Abstract
Specific-pathogen-free (SPF) swine appear to be the most appropriate candidate for pig to human xenotransplantation. Still, the risk of endogenous retrovirus transmission represents a major obstacle, since two human-tropic porcine endogenous retroviruses (PERVs) had been characterized in vitro (P. Le Tissier, J. P. Stoye, Y. Takeuchi, C. Patience, and R. A. Weiss, Nature 389:681–682, 1997). Here we addressed the question of PERV distribution in a French Large White SPF pig herd in vivo. First, PCR screening for previously described PERV envelope genes envA, envB, and envC (D. E. Akiyoshi, M. Denaro, H. Zhu, J. L. Greenstein, P. Banerjee, and J. A. Fishman, J. Virol. 72:4503–4507, 1998; Le Tissier et al., op. cit.). demonstrated ubiquity of envA and envB sequences, whereas envC genes were absent in some animals. On this basis, selective out-breeding of pigs of remote origin might be a means to reduce proviral load in organ donors. Second, we investigated PERV genome carriage in envC negative swine. Eleven distinct full-length PERV transcripts were isolated. The sequence of the complete envelope open reading frame was determined. The deduced amino acid sequences revealed the existence of four clones with functional and five clones with defective PERV PK-15 A- and B-like envelope sequences. The occurrence of easily detectable levels of PERV variants in different pig tissues in vivo heightens the need to assess PERV transmission in xenotransplantation animal models.
For a number of anatomical, physiological, and ethical reasons, pigs are considered the most adequate organ source for xenotransplantation, an alternative therapy to alleviate the chronic human transplant shortage (15, 30, 39). To overcome immunological barriers, transgenic pigs bearing human complement-inhibiting proteins have been developed, leading to increased control of hyperacute rejection in primate models (8, 13, 19, 24, 31, 41, 47; M. Winkler, M. Loss, M. Przemeck, J. Schmidtko, H. Arends, R. Kunz, A. Jalali, J. Klempnauer, E. Cozzi, and D. J. G. White, Abstr. 5th Int. Congr. Xenotransplant. abstr. 184, p. 64). However, xenotransplantation circumvents the natural barriers against infection, raising the risk of cross-species transmission of pathogens after intimate and prolonged contact of living pig and human cells (2, 4, 5, 27, 32). Whereas most pathogens liable to be transmitted from a pig graft to a human recipient can be ruled out by specific-pathogen-free (SPF) rearing conditions, this might not apply to new (14, 17, 23, 40) or hitherto unknown organisms and definitively does not apply to pathogens that are inherited as part of the germ line, i.e., porcine endogenous retroviruses (PERVs) (42). In theory, PERVs share the pathogenic potential of retroviruses in general, which includes insertional mutagenesis and immunosuppression by themselves or after recombination with human retroviruses (12, 34).
Recent findings showed that human-tropic type C PERVs are released in vitro from porcine pig kidney cell line PK-15 (PERV-PK15), from stimulated miniature swine primary peripheral blood mononuclear cells (PERV-MSL), and spontaneously from porcine aortic endothelial cells (PERV-PK15-like) (1, 18, 22, 26, 37, 45). Analysis of these PERVs revealed genomes of about 8.1 kb with close amino acid sequence similarities (>95%) for the gag and pol open reading frames (ORFs). In contrast, sequence discrepancies occurring in envelope ORFs led to the distinction of three envelope genes, termed envA, envB, and envC (1, 18). gag, pol, and env transcripts and close variants of the last have been discovered in many different pig herds (9; D. Cunningham et al., Abstr. 5th Int. Congr. Xenotranspl., abstr. 1154, 1999; C. Herring et al., Abstr. 5th Int. Congr. Xenotranspl., abstr. 1153, 1999; J. H. Lee et al., Abstr. 5th Int. Congr. Xenotranspl., abstr. 0164, 1999). In spite of these observations, until now, no case of PERV infection has been reported for patients and primates who had been treated with living pig tissues (16, 21, 25, 28). Albeit promising, the apparent absence of PERV transmission in these assays needs to be relativized with respect to the short period of exposure to pig cells before xenograft rejection, the number of cells introduced, the degree of cell contact (extracorporal versus in situ), and the genetically unmodified donor material used (35–37, 44). Compared to future xenotransplantation settings, these conditions might have minimized the viral load, a factor which determines successful PERV transmission in vitro (C. Patience, B. Oldmixon, T. Ericsson, and G. Andersson, Abstr. 5th Int. Congr. Xenotranspl., abstr. 1109, 1999). For all these reasons, it will be important to learn more about the incidence of PERV particles in vivo. Aside from work done by Akiyoshi et al. on PERV-MSL (envC) genomes in miniature swine (1), many unknowns remain concerning the type and level of transcription of replication-competent PERVs in vivo. Indeed, general observations made for endogenous retroviruses as for other nonessential genes show that multiple mutations accumulate during evolution (20). Among the 50 proviral loci estimated in the pig genome using a protease probe (26), only a small subset are expected to be infectious. Characterization of active full-length PERVs is of special interest with regard to localization of intact proviruses, a prerequisite for eliminatory cross-breeding or gene knock-out technology.
This study focused on proviral PERV distribution and characterization of transcriptionally active full-length type C PERVs in a specific-pathogen-free (SPF) Large White pig herd. We recorded absence of envelope envC genes in the genome of some animals of the herd. Results for long reverse transcriptase (RT)-PCR conducted on envC-negative swine revealed persistent high levels of full-length type C PERV genomes in seven pig organs in vivo. There is compelling evidence for at least 11 distinct biologically active proviral loci in three animals analyzed from this herd. Envelope data analysis showed distinct and intact ORFs for four PERVs with close homologies with PERV PK-15 A- and B-like sequences. On the contrary, five other PERVs with truncated envelope ORFs will constitute no infectious risk in xenotransplantation.
MATERIALS AND METHODS
Viruses and cell cultures.
The production of PERV PK-15 and Tsukuba-1 particles by, respectively, pig kidney PK-15 and pig spleen Schimozuma cell lines has been described previously (39). Schimozuma substrain G2 was a gift from B. Kaeffer (INRA, Nantes, France). Porcine PK-15, human K562, and green monkey kidney MARC cell lines were kindly provided by E. Albina (AFSSA, Ploufragan, France). These cells, except for K562, were grown in minimal essential medium (MEM) supplemented with 10% fetal calf serum (FCS), 100 U of penicillin per ml, and 100 mg of streptomycin per ml K562 cells were kept in RPMI medium, chicken hepatocellular carcinoma LMH cells (ATCC CRL-2117) were kept in Williams medium, and equine fibroblast ED cells (ATCC CCL-57) were kept in MEM with FCS and antibiotics as above.
SPF pigs.
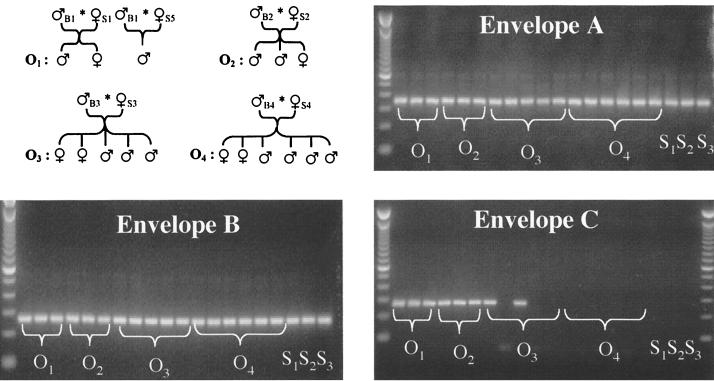
All samples were from Large White pigs raised under SPF conditions (6). Periodic controls ensure the maintenance of well-defined health status for this herd (C. Cariolet, personal communication). For the maintenance of the Ploufragan SPF pig out-bred herd, females are selected among the descendants. In order to avoid consanguinity, two boars delivered by hysterectomy and raised under sterile conditions until weaning are introduced each year. Hence, pig families were defined as the descendants of one boar. Tissue samples were harvested shortly after slaughter and stored in liquid nitrogen. Genomic full-length transcripts were cloned from pigs 6309, 6282, and 6407, weighing about 100 kg and 132 to 140 days of age at sacrifice. In order to avoid RNA degradation, a sample of pancreas was snap-frozen in liquid nitrogen, and RNA was extracted immediately after resection. Whole blood samples were collected from three females (sows 1 to 3 [S1 to S3]) and 17 offspring of four boars (O1 to O4). For practical reasons, no blood could be obtained from boars.
Isolation of DNA.
DNA was extracted from whole blood samples using the Qiagen tissue kit (Qiagen).
Isolation of mRNAs.
From 20 to 30 mg of frozen tissue sample was ground in liquid nitrogen. The frozen powder was lysed (LiDS), homogenized (Qiashredder; Qiagen) and fixed for 4 min on 1.25 mg of paramagnetic oligo(dT)25 beads (Dynal) according to the manufacturer's instructions. mRNA was eluted in Tris-HCl (10 mM [pH 7.5]) and stored at −80°C until use. For the extraction of mRNA from cell lines, cells were rinsed in phosphate-buffered saline (PBS) and directly subjected to lysis as stated here above.
Generation of genomic full-length PERV clones.
mRNA derived from 125 μg (375 μg for pancreas) of oligo(dT)25 beads was reverse transcribed in 50 μl of Thermoscript RT-PCR system reaction mixture with oligo(dT) primers as specified by the vendor (Gibco BRL). Negative controls without reverse transcriptase were prepared for all samples. RT conditions consisted of 5 min at 50°C, 60 min at 55°C, and 10 min at 60°C. Template mRNA was removed by addition of 5 U of Escherichia coli RNase H (Gibco BRL) at 37°C for 20 min. Then 5 μl of cDNAs or 2 ng of plasmid Tsukuba-1 DNA (kindly provided by J. P. Stoye) was subjected to PCR amplification with primers conserved in all three type C PERVs. The forward primer was designed from the leader region (L-fov, 5′-ACGTGCTAGGAGGATCACAGGCTGC-3′, nucleotides [nt] 342 to 356 in Tsukuba-1 or 347 to 372 in PK-15) and backward primer from the untranslated region downstream of the envelope gene (L-rev, 5′-GTTGTCTAAGTACCATGATCTGGACTGCAC-3′, nt 7476 to 7506 in Tsukuba-1 or 6665 to 6684 in PK-15). PCR with these primers should generate a 7.2-kb product for genomic porcine PERV type C RNA and products in the ∼3-kb range for spliced viral mRNAs. Amplification was carried out in 50 μl of Platinum Taq High Fidelity reaction mixture (Gibco BRL) with dimethyl sulfoxide added immediately prior to cycling to a final concentration of 10%. The initial denaturation step was 1 min at 94°C, followed by 10 cycles of 10 s at 94°C, 30 s at 66°C, ramp for 1 min to 68°C, 8 min at 68°C, and 25 cycles of 10 s at 94°C, 30 s at 66°C, ramp for 1 min to 68°C, 9 min at 68°C, and 20 min of final extension at 68°C. Full-length (approximately 7.2 kb) PCR products were gel purified (QIAEXII; Qiagen), ligated into the Topo XL cloning vector, and introduced into Top10 electrocompetent cells (Invitrogen). Eighty colonies with the 7.2-kb insert were isolated. Plasmid DNA was prepared with the Qiaprep 8 miniprep kit (Qiagen).
PCR.
PCR with primers specific for gag (1), pol (45), and envelope genes envA, envB, and envC (18, 38) was carried out using the primers described elsewhere but adapted here to higher melting temperatures: gag-fov (5′-CCCGATCAGGAGCCCTATATCCTTACGTG-3′), gag-rev (5′-CGCAGCGGTAATATCGCGATCTCGT-3′), pol-fov (5′-GACGGGTAACCCACTCGTTTCTGGT-3′), pol-rev (5′-ACGTACTGGAGGAGGGTCACCTAG-3′), envA-fov (5′-GAGATGGAAAGATTGGCAACAGCG-3′), envA-rev (5′-AGTGATGTTAGGCTCAGTGGGGAC-3′), envB-fov (5′-AATTCTCCTTTGTCAATTCCGGCCC-3′), envB-rev (5′-CCAGTACTTTATCGGGTCCCACTG-3′), envC-fov (5′-CTGACCTGGATTAGAACTGGAAGC-3′), and envC-rev (5′-GTTATGTTAGAGGATGGTCCTGGTC-3′). Reaction mixture (20 μl) containing 3 U of recombinant Tetrahymenis thermus (rTth) polymerase (Roche) bound to rTth antibody (Ozyme) and 1 U of uracil desoxynucleotide glycosylase (Gibco BRL) was subjected to 15 min at 37°C, 10 min at 94°C, followed by 30 rounds of thermocycling of 30 s at 94°C, 45 s at 65°C, 40 s at 72°C, and 10 min of final extension at 72°C.
RFLP.
For the restriction fragment length polymorphism (RFLP) assay, 80 full-length clones were digested with EcoRI (NEB).
Sequence analysis of PERV elements.
Clones were cycle sequenced on a 373 DNA sequencing system (Applied Biosystems) with the dye terminator cycle sequencing kit and Ampli Taq DNA polymerase FS (Applied Biosystems).
Nucleotide sequence accession numbers.
The complete envelope coding region was determined for one clone out of each profile group and deposited in the EMBL database (AJ288584 to AJ288592). Infobiogen's software was used to deduce the putative amino acid sequence. All sequence alignments were carried out using the INRA multialignment software (11), and sequence comparisons were carried out with NCI's BLAST (3) and LFASTA (7) software.
RESULTS
Distribution of proviral elements in SPF pigs in Ploufragan.
DNA extracted from blood samples of three sows and 17 offspring derived from four boars were screened by PCR for the distribution of gag, pol (data not shown), and envelope envA, envB, and envC gene sequences (Fig. 1). gag, pol, and envelope envA and envB genes were ubiquitously present in all animals. In contrast, our herd was heterogeneous for envC, which was absent in all mother sows tested, in offspring of boar B4, and in three of five offspring of boar B3. In all subsequent studies on transcriptionally active PERVs, only envC-negative swine, presumably cleaner in terms of proviral load, were used.
FIG. 1.
Distribution of proviral elements in SPF pigs in Ploufragan. PCR with primer pairs specific for envA, envB, and envC resulted in amplification products of 359 bp, 263 bp, and 281 bp, respectively. Here, a pig family was defined as the descendants of one boar. S1 to S3, mother sows; O1 to O4, piglets sired by boars 1 to 4, respectively.
Isolation of genome-length viral RNAs in pig tissues.
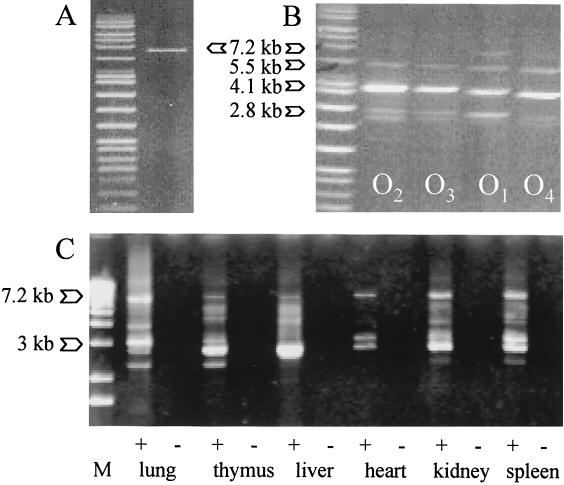
The specificity of long RT-PCR for PERV versus other ERV genomes was tested on RNA derived from human, simian, equine, avian, and porcine cell lines. Only porcine RNA yielded amplification products (data not shown). Long RT-PCR efficiently amplified full-length and additional shorter PERV type C transcripts from all pig tissues studied (Fig. 2). Faint bands larger than 8 kb likely correspond to either PCR artifacts or readthrough transcription of viral into cellular genes (20). Subgenomic fragments, derived from either truncated proviruses or spliced viral mRNAs, were observed in the 2.3-kb to 6-kb range. The intensity of these bands varied in a tissue-specific manner, but three times showed reproducibly consistent patterns in four different pigs: ∼2.3-kb, 2.8-kb, 3-kb, and 3.8-kb transcripts in the lung; ∼2.3-kb, 2.8-kb, 3-kb, 4-kb, and 5-kb transcripts in the thymus; a strong 3-kb transcript band in the liver; ∼3-kb and 3.3-kb transcripts in the heart; ∼3-kb, 4-kb, and 6-kb transcripts in the kidney; ∼3-kb and strong 3.3-kb bands in the spleen; and a ∼3.3-kb band in the pancreas. These patterns are more complex than those observed by Akiyoshi et al. for transcripts in miniature swine hybridized with an envC (PERV-MSLenv) probe (1). The multitude of products found here underscores the great number of PERV type C proviruses fixed in the germ line of our and probably most other pig herds.
FIG. 2.
Characterization of full-length PERV RNA and DNA. Long PCR performed on (A) plasmid Tsukuba-1 DNA, (B) porcine DNA extracted from whole blood of one offspring in each pig family, and (C) cDNA derived from major pig organs (lanes +) and negative controls without reverse transcriptase (lanes −). A specific amplification product was obtained for cloned PERV sequence, whereas porcine DNA revealed multiple products of viral origin. RT-PCR demonstrated the existence of full-length and spliced viral RNA in all pig tissues studied.
Subsequent PCR screening of 80 cloned full-length transcripts derived from heart, liver, lung, spleen, thymus, and kidney from two pigs (6309 and 6282) and pancreas from one pig (6407) ascertained retroviral origin: all clones presented gag elements and all except two clones showed pol elements. The latter, assumed to contain deletions or rearrangements which might render them noninfectious, were sorted out. The remaining clones were divided into 11 categories according to PCR results obtained for envelope genes (envA plus envB, or double positives, noted envAB) and EcoRI digestion profile patterns (1 to 8) (Table 1). These categories indicate the existence of at least 11 distinct full-length PERV genome variants. In consequence, there are at least 11 different transcriptionally active proviral insertion sites in the genome of these pigs. All tissues except lung transcribed three or more different variants. In particular, members of group envB1, which present a profile pattern identical to PERV PK-15 B (GenBank Y17013), and of group envA1 were prominent, i.e., all three pigs presented one or both types. Clones belonging to group envA1 were observed in all tissues except liver. However, we only isolated seven clones from liver and lung. This small sample size might not fully reflect the retroviral diversity in these tissues.
TABLE 1.
Characterization of full-length PERV genomes transcribed in three SPF pigs in vivoa
| Group | No. of clones | No. of pigs | Occurrence in organs | Envelope sequence | Envelope (amino acids) |
|---|---|---|---|---|---|
| envA1 | 27 | 3 | Heart, kidney, lung, pancreas, spleen, thymus | PERV-A1 | 661 |
| envA2 | 2 | 2 | Heart, thymus | PERV-A2 | 661 |
| envB1 | 25 | 3 | Kidney, liver, pancreas, spleen | PERV-B1 | 655 |
| envB4 | 1 | 1 | Spleen | PERV-B4 | 404 |
| envB5 | 1 | 1 | Kidney | NDb | |
| envB6 | 9 | 3 | Heart, kidney, liver, pancreas, thymus | PERV-B6 | 288 |
| envB7 | 1 | 1 | Pancreas | PERV-B7 | 632 |
| envB8 | 2 | 2 | Liver, kidney | ND | |
| envAB3 | 5 | 1 | Heart, liver, thymus | PERV-AB3 | 280 |
| envAB4 | 2 | 1 | Liver, thymus | PERV-AB4 | No protein |
| envAB7 | 3 | 2 | Heart, kidney | PERV-AB7 | 202 |
Each group includes clones with identical envelope PCR and RFLP profile (I to VIII) results.
ND, not determined.
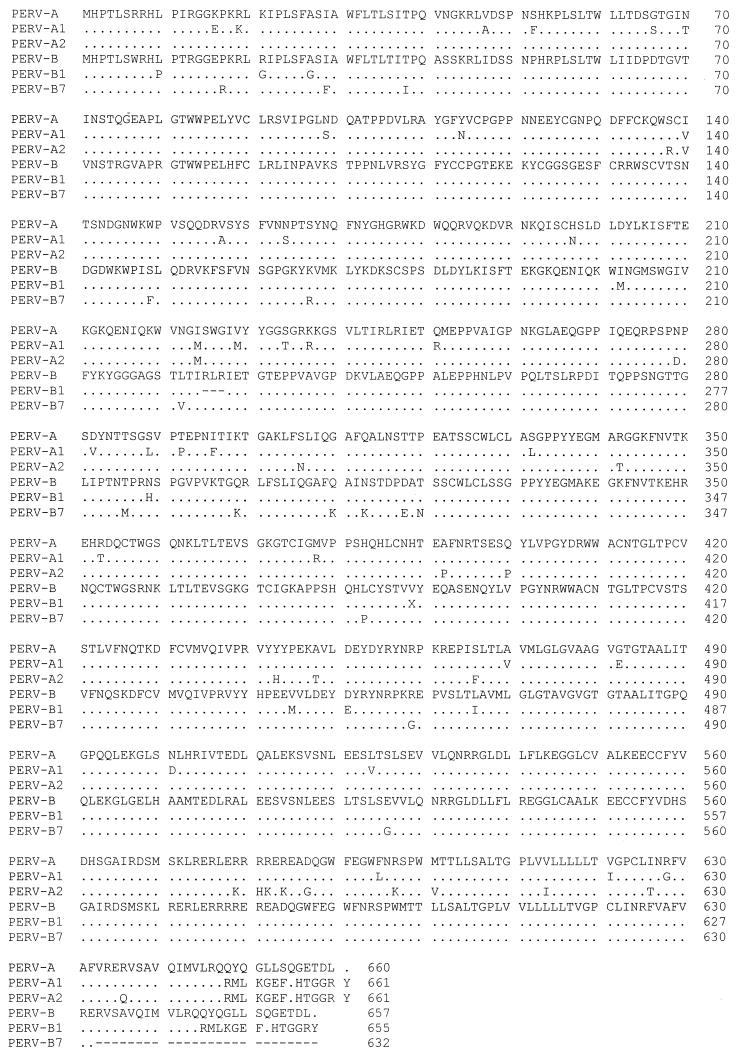
Envelope sequence data.
Alignment of the deduced amino acid sequence (Fig. 3) revealed intact envelope ORFs for four restriction groups: variants PERV-A1 and PERV-A2 shared homologies greater than 95 and 96%, respectively, with sequence PERV PK-15A, and variants PERV-B1 and PERV-B7 showed homologies greater than 97% to sequence PERV PK-15 B. The main sequence discrepancies occurred in a small stretch of 14 amino acid residues in the C-terminal regions of PERV-A1, PERV-A2, and PERV-B1, whereas 24 amino acid residues were deleted at the C-terminal region of PERV-B7. Five other clones, PERV-B4, PERV-B6, PERV-AB3, PERV-AB4, and PERV-AB7, presented insertions or deletions causing frameshifts and consecutive disruption of the translation process.
FIG. 3.
Amino acid sequences of untruncated envelope ORFs deduced from PERV transcripts. Dashes indicate gaps, and dots indicate identical amino acids.
DISCUSSION
We used Large White outbred SPF pigs to assess PERV carriage in vivo and suitability as an organ source for xenotransplantation. Consistent with observations made for other pig herds (37), PERV envC genes were absent in some of the animals, i.e., in all three mother sows tested, in the offspring of boar B4, and in three of five offspring of boar B3. Matched to the genealogical tree, these results sustain the hypothesis that our envelope envC primers are complementary to a single proviral insertion site on a somatic chromosome in the heterozygous boar B3 and its positive offspring. Hence, donor screening will permit us to preclude infectious risk for this subtype of PERVs in xenotransplantation. However, we cannot rule out that distantly related PERV envC viruses have not been recognized by the PCR primers used here. In the future, determination of chromosomal locations of PERVs in herds of remote origin might prove to be an efficient means of reducing PERV carriage by selective outbreeding.
Long PCR with primer pairs conserved in all three PERV sequences (PERV PK-15 A and B and PERV-MSL) confirmed predictions of whole and partial inserts in the genome of our pig herd (Fig. 2). Since proviral DNA load does not necessarily correlate with viral RNA load, we investigated whether the intact proviruses are dormant or biologically active. In contrast to other endogenous retroviruses which are transcribed at extremely low levels and require nested RT-PCR for their detection (43), simple RT-PCR was sufficient to detect constitutive transcription of full-length PERV genomes in all tissues tested. There are several implications to these findings. (i) Obviously, these viral genomic RNAs stem from long terminal repeats acting as strong promoters in porcine cells, which invalidates earlier assumptions that PERV expression might be triggered by in vitro culture conditions. (ii) Among the 11 different PERV genomes recognized here, a high percentage were of the envB1 type, which displays the restriction pattern expected for PERV PK-15 B (GenBank Y17013). This apparent similarity indicates replication competence for this group. (iii) Finally, the sample size and experimental conditions used might not account for minority or distantly related PERV genomes, thereby understating the viral diversity. A conceivable consequence of the presence of multiple variants is that earlier in vitro studies on the host range and interference of PERVs (37, 46) might not fully reflect the viral population encountered in different pig herds in vivo.
To assess infectious genomes, we sequenced the complete envelope coding region for one clone in each profile group. The deduced amino acid sequence revealed four variants with intact envelope ORFs and close homologies with sequences PERV PK-15 A and B. The main sequence discrepancies were located in a small stretch of amino acids at the C terminus. While amino acid residues 632 to 657 were missing in PERV-B7, the three other ORF variants showed C-terminal sequences markedly different from those of PERV-PK-15 but similar to each other. A likely reason for this phenomenon is that a predecessor PERV, after having acquired the C-terminal sequence, reintegrated into the pig genome. Indeed, ERVs (20) are prone to amplify in the genome in a retrotransposable fashion, thereby introducing genetic diversity and rapid genetic drift between separate strains. As a 16-amino-acid C-terminal fragment is removed from the end of the transmembrane envelope protein in murine leukemia virus-related viruses before budding (10), we suppose that this variation from PERV PK-15 A and B sequences does not impede replication competence in these clones. Nonetheless, downstream infectious particle formation remains to be demonstrated. Sequence analysis of PERV variants B4, B5, B6, AB4, and AB7 revealed multiple frameshifts with introduction of stop codons leading to truncated envelope proteins. Taken together with possible changes in other regions of the genome, the apparent envelope ORF alterations will not support replication in these genomes. The presence of multiple defective genomes in pig cells in vivo which are susceptible of virion assembly through complementation by helper viruses and interfere with replication-competent viruses is in agreement with the observation that a single in vitro passage of PERVs through human cells selects virus populations with increased infectious titers (46).
PCR analysis of envelope genes revealed double positive envAB in three of eight RFLP profile groups, all of which proved to code for defective envelope proteins. Since envA- and envB-specific primers bind to analogous but disparate regions of envelope gene sequences of two PK-15 PERVs (18), these findings suggest the presence of mutations or crossovers yielding novel mosaic genotypes. Evidence for envAB recombinant PERVs had been reported for two of sixty-four proviral envelope sequences derived from Australian Westran pigs (J. H. Lee et al., Abstr. 5th Int. Congr. Xenotranspl., abstr. 0164, 1999), and more recently, envAC recombinant viral RNAs were observed in U.S. minipig peripheral blood mononuclear cell cultures (46). Paradoxically, envelope sequence analysis of our envAB clones did not show recombination between envA and envB sequences but numerous point mutations. In spite of these alterations, we were unable to identify primer binding sites for envA amplification. As all PCR experiments were repeated twice at 3 month intervals on freshly prepared plasmid material in a PERV-free atmosphere, we suppose that envAB double-positive PCR results are due to crossovers of PERV PK-15 A- and B-like sequences upstream of the envelope ORF sequenced here rather than to PCR contamination.
This work demonstrates that full-length PERV genomes actively replicate to easily detectable levels in French Large White SPF pigs in vivo. Envelope analysis revealed the existence of five PERV variants coding for truncated envelope proteins, indicating that these genomes are devoid of infectious risk in a xenotransplantation setting. On the contrary, all tissues examined presented at least one of four PERVs with functional PERV PK-15-like envelope ORFs. Experiments trying to map potentially replication-competent variants to a Large White bacterial chromosome library (29) are currently under way. In the light of our findings, it seems illusory to control expression of PERVs in transplanted tissues. We therefore hope that the sequence data presented here will be valuable for the comparison of PERV distribution in different pig herds in an endeavor to rule out potentially infectious proviruses.
ACKNOWLEDGMENTS
We are grateful to R. Cariolet and P. Julou for production of SPF pigs and helpful advice. We also thank C. Rogel-Gaillard and P. Chardon for critical reading of the manuscript.
This work was supported by a grant from the Conseil Régional of Bretagne and Pays de Loire, France.
REFERENCES
- 1.Akiyoshi D E, Denaro M, Zhu H, Greenstein J L, Banerjee P, Fishman J A. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J Virol. 1998;72:4503–4507. doi: 10.1128/jvi.72.5.4503-4507.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allan J S, Broussaard S R, Michaels M G, Starzl T E, Leighton K L, Whitehead E M, Comuzzie A G, Lanford R E, Leland M M, Switzer W M, Heneine W. Amplification of simian retroviral sequences from human recipients of baboon liver transplants. Aids Res Hum Retroviruses. 1998;14:821–824. doi: 10.1089/aid.1998.14.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borie D C, Cramer D V, Phan-Thanh L, Vaillant J C, Bequet J L, Makowka L, Hannoun L. Microbiological hazards related to xenotransplantation of porcine organs into man. Infect Control Hosp Epidemiol. 1998;19:355–365. doi: 10.1086/647830. [DOI] [PubMed] [Google Scholar]
- 5.Brown J, Matthews A L, Sandstrom P A, Chapman L E. Xenotransplantation and the risk of retroviral zoonosis. Trends Microbiol. 1998;6:411–415. doi: 10.1016/s0966-842x(98)01347-x. [DOI] [PubMed] [Google Scholar]
- 6.Cariolet R. Bilan de 10 années d'utilisation de porcs exempts d'organismes pathogènes spécifiques (EOPS) à la station de pathologie porcine de Ploufragan. J Rech Porcine France. 1986;18:321–330. [Google Scholar]
- 7.Chao K M, Pearson W R, Miller W. Aligning two sequences within a specified diagonal band. Comput Appl Biosci. 1992;8:481–487. doi: 10.1093/bioinformatics/8.5.481. [DOI] [PubMed] [Google Scholar]
- 8.Chen R H, Naficy S, Logan J S, Diamond L E, Adams D H. Hearts from transgenic pigs constructed with CD59/DAF genomic clones demonstrate improved survival in primates. Xenotransplantation. 1999;6:194–200. doi: 10.1034/j.1399-3089.1999.00017.x. [DOI] [PubMed] [Google Scholar]
- 9.Clémenceau B, Lalain S, Martignat L, Saï P. Porcine endogenous retroviral mRNAs in pancreas and a panel of tissues from specific-pathogen-free pigs. Diabet Metab. 1999;25:518–525. [PubMed] [Google Scholar]
- 10.Coffin J M. Retroviridae: the viruses and their replication. In: Fields B N, editor. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1767–1847. [Google Scholar]
- 11.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denner J. Immunosuppression by retroviruses: implications for xenotransplantation. Ann NY Acad Sci. 1998;862:76–86. doi: 10.1111/j.1749-6632.1998.tb09119.x. [DOI] [PubMed] [Google Scholar]
- 13.Diamond L E, McCurry K R, Martin M J, McClellan S B, Oldham E R, Platt J L, Logan J S. Characterization of transgenic pigs expressing functional active human CD59 on cardiac endothelium. Transplantation. 1996;61:1241–1249. doi: 10.1097/00007890-199604270-00021. [DOI] [PubMed] [Google Scholar]
- 14.Ehlers B, Ulrich S, Goltz M. Detection of two novel porcine herpesviruses with high similarity to gammaherpesviruses. J Gen Virol. 1999;80:971–978. doi: 10.1099/0022-1317-80-4-971. [DOI] [PubMed] [Google Scholar]
- 15.French A J, Greenstein J L, Loveland B E, Mountford P S. Current and future prospects for xenotransplantation. Reprod Fertil Dev. 1998;10:683–696. doi: 10.1071/rd98112. [DOI] [PubMed] [Google Scholar]
- 16.Heneine W, Tibell A, Switzer W M, Sandstrom P, Rosales G V, Mathews A, Korsgren O, Chapman L E, Folks T M, Groth C G. No evidence of infection with porcine endogenous retrovirus in recipients of porcine islet-cell xenografts. Lancet. 1998;352:695–699. doi: 10.1016/S0140-6736(98)07145-1. [DOI] [PubMed] [Google Scholar]
- 17.Kroneman A, Cornelissen L A, Horzinek M C, de Groot R J, Egberink H F. Identification and characterization of a porcine torovirus. J Virol. 1998;72:3507–3511. doi: 10.1128/jvi.72.5.3507-3511.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Tissier P, Stoye J P, Takeuchi Y, Patience C, Weiss R A. Two sets of human-tropic pig retrovirus. Nature. 1997;389:681–682. doi: 10.1038/39489. [DOI] [PubMed] [Google Scholar]
- 19.Logan J S, Sharma A. Potential use of genetically modified pigs as organ donors for transplantation into humans. Clin Exp Pharmacol Physiol. 1999;26:1020–1025. doi: 10.1046/j.1440-1681.1999.03185.x. [DOI] [PubMed] [Google Scholar]
- 20.Löwer R, Löwer J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin U, Steinhoff G, Kiessig V, Chikobava M, Anssar M, Morschheuser T, Lapin B, Haverich A. Porcine endogenous retrovirus (PERV) was not transmitted from transplanted porcine endothelioal cells to baboons in vivo. Transplant Int. 1998;11:247–251. doi: 10.1007/s001470050136. [DOI] [PubMed] [Google Scholar]
- 22.Martin U, Kiessig V, Blusch J H, Haverich A, von Helm K, Herden T, Steinhoff G. Expression of pig endogenous retrovirus by primary porcine endothelial cells and infection of human cells. Lancet. 1998;352:692–694. doi: 10.1016/S0140-6736(98)07144-X. [DOI] [PubMed] [Google Scholar]
- 23.Meng X J, Purcell R H, Halbur P G, Lehman J R, Webb D M, Tsareva T S, Haynes J S, Thacker B J, Emerson S U. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci USA. 1997;94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mollnes T E, Fiane A E. Xenotransplantation: how to overcome the complement obstacle. Mol Immunol. 1999;36:269–276. doi: 10.1016/s0161-5890(99)00050-4. [DOI] [PubMed] [Google Scholar]
- 25.Paradis K, Langford G, Long Z, Heneine W, Sandstrom P, Switzer W M, Chapman L E, Lockey C, Onions D, Otto E XEN 111 Study Group. Search for cross-species transmission of porcine endogenous retrovirus in patients treated with living pig tissues. Science. 1999;285:1236–1241. doi: 10.1126/science.285.5431.1236. [DOI] [PubMed] [Google Scholar]
- 26.Patience C, Takeuchi Y, Weiss R A. Infection of human cells by an endogenous retrovirus of pigs. Nat Med. 1997;3:282–286. doi: 10.1038/nm0397-282. [DOI] [PubMed] [Google Scholar]
- 27.Patience C, Takeuchi Y, Weiss R A. Zoonosis in xenotransplantation. Curr Opin Immunol. 1998;10:539–542. doi: 10.1016/s0952-7915(98)80220-3. [DOI] [PubMed] [Google Scholar]
- 28.Pitkin Z, Mullon C. Evidence of absence of porcine endogenous retrovirus (PERV) infection in patients treated with a bioartificial liver support system. Artif Organs. 1999;23:829–833. doi: 10.1046/j.1525-1594.1999.06444.x. [DOI] [PubMed] [Google Scholar]
- 29.Rogel-Gaillard C, Bourgeaux N, Billault A, Vaiman M, Chardon P. Construction of a swine BAC library: application to the characterization and mapping of porcine type C endoviral elements. Cytogenet Cell Genet. 1999;85:205–211. doi: 10.1159/000015294. [DOI] [PubMed] [Google Scholar]
- 30.Sachs D H. The pig as a potential xenograft donor. Vet Immunol Immunopathol. 1994;43:185–191. doi: 10.1016/0165-2427(94)90135-x. [DOI] [PubMed] [Google Scholar]
- 31.Sandrin M S, McKenzie F C. Recent advances in xenotransplantation. Curr Opin Immunol. 1999;11:527–531. doi: 10.1016/s0952-7915(99)00011-4. [DOI] [PubMed] [Google Scholar]
- 32.Stoye J P, Le Tissier P, Takeuchi Y, Patience C, Weiss R A. Endogenous retroviruses: a potential problem for xenotransplantation? Ann NY Acad Sci. 1998;862:66–74. doi: 10.1111/j.1749-6632.1998.tb09118.x. [DOI] [PubMed] [Google Scholar]
- 33.Suzuka I, Shimizu N, Sekiguchi K, Hoshino H, Kodama M, Shimotohno K. Molecular cloning of unintegrated closed circular DNA of porcine retrovirus. FEBS Lett. 1986;198:339–343. doi: 10.1016/0014-5793(86)80432-x. [DOI] [PubMed] [Google Scholar]
- 34.Tacke S J, Kurth R, Denner J. Porcine endogenous retroviruses inhibit human immune cell function: risk for xenotransplantation? Virology. 2000;268:87–93. doi: 10.1006/viro.1999.0149. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi Y, Cosset F-L, Lachmann P J, Okada H, Weiss R A, Collins M K L. Type C retrovirus inactivation by human complement is determined by both the viral genome and the producer cell. J Virol. 1994;68:8001–8007. doi: 10.1128/jvi.68.12.8001-8007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeuchi Y, Porter C D, Straha K M, Preece A F, Gustafsson K, Cosset F-L, Weiss R A, Collins M K L. Sensitization of cells and retroviruses to human serum by (α1-3) galactosyltransferase. Nature. 1996;379:85–88. doi: 10.1038/379085a0. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi Y, Liong S-H, Bieniasz P D, Jäger U, Porter C D, Friedmann T, McClure M O, Weiss R A. Sensitization of rhabdo-, lenti-, and spumaviruses to human serum by galactosyl(α1-3)galactosylation. J Virol. 1997;71:6174–6178. doi: 10.1128/jvi.71.8.6174-6178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeuchi Y, Patience C, Magre S, Weiss R A, Banerjee P T, Le Tissier P, Stoye J P. Host range and interference studies of three classes of pig endogenous retrovirus. J Virol. 1998;72:9986–9991. doi: 10.1128/jvi.72.12.9986-9991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taniguchi S, Cooper D K C. Clinical xenotransplantation: past, present and future. Ann R Coll Surg Engl. 1997;79:13–19. [PMC free article] [PubMed] [Google Scholar]
- 40.Ulrich S, Goltz M, Ehlers B. Characterization of the DNA polymerase loci of the novel porcine lymphotropic herpesviruses 1 and 2 in domestic and feral pigs. J Gen Virol. 1999;80:3199–3205. doi: 10.1099/0022-1317-80-12-3199. [DOI] [PubMed] [Google Scholar]
- 41.Vial C M, Ostlie D J, Bhatti F N, Cozzi E, Goddard M, Chavez G P, Wallwork J, White D J, Dunning J J. Life supporting function for over one month of a transgenic porcine heart in a baboon. J Heart Lung Transplant. 2000;19:224–229. doi: 10.1016/s1053-2498(99)00099-6. [DOI] [PubMed] [Google Scholar]
- 42.Weiss R A, Griffiths D, Takeuchi Y, Patience C, Venables P J W. Retroviruses: ancient and modern. Arch Virol Suppl. 1999;15:171–177. doi: 10.1007/978-3-7091-6425-9_12. [DOI] [PubMed] [Google Scholar]
- 43.Weissmahr N R. Development and evaluation of a highly sensitive method for the identification of particle-associated retroviral sequences. Ph.D. thesis. Zürich, Switzerland: University of Zürich; 1995. [Google Scholar]
- 44.Welsh R M, O'Donnell C L, Reed D J, Rother R P. Evaluation of the Galα1-3Gal epitope as a host modification factor eliciting natural humoral immunity to enveloped viruses. J Virol. 1998;72:4650–4656. doi: 10.1128/jvi.72.6.4650-4656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson C A, Wong S, Muller J, Davidson C E, Rose T M, Burd P. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J Virol. 1998;72:3082–3087. doi: 10.1128/jvi.72.4.3082-3087.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson C A, Wong S, Van Brocklin M, Federspiel M J. Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J Virol. 2000;74:49–56. doi: 10.1128/jvi.74.1.49-56.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yannoutsos N, Langford G A, Cozzi E, Lancaster R, Elsome K, Chen P, White D J G. Production of pigs transgenic for human regulators of complement activation. Transplant Proc. 1995;27:324–325. [PubMed] [Google Scholar]