Abstract
Using subfragments of the simian virus 40 (SV40) core origin, we demonstrate that two alternative modules exist for the assembly of T-antigen (T-ag) double hexamers. Pentanucleotides 1 and 3 and the early palindrome (EP) constitute one assembly unit, while pentanucleotides 2 and 4 and the AT-rich region constitute a second, relatively weak, assembly unit. Related studies indicate that on the unit made up of pentanucleotide 1 and 3 and the EP assembly unit, the first hexamer forms on pentanucleotide 1 and that owing to additional protein-DNA and protein-protein interactions, the second hexamer is able to form on pentanucleotide 3. Oligomerization on the unit made up of pentanucleotide 2 and 4 and the AT-rich region is initiated by assembly of a hexamer on pentanucleotide 4; subsequent formation of the second hexamer takes place on pentanucleotide 2. Given that oligomerization on the SV40 origin is limited to double-hexamer formation, it is likely that only a single module is used for the initial assembly of T-ag double hexamers. Finally, we discuss the evidence that nucleotide hydrolysis is required for the remodeling events that result in the utilization of the second assembly unit.
A thorough understanding of the initiation of DNA replication and its regulation, will require a detailed description of the protein-DNA and protein-protein interactions that take place at origins of replication. Since origins of replication in higher eukaryotic organisms are poorly characterized (8, 21), little is known about the molecular interactions that take place at these sites. Therefore, well-defined viral model systems are being used in an effort to establish the molecular interactions required to initiate DNA replication. One of the best-characterized viral model systems is that based on simian virus 40 (SV40). This virus encodes a single protein, termed T antigen (T-ag) (68), that binds in a site-specific manner to the viral origin of replication, a necessary step for the initiation of DNA replication (72). Several reviews have been published that cover the SV40 origin of replication, T-ag, and the interactions that take place between these molecules (4, 7, 26). Therefore, only a brief introduction is provided that stresses recent observations in this field.
A 64-bp region of the SV40 genome, termed the core origin, is necessary and sufficient for viral replication (19, 22, 39, 52, 66). The core origin consists of a central region, termed site II, that is flanked by an AT-rich domain (AT) and a second region, termed the early palindrome (EP) (17). Site II contains four GAGGC pentanucleotides, arranged as inverted pairs, that serve as binding sites for T-ag (20, 43, 69, 71). All three regions of the core origin are required for DNA unwinding and initiation of DNA replication (13, 17, 32).
T-ag is a 708-amino-acid phosphoprotein that contains several structural and functional domains (for reviews, see references 7 and 26). One domain of T-ag, the T-ag origin binding domain (T-ag-obd131–260) has been extensively studied. The purified T-ag-obd131–260 can locate and bind site II within the core origin (reviewed in references 7 and 26). To better understand origin recognition, this domain was purified (33), and the solution structure of this polypeptide was determined (43). When viewed in terms of extensive mutagenesis studies (e.g., references 61 and 81), the structure of the T-ag-obd131–260 provided several important insights into pentanucleotide recognition (7). For instance, these studies established that GAGGC binding is mediated largely by a pair of loops (43).
Following origin recognition by T-ag monomers, a poorly understood oligomerization process takes place that results in the formation of two hexameric rings that encircle the core origin (12, 16, 30, 45) (reviewed in references 4 and 7). Previous studies demonstrated that the assembly of T-ag double hexamers on the core origin is a cooperative process (50, 55, 75, 76). Furthermore, transmission electron microscopy studies have provided images of T-ag hexamers (58) and double hexamers assembled on the core origin (73). The double-hexamer complex is 24 nm long and 8 to 12 nm wide, and the hexamers are oriented in a head-to-head manner (32, 73). Recent experiments indicate that hexamer-hexamer interactions are mediated by the T-ag-obd (73, 76), an observation consistent with previous studies of the T-ag-obd (33, 62).
Our long-term goal is to describe, in molecular terms, the mechanism of DNA unwinding. Since this process is initiated by T-ag assembly on the core origin, we have analyzed in detail the sequence requirements for hexamer and double-hexamer formation. Using mutant forms of the 64-bp core origin, we previously reported that individual pentanucleotides support hexamer formation while oligonucleotides containing active pairs of pentanucleotides, particularly pentanucleotides 1 and 3, support double-hexamer formation (32). The structural consequences of double-hexamer formation on pentanucleotides 1 and 3 are known to include melting of the EP and structural alterations of the AT (6, 32, 54). Properly arranged pairs of pentanucleotides were also sufficient for stable binding of T-ag-obd131–260 dimers to the core origin (33). In a subsequent study, we examined the role of the AT and EP regions in T-ag assembly events. These studies revealed that T-ag binding to the core origin requires not only interactions between the T-ag-obd and site II, but additional poorly defined interactions between non-T-ag-obd residues and either of the flanking sequences (7, 9, 35). They also demonstrated that in the presence of a nonhydrolyzable analogue of ATP, subfragments of the core origin containing site II and one additional flanking sequence that may be positioned on either side of site II are sufficient for double-hexamer formation (35, 55). However, in the presence of ATP, assembly events require a larger region of the core origin (35).
In view of these studies, we elected to establish which combination of pentanucleotide pairs and flanking sequences support T-ag assembly on the subfragments of the core origin. Results from these studies, presented herein, have refined our understanding of the sequence requirements for assembly of T-ag hexamers and double hexamers on the core origin. These observations are discussed in terms of how this viral helicase (14, 29, 65) is assembled and how subsequent remodeling events (2) are initiated.
MATERIALS AND METHODS
Commercial supplies of enzymes, DNA, reagents, and oligonucleotides.
T4 polynucleotide kinase was purchased from Promega, and HaeIII was purchased from Gibco-BRL/Life Technologies. Yeast hexokinase was obtained from Boehringer Mannheim, and glucose was obtained from Sigma Chemical Company.
Oligonucleotides were synthesized on an Applied Biosystems 394 DNA synthesizer at the Protein Chemistry Facility at Tufts University. The oligonucleotides were purified by electrophoresis through urea–10% or 15% polyacrylamide gels and isolated by standard methods (57, 64). HaeIII-digested plasmid pBR322, used as non-sequence-specific competitor DNA, was purified by conventional procedures (57).
Purification of T-ag.
SV40 T-ag was expressed in Sf9 cells with a baculovirus expression vector containing the T-ag-encoding SV40 A gene (53) and purified by immunoaffinity techniques with the PAb 419 monoclonal antibody as previously described (23, 60, 79). Purified T-ag was dialyzed against T-ag storage buffer (20 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 0.1 mM phenylmethylsulfonyl fluoride, 0.2 μg of leupeptin per ml, 0.2 μg of antipain per ml, and 10% glycerol) and stored at −80°C until use.
Band shift assays.
Double-stranded oligonucleotides, used as substrates in gel shift assays (27, 28), were formed by annealing complementary pairs of oligonucleotides in hybridization buffer (34). The double-stranded oligonucleotides were labeled at their 5′ ends with 32P by standard procedures (57). The labeled oligonucleotides were purified via electrophoresis on neutral 10% polyacrylamide gels (run in 1× Tris-borate-EDTA buffer at 10 W, ∼21 mA, and ∼465 V), and the bands of interest were removed. The DNA was eluted in oligonucleotide elution buffer (57). After being extracted with phenol-chloroform-isoamyl alcohol (25:24:1) and chloroform-isoamyl alcohol (24:1), the labeled oligonucleotides were precipitated with 100% ethanol, washed with 80% (vol/vol) ethanol, and dissolved in deionized H2O (25 fmol/μl).
Band shift reactions, using T-ag and the indicated double-stranded oligonucleotides, were conducted under replication conditions (79) as previously described (15, 44, 51, 64). The reaction mixtures (20 μl) contained 7 mM MgCl2, 0.5 mM DTT, 4 mM AMP-p-nitrophenyl (PNP) (or, where indicated, ADP, ATP, or no exogenous nucleotide), 40 mM creatine phosphate (pH 7.6), 0.48 μg of creatine phosphate kinase, 5 μg of bovine serum albumin, 0.8 μg of HaeIII-digested pBR322 DNA (∼2.5 pmol; used as a non-sequence-specific competitor), 25 fmol of labeled double-stranded oligonucleotide, and 6 pmol of T-ag (T-ag was the last component added to the reaction mixture). After a 20-min incubation at 37°C, glutaraldehyde was added (0.1% final concentration), and the reaction mixtures were incubated at 37°C for an additional 5 min. At the end of the incubation period, 5 μl of 6× gel loading dye II (15% Ficoll, 0.25% bromophenol blue, and 0.25% xylene cyanol [57]) was added to the reaction mixtures. The samples were then loaded on a 4 to 12% gradient polyacrylamide gel (19:1 acrylamide-to-bisacrylamide ratio) and electrophoresed in 0.5× Tris-borate-EDTA buffer for ∼95 min (∼500 V, 20 mA). The gels were dried, subjected to autoradiography, and subsequently placed in a PhosphorImager cassette. Products of gel shift reactions were quantitated with a Molecular Dynamics PhosphorImager. In an effort to remove trace amounts of ATP, the reactions conducted in the presence of ADP were incubated at 37°C for 5 min in the presence of 1 U of hexokinase and 10 mM glucose prior to the addition of T-ag (67). Furthermore, the creatine phosphate kinase, dissolved in 50 mM imidazole buffer, was replaced with a corresponding amount of imidazole buffer.
Nitrocellulose filter binding of SV40 T-ag–DNA complexes.
The nitrocellulose filter binding assay for T-ag binding was based on previously described methods (5, 42, 46, 64). Reaction mixtures (20 μl each) contained 7 mM MgCl2, 0.5 mM DTT, 40 mM creatine phosphate (di-Tris salt [pH 7.6]), 0.48 μg of creatine phosphate kinase, 0.2 mg of bovine serum albumin per ml, 0.8 μg of HaeIII-digested pBR322 DNA, 25 fmol of a given radioactively labeled oligonucleotide (∼106 cpm/pmol), the indicated amount of T-ag, and 4 mM either AMP-PNP or ATP.
RESULTS
Requirements for T-ag hexamer and double-hexamer formation on the site II + EP assembly unit.
A diagram of the SV40 core origin is presented in Fig. 1A. To establish which pairs of pentanucleotides are required for double-hexamer formation on the core origin subfragments (35), oligonucleotides were synthesized that contained all possible combinations of two pentanucleotides and one of the flanking sequences. A representative member of the set of oligonucleotides made up of two pentanucleotides plus the EP, designated the two pentanucleotide + EP set, is shown in Fig. 1B, diagram 2. This 48-bp oligonucleotide contains pentanucleotides 1 and 3 and the EP but contains transition mutations in pentanucleotides 2 and 4 and lacks the AT-rich region. Although not depicted in Fig. 1B, similar oligonucleotides containing pentanucleotides 1 and 2, 1 and 4, 2 and 3, 2 and 4, 3 and 4 and the EP were also synthesized. As a positive control, we also synthesized the 48-bp site II + EP oligonucleotide (Fig. 1B, diagram 1). The previously described 47-bp control oligonucleotide (35) served as a negative control (Fig. 1C).
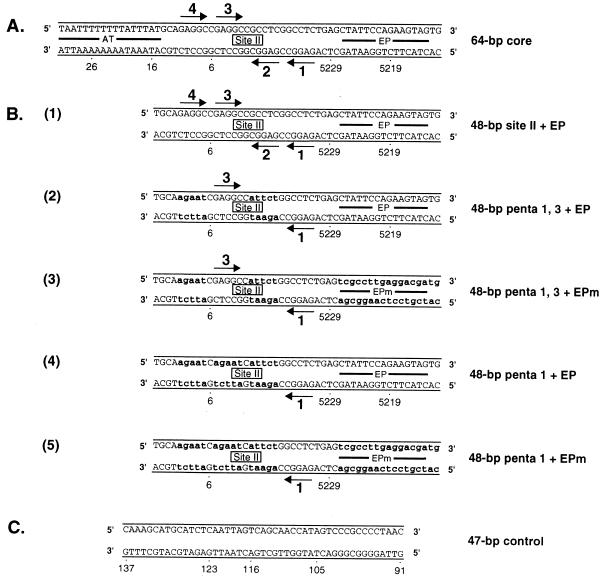
FIG. 1.
Sequences of the 64-bp core oligonucleotide and a set of oligonucleotides derived from the site II + EP oligonucleotide. (A) The 64-bp core origin; locations of the AT, site II, and the EP regions are indicated. Arrows depict the four GAGGC pentanucleotides within site II that serve as recognition sites for T-ag; pentanucleotides are numbered as previously described (38). SV40 sequences are numbered as described elsewhere (72). (B) Sequences of a set of oligonucleotides based on the 48-bp site II + EP oligonucleotide. The sequence of the 48-bp site II + EP oligonucleotide is presented in diagram 1. Diagram D2 presents the sequence of the 48-bp penta 1, 3 + EP oligonucleotide, a representative member of the two pentanucleotide + EP set of oligonucleotides. During the synthesis of the 48-bp penta 1, 3 + EP oligonucleotide, transition mutations were introduced at pentanucleotides 2 and 4. These mutations are indicated by the lowercase bold letters. Although not depicted, the 48-bp penta 1, 2 + EP, penta 1, 4 + EP, penta 2, 3 + EP, penta 2, 4 + EP, and penta 3, 4 + EP oligonucleotides were also synthesized. Diagram D3 presents the sequence of the 48-bp penta 1, 3 + EPm oligonucleotide. This oligonucleotide is similar to the 48-bp penta 1, 3 + EP oligonucleotide except that additional transition mutations have replaced the EP region (symbolized by lowercase bold letters). A representative member of the single pentanucleotide + EP set of oligonucleotides, the 48-bp penta 1 + EP oligonucleotide, is presented in diagram 4. During synthesis of this oligonucleotide, transition mutations were introduced at pentanucleotides 2, 3, and 4 (indicated by lowercase bold letters). The sequence of the 48-bp penta 1 + EPm oligonucleotide is presented in diagram 5; this molecule is similar to the 48-bp penta 1 + EP oligonucleotide except that additional transition mutations have replaced the EP region (indicated by lowercase bold letters). Although not depicted, the 48-bp penta 3 + EP and 48-bp penta 3 + EPm oligonucleotides were also synthesized. (C) Finally, the sequence of the 47-bp control oligonucleotide is presented; this molecule served as a control for non-sequence-specific binding events.
In a preliminary study, the two pentanucleotide + EP set of oligonucleotides was used in a series of band shift experiments (Fig. 2A). This set of reactions was conducted at a T-ag-to-oligonucleotide ratio of 240:1 in the presence of AMP-PNP. As positive controls, reactions were conducted with the 64-bp core and 48-bp site II + EP oligonucleotides (Fig. 2A, lanes 2 and 4); the products include T-ag hexamers and double hexamers (12, 55, 75). The reaction products formed with the two pentanucleotide + EP set of oligonucleotides are shown in lanes 6, 8, 10, 12, 14 and 16. To measure nonspecific binding to DNA, a reaction was conducted with the 47-bp control oligonucleotide (lane 18). Reactions shown in odd-numbered lanes were conducted in the absence of T-ag. Inspection of the even-numbered lanes reveals that while all of the two pentanucleotide + EP set of oligonucleotides formed hexamers, only the 48-bp oligonucleotide containing pentanucleotides 1 and 3 plus the EP (termed the penta 1, 3 + EP oligonucleotide) formed significant levels of double hexamers (lane 8). It is concluded that only pentanucleotides 1 and 3 are arranged in the correct orientation, relative to the EP, to support double-hexamer formation.
FIG. 2.
Representative gel mobility shift assay used to determine the sequence requirements for double hexamer formation on the two pentanucleotide + EP-based set of oligonucleotides. (A) Experiments were performed in the presence of AMP-PNP with 6 pmol of T-ag and 25 fmol of the indicated oligonucleotide. As positive controls, the reaction displayed in lane 2 was conducted with the 64-bp core oligonucleotide while that displayed in lane 4 was performed with the 48-bp site II + EP oligonucleotide. Reactions displayed in lanes 6, 8, 10, 12, 14, and 16 were conducted with the indicated members of the two pentanucleotide + EP set of oligonucleotides. As a negative control, the reaction in lane 18 was conducted with the 47-bp control oligonucleotide. Reactions in the odd-numbered lanes were conducted with the indicated oligonucleotides in the absence of T-ag. The arrows indicate the positions of T-ag hexamers (H) and T-ag double hexamers (DH). The position of input or free DNA is indicated by a bracket. (B) The reactions in Fig. 2A and similar reactions conducted in the presence of ADP, ATP, and no exogenous nucleotides (data not shown) were quantitated with a Molecular Dynamics PhosphorImager in order to determine the percentage of input DNA shifted into double hexamers. The nucleotide cofactor used in a given reaction is shown to the right of the figure, and the names of the individual oligonucleotides are presented along the x axis, while the percentage of input DNA present in double hexamers is listed on the y axis.
The experiments presented in Fig. 2A were repeated in the presence of ADP, ATP, and no exogenous nucleotide (data not shown). These experiments were quantitated with a Molecular Dynamics PhosphorImager. Results from these analyses are presented in Fig. 2B. Inspection of this figure confirms that in the presence of AMP-PNP, the penta 1, 3 + EP oligonucleotide supported DH formation at levels equivalent to those formed on the four-pentanucleotide-containing 48-bp site II + EP oligonucleotide. Furthermore, in the presence of ADP, the level of interactions of T-ag with the two pentanucleotide + EP set of oligonucleotides is similar, albeit somewhat lower in the case of the 48-bp penta 1, 3 + EP oligonucleotide, than that of the interactions taking place in the presence of AMP-PNP. It is also clear that in the absence of exogenous nucleotides, the two pentanucleotide + EP set of oligonucleotides supported limited double-hexamer formation, a result consistent with previous studies (5, 15, 18). In experiments conducted in the presence of ATP, it was previously reported (35) that the site II + EP oligonucleotide does not form double hexamers in the presence of ATP, a result confirmed by the data shown in Fig. 2B. Likewise, Fig. 2B demonstrates that all members of the two pentanucleotide + EP set of oligonucleotides were unable to support double-hexamer formation in the presence of ATP. In contrast, in the presence of ATP, hexamer formation was supported by these same oligonucleotides (data not shown). Quantitation of hexamer formation on the penta 1, 3 + EP assembly unit in the presence of ATP is presented in Fig. 3B).
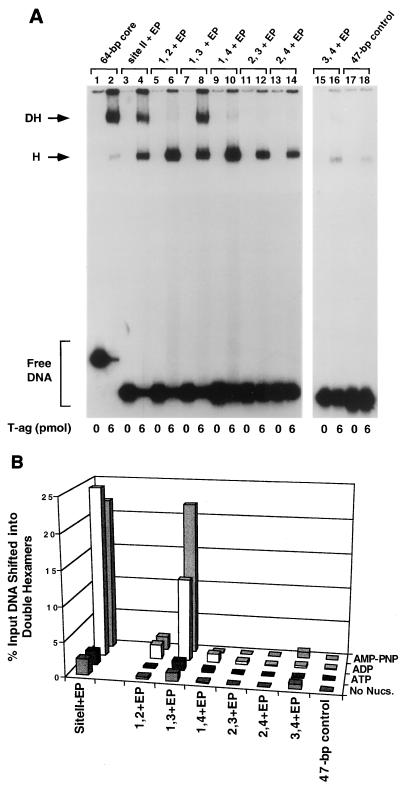
FIG. 3.
Representative gel mobility shift assay used to establish which components of the 48-bp penta 1, 3 + EP assembly unit are required for hexamer and double-hexamer formation. (A) Reactions were conducted in the presence of AMP-PNP with 6 pmol of T-ag and 25 fmol of the indicated oligonucleotide. As a positive control, one reaction was conducted with the 48-bp penta 1, 3 + EP oligonucleotide (lane 2). To test the role of the EP in these assembly events, a reaction was conducted with the 48-bp penta 1, 3 + EPm oligonucleotide (lane 4). Reactions conducted with the single pentanucleotide containing 48-bp penta 1 + EP and 48-bp penta 1 + EPm oligonucleotides are presented in lanes 6 and 8, respectively. Similar reactions, conducted with the 48-bp penta 3 + EP and 48-bp penta 3 + EPm oligonucleotides, are presented in lanes 10 and 12. To assay for non-sequence-specific binding events, a reaction was conducted with the 47-bp control oligonucleotide (lane 14). The reactions in the odd-numbered lanes were conducted in the absence of protein. The positions of T-ag hexamers (H) and double hexamers (DH) are indicated by arrows. The location of input or free DNA is indicated by a bracket. (B) The reactions in Fig. 3A and similar reactions conducted in the presence of ADP, ATP, and no exogenous nucleotides (data not shown) were quantitated with a Molecular Dynamics Phosphor- Imager. The percentage of input DNA, containing single pentanucleotides, that was shifted into hexamers is presented in histogram 2. The nucleotide cofactor used in a given set of reactions is shown to the right of the figure, and the names of the individual oligonucleotides are presented along the x axis, while the percentage of input DNA present in the hexamer species is listed on the y axis. Histogram 1 indicates the quantitative impact of mutating the EP region on T-ag oligomerization into hexamers and double hexamers.
Further characterization of hexamer and double-hexamer formation on the penta 1, 3 + EP assembly unit.
Given that in the presence of AMP-PNP, the 48-bp penta 1, 3 + EP oligonucleotide supported hexamer and double-hexamer formation, we elected to establish which of the three components were required for hexamer, as opposed to double-hexamer, assembly. We were also interested in characterizing the role of the EP in T-ag assembly events. Therefore, the penta 1, 3 + EP derivative set of oligonucleotides was synthesized (i.e., the 48-bp penta 1, 3 + EP mutant [EPm] oligonucleotide [Fig. 1B, diagram 3], the 48-bp penta 1 + EP and the 48-bp penta 1 + EPm oligonucleotides [Fig. 1B, diagrams 4 and 5], and the 48-bp penta 3 + EP and the 48-bp penta 3 + EPm oligonucleotides [diagrams not shown]).
One series of experiments conducted with the penta 1, 3 + EP derivative set of oligonucleotides, in the presence of AMP-PNP, is presented in Fig. 3A. As a positive control, a band shift experiment was conducted with the 48-bp penta 1, 3 + EP oligonucleotide (Fig. 1B, diagram 2); as previously demonstrated, this two-pentanucleotide-containing subfragment of the core origin supported hexamer and double-hexamer formation (Fig. 3A, lane 2). However, when these experiments were repeated with an oligonucleotide containing transition mutations in the EP, the penta 1, 3 + EPm oligonucleotide, T-ag assembly was greatly reduced (lane 4). Thus, as with assembly on the site II + EP oligonucleotide (35), T-ag assembly on the penta 1, 3 + EP assembly unit is promoted by sequence-specific, or perhaps conformation-dependent, interactions with the EP, an observation supported by previous studies (54, 59). Identical band shift experiments conducted with the 48-bp penta 1 + EP or 48-bp penta 3 + EP oligonucleotide, are presented in lanes 6 and 10, respectively. It is apparent that while the 48-bp penta 1 + EP oligonucleotide supported the formation of relatively high levels of T-ag hexamers, the 48-bp penta 3 + EP oligonucleotide did not. Confirmation that hexamer formation on pentanucleotide 1 requires an interaction with the EP region was demonstrated by additional experiments conducted with the 48-bp penta 1 + EPm oligonucleotide. Relative to the molecule containing the wild-type EP, this molecule was clearly defective in its ability to support hexamer formation (compare lanes 6 and 8). As expected, hexamer formation on the 48-bp penta 3 + EPm oligonucleotide was defective (lane 12); indeed, it formed hexamers at levels approaching those of background assembly on the 47-bp control oligonucleotide (lane 14). Reactions conducted in the absence of T-ag are presented in the odd-numbered lanes of Fig. 3A. It is concluded that in the presence of the EP, pentanucleotide 1, but not pentanucleotide 3, is capable of independently supporting hexamer formation.
The experiments presented in Fig. 3A and similar experiments conducted in the presence of ATP, ADP, or no exogenous nucleotides (data not shown) were quantitated with a PhosphorImager. Results from these studies are presented in Fig. 3B). Inspection of histogram 2 demonstrates that the pattern of hexamer formation on the penta 1, 3 + EP derivative set of oligonucleotides is similar in the presence of AMP-PNP and ADP. This histogram also confirms that while the 48-bp penta 1 + EP oligonucleotide supports hexamer formation, the 48-bp penta 3 + EP oligonucleotide is a poor substrate for hexamer assembly. Moreover, reactions conducted in the absence of nucleotide support very low levels of hexamer formation. Likewise, in the presence of ATP, very limited amounts of hexamer formed on this set of oligonucleotides. However, the 48-bp penta 1 + EP oligonucleotide was the single exception in that in the presence of ATP, it supported hexamer formation in this and several identical experiments (data not shown). Additional evidence that the 48-bp penta 1 + EP oligonucleotide supported T-ag binding in the presence of ATP is provided by other experiments (see Fig. 7B). Inspection of histogram 1 of Fig. 3B reveals that T-ag assembly on the 48-bp penta 1, 3 + EP oligonucleotide, in the presence of either AMP-PNP or ADP, is at least fivefold greater than on an oligonucleotide containing transition mutations in this flanking sequence.
FIG. 7.
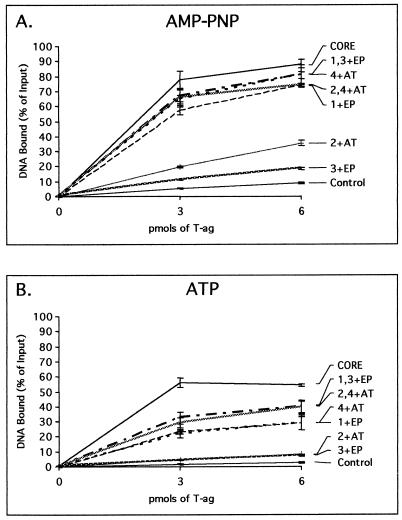
Filter binding assays used to measure the ability of T-ag to bind to oligonucleotides derived from the site II + EP- and site II + AT-based oligonucleotides. (A) Reactions were performed in the presence of AMP-PNP and the indicated amounts of T-ag (0, 3, and 6 pmol). The percentage of oligonucleotide bound to T-ag was established by nitrocellulose filter binding assays and scintillation counting. (B) The same set of oligonucleotides were used in additional nitrocellulose filter binding assays conducted in the presence of ATP. Following the addition of T-ag (0, 3, 6 pmol), the amount of bound oligonucleotide was determined. It is noted that a larger percentage of the substrates are bound in the filter binding assays than in the gel shift assays (compare the data in Fig. 7 with those in Fig. 3B and 6B). This may reflect that the T-ag–DNA complexes are subjected to harsher conditions during gel electrophoresis than during filter binding.
Requirements for T-ag hexamer and double-hexamer formation on the site II + AT assembly unit.
To establish the sequence requirements for double-hexamer assembly on the site II + AT assembly unit (35), a second series of band shift experiments was conducted with the two pentanucleotide + AT set of oligonucleotides. A representative member of this set of oligonucleotides is depicted in Fig. 4, diagram 2. This 47-bp oligonucleotide contains pentanucleotides 2 and 4 and the AT but contains transition mutations at pentanucleotides 1 and 3 and lacks the EP region. Similar oligonucleotides containing pentanucleotides 1 and 2, 1 and 3, 1 and 4, 2 and 3, 3 and 4 and the AT were synthesized. To serve as positive controls, the 47-bp site II + AT (Fig. 4, diagram 1) and the 64-bp core (Fig. 1A) oligonucleotides were also synthesized. The previously described 47-bp control oligonucleotide (Fig. 1B, diagram 6) served as a negative control.
FIG. 4.
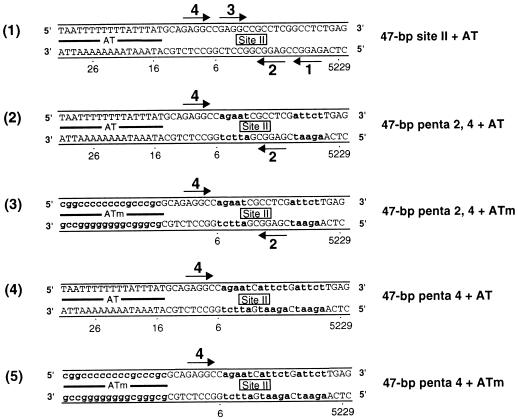
Sequences of the site II + AT-based oligonucleotides. Diagram D1 provides the sequence of the 47-bp site II + AT oligonucleotide. As in previous examples, the arrows depict the four GAGGC pentanucleotides that serve as recognition sites for T-ag. A representative member of the two pentanucleotide + AT set of oligonucleotides is presented in diagram 2. During the synthesis of the 47-bp penta 2, 4 + AT oligonucleotide, transition mutations were introduced at pentanucleotides 1 and 3. These mutations are indicated by lowercase bold letters. Although not depicted, the 47-bp penta 1, 2 + AT, penta 1, 3 + AT, penta 1, 4 + AT, penta 2, 3 + AT, and penta 3, 4 + AT oligonucleotides were also synthesized. Diagram D3 presents the sequence of the 47-bp penta 2, 4 + ATm oligonucleotide; this molecule is similar to the 47-bp penta 2, 4 + AT oligonucleotide except that additional transition mutations have replaced the AT (indicated by the lowercase bold letters). A representative member of the single pentanucleotide + AT set of oligonucleotides, the 47-bp penta 4 + AT oligonucleotide, is presented in diagram 4. During the synthesis of the 47-bp penta 4 + AT oligonucleotide, transition mutations were introduced at pentanucleotides 1, 2, and 3 (indicated by the lowercase bold letters). Diagram D5 presents the 47-bp penta 4 + ATm oligonucleotide; this molecule is similar to the 47-bp penta 4 + AT oligonucleotide except that the AT-rich region has been replaced by additional transition mutations (indicated by lowercase bold letters). Finally, although not depicted, the 47-bp penta 2 + AT and 47-bp penta 2 + ATm oligonucleotides were also synthesized.
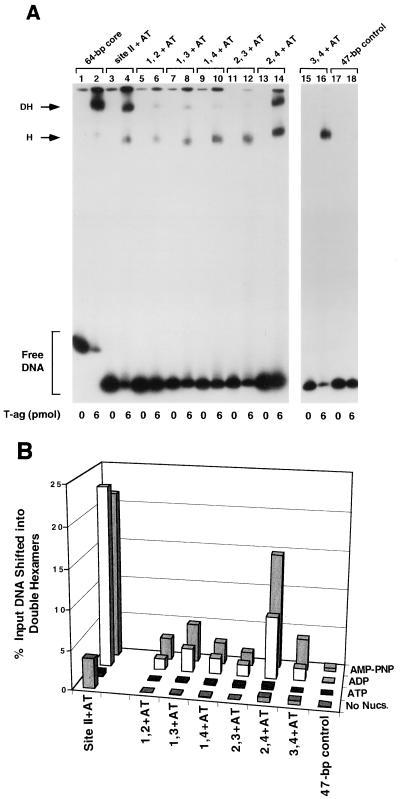
In an initial set of experiments, the two pentanucleotide + AT set of oligonucleotides were used in a series of band shift experiments that were conducted in the presence of AMP-PNP at a T-ag-to-oligonucleotide ratio of 240:1 (Fig. 5A). As positive controls, reactions were conducted with the 64-bp core origin oligonucleotide (Fig. 5, lane 2) and the 47-bp site II + AT oligonucleotide (lane 4); as expected, these molecules supported hexamer and double-hexamer formation. The reaction products formed with the two pentanucleotide + AT set of oligonucleotides are shown in lanes 6, 8, 10, 12, 14, and 16. Inspection of the protein-DNA complexes in these lanes demonstrates that while all of the two pentanucleotide + AT sets of oligonucleotides formed hexamers, only the penta 2, 4 + AT oligonucleotide formed double hexamers at levels approaching that of assembly on the 47-bp site II + AT oligonucleotide (compare lanes 14 and 4). As in previous reactions (Fig. 2A and 3A), the 47-bp control oligonucleotide supported very low levels of hexamer formation (Fig. 5, lane 18). Reactions in odd-numbered lanes were conducted in the absence of T-ag.
FIG. 5.
Representative gel mobility shift assay used to determine the sequence requirements for double hexamer formation on the two pentanucleotide + AT-based set of oligonucleotides. (A) Experiments were performed in the presence of AMP-PNP with 6 pmol of T-ag and 25 fmol of the indicated oligonucleotide. As positive controls, reactions were conducted with the 64-bp core oligonucleotide (lane 2) and the 47-bp site II + AT oligonucleotide (lane 4). The reactions displayed in lanes 6, 8, 10, 12, 14, and 16 were conducted with the indicated members of the two pentanucleotide + AT set of oligonucleotides. The reaction shown in lane 18 was conducted with the 47-bp control oligonucleotide. Reactions in the odd-numbered lanes were conducted in the absence of T-ag. The arrows indicate the positions of T-ag hexamers (H) and double hexamers (DH). The position of input or free DNA is indicated by a bracket. (B) The reactions shown in Fig. 5A and similar reactions conducted in the presence of ADP, ATP, and no exogenous nucleotides (data not shown) were quantitated with a Molecular Dynamics PhosphorImager to determine the percentage of input DNA shifted into double hexamers. The nucleotide cofactor is indicated to the right of the figure, and the names of the individual oligonucleotides are presented along the x axis, while the percentage of input DNA present in the double hexamers is indicated on the y axis.
The experiments shown in Fig. 5A were repeated in the presence of ADP, ATP, and no exogenous nucleotides (data not shown). These studies were quantitated with a Molecular Dynamics PhosphorImager; results from these analyses are presented in Fig. 5B. In the presence of AMP-PNP, the penta 2, 4 + AT oligonucleotide supported double-hexamer formation, albeit at a level somewhat lower than that on the 47-bp site II + AT oligonucleotide. It is also apparent that in the presence of ADP, T-ag's ability to form double hexamers on the two pentanucleotide + AT set of oligonucleotides is similar to, although somewhat less than, double-hexamer formation in the presence of AMP-PNP. As with the two pentanucleotide + EP set of oligonucleotides (Fig. 2B), T-ag's ability to form double hexamers on the two pentanucleotide + AT set of oligonucleotides was reduced to near-background levels in the presence of ATP or no exogenous nucleotides.
Further characterization of hexamer and double-hexamer formation on the penta 2, 4 + AT assembly unit.
As with the penta 1, 3 + EP assembly unit, we elected to establish which combinations of the penta 2, 4 + AT assembly unit are necessary for hexamer and double-hexamer formation. We also wanted to further characterize the role of the AT-rich region in assembly. Therefore, the penta 2, 4 + AT derivative set of oligonucleotides was synthesized (i.e., the 47-bp penta 2, 4 + ATm oligonucleotide [Fig. 4, diagram 3], the 47-bp penta 4 + AT and the 47-bp penta 4 + ATm oligonucleotides [Fig. 4, diagrams 4 and 5], and the 47-bp penta 2 + AT and 47-bp penta 2 + ATm oligonucleotides [diagrams not shown]).
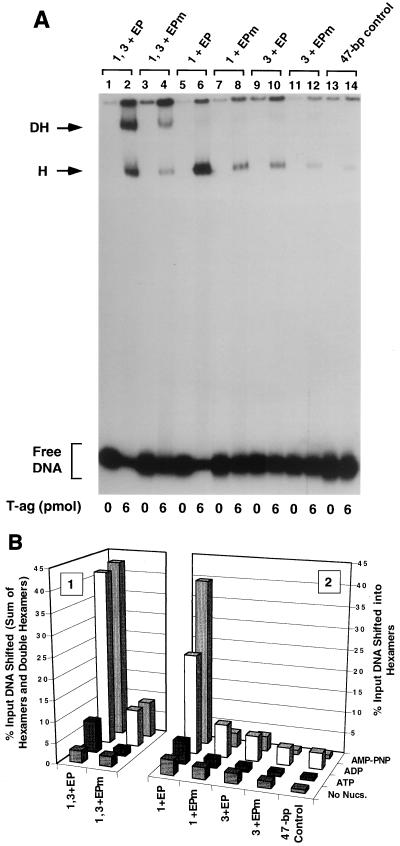
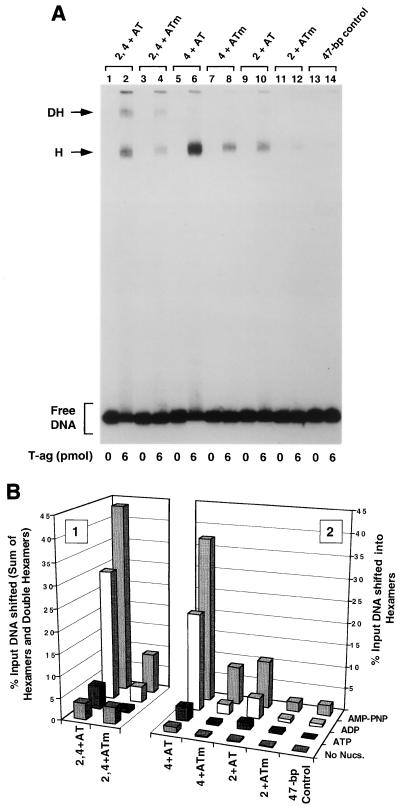
One series of experiments performed with this set of oligonucleotides, conducted in the presence of AMP-PNP, is presented in Fig. 6A. As a positive control, a band shift experiment was conducted with the 47-bp penta 2, 4 + AT oligonucleotide (Fig. 6A, lane 2). The products formed in this reaction, hexamers and double hexamers, are indicated. When these reactions were repeated with the 47-bp penta 2, 4 + ATm oligonucleotide, a molecule containing transition mutations in the AT sequences, the level of T-ag assembly was reduced (lane 4). Thus, as with assembly on the site II + AT oligonucleotide (35), T-ag assembly on the penta 2, 4 + AT assembly unit is promoted by sequence-specific, or conformation-dependent, interactions with the AT. Band shift reactions conducted with the 47-bp penta 4 + AT or 47-bp penta 2 + AT oligonucleotide are presented in lanes 6 and 10, respectively. It is apparent that the penta 4 + AT oligonucleotide supported a higher level of hexamer formation than the penta 2 + AT oligonucleotide. An additional reaction, conducted with the 47-bp penta 4 + ATm, is presented in lane 8. As in previous examples, a relatively low level of binding to this oligonucleotide indicates that sequence-specific, or perhaps conformation-dependent, interactions with the AT-rich region are important for hexamer assembly on pentanucleotide 4. As expected, the level of hexamer formation on the 47-bp penta 2 + ATm oligonucleotide was very low (lane 12). As in previous examples, the 47-bp control oligonucleotide was used to measure non-sequence-specific hexamer formation (lane 14). Reactions conducted in the absence of T-ag are presented in the odd-numbered lanes. It is concluded that in the presence of the AT-rich region, pentanucleotide 4 is the preferred substrate for hexamer formation.
FIG. 6.
Representative gel mobility shift assay used to establish which components of the 47-bp penta 2, 4 + AT assembly unit are required for hexamer and double-hexamer formation. (A) Reactions were conducted in the presence of AMP-PNP with 6 pmol of T-ag and 25 fmol of the indicated oligonucleotide. As a positive control, one reaction was conducted with the 47-bp penta 2, 4 + AT oligonucleotide (lane 2). To test the role of the AT in these assembly events, a reaction was conducted with the 47-bp penta 2, 4 + ATm oligonucleotide (lane 4). Reactions conducted with the single pentanucleotide containing 47-bp penta 4 + AT and 47-bp penta 4 + ATm oligonucleotides are presented in lanes 6 and 8, respectively. Similar reactions, conducted with the 47-bp 2 + AT and 47-bp penta 2 + ATm oligonucleotides, are presented in lanes 10 and 12, respectively. To assay for non-sequence-specific binding events, an additional reaction was conducted with the 47-bp control oligonucleotide (lane 14). Reactions in the odd-numbered lanes were conducted in the absence of protein. The positions of T-ag hexamers (H) and double hexamers (DH) are indicated by arrows, while the position of free DNA is indicated by a bracket. (B) The reactions shown in Fig. 6A and similar reactions conducted in the presence of ADP, ATP, and no exogenous nucleotides (data not shown) were quantitated with a Molecular Dynamics PhosphorImager. Histogram 2 displays the percentage of input DNA, containing single pentanucleotides, that is shifted into hexamers. The nucleotide cofactor used in a given set of reactions is shown to the right of the figure, the names of the individual oligonucleotides are shown on the x axis, and the percentage of input DNA shifted into hexamers is shown on the y axis. Histogram 1 reveals the quantitative impact of mutating the AT on T-ag oligomerization on the penta 2, 4 + AT assembly unit.
The experiments in Fig. 6A and similar experiments conducted in the presence of ATP, ADP, or no exogenous nucleotides (data not shown) were quantitated with a PhosphorImager; results from these studies are presented in Fig. 6B. Inspection of histogram 2 of Fig. 6B reveals that ADP supported similar, albeit somewhat lower, levels of hexamer formation on the penta 2, 4 + AT derivative set of oligonucleotides, as did AMP-PNP. In the absence of exogenous nucleotides, this set of oligonucleotides supported very low levels of hexamer formation. Moreover, in the presence of ATP, only the 47-bp penta 4 + AT oligonucleotide supported low, but detectable, levels of hexamer formation. (Additional evidence that the 47-bp penta 4 + AT oligonucleotide supports T-ag binding in the presence of ATP is provided by the experiments presented in Fig. 7B). In the presence of AMP-PNP, the 47-bp penta 4 + AT oligonucleotide supported a level of T-ag assembly approximately 3.5-fold higher than that of the 47-bp penta 2 + AT oligonucleotide (Fig. 7B). Finally, inspection of histogram 1 of Fig. 7B reveals that the level of T-ag assembly on the 47-bp penta 2, 4 + AT oligonucleotide, in the presence of AMP-PNP, is approximately fourfold higher than that on an oligonucleotide containing transition mutations in the AT.
Further characterization of T-ag assembly via nitrocellulose filter binding assays.
The T-ag assembly studies conducted in the preceding sections require cross-linking with glutaraldehyde (see Materials and Methods). Therefore, nitrocellulose filter binding assays were used to confirm that particular oligonucleotides (e.g., the 48-bp penta 1, 3 + EP, 47-bp penta 2, 4 + AT, 48-bp penta 1 + EP, and 47-bp penta 4 + AT) are preferred substrates for T-ag binding. Results from these assays, conducted with T-ag, AMP-PNP, and various oligonucleotides, are presented in Fig. 7A. As expected, T-ag bound the 48-bp penta 1, 3 + EP and 47-bp penta 2, 4 + AT oligonucleotides at levels only slightly lower than that of the 64-bp core origin oligonucleotide. Moreover, oligonucleotides containing a single pentanucleotide proximal to a given flanking sequence (i.e., the 48-bp penta 1 + EP and 47-bp penta 4 + AT oligonucleotides) bound T-ag at elevated levels, while those containing the flanking sequences distal to the single pentanucleotide (i.e., the 48-bp penta 3 + EP and 47-bp penta 2 + AT oligonucleotides) supported T-ag binding at reduced levels. Thus, results from both band shift and nitrocellulose filter binding assays are in agreement about which combinations of pentanucleotides and flanking sequences support T-ag assembly.
The same set of oligonucleotides was used in additional nitrocellulose filter binding assays conducted in the presence of ATP (Fig. 7B). In contrast to studies conducted in the presence of AMP-PNP (Fig. 7A), there was a considerable reduction in the level of binding to all oligonucleotides in the presence of ATP (Fig. 7B). Qualitatively, however, the results obtained with these two nucleotides are quite similar; those oligonucleotides containing pentanucleotides proximal to the flanking sequences support T-ag binding while those distal to the flanking sequences bound T-ag at levels comparable to that of the 47-bp control oligonucleotide.
DISCUSSION
The protein-DNA interactions that take place at the SV40 origin are of interest for a number of reasons. By studying T-ag assembly on the core origin, one is simultaneously characterizing origin recognition (reviewed in references 7 and 26) and the formation of a eukaryotic helicase (14, 29, 65). Moreover, it is likely that a clearer understanding of how T-ag assembles on the core origin will aid in efforts to establish the mechanism of T-ag-catalyzed DNA unwinding (14, 24, 80). A further motivation is that studies of T-ag–origin interactions continue to provide significant insights into how initiation of DNA replication is regulated by the cell cycle machinery (references 3, 47, 49, 75, 76 and references therein). Therefore, we have conducted a series of experiments designed to further refine the core origin sequence requirements for T-ag assembly events.
Our studies demonstrate that in the presence of SV40 core origin subfragments containing two pentanucleotides and the EP and the nonhydrolyzable analogue of ATP (AMP-PNP), double hexamers form only in the presence of pentanucleotides 1 and 3. In the presence of core origin subfragments containing two pentanucleotides and the AT, double hexamers form only when pentanucleotides 2 and 4 are present. Identical conclusions were drawn from experiments conducted in the presence of ADP, an indication that T-ag adopts a similar conformation upon binding of AMP-PNP or ADP. These and related studies (32, 33, 35) demonstrate that the 64-bp SV40 core origin contains two separate modular units for double-hexamer assembly. This observation raises the possibility that previous chemical and enzymatic studies of T-ag's interactions with the core origin (reviewed in reference 7) are composites of two signals; one signal is due to occupancy of the EP module, and the second weaker signal is due to occupancy of the AT module.
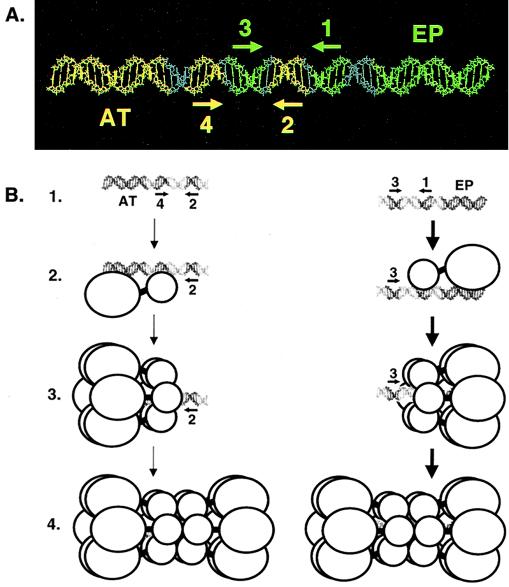
The relative positions of the penta 1, 3 + EP and penta 2, 4 + AT assembly units within the core origin are depicted in Fig. 8A. As previously noted, pentanucleotide pair 1 and 3 is on the same surface of B-DNA, as is pentanucleotide pair 2 and 4 (33). Based on these spatial arrangements, one would predict that T-ag contacts with the pentanucleotides are clustered along one face of B-DNA, a hypothesis confirmed by previous studies (31, 59). Moreover, a single nucleotide insertion between pentanucleotides 2 and 3 disrupts cooperative interactions between hexamers on the core origin (10, 75). Thus, separation of pentanucleotide pairs 1 and 3 and 2 and 4 by 3.4 Å is sufficient to disrupt the protein-protein interactions necessary for efficient double-hexamer formation. It is clear from Fig. 8 and previous studies that a very precise arrangement of sequences is necessary for efficient double-hexamer formation. These observations suggest that the two hexameric rings that assemble on these sequences have an equally precise spatial arrangement, a hypothesis consistent with recent images of T-ag double hexamers assembled on the core origin (73).
FIG. 8.
A model illustrating the relative positions of the penta 1, 3 + EP and penta 2, 4 + AT assembly units on the core origin and the formation of hexamers and double hexamers on oligonucleotides containing single assembly sites. (A) To depict the relative positions of the penta 1, 3 + EP and penta 2, 4 + AT assembly units, the 64-bp core origin is shown as a B DNA helix. The penta 1, 3 + EP assembly unit is shown in green, while the penta 2, 4 + AT assembly unit is shown in yellow. The locations of individual GAGGC pentanucleotides are indicated by arrows; the positions of the flanking sequences are also indicated. (B) Models for T-ag assembly events on the penta 2, 4 + AT (left) and 1, 3 + EP (right) assembly units (line 1). The structures used to depict T-ag monomers and hexamers are based on transmission electron microscopy studies reported by Valle et al. (73). Smaller circles represent the T-ag-obd, while the remaining residues of T-ag are represented by larger circles. Pentanucleotides proximal to the flanking sequences are recognized by the T-ag-obd, while non-T-ag-obd residues make both sequence-specific and non-sequence-specific interactions with the flanking sequences (this study; reference 35 and references therein). Following monomer binding (line 2), protein-protein interactions give rise to hexamer formation (line 3). Hexamer formation enables additional protein-protein and protein-DNA interactions to take place and subsequent formation of double hexamers to occur (line 4). Boldface arrows show that assembly on the penta 1, 3 + EP assembly unit is preferred over the penta 2, 4 + AT assembly unit.
A model is presented in Fig. 8B depicting double-hexamer formation on the two assembly units present within the SV40 core origin that incorporates recent insights into the structures of T-ag hexamers and double hexamers derived from studies by Valle et al. (73). On the penta 1, 3 + EP assembly unit, it is proposed that the initial hexamer forms on pentanucleotide 1 (Fig. 8B, lines 1 to 3). This oligomerization event is dependent upon both sequence-specific interactions between the T-ag-obd and pentanucleotide 1, as well as poorly characterized interactions between non-T-ag-obd regions of T-ag and the EP (this study; references 9, 35, 43, and 73 and references therein). Regions of T-ag required for hexamer formation (line 3) are known to include the T-ag-obd and Zn finger domains (41, 62, 76; reviewed in reference 7). Phosphorylation and ATP hydrolysis are not required for hexamer formation (3, 48, 49, 56, 76). The experiments shown in Fig. 2, 3, and 7 indicate that once the initial hexamer has formed on pentanucleotide 1, additional protein-protein and protein-DNA interactions promote the cooperative assembly of the second hexamer on pentanucleotide 3 (Fig. 8B, line 4). Regions of T-ag involved in the hexamer-hexamer interactions include the T-ag-obd and the N-terminal cluster of phosphorylation sites (references 3, 73, 75 and 76 and references therein). On the penta 2, 4 + AT assembly unit, it is proposed that the initial hexamer forms on pentanucleotide 4 (lines 1 to 3), presumably due to many of the same protein-protein and protein-DNA interactions described above for hexamer formation on the penta 1, 3 + EP assembly unit. The experiments shown in Fig. 5, 6, and 7 indicate that once the initial hexamer forms over pentanucleotide 4 (Fig. 8B, line 3), cooperative protein-protein interactions enable the second hexamer to form over pentanucleotide 2 (line 4).
Additional support for the model presented in Fig. 8B and especially for the hypothesis that the penta 1, 3 + EP unit is preferentially utilized is provided by previous studies of T-ag-obd131–260 and T-ag assembly events on oligonucleotides derived from the full-length core origin. For instance, purified T-ag-obd131–260 was shown to bind as a dimer to the four-pentanucleotide-containing 64-bp core origin (33). Pentanucleotides 1 and 3 were the preferred substrates for T-ag-obd131–260 dimer formation; only if pentanucleotides 1 and 3 were mutated were pentanucleotides 2 and 4 used to support T-ag-obd131–260 binding (33). To account for the observation that only two of the four pentanucleotides were bound by T-ag-obd131–260, we speculated that upon T-ag-obd131–260 binding to a given pentanucleotide (e.g., pentanucleotide 1), it obscured the neighboring pentanucleotide (e.g., pentanucleotide 2) from subsequent binding events. Similar observations were made with T-ag. For example, we demonstrated that only two pentanucleotides are required for T-ag double-hexamer formation on the 64-bp core origin (32) and that pentanucleotides 1 and 3 were the preferred substrates for T-ag assembly. Consistent with these observations, dimethyl sulfate studies revealed that T-ag protected pentanucleotides 1 and 3 more than 2 and 4 (6, 31), and preferential binding to the early half of the core origin has been reported by several investigators (55, 70, 75). Of particular interest, 1,10-phenanthroline-copper ion footprinting studies revealed that on the core origin, T-ag double hexamers and T-ag-obd131–260 dimers have virtually identical footprints that extend between pentanucleotides 1 and 3 (32). In the absence of pentanucleotides 1 and 3, pentanucleotides 2 and 4 supported T-ag double-hexamer formation but at relatively low levels when compared to pentanucleotides 1 and 3 (32).
It is noted that under certain conditions, double hexamers can form on the core origin at positions other than the penta 1, 3 + EP and penta 2, 4 + AT assembly units. For example, we previously reported that a 64-bp duplex molecule containing pentanucleotides 1 and 4, the 64-bp AT, penta 1, 4 + EP oligonucleotide, supported formation of two hexamers (32). In view of the experiments presented herein, we now suspect that these assembly events are due to aberrant formation, at relatively high protein-to-DNA ratios, of two hexamers at this distal pair of pentanucleotides. This observation may also be relevant to studies indicating that the SV40 core origin is organized into two complementary halves (i.e., penta 1, 2 + EP and penta 3, 4 + AT) (55). It is possible that Parsons et al. (55) detected EP-dependent assembly on pentanucleotide 1 and AT-dependent assembly on pentanucleotide 4.
While subfragments of the SV40 core origin support the assembly of T-ag double hexamers (this study and references 32, 33, and 35), the entire core origin region is needed for initiation of DNA unwinding (13, 32). These observations imply that certain core origin sequences associate with T-ag at a stage subsequent to double-hexamer formation. Regarding this remodeling process, it is noted that once the double hexamer is formed, two T-ag molecules are predicted to be bound to DNA. The remaining ten molecules are likely to be unbound and available for additional DNA binding events, such as interacting with the penta 2, 4 + AT assembly unit. An obvious question is what causes the double hexamers formed on one assembly unit to engage the second assembly unit? Given that T-ag is known to translocate along leading-strand templates in a 3′-to-5′ direction (29, 65, 78), it is unlikely that both hexamers simply shift to the unoccupied pair of pentanucleotides in the second unit. For example, a hexamer formed on pentanucleotide 1 could not directly translocate to pentanucleotide 2, although a hexamer formed on pentanucleotide 3 could migrate to pentanucleotide 4. Therefore, we suspect that the protein-DNA interactions used to assemble the hexamers are initially maintained and that under replication conditions (i.e., ATP and ATP hydrolysis), DNA present in the penta 2, 4 + AT assembly unit is reeled on to unbound T-ag molecules present in the double hexamers. This hypothesis is consistent with studies indicating that T-ag is immobilized on the origin and the DNA is pulled through during origin unwinding (63, 77) and replication events (reviewed in reference 11). Previous reports indicating that interactions between T-ag and the AT tract activate origin unwinding (40, 54) are consistent with the hypothesis that postassembly DNA-protein interactions are important for initiation of replication.
Several observations indicate that nucleotide hydrolysis may be involved in postassembly remodeling events. For instance, in contrast to double-hexamer formation in the presence of AMP-PNP or ADP, ATP supports hexamer, but not double-hexamer, formation on many of the core origin subfragments; similar observations were made by Kim et al. (35). Given that hexamers are detected at the pentanucleotides proximal to the flanking sequences in the presence of ATP (Fig. 3B, 6B, and 7B), it is possible that ATP or ATP hydrolysis causes preferential lability of the hexamers formed at the distal pentanucleotides. Related observations raise the possibility that this selective destabilization of one hexamer may be related to origin remodeling. For instance, on the penta 1, 3 + EP assembly unit, formation of the second hexamer, presumably on pentanucleotide 3, was not supported by ATP. However, in the presence of ATP, double-hexamer formation was supported by a 64-bp core origin oligonucleotide containing pentanucleotides 1 and 3 and the AT and EP regions (32). Thus in the presence of ATP, the AT is necessary for detection of the second hexamer. One interpretation of this observation is that ATP or ATP hydrolysis results in conformational changes that increase the repertoire of core origin sequences necessary for stable binding of the second hexamer. Consistent with this proposal, DNase I footprints of T-ag bound to the core origin were extended into the flanking regions upon the addition of ATP (6, 18). Moreover, many nucleotide-dependent conformational changes in prokaryotic helicases have been noted (e.g., in references 37 and 74).
It has been suggested that complicated biochemical processes do not change radically once solved by evolution (1, 25). Therefore, it is interesting that one architectural feature shared by many prokaryotic and viral origins of replication is the presence of multiple binding sites for initiators—those proteins that recognize origins in trans (36). In view of the similarity between the architectural features of these different replication origins, it is possible that subregions of many origins will support initiator assembly. If this hypothesis is proven to be correct, then additional evidence will have been obtained that the mechanisms operating during the initiation of DNA replication have been conserved during evolution.
ACKNOWLEDGMENTS
The first three authors contributed equally to this study.
We thank D. G. Sanford for help with computer modeling and A. J. Bullock for comments on the manuscript.
This study was supported by a grant from the NIH (9RO1GM55397).
REFERENCES
- 1.Alberts B. The DNA enzymology of protein machines. Cold Spring Harbor Symp Quant Biol. 1984;49:1–12. doi: 10.1101/sqb.1984.049.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Baker T A, Bell S P. Polymerases and the replisome: machines within machines. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 3.Barbaro B A, Sreekumar K R, Winters D R, Prack A E, Bullock P A. Phosphorylation of simian virus 40 T antigen on Thr124 promotes double-hexamer formation on subfragments of the viral core origin. J Virol. 2000;74:8601–8613. doi: 10.1128/jvi.74.18.8601-8613.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borowiec J A, Dean F B, Bullock P A, Hurwitz J. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell. 1990;60:181–184. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- 5.Borowiec J A, Hurwitz J. ATP stimulates the binding of the simian virus 40 (SV40) large tumor antigen to the SV40 origin of replication. Proc Natl Acad Sci USA. 1988;85:64–68. doi: 10.1073/pnas.85.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borowiec J A, Hurwitz J. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J. 1988;7:3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bullock P A. The initiation of simian virus 40 DNA replication in vitro. Crit Rev Biochem Mol Biol. 1997;32:503–568. doi: 10.3109/10409239709082001. [DOI] [PubMed] [Google Scholar]
- 8.Burhans W C, Huberman J A. DNA replication origins in animal cells: a question of context? Science. 1994;263:639–640. doi: 10.1126/science.8303270. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Joo W S, Bullock P A, Simmons D T. The N-terminal side of the origin-binding domain of simian virus 40 large T antigen is involved in A/T untwisting. J Virol. 1997;71:8743–8749. doi: 10.1128/jvi.71.11.8743-8749.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen G L, Wright P J, DeLucia A L, Lewton B A, Anderson M E, Tegtmeyer P. Critical spatial requirement within the origin of simian virus 40 DNA replication. J Virol. 1984;51:91–96. doi: 10.1128/jvi.51.1.91-96.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook P R. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 12.Dean F B, Borowiec J A, Eki T, Hurwitz J. The simian virus 40 T antigen double hexamer assembles around the DNA at the replication origin. J Biol Chem. 1992;267:14129–14137. [PubMed] [Google Scholar]
- 13.Dean F B, Borowiec J A, Ishimi Y, Deb S, Tegtmeyer P, Hurwitz J. Simian virus 40 large tumor antigen requires three core replication origin domains for DNA unwinding and replication in vitro. Proc Natl Acad Sci USA. 1987;84:8267–8271. doi: 10.1073/pnas.84.23.8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean F B, Bullock P, Murakami Y, Wobbe C R, Weissbach L, Hurwitz J. Simian virus 40 (SV40) DNA replication: SV40 large T antigen unwinds DNA containing the SV40 origin of replication. Proc Natl Acad Sci USA. 1987;84:16–20. doi: 10.1073/pnas.84.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean F B, Dodson M, Echols H, Hurwitz J. ATP-dependent formation of a specialized nucleoprotein structure by simian virus 40 (SV40) large tumor antigen at the SV40 replication origin. Proc Natl Acad Sci USA. 1987;84:8981–8985. doi: 10.1073/pnas.84.24.8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dean F B, Lee S H, Kwong A D, Bullock P, Borowiec J A, Kenny M K, Seo Y S, Eki T, Matsumoto T, Hodgins G, Hurwitz J. SV40 DNA replication in vitro. UCLA Symp Mol Cell Biol New Ser. 1989;127:315–326. [Google Scholar]
- 17.Deb S, DeLucia A L, Baur C-P, Koff A, Tegtmeyer P. Domain structure of the simian virus 40 core origin of replication. Mol Cell Biol. 1986;6:1663–1670. doi: 10.1128/mcb.6.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deb S P, Tegtmeyer P. ATP enhances the binding of simian virus 40 large T antigen to the origin of replication. J Virol. 1987;61:3649–3654. doi: 10.1128/jvi.61.12.3649-3654.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLucia A L, Deb S, Partin K, Tegtmeyer P. Functional interactions of the simian virus 40 core origin of replication with flanking regulatory sequences. J Virol. 1986;57:138–144. doi: 10.1128/jvi.57.1.138-144.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeLucia A L, Lewton B A, Tjian R, Tegtmeyer P. Topography of simian virus 40 A protein-DNA complexes: arrangement of pentanucleotide interaction sites at the origin of replication. J Virol. 1983;46:143–150. doi: 10.1128/jvi.46.1.143-150.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 22.DiMaio D, Nathans D. Cold-sensitive regulatory mutants of simian virus 40. J Mol Biol. 1980;140:129–146. doi: 10.1016/0022-2836(80)90359-9. [DOI] [PubMed] [Google Scholar]
- 23.Dixon R A F, Nathans D. Purification of simian virus 40 large T antigen by immunoaffinity chromatography. J Virol. 1985;53:1001–1004. doi: 10.1128/jvi.53.3.1001-1004.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dodson M, Dean F B, Bullock P, Echols H, Hurwitz J. Unwinding of duplex DNA from the SV40 origin of replication by T antigen. Science. 1987;238:964–967. doi: 10.1126/science.2823389. [DOI] [PubMed] [Google Scholar]
- 25.Echols H. Molecular mechanisms in DNA replication and recombination. 1989. [Google Scholar]
- 26.Fanning E, Knippers R. Structure and function of simian virus 40 large tumor antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 27.Fried M, Crothers D M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garner M M, Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981;9:3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goetz G S, Dean F B, Hurwitz J, Matson S W. The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J Biol Chem. 1988;263:383–392. [PubMed] [Google Scholar]
- 30.Huang S G, Weisshart K, Gilbert I, Fanning E. Stoichiometry and mechanisms of assembly of SV40 T antigen complexes with the viral origin of DNA replication and DNA polymerase α-primase. Biochemistry. 1998;37:15345–15352. doi: 10.1021/bi9810959. [DOI] [PubMed] [Google Scholar]
- 31.Jones K A, Tjian R. Essential contact residues within SV40 large T antigen binding sites I and II identified by alkylation-interference. Cell. 1984;36:155–162. doi: 10.1016/0092-8674(84)90084-9. [DOI] [PubMed] [Google Scholar]
- 32.Joo W S, Kim H Y, Purviance J D, Sreekumar K R, Bullock P A. Assembly of T-antigen double hexamers on the simian virus 40 core origin requires only a subset of the available binding sites. Mol Cell Biol. 1998;18:2677–2687. doi: 10.1128/mcb.18.5.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joo W S, Luo X, Denis D, Kim H Y, Rainey G J, Jones C, Sreekumar K R, Bullock P A. Purification of the SV40 T-antigen DNA binding domain and characterization of its interactions with the SV40 origin. J Virol. 1997;71:3972–3985. doi: 10.1128/jvi.71.5.3972-3985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadonaga J T, Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci USA. 1986;83:5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H Y, Barbaro B A, Joo W S, Prack A, Sreekumar K R, Bullock P A. Sequences requirements for the assembly of simian virus 40 T antigen and T-antigen origin binding domain on the viral core origin of replication. J Virol. 1999;73:7543–7555. doi: 10.1128/jvi.73.9.7543-7555.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kornberg A, Baker T A. DNA replication. 2nd ed. New York, N.Y: W. H. Freeman and Co.; 1992. [Google Scholar]
- 37.Korolev S, Hsieh J, Gauss G H, Lohman T M, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 38.Lewton B A, DeLucia A L, Tegtmeyer P. Binding of simian virus 40 A protein to DNA with deletions at the origin of replication. J Virol. 1984;49:9–13. doi: 10.1128/jvi.49.1.9-13.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J J, Peden K W C, Dixon R A F, Kelly T. Functional organization of the simian virus 40 origin of DNA replication. Mol Cell Biol. 1986;6:1117–1128. doi: 10.1128/mcb.6.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Li B L, Hock M, Wang E, Folk W R. Sequences flanking the pentanucleotide T-antigen binding sites in the polyomavirus core origin help determine selectivity of DNA replication. J Virol. 1995;69:7570–7578. doi: 10.1128/jvi.69.12.7570-7578.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loeber G, Stenger J E, Ray S, Parsons R E, Anderson M E, Tegtmeyer P. The zinc finger region of simian virus 40 large T antigen is needed for hexamer assembly and origin melting. J Virol. 1991;65:3167–3174. doi: 10.1128/jvi.65.6.3167-3174.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lorimer H E, Wang E H, Prives C. The DNA-binding properties of polyomavirus large T antigen are altered by ATP and other nucleotides. J Virol. 1991;65:687–699. doi: 10.1128/jvi.65.2.687-699.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo X, Sanford D G, Bullock P A, Bachovchin W W. Structure of the origin specific DNA binding domain from simian virus 40 T-antigen. Nat Struct Biol. 1996;3:1034–1039. doi: 10.1038/nsb1296-1034. [DOI] [PubMed] [Google Scholar]
- 44.Lusky M, Hurwitz J, Seo Y-S. Cooperative assembly of the bovine papilloma virus E1 and E2 proteins on the replication origin requires an intact E2 binding site. J Biol Chem. 1993;268:15795–15803. [PubMed] [Google Scholar]
- 45.Mastrangelo I A, Hough P V C, Wall J S, Dodson M, Dean F B, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature (London) 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 46.McEntee K, Weinstock G M, Lehman I R. recA protein-catalyzed strand assimilation: stimulation by Escherichia coli single-stranded DNA-binding protein. Proc Natl Acad Sci USA. 1980;77:857–861. doi: 10.1073/pnas.77.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McVey D, Brizuela L, Mohr I, Marshak D R, Gluzman Y, Beach D. Phosphorylation of large tumor antigen by cdc2 stimulates SV40 DNA replication. Nature (London) 1989;341:503–507. doi: 10.1038/341503a0. [DOI] [PubMed] [Google Scholar]
- 48.McVey D, Ray S, Gluzman Y, Berger L, Wildeman A G, Marshak D R, Tegtmeyer P. cdc2 phosphorylation of threonine 124 activates the origin-unwinding functions of simian virus 40 T antigen. J Virol. 1993;67:5206–5215. doi: 10.1128/jvi.67.9.5206-5215.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moarefi I F, Small D, Gilbert I, Hopfner M, Randall S K, Schneider C, Russo A A R, Ramsperger U, Arthur A K, Stahl H, Kelly T J, Fanning E. Mutation of the cyclin-dependent kinase phosphorylation site in simian virus 40 (SV40) large T antigen specifically blocks SV40 origin DNA unwinding. J Virol. 1993;67:4992–5002. doi: 10.1128/jvi.67.8.4992-5002.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murakami Y, Hurwitz J. DNA polymerase α stimulates the ATP-dependent binding of simian virus tumor T antigen to the SV40 origin of replication. J Biol Chem. 1993;268:11018–11027. [PubMed] [Google Scholar]
- 51.Murakami Y, Hurwitz J. Functional interactions between SV40 T antigen and other replication proteins at the replication fork. J Biol Chem. 1993;268:11008–11017. [PubMed] [Google Scholar]
- 52.Myers R M, Tjian R. Construction and analysis of simian virus 40 origins defective in tumor antigen binding and DNA replication. Proc Natl Acad Sci USA. 1980;77:6491–6495. doi: 10.1073/pnas.77.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Reilly D R, Miller L K. Expression and complex formation of simian virus 40 large T antigen and mouse p53 in insect cells. J Virol. 1988;62:3109–3119. doi: 10.1128/jvi.62.9.3109-3119.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parsons R, Anderson M E, Tegtmeyer P. Three domains in the simian virus 40 core origin orchestrate the binding, melting, and DNA helicase activities of T antigen. J Virol. 1990;64:509–518. doi: 10.1128/jvi.64.2.509-518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parsons R E, Stenger J E, Ray S, Welker R, Anderson M E, Tegtmeyer P. Cooperative assembly of simian virus 40 T-antigen hexamers on functional halves of the replication origin. J Virol. 1991;65:2798–2806. doi: 10.1128/jvi.65.6.2798-2806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reynisdottir I, Lorimer H E, Friedman P N, Wang E H, Prives C. Phosphorylation and active ATP hydrolysis are not required for SV40 T antigen hexamer formation. J Biol Chem. 1993;268:24647–24654. [PubMed] [Google Scholar]
- 57.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 58.San Martin M C, Gruss C, Carazo J M. Six molecules of SV40 large T antigen assemble in a propeller-shaped particle around a channel. J Mol Biol. 1997;268:15–20. doi: 10.1006/jmbi.1997.0952. [DOI] [PubMed] [Google Scholar]
- 59.SenGupta D J, Borowiec J A. Strand and face: the topography of interactions between the SV40 origin of replication and T-antigen during the initiation of replication. EMBO J. 1994;13:982–992. doi: 10.1002/j.1460-2075.1994.tb06343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simanis V, Lane D P. An immunoaffinity purification procedure for SV40 large T antigen. Virology. 1985;144:88–100. doi: 10.1016/0042-6822(85)90308-3. [DOI] [PubMed] [Google Scholar]
- 61.Simmons D T, Loeber G, Tegtmeyer P. Four major sequence elements of simian virus 40 large T antigen coordinate its specific and nonspecific DNA binding. J Virol. 1990;64:1973–1983. doi: 10.1128/jvi.64.5.1973-1983.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simmons D T, Upson R, Wun-Kim K, Young W. Biochemical analysis of mutants with changes in the origin-binding domain of simian virus 40 tumor antigen. J Virol. 1993;67:4227–4236. doi: 10.1128/jvi.67.7.4227-4236.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smelkova N V, Borowiec J A. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J Virol. 1997;71:8766–8773. doi: 10.1128/jvi.71.11.8766-8773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sreekumar K R, Barbaro B A, Prack A, Bullock P A. Methods for studying interactions between simian virus 40 T-antigen and the viral origin of replication. In: Raptis L, editor. Methods in molecular biology: SV40 protocols. Totowa, N.J: Humana Press, Inc.; 2000. [DOI] [PubMed] [Google Scholar]
- 65.Stahl H, Droge P, Knippers R. DNA helicase activity of SV40 large tumor antigen. EMBO J. 1986;5:1939–1944. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stillman B, Gerard R D, Guggenheimer R A, Gluzman Y. T antigen and template requirement for SV40 DNA replication in vitro. EMBO J. 1985;4:2933–2939. doi: 10.1002/j.1460-2075.1985.tb04026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Studwell P S, O'Donnell M. Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J Biol Chem. 1990;265:1171–1178. [PubMed] [Google Scholar]
- 68.Tegtmeyer P. Simian virus 40 deoxyribonucleic acid synthesis: the viral replicon. J Virol. 1972;10:591–598. doi: 10.1128/jvi.10.4.591-598.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tegtmeyer P, Lewton B A, DeLucia A L, Wilson V G, Ryder K. Topography of simian virus 40 A protein-DNA complexes: arrangement of protein bound to the origin of replication. J Virol. 1983;46:151–161. doi: 10.1128/jvi.46.1.151-161.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tenen D G, Taylor T S, Haines L L, Bradley M K, Martin R G, Livingston D M. Binding of simian virus 40 large T antigen from virus-infected monkey cells to wild-type and mutant viral replication origins. J Mol Biol. 1983;168:791–808. doi: 10.1016/s0022-2836(83)80075-8. [DOI] [PubMed] [Google Scholar]
- 71.Tjian R. The binding site on SV40 DNA for a T-antigen related protein. Cell. 1978;13:165–179. doi: 10.1016/0092-8674(78)90147-2. [DOI] [PubMed] [Google Scholar]
- 72.Tooze J. DNA tumor viruses. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. [Google Scholar]
- 73.Valle M, Gruss C, Halmer L, Carazo J M, Donate L E. Large T-antigen double hexamers imaged at the simian virus 40 origin of replication. Mol Cell Biol. 2000;20:34–41. doi: 10.1128/mcb.20.1.34-41.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Velankar S S, Soultanas P, Dillingham M S, Subramanya H S, Wigley D B. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 75.Virshup D M, Russo A A R, Kelly T J. Mechanism of activation of simian virus 40 DNA replication by protein phosphatase 2A. Mol Cell Biol. 1992;12:4883–4895. doi: 10.1128/mcb.12.11.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weisshart K, Taneja P, Jenne A, Herbig U, Simmons D T, Fanning E. Two regions of simian virus 40 T antigen determine cooperativity of double-hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. J Virol. 1999;73:2201–2211. doi: 10.1128/jvi.73.3.2201-2211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wessel R, Schweizer J, Stahl H. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wiekowski M, Schwarz M W, Stahl H. Simian virus 40 large T antigen DNA helicase. J Biol Chem. 1988;263:436–442. [PubMed] [Google Scholar]
- 79.Wobbe C R, Dean F, Weissbach L, Hurwitz J. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc Natl Acad Sci USA. 1985;82:5710–5714. doi: 10.1073/pnas.82.17.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wold M S, Li J J, Kelly T J. Initiation of simian virus 40 DNA replication in vitro: large-tumor-antigen- and origin-dependent unwinding of the template. Proc Natl Acad Sci USA. 1987;84:3643–3647. doi: 10.1073/pnas.84.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wun-Kim K, Upson R, Young W, Melendy T, Stillman B, Simmons D T. The DNA-binding domain of simian virus 40 tumor antigen has multiple functions. J Virol. 1993;67:7608–7611. doi: 10.1128/jvi.67.12.7608-7611.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]