Abstract
Human immunodeficiency virus type 1 (HIV-1) resistance to protease inhibitors (PI) is a major obstacle to the full success of combined antiretroviral therapy. High-level resistance to these compounds is the consequence of stepwise accumulation of amino acid substitutions in the HIV-1 protease (PR), following pathways that usually differ from one inhibitor to another. The selective advantage conferred by resistance mutations may depend upon several parameters: the impact of the mutation on virus infectivity in the presence or absence of drug, the nature of the drug, and its local concentration. Because drug concentrations in vivo are subject to extensive variation over time and display a markedly uneven tissue distribution, the parameters of selection for HIV-1 resistance to PI in treated patients are complex and poorly understood. In this study, we have reconstructed a large series of HIV-1 mutants that carry single or combined mutations in the PR, retracing the accumulation pathways observed in ritonavir-, indinavir-, and saquinavir-treated patients. We have then measured the phenotypic resistance and the drug-free infectivity of these mutant viruses. A deeper insight into the evolutionary value of HIV-1 PR mutants came from a novel assay system designed to measure the replicative advantage of mutant viruses as a function of drug concentration. By tracing the resultant fitness profiles, we determined the range of drug concentrations for which mutant viruses displayed a replicative advantage over the wild type and the extent of this advantage. Fitness profiles were fully consistent with the order of accumulation of resistance mutations observed in treated patients and further emphasise the key importance of local drug concentration in the patterns of selection of drug-resistant HIV-1 mutants.
Protease inhibitors (PIs) are widely used in the highly active antiretroviral therapy regimens currently prescribed for the treatment of human immunodeficiency virus type 1 (HIV-1) infection. These compounds block the activity of the HIV protease (PR) (1, 12, 17–19) and exert a profound inhibitory effect on HIV infectivity in vitro and in vivo, yielding long-term suppression of detectable HIV replication in treated patients and spectacular stabilization of the evolution of HIV disease (22, 24). However, when antiretroviral therapy fails to be fully suppressive of HIV replication, viral variants with decreased susceptibility to PIs can emerge (10, 20, 29, 31, 33, 37, 42, 45). HIV-1 resistance to PIs is the result of the accumulation of amino acid substitutions in HIV-1 PR, following a stepwise process that leads to increasing levels of resistance (4, 9, 20, 33). Most of the residues involved in PI resistance are highly conserved within the different clades of HIV-1 (3, 49). It is therefore assumed that these residues are essential for optimal PR function, ensuring optimal infectivity of HIV particles. Correspondingly, it has been shown by several laboratories that a number of HIV-1 mutants carrying PI resistance mutations, whether selected in vitro or in treated patients failing PI therapy, display significantly reduced infectivity, related to incomplete processing of the structural and enzymatically active proteins of HIV by PR (5, 7, 11, 29, 32, 43, 51). Particular substitutions or combinations of substitutions appear to exert more profound enzymatic and virus replicative defects: this is often the case for substitutions that are located within the active site of the enzyme and are directly involved in inhibitor and substrate binding (20, 23, 30, 41, 43). Interestingly, the enzymatic and replicative defects induced by such mutations can be partially, or even in some instances completely, compensated for by the emergence of secondary mutations located outside of the substrate-binding region of the enzyme (5, 20, 27, 29, 35).
Virus resistance is usually calculated by measuring the concentration of drug that is required to inhibit 50% (IC50) or 90% (IC90) of virus infectivity. For each virus variant, the level of resistance is therefore calculated relative to its own infectivity in drug-free conditions, regardless of whether this infectivity is affected by the presence of resistance mutations. The selection of any resistance mutation, however, is a function of both its impact in terms of resistance, as expressed by the IC50 and IC90 values for the virus, and its effect on virus infectivity both in the presence and in the absence of inhibitors. In fact, the probability of selection of any resistance mutation is a function of the concentration of drug at the site of virus replication: at a low drug concentration, the selective pressure will be insufficient to ensure the emergence of mutations that induce high levels of resistance but may significantly reduce drug-free virus infectivity, while at high drug concentrations, the pressure will be to high for selection of mutations that confer only low-level resistance. Therefore, each HIV variant carrying one or several PI resistance mutations should be best characterized by describing the range of drug concentrations for which it is advantaged relative to its parent strain and the extent of this selective advantage as a function of drug concentration.
In this study, we have examined the effect of single and combined amino acid substitutions in HIV-1 PR both in terms of resistance to PIs and in terms of drug-free infectivity of the virus. These mutants are representative of the described pathways of in vivo selection for HIV resistance to indinavir (IDV), ritonavir (RTV), and saquinavir (SQV) (9, 31, 33, 42, 47), three PIs often used in current antiretroviral regimens. Additionally, for each mutant we have determined the level of its selective advantage relative to wild-type virus over a range of drug concentrations, thus defining a unique and characteristic “fitness profile.” The fitness profiles that were calculated for viruses representing each of the mutational pathways studied were fully consistent with the observations made in vivo regarding the order of appearance of the mutations in treated patients. Therefore, we show that by integrating in vitro the main parameters of the selection for drug resistance, drug-free infectivity, resistance, and drug concentration, it is possible to anticipate the pattern of accumulation of resistance mutations in HIV-1 PR.
MATERIALS AND METHODS
Plasmid construction.
To construct a convenient vector for site-directed mutagenesis of HIV-1 PR, we cloned the fragment encompassing the entire PR sequence of pNL-4.3XCS into pBlueScript-SKII+ (Stratagene), generating plasmid SK-PR. The HIV-1 proviral clone pNL-4.3XCS is a modification of the molecular clone pNL-4.3 in which an XbaI site has been inserted immediately upstream of the PR coding sequence together with a ClaI site immediately downstream (generating pNL-4.3CX) (38) and which carries a SnaB1 site inserted by silent mutagenesis at position 3872. The Quick-change site-directed mutagenesis kit (Stratagene) was used to alter residues in the PR coding region of SK-PR, using for each mutation a positive- and a negative-strand oligonucleotide, according to the manufacturer's instructions. The mutated PR sequences were used to replace the corresponding fragment of pNL-4.3XCS, generating full-length mutant clones carrying typical RTV, IDV, and SQV resistance mutations. Mutant clones contained single PR mutations or combinations of two, three, and four mutations, retracing the accumulation pathways observed in treated patients.
The positive-strand oligonucleotides used in the mutagenesis procedure were as follows: L10-I+, 5′-CTCTTTGGCAGCGACCCATCGTCACAATAAAGATAG-3′; M36-I+, 5′-CAGTATTAGAAGAAATTAATTTGCCAGGAAGATGG-3′; M46-I+, 5′-GAAGATGGAAACCTAAGATAATAGGGGGAATTG-3′; G48-V+, 5′-CAAAACCAAAAATGATAGTGGGGATCGGAGGTTTTATCAAAC-3′; I54-V+, 5′-GAATTGGAGGTTTTGTCAAAGTGAGACAGTATGATCAG-3′; A71-V+, 5′-GAAATCTGCGGACATAAAGTTATAGGTACAGTATTAG-3′; V82-A+, 5′-GGACCTACACCTGCCAACATAATTGG-3′; and L90-M+, 5′-CAACATAATTGGAAGAAATCTCATGACTCAGATTGGCTGCAC-3′.
Negative-strand oligonucleotide sequences were antiparallel to those of the positive-strand oligonucleotides.
Cell cultures and PI resistance assay.
HeLa cells and P4 cells (HeLa-CD4, LTR-lacZ) (8) were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics. P4 cells were cultured in the presence of geneticin (500 μg/ml).
Subconfluent HeLa cells in 25-cm2 flasks were transfected with 8 μg of HIV proviral plasmid DNA by the calcium phosphate precipitation method. After 18 h, the transfected HeLa cells were trypsinized and split into 200-μl subcultures in triplicate in 96-well plates in the presence of increasing concentrations of protease inhibitor (0, 1, 5, 25, 125, 625, and 3,125 nM for RTV and IDV and 0, 0.064, 0.32, 1.6, 8, 40, 200, and 1,000 nM for SQV). After 30 h of treatment, viral supernatants containing equivalent amounts of p24 antigen from each subculture were used to infect subconfluent P4 cells cultures in 96-well plates in the presence of DEAE-dextran (20 μg/ml). The p24 concentration was measured for PI-naive subcultures and extrapolated for treated subcultures originating from the same transfection experiment. Forty hours after infection of P4 cells, the single-cycle titer of viruses produced in the presence of the inhibitor was determined by quantification of the β-galactosidase activity in P4 lysates, using a colorimetric assay (termed here the CPRG assay) based on the cleavage of chlorophenolred-β-d-galactopyranoside (CPRG) by β-galactosidase (adapted from Eustice et al. [21]). Briefly, following elimination of the supernatant, the P4 cells were lysed in 100 μl of lysis buffer (MgCl2, 5 mM; NP-40, 0.1% in phosphate-buffered saline). After incubation for 5 min at room temperature, 100 μl of reaction buffer (CPRG [6 mM] in lysis buffer) was added to the cell lysates and incubated for between 5 min and 2 h at 37°C. Optical densities in the reaction wells were read at 570 nm with a reference filter set at 690 nm. The susceptibility of the different viruses to PIs was expressed as the concentration of inhibitor that inhibited 50 or 90% of infectious events (IC50 and IC90, respectively). Fold change in susceptibility to PI was calculated as the ratio of the IC90 values for mutant viruses to the corresponding value for wild-type virus.
The 40-h infection time was adopted after careful assessment that the CPRG signal corresponded to single-cycle infections. This was established by comparison with the signal obtained when zidovudine was added 6 h after infection to prevent subsequent virus replication cycles. Expression and accumulation of β-galactosidase in infected P4 cells requires several hours, and at 40 h the signal increases linearly with the infectious titer.
Infectivity assays.
The single-cycle titer of the recombinant viruses was measured on indicator P4 cells. Briefly, triplicate subconfluent P4 cells in 96-well plates were infected with the equivalent of 5 and 10 ng of HIV-1 p24 of the different viruses obtained by transfection of HeLa cells in the presence of 20 μg of DEAE-dextran per ml. The infectious titer was measured using the CPRG assay and expressed as a percentage of wild-type infectivity.
Fitness profile assay.
To determine the replicative advantage of mutant viruses as a function of PI concentration, we performed resistance assays as described above, except that instead of calculating IC90 values, we calculated the ratio of mutant to wild-type infectivity (in CPRG units) for each drug concentration and for each mutant. The ratios were then interpolated as a continuous profile across the range of different drug concentrations tested using Microsoft Excel. A minimum of three independent experiments were performed for each of the mutants, and the curves shown in Fig. 3, 4, and 5 represent the averages of the values obtained for each drug concentration. With the wild-type infectivity set as the reference, the curve representing a mutant virus will be above the wild-type reference line for drug concentrations at which the mutant displayed a replicative advantage. The height of the peak is proportional to the extent of the replicative advantage.
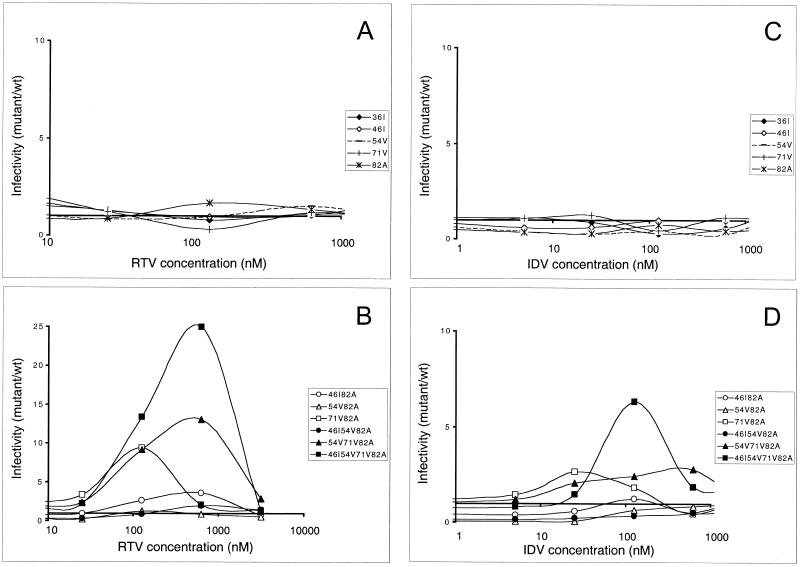
FIG. 3.
Fitness profiles: virus infectivity as a function of RTV and IDV concentration. Infectivity was measured for wild-type and PR mutant viruses in a range of PI concentrations. The ratio of mutant to wild-type infectivity (in CPRG units) was determined in the presence of various RTV (A and B) and IDV (C and D) concentrations. Viruses carrying single PR mutations are reported in panels A and C, while mutants carrying multiple resistance mutations are reported in panels B and D. The profiles shown correspond to the average values of at least three independent experiments. In panel B we used a different infectivity scale because of the high-level resistance to RTV reached by some mutant viruses.
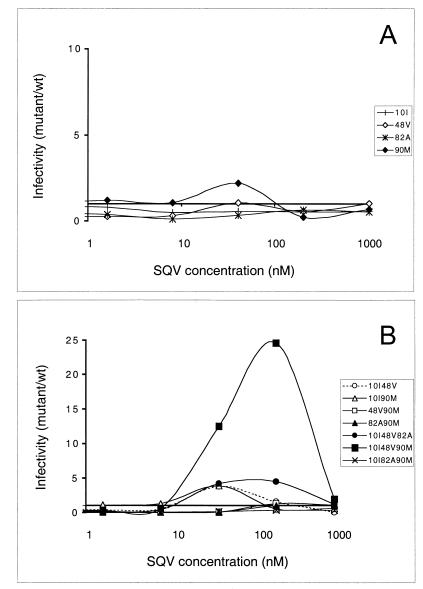
FIG. 4.
Fitness profiles: virus infectivity as a function of SQV concentration. As in Fig. 3, the ratio of mutant to wild-type infectivity (in CPRG units) was determined in the presence of various SQV concentrations. Viruses carrying single PR mutations are shown in panel A, while mutants carrying multiple resistance mutations are shown in panel B (note different scales). The profiles shown correspond to the average values of at least three independent experiments.
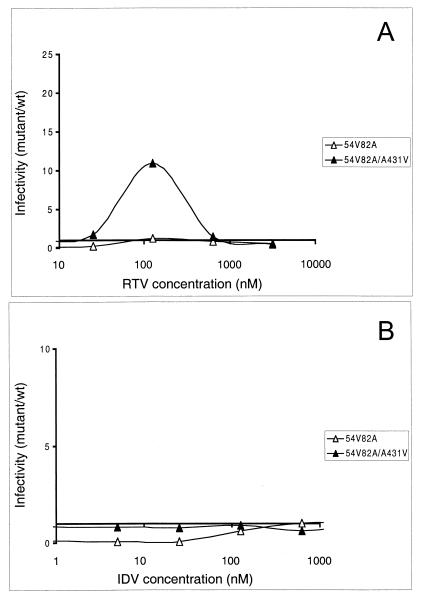
FIG. 5.
Effect of compensatory mutations in Gag cleavage sites. Comparison of fitness profiles for PR mutant I54V-V82A with and without compensatory changes in a Gag cleavage site (mutation A431V) in the presence of various concentrations of RTV (A) and IDV (B).
RESULTS
Effect of mutations in PR on resistance to PIs.
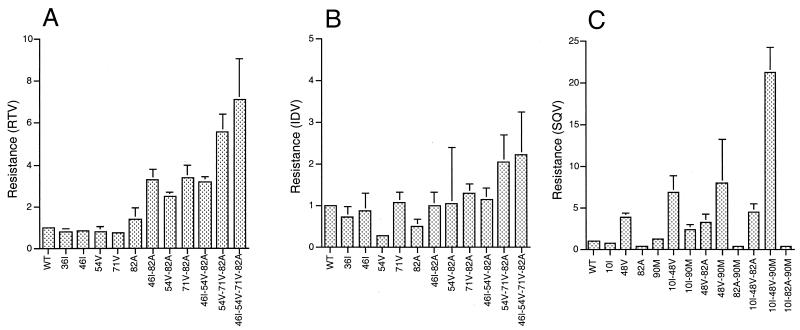
A large series of virus mutants were reconstructed in a variant of the pNL4-3 HIV-1 molecular clone according to the combinations of mutations typically observed in patients treated with RTV, IDV, or SQV (9, 31, 42, 47). We first determined the impact of amino acid substitutions in the HIV-1 PR domain on resistance to RTV, IDV, and SQV. For each mutant we calculated the fold change in susceptibility to the inhibitors as the ratio of their IC50 or IC90 values to those for wild-type virus. Mean fold changes based on IC90 values for the different mutants are reported in Fig. 1. None of the single mutants displayed a significant increase in resistance to RTV except V82A, which was slightly but reproducibly less sensitive than wild-type virus (Fig. 1A). Combinations of two or more mutations were required to attain significant resistance, the level of which generally increased with the number of mutations, as expected. However, some combinations of mutations clearly conferred higher levels of resistance than others. This trend was conserved when resistance based on IC50 values was compared (not shown).
FIG. 1.
PI resistance conferred by mutations in HIV-1 PR. Resistance to PIs was calculated for each PR mutant on the basis of IC90 values as fold increase with respect to wild-type (WT) NL-4.3 virus. Average values with standard deviation are shown for RTV (A), IDV (B), and SQV (C). Note that different scales are used for different inhibitors.
The same mutations in PR are usually observed in patients treated with RTV and in those treated with IDV, but while the accumulation of resistance mutations to RTV in vivo follows a conserved pathway, most often starting with the substitution at position 82 (33), evolution of resistance to IDV lacks such a landmark (9, 10). Analysis of resistance to IDV for the same series of mutant viruses showed that all single mutants were at least as sensitive as wild-type virus to IDV (Fig. 1B), as previously reported (9, 10). Only one of the three double mutants analyzed (mutant A71V-V82A) displayed a small but reproducible increase in resistance. Significant resistance could be observed only with the clone carrying four resistance mutations. Overall, the fold changes in susceptibility measured with IDV were lower than those obtained with RTV, indicating particular constraints to the development of resistance to IDV.
Resistance to SQV is characterized by the appearance of mutations G48V, L90M, and V82A, with the addition of mutations, like L10I, proposed to compensate for the structural modifications induced by primary changes. Different combinations of these mutations were frequently observed in SQV-treated patients except for mutations V82A and L90M, which have been previously reported as often being mutually exclusive. The reduced SQV susceptibility measured with different PR mutants (Fig. 1C) justifies previous observations made in treated patients and in in vitro virus cultures. The mutation G48V alone is sufficient for significant SQV resistance and was found in all combinations of mutations that conferred high-level resistance. This mutation is more frequently observed in patients in whom virus exposure to SQV is high due to more bioavailable formulations of the drug than in patients in whom SQV pressure is lower and in whom HIV-1 often displays L90M as a genetic marker of SQV resistance (6, 42, 46, 47, 50). In our analysis, L90M conferred a small but reproducible increase in SQV resistance. Viruses carrying both G48V and L90M mutations were markedly resistant, and if the L10I substitution was added, the fold increase in resistance was the highest observed in our study. Surprisingly, in the NL-4.3 background, the V82A substitution did not confer significant resistance to SQV, and its addition to different combinations of mutations did not augment the resistance level. Finally, in our system, mutants carrying both V82A and L90M displayed even higher sensitivity to SQV than wild-type virus. The resistance levels measured with our assay are somewhat lower than those obtained in systems based on multiple virus replication cycles using primary virus isolates (33). Nonetheless, the high reproducibility of our results allows accurate detection of small differences between individual clones. Both the resistance impact of the individual mutations described here and the finding that resistance to PIs increases with the number of PR mutations are in agreement with previous reports in which different techniques and target cells were used (4, 9, 10, 26, 33, 36, 39).
Impact of resistance mutations on viral infectivity.
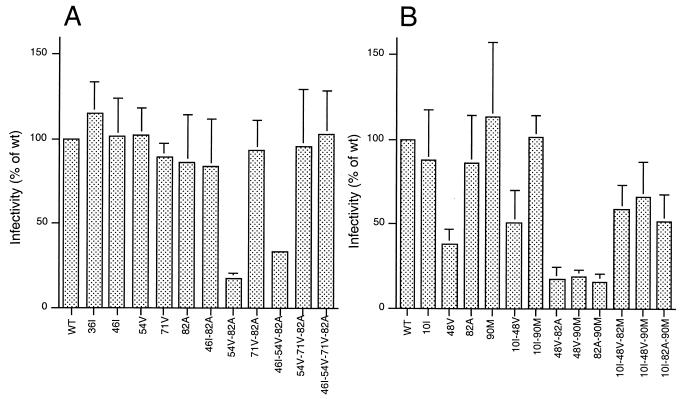
We and others have previously observed that viral variants carrying PI resistance mutations display a variable reduction in drug-free virus infectivity, often termed viral fitness (11, 29, 32, 43, 51). Here we determined the impact of single and combined mutations in the PR on drug-free virus infectivity, measured in a highly reproducible single-cycle infectivity assay (7, 8, 30, 51). For each of the mutants described above, we measured the infectivity of viral particles produced in drug-free cultures as a percentage of that of wild-type NL-4.3XCS virus. Most viral clones carrying single RTV or IDV resistance mutations were characterized by wild-type levels of infectivity (Fig. 2A). Interestingly, mutants with two mutations could be as infectious as wild-type virus (mutant A71V-V82A) or markedly impaired (mutant I54V-V82A). The same was true for the different combinations of three mutations. Wild-type infectivity was also observed for the clone carrying four mutations. Comparison of the infectivity of the different clones suggests that mutation I54V, which had no major impact on drug-free virus fitness when expressed alone, markedly decreased the infectivity of viral clones when other resistance mutations were present. This effect seemed to be neutralized to some extent by the addition of the substitution A71V.
FIG. 2.
Resistance-associated loss of viral infectivity. Drug-free infectivity of PR mutants of the RTV/IDV series (A) and SQV series (B) is shown as a percentage of wild-type (WT) NL-4.3 virus. Average values with standard deviation are illustrated.
The infectivities of viral clones carrying SQV resistance mutations are shown in Fig. 2B. The G48V mutation associated with high-level SQV resistance markedly affected viral infectivity whether expressed alone or in combination with other mutations. We previously described this phenomenon working on viral clones carrying patient-derived viral PR alleles (51). In line with the observations of other authors, the rare combination of V82A and L90M determined a marked reduction in viral infectivity, which could be only partially rescued by the compensatory mutation L10I. Addition of mutation L10I had a similar effect on the infectivity of clones G48V-V82A and G48V-L90M.
Selective advantage as a function of drug concentration: the fitness profiles.
To determine the range of drug concentrations for which each combination of mutations conferred a replicative advantage and the extent of this advantage, we traced fitness profiles for each of the mutants, representing the ratio of infectivity (in CPRG units) with respect to wild-type virus in variable drug concentrations (Fig. 3 and 4). From these comparisons, we could determine that mutation V82A conferred a small but reproducible replicative advantage in the presence of RTV concentrations ranging from 20 to 400 nM (Fig. 3A). For lower drug concentrations, this mutant displayed wild-type infectivity, while for higher drug concentrations, both wild-type and V82A mutant viruses were noninfectious. Again, none of the other single mutants analyzed displayed a replicative advantage with respect to wild-type virus. Caution should be used in interpreting minor differences in profiles at relatively high drug concentrations, at which wild-type virus infectivity is close to 0 CPRG units. Viruses carrying multiple mutations in the PR generally showed significant differences from wild-type virus (Fig. 3B; note the different scale with respect to Fig. 3A). A marked replicative advantage was observed for mutant A71V-V82A in the presence of drug concentrations of up to 1,000 nM. Mutant M46I-V82A replicated to a lower extent than V71A-V82A in low drug concentrations, but it was infectious even when produced in medium containing high concentration of RTV. The two mutants carrying a combination of three mutations in the PR gene had remarkably different phenotypes, showing that A71V was a more advantageous addition to I54V-V82A than was M46I, in terms of both extent of replication and range of inhibitor concentrations at which some infectivity was preserved. The mutant virus carrying four substitutions in the PR was characterized by a very high infectivity titer over a wide range of RTV concentrations, confirming that high-level resistance to PIs relies on the accumulation of several mutations.
Two main characteristics distinguished the curves describing resistance to IDV for the same series of mutant viruses (Fig. 3C and D): lower peaks, indicating that mutations conferred a smaller advantage with respect to RTV, and limitation of mutant virus infectivity at low IDV concentrations. Both of these findings reflect the difficulty that HIV-1 encounters in developing high-level resistance to IDV. Among the single mutants, only A71V surfaced over the wild-type infectivity threshold, while A71V-V82A seemed preferable to M46I-V82A in terms of both infectivity and range of IDV resistance, although limited to very low drug concentrations. As in the analysis performed with RTV, mutant I54V-A71V-V82A showed significant infectivity in the presence of IDV, and a marked advantage could be determined for the mutant virus carrying four mutations in the PR. The resistance-associated loss of viral fitness seems to be a limiting factor and may be responsible for the replicative disadvantage of virus M46I-I54V-V82A even in the presence of IDV.
Analysis of curves from SQV-resistant viruses (Fig. 4) clearly showed the advantage conferred by mutation L90M in low drug concentrations and the requirement for G48V to resist high concentrations of SQV. Less-expected features were the remarkable level of infectivity of mutant L10I-G48V-L90M in different SQV concentrations and the resistance of mutant L10I-G48V-V82A to a wide range of inhibitor concentrations, although for this virus the flat shape of the fitness profile indicated only a limited advantage. The phenotype of mutant L10I-G48V-L90M was even more impressive when compared to the profiles of mutants carrying combinations of two of the three mutations involved, which at the most showed a threefold advantage over wild-type virus. Finally, under no condition did mutants carrying both V82A and L90M present an advantage, reflecting the rare observation of such a combination in SQV resistance pathways both in patients and in virus culture.
Role of compensatory mutations in Gag.
We and others have previously reported that mutations in Gag cleavage sites can partially restore the viral infectivity of an HIV-1 PR mutant (16, 30, 52). In particular, we described an RTV-resistant patient-derived virus that, besides developing PR mutations I54V and V82A (one of the combinations of mutations analyzed here), displayed a Gag cleavage site amino acid change (A431V) associated with a significant rescue of drug-free infectivity. Here we measured the impact of this Gag cleavage site mutation on the reconstructed clone carrying the PR mutations I54V and V82A in different RTV concentrations (Fig. 5). Mutant I54V-V82A failed to display significant replicative advantage with respect to wild-type virus at any RTV concentration (Fig. 4B and 5A), despite the relatively frequent detection of this combination of mutations in treated patients (47). The addition of the A431V Gag mutation significantly increased the infectivity of this mutant virus over a wide range of RTV concentrations (Fig. 5A). Interestingly, in the presence of variable IDV concentrations (Fig. 5B), the presence of the Gag A431V substitution largely compensated for the resistance-associated loss of viral fitness of mutant I54V-V82A, producing a fitness profile similar to that of the wild type. Although this clone does not display a replicative advantage over the wild type in the presence of IDV, it may represent a viable intermediate for the subsequent accumulation of mutations in PR, in agreement with the reported high frequency of Gag cleavage site mutations in viruses from IDV-treated patients (52).
From these data, one can speculate that the emergence of combinations of mutations that markedly decrease PR function and HIV fitness in treated patients may depend on the presence of compensatory mutations in Gag.
DISCUSSION
When HIV-1 escapes PI therapy, viral replication under the selective pressure of these compounds leads to the emergence of amino acid substitutions that reduce inhibitor affinity for the mutated PR and thereby promote resistance. The selection of resistance mutations is a function of three main parameters: (i) the frequency of their introduction in the viral genome during replication; (ii) the concentration of inhibitor at the site of selection; and (iii) the impact of the mutations on the enzymatic performance of PR and therefore on the replicative fitness of the virus as a function of drug concentration. Here, we will only consider the last two of these parameters and will disregard the frequency of nucleotide misincorporation events during viral DNA synthesis by reverse transcriptase (40). Thus, we will assume that before introduction of therapy, the heterogeneous population of HIV-1 quasispecies that is characteristic of RNA viruses in vivo (15) potentially generates equal proportions of any variant bearing a single PI resistance mutation. All the amino acid substitutions analyzed here may be generated by a single nucleotide change in the corresponding codon.
Early stages of the selection for PI resistance.
During the process of selection for HIV drug resistance in vivo, each mutant quasispecies confronts different conditions of competitive growth relative to its parental wild-type counterpart and relative to other mutants, in tissue compartments where the concentration of inhibitor can vary. Resistance per se, as usually expressed by the IC50 and/or IC90 values for a virus, merely reflects the fraction of the viral replicative capacity that is reduced by a particular concentration of drug, regardless of the drug-free replicative capacity of the virus. On the other hand, drug-free infectivity, often termed viral fitness, does not take into account the selective advantage of a mutant in the presence of inhibitor. Here, we have devised a novel method of evaluation of the selective value of HIV-1 variants carrying mutations associated with resistance and viral escape to PIs, which is based on the assessment of the replicative advantage of the mutant relative to wild-type virus in the presence of different concentrations of inhibitors. Although fitness was not assessed using traditional growth competition experiments, the rapid single-cycle infectivity assay used here was reproducible and sensitive enough to allow fitness comparisons involving multiple HIV-1 variants and multiple drug concentrations.
Using the separate methods of assessment of resistance and infectivity, but more precisely using the novel fitness profiles method, we found that when present as single mutations, most of the amino acid substitutions that are part of the combinations known to mediate HIV-1 resistance to PIs do not confer any selective advantage to the virus, whatever the concentration of inhibitor. There are two notable exceptions: mutation V82A, which appears to confer a reproducible selective advantage in the presence of relatively low concentrations of RTV, and mutation L90M, which confers significant selective advantage in the presence of SQV. Interestingly, these two mutations are consistently found to be the first to emerge during in vivo HIV-1 escape to RTV and SQV therapy, respectively (6, 33). As for mutant G48V, for which the traditional evaluation of resistance by IC90 measurement revealed a significant level of resistance, it did not appear to be significantly advantaged even under strong SQV pressure in the absence of an accessory mutation such as L10I. This finding must relate to the fact that mutant G48V consistently displays a marked reduction in drug-free replicative capacity, which is likely to prevent its efficient selection in spite of significant resistance. Regarding resistance to IDV, the multiple genetic pathways leading to resistance to this drug in treated patients consistently involve mutations at position 46 and/or 82, with further accumulation of substitutions at position 71 and/or 54, among others (9, 52). Such disordered development of resistance is fully justified by the lack of selective advantage for any of the single mutants analyzed here (Fig. 3C).
Evolution toward higher levels of resistance.
The gradual accumulation of resistance mutations resulted in a notable increase in the extent and the range of the selective advantage displayed by the corresponding viruses. In the presence of RTV, we found that variants carrying combinations of A71V with V82A among other mutations were clearly the most efficient viruses, with the maximal advantage obtained for the mutant carrying all four of the tested substitutions in combination. This mutant, which appeared as fit as wild-type virus when tested in drug-free conditions, was considerably more efficient than wild-type virus and than viruses carrying fewer mutations over a wider range of both RTV and IDV concentrations. It has to be emphasized that although the same substitutions have been described in viruses escaping IDV or RTV therapy, resistance to RTV, whether expressed as traditional IC90 values or by the viral fitness profile, was always markedly more pronounced than resistance to IDV. In the presence of SQV, the most favorable combination associated mutations L10I, G48V, and L90M, a combination that is often observed in viruses escaping SQV in vivo. Similar to what was seen with RTV, this “optimal” combination markedly outperformed the other mutants both in the extent of the replicative performance and in the range of drug concentrations over which this advantage could be perceived. Overall, our findings explain why the selection for resistance to PIs is a gradual process, with only a marginal advantage conferred by the first mutations selected. Unlike mutations that mediate resistance to other compounds, such as lamivudine (3TC) and nonnucleosidic reverse transcriptase inhibitors, single mutations in PR are unable to lead to rapid outgrowth of resistant virus selected from quasispecies present even before therapy. Resistance to PIs can only be detected following the accumulation of two or more mutations, leading to a gradual increase in the range and extent of the replicative advantage. Because this process requires that HIV-1 continue to replicate in the presence of treatment, it is crucial that treatment with PIs achieve nearly complete suppression of viral replication in order to avoid the emergence of resistance.
Importance of drug levels for the development of resistance.
In individuals receiving antiretroviral therapy, the concentration of drug in peripheral blood can vary greatly over time during a single day, the result of discontinuous oral drug intake by patients and of more or less rapid drug inactivation by the natural clearance systems of the body (28, 34, 44). Drug concentrations are also presumed to vary widely from one anatomical compartment to another, with some compartments often considered possible sanctuaries for virus replication in spite of therapy (2, 13, 48). Virus variants bearing one particular mutation will therefore encounter different conditions of drug selective pressure within an infected individual. Upon analysis of the fitness profile of any given mutant, it is easy to determine which conditions of drug concentration will allow its emergence in competition with its parental wild-type counterpart. Therefore, even if these conditions are met only at certain periods or within particular anatomical compartments, one can envision that selection will follow successive fitness leaps depending on the virus phenotype and on its environment. In this respect, it is striking to observe that in some treated patients escaping a first line of therapy with a PI, resistance mutations do not appear to accumulate in spite of a high level of virus replication, as reflected by high amounts of virus in plasma (14, 25). In these patients, we propose that during peaks of high drug concentration or within compartments where drug concentration is high, the fitness of single PR mutants is insufficient to allow their initial selection, accounting for the subsequent accumulation of mutations. On the other hand, during troughs of low drug concentration or in compartments poorly permeated by the drug, the mutants are outgrown by wild-type virus, ensuring high viral load. It is remarkable that this phenomenon appears to have been described mostly in patients treated with IDV, a drug for which we clearly show here the fitness margin for selection of resistant mutants is strikingly narrower than for RTV and SQV. Overall, we believe that the examination of virus behavior using the fitness profile method will allow further understanding of the mechanisms of selection of HIV-1 drug resistance and may prove useful for the management of antiretroviral therapy in HIV-infected patients.
ACKNOWLEDGMENTS
This work was supported in part by a grant from the Agence Nationale de Recherche sur le SIDA (ANRS). F.M. was the recipient of a SIDACTION fellowship.
REFERENCES
- 1.Ashorn P, McQuade T J, Thaisrivongs S, Tomasselli A G, Tarpley W G, Moss B. An inhibitor of the protease blocks maturation of human and simian immunodeficiency viruses and spread of infection. Proc Natl Acad Sci USA. 1990;87:7472–7476. doi: 10.1073/pnas.87.19.7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aweeka F, Jayewardene A, Staprans S, Bellibas S E, Kearney B, Lizak P, Novakovic-Agopian T, Price R W. Failure to detect nelfinavir in the cerebrospinal fluid of HIV-1-infected patients with and without AIDS dementia complex. J Acquired Immune Defic Syndr Hum Retrovirol. 1999;20:39–43. doi: 10.1097/00042560-199901010-00006. [DOI] [PubMed] [Google Scholar]
- 3.Barrie K A, Perez E E, Lamers S L, Farmerie W G, Dunn B M, Sleasman J W, Goodenow M M. Natural variation in HIV-1 protease, Gag p7 and p6, and protease cleavage sites within Gag/Pol polyproteins: amino acid substitutions in the absence of protease inhibitors in mothers and children infected by human immunodeficiency virus type 1. Virology. 1996;219:407–416. doi: 10.1006/viro.1996.0266. [DOI] [PubMed] [Google Scholar]
- 4.Boden D, Markowitz M. Resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob Agents Chemother. 1998;42:2775–2783. doi: 10.1128/aac.42.11.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borman A M, Paulous S, Clavel F. Resistance of human immunodeficiency virus type 1 to protease inhibitors: selection of resistance mutations in the presence and absence of the drug. J Gen Virol. 1996;77:419–426. doi: 10.1099/0022-1317-77-3-419. [DOI] [PubMed] [Google Scholar]
- 6.Boucher C. Rational approaches to resistance: using saquinavir. AIDS. 1996;10(Suppl. 1):S15–S19. [PubMed] [Google Scholar]
- 7.Carron de la Carriere L, Paulous S, Clavel F, Mammano F. Effects of human immunodeficiency virus type 1 resistance to protease inhibitors on reverse transcriptase processing, activity, and drug sensitivity. J Virol. 1999;73:3455–3459. doi: 10.1128/jvi.73.4.3455-3459.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charneau P, Mirambeau G, Roux P, Paulous S, Buc H, Clavel F. HIV-1 reverse transcription: a termination step at the center of the genome. J Mol Biol. 1994;241:651–662. doi: 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
- 9.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E A. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Hobbins H L, Roth E, Shivaprakash M, Titus D L, Yang T, Teppler H, Squires K E, Deutsch P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 11.Croteau G, Doyon L, Thibeault D, McKercher G, Pilote L, Lamarre D. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J Virol. 1997;71:1089–1096. doi: 10.1128/jvi.71.2.1089-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debouck C, Metcalf B W. Human immunodeficiency virus protease—a target for AIDS therapy. Drug Dev Res. 1990;21:1–17. [Google Scholar]
- 13.Denissen J F, Grabowski B A, Johnson M K, Buko A M, Kempf D J, Thomas S B, Surber B W. Metabolism and disposition of the HIV-1 protease inhibitor ritonavir (ABT-538) in rats, dogs, and humans. Drug Metab Dispos. 1997;25:489–501. [PubMed] [Google Scholar]
- 14.Descamps D, Flandre P, Calvez V, Peytavin G, Meiffredy V, Collin G, Delaugerre C, Robert-Delmas S, Bazin B, Aboulker J P, Pialoux G, Raffi F, Brun-Vezinet F. Mechanisms of virologic failure in previously untreated HIV-infected patients from a trial of induction-maintenance therapy. Trilege (Agence Nationale de Recherches sur le SIDA 072) Study Team. JAMA. 2000;283:205–211. doi: 10.1001/jama.283.2.205. [DOI] [PubMed] [Google Scholar]
- 15.Domingo E, Escarmis C, Sevilla N, Baranowski E. Population dynamics in the evolution of RNA viruses. Adv Exp Med Biol. 1998;440:721–727. doi: 10.1007/978-1-4615-5331-1_93. [DOI] [PubMed] [Google Scholar]
- 16.Doyon L, Poulin F, Pilote L, Clouette C, Thibeault D, Croteau G, Lamarre D. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J Virol. 1996;70:3763–3769. doi: 10.1128/jvi.70.6.3763-3769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreyer G B, Metcalf B W, Tomaszek T A, Jr, Carr T J, Chandler A C d, Hyland L, Fakhoury S A, Magaard V W, Moore M L, Strickler J E, et al. Inhibition of human immunodeficiency virus 1 protease in vitro: rational design of substrate analogue inhibitors. Proc Natl Acad Sci USA. 1989;86:9752–9756. doi: 10.1073/pnas.86.24.9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erickson J, Kempf D. Structure-based design of symmetric inhibitors of HIV-1 protease. Arch Virol Suppl. 1994;9:19–29. doi: 10.1007/978-3-7091-9326-6_3. [DOI] [PubMed] [Google Scholar]
- 19.Erickson J, Neidhart D, VanDrie J, Kempf D, Wang X, Norbeck D, Plattner J, Rittenhouse J, Turon M, Wideburg N, Kohlbrenner W, Simmer R, Helfrick R, Paul D, Knigge M. Design, activity, and 2.8 A crystal structure of a C2 symmetric inhibitor complexed to HIV-1 protease. Science. 1990;249:527–533. doi: 10.1126/science.2200122. [DOI] [PubMed] [Google Scholar]
- 20.Erickson J W. The not-so-great escape. Nat Struct Biol. 1995;2:523–529. doi: 10.1038/nsb0795-523. [DOI] [PubMed] [Google Scholar]
- 21.Eustice D, Feldman P, Colberg-Poley A, Buckery R, Neubauer R. A sensitive method for the detection of β-galactosidase in transfected mammalian cells. BioTechniques. 1991;11:739–743. [PubMed] [Google Scholar]
- 22.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 23.Gulnick S V, Suvorov L I, Liu B, Yu B, Anderson B, Mitsuya H, Erickson J W. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry. 1995;34:9282–9287. doi: 10.1021/bi00029a002. [DOI] [PubMed] [Google Scholar]
- 24.Hammer S M, Squires K E, Hughes M D, Grimes J M, Demeter L M, Currier J S, Eron J J, Jr, Feinberg J E, Balfour H H, Jr, Deyton L R, Chodakewitz J A, Fischl M A. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 25.Havlir D V, Hellmann N S, Petropoulos C J, Whitcomb J M, Collier A C, Hirsch M S, Tebas P, Sommadossi J P, Richman D D. Drug susceptibility in HIV infection after viral rebound in patients receiving indinavir-containing regimens. JAMA. 2000;283:229–234. doi: 10.1001/jama.283.2.229. [DOI] [PubMed] [Google Scholar]
- 26.Hertogs K, de Bethune M P, Miller V, Ivens T, Schel P, Van Cauwenberge A, Van Den Eynde C, Van Gerwen V, Azijn H, Van Houtte M, Peeters F, Staszewski S, Conant M, Bloor S, Kemp S, Larder B, Pauwels R. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob Agents Chemother. 1998;42:269–276. doi: 10.1128/aac.42.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho D, Toyoshima T, Mo H, Kempf D, Norbeck D, Chen C, Wideburg N, Burt S, Erickson J, Singh M. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a C2-symmetric protease inhibitor. J Virol. 1994;68:2016–2020. doi: 10.1128/jvi.68.3.2016-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu A, Granneman G R, Bertz R J. Ritonavir: clinical pharmacokinetics and interactions with other anti-HIV agents. Clin Pharmacokinet. 1998;35:275–291. doi: 10.2165/00003088-199835040-00002. [DOI] [PubMed] [Google Scholar]
- 29.Kaplan A, Michael S, Wehbie R, Knigge M, Paul D, Everitt L, Kempf D, Norbeck D, Erickson J, Swanstrom R. Selection of multiple human immunodeficiency virus type 1 variants that encode viral proteases with decreased sensitivity to an inhibitor of the viral protease. Proc Natl Acad Sci USA. 1994;91:5597–5601. doi: 10.1073/pnas.91.12.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mammano F, Petit C, Clavel F. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J Virol. 1998;72:7632–7637. doi: 10.1128/jvi.72.9.7632-7637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markowitz M, Mo H, Kempf D, Norbeck D, Bhat T, Erickson J, Ho D. Selection and analysis of human immunodeficiency virus type 1 variants with increased resistance to ABT-538, a novel protease inhibitor. J Virol. 1995;69:701–706. doi: 10.1128/jvi.69.2.701-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Picado J, Savara A V, Sutton L, D'Aquila R T. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J Virol. 1999;73:3744–3752. doi: 10.1128/jvi.73.5.3744-3752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molla A, Kempf D, Korneyeva M, Gao Q, Shipper P, Mo H, Markowitz M, Vasavanonda S, Chernyavskyi T, Niu P, Lyons N, Hsu A, Granneman G, Ho D, Boucher C, Leonard J, Norbeck D. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 34.Moyle G J, Youle M, Higgs C, Monaghan J, Prince W, Chapman S, Clendeninn N, Nelson M R. Safety, pharmacokinetics, and antiretroviral activity of the potent, specific human immunodeficiency virus protease inhibitor nelfinavir: results of a phase I/II trial and extended follow-up in patients infected with human immunodeficiency virus. J Clin Pharmacol. 1998;38:736–743. doi: 10.1002/j.1552-4604.1998.tb04814.x. [DOI] [PubMed] [Google Scholar]
- 35.Nijhuis M, Schuurman R, de Jong D, Erickson J, Gustchina E, Albert J, Schipper P, Gulnik S, Boucher C A. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS. 1999;13:2349–2359. doi: 10.1097/00002030-199912030-00006. [DOI] [PubMed] [Google Scholar]
- 36.Parkin N T, Lie Y S, Hellmann N, Markowitz M, Bonhoeffer S, Ho D D, Petropoulos C J. Phenotypic changes in drug susceptibility associated with failure of human immunodeficiency virus type 1 (HIV-1) triple combination therapy. J Infect Dis. 1999;180:865–870. doi: 10.1086/314928. [DOI] [PubMed] [Google Scholar]
- 37.Patick A, Mo H, Markowitz M, Appelt K, Wu B, Musick L, Kalish V, Kaldor S, Reich S, Ho D, Webber S. Antiviral and resistance studies of AG1343, an orally bioavailable inhibitor of human immunodeficiency virus protease. Antimicrob Agents Chemother. 1996;40:292–297. doi: 10.1128/aac.40.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patick A, Rose R, Greytock J, Bechtold C, Hermsmeier M, Chen P, Barrish J, Zahler R, Colonno R, Lin P. Characterization of a human immunodeficiency virus type 1 variant with reduced sensitivity to an aminodiol protease inhibitor. J Virol. 1995;69:2148–2152. doi: 10.1128/jvi.69.4.2148-2152.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petropoulos C J, Parkin N T, Limoli K L, Lie Y S, Wrin T, Huang W, Tian H, Smith D, Winslow G A, Capon D J, Whitcomb J M. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44:920–928. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Preston B D, Poiesz B J, Loeb L A. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 41.Ridky T W, Kikonyogo A, Leis J, Gulnik S, Copeland T, Erickson J, Wlodawer A, Kurinov I, Harrison R W, Weber I T. Drug-resistant HIV-1 proteases identify enzyme residues important for substrate selection and catalytic rate. Biochemistry. 1998;37:13835–13845. doi: 10.1021/bi980612k. [DOI] [PubMed] [Google Scholar]
- 42.Roberts N A. Drug-resistance patterns of saquinavir and other HIV proteinase inhibitors. AIDS. 1995;9:S27–S32. [PubMed] [Google Scholar]
- 43.Rose R, Gong Y, Greytok J, Bechtold C, Terry B, Robinson B, Alam M, Colonno R, Lin P. Human immunodeficiency virus type 1 viral background plays a major role in development of resistance to protease inhibitors. Proc Natl Acad Sci USA. 1996;93:1648–1653. doi: 10.1073/pnas.93.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadler B M, Hanson C D, Chittick G E, Symonds W T, Roskell N S. Safety and pharmacokinetics of amprenavir (141W94), a human immunodeficiency virus (HIV) type 1 protease inhibitor, following oral administration of single doses to HIV-infected adults. Antimicrob Agents Chemother. 1999;43:1686–1692. doi: 10.1128/aac.43.7.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sardana V V, Schlabach A J, Graham P, Bush B L, Condra J H, Culberson J C, Gotlib L, Graham D J, Kohl N E, LaFemina R L, Schneider C L, Wolanski B S, Wolfgang J A, Emini E A. Human immunodeficiency virus type 1 protease inhibitors: evaluation of resistance engendered by amino-acid substitutions in the enzyme substrate binding site. Biochemistry. 1994;33:2004–2010. doi: 10.1021/bi00174a005. [DOI] [PubMed] [Google Scholar]
- 46.Schapiro J M, Winters M A, Lawrence J, Merigan T C. Clinical cross-resistance between the HIV-1 protease inhibitors saquinavir and indinavir and correlations with genotypic mutations. AIDS. 1999;13:359–365. doi: 10.1097/00002030-199902250-00008. [DOI] [PubMed] [Google Scholar]
- 47.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance. Int Antiviral News. 1997;5:129–137. [Google Scholar]
- 48.Shetty B V, Kosa M B, Khalil D A, Webber S. Preclinical pharmacokinetics and distribution to tissue of AG1343, an inhibitor of human immunodeficiency virus type 1 protease. Antimicrob Agents Chemother. 1996;40:110–114. doi: 10.1128/aac.40.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winslow D L, Stack S, King R, Scarnati H, Bincsik A, Otto M J. Limited sequence diversity in the HIV type 1 protease gene from clinical isolates and in vivo susceptibility to HIV protease inhibitors. AIDS Res Hum Retroviruses. 1995;11:107–113. doi: 10.1089/aid.1995.11.107. [DOI] [PubMed] [Google Scholar]
- 50.Winters M A, Schapiro J M, Lawrence J, Merigan T C. Human immunodeficiency virus type 1 protease genotypes and in vitro protease inhibitor susceptibilities of isolates from individuals who were switched to other protease inhibitors after long-term saquinavir treatment. J Virol. 1998;72:5303–5306. doi: 10.1128/jvi.72.6.5303-5306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zennou V, Mammano F, Paulous S, Mathez D, Clavel F. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J Virol. 1998;72:3300–3306. doi: 10.1128/jvi.72.4.3300-3306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Imamichi H, Imamichi T, Lane H, Falloon J, Vasudevachari M, Salzman N. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J Virol. 1997;71:6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]