Abstract
The 165-kb circularized chromosome of Epstein-Barr virus (EBV) is replicated in latently infected cells once per cell cycle by host proteins during S phase. Replication initiates at multiple sites on latent EBV chromosomes, including within a 1.8-kb region called oriP, which can provide both replication and stabilization for recombinant plasmids in the presence of the EBV-encoded protein, EBNA-1. Replication initiates at or near the dyad symmetry component (DS) of oriP, which depends on multiple EBNA-1 binding sites for activity. To test the importance of the replication function of oriP, the DS was deleted from the viral genome. EBV mutants lacking the DS and carrying a selectable gene could establish latent infections in BL30 cells, in which circular, mutant viral chromosomes were stably maintained. Analysis of replication fork movement using two-dimensional gel electrophoresis showed that the deletion of the DS reduced the initiation events to an undetectable level within the oriP region so that this segment was replicated exclusively by forks entering the region from either direction. A significant slowing or stalling of replication forks that occurs normally at the approximate position of the DS was also eliminated by deletion of the DS. The results confirm the DS as both a replication origin and a place where replication forks pause. Since the replication function of oriP is dispensable at least in certain cell lines, the essential role of EBNA-1 for infection of these cell lines is likely to be that of stabilizing the EBV chromosome by associating with the 30-bp repeats of oriP. The results also imply that in established cell lines, the EBV chromosome can be efficiently replicated entirely from origins that are activated by cellular factors. Presumably, initiation of replication at the DS, mediated by EBNA-1, is important for the natural life cycle of EBV, perhaps in establishing latent infections of normal B cells.
The life cycle of Epstein-Barr virus (EBV) includes the latent infection of B cells which the virus induces to proliferate (20). During latent infection, the viral genes that support viral DNA replication for virus production are inactive, so in order for the 165-kb EBV chromosome to be maintained in a circular, autonomous form, it must be replicated by the host cell during each cycle of division. In latently infected cells, the circular EBV chromosome is replicated during S phase once, and only once, per cell cycle (1, 54), suggesting that its replication is governed by “licensing,” the mechanism that is believed to control initiation of replication for the nuclear chromosomes of all eukaryotes (26).
We do not yet have a clear understanding of the structure of replication origins of mammalian chromosomes. A common feature that has emerged from origin-mapping studies is the existence of initiation zones, which may span only a few kilobases, as in the case of the human β-globin locus (3), or as much as 55 kb, in the cases of the hamster dihydrofolate reductase (DHFR) gene origin (7) or the murine β-globin locus (L. H. Nguyen, O. Ermakova, and C. Schildkraut, unpublished data), or 30 kb for the human ribosomal RNA gene locus (31). It is not clear how many individual sites of initiation may contribute to an initiation zone, but it is likely that in all cases, certain sites will be preferred over others (23). EBV reflects this complexity by having a broad initiation zone as well as a discrete replication origin, termed oriP, both of which function during latent infection.
oriP is a 1.8-kb region of the EBV genome that was identified based on its ability to support the stable maintenance of recombinant plasmids that were introduced into cells (52), an activity which requires a single EBV-encoded protein, EBNA-1 (34, 55). At the time that oriP was given its name, it was not appreciated that replication alone would be insufficient to maintain plasmids in mammalian cells (6). oriP depends on two cis-acting components, the DS and the FR (38), that function in different ways (Fig. 1). The DS, named for a dyad symmetry within it, is a cluster of four EBNA-1 binding sites that functions as a replicator in the presence of EBNA-1 (16, 43, 53). On plasmids supported by oriP, replication appears to initiate at or near the DS (11). The FR, named for a family of repeats, contains 20 EBNA-1 binding sites within 20 copies of a 30-bp sequence. In the presence of EBNA-1, the FR prevents plasmids from being rapidly lost from mitotically active cells (2, 25), apparently by allowing them to be tethered by EBNA-1 to condensed human chromosomes during mitosis (44). In addition, interactions between EBNA-1 molecules bound to the FR and to the DS bring the two components of oriP together, forming a DNA loop (10, 45), and this might contribute to initiation at the DS, since the FR might contribute to the efficiency of replication under some conditions (38, 51).
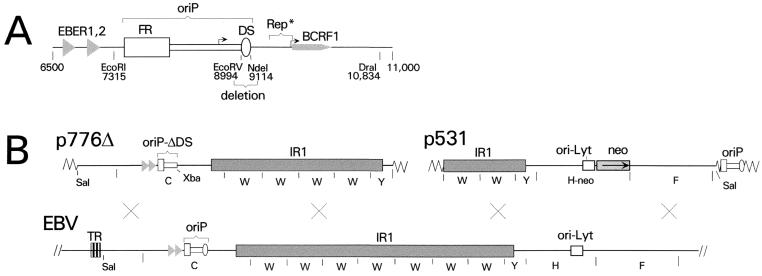
FIG. 1.
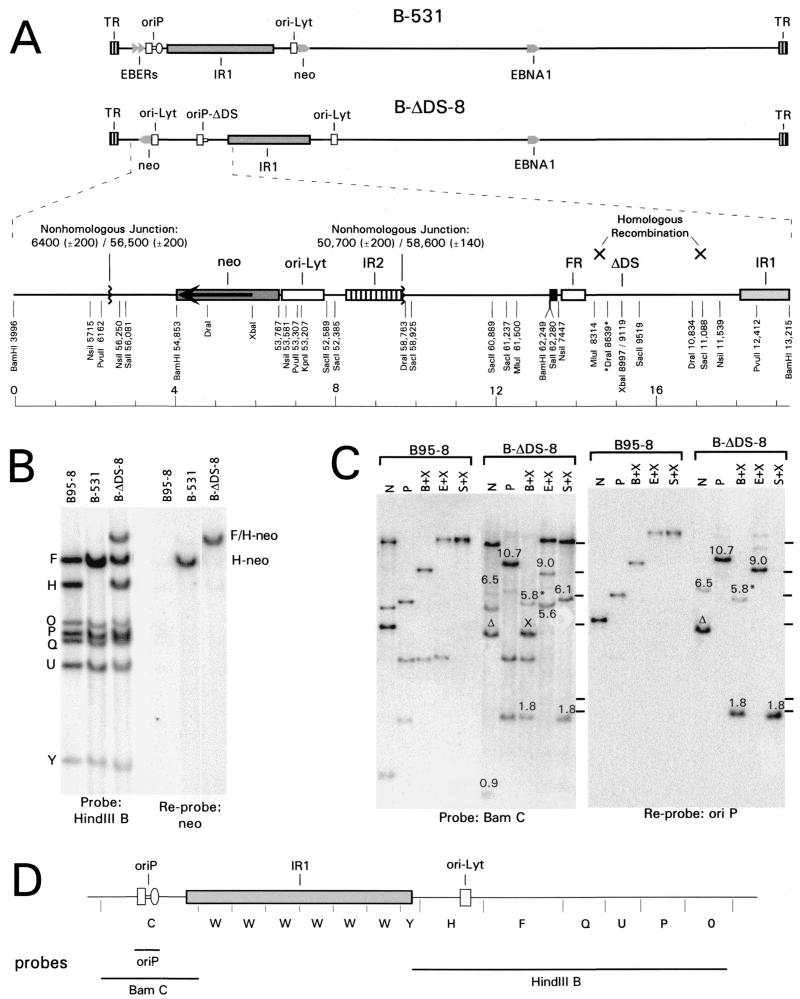
Deletion of the DS of oriP from EBV. (A) Map of the EBV chromosome in the vicinity of oriP, from position 6500 to 11000 (B95-8 coordinates). The two functional components of oriP, FR and DS, are indicated. The DS was deleted using the EcoRV and NdeI sites, as shown. Also indicated are the genes for EBER RNAs 1 and 2 and for the interleukin-10 homologue, BCRF1. Rep*, located between positions 9370 and 9668, is a region that was reported to have weak replication activity (22) and overlaps the proximal promoter of the BCRF1 gene. The two bent arrows indicate the BCRF1 gene promoters, which are active during lytic infection (27). (B) Linear maps of the plasmids p776Δ and p531 are shown above a map of the relevant part of the EBV chromosome. Homologous recombination in the regions indicated by crosses would transfer the DS deletion from p776Δ and the neo marker from p531 to the EBV chromosome. p776Δ contains 26 kb of EBV sequences, from a SalI site at position 644 to a BamHI site at position 48848, except that only four complete copies of the IR1 repeat (BamW segment) are present. The DS of oriP was deleted by removing 120 bp between EcoRV and NdeI sites (shown in panel A) and placing an XbaI linker at the site. BamHI restriction fragments C, W, and Y are indicated. The triangles represent the two EBER genes. The vector portions of the plasmids are indicated only by zigzag lines. p531 carries 21 kb of EBV DNA, from the BamHI site at position 40863 to the SalI site at position 62280, with the selectable CMVIE-neo construct (neo) replacing the nonessential EBV gene, BHRF1 (29). The BamHI fragments W, Y, H-neo, and F are indicated. oriP is also present, within the vector portion of the plasmid as indicated.
Analysis of the movements of replication forks on the EBV chromosome by using a two-dimensional (2D) electrophoretic technique revealed that oriP is only partly responsible for replicating the EBV chromosome. In contrast to small plasmids supported by oriP, where replication was shown to initiate at or near the DS essentially all of the time, on the EBV chromosome, oriP was found to be replicated passively most of the time (11, 32). For one EBV strain, Raji, which was investigated in the most detail, oriP was observed to be replicated primarily by forks progressing from the left, where a broad zone of initiation was located. Initiation was detected within every restriction fragment examined in a region extending leftward from oriP for 45 kb (32). As with initiation zones on human chromosomes, initiation at some regions occurred at higher frequencies than at others, but every region, like oriP, was replicated passively most of the time. It is not known how such large zones of initiation are determined, but for EBV, it is not likely to involve any viral protein directly. EBNA-1 is the only EBV protein that appears to be expressed in all latently infected cell lines, and a search for sites of EBNA-1 binding over the EBV genome (37) revealed only its sites at oriP and weaker-affinity sites at one distant locus, its autoregulated promoter (40, 41).
In this study, we asked the following: what might be the result of deleting the DS from the EBV genome? While the replication pattern of the EBV genome would suggest that the replication function of oriP is redundant, oriP might be the most active initiation site among partially active initiation sites in strains other than Raji (32). The DS has been conserved during evolution since the divergence between EBV and the related virus that infects baboons (33), indicating that it is important in the life cycles of these viruses. EBNA-1 has been shown to be essential in order for EBV to establish and maintain latent infection efficiently (28). Although this might be attributed to the plasmid stabilization function of the FR and EBNA-1, it could not be known without testing whether this maintenance function were itself a redundant feature of EBV.
Another issue that we sought to address is whether replication initiates at oriP on the EBV chromosome as part of a delocalized initiation region, in which several sites in the vicinity might contribute, or whether initiation at oriP and its vicinity is dependent upon the DS. In studies involving transient transfections with plasmids, relatively weak replication activity has been attributed to a short region named Rep* located close to the DS and just outside of oriP (22) (Fig. 1). The result is reminiscent of earlier studies of Calos and coworkers showing that most fragments of human DNA that are ∼10 kb and larger can be used to substitute for the DS of oriP to support rather stable replication of plasmids, on which replication appeared to initiate randomly within the cloned human DNA (24, 25). The results are most easily explained if a fairly large number of sites can each support initiation inefficiently in this context. This raised the question as to whether initiation of replication at oriP on the EBV chromosome might depend on the DS or, instead, on multiple sequences in the vicinity.
In this study, we found that the DS could be deleted from the EBV genome without appearing to hinder the ability of the virus to establish a latent infection. Thus, the essential role of EBNA-1 is likely to be that of stabilizing the EBV chromosome by acting in concert with the FR of oriP. Analysis of replication of the mutant EBV chromosomes using 2D gels showed that initiation of replication at oriP at significant levels requires the DS and provided additional clues as to how the EBV chromosome is replicated.
MATERIALS AND METHODS
Cell lines.
BL30 is an EBV-negative B-cell line derived from a Burkitt's lymphoma; it can be infected by EBV (9) but is not permissive for lytic EBV replication. P3HR1 clone 16 is a Burkitt's lymphoma cell line that carries a nontransforming EBV strain and in which lytic EBV replication can be induced (17). B95-8 is an EBV-transformed marmoset B-cell line that is semipermissive for EBV lytic replication (35). Cells were cultured in RPMI 1640 medium supplemented with 9% fetal bovine serum, penicillin, and streptomycin (Gibco-BRL). BL30 cells infected with EBV mutants conferring G418 resistance were grown in the presence of G418 (Gibco-BRL) at 1,700 μg/ml having a stated activity of 74%, for an effective concentration of 1,260 μg/ml. G418 was usually omitted during the last few doublings of cells before DNA was harvested.
Construction of p776Δ.
p776Δ (Fig. 1B) was constructed from pCΔJ, which contains 5.9 kb of EBV DNA between the EcoRI site (B95-8 coordinate, 7315), just to the left of oriP, and the BamHI site at the right end of BamC (13215). The plasmid was cut at this BamHI site and at a SalI site adjacent within the vector, and into it was inserted EBV DNA spanning four copies of BamW and BamY, excised from pBamW4Y (46) by cutting partially with BamHI and with SalI at its adjacent vector site, to construct p138.8. A ClaI linker was then inserted at the AatII site within the vector portion of p138.8, between the EcoRI side of the EBV insert and the bla gene; then, oriP was deleted between NdeI sites and replaced with an XbaI linker, yielding p775Δ. Next, p564, a plasmid built for a previous study (21) and which carries EBV DNA between the SalI site at position 644 and the SacI site at position 11088, was modified by introducing a ClaI linker into the vector adjacent to the SalI site and inserting an XbaI linker at the EcoRV site at position 8994, adjacent to the DS of oriP. The ClaI-XbaI fragment from this plasmid, containing EBV sequences 644 to 8994, was then inserted between the ClaI and XbaI sites of p775Δ to yield p776Δ.
Isolation of EBV mutants lacking the DS.
An amount of 8 × 106 P3HR1 cells was electroporated in 0.41 ml of complete growth medium with 24 μg of p776Δ mixed with 12 μg of p531 and 6 μg of pCMV-BZLF1 (15), the latter to induce lytic EBV replication, using a 0.4-cm chamber with a Bio-Rad Gene Pulser set at 220 V and 960 μF. The cells were cultured in 10 ml of medium for three days, and then the medium was collected, passed through a filter with a 0.4-μm pore size, and stored at 4°C. BL30 cells were infected in 48-well Falcon culture dishes by adding 50,000 cells, 25 μl of virus stock, and 0.3 ml of medium to each well. After 2 days, the medium was partially aspirated out of each well and replaced with 0.4 ml of medium containing 1,700 μg of G418 per ml (74% active; effective concentration, 1,260 μg/ml), and this was repeated two or three times at intervals of 2 days until all sensitive cells had died. In the two infections used in this study, G418-resistant clones emerged in 36 and 22 of the 48 wells. Similarly, to isolate mutants of B95-8 EBV, 107 cells were electroporated with the same plasmids in 0.45 ml of medium at 340 V and 500 μF. Cells were cultured for 4 days before medium was filtered. Volumes of 10 and 30 μl were used per well to infect BL30 cells, and drug-resistant clones emerged in 10 and 13 of 48 wells, respectively. G418-resistant BL30 clones were expanded in medium containing G418, and DNA was extracted from 0.5 × 106 to 1.0 × 106 cells by using sodium dodecyl sulfate and proteinase K (39), treated with phenol, and precipitated with ethanol after 20 μg of glycogen was added as carrier. A volume of 2% of each sample was analyzed by PCR using the primers 5′-GGGGGCGTCACCTGAAACCTTGTTTT and 5′-ACCGATAAGCGGACCCTCAAGAGGGC, with 5′ ends at EBV sequence positions 8758 and 9192, respectively, using Taq polymerase with 1.5 mM MgCl2 for 35 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 1.5 min. For mutants B-ΔDS-8 and P3-ΔDS-33, the PCR products carrying the DS deletion were purified from agarose gels and shown by DNA sequencing to be missing bases 8995 to 9114 with the XbaI linker sequence, GCTCTAGAGC, inserted. In the case of B-ΔDS-8, amplification was also done using a more distant left-side primer (5′-GTCGGCGTCCACTCTCTTTCCCCT, beginning at position 8440), which allowed the sequence to be determined across a novel DraI site at position 8636, which exists in this mutant due to the loss of the G at position 8637 of B95-8.
2D gel analysis of replication intermediates.
The procedures for enrichment of replication intermediates, 2D gel electrophoresis, and Southern analysis were essentially as described previously (30). Cells were harvested at densities no greater than 6 × 105/ml. Approximately 100 μg of EcoRI-digested nuclear matrix-associated DNA (8) was digested further with appropriate restriction enzymes and put through a BND cellulose column, and the entire caffeine eluate was applied to the first dimension of each 2D gel. DNA was transferred to Hybond N+ nylon membranes (Amersham) according to the manufacturer's recommendations and hybridized with specific probes as previously described (30).
For the experiment reported in Fig. 6B, cells were enriched for those within S phase by using centrifugal elutriation. Briefly, 3 × 109 to 7 × 109 cells were collected by centrifugation at 4°C, resuspended in 60 ml, passed through an 18-gauge needle to disrupt any aggregates, and injected into a Beckman JE-10X rotor spinning at 1,340 rpm with an initial flow rate of 30 ml/min. Fractions were collected at flow rates increasing by intervals of 5 ml/min, and cells were pelleted and frozen at −20°C, with aliquots taken to assess cell cycle position by fluorescence-activated cell sorter analysis of DNA content by propidium iodide fluorescence. Fractions corresponding to four intervals within S phase, from early to late, were analyzed for replication intermediates independently; replication intermediates were enriched in the early two S-phase fractions.
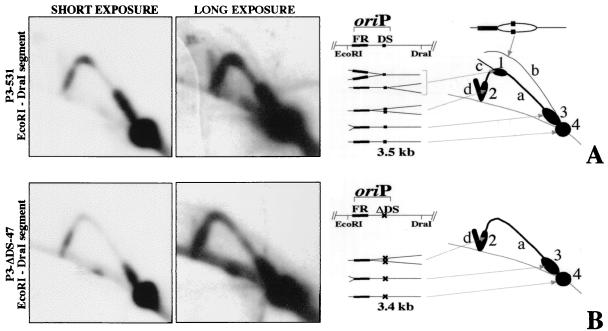
FIG. 6.
2D-gel analysis of replication in the vicinity of oriP on the EBV chromosome of strains P3-531 (A), in which oriP is wild type, and P3-ΔDS-47 (B), which lacks the DS of oriP. For the analysis (see Materials and Methods), DNA was digested with EcoRI and DraI and the membranes were probed with a 1-kb EcoRI-MluI segment (position 7315 to 8314) containing the FR. Short and long autoradiographic exposures are shown (left) together with diagrams summarizing the 2D gel patterns generated from replication intermediates (right). (A) With wild-type oriP (P3-531), four classes of replication intermediates can be observed: a, segments containing single replication forks generated from an external initiation site (Y arc); b, segments containing internal initiation sites (bubble arc); c, segments containing two opposing replication forks converging at the DS (this signal is barely detectable here but is stronger with other EBV strains); d, segments containing two opposing replication forks converging at the FR. As a result of fork pausing, replication intermediates accumulate at positions on the arcs to produce the spots indicated as 1, 2, and 3 in the diagram. Spot 1 is generated by forks coming from outside the EcoRI-DraI segment and pausing at the DS. Since the DS is in the middle of the segment, the two spots that would derive from the pausing of forks entering from either end of the segment overlap. Spot 2 is generated from forks entering from the right side of the oriP EcoRI-DraI segment and pausing at the FR. Spot 3 is generated from forks entering from the left and pausing at the FR. Finally, the EcoRI-DraI segment that does not contain a replication fork yields the 1N spot (spot 4 in the diagram). (B) With P3-ΔDS-47, which lacks the DS, only two classes or replication intermediates are visible, a and d. For P3-531, oriP is of the parental P3HR1 strain and the FR appears to be 200 to 300 bp longer than it is for P3-ΔDS-47 and the other DS mutants, for which the size of the FR matches that of strain B95-8, which was the source of oriP DNA used to generate the mutants. This could explain why the pauses along the Y arc (spots 2 and 3) appear longer with P3-531 than with P3-ΔDS-47.
RESULTS
Isolation of mutants of EBV strain P3HR1 that lack the DS.
A 120-bp deletion removing all four EBNA-1 binding sites of the DS of oriP was built into a 32-kb plasmid, p776Δ, which includes 27 kb of the EBV genome in the vicinity of oriP (Fig. 1). We tested whether the DS deletion could be transferred from p776Δ to the EBV chromosome by homologous recombination to yield mutant EBV capable of establishing a stable latent infection. To do this, p776Δ was introduced into P3HR1 clone 16 cells, which carry the inducible EBV strain P3HR1 along with a second plasmid, p531, which carries an overlapping region of the EBV genome with a CMVIE-neo (G418 resistance) gene construct replacing a nonessential EBV gene, BHRF1 (Fig. 1). EBV recombinants that have acquired the G418 resistance gene from p531 are phenotypically normal in culture and give rise to G418-resistant, latently infected cell clones after infection of susceptible cells (29). Virus released from the transfected P3HR1 cells was used to infect BL30 cells, which were then grown in multiwell plates in the presence of G418. We expected that a detectable fraction of the EBV chromosomes that had acquired the G418 resistance marker by recombination with p531 would also have acquired the deletion at oriP by recombining with p776Δ, based on previous studies involving recombination between EBV and transfected plasmids (49, 50).
To test for the presence of EBV chromosomes carrying the DS deletion, G418-resistant BL30 cells from 38 individual wells were expanded, and their DNA was analyzed by PCR using primers that flank the DS (Fig. 2A). Five clones appeared to carry only EBV chromosomes harboring the deletion within oriP, and 26 clones contained only intact oriP. In the remaining seven cases, both wild-type and mutant oriP genes were detected, indicating either that two infected clones had emerged in one well, that a clone was coinfected with parental P3HR1 virus, or that oriP DNA including the DS deletion was transferred onto the EBV chromosome without replacing the normal DS region. Despite some numerical uncertainty resulting from the mixed cases, it could be estimated that roughly one-sixth of the recovered virus chromosomes that had acquired the selective gene by recombining with p531 had also lost the DS at oriP by recombining with the nonselected plasmid, p776Δ, and were able to establish stable latent infections. Frequencies similar to this (49, 50) and much lower than this (56) have been reported for the introduction of neutral mutations into the P3HR1 genome by homologous recombination from a plasmid when selecting for incorporation of a selectable gene from a different plasmid. This indicated that loss of the DS did not greatly impair the ability of EBV to establish a stable, latent infection in BL30 cells.
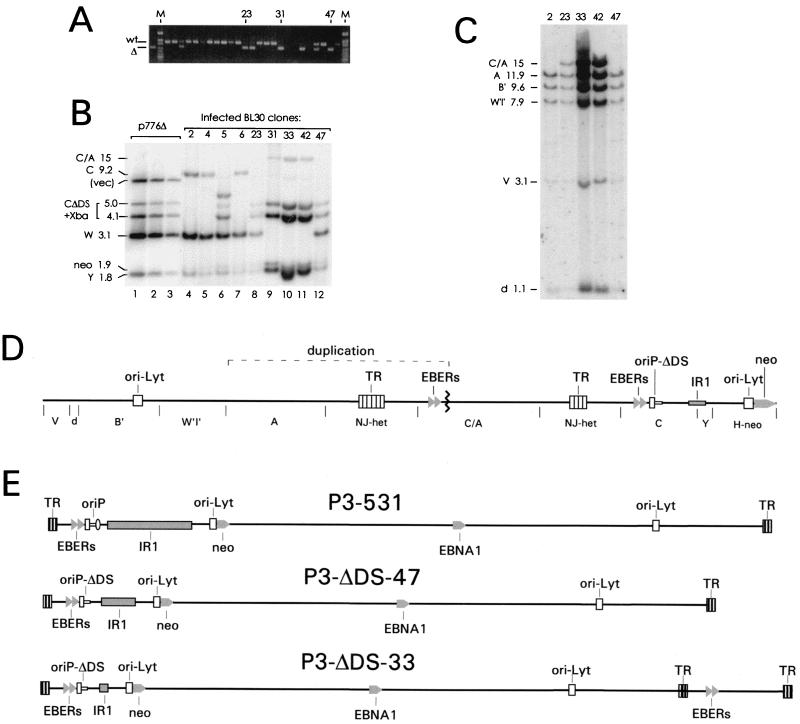
FIG. 2.
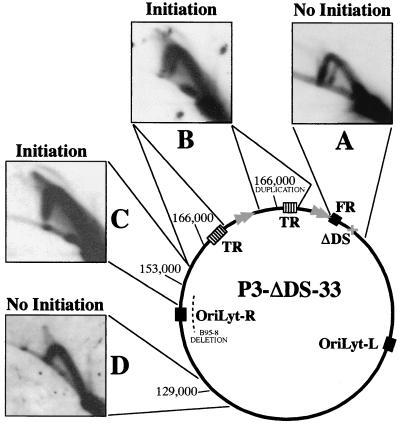
Isolation and characterization of mutants of EBV strain P3HR1 which lack the DS of oriP. (A) Screening BL30 clones by PCR for EBV mutants lacking the DS. An agarose gel stained with ethidium bromide is shown. A 435-bp DNA containing the DS was amplified from clones with wild-type oriP (wt), and a 315-bp product was detected with mutants carrying the deletion (Δ), as indicated. The leftmost lane shows a control PCR amplification of 1 pg of p776Δ DNA. Some clones that were studied further are indicated. Lane M, size standards. (B) Southern analysis of the BamC region of several clones. A volume of 5 μg of DNA from each clone was digested with BamHI plus XbaI. The probe was labeled p138.8, which contains EBV DNA extending from the EcoRI site at the left of oriP through four copies of BamW to the end of BamY (Fig. 1B). The 1.9-kb fragment containing the neo gene was detected partially by the probe (p138.8) which also contains the neo coding region. The fragments and their sizes (in kilobases) are indicated at the left. With five clones, the 9.2-kb BamC fragment was cleaved by XbaI to yield 5.0- and 4.1-kb bands (CΔDS + Xba), revealing the presence of the XbaI site at the DS deletion (lanes 8 to 12). Clone 5, which contained both normal and DS-deleted oriP genes based on PCR analysis, contained an additional band, most likely reflecting an insertion of sequences from one of the plasmids into the EBV chromosome. In lanes 1 to 3, 200, 100, and 25 pg of p776Δ, corresponding to 8, 4, and 1 copy per diploid human genome equivalent, respectively, were analyzed for comparison. DNA from clones 23 and 31 was slightly underloaded, and DNA from clone 33 was somewhat overloaded. (C) Southern analysis of the right end of the EBV genome of several clones. A volume of 5 μg of DNA from each clone was cut with BamHI, and the blot was probed with the 19-kb HindIII fragment D of B95-8, which detects BamHI fragments A, B′, W′I′, V, and d of strain P3HRI, as indicated. (D) A map showing 87 kb of the circular chromosome of mutants, such as P3-ΔDS-33, which carry a 26-kb duplication spanning the TR junction. The BamHI fragments are indicated below the line. The duplication involved joining (at the zigzag line) the leftmost 3 kb of BamC, including both EBER genes, to BamA near its left end, creating a 15-kb BamC/A fragment. (E) Structures of the chromosomes of three EBV strains derived from P3HR1, shown in their linearized forms. In the generation of each virus, recombination with p531 (Fig. 1B) introduced the G418 resistance gene (neo) adjacent to the left copy of oriLyt and at the same time restored the sequences to the right of IR1 that are missing in P3HR1. P3-ΔDS-47 and P3-ΔDS-33 each acquired the 120-bp deletion of the DS by recombining with p776Δ. P3-ΔDS-47 has only two complete copies of the IR1 repeat (BamW), while P3-ΔDS-33 has only a partial copy of the IR1 repeat (no copies of BamW). P3-ΔDS-33 also has the 26-kb duplication spanning the TR junction.
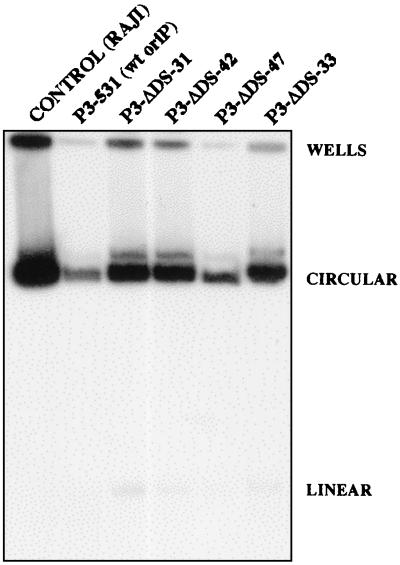
The EBV genomes of the five clones that lacked the DS were shown by Southern analysis to have the expected DNA structure in the vicinity of oriP. An XbaI linker that was placed at the site of the DS deletion allowed the 9.2-kb BamC fragment to be cut by XbaI, yielding fragments of 5.0 and 4.1 kb (Fig. 2B, lanes 8 to 12). All five clones also were found to carry the G418 resistance gene at the expected location in the EBV genome (data not shown). The EBV chromosomes of each of the clones were shown to be circular by electrophoresis into in situ lysis gels, as shown for four of the DS-deleted mutants in Fig. 3. The EBV mutants that lack the DS are designated with P3-ΔDS- followed by a number designating each clonal isolate; those that acquired only the G418 resistance gene (from p531) and were not altered at oriP are designated P3-531.
FIG. 3.
Autonomous maintenance of the circular chromosome of EBV mutants lacking the DS of oriP. BL30 clones carrying P3HR1 derivatives, 106 cells each, were lysed in the wells of a horizontal agarose gel as previously described (12). During electrophoresis, circular EBV chromosomes migrate more slowly than linear chromosomes, as indicated. The Southern blot shown was probed with radiolabeled EBV sequences from position 109802 (SacI) to 110760 (XbaI). Raji cells, which carry approximately 60 circular copies of the EBV chromosome per diploid human genome, were included for comparison.
The structure of the entire viral genome of each of the five P3-ΔDS mutants was examined by Southern analysis, and two unexpected differences were found. First, it was clear from the Southern analysis shown in Fig. 2B that the mutants that lacked the DS (lanes 8 to 12) also had fewer copies of the 3.1-kb BamW fragment, which is cleaved by BamHI from the internal repeat 1 (IR1) repeat array. Clones 31, 33, and 42 lacked BamW entirely and thus retained less than one complete copy of the IR1 repeat. Clones 23 and 47 had only two copies of BamW per viral chromosome, based on relative hybridization intensity and the size of restriction fragments spanning IR1 (data not shown). It is not clear why the recombination strategy that was used to introduce the DS deletion might have favored a reduction in the number of IR1 repeats. However, Yoo et al., who used a similar strategy to delete a promoter element near oriP from strain P3HR1, found that their recombinants contained only two or three copies of BamW (56). In that study, the EBV mutants were isolated by immortalizing human B cells, and it is likely that some BamW repeats are required in order for EBV to immortalize B cells.
The other difference was the presence in four of the five mutants of a duplication of 25 kb of the EBV genome that included the terminal repeat (TR) junction (Fig. 2D). At the time the mutants were being made, we were unaware that this duplication was present in most of the EBV genomes carried in the particular passage of P3HR1 clone 16 cells that was used. The duplication involved joining the leftmost 3.7 kb of the BamC segment to the left end of the BamA segment, resulting in a 15-kb junction segment designated BamC/A and shown in Fig. 2D. By Southern analysis, the BamC/A fragment was detected strongly by a probe covering 20 kb near the right end of the EBV genome (Fig. 2C), as well as by a probe made from the 3.1-kb EcoRI fragment that is just to the left of oriP (not shown), and it was detected weakly by the probe used in Fig. 2B, which included DNA from the right end of BamC extending past oriP to the EcoRI site at position 7315. The region containing the BamC/A junction was amplified by PCR and sequenced. Nucleotides at the positions equivalent to 7663 and 154913 of the B95-8 sequence form the junction. The duplication thus includes about one-third of the 30-bp repeats of oriP and contains seven EBNA-1 binding sites. What appears to be the same duplication, in that it contained the left end of BamC, was detected in P3HR1 EBV by Yoo et al. (56).
Three of the mutants, P3-ΔDS-31, -33, and -42, were maintained at the level of 20 EBV chromosomes per human diploid genome, compared to only 3 or fewer for the two other mutants lacking the DS or for the clones with wild-type oriP (Fig. 2B and C and data not shown). Clones 31, 33, and 42, which lack all copies of BamW, might have accumulated to higher-copy-number levels in BL30 cells because of the elimination of EBV gene expression that is unfavorable to the cells with a higher number of EBV genome copies. The 25-kb duplication, which these three clones also carry, is unlikely to have affected the copy level of the EBV genome. Clones P3-ΔDS-23 and -47 each have two copies of BamW and were each maintained at a low copy level, and clone 23 contains the 25-kb duplication while clone 47 does not (Fig. 2B and C). The absence of the DS itself had no apparent effect on the number of copies to which the EBV chromosome accumulated under selection in BL30 cells.
The fact that the duplication usually accompanied a large reduction in size of IR1 and was never seen otherwise is easily explained by the known size constraints under which the EBV chromosome is packaged into its viral capsid. The parental P3HR1 chromosome carrying the 25-kb duplication should be too large to be packaged efficiently (4), but because the duplication includes the TRs, which carry the processing and packaging signals (14), the duplication should be removed during packaging. However, for EBV recombinants that have lost most copies of BamW repeats, inclusion of the duplication should be favored during packaging to restore the chromosome size (4).
Deletion of the DS from strain B95-8.
In a comparison of DNA replication among several EBV strains by 2D gel analysis, strain B95-8 showed the most initiation at oriP (32). The B95-8 strain lacks 12 kb of sequence that overlap the region where the strongest initiation outside of oriP was detected in this study. Because of this, we reasoned that DS-deleted mutants of B95-8 might have a more severe phenotype (if any) than the P3HR1 mutants. We attempted to delete the DS from strain B95-8 by using the strategy that had succeeded with P3HR1. Of 22 G418-resistant BL30 clones that were screened, 5 clones had acquired the DS deletion, but 4 of the clones also carried a copy of intact oriP and through Southern analysis were found to have acquired additional sequences due to the insertion of one or both of the plasmids into the viral genome. One clone carried only oriP lacking the DS and did not contain significant vector sequences. The detailed structure of this EBV isolate, called B-ΔDS-8, was determined by Southern analysis and is presented in Fig. 4.
FIG. 4.
The chromosome structures of EBV strains B-531 and B-ΔDS-8 shown linearly (A), Southern analysis (B and C), and probes used (D). (A) B-ΔDS-8 acquired the DS deletion (ΔDS) at oriP by homologous recombination with p776Δ (Fig. 1B), but it acquired the G418 resistance gene and EBV sequences flanking it on p531 (Fig. 1B) through a complex insertion to the left of oriP. An expanded map of this region of its chromosome is shown. The EBV DNA to the left of oriP in B-ΔDS-8 is from the BamF region of the EBV genome, in the same arrangement as in p531 (Fig. 1B), separated from oriP by 124 bp of vector sequence, indicated by the small black box. This indicates that p531 recombined with EBV at oriP to the left of the DS or with p776Δ at oriP before the joined plasmids recombined with EBV. The G418 resistance gene and some flanking sequences of p531 then were inverted and inserted into the EBV chromosome through two nonhomologous events, as indicated. The map was confirmed by Southern analysis for every restriction site that is shown along with its inferred EBV sequence position. (B) A Southern blot analysis detecting the BamHI restriction fragments in the region of the EBV chromosome to the right of IR1, which was probed using the 29-kb HindIII B fragment (left) and reprobed with the neo gene (right). With B-531, the 6.0-kb BamH fragment was replaced by a 7.6-kb H-neo fragment (comigrating with the 7.5-kb BamF fragment). With B-ΔDS-8, the BamH fragment was unaltered, but the neo insert was present on a rearranged, 9.4-kb F/H-neo fragment. (C) Southern analysis of B-ΔDS-8 in the vicinity of oriP, compared to B95-8. DNAs were digested with NsiI (N), PvuII (P), and XbaI (X) plus either BamHI (B), EcoRI (E), or SalI (S). The blot was probed with the BamC fragment of EBV (position 3994 to 13215) (left) and then reprobed with the oriP region of p531 (a 1.8-kb SalI-EcoRV fragment) (right). Novel junction fragments, detected with B-ΔDS-8 but not with B95-8 and which verify the map shown in panel A, are indicated by their sizes (in kilobases). Δ, the NsiI fragment of B-ΔDS-8 that includes the DS deletion; X, the 4.1-kb BamHI-XbaI fragment that results from the XbaI site introduced at the DS deletion; 5.8*, the 5.8-kb BamHI fragment in the same lane which resulted from incomplete digestion of the sample by XbaI. For panels B and C, 5 μg of DNA from BL30 cells carrying either B-531 or B-ΔDS-8 or 0.2 μg of B95-8 DNA was analyzed. (D) BamHI restriction map of part of the EBV genome and the probes that were used in panels B and C.
During the generation of B-ΔDS-8, the DS deletion was transferred from p776Δ to the EBV chromosome at oriP in the expected manner by homologous recombination. However, the G418 resistance gene was not transferred from p531 to the EBV chromosome by homologous recombination at the expected position to the right of IR1 repeats (Fig. 1 and 4). Instead, recombination occurred between a copy of oriP that had been placed within the vector portion of p531 (for reasons unrelated to this study) and oriP of the EBV chromosome (or oriP of p776Δ before it recombined with the EBV chromosome). Several kilobases of EBV sequences from p531 that surround the selective marker were then inverted and recombined with the EBV chromosome near coordinate 6400 (±200 bp), resulting in the replacement of about 1,100 bp of sequence to the left of oriP, including the two EBER genes. Deletion of the EBER genes, which encode two small, abundant RNAs, was found previously to have no apparent effect on EBV infection (47, 48). B-ΔDS-8 was maintained in BL30 cells at approximately five circular copies per cell, as was B-531, a derivative of B95-8 that acquired the selective marker from p531 by homologous recombination and has wild-type oriP (data not shown).
EBV chromosomes that lack the DS are maintained stably without selection.
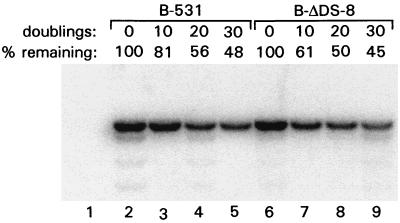
To test how stably the recombinant EBV genomes lacking the DS were being maintained, BL30 cells carrying B-ΔDS-8 or B-531 were grown in the absence of G418. After 10, 20, and 30 doublings of the cell populations, DNA was extracted to determine relative EBV genome levels. The EBV genomes carrying or lacking the DS were lost slowly from cells at similar rates, with just under half of the genomes remaining within each population after 30 doublings (Fig. 5), corresponding to exponential loss rates of less than 3% per doubling. BL30 cells carrying one of the P3HR1-derived mutants, P3-ΔDS-33, were grown for 2 months without drug selection or for over 80 population doublings, during which time the EBV genome decreased in copy level by less than 50% (data not shown). The results indicate that the EBV chromosome can be replicated very efficiently in the absence of the DS of oriP.
FIG. 5.
Comparison of the rates of loss of EBV chromosomes containing or lacking the DS. DNA was isolated from BL30 cells carrying either B-531 or B-ΔDS-8 after 0, 10, 20, and 30 doublings of the cell populations in the absence of G418. A volume of 5 μg of DNA from each sample was cut with EcoRI plus DraI and analyzed by Southern blotting. Staining of the gel by ethidium bromide revealed that equal amounts of DNA were loaded into each lane. A 3.1-kb EcoRI-DraI fragment was detected by probing with EBV sequences from position 125316 (EcoRI) to 125412 (EcoRV). The hybridization signals were quantitated using a beta imager (Molecular Dynamics) and are presented above each lane relative to the signal at 0 doublings. The weaker bands, most prominent in lanes 2, 4, and 9, most likely arose from cutting at “star” sites and were included in the quantitation. Lane 1, DNA from uninfected BL30 cells.
From these data, we conclude that the DS is not required for stable replication and episomal maintenance of the EBV chromosome during latent infection of BL30 cells.
Characterization of the replication of P3HR1-derived EBV with and without the DS.
To determine whether replication initiates at oriP in the absence of the DS, we examined the structure of the replicating molecules using 2D gel analysis. This method electrophoretically resolves different types of branched DNA molecules from each other and from linear ones based on their structure and size (5). We have applied the method to EBV in the past and have identified at oriP structures that arise from the following: initiation of replication at or near the DS, the “passive” replication of oriP by replication forks entering from either direction, the pausing or stalling of forks at the FR and the DS of oriP, and the convergence of forks into termination structures (11, 32). The right part of Fig. 6 depicts the electrophoretic patterns of the 2D gels and the structure of the branched DNA molecules corresponding to each pausing site (see the figure legend for a detailed description).
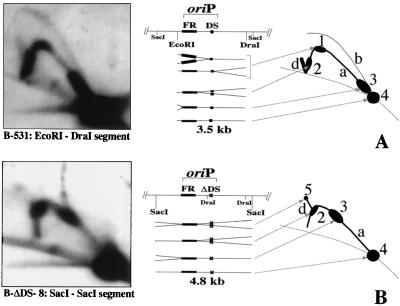
In Fig. 6, a 2D gel analysis of replication at oriP is shown for a derivative of P3HR1, P3-531, in which oriP is wild type, and for a similar virus, P3-ΔDS-47, which lacks the DS. In both cases, the number of copies of viral chromosomes per cell was low, at about three copies per diploid human genome (Fig. 2B and 3). A 3.5-kb restriction fragment, produced by cleavage with EcoRI and DraI, was chosen for analysis because the DS is at its center. The prominent arc generated by DNA molecules containing a single fork (the “Y arc”) indicates that most of the time oriP is replicated passively by replication forks that enter from either direction, as was found to be the case with most EBV strains previously (32). With P3-531, which has wild-type oriP, a weaker “bubble arc” is apparent from the longer exposure of the autoradiogram, indicating the movement of replication forks away from one or more points of initiation within the DNA segment. (A very faint arc is also detectable in the short-exposure film but it is not visible in its copy.) Because the bubble arc is full length, it reveals initiation of replication within the central one-third of the restriction fragment, presumably at the DS. A bubble arc could not be detected with P3-ΔDS-47, indicating that the DS is either important or required for initiation of replication at oriP. The experiment reported for P3-531 was repeated twice with two different matrix preps. In both cases a clear bubble arc is visible. The P3-DS-47 assay was repeated three times with three different matrix preps. Two matrix preps were made from asynchronous populations of cells while a third matrix prep was made starting from elutriated cells, pooling the early S and the late S fractions separately and running two different 2D gels. Figure 6B shows the result of the blot obtained from the early fraction of the elutriated cells, but in all cases bubble arcs were not detectable. The experiment with elutriated cells was performed in order to have a signal for P3-ΔDS-47 of intensity sufficiently high to exclude initiation events at oriP.
Intense regions of the Y arcs shown in Fig. 6 reflect the accumulation of replicating molecules for which the fork moves slowly or stalls, at the FR or at the DS of oriP (11, 32). These are most clearly appreciated by viewing the shorter exposures in the left panels. The accumulation of forks at the apex of the Y arc indicate pausing at or near the DS in the case of P3-531. With P3-ΔDS-47, this pause was virtually eliminated, implicating the DS in causing replication forks to stall.
This experiment strongly suggests that the DS element is required to specify initiation of DNA replication within oriP and, as a consequence, that initiation of DNA replication within oriP is not required for EBV infection and stable replication of the EBV genome in BL30 cells.
Characterization of the replication of a DS-deleted mutant of P3HR1 EBV maintained at a high copy number.
We also examined the replication of a DS-deleted mutant (P3-ΔDS-33) that lacked the BamW repeats. Because this mutant was maintained in BL30 cells at 20 to 30 copies per cell, stronger signals could be obtained from replication intermediates on 2D gels. As shown in Fig. 7A, an intense Y arc was apparent at oriP, without any detectable bubble arc. (A weaker Y arc of a smaller size seen in Fig. 7A corresponds to the 2.9-kb EcoRI-DraI junction fragment of the duplication, described above. It was detected because about 350 bp at the EcoRI end of the fragment is from the BamC side of the junction and overlaps the EcoRI-MluI fragment oriP that was used as a probe.) Because of the high number of copies of the EBV chromosome in this strain, replication intermediates were much easier to detect than with P3-531 and P3-ΔDS-47 (Fig. 6), as is apparent by comparing the intensities of Y arcs at the region where the forks are not stalled (the less intense region on the right half of the arcs.) Since a bubble arc was readily detected with P3-531 (Fig. 6), revealing initiation at or near the DS on some EBV chromosomes, initiation at oriP should have been detected with P3-ΔDS-33 (Fig. 7) even if it occurred at only a small fraction of the frequency that applies when the DS is present. We conclude from this that the DS is required for replication to initiate at oriP at any significant frequency, at least in strain P3HR1.
FIG. 7.
2D gel analysis of DNA replication at different regions of the EBV chromosome of the strain, P3-ΔDS-33, which lacks the DS of oriP. (A) A 3.5-kb segment between EcoRI and DraI sites, centered at the DS deletion of oriP, showed no evidence of initiation. (B) The EcoRI fragment (∼13 kb) spanning the TR junction, duplicated in this strain, with one copy containing two fewer 0.5-kb TR units due to a cycle of cleavage during packaging in P3HR1 cells and recircularization upon infection of BL30 cells. This fragment, its center near B95-8 coordinate 166000, produced a bubble arc, indicating initiation. (C) A 14-kb XbaI-EcoRI segment, centered near B95-8 position 153000, with its left half within the region that is deleted from B95-8, also produced a bubble arc. (D) A 7.8-kb EcoRI-XbaI centered near position 129000 did not reveal initiation. Probes for hybridization were as follows: A, DraI-MluI (position 7335 to 8314); B, the 5.3-kb BamI′ segment of strain AG876 (36); C, HpaI-XbaI (position 131959 to 133151); D, EcoRI-XbaI (position 125316 to 133151).
As with P3-ΔDS-47, there appeared to be little or no pausing at the position of the DS deletion in P3-ΔDS-33, while replication pausing at the FR suggests that, also in this case, oriP is replicated passively by replication forks that enter from either direction. This was more apparent with shorter exposures of the autoradiogram (data not shown).
Initiation of replication within the Raji EBV genome was most readily detected within a 14-kb EcoRI-XbaI fragment that is centered 18 kb from the TRs (32). We analyzed this fragment of P3-ΔDS-33 and detected initiation (Fig. 7C). We also detected initiation closer to the terminal repeats within a nearby XbaI-EcoRI fragment (Fig. 7B). Because this region of the P3-ΔDS-8 was duplicated and one TR junction had two more 0.5-kb terminal repeats than the other, a double Y arc was detected (Fig. 7B). It is worth noting that the bubble arcs detected (Fig. 7B and C) show a stronger signal in the upper part of the arc and a weaker signal (that practically disappears) at the lower part of the arc approaching the unreplicated restriction fragment (the 1N spot). This is the expected pattern for delocalized initiation events (42) in which replication initiated at different sites within the same restriction fragment. In this case, each initiation site generated a slightly different bubble arc that overlapped only in the upper part of the arc as a result of the “compression effect” that occurs in the gel for the higher-molecular-weight molecules. This confirms the presence of a delocalized initiation region in a P3HR1-derived clone as was previously described for EBV in Raji cells (32).
On the other hand, not all of the restriction fragments examined with P3-DS-33 showed initiation events. Initiation of DNA replication was not detected in a 7.8-kb EcoRI-XbaI segment upstream from the delocalized initiation region (D) where termination events were instead detected. Since the oriP region is replicated passively by replication forks that enter from either direction and that termination events were detected at specific sites (at the FR) or at random sites within all the other restriction segments analyzed (B, C, and D), it is possible that further initiation sites might be present in the EBV genome region between oriP and the 7.8-kb EcoRI-XbaI segment.
All gels shown in Fig. 7 were made from the same matrix prep. B, C, and D represent rehybridization of the same filter after stripping of the probe.
The results obtained for P3-DS-33 confirm that the DS is required to specify initiation of DNA replication within oriP. The results also show that a wide delocalized initiation zone exists in P3HR1-derived EBV, similar to what was observed with Raji EBV, which suggests that initiation sites might also be present in other regions of the EBV genome.
Characterization of the replication of B95-8 derived EBV clones with or without the DS.
With the B95-8 strain of EBV, initiation was observed to occur more frequently at oriP than it does with other EBV strains (32), as discussed above. The same higher frequency of activation was also detected for the B95-8 derivative, B-531, carried in BL30 cells. In the 2D gel analysis of replication at oriP of B-531 shown in Fig. 8, a bubble arc was clearly visible and closer in intensity to that of the Y arc than was the case with the comparable strain derived from P3HR1, P3-531, as seen in Fig. 6A. (Comparisons should be made between the bubble arc and the part of the Y arc that lacks pause sites, because replication fork pausing is not observed in a bubble arc as long as one fork is free to move; when one fork moves past the restriction site used to define the segment, the bubble structure will convert to a paused Y structure.)
FIG. 8.
DNA replication at oriP of B95-8-derived strains B-531, which contains wild-type oriP, and B-ΔDS-8, which lacks the DS. The results of 2D gel analysis are shown for each (left), along with a diagram summarizing the gel patterns generated by different replication intermediates (right). (A) For B-531, a 3.5-kb EcoRI-DraI segment was analyzed as performed for P3-531 (Fig. 6). With oriP intact, the same replication intermediates as those shown in Fig. 6 were observed, but note the increased initiation activity at oriP, indicated by the higher ratio of intensity of the bubble arc (b) to the Y arc (a). (B) For B-ΔDS-8, a 4.8-kb SacI segment was analyzed. Initiation was not detected. Spots 2 and 3, reflecting pausing at the FR by forks moving in each direction, were shifted to higher positions on the Y arc because the FR is closer to the center of the SacI fragment.
Comparable analysis of the B95-8 derivative B-ΔDS-8 did not reveal a detectable bubble arc (Fig. 8, right). In the process of this analysis it was learned that B-ΔDS-8 contains a novel DraI site 358 bp from the DS deletion due to a deletion of a single base pair at B95-8 coordinate 8637 (Fig. 4A). This prevented us from using the same EcoRI-DraI restriction fragment that had been used in the 2D gel analysis of the other strains, so instead, we analyzed a 4,800-bp SacI fragment that is roughly centered over oriP and which includes the EcoRI-DraI segment used in the analysis of B-531. The somewhat larger SacI fragment allowed us to exclude initiation of DNA replication within a 2-kb region surrounding the site of the DS deletion. The right part of Fig. 8 depicts the structure of the two segments analyzed, the electrophoretic patterns of the 2D gels, and the structure of the branched DNA molecules. The intense regions of the Y arc, which are due to the pausing of forks moving in either direction at the FR of oriP, appear closer to the apex of the Y arc of the SacI fragment of B-ΔDS-8 rather than at the bases of the arc of the EcoRI-DraI fragment. This is because the FR of oriP is located between 1,250 and 1,850 bp from the left end of the SacI fragment, rather than just 150 to 750 bp from the left end of the EcoRI-DraI fragment.
The central one-third of the SacI fragment of B-ΔDS-8 extends well into the FR at the left and 400 bp past the DS at the right. Thus, if replication initiated anywhere within oriP or within several hundred bases to the right of it at any significant frequency in this DS-deleted strain, a bubble arc should have been seen. Since oriP is used more frequently for initiation in B95-8 than in other EBV strains, as previously mentioned, if initiation could occur within or near oriP in the absence of the DS, we would have been most likely to detect it in this strain. Therefore, the replication initiation function of oriP does not appear to be required even by the B95-8 strain in order for its chromosome to be replicated efficiently during latent infection.
DISCUSSION
The main conclusion of this work is that the circular chromosome of EBV can be replicated stably during latent infection in the absence of the replication function of oriP, its only characterized replication origin that functions during latent infection. EBV mutants lacking the DS of oriP were readily isolated within latently infected clones of BL30 cells, where the mutant viral chromosomes were maintained autonomously. 2D gel analysis of the replication of the mutant EBV chromosomes showed that the DS provides the replication initiation function of oriP in the context of the viral chromosome. In the absence of the DS, oriP was found to be replicated entirely passively on the EBV chromosome by forks that move through the region in either direction.
The EBV-encoded protein EBNA-1 is essential for the functions of both components of oriP, the replication function of the DS and the episome maintenance function of the FR (see Introduction). EBV mutants lacking a functional EBNA-1 gene were shown in a previous study to be incapable of establishing latent infections in BL30 cells (28). Since the results of this study indicate that the DS is dispensable for replication of the latent EBV chromosome in BL30 cells, the essential function of EBNA-1 is likely to be that of stabilizing the EBV episome through its interaction with the FR of oriP.
The replication initiation function of oriP is dispensable for latent infection of BL30 and other established cell lines.
The previous 2D gel analysis of EBV replication during latent infection by Little and Schildkraut revealed that replication initiates at oriP only a fraction of the time (32), so it was not unreasonable to expect that the replication function of oriP might be dispensable under some experimental conditions. For the present study, EBV mutants lacking the DS were used to infect one cell line, BL30, which was derived from a Burkitt's lymphoma. In similar studies not presented here, we have also found that the EBV mutants lacking the DS can establish latent infections of other Burkitt's lymphoma lines, Raji and EBV-negative Akata, and an adenovirus-transformed epithelial cell line, 293 (13), with episomal maintenance of the viral chromosome. These results are also consistent with the fact that the EBV chromosome was stably maintained after being transferred into the mouse A9 cell line (19), a cellular environment in which oriP is not expected to be active. In rodent cell lines, plasmids carrying oriP and expressing EBNA-1 do not replicate detectably above background levels and cannot be maintained (51, 55). Initiation of replication at oriP was not detectable by 2D gel analysis in the mouse A9 cells carrying EBV episomes (M. A. Gilson, P. Norio, H. Fu, J.-M. Vos, and C. L. Schildkraut, unpublished data), implying that origins of replication outside of oriP were responsible for maintaining the EBV episomes.
Yet the presence of a functional homologue of the DS within oriP of herpesvirus papio, an EBV-like virus that infects baboons (33), indicates that the DS has been conserved during evolution and suggests that it is important in nature. It is conceivable that the replication function of oriP could be more important for latent infection of native B cells than for latent infection of established cell lines, and this is under investigation. Another possibility is that the DS might be more important for establishing latent infection than for maintaining it. For example, it is conceivable that the DS could be more important as chromatin structure is being established soon after infection than in subsequent cell division cycles.
From the present study we know that the EBV chromosome lacking the DS can replicate stably in BL30 cells once a latent infection has been established, but we did not measure directly how efficiently the EBV mutants lacking the DS establish a stable episomal state following infection. The efficiency with which the DS deletion mutants were isolated as clones of latently infected cells, however, suggests that the mutants must establish latent infections with an efficiency that is within the same order of magnitude as that of wild-type EBV. The mutants lacking the DS were isolated by allowing two plasmids to recombine with the EBV chromosome, one plasmid to transfer a G418 resistance gene to the viral chromosome by homologous recombination at a site distant from oriP and a second plasmid to transfer the DS deletion onto the viral chromosome at oriP. For the isolation of mutants using the P3HR1 strain, it was estimated that one-sixth of the EBV chromosomes that acquired the G418 resistance marker and were able to establish latent infection also had acquired the deletion that eliminated the DS. This frequency is within the range reported previously for the transfer of harmless mutations into this strain of EBV using similar genetic methods (49, 50, 56). If the EBV recombinants lacking the DS established latent infections in BL30 cells only one-tenth as efficiently, for example, as EBV containing the DS, then the mutants should have appeared at a much lower frequency. A reduction of efficiency by 50%, on the other hand, would not have been detected.
The ease with which the DS deletion was introduced also argues against the possibility that compensating mutations elsewhere in the EBV genome, which might have restored a function similar to that of the DS, were being selected. For this to have been the case, a compensating mutation would have had to preexist in a large portion of the genomes of the parental P3HR1 strain. Because we also were able to isolate at least one mutant lacking the DS using a different EBV strain, B95-8, it is unlikely that any feature peculiar to strain P3HR1 made the DS redundant.
Initiation of replication at oriP requires the DS.
While considerable evidence implicated the DS as a replication origin, it could not have been predicted that origin activity in its vicinity on the EBV chromosome would appear to cease in its absence. The large zone of initiation detected with Raji EBV, within which initiation events were detected in every restriction fragment that was examined, seemed to extend to oriP (32). In addition, Kirchmaier and Sugden identified weak replication activity within a 300-bp region, which they named Rep*, located to the right of the DS (22). For the 2D gel analysis of the P3HR1 mutants shown in Fig. 6 and 7, Rep* was positioned within the central one-third of the restriction fragment that was examined, where initiation of bidirectional replication would be most clearly detected as a full bubble arc. For the 2D analysis of the B95-8 mutant shown in Fig. 8 (B-ΔDS-8), the restriction fragment was positioned such that origin activity anywhere within oriP lacking the DS or at Rep* would have been detected. The results indicated that these other potential elements were not sufficient, in the absence of the DS, to support initiation from this region of the EBV chromosome at a detectable frequency.
It should be pointed out that the site(s) of initiation that must occur within a few hundred base pairs of the DS (11) has not been determined precisely, so it is still possible that the DS, although required for initiation of DNA replication, is not the primary initiation site. In this case, its function could be to increase the origin efficiency or to keep the initiation site active (for example, by helping to recruit initiation factors to the region or by excluding nucleosomes from the true initiation site). Similarly, Rep* or other sequences within or near oriP might play indirect, ancillary roles in supporting replication initiation at the DS (or its vicinity) on the EBV chromosome, a possibility which could be tested by deleting these regions from the EBV chromosome.
Initiation of replication on the EBV chromosome at sites distant from oriP.
Where replication initiates on the EBV chromosome outside of oriP has been determined only in a limited way, largely because the answer is complex. DNA replication appears to initiate at multiple sites within a zone spread across tens of kilobases to the left of oriP on the EBV chromosome, perhaps without any single site serving as an origin the majority of the time (32). In the EBV mutant derived from strain P3HR1 that we examined here, replication also was found to initiate within the broad zone, both within a 14-kb XbaI-EcoRI fragment centered at coordinate 153000, as found previously (32), and also within a 13-kb EcoRI fragment spanning the terminal repeats and centered at 166000, a region which had not been examined before. With EBV of the cell line Raji, oriP was found to be replicated primarily from left to right, which was consistent with replication being initiated within the zone to the left most of the time (32). However, in the present study involving EBV genomes that were derived from strains P3HR1 and B95-8, the FR of oriP was found to be replicated close to half of the time by forks entering from the right, even when the DS was deleted and unable to support initiation. This suggests that replication is likely to initiate somewhere to the right of oriP much of the time in these EBV strains.
It may be worth noting that diffuse termination structures were detected in the analysis of the EcoRI-XbaI fragment centered at position 129000 on the EBV map (Fig. 7). This could be explained most easily by the initiation of replication at one or more sites located between oriP and position 129000, since forks coming from this region could converge here with forks coming from the direction of the TRs (Fig. 7). Prominent, diffuse termination structures were also detected in the analysis of the 14-kb XbaI-EcoRI fragment centered at position 153000 (Fig. 7), which could be due to convergence of two forks that initiated at two flanking sites on the same viral chromosome. Evidence that this occurs was discussed previously (32).
The functional redundancy of replication origins.
The main conclusion of this report is that the replication function of oriP is redundant, at least for the latent infection of laboratory cell lines. Replication initiates on the EBV chromosome at sites other than oriP most of the time. Nevertheless, initiation has been detected more efficiently at oriP than at any other single site with most EBV strains, the only exception being Raji EBV, for which the 14-kb XbaI-EcoRI fragment, centered at position 153000 and within the broad initiation zone, was the most active (32). The broad initiation zone that exists mostly to the left of the TRs of EBV (Fig. 7) resembles the initiation zones that occur at some mammalian chromosomal loci, and initiation at sites within the zone is likely to be determined entirely by interactions with cellular proteins (see Introduction). Recent analysis of origin activity 3′ of the hamster DHFR gene suggests that large initiation zones may be comprised of discrete sites of initiation (23). Interestingly, the most active origin in the DHFR gene origin cluster, oriβ, can be deleted without affecting the timing of DNA replication for the locus during S phase or the apparent frequency of initiation at other sites within the locus (18). A functional redundancy of sites that have the potential to serve as replication origins may be a common feature of DNA replication in the mammalian nucleus.
ACKNOWLEDGMENTS
We thank Sarah Camiolo for constructing p776Δ and for the initial PCR and Southern analyses; Jaqueline Bashaw for the results of Fig. 2 and 4, for preparing figures, and for PCR analysis; and Prasad Kularni and M. Vogel for PCR amplification and DNA sequencing of EBV mutants.
This work was supported by NIH grants CA4312212 to J.L.Y. and GM45751 to C.L.S. and Cancer Center Core Grants CA16056 to the RPCI Biopolymer Facility and SP30-CA13330 to the AECOM FACS facility.
REFERENCES
- 1.Adams A. Replication of latent Epstein-Barr virus genomes in Raji cells. J Virol. 1987;61:1743–1746. doi: 10.1128/jvi.61.5.1743-1746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiyar A, Tyree C, Sugden B. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aladjem M I, Groudine M, Brody L L, Dieken E S, Fournier R E, Wahl G M, Epner E M. Participation of the human beta-globin locus control region in initiation of DNA replication. Science. 1995;270:815–819. doi: 10.1126/science.270.5237.815. [DOI] [PubMed] [Google Scholar]
- 4.Bloss T A, Sugden B. Optimal lengths for DNAs encapsidated by Epstein-Barr virus. J Virol. 1994;68:8217–8222. doi: 10.1128/jvi.68.12.8217-8222.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewer B J, Fangman W L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 6.Calos M P. Stability without a centromer. Proc Natl Acad Sci USA. 1998;95:4084–4085. doi: 10.1073/pnas.95.8.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dijkwel P A, Hamlin J L. The Chinese hamster dihydrofolate reductase origin consists of multiple potential nascent-strand start sites. Mol Cell Biol. 1995;15:3023–3031. doi: 10.1128/mcb.15.6.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dijkwel P A, Vaughn J P, Hamlin J L. Mapping of replication initiation sites in mammalian genomes by two-dimensional gel analysis: stabilization and enrichment of replication intermediates by isolation on the nuclear matrix. Mol Cell Biol. 1991;11:3850–3859. doi: 10.1128/mcb.11.8.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Favrot M C, Maritaz O, Suzuki T, Cooper M, Philip I, Philip T, Lenoir G. EBV-negative and -positive Burkitt cell lines variably express receptors for B-cell activation and differentiation. Int J Cancer. 1986;38:901–906. doi: 10.1002/ijc.2910380618. [DOI] [PubMed] [Google Scholar]
- 10.Frappier L, O'Donnell M. Epstein-Barr nuclear antigen 1 mediates a DNA loop within the latent replication origin of Epstein-Barr virus. Proc Natl Acad Sci USA. 1991;88:10875–10879. doi: 10.1073/pnas.88.23.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gahn T A, Schildkraut C L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- 12.Gardella T, Medveczky P, Sairenji T, Mulder C. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J Virol. 1984;50:248–254. doi: 10.1128/jvi.50.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 14.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 15.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 16.Harrison S, Fisenne K, Hearing J. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J Virol. 1994;68:1913–1925. doi: 10.1128/jvi.68.3.1913-1925.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heston L, Rabson M, Brown N, Miller G. New Epstein-Barr virus variants from cellular subclones of P3J-HR-1 Burkitt lymphoma. Nature. 1982;295:160–163. doi: 10.1038/295160a0. [DOI] [PubMed] [Google Scholar]
- 18.Kalejta R F, Li X, Mesner L D, Dijkwel P A, Lin H B, Hamlin J L. Distal sequences, but not ori-beta/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol Cell. 1998;2:797–806. doi: 10.1016/s1097-2765(00)80294-4. [DOI] [PubMed] [Google Scholar]
- 19.Kelleher Z T, Fu H, Livanos E, Wendelburg B, Gulino S, Vos J M. Epstein-Barr-based episomal chromosomes shuttle 100 kb of self-replicating circular human DNA in mouse cells. Nat Biotechnol. 1998;16:762–768. doi: 10.1038/nbt0898-762. [DOI] [PubMed] [Google Scholar]
- 20.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2324–2396. [Google Scholar]
- 21.Kim O J, Yates J L. Mutants of Epstein-Barr virus with a selective marker disrupting the TP gene transform B cells and replicate normally in culture. J Virol. 1993;67:7634–7640. doi: 10.1128/jvi.67.12.7634-7640.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirchmaier A L, Sugden B. Rep*: a viral element that can partially replace the origin of plasmid DNA synthesis of Epstein-Barr virus. J Virol. 1998;72:4657–4666. doi: 10.1128/jvi.72.6.4657-4666.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi T, Rein T, DePamphilis M L. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol Cell Biol. 1998;18:3266–3277. doi: 10.1128/mcb.18.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krysan P J, Calos M P. Replication initiates at multiple locations on an autonomously replicating plasmid in human cells. Mol Cell Biol. 1991;11:1464–1472. doi: 10.1128/mcb.11.3.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krysan P J, Haase S B, Calos M P. Isolation of human sequences that replicate autonomously in human cells. Mol Cell Biol. 1989;9:1026–1033. doi: 10.1128/mcb.9.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laskey R, Madine M. Roles of nuclear structure in DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 119–130. [Google Scholar]
- 27.Lau R, Middeldorp J, Farrell P J. Epstein-Barr virus gene expression in oral hairy leukoplakia. Virology. 1993;195:463–474. doi: 10.1006/viro.1993.1397. [DOI] [PubMed] [Google Scholar]
- 28.Lee M A, Diamond M E, Yates J L. Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus. J Virol. 1999;73:2974–2982. doi: 10.1128/jvi.73.4.2974-2982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee M-A, Yates J L. BHRF1 of Epstein-Barr virus, which is homologous to human proto-oncogene bcl2, is not essential for transformation of B cells or for virus replication in vitro. J Virol. 1992;66:1899–1906. doi: 10.1128/jvi.66.4.1899-1906.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Little R, Schildkraut C. Methods in molecular genetics. Vol. 4. New York, N.Y: Academic Press, Inc.; 1994. Mapping sites of replication initiation and termination in circular viral genomes using two-dimensional gel electrophoresis. [Google Scholar]
- 31.Little R D, Platt T H, Schildkraut C L. Initiation and termination of DNA replication in human rRNA genes. Mol Cell Biol. 1993;13:6600–6613. doi: 10.1128/mcb.13.10.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Little R D, Schildkraut C L. Initiation of latent DNA replication in the Epstein-Barr virus genome can occur at sites other than the genetically defined origin. Mol Cell Biol. 1995;15:2893–2903. doi: 10.1128/mcb.15.5.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loeb D D, Sung N S, Pesano R L, Sexton C J, Hutchison C D, Pagano J S. Plasmid origin of replication of herpesvirus papio: DNA sequence and enhancer function. J Virol. 1990;64:2876–2883. doi: 10.1128/jvi.64.6.2876-2883.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lupton S, Levine A J. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol Cell Biol. 1985;5:2533–2542. doi: 10.1128/mcb.5.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller G, Lipman M. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc Natl Acad Sci USA. 1973;70:190–194. doi: 10.1073/pnas.70.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raab-Traub N, Dambaugh T, Kieff E. DNA of Epstein-Barr virus VIII: B95-8, the previous prototype, is an unusual deletion derivative. Cell. 1980;22:257–267. doi: 10.1016/0092-8674(80)90173-7. [DOI] [PubMed] [Google Scholar]
- 37.Rawlins D R, Milman G, Hayward S D, Hayward G S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 38.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sample J, Henson E B, Sample C. The Epstein-Barr virus nuclear protein 1 promoter active in type I latency is autoregulated. J Virol. 1992;66:4654–4661. doi: 10.1128/jvi.66.8.4654-4661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaefer B C, Strominger J L, Speck S H. Redefining the Epstein-Barr virus-encoded nuclear antigen EBNA-1 gene promoter and transcription initiation site in group I Burkitt lymphoma cell lines. Proc Natl Acad Sci USA. 1995;92:10565–10569. doi: 10.1073/pnas.92.23.10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schvartzman J B, Martinez-Robles M L, Hernandez P. The migration behaviour of DNA replicative intermediates containing an internal bubble analyzed by two-dimensional agarose gel electrophoresis. Nucleic Acids Res. 1993;21:5474–5479. doi: 10.1093/nar/21.23.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shirakata M, Hirai K. Identification of minimal oriP of Epstein-Barr virus required for DNA replication. J Biochem (Tokyo) 1998;123:175–181. doi: 10.1093/oxfordjournals.jbchem.a021907. [DOI] [PubMed] [Google Scholar]
- 44.Simpson K, McGuigan A, Huxley C. Stable episomal maintenance of yeast artificial chromosomes in human cells. Mol Cell Biol. 1996;16:5117–5126. doi: 10.1128/mcb.16.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su W, Middleton T, Sugden B, Echols H. DNA looping between the origin of replication of Epstein-Barr virus and its enhancer site: stabilization of an origin complex with Epstein-Barr nuclear antigen 1. Proc Natl Acad Sci USA. 1991;88:10870–10874. doi: 10.1073/pnas.88.23.10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugden B, Yates J, Mark W. Transforming functions associated with Epstein-Barr virus. J Investig Dermatol. 1984;83(Suppl. 1):82s–87s. doi: 10.1111/1523-1747.ep12281491. [DOI] [PubMed] [Google Scholar]
- 47.Swaminathan S, Huneycutt B S, Reiss C S, Kieff E. Epstein-Barr virus-encoded small RNAs (EBERs) do not modulate interferon effects in infected lymphocytes. J Virol. 1992;66:5133–5136. doi: 10.1128/jvi.66.8.5133-5136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci USA. 1991;88:1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomkinson B, Kieff E. Second-site homologous recombination in Epstein-Barr virus: insertion of type 1 EBNA 3 genes in place of type 2 has no effect on in vitro infection. J Virol. 1992;66:780–789. doi: 10.1128/jvi.66.2.780-789.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomkinson B, Robertson E, Yalamanchili R, Longnecker R, Kieff E. Epstein-Barr virus recombinants from overlapping cosmid fragments. J Virol. 1993;67:7298–7306. doi: 10.1128/jvi.67.12.7298-7306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wysokenski D A, Yates J L. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J Virol. 1989;63:2657–2666. doi: 10.1128/jvi.63.6.2657-2666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yates J L, Camiolo S M, Bashaw J M. The minimal replicator of Epstein-Barr virus oriP. J Virol. 2000;74:4512–4522. doi: 10.1128/jvi.74.10.4512-4522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yates J L, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 56.Yoo L I, Mooney M, Puglielli M T, Speck S H. B-cell lines immortalized with an Epstein-Barr virus mutant lacking the Cp EBNA2 enhancer are biased toward utilization of the oriP-proximal EBNA gene promoter Wp1. J Virol. 1997;71:9134–9142. doi: 10.1128/jvi.71.12.9134-9142.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]