Abstract
Human herpesvirus 8 (HHV-8; also known as Kaposi's sarcoma-associated herpesvirus) is the causative agent of Kaposi's sarcoma and certain B-cell lymphomas. In most infected cells, HHV-8 establishes a latent infection characterized by the expression of latency-associated nuclear antigen (LANA) encoded by open reading frame 73. Although unrelated by sequence, there are functional similarities between LANA and the EBNA-1 protein of Epstein-Barr virus. Both accumulate as subnuclear speckles and are required for maintenance of the viral episome. EBNA-1 also regulates viral gene expression and is required for cell immortalization, suggesting that LANA performs similar functions in the context of HHV-8 infection. Here we show that LANA forms stable dimers, or possibly higher-order multimers, and that this is mediated by a conserved region in the C terminus. By expressing a series of truncations, we show that both the N- and C-terminal regions localize to the nucleus, although only the C terminus accumulates as nuclear speckles characteristic of the intact protein. Lastly, we show that LANA can function as a potent transcriptional repressor when tethered to constitutively active promoters via a heterologous DNA-binding domain. Domains in both the N and C termini mediate repression. This suggests that one function of LANA is to suppress the expression of the viral lytic genes or cellular genes involved in the antiviral response.
Human herpesvirus 8 (HHV-8) is a recently identified gamma-2 herpesvirus (rhadinovirus) involved in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma (reviewed in references 4, 6 and 30). In most infected cells, HHV-8 establishes a latent infection in which the circularized genome resides as a multicopy episome and expresses a limited repertoire of viral genes. Foremost among these is latency-associated nuclear antigen (LANA; also referred to as LNA or LNA1), encoded by viral open reading frame 73 (ORF73) (17, 18, 33).
Although the role of LANA during HHV-8 latency and tumor formation is poorly understood, there are interesting functional parallels to the EBNA-1 protein of Epstein-Barr virus. LANA and EBNA-1 are unrelated by sequence; however, both accumulate in the infected cell nucleus as characteristic speckles, associate with host mitotic chromosomes, contain repetitive amino acid sequences, and are required for maintenance of the viral chromosome in the dividing host cell (3, 16, 19, 41, 42). EBNA-1 is a DNA-binding protein that initiates viral replication through recognition of multiple EBNA-1-binding sites within the viral origin of replication (34). In addition, EBNA-1 regulates expression of a number of viral promoters, including its own (23). EBNA-1 forms a stable dimer, and this is required for DNA binding, plasmid maintenance, and transcription regulation (2, 12).
Here we show that LANA is also capable of dimerization and that this is independent of other HHV-8 gene products or DNA. The dimerization domain lies between residues 884 and 1089 within the C terminus and is thus distinct from the putative leucine zipper motif located in the central repetitive domain (37). Both the N- and C-terminal regions of the protein contain nuclear localization sequences; however, only the C terminus accumulates as nuclear speckles similar to those observed with the full-length protein. Lastly, we show that LANA can function as a potent transcriptional repressor when tethered to constitutively active promoters using a heterologous DNA-binding domain. Independent repression domains were identified in both N- and C-terminal domains.
MATERIALS AND METHODS
Plasmids.
ORF73 was amplified from cloned HHV-8 genomic DNA (GenBank accession number AAB62657, phage sy3-7) by high-fidelity PCR as two segments using primers 5′-GCTCTAGAGCGCCCCCGGGAATGCGCCTG-3′ with 5′-CTCAACGTTTTGTTTCCATC-3′ and 5′-GATGGAAACAAAACGTTGAG-3′ with 5′-CAGGATCCTATGTCATTTCCTGTGGAGA-3′ (added restriction sites are underlined). Each fragment was subcloned into the pGEM-T Easy vector (Promega, Inc.), released by digestion with XbaI and Psp1406I or Psp1406I and BamHI, and ligated into the mammalian expression vectors pCGN and pCGT using the unique XbaI and BamHI sites. The resulting constructs, pCGNLANAFL and pCGTLANAFL, respectively, express full-length LANA bearing N-terminal influenza virus hemagglutinin (HA) or bacteriophage T7 gene 10 (T7) epitope tags, respectively (44, 47).
The isolated N- and C-terminal domains were also generated by PCR using the following primer combinations for the N-terminal region (residues 2 to 329), 5′-GCTCTAGAGCGCCCCCGGGAATGCGCCTG-3′ with 5′-CGGATCCAAAGCTTATTGTCATTGTCATCCTTGTC-3′, and for the C-terminal region (residues 863 to 1089), 5′-GTCTAGAAAGCTTCCCATAATCTTGCACGGGTCG-3′ with 5′-CAGGATCCTATGTCATTTCCTGTGGAGA-3′ (added XbaI, BamHI, and HindIII restriction sites are underlined). The internal deletion (LANANC) was constructed by releasing the N-terminal coding fragment using XbaI and HindIII and the C-terminal encoding fragment using HindIII and BamHI and then simultaneously ligating both fragments into pCGN or pCGT.
The Gal4 fusion proteins were constructed by cloning appropriate XbaI-BamHI fragments into the expression vector pCGNGal(1–94), which fuses the first 94 residues of the yeast Gal4 DNA-binding domain to the N terminus of the LANA fragments. Luciferase reporter constructs p5xGal4SV40-luc and p5xGal4tk-luc were a generous gift of Milo Vassallo and Naoko Tanese. Green fluorescent protein (GFP)-LANA fusions were constructed by subcloning appropriate XbaI-BamHI fragments into a modified polylinker that added a unique EcoRI site immediately upstream of the XbaI site. Each ORF was then released as an EcoRI-BamHI fragment and cloned into the pEGFP-C2 vector (Clontech). GFP-HCF-1C expresses residues 1756 to 2035 of human HCF-1, which include the C-terminal nuclear localization signal.
Sequence analysis.
Alignments of LANA-like protein sequences were compiled using ZEGA and CLUSTALW (Molecular Modeling and Bioinformatics Group, Skirball Institute). Secondary-structure predictions were performed for each viral sequence using the PHD algorithm (36).
Transfections, coimmunoprecipitations, and immunoblotting.
293T cells were transfected with lipofectamine (Life Technologies) using 20 μl of lipid reagent per 6-cm dish. Whole cell/nuclear extracts were prepared after 40 h by lysing cells in high-salt lysis buffer (420 mM KCl, 10 mM Tris-HCl [pH 7.9], 5% glycerol, 0.25% Nonidet P-40, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 0.2 mM sodium vanadate, 50 μM sodium fluoride, 1 mM dithiothreitol). Nuclei were extracted at 4°C for 30 min and removed by centrifugation. For immunoprecipitations, 100 μl of extract was incubated with 2.5 μl of anti-HA (αHA) antibody 12CA5-coupled protein G-agarose beads at 4°C for 1 h. The beads were washed four times in 1 ml of wash buffer (200 mM KCl, 10 mM Tris-HCl [pH 7.9], 5% glycerol, 0.5 mM EDTA), before separation by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Immunoblotting was performed by semidry transfer and detected by enhanced chemiluminescence (SuperSignal; Pierce). The αHA antibody and αT7 antibody (Novagen) were diluted 1:5,000 and 1:10,000, respectively.
Epifluorescence microscopy.
Cos-1 cells were plated into 35-mm dishes containing a glass coverslip-covered 15-mm cutout (MatTek Corporation) and transfected the next day with 500 ng of GFP expression plasmid. Cells were examined 24 h posttransfection with a Zeiss Axiovert epifluorescence microscope equipped with a Princeton Instruments cooled charge-coupled device camera and MetaMorph digital imaging software (Universal Imaging).
Luciferase reporter assays.
For transcriptional repression assays, 293T cells were transfected by electroporation (106 cells/assay) using a Bio-Rad Genepulser with capacitance extendor set at 0.22 kV and 950 μF, and extracts were prepared after 30 h by adding 300 μl of LCCLR buffer (Promega, Inc.) to each 6-cm dish. For the luciferase activity assay, 50 μl of cell extract and 300 μl of reaction buffer (25 mM glycineglycine [pH 7.8], 15 mM MgSO4, 1 mM ATP [pH 7.0], 0.1 mg of bovine serum albumin per ml, and 1 mM dithiothreitol) were mixed, added to 1 mM D-luciferin substrate (Analytical Luminescence Laboratory), and immediately assayed using an LB9507 luminometer (EG&G Berthold, Inc.). Values represent the means of three independent transfections, and error bars indicate standard deviation from the mean.
RESULTS
LANA can be divided into three regions.
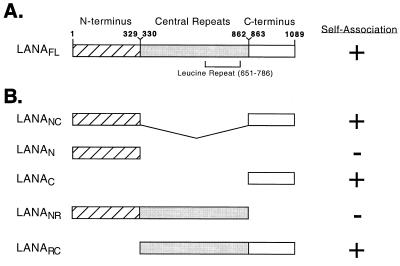
LANA can be divided into three general regions based on amino acid composition (Fig. 1A): a 329-amino-acid N-terminal portion, a 534-amino-acid highly repetitive central domain, and a 227-amino-acid C-terminal domain. The N terminus is rich in serine/threonine, proline, and basic residues. The large central repetitive region is highly enriched for four residues, glutamic acid, aspartic acid, glutamine, and leucine, arranged as imperfect reiterations of several different repeat units, including 19 copies of a leucine heptad repeat (LEEQEQEL) predicted to form a coiled-coil structure or leucine zipper (37). The calculated pI of the entire central region is 2.97, reflecting the extremely acidic composition (51% Glu or Asp). The C terminus is rich in charged and bulky hydrophobic residues.
FIG. 1.
Schematic representation of the primary structure of recombinant LANA and derivatives. (A) LANA can be divided into three regions by virtue of amino acid composition: the 329-amino-acid N terminus (residues 1 to 329); a variable central region (residues 330 to 862) consisting of multiple repeat elements rich in leucine residues as well as the charged polar amino acids glutamine, glutamic acid, and aspartic acid; and the 227-amino-acid C terminus (residues 863 to 1089). Amino acid numbers follow the sequence determined by Neipel et al. (GenBank accession number AAB62657) (29). (B) Fragments used in this study. The ability to self-associate is indicated with a plus.
LANA associates with itself in the absence of viral DNA or other viral gene products.
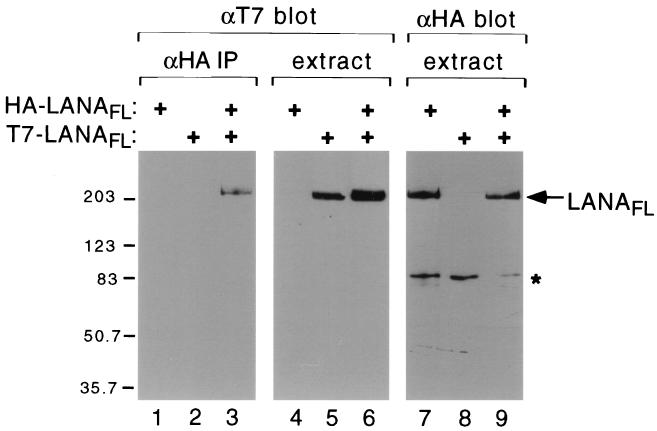
To determine whether LANA is capable of dimerization, we transiently expressed a mixture of HA and T7 epitope-tagged versions of the full-length protein in transiently transfected 293T cells (Fig. 2). Extracts were prepared and immunoprecipitated using αHA antibody-coupled agarose beads. The resulting immunocomplexes were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted using an αT7 monoclonal antibody (Fig. 2, lanes 1 to 3). T7-tagged LANA was recovered only in the presence of HA-tagged LANA (compare lanes 2 and 3), suggesting that HA- and T7-tagged LANA polypeptides exist within a complex. Direct immunoblotting of the starting extracts with the αT7 antibody (lanes 4 to 6) or αHA antibody (lanes 7 to 9) showed that similar amounts of recombinant protein (either T7- or HA-LANAFL) were expressed in each sample. Consistent with previous studies, recombinant LANA migrated with an apparent mass of greater than 200 kDa, perhaps reflecting the unusual amino acid composition or presence of extensive posttranslational modifications (18, 19, 33, 48). Association of HA- and T7-tagged polypeptides was maintained in the presence of the DNA intercalator ethidium bromide (400 μg/ml) (22), indicating that it is not mediated by fragments of DNA present in the extract (data not shown).
FIG. 2.
LANA interacts with itself in solution. Human 293T cells were transfected with expression plasmids (2 μg each) encoding HA-tagged LANAFL (lanes 1, 3, 4, 6, 7, and 9) and T7-tagged LANAFL (lanes 2, 3, 5, 6, 8, and 9). After 40 h, whole-cell extracts were prepared and immunoprecipitated (IP) using an αHA monoclonal antibody (12CA5) coupled to agarose beads. Immunoprecipitates were resolved on an SDS–8% polyacrylamide gel, transferred to nitrocellulose, and blotted with the αT7.tag monoclonal antibody (Invitrogen). Approximately 1/20 of the starting extracts were electrophoresed in parallel and immunoblotted with the αT7 or αHA antibodies. Nonspecific cross-reacting polypeptides are indicated with an asterisk. Sizes are shown in kilodaltons.
Self-association is mediated by the C terminus of LANA.
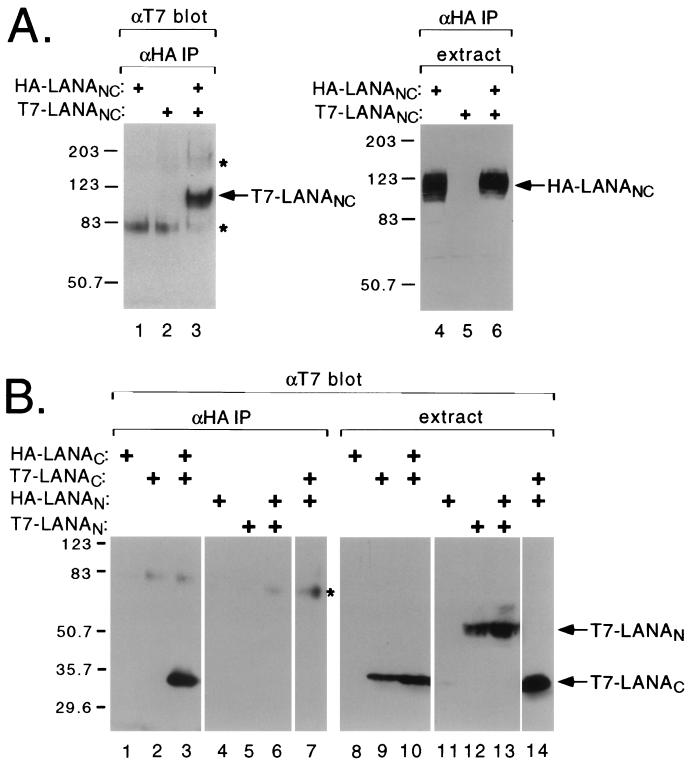
To map the sequences necessary for self-association, we generated an internal deletion variant lacking the central repetitive domain (LANANC, shown schematically in Fig. 1B) and examined self-association by coimmunoprecipitation (Fig. 3A). HA-epitope tagged LANANC migrates as a cluster of bands of 120 to 130 kDa (Fig. 3A, lanes 4 and 6). Following immunoprecipitation with the αHA antibody, T7-LANANC was recovered when coexpressed with HA-LANANC (lane 3). This result shows that the central domain is not required for self-association.
FIG. 3.
LANA associates with itself through the C-terminal domain. (A) The central repetitive domain is dispensable for self-association. Human 293T cells were transfected with expression plasmids (2 μg each) encoding HA-tagged LANANC (lanes 1, 3, 4, and 6) and/or T7-tagged LANANC (lanes 2, 3, 5, and 6), and coassociation was monitored by immunoprecipitation (IP) using αHA antibody beads followed by SDS–7% PAGE and immunoblotting with αT7 antibody (lanes 1 to 3). One-twentieth of the starting extract was resolved in parallel and immunoblotted with the αHA antibody (lanes 4 and 5). Nonspecific cross-reacting polypeptides are indicated with an asterisk. (B) The C-terminal domain interacts with itself. HA- and T7-tagged polypeptides corresponding to the N- and C-terminal regions of LANA were assayed for coassociation as described above. 293T cells were transfected with plasmids encoding HA-LANAC (lanes 1, 3, 8, and 9), T7-LANAC (lanes 2, 3, 7, 9, 10, and 14), HA-LANAN (lanes 4, 6, 7, 11, 13, and 14), and T7-LANAN (lanes 5, 6, 12, and 13). Extracts were immunoprecipitated using αHA antibody beads and probed with the αT7 antibody (lanes 1 to 7) or probed directly with the αT7 antibody (lanes 8 to 14). Nonspecific cross-reacting polypeptides are indicated with an asterisk. Sizes are shown in kilodaltons.
Next we asked whether individual N- or C-terminal subdomains were capable of interaction (Fig. 3B). The T7-LANAC polypeptide was efficiently coprecipitated when coexpressed with HA-LANAC (Fig. 3B, lane 3) but not with HA-LANAN (lane 7). Expression and recovery of the HA-tagged fusion proteins was confirmed by immunoblotting (not shown). This suggests that self-association is mediated through the C-terminal domain alone and does not involve an “end-to-end” interaction. T7-LANAN could not be recovered with HA-LANAN (Fig. 3B, lane 6), indicating that the association between C-terminal domains is likely to be the predominant interaction.
We were unable to detect expression of the isolated central region (data not shown); however, we were able to express T7-tagged fusions containing the N terminus linked to the central region (LANANR) or the central region linked to the C terminus (LANARC). Consistent with the previous results, only the LANARC fusion could be coimmunoprecipitated using LANAC or LANAFL (summarized in Fig. 1B).
Sequences required for self-association are conserved in LANA-like proteins.
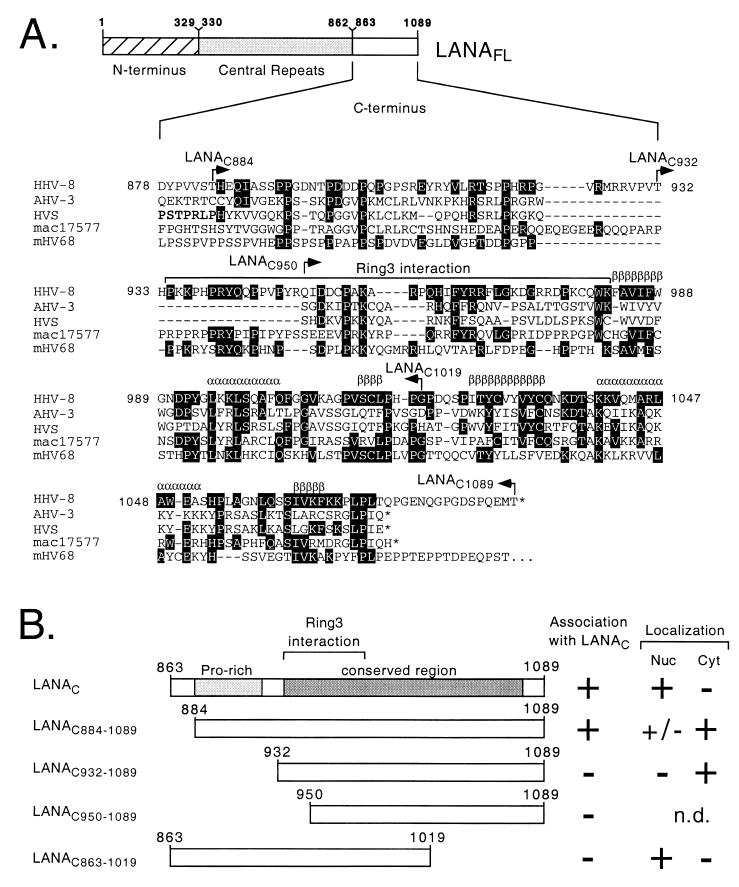
Figure 4A compares the amino acid sequence of the C terminus of HHV-8 LANA with the equivalent regions of LANA-like proteins encoded by four other rhadinoviruses: Ateline herpesvirus-3 (11), herpesvirus saimiri (1), macaque rhadinovirus 17577 (38), and murine herpesvirus 68 (45). Within this region, the area of greatest conservation corresponds to residues 934 to 1072 in the HHV-8 LANA sequence, with a more limited conservation (notably three invariant prolines residues) between residues 885 and 922. To localize sequences required for self-association more precisely, we generated four additional truncations within the C terminus and assayed these for coimmunoprecipitation with the full C terminus (LANAC). These results are summarized in Fig. 4B. The truncation preserving the entire conserved region (LANAC884–1089) was coprecipitated with similar efficiency to LANAC, while further N-terminal truncation (LANAC932–1089 and LANAC950–1089) abolished the association entirely. A relatively large deletion from the C-terminal end of the fragment (LANAC803–1019) also prevented association. Thus, almost the entire C terminus of LANA (residues 884 to 1089) is required for self-association and encompasses the majority of residues conserved between rhadinovirus family members.
FIG. 4.
Mapping of the self-association domain within the C terminus. (A) Alignment of the C-terminal domains from LANA-like proteins encoded by HHV-8, Ateline herpesvirus-3 (AHV-3), herpesvirus saimiri (HVS), macaque rhadinovirus 17577 (mac17577), and murine herpesvirus 68 (mHV68). Residues identical to the HHV-8 sequence are shown in black, and introduced gaps are represented by dashes. An asterisk signifies the carboxy terminus of the polypeptide. Structural predictions (shown above the HHV-8 sequence) suggest a conserved arrangement of interspersed β-sheets (β) and α-helices (α). The minimal RING3-binding peptide (residues 934 to 982) defined by Platt et al. (32) is indicated with a bracket. (B) HA-tagged LANAC863–1089 was coexpressed in Cos-1 cells with a the following T7-tagged C-terminal derivatives: LANAC863–1089, LANAC884–1089, LANAC932–1089, LANAC950–1089, and LANAC863–1019. Plus signs indicate a positive interaction with the HA-tagged LANAC fragment or specific localization to the nucleus (Nuc) or cytoplasm (Cyt). n.d., not determined.
Both the N and C termini localize to the nucleus.
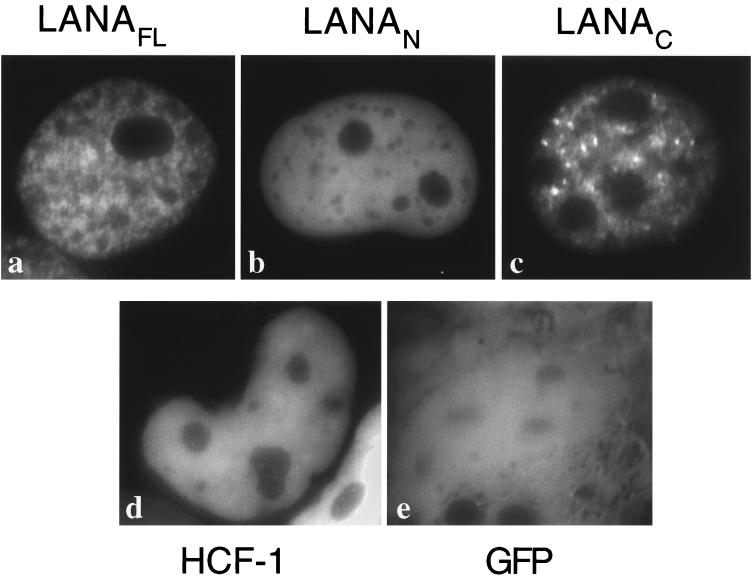
Immunolocalization studies in latently infected cell lines, tumor biopsies, and transfected tissue culture cells have shown that LANA accumulates in discrete spots or speckles within the interphase nucleus (16, 19, 33, 41, 42). To map the regions of LANA required for the punctate nuclear distribution, selected LANA polypeptides were fused to GFP, and the subcellular localization was determined in live Cos-1 cells using epifluorescence microscopy (Fig. 5). Fusions containing full-length LANA (GFP-LANAFL, Fig. 5A) or the C-terminal domain (GFP-LANAC, Fig. 5C) accumulated in the majority of transfected nuclei as discrete speckles, strongly reminiscent of the staining pattern described for HHV-8-infected cells. Interestingly, the N terminus (GFP-LANAN, Fig. 5B) also localized to the nucleus, but with a more uniform distribution similar to that shown by HCF-1 (GFP-HCF-1, Fig. 5D), a cellular transcription factor that contains a bipartite nuclear localization signal (21, 46). Expressed on its own, the GFP moiety was distributed evenly throughout both cytoplasm and nucleoplasm (Fig. 5E), indicating that the nuclear accumulation of all three LANA derivatives was specific.
FIG. 5.
N- and C-terminal domains of LANA contain nuclear localization signals. Cos-1 cells were transiently transfected with expression plasmids encoding (a) GFP-LANAFL, (b) GFP-LANAN, (c) GFP-LANAC, (d) GFP-HCF-1, and (e) GFP alone, and GFP localization was visualized in living cells by epifluorescence microscopy.
To further localize the determinants for nuclear localization and speckling, we assayed the truncations of the C-terminal region described above. The results are summarized in Fig. 4B. Removal of the first 22 residues from the N terminus (GFP-LANAC884–1089) resulted in a predominantly cytoplasmic distribution with a weak signal in the nuclei of some but not all cells. Nuclear localization was lost entirely by further truncation. In contrast, deletion from residue 1020 to the end of the polypeptide (GFP-LANAC863–1019) did not prevent nuclear localization, although the GFP fusion protein accumulated in large granules that were clearly distinct from the more numerous speckles seen with the intact C terminus. These results show that residues N-terminal to the minimal self-association fragment (residues 884 to 1089) are required for nuclear localization. Deletion of residues 1020 to 1089 prevents formation of nuclear speckles and also disrupts self-association, suggesting that the two functions might be related.
LANA functions as a transcriptional repressor.
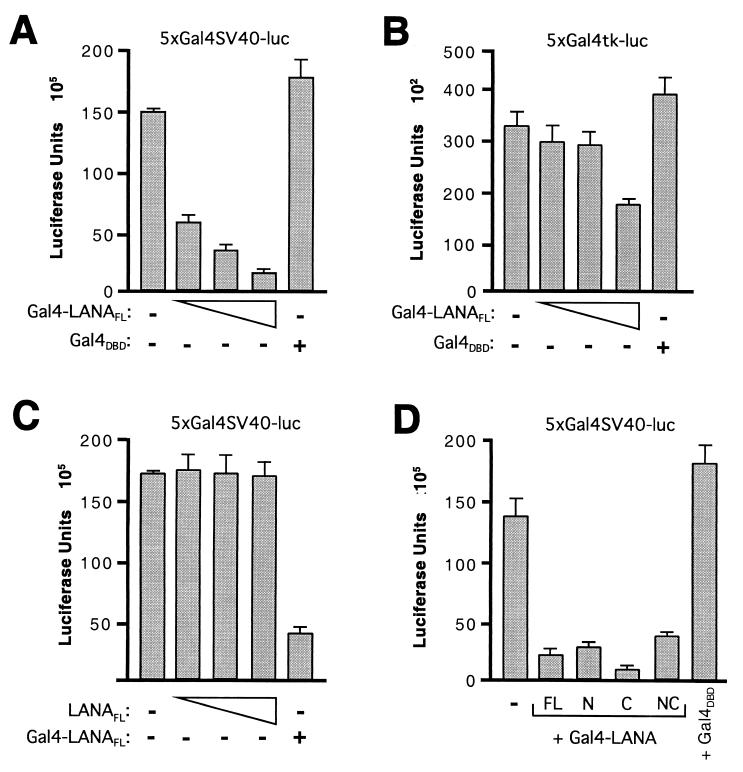
EBNA-1 acts as both a transcriptional activator and repressor (reviewed in reference 23). To determine whether LANA is capable of modulating gene expression, we fused LANA to the Gal4 DNA-binding domain (Gal4-LANAFL) and assayed two reporter genes in which five Gal4-binding sites had been placed upstream of either the simian virus 40 (SV40) early promoter (5xGal4SV40-luc, Fig. 6A) or the herpes simplex virus thymidine kinase promoter (5xGal4tk-luc, Fig. 6B). The constitutive expression of both reporters was repressed in a dose-dependent manner by cotransfection with Gal4-LANAFL. This is specific to the fusion protein, as no repression was observed using the Gal4 DNA-binding domain alone (Gal4DBD). The SV40 promoter was significantly more sensitive to LANA-mediated repression than the thymidine kinase promoter. For example, 50 ng of Gal4-LANAFL reduced SV40 promoter activity by 60%, compared to a 10% reduction for the thymidine kinase promoter. The reasons for this interesting difference are unclear but may be related to the much higher activity of the SV40 promoter. Although constitutively active, both promoters could be further stimulated using activator fusions such as Gal4-VP16 or Gal4-LZIP (data not shown).
FIG. 6.
LANA represses transcription when tethered to a promoter by a heterologous DNA-binding domain. 293T cells were cotransfected with (A) p5xGal4SV40-luc (25 ng) or (B) p5xGal4tk-luc (250 ng) luciferase reporter plasmids together with expression vectors encoding Gal4DBD alone (500 ng) or increasing amounts of Gal4-LANAFL (50, 100, and 500 ng). (C) The p5xGal4SV40-luc reporter (25 ng) was cotransfected with increasing amounts of T7-tagged LANAFL (50, 100, and 500 ng) or Gal4-LANAFL (500 ng) expression vectors. (D) The p5xGal4SV40-luc reporter (25 ng) was cotransfected with vectors (500 ng) encoding Gal4-LANAFL, Gal4-LANAN, Gal4-LANAC, Gal4-LANANC, and Gal4DBD alone. Values are the means of three independent assays, and the error bars indicate standard deviation.
In addition, we measured the activity of the SV40 promoter in the presence of full-length LANA lacking the Gal4 DNA-binding domain (LANAFL, Fig. 6C). Increasing amounts of LANAFL had no effect on promoter activity, showing that LANA must be bound to the target promoter to mediate repression. Immunoblotting confirmed that LANAFL and Gal4-LANAFL were expressed at comparable levels (data not shown).
Both the N and C termini contain repression domains.
To map the regions of LANA that are required for repression more precisely, we fused the N- and C-terminal regions of LANA to the Gal4 DNA-binding domain and monitored expression of the SV40-derived reporter (Fig. 6D). Both fusion proteins were able to repress transcription with similar efficiency to Gal4-LANAFL or a derivative lacking the central repetitive domain (Gal4-LANANC). Because protein expression was not normalized in this experiment, we are unable to draw meaningful conclusions from the small differences in repression activity between the various fusion proteins. As with full-length LANA, repression by the isolated N- and C-terminal domains was only observed when tethered to the promoter via the Gal4 DNA-binding domain (data not shown). In summary, these results show that LANA contains at least two repression domains, one located in the N terminus and one in the C terminus.
DISCUSSION
This study presents a structure-function analysis of LANA, the major latency-associated nuclear antigen of HHV-8-infected cells. Using epitope-tagged proteins, we show that LANA is capable of multimerization and that this occurs in the absence of other HHV-8 gene products. The self-association function was mapped to a 206-amino-acid region (residues 884 to 1089) within the C terminus. This corresponds to region that is conserved in other LANA-like proteins, suggesting that self-association is a general property of these proteins. Additional studies are needed to determine whether LANA forms dimers, as with EBNA-1 (2), or higher-order multimers, as described for a number of viral replication proteins, including SV40 large T antigen (27, 31). While it is not known whether LANA is able to bind DNA directly, it would be consistent with its role in maintenance of the viral latent genome (3) and association with both viral and cellular chromosomes (3, 7, 42). Dimerization is a common feature of DNA-binding proteins and may be critical for the DNA-binding activity of LANA.
C terminus of LANA specifies punctate nuclear distribution.
Using GFP-tagged polypeptides, we have found that both the N- and C-terminal regions of LANA localize to the nucleus. While the N terminus gives a diffuse pattern in interphase nuclei typical of many transcription factors, the C terminus accumulates as discrete nuclear speckles reminiscent of the punctate pattern shown by full-length LANA (7, 16, 19, 42). The primary sequence of the N terminus contains several short stretches of basic residues which might function as simple nuclear localization sequences (9, 25), and similar sequences occur in the N termini of all the known LANA-like proteins (1, 11, 24, 38, 45). Using truncations within the C-terminal region, we show that residues amino-terminal to the minimal self-association domain are required for nuclear localization. Deletion of residues 1020 to 1089 at the C terminus does not affect nuclear localization but disrupts speckle formation and self-association. While this implies a causal relationship, additional mutations will need to be tested.
Why LANA accumulates at specific locations within the nucleus is not known. Similar behavior has been described for a variety of viral regulatory proteins, and these structures may act as viral replication centers (28, 40). LANA does not associate with ND10 bodies, which contain PML and Sp100 proteins, and does not colocalize with EBNA-1 in spite of the functional similarities outlined above (5, 42). The absence of a recognizable nuclear localization signal in the C terminus suggests that it is targeted through protein-protein interactions. One candidate is RING3, a nuclear protein related to the female sterile homeotic protein of Drosophila melanogaster (32). RING3 contains two bromodomains, which may be involved in association with chromatin, and an extraterminal (ET) domain of unknown function (35). The C terminus of LANA interacts directly with the ET domain, and in vivo this results in phosphorylation of the LANA C terminus. RING3 does not appear to be a kinase itself but is thought to recruit another uncharacterized activity (32, 35). Our deletion mapping suggests that interaction with RING3 is not sufficient for speckle formation or LANA multimerization. By immunofluorescence, RING3 is distributed evenly throughout the interphase nucleus (8), suggesting that other factors specify LANA's pattern distribution. Multimerization requires a larger region (residues 884 to 1089) than is needed to recruit RING3 (corresponds to residues 934 to 982 in Fig. 4A) (32), again implying that these are separate functions.
LANA represses transcription.
We have shown that LANA acts as a transcriptional repressor utilizing independent repression domains located in both the N- and C-terminal regions. The fact that LANA can suppress two heterologous promoters suggests that it acts on a generalized target such as chromatin structure or some aspect of the general transcription machinery (reviewed in reference 20). Alternatively, one or both repression domains may direct the reporter gene to a transcriptionally silent compartment within the nucleus, such as heterochromatin (42). This might provide an efficient mechanism for silencing the entire repertoire of lytic cycle genes, provided the handful of latency-associated promoters manage to escape repression.
Repression by LANA may also play an important role in preventing apoptosis of the latently infected cell. Friborg and colleagues have shown that LANA interacts with the cellular transcription factor p53, preventing transcriptional activation of genes containing p53 binding sites (13). p53 plays a unique role in protecting cells against DNA damage and in triggering programmed cell death in response to viral infection. Loss of p53 function is also an important contributor to cellular transformation (10, 14). It is perhaps not surprising that HHV-8, like many other DNA tumor viruses, has evolved a mechanism to interfere with p53 function (15, 39, 43). LANA binds to directly to p53; however, this does not prevent DNA binding or alter p53 stability (13). Instead, LANA may selectively repress p53-responsive promoters through a “piggybacking” mechanism involving the repression domains identified in this study. A similar mechanism is employed by the adenovirus E1B 55K protein, which binds to the N-terminal activation domain of p53 and inhibits transcription via a separate repression domain (reviewed in reference 26).
By analogy to EBNA-1, we suspect that LANA performs multiple functions required for latent infection. The list may include regulation of viral gene expression, replication of the viral genome, tethering to the cellular chromosome, and suppression of the antiviral response (3, 13). Because multimerization of LANA is likely to be important for several or all of these functions, this virus-specific interaction provides an attractive therapeutic target.
ACKNOWLEDGMENTS
We offer special thanks to Shaun Walters and Ellen Choy for initiating this project and to Naoko Tanese for many helpful discussions, reagents, and continued interest in the project. Mark Philips graciously provided access to the epifluorescence microscope and image capturing facilities. Michael Garabedian and Naoko Tanese offered many useful comments on the manuscript, and Milo Vassallo, Andy Shih, Alvin Friedman-Kien, and Frank Neipel provided essential reagents.
This work was supported by funds from the Center for AIDS Research, Kaplan Comprehensive Cancer Center, and Sackler Institute for Graduate Studies. D.R.S. was supported by NIH training grant 5T32 GM 07308 from the NIGMS.
REFERENCES
- 1.Albrecht J C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, et al. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambinder R F, Mullen M A, Chang Y N, Hayward G S, Hayward S D. Functional domains of Epstein-Barr virus nuclear antigen EBNA-1. J Virol. 1991;65:1466–1478. doi: 10.1128/jvi.65.3.1466-1478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 4.Boshoff C, Weiss R A. Kaposi's sarcoma-associated herpesvirus. Adv Cancer Res. 1998;75:57–86. doi: 10.1016/s0065-230x(08)60739-3. [DOI] [PubMed] [Google Scholar]
- 5.Callahan J, Pai S, Cotter M, Robertson E S. Distinct patterns of viral antigen expression in Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus coinfected body-cavity-based lymphoma cell lines: potential switches in latent gene expression due to coinfection. Virology. 1999;262:18–30. doi: 10.1006/viro.1999.9876. [DOI] [PubMed] [Google Scholar]
- 6.Cesarman E, Knowles D M. The role of Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in lymphoproliferative diseases. Semin Cancer Biol. 1999;9:165–174. doi: 10.1006/scbi.1998.0118. [DOI] [PubMed] [Google Scholar]
- 7.Cotter M A, 2nd, Robertson E S. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology. 1999;264:254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- 8.Denis G V, Green M R. A novel, mitogen-activated nuclear kinase is related to a Drosophila developmental regulator. Genes Dev. 1996;10:261–271. doi: 10.1101/gad.10.3.261. [DOI] [PubMed] [Google Scholar]
- 9.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 10.el-Deiry W S. Regulation of p53 downstream genes. Semin Cancer Biol. 1998;8:345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 11.Ensser A, Pflanz R, Fleckenstein B. Primary structure of the alcelaphine herpesvirus 1 genome. J Virol. 1997;71:6517–6525. doi: 10.1128/jvi.71.9.6517-6525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frappier L, O'Donnell M. Overproduction, purification, and characterization of EBNA1, the origin binding protein of Epstein-Barr virus. J Biol Chem. 1991;266:7819–7826. [PubMed] [Google Scholar]
- 13.Friborg J, Jr, Kong W, Hottiger M O, Nabel G J. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 14.Gottlieb T M, Oren M. p53 and apoptosis. Semin Cancer Biol. 1998;8:359–368. doi: 10.1006/scbi.1998.0098. [DOI] [PubMed] [Google Scholar]
- 15.Kannabiran C, Morris G F, Mathews M B. Dual action of the adenovirus E1A 243R oncoprotein on the human proliferating cell nuclear antigen promoter: repression of transcriptional activation by p53. Oncogene. 1999;18:7825–7833. doi: 10.1038/sj.onc.1203294. [DOI] [PubMed] [Google Scholar]
- 16.Katano H, Sato Y, Kurata T, Mori S, Sata T. High expression of HHV-8-encoded ORF73 protein in spindle-shaped cells of Kaposi's sarcoma. Am J Pathol. 1999;155:47–52. doi: 10.1016/S0002-9440(10)65097-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J Clin Investig. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kellam P, Boshoff C, Whitby D, Matthews S, Weiss R A, Talbot S J. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J Hum Virol. 1997;1:19–29. [PubMed] [Google Scholar]
- 19.Kellam P, Bourboulia D, Dupin N, Shotton C, Fisher C, Talbot S, Boshoff C, Weiss R A. Characterization of monoclonal antibodies raised against the latent nuclear antigen of human herpesvirus 8. J Virol. 1999;73:5149–5155. doi: 10.1128/jvi.73.6.5149-5155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knoepfler P S, Eisenman R N. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 21.La Boissiere S, Hughes T, O'Hare P. HCF-dependent nuclear import of VP16. EMBO J. 1999;18:480–489. doi: 10.1093/emboj/18.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai J S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leight E R, Sugden B. EBNA-1: a protein pivotal to latent infection by epstein-barr virus. Rev Med Virol. 2000;10:83–100. doi: 10.1002/(sici)1099-1654(200003/04)10:2<83::aid-rmv262>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 24.Lomonte P, Bublot M, van Santen V, Keil G M, Pastoret P-P, Thiry E. Analysis of bovine herpesvirus 4 genomic regions located outside the conserved gammaherpesvirus gene blocks. J Gen Virol. 1995;76:1835–1841. doi: 10.1099/0022-1317-76-7-1835. [DOI] [PubMed] [Google Scholar]
- 25.Makkerh J P S, Dingwall C, Laskey R A. Comparative mutagenesis of nuclear localization signals reveals the importance of neutral and acidic amino acids. Curr Biol. 1996;6:1025–1027. doi: 10.1016/s0960-9822(02)00648-6. [DOI] [PubMed] [Google Scholar]
- 26.Martin M E, Berk A J. Corepressor required for adenovirus E1B 55,000-molecular-weight protein repression of basal transcription. Mol Cell Biol. 1999;19:3403–3414. doi: 10.1128/mcb.19.5.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mastrangelo I A, Hough P V, Wall J S, Dodson M, Dean F B, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature. 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 28.Maul G G. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 29.Neipel F, Albrecht J C, Fleckenstein B. Human herpesvirus 8—the first human rhadinovirus. J Natl Cancer Inst Monogr. 1998;23:73–77. doi: 10.1093/oxfordjournals.jncimonographs.a024178. [DOI] [PubMed] [Google Scholar]
- 30.Neipel F, Fleckenstein B. The role of HHV-8 in Kaposi's sarcoma. Semin Cancer Biol. 1999;9:151–164. doi: 10.1006/scbi.1999.0129. [DOI] [PubMed] [Google Scholar]
- 31.Parsons R E, Stenger J E, Ray S, Welker R, Anderson M E, Tegtmeyer P. Cooperative assembly of simian virus 40 T-antigen hexamers on functional halves of the replication origin. J Virol. 1991;65:2798–2806. doi: 10.1128/jvi.65.6.2798-2806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Platt G M, Simpson G R, Mittnacht S, Schulz T F. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J Virol. 1999;73:9789–9795. doi: 10.1128/jvi.73.12.9789-9795.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rawlins D R, Milman G, Hayward S D, Hayward G S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 35.Rhee K, Brunori M, Besset V, Trousdale R, Wolgemuth D J. Expression and potential role of Fsrg1, a murine bromodomain-containing homologue of the Drosophila gene female sterile homeotic. J Cell Sci. 1998;111:3541–3550. doi: 10.1242/jcs.111.23.3541. [DOI] [PubMed] [Google Scholar]
- 36.Rost B. PHD: predicting 1D protein structure by profile-based neural networks. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 37.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Searles R P, Bergquam E P, Axthelm M K, Wong S W. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steegenga W T, Shvarts A, Riteco N, Bos J L, Jochemsen A G. Distinct regulation of p53 and p73 activity by adenovirus E1A, E1B, and E4orf6 proteins. Mol Cell Biol. 1999;19:3885–3894. doi: 10.1128/mcb.19.5.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sternsdorf T, Grotzinger T, Jensen K, Will H. Nuclear dots: actors on many stages. Immunobiology. 1997;198:307–331. doi: 10.1016/S0171-2985(97)80051-4. [DOI] [PubMed] [Google Scholar]
- 41.Szekely L, Chen F, Teramoto N, Ehlin-Henriksson B, Pokrovskaja K, Szeles A, Manneborg-Sandlund A, Lowbeer M, Lennette E T, Klein G. Restricted expression of Epstein-Barr virus (EBV)-encoded, growth transformation-associated antigens in an EBV- and human herpesvirus type 8-carrying body cavity lymphoma line. J Gen Virol. 1998;79:1445–1452. doi: 10.1099/0022-1317-79-6-1445. [DOI] [PubMed] [Google Scholar]
- 42.Szekely L, Kiss C, Mattsson K, Kashuba E, Pokrovskaja K, Juhasz A, Holmvall P, Klein G. Human herpesvirus-8-encoded LNA-1 accumulates in heterochromatin-associated nuclear bodies. J Gen Virol. 1999;80:2889–2900. doi: 10.1099/0022-1317-80-11-2889. [DOI] [PubMed] [Google Scholar]
- 43.Szekely L, Selivanova G, Magnusson K P, Klein G, Wiman K G. EBNA-5, an Epstein-Barr virus-encoded nuclear antigen, binds to the retinoblastoma and p53 proteins. Proc Natl Acad Sci USA. 1993;90:5455–5459. doi: 10.1073/pnas.90.12.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 45.Virgin H W, 4th, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson, A. C., M. J. Boutros, K. M. Johnson, and W. Herr. HCF-1 amino- and carboxy-terminal subunit association through two separate sets of interaction modules: involvement of fibronectin type 3 repeats. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 47.Wilson A C, Freiman R N, Goto H, Nishimoto T, Herr W. VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol Cell Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu L, Wang R, Sweat A, Goldstein E, Horvat R, Chandran B. Comparison of human sera reactivities in immunoblots with recombinant human herpesvirus (HHV)-8 proteins associated with the latent (ORF73) and lytic (ORFs 65, K8.1A, and K8.1B) replicative cycles and in immunofluorescence assays with HHV-8-infected BCBL-1 cells. Virology. 1999;256:381–392. doi: 10.1006/viro.1999.9674. [DOI] [PubMed] [Google Scholar]