456. doi: 10.1016/j.cell.2022.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Anderson CJ, Medina CB, Barron BJ, Karvelyte L, Aaes TL, Lambertz I, Perry JSA, Mehrotra P, Gonçalves A, Lemeire K, Blancke G, Andries V, Ghazavi F, Martens A, van Loo G, Vereecke L, Vandenabeele P, Ravichandran KS. 2021. Microbes exploit death-induced nutrient release by gut epithelial cells. Nature 596:262–267. doi: 10.1038/s41586-021-03785-9 [DOI] [PubMed] [Google Scholar]
- 59. Fuller MF, Reeds PJ. 1998. Nitrogen cycling in the gut. Annu Rev Nutr 18:385–411. doi: 10.1146/annurev.nutr.18.1.385 [DOI] [PubMed] [Google Scholar]
- 60. Sung J, Kim S, Cabatbat JJT, Jang S, Jin Y-S, Jung GY, Chia N, Kim P-J. 2017. Global metabolic interaction network of the human gut microbiota for context-specific community-scale analysis. Nat Commun 8:15393. doi: 10.1038/ncomms15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pérez Escriva P, Fuhrer T, Sauer U. 2022. Distinct N and C cross-feeding networks in a synthetic mouse gut consortium. mSystems 7:e0148421. doi: 10.1128/msystems.01484-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cohen PS, Conway T. 2015. Applying the restaurant hypothesis to intestinal microbiota. Microbe Magazine 10:324–328. doi: 10.1128/microbe.10.324.1 [DOI] [Google Scholar]
- 63. Wong JMW, Jenkins DJA. 2007. Carbohydrate digestibility and metabolic effects. J Nutr 137:2539S–2546S. doi: 10.1093/jn/137.11.2539S [DOI] [PubMed] [Google Scholar]
- 64. Jakubovics NS. 2015. Saliva as the sole nutritional source in the development of multispecies communities in dental plaque. Microbiol Spectr 3. doi: 10.1128/microbiolspec.MBP-0013-2014 [DOI] [PubMed] [Google Scholar]
- 65. Conway T, Krogfelt KA, Cohen PS. 2007. Escherichia coli at the intestinal mucosal surface, p 175–196. In Brogden KA MF, Cornick N, Stanton TB, Zhang Q, Nolan LK, Wannemuehler MJ (ed), Virulence mechanisms of bacterial pathogens, 4 ed. ASM Press, Washington, DC. [Google Scholar]
- 66. Kim YS, Ho SB. 2010. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep 12:319–330. doi: 10.1007/s11894-010-0131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Camilleri M. 2006. Integrated upper gastrointestinal response to food intake. Gastroenterology 131:640–658. doi: 10.1053/j.gastro.2006.03.023 [DOI] [PubMed] [Google Scholar]
- 68. Schubert C, Winter M, Ebert-Jung A, Kierszniowska S, Nagel-Wolfrum K, Schramm T, Link H, Winter S, Unden G. 2021. C4-dicarboxylates and L-aspartate utilization by Escherichia coli K-12 in the mouse intestine: L-aspartate as a major substrate for fumarate respiration and as a nitrogen source. Environ Microbiol 23:2564–2577. doi: 10.1111/1462-2920.15478 [DOI] [PubMed] [Google Scholar]
- 69. Ruddle SJ, Massis LM, Cutter AC, Monack DM. 2023. Salmonella-liberated dietary L-arabinose promotes expansion in superspreaders. Cell Host Microbe 31:405–417. doi: 10.1016/j.chom.2023.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502:96–99. doi: 10.1038/nature12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Eberl C, Weiss AS, Jochum LM, Durai Raj AC, Ring D, Hussain S, Herp S, Meng C, Kleigrewe K, Gigl M, Basic M, Stecher B. 2021. E. coli enhance colonization resistance against Salmonella Typhimurium by competing for galactitol, a context-dependent limiting carbon source. Cell Host Microbe 29:1680–1692. doi: 10.1016/j.chom.2021.09.004 [DOI] [PubMed] [Google Scholar]
- 72. Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307:1955–1959. doi: 10.1126/science.1109051 [DOI] [PubMed] [Google Scholar]
- 73. Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, Young VB, Henrissat B, Wilmes P, Stappenbeck TS, Núñez G, Martens EC. 2016. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167:1339–1353. doi: 10.1016/j.cell.2016.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Martens EC, Chiang HC, Gordon JI. 2008. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4:447–457. doi: 10.1016/j.chom.2008.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jimenez AG, Ellermann M, Abbott W, Sperandio V. 2020. Diet-derived galacturonic acid regulates virulence and intestinal colonization in enterohaemorrhagic Escherichia coli and citrobacter rodentium. Nat Microbiol 5:368–378. doi: 10.1038/s41564-019-0641-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Luis AS, Briggs J, Zhang X, Farnell B, Ndeh D, Labourel A, Baslé A, Cartmell A, Terrapon N, Stott K, Lowe EC, McLean R, Shearer K, Schückel J, Venditto I, Ralet M-C, Henrissat B, Martens EC, Mosimann SC, Abbott DW, Gilbert HJ. 2018. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol 3:210–219. doi: 10.1038/s41564-017-0079-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, Cantarel BL, Coutinho PM, Henrissat B, Crock LW, Russell A, Verberkmoes NC, Hettich RL, Gordon JI. 2009. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci U S A 106:5859–5864. doi: 10.1073/pnas.0901529106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wu M, McNulty NP, Rodionov DA, Khoroshkin MS, Griffin NW, Cheng J, Latreille P, Kerstetter RA, Terrapon N, Henrissat B, Osterman AL, Gordon JI. 2015. Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut bacteroides. Science 350:aac5992. doi: 10.1126/science.aac5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pultz NJ, Hoskins LC, Donskey CJ. 2006. Vancomycin-resistant enterococci may obtain nutritional support by scavenging carbohydrate fragments generated during mucin degradation by the anaerobic microbiota of the colon. Microb Drug Resist 12:63–67. doi: 10.1089/mdr.2006.12.63 [DOI] [PubMed] [Google Scholar]
- 80. Falony G, Vlachou A, Verbrugghe K, De Vuyst L. 2006. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl Environ Microbiol 72:7835–7841. doi: 10.1128/AEM.01296-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lendenmann U, Snozzi M, Egli T. 1996. Kinetics of the simultaneous utilization of sugar mixtures by Escherichia coli in continuous culture. Appl Environ Microbiol 62:1493–1499. doi: 10.1128/aem.62.5.1493-1499.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Münster U. 1993. Concentrations and fluxes of organic carbon substrates in the aquatic environment. Antonie Van Leeuwenhoek 63:243–274. doi: 10.1007/BF00871222 [DOI] [PubMed] [Google Scholar]
- 83. Leatham-Jensen MP, Frimodt-Møller J, Adediran J, Mokszycki ME, Banner ME, Caughron JE, Krogfelt KA, Conway T, Cohen PS. 2012. The streptomycin-treated mouse intestine selects Escherichia coli envZ missense mutants that interact with dense and diverse intestinal microbiota. Infect Immun 80:1716–1727. doi: 10.1128/IAI.06193-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Adediran J, Leatham-Jensen MP, Mokszycki ME, Frimodt-Møller J, Krogfelt KA, Kazmierczak K, Kenney LJ, Conway T, Cohen PS. 2014. An Escherichia coli Nissle 1917 missense mutant colonizes the streptomycin-treated mouse intestine better than the wild type but is not a better probiotic. Infect Immun 82:670–682. doi: 10.1128/IAI.01149-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. McCormick BA, Franklin DP, Laux DC, Cohen PS. 1989. Type 1 pili are not necessary for colonization of the streptomycin-treated mouse large intestine by type 1-piliated Escherichia coli F-18 and E. coli K-12. Infect Immun 57:3022–3029. doi: 10.1128/iai.57.10.3022-3029.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Meador JP, Caldwell ME, Cohen PS, Conway T. 2014. Escherichia coli pathotypes occupy distinct niches in the mouse intestine. Infect Immun 82:1931–1938. doi: 10.1128/IAI.01435-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, Anderson AB, Grissom JE, Laux DC, Cohen PS, Conway T. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A 101:7427–7432. doi: 10.1073/pnas.0307888101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JCD. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol 47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x [DOI] [PubMed] [Google Scholar]
- 89. Bowden SD, Rowley G, Hinton JCD, Thompson A. 2009. Glucose and glycolysis are required for the successful infection of macrophages and mice by Salmonella enterica serovar typhimurium. Infect Immun 77:3117–3126. doi: 10.1128/IAI.00093-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jones SA, Jorgensen M, Chowdhury FZ, Rodgers R, Hartline J, Leatham MP, Struve C, Krogfelt KA, Cohen PS, Conway T. 2008. Glycogen and maltose utilization by Escherichia coli O157:H7 in the mouse intestine. Infect Immun 76:2531–2540. doi: 10.1128/IAI.00096-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Boos W, Shuman H. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev 62:204–229. doi: 10.1128/MMBR.62.1.204-229.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Preiss J. 1984. Bacterial glycogen synthesis and its regulation. Annu Rev Microbiol 38:419–458. doi: 10.1146/annurev.mi.38.100184.002223 [DOI] [PubMed] [Google Scholar]
- 93. Svanborg C. 1995. Re.: oral administration of a certain strain of live Escherichia coli for intestinal disorders. Infection 23:251–251. doi: 10.1007/BF01781214 [DOI] [PubMed] [Google Scholar]
- 94. Sartor RB. 2005. Probiotic therapy of intestinal inflammation and infections. Curr Opin Gastroenterol 21:44–50. [PubMed] [Google Scholar]
- 95. Kruis W, Fric P, Pokrotnieks J, Lukás M, Fixa B, Kascák M, Kamm MA, Weismueller J, Beglinger C, Stolte M, Wolff C, Schulze J. 2004. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53:1617–1623. doi: 10.1136/gut.2003.037747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Schultz M. 2008. Clinical use of E. coli Nissle 1917 in inflammatory bowel disease. Inflamm Bowel Dis 14:1012–1018. doi: 10.1002/ibd.20377 [DOI] [PubMed] [Google Scholar]
- 97. Levine MM, Bergquist EJ, Nalin DR, Waterman DH, Hornick RB, Young CR, Sotman S. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet 1:1119–1122. doi: 10.1016/s0140-6736(78)90299-4 [DOI] [PubMed] [Google Scholar]
- 98. Formal SB, Dammin GJ, Labrec EH, Schneider H. 1958. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J Bacteriol 75:604–610. doi: 10.1128/jb.75.5.604-610.1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. O’Brien AD, Melton AR, Schmitt CK, McKee ML, Batts ML, Griffin DE. 1993. Profile of Escherichia coli O157:H7 pathogen responsible for hamburger-borne outbreak of hemorrhagic colitis and hemolytic uremic syndrome in Washington. J Clin Microbiol 31:2799–2801. doi: 10.1128/jcm.31.10.2799-2801.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bertin Y, Chaucheyras-Durand F, Robbe-Masselot C, Durand A, de la Foye A, Harel J, Cohen PS, Conway T, Forano E, Martin C. 2013. Carbohydrate utilization by enterohaemorrhagic Escherichia coli O157:H7 in bovine intestinal content. Environ Microbiol 15:610–622. doi: 10.1111/1462-2920.12019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Alteri CJ, Smith SN, Mobley HLT. 2009. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog 5:e1000448. doi: 10.1371/journal.ppat.1000448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Levine MM, Nataro JP, Karch H, Baldini MM, Kaper JB, Black RE, Clements ML, O’Brien AD. 1985. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J Infect Dis 152:550–559. doi: 10.1093/infdis/152.3.550 [DOI] [PubMed] [Google Scholar]
- 103. Taylor SJ, Winter MG, Gillis CC, Silva L da, Dobbins AL, Muramatsu MK, Jimenez AG, Chanin RB, Spiga L, Llano EM, Rojas VK, Kim J, Santos RL, Zhu W, Winter SE. 2022. Colonocyte-derived lactate promotes E. coli fitness in the context of inflammation-associated gut microbiota dysbiosis. Microbiome 10:200. doi: 10.1186/s40168-022-01389-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Garzetti D, Eberl C, Stecher B. 2018. Complete genome sequencing of the mouse intestinal isolate Escherichia coli Mt1B1. Genome Announc 6:e00426-18. doi: 10.1128/genomeA.00426-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Viola RE. 2000. L-aspartase: new tricks from an old enzyme. Adv Enzymol Relat Areas Mol Biol 74:295–341. doi: 10.1002/9780470123201.ch7 [DOI] [PubMed] [Google Scholar]
- 106. Schubert C, Zedler S, Strecker A, Unden G. 2021. L-aspartate as a high-quality nitrogen source in Escherichia coli: regulation of L-aspartase by the nitrogen regulatory system and interaction of L-aspartase with GlnB. Mol Microbiol 115:526–538. doi: 10.1111/mmi.14620 [DOI] [PubMed] [Google Scholar]
- 107. Kitamoto S, Alteri CJ, Rodrigues M, Nagao-Kitamoto H, Sugihara K, Himpsl SD, Bazzi M, Miyoshi M, Nishioka T, Hayashi A, et al. 2020. Dietary L-serine confers a competitive fitness advantage to enterobacteriaceae in the inflamed gut. Nat Microbiol 5:116–125. doi: 10.1038/s41564-019-0591-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bertin Y, Girardeau JP, Chaucheyras-Durand F, Lyan B, Pujos-Guillot E, Harel J, Martin C. 2011. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol 13:365–377. doi: 10.1111/j.1462-2920.2010.02334.x [DOI] [PubMed] [Google Scholar]
- 109. Doranga S, Conway T, Kaspar JR. 2023. OmpC-dependent bile tolerance contributes to E. coli colonization of the mammalian intestine. Microbiol Spectr 11:e0524122. doi: 10.1128/spectrum.05241-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Leatham MP, Banerjee S, Autieri SM, Mercado-Lubo R, Conway T, Cohen PS. 2009. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine . Infect Immun 77:2876–2886. doi: 10.1128/IAI.00059-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Segura A, Bertoni M, Auffret P, Klopp C, Bouchez O, Genthon C, Durand A, Bertin Y, Forano E. 2018. Transcriptomic analysis reveals specific metabolic pathways of enterohemorrhagic Escherichia coli O157:H7 in bovine digestive contents. BMC Genomics 19:766. doi: 10.1186/s12864-018-5167-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Miranda RL, Conway T, Leatham MP, Chang DE, Norris WE, Allen JH, Stevenson SJ, Laux DC, Cohen PS. 2004. Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDl933) and E. coli K-12 (MG1655) in the mouse intestine. Infect Immun 72:1666–1676. doi: 10.1128/IAI.72.3.1666-1676.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Schinner SAC, Mokszycki ME, Adediran J, Leatham-Jensen M, Conway T, Cohen PS. 2015. Escherichia coli EDL933 requires gluconeogenic nutrients to successfully colonize the intestines of streptomycin-treated mice precolonized with E. coli Nissle 1917. Infect Immun 83:1983–1991. doi: 10.1128/IAI.02943-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Bertin Y, Deval C, de la Foye A, Masson L, Gannon V, Harel J, Martin C, Desvaux M, Forano E. 2014. The Gluconeogenesis pathway is involved in maintenance of enterohaemorrhagic Escherichia coli O157:H7 in bovine intestinal content. PLoS One 9:e98367. doi: 10.1371/journal.pone.0098367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Leatham MP, Stevenson SJ, Gauger EJ, Krogfelt KA, Lins JJ, Haddock TL, Autieri SM, Conway T, Cohen PS. 2005. Mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect Immun 73:8039–8049. doi: 10.1128/IAI.73.12.8039-8049.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sim M, Koirala S, Picton D, Strahl H, Hoskisson PA, Rao CV, Gillespie CS, Aldridge PD. 2017. Growth rate control of flagellar assembly in Escherichia coli strain RP437. Sci Rep 7:41189. doi: 10.1038/srep41189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Gauger EJ, Leatham MP, Mercado-Lubo R, Laux DC, Conway T, Cohen PS. 2007. Role of motility and the flhDC operon in Escherichia coli MG1655 colonization of the mouse intestine. Infect Immun 75:3315–3324. doi: 10.1128/IAI.00052-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. De Paepe M, Gaboriau-Routhiau V, Rainteau D, Rakotobe S, Taddei F, Cerf-Bensussan N. 2011. Trade-off between bile resistance and nutritional competence drives Escherichia coli diversification in the mouse gut. PLoS Genet 7:e1002107. doi: 10.1371/journal.pgen.1002107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Giraud A, Arous S, De Paepe M, Gaboriau-Routhiau V, Bambou J-C, Rakotobe S, Lindner AB, Taddei F, Cerf-Bensussan N. 2008. Dissecting the genetic components of adaptation of Escherichia coli to the mouse gut. PLoS Genet 4:e2. doi: 10.1371/journal.pgen.0040002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol 46:281–291. doi: 10.1046/j.1365-2958.2002.03170.x [DOI] [PubMed] [Google Scholar]
- 121. Guillier M, Gottesman S. 2006. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol 59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x [DOI] [PubMed] [Google Scholar]
- 122. Weickert MJ, Adhya S. 1993. The galactose regulon of Escherichia coli. Mol Microbiol 10:245–251. doi: 10.1111/j.1365-2958.1993.tb01950.x [DOI] [PubMed] [Google Scholar]
- 123. Pal RR, Baidya AK, Mamou G, Bhattacharya S, Socol Y, Kobi S, Katsowich N, Ben-Yehuda S, Rosenshine I. 2019. Pathogenic E. coli extracts nutrients from infected host cells utilizing Injectisome components. Cell 177:683–696. doi: 10.1016/j.cell.2019.02.022 [DOI] [PubMed] [Google Scholar]
- 124. Bäumler AJ, Sperandio V. 2016. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535:85–93. doi: 10.1038/nature18849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Pacheco AR, Sperandio V. 2015. Enteric pathogens exploit the microbiota-generated nutritional environment of the gut. Microbiol Spectr 3. doi: 10.1128/microbiolspec.MBP-0001-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Khan I, Bai Y, Zha L, Ullah N, Ullah H, Shah SRH, Sun H, Zhang C. 2021. Mechanism of the gut microbiota colonization resistance and enteric pathogen infection. Front Cell Infect Microbiol 11:716299. doi: 10.3389/fcimb.2021.716299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wotzka SY, Kreuzer M, Maier L, Arnoldini M, Nguyen BD, Brachmann AO, Berthold DL, Zünd M, Hausmann A, Bakkeren E, et al. 2019. Escherichia coli limits Salmonella Typhimurium infections after diet shifts and fat-mediated microbiota perturbation in mice. Nat Microbiol 4:2164–2174. doi: 10.1038/s41564-019-0568-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt W-D. 2007. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5:2177–2189. doi: 10.1371/journal.pbio.0050244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Ferreira RBR, Gill N, Willing BP, Antunes LCM, Russell SL, Croxen MA, Finlay BB. 2011. The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLoS One 6:e20338. doi: 10.1371/journal.pone.0020338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, Finlay BB. 2008. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun 76:4726–4736. doi: 10.1128/IAI.00319-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Stecher B. 2015. The roles of inflammation, nutrient availability and the commensal microbiota in enteric pathogen infection. Microbiol Spectr 3. doi: 10.1128/microbiolspec.MBP-0008-2014 [DOI] [PubMed] [Google Scholar]
- 132. Buffie CG, Pamer EG. 2013. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 13:790–801. doi: 10.1038/nri3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Osbelt L, Wende M, Almási É, Derksen E, Muthukumarasamy U, Lesker TR, Galvez EJC, Pils MC, Schalk E, Chhatwal P, Färber J, Neumann-Schaal M, Fischer T, Schlüter D, Strowig T. 2021. Klebsiella oxytoca causes colonization resistance against multidrug-resistant K. pneumoniae in the gut via cooperative carbohydrate competition. Cell Host Microbe 29:1663–1679. doi: 10.1016/j.chom.2021.09.003 [DOI] [PubMed] [Google Scholar]
- 134. Kamada N, Kim Y-G, Sham HP, Vallance BA, Puente JL, Martens EC, Núñez G. 2012. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336:1325–1329. doi: 10.1126/science.1222195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Osbelt L, Thiemann S, Smit N, Lesker TR, Schröter M, Gálvez EJC, Schmidt-Hohagen K, Pils MC, Mühlen S, Dersch P, Hiller K, Schlüter D, Neumann-Schaal M, Strowig T, Baumler AJ. 2020. Variations in microbiota composition of laboratory mice influence Citrobacter rodentium infection via variable short-chain fatty acid production. PLoS Pathog 16:e1008448. doi: 10.1371/journal.ppat.1008448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Sorbara MT, Dubin K, Littmann ER, Moody TU, Fontana E, Seok R, Leiner IM, Taur Y, Peled JU, van den Brink MRM, Litvak Y, Bäumler AJ, Chaubard J-L, Pickard AJ, Cross JR, Pamer EG. 2019. Inhibiting antibiotic-resistant enterobacteriaceae by microbiota-mediated intracellular acidification. J Exp Med 216:84–98. doi: 10.1084/jem.20181639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Corr SC, Li Y, Riedel CU, O’Toole PW, Hill C, Gahan CGM. 2007. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci U S A 104:7617–7621. doi: 10.1073/pnas.0700440104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Fang K, Jin X, Hong SH. 2018. Probiotic Escherichia coli inhibits biofilm formation of pathogenic E. coli via extracellular activity of DegP. Sci Rep 8:4939. doi: 10.1038/s41598-018-23180-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Hecht AL, Casterline BW, Earley ZM, Goo YA, Goodlett DR, Bubeck Wardenburg J. 2016. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep 17:1281–1291. doi: 10.15252/embr.201642282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Cash HL, Whitham CV, Behrendt CL, Hooper LV. 2006. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313:1126–1130. doi: 10.1126/science.1127119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Zhan Z, Tang H, Zhang Y, Huang X, Xu M. 2022. Potential of gut-derived short-chain fatty acids to control enteric pathogens. Front Microbiol 13:976406. doi: 10.3389/fmicb.2022.976406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3:777–788. doi: 10.1038/nrmicro1273 [DOI] [PubMed] [Google Scholar]
- 143. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MRM, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Hancock V, Dahl M, Klemm P. 2010. Probiotic Escherichia coli strain Nissle 1917 outcompetes intestinal pathogens during biofilm formation. J Med Microbiol 59:392–399. doi: 10.1099/jmm.0.008672-0 [DOI] [PubMed] [Google Scholar]
- 145. Serapio-Palacios A, Woodward SE, Vogt SL, Deng W, Creus-Cuadros A, Huus KE, Cirstea M, Gerrie M, Barcik W, Yu H, Finlay BB. 2022. Type VI secretion systems of pathogenic and commensal bacteria mediate niche occupancy in the gut. Cell Rep 39:110731. doi: 10.1016/j.celrep.2022.110731 [DOI] [PubMed] [Google Scholar]
- 146. Kim S, Covington A, Pamer EG. 2017. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol Rev 279:90–105. doi: 10.1111/imr.12563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Sorbara MT, Pamer EG. 2019. Interbacterial mechanisms of colonization resistance and the strategies pathogens use to overcome them. Mucosal Immunol 12:1–9. doi: 10.1038/s41385-019-0151-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Pereira FC, Berry D. 2017. Microbial nutrient niches in the gut. Environ Microbiol 19:1366–1378. doi: 10.1111/1462-2920.13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Michino H, Araki K, Minami S, Takaya S, Sakai N, Miyazaki M, Ono A, Yanagawa H. 1999. Massive outbreak of Escherichia coli O157:H7 infection in school children in Sakai city, Japan, associated with consumption of white radish sprouts. Am J Epidemiol 150:787–796. doi: 10.1093/oxfordjournals.aje.a010082 [DOI] [PubMed] [Google Scholar]
- 150. Caballero-Flores G, Pickard JM, Fukuda S, Inohara N, Núñez G. 2020. An enteric pathogen subverts colonization resistance by evading competition for amino acids in the gut. Cell Host Microbe 28:526–533. doi: 10.1016/j.chom.2020.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Theriot CM, Koenigsknecht MJ, Carlson PE Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, Z Li J, Young VB. 2014. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to clostridium difficile infection. Nat Commun 5:3114. doi: 10.1038/ncomms4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Olsen SJ, Miller G, Breuer T, Kennedy M, Higgins C, Walford J, McKee G, Fox K, Bibb W, Mead P. 2002. A waterborne outbreak of Escherichia coli O157:H7 infections and hemolytic uremic syndrome: implications for rural water systems. Emerg Infect Dis 8:370–375. doi: 10.3201/eid0804.000218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Anderson JD, Gillespie WA, Richmond MH. 1973. Chemotherapy and antibiotic-resistance transfer between enterobacteria in the human gastro-intestinal tract. J Med Microbiol 6:461–473. doi: 10.1099/00222615-6-4-461 [DOI] [PubMed] [Google Scholar]
- 154. Zhang W, Qi W, Albert TJ, Motiwala AS, Alland D, Hyytia-Trees EK, Ribot EM, Fields PI, Whittam TS, Swaminathan B. 2006. Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms. Genome Res 16:757–767. doi: 10.1101/gr.4759706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Rasko DA, Rosovitz MJ, Myers GSA, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. 2008. The Pangenome structure of Escherichia coli: Comparative Genomic analysis of E. coli Commensal and pathogenic isolates. J Bacteriol 190:6881–6893. doi: 10.1128/JB.00619-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Oliveira RA, Ng KM, Correia MB, Cabral V, Shi H, Sonnenburg JL, Huang KC, Xavier KB. 2020. Klebsiella michiganensis transmission enhances resistance to enterobacteriaceae gut invasion by nutrition competition. Nat Microbiol 5:630–641. doi: 10.1038/s41564-019-0658-4 [DOI] [PubMed] [Google Scholar]
- 157. Brugiroux S, Beutler M, Pfann C, Garzetti D, Ruscheweyh H-J, Ring D, Diehl M, Herp S, Lötscher Y, Hussain S, Bunk B, Pukall R, Huson DH, Münch PC, McHardy AC, McCoy KD, Macpherson AJ, Loy A, Clavel T, Berry D, Stecher B. 2016. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat Microbiol 2:16215. doi: 10.1038/nmicrobiol.2016.215 [DOI] [PubMed] [Google Scholar]
- 158. Isaac S, Flor-Duro A, Carruana G, Puchades-Carrasco L, Quirant A, Lopez-Nogueroles M, Pineda-Lucena A, Garcia-Garcera M, Ubeda C. 2022. Microbiome-mediated fructose depletion restricts murine gut colonization by vancomycin-resistant Enterococcus. Nat Commun 13:7718. doi: 10.1038/s41467-022-35380-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Freter R, Hentges D.. 1983. Human intestinal microflora in health and disease. Mechanisms that control the microflora in the large Intestine Academic Press, Inc, San Diego, Calif:33-54. [Google Scholar]
- 160. Freter R, Stauffer E, Cleven D, Holdeman LV, Moore WE. 1983. Continuous-flow cultures as in vitro models of the ecology of large intestinal flora. Infect Immun 39:666–675. doi: 10.1128/iai.39.2.666-675.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Keeney KM, Finlay BB. 2011. Enteric pathogen exploitation of the microbiota-generated nutrient environment of the gut. Curr Opin Microbiol 14:92–98. doi: 10.1016/j.mib.2010.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Gillilland MG III, Erb-Downward JR, Bassis CM, Shen MC, Toews GB, Young VB, Huffnagle GB. 2012. Ecological succession of bacterial communities during conventionalization of germ-free mice. Appl Environ Microbiol 78:2359–2366. doi: 10.1128/AEM.05239-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Marino S, Baxter NT, Huffnagle GB, Petrosino JF, Schloss PD. 2014. Mathematical modeling of primary succession of murine intestinal microbiota. Proc Natl Acad Sci U S A 111:439–444. doi: 10.1073/pnas.1311322111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Palestrant D, Holzknecht ZE, Collins BH, Parker W, Miller SE, Bollinger RR. 2004. Microbial biofilms in the gut: visualization by electron microscopy and by acridine orange staining. Ultrastruct Pathol 28:23–27. [PubMed] [Google Scholar]
- 165. Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. 2005. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol 43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Rolhion N, Carvalho FA, Darfeuille-Michaud A. 2007. OmpC and the sigma(E) regulatory pathway are involved in adhesion and invasion of the Crohn’s disease-associated Escherichia coli strain LF82. Mol Microbiol 63:1684–1700. doi: 10.1111/j.1365-2958.2007.05638.x [DOI] [PubMed] [Google Scholar]
- 167. Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J Bacteriol 181:5993–6002. doi: 10.1128/JB.181.19.5993-6002.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Shelton CD, Yoo W, Shealy NG, Torres TP, Zieba JK, Calcutt MW, Foegeding NJ, Kim D, Kim J, Ryu S, Byndloss MX. 2022. Salmonella enterica serovar Typhimurium uses anaerobic respiration to overcome propionate-mediated colonization resistance. Cell Rep 38:110180. doi: 10.1016/j.celrep.2021.110180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Byrne BM, Dankert J. 1979. Volatile fatty acids and aerobic flora in the gastrointestinal tract of mice under various conditions. Infect Immun 23:559–563. doi: 10.1128/iai.23.3.559-563.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM. 2013. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54:2325–2340. doi: 10.1194/jlr.R036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Horswill AR, Dudding AR, Escalante-Semerena JC. 2001. Studies of propionate toxicity in Salmonella enterica identify 2-methylcitrate as a potent inhibitor of cell growth. J Biol Chem 276:19094–19101. doi: 10.1074/jbc.M100244200 [DOI] [PubMed] [Google Scholar]
- 172. Jacobson A, Lam L, Rajendram M, Tamburini F, Honeycutt J, Pham T, Van Treuren W, Pruss K, Stabler SR, Lugo K, Bouley DM, Vilches-Moure JG, Smith M, Sonnenburg JL, Bhatt AS, Huang KC, Monack D. 2018. A gut commensal-produced metabolite mediates colonization resistance to salmonella infection. Cell Host Microbe 24:296–307. doi: 10.1016/j.chom.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Hudson AW, Barnes AJ, Bray AS, Ornelles DA, Zafar MA. 2022. Klebsiella pneumoniae L-fucose metabolism promotes gastrointestinal colonization and modulates its virulence determinants. Infect Immun 90:e0020622. doi: 10.1128/iai.00206-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Winter SE, Bäumler AJ. 2014. “Why related bacterial species bloom simultaneously in the gut: principles underlying the 'like will to like' concept”. Cell Microbiol 16:179–184. doi: 10.1111/cmi.12245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. doi: 10.1038/nature09415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485. doi: 10.1073/pnas.1107857108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Nedialkova LP, Denzler R, Koeppel MB, Diehl M, Ring D, Wille T, Gerlach RG, Stecher B. 2014. Inflammation fuels colicin Ib-dependent competition of Salmonella serovar typhimurium and E. coli in enterobacterial blooms. PLoS Pathog 10:e1003844. doi: 10.1371/journal.ppat.1003844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Fast D, Kostiuk B, Foley E, Pukatzki S. 2018. Commensal pathogen competition impacts host viability. Proc Natl Acad Sci U S A 115:7099–7104. doi: 10.1073/pnas.1802165115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Zhao W, Caro F, Robins W, Mekalanos JJ. 2018. Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence . Science 359:210–213. doi: 10.1126/science.aap8775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Sana TG, Flaugnatti N, Lugo KA, Lam LH, Jacobson A, Baylot V, Durand E, Journet L, Cascales E, Monack DM. 2016. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci U S A 113:E5044–51. doi: 10.1073/pnas.1608858113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Aurass P, Prager R, Flieger A. 2011. EHEC/EAEC O104:H4 strain linked with the 2011 german outbreak of haemolytic uremic syndrome enters into the viable but non-culturable state in response to various stresses and resuscitates upon stress relief. Environ Microbiol 13:3139–3148. doi: 10.1111/j.1462-2920.2011.02604.x [DOI] [PubMed] [Google Scholar]
- 182. Dinu L-D, Bach S. 2011. Induction of viable but nonculturable Escherichia coli O157:H7 in the phyllosphere of lettuce: a food safety risk factor. Appl Environ Microbiol 77:8295–8302. doi: 10.1128/AEM.05020-11 [DOI] [PMC free article] [PubMed] [Google Scholar]