Abstract
The lack of a susceptible cell line and an animal model for Norwalk virus (NV) infection has prompted the development of alternative strategies to generate in vitro RNAs that approximate the authentic viral genome. This approach has allowed the study of viral RNA replication and gene expression. In this study, using mobility shift and cross-linking assays, we detected several cellular proteins from HeLa and CaCo-2 cell extracts that bind to, and form stable complexes with, the first 110 nucleotides of the 5′ end of NV genomic RNA, a region previously predicted to form a double stem-loop structure. These proteins had molecular weights similar to those of the HeLa cellular proteins that bind to the internal ribosomal entry site of poliovirus RNA. HeLa proteins La, PCBP-2, and PTB, which are important for poliovirus translation, and hnRNP L, which is possibly implicated in hepatitis C virus translation, interact with NV RNA. These protein-RNA interactions are likely to play a role in NV translation and/or replication.
Norwalk virus (NV) is the prototype strain of human caliciviruses and has been implicated in outbreaks of nonbacterial acute gastroenteritis in the U.S. and many other countries (2, 15, 30). The virus is small (27 to 35 nm in diameter), round, nonenveloped, and with an amorphous surface structure (29, 40). The virion contains a 7.7-kb single-stranded, positive RNA genome; the RNA is polyadenylated and attaches with a VPg at its 5′ end (6, 10). Genome sequence analysis has revealed three open reading frames (ORFs). ORF 1 encodes a polyprotein that is processed into nonstructural proteins required for virus replication and has sequence homology to picornavirus 2C helicase, 3C protease, and 3D RNA-dependent RNA polymerase (12). ORF 2 encodes the viral capsid protein, and ORF 3 encodes a small basic protein with an unknown function (27). NV also produces a 2.3-kb subgenomic RNA containing ORFs 2 and 3, each of them having a strong AUG initiation codon, suggesting that they may be expressed independently (27).
The conserved sequence identified at the 5′ end of the genomic and subgenomic RNAs of NV suggests that it might be important for virus replication; 23 (88%) of the first 26 nucleotides (nt) of the two RNAs are identical (20). This element is also present in other caliciviruses, including the rabbit hemorrhagic disease virus and the feline calicivirus (9, 20, 34, 42). Sequence analysis of the NV RNA predicted a double stem-loop structure at the 5′ end of the genomic (nt 1 to 110) and subgenomic (nt 5280 to 5356) RNAs (27). A similar double stem loop was also predicted upstream of ORF 3 (nt 6848 to 6941). However, the role of these predicted structures in viral RNA replication remains unknown.
Highly conserved secondary RNA structures are known to be present at the 5′ and 3′ ends or in the internal regions of the genomes of picornaviruses, hepatitis C virus, dengue virus, Japanese encephalitis virus, and simian hemorrhagic fever virus (7, 8, 11, 13, 22, 24, 28, 34, 38, 43, 47). Studies of viral RNA interaction with cellular proteins have identified several elements in the viral RNAs that are important for viral replication (1, 3, 13). RNA-protein complexes are formed when authentic viral RNA or in vitro synthesized viral RNA transcripts are incubated with cell extracts. These complexes are involved in viral RNA replication and translation (1, 7, 8, 13, 17, 19, 22, 23, 24, 25, 28, 33, 43, 47, 48).
The absence of a permissive cell line and a susceptible animal model for NV infection has made it difficult to study the biology of the virus. The successful cloning and sequencing of the NV genome and other human calicivirus genomes has allowed much progress in our knowledge of gene coding strategies, genomic organization, viral RNA replication, and gene expression (20, 26, 27, 34). However, in the case of NV little is known about the mechanisms of viral replication. In this study, we performed binding experiments of in vitro synthesized NV RNA and HeLa and CaCo-2 cell extracts. Our results demonstrate that the 5′ end of the NV genome contains elements that bind specifically to different cellular proteins, some of which include HeLa proteins, such as La, hnRNPL, PTB, and PCBP-2, that are known to be involved in the poliovirus internal ribosomal entry site (IRES)-associated translation (3, 16, 17, 19, 21, 28, 34, 35, 36) and hepatitis C virus translation (18, 23).
MATERIALS AND METHODS
Cells.
HeLa cells were grown in Dulbecco's minimal essential medium supplemented with 10% newborn calf serum, 5,000 U of penicillin, and 5 μg of streptomycin. CaCo-2 cells (a human colon adenocarcinoma cell line) were grown in Dulbecco's minimal essential medium containing 0.11% glutamine, 0.02% sodium pyruvate, 0.47% NaCl, 1× nonessential amino acids, 5,000 U of penicillin, 5 μg of streptomycin, and 10% fetal bovine serum. Both cell lines were grown in a 5% CO2 incubator at 37°C. The culture medium was changed every other day until the cells reached confluency.
Subcloning of the 5′ end of NV genomic cDNA.
A cDNA clone of nt 1 to 1810 of the NV genome was constructed from the cDNA clone 5030 (27) by PCR using a 5′ primer containing the first 12 nt of the NV genome that were missed in cDNA 5030 and a 3′ primer containing the sequence around the EcoRV site at nt 1810 of the NV genome. To facilitate synthesis of RNA using this cDNA as a template, a bacteriophage T7 RNA promoter sequence and a MluI site were included in the 5′ primer. The PCR-amplified 1.8-kb cDNA was subsequently cloned into a pGEM-T vector (Promega).
In vitro transcription of 5′ NV genomic RNAs.
NV genomic RNAs containing nt 1 to 191 were prepared by in vitro transcription using T7 RNA polymerase with the NV 1.8-kb cDNA predigested with MluI and HaeII as a template, following a method previously described (38). After the transcription reaction, the DNA template was removed by treating the samples with DNase RQ1 (Promega) in the presence of RNase inhibitors (Promega). Unincorporated nucleotides in the reaction mixture were removed by gel filtration. For synthesis of radiolabeled RNA transcripts, [α-32P]UTP (Dupont) was included in the transcription reaction.
Two additional RNAs containing nt 1 to 110 and 111 to 191 of the NV genome were produced by in vitro transcription using T7 RNA polymerase from the two PCR-amplified cDNAs containing the respective regions. The PCR was performed using the NV 1.8-kb cDNA as the template. The 5′ primers of the two pairs used in the PCR also contained the bacteriophage T7 promoter sequence. The PCR was performed for 35 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 30 s, using a Perkin-Elmer Cetus DNA thermocycler. The resulting PCR products were purified by a QIAquiq gel extraction G-50 kit (Qiagen) before they were used as templates for RNA synthesis.
Preparation of HeLa and CaCo-2 cell extracts.
HeLa and CaCo-2 cells were washed twice with cold phosphate-buffered saline and once with 5 volumes of washing buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl). After the final wash, the cells were resuspended in 2 cell volumes of washing buffer. HeLa cells were homogenized with 20 strokes and CaCo-2 cells were homogenized with 60 strokes in a glass Dounce homogenizer. The cell homogenates were centrifuged at 10,000 rpm for 30 min in a Sorvall GSA rotor. The supernatant, called S10 extract, was divided into aliquots, and the concentration of proteins in each extract was determined by the Bradford assay (5).
Mobility shift electrophoresis assay.
Variable amounts (4 to 20 μg) of S10 extracts from HeLa or CaCo-2 cells were preincubated for 15 min at 4°C with equal amounts of tRNA in a buffer containing 10 mM HEPES (pH 7.4), 0.1 mM EDTA, 0.2 mM dithiothreitol, 8 mM MgCl2, 4 mM spermidine, 3 mM ATP, 2 mM GTP, and 10% (vol/vol) glycerol in a final volume of 10 μl. For each RNA binding experiment, 1 × 106 cpm of 32P-labeled NV RNA was added to the reaction mixture and incubated for 15 min at 4°C. Before loading the gels, a final incubation with 20 units of RNase A and 20 μg of RNase T1 was performed for 15 min at room temperature. RNA-protein complexes were analyzed by electrophoresis on a 6% polyacrylamide (acrylamide-bisacrylamide, 80:1) gel in 0.5× TBE buffer (90 mM Tris, 64.6 mM boric acid, 2.5 mM EDTA [pH 8.3]) run at 20 mA for 4 h. Gels were dried and autoradiographed. To determine the stability of the RNA-protein complexes, different concentrations of KCl were included in the preincubation reaction. For competition experiments, unlabeled RNAs were added to the preincubation reaction mixture.
Mobility supershift electrophoresis assay.
Twelve micrograms of monoclonal antibodies to HeLa PCBP-2 protein were incubated with 12 μg of S10 extracts. The antigen-antibody reaction was allowed to proceed for 30 min on ice before addition of labeled RNA. The antibody-RNA-protein supercomplex was further processed under the same conditions described for the RNA-protein complex. The monoclonal antibodies were generously provided by B. Semler, University of California, Irvine.
UV cross-linking of RNA-protein complex.
UV cross-linking of RNA-protein complexes was performed using a method described previously (16, 17) in a reaction mixture containing 200 pmol of α-32P-labeled RNA and 60 μg of S10 extract or 500 ng of the recombinant polypyrimidine tract-binding (PTB) protein. The recombinant PTB protein was kindly provided by Monica DeNova, Centro de Investigación y de Estudios Avanzados del IPN, Mexico City, Mexico. The sequence of the human PTB was obtained from plasmid PTB-pRC/CMV (kindly provided by Stanley Lemon, University of North Carolina). Following cross-linking, the samples were fractionated in 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels. The gels were fixed, dried, and autoradiographed.
Immunoprecipitation of La-RNA and hnRNP-L-RNA complexes.
After UV cross-linking of CaCo-2 cytoplasmic extracts to nt 1 to 110 of NV RNA and RNase treatment were done as described above, the samples were incubated with 10 μl of protein G-Sepharose 4B beads for 2 h at 4°C and centrifuged at 12,000 rpm for 5 min in a microcentrifuge. Supernatants were incubated overnight at 4°C with monoclonal anti-La or anti-hnRNP-L antibodies (kindly provided by N. Sonenberg, McGill University, Montreal, Quebec, Canada, and G. Dreyfuss, University of Pennsylvania School of Medicine, Philadelphia, respectively) overnight at 4°C. The immunocomplexes were immobilized on protein G-Sepharose 4B beads saturated with 2% bovine serum albumin for another 2 h at 4°C. Unbound materials were washed five times with NETS buffer (50 mM Tris-HCl [pH 7.4], 5 mM EDTA, 1 mM dithiothreitol, 100 mM NaCl, 0.05% Nonidet P-40). Bound proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) followed by autoradiography. Parallel reaction mixtures were made with an unrelated antiactin monoclonal antibody as a control.
RESULTS
Interaction of the 5′ end of NV RNAs with cell extracts.
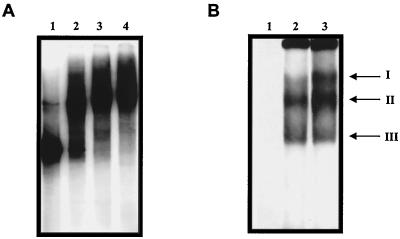
To determine whether the 5′ end of the NV genome was recognized by cellular proteins, 32P-labeled RNA transcripts from nt 1 to 191 (including the predicted double stem loop of nt 1 to 110) were incubated with the S10 extract from HeLa cells. A major RNA-protein complex was observed following electrophoresis (Fig. 1). Similar results were obtained when CaCo-2 extracts were used (data not shown). The migration of the complex was retarded when increasing concentrations of S10 extracts were used (Fig. 1A, lanes 2 to 4). Electrophoresis performed following RNase treatment showed the formation of three complexes with different migration patterns in both HeLa cells (Fig. 1B, lane 2) and CaCo-2 cells (lane 3). Complexes II and III revealed stronger RNA labeling than complex I in HeLa S10 extracts, while complexes I and II showed stronger signals in CaCo-2 extracts.
FIG. 1.
Mobility shift analysis of RNA consisting of nt 1 to 191 from the NV 5′ end. (A) Labeled RNA incubated in the absence (lane 1) or presence of 2, 6, and 8 μg of HeLa S10 extracts (lanes 2, 3, and 4, respectively). (B) Labeled RNA incubated in the absence (lane 1) or presence of 12 μg of HeLa (lane 2) or CaCo-2 S10 extracts (lane3), followed by RNase treatment. Complex formation was assayed by electrophoresis on native polyacrylamide gels and detected by autoradiography. Mobility of complexes I, II, and III is indicated on the right side of the figure.
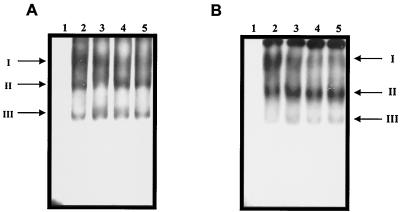
In order to determine the stability of the RNA-protein complexes, the samples were incubated with variable KCl concentrations. The three complexes formed with the HeLa S10 extract and nt 1 to 191 of NV RNA were not altered in a range of 0 to 1.2 M KCl (Fig. 2A, lanes 2 to 5). However, the intensity of complex I formed with the CaCo-2 S10 extract was reduced at 0.9 and 1.2 M of KCl (Fig. 2B, lanes 4 and 5).
FIG. 2.
Stability of RNA-protein complexes formed with nt 1 to 191 of NV. Labeled RNA was incubated in the absence (lanes 1) or presence of 0.6, 0.9, and 1.2 M KCl (lanes 3 to 5, respectively) with HeLa (A) or CaCo-2 (B) S10 extracts. Complex formation was assayed by electrophoresis on native polyacrylamide gels and detected by autoradiography.
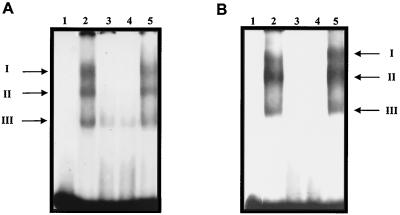
The specificity of the RNA-protein binding was further demonstrated in competition experiments using HeLa (Fig. 3A) or CaCo-2 extracts (Fig. 3B) incubated with a 20-fold-molar excess of cold homologous or heterologous RNAs as competitors. Significant reduction of the three RNA-protein complexes was observed in samples incubated with homologous (nt 1 to 191 from NV) (lanes 3) but not heterologous RNA transcripts from pBluescript plasmid DNA (lanes 5). Most interestingly, a heterologous RNA (nt 275 to 628) from poliovirus (PV) efficiently competed with the NV RNA in the formation of complexes (lanes 4), suggesting that both viruses could share the same cellular machinery for replication.
FIG. 3.
Specificity of RNA-protein complexes formed with nt 1 to 191 of NV. Labeled RNA was incubated in the absence (lanes 2) or presence of a 20-fold-molar excess of a homologous (lanes 3) or heterologous competitor consisting of poliovirus nt 275 to 628 RNA (lanes 4) or pBluescript RNA (lanes 5). Free RNA was loaded on lanes 1. HeLa S10 extracts (A) and CaCo-2 S10 extracts (B) are shown. Complex formation was assayed by electrophoresis through native polyacrylamide gels and detected by autoradiography.
Identification of cellular proteins present in the RNA-protein complexes.
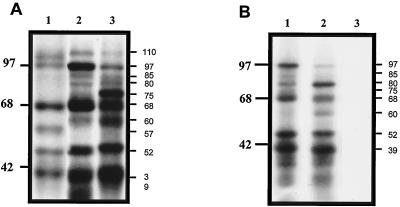
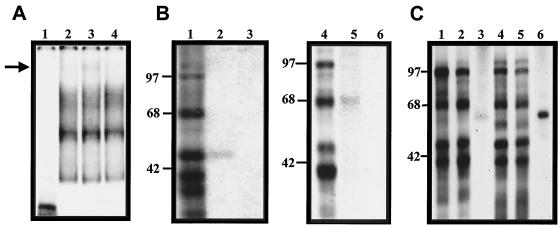
To identify the cellular proteins present in the RNA-protein complexes, labeled RNA containing nt 1 to 191 of NV RNA was incubated with HeLa or CaCo-2 extracts, followed by UV-induced cross-linking, and was analyzed on denaturing SDS-PAGE. Nine proteins with apparent molecular masses of 110, 97, 85, 80, 75, 68, 60, 52, and 39 kDa were identified from both HeLa and CaCo-2 cells (Fig. 4A, lanes 2, and 3, respectively). Although similar molecular weights were observed for both cell lines, the relative intensities of individual proteins differed. The 97- and 68-kDa proteins were more intense in HeLa cell extracts (lane 2), while the 75- and 39-kDa proteins were stronger in the CaCo-2 cell extracts (lane 3). Cellular proteins with molecular masses similar to those of proteins cross-linked to NV RNA also bound to the PV 5′ untranslated region (nt 275 to 628) RNA (Fig. 4A, lane 1), except for a 60-kDa protein that was cross-linked to the NV RNA but not to the PV RNA. However, the intensity of labeled proteins bound to the PV RNA was generally weaker.
FIG. 4.
UV-induced cross-linking of HeLa and CaCo-2 S10 extracts to labeled RNAs from nt 1 to 191, 1 to 110, and 111 to 191 of NV. (A) Labeled RNA of nt 1 to 191 of NV (lanes 2 and 3) and nt 275 to 628 of the PV 5′ untranslated region (lane 1) were UV cross-linked with 60 μg of HeLa extracts (lanes 1 and 2) or CaCo-2 S10 extracts (lane 3). (B) Labeled RNA consisting of nt 1 to 110 (lanes 1 and 2) and nt 111 to 191 (lane 3) were UV cross-linked with 60 μg of HeLa extracts (lane1) or CaCo-2 S10 extracts (lanes 2 and 3). Proteins were separated by SDS–12.5% PAGE and detected by autoradiography. Numbers to the right of panels indicate molecular masses in kilodaltons.
To locate the specific region of the 5′ end of the NV genome that is responsible for the RNA-protein interactions, two RNA probes consisting of nt 1 to 110 (double stem loop) and nt 111 to 191 were used in the UV cross-linking assays. The RNA probe from nt 1 to 110 cross-linked with the same nine proteins from both HeLa (Fig. 4B, lane 1) and CaCo-2 (Fig. 4B, lane 2) cell extracts that bound to the probe from nt 1 to 191 (Fig. 4A). In contrast, the RNA probe from nt 111 to 191 failed to cross-link with any cellular proteins (Fig. 4B, lane 3), indicating that the protein interaction occurs within nt 1 to 110 of NV.
Proteins of 97, 68, 57/60, 52, and 39 kDa bound to the PV IRES element have been previously identified as unr, hnRNP L, PTB, La, and PCBP2, respectively. All of them are involved in picornavirus and hepatitis C virus translation (3, 18, 21, 22, 23, 24, 28, 35, 36, 44). Due to the similarities in molecular mass to the proteins detected in this study, the possibility of the presence of PCBP-2, La, hnRNP-L, and PTB was investigated. To determine whether PCBP-2 binds to nt 1 to 110 of NV RNA, a supershift assay was performed. Incubation with anti-PCBP-2 antibodies resulted in the formation of an additional complex with less mobility (Fig. 5A, lane 2) than the three complexes observed previously (Fig. 5A, lane 1). This additional complex was not formed when the reaction was performed in the presence of an unrelated antibody (anti-actin) (Fig. 5A, lane 3).
FIG. 5.
Identification of proteins bound to nt 1 to 110 of NV RNA. (A) Mobility supershift analysis of complexes formed with nt 1 to 110 of NV. Labeled RNA was incubated with 10 μg of HeLa S10 extracts in the absence (lane 1) and presence of antibodies to PCBP-2 (lane 2) or an antiactin antibody (lane 3). Complex formation was assayed by electrophoresis through native polyacrylamide gels and detected by autoradiography. The arrow indicates the complex formed in the presence of the specific antibody. (B) Immunoprecipitation assay of La and hnRNP-L proteins bound to nt 1 to 110 of NV RNA. Labeled RNA was cross-linked with 60 μg of CaCo-2 S10 proteins (lanes 1 and 4) and immunprecipitated with anti-La (lane 2), anti-hnRNP-L (lane 5) or anti-actin antibodies (lanes 3 and 6). (C) UV cross-linking of nt 1 to 110 of NV RNA and nt 275 to 628 of PV RNA with S10 extracts from HeLa and CaCo-2 cells and with the recombinant PTB protein. Labeled RNAs of nt 1 to 110 of NV (lanes 1 to 3) and nt 275 to 628 of the PV 5′ UTR (lanes 4 to 6) were UV cross-linked with 60 μg of HeLa (lanes 1 and 4) or CaCo-2 (lane 2 and 5) S10 extract or 500 μg of recombinant PTB protein (lanes 3 and 6). Samples were loaded on an SDS–10% polyacrylamide gel followed by electrophoresis and were detected by autoradiography.
To analyze if the 52- and 68-kDa cross-linked proteins correspond to the La and hnRNP L proteins, an immunoprecipitation assay using monoclonal antibodies against each protein was carried out (Fig. 5B). A protein of 52 kDa that comigrates with the 52-kDa cross-linked protein was immunoprecipitated by anti-La antibodies (Fig. 5B, lane 2), while a 68-kDa protein, which also migrates with the 68-kDa cross-linked protein, was identified by the anti-hnRNP L antibodies (Fig. 5B, lane 5). Antiactin antibodies were unable to immunoprecipitate any labeled proteins (Fig. 5B, lanes 3 and 6).
Finally, a cross-linking assay of nt 1 to 110 of NV RNA with a recombinant PTB protein was performed (Fig. 5C). The recombinant PTB was able to bind to nt 1 to 110 of NV RNAs and to nt 275 to 628 of PV RNAs. However, this recombinant PTB showed a migration slightly different from that of the 57/60-kDa proteins cross-linked with HeLa and CaCo-2 cells with NV (Fig. 5C, lanes 1 and 2, respectively) or PV (Fig. 5C, lanes 4 and 5, respectively). The amount of labeled PTB detected when PV RNA was used was larger than the amount of PTB bound to NV RNA.
DISCUSSION
This is the first report to show binding of NV RNA to proteins from eukaryotic cells. Two different cell extracts were used. HeLa cells were included in the study because they have been widely used for cultivation of several viruses and contain the proteins and factors required for their translation either in cell culture or in cell-free systems (45). On the other hand, CaCo-2 cells (derived from a human colon adenocarcinoma) were selected for their resemblance to human enterocytes of the small intestine, where NV might replicate during human infection (39, 40). Moreover, recombinant NV-like particles, which are morphologically and antigenically similar to native NV, bind and penetrate CaCo-2 cells (46).
In the absence of RNase, the mobility shift assays demonstrated the formation of a large ribonucleoprotein complex with the 5′ end of the NV genome using HeLa and CaCo-2 S10 extracts. After RNase treatment with HeLa or CaCo-2 S10 extracts, three ribonucleoprotein complexes were distinguished. There are several possibilities to explain the presence of these bands: they might reflect (i) different ribonucleoprotein complexes, (ii) the same complex with different conformations, or (iii) three stages of the same complex with different amounts of bound proteins. It is important to emphasize that these complexes were not disrupted by increasing concentrations of KCl, indicating that the RNA-protein interactions are stable.
The specificity of the complex formation was confirmed through the competition with homologous but not with heterologous pBluescript RNA. The competition observed in the presence of PV RNA suggests that NV and PV probably use the same cellular machinery for replication. Although the primary 5′ end sequences of the two viral RNAs are different, the specific competition suggests that they might share similar secondary structures that are recognized by the same panel of proteins in these cells.
Since one of the three double stem-loop structures is predicted to be located between nt 1 and 110 of NV RNA (27), we determined which of the cross-linked proteins that bound to nt 1 to 191 also bound to this region. All HeLa and CaCo-2 cell proteins that bind to nt 1 to 191 formed complexes with NV nt 1 to 110, while no binding was observed to nt 111 to 191, suggesting that the region from nt 1 to 110 is responsible for the RNA-protein interaction. Similar double stem loops are also predicted upstream of ORF 2 (nt 5280 to 5356) and ORF 3 (nt 6848 to 6941). Preliminary cross-linking assays performed with these two regions suggest a protein binding pattern similar to the one observed with nt 1 to 110 of NV RNA.
The cross-linking assays demonstrated the presence of nine proteins in the NV RNA-protein complexes formed with the HeLa and CaCo-2 S10 extracts. Eight of these proteins had the same molecular mass as those bound to nt 275 to 628 of PV RNA, providing further evidence that NV and PV may use similar replication or translation mechanisms. The 57-kDa protein bound to PV RNA and identified as PTB (4, 14, 21) was detected in NV with a slightly higher migration. It is then possible that the 60-kDa protein cross-linked to NV RNA could be an isoform of PTB, as has been previously described (4, 14). The recombinant PTB protein bound to PV RNA and NV RNA showed the same migration, but the amount of protein bound to PV RNA was greater. Although the reported PTB-binding consensus sequence is not present within the first 110 nt of NV RNA, multiple binding sites of PTB have been identified in PV and encephalomyocarditis virus IRES, where this consensus sequence does not exist (24). It could be speculated that PTB can bind to similar structures present in both RNAs but with different affinity.
The presence of five favored initiation codons based on Kozak's rule (31) has been reported within the first 1,106 nt of NV. The first favored AUG is located at nt 11 and has been suggested to be the translation initiation site (9). This site is very close to the 5′ end and might be inefficiently recognized or ignored by the ribosome, as has been described previously (32). In addition, the absence of the cap structure in NV RNA could suggest that the ribosome requires additional elements to initiate translation. One possibility is that the observed ribonucleoprotein complex formed with the double stem-loop structure located at nt 1 to 110 allows entry to the ribosome, as occurs during picornavirus translation. In that case, the next favored initiation codon located at nt 167 could be used as the translation initiation site. La, hnRNP L, PTB, and PCBP-2 were shown to interact with nt 1 to 110 of NV RNA by immunoprecipitation, cross-linking, or mobility gel shift assays. La protein is responsible for the selection of the correct translation initiation site and the enhancement of PV translation (36). On the other hand, PTB protein has been proposed to promote the correct folding of encephalomyocarditis virus IRES in order to present the critical primary nucleotide sequence motifs in the correct three-dimensional organization to allow internal ribosome entry (28). For NV, interaction with La and PTB could play a similar role, promoting the correct folding of the RNA and selecting the correct site for ribosomal entry.
Although at present we do not know how NV RNAs interact with cellular proteins, the formation of stable complexes with the 5′ end of NV and the identification of proteins related to PV replication and translation in these complexes suggest that the RNA-protein interactions could play a significant role in the translation of NV.
Further analysis aimed at understanding the role of the RNA-protein interactions in viral translation or replication is currently being performed in our laboratory using deletion and site-directed mutagenesis. Since HeLa cells contain translation factors that bind to NV RNA, these cells could be used to analyze whether they allow NV translation and shed some light on one of the processes of viral replication. However, if NV translation can be demonstrated, this does not mean that the complete viral cycle could take place in this cell line. It is possible that other stages of the cycle, such as entrance, RNA replication, or assembly, are blocked in these cells, thus explaining their nonpermissive nature to NV.
ACKNOWLEDGMENTS
We thank Fernando Medina for the cell cultures, Jaime Escobar for technical assistance, and Monica DeNova for the recombinant PTB protein. We gratefully acknowledge Stanley Lemon for providing the PTB sequence, B. Semler (University of California, Irvine) for the anti-PCBP2 antibodies, N. Sonenberg (McGill University, Montreal, Quebec, Canada) for the anti-La antibodies, and G. Dreyfuss (University of Pennsylvania School of Medicine, Philadelphia) for the anti-hnRNP-L antibodies. We also thank Martha Espinosa-Cantellano for critical comments on the manuscript.
This work was supported by grants from the Consejo Nacional de Ciencia y Tecnología.
REFERENCES
- 1.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′ end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blacklow N R, Greenberg H B. Viral gastroenteritis. N Engl J Med. 1991;325:252–264. doi: 10.1056/NEJM199107253250406. [DOI] [PubMed] [Google Scholar]
- 3.Blyn L B, Towner J S, Semler B L, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bothwell A L M, Ballard D W, Philbrick W M, Lindwall G, Maher S E, Bridgett M M, Jamison S F, Garcia-Blanco M A. Purine polypyrimidine tract binding protein. J Biol Chem. 1991;25:24657–24663. [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Burroughs J N, Brown F. Presence of a covalently linked protein on calicivirus RNA. J Gen Virol. 1978;41:443–446. doi: 10.1099/0022-1317-41-2-443. [DOI] [PubMed] [Google Scholar]
- 7.Chang K H, Brown E A, Lemon S M. Cell type-specific proteins which interact with the 5′ nontranslated region of hepatitis A virus RNA. J Virol. 1993;67:6716–6725. doi: 10.1128/jvi.67.11.6716-6725.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C J, Kuo M D, Chen L J, Hsu S L, Wang Y M, Lin J H. RNA-protein interactions: involvement of NS3, NS5, and 3′ coding regions of Japanese encephalitis virus genomic RNA. J Virol. 1997;71:3466–3473. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke I N, Lambden P R. The molecular biology of caliciviruses. J Gen Virol. 1997;78:291–301. doi: 10.1099/0022-1317-78-2-291. [DOI] [PubMed] [Google Scholar]
- 10.Dunham D M, Jiang X, Berke T, Smith A W, Matson D O. Genomic mapping of a calicivirus VPg. Arch Virol. 1998;143:2421–2430. doi: 10.1007/s007050050471. [DOI] [PubMed] [Google Scholar]
- 11.Ehrenfeld E, Semler B. Anatomy of the poliovirus internal ribosome entry site. Curr Top Microbiol Immunol. 1995;203:65–83. doi: 10.1007/978-3-642-79663-0_3. [DOI] [PubMed] [Google Scholar]
- 12.Ehresmann D W, Schaffer F L. RNA synthesized in calicivirus-infected cells is atypical of picornaviruses. J Virol. 1977;22:572–576. doi: 10.1128/jvi.22.2.572-576.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamarnik A V, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gil A, Sharp P A, Jamison S F, Garcia-Blanco M A. Characterization of cDNAs encoding the polypyrimidine tract binding protein. Genes Dev. 1991;5:1224–1236. doi: 10.1101/gad.5.7.1224. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg H B, Valdesuso J, Kapikian A Z, Chanock R M, Wyatt R G, Szmuness W, Larrick J, Kaplan J, Gilman H, Sack D A. Prevalence of antibodies to the Norwalk virus in various countries. Infect Immun. 1979;26:270–273. doi: 10.1128/iai.26.1.270-273.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutiérrez-Escolano A L, del Angel R M. Nuclear proteins bind to poliovirus 5′ untranslated region. Arch Med Res. 1996;27:413–419. [PubMed] [Google Scholar]
- 17.Gutiérrez-Escolano A L, Denova M A, Racaniello V R, del Angel R M. Attenuating mutations in the poliovirus 5′ untranslated region alter its interaction with polypyrimidine tract-binding protein. J Virol. 1997;71:3826–3833. doi: 10.1128/jvi.71.5.3826-3833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahm B, Kim Y K, Kim J H, Kim T Y, Jang S K. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J Virol. 1998;72:8782–8788. doi: 10.1128/jvi.72.11.8782-8788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haller A A, Semler B L. Stem-loop structure synergy in binding cellular proteins to the 5′ noncoding region of poliovirus RNA. Virology. 1995;206:923–934. doi: 10.1006/viro.1995.1015. [DOI] [PubMed] [Google Scholar]
- 20.Hardy M E, Estes M K. Completion of the Norwalk virus genome sequence. Virus Genes. 1996;12:287–290. doi: 10.1007/BF00284649. [DOI] [PubMed] [Google Scholar]
- 21.Hellen C U, Witherella G W, Schmid M, Shin H S, Pestova T V, Gil A, Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc Natl Acad Sci USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunt S L, Hsuan J J, Totky N, Jackson R. Unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 1998;13:437–448. doi: 10.1101/gad.13.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang Y K, Brinton M A. A 68-nucleotide sequence within the 3′ noncoding region of simian hemorrhagic fever virus negative-strand RNA binds to four MA104 cell proteins. J Virol. 1998;72:4341–4351. doi: 10.1128/jvi.72.5.4341-4351.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito T, Lai M. An internal polypyrimidine-tract-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′ untranslated sequence. Virology. 1999;254:288–296. doi: 10.1006/viro.1998.9541. [DOI] [PubMed] [Google Scholar]
- 25.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang X, Wang M, Graham D Y, Estes M K. Expression, self-assembly and antigenicity of the Norwalk virus capsid protein. J Virol. 1992;66:6527–6532. doi: 10.1128/jvi.66.11.6527-6532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang X, Wang M, Wang K, Estes M K. Sequence and genomic organization of Norwalk virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- 28.Kaminski A, Hunt S L, Patton J G, Jackson R J. Direct evidence that polypyrimidine tract-binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA. 1995;1:924–938. [PMC free article] [PubMed] [Google Scholar]
- 29.Kapikian A Z, Wyatt R G, Dolin R, Thornhill T S, Kalika A R, Chanock R M. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J Virol. 1972;10:1075–1081. doi: 10.1128/jvi.10.5.1075-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaplan J E, Gary G W, Baron R C. Epidemiology of Norwalk gastroenteritis and the role of Norwalk virus in outbreaks of acute nonbacterial gastroenteritis. Ann Intern Med. 1982;96:756–761. doi: 10.7326/0003-4819-96-6-756. [DOI] [PubMed] [Google Scholar]
- 31.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak M. A short leader sequence impairs the fidelity of initiation by eucaryotic ribosomes. Gene Expr. 1991;1:111–115. [PMC free article] [PubMed] [Google Scholar]
- 33.Lai M M C. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 34.Lambden P R, Lui B, Clarke I N. A conserved sequence motif at the 5′ terminus of the Southampton virus genome is characteristic of the caliciviridae. Virus Genes. 1995;10:149–152. doi: 10.1007/BF01702595. [DOI] [PubMed] [Google Scholar]
- 35.Luz N, Beck E. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J Virol. 1991;65:6486–6494. doi: 10.1128/jvi.65.12.6486-6494.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meerovitch K, Svitkin Y V, Lee H S, Lejbkowics F, Kenan D J, Chan E K L, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmenberg A C. Sequence alignments of picornaviral capsid proteins. In: Semler B L, Ehrenfeld E, editors. Molecular aspects of picornavirus infection and detection. Washington, D.C.: American Society for Microbiology; 1989. pp. 211–241. [Google Scholar]
- 38.Pelletier J, Kaplan G, Racaniello V R, Sonenberg N. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol Cell Biol. 1988;8:1103–1112. doi: 10.1128/mcb.8.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pintó R M, Diez J M, Bosch A. Use of the colonic carcinoma cell line CaCo-2 for in vivo amplification and detection of enteric viruses. J Med Virol. 1994;44:310–314. doi: 10.1002/jmv.1890440317. [DOI] [PubMed] [Google Scholar]
- 40.Pintó R M, Robine-Leon S, Appay M D, Kedinger M, Triadou N, Dussaulx E, Lacroix B, Simon-Assmann P, Haffen K, Fogh J, Zweibaum A. Enterocyte-like differentiation and polarization of the human colon carcinoma cell line Caco-2 in culture. Biol Cell. 1983;47:323–330. [Google Scholar]
- 41.Prasad B V V, Rothnagel R, Jiang X, Estes M K. Three-dimensional structure of the baculovirus-expressed Norwalk virus capsid. J Virol. 1994;68:5117–5125. doi: 10.1128/jvi.68.8.5117-5125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rinehart-Kim J E, Zhong W M, Jiang X, Smith A W, Matson D O. Complete nucleotide sequence and genomic organization of a primate calicivirus, Pan-1. Arch Virol. 1998;144:199–208. doi: 10.1007/s007050050497. [DOI] [PubMed] [Google Scholar]
- 43.Roehl H H, Semler B L. Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3′ end of viral negative-strand RNA. J Virol. 1995;69:2954–2961. doi: 10.1128/jvi.69.5.2954-2961.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svitkine Y V, Meerovitch K, Lee H S, Dholakia J N, Kenan D J, Agol V I, Sonenberg N. Internal initiation on poliovirus RNA: further characterization of La function in poliovirus translation in vitro. J Virol. 1994;68:1544–1550. doi: 10.1128/jvi.68.3.1544-1550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villa-Komaroff L, Gutteman N, Baltimore D, Lodish H F. Complete translation of poliovirus RNA in a eukaryotic cell-free system. Proc Natl Acad Sci USA. 1975;72:4157–4161. doi: 10.1073/pnas.72.10.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White I J, Ball J M, Hardy M E, Tanaka T N, Kitamoto N, Estes M K. Attachment and entry of recombinant Norwalk virus capsid to cultured human and animal cell lines. J Virol. 1996;70:6589–6597. doi: 10.1128/jvi.70.10.6589-6597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Witherell G W, Gil A, Wimmer E. Interaction of polypyrimidine tract binding protein with the encephalomyocarditis virus mRNA internal ribosomal entry site. Biochemistry. 1993;32:8268–8275. doi: 10.1021/bi00083a030. [DOI] [PubMed] [Google Scholar]
- 48.Zeng L, Falgout B, Markoff L. Identification of specific nucleotide sequences within the conserved 3′-SL in the dengue type 2 virus genome required for replication. J Virol. 1998;72:7510–7522. doi: 10.1128/jvi.72.9.7510-7522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]