Abstract
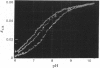
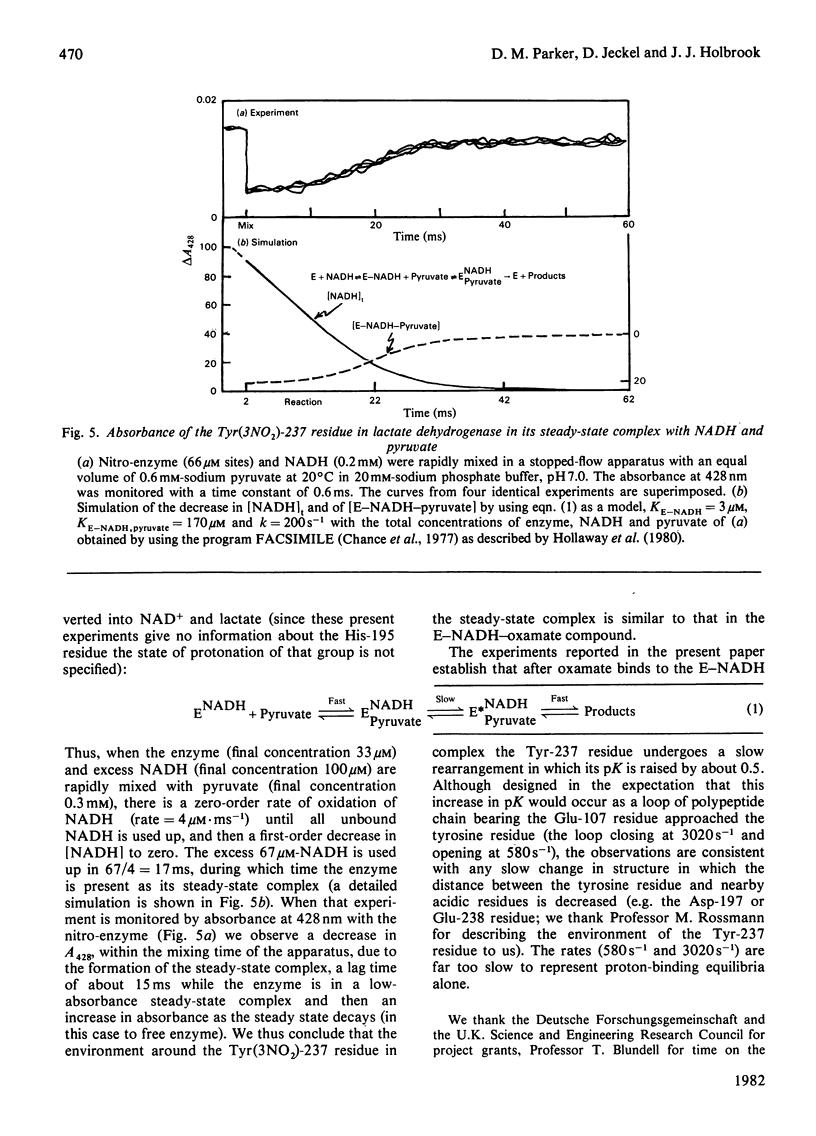
1. The pKa of the phenolic hydroxy group of the Tyr(3NO2)-237 residue in pig heart [Tyr(3NO2)237]lactate dehydrogenase is 7.2 in the apoenzyme, 7.4 in the enzyme-NADH complex and 7.8 in the enzyme-NADH-oxamate complex. The alkaline shift from apoenzyme to ternary complex is ascribed to the approach of the Glu-107 residue during the movement of the polypeptide loop residues 98-110. 2. The affinities of the nitrated enzyme for NADH and for oxamate (in the presence of NADH) are slightly less than those of the native enzyme. The turnover number for the nitrated enzyme in the pyruvate-to-lactate direction is about 0.75 of the value for the native enzyme. 3. Temperature-jump relaxation experiments of the enzyme saturated with NADH but fractionally saturated with oxamate are interpreted to show that the pKa of the nitrotyrosine residue responds to a protein rearrangement after oxamate binds to the binary enzyme-NADH complex. 4. Transient-kinetic experiments show the environment of the Tyr(3NO2)-237 residue in the enzyme-NADH-pyruvate complex of the steady state to be similar to that in the enzyme-NADH-oxamate inhibitor complex.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Eventoff W., Rossmann M. G., Taylor S. S., Torff H. J., Meyer H., Keil W., Kiltz H. H. Structural adaptations of lactate dehydrogenase isozymes. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2677–2681. doi: 10.1073/pnas.74.7.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau U., Kapmeyer H., Trommer W. E. Combined coenzyme-substrate analogues of various dehydrogenases. Synthesis of (3S)- and (3R)-5-(3-carboxy-3-hydroxypropyl)nicotinamide adenine dinucleotide and their interaction with (S)- and (R)-lactate-specific dehydrogenases. Biochemistry. 1978 Oct 31;17(22):4621–4626. doi: 10.1021/bi00615a007. [DOI] [PubMed] [Google Scholar]
- Heck H. de A. Porcine heart lactate dehydrogenase. Optical rotatory dispersion, thermodynamics, and kinetics of binding reactions. J Biol Chem. 1969 Aug 25;244(16):4375–4381. [PubMed] [Google Scholar]
- Holbrook J. J. Protein fluorescence of lactate dehydrogenase. Biochem J. 1972 Jul;128(4):921–931. doi: 10.1042/bj1280921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook J. J., Stinson R. A. The use of ternary complexes to study ionizations and isomerizations during catalysis by lactate dehydrogenase. Biochem J. 1973 Apr;131(4):739–748. doi: 10.1042/bj1310739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook J. J., Wolfe R. G. Malate dehydrogenase. X. Fluorescence microtitration studies of D-malate, hydroxymalonate, nicotinamide dinucleotide, and dihydronicotinamide-adenine dinucleotide binding by mitochondrial and supernatant porcine heart enzymes. Biochemistry. 1972 Jun 20;11(13):2499–2502. doi: 10.1021/bi00763a018. [DOI] [PubMed] [Google Scholar]
- Jeckel D., Anders R., Pfleiderer G. Zum Wirkungsmechanismus der Lactat-Dehydrogenase. VI. Anderung der biochemischen Eigenschaften von Lactat-Dehydrogenase aus Schweineherzmuskel nach Nitrierung mit Tetranitromethan. Hoppe Seylers Z Physiol Chem. 1971 Jun;352(6):769–779. [PubMed] [Google Scholar]
- Lodola A., Parker D. M., Jeck R., Holbrook J. J. Malate dehydrogenase of the cytosol. Ionizations of the enzyme-reduced-coenzyme complex and a comparison with lactate dehydrogenase. Biochem J. 1978 Aug 1;173(2):597–605. doi: 10.1042/bj1730597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D. M., Holbrook J. J., Birdsall B., Roberts G. C. H NMR studies of Tyr-237 of lactate dehydrogenase. FEBS Lett. 1981 Jun 29;129(1):33–35. doi: 10.1016/0014-5793(81)80748-x. [DOI] [PubMed] [Google Scholar]
- Parker D. M., Holbrook J. J. The oxaloacetate reductase activity of vertebrate lactate dehydrogenase. Int J Biochem. 1981;13(10):1101–1105. doi: 10.1016/0020-711x(81)90173-7. [DOI] [PubMed] [Google Scholar]
- Trommer W. E., Glöggler K. Solution conformation of lactate dehydrogenase as studied by saturation transfer ESR spectroscopy. Biochim Biophys Acta. 1979 Dec 7;571(2):186–194. doi: 10.1016/0005-2744(79)90089-5. [DOI] [PubMed] [Google Scholar]
- WINER A. D., SCHWERT G. W. Lactic dehydrogenase. VII. Fluorescence spectra of ternary complexes of lactic dehydrogenase, reduced diphosphopyridine nucleotide, and carboxylic acids. J Biol Chem. 1959 May;234(5):1155–1161. [PubMed] [Google Scholar]