Abstract
The Kaposi's sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8, open reading frame (ORF) K9 encodes a viral interferon regulatory factor (vIRF) that functions as a repressor for interferon-mediated signal transduction. Consequently, this gene is thought to play an important role in the tumorigenicity of KSHV. To understand the molecular mechanisms underlying vIRF expression, we studied the transcriptional regulation of this gene. Experiments using 5′ rapid amplification of cDNA ends and primer extension revealed that vIRF had different transcriptional patterns during the latent and lytic phases. The promoter region of the minor transcript, which was mainly expressed in uninduced BCBL-1 cells, did not contain a canonical TATA box, but a cap-like element and an initiator element flanked the transcription start site. The promoter of the major transcript, which was mainly expressed in tetradecanoyl phorbol acetate-induced BCBL-1 cells, contained a canonical TATA box. A luciferase reporter assay using a deletion mutant of the vIRF promoter and a mutation in the TATA box showed that the TATA box was critical for the lytic activity of vIRF. The promoter activity in the latent phase was eight times stronger than that of the empty vector but was less than 10% of the activity in the lytic phase. Therefore, KSHV may use different functional promoter elements to regulate the expression of vIRF and to antagonize the cell's interferon-mediated antiviral activity. We have also identified a functional domain in the ORF 50 protein, an immediate-early gene product that is mainly encoded by ORF 50. The ORF 50 protein transactivated the vIRF and DNA polymerase promoters in BCBL-1, 293T, and CV-1 cells. Deleting one of its two putative nuclear localization signals (NLSs) resulted in failure of the ORF 50 protein to localize to the nucleus and consequently abrogated its transactivating activity. We further confirmed that the N-terminal region of the ORF 50 protein included an NLS domain. We found that this domain was sufficient to translocate β-galactosidase to the nucleus. Analysis of deletions within the vIRF promoter suggested that two sequence domains were important for its transactivation by the ORF 50 protein, both of which included putative SP-1 and AP-1 binding sites. Competition gel shift assays demonstrated that SP-1 bound to these two domains, suggesting that the SP-1 binding sites in the vIRF promoter are involved in its transactivation by ORF 50.
Kaposi's sarcoma associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV-8), was first discovered in tissues obtained from Kaposi's sarcomas (9, 23) and later from a patient with multicentric Castleman's disease (53) and from a body cavity-based lymphoma (BCBL) (primary effusion lymphoma) (7). Although the scope of its etiology and pathogenic mechanisms is yet to be elucidated, the available evidence strongly suggests that KSHV promotes certain types of cell proliferation (38, 52). Although KSHV is difficult to transmit to other cells, it can be maintained in tissues, and viral production can be induced in some cell lines by treatment with certain reagents (1, 8, 12, 14, 39).
KSHV is a new member of the gammaherpesvirus family and has genetic similarity to herpesvirus saimiri and Epstein-Barr virus (EBV) (39, 48). The KSHV genome is double stranded and consists of a long unique DNA sequence of 140.5 kb flanked by multiple GC-rich terminal-repeat sequences (48). The complete nucleotide sequence of KSHV has revealed that KSHV contains 80 complete open reading frames (ORFs), some of which have similarity to those of herpesvirus saimiri; however, 15 ORFs (designated ORF K1 to 15) are unique to KSHV (41, 48). Interestingly, KSHV encodes several homologues to human genes that are associated with the immune response and cell-cycle regulation: i.e., ORF 16 (viral Bcl-2) (10, 50), ORF 72 (viral cyclin D) (16, 30, 45), ORF 74 (viral interleukin 8 [vIL-8] receptor) (2, 19), ORF K2 (vIL-6) (5, 36, 40, 42), ORF K4 and K6 (viral MIP-I and -II) (3, 27, 42), ORF K9 (viral interferon regulatory factor [vIRF]) (29, 63), and ORF K13 (viral FLICE [caspase-8]-inhibitory protease) (57). Previous reports suggested that some of these genes indeed initiate cell proliferation and thereby promote tumor progression.
Herpesvirus genes can be classified as latent, immediate-early (IE), early, and late. The expression of IE and early genes is independent of viral DNA replication, and some of these genes are involved in gene regulation and DNA replication. The late genes are dependent on viral DNA replication and mainly encode structural proteins (22). In cells infected with herpesviruses, the regulation of gene expression generally follows an ordered cascade, with the IE genes being transcribed first, following penetration of the virus. The early and late genes are transcribed thereafter. However, viruses often have a more-complicated pattern of expression that depends on cellular as well as viral transactivator regulation. KSHV transcripts in cell lines derived from primary effusion lymphomas, like BCBL-1, can also be divided into the four classes of latent, IE, early, and late, based on their responsiveness to phorbol ester treatment (49, 55, 62). Based on their sequence homology with other gammaherpesviruses or the kinetics of their expression, several IE candidate genes in KSHV have been proposed, including ORF K3, ORF K5, ORF K8, ORF 50, ORF 57 (48), ORF K4.2, and ORF 45 (62). Among the products of these genes, ORF 50 protein, which is mainly encoded by ORF 50, can activate the expression of several early genes (32, 54).
KSHV vIRF is a homologue of the cellular IRF. Previous studies indicated that vIRF repressed cellular IFN-mediated signal transduction and induced the transformation of NIH 3T3 cells (6, 15, 29, 43, 63). vIRF may also play an important role in the transcriptional activation of KSHV gene expression and cell cycle regulation (26, 46). These facts strongly suggest that the vIRF gene product augments the tumorigenicity of KSHV. In this report, we investigated the molecular mechanisms of the transcriptional regulation of vIRF in the latent and lytic phases of the viral life cycle. We found that an IE gene product, ORF 50 protein, could transactivate vIRF and DNA polymerase, and we characterized one of its nuclear localization signal (NLS) domains. We also identified the response elements to ORF 50 protein in the vIRF promoter region. These results demonstrate an elaborate molecular regulation of vIRF gene expression that has been developed by KSHV to antagonize the IFN-mediated cellular antiviral activity, and that subsequently promotes tumor formation.
MATERIALS AND METHODS
Cell lines and cell culture.
BCBL-1 cells, which are latently infected with KSHV, were grown in RPMI 1640 (Nissui, Tokyo, Japan) supplemented with 10% heat-inactivated fetal bovine serum (GIBCO BRL, Gaithersburg, Md.) at 37°C in a 5% CO2 incubator. When necessary, cells were treated with 25 ng of phorbol ester (12-O-tetradecanoyl phorbol-13 acetate [TPA]) per ml (Sigma, St. Louis, Mo.) to induce the viral lytic cycle. Raji (KSHV-negative, EBV-positive) and Ramos cells (KSHV-negative, EBV-negative) were cultured under the same conditions as BCBL-1 cells. 293T and CV-1 cells were grown in Dulbecco's modified Eagle medium (Nissui) with 10% fetal bovine serum and used for transient transfections.
5′ RACE.
All primers used for cDNA synthesis and PCR were designed on the basis of the published vIRF cDNA sequence data, which were drawn from a KSHV cDNA library (25). 5′ rapid amplification of cDNA ends (5′-RACE) was performed using an amplification kit for cDNA ends (GIBCO BRL) with poly(A)+ RNA extracted from uninduced and TPA-induced BCBL-1 cells. For the 5′-RACE, the first cDNA strand was synthesized using a KSHV-specific primer, K9F6 (5′-GACTCCACATTCCACGCATT-3′) and then tailed with an oligo(dC). The cDNA was amplified by PCR using an adapter-specific [poly(G)] primer provided in the kit and with a second KSHV-specific primer K9F7 (5′-CCACTGTTGACTTGACATACTATCCAATCC-3′). A nested PCR was performed using a nested adapter primer provided in the kit and a third KSHV-specific primer, K9F9 (5′-CACCGGTAGATATTCTTAGTTGTCGTGTCC-3′). The PCR products were inserted into the pCR2.1 vector using a TA cloning kit (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions and analyzed by sequencing using a SequiTherm EXCEL sequencing kit (Epicentre Technologies, Madison, Wis.), which is based on the dideoxynucleotide termination method. Sequence analysis was carried out by comparison with the published KSHV sequence data (48).
Primer extension.
The oligonucleotide primer pK9-Rseq (see Fig. 4) (5′-GTCCCGCAACCAGACTAGCT-3′) was 5′ end labeled with IRD41 (Aloka, Tokyo, Japan). One microgram of poly(A)+ RNA extracted from TPA-treated BCBL-1 cells and 10 μg of poly(A)+ RNA extracted from untreated BCBL-1 cells were mixed with 5 μl of the 1 μM dye primer pK9-Rseq, heated at 70°C for 10 min, and then immediately transferred to 50°C. The extension reaction was carried out at 50°C for 50 min in a total volume of 50 μl, with 200 U of Superscript II reverse transcriptase (GIBCO BRL). After digestion with RNase A for 30 min at 37°C, the sample was precipitated with ethanol, dissolved in Tris-EDTA, and then analyzed on a 6% polyacrylamide sequencing gel containing 8 M urea, on a Li-Cor DNA sequencer (model 4000). A sequencing reaction using the same labeled primer was run alongside to determine the size of the primer extension product. The vIRF promoter-reporter clone was used as a template for the sequencing reactions.
FIG. 4.
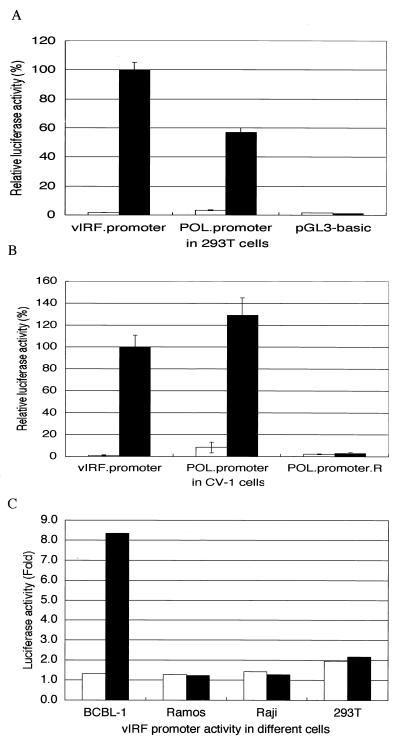
ORF50-cDNA clone transactivated the vIRF promoter and the DNA polymerase promoter in 293T (A) and CV-1 cells (B). The expression constructs for the ORF50-cDNA clone (solid bars) or the ORF50-genomic clone (open bars) were cotransfected with the luciferase reporter constructs driven by the vIRF promoter or the DNA polymerase promoter into 293T cells or CV-1 cells. As a negative control, a promoterless vector, pGL3.basic (A), or a DNA polymerase promoter orientation reverse construct, POL.promoter.R (B), was also included. The relative luciferase activities were normalized to the β-Gal activity. The luciferase activities are shown as percentages compared with the vIRF promoter value, which was defined as 100%. The average (with standard deviation) of duplicate experiments is represented. (C) vIRF promoter activities in different cell lines with (solid bars) or without (open bars) TPA treatment. The luciferase activities were normalized to the β-Gal activity and are shown as fold increases compared with the activity of the promoterless pGL3.basic vector.
Construction of an ORF 50 genomic clone, a full-length cDNA clone, and a β-galactosidase (β-Gal)–NLS-1 fusion protein clone.
Total cellular DNA was extracted from BCBL-1 cells treated with TPA for 48 h, using a QIAGEN (Hilden, Germany) genomic DNA purification kit according to the manufacturer's protocol. The ORF 50 sequence was predicted from the published sequence data (48) and was amplified from total BCBL-1 DNA by PCR using the primers ORF50F-Eco (5′-CGGAATTCATGAAAGAATGTTCCAAGCTTG-3′) and ORF50R-Eco (5′-CTGAATTCTCGGAAGTAATTACGCCATTGG-3′). The amplified DNA fragment was digested with EcoRI and inserted into the pcDNA3.1(−)B expression vector at the EcoRI site (Invitrogen) in-frame with the myc epitope and the polyhistidine tag at the tail.
Because the ORF 50 transcript is generated by a splicing event (54), a full-length cDNA clone was obtained using RT-PCR. Total RNA was isolated from BCBL-1 cells that had been treated with TPA for 24 h, using a QIAGEN RNeasy kit according to the manufacturer's protocol. Poly(A)+ RNA was isolated from total RNA using an mRNA purification kit (TAKARA, Kyoto, Japan) according to the manufacturer's instructions. The first-strand cDNA was synthesized with Superscript II reverse transcriptase and an oligo(dT) primer (GIBCO BRL). The full cDNA fragment was generated by PCR using primers RtaF-Xba (5′-GCTCTAGAAAAATGGCGCAAGATGACAAGG-3′) and RtaR-Xba (5′-GCTCTAGACAGTCTCGGAAGTAATTACGCC-3′). The cDNA fragment was digested with XbaI and inserted into the XbaI site of pcDNA3.1(−)B, in-frame with the myc epitope and the polyhistidine tag. This construct was named ORF50-cDNA to distinguish it from the ORF50-genomic clone. The two constructs were sequenced to confirm that they carried no mutations and were in frame with the myc epitope and polyhistidine tag.
pRSV-lacZ (28) was used for the construction of the β-Gal–NLS-1 fusion protein clone. The first 39-bp oligonucleotide of ORF50-cDNA, which included the first putative NLS consensus sequence, was chemically synthesized to be flanked by KpnI sites and was inserted into the KpnI site of the vector, in frame with the lacZ gene. This construct was named β-Gal–NLS-1.
Transient expression of ORF50-genomic, ORF50-cDNA, and β-Gal–NLS-1 clones in 293T cells.
The pcDNA-ORF50-genomic, pcDNA-ORF50-cDNA, and β-Gal–NLS-1 constructs were used for transient expression. One day before transfection, 293T cells (105 cells) were plated on poly-d-lysine–laminin-coated coverslips (Becton Dickinson Labware) and incubated at 37°C in a 5% CO2 incubator. They were transfected with the expression vectors using SuperFect Transfection Reagent (QIAGEN) according to the manufacturer's instructions.
Indirect immunofluorescence assay.
Forty-eight hours after transfection, the transfected cells were fixed with acetone (for ORF 50) or 4% paraformaldehyde in phosphate-buffered saline (PBS) (for β-Gal) for 15 min, washed with PBS supplemented with 0.1% Triton X-100 for 10 min, and incubated with an appropriate dilution (1:100) of anti-myc monoclonal antibody (MAb) (Invitrogen) (for ORF 50), which recognizes the myc epitope encoded by the pcDNA 3.1 vector or anti-β-Gal monoclonal antibody (GIBCO BRL) (for β-Gal). After incubation for 1 h at 37°C, the cells were washed with PBS and incubated with fluorescein isothiocyanate-conjugated rabbit anti-mouse immunoglobulin G (DAKO, Copenhagen, Denmark) diluted 1:30 for 1 h at 37°C. After being washed as described above, the coverslips were then mounted on slides with 50% glycerol in PBS and examined using fluorescence microscopy.
Construction of the promoter-luciferase reporter clones and a series of vIRF promoter deletion mutants.
The pGL3-Basic promoterless plasmid (Promega, Madison, Wis.) containing the luciferase gene was used for construction of the vIRF and DNA polymerase gene promoters, using a PCR-based strategy. The primers used to construct the vIRF promoter were K9P-F (5′-GCCCATGGTCATATATGTGAATTATAAAAC-3′) and K9P-R (5′-GGGTCCATGGTCCCGCAACCAGACTAGCTC-3′), and the primers for construction of the DNA polymerase promoter were POL.P-F (5′-CCCCATGGTGACGTTTAAGTTTTTGA-3′) and POL.P-R (5′-ATCCATGGTCTGCGGACGGTAATTTG-3′). All of these primers were carefully designed so that the native initiating ATGs for the vIRF and DNA polymerase genes were used to control the translation of the luciferase gene. A mutant vIRF promoter with its 3′ end deleted was constructed using primers K9P-F (5′-GCCCATGGTCATATATGTGAATTATAAAAC-3′) and K9P-R2 (5′-ACATGTGACGTCCCATGGAAAACCAGCGTTTCTCAAAT-3′). In this mutant, part of the putative vIRF lytic promoter sequence was removed, but all of the latent promoter sequence was retained.
A series of deletion mutants of the vIRF promoter was generated using a deletion kit for kilo-sequences (TAKARA). Briefly, 10 μg of pGL3-K9P was digested with KpnI and HindIII to generate a deletion starting site in the promoter region. After purification, the DNA was treated with exonuclease III at 37°C in a total volume of 200 μl, which deletes nucleotides at a speed of about 300 bp/min. At 10-s intervals, 10-μl aliquots of reaction mixture were removed and incubated at 65°C for 5 min to stop the reaction. After removing the single-stranded DNA by treatment with mung bean nuclease and blunting the ends of the double-stranded DNA with the Klenow fragment, the DNAs were self-ligated and transformed into the DH5α bacterial strain. A series of deletion mutants was selected by colony PCR.
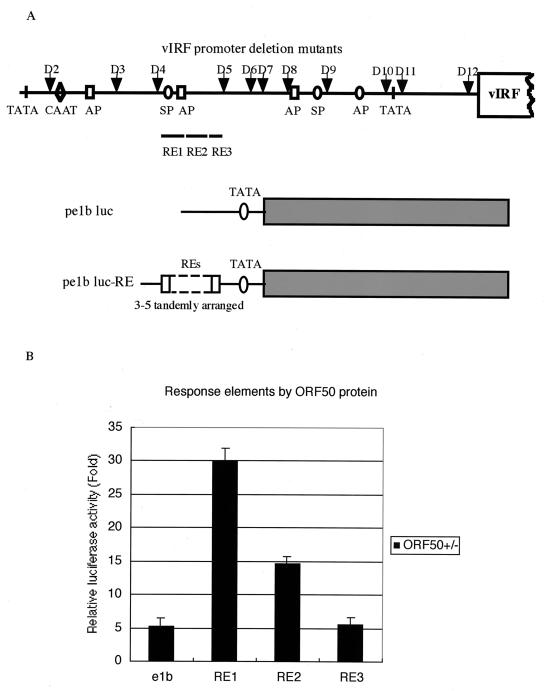
The ORF 50 response element (RE) reporter clones were constructed as follows. First, the reporter plasmid pe1b luc was generated by inserting a minimal TATA box upstream of the firefly luciferase gene in the pSP72 plasmid (Clontech, Palo Alto, Calif.). The putative KSHV ORF 50 REs were chemically synthesized and inserted in a 3-5 tandem arrangement upstream of the e1b minimal TATA element. Three clones were constructed and assigned the following names: pe1b luc-RE1, pe1b luc-RE2, and pe1b luc-RE3. The first clone contained the first, 56-bp putative RE (nucleotides [nt] 85769 to 85714 [GenBank accession no. U75698]) (48), the second clone contained the second, 60-bp element (nt 85713 to 85654), and the third clone contained the last, 29-bp element (nt 85653 to 85625).
Site-directed mutagenesis.
A specific mutation in the TATA box at −30 bp upstream of the major transcription start site (+1) of the vIRF gene was generated using the Quickchange site-directed mutagenesis kit (Strategene, La Jolla, Calif.) following the manufacturer's procedure. The primers used in the PCR to generate this mutation were K9PM1 (5′-CCTAGCCGTGATAGCTCGAGAGCCTGTC-3′) and K9PM2 (5′-GACAGGCTCTCGAGCTATCACGGCTAGG-3′). This mutation changed the TATA box from TATATA to TAGCTC, which should not be functional based on the consensus TATA sequence. The mutations were confirmed by sequencing.
DNA transfection and reporter assays.
DNA transfections into 293T and CV-1 cells were carried out with the SuperFect Transfection Reagent (QIAGEN) according to the manufacturer's instructions. Twelve-well plastic plates were used for the reporter assays. For each transfection into 293T and CV-1 cells, 105 cells and 3 μg of DNA were used. DNA transfections into BCBL-1, Ramos, and Raji cells were carried out by electroporation. Briefly, cells were washed and resuspended in serum-free RPMI 1640 medium at a density of 107 cells/ml. Cell suspensions (0.8 ml) were placed in electroporation cuvettes (diameter, 0.4 cm) with 20 μg of DNA. Cells were electroporated at 960 μF and 250 mV and then transferred to 8 ml of complete RPMI medium. For TPA-treated cells, TPA was added immediately after transfection to a final concentration of 25 ng/ml. Transfection efficiency was normalized by cotransfection of an internal control plasmid, pCMV-β-galactosidase, driven by the human cytomegalovirus IE promoter (Promega) because of its low responsiveness to TPA (J. Chen, unpublished observation). Cell lysates were prepared 48 h after transfection using a luciferase assay kit (Promega) and a galactosidase assay kit (Clontech). The luciferase and β-Gal activities were measured using a Lumat LB9507 photon counter (EG&G Berthold, Germany). In all cases, three or more separate transfections were performed, and the mean with standard deviations was calculated.
Gel-shift assay.
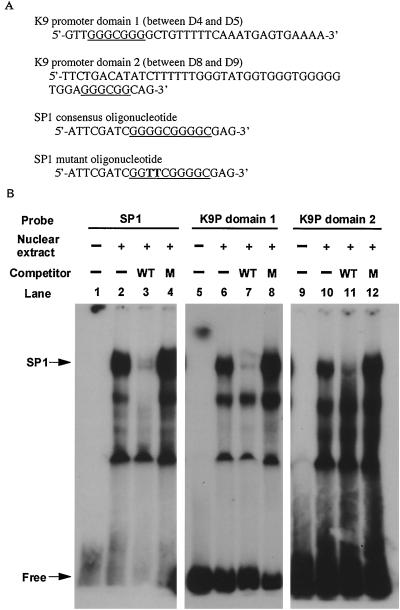
The SP1 consensus oligonucleotide and SP-1 mutant oligonucleotide were commercial products (Santa Cruz Biotechnology, Inc. Santa Cruz, Calif.). They were labeled at the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase (GIBCO BRL). The assay was performed according to the supplier's instructions. To prepare 32P-labeled probe for the assay, K9 promoter domains 1 (between D4 and D5) and 2 (between D8 and D9) were chemically synthesized and 3′ end labeled with [α-32P]dCTP and the Klenow fragment (GIBCO BRL). Nuclear extracts were prepared from BCBL-1 cells treated with TPA for 24 h.
Nucleotide sequence accession numbers.
The nucleotide sequences reported here have been deposited in the GenBank database and assigned the accession numbers AF145700 (for the major transcript) and AF145701 (for the minor transcript).
RESULTS
vIRF has different transcriptional patterns during the latent and lytic phases.
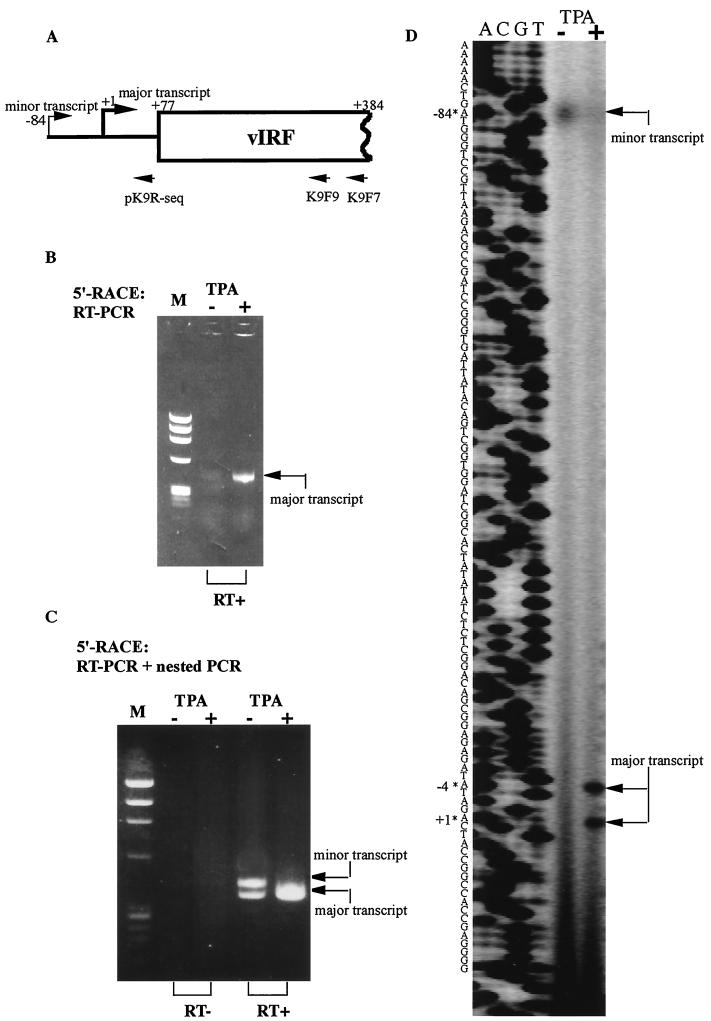
Northern blot analysis showed that vIRF mRNA is 1.5 kb long and expressed during latency, but that it is induced to higher transcription levels by TPA treatment (37, 49). We previously developed a MAb, B291, that reacts specifically with the ORF K9 gene product, vIRF. ORF K9 encodes a 450-amino acid (aa) product that is recognized by MAb B291 as a 50-kDa protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (25). Here we attempted to further analyze the expression kinetics of vIRF using 5′-RACE and primer extension analysis. After the reverse transcription (RT)-PCR in the 5′-RACE analysis (Fig. 1B), an intense band was seen during the lytic phase (TPA treated) and a very weak band could also be seen in untreated BCBL-1 cells. Further analysis of the RT-PCR products by nested PCR (Fig. 1C) showed that the vIRF transcription patterns were different before and after TPA treatment. A higher-molecular-weight band could be observed in untreated BCBL-1 cells. A primer extension experiment (Fig. 1D) showed similar results to 5′-RACE, but there was only one band (the minor transcript) in the latent phase. This could be explained by the different sensitivities of these two methods. The low-molecular-weight band (the major transcript) was also observed in the latent phase in the 5′-RACE experiment, probably because the virus in some uninduced BCBL-1 cells reactivated spontaneously. Although in uninduced cells the spontaneous expression (major transcript) was very low compared with the bona fide latent expression (minor transcript) as shown in primer extension assay, in the PCR system, and especially in the nested PCR system, the low-molecular-weight transcript (spontaneous major transcript) tended to be amplified more efficiently, causing the two bands in Fig. 1C to appear to be equally abundant in the pre-TPA sample. The minor transcription start site was 84 bp upstream of the major transcription start site (defined as +1) (Fig. 1A and D). The minor transcript was less abundant and ran as a wider, blurrier band than did the major transcript, which was strongly expressed in the lytic phase (Fig. 1D). These results indicated that vIRF had different transcriptional patterns during the latent and lytic phases.
FIG. 1.
Mapping of the vIRF transcription start sites identified by 5′-RACE and primer extension. (A) Schematic diagram of the vIRF transcripts and the primers used for 5′-RACE (K9F7 and K9F9) and primer extension (pK9R-seq). (B) RT-PCR result of 5′-RACE. The expression of vIRF mRNA in untreated BCBL-1 cells was very low compared with that in TPA-treated cells. RT+, reverse transcriptase treated. (C) Nested-PCR result of 5′-RACE. Different transcription patterns are seen in untreated versus TPA-treated BCBL-1 cells. RT−, control without reverse transcriptase. (D) Primer extension result. Minor and major transcripts had different start sites. Please note that the amounts of mRNA were different (10 μg of RNA for the lane without TPA [TPA−] and 1 μg of RNA for the lane with TPA [TPA+]; see Materials and Methods). The size of the primer extension product was indicated by a sequencing ladder initiated with the same primer. The arrows indicate the positions of the transcription start sites.
vIRF uses different promoter domains during latent and lytic phases.
Analysis of the sequence of the vIRF promoter showed a putative TATA box located −30 bp upstream of the major transcription start site but downstream of the minor transcription start site (Fig. 2). Additionally, a putative cap site, CTGGT (−96 to −92 bp), and a conserved initiator element, CAATTC (−76 to −71 bp), flanked the minor transcription start site. Several predicted binding sites for cellular transcriptional factors were identified in this region that might be involved in the transcriptional regulation of vIRF (Fig. 2).
FIG. 2.
Nucleotide sequence of the vIRF promoter region. The sequence was taken from GenBank KSU75698, nt 86073 to 85207 (48). The boldface letters ATG represent the vIRF translation start codon. The TATA element is indicated in boldface type and boxed. Four potential AP-1 sites and two potential SP-1 sites are boxed. The putative minor and major transcription start sites are indicated with arrows. One of the major transcription start sites was defined as the first nucleotide, based on the results of both 5′-RACE and primer extension. In addition, a conserved cap site and an initiator element flanking the minor transcription start site are indicated.
We next compared the relative activity of the vIRF promoter during the latent (uninduced) and lytic (TPA-treated) phases (Fig. 3). The activity of the vIRF promoter in the latent phase was eight times higher than that of the promoterless pGL3-basic vector, which was used as a control. However, its activity was less than 10% of the activity observed after TPA treatment. This result was compatible with the result we obtained by immunofluorescence staining of BCBL-1 cells with MAb B-291 (25). During the latent phase (uninduced), the expression of vIRF was detectable in about 1% of the cells, while during the lytic phase (TPA treated), approximately 10 to 20% of the cells had detectable vIRF expression.
FIG. 3.
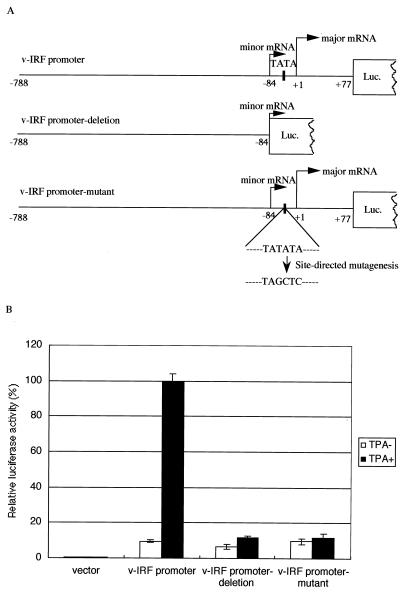
Activation of the vIRF promoter 3′-end deletion mutant and TATA element site-directed mutant. (A) Schematic diagram of the vIRF promoter and two mutant constructs. The vIRF promoter deletion mutant lacked part of the putative vIRF lytic promoter sequence but retained all of the latent promoter sequence. The TATA box was located between the minor and major transcript start sites. In the vIRF promoter TATA mutant, TATATA was changed to TAGCTC. (B) Relative luciferase activity of the vIRF promoter deletion mutant and the TATA element mutant compared with the wild-type promoter. Neither mutant construct was responsive to TPA induction. Note that the vIRF promoter still had low activity in the latent phase (uninduced). The wild-type vIRF promoter and the two mutant promoter constructs were transfected into BCBL-1 cells that were not induced or were induced with TPA immediately after transfection. Cells were harvested 48 h later and assayed for luciferase activity. Luciferase activity values were normalized by cotransfecting an internal control plasmid (pCMV-β-Gal). Activities were expressed relative to the wild-type promoter activity induced by TPA, which was defined as 100%. The average of duplicate experiments (with standard deviation [error bar]) is represented.
To investigate whether vIRF uses different promoter elements to drive its expression during the latent and lytic phases, we generated a promoter deletion mutant (vIRF promoter deletion) in which the 3′ end of the promoter was deleted downstream of the minor transcription start site (Fig. 3A). The luciferase assay showed that this construct lost its responsiveness to TPA induction (Fig. 3B). Furthermore, we investigated which transcriptional element was critical for the lytic activity. In vIRF, the promoter for the minor transcription should not include the TATA element that is located downstream of the minor transcription start site and upstream of the major transcription start site. To determine if the TATA box is active during the lytic phase, we constructed a nonfunctional, site-directed mutant of the TATA box, based on the TATA consensus sequence, in which TATATA was changed to TAGCTC (Fig. 3A). The luciferase activity of this promoter (vIRF promoter mutant) was measured and compared with that of the wild-type promoter. The vIRF promoter TATA mutant also lost its responsiveness to TPA induction, whereas the basal level of activity was retained in the presence and absence of TPA. This result demonstrated that the TATA box was critical for the lytic expression of vIRF upon TPA induction, while in the latent phase, the TATA box was dispensable.
ORF50-cDNA clone transactivates the vIRF and DNA polymerase promoters.
Previous studies showed that the ORF50 protein activates early lytic genes, including those coding for vIL-6, polyadenylated nuclear RNA (nut-1), thymidine kinase, kaposin, and DNA binding protein, and a late gene coding for a small viral capsid antigen (32, 54). The KSHV ORF 50 protein is a homologue of EBV Rta, which is an IE gene of EBV and transactivates many EBV viral promoters (17, 21, 33, 35, 44, 58, 59). To determine whether the KSHV ORF 50 protein, like EBV Rta, is also a transactivator, its ability to transactivate the vIRF and DNA polymerase promoters was tested. A genomic clone (ORF50-genomic) and a cDNA clone (ORF50-cDNA) were constructed to test whether the splicing event was important for the transactivating ability of the ORF50 protein (see Fig. 5A).
FIG. 5.
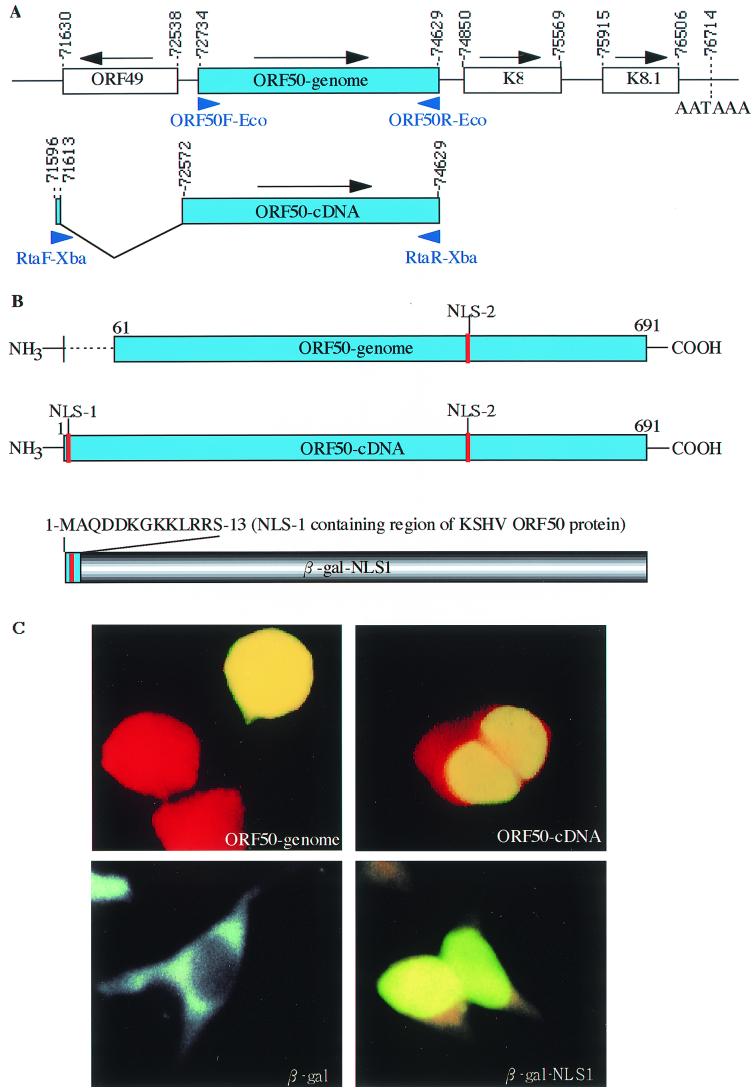
An NLS was critical for the nuclear translocation of ORF 50 protein, an IE gene product of KSHV, which is mainly encoded by ORF50. (A) Diagram of the genomic region containing the ORF50-genomic clone and the ORF50-cDNA clone encoding the ORF 50 gene product. ORFs are shown in open boxes. The direction of the ORF is indicated with an arrow above the box. The numbers indicate nucleotide positions in the KSHV genome (48). The position of the ORF50-cDNA clone used for transfection studies is shown, with the splice site between 71,613 and 72,572 indicated. The two open boxes indicate the fused ORF50-cDNA ORF. The positions of the primers used for PCR amplification of the ORF50-genomic and ORF50-cDNA clones are indicated by arrows below the boxes. (B) Construction of ORF50-genomic, ORF50-cDNA, and β-Gal–NLS-1. The ORF50-genomic clone lacks the N-terminal 60 aa that was added to the ORF50-cDNA clone through a splicing event. This region also includes a putative NLS (NLS-1). To test the function of this NLS, the first 13 aa of ORF50-cDNA was fused to β-Gal and its nuclear localization ability was tested. (C) Immunofluorescence staining results. The ORF50-cDNA clone localized to the nucleus, while most of the ORF50-genomic clone localized to both the nucleus and the cytoplasm. The upper two panels show the localization of the ORF50-genomic and ORF50-cDNA clones. The lower two panels show that β-Gal was able to localize to the nucleus when fused with NLS-1 of KSHV ORF 50 protein.
ORF50-genomic and ORF50-cDNA clones were tested for their effects on the luciferase activity of vIRF and DNA polymerase promoters in 293T, CV-1, and BCBL-1 cells. ORF50-cDNA transactivated the vIRF and DNA polymerase promoters in 293T cells (Fig. 4A), CV-1 cells (Fig. 4B), and BCBL-1 cells (data not shown), while ORF50-genomic did not transactivate these two promoters (Fig. 4A and B and data not shown). Because ORF50-cDNA transactivated the vIRF and DNA polymerase promoters in 293T and CV-1 cells, we speculated that this transactivation did not require other viral factors. These results indicated that KSHV ORF 50 protein, like its homologue, EBV Rta, was a transactivator.
To determine whether TPA could activate vIRF expression directly, without the expression of other KSHV genes, we compared the activity of the vIRF promoter in several cell lines with or without TPA treatment. TPA transactivated the vIRF promoter in KSHV-infected BCBL-1 cells but had no effect on the promoter in Ramos (KSHV-negative, EBV-negative) Raji (KSHV-negative, EBV-positive), and 293T cells (Fig. 4C). These results indicated that the ORF 50 product was required for the activation of vIRF. We confirmed these results by transfecting the ORF50-cDNA clone into BCBL-1 cells and observing the vIRF expression using a vIRF-specific MAb (25). The results of this experiment also suggested that ORF 50 activated endogenous vIRF expression.
An NLS domain in the N terminus of the KSHV ORF 50 protein is critical for its nuclear localization and consequently for its transactivating ability.
To better understand the differences between the transactivating abilities of the ORF50-genomic and ORF50-cDNA clones, we compared their sequences. The ORF50-cDNA sequence is in frame with ORF50-genomic and is encoded almost entirely by ORF50, but the first 958 bp of the genomic sequence, which encode ORF49, are deleted in a splicing event (Fig. 5A). This splicing event provides a new start site for ORF50-cDNA, 60 aa upstream of the ORF50-genomic start site, and also introduces a putative NLS sequence (Fig. 5B). We compared the expression patterns of ORF50-genomic and ORF50-cDNA in 293T cells using an immunofluorescence assay to detect the myc epitope tag, which was encoded in frame with both ORF50-cDNA and ORF50-genomic (Fig. 5C). The results showed that ORF50-cDNA was localized to the nucleus. In contrast, ORF50-genomic was unstable and tended to localize to both the nucleus and the cytoplasm.
To confirm that there is an NLS in the N terminus of the ORF50 protein, a clone was constructed (Fig. 5B) in which a fragment containing the first 13 aa of the protein encoded by ORF50-cDNA was fused with the N-terminal region of a β-Gal protein, and the nuclear localization ability of this fusion was tested. As shown in Fig. 5C, this 13-aa fragment conferred its nuclear localization capability to the fusion protein.
Another putative nuclear targeting sequence was located in the C-terminal region of the ORF50 protein. This NLS seemed to be less effective, since deletion of the N-terminal NLS resulted in unstable expression and both nuclear and cytoplasmic location of the ORF 50 protein (Fig. 5C, first panel). Compared with ORF50-cDNA, ORF50-genomic, which was unstable and was not strictly localized to the nucleus, had no transactivator function (Fig. 4A and B). These data suggest that the NLS in the first 60 aa of the ORF 50 protein is essential for its translocation to the nucleus and that this translocation is in turn required for its transactivating activity.
Identification of vIRF promoter elements required for promoter activity.
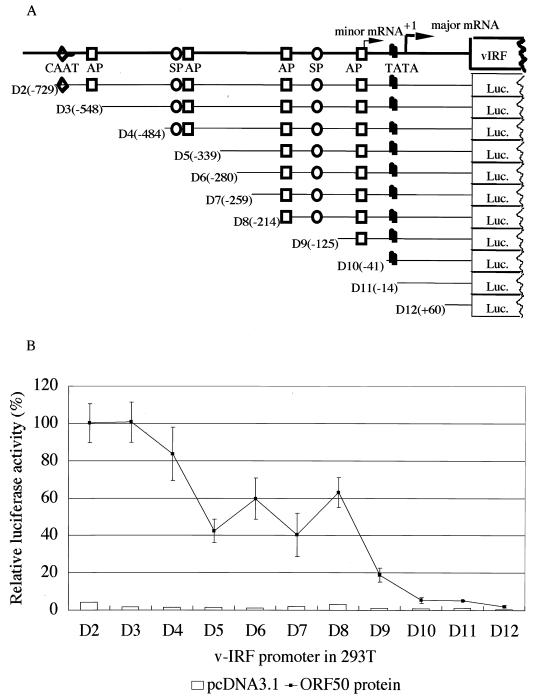
An 867-bp sequence encompassing the region from the stop codon of ORF K10 to the start codon of ORF K9 (vIRF) was examined for potential regulatory sites (Fig. 2). Analysis of the vIRF promoter sequence demonstrated the presence of several putative binding sites for transcription factors that could be important for promoter activity. This sequence contained the TATA box −30 bp upstream of the major transcription start site, four putative AP-1 recognition sequences, and two GC-rich SP-1 binding sites. The AP-1 transcription factor could be responsible for the induction of transcription by phorbol esters (13, 24). We believed the responsiveness of the vIRF promoter to TPA to be mediated through the activation of the ORF 50 protein, since TPA could not induce vIRF promoter activity directly in 293T cells, nor in KSHV-negative lymphoid cells such as Raji and Ramos cells (Fig. 4C). To assess the REs in the vIRF promoter, we generated a series of deletion mutants within the vIRF promoter (Fig. 6A) and analyzed their effects on vIRF promoter activity in the presence of the viral transactivator ORF 50 protein (Fig. 6B). A luciferase reporter assay indicated that there were two sequence domains that were critical for the activation of the vIRF promoter. One was located between −548 bp (D3) and −339 bp (D5) upstream of the major transcription start site and included an SP-1 and an AP-1 site. Another was located between −214 bp (D8) and −41 bp (D10) upstream of the major transcription start site and included two AP-1 sites and an SP-1 site. Deletion of the first activation domain (D3 to D5) reduced the promoter activation by half. The sequence between −339 bp (D5) and −214 bp (D8) seemed not to contain major activation elements, since there was no significant difference in the activity among mutants bearing deletions in this region. Further deletion of an AP-1 site and an SP-1 site (D8 to D9) reduced the promoter activation to 20% of its full activity. The additional deletion of an AP-1 site (D9 to D10) completely abolished the vIRF promoter's activity in response to ORF 50 protein transactivation.
FIG. 6.
(A) Schematic diagram of the vIRF promoter and a series of nested deletion mutants. Putative transcription elements located within the vIRF promoter are shown. AP, activator protein; SP, stimulating protein; TATA, TATA box. The numbers indicate the positions relative to the major transcription start site, which was defined as +1. (B) Transactivation of the vIRF promoter by the ORF50-cDNA clone in 293T cells. The indicated deletion mutants of the vIRF promoter were cotransfected into 293T cells with a transactivator (pcDNA3.1-ORF50-cDNA) or empty vector (pcDNA3.1) as a negative control. Cells were harvested 48 h after transfection and assayed for luciferase activity. The luciferase activity values were normalized to a cotransfected internal control plasmid (pCMV-β-gal). The activation was indicated as a percentage of that of the full-length vIRF promoter, which was defined as 100%. Each experiment was repeated at least three times with similar results. The average value (with standard deviation) of duplicate experiments is shown.
The results of the luciferase reporter assay implied the involvement of AP-1 or SP-1 in ORF50 response activity. We constructed three luciferase reporter clones driven by different REs: RE1, RE2, and RE3 (Fig. 7A). The relative luciferase activities of these three constructs indicated that RE1, which included an AP-1 and an SP-1 binding site, had the greatest transactivating capability (Fig. 7B), suggesting that AP-1 or SP-1 is indeed involved in the transactivation of vIRF by ORF 50. Together with the results shown in Fig. 6, these results demonstrated that multiple REs might be important in the transactivation of the vIRF promoter.
FIG. 7.
Luciferase reporter assay of the response elements to the ORF 50 protein in the vIRF promoter region. (A) Schematic diagram of the vector (p e1b luc) and the constructs (p e1b luc-RE). The locations of RE1, RE2, and RE3 are shown. The arrows show the locations of the vIRF promoter deletion mutants. (B) The relative luciferase activity of the RE reporter constructs in response to the ORF 50 protein. The mean luciferase activities in response to the ORF 50 protein obtained from three transfections after normalization for β-Gal expression are indicated as fold increases relative to the pcDNA3.1 vector control.
To confirm this result, we performed a competition gel-shift assay (Fig. 8A), using an SP-1 consensus oligonucleotide and an SP-1 mutant oligonucleotide as the competitor. As shown in Fig. 8B, the binding of SP-1 by K9 promoter domain 1 (between D4 and D5), which included RE1, RE2, and RE3, could be inhibited by wild-type SP-1 but not by mutant SP-1 (lanes 7 and 8). Another activation domain (between D8 and D9), identified by the promoter deletion mutant reporter assay, was similarly inhibited, although in this case the competition was partial (lane 11). These results suggested that these SP-1-like motifs were functional and that SP-1 might be involved in the transactivation of the vIRF promoter by the ORF 50 protein. We also noticed an additional lower band that was competed out with SP-1 in lane 3 but that was not competed with K9P domains 1 and 2 (lanes 7 and 11). We believe that this additional lower band was not SP-1, but another cellular DNA binding factor, since both K9P domains 1 and 2 contained consensus sequences for both SP-1 (matrix similarity, 95.9 and 88.9%, respectively) and AP-1 (matrix similarity, 86.2 and 85.4%, respectively).
FIG. 8.
Competition gel shift assay. (A) Oligonucleotide sequences used in the assay. The SP1 consensus sequences are underlined. The mutant nucleotides are shown in boldface type. (B) Lanes 1, 5, and 9, 32P-labeled probe only; lanes 2, 6, and 10, labeled probe with 2 μg of nuclear extract prepared from BCBL-1 cells treated with TPA for 24 h; lanes 3, 7, and 11, labeled probe with nuclear extract and 100 ng of unlabeled SP1 oligonucleotide (WT) as a competitor; lanes 4, 8, and 12, labeled probe with nuclear extract and 100 ng of unlabeled SP1 mutant oligonucleotide (M) as a competitor. The SP1 consensus oligonucleotide and K9 promoter domain 1 (D4 to D5) and domain 2 (D8 to D9) were labeled with 32P and used as probes in lanes 1 to 4, 5 to 8, and 9 to 12, respectively.
DISCUSSION
Like most of the virus-encoded cytokines and signal transduction genes, vIRF can be expressed in latency but is induced to higher transcription levels by TPA treatment (37). The vIRF gene is therefore defined as a class II gene, differing from class I genes (constitutively transcribed both in presence and absence of TPA) and class III genes (transcribed only following TPA treatment) (49). In a previous report, we showed that the vIRF mRNA was expressed during the early stage of viral infection (25). In uninduced BCBL-1 cells, about 1% of the cells expressed vIRF as examined using an immunofluorescence assay, while after TPA treatment, the percentage of vIRF-positive cells increased to 10 to 20%. In this report, we further investigated the molecular mechanisms underlying the differential expression of vIRF during the latent and lytic phases. We found that there were different transcriptional patterns during the latent and lytic phases, which suggested that the differential expression is due to the use of different promoters. In untreated cells, we observed weak expression of vIRF mRNA, which was greatly elevated in cells treated with TPA. However, it was difficult to distinguish the minor and major mRNAs by Northern blot, since the minor mRNA was only 84 bp longer than the major mRNA (based on the results of 5′-RACE and primer extension).
Expression of eukaryotic genes is often regulated through a mechanism that depends on alternative promoter use. This mechanism may also be common in viruses. In KSHV, we found vIRF expression to be regulated through different promoter elements. The promoter for the major transcript included a TATA box element, while the promoter for the minor transcript lacked a TATA-like sequence but included a cap site and a conserved initiator element. Initiator elements are often present in TATA-less promoters (47). In such cases, initiator elements bind the transcription factor TFIID and can replace the TATA box as the initiation site for RNA polymerase II-dependent transcription. Typically, this transcriptional activity is weak and the transcript start site is less precisely positioned (4, 34). In our 5′-RACE experiment, there was a major as well as a minor transcript band in the latent phase (uninduced), but in our primer extension experiment only the minor transcript band was seen. This difference was likely to be due to the different sensitivities of these two methods. In the latent phase, less than 1% of the cells could be spontaneously activated, and the resultant low-molecular-weight transcript was amplified more efficiently after two rounds of PCR than the high-molecular-weight transcript, resulting in equally abundant bands in pre-TPA-treated cells in the 5′-RACE–nested-PCR experiment.
By using a different promoter element during the latent and lytic phases, KSHV may be able to regulate vIRF expression and subsequently to contribute to viral latency by repressing cellular IFN-mediated signal transduction. Although the promoter activity in the latent phase was weak (only 8% of that of the lytic phase), vIRF expression in latency, like that of other virus-encoded cytokines and signal-transduction genes, should also be essential for KSHV's escape from the host cell's defense system against virus invasion.
The transport of proteins into the nucleus is dependent on the NLS, although many nuclear targeting sequences appear to be quite complex. A consensus NLS bipartite motif, (K/R)(K/R)XXXXXXXXXXKXXKK, was identified in the simian virus 40 large T antigen (11). Zhang et al. (61) showed that the last portion of this sequence (KXXKK) was sufficient for its localization to the nucleus. In many cases, localization to the nucleus is essential for the activity of a putative transactivator. The ORF 50 protein acquires a new nuclear targeting sequence at its N terminus through a splicing event. The fact that it simultaneously obtains transactivation activity showed that the splicing event is very important for the function of this gene. Our data indicated that the N-terminal NLS of the ORF 50 protein is critical for its localization to the nucleus. As with many other transactivators, the localization of ORF 50 protein to the nucleus is probably necessary for its transactivating activity.
The ORF 50 protein has homologues in other gammaherpesviruses that function as transcriptional activators and activate lytic cycle gene expression (21, 33, 35, 58, 59). The gammaherpesvirus Rta protein appears to be unique in that the region involved in DNA binding does not contain any well-characterized DNA-binding motifs, and Rta is not homologous with any known cellular transcriptional activators. EBV Rta is known to bind DNA directly in some cases, but it may also interact with DNA via other proteins or transcriptional factors (20). The N-terminal 238 aa of the KSHV ORF 50 protein contains a region that is 20 to 32% identical and 41 to 52% similar to the Rta proteins of EBV, herpesvirus saimiri, bovine herpesvirus 4, equine herpesvirus, and mouse herpesvirus 68 (32). In EBV Rta, this domain mediates dimerization and DNA binding. This functional domain, as well as an activation domain located near the carboxyl terminus, is conserved in the ORF 50 protein, suggesting that the ORF 50 protein may function similarly to its EBV counterpart in the viral life cycle (18, 21). The ORF 50 protein transactivated the vIRF and DNA polymerase promoters in 293T cells and in CV-1 cells. This result suggests that the ORF 50 protein may interact with DNA directly or through other cellular factors without requiring additional virally encoded factors.
Transcriptional regulation of KSHV genes relies on a complex interaction between cellular and viral transactivators. Previous studies have implicated the cellular transcription factors AP-1 and SP-1 to be involved in ORF 50 protein-directed early gene regulation (51, 56, 60). A study on the activation of another early gene, the thymidine kinase gene of KSHV, by the ORF 50 gene product suggests that it is SP-1 dependent (60). Although the AP-1 transcription factor could be responsible for the transcription induced by treatment with phorbol esters (TPA), the induction of vIRF by TPA treatment in BCBL-1 cells is probably mediated through the activation of the ORF50 protein, since TPA could not transactivate the vIRF promoter directly in 293T, Raji, or Ramos cells. We confirmed this idea by directly transfecting the ORF50-cDNA clone into BCBL-1 cells and testing vIRF expression by an immunofluorescence assay using a MAb that reacts specifically with the vIRF protein (25). A significant increase in the expression of vIRF was observed. This increase in vIRF expression was comparable to that obtained with TPA treatment (data not shown), suggesting that ORF 50 indeed activated endogenous vIRF expression.
The KSHV ORF 50 gene product plays an essential role in KSHV lytic replication and is a putative molecular switch controlling the induction of virus from latency (31, 32, 54). The ORF 50 protein can activate the expression of many early and some late genes. However, it is not yet clear whether ORF 50 expression can ultimately lead to viral DNA synthesis or viral particle formation, although EBV Rta can activate the EBV lytic cascade in epithelial cells and disrupts latency in several B-lymphoid cell lines (44, 59). Several putative IE genes of KSHV, including ORF K5, ORF K8, ORF K3, ORF 45, and ORF 4.2 are under investigation in our laboratory to identify their functional characteristics.
REFERENCES
- 1.Arvanitakis L, Mesri E A, Nador R G, Said J W, Asch A S, Knowles D M, Cesarman E. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–2654. [PubMed] [Google Scholar]
- 2.Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka E G, Gutkind J S, Asch A S, Cesarman E, Gershengorn M C, Mesri E A, Gerhengorn M C. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391:86–89. doi: 10.1038/34193. . (Erratum, 392:210.) [DOI] [PubMed] [Google Scholar]
- 3.Boshoff C, Endo Y, Collins P D, Takeuchi Y, Reeves J D, Schweickart V L, Siani M A, Sasaki T, Williams T J, Gray P W, Moore P S, Chang Y, Weiss R A. Angiogenic and HIV-inhibitory functions of KSHV-encoded chemokines. Science. 1997;278:290–294. doi: 10.1126/science.278.5336.290. [DOI] [PubMed] [Google Scholar]
- 4.Bulfone-Paus S, Dempsey L A, Maizels N. Host factors LR1 and Sp1 regulate the Fp promoter of Epstein-Barr virus. Proc Natl Acad Sci USA. 1995;92:8293–8297. doi: 10.1073/pnas.92.18.8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burger R, Neipel F, Fleckenstein B, Savino R, Ciliberto G, Kalden J R, Gramatzki M. Human herpesvirus type 8 interleukin-6 homologue is functionally active on human myeloma cells. Blood. 1998;91:1858–1863. [PubMed] [Google Scholar]
- 6.Burysek L, Yeow W S, Lubyova B, Kellum M, Schafer S L, Huang Y Q, Pitha P M. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J Virol. 1999;73:7334–7342. doi: 10.1128/jvi.73.9.7334-7342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS- related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 8.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 9.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 10.Cheng E H, Nicholas J, Bellows D S, Hayward G S, Guo H G, Reitz M S, Hardwick J M. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci USA. 1997;94:690–694. doi: 10.1073/pnas.94.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 12.Foreman K E, Friborg J, Jr, Kong W P, Woffendin C, Polverini P J, Nickoloff B J, Nabel G J. Propagation of a human herpesvirus from AIDS-associated Kaposi's sarcoma. N Engl J Med. 1997;336:163–171. doi: 10.1056/NEJM199701163360302. [DOI] [PubMed] [Google Scholar]
- 13.Franklin C C, Sanchez V, Wagner F, Woodgett J R, Kraft A S. Phorbol ester-induced amino-terminal phosphorylation of human JUN but not JUNB regulates transcriptional activation. Proc Natl Acad Sci USA. 1992;89:7247–7251. doi: 10.1073/pnas.89.15.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaidano G, Cechova K, Chang Y, Moore P S, Knowles D M, Dalla-Favera R. Establishment of AIDS-related lymphoma cell lines from lymphomatous effusions. Leukemia. 1996;10:1237–1240. [PubMed] [Google Scholar]
- 15.Gao S J, Boshoff C, Jayachandra S, Weiss R A, Chang Y, Moore P S. KSHV ORF K9 (vIRF) is an oncogene, which inhibits the interferon signaling pathway. Oncogene. 1997;15:1979–1985. doi: 10.1038/sj.onc.1201571. [DOI] [PubMed] [Google Scholar]
- 16.Godden-Kent D, Talbot S J, Boshoff C, Chang Y, Moore P, Weiss R A, Mittnacht S. The cyclin encoded by Kaposi's sarcoma-associated herpesvirus stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J Virol. 1997;71:4193–4198. doi: 10.1128/jvi.71.6.4193-4198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruffat H, Duran N, Buisson M, Wild F, Buckland R, Sergeant A. Characterization of an R-binding site mediating the R-induced activation of the Epstein-Barr virus BMLF1 promoter. J Virol. 1992;66:46–52. doi: 10.1128/jvi.66.1.46-52.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruffat H, Sergeant A. Characterization of the DNA-binding site repertoire for the Epstein-Barr virus transcription factor R. Nucleic Acids Res. 1994;22:1172–1178. doi: 10.1093/nar/22.7.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo H G, Browning P, Nicholas J, Hayward G S, Tschachler E, Jiang Y W, Sadowska M, Raffeld M, Colombini S, Gallo R C, Reitz M S., Jr Characterization of a chemokine receptor-related gene in human herpesvirus 8 and its expression in Kaposi's sarcoma. Virology. 1997;228:371–378. doi: 10.1006/viro.1996.8386. [DOI] [PubMed] [Google Scholar]
- 20.Gutsch D E, Marcu K B, Kenney S C. The Epstein-Barr virus BRLF1 gene product transactivates the murine and human c-myc promoters. Cell Mol Biol (Noisy-le-Grand) 1994;40:747–760. [PubMed] [Google Scholar]
- 21.Hardwick J M, Tse L, Applegren N, Nicholas J, Veliuona M A. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J Virol. 1992;66:5500–5508. doi: 10.1128/jvi.66.9.5500-5508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y Q, Li J J, Kaplan M H, Poiesz B, Katabira E, Zhang W C, Feiner D, Friedman-Kien A E. Human herpesvirus-like nucleic acid in various forms of Kaposi's sarcoma. Lancet. 1995;345:759–761. doi: 10.1016/s0140-6736(95)90641-x. [DOI] [PubMed] [Google Scholar]
- 24.Imagawa M, Chiu R, Karin M. Transcription factor AP-2 mediates induction by two different signal-transduction pathways: protein kinase C and cAMP. Cell. 1987;51:251–260. doi: 10.1016/0092-8674(87)90152-8. [DOI] [PubMed] [Google Scholar]
- 25.Inagi R, Okuno T, Ito M, Chen J, Mori Y, Haque M, Zou P, Yagi H, Kiniwa S, Saida T, Ueyama Y, Hayashi K, Yamanishi K. Identification and characterization of human herpesvirus 8 open reading frame K9 viral interferon regulatory factor by a monoclonal antibody. J Hum Virol. 1999;2:63–71. [PubMed] [Google Scholar]
- 26.Jayachandra S, Low K G, Thlick A E, Yu J, Ling P D, Chang Y, Moore P S. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc Natl Acad Sci USA. 1999;96:11566–11571. doi: 10.1073/pnas.96.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Kledal T N, Rosenkilde M M, Coulin F, Simmons G, Johnsen A H, Alouani S, Power C A, Luttichau H R, Gerstoft J, Clapham P R, Clark-Lewis I, Wells T N C, Schwartz T W. A broad-spectrum chemokine antagonist encoded by Kaposi's sarcoma-associated herpesvirus. Science. 1997;277:1656–1659. doi: 10.1126/science.277.5332.1656. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi T, Shoya Y, Koda T, Takashima I, Lai P K, Ikuta K, Kakinuma M, Kishi M. Nuclear targeting activity associated with the amino terminal region of the Borna disease virus nucleoprotein. Virology. 1998;243:188–197. doi: 10.1006/viro.1998.9049. [DOI] [PubMed] [Google Scholar]
- 29.Li M, Lee H, Guo J, Neipel F, Fleckenstein B, Ozato K, Jung J U. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor. J Virol. 1998;72:5433–5440. doi: 10.1128/jvi.72.7.5433-5440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Lee H, Yoon D W, Albrecht J C, Fleckenstein B, Neipel F, Jung J U. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukac D M, Kirshner J R, Ganem D. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukac D M, Renne R, Kirshner J R, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 33.Manet E, Gruffat H, Trescol-Biemont M C, Moreno N, Chambard P, Giot J F, Sergeant A. Epstein-Barr virus bicistronic mRNAs generated by facultative splicing code for two transcriptional trans-activators. EMBO J. 1989;8:1819–1826. doi: 10.1002/j.1460-2075.1989.tb03576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manzano-Winkler B, Novina C D, Roy A L. TFII is required for transcription of the naturally TATA-less but initiator-containing Vbeta promoter. J Biol Chem. 1996;271:12076–12081. doi: 10.1074/jbc.271.20.12076. [DOI] [PubMed] [Google Scholar]
- 35.Marschall M, Leser U, Seibl R, Wolf H. Identification of proteins encoded by Epstein-Barr virus trans-activator genes. J Virol. 1989;63:938–942. doi: 10.1128/jvi.63.2.938-942.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molden J, Chang Y, You Y, Moore P S, Goldsmith M A. A Kaposi's sarcoma-associated herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 receptor subunit. J Biol Chem. 1997;272:19625–19631. doi: 10.1074/jbc.272.31.19625. [DOI] [PubMed] [Google Scholar]
- 37.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 38.Moore P S, Chang Y. Antiviral activity of tumor-suppressor pathways: clues from molecular piracy by KSHV. Trends Genet. 1998;14:144–150. doi: 10.1016/s0168-9525(98)01408-5. [DOI] [PubMed] [Google Scholar]
- 39.Moore P S, Gao S J, Dominguez G, Cesarman E, Lungu O, Knowles D M, Garber R, Pellett P E, McGeoch D J, Chang Y. Primary characterization of a herpesvirus agent associated with Kaposi's sarcomae. J Virol. 1996;70:549–558. doi: 10.1128/jvi.70.1.549-558.1996. . (Erratum, 70:9083.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neipel F, Albrecht J C, Ensser A, Huang Y Q, Li J J, Friedman-Kien A E, Fleckenstein B. Human herpesvirus 8 encodes a homolog of interleukin-6. J Virol. 1997;71:839–842. doi: 10.1128/jvi.71.1.839-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neipel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicholas J, Ruvolo V R, Burns W H, Sandford G, Wan X, Ciufo D, Hendrickson S B, Guo H G, Hayward G S, Reitz M S. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 43.Pitha P M. Reflections on the years in interferon research. J Interferon Cytokine Res. 1997;17:181–184. doi: 10.1089/jir.1997.17.181. [DOI] [PubMed] [Google Scholar]
- 44.Ragoczy T, Heston L, Miller G. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J Virol. 1998;72:7978–7984. doi: 10.1128/jvi.72.10.7978-7984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reed J A, Nador R G, Spaulding D, Tani Y, Cesarman E, Knowles D M. Demonstration of Kaposi's sarcoma-associated herpes virus cyclin D homolog in cutaneous Kaposi's sarcoma by colorimetric in situ hybridization using a catalyzed signal amplification system. Blood. 1998;91:3825–3832. [PubMed] [Google Scholar]
- 46.Roan F, Zimring J C, Goodbourn S, Offermann M K. Transcriptional activation by the human herpesvirus-8-encoded interferon regulatory factor. J Gen Virol. 1999;80:2205–2209. doi: 10.1099/0022-1317-80-8-2205. [DOI] [PubMed] [Google Scholar]
- 47.Roy A L, Malik S, Meisterernst M, Roeder R G. An alternative pathway for transcription initiation involving TFII-I. Nature. 1993;365:355–359. doi: 10.1038/365355a0. [DOI] [PubMed] [Google Scholar]
- 48.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarid R, Sato T, Bohenzky R A, Russo J J, Chang Y. Kaposi's sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat Med. 1997;3:293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- 51.Sarid R, Wiezorek J S, Moore P S, Chang Y. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol. 1999;73:1438–1446. doi: 10.1128/jvi.73.2.1438-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schalling M, Ekman M, Kaaya E E, Linde A, Biberfeld P. A role for a new herpes virus (KSHV) in different forms of Kaposi's sarcoma. Nat Med. 1995;1:707–708. doi: 10.1038/nm0795-707. [DOI] [PubMed] [Google Scholar]
- 53.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 54.Sun R, Lin S F, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun R, Lin S F, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol. 1999;73:2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Talbot S J, Weiss R A, Kellam P, Boshoff C. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology. 1999;257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- 57.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J L, Schroter M, Scaffidi C, Krammer P H, Peter M E, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 58.van Santen V L. Characterization of a bovine herpesvirus 4 immediate-early RNA encoding a homolog of the Epstein-Barr virus R transactivator. J Virol. 1993;67:773–784. doi: 10.1128/jvi.67.2.773-784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zalani S, Holley-Guthrie E, Kenney S. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc Natl Acad Sci USA. 1996;93:9194–9199. doi: 10.1073/pnas.93.17.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang L, Chiu J, Lin J C. Activation of human herpesvirus 8 (HHV-8) thymidine kinase (TK) TATAA-less promoter by HHV-8 ORF50 gene product is SP1 dependent. DNA Cell Biol. 1998;17:735–742. doi: 10.1089/dna.1998.17.735. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Q, Holley-Guthrie E, Dorsky D, Kenney S. Identification of transactivator and nuclear localization domains in the Epstein-Barr virus DNA polymerase accessory protein, BMRF1. J Gen Virol. 1999;80:69–74. doi: 10.1099/0022-1317-80-1-69. [DOI] [PubMed] [Google Scholar]
- 62.Zhu F X, Cusano T, Yuan Y. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J Virol. 1999;73:5556–5567. doi: 10.1128/jvi.73.7.5556-5567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zimring J C, Goodbourn S, Offermann M K. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J Virol. 1998;72:701–707. doi: 10.1128/jvi.72.1.701-707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]