Abstract
This article offers a new method of clustering data. This method is constructed by combining the multivariate adaptive regression spline biresponse continuous model (MARSBC) with the fuzzy clustering means (FCM) approach, called the multivariate adaptive biresponse fuzzy clustering means regression splines (MABFCMRS) model. This method uses patterns obtained from the MARSBC model to separate data into specific groups. Observing unobserved heterogeneity that has not been obtained from previous models. Unlike the classic fuzzy clustering methods that use euclid distances to determine the weight of the object, this method uses the total square of the massed residual distance generated by the MARSBC model. Theoretical studies were conducted to obtain predictions for the MABFCMRS model parameters. Furthermore, this method was applied to stunting and wasting cases in southeastern Sulawesi province. The results of the research show that in the case of stunting modeling and wasting in southeast Sulawesi province, the best clusters were obtained based on the criteria of partition coefficient (PC) and modification of PC (MPC). This research is able to show that the clustering process using the MABFCMRS model has been able to improve generalized cross-validation (GCV) values and determination coefficients.
-
•
This paper presents a new, effective method for clustering data based on unobserved heterogeneity.
-
•
This is applicable to sizable samples with 3 to 20 predictors.
Keywords: MARSBC, FCM, MABFCMRS, Stunting, Wasting
Method name: Multivariate adaptive biresponse fuzzy clustering means regression splines (MABFCMRS)
Graphical abstract
Specifications table
| Subject area: | Mathematics |
| More specific subject area: | Statistics: Non-parametric modeling |
| Name of your method: | Multivariate adaptive biresponse fuzzy clustering means regression splines (MABFCMRS) |
| Name and reference of original method: |
|
| Resource availability: | Stunting and wasting prevalence data in southeastern Sulawesi province, along with predictor variables for 2021, came from the health service of Southeast Sulawesi province. |
Background
One of the most popular mathematical models for explaining the relationship between a predictor and a response variable is regression. Regression curves describe these patterns of relationships and can be predicted using three types of approaches. The approaches used are parametric, non-parametric and semi-parameter regression approaches. A parametric regression approach is used when the shape of a regression curve is linear, quadratic, or cubic [1]. In cases where the regression curve's shape is unknown or nonexistent, the non-parametric regression approach applies [2]. If some forms of the regression pattern are known and some are unknown, then the semiparametric approach is used.
Non-parametric regression provides a versatile option for data modeling, particularly in the presence of non-linear and high-dimensional patterns. Typically, parametric regression is more inflexible than non-parametric regression because it is used when regression curves are predetermined. Some of the non-parametric regression approaches are spline methods [3], Fourier series estimator [2], kernel [4] and multivariate adaptive regression splines (MARS) [5]. MARS is a complex combination of truncated spline and recursive partition regression (RPR), capable of accommodating additive effects as well as predictor interactions, unlike truncated spline, which deals only with additive effects. With this flexibility, MARS is effective for regression analysis that includes the response variable of continuous data [6], categorical [7,8] and count data [[9], [10], [11], [12]]. MARS is capable of handling large-scale data and large data samples differently with a truncated spline, which is more suited for continuous response data. In addition, MARS produces a smooth model around the node based on the minimalization value of generalized cross-validation (GCV), showing his ability to reduce forecasting mistakes. The MARS model facilitates providing continuous answers with multiple good responses [13] neither category [14].
This research present the development of a multivariate adaptive regression splines biresponse continuous (MARSBC) model enchanced by a fuzzy clustering means (FCM) approach, termed the multivariate adaptive bisresponse fuzzy clustering means regression splines (MABFCMRS). The main objective of this model is to account for heterogeneity that was not included in the prior model and was not seen. This method differs from conventional fuzzy clustering methods that use Euclid distance since it bases cluster creation on the total square of the residual distance given the weight derived from the MARSBC model. A modification of these two processes offers results that differ from the prior model. Compared to the prior model, the resulting one has more complex basis functions and parameters. Based on the theoretical framework of information, this study suggests a total number of weighted sum-of-squared-errors criteria to determine an estimate of the MABFCMRS model.
Method details
Model specifications and estimation procedures
MARS Model
The model was first introduced by Friedman in 1991. MARS is a non-parametric regression model that incorporates many different types of regression, including recursive partitioning regression (RPR) and truncated spline regression, in a sophisticated way [5]. The modeling features of MARS allow it to consider factors in additive effects and interactions between predictors, which are challenges that spline cannot overcome. MARS versatility extends to a wide range of applications, encompassing both categorical [7,14,15] and continuous responses [[16], [17], [18], [19]], making it more powerful than a truncated spline. MARS, using an approach that is both effective and useful, can estimate models of high-dimensional datasets [5]. The model is produced in a continuous MARS on a node that cannot be obtained from RPR. Model formation is conducted in two stages, with stepwise procedures for forming basis functions (BF), maximum interactions (MI), and minimum observations. (MO). The first stage is a forward selection, which involves selecting all of the available basis functions derived with the highest feasible complexity. The second stage involves a backward selection, where the complexity of the formed basis function is reduced based on the smallest GCV value [5,6]. The form of the MARS model is seen in Eq. (1). Given the response variable:
| (1) |
where represent the vector of predictor variables, with p indicating the total number of predictors, represents a random error that has a mean value of zero and a variance that remains constant, is the total number of observations. M denotes the number of non-constant basis functions. is a function parameter of a constant basis, is a non-constant-m basis function. is mth basis function that declared in Eq. (2).
| (2) |
with , is the maximum interaction of mth basis function, is the computation that involves multiplying the independent variables of the kth and mth basis function, is the value of the kth knot on interaction, the mth basis function, and is the sign of the basis function.
MARSBC model
MARSBC is multivariate adaptive regression splines biresponse continuous, which builds upon the MARS paradigm [5] that was initially proposed in 2015 by Ampulembang [13]. A non-parametric regression model with biresponse continuous can get close to an unknown regression function of type . This lets the relationship be written as an Eq. (3)
| (3) |
is the number of responses. When written in matrix format,
| (4) |
where is parameter of MARSBC, and
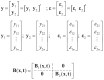 |
(5) |
assuming . The is a covariance error matrix. The random errors in MARSBC equation follow a continuous normal distribution with mean dan and covariance error matrices . is a weight matrix used to accommodate the correlation between responses through an estimation of the model parameters. Model parameter estimates assume the matrix is known (fixed variable). Eq. (6) illustrated the use of the maximum likelihood method to obtain parameter estimate for the MARSBC [13].
| (6) |
Estimate the matrix covariance error using the provided Eq. (7).
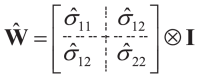 |
(7) |
with the Eq. (8):
| (8) |
Fuzzy clustering means
Fuzzy clustering means (FCM) is a non-hierarchical clustering method within fuzzy clusters, originally invented by Jim Bezdek in 1981 [20]. FCM is a method of clustering data in which the assigned position of each data point in a cluster is based on a membership value that fluctuates between 0 and 1 [21]. The iterative process known as FCM clustering uses the restrictions necessary to minimize objective functions. The grouping begins with the initialization of the center of the cluster, which is subsequently updated through iterations based on the objective function minimization in Eq. (9):
| (9) |
let is the data object, where denotes the value of the jth predictor variable, with j ranging from 1 to p. is the center of the cluster-, is the degree of membership i on the data cluster-. m is the weighted exponent of the fuzzifier, has the potential to influence FCM clustering performance. The membership value of an object achieve each aspect of the Eq. (10).
| (10) |
Optimizing functions using iterative approaches. The Lagrange multiplier examination was used to derive the formula for minimizing Eq. (9), subject to the constraint in Eq. (10). This resulted in the iteration update formula for the prototype and the partition matrix [22]:
| (11) |
| (12) |
Continue implementing the method outlined in Eqs. (11) and (12) to attain a value lower than the previous iteration, resulting in the lowest achievable value. The goal is to iteratively improve a series of sets on fuzzy clusters, and it should continue until no further improvements in are possible [23].
Multivariate fuzzy clustering means membership
The degree of membership in the Multivariate Fuzzy Clustering Means (MFCM) algorithm varies between one response and another, as well as between different clusters. With the goal of performing this, it is essential to ascertain the proper representation of membership, in addition to the methodology for computing the effective distance between the cluster and its prototype [24]. Known multivariate membership degree matrix , is the degree of membership from object-i to cluster-c, in respon tu- . Now suppose that there is a distance matrix of n where is the distance between patterns and prototype the cluster with a goal to, in response to- . According to MFCM, the goal is to observe multivariate fuzzy partitions based on the data set , during the process of minimizing the objective function using Eq. (13).
| (13) |
where is the center of the cluster seen in Eq. (14):
| (14) |
Index validity
Researchers primarily focus on identifying the optimal criterion for c that can reveal the data's structure. Most cluster methods require calculating c from the cluster. Working with the correct cluster size as an approach to determining the validity of the cluster is necessary to evaluate the size of a separate cluster [25]. The Partition Coefficient (PE) [[26], [27], [28]] were the first index validations linked to the FCM and Modification of PC (MPC) index, which Dave 1996 [29] presented. Eqs. (15) and (16) show the PC and MPC formulations appropriately.
| (15) |
where . Breaking determines the optimal number of c* clusters to optimize the cluster's efficacy for X data sets. The performance of the MPC can reduce monotony and is distinguished by
| (16) |
with . Generally, an optimal c* cluster will be obtained by breaking to produce the best cluster performance for the X data set.
MABFCMRS model
The MABFCMRS model is the development of the MARS Continuous Response model using the FCM approach. The MABFCMRS model can account for the variability of the process, resulting in a unique model for each cluster. The MABFCMRS model can be seen in the Eq. (17).
| (17) |
The Eq. (17) is represented as a matrix. The expression for the set of n observations is in Eq. (18).
| (18) |
where
by assume . is the parent basis's function parameter depending on the cluster's initial response and is a non-constant parameter to m in the initial response of the cluster to (c).
Parameter Estimation of MABFCMRS
The MBFCMRS parameter assumption determines the number of clusters in a data set using an objective function of the total number of square errors. This function is designed to identify a cluster partition that minimizes the objective function value, indicating the optimal cluster based on the provided parameters. The following theorem illustrates the MABFCMRS parameter.
Theorem 1
Given MABFCMRS model onEq. (18)withis the normal distribution, having a mean of 0 and a variance of. The FCM goal function is defined byEq. (13)which uses the approach weighted least squares to define the FCM goal function and then determines a model parameter MABFCMRS based on theEq. (19).
(19)
Proof of Theorem 1. The estimate of the MABFCMRS parameter is obtained by using the objective function of the total number of weighted sum-of-squared- errors criterion to find the number of clusters in the data set [30], as follows:
| (20) |
is the objective function of MABFCMRS model and for every i. is residual of the MABFCMRS model associated with cluster observations ith in cluster cth. Objective function Eq. (20) if written in vector notation, the matrix will be Eq. (21):
| (21) |
where
from Eq. (18) known that so
by
Where vector of size , matrix of size . is a basis function matrix having size and vector has a size of . The parameter predictor for the MABFCMRS model uses the Lagrange multiplier approach. Eq. (22) represents the objective function:
| (22) |
To find the parameter of the MABFCMRS model, the Eq. (22) is the derivative against the where the function is equated with zero, until obtained.
| (23) |
with
Fuzzy membership values
Determines fuzzy membership values using the langrange multiplier method. Deduct the assumption from the Eq. (24):
| (24) |
With a formula of for by . First derivative of relative to then equate zero until obtained for and specific:
| (25) |
In contrast, following the summation of Eq. (25) for every c and specific i then
| (26) |
Modify Eq. (26) with Eq. (25) and then acquire fuzzy membership values.
| (27) |
In order to obtain the MABFCMRS function meter, first it is necessary to substitute Eq. (23) into Eq. (18) then obtained function measurement MABFCMRS in Eq. (28).
| (28) |
where
GCV for MABFCMRS
GCV is associated with the concept of reducing the residual sum of squares [31]. The most powerful MARS type is the one with the lowest GCV. GCV is employed to choose the most optimal basis function for constructing the MARS model [5]. GCV formula can be seen in the Eq. (29).
| (29) |
with
is complex function, is the number of estimable parameters, d is the degree of interaction, indicating the additional contribution of each basis function to the overall complexity of the model resulting from the nonlinearity of the basis function parameters has been identified [32]. The optimum d value is in the interval , is the number of maximum nonconstant basis function and is the estimated value of response variable. The GCV MABFCMRS formula can be seen based on the Eq.(30).
| (30) |
Method validation
Application of MABFCMRS model
The MABFCMRS model obtained was applied to stunting and wasting prevalence data in Southeast Sulawesi province by 2021. Stunting is a length or lower height based on age, while wasting is a type of malnutrition that occurs when a child's weight is below the appropriate level of their height, according to the z-score according to the WHO nutritional status table [33]. The information that was collected comes from the health department of the southeastern province of Sulawesi, which has 222 subdistrict which is spread out in 17 districts and cities. There are two response variables and five predictor factors. The variable responses to this study are the prevalence of stunting and the prevalence of wasting . The predictor variable used is: is the percentage of babies with a low birth weight, is the percentage of pregnant mothers who are anemic, is the percentage of infants exclusively receiving breast milk at six months of age, is the percentage of babies getting a complete basic immunization, is the percentage of toddler getting vitamin A and is the percentage of mothers getting vitamin A. Table 1 shows the descriptive statistics of each variable. The relationships between each predictor variable and each responder variable displayed in Fig. 1, Fig. 2. Data pairs illustrate the geometric structure of the unidentified forms. Non-parametric regression is the appropriate approach to use in this research. This study contrasts two non-parametric regression methodologies: MARSBC and MABFCMRS.
Table 1.
Descriptive statistics based on research variables.
| Variable | Mean | StDev | Minimum | Maximum |
|---|---|---|---|---|
| 15.01 | 11.85 | 0 | 51.91 | |
| 4.34 | 5.07 | 0 | 45.09 | |
| 3.53 | 3.35 | 0 | 23.1 | |
| 43.68 | 37.4 | 0 | 101.4 | |
| 52 | 26.12 | 0 | 98.6 | |
| 85.82 | 30.48 | 0 | 201.7 | |
| 74.13 | 25.19 | 0 | 168.1 | |
| 86.51 | 18.89 | 17.9 | 157.1 |
Fig. 1.
Visualization of research variables of respon .
Fig. 2.
Visualization of research variables of respon .
Table 1 presents the descriptive statistics for the dependent variables, prevalence of stunting and prevelance of wasting and the independent variables (to ) employed in this research. The average value for is 15.01, with a standard deviation of 11.85, suggesting a moderate degree of variability from the average. This variable ranges from a minimum value of 0 to a maximum value of 51.91. , the second dependent variable, has a mean of 4.34 and a standard deviation of 5.07. It also has a range of values from 0 to 45.09, suggesting a more uniform distribution in comparison to . The independent variables through have significantly different mean values, ranging from 3.53 for to 86.51 for . These standard deviations also indicate different levels of dispersion, suggesting that there is variation in the spread of data for each variable. Some variables, like , show a much greater range of values, whereas shows limited oscillations in the range of minimum and maximum values, indicating significant disparities in the scale of measurement.
Fig. 1, Fig. 2 illustrate the distribution of predictor variables against response variables. The emerging pattern from these distributions lacks a clear form, suggesting that the Multivariate Adaptive Biresponse Fuzzy Clustering Means Regression Splines (MABFCMRS) model is suitable for further data analysis. Estimates of MARSBC models are shown in Table 2. The best model is obtained when the optimal BF= 22 MO =2 and MI= 3. Estimation of the MARSBC model resulted in 22 basis functions obtained for response 1 (prevelence of stunting) and response 2 (prevalence of wasting).
Table 2.
Model MARSBC.
| Model in response 1 |
Model in response 2 |
||
|---|---|---|---|
| Variables | Estimate | Variables | Estimate |
| (Intercept) | 0.1317 | (Intercept) | 0.1264 |
| bx_marsbc [, -1]h(X4-66.4) | 0.0421 | bx_marsbc [, -1]h(X3-56.9) | 0.0352 |
| bx_marsbc [, -1]h(66.4-X4) | -0.2614 | bx_marsbc [, -1]h(56.9-X3) | -0.0281 |
| bx_marsbc [, -1]h(X6-100) | 0.0214 | bx_marsbc [, -1]h(X4-124.1) | -0.2482 |
| bx_marsbc [, -1]h(100-X6) | 0.0056 | bx_marsbc [, -1]h(124.1-X4) | 0.2573 |
| bx_marsbc [, -1]h(X2-97.2) *h(X6-100) | 0.0107 | bx_marsbc [, -1]h(X3-56.9) *h(X6-109.6) | 0.6039 |
| bx_marsbc [, -1]h(97.2-X2) *h(X6-100) | 0.0010 | bx_marsbc [, -1]h(X3-56.9) *h(109.6-X6) | 0.0078 |
| bx_marsbc [, -1]h(X1-1.1) | 0.0433 | bx_marsbc [, -1]h(X2-100) *h(56.9-X3) | 0.8678 |
| bx_marsbc [, -1]h(1.1-X1) | 0.0042 | bx_marsbc [, -1]h(100-X2) *h(56.9-X3) | 0.0517 |
| bx_marsbc [, -1]h(X1-3.8) *h(X4-66.4) | -0.0002 | bx_marsbc [, -1]h(56.9-X3) *h(X5-42.7) | -0.0034 |
| bx_marsbc [, -1]h(3.8-X1) *h(X4-66.4) | -0.0014 | bx_marsbc [, -1]h(56.9-X3) *h(42.7-X5) | 0.0275 |
| bx_marsbc [, -1]h(1.1-X1) *h(X3-43) | 0.3286 | bx_marsbc [, -1]h(X1-3.3) | 0.2782 |
| bx_marsbc [, -1]h(1.1-X1) *h(43-X3) | 0.5103 | bx_marsbc [, -1]h(3.3-X1) | -0.0219 |
| bx_marsbc [, -1]h(X2-81.8) *h(100-X6) | 0.0319 | bx_marsbc [, -1]h(X1-3.3) *h(X6-100) | -0.0026 |
| bx_marsbc [, -1]h(81.8-X2) *h(100-X6) | -0.0012 | bx_marsbc [, -1]h(X1-3.3) *h(100-X6) | -0.0007 |
| bx_marsbc [, -1]h(X3-94.1) *h(100-X6) | 0.1700 | bx_marsbc [, -1]h(X2-3.4) | -0.1261 |
| bx_marsbc [, -1]h(94.1-X3) *h(100-X6) | 0.0698 | bx_marsbc [, -1]h(3.4-X2) | -0.0019 |
| bx_marsbc [, -1]h(1.1-X1) *h(X2-89.5) | -0.0004 | bx_marsbc [, -1]h(X2-3.4) *h(X6-63.3) | 0.7803 |
| bx_marsbc [, -1]h(1.1-X1) *h(89.5-X2) | 0.0000 | bx_marsbc [, -1]h(X2-3.4) *h(63.3-X6) | -0.0712 |
| bx_marsbc [, -1]h(1.1-X1) *h(X5-96.8) | -0.0021 | bx_marsbc [, -1]h(X2-3.4) *h(X3-8.3) | 0.4917 |
| rbx_marsbc [, -1]h(X3-75.9) | -0.0064 | bx_marsbc [, -1]h(3.3-X1) *h(X3-44.2) | 0.3072 |
| bx_marsbc [, -1]h(75.9-X3) | -0.0024 | bx_marsbc [, -1]h(3.3-X1) *h(44.2-X3) | 0.2318 |
The model for response 1, derived from Table 2, is presented in Eq. (31).
| (31) |
where
| (32) |
The Eq. (31) shows that all predictor variables have an impact on the response . The interpretation of the model using basis function bf1 indicates that for values of greater than 66.4, there will be an increase in the prevalence of stunting by 0.0421, assuming all other predictor variables are held constant. But, if is less than 1.1 and is less than 96.8, the product of their interactions will make stunting more common by 0.0036, assuming that all other predictors stay the same. This is shown by the interaction term in basis function bf20. It is important to note that represents the percentage of babies born with low birth weight, x4 indicates the percentage of babies receiving complete basic immunization, and corresponds to the percentage of toddlers receiving vitamin A supplementation. These variables are critical because the model assumes linearity and interaction effects are specified.
| (33) |
where
| (34) |
The Eq. (33) displays that all predictor variables have an impact on the response . In the model utilizing basis function bf1, when exceeds 56.9, the prevalence of wasting increases by 0.0352, assuming all other predictor variables remain constant. On the other hand, if is less than 33.1 and is greater than 44.2, their interaction within the basis function bf21 causes the prevalence of stunting to rise by 0.3072, assuming that all other variables stay the same. It is crucial to note that represents the percentage of babies born with low birth weight, while indicates the percentage of infants exclusively breastfed at six months of age. Both variables are essential, as the model assumes linearity and interaction effects among the variables.
After establishing the MARSBC model based on the optimal basis functions determined using the FCM approach, the MABFCMRS model was subsequently created. During this development phase, we validated the indices using PC and MPC, as outlined in Eq. (24). Table 3 displays the results of the clustering index validation using PC and MPC.
Table 3.
Validation index number of clusters.
| Number of cluster | PC | MPC |
|---|---|---|
| 2 | 0.76115117 | 0.52230234 |
| 3 | 0.51952332 | 0.27928498 |
| 4 | 0.39922768 | 0.19897024 |
| 5 | 0.33300297 | 0.16625371 |
The validation index calculation using PC and MPC indicates that the optimal number of clusters is 2. This is derived from the optimum PC and MPC values. The value of the fuzzy membership degree is shown in Appendix A. Appendix A displays the distribution of the incidence spreads that enter each cluster. Out of all the existing incidences, 102 were included in cluster 1 and 120 were included in cluster 2. The MABFCMRS model designed for cluster 1 is shown in Table 4. The best model is achieved when BF = 20, MO = 1, and MI = 2.
Table 4.
MABFCMRS Cluster 1 model.
| Response 1 | |
|---|---|
| Model | Estimate |
| (Intercept) | 30.0914 |
| bx_mabfcmrs [, -1]h(X3-80) | -1.7595 |
| bx_mabfcmrs [, -1]h(110.2-X4) | -0.3997 |
| bx_mabfcmrs [, -1]h(X1-1.2) *h(X4-75.8) *h(X6-96.8) | 0.0039 |
| bx_mabfcmrs [, -1]h(X1-1.2) *h(75.8-X4) *h(X6-96.8) | 0.0183 |
| bx_mabfcmrs [, -1]h(1.2-X1) *h(29.7-X2) | -0.5832 |
| bx_mabfcmrs [, -1]h(1.2-X1) *h(29.7-X2) *h(X3-68.2) | 0.0270 |
| bx_mabfcmrs [, -1]h(X3-79.1) *h(110.2-X4) | 0.0240 |
| bx_mabfcmrs [, -1]h(79.1-X3) *h(110.2-X4) | 0.0049 |
| Response 2 | |
| Model | Estimate |
| (Intercept) | 7.2643 |
| bx_mabfcmrs [, -1]h(X3-54.7) | -0.6118 |
| bx_mabfcmrs [, -1]h(X4-91.3) | -0.1633 |
| bx_mabfcmrs [, -1]h(63.3-X6) | 0.5623 |
| bx_mabfcmrs [, -1]h(39.1-X2) *h(54.7-X3) | 0.0061 |
| bx_mabfcmrs [, -1]h(99-X2) *h(91.3-X4) | -0.0017 |
| bx_mabfcmrs [, -1]h(X2-94) *h(X3-54.7) | -0.0117 |
| bx_mabfcmrs [, -1]h(94-X2) *h(X3-54.7) | 0.0025 |
| bx_mabfcmrs [, -1]h(X3-57.1) | -2.3486 |
| bx_mabfcmrs [, -1]h(X2-4.2) *h(X3-54) | 0.0279 |
| bx_mabfcmrs [, -1]h(4.2-X2) *h(X3-54) | -0.0485 |
| bx_mabfcmrs [, -1]h(94.6-X2) *h(X3-57.1) | 0.0313 |
Table 4 MABFCMRS cluster 1 model for response 1 yields a model as depicted in Eq.(35). It is observed that out of the six predictor variables utilized, only five significantly influence the prevalence of stunting. These impactful variables are , , , and Eq. (35) describes the model where bf6 represents the interaction among three variables: , , . This suggests that the prevalence of stunting will increase by 0.0270 if is less than 1.2, is less than 29.7, and is greater than 68.2, assuming all other variables remain constant.
| (35) |
where
| (36) |
Table 4 features the MABFCMRS model for response 2 in cluster 1 presents a model as shown in Eq. (37). Of the six predictor variables used, only four (, , and ) have a significant impact. The interpretation of basis function bf11 indicates that if is less than 94 and is greater than 57.1, the prevalence of wasting will increase by 0.0313, assuming all other variables are held constant.
| (37) |
where
| (38) |
The MABFCMRS model designed for cluster 2 is shown in Table 5. The best model is achieved when BF = 22, MO = 3, and MI = 1.
Table 5.
MABFCMRS Cluster 2 model.
| Response 1 | |
|---|---|
| Model | Estimate |
| (Intercept) | 0.0008 |
| bx_mabfcmrs [, -1]h(1.3-X1) *h(X3-47.4) | 0.0008 |
| bx_mabfcmrs [, -1]h(1.3-X1) *h(X2-89.5) | -0.0005 |
| bx_mabfcmrs [, -1]h(1.3-X1) *h(89.5-X2) | 0.0924 |
| bx_mabfcmrs [, -1]h(X1-1.3) *h(X3-73.3) | -0.0008 |
| bx_mabfcmrs [, -1]h(X1-1.3) *h(73.3-X3) | 0.0412 |
| bx_mabfcmrs [, -1]h(X1-1.3) *h(X2-57.1) *h(X3-73.3) | 0.0030 |
| bx_mabfcmrs [, -1]h(X1-1.3) *h(57.1-X2) *h(X3-73.3) | 0.0027 |
| bx_mabfcmrs [, -1]h(1.3-X1) *h(47.4-X3) *h(X4-95.5) | 0.0032 |
| bx_mabfcmrs [, -1]h(1.3-X1) *h(47.4-X3) *h(95.5-X4) | 0.0093 |
| bx_mabfcmrs [, -1]h(X6-100) | 0.0075 |
| bx_mabfcmrs [, -1]h(100-X6) | -0.0054 |
| bx_mabfcmrs [, -1]h(1.3-X1) *h(X3-47.4) *h(X4-64.3) | 0.0123 |
| bx_mabfcmrs [, -1]h(1.3-X1) *h(X3-47.4) *h(64.3-X4) | 0.0077 |
| bx_mabfcmrs [, -1]h(X1-1.3) *h(73.3-X3) *h(111.3-X6) | -0.0005 |
| bx_mabfcmrs [, -1]h(136.3-X4) *h(X6-111.7) | 0.0480 |
| bx_mabfcmrs [, -1]h(136.3-X4) *h(111.7-X6) | 0.0014 |
| Response 2 | |
| Model | Estimate |
| (Intercept) | 0.0029 |
| bx_mabfcmrs [, -1]h(X2-98.8) *h(X5-77) | 0.0025 |
| bx_mabfcmrs [, -1]h(X6-98) | -0.0333 |
| bx_mabfcmrs [, -1]h(98-X6) | 0.0649 |
| bx_mabfcmrs [, -1]h(X6-49.3) | 0.0443 |
| bx_mabfcmrs [, -1]h(X3-87.4) *h(X6-98) | 0.0784 |
| bx_mabfcmrs [, -1]h(X3-71) *h(98-X6) | -0.0003 |
| bx_mabfcmrs [, -1]h(71-X3) *h(X4-50.6) *h(98-X6) | 0.0002 |
| bx_mabfcmrs [, -1]h(3.7-X2) *h(X6-49.3) | -0.0076 |
Table 5 for response 1 presents the parameter estimates of the MABFCMRS model for cluster 2, as shown in Eq. (39) Of the six predictor variables used, only four significantly influence the prevalence of stunting: , , , , and . The interpretation for basis function bf11 suggests that if is less than 100, the prevalence of stunting will decrease by 0.0054, assuming all other variables are held constant.
| (39) |
where
| (40) |
Table 5 for response 2 displays the parameter estimates of the MABFCMRS model for Cluster 2, as detailed in Eq. (41). Among the 6 predictor variables employed, only five significantly affect the prevalence of stunting: , , , and . If does not exceeds 98, the basis function bf3 interpretation indicates a 0.0649 increase in the prevalence of wasting, assuming all other variables remain constant
| (41) |
where
| (42) |
Comparison of the Goodness of fit model can be seen in Table 6. The MABFCMRS model in cluster 1 and cluster 2 has the smallest GCV and MSE value. Similarly, the R-square value shows a substantial rise from the MARSBC model to the MABFCMRS model.
Table 6.
Comparison of the goodness of fit model.
| Model | Criteria |
N | ||
|---|---|---|---|---|
| MSE | R2 | GCV | ||
| MARSBC | 187.214596 | 0.32065947 | 211.099929 | 222 |
| MABFCMRS Cluster1 | 54.9758835 | 0.67400923 | 134.91909 | 102 |
| MABFCMRS Cluster 2 | 19.9363767 | 0.74444143 | 41.5878378 | 120 |
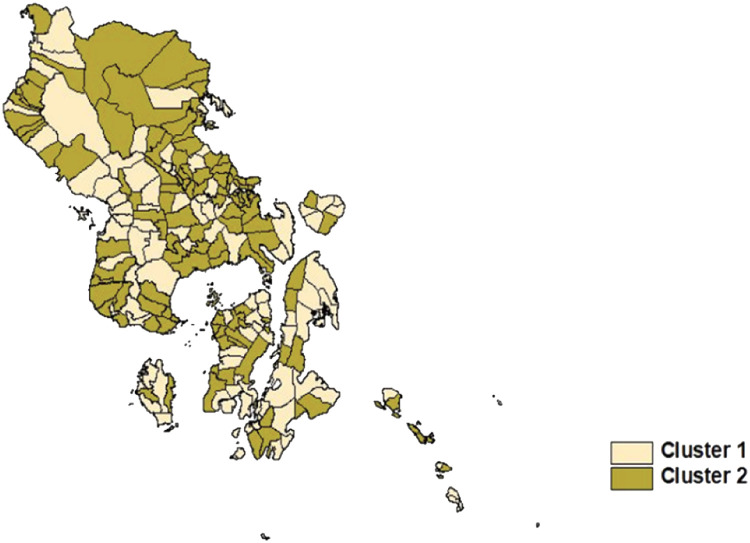
The MABFCMRS model in Fig. 3 categorizes the subdistricts based on the maximum validation values of the PC and MPC indices. The 222 subdistricts in Southeast Sulawesi province are divided into two clusters: the lighter color represents cluster 1, while the darker color represents cluster 2.
Fig. 3.
Southeast Sulawesi cluster distribution map.
Conclusion
This study developed a new MABFCMRS model that combines the MARSBC model and the FCM method. The MABFCMRS model is designed to involve unobserved heterogeneity. Creates a data cluster based on a previously established model cluster center. This makes the MABFCMRS model more complicated than the MARSBC model. The MABFCMRS model was applied to the prevalence of stunting and wasting in 222 subdistricts in the southeastern province of Sulawesi. Based on PC and MPC index validation values, there are two clusters. Cluster 1 groups 102 clusters, and cluster 2 groups 120. Based on the goodness of fit values, MSE, R2, and GCV show that the MABFCMRS model is better than the MARSBC model.
Limitations
In order to avoid the increased complexity and challenges associated with interpreting more extensive interactions, this study identified a limitation in the MABFCMRS model, restricting it to only include up to three interactions. Additionally, the current article does not include hypothesis testing for the proposed model, presenting an opportunity for future research to explore this aspect. The model's adaptability also allows for the inclusion of categorical and count data in subsequent studies.
CRediT authorship contribution statement
Mira Meilisa: Conceptualization, Methodology, Formal analysis, Writing – original draft, Visualization. Bambang Widjanarko Otok: Conceptualization, Methodology, Writing – review & editing, Supervision. Jerry Dwi Trijoyo Purnomo: Methodology, Validation, Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Ethics statements
The proposed study uses secondary and primary data. Secondary data was obtained from the Southeast Sulawesi Provincial Health Service for 2021. Primary data was retrieved from focus group discussions (FGDs) to confirm and validate research data with the Southeast Sulawesi Provincial Health Office.
Acknowledgments
The authors expressed their gratitude to Kementrian Pendidikan, Kebudayaan, Risetdan Teknologi (Center for Higher Education Funding - BPPT) and Lembaga Pengelolaan Dana Pendidikan (LPDP) for their support in implementing this research.
Appendix A
Table A1. Degree of membership value for MABFCMRS to determine the sub-districts in Southeast Sulawesi province that belong to each cluster.
Table A1.
Degree of membership value for MABFCMRS.
| No. | Subdistrict | Cluster 1 | Cluster 2 | Number of clusters | No. | Subdistrict | Cluster 1 | Cluster 2 | Number of clusters |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Kapontori | 0.7039 | 0.2961 | 1 | 112 | Poleang Utara | 0.9411 | 0.0589 | 1 |
| 2 | Lasalimu | 0.6803 | 0.3197 | 1 | 113 | Rarowatu | 0.0513 | 0.9487 | 2 |
| 3 | Lasalimu Selatan | 0.7895 | 0.2105 | 1 | 114 | Rarowatu Utara | 0.1195 | 0.8805 | 2 |
| 4 | Pasar Wajo | 0.9253 | 0.0747 | 1 | 115 | Rumbia | 0.0027 | 0.9973 | 2 |
| 5 | Siontapina | 0.4932 | 0.5068 | 2 | 116 | Rumbia Tengah | 0.1884 | 0.8116 | 2 |
| 6 | Wabula | 0.8059 | 0.1941 | 1 | 117 | Tontonunu | 0.0397 | 0.9603 | 2 |
| 7 | Wolowa | 0.0531 | 0.9469 | 2 | 118 | Binongko | 0.6248 | 0.3752 | 1 |
| 8 | Batalaiworu | 0.1612 | 0.8388 | 2 | 119 | Kaledupa | 0.2647 | 0.7353 | 2 |
| 9 | Batukara | 0.7274 | 0.2726 | 1 | 120 | Kaledupa Selatan | 0.0023 | 0.9977 | 2 |
| 10 | Bone | 0.1785 | 0.8215 | 2 | 121 | Togo Binongko | 0.6005 | 0.3995 | 1 |
| 11 | Duruka | 0.1249 | 0.8751 | 2 | 122 | Tomia | 0.9319 | 0.0681 | 1 |
| 12 | Kabangka | 0.2495 | 0.7505 | 2 | 123 | Tomia Timur | 0.4765 | 0.5235 | 2 |
| 13 | Kabawo | 0.9619 | 0.0381 | 1 | 124 | Wangi-Wangi | 0.9375 | 0.0625 | 1 |
| 14 | Katobu | 0.8793 | 0.1207 | 1 | 125 | Wangi-Wangi Selatan | 0.1604 | 0.8396 | 2 |
| 15 | Kontukowuna | 0.1144 | 0.8856 | 2 | 126 | Uluiwoi | 0.9999 | 0.0001 | 1 |
| 16 | Kontunaga | 0.9010 | 0.0990 | 1 | 127 | Batu Putih | 0.7517 | 0.2483 | 1 |
| 17 | Lasalepa | 0.7625 | 0.2375 | 1 | 128 | Katoi | 0.1286 | 0.8714 | 2 |
| 18 | Lohia | 0.8736 | 0.1264 | 1 | 129 | Kodeoha | 0.0074 | 0.9926 | 2 |
| 19 | Maligano | 0.0167 | 0.9833 | 2 | 130 | Lambai | 0.1037 | 0.8963 | 2 |
| 20 | Marobo | 0.0844 | 0.9156 | 2 | 131 | Lasusua | 0.2740 | 0.7260 | 2 |
| 21 | Napabalano | 0.9643 | 0.0357 | 1 | 132 | Ngapa | 0.0668 | 0.9332 | 2 |
| 22 | Parigi | 0.9334 | 0.0666 | 1 | 133 | Pakue | 0.5195 | 0.4805 | 1 |
| 23 | Pasi Kolaga | 0.9682 | 0.0318 | 1 | 134 | Pakue Tengah | 0.2492 | 0.7508 | 2 |
| 24 | Pasir Putih | 0.1287 | 0.8713 | 2 | 135 | Pakue Utara | 0.9225 | 0.0775 | 1 |
| 25 | Tongkuno | 0.0417 | 0.9583 | 2 | 136 | Porehu | 0.1555 | 0.8445 | 2 |
| 26 | Tongkuno Selatan | 0.9118 | 0.0882 | 1 | 137 | Ranteangin | 0.0020 | 0.9980 | 2 |
| 27 | Towea | 0.5925 | 0.4075 | 1 | 138 | Tiwu | 0.2740 | 0.7260 | 2 |
| 28 | Wakorumba Selatan | 0.8681 | 0.1319 | 1 | 139 | Tolala | 0.8487 | 0.1513 | 1 |
| 29 | Watopute | 0.9316 | 0.0684 | 1 | 140 | Watunohu | 0.7360 | 0.2640 | 1 |
| 30 | Abuki | 0.0837 | 0.9163 | 2 | 141 | Wawo | 0.7581 | 0.2419 | 1 |
| 31 | Amonggedo | 0.1121 | 0.8879 | 2 | 142 | Bonegunu | 0.2203 | 0.7797 | 2 |
| 32 | Anggaberi | 0.0016 | 0.9984 | 2 | 143 | Kalisusu | 0.7896 | 0.2104 | 1 |
| 33 | Anggalomoare | 0.2332 | 0.7668 | 2 | 144 | Kambowa | 0.9032 | 0.0968 | 1 |
| 34 | Anggotoa | 0.0238 | 0.9762 | 2 | 145 | Kulisusu Barat | 0.8953 | 0.1047 | 1 |
| 35 | Asinua | 0.6343 | 0.3657 | 1 | 146 | Kulisusu Utara | 0.0142 | 0.9858 | 2 |
| 36 | Bandoala | 0.5237 | 0.4763 | 1 | 147 | Wakorumba Utara | 0.1160 | 0.8840 | 2 |
| 37 | Besulutu | 0.0663 | 0.9337 | 2 | 148 | Andowia | 0.0762 | 0.9238 | 2 |
| 38 | Kapoiala | 0.4619 | 0.5381 | 2 | 149 | Asera | 0.0376 | 0.9624 | 2 |
| 39 | Konawe | 0.1399 | 0.8601 | 2 | 150 | Landawe | 0.0117 | 0.9883 | 2 |
| 40 | Lalonggasumeeto | 0.0676 | 0.9324 | 2 | 151 | Langgikima | 0.4067 | 0.5933 | 2 |
| 41 | Lambuya | 0.0397 | 0.9603 | 2 | 152 | Lasolo | 0.9994 | 0.0006 | 1 |
| 42 | Latoma | 0.0063 | 0.9937 | 2 | 153 | Lasolo Kepulauan | 0.2913 | 0.7087 | 2 |
| 43 | Meluhu | 0.8239 | 0.1761 | 1 | 154 | Lembo | 0.0500 | 0.9500 | 2 |
| 44 | Morosi | 0.2426 | 0.7574 | 2 | 155 | Molawe | 0.2367 | 0.7633 | 2 |
| 45 | Onembute | 0.0175 | 0.9825 | 2 | 156 | Motui | 0.8340 | 0.1660 | 1 |
| 46 | Padangguni | 0.2026 | 0.7974 | 2 | 157 | Oheo | 0.0838 | 0.9162 | 2 |
| 47 | Pondidaha | 0.4879 | 0.5121 | 2 | 158 | Sawa | 0.8952 | 0.1048 | 1 |
| 48 | Puriala | 0.9915 | 0.0085 | 1 | 159 | Wawolesea | 0.0720 | 0.9280 | 2 |
| 49 | Routa | 0.0109 | 0.9891 | 2 | 160 | Wiwirano | 0.7769 | 0.2231 | 1 |
| 50 | Sampara | 0.2058 | 0.7942 | 2 | 161 | Aere | 0.0292 | 0.9708 | 2 |
| 51 | Soropia | 0.0723 | 0.9277 | 2 | 162 | Dangia | 0.1693 | 0.8307 | 2 |
| 52 | Tongauna | 0.5509 | 0.4491 | 1 | 163 | Ladongi | 0.0085 | 0.9915 | 2 |
| 53 | Tongauna Utara | 0.7730 | 0.2270 | 1 | 164 | Lalolae | 0.9978 | 0.0022 | 1 |
| 54 | Uepai | 0.2092 | 0.7908 | 2 | 165 | Lambandia | 0.7515 | 0.2485 | 1 |
| 55 | Unaaha | 0.5181 | 0.4819 | 1 | 166 | Loea | 0.3561 | 0.6439 | 2 |
| 56 | Wawotobi | 0.0142 | 0.9858 | 2 | 167 | Mowewe | 0.6602 | 0.3398 | 1 |
| 57 | Wonggeduku | 0.0725 | 0.9275 | 2 | 168 | Poli-Polia | 0.9493 | 0.0507 | 1 |
| 58 | Wonggeduku Barat | 0.2465 | 0.7535 | 2 | 169 | Tinondo | 0.7566 | 0.2434 | 1 |
| 59 | Baula | 0.7151 | 0.2849 | 1 | 170 | Tirawuta | 0.9861 | 0.0139 | 1 |
| 60 | Iwoimendaa | 0.0410 | 0.9590 | 2 | 171 | Uesi | 0.7781 | 0.2219 | 1 |
| 61 | Kolaka | 0.9697 | 0.0303 | 1 | 172 | Wawonii Barat | 0.1286 | 0.8714 | 2 |
| 62 | Latambaga | 0.7456 | 0.2544 | 1 | 173 | Wawonii Selatan | 0.6112 | 0.3888 | 1 |
| 63 | Polinggona | 0.6074 | 0.3926 | 1 | 174 | Wawonii Tengah | 0.9911 | 0.0089 | 1 |
| 64 | Pomalaa | 0.6024 | 0.3976 | 1 | 175 | Wawonii Tenggara | 0.0065 | 0.9935 | 2 |
| 65 | Samaturu | 0.6327 | 0.3673 | 1 | 176 | Wawonii Timur | 0.6458 | 0.3542 | 1 |
| 66 | Tanggetada | 0.2852 | 0.7148 | 2 | 177 | Wawonii Timur Laut | 0.9826 | 0.0174 | 1 |
| 67 | Toari | 0.1782 | 0.8218 | 2 | 178 | Wawonii Utara | 0.9974 | 0.0026 | 1 |
| 68 | Watubangga | 0.3969 | 0.6031 | 2 | 179 | Barangka | 0.0946 | 0.9054 | 2 |
| 69 | Wolo | 0.2452 | 0.7548 | 2 | 180 | Kusambi | 0.0303 | 0.9697 | 2 |
| 70 | Wundulako | 0.5091 | 0.4909 | 1 | 181 | Lawa | 0.8350 | 0.1650 | 1 |
| 71 | Andoolo | 0.3676 | 0.6324 | 2 | 182 | Maginti | 0.1734 | 0.8266 | 2 |
| 72 | Andoolo Barat | 0.9353 | 0.0647 | 1 | 183 | Napano Kusambi | 0.7787 | 0.2213 | 1 |
| 73 | Angata | 0.0093 | 0.9907 | 2 | 184 | Sawerigadi | 0.0975 | 0.9025 | 2 |
| 74 | Baito | 0.6736 | 0.3264 | 1 | 185 | Tiworo Kepulauan | 0.9882 | 0.0118 | 1 |
| 75 | Basala | 0.0208 | 0.9792 | 2 | 186 | Tiworo Selatan | 0.4487 | 0.5513 | 2 |
| 76 | Benua | 0.5793 | 0.4207 | 1 | 187 | Tiworo Tengah | 0.1652 | 0.8348 | 2 |
| 77 | Buke | 0.1688 | 0.8312 | 2 | 188 | Tiworo Utara | 0.0466 | 0.9534 | 2 |
| 78 | Kolono | 0.0690 | 0.9310 | 2 | 189 | Wa Daga | 0.0107 | 0.9893 | 2 |
| 79 | Kolono Timur | 0.5351 | 0.4649 | 1 | 190 | Gu | 0.6983 | 0.3017 | 1 |
| 80 | Konda | 0.1088 | 0.8912 | 2 | 191 | Lakudo | 0.8179 | 0.1821 | 1 |
| 81 | Laeya | 0.9764 | 0.0236 | 1 | 192 | Mawasangka | 0.3564 | 0.6436 | 2 |
| 82 | Lainea | 0.3826 | 0.6174 | 2 | 193 | Mawasangka Tengah | 0.6652 | 0.3348 | 1 |
| 83 | Lalembuu | 0.1031 | 0.8969 | 2 | 194 | Mawasangka Timur | 0.9583 | 0.0417 | 1 |
| 84 | Landono | 0.8695 | 0.1305 | 1 | 195 | Sangia Wambulu | 0.0027 | 0.9973 | 2 |
| 85 | Laonti | 0.6962 | 0.3038 | 1 | 196 | Talaga Raya | 0.7931 | 0.2069 | 1 |
| 86 | Moramo | 0.0039 | 0.9961 | 2 | 197 | Batauga | 0.0358 | 0.9642 | 2 |
| 87 | Moramo Utara | 0.0039 | 0.9961 | 2 | 198 | Batuatas | 0.9899 | 0.0101 | 1 |
| 88 | Mowila | 0.6002 | 0.3998 | 1 | 199 | Kadatua | 0.9357 | 0.0643 | 1 |
| 89 | Palangga | 0.2098 | 0.7902 | 2 | 200 | Lapandewa | 0.9990 | 0.0010 | 1 |
| 90 | Palangga Selatan | 0.0733 | 0.9267 | 2 | 201 | Sampolawa | 0.2096 | 0.7904 | 2 |
| 91 | Ranomeeto | 0.8837 | 0.1163 | 1 | 202 | Siompu | 0.5026 | 0.4974 | 1 |
| 92 | Ranometoo Barat | 0.0695 | 0.9305 | 2 | 203 | Siompu Barat | 0.9971 | 0.0029 | 1 |
| 93 | Sabulakoa | 0.8121 | 0.1879 | 1 | 204 | Abeli | 0.1991 | 0.8009 | 2 |
| 94 | Tinanggea | 0.0211 | 0.9789 | 2 | 205 | Baruga | 0.0684 | 0.9316 | 2 |
| 95 | Wolasi | 0.0580 | 0.9420 | 2 | 206 | Kadia | 0.8064 | 0.1936 | 1 |
| 96 | Kabaena | 0.0537 | 0.9463 | 2 | 207 | Kambu | 0.0920 | 0.9080 | 2 |
| 97 | Kabaena Barat | 0.7824 | 0.2176 | 1 | 208 | Kendari | 0.2313 | 0.7687 | 2 |
| 98 | Kabaena Selatan | 0.7598 | 0.2402 | 1 | 209 | Kendari Barat | 0.1846 | 0.8154 | 2 |
| 99 | Kabaena Tengah | 0.9589 | 0.0411 | 1 | 210 | Mandonga | 0.3307 | 0.6693 | 2 |
| 100 | Kabaena Timur | 0.1982 | 0.8018 | 2 | 211 | Nambo | 0.2062 | 0.7938 | 2 |
| 101 | Kabaena Utara | 0.9564 | 0.0436 | 1 | 212 | Poasia | 0.2365 | 0.7635 | 2 |
| 102 | Kep. Masaloka Raya | 0.9776 | 0.0224 | 1 | 213 | Puuwatu | 0.8947 | 0.1053 | 1 |
| 103 | Lantari Jaya | 0.9652 | 0.0348 | 1 | 214 | Wua-Wua | 0.2462 | 0.7538 | 2 |
| 104 | Mataoleo | 0.0350 | 0.9650 | 2 | 215 | Batupoaro | 0.1842 | 0.8158 | 2 |
| 105 | Matausu | 0.8621 | 0.1379 | 1 | 216 | Betoambari | 0.6757 | 0.3243 | 1 |
| 106 | Poleang | 0.1038 | 0.8962 | 2 | 217 | Bungi | 0.7136 | 0.2864 | 1 |
| 107 | Poleang Barat | 0.4822 | 0.5178 | 2 | 218 | Kokalukuna | 0.6965 | 0.3035 | 1 |
| 108 | Poleang Selatan | 0.1768 | 0.8232 | 2 | 219 | Lea-Lea | 0.8083 | 0.1917 | 1 |
| 109 | Poleang Tengah | 0.9368 | 0.0632 | 1 | 220 | Murhum | 0.2156 | 0.7844 | 2 |
| 110 | Poleang Tenggara | 0.1081 | 0.8919 | 2 | 221 | Sorawolio | 0.0256 | 0.9744 | 2 |
| 111 | Poleang Timur | 0.7637 | 0.2363 | 1 | 222 | Wolio | 0.9955 | 0.0045 | 1 |
Data availability
The authors do not have permission to share data.
References
- 1.Draper N.R., Smith H. 3rd ed. John Wiley & Sons, Inc; 1998. Applied Regression Analysis. [Google Scholar]
- 2.Eubank R.L. 2nd ed. Marcel Dekker Inc; 1999. Nonparametric Regression and Spline Smoothing. [Google Scholar]
- 3.Wahba G. Society for Industrial and Applied Mathematics; 1990. Spline Models for Observational Data. [Google Scholar]
- 4.Hardle W. Applied nonparametric regression. J. R. Stat. Soc. Ser. A. 1994:433. doi: 10.2307/2982873. [DOI] [Google Scholar]
- 5.Friedman J.H. Multivariate adaptive regression splines. Ann. Stat. 1991;19:1–141. [Google Scholar]
- 6.Bekar Adiguzel M., Cengiz M.A. Model selection in multivariate adaptive regressions splines (MARS) using alternative information criteria. Heliyon. 2023;9 doi: 10.1016/j.heliyon.2023.e19964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meilisa M., Otok B.W., Purnomo J.D.T. Factor affecting the severity of the dengue fever using multivariate adaptive regression spline (Mars) method. AIP Conf. Proc. 2023:2556. doi: 10.1063/5.0131508. [DOI] [Google Scholar]
- 8.Leathwick J.R., Elith J., Hastie T. Comparative performance of generalized additive models and multivariate adaptive regression splines for statistical modelling of species distributions. Ecol. Modell. 2006;199:188–196. doi: 10.1016/j.ecolmodel.2006.05.022. [DOI] [Google Scholar]
- 9.Sriningsih R., Otok B.W. Sutikno, determination of the best multivariate adaptive geographically weighted generalized poisson regression splines model employing generalized cross-validation in dengue fever cases. MethodsX. 2023;10 doi: 10.1016/j.mex.2023.102174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sriningsih R., Otok B.W. Sutikno, factors affecting the number of dengue fever cases in West Sumatra province using the Multivariate Adaptive Regression Splines (MARS) approach. J. Phys. Conf. Ser. 2021 doi: 10.1088/1742-6596/1722/1/012094. [DOI] [Google Scholar]
- 11.Yasmirullah S.D.P., Otok B.W., Purnomo J.D.T., Prastyo D.D. Parameter estimation of spatial error model – multivariate adaptive generalized poisson regression spline. Eng. Lett. 2023;31:1265–1272. doi: 10.5267/j.dsl.2023.7.004. [DOI] [Google Scholar]
- 12.Yasmirullah S.D.P., Otok B.W., Trijoyo Purnomo J.D., Prastyo D.D. A hybrid model of spatial autoregressive-multivariate adaptive generalized Poisson regression spline. Decis. Sci. Lett. 2023;12:721–728. doi: 10.5267/dsl.2023.7.004. [DOI] [Google Scholar]
- 13.Ampulembang A.P., Otok B.W., Rumiati A.T. Budiasih, Bi-responses nonparametric regression model using MARS and its properties. Appl. Math. Sci. 2015;9:1417–1427. doi: 10.12988/ams.2015.5127. [DOI] [Google Scholar]
- 14.Ramadany R., Otok B.W. 3rd Int. Conf. Sci. Math. Environmet Educ. Flex. Res. Innov. Sci. Math. Environ. Educ. Sustain. Dev. 2023. Purhadi, parameter estimation and hypothesis testing of multivariate adaptive bivariate generalized poisson regression spline. [Google Scholar]
- 15.Yasmirullah S.D.P., Otok B.W., Purnomo J.D.T., Prastyo D.D. Parameter estimation of Multivariate Adaptive Regression Spline (MARS) with stepwise approach to Multi Drug-Resistant Tuberculosis (MDR-TB) modeling in lamongan regency. J. Phys. Conf. Ser. ICSMTR. 2019:2021. doi: 10.1088/1742-6596/1752/1/012017. [DOI] [Google Scholar]
- 16.Otok B.W., Rumiati A.T., Ampulembang A.P., Al Azies H. Anova decomposition and importance variable process in multivariate adaptive regression spline model. Int. J. Adv. Sci. Eng. Inf. Technol. 2023;13:928–934. doi: 10.18517/ijaseit. [DOI] [Google Scholar]
- 17.Devi S., Yasmirullah P., Otok B.W., Dwi J., Purnomo T., Prastyo D.D. Parameter estimation and hypothesis testing of the modified multivariate adaptive regression spline for modeling the number of diseases. IOSR J. Math. 2021;17:35–47. doi: 10.9790/5728-1705033547. [DOI] [Google Scholar]
- 18.Yasmirullah S.D.P., Otok B.W., Purnlmo J.D.T., Prastyo D.D. AIP Conf. Proc. 2021. Multivariate adaptive regression spline (MARS) methods with application to multi drug-resistant tuberculosis (mdr-tb) prevalence. [DOI] [Google Scholar]
- 19.Prihastuti Yasmirullah S.D., Otok B.W., Trijoyo Purnomo J.D., Prastyo D.D. Modification of Multivariate Adaptive Regression Spline (MARS) J. Phys. Conf. Ser. 2021;1863 doi: 10.1088/1742-6596/1863/1/012078. [DOI] [Google Scholar]
- 20.Bezdek J.C. Plenum Press New York and London; 1981. Pattern Recognition with Fuzzy Objective Function Algorithms. [Google Scholar]
- 21.Wang J., Yang Z., Liu X., Li B., Yi J., Nie F. Projected fuzzy C-means with probabilistic neighbors. Inf. Sci. 2022;607:553–571. doi: 10.1016/j.ins.2022.05.097. [DOI] [Google Scholar]
- 22.Pang Y., Shi M., Zhang L., Song X., Sun W. PR-FCM: a polynomial regression-based fuzzy C-means algorithm for attribute-associated data. Inf. Sci. 2022;585:209–231. [Google Scholar]
- 23.Wang W., Zhang Y. On Fuzzy cluster validity indices. Fuzzy Sets Syst. 2007;158 doi: 10.1016/j.fss.2007.03.004. [DOI] [Google Scholar]
- 24.Pimentel B.A., De Souza R.M.C.R. A multivariate fuzzy c-means method. Appl. Soft Comput. J. 2013;13:1592–1607. doi: 10.1016/j.asoc.2012.12.024. [DOI] [Google Scholar]
- 25.Bezdek J.C., Ehrlich R., Full W. FCM: the fuzzy c-means clustering algorithm. Comput. Geosci. 1984;10:191–203. doi: 10.1016/0098-3004(84)90020-7. [DOI] [Google Scholar]
- 26.Bezdek J.C. Plenum Press New York and London; 1981. Pattern Recognition With Fuzzy Objective Function Algorithms. [Google Scholar]
- 27.Trauwaert E. On the meaning of dunn's partition coefficient for fuzzy clusters. Fuzzy Sets Syst. 1988;25 [Google Scholar]
- 28.Wu K.L., Yang M.S. A cluster validity index for fuzzy clustering. Pattern Recognit. Lett. 2005;26:1275–1291. doi: 10.1016/j.patrec.2004.11.022. [DOI] [Google Scholar]
- 29.Dave R.N. Validating fuzzy partitions obtained through c-shells clustering. Pattern Recognit. Lett. 1996;17:613–623. doi: 10.1016/0167-8655(96)00026-8. [DOI] [Google Scholar]
- 30.Wedel M., Steenkamp J.B.E.M. A fuzzy clusterwise regression approach to benefit segmentation. Int. J. Res. Mark. 1989;6:241–258. doi: 10.1016/0167-8116(89)90052-9. [DOI] [Google Scholar]
- 31.Craven P., Wahba G. Smoothing noisy data with spline functions. Numer. Math. 1979;31:377–403. [Google Scholar]; https://pdfs.semanticscholar.org/e08c/75c7047d9bb2af6a339428c8bf4c11ad01d5.pdf.
- 32.Friedman J.H., Roosen C.B. An introduction to multivariate adaptive regression splines. Stat. Methods Med. Res. 1995;4:197–217. doi: 10.1177/096228029500400303. [DOI] [PubMed] [Google Scholar]
- 33.WHO . 2014. World Health Statistics 2014, Italy. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors do not have permission to share data.